Abstract
Many open questions in RNA biology relate to the kinetics of gene expression and the impact of RNA binding regulatory factors on processing or decay rates of particular transcripts. Steady state measurements of RNA abundance obtained from RNA-seq approaches are not able to separate the effects of transcription from those of RNA decay in the overall abundance of any given transcript, instead only giving information on the (presumed steady-state) abundances of transcripts. Through the combination of metabolic labeling and high-throughput sequencing, several groups have been able to measure both transcription rates and decay rates of the entire transcriptome of an organism in a single experiment. This review focuses on the methodology used to specifically measure RNA decay at a global level. By comparing and contrasting approaches and describing the experimental protocols in a modular manner, we intend to provide both experienced and new researchers to the field the ability to combine aspects of various protocols to fit the unique needs of biological questions not addressed by current methods.
Keywords: RNA decay, metabolic labeling, high-throughput sequencing, 4sU, EU, BrU
1. Introduction
Gene expression is modulated at multiple stages including transcription and processing of nascent transcripts, regulation of translation efficiency and intracellular localization, and control of the rate of RNA degradation. This chapter focuses on advanced methods to measure mRNA decay rates on a transcriptome-wide basis. Multiple RNA decay pathways that degrade RNAs have been discovered and specific regulatory factors that control the rates of RNA decay have been reported [1, 2, 3]. These include short regulatory RNAs (siRNA and microRNAs) [4] and a plethora of RNA binding proteins [5]. The key challenge now is to determine the impact of each of these factors on the transcriptome using facile quantitative approaches. Early approaches to measuring mRNA decay involved shutting off transcription and measuring RNA abundance over time using either northern blots, dot blots, or radioactively labeled RNA [6, 7, 8, 9, 10]. However, concerns over the impact on the underlying biology for cells undergoing transcription shutoff have led to the development of various methods to metabolically label RNA to measure RNA decay in a less intrusive manner. By incorporating a chemically modified nucleobase into the cellular pool of ribonucleotide triphosphates (NTPs), RNAs can be labeled without disrupting gene expression, thereby minimally perturbing the underlying biology. Additionally, the indiscriminate nature of metabolic labeling combined with label-based purification methods and modern RNA sequencing allows for transcriptome-wide determinations of both transcription rates and RNA decay in a single experiment. Here we review both historical and recent advances in methods using metabolic labeling to quantitatively measure RNA decay in living cells. We take a modular approach, by describing individual aspects of the methods that have been developed in such a way that each step can be mixed and matched with later steps, so that unique experimental designs can be developed to answer challenging biological questions using an optimal combination of approaches.
2. Metabolic Labels
The cornerstone of most modern sequencing-based workflows for measuring RNA decay is the use of metabolic labeling, via the incorporation of nucleotide analogs into RNA, which are used to separate or distinguish the labeled RNA from the rest of the cellular RNA pool. Here we review the development and characteristics of several of the most frequently used metabolic labels in modern practice.
2.1. Thiol-containing uracil analogs
A variety of different modified uracil labels have been used to measure both mRNA decay and mRNA synthesis. The most basic requirements for such a label are that it be cell permeable, readily incorporated into RNA, minimally perturb cellular physiology, and permit either the purification or specific detection of RNA molecules containing the label. Several commonly used metabolic labels meeting these criteria are shown in Figure 1. The most widely used label is 4-thiouridine in either its nucleoside (4sU) or nucleobase (4tU) forms (Figure 1A). Both 4tU and 4sU are readily taken up by yeast [11, 12], archaea [13], and higher eukaryotes including human cells [14, 15, 16]. In contrast to other thiol-modified nucleotides, incorporation of 4sU at concentrations of up to 100 μM in cell culture does not have a discernible impact on the synthesis of RNA or protein degradation rates indicating limited perturbation of transcription and translation following incorporation of the label [17]. In contrast, 6-thioguanine (6sG) and related compounds are still at times used for metabolic labeling of RNA [18], but as 6sG has been shown to perturb both transcription and translation [17], 6sG is of substantially less utility for the long term labeling required for RNA stability experiments. Although long term culture (48 hr) in the presence of 4sU has been associated with a decrease in cell viability [19], short term labeling (10 hrs) of up to 4 mM 4tU does not appear to have a discernible impact on cell growth in yeast [12]. However, in vitro translation assays have revealed that 4sU-containing mRNAs can decrease ribosomal elongation processivity and increase downstream initiation rates [20]. For organisms such as S. cerevisiae and E. coli that express a functional Uracil Phosphoribosyltransferase (UPRT), 4-thiouracil can be used in place of 4-thiouridine as it is readily converted to 4sU as needed by the cells. However, in both mouse and human cells incorporation of 4tU into cellular RNA does not readily occur and coexpression of UPRT from another organism is needed in order to incorporate 4tU into nascent RNA [21, 11]. Expression of the well-characterized Toxoplasma gondii UPRT has been used to successively label RNA with 4tU in human foreskin fibroblasts [11] and, subsequently, in a variety of other cell types [22, 23]. The requirement for UPRT activity in labeling with 4tU has also led to the development of “TU-tagging”, a method to selectively label mRNAs in only one cell type in the context of a mixed population of cells. By expressing UPRT only in the cell type of interest, one can determine both the identity and mRNA decay rates of the mRNAs from that cell type [24]. Additionally, 4tU [Sigma Aldrich Cat. No. T4509] is substantially cheaper than 4sU [Sigma Aldrich Cat. No. 440736] and is more economical to use for organisms that already have robust endogenous UPRT activity (please note that throughout this review we indicate product numbers merely as examples, and not as a reflection of endorsement of any particular product or manufacturer). In whatever form it is introduced, RNA-incorporated 4sU readily crosslinks to both RNA and protein upon exposure to 365 nm UV light, a feature that is taken advantage of for the analysis of RNA-protein interactions but should be minimized in the analysis of mRNA decay [25, 26]
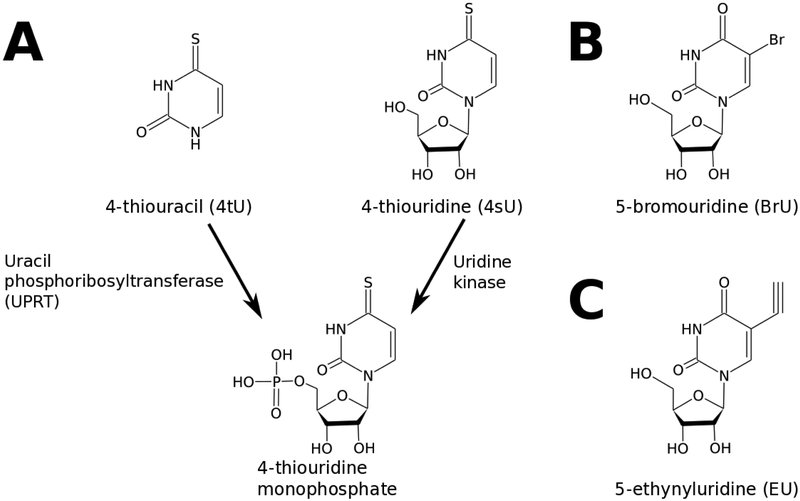
Figure 1:
Structures and inclusion chemistries of common RNA metabolic labels. A) 4-thiouracil variants and pathways for incorporation into nucleotide metabolism; once the nucleotide monophosphate is formed, the resulting compound is readily incorporated into cellular RNA. B) Structure of 5-bromouridine, which can be assimilated through the uridine kinase pathway as on the right side of panel A. C) Structure of the click chemistry substrate 5-ethynyluridine, again typically incorporated into the cellular nucleotide pool via uridine kinase activity.
2.2. Halogen-containing uracil analogs
Incorporation of 5-bromodeoxyuridine (BrdU) into cellular DNA was first described in the 1950s [27, 28] and the development of an anti-BrdU antibody allowed for visualization of DNA within a living cell [29]. Some BrdU antibodies cross-react with 5-bromouridine (BrU) and labeling with BrU can be used to selectively purify BrU labeled RNA with an anti-BrdU antibody [30]. Like 4sU, BrU (Figure 1B) is readily taken up by mammalian cells [31] and BrU does not appear to have the same general toxicity effects that 4sU has under long exposure [19], making it an attractive reagent to use for measuring mRNA decay over a longer time course. However, in vitro translation assays have revealed that BrU containing mRNAs have a modest negative impact on both ribosomal elongation and initiation, but not as large in magnitude as the effects seen from 4sU [20]. Additionally, BrU [Sigma Aldrich Cat. No. 850187] is comparable in price to 4tU and does not require UPRT activity for incorporation into mammalian cellular RNA.
2.3. Alkyne-containing uracil analogs
First described as a labeling reagent for fixed cells, 5-Ethynyluridine (EU; Figure 1C) is a uracil derivative capable of performing “click” chemistry (reviewed [32]) both in vivo and in vitro [33]. Like BrU, 4sU and 4tU, EU is rapidly taken up into the cellular pool of NTPs and incorporated into transcribed RNAs. Similar to 4sU and 4tU, short-term labeling with EU does not appear to have negative effects on cellular health, but longer incubation times do negatively impact growth rates [19]. Although EU could be used for high throughput determinations of RNA synthesis and decay rates, most studies have been primarily focused on targeted measurements of select RNAs through the use of qRT-PCR [34, 35]. Recent development of 5-Ethynylcytosine (EC) [36] in conjunction with expression of cytidine deaminase and UPRT in Drosophila has led to the development of “EC-tagging”, a method to purify cell-type specific RNAs with higher specificity than “TU-tagging” with 4tU as described above [37]: EU is generated in situ by target cells through the combined activities of ectopically expressed cytidine deaminase (to generate 5-ethynyluracil) and UPRT (to generate EU, analogous to the reaction in Figure 1A). Additionally, EU has recently been used to determine the nascent-RNA “interactome” through a combination of EU labeling and UV crosslinking coupled with RNA-seq and proteome analysis, indicating that EU labeling can be successively used with high throughput methods [38]. EU is significantly more costly than 4sU, 4tU or BrU, and can be purchased either stand alone [Invitrogen Cat. No. E10345] or in the Click-iT Nascent RNA Capture Kit [Invitrogen Cat. No. C10365] along with buffers and protocols for its use.
2.4. Impact of exogenous labels on RNA decay rates
A growing body of evidence has suggested a role for RNA modifications in the post-transcriptional control of gene expression, including control of RNA processing, binding of RNA binding proteins, changes in secondary structure, and stop-codon readthrough (reviewed in [39]). Although none of the labels introduced above perfectly match the modifications that have been found naturally in eukaryotic cells, in principle, these exogenous labels could still disrupt RNA decay rates through similar mechanisms. While this represents an important consideration, to the best of our knowledge there have been no targeted experiments designed to test the impact of any of the labels above on RNA decay rates themselves. Genome-wide comparisons between 4sU labeling and transcriptional shutoff experiments in yeast have shown that RNA decay rates determined from transcriptional shutoff experiments have greater agreement with one another than they do with RNA decay rates determined using metabolic labeling [40]. However, Sun et al. also show that decay rates determined from transcriptional shutoff experiments correlate well with genome-wide measurements of mRNA decay made using metabolically labeled RNA in cells displaying a transcriptional shutoff phenotype. They further show, by using measurements of metabolically labeled RNA, that RNA decay in cells under osmotic stress or heat shock also correlate well with RNA decay rates determined in transcriptional shutoff experiments, suggesting that perturbations of RNA abundances from cellular responses to transcriptional shutoff may mimic stress responses and confound measurements of RNA decay [40]. On the other hand, comparisons of RNA decay rates determined from seperate labs using different experimental strategies with the same metabolic label do not correlate well with one another, suggesting that there may be sources of experimental error in labeling experiments that are poorly understood [40]. One possible source of error could be attributed to differences in normalization between RNA abundance measurements. For example, Lugowski et al. report better replicate to replicate correlation, as well as better agreement to transcriptional shutoff experiments and metabolic labeling experiments from other labs, using an internal normalization method (normalize to introns) as opposed to an external method (normalize to spike-in) [41]. Further discussion on the impact of normalization methods on measurements of RNA decay can be found in Section 7. As it stands, it is unclear if there is a single major source of discrepancy that results in disagreement between measurements of RNA decay between different labs and experimental approaches.
3. Selection and purification of labeled RNAs
In the majority of metabolic labeling experiments at present, labeled RNA molecules are physically isolated from the total RNA pool prior to analysis (one notable exception, SLAM-Seq, is described below). After purification of total RNA from the cell lysate, the newly labeled RNAs must be separated and purified using methods specific for the incorporated label. Each label discussed above uses different chemistry for selection, but the general principle is the same: select the label with as high affinity as possible thereby minimizing the amount of starting material needed and maximizing capture specificity. Once purified, the labeled RNAs are then quantified using standard RNA-seq methods (Figure 2) [16, 42, 43, 44].
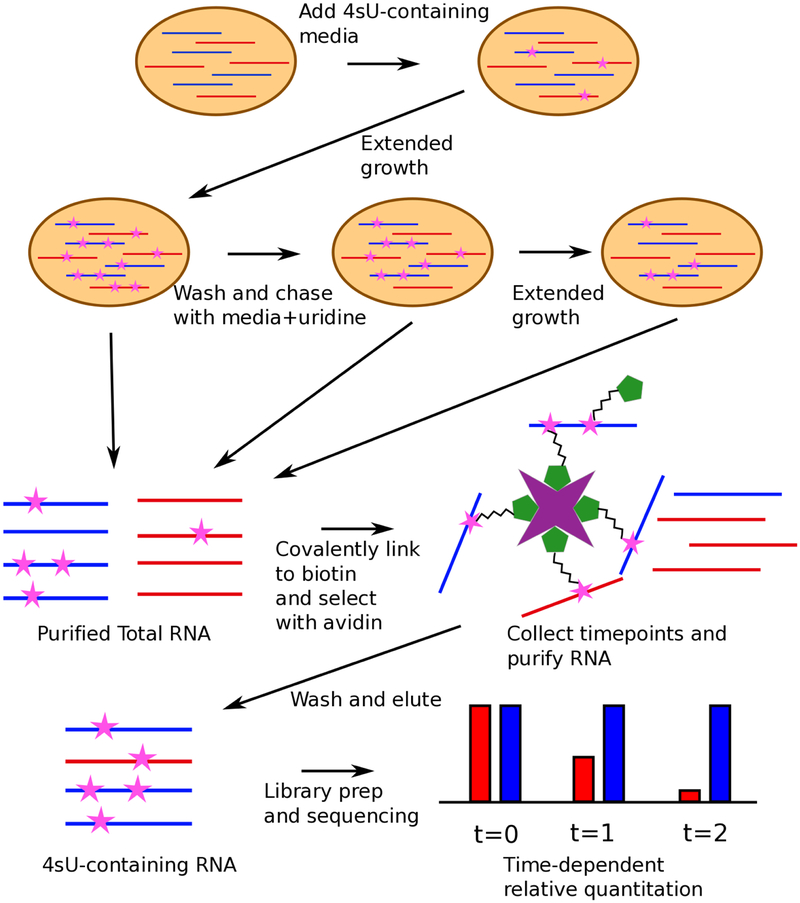
Figure 2:
Workflow of a 4sU chase experiment to measure the stabilities of different RNA species. Shown is a hypothetical cell containing two types of transcript (blue and red), with similar equilibrium levels but differing stabilities. Cells are grown in media with 4sU added to label transcripts, and then washed and chased with media containing unlabeled uridine, with samples harvested for RNA extraction at two or more time points during the pulse/chase. 4sU-containing transcripts are then covalently linked biotin and purified using streptavidin, and the enriched RNA prepared for sequenced using standard methods. Note that the RNA purification and 4sU enrichment steps are performed separately for each time point.
3.1. HDPD-biotin
For 4sU and 4tU, purification is performed by chemically linking the labeled RNA to biotin and using the well-studied affinity between biotin and streptavidin to purify the RNA-biotin complex [45]. 4sU labeled RNA can be covalently linked to biotin by taking advantage of the thiol-containing uridine and forming a disulfide bond to modified biotin molecules. The most commonly used modification to biotin is N-[6-(Biotinamido]hexyl]-3’-(2’-pyridyldithio)-propionamide (HPDP-biotin) [11, 14, 12, 16] and HPDP-biotin is readily available in the form of the EZ-link HPDP-Biotin kit [Thermo Scientific Cat. No. 21341]. The covalent link between 4sU and HPDP-Biotin is completely reversible and elution is performed through the reduction of disulfide bonds with a reducing agent such as DTT, which results in RNA without covalently bound adducts as input into downstream sequencing.
3.2. MTS-biotin
While the HDPD-biotin based procedure described above has been widely used, the formation of a disulfide bond between 4sU and HPDP-biotin is inefficient; disulfide exchange reactions between 4sU and HPDP-biotin indicate that less than 20% of free 4sU is converted to 4sU-HPDP-biotin in reactions as long as 120 minutes. Recent developments using methylthiosulfonate-biotin (MTS-biotin) have indicated greater than 95% conversion of free 4sU to 4sU-MTS-biotin in as little as five minutes, indicating a fast and efficient reaction resulting in capture of labeled RNA without the need for as much starting material [44]. The MTS-biotin purification protocol has been used to study miRNA turnover [46], response to viral infection [47], and transcription rates in yeast [48], but it has not enjoyed as much widespread use as HPDP-biotin, possibly because of MTS-biotin’s relatively recent introduction as a viable alternative to HPDP-biotin. Additionally, MTSEA-biotin [Biotium Cat. No. 90064] is less costly than HPDP-biotin, making it a more economical alternative.
3.3. Anti-BrdU antibody
Unlike 4sU, BrU does not have a chemical group that can be easily used to create reversible crosslinks with modified biotin. Thus, purification of BrU-containing RNAs must proceed with non-covalent interactions mediated through well-established anti-BrdU antibodies (which frequently also bind BrU). Many commercially available Anti-BrdU antibodies have been used for the quantification of mRNA synthesis or decay through BrU labels: mouse anti-BrdU [Roche 11170376001][31], BrdU Antibody (IIB5) [Santa Cruz sc-32323] for GRO-Seq [49], Anti-BrdU mAb 2B1 [MBL International Corporation, cat. No. MI-11-3] for BRIC-seq [50], and mouse anti-BrdU [BD Pharmingen, 555627] for Bru-Seq and BruChase-Seq [42]. Imamchi et al. [50] indicated that they have tried multiple anti-BrdU antibodies and the reported 2B1 antibody resulted in the highest yields, but to the best of our knowledge, no extensive comparison of antibody purification efficiencies has been published.
3.4. Click chemistry
As with 4sU, purifying RNAs labeled with EU usually relies on a covalent linkage with biotin and selection using streptavidin beads, in this case using the bio-orthogonal copper-catalyzed azide-alkyne cycloaddition reaction typical of modern ‘click’ chemistry. Most uses of EU to purify RNA follow the Click-iT Nascent RNA Capture Kit protocol, which involves the use of PEG4 carboxamide-6-azidohexanyl biotin (azide-biotin) with a copper (I) catalyst (generated in situ in the reaction by reduction of copper (II)) to covalently link the EU to biotin [34, 35, 51]. Unlike 4sU, this covalent bond is not easily reversed and generation of cDNA libraries for sequencing or qRT-PCR for direct quantification has to be done while linked to the streptavidin-beads [52]. It is not clear what effect, if any, this has on the error rate of the reverse transcriptase.
It may be possible to take advantage of the ability of very low-salt solutions to cause surprisingly rapid dissociation of the streptavidin-biotin interaction [53] prior to quantitation or sequencing library preparation. To our knowledge, this strategy has not been employed to date in the published literature.
3.5. Purification-free detection through enhanced T→C mutation rates
The use of 4sU-containing RNAs for cDNA synthesis results in the reverse transcriptase misincorporating a guanine residue opposite the 4sU at a low level that is exacerbated when cross-linked to protein [26]. Substituting iodoacetamide (IAA) in place of cross-linked protein allows for non-specific enhancement of T→C conversion rates in the reverse transcriptase reaction for all 4sU sites in a library through disulfide bond formation between the IAA and 4sU. T→C mutation rates increase from 10% without IAA to 94% with IAA. SLAM-Seq takes advantage of this increase in mutation rates to quantify mRNA synthesis and decay rates without a purification step. By labeling with 4sU and treating with IAA before library preparation, SLAM-Seq can differentiate labeled RNA from unlabeled RNA strictly through quantification of T→C mutation rates of the final library. Removal of a purification step vastly decreases the amount of input RNA needed and greatly simplifies the mRNA decay protocol [54].
3.6. Impact of pulldown efficiency and label incorporation rates on experimental measurements
Two additional parameters that could introduce noise into measurements of RNA decay using metabolic labeling include the incorporation rate of the label into newly synthesized RNA, and the efficiency of pulling down labeled RNA from the total purified RNA. We are unaware of any systematic characterization of the differences in label incorporation between the different metabolic labels discussed in Section 2. In some sense, differences in incorporations rates, so long as they are consistent across timepoints, are of no consequence in the experimental designs discussed below since quantification of RNA abundance is either relative to the total amount of RNA pulled down or is normalized by sequencing both the unlabeled and labeled RNAs for each time point (see Section 7 for details). However, incorporation rates may be a crucial parameter for measurements of either fast-decaying or slow-decaying RNAs, as they may limit detection. In such cases, optimization of the amount of label added to the cells, incubation times with label, and/or choice of time points may allow for detection of difficult transcripts. As with incorporation rates, a systematic comparison of pulldown efficiencies between different labels and selection strategies has also not been performed. In a typical RNA decay experiment, differences in pulldown efficiency within a single experiment will be controlled for through the use of spike-ins or internal normalization (as discussed in Section 7), thereby largely eliminating pulldown efficiencies as a major source of experimental error as long as saturation is not reached. However, improvements in pulldown efficiency can result in less needed biological material for a given experiment. Furthermore, many of the computational methods used to analyze RNA decay experiments operate under the implicit assumption that the sequenced pool of labeled RNA contains no contaminating unlabeled RNA, which may not be accurate to actual experimental conditions but will be closer approximated with better pulldown efficiencies. As discussed above, some improvements have been made to biotin based pulldown strategies for experiments using 4sU as a label through changing the identity of the chemical crosslinker [44]. Additionally, the use of mutation rates induced by the metabolic label removes the need for a pulldown step but introduces a separate source of experimental error related to modification efficiencies of the label itself and misincorporation rates of the reverse transcriptase [54].
4. Experimental Design for measuring RNA decay
With a label and purification method in hand, an experimental design must be chosen that maximizes the amount of information to be gained per unit cost. Different considerations must be made if both synthesis rates and decay rates are to be determined. Additionally, it is critical to decide whether precise RNA half-lives are to be measured or if end-point abundance estimations are sufficient for the biological question of interest. A comparison of different experimental designs frequently used for the determination of RNA decay is shown in Figure 3.
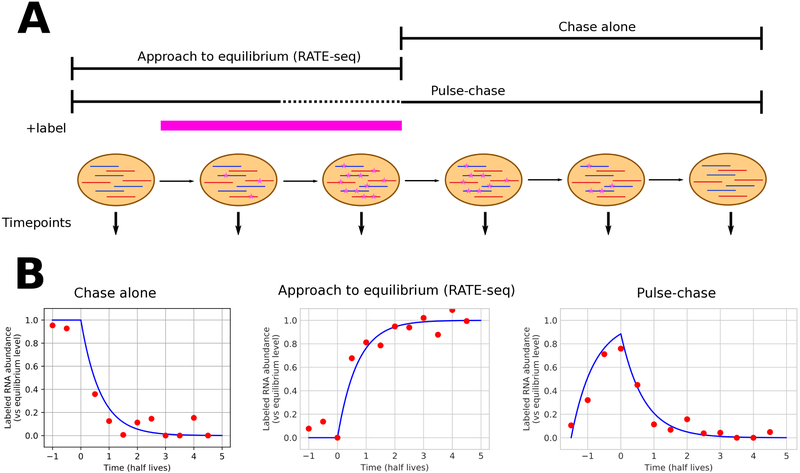
Figure 3:
Overview of different metabolic labeling time strategies, as discussed in detail in the text. A) Schematic of the timing of labeling and sample harvest for three different methods; n.b. labeling in a pulse-chase experiment is typically too short for equilibrium levels to be reached. The pink bar (+label) indicates the time period during which labeled nucleotide is present. B) Expected abundance curves (blue) and hypothetical experimental data (red) for the fractional abundance of labeled transcript for any particular RNA under each experimental procedure shown in panel A. Time is relative to a zero point at the time of labeled nucleotide removal/washout (chase-alone and pulse-chase) or addition (RATE-seq).
4.1. Chase alone
To determine RNA decay alone, cells can be grown for an extended period of time, often 24 hours, in the presence of a label. At time zero, the growth media is replaced with identical media containing the same concentration of unlabeled uridine and the labeled RNAs are tracked via purification and sequencing. If determining RNA half-lives, several time points are taken and used for fitting a single exponential decay model [50]. For a more coarse-grained determination of decay, a single time point can be taken after the switch to unlabeled media and compared to a sample taken at time zero. There are major trade offs to consider between these two approaches. By taking only two time points, one drastically cuts down on the costs of sequencing and the labor to prepare the samples. This can be particularly useful when comparing the difference in RNA decay between two biological conditions where the exact half-life is not as useful as as the relative change in decay between the two conditions is. On the other hand, taking several time points allows one to capture both short-lived and long-lived transcripts that may be missed with a single time point. In cultured mammalian cells, the average mRNA half-life is 7–9 hours [55, 56, 16] and it is critical to choose time points that capture the decay of mRNA transcripts of interest. Furthermore, many time points are needed to accurately fit the exponential models used for half-life determination. Thus, selection of the duration and number of time points to be analyzed typically needs to be optimized (left side of Figure 3B).
4.2. Approach to equilibrium
The converse of the chase-alone experimental methodology, approach to equilibrium, allows for RNA decay rates to be determined from measuring time points after the addition of the labeled uridine to the media. Although cells harvested after a short incubation time can be used to measure transcription rates [57], taking several time points over an extended time course in the presence of the labeled uridine can allow for the mRNA decay rates to be determined instead. The biological motivation behind approach to equilibrium is the concern that labeled nucleotides can be recycled within a cell leading to an ineffective chase with unlabeled nucleotides [58]. To see the quantitative motivation for the approach to equilibrium method, it is useful to consider the overall dynamics of a given transcript. Assuming a constant rate of transcription, the concentration of any particular RNA species, X, will generally follow the equation
| (1) |
Here, τ represents the rate of transcription under the condition of interest, δ is the decay rate of the RNA (typically the quantity of interest), and γ is a dilution term dependent on the growth rate of the cell (if not explicitly accounted for, dilution effects will be incorporated into the inferred value of δ, which for the slow-growing cells of higher eukaryotes is typically a negligible correction)[59]. If one considers the labeled form of an RNA of interest as a separate species, X*, then Eq. 1 will likewise be followed for the labeled species, except that the synthesis rate will be proportional to τ when the label is present, and equal to zero when the label is not. As the steady state level is defined by the point at which the synthesis and decay rates are perfectly balanced, the steady state concentration requires [X]′ = 0, or τ = (δ + γ)[X]eq. From this equation it immediately follows that knowledge of any two of the equilibrium concentration, overall decay rate, and synthesis rate are sufficient to specify the third.
By growing cells in a constant amount of label, the fraction of each RNA that is labeled will increase at a rate that is determined only by its degradation rate and the growth rate of the cells until it reaches a steady state level [60]. By measuring time points along this increase, one can capture the decay rate of any given RNA molecule [43], as the equilibrium value will be known from a very late time point and a curve fit can then reveal the decay parameters (see Figure 5B and Figure 3B (middle)). However, approach to equilibrium requires cells to grow in the presence of the label for an extended period of time, which may be problematic for labels that have demonstrated toxicity under longer exposure, such as 4sU.
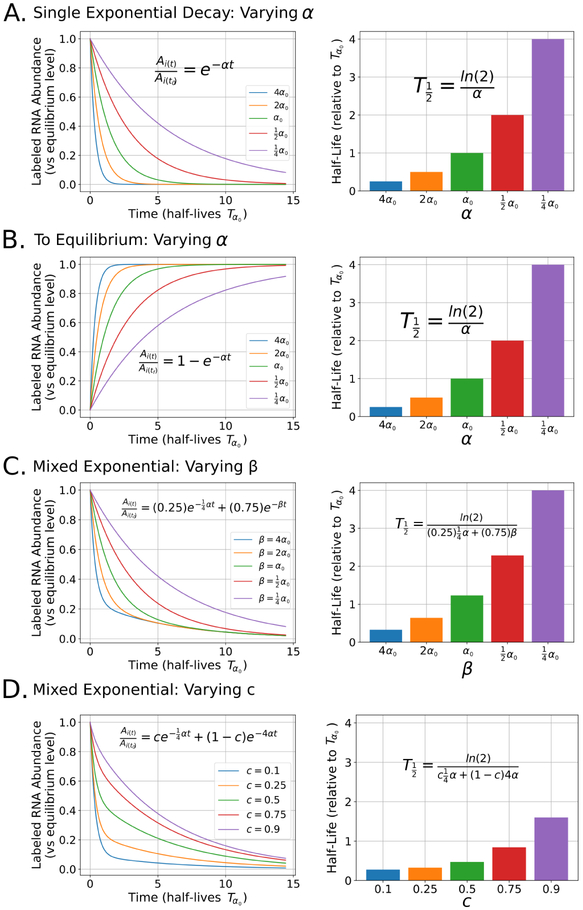
Figure 5:
Impact of varying parameters in the equations used for modeling RNA decay: For each panel, the left graph represents the shapes of several arbitrary exponential curves after perturbing a single parameter in each model. Here, the y-axis represents labeled RNA abundance relative to the equilibrium level of labeled RNA in label-containing media. The x-axis represents time in the number of half-lives for a single exponential curve with a decay rate of α0, indicated with Tα0. The right graph in each panel represents half-lives calculated from each of the curves from the corresponding left graph in the same panel and relative to Tα0. In each graph, the equation used to either model the decay or determine the half-life is displayed. A) The effect of varying α on relative RNA abundance (left) and half-life (right) when modeling RNA decay with a single exponential. B) The effect of varying α when modeling RNA decay in a to equilibrium experimental design. C) The effect of varying β with a fixed c and a fixed α when modeling RNA decay with a two component mixed exponential. D) The effect of varying c with a fixed α and β when modeling RNA decay with a two component mixed exponential.
4.3. Pulse-chase
It is often advantageous to determine both the synthesis and decay rates of an RNA molecule within the bounds of a single experiment. By incubating with a short “pulse” of label and “chasing” with unlabeled media one can both minimize exposure of the cells to the label and determine both synthesis and decay rates separately [11, 16, 42, 61, 62]. Through taking time points at the initial addition of the label, the switch to unlabeled media, and throughout the “chase” period, the lifespan of all nascent labeled RNAs can be tracked (Figure 3B (right)). Pulse-chase methods have the advantage of subjecting the cells to short-exposures of the label thereby mitigating any potential toxicity.
5. Quantification of RNA abundance
Although specialized DNA microarrays have been used previously [55, 14, 56], global analysis of RNA decay is more recently measured through the use of high throughput sequencing and well-established bioinformatics tools are used to analyze the resulting sequencing reads. Library preparation for RNA sequencing experiments is available through several commercial kits or custom methods that are specific to the experiment of interest. As a general rule, paired-end and stranded sequencing is preferred, particularly for organisms that perform splicing or have transcripts regulated by antisense RNAs. Additionally, several strategies exist to remove highly abundant ribosomal RNAs (rRNA) from samples prior to library preparation, including rRNA depletion with custom oligos or selection of poly-adenylated mRNAs. Because poly(A) metabolism plays an important role in mRNA decay pathways, it is advisable to avoid poly(A) selection when analyzing mRNA decay kinetics. [1, 3] After sequencing, several data processing steps must occur to take the raw sequencing reads to a measurement of RNA abundance. Many of the programs and tools written for the analysis of high throughput sequencing data are driven by a text interface, so it is expected that users have some familiarity with the Unix command line. Many institutions have workshops designed to teach new users both familiarity and comfort with the command line, and readers who feel uncomfortable working with command line programs can find help both online and locally. For most applications, the RNA sequencing reads obtained from the methods above can be treated like data from any other RNA-seq experiment. Typically, sequencing reads are stored in the fastq file format where both sequence and base-calling quality information can be stored. Here we will briefly outline the set of steps needed to analyze RNA-sequencing data for RNA decay experiments with extra commentary on possible locations in the analysis that may differ for RNA decay-type experiments as compared to standard RNA-seq workflows. For more information on best practices concerning RNA-seq data we point the reader to recent reviews in the literature [63, 64].
5.1. Adapter removal and quality control
As with any sequencing analysis, standard quality control must be employed. Several steps must be taken to remove adapters needed for Illumina sequencing as well as reads containing low confidence base calls. For the removal of adapters and low quality sequences, several programs exist including cutadapt [65], fastx toolkit [http://hannonlab.cshl.edu/fastx_toolkit/], and trimmomatic [66]. Several key statistics about the quality of the sequencing reads can be calculated both before and after adapter and quality trimming using FastQC [67] (Figure 4A). Next the reads must be aligned to a reference transcriptome which is available from either NCBI or the UCSC Genome Browser for most model organisms. Several different aligners have been developed for processing RNA sequencing reads including bowtie2 [68], tophat2 [69], STAR [70], kallisto [71], and many others. A comparison of the most commonly used aligners indicates tradeoffs between each tool and the specific aligner used will depend on the question being asked [72]. However, if one is using the SLAM-seq methodology that is dependent on T→C mutations then it is recommended to use the T→C mutation aware aligner NextGenMap [73] with special settings designed to weaken the penalty for mismatches resulting from a T→C mutation event [54] (Figure 4B).
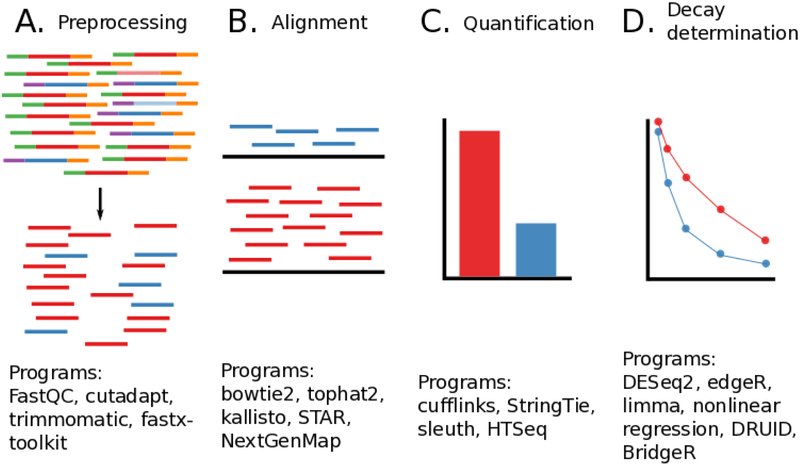
Figure 4:
Overall diagram of data analysis steps needed to process high throughput sequencing reads from RNA decay experiments. Widely used example software packages are noted underneath each step. A) Preprocessing and quality control, here adapters and low quality reads are removed from analysis. B) Alignment of reads to a reference genome or transcriptome. Several key considerations are highlighted in the text below. C) Quantification of each transcript or feature of interest. Several different programs can be used to convert alignment information into a measure of RNA abundance that is comparable between experiments. D) Modeling of RNA decay. Many different models can be used to determine the decay rates of each transcript of interest.
5.2. Reference-based alignment, transcriptome assembly or pseudoalignment?
Several additional considerations need to be made when choosing both the aligner and the downstream quantification software for processing the data from a high-throughput RNA decay experiment. For single-celled organisms such as bacteria or archaea where a high quality reference transcriptome is known for the organism and that organism does not process RNAs through splicing, a simple aligner such as bowtie2 will perform well. However, most higher eukaryotes do process RNAs through splicing and thus splice-aware aligners, such as hisat2 [74] and STAR, are recommended. Under some biological conditions, novel transcripts are may be expected and have not yet been characterized and logged in the reference transcriptome of the organism under study. Here, downstream software will be needed to infer the presence of novel transcripts and assemble a transcriptome either de novo or through assistance of an existing reference transcriptome. However, many experiments are not designed to look for new transcripts and are instead concerned with the abundance of well-characterized transcripts annotated in a reference transcriptome. Pseudoaligners such as kallisto [71] and salmon [75] are designed to deal efficiently with this latter case. Rather than do a full alignment, pseudoaligners allow for RNA quantification without needed to fully align the reads to the reference transcriptome. Pseudoaligners have the advantage of being substantially faster than traditional alignment methods, but will not be able to detect any novel transcripts and are wholly reliant on the quality of the reference transcriptome. Unlike the pseudoaligners, most major aligners will output a sequence alignment map (SAM) file or its binary equivalent (BAM) that contains several details of where a particular sequence aligned and the quality of that alignment. Key statistics and simple manipulations of this file format can be obtained using samtools [76]. After alignment, downstream tools are needed to convert the sequence alignment information into some form of quantification of RNA abundance. The most commonly used software suite that performs this quantification is cufflinks [77] however, StringTie has shown better performance than cufflinks and is currently recommended as a replacement [78]. Both cufflinks and StringTie (as well as other related tools) perform novel transcript discovery and transcriptome assembly, which is useful under conditions where new transcripts are expected and informative but is not always necessary. If transcriptome assembly is not needed due to the existence of an already annotated, high quality reference transcriptome, or if the investigators biological question is not concerned with novel transcripts, then a simple feature level quantification can be obtained using HTSeq [79] instead (Figure 4C).
5.3. Gene level or exon level?
Another key consideration when quantifying data from RNA decay experiments is to determine whether to quantify at the gene level (where all reads for a gene are pooled together regardless of transcript isoforms) or exon level (where each exon is quantified separately). Most reports for determining RNA decay have focused on gene level quantification, but exon level information may be needed if one is tracking decay of specific transcript isoforms.
5.4. Count level or TPM?
When considering differences between two experimental conditions, another major consideration to make is how to quantify the amount of change in RNA decay between the two conditions. Without proper statistical analysis, differences in sequencing depth, the efficiency of labeled RNA recovery, and biological variability between replicates can confound any true biological difference that is being measured. Fragments Per Kilobase per Million (FPKM) or Reads Per Kilobase per Million (RPKM) are two measures that were designed to correct for both sequencing depth and transcript length bias between different samples and genes (or exons). However, the Transcripts Per Million (TPM) unit has superseded RPKM and FPKM as the preferred value for reporting RNA expression, since TPM values can more accurately be directly compared between experiments [80]. TPM is commonly reported as measure of relative RNA abundance under a particular experimental condition for a feature of interest, but more sophisticated statistical models have arisen that better account for the biological variability seen in the quantification of RNA-seq data. The use of negative binomial models based on count-level data instead of FPKM or TPM for each feature of interest allow for better estimation of biological variability and thus more accurate and reproducible results. Negative binomial models are implemented in all of the major differential expression packages currently used in RNA-seq analysis and are applicable to RNA decay analysis. Some of the key differential expression software packages include DESeq2 [81], edgeR [82], limma [83], cufflinks [77], and StringTie [78]. These packages will take count-level data for each feature (at the gene or exon level) of interest and use negative binomial-based statistical models to properly account for variability between conditions. Additionally, the kallisto pseudo-aligner has a downstream package, sleuth [84] designed specifically for use with kallisto, and uses the same general principles as the packages mentioned above (Figure 4D).
6. Modeling RNA Decay
The ultimate goal for most RNA decay experiments is to quantitatively measure the kinetics of RNA abundance over time. For some research questions, a measure of relative changes in RNA decay between two conditions or two transcripts may suffice. However, for another subset of research questions, the determination of a quantitative rate constant with meaningful units is the object of interest. A careful consideration of normalization procedures for measurements of RNA abundance using high-throughput sequencing techniques is essential for this latter class of experiments (and still useful for the former class), as discussed in Section 7. However, a discussion of the theory that underlies models used for the determination of RNA decay as applied to perfect measurements of RNA abundance and discussed below is, nevertheless, instructive.
6.1. Single exponential decay
Guided by historical transcription shutoff experiments, most chase experimental designs use a single exponential equation to determine RNA decay half-lives. A single exponential model assumes that RNA decays at a rate proportional to its instantaneous concentration over the measurement time of the experiment:
| (2) |
Where is the relative abundance for labeled RNA i at time t as compared to time t0: the initial time point taken when the labeled RNA has reached equilibrium. Here αi represents the constant decay rate for RNA i. Note that the exponential form for RNA abundance is obtained directly from integration of Eq. 1 with the production term set to zero and growth term omitted. Thus the half-life of the RNA can be determined by fitting the data with the following equation (Figure 5A):
| (3) |
It is important to note that both the approach given here, and the more sophisticated variations below, work under the assumption that the fitted parameters (e.g., decay rate) do not vary throughout the experimental time course. An additional modification for the half-life determination to account for dilution due to cell growth has also been suggested by several groups. [12, 43]:
| (4) |
Where kgrowth is the same for all RNAs and is determined by the growth rate of the culture; again this equation arises directly from the presumed time-dependent change in RNA abundance stated in Eq. 1. Note that in the context of Eq. 4 the “half life” so calculated yields a half life for the individual RNA molecules themselves, rather than the bulk half life that would be observed for a population of molecules (the latter ought to include dilution effects, while the former should not).
6.2. Mixed exponential decay
Imamachi et al. [50] have noted that a subset of RNAs do not decay in a manner that is easily described by a single exponential and have suggested fitting the data with a model that considers a mixed population of RNAs with different decay rates:
| (5) |
Where c indicates a weight for one subpopulation vs. the other subpopulation and βi is the decay rate for a second population for a particular RNA. In principle, even more complex functional forms could be considered, such as adding additional exponential terms or using a stretched exponential, which might better account for data where multiple subpopulations decayed on different timescales. Precisely such a situation might easily emerge if multiple different subpopulations of cells were present in the measurements, or if gene-level quantification was in use but multiple transcript isoforms existed with differing stabilities. Using more complicated models can be prone to overfitting and appropriate model selection criteria [85] must be made when choosing between models with more or fewer parameters (Figure 5C-D).
6.3. Approach to equilibrium
For approach to equilibrium experimental designs, several assumptions and considerations must be made to properly model the RNA half-lives. Neymotin et al. determine RNA half-lives by considering the decay of unlabeled RNAs and also taking into account the cell growth rates [43]. They ultimately model the abundance of any given labeled RNA at time t as the following:
| (6) |
Where tf is the final time point where the labeled RNA has reached steady state levels (at the end of the time course) and td is the time between the addition of the label and the first measurement of labeled RNA. Both the overall α, which is equal to the αi–kgrowth, and the Yeq for each RNA can then be estimated from the experimental data, here assuming the td is fixed for all RNAs based on experimental measurements for when RNA first appears in after label selection (Figure 5B). Half-lives can then be calculated as above using the growth rate-corrected half-life formula above (Eq. 4). DRUID, an automated pipeline for approach-to-equilibrium experiments, has been developed to deal to help analyze data from this type of experimental design without the need to have complicated spike-ins or sophisticated ways to deal with normalization [41].
6.4. Pulse-chase considerations
Pulse-chase experimental designs have the advantage of allowing the experimenter to separately determine both transcription rates and decay rates from a single experiment. By comparing labeled RNA abundances to unlabeled RNAs (or labeled RNAs between two different experimental conditions) at the beginning of the chase, one can have a general idea of nascent transcription rates [54] or condition-specific effects on transcription [42]. By taking several time points throughout the chase part of the experiment, one can use the single exponential equations described above to fit half-lives of each RNA of interest. Alternatively, one can take a single time point after a chase and measure differences between labeled RNA abundances in two different experimental conditions or between the start and end of the chase to determine relative decay rates without determining the half-life of the RNAs. It is worth noting that many of the methods mentioned above have focused on following a single species: the labeled RNAs. Whether through following the decay of the labeled species over time (chase and pulse-chase), or through measuring the decay of the unlabeled species over time indirectly by measuring the approach to equilibrium of the labeled species, these methods allow for accurate determinations of mRNA decay rates. However, valuable information that may also be gained by sequencing both the labeled and unlabeled pools. More sophisticated methods that take into account both transcription rates, RNA decay rates and measurement of both the pool of unlabeled RNAs and the pool of labeled RNAs have also been reported throughout the literature [16, 15, 86] and can give deeper insight into the full kinetics of individual mRNA transcripts but are outside the scope of this particular review.
6.5. Half-lives vs. differential abundance
The exponential equations above are typically fit using non-linear least squares methods to determine αi by minimizing the squared sum of the errors between the model and the data for each RNA. Although half-lives can be determined with as few as three time points, it has been recommended to use at least 5 time points [87] in order to accurately determine half-lives. The Akimitsu lab has developed a custom R package [https://github.com/AkimitsuLab/BridgeR] for determining the difference in mRNA half-lives between two different conditions of interest. However, determining the RNA half lives for many different replicates and experimental conditions can be incredibly costly due to the amount of sequencing samples needed in order to properly fit the exponential equations. Instead of determining full half-lives for every RNA of interest, one could consider capturing an initial and final time point and using differential expression software to measure the impact of a particular condition on the relative abundance of RNA in the final time point compared to the initial time point. Simple models designed to measure the condition-specific effects on RNA abundance can be specified easily in differential expression analysis software such as DEseq2 [81], which at least permits determination of whether or not the decay of a particular transcript changes between a pair of conditions.
7. Normalization and the use of spike-ins for estimation of labeled RNA abundance
It is important to note that high-throughput sequencing reactions only give relative abundance measurements of RNA. Any comparison of two separate RNA sequencing reactions will thus require some sort of normalization in order to put RNA abundance estimations on the same numerical scale relative to one another. The most commonly reported normalization schemes for RNA-seq type experiments include RPKM and TPM, which act to normalize the count data obtained from a typical RNA seq workflow to both the length of the genomic feature of interest as well as the sequencing depth for that particular sample as discussed above. As most metabolic labeling experiments described here involve a pulldown step, however, the normalization provided by TPM-type measurements is insufficient, because the resulting abundance measurements are still only known relative to the total set of labeled RNA. Comparison of different time points, essential for calculation of RNA stability, is thus impossible without some sort of normalization that allows for proper scaling of the observed abundances relative to the total (and not only labeled) RNA present in the sample.
7.1. Rationale for the use of spike-ins
To more clearly demonstrate the necessity and utility of a constant reference value for normalization of RNA abundance, we must consider what is actually being measured when one performs an RNA decay experiment where only the labeled RNA is sequenced. Let us represent the abundance of labeled RNA for any given transcript i as Xi,L(t) and the corresponding abundance of unlabeled RNA in the same experiment as Xi,U(t). We can then consider the entire abundance of labeled RNA for all genes at any given time point t to be:
| (7) |
Likewise, the entire abundance of unlabeled RNA for all genes can be represented as:
| (8) |
Thus the total RNA abundance A is simply:
| (9) |
The quantity of interest for the purposes of fitting RNA decay equations is the overall abundance of a labeled RNA of interest relative to total RNA over time, that is,
| (10) |
What is actually measured, however, for any given gene in an RNA decay experiment when only the labeled RNA pool is sequenced is:
| (11) |
Where Ri(t) is the relative abundance of RNA i in the total labeled RNA pool at time point t. Since both Γ(t) and Xi,L are changing throughout the course of an experiment, attempting to fit the RNA decay equations we have described here to raw RPKM measurements is not physically meaningful. However, if one is able to change the variable quantity Γ(t) in the denominator of Eq. 11 into something that is known to be constant throughout the experiment, then R(t) can be transformed into a reliable estimator of RNA abundance on an arbitrary scale. One approach to add a constant to any RNA decay experiment is to add a labeled spike-in RNA at a known ratio 1/d of labeled spike-in to total RNA and normalize RPKM values to that of the measured RPKM of the spike-in. Thus, the spike-in will be added at some constant value S(t) that is a function of A(t):
| (12) |
Now we can modify Eq. 11 to include a known constant amount of spiked-in label S:
| (13) |
Likewise, we can represent the relative abundance of the spike in by Rs(t):
| (14) |
By normalizing the fractional abundance of labeled RNA in the total labeled pool with the spike-in Ri(t) (Eq. 13) to the relative abundance of the spike-in Rs(t) (Eq. 14) we can see that the denominator Γ(t) + S(t) will cancel, resulting in a spike-in normalized estimation of labeled RNA abundance, Ni(t).
| (15) |
Furthermore, upon substitution of S(t) with Eq. 12 we can see that Ni(t) in Eq. 15 is a reliable estimator for the fractional abundance of labeled RNA i in the total RNA A(t) (as sought in Eq. 10) rather than just the labeled RNA pool Γ(t), clearly demonstrating the need for normalization to some source of constant labeled RNA when determining RNA decay rates and half-lives. It is important to note, that any error in the fraction of added spike-in 1/d will add additional noise to the normalized RNA abundance estimate described by Eq. 15.
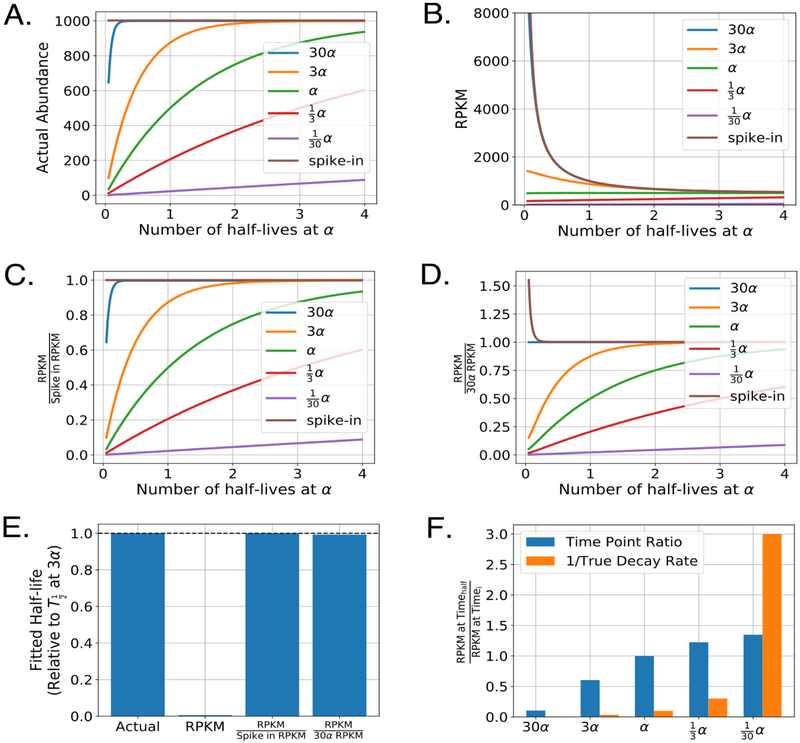
To further illustrate the impact of normalization on the determination of RNA decay rates, we simulated an approach to equilibrium experiment where the bulk RNA representing 99% of the sequences decayed at a rate of α. We then considered several different RNA transcripts each at the same steady state level of overall abundance but at several different multiples of the overall bulk RNA decay rate. The actual abundances of labeled RNA from this simulation can be seen in Figure 6A. We then added in a spike-in RNA at 0.5% of the total RNA for each time point and determined what the resulting RPKM values for each of these transcripts would be under a scenario where labeled RNA is pulled-down with perfect efficiency (Figure 6B). In this simulation, it is evident that the raw RPKM values do not represent the actual RNA abundances. Note that the spike-in RNA rapidly decays in RPKM abundance throughout the time course even though it is added at a constant amount relative to the total RNA. This is to be expected, as in early time points the spiked-in RNA represents the only labeled RNA species in the reaction. As more labeled RNA is created in the cells, the relative fraction of spike-in RNA drops precipitously. However, if we normalize the RPKM traces to the spike-in RPKM (Figure 6C) the actual RNA abundances are exactly reproduced, demonstrating both the utility and necessity of a constant reference value in RNA decay experiments. Therefore, it should not be surprising that many groups advocate for the use of labeled RNA spike-ins when determining RNA half-lives across a variety of model organisms [19, 50, 43, 44, 88, 41].
Figure 6:
Impact of common normalization procedures on the determination of RNA half-lives. A) Simulated labeled mRNA counts for several transcripts decaying at the indicated rates in an approach to equilibrium experiment. For this simulation, bulk RNA (not plotted) decayed at a rate of α and represented 99% of the total RNA sample. Spike-ins were added at 0.5% of the total RNA (that is, sum of labeled and unlabeled). B) Raw RPKM values for each transcript and spike-in RNA. For simplicity, each simulated time point was sequenced to the same depth of 5,000,000 reads and each transcript and spike-in RNA was considered to be the same exact length. Time is indicated in number of half-lives of the bulk RNA, which decays at a rate of α. C) As in B but RPKM values are normalized to the spike-in RPKM values for each sample. D) As in B-C but RPKM values are normalized to a transcript that decays at a rate of 30α. E) Calculated half-lives for the transcript with a decay rate of α. Each half-life was determined by fitting the the approach to equilibrium equation indicated in Figure 5B using non-linear least squares on five evenly spaced time points from the indicated simulated traces in panels A-D. F) Relative mRNA decay as determined by the change in raw RPKM from two time points. Timehalf was chosen to be the time point at exactly one half-life for the bulk RNA. For comparison, orange bars represent the inverse decay rate for each of the indicated transcripts.
7.2. Practical use of labeled spike-ins for RNA decay experiments
For standard quantification of RNA in RNA-seq experiments, a set of agreed upon standards have been adopted and maintained by the External RNA Controls Consortium (ERCC) [89, 90, 91]. Furthermore, spike-ins are seeing widespread use throughout most high-throughput sequencing technologies (reviewed in [92]). However, unlike RNA-seq, no agreed upon set of labeled RNA spike-in standards have been established for RNA decay experiments and the ERCC collection is not available in labeled form. Instead, each lab has developed their own set of standards to use as spike-ins for their system. Tani et al. have established the use of the exogenous luciferase RNA, in vitro transcribed with a known quantity of label, and added to the total purified RNA directly before label selection [19, 50]. Russo et al. use an expensive synthetic labeled positive control that is not reliant upon the efficiency of labeling within an in vitro transcription reaction [88]. Neymotin et al. used a combination of three spike-ins with different lengths from a different organism but with matched GC content to their organism of interest [43]. Likewise, Duffy et al. also use a mix of RNAs from a different organism as a spike in [44]. Finally, Lugowski et al. use two sets of spike-ins, a labeled spike-in of whole genome reads from one organism and an unlabeled spike-in of whole genome reads from a second organism, where both spike-in species originate from organisms that are sufficiently different from the organism of interest [41]. There are several advantages and disadvantages to each of the approaches used above. Spike-ins labeled by in vitro transcription are much cheaper than buying synthetic spike-ins but are also sensitive to variations in the in vitro transcription reaction itself. To mitigate this effect experiments using in vitro transcription to create labeled spike-in RNAs should use RNAs from the same transcription reaction for all samples that are to be compared. From the ERCC experiments, it is evident that sequence bias can have a major impact on measurements from high throughput sequencing experiments [93]. Thus, the use of a single spike-in may not be sufficient for precise measurements of mRNA half-lives. The use of whole-genome labeled RNAs from a non-target organism may help alleviate some of these concerns since a variety of length distributions and sequence compositions are present from those samples, but mismatches between sequence bias in two different organisms can add additional source of noise to the experiment. Additionally, any spike-in is particularly subject to pipetting errors as any mis-quantification of the precise amount of spike-in added to a reaction will add a considerable amount of noise to the quantification procedure, as the spike-in provides the sole normalizing factor for recovering proper decay rates (Eq. 15).
7.3. Spike-in free approaches
Despite the clear utility of a spike-in in estimating RNA abundance, several groups have found additional ways to accurately estimate RNA abundance in RNA decay experiments without using a spike-in RNA [14, 16, 42, 54, 41]. Both Dolken et al. and Schwanhausser et al. use a procedure in which they determine the abundance of both the labeled and unlabeled RNA species by sequencing both the selected labeled RNAs and the unlabeled RNAs found in the unbound fraction, which allows them to determine absolute RNA abundance and decay rates for each transcript, albeit at greater cost than a typical RNA decay experiment [14, 16]. Similarly, Herzog et al. have the ability to measure both labeled and unlabeled pools of RNA abundance with a single sequencing reaction since their method relies on the determination of T to C mutations to determine labeled RNAs and they are able to internally normalize to the total abundance of RNA through this method [54]. Lugowski et al. developed an entirely new pipeline (DRUID) that uses rapidly decaying RNA introns as a constant internal normalization in approach to equilibrium experiments, which they found to be superior to the spike-in based normalization that they attempted in parallel [41]. To illustrate how the DRUID procedure works, we simulated normalization to a rapidly decaying transcript in an approach to equilibrium experiment (Figure 6D). Here it is evident that the rapidly decaying transcript approaches a constant labeled value quickly within the experimental procedure and can be used, instead of a constant spike-in, to normalize the RPKM abundances and recover a true estimator of RNA abundance. We also demonstrate that RNA half-lives determined using the DRUID approach vs. the spike-in approach are able to easily recover the true half-life for a transcript in our simulations (Figure 6E). Lugowski et al. directly compare their DRUID approach to a spike-in approach and find that half-lives determined from the DRUID approach have higher replicate-replicate agreement and also outperformed both spike-in normalization and transcription shutoff experiments when compared against a benchmark dataset [41], possibly due to the pipetting error inherent in the use of spike-ins. In theory, a similar approach could be used for pulse-chase and chase-alone experimental set-ups. However, instead of normalizing to a highly unstable transcript, one would need to normalize to an extremely stable transcript (after sufficient labeling time) as has been suggested by some groups [15, 50]. All such internal-reference approaches provide a potentially simpler workflow than spike-in based methods, and avoid concerns such as pipetting and RNA quantitation errors, but necessitate the identification of extremely unstable or stable pieces of RNA that can be relied upon to have far longer or shorter half lives than any transcripts of biological interest.
As a simpler alternative, Paulsen et al. 2014 suggest measuring the labeled RNA species at just two time points, one time point after a short labeling period, and a second time point chosen at the average half-life of RNA in the organism of interest [42]. A comparison between these two time points can then be made using differential expression software to get a semi-quantitative view of RNA decay at a much lower cost. To illustrate this approach we compared the RPKMs of two different time points in our simulation and compared these ratios with the true decay rates of the transcripts (Figure 6F). Here, it is clear that the rank ordering of the transcript stabilities is preserved, but no interpretation can be given as far as the magnitude change between each of the transcripts, and any attempts to fit a decay rate using such data would fail even if many timepoints were collected. However, the true utility of this approach can be seen when comparing these relative measurements of RNA decay between two different experimental conditions. We further illustrate this approach with a case study in Section 8.
8. Interpretation and follow-up
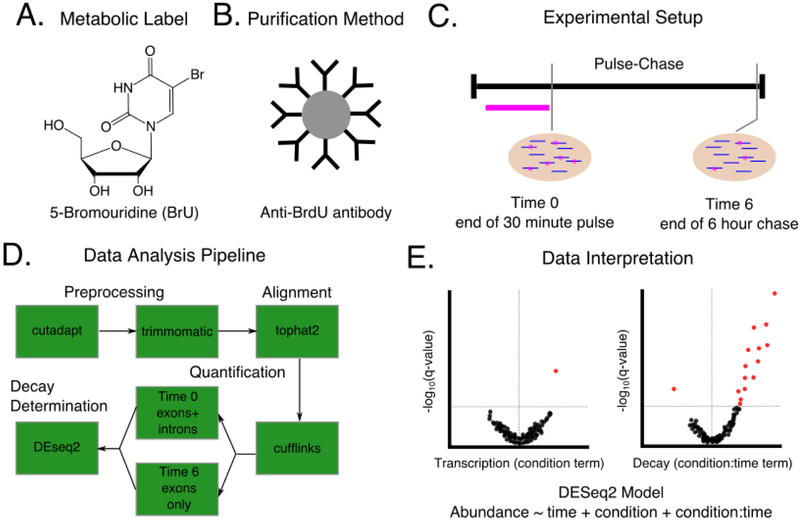
A careful consideration of each aspect of a successful RNA decay experiment can be best described through a sample case study. Consider the scenario where one wants to identify the set of mRNA targets for which RNA decay is primarily mediated by a particular RNA binding protein of interest. To determine possible targets, mRNA decay is measured transcriptome-wide in both mock-treated cells and cells in which the RNA binding protein of interest is knocked down with a silencing RNA. Possible targets will include RNAs that have differential mRNA decay in the knockout genotype compared to the wild-type cells. For this case study, we select an experimental procedure designed to minimize both the cost and cellular manipulations needed to conduct the experiment. Given these constraints, BrU is chosen as the labeling reagent for its low toxicity, cost, and the avoidance of any requirement to incorporate a functional UPRT into the human cell line used for the experiment (Figure 7A). With BrU, an anti-BrdU antibody with known cross-reactivity to BrU is chosen as the selection reagent (Figure 7B). For this particular experiment we are not interested in the exact half-lives of expressed RNAs, but rather the effect of the RNA-binding protein of interest on mRNA decay. Since the RNA binding protein of interest is hypothesized to be involved only in post-transcriptional regulation and not in transcriptional regulation, we are also interested in differentiating transcriptional effects from stability effects. Thus, the pulse-chase experimental design is chosen in order to be able to determine effects on both processes. In this case, we take a single time point at the start of the chase after 30 minutes of labeling and take a second time point at the end of the chase several hours later (Figure 7C). The end of the chase was chosen to coincide with the average mRNA half-life in cultured mammalian cells [55, 56, 16]. To assess biological reproducibility, three replicates for each time point and genotype are performed and analyzed. Three replicates were chosen to be consistent with long RNA-seq ENCODE guidelines which suggest that at least two biological replicates should be used to assess biological reproducibility [94]. Furthermore, the ENCODE ChIP-Seq guidelines suggest that more than two replicates are not absolutely necessary as experiments with RNA pol II indicated that more than two replicates did not increase the number of sites discovered [95, 96]. Since RNA decay experiments have aspects in common with both ChIP-Seq (with an immunoprecipitation step) and RNA-seq (with quantification of RNA abundance), elements of both recommendations are likely applicable here. After RNA quantification, replicate agreement among a single time point can be assessed using rank-based statistics, such as Spearman correlation coefficients. However, correlation coefficients between samples at different timepoints are not meaningful as the RNA abundances are expected to decay at different rates throughout the experiment. Major disagreements between replicates at the same time point can indicate a need for more replicates to better assess variability or a need to repeat the experiment and obtain higher quality samples. It is important to note that this experimental design disfavors detection of regulation of mRNAs with very short or very long half lives. After preparing stranded paired-end libraries for each sample and sending them for sequencing, we perform quality control and clean-up of the sequencing reads using a combination of FastQC, cutadapt, and trimmomatic. Since we want to differentiate transcription effects from decay effects of the RNA binding protein on the transcriptome, we choose to use the splice-aware aligner tophat2 and associated analysis suite cufflinks to assign read counts at both the exon and gene level. We follow the recommendation of Paulsen et al. [42] and use full gene level counts (including exons and introns) at the early time point to measure nascent RNA abundance and use the sum of all possible exons (but not introns) at the late time point to measure mature RNA abundance (Figure 7D). We then take this count data and use a simple model to determine changes in transcription and stability resulting from the RNA binding protein knock down with DEseq2:
| (16) |
Figure 7:
A hypothetical experimental design for determining changes in RNA decay between a WT and RBP knockdown condition. A)-C). The choices to be made in designing the experiment including which metabolic label to use (A), how to purify labeled RNA (B), and the timing of label introduction and sample harvest (C). Note that this experimental design is done in parallel for knockdown and control cells. D) A sample data analysis pipeline to be used to analyze sequencing results from the experiment described in A. Choices must be made at the preprocessing, alignment, quantification, and decay determination stages as indicated in Figure 4. E) Hypothetical volcano plots to visualize the results from the experiment in A-C as analyzed by the pipeline in D. A generalized linear model is used with DEseq2 to determine knockdown specific changes in both transcription and decay. Red dots indicate significant genes as measured by a FDR corrected p-value < 0.1. The vertical gray line in each plot indicates a log2 fold change of zero. For this hypothetical experiment, few genes had a significant change in transcription under the knockdown condition (left plot), but many genes were stabilized in the knockdown condition (right plot), suggesting that experiment identified several genes that can be considered putative targets for the RBP of interest and represent good candidates for targeted experimental follow-ups.
Where A is the abundance of any particular transcript, time is a binary term for the time point (start or end of the chase), condition is a binary term for which condition the RNA is in (knockdown or control) and condition : time is an interaction term between the time and knockdown information. Here, the magnitude and direction of the condition term is interpreted as the knockdown effect on RNA abundance after 30 minutes transcription during the pulse. The magnitude and direction of the interaction term condition : time is interpreted as the knockdown effect on the change in RNA abundance from the start to the end of the chase. After false discovery rate correction using the Benjamini-Hochberg procedure [97], several high confidence targets for the RNA binding protein of interest can be identified and followed up with targeted experiments (Figure 7E).
9. Concluding Remarks
This review provides a general overview of the decisions to be made when planning experiments to globally analyze RNA decay using metabolic labeling coupled with high throughput sequencing. We hope this article will serve as a resource for new and experienced researchers in the field. For additional information, we refer readers to several recent reviews that provide more depth on each topic presented above [98, 99, 87]. With the advent of low cost high-throughput sequencing, measurements of RNA decay at a global scale are broadly achievable. Metabolic labeling of RNA has allowed for the measurement of both transcription rates and decay rates with minimal perturbation of the underlying biology. Recent advances in chemistry have allowed for enhanced selection of labeled RNAs from the pool of total RNA in the cell [44] or removal of the need to select the labeled species from the pool of RNA altogether [54], greatly reducing the amount of starting material needed for these experiments and lowering the overall cost. Additionally, new experimental approaches using the metabolic labels and methods described here have allowed for novel insights into RNA biology including the identification of RNA binding proteins involved in nascent transcription [38], the impact of a single RNA binding protein in amyotrophic lateral sclerosis [62], and the discovery of antisense RNAs expressed during herpes infection [47], to name a few. Future applications can include analysis of RNA metabolism during development, differentiation, the course of the cell cycle, and in response to external cues, stresses and infections. Many open questions in RNA biology involve the kinetics of RNA abundance and the effect of various players on RNA synthesis and degradation, rather than the steady-state abundance of RNA alone. The combination of metabolic labeling with high-throughput sequencing has allowed researchers to address these questions at a global level and will prove to be a valuable asset in the RNA biologist’s toolkit.
Acknowledgments
This work was supported in part by the National Institute of General Medical Sciences, National Institutes of Health grant R35 GM128637 to P.L.F. and grant R01 GM105707 to A.C.G. Additionally M.B.W. is supported by an National Science Foundation Graduate Research Fellowship DGE1256260.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Garneau NL, Wilusz J, Wilusz CJ, The highways and byways of mRNA decay, Nature Reviews Molecular Cell Biology 8 (2) (2007) 113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- [2].Jonas S, Izaurralde E, Towards a molecular understanding of microRNA-mediated gene silencing, Nature Reviews Genetics 16 (7) (2015) 421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- [3].Houseley J, Tollervey D, The Many Pathways of RNA Degradation, Cell 136 (4) (2009) 763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- [4].Bartel DP, Metazoan MicroRNAs, Cell 173 (1) (2018) 20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gerstberger S, Hafner M, Tuschl T, A census of human RNA-binding proteins, Nature Reviews Genetics 15 (12) (2014) 829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Santiago TC, Purvis IJ, Bettany AJ, Brown AJ, The relationship between mRNA stability and length in Saccharomyces cerevisiae., Nucleic Acids Research 14 (21) (1986) 8347–8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nonet M, Scafe C, Sexton J, Young R, Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis., Molecular and Cellular Biology 7 (5) (1987) 1602–1611. doi: 10.1128/MCB.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Herrick D, Parker R, Jacobson A, Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae., Molecular and Cellular Biology 10 (5) (1990) 2269–2284. doi: 10.1128/MCB.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ross J, mRNA stability in mammalian cells., Microbiological Reviews 59 (3) (1995) 423–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Brown AJP, Sagliocco FA, mRNA Abundance and Half-Life Measurements, in: Yeast Protocols, Methods in Molecular Biology™, Humana Press, 1996, pp. 277–295. doi: 10.1385/0-89603-319-8:277. [DOI] [PubMed] [Google Scholar]
- [11].Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC, Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay, Nature Biotechnology 23 (2) (2005) 232–237. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- [12].Munchel SE, Shultzaberger RK, Takizawa N, Weis K, Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay, Molecular Biology of the Cell 22 (15) (2011) 2787–2795. doi: 10.1091/mbc.e11-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Knüppel R, Kuttenberger C, Ferreira-Cerca S, Toward Time-Resolved Analysis of RNA Metabolism in Archaea Using 4-Thiouracil., Frontiers in microbiology 8 (2017) 286–286. doi: 10.3389/fmicb.2017.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, Koszinowski UH, High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay, RNA 14 (9) (2008) 1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, Amit I, Regev A, Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells, Nature Biotechnology 29 (5) (2011) 436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M, Global quantification of mammalian gene expression control, Nature 473 (7347) (2011) 337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- [17].Melvin WT, Milne HB, Slater AA, Allen HJ, Keir HM, Incorporation of 6-Thioguanosine and 4-Thiouridine into RNA, European Journal of Biochemistry 92 (2) (1978) 373–379. doi: 10.1111/j.1432-1033.1978.tb12756.x. [DOI] [PubMed] [Google Scholar]
- [18].Spitzer J, Hafner M, Landthaler M, Ascano M, Farazi T, Wardle G, Nusbaum J, Khorshid M, Burger L, Zavolan M, Tuschl T, Chapter Eight - PAR-CLIP (Photoactivatable Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation): A Step-By-Step Protocol to the Transcriptome-Wide Identification of Binding Sites of RNA-Binding Proteins, in: Lorsch J (Ed.), Methods in Enzymology, Vol. 539 of Laboratory Methods in Enzymology: Protein Part B, Academic Press, 2014, pp. 113–161. doi: 10.1016/B978-0-12-420120-0.00008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, Akimitsu N, Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals, Genome Research 22 (5) (2012) 947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Aspden JL, Jackson RJ, Differential effects of nucleotide analogs on scanning-dependent initiation and elongation of mammalian mRNA translation in vitro, RNA 16 (6) (2010) 1130–1137. doi: 10.1261/rna.1978610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pfefferkorn ER, Pfefferkorn LC, Specific labeling of intracellular Toxoplasma gondii with uracil, The Journal of Protozoology 24 (3) (1977) 449–453. [DOI] [PubMed] [Google Scholar]
- [22].Gay L, Karfilis KV, Miller MR, Doe CQ, Stankunas K, Applying thiouracil tagging to mouse transcriptome analysis, Nature Protocols 9 (2) (2014) 410–420. doi: 10.1038/nprot.2014.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chatzi C, Zhang Y, Shen R, Westbrook GL, Goodman RH, Transcriptional Profiling of Newly Generated Dentate Granule Cells Using TU Tagging Reveals Pattern Shifts in Gene Expression during Circuit Integration,, eNeuro 3 (1). doi: 10.1523/ENEURO.0024-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cleary MD, Chapter 19 Cell Type–Specific Analysis of mRNA Synthesis and Decay In Vivo with Uracil Phosphoribosyltransferase and 4-thiouracil, in: Methods in Enzymology, Vol. 448, Elsevier, 2008, pp. 379–406. doi: 10.1016/S0076-6879(08)02619-0. [DOI] [PubMed] [Google Scholar]
- [25].Sontheimer EJ, Site-specific RNA crosslinking with 4-thiouridine, Molecular Biology Reports 20 (1) (1994) 35–44. doi: 10.1007/BF00999853. [DOI] [PubMed] [Google Scholar]
- [26].Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp A-C, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T, Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP, Cell 141 (1) (2010) 129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zamenhof S, Griboff G, Incorporation of Halogenated Pyrimidines into the Deoxyribonucleic Acids of Bacterium Coli and its Bacteriophages, Nature 174 (4424) (1954) 306–307. doi: 10.1038/174306a0. [DOI] [PubMed] [Google Scholar]
- [28].Eidinoff ML, Cheong L, Rich MA, Incorporation of Unnatural Pyrimidine Bases into Deoxyribonucleic Acid of Mammalian Cells, Science 129 (3362) (1959) 1550–1551. doi: 10.1126/science.129.3362.1550. [DOI] [PubMed] [Google Scholar]
- [29].Gratzner HG, Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication, Science 218 (4571) (1982) 474–475. doi: 10.1126/science.7123245. [DOI] [PubMed] [Google Scholar]
- [30].Haider S, Juan G, Traganos F, Darzynkiewicz Z, Immunoseparation and Immunodetection of Nucleic Acids Labeled with Halogenated Nucleotides, Experimental Cell Research 234 (2) (1997) 498–506. doi: 10.1006/excr.1997.3644. [DOI] [PubMed] [Google Scholar]
- [31].Ohtsu M, Kawate M, Fukuoka M, Gunji W, Hanaoka F, Utsugi T, Onoda F, Murakami Y, Novel DNA Microarray System for Analysis of Nascent mRNAs, DNA Research 15 (4) (2008) 241–251. doi: 10.1093/dnares/dsn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Best MD, Click Chemistry and Bioorthogonal Reactions: Unprecedented Selectivity in the Labeling of Biological Molecules, Biochemistry 48 (28) (2009) 6571–6584. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- [33].Jao CY, Salic A, Exploring RNA transcription and turnover in vivo by using click chemistry, Proceedings of the National Academy of Sciences 105 (41) (2008) 15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Abe K, Ishigami T, Shyu A-B, Ohno S, Umemura S, Yamashita A, Analysis of interferon-beta mRNA stability control after poly(I:C) stimulation using RNA metabolic labeling by ethynyluridine, Biochemical and Biophysical Research Communications 428 (1) (2012) 44–49. doi: 10.1016/j.bbrc.2012.09.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ideue T, Adachi S, Naganuma T, Tanigawa A, Natsume T, Hirose T, U7 small nuclear ribonucleoprotein represses histone gene transcription in cell cycle-arrested cells, Proceedings of the National Academy of Sciences 109 (15) (2012) 5693–5698. doi: 10.1073/pnas.1200523109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Qu D, Zhou L, Wang W, Wang Z, Wang G, Chi W, Zhang B, 5-Ethynylcytidine as a new agent for detecting RNA synthesis in live cells by “click” chemistry, Analytical Biochemistry 434 (1) (2013) 128–135. doi: 10.1016/j.ab.2012.11.023. [DOI] [PubMed] [Google Scholar]
- [37].Hida N, Aboukilila MY, Burow DA, Paul R, Greenberg MM, Fazio M, Beasley S, Spitale RC, Cleary MD, EC-tagging allows cell type-specific RNA analysis, Nucleic Acids Research 45 (15) (2017) e138–e138. doi: 10.1093/nar/gkx551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bao X, Guo X, Yin M, Tariq M, Lai Y, Kanwal S, Zhou J, Li N, Lv Y, Pulido-Quetglas C, Wang X, Ji L, Khan MJ, Zhu X, Luo Z, Shao C, Lim D-H, Liu X, Li N, Wang W, He M, Liu Y-L, Ward C, Wang T, Zhang G, Wang D, Yang J, Chen Y, Zhang C, Jauch R, Yang Y-G, Wang Y, Qin B, Anko M-L, Hutchins AP, Sun H, Wang H, Fu X-D, Zhang B, Esteban MA, Capturing the interactome of newly transcribed RNA, Nature Methods 15 (3) (2018) 213–220. doi: 10.1038/nmeth.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Roundtree IA, Evans ME, Pan T, He C, Dynamic RNA Modifications in Gene Expression Regulation, Cell 169 (7) (2017) 1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sun M, Schwalb B, Schulz D, Pirkl N, Etzold S, Larivière L, Maier KC, Seizl M, Tresch A, Cramer P, Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation, Genome Research 22 (7) (2012) 1350–1359. doi: 10.1101/gr.130161.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lugowski A, Nicholson B, Rissland OS, DRUID: A pipeline for transcriptome-wide measurements of mRNA stability, RNA (2018) rna.062877.117doi: 10.1261/rna.062877.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Paulsen MT, Veloso A, Prasad J, Bedi K, Ljungman EA, Magnuson B, Wilson TE, Ljungman M, Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA, Methods 67 (1) (2014) 45–54. doi: 10.1016/j.ymeth.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Neymotin B, Athanasiadou R, Gresham D, Determination of in vivo RNA kinetics using RATE-seq, RNA 20 (10) (2014) 1645–1652. doi: 10.1261/rna.045104.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Duffy EE, Rutenberg-Schoenberg M, Stark CD, Kitchen RR, Gerstein MB, Simon MD, Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry, Molecular Cell 59 (5) (2015) 858–866. doi: 10.1016/j.molcel.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chaiet L, Frank J. W, The properties of streptavidin, a biotin-binding protein produced by Streptomycetes, Archives of Biochemistry and Biophysics 106 (1964) 1–5. doi: 10.1016/0003-9861(64)90150-X. [DOI] [PubMed] [Google Scholar]
- [46].Duffy EE, Simon MD, Enriching s4U-RNA Using Methane Thiosulfonate (MTS) Chemistry, Current Protocols in Chemical Biology 8 (4) (2016) 234–250. doi: 10.1002/cpch.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wyler E, Menegatti J, Franke V, Kocks C, Boltengagen A, Hennig T, Theil K, Rutkowski A, Ferrai C, Baer L, Kermas L, Friedel C, Rajewsky N, Akalin A, Dölken L, Grässer F, Landthaler M, Widespread activation of antisense transcription of the host genome during herpes simplex virus 1 infection, Genome Biology 18 (1). doi: 10.1186/s13059-017-1329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Warfield L, Ramachandran S, Baptista T, Devys D, Tora L, Hahn S, Transcription of Nearly All Yeast RNA Polymerase II-Transcribed Genes Is Dependent on Transcription Factor TFIID, Molecular Cell 68 (1) (2017) 118–129.e5. doi: 10.1016/j.molcel.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Core LJ, Waterfall JJ, Lis JT, Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters, Science (New York, N.Y.) 322 (5909) (2008) 1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Imamachi N, Tani H, Mizutani R, Imamura K, Irie T, Suzuki Y, Akimitsu N, BRIC-seq: A genome-wide approach for determining RNA stability in mammalian cells, Methods 67 (1) (2014) 55–63. doi: 10.1016/j.ymeth.2013.07.014. [DOI] [PubMed] [Google Scholar]
- [51].Ierusalimsky VN, Balaban PM, Long-living RNA in the CNS of terrestrial snail, RNA Biology 15 (2) (2018) 207–213. doi: 10.1080/15476286.2017.1411460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Palozola KC, Donahue G, Liu H, Grant GR, Becker JS, Cote A, Yu H, Raj A, Zaret KS, Mitotic transcription and waves of gene reactivation during mitotic exit, Science 358 (6359) (2017) 119–122. doi: 10.1126/science.aal4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Holmberg A, Blomstergren A, Nord O, Lukacs M, Lundeberg J, Uhlén M, The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures, ELECTROPHORESIS 26 (3) (2005) 501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- [54].Herzog VA, Reichholf B, Neumann T, Rescheneder P, Bhat P, Burkard TR, Wlotzka W, von Haeseler A, Zuber J, Ameres SL, Thiol-linked alkylation of RNA to assess expression dynamics, Nature Methods 14 (12) (2017) 1198–1204. doi: 10.1038/nmeth.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, D. JE Jr, Decay Rates of Human mRNAs: Correlation With Functional Characteristics and Sequence Attributes, Genome Research (2003) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS, Database for mRNA Half-Life of 19 977 Genes Obtained by DNA Microarray Analysis of Pluripotent and Differentiating Mouse Embryonic Stem Cells, DNA Research 16 (1) (2009) 45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Paulsen MT, Veloso A, Prasad J, Bedi K, Ljungman EA, Tsan Y-C, Chang C-W, Tarrier B, Washburn JG, Lyons R, Robinson DR, Kumar-Sinha C, Wilson TE, Ljungman M, Coordinated regulation of synthesis and stability of RNA during the acute TNF-induced proinflammatory response, Proceedings of the National Academy of Sciences 110 (6) (2013) 2240–2245. doi: 10.1073/pnas.1219192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Nikolov EN, Dabeva MD, Re-utilization of pyrimidine nucleotides during rat liver regeneration., Biochemical Journal 228 (1) (1985) 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Phillips R, Kondev J, Theriot J, Garcia HG, Orme N, Physical Biology of the Cell, London ; New York, NY: : Garland Science, [2013], 2013. [Google Scholar]
- [60].Greenberg JR, High Stability of Messenger RNA in Growing Cultured Cells, Nature 240 (5376) (1972) 102–104. doi: 10.1038/240102a0. [DOI] [PubMed] [Google Scholar]
- [61].Lefkofsky HB, Veloso A, Ljungman M, Transcriptional and post-transcriptional regulation of nucleotide excision repair genes in human cells, Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 776 (2015) 9–15. doi: 10.1016/j.mrfmmm.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tank EM, Figueroa-Romero C, Hinder LM, Bedi K, Archbold HC, Li X, Weskamp K, Safren N, Paez-Colasante X, Pacut C, Thumma S, Paulsen MT, Guo K, Hur J, Ljungman M, Feldman EL, Barmada SJ, Abnormal RNA stability in amyotrophic lateral sclerosis, Nature Communications 9 (1) (2018) 2845. doi: 10.1038/s41467-018-05049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, Mortazavi A, A survey of best practices for RNA-seq data analysis, Genome Biology 17. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hrdlickova R, Toloue M, Tian B, RNA-Seq methods for transcriptome analysis, Wiley interdisciplinary reviews. RNA 8 (1). doi: 10.1002/wrna.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads, EMB-net.journal 17 (1) (2011) pp. 10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- [66].Bolger AM, Lohse M, Usadel B, Trimmomatic: A flexible trimmer for Illumina Sequence Data, Bioinformatics (2014) btu170doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Andrews S, FastQC: A quality control tool for high throughput sequence data (2010). [Google Scholar]
- [68].Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2, Nature Methods 9 (4) (2012) 357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL, TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions, Genome Biology 14 (4) (2013) R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: Ultrafast universal RNA-seq aligner, Bioinformatics 29 (1) (2013) 15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification, Nature Biotechnology 34 (5) (2016) 525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- [72].Baruzzo G, Hayer KE, Kim EJ, Di Camillo B, FitzGerald GA, Grant GR, Simulation-based comprehensive benchmarking of RNA-seq aligners, Nature Methodsdoi: 10.1038/nmeth.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sedlazeck FJ, Rescheneder P, von Haeseler A, NextGenMap: Fast and accurate read mapping in highly polymorphic genomes, Bioinformatics 29 (21) (2013) 2790–2791. doi: 10.1093/bioinformatics/btt468. [DOI] [PubMed] [Google Scholar]
- [74].Kim D, Langmead B, Salzberg SL, HISAT: A fast spliced aligner with low memory requirements, Nature Methods 12 (4) (2015) 357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, Salmon: Fast and bias-aware quantification of transcript expression using dual-phase inference, Nature methods 14 (4) (2017) 417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, The Sequence Alignment/Map format and SAMtools, Bioinformatics 25 (16) (2009) 2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks, Nature Protocols 7 (3) (2012) 562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL, StringTie enables improved reconstruction of a transcriptome from RNA-seq reads, Nature biotechnology 33 (3) (2015) 290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Anders S, Pyl PT, Huber W, HTSeq—a Python framework to work with high-throughput sequencing data, Bioinformatics 31 (2) (2015) 166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Li B, Dewey CN, RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome, BMC Bioinformatics 12 (1) (2011) 323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biology 15 (2014) 550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Robinson MD, McCarthy DJ, Smyth GK, edgeR: A Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics 26 (1) (2010) 139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, Limma powers differential expression analyses for RNA-sequencing and microarray studies, Nucleic Acids Research 43 (7) (2015) e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Pimentel H, Bray NL, Puente S, Melsted P, Pachter L, Differential analysis of RNA-seq incorporating quantification uncertainty, Nature Methods 14 (7) (2017) 687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- [85].Friedman J, Hastie T, Tibshirani R, The Elements of Statistical Learning, Vol. 1, Springer series in statistics Springer, Berlin, 2001. [Google Scholar]
- [86].Schwalb B, Schulz D, Sun M, Zacher B, Dümcke S, Martin DE, Cramer P, Tresch A, Measurement of genome-wide RNA synthesis and decay rates with Dynamic Transcriptome Analysis (DTA), Bioinformatics 28 (6) (2012) 884–885. doi: 10.1093/bioinformatics/bts052. [DOI] [PubMed] [Google Scholar]
- [87].Lugowski A, Nicholson B, Rissland OS, Determining mRNA half-lives on a transcriptome-wide scale, Methods 137 (2018) 90–98. doi: 10.1016/j.ymeth.2017.12.006. [DOI] [PubMed] [Google Scholar]
- [88].Russo J, Heck AM, Wilusz J, Wilusz CJ, Metabolic labeling and recovery of nascent RNA to accurately quantify mRNA stability, Methods 120 (2017) 39–48. doi: 10.1016/j.ymeth.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Cronin M, Ghosh K, Sistare F, Quackenbush J, Vilker V, O’Connell C, Universal RNA Reference Materials for Gene Expression, Clinical Chemistry 50 (8) (2004) 1464–1471. doi: 10.1373/clinchem.2004.035675. [DOI] [PubMed] [Google Scholar]
- [90].External RNA Controls Consortium, Proposed methods for testing and selecting the ERCC external RNA controls, BMC Genomics 6 (1) (2005) 150. doi: 10.1186/1471-2164-6-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].The External RNA Controls Consortium, Baker SC, Bauer SR, Beyer RP, Brenton JD, Bromley B, Burrill J, Causton H, Conley MP, Elespuru R, Fero M, Foy C, Fuscoe J, Gao X, Gerhold DL, Gilles P, Goodsaid F, Guo X, Hackett J, Hockett RD, Ikonomi P, Irizarry RA, Kawasaki ES, Kaysser-Kranich T, Kerr K, Kiser G, Koch WH, Lee KY, Liu C, Liu ZL, Lucas A, Manohar CF, Miyada G, Modrusan Z, Parkes H, Puri RK, Reid L, Ryder TB, Salit M, Samaha RR, Scherf U, Sendera TJ, Setterquist RA, Shi L, Shippy R, Soriano JV, Wagar EA, Warrington JA, Williams M, Wilmer F, Wilson M, Wolber PK, Wu X, Zadro R, The External RNA Controls Consortium: A progress report, Nature Methods 2 (2005) 731–734. doi: 10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- [92].Hardwick SA, Deveson IW, Mercer TR, Reference standards for next-generation sequencing, Nature Reviews Genetics 18 (8) (2017) 473–484. doi: 10.1038/nrg.2017.44. [DOI] [PubMed] [Google Scholar]
- [93].Seqc/Maqc-Iii Consortium, Su Z, Łabaj PP, Li S, Thierry-Mieg J, Thierry-Mieg D, Shi W, Wang C, Schroth GP, Setterquist RA, Thompson JF, Jones WD, Xiao W, Xu W, Jensen RV, Kelly R, Xu J, Conesa A, Furlanello C, Gao H, Hong H, Jafari N, Letovsky S, Liao Y, Lu F, Oakeley EJ, Peng Z, Praul CA, Santoyo-Lopez J, Scherer A, Shi T, Smyth GK, Staedtler F, Sykacek P, Tan X-X, Thompson EA, Vandesompele J, Wang MD, Wang J, Wolfinger RD, Zavadil J, Auerbach SS, Bao W, Binder H, Blomquist T, Brilliant MH, Bushel PR, Cai W, Catalano JG, Chang C-W, Chen T, Chen G, Chen R, Chierici M, Chu T-M, Clevert D-A, Deng Y, Derti A, Devanarayan V, Dong Z, Dopazo J, Du T, Fang H, Fang Y, Fasold M, Fernandez A, Fischer M, Furió-Tari P, Fuscoe JC, Caimet F, Gaj S, Gandara J, Gao H, Ge W, Gondo Y, Gong B, Gong M, Gong Z, Green B, Guo C, Guo L, Guo L-W, Hadfield J, Hellemans J, Hochreiter S, Jia M, Jian M, Johnson CD, Kay S, Kleinjans J, Lababidi S, Levy S, Li Q-Z, Li L, Li L, Li P, Li Y, Li H, Li J, Li S, Lin SM, López FJ, Lu X, Luo H, Ma X, Meehan J, Megherbi DB, Mei N, Mu B, Ning B, Pandey A, Pérez-Florido J, Perkins RG, Peters R, Phan JH, Pirooznia M, Qian F, Qing T, Rainbow L, Rocca-Serra P, Sambourg L, Sansone S-A, Schwartz S, Shah R, Shen J, Smith TM, Stegle O, Stralis-Pavese N, Stupka E, Suzuki Y, Szkotnicki LT, Tinning M, Tu B, van Delft J, Vela-Boza A, Venturini E, Walker SJ, Wan L, Wang W, Wang J, Wang J, Wieben ED, Willey JC, Wu P-Y, Xuan J, Yang Y, Ye Z, Yin Y, Yu Y, Yuan Y-C, Zhang J, Zhang KK, Zhang W, Zhang W, Zhang Y, Zhao C, Zheng Y, Zhou Y, Zumbo P, Tong W, Kreil DP, Mason CE, Shi L, A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium, Nature Biotechnology 32 (9) (2014) 903–914. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].ENCODE Project Consortium, An integrated encyclopedia of DNA elements in the human genome, Nature 489 (7414) (2012) 57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, Chen Y, DeSalvo G, Epstein C, Fisher-Aylor KI, Euskirchen G, Gerstein M, Gertz J, Hartemink AJ, Hoffman MM, Iyer VR, Jung YL, Karmakar S, Kellis M, Kharchenko PV, Li Q, Liu T, Liu XS, Ma L, Milosavljevic A, Myers RM, Park PJ, Pazin MJ, Perry MD, Raha D, Reddy TE, Rozowsky J, Shoresh N, Sidow A, Slattery M, Stamatoyannopoulos JA, Tolstorukov MY, White KP, Xi S, Farnham PJ, Lieb JD, Wold BJ, Snyder M, ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia, Genome Research 22 (9) (2012) 1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB, PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls, Nature Biotechnology 27 (1) (2009) 66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Benjamini Y, Hochberg Y, Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing, J. R. Statist. Soc. B 57 (1) (1995) 289–300. [Google Scholar]
- [98].Tani H, Akimitsu N, Genome-wide technology for determining RNA stability in mammalian cells: Historical perspective and recent advantages based on modified nucleotide labeling, RNA Biology 9 (10) (2012) 1233–1238. doi: 10.4161/rna.22036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wada T, Becskei A, Impact of Methods on the Measurement of mRNA Turnover, International Journal of Molecular Sciences 18 (12) (2017) 2723. doi: 10.3390/ijms18122723. [DOI] [PMC free article] [PubMed] [Google Scholar]