Abstract
In synthetic biology approaches, lipid vesicles are widely used as protocell models. While many compounds have been encapsulated in vesicles (e.g. DNA, cytoskeleton and enzymes), the incorporation of glycocalyx components in the lipid bilayer has attracted much less attention so far. In recent years, glycoconjugates have been integrated in the membrane of giant unilamellar vesicles (GUVs). These minimal membrane systems have largely contributed to shed light on the molecular mechanisms of cellular processes. In this review, we first introduce several preparation and biophysical characterization methods of GUVs. Then, we highlight specific applications of protocells investigating glycolipid-mediated endocytosis of toxins, viruses and bacteria. In addition, we delineate how prototissues have been assembled from glycan-decorated protocells by using lectin-mediated cross-linking of opposed glycoreceptors (e.g. glycolipids and glycopeptides). In future applications, glycan-decorated protocells might be useful for investigating cell–cell interactions (e.g. adhesion and communication). We also speculate about the implication of lectin–glycoreceptor interactions in membrane fusion processes.
Keywords: synthetic glycobiology, lectins, glycosphingolipids, endocytosis, fluorescence microscopy, atomic force microscopy
1. Introduction
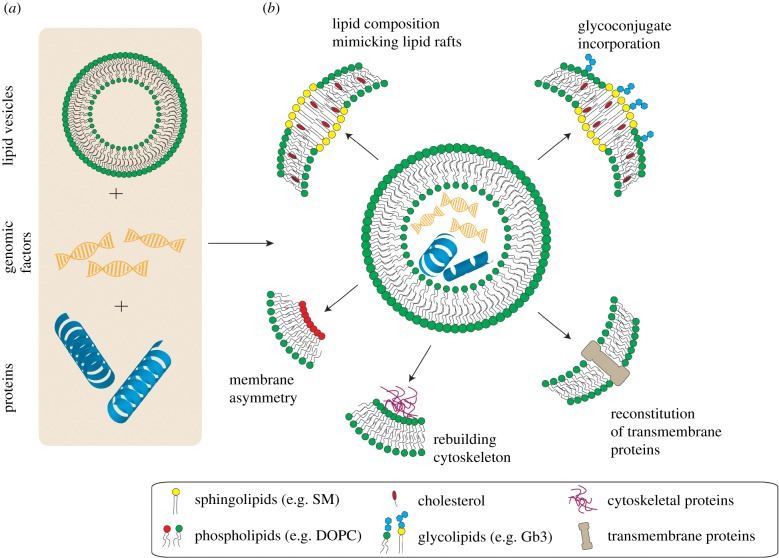
Since the mid-twentieth century, scientists from different disciplines such as chemistry, physics and biology have devoted enormous efforts to build an artificial cell, which would ideally present several features of cellular life, e.g. self-maintenance, self-reproduction and evolvability [1–3]. Creating an artificial cell, also termed protocell [4–6], which provides most of the above-mentioned properties, is extremely difficult and has not been accomplished yet. In a bottom-up approach, non-living components are assembled together to form an ensemble that can replicate the cellular life [7,8]. The concept is to start with a simple and limited number of constituents and then increase the complexity of the ensemble by integrating more and more natural or synthetic building blocks in order to fill the gap between non-living and living worlds [7,9]. Extensively used building blocks for protocell construction are lipid vesicles, also called liposomes (figure 1a).
Figure 1.
The protocell concept. (a) Assembly of non-living matter such as lipid vesicles, genomic factors (e.g. DNA/RNA) and other cellular components (e.g. proteins) in a bottom-up approach to build a protocell. (b) Protocells can be varied with respect to the membrane composition to reconstitute different aspects of native cell membranes such as the coexistence of different domains (lipid rafts) or leaflet asymmetry. Moreover, membrane components such as glycoconjugates or transmembrane proteins as well as cytosolic components (e.g. actin filaments) can be incorporated into protocells. (Online version in colour.)
In the aqueous environment, distinct lipids such as phospholipids self-assemble into free-standing lipid bilayer spheres due to their geometric shape (in particular, cylinder-shaped) and amphiphilic nature (consisting of a polar water-soluble head group attached to a water-insoluble hydrocarbon tail) [10,11]. Since vesicles possess physical and chemical properties related to cellular membranes, such as interaction with the surrounding environment (permeability) and physical integrity (stability), they are well suited as an artificial membrane model to rebuild a native cell [4,12]. Moreover, cellular constituents such as RNA/DNA or enzymes, which are reproduction and evolutionary factors in living cells, can be incorporated in vesicles (figure 1a). A more detailed review about experimental endeavours for encapsulating life encoders into vesicles can be found in [1,7].
Cellular membranes are selective barriers, which allow the uptake of essential nutrients and the transfer of ions necessary for cellular metabolism and maintenance. As they also protect the cellular body from foreign matter, membranes play a vital role in cellular life. The cell membrane was first postulated as a bi-dimensional fluid formed as a lipid bilayer in which proteins are homogeneously distributed [13]. Over time, several experimental evidences mostly based on in vitro assays of detergent solubilization supported the idea that distinct lipid domains may exist in biological cell membranes [14,15]. These findings were finally concluded in the ‘lipid raft’ hypothesis, which proposes the existence of membrane domains that are enriched in specific lipid species, in particular in sphingomyelin (SM) and cholesterol, and associated with special types of proteins [16]. It is supposed that raft domains dynamically form and dissociate in the exoplasmic leaflet and function as a platform for membrane signalling and trafficking [17–19].
Glycosphingolipids (GSLs) are components of biological membranes, which consist of a ceramide backbone covalently attached to carbohydrate moieties. In the exoplasmic leaflet, GSLs are mostly associated with ordered membrane domains (e.g. lipid rafts or caveolae), together with SM and cholesterol [16,20–22]. The carbohydrate parts of GSLs are exposed to the extracellular environment and act as receptors for carbohydrate-binding proteins, so-called lectins. GSL-enriched domains are involved in cell–cell communication and cell adhesion [16,23]. In addition to these functions, host GSLs can be hijacked by several pathogens and pathogenic products (e.g. toxins) in order to gain access to cells [24,25].
Decorating protocells with natural and synthetic glycoconjugates represents a novel direction in the emerging field of synthetic glycobiology and will allow scientists to enlighten their understanding of several biological processes such as endocytosis and cell adhesion. In this review, we briefly explain some production and characterization methods of lipid-based protocells and then highlight recent achievements using glycan-decorated protocells in basic science.
2. Preparation and characterization of protocells
A protocell can be described as a confined object comparable in size to cells. Vesicles can be produced in various sizes from tens of nanometres to tens of micrometres. Giant unilamellar vesicles (GUVs) with sizes ranging from 5 to 50 µm represent the most popular biomimetic systems that resemble mammalian cells. The hydration method [26] is a conventional and relatively simple technique to form vesicles; however, it usually yields multilamellar vesicles. Over time, several other techniques like electroformation [27,28], gel- [29] or paper-assisted hydration [30], inverted emulsion [31] and microfluidics [32,33] have been developed to produce vesicles in a more efficient and controllable manner, and to assemble more complex GUVs. Figure 1b depicts various lipid-based protocells that can be established by modifying the membrane composition and/or encapsulating biological or chemical components. Liquid-ordered domains (Lo), which are considered as artificial structures resembling lipid rafts in cell membranes, can be reconstituted in protocells by mixing three lipid components, a low melting temperature lipid (e.g. dipalmitoylphosphatidylcholine, DOPC), cholesterol and SM as high melting temperature lipid. The fabrication of GUVs with an asymmetric lipid bilayer (containing different lipid species in the inner and the outer membrane leaflets) [34,35] or the encapsulation of cytoskeletal proteins [36,37] are other cases of increased complexity. Incorporation of natural or synthetic glycolipids [38,39] to GUVs can be achieved with all common GUV preparation techniques by usually mixing the glycolipid with other lipid species. At low concentrations of glycolipids, they are normally fully dispersed in the GUV membrane. The reconstitution of functional membrane proteins into GUVs is yet more challenging [40,41]. High yield GUV production in more relevant physiological solutions [42–45] and recently in phosphate-buffered saline [46] represents other attempts to bridge from protocells to native cells.
Since the first establishment of GUVs, their basic properties such as size, shape and uniformity could be non-destructively measured by conventional optical microscopy techniques like phase contrast microscopy [45]. The lamellarity of GUVs is generally judged based on the fluorescence intensity measurements of fluorescently labelled GUVs [45], although a label-free optical method based on differential interference contrast was proposed to measure the GUV lamellarity [47]. Taking advantage of fluorescent labelling of lipids and proteins, many studies, including most of those cited in §3, have been conducted using different fluorescence microscopy techniques.
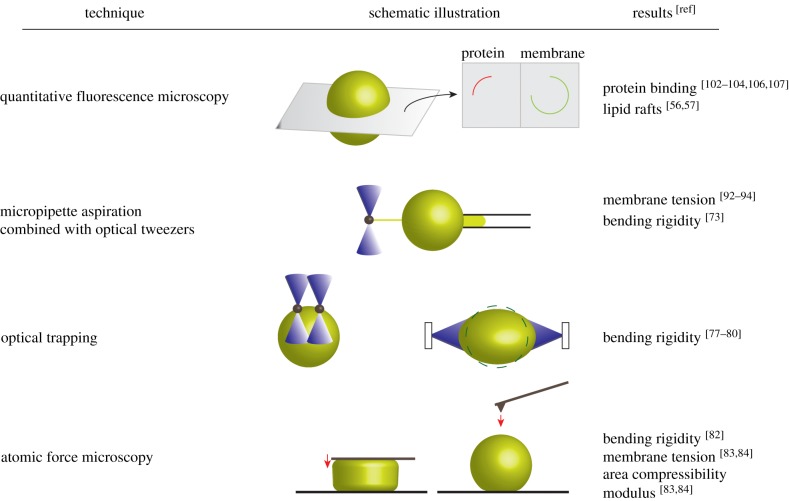
A small portion of fluorescently labelled lipids can be easily added to the lipid composition as a membrane marker. By optical sectioning microscopies, such as confocal microscopy, the dynamics and binding efficiencies of fluorescently labelled proteins (with different excitation/emission wavelengths compared with the membrane marker) can be quantified by measuring the (mean) intensity of fluorescent signals from cross sections of GUVs (figure 2). Automated programmes for detection, tracking and computation of protein binding to GUVs have facilitated the quantitative analysis of a huge number of microscopy datasets (as images or movies) [48,49].
Figure 2.
Selected characterization methods of GUVs. Fluorescence microscopy techniques can be used to visualize and quantify protein binding to a lipid vesicle in different cross sections. Preferential partitioning of fluorescently labelled molecules into different lipid phases (liquid-disordered or liquid-ordered) can be monitored. Different physical properties of membranes can be assessed by several micromanipulation techniques, e.g. micropipette aspiration (solely or in combination with optical tweezers), optical trapping of beads or entire GUVs and atomic force microscopy (as parallel plate compression or local indentation measurement. (Online version in colour.)
Despite notable advancements in experimental techniques, in particular in high-resolution fluorescence microscopy, the direct observation of membrane domains in native cellular membranes remains challenging, and thus the lipid raft hypothesis is still a topic of debate [50,51]. Nowadays, lipid rafts in cell membranes are defined as dynamic, nanometre-sized domains [19]. In membrane model systems, such as GUVs, the demixing of membrane components in more ordered (liquid-ordered, Lo) and less ordered (liquid-disordered, Ld) domains has been reported about two decades ago [52]. The coexistence of different phases with sizes above the optical resolution (greater than 200 nm) can be perceived due to preferential partitioning of different membrane markers in different phases [52–55]. Even though liquid-ordered domains in model membranes are considered as simplified equivalents of native lipid rafts, we feel obliged to remark the discrepancy in size and in composition. The dynamics of ordered, ‘raft-like’ domains in GUVs can be studied by various fluorescence microscopy techniques, e.g. fluorescence recovery after photobleaching (FRAP) or, in molecular detail, by fluorescence correlation spectroscopy (FCS) [56,57]. FCS offers a less destructive method with limited photobleaching and phototoxicity compared to FRAP [57]. Partitioning of GSLs in different phases can also be studied in GUVs. Fricke & Dimova [58] demonstrated the coexistence of liquid-disordered and gel-like phases in GUVs at room temperature in the presence of monosialotetrahexosylganglioside (GM1) (above 5 mol%) by fluorescence microscopy. In accordance, liquid-ordered/liquid-disordered coexistence induced by the GM1 receptor was reported in [59].
In many cellular processes, e.g. cell division, autophagy and endocytosis, the cell and, particularly, the cell membrane undergo drastic morphological changes that are inevitably linked to membrane elastic properties. As a pioneering work [60], Helfrich proposed that the total elastic energy of a vesicle is a combination of stretching (i.e. change in area) and bending (i.e. change in curvature) energies. The unknown mechanical parameters in Helfrich's theory are membrane tension (for stretching energy term), bending rigidity and membrane spontaneous curvature (for bending energy term). Afterwards, a significant number of theoretical [61,62] and experimental [63–65] studies have been inspired by Helfrich's theory to measure the above-mentioned properties for fluid membranes. The free-standing giant vesicles with no steric constraint were used as a popular protocell model.
The membrane spontaneous curvature is influenced by any kind of asymmetry across the lipid bilayer. Ideally, when a membrane has perfectly symmetric inner and outer leaflets, symmetric transmembrane compositions and, moreover, the inner and outer solutions have a symmetric distribution of ions or particles, the membrane spontaneous curvature is zero [66]. Although the membrane spontaneous curvature is clearly not zero in biological membranes, most experimental works on GUVs have focused on measuring bending rigidity and membrane tension [64]. The reason was the limitation of experimental techniques that cannot simultaneously assess the bending rigidity and spontaneous curvature. The micropipette aspiration (MA) approach [67,68], where the vesicle surface is aspirated into a micropipette by applying a defined suction pressure [69,70], is the most common micromanipulation technique to measure membrane tension and bending rigidity of vesicles. The membrane tension and area expansion modulus can be determined from the vesicle radius, pipette radius and length of the aspirated part for an applied pressure (figure 2). At low tension (less than 0.5 mN m−1), the bending rigidity can be computed from the slope of the logarithm of the tension versus area expansion modulus [65,71]. Using the MA method, Lu et al. [71] interestingly reported that the bending modulus and area expansion modulus of synthetic asymmetric lipid bilayers were up to 50% larger than the values acquired for symmetric bilayers. Pulling outward membrane tubules from aspirated GUVs (figure 2) by trapped beads with electromagnetic field [72] or optical tweezers (OTs) [73] has enabled the direct measurement of bending rigidity independent of area expansion and, consequently, can be used to characterize stiffer GUVs (e.g. multicomponent fluid vesicles displaying higher membrane tension) [72,73]. Combinations of MA and OT techniques have been applied to glycan-decorated vesicles to study the relation between membrane tension and endocytosis events (membrane invagination, tubulation; further detailed in the next section). Recently, Dasgupta et al. [74] presented a combined MA/OT technique to pull inward and outward tubules from GM1-containing GUVs to assess the spontaneous curvature parameter generated by GM1 molecules. In a special application of micromanipulation by OT [75,76], two trapped micrometre-sized beads were attached to the vesicle surface (figure 2). While one bead was kept fixed, the other bead was moved to bend the membrane. Neglecting the membrane stretching and knowing the trap stiffness and the displacement between two beads, one can compute the membrane-bending rigidity. With this approach, Dimova et al. [77] could measure the elastic properties of dimyristoylphosphatidylcholine containing vesicles during transition from fluid to solid (gel) phases. The whole GUV can be trapped and stretched in a dual-beam optical trap to measure the bending rigidity [78,79] and even the viscoelastic response of a vesicle [80] (figure 2).
Even though less frequently applied than MA, atomic force microscopy (AFM) has been employed in combination with GUVs to study the mechanical properties of a lipid bilayer [81]. Here, a micrometre-sized beam called a cantilever (tipless or with a small tip) compresses the membrane (figure 2). The deflection of the cantilever (which later converts to force) and the indentation depth are measured. By applying contact mechanic theories to measured parameters, elastic properties can be quantified. Dieluweit et al. [82] implemented AFM force measurements on protein-coated vesicles to determine the bending rigidity, although they reported unsuccessful efforts for uncoated phosphatidylcholine (SOPC) containing vesicles. Interestingly, the immobilization of GUVs on substrates can assist the AFM force measurement. The membrane tension and area compressibility modulus of empty and actin-encapsulated phosphatidylcholine (DOPC) vesicles have been measured by performing AFM force measurements on sessile GUVs [83,84]. To our knowledge, there exists no AFM study to examine the mechanics of glycan-decorated protocells, despite of the fact that AFM-based theoretical [85] and experimental [86] works have investigated the role of the glycocalyx layer in cell mechanics.
For more detailed literature and further characterization methods like optical microscopy-based techniques (e.g. fluctuation spectroscopy or membrane deformation induced by an electric or magnetic field), readers are referred to comprehensive reviews [64,65] and references therein.
3. Application of glycan-decorated protocells in endocytosis
Multiple endocytic pathways, such as phagocytosis, macro- and micropinocytosis (including clathrin-dependent and caveolin-dependent endocytosis, CLIC/GEEC pathway and others), have been proposed to function in eukaryotic cells [87–90], whereby extracellular molecules/particles (e.g. nutrients, growth factors and also pathogens) as well as plasma membrane constituents are internalized. In general, an endocytic process starts with the segregation and concentration of molecules at the plasma membrane, which leads to a local change of membrane shape. For clathrin-dependent pathways, it is well known that the recruitment of several molecular machineries (e.g. clathrin coats and dynamin) induces inward membrane bending to form a clathrin-coated pit. However, membrane bending and formation of membrane invaginations can also occur in the absence of molecular regulators as proposed in clathrin-independent mechanisms [91]. Finally, the membrane pit/invagination is detached from the membrane and specifically transported as vesicle to intracellular destinations. Saleem et al. [92] reconstituted clathrin assembly by employing GUVs and observed that clathrin polymerization and membrane budding are counteracted by membrane tension. In another example of using GUVs to study endocytosis processes, Roux et al. [93] showed that polymerization of dynamin, which plays a role in the formation of endocytic vesicles through membrane fission, generates enough force to deform GUV membranes at low membrane tension. The BAR domain protein endophilin A1, which is another protein involved in clathrin-dependent endocytosis, induced the growth of membrane buds and tubulation on flaccid GUVs [94]. Above-mentioned studies, which have been performed on GUVs, highlight the role of proteins that are involved in the well-known clathrin-dependent endocytic pathway. But could GUVs be employed in studies, which aim at elucidating clathrin-independent mechanisms, for instance lipid-driven endocytosis?
The discovery of gangliosides (i.e. GSLs containing one or more sialic acids) as receptors of several bacterial toxins dates back to 40 years ago [95,96]. For instance, GM1 and GD1b are specifically bound by cholera toxin (CT) and tetanus toxin, respectively [95,97]. The specific binding of these toxins to gangliosides associated with lipid raft domains activates a cascade of signalling events, which leads to toxin internalization into host cells. Also in this field, bottom-up synthetic approaches have been proved useful for elucidating the molecular mechanisms of toxin uptake. Preparation of vesicles containing the glycolipid of interest made it possible to investigate the role of toxin–receptor interactions leading to uptake. The ganglioside GM1 is targeted by CT, which is secreted by the bacterium Vibrio cholera, one of the main responsible for diarrhoea disease [98,99]. The fully assembled CT consists of a catalytic A-subunit (CTA) and a non-toxic, pentameric B-subunit (CTB), which mediates the internalization of the toxin into intestinal epithelial cells, probably through multivalent binding of (up to five) GM1 receptors [99].
Using fluorescently labelled GUVs, Hammond et al. [100] demonstrated that the binding of CTB to GM1 receptors separates the membrane into raft-like (liquid-ordered, Lo) and non-raft (liquid-disordered, Ld) phases even though the vesicle membrane had initially one uniform phase (either Lo or Ld). The authors also observed that CTB–GM1 complexes partitioned into Lo domains, which was consistent with the belief that GM1 receptors are associated with raft domains [100]. It has been reported that GM1 also functions as the natural receptor for simian virus 40 (SV40), which binds via its pentameric VP1 capsid proteins [101]. Ewers et al. showed that SV40-like particles (composed of 72 VP1 proteins) induce the formation of tubular membrane invaginations in energy-depleted GM1-expressing cells [102]. To prove the sufficiency of SV40–GM1 interactions for the formation of membrane invaginations, the authors prepared GUVs containing different GM1 receptor species (i.e. natural and synthetic ones mainly differing in the fatty acyl chain length and degree of saturation) and observed membrane tubulation only for specific GM1 species. Membrane-phase separation and Lo/Ld domain formation occurred shortly after SV40 binding to GM1 [102], as observed for CT [100]. Finally, a clathrin-independent, lipid-mediated mechanism was proposed as SV40 initiates its internalization process by inducing membrane curvature through multivalent binding of its VP1 pentamers to host cell GM1 receptors [102].
Shiga toxin (Stx), which is produced by Shigella dysenteriae and enterohaemorrhagic Escherichia coli strains, is another prominent example, where the application of protocells has largely contributed to a better understanding of toxin uptake. Similar to CT, Stx is composed of a toxic A-subunit and a receptor-binding B-subunit. The homopentameric B-subunit of Shiga toxin (STxB) specifically binds up to 15 globotriaosyl ceramide (Gb3; also known as CD77 or the Pk blood group antigen) molecules [38,103]. Römer et al. studied StxB-induced tubular membrane invaginations in cells and GUVs. StxB-triggered Gb3 clustering led to the formation of membrane tubules in vesicles with lower membrane tension. By contrast, StxB formed micrometre-sized domains when highly tensed vesicles were used [104]. By probably similar mechanisms, the fucose-binding bacterial lectin RSL from Ralstonia solanacearum is able to induce tubular membrane invaginations in GUVs containing fucosylated glycolipids (figure 3) [106].
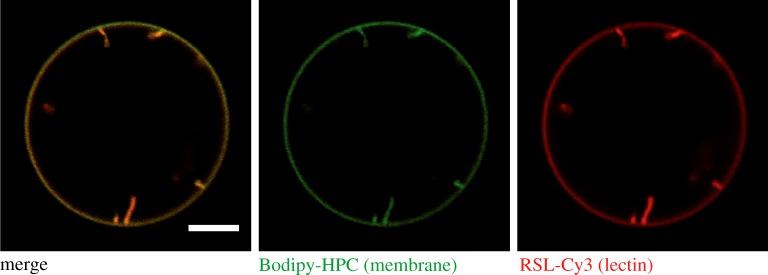
Figure 3.
Lectin-induced membrane invaginations. GUVs containing the synthetic fucosylated glycolipid FSL-Lewis a were incubated with Cy3-labelled RSL (200 nM, red colour). Confocal microscopy images from the equatorial plane of GUVs show lectin binding to the membrane (labelled with beta-BODIPY FL C5-HPC, green colour) as well as inward membrane tubulation. GUVs were composed of DOPC/cholesterol/FSL-Lewis a/beta-BODIPY FL C5-HPC (64 : 30 : 5 : 1 mol%) and prepared by the electroformation [105] technique. Scale bar: 5 µm. (Online version in colour.)
When investigating the interactions of the StxB-binding site mutant W34A with GUVs containing a natural mixture of Gb3 receptors, neither Gb3 clustering nor domain formation could be observed, despite adequate StxB binding to GUVs. The result was similar for wild-type StxB binding when the saturated Gb3 species Gb3-C22 : 0 was used as single receptor molecules, demonstrating that the formation of membrane invaginations crucially depends on toxin structure and valency as well as on receptor structure [104]. The impact of the configuration of the 2OH group of the fatty acyl chain in Gb3 receptors on StxB-induced membrane tubulation was demonstrated in [107]. The subtle change in chirality of the receptor altered membrane organization and consequently the propensity of tubule formation [107]. Similarly, it has been shown that engineered RSLs (neoRSLs) with controlled number (0–6) and position of binding sites bound to glycolipid-containing GUVs, but differed in inducing tubular membrane invaginations [106,108].
Furthermore, the impact of actin polymerization on the scission of STxB-induced endocytic membrane tubules in the absence of dynamin was demonstrated using GUVs [109]. Polymerizing actin supplies additional mechanical stress in synergy with already generated stresses at the boundary region of membrane domains (formed by StxB) and favours the scission process [109].
As the formation of membrane invaginations has been observed for several bacterial toxins and viruses, the question arose if also the much bigger bacteria exploit this concept of glycolipid-mediated entry into host cells by receptor clustering. Eierhoff et al. [110] showed the engulfment of the bacterium Pseudomonas aeruginosa into Gb3-containing GUVs when the bacterium expresses the tetrameric, Gb3-binding lectin LecA.
In this section we have highlighted some applications of protocells to gain a more detailed understanding of endocytosis processes. For more details, especially about lipid-driven endocytosis, we refer readers to previously published reviews [105,111,112]. Except for the combined MA/OT techniques, which have been used in above-mentioned examples [92,93,109], other micromanipulation techniques can be applied to study, for example, phagocytosis in artificial membrane systems. Meinel et al. [113] investigated energy and force profiles during the uptake of an optically trapped bead into GUVs. In this study, the authors used uncoated beads, and GUVs were made from egg-PC lipids. In future studies, the physical interactions of lectin-coated beads (to mimic a bacterium) with glycolipid-containing GUVs (to mimic the host plasma membrane) would be a logical extension of this work. Similarly, AFM cantilevers can be functionalized with lectin-coated beads and adhesion forces with glycan-containing vesicles can be recorded. These studies would reveal further insights into the role of bacterial lectins and host cell glycans in the attachment and uptake of bacteria.
4. Assembly of prototissues by protocell cross-linking and adhesion
In multicellular organisms, cell adhesion to neighbouring cells or the extracellular matrix is very crucial for the integrity and maintenance of tissues and organs as well as for cellular processes such as migration or tissue development. In several diseases like cancer [114,115] or arthritis [116], cell adhesion is aberrantly transformed. Moreover, to initiate bacterial and viral infection, bacteria or viruses first need to adhere to host cells, which is very often established by lectin–carbohydrate interactions [117]. The cell adhesion phenomenon is mainly mediated by the interaction of transmembrane proteins called cell adhesion molecules (CAMs), which are classified into four groups: cadherins (Ca2+-dependent adhesion molecules), immunoglobulin-like CAM superfamily, integrins and selectins [118]. Selectins are carbohydrate-binding molecules, which facilitate the attachment of two different cell types (heterotypic adhesion). For instance, during an inflammatory response, activated endothelial cells express E-selectin molecules on their surface. The interaction of E-selectin with glycoproteins of circulating leucocytes in the blood slows down the leucocyte motion. Hence, more leucocytes attach to the endothelial layer of blood vessels and therefore leave the blood stream towards inflammatory locations [116]. The heterodimeric integrins represent another type of cell adhesion molecules that function as attachment regulators to the extracellular matrix and as a signal transmitter from the extracellular environment into the cell, and vice versa [119]. For instance, integrins are involved in cell migration, extracellular matrix assembly, cell growth and proliferation [120,121]. They specifically interact with the RGD peptide of extracellular macromolecules such as fibrinogen or fibronectin [122].
Studying the interaction of receptor-containing giant vesicles with ligands immobilized on a surface or diffusing in a supported lipid bilayer can mimic cell–matrix and cell–cell adhesion processes, respectively [123]. The integrin–RGD interaction was reconstituted by incorporating a fluorescently labelled RGD peptide into vesicles, which then adhered to an integrin-functionalized substrate [124]. Reflection interference contrast microscopy (RICM) and its modifications represent techniques to quantify inter-surface distances and membrane fluctuations [123,125,126], and therefore provide quantitative information about different stages of adhesion (nucleation, spreading until saturation). Simple membrane models like GUVs with a controllable lipid composition are well-suited samples for RICM to measure adhesion kinetics of strong (e.g. biotin–streptavidin [127]) or weak (e.g. E-selectin to a glycolipid carrying sialyl-LewisX (SLex) [128]) interactions. For instance, growth kinetics of the adhesion area of integrin-containing GUVs on a fibrinogen-coated substrate have been measured by RICM [122]. In a recent publication, preferential partitioning of platelet integrin αIIbβ3 into Ld domains of phase-separated GUVs has been observed in both inactivated and activated (presence of DTT and Mn2+) states. Fibrinogen binding to activated integrins did not change the preference of the integrin αIIbβ3 heterodimer to Ld domains [129]. Moreover, homotypic cell–cell adhesion was modelled by incorporation of epithelial cadherin (E-cadherin) from the cadherin family into protocells and supported lipid bilayers [130,131]. In the framework of another experimental approach, spreading dynamics of GUVs comprising an actin cortex on a substrate exhibited dependency on the density of the cortical shell. Vesicles with sparse actin shell density behaved like an empty vesicle, while the vesicle behaviour was reminiscent of cells with high actin shell density [132]. The spreading behaviour of a novel type of protocells called droplet-stabilized GUVs (dsGUVs) on different matrices was investigated. The integrin αIIbβ3-functionalized dsGUVs spread better on fibrinogen compared to fibronectin or collagen matrices [133]. Not only through molecular binding, vesicles can also adhere to the substrate by the electrostatic interaction [134].
Furthermore, and without any doubt, multiple glycocalyx components are involved in cell recognition and adhesion [135]. To investigate the adhesion process, glycan-decorated vesicles can be implemented to mimic the glycocalyx layer of the cell surface. Stuhr-Hansen et al. [136] reconstituted another component of the glycocalyx layer by integrating synthesized, cholesterol-anchored glycopeptides into GUVs and checked their accessibility with lectin binding.
The cooperativity between multiple eukaryotic cells and the extracellular matrix in a tissue carries out a specific function that a single isolated cell is unable to fulfil. For years, studies about minimal cell models have focused on single protocells, and the cooperativity has not been addressed. As a model for primitive cellular assemblies, a colony of giant vesicles has been assembled by polypeptide-mediated electrostatic attraction. Enhanced vesicle fusion and solute capture on the colony surface, which are two principal mechanisms in living cells, have been observed compared to isolated vesicles [137,138]. In nature, however, this cellular cooperativity and communication are conducted by cell adhesion molecules. For the first time, a network of protocells has been recently assembled based on lectin–glycan interactions. Using multivalent lectins with opposing binding sites for carbohydrate moieties, Villringer et al. could cross-link giant vesicles in a controlled (and reversible) manner forming a mimic of cell–cell junctions (protocellular junctions). Furthermore, cell–matrix adhesion was mimicked by adhering cross-linked vesicles to a substrate via biotin–streptavidin interactions. Hence, the authors succeeded in engineering a stable prototissue from giant vesicles presenting both cell–cell and cell–substrate adhesion properties as an important step towards synthetic minimal tissues. The competition of repulsion forces (originating from the glycocalyx coat that covers the cell surface) and specific ligand–receptor attraction forces (lectin–glycan interactions in this example) was mimicked by inserting different concentrations of lipopolymers into GUVs [139]. Figure 4 displays a prototissue, which was formed from GUVs containing the GSL receptor Gb3 by cross-linking GUVs with the tetrameric lectin LecA. The vesicles were carrying a small quantity of biotinylated lipids in order to adhere to a streptavidin-functionalized surface (figure 4b,c). In another study, Ribeiro et al. engineered the first bispecific lectin (termed Janus lectin) with two distinct recognition surfaces for fucosylated and sialylated glycoconjugates, respectively. This bispecific lectin could bridge protocells carrying two different types of glycoconjugates and form a network of heterogeneous protocells [140].
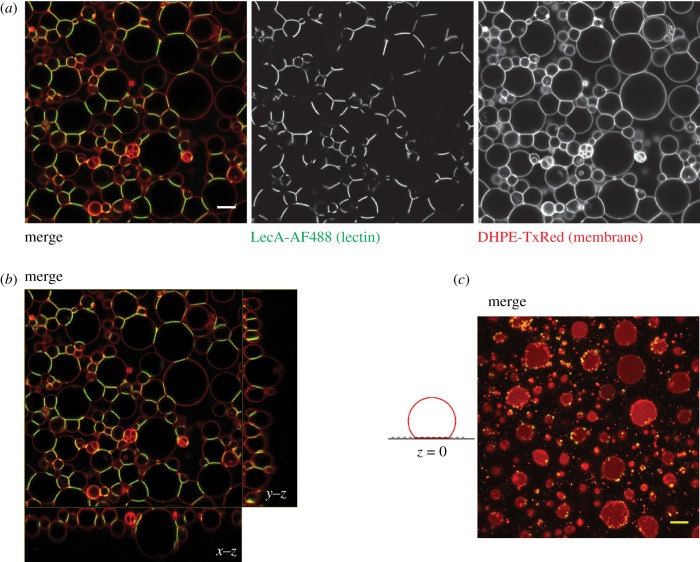
Figure 4.
Assembly of prototissues by lectin–glycan interactions. (a) Confocal sections of an arbitrary plane of a prototissue made by cross-linking Gb3-containing GUVs (membrane marker: DHPE-TxRed, red colour) in the presence of Alexa488-labelled lectin LecA (200 nM, green colour). (b) Representative x–y, x–z and y–z cross sections of the merge image of (a), demonstrating the attachment of most GUVs to the streptavidin-coated coverslip. (c) For better clarity, the saturated adhesion patches of GUVs (same image as in (a) and (b)) are shown at z = 0, as depicted in the schematic illustration. GUVs were prepared by electroformation [105] with a lipid composition of DOPC/cholesterol/Gb3/DOPE-biotin/DHPE-TxRed (64 : 30 : 5:0.5 : 0.5 mol%). Scale bar: 5 µm. (Online version in colour.)
As discussed in §2, membrane physical parameters can be investigated in single isolated vesicles using micromanipulation techniques. We suppose that those techniques, for instance AFM, can also be technically applicable to characterize single vesicles in prototissues. Comparison of measured membrane physical parameters in prototissues to those in single isolated vesicles might be used to study the effect of adhesion (due to protocell–protocell or protocell–substrate attachment) on lipid bilayer properties. Figure 5 displays a schematic illustration of a prototissue formed from cross-linked protocells. As in the experimental design, vesicles are cross-linked to each other through binding of the tetrameric lectin LecA to the GSL Gb3 receptors from adjacent vesicles. In cross-linked regions, clustering of lectin–receptor complexes occurs and deforms the membrane. Attachment to the substrate is provided by biotin–streptavidin interaction (not all vesicles can adhere to the substrate due to the limited space). The AFM cantilever can indent individual protocells and provide quantitative information about membrane mechanical properties.
Figure 5.
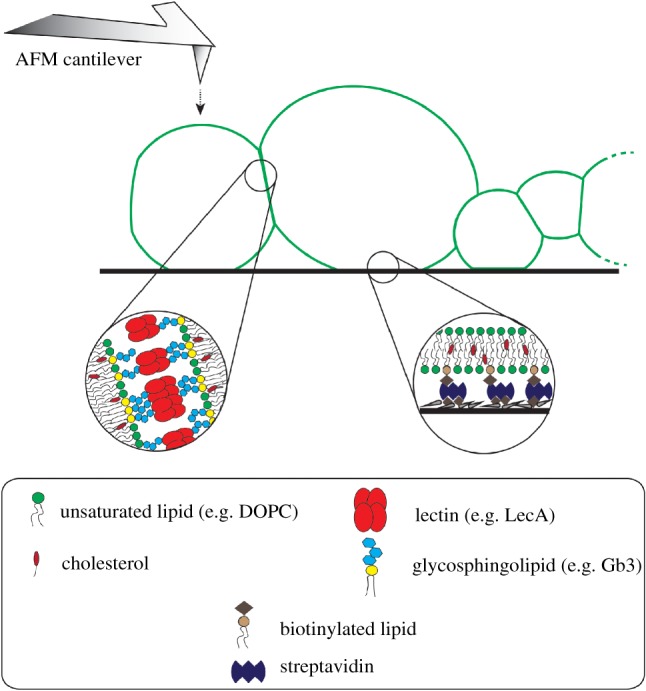
Schematic illustration of prototissues and possible application of micromanipulation techniques on prototissues. The schematic illustration depicts protocell–protocell junctions and protocell–substrate adhesion in a prototissue (not to scale). The protocell–protocell adhesion is regulated by lectin–glycan interactions, while the protocell–substrate is controlled by biotin–streptavidin attachment. The atomic force microscopy technique might be applied to prototissues to measure membrane physical parameters as discussed in §2 for single vesicles. (Online version in colour.)
Very recently, Bartelt et al. [141] were able to move GUVs across a substrate by introducing asymmetry in the GUV–substrate adhesion. This study represents a first step towards the understanding of cell motility by using minimal protocells.
5. Application of protocells in membrane fusion studies
As a simple description, membrane fusion is a process whereby two separate lipid bilayers merge to become one. This controlled membrane fusion process is essential for basic cellular functions like uptake and transport of nutrients or hormones (endo- and exocytosis), fertilization (fusion of sperm and oocyte) and signalling in nerve cells (synaptic vesicle exocytosis) [142,143]. Membrane fusions of two mitochondria or endosome membranes are some examples of ubiquitous and continuous fusion processes between intracellular compartments. Moreover, several viruses such as HIV enter the cytosol and infect host cells by fusing their viral envelope to the host's membrane. Despite the diversity of biological membrane fusion processes with respect to the involved fusogenic proteins (SNARE superfamily [144], haemagglutinin [145]), the location (mitochondria, endosome or cell plasma membrane) and the spatial (nano- to micro-sized contact zone between two membranes) and temporal (milliseconds to minutes) scale, it has been postulated that most membrane fusion processes follow a similar order of events nicely explained by the stalk model: first, two membranes are brought into contact by specific tethering proteins. After overcoming electrostatic forces, the lipids of the contact zone are at a very close proximity and a local perturbation of bilayer structure emerges. It is followed by the generation of an intermediate neck (either hemifusion stalk or hemifusion diaphragm, HD) and finally a fusion pore opens and expands until two membranes become united [143,146,147].
To investigate protein–lipid interactions involved in the membrane fusion process, synthetic membrane models have been extensively implemented. The fusion of single large unilamellar vesicles (LUVs, diameter of approx. 100 nm), which carry fusogenic proteins, either with supported lipid bilayers [148,149], the plasma membrane [150] or other LUVs [151], has been studied with different analytical techniques. The fluorescence resonance energy transfer (FRET) technique is frequently applied in vesicle fusion assays. The principle is based on the energy transfer level between a fluorophore called donor and another one termed acceptor. The energy transfer efficiency is inversely related to the distance between them. The FRET efficiency will decrease if a liposome carrying both donor and acceptor fluorophore fuses with a non-labelled liposome indicating the increase of distance between donor and acceptor and fusion-associated lipid mixing [152]. Mixing of the content of liposomes is another indirect evidence for membrane fusion. Two distinct vesicle populations are differently loaded, each with one part of a complex (e.g. TbCl3 and dipicolinic acid (DPA), respectively). Upon vesicle fusion the Tb3+/DPA complex is assembled, which results in a much higher fluorescence intensity than can be detected for Tb3+ alone [152].
However, due to their small size, LUVs could not provide direct observation of intermediate steps of membrane fusion processes in order to experimentally demonstrate the stalk/hemifusion hypothesis. In contrast, fusion steps can be directly observed and monitored by using GUVs in combination with optical (fluorescence) microscopy techniques [153–155]. With the combination of micropipette aspiration and optical video microscopy, two trapped GUVs were brought into contact and the formation of the fusion neck was imaged in a ligand-mediated fusion process [156]. The formation and expansion dynamics of the intermediate neck were also monitored in electroporation-based fusion [156]. Moreover, the micrometre-sized HD formation in viral peptide-induced fusion was observed in giant vesicles [157]. Fusion of LUVs with sessile GUVs (i.e. immobilized on the substrate) of different membrane tension in the presence of SNARE proteins showed an increase in fusion efficiency for more tensed vesicles [158].
As mentioned in previous sections, glycans are important in initial adhesion events between cells or cells and pathogens. Pathogenic virulence factors such as lectins that bind to host cell glycans have not yet been shown to be implicated in membrane fusion processes. However, modifying their structure might turn them into fusogenic molecules.
6. Conclusion
Synthetic membrane models, such as GUVs, have been proved in the past to be fairly helpful tools in mimicking cellular membranes and hence in understanding cellular processes. Nevertheless, we should be aware of and not ignore that native membranes are highly complex systems consisting of a multitude of lipid species and proteins that exhibit specific functions. By integrating diverse glycoconjugates (from glycolipids, over glycopeptides and glycoproteins to proteoglycans) into the lipid bilayer of protocells, an environment resembling the glycocalyx layer of native cells is created. These novel features will allow mimicking and grasping more complex cell adhesion and cell–cell communication processes in future studies and may finally lead to the development of bioinspired applications and therapies.
Acknowledgements
We thank Dr Sarah Villringer for providing the raw image data of figure 4.
Data accessibility
This article has no additional data.
Authors' contributions
R.O. and W.R. designed and wrote the manuscript.
Competing interests
The authors declare no conflict of interest.
Funding
This work has been supported by the Excellence Initiative of the German Research Foundation (EXC 294) and the German Research Foundation (grant RO 4341/3-1).
References
- 1.Luisi PL, Ferri F, Stano P. 2006. Approaches to semi-synthetic minimal cells: a review. Naturwissenschaften 93, 1–13. ( 10.1007/s00114-005-0056-z) [DOI] [PubMed] [Google Scholar]
- 2.Pohorille A, Deamer D, Pohorille A. 2002. Artificial cells: prospects for biotechnology. Trends Biotechnol. 20, 123–128. ( 10.1016/S0167-7799(02)01909-1) [DOI] [PubMed] [Google Scholar]
- 3.Hammer DA, Kamat NP. 2012. Towards an artificial cell. FEBS Lett. 586, 2882–2890. ( 10.1016/j.febslet.2012.07.044) [DOI] [PubMed] [Google Scholar]
- 4.Sakuma Y, Imai M. 2015. From vesicles to protocells: the roles of amphiphilic molecules. Life 5, 651–675. ( 10.3390/life5010651) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen S, Chen L, Deamer D, Krakauer DC, Packard NH, Stadler PF, Rasmussen S. 2004. Transitions from nonliving to living matter. Science 303, 963–965. ( 10.1126/science.1093669) [DOI] [PubMed] [Google Scholar]
- 6.Hest JCM. 2017. Artificial cells: synthetic compartments with life-like functionality and adaptivity. Acc. Chem. Res. 50, 769–777. ( 10.1021/acs.accounts.6b00512) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwille P. 2015. Jump-starting life? Fundamental aspects of synthetic biology. J. Cell Biol. 210, 687–690. ( 10.1083/jcb.201506125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noireaux V, Maeda YT, Libchaber A. 2011. Development of an artificial cell, from self-organization to computation and self-reproduction. Proc. Natl Acad. Sci. USA 108, 3473–3480. ( 10.1073/pnas.1017075108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwille P. 2011. Bottom-up synthetic biology: engineering in a tinkerer's world. Science 333, 1252–1254. ( 10.1126/science.1211701) [DOI] [PubMed] [Google Scholar]
- 10.Israelachvili JN, Mitchell DJ, Ninham BW. 1977. Theory of self-assembly of lipid bilayers and vesicles. Biochim. Biophys. Acta 470, 185–201. ( 10.1016/0005-2736(77)90099-2) [DOI] [PubMed] [Google Scholar]
- 11.Israelachvili JN. 2011. Intermolecular and surface forces. New York, NY: Academic Press. [Google Scholar]
- 12.Monnard PA, Deamer DW. 2011. Membrane self-assembly processes: steps toward the first cellular life. Minimal Cell Biophys. Cell Compart. Orig. Cell Funct. 207, 123–151. [DOI] [PubMed] [Google Scholar]
- 13.Singer SJ, Nicolson GL. 1972. The fluid mosaic model of the structure of cell membranes. Science 175, 720–731. ( 10.1126/science.175.4023.720) [DOI] [PubMed] [Google Scholar]
- 14.Seddon AM, Curnow P, Booth PJ. 2004. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta Biomembr. 1666, 105–1617. [DOI] [PubMed] [Google Scholar]
- 15.Lingwood D, Simons K. 2007. Detergent resistance as a tool in membrane research. Nat. Protoc. 2, 2159–2165. ( 10.1038/nprot.2007.294) [DOI] [PubMed] [Google Scholar]
- 16.Simons K, Ikonen E. 1997. Functional rafts in cell membranes. Nature 387, 569–572. ( 10.1038/42408) [DOI] [PubMed] [Google Scholar]
- 17.Simons K, Toomre D. 2000. lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–41. ( 10.1038/35036052) [DOI] [PubMed] [Google Scholar]
- 18.Brown DA, London E. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14, 111–136. ( 10.1146/annurev.cellbio.14.1.111) [DOI] [PubMed] [Google Scholar]
- 19.Lingwood D, Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science 46, 46–51. ( 10.1126/science.1174621) [DOI] [PubMed] [Google Scholar]
- 20.Sych T, Mély Y, Römer W. 2018. Lipid self-assembly and lectin-induced reorganization of the plasma membrane. Phil. Trans. R. Soc. B 373, 20170117 ( 10.1098/rstb.2017.0117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooper NM. 1999. Detergent-insoluble glycosphingolipid/cholesterol-rich membrane domains, lipid rafts and caveolae. Mol. Membr. Biol. 16, 145–156. ( 10.1080/096876899294607) [DOI] [PubMed] [Google Scholar]
- 22.Fra AM, Williamson E, Simons K. 1994. Detergent-insoluble glycolipid microdomains in lymphocytes in the absence of caveolae. Biochemistry 269, 30745–30748. [PubMed] [Google Scholar]
- 23.Kasahara K, Sanai Y. 1999. Possible roles of glycosphingolipids in lipid rafts. Biophys. Chem. 82, 121–127. ( 10.1016/S0301-4622(99)00111-8) [DOI] [PubMed] [Google Scholar]
- 24.Riethmüller J, Riehle A, Grassmé H, Gulbins E. 2006. Membrane rafts in host-pathogen interactions. Biochim. Biophys. Acta Biomembr. 1758, 2139–2147. [DOI] [PubMed] [Google Scholar]
- 25.Pizarro-Cerdá J, Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell 124, 715–727. ( 10.1016/j.cell.2006.02.012) [DOI] [PubMed] [Google Scholar]
- 26.Reeves JP, Dowben RM. 1969. Formation and properties of thin-walled phospholipid vesicles. J. Cell. Physiol. 73, 49–60. ( 10.1002/jcp.1040730108) [DOI] [PubMed] [Google Scholar]
- 27.Angelova MI, Dimitrov DS. 1986. Liposome electro-formation. Faraday Discuss. Chem. Soc. 81, 303–311. ( 10.1039/dc9868100303) [DOI] [Google Scholar]
- 28.Angelova MI, Soléau S, Méléard P, Faucon F, Bothorel P. 1992. Preparation of giant vesicles by external AC electric fields. Kinetics and applications. In Trends in colloid and interface science VI, pp. 127–131. Berlin, Germany: Springer. [Google Scholar]
- 29.Weinberger A, et al. 2013. Gel-assisted formation of giant unilamellar vesicles. Biophys. J. 105, 154–164. ( 10.1016/j.bpj.2013.05.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kresse KM, Xu M, Pazzi J, García-Ojeda M, Subramaniam AB. 2016. Novel application of cellulose paper as a platform for the macromolecular self-assembly of biomimetic giant liposomes. ACS Appl. Mater. Interfaces 8, 32 102–32 107. ( 10.1021/acsami.6b11960) [DOI] [PubMed] [Google Scholar]
- 31.Pautot S, Frisken BJ, Weitz DA. 2003. Production of unilamellar vesicles using an inverted emulsion. Langmuir 19, 2870–2879. ( 10.1021/la026100v). [DOI] [Google Scholar]
- 32.Karamdad K, Law RV, Seddon JM, Brooks NJ, Ces O. 2015. Preparation and mechanical characterisation of giant unilamellar vesicles by a microfluidic method. Lab Chip 15, 557–562. ( 10.1039/C4LC01277A) [DOI] [PubMed] [Google Scholar]
- 33.Van Swaay D, Demello A.. 2013. Microfluidic methods for forming liposomes. Lab Chip 13, 752–767. ( 10.1039/c2lc41121k) [DOI] [PubMed] [Google Scholar]
- 34.Karamdad K, Law RV, Seddon JM, Brooks NJ, Ces O. 2016. Studying the effects of asymmetry on the bending rigidity of lipid membranes formed by microfluidics. Chem. Commun. 52, 5277–5280. ( 10.1039/C5CC10307J) [DOI] [PubMed] [Google Scholar]
- 35.Hu PC, Li S, Malmstadt N. 2011. Microfluidic fabrication of asymmetric giant lipid vesicles. ACS Appl. Mater. Interfaces 3, 1434–1440. ( 10.1021/am101191d) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai FC, Stuhrmann B, Koenderink GH. 2011. Encapsulation of active cytoskeletal protein networks in cell-sized liposomes. Langmuir 27, 10 061–10 071. ( 10.1021/la201604z) [DOI] [PubMed] [Google Scholar]
- 37.Pontani LL, Van Der Gucht J, Salbreux G, Heuvingh J, Joanny JF, Sykes C.. 2009. Reconstitution of an actin cortex inside a liposome. Biophys. J. 96, 192–198. ( 10.1016/j.bpj.2008.09.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johannes L, Römer W. 2010. Shiga toxins from cell biology to biomedical applications. Nat. Rev. Microbiol. 8, 105–116. ( 10.1038/nrmicro2279) [DOI] [PubMed] [Google Scholar]
- 39.Migas UM, Abbey L, Velasco-Torrijos T, McManus JJ. 2014. Adding glycolipid functionality to model membranes—phase behaviour of a synthetic glycolipid in a phospholipid membrane. Soft Matter 10, 3978–3983. ( 10.1039/C4SM00147H) [DOI] [PubMed] [Google Scholar]
- 40.Dezi M, Di Cicco A, Bassereau P, Levy D.. 2013. Detergent-mediated incorporation of transmembrane proteins in giant unilamellar vesicles with controlled physiological contents. Proc. Natl Acad. Sci. USA 110, 7276–7281. ( 10.1073/pnas.1303857110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rigaud J-L, Levy D. 2003. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 372, 65–86. ( 10.1016/S0076-6879(03)72004-7) [DOI] [PubMed] [Google Scholar]
- 42.Montes LR, Alonso A, Goñi FM, Bagatolli LA. 2007. Giant unilamellar vesicles electroformed from native membranes and organic lipid mixtures under physiological conditions. Biophys. J. 93, 3548–3554. ( 10.1529/biophysj.107.116228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein H, Spindler S, Bonakdar N, Wang C, Sandoghdar V. 2017. Production of isolated giant unilamellar vesicles under high salt concentrations. Front. Physiol. 8, 63 ( 10.3389/fphys.2017.00063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q, Wang X, Ma S, Zhang Y, Han X. 2016. Electroformation of giant unilamellar vesicles in saline solution. Colloids Surf. B Biointerfaces 147, 368–375. ( 10.1016/j.colsurfb.2016.08.018) [DOI] [PubMed] [Google Scholar]
- 45.Akashi K, Miayata H, Itoh H, Kazuhika K. 1996. Preparation of giant liposomes in physiological conditions and their characterization under an optical microsocpe. Biophys. J. 71, 3242–3250. ( 10.1016/S0006-3495(96)79517-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lefrançois P, Goudeau B, Arbault S. 2018. Electroformation of phospholipid giant unilamellar vesicles in physiological phosphate buffer. Integr. Biol. 10, 429–434. ( 10.1039/C8IB00074C) [DOI] [PubMed] [Google Scholar]
- 47.McPhee CI, Zoriniants G, Langbein W, Borri P. 2013. Measuring the lamellarity of giant lipid vesicles with differential interference contrast microscopy. Biophys. J. 105, 1414–1420. ( 10.1016/j.bpj.2013.07.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hermann E, Bleicken S, Subburaj Y, García-Sáez AJ. 2014. Automated analysis of giant unilamellar vesicles using circular Hough transformation. Bioinformatics 30, 1747–1754. ( 10.1093/bioinformatics/btu102) [DOI] [PubMed] [Google Scholar]
- 49.Sych T, et al. In press. GUV-AP: multifunctional FIJI-based tool for quantitative image analysis of giant unilamellar vesicles. Bioinformatics. ( 10.1093/bioinformatics/bty962) [DOI] [PubMed] [Google Scholar]
- 50.Munro S. 2003. Lipid rafts: elusive or illusive? Cell 115, 377–388. ( 10.1016/S0092-8674(03)00882-1) [DOI] [PubMed] [Google Scholar]
- 51.Yethiraj A, Weisshaar JC. 2007. Why are lipid rafts not observed in vivo? Biophys. J. 93, 3113–3119. ( 10.1529/biophysj.106.101931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dietrich C, Bagatolli LA, Volovyk ZN, Thompson NL, Levi M, Jacobson K, Gratton E. 2001. Lipid rafts reconstituted in model membranes. Biophys. J. 80, 1417–1428. ( 10.1016/S0006-3495(01)76114-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumgart T, Hunt G, Farkas ER, Webb WW, Gerald W. 2009. Fluorescence probe partitioning between Lo/Ld phases in lipid membranes. Biochim. Biophys. Acta 1768, 2182–2194. ( 10.1016/j.bbamem.2007.05.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kilin V, Glushonkov O, Herdly L, Klymchenko A, Richert L, Mely Y. 2015. Fluorescence lifetime imaging of membrane lipid order with a ratiometric fluorescent probe. Biophys. J. 108, 2521–2531. ( 10.1016/j.bpj.2015.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baumgart T, Hess ST, Webb WW. 2003. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature 425, 821–824. ( 10.1038/nature02013) [DOI] [PubMed] [Google Scholar]
- 56.Kahya N, Scherfeld D, Bacia K, Poolman B, Schwille P. 2003. Probing lipid mobility of raft-exhibiting model membranes by fluorescence correlation spectroscopy. J. Biol. Chem. 278, 28 109–28 115. ( 10.1074/jbc.M302969200) [DOI] [PubMed] [Google Scholar]
- 57.Kahya N, Scherfeld D, Bacia K, Schwille P. 2004. Lipid domain formation and dynamics in giant unilamellar vesicles explored by fluorescence correlation spectroscopy. J. Struct. Biol. 147, 77–89. ( 10.1016/j.jsb.2003.09.021) [DOI] [PubMed] [Google Scholar]
- 58.Fricke N, Dimova R. 2016. GM1 softens POPC membranes and induces the formation of micron-sized domains. Biophys. J. 111, 1935–1945. ( 10.1016/j.bpj.2016.09.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puff N, Watanabe C, Seigneuret M, Angelova MI, Staneva G. 2014. Lo/Ld phase coexistence modulation induced by GM1. Biochim. Biophys. Acta Biomembr. 1838, 2105–2114. [DOI] [PubMed] [Google Scholar]
- 60.Helfrich W. 1973. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C 28, 693–703. ( 10.1515/znc-1973-11-1209) [DOI] [PubMed] [Google Scholar]
- 61.Campelo F, Arnarez C, Marrink SJ, Kozlov MM. 2014. Helfrich model of membrane bending: from Gibbs theory of liquid interfaces to membranes as thick anisotropic elastic layers. Adv. Colloid Interface Sci. 208, 25–33. ( 10.1016/j.cis.2014.01.018) [DOI] [PubMed] [Google Scholar]
- 62.Achim G, Stephen G. 2017. Theory and algorythms to compute Helfrish bending forces: a review. J. Phys. Condens. Matter 29, 203001 ( 10.1088/1361-648X/aa6313) [DOI] [PubMed] [Google Scholar]
- 63.Pontes B, Monzo P, Gauthier NC. 2017. Membrane tension: a challenging but universal physical parameter in cell biology. Semin. Cell Dev. Biol. 71, 30–41. ( 10.1016/j.semcdb.217.08.030) [DOI] [PubMed] [Google Scholar]
- 64.Bassereau P, Sorre B, Lévy A. 2014. Bending lipid membranes: experiments after W. Helfrich's model. Adv. Colloid Interface Sci. 208, 47–57. ( 10.1016/j.cis.2014.02.002) [DOI] [PubMed] [Google Scholar]
- 65.Dimova R. 2014. Recent developments in the field of bending rigidity measurements on membranes. Adv. Colloid Interface Sci. 208, 225–234. ( 10.1016/j.cis.2014.03.003) [DOI] [PubMed] [Google Scholar]
- 66.Bassereau P, et al. 2018. The 2018 biomembrane curvature and remodeling roadmap. J. Phys. D: Appl. Phys. 51, 343001 ( 10.1088/1361-6463/aacb98) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Evans EA. 1983. Bending elastic modulus of red blood cell membrane derived from buckling instability in micropipet aspiration tests. Biophys. J. 43, 27–30. ( 10.1016/S0006-3495(83)84319-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Evans EA, Skalak R. 1979. Mechanics and thermodynamics of biomembranes: part 1. CRC Crit. Rev. Bioeng. 3, 181–330. [PubMed] [Google Scholar]
- 69.Tierney KJ, Block DE, Longo ML. 2005. Elasticity and phase behavior of DPPC membrane modulated by cholesterol, ergosterol, and ethanol. Biophys. J. 89, 2481–2493. ( 10.1529/biophysj.104.057943) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tian A, Baumgart T. 2009. Sorting of lipids and proteins in membrane curvature gradients. Biophys. J. 96, 2676–2688. ( 10.1016/j.bpj.2008.11.067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu L, Doak WJ, Schertzer JW, Chiarot PR. 2016. Membrane mechanical properties of synthetic asymmetric phospholipid vesicles. Soft Matter 12, 7521–7528. ( 10.1039/C6SM01349J) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heinrich V, Waugh RE. 1996. A piconewton force transducer and its application to measurement of the bending stiffness of phospholipid membranes. Ann. Biomed. Eng. 24, 595–605. ( 10.1007/BF02684228) [DOI] [PubMed] [Google Scholar]
- 73.Cuvelier D, Derényi I, Bassereau P, Nassoy P. 2005. Coalescence of membrane tethers: experiments, theory, and applications. Biophys. J. 88, 2714–2726. ( 10.1529/biophysj.104.056473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dasgupta R, Miettinen MS, Fricke N, Lipowsky R, Dimova R.. 2018. The glycolipid GM1 reshapes asymmetric biomembranes and giant vesicles by curvature generation. Proc. Natl Acad. Sci. USA 115, 5756–5761. ( 10.1073/pnas.1722320115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chu S, Bjorkholm JE, Ashkin A, Cable A. 1986. Experimental observation of optically trapped atoms. Phys. Rev. Lett. 57, 314–317. ( 10.1103/PhysRevLett.57.314) [DOI] [PubMed] [Google Scholar]
- 76.Ashkin A, Dziedzic JM, Bjorkholm JE, Chu S. 1986. Observation of a single-beam gradient force optical trap for dielectric particles. Opt. Lett. 11, 288–290. ( 10.1364/OL.11.000288) [DOI] [PubMed] [Google Scholar]
- 77.Dimova R, Pouligny B, Dietrich C. 2000. Pretransitional effects in dimyristoylphosphatidylcholine vesicle membranes: optical dynamometry study. Biophys. J. 79, 340–356. ( 10.1016/S0006-3495(00)76296-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Solmaz ME, Biswas R, Sankhagowit S, Thompson JR, Mejia CA, Malmstadt N, Povinelli ML. 2012. Optical stretching of giant unilamellar vesicles with an integrated dual-beam optical trap. Biomed. Opt. Express 3, 2419 ( 10.1364/BOE.3.002419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Delabre U, Feld K, Crespo E, Whyte G, Sykes C, Seifert U, Guck J. 2015. Deformation of phospholipid vesicles in an optical stretcher. Soft Matter 11, 6075–6088. ( 10.1039/C5SM00562K) [DOI] [PubMed] [Google Scholar]
- 80.Wu SH, Sankhagowit S, Biswas R, Wu S, Povinelli ML, Malmstadt N. 2015. Viscoelastic deformation of lipid bilayer vesicles. Soft Matter 11, 7385–7391. ( 10.1039/C5SM01565K) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Binnig G, Quate CF, Gerber C. 1986. Atomic force microscope. Phys. Rev. Lett. 56, 930–933. ( 10.1103/PhysRevLett.56.930) [DOI] [PubMed] [Google Scholar]
- 82.Dieluweit S, Csiszár A, Rubner W, Fleischhauer J, Houben S, Merkel R. 2010. Mechanical properties of bare and protein-coated giant unilamellar phospholipid vesicles. A comparative study of micropipet aspiration and atomic force microscopy. Langmuir 26, 11 041–11 049. ( 10.1021/la1005242) [DOI] [PubMed] [Google Scholar]
- 83.Schäfer E, Vache M, Kliesch T-T, Janshoff A. 2015. Mechanical response of adherent giant liposomes to indentation with a conical AFM-tip. Soft Matter 11, 4487–4495. ( 10.1039/C5SM00191A) [DOI] [PubMed] [Google Scholar]
- 84.Schäfer E, Kliesch T-T, Janshoff A. 2013. Mechanical properties of giant liposomes compressed between two parallel plates: impact of artificial actin shells. Langmuir 29, 10 463–10 474. ( 10.1021/la401969t) [DOI] [PubMed] [Google Scholar]
- 85.Guz N, Dokukin M, Kalaparthi V, Sokolov I. 2014. If cell mechanics can be described by elastic modulus: study of different models and probes used in indentation experiments. Biophys. J. 107, 564–575. ( 10.1016/j.bpj.2014.06.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dokukin M, Ablaeva Y, Kalaparthi V, Seluanov A, Gorbunova V, Sokolov I. 2016. Pericellular brush and mechanics of guinea pig fibroblast cells studied with AFM. Biophys. J. 111, 236–246. ( 10.1016/j.bpj.2016.06.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaksonen M, Roux A. 2018. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 19, 313–326. ( 10.1038/nrm.2017.132) [DOI] [PubMed] [Google Scholar]
- 88.Johannes L, Mayor S. 2010. Induced domain formation in endocytic invagination, lipid sorting, and scission. Cell 142, 507–510. ( 10.1016/j.cell.2010.08.007) [DOI] [PubMed] [Google Scholar]
- 89.Howes MT, Mayor S, Parton RG. 2010. Molecules, mechanisms, and cellular roles of clathrin-independent endocytosis. Curr. Opin Cell Biol. 22, 519–527. ( 10.1016/j.ceb.2010.04.001) [DOI] [PubMed] [Google Scholar]
- 90.Johannes L, Parton RG, Bassereau P, Mayor S. 2015. Building endocytic pits without clathrin. Nat. Rev. Mol. Cell Biol. 16, 311–321. ( 10.1038/nrm3968) [DOI] [PubMed] [Google Scholar]
- 91.Johannes L, Wunder C, Shafaq-Zadah M. 2016. Glycolipids and lectins in endocytic uptake processes. J. Mol. Biol. 428, 4792–4818. ( 10.1016/j.jmb.2016.10.027) [DOI] [PubMed] [Google Scholar]
- 92.Saleem M, Morlot S, Hohendahl A, Manzi J, Lenz M, Roux A. 2015. A balance between membrane elasticity and polymerization energy sets the shape of spherical clathrin coats. Nat. Commun. 6, 6249 ( 10.1038/ncomms7249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roux A, Koster G, Lenz M, Sorre B, Manneville J-B, Nassoy P, Bassereau P. 2010. Membrane curvature controls dynamin polymerization. Proc. Natl Acad. Sci. USA 107, 4141–4146. ( 10.1073/pnas.0913734107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi Z, Baumgart T. 2015. Membrane tension and peripheral protein density mediate membrane shape transitions. Nat. Commun. 6, 5974 ( 10.1038/ncomms6974) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Heyningen WE. 1974. Gangliosides as membrane receptors for tetanus toxin, cholera toxin and serotonin. Nature 249, 415 ( 10.1038/249415a0) [DOI] [Google Scholar]
- 96.Aigal S, Claudinon J, Römer W. 2015. Plasma membrane reorganization: a glycolipid gateway for microbes. Biochim. Biophys. Acta Mol. Cell Res. 1853, 858–1871. [DOI] [PubMed] [Google Scholar]
- 97.Yamakawa T, Nagai Y. 1978. Glycolipids at the cell surface and their biological functions. Trends Biochem. Sci. 3, 128–131. ( 10.1016/S0968-0004(78)80031-0) [DOI] [Google Scholar]
- 98.Holmgren J, Lonnroth I, Svennerholm L. 1973. Tissue receptor for cholera exotoxin: postulated structure from studies with G(M1) ganglioside and related glycolipids. Infect. Immun. 8, 208–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sanchez J, Holmgren J. 2011. Cholera toxin- afoe and a friend. Indian J. Med. Res. 133, 153–163. [PMC free article] [PubMed] [Google Scholar]
- 100.Hammond AT, Heberle FA, Baumgart T, Holowka D, Baird B, Feigenson GW. 2005. Crosslinking a lipid raft component triggers liquid ordered–liquid disordered phase separation in model plasma membranes. Proc. Natl Acad. Sci. USA 98, 9471–9473. ( 10.1073/pnas.0405654102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Campanero-Rhodes MA, et al. 2007. N-glycolyl GM1 ganglioside as a receptor for Simian Virus 40. J. Virol. 81, 12 846–12 858. ( 10.1128/JVI.01311-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ewers H, et al. 2010. GM1 structure determines SV40-induced membrane invagination and infection. Nat. Cell Biol. 12, 11–18. ( 10.1038/ncb1999) [DOI] [PubMed] [Google Scholar]
- 103.Juillot S, Römer W. 2014. Shiga toxins. In Pathogenic Escherichia coli: molecular and cellular microbiology, pp. 79–101. Poole, UK: Caister Academic Press. [Google Scholar]
- 104.Römer W, et al. 2007. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 450, 670–675. ( 10.1038/nature05996) [DOI] [PubMed] [Google Scholar]
- 105.Madl J, Villringer S, Römer W. 2016. Delving into lipid-driven endocytic mechanisms using biomimetic membranes. In Chemical and synthetic approaches in membrane biology (ed. Shukla AK.). Springer Protocols Handbooks, pp. 17–36. New York, NY: Humana Press; ( 10.1007/8623_2016_7) [DOI] [Google Scholar]
- 106.Arnaud J, Tröndle K, Claudinon J, Audfray A, Varrot A, Römer W, Imberty A. 2014. Membrane deformation by neolectins with engineered glycolipid binding sites. Angew. Chem. Int. Ed. 53, 9267–9270. ( 10.1002/anie.201404568) [DOI] [PubMed] [Google Scholar]
- 107.Schütte OM, Patalag LJ, Weber LMC, Ries A, Römer W, Werz DB, Claudia S. 2015. 2-Hydroxy fatty acid enantiomers of Gb3 impact shiga toxin binding and membrane organization. Biophys. J. 108, 2775–2778. ( 10.1016/j.bpj.2015.05.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arnaud J, et al. 2013. Reduction of lectin valency drastically changes glycolipid dynamics in membranes but not surface avidity. ACS Chem. Biol. 8, 1918–1924. ( 10.1021/cb400254b) [DOI] [PubMed] [Google Scholar]
- 109.Römer W, et al. 2010. Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell 140, 540–553. ( 10.1016/j.cell.2010.01.010) [DOI] [PubMed] [Google Scholar]
- 110.Eierhoff T, et al. 2014. A lipid zipper triggers bacterial invasion. Proc. Natl Acad. Sci. USA 111, 12 895–12 900. ( 10.1073/pnas.1402637111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schubert T, Römer W. 2015. How synthetic membrane systems contribute to the understanding of lipid-driven endocytosis. Biochim. Biophys. Acta Mol. Cell Res. 1853, 2992–3005. ( 10.1016/j.bbamcr.2015.07.014) [DOI] [PubMed] [Google Scholar]
- 112.Eierhoff T, Stechmann B, Römer W. 2012. Pathogen and toxin entry—how pathogens and toxins induce and harness endocytotic mechanisms. In Molecular regulation of endocytosis (ed. Ceresa B.), pp. 249–276. London, UK: InTech. [Google Scholar]
- 113.Meinel A, Tränkle B, Römer W, Rohrbach A. 2014. Induced phagocytic particle uptake into a giant unilamellar vesicle. Soft Matter 10, 3667–3678. ( 10.1039/C3SM52964A) [DOI] [PubMed] [Google Scholar]
- 114.Hirohashi S, Kanai Y. 2003. Cell adhesion system and human cancer morphogenesis. Cancer Sci. 94, 575–581. ( 10.1111/j.1349-7006.2003.tb01485.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Berx G, Nollet F, Leeuwl WJF, Vijverl MJ, Cornelisse C, Roy FV. 1995. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J. 14, 6107–6115. ( 10.1002/j.1460-2075.1995.tb00301.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ley K. 2003. The role of selectins in inflammation and disease. Trends Mol. Med. 9, 263–268. ( 10.1016/S1471-4914(03)00071-6) [DOI] [PubMed] [Google Scholar]
- 117.Sharon N. 2007. Lectins: carbohydrate-specific reagents and biological recognition molecules. J. Biol. Chem. 282, 2753–2764. ( 10.1074/JBC.X600004200) [DOI] [PubMed] [Google Scholar]
- 118.Chothia C, Jones EY. 1997. The molecular structure of cell adhesion molecules. Annu. Rev. Biochem. 66, 823–862. ( 10.1146/annurev.biochem.66.1.823) [DOI] [PubMed] [Google Scholar]
- 119.Michaud S, Marin R, Westwood JT, Tanguay RM. 2011. The tail of integrin activation. Trends Biochem. Sci. 36, 191–198. ( 10.1016/j.tibs.2010.11.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huttenlocher A, Sandborg RR, Horwitz AF. 1995. Adhesion in cell migration. Curr. Opin. Cell Biol. 7, 697–706. ( 10.1016/0955-0674(95)80112-X) [DOI] [PubMed] [Google Scholar]
- 121.Rosso F, Giordano A, Barbarisi M, Barbarisi A. 2004. From cell-ECM interactions to tissue engineering. J. Cell. Physiol. 199, 174–180. ( 10.1002/jcp.10471) [DOI] [PubMed] [Google Scholar]
- 122.Streicher P, et al. 2009. Integrin reconstituted in GUVs: a biomimetic system to study initial steps of cell spreading. Biochim. Biophys. Acta Biomembr. 1788, 2291–2300. [DOI] [PubMed] [Google Scholar]
- 123.Fenz SF, Sengupta K. 2012. Giant vesicles as cell models. Integr. Biol. 4, 982–995. ( 10.1039/c2ib00188h) [DOI] [PubMed] [Google Scholar]
- 124.Marchi-Artzner V, Lorz B, Gosse C, Jullien L, Merkel R, Kessler H, Sackmann E. 2003. Adhesion of Arg-Gly-Asp (RGD) peptide vesicles onto an integrin surface: visualization of the segregation of RGD ligands into the adhesion plaques by fluorescence. Langmuir 19, 835–841. ( 10.1021/la026227k) [DOI] [Google Scholar]
- 125.Monzel C, Fenz SF, Merkel R, Sengupta K. 2009. Probing biomembrane dynamics by dual-wavelength reflection interference contrast microscopy. ChemPhysChem 10, 2828–2838. ( 10.1002/cphc.200900645) [DOI] [PubMed] [Google Scholar]
- 126.Limozin L, Sengupta K. 2009. Quantitative reflection interference contrast microscopy (RICM) in soft matter and cell adhesion. ChemPhysChem 10, 2752–2768. ( 10.1002/cphc.200900601) [DOI] [PubMed] [Google Scholar]
- 127.Fenz SF, Smith AS, Merkel R, Sengupta K. 2011. Inter-membrane adhesion mediated by mobile linkers: effect of receptor shortage. Soft Matter 7, 952–962. ( 10.1039/C0SM00550A) [DOI] [Google Scholar]
- 128.Reister-Gottfried E, Sengupta K, Lorz B, Sackmann E, Seifert U, Smith AS. 2008. Dynamics of specific vesicle-substrate adhesion: from local events to global dynamics. Phys. Rev. Lett. 101, 208103 ( 10.1103/PhysRevLett.101.208103) [DOI] [PubMed] [Google Scholar]
- 129.Gaul V, Lopez SG, Lentz BR, Moran N, Forster RJ, Keyes TE. 2015. The lateral diffusion and fibrinogen induced clustering of platelet integrin αiIbβ3 reconstituted into physiologically mimetic GUVs. Integr. Biol. 7, 402–411. ( 10.1039/C5IB00003C) [DOI] [PubMed] [Google Scholar]
- 130.Puech PH, Feracci H, Brochard-Wyart F. 2004. Adhesion between giant vesicles and supported bilayers decorated with chelated E-cadherin fragments. Langmuir 20, 9763–9768. ( 10.1021/la048682h) [DOI] [PubMed] [Google Scholar]
- 131.Fenz SF, Merkel R, Sengupta K. 2009. Diffusion and intermembrane distance: case study of avidin and E-cadherin mediated adhesion. Langmuir 25, 1074–1085. ( 10.1021/la803227s) [DOI] [PubMed] [Google Scholar]
- 132.Murrell M, Pontani LL, Guevorkian K, Cuvelier D, Nassoy P, Sykes C. 2011. Spreading dynamics of biomimetic actin cortices. Biophys. J. 100, 1400–1409. ( 10.1016/j.bpj.2011.01.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Weiss M, et al. 2018. Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nat. Mater. 17, 89–96. ( 10.1038/nmat5005) [DOI] [PubMed] [Google Scholar]
- 134.Steinkühler J, Agudo-Canalejo J, Lipowsky R, Dimova R. 2016. Modulating vesicle adhesion by electric fields. Biophys. J. 111, 1454–1464. ( 10.1016/j.bpj.2016.08.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Weinbaum S, Tarbell JM, Damiano ER. 2007. The structure and function of the endothelial glycocalyx layer. Annu. Rev. Biomed. Eng. 9, 121–167. ( 10.1146/annurev.bioeng.9.060906.151959) [DOI] [PubMed] [Google Scholar]
- 136.Stuhr-Hansen N, Madl J, Villringer S, Aili U, Römer W, Blixt O. 2016. Synthesis of cholesterol-substituted glycopeptides for tailor-made glycocalyxification of artificial membrane systems. ChemBioChem 17, 1403–1406. ( 10.1002/cbic.201600258) [DOI] [PubMed] [Google Scholar]
- 137.Carrara P, Stano P, Luisi PL. 2012. Giant vesicles ‘colonies’: a model for primitive cell communities. ChemBioChem 13, 1497–1502. ( 10.1002/cbic.201200133) [DOI] [PubMed] [Google Scholar]
- 138.De Souza TP, et al. 2017. Vesicle aggregates as a model for primitive cellular assemblies. Phys. Chem. Chem. Phys. 19, 20 082–20 092. ( 10.1039/C7CP03751A) [DOI] [PubMed] [Google Scholar]
- 139.Villringer S, Madl J, Sych T, Manner C, Imberty A, Römer W. 2018. Lectin-mediated protocell crosslinking to mimic cell-cell junctions and adhesion. Sci. Rep. 8, 1932 ( 10.1038/s41598-018-20230-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ribeiro JP, Villringer S, Goyard D, Coche-Guerente L, Höferlin M, Renaudet O, Römer W, Imberty A. 2018. Tailor-made Janus lectin with dual avidity assembles glycoconjugate multilayers and crosslinks protocells. Chem. Sci. 9, 7634–7641. ( 10.1039/C8SC02730G) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bartelt SM, Steinkuehler J, Dimova R, Wegner SV. 2018. Light guided motility of a minimal synthetic cell. Nano Lett. 18, 7268–7274. ( 10.1021/acs.nanolett.8b03469) [DOI] [PubMed] [Google Scholar]
- 142.Martens S, McMahon HT. 2008. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 9, 543–556. ( 10.1038/nrm2417) [DOI] [PubMed] [Google Scholar]
- 143.Jahn R. 2011. Membrane fusion. Ann. Plast. Surg. 67, 98 ( 10.1097/SAP.0b013e31820dc0e3) [DOI] [Google Scholar]
- 144.Wickner W, Schekman R. 2008. Membrane fusion. Nat. Struct. Mol. Biol. 15, 658–664. ( 10.1038/nsmb.1451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tamm LK, Han X. 2000. Viral fusion peptides: a tool set to disrupt and connect biological membranes. Biosci. Rep. 20, 501–518. ( 10.1023/A:1010406920417) [DOI] [PubMed] [Google Scholar]
- 146.Kozlovsky Y, Kozlov MM. 2002. Stalk model of membrane fusion: solution of energy crisis. Biophys. J. 82, 882–895. ( 10.1016/S0006-3495(02)75450-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Markin VS, Albanesi JP. 2002. Membrane fusion: stalk model revisited. Biophys. J. 82, 693–712. ( 10.1016/S0006-3495(02)75432-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fix M, Melia TJ, Jaiswal JK, Rappoport JZ, You D, Sollner TH, Rothman JE, Simon SM. 2004. Imaging single membrane fusion events mediated by SNARE proteins. Proc. Natl Acad. Sci. USA 101, 7311–7316. ( 10.1073/pnas.0401779101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ma M, Gong Y, Bong D. 2009. Lipid membrane adhesion and fusion driven by designed, minimally multivalent hydrogen-bonding lipids. J. Am. Chem. Soc. 131, 16 919–16 926. ( 10.1021/ja9072657) [DOI] [PubMed] [Google Scholar]
- 150.Melikyan GB, Barnard RJO, Abrahamyan LG, Mothes W, Young JAT. 2005. Imaging individual retroviral fusion events: from hemifusion to pore formation and growth. Proc. Natl Acad. Sci. USA 102, 8728–8733. ( 10.1073/pnas.0501864102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yoon T-Y, Okumus B, Zhang F, Shin Y-K, Ha T. 2006. Multiple intermediates in SNARE-induced membrane fusion. Proc. Natl Acad. Sci. USA 103, 19 731–19 736. ( 10.1073/pnas.0606032103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Marsden HR, Tomatsu I, Kros A. 2011. Model systems for membrane fusion. Chem. Soc. Rev. 40, 1572–1585. ( 10.1039/C0CS00115E) [DOI] [PubMed] [Google Scholar]
- 153.Pantazatos DP, MacDonald RC. 1999. Directly observed membrane fusion between oppositely charged phospholipid bilayers. J. Membr. Biol. 170, 27–38. ( 10.1007/s002329900535) [DOI] [PubMed] [Google Scholar]
- 154.Lei G, MacDonald RC. 2003. Lipid bilayer vesicle fusion: intermediates captured by highspeed microfluorescence spectroscopy. Biophys. J. 85, 1585–1599. ( 10.1016/S0006-3495(03)74590-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Heuvingh J, Pincet F, Cribier S. 2004. Hemifusion and fusion of giant vesicles induced by reduction of inter-membrane distance. Eur. Phys. J. E 14, 269–276. ( 10.1140/epje/i2003-10151-2) [DOI] [PubMed] [Google Scholar]
- 156.Haluska CK, Riske KA, Marchi-Artzner V, Lehn J-M, Lipowsky R, Dimova R. 2006. Time scales of membrane fusion revealed by direct imaging of vesicle fusion with high temporal resolution. Proc. Natl Acad. Sci. USA 103, 15 841–15 846. ( 10.1073/pnas.0602766103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nikolaus J, Stöckl M, Volkmer R, Herrmann A. 2010. Direct visualization of large and protein-free hemifusion diaphragms. Biophys. J. 98, 1192–1199. ( 10.1016/j.bpj.2009.11.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kliesch TT, Dietz J, Turco L, Halder P, Polo E, Tarantola M, Jahn R, Janshoff A. 2017. Membrane tension increases fusion efficiency of model membranes in the presence of SNAREs. Sci. Rep. 7, 12070 ( 10.1038/s41598-017-12348-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.