Abstract
The initial part of this review details the controversy behind the use of a serological level of prostate-specific antigen (PSA) for the diagnostics of prostate cancer (PCa). Novel biomarkers are in demand for PCa diagnostics, outperforming traditional PSA tests. The review provides a detailed and comprehensive summary that PSA glycoprofiling can effectively solve this problem, thereby considerably reducing the number of unnecessary biopsies. In addition, PSA glycoprofiling can serve as a prognostic PCa biomarker to identify PCa patients with an aggressive form of PCa, avoiding unnecessary further treatments which are significantly life altering (incontinence or impotence).
Keywords: prostate cancer, diagnostics, biomarkers, lectins, glycans, prostate-specific antigen
1. Introduction
1.1. Prostate cancer
Cancer is a major cause of deaths in both developing and developed countries [1]. About 14.1 million new cancer cases and 8.2 million deaths from cancer were estimated to have occurred worldwide in 2012 [2]. Prostate cancer (PCa) is the most frequently diagnosed cancer among men and lung cancer is the leading cause of cancer death among women in the more developed countries. PCa is diagnosed in 1.1 million men with the number of deaths estimated at 307 500 per annum [1].
In the nineteenth century, PCa was described as ‘a very rare disease’, but in the twenty-first century it has become the major cancer diagnosed in men with a projected increase in incidence to 2.1 million by 2035 with up to 633 328 associated deaths [3]. The survival rate is proportional to the stage reached at diagnosis, hence early-stage diagnosis using disruptive and effective diagnostic tools is a key to reducing mortality.
1.2. Prostate-specific antigen as prostate cancer biomarker
The gold standard in early-stage PCa diagnosis/screening is analysis of the level of prostate-specific antigen (PSA) in serum. PSA is a glycoprotein and an enzyme produced by the prostate, responsible for seminal fluid liquefaction. The serological PSA level is low in healthy men with an increased level observed after disruption of the basement membrane of the prostate gland (i.e. as a result of PCa and other conditions). A PSA level of up to 4 ng ml−1 is considered ‘normal’ [4]. A PSA level in the range of 4–10 ng ml−1 (also often defined as a grey zone) is considered ‘intermediate’, with cancer present in 30–35% of patients [4]. A PSA level of over 10 ng ml−1 is considered to be ‘high’ with a 67% probability of advanced disease [4]. The pathologist Richard Ablin, who discovered PSA, wrote The Great Prostate Hoax: How Big Medicine Hijacked the PSA Test and Caused a Public Health Disaster. He listed the following major concerns about using PSA as a diagnostic PCa biomarker [5,6]:
-
—
PSA is a prostate- and not a PCa-specific biomarker, hence it cannot be used as a diagnostic PCa biomarker.
-
—
The serological PSA level does not provide information on whether the PCa is indolent or aggressive.
-
—
PSA should only be used as an indicator of PCa recurrence.
The controversy behind using PSA for PCa diagnosis can be documented as follows:
-
—
The US Food and Drug Administration (FDA) agency approved PSA tests together with digital rectal examination (DRE) for PCa diagnostics in 1994 [3].
-
—
Owing to PCa over-diagnosis, the US Preventative Services Task Force (USPSTF) published a recommendation against the use of PSA for PCa diagnosis in 2012 [7].
-
—
Since the publication of this recommendation, a more advanced PCa with a higher proportion of tumours of higher grade and stage was detected [8], hence in 2017 the USPSTF recommended selective use of PSA tests for men aged from 55 to 70 [9,10].
The debate continues about the clinical value of PSA testing, because the application of such screening leads to unnecessary biopsies (up to 75% of biopsies are avoidable) [11] and other treatments leading to life-altering issues, such as incontinence or impotence [3]. A biopsy is performed in the event of suspicious results from PSA tests and DRE to validate the presence of PCa; however, a biopsy provides substantially false-negative results. Here is a summary of the use of PSA tests [11]:
-
—
Screening does not reduce PCa risk but increases the chance of finding it.
-
—
PSA tests can identify early-stage PCa that a DRE would miss.
-
—
A ‘normal’ PSA level of up to 4 ng ml−1 is not a guarantee of health, because a biopsy in 15% of men with this PSA level reveals PCa.
-
—
A high PSA level may prompt the patient to seek treatment, resulting in possible urinary and sexual side-effects, even in cases when active surveillance without any treatment would be the preferred option for indolent localized PCa disease.
-
—
An elevated PSA level could indicate other prostate diseases, such as benign prostatic hyperplasia (BPH, a benign prostate disease) and prostatitis, besides PCa.
As a result of the suboptimal clinical performance of PSA tests, numerous other PCa diagnostic tests are currently under development with clinical validation of a few of them already approved by regulatory agencies such as the prostate health index (PHI) and 4 K score tests [3,4,12–14]. In order to successfully validate PCa biomarkers, the biomolecule of interest needs to meet the following requirements [3,15]:
-
—
Is cancer-specific;
-
—
Can distinguish between healthy individuals and cancer patients (a diagnostic biomarker);
-
—
Distinguishes between indolent and aggressive prostate cancer (a prognostic biomarker);
-
—
Monitors disease progression;
-
—
Predicts recurrence of disease;
-
—
Monitors response of disease to treatment (a predictive biomarker);
-
—
Is stable and readily detectable;
-
—
Can be applied as a target for therapy (a therapeutic biomarker).
1.3. Prostate-specific antigen structure
PSA (also denoted as human kallikrein 3, hk3 or KLK3) is present in serum in a complex (cPSA) that forms a covalent bond with proteins such as α1-antichymotrypsin and α2-macroglobulin [13,16], while some PSA is present in a free form (fPSA). fPSA consists of several isoforms and the detection of some of these isoforms forms an integral part of PCa diagnostic kits such as the PHI test (detection of total PSA, fPSA and [−2] proPSA) and 4 K score (fPSA, intact PSA, total PSA and human kallikrein hk2) [13]. The sum of all PSA forms is referred to as the total PSA (tPSA). fPSA can be released from cPSA upon prolonged incubation with ethanolamine (see §5.1.2). N-glycan on PSA stabilizes the structure of the enzyme while to some extent changing the activation profile and enzymatic activity [17]. A detailed description of the PSA's glycan structure can be found elsewhere [18,19].
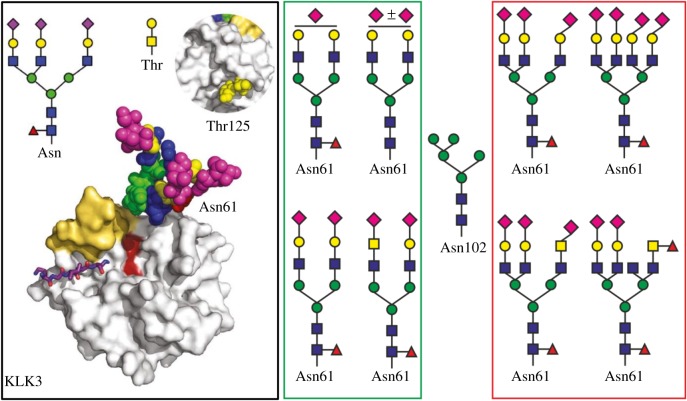
Mechref and co-workers discovered a possible additional glycosylation site Asn102 besides the one at Asn69 [20] (according to another assignment Asn69 = Asn61 [18]) (figure 1). This novel glycosylation site is a result of unusual missense mutation (the rs61752561 in KLK3 genes) resulting in the conversion of Asp102 to Asn102. Such a mutation is closely associated with PCa patient survival [23]. Of the three commercially available PSA standards investigated in the study, the predominant glycan forms at Asn102 and at Asn69 (Asn61) are shown (figure 1) [20]. The structure of glycan present on Asn61 of PSA when isolated from healthy men (BPH patients) and men with PCa is shown in figure 1, indicating that glycan from PCa contains α-2,3-sialic acid, an increased amount of α-2,6-sialic acid, one or two branching points (tri- and tetra-antennary glycans), antennary and core fucose and LacdiNac (GalNAc-GlcNAc) (figure 1).
Figure 1.
Black box (left) image: natural PSA (KLK3, hk3; PDB code 3QUM) exhibiting a complex tri-antennary glycan at Asn61, with terminal sialic acid units, often N-acetyl neuraminic acid (Neu5Ac) (magenta). The active site is in an open state permitting access of substrates, e.g. shown as a stick model bound to the protein. At the opposite side to the active site, an O-glycan is linked to Thr125, which consists of GalNAc-Gal (insert). The figure is taken from an open access publication [17]. Green box (middle) image: typical glycan structures present at Asn61 on PSA from healthy men or men with BPH. Red box (right) image: typical glycan structures present at Asn61 of PSA from PCa patients. Glycans present at position Asn61 were compiled from various sources [17,19–22]. A typical glycan structure present at Asn102 on PSA as a result of an unusual missense mutation (the rs61752561 in KLK3 genes) resulting in the conversion of Asp102 to Asn102 is positioned between the green and red boxes. (Online version in colour.)
2. Glycans as prostate cancer biomarkers
2.1. Glycans in cancer development and progression
Glycomics studies the glycan (a complex carbohydrate attached to a protein or lipid) structure and function. There are reasons why glycomics lags behind genomics and proteomics:
-
—
glycans are more structurally complex than proteins and DNA/RNA;
-
—
determination of glycan sequences using traditional instrumental techniques is challenging [24];
-
—
glycan biosynthesis cannot be predicted from a template, as in the case of DNA and proteins [25].
The role of glycans in cancer development and progression is recognized [26–30]. A changed glycosylation in cancer cells, in comparison with healthy cells, can result from the following processes [15,31,32]:
-
—
Expression levels of glycosyltransferases (GTs);
-
—
Localization of GTs in the cellular compartments (i.e. nucleus and mitochondria);
-
—
Expression of chaperones responsible for proper folding of glycoproteins and GTs;
-
—
Site- and protein-specific enzymatic preference of GTs;
-
—
Expression levels of glycosidases during glycoprotein processing;
-
—
Expression levels of hydrolases in lysosomes and cellular secretions;
-
—
Availability of protein substrates;
-
—
Availability, level and activity of activated nucleotide sugars;
-
—
Activity of monosaccharide transporters;
-
—
Glycoprotein turnover kinetics;
-
—
pH of endoplasmic reticulum (ER) and Golgi apparatus;
-
—
Competition reactions between GTs for similar glycan acceptors.
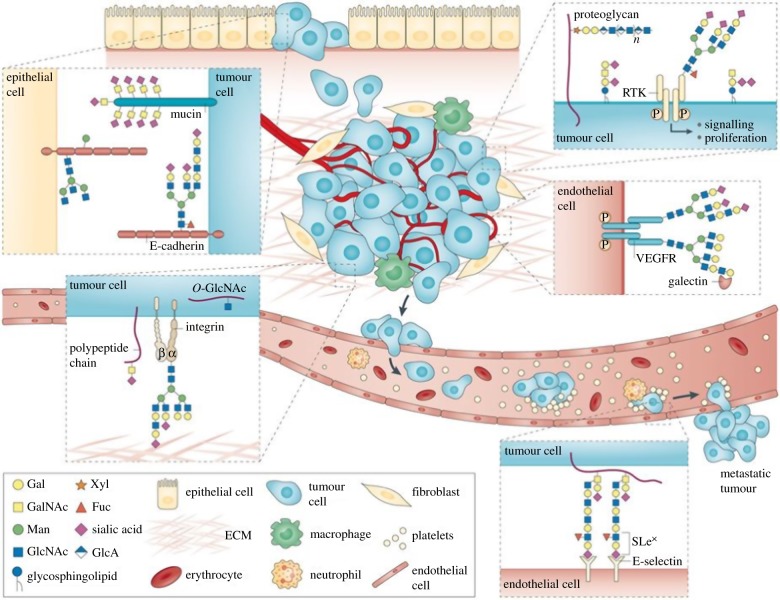
Cancer-associated changes in the glycan structures involve: α2,3-sialylation; core-fucosylation (i.e. the addition of fucose to the innermost GlcNAc residue in the vicinity of the protein backbone); O-glycan truncation (short O-glycans); presence of polysialic acid, N-acetylglucosamine and Lewis antigens; and N- and O-glycan branching (figure 2) [15,31,32,34,35]. Accordingly, changes in the glycan types can differ between cells from the same tissue/organ and also between organs [36]. Changes in the glycan profile are often cited as a ‘hallmark of cancer’ [37]. ‘Rather than aberrant glycosylation being itself a hallmark of cancer [38], another perspective is that glycans play a role in every recognized cancer hallmark’ [37].
Figure 2.
In the process of tumour cell dissociation/invasion, glycans interfere with cell–cell adhesion via electrostatic repulsion mediated by negatively charged sialic acids and by branched N-glycans present on E-cadherin (upper left). Expression of gangliosides in the cancer cell membrane can also modulate signal transduction, activating various cellular pathways that induce tumour growth and progression (upper right). In the process of tumour cell migration, integrins show altered glycosylation in both O-linked and N-linked glycans. Terminal sialylation interferes with cell-extracellular matrix (ECM) interactions, promoting an increased migratory and invasive phenotype (lower left). The aberrant glycosylation of vascular endothelial growth factor receptor (VEGFR) modulates its interaction with galectins and is associated with tumour angiogenesis. The tumour-associated carbohydrate determinants sialyl Lewis x (SLex) and SLea serve as ligands for the adhesion receptors expressed in activated endothelial cells (E-selectin), promoting cancer cell adhesion/metastasis. Reprinted with permission from Pinho & Reis [33] (copyright © 2015 Nature). (Online version in colour.)
2.2. Glycans as a novel type of biomarkers
In addition to already approved diagnostic procedures (DRE, PSA test and magnetic resonance imaging (MRI)), changes in the marker's glycan structure as biomarkers have not yet been approved for PCa diagnostics [12]. A paper recently published in Science suggests that, for reliable and accurate early-stage cancer diagnostics, multi-analyte blood tests need to be performed, i.e. serological analysis of the levels of several proteins with cell-free DNA [39]. However, in the pioneering study published in Science [39], glycan analysis was not included for cancer diagnostics. In another study, an integrated approach using six different types of biomarker blocks (clinical data, DNA methylation, coding and non-coding transcripts, proteins and glycans) including glycans (i.e. glycans released from the glycoproteins by the enzymatic action) to distinguish indolent localized PCa from aggressive non-localized PCa was described for the first time [40]. By analysis of 61 different biomolecules including four clinical parameters, it was possible to distinguish indolent PCa from the aggressive PCa form with AUC = 0.91 (AUC = area under the curve in the receiver operating characteristic curve). A controlled clinical data evaluation using only four clinical parameters (age, PSA level, Gleason score (GS) and DRE results) affords a lower AUC = 0.67 [40].
The introduction of new PCa biomarkers into clinical practice is slow owing to the strict clinical requirements for the accuracy, specificity (true negative rate, i.e. correct identification of healthy individuals) and sensitivity (true positive rate, i.e. correct identification of patients with disease) of such tests [41]. In addition, the biomarkers need to be validated on large patient cohorts with a proper selection of control samples. The co-morbidities present in the patient and control groups, which can affect clinical validation, need to be provided [42]. The biomarker variability depending on gender, age, habits or environmental factors also has to be investigated. Recent studies really suggest that glycosylation is age related and that the control group needs to age match the group with disease [43]. Clinical validation is required using a multi-centre study to address inter-laboratory variability (i.e. storage and processing conditions).
Although not yet approved by the regulatory bodies, the application of glycans into accurate and robust PCa diagnostics is gaining momentum. Specific PSA glycoprofiling, applied as a diagnostic or prognostic biomarker, has already been described, albeit marginally, in a series of review papers from O'Kennedy's laboratory [3,4,13,16] and also from other laboratories [11,12,21,44,45].
There is another very important issue to consider while implementing glycan assays for PCa diagnostics—the assay format. Only a limited number of techniques available for glycan analysis can provide the quantitative data required in clinical practice, including lectin- and antibody-based ELISA assays [15,41]. Such assays, however, entail drawbacks such as the need for labelling and external standards, as well as the limited repertoire of lectin and anti-carbohydrate antibodies recognizing glycans and, commonly, their low binding affinity [41]. This review seeks to provide a detailed and comprehensive overview of PSA glycoprofiling applied as a diagnostic or prognostic PCa biomarker using instrumental-based and lectin-integrating approaches.
3. Instrumental-based approaches for prostate-specific antigen glycoprofiling
Because of either a lack of sensitivity or insufficient resolution for quantifying minor glycoforms by employing mass spectrometry (MS) or chromatographic methods [41], the instrument-based approach is only infrequently applied to PSA glycoprofiling. These techniques are constantly evolving, with the advances comprehensively summarized by Mechref's team [46].
PSA from human serum was enriched using anti-PSA antibodies immobilized on magnetic beads [47]. In the two subsequent steps performed on beads, PSA still affinity-captured to the antibody was deglycosylated with endoglycosidase F3 and subsequently digested by trypsin prior to liquid chromatography (LC)–MS/MS analysis. A core-fucosylated form of PSA was investigated down to 1 ng ml−1. The method was applied to the analysis of three spiked female human sera, hence no relevant clinical parameters are available [47]. In the other study, PSA was immunoprecipitated from expressed prostate secretion (EPS) urine [48]. PSA was excised from the band on the polyacrylamide gel electrophoresis (PAGE) gel and the N-glycan was released from PSA using PNGase F enzyme and further analysed using LC–MS. Two glycans were identified as being significantly different when comparing PCa and BPH patients and no significant difference was observed between PCa and BPH patients when considering overall sialylation and fucosylation [48].
α-2,3-sialylation of PSA is one of the dominant glycan forms of PSA, present in serum samples from PCa patients, hence there is a requirement for instrument-based methods for selective and specific detection of this glycan form. Wuhrer and co-workers [24] developed a method for the specific detection of α-2,3-sialic acid (α-2,3-SA) and α-2,6-sialic acid (α-2,6-SA) containing glycopeptides using a combination of capillary electrophoresis (CE) with electrospray ionization MS. While the fragmentation pattern (collision-induced dissociation) of both glycopeptides is identical, these two isomers are separated using a different electrophoretic mobility. This difference in electrophoretic mobility results from a minute difference in pKa (ΔpKa = 0.034) between these two isomers. The method is quite complex, including PSA denaturation, reduction of disulfide bonds, sulfide alkylation, tryptic digest, glycan release using PNGase and glycan enrichment using hydrophilic interaction LC. The analysis has not been deployed to an analysis of real samples and the method requires a microgram quantity of the PSA for analysis [24]. The group led by Professor Paleček developed an electrochemical method for distinguishing between terminal α-2,3-SA and α-2,6-SA using Os(VI) complexes [49]. The optimal experimental conditions could then be applied for simple discrimination between these two isomers on two types of electrodes. The method requires a relatively high glycan concentration (500 nM) for the analysis [49].
An increased sialylation of glycans on PSA isolated from a tissue and detected using an instrumental approach could distinguish PCa from BPH patients with a good statistical significance (p = 0.051) [50]. Serum samples were initially digested using proteinase and glycopeptides were purified using desalting columns. The glycan part of the glycopeptides was oxidized, and such oxidized glycopeptides were attached to a hydrazide resin. Unconjugated peptides were removed by washing and glycopeptides were subsequently released from the resin using PNGase F with subsequent MS analysis [50].
LC–MS/MS analysis of four distinct PSA glycoforms was applied to PCa diagnostics using urine [51]. Urinary PSA was immunoprecipitated using magnetic beads, PSA was denatured and then applied on the PAGE gel. Next, the protein was digested in the gel by chymotrypsin with subsequent analysis of peptides and glycopeptides by LC–MS. The most successful discrimination between BPH patients (PSA level up to 78 ng ml−1) and PCa patients (PSA level up to 27 µg ml−1) was obtained when one glycan form was analysed with an AUC of 0.74 (see table 3, no. 20) [51].
4. Lectin-based methods for prostate-specific antigen glycoprofiling
The detection of any type of biomarker directly in urine is considered to be a non-invasive method, while blood/serum sampling is a mildly invasive procedure. Accordingly, a biomarker assay in urine might appear to have advantages over the use of serum samples. There remain, however, a few important issues related to biomarker analysis in urine. Some aberrant glycans do not necessarily need to reach the urine and can be detected only in serum (i.e. α-2,3-SA PSA form) [52] and a highly glycosylated Tamm–Horsfall protein forming large aggregates in urine should be removed prior to glycan analysis [53].
The clinical performance of glycans as diagnostic PCa biomarkers should be compared with the clinical performance of PSA and PHI tests [14]. For example, the analysis of PSA in serum provides the following diagnostic performance with AUC = 0.68: (i) at a cut-off value of 4.1 ng ml−1: sensitivity 20%, specificity 94%; (ii) at a cut-off value of 2.6 ng ml−1: sensitivity 40%, specificity 81% [14].
4.1. Analysis in serum
4.1.1. Diagnostic prostate cancer biomarkers
4.1.1.1. Performance of PHI test
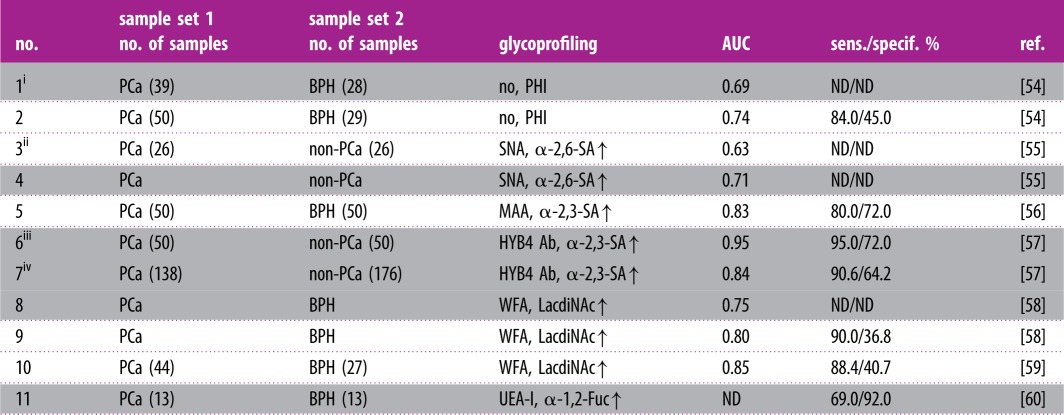
The diagnostic performance of PHI to discriminate between PCa patients (tPSA of 2.6–40.6 ng ml−1) and BPH patients (tPSA of 3.9–14.5 ng ml−1) showed an AUC of 0.74; sensitivity 84% and specificity 45% (table 1, no. 2). Thus, the diagnostic performance was better than in the tPSA test with AUC = 0.51 and %fPSA test with AUC = 0.63. When the sub-cohort of patients with tPSA lower than 13 ng ml−1 (28 BPH versus 39 PCa patients) was investigated using the PHI test, the AUC was 0.69 (table 1, no. 1) [54].
Table 1.
Clinical performance of PSA glycoprofiling in serum samples as a diagnostic PCa biomarker. PCa, prostate cancer patients; BPH, patients with benign prostatic hyperplasia; AUC, area under the curve in a receiver operating curve; sens., sensitivity; specif., specificity; i, not exactly the grey zone, but tPSA was below 13 ng ml−1; ii, %fPSA level in the range 10–20%; PHI, prostate health index; SA, sialic acid; ND, not determined, SNA, Sambucus nigra agglutinin; MAA, Maackia amurensis agglutinin; iii, training test using 100 serum samples; iv, validation test using 314 serum samples; HYB4 Ab, antibody specific towards α-2,3 sialic acid; WFA, Wisteria floribunda agglutinin; UEA-I, Ulex europaeus agglutinin I; Fuc, fucose; highlights in grey: studies performed with serum samples having tPSA level in a grey zone.
 |
4.1.1.2. Sialylation changes
From five different lectins investigated in an ELISA format of analysis with electrochemiluminescent detection, Sambucus nigra agglutinin (SNA), in particular, displayed the best discrimination power (PCa patients versus non-PCa patients) with p = 0.47 [55]. PSA was attached to the special ELISA wells using adsorbed anti-PSA antibodies and PSA was glycoprofiled using biotinylated lectins and streptavidin modified by a fluorescent tag. The PSA level in both sets of samples was up to 10.4 ng ml−1. Although the SNA-based analysis did not perform better than a %fPSA test using the whole set of samples (AUC = 0.63 versus AUC = 0.85) (table 1, no. 3), the SNA-based assay performed very well in the discrimination of a subset of samples (i.e. AUC = 0.71 versus AUC = 0.54) (table 1, no. 4), which could not be distinguished using a %fPSA test (%fPSA level in the range 10–20%) [55].
An automated micro-total immunoassay system using microchip CE and a liquid-phase binding assay was applied to measuring the percentage of PCa-associated α-2,3-SA on PSA in serum from BPH and PCa patients [56]. The assay entails the use of Maackia amurensis agglutinin (MAA) for affinity separation of α-2,3-SA containing PSA. Such assays outperform (sensitivity = 80%, specificity = 72%, AUC = 0.83, table 1, no. 5) the clinical performance of a PSA analysis (sensitivity = 80%, specificity = 14%, AUC = 0.51). The PSA level in both groups with a median value of 6.45 ng ml−1 (1.9–20.4 ng ml−1) for BPH and 6.60 ng ml−1 for PCa (1.5–21.4 ng ml−1) was very similar [56]. Another study also confirmed the practical utility of detecting α-2,3-SA on PSA for PCa diagnostics using only a limited number of samples [61]. The method involves immobilization of anti-PSA antibodies on the modified gold surface with application of serum samples and subsequent use of lectins to form a sandwich configuration. Since the method is based on a label-free electrochemical technique the assay worked without the need to use a label [61].
The Luminex flow-through system with anti-PSA antibodies immobilized on magnetic beads was applied to the detection of α-2,3-SA on PSA using a monoclonal antibody (HYB4) instead of MAA for the detection of α-2,3-SA [57]. The method was initially tested using a training dataset (50 PCa samples and 50 BPH samples) with the PSA level in the grey zone with a significant clinical difference between these two sets represented by AUC = 0.95 (table 1, no. 6). When 314 serum samples were applied to a validation sample set (grey zone), the AUC value dropped to 0.84 (table 1, no. 7), but still outperformed the PSA test with AUC = 0.61 [57].
4.1.1.3. LacdiNAc (GalNAc-GlcNAc) detection
An automated lectin-based immunoassay using Wisteria floribunda agglutinin (WFA) displayed a promising PCa diagnostic performance [58]. Anti-PSA was immobilized on the gold thin film, the sample was applied in a flow system and finally fluorescently labelled lectin-recognizing LacdiNAc was injected over the surface forming a sandwich. The signal was read as fluorescence intensity with a surface plasmon field-enhanced fluorescence spectrometer. The change in the PSA's glycan outperformed the PSA level in terms of clinical performance for samples with a PSA level up to 20 ng ml−1 (AUC of 0.80 versus 0.64) (table 1, no. 9) and with the PSA level up to 10 ng ml−1 (AUC of 0.75 versus 0.55) (table 1, no. 8) [58]. When the above-described assay system was further optimized, a higher clinical performance was achieved with an immobilized anti-PSA antibody and employing WFA to the analysis of LacdiNAc on PSA [59]. The method was tested to reveal the diagnostic potential of the approach by comparing serum samples from PCa patients and BPH patients with a PSA level in the range 4–20 ng ml−1 in both groups. The results indicate the great potential of the methods with AUC = 0.85 (table 1, no. 10), much higher than the performance of a PSA test with AUC = 0.56 [59].
4.1.1.4. Fucose detection
An ELISA-like assay format with anti-PSA immobilized within a 96-well plate was applied for the analysis of PSA's fucosylation integrating Ulex europaeus agglutinin (UEA-I) lectin. The method was used to distinguish PCa patients (PSA up to 10.7 ng ml−1) from BPH patients (PSA level up to 9.1 ng ml−1) [60]. The results showed a significant statistical difference in the signal obtained using these two sets of samples (p < 0.002) with 69% sensitivity and 92% specificity. The PSA test afforded 56% sensitivity and 70% specificity [60]. Two different lectin-based fluorescent immunoassays were applied to the glycoprofiling of PSA isolated from tissues, serum samples and urine [62]. Both detection methods rely on anti-PSA immobilized in wells within an ELISA plate, followed by incubation with the sample and finally with labelled Aleuria aurantia lectin (AAL). In the first assay protocol, AAL (fucose-specific) was labelled with europium chelate and in the second assay protocol the lectin was labelled with polystyrene nanoparticles doped with europium. The second method was much more sensitive than the first one and applied for real sample analysis using time-resolved fluorescence assays. In the serum/plasma samples, a high background signal rendered it not possible to glycoprofile PSA. However, urine was a proper matrix, making it possible to glycoprofile PSA and the results showed a statistically significant (p = 0.03) increase in PSA fucosylation in PCa patients compared with BPH patients. Unfortunately, there was no statistically significant difference in urine PSA fucosylation between the PCa patients and healthy individuals [62].
The homogeneous lectin-based assay was based on incubation of samples with Sepharose-bound lectins [22]. After washing, glycoproteins were eluted from the columns and the PSA level was determined in the pre-column and bound fraction for calculation of the relative amount of the lectin-bound fraction. Trichosanthes japonica agglutinin-II (recognizing α-1,2-Fuc) showed the best clinical performance (p < 0.05) as a diagnostic PCa biomarker (PCa versus BPH) out of eight lectins tested, when applied to PSA glycoprofiling [22].
4.1.1.5. Mannose detection
Two studies have detailed the usefulness of Concanavalin A in the glycoprofiling of PSA (a decreased mannose level with PCa development) with the potential to be a diagnostic PCa biomarker [63,64]. The first was a lectin-based assay in which samples were incubated with Sepharose-bound lectins with a final calculation of the relative amount of the lectin-bound fraction, which was statistically lower in the case of PCa patients than in BPH patients (p < 0.05) [63]. The second method relied on precipitation of serum samples using lectin, with a statistically lower binding of lectin to PSA from PCa patients than from BPH patients with p<0.001 [64].
4.1.2. Prognostic prostate cancer biomarkers
4.1.2.1. Sialylation changes
Sophisticated glycoprofiling of PSA to investigate the α-2,3-SA percentage of PSA as a prognostic PCa biomarker was compared with the performance of a PHI test [54]. The method involved the release of PSA from the complex with α1-antichymotrypsin and α2-macroglobulin proteins using ethanolamine. Immunopurified PSA was then applied to a lectin chromatography column using SNA lectin. Eluted and bound fractions were collected and fPSA was detected in both fractions to calculate the α-2,3-SA percentage of PSA and α-2,6-SA percentage of PSA. The results indicate that the α-2,3-SA percentage cannot be applied to PCa diagnostics because no significant differences were observed between BPH (PSA level in the range 3.9–14.5 ng ml−1) and low- and intermediate-risk PCa (PSA level in the range 2.6–12.4 ng ml−1) patients. This is why such assays were applied as a prognostic PCa biomarker, i.e. to distinguish high-risk PCa patients from BPH, low- and intermediate-risk PCa patients with AUC = 0.97, sensitivity of 81.8% and specificity of 96.5% (table 2, no. 13). Analysis of the tPSA revealed AUC of 0.81 and a PHI test value of 0.84 (table 2, no. 12). The combination of PHI with the α-2,3-SA percentage of PSA displayed a high prognostic potential with the following parameters: AUC = 0.985, sensitivity 100% and specificity 94.7% (table 2, no. 14) [54]. Out of five different Sepharose-bound lectins investigated for glycoprofiling of fPSA, MAA, in particular, was capable of distinguishing between PCa patients and BPH patients with a high statistical significance (p < 0.001) [63].
Table 2.
Clinical performance of PSA glycoprofiling in serum samples as a prognostic PCa biomarker. HR, high-risk PCa patients; LR, low-risk PCa patients; IR, intermediate-risk PCa patients; Pbx GG, prostate biopsy grade group; GS, Gleason score; AAL, Aleuria aurantia lectin; PhoSL, Pholiota squarrosa lectin; for other abbreviations see table 1.
| no. | sample set 1 no. of samples |
sample set 2 no. of samples |
glycoprofiling | AUC | sens./specif. % | ref. |
|---|---|---|---|---|---|---|
| 12 | HR (22) | BPH + LR + IR (57) | no, PHI | 0.84 | 81.8/84.2 | [54] |
| 13 | HR (22) | BPH + LR + IR (57) | SNA, α-2,3-SA↑ | 0.97 | 81.8/96.5 | [54] |
| 14 | HR (22) | BPH + LR + IR (57) | PHI + SNA, α-2,3-SA↑ | 0.985 | 100/94.7 | [54] |
| 15 | HR (22) | BPH + LR + IR (51) | SNA, α-2,3-SA↑ | 0.97 | 85.7/95.5 | [65] |
| 16 | PCa, Pbx GG ≥ 3 (159) | PCa, Pbx GG ≤ 2 (85) | WFA, LacdiNAc↑ | 0.65 | 57.1/80.8 | [58] |
| 17 | PCa, GS > 6 (33) | PCa, GS = 6 (14) | AAL, α-1,6(3)-Fuc↑ | 0.71 | ND/ND | [66] |
| 18 | PCa, GS>8 (20) | PCa, GS = 6 (14) | AAL, α-1,6(3)-Fuc↑ | 0.86 | ND/ND | [66] |
| 19 | HR (22) | BPH + LR + IR (51) | PhoSL, core α-1,6-Fuc↓ | 0.94 | 90.0/95.0 | [65] |
The sophisticated study for the glycoprofiling of PSA from serum was performed by Peracaula's group [65]. In their approach, PSA was released from a complex with α1-antichymotrypsin and α2-macroglobulin proteins using ethanolamine, then a free form of PSA was glycoprofiled using two lectins. These two proteins are glycoproteins [22], so the lectins can interact with the glycans of these two proteins in addition to the glycan of PSA when working with tPSA. The procedure for the preparation of fPSA from tPSA required a relatively long incubation time of 72 h and a large volume of the sample of 1.5 ml. When the core fucosylation ratio determined using Pholiota squarrosa lectin (PhoSL) was applied (table 2, no. 19) to distinguish high-risk PCa patients (PSA level up to 110 ng ml−1) from BPH patients and low- and intermediate-risk PCa patients (PSA level up to 18.2 ng ml−1), the assay afforded a sensitivity of 90%, specificity of 95% and AUC = 0.94. When the α-2,3-SA percentage of PSA was applied as a biomarker, the following clinical outcomes were revealed: sensitivity 85.7%, specificity 95.5% and AUC = 0.97 (table 2, no. 15) [65].
4.1.2.2. LacdiNAc detection
The prognostic potential of the application of WFA for PSA glycoprofiling within an automated lectin-based flow-through immunoassay can be affirmed by the ability to discriminate between PCa patients with a prostate biopsy grade group of 2 or higher and PCa patients with a prostate biopsy grade group of greater than or equal to 3, displaying sensitivity 57.1%, specificity 80.8% and AUC = 0.65 (table 2, no. 16). A PSA test afforded AUC of only 0.52 [58]. The application of WFA to the glycoprofiling of various glycoproteins as potential disease biomarkers including PSA is duly summarized in a recent review paper [67].
4.1.2.3. Fucose detection
The lectin-based fluorescent immunoassay method employing time-resolved fluorescence assays for the analysis of PSA's fucosylation was capable of distinguishing low-grade PCa tissues from tissue samples from men with an aggressive form of PCa (GS 4–5) [62]. A significant increase (p = 0.001) in the PSA fucosylation was observed in the PCa tissue compared with the benign tissue [62].
The prognostic utility of PSA's fucosylation was tested using AAL [66]. Agarose bead-bound AAL was incubated with the samples and, after incubation, the glycoproteins were eluted from the beads. Then, this eluate was incubated with anti-PSA antibodies immobilized on magnetic beads and the multiplex assay was completed using the Bioplex200 system. In this case, the authors showed that, with increased GS, there was an increase in fucosylation of PSA (i.e. % of PSA fucosylation) and this increase statistically discriminated between aggressive and indolent PCa (GS > 6 versus GS = 6) with p = 0.0053. Analysis of the results in receiver operating characteristic form showed AUC = 0.71 for the percentage of fucosylated PSA (table 2, no. 17), which is much higher than the PSA level with AUC = 0.66 (GS > 6 versus GS = 6). If the percentage of fucosylated PSA was applied in order to discriminate between GS ≥ 8 and GS = 6, the AUC increased to 0.86 (table 2, no. 18) and the PSA test showed AUC = 0.81 [66].
Eight different lectins immobilized on Sepharose columns were applied to the PSA glycoprofiling [22]. The discrimination potential of PSA's glycosylation status was determined using 20 samples from PCa patients and 20 samples from BPH patients. The results suggest that the best performance was that of Trichosanthes japonica agglutinin-II (TJA-II, specific for α1,2-fucose and GalNAc), but other lectins such as WFA (binding to LacdiNAc or GalNAc), UEA-I (Ulex europaeus agglutinin, binding to α1,2-fucose), MAA and DSA (Datura stramonium agglutinin, binding to tri- and tetra-antennary glycans and GlcNAc) also showed promising results. When using TJA-II, a significant clinical significance in distinguishing PCa patients from BPH patients was determined (p < 0.05) [22].
4.2. Analysis in urine
4.2.1. Diagnostic prostate cancer biomarkers
Peracaula's group with an assay approach described in [65] showed that PSA glycoprofiling using two lectins, PhoSL (specific for core fucose) and SNA (specific for α-2,6-SA), in ELISA format could not be applied to PCa diagnostics [52]. This is a surprising outcome because the same assay format worked well using serum samples [65]. The explanation is that the level of PSA from a tumour tissue cannot reach the urine, with only a minor level of aberrantly glycosylated PSA present in urine [52]. The low level of PSA with α-2,3-SA can also be explained by the preferential presence of the α-2,6-SA form on PSA in seminal plasma [68] and this form of PSA can be released into urine [52]. The AAL labelled with polystyrene nanoparticles doped with europium applied to the fluorescent ELISA format of the analysis has the potential for use in PSA glycoprofiling as a diagnostic PCa biomarker (PCa versus BPH) with p = 0.03 [62]. The performance of glycans as diagnostic PCa biomarkers is shown in table 3.
Table 3.
Clinical performance of PSA glycoprofiling in urine samples as a diagnostic PCa biomarker. For abbreviations see table 1. H2N4S1F1 = glycan containing eight monosaccharides, i.e. two hexoses (H2), four N-acetyl hexoses (N4), one sialic acid (S1) and one fucose (F1).
| no. | sample set 1 no. of samples |
sample set 2 no. of samples |
glycoprofiling | AUC | sens./specif. % | ref. |
|---|---|---|---|---|---|---|
| 20 | PCa (38) | BPH (61) | H2N4S1F1↑ (a) | 0.74 | 92.9/59.0 | [51] |
| 21 | PCa (38) | BPH (61) | mono-SA↑ (b) | 0.71 | 100/47.5 | [51] |
| 22 | PCa (38) | BPH (61) | SA↑ (c) | 0.69 | 68.8/75.0 | [51] |
| 23 | PCa (38) | BPH (61) | unfucosylated↑ (d) | 0.68 | 87.5/60.0 | [51] |
| 24 | PCa (38) | BPH (61) | a + b+c + d | 0.72 | 87.5/60.0 | [51] |
4.2.2. Prognostic prostate cancer biomarkers
The application of AAL and PhoSL in a sandwich ELISA-like format of analysis to PSA glycoprofiling was successfully used in the prognosis of PCa [69]. When PCa patients with GS ≥ 7 were discriminated against patients with a lower GS, AUC = 0.69 (AAL) and AUC = 0.72 (PhoSL) were obtained. By contrast, the serum level of PSA afforded only AUC = 0.59. When multivariate analysis was performed taking into account both the glycosylation changes and the density of PSA, the AUC value increased to 0.82 [69] (table 4). The AAL labelled with polystyrene nanoparticles doped with europium applied in the fluorescent ELISA format of the analysis also showed the potential to be applied as a prognostic PCa biomarker (PCa with GS ≥ 7 versus BPH) with p = 0.01 in an additional study [62].
Table 4.
Clinical performance of PSA glycoprofiling in urine samples as a prognostic PCa biomarker. PSAD, PSA density; AAL, Aleuria aurantia lectin; PhoSL, Pholiota squarrosa lectin; for other abbreviations see table 1 and table 2.
| no. | sample set 1 no. of samples |
sample set 2 no. of samples |
glycoprofiling | AUC | sens./specif. % | ref. |
|---|---|---|---|---|---|---|
| 25 | PCa, GS ≥ 7 (31) | PCa, GS < 7 (18) | AAL, α-1,6(3)-Fuc↓ | 0.69 | 90.3/47.3 | [69] |
| 26 | PCa, GS ≥ 7 (31) | PCa, GS < 7 (18) | PhoSL, core α-1,6-Fuc↓ | 0.72 | 90.0/39.5 | [69] |
| 27 | PCa, GS ≥ 7 (31) | PCa, GS < 7 (18) | Fuc, core Fuc and PSAD | 0.82 | 74.1/81.5 | [69] |
4.3. Analysis in other types of samples
Tissue samples from 10 PCa patients were divided into two sets, i.e. a set from healthy prostates and a set from prostates with a tumour [70]. Anti-PSA immobilized within wells of the plate was applied for PSA capture from these sample sets with subsequent O-glycan analysis by addition of an antibody against core 2 sialyl Lewis X (sLex) glycan. This is the only published study focusing on the glycoprofiling of O-glycan on PSA. The results indicate a significant difference in the O-glycan profile of PSA when these two sets of samples were compared with p = 0.03 [70]. Moreover, the O-glycoprofiling of two other proteins, such as MUC1 and prostatic acid phosphatase (PAP), could also be applied to distinguish between these two sets of samples (p = 0.01 or p = 0.06, respectively) [70]. Three different lectins proved to be useful for the glycoprofiling of PSA isolated from a tissue to distinguish aggressive from indolent PCa: p = 0.0049 for SNA [71], p = 0.006 for Jacalin [71] and p = 0.001 for AAL [62].
Drake's group used seminal plasma to glycoprofile PSA (identification of 40 unique glycans) and PAP (identification of 21 unique glycans on three glycosylation sites—Asn62, Asn188 and Asn301) using a combination of high-performance liquid chromatography and matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) MS. A PSA was isolated from the samples using thiophilic adsorption chromatography. A PSA gel slice from PAGE was digested by PNGase, glycans were labelled and subsequently analysed using LC. The study, however, did not reveal a statistical significance of glycan analysis for PCa diagnostics [72].
5. Conclusion
It may be concluded that, when the analysis of a biomarker is applied to a serum sample from a grey zone, the AUC value drops by 0.05 compared with the analysis performed on samples with PSA levels across a wide range. This pertains to the use of a PHI test (0.694 versus 0.735) [54] and also to PSA glycoprofiling using WFA lectin (0.75 versus 0.80) [58].
Another conclusion can be drawn by evaluating the clinical performance of a glycan biomarker when a moderate number (100) of samples is compared with a wider number of samples (314). The AUC dropped from a value of 0.95 (100 samples) to 0.84 when 314 serum samples were assayed [57].
In general, it may be concluded that PSA glycoprofiling is applied more frequently as a diagnostic PCa biomarker in serum (seven studies with relevant clinical data) than in urine (one study with relevant clinical data). The same trend is observed for PSA glycoprofiling applied as a prognostic PCa biomarker (4 versus 1). The explanation might be that the level of PSA from a tumour is very low in urine samples with only a minor level of aberrantly glycosylated PSA present in urine [52].
The dominant detection platform for specific PSA glycoprofiling is based on the integration of lectins either into an ELISA format or using other sophisticated protocols rather than in an instrument-based approach. This can be explained by the low sensitivity of instrumental techniques to working with biomarkers that are present in the blood in very low levels (i.e. down to several ng ml−1).
It is worth comparing the clinical performance of PSA glycoprofiling in serum applied to PCa diagnostics with the performance of PSA and PHI tests. The PSA tests afford AUC = 0.68 [14] and the PHI test could discriminate between PCa and BPH patients with AUC = 0.74 [54]. The detection of increased levels of α-2,6-SA by the SNA lectin does not offer any advantage in terms of clinical performance (AUC = 0.63) [55] over the PSA and PHI tests. An analysis of increased levels of α-2,3-SA by MAA (AUC = 0.83) [56] or HYB4 Ab (AUC = 0.95) [57] and of increased levels of LacdiNAc by the WFA lectin (AUC = 0.80–0.85) [58,59] offers a significant advantage over the PSA and PHI tests. Moreover, PSA glycoprofiling exhibits a significant advantage over PSA and PHI tests when serum samples with a PSA level in a grey zone are analysed, which is important from a clinical perspective. Accordingly, PSA glycoprofiling (especially analysis of α-2,3-SA) could be applied as a diagnostic PCa biomarker to better distinguish those men who need to undergo a biopsy, so as to avoid unnecessary biopsies.
PSA glycoprofiling can also be useful for distinguishing PCa patients with an aggressive form of the disease from those with only indolent disease when analysing α-2,3-SA (AUC = 0.97) [54,65] and α-1,6-Fuc (AUC = 0.94) [65] over the PHI test (AUC = 0.84) [54]. The detection of LacdiNAc (AUC = 0.65) [58] and α-1,6(3)-Fuc (AUC = 0.71–0.86) [66] does not provide any advantage over the PHI test.
Our results also suggest that antibody fragments for the affinity purification of PSA have distinct advantages over the application of full antibodies, including the absence of any glycan moiety within antibody fragments and the possibility of immobilizing a high density of antibody fragments on surfaces and a higher stability of antibody fragments than antibodies. Full-length antibodies need to be blocked in order to suppress the possible interference of the antibody's glycan with the lectin-based glycoprofiling [73]. Besides the antibody fragments, DNA aptamers with advantages over antibodies similar to those described for antibody fragments could also replace antibodies in the future [74].
From the authors’ perspective, the most exciting area is the use of exosomes as a precious source of cancer-related glycan-based biomarkers. It can be anticipated that some glycan biomarkers could be enriched with exosomes resulting in simplified assay formats [15]. Unfortunately, PSA present in the exosomes has not been glycoprofiled yet.
Data accessibility
No new data were collected in the course of this research. All the data are presented from the published literature with permission.
Authors' contributions
All authors contributed to writing.
Competing interests
We declare we have no competing interests.
Funding
The authors wish to acknowledge the financial support received from the Slovak Scientific Grant Agency VEGA 2/0137/18 and 2/0090/16 from the Slovak Research and Development Agency APVV 17-0300. We would like to acknowledge the support received from the ERC Proof of Concept grant (no. 825586) and from the Innovative Training Network (no. 813120). This publication is the result of the project implementation: Centre for materials, layers and systems for applications and chemical processes under extreme conditions – stage I, ITMS no.: 26240120007, supported by the ERDF.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. 2015. Global cancer statistics, 2012. CA: Cancer J. Clin. 65, 87–108. ( 10.3322/caac.21262) [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. 2015. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386. ( 10.1002/ijc.29210) [DOI] [PubMed] [Google Scholar]
- 3.O'Reilly J-A, O'Kennedy RJ. 2017. Prostate cancer detection: complexities and strategies. J. Cancer Treat. Diagn. 2, 18–25. [Google Scholar]
- 4.Rodríguez JZ, O'Kennedy R. 2017. New approaches for the development of diagnostic systems for prostate cancer. Asian Hosp. Healthc. Manag. 36, 18–23. [Google Scholar]
- 5.Ablin RJ, Piana R. 2014. The great prostate hoax: how big medicine hijacked the PSA test and caused a public health disaster. New York, NY: Palgrave McMillian. [Google Scholar]
- 6.Ablin RJ. 2014. Prostate cancer test has been misused for money. In The scientist (internet version). See https://www.newscientist.com/article/mg22129564-400-prostate-cancer-test-has-been-misused-for-money/. [Google Scholar]
- 7.Moyer VA. 2012. Screening for prostate cancer: US Preventive Services Task Force recommendation statement. Ann. Intern. Med. 157, 120–134. ( 10.7326/0003-4819-157-2-201207170-00459) [DOI] [PubMed] [Google Scholar]
- 8.Fleshner K, Carlsson SV, Roobol MJ. 2017. The effect of the USPSTF PSA screening recommendation on prostate cancer incidence patterns in the USA. Nat. Rev. Urol. 14, 26–37. ( 10.1038/nrurol.2016.251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Der Kwast TH, Roobol MJ.. 2017. Prostate cancer: draft USPSTF 2017 recommendation on PSA testing—a sea-change? Nat. Rev. Urol. 14, 457–458. ( 10.1038/nrurol.2017.89) [DOI] [PubMed] [Google Scholar]
- 10.Grossman DC, et al. 2018. Screening for prostate cancer: US Preventive Services Task Force recommendation statement. JAMA 319, 1901–1913. ( 10.1001/jama.2018.3710) [DOI] [PubMed] [Google Scholar]
- 11.Garnick MB, et al. 2018. Harvard Medical School 2018 annual report on prostate diseases. Boston, MA: Harvard Health Publishing. [Google Scholar]
- 12.Umberto A, et al. 2018. Novel diagnostic biomarkers of prostate cancer: an update. Curr. Med. Chem. 25, 1–14. ( 10.2174/0929867325666180914115416) [DOI] [PubMed] [Google Scholar]
- 13.Sharma S, Zapatero-Rodríguez J, O'Kennedy R. 2017. Prostate cancer diagnostics: clinical challenges and the ongoing need for disruptive and effective diagnostic tools. Biotechnol. Adv. 35, 135–149. ( 10.1016/j.biotechadv.2016.11.009) [DOI] [PubMed] [Google Scholar]
- 14.Hatakeyama S, Yoneyama T, Tobisawa Y, Ohyama C. 2017. Recent progress and perspectives on prostate cancer biomarkers. Int. J. Clin. Oncol. 22, 214–221. ( 10.1007/s10147-016-1049-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tkac J, Bertok T, Hires M, Jane E, Lorencova L, Kasak P. 2019. Glycomics of prostate cancer: updates. Exp. Rev. Proteomics. 16, 65–76. ( 10.1080/14789450.2019.1549993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilgunn S, Conroy PJ, Saldova R, Rudd PM, O'Kennedy RJ. 2013. Aberrant PSA glycosylation—a sweet predictor of prostate cancer. Nat. Rev. Urol. 10, 99–107. ( 10.1038/nruro1.2012.258) [DOI] [PubMed] [Google Scholar]
- 17.Guo S, Briza P, Magdolen V, Brandstetter H, Goettig P. 2018. Activation and activity of glycosylated KLKs 3,4 and 11. Biol. Chem. 399, 1009–1022. ( 10.1515/hsz-2018-0148) [DOI] [PubMed] [Google Scholar]
- 18.Stura EA, Muller BH, Bossus M, Michel S, Jolivet-Reynaud C, Ducancel F. 2011. Crystal structure of human prostate-specific antigen in a sandwich antibody complex. J. Mol. Biol. 414, 530–544. ( 10.1016/j.jmb.2011.10.007) [DOI] [PubMed] [Google Scholar]
- 19.Guo S, Skala W, Magdolen V, Brandstetter H, Goettig P. 2014. Sweetened kallikrein-related peptidases (KLKs): glycan trees as potential regulators of activation and activity. Biol. Chem. 395, 959–976. ( 10.1515/hsz-2014-0140) [DOI] [PubMed] [Google Scholar]
- 20.Song E, Hu Y, Hussein A, Yu C-Y, Tang H, Mechref Y. 2015. Characterization of the glycosylation site of human PSA prompted by missense mutation using LC-MS/MS. J. Proteome Res. 14, 2872–2883. ( 10.1021/acs.jproteome.5b00362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drake RR, Jones EE, Powers TW, Nyalwidhe JO. 2015. Altered glycosylation in prostate cancer. In Glycosylation and cancer (eds Drake RR, Ball LE), pp. 345–382. London, UK: Academic Press. [DOI] [PubMed] [Google Scholar]
- 22.Fukushima K, Satoh T, Baba S, Yamashita K. 2010. Alpha 1,2-fucosylated and beta-N-acetylgalactosaminylated prostate-specific antigen as an efficient marker of prostatic cancer. Glycobiology 20, 452–460. ( 10.1093/glycob/cwp197) [DOI] [PubMed] [Google Scholar]
- 23.Scorilas A, Mavridis K. 2014. Predictions for the future of kallikrein-related peptidases in molecular diagnostics. Exp. Rev. Mol. Diagn. 14, 713–722. ( 10.1586/14737159.2014.928207) [DOI] [PubMed] [Google Scholar]
- 24.Kammeijer GSM, et al. 2017. Sialic acid linkage differentiation of glycopeptides using capillary electrophoresis–electrospray ionization–mass spectrometry. Sci. Rep. 7, 3733 ( 10.1038/s41598-017-03838-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dosekova E, Filip J, Bertok T, Both P, Kasak P, Tkac J. 2017. Nanotechnology in glycomics: applications in diagnostics, therapy, imaging, and separation processes. Med. Res. Rev. 37, 514–626. ( 10.1002/med.21420) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landhuis E. 2017. Glycobiology: sweet success. Nature 547, 127–129. ( 10.1038/nj7661-127a) [DOI] [Google Scholar]
- 27.RodrÍguez E, Schetters STT, van Kooyk Y. 2018. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 18, 204–211. ( 10.1038/nri.2018.3) [DOI] [PubMed] [Google Scholar]
- 28.Schneider M, et al. 2017. Inhibition of delta-induced notch signaling using fucose analogs. Nat. Chem. Biol. 14, 65–71. ( 10.1038/nchembio.2520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, et al. 2017. Selective in vivo metabolic cell-labeling-mediated cancer targeting. Nat. Chem. Biol. 13, 415–424. ( 10.1038/nchembio.2297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beatson R, et al. 2016. The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat. Immunol. 17, 1273–1281. ( 10.1038/ni.3552) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinho SS, Reis CA. 2015. Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555. ( 10.1038/nrc3982) [DOI] [PubMed] [Google Scholar]
- 32.Stowell SR, Ju T, Cummings RD. 2015. Protein glycosylation in cancer. Annu. Rev. Pathol. 10, 473–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinho SS, Reis CA. 2015. Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540. [DOI] [PubMed] [Google Scholar]
- 34.Teoh ST, Ogrodzinski MP, Ross C, Hunter KW, Lunt SY. 2018. Sialic acid metabolism: a key player in breast cancer metastasis revealed by metabolomics. Front. Oncol. 8, 174 ( 10.3389/fonc.2018.00174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanas A, Sahasrabudhe NM, Rodríguez E, van Kooyk Y, van Vliet SJ. 2018. Fucosylated antigens in cancer: an alliance toward tumor progression, metastasis, and resistance to chemotherapy. Front. Oncol. 8, 39 ( 10.3389/fonc.2018.00039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kailemia MJ, Xu GG, Wong M, Li QY, Goonatilleke E, Leon F, Lebrilla CB. 2018. Recent advances in the mass spectrometry methods for glycomics and cancer. Anal. Chem. 90, 208–224. ( 10.1021/acs.analchem.7b04202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munkley J, Elliott DJ. 2016. Hallmarks of glycosylation in cancer. Oncotarget 7, 35 478–35 489. ( 10.18632/oncotarget.8155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Wuhrer M, Holst S. 2018. Serum sialylation changes in cancer. Glycoconjugate J. 35, 139–160. ( 10.1007/s10719-018-9820-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen JD, et al. 2018. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930. ( 10.1126/science.aar3247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy K, et al. 2018. Integrating biomarkers across omic platforms: an approach to improve stratification of patients with indolent and aggressive prostate cancer. Mol. Oncol. 12, 1513–1525. ( 10.1002/1878-0261.12348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Etxebarria J, Reichardt N-C. 2016. Methods for the absolute quantification of N-glycan biomarkers. Biochim. Biophys. Acta-Gen. Subj. 1860, 1676–1687. ( 10.1016/j.bbagen.2016.03.003) [DOI] [PubMed] [Google Scholar]
- 42.Chocholova E, et al. 2018. Glycomics meets artificial intelligence—potential of glycan analysis for identification of seropositive and seronegative rheumatoid arthritis patients revealed. Clin. Chim. Acta 481, 49–55. [DOI] [PubMed] [Google Scholar]
- 43.Bertok T, et al. 2015. Carboxybetaine modified interface for electrochemical glycoprofiling of antibodies isolated from human serum. Langmuir 31, 7148–7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vermassen T, Speeckaert MM, Lumen N, Rottey S, Delanghe JR. 2012. Glycosylation of prostate specific antigen and its potential diagnostic applications. Clin. Chim. Acta 413, 1500–1505. ( 10.1016/j.cca.2012.06.007) [DOI] [PubMed] [Google Scholar]
- 45.Damborska D, Bertok T, Dosekova E, Holazova A, Lorencova L, Kasak P, Tkac J. 2017. Nanomaterial-based biosensors for detection of prostate specific antigen. Microchim. Acta 184, 3049–3067. ( 10.1007/s00604-017-2410-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banazadeh A, Veillon L, Wooding KM, Zabet-moghaddam M, Mechref Y. 2017. Recent advances in mass spectrometric analysis of glycoproteins. Electrophoresis 38, 162–189. ( 10.1002/elps.201600357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lang R, Leinenbach A, Karl J, Swiatek-de Lange M, Kobold U, Vogeser M. 2018. An endoglycosidase-assisted LC-MS/MS-based strategy for the analysis of site-specific core-fucosylation of low-concentrated glycoproteins in human serum using prostate-specific antigen (PSA) as example. Clin. Chim. Acta 480, 1–8. ( 10.1016/j.cca.2018.01.040) [DOI] [PubMed] [Google Scholar]
- 48.Jia G, et al. 2017. Alterations in expressed prostate secretion—urine PSA N-glycosylation discriminate prostate cancer from benign prostate hyperplasia. Oncotarget 8, 76 987–76 999. ( 10.18632/oncotarget.20299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trefulka M, Paleček E. 2017. Distinguishing glycan isomers by voltammetry. Modification of 2,3-sialyllactose and 2,6-sialyllactose by osmium(VI) complexes. Electrochem. Commun. 85, 19–22. ( 10.1016/j.elecom.2017.10.014) [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Tian Y, Rezai T, Prakash A, Lopez MF, Chan DW, Zhang H. 2011. Simultaneous analysis of glycosylated and sialylated prostate-specific antigen revealing differential distribution of glycosylated prostate-specific antigen isoforms in prostate cancer tissues. Anal. Chem. 83, 240–245. ( 10.1021/ac102319g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsiao C-J, Tzai T-S, Chen C-H, Yang W-H, Chen C-H. 2016. Analysis of urinary prostate-specific antigen glycoforms in samples of prostate cancer and benign prostate hyperplasia. Dis. Markers 2016, 8915809 ( 10.1155/2016/8915809) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barrabes S, Llop E, Ferrer-Batalle M, Ramirez M, Aleixandre RN, Perry AS, de Llorens R, Peracaula R.. 2017. Analysis of urinary PSA glycosylation is not indicative of high-risk prostate cancer. Clin. Chim. Acta 470, 97–102. ( 10.1016/j.cca.2017.05.009) [DOI] [PubMed] [Google Scholar]
- 53.Kosanovic M, Jankovic M. 2014. Isolation of urinary extracellular vesicles from Tamm-Horsfall protein-depleted urine and their application in the development of a lectin-exosome-binding assay. Biotechniques 57, 143–149. ( 10.2144/000114208) [DOI] [PubMed] [Google Scholar]
- 54.Ferrer-Batallé M, Llop E, Ramírez M, Aleixandre RN, Saez M, Comet J, de Llorens R, Peracaula R.. 2017. Comparative study of blood-based biomarkers, α2, 3-sialic acid PSA and PHI, for high-risk prostate cancer detection. Int. J. Mol. Sci. 18, 845 ( 10.3390/ijms18040845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meany DL, Zhang Z, Sokoll LJ, Zhang H, Chan DW. 2009. Glycoproteomics for prostate cancer detection: changes in serum PSA glycosylation patterns. J. Proteome Res. 8, 613–619. ( 10.1021/pr8007539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishikawa T, et al. 2017. An automated micro-total immunoassay system for measuring cancer-associated 2,3-linked sialyl N-glycan-carrying prostate-specific antigen may improve the accuracy of prostate cancer diagnosis. Int. J. Mol. Sci. 18, 470 ( 10.3390/ijms18020470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoneyama T, et al. 2014. Measurement of aberrant glycosylation of prostate specific antigen can improve specificity in early detection of prostate cancer. Biochem. Biophys. Res. Commun. 448, 390–396. ( 10.1016/j.bbrc.2014.04.107) [DOI] [PubMed] [Google Scholar]
- 58.Hagiwara K, et al. 2017. Wisteria floribunda agglutinin and its reactive-glycan-carrying prostate-specific antigen as a novel diagnostic and prognostic marker of prostate cancer. Int. J. Mol. Sci. 18, 261 ( 10.3390/ijms18020261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaya T, Kaneko T, Kojima S, Nakamura Y, Ide Y, Ishida K, Suda Y, Yamashita K. 2015. High-sensitivity immunoassay with surface plasmon field-enhanced fluorescence spectroscopy using a plastic sensor chip: application to quantitative analysis of total prostate-specific antigen and GaINAc beta 1–4GIcNAc-linked prostate-specific antigen for prostate cancer diagnosis. Anal. Chem. 87, 1797–1803. ( 10.1021/ac503735e) [DOI] [PubMed] [Google Scholar]
- 60.Dwek MV, Jenks A, Leathem AJC. 2010. A sensitive assay to measure biomarker glycosylation demonstrates increased fucosylation of prostate specific antigen (PSA) in patients with prostate cancer compared with benign prostatic hyperplasia. Clin. Chim. Acta 411, 1935–1939. ( 10.1016/j.cca.2010.08.009) [DOI] [PubMed] [Google Scholar]
- 61.Pihikova D, Kasak P, Kubanikova P, Sokol R, Tkac J. 2016. Aberrant sialylation of a prostate-specific antigen: electrochemical label-free glycoprofiling in prostate cancer serum samples. Anal. Chim. Acta 934, 72–79. ( 10.1016/j.aca.2016.06.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kekki H, Peltola M, van Vliet S, Bangma C, van Kooyk Y, Pettersson K. 2017. Improved cancer specificity in PSA assay using Aleuria aurantia lectin coated Eu-nanoparticles for detection. Clin. Biochem. 50, 54–61. ( 10.1016/j.clinbiochem.2016.06.015) [DOI] [PubMed] [Google Scholar]
- 63.Hatakeyama S, et al. 2006. Carbohydrate structure of prostate-specific antigen and its distinct affinity to Maackia amurensis lectin between cancer and non-cancer source. Glycobiology 16, 1152 ( 10.1093/glycob/cwh071) [DOI] [Google Scholar]
- 64.Basu PS, Majhi R, Batabyal SK. 2003. Lectin and serum-PSA interaction as a screening test for prostate cancer. Clin. Biochem. 36, 373–376. ( 10.1016/s0009-9120(03)00050-x) [DOI] [PubMed] [Google Scholar]
- 65.Llop E, et al. 2016. Improvement of prostate cancer diagnosis by detecting PSA glycosylation-specific changes. Theranostics 6, 1190–1204. ( 10.7150/thno.15226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li QK, Chen L, Ao M-H, Chiu JH, Zhang Z, Zhang H, Chan DW. 2015. Serum fucosylated prostate-specific antigen (PSA) improves the differentiation of aggressive from non-aggressive prostate cancers. Theranostics 5, 267–276. ( 10.7150/thno.10349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narimatsu H, Sato T. 2018. Wisteria floribunda agglutinin positive glycobiomarkers: a unique lectin as a serum biomarker probe in various diseases. Exp. Rev. Proteomics. 15, 183–190. ( 10.1080/14789450.2018.1419066) [DOI] [PubMed] [Google Scholar]
- 68.Bhanushali PB, Badgujar SB, Tripathi MM, Gupta S, Murthy V, Krishnasastry MV, Puri CP. 2016. Development of glycan specific lectin based immunoassay for detection of prostate specific antigen. Int. J. Biol. Macromol. 86, 468–480. ( 10.1016/j.ijbiomac.2016.01.110) [DOI] [PubMed] [Google Scholar]
- 69.Fujita K, et al. 2016. Decreased fucosylated PSA as a urinary marker for high Gleason score prostate cancer. Oncotarget 7, 56 643–56 649. ( 10.18632/oncotarget.10987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Z, Gulzar ZG, St Hill CA, Walcheck B, Brooks JD. 2014. Increased expression of GCNT1 is associated with altered o-glycosylation of PSA, PAP, and MUC1 in human prostate cancers. Prostate 74, 1059–1067. ( 10.1002/pros.22826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Y, Tao S-C, Bova GS, Liu AY, Chan DW, Zhu H, Zhang H. 2011. Detection and verification of glycosylation patterns of glycoproteins from clinical specimens using lectin microarrays and lectin-based immunosorbent assays. Anal. Chem. 83, 8509–8516. ( 10.1021/ac201452f) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.White KY, et al. 2009. Glycomic characterization of prostate-specific antigen and prostatic acid phosphatase in prostate cancer and benign disease seminal plasma fluids. J. Proteome Res. 8, 620–630. ( 10.1021/pr8007545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen S, LaRoche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, Brand RE, Haab BB. 2007. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat. Methods 4, 437–444. ( 10.1038/NMETH1035) [DOI] [PubMed] [Google Scholar]
- 74.Jolly P, Damborsky P, Madaboosi N, Soares RRG, Chu V, Conde JP, Katrlik J, Estrela P. 2016. DNA aptamer-based sandwich microfluidic assays for dual quantification and multi-glycan profiling of cancer biomarkers. Biosens. Bioelectron. 79, 313–319. ( 10.1016/j.bios.2015.12.058) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were collected in the course of this research. All the data are presented from the published literature with permission.