Abstract
Background
Approximately 10 million people in Pakistan are infected with the hepatitis C virus (HCV). Most patients develop chronic hepatitis, with rare cases of spontaneous clearance. However, little is known about multidrug resistant viral variants in Pakistan.
Findings
This case study describes a 47-year-old male diagnosed with chronic HCV genotype 3a infection in 2003. After an initial diagnosis of viral infection, the patient remained treatment naïve for 5 years. He received two therapy cycles of interferon (IFN) plus ribavirin (RBV) in 2007 and 2010, however, he was non-responsive to the therapy. The patient then received an additional two treatment cycles of pegylated IFN α-2b plus RBV (in 2011 and 2013); he was still non-responsive. In 2016, the patient underwent sofosbuvir plus RBV combination therapy, however, the sustained virological response was still not achieved. The host genetic factor was found to be heterozygous guanine and thymine (GT) and cytosine and thymine (CT) genotypes of rs8099917 and rs12979860 polymorphism of IL28B, respectively. Phylogenetic analysis suggests that the resistant variant belong to an out-group and may require triple therapy.
Conclusions
This is the first case that reports on a HCV-infected individual who was a non-responder to multiple IFN therapies in Pakistan. Further studies are needed to understand multidrug-resistant HCV variants in the Pakistani population.
Electronic supplementary material
The online version of this article (10.1186/s40249-018-0386-7) contains supplementary material, which is available to authorized users.
Multilingual abstracts
Please see additional file 1 for translations of the abstract into the five official working languages of the United Nations.
Background
The hepatitis C virus (HCV) is a single-stranded positive ribonucleic acid (RNA) virus with a high genetic variability rate. A single infected HCV patient can have a mixture of closely related viral genomes, either as quasispecies or separate groups referred to as genotypes [1]. Genotypes strongly affect outcomes of the standard interferon (IFN) treatment regimen [2, 3]. Different studies have reported that close relationships exist between genotyping, disease behaviour, and response to IFN treatment [4–7]. Patients infected with HCV genotypes 2 and 3 respond more efficiently to IFN-based therapies than those infected with a genotype 1 infection [8]. Sustained virological response (SVR) rates to standard IFN plus ribavirin (RBV) treatment regimens in the HCV-positive population of Pakistan have been reported to be between 27.8% and 62% [9]. Higher SVR rates have been reported in relation to pegylated interferon (PEG-IFN) in HCV-2a infected patients as compared to patients infected with HCV-3a [10–12]. Recently, more effective oral treatment options have been introduced with reported SVR rates of about 90% (sometimes > 90%) and minimal adverse effects.
Sofosbuvir (SOF) is one of the direct-acting antivirals (DAAs) approved by the U.S. Food and Drug Administration (US-FDA). It was approved on December 6, 2013 [13, 14]. In treatment-naïve patients infected with genotype 1, phase 2 trials of 400 mg/day of PEG-IFN, RBV, and SOF for 12 or 24 weeks resulted in SVRs of 87–92% [15, 16].
Although SOF has been available in Pakistan at a subsidized price since December 2014, there is limited literature on treatment response rates [17, 18]. This paper reports on a HCV-infected patient who was non-responsive to multiple combination antiviral therapies.
Case presentation
A 47-year-old male with a body weight of 84 kg and height of 1.65 m living in underprivileged part of Islamabad has a history of generalized pain, fatigue, and fever. He was diagnosed with chronic HCV (3a genotype) infection in 2003. The patient’s medical history was not significant except for dental surgery and few surgical stitches. He remained treatment naïve for 5 years. In 2007, he received the first IFN (100 mg/week) plus RBV (400 mg/day) combination therapy and was on this treatment for 6 months. However, SVR was not achieved. He remained without treatment for the next 2 years (2008–2009). In 2010, he again underwent the same combination therapy. After 6 months of treatment, he, remained positive for HCV RNA. These treatments were not only expensive but also resulted in adverse effects, including stomach burning, loss of appetite, nausea, fever, fatigue, and anxiety.
In 2011, Patient was advised to undergo PEG-IFN plus RBV combination therapy. However, he remained a non-responder. After one and a half year, in 2013, patient received the same (PEG-IFN plus RBV) combination therapy for 6 months. Yet patient’s serum was still positive for HCV RNA. Eventually, in 2015, he underwent SOF (400 mg/day) plus RBV combination therapy for 6 months. Still SVR was not achieved and surprisingly a high viral load of 5.2 × 105 IU/ml was reported by real-time polymerase chain reaction (PCR) diagnosis. An ultrasound revealed that his liver was of normal shape, size, and echotexture; he had a mildly fatty liver with no fibrosis or lesion.
The patient’s diagnostic and treatment history are summarized in Tables 1 and 2, respectively. Viral genotype remained undetermined/untypable for the years 2012, 2014, and early 2016 (Table 1) following 6 months of combination therapy (Table 2), perhaps due to the detection method’s incapability or the detection limit as it was performed on conventional PCR-based method followed by detection on the agarose gel. In late 2016, the viral load was found to be 5.2 × 105 IU/ml and the patient was found positive for genotype 3a (Table 1).
Table 1.
Patient diagnostic history
| Diagnostic tests | Method | Results | Date/Year |
|---|---|---|---|
| HCV | Qualitative PCRa | Detectable RNA | 2003 |
| HCV | Qualitative PCR | Detectable RNA | 12–03-2008 |
| Ultrasound | Ultrasound (liver) | Liver is normal shape, size and echotexture, mild fatty liver | 2009 |
| HCV | Qualitative PCR | Detectable RNA | 06–05-2011 |
| HCV | Qualitative PCR | Detectable RNA | 12–12-2011 |
| Ultrasound | Ultrasound (liver) | Liver is normal size, shape and echotexture, no focal lesion, mild fatty liver | 23–12-2011 |
| HCV | Qualitative PCR | Non-Detectable | 18–04-2012 |
| HCV | PCR Genotyping | Un-typable | 24–4- 2012 |
| Liver enzymes | ALT (NR 10–50) | 40 IU | 8–7-2013 |
| HCV | Quantitative PCR | Detectable RNA 77874 IU/ml | 21–5-2014 |
| HCV | PCR Genotyping | Un-typable | 27–6-2014 |
| Liver enzymes | ALT (NR 10–50) | 36 IU | 18–09-2015 |
| Liver enzymes | ALT (NR 10–50) | 27 IU | 14–12-2015 |
| HCV | Qualitative PCR | Detectable RNA | 23–4-2016 |
| Ultrasound | Ultrasound (liver) | Liver is normal size, shape and echotexture, no focal lesion | 8–05–2016 |
| HCV | PCR Genotyping | Untypable | 28–5-2016 |
| HCV | PCR Genotyping | 3a | 11–8-2016 |
| HCV | Quantitative PCR | Detectable RNA 5.2 × 105 IU/ml | 11–8-2016 |
Abbreviations: HCV hepatitis C virus, PCR polymerase chain reaction, ALT alanine aminotransferase, NR normal range
aThere was no trend of prescribing quantitative PCR before treatment in Pakistan
Table 2.
Patient treatment history
| Therapy used | Mode of treatment | Start date | End date | Duration of therapy | Result | Side effects/complications |
|---|---|---|---|---|---|---|
| interferon alpha-2b plus RBV | Interferon | 9–07-2007 | 5–01-2008 | 6 months | Resistant to treatment/Non responder | Stomach burning, Loss of appetite, Nausea, Fever, Fatigue, feeling anxious or aggressive |
| interferon alpha-2b plus RBV | Interferon | 11–07-2010 | 5–01-2011 | 6 months | Resistant to treatment/Non responder | Stomach burning, Loss of appetite, Nausea, Fever, Fatigue, feeling anxious or aggressive |
| PEG-Interferon alpha-2b plus RBV | Interferon | 24–08-2011 | 20–02-2012 | 6 months | Resistant to treatment/Non responder | Stomach burning, Loss of appetite, Nausea, Fever, Fatigue, feeling anxious or aggressive |
| PEG-interferon alpha-2b plus RBV | Interferon | 11–08-2013 | 08–02-2014 | 6 months | Resistant to treatment/Non responder | Stomach burning, Loss of appetite, Nausea, Fever, Fatigue, feeling anxious or aggressive |
| Sofobuvir plus RBV | Polymerase inhibitor | 1–08-2015 | 1–02-2016 | 6 months | Resistant to treatment/Non responder | Loss of appetite |
After partial genome sequencing of NS5B, BLAST analysis showed 93% similarity to the already existing NS5B nucleotide sequences in the GenBank database (Fig. 2). This shows virus (accession number KY971494; variant ‘Pk1-RV’) is of genotype 3a. Analysis further confirmed that the variant (Pk1-RV) is distinct from HCV genotypes 3 k, 3b, 1a, and 1b (Fig. 2).
Fig. 2.
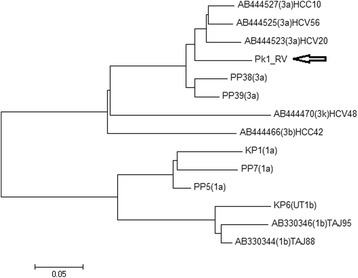
Nucleotide sequence-based genotyping of HCV. The resistant variant is labeled as Pk1-RV (GenBank accession no. KY971494) and clustered with genotype 3a reference sequences. The fragment length of target sequence is 353 nucleotides of NS5B gene. Analysis was carried out using 14 reference viral genome sequences. Sequences included genotype 3a (accession no. AB44523, AB44525, AB44527, PP3, PP39, and KPK2); 3b (accession no. AB444466); 3 k (accession no. AB444470); 1a (KPK1, PP5, PP7); and 1b (KPK6, accession no. AB330344, AB330346)
Taking into account the medical history of the patient, we performed restriction fragment length polymorphism (RFLP) for interleukin 28B (IL28B) at rs8099917 and rs12979860. The present study showed polymorphism cytosine and thymine (CT) and guanine and thymine (GT) at (rs12978960, (rs8099917) respectively, as in (Fig. 1a and b). Same polymorphism was reported by Yang et al. who has linked it with successful treatment outcome (SVR) [19].
Fig. 1.
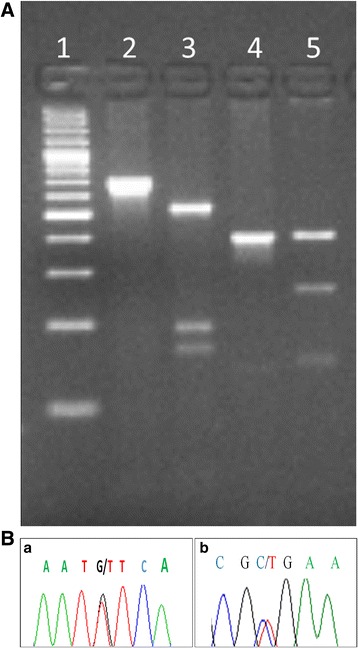
a IL-28B polymorphism showing host genotype CT (rs12978960) and TG (rs8099917) in the patient. Line 1: DNA ladder; Line 2: Amplified uncut product of rs12978960; Line 3: CT genotype by RFLP; Line 4: Amplified uncut product of rs8099917; Line 5: GT genotype by Restriction Fragment Length Polymorphism (RFLP). b Chromatogram showing (a) heterozygous G/T (rs8099917) and (b) heterozygous C/T (rs12979860) polymorphism of IL28b
Materials and methods
Patient history and blood sample collection
Patient’s medical record showed that he remained non-responding to multiple antiviral therapies (i.e. IFN + RBV and PEG-IFN + RBV) during 2003–2015 (as stated in Tables 1 and 2). In 2016, patient was found also non-responsive to sovaldi + RBV combination therapy. His blood sample was analysed for viral and host factors such as viral genotyping, viral load, IL28B polymorphism and partial genome sequencing of NS5B.
Biochemical test
Blood level of liver functional enzyme alanine aminotransferase (ALT) was analysed using the Abcam® Alanine Transaminase Activity Assay Kit (ab105134), (Cambridge, UK) as according to the manufacturer’s instructions.
RNA and DNA extraction
For viral load determination, genotyping and partial genome sequencing of NS5B, viral RNA was isolated using viral RNA extraction kit (Instant Virus RNA Kit, AJ Roboscreen). The human genomic DNA for IL28B typing is extracted (Phenol chloroform method).
Viral load
The viral load was determined using a previously reported method [20] through real-time PCR machine (Rotor-Gene 3 000™, Corbett Research, Sydney, Australia). The lower limit of detection of the assay was set at 50 IU/ml [21].
HCV genotyping
Extracted viral RNA was processed for viral genotyping using a method previously reported by Aziz et al. [22].
IL28B typing
Polymorphism in the IL28B gene (rs8099917, rs12979860) was detected by amplifying genomic DNA using the primer sets, as described previously [23, 24]. Restriction digestion was performed using BseMI (BsrDI) and Hpy166II and DNA fragments were detected on 2% agarose gel. The gel image pattern confirmed GT and CT genotypes for IL28B (rs8099917 and rs12979860, respectively).
Phylogenetic analysis of HCV isolate obtained from target patient
Sequencing-based viral genotyping was assessed using NS5B genome sequencing, as reported previously by Ali et al. [25]. Sequencing results showed that the genotype was 3a.
MEGA 7 software [26] was used for assessing the phylogenetic relationship of the partial genome sequence (Pk1-RV; GenBank accession no. KY971494), with the reference sequences obtained from GenBank. HCV NS5B reference sequences from different regions of Pakistan and Tajikistan were compared with the partial viral genome sequence obtained from the patient. A few reference (genotype 1b obtained from GenBank database) sequences were from Tajikistan.
The resistant variant clustered with out-group, which need further research to get in site of the phenomenon. These sequences include different genotypes and subtypes (3a, 3b, and 3 k, and 1a and 1b). Evolutionary history was inferred using the neighbour-joining method [27]. The optimal tree with the sum of branch length = 1.30808452 was constructed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches [28]. The tree was constructed using a neighbor-joining algorithm (Fig. 2).
Discussion
In present study, we reported a multidrug-resistant HCV-infected patient with genotype 3a and its association with IL28B polymorphism. Ultrasound examination showed that patient’s liver was normal in size, shape and echo texture with no focal lesion. In spite of having favourable virus and host genetic factor i.e. IL28B polymorphism CT and GT at rs12978960 and rs8099917, respectively, and ALT in normal range, we observed higher viremia in blood at the end of sovaldi plus ribavirin treatment.
Sezaki et al. conducted a study to determine the impact of IL28B polymorphism on SVR in patients on a ledipasvir/SOF regimen and observed that genotypes GT and CT were associated with lower response rates [29]. It has been reported that polymorphism in IL28B (rs12979860) is strongly associated with the response rate to the antiviral treatment regimen. The most favourable polymorphism in terms of responsiveness to therapy is cytosine-cytosine (CC) (rs12979860), which has been reported to have an almost two-fold higher likelihood of attaining a SVR as compared to patients with a cytosine-thymine (CT) or thymine-thymine (TT) genotype at the same locus [30].
Previous studies have shown that the TT (rs8099917) and CC (rs12978960) host-genotype population is associated with higher SVR rates as a result of antiviral therapies [31–33]. However, a detailed study needs to be performed to investigate host genotypes in HCV-infected individuals in Pakistan to better understand the pattern of viral response to antiviral therapies.
In the current study, the patient tolerated the SOF and RBV combination therapy well, however, he failed to achieve SVR. Ali et al. recently conducted a systematic review of treatment response rates in Pakistani HCV-infected patients and concluded that IFN plus RBV combination therapies have SVR rates of 64.38–68.38% [31]. Oral treatment against all HCV genotypes, in the majority of patients, is now possible due to the availability of a number of highly effective IFN-free regimens [34]. Kowdley et al. performed a multicenter study and reported that SOF was relatively safe, with rare viral breakthrough during treatment and fewer drug interactions [15]. Akhter et al. conducted the first study in Pakistan that reported SVR rates of 85.5% to RBV plus SOF combination therapy (n = 502; genotype 3a) [17]. Patients infected with HCV-3 have been reported to have lower response rates to SOF and RBV therapy than those infected with genotype 2 [16]. Therefore, a combined therapy of RBV and SOF for 24 weeks is an effective treatment regimen for post-transplantation HCV infection [35].
Mutations associated with resistance to DAAs especially SOF have been reported in several studies [36, 37]. However, there is limited information from Pakistan about the selection of DAA-resistant viral mutations [23]. Viral sequences from Pakistan need to be investigated for mutations and/or amino acid substitutions associated with possible non-responsiveness to/relapse associated with antiviral therapies.
Conclusions
In this case, viral genetic factor (genotype 3a) and host genetic factors (CT at locus rs12978960 and GT at rs8099917) were mildly favourable in terms of responsiveness to therapies. The phylogenetic analysis showed a distinct 3a genotype, which cluster with the 3a genotype from Pakistan however, it was observed that the sequence is more evolutionary diverse on the basis of an increased branched length of the node in the phylogenetic tree. Therefore, it is concluded that mutation in the viral genome and host genetic factor could be responsible for the patient’s non-responsiveness to the therapies. We recommend administration of triple therapies (RBV + INF + SOF/ Boceprevir or RBV + BRF + SOF), which could be effective in non-responding patients. It is also suggested to use a combined therapy of Olysio™ and SOF. In addition, further studies should be conducted to understand the possible mechanism(s) of viral non-responsiveness to therapy regimens, with a special focus on Pakistani patients.
Additional file
Multilingual abstract in the five official working languages of the United Nations. (PDF 460 kb)
Acknowledgements
The authors are thankful to the participant of the study for his cooperation. Financial support from the Higher Education Commission (HEC) of Pakistan is highly acknowledged.
Funding
AZ is supported by “Indigenous Ph.D. fellowship program” from the Higher Education Commission (HEC) of Pakistan.
Availability of data and materials
All data generated or analysed during this study are included in the published paper (and its supplementary information files).
Sequence submitted to GenBank
Partial NS5B sequence of the current viral isolate has been submitted to GenBank under accession no. KY971494 (isolate Pk1_RV).
Abbreviations
- BRF
Boceprevir
- CT
Cytosine and thymine
- DAA
Direct-acting antiviral
- GT
Guanine and thymine
- HCV
Hepatitis C Virus
- IFN
Interferon
- IL28B
Interleukin 28B
- PCR
Polymerase chain reaction
- PEG-IFN
Pegylated interferon
- RBV
Ribavirin
- RFLP
Restriction Fragment Length Polymorphism
- RNA
Ribonucleic acid
- SOF
Sofosbuvir
- SVR
Sustained virological response
Authors’ contributions
AZ and AR conceived the study. AZ collected the samples and data. AZ, HA and MA performed the experiments, analysed the data, and drafted the paper. AR, ZKS, MZ, and MA critically revised the paper. All authors read and approved the paper for publication.
Ethics approval and consent to participate
The study has been approved by the departmental ethical committee of Quaid-i-Azam University Islamabad under the number biotech-F-15-657. Informed consent was obtained from the patient before the blood samples were collected.
Consent for publication
Authors have obtained consent from the study participant to report the data without disclosing his identity.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Asad Zia, Email: asadzia96@gmail.com.
Muhammad Ali, Email: ali_biotech01@yahoo.com.
Hafsa Aziz, Email: hafsa.aziz@gmail.com.
Muhammad Zia, Email: ziachaudhary@gmail.com.
Zabta Khan Shinwari, Email: shinwari2008@gmail.com.
Abida Raza, Phone: +92 51 9248671-6, Email: abida_rao@yahoo.com.
References
- 1.Echeverría N, Moratorio G, Cristina J, Moreno P. Hepatitis C virus genetic variability and evolution. World J Hepatol. 2015;7(6):831–845. doi: 10.4254/wjh.v7.i6.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeuzem S, Feinman SV, Rasenack J, Heathcote EJ, Lai MY, Gane E, et al. Peginterferon alfa-2a in patients with chronic hepatitis C. N Engl J Med. 2000;343(23):1666–1672. doi: 10.1056/NEJM200012073432301. [DOI] [PubMed] [Google Scholar]
- 3.Heathcote EJ, Shiffman ML, WGE C, Dusheiko GM, Lee SS, Balart L, et al. Peginterferon alfa-2a in patients with chronic hepatitis C and cirrhosis. N Engl J Med. 2000;343(23):1673–1680. doi: 10.1056/NEJM200012073432302. [DOI] [PubMed] [Google Scholar]
- 4.Trepo C, Habersetzer F, Bailly F, Berby F, Pichoud C, Berthillon P, et al. Interferon therapy for hepatitis C. Antivir Res. 1994;24(2–3):155–163. doi: 10.1016/0166-3542(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 5.Akram M, Idrees M, Zafar S, Hussain A, Butt S, Afzal S, et al. Effects of host and virus related factors on interferon-α+ ribavirin and pegylated-interferon+ ribavirin treatment outcomes in chronic hepatitis C patients. Virol J. 2011;8:234. doi: 10.1186/1743-422X-8-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inamullah I, Idrees M, Ahmed H, Ali M, Ali L, Ahmed A. Hepatitis C virus genotypes circulating in district swat of Khyber Pakhtoonkhaw. Pakistan Virol J. 2011;8:16. doi: 10.1186/1743-422X-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rehman I-u, Idrees M, Ali M, Ali L, Butt S, Hussain A, et al. Hepatitis C virus genotype 3a with phylogenetically distinct origin is circulating in Pakistan. Genet Vaccines Ther. 2011;9:2. doi: 10.1186/1479-0556-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dusheiko G, Main J, Thomas H, et al. Ribavirin treatment for patients with chronic hepatitis C: results of a placebo-controlled study. J Hepatol. 1996;25(5):591–598. doi: 10.1016/S0168-8278(96)80225-X. [DOI] [PubMed] [Google Scholar]
- 9.Idrees M. Development of an improved genotyping assay for the detection of hepatitis C virus genotypes and subtypes in Pakistan. J Virol Methods. 2008;150(1–2):50–56. doi: 10.1016/j.jviromet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Shiffman ML, Suter F, Bacon BR, Nelson D, Harley H, Solá R, et al. Peginterferon alfa-2a and ribavirin for 16 or 24 weeks in HCV genotype 2 or 3. N Engl J Med. 2007;357(2):124–134. doi: 10.1056/NEJMoa066403. [DOI] [PubMed] [Google Scholar]
- 11.Alberti A. Optimizing PEG-interferon and ribavirin combination therapy for patients infected with HCV-2 or HCV-3: is the puzzle completed? J Hepatol. 2004;40(6):1032–1035. doi: 10.1016/j.jhep.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Marcellin P, Cheinquer H, Curescu M, Dusheiko GM, Ferenci P, Horban A, et al. High sustained virologic response rates in rapid virologic response patients in the large real world prophesys cohort confirm results from randomized clinical trials. Hepatology. 2012;56(6):2039–2050. doi: 10.1002/hep.25892. [DOI] [PubMed] [Google Scholar]
- 13.Lawitz E, Sulkowski MS, Ghalib R, Rodriguez-Torres M, Younossi ZM, Corregidor A, et al. Simeprevir plus sofosbuvir, with or without ribavirin, to treat chronic infection with hepatitis C virus genotype 1 in non-responders to pegylated interferon and ribavirin and treatment-naive patients: the COSMOS randomised study. Lancet. 2014;384(9956):1756–1765. doi: 10.1016/S0140-6736(14)61036-9. [DOI] [PubMed] [Google Scholar]
- 14.Lawitz E, Gane EJ. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;369(7):678–679. doi: 10.1056/NEJMc1307641. [DOI] [PubMed] [Google Scholar]
- 15.Kowdley KV, Lawitz E, Crespo I, Hassanein T, Davis MN, DeMicco M, et al. Sofosbuvir with pegylated interferon alfa-2a and ribavirin for treatment-naive patients with hepatitis C genotype-1 infection (ATOMIC): an open-label, randomised, multicentre phase 2 trial. Lancet. 2013;381(9883):2100–2107. doi: 10.1016/S0140-6736(13)60247-0. [DOI] [PubMed] [Google Scholar]
- 16.Lawitz E, Lalezari JP, Hassanein T, Kowdley KV, Poordad FF, Sheikh AM, et al. Sofosbuvir in combination with peginterferon alfa-2a and ribavirin for non-cirrhotic, treatment-naive patients with genotypes 1, 2, and 3 hepatitis C infection: a randomised, double-blind, phase 2 trial. Lancet Infect Dis. 2013;13(5):401–408. doi: 10.1016/S1473-3099(13)70033-1. [DOI] [PubMed] [Google Scholar]
- 17.Akhter TS, Umar M, Aslam F, Nisar G, Naseer A, Ahmad S, et al. Sofosbuvir for the treatment of hepatitis c genotype 3 infected patients in pakistan. J Ayub Med Coll Abbottabad. 2017;28((4) Suppl 1):S884–S889. [PubMed] [Google Scholar]
- 18.Mansoor VB, Ahmed U, Jahanzaib M, Ali Z, Haroon MA, Ahmed H, et al. End treatment response in chronic hepatitis C patients to Sofosbuvir and Ribavirin. Ann of PIMS. 2016;127:127–30.
- 19.Yang M, Rao HY, Feng B, Zhang W, Wei L. Impact of interleukin 28B polymorphisms on spontaneous clearance of hepatitis C virus infection: a meta analysis. J Gastroenterol Hepatol. 2013;28(7):1114–1121. doi: 10.1111/jgh.12233. [DOI] [PubMed] [Google Scholar]
- 20.Aziz H, Raza A, Irfan J. Optimum predictors of therapeutic outcome in HCV patients in Pakistan. J Med Virol. 2016;88(1):100–108. doi: 10.1002/jmv.24305. [DOI] [PubMed] [Google Scholar]
- 21.Raza A, Ali Z, Irfan J, Murtaza S, Shakeel S. Analytical variables influencing the HCV RNA determination by TaqMan real-time PCR in routine clinical laboratory practice. Mol Biol Rep. 2012;39(7):7421–7427. doi: 10.1007/s11033-012-1574-3. [DOI] [PubMed] [Google Scholar]
- 22.Aziz H, Raza A, Murtaza S, Waheed Y, Khalid A, Irfan J, et al. Molecular epidemiology of hepatitis C virus genotypes in different geographical regions of Punjab Province in Pakistan and a phylogenetic analysis. Int J Infect Dis. 2013;17(4):e247–e253. doi: 10.1016/j.ijid.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 23.Moreira S, Garcia RF, Gutberlet A, Bertol BC, Ferreira LE, Pinho MD, et al. A straightforward genotyping of the relevant IL28B SNPs for the prediction of hepatitis C treatment outcome. J Virol Methods. 2012;184(1–2):93–97. doi: 10.1016/j.jviromet.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 24.Aziz H, Raza A, Ali K, JZK K, Irfan J, Gill ML. Polymorphism of the IL28B gene (rs8099917, rs12979860) and virological response of Pakistani hepatitis C virus genotype 3 patients to pegylated interferon therapy. Int J Infect Dis. 2015;30:91–97. doi: 10.1016/j.ijid.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 25.Ali M, Rehman IU, Idrees M. Emergence of genetically variant hepatitis C virus population in response to increased antiviral drug pressure, Pakistan. Virus Genes. 2014;48(3):543–549. doi: 10.1007/s11262-014-1047-y. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 29.Sezaki H, Suzuki F, Fujiyama S, Kawamura Y, Hosaka T, Akuta N, et al. The impact of IL28B polymorphism and HCV NS5A resistance associated variants on treatment response with ledipasvir and sofosbuvir regimen in Japanese real-life settings. Hepatology. 2016;64(Suppl S1):968A–9A. https://liverlearning.aasld.org/aasld/2016/thelivermeeting/144845/hitomi.sezaki.the.impact.of.il28b.polymorphism.and.hcv.ns5a.resistance.html.
- 30.Muir AJ, Gong L, Johnson SG, Lee MM, Williams MS, Klein TE, et al. Clinical Pharmacogenetics implementation consortium (CPIC) guidelines for IFNL3 (IL28B) genotype and PEG interferon- α–based regimens. Clin Pharmacol Ther. 2014;95(2):141–146. doi: 10.1038/clpt.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali M, Afzal S, Zia A, Hassan A, Khalil AT, Ovais M, et al. A systematic review of treatment response rates in Pakistani hepatitis C virus patients; current prospects and future challenges. Medicine (Baltimore) 2016;95(50):e5327. doi: 10.1097/MD.0000000000005327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, et al. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;368(20):1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 33.Poordad F, Bronowicki JP, Gordon SC, Zeuzem S, Jacobson IM, Sulkowski MS, et al. Factors that predict response of patients with hepatitis C virus infection to boceprevir. Gastroenterology. 2012;143(3):608–618.e5. doi: 10.1053/j.gastro.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 34.Alghamdi AS, Alqutub A, Abaalkhail F, Sanai FM, Alghamdi H, Altraif I, et al. SASLT position statement on the direct-acting antiviral agents for the treatment of hepatitis C virus infection. Saudi J Gastroenterol. 2015;21(2):60–63. doi: 10.4103/1319-3767.153810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charlton M, Gane E, Manns MP, Brown RS, Curry MP, Kwo PY, et al. Sofosbuvir and ribavirin for treatment of compensated recurrent hepatitis C virus infection after liver transplantation. Gastroenterology. 2015;148(1):108–117. doi: 10.1053/j.gastro.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Costantino A, Spada E, Equestre M, Bruni R, Tritarelli E, Coppola N, et al. Naturally occurring mutations associated with resistance to HCV NS5B polymerase and NS3 protease inhibitors in treatment-naïve patients with chronic hepatitis C. Virol J. 2015;12:186. doi: 10.1186/s12985-015-0414-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donaldson EF, Harrington PR, O'Rear JJ, Naeger LK. Clinical evidence and bioinformatics characterization of potential hepatitis C virus resistance pathways for sofosbuvir. Hepatology. 2015;61(1):56–65. doi: 10.1002/hep.27375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multilingual abstract in the five official working languages of the United Nations. (PDF 460 kb)
Data Availability Statement
All data generated or analysed during this study are included in the published paper (and its supplementary information files).


