Abstract
Quantitative systems pharmacology (QSP) is a rapidly emerging discipline with application across a spectrum of challenges facing the pharmaceutical industry, including mechanistically informed prioritization of target pathways and combinations in discovery, target population, and dose expansion decisions early in clinical development, and analyses for regulatory authorities late in clinical development. QSP's development has influences from physiologic modeling, systems biology, physiologically‐based pharmacokinetic modeling, and pharmacometrics. Given a varied scientific heritage, a variety of tools to accomplish the demands of model development, application, and model‐based analysis of available data have been developed. We report the outcome from a community survey and resulting analysis of how modelers view the impact and growth of QSP, how they utilize existing tools, and capabilities they need improved to further accelerate their impact on drug development. These results serve as a benchmark and roadmap for advancements to the QSP tool set.
Executive Summary
Quantitative systems pharmacology (QSP) is a growing discipline focused on the development of mechanistic models of biological and/or physiological processes and pharmacology to integrate knowledge and data into a predictive, testable framework. A web‐based survey sponsored by the International Society of Pharmacometrics’ (ISoP) QSP Special Interest Group (SIG) was conducted to assess the perceived adequacy and capabilities of software packages currently used for QSP model development. The survey was divided into three sections: demographic information, a set of detailed questions about the capabilities for QSP‐centric modeling tasks for a single software package, and a listing of potential features for an “ideal” QSP software platform that the respondents were asked to prioritize. The survey garnered 105 unique responses, with the majority of respondents being primarily QSP modelers and the rest identifying as either other types of modelers in the pharmaceutical industry (e.g., pharmacokinetic (PK), PK/pharmacodynamic (PK/PD), physiologically based pharmacokinetic (PBPK), etc.) or nonmodeling pharmaceutical professionals that oversee or collaborate with modelers (Figure 1). Results suggest that QSP modelers primarily are building deterministic systems of ordinary differential equations (ODEs) and tend to prefer tools with extensive scripting capabilities (Figures 4 and 5). However, the community is divided over the utility of modularity and object‐oriented principles in model construction and the utility of graphical‐based model design suites, indicating potential opportunities for improvements in these feature domains. Two other definitive areas for improvement identified among the platforms surveyed were intrinsic documentation capabilities (Figure 5) and organization of parameters. Despite the lack of structure for maintaining parameters, almost all modelers utilize some form of parameter estimation algorithm that is associated with the software platforms, with gradient‐based, nonlinear mixed‐effects, and global optimization algorithms approximately equally utilized.
Prioritization of new features was analyzed collectively among respondents and then stratified for self‐reported experience in QSP modeling (Table 2). Feature requests were generally aligned with the results of the capabilities assessed in section 2, with the need for scripting tools, high performance computing, visualization capacity, and multiple parameter estimation techniques highlighted strongly in the collective population and across experience groups. In addition, across experience groups, modularity of model components and integration with additional external tools from related disciplines (e.g., bioinformatics) were seen as less important features. Of note, several trends emerged showing divergent opinions of feature utility as a function of experience. Less experienced QSP modelers preferred packages with graphical model design capability and access to pre‐existing models in a compatible format and were less interested in the ability to generate large models. Conversely, more experienced modelers were less interested in the graphical design and model‐sharing features and favored platforms capable of large model design, simulation, and management.
The results of this survey identify some intriguing relationships between modeler experience/role and preferred software platform/features and offer potential for future development efforts. Modelers new to QSP seem to find value in graphical model design and availability of pre‐existing models. A question that arises is: Why are those capabilities ranked important less frequently by experienced QSP modelers? Is it because in their training those features were not readily available, so they adapted and now see no need to use them? Is it that, as models become larger and more complex, the available graphical design tools become less effective in their stated task? Perhaps a renewed platform development effort is needed to improve the ability of graphical suites to implement self‐contained submodels, thus reducing the complexity of model diagrams at each scale of the model; however, the feature request for such modularity was low. Again, is this due to such features having been unavailable for most modelers or only implemented by those with considerable object‐oriented programming experience? These are only a few of the questions that can be mined from the survey results, and the community is invited to analyze and interpret the results on their own and to find additional relationships that describe how QSP modelers are using current software tools. The final question, however, must be centered on how these results and requests are acknowledged by the software platform developers to enable continued and sustained growth for QSP modeling applications.
Over the last few decades, the pharmaceutical industry has increasingly relied on mathematical modeling as part of model‐informed drug discovery and development (MID3).1 Mathematical modeling and simulation is used for rigorous experimental data analysis, drug candidate screening, dose selection, design, and optimization of preclinical and clinical studies and other purposes.2, 3 PK, PD, and population PK (PopPK) modeling have become routine practice in drug evaluation and approval by drug regulatory agencies.4, 5 Recently, a more comprehensive approach termed QSP modeling has been gaining popularity as a way to accelerate, improve, and reduce costs of MID3.6, 7 QSP has been described as a “quantitative analysis of the dynamic interactions between drug(s) and a biological system” that “…aims to understand the behavior of the system as a whole.”8 In 2011, a National Institutes of Health White Paper published by the QSP Workshop Group9 provided an authoritative and comprehensive summary of what QSP is and how it will promote and accelerate drug discovery and development. In contrast to traditional PK/PD approaches, QSP models rely on more detailed representations of a drug's mechanism of action and the underlying physiology, which lead to more complex model structures, often with a larger number of equations and parameters. With sufficient project scope and time frame, QSP modeling may attempt to achieve a broader goal of creating comprehensive disease models covering multiple pharmacological targets. QSP also may be used to answer more focused and specific questions. Naturally, QSP modeling requires an advanced set of software tools to accomplish these goals. Given these diverse and potentially demanding tasks, QSP modeling software ideally should be capable of doing the following:
Create and run reusable, physiologically based models that include a quantitative description of a drug's pharmacological effects on disease pathophysiology and/or progression.
Offer comprehensive model documentation tools.
Provide capabilities to represent individual and population level variability in PK and PD responses.
Execute computationally intensive simulation tasks for modeling clinical trials with multiple therapies and different drug regimens and provide a means to store, document, and analyze large numbers of simulation results.
The majority of software tools currently used for QSP modeling were not originally designed for this purpose. Existing PK, PopPK, or PK/PD software packages (e.g., NONMEM,10 Phoenix11); systems biology software (e.g., JDesigner, Systems Biology Workbench12, 13); or general purpose modeling software (e.g., Mathworks MATLAB/Mathworks SimBiology,14 R and its packages,15, 16 Wolfram Mathematica and SystemModeler,17 and Berkeley Madonna18) are often used. All have advantages and disadvantages and have limits in their applicability for QSP modeling. Several software tools (e.g., PhysioLab19 by Entelos, Aegis by Immunetrics,20 PKSim, and MoBi21 by Bayer Technologies) were developed specifically for QSP modeling as a primary objective. There have been additional QSP software initiatives, including DBSolve,22 ViSP,23 and Open Systems Pharmacology,24 as well as specialized MATLAB and R packages including KroneckerBio,25 the QSP Toolbox,26 MatVPC,27 and mrgsolve.28 However, these tools have yet to gain a large following in the QSP community, as they are used primarily by end users within or in collaboration with their originating institutions. With a growing popularity and acceptance of QSP modeling, there is an ever‐increasing demand for a comprehensive QSP software suite that can satisfy the larger requirements. Developing such software will not be an easy task, as it requires a thorough understanding of the current and future needs and challenges of QSP modeling.
ISoP is a nonprofit organization whose mission is the promotion and advancement of the discipline of pharmacometrics, which encompasses increasingly diverse mathematical and statistical specializations, including mechanistic modeling and QSP. A QSP SIG has been established within ISoP, with goals that include advancing knowledge and practice through timely communication and resources, including publications. Feedback from the QSP community provides a good starting point for guidance on features and requirements for effective and successful QSP software. We conducted a detailed survey of the QSP community to identify and emphasize the software tool requirements, as well as to highlight the ongoing trends in this developing field as part of recent activities endorsed by the ISoP QSP SIG. This survey complements other efforts seeking feedback on existing competencies and modeling approaches (e.g., by Drug Disease Model Resources consortium education and training working group29, 30) as well as feedback on practices in preclinical PK/PD and QSP analysis adopted across pharmaceutical companies.31, 32
We established a web‐based survey with the goals to:
Gain a comprehensive view of how organizations and individual scientists are implementing QSP modeling and how software tools are currently used by the QSP community for MID3.
Identify and prioritize software and modeling capabilities that are considered necessary for successful application of QSP modeling in MID3.
Provide necessary information and feedback for software developers on how QSP software could be improved.
Provide an objective evaluation to inform the community of the capabilities of popular QSP modeling tools.
The emphasis of this survey was on tools and capabilities available now and those expected in the near future, approximately a 3‐year period. This article describes and analyzes the results of the survey with the hope that its conclusions will be informative for the QSP community in general and helpful for the software developers to deliver tools to better serve QSP modelers’ needs.
Methods
A web‐based survey format was chosen to provide flexibility and to reach a broad respondent pool that uses mathematical and computer modeling in MID3. The survey was designed using Google Forms technology and contained a set of questions with answer options. Snapshots of the web pages, including questions and suggested answers, can be viewed in Supplementary File S1 . The survey was advertised via multiple internet channels (websites) and by sending email announcements to members registered with the ISoP. Survey participants registered with their email addresses, which were used solely for the purpose of returning the survey results, if requested, and for identifying duplicate responses. The survey was opened in mid‐October 2016 and closed on April 1, 2017. Once closed, the survey results were collected and saved in a Microsoft Excel spreadsheet.
The survey was divided into several sections. The introductory section stated the goals of the survey and provided instructions and was followed by three question‐based sections. First, the “general” section asked broad scope questions about the respondent's background and experience with QSP modeling, as well as her/his organization's application of and commitment to QSP modeling. Second, the “technical” section queried specific questions about respondent's experience with a particular QSP software of choice. The final “preferred features” section aimed to collect the respondent's feedback on features of a “dream tool” for QSP modeling work. Sections 1 through 3 contained 16, 36, and 18 questions, respectively. For ease of navigation, the first and second sections were divided into subsections. Each section provided an approximate time for completion, with an estimated total time to complete the survey of 30–35 minutes. Depending on the question, users were allowed to select single or multiple answers from provided choices. Some questions had a text box for customized feedback not covered by the available options.
The data analysis presented below, with the exception of section 3, is not intended to be a comprehensive statistical evaluation of the results for significant trends in respondents’ answers. Instead, it is aimed at providing a snapshot of available QSP software tools, their utilization, and users’ opinions on how these tools could be improved in the future. In section 3 of the survey, respondents were asked to rate 18 selected QSP software features as to their importance in a hypothetical new QSP modeling platform. Features could be rated in the order of importance and placed into three groups—(i) “most important,” (ii) “somewhat important,” and (iii) “least important”—with each group containing six features. When a feature was selected, a score of 3, 2, or 1 was assigned to indicate most (3) to least (1) level of importance. We analyzed the mean and rank‐order trends for the entire response set and also categorized responses based on respondents’ self‐reported experience in QSP. Respondents were divided into three categories, those with “low” experience in QSP (< 1 year), “medium” experience in QSP (1–3 years), and “high” experience in QSP (> 3 years). As a result, a 3 × 3 contingency table was formed for each question. The responses were tested by Fisher's exact method to determine significant differences in the pattern of response based on experience category. Trends were deemed significant if the P value was < 0.05.
Results
We received a total of 109 records (participants), of which 4 were deemed duplicates submitted by participants twice at different times. For each duplicate pair, we excluded the record that had the earlier time stamp. Not all respondents answered all questions; some questions were answered in an incorrect manner, which made unique interpretation impossible. Consequently, when calculating percent distributions, only nonblank, unambiguous answers were included. In general, we did not attempt a rigorous statistical analysis of the results. Instead, we chose to present the observations and conclusions from the survey straightforwardly to highlight apparent trends in the QSP community's response. Below we present the results of the survey by section. Additional details can be found in the Supplementary Materials . An anonymized version of the survey results is provided in Supplementary File S4 , in case the reader wants to perform more detailed analyses of the responses.
Information about survey participants
Affiliation and experience
Figure 1 presents composition of survey participants by (i) affiliation and (ii) level of involvement and experience in QSP modeling. It is worth noting that the majority of participants (72%) are employed by for‐profit organizations, biopharma, contract research organizations (CROs), consulting, and QSP service providers (Figure 1 a), 71% of whom are doing QSP modeling. This suggests that modeling, and particularly QSP modeling (~40% of participants from the for‐profit segment are dedicated to QSP modeling), is considered a valuable activity deserving investment of time and resources. The overall distribution of QSP modelers follows the composition of the survey participants by affiliation, indicating that QSP modeling has similar representation in various sectors.
Figure 1.
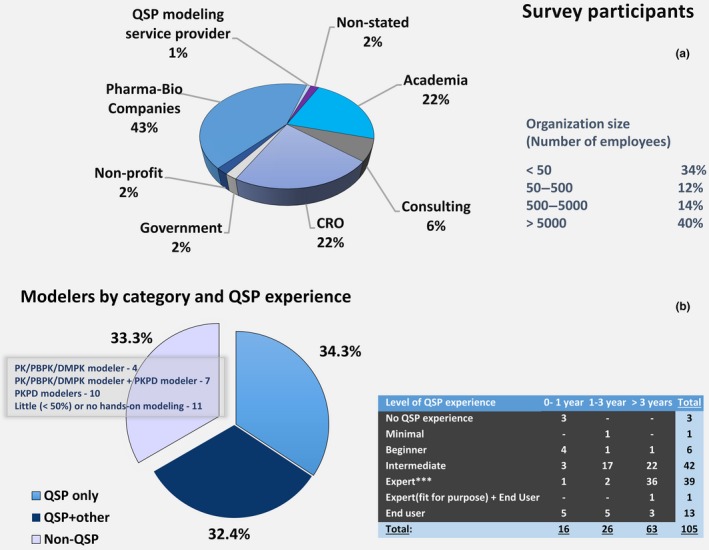
Composition of survey participants by (a) affiliation and (b) experience. DMPK, drug metabolism and pharmacokinetics; PBPK, physiologically based pharmacokinetic; PK, pharmacokinetic; PKPD, pharmacokinetic/pharmacodynamic; QSP, quantitative systems pharmacology.
With regard to QSP modeling experience, almost all participants (97%) have some, and the majority (60%) have three or more years of experience (table inset in Figure 1 b). According to the poll, it takes ≥ 3 years to become a QSP expert as evidenced by the people who self‐classified as QSP experts, and > 1 year to reach an intermediate level. Expert modelers tend to be in higher numbers in for‐profit organizations (pharma, 45%; CROs, 30%; and consulting, 10%), whereas only 15% were reported to be in academia. This may reflect the fact that a significant proportion of QSP modelers in academia are students and post‐docs, who leave academia before reaching an expert level.
Within organizations, QSP modeling is conducted primarily in relatively small (< 50 people) departments. Within these departments, in 54% of the cases, it is a group of 1–5 scientists that perform QSP modeling and use QSP in research (Table 1), whereas only in 10% of cases these groups are larger than 10 scientists. Similar proportions hold when considering the number of “dedicated” QSP scientists that spend more than half of their time on QSP modeling. At the same time, in ~9.5% of all cases surveyed, departments have no scientists who conduct QSP modeling. However, according to respondents, the situation will change within the next 3 years, as there will be more scientists working primarily in QSP research. In addition, a shift toward larger QSP groups is anticipated within departments. The number of people for whom QSP modeling is not a primary function will stay approximately unchanged.
Table 1.
Allocation of resources for QSP modeling
| Size of department | No. of employees using QSP in research | Dedicated scientists with > 50% of their time on QSP | Dedicated scientists with < 50% of their time on QSP | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1–5 | 5–10 | > 10 | 0 | 1–5 | 5–10 | > 10 | 0 | 1–5 | 5–10 | > 10 | |
| < 50 | 9 | 43 | 20 | 8 | 17 (11) | 42 (36) | 12 (18) | 8 (15) | 27 (26) | 42 (39) | 5 (9) | 6 (6) |
| 50–500 | 1 | 15 | 4 | 3 | 5 (2) | 14 (13) | 2 (6) | 2 (2) | 7 (7) | 11 (10) | 3 (4) | 2 (1) |
| 500–5,000 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 (0) | 0 | 0 | 0 |
| > 5,000 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| (%) – distribution | ||||||||||||
| < 50 | 11% | 54% | 25% | 10% | 22 (13)% | 53 (45)% | 15 (22)% | 10 (20)% | 34 (33)% | 53 (49)% | 6 (11)% | 8 (8)% |
| 50–500 | 4% | 65% | 17% | 13% | 22 (9)% | 61 (57)% | 9 (26)% | 9 (9)% | 30 (32)% | 48 (45)% | 13 (18)% | 9 (5)% |
Resources for QSP modeling inside departments where it is conducted. The upper part of the table provides the absolute number of responses; the lower part reports responses as percent of total. Numbers in parenthesis show values expected in 3 years. QSP, quantitative systems pharmacology.
Utilization of QSP modeling
A large proportion of respondents (42%) stated that their organizations rely entirely on internal resources when doing QSP modeling; almost half (48%) of them work in a CRO. Pharma companies, academic institutions, government agencies, and nonprofit organizations rely less on internal resources, as reported in ~16%, 27%, 2%, and 2% cases, correspondingly. In 41% of cases, a combination of outsourcing and internal work is reported; 70% of these are in pharma companies. At the opposite end of the spectrum, < 3% of responses claimed that their organizations outsource QSP modeling work entirely, and in 13% of cases no resources are dedicated to QSP modeling. Thus, ~84% of respondents reported at least some level of involvement with QSP modeling in their organizations.
QSP modeling is expected to have a broader scope and impact in MID3. It provides valuable insights and has the potential to influence decisions throughout the entire drug discovery and development process. This is reflected in answers to questions related to where QSP modeling has demonstrated significant impact. About 60% of respondents placed evaluating drug combinations, dosing regimens, and preclinical‐clinical translational studies at the top of their lists (Figure 2 a). Other questions belonging to use of QSP modeling in early clinical development, such as evaluating drug safety, go/no go decisions, and phase I and II study design and data analyses, rated highly (45–50% of responses). Late clinical development is generally regarded as less impacted by QSP work (33% of responses); however, patient stratification and biomarker evaluation, which are relevant to phase III studies, are considered important use‐cases by 56% of respondents. In addition, late‐stage development can be influenced by QSP modeling by comparing drug candidates to competitors and standards of care, as indicated by 42% of respondents. QSP modeling plays an important role throughout the discovery phase and in target prioritization (both 56% cases), followed by compound optimization (47%). QSP modeling in the preclinical phase also has more immediate priorities. When asked about near‐term (1 year) deliverables, the majority of modelers (78%) expect to apply QSP to understand underlying biology and a drug's mechanism of action (Figure 2 b). Other frequent and significant questions listed as immediate QSP goals are data analysis and parameter estimation (61%), simulation and optimization of experiments, clinical studies (58–60%), and simulating disease progression (47%). Strikingly, issues, such as predicting drug toxicity, were ranked much lower, despite being recognized by 45% of respondents as one of the areas influenced by QSP modeling (Figure 2 a).
Figure 2.
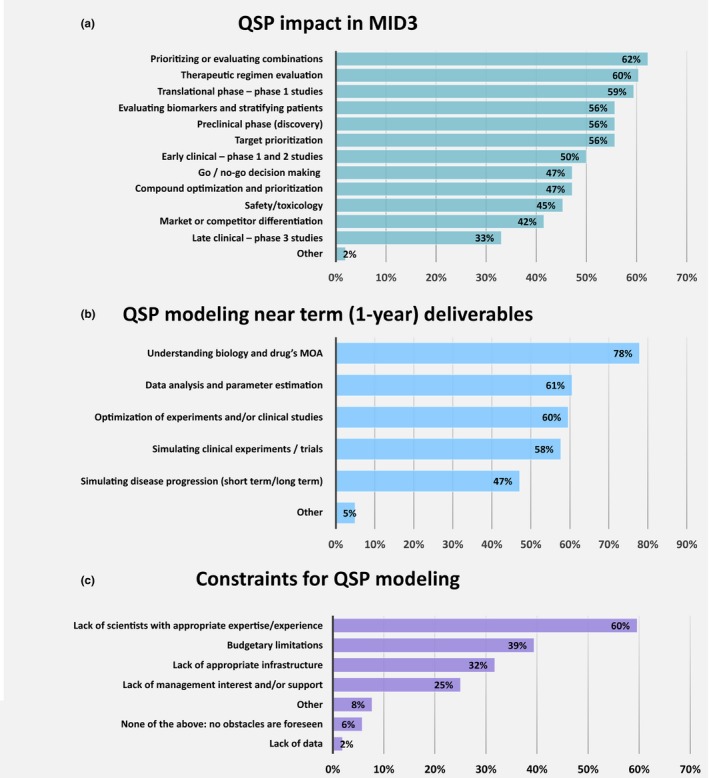
(a) Effect of quantitative systems pharmacology (QSP) modeling in drug discovery and development, (b) expected near term QSP modeling goals and deliverables, and (c) major obstacles to further progress in QSP modeling. MID3, model‐informed drug discovery and development; MOA, mechanism of action.
Adoption and widespread utilization of QSP modeling faces multiple obstacles. In most occasions, there is more than one factor that reduces the efficiency and impact of QSP modeling from its full potential. The lack of scientists with appropriate expertise is considered the biggest impediment according to 60% of respondents, followed by budgetary limitations (39%) and lack of infrastructure (32%; Figure 2 c). Lack of management interest and support is highlighted by 25% of respondents (69% of them in the pharmaceutical industry), whereas only 6% of all respondents see no obstacles. Other barriers not stated specifically in the survey but submitted by respondents were lack of data to inform models, organizational issues that lead to a disconnect from project teams, lack of demonstrated impact, and insufficient time for development of QSP models to produce such impact. Although much less common, there were opinions about more serious barriers for QSP modeling, including a lack of clinical buy‐in, absence of modeling standards, challenges to measure the value of QSP modeling, and quite skeptical views of QSP modeling resulting from oversold and unrealistic expectations (~4% combined).
When asked about types of models developed and utilized, 92% of respondents rely on deterministic ODE‐based models as part of their work. Stochastic models and frameworks that combine different types of models are used equally by 30% of respondents, followed by deterministic partial differential equation models with 23% and agent‐based models with 18%. Less used approaches included delayed differential equations, Boolean network‐based models, and machine learning (5% combined).
QSP software analysis
Evaluated tools
Section 2 of the survey comprised questions evaluating the current state of software tools as well as the features modelers considered necessary for their work. Respondents were asked to select the software that is most familiar to her/him and to provide an assessment of the selected tool. Rather than presenting a comprehensive breakdown of responses for each tool here, the responses have been combined to give a better general description of the features available in current software and where new features may be required to address the needs of the QSP community. Each question in section 2 received between 96 and 103 responses.
At the beginning of this section, the survey form listed 11 software tools to evaluate, and the user had an option to specify an additional tool, if such was missing. Sixteen were identified as QSP tools of choice, which are shown in Figure 3. The popularity and the original scope of these tools vary substantially. Three quarters of respondents preferred four tools: (i) MathWorks MATLAB, (ii) MathWorks SimBiology, (iii) R and R packages, and (iv) NONMEM. MATLAB and R were the most prevalent and belong to a category of multipurpose computing software platforms, whereas NONMEM was developed as a standard tool for pharmacometric analysis. Following the top four tools were the JDesigner and Entelos PhysioLab Modeler (~5% share each). Both of these software tools have visual modeling environments. However, the PhysioLab Modeler was specifically designed for the purpose of physiologic modeling as opposed to modeling chemical reaction networks. Next in popularity, the Berkeley Madonna and DBSolve were developed to quickly solve ODEs resulting from systems of biochemical reactions. Together, these eight software tools represented 90% of responses. The remaining eight tools have smaller footprints; each was selected by users only once or twice. Like the above tools, these were not necessarily designed for QSP modeling. Thus, specialized QSP modeling software represents a relatively small portion of the tools surveyed, showing a clear opportunity for developing novel tools for the QSP community. This fact also underscores that QSP modeling is a relatively new discipline; existing tools have filled the void, wheres the development of available customized tools is just beginning.
Figure 3.
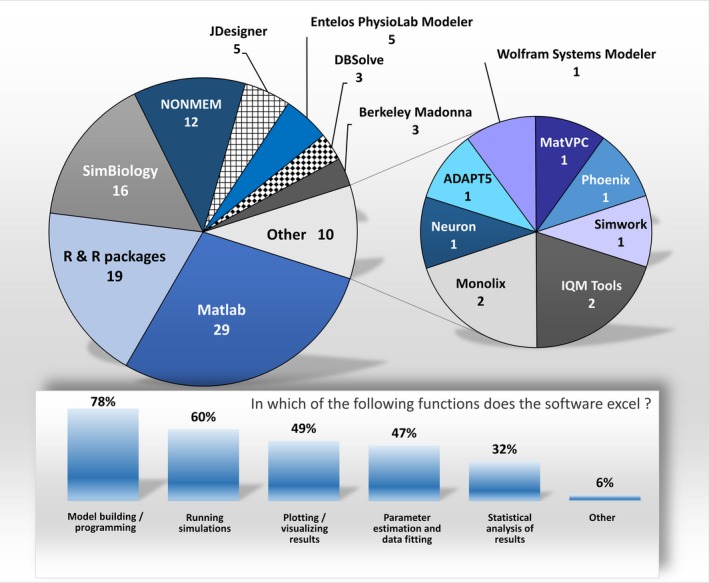
Upper part shows the software tools selected by survey participants for their evaluation; 102 responses total. Lower plot indicates the feature(s) in which the selected software tool excels, as perceived by its user; responses combined for all tools evaluated.
Software basics
Once a software tool was selected, respondents were asked which tasks their tool excelled at. QSP software tended to perform well at model building tasks (78%), running simulations (60%), plotting (49%), parameter estimation (47%), and statistical analysis (32%; Figure 3). Indeed, the features that are used most frequently are also praised the most, and likely the software tools are selected based on their capabilities for these specific features. Few users gave software high marks for other features, such as model visualization (1%), knowledge and data integration (1%), defined precision (1%), model generalizability and transparent syntax (1%), and hierarchical modeling capabilities (1%).
The first series of questions was focused on basic software features and use, such as software and model development, deployment, and collaboration environment (for results see Supplementary File S2 and Figures S2–S10 ). According to responses, most software tools are run on Windows operating systems (80%), although a substantial portion used Linux (27%) and MacOS (27%; Figure S3 ). More than half (59%) used parallel computing capabilities ( Figure S4 ), whereas the remainder did not use or were unaware of parallel capabilities in their software tool (32% and 9%, respectively). As for hardware, surprisingly, the majority of respondents run jobs on their laptops (77%), although many also ran simulations on a workstation (42%), cluster (37%), and cloud (20%; Figure S5 ). Respondents reported using a mix of software deployment and IT software options for their QSP software tool: in 65% of cases it was installed as a standalone application, 51% used server‐based, and 19% used web‐based deployment ( Figure S8 ). Utilization of collaboration capabilities and version control to develop models with colleagues was varied. Many (47%) did not use such capabilities, 26% used them directly within the tool, 21% used alternative version control software, and the rest were not aware of such capabilities ( Figure S7 ). Collaboration can take place through joint model use; however, the majority of respondents reported their models were primarily deployed on a standalone basis (85%), with less server‐based and web‐based deployment (27% and 13%, respectively; Figure S9 ). About half (53%) of respondents also indicated developing standalone applications with their software tool ( Figure S6 ). When it comes to exporting models for use with a different application, at least two options exist: (i) saving a model in a format suitable for import by a different application and, less convenient, (ii) saving a model in the form of equations to be later reprogrammed in an alternative software. For the first option, Systems Biology Markup Language (SBML) is one of the most widespread model export formats available today. However, it has not become a dominant approach in QSP, as 53% of respondents do not use it, and only 24% indicated that they need it for work ( Figure S2 ). The remaining respondents either did not know about SBML (15%) or were not aware of SBML support by their tool (9%). The ability to export models as equations was used by 46% of respondents, whereas an equal percent indicated they did not use such capabilities, and 9% were not aware of the capability ( Figure S10 ).
Model development capabilities
The next set of questions concentrated on tasks, such as model development and documentation. Although broad descriptions have been proposed,9, 33 currently there is no clear and universally accepted definition of what a QSP model is, and different mathematical approaches are used in building QSP models. However, respondents reported overwhelmingly using their QSP tool for developing ODE‐based models (95%), followed by statistical models (44%), stochastic models (29%), partial differential equation–based models (19%), and agent‐based models (14%) (Figure 4). All other types of models fell below the 5% threshold (e.g., 2% of respondents reported developing delayed differential equation models, whereas network‐based models were used by 1%).
Figure 4.
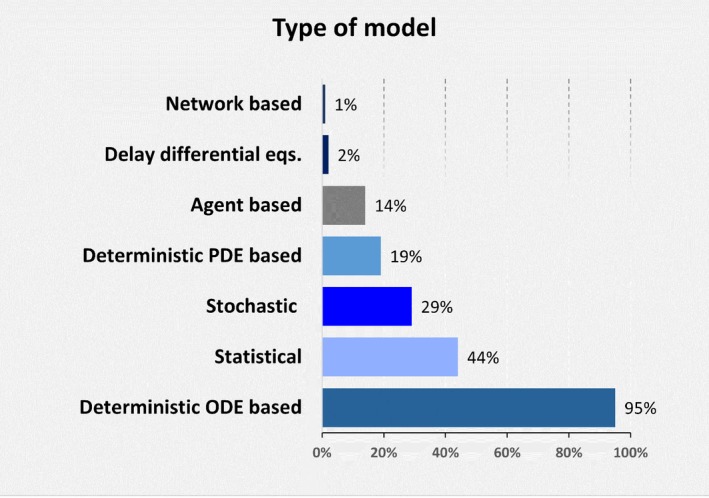
Quantitative systems pharmacology models most frequently developed by users. ODE, ordinary differential equation; PDE, partial differential equation.
The respondents’ evaluation of specific software capabilities is shown in Figure 5. One feature that scored highly is the scripting capability provided by software. Scripting becomes important when options available for model customization and model‐based analyses, for example, through a graphical user interface (GUI), are limited. The majority of respondents (57%) reported extensively using scripting tools during model development to expand capabilities of the tool, 22% reported using limited scripting, 16% reported that they did not use scripting, and 5% reported extensively using scripting but by a different software platform. Two thirds (66%) were satisfied with the available scripting tool, reporting it offered adequate scripting capabilities, languages, and editors. Remaining respondents indicated some desire for improvement: 7% pointed out they would like a more comprehensive language, 6% wished they would have a better text editor, and the other 6% indicated both need improvement. In 15% of cases, users revealed they did not use scripting. It is worth noting that for some QSP tools (e.g., MATLAB and R), the separation between the scripting and nonscripting aspects of model development and application is rather ambiguous or not relevant.
Figure 5.
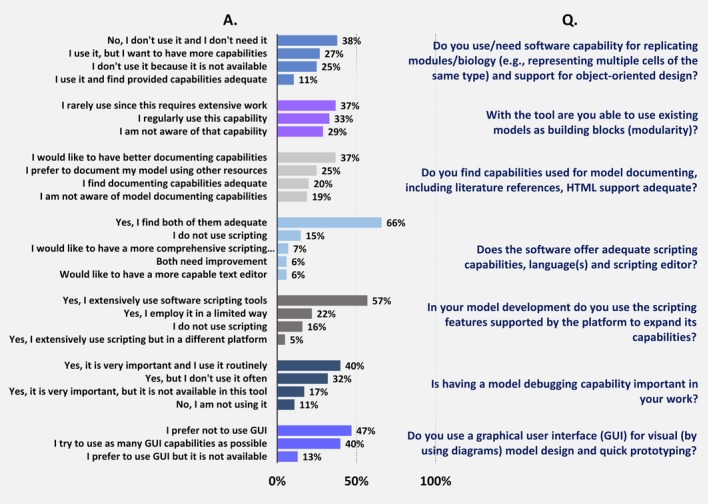
Quantitative systems pharmacology software features and their importance as evaluated by users. Survey questions are given on the right side of the plot, answer options are presented on the left against corresponding bar plots. Percent values show numbers calculated with respect to the total number of answers (survey questions 2.12–2.18).
A GUI is now a standard feature in the majority of software applications. For QSP software tools, it provides a convenient means for rapid model development and facilitates running simulations and analyzing results. GUI may also be useful for communication and collaboration, especially with nonmodelers. The majority of respondents indicated some interest in the availability and implementation of a GUI for model design and quick prototyping: 40% indicated they used as many GUI capabilities as possible, and 13% indicated they would prefer to use GUI but it is not available (Figure 5). At the same time, a substantial portion (47%) indicated they preferred not to use a GUI, presumably due to the lack of functionality and efficiency in the currently existing graphical tools.
An essential and widespread feature in traditional computing software is the availability of a debugging tool. The need for such capability during QSP model development is clearly supported by the survey results. Almost 90% of respondents acknowledge in some way the importance of debugging tools as valuable software components. Having a model debugging capability was reported as very important and regularly used by 40% of respondents, important but not used often by 32%, and very important but not available by 17%. Only 11% of survey participants did not use debugging, more than half of which are either beginners or those who are primarily interested in the results of modeling rather than model development itself.
A QSP model is often designed for multiple applications, capable of answering more questions and to be extended and reused, as compared with traditional fit‐for‐purpose PK/PD models. In order to fulfill this promise, QSP models must be well‐documented to be used by various scientists and not only by those who originally developed them. In the survey, when asked about documentation capabilities, including references and HTML support, only 20% of respondents agreed that the documentation capabilities were adequate (Figure 5). Many (37%) indicated they would like to have better capabilities, 25% preferred to document the model with other resources, and 19% were not aware of documentation capabilities. Although documentation per se is not directly connected to modeling results and predictions, there is an apparent gap in existing documentation tools, which may negatively affect a model's overall value and reusability.
Rapid model development and reliability greatly benefit from modular architecture and incorporation of pre‐existing mathematical models of organ, tissue, or cellular processes into a new model. Modular model development is widely employed in traditional engineering applications but currently is used to a limited extent in QSP model development. When asked whether QSP modelers were able to use existing models as building blocks, 29% indicated they were not aware of the capability, 37% indicated they rarely resort to this option because it requires extensive work, and only 33% indicated they regularly used the capability in their software (Figure 5). Similarly, when asked whether respondents needed a software capability for replicating modules and for supporting object‐oriented design, although 38% indicated they did not need such capability, the majority of users expressed interest: 27% replied they use it but want more capabilities, 25% indicated they did not use it because it is not available, and 11% stated they use it and find the capabilities adequate.
Use of model parameters
QSP models usually include a large number of parameters, often running into hundreds and even thousands. QSP model parameterizations are often referred to as virtual patients (VPs),26, 34 or virtual subjects,35 and can span alternate clinical phenotypes.36 Variability in model parameters may increase with multiple virtual patients, multiple patient phenotypes, and various therapies. Consequently, handling, storing, and organizing parameters can be a nontrivial task in itself. Responses to the set of questions related primarily to model parameters are summarized by Figures S19–S24 in Supplementary File S2 . A minority of respondents (20%) answered that the parameters in their selected software were organized in a flexible structure with powerful handling features. Many respondents indicated room for improvement in this area: 49% stated that parameters were organized but with limited features, and 31% replied they were not organized ( Figure S19 ). When asked about parameter manipulation and export/import to/from different software packages, 43% responded that they use this feature but options are limited, 24% found the export/import capabilities adequate, and equal fractions of respondents (17%) did not use export/import, either because parameter manipulation provided by the software is adequate or because they were not aware of such a capability ( Figure S20 ). This highlights a need for improvement in parameter organization and handling, especially in the view that, in the future, models may grow (e.g., due to extension and reuse), and, therefore, will have even more parameters.
As has been identified earlier (Figure 3), parameter estimation is a major and challenging task for QSP modelers. Accurate and efficient parameter estimation can be crucial for success of a project; therefore, QSP software should offer adequate capabilities. When asked whether parameter estimation methods provided by the software were overall sufficient, 55% of survey participants answered positively; however, 36% indicated capabilities are very limited, and 9% were not aware of parameter estimation capabilities within their software ( Figure S21 ). When asked which parameter estimation methods they used most often, 53% preferred global optimization methods, 49% used nonlinear mixed effects modeling, 47% utilized gradient‐based algorithms, and 25% applied simplex algorithms (Figure 6). About 4% of respondents tended to use other methods or a combination of methods, and 4% of respondents indicated they did not use parameter estimation. Accuracy and reliability of parameter estimation and/or model qualification also depends on the quality of available data. Half of respondents (51%) indicated they had access and used rich data types and sources (e.g., individual time courses, ‘omics data), whereas a slightly smaller percentage (45%) relied on simpler data (e.g., summary statistics, mean time courses, etc.) whether intentionally or due to lack of a better option ( Figure S24 ).
Figure 6.
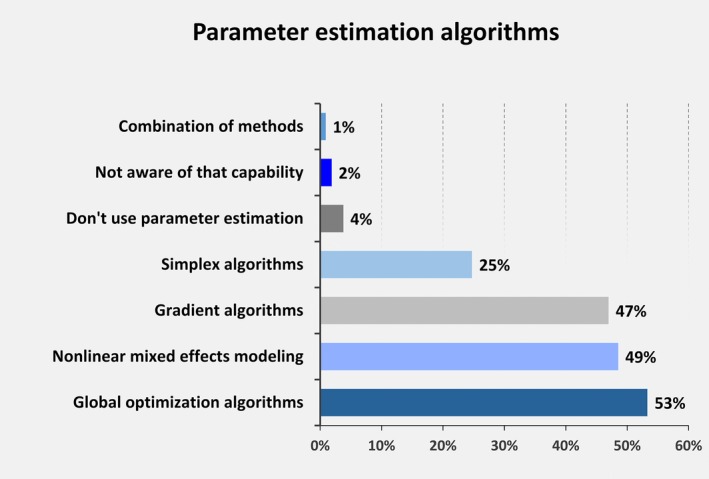
Prevalence of parameter estimation algorithms used for quantitative systems pharmacology modeling.
Parameter sensitivity analysis (SA) is another task routinely performed by QSP modelers. Like parameter estimation, SA depends on specialized algorithms and significant computational resources. When questioned whether respondents used their software for parameter SA, 39% answered affirmatively and were satisfied with existing options, whereas the other 48% who use SA stated the software had very limited capabilities. About 7% of users replied they did not use the software for SA, and 6% of users were not aware of SA capabilities ( Figure S22 ). When it comes to satisfaction with available equation solvers and algorithms, respondents were generally content with what is available (78%); no more than 21% expressed that one or another solver is missing for their work ( Figures S25 and S26 ).
Analysis of simulation results
This section of the survey examined how QSP modelers use their software for visualizing, analyzing, and exporting simulation results, and which features are represented adequately or are missing. Quite often, analysis of simulation results requires a substantial amount of time. The same can be said about achieving desired visual appearance suitable for presenting results and delivering convincing messages to diverse audiences. The survey revealed a mixed picture with regard to the efficiency and visual effectiveness generating plots within QSP software. Here, although 38% of respondents stated that their software was highly customizable and met all their needs, 28% indicated it was easy and simple but they wished it were more flexible. In 23% of cases, respondents faced a lot of repetitive work, and 11% of respondents reported plotting capabilities were absent or very limited ( Figure S27 ). When it comes to what plotting/visualization features users value the most, flexibility in data visualization and ability to create custom plots was at the top of the list (65%), whereas 52% praised the ability to overlay simulation results with external data. Among other features, 43% of responses valued the ability to quickly visualize results with prebuilt plot templates, and 36% appreciated a large selection of visualization/plot types. A quarter of the users were presumably dissatisfied with their software, as they preferred a different tool for doing most of their plotting tasks ( Figure S28 ). There is a clear opportunity for improvement in the statistical analysis tools provided by the software, because less than half (43%) of respondents were satisfied with provided features. A substantial portion (28%) found the available selection limited, and 21% of modelers found specialized statistical tools more attractive ( Figure S29 ). Using external tools for data visualization and statistical analysis is not unusual, but it also requires data export capabilities. The survey results showed this feature is largely available as 95% of modelers use it in some form. A majority of modelers (60%) use the simplest data export in the form of a text file, whereas 31% export data to spreadsheets or using a proprietary format, and 4% upload results directly to a database ( Figure S30 ). Only 6% do not use data export as they are not aware of their software's export capability.
Virtual patients
The concept of a VP in QSP modeling offers an attractive way to project data and behaviors of real world patients into the world of simulated data and predictions. Not only does it bring a structure to the massive amount of information QSP models use and create, the concepts of VP and virtual population (VPop) facilitate communication of modeling results outside of the QSP modeling community (e.g., to biologists and clinicians). Creating and qualifying VPs and VPops can be computationally costly and time‐consuming and requires considerable software capabilities to accomplish the task. The survey results suggest that this feature represents an opportunity for future software development and improvement. Specifically, when questioned about software capabilities for creating VPs and VPops, the majority of respondents indicated they used VPs and VPops in some capacity, but only 21% indicated that their software fully covers all needs in VP and/or VPop development and running virtual experiments. For the rest, 45% of respondents used the software to create VPs and VPops but found the capabilities limited. At the same time, 27% indicated they do not use the concept of VPs and VPops, and 8% were not aware of any tool for creating VPs and/or VPops ( Figure S31 ). As the numbers of VPs required to explore parametric and response variability can be quite high, the structure and the organization of VPs, VPops, and simulation experiments become very important. It is expected that QSP software should provide these features together with convenient means for running many computationally intensive simulations and storing the results. Currently, only 25% of survey participants indicated they find their existing capabilities scalable, flexible, and easy to use. A large proportion of users (44%) answered that available capabilities were limited and not easily scalable, and 31% indicated they are not aware of any feature or tool that will do the job ( Figure S32 ).
Software ownership and maintenance
The final set of questions in the second section was focused on additional aspects of the software, including cost, support, and the use of pre‐existing models. When asked whether the cost of software ownership, including the cost of the license, add‐on packages, and/or customer support and annual maintenance fees, plays an important/definitive role, the majority of respondents indicated that cost played an important but not decisive role (58%). Still, 33% responded they would use a different software tool if there were no budgetary constraints. A smaller percentage of respondents indicated that specified capabilities but not cost plays a decisive role (9%) ( Figure S33 ). To the question whether they find the software's customer support adequate and useful, 45% answered they find it somewhat useful, 28% were not aware of software support or it was not provided, and 27% found it helpful most of the time ( Figure S34 ). When it comes to exploiting other sources of information about their software, respondents indicated using a variety of resources: 79% used the documentation and help system, 69% used community help forums, 65% used other online resources, and 61% used peer‐to‐peer communication ( Figure S35 ). One possibility to expand the utility of the user's software is to obtain and use existing QSP models, either free or commercially available. This option was used by 39% of respondents, 30% did not find existing models very useful, and 27% were not aware of existing models. A small fraction of participants (4%) indicated they do not use existing models or platforms due to cost constraints ( Figure S36 ).
QSP software defined by users
Analysis of preferred features
After surveying existing software tools, we asked respondents to consider the features of an ideal tool for their QSP modeling work. The intent for this section was to obtain respondents’ feedback as to which out of 18 selected features they consider more or less important to have in QSP software (Table 2). Means and ranks were computed for each question, both for the full population and split by experience category, as explained in the Methods section. In this stratification, 61% of respondents fell into the most experienced category, 23% into the intermediate experience category, and 16% into the least or no experience category.
Table 2.
Relative importance of QSP software features
| Feature | Rank | Mean | P value | ||||||
|---|---|---|---|---|---|---|---|---|---|
| All | Low | Med | High | All | Low | Med | High | ||
| 3.1 Ease of large model development (> 20 state variables) | 1 | 11 | 1 | 1 | 2.68 | 2.17 | 2.70 | 2.82 | 0.0004 |
| 3.7 Support for multiple parameter estimation algorithms | 2 | 1 | 3 | 2 | 2.58 | 2.63 | 2.54 | 2.59 | 0.9884 |
| 3.6 Support for scripting tasks that extend the tool's capabilities | 3 | 6.5 | 4 | 4 | 2.51 | 2.38 | 2.50 | 2.55 | 0.7578 |
| 3.12 Built‐in support for flexible visualization of simulation results | 4 | 6.5 | 6 | 3 | 2.51 | 2.38 | 2.46 | 2.56 | 0.3728 |
| 3.8 Handling a large number of parameters including export/import | 5 | 3 | 6 | 5 | 2.49 | 2.50 | 2.46 | 2.49 | 0.9254 |
| 3.3 High‐performance parallel computing enabled | 6 | 6 | 2 | 8 | 2.35 | 2.42 | 2.65 | 2.24 | 0.2103 |
| 3.5 Availability of multiple numerical solvers | 6 | 2 | 11 | 6 | 2.35 | 2.58 | 2.26 | 2.35 | 0.8951 |
| 3.4 Support for flexible hardware/software architecture (cluster, cloud, different OS) | 8 | 6 | 8 | 7 | 2.29 | 2.42 | 2.39 | 2.26 | 0.6782 |
| 3.14 Support for VPops manipulation, sampling, and clinical trial simulation | 9 | 11 | 5 | 10 | 2.26 | 2.17 | 2.43 | 2.18 | 0.6951 |
| 3.2 Support for export to SBML or other language | 10 | 1 | 11 | 14 | 2.22 | 2.67 | 2.26 | 2.11 | 0.1321 |
| 3.13 Tools for VPs and VPops creation | 11 | 15 | 9 | 11 | 2.21 | 2.08 | 2.35 | 2.15 | 0.706 |
| 3.15 Low cost of ownership and maintenance | 12 | 9 | 10 | 11 | 2.19 | 2.25 | 2.32 | 2.15 | 0.7344 |
| 3.16 Customer support | 13 | 17 | 13 | 9 | 2.18 | 2.00 | 2.17 | 2.21 | 0.7833 |
| 3.11 Ease of creation of replicated features (e.g., array of cells, similar compounds) | 14 | 11 | 13 | 13 | 2.15 | 2.17 | 2.17 | 2.13 | 0.9902 |
| 3.9 Visual diagrammatic model development (in contrast to purely text‐based) | 15 | 8 | 16 | 15 | 2.08 | 2.33 | 2.13 | 1.97 | 0.0265 |
| 3.10 Modular (plug‐and‐play) model architecture | 16 | 15 | 13 | 16 | 2.03 | 2.08 | 2.17 | 1.95 | 0.7437 |
| 3.18 Integration with additional external tools (e.g., bioinformatics) | 17 | 18 | 18 | 17 | 1.87 | 1.92 | 1.91 | 1.82 | 0.6384 |
| 3.17 Selection of available disease models/platforms for this particular software | 18 | 11 | 17 | 18 | 1.81 | 2.17 | 2.04 | 1.60 | 0.0005 |
Categorical breakdown of feature importance in a hypothetical QSP modeling software platform. The respondents were asked to place features into one of three categories by the order of importance and to assign a score as follows: 3 = most important, 2 = somewhat important, and 1 = least important. Right part of the table presents combined average scores given by the respondents (All) as well as split between groups of respondents based on their experience: low = < 1 year experience, medium = 1–3 years of experience, high = > 3 years of experience. Based on the scores given by all respondents and each group separately features are ranked as shown in middle part of the table.
OS, operating system; QSP, quantitative systems pharmacology; SBML, Systems Biology Markup Language; VPops, virtual populations; VPs, virtual patients.
In Table 2, features were sorted according to their rank, with the highest‐ranking feature placed at the top. Three of the top six, features 3.6, 3.7, and 3.8, were highly regarded irrespective of level of experience. They included management and estimation of parameters and the need for programming/scripting capabilities beyond pure model‐building. Several design options showed statistically significant differences according to the experience levels of the respondents. Feature 3.1, which is the ability to easily develop and simulate large (> 20 state variable) models, ranked first for the full population but was of low importance for the least experienced users. This feature showed sharply increasing importance for users with experience (P < 0.001). For two other features, importance decreased with experience: Features 3.9 (visual diagrammatic model creation/editing capability…) (P < 0.05) and 3.17 (selection of available disease models (platforms) …) (P < 0.001). This may be indicative of the utility of graphical interfaces and open‐access model repositories as sources of training material for new QSP scientists and engineers, whereas more experienced modelers prefer to construct their own models without need for a GUI. One other question, 3.2 (support for SBML export or export to any other widely used language (R, MATLAB, Python, etc.)) approached significance (P = 0.13), with the trend also showing decreased importance with increasing experience. Unexpectedly, features, such as the ability to construct models in a modular style (3.10 and 3.11), which should accelerate model development, were generally not selected as important across all experience groups. Similarly, integration with other data/modeling platforms (3.18) was at the bottom of users’ priority lists.
In stratifying the responses, it was also noted that 32 respondents completed section 3 of the survey without self‐identifying as a current QSP modeler (survey question 1.3). To determine whether there was a change in software requirements by considering only current QSP modelers, we performed a similar analysis on the 69 QSP‐labeled responses. With significantly fewer numbers, we lumped the intermediate and least experienced classes together to yield 50 more experienced respondents compared with 19 less experienced respondents. The tabulated results are given in Supplementary File S3 and Table S1 . Overall, five of the top six features and all seven of the top seven features were common between the analyses; features 3.3 and 3.4 were rated 6 and 7 (respectively, 7 and 6) in the complete and restricted data sets. Two differences stood out from this analysis. First, there was more agreement between experience categories about the most important features; five of the top six overall features were common between the groups. There was only one statistically significant difference between experience groups in the reduced data set, with feature 3.9 showing decreased importance to the more experienced set (P < 0.05). Features 3.1 (P = 0.08) and 3.17 (P = 0.0572) approached significance. No other features even approached a statistical difference between the groups. However, given the limited number of self‐identified less‐experienced QSP modelers in our dataset, it should not be surprising that statistical differences were not found.
Discussion
In 2011, a National Institutes of Health White Paper by the QSP Workshop Group9 formally endorsed QSP as a novel approach to accelerate and advance the drug discovery and development process, in which mathematical modeling and simulation plays an important role. Creation and application of QSP models relies on the availability of appropriate software tools, which in turn requires a clear understanding of the capabilities such tools need to possess. This survey provides a representative picture of the QSP community, with over 100 respondents from a variety of institutions and experiences. The survey highlights challenges facing the QSP community, as well as the community's opinion about important features for consideration during new QSP software tool development. Results of the survey, particularly those obtained from questions in sections 2 and 3, may provide improved guidelines for software developers, for organizations using QSP modeling, and for the QSP modelers themselves.
Analysis of the survey data led to several questions on the scope and features required of QSP software: What kind of tool, one focused and specifically developed for QSP modeling, or a platform with a more general scope, will be the preferred choice of QSP modelers? Do we need a single integrated tool covering all aspects of QSP modeling, or will a suite of specialized tools be a better solution? Of 16 tools assessed, two general purpose computing software platforms, MATLAB and R, were favored by almost half of respondents (Figure 3). Of the remaining 14 software tools, perhaps only the PhysioLab Modeler was designed specifically for QSP modeling, and others were developed either for PK, PK/PD, or PopPK modeling, or came from disciplines not necessarily associated with pharmacology. Preference for general purpose tools might be attributed to (i) a lack of capabilities and/or flexibility in existing specialized tools required for developing and exploiting QSP models and (ii) widespread use and accessibility of general purpose software, which are often used in training programs and relatively inexpensive to acquire or freely available. In support of the first assertion, QSP modelers placed “capable of building large models” at the top of the list for desired/must‐have features (see feature 3.1 in Table 2 and Figure S1 in Supplementary File S2 ). General purpose computing software packages typically impose relatively few limitations on the size of the model and the number of equations, which is not always the case for specialized software. General purpose software also provides scripting support, which was ranked highly by respondents ( Figure S14 , feature 3.6 in Table 2). General purpose tools also offer flexibility to implement many aspects of QSP workflows that may not be available in fully commercial solutions, such as VPops or multivariable SA,34, 35, 36, 37, 38 and normally offer a debugging capability ( Figure S13 ). Although valued less by the respondents, debugging capabilities are rarely found in more specialized tools. Other highly ranked features, such as multiple parameter estimation algorithms and flexible visualization of results, are usually well‐supported by general purpose tools. Finally, tools are being designed with general scope in mind in order to appeal to as many users as possible, and their use becomes widespread. Making software less expensive is another attractive feature which, with economy of scale, is easier to achieve with a large user base compared with a specialized tool used by few.
On the downside, the use of general purpose tools necessitates good programming skills and substantial time for code writing during model development, whereas specialized software automates and performs certain tasks itself. Nonetheless, many QSP modelers either are not ready to trade flexibility for convenience or have not found a specialized tool that adequately addresses their needs. Perhaps surprisingly, QSP modelers ranked low the use of GUIs and visual model development, features that are intended to facilitate large model development (feature 3.9 in Table 2). Models designed visually as diagrams may also help to communicate model scope to scientists without a mathematical background. Nonetheless, more experienced users tended to assign less importance to this feature.
The opinion has been expressed that familiarity with QSP models and their assumptions beyond the research phase of drug development may improve their acceptance.39 Making models available publicly in a common exchange format may help to facilitate critical review and ultimate acceptance by the broader community. Furthermore, such a public model exchange would facilitate reuse and refinement, increasing the efficiency and quality of QSP model development. To date, SBML seems to be the most widely used and perhaps most advanced format for this purpose. Still, three in four respondents either do not use, know nothing about, or were not aware of SBML support in their software of choice. Support for model export to SBML or other formats was also ranked low for an “ideal” QSP software (feature 3.2 in Table 2). Furthermore, results of the survey suggest that collaboration capabilities and availability of pre‐existing models are not of great importance, the latter especially to very experienced QSP modelers (feature 3.17 in Table 2).
The combination of low interest in existing models and collaboration capabilities of software tools may be a reflection of QSP modeling being in a nascent stage. The majority of models currently available with associate computer code and proper documentation typically has limited scope and, therefore, has limited application. Publicly available disease‐scale QSP platforms with detailed representations of biological processes are relatively rare. Fit‐for‐purpose models may be developed without need for collaboration, so they may be favored by time and resource constraints routinely faced by modelers in the pharmaceutical industry. However, in the future, this may change as we see increased efforts to promote efficient model and model‐based information exchange.40, 41 Furthermore, collaboration environments for improved integration are forming, such as in the Garuda Alliance,42 and efforts to collect, organize, and make available large biomedical datasets are ongoing in a number of different fields.43, 44 However, any similar attempt in the QSP community will require a critical mass of talented modelers and developers to join the initiative before it becomes a common practice.
In conclusion, the choice of software is a critical element to the successful application of QSP modeling in the drug discovery and development process. As the survey results indicated, the search for adequate QSP software tools is still in its early phase. Currently, there is no single QSP software accepted by a majority of modelers; instead, we often resort to general purpose computing tools or to PK/PD simulation software. QSP modeling remains primarily in the hands of individuals rather than teams due to lack of software collaboration capabilities and a shortage of dedicated resources. QSP software development faces multiple challenges, as the set of features required by modelers is extensive and evolves together with the field. We hope the survey results and interpretation offered here will help to set priorities for software development and will provide guidelines for those who enter this rapidly advancing and changing field.
Funding
All authors are employees of their respective companies.
Conflict of Interest
The authors declared no competing interests for this work.
Author Contributions
S.E., B.J.S., C.J.T., and C.J.M. designed the survey. S.E., B.J.S., and C.J.T. analyzed the data. S.E., B.J.S., C.J.T., and C.J.M. wrote the manuscript.
Supporting information
Supplementary File S1. Survey web forms.
Supplementary File S2. Figures S1–S36 (section 2 questions).
Supplementary File S3. Table S1 (section 3 questions).
Supplementary File S4. Survey results data.
Acknowledgments
The authors thank the International Society of Pharmacometrics and the QSP Special Interest Group for their endorsement of this effort and Brian Corrigan, Valeriu Damian, Marko Miladinov, Saroja Ramanujan, Enrico Smith, Michael Weis, and Michael Zager for their support of the survey, from concept through implementation, including valuable feedback on its content. Last but not least, we thank survey participants for their contributions and willingness to share their thoughts and ideas.
References
- 1. Lalonde, R.L. et al Model‐based drug development. Clin. Pharmacol. Ther. 82, 21–32 (2007). [DOI] [PubMed] [Google Scholar]
- 2. Sliwoski, G. , Kothiwale, S. , Meiler, J. & Lowe, E.W. Computational methods in drug discovery. Pharmacol. Rev. 66, 334–395 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bleicher, K.H. , Bohm, H.J. , Muller, K. & Alanine, A.I. Hit and lead generation: beyond high‐throughput screening. Nat. Rev. Drug Discov. 2, 369–378 (2003). [DOI] [PubMed] [Google Scholar]
- 4. Tuntland, T. et al Implementation of pharmacokinetic and pharmacodynamic strategies in early research phases of drug discovery and development at Novartis Institute of Biomedical Research. Front. Pharmacol. 5, 174 (2014). 10.3389/fphar.2014.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang, S.‐M. , Abernethy, D.R. , Wang, Y. , Zhao, P. & Zineh, I. The utility of modeling and simulation in drug development and regulatory review. J. Pharm. Sci. 102, 2912–2923 (2013). [DOI] [PubMed] [Google Scholar]
- 6. Leil, T.A. & Bertz, R.J. Quantitative systems pharmacology can reduce attrition and improve productivity in pharmaceutical research and development. Front. Pharmacol. 5, 247 (2014). 10.3389/fphar.2014.00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Visser, S.A.G. , de Alwis, D.P. , Kerbusch, T. , Stone, J.A. & Allerheiligen, S.R.B. Implementation of quantitative and systems pharmacology in large pharma. CPT Pharmacometrics Syst. Pharmacol. 3, e142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Van Der Graaf, P.H. & Gabrielsson, J. Pharmacokinetic‐pharmacodynamic reasoning in drug discovery and early development. Future Med. Chem. 1, 1371–1374 (2009). [DOI] [PubMed] [Google Scholar]
- 9. Sorger, P.K. et al Quantitative and systems pharmacology in the post‐genomic era: new approaches to discovering drugs and understanding therapeutic mechanisms. NIH White Paper by the QSP Workshop Group: (National Institutes of Health, Bethesda, MD, 2011). [Google Scholar]
- 10. NONMEM . <http://www.iconplc.com/innovation/nonmem/>.
- 11. Phoenix. Version 8.0 (Certara USA, Inc., Princeton, NJ: ). [Google Scholar]
- 12. Hucka, M. et al The ERATO Systems Biology Workbench: enabling interaction and exchange between software tools for computational biology. Pac. Symp. Biocomput. 7, 450–461 (2002). [DOI] [PubMed] [Google Scholar]
- 13. Sauro, H.M. et al Next generation simulation tools: the Systems Biology Workbench and BioSPICE integration. OMICS 7, 355–372 (2003). [DOI] [PubMed] [Google Scholar]
- 14. MathWorks . SimBiology by MathWorks. <http://www.mathworks.com/products/simbiology/>.
- 15. R_language . Open source software tools. <https://www.r-project.org/about.html>.
- 16. Wojciechowski, J. , Hopkins, A.M. & Upton, R.N. Interactive pharmacometric applications using R and the Shiny package. CPT Pharmacometrics Syst. Pharmacol. 4, 146–159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolfram . System Modeler, Mathematica. <http://www.wolfram.com/system-modeler/?source=nav>.
- 18. Macey, R. , George, O. & Zahnley, T. Berkeley Madonna User's Guide (Department of Molecular and Cellular Biology, University of California, Berkeley, CA, 2003). <https://mcb.berkeley.edu/courses/mcb137/exercises/madonnamanual.pdf>. [Google Scholar]
- 19. Shoda, L. et al The Type 1 Diabetes PhysioLab® Platform: a validated physiologically based mathematical model of pathogenesis in the non‐obese diabetic mouse. Clin. Exp. Immunol. 161, 250–267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Immunetrics . Aegis Modeling Platform. <https://www.immunetrics.com/products/modeling-platform/>.
- 21. Eissing, T. et al A computational systems biology software platform for multiscale modeling and simulation: integrating whole‐body physiology, disease biology, and molecular reaction networks. Front. Physiol. 2, 4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gizzatkulov, N.M. et al DBSolve Optimum: a software package for kinetic modeling which allows dynamic visualization of simulation results. BMC Syst. Biol. 4, 109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ermakov, S. et al Virtual Systems Pharmacology (ViSP) software for mechanistic system‐level model simulations. Front. Pharmacol. 5, 232 (2014). 10.3389/fphar.2014.00232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Open‐Systems‐Pharmacology . <https://github.com/Open-Systems-Pharmacology>.
- 25. Kroneckerbio . <https://github.com/kroneckerbio/kroneckerbio>.
- 26. Cheng, Y. et al QSP toolbox: computational implementation of integrated workflow components for deploying multi‐scale mechanistic models. AAPS J. 19, 1002–1016 (2017). [DOI] [PubMed] [Google Scholar]
- 27. Biliouris, K. , Lavielle, M. & Trame, M.N. MatVPC: a user‐friendly MATLAB‐based tool for the simulation and evaluation of systems pharmacology models. CPT Pharmacometrics Syst. Pharmacol. 4, 547–557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baron, K.T. & Gastonguay, M.R. Simulation from ODE‐based population PK/PD and systems pharmacology models in R with mrgsolve. J. Pharmacokinet. Pharmacodyn. 42, S84–S85 (2015). [Google Scholar]
- 29. DDMoRe . The Drug Disease Model Resources (DDMoRe) consortium. <www.ddmore.eu>.
- 30. Vlasakakis, G. et al White Paper: Landscape on technical and conceptual requirements and competence framework in drug/disease modeling and simulation. CPT Pharmacometrics Syst. Pharmacol. 2, 1–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schuck, E. et al Preclinical pharmacokinetic/pharmacodynamic modeling and simulation in the pharmaceutical industry: an IQ consortium survey examining the current landscape. AAPS J. 17, 462–473 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nijsen, M.J.M.A. et al Preclinical QSP modeling in the pharmaceutical industry: an IQ consortium survey examining the current landscape. CPT Pharmacometrics Syst. Pharmacol. 7, 135–146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van der Graaf, P.H. & Benson, N. Systems pharmacology: bridging systems biology and pharmacokinetics‐pharmacodynamics (PKPD) in drug discovery and development. Pharm. Res. 28, 1460–1464 (2011). [DOI] [PubMed] [Google Scholar]
- 34. Allen, R.J. , Rieger, T.R. & Musante, C.J . Efficient generation and selection of virtual populations in quantitative systems pharmacology models. CPT Pharmacometrics Syst. Pharmacol. 5, 140–146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gadkar, K. , Kirouac, D.C. , Mager, D.E. , van der Graaf, P.H. & Ramanujan, S. A six‐stage workflow for robust application of systems pharmacology. CPT Pharmacometrics Syst. Pharmacol. 5, 235–249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klinke, D.J. 2nd . Integrating epidemiological data into a mechanistic model of type 2 diabetes: validating the prevalence of virtual patients. Ann. Biomed. Eng. 36, 321–334 (2008). [DOI] [PubMed] [Google Scholar]
- 37. Schmidt, B.J. , Casey, F.P. , Paterson, T. & Chan, J.R. Alternate virtual populations elucidate the type I interferon signature predictive of the response to rituximab in rheumatoid arthritis. BMC Bioinformatics 14, 221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang, X.Y. , Trame, M.N. , Lesko, L.J. & Schmidt, S. Sobol sensitivity analysis: a tool to guide the development and evaluation of systems pharmacology models. CPT Pharmacometrics Syst. Pharmacol. 4, 69–79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peterson, M.C. & Riggs, M.M. FDA advisory meeting clinical pharmacology review utilizes a quantitative systems pharmacology (QSP) model: a watershed moment? CPT Pharmacometrics Syst. Pharmacol. 4, 189–192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harnisch, L. , Matthews, I. , Chard, J. & Karlsson, M.O. Drug and disease model resources: a consortium to create standards and tools to enhance model‐based drug development. CPT Pharmacometrics Syst. Pharmacol. 2, e34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Le Novere, N. et al BioModels database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 34, D689–D691 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garuda Alliance . <http://www.garuda-alliance.org/>.
- 43. NHANES . National Health and Nutrition Examination Survey Data. Centers for Disease Control and Prevention 2011‐2012. <http://www.cdc.gov/nchs/nhanes.htm>.
- 44. Vivian, J. et al Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 35, 314–316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File S1. Survey web forms.
Supplementary File S2. Figures S1–S36 (section 2 questions).
Supplementary File S3. Table S1 (section 3 questions).
Supplementary File S4. Survey results data.


