Abstract
Objective:
To review observational human, murine, and interventional trial studies that have examined the gut microbiome in food allergy, and to provide perspective on future investigations in this field.
Data Sources:
A review of the published literature was performed with PubMed, and clinical studies catalogued at ClinicalTrials.gov were also reviewed.
Study Selections:
The most recent relevant studies, seminal works, and topical clinical trials were selected.
Results:
Gut dysbiosis likely precedes the development of food allergy, and the timing of such dysbiosis is critical. Gut microbiota associated with individual food allergies may be distinct. Murine models support the importance of gut microbiota in shaping immune maturation and tolerance. Gut microbiota may affect food allergy susceptibility by modulating type 2 immunity, influencing immune development and tolerance, regulating basophil populations, and promoting intestinal barrier function. Ongoing and future interventional trials of probiotics, prebiotics, synbiotics, and fecal microbiota transfer will help translate our understanding of the gut microbiome in food allergy to clinical practice. Future work in this area will include deepening of current research foci, as well as expansion of efforts to include the virome, mycobiome, and interactions between the microbiome, host, and environment. Robust and consistent study designs, multi-dimensional profiling, and systems biology approaches will enable this future work.
Conclusion:
By advancing research on the microbiome in food allergy, we can further our understanding of food allergy and derive new approaches for its prevention and therapy.
Keywords: microbiome, food allergy, gut, dysbiosis, microbiota, milk, egg, peanut, probiotic, prebiotic, synbiotic, systems biology
Introduction
Food allergy is a clinical and public health problem that affects up to 10% of the US population.1 Defined as an adverse health effect arising from a specific immune response that occurs reproducibly on exposure to a food, IgE-mediated food allergy encompasses relatively immediate symptoms affecting the skin, respiratory, gastrointestinal, and/or cardiovascular systems.2 The etiology of food allergy involves deviation from a default state of immune tolerance that is likely driven by antigen exposure, commensal microbiota, and their interactions.3
Resident microbial communities vastly outnumber human cells and genes, motivating interest in how their dysregulation (i.e. dysbiosis) may influence host immunologic development and risk for allergic disorders.4 The sum of microbes, their genomic elements, and interactions in a given ecologic niche (i.e. microbiome) differs by body site.5 Growing evidence supports a potential role for the gut microbiome in the pathogenesis and course of food allergy.3–14 Next-generation sequencing, including 16S rRNA sequencing and shotgun metagenomic sequencing, has advanced microbiome research in recent years by enabling more comprehensive and culture-free profiling of taxa in a given sample.4
Building on our previous address of this topic3, here we review the potential role of the gut microbiome in the development and course of food allergy through a targeted discussion of results from observational human, murine, and interventional trial studies. We additionally discuss the directions in which we envision future investigations of this exciting area will move.
Observational human studies
Studies of the microbiome in human subjects with food sensitization and food allergy thus far have yielded variable findings, likely due to several factors including heterogeneity of study populations, variable definitions of food sensitization and food allergy, differences in profiling and study designs, and varying sample sizes.
Early studies of gut microbiota and food allergy were culture-based, targeting groups of and individual bacteria of interest to explore hypotheses regarding the relationship between gut microbial characteristics and food allergy in early childhood. For example, a culture-based study of Spanish children with milk allergy (based on sensitization by skin prick test (SPT) > 3 mm, sIgE ≥ 0.35 kUA/L, and milk formula challenge) showed that milk allergic infants had higher total bacteria and anaerobic counts compared to healthy controls; after 6 months of differential formula intake, the 46 milk allergic infants had higher proportions of Lactobacilli and lower proportions of Enterobacteria and Bifidobacteria observed in bacterial cultures.6 While culture-based methods can quantify counts of specific bacteria, the approach is tied to the specific culture conditions of individual bacteria of a priori interest, and the majority of bacteria cannot be cultured. Culture-based approaches are therefore limited in the number and diversity of microbes that can be profiled.
More recent studies of gut microbiota in food allergic individuals have employed sequencing of the 16S rRNA gene, which encodes a component of the prokaryotic ribosome.15 The 16S rRNA gene has highly conserved primer binding sties and contains hypervariable regions that can provide species-specific signature sequences for bacterial identification, thus enabling more comprehensive bacterial identification without the constraints of culture-based methods. 15
Findings from a 16S rRNA sequencing-based studies of food sensitization and food allergy defined broadly suggest that gut dysbiosis may precede the development of food allergy. Among a cohort of 225 US children, lower relative abundances of Haemophilus, Dialister, Dorea, and Clostridium in stool samples collected at age 3–6 months was associated with sensitization (sIgE ≥ 0.10 kUA/L) to at least one food allergen among milk, egg, peanut, soy, and wheat.7 Additionally, the investigators found lower relative abundances of Citrobacter, Oscillospira, Lactococcus, and Dorea in stool collected at age 3–6 months in children who had food allergy by age 3 years, defined by sensitization and convincing allergic symptom.7 Separately, a longitudinal study of 166 infants at age 3 and 12 months from the Canadian Healthy Infant Longitudinal Development (CHILD) study showed that lower gut microbial richness at age 3 months was associated with increased likelihood of food sensitization (SPT wheal ≥ 2mm than negative control) by age 12 months.16 Each quartile increase in richness at 3 months was associated with a 55% reduced risk for food sensitization by age 12 months (adjusted OR, 0.45; 95% CI, 0.23, 0.87).16 Enterobacteria were overrepresented and Bacteroidaceae were underrepresented in food-sensitized infants at 3 months and 1 year.16 These associations between early gut microbiome composition and later food allergen sensitization and/or food allergy suggest a possible role for gut dysbiosis in the development of food allergy.
Studies of the gut microbiome in food allergy and related phenotypes additionally suggest a critical role of timing. The gut microbiome is known to change with time, with the most rapid change occurring in early life.17 In the CHILD cohort, gut microbial richness at age 3 months was associated with increased likelihood of food sensitization by age 1 year, while there was no association between gut microbial characteristics at age 12 months and food sensitization.16 Beyond food allergen sensitization, a multi-center, longitudinal study of 226 milk allergic children in the United States from the Consortium for Food Allergy Research (CoFAR) examined the relationship between infant gut microbiome and food allergy resolution, with milk allergy defined based on oral food challenge, convincing history and positive allergy testing, or flare of atopic dermatitis associated with milk ingestion and milk sIgE ≥ 5 kUA/L. The investigators found that Firmicutes including Clostridia were enriched in the gut microbiome of infants age 3–6 months whose milk allergy resolved by age 8 years.18 However, in older infants, there was no association between gut microbiome composition and milk allergy resolution.18 Findings from murine models also support age-sensitive contact with microbiota.19 Colonization of gnotobiotic mice with a diverse microbial population early, but not late, in life suppresses IgE and prevents mice from development of food allergy.20 These collective findings support the notion that microbial effects on early immune system development play a role in subsequent food allergy development.
Given differences in the presentations and natural histories of specific food allergies, it is possible that gut microbiota associated with individual food allergies are also distinct. In a study of 141 children, genera from Lachnospiraceae, Streptococcaceae, and Leuconostocaceae were differentially abundant in the gut microbiome of US children with egg allergy vs. non food allergic controls from the multi-site Consortium for Food Allergy Research (CoFAR). In this study, egg allergy was defined based on oral food challenge, convincing history and positive allergy testing, or flare of atopic dermatitis associated with egg ingestion and egg sIgE ≥ 2 kUA/L (n=141).9 Studies of gut microbiota in milk allergic individuals demonstrate some overlapping but also contrasting gut microbial compositions. For example, a study of 39 Italian children showed that compared to healthy controls, milk allergic infants (diagnosed based on history, oral food challenge, and milk sIgE level) had gut microbial community structures dominated by Lachnospiraceae and Ruminococcaceae.12 While differences in the implicated taxa reported in these studies may have been due to the specific food allergy targeted, additional explanations include differences in other population characteristics, phenotyping details, and/or analytic strategy including assessment of microbiota at varying taxonomic levels. The characterization and comparison of microbial signatures of different food allergies remains an area that merits further investigation.
Murine Studies
Murine models of food allergy have provided insightful experimental dimensions to understanding the role of the microbiome in food allergy, with intriguing results to suggest that risk for food allergy can be altered by gut microbiota. For example, food allergy-prone mice with a gain-of-function mutation in the IL-4 receptor-a chain have differential gut abundances of Lachnospiraceae, Lactobacillaceae, Rikenallaceae, and Porphyromononadaceae compared to wild type mice.14 Transfer of gut microbiota from these food allergy-prone mice to germ-free mice seemed to transfer disease susceptibility, with reconstituted mice demonstrating OVA-specific IgE responses and symptoms consistent with anaphylaxis upon OVA challenge.14 Interestingly, although therapy of these food allergy-prone mice with Treg cells specific for the immunodominant OVA peptide suppressed allergen sensitization and correlates of anaphylaxis, and this Treg therapy altered the gut microbiota, their gut microbiota did not shift back to baseline, suggesting that immunomodulatory mechanisms beyond Treg cells are involved.14 Another study demonstrated the converse—that transfer of gut microbiota from healthy mice to allergy-prone mice is protective. Specifically, colonization of milk sensitized, germ-free mice with bacteria from healthy infant stool, which contains abundant taxa from Bifidobacterium and Bacteroides, reduced signs of systemic allergic response (e.g. lower magnitude of rectal temperature drop) upon challenge to cow’s milk protein compared to their uncolonized murine counterparts.21 Additionally, wild-type mice induced to have altered gut flora by antibiotic treatment also become more susceptible to systemic anaphylaxis upon oral challenge compared to mice not treated with antibiotics.22 When the gut flora of these mice are allowed to repopulate, allergen-specific IgE levels in these mice decrease.22 Collectively, these findings from murine models suggest that microbial dysbiosis plays a role in food allergy pathogenesis.
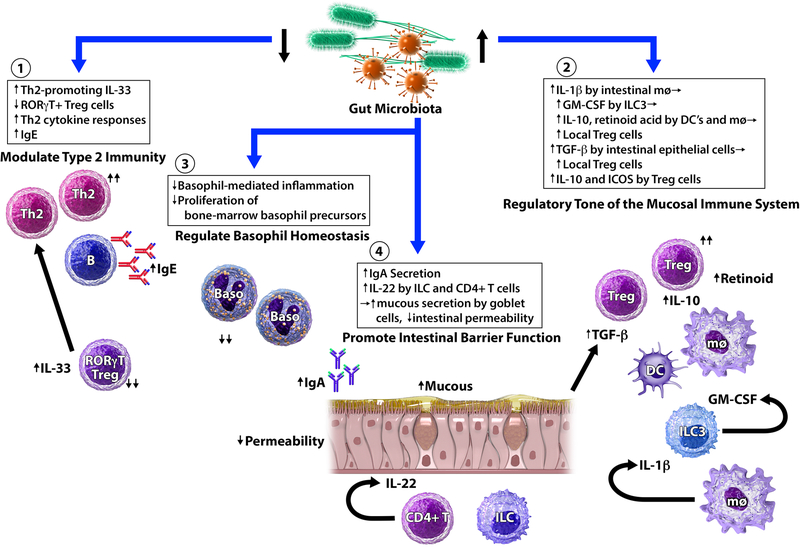
The mechanisms by which gut microbiota contribute to food allergy risk are not yet completely understood. Based on observations from murine studies, we have previously described the potential mechanisms by which gut microbiota may influence allergic diathesis,3 which we summarize in Figure 1 and below under the following categories: (1) modulation of type 2 immunity, (2) influence on immune maturation and tolerance, (3) regulation of basophil populations, and (4) promotion of intestinal barrier function.
Figure 1. Mechanisms by which gut microbiota may affect food allergy susceptibility.
Murine models of food allergy have shown that gut microbiota interact with multiple aspects of the intestinal mucosa, innate immunity, and adaptive immunity. (1) Gut microbiota modulate type 2 immunity. In germ-free mice, expression of Th2-promoting cytokine IL-33 by epithelial cells is increased. In contrast, Th2-inhibiting RORγt+ Treg cells are profoundly reduced in both germ-free and antibiotic-treated mice. As a result, Th2-associated pathology is exacerbated and IgE levels are elevated in the absence of key microbial signals. (2) Gut microbiota influence immune development and tolerance. Microbial colonization promotes the expansion of Treg cells. Microbial signals promote IL-1β secretion by intestinal macrophages (mφ), which leads to GM-CSF release by type 3 innate lymphoid cells (ILC3). As a result, IL-10 and retinoid acid secretions by dendritic cells (DC) and mφ are elevated, which leads to the expansion of local Treg cells. Strain-specific Clostridia colonization of gnotobiotic mice stimulates the secretion of TGF- β from intestinal epithelial cells, leading to the expansion of colonic Treg cells. Strain-specific Clostridia colonization also induces key anti-inflammatory molecules (IL-10 and ICOS) in Treg cells. (3) Gut microbiota regulate basophil populations. Circulating basophil levels increase in antibiotic-treated mice. Conversely, the presence of microbial signals limits the proliferation of bone-marrow basophil precursors, and reduces basophil-mediated allergic inflammation. (4) Gut microbiota promote intestinal barrier function. Selective colonization of germ-free mice with certain strains of Clostridia and Bacteroides promotes intestinal IgA secretion, which can contribute to immune exclusion and reduce allergen uptake. Strain-specific Clostridia colonization of germ-free mice also induces IL-22 production by innate lymphoid cells (ILC) and CD4+ T cells. IL-22, in turn, promotes mucous secretion by goblet cells and reduces intestinal permeability to dietary allergens. Adapted from Ho and Bunyavanich, Curr Allergy Asthma Rep. 2018 Apr 5;18(4):27.
Modulation of type 2 immunity
Evidence from germ-free mouse models supports a role for microbiota in modulating type 2 immunity. Compared to wild-type mice, germ-free mice exhibit elevated serum IgE levels at baseline. This is thought to be due to isotype switching of B cells to IgE at mucosal sites in a CD4 T-cell and IL-4 dependent manner.20 Germ-free mice are also more likely to become sensitized to cow’s milk protein and ovalbumin through oral exposure.20, 21 When challenged, milk-sensitized germ-free mice exhibit exacerbated systemic anaphylaxis with reduced rectal temperatures, higher levels of blood mouse mast cell protease-1 (mMCP-1), beta lactoglobulin-specific IgG1, and a systemic Th2-skewed response compared to sensitized wild-type controls.21 In the absence of microbial signals, increased expression of IL-33, a type 2 cytokine, has also been observed in the intestinal epithelium.23 The presence of microbiota can induce IL6 and IL23 production toward a type 3 response that suppresses type 2 responses.23 Microbiota may also regulate type 2 responses by inducing RORγt+ Tregs in a retinoic acid (RA)-dependent manner. RORγt+ Tregs regulate coactivator functions of dendritic cells by expressing high levels of CTLA4, thereby regulating the generation of Th2 cells in the intestine. Mice deficient in RORγt+ Tregs produced higher amounts of the type 2 cytokines IL-4 and IL-5.23
Immune maturation and tolerance
Conventional immune maturation in mice requires colonization by mouse-specific microbiota.24 In contrast, germ-free mice and mice populated with human microbiota have low levels of CD4+ and CD8+ T cells, low rates of T cell proliferation, few dendritic cells, and low antimicrobial peptide expression.24 These differences in immune maturation are possibly due to the decreased ability of human microbiota in mice to induce antigen uptake from the gut lumen by dendritic cells, thereby decreasing downstream T cell activation and proliferation. 24 Colonizing germ-free mice with segmented filamentous bacteria (SFB), which are commensal taxa in mice, partially restores CD4+ RORγt+ T cell counts, while failing to increase CD4+ Foxp3+ cell numbers.24 These results suggest that individual components of host-specific microbiota may be responsible for specific aspects of immune maturation.
In murine models, microbial colonization promotes the expansion of Treg cells, which are essential for immune tolerance and modulation of adaptive immunity.25 Oral colonization of mice with certain Clostridia strains promotes colonic Treg cell population by stimulating the secretion of TGF-β from intestinal epithelial cells.26 In addition, strain-specific Clostridia colonization induced key anti-inflammatory molecules (IL-10 and ICOS) in Treg cells, leading to an attenuated phenotype in an allergic disease model. 26 Bacteroides strains similarly promoted Treg cells and functions, although to a lesser degree compared to strains within Clostridia.25
The presence of microbiota overall also promotes IL-1β secretion by intestinal macrophages, which leads to GM-CSF release by type 3 innate lymphoid cells and subsequent release of retinoic acid (RA) and IL-10 by dendritic cells and macrophages. RA and IL-10, in turn, promote the expansion of local Treg cells.27 In one study aimed at identifying CD4+ FOXP3+ Treg-inducing bacterial strains from human microbiota, 17 strains of bacteria from human stool were identified as enhancing Treg cell abundance, with genome sequencing revealing that all 17 strains were within Clostridia.26 In another study, colonization of gnotobiotic mice with Clostridium clusters was found to be protective against sensitization to peanut/cholera toxin. Compared to germ-free controls, the mice colonized with Clostridium clusters showed reduced levels of peanut-specific and total IgE and no drop in rectal temperature upon allergen challenge.28 While many studies in murine models have found that taxa from Clostridia confer an overall protective effect on the host, their association with atopic risk has been more variable in human studies5, 9, 11, 18, 29, 30
Regulation of basophil populations
Gut microbiota may modulate allergic responses by modulating basophil populations. In mice treated with broad-spectrum antibiotics, increased numbers of steady-state circulating basophil populations were observed along with exaggerated Th2 cell responses and elevated IgE concentrations.31 Depletion of basophils in these mice attenuated Th2 cell responses, suggesting that basophils contributed to the observed allergic inflammation. Microbially-derived signals were also observed to influence proliferation of bone-marrow resident precursor populations for basophil development.31
Promotion of intestinal barrier function
Gut microbiota can promote IgA secretion, contribute to immune exclusion, and reduce allergen uptake.28 For example, fecal IgA levels in germ-free mice can be restored with colonization by Clostridia clusters (predominantly clusters XIVa, XIVb, and IV), and partially restored with Bacteroides uniformis colonization.28 Gene expression of IL-22, a cytokine that protects intestinal epithelial barrier by promoting mucous secretion by goblet cells, is also upregulated in lamina propria lymphocytes of germ-free mice colonized with Clostridia clusters (but not B. uniformis) with concomitant increases in the numbers of mucus-producing goblet cells.28 This is thought to affect permeability of the intestinal barrier to dietary allergens, such as peanut Arah2 and Arah6 proteins, as circulating concentrations of these proteins after intragastric peanut gavage were reduced in Clostridia-colonized (but not B. uniformis colonized) mice.28
Although murine models provide insight into potential mechanisms that characterize microbiome interactions with immune development and tolerance in humans, such mechanisms may be distinct in human beings, and it remains to be seen the extent to which findings from murine models can be observed in human beings. In human beings, evidence supporting a causal role for gut microbiota in the development and course of food allergy remains largely unexplored, in large part because of the challenges of performing interventional trials in this area.
Interventional trials
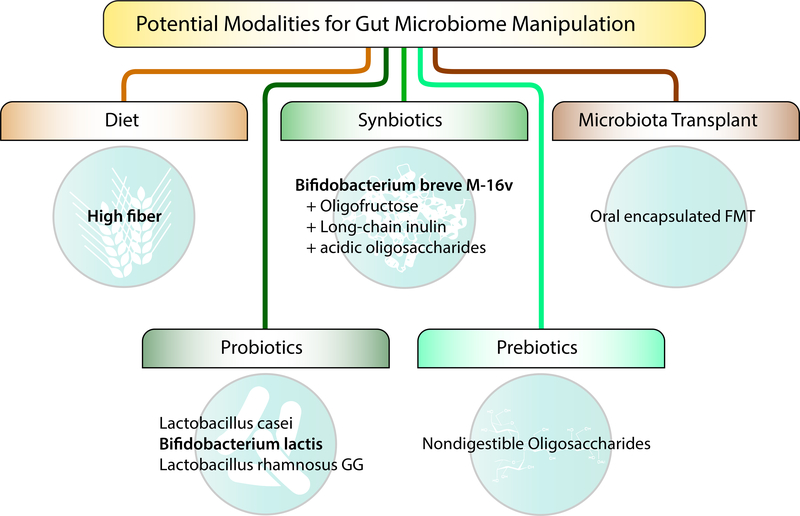
Findings that gut microbiota can modulate food allergy susceptibility suggest potential therapeutic utility from the manipulation of the gut microbiome to the host’s advantage. This is an area of great interest given the potential benefits and possible opportunities to shape food allergy development and treatment. Potential modalities for gut microbiome manipulation include diet, probiotics, prebiotics, synbiotics, and fecal microbiota transplantation (Figure 2).
Figure 2. Potential modalities for gut microbiome manipulation.
Potential modalities for gut microbiome manipulation include diet, probiotics, prebiotics, synbiotics, and fecal microbiota transplantation. Probiotics are live microorganisms that, when administered, confer a health benefit to the host. Prebiotics are non-digestible substrates that can be selectively utilized by host microorganisms. Synbiotics are products that contain both prebiotics and probiotics. Fecal microbiota transplantation (FMT) is the administration of fecal matter from a donor to a recipient. Examples of each based on existing or planned studies are provided.
Probiotics
A limited number of clinical trials of probiotics for the prevention or treatment of challenge-proven food allergies have been published. Probiotics refer to live micro-organisms that can confer benefits to the host when administered. Clinical trials of probiotic supplementation with Lactobacillus casei and Bifidobacterium lactis in children with milk allergy for 12 months showed no effect on milk allergy resolution.13, 32 However, another trial of milk allergic children studying supplementation with Lactobacillus rhamnosus GG combined with extensively hydrolyzed casein formula demonstrated increased rates of milk allergy resolution after 6 and 12 months compared to a control group receiving the formula alone.13, 32 At 12 months, the absolute risk difference in cow’s milk tolerance was 0.20 (95% CI 0.05–0.35) between the trial arms. A subsequent study comparing stool from healthy infants and from cow’s milk allergic infants before and after treatment with extensively hydrolyzed formula with or without Lactobacillus rhamnosus GG showed that Blautia and Roseburia were enriched in the gut microbiome of tolerant infants with higher concentrations of the short chain fatty acid butyrate, leading the investigators to hypothesize that the acquisition of specific strains in these genera is associated with tolerance.12 Consistent with observations from other studies of gut microbiota, the effects of probiotics are likely strain-specific, and thus careful attention to strain-level effects and interventions is merited.
Much of the work on probiotics and food allergy has focused on individuals with cow’s milk allergy, with far fewer studies of other food allergies. However, the rationale for an effect of probiotics on other food allergies is analogous, and Lactobacillus rhamnosus GG has also been studied in the context of peanut allergy. In a clinical trial, Lactobacillus rhamnosus GG was administered with peanut oral immunotherapy for 18 months. Subjects receiving the combination treatment had higher rates of desensitization to peanut compared to placebo (82.1% vs. 3.6%, respectively).33 However, as there was no OIT-only or probiotic-only group, efficacy due to the probiotic itself was unclear. A follow up study of 48 of the 56 children who participated in this combined probiotic and oral immunotherapy trial four years after treatment cessation found that individuals who had been treated were more likely to have continued eating peanut compared to those who had taken placebo (67% vs. 4%, p = 0.001), and more participants from the treated group had smaller peanut skin prick test size and higher peanut sIgG4:sIgE ratios compared to placebo-treated controls.34
Given the limited aggregate literature on probiotics and food allergy thus far, the most recent Cochrane review35 states that more data are needed before recommendations can be made to support probiotic supplementation for food allergy. This is supported by more recent meta-analyses on the subject36 as well as recognition by the World Allergy Organization that there is “very low quality evidence” on this topic at present.37
Prebiotics
Prebiotics are nondigestible food components that selectively foster the growth and activity of selected commensal microbiota in the host. For example, fiber is a prebiotic that many gut bacteria use as a nutrient and in turn produce short chain fatty acids, which are thought to beneficially inhibit allergic inflammation.38, 39 Prebiotics are frequently added to infant formulas. Effective prebiotics are not digested or absorbed in the upper gastrointestinal tract and reach the large intestines, where they are selectively used by microorganisms with health-promoting effects.40 Clinical trials of prebiotic supplementation in infants have shown no effect on the development of food allergy.40–42 However, risk of asthma and eczema was reduced in some individual studies.40–42
Synbiotics
Combinations of probiotics and prebiotics are termed synbiotics, and clinical trials of these for the prevention of allergy are underway. A prospective, randomized, double-blind controlled study of 110 full term infants with cow’s milk allergy receiving either amino acid-based formula (AAF) or AAF with synbiotics showed that subjects in both arms have normal and similar growth.43 The synbiotics used in this trial included Bifidobacterium breve M-16V, oligofructose, long-chain inulin, and acidic oligosaccharides. A planned but not yet recruiting randomized, double-blind clinical trial of children at high risk for allergy sponsored by the same company will compare partially hydrolyzed infant formula with synbiotics vs. standard infant formula (NCT03067714) for the primary outcome of doctor-diagnosed IgE-mediated allergic manifestations.
Fecal microbiota transplantation
Fecal microbiota transplantation (FMT) refers to the transfer of communities of microbes from a donor to a recipient, such as has been used for patients with Clostridium difficile colitis.44, 45 This is an intriguing option to consider, given findings from murine models supporting that fecal transfer can be an effective mode to shape allergy outcomes. A small Phase I open label trial to evaluate the safety and efficacy of oral encapsulated FMT for the treatment of peanut allergy is underway (NCT02960074). However, until further work is done in this area, significant questions will remain about FMT for food allergy treatment, including the optimal community to transfer, how best to ensure establishment of the donated community, and safety of the procedure.
Future directions
Knowledge about the gut microbiome’s role in food allergy is an emerging area with ample opportunities to deepen current foci as well as develop new directions. Findings in this area of research have been relatively heterogeneous, reflecting the variability of study designs, diverse phenotyping criteria, and differences in profiling and analytic approaches. Especially for human studies of the gut microbiome of food allergy, thoughtful prospective planning of study design and execution in robust sample sizes are needed to enable reliable findings that allow for valid cross-study comparisons. Attention to clinical phenotyping will ensure rigorous and reproducible definitions of food sensitization and food allergy, and more studies should be conducted to assess the extent to which associations between the gut microbiome and food allergy differ by specific food allergy. While murine studies have yielded many insights, additional work will be needed to assess the degree to which murine findings are translatable to the human context. Longitudinal studies of the gut microbiome in human populations, involving sampling and phenotyping at multiple time points, will also further our understanding of temporal sequence, causality, and how host-microbe interactions may shape the development and course of food allergy.
As most current investigations of the microbiome in food allergy have focused on bacterial microbiota, future research could assess contributions from the virome and mycobiome. Sequencing methods, references databases, and analytic tools to assess the virome and mycobiome are less developed than those for the bacterial microbiome.5 However, increasingly available tools for the analysis of data from shotgun metagenomic sequencing (i.e. whole genome sequencing performed on genomic DNA from a mixed microbial community) are enabling microbiome research efforts beyond the bacterial microbiome.4, 46 For characterizing the bacterial microbiome, shotgun metagenomic sequencing can also offer better resolution than 16S rRNA sequencing at the species and strain level and more accurate community diversity estimates by avoiding PCR biases4, 46 For these reasons, shotgun sequencing is increasingly used in prominent studies of the microbiome,47–49 although its relative higher cost is a consideration. Because both human and murine studies have shown that strains within the same bacterial species can have differing immunologic effects,26, 28 the capacity to decipher microbiota at the strain level will likely become increasingly desired.
Better characterization of interactions between microbiota and host, and the specific pathways and effects by which microbiota exert their influence, is another worthwhile research direction that will be enabled by multidimensional profiling and systems biology.4, 50, 51 Because food allergy is a complex and heterogeneous disease, it is unlikely that the microbiota implicated in its pathogenesis and disease course, or even the microbiome in its entirety, can capture the interdependent dynamics of the molecular networks involved in food allergy.4 Our understanding of food allergy has been advanced not only by studies of the microbiome, but also by data generated through genome-wide association,52–55 transcriptome,56 epigenome,57, 58 and metabolome studies.4, 59, 60 Integrating these types of system-wide data is critical if we are to construct models that are predictive of complex biological interactions and systems, a necessary step to developing a more complete understanding of food allergy.4, 11, 50, 51
It is likely that bacterial, viral, and fungal biomes interact with the human genome in complex ways to influence food allergy.4, 37, 50, 51 System biology approaches have been used to examine relationships between microbiota and host genomic profiles in other disease areas such as inflammatory bowel disease.61, 62 To make accurate disease predictions, we need to apply a systems biology approach to create the networks capable of representing causal relationships among molecular features within a given cell or tissue type,56 between different tissues63, and between host and environment, including the microbiome.4
Conclusion
Our understanding of the role of the gut microbiome in food allergy has evolved with knowledge gained form observational human, murine, and interventional trial studies. The evidence thus far suggests that gut dysbiosis likely precedes the development of food allergy, and the timing of such dysbiosis is critical. Gut microbiota associated with individual food allergies may be distinct, but further research in this area is needed. Murine models have provided important experimental dimensions to our study of the microbiome in food allergy. Mechanistically, gut microbiota may affect food allergy susceptibility by modulating type 2 immunity, influencing immune maturation and tolerance, regulating basophil populations, and promoting intestinal barrier function. Ongoing and future clinical trials of probiotics, prebiotics, synbiotics, and fecal microbiota transfer will help translate our enhanced understanding of the gut microbiome in food allergy to clinical practice. Future work in this area will include the deepening of current research foci as well as expansion to considerations of the virome, mycobiome, and interactions between the microbiome, host, and environment through systems biology approaches. By moving forward research on the microbiome in food allergy, we can further our understanding of food allergy and derive new approaches for its prevention and therapy.
Key messages.
Growing evidence supports a role for the gut microbiome in the pathogenesis and course of food allergy
Gut dysbiosis may precede the development of food allergy, and the timing of such dysbiosis is critical.
Gut microbiota associated with individual food allergies may be distinct.
Murine models suggest that gut microbiota affect food allergy susceptibility by modulating type 2 immunity, influencing immune maturation and tolerance, regulating basophil populations, and promoting intestinal barrier function.
Ongoing and future interventional clinical trials of probiotics, prebiotics, synbiotics, and fecal microbiota transfer can help translate our understanding of the gut microbiome in food allergy to clinical practice.
Robust and consistent study design, multi-dimensional profiling, and systems biology approaches will further advance our understanding of the microbiome in food allergy.
Acknowledgments
SB is supported by the National Institutes of Health (R01AI118833, U19AI136053)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sicherer SH, Sampson HA. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J Allergy Clin Immunol 2018; 141:41–58. [DOI] [PubMed] [Google Scholar]
- 2.Boyce JA, Assa’ad A, Burks AW, Jones SM, Sampson HA, Wood RA, et al. Guidelines for the diagnosis and management of food allergy in the United States: report of the NIAID-sponsored expert panel. J Allergy Clin Immunol 2010; 126:S1–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho HE, Bunyavanich S . Role of the Microbiome in Food Allergy. Curr Allergy Asthma Rep 2018; 18:27. [DOI] [PubMed] [Google Scholar]
- 4.Bunyavanich S, Schadt EE. Systems biology of asthma and allergic diseases: a multiscale approach. J Allergy Clin Immunol 2015; 135:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang YJ, Marsland BJ, Bunyavanich S, O’Mahony L, Leung DY, Muraro A, et al. The microbiome in allergic disease: Current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J Allergy Clin Immunol 2017; 139:1099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson-Chagoyan OC, Vieites JM, Maldonado J, Edwards C, Gil A. Changes in faecal microbiota of infants with cow’s milk protein allergy--a Spanish prospective case-control 6-month follow-up study. Pediatr Allergy Immunol 2010; 21:e394–400. [DOI] [PubMed] [Google Scholar]
- 7.Savage JH, Lee-Sarwar KA, Sordillo J, Bunyavanich S, Zhou Y, O’Connor G, et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunyavanich S, Shen N, Grishin A, Wood R, Burks W, Dawson P, et al. Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol 2016; 138:1122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fazlollahi M, Chun Y, Grishin A, Wood RA, Burks AW, Dawson P, et al. Early-life gut microbiome and egg allergy. Allergy 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee-Sarwar K, Kelly RS, Lasky-Su J, Moody DB, Mola AR, Cheng TY, et al. Intestinal microbial-derived sphingolipids are inversely associated with childhood food allergy. J Allergy Clin Immunol 2018; 142:335–8 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol 2001; 107:129–34. [DOI] [PubMed] [Google Scholar]
- 12.Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J 2016; 10:742–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berni Canani R, Nocerino R, Terrin G, Coruzzo A, Cosenza L, Leone L, et al. Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy: a randomized trial. J Allergy Clin Immunol 2012; 129:580–2, 2 e1–5. [DOI] [PubMed] [Google Scholar]
- 14.Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, Garcia Lloret M, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol 2013; 131:201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinstock GM. Genomic approaches to studying the human microbiota. Nature 2012; 489:250–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy 2015; 45:632–43. [DOI] [PubMed] [Google Scholar]
- 17.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature 2012; 486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bunyavanich S, Shen N, Grishin A, Wood R, Burks W, Dawson P, et al. Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012; 336:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe 2013; 14:559–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez B, Prioult G, Hacini-Rachinel F, Moine D, Bruttin A, Ngom-Bru C, et al. Infant gut microbiota is protective against cow’s milk allergy in mice despite immature ileal T-cell response. FEMS Microbiol Ecol 2012; 79:192–202. [DOI] [PubMed] [Google Scholar]
- 22.Bashir ME, Louie S, Shi HN, Nagler-Anderson C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol 2004; 172:6978–87. [DOI] [PubMed] [Google Scholar]
- 23.Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 2015; 349:989–93. [DOI] [PubMed] [Google Scholar]
- 24.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 2012; 149:1578–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Round JL, Mazmanian SK. Inducible Foxp(3+) regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America 2010; 107:12204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500:232–6. [DOI] [PubMed] [Google Scholar]
- 27.Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, et al. Microbiota-Dependent Crosstalk Between Macrophages and ILC3 Promotes Intestinal Homeostasis. Science 2014; 343:1477–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A 2014; 111:13145–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sepp E, Julge K, Mikelsaar M, Bjorksten B. Intestinal microbiota and immunoglobulin E responses in 5-year-old Estonian children. Clin Exp Allergy 2005; 35:1141–6. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka M, Korenori Y, Washio M, Kobayashi T, Momoda R, Kiyohara C, et al. Signatures in the gut microbiota of Japanese infants who developed food allergies in early childhood. FEMS Microbiol Ecol 2017; 93. [DOI] [PubMed] [Google Scholar]
- 31.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nature Medicine 2012; 18:538–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hol J, van Leer EH, Elink Schuurman BE, de Ruiter LF, Samsom JN, Hop W, et al. The acquisition of tolerance toward cow’s milk through probiotic supplementation: a randomized, controlled trial. J Allergy Clin Immunol 2008; 121:1448–54. [DOI] [PubMed] [Google Scholar]
- 33.Tang ML, Ponsonby AL, Orsini F, Tey D, Robinson M, Su EL, et al. Administration of a probiotic with peanut oral immunotherapy: A randomized trial. J Allergy Clin Immunol 2015; 135:737–44 e8. [DOI] [PubMed] [Google Scholar]
- 34.Hsiao KC, Ponsonby AL, Axelrad C, Pitkin S, Tang MLK, Team PS. Long-term clinical and immunological effects of probiotic and peanut oral immunotherapy after treatment cessation: 4-year follow-up of a randomised, double-blind, placebo-controlled trial. Lancet Child Adolesc Health 2017; 1:97–105. [DOI] [PubMed] [Google Scholar]
- 35.Osborn DA, Sinn JK. Probiotics in infants for prevention of allergic disease and food hypersensitivity. Cochrane Database Syst Rev 2007:CD006475. [DOI] [PubMed] [Google Scholar]
- 36.Cuello-Garcia CA, Brozek JL, Fiocchi A, Pawankar R, Yepes-Nunez JJ, Terracciano L, et al. Probiotics for the prevention of allergy: A systematic review and meta-analysis of randomized controlled trials. J Allergy Clin Immunol 2015; 136:952–61. [DOI] [PubMed] [Google Scholar]
- 37.Fiocchi A, Pawankar R, Cuello-Garcia C, Ahn K, Al-Hammadi S, Agarwal A, et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ J 2015; 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013; 504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504:446–50. [DOI] [PubMed] [Google Scholar]
- 40.Osborn DA, Sinn JK. Prebiotics in infants for prevention of allergy. Cochrane Database Syst Rev 2013:CD006474. [DOI] [PubMed] [Google Scholar]
- 41.Grimshaw K, Logan K, O’Donovan S, Kiely M, Patient K, van Bilsen J, et al. Modifying the infant’s diet to prevent food allergy. Arch Dis Child 2017; 102:179–86. [DOI] [PubMed] [Google Scholar]
- 42.Wopereis H, Sim K, Shaw A, Warner JO, Knol J, Kroll JS. Intestinal microbiota in infants at high risk for allergy: Effects of prebiotics and role in eczema development. J Allergy Clin Immunol 2018; 141:1334–42 e5. [DOI] [PubMed] [Google Scholar]
- 43.Burks AW, Harthoorn LF, Van Ampting MT, Oude Nijhuis MM, Langford JE, Wopereis H, et al. Synbiotics-supplemented amino acid-based formula supports adequate growth in cow’s milk allergic infants. Pediatr Allergy Immunol 2015; 26:316–22. [DOI] [PubMed] [Google Scholar]
- 44.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368:407–15. [DOI] [PubMed] [Google Scholar]
- 45.Youngster I, Russell GH, Pindar C, Ziv-Baran T, Sauk J, Hohmann EL. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA 2014; 312:1772–8. [DOI] [PubMed] [Google Scholar]
- 46.Tessler M, Neumann JS, Afshinnekoo E, Pineda M, Hersch R, Velho LFM, et al. Large-scale differences in microbial biodiversity discovery between 16S amplicon and shotgun sequencing. Sci Rep 2017; 7:6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018; 359:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yassour M, Jason E, Hogstrom LJ, Arthur TD, Tripathi S, Siljander H, et al. Strain-Level Analysis of Mother-to-Child Bacterial Transmission during the First Few Months of Life. Cell Host Microbe 2018; 24:146–54 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe 2018; 24:133–45 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knight R, Callewaert C, Marotz C, Hyde ER, Debelius JW, McDonald D, et al. The Microbiome and Human Biology. Annu Rev Genomics Hum Genet 2017; 18:65–86. [DOI] [PubMed] [Google Scholar]
- 51.Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, et al. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 2016; 535:94–103. [DOI] [PubMed] [Google Scholar]
- 52.Marenholz I, Grosche S, Kalb B, Ruschendorf F, Blumchen K, Schlags R, et al. Genome-wide association study identifies the SERPINB gene cluster as a susceptibility locus for food allergy. Nat Commun 2017; 8:1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asai Y, Eslami A, van Ginkel CD, Akhabir L, Wan M, Ellis G, et al. Genome-wide association study and meta-analysis in multiple populations identifies new loci for peanut allergy and establishes C11orf30/EMSY as a genetic risk factor for food allergy. J Allergy Clin Immunol 2018; 141:991–1001. [DOI] [PubMed] [Google Scholar]
- 54.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, et al. letters Genomewide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nature Genetics 2012; 44:1222–6. [DOI] [PubMed] [Google Scholar]
- 55.Hong X, Hao K, Ladd-Acosta C, Hansen KD, Tsai HJ, Liu X, et al. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children. Nat Commun 2015; 6:6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watson CT, Cohain AT, Griffin RS, Chun Y, Grishin A, Hacyznska H, et al. Integrative transcriptomic analysis reveals key drivers of acute peanut allergic reactions. Nat Commun 2017; 8:1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong X, Ladd-Acosta C, Hao K, Sherwood B, Ji H, Keet CA, et al. Epigenome-wide association study links site-specific DNA methylation changes with cow’s milk allergy. J Allergy Clin Immunol 2016; 138:908–11 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martino D, Joo JE, Sexton-Oates A, Dang T, Allen K, Saffery R, et al. Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy. Epigenetics 2014; 9:998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steinke JW, Pochan SL, James HR, Platts-Mills TAE, Commins SP. Altered metabolic profile in patients with IgE to galactose-alpha-1,3-galactose following in vivo food challenge. J Allergy Clin Immunol 2016; 138:1465–7 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kong J, Chalcraft K, Mandur TS, Jimenez-Saiz R, Walker TD, Goncharova S, et al. Comprehensive metabolomics identifies the alarmin uric acid as a critical signal for the induction of peanut allergy. Allergy 2015; 70:495–505. [DOI] [PubMed] [Google Scholar]
- 61.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012; 491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Virgin HW, Todd JA. Metagenomics and personalized medicine. Cell 2011; 147:44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobrin R, Zhu J, Molony C, Argman C, Parrish ML, Carlson S, et al. Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol 2009; 10:R55. [DOI] [PMC free article] [PubMed] [Google Scholar]