Abstract
Background
The plastid is a semiautonomous organelle with its own genome. Plastid genomes have been widely used as models for studying phylogeny, speciation and adaptive evolution. However, most studies focus on comparisons of plastid genome evolution at high taxonomic levels, and comparative studies of the process of plastome evolution at the infrageneric or intraspecific level remain elusive. Holcoglossum is a small genus of Orchidaceae, consisting of approximately 20 species of recent radiation. This made it an ideal group to explore the plastome mutation mode at the infrageneric or intraspecific level.
Results
In this paper, we reported 15 complete plastid genomes from 12 species of Holcoglossum and 1 species of Vanda. The plastid genomes of Holcoglossum have a total length range between 145 kb and 148 kb, encoding a set of 102 genes. The whole set of ndh-gene families in Holcoglossum have been truncated or pseudogenized. Hairpin inversion in the coding region of the plastid gene ycf2 has been found.
Conclusions
Using a comprehensive comparative plastome analysis, we found that all the indels between different individuals of the same species resulted from the copy number variation of the short repeat sequence, which may be caused by replication slippage. Annotation of tandem repeats shows that the variation introduced by tandem repeats is widespread in plastid genomes. The hairpin inversion found in the plastid gene ycf2 occurred randomly in the Orchidaceae.
Electronic supplementary material
The online version of this article (10.1186/s12862-019-1384-5) contains supplementary material, which is available to authorized users.
Keywords: Holcoglossum, Plastid genome, NDH complex, Divergence hotspot, Intraspecific variation, Tandem repeat, Hairpin inversion
Background
The plastid is a semiautonomous organelle that evolved from cyanobacteria by endosymbiosis [1]. During the course of evolution, the coding capacity of plastid genomes (plastomes) has experienced drastic reductive evolution with gene loss or transfer to the nucleus [2–4]. The genes reserved in plastomes are usually necessary for the chloroplast to perform its normal functions, including approximately 80 unique protein-coding genes, 30 tRNA genes and 4 rRNA genes. In addition to highly conserved gene content, the organization of the plastome in higher plants is remarkably conserved, which is characterized by two large inverted repeat regions (IRA and IRB) separated by two single copy regions with different lengths, known as a large single copy region (LSC) and a small single copy region (SSC) [3, 5–7].
Benefiting from the advances in next-generation sequencing, more plastid genomes have been sequenced, and there are more than 2800 records of eukaryotic plastid genomes available in the NCBI database (https://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?opt=plastid&taxid=2759 last accessed May 30, 2018). Due to their frequent sequencing and wide availability, plastid genomes have been used as models in genetic variation studies, encompassing both micro- and macro-evolutionary events across all lineages of plants [8–14]. However, previous studies have mostly focused on comparisons of plastid genome evolution at higher taxonomic levels (e.g., across genera or families or orders) or between autotrophic and heterotrophic plants, which may have phylogenetic sampling ‘gaps’ or evolutionary route ‘gaps’ [15]. This may cause key steps in the process of plastome evolution at the infrageneric or intraspecific level to remain elusive.
The genus Holcoglossum Schltr. (Vandeae, Orchidaceae) consists of approximately 20 species that are mainly distributed in southwestern China and neighbouring regions [16–24]. Holcoglossum has two diversity centres, one in the tropical region and the other in the temperate alpine region of the Hengduan Mountains (HDM), with an elevation of over 2000 m [20, 23, 25]. At least six species of Holcoglossum are distributed in the HDM, five of which are restricted to this area [23]. Biogeographic analyses and molecular phylogeny suggest that Holcoglossum dispersed from tropical regions to the HDM and then radiated there [23]. Previous results indicated that the pendent growing pattern [23] and laterocytic and polarcytic stomata are perhaps ecological adaptations to the strong winds and ample rains in the alpine region of the HDM [26]. Rapid changes in temperature and weather conditions are major challenges for the species living in temperate alpine regions in the HDM. Previous results indicated that plastid genes of Cardamine resedifolia (Brassicaceae) experienced more intense positive selection than those of the low altitude C. impatiens, possibly as a consequence of adaptation to high altitude environments [12].
Here, using comparative plastid genomes of 15 complete plastome sequences of 12 species of Holcoglossum and 1 species of Vanda, we aim to (1) understand the evolution of the plastid genome in Holcoglossum and (2) investigate the evolutionary pattern of the plastid genome at infrageneric and intraspecific levels.
Methods
Taxa sampling, DNA isolation, library preparation, and sequencing
In this study, we sampled and sequenced 12 species of Holcoglossum, including 2 individuals of H. flavescens and H. nujiangense, and 1 species of Vanda. Two plastomes of Neofinetia were downloaded from NCBI (Table 1) as outgroups. Fresh leaves, stems and flowers were collected in the field and preserved in silica gel as well as frozen at − 20 °C. Total DNA was isolated using a modified cetyltrimethyl ammonium bromide (CTAB) protocol [27]. DNA with concentrations greater than 100 ng/ml was sheared to 500 bp using Covaris M220. Sequencing libraries were prepared using the NEBNext Ultra DNA Library Prep Kit (according to the manufacturer’s protocol) for subsequent paired-end sequencing on an Illumina HiSeq 2500 at the Institute of Botany, Chinese Academy of Sciences.
Table 1.
Basic information of plastid genomes used in this study
| Species | Mean coverage | Length (bp) | GC Content (%) | No. vouchers specimen or NCBI accession | |||
|---|---|---|---|---|---|---|---|
| Total | LSC | SSC | IR | ||||
| Holcoglossum nujiangense_S1_S16 | 220 | 146,487 | 82,955 | 11,916 | 25,808 | 35.4 | Jin Xiaohua 6930 |
| Holcoglossum nujiangense_S5_S9 | 539 | 146,395 | 82,873 | 11,906 | 25,808 | 35.4 | Jin Xiaohua 10,897 |
| Holcoglossum weixiense | 205 | 146,597 | 82,981 | 12,000 | 25,808 | 35.4 | HK Kadoorie Program Team 3490 |
| Holcoglossum sinicum | 507 | 145,909 | 82,658 | 11,635 | 25,808 | 35.4 | Jin Xiaohua 14,683 |
| Holcoglossum flavescens _S2_S18 | 183 | 146,863 | 83,288 | 11,959 | 25,808 | 35.3 | Jin Xiaohua 8943 |
| Holcoglossum flavescens_S5_S10 | 489 | 146,763 | 83,188 | 11,959 | 25,808 | 35.4 | Jin Xiaohua 15,165 |
| Holcoglossum rupestre | 75 | 147,163 | 83,575 | 11,936 | 25,826 | 35.3 | Jin Xiaohua 9015 |
| Holcoglossum quasipinifolium | 131 | 147,063 | 83,440 | 12,079 | 25,772 | 35.4 | JXH028 |
| Holcoglossum lingulatum | 151 | 146,525 | 83,713 | 11,274 | 25,769 | 35.5 | Jin Xiaohua 9491 |
| Holcoglossum nagalandensis | 357 | 146,826 | 83,763 | 11,477 | 25,793 | 35.3 | Jin Xiaohua 10,522 |
| Holcoglossum amesianum | 425 | 148,074 | 84,250 | 12,026 | 25,899 | 35.3 | Jin Xiaohua 9419 |
| Holcoglossum himalaicum | 752 | 145,207 | 83,712 | 11,413 | 25,041 | 35.3 | Jin Xiaohua 9496 |
| Holcoglossum wangii | 209 | 147,170 | 83,846 | 11,594 | 25,866 | 35.4 | Jin Xiaohua 13,881 |
| Holcoglossum subulifolium | 360 | 146,930 | 83,398 | 11,802 | 25,865 | 35.5 | Jin Xiaohua 13,614 |
| Neofinetia falcata | _ | 146,497 | 83,809 | 11,775 | 25,456 | 35.3 | NC_036372 |
| Neofinetia richardsiana | _ | 146,498 | 83,809 | 11,775 | 25,457 | 35.3 | NC_036373 |
| Vanda brunnea | 191 | 149,216 | 85,783 | 11,713 | 25,860 | 35.3 | Jin Xiaohua 13,059 |
Plastome assembly and annotation
Plastome assembly and annotation followed the methods of Feng et al. (2016) [10]. In short, raw reads were trimmed and filtered with NGSQCTOOLKIT v 2.3.3 [28], and bases with a PHRED quality lower than 20 were trimmed. All trimmed reads shorter than 70 bp were discarded. The filtered reads were mapped to the plastome of Calanthe triplicata (https://www.ncbi.nlm.nih.gov/nuccore/NC_024544.1) in Geneious v10.2.2 (http://www.geneious.com, last accessed June 4, 2017) to filter reads matching the reference genomes. De novo assemblies were constructed in VELVET [29] with several K-mer values, and contigs from each assembly were merged in Geneious and combined into scaffolds using the default parameters (minimum overlap 20 bp, minimum similarity 70%). Alternatively, contigs from both assemblies (Geneious or Velvet) were merged in SSPACE [30] to form scaffolds/draft genomes. IR boundaries for each draft plastome were confirmed by BLAST [31], with the first and last sequences (approximately 50 bp) of the draft plastome used as search terms. The finished plastomes were annotated by using DOGMA with an e value of 5% and identity thresholds of 60 and 80% for protein-coding genes and tRNAs, respectively [32]. Smaller exons (< 30 bp) were manually annotated by local BLAST in Geneious. The initiation codon, termination codon, and other annotation errors for each gene were revised in Sequin and exported as GenBank files.
DNA alignment and phylogenetic analysis
We generated multiple sequence alignments of whole plastid genomes using MAFFT under the automatic model selection option with some manual adjustments [33]. At the same time, 68 protein-coding sequences were exported from plastomes in Geneious. The protein-coding sequences were aligned at the codon level with the option “-codon” using MUSCLE [34] in MEGA v7.0.2 [35]. Stop codons were removed from the sequences prior to alignment. The phylogenetic trees were reconstructed based on the nucleotide data of whole plastid genomes with the GTRGAMMA model using RAxML v8.0.9 [36] in the CIPRES Science Gateway [37], and branch support was assessed using 1000 standard bootstrap replicates.
Sequence divergence analysis
We compared the overall similarities among different plastomes in Holcoglossum using H. subulifolium with one IR region removed as a reference. The sequence identity of the Holcoglossum plastid genomes was plotted using the mVISTA program with the LAGAN mode [38]. To screen variable characters within Holcoglossum, the average number of nucleotide differences (K) and total number of mutations (Eta) were determined to analyse nucleotide diversity (Pi) using DnaSP v6.10.04 [39]. The step size was set to 200 bp, with a 500 bp window length.
The complete plastomes of two H. flavescens individuals and two H. nujiangense individuals were aligned in Geneious with the MAFFT algorithm, and differences were identified by using the “Find Variations/SNPs” function and checked individually. We recorded substitutions and indels separately, as well as their location in the chloroplast genome.
Since all of the indels in intraspecific variation are caused by the copy number variation of the short repeat sequence, as shown in our results, we further explored whether the tandem repeat also contributed to interspecific plastid genome variation. We located and annotated the tandem repeats on the multiple sequence alignment matrix of Holcoglossum plastome with Phobos [40] in Geneious.
Molecular evolutionary pattern analysis of plastid genes
To explore the selection patterns and identify positive selection on the protein-coding genes, we use two models, model M0 and a branch-site model, implemented in the PAML Codeml program [41]. The codon frequencies were determined by the F3 × 4 model. Twenty-eight genes with too few variable sites were not examined (Additional file 1: Table S2). Alignment gaps and uncertainties were deleted to avoid false positives [42].
The model M0 (model = 0, Nsites = 0, which assumes no site-wise or branch-wise dN/dS variation) estimates the rates of synonymous (dS) and non-synonymous substitutions (dN) and the dN/dS value of each gene, which can be an indication of the selection pattern.
The branch-site model (model = 2, Nsites = 2, fixed omega = 0, omega = 2) was used to detect evidence of positive selection on specific sites along a specific lineage. The goal of our study was to explore the role of positive selection in the adaptive patterns of Holcoglossum adapted to tropical regions and temperate alpine regions; thus, the tropical clade and alpine clade were used to perform the selection analyses. The likelihood ratio test (LRT) with a χ2 distribution was used to determine which models were significantly different from the null model (model = 2, Nsites = 2, fixed omega = 1, omega = 1) at a threshold of P < 0.05. The Bayes empirical Bayes (BEB) method was used to statistically identify sites under positive selection with posterior probabilities ≥0.95 [43].
Results
Plastome structure and phylogenomics of Holcoglossum
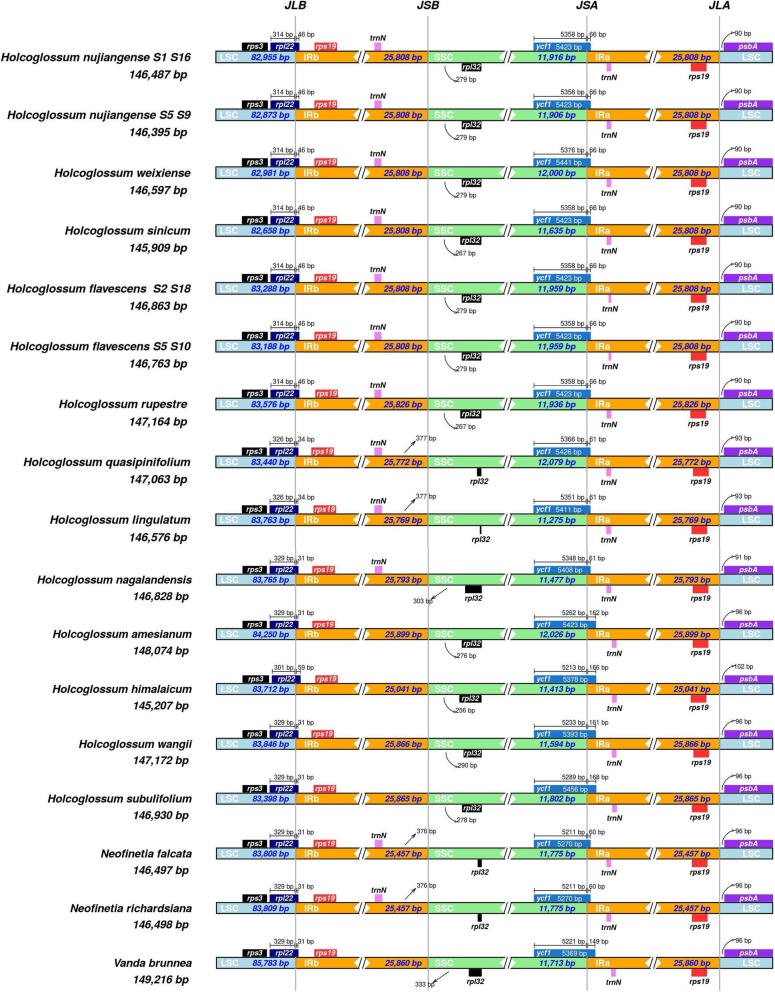
In the present study, 14 complete plastomes of 12 species of Holcoglossum and 1 species of Vanda were obtained for the first time. These plastomes showed the typical quadripartite structure of most angiosperms. The plastomes of Holcoglossum had a total length range between 145,207 bp in H. himalaicum and 148,074 bp in H. amesianum. The length variation of the Holcoglossum plastomes observed here was low (145–148 kb). The expansion and contraction of the inverted repeat regions usually contribute to variation in the length of plastomes. In this study, we found that the IR/SSC boundary was located differently among the 12 Holcoglossum species, but the location of the boundary and length of the IR regions only showed moderate variation (Table 1), and there was no obvious phylogenetic implication of extension/contraction of IRs among the Holcoglossum plastomes (Fig. 1).
Fig. 1.
Comparison of the LSC, IR, and SSC junction positions in Holcoglossum and three outgroup plastid genomes
All of the sequenced Holcoglossum plastomes are highly conserved in structure compared to most angiosperms, sharing the common typical quadripartite structure comprising two copies of IR (25,041–25,899 bp) separated by the LSC (82,658–84,250 bp) and SSC (11,275–12,079 bp) regions (Table 1). The overall GC content was between 35.3–35.5% (Table 1), which is similar to the other Orchidaceae plastomes sequenced thus far [44, 45]. The Holcoglossum plastomes encoded an identical set of 102 genes, of which 85 were unique and 17 were duplicated in the IR regions. The 102 genes contained 68 protein-coding genes, 30 tRNA genes, and 4 rRNA genes (Additional file 2: Table S1). Functional cp-ndh genes have been lost or pseudogenized in all Holcoglossum species.
Phylogenetic analyses indicated that Holcoglossum is monophyletic and subdivided into three strongly supported clades (ML bootstrap =100%): the tropical clade (TC) with five species, the alpine clade (AC) with five species and the HC clade with two species (Additional file 3: Figure S1). All of the nodes among the lineages in our tree were strongly supported by ML bootstrap values ≥94% (Additional file 3: Figure S1). Our results indicated that H. amesianum and H. naglandensis are sister groups forming a sister clade to H. himalaicum and H. wangii.
Intraspecific plastome variation and mutation hotspots of Holcoglossum plastomes
Comparing plastomes of two individuals of H. flavescens, we found 17 SNPs, 1 single nucleotide indel and 3 multi-nucleotide indels ranging from 14 to 57 bp in H. flavescens. Between the two individuals of H. nujiangense, 8 SNPs, 3 single nucleotide indels and 5 multi-nucleotide indels of 3–36 bp length have been found (Table 2). All of the SNPs and indels are located in the LSC and SSC regions, and all of the indels contributing to intraspecific variation are caused by the copy number variation of short repeat sequences.
Table 2.
Intraspecific variation between two individuals of H. flavenscens and H. nujiangense
| Holcoglossum flavenscens | Holcoglossum nujiangense | ||||||
|---|---|---|---|---|---|---|---|
| Position | Varitation type | Location | Location type | Position | Varitation type | Location | Location type |
| 895 | G/A | psbA | coding | 4271 | Indel(3 bp) | trnK | IGS |
| 9882 | A/G | trnG | IGS* | 8011 | ./T | psbK_psbI | IGS |
| 10,370 | T/C | trnR_atpA | IGS | 20,023 | A/G | rpoC2 | coding |
| 17,761 | T/G | rpoC2 | coding | 21,021 | A/C | rpoC1 | coding |
| 33,914 | C/A | trnT_psbD | IGS | 21,031 | G/A | rpoC1 | coding |
| 36,702 | A/C | trnS_psbZ | IGS | 28,263 | Indel(20 bp) | rpoB_trnC | IGS |
| 43,116 | C/T | psaA_ycf3 | IGS | 32,903 | ./A | trnE_trnT | IGS |
| 49,010 | C/A | trnL_trnF | IGS | 33,559 | A/G | trnT_psbD | IGS |
| 49,020 | Indel(28 bp) | trnL_trnF | IGS | 33,845 | Indel(19 bp) | trnT_psbD | IGS |
| 49,378 | Indel(57 bp) | trnF_trnV | IGS | 49,670 | Indel(36 bp) | trnF_trnV | IGS |
| 49,514 | Indel(14 bp) | trnF_trnV | IGS | 56,132 | G/T | rbcL_accD | IGS |
| 49,981 | C/A | trnV | IGS | 79,512 | T/C | rps8_rpl14 | IGS |
| 56,483 | A/G | accD | coding | 81,511 | ./T | rpl14_rps3 | IGS |
| 64,283 | A/C | psbE_petL | IGS | 111,832 | Indel(10 bp) | ccsA_psaC | IGS |
| 67,306 | T/C | rpl20 | coding | 112,042 | T/C | ccsA_psaC | IGS |
| 69,392 | T/. | clpP | IGS | 119,788 | G/A | ycf1 | coding |
| 73,253 | G/A | psbB/psbT | IGS | ||||
| 73,948 | A/G | psbH/petB | IGS | ||||
| 110,107 | G/T | rpl32_trnL | IGS | ||||
| 112,694 | A/C | ccsA_psaC | IGS | ||||
| 120,678 | T/C | ycf1 | coding | ||||
*IGS: Inter-Genic Sequence
The border regions of LSC/IRB, IRB/SSC, SSC/IRA, and IRA/LSC are usually highly variable even between closely related species [46, 47]. Therefore, we compared and visualized the exact IR border positions and their adjacent genes among the Holcoglossum chloroplast genomes and the reference genome using the IRscope online program [48]. The results showed that the genes trnN-rpl32-ycf1 and rpl22-rps19-psbA were located in the junctions of the SSC/IR and LSC/IR regions. The ycf1 gene spans the SSC/IRA region and extends to the IR region from 61 to 168 bp (Fig. 1). The mVISTA percent identity plot and slide window analysis show that the most divergent regions are located in the trnS-trnG, trnE-trnT, trnL-trnV, clpP-psbB and psaC-rps15 regions in the Holcoglossum plastome (Figs. 2 and 3).
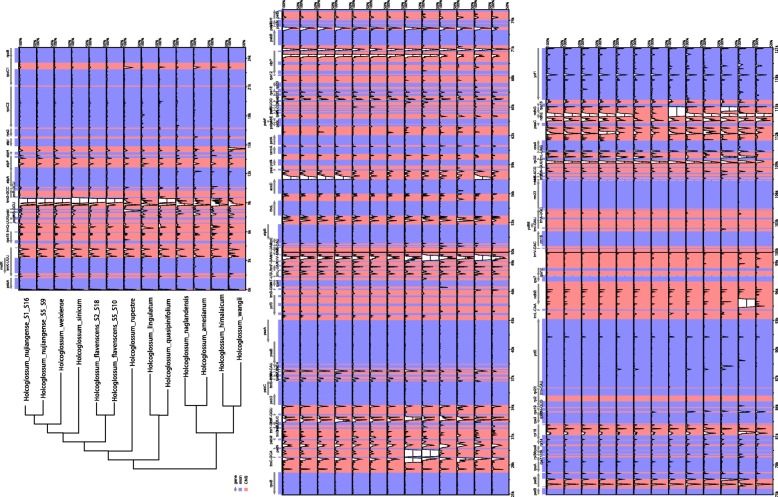
Fig. 2.
mVISTA percent identity plot of available Holcoglossum plastomes using H. subulifolium as a reference. The vertical scale indicates the percentage of identity ranging from 50 to 100%. Coding regions are in blue, and noncoding regions are in red. Cladogram redrawn from Additional file 3: Figure S1, branch lengths are not representative of evolutionary changes
Fig. 3.
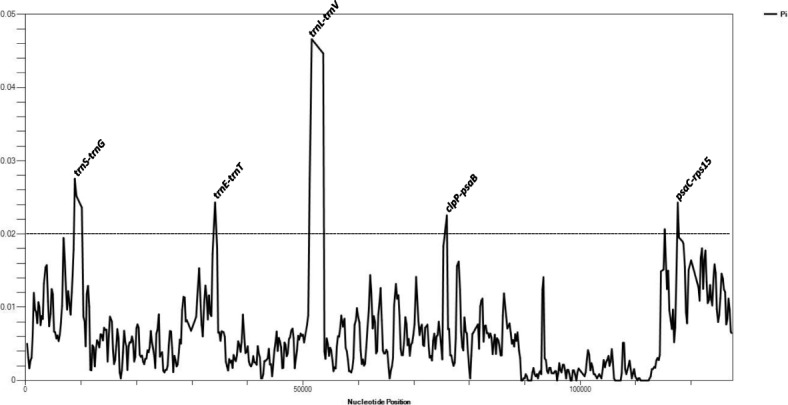
Sliding window analysis of the whole plastid genomes of Holcoglossum taxa. The 5 mutation hotspot regions (Pi > 0.02) are annotated
Molecular evolutionary pattern of Holcoglossum plastid genes
Most of the plastid genes in Holcoglossum are under strongly negative selection with a very low ω value (ω < 0.5), yet the genes ycf2 and ycf1 of uncertain function are under neutral selection with a ω value near to 1.0; the only gene found under positive selection is psbK with a high ω value (ω = 1.92088) (Additional file 1: Table S2). The branch-site model analysis does not detect any site under positive selection when the alpine clade is set as the foreground branch, while there are 14 sites in ycf2 and 2 sites in the ycf1 gene have been detected theoretically under positive selection (as the Bayes Empirical Bayes probability > 0.95) when the tropical clade is set as the foreground branch (Additional file 4: Table S3).
Discussion
Phylogeny of Holcoglossum
The phylogenetic relationships among the major lineages of Holcoglossum based on plastomes were essentially in agreement with the results of Xiang et al. [24] based on four markers (matK, trnH-psbA, trnL-F, and nuclear ITS sequences) with the exception of the placement of H. amesianum. Our results indicated that H. amesianum and H. naglandensis are sister groups forming a sister clade with H. himalaicum and H. wangii. However, H. amesianum had been placed in a sister clade to the clade formed by H. naglandensis, H. himalaicum and H. wangii but with low support (PP = 0.78, BS < 50) in previous results [24]. The difference may be due to the different taxonomic sampling in the two studies or the markers used in the previous study being unable to resolve the phylogenetic relationships in Holcoglossum.
Hairpin inversion in plastid gene ycf2
The plastid gene ycf2 is a large yet functionally undefined ORF in land plants. Nucleotide sequence similarity among land plant ycf2 is extraordinarily low compared to other plastid-encoded genes, being less than 50% across bryophytes, ferns, and seed plants [5]. When we aligned the protein coding gene ycf2 of Holcoglossum, we found a short inversion mediated by a 17 bp inverted repeat sequence located down- and up-stream in H. flavescens, H. quasipinifolium, H. amesianum and H. naglandensis (Additional file 5: Figure S2). To understand whether this inversion occurred randomly, we analysed it across the Orchidaceae family. We found that this motif is conserved at the sequence level in Orchidaceae but is inversely randomly mediated by the hairpin structure. In some species, this motif has been lost or disrupted (Additional file 6: Figure S3).
Previous studies show that most stem-loop structures involving small inversions occur in close proximity to the stop codons of genes and have the function of stabilizing the corresponding mRNA molecules [49], and the majority of the small inversions were located downstream of adjacent genes with a tail-to-tail orientation [50]. However, the hairpin inversion in the plastid gene ycf2 found in this study is located in the coding region, occurring randomly and being disrupted in some species. These results indicated that this motif may not be pivotal for ycf2 to exercise its function, and this needs to be revised with a broader sample.
Intraspecific variation of plastomes
Most of the SNPs found between the two different individuals of Holcoglossum are located in intergenic regions. We found 5 SNPs located in the coding region of psbA, rpoC2, accD, rpl20 and ycf1 in H. flavescens, among which the SNPs located in rpoC2 and accD lead to a nonsynonymous mutation between these two individuals. In H. nujiangense, we found 1 synonymous mutation SNP in rpoC2, 2 nonsynonymous mutation SNPs in rpoC1, and 1 nonsynonymous mutation SNP in ycf1. Interestingly, all of these intraspecific variation sites in coding regions are usually conserved between species. All 3 indels found in H. flavescens are located in the intergenic region (1 in trnL-trnF, 2 in trnF-trnV); the 5 indels found in H. nujiangense are located in the intron region of trnK, the intergenic region of rpoB-trnC, trnT-psbD, trnF-trnV and ccsA-psaC. Comparative analysis found that all indels are caused by the copy number variation of the short repeat sequence, which may be caused by replication slippage (Additional file 7: Figure S4). This is in line with a previous study that found that the intraspecific variation in the chloroplast genome of Astragalus membranaceus was due to an extra copy of the “TATATATTTA” repeat [51], and the vast majority of mutations in the spontaneous plastome mutants of Oenothera are indels originating from DNA replication slippage events [52]. Furthermore, the location of intraspecific variation loci shows that most variations in these two species are species-specific except for the variation in the mutation hotspot region trnL-trnV. These intraspecific loci represent potential markers that can be used to distinguish closely related varieties of specific taxa. However, further population genetic studies are still needed to determine whether intraspecific genetic diversity is linked to geographic ranges or the intrinsic characteristics of the taxonomic group.
Tandem repeat sequences contribute to plastid genome evolution
DNA tandem repeats (TRs) are not just popular molecular markers but are also important genomic elements from an evolutionary and functional perspective [53–56]. Because all the indels found in intraspecific variation are caused by the copy number variation of the short repeat sequence, as shown in our results, we further explored whether the tandem repeat also contributed to interspecific plastid genome variation. We located and annotated the tandem repeats on the multiple sequence alignment matrix of the Holcoglossum plastome with Phobos [40] in Geneious. Our results indicated that the mutation hotspot regions are always accompanied by densely distributed tandem repeats (Additional file 8: Figure S5), which indicates that the tandem repeat sequences play an important role in plastid genome variation between closely related species. This finding is consistent with the observation that nearly all detected mutations in the spontaneous plastome mutants of Oenothera could be associated with repetitive elements [52].
Furthermore, we found that in the plastid gene ycf2, a 15 bp extra copy of “TCGATATTGATGATA” is synapomorphic for the TC clade, whereas the possession of the 9 bp duplication of “ATGATAGTA” is synapomorphic for the HC plus AC clade, with a reversal (secondary loss) in H. lingulatum (Additional file 9: Figure S6). Therefore, the HC clade can be referred to as the “intermediate clade” as suggested by Xiang et al. [24]. However, whether these repeat regions have contributed to the adaption to different habitats (here referring to tropical and temperate alpine regions) remains to be verified.
Positive selection on photosynthetic chloroplast genes
Understanding the patterns of divergence and adaptation among the members of a specific phylogenetic clade can offer important clues about the forces driving its evolution [12, 57–59]. In this study, we detected some positive selective signals in the tropical clade, but sites under positive selection are quite rare and mainly detected in the ycf1 and ycf2 genes. This may be because adaptive modifications to other abiotic stresses targeting genes in the nucleus were sufficient to maintain homeostasis for photosynthesis since there are a variety of strategies for plants to adapt to the environment, so there is no need for adaptive evolution of chloroplast-encoded genes [60, 61].
NDH complex coding genes lost in Holcoglossum plastome
The chloroplast NAD(P)H-dehydrogenase-like (NDH) complex is located in the thylakoid membrane and plays an important role in mediating photosystem I cyclic electron transport (PSI-CET) and facilitating chlororespiration [62, 63]. Loss of the cp-ndh genes is widely reported in heterotrophic species because they do not need to synthesize organic carbon through photosynthesis by themselves [10, 11, 13, 64, 65]. However, as more plastid genomes have been sequenced, some autotrophic plants, such as some species of Pinales, Geraniaceae and Orchidaceae, have also been reported to lose almost the entire set of cp-ndh genes [66–70]. In our study, we also found that all of the cp-ndh genes were truncated or pseudogenized in the Holcoglossum plastid genome.
The loss of plastome genes may be due to transfer to the nucleus, substitution of a nuclear encoded mitochondrial targeted gene or substitution of a nuclear gene for a plastid gene. Translocation of ndh genes to the chondriome in Cymbidium has been reported, and different levels of ndh gene degradation among even closely related species in Cymbidium may be due to multiple bidirectional intracellular gene transfers between two organellar genomes [71]. As there is an alternative PSI cyclic electron transport pathway: the proton gradient regulation 5 (PGR5)/PGR5-like photosynthetic phenotype 1 (PGRL1)-dependent antimycin A-sensitive pathway [72–74], especially under high light conditions, the NDH1 pathway would be minor, while the PGR5 pathway would be dominant [63, 75]. The NDH complex may not be necessary for some plants. Using comparative genome analyses, Lin et al. found that nuclear NDH-related genes are also lost in orchids without cp-ndh genes [76].
Conclusions
In this study, we reported 15 completed plastid genomes using Illumina sequencing technology via a reference-guided assembly. These plastid genomes were highly conserved, and the whole set of ndh-gene families was truncated or pseudogenized. The five mutation hotspot regions were identified across the Holcoglossum plastid genomes, which could serve as potential markers for phylogenetic and population genetic studies. We further investigated the intraspecific variation of indels and substitutions in two species, and potentially diagnostic variations have been found in the plastomes of different individuals. A hairpin inversion in the coding region of the plastid gene ycf2, which occurred randomly in Orchidaceae, was found in this study. We additionally found evidence that the tandem repeat sequences contribute to the evolution of the plastid genome not only in the intergenic region but also in the coding region.
Additional files
Table S2. Statistic of substitution sites and ω values of Holcoglossum plastid genes. (XLSX 15 kb)
Table S1. List of genes identified in the plastid genomes of Holcoglossum. (DOCX 26 kb)
Figure S1. Maximum Likelihood phylogenetic tree of Holcoglossum based on the whole plastid genome except for one invert repeat region. Bootstrap support is indicated on the nodes. (PDF 171 kb)
Table S3. Detected positive selection sites in the plastid genes of TC clade Holcoglossum species. (DOCX 19 kb)
Figure S2. Hairpin inversion of ycf2 in Holcoglossum. (PNG 236 kb)
Figure S3. Hairpin inversion of ycf2 in Orchidaceae. (PDF 316 kb)
Figure S4. Intraspecific variation resulting from tandem repeats in H. flavenscens. (PNG 201 kb)
Figure S5. Tandem repeat annotated to the whole plastid genome (with only one invert repeat region) alignment. The brown triangles represent the tandem repeat regions. (PDF 1013 kb)
Figure S6. Aligned sequence matrix of ycf2 gene shows the duplication of tandem repeat in Holcoglossum. (JPG 727 kb)
Acknowledgements
We would like to thank Yan-Lei Feng for helping in plastid genome assembly, Yi-Zhen Sun for DNA sequencing, and American Journal Experts for language editing.
Funding
This study was financially supported by Strategic Priority Research Program, Chinese Academy of Sciences (XDA19050201), National Natural Science Foundation of China (31670194, 31470299, 41672018), Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences (Y4ZK111B01 to X.H.J).
Availability of data and materials
All annotated plastid genomes generated in this study have been submitted to NCBI with accession of MK442924 - MK442937, MK460222.
Abbreviations
- HDM
The Hengduan Mountains
- IRA/ IRB
Inverted repeat regions A/B
- LSC
Large single copy region
- ML
Maximum likelihood
- ORF
Open Reading Frame
- SSC
Small single copy region
Authors’ contributions
X.H.J designed the study. X. M, Z.H.L, Y.X.L and C.W.W performed the experiments. X. M and Z.H.L assembled the plastid genome. Z.H.L and X. M analysed the data. X.H.J, Z.H.L and D.Y.W wrote the initial manuscript. All authors contributed to and approved the final version.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhang-Hai Li, Email: zhanghailee@ibcas.ac.cn.
Xiao Ma, Email: maxiao@ibcas.ac.cn.
De-Yi Wang, Email: deyiwang@ibcas.ac.cn.
Yun-Xia Li, Email: yysansi@vip.qq.com.
Cheng-Wang Wang, Email: wangchengwang@ibcas.ac.cn.
Xiao-Hua Jin, Phone: +86 10-62836582, Email: xiaohuajin@ibcas.ac.cn.
References
- 1.Mereschkowsky C. Uber natur und ursprung der chromatophoren im pflanzenreiche. Biol Centralblatt. 1905;25:293–604. [Google Scholar]
- 2.Archibald JM. Genomic perspectives on the birth and spread of plastids. Proc Natl Acad Sci U S A. 2015;112(33):10147–10153. doi: 10.1073/pnas.1421374112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green BR. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66(1):34–44. doi: 10.1111/j.1365-313X.2011.04541.x. [DOI] [PubMed] [Google Scholar]
- 4.Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegawa M, Kowallik KV. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393(6681):162. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- 5.Wicke S, Schneeweiss GM, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011;76(3–5):273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravi V, Khurana J, Tyagi A, Khurana P. An update on chloroplast genomes. Plant Syst Evol. 2008;271(1–2):101–122. [Google Scholar]
- 7.Sugiura M. The chloroplast genome. Plant Mol Biol. 1992;19(1):149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- 8.Gitzendanner MA, Soltis PS, Wong GK, Ruhfel BR, Soltis DE. Plastid phylogenomic analysis of green plants: a billion years of evolutionary history. Am J Bot. 2018;105(3):291–301. doi: 10.1002/ajb2.1048. [DOI] [PubMed] [Google Scholar]
- 9.Lam VKY, Darby H, Merckx V, Lim G, Yukawa T, Neubig KM, et al. Phylogenomic inference in extremis: a case study with mycoheterotroph plastomes. Am J Bot. 2018;105(3):480–494. doi: 10.1002/ajb2.1070. [DOI] [PubMed] [Google Scholar]
- 10.Feng YL, Wicke S, Li JW, Han Y, Lin CS, Li DZ, et al. Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol Evol. 2016;8(7):2164–2175. doi: 10.1093/gbe/evw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wicke S, Müller KF, Quandt D, Bellot S, Schneeweiss GM. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc Natl Acad Sci U S A. 2016;113(32):9045–9050. doi: 10.1073/pnas.1607576113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu S, Sablok G, Wang B, Qu D, Barbaro E, Viola R, et al. Plastome organization and evolution of chloroplast genes in Cardamine species adapted to contrasting habitats. BMC Genomics. 2015;16(1):306. doi: 10.1186/s12864-015-1498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett CF, Freudenstein JV, Li J, Mayfield-Jones DR, Perez L, Pires JC, et al. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 2014;31(12):3095–3112. doi: 10.1093/molbev/msu252. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Liu B, Cheng F, Ramchiary N, Choi S-R, Lim YP, et al. Sequencing of chloroplast genome using whole cellular DNA and Solexa sequencing technology. Front Plant Sci. 2012;3:243. doi: 10.3389/fpls.2012.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett CF, Wicke S, Sass C. Dense infraspecific sampling reveals rapid and independent trajectories of plastome degradation in a heterotrophic orchid complex. New Phytol. 2018;218(3):1192–204. doi: 10.1111/nph.15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seidenfaden G. Orchid genera in Thailand: 14. Fifty-nine vandoid genera. Op Bot. 1988;95:1–398. [Google Scholar]
- 17.Christenson E. Two new species of Holcoglossum Schltr.(Orchidaceae: Aeridinae) from China. Lindleyana. 1998;13(2):121–124. [Google Scholar]
- 18.Jin XH, Qin HN, Chen SC. A new species of Holcoglossum (Orchidaceae: Aeridinae) from China. Kew Bull. 2004;59:633–5. [Google Scholar]
- 19.Jin XH, Chen SC, Qin HN, Zhu GH, Laiping GS. A new species of Holcoglossum (Orchidaceae) from China. Novon. 2004;14(2):178–179. [Google Scholar]
- 20.JIN XH. Generic delimitation and a new infrageneric system in the genus Holcoglossum (Orchidaceae: Aeridinae) Bot J Linean Soc. 2005;149(4):465–468. [Google Scholar]
- 21.Jin XH, Chen SC, Li DZ. Holcoglossum nujiangense (Orchidaceae: Aeridinae)–a new species and its pollination system. Nord J Bot. 2007;25(1–2):125–128. [Google Scholar]
- 22.Jin XH, Zhang T, Gu ZJ, Li DZ. Cytological studies on the genus Holcoglossum (Orchidaceae) Bot J Linean Soc. 2007;154(2):283–288. [Google Scholar]
- 23.Fan J, Qin HN, Li DZ, Jin XH. Molecular phylogeny and biogeography of Holcoglossum (Orchidaceae: Aeridinae) based on nuclear ITS, and chloroplast trnL-F and matK. Taxon. 2009;58(3):849–861. [Google Scholar]
- 24.Xiang XG, Li DZ, Jin XH, Hu H, Zhou HL, Jin WT, et al. Monophyly or paraphyly–the taxonomy of Holcoglossum (Aeridinae: Orchidaceae) PLoS One. 2012;7(12):e52050. doi: 10.1371/journal.pone.0052050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aver’janov LV, Averyanova AL. Updated checklist of the orchids of Vietnam. Hanoi: Vietnam National University Publishing House; 2003. [Google Scholar]
- 26.Fan J, He R, Zhang Y, Jin X. Systematic significance of leaf epidermal features in Holcoglossum (Orchidaceae) PLoS One. 2014;9(7):e101557. doi: 10.1371/journal.pone.0101557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Wang S, Jing Y, Wang L, Zhou SL. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 2013;48(1):72–78. [Google Scholar]
- 28.Patel RK, Jain M. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7(2):e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2010;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 31.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32(suppl_2):W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20(17):3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 33.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller MA, Pfeiffer W, Schwartz T. Gateway Computing Environments Workshop (GCE), 2010. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; pp. 1–8. [Google Scholar]
- 38.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32(suppl_2):W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34(12):3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 40.Phobos 3.3.11 [http://www.rub.de/ecoevo/cm/cm_phobos.htm].
- 41.Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 42.Fletcher W, Yang ZH. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol. 2010;27(10):2257–2267. doi: 10.1093/molbev/msq115. [DOI] [PubMed] [Google Scholar]
- 43.Zhang JZ, Nielsen R, Yang ZH. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22(12):2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 44.Yang JB, Tang M, Li HT, Zhang ZR, Li DZ. Complete chloroplast genome of the genus cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol Biol. 2013;13(1):84. doi: 10.1186/1471-2148-13-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niu ZT, Zhu SY, Pan JJ, Li LD, Sun J, Ding XY. Comparative analysis of dendrobium plastomes and utility of plastomic mutational hotspots. Sci Rep. 2017;7(1):2073. doi: 10.1038/s41598-017-02252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Long HX, Zhang L, Liu ZM, Cao HP, Shi MW, et al. The complete chloroplast genome sequence of tung tree (Vernicia fordii): organization and phylogenetic relationships with other angiosperms. Sci Rep. 2017;7(1):1869. doi: 10.1038/s41598-017-02076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu ZT, Pan JJ, Zhu SY, Li LD, Xue QY, Liu W, et al. Comparative analysis of the complete Plastomes of Apostasia wallichii and Neuwiedia singapureana (Apostasioideae) reveals different evolutionary dynamics of IR/SSC boundary among photosynthetic orchids. Front Plant Sci. 2017;8:1713. doi: 10.3389/fpls.2017.01713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amiryousefi A, Hyvönen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34(17):3030–1. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 49.Shinozaki K, Hayashida N, Sugiura M. Molecular Biology of Photosynthesis. Dordrecht: Springer; 1988. Nicotiana chloroplast genes for components of the photosynthetic apparatus; pp. 7–31. [DOI] [PubMed] [Google Scholar]
- 50.Kim KJ, Lee HL. Widespread occurrence of small inversions in the chloroplast genomes of land plants. Mol Cells. 2005;19(1):104–13. [PubMed] [Google Scholar]
- 51.Lei W, Ni D, Wang Y, Shao J, Wang X, Yang D, et al. Intraspecific and heteroplasmic variations, gene losses and inversions in the chloroplast genome of Astragalus membranaceus. Sci Rep. 2016;6:21669. doi: 10.1038/srep21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Massouh A, Schubert J, Yaneva-Roder L, Ulbricht-Jones ES, Zupok A, Johnson MT, et al. Spontaneous chloroplast mutants mostly occur by replication slippage and show a biased pattern in the Plastome of Oenothera. Plant Cell. 2016;28(4):911–929. doi: 10.1105/tpc.15.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jernigan KK, Bordenstein SR. Tandem-repeat protein domains across the tree of life. PeerJ. 2015;3:e732. doi: 10.7717/peerj.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S, et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet. 2016;48(1):22. doi: 10.1038/ng.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hannan AJ. Tandem repeats mediating genetic plasticity in health and disease. Nature Reviews Genetics. 2018;19(5):286–98. doi: 10.1038/nrg.2017.115. [DOI] [PubMed] [Google Scholar]
- 56.Zhao X, Su L, Schaack S, Sadd BM, Sun C. Tandem repeats contribute to coding sequence variation in bumblebees (hymenoptera: Apidae) Genome Biol Evol. 2018;10(12):3176–3187. doi: 10.1093/gbe/evy244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duchene D, Bromham L. Rates of molecular evolution and diversification in plants: chloroplast substitution rates correlate with species-richness in the Proteaceae. BMC Evol Biol. 2013;13(1):65. doi: 10.1186/1471-2148-13-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wicke S, Schäferhoff B, de Pamphilis CW, Müller KF. Disproportional plastome-wide increase of substitution rates and relaxed purifying selection in genes of carnivorous Lentibulariaceae. Mol Biol Evol. 2013;31(3):529–545. doi: 10.1093/molbev/mst261. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Z, An M, Miao J, Gu Z, Liu C, Zhong B. The Antarctic sea ice alga Chlamydomonas sp. ICE-L provides insights into adaptive patterns of chloroplast evolution. BMC Plant Biol. 2018;18(1):53. doi: 10.1186/s12870-018-1273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dolhi JM, Maxwell DP, Morgan-Kiss RM. The Antarctic Chlamydomonas raudensis: an emerging model for cold adaptation of photosynthesis. Extremophiles. 2013;17(5):711–722. doi: 10.1007/s00792-013-0571-3. [DOI] [PubMed] [Google Scholar]
- 61.Hirooka S, Hirose Y, Kanesaki Y, Higuchi S, Fujiwara T, Onuma R, et al. Acidophilic green algal genome provides insights into adaptation to an acidic environment. Proc Natl Acad Sci U S A. 2017;114(39):E8304–E8313. doi: 10.1073/pnas.1707072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peltier G, Aro E-M, Shikanai T. NDH-1 and NDH-2 plastoquinone reductases in oxygenic photosynthesis. Annu Rev Plant Biol. 2016;67:55–80. doi: 10.1146/annurev-arplant-043014-114752. [DOI] [PubMed] [Google Scholar]
- 63.Yamori W, Shikanai T. Physiological functions of cyclic electron transport around photosystem I in sustaining photosynthesis and plant growth. Annu Rev Plant Biol. 2016;67:81–106. doi: 10.1146/annurev-arplant-043015-112002. [DOI] [PubMed] [Google Scholar]
- 64.Graham SW, Lam VK, Merckx VS. Plastomes on the edge: the evolutionary breakdown of mycoheterotroph plastid genomes. New Phytol. 2017;214(1):48–55. doi: 10.1111/nph.14398. [DOI] [PubMed] [Google Scholar]
- 65.Niu ZT, Xue QY, Zhu SY, Sun J, Liu W, Ding XY. The complete Plastome sequences of four orchid species: insights into the evolution of the Orchidaceae and the utility of Plastomic mutational hotspots. Front Plant Sci. 2017;8:715. doi: 10.3389/fpls.2017.00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wakasugi T, Tsudzuki J, Ito S, Nakashima K, Tsudzuki T, Sugiura M. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc Natl Acad Sci U S A. 1994;91(21):9794–9798. doi: 10.1073/pnas.91.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang CC, Lin HC, Lin IP, Chow TY, Chen HH, Chen WH, et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 2005;23(2):279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- 68.Wu FH, Chan MT, Liao DC, Hsu CT, Lee YW, Daniell H, et al. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 2010;10(1):68. doi: 10.1186/1471-2229-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bellot S, Renner SS. The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biol Evol. 2015;8(1):189–201. doi: 10.1093/gbe/evv251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin CS, Chen JJ, Huang YT, Chan MT, Daniell H, Chang WJ, et al. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci Rep. 2015;5:9040. doi: 10.1038/srep09040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim HT, Chase MW. Independent degradation in genes of the plastid ndh gene family in species of the orchid genus cymbidium (Orchidaceae; Epidendroideae) PLoS One. 2017;12(11):e0187318. doi: 10.1371/journal.pone.0187318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schünemann D, Finazzi G, et al. A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell. 2008;132(2):273–285. doi: 10.1016/j.cell.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 73.Munekage Y, Hashimoto M, Miyake C, Tomizawa KI, Endo T, Tasaka M, et al. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature. 2004;429(6991):579. doi: 10.1038/nature02598. [DOI] [PubMed] [Google Scholar]
- 74.Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T. PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell. 2002;110(3):361–371. doi: 10.1016/s0092-8674(02)00867-x. [DOI] [PubMed] [Google Scholar]
- 75.Alric J, Johnson X. Alternative electron transport pathways in photosynthesis: a confluence of regulation. Curr Opin Plant Biol. 2017;37:78–86. doi: 10.1016/j.pbi.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 76.Lin CS, Chen JJ, Chiu CC, Hsiao HC, Yang CJ, Jin XH, et al. Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 2017;90(5):994–1006. doi: 10.1111/tpj.13525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Statistic of substitution sites and ω values of Holcoglossum plastid genes. (XLSX 15 kb)
Table S1. List of genes identified in the plastid genomes of Holcoglossum. (DOCX 26 kb)
Figure S1. Maximum Likelihood phylogenetic tree of Holcoglossum based on the whole plastid genome except for one invert repeat region. Bootstrap support is indicated on the nodes. (PDF 171 kb)
Table S3. Detected positive selection sites in the plastid genes of TC clade Holcoglossum species. (DOCX 19 kb)
Figure S2. Hairpin inversion of ycf2 in Holcoglossum. (PNG 236 kb)
Figure S3. Hairpin inversion of ycf2 in Orchidaceae. (PDF 316 kb)
Figure S4. Intraspecific variation resulting from tandem repeats in H. flavenscens. (PNG 201 kb)
Figure S5. Tandem repeat annotated to the whole plastid genome (with only one invert repeat region) alignment. The brown triangles represent the tandem repeat regions. (PDF 1013 kb)
Figure S6. Aligned sequence matrix of ycf2 gene shows the duplication of tandem repeat in Holcoglossum. (JPG 727 kb)
Data Availability Statement
All annotated plastid genomes generated in this study have been submitted to NCBI with accession of MK442924 - MK442937, MK460222.