Abstract
Human and ecological hazard assessment of chemicals requires the extrapolation of toxicities measured in a small number of laboratory model species to species of concern. The Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS) tool was developed as a rapid, cost effective method to aid cross-species extrapolation of susceptibility to chemicals acting on specific protein targets through evaluation of protein structural similarities and differences. The greatest resolution for extrapolation of chemical susceptibility across species involves comparisons of individual amino acid residues at key positions involved in protein-chemical interactions. However, a lack of understanding of whether specific amino acid substitutions among species at key positions in proteins affect interaction with chemicals made manual interpretation of alignments time consuming and potentially inconsistent. Therefore, this study used in silico site-directed mutagenesis coupled with docking simulations of computational models for acetylcholinesterase (AChE) and ecdysone receptor (EcR) to investigate how specific amino acid substitutions impact protein-chemical interaction. This study found that computationally derived substitutions in identities of key amino acids caused no change in protein-chemical interaction if residues share the same side chain functional properties and have comparable molecular dimensions, while differences in these characteristics can change protein-chemical interaction. These findings were considered in the development of capabilities for automatically generated species-specific predictions of chemical susceptibility in SeqAPASS. These predictions for AChE and EcR were shown to agree with less robust SeqAPASS predictions comparing the primary sequence and functional domain sequence of proteins for more than 90 % of the investigated species, but also identified dramatic species-specific differences in chemical susceptibility that align with results from standard toxicity tests. These results provide a compelling line-of-evidence for use of SeqAPASS in deriving screening level, species-specific, susceptibility predictions across broad taxonomic groups for application to human and ecological hazard assessment.
Keywords: docking simulations, acetylcholinesterase, ecdysone receptor, sequence similarity
INTRODUCTION
Human and ecological hazard assessment of chemicals requires the extrapolation of toxicities measured in a small number of laboratory model species to species of concern. In the case of ecological hazard assessment, this could involve extrapolation to thousands of species representing taxonomic groups as diverse as mammals, fishes, invertebrates, and plants. Extrapolation of chemical toxicity data across species is a complex challenge for risk assessors because potential differences in species sensitivity can range from a few fold to more than a hundred- or thousand-fold (Cohen-Barnhouse et al 2011; Doering et al 2013; Russom et al 2014; Song et al 1997; Song et al 2016; Thomas & Janz 2015; Vardy et al 2013; Wang et al 2013). These documented differences in chemical sensitivities across species and the fact that all species of concern cannot be tested in standard toxicity tests has elevated the need for rapid, cost-effective methods for species extrapolation. To begin to address this challenge, the U.S. Environmental Protection Agency (US EPA) developed the Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS; https://seqapass.epa.gov/seqapass/) tool and made it publicly available in 2016 (LaLone et al 2016). Based on the knowledge that proteins are common targets of chemical perturbation which lead to adverse effects across species, the SeqAPASS tool was designed to rapidly and computationally predicts chemical susceptibility across phylogenetically diverse species through evaluation of protein structural similarities and differences (Figure 1) (LaLone et al 2016). Specifically, the SeqAPASS tool compares the amino acid sequence of the protein target of a chemical in a known sensitive species to the sequences of more than 95 million proteins available in the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/refseq/ accessed November 2017) protein database and calculates sequence similarity metrics that are used as a basis for inferring potential cross-species chemical susceptibility (LaLone et al 2013; 2016). Several case studies have been developed demonstrating the utility of the SeqAPASS tool in predicting cross-species chemical susceptibility to insecticides and pharmaceuticals, as well as for extrapolation of high throughput screening data (Ankley et al 2016; Fay et al 2017; LaLone et al 2013; 2016; 2017; Martinovic-Weigelt et al 2017; Russom et al 2014).
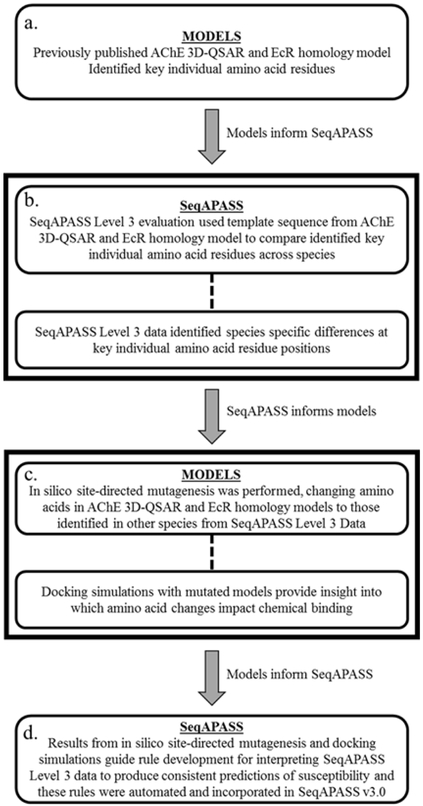
Figure 1.
Schematic of approach using in silico site-directed mutagenesis coupled with docking simulations of computational models for acetylcholinesterase (AChE) and ecdysone receptor (EcR) to investigate how specific amino acid substitutions impact protein-chemical interaction to develop automated Level 3 susceptibility predictions for incorporation into SeqAPASS v.3.0.
The SeqAPASS tool allows for the evaluation of protein targets at three levels of complexity depending on how well the protein-chemical interaction has been characterized (LaLone et al 2016). Results from each level of the SeqAPASS evaluation provide an additional line-of-evidence for predicting the likelihood of a chemical, or chemical class, to act on that same protein target in another species based on comparison to a known sensitive species (LaLone et al 2016). Briefly, Level 1 of the SeqAPASS analysis allows for cross-species comparisons of the primary amino acid sequence (including ortholog detection) (LaLone et al 2016). Level 2 provides a means to examine similarity of functional domains (such as ligand binding domains) within a protein sequence (LaLone et al 2016). With either Level 1 or Level 2 analyses, a susceptibility cut-off is automatically determined by the tool. The cut-off is based on ortholog determinations where it is assumed that orthologous proteins, which share a common genetic ancestry and diverged through a speciation event, are likely to share similar function (LaLone et al 2016). The Level 1 and 2 evaluations of sequence similarity provide broad predictions of susceptibility across taxonomic groups. For example, it is anticipated that Level 1 data might distinguish differences between vertebrate and invertebrate susceptibility and that Level 2 data might be slightly more specific in predicting susceptibilities of specified taxonomic groups. However, the Level 3 evaluation integrates knowledge of protein structure and protein-chemical interaction to allow for more precise, higher resolution susceptibility predictions across specific species.
Level 3 of the SeqAPASS tool compares the identities of individual amino acids at specific positions in a protein target that have been identified as important for chemical binding, maintaining protein conformation, transcriptional activation, or other key functions (Figure 1) (LaLone et al 2016). Increasing numbers of investigations have demonstrated the importance of identities of amino acids at key positions of a protein in determining protein interaction with chemicals. Species-, strain-, or population-specific additions, deletions, or substitutions of amino acids at key positions can alter or even abolish the interaction of the protein with certain chemicals and dramatically alter chemical sensitivity of the organism (Doering et al 2015; Farmahin et al 2012; 2013; Ffrench-Constant et al 1993; Karchner et al 2006; Liu et al 2005; Martinez-Torres 1999; Mutero et al 1994; Wirgin et al 2011). Previous published case studies using early versions (v.1.0 and v.2.0) of the SeqAPASS Level 3 analysis were conducted based on the assumption that all identified key amino acid residues must be identical across species or contain a similar side chain (e.g. acidic, aromatic) compared to the template amino acid residue to be predicted susceptible. The interpretation of Level 3 data was conducted manually by the user based on the identity of the amino acids automatically aligned with selected species in SeqAPASS (Ankley et al 2016) which makes this effort relatively time consuming and potentially inconsistent among users.
Recent advances in the capabilities and accuracy of computational docking simulations allows for rapid, cost-effective, and comprehensive investigations of protein-chemical interactions using computers (i.e. in silico). For such simulations, 3-dimensional (3-D) protein models can be built by aligning protein sequence data with available protein crystal structures bound to natural or synthetic ligands. Key amino acids can then be identified by computationally docking (i.e. binding) a variety of ligands to identify chemical properties and protein conformations necessary for proper binding. To further probe the essentiality of key amino acids in protein-chemical interactions, computer generated or in silico site-directed mutagenesis (i.e. specific and intentional changes to the amino acid residues at key positions in computer models of a protein) can be used to simulate substitutions in identities of key amino acid residues with subsequent docking simulations (Dow et al 2016). The present study utilized in silico site-directed mutagenesis with docking simulations as an initial step towards developing consistent rules for interpreting SeqAPASS Level 3 individual amino acid residue alignments across species.
Development of consistent rules for SeqAPASS Level 3 predictions was achieved through investigation of two case studies that focused on two different classes of pesticides with well characterized target proteins for which in silico models currently exist, namely inhibitors of acetylcholinesterase (AChE) and agonists of the ecdysone receptor (EcR) (Evenseth 2014; Lee & Barron 2015; 2016). Acetylcholinesterase is a key enzyme involved in neurotransmission in animals and functions in the termination of synaptic transmission at the synapse through the rapid hydrolysis of the neurotransmitter, acetylcholine (Russom et al 2014). Inhibition of AChE can result in accumulation of acetylcholine in synapses causing an excitatory response in muscle and brain leading to neurotoxic symptomology causing death (Russom et al 2014). Several pesticides have been designed to inhibit AChE, including organophosphates and carbamates. These compounds can inhibit AChE resulting in mortality among a wide range of invertebrate and vertebrate species which has resulted in their use as insecticides (insects), nematocides (nematodes), acaricides (ticks and mites), rodenticides (rodents), and avicides (birds) (Glaser 1999). However, this broad susceptibility among animals can result in toxicities to a wide range of non-target species (Beketov et al 2008; Mineau 2002; Reinecke & Reinecke 2007; Webber et al 2010). In contrast to the broad taxonomic susceptibility to AChE inhibitors, the EcR mediates transcriptional regulation of molting through activation by the hormone 20-hydroxyecdysone (20E) in a process that is unique to arthropods (Song et al 2017). Molting is the process in arthropods of generating a new exoskeleton and shedding the old exoskeleton, which is required for growth and development (Song et al 2017). The EcR has a key role in invertebrate endocrine regulation of molting and has been shown to be well conserved among all arthropods, which includes the Hexapoda (insects and springtails), Crustacea (shrimps, crabs, etc.), Myriapoda (centipedes, millipedes), and Chelicerata (spiders, scorpions, etc.) (Fay et al 2017; Song et al 2017). Insecticides that act as agonists of the EcR can cause premature molting disruption that leads to mortality (Song et al 2017). Therefore, certain chemicals that disrupt EcR have been developed for use as arthropod pest-specific pesticides, including RH-5849, tebufenozide, and methoxyfenozide.
The specific objectives of this study were to utilize in silico site-directed mutagenesis of existing computational models for AChE and EcR proteins and docking simulations to investigate how substitutions in identities of amino acids affect protein-chemical interactions. This knowledge could then be utilized to develop general rules for automating the SeqAPASS Level 3 evaluation to predict chemical susceptibility and incorporated into SeqAPASS v.3.0 to improve the utility of the tool for application to human and ecological screening-level hazard assessment.
MATERIALS AND METHODS
Case Study: Acetylcholinesterase inhibition
Acetylcholinesterase 3D-QSAR informs SeqAPASS (Figure 1A)
A 3-D quantitative structure activity relationship (3D-QSAR) model for AChE inhibitors was previously constructed and validated for quantitative understanding of protein-chemical interactions (Lee & Barron 2015; 2016). Briefly, protein-chemical complex structures from molecular docking were mapped onto a structure-based pharmacophore and transformed into a 3D-fingerprint descriptor encoding key protein-chemical interactions to evaluate the structural requirements responsible for chemical activity (Lee & Barron 2015; 2016). Four native and optimized AChE structures of two different species (Mus musculus and Torpedo californica) were used for molecular docking (Lee & Barron 2015). Validation of selectivity of these protein models had been performed through in silico docking simulations of a training set of 63 compounds and an external validation set of 26 compounds (Lee & Barron 2015). Ten key amino acid residue positions were selected for evaluation in SeqAPASS Level 3 based on their importance in the 3D-QSAR model for AChE, namely tryptophan 84, glycine 118, glycine 119, serine 200, alanine 201, phenylalanine 288, glutamic acid 327, phenylalanine 330, phenylalanine 331, and histidine 440.
SeqAPASS analyses to inform in silico site-directed mutagenesis (Figure 1B)
Predictions of cross-species chemical susceptibility for AChE were determined using the SeqAPASS tool (LaLone et al 2016). House mouse (Mus musculus) AChE (NCBI accession: NP_033729.1) was used as the SeqAPASS Level 1 query sequence as it aligned with the crystal structure employed in developing the AChE 3D-QSAR model (Lee & Barron 2015; 2016). Level 2 functional domain analysis was conducted in SeqAPASS using the esterase_lipase domain of AChE (NCBI conserved domain accession: cd00312) which contains the catalytic triad described as a key functional component of the enzyme (Lee & Barron 2016). The template sequence selected for alignments in the Level 3 evaluation was also the house mouse AChE (NCBI accession: NP_033729.1). Select species from taxonomic groups identified as susceptible (i.e. those taxonomic groups above the susceptibility cut-off) along with a few species from taxonomic groups predicted less likely to be susceptible (i.e. first four taxonomic groups below the susceptibly cut-off) in the Level 1 and 2 evaluations were aligned with the template sequence in Level 3. From SeqAPASS data, identification of differences in individual amino acids at the ten key amino acid residue positions selected in the 3D-QSAR model for AChE were then used to inform in silico site-directed mutagenesis.
In silico site-directed mutagenesis and docking simulations (Figure 1C)
In silico site-directed mutagenesis and docking simulations were performed with the 3D-QSAR model for AChE. In silico site-directed mutations were performed by use of the “build model” function in the Internal Coordinate Mechanics (ICM) software package v.3.7.3c (Abagyan et al 1994) by changing selected amino acids at positions in the active site of the models of the AChE to the amino acid in the corresponding position of the AChE of other species identified by SeqAPASS. External validation of the in silico site-directed mutagenesis process was performed with six mutations for ten organophosphates (Figure S1) and ten carbamates (Figure S2) and validated against previously published experimental information (Table S1; Table S2). For the docking simulations, binding affinity of the inhibitors with the mutant or wild-type protein structures was estimated as the sum of contributions of each protein-chemical interaction predicted by the 3D-QSAR model.
Case Study: Ecdysone receptor activation
Ecdysone receptor homology model informs SeqAPASS (Figure 1A)
A homology model for EcR was previously constructed and validated for quantitative understanding of protein-chemical interactions (Evenseth 2014). Briefly, this model was derived based on a multiple sequence alignment between the ligand binding domain (LBD) of the EcR of common water flea and the top eight most similar structures found in the protein data bank (PDB; http://www.rcsb.org/pbd/home/home.do). The crystal structure of the EcR of tobacco budworm (Heliothis virescens; PBD ID 2R40_D) was used as a template for the homology model (Evenseth 2014). The homology model was constructed by use of the “build model” function in the ICM software package (Abagyan et al 1994). This model had been evaluated for stereochemical and geometric quality in the Structural Analysis and Verification Server (SAVES) by use of PROCHECK (Laskowski et al 1993), ERRAT (Colovos & Yeates 1993), and VERTIFY_3D (Bowie et al 1991; Luthy et al 1992). Validation of selectivity of this model had been performed through in silico docking simulations of a training set of 9 known agonists and 155 decoys (Evenseth 2014). The evaluation of the stereochemical and geometrical quality indicated a structurally sound homology model, while the docking simulations indicated that the model had the ability to separate known binders from decoys. Four key amino acid residue positions were selected for evaluation in SeqAPASS Level 3 based on their importance in the homology model for EcR, namely aspartic acid 384, threonine 415, alanine 470, and asparagine 573.
SeqAPASS analyses to inform site-directed mutagenesis (Figure 1B)
Predictions of cross-species chemical susceptibility for EcR were determined using the SeqAPASS tool (LaLone et al 2016). Common water flea (Daphnia magna) EcR (NCBI accession: BAF49029.1) was used as the SeqAPASS Level 1 query sequence because it aligned with the crystal structures used to develop the EcR homology model (Evenseth 2014). Level 2 SeqAPASS analysis was conducted on the LBD of EcR which directly interacts with agonists (NCBI conserved domain accession: cd06938). The template protein sequence selected for the Level 3 individual amino acid residue alignments was also the common water flea EcR (NCBI accession: BAF49029.1). Select species from taxonomic groups identified as susceptible along with a few species from taxonomic groups predicted less likely susceptible in the Level 1 and 2 evaluations were aligned with the template sequence in Level 3. From SeqAPASS data, identification of differences in individual amino acids at the four key amino acid residue positions selected in the homology model for EcR were then used to inform in silico site-directed mutagenesis.
In silico site-directed mutagenesis and docking simulations (Figure 1C)
In silico site-directed mutagenesis docking simulations were performed with the homology model of common water flea EcR (Evenseth 2014). In silico site-directed mutagenesis and docking simulations were performed with 20-Hydroxyecdysone (20E), the natural ligand, docked at the active site. The molecular structure of 20E was downloaded from the ChEMBL database (Harada et al 2009). Docking of 20E was performed with the ICM-software (Abagyan et al 1994). Homology model mutations were performed by use of the “build model” function in the ICM software package (Abagyan et al 1994) by changing amino acids at the four selected amino acid positions in the homology model to the desired amino acid based on SeqAPASS data. After the mutation, structural refinements were performed to all amino acids within 5 A of 20E by use of the “refineModel” function in the ICM software package (Abagyan et al 1994). For docking simulations, binding energy of 20E interacting with the computationally manipulated protein models were scored by use of the “Virtual Ligand Screening” function in the ICM software package for evaluation of properties of amino acids that allowed binding (Abagyan et al 1994).
Development of automated Level 3 susceptibility predictions for SeqAPASS v.3.0 (Figure 1D)
The assumptions used for SeqAPASS Level 3 evaluations from earlier versions of the tool relied on the user’s interpretation of the data to predict susceptibility between the query species and other species of interest. This analysis was expanded through consistent comparison of side chain functional properties and molecular dimension of amino acid residues between the query species and other species of interest based on two case studies using in silico site-directed mutagenesis and docking simulations. For comparisons of side chain functional properties, each of the twenty common amino acids were grouped by their standard side chain class as 1) acidic, 2) aliphatic, 3) amidic, 4) aromatic, 5) basic, 6) hydroxylic, or 7) sulfur-containing (Table 1). For comparison of molecular dimensions, standard molar mass in grams per mol (g/mol) were assigned to each of the twenty common amino acids and which is used as a surrogate for molecular dimensions as amino acids with larger side chains have greater molar mass relative to amino acids with smaller side chains (Table 1). Based on these two amino acid classification schemes, the SeqAPASS v.3.0 tool was developed to automatically compare the identity of amino acids for each selected species at selected protein sequence positions against the template species sequence for common side chain classification and molar mass. Therefore, species that have key amino acid residues that share the same side chain classification and/or have a molar mass within an absolute value of 30 g/mol based on results obtained in this study for incidence of steric hindrance (Table 2; Table 3) are identified as similar and predicted to share susceptibility to chemicals with the template species (Figure 1). Species that have one or more key amino acid residues that do not share the same side chain classification and have a difference in molar mass of 30 g/mol or greater relative to the template sequence are predicted as less likely of sharing susceptibility to chemicals with the template species (Figure 1). Requiring one or more key amino acids to share neither the same side chain classification or molar mass as the template species was used for determination of chemical susceptibility in order to produce conservative predictions, as dramatic differences among amino acid residues are more likely to change the protein-chemical interaction relative to minor differences. Based on the summation of predictions for all of the identified key amino acid residues, an overall chemical susceptibility prediction of “yes” or “no” is generated for each aligned species (Figure 1). The improved Level 3 analysis has been incorporated into SeqAPASS v.3.0 which is available online and includes a user guide on performing this analysis in the tool (https://seqapass.epa.gov/seqapass/).
Table 1.
Amino acid classification system.
| Amino Acid | 1-Letter | Side Chain Class | Molecular Weight (g/mol) |
|---|---|---|---|
| Aspartic Acid | D | Acidic | 133.104 |
| Glutamic Acid | E | Acidic | 147.131 |
| Alanine | A | Aliphatic | 89.094 |
| Glycine | G | Aliphatic | 75.067 |
| Isoleucine | I | Aliphatic | 131.175 |
| Leucine | L | Aliphatic | 131.175 |
| Proline | P | Aliphatic | 115.132 |
| Valine | V | Aliphatic | 117.148 |
| Asparagine | N | Amidic | 132.119 |
| Glutamine | Q | Amidic | 146.146 |
| Phenylalanine | F | Aromatic | 165.192 |
| Tryptophan | W | Aromatic | 204.228 |
| Tyrosine | Y | Aromatic | 181.191 |
| Histidine | H | Basic | 155.156 |
| Lysine | K | Basic | 146.189 |
| Arginine | R | Basic | 174.203 |
| Serine | S | Hydroxylic | 105.093 |
| Threonine | T | Hydroxylic | 119.119 |
| Methionine | M | Sulfur-Containing | 149.208 |
| Cysteine | C | Sulfur-Containing | 121.154 |
Table 2.
| Mutation | Organophosphate Bindingc | Carbamate Bindingc | Predicted Mechanism |
Same Side Chain Class |
Molecular Weight Difference (g/mol) |
Susceptibility Prediction |
|---|---|---|---|---|---|---|
| W117Y | No change | No change | None | Yes | 23 | Yes |
| W117A | No change | No change | None | No | 115 | Yes |
| G152A | No change | No change | None | Yes | 14 | Yes |
| G152S | Decrease | Decrease | Steric hindrance | No | 30 | No |
| A235S | No change | No change | None | No | 16 | Yes |
| F326C | No change | No change | None | No | 44 | No |
| F368A | Decrease | No change | Electrostatic | No | 76 | No |
| F368W | Decrease | No change | Electrostatic | Yes | 39 | Yes |
| F369L | Decrease | Decrease | Steric hindrance | No | 34 | No |
| F369S | Decrease | Decrease | Steric hindrance | No | 60 | No |
| F369W | Decrease | Decrease | Steric hindrance | Yes | 39 | Yes |
Raw data results are presented (Supplementary Data File).
Amino acids represented by acronyms is listed (Table 1).
Binding -log of the 50 % inhibition concentration (pIC50) of mutant model relative to binding pIC50 of wild-type model.
Table 3.
| Mutant | Binding b | Predicted Mechanism |
Same Side Chain Class |
Molecular Weight Difference (g/mol) |
Susceptibility Prediction |
|---|---|---|---|---|---|
| D506E | No change | None | Yes | 14 | Yes |
| D506P | Decrease | Electrostatic | No | 18 | Yes |
| T537A | No change | None | No | 30 | Yes |
| A592G | No change | None | Yes | 14 | Yes |
| A592F | Decrease | Steric hindrance | No | 76 | No |
| A592V | No change | None | Yes | 28 | Yes |
| N695A | Decrease | Steric hindrance | No | 43 | No |
Raw data results are presented (Supplementary Data File).
Amino acids represented by acronyms is listed (Table 1).
Binding free energy of mutant model relative to binding free energy of wild-type model.
RESULTS
Case Study: Acetylcholinesterase
SeqAPASS Level 1 analysis:
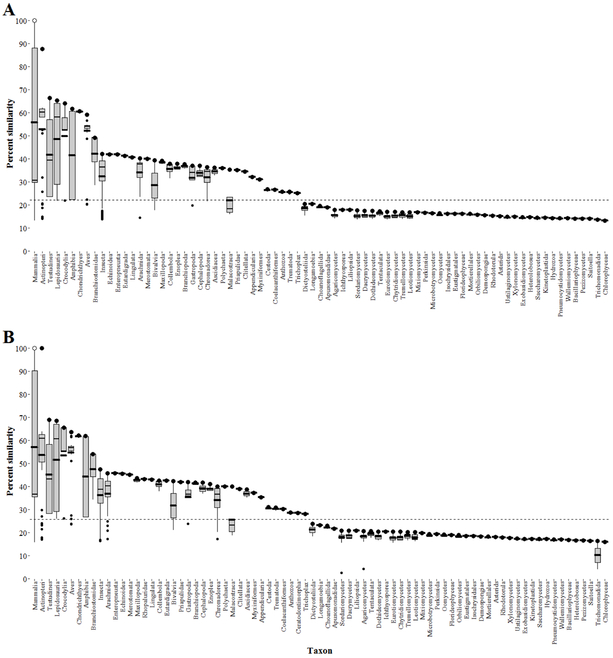
Level 1, primary amino acid sequence alignments predicted susceptibility to inhibitors of AChE among animals of the clade Bilateria, including Mammalia (mammals), Actinopteri (bony fishes), Aves (birds), Amphibia (frogs, salamanders, newts), Chondrichthyes (sharks, rays, skates, chimaeras), Arachnida (spiders, scorpions, ticks, mites), Insecta (insects), Gastropoda (snails, slugs), and Polychaeta (bristle worms), among others (Figure 2A). Further, Anthozoa (corals, sea anemones) of the clade Radiata are also predicted as likely to be susceptible to inhibitors of AChE (Figure 2A). Level 1 analysis predicted taxon represented by fungi, plants, unicellular eukaryotes, and simple multicellular animals such as Demospongiae (sea sponges), Tentaculata (comb jellies), and Hydrozoa (hydrozoans) as less likely to be susceptible to AChE inhibitors (Figure 2A).
Figure 2.
Box-plots illustrating Level 1 primary amino acid sequence similarity (A) and Level 2 functional domain sequence similarity (B) to the house mouse (Mus musculus) acetylcholinesterase (AChE) by use of the SeqAPASS tool. The open dot represents the house mouse AChE and the filled black dots represent the individual species within the specified taxonomic group with the greatest percent similarity. Within a given taxonomic box, the thick and thin horizontal solid lines represent the mean and median percent similarity, respectively. The dashed horizontal line represents the susceptibility cut-off. Primary amino acid sequence similarity (A) used a susceptibility cut-off of 22.1 %. Functional domain sequence similarity used a susceptibility cut-off of 25.7 % (B).
SeqAPASS Level 2 analysis:
Level 2 SeqAPASS analysis predicted likelihood for susceptibility to inhibitors of AChE among taxonomic groups comparable to those predicted by the Level 1 analysis (Figure 2B). Level 2 analysis predicted taxon represented by fungi, plants, unicellular eukaryotes, and simple multicellular animals as less likely of being susceptible, in common with results of Level 1 analysis (Figure 2B).
SeqAPASS Level 3 analysis:
Ten amino acid positions associated with house mouse AChE, which were identified in 3D crystal structures as contributing to the reactive site, were selected for investigation (Lee et al 2015; 2016). Level 3 analysis in SeqAPASS indicates significant conservation in identities of amino acids at these ten positions among phylogenetically diverse species that were predicted to be susceptible in Levels 1 and 2 of the analyses (Table 4; Supplementary Data File). All investigated species that were predicted to be susceptible shared amino acid identities at the equivalent positions to glycine 152, serine 234, glutamic acid 365, and histidine 478 of house mouse (Table 4; Supplementary Data File). However, amino acid substitutions were detected at six of the ten investigated positions (117, 153, 235, 326, 368, and 369) among some species that were predicted susceptible in Levels 1 and 2 (Table 4; Supplementary Data File). House mouse AChE has tryptophan 117 in common with all species except for Farrer’s scallop (Bivalvia) which has tyrosine (Table 4; Supplementary Data File). Additionally, house mouse AChE has glycine 153 in common with numerous species (Table 4; Supplementary Data File). However, other species have serine at the corresponding position, including species of Insecta (insects), Arachnida (spiders, scorpions, ticks, mites), Bivalvia (clams, oysters, cockles, mussels, scallops), Ascidiacea (sea squirts), and Trematoda (flukes) (Table 4; Supplementary Data File). One species, the common octopus (Cephalopoda), has alanine at the corresponding position (Table 4). Furthermore, house mouse AChE has alanine 235 in common with most species (Table 4; Supplementary Data File). However, some species of Insecta (insects), Bivalvia (clams, oysters, cockles, mussels, scallops), and Enoplea (nematodes) have serine at the equivalent position (Table 4; Supplementary Data File). One species, the mygalomorph spider (Arachnida), has valine at the corresponding position (Table 4). House mouse AChE has phenylalanine 326 in common with all investigated species of the phylum Chordata as well as the Cestoda (tapeworms) and Trematoda (flukes) (Table 4; Supplementary Data File). However, invertebrates have variable amino acid identities that align, including substitutions with cysteine, valine, leucine, isoleucine, or tyrosine (Table 4; Supplementary Data File). House mouse AChE has tyrosine 368 (Table 4). Again, variability in amino acid identities exists at the equivalent position among species from most taxonomic groups, including substitutions with phenylalanine, serine, tryptophan, alanine, isoleucine, or valine (Table 4; Supplementary Data File). Finally, house mouse AChE has phenylalanine 369 in common with most other species (Table 4; Supplementary Data File). However, several species have tryptophan at the equivalent position, including mosquitos (Insecta), red spider mite (Arachnida), Atlantic horseshoe crab (Merostomata), roundworms (Chromadorea), and tapeworms (Cestoda) (Table 4; Supplementary Data File). In addition, several aphid spp. (Insecta) have serine, while the mygalomorph spider (Arachnida) has leucine, and the Farrer’s scallop (Bivalvia) has threonine (Table 4; Supplementary Data File).
Table 4.
Level 3 comparison of key amino acid residues in house mouse (Mus musculus) acetylcholinesterase (AChE) relative to other species.a, b
| Class Name | Scientific Name | Common Name | W117 | G152 | G153 | S234 | A235 | F326 | E365 | Y368 | F369 | H478 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mammalia | Mus musculus | house mouse | W | G | G | S | A | F | E | Y | F | H |
| Mammalia | Homo sapiens | human | W | G | G | S | A | F | E | Y | F | H |
| Actinopteri | Electrophorus electricus | electric eel | W | G | G | S | A | F | E | Y | F | H |
| Lepidosauria | Echis coloratus | vipers | W | G | G | S | A | F | E | Y | F | H |
| Aves | Pseudopodoces humilis | Tibetan ground-tit | W | G | G | S | A | F | E | Y | F | H |
| Chondrichthyes | Torpedo californica | Pacific electric ray | W | G | G | S | A | F | E | F | F | H |
| Amphibia | Xenopus tropicalis | western clawed frog | W | G | G | S | A | F | E | Y | F | H |
| Branchiostomidae | Branchiostoma belcheri | Belcher’s lancelet | W | G | G | S | A | F | E | F | F | H |
| Insecta | Aedes aegypti | yellow fever mosquito | W | G | G | S | A | C | E | Y | F | H |
| Insecta | Phlebotomus papatasi | sandflies | W | G | G | S | A | C | E | Y | F | H |
| Insecta | Culex tritaeniorhynchus | mosquitos | W | G | G | S | A | C | E | Y | W | H |
| Insecta | Rhopalosiphum padi | bird cherry-oat aphid | W | G | G | S | A | C | E | Y | S | H |
| Insecta | Myzus persicae | green peach aphid | W | G | G | S | A | C | E | Y | S | H |
| Insecta | Operophtera brumata | winter moth | W | G | G | S | A | C | E | Y | F | H |
| Insecta | Culex quinquefasciatus | southern house mosquito | W | G | S | S | S | C | E | Y | F | H |
| Arachnida | Trittame loki | mygalomorph spiders | W | G | G | S | V | I | E | A | L | H |
| Arachnida | Oligonychus coffeae | spider mites | W | G | S | S | A | V | E | Y | F | H |
| Arachnida | Rhipicephalus microplus | southern cattle tick | W | G | G | S | A | V | E | W | F | H |
| Enteropneusta | Saccoglossus kowalevskii | acorn worms | W | G | G | S | A | Y | E | F | F | H |
| Merostomata | Limulus polyphemus | Atlantic horseshoe crab | W | G | G | S | A | I | E | Y | W | H |
| Maxillopoda | Tigriopus japonicus | copepods | W | G | G | S | A | C | E | F | F | H |
| Bivalvia | Crassostrea gigas | Pacific oyster | W | G | S | S | S | L | E | Y | F | H |
| Bivalvia | Azumapecten farreri | Farrer’s scallop | Y | G | S | S | A | F | E | I | T | H |
| Gastropoda | Ambigolimax valentianus | land snails | W | G | G | S | A | V | E | Y | F | H |
| Cephalopoda | Octopus vulgaris | common octopus | W | G | A | S | A | L | E | Y | F | H |
| Enoplea | Trichuris trichiura | human whipworm | W | G | G | S | S | L | E | Y | F | H |
| Chromadorea | Ascaris suum | pig roundworm | W | G | G | S | A | L | E | F | W | H |
| Ascidiacea | Ciona intestinalis | vase tunicate | W | G | S | S | A | F | E | Y | F | H |
| Cestoda | Echinococcus multilocularis | tapeworms | W | G | G | S | A | F | E | Y | W | H |
| Anthozoa | Exaiptasia pallida | sea anemones | - | G | G | S | A | - | E | V | F | H |
Listing of all species investigated in Level 3 is provided (Supplementary Data File).
Amino acids represented by acronyms is listed (Table 1).
AChE in silico site-directed mutagenesis and docking simulations:
The six positions with differences in identities of amino acids among species (117, 153, 235, 326, 368, and 369) were investigated by use of in silico site-directed mutagenesis and docking simulations. However, all amino acid substitutions at these positions could not be pragmatically evaluated due to the required computational effort. Therefore, certain amino acid substitutions were selected based on incidence among species and those common to key taxonomic groups. No effect on in silico docking of organophosphates or carbamates was detected as a result of substitutions of tryptophan for tyrosine at position 117, glycine for alanine at position 153, alanine for serine at position 235, or phenylalanine for cysteine at position 326 (Table 2). Docking of organophosphates and carbamates to AChE was decreased in silico when glycine was substituted for serine at position 153 and phenylalanine for leucine, serine, or tryptophan at position 369 (Table 2). Decreased in silico docking of organophosphates, but not carbamates, was identified when phenylalanine was substituted for alanine or tryptophan at position 368 (Table 2). Among the amino acids substitutions that were investigated, complete abolishment of in silico docking was only observed as a result of substitution of glycine for serine at position 153, and only for three chemicals (Table S3; Table S4).
Application of rules for Level 3 susceptibility predictions
Of the eleven amino acid substitutions at six different positions investigated here for AChE, the automated Level 3 predictions developed here supported results of in silico site-directed mutagenesis and docking simulations for nine of eleven amino acid residue substitutions (82 %) with the other two not present in other species (Table 2). Side chain functional classification and molar mass of the key amino acid positions in AChE were compared across 376 species from 38 taxonomic groups predicted susceptible by Level 1 and 2 analyses and 11 species from four taxonomic groups were predicted as less likely to be susceptible (Figure 2; Supplementary Data File 1). Automated species-specific Level 3 predictions of chemical susceptibility developed here agree with results of Level 1 and 2 predictions for 350 of the 387 (90 %) aligned species (Supplementary Data File).
Case Study: Ecdysone Receptor
SeqAPASS Level 1 analysis:
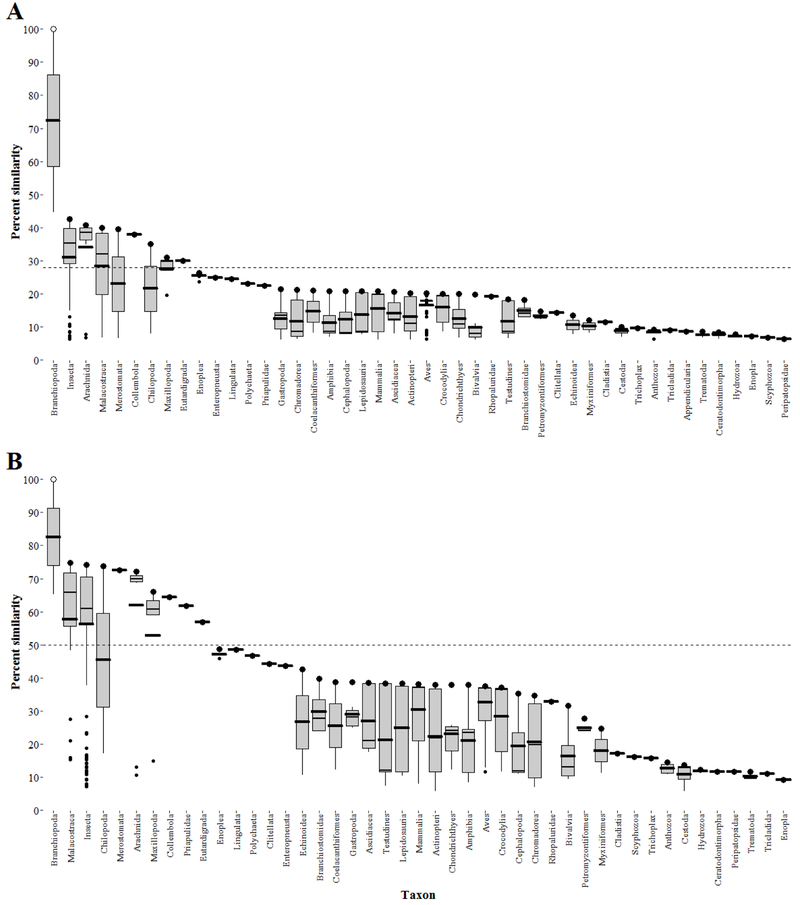
Level 1 primary amino acid alignments predicted susceptibility to agonists of the EcR among animals of the phylum Arthropoda, namely Branchiopoda (fairy shrimps, clam shrimps, water fleas, shield shrimps), Insecta (insects), Arachnida (spiders, scorpions, ticks, mites), Malacostraca (crabs, lobsters, crayfish, shrimp, krill, woodlice, amphipods, mantis shrimps), Merostomata (horseshoe crab), Collembola (springtails), Chilopoda (centipedes), and Maxillopoda (barnacles, copepods) (Figure 3A). However, Eutardigrada (water bears; phylum Tardigrada) are also predicted to be susceptible to agonists of the EcR (Figure 3A). Level 1 analysis predicted taxon represented by all vertebrates, all invertebrates not of the superphylum Ecdysozoa, and all fungi, plants, and unicellular eukaryotes as less likely to be susceptible to agonists of the EcR (Figure 3A).
Figure 3.
Box-plots illustrating Level 1 primary amino acid sequence similarity (A) and Level 2 functional domain sequence similarity (B) to the common water flea (Daphnia magna) ecdysone receptor (EcR) by use of the SeqAPASS tool. The open dot represents the water flea EcR and the filled black dots represent the individual species within the specified taxonomic group with the greatest percent similarity. Within a given taxonomic box, the thick and thin horizontal solid lines represent the mean and median percent similarity, respectively. The dashed horizontal line represents the susceptibility cut-off. Primary amino acid sequence similarity (A) used a susceptibility cut-off of 27.9 %. Functional domain sequence similarity used a susceptibility cut-off of 50.0 %.
SeqAPASS Level 2 analysis:
SeqAPASS Level 2 analysis predicted susceptibility to agonists of the EcR among taxonomic groups comparable to those predicted by the Level 1 analysis, namely arthropods (Figure 3B). However, level 2 analysis also predicted susceptibility of Priapulidae (priapulid worms; phylum Priapulida), while predicting lesser likelihood of susceptibility of Eutardigrada (water bears; phylum Tardigrada) (Figure 3B). Additionally, taxon predicted as less likely of being susceptible are again represented by all vertebrates, all invertebrates not of the superphylum Ecdysozoa, and all fungi, plants, and unicellular eukaryotes (Figure 3B).
SeqAPASS Level 3 analysis:
Seven amino acid positions were selected for investigation with SeqAPASS Level 3 individual amino acid residue comparison which were identified in 3-D crystal structures of EcR as contributing to the binding pocket (Amor et al 2012; Evenseth 2014). Level 3 demonstrates significant amino acid conservation at these seven positions among phylogenetically diverse species that were predicted to be susceptible in SeqAPASS Levels 1 and 2 analysis (Table 5). However, little conservation is present in identities of amino acids at these seven positions among species representing taxa predicted less likely to be susceptible, but which possess identified ortholog candidates for EcR (Table 5). All investigated species that were predicted to be susceptible by both Level 1 and 2 analyses shared amino acid identities at the equivalent positions to threonine 540, arginine 577, tyrosine 602, and asparagine 695 of common water flea (Table 5; Supplementary Data File). However, three of the seven investigated positions (506, 537, 592) have substitutions in identities of amino acids among some species that were predicted to be susceptible in Levels 1 and 2 (Table 5; Supplementary Data File). Common water flea EcR has aspartic acid 506 in contrast to all other investigated arthropods, with the exception of other species of common water fleas (Branchiopoda) and the squinting bush brown (Insecta) (Table 5; Supplementary Data File). The American cockroach (Insecta) has proline at the corresponding position (Table 5). All other species of arthropods have glutamic acid at the corresponding position (Table 5; Supplementary Data File). Common water flea has threonine 537 in common with all species except for opossum shrimps (Malacostraca) and coleseed sawfly (Insecta), which have serine (Table 5; Supplementary Data File). Common water flea has alanine 592 in common with most species (Table 5; Supplementary Data File). However, Malacostraca (crustaceans) have glycine, serine, or phenylalanine (Table 5; Supplementary Data File). Several species of Insecta (insects) have valine at the equivalent position (Table 5; Supplementary Data File). Priapulids (Priapulidae) have glycine at the equivalent position (Table 5).
Table 5.
Level 3 comparison of key amino acid residues in common water flea (Daphnia magna) ecdysone receptor (EcR) relative to other species. a, b
| Class Name | Scientific Name | Common Name | D506 | T537 | T540 | R577 | A592 | Y602 | N695 |
|---|---|---|---|---|---|---|---|---|---|
| Branchiopoda | Daphnia magna | common water fleas | D | T | T | R | A | Y | N |
| Branchiopoda | Daphnia pulex | common water flea | D | T | T | R | A | Y | - |
| Malacostraca | Crangon crangon | crustaceans | E | T | T | R | G | Y | N |
| Malacostraca | Portunus trituberculatus | swimming crab | E | T | T | R | G | Y | N |
| Malacostraca | Procambarus clarkii | red swamp crayfish | E | T | T | R | G | Y | N |
| Malacostraca | Scylla paramamosain | green mud crab | E | T | T | R | G | Y | N |
| Malacostraca | Neomysis integer | opossum shrimps | E | S | T | R | F | Y | N |
| Insecta | Bicyclus anynana | squinting bush brown | D | T | T | R | A | Y | N |
| Insecta | Locusta migratoria | migratory locust | E | T | T | R | A | Y | N |
| Insecta | Heliothis virescens | tobacco budworm | E | T | T | R | A | Y | N |
| Insecta | Aedes aegypti | yellow fever mosquito | E | T | T | R | A | Y | N |
| Insecta | Harmonia axyridis | ladybird beetles | E | T | T | R | V | Y | N |
| Insecta | Colaphellus bowringi | leaf beetles | E | T | T | R | V | Y | N |
| Insecta | Periplaneta americana | American cockroach | P | T | T | R | A | Y | N |
| Chilopoda | Lithobius peregrinus | centipedes | E | T | T | R | A | Y | N |
| Merostomata | Limulus polyphemus | Atlantic horseshoe crab | E | T | T | R | A | Y | N |
| Arachnida | Liocheles australasiae | scorpions | E | T | T | R | A | Y | N |
| Arachnida | Parasteatoda tepidariorum | common house spider | E | T | T | R | A | Y | N |
| Arachnida | Varroa destructor | honeybee mite | E | T | T | R | A | Y | N |
| Maxillopoda | Tigriopus japonicus | copepods | E | T | T | R | A | Y | N |
| Maxillopoda | Paracyclopina nana | copepods | E | T | T | R | A | Y | N |
| Collembola | Orchesella cincta | slender springtails | E | T | T | R | A | Y | - |
| Priapulidae | Priapulus caudatus | priapulids | E | T | T | R | G | Y | H |
| Eutardigrada | Ramazzottius varieornatus | water bears | E | T | T | R | A | Y | W |
| Enopleac | Trichuris trichiura | human whipworm | D | T | N | K | G | Y | H |
| Enopleac | Trichinella papuae | roundworms | D | T | N | K | G | Y | H |
| Lingulatac | Lingula anatina | lampshells | E | T | V | R | G | L | H |
Listing of all species investigated in Level 3 is provided (Supplementary Data).
Amino acids represented by acronyms is listed (Table 1).
Species predicted lesser likelihood of susceptibility to agonists of the EcR, but which possess identified ortholog candidates for EcR.
In silico site-directed mutagenesis and docking simulations:
From the SeqAPASS Level 3 alignment, three positions were found in the EcR with differences in identities of amino acids among species (506, 537, and 592) as well as hypothetical differences at position 695 (Supplementary Data File). Therefore, a total of four amino acid positions were investigated using in silico site-directed mutagenesis and docking simulations (Table S5). As for AChE, all amino acid substitutions could not be pragmatically evaluated due to the required computational effort. Therefore, certain amino acid substitutions were selected. No effect on in silico docking of the natural ligand 20E was detected as a result of substitutions of aspartic acid for glutamic acid at position 506, threonine for alanine at position 537, or alanine for glycine or valine at position 592 (Table 3). However, in silico docking of 20E was decreased as a result of substitution of aspartic acid for proline at position 506 and alanine for phenylalanine at position 592 (Table 3). A hypothetical substitution of asparagine for alanine at position 695 also decreased in silico docking of 20E (Table 3). Complete abolishment of in silico docking of 20E was not observed as a result of any substitution in identities of amino acids that were investigated (Table 3).
Application of rules for Level 3 susceptibility predictions
Of the seven amino acid substitutions at four different positions investigated in the EcR sequence, the automated Level 3 predictions supported results of in silico site-directed mutagenesis and docking simulations based on the presence of such substitutions across species for six of the seven (86 %) substitutions (Table 3). Side chain functional classification and molar mass of the investigated key amino acid positions in EcR were compared across 178 species from 10 taxonomic groups predicted by Level 1 and 2 analyses to be susceptible and 8 species from 5 taxonomic groups predicted as less likely to be susceptible (Figure 3; Supplementary Data File). Automated species-specific Level 3 predictions of chemical susceptibility developed here agree with the results of Level 1 susceptibility predictions for 176 of 186 (95 %) species and Level 2 predictions for 178 of 186 (96 %) of species (Supplementary Data File).
Supplementary Data
Data available from the Dryad Digital Repository: 10.5061/dryad.2tg6967.
DISCUSSION
The SeqAPASS tool was developed by the US EPA to address needs for rapid, cost effective methods for species extrapolation to understand chemical susceptibility for human and ecological hazard assessment (LaLone et al 2016). The approaches utilized by Level 1 (cross-species primary sequence comparison) and Level 2 (cross-species functional domain sequence comparison) analyses in SeqAPASS offers an automated means for inferring probable susceptibility to chemicals that act on well defined protein targets. The primary objective of this investigation utilizing in silico site-directed mutagenesis and docking simulations was to better understand the role of specific amino acid substitutions present in proteins of phylogenetically diverse species and how these substitutions affect protein-chemical interactions. The intent was then to use this information to develop a standard set of rules to advance and automate the interpretation of SeqAPASS Level 3 data for improved species-specific predictions of chemical susceptibility. The SeqAPASS Level 3 analyses initially utilized a set of assumptions for manual interpretation of data where susceptibility of the species of interest required residues from other species to have either 1) the identical amino acid residue as the template species, 2) a similar side chain as the amino acid residue in the template sequence, or 3) specific information regarding amino acid differences in the published literature (LaLone et al 2016). However, a standardized method of amino acid groupings was not defined (LaLone et al 2016). Both, SeqAPASS Level 1 and 2 analyses predicted susceptibility among animals of the clade Bilateria to inhibitors of AChE and among animals of the phylum Arthropoda to agonists of the EcR (Figure 2; Figure 3). However, numerous differences in the amino acid sequences of these and other protein targets are present among phylogenetically diverse species where point mutations could affect protein-chemical interactions and therefore susceptibility of the species. However, complete abolishment of in silico chemical docking was only observed as a result of a single investigated substitution of glycine for serine at position 153 of AChE and only for three of the ten investigated chemicals (Table 2; Supplementary Data File). No other differences in identities of amino acids were found to completely abolish binding of any investigated chemicals among the more than 1,000 different species from up to 38 different taxonomic groups investigated through in silico site-directed mutagenesis and docking simulations (Table 2; Table3; Supplementary Data File). Therefore, for both AChE and EcR, the use of in silico site-directed mutagenesis docking simulations broadly support the cross-species susceptibility predictions generated from Level 1 and Level 2 analyses in SeqAPASS.
Increasing numbers of investigations demonstrate the importance of identities of amino acids at key positions of a protein in determining interaction with chemicals. Substitutions at key positions can affect protein-chemical interactions by 1) altering electrostatic properties or reactivity or 2) through the process of steric hindrance (Lee & Baron 2016). Altered electrostatic properties or reactivity can occur when an amino acid with a particular side chain functional property (e.g. negative charge) is substituted for an amino acid with a different side chain functional property (e.g. positive charge). In a previously developed case study that focused on predicting pollinator susceptibility to neonicotinoids, which act on the nicotinic acetylcholine receptor, considerations of altered electrostatic properties or reactivity were manually accounted for when evaluating SeqAPASS Level 3 data (LaLone et al 2016). In that case study, arachnids known to be insensitive to neonicotinoids and aphids that had developed resistance to neonicotinoids contained glutamine or threonine residues (both uncharged) in a critical position where targeted pest insects and beneficial insects contained an arginine amino acid (positively charged) (LaLone et al 2016). Arginine was shown to optimally interact electrostatically with the negatively charged components of neonicotinoids, whereas glutamine or threonine did not (LaLone et al 2016). Steric hindrance can occur when an amino acid with a smaller sized side chain (or lesser molecular dimensions) is replaced by an amino acid with a larger sized side chain (or greater molecular dimensions). This substitution could change the shape of the protein or the size of the ligand binding pocket, thereby altering its interaction with chemicals. Previously, steric hindrance was manually considered when evaluating SeqAPASS Level 3 data in a case study focused on predicting susceptibility of beneficial insects to agonists of the EcR (LaLone et al 2016). In this case study, Hemiptera (cicadas, aphids, planthoppers, leafhoppers, shieldbugs) and Hymenoptera (sawflies, wasps, bees, ants) known to be insensitive to molt-accelerating compounds that target the EcR, contained an isoleucine residue in a critical position of the EcR (LaLone et al 2016). In contrast, targeted pest insects of the Lepidoptera (bufferflies, moths) contained a methionine residue (LaLone et al 2016). Specific information in the published literature demonstrate that the presence of isoleucine at this critical position, along with other amino acid substitutions, generates steric hindrance between the EcR and the molt-accelerating compound, tebufenozide (Amor et al 2012).
Six of the eleven investigated mutations for AChE and four of the seven investigated mutations for EcR caused no detectable change in docking simulations (Table 2; Table 3). For example, substitution of tryptophan (aromatic; large) at position 117 of AChE for tyrosine (also aromatic; large) or substitution of aspartic acid (acidic; large) at position 506 of EcR for glutamic acid (also acidic; large) caused no detectable change in docking (Table 2; Table 3). However, other substitutions in identities of amino acids at certain key positions resulted in changes in docking simulations. For example, substitution of phenylalanine (aromatic; larger) at position 369 of AChE for serine (hydroxylic; smaller) and substitution of alanine (aliphatic; smaller) at position 592 of EcR for phenylalanine (aromatic; larger) decreased docking (Table 2; Table 3). In one case, the substitution of glycine (aliphatic; smaller) at position 153 of AChE for serine (hydroxylic; larger) decreased or even abolished docking (Table 2; Supplementary Data File). All substitutions in identities of amino acids investigated through in silico site-directed mutagenesis and docking simulations that caused decreased docking were the result of substitutions that caused either 1) altered electrostatic properties or reactivity or 2) steric hindrance (Table 2; Table 3).
Results of the present study make three key contributions that facilitated the automated interpretation of Level 3 analyses now integrated in SeqAPASS v.3.0. 1) the most common substitutions in amino acids at key positions of proteins among species cause indiscernible changes in the ability of the protein to interact with chemicals and therefore appear to act as “silent” substitutions, 2) substitutions in amino acids that cause indiscernible changes in chemical interaction with proteins in silico share the same electrostatic properties or reactivity and comparable molecular dimensions, and 3) substitutions in amino acids that cause a change in chemical interaction with proteins in silico differ in electrostatic properties or reactivity and/or have different molecular dimensions. This allows for a simple set of rules derived from basic descriptors of same side chain functional properties and molecular dimensions to be integrated into the SeqAPASS tool. Based on the present study, in most cases application of these SeqAPASS level 3 rules will yield conclusions similar to those derived from more sophisticated, computationally intensive, and more difficult to implement in silico docking approaches, provided amino acid positions critical for function are known a priori. Therefore, this work supports the similar side chain assumption manually applied for previous Level 3 analyses, but further defined the side chain classifications and incorporated considerations of molecular dimensions in an automated fashion.
Rules for assigning susceptibility predictions from Level 3 individual amino acid residue alignments based on these three key findings were incorporated in SeqAPASS v.3.0. For comparisons of side chain functional properties and molecular dimensions, each of the twenty common amino acids were assigned to their standard side chain classification and standard molar mass, respectively (Table 1). Using these rules, the improved Level 3 analysis can automatically generate species-specific predictions of chemical susceptibility which in these two case studies agree with Level 1 and 2 predictions for more than 90 % of investigated species for AChE and EcR (Supplementary Data File). Therefore, the considerable consistency of automated predictions across Levels 1, 2 and 3 of SeqAPASS provides a compelling line-of-evidence for species-specific chemical susceptibilities across broad taxonomic groups. This line-of-evidence uses publicly available data, is quick and consistent to interpret by regulators and researchers from a wide variety of fields, and has immediate applications to human and ecological screening-level hazard assessments.
The automated Level 3 predictions of chemical susceptibility developed here identified some species-specific differences relative to predictions of Level 1 and 2 analyses. The majority of these differences involved either poor quality sequences or proteins that might not be orthologs of the query protein (Supplementary Data File). Protein sequences that are of poor quality or not homologous to the query protein cannot be interpreted with certainty for predictions of chemical susceptibility. However, some species-specific predictions of chemical susceptibility differed from Level 1 and 2 predictions but included high quality protein sequences (Supplementary Data File). A vast literature exists on dramatic species-specific differences in chemical susceptibility, including complete resistance (Karchner et al 2006; McKenzie 1996; Shaw et al 1999; Van Leeuwen et al 2010; Wirgin & Waldman 2004). Automated Level 3 analyses predicted the southern house mosquito (Insecta), Colorado potato beetle (Insecta), several aphid spp. (Insecta), and several mite spp. (Arachnida) to be less likely of being susceptible to organophosphates and carbamates (Supplementary Data File). Lack of susceptibility to organophosphates and carbamates has been demonstrated for these species in standard toxicity tests and is known to result from reduced binding affinity of AChE (Alyokhin et al 2008; Fournier 2005; Fournier & Mutero 1994; Li & Han 2004; Moores et al 1996; Naqqash et al 2016; Osta et al 2012; Zahavi & Tahori 1970). Similarly, automated Level 3 analyses predicted the opossum shrimp (Malacostraca) and slender springtail (Collembola) to be less likely of being susceptible to agonists of the EcR (Supplementary Data File). Lack of susceptibility to agonists of EcR has been demonstrated in standard toxicity tests for opossum shrimp (Malacostraca) (De Wilde et al 2013). Susceptibility of slender springtail (Collembola) has not been investigated in standard toxicity tests, but lack of susceptibility to agonists of EcR has been demonstrated in standard toxicity tests for another springtail sp. (Collembola) (Campiche et al 2006). Further, the automated Level 3 analyses can provide predictions of susceptibility for other groups within the Superphylum Ecdysozoa (animals that shed their exoskeletons), which also includes the Priapulidae (priapulid worms), Eutardigrada (water bears), and Enoplea (nematodes), among others. Level 1 of SeqAPASS predicts susceptibility of Eutardigrada (water bears), but not Enoplea (nematodes) or Priapulidae (priapulid worms), while Level 2 predicts susceptibility of Eutardigrada (water bears) and Priapulidae (priapulid worms), but not Enoplea (nematodes) (Figure 3). In contrast, Level 3 analyses based on identities of key amino acid residues predict susceptibility of Priapulidae (priapulid worms) and Enoplea (nematodes), but not Eutardigrada (water bears) (Supplementary Data File). Results of standard toxicity tests support Level 3 predictions of susceptibility for other nematodes (Secernentea), but no toxicity information is available for Enoplea (nematodes), Priapulidae (priapulid worms), or Eutardigrada (water bears) (Graham et al 2010). Therefore, the automated Level 3 analyses incorporated into SeqAPASS v.3.0 can identify dramatic species-specific differences in chemical susceptibility to inhibitors of AChE and agonists of EcR that differ from Level 1 and 2 predictions, but which align with results of standard toxicity tests. This agreement with results of standard toxicity tests supports the usefulness of Level 3 SeqAPASS predictions for these and other protein targets allowing for comparisons of numerous species from diverse taxonomic groups for which toxicity data has not been or cannot be generated, such as the Enoplea (nematodes), Priapulidae (priapulid worms), and Eutardigrada (water bears). Further, in cases where Level 1 and 2 susceptibility predictions differ, Level 3 analysis can be used as a deciding factor in order to make a susceptibility prediction. However, despite major improvements in the automation of Level 3 predictions of chemical susceptibility incorporated into SeqAPASS v.3.0, there are still challenges in evaluating the data. The analyses assume accurate identification of all key amino acid positions in a protein and that the general rules developed here are readily applicable to predicting other protein-chemical interactions. As knowledge of protein-chemical interactions expand, refinement of the rules for predicting susceptibility will continue to evolve with future versions of SeqAPASS.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Villeneuve for providing thoughtful review comments on an earlier version of the paper, J. Swintek for R software support, and T. Transue and C. Simmons for incorporating the improve Level 3 analysis into SeqAPASS v.3.0. This manuscript has been reviewed in accordance with the requirements of the U.S. EPA Office of Research and Development; however, the recommendations made herein do not represent U.S. EPA policy. Mention of products or trade names does not indicate endorsement by the U.S. EPA.
FUNDING INFORMATION
This work was supported by the U.S. Environmental Protection Agency.
REFERENCES
- Abagyan RA, Totrov MM, Kuznetsov DA (1994). ICM: A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J. Com. Chem. 15, 488–506. [Google Scholar]
- Alyokhin A, Baker M, Mota-Sanchez D, Dively G, Grafius E (2008). Colorado potato beetle resistance to insecticides. American J. Potato Res. 85(6), 395–413. [Google Scholar]
- Amor F, Christiaens O, Bengochea P, Medina P, Rouge P, Vinuela E, Smagghe G (2012). Selectivity of diaclhydrazine insecticides to the predatory bug Orius laevigatus: In vivo and modelling/docking experiments. Pest Manag. Sci. 68, 1586–1594. [DOI] [PubMed] [Google Scholar]
- Ankley GT, LaLone CA, Gray LE, Villeneuve DL, Hornung MW (2016). Evaluation of the scientific underpinnings for identifying estrogenic chemicals in nonmammalian taxa using mammalian test systems. Environ. Toxicol. Chem. 35(11), 2806–2816. [DOI] [PubMed] [Google Scholar]
- Beketov M, Schafer RB, Marwitz A, Paschke A, Liess M (2008). Longterm stream invertebrate community alterations induced by the insecticide thiacloprid: effect concentrations and recovery dynamics. Sci Total Environ. 405, 96–108. [DOI] [PubMed] [Google Scholar]
- Bowie JU, Luthy R, Eisenberg DA (1991). Method to identify protein sequences that fold into a known three-dimensional structure. Science. 253(5016), 164–170. [DOI] [PubMed] [Google Scholar]
- Campiche S, Becker-van Slooten K, Ridreau C, Tarradellas J (2006). Effects of insect growth regulators on the nontarget soil arthropod Folsomia candida (Collembola). Ecotox. Environ. Saft. 63, 216–225. [DOI] [PubMed] [Google Scholar]
- Cohen-Barnhouse AM, et al. (2011). Sensitivity of Japanese quail (Coturnixjaponica), common pheasant (Phasianus colchicus), and white leghorn chicken (Gallus gallus domesticus) embryos to in ovo exposure to TCDD, PeCDF, and TCDF. Toxicol. Sci. 119, 93–102. [DOI] [PubMed] [Google Scholar]
- Colovos C, Teates TO (1993). Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2(9), 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wilde R, Swevers L, Soin T, Christiaens O, Rouge P, Cooreman K, Janssen CR, Smagghe G (2013). Cloning and functional analysis of the ecdysteroid receptor complex in the opossum shrimp Neomysis integer (Leach, 1814). Aquat. Toxicol. 130–131(15), 31–40. [DOI] [PubMed] [Google Scholar]
- Doering JA, Farmahin R, Wiseman S, Beitel SC, Kennedy SW, Giesy JP, Hecker M (2015). Differences in activation of aryl hydrocarbon receptors of white sturgeon relative to lake sturgeon are predicted by identities of key amino acids in the ligand binding domain. Environ. Sci. Technol. 49, 4681–4689. [DOI] [PubMed] [Google Scholar]
- Doering JA, Giesy JP, Wiseman S, Hecker M (2013). Predicting the sensitivity of fishes to dioxin-like compounds: possible role of the aryl hydrocarbon receptor (AhR) ligand binding domain. Environ. Sci. Pollut. Res. 20(3), 1219–1224. [DOI] [PubMed] [Google Scholar]
- Dow BA, Sehanobish E, Davidson VL (2016). In silico approaches to identify mutagenesis targets to probe and alter protein-cofactor and protein-protein functional relationships In Vitro Mutagenesis, In Methods in Molecular Biology. Springer. [DOI] [PubMed] [Google Scholar]
- Evenseth LM (2014). Structure, function and ligand interactions of the ecdysone receptor from Daphnia magna. Master thesis in Molecular Biotechnology, Faculty of Health Sciences, UiT – The Arctic University of Norway. https://munin.uit.no/handle/10037/9290. [Google Scholar]
- Farmahin R, et al. (2013). Amino acid sequence of the ligand-binding domain of the aryl hydrocarbon receptor 1 predicts sensitivity of wild birds to effects of dioxin-like compounds. Toxicol. Sci. 131(1), 139–152. [DOI] [PubMed] [Google Scholar]
- Farmahin R, et al. (2012). Sequence and in vitro function of chicken, ring-necked pheasant, and Japanese quail AHR1 predict in vivo sensitivity to dioxins. Environ. Sci. Toxicol. 46(5), 2967–2975. [DOI] [PubMed] [Google Scholar]
- Fay KA, Villeneuve DL, LaLone CA, Song Y, Tollefsen KE, Ankley GT (2017). Practical approaches to adverse outcome pathway development and weight-of-evidence evaluation as illustrated by ecotoxicological case studies. Environ. Toxicol. Chem. 36(6), 1429–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench-Constant RH, Rocheleau TA, Stelchen JC, Chalmers AE (1993). A point mutation in a Drosophilia GABA receptor confers insecticide resistance. Nature; London 3636428, 449–451. [DOI] [PubMed] [Google Scholar]
- Fournier D (2005). Mutations of acetylcholinesterase which confer insecticide resistance in insect populations. Chemico-Biol Inter. 157–158, 257–261. [DOI] [PubMed] [Google Scholar]
- Fournier D, Mutero A (1994). Modification of acetylcholinesterase as a mechanism of resistance to insecticides. Comp. Biochem. Physiol. C 108(1), 19–31. [Google Scholar]
- Glaser LC Organophosphorus and Carbamate Pesticides In Friend M; Franson JC eds, Field Manual of Wildlife Diseases, USGS Biological Resource Division, National Wildlife Health Center, Madison, WI, USA, pp 287–293. [Google Scholar]
- Graham LD, Kotze AC, Fernley RT, Hill RJ (2010). An ortholog of the ecdysone receptor protein (EcR) from the parasitic nematode Haemonchus contortus. Molec. Biochem. Parasit. 171(2), 104–107. [DOI] [PubMed] [Google Scholar]
- Harada T, Nakagawa Y, Akamatsu M, Miyagawa H (2009). Evaluation of hydrogen bonds of ecdysteroids in the ligand-receptor interactions using a protein modeling system. Bioorg. Med. Chem. 17, 5868–5873. [DOI] [PubMed] [Google Scholar]
- Karchner SI, Franks DG, Kennedy SW, Hahn ME (2006). The molecular basis for differential dioxin sensitivity in birds: role of the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA. 103(16), 6252–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalone CA, Villeneuve DL, Burgoon LD, Russom CL, Helgen HW, Berninger JP, Tietge JE, Severson MN, Cavallin JE, Ankley GT (2013). Molecular target sequence similarity as a basis for species extrapolation to assess the ecological risk of chemicals with known modes of action. Aquat. Toxicol. 144, 141–154. [DOI] [PubMed] [Google Scholar]
- LaLone CA, Villeneuve DLDL, Lyons HWD, Helgen SLHW, Robinson JASL, Swintek TWJA, Saari TW, Ankley GT (2016). Sequence alignment to predict across species susceptibility (SeqAPASS): A web-based tool for addressing the challenges of species extrapolation of chemical toxicity. Toxicol. Sci. 153(2), 228–245. [DOI] [PubMed] [Google Scholar]
- LaLone CA, Villeneuve DL, Wu-Smart J, Milsk RY, Sappington K, Garber KV, Housenger J, Ankley GT (2017). Weight of evidence evaluation of a network of adverse outcome pathways linking activation of the nicotinic acetylcholine receptor in honey bees to colony death. Sci. Total Environ. 15, 584–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993). PROCHECK – a program to check the stereochemical quality of protein structures. J. App. Cryst. 26, 283–291. [Google Scholar]
- Lee S, Barron MG (2015). Development of a 3D-QSAR model for acetylcholinesterase inhibitors using a combination of fingerprint, molecular docking, and structure-based pharmacophore approaches. Toxicol. Sci. 148(1), 60–70. [DOI] [PubMed] [Google Scholar]
- Lee S, Barron MG (2016). A mechanism-based 3D-QSAR approach for classification and prediction of acetylcholinesterase inhibitory potency of organophosphate and carbamate analogs. J. Comput. Aided Mol. Des. 30, 347–363. [DOI] [PubMed] [Google Scholar]
- Li F, Han Z (2004). Mutations in acetylcholinesterase associated with insecticide resistance in the cotton aphid, Aphis gossypii Glover. Insect Biochem. Molec. Biol. 34(4), 397–405. [DOI] [PubMed] [Google Scholar]
- Liu Z; Williamson MS; Lansdell SJ; Denholm I; Han Z; Millar NS (2005). A nicotinic acetylcholine receptor mutation conferring target-site resistance to imidacloprid in Nilaparvata lugens (brown planthopper). Proc. Natl. Acad. Sci. USA. 102 (24), 8420–8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthy R, Bowie JU, Eisenberg D (1992). Assessment of protein models with three-dimensional profiles. Nature. 356(6364), 83–85. [DOI] [PubMed] [Google Scholar]
- Martinez-Torres D, Foster SP, Field LM, Devonshire AL, Williamson MS (1999). A sodium channel point mutation is associated with resistance to DDT and pyrethroid insecticides in the peach-potato aphid, Myzus persicae (Sulzer) (Hemiptera: Amphididae). Insect Mol. Biol. 8(3), 339–346. [DOI] [PubMed] [Google Scholar]
- Martinovic-Weigelt D, et al. (2017). Derivation and evaluation of putative adverse outcome pathways for the effects of cyclooxygenase inhibitors on reproductive processes in female fish. Toxicol. Sci. 156(2), 344–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie JA (1996). Ecological and Evolutionary Aspects of Insecticide Resistance, R.G. Landes Company, Austin, TX. [Google Scholar]
- Mineau P (2002). Estimating the probability of bird mortality from pesticides sprays on the basis of the field study record. Environ. Toxicol. Chem. 21, 1497–1506. [PubMed] [Google Scholar]
- Moores GD, Gao X, Denholm I (1996). Characterization of insensitive acetylcholinesterase in insecticide-resistant cotton aphids, aphis gossypiiglover (homoptera: Aphididae). Pest. Biochem. Physiol. 56(2), 102–110. [Google Scholar]
- Mutero A, Pralavoria M, Bride JM, Fournier D (1994). Resistance-associated point mutations in insecticide-insensitive acetylcholinesterase. Proc. Natl. Acad. Sci. USA. 91, 5922–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqqash MN, Gokce A, Bakhsh A, Salim M (2016). Insecticide resistance and its molecular basis in urban insect pests. Parasitology Res. 115(4), 1363–1373. [DOI] [PubMed] [Google Scholar]
- Osta MA, Rizk ZJ, Labbe P, Weill M, Knio K (2012). Insecticide resistance to organophosphates in Culex pipiens complex from Lebanon. Parasit. Vectors. 5, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke S, Reinecke A (2007). The impact of organophosphate pesticides in orchards on earthworms in the Western Cape, South Africa. Ecotoxicol. Environ. Saf. 66, 244–251. [DOI] [PubMed] [Google Scholar]
- Russom CL, LaLone CA, Villeneuve DL, Ankley GT (2014). Development of an adverse outcome pathway for acetylcholinesterase inhibition leading to acute mortality. Enviro. Toxicol. Chem. 33(10), 2157–2169. [DOI] [PubMed] [Google Scholar]
- Shaw AJ (1999). The evolution of heavy metal tolerance in plants: adaptations, limits, and costs, in: Forbes VE (Ed.), Genetics and Ecolotoxicology, Taylor and Francis, Philadelphia, PA, pp. 9–30. [Google Scholar]
- Song T, Doering JA, Sun J, Beitel SC, Shekh K, Patterson S, Crawford S, Giesy JP, Wiseman SB, Hecker M (2016). Linking oxidative stress and magnitude of compensatory responses with life-stage specific differences in sensitivity of white sturgeon (Acipenser transmontanus) to copper or cadmium. Enviro. Sci. Technol. 50(17), 9717–9726. [DOI] [PubMed] [Google Scholar]
- Song MY, Stark JD, Brown JJ (1997). Comparative toxicity of four insecticides, including imidacloprid and tebufenozide, to four aquatic arthropods. Enviro. Toxicol. Chem. 16(12), 2494–2500. [Google Scholar]
- Song Y, Villeneuve DL, Toyota K, Iguchi T, Tollefsen KE (2017). Ecdysone receptor agonism leading to lethal molting disruption in arthropods: Review and adverse outcome pathway development. Enviro. Sci. Technol. 51, 4142–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JK, Janz DM (2015). Developmental and persistent toxicities of maternally deposited selenomethionine in zebrafish (Danio rerio). Enviro. Sci. Technol. 49(16), 10182–10189. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen T, Vontas J, Tsagkarakou A, Dermauw W, Tirry L (2010). Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: A review. Insect Biochem. Molec. Biol. 40(8), 563–572. [DOI] [PubMed] [Google Scholar]
- Vardy DW, Oellers J, Doering JA, Hollert H, Giesy JP, Hecker M (2013). Sensitivity of early life stages of white sturgeon, rainbow trout, and fathead minnow to copper. Ecotoxicology. 22(1), 139–147. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Q, Wu B, Li Y, Lu G (2013). Correlation between TCDD acute toxicity and aryl hydrocarbon receptor structure for different mammals. Ecotox. Enviro. Soft. 89, 84–88. [DOI] [PubMed] [Google Scholar]
- Webber NR, Boone MD, Distel CA (2010). Effects of aquatic and terrestrial carbaryl exposure on feeding ability, growth, and survival of American toads. Environ. Toxicol. Chem. 29, 2323–2327. [DOI] [PubMed] [Google Scholar]
- Wirgin I, Roy NK, Loftus M, Chambers RC, Franks DG, Hahn ME (2011). Mechanistic basis of resistance to PCBs in Atlantic tomcod from the Hudson River. Science. 331, 1322–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirgin I, Waldman JR (2004). Resistance to contaminants in North American fish populations. Mut. Res. Fund. Molec. Mech. Mutagen. 552(1–2), 73–100. [DOI] [PubMed] [Google Scholar]
- Zahavi M, Tahori AS (1970). Sensitivity of acetylcholinesterase in spider mites to organo- phosphorus compounds. Biochem. Pharmacol. 19(1), 219–225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.