Abstract
It is widely believed that protection against acquisition of HIV or SIV infection requires anti-envelope (anti-Env) antibodies, and that cellular immunity may affect viral loads but not acquisition, except in special cases. Here we provide evidence to the contrary. Mucosal immunization may enhance HIV vaccine efficacy by eliciting protective responses at portals of exposure. Accordingly, we vaccinated macaques mucosally with HIV/SIV peptides, modified vaccinia Ankara–SIV (MVA-SIV), and HIV-gp120–CD4 fusion protein plus adjuvants, which consistently reduced infection risk against heterologous intrarectal SHIVSF162P4 challenge, both high dose and repeated low dose. Surprisingly, vaccinated animals exhibited no anti-gp120 humoral responses above background and Gag- and Env-specific T cells were induced but failed to correlate with viral acquisition. Instead, vaccine-induced gut microbiome alteration and myeloid cell accumulation in colorectal mucosa correlated with protection. Ex vivo stimulation of the myeloid cell–enriched population with SHIV led to enhanced production of trained immunity markers TNF-α and IL-6, as well as viral coreceptor agonist MIP1α, which correlated with reduced viral Gag expression and in vivo viral acquisition. Overall, our results suggest mechanisms involving trained innate mucosal immunity together with antigen-specific T cells, and also indicate that vaccines can have critical effects on the gut microbiome, which in turn can affect resistance to infection. Strategies to elicit similar responses may be considered for vaccine designs to achieve optimal protective efficacy.
Keywords: Vaccines
Keywords: AIDS vaccine
Introduction
The protective immune correlates of the RV144 vaccine trial were non-neutralizing anti-envelope (anti-Env) antibodies (1, 2). It is widely believed that protection against HIV or SIV acquisition requires anti-Env antibodies, and that cellular immunity may affect viral load (VL) but not prevent acquisition. Here, we provide contrary evidence that antibodies are not always necessary for protection. We have been developing a mucosal SIV vaccine, with components analogous to RV144 (including poxvirus vectors and protein or peptides), with which we achieved reduction of VLs in an SIVmac251 infection model (3). In that model, the magnitudes of Gag-specific polyfunctional CD8+ T cells and the expression levels of innate factor APOBEC3G inversely correlated with VL (4). However, Env-specific antibody responses were not elicited (4). In light of RV144, we asked whether the combination of mucosal T cell– and antibody-based vaccines would generate better immunity to mediate protection. In the current studies, we have included full-length single chain (FLSC), which is a chimeric protein containing HIV-1BaLgp120 and the D1 and D2 domains of macaque CD4, in the mucosal vaccine regimen. Recombinant FLSC containing rhesus CD4 (rhFLSC) has been shown to induce cross-reactive antibodies and protective immune responses in macaques (5, 6), including V2 loop antibodies that correlated with protection (7). We hypothesized that the inclusion of rhFLSC would help to induce anti-Env antibody responses. However, in the first study, we found that even with rhFLSC in the mucosal vaccine regimen, anti-Env antibodies were not elicited above background levels. The second study confirmed that antibodies against other non-HIV components of the mucosal vaccine, but not Env, were induced. Since most HIV and SIV vaccination regimens elicit virus-specific cellular and antibody responses, and not all vaccine-induced immunity mediates protection, it is difficult to dissect the protective mechanisms. Our mucosal vaccine provided an opportunity to address this question in the absence of anti-Env antibodies.
In the third cohort, which was an expansion cohort of the first one, we immunized 21 macaques using a similar regimen, and challenged them with repeated low-dose HIV/SIV (SHIV). As predicted, anti-Env antibody responses were not elicited in blood or mucosa. Nevertheless, the vaccinated animals demonstrated significantly reduced risk of viral acquisition, with 44% vaccine efficacy. Therefore, the reduced risk of viral acquisition was achieved solely through antigen-specific T cell responses and/or innate immunity. Though indeed, Gag- and Env-specific cellular responses were elicited, the magnitude of the responses did not correlate with the reduced risk of viral acquisition. Instead, the mucosal vaccine induced significant increases in myeloid cell subsets in the PBMCs (including CD15+ myeloid-derived suppressor cells [MDSCs]) and colorectal intraepithelial compartment of the vaccinated animals, both correlating with viral acquisition. Upon ex vivo stimulation with SHIV, the myeloid cell–enriched population from the vaccinated animals produced more TNF-α and IL-6, which are the markers of trained immunity, as well as virus coreceptor agonist MIP1α, than those from the naive ones. The rapidly produced TNF-α, IL-6, and MIP1α correlated with the reduced viral Gag expression in the ex vivo system, and/or reduced in vivo infection risk, suggesting the involvement of trained immunity. Furthermore, the composition of the gut microbiome was altered in the vaccinated animals. The latter correlated with viral acquisition and colorectal monocytes. Overall, our data suggested that the current mucosal vaccine might induce trained innate immunity (8, 9), which affected vaccine efficacy against heterologous SHIV challenge. Thus, a combination of T cell and/or trained immunity appears capable of mediating protection without anti-Env antibody responses. Antigen-specific T cells may confer the necessary specificity to guide the trained innate immune cells to the relevant targets. Since the mucosal vaccines had different correlates of protection from those of RV144, the combination might lead to increased vaccine efficacy.
Results
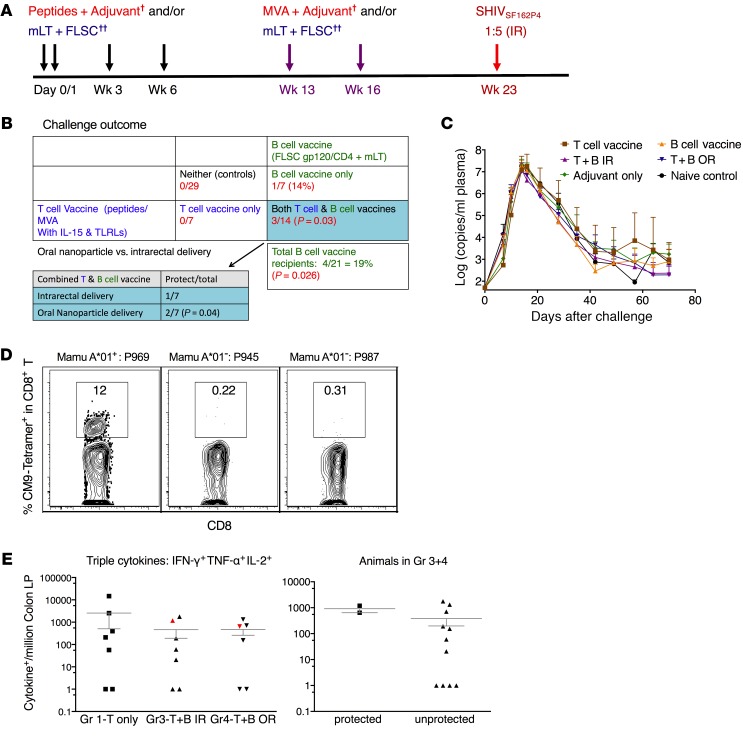
In the absence of anti-Env antibody responses, partial protection against a single high-dose SHIVSF162P4challenge was achieved in the combination group. To test whether combination of a T cell–based mucosal HIV vaccine with an Env antibody–based vaccine will mediate better protection against intrarectal SHIV challenge, we vaccinated 4 groups of macaques in the first study (Supplemental Tables 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/JCI122110DS1). Each group had as close to an equal number of Mamu-A*01–positive and -A*02–positive animals as possible, and very few B*08 and B*17 animals, distributed evenly. Group-1 macaques were vaccinated with a T cell–based mucosal vaccine, which included HIV/SIV peptides adjuvanted with TLR agonists and IL-15 as a priming immunization, and modified vaccinia Ankara (MVA) expressing the SIV Env, Gag, pol, Rev, Tat, and Nef with the same adjuvants as a boost, while group-2 macaques were vaccinated with rhFLSC adjuvanted with mutant heat-labile E. coli toxin (mLT). The vaccine components were delivered either intrarectally or orally, all targeting the colorectal tissues. We developed a Eudragit-coated microparticle/nanoparticle formulation oral delivery system to induce immunity in the colorectal mucosa in mice (10) and translated that here to macaques. The Eudragit-coated microparticle/nanoparticle formulation was optimized for oral delivery in macaques (Supplemental Figure 1) based on our previous murine study (10). Groups 3 and 4 were given the combination of both vaccines with the peptides/adjuvants either intrarectally (group 3) or orally (group 4), but were otherwise identical. Seven weeks after the last boost, the animals were challenged intrarectally with a single high-dose SHIVSF162P4, which infected all 29 control animals that were part of a large set of collaborative studies in the same facility with animals from the same source (including 6 adjuvant, and 4 mock controls) (Figure 1, A and B). Seven animals in the T cell–based vaccine were all infected, while 1 of 7 was uninfected in the rhFLSC-alone group. In the combination of groups 3 and 4, three of 14 animals were protected, which was significantly different from the 29 control animals (P = 0.03), indicating the protection was significant. After SHIV infection, there was no difference among the groups in the VLs of those animals that were infected (Figure 1C).
Figure 1. Partial protection against a single high-dose SHIVSF162P4 challenge was achieved in the first cohort study.
The 3 protected macaques that were vaccinated with the combined mucosal vaccines had Gag-specific CD8+ T cell responses in the rectal mucosa. (A) Schematic illustration of mucosal vaccination and challenge protocol of the first study. (B) Challenge outcome. Fisher’s exact test was used to calculate the P values. TLRLs, TLR ligands. (C) Geometric mean of the viral load (VL) in the plasma of the infected animals. (D) Dominant CM9-tetramer+CD8+ T cell responses were induced in one of the protected animals, which was Mamu-A*01+ in the rectal lamina propria (LP). The 2 other protected animals were Mamu-A*01–. (E) Intracellular cytokine+ CD8+ T cell responses against SIV Gag were induced in the rectal LP of the 2 Mamu-A*01– animals. †MVA, modified vaccinia Ankara, plus adjuvant (triple TLR [TLR2, -3, and -9] agonists) plus IL-15. ††FLSC, full-length single chain, plus cross-linked gp120-CD4 complexes. IR, intrarectal; mLT, mutant heat-labile E. coli toxin (R192G); OR, oral.
We initially hypothesized that the protection against acquisition was mediated by anti-Env antibody responses, based on the fact that all the protected animals were in the groups including rhFLSC. However, when we measured the humoral immunity against Env, we were surprised to find that there were no or extremely low levels of binding antibodies against gp120 of either the vaccine strain (BaL) or the challenge strain (SF162P4), rhFLSC, or CD4, let alone neutralizing antibodies against SHIV. There were also no CD4-inducible antibodies or antibody-dependent cellular cytotoxicity activity (ADCC) in the plasma. Moreover, no mucosal antibodies in the rectal mucosa or Env-specific B cell responses in mesenteric lymph nodes (MLNs) were observed (Supplemental Table 3 and Supplemental Figure 2, A–D). Taken together, these results show that the mucosal vaccines induced negligible to very low levels of systemic or mucosal Env-specific humoral immune responses (not significantly different from prevaccination background levels), which showed no correlations with acquisition.
We then examined the Gag-specific CD8+ T cell responses in the colorectal tissues. Among the 3 protected animals exposed to Gag, high Gag CM9-tetramer+CD8+ T cell responses were induced in the only Mamu-A*01–positive animal (Figure 1D), and Gag-specific polyfunctional CD8+ T cell responses were induced in the 2 Mamu-A*01–negative animals in the colorectal tissues (Figure 1E). However, the magnitudes of the Gag-specific responses did not correlate with protection. The animals in group 1 had the 2 highest Gag-specific CD8+ T cell responses, but none of them was protected (Figure 1E).
Anti–cholera toxin, but not anti-rhFLSC, responses were effectively elicited after intrarectal immunization of rhFLSC with cholera toxin in the second study.
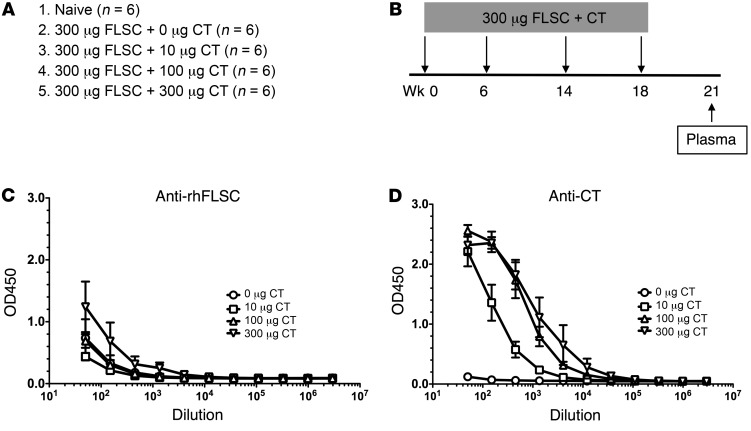
The finding that the mucosal vaccine did not induce anti-Env antibodies is consistent with what we have found in the second study, where we explored whether cholera toxin (CT) coformulated with rhFLSC could elicit long-lived anti-rhFLSC responses after intrarectal immunization (Figure 2, A and B). We have had a long-standing interest in overcoming the poor persistence of anti-Env antibody responses, a major problem confronting HIV-1 vaccine development (11–14). Previous studies from our group in mice suggested that mucosal immunization with a gp140 Env immunogen formulated with wild-type CT, enzymatically inactivated K63 mutant of CT (CT-K63), or wild-type A1 domain of CT efficiently elicited persistent antibody responses to Env (11, 12, 15, 16). Based on these observations, we immunized the macaques intrarectally with 300 μg of rhFLSC and with varying doses of wild-type CT with the idea of ultimately determining whether persistent antibody responses to rhFLSC could provide protection against a rectal challenge with SHIV. After immunization of 30 macaques intrarectally with 300 μg of rhFLSC plus varying doses of CT, surprisingly, only marginal anti-rhFLSC responses were elicited at the highest CT dose and no detectable responses at lower CT doses (Figure 2C). By contrast, anti-CT antibody responses were elicited in a dose-dependent fashion (Figure 2D). It should be noted that macaques immunized with 100–300 μg rhFLSC by various routes and adjuvants routinely elicit anti-rhFLSC antibody responses (5, 6, 17), suggesting that a factor other than the immunogen itself accounts for the poor rectal immunogenicity observed here.
Figure 2. Anti-rhFLSC responses were not effectively elicited 21 weeks (3 weeks after the last vaccination) after intrarectal immunization with rhFLSC plus cholera toxin (CT) in the plasma of the macaques in the second study.
(A and B) The immunization groups (A) and protocol (B) of the 30 macaques. (C) Only marginal anti-rhFLSC responses (IgG) were induced at the highest CT dose and no detectable responses at the lower doses of CT. (D) Anti-CT antibody responses (IgG) were elicited in a dose-dependent fashion.
The mucosal vaccine mediated delay of viral acquisition without Env-specific antibody responses in the expansion (third) cohort.
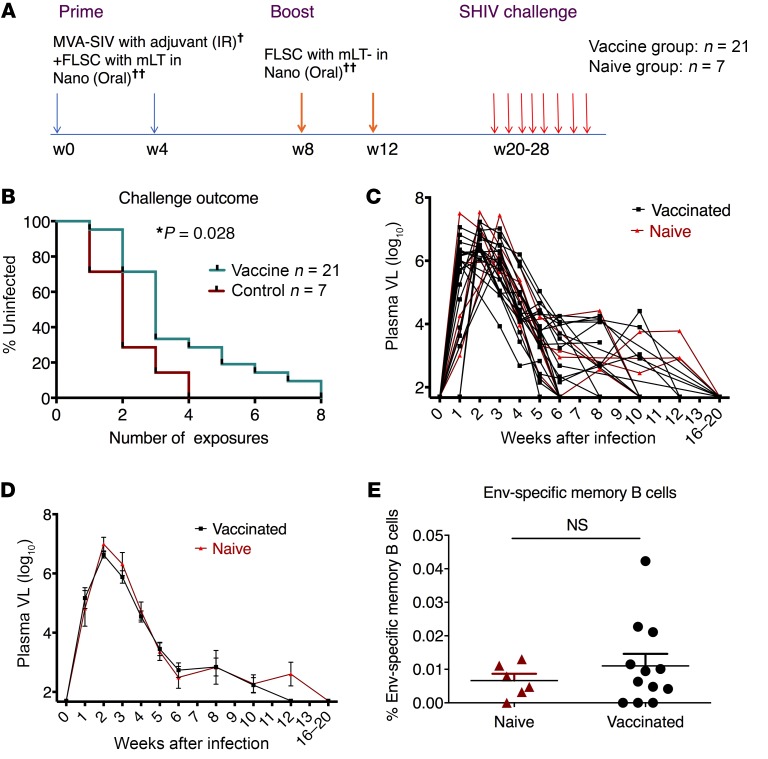
Most vaccine platforms elicit a myriad of innate, cellular, and humoral responses, not all of which contribute to protective immunity. The finding that our mucosal vaccine did not induce Env-specific antibody responses gave us a unique opportunity to evaluate the role of virus-specific T cell responses and/or innate immunity in mediating protection against viral acquisition. We therefore initiated an expansion cohort to examine protective mechanisms (Supplemental Table 4). Twenty-one macaques were primed with MVA-SIV plus rhFLSC, and boosted with rhFLSC, in both cases with adjuvants (TLR ligands and IL-15 for the MVA and mLT for the rhFLSC boost, the latter delivered in Eudragit-coated microparticle/nanoparticle formulation). Peptides were omitted from this vaccine, as we had noted protection with the animals that received rhFLSC in the first cohort. Eight weeks after the last boost, we intrarectally challenged the animals with 8 repeated weekly low doses of SHIVSF162P4 (Figure 3A). Compared with naive controls, the vaccinated animals demonstrated significant delay of viral acquisition, with vaccine efficacy of 44% (P = 0.028, Figure 3B). We did not observe any VL difference between the vaccinated and naive groups once the animals were infected by SHIVSF162P4 (Figure 3, C and D).
Figure 3. The mucosal vaccine mediated delay of viral acquisition against repeated low-dose SHIV challenges in the absence of anti-Env antibody responses in the third cohort study.
(A) Schematic illustration of mucosal vaccination and challenge protocol of the third cohort study. Twenty-eight macaques were distributed into either a vaccine group (n = 21) or naive group (n = 7). MVA-SIV with adjuvant combination of triple TLR (TLR2, -3, and -9) agonists plus IL-15 was given intrarectally, while FLSC with mLT in nanoparticle format was given orally. (B) Infection-free curves for the vaccinated and naive groups. A Kaplan-Meier curve analysis was performed after a series of 8 weekly intrarectal challenges. (C and D) The plasma VLs of each of the infected animals (C) and the geometric mean and SEM of the vaccinated and naive groups. (E) Anti-HIV humoral responses in the colorectal samples (rectal swab, rectal pinches, MLNs) 4 weeks after the last vaccination were measured. No gp120-specific IgG or IgA was detected. HIV Env–specific B cell responses in the rectal pinches 4 weeks after the last vaccination did not change. Mann-Whitney test was used to compare the 2 groups (n = 6, and 12 for naive and vaccinated groups). †MVA, modified vaccinia Ankara, plus adjuvant (triple TLR [TLR2, -3, and -9] agonists) plus IL-15. ††FLSC, full-length single chain, cross-linked gp120-CD4 complexes. IR, intrarectal; mLT, mutant heat-labile E. coli toxin (R192G); NS, not significant.
To explore the protective mechanisms, we first examined the humoral immune responses. Consistent with the previous cohorts, we did not find significant titers of binding antibody responses against gp120 (either BaL or SF162) in either the plasma or the rectal mucosa, including IgM, IgG, and IgA (Supplemental Table 5). Thus, surprisingly, in the absence of measurable anti-Env antibody, we have achieved significantly reduced risk of acquisition in this repeated low-dose SHIV challenge model, consistent with our finding in the earlier high-dose challenge study. Similarly to the second cohort, we found that the antibody responses against adjuvant mLT were induced (Supplemental Figure 3, A and B), suggesting different antigens may have different antibody induction mechanisms.
We further evaluated B cell responses in the rectal mucosa after vaccination. For the Env-specific memory B cells, the vaccinated animals did not differ from the naive animals (Figure 3E). Only very weak responses in several animals were found, concordant with the lack of anti-Env antibodies in the rectal swab. The total plasmablasts were also not affected by the mucosal vaccine (Supplemental Figure 3C).
In the present study, no antibodies were detected against CD4, as shown in Supplemental Figure 2A, consistent with the observations that rhFLSC did not raise anti-CD4 autoantibodies in any previous macaque studies, including a formal immunotoxicological study published by Schwartz et al. (18). Other components, e.g., MVA, did not contain any xeno antigens. Furthermore, the SHIV stock was grown in rhesus cells, which is different from some of the studies using virus stocks grown in human cells.
The mucosal vaccine induced Gag- and Env-specific T cell responses in the colorectal mucosal tissues and MLNs.
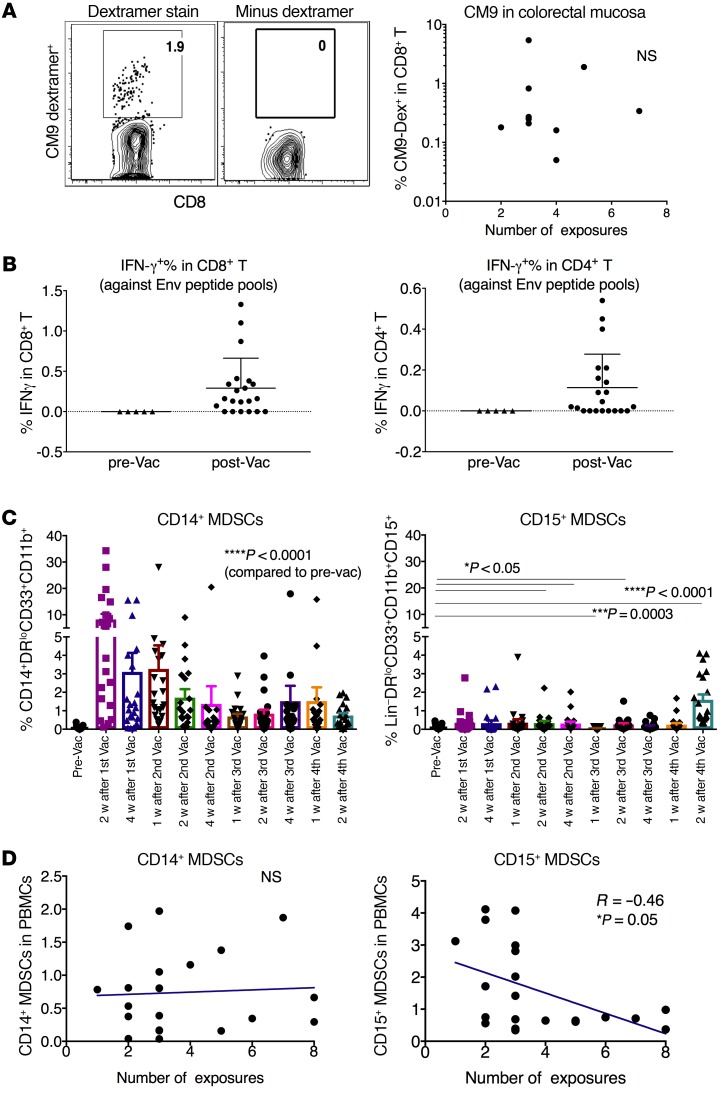
In the first cohort, we found that the protected animals had vaccine-induced Gag-specific T cell responses in the colorectal lamina propria. In this cohort, we focused on both Gag- and Env-specific T cell responses. As 10 animals were Mamu-A*01 positive, we monitored the dominant Gag-specific CM9-dextramer+ CD8+ T cell responses in colorectal intraepithelial lymphocytes (IELs) during the vaccination, since this is the compartment most closely exposed to the colorectal luminal contents. The frequency was increased after priming, but decreased after boosting. After boosting, some animals demonstrated Gag-CM9 dextramer+ T cell responses in IELs, but they did not correlate with delay of viral acquisition (Figure 4A). In MLNs and PBMCs, the CM9-dextramer+ T cell responses were either low or absent (Supplemental Figures 4 and 5). Overall, none of them correlated with viral acquisition.
Figure 4. The mucosal vaccine induced Gag- and Env-specific T cell responses in the colorectal mucosal tissues and MLNs, and MDSCs in PBMCs, which inversely correlated with viral acquisition in the third cohort.
(A) Gag-specific CM9 dextramer+CD8+ T cell responses were induced in the colorectal tissues of the vaccinated Mamu-A*01+ macaques 4 weeks after the last vaccination. Left panel shows a representative staining, while the right panel shows a negative control without the dextramer. (B) Env-specific CD8+ and CD4+ T cell responses were induced in the MLNs of the vaccinated animals 4 weeks after the last vaccination. (C) The kinetics of the MDSCs in the PBMCs upon each vaccination. Wilcoxon’s signed-rank test was used to compare each time point with prevaccination. (D) CD15+, but not CD14+, MDSCs in the PBMCs at 2 weeks after the last vaccination inversely correlated with number of viral exposures necessary for viral acquisition. Spearman’s r and exact Jonckheere-Terpstra test P value of the correlation are indicated. NS, not significant.
The HIV Env–specific T cell responses were evaluated in the MLNs of all the vaccinated animals 4 weeks after the last vaccination (Supplemental Figure 6A). Fifteen of 21 animals showed positive IFN-γ+CD8+ T cell responses against Env, and similar responses were found for CD4+ T cells (Figure 4B). However, just as in the case of the Gag-specific T cell responses, the magnitude of Env-specific T cell responses did not correlate with delay of viral acquisition (Supplemental Figure 6B).
The mucosal vaccine–induced MDSCs in PBMCs.
The fact that neither Gag- nor Env-specific T cell responses correlated with viral acquisition suggested that regulatory factors might affect the challenge outcome besides a possible role for T cells. Our previous study demonstrated that the vaccine-induced MDSCs could dampen protective immunity to SIV (19). In the third cohort, we asked whether this mucosal regimen could induce MDSCs; if so, what role did the induced MDSCs play in this repeated low-dose SHIV challenge model? MDSCs act as a double-edged sword in HIV and SIV infection, depending on which type of immunity the MDSCs suppress more, the immune activation of CD4+ T cells as viral targets, or the adaptive antiviral immunity that suppresses the virus. To answer these questions, we measured the kinetics of the MDSCs in PBMCs. Two subsets of MDSCs, namely, CD14+ (monocytic) and CD15+ (granulocytic), were investigated. In the naive animals, the frequencies of both MDSC subsets were consistently low over time (the mean of the sum of both types of MDSCs <0.5%). For the CD14+ subset, compared with the low level at prevaccination, the first MVA prime induced a sharp increase, which then gradually declined (Figure 4C). Throughout the whole course of vaccination, CD14+ MDSCs persisted in the blood, at significantly higher levels than the prevaccination baseline levels, which were very low (Figure 4C). In contrast, the CD15+ MDSCs were mostly unchanged from prevaccination through 1 week after the last vaccine (except a few time points), after which they sharply increased 2 weeks after the last boost (Figure 4C). Thus, the mucosal vaccine induced a significant quantity of both MDSCs in the PBMCs. In the previous study, we found expansions of MDSCs in the vaccinated animals, but not in the adjuvant-only group, suggesting that the vaccine components such as vaccinia virus or HIV proteins might play roles in MDSC inductions (19).
We then used the data from the 2-week time point after the last boost to assess the role of MDSCs in viral acquisition. CD15+, but not CD14+, MDSCs inversely correlated with viral acquisition (Figure 4D). Animals with higher CD15+ MDSCs were more likely to be infected early, suggesting the detrimental role of the induced CD15+ MDSCs. This might be mediated by suppression of virus-specific T cell responses.
Mucosal vaccine induced newly migrated monocytes in the colorectal IELs, which displayed a biphasic positive correlation with viral exposure.
We next checked the viral target cells (defined as Ki67+CCR5+CD4+ T cells) in the rectal mucosa, which have been shown to be able to influence the challenge outcome (21, 22). The frequencies of viral target cells in the rectal IELs 4 weeks after vaccination did not significantly differ from those prevaccination, or those in the naive controls (Supplemental Figure 7).
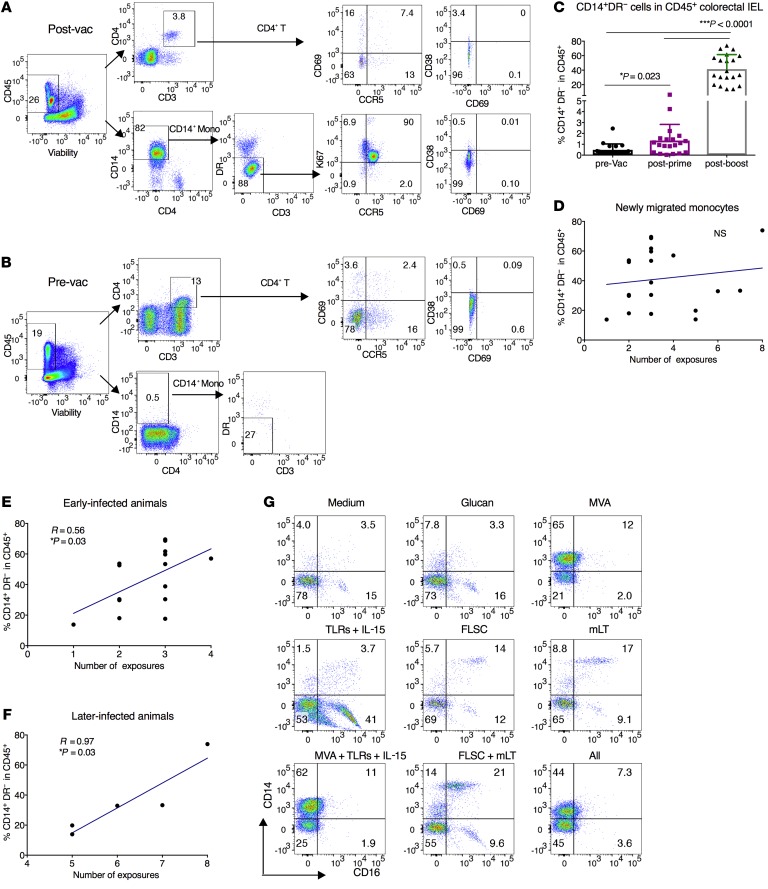
However, the CD14+ monocytes in the rectal IELs were dramatically increased after the boost (Figure 5, A–C). The majority of the CD14+ monocytes accumulating in the colorectal IELs after vaccination were HLA-DR negative/low, resembling the phenotype of newly migrated monocytes from blood (23). Most of these cells were phenotypically CCR5+Ki67+CD69–CD38–CD4–.
Figure 5. Mucosal vaccine induced newly migrated monocytes in the IELs of the colorectal tissues, which demonstrated a biphasic positive correlation with viral exposure in the third study.
(A and B) In the CD45+ live mononuclear cell gating of one representative animal, the CD14+DR– subset was dramatically increased after vaccination. The majority of the CD14+DR– subset was CCR5+Ki67+CD69–CD38–CD4–. (C) The kinetics of the CD14+DR– subset before and after prime/boost in the vaccinated animals. Wilcoxon’s signed-rank test was used for comparison. (D–F) The CD14+DR– subset did not correlate with the number of viral exposures in all the vaccinated animals. If the animals were split into early- and later-infected animals, in both subpopulations of the animals there were positive associations (trends) between the CD14+DR– subset and the number of viral exposures. Spearman’s test was used to calculate the r and P values of the correlations. (G) The adherent cells from naive PBMCs were treated with vaccine components or medium/glucan (as indicated in the figure) for 24 hours. After washout, the cells were cultured for 6 days in R10 medium. On day 6, the cells were detached and analyzed by flow cytometry. NS, not significant.
To determine whether these monocytes affected viral acquisition, we performed a correlation analysis. If all the animals were included, the frequencies of CD14+ monocytes in the colorectal IELs did not correlate with viral acquisition (Figure 5D). However, if we split the animals into the early (N ≤ 4) and later-infected groups (N > 4), both the early- and later-infected animals showed positive correlations (P = 0.03) (Figure 5, E and F). To investigate which vaccine component drove the biggest shifts in monocytes and MDSCs, we did an in vitro stimulation study, and found that MVA or any combination with MVA induced a high accumulation of CD14+ monocytes (Figure 5G). Considering the immature nature of these monocytes, the biphasic correlation may indicate the existence of a transition point where the monocytes either matured or differentiated to other cell types dependent on the gut microenvironment. Since myeloid cells respond to microbial signals to initiate innate and adaptive immune responses, the dramatic increase of these myeloid cells suggested that the gut microbiome might also be altered in the vaccinated animals.
The gut microbiome correlated with viral exposure, and CD14+HLA-DR–/lo monocytes in the colorectal IELs.
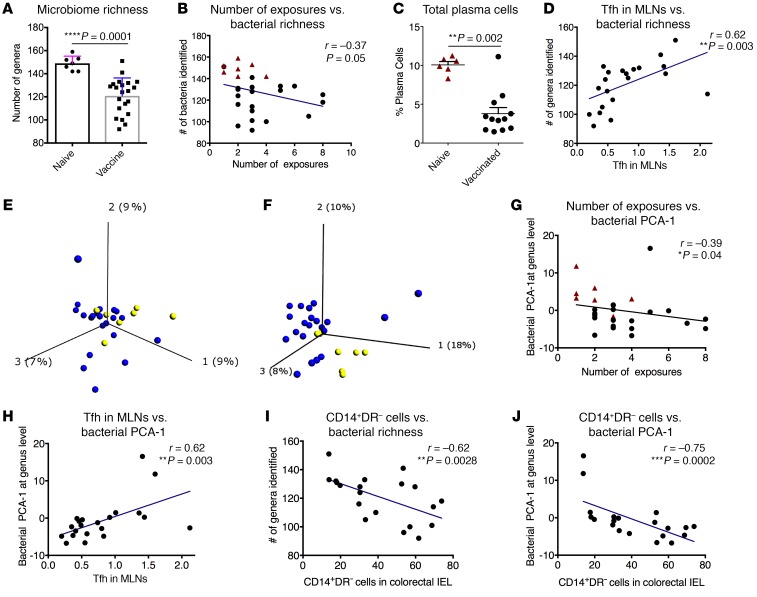
To test this hypothesis, we sequenced the 16S rRNA using fecal samples collected 1 week before the serial viral challenges (Supplemental Table 6). We first compared the number of species identified in the vaccinated and naive animals. Notably, the bacterial richness was significantly lower in the vaccinated group than the naive group at both the genus (Figure 6A) and species (Supplemental Figure 8A) levels. Furthermore, the bacterial richness at both levels inversely correlated with viral acquisition, but this may be an indirect association due to the differing group means (Figure 6B and Supplemental Figure 8B). Commensal microbes have been shown to promote the Tfh and germinal center B cell responses, which led to IgA production against commensals (24). We found that the frequency of total plasma cells among CD45+ cells was significant lower in the rectal mucosa of the vaccinated animals than that of the naive ones (Figure 6C). Given the lack of mucosal virus-specific antibody, this could also relate to the alteration of gut microbial microenvironment. One likely explanation was that the alteration of the gut microbiome, especially the reduction of the bacterial richness, might lead to these changes. Future studies are needed to confirm this interpretation. Total Tfh in the MLNs, which were upstream of the plasma cells, also correlated with bacterial richness (Figure 6D and Supplemental Figure 8C).
Figure 6. Gut microbiome correlated with viral exposure, Tfh in MLNs, and newly migrated monocytes in the IELs of the colorectal tissues in the third study.
(A) The diversity of the gut microbiome at the genus level was lower in the vaccinated animals compared with that of the naive controls. Mann-Whitney test was used for comparison. (B) Bacterial richness inversely correlated with viral acquisition. (C) The total plasma cells were lower in the rectal pinches 4 weeks after the last vaccination compared with those of the naive controls. (D) Bacterial richness positively correlated with Tfh cells in the MLNs. (E and F) Principal component analysis (PCA) plot of the gut microbiome at the genus level 1 month before vaccination (E) and 7 weeks after the last vaccination (F). The blue dots indicate vaccinated animals, while the yellow ones indicate the naive animals. (G and H) PCA-1 of the gut microbiome at the genus level was exported for each animal. The bacterial PCA-1 showed associations with the number of viral exposures (G), and Tfh in the MLNs (H). (I and J) CD14+DR– cells in the IELs of the colorectal tissues correlated with bacterial richness (I) and bacterial PCA-1 at the genus level (J). Spearman’s test was used to calculate the r and P values of the correlations.
Further analysis revealed that the vaccinated groups had different gut microbiome compositions. We visualized the gut microbiome data using principal component analysis (PCA, Supplemental Figures 8D and 9). The naive and the vaccinated animals displayed different bacterial compositions after (Figure 6F and Supplemental Figures 8E and 9) but not before vaccination (Figure 6E and Supplemental Figure 8D). Bacterial PCA-1 did not correlate with the viral target cells in the rectal mucosa (Supplemental Figure 10), but it did correlate with viral acquisition (Figure 6G and Supplemental Figure 8F), suggesting that the gut bacteria might associate with SHIV susceptibility. The bacterial composition correlated with total Tfh in the MLNs (Figure 6H and Supplemental Figure 8G).
The mucosal vaccine induced a dramatic increase of CD14+DR– monocytes in the colorectal IELs. The accumulation of these cells correlated with the bacterial richness and bacterial PCA-1 (Figure 6, I and J, and Supplemental Figure 8, H and I). It is possible that the gut microbiome attracted the CD14+ monocytes from blood to the gut, as this was concurrent with the disappearance of CD14+ MDSCs in the blood. Alternatively, the alteration of the gut microbiome caused the local monocyte proliferation. Regardless, the gut microbiome might interact with the CD14+DR– monocytes in the colorectal mucosa, and further studies are needed to determine the exact mechanisms mediating the accumulation of and roles of these myeloid cells.
Although the frequency of viral target cells (Ki67+CCR5+CD4+ T cells) did not change after vaccination (Supplemental Figure 7), it inversely correlated with CD14+DR– cells in the colorectal IELs, suggesting that the CD14+DR– cells might be suppressive to viral target T cells (Supplemental Figure 11A). Furthermore, the other markers of immune activation on the T cells did shift after vaccination (Supplemental Figure 11, B and C). To determine whether the shift in immune activation markers with vaccination had any association with microbiome complexity, we did correlation analyses. We found that before vaccination, the gut microbiome PCAs correlated with HLA-DR on T cells and CCR5 on CD14+ monocytes; after vaccination, the gut microbiome PCAs correlated with CD38, CCR5, and CD69 on T cells, and CD69 and CCR5 on CD14+ monocytes (Supplemental Figure 11D). This cross-sectional study of the immune activation and gut microbiome suggested a possible link between immune activation and gut microbiome. This is consistent with our recent ex vivo study, where we found that the gut microbiome can shape the immune activation of the T cells (25). Nevertheless, a longitudinal study will provide a better picture of the shift.
Enhanced expression of TNF-α, IL-6, and MIP1α by monocytes was inversely correlated with viral Gag expression upon SHIV stimulation ex vivo, and associated with reduced viral infection risk in vivo.
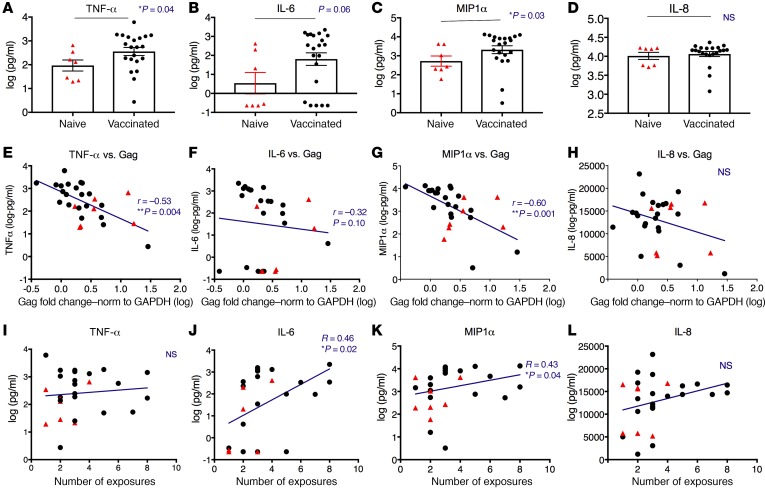
The reduced risk of viral acquisition was achieved in the absence of anti-Env antibody responses and did not correlate with virus-specific T cell responses, suggesting a key role of innate immunity, especially memory monocyte–mediated trained immunity (8). Trained immunity previously has been described in bacillus Calmette-Guérin (BCG) vaccination and in vitro restimulations with TLR agonists on monocytes (9, 26). Monocytes and macrophages can respond with an increased cytokine production upon restimulation after priming with fungal cell wall component β-glucan, TLR agonists, or BCG, with TNF-α and IL-6 as the primary markers of trained immunity (26, 27). Furthermore, BCG-educated myeloid cells could generate protective innate immunity against tuberculosis in vivo (28). We hypothesized that CD14+ myeloid cells induced by our mucosal vaccine might constitute trained immunity and have contributed to control of SHIV acquisition. To test that, we thawed PBMCs from vaccinated (1 week after the last boost) and naive animals and enriched the adherent myeloid cells by washout of the suspension cells after 2 hours of culture. Cytokine indicators of trained immunity (TNF-α, IL-6), an agonist of viral coreceptor CCR5 (MIP1α), and a cytokine unrelated to trained immunity (IL-8) were measured after 18 hours of stimulation with SHIV. We found that the expression of TNF-α, IL-6, and MIP1α (but not IL-8, an internal negative control) was higher in the vaccinated group than in the naive group upon SHIV stimulation (Figure 7, A–D). We also evaluated the SHIV Gag RNA expression in the cells exposed to SHIV as a measure of viral replication. The median relative Gag level was 3.54 in the naive controls and only 1.99 in the vaccinated group (P = 0.1 by nonparametric Mann-Whitney test). Interestingly, TNF-α, IL-6, and MIP1α showed inverse correlations (or trends) with viral Gag expression (Figure 7, E–G), suggesting the enhanced production of TNF-α, IL-6, and MIP1α by monocytes was associated with inhibition of viral replication. These inverse correlations also suggested that the cytokine/chemokine difference cannot be simply related to infection levels across these samples, or Gag would have been expected to positively correlate. IL-8 as a negative control did not show any association with Gag expression (Figure 7H). Moreover, the expression of IL-6 and MIP1α was positively correlated with numbers of viral exposures required for the animals to become infected in vivo (Figure 7, J and K), but not TNF-α and IL-8 (Figure 7, I and L). Besides the original trained-immunity markers, TNF-α and IL-6, MIP1α might be another marker of trained immunity against HIV infection. Overall, increased expression of 2 indicators of trained immunity (TNF-α and IL-6) were found in the monocytes from vaccinated animals compared with those from the naive animals. The enhanced expression of TNF-α and IL-6 demonstrated associations with decreased viral Gag expression as a measure of virus replication in the ex vivo restimulation system, and/or with reduced viral acquisition in vivo. Though further supporting data are needed, our results support the hypothesis that trained immunity mediated by monocytes might be involved in mediating reduction of viral acquisition.
Figure 7. Enhanced expression of trained-immunity markers was inversely correlated with SHIV Gag expression ex vivo and positively with number of viral exposures in vivo.
PBMC samples collected from the vaccinated (1 week after the last boost) and naive animals were thawed, and the monocytes were enriched by 3 washes with warm PBS to remove the suspended cells after 2 hours of adherence to 48-well plates. The adherent monocyte-enriched cell populations were then cocultured with SHIV (1:100) for 18 hours. The supernatant was collected and the production of TNF-α, IL-6 (both as classical trained-immunity markers), MIP1α (SHIV coreceptor agonist), and IL-8 (negative control unrelated to trained immunity) were measured. The cells were further cultured for 2 more days before the expression levels of SHIV Gag in the cells of each sample were measured by qPCR. (A–D) The protein levels of TNF-α, IL-6, MIP1α, and IL-8 were compared between the naive and vaccinated animals. Mann-Whitney test was used for comparisons. Mean ± SEM are shown. (E–H) The cellular expression of SHIV Gag RNA was inversely correlated with TNF-α, IL-6 (trend), and MIP1α (but not with IL-8) production in the supernatant. Spearman’s r and P values of the correlations are indicated. (I–L) IL-6 and MIP1α (but not TNF-α or IL-8) were positively correlated with the number of viral exposures required for the animals to become infected in vivo. Jonckheere-Terpstra test was used to calculate the r and P values. Naive n = 7, shown in red triangles; vaccinated n = 21, shown in black dots. NS, not significant.
Discussion
Antibody against HIV/SIV Env, whether it be neutralizing or V2-binding, has been widely thought to be essential to prevent acquisition of HIV or SIV infection. In contrast, we demonstrated here that the mucosal vaccine composed of MVA and rhFLSC coinjected with TLR agonists, IL-15, and mLT mediated significant protection, or reduced risk of acquisition against SHIV intrarectal viral challenges. These 2 vaccine-challenge studies conclusively demonstrated that SHIV vaccine efficacy could be achieved without anti-Env antibody responses, and therefore, could be mediated by adaptive cellular and/or innate immunity. Further delineating the immune correlates will inform future HIV vaccine design.
HIV-specific T cell responses have been detected in a significant proportion of exposed but uninfected individuals, and certain T cell response patterns correlate with infection risk (29). Rectal virus-specific CD8+ T cells, positioned at the mucosal site of viral entry, play critical roles in preventing and/or clearing of viral infection (30–33). Here, we found that mucosal Gag- and Env-specific T cell responses were elicited in both cohorts, although the magnitude of the responses did not correlate with viral acquisition, indicating mucosal T cells alone did not directly prevent virus transmission. This is consistent with the recent finding that rectal CD8+ T cells have lower expression levels of perforin and GrzB compared with blood counterparts, and therefore are not programmed primarily for cytotoxicity (34). Since mucosal virus-specific CD8+ T cells are intrinsically biased toward chemokine/cytokine production rather than cytotoxic responses (35, 36), it is likely that upon viral challenge, the vaccine-induced virus-specific T cells released chemokines and cytokines, which either directly blocked the viral infection (such as MIP-1α and 1β) or initiated innate immune responses to limit viral acquisition. In addition, correlation of T cell responses with protection may be masked by negative regulatory factors such as MDSCs that affect T cell efficacy, requiring both to be considered. However, a simple ratio of T cell to MDSC numbers was not predictive of protection. Nevertheless, infection of all 6 adjuvant-only controls suggested that there was some role for adaptive immunity. Thus, a combination of T cell immunity and innate immunity may determine protection.
In both cohorts, the viral challenges were performed 7–8 weeks after the last boost. The long interval between vaccination and challenge suggested the involvement of adaptive immunity. However, the lack of associations of anti-Env antibody and virus-specific T cell responses with viral acquisition argued against this notion, suggesting a key role of innate immunity. We have shown in our previous study that APOBEC3G, combined with virus-specific polyfunctional T cell responses, correlated with viral control after SIV infection (4). However, innate immunity alone traditionally was considered to lack antigen specificity and long-term memory. Recently, this concept was challenged by studies showing that certain stimuli can prime innate immune cells, especially the myeloid cells, to respond to future stimuli more strongly (8). Experimental and proof-of-concept clinical studies have demonstrated that protection against infections could be achieved in vaccination models independently of lymphocytes (37). This phenomenon of mammalian innate immunity exhibiting an immunological memory of past insults is termed trained (innate) immunity (8), which is regulated through epigenetic programming (38, 39). These epigenetic and associated metabolic changes led to enhanced (training) or decreased (tolerance) cytokine production, depending on the insults (26, 38, 39). Since myeloid cells, including monocytes and macrophages, are among the main innate immune cells that can be trained (26, 38, 40), we examined these cells in our studies. We found that 2 subsets of myeloid cells were associated with viral acquisition. One was the MDSCs, which we have shown in our previous study to dampen the protective immunity against VL (19). Both CD14+ and CD15+ MDSCs were increased in the PBMCs of the vaccinated animals, and only CD15+ MDSCs inversely correlated with reduced risk of viral acquisition. As MDSCs can be infected by SIV in vivo (41), the correlation could be the result of MDSCs suppressing the protective immunity, or MDSCs directly acting as viral target cells. The other subset of myeloid cells was the CD14+DR– monocytes in the colorectal intraepithelial compartment, which was dramatically increased after vaccination. These cells had possibly newly migrated from blood, as the increase of these cells was concurrent with the disappearance of CD14+ MDSCs in the PBMCs. The chemokines released by antigen-specific T cells in the colorectal mucosa might contribute to the recruitment. Once these myeloid cells entered the gut, they could differentiate into different cell types, such as DCs, or M1/M2 macrophages depending on the gut microenvironment, and differentially influence host defense (38). Notably, another poxvirus vaccine has just been found to induce protection associated with CD14+ monocytes, which were hypothesized to provide trained immunity working in combination with anti-V2 antibodies to protect (42). Immunological imprinting of either tolerance or trained immunity determines the functional fates of the macrophages and susceptibility to secondary infections (38). It is tempting to hypothesize that these myeloid cells might constitute the trained innate immunity, and mediate protection in these 2 cohorts. Since trained immunity was mostly studied in BCG and Candida infections, LPS or glucans were used as surrogate stimulation reagents for bacteria or fungi. When we investigated the roles of trained immunity in protecting against SHIV infections, stimulation with SHIV would better predict outcomes of the in vivo challenges. Therefore, we stimulated with SHIV instead of LPS or glucan, and measured MIP1α, and viral Gag expression besides traditional trained-immunity markers TNF-α and IL-6. We found in our ex vivo study that upon SHIV stimulation, the monocyte/macrophage–enriched cell population in the vaccinated animals produced more TNF-α, IL-6, and MIP1α than those of the naive ones, and most importantly, the enhanced expression of cytokines/chemokines also correlated with the control of ex vivo SHIV replication and/or reduced in vivo viral infection risks. This suggested that myeloid cell–mediated trained immunity might be involved in protection. However, whether this was through the vaccine-mediated epigenetic modification of the monocyte/macrophage, and whether part of the vaccine and/or the whole vaccine platform is necessary for the enhanced production of cytokines/chemokines upon SHIV stimulation, remain unknown. Further vaccine component breakdown studies and characterization of the epigenetic modification of these myeloid cells are underway and will provide more information on the involvement of trained immunity. Nevertheless, we did observe that induction of CD14+ monocytes was dependent on the MVA component of the vaccine (Figure 5G). Whether MVA alone was sufficient for the whole process of training is unknown and will be determined by further epigenetic studies. Because trained immunity is less specific than adaptive immunity, we expect that training by one virus could provide innate protection against some other viruses.
We used proportional hazards modeling to investigate the number of viral exposures before infection as a function of multiple factors. Because of the size of the challenge study (28 animals with the primary outcomes), we limited the models to at most 3 factors. The association with IL-6 seen in Figure 7J was a component of all the most significant models, and the association with CD15+ MDSCs seen in Figure 4D made a secondary, weaker contribution while reducing the study size to 21. No other factors added substantial independent improvements or interactions to the models.
Bacteria, viruses, and proinflammatory cytokines induce trained immunity in myeloid cells (26). Notably, commensal microbiota are required to program the innate immune cells, and the H3K4m3 histone modification of inflammatory response genes in NK cells is increased in normal mice compared with germ-free ones (43, 44). In line with this, gut microbiota have been shown to impact immune cell development and differentiation, and thus affect the immunogenicity and clinical efficacy of some vaccines (45, 46). Our mucosal vaccine, composed of poxvirus, rhFLSC, TLR ligands, mLT, and IL-15, had the capacity to potentially train innate immunity, and thus influence the vaccine efficacy. Moreover, since the mucosal vaccine was either delivered directly to the colorectal lumen or given orally but also targeting the colorectal mucosa, the vaccine itself could physically interact with the gut microbiome, and alter the bacterial composition. Trained monocytes, in turn, have been shown to cause metabolic changes including the induction of HIF1α (47), which promoted MDSC accumulation (48), and enhanced aerobic glycolysis (47, 49), which might affect the gut microbiome composition. Indeed, the richness and the composition of the gut microbiome were significantly different between the vaccinated animals and the naive ones, in contrast to no difference before vaccination. Interestingly, CD14+DR– monocytes in the colorectal mucosa also correlated with bacterial PCA-1 and richness at both genus and species levels. Determining whether the vaccine-altered gut microbiota functionally reprogrammed the myeloid cells and/or NK cells will help to evaluate the relative contribution of trained immunity to protection. Overall, the mucosal vaccination had a profound impact on the gut microenvironments, and caused the alteration of the gut microbiome, total plasma cells (possibly related to microbiome changes), Tfh, and CD14+DR– monocytes, which altogether might contribute to reduced risk of viral acquisition.
Since Env-specific non-neutralizing antibodies (nnAbs) have been identified as a protective immune correlate in the RV144 trial, great efforts have been made to test whether such nnAbs could mediate protection/delay against SHIV acquisition using adoptive transfer macaque models. However, so far, there is no proof of concept that such nnAbs mediated protection against viral acquisition (50–52). One study showed stronger antibody responses associated with less or no protection (53). In other studies the nnAbs were found to limit the number of founder viruses (54), and reduce (55) or not reduce VL after infection (56). The mechanistic role of nnAbs in protection against HIV infection remains undefined.
In the absence of detectable anti-Env antibodies, viral acquisition in the vaccinated macaques was significantly delayed or prevented. This opens a new avenue of HIV vaccine development. Mechanistically, this mucosal vaccine platform is complementary to the RV144 vaccine platform, so it is tempting to speculate that the combination of both might lead to enhanced vaccine efficacy, although an RV144-like regimen may also induce protection through monocytes and antibodies (42). Even more interesting is that our data suggested that the mucosal vaccine might induce trained innate immunity, which, alone or together with T cells, mediated the protection/reduced risk of viral acquisition against SHIV challenges. Adaptive HIV- and SIV-specific T cells may help confer specificity to trained immunity to target relevant HIV- and SIV-infected cells or virus. To our knowledge, this and a concurrent study (42) (but which also required antibodies) are the first demonstrations that trained immunity against HIV is possible via vaccination, in the present case even without antibodies. Here we also provide more evidence to indicate induction of trained innate immunity (Figure 7). Moreover, our findings provide insights for interpreting other reported vaccine effects achieved with complex immunization regimens. Overall, the induced trained immunity may be a new opportunity for innovative HIV vaccine design.
Methods
Animals.
Two cohorts of a total of 66 adult Indian rhesus macaques (Macaca mulatta) were housed in the NCI animal facility. All macaques were free of SIV, simian retroviruses 1, 2, and 5, and simian T cell leukemia/lymphotropic virus type 1 before the study. The basic information on sex, age, weight, and major histocompatibility complex alleles of the animals is shown in Supplemental Tables 1 and 4. Twenty-nine control animals (including the control animals in the first cohort), which were challenged with intrarectal inoculation of 1 high dose (1:5 diluted) of SHIVSF162P4 stock, were used for analysis in the first cohort. Twenty-two of them were housed in the NCI animal facility, while 7 of them were maintained in Advanced BioScience Laboratories. The second cohort of 30 adult Indian rhesus macaques was housed in, and approved by, the University of Maryland animal facility.
Development of Eudragit-coated microparticle/nanoparticle formulation oral delivery system.
We had previously shown in mice that to protect against rectal mucosal challenge with a virus without antibody neutralization, a local mucosal T cell immune response was necessary (30), and that the most effective way to induce that in mice and macaques was by direct intrarectal immunization (31, 32). However, intrarectal immunization has not been considered practical for widespread human use. Oral administration would be much more practical but is hindered by the need to protect the vaccine against acid and enzymes in the stomach and small intestine and successfully deliver it to the large intestine or colon. To accomplish this, we have designed and developed a new mucosal vaccine delivery strategy that allows colorectal immunization through the oral route (10). We incorporated antigen and adjuvants into nanoparticles of the appropriate size for uptake by DCs, and then coated these with a Eudragit coating (forming microparticles) that protects them from stomach acid and releases them selectively at the pH in the small or large intestine. Using the formulation to deliver the vaccine contents to the large intestine, we found that we could mimic the efficacy of intrarectal delivery to protect against intrarectal challenge with a recombinant vaccinia virus expressing the HIV Env protein in the mouse model (10).
To translate this approach to rhesus macaques, we have optimized 2 formulations: Eudragit FS-30D single-coated BSA-FITC and double-coated BSA–Alexa 647 microparticles. The 2 types of microparticles were given orally to the same macaque, and followed by necropsy after 24 hours. Single-cell suspensions from the duodenum, jejunum, ileum, caecum; ascending, transverse, and descending colons; and rectum were collected. Flow cytometry was run to identify the fluorescently labeled particles in live cells. We found that the double-coated rather than the single-coated microparticles mainly released their contents for uptake in the large intestine (Supplemental Figure 1). Therefore, we used the double-coated formula for oral delivery of our mucosal SHIV vaccines. The double-coated formula was manufactured at Nanotherapeutics, Inc. under a Collaborative Research and Development Agreement. PLGA nanoparticles were manufactured with NanoDRY technology (US Patent 8,501,232). Chloroform solution (0.9 ml) containing 9 mg of PLGA was prepared, followed by addition of 0.1–0.2 mg of antigen in 0.1 ml of water to form the primary emulsion. The PLGA mixture was emulsified with a vortex mixer for 30 seconds to form the water-in-oil emulsion. The mixture emulsion was added dropwise into 9 ml of isopropanol in a small stainless-steel bowl agitated at low-frequency sonication (60 hertz, 60% max) with 4 MOJO beads (1-cm ceramic marbles). Nanoparticles were precipitated at 300 to 400 Torr for 15 minutes at 25°C. The suspension was allowed to settle for 1 hour in a freezer followed by centrifugation and the supernatant removed. EUDRAGIT FS30D (Evonik) in isopropanol was used to coat PLGA nanoparticles, with 20 mg of FS30D microparticles containing 200 μg of antigen (1% loading).
Immunization of the first and third cohorts.
Rhesus macaques were immunized with a combination of a peptide vaccine, MVA expressing SIV antigens, and a protein vaccine consisting of the macaque FLSC fusion protein of HIV-gp120 fused to 2 domains of macaque CD4 to expose CD4-induced epitopes (5). The SIV/HIV peptide vaccine has been described in previous studies (4, 19). In both cohorts, recombinant MVA viruses expressing SIVmac239 Gag, Pol, Env, Rev, Tat, and Nef (5 × 108/immunization for each) (57) were administered intracolorectally with different adjuvants. The adjuvants included a combination of 500 μg per dose of D-type CpG oligodeoxynucleotide (TLR9 agonist), 10 μg per dose of MALP2 (TLR2/6 agonist), 1 mg per dose of PolyI:C (Invivogen) (TLR3 agonist), and 300 μg per dose of recombinant human IL-15 (Peprotech). The peptide vaccine incorporated 0.5 mg of each peptide mixed with DOTAP (Roche, 100 μl per dose), with a mixture of molecular adjuvants consisting of IL-15 and ligands cited above for TLR2/6, -3, and -9 that induce innate as well as adaptive immunity against SIV (3). The rhFLSC vaccine contained rhFLSC (at 100 μg per dose) mixed with mutant E. coli labile toxin R192G (mLT) (at 100 μg per dose) (58). The peptide and rhFLSC vaccines were prepared in nanoparticle format or not as indicated in Figures 1 and 3, and Supplemental Table 2. Intracolorectal inoculations were performed as previously described (4).
The details of the immunization protocol and groups of the first cohort are shown in Figure 1 and Supplemental Tables 1 and 2. Group 1 (T cell vaccine): Pep/MVA + TLRLs + IL-15; group 2 (FLSC vaccine): rhFLSC + mLT; group 3 and 4: the combination of group 1 and 2 vaccines in oral nanoparticle format (group 4) or direct intrarectal delivery (group 3). Adjuvant-only and naive animals were designated as groups 5 and 6, respectively. The animals were primed at week 0, 3, and 6, and boosted at week 13 and 16.
For the immunization of the third cohort, 7 macaques served as naive control, while 21 animals were the vaccinees (Supplemental Table 4 and Figure 2). The same doses of vaccine were given in the third cohort except for rhFLSC, in which 200 μg instead of 100 μg per dose was mixed with mLT (at 100 μg mLT per dose). MVA with adjuvant intrarectally along with rhFLSC plus mLT in oral nanoparticles were used for priming at week 0 and 4, followed by boosting with rhFLSC plus mLT in oral nanoparticles at week 8 and 12.
SHIV challenge of the first and third cohorts.
Seven weeks after the last boost, all animals in the first cohort received an intrarectal inoculation of 1 high dose (1:5 diluted) of SHIVSF162P4 stock (M661-derived harvest 1 dated October 5, 2006), which was provided by Nancy Miller, National Institute of Allergy and Infectious Diseases (NIAID), NIH.
Eight weeks after the last boost, all animals in the third cohort were intrarectally exposed to 8 weekly low-dose SHIVSF162P4 (1:35 diluted) from the same stock until they were infected. Once the animals were confirmed infected, they were not challenged further. SIV RNA levels determined by nucleic acid sequence–based amplification (NASBA) assays, and absolute CD4+ T cell counts were monitored for 6–14 months after infection by Advanced BioScience Laboratories. The cutoff threshold for viral RNA detection was 50 copies/ml.
Immunization of the second cohort.
A cohort of 30 Indian rhesus macaques was divided into 5 groups (each with n = 6 macaques). Group 1 was the naive group, while groups 2–5 were immunized intrarectally with 300 μg of rhFLSC, and with varying doses of wild-type CT (0, 10, 100, and 300 μg). Four immunizations were given at week 0, 6, 14, and 18. Three weeks after the last immunization, plasma was collected to measure the antibody titers against rhFLSC and CT. Because we did not observe anti-rhFLSC antibodies in the CT titration study we did not challenge the macaques.
Flow cytometric analysis of virus-specific T cell responses, Env-specific B cell responses, Tfh, MDSCs, and immune activation in PBMCs, MLNs, and colorectal tissues.
Virus-specific T cells were measured in mononuclear cells isolated from colorectal lamina propria and MLNs by flow cytometric intracellular cytokine analysis, as previously described in detail (4, 59). Tfh cells were measured in MLNs 4 weeks after the last vaccination. For dextramer staining, we used Mamu-A*01–positive CM9 dextramer obtained from ImmuDex. MDSCs in PBMCs were measured freshly using blood processed with Lympholyte-H Cell Separation Media (Cedarlane) as previously described (41). The immune activation in the colorectal tissues was also measured freshly after processing the tissues as described previously (4, 59). The details of the processing of the colorectal tissues is described in the Supplemental Methods. For MDSC and immune activation measurements, cells were first incubated with Fc receptor (FcR) blocking reagent (Miltenyi Biotec) to saturate FcR and then labeled with monoclonal antibodies. The gating strategy and the definition of CD14+ (CD33+CD11b+DR–CD14+) and CD15+ (CD33+CD11b+DR–CD15+Lin–) MDSCs were as described in a previous study (41). Yellow viability dye was used to distinguish the live/dead cells (Invitrogen). For Env-specific memory B cell, plasmablast (PB), and plasma cell (PC) flow cytometry assays, details are provided in the Supplemental Methods. Mucosal PBs were identified as CD19+CD20+/–HLA-DR+Ki67+IRF4+CD138+/– and mucosal PCs as CD19+CD20–HLA-DR–Ki67–IRF4+CD138+ as previously described in mucosal tissues (60). The antibody information is listed in Supplemental Table 7. Data acquisition was performed with a BD LSR II flow cytometer, and data were analyzed with FlowJo software (Tree Star Inc). Env-specific memory B cells were defined as CD19+CD20+CD21+/−IgD−gp120+ B cells, as described previously (61).
Measurements of rhFLSC-binding antibodies in rectal secretions and rectal tissue explants.
Rectal sample processing has been described previously and is provided in the Supplemental Methods (62, 63). rhFLSC-specific IgA and IgG antibody titers were measured by ELISA using recombinant rhFLSC protein (17). The titer was defined as the reciprocal of the sample dilution at which the optical density (OD) of the test serum was 2 times greater than that of the negative-control samples obtained from naive unvaccinated macaques (64). To account for varying amounts of total immunoglobulin in samples, total concentrations of IgA and IgG were measured in each sample by ELISA and used to normalize rhFLSC-specific IgA and IgG titers. Details of the protocol are provided in the Supplemental Methods. rhFLSC-specific IgA and IgG are expressed as titer of rhFLSC-specific IgA/G per μg of total IgA/G (65).
Gut microbiome analysis.
In the third cohort, fecal samples were collected 1 week before the SHIV challenges. A Qiagen DNA Stool Mini Kit was used for DNA extraction from stool samples. The sequencing was performed on the V4 region of the 16S rDNA gene (515F–806R) using the Illumina MiSeq platform (66). Chimeric sequences were filtered out of the FASTQ files containing the 16S rRNA gene sequences using the USEARCH (version 8.1.1831) utility’s UCHIME implementation and the ‘gold’ database (version microbiomeutil-r20110519) (67, 68). The reads, thus filtered, were then binned into operational taxonomic units (OTUs) at 97% similarity using USEARCH’s cluster_otus command. QIIME (1.9.1) scripts were used to classify and align the obtained OTUs (69). The assign_taxonomy.py script was used to assign taxonomy using the default RDP method and GreenGenes database (http://qiime.org/home_static/dataFiles.html).
In vitro/ex vivo SHIV stimulation experiments.
Frozen PBMCs collected from vaccinated (1 week after last boost) and naive animals were thawed, washed, and diluted to 5 × 106/ml. PBMCs (400 μl/well) were added to 48-well plates and incubated for 2 hours before washing with warm PBS to remove the cells in suspension. For in vitro assays, PBMCs from naive animals were used. Different components of the vaccine, or medium, or glucan were added to stimulate the cells for 18 hours. After 3 washes to eliminate the stimulators, the cells were cultured in R10 medium with 10% human AB serum, with medium changed every 3 days. On day 6, the monocytes/macrophages were collected and stained with antibody mix after treatment with Versene for 1 hour to detach adherent cells. The cells were then analyzed by flow cytometry. For ex vivo study, the monocyte-enriched cells were then stimulated with SHIV (1:100 diluted, in vivo challenge stock virus) for 18 hours. Supernatants were collected and TNF-α, IL-6, IL-8, and MIP1α were measured using ELISA kits from R&D Systems. The cells were washed 3 times with warm PBS to remove SHIV on the cell surface. The cells were cultured for another 48 hours with fresh R10 medium added before the cells were lysed, and RNA was isolated using Qiagen RNA isolation kits. For measuring Gag expression and GAPDH, TaqMan probe and primer sets were used (Thermo Fisher Scientific). RNA transcription kits and qPCR mixture were from Bioline USA Inc. Relative mRNA expression levels were compared by the comparative threshold cycle (Ct) method of relative quantification (PerkinElmer User Bulletin no. 2) as described previously (41). After normalization to GAPDH, Gag expression in one animal was used as the reference to calculate the Gag fold changes in the rest of the animals.
Statistics.
We performed statistical analyses with Prism version 6 (GraphPad). Mann-Whitney and Wilcoxon tests were used as shown in the figures. Spearman’s analysis and Jonckheere-Terpstra tests were used for correlations. A 2-sided significance level of 0.05 was used for all analyses. PCA was performed with Qlucore Omics Explorer or SAS.
Study approval.
All animals were maintained in accordance with guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International and in protocols approved by the NCI Animal Care and Use Committee, or the Animal Care and Use Committee of the Institute of Human Virology, University of Maryland.
Author contributions
YS, JAB, and RCG conceived and designed the projects, and interpreted the data for the first and third studies. GKL, A. DeVico, and RCG designed, ran experiments, and/or interpreted the data for the second study. For the first and third studies, YS, YW, KB, and BF processed samples. YS, A. Dzutsev, GT, DVI, VM, TM, MRG, XS, GDT, and JAB performed and interpreted experiments. GKL, A. DeVico, and TF provided the rhFLSC protein. JK, RCW, and JT optimized and produced the nanoparticle vaccine. DK synthesized and provided D-type CpG oligonucleotides and contributed to experimental planning. JC provided the mLT. GF provided the seeds of the MVA-SIV. YS and DV performed statistical analyses. YS and JAB wrote the manuscript with input from all the authors.
Supplementary Material
Acknowledgments
We thank H.K. Chung, R. Pal, and S. Orndorff from Advanced BioScience Laboratories, Inc. for the viral load and CD4 assays; A. Hogg and H. Yu for tissue processing; H.X. Liao and the Duke Human Vaccine Institute Protein Production Facility for providing Rh CH38 IgA; D. Watkins (University of Miami) for MHC typing; Nancy Miller (NIAID) for providing SHIVSsf162P4 challenge stock; and Chris Miller (UC Davis) for helpful discussion. This work was supported by the Intramural Program of the NIH/NCI Center for Cancer Research funding Z01 SC 004020 (to JAB), NIH NIAID HHSN 272201100016C, NIAID/NIH P01AI124912 (to RCG, GKL, and A. DeVico), and NCI innovation award (to YS).
Version 1. 02/18/2019
Electronic publication
Version 2. 03/01/2019
Print issue publication
Funding Statement
This work was supported by the Intramural Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research funding Z01 SC 004020 to Jay B. Berzofsky and NIH NIAID HHSN 272201100016C.
Footnotes
RCW’s present address is: Ology Bioservices, Inc.
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(3):1314–1328.https://doi.org/10.1172/JCI122110.
Contributor Information
Yongjun Sui, Email: suiy@mail.nih.gov.
George K. Lewis, Email: glewis@ihv.umaryland.edu.
Yichuan Wang, Email: yichuan.wang@nih.gov.
Kurt Berckmueller, Email: keberckmueller@email.wm.edu.
Amiran Dzutsev, Email: Dzutsev@pop.nci.nih.gov.
Venkatramanan Mohanram, Email: venkatramanan81@gmail.com.
Thomas Musich, Email: musichta@hiv-th.org.
Xiaoying Shen, Email: sxshen@duke.edu.
Anthony DeVico, Email: adevico@ihv.umaryland.edu.
Timothy Fouts, Email: timothy.fouts@ablinc.com.
James Kirk, Email: jkirk99@msn.com.
Dennis Klinman, Email: klinmand@mail.nih.gov.
Georgia D. Tomaras, Email: georgia.tomaras@duke.edu.
Genoveffa Franchini, Email: franchig@mail.nih.gov.
Marjorie Robert-Guroff, Email: guroffm@mail.nih.gov.
Giorgio Trinchieri, Email: trinchig@mail.nih.gov.
Jay A. Berzofsky, Email: berzofsk@helix.nih.gov.
References
- 1.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361(23):2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366(14):1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sui Y, et al. TLR agonists and/or IL-15 adjuvanted mucosal SIV vaccine reduced gut CD4+ memory T cell loss in SIVmac251-challenged rhesus macaques. Vaccine. 2011;30(1):59–68. doi: 10.1016/j.vaccine.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sui Y, et al. Innate and adaptive immune correlates of vaccine and adjuvant-induced control of mucosal transmission of SIV in macaques. Proc Natl Acad Sci U S A. 2010;107(21):9843–9848. doi: 10.1073/pnas.0911932107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeVico A, et al. Antibodies to CD4-induced sites in HIV gp120 correlate with the control of SHIV challenge in macaques vaccinated with subunit immunogens. Proc Natl Acad Sci U S A. 2007;104(44):17477–17482. doi: 10.1073/pnas.0707399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouts TR, et al. Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. Proc Natl Acad Sci U S A. 2015;112(9):E992–E999. doi: 10.1073/pnas.1423669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon SN, et al. Boosting of ALVAC-SIV vaccine-primed macaques with the CD4-SIVgp120 fusion protein elicits antibodies to V2 associated with a decreased risk of SIVmac251 acquisition. J Immunol. 2016;197(7):2726–2737. doi: 10.4049/jimmunol.1600674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe. 2011;9(5):355–361. doi: 10.1016/j.chom.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Netea MG, et al. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352(6284):aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Q, et al. Large intestine-targeted, nanoparticle-releasing oral vaccine to control genitorectal viral infection. Nat Med. 2012;18(8):1291–1296. doi: 10.1038/nm.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagley KC, et al. Immunogenicity of DNA vaccines that direct the coincident expression of the 120 kDa glycoprotein of human immunodeficiency virus and the catalytic domain of cholera toxin. Vaccine. 2003;21(23):3335–3341. doi: 10.1016/S0264-410X(03)00038-0. [DOI] [PubMed] [Google Scholar]
- 12.Fouts TR, et al. Progress toward the development of a bacterial vaccine vector that induces high-titer long-lived broadly neutralizing antibodies against HIV-1. FEMS Immunol Med Microbiol. 2003;37(2–3):129–134. doi: 10.1016/S0928-8244(03)00067-1. [DOI] [PubMed] [Google Scholar]
- 13.Bagley KC, Lewis GK, Fouts TR. Adjuvant activity of the catalytic A1 domain of cholera toxin for retroviral antigens delivered by GeneGun. Clin Vaccine Immunol. 2011;18(6):922–930. doi: 10.1128/CVI.05019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis GK, DeVico AL, Gallo RC. Antibody persistence and T-cell balance: two key factors confronting HIV vaccine development. Proc Natl Acad Sci U S A. 2014;111(44):15614–15621. doi: 10.1073/pnas.1413550111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douce G, et al. Mutants of Escherichia coli heat-labile toxin lacking ADP-ribosyltransferase activity act as nontoxic, mucosal adjuvants. Proc Natl Acad Sci U S A. 1995;92(5):1644–1648. doi: 10.1073/pnas.92.5.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontana MR, et al. Construction of nontoxic derivatives of cholera toxin and characterization of the immunological response against the A subunit. Infect Immun. 1995;63(6):2356–2360. doi: 10.1128/iai.63.6.2356-2360.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas MA, et al. HIV-1 CD4-induced (CD4i) gp120 epitope vaccines promote B and T-cell responses that contribute to reduced viral loads in rhesus macaques. Virology. 2014;471-473:81–92. doi: 10.1016/j.virol.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz JA, et al. An HIV gp120-CD4 immunogen does not elicit autoimmune antibody responses in cynomolgus macaques. Clin Vaccine Immunol. 2016;23(7):618–627. doi: 10.1128/CVI.00115-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sui Y, et al. Vaccine-induced myeloid cell population dampens protective immunity to SIV. J Clin Invest. 2014;124(6):2538–2549. doi: 10.1172/JCI73518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Permar SR, Staats HF. Which comes first: the antigen or the adjuvant? J Clin Invest. 2014;124(6):2364–2365. doi: 10.1172/JCI76263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carnathan DG, et al. Activated CD4+CCR5+ T cells in the rectum predict increased SIV acquisition in SIVGag/Tat-vaccinated rhesus macaques. Proc Natl Acad Sci U S A. 2015;112(2):518–523. doi: 10.1073/pnas.1407466112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sui Y, et al. Early SIV dissemination after intrarectal SIVmac251 challenge was associated with proliferating virus-susceptible cells in the colorectum. J Acquir Immune Defic Syndr. 2016;71(4):353–358. doi: 10.1097/QAI.0000000000000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bain CC, et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013;6(3):498–510. doi: 10.1038/mi.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubinak JL, et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe. 2015;17(2):153–163. doi: 10.1016/j.chom.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sui Y, et al. Influence of gut microbiome on mucosal immune activation and SHIV viral transmission in naive macaques. Mucosal Immunol. 2018;11(4):1219–1229. doi: 10.1038/s41385-018-0029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ifrim DC, et al. Trained immunity or tolerance: opposing functional programs induced in human monocytes after engagement of various pattern recognition receptors. Clin Vaccine Immunol. 2014;21(4):534–545. doi: 10.1128/CVI.00688-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinnijenhuis J, et al. Long-lasting effects of BCG vaccination on both heterologous Th1/Th17 responses and innate trained immunity. J Innate Immun. 2014;6(2):152–158. doi: 10.1159/000355628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufmann E, et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell. 2018;172(1–2):176–190.e19. doi: 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 29.Kuebler PJ, et al. Cellular immune correlates analysis of an HIV-1 preexposure prophylaxis trial. Proc Natl Acad Sci U S A. 2015;112(27):8379–8384. doi: 10.1073/pnas.1501443112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belyakov IM, et al. The importance of local mucosal HIV-specific CD8(+) cytotoxic T lymphocytes for resistance to mucosal viral transmission in mice and enhancement of resistance by local administration of IL-12. J Clin Invest. 1998;102(12):2072–2081. doi: 10.1172/JCI5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belyakov IM, et al. Mucosal immunization with HIV-1 peptide vaccine induces mucosal and systemic cytotoxic T lymphocytes and protective immunity in mice against intrarectal recombinant HIV-vaccinia challenge. Proc Natl Acad Sci U S A. 1998;95(4):1709–1714. doi: 10.1073/pnas.95.4.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belyakov IM, et al. Mucosal AIDS vaccine reduces disease and viral load in gut reservoir and blood after mucosal infection of macaques. Nat Med. 2001;7(12):1320–1326. doi: 10.1038/nm1201-1320. [DOI] [PubMed] [Google Scholar]
- 33.Xu H, et al. Mucosal vaccination with heterologous viral vectored vaccine targeting subdominant SIV accessory antigens strongly inhibits early viral replication. EBioMedicine. 2017;18:204–215. doi: 10.1016/j.ebiom.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiniry BE, et al. Predominance of weakly cytotoxic, T-betLowEomesNeg CD8+ T-cells in human gastrointestinal mucosa: implications for HIV infection. Mucosal Immunol. 2017;10(4):1008–1020. doi: 10.1038/mi.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shacklett BL, Anton PA. HIV Infection and gut mucosal immune function: updates on pathogenesis with implications for management and intervention. Curr Infect Dis Rep. 2010;12(1):19–27. doi: 10.1007/s11908-009-0072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shacklett BL, Ferre AL. Mucosal immunity in HIV controllers: the right place at the right time. Curr Opin HIV AIDS. 2011;6(3):202–207. doi: 10.1097/COH.0b013e3283453e2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintin J, Cheng SC, van der Meer JW, Netea MG. Innate immune memory: towards a better understanding of host defense mechanisms. Curr Opin Immunol. 2014;29:1–7. doi: 10.1016/j.coi.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Saeed S, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345(6204):1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Heijden CDCC, Noz MP, Joosten LAB, Netea MG, Riksen NP, Keating ST. Epigenetics and trained immunity. Antioxid Redox Signal. 2018;29(11):1023–1040. doi: 10.1089/ars.2017.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Meer JW, Joosten LA, Riksen N, Netea MG. Trained immunity: A smart way to enhance innate immune defence. Mol Immunol. 2015;68(1):40–44. doi: 10.1016/j.molimm.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 41.Sui Y, et al. Paradoxical myeloid-derived suppressor cell reduction in the bone marrow of SIV chronically infected macaques. PLoS Pathog. 2017;13(5):e1006395. doi: 10.1371/journal.ppat.1006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaccari M, et al. HIV vaccine candidate activation of hypoxia and the inflammasome in CD14+ monocytes is associated with a decreased risk of SIVmac251 acquisition. Nat Med. 2018;24(6):847–856. doi: 10.1038/s41591-018-0025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarke TB. Microbial programming of systemic innate immunity and resistance to infection. PLoS Pathog. 2014;10(12):e1004506. doi: 10.1371/journal.ppat.1004506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganal SC, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37(1):171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 45.Valdez Y, Brown EM, Finlay BB. Influence of the microbiota on vaccine effectiveness. Trends Immunol. 2014;35(11):526–537. doi: 10.1016/j.it.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Oh JZ, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41(3):478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng SC, et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345(6204):1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiu DK, et al. Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat Commun. 2017;8(1):517. doi: 10.1038/s41467-017-00530-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kleinnijenhuis J, et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci U S A. 2012;109(43):17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burton DR, et al. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc Natl Acad Sci U S A. 2011;108(27):11181–11186. doi: 10.1073/pnas.1103012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dugast AS, et al. Lack of protection following passive transfer of polyclonal highly functional low-dose non-neutralizing antibodies. PLoS ONE. 2014;9(5):e97229. doi: 10.1371/journal.pone.0097229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bruel T, et al. Lack of ADCC breadth of human nonneutralizing anti-HIV-1 antibodies. J Virol. 2017;91(8):e02440-16. doi: 10.1128/JVI.02440-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaccari M, et al. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat Med. 2016;22(7):762–770. doi: 10.1038/nm.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santra S, et al. Human non-neutralizing HIV-1 envelope monoclonal antibodies limit the number of founder viruses during SHIV mucosal infection in rhesus macaques. PLoS Pathog. 2015;11(8):e1005042. doi: 10.1371/journal.ppat.1005042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hessell AJ, et al. Reduced cell-associated DNA improved viral control in macaques following passive transfer of a single anti-V2 monoclonal antibody and repeated SHIV challenges [published online ahead of print March 7, 2018] J Virol. doi: 10.1128/JVI.02198-17. https://jvi.asm.org/content/92/11/e02198-17.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakane T, et al. Limited impact of passive non-neutralizing antibody immunization in acute SIV infection on viremia control in rhesus macaques. PLoS ONE. 2013;8(9):e73453. doi: 10.1371/journal.pone.0073453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaccari M, et al. Reduced protection from simian immunodeficiency virus SIVmac251 infection afforded by memory CD8+ T cells induced by vaccination during CD4+ T-cell deficiency. J Virol. 2008;82(19):9629–9638. doi: 10.1128/JVI.00893-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chong C, Friberg M, Clements JD. LT(R192G), a non-toxic mutant of the heat-labile enterotoxin of Escherichia coli, elicits enhanced humoral and cellular immune responses associated with protection against lethal oral challenge with Salmonella spp. Vaccine. 1998;16(7):732–740. doi: 10.1016/S0264-410X(97)00255-7. [DOI] [PubMed] [Google Scholar]
- 59.Lamoreaux L, Roederer M, Koup R. Intracellular cytokine optimization and standard operating procedure. Nat Protoc. 2006;1(3):1507–1516. doi: 10.1038/nprot.2006.268. [DOI] [PubMed] [Google Scholar]
- 60.Demberg T, Mohanram V, Venzon D, Robert-Guroff M. Phenotypes and distribution of mucosal memory B-cell populations in the SIV/SHIV rhesus macaque model. Clin Immunol. 2014;153(2):264–276. doi: 10.1016/j.clim.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mohanram V, et al. Improved flow-based method for HIV/SIV envelope-specific memory B-cell evaluation in rhesus macaques. J Immunol Methods. 2014;412:78–84. doi: 10.1016/j.jim.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas MA, et al. Rhesus macaque rectal and duodenal tissues exhibit B-cell sub-populations distinct from peripheral blood that continuously secrete antigen-specific IgA in short-term explant cultures. Vaccine. 2014;32(7):872–880. doi: 10.1016/j.vaccine.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patterson LJ, et al. Rapid SIV Env-specific mucosal and serum antibody induction augments cellular immunity in protecting immunized, elite-controller macaques against high dose heterologous SIV challenge. Virology. 2011;411(1):87–102. doi: 10.1016/j.virol.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Demberg T, et al. Dynamics of memory B-cell populations in blood, lymph nodes, and bone marrow during antiretroviral therapy and envelope boosting in simian immunodeficiency virus SIVmac251-infected rhesus macaques. J Virol. 2012;86(23):12591–12604. doi: 10.1128/JVI.00298-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vargas-Inchaustegui DA, et al. Humoral immunity induced by mucosal and/or systemic SIV-specific vaccine platforms suggests novel combinatorial approaches for enhancing responses. Clin Immunol. 2014;153(2):308–322. doi: 10.1016/j.clim.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 68.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.