Abstract
The prognosis for patients with bladder cancer (BCa) with lymph node (LN) metastasis is poor, and it is not improved by current treatments. Long noncoding RNAs (lncRNAs) are involved in the pathology of various tumors, including BCa. However, the role of Differentiation antagonizing non-protein coding RNA (DANCR) in BCa LN metastasis remains unclear. In this study, we discover that DANCR was significantly upregulated in BCa tissues and cases with LN metastasis. DANCR expression was positively correlated with LN metastasis status, tumor stage, histological grade, and poor patient prognosis. Functional assays demonstrated that DANCR promoted BCa cell migration, invasion, and proliferation in vitro and enhanced tumor LN metastasis and growth in vivo. Mechanistic investigations revealed that DANCR activated IL-11-STAT3 signaling and increased cyclin D1 and PLAU expression via guiding leucine-rich pentatricopeptide repeat containing (LRPPRC) to stabilize mRNA. Moreover, oncogenesis facilitated by DANCR was attenuated by anti-IL-11 antibody or a STAT3 inhibitor (BP-1-102). In conclusion, our findings indicate that DANCR induces BCa LN metastasis and proliferation via an LRPPRC-mediated mRNA stabilization mechanism. DANCR may serve as a multi-potency target for clinical intervention in LN-metastatic BCa.
Keywords: DANCR, bladder cancer, lymph node metastasis, IL-11-STAT3 signaling, proliferation
Graphical Abstract
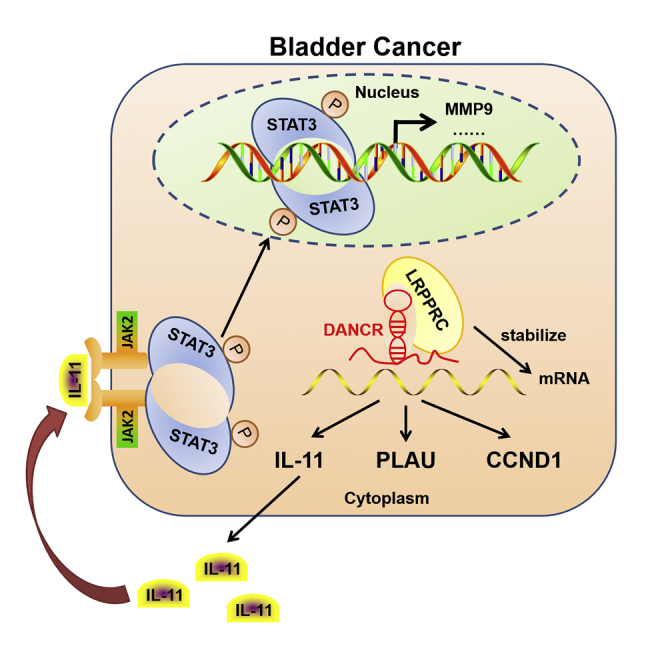
Chen et al. demonstrate that DANCR activates IL-11-STAT3 signaling and increases cyclin D1 and PLAU expression via guiding LRPPRC to stabilize mRNA, leading to LN metastasis and proliferation of bladder cancer. Therefore, DANCR may serve as a multi-potency target for clinical intervention in LN-metastatic bladder cancer.
Introduction
Bladder cancer (BCa) is the most common malignancy of the urinary system in China1 and worldwide,2 and it is the number one cause of death in patients with urinary tract disease.3 Muscle-invasive BCa (MIBC) represents 25%–40% of all BCa, and it can spread from the bladder to the pelvic lymph nodes (LNs) and then to visceral organs.4 The probability of death from MIBC with LN metastasis is significantly higher than that from MIBC without LN metastasis. The death rate increases from 18.6% to 77.6% within 5 years, even when the MIBC is treated with radical cystectomy.5, 6 LN metastasis is a complex multistep process that involves dissemination of cancer cells to lymphatic vessels, transport, settlement, and colonization expansion of cancer cells in the LNs.7, 8 However, the biological character and molecular mechanism of BCa cell invasion and metastasis to the LNs remain largely unknown.
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nt with no or weak protein-coding capacity.9 Accumulating evidence indicates that lncRNAs play diverse roles in the initiation and progression of human cancers.10, 11 For example, lncRNAs HOTAIR, CYTOR, and SNHG14 participate in the metastatic cascade by regulating cell migration and invasion,12, 13, 14 and lncRNAs ATB and CILA1 promote metastasis by inducing the epithelial-mesenchymal transition (EMT).15, 16 Our recent study discovered that lncRNA LBCS inhibits self-renewal and chemoresistance of BCa stem cells through the epigenetic silencing of SOX2.17 Our previous study found that lncRNA BLACAT2 promoted BCa-associated lymphangiogenesis and lymphatic metastasis by enhancing vascular endothelial growth factor C (VEGF-C) signaling.18 However, the biological function and molecular mechanism of lncRNAs in BCa LN metastasis remain to be fully clarified.
Recently, the lncRNA DANCR was reported to play critical roles in diverse biological processes, including stem cell differentiation, cell proliferation, and cancer progression.19, 20, 21 DANCR regulates the differentiation of mesenchymal tissues, such as in chondrogenic and osteogenic differentiation.22, 23 In addition, DANCR increased the stemness features of hepatocellular carcinoma cells and promoted the invasion of prostate cancer cells.21, 24 Moreover, DANCR is associated with tumor progression and poor prognosis in colorectal cancer.25 However, whether DANCR has a functional role in BCa and, if so, what is the underlying molecular mechanism are unknown.
In the present study, we identified that DANCR was significantly overexpressed in LN-metastatic BCa and correlated closely with poor prognosis. Through gain or loss of function, we demonstrated that DANCR promoted migration, invasion, and proliferation in BCa cells in vitro and enhanced tumor LN metastasis and growth in vivo. Mechanistically, DANCR guided leucine-rich pentatricopeptide repeat containing (LRPPRC) to stabilize mRNA of interleukin 11 (IL-11), CCND1, and plasminogen activator urokinase (PLAU), contributing to activate IL-11-STAT3 signaling and increase CCND1 and PLAU expression. Therefore, targeting DANCR could be a potential therapeutic strategy leading to less metastasis and proliferation in BCa.
Results
DANCR Overexpression Correlates with LN Metastasis and Poor Prognosis in BCa
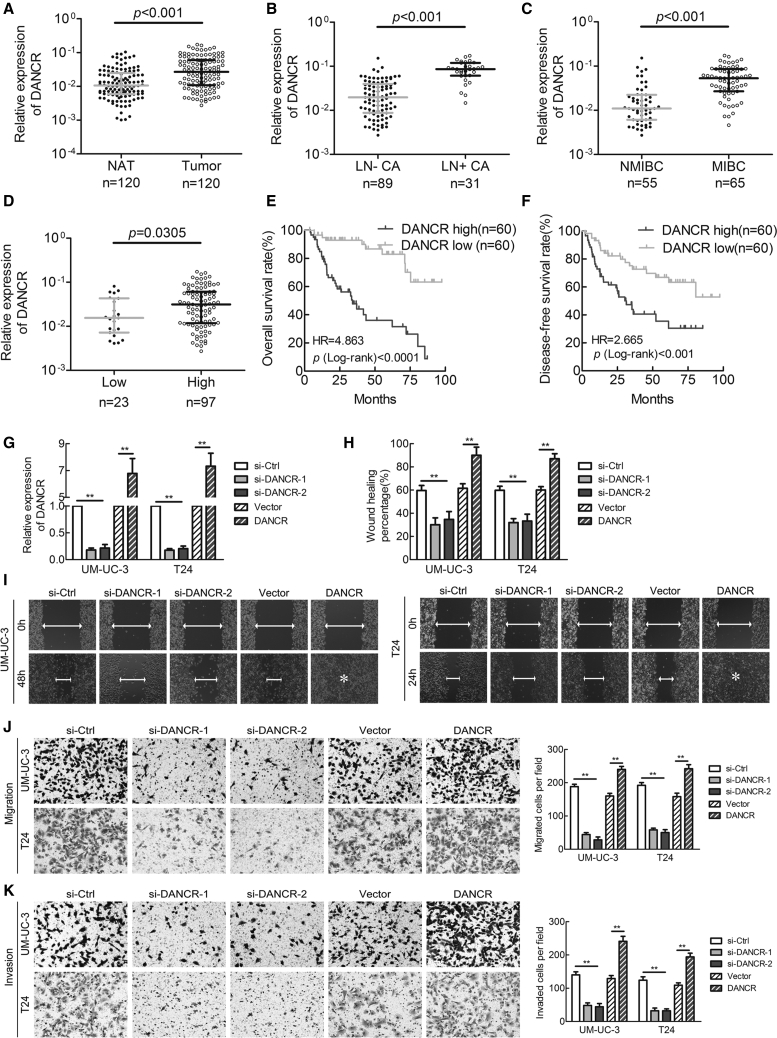
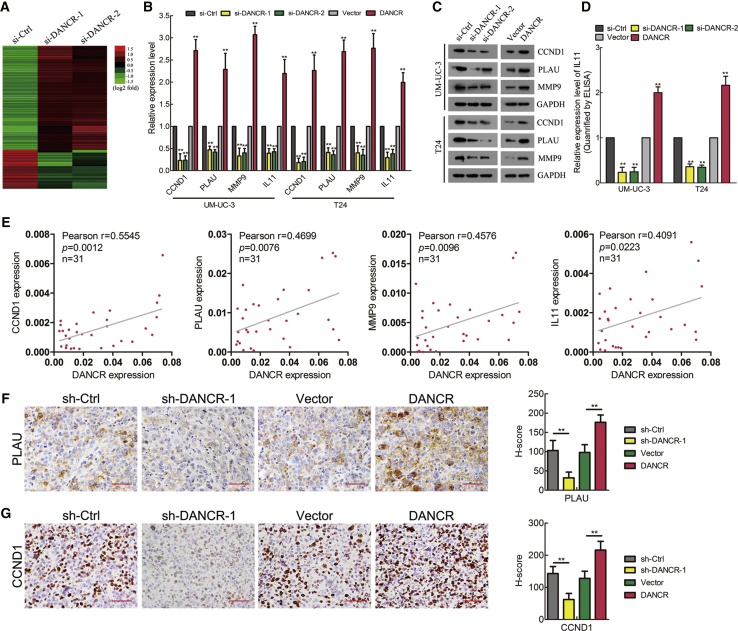
To evaluate whether DANCR is involved in BCa progression, DANCR expression was investigated in a large 120-case cohort of BCas using qRT-PCR. The results showed that DANCR was overexpressed in BCa tissues compared with normal adjacent tissues and in LN-positive bladder tumors compared with LN-negative tumors (Figures 1A and 1B). Additionally, DANCR was upregulated in MIBC compared with non-MIBC (NMIBC), as well as in high-grade BCa compared with lower-grade tumors (Figures 1C and 1D). Moreover, clinicopathological correlation analysis revealed that DANCR expression correlated strongly with pathological stage, grade, and LN metastasis status of BCa (Table S1). Furthermore, patients with high DANCR-expressing BCas had shorter overall survival (OS) and disease-free survival (DFS) (Figures 1E and 1F), suggesting a potential link between high DANCR expression and human BCa progression. In addition, univariate analysis indicated that DANCR expression was significantly associated with OS and DFS (Table 1; Table S2). The multivariate Cox regression analysis demonstrated that high DANCR expression in BCa tissues was an independent prognostic factor for shorter OS (Table 1). Collectively, these data demonstrate that DANCR is associated with LN metastasis status, tumor stage, and histological grade and may serve as a marker of poor prognosis in BCa.
Figure 1.
DANCR Expression Correlates with Bladder Cancer LN Metastasis and Predicts Poor Prognosis, and It Promotes Metastasis of Bladder Cancer Cells In Vitro
(A) DANCR expression was detected by qRT-PCR in 120 cases of BCa tissues paired with normal adjacent tissues (NAT). (B) DANCR expression was detected in LN-negative and LN-positive BCa tissues. (C) DANCR expression was detected in NMIBC and MIBC. (D) DANCR expression was detected in high-grade compared with lower-grade bladder cancer. (E and F) Kaplan-Meier curves for OS (E) and DFS (F) of patients with bladder cancer with high versus low expression of DANCR. The patients were divided into DANCR-low (n = 60) and DANCR-high groups (n = 60). (G) qRT-PCR analysis of DANCR expression levels in DANCR-silenced cells, DANCR-overexpressing cells, and control cells. (H) A histogram analysis of cell migration distances is shown. (I) Representative images of wound-healing assays using UM-UC-3 and T24 cells, showing cell motility after knockdown or overexpression of DANCR. (J and K) Representative images of migration (J) and invasion (K) assays using UM-UC-3 and T24 cells (left panels), showing cell migration and invasion after knockdown or overexpression of DANCR. A histogram analysis of migrated or invaded cell counts is shown (right panels). Statistical significance was assessed using two-tailed t tests or ANOVA. *p < 0.05 and **p < 0.01.
Table 1.
Univariate and Multivariate Analysis of Factors Associated with Overall Survival in Bladder Cancer
| Variable | Univariate |
Multivariate |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p Value | HR | 95% CI | p Value | |
| Age, years (>65/≤65) | 1.980 | 1.090–3.597 | 0.025* | 1.283 | 0.676–2.437 | 0.446 |
| Gender (female/male) | 0.478 | 0.188–1.215 | 0.121 | – | – | NA |
| Histological grade (high/low) | 3.352 | 1.194–9.409 | 0.022* | 1.640 | 0.515–5.225 | 0.403 |
| Tumor stage (T2–T4/Ta–T1) | 5.156 | 2.392–11.113 | <0.001* | 1.603 | 0.539–4.770 | 0.396 |
| Nodal metastasis (N1–N2/N0) | 3.802 | 2.009–7.194 | <0.001* | 2.175 | 1.042–4.542 | 0.039* |
| Tumor size (>3 cm/≤3 cm) | 1.287 | 0.705–2.353 | 0.411 | – | – | NA |
| Tumor number (multiple/single) | 0.780 | 0.412–1.476 | 0.445 | – | – | NA |
| DANCR (high/low) | 5.290 | 2.589–10.809 | <0.001* | 2.945 | 1.200–7.232 | 0.018* |
Univariate and multivariate analysis. Cox proportional hazards regression model. Variables associated with survival by univariate analyses were adopted as covariates in multivariate analyses. HR > 1, risk for death increased; HR < 1, risk for death reduced. *Significant p value.
DANCR Enhances the Metastatic Behavior of Bladder Cancer Cells In Vitro and LN Metastasis In Vivo
To investigate the role of DANCR in BCa progression, we silenced or overexpressed DANCR in BCa cells using two small interfering RNAs (siRNAs) or lentiviral infection, respectively. qRT-PCR showed that DANCR was remarkably downregulated or increased in UM-UC-3 and T24 cells (Figure 1G). Overexpression of DANCR was significantly associated with tumor invasion and LN metastasis in patients with BCa; therefore, the effects of DANCR on cell motility and tumor metastasis were studied using wound-healing, cell migration, and invasion assays. Wound-healing assays showed that the downregulation of DANCR decreased, whereas the upregulation of DANCR increased, the migratory speed of T24 and UM-UC-3 cells (Figures 1H and 1I). Moreover, DANCR knockdown inhibited migration and invasion of BCa cells, whereas the opposite outcome was observed after DANCR overexpression (Figures 1J and 1K). Taken together, our results indicated that DANCR is required for the migration and invasiveness of BCa cells in vitro.
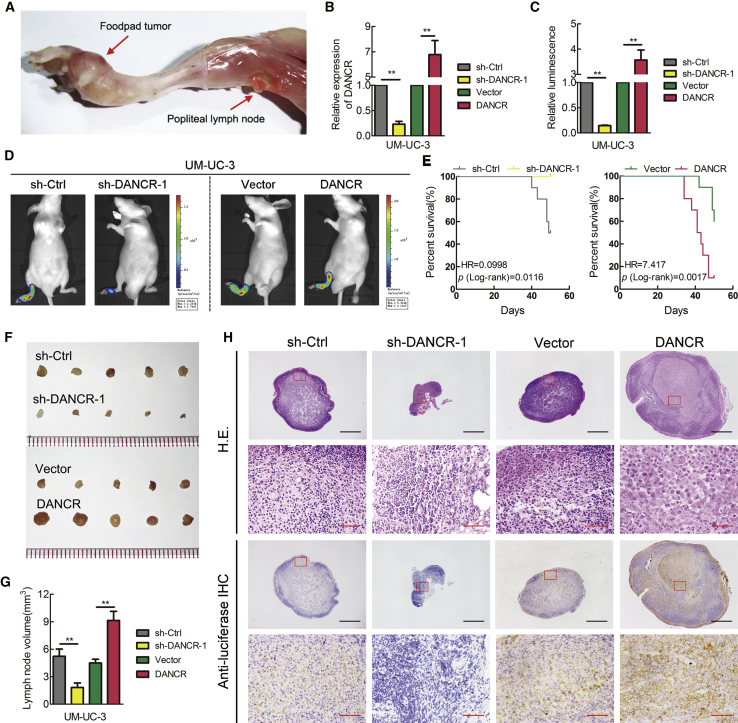
To further evaluate the effects of DANCR in LN metastasis of BCa, an in vivo nude mouse popliteal LN metastasis model was constructed (Figure 2A). UM-UC-3/luc BCa cell lines, which stably expressed a short hairpin RNA (shRNA) targeting DANCR or overexpressed DANCR, were inoculated into the footpads of nude mice (n = 10/group) (Figure 2B). The primary footpad tumors and popliteal LNs were dissected after 6 weeks. Strikingly, DANCR knockdown significantly inhibited metastasis of BCa cells to the LNs. In contrast, overexpression of DANCR promoted LN metastasis, as determined by in vivo imaging systems (IVISs) (Figures 2C and 2D). Additionally, the mice bearing DANCR-overexpressing tumors had shorter survival times, while mice bearing DANCR-knockdown tumors had longer survival times compared with those in the corresponding control groups (Figure 2E). Moreover, the volumes of the popliteal LNs were markedly smaller in the DANCR shRNA mice but larger in the DANCR-overexpressing mice than in the corresponding control mice (Figures 2F and 2G). BCa cell metastatic LNs were confirmed by H&E staining and immunostaining (immunohistochemistry [IHC]) for luciferase (Figure 2H; Table S3). Collectively, these results indicate that DANCR promotes BCa cells’ LN metastasis in vivo via augmenting their invasive ability, suggesting that DANCR is a promising target for preventing LN metastasis in BCa.
Figure 2.
DANCR Facilitates LN Metastasis of Bladder Cancer Cells In Vivo
(A) Representative images of the nude mouse model of popliteal LN metastasis. The indicated UM-UC-3 cells were injected into the footpads of the nude mice, and the popliteal LNs were enucleated and analyzed. (B) qRT-PCR analysis of DANCR expression levels in stably DANCR-silenced or DANCR-overexpressing cells and control cells. (C and D) Histogram analysis of popliteal LN metastasis (C) and representative images of bioluminescence (D) (popliteal LNs) in the indicated cell groups (n = 10 per group). (E) Kaplan-Meier survival analysis of the mice (n = 10 per group) that were inoculated with DANCR-knockdown or -overexpressing UM-UC-3 cells compared with the corresponding control cells. (F and G) Representative images of dissected popliteal LNs (F) and histogram analysis (H) of the LN volume. (H) Representative images of H&E and IHC staining confirming the LN status (n = 10). Scale bars, 500 μm (black) and 50 μm (red). *p < 0.05 and **p < 0.01.
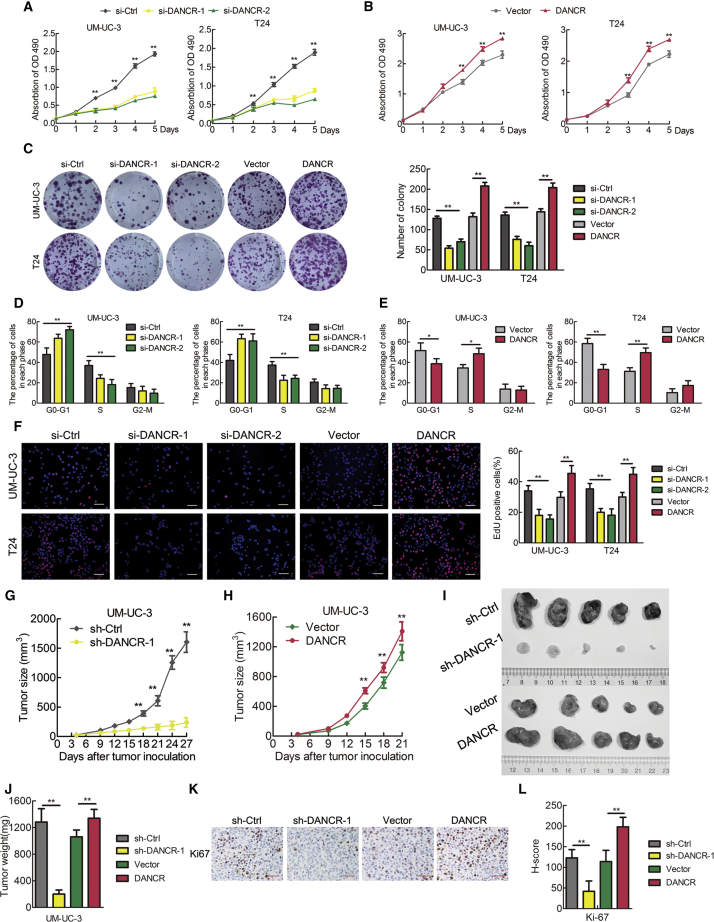
DANCR Promotes the Proliferation of Bladder Cancer Cells In Vitro and Tumor Growth In Vivo
We next investigated the function of DANCR on the proliferation of BCa cells using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and colony formation assays. We found that DANCR knockdown significantly reduced the viability and colony formation ability of UM-UC-3 and T24 cells (Figures 3A and 3C). In contrast, overexpression of DANCR enhanced BCa cell viability and colony formation ability (Figures 3B and 3C). However, DANCR had no obvious effect on the apoptosis of BCa cells (Figure S1A). To characterize whether DANCR is involved in the cell cycle, we performed flow cytometry and ethynyl deoxyuridine (EdU) assays. Interestingly, DANCR silencing dramatically increased the cell population in the G0/G1 phase, whereas it reduced the cell population in the S phase; DANCR overexpression had the opposite effect (Figures 3D and 3E; Figure S1B). Similarly, the EdU assay showed that DANCR knockdown significantly decreased the cell population in the S phase, whereas DANCR overexpression had the opposite effect (Figure 3F; Figure S1C). Moreover, DANCR knockdown markedly reduced, but DANCR overexpression increased, the Ki67 expression in vitro (Figure S1D). Therefore, these data show that DANCR promotes BCa cell proliferation via regulating the G1/S phase transition.
Figure 3.
DANCR Enhances the Proliferation of Bladder Cancer Cells In Vitro and Tumor Growth In Vivo
(A and B) Cell viability was evaluated in DANCR-knockdown (A) or -overexpressing (B) UM-UC-3 and T24 cells. (C) Colony formation assays were performed in DANCR-knockdown or -overexpressing UM-UC-3 and T24 cells. (D and E) Flow cytometry analysis of UM-UC-3 and T24 cells transfected with DANCR siRNA (D) or stable DANCR-overexpressing (E) cells, compared with the corresponding control cells. The percentages (%) of cell populations at different stages of the cell cycle are listed in the panels. (F) EdU assay measurement of the cell population in the S phase and a histogram analysis of EdU-positive cell counts are shown. Blue, nucleus; red, S-phase cells. Scale bars, 100 μm (white). (G and H) Tumor growth curves of DANCR knockdown (G) or overexpression groups (H) are summarized in the line chart. The average tumor volume is expressed as the mean ± SD of five mice. (I) Representative images of the tumors of DANCR-knockdown or -overexpression groups and their respective controls. (J) Tumor weights were measured after the tumors were surgically dissected. (K) IHC examination of xenograft tumor Ki67 expression. (L) Histogram shows the H-score in DANCR-knockdown or -overexpression groups and the control group. Scale bars, 50 μm (red). *p < 0.05 and **p < 0.01.
To further explore the effects of DANCR in BCa tumorigenesis in vivo, stable DANCR-silenced, DANCR-overexpressing, or control UM-UC-3 cells were subcutaneously injected into BALB/c nude mice, and the tumor growth activity was measured (Figures 3G and 3H). Strikingly, the growth, size, and weight of tumors derived from the DANCR-knockdown group were markedly reduced compared with those in the control group (Figures 3I and 3J). Conversely, DANCR overexpression promoted tumor growth of BCa cells (Figures 3I and 3J). Moreover, the tumors derived from the DANCR-knockdown group exhibited lower expression of the proliferation marker Ki67 than the control group. However, the upregulation of DANCR was associated with a higher Ki67 expression in the DANCR-overexpressing group compared with that in the control group (Figures 3K and 3L). Interestingly, we observed that the tumors formed by the DANCR-overexpressing BCa cells grown in the nude mice displayed spike-like structures that invaded the surrounding muscle tissues, while the control tumors exhibited sharp edges (Figure S2), further supporting that DANCR enhanced BCa cell invasion. Collectively, these data strongly suggest that targeting DANCR could inhibit tumorigenesis and tumor growth of BCa cells in vivo.
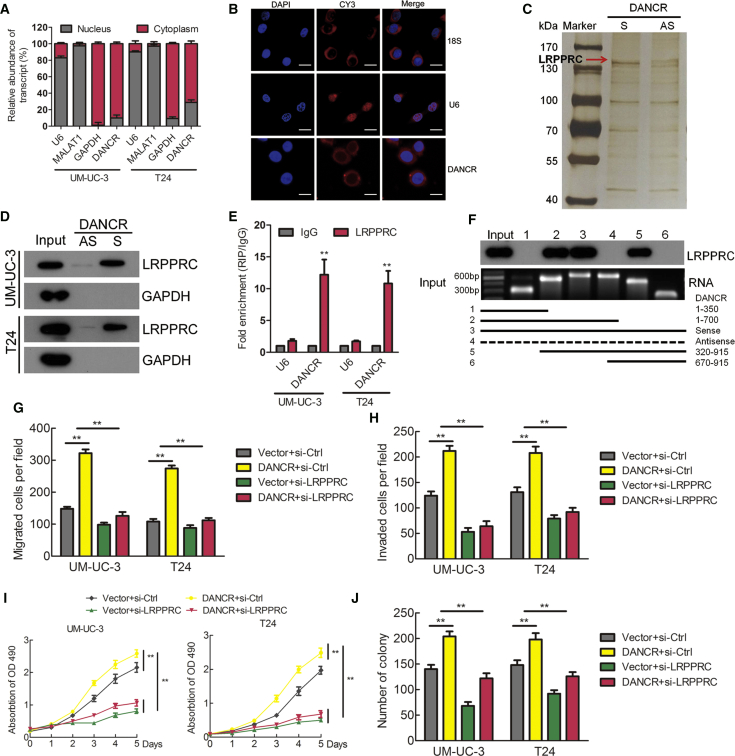
DANCR Directly Interacts with LRPPRC to Play Key Roles in BCa
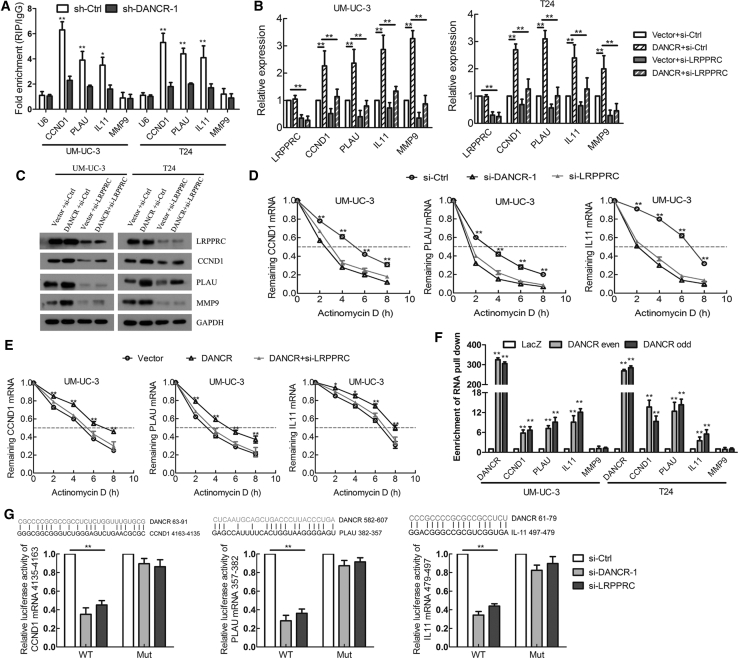
The subcellular localization of an lncRNA is associated closely with its biological mechanism. Cellular fractionation assays and RNA fluorescence in situ hybridization (RNA-FISH) showed that DANCR was distributed mainly in the cytoplasm in BCa cells (Figures 4A and 4B). lncRNAs located in the cytoplasm are usually associated with post-transcriptional regulation.20, 26 To identify DANCR-interacting proteins in BCa cells, we performed RNA pull-down assays using in vitro-transcribed biotinylated DANCR. One overtly differential band between 130 and 150 kDa appeared after silver staining, and it was identified as LRPPRC by mass spectrometry (Figure 4C; Figure S3A). Consistent with DANCR localization, LRPPRC was also distributed mainly in the cytoplasm in BCa cells (Figure S3B). We confirmed the special interaction between DANCR and LRPPRC using western blotting (Figure 4D). We also verified this result using RNA immunoprecipitation (RIP), and we found that DANCR, but not U6, was enriched in LRPPRC precipitates (Figure 4E). Moreover, a serial deletion analysis revealed that 350–670 nt in the DANCR transcript, forming a stem-loop structure, are critical for the interaction with LRPPRC (Figure 4F; Figure S3C).
Figure 4.
DANCR Directly Interacts with LRPPRC to Play Key Roles in Bladder Cancer
(A) Nuclear fraction experiment and qRT-PCR detected the abundance of DANCR in the nucleus and cytoplasm. GAPDH was the positive control for cytoplasm, and MALAT1 and U6 were the positive controls for the nucleus. (B) The subcellular distribution of DANCR was visualized by RNA fluorescence in situ hybridization (RNA-FISH) in UM-UC-3 cells. 18S was the positive control for the cytoplasm, and U6 was the positive control for the nucleus. Scale bars, 10 μm (white). (C) RNA pull-down assay was performed using DANCR sense and antisense RNAs incubated with cytoplasm extracts of UM-UC-3 cells, followed by silver staining. A red arrow indicates LRPPRC. (D) The interaction between DANCR and LRPPRC was confirmed by RNA pull-down and western blotting. GAPDH served as the negative control. (E) RIP was performed using anti-LRPPRC and control IgG antibodies, followed by qRT-PCR to examine the enrichment of DANCR and U6. U6 served as the negative control. (F) Serial deletions of DANCR were used in the RNA pull-down assays to identify the core regions of DANCR that were required for the physical interaction with LRPPRC. (G and H) Cell migration (G) and invasion (H) were analyzed using DANCR overexpression or control cells combined with LRPPRC knockdown. (I and J) Cell viability (I) and colony formation (J) were analyzed using DANCR overexpression or control cells combined with LRPPRC knockdown. *p < 0.05 and **p < 0.01.
To explore the role of the DANCR-LRPPRC association in DANCR’s function, we overexpressed DANCR, and then we knocked down LRPPRC in BCa cells. Interestingly, knockdown of LRPPRC eliminated the DANCR-mediated enhancement of the metastasis and proliferation of BCa cells in vitro (Figures 4G–4J; Figures S4A–S4C). Therefore, these data strongly suggest that DANCR regulates metastasis and proliferation of BCa cells in an LRPPRC-dependent manner.
DANCR Regulates CCND1, PLAU, and IL-11 Expression
To explore the molecular mechanism underlying DANCR-induced metastasis and proliferation in BCa, a genome-wide mRNA expression profile screen was used to compare gene expression profiles between DANCR-silenced UM-UC-3 cells and their control cells. Among 236 genes that were regulated by DANCR (fold change >2.0), many genes that play critical roles in cell metastasis and proliferation, such as IL-11, PLAU, Matrix metalloproteinase 9 (MMP9), and CCND1,27, 28, 29 were significantly downregulated in DANCR-silenced cells (Figure 5A). Furthermore, qRT-PCR, western blotting, and ELISAs revealed that the expression levels of CCND1, PLAU, MMP9, and IL-11, at both the mRNA and protein levels, were decreased in DANCR-silenced cells and increased in DANCR-overexpressing cells (Figures 5B–5D). Importantly, IL-11, PLAU, and MMP9 were upregulated in BCa tissues compared with normal tissues, and they correlated positively with poor OS in BCa from The Cancer Genome Atlas (TCGA) cohort (Figures S5A and S5B). Moreover, positive correlations between DANCR expression and the CCND1, PLAU, MMP9, and IL-11 levels were also observed in a 31-case cohort of BCa specimens (Figure 5E). Additionally, the protein expression levels of PLAU and CCND1 were markedly decreased in the DANCR-silenced xenograft tumors, but they were markedly increased in the DANCR-overexpressing xenograft tumors (Figures 5F and 5G). Taken together, these results suggest that DANCR upregulates the expression levels of IL-11, PLAU, MMP9, and CCND1, which contribute to cancer cell lymphatic metastasis and proliferation in BCa.
Figure 5.
DANCR Regulates CCND1, PLAU, and IL-11 Expression Levels
(A) A heatmap representing mRNA expression levels in the UM-UC-3 cells transfected with control or DANCR siRNA for 48 h. (B) The differentially expressed genes in the microarray were verified in UM-UC-3 and T24 cells by qRT-PCR. (C) The expression of DANCR target genes was detected by western blotting. GAPDH was used as the internal control. (D) The secreted IL-11 expression was detected using ELISA. (E) Pearson correlations between the expression levels of DANCR and CCND1, PLAU, MMP9, and IL-11, as assessed by qRT-PCR analysis, from 31 bladder cancer patients. (F and G) The PLAU expression in xenograft footpad tumor (F) and CCND1 expression in xenograft subcutaneous tumor (G) were detected by IHC. Histogram shows the H-score in DANCR-knockdown or -overexpression groups and control group. Scale bars, 50 μm (red). *p < 0.05 and **p < 0.01.
DANCR Regulates mRNA Stability by Guiding LRPPRC to Target Genes
LRPPRC was reported to regulate mRNA stability;30, 31 therefore, we investigated whether DANCR guided LRPPRC to stabilize target gene mRNAs. First, we found that CCND1, PLAU, and IL-11, but not MMP9, mRNAs were enriched in LRPPRC precipitates using a RIP assay (Figure 6A). However, the enrichment of IL-11, PLAU, and CCND1 mRNAs in LRPPRC precipitates was markedly decreased in DANCR-knockdown cells compared with control cells (Figure 6A), suggesting that LRPPRC interacted with these mRNAs in a DANCR-dependent manner. In addition, knockdown of LRPPRC eliminated the DANCR-mediated increase in the target genes’ mRNA and protein expression levels in BCa cells (Figures 6B and 6C; Figure S6A). Furthermore, we treated BCa cells with actinomycin D, which allowed us to measure the decay of pre-existing mRNA. The data showed that knockdown of DANCR or LRPPRC resulted in decreased half-lives of CCND1, PLAU, and IL-11 mRNAs in BCa cells, whereas overexpression of DANCR increased their half-lives, but not that of MMP9 mRNA (Figure 6E; Figures S6C and S6E). Furthermore, knockdown of LRPPRC eliminated the enhanced effect of DANCR on mRNA stability, indicating that DANCR stabilizes IL-11, PLAU, and CCND1 mRNAs in an LRPPRC-dependent manner (Figures 6D and 6E; Figures S6B–S6E).
Figure 6.
DANCR Regulates mRNA Stability by Guiding LRPPRC to Target Genes
(A) RIP was performed using anti-LRPPRC and control IgG antibodies in DANCR-knockdown or control cells, followed by qRT-PCR to examine the enrichment of CCND1, PLAU, IL-11, and MMP9 mRNA and U6. (B and C) The mRNA (B) and protein (C) expression levels of DANCR target genes were detected in DANCR-overexpression or control cells combined with LRPPRC knockdown. (D) UM-UC-3 cells expressing control siRNA, DANCR siRNA-1, or LRPPRC siRNA were treated with actinomycin D (5 μg/mL) for the indicated periods of time. (E) UM-UC-3 cells stably expressing control, DANCR, or DANCR + LRPPRC siRNA were treated with actinomycin D (5 μg/mL) for the indicated periods of time. Total RNA was purified and then analyzed using qRT-PCR to examine the mRNA half-lives of CCND1, PLAU, and IL-11. (F) UM-UC-3 cell lysates were incubated with in vitro-synthesized, biotin-labeled control LacZ DNA probes or antisense DNA probes against DANCR for the biotinylated oligonucleotide pull-down assay. The precipitates from the pull-down were analyzed by qRT-PCR to detect the interacting mRNAs. (G) The putative CCND1-, PLAU-, and IL-11-binding sites were identified in DANCR. Indicated CCND1, PLAU, and IL-11 wild-type or mutated DANCR-binding sites were constructed in the psiCHECK2 vector and subjected to luciferase reporter assays in DANCR- or LRPPRC-silenced and control cells. *p < 0.05 and **p < 0.01.
To determine whether DANCR directly interacted with CCND1, PLAU, and IL-11 mRNAs, we performed a biotinylated oligonucleotide pull-down assay. We found that endogenous CCND1, PLAU, and IL-11 mRNAs, but not MMP9 mRNA, co-precipitated with DANCR (Figure 6F), indicating an association between DANCR and these mRNAs. In addition, we found there were some complementary base pairings between DANCR and CCND1, PLAU, and IL-11 mRNAs (Figure 6G). For further validation, the probable binding sites of CCND1, PLAU, and IL-11 were cloned into psiCHECK2 vectors for luciferase reporter assays. We found that knockdown of DANCR or LRPPRC decreased the luciferase activity of wild-type 4,135–4,163 nt CCND1. Similar results were observed for 357–382 nt PLAU and 479–497 nt IL-11 (Figure 6G). Moreover, the luciferase activity of mutated binding sites of these genes remained unaltered between control and DANCR- or LRPPRC-knockdown cells (Figure 6G). Taken together, these data indicate that DANCR guides LRPPRC to stabilize CCND1, PLAU, and IL-11 mRNAs, contributing to their increased expression.
DANCR Activates the IL-11-STAT3-Signaling Pathway to Promote BCa Metastasis
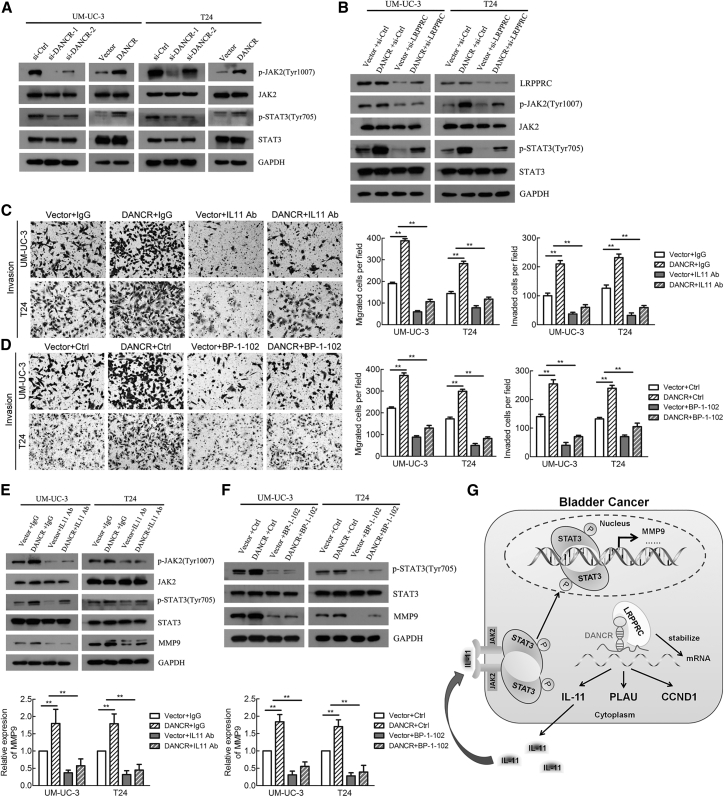
To investigate whether DANCR activated STAT3 signaling in BCa cells by increasing IL-11 secretion, we detected total and phosphorylated JAK2 and STAT3 in DANCR-knockdown and -overexpressing BCa cells using western blotting. As shown in Figure 7A, p-JAK2(Tyr1007) and p-STAT3(Tyr705) levels were decreased in DANCR-knockdown cells, whereas they were increased in DANCR-overexpressing cells. Furthermore, knockdown of LRPPRC attenuated the STAT3 signaling activated by DANCR overexpression in BCa cells (Figure 7B).
Figure 7.
DANCR Activates IL-11-STAT3-Signaling Pathway to Promote BCa Metastasis
(A) The expression levels of total and phosphorylated JAK2 and STAT3 were detected by western blotting in DANCR-knockdown or -overexpression groups. (B) The levels of total and phosphorylated JAK2 and STAT3 were detected by western blotting in DANCR overexpression combined with knockdown of LRPPRC cells. (C and D) Cell migration and invasion were analyzed using DANCR-overexpression or control cells combined with anti-IL-11 antibody (C) and STAT3 inhibitor (BP-1-102) (D). (E) The levels of total and phosphorylated JAK2 and STAT3 and MMP9 were detected by western blotting in DANCR overexpression combined with anti-IL-11 antibody. (F) The levels of total and phosphorylated STAT3 and MMP9 were detected by western blotting in DANCR-overexpressing cells combined with STAT3 inhibitor (BP-1-102). The mRNA expression of MMP9 was detected by qRT-PCR in DANCR-overexpressing or control cells combined with anti-IL-11 antibody and STAT3 inhibitor (BP-1-102). (G) A schematic model of the mechanism underlying the role of DANCR in BCa metastasis and proliferation. *p < 0.05 and **p < 0.01.
To verify that IL-11-STAT3 signaling activation was critical to BCa metastasis, we used anti-IL-11 antibody and a STAT3 inhibitor (BP-1-102) to block STAT3 signaling in DANCR-overexpression BCa cells. Interestingly, either the anti-IL-11 antibody or the STAT3 inhibitor (BP-1-102) not only suppressed BCa cell migration and invasion in control BCa cells but also eliminated the majority of DANCR overexpression-mediated enhancements in BCa cells (Figures 7C and 7D; Figure S7A). Furthermore, the anti-IL-11 antibody reduced p-JAK2, p-STAT3, and MMP9 levels, but not total JAK2 and STAT3 levels (Figure 7E). Similarly, the STAT3 inhibitor downregulated p-STAT3 and MMP9 levels (Figure 7F). Either the anti-IL-11 antibody or the STAT3 inhibitor attenuated STAT3 signaling and MMP9 activation by DANCR overexpression in BCa cells (Figures 7E and 7F). Overall, the data indicate that DANCR promoted metastasis and increased MMP9 expression in an IL-11-JAK-STAT3 signaling-dependent manner in BCa cells, suggesting that the anti-IL-11 antibody and STAT3 inhibitor could be promising targeted therapies to block metastasis in patients with BCa that overexpresses DANCR.
Discussion
LN metastasis leads to a poor prognosis for patients with BCa, and currently it has limited treatment options in the clinic.32, 33 Our previous studies established an effective method to diagnose LN metastasis in BCa.34 However, determination of the molecular mechanisms underlying LN metastasis and the identification of novel targets are urgently needed for prevention and therapy. Accumulating evidence indicates that lncRNAs play critical roles in the initiation and progression of human cancers.35, 36, 37, 38 However, the biological function and molecular mechanisms of lncRNAs in BCa LN metastasis are largely unknown. To the best of our knowledge, this is the first study to systematically evaluate the role of DANCR in LN metastasis of BCa. In this study, we reported that DANCR is overexpressed in LN-metastatic BCa and is associated with poor clinical prognosis. Moreover, overexpression of DANCR promoted metastasis and proliferation of BCa cells in vitro and in vivo. Mechanistically, DANCR activates IL-11-STAT3 signaling and increases CCND1 and PLAU expression levels by guiding LRPPRC to stabilize their mRNAs.
lncRNAs are reported to regulate biological functions via diverse mechanisms: guider; decoy; scaffold effect on DNA, RNA, or protein; and post-transcriptional effects.39 Previous studies showed that DANCR mainly exerted oncogenic roles in cancers by serving as a microRNA (miRNA) sponge.20, 40, 41 DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mammalian target of rapamycin (mTOR) expression.20 DANCR promotes rho-associated coiled-coil-containing protein kinase 1 (ROCK1)-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma.40 However, some reports showed that DANCR could directly bind to some proteins or mRNAs. Yuan et al.21 found that DANCR could bind to CTNNB1 mRNA and then block the repressing effect of microRNA miR-2214, miR-320a, and miR-199a on CTNNB1. Li et al.42 found that DANCR interacted with EZH2 and then modulated the stability of EZH2. Additionally, one study showed that DANCR recruited the PRC2 complex to inhibit TIMP2/3 expression.24
In this study, we found that DANCR directly interacted with LRPPRC. LRPPRC, an RNA-binding protein, regulates mRNA stability and polyadenylation mainly in mitochondria but also in the cytoplasm and nucleus.30, 31, 43, 44 We found that both DANCR and LRPPRC were distributed mainly in the cytoplasm in BCa cells. Knockdown of DANCR or LRPPRC resulted in decreases in the half-lives of IL-11, PLAU, and CCND1 mRNAs in BCa cells. Furthermore, we found that DANCR directly interacted with IL-11, PLAU, and CCND1 mRNAs. Knockdown of DANCR attenuated the interaction between LRPPRC and its target mRNAs. Overall, we identified a novel mechanism by which DANCR guides LRPPRC to stabilize IL-11, PLAU, and CCND1 mRNAs, which activates IL-11-STAT3 signaling and enhances CCND1 and PLAU expression levels.
LN metastasis is a complex multistep process, in which enhanced migration and invasion are key initial steps of cancer metastasis.7, 8 We found that DANCR was necessary for BCa cell migration and invasion in vitro and LN metastasis in vivo. Further investigations revealed that DANCR increased IL-11 expression. IL-11 is implicated in various malignancies, including gastric, colorectal, pancreatic, and prostate cancers.45, 46, 47 Many of the pro-oncogenic effects of IL-11 have been shown to be associated with the JAK-STAT3-signaling pathway.27, 45 We observed that JAK-STAT3 signaling was activated in DANCR-overexpressing cells, whereas it was decreased in DANCR-silenced cells. JAK-STAT3 signaling acts to drive the proliferation, survival, invasiveness, and metastasis of tumor cells while strongly suppressing the anti-tumor immune response in many cancers.48, 49 In this study, blocking STAT3 signaling using anti-IL-11 antibody or a STAT3 inhibitor (BP-1-102) showed promising anti-tumor effects in DANCR-overexpressing BCa by inhibiting migration and invasion. Recently, treatments that target the JAK-STAT3 pathway have shown promising therapeutic benefit in mice and patients with cancer by directly inhibiting tumor cell growth and metastasis.50, 51, 52 Taken together, DANCR is critical for IL-11-JAK-STAT3 signaling activation and metastasis in BCa, suggesting that an anti-IL-11 antibody or STAT3 inhibitor would serve as an effective therapeutic approach for patients with BCa that overexpresses DANCR.
In this study, we found that DANCR upregulated PLAU, MMP9, and CCND1 expression levels in BCa. PLAU, urokinase-type plasminogen activator protease (uPA), is a secreted factor that binds to the outer cell surface, remodels the extracellular matrix, and then allows cells to move inside tissues.53, 54 Furthermore, PLAU is overexpressed in many cancer cells and is important for cancer cell metastatic spread.28, 55 MMP9 is involved in the breakdown of the extracellular matrix and promotes cancer cell metastasis.56 Importantly, PLAU and MMP9 were upregulated in BCa tissues compared with normal tissues, and they are positively correlated with poor OS in BCa from TCGA cohort, suggesting their important roles in BCa. CCND1, a key regulator in G1-to-S phase transition, is overexpressed in BCa and is associated with poor prognosis.29, 57, 58 Collectively, these results indicated that DANCR has great potential as a multi-potency therapeutic target to inhibit metastasis and proliferation in BCa.
Recently, small molecules designed to target the folded structure of oncogenic noncoding RNA have shown significant anti-tumor effects in vivo by selectively modulating noncoding RNAs in cancer cells, with no effects in normal cells.59 Additionally, siRNA or locked nucleic acids can serve as therapeutic agents that specifically target lncRNAs in vivo.60 In the future, the inhibition of cancer invasiveness and proliferation via small molecules that specifically target DANCR might represent potential therapeutics for human cancer.
In summary, we report the discovery that DANCR clinically and functionally participates in LN metastasis and proliferation of BCa via IL-11-mediated activation of JAK-STAT3 signaling (Figure 7G). Uncovering the precise role of DANCR in the progression of BCa will not only increase our knowledge of lncRNA-induced LN metastasis and proliferation but also enable the development of novel therapeutic strategies to treat BCa LN metastasis.
Materials and Methods
Human Tissue Samples
A total of 120 snap-frozen fresh BCa tissues and 120 normal adjacent tissues (NATs) were obtained with the written consent of patients who underwent surgery at Sun Yat-sen Memorial Hospital, Sun Yat-sen University (Guangzhou, China) between January 2010 and January 2017. All samples were pathologically confirmed as BCa by two independent pathologists. Ethical approval was obtained from Sun Yat-sen University’s Committees for Ethical Review of Research Involving Human Subjects. Table S1 lists the patient and tumor demographics.
Cell Culture
The cell lines used in this study included the human BCa cells UM-UC-3 and T24 and SV40-transformed kidney cell line 293T (ATCC, Manassas, VA, USA). UM-UC-3 and 293T cells were cultured in DMEM (Gibco, Shanghai, China), whereas T24 cells were cultured in RPMI 1640 (Gibco). All media were supplemented with 10% FBS (fetal bovine serum; Shanghai ExCell Biology, Shanghai, China) and 1% penicillin and streptomycin. The STAT3 inhibitor (BP-1-102) was purchased from Selleck (Houston, TX, USA) and used at 15 μM for 48 h. The anti-IL-11 antibody was purchased from R&D Systems (Minneapolis, MN, USA; catalog MAB218) and used at 15 μg/mL for 48 h. Cells were grown in a humidified atmosphere of 5% CO2 at 37°C. The cell lines used in this study were not contaminated by mycoplasma.
RNA Isolation and qRT-PCR
RNA isolation and qRT-PCR analysis were performed following standard protocols, as previously reported.61 The transcription level of GAPDH was used as an internal control. The specific primers used are listed in Table S4.
RNAi
siRNA oligonucleotides targeting DANCR, LRPPRC, and negative control siRNAs were purchased from GenePharma (Shanghai, China). The siRNA sequence is listed in Table S4. siRNA transfections were performed using 75 nM siRNA and Lipofectamine RNAimax (Life Technologies, Waltham, MA, USA), as described previously.57
Lentivirus Transduction
To establish stable overexpression and knockdown cell lines, full-length DANCR or shRNA sequences that specifically target DANCR were cloned into vectors pCDH-CMV-MCS-EF1-Puro or pLKO.1-Puro. Bidirectional sequencing was performed to verify the correct sequences. The sequences of the shRNAs are listed in Table S5. Lentivirus production and infection were conducted as described previously.62
In Vitro Cell Wound-Healing, Migration, and Invasion Assays
Wound-healing assays and Transwell assays were performed to detect cell migration and invasion. The details were described in our previous study.63
Cell Proliferation Assay
The MTT assay and the colony formation assay were performed to detect cell viability. Cell cycle analysis and EdU assays were used to detect cell populations at different phases. The details were described in our previous study.57, 62
In Vivo Popliteal LN Metastasis and Tumorigenesis Assays
All procedures involving animals were approved by the Institute Animal Care and Use Committee of Sun Yat-sen University. Male BALB/c nude mice (4–5 weeks old) were purchased from the Experimental Animal Center of Sun Yat-sen University and housed in specific pathogen-free (SPF) barrier facilities. The popliteal LN metastasis assay was described previously.18 Ten mice were included in each group, and lentivirus-transduced UM-UC-3 cells (3 × 106 cells) that stably expressed firefly luciferase were inoculated into the mice’s footpads. Lymphatic metastasis was monitored and imaged using a bioluminescence imaging system (PerkinElmer, IVIS Spectrum Imaging System, Waltham, MA, USA) 6 weeks after the injections. The primary tumors and popliteal LNs were enucleated and embedded in paraffin. The LN volumes were calculated using the following formula: LN volume (mm3) = (length [mm]) × (width [mm])2 × 0.52. The formalin-fixed paraffin-embedded (FFPE) samples were analyzed using IHC with anti-luciferase antibodies (Abcam, Cambridge, MA, USA). Images were captured using a Nikon Eclipse 80i system with NIS-Elements software (Nikon, Tokyo, Japan).
The tumorigenesis assay was performed as described previously.63, 64 A total of 1 × 106 cells were injected subcutaneously into the right or left side of the dorsum, and five mice were used. At 4 weeks post-implantation, the mice were euthanized and tumors were surgically dissected. The tumor specimens were fixed in 4% paraformaldehyde.
Microarray Analysis
Total RNA was extracted using the Trizol reagent and further purified using a QIAGEN RNeasy Mini Kit, according to the manufacturer’s instructions (QIAGEN, Hilden, Germany). To detect the expression of genes after DANCR knockdown, the Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA) was used, and the analysis was performed by CapitalBio (Beijing, China), according to the manufacturer’s instructions. All primary data in the microarray analysis have been uploaded to GEO: GSE119639.
Western Blotting
Western blotting was performed as previously described.64, 65 Primary antibodies specific to MMP9, CCND1, STAT3, phospho (p)-STAT3(Tyr705), JAK2, p-JAK2(Tyr1007), GAPDH (1:1,000, Cell Signaling Technology, Danvers, MA, USA), PLAU, Ki-67 (1:1,000, Abcam,), and LRPPRC (1:200, Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used. The blots were then incubated with goat anti-rabbit or anti-mouse secondary antibody (Cell Signaling Technology), and they were visualized using enhanced chemiluminescence.
ELISA-Based Quantification of Secreted IL-11
The cell culture supernatant was collected, and the secreted IL-11 was quantified using a Human IL-11 Quantikine ELISA Kit (D1100, R&D Systems), according to the manufacturer’s instructions.
IHC Staining and Scoring Analyses
IHC staining and score calculation were conducted as described previously.57, 62 Anti-Ki67 antibodies (1:500, Zhongshan Bio-Tech, Beijing, China), CCND1 (1:100, Cell Signaling Technology), and PLAU (1:100, Abcam) were used to detect their expression levels in mouse tumors. Images were visualized using a Nikon ECLIPSE Ti (Tokyo, Japan) microscope system and processed using Nikon software.
Cytosolic and Nuclear Fraction and RNA-FISH
The cellular fraction was isolated as described previously.11 Briefly, 107 cells were harvested, resuspended in 1 mL ice-cold RNase-free PBS, 1 mL buffer C1 (1.28 M Sucrose, 40 mM Tris-HCl [pH 7.5], 20 mM MgCl2, and 4% Triton X-100), and 3 mL RNase-free water; and incubated for 15 min on ice. The cells were then centrifuged for 15 min at 3,000 × g, and the supernatant containing the cytoplasmic constituents and the nuclear pellet was retained for RNA extraction.
The FISH was performed as previously described.11 Briefly, UM-UC-3 cells were seeded and fixed with 4% paraformaldehyde, treated with 0.5% Triton in PBS, followed by pre-hybridization. They were then hybridized with the probe (5 μM) overnight. The CY3-labeled U6 and 18S probes were provided by Ribo Bio (Guangzhou, China), and the DANCR probes were synthesized by Sangon (Shanghai, China). The cells were visualized under a confocal microscope (Zeiss, Munich, Germany). The sequences of the probes are listed in Table S6.
RNA Pull-Down
DANCR full-length sense, antisense, and serial deletion sequences were prepared via in vitro transcription using a TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific, Waltham, MA, USA), treated with RNase-free DNase I, and purified using a GeneJET RNA purification kit (Thermo Scientific). The RNA pull-down assay was performed using a Magnetic RNA-Protein Pull-down Kit (Thermo Scientific), according to the manufacturer’s instructions and as described previously.17 The samples were separated by electrophoresis, and DANCR-specific bands were identified using mass spectrometry and retrieved from a human proteome library.
The method to pull down the RNAs interacting with DANCR is shown below. Briefly, 3′ end Biotin-TEG (15 atom triethylene glycol spacer) modified-DNA probes against DANCR and LacZ were synthesized by Sangon. The sequences of the probes are shown in Table S6. UM-UC-3 and T24 cells (2 × 107) were cross-linked for each hybridization reaction. The cell lysates were hybridized with a mixture of biotinylated DNA probes for 4 h at 37°C. The binding complexes were then recovered using streptavidin-conjugated magnetic beads. Finally, RNA was eluted and purified from the beads for qRT-PCR analyses. The LacZ probe set was provided along with the kit to serve as a negative control probe.
RIP Assay
The RIP was performed using the EZ-Magna RIP kit (Millipore, Burlington, MA, USA), according to the manufacturer’s instructions and as described previously.11 Briefly, 107 cells were lysed with RIP lysis buffer using one freeze-thaw cycle. Cell extracts were coimmunoprecipitated with anti-LRPPRC (Abcam) antibodies, and the retrieved RNA was subjected to qRT-PCR analysis. Normal immunoglobulin G (IgG) was used as a negative control. For qRT-PCR analysis, U6 was used as a non-specific control.
Actinomycin D Treatment for mRNA Stability Assay
BCa cells were treated with 5 μg/mL actinomycin D (Sigma-Aldrich, St. Louis, MO, USA) to inhibit gene transcription, and cells were collected at various time intervals. CCND1, PLAU, IL-11, and MMP9 mRNA levels were assessed using qRT-PCR.
Luciferase Assay
The indicated regions of the IL-11, CCND1, and PLAU promoters were cloned into psiCHECK2 luciferase reporter plasmid and transfected into 293T cells, separately. The DANCR siRNA-1 or LRPPRC siRNA was co-transfected into cells, separately. The luciferase activity was detected according to the manual of the Dual-Luciferase Reporter Assay system (Promega, Madison, WI, USA) at 48 h after transfection. The Renilla luciferase activity was normalized against the Firefly luciferase activity.
Statistical Analyses
Quantitative data are presented as the means ± the SD of three independent experiments. Differences between two groups were analyzed using the unpaired or paired Student’s t test (two-tailed tests). When more than two groups were compared, one-way ANOVA followed by Dunnett’s multiple comparison test were performed. Data from the clinical analysis were shown as the median with the interquartile range. The Mann-Whitney U test was used for independent samples when the population could not be assumed to be normally distributed. Pearson’s chi-square test was used to analyze the clinical variables. Cumulative survival time was calculated using the Kaplan-Meier method and analyzed by the log-rank test. A multivariate Cox proportional hazards model was used to estimate the adjusted hazard ratios (HRs) and 95% confidence intervals (CIs) and to identify independent prognostic factors. Spearman’s correlation analysis was performed to determine the correlation between two variables. All statistical analyses in this study were performed using SPSS 19.0 software (SPSS, Armonk, NY, USA) or GraphPad Prism 5.0 (GraphPad, La Jolla, CA, USA). A p value < 0.05 was considered significant.
Author Contributions
X.C., Z.C., and T.L. designed this study. Z.C., X.C., and R.X. conducted experiments and performed data analysis. W.D. and J. Han performed clinical data analysis. H.L. performed bioinformatic analysis. J.Z. and Q.Z. performed RIP and RNA pull-down assay. X.C., J. Huang, and T.L. wrote and reviewed the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grants 81702523, 81825016, 81772719, 81772728, 81572514, and 81472384), the National Natural Science Foundation of Guangdong (grants 2016A030313321, 2016A030313244, and 2015A030311011), the Science and Technology Program of Guangzhou (grants 201804010041, 201604020156, and 201604020177), the Science and Technology Planning Project of Guangdong Province (grant 2017B020227007), the Guangdong Special Support Program (2017TX04R246), the Fundamental Research Funds for the Central Universities (for X.C., 18ykpy18), the Project Supported by Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (for T.L.), the Yat-Sen Scholarship for Young Scientist (for X.C.), Sun Yat-sen Initiative Program for Scientific Research (for X.C., YXQH201708), the Cultivation of Major Projects and Emerging, Interdisciplinary Fund, Sun Yat-Sen University (grant 16ykjc18), and the National Clinical Key Specialty Construcion Project for Department of Urology and Department of Oncology, as well as grant KLB09001 from the Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education Institutes, Sun-Yat-Sen University and grant [2013]163 from the Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology.
Footnotes
Supplemental Information includes seven figures and six tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.12.015.
Contributor Information
Xu Chen, Email: chenx457@mail.sysu.edu.cn.
Jian Huang, Email: urolhj@sina.com.
Tianxin Lin, Email: lintx@mail.sysu.edu.cn.
Supplemental Information
References
- 1.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Van Batavia J., Yamany T., Molotkov A., Dan H., Mansukhani M., Batourina E., Schneider K., Oyon D., Dunlop M., Wu X.R. Bladder cancers arise from distinct urothelial sub-populations. Nat. Cell Biol. 2014;16:982–991. doi: 10.1038/ncb3038. [DOI] [PubMed] [Google Scholar]
- 4.Youssef R.F., Raj G.V. Lymphadenectomy in management of invasive bladder cancer. Int. J. Surg. Oncol. 2011;2011:758189. doi: 10.1155/2011/758189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X.R. Urothelial tumorigenesis: a tale of divergent pathways. Nat. Rev. Cancer. 2005;5:713–725. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- 6.Hautmann R.E., de Petriconi R.C., Pfeiffer C., Volkmer B.G. Radical cystectomy for urothelial carcinoma of the bladder without neoadjuvant or adjuvant therapy: long-term results in 1100 patients. Eur. Urol. 2012;61:1039–1047. doi: 10.1016/j.eururo.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 7.Cao Y. Opinion: emerging mechanisms of tumour lymphangiogenesis and lymphatic metastasis. Nat. Rev. Cancer. 2005;5:735–743. doi: 10.1038/nrc1693. [DOI] [PubMed] [Google Scholar]
- 8.Karaman S., Detmar M. Mechanisms of lymphatic metastasis. J. Clin. Invest. 2014;124:922–928. doi: 10.1172/JCI71606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitt A.M., Chang H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu P., Chen X., Xie R., Han J., Xie W., Wang B., Dong W., Chen C., Yang M., Jiang J. lncRNA HOXD-AS1 Regulates Proliferation and Chemo-Resistance of Castration-Resistant Prostate Cancer via Recruiting WDR5. Mol. Ther. 2017;25:1959–1973. doi: 10.1016/j.ymthe.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu G., Ye Z., Zhao X., Ji Z. SP1-induced up-regulation of lncRNA SNHG14 as a ceRNA promotes migration and invasion of clear cell renal cell carcinoma by regulating N-WASP. Am. J. Cancer Res. 2017;7:2515–2525. [PMC free article] [PubMed] [Google Scholar]
- 14.Yue B., Liu C., Sun H., Liu M., Song C., Cui R., Qiu S., Zhong M. A Positive Feed-Forward Loop between LncRNA-CYTOR and Wnt/β-Catenin Signaling Promotes Metastasis of Colon Cancer. Mol. Ther. 2018;26:1287–1298. doi: 10.1016/j.ymthe.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan J.H., Yang F., Wang F., Ma J.Z., Guo Y.J., Tao Q.F., Liu F., Pan W., Wang T.T., Zhou C.C. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Lin Z., Sun L., Xie S., Zhang S., Fan S., Li Q., Chen W., Pan G., Wang W., Weng B. Chemotherapy-Induced Long Non-coding RNA 1 Promotes Metastasis and Chemo-Resistance of TSCC via the Wnt/β-Catenin Signaling Pathway. Mol. Ther. 2018;26:1494–1508. doi: 10.1016/j.ymthe.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X., Xie R., Gu P., Huang M., Han J., Dong W., Xie W., Wang B., He W., Zhong G. Long noncoding RNA LBCS inhibits self-renewal and chemoresistance of bladder cancer stem cells through epigenetic silencing of SOX2. Clin. Cancer Res. 2018 doi: 10.1158/1078-0432.CCR-18-1656. Published online November 5, 2018. [DOI] [PubMed] [Google Scholar]
- 18.He W., Zhong G., Jiang N., Wang B., Fan X., Chen C., Chen X., Huang J., Lin T. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J. Clin. Invest. 2018;128:861–875. doi: 10.1172/JCI96218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kretz M., Webster D.E., Flockhart R.J., Lee C.S., Zehnder A., Lopez-Pajares V., Qu K., Zheng G.X., Chow J., Kim G.E. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Q.C., Rui Z.H., Guo Z.L., Xie W., Shan S., Ren T. LncRNA-DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mTOR expression. J. Cell. Mol. Med. 2018;22:1527–1537. doi: 10.1111/jcmm.13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan S.X., Wang J., Yang F., Tao Q.F., Zhang J., Wang L.L., Yang Y., Liu H., Wang Z.G., Xu Q.G. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L., Yang C., Chen S., Wang G., Shi B., Tao X., Zhou L., Zhao J. Long Noncoding RNA DANCR Is a Positive Regulator of Proliferation and Chondrogenic Differentiation in Human Synovium-Derived Stem Cells. DNA Cell Biol. 2017;36:136–142. doi: 10.1089/dna.2016.3544. [DOI] [PubMed] [Google Scholar]
- 23.Zhu L., Xu P.C. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem. Biophys. Res. Commun. 2013;432:612–617. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 24.Jia J., Li F., Tang X.S., Xu S., Gao Y., Shi Q., Guo W., Wang X., He D., Guo P. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget. 2016;7:37868–37881. doi: 10.18632/oncotarget.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y., Zhang M., Liang L., Li J., Chen Y.X. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int. J. Clin. Exp. Pathol. 2015;8:11480–11484. [PMC free article] [PubMed] [Google Scholar]
- 26.Xie S., Yu X., Li Y., Ma H., Fan S., Chen W., Pan G., Wang W., Zhang H., Li J., Lin Z. Upregulation of lncRNA ADAMTS9-AS2 Promotes Salivary Adenoid Cystic Carcinoma Metastasis via PI3K/Akt and MEK/Erk Signaling. Mol. Ther. 2018;26:2766–2778. doi: 10.1016/j.ymthe.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst M., Putoczki T.L. Molecular pathways: IL11 as a tumor-promoting cytokine-translational implications for cancers. Clin. Cancer Res. 2014;20:5579–5588. doi: 10.1158/1078-0432.CCR-13-2492. [DOI] [PubMed] [Google Scholar]
- 28.Moquet-Torcy G., Tolza C., Piechaczyk M., Jariel-Encontre I. Transcriptional complexity and roles of Fra-1/AP-1 at the uPA/Plau locus in aggressive breast cancer. Nucleic Acids Res. 2014;42:11011–11024. doi: 10.1093/nar/gku814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watters A.D., Latif Z., Forsyth A., Dunn I., Underwood M.A., Grigor K.M., Bartlett J.M. Genetic aberrations of c-myc and CCND1 in the development of invasive bladder cancer. Br. J. Cancer. 2002;87:654–658. doi: 10.1038/sj.bjc.6600531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chujo T., Ohira T., Sakaguchi Y., Goshima N., Nomura N., Nagao A., Suzuki T. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 2012;40:8033–8047. doi: 10.1093/nar/gks506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuchiya N., Fukuda H., Nakashima K., Nagao M., Sugimura T., Nakagama H. LRP130, a single-stranded DNA/RNA-binding protein, localizes at the outer nuclear and endoplasmic reticulum membrane, and interacts with mRNA in vivo. Biochem. Biophys. Res. Commun. 2004;317:736–743. doi: 10.1016/j.bbrc.2004.03.103. [DOI] [PubMed] [Google Scholar]
- 32.May M., Herrmann E., Bolenz C., Tiemann A., Brookman-May S., Fritsche H.M., Burger M., Buchner A., Gratzke C., Wülfing C. Lymph node density affects cancer-specific survival in patients with lymph node-positive urothelial bladder cancer following radical cystectomy. Eur. Urol. 2011;59:712–718. doi: 10.1016/j.eururo.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 33.Shariat S.F., Ehdaie B., Rink M., Cha E.K., Svatek R.S., Chromecki T.F., Fajkovic H., Novara G., David S.G., Daneshmand S. Clinical nodal staging scores for bladder cancer: a proposal for preoperative risk assessment. Eur. Urol. 2012;61:237–242. doi: 10.1016/j.eururo.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 34.Wu S., Zheng J., Li Y., Yu H., Shi S., Xie W., Liu H., Su Y., Huang J., Lin T. A Radiomics Nomogram for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. Clin. Cancer Res. 2017;23:6904–6911. doi: 10.1158/1078-0432.CCR-17-1510. [DOI] [PubMed] [Google Scholar]
- 35.Lin C., Zhang S., Wang Y., Wang Y., Nice E., Guo C., Zhang E., Yu L., Li M., Liu C. Functional Role of a Novel Long Noncoding RNA TTN-AS1 in Esophageal Squamous Cell Carcinoma Progression and Metastasis. Clin. Cancer Res. 2018;24:486–498. doi: 10.1158/1078-0432.CCR-17-1851. [DOI] [PubMed] [Google Scholar]
- 36.Hou Z., Xu X., Fu X., Tao S., Zhou J., Liu S., Tan D. HBx-related long non-coding RNA MALAT1 promotes cell metastasis via up-regulating LTBP3 in hepatocellular carcinoma. Am. J. Cancer Res. 2017;7:845–856. [PMC free article] [PubMed] [Google Scholar]
- 37.Ma M., Zhang Y., Weng M., Hu Y., Xuan Y., Hu Y., Lv K. lncRNA GCAWKR Promotes Gastric Cancer Development by Scaffolding the Chromatin Modification Factors WDR5 and KAT2A. Mol. Ther. 2018;26:2658–2668. doi: 10.1016/j.ymthe.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C., He W., Huang J., Wang B., Li H., Cai Q., Su F., Bi J., Liu H., Zhang B. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat. Commun. 2018;9:3826. doi: 10.1038/s41467-018-06152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Zeng X., Wang N., Zhao W., Zhang X., Teng S., Zhang Y., Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol. Cancer. 2018;17:89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Lu Z., Wang N., Feng J., Zhang J., Luan L., Zhao W., Zeng X. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp. Mol. Med. 2018;50:57. doi: 10.1038/s12276-018-0082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z., Hou P., Fan D., Dong M., Ma M., Li H., Yao R., Li Y., Wang G., Geng P. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24:59–71. doi: 10.1038/cdd.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruzzenente B., Metodiev M.D., Wredenberg A., Bratic A., Park C.B., Cámara Y., Milenkovic D., Zickermann V., Wibom R., Hultenby K. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volpon L., Culjkovic-Kraljacic B., Sohn H.S., Blanchet-Cohen A., Osborne M.J., Borden K.L.B. A biochemical framework for eIF4E-dependent mRNA export and nuclear recycling of the export machinery. RNA. 2017;23:927–937. doi: 10.1261/rna.060137.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu D.H., Zhu Z., Wakefield M.R., Xiao H., Bai Q., Fang Y. The role of IL-11 in immunity and cancer. Cancer Lett. 2016;373:156–163. doi: 10.1016/j.canlet.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Zurita A.J., Troncoso P., Cardó-Vila M., Logothetis C.J., Pasqualini R., Arap W. Combinatorial screenings in patients: the interleukin-11 receptor alpha as a candidate target in the progression of human prostate cancer. Cancer Res. 2004;64:435–439. doi: 10.1158/0008-5472.can-03-2675. [DOI] [PubMed] [Google Scholar]
- 47.Onnis B., Fer N., Rapisarda A., Perez V.S., Melillo G. Autocrine production of IL-11 mediates tumorigenicity in hypoxic cancer cells. J. Clin. Invest. 2013;123:1615–1629. doi: 10.1172/JCI59623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tao L., Huang G., Wang R., Pan Y., He Z., Chu X., Song H., Chen L. Cancer-associated fibroblasts treated with cisplatin facilitates chemoresistance of lung adenocarcinoma through IL-11/IL-11R/STAT3 signaling pathway. Sci. Rep. 2016;6:38408. doi: 10.1038/srep38408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng J., Liu Y., Lee H., Herrmann A., Zhang W., Zhang C., Shen S., Priceman S.J., Kujawski M., Pal S.K. S1PR1-STAT3 signaling is crucial for myeloid cell colonization at future metastatic sites. Cancer Cell. 2012;21:642–654. doi: 10.1016/j.ccr.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X., Yue P., Page B.D., Li T., Zhao W., Namanja A.T., Paladino D., Zhao J., Chen Y., Gunning P.T., Turkson J. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc. Natl. Acad. Sci. USA. 2012;109:9623–9628. doi: 10.1073/pnas.1121606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Canesin G., Evans-Axelsson S., Hellsten R., Sterner O., Krzyzanowska A., Andersson T., Bjartell A. The STAT3 Inhibitor Galiellalactone Effectively Reduces Tumor Growth and Metastatic Spread in an Orthotopic Xenograft Mouse Model of Prostate Cancer. Eur. Urol. 2016;69:400–404. doi: 10.1016/j.eururo.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 52.Johnson D.E., O’Keefe R.A., Grandis J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blasi F., Sidenius N. The urokinase receptor: focused cell surface proteolysis, cell adhesion and signaling. FEBS Lett. 2010;584:1923–1930. doi: 10.1016/j.febslet.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 54.Resmini G., Rizzo S., Franchin C., Zanin R., Penzo C., Pegoraro S., Ciani Y., Piazza S., Arrigoni G., Sgarra R., Manfioletti G. HMGA1 regulates the Plasminogen activation system in the secretome of breast cancer cells. Sci. Rep. 2017;7:11768. doi: 10.1038/s41598-017-11409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banyard J., Chung I., Migliozzi M., Phan D.T., Wilson A.M., Zetter B.R., Bielenberg D.R. Identification of genes regulating migration and invasion using a new model of metastatic prostate cancer. BMC Cancer. 2014;14:387. doi: 10.1186/1471-2407-14-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sasano T., Mabuchi S., Kozasa K., Kuroda H., Kawano M., Takahashi R., Komura N., Yokoi E., Matsumoto Y., Hashimoto K. The Highly Metastatic Nature of Uterine Cervical/Endometrial Cancer Displaying Tumor-Related Leukocytosis: Clinical and Preclinical Investigations. Clinical Cancer Res. 2018;24:4018–4029. doi: 10.1158/1078-0432.CCR-17-2472. [DOI] [PubMed] [Google Scholar]
- 57.Chen X., Gu P., Xie R., Han J., Liu H., Wang B., Xie W., Xie W., Zhong G., Chen C. Heterogeneous nuclear ribonucleoprotein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J. Cell. Mol. Med. 2017;21:1266–1279. doi: 10.1111/jcmm.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malumbres M., Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 59.Velagapudi S.P., Cameron M.D., Haga C.L., Rosenberg L.H., Lafitte M., Duckett D.R., Phinney D.G., Disney M.D. Design of a small molecule against an oncogenic noncoding RNA. Proc. Natl. Acad. Sci. USA. 2016;113:5898–5903. doi: 10.1073/pnas.1523975113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Disney M.D., Angelbello A.J. Rational Design of Small Molecules Targeting Oncogenic Noncoding RNAs from Sequence. Acc. Chem. Res. 2016;49:2698–2704. doi: 10.1021/acs.accounts.6b00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen X., Gu P., Li K., Xie W., Chen C., Lin T., Huang J. Gene expression profiling of WDR5 regulated genes in bladder cancer. Genom. Data. 2015;5:27–29. doi: 10.1016/j.gdata.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen X., Xie W., Gu P., Cai Q., Wang B., Xie Y., Dong W., He W., Zhong G., Lin T., Huang J. Upregulated WDR5 promotes proliferation, self-renewal and chemoresistance in bladder cancer via mediating H3K4 trimethylation. Sci. Rep. 2015;5:8293. doi: 10.1038/srep08293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang J., Chen X., Liu H., Shao J., Xie R., Gu P., Duan C. Polypyrimidine Tract-Binding Protein 1 promotes proliferation, migration and invasion in clear-cell renal cell carcinoma by regulating alternative splicing of PKM. Am. J. Cancer Res. 2017;7:245–259. [PMC free article] [PubMed] [Google Scholar]
- 64.Fan X., Chen X., Deng W., Zhong G., Cai Q., Lin T. Up-regulated microRNA-143 in cancer stem cells differentiation promotes prostate cancer cells metastasis by modulating FNDC3B expression. BMC Cancer. 2013;13:61. doi: 10.1186/1471-2407-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen X., Wu J., Liu H., He Z., Gu M., Wang N., Ma J., Hu J., Xia L., He H. Approaches to efficient production of recombinant angiogenesis inhibitor rhVEGI-192 and characterization of its structure and antiangiogenic function. Protein Sci. 2010;19:449–457. doi: 10.1002/pro.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.