Abstract
Significance: Cellular reactive oxygen species (ROS) mediate redox signaling cascades that are critical to numerous physiological and pathological processes. Analytical methods to monitor cellular ROS levels and proteomic platforms to identify oxidative post-translational modifications (PTMs) of proteins are critical to understanding the triggers and consequences of redox signaling.
Recent Advances: The prevalence and significance of redox signaling has recently been illuminated through the use of chemical probes that allow for sensitive detection of cellular ROS levels and proteomic dissection of oxidative PTMs directly in living cells.
Critical Issues: In this review, we provide a comprehensive overview of chemical probes that are available for monitoring ROS and oxidative PTMs, and we highlight the advantages and limitations of these methods.
Future Directions: Despite significant advances in chemical probes, the low levels of cellular ROS and low stoichiometry of oxidative PTMs present challenges for accurately measuring the extent and dynamics of ROS generation and redox signaling. Further improvements in sensitivity and ability to spatially and temporally control readouts are essential to fully illuminate cellular redox signaling.
Keywords: oxidative stress, cysteine oxidation, redox proteomics, redox sensors
Introduction
Reactive oxygen species (ROS) are produced endogenously during a variety of cellular processes, and primary among these is aerobic respiration. In eukaryotic systems, the majority of cellular ROS is believed to be generated during the transfer of electrons through components of the electron transport chain localized in the inner mitochondrial membrane. This electron transport process results in the reduction of molecular oxygen (O2) to water, and it drives adenosine triphosphate (ATP) synthesis to fuel key physiological processes. However, partial reduction of O2 can result in the production of numerous ROS, including superoxide (O2•−) and hydrogen peroxide (H2O2) (Fig. 1A). Further, controlled ROS production also occurs at reduced nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX) enzymes, which oxidize NADPH to nicotinamide adenine dinucleotide phosphate (NADP+), and reduce O2 to produce O2•− as a primary product. In various physiological processes, including cellular proliferation and migration, cytosolic regulatory subunits are assembled on membrane catalytic NOX proteins to activate ROS production (60, 118). Importantly, localization of NOX subunits provides spatial regulation within cells, such as the assembly of regulatory and catalytic subunits at the leading edge of migrating cells (125).
FIG. 1.
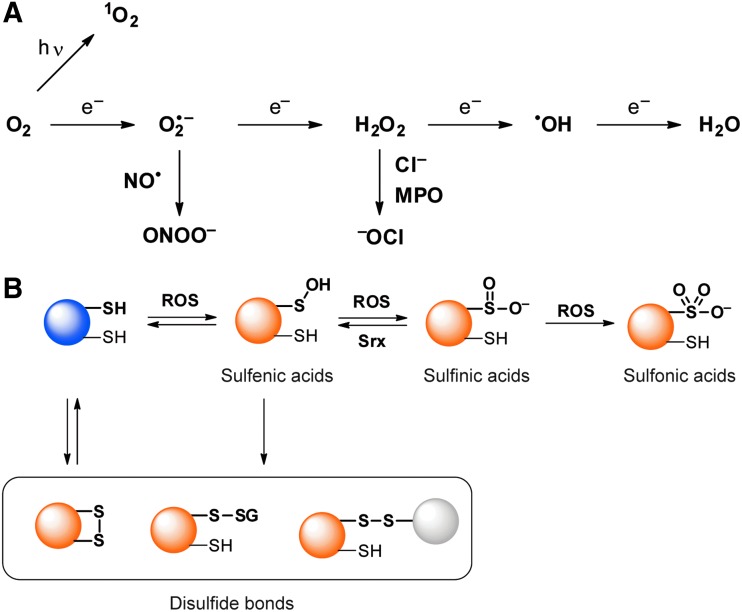
ROS and oxidative PTMs of cysteine. (A) ROS are derived from molecular oxygen through excitation and reduction. (B) Oxidative PTMs of cysteine highlighted in this review. PTMs, post-translational modifications; ROS, reactive oxygen species. Color images are available online.
To counteract the damaging effects of excessive ROS generation, organisms possess a system of small-molecule antioxidants, including ascorbic acid and glutathione, as well as redox enzymes, including glutaredoxins (Grxs), thioredoxins (Trxs), and peroxiredoxins (Prxs) (126). In unicellular organisms, ROS sensors trigger activation of antioxidant systems when ROS concentrations exceed physiological levels. For example, the H2O2 sensors OxyR (e.g., prokaryotes) and Orp1 (e.g., yeast) directly sense peroxide levels and activate the production of antioxidant enzymes such as catalase (18). In multicellular organisms, antioxidant enzymes play important roles in spatiotemporal regulation of redox signaling. Evidence suggests that the subcellular localization of antioxidant enzymes restricts ROS to a particular cellular compartment (126). In addition, antioxidant enzymes such as Prxs are inactivated by oxidation or phosphorylation, and they form temporal “hot spots” for redox signaling through feedback inhibition (102, 133).
These highly controlled systems of cellular ROS generation and attenuation result in strict spatiotemporal regulation of cellular ROS levels. It was initially believed that the high reactivity of ROS resulted mainly in irreversible damage to biomolecules; however, it is now realized that ROS can play critical roles in cell signaling and regulation. This process, known as redox signaling, is a key component of numerous physiological and pathological processes, including growth-factor signaling and inflammation. One of the major effectors of ROS in redox signaling are thiol groups of cysteine residues in proteins, which can be transformed to form distinct oxidative post-translational modifications (PTMs) such as S-sulfenylation, S-sulfinylation, and S-sulfonylation, as well as disulfide-bond formation (Fig. 1B). S-sulfenylation (sulfenic acid [RSOH]) is a reversible PTM under cellular redox potentials, at which sulfenic acids can readily be reduced to the corresponding thiol by cellular antioxidants. Sulfenic acids can also be converted to intra- or intermolecular disulfide bonds with thiol groups of redox-active small molecules and proteins. Further oxidation of sulfenic acids results in S-sulfinylation and S-sulfonylation. Sulfinic acids (RSO2H) are only reversed by sulfiredoxin (Srx) (14), and sulfonic acids (RSO3H) are considered irreversible in cells. Among the oxidative PTMs of cysteine (Cys), the reversibility of S-sulfenylation renders it suitable for temporal signal transductions akin to phosphorylation. Well-studied examples of S-sulfenylation include protein tyrosine phosphatase 1B (PTP1B) and epidermal growth factor receptor (EGFR). PTP1B is inhibited by H2O2 through S-sulfenylation of the catalytic Cys215 on growth-factor stimulation (63), and EGFR is activated through S-sulfenylation of Cys797 (93). Both these S-sulfenylation events serve to further magnify phosphorylation events that are key to growth-factor signaling.
To elucidate the ubiquity and functional consequences of redox signaling resulting from cellular ROS generation, it is critical to be able to visualize ROS production and analyze subsequent oxidative PTMs of cysteine. ROS detection has been enabled by small-molecule and protein-based probes, which are coupled to spectroscopic readouts that provide sensitive ROS detection in living cells (in situ). Oxidative PTMs resulting from increased ROS levels can be interrogated by applying chemical probes coupled with proteomic technologies such as gel electrophoresis and mass spectrometry (MS). In this review, we summarize the available array of chemical probes for analyzing both ROS generation and downstream oxidative cysteine PTMs. Detailed mechanisms, advantages, and limitations of each method will be discussed where relevant.
Chemical Probes for ROS
ROS and the related reactive nitrogen species (RNS) typically display high reactivity with biomolecules (Fig. 1A). ROS include O2•−, H2O2, hydroxyl radical (•OH), hypochlorite (−OCl), and singlet oxygen (1O2). RNS include peroxynitrite (ONOO−), which is generated via the reaction of nitric oxide (•NO) with O2•−. Due to the distinct reactivity characteristics of each ROS and RNS, these species can display unique effects on physiological processes. For example, O2•− and H2O2 are relatively mild reactants and are typically implicated in redox signaling that is mediated through oxidative PTMs of cysteine. In contrast, −OCl is a stronger oxidant that is produced by myeloperoxidase (MPO) in phagosomes, and it attacks invading microbes that are engulfed by neutrophils and macrophages. Due to the unique reactivity characteristics of each ROS, it is imperative to develop methods that are specific for each species. To this end, a number of chemical probes that are specific to distinct ROS have been developed and utilize electron paramagnetic resonance (EPR), chemiluminescence, fluorescence spectroscopy, and microscopy platforms to monitor ROS levels. In this review, we focus on chemical probes for O2•− and H2O2, which are considered the most relevant to redox signaling (18, 102).
Chemical Probes for O2•−
EPR probes for O2•−
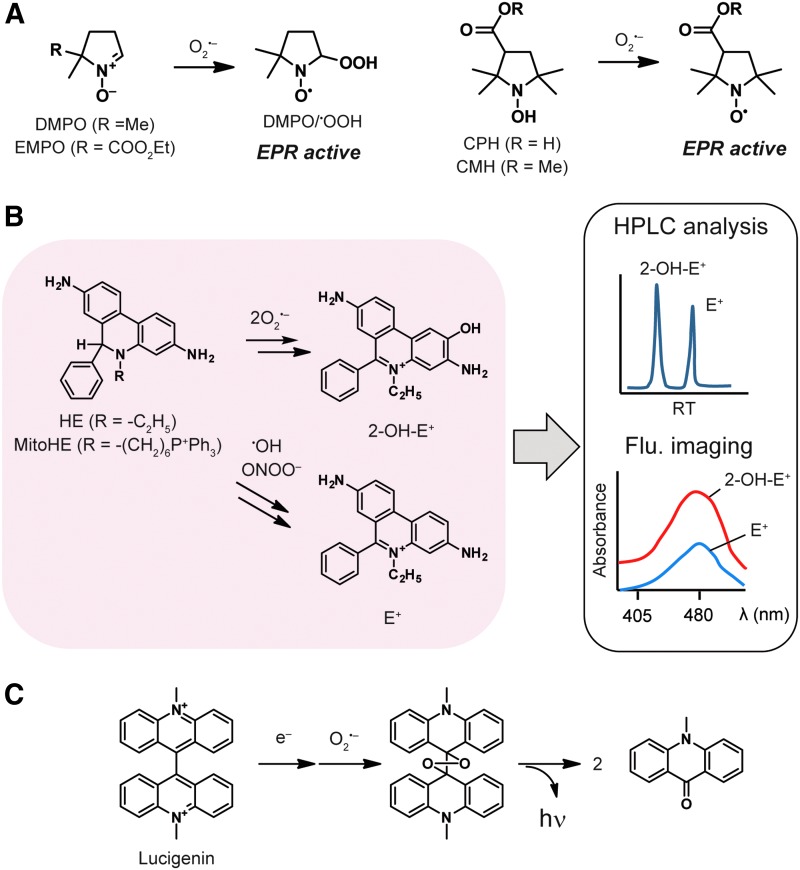
EPR detects the absorbance of microwave energy by radicals with unpaired electrons in an applied magnetic field. Because ROS are very short lived and unable to be directly detected in biological samples, chemical probes that form stable adducts with ROS, termed spin traps, are used for biological ROS detection by EPR. A nitrone-based probe, 5,5-dimethyl-1-pyrroline-N-oxide (DMPO), is the most widely used EPR probe for O2•−. DMPO reacts with unstable free radicals, such as O2•− and •OH, to form relatively stable radical adducts, DMPO/•OOH and DMPO/•OH, respectively, which can be characterized by EPR (Fig. 2A). The DMPO probe has been applied to detect free radicals in a wide variety of cells, including endothelial cells (105, 144) and myocytes (128). One of the limitations of DMPO is the short half-life of DMPO/•OOH (t1/2 ∼45 s), which lowers the detection sensitivity in EPR measurements. To overcome this limitation, a derivative of DMPO, 5-(diethoxyphosphoryl)-5-methyl-1-pyrroline-N-oxide (DEPMPO), has been developed, which generates a more stable adduct with O2•− (t1/2 ∼15 min) (29). Another limitation of DMPO is the potential decomposition of DMPO/•OOH to DMPO/•OH through reductive degradation by biological antioxidants. The susceptibility to reductive degradation hampers sensitive detection in situ, resulting in false negatives when O2•− levels are low (9). The possibility of degradation also requires control experiments with superoxide dismutase (SOD) mimics to distinguish O2•− from •OH (119). Further, nitrone-based probes are limited by the slow reaction rates displayed with O2•− (e.g., k = 74 M−1·s−1 for a nitrone-based derivative, EMPO) (22).
FIG. 2.
Chemical probes for superoxide. (A) EPR-based spin traps form stable radicals with superoxide. (B) HE and MitoHE react with superoxide to form fluorescent products. (C) Lucigenin releases a photon on reaction with superoxide. EPR, electron paramagnetic resonance; HE, hydroethidine. Color images are available online.
Cyclic hydroxylamine-based probes, including 1-hydroxy-3-carboxy-2, 2, 5-tetramethyl-pyrrolidine hydrochloride (CPH), provide an alternative method for O2•− detection with high reliability and sensitivity (59) (Fig. 2A). CPH reacts with O2•− with a fast reaction rate (k = 3.2 × 103 M−1·s−1) to give a stable radical product (t1/2 = 330 min in smooth muscle cells) (23). CMH (1-hydroxy-3-methoxycarbonyl-2,2,5,5-tetramethylpyrrolidine), a plasma membrane-permeable derivative of CPH, has been applied to O2•− detection in cultured cells and tissue samples (24, 59). Cyclic hydroxylamine-based probes have overcome drawbacks of nitrone-based probes, such as slow reaction with O2•− and reductive degradation in living organisms. However, the reaction rates of hydroxylamine-based probes are still slower than that of spontaneous dismutation of O2•− under physiological conditions (k = 105–106 M−1·s−1) (113). In addition, these probes can be oxidized by multiple ROS and auto-oxidation as well, which necessitates control experiments with specific ROS-scavengers, such as SOD and catalase.
Triarylmethyl (TAM) radical-based probes have also been reported; the TAM radicals are EPR active and lose the EPR signal through the reaction with O2•− at fast reaction rates (k = 3.1 × 103 M−1·s−1 for a TAM derivative, OXO63) (103). The TAM radical-based probe, CT-02H, showed selectivity to O2•− and ROO• among ROS and was applied to the detection of cellular O2•− release on menadione stimulation (70). A major limitation of TAM radical-based probes is the signal-decreasing (turn-off) response to O2•−, which lowers sensitivity of detection due to high background signals and hampers the wide application of TAM probes in living cells.
Fluorescent and chemiluminescent probes for O2•−
Fluorescence imaging is a facile and sensitive methodology to detect ROS generation by coupling ROS-reactive chemical probes with fluorescence spectroscopy. Hydroethidine (HE) is a widely used fluorescence probe for O2•− both in vitro and in situ (Fig. 2B). HE is oxidized by O2•−, to form the highly fluorescent 2-hydroxyethidium (2-OH-E+) with a high reaction rate (k = 2.6 × 106 M−1·s−1) (138, 139). However, other ROS such as ONOO−, OH•, and H2O2 oxidize HE to give a fluorescent product, ethidium (E+). High-performance liquid chromatography can be used to separate 2-OH-E+ from E+ and provide a quantitative and specific measure of O2•− in the system (28). E+ and 2-OH-E+ can also be distinguished by fluorescence spectroscopy by excitation at 405 nm instead of 480 nm, which significantly reduces the fluorescence signal from E+ (85). However, control experiments utilizing SOD are still required to confirm specific detection of O2•−. A mitochondria-targeted HE derivative, Mito-HE (also known as MitoSOX) contains a positively charged triphenylphosphonium (TPP) group, which allows Mito-HE to accumulate in mitochondria (142). Mito-HE has been applied to the detection of mitochondrial O2•− production in a wide range of cells, including aortic endothelial cells (86) and cultured cerebellar granule neurons (48).
Hydrocyanines are reported as probes for several ROS, including O2•− (57). The reduced cyanines are readily prepared by reduction of cyanine dyes with sodium borohydride, resulting in a drastic change in cyanine absorption and fluorescence. One derivative, hydro-carbocyanine 3 (Cy3), displays a fluorescence increase on treatment with O2•−, •OH, and t-butyl peroxyl radical (TBO•). One advantage of hydrocyanines over HE is the increased stability in aqueous solutions; the half-lives of hydro-Cy3 and hydro-carbocyanine 7 (Cy7) in neutral aqueous solutions are ∼3 days, whereas that of HE is 30 min. Deuterated hydrocyanines have been reported to be more stable against auto-oxidation, which reduces background signals and allows for more sensitive detection of ROS (56). Hydro-Cy3 has been applied to live-cell imaging of rat aortic smooth muscle cells stimulated by angiotensin II. Another advantage of hydrocyanines is near-infrared absorption and fluorescence, which enables applications of hydrocyanines for in vivo imaging of ROS production in mice (35, 57).
Sulfonate-based protecting groups are utilized as a design strategy for O2•− probes. Screening a library of benzenesulfonyl fluorescein (BES) derivatives identified BES-So and BES-H2O2 as fluorescent probes for O2•− and H2O2, respectively (74, 75). In addition, HKSOX-1, a fluorescein protected by triflate groups, is reported to be responsive to O2•− and applied to in vivo O2•− imaging in zebrafish (44).
Chemiluminescent probes are widely used for O2•− detection. The most common chemiluminescent probe for O2•− is lucigenin (Fig. 2C). In the initial step, lucigenin is reduced by O2•− to a cation radical, which then reacts with a second O2•− molecule to form a dioxetane structure. A photon is emitted when the high-energy dioxetane structure is cleaved. Although lucigenin-based chemiluminescence is facile to monitor, lucigenin itself can produce O2•− through a process known as redox cycling. This redox cycling phenomenon leads to the overestimation of O2•− levels in the sample (120), and it is important to minimize artifactual production of O2•− from lucigenin by choosing appropriate experimental conditions (67). Another class of chemiluminescent probes for O2•− are derived from coelenterazine, which is a luciferin that is responsible for the bioluminescence of marine organisms. Coelenterazine has been reported as a chemiluminescent probe for O2•− with enhanced light emission relative to lucigenin (52). One advantage of coelenterazine is that it does not produce artificial O2•− through redox cycling (120). Derivatization of coelenterazine has resulted in the improved probes, 2-methyl-6-phenylimidazo[1,2-a]pyrazin-3(7H)-one (CLA) (116) and 6-(4-methoxyphenyl)-2-methyl-imidazo[1,2-a]pyrazin-3(7H)-one (MCLA) (89); the optimized derivative MCLA has a significantly enhanced emission (>100-fold) over coelenterazine. CLA and MCLA have been applied to O2•− detection in a variety of biological samples such as living cells (53, 54) and tissues (58, 130). It is reported that MCLA can emit light by the reaction with ROS other than O2•− (49, 90); thus, control experiments with SOD and other specific quenchers of ROS are required.
Chemical Probes for Hydrogen Peroxide
Small-molecule-based probes for H2O2
The most widely used chemical probes for H2O2 are reduced dyes such as dihydrodichlorofluorescein (DCFH2) and dihydrorhodamine123. Oxidation of these reduced dyes by ROS generates a fluorescence increase. However, these reduced dyes have several critical drawbacks when considered as fluorescence probes for H2O2. First, DCFH2 has poor reactivity and specificity to H2O2; DCFH2 is even more responsive to highly ROS, including •OH, ONOO−, and −OCl, than H2O2 (112). Thus, DCFH2 should be regarded as an indicator of gross ROS amount rather than an H2O2-specific probe. In addition, DCFH2 can be oxidized by light irradiation, which requires intensive protection from light throughout the experimental workflow. In addition, the irradiation during fluorescence readings can also trigger increased DCFH2 fluorescence, rendering the interpretation of imaging results complicated and unreliable (5, 112).
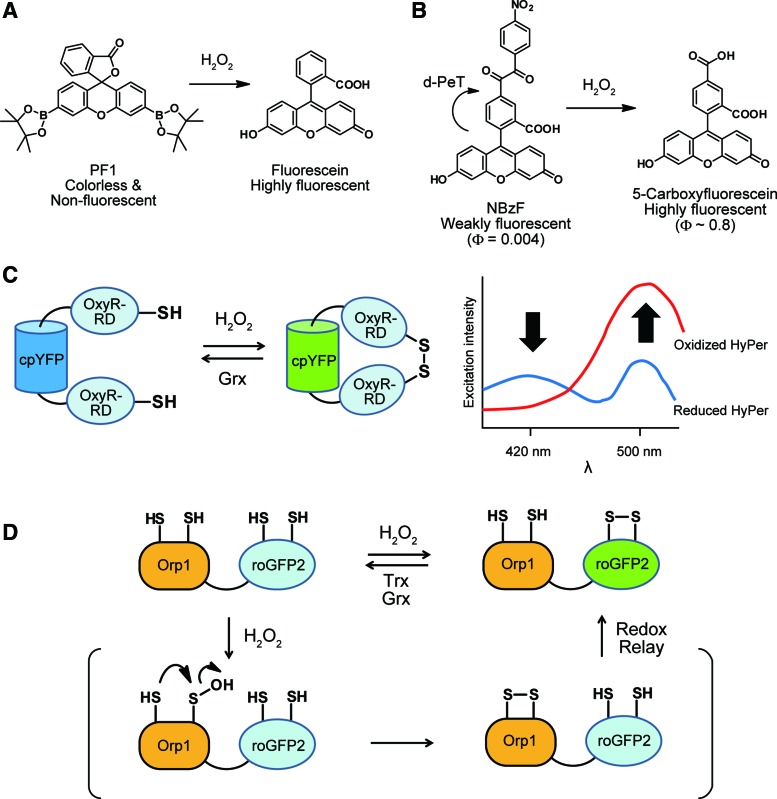
In attempts to overcome these drawbacks, chemical reactions that are specific for H2O2 have been sought as the underlying basis for fluorescence-based H2O2 probes. One example is the deprotection of boronate esters through oxidation by H2O2. Peroxyfluor-1 (PF1) and peroxyresorufin-1 (PR1) contain boronate ester groups, which can be converted to phenols by H2O2 to produce the highly fluorescent molecules, fluorescein (λabs = 488 nm, λem = 520 nm) and resorufin (λabs = 571 nm, λem = 585 nm), respectively (82) (Fig. 3A). PF1 and PR1 show a large fluorescence increment (>1000-fold) with reaction rate constants k = 0.54 M−1·s−1, and 1.0 M−1·s−1, respectively. The response of PF1 is reported to be selective for H2O2, with relatively small fluorescence increments observed for •NO treatment in vitro. Peroxy Green 1 (PG1), a derivative based on TokyoGreen (124), has also been applied for fluorescence imaging of endogenous H2O2 produced by A431 cells and cultured rat hippocampal neurons under epidermal growth factor (EGF) stimulation (83). Similarly, a wide variety of fluorophores have been protected with boronates; for example, rhodol-based derivatives, Peroxy Yellow 1 (PY1) and Peroxy Orange 1 (PO1), were developed for multi-color imaging, generating products that emit yellow (λabs = 519 nm, λem = 548 nm) and orange fluorescence (λabs = 540 nm, λem = 565 nm), respectively (21). PY1 and PO1 have been applied to live-cell imaging of endogenous H2O2 in multiple types of cells, including RAW264.7 macrophages stimulated by phorbol 12-myristate 13-acetate (PMA), and EGF-stimulated A431 cells. A major limitation of boronate-based probes is the slow reaction rate with H2O2 (k = 0.1–1 M−1·s−1). This slow reaction rate results in low sensitivity under physiological conditions where H2O2 is rapidly removed by antioxidant enzymes such as Prxs (132). Another limitation is that boronate ester-based probes have a fluorescence response to ONOO− as well. Multiple reports show the application of boronate-based probes to the detection of ONOO− (114, 137, 143); thus, control experiments with ONOO− scavengers are required to differentiate H2O2 and ONOO− when using boronate-based probes.
FIG. 3.
Fluorescent probes for H2O2. (A) Boronate-protected fluorophores are deprotected by H2O2. (B) The benzil group is cleaved by H2O2 to form highly fluorescent 5-carboxyfluorescein. (C) HyPer enables ratiometric fluorescent measurements of H2O2 in situ. (D) Orp1 forms a disulfide bond on reaction with H2O2, which is relayed to the fused roGFP2, leading to a fluorescence change. H2O2, hydrogen peroxide; roGFP2, redox-sensitive green fluorescent protein 2. Color images are available online.
Benzil chemistry can also be used as a design strategy for H2O2 probes. Benzil (α-dibenzoyl) has been reported to generate benzoic acid by H2O2 in alkaline conditions through an acyclic (Baeyer-Villiger) mechanism (109). To exploit this reaction for H2O2 sensing, benzil groups were appended to fluorescein to generate 5-benzoylcarbonylfluorescein derivatives (3) (Fig. 3B). The benzil group acts as an H2O2-reactive group while also quenching fluorescence through a donor-excited photoinduced electron transfer (d-PeT) mechanism (123, 124). In the d-PeT process, an electron from the excited fluorophore transfers to the lowest unoccupied molecular orbital of the benzil group, which results in non-radiative relaxation of the excitation energy (123). A derivative with a nitro group on the benzil moiety, 5-(4-nitrobenzoyl)carbonylfluorescein (NBzF), displays a 150-fold fluorescence increase (λabs = 495 nm, λem = 519 nm) in the presence of H2O2 with a reaction rate constant k = 3.2 M−1·s−1. In addition to H2O2, NBzF shows a relatively small fluorescence increment in the presence of t-butyl hydroperoxide (t-BuOOH) and ONOO−, but it does not respond to •NO in vitro. NBzF was applied for live-cell imaging of endogenous H2O2 in PMA-stimulated RAW264.7 cells and EGF-stimulated A431 cells. However, NBzF has limitations that are similar to boronate-based probes, including the relatively slow reaction rate with H2O2 and the necessity of control experiments with ROS scavengers.
The enzyme-mediated reaction of a small molecule with H2O2 can provide a robust and sensitive strategy for H2O2 detection. For example, Amplex Red is oxidized by H2O2 in the presence of horseradish peroxidase (HRP) to produce the fluorescent product, resorufin (140). This Amplex Red assay is a reliable method to detect extracellular H2O2 with a limit of detection of ∼5 pmol. However, HRP is inactivated in the highly reducing cytosolic environment, likely due to the reduction of functionally essential disulfide bonds on HRP (43), rendering Amplex Red not applicable to cytosolic H2O2 detection.
Due to the distinct physiological roles of H2O2 in different subcellular locations, it is important to visualize H2O2 production in a particular organelle. To this end, organelle-targeted probes for H2O2 have been developed. One strategy to target small-molecule-based probes to a particular organelle is to utilize organelle-directing groups. Mitochondria peroxy yellow 1 (MitoPY1) is a boronate-based probe conjugated with a TPP group, which is well known to localize in mitochondria due to its positive charge (20). MitoPY1 successfully detected exogenously added H2O2 in the mitochondria. Another strategy for directing probes to distinct organelles is through the use of protein tags such as the SNAP-tag. The SNAP-tag is an engineered human DNA repair protein, human O6-alkylguanine-DNA alkyltransferase (hAGT), which covalently binds to small molecules with an O6-benzylguanine group (51). Boronate-based probes are conjugated with SNAP-tag substrates (SNAP PG1 and PG2) (115) and directed to SNAP-tagged proteins with distinct subcellular localizations. This approach has been used to target boronate probes to the cytosol, nucleus, the extracellular face of the plasma membrane, as well as the endoplasmic reticulum and mitochondrial inner membrane, for exogenous H2O2 detection in each specific subcellular location. Similarly, organelle-specific imaging of endogenous H2O2 has been achieved. An organelle-targeted benzil-based probe, NBzF-BG, which has an O6-benzylguanine group conjugated with NBzF, has been applied to detect endogenous H2O2 produced during phagocytosis (2). NBzF-BG has been presented on the extracellular side of the plasma membrane of RAW264.7 macrophages, localizing NBzF-BG on the inside of the phagosomal membrane on phagocytosis of opsonized microbeads. NBzF-BG localized inside phagosomes has enabled the visualization of H2O2 production in phagosomes.
Genetically encoded fluorescence probes for H2O2
Genetically encoded fluorescent protein-based H2O2 probes, such as HyPer and redox-sensitive green fluorescent protein 2 (roGFP2)-Orp1, are powerful imaging tools with several advantages such as high sensitivity, ratiometric measurements, and ability to localize to a particular organelle. HyPer consists of a circularly permutated yellow fluorescent protein (cpYFP) inserted into the regulatory domain of OxyR, a prokaryotic H2O2 sensor (11) (Fig. 3C). H2O2 oxidizes Cys199 of the OxyR regulatory domain to form a sulfenic acid, followed by disulfide-bond formation with Cys208. The disulfide formation changes the conformation and excitation properties of HyPer, which enables ratiometric measurements with two excitation wavelengths (488/405 nm). The fluorescence response of HyPer is relatively sensitive to H2O2 concentrations, and HyPer is not responsive to other ROS including O2•−, •NO, and ONOO−, and oxidized glutathione (GSSG). Several variants of HyPer have been developed with larger signal enhancement (HyPer-2 and HyPer-3) (13, 77) and different excitation and emission wavelengths (HyPer-Red) (27). The fast reaction rates (k = 1.2–5.0 × 105 M−1·s−1) with H2O2 and the ratiometric response confer HyPer probes that are suitable for various biological applications. HyPer has been applied for live-cell imaging in bacterial and mammalian cells, and it has also been targeted to organelles such as the cytoplasm, mitochondria, nucleus, endoplasmic reticulum, peroxisomes, and plasma membrane (12). The HyPer probes have also been applied to imaging of whole animals, including Caenorhabditis elegans (55) and zebrafish (88). A limitation of HyPer probes is that the fluorescence signal of HyPer can be affected by pH; thus, control experiments with pH sensors such as SypHer (79) are required to interpret HyPer-based imaging results.
Another strategy for genetically encoded H2O2 probes is based on roGFPs (25, 41) coupled with a yeast peroxidase Orp1 (38). Orp1 is oxidized to form a sulfenic acid by H2O2 at the catalytic Cys36, which rapidly forms an intramolecular disulfide bridge with the resolving Cys82. When fused to roGFP2, the disulfide bridge of Orp1 is efficiently relayed to roGFP2, which results in a ratiometric fluorescence signal change (390/480 nm) (Fig. 3D). The response of roGFP2-Orp1 is specific for H2O2 among oxidants such as GSSG, cystine, and dehydroascorbic acid (38). One advantage of roGFP2-Orp1 over HyPer is insensitivity to pH changes within the physiological range. Applications of roGFP2-Orp1 include the visualization of physiological H2O2 production in Drosophila larvae and tissues (6, 8, 122). Although roGFP2-Orp1 and HyPer have been successfully applied to H2O2 imaging in vivo, reaction rates of their sensing moiety are still >100-fold slower than that of typical 2-Cys Prxs (k = ∼107 M−1·s−1), which are abundant in the cytosol. Recently, improved roGPF2-based H2O2 probes, roGFP2-Tsa2ΔCR and roGFP2-Tsa2ΔCPΔCR, have been reported (84). These probes utilize a typical 2-Cys Prx Tsa2, with mutations at the peroxidatic cysteine (CP) and/or the resolving cysteine (CR), as an H2O2-sensitive moiety. These probes are ∼50% oxidized under unstressed conditions, which allows for dynamic monitoring of both increases and decreases in basal H2O2. By using the roGFP2-Tsa2ΔCR probe, it has been shown that cellular growth rate correlates with basal mitochondrial H2O2 levels in yeast cells.
Chemical Probes for Oxidative PTMs of Cysteine
Cysteine residues have unique chemical reactivity, including high nucleophilicity and redox sensitivity, and play important roles in many protein functions, such as nucleophilic and redox catalysis, structural-disulfide formation, and binding to metal ions (91). Oxidative PTMs of cysteine play central roles in redox signaling by regulating the activity of key proteins within signaling pathways. One well-characterized example of cysteine-mediated redox signaling is in EGF signaling. In A431 epidermoid carcinoma cells, PTP1B is inhibited (63) and EGFR is activated (93) through S-sulfenylation, resulting in augmentation of the phosphorylation signals resulting from EGF binding to EGFR. Therefore, methods for identifying and quantifying oxidative cysteine PTMs are critical to understanding redox signaling. Numerous chemical probes with a variety of reactive handles have been developed and applied to globally monitor oxidative cysteine PTMs.
Chemical probes for oxidative cysteine PTMs can be categorized into two groups: (i) cysteine-reactive probes to visualize general cysteine PTMs and (ii) probes that are specific for a particular oxidative PTM of cysteine. Probes in the first category are based on monitoring the intrinsic cysteine reactivity with electrophilic chemical probes that form stable covalent adducts with reduced cysteines. Oxidation of a particular cysteine residue is reflected by a loss in nucleophilicity and subsequent decrease in probe labeling. These methods allow for identification of any cysteine PTM that reduces nucleophilicity, and therefore can be applied to all oxidative cysteine modifications. However, secondary methods need to be employed to identify the exact modification that is causing the measured loss in cysteine reactivity. In the second category, PTM-specific reactions are harnessed to design chemical probes that are specific for each oxidative cysteine PTM, including S-sulfenylation, S-sulfinylation, and S-glutathionylation. Contrary to the general cysteine-reactive probes, PTM-specific probes can focus on a particular PTM, lower the complexity of proteome samples, and enhance sensitivity and reliability of analysis. However, probes to directly monitor inter- and intra-molecular disulfides, and sulfonic acids, are currently lacking, rendering these PTMs intractable to existing PTM-specific methods. To date, both general and specific methods rely on two-dimensional electrophoresis and MS-based analytical methods for identification and quantification of labeled proteins.
Chemical-Proteomic Methods for General Oxidative PTMs of Cysteine
Two-dimensional gel electrophoresis
Two-dimensional gel electrophoresis (2DE)-based methods have been commonly used in proteomic research. 2DE methods are also frequently applied to the analysis of oxidative cysteine PTMs for visualization with radioactive or fluorescent reporters (17). In typical procedures for autoradiography, non-oxidized cysteine residues are capped by iodoacetamide (IAM) or N-ethylmaleimide (NEM). Then, oxidized cysteines are reduced by reducing agents such as dithiothreitol (DTT) or tris(2-carboxyethyl)phosphine (TCEP). Subsequently, the newly reduced cysteine residues are labeled with radioactive 14C-IAM or 14C-NEM for visualization by autoradiography (61, 65). Due to non-specific reduction by DTT or TCEP (34), these methods visualize multiple forms of oxidative cysteine PTMs, including disulfide bonds, sulfenic acids, and nitrosothiols. To achieve more facile visualization of 2D gels, radioactive probes have been replaced by fluorescent cysteine-reactive probes such as 5-iodoacetamidefluorescein (10), monobromobimane (136), and boron dipyrromethene (BODIPY) FL C1-IA (42). The signal intensity of radiation or fluorescence from proteins can be normalized by the protein abundance estimated from Coomassie Brilliant Blue (CBB) staining to provide a qualitative estimate of the stoichiometry of oxidation (42, 65).
2DE-based methods can be applied to directly compare the degree of cysteine oxidation between two samples. In a method termed redox differential in-gel electrophoresis (redox-DIGE), a pair of cysteine-reactive probes conjugated with orthogonal fluorophores such as Cy3-maleimide and carbocyanine 5 (Cy5)-maleimide are used for labeling two samples, respectively (16, 30, 46). After blocking reduced cysteines, oxidized cysteines in two distinct cell lysates are labeled with two different fluorophores. The two labeled samples are combined and analyzed on the same 2D gel, enabling direct visualization of differences in oxidative cysteine PTMs across two samples. Redox-DIGE has been applied to identify mitochondrial proteins that are sensitive to oxidative modifications by endogenous ROS (46). However, this redox-DIGE analysis lacks information about protein abundance; thus, fluorescence signals cannot be normalized with the protein amounts. To overcome this limitation, another differential labeling strategy was developed, whereby both oxidized and reduced cysteine residues are differentially labeled with two distinct fluorophores (62). A ratio between the two fluorescence signals gives a quantitative measure of the stoichiometry of oxidation by normalizing for protein abundance.
The 2DE-based methods described earlier commonly have major drawbacks. The limited resolution of 2DE results in insufficient separation of proteins with similar physicochemical properties. Further, limited detection sensitivity of gel-imaging methods prohibits the detection of low abundant proteins. In addition, even if protein spots are found to be oxidized by 2DE, the identification of each protein has to be performed individually by MS. Time-consuming 2DE procedures and subsequent individual identification of each protein by MS make 2DE methods low-throughput. Further, given that a protein has multiple cysteines, a signal of the protein on 2DE can represent an average of multiple cysteine residues, and identifying the exact cysteine(s) that are oxidized can be challenging.
Isotope-coded affinity tags
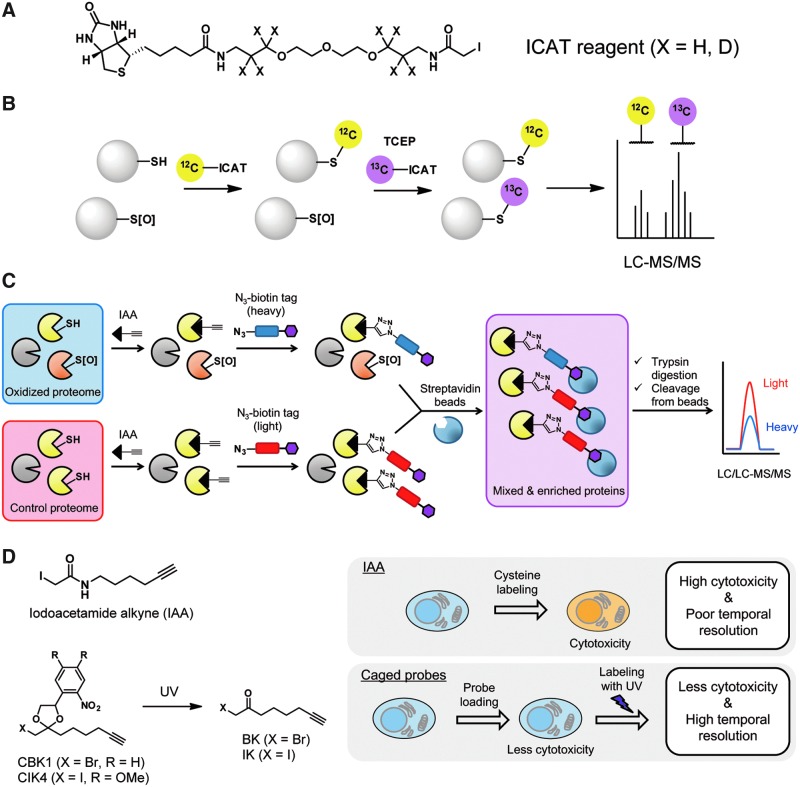
To overcome the multiple drawbacks of 2DE, a method based on isotope-coded affinity tags (ICAT) has been developed. The ICAT method utilizes biotinylated IAM derivatives that are labeled with isotopically light and heavy linkers (39). Peptides tagged with the ICAT probes were enriched by streptavidin beads and analyzed by quantitative MS (Fig. 4A). Isotopically labeled ICAT probes enable relative quantification of oxidized cysteines between the light- and heavy-labeled samples. The ICAT strategy was applied to identify cysteines that are susceptible to oxidation on H2O2 treatment (111). A modified ICAT strategy termed OxICAT (oxidative isotope-coded affinity tag) has been developed for differential labeling of oxidized and reduced cysteine in the same sample (64). In this OxICAT strategy, reduced cysteines are labeled with a light ICAT reagent, and oxidized cysteine residues are reduced with TCEP and labeled with a heavy ICAT reagent (Fig. 4B). This differential labeling enables quantification of the ratio between reduced and oxidized forms of each cysteine, and this ratio can then be compared across multiple samples. To better monitor protein abundance changes in tandem with oxidation variations, isotopic dimethyl labeling has been coupled with ICAT, enabling protein abundance measurements before cysteine enrichment (32, 33). Another strategy for protein quantification is a method termed GELSILOX in which a stable O2 isotope is incorporated during the trypsin digestion step (78). These ICAT, OxICAT, and GELSILOX methods overcome major limitations of 2DE by achieving both the identification and quantification of multiple oxidized proteins in a single experiment, and, in addition, provide higher sensitivity than gel-based methods for the detection of low abundance proteins. However, detection sensitivity can be decreased by low ionization efficiency of certain peptides, multiple cysteines on the same peptide can complicate site identifications, and a cysteine cannot be identified if the residue is on a very short or long tryptic peptide.
FIG. 4.
Chemical-proteomic platforms for general cysteine PTMs. (A) Structures of ICAT reagents. (B) OxICAT is a modified ICAT technology for oxidative cysteine PTMs. (C) A quantitative proteomic platform, isoTOP-ABPP, enables comprehensive identification and quantification of cysteine PTMs. (D) Caged electrophilic probes are developed to achieve in situ labeling of cysteine and analysis of cysteine PTMs within a physiological context. CBK1, caged α-bromomethylketone alkyne; IAA, idoacetamide alkyne; ICAT, isotope-coded affinity tags; isoTOP-ABPP, isotopic tandem orthogonal proteolysis activity-based protein profiling; LC-MS/MS, liquid chromatography-tandem mass spectrometry; OxICAT, oxidative isotope-coded affinity tag; TCEP, tris(2-carboxyethyl)phosphine; UV, ultraviolet. Color images are available online.
Isotopic tandem orthogonal proteolysis activity-based protein profiling
ICAT probes have a bulky biotin reporter that may inhibit labeling of sterically hindered cysteines. An alternative MS-based method, termed isotopic tandem orthogonal proteolysis activity-based protein profiling (isoTOP-ABPP), relies on a simpler iodoacetamide alkyne (IAA) probe (131). In isoTOP-ABPP, proteome samples are labeled with IAA, before incorporation of a biotin-azide tag via copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC) (Fig. 4C). The biotin moiety facilitates subsequent enrichment of labeled proteins by streptavidin beads, after which proteins then undergo an on-bead tryptic digestion. The biotin-azide tag has cleavable linkers that are susceptible to cleavage by treatment with tobacco etch virus protease or sodium dithionite (98, 131), which releases the enriched peptides for MS analysis. By using isotopically light- or heavy-labeled biotin-azide tags, relative quantification across two samples is achieved. An isoTOP-ABPP ratio, R, is calculated from signal intensities of light- and heavy-tagged peptides for each identified cysteine. The R value provides a quantitative measure of changes in cysteine reactivity, and the isoTOP-ABPP method can identify and quantify cysteine reactivity changes of ∼1000 to 1500 cysteine residues. The isoTOP-ABPP method and its derivatives have been applied for identification of bacterial proteins that are susceptible to oxidation by H2O2 (19). This method was also applied to identify S-nitrosation sites on numerous proteins on treatment with transnitrosation donors such as S-nitrosoglutathione (GSNO), identifying S-nitrosation sites on proteins including 3-hydroxyacyl-CoA dehydrogenase type 2 (HADH2) and cathepsin D (CTSD) (141).
Although isoTOP-ABPP can quantify and identify more than 1000 cysteines in a single tandem liquid chromatography-tandem mass spectrometry (LC/LC-MS/MS) run, coverage of the whole proteome by this method can still be limited due to the high complexity and heterogeneity of the proteome samples. Organelle isolation strategies can be coupled with isoTOP-ABPP to improve coverage of reactive cysteines in a particular organelle such as mitochondria (7). Another limitation of isoTOP-ABPP has been the usage of cytotoxic IAA probe, which is not suitable for live-cell applications. Thus, applications of isoTOP-ABPP have been primarily performed in lysate samples. However, because oxidative PTMs of cysteine are highly labile to environmental redox changes caused by cell lysis, it is ideal to perform the initial labeling step directly in living cells. Photolabile caged electrophilic probes have been developed to achieve cysteine labeling in situ (1, 4). A caged electrophile named caged α-bromomethylketone alkyne, or CBK1, is not cysteine reactive and, therefore, can be loaded into live cells with negligible cytotoxicity (Fig. 4D). CBK1 is activated to be cysteine reactive by ultraviolet (UV) light irradiation in situ to trigger cysteine labeling, and the labeled cysteine residues are identified by LC/LC-MS/MS. An additional advantage of this labeling strategy with caged probes is the high temporal resolution of cysteine labeling, which is required for accurate analysis of transient and reversible cysteine oxidations in redox signaling. The practical usability of CBK1 has been demonstrated by analyzing redox signaling in A431 epidermoid carcinoma cells stimulated by EGF. CBK1 coupled with isoTOP-ABPP identified hundreds of cysteine residues with R values, indicating the degree of oxidative modification for each cysteine under EGF stimulation. More recently, an optimized caged probe, caged α-iodoketone alkyne 4 (CIK4), has been reported with improved cysteine reactivity and uncaging efficiency, which enables more efficient detection of cysteine residues by LC/LC-MS/MS (1). Though caged electrophilic probes enable cysteine labeling in situ with short exposure time to cytotoxic agents and high temporal resolution, the limitation is the usage of UV light, which may cause cytotoxic effects on live cells.
Chemical Proteomic Methods for a Specific Oxidative PTM of Cysteine
Chemical probes for sulfenic acids
The 2DE, ICAT, and isoTOP-ABPP methods discussed earlier all rely on changes to the intrinsic reactivity of a cysteine induced by oxidation, and, therefore, do not differentiate cysteine PTMs such as sulfenic acids, sulfinic acids, and disulfides. Complementary strategies focus on a specific cysteine PTM by utilizing chemical probes that selectively react with a single PTM. The reversibility of sulfenic acids renders this PTM the ability to mediate cellular signal transduction events that are akin to phosphorylation and acetylation. One strategy for the specific labeling of sulfenylation is based on the selective reduction of sulfenic acids by arsenite, which is analogous to the differential tagging strategies described earlier (108). In this method, sulfenic acids are reduced by arsenite under denaturing conditions, then labeled with a biotin-maleimide probe. Another strategy is the direct labeling with chemical probes that are specific for sulfenic acids, including 5, 5-dimethyl-1, 3-cyclohexanedione, also known as dimedone, as a reactive moiety (Fig. 5A). The enolate form of dimedone facilitates nucleophilic attack onto sulfenic acids to form a covalent adduct. Dimedone or other 1,3-diketone compounds are conjugated with fluorophores or biotin analogues for in-gel visualization and enrichment with avidin for MS-based proteomic experiments (37, 87, 95, 96).
FIG. 5.
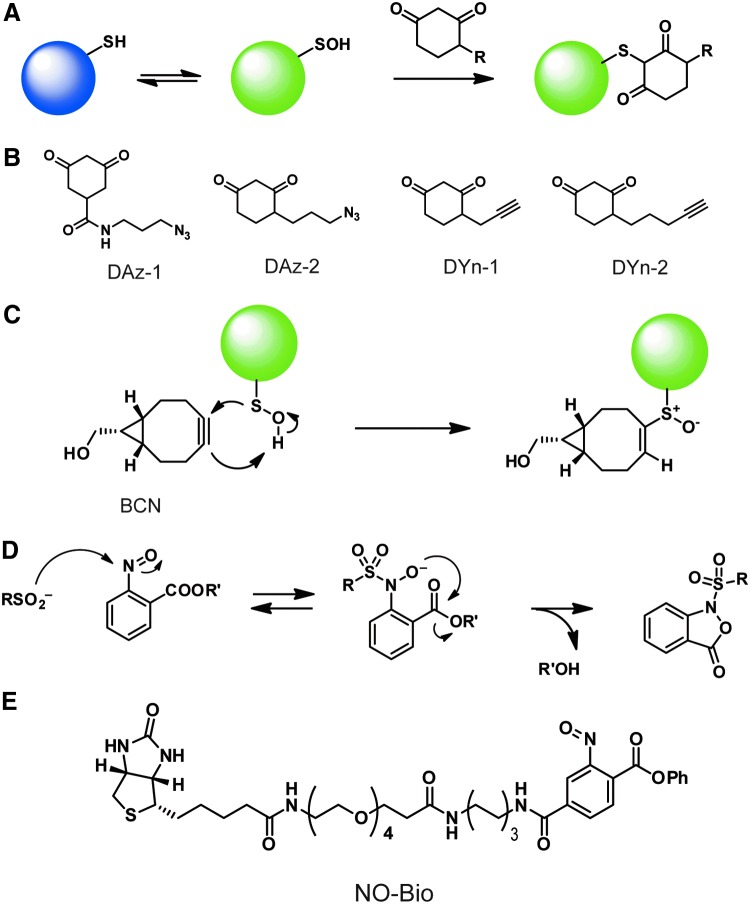
Chemical probes for sulfenic acids and sulfinic acids. (A) The reaction between sulfenic acids and dimedone. (B) Structures of dimedone-based probes. (C) A strained alkyne-based probe, BCN, can be conjugated to sulfenic acids. (D) The reaction between sulfinic acids and 2-nitroso benzoic acid derivatives. (E) NO-Bio is a nitroso-based chemical probe for sulfinic acids. BCN, bicycle[6.1.0]nonyne; DAz, azide-tagged dimedone; DYn, alkyne-tagged dimedone; NO-Bio, biotinylated 2-nitroso terepththalic acid. Color images are available online.
The presence of bulky reporter groups such as fluorophores and biotin analogues are not ideal because they result in low labeling efficiency by blocking access to buried sulfenic acids, as well as limiting the plasma membrane permeability of the probes. Sulfenic acids are highly labile to redox changes, and, therefore, labeling in situ is important for accurate analysis. To this end, dimedone derivatives with minimal modifications have been developed; azide-tagged probes (azide-tagged dimedone [DAz]-1 and DAz-2) (66, 100) and alkyne-tagged probes (alkyne-tagged dimedone [DYn]-1 and DYn-2) (93) have chemical handles for incorporation of reporters via a Staudinger reaction or CuAAC in subsequent steps (Fig. 5B). These probes are membrane permeable and can be applied to the labeling of sulfenic acids directly in living cells.
A strategy for differential labeling of sulfenic acids has been developed by utilizing isotopically coded dimedone- and iododimedone-bearing probes, which are reactive with sulfenic acids and reduced cysteines, respectively (110). Based on this strategy, the degree of sulfenylation at a particular cysteine residue can be accurately quantified. In a similar manner, dimedone and IAM conjugated with lantanide-1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) have been applied to quantify sulfenylation using inductively coupled plasma-MS (26).
Dimedone-based probes have been applied to various biological systems to uncover sulfenylation sites. For example, a biotinylated dimedone probe has been utilized to reveal that Cys124 of protein kinase B β (Akt2) is sulfenylated under physiologically relevant platelet-derived growth factor (PDGF) stimulation (129). The sulfenylation of Cys124 inhibits the activity of Akt2, showing a novel regulatory mechanism for kinase regulation in addition to phosphorylation. As another example, a biotinylated dimedone probe has been applied to T cells to identify increased sulfenylation on protein tyrosine phosphatases such as Src homology domain 2-containing protein tyrosine phosphatase (SHP)-1 and SHP-2, and actin on T cell activation (81). This study demonstrated that reversible sulfenylation is an important regulatory factor for proliferation and function of cluster of differentiation 8 (CD8+) T cells. An alkyne-functionalized dimedone probe, DYn-2, revealed that Cys797 of EGFR is sulfenylated under EGF stimulation and sulfenylation augments the kinase activity of EGFR (93). Lastly, isotopically light- and heavy-labeled DYn-2 probes were coupled to advanced LC-MS/MS-based methods to globally identify and quantify the exact sites of sulfenylation within proteomes (134).
Although dimedone-based probes have been successfully applied to proteomic experiments, one limitation of dimedone is the demonstrated slow reaction rate with sulfenic acids, which requires the use of the chemical probes at high concentrations to achieve efficient labeling. To overcome this limitation, a library of ∼100 cyclic C-nucleophiles have been evaluated for reactivity with sulfenic acids to find several novel compounds with more than 200-fold enhanced reactivity than that of dimedone (36). Strained bicycle[6.1.0]nonyne (BCN) derivatives were also reported as a new reactive group with sulfenic acids (Fig. 5C). The BCN derivatives can react with sulfenic acids with a faster reaction rate than dimedone by over two orders of magnitude (97). It is noteworthy that BCN is reported to undergo thiol-yne addition and can be non-selectively consumed by thiols (121). These new chemistries can offer an alternative strategy for monitoring sulfenylation in addition to the widely used dimedone-based probes.
Chemical probes for sulfinic acids
S-sulfinylation (RSO2H) was regarded an artifactual hyper-oxidation of cysteine that occurs during sample handling, but increasing evidence indicates that S-sulfinylation is not only physiologically relevant but also important in redox signaling (47, 71). It was revealed that ∼5% of cysteine residues are oxidized to sulfinic acids in a soluble fraction of rat liver (40). Among S-sulfinylated proteins are Prxs, which are a family of H2O2-scavanging enzymes. Due to high abundance in the cytosol and very fast reaction rates with H2O2 (k = ∼107 M−1·s−1), Prxs remove H2O2 very rapidly to keep the concentration of cytosolic H2O2 relatively low (132). Therefore, it has been proposed that Prxs activity may be locally regulated under redox signaling through PTMs such as S-sulfinylation (135) and phosphorylation (133). Although sulfinic acids are not reduced by most biological antioxidants, the S-sulfinylation on Prxs can be reduced through an ATP-dependent reduction by Srx (14).
To capture sulfinic acids in a specific manner, the reported reaction between 2-nitroso benzoic acid derivatives and sulfinic acids has been adapted (72) (Fig. 5D). The reactive nitroso warhead covalently modifies sulfinic acids to form stable nitroso-benzene derivatives. This ligation reaction is shown to be selective to sulfinic acids among various nucleophilic and/or redox-active small molecules. An observed side reaction involves the oxidation of cysteine to cystine concomitant with the reduction of nitroso compounds. A nitroso compound, 2-nitroso terepththalic acid (NO-Ph), was shown to successfully label a sulfinic acid on a recombinant Gpx3 mutant; however, NO-Ph also formed a stable sulfonamide adduct with a reduced cysteine on Gpx3. Thus, the blocking of reduced cysteine with NEM before the labeling of sulfinic acids is required for efficient MS analysis. The reaction termed sulfinic-acid-nitroso ligation (SNL) has been utilized to design a sulfinic acid-specific probe, biotinylated 2-nitroso terepththalic acid (NO-Bio), in which a 2-nitroso benzoic acid derivative is conjugated with a biotin analogue (73) (Fig. 5E). The NO-Bio probe has been applied to the labeling of sulfinic acids on recombinant Parkinson disease protein 7 (DJ-1) oxidized by H2O2 and in cell lysates induced by 2,2′-dipyridyl disulfide (DPS), and to profile oxidation among normal and matched tumor cells. The SNL reaction can also be utilized to monitor S-nitrosation (76). Biotinylated probes with sulfinic acid (Biotin-SO2H) and GSNO (Biotin-GSNO) have been applied to quantitative MS using stable isotope labeling with amino acids in cell culture (SILAC), revealing relative occupancy ratios of S-sulfinylation and S-nitrosation in 293T cells. The SNL chemistry has not yet been applied to a wide variety of biological samples; however, it opens the door to proteome-wide analyses focusing on S-sulfinylation and S-nitrosation.
Chemical probes for S-glutathionylation
S-glutathionylation is known to regulate important protein functions that are involved in various physiological processes such as gene expression, signaling, energy metabolism, and cell survival (92). For example, glutathionylation of nuclear factor-κB (NF-κB) inhibits DNA-binding activity and downstream transcriptional activation (94). Another key protein in this pathway, inhibitor of κB kinase (IKK), is also glutathionylated, rendering the NF-κB pathway highly susceptible to redox changes in the cellular environment (101). The extent of modification with glutathione depends on the ratio of reduced (GSH) and oxidized forms (GSSG) of glutathione.
Various chemical-proteomic strategies have been developed to analyze S-glutathionylation within a biological context. In early methods based on autoradiography, GSH tagged with 3H or 35S was utilized for labeling in vitro, and 35S-cysteine was applied for metabolic incorporation in situ (31). GSH is synthesized from glutamate, cysteine, and glycine through two subsequent ATP-dependent reactions catalyzed by γ-glutamylcysteine synthetase and glutathione synthetase (GS) (80). The method for metabolic incorporation of 35S-cysteine requires treatment with cycloheximide to inhibit incorporation of 35S-cysteine into proteins, ensuring that 35S-cysteine is only incorporated into GSH. Proteome samples modified with 35S-labeled GSH are analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and imaged by autoradiography. This method was applied to monitor S-glutathionylation in human monocytes (104), revealing that glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is S-glutathionylated during a respiratory burst (99). However, this method has major drawbacks, including the necessity to inhibit protein synthesis, which greatly affects cellular metabolism, and the lack of a handle to enrich modified proteins for global MS identification.
To overcome these drawbacks of radioactive GSHs, biotinylated GSH (BioGSH) analogues have been developed. GSH and GSSG can be conjugated with a biotin group at the primary amine by treatment with sulfosuccinimidyl-6-(biotinamido)-hexanoate (sulfo-NHS-biotin) (15). A membrane-permeable precursor of GSH, glutathione ethyl ester (GEE), can also be modified by sulfo-NHS-biotin to form biotinylated glutathione ethyl ester (BioGEE) (117) (Fig. 6A). The biotin groups can be utilized for enrichment of glutathionylated proteins by streptavidin beads, followed by reductive cleavage of the enriched proteins from the beads and analysis by gel electrophoresis and MS. Non-reducing Western blotting enables sensitive detection of glutathionylated proteins in combination with streptavidin-HRP or anti-biotin antibodies. These BioGSH derivatives have been applied to the identification of glutathionylation in vitro (45, 127) and in cells. BioGEE was applied to live HeLa cells under oxidative stress by exogenous H2O2 or tumor necrosis factor-α (TNF-α) treatment to identify annexin A2 and Trx peroxidase 2 as glutathionylated proteins (117). Limitations of this method include the modification with a bulky biotin group that may perturb a subset of proteins tagged with these reagents, and the exogenous addition of redox-active GSH derivatives that affect the native GSH/GSSG ratio.
FIG. 6.
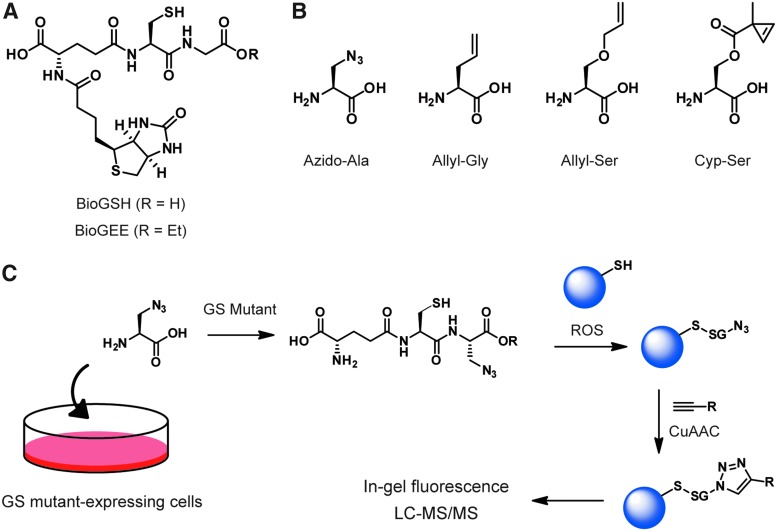
Chemical probes for S-glutathionylation. (A) Structures of biotinylated glutathione derivatives, BioGSH and BioGEE. (B) Glycine surrogates with bioorthogonal handles for metabolic incorporation. (C) The glycine surrogates can be incorporated into GSH catalyzed by mutated GSs in living cells, which enables proteomic analyses of native glutathionylation. BioGEE, biotinylated glutathione ethyl ester; BioGSH, biotinylated GSH; CuAAC, copper(I)-catalyzed alkyne-azide cycloaddition; GS, glutathione synthetase; GSH, reduced glutathione. Color images are available online.
An elegant method to overcome the limitations of BioGSH derivatives has been developed by utilizing a mutated GS that catalyzes coupling of an azide-tagged alanine derivative (azido-Ala) to γ-glutamylcysteine (γGlu-Cys) (106) (Fig. 6B, C). The GSH derivative containing an azide handle (azide-GSH) for CuAAC allows for identification of glutathionylated proteins by gel and MS analysis. The strategy was utilized for analysis of glutathionylation in living cells under exogenous H2O2 stimulation and glucose depletion (107). Alkene-bearing Gly surrogates (ally-containing glycine [Allyl-Gly], allyl-containing serine [Allyl-Ser], and cyclopropane-containing serine [Cyp-Ser]) can also be enzymatically incorporated into GSH by mutated GSs, which enable bioorthogonal tagging by tetrazine-based reporters (50) (Fig. 6B).
Selective reduction of glutathione-containing disulfides mediated by Grx has been applied to proteomic analysis of glutathionylation (68). In the Grx-based method, on blocking non-oxidized cysteine residues with NEM, glutathione-containing disulfide bonds are reduced by Grx. Freshly formed cysteines are labeled with a biotin-tagged NEM probe, resulting in subsequent enrichment of labeled proteins. The Grx-based method has been applied to analyze glutathionylation under oxidative stress induced by diamide (69).
Summary
Redox signaling is involved in various physiological processes such as antioxidant induction, cellular proliferation, and migration. Due to the highly reactive and unstable nature of ROS, it has been challenging to measure physiological ROS generation in a reliable and quantitative manner. Taking advantage of the distinct reactivities of different ROS, chemical probes have been developed and applied to detect and quantify biological ROS production. To date, various detection methods for O2•− are available and widely used with EPR, fluorescence, and chemiluminescence instrumentation. Fluorescence probes for H2O2 have been designed by utilizing H2O2-specific chemical reactions and engineered H2O2-sensitive proteins. Despite the low reactivity of H2O2, fluorescent protein-based H2O2 probes are highly sensitive to H2O2. Currently available ROS probes have several advantages and limitations in terms of sensitivity, selectivity, and usability; thus, it is important to choose the appropriate methods for a given application. Control experiments with ROS scavengers and a combination of multiple ROS-detection methods can increase the reliability of the measurements. Besides the detection of ROS generation, proteome-wide analysis of oxidative cysteine PTMs is also critical to uncover the mechanisms and downstream consequences of redox signaling. Increased evidence shows that cysteine PTMs such as sulfenylation, sulfinylation, and glutathionylation can transiently regulate protein activity with critical downstream consequences. The dynamic nature of oxidative PTMs of cysteine has made it a challenge to perform comprehensive and quantitative analysis of these PTMs within native biological environments. Recent advances in chemical probes for in situ labeling of cysteine and its PTMs will provide facile and reliable approaches to capture native PTMs within a physiological context. In parallel, advances in MS instrumentation and quantitative proteomic strategies will enhance the efficiency of identification and quantification of cysteine PTMs. Analytical platforms for ROS and cysteine PTMs highlighted in this review are complementary to each other, and their combined use will offer a robust and comprehensive strategy to dissect redox signaling. The detection of ROS and oxidative cysteine PTMs will benefit from analytical advances that can improve the sensitivity of fluorescence and MS-based analyses, and further, methods to provide spatial and temporal control over measurements will be instrumental in furthering our understanding of redox signaling.
Acknowledgments
M.A. is supported by the Postdoctoral Fellowship for Research Abroad from Japan Society for the Promotion of Science. This work was funded by NIH grants 1R01GM117004 and 1R01GM118431-01A1 to E.W.
Abbreviations Used
- 2DE
two-dimensional gel electrophoresis
- 2-OH-E+
2-hydroxyethidium
- Akt2
protein kinase B β
- ATP
adenosine triphosphate
- BCN
bicycle[6.1.0]nonyne
- BES
benzenesulfonyl fluorescein
- BioGEE
biotinylated glutathione ethyl ester
- BioGSH
biotinylated GSH
- CBK1
caged α-bromomethylketone alkyne
- CLA
2-Methyl-6-phenylimidazo[1,2-a]pyrazin-3(7H)-one
- CP
peroxidatic cysteine
- CPH
1-hydroxy-3-carboxy-2, 2, 5-tetramethyl-pyrrolidine hydrochloride
- CR
resolving cysteine
- CuAAC
copper(I)-catalyzed alkyne-azide cycloaddition
- Cy3
carbocyanine 3
- Cys
cysteine
- DAz
azide-tagged dimedone
- DCFH2
dihydrodichlorofluorescein
- DMPO
5,5-dimethyl-1-pyrroline-N-oxide
- d-PeT
donor-excited photoinduced electron transfer
- DTT
dithiothreitol
- DYn
alkyne-tagged dimedone
- E+
ethidium
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EPR
electron paramagnetic resonance
- Grx
glutaredoxin
- GS
glutathione synthetase
- GSH
reduced glutathione
- GSNO
S-nitrosoglutathione
- GSSG
oxidized glutathione
- H2O2
hydrogen peroxide
- HE
hydroethidine
- HRP
horseradish peroxidase
- IAA
iodoacetamide alkyne
- IAM
iodoacetamide
- ICAT
isotope-coded affinity tag
- isoTOP-ABPP
isotopic tandem orthogonal proteolysis activity-based protein profiling
- LC/LC-MS/MS
tandem liquid chromatography-tandem mass spectrometry
- MCLA
6-(4-methoxyphenyl)-2-methyl-imidazo[1,2-a]pyrazin-3(7H)-one
- MitoPY1
mitochondria peroxy yellow 1
- MS
mass spectrometry
- NADPH
reduced nicotinamide adenine dinucleotide phosphate
- NBzF
5-(4-nitrobenzoyl)carbonylfluorescein
- NBzF-BG
O6-benzylguanine-conjugated NBzF
- NEM
N-ethylmaleimide
- NF-κB
nuclear factor-κB
- •NO
nitric oxide
- NO-Bio
biotinylated 2-nitroso terepththalic acid
- NO-Ph
2-nitroso terepththalic acid
- NOX
NADPH oxidase
- O2
oxygen
- O2•−
superoxide
- −OCl
hypochlorite
- •OH
hydroxyl radical
- ONOO−
peroxynitrite
- OxICAT
oxidative isotope-coded affinity tag
- PF1
Peroxyfluor-1
- PG1
Peroxy Green 1
- PMA
phorbol 12-myristate 13-acetate
- PO1
Peroxy Orange 1
- PR1
Peroxyresorufin-1
- Prx
peroxiredoxin
- PTM
post-translational modification
- PTP1B
protein tyrosine phosphatase 1B
- PY1
Peroxy Yellow 1
- redox-DIGE
redox differential in-gel electrophoresis
- RNS
reactive nitrogen species
- roGFP
redox-sensitive green fluorescent protein
- ROS
reactive oxygen species
- SHP
Src homology domain 2-containing protein tyrosine phosphatase
- SNL
sulfinic-acid-nitroso ligation
- SOD
superoxide dismutase
- Srx
sulfiredoxin
- Sulfo-NHS-biotin
sulfosuccinimidyl-6-(biotinamido)-hexanoate
- TAM
triarylmethyl
- TCEP
tris(2-carboxyethyl)phosphine
- TPP
triphenylphosphonium
- Trx
thioredoxin
- UV
ultraviolet
References
- 1. Abo M, Bak DW, and Weerapana E. Optimization of caged electrophiles for improved monitoring of cysteine reactivity in living cells. Chembiochem 18: 81–84, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abo M, Minakami R, Miyano K, Kamiya M, Nagano T, Urano Y, and Sumimoto H. Visualization of phagosomal hydrogen peroxide production by a novel fluorescent probe that is localized via SNAP-tag labeling. Anal Chem 86: 5983–5990, 2014 [DOI] [PubMed] [Google Scholar]
- 3. Abo M, Urano Y, Hanaoka K, Terai T, Komatsu T, and Nagano T. Development of a highly sensitive fluorescence probe for hydrogen peroxide. J Am Chem Soc 133: 10629–10637, 2011 [DOI] [PubMed] [Google Scholar]
- 4. Abo M. and Weerapana E. A caged electrophilic probe for global analysis of cysteine reactivity in living cells. J Am Chem Soc 137: 7087–7090, 2015 [DOI] [PubMed] [Google Scholar]
- 5. Afzal M, Matsugo S, Sasai M, Xu B, Aoyama K, and Takeuchi T. Method to overcome photoreaction, a serious drawback to the use of dichlorofluorescin in evaluation of reactive oxygen species. Biochem Biophys Res Commun 304: 619–624, 2003 [DOI] [PubMed] [Google Scholar]
- 6. Albrecht SC, Barata AG, Grosshans J, Teleman AA, and Dick TP. In vivo mapping of hydrogen peroxide and oxidized glutathione reveals chemical and regional specificity of redox homeostasis. Cell Metab 14: 819–829, 2011 [DOI] [PubMed] [Google Scholar]
- 7. Bak DW, Pizzagalli MD, and Weerapana E. Identifying functional cysteine residues in the mitochondria. ACS Chem Biol 17: 947–957, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barata AG. and Dick TP. In vivo imaging of H2O2 production in Drosophila. Methods Enzymol 526: 61–82, 2013 [DOI] [PubMed] [Google Scholar]
- 9. Bardelang D, Rockenbauer A, Karoui H, Finet JP, Biskupska I, Banaszak K, and Tordo P. Inclusion complexes of EMPO derivatives with 2,6-di-O-methyl-beta-cyclodextrin: synthesis, NMR and EPR investigations for enhanced superoxide detection. Org Biomol Chem 4: 2874–2882, 2006 [DOI] [PubMed] [Google Scholar]
- 10. Baty JW, Hampton MB, and Winterbourn CC. Detection of oxidant sensitive thiol proteins by fluorescence labeling and two-dimensional electrophoresis. Proteomics 2: 1261–1266, 2002 [DOI] [PubMed] [Google Scholar]
- 11. Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, and Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods 3: 281–286, 2006 [DOI] [PubMed] [Google Scholar]
- 12. Bilan DS. and Belousov VV. HyPer family probes: state of the art. Antioxid Redox Signal 24: 731–751, 2016 [DOI] [PubMed] [Google Scholar]
- 13. Bilan DS, Pase L, Joosen L, Gorokhovatsky AY, Ermakova YG, Gadella TW, Grabher C, Schultz C, Lukyanov S, and Belousov VV. HyPer-3: a genetically encoded H2O2 probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chem Biol 8: 535–542, 2013 [DOI] [PubMed] [Google Scholar]
- 14. Biteau B, Labarre J, and Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425: 980–984, 2003 [DOI] [PubMed] [Google Scholar]
- 15. Brennan JP, Miller JI, Fuller W, Wait R, Begum S, Dunn MJ, and Eaton P. The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Mol Cell Proteomics 5: 215–225, 2006 [DOI] [PubMed] [Google Scholar]
- 16. Bruschi M, Grilli S, Candiano G, Fabbroni S, Della Ciana L, Petretto A, Santucci L, Urbani A, Gusmano R, Scolari F, and Ghiggeri GM. New iodo-acetamido cyanines for labeling cysteine thiol residues. A strategy for evaluating plasma proteins and their oxido-redox status. Proteomics 9: 460–469, 2009 [DOI] [PubMed] [Google Scholar]
- 17. Chiappetta G, Ndiaye S, Igbaria A, Kumar C, Vinh J, and Toledano MB. Proteome screens for Cys residues oxidation: the redoxome. Methods Enzymol 473: 199–216, 2010 [DOI] [PubMed] [Google Scholar]
- 18. D'Autreaux B. and Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol 8: 813–824, 2007 [DOI] [PubMed] [Google Scholar]
- 19. Deng X, Weerapana E, Ulanovskaya O, Sun F, Liang H, Ji Q, Ye Y, Fu Y, Zhou L, Li J, Zhang H, Wang C, Alvarez S, Hicks LM, Lan L, Wu M, Cravatt BF, and He C. Proteome-wide quantification and characterization of oxidation-sensitive cysteines in pathogenic bacteria. Cell Host Microbe 13: 358–370, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dickinson BC. and Chang CJ. A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells. J Am Chem Soc 130: 9638–9639, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dickinson BC, Huynh C, and Chang CJ. A palette of fluorescent probes with varying emission colors for imaging hydrogen peroxide signaling in living cells. J Am Chem Soc 132: 5906–5915, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dikalov S, Griendling KK, and Harrison DG. Measurement of reactive oxygen species in cardiovascular studies. Hypertension 49: 717–727, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dikalov S, Skatchkov M, Fink B, and Bassenge E. Quantification of superoxide radicals and peroxynitrite in vascular cells using oxidation of sterically hindered hydroxylamines and electron spin resonance. Nitric Oxide 1: 423–431, 1997 [DOI] [PubMed] [Google Scholar]
- 24. Dikalova A, Clempus R, Lassegue B, Cheng G, McCoy J, Dikalov S, San Martin A, Lyle A, Weber DS, Weiss D, Taylor WR, Schmidt HH, Owens GK, Lambeth JD, and Griendling KK. Nox1 overexpression potentiates angiotensin II-induced hypertension and vascular smooth muscle hypertrophy in transgenic mice. Circulation 112: 2668–2676, 2005 [DOI] [PubMed] [Google Scholar]
- 25. Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, and Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem 279: 22284–22293, 2004 [DOI] [PubMed] [Google Scholar]
- 26. El-Khatib AH, Esteban-Fernandez D, and Linscheid MW. Inductively coupled plasma mass spectrometry-based method for the specific quantification of sulfenic acid in peptides and proteins. Anal Chem 86: 1943–1948, 2014 [DOI] [PubMed] [Google Scholar]
- 27. Ermakova YG, Bilan DS, Matlashov ME, Mishina NM, Markvicheva KN, Subach OM, Subach FV, Bogeski I, Hoth M, Enikolopov G, and Belousov VV. Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nat Commun 5: 5222, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fink B, Laude K, McCann L, Doughan A, Harrison DG, and Dikalov S. Detection of intracellular superoxide formation in endothelial cells and intact tissues using dihydroethidium and an HPLC-based assay. Am J Physiol Cell Physiol 287: 895, 2004 [DOI] [PubMed] [Google Scholar]
- 29. Frejaville C, Karoui H, Tuccio B, Le Moigne F, Culcasi M, Pietri S, Lauricella R, and Tordo P. 5-(Diethoxyphosphoryl)-5-methyl-1-pyrroline N-oxide: a new efficient phosphorylated nitrone for the in vitro and in vivo spin trapping of oxygen-centered radicals. J Med Chem 38: 258–265, 1995 [DOI] [PubMed] [Google Scholar]
- 30. Fu C, Hu J, Liu T, Ago T, Sadoshima J, and Li H. Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J Proteome Res 7: 3789–3802, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao XH, Bedhomme M, Veyel D, Zaffagnini M, and Lemaire SD. Methods for analysis of protein glutathionylation and their application to photosynthetic organisms. Mol Plant 2: 218–235, 2009 [DOI] [PubMed] [Google Scholar]
- 32. Garcia-Santamarina S, Boronat S, Domenech A, Ayte J, Molina H, and Hidalgo E. Monitoring in vivo reversible cysteine oxidation in proteins using ICAT and mass spectrometry. Nat Protoc 9: 1131–1145, 2014 [DOI] [PubMed] [Google Scholar]
- 33. Garcia-Santamarina S, Boronat S, Espadas G, Ayte J, Molina H, and Hidalgo E. The oxidized thiol proteome in fission yeast—optimization of an ICAT-based method to identify H2O2-oxidized proteins. J Proteomics 74: 2476–2486, 2011 [DOI] [PubMed] [Google Scholar]
- 34. Getz EB, Xiao M, Chakrabarty T, Cooke R, and Selvin PR. A comparison between the sulfhydryl reductants tris(2-carboxyethyl)phosphine and dithiothreitol for use in protein biochemistry. Anal Biochem 273: 73–80, 1999 [DOI] [PubMed] [Google Scholar]
- 35. Goodson P, Kumar A, Jain L, Kundu K, Murthy N, Koval M, and Helms MN. NADPH oxidase regulates alveolar epithelial sodium channel activity and lung fluid balance in vivo via O2− signaling. Am J Physiol Lung Cell Mol Physiol 302: 410, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gupta V. and Carroll KS. Profiling the reactivity of cyclic C-nucleophiles towards electrophilic sulfur in cysteine sulfenic acid. Chem Sci 7: 400–415, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gupta V. and Carroll KS. Sulfenic acid chemistry, detection and cellular lifetime. Biochim Biophys Acta 1840: 847–875, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gutscher M, Sobotta MC, Wabnitz GH, Ballikaya S, Meyer AJ, Samstag Y, and Dick TP. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J Biol Chem 284: 31532–31540, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, and Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 17: 994–999, 1999 [DOI] [PubMed] [Google Scholar]
- 40. Hamann M, Zhang T, Hendrich S, and Thomas JA. Quantitation of protein sulfinic and sulfonic acid, irreversibly oxidized protein cysteine sites in cellular proteins. Methods Enzymol 348: 146–156, 2002 [DOI] [PubMed] [Google Scholar]
- 41. Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, and Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem 279: 13044–13053, 2004 [DOI] [PubMed] [Google Scholar]
- 42. Hochgrafe F, Mostertz J, Albrecht D, and Hecker M. Fluorescence thiol modification assay: oxidatively modified proteins in Bacillus subtilis. Mol Microbiol 58: 409–425, 2005 [DOI] [PubMed] [Google Scholar]
- 43. Hopkins C, Gibson A, Stinchcombe J, and Futter C. Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope. Methods Enzymol 327: 35–45, 2000 [DOI] [PubMed] [Google Scholar]
- 44. Hu JJ, Wong NK, Ye S, Chen X, Lu MY, Zhao AQ, Guo Y, Ma AC, Leung AY, Shen J, and Yang D. Fluorescent probe HKSOX-1 for imaging and detection of endogenous superoxide in live cells and in vivo. J Am Chem Soc 137: 6837–6843, 2015 [DOI] [PubMed] [Google Scholar]
- 45. Huang Z, Pinto JT, Deng H, and Richie JP. Inhibition of caspase-3 activity and activation by protein glutathionylation. Biochem Pharmacol 75: 2234–2244, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hurd TR, Prime TA, Harbour ME, Lilley KS, and Murphy MP. Detection of reactive oxygen species-sensitive thiol proteins by redox difference gel electrophoresis: implications for mitochondrial redox signaling. J Biol Chem 282: 22040–22051, 2007 [DOI] [PubMed] [Google Scholar]
- 47. Jacob C, Holme AL, and Fry FH. The sulfinic acid switch in proteins. Org Biomol Chem 2: 1953–1956, 2004 [DOI] [PubMed] [Google Scholar]
- 48. Johnson-Cadwell LI, Jekabsons MB, Wang A, Polster BM, and Nicholls DG. “Mild uncoupling” does not decrease mitochondrial superoxide levels in cultured cerebellar granule neurons but decreases spare respiratory capacity and increases toxicity to glutamate and oxidative stress. J Neurochem 101: 1619–1631, 2007 [DOI] [PubMed] [Google Scholar]
- 49. Kambayashi Y. and Ogino K. Reestimation of Cypridina luciferin analogs (MCLA) as a chemiluminescence probe to detect active oxygen species—cautionary note for use of MCLA. J Toxicol Sci 28: 139–148, 2003 [DOI] [PubMed] [Google Scholar]
- 50. Kekulandara DN, Samarasinghe KT, Munkanatta Godage DN, and Ahn YH. Clickable glutathione using tetrazine-alkene bioorthogonal chemistry for detecting protein glutathionylation. Org Biomol Chem 14: 10886–10893, 2016 [DOI] [PubMed] [Google Scholar]
- 51. Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, and Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol 21: 86–89, 2003 [DOI] [PubMed] [Google Scholar]
- 52. Kervinen M, Patsi J, Finel M, and Hassinen IE. Lucigenin and coelenterazine as superoxide probes in mitochondrial and bacterial membranes. Anal Biochem 324: 45–51, 2004 [DOI] [PubMed] [Google Scholar]
- 53. Kimura C, Cheng W, Hisadome K, Wang YP, Koyama T, Karashima Y, Oike M, and Ito Y. Superoxide anion impairs contractility in cultured aortic smooth muscle cells. Am J Physiol Heart Circ Physiol 283: 382, 2002 [DOI] [PubMed] [Google Scholar]
- 54. Kimura C, Oike M, and Ito Y. Hypoxia-induced alterations in Ca2+ mobilization in brain microvascular endothelial cells. Am J Physiol Heart Circ Physiol 279: 2310, 2000 [DOI] [PubMed] [Google Scholar]
- 55. Knoefler D, Thamsen M, Koniczek M, Niemuth NJ, Diederich AK, and Jakob U. Quantitative in vivo redox sensors uncover oxidative stress as an early event in life. Mol Cell 47: 767–776, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kundu K, Knight SF, Lee S, Taylor WR, and Murthy N. A significant improvement of the efficacy of radical oxidant probes by the kinetic isotope effect. Angew Chem Int Ed Engl 49: 6134–6138, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kundu K, Knight SF, Willett N, Lee S, Taylor WR, and Murthy N. Hydrocyanines: a class of fluorescent sensors that can image reactive oxygen species in cell culture, tissue, and in vivo. Angew Chem Int Ed Engl 48: 299–303, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kuwahara K, Oizumi N, Fujisawa S, Tanito M, and Ohira A. Carteolol hydrochloride protects human corneal epithelial cells from UVB-induced damage in vitro. Cornea 24: 213–220, 2005 [DOI] [PubMed] [Google Scholar]
- 59. Kuzkaya N, Weissmann N, Harrison DG, and Dikalov S. Interactions of peroxynitrite, tetrahydrobiopterin, ascorbic acid, and thiols: implications for uncoupling endothelial nitric-oxide synthase. J Biol Chem 278: 22546–22554, 2003 [DOI] [PubMed] [Google Scholar]
- 60. Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol 4: 181–189, 2004 [DOI] [PubMed] [Google Scholar]
- 61. Le Moan N, Clement G, Le Maout S, Tacnet F, and Toledano MB. The Saccharomyces cerevisiae proteome of oxidized protein thiols: contrasted functions for the thioredoxin and glutathione pathways. J Biol Chem 281: 10420–10430, 2006 [DOI] [PubMed] [Google Scholar]
- 62. Le Moan N, Tacnet F, and Toledano MB. Protein-thiol oxidation, from single proteins to proteome-wide analyses. Methods Mol Biol 476: 181–198, 2008 [PubMed] [Google Scholar]
- 63. Lee SR, Kwon KS, Kim SR, and Rhee SG. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J Biol Chem 273: 15366–15372, 1998 [DOI] [PubMed] [Google Scholar]
- 64. Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, and Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A 105: 8197–8202, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Leichert LI. and Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol 2: e333, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Leonard SE, Reddie KG, and Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem Biol 4: 783–799, 2009 [DOI] [PubMed] [Google Scholar]
- 67. Li Y, Zhu H, Kuppusamy P, Roubaud V, Zweier JL, and Trush MA. Validation of lucigenin (bis-N-methylacridinium) as a chemilumigenic probe for detecting superoxide anion radical production by enzymatic and cellular systems. J Biol Chem 273: 2015–2023, 1998 [DOI] [PubMed] [Google Scholar]
- 68. Lillig CH. and Berndt C. Glutaredoxins in thiol/disulfide exchange. Antioxid Redox Signal 18: 1654–1665, 2013 [DOI] [PubMed] [Google Scholar]
- 69. Lind C, Gerdes R, Hamnell Y, Schuppe-Koistinen I, von Lowenhielm HB, Holmgren A, and Cotgreave IA. Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch Biochem Biophys 406: 229–240, 2002 [DOI] [PubMed] [Google Scholar]
- 70. Liu Y, Song Y, De Pascali F, Liu X, Villamena FA, and Zweier JL. Tetrathiatriarylmethyl radical with a single aromatic hydrogen as a highly sensitive and specific superoxide probe. Free Radic Biol Med 53: 2081–2091, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lo Conte M. and Carroll KS. The redox biochemistry of protein sulfenylation and sulfinylation. J Biol Chem 288: 26480–26488, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lo Conte M. and Carroll KS. Chemoselective ligation of sulfinic acids with aryl-nitroso compounds. Angew Chem Int Ed Engl 51: 6502–6505, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lo Conte M, Lin J, Wilson MA, and Carroll KS. A chemical approach for the detection of protein sulfinylation. ACS Chem Biol 10: 1825–1830, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Maeda H, Fukuyasu Y, Yoshida S, Fukuda M, Saeki K, Matsuno H, Yamauchi Y, Yoshida K, Hirata K, and Miyamoto K. Fluorescent probes for hydrogen peroxide based on a non-oxidative mechanism. Angew Chem Int Ed Engl 43: 2389–2391, 2004 [DOI] [PubMed] [Google Scholar]
- 75. Maeda H, Yamamoto K, Nomura Y, Kohno I, Hafsi L, Ueda N, Yoshida S, Fukuda M, Fukuyasu Y, Yamauchi Y, and Itoh N. A design of fluorescent probes for superoxide based on a nonredox mechanism. J Am Chem Soc 127: 68–69, 2005 [DOI] [PubMed] [Google Scholar]
- 76. Majmudar JD, Konopko AM, Labby KJ, Tom CTMB, Crellin JE, Prakash A, and Martin BR. Harnessing redox cross-reactivity to profile distinct cysteine modifications. J Am Chem Soc 138: 1852–1859, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Markvicheva KN, Bilan DS, Mishina NM, Gorokhovatsky AY, Vinokurov LM, Lukyanov S, and Belousov VV. A genetically encoded sensor for H2O2 with expanded dynamic range. Bioorg Med Chem 19: 1079–1084, 2011 [DOI] [PubMed] [Google Scholar]
- 78. Martinez-Acedo P, Nunez E, Gomez FJ, Moreno M, Ramos E, Izquierdo-Alvarez A, Miro-Casas E, Mesa R, Rodriguez P, Martinez-Ruiz A, Dorado DG, Lamas S, and Vazquez J. A novel strategy for global analysis of the dynamic thiol redox proteome. Mol Cell Proteomics 11: 800–813, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Matlashov ME, Bogdanova YA, Ermakova GV, Mishina NM, Ermakova YG, Nikitin ES, Balaban PM, Okabe S, Lukyanov S, Enikolopov G, Zaraisky AG, and Belousov VV. Fluorescent ratiometric pH indicator SypHer2: applications in neuroscience and regenerative biology. Biochim Biophys Acta 1850: 2318–2328, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Meyer AJ. and Hell R. Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth Res 86: 435–457, 2005 [DOI] [PubMed] [Google Scholar]
- 81. Michalek RD, Nelson KJ, Holbrook BC, Yi JS, Stridiron D, Daniel LW, Fetrow JS, King SB, Poole LB, and Grayson JM. The requirement of reversible cysteine sulfenic acid formation for T cell activation and function. J Immunol 179: 6456–6467, 2007 [DOI] [PubMed] [Google Scholar]
- 82. Miller EW, Albers AE, Pralle A, Isacoff EY, and Chang CJ. Boronate-based fluorescent probes for imaging cellular hydrogen peroxide. J Am Chem Soc 127: 16652–16659, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Miller EW, Tulyathan O, Isacoff EY, and Chang CJ. Molecular imaging of hydrogen peroxide produced for cell signaling. Nat Chem Biol 3: 263–267, 2007 [DOI] [PubMed] [Google Scholar]
- 84. Morgan B, Van Laer K, Owusu TN, Ezerina D, Pastor-Flores D, Amponsah PS, Tursch A, and Dick TP. Real-time monitoring of basal H2O2 levels with peroxiredoxin-based probes. Nat Chem Biol 12: 437–443, 2016 [DOI] [PubMed] [Google Scholar]
- 85. Nazarewicz RR, Bikineyeva A, and Dikalov SI. Rapid and specific measurements of superoxide using fluorescence spectroscopy. J Biomol Screen 18: 498–503, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nazarewicz RR, Dikalova AE, Bikineyeva A, and Dikalov SI. Nox2 as a potential target of mitochondrial superoxide and its role in endothelial oxidative stress. Am J Physiol Heart Circ Physiol 305: 1131, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nelson KJ, Klomsiri C, Codreanu SG, Soito L, Liebler DC, Rogers LC, Daniel LW, and Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection methods to visualize and identify labeled proteins. Methods Enzymol 473: 95–115, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Niethammer P, Grabher C, Look AT, and Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 459: 996–999, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nishida A, Kimura H, Nakano M, and Goto T. A sensitive and specific chemiluminescence method for estimating the ability of human granulocytes and monocytes to generate O2−. Clin Chim Acta 179: 177–181, 1989 [DOI] [PubMed] [Google Scholar]
- 90. Oosthuizen MM. and Greyling D. Hydroxyl radical generation: the effect of bicarbonate, dioxygen and buffer concentration on pH-dependent chemiluminescence. Redox Rep 6: 105–116, 2001 [DOI] [PubMed] [Google Scholar]
- 91. Pace NJ. and Weerapana E. Diverse functional roles of reactive cysteines. ACS Chem Biol 8: 283–296, 2013 [DOI] [PubMed] [Google Scholar]
- 92. Pastore A. and Piemonte F. S-glutathionylation signaling in cell biology: progress and prospects. Eur J Pharm Sci 46: 279–292, 2012 [DOI] [PubMed] [Google Scholar]
- 93. Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, and Carroll KS. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol 8: 57–64, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pineda-Molina E, Klatt P, Vazquez J, Marina A, Garcia de Lacoba M, Perez-Sala D, and Lamas S. Glutathionylation of the p50 subunit of NF-kappaB: a mechanism for redox-induced inhibition of DNA binding. Biochemistry 40: 14134–14142, 2001 [DOI] [PubMed] [Google Scholar]
- 95. Poole LB, Klomsiri C, Knaggs SA, Furdui CM, Nelson KJ, Thomas MJ, Fetrow JS, Daniel LW, and King SB. Fluorescent and affinity-based tools to detect cysteine sulfenic acid formation in proteins. Bioconjug Chem 18: 2004–2017, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Poole LB, Zeng BB, Knaggs SA, Yakubu M, and King SB. Synthesis of chemical probes to map sulfenic acid modifications on proteins. Bioconjug Chem 16: 1624–1628, 2005 [DOI] [PubMed] [Google Scholar]
- 97. Poole TH, Reisz JA, Zhao W, Poole LB, Furdui CM, and King SB. Strained cycloalkynes as new protein sulfenic acid traps. J Am Chem Soc 136: 6167–6170, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Qian Y, Martell J, Pace NJ, Ballard TE, Johnson DS, and Weerapana E. An isotopically tagged azobenzene-based cleavable linker for quantitative proteomics. Chembiochem 14: 1410–1414, 2013 [DOI] [PubMed] [Google Scholar]
- 99. Ravichandran V, Seres T, Moriguchi T, Thomas JA, and Johnston RB. S-thiolation of glyceraldehyde-3-phosphate dehydrogenase induced by the phagocytosis-associated respiratory burst in blood monocytes. J Biol Chem 269: 25010–25015, 1994 [PubMed] [Google Scholar]
- 100. Reddie KG, Seo YH, Muse WB, III, Leonard SE, and Carroll KS. A chemical approach for detecting sulfenic acid-modified proteins in living cells. Mol Biosyst 4: 521–531, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Reynaert NL, van der Vliet A, Guala AS, McGovern T, Hristova M, Pantano C, Heintz NH, Heim J, Ho YS, Matthews DE, Wouters EF, and Janssen-Heininger YM. Dynamic redox control of NF-kappaB through glutaredoxin-regulated S-glutathionylation of inhibitory kappaB kinase beta. Proc Natl Acad Sci U S A 103: 13086–13091, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science 312: 1882–1883, 2006 [DOI] [PubMed] [Google Scholar]
- 103. Rizzi C, Samouilov A, Kutala VK, Parinandi NL, Zweier JL, and Kuppusamy P. Application of a trityl-based radical probe for measuring superoxide. Free Radic Biol Med 35: 1608–1618, 2003 [DOI] [PubMed] [Google Scholar]
- 104. Rokutan K, Thomas JA, and Johnston RB. Phagocytosis and stimulation of the respiratory burst by phorbol diester initiate S-thiolation of specific proteins in macrophages. J Immunol 147: 260–264, 1991 [PubMed] [Google Scholar]
- 105. Rosen GM. and Freeman BA. Detection of superoxide generated by endothelial cells. Proc Natl Acad Sci U S A 81: 7269–7273, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Samarasinghe KT, Munkanatta Godage DN, VanHecke GC, and Ahn YH. Metabolic synthesis of clickable glutathione for chemoselective detection of glutathionylation. J Am Chem Soc 136: 11566–11569, 2014 [DOI] [PubMed] [Google Scholar]
- 107. Samarasinghe KT, Munkanatta Godage DN, Zhou Y, Ndombera FT, Weerapana E, and Ahn YH. A clickable glutathione approach for identification of protein glutathionylation in response to glucose metabolism. Mol Biosyst 12: 2471–2480, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Saurin AT, Neubert H, Brennan JP, and Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc Natl Acad Sci U S A 101: 17982–17987, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sawaki Y. and Foote CS. Acyclic mechanism in the cleavage of benzils with alkaline hydrogen peroxide. J Am Chem Soc 101: 6292–6296, 1979 [Google Scholar]
- 110. Seo YH. and Carroll KS. Quantification of protein sulfenic acid modifications using isotope-coded dimedone and iododimedone. Angew Chem Int Ed Engl 50: 1342–1345, 2011 [DOI] [PubMed] [Google Scholar]
- 111. Sethuraman M, McComb ME, Heibeck T, Costello CE, and Cohen RA. Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol Cell Proteomics 3: 273–278, 2004 [DOI] [PubMed] [Google Scholar]
- 112. Setsukinai K, Urano Y, Kakinuma K, Majima HJ, and Nagano T. Development of novel fluorescence probes that can reliably detect reactive oxygen species and distinguish specific species. J Biol Chem 278: 3170–3175, 2003 [DOI] [PubMed] [Google Scholar]
- 113. Sheng Y, Abreu IA, Cabelli DE, Maroney MJ, Miller AF, Teixeira M, and Valentine JS. Superoxide dismutases and superoxide reductases. Chem Rev 114: 3854–3918, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Sikora A, Zielonka J, Lopez M, Joseph J, and Kalyanaraman B. Direct oxidation of boronates by peroxynitrite: mechanism and implications in fluorescence imaging of peroxynitrite. Free Radic Biol Med 47: 1401–1407, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Srikun D, Albers AE, Nam CI, Iavarone AT, and Chang CJ. Organelle-targetable fluorescent probes for imaging hydrogen peroxide in living cells via SNAP-Tag protein labeling. J Am Chem Soc 132: 4455–4465, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sugioka K, Nakano M, Kurashige S, Akuzawa Y, and Goto T. A chemiluminescent probe with a Cypridina luciferin analog, 2-methyl-6-phenyl-3,7-dihydroimidazo[1,2-a]pyrazin-3-one, specific and sensitive for O2- production in phagocytizing macrophages. FEBS Lett 197: 27–30, 1986 [DOI] [PubMed] [Google Scholar]
- 117. Sullivan DM, Wehr NB, Fergusson MM, Levine RL, and Finkel T. Identification of oxidant-sensitive proteins: TNF-alpha induces protein glutathiolation. Biochemistry 39: 11121–11128, 2000 [DOI] [PubMed] [Google Scholar]
- 118. Sumimoto H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J 275: 3249–3277, 2008 [DOI] [PubMed] [Google Scholar]
- 119. Tamura Y, Chi LG, Driscoll EM, Hoff PT, Freeman BA, Gallagher KP, and Lucchesi BR. Superoxide dismutase conjugated to polyethylene glycol provides sustained protection against myocardial ischemia/reperfusion injury in canine heart. Circ Res 63: 944–959, 1988 [DOI] [PubMed] [Google Scholar]
- 120. Tarpey MM, White CR, Suarez E, Richardson G, Radi R, and Freeman BA. Chemiluminescent detection of oxidants in vascular tissue. Lucigenin but not coelenterazine enhances superoxide formation. Circ Res 84: 1203–1211, 1999 [DOI] [PubMed] [Google Scholar]
- 121. Tian H, Sakmar TP, and Huber T. A simple method for enhancing the bioorthogonality of cyclooctyne reagent. Chem Commun (Camb) 52: 5451–5454, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Tormos KV. and Chandel NS. Seeing the light: probing ROS in vivo using redox GFP. Cell Metab 14: 720–721, 2011 [DOI] [PubMed] [Google Scholar]
- 123. Ueno T, Urano Y, Setsukinai K, Takakusa H, Kojima H, Kikuchi K, Ohkubo K, Fukuzumi S, and Nagano T. Rational principles for modulating fluorescence properties of fluorescein. J Am Chem Soc 126: 14079–14085, 2004 [DOI] [PubMed] [Google Scholar]
- 124. Urano Y, Kamiya M, Kanda K, Ueno T, Hirose K, and Nagano T. Evolution of fluorescein as a platform for finely tunable fluorescence probes. J Am Chem Soc 127: 4888–4894, 2005 [DOI] [PubMed] [Google Scholar]
- 125. Ushio-Fukai M. Localizing NADPH oxidase-derived ROS. Sci STKE 2006: re8, 2006 [DOI] [PubMed] [Google Scholar]
- 126. Veal EA, Day AM, and Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell 26: 1–14, 2007 [DOI] [PubMed] [Google Scholar]
- 127. Velu CS, Niture SK, Doneanu CE, Pattabiraman N, and Srivenugopal KS. Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry 46: 7765–7780, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wang P, Chen H, Qin H, Sankarapandi S, Becher MW, Wong PC, and Zweier JL. Overexpression of human copper, zinc-superoxide dismutase (SOD1) prevents postischemic injury. Proc Natl Acad Sci U S A 95: 4556–4560, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wani R, Qian J, Yin L, Bechtold E, King SB, Poole LB, Paek E, Tsang AW, and Furdui CM. Isoform-specific regulation of Akt by PDGF-induced reactive oxygen species. Proc Natl Acad Sci U S A 108: 10550–10555, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Warnholtz A, Nickenig G, Schulz E, Macharzina R, Brasen JH, Skatchkov M, Heitzer T, Stasch JP, Griendling KK, Harrison DG, Bohm M, Meinertz T, and Munzel T. Increased NADH-oxidase-mediated superoxide production in the early stages of atherosclerosis: evidence for involvement of the renin-angiotensin system. Circulation 99: 2027–2033, 1999 [DOI] [PubMed] [Google Scholar]
- 131. Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, and Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468: 790–795, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol 4: 278–286, 2008 [DOI] [PubMed] [Google Scholar]
- 133. Woo HA, Yim SH, Shin DH, Kang D, Yu DY, and Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell 140: 517–528, 2010 [DOI] [PubMed] [Google Scholar]
- 134. Yang J, Gupta V, Carroll KS, and Liebler DC. Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat Commun 5: 4776, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Yang KS, Kang SW, Woo HA, Hwang SC, Chae HZ, Kim K, and Rhee SG. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem 277: 38029–38036, 2002 [DOI] [PubMed] [Google Scholar]
- 136. Yano H. Fluorescent labeling of disulfide proteins on 2D gel for screening allergens: a preliminary study. Anal Chem 75: 4682–4685, 2003 [DOI] [PubMed] [Google Scholar]
- 137. Yu F, Song P, Li P, Wang B, and Han K. A fluorescent probe directly detect peroxynitrite based on boronate oxidation and its applications for fluorescence imaging in living cells. Analyst 137: 3740–3749, 2012 [DOI] [PubMed] [Google Scholar]
- 138. Zhao H, Joseph J, Fales HM, Sokoloski EA, Levine RL, Vasquez-Vivar J, and Kalyanaraman B. Detection and characterization of the product of hydroethidine and intracellular superoxide by HPLC and limitations of fluorescence. Proc Natl Acad Sci U S A 102: 5727–5732, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Zhao H, Kalivendi S, Zhang H, Joseph J, Nithipatikom K, Vasquez-Vivar J, and Kalyanaraman B. Superoxide reacts with hydroethidine but forms a fluorescent product that is distinctly different from ethidium: potential implications in intracellular fluorescence detection of superoxide. Free Radic Biol Med 34: 1359–1368, 2003 [DOI] [PubMed] [Google Scholar]
- 140. Zhou M, Diwu Z, Panchuk-Voloshina N, and Haugland RP. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal Biochem 253: 162–168, 1997 [DOI] [PubMed] [Google Scholar]
- 141. Zhou Y, Wynia-Smith SL, Couvertier SM, Kalous KS, Marletta MA, Smith BC, and Weerapana E. Chemoproteomic strategy to quantitatively monitor transnitrosation uncovers functionally relevant S-nitrosation sites on cathepsin D and HADH2. Cell Chem Biol 23: 727–737, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zielonka J. and Kalyanaraman B. Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic Biol Med 48: 983–1001, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Zielonka J, Zielonka M, Sikora A, Adamus J, Joseph J, Hardy M, Ouari O, Dranka BP, and Kalyanaraman B. Global profiling of reactive oxygen and nitrogen species in biological systems: high-throughput real-time analyses. J Biol Chem 287: 2984–2995, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Zweier JL, Kuppusamy P, and Lutty GA. Measurement of endothelial cell free radical generation: evidence for a central mechanism of free radical injury in postischemic tissues. Proc Natl Acad Sci U S A 85: 4046–4050, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]