Abstract
Protein synthesis is a central and dynamic process, frequently deregulated in cancer through aberrant activation or expression of translation initiation factors and tRNAs. The discovery of tRNA-derived fragments, a new class of abundant and, in some cases stress-induced, small Noncoding RNAs has perplexed the epigenomics landscape and highlights the emerging regulatory role of tRNAs in translation and beyond. Skin is the biggest organ in human body, which maintains homeostasis of its multilayers through regulatory networks that induce translational reprogramming, and modulate tRNA transcription, modification and fragmentation, in response to various stress signals, like UV irradiation. In this review, we summarize recent knowledge on the role of translation regulation and tRNA biology in the alarming prevalence of skin cancer.
Keywords: : Noncoding RNAs, ribosome, skin cancer, translation regulation, tRFs, tRNA
Translation regulation, tRNAs & tRFs: a dynamic balance
Protein synthesis is a central and essential biological process during which the genetic information imprinted on mRNAs is decoded into the necessary protein context required for cell's viability by tRNAs carrying cognate amino acids onto ribosomes [1–3]. Uninterrupted and flowless translation depends on the transcription, folding, maturation and posttranscriptional modification of rRNAs and tRNAs, in a pipeline of events tightly controlled and mediated by many important enzymes and scaffold proteins [4]. The order and the accuracy of the reactions that occur during translation is well-defined and consists of several steps, some of which are highly energy-consuming, and ensure the quality of the produced proteins for every cell type [5]. During cell's life, translational rates reflect the metabolic rewiring that occurs in response to various intra- or extracellular signals, through pathways which converge to target translation initiation and affect directly or indirectly the transcription of tRNAs and rRNAs [6]. Regulation of translation rates is cell- or tissue- specific and defines cell's life cycle, providing the means of cross-talk for virtually every known biochemical pathway [7]. Therefore, translation is considered a key process for the regulation of gene expression which maintains cellular and tissue homeostasis [8]. The rapid development of powerful high-throughput methodologies, such as next generation RNA sequencing, mass spectrometry and bioinformatics tools, combined with ribosomal profiling studies have unveiled an unprecedented complexity of several RNA–protein networks that control translation pace, especially under stress conditions [9,10]. These interactions often appear derailed and therefore, translation deregulation leading to aberrant production of tumor promoting proteins has been considered a hallmark of cancer [11,12].
The complex landscape of translation regulation is further enriched by several events that occur either at the post-transcriptional or at the post-translational level [13]. Recent studies indicated ribosomal assembly and modifications as important modulators during early development, thus emphasizing the spatial and temporal organization and regulation of translation during stem cell self-renewal and differentiation [14,15]. Ribosomes exhibit considerable variation even within the same cell and contribute to gene expression regulation through selective translation of distinct subpools of mRNAs [16,17]. Moreover, specific ribosomal proteins play role in embryonic development and exhibit tissue specific patterns [18]. Similarly, specific ribosomal protein patterns can distinguish normal from malignant human cells and in addition, aberrant overexpression of rRNA may also occur in several tumors [19–21]. In addition, several important rRNA modifications contribute significantly to the heterogeneity of ribosomal population within a cell and several studies associate modifications deregulation of specific ribosomal constituents to carcinogenesis [22,23]. Defects in translation machinery directly affect the expression of proto-oncogenes such as KRAS, mTOR and MYC which in turn, control transcription of rRNAs and tRNAs by RNA polymerase I and III (RNAP I and III) [24–26].
Since their discovery, tRNA molecules were considered ‘housekeeping’ RNAs and passive carriers of amino acids, representing adaptor molecules between the RNA and protein world [27]. In human, an unexpectedly high number of genes (>600) and an even more surprising copy number variation suggest additional roles for these ancient molecules [28,29]. Advances in RNA sequencing allowed the quantitation of tRNA gene expression in various cell types and tissues which revealed the complexity of the human tRNAome [30]. Several recent studies have shown that translation of specific mRNAs can occur as a mechanism of adaptation in response to stress and interestingly, distinct tRNA expression patterns between different cell types and differences in tRNA content and usage in response to various signals and conditions affect translational regulation and promote adaptive translation in a very dynamic way [31]. A closer look on the codon context of different cell types suggests a codominant expression of their codon-enriched tRNAs and clearly indicate coordination between transcription and translation, in support of the notion that translation regulation relies on distinct transcriptional programs coordinating tRNA supply and demand [32,33]. In addition, the fact that specific tRNAs and aminoacyl-tRNA synthetases are modulated in several cancers opens new avenues and provides novel comprehensive and specific molecular signatures that could be used for diagnosis [34,35]. Finally, numerous posttranscriptional modifications on tRNAs have been associated with a broad spectrum of diseases, including cancer, thus highlighting the importance of correctly synthesized and fully active tRNAs for optimization of translation rates and proteome integrity [36,37]. It is evident from many reports that alterations of tRNA expression, modification and usage represent a mechanism of stress response and these changes affect translation rates [38]. Upregulation of tRNAs can drive cancer metastasis by enhancing the stability of translation of nodal mRNAs enriched in rare codons. In addition, mischarged tRNAs can incorporate amino acids erroneously into proteins to promote dynamic translational adaptations under stress [39]. Therefore, codon bias, a term which describes the uneven use of synonymous codons, codon usage and codon optimality has been proposed as an additional epigenetic code which fine-tunes the networks of translation machinery [40].
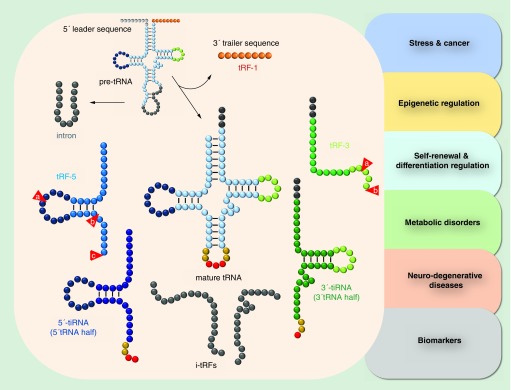
The regulatory roles of tRNAs beyond translation were further elaborated with the recent discovery of tRNA-derived fragments (tRFs), which represent a new class of small noncoding RNAs, first detected decades ago in the urine samples of cancer patients [27,41]. Although at the beginning they were considered tRNA degradation products, recent meta-analyses of several datasets showed that tRFs are not produced randomly [42–44]. Instead, they are present in all kingdoms of life and are associated with a vast array of biological functions, spanning from stem cell self-renewal and epigenetic inheritance to translation control and cancer [45–50]. tRNA fragments can derive from either pre- or mature tRNAs and, depending on the origin and length (14–40 nt), are classified as tRF-5 (type a, b and c, mainly located in the nucleus), tRF-3 (type a and b, mainly located in the cytoplasm), tRF-1 (3′ maturation products, located in the cytoplasm) and i-tRFs (or tiRs, of intronic origin) [51,52] (Figure 1). Biogenesis of tRFs depends on cell type, condition or developmental stage and specific tRF signatures have been reported for various cancer types suggesting that tRFs can serve as novel auxiliary biomarkers for several pathological phenotypes [53]. Moreover, fragmentation of tRNAs is affected by the presence or absence of post-transcriptional modifications and is mediated by different endonucleases, including tRNase Z and Dicer, while the existence of additional enzymes that produce tRFs still remain elusive [54]. In eukaryotes, tRNA cleavage is a conserved response to oxidative stress and angiogenin (ANG), an essential ribonuclease known to promote angiogenesis, is responsible for the production of stress-derived tRFs known as 5′- or 3′-tiRNAs (or tRNA halves; stress induced tRFs), in response to various external stress signals, like UV radiation (UVR), heat shock, oxidative and nutrition stress, which can repress translation similarly to miRNAs [55–57].
Figure 1. . Classification of tRNA-derived fragments.
tRNA fragments can derive from either pre- or mature tRNAs and, depending on the origin and length (14–40 nt), are classified as tRNA-derived fragment (tRF)-5 (type a, b and c), tRF-3 (type a and b), tRF-1 (3′ maturation products) and i-tRFs (of intronic origin). In eukaryotes, tRNA cleavage by angiogenin (ANG) in response to stress signals leads to the production of stress-derived tRFs known as 5′- or 3′-tiRNAs (or tRNA halves; stress induced tRFs). tRFs and tiRNAs have been implicated in several conditions, including cancer, stress, epigenetic regulation, self-renewal and differentiation, various metabolic disorders and neurodegenerative diseases, while their use as biomarkers has only recently began to emerge.
Although deregulation of translation initiation, tRNA expression and modification, and tRF-mediated regulation have been reported individually for several cancer types, a comprehensive picture of the dynamic balance between all has emerged only recently, regarding translation reprogramming in skin stem cells and melanoma [58–60]. Skin cancer represents a group of heterogeneous nonmelanoma and melanoma tumors, which are highly correlated to environmental stress signals such as UV irradiation and exposure to chemical pollutants, resulting in excessive oxidative stress and mutagenesis and leading in response, to deregulated or adaptive translation [61]. During the last decades climate change and prolonged exposure to UV irradiation has brought skin cancer in the spotlight as the most common type of cancer, worldwide [62,63]. Subsequently, the development and analysis of genetically engineered mouse models was necessary and has helped significantly towards the identification of the molecular and microenvironmental mechanisms underlying UV induction of aggressive skin cancer such as melanoma, as well as its metastatic spread [64,65]. Moreover, it allowed a better understanding of the inherent and acquired resistance to targeted and immune-based therapeutic and helped to improve preclinical models [66,67].
Herein, we summarize recent knowledge regarding the role of deregulated translation initiation, the biosynthesis and overexpression of tRNAs and their fragments in skin cancer mechanisms and we discuss the putative role of tRFs in keratinocytes.
The complexity & plasticity of skin anatomy
Skin is the largest organ in the human body and accounts for almost 15% of the total adult bodyweight [68]. It represents an effective barrier between the organism and the environment and provides the primary protection against several external dangers, like sunlight exposure, injuries, microbial infections and pollutants, that cause allergies and/or inflammation. In addition, skin layers prevent excessive dehydration from heat, thus significantly contributing to thermoregulation [69]. The essential functions of skin rely on its stratification, the adhesion level between layers and different cell types and various signals that fine-tune gene expression which maintains skin homeostasis [70]. Skin consists of three major layers: the epidermis, the dermis and subcutaneous tissue (or hypodermis), each well-defined and with a content of certain differentiated cells and niches of pluripotent stem cells (Figure 2). Skin stem cells of epidermis and other layers allow their continuous renewal, a characteristic of skin which makes it an ideal model for studying mechanisms of differentiation and tissue regeneration [71].
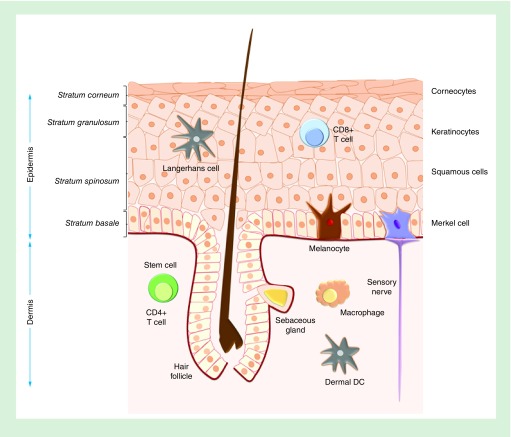
Figure 2. . The layers and cell types of human epidermis.
Epidermal skin layers include the basal layer (stratum basale), the squamous cell layer (stratum spinosum), the granular layer (stratum granulosum) and the cornified layer (stratum corneum). Skin cells include keratinocytes (majority of cells), skin stem cells, melanocytes, Langerhans cells and Merkel cells.
Epidermis is the outer layer of skin; a stratified epithelium composed of keratinocytes (ectodermally-derived representing almost 80% of all cells), melanocytes of neural crest origin responsible for melanin production and skin color and finally, Merkel and Langerhans cells, which are responsible for immune protection against infections and certain chemicals. Keratinocytes move along the four layers of epidermis in repeating cycles of renewal: the basal layer (stratum basale) which contains mitotically active cells forming a single layer, the squamous cell layer (stratum spinosum), the granular layer (stratum granulosum) and the cornified layer (stratum corneum) which provides the mechanical protection due to an extensive network of keratin filaments (Figure 2). The largest pool of epidermal keratinocytes differentiates during migration from the basal layer to the surface, in a complex process known as terminal differentiation or keratinization [72]. During keratinization, the expression, alternative splicing and translation of different keratin types is essential for the regulation and promotion of the proliferative process [73]. In addition, the basal layer harbors stem cells dividing in a relatively low rate which contribute to the continuous renewal of outer epidermis by losing their self-renewal ability and proliferating as they move to the surface [74]. It has been proposed that epidermal stem cells contribute to the dynamic homeostasis of epidermis, only when intense external stimuli (i.e., wounding, sunburns, etc.) require extensive recruitment of proliferating and differentiating cells for effective and fast tissue reconstitution, in response to stress [75]. Recently the remarkable existence of long-lived stem cells that produce progenitors that replenish terminally differentiated cells was described, providing new perspectives regarding skin healing and regeneration [76].
The dermis (ten-times thicker) is separated from epidermis by the basal membrane which provides mechanical support and polarity coordination for epidermal cells and serves as an important gatekeeper for exchange of cells and molecules. Basal membrane regulates the controlled release of important cytokines and growth factors, during tissue development or repair and provides the niche for important biochemical events. Epidermal morphogenesis and differentiation depend to a great extent on dermis and their interaction is crucial for the development of the appendages such as the hair follicles, apocrine and eccrine glands [77]. In addition, multiple stem cell niches contribute to dermal regeneration when necessary [78]. Finally, the subcutaneous tissue layer varies in size and buoyancy and consists of lipocytes which store and provide energy for the body [68]. The plasticity and nature of skin layers and their frequent exposure to various stress signals that lead to cell damage have contributed to the alarming increase of various types of skin cancer in human.
Molecular mechanisms involved in skin cancer
Skin cancer is the most frequent and common type of cancer globally accounting for at least 40% of cases and the most widespread cancer among Caucasian population [62]. It arises from a combination of genetic and environmental factors, with DNA damage caused by exposure to UVR being the major effector [79]. Both UVR and environmental pollution lead to damaging oxidative stress which triggers aging and associates with increasing incidents of skin cancer [80].
The three major types of skin cancer are the basal-cell skin cancer (BCC), the cutaneous squamous-cell skin cancer (SCC) and, melanoma, the deadliest type of a severe malignancy deriving from melanocytes of the basal layer [81]. BCCs and SCCs are known as nonmelanoma skin cancers (NMSCs). They account for the 40% of all cancers and more than 90% of the skin cancers, mostly found in body parts frequently exposed to sunlight. The term ‘keratinocyte carcinomas’ is also used to distinguish them from less common skin cancers, such as Merkel-cell carcinoma (MCC), adnexal carcinoma, dermatofibrosarcoma protuberans (DFSB) and cutaneous lymphoma [82,83]. BCCs and SCCs are found with a 4:1 ratio in keratinocyte carcinomas. Exposure to UVR, age, skin pigmentation and lifestyle are the major factors for developing NMSC [84]. Intermittent and childhood sun exposure seems to increase the risk of BCC, while SCC appears more related to chronic UV exposure. Although BCC is generally considered not life-threating, there is a growing concern for SCC mortality especially among the elderly and immunosuppressed patients [85].
Skin cancer development can be affected by a combination of gene expression fluctuations and cell communication in the cells surrounding a tumorigenic lesion containing epithelial and mesenchymal cells. Until recently, it was unclear whether changes in the different layers of the skin were associated to more widespread alterations [86]. Notch signaling is an important cell–cell communication pathway with a key role in skin cells and contributes to development and tissue homeostasis [87]. Mice with inactivated Notch signaling develop spontaneous, multifocal squamous carcinomas of diverse cellular origin and the formation of these lesions can be prevented by administration of anti-inflammatory drugs. It was proposed that human skin fibroblasts with dysfunctional Notch signaling could also promote the neoplastic transformation of epithelial cells through transcriptional activation by AP-1 transcription factor, which plays important role in the regulation of keratinocytes differentiation and proliferation during renewal of epidermis. In an important study, the stroma surrounding human preneoplastic skin lesions showed similar alterations in Notch signaling to those in the mouse model, and these changes could be induced by exposure to UVA, highlighting the role of mesenchymal components as cancer drivers [88].
Although UVA is more effective in skin penetration, the most biologically severe effects are attributed to UVB and include mainly DNA damage, inflammatory response leading to immunosuppression, production of reactive oxygen species (ROS) and activation of receptor tyrosine kinases-mediated (RTK) signaling [89]. In keratinocytes, DNA damage can activate cell cycle checkpoints and induce mechanisms which can initiate apoptosis, while DNA repair defects can lead to carcinogenesis [90]. In NMSC, ATM and ATR kinases are critical in maintaining and regulating DNA damage checkpoints and activating downstream signals that either repair DNA or induce apoptosis, involving targeting or activation of tumor suppressor protein p53 (for more extensive review see [86]).
Inflammatory response can drive keratinocyte proliferation and induce invasion and metastasis of cancer cells. The most important pathways involved are the NFκB pathway (including RTKs, TNF and TLR receptors) and the EGFR-dependent activation of STAT3, an important transcription factor with role in survival and proliferation of keratinocytes after UVB exposure [91]. STAT3 regulates expression of inflammatory interleukins and nodal genes including c-MYC, c-FOS and BCL-2 with role in modulation of tumor progression, survival and metastasis, respectively [92–95].
UVR induces ROS production which directly affect the nuclear and mitochondrial DNA and RNA of keratinocytes and results in deregulation of central signaling pathways, through activation of members of the MAPK family, p38 and JNK signaling pathways [96,97]. Activation of p38 pathway promotes p53-dependent or independent (through increased expression of HIF1α) apoptosis and recent studies in mice suggest that survival of keratinocytes can occur through p38 activation and subsequent upregulation of Bcl-XL and COX2 which in turn, modulate mitochondrial apoptosis. As a result p38-deficient mice are resistant to skin carcinogenesis induced after prolonged UVR [98,99]. In the JNK pathway the main target is again the transcription factor AP-1 which has oncogenic capacity, as mentioned above [100].
The RTK receptors (such as IR, IGF-1-R and EGFR) are also rapidly activated in response to UVR exposure in NMSC. The main affected downstream cascades are the RAS/RAF/MEK/ERK and the PI3K/PTEN/AKT/mTOR signaling pathways. Both pathways converge to the translation initiation, stimulate translation of specific oncogenes (i.e., c-MYC) and affect global protein synthesis rates [101]. Finally, all the above pathways modulate UVB-induced keratinocyte apoptosis via suppression of Bcl-2 expression (intrinsic pathway) and activation of Caspase 8 (extrinsic pathway) [102].
Essentially the same pathways are deregulated in melanoma which represents the most aggressive type of skin cancer, due to its metastatic potential and resistance to available therapies. Melanomas are heterogenous tumors which usually develop de novo in normal skin (70%) or within a pre-existing acquired or congenital melanocytic nevus (to a much lower percentage) [103]. DNA damage caused by UV irradiation is the main trigger, with genetic predisposition to account for 10% of melanoma cases [104]. The oncogenic transformation is triggered by a combination of senescence bypass, such as PTEN deletion or inactivation of the CDKN2A locus, combined with deregulated MAPK signaling primarily driven by activating mutations in BRAF (∼50%) or NRAS (∼20%), which rarely co-exist [105]. The most common mutation found in >90% of melanoma cases is BRAFV600E, resulting in activation of the MAPK pathway, secretion of VEGF and vascular development [106]. Interestingly, the risk of cutaneous melanoma is primarily associated with intermittent rather than cumulative UVR exposure or severe episodes of sunburn, suggesting involvement of mechanisms different to those of NMSC [107].
Whole-genome sequencing analyses showed that melanomas of different origin (cutaneous, mucosal and acral) exhibit different mutational signatures, an observation which explains differences in melanoma epidemiology and genetic cancer drivers in different body parts that are not susceptible to UV irradiation. Most cutaneous melanomas have a distinct UV-associated signature of mutations, with C > T nucleotide transitions being the most dominant. TERT promoter mutations were the most frequently observed alterations, followed by driver mutations in TP53, PTEN or RB1 and mutations in components of the MAPK or PI3K pathways [108]. Existing reports suggest a synergistic effect of essential genes expression induced by microphthalmia-associated transcription factor (MITF), a master regulator of melanocyte development, function and survival. Interestingly, a short isoform (419 aa) of MITF transcription factor (MITF-M) is expressed exclusively in melanocytes and melanoma cells [109,110]. MITF expression at transcription and translation levels differs by many folds but it is imperative for the fate of melanocytes to remain between tightly regulated margins, especially given that MITF is associated with oncogenic programs and is affected by BRAF- or NRAS-derived deregulation of MAPK pathway, together with HIF1α and MYC [111]. Moreover, MITF regulates the metabolic rewiring in melanoma and switch from oxidative phosphorylation to glycolysis [112].
MITF amplicon harbors the melanoma specific long noncoding RNA SAMMSON. SAMMSON is transcriptionally cogained with MITF in 10% of the melanomas and the expression levels of MITF and SAMMSON were not previously correlated, although SAMMSON is expressed in more than 90% of melanomas. Recently it was shown that SAMMSON is an important regulatory long noncoding RNA and its silencing can reduce melanoma cell growth and survival, independent of the transcriptional state of BRAF, NRAS or TP53. Moreover, cells that are intrinsically resistant to BRAF inhibition were found to be sensitive to SAMMSON silencing which enhanced the effects of BRAF and MEK inhibitors. Finally, cells with acquired resistance to BRAF inhibition remained sensitive to SAMMSON targeting [113]. Interestingly, SAMMSON regulates mitochondrial translation through interaction with p32 (also known as C1QBP/gC1qR/HABP1) a widely distributed, multiligand-binding and multifunctional protein, which contributes to maintenance of mitochondrial membrane potential and oxidative phosphorylation [114]. It was observed that depletion of SAMMSON decreases the fraction of p32 associated with mitochondria, diminishes mitochondrial membrane potential and ultimately, triggers the p53-independent apoptotic response [115]. The fact that a SAMMSON-specific antisense oligonucleotide decreases tumor growth and synergizes with dabrafenib, a BRAF inhibitor, to induce apoptosis in vivo, is likely to arise because BRAF inhibition activates a MITF-PGC1α axis to elevate mitochondrial oxidative phosphorylation [116]. Most recently, it was demonstrated that SAMMSON promotes increased ribosomal RNA maturation and translation both in the cytosol and mitochondria. This is achieved through modulation of the localization of CARF, an RNA-binding protein. CARF binds and sequesters the exo-ribonuclease XRN2 in the nucleoplasm, which under normal conditions limits nucleolar rRNA maturation. SAMMSON interferes with XRN2 binding to CARF in the nucleus and favors the formation of an aberrant cytoplasmic RNA–protein complex containing CARF and p32 which is required for the processing of the mitochondrial rRNAs. Therefore, SAMMSON represents an oncogenic lncRNA that can simultaneously modulate RNA–protein complex formation in two distinct cellular compartments to promote cell growth [117]. These recent findings illuminate the multiple levels of epigenetic regulation that exist and contribute to skin carcinogenesis (especially in melanoma) and include long noncoding RNAs and many miRNAs, which are either used as predictive biomarkers in skin cancer or represent regulators of the MAPK or the PI3K/mTOR pathways which in turn, adjust the potential of their signals to fine-tune translation initiation [118]. Finally, exosomes have emerged as important mediators of tumorigenesis which facilitate cell-to-cell cross-talk distantly from the primary tumor, preparing metastasis. Exosomes are small circulating vesicles which carry exosomal-specific DNA, mRNAs, tumor neo-antigen peptides and miRNAs. They are secreted from most human cell types, including melanoma cells, and the surrounding cells of the tumor microenvironment. In melanoma, exosomes secretion is regulated by the Ras-related Rab27a protein and they carry as cargo the oncoprotein MET to educate bone marrow progenitor cells towards a pro-metastatic phenotype [119].
Translation regulation in skin tissue homeostasis & neoplastic transformation
The ability of skin tissue to maintain homeostasis resides mainly in the stem cells of epidermis [120]. For example, in the case of an injured epidermis layer, tissue damage is repaired by surrounding epidermal cells that migrate in and proliferate [121]. During this process, a new self-renewing epidermal stem cells population is established and adjusts its numbers to fit the available niche and maintain its stem cell character. This is highly dependent on the contact with the basal membrane, which preserves stem cell potential [74]. Loss of contact for any reason is transmitted through the major signaling pathways that target and accelerate translation or promote selective translation which triggers terminal differentiation [122]. Moreover, recent studies showed that epidermis is compartmentalized during homeostasis and the autonomous compartments are maintained by distinct stem cell populations, which during wound healing acquire lineage plasticity and contribute to tissue regeneration, suggesting the existence of mechanisms which require fine-tuned metabolic adaptations [123].
The dynamic equilibrium between stem cell self-renewal and stem cell differentiation reflects on gene expression levels and activation or suppression of the components that modulate translation initiation and protein synthesis rates [58]. Self-renewal without differentiation of at least some of the cells leads to tumorigenesis, while the opposite results (on a long term) in decreased skin regeneration ability due to insufficient stem cell population [75]. Epidermal stem cells exhibit low rate of division and protein synthesis which accelerate during differentiation. Terminal differentiation is an example of these dynamic adaptations of the translation machinery and involves orchestrated expression, alternative mRNA splicing and translation of different types of keratins which mark each cell status [124,125].
Protein synthesis in stem cells is regulated mainly through the mTOR complex, an important target of PI3K/AKT pathway and sensor of diverse signals, including nutritional and oxidative stress [126]. In response, mTOR modulates translation directly or through ribosomal protein S6K1, thus affecting the levels of translation initiation [127]. In addition, mTOR affects transcription of RNAP I (via TIF-1A) and III (via Maf1) which are responsible for rRNA and tRNA transcription [128,129]. Many tissues, including skin, depend on mTOR signaling to regulate stem cell self-renewal, proliferation and tumor suppression [130]. Recently, both mTORC1 (with its associated regulatory protein RAPTOR) and mTORC2 complexes (with RICTOR) were found essential for skin morphogenesis and epidermal barrier formation in mice, a finding which underlines the essential role of translation regulation. Downstream phosphorylation of S6K, RPS6 and 4E-BP1 in mTOR-deficient mice was noticeably decreased in the epidermis. Interestingly, each complex was found dedicated to the control of different cellular processes, with mTORC1 being involved in the regulation of early phase epidermal differentiation and stratification (including keratinocyte proliferation and hair follicle formation), while mTORC2 acting at a later stage, controlling interfollicular epidermis stratification, barrier formation and asymmetric cell division [131].
Ribosomes play a central role in translation and their biogenesis is a well-orchestrated and a fine-tuned process and ribosome inactivating proteins can target melanoma cells both in vitro and iv vivo [132]. Heterogenous ribosomes exhibit selectivity for distinct pools of mRNAs and under specific conditions may have different protein composition that could explain differential translation [133]. In addition, rRNA is modified by several noncoding RNAs (mostly snoRNAs) and enzymes, which contribute further to the complexity of ribosome content [23].
Although the contribution of components from MAPK and PI3K pathways on RNAP I transcription has been extensively studied in many types of cancer, only few studies exist regarding skin cancer [6,134]. Both signaling pathways are coupled through UBF, TBP and TIF-IA and the overactivation of the MAPK pathway, which is observed in melanoma, could lead in enhanced rRNA transcription [135,136]. Similarly, PI3K/AKT/mTOR pathway activation, which in many melanoma cases co-exists with PTEN loss, was found to enhance RNAP I transcription through stabilization of the SL1-mediated pre-initiation transcription complex, resulting in elevated rRNA transcription and ribosomal ribonucleoprotein subunits assembly [137]. Only few studies exist regarding the involvement of small (RPSs) or large (RPLs) ribosomal subunit proteins in melanoma. A screening of all RPLs showed variable effect in melanoma cells, with many of them found to facilitate survival. Knockdown of RPL13 levels caused increased p53 stability, through binding of RPL5 and RPL11 to MDM2. This interaction prevented p53 degradation, induced p53-dependent cell cycle arrest and decreased translation rates [138]. Similarly, knockdown of RPS3 suppressed cell growth and induced apoptosis. RPS3 was found highly expressed in melanoma cell lines and melanoma tumor tissues and its overexpression is associated with poor prognosis of melanoma patients. In addition, RPS3 knockdown triggered apoptosis through the release of cytochrome c and cleavage of pro-apoptotic proteins (PARP, caspase-3 and -9) resulting in tumor growth inhibition in a melanoma xenograft mouse model [139]. Finally, mutations in Rps19 and Rps20 in mice causing epidermal melanocytosis, were associated with p53 stabilization and stimulation of stem cell factor expression (SCF or KIT ligand), an important cytokine that plays role in melanogenesis [140].
Protein synthesis in every cell type is a constant process, initiated through interactions of translation initiation factors, ribosomal subunits and uninterrupted delivery of aminoacyl-tRNAs for amino acid incorporation. The majority of translation in eukaryotic cells (∼90%) is cap-dependent, requiring recognition of the 5′ 7-methyl-guanosine of mRNAs and spatial and temporal assembly of the initiation complex that recruits the small and the large ribosomal subunits to form functional ribosomes [141]. A small fraction (∼10%) of the mRNAs however initiate cap-independent translation and, especially under stress conditions, interactions with 5′cap are diminished. Instead, internal ribosome entry sites (IRES) serve for binding of a subset of translation initiation factors, which are recruited by IRES-trans acting factors and attract ribosomal subunits binding and ribosome assembly [142]. The switch between cap-dependent and IRES-mediated translation is, in many cases, crucial for the cellular decisions and fate and is profoundly affected by major transcription factors such as MYC and perturbations affecting this switch can lead to false mitotic progression, increased chromosome instability and cancer [24,143].
Translation initiation is considered the rate-limiting step of the overall protein synthesis and many key-factors participating in this process are found deregulated in cancer, including skin cancer [4,144]. Many of them are targeted by either the MAPK or the PI3K/AKT/mTOR signaling pathways or both and in melanoma, some of them are used as prognostic markers for the survival of patients (Figure 3) [145]. A detailed list of the translation initiation factors that contribute to skin cancer are presented in Table 1. Cap-dependent translation initiation requires the recognition of the 5′ cap by eIF4E, which is attached to the scaffold protein eIF4G. Attached to eIF4G is also eIF4A, an ATP-dependent helicase of the DEAD box family, which unwinds perplexed RNA secondary structures and facilitates translation. Together, they constitute the eIF4F complex, which is further stabilized by poly(A)-binding protein (PABP) which binds both the 3′ poly(A) tail of mRNA and eIF4G [141]. A nodal initiation factor is eIF4B, which binds on eIF4G to activate eIF4A. These interactions are necessary to circularize the mRNA and the gradual formation of the ternary complex is committed to the recruitment of 40S ribosomal subunit along with eIF2, an essential heterotrimeric GTP-binding protein (comprised of α-, β- and γ- subunits) which carries the first initiator Met-tRNAi on the P-site [128] (Figure 3). The activity of eIF2 depends on recycling of GTP to GDP and is regulated by eIF2B. Only the GTP-bound form of eIF2 is active and allows the 40S ribosomal subunit, loaded with the eIF1, eIF1A and eIF3 factors (also known as the 43S pre-initiation complex; PIC), to initiate translation (Figure 3) [146]. Both eIF1 and eIF1A are important for the fidelity of recognition of the first codon and several mutations in eIF1A are associated with uveal melanoma, while specific residues were recently implicated in stabilization of pre-initiation complexes [147–149]. Phosphorylation of eIF2 is critical for translation repression and reprogramming and is regulated by the opposing activities of the eIF2 kinases (PKR, PERK, GCN2) and the eIF2 phosphatase GADD34 [150,151]. Subsequent phosphorylation of eIF2 on serine 51 of the α subunit converts eIF2 to a competitive inhibitor of eIF2B, while GADD34 waives its phosphorylation (Figure 3). These activities are essential in early stress response and possibly contribute in translation rewiring by reducing protein synthesis to facilitate cell survival or at a later stage to regain and sustain higher protein synthesis rates [152]. In agreement with the above, it has been reported that translational repression, which represents an immediate response to stress, protects human keratinocytes from UVB-induced apoptosis through phosphorylation of eIF2α [153,154]. During differentiation of human keratinocytes the translation of specific mRNAs is regulated by GCN2 and microarray analysis has shown differential expression of eIF2γ subunit that could discriminate melanoma from other malignant and nonmalignant specimens [155,156]. The essential role of eIF2α-eIF2B control through repetitive phosphorylation‐dephosphorylation cycles was recently exemplified in an important report suggesting that translation reprogramming in response to starvation is mediated by eIF2B and can drive, under specific conditions, the phenotypic plasticity and therapeutic resistance which is observed in many melanoma cases. Microenvironmental stress signals can inhibit eIF2B, resulting in transcriptional repression of MITF via ATF4, which is a key transcription factor and activator of the integrated stress response (ISR) implicated in drug resistance. Given that MITF levels are critical for maintaining normal differentiation and low MITF levels are correlated with resistance to MEK and BRAF inhibitors, it was observed that translational reprogramming dramatically enhances tumorigenesis and is linked to a previously unexplained gene expression reprogramming associated with anti-PD-1 immunotherapy resistance [59].
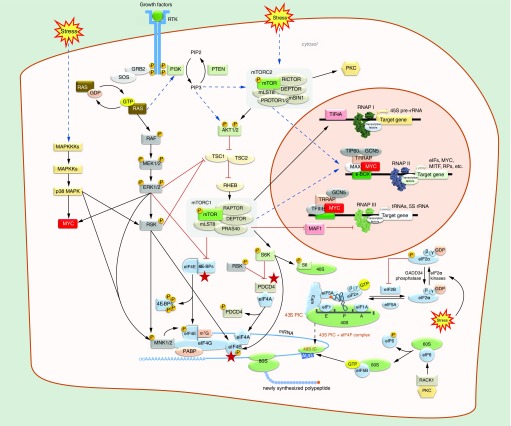
Figure 3. . Signal transduction pathways that affect the cap-dependent translation initiation.
Dimerization of receptor tyrosine kinases (RTKs) by mitogenic signals activate RAS GTPases, which act as molecular switches that transduce extracellular signals through the RAF/MEK/ERK and PI3K/AKT/mTOR signaling pathways to accordingly regulate translation at the initiation step. Kinases ERK1/2 and the stress-induced p38 MAPK stimulate cap-dependent translation by activating directly or through p90RSK (RSK) the eIF4G-associated kinases MNK1 and MNK2, which phosphorylate the cap-binding factor eIF4E on Ser209. Both ERK1/2 and AKT1/2 activate indirectly mTORC1 which phosphorylates the 4E-binding proteins (4E-BPs) at multiple sites and causes their dissociation from eIF4E. 4E-BPs can also be phosphorylated by ERK1/2. Activated mTORC1 also activates via phosphorylation the p70S6K (S6K), a kinase that affects translation by phosphorylating, among others, the ribosome protein S6. S6K and RSK also phosphorylate PDCD4, leading to release of eIF4A, and eIF4B, increasing its association and enhancing the helicase activity of eIF4A. Stress signals also induce the eIF2α kinases which phosphorylate the eIF2α subunit thus inhibiting translation initiation by preventing eIF2-bound GTP recycling by eIF2B. Stress-induced activation of mTORC2 complex leads to activation of PKC, which phosphorylates eIF6, leading to its release from free 60S subunits, which then become available for 80S ribosome assembly by eIF5B. Translation is also regulated at the transcriptional level. mTORC1/2- and c-MYC-mediated regulation of all three RNA polymerases leads to reprogramming of protein synthesis via modulation of ribosome biogenesis, expression of initiation factors and several oncoproteins, including MYC and MITF, and tRNA transcription. Arrows indicate activation and bar-headed lines indicate inhibition. Indirect regulation is indicated by dashed arrows. Stars show convergence of signaling pathways.
Table 1. . Deregulation of eukaryotic initiation factors in skin cancer.
| Factor | Deregulation in skin cancer | Ref. |
|---|---|---|
| eIF1 | N/A | N/A |
| eIF1A | Mutations associated with metastasis in uveal melanoma | [147–149] |
| eIF2α | Inhibition via phosphorylation protects from transformation | [150,153–155] |
| eIF2β | N/A | N/A |
| eIF2γ | Transiently upregulated | [156] |
| eIF2B | Inhibition by p-eIF2α | [59] |
| eIF3a | Increased expression | [157] |
| eIF3b | N/A | N/A |
| eIF3c | Decreased expression | [158] |
| eIF3d | KD suppresses proliferation | [159] |
| eIF3e | N/A | N/A |
| eIF3f | Decreased expression | [160,161] |
| eIF3g | N/A | N/A |
| eIF3h | N/A | N/A |
| eIF3i | Increased expression | [162] |
| eIF3j | N/A | N/A |
| eIF3k | N/A | N/A |
| eIF3l | N/A | N/A |
| eIF3m | N/A | N/A |
| eIF4A1 | Increased expression | [163] |
| PDCD4 | Decreased expression - Inhibition via phosphorylation | [164–167] |
| eIF4B | Enhancement via phosphorylation | [168,169] |
| eIF4E | Increased expression & phosphorylation | [170–174] |
| 4E-BPs | Hyperphosphorylation | [175–178] |
| eIF4G | N/A | N/A |
| eIF4F complex | Persistent formation leads to resistance to treatment | [178] |
| eIF5A1 | Inhibition impairs melanoma growth | [179] |
| eIF5A2 | Increased expression | [180] |
| eIF5B | N/A | N/A |
| eIF6 | Increased expression | [181] |
Binding of eIF3, an important mediator protein of 13 subunits, to eIF4G scaffold completes the translation initiation binding events. Subsequently, the 40S ribosomal subunit scans the mRNA until an AUG start codon is recognized and enters the P-site to base-pair with the tRNAi Met, thus forming the 48S initiation complex [128]. The events are promoted by the scaffold protein RAPTOR of the mTORC1 complex, which phosphorylates the 4E-BP1 and the ribosomal S6K1 which is associated with eIF3. The phosphorylated 4E-BP1 dissociates from eIF4E and cannot longer serve as translational repressor. At the same time, S6K1 targets with phosphorylation S6 protein, a constituent of the 40S subunit and eIF4B which associates with eIF3 and indirectly enhances the helicase activity of eIF4A [182].
The role of the multiple eIF3 subunits have been studied in melanoma and EIF3F loss of heterozygosity (LOH) was found in 75% to 92% of melanomas [160]. The eIF3a (p170) and eIF3i (Trip-1) subunits are overexpressed in melanoma cell lines and the eIF3c subunit (p110) exhibits decreased expression during melanoma progression, while knockdown of eIF3d suppresses proliferation of human melanoma cells [157–159,162]. In addition, downregulation of eIF3c in mouse melanoma cells causes cell cycle arrest, reduced cell proliferation and cell death [183]. The eIF3f subunit is considered a negative regulator of translation and is decreased in many melanomas, while its overexpression induces apoptosis and inhibits cell proliferation, likely by enhancing ribosome degradation [161]. Subsequent recruitment of eIF5 (also known as eIF5A or eIF5A1, exhibiting GTPase activity) triggers hydrolysis of GTP bound on eIF2, release of translation initiation factors from the 40S ribosomal subunits and finally eIF5B guides the 60S ribosomal subunit for assembly and formation of 80S elongating ribosome [141]. eIF5A is a ubiquitous protein and bears an unusual posttranslational modification on lysine 50 with the polyamine hydroxyputrescine, known as hypusine, which is required for functionality [184]. Interestingly, inhibition of hypusination has been shown to impair melanoma growth [179]. A recent study revealed that eIF5A2, a paralogue of eIF5A, which is present only in testis, parts of the brain and in tumors, was found overexpressed after extensive screening of 459 melanocytic lesions and several melanoma cell lines [180]. In a parallel role, the anti-association factor eIF6 binds to 60S ribosomal subunit to prevent 80S assembly when necessary. Interestingly, this overlooked factor is transcribed after Ras activation of Notch-1 and eIF6 overexpression was recently reported to indirectly affect melanoma cells motility through regulation of cdc42 (cell division control protein 42) [181,185].
The evidence that global protein synthesis rates regulate almost all aspects of gene expression are compelling for both normal and cancer cells under various stages of differentiation and growth conditions, as a general mechanism that monitors cellular homeostasis. Over 2 decades ago, it was observed that eIF4E is present in limiting amounts in cells and overexpression of eIF4E caused the transformation of fibroblasts [186]. Furthermore, its overexpression can selectively alter the translation of cancer-related mRNA transcripts [187]. Of note, several mRNAs like those of cyclins, MYC and VEGF are considered eIF4E-sensitive, and contain highly structured or unusually long 5′ UTRs which require helicase activity from eIF4A, which is 20-fold higher when eIF4A is in complex with eIF4F, than in its free form [188]. Moreover, stimulation of eIF4F-mediated translation initiation is modulated by either directly altering the expression of phosphorylation of its components or through regulation of its formation and activity which is tightly controlled to maintain cellular and tissue homeostasis. Finally, the dynamic equilibrium between eIF4E and 4E-BP1 is important because hyperphosphorylated 4E-BP1 favors eIF4E stimulated cap-dependent translation, while 4E-BP1 hypophosphorylation stabilizes the binding to eIF4E and suppression of translation, thus favoring IRES-dependent translation initiation [189].
The MAPK pathway phosphorylates through ERK1/2 kinases, the MAPK-interacting serine/threonine kinases 1 and 2 (MNK1/2) as well as the p90 ribosomal protein S6 kinase (RSK; member of the AGC protein kinase family). Alternatively, MNKs can be also activated by the stress-induced JNK and p38 pathway that play important role in melanoma [190,191]. In all cases, the major target is eIF4E phosphorylation on serine 209 which has been associated with tumor progression and reduced survival in malignant melanoma [170,171]. Activation of RSK phosphorylates serine 422 of eIF4B which is considered an important point of convergence for the two major signaling pathways, since the same phosphorylation is also mediated by the mTOR-activated S6K1 [168,169,192]. Recent reports pinpoint the role of RSK1 as promoter of melanoma growth and proliferation and its constitutive activation leads to increased melanoma invasion [193]. Of note, RSK1 was found constitutively phosphorylated at serine 380 in nodular but not superficial spreading melanoma and the tumor suppressor PDCD4 which inhibits eIF4A upon binding, was recently identified as substrate of RSK in a phosphoproteomic analysis [164]. Interestingly, PDCD4 downregulation has been linked in many studies with melanoma progression [165–167]. Accordingly, the reported eIF4A1 overexpression in melanoma is associated with the levels of PDCD4 which can either promote or suppress cancer progression [163]. The kinases that mediate the MAPK signal downstream of ERK are considered important therapeutic targets and inhibition of MNK1/2 by carcosporamide in B16 melanoma pulmonary metastases or in KIT-mutant melanoma are effective in reducing cancer progression [194,195]. Finally, inhibition of eIF4A acts synergistically with MEK inhibitors against NRAS-mutant melanoma cell lines [196].
On the other hand, mTOR-mediated S6K1 phosphorylation targets S6 ribosomal protein and 4E-BPs [175]. 4E-BPs are considered translation inhibitors and are also activated by the MAPK pathway through phosphorylation of serine 65 by ERKs under ionizing irradiation (Figure 3) [197]. The fact that 4E-BP1 phosphorylation is hierarchical and the suggestion that additional, but elusive so far, kinases might be involved, could account for the potency level of translation initiation regulation under different conditions [198]. The two pathways phosphorylate 4E-BPs in different residues and overall, affect the availability of eIF4E for 5′ cap binding [176]. They are also considered as the second important point of convergence for the two pathways in addition to eIF4B. Subsequently, phosphorylation cross-talk between 4E-BPs and eIF4E regulate excessive levels of translation initiation and eIF4F complex assembly. Accumulating evidence suggest that the levels of 4E-BPs phosphorylation, the expression levels of eIF4E and their ratio in their complex drives resistance against inhibitors that target both signaling pathways, while phosphorylated 4E-BP1 has been associated with poor survival in melanoma [172,177,199,200]. Recent reports have shown that this is the case for the natural polyphenol Rottlerin which in SK-Mel-28 melanoma cells inhibits mTORC1 and 4E-BP1 [201].
Although early studies have established the role of eIF4E deregulation in cancer, its contribution in skin cancer was, until recently, unclear and not well studied. In multiple melanoma cell lines, the activation of the MAPK pathway downregulates miR-768-3p leading to eIF4E upregulation and increased protein synthesis [202]. Recently, persistent formation of the eIF4F complex was associated with resistance to anti-BRAF, anti-MEK and combination therapies in melanoma [178]. Furthermore, expression of BRAFV600E in melanocytes increased eIF4E phosphorylation and protein synthesis [203]. DNA damage, oncogene induced senescence, levels of oxygen, nutritional or energetic stress (through impairment in mitochondria), drug resistance (including eIF4E-mediated resistance to irradiation) and metastasis are related to deregulation of eIF4E. The deregulation is linked to enhanced transcription, phosphorylation of eIF4E at serine 209 by the eIF4G-associated MNK1 and MNK2 kinases and through mTORC1-dependent phosphorylation and inactivation of the 4E-BPs. Recent reports showed that increased expression of eIF4E is observed in several melanoma cell lines and proliferation rates were significantly restrained after knockdown of eIF4E. The results were correlated with decreased expression and activity of oncogenes such as c-MYC and BCL2, suggesting that eIF4E is a prognostic marker of melanoma patient survival [170,173]. An independent study proposed that in BRAFV600E melanoma cell lines, vemurafenib inhibits 4E-BP1 phosphorylation and promotes eIF4E-4E-BP1 binding that prevents translation initiation [174]. Moreover, a study of surgical specimens from 114 patients and immunohistochemistry analysis showed that phosphorylation of eIF4E on serine 209 is essential and associated with metastatic potential, reduced survival and increased risk of death, suggesting that eIF4E can be used as a prognostic marker in melanoma [171].
Finally, in skin cancer and especially in melanoma, ROS induce rewiring of translation and dynamic metabolic changes [112]. For example, oxidative stress can inhibit distant metastasis by human melanoma cells, however, few cells can escape and successfully metastasize by increasing their dependence on NADPH-generating enzymes [204]. Therefore, the metastatic potential of melanoma, which in a mouse model is promoted by cooperation of BRAFV600E with PTEN loss and disrupted by mTORC1 or MEK1/2 inhibitors, can be increased after administration of antioxidants [205,206]. These reports underline the important role of translational regulation on metabolic homeostasis in response to various stress signals. In agreement, a very recent and elegant study identified in mice the ribosome rescue factor Pelota (Pelo), which is expressed in skin dermis and epidermis, as an evolutionary conserved mechanism which besides rescuing active translation from stalled ribosomes, it also maintains epidermal homeostasis via mTOR. Interestingly, deletion of Pelo from dermis resulted in mice with no obvious dermal abnormalities. However, when the same deletion was performed in epidermal cells, the resulted mice exhibited severe epidermal defects. Deletion of Pelo in cultures of a mixed population of human epidermal keratinocytes expressing keratin 14 and epidermal stem cells resulted in mice with scaly skin, epidermal thickening, increased water loss and loss of some hair follicle stem cells. The same defects could be also observed when Pelo loss was induced in adult mice. Surprisingly, the result of Pelo loss in epidermis was an increase in global translation efficiency in intrafollicular epidermal cells and bioinformatics analysis showed notably translation efficiency for keratins and ribosomal proteins and upregulation of mTOR, which can sense ribosome stalling [207]. The results of this study indicate that skin tissue homeostasis relies on translational regulation and suggest that the mTOR pathway serves as sensor of impaired ribosomes and counteracts as stimulator of translation initiation and elongation factors, acting either directly by mediating stimulation of differentiation in the basal membrane or indirectly, by promoting proliferation [208].
The emerging role of tRNA biology & tRFs in skin cancer
Since their discovery, tRNAs have been considered as passive carriers of amino acids during translation and their putative regulatory role had been overlooked, until recently [2,209]. Nowadays, tRNAs represent a dynamic population of essential noncoding RNAs with diverse regulatory roles and profound impact on human health [31].
In all cell types, tRNAs are abundant, representing up to the 15% of the total RNA content. It is therefore not surprising that RNAP III, which is responsible for transcription of more than 600 tRNA genes in human, accomplishes approximately 20% of the overall cellular transcription [33,210]. Transcription by RNAP III is guided by a small number of transcription factors which form initiation complexes onto three major promoter types [211,212]. Besides tRNAs and 5S rRNAs, RNAP transcribes several important noncoding RNAs such as miRNAs, U6 RNA, 7SL, 7SK, Y RNA, vault RNA, RNase P (H1) and RNase MRP RNA subunits, affecting collectively the expression of more than 1000 genes in human and mouse [213,214]. Interestingly, many recent studies revealed differences of RNAP III binding among eukaryotes and, in human cells, a large fraction of RNAP III genes are permanently repressed (reviewed in [30]). The recently published crystal structure of RNAP III revealed a complex of 17 subunits (compared with 14 and 12 for RNA pol I and II respectively), implying that regulation of RNAP III can occur through multiple protein-protein interactions [215]. For example, BRF2, a core RNAP III transcription factor, can directly sense redox signaling and couples cellular responses to oxidative stress with RNAP III output. BRF2 knockdown in A549 lung adenocarcinoma cells resulted in diminished levels of SecCys tRNAs and consequently, low levels of the TrXR1 and Gpx2 selenoproteins [216]. The observation that BRF2 has redox-sensing properties and contributes to the ability of cells to escape apoptosis induced by ROS could be also explored regarding the skin cancer etiology, since a bioinformatics analysis identified overexpression of BRF2 in melanoma, among several cancer types [217]. Moreover, MOAG-2 which promotes cytotoxicity in Huntington's disease by causing protein aggregation in the cytoplasm, was unexpectedly found to associate with RNAP III complex in the nucleus regulating transcription [218].
RNAP III is regulated similarly to translation initiation, by the opposing targeting of oncogenes and tumor suppressors (mTORC1, MYC, RAS, p53 and Rb) and enhanced transcription contributes to oncogenic transformation and cancer development [219]. In human, the transcription factor MAF1 is a central repressor of RNAP III and is considered as a tumor suppressor because it restricts transcription in response to nutrient deprivation, oxidative stress and DNA damage [220]. During transcription, mTOR directly associates with TFIIIC complex and waives tRNA and 5S rRNA gene MAF1-mediated transcription suppression [221]. Moreover, MAF1 has been identified as a new target of PTEN, an important tumor suppressor which inhibits signal transduction though the PI3K/mTOR axis and is absent in many BRAF-associated melanomas [222]. When MAF1 is nonfunctional, cells increase transcription by RNAP III resulting in enhanced energy waste and highlighting the key role of MAF1 in metabolic economy [223]. Finally, a recent study implicating RNAP III activity in age-related stem cell dysfunction exemplifies the role of RNAP III/mTORC1 interconnection on the regulation of translational machinery or through effects on organization of chromatin loci containing tRNA genes clusters [26,224].
Correct folding of tRNAs is facilitated by La protein (Lupus antigen, SSB) an important phosphoprotein chaperone, first identified as the major antigen in the serum of patients suffering from systemic lupus erythematosus [225]. La belongs to a broad and evolutionary conserved family of La-related proteins (LARPS) bearing the characteristic La motif (LAM), which facilitates folding of all RNAP III transcripts and has been found at genes transcribed by RNAP III, in vivo [226]. Although many of the LARP members are involved in several cancer types, the role of La in cancer is essentially unexplored with only few studies reporting that La overexpression affects regulation of MDM2-mediated p53 stability and degradation, which could be correlated to various skin cancer types [227]. Upon phosphorylation, La is localized in the nucleus and is associated with tRNAs, while dephosphorylation guides La to the cytoplasm where it associates with 5′-terminal oligopyrimidine (5′TOP) mRNAs, known to control protein synthesis [228]. The repertoire of La was recently expanded as modifier of Bcl2 mRNA structure around the translational start thus, stimulating translation of Bcl2 in cancer cells and as a gatekeeper of tRNA misfolding and processing through the miRNA biogenesis pathway [229,230]. The latter observation suggests cross-talk of tRFs and miRNA biogenesis pathways, with possible implication in translation regulation [231]. Finally, La guides removal of the 5′ leader by RNase P, an essential ribonucleoprotein with a catalytic RNA and ten protein subunits in human, as well as removal of 3′ trailer sequences by RNase ZL (ELAC2), from premature tRNA transcripts [232]. Interestingly, RNase P in human mitochondria is a protein-only holoenzyme of three subunits, representing an unusual RNase P type which is widespread among eukaryotes [233]. Individual RNase P protein subunits can interact with RNAP III components, can repress histone H3.3 recruitment and have a role in Double Strand Brake (DSB) repair via Homology Directed Repair (HDR) [234]. Similarly, ELAC2 is required not only for 3′ tRNA processing but also for the maintenance of C/D box snoRNAs, miRNAs and tRFs [235]. All these reports highlight a new dynamic regulatory network which requires detailed investigation, regarding coupling RNAP III transcription and translation regulation of key-proteins.
Mature tRNA biogenesis concludes with the addition of the universally conserved 3′ CCA end by the CCA tRNA nucleotidyl-transferase which has several functions in tRNA repair, protection, degradation and aminoacylation in stress response and excision of introns in the nucleus (in human) prior to export by the RAN GTPase exportin-t (XPOT), aminoacylation by aminoacyl-tRNA synthetases and delivery to the elongating ribosome by eEF1α [236,237]. In melanoma, lysyl-tRNA synthetase, a member of the mammalian multi-synthetase complex, translocate to the nucleus and binds to MITF to drive oncogenic transcriptional activation [34]. In addition, eEF1α has been reported as a molecular target of plant-derived isocarbostyrils in mouse and human melanoma cell cultures [238].
Modulation of RNAP III transcription can lead to alterations in the levels of specific tRNAs which could serve codon-usage demand for specific mRNAs. The reported overexpression of tRNAs in cancer verified this notion and raised questions on the role of tRNA levels in development and cancer [239,240]. Analyses of tRNA pools showed that there are marked differences in types and amounts of tRNAs on the genomic scale among cancerous and noncancerous cells, as well as among cells form different tissues [241,242]. The content on tRNAs corresponds to specific mRNA codon-usage signatures depending on the cell type and status thus providing a coordination between transcription and translation [32]. Interestingly, screening of cancer lines (including three melanoma lines; BLM, G361, LOX-IMVI) and stem cell lines showed that reactivation of TERT promoter driven by mutations or oncogenes results in increased TERT levels which, in turn, can directly associate with RNAP III subunit RPC32 to increase tRNA transcription [243]. The same study suggested that TERT promotes aberrant cell proliferation by augmenting expression of several tRNA species decoding for Ala, Arg, Asn, Lys, Glu and Cys. Moreover, overexpression of tRNAi Met can induce significant changes in tRNA expression profiles, suggesting that deregulation in translation initiation affects RNAP III transcription [244]. Similarly, tumor growth and angiogenesis were progressing faster in mice expressing additional copies of the tRNAi Met gene (termed 2+ tRNAi Met). Stromal fibroblasts overexpressing tRNAi Met could increase synthesis of specific collagen types (in particular type II) thus, contributing to tumor progression by enhancing the ability to synthesize and secrete a type II collagen extracellular matrix that supports endothelial cell migration and angiogenesis [245]. A similar study in mice suggested that tRNAi Met drives cell migration and invasion through modulation of α5β1 integrin and translation initiation levels leading to increased metastatic potential in melanoma without affecting cell proliferation and primary tumor growth. In addition, the expression of RNAP III associated genes are elevated in metastatic compared with primary tumors [246].
Advances in ribosome profiling made evident that codon bias serves as an auxiliary code that ensures the efficiency and fidelity of translation and, under various conditions, affects mRNA stability and shapes the proteome, allowing swift translation reprogramming to maintain homeostasis [40,247]. It has been proposed that synthesis of mutated proteins when cells are stressed or during adaptation to stressful environmental changes can be beneficial for cancer cells and can further drive tumorigenesis [248]. Therefore, adaptive translation has been proposed as a mechanism of response to various stress and environmental adaptation signals and in the case of cancer cells, translational errors can be tolerated and regulated. For example, rare codon bias affects Kras-driven tumorigenesis in mice [249,250]. Moreover, alteration of single tRNA levels can cause specific phenotypic alterations like in the case of breast cancer cells, where upregulation of tRNAArg CCG and tRNAGlu UUC promote metastatic progression by enhancing the stability and translation efficiency of mRNAs such as EXOSC2 and GRIPP1 [39]. Of note, loss of function of one of the tRNAArg UCU isodecoders specifically expressed in the mouse central nervous system is associated with increased ribosome stalling and leads to neurodegeneration in GTPBP2-deficient mice [251]. GTPBP2 is a binder to Pelota (Pelo) which as described previously is important for skin homeostasis, indicating that ribosome stalling induced either through Pelo or tRNA deficiency, share common regulatory mechanisms that affect translation regulation [207].
Maturation of tRNAs is considered complete, only when post-transcriptional modifications are introduced to specific tRNA positions. So far, numerous modifications are known to be crucial for tRNA structure, stability, recognition and function with role on severe pathological conditions including cancer [36,252]. For example, NSUN2 an important enzyme that introduces cytosine-5 methylation (m5C) in tRNA's wobble position 34, is a downstream target of MYC and responsible for proliferation and cell cycle progression of keratinocytes induced by MYC [253]. Lack of modifications can lead to rapid tRNA decay or fragmentation [54]. Moreover, it is known that oxidative stress induces several functional and structural changes to tRNAs and that specific modifications under these conditions can promote tRNA stability and protein synthesis [38,254]. Response to oxidative stress is also controlled by reprogramming of tRNA modifications and codon-biased selective translation of survival proteins [255]. Most recently, it was shown that tRNA modification of wobble U34 optimizes translation rates and maintains proteome integrity [256]. Finally, as mentioned above oxidative stress affects the integrity of mitochondrial DNA and induces mutations in mitochondrial tRNA genes. Recent attempts to identify somatic mtDNA mutations across 527 tumors and 14 cancer types revealed a selective pressure against deleterious coding mutations, supporting that functional mitochondria are required in tumor cells. In addition, a strong mutational strand bias was observed, compatible with endogenous replication-coupled errors as the major source of mutations. Interestingly, some mutations in tRNAs were caused by accumulation of unprocessed tRNA precursors due to incorrect tRNA folding [257]. It is well established that impaired mitochondrial tRNA biosynthesis has been linked to severe pathological syndromes. Moreover, it has been proposed that interaction of mitochondrial tRNAs with proteins ensures not only tRNA synthesis, maturation and function, but also protection from degradation. Therefore, critical mutations perturbing this interaction could lead to decreased tRNA stability [258].
During recent years, numerous quality control and surveillance pathways have been discovered and studied thoroughly [259]. Therefore, conformational changes in tRNA have been proposed as an early indicator of acute cellular damage and could be used as useful tool of prognosis [260]. The recent discovery of tRFs exemplified the regulatory role of tRNAs however, still baffles the field, since it seems that tRFs exist in cells under a normal lifecycle and even stress-induced tRFs (tiRNAs or tRNA halves) are present also under nonstress conditions [261]. Undoubtedly, the discovery of tRFs made evident that tRNA fragmentation either due to various stress signals or due to insufficient modifications is a first line of the cell's response to slow down translation. Stress-induced tRFs produced by ANG represent less than 5% of total tRNAs in a cell, and does not seem to affect essentially the overall tRNA levels [262]. Of note, selected 5′-tiRNAs (5′-tiRNAAla and 5′-tiRNACys) containing 5′-TOG motifs and assembling G-quadruplex-like structures can interfere with the assembly of the cap-binding eIF4F complex to inhibit translation initiation suggesting a broader role of tRFs in translation regulation [263]. On the cellular level, tiRNAs promote the assembly of cytoprotective stress granules (SGs) through binding to Y-Box protein 1 (YB-1 or YBX-1) which is highly overexpressed in many cancer types [264,265]. Interestingly, YB-1 is activated by the MAPK and the PI3K/AKT/mTOR pathways to promote melanoma cell proliferation [266]. Moreover, it was shown recently that YB-1 is overexpressed in melanoma and its nonphosphorylated cytoplasmic form enhances the migratory and invasive potential and promotes endothelial-to-mesenchymal transition (EMT) [267]. A previous report had also implicated tiRNA-mediated sequestration of YBX-1 from binding to pro-oncogenic transcripts as a mechanism of decreasing the hypoxia-stimulated highly metastatic potential of breast cancer cells, indicating a tumor-suppressor role for tiRNAs [268]. Interestingly, tRFs were found associated with Ago proteins 1, 3 and 4 but not 2 and recent bioinformatics analyses suggested that tRFs can mediate translation repression through a mechanism similar to miRNA-mediated interference [269]. In addition, tRF-3 can repress target genes in a Dicer-independent manner through complementarity to the 3′UTR of the target mRNAs and recruitment of an Ago-GW182 RISC [49]. In either case, the regulatory repertoire of tRFs is constantly expanding and tRFs have been reported to block long terminal repeats retrotransposons or to associate with human multisynthetase complex, to modulate translation and together with their tRNAs can affect decisions in stem or tumor cells (Figure 1) [270,271].
Although the relationship between translation regulation and stress response in controlling stem cells’ fate is unclear, recent breakthrough studies revealed a very delicate balance between tRNA modification, translation reprogramming and tRF production. Skin stem cells have low translation rates and their differentiation is regulated by the important NSUN2 methylase. The methylation activity is low in epidermal stem cells but is increased upon commitment to differentiation [272]. Deletion of NSUN2 delays differentiation of hair follicle stem cells in mice and triggers ANG-mediated tiRNA production, in an effort to slow down global translation [58]. In a most recent study, a similar effect was shown in human, where NSUN2 depletion triggered again tRNA cleavage into tRNA-halves to decrease protein synthesis levels. The abundance of proteins enriched in codons such as Gln, Glu and Lys, known to require thiolated tRNAs, were significantly increased in BRAFV600E-driven melanoma. The authors showed that H1F1A translation depends on specific tRNA modifications and affect HIF1α synthesis [60,273]. HIF1α is important for skin physiology and controls angiogenesis and tumorigenesis when increased in the hypoxic microenvironment of human epidermis [274]. Finally, translational control in stem cells is modulated by pseudouridylation of specific tRFs and stem cells are enriched with PUS7 Ψ synthase which governs protein synthesis and cell growth [46].
Putative roles of tRFs in keratinocytes
The discovery of tRFs has perplexed the epigenetic landscape of gene expression regulation and highlighted the regulatory role of tRNAs. However, although hundreds of tRFs sequences pile up in databases, their exact role in translation regulation and possibly in numerous other pathways remains elusive to a great extend [275,276]. In addition, although tRFs emerge as novel cancer biomarkers, information on skin-derived tRFs is restricted only in samples from skin cutaneous melanoma [277]. Therefore, the need for further studies regarding the existence and the expression levels and role of tRFs and tRNAs in various cell niches of human epidermis is imperative. Interestingly, the observation that few miRNAs are generated by pre-existing tRNAs has led to missannotation of some miRNAs which overlap with tRFs in miRbase [278].
To get novel information on skin-derived tRFs we provide herein, preliminary data showing the existence of tRFs in unstressed HaCaT cells, which represent immortalized keratinocytes resembling human epidermis. Although the scope of this review is to highlight the emerging role of various aspects of tRNA biology (including tRFs) in skin cancer, and its implications in translation regulation rather than the presentation of descriptive data, the detection of all tRFs types (including some of those that are classified as stress-derived) in unstressed cells suggests that all types of tRFs are constitutively produced in human keratinocytes (see detailed experimental description in Figure 4 legend). As mentioned above, keratinocytes represent the predominant cell type (90%) of epidermis and are subjected to various types of stress that could induce production of additional tRFs or alter their levels and types that can be detected. Moreover, tRFs in keratinocytes could represent a ‘stress surveillance’ mechanism that could help translation to adjust its rates depending on the conditions sensed. The results from the sequencing reads were mapped on the tRF database and revealed an uneven distribution of tRFs with tRF-3 being the most abundant group (Figure 4; raw and summarized data are available under the GEO repository accession number GSE119765). As mentioned previously tRF-5 are mainly located in the nucleus, while tRF-1 and tRF-3 are mainly located in the cytoplasm with the latter having the ability to suppress gene expression in a Dicer-independent and Ago-GW182RISC-dependent manner, through complementarity to the 3′UTR of the target mRNAs in a fashion similar to miRNAs [49]. Since, biogenesis of tRFs depends on cell type, condition or developmental stage, the specific tRF pattern presented in Table 2 could be considered representative of keratinocytes under normal conditions. The existence of tRFs in unstressed cells has been demonstrated for several cell lines, indicating that production of tRFs, including stress induced tRFs (i-tRFs), represents a global epigenetic mechanism which most likely couples tRNA transcription, modification and cleavage, with translation regulation. Interestingly, some of the most abundant tRFs identified have been previously identified as tRNA-derived miRNAs, an observation that verifies previous reports (Table 2) [278].
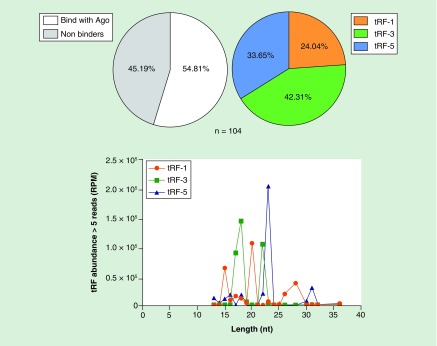
Figure 4. . Detection and distribution of tRF-1, tRF-3 and tRF-5 in human immortalized keratinocytes.
HaCaT cells (Thermo Fisher Scientific) were cultured under normal conditions in DMEM supplemented with 10% FBS, at 37°C under 5% CO2. Cells (∼2 × 106) were harvested and lysed immediately using the lysis buffer of the mirVANA isolation kit (Ambion) according to the manufacturer's instructions. Small RNAs (<200 nt) were isolated according to the manufacture's protocol and the size distribution quantity of the isolated RNAs was assessed using the RNA 6000 Nano Kit (Agilent) on an Agilent 2100 bioanalyzer. Subsequently, cDNA libraries were prepared using the Ion Total RNA-Seq Kit v2 protocol for small RNA sequencing according to the manufacturer's instructions. Templated ion sphere particles were generated using the Ion OneTouch 200 template kit v2. Sequencing was performed by semiconductor sequencing on the Ion Torrent PGM platform, using an Ion Torrent 318 chip and the Ion PGM 200 sequencing kit (Thermo Fisher Scientific). Annotated tRFs were quantified using an in-house bash script cucullated directly from the fastq files without mapping to the genome. Raw sequencing, summarized data and the current protocol are available in the GEO repository with accession number GSE119765. Out of 133 tRNA-derived fragments (tRFs) unique sequences annotated in the tRF database (http://genome.bioch.virginia.edu/trfdb/), we identified 104 (>5 reads). The detected tRFs sequences were compared with data from CLASH analyses (Crosslinking, Ligation and Sequencing of Hybrids) of AGO proteins as described in the text [269]. The analysis detects known tRFs that have been previously found bound on AGO proteins and could probably act through a mechanism similar to that of miRNAs. The analysis shows that more than half of the tRFs population (54.81%) that was detected in HaCaT cells has been previously reported to associate with AGO proteins. In addition, some of the HaCaT tRFs were found to overlap with known miRNAs as has been previously reported (Table 2) [278]. The most abundant group of detected tRFs were tRF-3 (42.31%) and length distribution analysis showed that their length varies in comparison with tRF-5. The existence of tRFs in keratinocytes and their possible roles are discussed in the text.
Table 2. . The 25 most abundant tRNA-derived fragments from HaCaT (>100 reads) are presented according to their sequence, ability to bind with Ago and similarity with known miRNAs.
| tRF ID | Type | Codon | Length | Sequence | Reads | Overlapping miRNAs | Bound with Ago | Predicted targets | Molecular function |
|---|---|---|---|---|---|---|---|---|---|
| 5030b | trf-5 | Glu GAG | 23 | TCCCTGGTGGTCTAGTGGTTAGG | 1593 | Yes | N/A | ||
| 1001 | trf-1 | Ser UCA | 20 | GAAGCGGGTGCTCTTATTTT | 810 | Yes | N/A | ||
| 3001a | trf-3 | Leu CUA, CUU | 18 | ATCCCACCGCTGCCACCA | 559 | miR-1260a | Yes | RPL4, USP9X, SAFB, E2F3, PITX1, UPF1, CDC37, PRDM10, HIST1H2AG, ZNF607, HIST3H2A, ENSA, AK2, HIST2H4B, PRPF31 | RNA binding, Structural constituent of ribosome, Receptor binding, Chaperone activity, DNA binding, Transcription factor activity |
| miR-1260b | |||||||||
| miR-4695-3p | |||||||||
| miR-6865-3p | |||||||||
| 3025b | trf-3 | Asp GAC | 22 | TCGATTCCCCGACGGGGAGCCA | 533 | Yes | N/A | ||
| 3003a | trf-3 | Cys UGC | 17 | TCCGGGTGCCCCCTCCA | 468 | Yes | PEPD, RPL23, PRPS2, MAZ, XBP1, COPA, C21orf91, SMEK1, ENO1, ETFB, SET, TAF15, RAB11A, LNPEP, PIBF1, ELL, TAF5, SLC35A3, DHX38, UBE2S, GPSM1, TRIAP1, MAP7D1, BIN3, TMSB10, NFYA, XPO4, GID4, GABARAPL1, MRPL39, PATL1, CAMKK2, TUBB, HNRNPR, PIK3R1, KLHDC10, PAX6, RNPS1, CIC, SNX25, DNMT3A, ZC3H13, RDH11, BAZ2A, SCAF11, MCC, SIPA1L2, LOC101930123 | GTPase activator activity, Phosphorylase activity, Enzyme inhibitor activity, DNA-methyltransferase activity, Transcription factor activity, RNA binding | |
| 1042 | trf-1 | Thr ACU | 28 | AACCGAGCGTCCAAGCTCTTTCCATTTT | 403 | Yes | N/A | ||
| 3014a | trf-3 | Ser UAC, UCU | 18 | ATCCTGCCGACTACGCCA | 389 | Yes | USP22 | A histone deubiquitinating component of the transcription regulatory histone acetylation (HAT) complex SAGA | |
| 5027b | trf-5 | Val GUG, GUU | 23 | GTTTCCGTAGTGTAGTGGTTATC | 362 | Yes | N/A | ||
| 3015b | trf-3 | Glu GAA | 22 | TCGACTCCCGGTGTGGGAACCA | 361 | Yes | N/A | ||
| 1037 | trf-1 | LeuTAG | 20 | ACCTCAGAAGGTCTCACTTT | 311 | miR-1285-3p | Yes | N/A | |
| 1030 | trf-1 | Ser UCU | 15 | GGAATGTCAGCTTTT | 298 | No | N/A | ||
| 1003 | trf-1 | Ser AGC | 26 | GCTAAGGAAGTCCTGTGCTCAGTTTT | 203 | Yes | N/A | ||
| 3020a | trf-3 | Ala GCA, GCU | 18 | TCCCCGGCACCTCCACCA | 194 | Yes | BCKDK, CNBP, ATP5B, TAF15, PA2G4, PRMT7, MINK1, FBL | Catalytic activity, Ubiquitin-specific protease activity, RNA binding, Protein serine/threonine kinase activity, Transcription regulator activity, Methyltransferase activity | |
| 3021a | trf-3 | Ala GCA, GCG | 18 | TCCCCGGCATCTCCACCA | 178 | Yes | RPS3, CSRP2, SLU7 | Involved in the repair of UV-induced DNA damage, development and cellular differentiation | |
| 5005c | trf-5 | Lys AAA | 31 | GCCCGGATAGCTCAGTCGGTAGAGCATCAGA | 162 | miR-6078 | Yes | N/A | |
| 5019a | trf-5 | Leu CUA, CUU | 16 | GGTAGCGTGGCCGAGC | 158 | Yes | N/A | ||
| 3009a | trf-3 | Leu CUU | 18 | ACCCCACTCCTGGTACCA | 157 | Yes | FECH, POLR2A, SERBP1, KDM5A, MICAL3, GHDC, GATSL1, DCTN5, SPHK2, CDKN1A, CHD4, TTC1, UBR5, FABP5, CTNNB1, MTAP, POLA2, SMARCB1, NIPSNAP1, PEA15, PDIA4, PCBP2, VTI1B, IPO7, AKAP13, RAB35, PTPN18, BAHD1, KCTD3, TRPM7, UBE2O, CCND1, DDOST, TGFBR1, ATXN2L, EIF3CL, GJC1, PTRF, ZNF790, KHSRP | DNA-directed DNA polymerase activity, Phosphorylase activity, Kinase binding, Transporter activity, Receptor signaling protein serine/threonine kinase activity, Ion channel activity | |
| 5004b | trf-5 | Gly GGC, GGG | 22 | GCATTGGTGGTTCAGTGGTAGA | 154 | Yes | N/A | ||
| 3025a | trf-3 | Asp GAC | 18 | TTCCCCGACGGGGAGCCA | 130 | Yes | RPL35A, SLC25A5, HCN2, ELP3, BTG1, PCBP2, ITPKC, RBM12B | Structural constituent of ribosome, Voltage-gated ion channel activity, Acyltransferase activity, Lipid kinase activity, ATP binding, Enzyme activator activity | |
| 3019a | trf-3 | Ala GCU | 18 | TCCCCAGTACCTCCACCA | 125 | Yes | GFI1, PRELID1, APMAP, GPATCH4 | Transription regulation activity | |
| 1004 | trf-1 | Asp GAC | 18 | GTGTGTAGCTGCACTTTT | 124 | No | N/A | ||
| 3008a | trf-3 | Val GUG, GUU | 17 | ACCGGGCGGAAACACCA | 115 | Yes | N/A | ||
| 1024 | trf-1 | Lys AAG | 15 | GCAACTGGTCGTTTT | 114 | Yes | N/A | ||
| 1026 | trf-1 | Ile AUU | 17 | TTCCGTGGGTTTGTTTT | 104 | No | N/A | ||
| 1041 | trf-1 | Gly GGU | 15 | TGCGGTACCACTTTT | 102 | No | N/A | ||
Each tRNA-derived fragment was corelated with mRNA targets using CLASH data meta-analysis and their predicted targets were also reported. Gene ontology enrichment analysis of the predicted gene targets shows putative participation in important molecular functions including ribosome structure and RNA binding, transcription regulator activity, DNA methyltransferase activity, phosphorylase activity, transcription factor activity and GTPase activity
MCC: Merkel-cell carcinoma.
In a further step, the list of the most abundant tRFs was filtered according to their ability to bind Ago proteins according to previous studies [269]. The predicted Ago binding tRFs represent more than half of the total tRFs identified (54.81% of total 104 detected tRFs and 134 total tRFs sequences in the tRF database). To get a picture of the putative targets of the AGO-binders tRFs, subsequent gene ontology enrichment analysis revealed putative target genes that are involved in the regulation of translation factors, ribosomal proteins and RNA binding proteins (Table 2). Although further and in-depth experimental verification is necessary to assess the role for some of the tRFs detected, the preliminary data presented herein coincide with previous reports and elaborate the complex epigenetic role of tRFs. Moreover, the analysis suggests that tRFs from keratinocytes may have the potential to play important role on transcriptional and translational regulation and may ultimately affect molecular mechanisms that define the fate of various types of skin cells under stress but also under normal conditions. The mechanisms by which they transmit their effect in each case remain under investigation and are expected to shed light in the expanding noncoding RNA world.
Conclusion & open questions
Skin is an elegant multilayer organ, ideal for studies of molecular networks that control stem cell renewal and differentiation under stress. Although modifications of protein synthesis patterns that occur in the epidermis following exposure to cellular stress have been known for more than 50 years, the exact underlying mechanisms and the contribution of tRNAs have only recently emerged [279]. Given the recent advances in analytical methodologies, a deep knowledge of the mechanisms that control the various skin cell niches could also apply to various other tissues that share common characteristics. Moreover, it can give answers regarding many molecular events that either maintain homeostasis or lead through deregulation to various diseases, including cancer. A major contribution in understanding skin malignant transformation has been provided by several recent studies on the role of central signaling pathways, related to the emerging prevalence of small noncoding RNAs, like miRNAs, as epigenetic regulators [280]. Nevertheless, the most important downstream targets that ultimately control the quality and quantity of the proteins produced for the cellular functions are the translation initiation factors and the tRNAs. In-depth studies on several components of translation regulation are either missing or are very limited and certainly require a closer attention (Table 1). Consequently, the events that affect tRNA transcription, maturation, modification and fragmentation are dynamic and converge in the regulation of transcription and translation to maintain skin tissue homeostasis and in the translational reprogramming which rewires important metabolic pathways in skin malignancies. More experimentation is needed to determine whether different tRNA pools may govern important differentiation processes such as keratinization and whether this is also strongly regulated through specific tRNA modifications, production of tRFs and finally altered translation rates of specific mRNAs.
Future perspective
The methodologies provided by combined ribosomal profiling and subsequent NGS and mass spectrometry analyses of active translation have already started to provide new information on novel RNAs or proteins that affect translation regulation, in a case-by-case basis. Moreover, the role of tRNA biology in melanoma has just begun to emerge suggesting that tRNAs and tRFs are important players in the regulation and/or the fine-tuning of expression of factors or pathways that are specifically expressed under either normal or stress conditions. Moreover, the presence of tRFs in human keratinocytes under normal conditions suggests possible roles in gene expression regulation, beyond stress response. The fact that keratinocytes are continuously exposed to various stress conditions suggests that, most likely, tRFs alter their levels and subsequently affect gene expression both at transcriptional and translational level, a notion that merits meticulous attention, in-depth investigation and experimental verification. Finally, the specific mechanism through which tRNA overexpression and tRFs transmit their effect, independently or in combination with other small or long noncoding regulatory RNAs in skin cancer, will provide a more comprehensive picture regarding the dynamic balance among various populations of noncoding RNAs in skin cancer, a better diagnostic value and possibly will pinpoint novel targets for specific and efficient anticancer therapy.
Executive summary.
Translation regulation, tRNAs & tRNA-derived fragments: a dynamic balance
Translation initiation is considered the rate-limiting step of the overall protein synthesis and many key-factors participating in this process are found deregulated in cancer, including skin cancer.
Translational rates reflect to the metabolic rewiring that occurs in response to various intra- or extracellular signals, through pathways which converge to targeting translation initiation and affect directly or indirectly the transcription of tRNAs and rRNAs.
Translation of specific mRNAs can occur as a mechanism of adaptation in response to stress and distinct tRNA expression patterns between different cell types.
tRNA-derived fragments represent a new class of small noncoding RNAs with role in the regulation of translation and possibly other nodal pathways and are found in both stressed and unstressed cells.
Differences in tRNA content and usage in response to various signals and conditions affect translational regulation and promote adaptive translation in a very dynamic way.
The complexity & plasticity of skin anatomy
Skin is the largest organ in the human body represents an effective barrier between the organism and the environment and provides the primary protection against several external dangers.
Skin consists of three major layers: the epidermis, the dermis and subcutaneous tissue (or hypodermis), each well-defined and with a content of certain differentiated cells and niches of pluripotent stem cells.
Skin stem cells of epidermis and other layers allow their continuous renewal, a characteristic of skin which makes it an ideal model for studying mechanisms of differentiation and tissue regeneration.
Molecular mechanisms involved in skin cancer
Skin cancer arises from a combination of genetic and environmental factors, with DNA damage caused by exposure to UV radiation being the major effector.
There are three major types of skin cancer: the basal-cell skin cancer (BCC), the cutaneous squamous-cell skin cancer (SCC) and, melanoma, the deadliest type of a severe malignancy deriving from melanocytes of the basal layer.
Skin cancer development can be affected by a combination of gene expression fluctuations and cell communication in the cells surrounding a tumorigenic lesion containing epithelial and mesenchymal cells.
Translation regulation in skin tissue homeostasis & neoplastic transformation
The dynamic equilibrium between stem cell self-renewal and stem cell differentiation reflects on gene expression levels and activation or suppression of the components that modulate translation initiation and protein synthesis rates.
Skin tissue homeostasis relies on translational regulation and suggest that the mTOR pathway serves as sensor of impaired ribosomes and counteracts as stimulator of translation initiation and elongation factors, acting either directly by mediating stimulation of differentiation in the basal membrane or indirectly, by promoting proliferation.
Deregulation of several key-factors involved in translation initiation have been reported in both melanoma and nonmelanoma skin cancers.
The emerging role of tRNA biology & tRNA-derived fragments in skin cancer
Distinct tRNA overexpression and/or tRNA modification patterns contribute to the dynamic regulation of translation in skin niches that contain different cell types.
UVR-induced DNA damage and oxidative stress impair tRNA integrity and decoding ability and induce production of specific tRNA-derived fragments.
In response, the presence of absence of various tRNA modifications contribute to tRNA stability and are involved in reprogramming of, and codon-biased selective translation.
Preliminary analysis of tRNA-derived fragments in keratinocytes suggests that tRNA-derived fragments possibly play role in the modulation of important molecular pathways.
Acknowledgments
The authors are indebted to our colleagues at the Departments of Biochemistry (RNA biology group) and Dermatology for helpful discussions. We apologize to those colleagues whose work was not cited due to space considerations. We thank D Chartoumpekis and I Habeos (School of Medicine, University of Patras) for the gift of HaCaT cells. I Skeparnias is supported by a PhD fellowship from the General Secretariat for Research and Technology (GSRT) and Hellenic Foundation for Research and Innovation (HFRI) and a Fulbright Doctoral Dissertation Visiting Research Student Fellowship (2018–2019). Constantinos Stathopoulos is a recipient of a Fulbright Visiting Research Scholar Fellowship (2018–2019). This work was supported in part by INSPIRED (MIS 5002550) which is implemented under the Action ‘Reinforcement of the Research and Innovation Infrastructure,’ funded by the Operational Program ‘Competitiveness, Entrepreneurship and Innovation’ (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund).
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Crick FH. On protein synthesis. Symp. Soc. Exp. Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- 2.Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamecnik PC. A soluble ribonucleic acid intermediate in protein synthesis. J. Biol. Chem. 1958;231(1):241–257. [PubMed] [Google Scholar]
- 3.Brenner S, Jacob F, Meselson M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature. 1961;190(4776):576–581. doi: 10.1038/190576a0. [DOI] [PubMed] [Google Scholar]
- 4.Hershey JWB, Sonenberg N, Mathews MB. Principles of translational control: an overview. Cold Spring Harb. Perspect. Biol. 2012;4(12) doi: 10.1101/cshperspect.a011528. pii: a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuller AP, Green R. Roadblocks and resolutions in eukaryotic translation. Nat. Rev. Mol. Cell Biol. 2018;19(8):526–541. doi: 10.1038/s41580-018-0011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tahmasebi S, Khoutorsky A, Mathews MB, Sonenberg N. Translation deregulation in human disease. Nat. Rev. Mol. Cell Biol. 2018;19(12):791–807. doi: 10.1038/s41580-018-0034-x. [DOI] [PubMed] [Google Scholar]
- 7.Roux PP, Topisirovic I. Signaling pathways involved in the regulation of mRNA translation. Mol. Cell. Biol. 2018;38(12) doi: 10.1128/MCB.00070-18. pii: e00070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buszczak M, Signer RAJ, Morrison SJ. Cellular differences in protein synthesis regulate tissue homeostasis. Cell. 2014;159(2):242–251. doi: 10.1016/j.cell.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol. Cell. 2010;40(2):228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 10.Gobet C, Naef F. Ribosome profiling and dynamic regulation of translation in mammals. Curr. Opin. Genet. Dev. 2017;43:120–127. doi: 10.1016/j.gde.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015;14(4):261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 12.Robichaud N, Sonenberg N. Translational control and the cancer cell response to stress. Curr. Opin. Cell Biol. 2017;45(Figure 1):102–109. doi: 10.1016/j.ceb.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer. 2016;16(5):288–304. doi: 10.1038/nrc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez CG, Teixeira FK, Czech B, et al. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell. 2016;18(2):276–290. doi: 10.1016/j.stem.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; • An important study showing that stem cell differentiation relies on pathways promoting global protein synthesis.
- 15.Bastide A, David A. The ribosome, (slow) beating heart of cancer (stem) cell. Oncogenesis. 2018;7(4) doi: 10.1038/s41389-018-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simsek D, Tiu GC, Flynn RA, et al. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell. 2017;169(6):1051–1065.e18. doi: 10.1016/j.cell.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The first extensive mammalian ribo-intercatome which revealed an unexpected diversity in the population of ribosomes.
- 17.Shi Z, Fujii K, Kovary KM, et al. Heterogeneous Ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell. 2017;67(1):71–83.e7. doi: 10.1016/j.molcel.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondrashov N, Pusic A, Stumpf CR, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145(3):383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guimaraes JC, Zavolan M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 2016;17(1):1–13. doi: 10.1186/s13059-016-1104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uemura M, Zheng Q, Koh CM, Nelson WG, Yegnasubramanian S, De Marzo AM. Overexpression of ribosomal RNA in prostate cancer is common but not linked to rDNA promoter hypomethylation. Oncogene. 2012;31(10):1254–1263. doi: 10.1038/onc.2011.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, Wang Y, Lv Q, et al. Overexpression of ribosomal RNA in the development of human cervical cancer is associated with rDNA promoter hypomethylation. PLoS ONE. 2016;11(10):e0163340. doi: 10.1371/journal.pone.0163340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruggero D, Grisendi S, Piazza F, et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science (80-.) 2003;299(5604):259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 23.Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DLJ, Bohnsack MT. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017;14(9):1138–1152. doi: 10.1080/15476286.2016.1259781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barna M, Pusic A, Zollo O, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456(7224):971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruggero D. Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 2013;4(2):a012336–a012336. doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arimbasseri AG, Maraia RJ. RNA polymerase III advances: structural and tRNA functional views. Trends Biochem. Sci. 2016;41(6):546–559. doi: 10.1016/j.tibs.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2017;19(1):45–58. doi: 10.1038/nrm.2017.77. [DOI] [PubMed] [Google Scholar]
- 28.Parisien M, Wang X, Pan T. Diversity of human tRNA genes from the 1000-genomes project. RNA Biol. 2013;10(12):1853–1867. doi: 10.4161/rna.27361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44(D1):D184–D189. doi: 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orioli A. tRNA biology in the omics era: stress signalling dynamics and cancer progression. BioEssays. 2017;39(3):1–11. doi: 10.1002/bies.201600158. [DOI] [PubMed] [Google Scholar]
- 31.Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015;16(2):98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 32.Gingold H, Tehler D, Christoffersen NR, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158(6):1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]; • The first detailed screening which revealed different patterns of tRNAs usage between proliferating and differentiating cells.
- 33.Grewal SS. Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochim. Biophys. Acta - Gene Regul. Mech. 2015;1849(7):898–907. doi: 10.1016/j.bbagrm.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, You S, Hwang D. Aminoacyl-tRNA synthetases and tumorigenesis: More than housekeeping. Nat. Rev. Cancer. 2011;11(10):708–718. doi: 10.1038/nrc3124. [DOI] [PubMed] [Google Scholar]
- 35.Huang SQ, Sun B, Xiong ZP, et al. The dysregulation of tRNAs and tRNA derivatives in cancer. J. Exp. Clin. Cancer Res. 2018;37(1):1–11. doi: 10.1186/s13046-018-0745-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014;20(6):306–314. doi: 10.1016/j.molmed.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Close P, Bose D, Chariot A, Leidel SA. Cancer and Noncoding RNAs. Elsevier; 2018. Chapter 10: Dynamic regulation of tRNA modifications in cancer [Internet] pp. 163–186. [Google Scholar]
- 38.Gu C, Begley TJ, Dedon PC. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014;588(23):4287–4296. doi: 10.1016/j.febslet.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016;165(6):1416–1427. doi: 10.1016/j.cell.2016.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanson G, Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2017;19(1):20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vallee BL, Riordan JF, Lobb RR, et al. Tumor-derived angiogenesis factors from rat Walker 256 carcinoma: an experimental investigation and review. Experientia. 1985;41(1):1–15. doi: 10.1007/BF02005853. [DOI] [PubMed] [Google Scholar]
- 42.Cole C, Sobala A, Lu C, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15(12):2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138(2):215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Pederson T. Regulatory RNAs derived from transfer RNA? RNA. 2010;16(10):1865–1869. doi: 10.1261/rna.2266510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588(23):4297–304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guzzi N, Cieśla M, Ngoc PCT, et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. 2018;173(5):1204–1216.e26. doi: 10.1016/j.cell.2018.03.008. [DOI] [PubMed] [Google Scholar]; • An important recent report linking tRNA-derived fragments (tRFs) modification and translational control in stem cells.
- 47.Sharma U, Conine CC, Shea JM, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351(6271):391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Q, Yan M, Cao Z, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 49.Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA. 2018;24(8):1093–1105. doi: 10.1261/rna.066126.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cristodero M, Polacek N. The multifaceted regulatory potential of tRNA-derived fragments. Non-Coding RNA Investig. 2017;1:7. [Google Scholar]
- 51.Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs) Trends Biochem. Sci. 2016;41(8):679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soares AR, Santos M. Discovery and function of transfer RNA-derived fragments and their role in disease. Wiley Interdiscip. Rev. RNA. 2017;8(5):1–13. doi: 10.1002/wrna.1423. [DOI] [PubMed] [Google Scholar]
- 53.Sun C, Fu Z, Wang S, et al. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 2018;414:16–25. doi: 10.1016/j.canlet.2017.10.031. [DOI] [PubMed] [Google Scholar]
- 54.Lyons SM, Fay MM, Ivanov P. The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett. 2018;592(17):2828–2844. doi: 10.1002/1873-3468.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14(10):2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamasaki S, Ivanov P, Hu G-F, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185(1):35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nawrot B, Sochacka E, Düchler M. tRNA structural and functional changes induced by oxidative stress. Cell. Mol. Life Sci. 2011;68(24):4023–4032. doi: 10.1007/s00018-011-0773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blanco S, Bandiera R, Popis M, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534(7607):335–340. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• An important study linking protein synthesis with stem cells fate during regeneration of a tissue or during tumorigenesis.
- 59.Falletta P, Sanchez-del-Campo L, Chauhan J, et al. Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Genes Dev. 2017;31(1):18–33. doi: 10.1101/gad.290940.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rapino F, Delaunay S, Rambow F, et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature. 2018;558(7711):605–609. doi: 10.1038/s41586-018-0243-7. [DOI] [PubMed] [Google Scholar]; •• Association between tRNA modification and expression and codon-specific translation reprogramming.
- 61.Ossio R, Roldán-Marín R, Martínez-Said H, Adams DJ, Robles-Espinoza CD. Melanoma: a global perspective. Nat. Rev. Cancer. 2017;17(7):393–394. doi: 10.1038/nrc.2017.43. [DOI] [PubMed] [Google Scholar]
- 62.Apalla Z, Nashan D, Weller RB, Castellsague X. Skin cancer: epidemiology, disease burden, pathophysiology, diagnosis, and therapeutic approaches. Dermatol. Ther. (Heidelb). 2017;7(Suppl. 1):5–19. doi: 10.1007/s13555-016-0165-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Michael HT, Merlino G. A topical solution to the sunless tanning problem. Trends Mol. Med. 2017;23(9):771–773. doi: 10.1016/j.molmed.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaidi MR, Day C-P, Merlino G. From UVs to metastases: modeling melanoma initiation and progression in the mouse. J. Invest. Dermatol. 2008;128(10):2381–2391. doi: 10.1038/jid.2008.177. [DOI] [PubMed] [Google Scholar]
- 65.Noonan FP, Recio JA, Takayama H, et al. Neonatal sunburn and melanoma in mice. Nature. 2001;413(6853):271–272. doi: 10.1038/35095108. [DOI] [PubMed] [Google Scholar]; •• Use of a genetically engineered mouse model showing that a single dose of burning ultraviolet radiation to neonates, but not adults, is sufficient to induce tumors that are reminiscent of human melanoma.
- 66.Zaidi MR, Davis S, Noonan FP, et al. Interferon-γ links ultraviolet radiation to melanomagenesis in mice. Nature. 2011;469(7331):548–553. doi: 10.1038/nature09666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Day C-P, Marchalik R, Merlino G, Michael H. Mouse models of UV-induced melanoma: genetics, pathology, and clinical relevance. Lab. Investig. 2017;97(6):698–705. doi: 10.1038/labinvest.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanitakis J. Anatomy, histology and immunohistochemistry of normal human skin. Eur. J. Dermatol. 2002;12(4):390–399. quiz 400–401. [PubMed] [Google Scholar]
- 69.Proksch E, Brandner JM, Jensen JM. The skin: an indispensable barrier. Exp. Dermatol. 2008;17(12):1063–1072. doi: 10.1111/j.1600-0625.2008.00786.x. [DOI] [PubMed] [Google Scholar]
- 70.Fuchs E, Horsley V. More than one way to skin. Genes Dev. 2008;22(8):976–985. doi: 10.1101/gad.1645908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clayton E, Doupé DP, Klein AM, Winton DJ, Simons BD, Jones PH. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446(7132):185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 72.Kolarsick PAJ, Kolarsick MA, Goodwin C. Anatomy and physiology of the skin. J. Dermatol. Nurses Assoc. 2011;3(4):203–213. [Google Scholar]
- 73.Suter MM, Schulze K, Bergman W, Welle M, Roosje P, Müller EJ. The keratinocyte in epidermal renewal and defence. Vet. Dermatol. 2009;20(5–6):515–532. doi: 10.1111/j.1365-3164.2009.00819.x. [DOI] [PubMed] [Google Scholar]
- 74.Fuchs E. Skin stem cells: rising to the surface. J. Cell Biol. 2008;180(2):273–284. doi: 10.1083/jcb.200708185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boehnke K, Falkowska-Hansen B, Stark HJ, Boukamp P. Stem cells of the human epidermis and their niche: composition and function in epidermal regeneration and carcinogenesis. Carcinogenesis. 2012;33(7):1247–1258. doi: 10.1093/carcin/bgs136. [DOI] [PubMed] [Google Scholar]
- 76.Hirsch T, Rothoeft T, Teig N, et al. Regeneration of the entire human epidermis using transgenic stem cells. Nature. 2017;551(7680):327–332. doi: 10.1038/nature24487. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Discovery of long-lived skin stem cells that produce progenitors.
- 77.Vapniarsky N, Arzi B, Hu JC, Nolta JA, Athanasiou KA. Concise review: human dermis as an autologous source of stem cells for tissue engineering and regenerative medicine. Stem Cells Transl. Med. 2015;4(10):1187–1198. doi: 10.5966/sctm.2015-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Toma JG, Akhavan M, Fernandes KJL, et al. Isolation of multipotent adult stem cells from the dermis of mammalian skin. Nat. Cell Biol. 2001;3(9):778–784. doi: 10.1038/ncb0901-778. [DOI] [PubMed] [Google Scholar]
- 79.Narayanan DL, Saladi RN, Fox JL. Ultraviolet radiation and skin cancer. Int. J. Dermatol. 2010;49(9):978–86. doi: 10.1111/j.1365-4632.2010.04474.x. [DOI] [PubMed] [Google Scholar]
- 80.Baudouin C, Charveron M, Tarroux R, Gall Y. Environmental pollutants and skin cancer. Cell Biol. Toxicol. 2002;18(5):341–348. doi: 10.1023/a:1019540316060. [DOI] [PubMed] [Google Scholar]
- 81.Gálvez JM, Castillo D, Herrera LJ, et al. Multiclass classification for skin cancer profiling based on the integration of heterogeneous gene expression series. PLoS ONE. 2018;13(5):e0196836. doi: 10.1371/journal.pone.0196836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nehal KS, Bichakjian CK. Update on keratinocyte carcinomas. N. Engl. J. Med. 2018;379(4):363–374. doi: 10.1056/NEJMra1708701. [DOI] [PubMed] [Google Scholar]
- 83.Harms PW, Harms KL, Moore PS, et al. The biology and treatment of Merkel cell carcinoma: current understanding and research priorities. Nat. Rev. Clin. Oncol. 2018;15(12):763–776. doi: 10.1038/s41571-018-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim IY, He YY. Ultraviolet radiation-induced non-melanoma skin cancer: regulation of DNA damage repair and inflammation. Genes Dis. 2014;1(2):188–198. doi: 10.1016/j.gendis.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Didona D, Paolino G, Bottoni U, Cantisani C. Non melanoma skin cancer pathogenesis overview. Biomedicines. 2018;6(1):6. doi: 10.3390/biomedicines6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feehan RP, Shantz LM. Molecular signaling cascades involved in nonmelanoma skin carcinogenesis. Biochem. J. 2016;473(19):2973–2994. doi: 10.1042/BCJ20160471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Watt FM, Estrach S, Ambler CA. Epidermal Notch signalling: differentiation, cancer and adhesion. Curr. Opin. Cell Biol. 2008;20(2):171–179. doi: 10.1016/j.ceb.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hu B, Castillo E, Harewood L, et al. Multifocal epithelial tumors and field cancerization from loss of mesenchymal CSL signaling. Cell. 2012;149(6):1207–1220. doi: 10.1016/j.cell.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu S-L, Lee S-K. Ultraviolet radiation: DNA damage, repair, and human disorders. Mol. Cell. Toxicol. 2017;13(1):21–28. [Google Scholar]
- 90.Lee C-H, Wu S-B, Hong C-H, Yu H-S, Wei Y-H. Molecular mechanisms of UV-induced apoptosis and its effects on skin residential cells: the implication in UV-based phototherapy. Int. J. Mol. Sci. 2013;14(3):6414–6435. doi: 10.3390/ijms14036414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maru GB, Gandhi K, Ramchandani A, Kumar G. The role of inflammation in skin cancer. Adv. Exp. Med. Biol. 2014;816:437–469. doi: 10.1007/978-3-0348-0837-8_17. [DOI] [PubMed] [Google Scholar]
- 92.Carpenter RL, Lo H-W. STAT3 target genes relevant to human cancers. Cancers (Basel) 2014;6(2):897–925. doi: 10.3390/cancers6020897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Watt FM, Frye M, Benitah SA. MYC in mammalian epidermis: how can an oncogene stimulate differentiation? Nat. Rev. Cancer. 2008;8(3):234–242. doi: 10.1038/nrc2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Briso EM, Guinea-Viniegra J, Bakiri L, et al. Inflammation-mediated skin tumorigenesis induced by epidermal c-Fos. Genes Dev. 2013;27(18):1959–1973. doi: 10.1101/gad.223339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nys K, Agostinis P. Bcl-2 family members: essential players in skin cancer. Cancer Lett. 2012;320(1):1–13. doi: 10.1016/j.canlet.2012.01.031. [DOI] [PubMed] [Google Scholar]
- 96.Sander CS, Hamm F, Elsner P, Thiele JJ. Oxidative stress in malignant melanoma and non-melanoma skin cancer. Br. J. Dermatol. 2003;148(5):913–922. doi: 10.1046/j.1365-2133.2003.05303.x. [DOI] [PubMed] [Google Scholar]
- 97.Son Y, Kim S, Chung H-T, Pae H-O. Reactive oxygen species in the activation of MAP kinases [Internet] Methods Enzymol. 2013;528:27–48. doi: 10.1016/B978-0-12-405881-1.00002-1. [DOI] [PubMed] [Google Scholar]
- 98.Bulavin DV. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 1999;18(23):6845–6854. doi: 10.1093/emboj/18.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nys K, Van Laethem A, Michiels C, et al. A p38MAPK/HIF-1 pathway initiated by UVB irradiation is required to induce noxa and apoptosis of human keratinocytes. J. Invest. Dermatol. 2010;130(9):2269–2276. doi: 10.1038/jid.2010.93. [DOI] [PubMed] [Google Scholar]
- 100.Silvers AL, Bachelor MA, Bowden GT. The role of JNK and p38 MAPK activities in UVA-Induced signaling pathways leading to AP-1 activation and c-Fos expression. Neoplasia. 2003;5(4):319–329. doi: 10.1016/S1476-5586(03)80025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nat. Rev. Genet. 2002;3(3):199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- 102.Knezevic D, Zhang W, Rochette PJ, Brash DE. Bcl-2 is the target of a UV-inducible apoptosis switch and a node for UV signaling. Proc. Natl Acad. Sci. USA. 2007;104(27):11286–11291. doi: 10.1073/pnas.0701318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shain AH, Bastian BC. From melanocytes to melanomas. Nat. Rev. Cancer. 2016;16(6):345–358. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
- 104.Soura E, Eliades PJ, Shannon K, Stratigos AJ, Tsao H. Hereditary melanoma: update on syndromes and management. J. Am. Acad. Dermatol. 2016;74(3):395–407. doi: 10.1016/j.jaad.2015.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jakob JA, Bassett RL, Ng CS, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2012;118(16):4014–4023. doi: 10.1002/cncr.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 107.von Thaler AK, Kamenisch Y, Berneburg M. The role of ultraviolet radiation in melanomagenesis. Exp. Dermatol. 2010;19(2):81–88. doi: 10.1111/j.1600-0625.2009.01025.x. [DOI] [PubMed] [Google Scholar]
- 108.Hayward NK, Wilmott JS, Waddell N, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545(7653):175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 109.Wang Y, Radfar S, Liu S, Riker AI, Khong HT. Mitf-Mdel, a novel melanocyte/melanoma-specific isoform of microphthalmia-associated transcription factor-M, as a candidate biomarker for melanoma. BMC Med. 2010;8(1):14. doi: 10.1186/1741-7015-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ennen M, Keime C, Gambi G, et al. MITF -High and MITF -low cells and a novel subpopulation expressing genes of both cell states contribute to intra- and intertumoral heterogeneity of primary melanoma. Clin. Cancer Res. 2017;23(22):7097–7107. doi: 10.1158/1078-0432.CCR-17-0010. [DOI] [PubMed] [Google Scholar]
- 111.Kawakami A, Fisher DE. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Investig. 2017;97(6):649–656. doi: 10.1038/labinvest.2017.9. [DOI] [PubMed] [Google Scholar]
- 112.Ratnikov BI, Scott DA, Osterman AL, Smith JW, Ronai ZA. Metabolic rewiring in melanoma. Oncogene. 2017;36(2):147–157. doi: 10.1038/onc.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leucci E, Vendramin R, Spinazzi M, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531(7595):518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 114.Fogal V, Zhang L, Krajewski S, Ruoslahti E. Mitochondrial/cell-surface protein p32/gC1qR as a molecular target in tumor cells and tumor stroma. Cancer Res. 2008;68(17):7210–7218. doi: 10.1158/0008-5472.CAN-07-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goding CR. Targeting the lncRNA SAMMSON reveals metabolic vulnerability in melanoma. Cancer Cell. 2016;29(5):619–621. doi: 10.1016/j.ccell.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 116.Haq R, Shoag J, Andreu-Perez P, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23(3):302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vendramin R, Verheyden Y, Ishikawa H, et al. SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat. Struct. Mol. Biol. 2018;25(11):1035–1046. doi: 10.1038/s41594-018-0143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Epigenetic regulation in human melanoma: past and future. Epigenetics. 2015;10(2):103–21. doi: 10.1080/15592294.2014.1003746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu. Rev. Cell Dev. Biol. 2006;22(1):339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li Y, Zhang J, Yue J, Gou X, Wu X. Epidermal stem cells in skin wound healing. Adv. Wound Care. 2017;6(9):297–307. doi: 10.1089/wound.2017.0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ojeh N, Pastar I, Tomic-Canic M, Stojadinovic O. Stem cells in skin regeneration, wound healing, and their clinical applications. Int. J. Mol. Sci. 2015;16(10):25476–25501. doi: 10.3390/ijms161025476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Page ME, Lombard P, Ng F, Göttgens B, Jensen KB. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell. 2013;13(4):471–482. doi: 10.1016/j.stem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim S, Wong P, Coulombe PA. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 2006;441(7091):362–365. doi: 10.1038/nature04659. [DOI] [PubMed] [Google Scholar]
- 125.Loschke F, Seltmann K, Bouameur J-E, Magin TM. Regulation of keratin network organization. Curr. Opin. Cell Biol. 2015;32:56–64. doi: 10.1016/j.ceb.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 126.Russell RC, Fang C, Guan K-L. An emerging role for TOR signaling in mammalian tissue and stem cell physiology. Development. 2011;138(16):3343–3356. doi: 10.1242/dev.058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009;10(5):307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 128.Blagden SP, Willis AE. The biological and therapeutic relevance of mRNA translation in cancer. Nat. Rev. Clin. Oncol. 2011;8(5):280–291. doi: 10.1038/nrclinonc.2011.16. [DOI] [PubMed] [Google Scholar]
- 129.Gentilella A, Kozma SC, Thomas G. A liaison between mTOR signaling, ribosome biogenesis and cancer. Biochim. Biophys. Acta. 2015;1849(7):812–820. doi: 10.1016/j.bbagrm.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Werzowa J, Koehrer S, Strommer S, et al. Vertical inhibition of the mTORC1/mTORC2/PI3K pathway shows synergistic effects against melanoma in vitro and in vivo . J. Invest. Dermatol. 2011;131(2):495–503. doi: 10.1038/jid.2010.327. [DOI] [PubMed] [Google Scholar]
- 131.Ding X, Bloch W, Iden S, et al. mTORC1 and mTORC2 regulate skin morphogenesis and epidermal barrier formation. Nat. Commun. 2016;7:13226. doi: 10.1038/ncomms13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cheung MC, Revers L, Perampalam S, et al. An evolved ribosome-inactivating protein targets and kills human melanoma cells in vitro and in vivo . Mol. Cancer. 2010;9(1):28. doi: 10.1186/1476-4598-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012;13(6):355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kardos GR, Robertson GP. Therapeutic interventions to disrupt the protein synthetic machinery in melanoma. Pigment Cell Melanoma Res. 2015;28(5):501–519. doi: 10.1111/pcmr.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stefanovsky VY, Pelletier G, Hannan R, Gagnon-Kugler T, Rothblum LI, Moss T. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell. 2001;8(5):1063–1073. doi: 10.1016/s1097-2765(01)00384-7. [DOI] [PubMed] [Google Scholar]
- 136.Zhao J, Yuan X, Frödin M, Grummt I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell. 2003;11(2):405–13. doi: 10.1016/s1097-2765(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 137.Zhang C, Comai L, Johnson DL. PTEN represses RNA polymerase I transcription by disrupting the SL1 complex. Mol. Cell. Biol. 2005;25(16):6899–6911. doi: 10.1128/MCB.25.16.6899-6911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kardos GR, Dai M-S, Robertson GP. Growth inhibitory effects of large subunit ribosomal proteins in melanoma. Pigment Cell Melanoma Res. 2014;27(5):801–812. doi: 10.1111/pcmr.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tian Y, Qin L, Qiu H, et al. RPS3 regulates melanoma cell growth and apoptosis by targeting Cyto C/Ca2+/MICU1 dependent mitochondrial signaling. Oncotarget. 2015;6(30):29614–29625. doi: 10.18632/oncotarget.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.McGowan KA, Li JZ, Park CY, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat. Genet. 2008;40(8):963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jackson RJ, Hellen CUTT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Walters B, Thompson SR. Cap-independent translational control of carcinogenesis. Front. Oncol. 2016;6:128. doi: 10.3389/fonc.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Malka-Mahieu H, Newman M, Désaubry L, Robert C, Vagner S. Molecular pathways: the eIF4F translation initiation complex – new opportunities for cancer treatment. Clin. Cancer Res. 2017;23(1):21–25. doi: 10.1158/1078-0432.CCR-14-2362. [DOI] [PubMed] [Google Scholar]
- 145.Ali MU, Ur Rahman MS, Jia Z, Jiang C. Eukaryotic translation initiation factors and cancer. Tumour Biol. 2017;39(6) doi: 10.1177/1010428317709805. 1010428317709805. [DOI] [PubMed] [Google Scholar]
- 146.Bogorad AM, Lin KY, Marintchev A. Novel mechanisms of eIF2B action and regulation by eIF2α phosphorylation. Nucleic Acids Res. 2017;45(20):11962–11979. doi: 10.1093/nar/gkx845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Martin M, Maßhöfer L, Temming P, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013;45(8):933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dono M, Angelini G, Cecconi M, et al. Mutation frequencies of GNAQ, GNA11, BAP1, SF3B1, EIF1AX and TERT in uveal melanoma: Detection of an activating mutation in the TERT gene promoter in a single case of uveal melanoma. Br. J. Cancer. 2014;110(4):1058–1065. doi: 10.1038/bjc.2013.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ewens KG, Kanetsky PA, Richards-Yutz J, et al. Chromosome 3 status combined with BAP1 and EIF1AX mutation profiles are associated with metastasis in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2014;55(8):5160–5167. doi: 10.1167/iovs.14-14550. [DOI] [PubMed] [Google Scholar]
- 150.Holcik M. Could the eIF2α-independent translation be the achilles heel of cancer? Front. Oncol. 2015;5:1–8. doi: 10.3389/fonc.2015.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Choy MS, Yusoff P, Lee IC, et al. Structural and functional analysis of the GADD34:PP1 eIF2α phosphatase. Cell Rep. 2015;11(12):1885–1891. doi: 10.1016/j.celrep.2015.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5(10):827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Collier AE, Wek RC, Spandau DF. Translational repression protects human keratinocytes from UVB-induced apoptosis through a discordant eIF2 Kinase stress response. J. Invest. Dermatol. 2015;135(10):2502–2511. doi: 10.1038/jid.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wengrod J, Wang D, Weiss S, Zhong H, Osman I, Gardner LB. Phosphorylation of eIF2α triggered by mTORC1 inhibition and PP6C activation is required for autophagy and is aberrant in PP6C-mutated melanoma. Sci. Signal. 2015;8(367):1–12. doi: 10.1126/scisignal.aaa0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Collier AE, Wek RC, Spandau DF. Human keratinocyte differentiation requires translational control by the eIF2α kinase GCN2. J. Invest. Dermatol. 2017;137(9):1924–1934. doi: 10.1016/j.jid.2017.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Becker B, Roesch A, Hafner C, et al. Discrimination of melanocytic tumors by cDNA array hybridization of tissues prepared by laser pressure catapulting. J. Invest. Dermatol. 2004;122(2):361–368. doi: 10.1046/j.0022-202X.2004.22240.x. [DOI] [PubMed] [Google Scholar]
- 157.Pincheira R, Chen Q, Zhang JT. Identification of a 170-kDa protein over-expressed in lung cancers. Br. J. Cancer. 2001;84(11):1520–1527. doi: 10.1054/bjoc.2001.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Baldi A, Battista T, De Luca A, et al. Identification of genes down-regulated during melanoma progression: a cDNA array study. Exp. Dermatol. 2003;12(2):213–218. doi: 10.1034/j.1600-0625.2003.00026.x. [DOI] [PubMed] [Google Scholar]
- 159.Li H, Zhou F, Wang H, et al. Knockdown of EIF3D suppresses proliferation of human melanoma cells through G2/M phase arrest. Biotechnol. Appl. Biochem. 2015;62(5):615–620. doi: 10.1002/bab.1305. [DOI] [PubMed] [Google Scholar]
- 160.Doldan A, Chandramouli A, Shanas R, et al. Loss of the eukaryotic initiation factor 3f in melanoma. Mol. Carcinog. 2008;47(10):806–813. doi: 10.1002/mc.20436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shi J, Kahle A, Hershey JWB, et al. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene. 2006;25(35):4923–4936. doi: 10.1038/sj.onc.1209495. [DOI] [PubMed] [Google Scholar]
- 162.Bernardini S, Melino G, Saura F, et al. Expression of Co-factors (SMRT and Trip-1) for retinoic acid receptors in human neuroectodermal cell lines. Biochem. Biophys. Res. Commun. 1997;234(1):278–282. doi: 10.1006/bbrc.1997.6626. [DOI] [PubMed] [Google Scholar]
- 163.Eberle J, Krasagakis K, Orfanos CE. Translation initiation factor eIF-4A1 mRNA is consistently overexpressed in human melanoma cells in vitro . Int. J. Cancer. 1997;71(3):396–401. doi: 10.1002/(sici)1097-0215(19970502)71:3<396::aid-ijc16>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 164.Galan JA, Geraghty KM, Lavoie G, et al. Phosphoproteomic analysis identifies the tumor suppressor PDCD4 as a RSK substrate negatively regulated by 14-3-3. Proc. Natl Acad. Sci. 2014;111(29):e2918–e2927. doi: 10.1073/pnas.1405601111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vikhreva PN, Korobko IV. Expression of Pdcd4 tumor suppressor in human melanoma cells. Anticancer Res. 2014;34(5):2315–2318. [PubMed] [Google Scholar]
- 166.Wang D, Guo S, Han SY, Xu N, Guo JY, Sun Q. Distinct roles of different fragments of PDCD4 in regulating the metastatic behavior of B16 melanoma cells. Int. J. Oncol. 2013;42(5):1725–1733. doi: 10.3892/ijo.2013.1841. [DOI] [PubMed] [Google Scholar]
- 167.Wan J, Yang J, Huang Y, Deng L. MicroRNA-150 inhibitors enhance cell apoptosis of melanoma by targeting PDCD4. Oncol. Lett. 2018;15(2):1475–1482. doi: 10.3892/ol.2017.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shahbazian D, Roux PP, Mieulet V, et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25(12):2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Degen M, Barron P, Natarajan E, Widlund HR, Rheinwald JG. RSK activation of translation factor eIF4B drives abnormal increases of Laminin γ2 and MYC protein during neoplastic progression to squamous cell carcinoma. PLoS ONE. 2013;8(10):2–10. doi: 10.1371/journal.pone.0078979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Khosravi S, Tam KJ, Ardekani GS, Martinka M, McElwee KJ, Ong CJ. EIF4E is an adverse prognostic marker of melanoma patient survival by increasing melanoma cell invasion. J. Invest. Dermatol. 2015;135(5):1358–1367. doi: 10.1038/jid.2014.552. [DOI] [PubMed] [Google Scholar]
- 171.Carter JH, Deddens JA, Spaulding IV NR, et al. Phosphorylation of eIF4E serine 209 is associated with tumour progression and reduced survival in malignant melanoma. Br. J. Cancer. 2016;114(4):444–453. doi: 10.1038/bjc.2015.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Salehi Z, Mashayekhi F, Shahosseini F. Significance of eIF4E expression in skin squamous cell carcinoma. Cell Biol. Int. 2007;31(11):1400–1404. doi: 10.1016/j.cellbi.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 173.Joyce CE, Yanez AG, Mori A, Yoda A, Carroll JS, Novina CD. Differential regulation of the melanoma proteome by eIF4A1 and eIF4E. Cancer Res. 2017;77(3):613–622. doi: 10.1158/0008-5472.CAN-16-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhan Y, Dahabieh MS, Rajakumar A, et al. The role of eIF4E in response and acquired resistance to vemurafenib in melanoma. J. Invest. Dermatol. 2015;135(5):1368–1376. doi: 10.1038/jid.2015.11. [DOI] [PubMed] [Google Scholar]
- 175.Lopez-Rivera E, Jayaraman P, Parikh F, et al. Inducible nitric oxide synthase drives mTOR pathway activation and proliferation of human melanoma by reversible nitrosylation of TSC2. Cancer Res. 2014;74(4):1067–1078. doi: 10.1158/0008-5472.CAN-13-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tiwari R, Sahu I, Soni BL, et al. Quantitative phosphoproteomic analysis reveals system-wide signaling pathways regulated by site-specific phosphorylation of Keratin-8 in skin squamous cell carcinoma derived cell line. Proteomics. 2017;17(7):1–17. doi: 10.1002/pmic.201600254. [DOI] [PubMed] [Google Scholar]
- 177.O'Reilly KE, Warycha M, Davies MA, et al. Phosphorylated 4E-BP1 is associated with poor survival in melanoma. Clin. Cancer Res. 2009;15(8):2872–2878. doi: 10.1158/1078-0432.CCR-08-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Boussemart L, Malka-Mahieu H, Girault I, et al. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature. 2014;513(7516):105–109. doi: 10.1038/nature13572. [DOI] [PubMed] [Google Scholar]
- 179.Jasiulionis MG, Luchessi AD, Moreira AG, et al. Inhibition of eukaryotic translation initiation factor 5A (eIF5A) hypusination impairs melanoma growth. Cell Biochem. Funct. 2007;25(1):109–114. doi: 10.1002/cbf.1351. [DOI] [PubMed] [Google Scholar]
- 180.Khosravi S, Wong RPC, Ardekani GS, et al. Role of EIF5A2, a downstream target of Akt, in promoting melanoma cell invasion. Br. J. Cancer. 2014;110(2):399–408. doi: 10.1038/bjc.2013.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pinzaglia M, Montaldo C, Polinari D, et al. eIF6 over-expression increases the motility and invasiveness of cancer cells by modulating the expression of a critical subset of membrane-bound proteins. BMC Cancer. 2015;15(1):1–15. doi: 10.1186/s12885-015-1106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 183.Emmanuel R, Weinstein S, Landesman-Milo D, Peer D. eIF3c: a potential therapeutic target for cancer. Cancer Lett. 2013;336(1):158–166. doi: 10.1016/j.canlet.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 184.Mathews MB, Hershey JWBB. The translation factor eIF5A and human cancer. Biochim. Biophys. Acta. 2015;1849(7):836–844. doi: 10.1016/j.bbagrm.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhu W, Li GX, Chen HL, Liu XY. The role of eukaryotic translation initiation factor 6 in tumors. Oncol. Lett. 2017;14(1):3–9. doi: 10.3892/ol.2017.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345(6275):544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 187.Larsson O, Li S, Issaenko OA, et al. Eukaryotic translation initiation factor 4E-induced progression of primary human mammary epithelial cells along the cancer pathway is associated with targeted translational deregulation of oncogenic drivers and inhibitors. Cancer Res. 2007;67(14):6814–6824. doi: 10.1158/0008-5472.CAN-07-0752. [DOI] [PubMed] [Google Scholar]
- 188.Truitt ML, Conn CS, Shi Z, et al. Differential requirements for eIF4E dose in normal development and cancer. Cell. 2015;162(1):59–71. doi: 10.1016/j.cell.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.She Q-B. 4E-BP1 as an oncotarget. Aging (Albany NY) 2015;7(8):517–518. doi: 10.18632/aging.100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Whitmarsh AJ. Regulation of gene transcription by mitogen-activated protein kinase signaling pathways. Biochim. Biophys. Acta. 2007;1773(8):1285–1298. doi: 10.1016/j.bbamcr.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 191.Lineham E, Spencer J, Morley SJ. Dual abrogation of MNK and mTOR: a novel therapeutic approach for the treatment of aggressive cancers. Future Med. Chem. 2017;9(13):1539–1555. doi: 10.4155/fmc-2017-0062. [DOI] [PubMed] [Google Scholar]
- 192.Yang H, Kircher DA, Kim KH, et al. Activated MEK cooperates with Cdkn2a and Pten loss to promote the development and maintenance of melanoma. Oncogene. 2017;36(27):3842–3851. doi: 10.1038/onc.2016.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Salhi A, Farhadian JA, Giles KM, et al. RSK1 activation promotes invasion in nodular melanoma. Am. J. Pathol. 2015;185(3):704–716. doi: 10.1016/j.ajpath.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Konicek BW, Stephens JR, McNulty AM, et al. Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res. 2011;71(5):1849–1857. doi: 10.1158/0008-5472.CAN-10-3298. [DOI] [PubMed] [Google Scholar]
- 195.Zhan Y, Guo J, Yang W, et al. MNK1/2 inhibition limits oncogenicity and metastasis of KIT -mutant melanoma. J. Clin. Invest. 2017;127(11):4179–4192. doi: 10.1172/JCI91258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Malka-Mahieu H, Girault I, Rubington M, et al. Synergistic effects of eIF4A and MEK inhibitors on proliferation of NRAS-mutant melanoma cell lines. Cell Cycle. 2016;15(18):2405–2409. doi: 10.1080/15384101.2016.1208862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Herbert TP, Tee AR, Proud CG. The extracellular signal-regulated kinase pathway regulates the phosphorylation of 4E-BP1 at multiple sites. J. Biol. Chem. 2002;277(13):11591–11596. doi: 10.1074/jbc.M110367200. [DOI] [PubMed] [Google Scholar]
- 198.Gingras AC, Raught B, Gygi SP, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15(21):2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Alain T, Morita M, Fonseca BD, et al. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res. 2012;72(24):6468–6476. doi: 10.1158/0008-5472.CAN-12-2395. [DOI] [PubMed] [Google Scholar]
- 200.Ducker GS, Atreya CE, Simko JP, et al. Incomplete inhibition of phosphorylation of 4E-BP1 as a mechanism of primary resistance to ATP-competitive mTOR inhibitors. Oncogene. 2014;33(12):1590–1600. doi: 10.1038/onc.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Daveri E, Maellaro E, Valacchi G, Ietta F, Muscettola M, Maioli E. Inhibitions of mTORC1 and 4EBP-1 are key events orchestrated by Rottlerin in SK-Mel-28 cell killing. Cancer Lett. 2016;380(1):106–113. doi: 10.1016/j.canlet.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 202.Jiang CC, Croft A, Tseng H-Y, et al. Repression of microRNA-768-3p by MEK/ERK signalling contributes to enhanced mRNA translation in human melanoma. Oncogene. 2014;33(20):2577–2588. doi: 10.1038/onc.2013.237. [DOI] [PubMed] [Google Scholar]
- 203.Croft A, Tay KH, Boyd SC, et al. Oncogenic activation of MEK/ERK primes melanoma cells for adaptation to endoplasmic reticulum stress. J. Invest. Dermatol. 2014;134(2):488–497. doi: 10.1038/jid.2013.325. [DOI] [PubMed] [Google Scholar]
- 204.Piskounova E, Agathocleous M, Murphy MM, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527(7577):186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dankort D, Curley DP, Cartlidge RA, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat. Genet. 2009;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Le Gal K, Ibrahim MX, Wiel C, et al. Antioxidants can increase melanoma metastasis in mice. Sci. Transl. Med. 2015;7(308):308re8–308re8. doi: 10.1126/scitranslmed.aad3740. [DOI] [PubMed] [Google Scholar]
- 207.Liakath-Ali K, Mills EW, Sequeira I, et al. An evolutionarily conserved ribosome-rescue pathway maintains epidermal homeostasis. Nature. 2018;556(7701):376–380. doi: 10.1038/s41586-018-0032-3. [DOI] [PubMed] [Google Scholar]
- 208.Baumann K. Smooth translation to maintain a healthy skin. Nat. Rev. Mol. Cell Biol. 2018;19(6):345–345. doi: 10.1038/s41580-018-0025-y. [DOI] [PubMed] [Google Scholar]
- 209.Rak R, Dahan O, Pilpel Y. Repertoires of tRNAs: the couplers of genomics and proteomics. Annu. Rev. Cell Dev. Biol. 2018;34(1) doi: 10.1146/annurev-cellbio-100617-062754. annurev-cellbio-100617–062754. [DOI] [PubMed] [Google Scholar]
- 210.Moir RD, Willis IM. Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim. Biophys. Acta. 2013;1829(3–4):361–375. doi: 10.1016/j.bbagrm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell. 2012;45(4):439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 212.Arimbasseri GA. Interactions between RNAP III transcription machinery and tRNA processing factors. Biochim. Biophys. Acta. 2018;1861(4):354–360. doi: 10.1016/j.bbagrm.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 213.White RJ. Transcription by RNA polymerase III: more complex than we thought. Nat. Rev. Genet. 2011;12(7):459–463. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 214.Dieci G, Conti A, Pagano A, Carnevali D. Identification of RNA polymerase III-transcribed genes in eukaryotic genomes. Biochim. Biophys. Acta. 2013;1829(3–4):296–305. doi: 10.1016/j.bbagrm.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 215.Hoffmann NA, Jakobi AJ, Moreno-Morcillo M, et al. Molecular structures of unbound and transcribing RNA polymerase III. Nature. 2015;528(7581):231–236. doi: 10.1038/nature16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gouge J, Satia K, Guthertz N, et al. Redox signaling by the RNA polymerase III TFIIB-related factor Brf2. Cell. 2015;163(6):1375–1387. doi: 10.1016/j.cell.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Cabarcas S, Schramm L. RNA polymerase III transcription in cancer: the BRF2 connection. Mol. Cancer. 2011;10:1–10. doi: 10.1186/1476-4598-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sin O, de Jong T, Mata-Cabana A, et al. Identification of an RNA polymerase III regulator linked to disease-associated protein aggregation. Mol. Cell. 2017;65(6):1096–1108.e6. doi: 10.1016/j.molcel.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Johnson SAS, Dubeau L, Johnson DL. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J. Biol. Chem. 2008;283(28):19184–19191. doi: 10.1074/jbc.M802872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Upadhya R, Lee JH, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol. Cell. 2002;10(6):1489–1494. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 221.Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc. Natl Acad. Sci. USA. 2010;107(26):11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Johnson DL, Stiles BL. Maf1, a new PTEN target linking RNA and lipid metabolism. Trends Endocrinol. Metab. 2016;27(10):742–750. doi: 10.1016/j.tem.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Bonhoure N, Byrnes A, Moir RD, et al. Loss of the RNA polymerase III repressor MAF1 confers obesity resistance. Genes Dev. 2015;29(9):934–947. doi: 10.1101/gad.258350.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Filer D, Thompson MA, Takhaveev V, et al. RNA polymerase III limits longevity downstream of TORC1. Nature. 2017;552(7684):263–267. doi: 10.1038/nature25007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Maraia RJ, Mattijssen S, Cruz-Gallardo I, Conte MR. The La and related RNA-binding proteins (LARPs): structures, functions, and evolving perspectives. Wiley Interdiscip. Rev. RNA. 2017;8(6):e1430. doi: 10.1002/wrna.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Fairley JA, Kantidakis T, Kenneth NS, Intine RV, Maraia RJ, White RJ. Human La is found at RNA polymerase III-transcribed genes in vivo . Proc. Natl Acad. Sci. USA. 2005;102(51):18350–18355. doi: 10.1073/pnas.0506415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Stavraka C, Blagden S. The La-related proteins, a family with connections to cancer. Biomolecules. 2015;5(4):2701–2722. doi: 10.3390/biom5042701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Intine RV, Tenenbaum SA, Sakulich AL, Keene JD, Maraia RJ. Differential phosphorylation and subcellular localization of La RNPs associated with precursor tRNAs and translation-related mRNAs. Mol. Cell. 2003;12(5):1301–1307. doi: 10.1016/s1097-2765(03)00429-5. [DOI] [PubMed] [Google Scholar]
- 229.Heise T, Kota V, Brock A, et al. The La protein counteracts cisplatin-induced cell death by stimulating protein synthesis of anti-apoptotic factor Bcl2. Oncotarget. 2016;7(20):29664–76. doi: 10.18632/oncotarget.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hasler D, Lehmann G, Murakawa Y, et al. The Lupus Autoantigen La prevents Mis-channeling of tRNA fragments into the human MicroRNA pathway. Mol. Cell. 2016;63(1):110–124. doi: 10.1016/j.molcel.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 231.Hasler D, Meister G. From tRNA to miRNA: RNA-folding contributes to correct entry into noncoding RNA pathways. FEBS Lett. 2016;590(15):2354–2363. doi: 10.1002/1873-3468.12294. [DOI] [PubMed] [Google Scholar]
- 232.Jarrous N. Roles of RNase P and its subunits. Trends Genet. 2017;33(9):594–603. doi: 10.1016/j.tig.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 233.Lechner M, Rossmanith W, Hartmann RK, et al. Distribution of ribonucleoprotein and protein-only RNase P in Eukarya. Mol. Biol. Evol. 2015;32(12):3186–3193. doi: 10.1093/molbev/msv187. [DOI] [PubMed] [Google Scholar]
- 234.Jarrous N, Reiner R. Human RNase P: a tRNA-processing enzyme and transcription factor. Nucleic Acids Res. 2007;35(11):3519–3524. doi: 10.1093/nar/gkm071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Siira SJ, Rossetti G, Richman TR, et al. Concerted regulation of mitochondrial and nuclear non-coding RNAs by a dual-targeted RNase Z. EMBO Rep. 2018;19(10):e46198. doi: 10.15252/embr.201846198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Hopper AK, Huang H-Y. Quality Control pathways for nucleus-encoded eukaryotic tRNA biosynthesis and subcellular trafficking. Mol. Cell. Biol. 2015;35(12):2052–2058. doi: 10.1128/MCB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Chatterjee K, Nostramo RT, Wan Y, Hopper AK. tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: location, location, location. Biochim. Biophys. Acta. 2018;1861(4):373–386. doi: 10.1016/j.bbagrm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Van Goietsenoven G, Hutton J, Becker J-P, et al. Targeting of eEF1A with amaryllidaceae isocarbostyrils as a strategy to combat melanomas. FASEB J. 2010;24(11):4575–4584. doi: 10.1096/fj.10-162263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37(21):7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Mahlab S, Tuller T, Linial M. Conservation of the relative tRNA composition in healthy and cancerous tissues. RNA. 2012;18(4):640–652. doi: 10.1261/rna.030775.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Dittmar KA, Mobley EM, Radek AJ, Pan T. Exploring the regulation of tRNA distribution on the genomic scale. J. Mol. Biol. 2004;337(1):31–47. doi: 10.1016/j.jmb.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 242.Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2(12):e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Khattar E, Kumar P, Liu CY, et al. Telomerase reverse transcriptase promotes cancer cell proliferation by augmenting tRNA expression. J. Clin. Invest. 2016;126(10):4045–4060. doi: 10.1172/JCI86042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Pavon-Eternod M, Gomes S, Rosner MR, Pan T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA. 2013;19(4):461–466. doi: 10.1261/rna.037507.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Clarke CJ, Berg TJ, Birch J, et al. The initiator methionine tRNA drives secretion of type II collagen from stromal fibroblasts to promote tumor growth and angiogenesis. Curr. Biol. 2016;26(6):755–765. doi: 10.1016/j.cub.2016.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Birch J, Clarke CJ, Campbell AD, et al. The initiator methionine tRNA drives cell migration and invasion leading to increased metastatic potential in melanoma. Biol. Open. 2016;5(10):1371–1379. doi: 10.1242/bio.019075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Presnyak V, Alhusaini N, Chen Y-H, et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160(6):1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Karpinets TV, Foy BD. Tumorigenesis: the adaptation of mammalian cells to sustained stress environment by epigenetic alterations and succeeding matched mutations. Carcinogenesis. 2005;26(8):1323–1334. doi: 10.1093/carcin/bgi079. [DOI] [PubMed] [Google Scholar]
- 249.Lampson BL, Pershing NLK, Prinz JA, et al. Rare codons regulate KRas oncogenesis. Curr. Biol. 2013;23(1):70–75. doi: 10.1016/j.cub.2012.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Pershing NLK, Lampson BL, Belsky JA, Kaltenbrun E, MacAlpine DM, Counter CM. Rare codons capacitate Kras-driven de novo tumorigenesis. J. Clin. Invest. 2015;125(1):222–233. doi: 10.1172/JCI77627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ishimura R, Nagy G, Dotu I, et al. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345(6195):455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Pan T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018;28(4):395–404. doi: 10.1038/s41422-018-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 2006;16(10):971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 254.Tuorto F, Liebers R, Musch T, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012;19(9):900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 255.Chan CTY, Pang YLJ, Deng W, et al. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nedialkova DD, Leidel SA. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161(7):1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Stewart JB, Alaei-Mahabadi B, Sabarinathan R, et al. Simultaneous DNA and RNA mapping of somatic mitochondrial mutations across diverse human cancers. PLoS Genet. 2015;11(6):e1005333. doi: 10.1371/journal.pgen.1005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Belostotsky R, Frishberg Y, Entelis N. Human mitochondrial tRNA quality control in health and disease. RNA Biol. 2012;9(1):33–39. doi: 10.4161/rna.9.1.18009. [DOI] [PubMed] [Google Scholar]
- 259.Megel C, Morelle G, Lalande S, Duchêne A-M, Small I, Maréchal-Drouard L. Surveillance and cleavage of eukaryotic tRNAs. Int. J. Mol. Sci. 2015;16(12):1873–1893. doi: 10.3390/ijms16011873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Mishima E, Inoue C, Saigusa D, et al. Conformational change in transfer RNA is an early indicator of acute cellular damage. J. Am. Soc. Nephrol. 2014;25(10):2316–2326. doi: 10.1681/ASN.2013091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Yang X-LL, Schimmel P. Functional expansion of the tRNA world under stress. Mol. Cell. 2011;43(4):500–502. doi: 10.1016/j.molcel.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wilusz JE. Controlling translation via modulation of tRNA levels. Wiley Interdiscip. Rev. RNA. 2015;6(4):453–470. doi: 10.1002/wrna.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43(4):613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Emara MM, Ivanov P, Hickman T, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010;285(14):10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lyons SM, Achorn C, Kedersha NL, Anderson PJ, Ivanov P. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res. 2016;44(14):6949–6960. doi: 10.1093/nar/gkw418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Sinnberg T, Sauer B, Holm P, et al. MAPK and PI3K/AKT mediated YB-1 activation promotes melanoma cell proliferation which is counteracted by an autoregulatory loop. Exp. Dermatol. 2012;21(4):265–270. doi: 10.1111/j.1600-0625.2012.01448.x. [DOI] [PubMed] [Google Scholar]
- 267.Kosnopfel C, Sinnberg T, Sauer B, et al. YB-1 expression and phosphorylation regulate tumorigenicity and invasiveness in melanoma by influencing EMT. Mol. Cancer Res. 2018;16(7):1149–1160. doi: 10.1158/1541-7786.MCR-17-0528. [DOI] [PubMed] [Google Scholar]
- 268.Goodarzi H, Liu X, Nguyen HCB, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-Derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161(4):790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12(1):78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017;170(1):61–71.e11. doi: 10.1016/j.cell.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Keam SP, Sobala A, Ten Have S, Hutvagner G. tRNA-derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation. J. Proteome Res. 2017;16(2):413–420. doi: 10.1021/acs.jproteome.6b00267. [DOI] [PubMed] [Google Scholar]
- 272.Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. The RNA–methyltransferase misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011;7(12):e1002403. doi: 10.1371/journal.pgen.1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.McMahon M, Ruggero D. A wobbly road to drug resistance in melanoma: tRNA-modifying enzymes in translation reprogramming. EMBO J. 2018;37(15):e99978. doi: 10.15252/embj.201899978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Rezvani HR, Ali N, Nissen LJ, et al. HIF-1α in epidermis: oxygen sensing, cutaneous angiogenesis, cancer, and non-cancer disorders. J. Invest. Dermatol. 2011;131(9):1793–1805. doi: 10.1038/jid.2011.141. [DOI] [PubMed] [Google Scholar]
- 275.Kumar P, Mudunuri SB, Anaya J, Dutta A. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015;43(D1):D141–D145. doi: 10.1093/nar/gku1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Pliatsika V, Loher P, Magee R, et al. MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects. Nucleic Acids Res. 2018;46(D1):D152–D159. doi: 10.1093/nar/gkx1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zheng LL, Xu WL, Liu S, et al. tRF2Cancer: a web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucleic Acids Res. 2016;44(W1):W185–W193. doi: 10.1093/nar/gkw414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Venkatesh T, Suresh PS, Tsutsumi R. tRFs: miRNAs in disguise. Gene. 2016;579(2):133–138. doi: 10.1016/j.gene.2015.12.058. [DOI] [PubMed] [Google Scholar]
- 279.Baden HP, Pearlman C. The effect of ultraviolet light on protein and nucleic acid synthesis in the epidermis. J. Invest. Dermatol. 1964;42:71–75. [PubMed] [Google Scholar]
- 280.Kaushik SB, Kaushik N. Non-coding RNAs in skin cancers: an update. Non-coding RNA Res. 2016;1(1):83–86. doi: 10.1016/j.ncrna.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]