Abstract
Dendritic cells (DCs) and macrophages (MΦs) are antigen-presenting phagocytic cells found in many peripheral tissues of the human body, including the blood, lymph nodes, skin, and lung. They are vital to maintaining steady-state respiration in the human lung based on their ability to clear airways while also directing tolerogenic or inflammatory responses based on specific stimuli. Over the past three decades, studies have determined that there are multiple subsets of these two general cell types that exist in the airways and interstitium. Identifying these numerous subsets has proven challenging, especially with the unique microenvironments present in the lung. Cells found in the vasculature are not the same subsets found in the skin or the lung, as demonstrated by surface marker expression. By transcriptional profiling, these subsets show similarities but also major differences. Primary human lung cells and/or tissues are difficult to acquire, particularly in a healthy condition. Additionally, surface marker screening and transcriptional profiling are continually identifying new DC and MΦ subsets. While the overall field is moving forward, we emphasize that more attention needs to focus on replicating the steady-state microenvironment of the lung to reveal the physiological functions of these subsets.
Keywords: innate immunity, monocytes, respiratory system, phagocytes, transcriptional profiling
I. INTRODUCTION
Dendritic cells (DCs) and macrophages (MΦs) are specialized mononuclear phagocytic cells that also present portions of antigens to other members of the immune system. As phagocytes, they process ingested microbes, cellular debris, and particulates; as antigen-presenting cells (APCs), they subsequently present epitopes of these antigens to other cell types to direct inflammatory or tolerogenic innate and adaptive immune responses. Both DCs and MΦs have for over two decades been known to exist throughout the human lung,1,2 although their exact identification and potential functions have changed during this time as scientific techniques and technologies have improved. Instead of just two cell types, it is now clear that a heterogenous collection of these cells works together to maintain respiratory function.1,3–8 Although DCs and MΦs are generally well studied, a large proportion of what we know, or assume to know, about their identification and biology has come from small animal work or in vitro models. Comparatively, the scientific community has been impeded in its investigations of resident phagocytes of the human lung by issues associated with acquiring cells/tissues from healthy donors.
The goal of this review is to describe what is currently known about DC and MΦ subsets of the lung, including their identification, characterization, and functional properties, based mainly on studies involving primary human cells. Where relevant and applicable, we will draw on comparative work from mice or other human tissues, with the overall goal to point out the unique microenvironment of the lung interstitium and airways, and to focus on how future studies should take into account these microenvironments when studying resident DCs and MΦs. As we describe previous work, we will try to reconcile discrepancies in subset identification as we show how the field has moved forward in recent years. As we will demonstrate, many groups have likely been studying the same subsets but merely calling them by different names based on certain marker expression profiles. To understand the importance of DCs and MΦs in the human lung, we must first understand the anatomical and physiological complexities of this essential organ.
II. HUMAN LUNG STRUCTURE, FUNCTION, AND CELLULAR COMPOSITION
Among human tissues, the lung is highly specialized for the vital function of respiration. Like the skin and gut, the lung is an open system continually exposed to the external environment. Structurally, the lung as a whole (Fig. 1) must be flexible to allow inspiration and expiration. Inspired air travels down the trachea before splitting into the primary, or main-stem, bronchi that enter the right and left lungs. Further branching results in secondary bronchi entering each of the three right and two left lobes of the lungs. Tertiary bronchi and subsequent smaller bronchioles continue this asymmetrical dichotomy several levels down within individual lobes.9,10 At the bronchiole level, the descending airways change in composition from mainly hyaline cartilage, meant for rigid support, to smooth muscle and elastin fibers.11 This latter framework allows rapid dilation or constriction to regulate airflow into the deeper regions of the lungs to occur. The bronchioles continue branching and decreasing in channel width to the point of the terminal bronchioles, the final level of the conducting airways (Fig. 1).
FIG. 1:
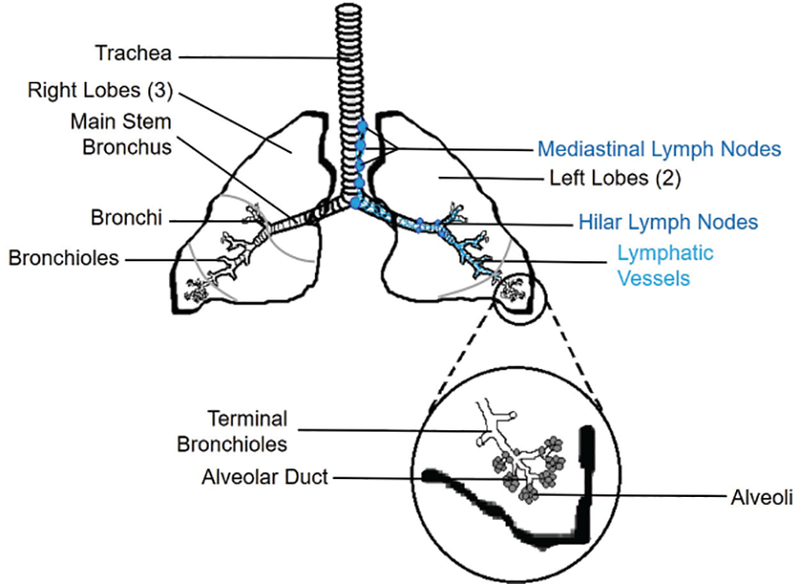
Human lung anatomy. Inhaled air travels down the trachea, enters the lungs, and follows a network of branching bronchi and bronchioles until it reaches the terminal bronchioles and the alveoli. An ascending network of lymphatic vessels (right side) carries lymph, particulates, and immune cells from the interstitial spaces to the hilar and eventually the mediastinal lymph nodes. Inset: The alveoli are grape-like structures responsible for exchange of oxygen and carbon dioxide.
Physiologically, a very complex network of airways and capillaries exists within the human lower respiratory tract that facilitates constant gas exchange between inhaled air and the bloodstream. The human lung develops and expands rapidly after birth, to the point that the adult version of this organ averages over 120 m2 of respiratory surface area.12 The actual structures involved in respiration are open-ended spherical sacs called alveoli, which form millions of clusters within the distal regions of the lung lobes (Figs. 1 and 2).13 The alveolar walls are only one epithelial cell layer thick, and each alveolus is covered in a dense network of blood capillaries that is similarly a single endothelial cell layer thick (Fig. 2). Thus, oxygen from inspired air can rapidly cross the alveolar epithelium and the capillary endothelium, and likewise carbon dioxide can cross both cell layers and be removed during expiration.
FIG. 2:
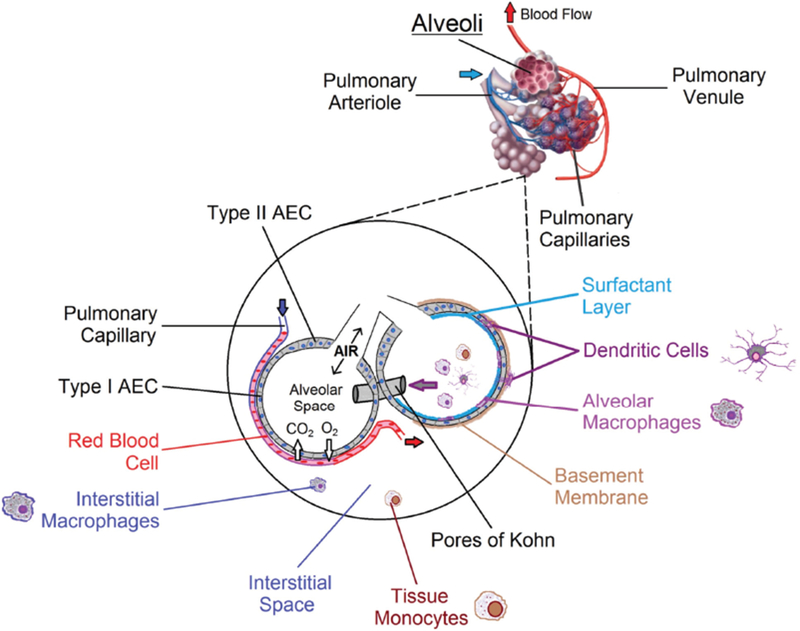
Human alveolar structure and cellular composition. At the alveolar level, oxygen diffuses into and carbon dioxide diffuses out of blood flowing through pulmonary capillaries. Inset: A wide variety of cell types are found in and around alveoli. Type I AECs make up the majority of the surface area involved in respiration. DCs are found embedded within the alveolar epithelium and just below it in the submucosa, from where they extend processes into the alveolar space to sample antigens. Immune cells transit between alveoli via pores of Kohn. Together, subsets of macrophages, dendritic cells, and tissue monocytes keep the airways clear of obstruction and direct interactions with cells of the adaptive immune system.
About 97% of the surface area of each alveolus is composed of large thin type I alveolar epithelial cells (AECs).14 More numerous but much smaller type II AECs can be found along the edges of an alveolus, where they serve to replenish type I AECs and to produce a layer of surfactant that coats the inner alveolar lining.14,15 Surfactant is a complex mixture of mainly lipids, but also about 10% proteins, which reduces the surface tension at the gas-liquid interface.16 Without surfactant, alveoli would collapse during expiration and require additional energy to inflate during inspiration. A thin basement membrane runs along both the blood capillaries and the alveoli, providing physical support for the delicate air-blood barrier. While each alveolus is a separate unit, alveoli are connected by septal holes known as pores of Kohn.17 These connections allow the flow of surfactant and the movement of immune cells such as DCs and MΦs between neighboring alveoli (Fig. 2).17,18
Due to the single-cell layers that separate inhaled air from the blood circulation, injury or inflammation in the lower airways can quickly affect respiration. As such, the alveolar spaces must be kept in natural anti-inflammatory conditions that allow tolerance to particles such as the dust and pollen we breathe in on a regular basis. Additionally, for decades we believed that the lower airways were completely sterile in the steady state and only contained microbes during infections. This theory has changed with recent findings of 16S ribosomal RNA from the lower airways of healthy volunteers, which suggest that these regions in fact house a low microbiome concentration that is distinct from merely upper airway contaminants.19–25 As would be expected, the density of lower airway microbes is much lower than that observed in high-nutrient locations like the intestines, skin, and nasal passages.26,27
Although there are some variations between individuals depending on age and environment, many bacterial phyla have been consistently demonstrated in healthy human lungs based on 16S ribosomal RNA sequencing, including Proteobacteria, Fusobacteria, Bacteroidetes, Firmicutes, and Actinobacteria.27,28 Considering that alveolar development occurs predominantly after birth and proceeds through the first three years of life,29 it is likely that the lower airways are progressively colonized by members of these phyla during this same time period. In mice, the presence and diversity of lower respiratory tract microbiota have been shown to positively correlate with smaller and more numerous alveoli.30 It is quite possible that development of the human respiratory structure is similarly affected by microbial exposure and possible colonization. While bacterial species establish themselves in the airways, they cannot be allowed to reach densities that would impair respiration. Cells residing along the airways must routinely keep bacterial numbers in check without stimulating inflammation. The epithelial cells of both the bronchi and the alveoli help maintain tolerance under steady-state conditions by constant production of the anti-inflammatory cytokines IL-10 and TGF-β.31,32 Alveolar and interstitial macrophages (IMs) may also contribute to the development of tolerance to nonpathogenic microbes and other inhaled particulates by decreasing antigen presentation by resident DC populations33,34 and by stimulating the production of Foxp3+ regulatory T-cells along the airways.35
Overall, DCs and MΦs must keep the airways free of obstructions caused by dead cells, microbes, and particulates without triggering inflammation and damage to the airways. Accordingly, we suggest that future work to elucidate the potential roles of human airway DCs and MΦs should culture them together during experimental stimulations. This way, any interactions between the cell subsets are exposed rather than just demonstrating a specific cell response that is not relevant to the airway environment. We have exposed human airway antigen-presenting cells (APCs) subsets, acquired by whole-lung lavage, to bacterial stimulation as a pooled batch prior to flow cytometric analysis to incorporate these types of cellular interactions.8 We have also studied the ability of airway APCs (HLA-DR+) to phagocytose Bacillus anthracis spores using a novel precision-cut lung-slice model that allows cells to remain in their normal anatomical locations and frequencies, and incorporates AECs into the culture conditions.36
Because DCs and MΦs are APCs, their functional roles are greater than merely the ingestion of antigens along the lower airways. They serve a vital role in directing adaptive immune responses either to tolerance after the majority of exposures or to inflammation in the rare instances in which pathogenic microbes invade. On most occasions, these interactions with the adaptive immune system (specifically B- and T-cells) occur in the mediastinal lymph nodes (LNs). The afferent lymphatic network of the lung allows APCs found in and around the airways to transit first to the smaller hilar LNs and then to the mediastinal LNs located along the trachea (Fig. 1).37 The primary role of lymphatics is to regulate interstitial fluid volume in conjunction with constantly changing volume and pressure in the blood vasculature.38,39
Lymphatic vessels are open-ended and unidirectional in their fluid flow. Lobed valves in the microlymphatic vessels open in response to increased interstitial fluid pressure, but close as pressure falls to prevent backflow. In the steady state, the fluid carries with it cellular debris and foreign antigens that have evaded internalization by phagocytic APCs. Smooth muscle rhythmically contracts along larger lymphatic collectors to aid in lymphatic flow toward regional LNs.40 Human lungs contain an afferent lymphatic network of decreasing density all the way down to the respiratory bronchioles and interalveolar interstitium,41 providing a direct conduit for migratory immune cells, specifically DCs and tissue monocytes (TMs) (and possibly MΦs under certain conditions, as discussed in a later section), to reach draining LNs.
Now that we have described the basic structure and function of the human lung, we will shift our attention to DC and MΦ subsets that reside within the lung, with particular emphasis placed on those present along the airways.
III. HUMAN DENDRITIC CELLS
A. Introduction to Human DCs
DCs are found in many different tissues of the human body, including blood,42 skin,43 LNs,44 and lung.2 Due to their strength in stimulating T-cell proliferation and cytokine production, they have been harnessed for immunotherapeutics, where donor-derived DCs are exposed to a specific antigen ex vivo and reintroduced to the patient.45–49 In future clinical trials, the efficacy of these treatments may be improved by the use of specific DC subsets that are particularly useful against certain cancers or pathogens. Just as DCs are important for directing immune responses against certain microbes, microbes have also evolved to utilize DCs to spread themselves throughout the body. This “Trojan horse” or “carrier cell” model has been linked to the spread of human pathogens such as Bacillus anthracis,50,51 Burkholderia pseudomallei,52 and Cryptococcus neoformans.53 When steady-state DC functions are perturbed, disease processes like chronic obstructive pulmonary disease54 and systemic lupus erythematosus55 may be initiated, worsen, or be prolonged. Additional mechanisms of these and similar diseases may be revealed with the identification and characterization of specific DC subsets, particularly if the microenvironment of the anatomical sources is also considered.
Studying primary human DCs is not without its barriers, which must be overcome to make progress in the field. DCs as a whole are naturally rare in human tissues and are rarer still when individual subsets are identified. For example, DCs are approximately 1 percent of peripheral blood mono-nuclear cells,56 so a large amount of source tissue is required to obtain enough cells to do substantial functional assays. These tissues, therefore, come at a high monetary cost, must be acquired as part of surgical resections of diseased tissues, or are just not regularly available for research purposes. While monocyte-derived DCs (and MΦs) have been used as stand-ins due to their ability to be generated in higher frequencies from human blood,57,58 our group has shown that transcriptionally monocyte-derived DCs and MΦs are weak models of several DC and MΦ subsets found in human lower airways.8 Another obvious limitation of human DC studies is that DC migration into or out of tissues cannot be followed in vivo. The human DC microenvironment can also dramatically affect expression of surface markers often used in their discrimination.59 With this in mind, we first describe some basics of DCs isolated from the microenvironment of the blood and skin and then transition to the lung for the majority of our commentary.
B. Basic Markers for Identifying Human DC Subsets
DCs were first identified based on their dendrites, unique membrane extensions that are used by this cell type to sample antigens from the surrounding environment.60 DCs are generally characterized as also having a high cytoplasmic–nuclear ratio, which is evident upon microscopic investigation.60 Unfortunately, such morphological characteristics are not enough to differentiate individual DC subsets.61 This is coupled with the fact that just isolating DCs from human tissue is enough to cause changes in their morphological appearance.62 Unlike other cell types, such as T-cells, DCs have no single surface or intracellular marker that is specific to them. MΦs, monocytes, and DCs all fit together as mononuclear phagocytes, and thus, are expected to be somewhat related. Therefore, it has become necessary to use several markers just to tell these cell types apart, with the addition of more markers to separate DC subsets. With technological advances such as transcriptional profiling, new markers are continuously being identified that may in the future divide currently known DC subsets into multiple subsets. An additional challenge then arises of identifying a functional role that correlates with a DC subset based on a specific set of markers.
Identification of standard human DCs often begins with exclusion of other cell types. Expression of specific markers for T-cells (CD3), B-cells (CD19, CD20), and natural killer cells (CD56) provides an initial framework that narrows cell identification.63 After these exclusions, often termed a “lineage dump” in flow cytometry, APCs are further identified based on their expression of the class II MHC molecule HLA-DR.61 At this point, the researcher has separated DCs, monocytes, and MΦs from the other common cell types found in most human tissues. An additional discriminatory surface marker is the integrin CD11c,64 as it is expressed on most myeloid cells61 but is not expressed by human plasmacytoid DCs (pDCs).42,57 DC subsets identified in human skin, including CD1a+ DCs, CD14+ DCs, and Langerhans cells (LCs) express CD11c,65 which we have also used as part of a schema to identify multiple DC subsets of the human airways.8
A major breakthrough in DC identification came in 2000, when Dzionek et al.42 performed a screen of monoclonal antibodies against various surface markers on human blood cells. Starting with the lineage exclusion described earlier, the blood dendritic cell antigens (BDCAs) were identified as markers that reliably and reproducibly separate the three major DC subsets in blood. BDCA-1 (CD1c) and BDCA-3 (CD141) identify two myeloid DC subsets, while BDCA-2 (CD303) specifically identify pDCs. Since this work, others have been able to characterize human pDCs as being BDCA-2+ and CD123+ (IL-3R) while being negative for CD11c.66–68
Plasmacytoid DCs are actually similar to T-cells in that they express the pre-T-cell receptor alpha chain and also CD4 found on T helper cells.67 Physiologically, pDCs express TLR7 and TLR9, which are both endosomal sensors for pathogen nucleic acids.69 Instead of primarily phagocytosis, it is hypothesized that the major function of pDCs is the production of type I interferon upon viral infection.70 Some work has suggested that BDCA-2 expression is important for regulating the amount of type I interferon actually secreted, as antibody ligation of this molecule dampens interferon production in response to known TLR7 and TLR9 stimuli.71,72
Under steady-state conditions, pDCs promote tolerance, at least in primary lymphoid tissue,73 by stimulating the differentiation of T-cells into regulatory T-cells.74 We have not isolated pDCs from human airways based on CD123 expression,8 but they may serve a tolerogenic role in the lung inter-stitium based on their identification by others at very low levels in bronchoalveolar lavage (BAL) fluid.75 Our inability to identify pDCs from human airways agrees with the suggestion that pDCs migrate into peripheral tissue from the bloodstream or lymphoid organs upon viral infection76 and then perhaps remain there short-term to reinitiate regulatory T-cell differentiation during the healing process. CD123 seems to be the more reliable marker for pDCs, as surface BDCA-2 levels decrease quickly with time in culture.42 While CD123+ HLA-DR+ lineage− cells are still accepted as pDCs,42,57 recent work has challenged the notion that CD123 is specific to pDCs in the blood77,78 given that precursors of BDCA-1+ and BDCA-3+ DCs (discussed next) also express this marker and may confound results that rely purely on CD123 to identify pDCs.
BDCA-1 and BDCA-3 have both been used as specific surface markers for two myeloid DC subsets found in human blood and several other human tissues.42,57 Unfortunately, BDCA-3 expression increases on pDCs and BDCA-1+ DCs soon after culturing,42,57 which affects subset identification if stimulation in vitro occurs first. Certain transcription factors can serve as a secondary means of identifying these two DC subsets. The classical BDCA-1+ DCs can also be identified by specific expression of the transcription factor IFN regulatory factor 4 (IRF4).79 In contrast, true BDCA-3+ DCs can be identified by their expression of both IFN regulatory factor 8 (IRF8) and the basic leucine zipper ATF-like transcription factor 3 (BATF3).80 We have also seen specific expression of IRF4 in BDCA-1+ CD14− DCs in the human airways based on whole-genome transcriptional profiling,8 so these transcription factor differences between the two subsets are likely conserved between different human tissues.
Interestingly, BDCA-1/CD1c has been shown to present nonpeptide antigens (as CD1a and CD1b also do) specifically to γδ T-cells.81,82 Further, BDCA-1 transits through early and late endosomes after phagocytosis, while CD1a and CD1b do not.83 Together, these results reveal a scenario in which BDCA-1+ DCs found in the periphery present a wide variety of nonpeptide epitopes to γδ T-cells, which then aid in mounting an overall immune response. With variations in BDCA expression, other markers have been identified that may prove to be more stable in identifying BDCA-3+ DCs. Examples include CLEC9A80 and XCR1.84 CLEC9A has been associated with BDCA-3high DCs that are functionally potent at cross-presentation of various antigens, including necrotic cells.85–88 Of similar importance, XCR1 is a known receptor for a ligand produced by CD8+ cytotoxic T-cells.89 Therefore, we suggest that both XCR1 and CLEC9A facilitate interactions of BDCA-3+ DCs with cytotoxic T-cells in LNs and in peripheral tissues during active tissue damage.
It should be noted that multiple levels of human DC progenitor cells have been identified for pDCs, BDCA-1+ DCs, and BDCA-3+ DCs.90–92 Subsequent to hematopoietic stem cells, these progenitors can be identified by specific sets of surface markers as they follow distinct differentiation pathways from the bone marrow through the blood and then to the periphery.90–92 Breton et al.77 revealed that CD172a expression can be used to distinguish bone marrow precursor cells committed to becoming either BDCA-1+ or BDCA-3+ DCs of human blood in adults. By single-cell RNA-seq, they showed that CD172a+ bone marrow cells become blood BDCA-1+ DCs, while CD172a− precursors become blood BDCA-3+ DCs.
It is possible that plasticity exists between DC subsets within specific microenvironments such as the skin or airways. Perhaps a DC expressing the surface markers of a BDCA-1+ DC can functionally perform like a BDCA-3+ DC under a specific set of microenvironmental conditions. While it has been proposed that BDCA-1+ DCs can become CD123+ pDCs in culture, this is unlikely without additional stimuli.93 Another possibility, supported by recent findings, is that precursors of myeloid BDCA-1+ and BDCA-3+ DCs are also CD123+.77,78 Therefore, pDCs can be separated from mature myeloid DCs based on CD123 expression, but not from precursors that exist in human blood. While it is unknown whether myeloid DC precursors migrate into the lung, we suspect that commitment of premyeloid DCs into becoming myeloid DCs occurs in the blood, followed by sensing of microenvironmental signals after emigrating into the tissue. As it is yet to be determined whether all steady-state peripheral DC subsets in humans, particularly those of the lung, are replenished from the bone marrow, discussion of DC developmental stages and their associated surface markers is beyond the scope of this review.
C. Identifying Human DC Subsets by Transcriptional Comparisons
Recent technological advances at the genomic level have allowed characterization of several rare cell populations by transcriptional profiling.79,94–97 Based on these comparisons, the relatedness of DC subsets can be determined, as can differentially expressed genes (DEGs) specific to individual DC subsets. Of course, major variations in identifying DEGs may exist based on the cutoff used for importance regarding fold expression. With rare genes, a low cutoff will result in little variation between signal and noise while a high cutoff may disregard true DEGs that are just naturally expressed at very low levels. For example, one group may use a 2-fold difference while another may use a 4-fold difference; based on this, the determined DEGs can be dramatically different in number and identity. For this reason, we have specifically ignored fold-difference when analyzing transcriptional data of lung phagocyte subsets and instead have compared results based on adjusted P values only.8
Multiple groups have used single-cell RNA-seq to identify precursor cells to blood DC subsets77 and even to split the accepted BDCA-1+ and BDCA-3+ DC subsets of human blood into further subpopulations.98 Future work using single-cell RNA-seq technology comparing cells isolated from human airways or lung digestion may in fact determine equivalency of these blood subsets to certain lung populations. Additionally, such comparisons will allow direct identification of genes that are specifically expressed by lung DC subsets as a result of the microenvironment of the airways or interstitial spaces rather than their DC identity. Such findings will aid researchers in developing more physiologically relevant in vitro models of the human lung. However, these recently defined blood DC subsets likely will not all have equivalents in the lung. For example, we have identified a specific DC subset found in human airways that expresses surface Langerin,8 and to our knowledge no such naturally Langerin-expressing DCs exist in the blood.
Work is continuously being done to identify additional markers of specific DC subsets in human blood,99 as this biological material is readily available. Some of these markers may turn out to be more applicable for identifying DC subsets in the lung under steady-state conditions. With our overall theme of this review, we continue to emphasize that the specific microenvironmental cues should be accounted for when performing transcriptional comparisons. Therefore, if transcriptomes of cell subsets from the airways are to be compared, the processing time (and likewise cell manipulation) should be minimized.
Our group has shown that the monocyte-derived DCs and MΦs often used in research are poor models for human lung DC and MΦ subsets based on whole-genome transcriptional profiling.8 The differences are so dramatic that primary lung DCs and MΦs are more closely related to each other than they are to their monocyte-derived counterparts. For example, BDCA1+ CD14− DCs isolated from human airways are more closely related to alveolar macrophages (AMs) than they are to monocyte-derived DCs. Therefore, future lung studies should focus on coculturing of cells obtained from primary human tissue, such as DCs, MΦs, and AECs, or conversely do in situ work with human tissue in which all cell types are present in their natural anatomic locations and frequencies. Studying cells in isolation may provide insight into a cell type’s potential surface markers and functionality but does not necessarily replicate what it does within the tissue in situ.
D. Techniques for Isolating DCs from the Human Lung
As described earlier, the human lung possesses two unique microenvironments for DC subsets: the airways themselves and the interstitial spaces (Fig. 2). For the purposes of APC characterization, we define the airways as predominantly the bronchioles and alveoli, whereas the interstitium encompasses the regions around the airways and pulmonary vasculature. In the steady state, DCs have been identified within the mucosa of the human bronchioles and alveoli.7,100,101 However, the exact identities of these DC subsets are just beginning to be understood based on surface markers and transcriptional profiling. In mice, DCs extend their dendrites through tight junctions between alveolar epithelial cells to allow direct internalization of any microbes or particles trapped within the surfactant layer, followed by their maturation and migration to the regional LNs.102 This same process likely occurs in humans, as DC subsets matching airway cells, based on surface marker expression, have been detected in the digests of mediastinal LNs (Fig. 2).6 Cells resembling DCs have been seen in human airways for over three decades,2 although individual subsets have just recently been discriminated.
Progress in the field of lung DC research has been slowed by a lack of availability of healthy lung, with the majority of our existing knowledge instead coming from digestion of surgical resection from cancer patients.3,103–105 Using single-cell suspensions from digestion of such resections, Nicod et al.106 were the first to identify a rare population of low-autofluorescence APCs that are strong stimulators of CD4+ T-cell proliferation. At the time, it was assumed that these cells, likely DCs, are present only in the interstitium and that AMs are the only APCs found in the airways.
While resected lung sections are deemed non-cancerous based on visual inspection, the effects of proximity to cancerous tissue are unknown. Others have observed that BDCA-2+ pDCs and CLEC9A+ DCs are absent from digestions of healthy lung donors but present in noncancerous tissue resections of non-small-cell lung carcinoma.6 These results suggest that bystander cells are affected by cancerous tissue in the lung. Another concern with isolating cells from whole-lung tissue is the requirement of digestion, which presents two separate constraints. The first is that digestion itself is a harsh process that can activate cells like DCs and MΦs. The second is that cells present in the vasculature and expressing the same surface markers mix with resident cells of the interstitium and/or airways. Pioneering work by Desch et al.6 minimized the latter problem by first perfusing the blood vessels of en bloc human lungs, which were deemed unsuitable for transplant. This allowed removal of a large fraction of blood cells that could contaminate the true lung cell pool.
Desch et al.6 further identified remaining vascular cells by labeling with an anti-CD45 antibody via the blood vessels, with separate CD45 labeling of cells through the airways. This allowed separation of intravascular and extravascular cells based on prestaining after tissue digestion. Indeed, BDCA-1+ and BDCA-3+ DC percentages decreased with preperfusion, and subsequently intravascular cells could be separated from extravascular cells after tissue digestion.
Other groups have attempted to circumvent the issues related to tissue digestion by isolating and characterizing DC subsets directly from the airways of volunteers by BAL.7,107–109 This technique is ideal for isolating cells directly from the microenvironment of the airways, but does not provide information about interstitial cell populations. Using BAL, resident DCs have been isolated not only from the distal airways but also as far proximally as the walls of the trachea.101,110 While using healthy volunteers elim inates the concern of underlying disease processes, the procedure has high variability in the number of total cells acquired. This can be troublesome, particularly when analyzing rare DC subsets. We recently combined these two procedures by acquiring en bloc lungs, perfusing the vasculature, and then performing repeated BAL of the airways.8 With this technique, we obtained large quantities of airway and alveolar resident phagocytes while circumventing the issues of blood cell contamination and harsh tissue digestion.
E. Surface Markers for Identifying Human Lung DC Subsets
Demedts et al.3 were the earliest investigators of DC subsets of the human lung based on the BDCA markers previously used to characterize blood DCs.42 By digestion of tissue resections and subsequent flow cytometry of the cell suspensions, they classified BDCA-1+ and BDCA-3+ myeloid DCs, along with BDCA-2+ CD123+ pDCs from an overall CD14− DC pool. Importantly, they included CD14 in their lineage dump, implying that if a separate population of CD14+ DCs exists in the lung inter-stitium or airways, it is distinct from both BDCA-1+ and BDCA-3+ DC subsets. Consistent with this, inclusion of CD14 positivity revealed a small population of BDCA-1+ CD14+ DCs. Others have identified BDCA-1+ CD14+ DCs from human blood, calling them CD1c+ monocytes, with the distinct stimulatory capacity of naïve CD4+ T-cells compared to conventional BDCA-1+ CD14− DCs.111 However, because these cells were isolated after tissue digestion, it cannot be determined whether these BDCA-1+ CD14+ DCs originate from the lungs or are a residual population from the vasculature. We recently isolated cells with the same marker expression pattern from human lung airways after whole-lung BAL.8 Transcriptional profiling of these cells showed that they group along with Langerin+ DCs and BDCA-1+ CD14− classical DCs in a “DC clade.” This grouping is quite distinct from a “MΦ/monocyte clade” that includes AMs, BDCA-1− CD14+ DCs (same as CD14+ DCs), and BDCA-1− CD14− cells.8 These results together imply two basic interpretations. First, while CD14 positivity has been used to exclude monocyte contamination,105 it is likely a marker for true resident cells of the human airways.
Masten et al.105 originally isolated a population of BDCA-1+ CD1a+ CD14− DCs and found that they are far more allostimulatory of T-cell proliferation compared to a subset of BDCA-1− CD14+ cells thought to be blood monocytes. However, a large portion of those cells were probably CD14+ DCs resident in the actual lung interstitium or airways. Second, these BDCA-1− CD14+ cells of the human lung are likely the equivalent subset as CD14+ dermal DCs.94,112 CD14+ DCs isolated from the skin do indeed have relatively poor ability to stimulate T-cell proliferation as compared with dermal CD1a+ DCs.112 In human skin, these CD14+ DCs express low levels of the activation marker CD83 relative to CD1a+ DCs from the same location.113 Similarly, we have observed that CD14+ DCs express low levels of surface CD83, even after exposure to heat-killed bacteria.8 While CD14+ dermal DCs express variable levels of CD163, a scavenger receptor associated with monocytes and macrophages,94,114 they also express DC-SIGN, which is expressed specifically by DCs.65,115
Desch et al.6 noted that of multiple DC/monocyte/monocyte-derived populations resident in the human lung, only BDCA-1+ CD1a+ “pulmonary DCs” completely lack CD14 expression. When tested in mixed leukocyte reactions (MLRs), this BDCA-1+ CD1a+ DC subset was a far better stimulator of T-cell proliferation in comparison with BDCA-1− CD14+ CD206+ “tissue monocytes.” Thus, both Masten et al.105 and Desch et al.6 had probably identified the same two cellular subsets of the lung based on slightly differing marker expression, as evidenced by MLRs with similar measured results.
Demedts et al.3 did verify that expression of the integrin CD11c can be translated from blood DC identification42,57 to lung DC discrimination. As in blood, both BDCA-1+ and BDCA-3+ DCs express CD11c, while pDCs (identified by BDCA-2 expression) lack this marker. Demedts et al. were also able to separate a small but definite subset of DCs that express CD1a and Langerin after lung digestion.3 These cells were not likely to have come from vasculature contamination, as freshly isolated BDCA −1+ and −3+ DCs from human blood do not express Langerin116 or CD1a6,42 at significant levels. According to immunohistochemistry, these double positive cells were observed along the epithelial lining of the large and small airways but not in the underlying interstitium.3
Such findings of CD1a+ Langerin+ lung DCs initially suggested that a skin LC equivalent population can exist in the human airways. Since the airways do not present with layering similar to epidermal and dermal layers of the skin, such an LC-like cell must anatomically occur within the human airway epithelium. However, Demedts did observe that most CD1a+ DCs also express BDCA-1,3 and thus can be confused with conventional BDCA-1+ DCs without the inclusion of Langerin staining.103 With the greater number of markers tested by Demedts et al.,3 the investigators also were able to identify a small fraction of BDCA-1+ CD1a− DCs that do not display a LC-like phenotypic appearance, and which therefore are conventional BDCA-1+ CD14− DCs. When comparing the BDCA-1+ and BDCA-3+ DC subsets, BDCA-1+ DCs show a stronger ability to stimulate T-cell proliferation, although it should be noted that both myeloid DC subsets are stronger stimulators compared to BDCA-2+ pDCs.4
F. Potential DC Subsets of the Human Airways
Using BAL, Ten Berge et al.108 were the first to look at the specific microenvironment of the lung airways, with the goal of trying to separate pDCs and myeloid DCs, which include both BDCA-1+ and BDCA-3+ DCs. With the basic discrimination schema of pDCs being CD123+ CD11c− HLA-DR+ cells and myeloid DCs being low-autofluorescence CD11c+ HLA-DR+ cells, they were able to show that both cell types can be identified in lavage samples and further showed that pooled myeloid cells can be obtained in quantities useful for functional assays. As expected, the majority of cells in BAL samples were easy to separate as high-autofluorescence AMs.
In MLRs, the pooled myeloid DCs stimulate CD4+ T helper cell proliferation at ratios as low as 1 APC to 20 T-cells. If the researchers pre-exposed myeloid DCs to lipopolysaccharide (LPS), they found that proliferation dramatically increased. The specific cytokines stimulated by the T-cells included a Th2-skewed pattern of IL-4, IL-5, and IL-13 expression. A similar pattern has been observed in MLRs involving epidermal LCs and CD1a+ dermal DCs found in human skin.65 Thus, while Ten Berge et al.108 were looking only for myeloid DCs, they would have included four potentially separate DC subsets: BDCA-1+, BDCA-3+, CD1a+, and Langerin+ DCs. van Haarst et al.117 identified a very small subset of CD1a+ DCs via BAL that trigger a high level of CD4+ T helper cell proliferation. So it is possible that these CD1a+ DCs are the major stimulant of the pooled myeloid DCs tested by Ten Berge et al.108 Unfortunately, van Haarst et al.117 did not use Langerin to further determine if the CD1a+ DCs are separate from Langerin+ DCs.
More recent work has actually shown that Langerin+ DCs of the human lung are definitively not equivalent to epidermal LCs but are closely related to BDCA-1+ Langerin− conventional DCs.116 These lung Langerin+ DCs express far lower levels of both Langerin and CD1a compared to true LCs, and have their counterparts in various human tissues, including dermis, liver, and tonsils.116,118 LCs are also likely functionally specialized compared to Langerin+ DCs. LCs are unique to the epidermal layer of human skin (as opposed to the dermis for Langerin+ DCs), and are identified by their high expression levels of both CD1a and Langerin, a C-type lectin.65,119,120 LCs can be further identified by rod-shaped structures in their cytoplasm called Birbeck granules (BGs).121 Langerin is expressed on the LC surface but is expressed at much higher levels in the LC BGs.121 Langerin specifically binds to sugar residues122; this binding likely plays a functional role in LCs being able to bind portions of various types of pathogens that may be encountered at the skin surface. Interestingly, Langerin+ DCs do not contain BGs, which may provide future clues as to the functionally distinct role of this DC subset in various tissue types.
Tsoumakidou et al.107 measured subsets of BDCA-1+, BDCA-2+, and BDCA-3+ DCs in BAL fluid from healthy volunteers, but they did not use CD1a or Langerin to further discriminate DCs subsets. Based on work by Segura et al.44 in human skin, all BDCA-1+ DCs fit the classification of either LCs from the epidermis or CD1a+ DCs from the dermis. The CD1a+ dermal DCs indeed also include Langerin+ DCs discussed earlier.116,118 Desch et al.6 actually identified three separate extravascular BDCA-1+ subsets from digestion of whole human lung: CD1a+ CD206−, CD1a+ CD206+, and CD1a− CD206+. However, of these subsets, only the CD1a+ CD206− cells exhibited the classical DC characteristics of dendritic extensions and lamellipodial movement. As such, they were identified as “pulmonary DCs” while the latter two subsets were distinguished as “monocyte-derived” based on morphology and CD206 positivity. It is thus likely that Langerin+ DCs of the human lung8,116 are a subset of the CD1a+ CD206− DCs identified by Desch et al.6 rather than monocyte-derived CD1a+ CD206+ cells.
Using the methodology employed by Desch et al., it was determined that a CD1a+ BDCA-1− DC subset does not exist in the lung. This finding agrees with the recent report by Baharom et al.7 that less than 20% of BDCA-1+ DCs expressed CD1a in BAL fluid from healthy volunteers. Additionally, even BDCA-1 and CD1a expression together were not sufficient to completely distinguish all DC subsets resident in the lung. Inclusion of CD206, or the man-nose receptor, did create another level of DC subset separation whereby CD206 positivity indicated monocyte-derived cells that had extravasated from the blood.6,7 However, CD206 expression has been observed on inflammatory DCs as well.123 Thus, it remains to be conclusively determined whether the two CD206+ cell subsets (CD1a+ and CD1a−) are resident in the lungs (specifically the airways) under true steady-state conditions, and if they are, what their function may be.
We used surface Langerin expression to separate a specific DC subset of the airways from two subsets negative for Langerin: BDCA-1+ CD14+ DCs and BDCA-1+ CD14− DCs.8 Further measurement of CD1a on these subsets showed that, while it is expressed on the surface of a large fraction (~75%) of Langerin+ DCs, it is not uniformly expressed on all of these cells. Of the other two DC subsets, approximately 60% express surface CD1a. These results suggest that (1) CD1a coexpression is not a defining characteristic of all Langerin+ DCs; (2) Langerin+ DCs, with or without CD1a expression, can be separated from BDCA-1+ CD14+ and BDCA-1+ CD14− DCs; and (3) BDCA-1 is not the only surface marker that identifies DC subsets of the lung. It is possible that further examination will reveal that the Langerin+ CD1a+ subset we examined has characteristics of LCs of the skin whereas the Langerin+ CD1a− DCs have characteristics of Langerin+ DCs of the dermis.65,116
Indeed, work by Bigley et al.116 determined that Langerin is not a specific marker for LCs in humans. They found that true LCs of the epidermis express very high levels of both Langerin and CD1a, while Langerin+ DCs of the dermis express both but at lower levels. A Langerin+ DC subset was also observed by this group in lung digests, but equivalent LCs were not seen. In agreement with these results, we did not identify any Langerinhi CD1ahi cells characteristic of LCs from human airways.8 However, although Bigley et al.116 did not observe a Langerin+ CD1a− subset from tissue digestion, we did observe such a population, though quite small, from fresh BAL.8 We suggest that the harshness of tissue digestion may promote surface expression of CD1a on these Langerin+ DCs but that BAL limits this procedural stimulation, rendering surface CD1a levels low to intermediate, not high as observed on true LCs.116 Similar Langerin+ cells have also been isolated from human tonsils and characterized as closely related to BDCA-1+ DCs but distinct from LCs of the epidermis.118 Future studies will need to determine how surface expression of these many markers, including BDCA, Langerin, CD1a, and CD123 relate to individual DC subsets in human airways, and how the microenvironments of the airways (and possibly the interstitium) affect their expression levels.
G. Correlating DC Subsets with Airway Anatomy
DC subsets of the airways have also been investigated based on their anatomical locations. Todate et al.101 used histology to examine the mucosal lining and submucosal layers of human bronchioles. The researchers were able to identify distinct CD1a+ and BDCA-1+ DCs embedded in the bronchial epithelium in similar frequencies. However, in the underlying submucosa, BDCA-1+ DCs were found to be far more numerous than CD1a+ DCs. The equal presence of both DC subsets in the epithelial lining suggests functional specialization of both, although we cannot conclude merely by their location that CD1a+ DCs in bronchioles are equivalent to LCs in the epidermis. Baharom et al.7 collected separate lavage fractions of the bronchial and bronchoalveolar regions of healthy volunteers and observed CD1a expression only on a small percentage of BDCA-1+ DCs.
The discrepancies between the two studies may derive from the methods of DC identification rather than the markers used. Histology alone, as used by Todate et al.,101 is not sensitive to low levels of BDCA-1 expression, particularly when trying to determine coexpression. Conversely, Baharom et al.7 used flow cytometry for detection of BDCA-1 and CD1a, which is far more sensitive in detecting low levels of surface marker expression. With the more sensitive assay, Baharom et al.7 were likely correct in their assessment of BDCA-1+ CD1a− and BDCA-1+ CD1a+ DC subsets from lavage, making their frequencies similar to those observed by Todate et al.101 Additionally, BDCA-1+ CD1a− DCs in and along the alveolar epithelium may be dissociated readily by BAL compared to BDCA-1+ CD1a+ DCs that are mainly located under the epithelium. This means that BDCA-1+ CD1a− and BDCA-1+ CD1a+ DCs identified by Baharom et al.7 are likely equivalent to “CD1a− monocyte–derived cells” and “pulmonary DCs,” respectively, as described by Desch et al.6
We acknowledge that our whole-lung lavage may have acquired mononuclear phagocytes from both bronchial and bronchoalveolar regions, although results were comparable, as far as subset frequencies, to results in BAL fluid obtained purely from the lower regions of the lungs.8 Thus, while the frequencies of our identified phagocyte subsets likely exist along the entire lower airways, the bronchial contribution is not that different from the subsets found in the alveolar regions. Staining of lung slices from different regions of the lung, as we have performed for other lung cell types,36 should be conducted to clarify this issue.
A recent study developed a protocol to identify BDCA-1+ DCs, BDCA-3+ DCs, and CD123+ pDCs from endobronchial biopsies of healthy volunteers.124 Cell frequencies were determined by the location of the biopsies, immunohistochemical staining of tissue, and digestion of the tissue samples for flow cytometry. However, alveolar regions cannot be sampled in such a procedure, while they can be sampled with our techniques using whole human lungs and staining of intact lung slices for identification and localization of phagocyte subsets.8,36
H. Transcriptional Profiling to Identify Lung DC Subsets
Finally, DC subsets of the lung cannot be identified and characterized by surface marker expression or their results in MLRs alone. For this reason, researchers have begun comparing DC subsets based on partial or complete transcriptional profiling.6,79,85,95 Haniffa et al.85 moved the DC field forward by first comparing skin DC subsets to those isolated from other organs, including the lung. While they focused on BDCA-3high DCs isolated from skin and lung tissue digests, they extended this work by identifying a DEG list specific to these cross-presenting cells that included the genes XCR1,84 CADMI,125 and CLEC9A.80 Desch et al.6 were not able to observe CLEC9A on any extravascular lung cell type expressing BDCA-3, regardless of expression levels. In agreement with these findings, we were unable to measure high BDCA-3 or CLEC9A expression on Langerin+, BDCA-1+ CD14+, or BDCA-1+ CD14− DCs isolated from BAL of healthy volunteers or of whole donor lungs.8 Thus, it is conceivable that Haniffa et al.85 had actually identified a signature set of genes for BDCA-3high cells from the blood rather than the lung interstitium or airways.
Langerin is expressed on a fraction of BDCA-1+ DCs of the lung,116 and we were able to further demonstrate that Langerin+ DCs can be separated from conventional BDCA-1+ CD14− and BDCA-1+ CD14+ DC subsets of the airways that lack this marker.8 Our analysis based on comparative transcriptomics identifies Langerin as a highly specific gene expressed by Langerin+ DCs.8 Therefore, while Langerin+ DCs also express BDCA-1, the converse is not true of all BDCA-1+ DCs, which emphasizes the importance of Langerin as a marker for DC separation in the airways. Interestingly, mRNA levels of Langerin were high in two separate DC subsets identified by Desch et al.: BDCA1+ CD1a+ CD206+ DCs and BDCA1+ CD1a+ CD206− DCs.6 While we only observed high levels of Langerin mRNA in our Langerin+ DCs, we did not attempt to further separate this population based on BDCA-1 or CD206 expression.8 Therefore, our Langerin+ DC subset, of which a substantial proportion was CD1a+, may indeed include both CD206+ and CD206− cells.
Although we were able to determine that Langerin RNA is not highly expressed in BDCA-1+ CD14− or BDCA-1+ CD14+ DCs of the airways, these were relative comparisons based on microar-rays rather than absolute values from RNA-seq. As Bigley et al.116 showed, blood BDCA-1+ CD14− DCs do not express surface Langerin immediately after isolation but can gain expression within 18 hours if cultured with medium containing serum or TGF-β. This rapid induction of surface Langerin suggests that the RNA is made at some level constantly in BDCA-1+ DCs, and agrees with findings by our group8 and others,116,118 suggesting a close relationship between Langerin+ and BDCA-1+ CD14− DCs in various tissues.
IV. HUMAN MACROPHAGES
A. Introduction to Lung MΦs and Their Phenotypic Plasticity
Macrophages (MΦs) are considered important for removal of dead cells and debris in the periphery in the steady state and also removal after injury or infection. They are generally regarded as nonmigra-tory and poor at antigen presentation. MΦs should be included when discussing DC functions due to their continuous interactions with DC subsets in the periphery. MΦs, like DCs, are found in various human peripheral tissues, including the skin94,114 and lung.1,5 Having previously described characterization of human DC subsets in the lung, we focus on this site overall for investigating MΦ subsets. Throughout our discussion of MΦ subsets, we observe the same future needs of incorporating micro-environmental differences into studies, as proposed for furthering DC research.
MΦs are traditionally thought of as sentinel cells, protecting the lung from bacterial, viral, and fungal infections. While lung-resident MΦs are indeed capable of rapid inflammatory responses to microbial threats, they are also vital for resolution of inflammation with minimal airway damage once these threats have been eliminated. MΦs carry out other functions, including clearance of dead cells, wound healing, and overall maintenance of homeostasis in the steady state through their interactions with other cell types (such as epithelial cells and DCs). The extent of MΦs’ capacities indicates that plasticity likely exists in this overall cell type so as to allow rapid response to a broad range of proinflammatory and inhibitory stimuli. Indeed, Stein et al.,126 working with murine models, suggested that MΦs can take on distinct pro- or anti-inflammatory phenotypes depending on exposure to IFN-γ or IL-4. Since these initial studies, the two phenotypes have been termed M1 (classically activated) and M2 (alternatively activated), respectively.127
Several groups have attempted to further split and/or reclassify M1 and M2 MΦs to encompass TLR signaling, endogenous cytokine/chemokine expression in different microenvironments, amino acid metabolism, and branches of the adaptive immune system.127–129 However, most of these investigations have been restricted to mice and thus their relevance to human tissue MΦ identification and function remains to be determined. Shaykhiev et al.130 performed whole-genome transcriptional profiling of healthy human alveolar MΦs (AMs) acquired via BAL to examine their activation states. Because the researchers’ goal was to elicit activation skewing caused by smoking, however, their study focused only on the few genes specifically induced by M1 or M2 stimuli (but not both) based on previous murine studies and limited work on human monocytes. Interestingly, these restricted gene sets implied that AMs are slightly M1-polarized at the transcriptional level, suggesting that, although steady-state AMs reside in an anti-inflammatory environment containing TGF-β and IL-1031,32 and contribute to tolerance by regulating DC antigen presentation33,34 and stimulating regulatory T-cells,35 they remain poised to shift toward an M1 phenotype if exposed to proinflammatory stimuli.
Future studies should take into account the many genes that are up- or downregulated by M1 and M2 stimuli, as the steady-state airways likely contain a milieu of both types at varying levels. For example, exposure of human AMs to IL-4 alone increases their production of the M2-related chemokines CCL17 and CCL22, but the addition of IL-10 inhibits this effect.131 The complex micro-environments of the human lung under steady-state conditions likely prevent distinct separation of tissue-resident MΦ subtypes based on specific M1 or M2 activation states, meaning that cells from this site may show mixed phenotypes along a spectrum in the absence of disease. Dysregulation of tissue MΦs has been linked to various diseases, including atopic dermatitis,132 psoriasis,133 lung cancer,134 COPD,135 and tuberculosis.136 Being able to identify and characterize steady-state MΦ subsets in human tissues will thus provide a foundation for determining their protective or harmful contributions to various disease states.
B. Strategies for Characterizing Human Lung MΦ Subsets by Functional Properties
Human lungs are known to contain two major subsets of MΦs: larger AMs found in the alveoli and smaller but more phenotypically heterogeneous interstitial MΦs (IMs) found in the parenchymal spaces outside of the airways and alveoli (Fig. 2).137,138 It should be noted that Gibbings et al.139 recently identified three subsets of IMs from murine lung digestions based on unique surface marker and gene expression profiles. In the future, equivalent subsets may be isolated from human lungs as well. Morphologically, AMs have a lower nuclear–cytoplasmic ratio and more euchromatin (versus heterochromatin) compared to IMs.137 Between these two subsets, most of our current knowledge relates to AMs, as they are the predominant cell type in BAL fluid. For this reason, BAL in healthy volunteers provides a technique, free of surgical procedures, in which AMs can be isolated in numbers practical for functional and transcriptional analyses (unlike rare DC subsets).
It should be mentioned that the ratio of AMs to alveoli is actually quite low, estimated to be one to three.140 Therefore, the large number of AMs acquired by BAL are the result of the millions of alveoli13 responsible for gas exchange in the lower airways of human lungs. We can also infer that AMs likely transit constantly between groups of alveoli in the human lung via the pores of Kohn17,141 to engulf inhaled particulates and microbes in the steady state. Physiologically, AMs must be rare within the alveolar spaces so as to not impair gas exchange with the capillary network.
IMs from human lungs are a more difficult target of investigation, largely due to their heterogeneity and their similarity to AMs. Lung resections can be lavaged for AM isolation, followed by tissue digestion for collection of IMs.137,138 Since human blood does not contain MΦs, digestion of lung re-sections can yield large numbers of this general cell type free of vascular contamination. However, as described for studies of DC subsets, lung resections are performed for underlying conditions that likely alter MΦ populations from their true steady-state phenotypes. For example, AMs collected from BAL fluid of patients with varying types of lung cancer showed distinct reductions in phagocytosis of polystyrene beads and proinflammatory cytokine secretion in response to LPS (as measured by levels of TNF-α, IL-1, and IL-6) when compared to healthy controls.134 Therefore, functional differences between human AMs and IMs may be confounded by diseases of the lung that affect these two MΦ subsets to varying degrees.
Limited studies attempting to compare human AMs and IMs have begun with cultures enriched for adherent cells.137,138 Intrinsic adherence by lung MΦs142 provides a means of washing away nonadherent lymphocytes and red blood cells from BAL or digest preparations. However, considering that AMs likely transit between alveoli in vivo, this adherent property in vitro may reflect activation during the isolation process, response to culture conditions, or a combination of both. AMs freshly isolated by BAL show a baseline proinflammatory transcriptional profile immediately after collection that decreases after 24 hours in culture.143 Thus, while Fathi et al.137 showed that AMs possess a greater potential for phagocytosis of yeast particles compared to IMs, they also perform their work within a 24-hour window postisolation. It is possible that had Fathi et al. allowed the MΦs to return to a true resting state, the differences would have diminished. More recently, Hoppstädter et al.138 observed comparable internalization levels of fluorescent beads between the two subsets after unstimulated culture for 72 hours.
Culture conditions, such as rigidity of the plating surface, can also skew measurements of phagocytosis by adherent MΦs. Standard tissue culture plates are rigid and promote adhesion. The steady-state microenvironment of the alveoli experiences constant perturbations based on fluctuating oxygen tension and a broad spectrum of inhaled particulates. However, physical perturbations are also constantly occurring as low-rigidity alveoli stretch and shrink during respiration. In vivo, a thin layer of surfactant coats the inner alveolar surfaces (Fig. 2). Akei et al.144 demonstrated that specific surfactant proteins decrease the overall surface tension of this film and thus allow AMs to reside below the surface of this layer but remain in a more globular (or less flattened) shape. They further showed that this rounded shape of AMs in mice correlates with increased phagocytosis of intratracheally instilled microbeads.
Other researchers have observed that lower rigidity substrates, comparable to that of human lung, allow more rounding of human AMs.145 Yet rhythmic stretching of AMs, as encountered in alveoli, decreases both cell elasticity and phagocytic potential.145 It is likely that, in vivo, a combination of surface tension, low rigidity of alveolar epithelia, and cyclical AM stretching determine the overall phagocytic ability of steady-state AMs for nonpathogenic particles. We suggest that AMs and IMs are initially activated in a proinflammatory manner during isolation, and that triggers strong adhesion to highly rigid tissue culture dishes on which most MΦ experiments are performed. Thus, MΦs remain stationary and in a more flattened form in vitro in the absence of rhythmic stretching. Without more focus on replicating in vivo microenvironments of the alveolar and parenchymal regions, differences measured between AMs and IMs, such as phagocytic ability or cytokine production, may not accurately convey their true functional phenotypes. Importantly, freshly isolated MΦs must be allowed to return to a resting condition prior to exposure to external stimuli. Standardization of isolation procedures and culture times for human lung MΦs can also help delineate functional differences between MΦ subsets.
Based on their higher frequency in the alveoli, AMs are expected to have a temporal advantage over DCs in interaction with inhaled particles. However, unlike peripheral DCs, human lung–resident MΦs are generally accepted as nonmigratory, performing most of their responsibilities in and around the airways. In murine models, the migratory potential of AMs to drain LNs in the steady state is unclear.146,147 While Kirby et al.147 found a small number of high-autofluorescence cells fitting the surface marker description of AMs in the regional LNs of mice, these cells did not express CCR7. Intranasal exposure to Streptococcus pneumoniae led to increased MΦ-like cells, containing bacteria, in the draining LNs. Yet these cells were not observed to express CCR7, even 24 hours postexposure.
Multiple physiological occurrences explain such contrasting results. It is possible that the cells identified in the LNs do not originate from the airways but rather are IMs that acquire bacteria passing from the apical to the basolateral sides of the alveolar epithelia via transcytosis. Additionally, signifi-cant migration of AMs and/or IMs may be pathogen dependent, where only certain host-pathogen interactions drive MΦ migration. Finally, if lung MΦs do migrate, the process may be via an uncharacterized mechanism that is not CCR7-dependent. How murine studies correlate with findings in humans is still unclear. Desch et al.6 surveyed steady-state phagocytic mononuclear cells, including MΦs, present in the mediastinal LNs of whole human lungs. There they only found DCs, specifically BDCA-3+ CD14+ and BDCA-1+ CD206− DCs, matching cell subsets seen in the lungs based on surface marker expression. Of these two DC subsets, only the BDCA-1+ CD206− DCs were shown to express CCR7, which implies that the BDCA-3+ CD14+ DCs either are resident in the LNs or migrate there via a CCR7-independent mechanism.
We did not observe upregulation of CCR7 in AMs after exposure to heat-killed Escherichia coli or Staphylococcus aureus.8 A lack of LN-directed migration by human tissue MΦs is not restricted to the lung, as dermal MΦs also do not migrate out of skin explants or upregulate CCR7, even after maturation with various proinflammatory cytokines.114 If lung MΦ subsets do not possess migratory capacity, this property can be used in future studies to discriminate them from DC subsets that are known to home to draining LNs under steady-state and inflammatory conditions.
With the significant uncertainty as to whether tissue MΦ migrate, the physiological relevance of measuring their stimulatory capacity in MLRs is questionable. However, AMs and IMs do interact with steady-state migratory DCs and also T-cells migrating to the airways during infection. These interactions must be tightly regulated to minimize tissue damage during inflammation, and more importantly to maintain tolerance to innocuous particles in the steady state. While they do express HLA-DR and present antigens, lung MΦs as a whole are weaker at stimulating T-cells compared to lung DCs,106,148,149 likely relating to this induction and preservation of tolerance. In addition to direct binding of the T-cell receptor to an MHC complex, APC-dependent cytokine signaling and costimulatory molecules dictate the strength and direction of T-cell responses. Lipscomb et al.149 demonstrated that AMs secrete low levels of a proinflammatory cytokine signal, as measured by IL-1 production after antigen exposure. However, cytokine production did not completely explain the weak T-cell activation phenomenon, as addition of exogenous cytokine did not dramatically increase T-cell proliferation. In a follow-up study, Lyons et al.150 observed that AMs express a low density of the integrin leukocyte function-associated antigen-1 (LFA-1) important for initial binding of APCs to T-cells. Finally, AMs express minimal levels of the costimulatory molecules CD80 and CD86, even after activation with IFN-γ.151
We have observed minimal upregulation of CD86 on AMs, even after exposure to heat-killed bacteria.8 APC–T-cell interactions without costimulation result in anergic T-cells and the subsequent development of peripheral tolerance. Blumenthal et al.152 further implicated physical interaction of AMs with T-cells as causing this effect, as peripheral blood monocytes strongly stimulated proliferation of CD4+ T-cells, and this response was not inhibited by mixing of AMs one to one with monocytes prior to MLRs. They also verified that these T-cells were not skewed towards a Th1 or Th2 response without proliferation, as they produced very low levels of IFN-γ and IL-4 in the presence of AMs. These results collectively suggest that AMs are indeed weak stimulators of T-cells, but that they do not confer a suppressive phenotype onto other APCs (like DCs) that would also interact with T-cells. However, the latter idea would need verification by co-culturing human AMs (or IMs) with the various lung DC subsets described earlier.
C. Strategies for Characterizing Human Lung MΦ Subsets by Origins and Specific Markers
Although MΦs were originally thought to be replenished by blood monocytes in steady-state conditions,153 this idea was based on monocytes having the potential to become MΦs in vitro under certain culture conditions or in vivo after whole-animal irradiation.154,155 Only recently have parabiotic studies in mice suggested that many tissue-resident MΦ populations, including microglia and AMs, are actually seeded early in embryonic development and are then maintained in the steady state by self-renewal within the tissue itself156–159 rather than by external replenishment by hematopoietic stem cells.160 This local renewal of some lung MΦ populations likely occurs in humans as well, as is supported by Nayak et al.161 and Eguíluz-Gracia et al.,162 who in their study of lung transplant patients found that the majority of AMs remain of donor origin years after surgery. It remains to be seen whether steady-state IMs are actually the precursor cells of AMs, whether each subset self-renews independently, or whether IMs self-renew at all.
Jakubzick et al.163 demonstrated that, at least in mice, monocytes in the steady state can enter the lung tissue from blood circulation without differentiating into DCs or MΦs. Instead, monocytes upregulate MHCII while crossing the endothelium, acquire antigen in the periphery, and migrate back to draining LNs without acquiring any phenotypic characteristics of DCs or MΦs.163 As expected, migrating DCs far outnumber monocytes in LNs, but Jakubzick et al.163 demonstrated that monocytes do not automatically become DCs or MΦs by entering the lungs. These findings do not exclude monocytes from being the natural source of IMs in the steady state. Differentiation of monocytes to IMs may occur at a low basal rate but then be strongly induced under inflammatory conditions in which resident IMs (as well as AMs) are depleted.
Guilliams and Scott164 proposed that the population(s) of tissue-resident MΦs in most tissues are replenished based on “niche competition.” According to this model, a certain frequency of MΦs, such as AMs and IMs, exist in a given tissue. As long as that niche for that cell type is filled, monocytes can enter and exit the tissue without differentiating into MΦs. However, if slots in the niche become vacant, as can occur during infection and inflammation, then remaining resident MΦs and infiltrating monocytes can rapidly replenish the niche frequency. In this case, microenvironmental signals allow monocytes to differentiate into self-renewing tissue macrophages. In the human lung, the interstitial spaces and the alveoli present two distinct niches. The niche competition model is complicated by the fact that a physical barrier exists at the alveolar epithelium and breakdown of this barrier during inflammation may also contribute to monocyte differentiation into AMs during tissue repair.
It is possible that a subset of IMs renews AMs in the steady state, but during inflammation monocytes also directly differentiate into AMs. The other possibility involves depletion of the IM niche first during inflammation. Monocytes would first exit the blood to differentiate into IMs until that niche is filled, followed by IM migration into the alveoli to replenish the AM niche. Finally, once both niches are refilled, monocytes no longer differentiate into either type of resident macrophage. Whether any of the true DC subsets discussed follow a similar means of replenishment based on the niche model is unknown and will be an active area of future study.
Unlike MΦs, DCs are thought to regularly migrate to draining LNs, even without exposure to antigens.165 Therefore, they need constant replenishment, even in the absence of tissue injury or infection. Monocytes may regularly enter the lungs, including the airways, and have the potential to differentiate into both DCs and MΦ subsets depending on vacancies in their respective niche and subsequent microenvironmental signals. Molawi et al.166 observed in mice that, while embryonic stem cells seed the resident macrophage population of the heart early in life, monocytes progressively contribute more to this population with increasing age. It is entirely possible that a similar transition from embryonic to bone marrow–derived macrophage sources occurs in human lungs, as embryonically derived resident macrophages (AMs or IMs) lose their ability to self-renew as the late life stages are reached. In humans, CD14+ DCs described previously may be the equivalent cell subset to these constant-turnover monocytes, but they are likely not the equivalent of IMs. Dermal CD14+ DCs show properties of both blood BDCA-1+ DCs and CD14+ monocytes.85 Complete transcriptional profiling of these dermal CD14+ DCs identifies them as closely related to dermal MΦs.94 We have determined that CD14+ DCs isolated from human airways are closely related to definite AMs but inversely related to members of the “DC-clade” isolated from human airways.8
The source cells for replenishment of steady-state dermal MΦs remain unclear in humans, although work in mice indicates that blood monocytes may contribute a significant fraction of the overall population.167 In contrast, studies of patients undergoing skin transplants indicate that only approximately 30% of dermal MΦs are of recipient origin in engraftments more than a year after the procedure.114 These results suggest that human dermal MΦs are long-lived, but they also imply that two subsets of dermal MΦs exist: one replenished by circulating monocytes and a second populated by self-renewal in the tissue.
As with DC subsets, identification of human lung MΦ subsets is likely confounded by the variety of surface markers used to separate them. MΦs as a general cell type can be identified by their high autofluorescence (AF) in mice and humans when analyzed by flow cytometry.106,168,169 While the main source of AF in MΦs seems to be phagolysosomes,168 the actual cellular fluorophores involved remain a mystery. AF can also be seen as a nuisance, as it can mask actual marker signals if the marker’s expression is low. For this reason, MΦs are often excluded from analyses of DCs by collecting only nonadherent cells106 or initially gating outside-scatterhigh (high-granularity), forward-scatterhigh (large) AFhigh cells by flow cytometry.7 Another concern is that, while AMs are high in AF, IMs are more heterogeneous in their AF levels.7
Studies have actually revealed three distinct subsets of IMs in the mouse lung,139 and it is yet to be determined whether equivalent subsets exist in the human lung. Several groups have worked to develop sets of surface markers that can specifically identify human AMs, IMs, and lung-immigrating monocytes,6,7,170,171 although the three cell subsets have not been investigated in depth concurrently. Desch et al.6 analyzed all HLA-DR+ extravascular cells from digestion of healthy lung and, aside from BDCA-1+ cells, only identified SSChigh AMs and a subset of SSCint cells that the investigators termed “tissue monocytes.” There are two possible explanations for this observation (of a lack of IM). First, BDCA-1 positivity may not automatically indicate a DC subset, and thus what Desch et al.6 called “CD1a− monocyte-derived cells” actually are or at least include IMs. Second, IMs were pooled together with the identified TMs as a single cell subset when in fact two separate subsets were present. Interestingly, both AMs and TMs were shown to express the C-type lectin CD206 (mannose receptor) and the Fc receptor CD64 (FcγRI), but could be differentiated by expression of the low-affinity Fc receptor CD16 specifically found on TMs.6
In direct contrast, Yu et al.170 and Bharat et al.171 independently used expression of CD206 by human lung MΦs as a specific means of separation from monocyte subsets. They identified the sialic acid receptor CD169 (Siglec-1) as a marker that can separate AMs from IMs, while all TMs were identified as CD206− and CD169−. Another sialic acid receptor, CD33 (Siglec-3), has been used to identify human AMs in place of the integrin CD11c, but this marker is also expressed by TMs and eosinophils.172 Thus, CD11c or CD33 expression can be included as a myeloid cell marker, but does not specifically separate out MΦs.
It appears physiologically logical for AMs to express CD169, as this marker has been associated with a proinflammatory phenotype173 that AMs can acquire (i.e., M1 skewing) upon exposure to pathogenic microbes in the airways. Confocal microscopy visually supports Yu et al.’s170 classifications, as the cells they termed IMs were localized outside the alveoli (they did not try to localize TMs). If CD206 is selectively expressed by lung MΦs relative to expression on monocytes, then the majority of TMs analyzed by Desch et al.6 were actually IMs. However, Desch et al. showed that, while CD14+ monocytes from the blood are CD206−, all TMs, AMs, and monocyte-derived cells in the lung (both CD1a+ and CD1a−) are CD206+. These results suggest that CD206 expression is induced by migration across the endothelium and that CD206 expression does not separate TMs from IMs in the lung. Perhaps the upregulation of CD206 on TMs emigrating from the blood is not an immediate event so that crossing the endothelium does signal the upregulation of CD206, but this process can take days to occur. Levels of CD206 cannot separate AMs from IMs, as this marker is expressed along a continuum rather than distinctly bimodal.172
The studies just mentioned may be reconciled if we characterize IMs as HLA-DR+ CD11c/CD33+ CD206+ CD16+ CD169−, and characterize AMs as AFhigh HLA-DR+ CD11c/CD33+ CD206+ CD16− CD169+. Baharom et al.7 performed BAL on healthy volunteers and identified CD14+ CD16− “classical monocytes” and a second subset of CD14+ CD16+ “intermediate monocytes” in BAL fluid after AM exclusion. These results strongly support the existence of separate IMs and TM subsets considering IMs are not collected by BAL. Staining these two monocytic subsets found in the airways for CD206 and CD169 expression further substantiates these levels of phagocyte discrimination. Also, the CD14+ DCs we discussed previously can be split into two separate monocytic subsets based on CD16 positivity. Interestingly, CD16 has been described as a marker used to identify and separate “migratory monocytes” that can leave the circulation and enter tissue from “patrolling monocytes” that remain along the capillary walls.174 In this case, the migratory monocytes are CD14high CD16− and the patrolling monocytes are CD14low CD16+.
Considering the close proximity of alveolar blood capillaries and the alveolar epithelium, it is possible that migratory monocytes migrate straight out of the capillaries and into the alveolar spaces. They can also migrate straight out of the capillaries and into the interstitial spaces. At the same time, since the thin capillary walls are in direct contact with the thin epithelium, patrolling monocytes may be sentinels of both barriers. That would explain how Baharom et al.7 isolated both classical and intermediate monocytes from human airway lavage. With the likelihood of isolating patrolling monocytes from the lung during tissue digestion, CD16 would not aid in their discrimination from IMs.
The function of one or more subsets of TMs in the human airways under steady-state conditions remains unclear, although we speculate that TMs can fully enter alveolar spaces, acquire antigens, and migrate back out, while AMs cannot. Once out of the alveoli, TMs may transit to the mediastinal LNs under a different time frame compared to steady-state DCs, or they may promote IMs tolerance of particulates and debris in the interstitial spaces by direct interactions.
Reports of monocytes in healthy human airways7 suggest that, in the “niche competition” model described earlier, availability of slots dictates replenishment of AMs rather than accessibility of the alveoli. For future studies, surface markers for MΦ/monocyte subsets in the lung should be standardized. Additionally, AMs, IMs, and TMs will need to be analyzed together for the true scope of subsets present and their functional specialties to be determined. A summary diagram of the markers described for human lung MΦs and monocyte subset identification can be found in Fig. 3.
FIG. 3:
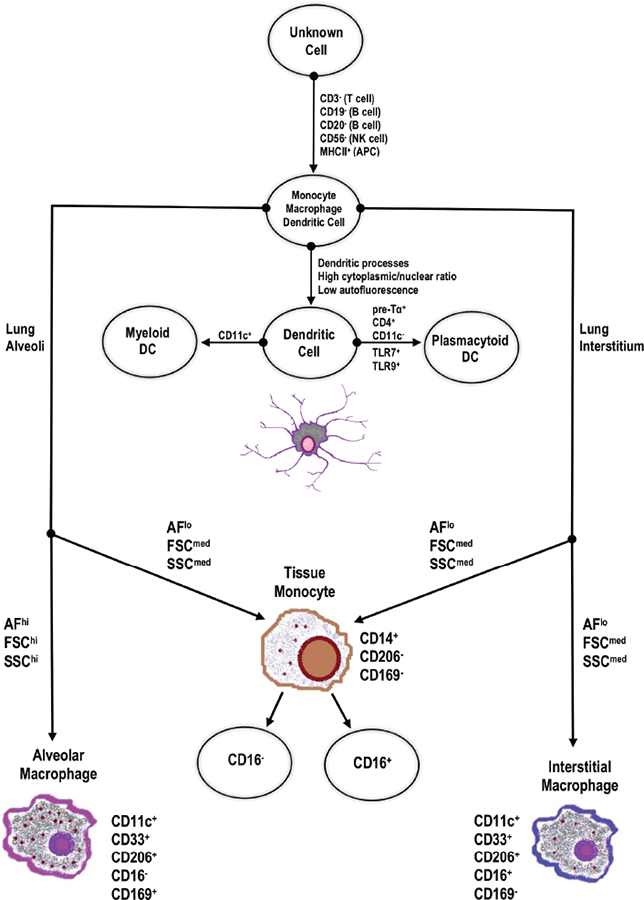
Summary diagram of stepwise human lung MΦs and monocyte subset identification, including markers and microenvironment. MHC, major histocompatibility complex; APC, antigen-presenting cell; AF, autofluorescence; FSC, forward scatter; SSC, side scatter.
D. Other Potential Human Lung MΦ Markers
Steady-state expression and activation-based induction of other surface markers, including toll-like receptors (TLRs), has been used to classify human lung MΦs. Specific expression of these pattern recognition receptors (PRRs) on AMs would be in line with their location in the alveolar spaces, where they are the first cells to encounter inhaled pathogens. However, TLR profiles are not specific to AMs or IMs, as there is high variability in expression between cell donors under both steady-state and inflammatory conditions.172 Hoppstädter et al.138 observed 3-fold higher expression of HLA-DR on IMs compared to AMs without stimulation, but did not observe any differences in mRNA levels of TLR 1–10. However, this group did not differentiate TMs from IMs, and thus could have been surveying AMs or a number of MΦ/monocyte subsets. Additionally, they did not measure TLR protein expression, so stimulation with TLR agonists may not have led to surface TLR upregulation. For example, separate studies have shown that less than 5% of AMs express surface TLR-2 and TLR-4 in the steady state, although the percentages increase slightly after 10 minutes of LPS exposure.175 While the researchers observed that LPS stimulation via TLR-4 led to increased production of TNF-α and IL-6 mRNA by both MΦ subsets, IMs produced less TNF-α and more IL-6 compared to AMs.
Baharom et al.7 also measured high TNF-α production by TMs isolated by BAL after exclusion of AFhigh AMs. Together, these results from this group and Hoppstädter et al.138 agree with the notion that phagocytic APCs, whether AMs or TMs, located within the alveoli are primed for faster proinflammatory responses compared to APC subsets in the interstitial spaces.
Asada et al.176 identified a transcription factor called peroxisome proliferator–activated receptor gamma (PPAR-γ), which is expressed at high levels by AMs and is vital for maintaining their anti-inflammatory phenotype in human airways. Whether this transcription factor is specific to AMs or is also expressed by TMs, IMs, or airway DCs is unknown. Expression of PPAR-γ may be specific to AMs and serve as another selective marker of this cell type, but it also may be inducible in multiple mononuclear phagocytes based on microenvironmental signals in the alveolar spaces. Cells like type I AECs likely stimulate expression of PPAR-γ by AMs in human airways; thus, without similar coculturing of AECs and AMs in vitro, expression levels of PPAR-γ decrease with time. We contend that a similar distinction may be seen between TMs in the two lung microenvironments. Whether identifying surface markers vary between TMs in the interstitium and those in the alveoli remains to be determined. Higher HLA-DR levels on IMs138 suggest that the function of this cell type involves constant interaction with any immigrating B- and T-cells in the interstitial spaces. Importantly, these interactions may be inhibitory due to poor T-cell stimulation, as discussed earlier regarding AMs.
V. CONCLUSIONS AND FUTURE DIRECTIONS
Over the past 30 years, researchers have progressed from determining that DCs and MΦs are distinct cell types to identifying numerous subsets of each in specific tissues. Such advances are especially evident in the human lung. Regardless of whether subsets are identified by comparing surface markers, MLR results, or transcriptional profiling, every study is a step forward in the field. Unfortunately, as we identify new markers for certain subsets, we may find it difficult to reconcile previous work with the most recent findings. Surface markers such as BDCA, CD1a, CD14, and CD206 are not specific to just one cell subset. Additionally, depending on the source material and means of cellular isolation, marker expression may rapidly increase or decrease. Methods to resolve these issues, in our opinion, will include additional emphasis on transcriptional profiling and hierarchical clustering. As we establish sets of DEGs for individual DC and MΦ subsets, we can rapidly determine correlative protein expression by methods such as flow cytometry.
Specific to the human lung, the difficulties in acquiring healthy tissue are clear and will continue to impose barriers to our understanding of resident DC and MΦ subsets. While some major developments have come from cells isolated from the digestion of surgical resections,3,103–105 these cells may be altered by the effects of diseased tissue nearby. Additionally, digestion itself is harsh on the cells of the lung, leading to their activation or causing changes in expression of identifying surface markers. Even with proper perfusion, contamination of resident lung cells with capillary cells is a concern unless the intravascular populations can be identified prior to digestion.6 Isolation of cells directly from the airways of healthy volunteers has shown promise, and several groups have isolated and characterized some DCs and MΦ subsets in this manner.107–109,117 These cells are acquired by minimal treatment and processing, and are free of vascular contamination. A drawback to this procedure is the lack of large numbers of cells.107 We have circumvented this issue by acquiring cells from whole donor lungs.8 Of course, this does not help the researcher obtain interstitial DC and MΦ subsets.
A major avenue of future research should be the replication of the microenvironments of the human airways and interstitium. Differences in microenvironments affect the marker expression profiles of DC and MΦ subsets as well as their responses to various tolerogenic or inflammatory stimuli. Instead of isolating individual DC and MΦ subsets and seeing what cytokines and chemokines they produce in response to certain antigens, more emphasis needs to be placed on how the subsets function together to maintain steady-state equilibrium in the microenvironments of the human lung. At the very least, subsets should be cocultured when in vitro assays are performed or, if possible, studied ex vivo using models like the precision-cut lung slice.36 In the alveolar regions of the lung, for example, DC subsets interact with both MΦ and monocyte subsets as well as with epithelial cells that make up the alveolar walls. Thus, studying a single cell subset in isolation may not be relevant to its behavior within the alveolar architecture.
Additionally, that a certain subset responds to a given stimulus in isolation does not mean it cannot respond in a different manner in the microenvironments of the lung. DC and MΦ subsets may express a fixed set of markers, but they have the potential to behave like other subsets if microenvironmental conditions change (i.e., during infection or in response to tissue damage). Plasticity may complicate subset studies, but it is nonetheless a physiological response vital for cellular responses to rapidly changing conditions in the airways or interstitium.
As we emphasize throughout this review, the microenvironments found in the human lung are unique and must be taken into account in future work to truly elucidate how DC and MΦ subsets protect the lung against constant environmental threats and allow it to carry on the respiration necessary for life. Physiological models of the human alveoli and underlying interstitium will be difficult to develop, but can begin with simple coculture systems in which two or more cell types are cultured together in close physical and chemical proximity.
ACKNOWLEDGMENTS
We would like to acknowledge the following funding sources for making this review possible: National Institutes of Health (Grant No. 5U19AI062629)–Mechanisms by Which B. anthracis Spores Escape the Lung (Jordan P. Metcalf); and U.S. Department of Veterans Affairs (Grant No. 5I01BX001937)–Antiviral Immunosuppression by Cigarette Smoke (Jordan P. Metcalf).
ABBREVIATIONS:
- AEC
alveolar epithelial cell
- AM
alveolar macrophage
- APC
antigen-presenting cell
- BAL
bronchoalveolar lavage
- BDCA
blood dendritic cell antigen
- DC
dendritic cell
- DEG
differentially expressed gene
- IM
interstitial macrophage
- LC
Langerhans cell
- LN
lymph node
- MΦ
macrophage
- MLR
mixed leukocyte reaction
- TM
tissue monocyte
REFERENCES
- 1.Krombach F, Gerlach JT, Padovan C, Burges A, Behr J, Beinert T, Vogelmeier C. Characterization and quantification of alveolar monocyte-like cells in human chronic inflammatory lung disease. Eur Respir J. 1996;9(5):984–91. [DOI] [PubMed] [Google Scholar]
- 2.Sertl K, Takemura T, Tschachler E, Ferrans VJ, Kaliner MA, Shevach EM. Dendritic cells with antigen-presenting capability reside in airway epithelium, lung parenchyma, and visceral pleura. J Exp Med. 1986;163(2):436–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demedts IK, Brusselle GG, Vermaelen KY, Pauwels RA. Identification and characterization of human pulmonary dendritic cells. Am J Respir Cell Mol Biol. 2005;32(3):177–84. [DOI] [PubMed] [Google Scholar]
- 4.Demedts IK, Bracke KR, Maes T, Joos GF, Brusselle GG. Different roles for human lung dendritic cell subsets in pulmonary immune defense mechanisms. Am J Respir Cell Mol Biol. 2006;35(3):387–93. [DOI] [PubMed] [Google Scholar]
- 5.Gant VA, Hamblin AS. Human bronchoalveolar macrophage heterogeneity demonstrated by histochemistry, surface markers and phagocytosis. Clin Exp Immunol. 1985;60(3):539–45. [PMC free article] [PubMed] [Google Scholar]
- 6.Desch AN, Gibbings SL, Goyal R, Kolde R, Bednarek J, Bruno T, Slansky JE, Jacobelli J, Mason R, Ito Y, Messier E, Randolph GJ, Prabagar M, Atif SM, Segura E, Xavier RJ, Bratton DL, Janssen WJ, Henson PM, Jakubzick CV. Flow cytometric analysis of mononuclear phagocytes in nondiseased human lung and lung-draining lymph nodes. Am J Respir Crit Care Med. 2016;193(6):614–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baharom F, Thomas S, Rankin G, Lepzien R, Pourazar J, Behndig AF, Ahlm C, Blomberg A, Smed-Sörensen A. Dendritic cells and monocytes with distinct inflammatory responses reside in lung mucosa of healthy humans. J Immunol. 2016;196(11):4498–509. [DOI] [PubMed] [Google Scholar]
- 8.Patel VI, Booth JL, Duggan ES, Cate S, White VL, Hutchings D, Kovats S, Burian DM, Dozmorov M, Metcalf JP. Transcriptional classification and functional characterization of human airway macrophage and dendritic cell subsets. J Immunol. 2017;198(3):1183–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horsfield K, Dart G, Olson DE, Filley GF, Cumming G. Models of the human bronchial tree. J Appl Physiol. 1971;31(2):207–17. [DOI] [PubMed] [Google Scholar]
- 10.Parker H, Horsfield K, Cumming G. Morphology of distal airways in the human lung. J Appl Physiol. 1971;31(3): 386–91. [DOI] [PubMed] [Google Scholar]
- 11.Pierce JA, Ebert RV. Fibrous network of the lung and its change with age. Thorax. 1965;20(5):469–76. [Google Scholar]
- 12.Burri PH. Development and growth of the human lung In: Comprehensive physiology. Hoboken, NJ: Wiley; 2011. [Google Scholar]
- 13.Ochs M, Nyengaard JR, Jung A, Knudsen L, Voigt M, Wahlers T, Richter J, Gundersen HJG. The number of alveoli in the human lung. Am J Respir Crit Care Med. 2004;169(1):120–24. [DOI] [PubMed] [Google Scholar]
- 14.Simionescu M Ultrastructural organization of the alveolar-capillary unit. Ciba Found Symp. 1980;78:113–6. [DOI] [PubMed] [Google Scholar]
- 15.Phelps DS, Floros J. Localization of surfactant protein synthesis in human lung by in situ hybridization. Am Rev Respir Dis. 1988;137(4):939–42. [DOI] [PubMed] [Google Scholar]
- 16.Schürch S Surface tension at low lung volumes: dependence on time and alveolar size. Respir Physiol. 1982;48(3): 339–55. [DOI] [PubMed] [Google Scholar]
- 17.Bastacky J, Goerke J. Pores of Kohn are filled in normal lungs: low-temperature scanning electron microscopy. J Appl Physiol. 1992;73(1):889–95. [DOI] [PubMed] [Google Scholar]
- 18.Takaro T, Parra SC, Peduzzi PN. Anatomical relationships between type II pneumonocytes and alveolar septal gaps in the human lung. Anat Rec. 1985;213(4):540–50. [DOI] [PubMed] [Google Scholar]
- 19.Cabrera-Rubio R, Garcia-Núñez M, Setó L, Antó JM, Moya A, Monsó E, Mira A. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J Clin Microbiol. 2012;50(11):3562–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184(8):957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charlson ES, Bittinger K, Chen J, Diamond JM, Li H, Collman RG, Bushman FD. Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PLoS One. 2012;7(9):e42786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN, Huffnagle GB. Analysis of culture-dependent versus culture-independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid. J Clin Microbiol. 2014;52(10):3605–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, Curtis JL. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12(6):821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, Martinez FJ, Huffnagle GB. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6(2):e16384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, Flores SC, Fontenot AP, Ghedin E, Huang L, Jablonski K, Kleerup E, Lynch SV, Sodergren E, Twigg H, Young VB, Bassis CM, Venkataraman A, Schmidt TM, Weinstock GM, Lung HIV Microbiome Project. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187(10):1067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt MF, Cookson WOC. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, Beck JM, Curtis JL, Huffnagle GB. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio. 2015;6(2):e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adami AJ, Cervantes JL. The microbiome at the pulmonary alveolar niche and its role in Mycobacterium tuberculosis infection. Tuberc. 2015;95(6):651–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burri PH. Fetal and postnatal development of the lung. Annu Rev Physiol. 1984;46:617–28. [DOI] [PubMed] [Google Scholar]
- 30.Yun Y, Srinivas G, Kuenzel S, Linnenbrink M, Alnahas S, Bruce KD, Steinhoff U, Baines JF, Schaible UE. Environmentally determined differences in the murine lung microbiota and their relation to alveolar architecture. PLoS One. 2014;9(12):e113466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonfield TL, Konstan MW, Burfeind P, Panuska JR, Hilliard JB, Berger M. Normal bronchial epithelial cells constitutively produce the anti-inflammatory cytokine interleukin-10, which is downregulated in cystic fibrosis. Am J Respir Cell Mol Biol. 1995;13(3):257–61. [DOI] [PubMed] [Google Scholar]
- 32.Mayer AK, Bartz H, Fey F, Schmidt LM, Dalpke AH. Airway epithelial cells modify immune responses by inducing an anti-inflammatory microenvironment. Eur J Immunol. 2008;38(6):1689–99. [DOI] [PubMed] [Google Scholar]
- 33.Holt PG, Oliver J, Bilyk N, McMenamin C, McMenamin PG, Kraal G, Thepen T. Downregulation of the antigen presenting cell function(s) of pulmonary dendritic cells in vivo by resident alveolar macrophages. J Exp Med. 1993;177(2):397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holt PG, Schon-Hegrad MA, Oliver J. MHC class II antigen-bearing dendritic cells in pulmonary tissues of the rat. Regulation of antigen presentation activity by endogenous macrophage populations. J Exp Med. 1988;167(2):262–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soroosh P, Doherty TA, Duan W, Mehta AK, Choi H, Adams YF, Mikulski Z, Khorram N, Rosenthal P, Broide DH, Croft M. Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J Exp Med. 2013;210(4):775–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Booth JL, Duggan ES, Patel VI, Langer M, Wu W, Braun A, Coggeshall KM, Metcalf JP. Bacillus anthracis spore movement does not require a carrier cell and is not affected by lethal toxin in human lung models. Microbes Infect. 2016;18(10):615–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lannes D, Monteiro AS, Toscano E, Cavalcanti A, Nascimento M, de Biasi P, Zamboni M. Transbronchial needle aspiration of hilar and mediastinal lymph nodes. Rev Port Pneumol. 2007;13(5):651–58. [DOI] [PubMed] [Google Scholar]
- 38.Guyton AC. A concept of negative interstitial pressure based on pressures in implanted perforated capsules. Circ Res. 1963;12:399–414. [DOI] [PubMed] [Google Scholar]
- 39.Elhay S, Casley-Smith JR. Mathematical model of the initial lymphatics. Microvasc Res. 1976;12(2):121–40. [DOI] [PubMed] [Google Scholar]
- 40.Benoit JN, Zawieja DC, Goodman AH, Granger HJ. Characterization of intact mesenteric lymphatic pump and its responsiveness to acute edemagenic stress. Am J Physiol. 1989;257(6 Pt 2):H2059–69. [DOI] [PubMed] [Google Scholar]
- 41.Kambouchner M, Bernaudin J-F. Intralobular pulmonary lymphatic distribution in normal human lung using D24–0 antipodoplanin immunostaining. J Histochem Cytochem. 2009;57(7):643–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S, Buck DW, Schmitz J. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol. 2000;165(11):6037–46. [DOI] [PubMed] [Google Scholar]
- 43.Richters CD, Hoekstra MJ, van Baare J, Du Pont JS, Hoefsmit EC, Kamperdijk EW. Isolation and characterization of migratory human skin dendritic cells. Clin Exp Immunol. 1994;98(2):330–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segura E, Valladeau-Guilemond J, Donnadieu M-H, Sastre-Garau X, Soumelis V, Amigorena S. Characterization of resident and migratory dendritic cells in human lymph nodes. J Exp Med. 2012;209(4):6536–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okamoto M, Furuichi S, Nishioka Y, Oshikawa T, Tano T, Ahmed SU, Takeda K, Akira S, Ryoma Y, Moriya Y, Saito M, Sone S, Sato M. Expression of toll-like receptor 4 on dendritic cells is significant for anticancer effect of dendritic cell-based immunotherapy in combination with an active component of OK-432, a streptococcal preparation. Cancer Res. 2004;64(15):5461–70. [DOI] [PubMed] [Google Scholar]
- 46.Kimura Y, Tsukada J, Tomoda T, Takahashi H, Imai K, Shimamura K, Sunamura M, Yonemitsu Y, Shimodaira S, Koido S, Homma S, Okamoto M. Clinical and immunologic evaluation of dendritic cell-based immuno-therapy in combination with gemcitabine and/or S-1 in patients with advanced pancreatic carcinoma. Pancreas. 2012;41(2):195–205. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi H, Okamoto M, Shimodaira S, Tsujitani S, Nagaya M, Ishidao T, Kishimoto J, Yonemitsu Y; DCVaccine Study Group at the Japan Society of Innovative Cell Therapy (J-SICT). Impact of dendritic cell vaccines pulsed with Wilms’ tumour-1 peptide antigen on the survival of patients with advanced non-small cell lung cancers. Eur J Cancer. 2013;49(4):852–59. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi M, Sakabe T, Abe H, Tanii M, Takahashi H, Chiba A, Yanagida E, Shibamoto Y, Ogasawara M, Tsujitani S, Koido S, Nagai K, Shimodaira S, Okamoto M, Yonemitsu Y, Suzuki N, Nagaya M; DC-Vaccine Study Group at the Japan Society of Innovative Cell Therapy (J-SICT). Dendritic cell-based immunotherapy targeting synthesized peptides for advanced biliary tract cancer. J Gastrointest Surg. 2013;17(9):1609–17. [DOI] [PubMed] [Google Scholar]
- 49.Sakai K, Shimodaira S, Maejima S, Udagawa N, Sano K, Higuchi Y, Koya T, Ochiai T, Koide M, Uehara S, Nakamura M, Sugiyama H, Yonemitsu Y, Okamoto M, Hongo K. Dendritic cell-based immunotherapy targeting Wilms’ tumor 1 in patients with recurrent malignant glioma. J Neurosurg. 2015;123(4):989–97. [DOI] [PubMed] [Google Scholar]
- 50.Brittingham KC, Ruthel G, Panchal RG, Fuller CL, Ribot WJ, Hoover TA, Young HA, Anderson AO, Bavari S. Dendritic cells endocytose Bacillus anthracis spores: implications for anthrax pathogenesis. J Immunol Baltim Md 1950. 2005;174(9):5545–52. [DOI] [PubMed] [Google Scholar]
- 51.Cleret A, Quesnel-Hellmann A, Vallon-Eberhard A, Verrier B, Jung S, Vidal D, Mathieu J, Tournier J-N. Lung dendritic cells rapidly mediate anthrax spore entry through the pulmonary route. J Immunol. 2007;178(12):7994–8001. [DOI] [PubMed] [Google Scholar]
- 52.Williams NL, Morris JL, Rush CM, Ketheesan N. Migration of dendritic cells facilitates systemic dissemination of Burkholderia pseudomallei. Infect Immun. 2014;82(10):4233–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Botts MR, Hull CM. Dueling in the lung: how Cryptococcus spores race the host for survival. Curr Opin Microbiol. 2010;13(4):437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Demedts IK, Bracke KR, Van Pottelberge G, Testelmans D, Verleden GM, Vermassen FE, Joos GF, Brusselle GG. Accumulation of dendritic cells and increased CCL20 levels in the airways of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2007;175(10):998–1005. [DOI] [PubMed] [Google Scholar]
- 55.Ding D, Mehta H, McCune WJ, Kaplan MJ. Aberrant phenotype and function of myeloid dendritic cells in systemic lupus erythematosus. J Immunol. 2006;177(9):5878–89. [DOI] [PubMed] [Google Scholar]
- 56.Hart DN. Dendritic cells: unique leukocyte populations which control the primary immune response. Blood. 1997;90(9):3245–87. [PubMed] [Google Scholar]
- 57.MacDonald KPA, Munster DJ, Clark GJ, Dzionek A, Schmitz J, Hart DNJ. Characterization of human blood dendritic cell subsets. Blood. 2002;100(13):4512–20. [DOI] [PubMed] [Google Scholar]
- 58.Tserel L, Runnel T, Kisand K, Pihlap M, Bakhoff L, Kolde R, Peterson H, Vilo J, Peterson P, Rebane A. MicroRNA expression profiles of human blood monocyte-derived dendritic cells and macrophages reveal miR-511 as putative positive regulator of Toll-like receptor 4. J Biol Chem. 2011;286(30):2648–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takenaka S, McCormick S, Safroneeva E, Xing Z, Gauldie J. Influence of the tissue microenvironment on Toll-like receptor expression by CD11c+ antigen-presenting cells isolated from mucosal tissues. Clin Vaccine Immunol CVI. 2009;16(11):1615–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J Exp Med. 1973;137(5): 1142–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hart DN, McKenzie JL. Isolation and characterization of human tonsil dendritic cells. J Exp Med. 1988; 168(1):157–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheicher C, Mehlig M, Zecher R, Reske K. Dendritic cells from mouse bone marrow: in vitro differentiation using low doses of recombinant granulocyte-macrophage colony-stimulating factor. J Immunol Methods. 1992; 154(2):253–64. [DOI] [PubMed] [Google Scholar]
- 63.Williams LA, Egner W, Hart DN. Isolation and function of human dendritic cells. Int Rev Cytol. 1994;153:41–103. [DOI] [PubMed] [Google Scholar]
- 64.Hogg N Human mononuclear phagocyte molecules and the use of monoclonal antibodies in their detection. Clin Exp Immunol. 1987;69(3):687–94. [PMC free article] [PubMed] [Google Scholar]
- 65.Klechevsky E, Morita R, Liu M, Cao Y, Coquery S, Thompson-Snipes L, Briere F, Chaussabel D, Zurawski G, Palucka AK, Reiter Y, Banchereau J, Ueno H. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity. 2008;29(3): 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rissoan MC, Soumelis V, Kadowaki N, Grouard G, Briere F, de Waal Malefyt R, Liu YJ. Reciprocal control of T helper cell and dendritic cell differentiation. Science. 1999;283(5405):1183–86. [DOI] [PubMed] [Google Scholar]
- 67.Bruno L, Res P, Dessing M, Cella M, Spits H. Identification of a committed T cell precursor population in adult human peripheral blood. J Exp Med. 1997;185(5):875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robinson SP, Patterson S, English N, Davies D, Knight SC, Reid CD. Human peripheral blood contains two distinct lineages of dendritic cells. Eur J Immunol. 1999; 29(9):2769–78. [DOI] [PubMed] [Google Scholar]
- 69.Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, Cella M, Lande R, Uzé G. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol. 2004;34(3):796–805. [DOI] [PubMed] [Google Scholar]
- 70.Cella M, Jarrossay D, Facchetti F, Alebardi O, Nakajima H, Lanzavecchia A, Colonna M. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat Med. 1999;5(8):919–23. [DOI] [PubMed] [Google Scholar]
- 71.Pellerin A, Otero K, Czerkowicz JM, Kerns HM, Shapiro RI, Ranger AM, Otipoby KL, Taylor FR, Cameron TO, Viney JL, Rabah D. Anti-BDCA2 monoclonal antibody inhibits plasmacytoid dendritic cell activation through Fc-dependent and Fc-independent mechanisms. EMBO Mol Med. 2015;7(4):464–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dzionek A, Sohma Y, Nagafune J, Cella M, Colonna M, Facchetti F, Günther G, Johnston I, Lanzavecchia A, Nagasaka T, Okada T, Vermi W, Winkels G, Yamamoto T, Zysk M, Yamaguchi Y, Schmitz J. BDCA-2, a novel plasmacytoid dendritic cell-specific type II C-type lectin, mediates antigen capture and is a potent inhibitor of interferon alpha/beta induction. J Exp Med. 2001;194(12):1823–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martín-Gayo E, Sierra-Filardi E, Corbí AL, Toribio ML. Plasmacytoid dendritic cells resident in human thymus drive natural Treg cell development. Blood. 2010; 115(26):5366–75. [DOI] [PubMed] [Google Scholar]
- 74.Ito T, Yang M, Wang Y-H, Lande R, Gregorio J, Perng OA, Qin X-F, Liu Y-J, Gilliet M. Plasmacytoid dendritic cells prime IL-10–producing T regulatory cells by inducible costimulator ligand. J Exp Med. 2007;204(1):105–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ten Berge B, Kleinjan A, Muskens F, Hammad H, Hoogsteden HC, Hendriks RW, Lambrecht BN, Van den Blink B. Evidence for local dendritic cell activation in pulmonary sarcoidosis. Respir Res. 2012;13:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cox K, North M, Burke M, Singhal H, Renton S, Aqel N, Islam S, Knight SC. Plasmacytoid dendritic cells (PDC) are the major DC subset innately producing cytokines in human lymph nodes. J Leukoc Biol. 2005;78(5):1142–52. [DOI] [PubMed] [Google Scholar]
- 77.Breton G, Zheng S, Valieris R, Tojal da Silva I, Satija R, Nussenzweig MC. Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs. J Exp Med. 2016;213(13):2861–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.See P, Dutertre C-A, Chen J, Günther P, McGovern N, Irac SE, Gunawan M, Beyer M, Händler K, Duan K, Sumatoh HRB, Ruffin N, Jouve M, Gea-Mallorquí E, Hennekam RCM, Lim T, Yip CC, Wen M, Malleret B, Low I, Shadan NB, Fen CFS, Tay A, Lum J, Zolezzi F, Larbi A, Poidinger M, Chan JKY, Chen Q, Rénia L, Haniffa M, Benaroch P, Schlitzer A, Schultze JL, Newell EW, Ginhoux F. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017;356(6342):eaag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, Low D, Ho AWS, See P, Shin A, Wasan PS, Hoeffel G, Malleret B, Heiseke A, Chew S, Jardine L, Purvis HA, Hilkens CMU, Tam J, Poidinger M, Stanley ER, Krug AB, Renia L, Sivasankar B, Ng LG, Collin M, Ricciardi-Castagnoli P, Honda K, Haniffa M, Ginhoux F. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38(5):970–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Poulin LF, Salio M, Griessinger E, Anjos-Afonso F, Craciun L, Chen J-L, Keller AM, Joffre O, Zelenay S, Nye E, Moine AL, Faure F, Donckier V, Sancho D, Cerundolo V, Bonnet D, Sousa CR. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8α+ dendritic cells. J Exp Med. 2010;207(6):1261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sköld M, Behar SM. The role of group 1 and group 2 CD1-restricted T cells in microbial immunity. Microbes Infect Inst Pasteur. 2005;7(3):544–51. [DOI] [PubMed] [Google Scholar]
- 82.Roy S, Ly D, Castro CD, Li N-S, Hawk AJ, Altman JD, Meredith SC, Piccirilli JA, Moody DB, Adams EJ. molecular analysis of lipid-reactive Vδ1 γδ T cells identified by CD1c tetramers. J Immunol. 2016;196(4):1933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sugita M, Cao X, Watts GFM, Rogers RA, Bonifacino JS, Brenner MB. Failure of trafficking and antigen presentation by CD1 in AP-3-deficient cells. Immunity. 2002;16(5):697–706. [DOI] [PubMed] [Google Scholar]
- 84.Crozat K, Guiton R, Contreras V, Feuillet V, Dutertre C-A, Ventre E, Vu Manh T-P, Baranek T, Storset AK, Marvel J, Boudinot P, Hosmalin A, Schwartz-Cornil I, Dalod M. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J Exp Med. 2010;207(6):1283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haniffa M, Shin A, Bigley V, McGovern N, Teo P, See P, Wasan PS, Wang X-N, Malinarich F, Malleret B, Larbi A, Tan P, Zhao H, Poidinger M, Pagan S, Cookson S, Dickinson R, Dimmick I, Jarrett RF, Renia L, Tam J, Song C, Connolly J, Chan JKY, Gehring A, Bertoletti A, Collin M, Ginhoux F. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 2012;37(1):60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sancho D, Joffre OP, Keller AM, Rogers NC, Martínez D, Hernanz-Falcón P, Rosewell I, Reis e Sousa C. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 2009;458(7240):899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, Angel CE, Chen C-JJ, Dunbar PR, Wadley RB, Jeet V, Vulink AJE, Hart DNJ, Radford KJ. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med. 2010;207(6):1247–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huysamen C, Willment JA, Dennehy KM, Brown GD. CLEC9A is a novel activation C-type lectin-like receptor expressed on BDCA3+ dendritic cells and a subset of monocytes. J Biol Chem. 2008;283(24):16693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bachem A, Güttler S, Hartung E, Ebstein F, Schaefer M, Tannert A, Salama A, Movassaghi K, Opitz C, Mages HW, Henn V, Kloetzel P-M, Gurka S, Kroczek RA. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J Exp Med. 2010;207(6):1273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Breton G, Lee J, Liu K, Nussenzweig MC. Defining human dendritic cell progenitors by multiparametric flow cytometry. Nat Protoc. 2015;10(9):1407–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee J, Breton G, Aljoufi A, Zhou YJ, Puhr S, Nussenzweig MC, Liu K. Clonal analysis of human dendritic cell progenitor using a stromal cell culture. J Immunol Methods. 2015;425:21–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee J, Breton G, Oliveira TYK, Zhou YJ, Aljoufi A, Puhr S, Cameron MJ, Sékaly R-P, Nussenzweig MC, Liu K. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J Exp Med. 2015;212(3):385–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Watanabe N, Narita M, Yamahira A, Nakamura T, Tochiki N, Saitoh A, Kaji M, Hashimoto S, Furukawa T, Toba K, Fuse I, Aizawa Y, Takahashi M. Transformation of dendritic cells from plasmacytoid to myeloid in a leukemic plasmacytoid dendritic cell line (PMDC05). Leuk Res. 2010;34(11):1517–24. [DOI] [PubMed] [Google Scholar]
- 94.McGovern N, Schlitzer A, Gunawan M, Jardine L, Shin A, Poyner E, Green K, Dickinson R, Wang X-N, Low D, Best K, Covins S, Milne P, Pagan S, Aljefri K, Windebank M, Miranda-Saavedra D, Saavedra DM, Larbi A, Wasan PS, Duan K, Poidinger M, Bigley V, Ginhoux F, Collin M, Haniffa M. Human dermal CD14+ cells are a transient population of monocyte-derived macrophages. Immunity. 2014;41(3):465–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lindstedt M, Lundberg K, Borrebaeck CAK. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J Immunol. 2005;175(8):4839–46. [DOI] [PubMed] [Google Scholar]
- 96.Lundberg K, Albrekt A-S, Nelissen I, Santegoets S, de Gruijl TD, Gibbs S, Lindstedt M. Transcriptional profiling of human dendritic cell populations and models—unique profiles of in vitro dendritic cells and implications on functionality and applicability. PLoS One. 2013;8(1):e52875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TC, Ulas T, Schultze JL. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40(2):274–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, Jardine L, Dixon D, Stephenson E, Nilsson E, Grundberg I, McDonald D, Filby A, Li W, De Jager PL, Rozenblatt-Rosen O, Lane AA, Haniffa M, Regev A, Hacohen N. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017. April 21;356(6335). pii: eaah4573. DOI: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fromm PD, Kupresanin F, Brooks AES, Dunbar PR, Haniffa M, Hart DNJ, Clark GJ. A multi-laboratory comparison of blood dendritic cell populations. Clin Transl Immunol. 2016;5(4):e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Veres TZ, Rochlitzer S, Shevchenko M, Fuchs B, Prenzler F, Nassenstein C, Fischer A, Welker L, Holz O, Müller M, Krug N, Braun A. Spatial interactions between dendritic cells and sensory nerves in allergic airway inflammation. Am J Respir Cell Mol Biol. 2007;37(5):5535–61. [DOI] [PubMed] [Google Scholar]
- 101.Todate A, Chida K, Suda T, Imokawa S, Sato J, Ide K, Tsuchiya T, Inui N, Nakamura Y, Asada K, Hayakawa H, Nakamura H. Increased numbers of dendritic cells in the bronchiolar tissues of diffuse panbronchiolitis. Am J Respir Crit Care Med. 2000;162(1):148–53. [DOI] [PubMed] [Google Scholar]
- 102.Thornton EE, Looney MR, Bose O, Sen D, Sheppard D, Locksley R, Huang X, Krummel MF. Spatiotemporally separated antigen uptake by alveolar dendritic cells and airway presentation to T cells in the lung. J Exp Med. 2012;209(6):1183–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cochand L, Isler P, Songeon F, Nicod LP. Human lung dendritic cells have an immature phenotype with efficient mannose receptors. Am J Respir Cell Mol Biol. 1999;21(5):547–54. [DOI] [PubMed] [Google Scholar]
- 104.Nicod LP, Lipscomb MF, Weissler JC, Toews GB. Mononuclear cells from human lung parenchyma support antigen-induced T lymphocyte proliferation. J Leukoc Biol. 1989;45(4):336–44. [DOI] [PubMed] [Google Scholar]
- 105.Masten BJ, Olson GK, Tarleton CA, Rund C, Schuyler M, Mehran R, Archibeque T, Lipscomb MF. Characterization of myeloid and plasmacytoid dendritic cells in human lung. J Immunol. 2006;177(11):7784–93. [DOI] [PubMed] [Google Scholar]
- 106.Nicod LP, Lipscomb MF, Toews GB, Weissler JC. Separation of potent and poorly functional human lung accessory cells based on autofluorescence. J Leukoc Biol. 1989;45(5):458–65. [DOI] [PubMed] [Google Scholar]
- 107.Tsoumakidou M, Tzanakis N, Papadaki HA, Koutala H, Siafakas NM. Isolation of myeloid and plasmacytoid dendritic cells from human bronchoalveolar lavage fluid. Immunol Cell Biol. 2006;84(3):267–73. [DOI] [PubMed] [Google Scholar]
- 108.Ten Berge B, Muskens F, Kleinjan A, Hammad H, Hoogsteden HC, Lambrecht BN, Van den Blink B. A novel method for isolating dendritic cells from human bronchoalveolar lavage fluid. J Immunol Methods. 2009; 351(12):132–3. [DOI] [PubMed] [Google Scholar]
- 109.Bratke K, Lommatzsch M, Julius P, Kuepper M, Kleine H-D, Luttmann W, Christian Virchow J. Dendritic cell subsets in human bronchoalveolar lavage fluid after segmental allergen challenge. Thorax. 2007;62(2):168–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tschernig T, de Vries VC, Debertin AS, Braun A, Walles T, Traub F, Pabst R. Density of dendritic cells in the human tracheal mucosa is age dependent and site specific. Thorax. 2006;61(11):986–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schrøder M, Melum GR, Landsverk OJB, Bujko A, Yaqub S, Gran E, Aamodt H, Bækkevold ES, Jahnsen FL, Richter L. CD1c-expression by monocytes—implications for the use of commercial CD1c+ dendritic cell isolation kits. PLoS One. 2016;11(6):e0157387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Angel CE, George E, Brooks AES, Ostrovsky LL, Brown TLH, Dunbar PR. Cutting edge: CD1a+ antigen-presenting cells in human dermis respond rapidly to CCR7 ligands. J Immunol. 2006;176(10):5730–4. [DOI] [PubMed] [Google Scholar]
- 113.Angel CE, Lala A, Chen C-JJ, Edgar SG, Ostrovsky LL, Dunbar PR. CD14+ antigen-presenting cells in human dermis are less mature than their CD1a+ counterparts. Int Immunol. 2007;19(11):1271–9. [DOI] [PubMed] [Google Scholar]
- 114.Haniffa M, Ginhoux F, Wang X-N, Bigley V, Abel M, Dimmick I, Bullock S, Grisotto M, Booth T, Taub P, Hilkens C, Merad M, Collin M. Differential rates of replacement of human dermal dendritic cells and macrophages during hematopoietic stem cell transplantation. J Exp Med. 2009;206(2):371–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ochoa MT, Loncaric A, Krutzik SR, Becker TC, Modlin RL. “Dermal dendritic cells” comprise two distinct populations: CD1+ dendritic cells and CD209+ macrophages. J Invest Dermatol. 2008;128(9):2225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bigley V, McGovern N, Milne P, Dickinson R, Pagan S, Cookson S, Haniffa M, Collin M. Langerin-expressing dendritic cells in human tissues are related to CD1c+ dendritic cells and distinct from Langerhans cells and CD141high XCR1+ dendritic cells. J Leukoc Biol. 2015;97(4):627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van Haarst JM, Hoogsteden HC, de Wit HJ, Verhoeven GT, Havenith CE, Drexhage HA. Dendritic cells and their precursors isolated from human bronchoalveolar lavage: immunocytologic and functional properties. Am J Respir Cell Mol Biol. 1994;11(3):344–50. [DOI] [PubMed] [Google Scholar]
- 118.De Monte A, Olivieri C-V, Vitale S, Bailleux S, Castillo L, Giordanengo V, Maryanski JL, Segura E, Doglio A. CD1c-related DCs that express CD207/Langerin, but are distinguishable from Langerhans cells, are consistently present in human tonsils. Front Immunol. 2016;7:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nestle FO, Zheng XG, Thompson CB, Turka LA, Nickoloff BJ. Characterization of dermal dendritic cells obtained from normal human skin reveals phenotypic and functionally distinctive subsets. J Immunol. 1993;151(11):6535–5. [PubMed] [Google Scholar]
- 120.Lenz A, Heine M, Schuler G, Romani N. Human and murine dermis contain dendritic cells. Isolation by means of a novel method and phenotypical and functional characterization. J Clin Invest. 1993;92(6):2587–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mc Dermott R, Ziylan U, Spehner D, Bausinger H, Lipsker D, Mommaas M, Cazenave J-P, Raposo G, Goud B, de la Salle H, Salamero J, Hanau D. Birbeck granules are subdomains of endosomal recycling compartment in human epidermal Langerhans cells, which form where Langerin accumulates. Mol Biol Cell. 2002;13(1):317–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feinberg H, Taylor ME, Razi N, McBride R, Knirel YA, Graham SA, Drickamer K, Weis WI. Structural basis for Langerin recognition of diverse pathogen and mammalian glycans through a single binding site. J Mol Biol. 2011;405(4):1027–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Segura E, Touzot M, Bohineust A, Cappuccio A, Chiocchia G, Hosmalin A, Dalod M, Soumelis V, Amigorena S. Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity. 2013;38(2):336–48. [DOI] [PubMed] [Google Scholar]
- 124.Baharom F, Rankin G, Scholz S, Pourazar J, Ahlm C, Blomberg A, Smed-Sörensen A. Human lung dendritic cells: spatial distribution and phenotypic identification in endobronchial biopsies using immunohistochemistry and flow cytometry. J Vis Exp JoVE. 2017;(119). DOI: 10.3791/55222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Galibert L, Diemer GS, Liu Z, Johnson RS, Smith JL, Walzer T, Comeau MR, Rauch CT, Wolfson MF, Sorensen RA, Van der Vuurst de Vries A-R, Branstetter DG, Koelling RM, Scholler J, Fanslow WC, Baum PR, Derry JM, Yan W. Nectin-like protein 2 defines a subset of T-cell zone dendritic cells and is a ligand for class-I-restricted T-cell-associated molecule. J Biol Chem. 2005;280(23):21955–64. [DOI] [PubMed] [Google Scholar]
- 126.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176(1):287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. 2000;164(12):6166–73. [DOI] [PubMed] [Google Scholar]
- 128.Edwards JP, Zhang X, Frauwirth KA, Mosser DM. Biochemical and functional characterization of three activated macrophage populations. J Leukoc Biol. 2006;80(6): 1298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stout RD, Jiang C, Matta B, Tietzel I, Watkins SK, Suttles J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J Immunol. 2005;175(1):342–9. [DOI] [PubMed] [Google Scholar]
- 130.Shaykhiev R, Krause A, Salit J, Strulovici-Barel Y, Harvey B-G, O’Connor TP, Crystal RG. Smoking-dependent reprogramming of alveolar macrophage polarization: implication for pathogenesis of chronic obstructive pulmonary disease. J Immunol. 2009;183(4):2867–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pechkovsky DV, Prasse A, Kollert F, Engel KMY, Dentler J, Luttmann W, Friedrich K, Müller-Quernheim J, Zissel G. Alternatively activated alveolar macrophages in pulmonary fibrosis-mediator production and intracellular signal transduction. Clin Immunol. 2010;137(1):89–101. [DOI] [PubMed] [Google Scholar]
- 132.Kasraie S, Niebuhr M, Kopfnagel V, Dittrich-Breiholz O, Kracht M, Werfel T. Macrophages from patients with atopic dermatitis show a reduced CXCL10 expression in response to staphylococcal α-toxin. Allergy. 2012;67(1):41–9. [DOI] [PubMed] [Google Scholar]
- 133.Fuentes-Duculan J, Suárez-Fariñas M, Zaba LC, Nograles KE, Pierson KC, Mitsui H, Pensabene CA, Kzhyshkowska J, Krueger JG, Lowes MA. A subpopulation of CD163-positive macrophages is classically activated in psoriasis. J Invest Dermatol. 2010;130(10):2412–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pouniotis DS, Plebanski M, Apostolopoulos V, McDonald CF. Alveolar macrophage function is altered in patients with lung cancer. Clin Exp Immunol. 2006;143(2):363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Berenson CS, Kruzel RL, Eberhardt E, Dolnick R, Minderman H, Wallace PK, Sethi S. Impaired innate immune alveolar macrophage response and the predilection for COPD exacerbations. Thorax. 2014;69(9):811–8. [DOI] [PubMed] [Google Scholar]
- 136.Choi H-S, Rai PR, Chu HW, Cool C, Chan ED. Analysis of nitric oxide synthase and nitrotyrosine expression in human pulmonary tuberculosis. Am J Respir Crit Care Med. 2002;166(2):178–86. [DOI] [PubMed] [Google Scholar]
- 137.Fathi M, Johansson A, Lundborg M, Orre L, Sköld CM, Camner P. Functional and morphological differences between human alveolar and interstitial macrophages. Exp Mol Pathol. 2001;70(2):77–82. [DOI] [PubMed] [Google Scholar]
- 138.Hoppstädter J, Diesel B, Zarbock R, Breinig T, Monz D, Koch M, Meyerhans A, Gortner L, Lehr C-M, Huwer H, Kiemer AK. Differential cell reaction upon Toll-like receptor 4 and 9 activation in human alveolar and lung inter-stitial macrophages. Respir Res. 2010;11:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gibbings SL, Thomas SM, Atif SM, McCubbrey AL, Desch AN, Danhorn T, Leach SM, Bratton DL, Henson PM, Janssen WJ, Jakubzick CV. Three unique interstitial macrophages in the murine lung at steady state. Am J Respir Cell Mol Biol. 2017;57(1):66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Westphalen K, Gusarova GA, Islam MN, Subramanian M, Cohen TS, Prince AS, Bhattacharya J. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature. 2014;506(7489):503–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Peão MN, Aguas AP, de Sá CM, Grande NR. Morphological evidence for migration of particle-laden macrophages through the interalveolar pores of Kohn in the murine lung. Acta Anat (Basel). 1993;147(4):227–32. [DOI] [PubMed] [Google Scholar]
- 142.Cohen AB, Cline MJ. The human alveolar macrophage: isolation, cultivation in vitro, and studies of morphologic and functional characteristics. J Clin Invest. 1971;50(7):1390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tomlinson GS, Booth H, Petit SJ, Potton E, Towers GJ, Miller RF, Chain BM, Noursadeghi M. Adherent human alveolar macrophages exhibit a transient pro-inflammatory profile that confounds responses to innate immune stimulation. PLoS One. 2012;7(6):e40348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Akei H, Whitsett JA, Buroker M, Ninomiya T, Tatsumi H, Weaver TE, Ikegami M. Surface tension influences cell shape and phagocytosis in alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2006;291(4):L572–9. [DOI] [PubMed] [Google Scholar]
- 145.Patel NR, Bole M, Chen C, Hardin CC, Kho AT, Mih J, Deng L, Butler J, Tschumperlin D, Fredberg JJ, Krishnan R, Koziel H. Cell elasticity determines macrophage function. PLoS One. 2012;7(9):e41024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jakubzick C, Tacke F, Llodra J, van Rooijen N, Randolph GJ. Modulation of dendritic cell trafficking to and from the airways. J Immunol. 2006;176(6):3578–84. [DOI] [PubMed] [Google Scholar]
- 147.Kirby AC, Coles MC, Kaye PM. Alveolar macrophages transport pathogens to lung draining lymph nodes. J Immunol. 2009;183(3):1983–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Toews GB, Vial WC, Dunn MM, Guzzetta P, Nunez G, Stastny P, Lipscomb MF. The accessory cell function of human alveolar macrophages in specific T cell proliferation. J Immunol. 1984;132(1):181–6. [PubMed] [Google Scholar]
- 149.Lipscomb MF, Lyons CR, Nunez G, Ball EJ, Stastny P, Vial W, Lem V, Weissler J, Miller LM. Human alveolar macrophages: HLA-DR-positive macrophages that are poor stimulators of a primary mixed leukocyte reaction. J Immunol. 1986;136(2):497–504. [PubMed] [Google Scholar]
- 150.Lyons CR, Ball EJ, Toews GB, Weissler JC, Stastny P, Lipscomb MF. Inability of human alveolar macrophages to stimulate resting T cells correlates with decreased antigen-specific T cell-macrophage binding. J Immunol. 1986;137(4):1173–80. [PubMed] [Google Scholar]
- 151.Chelen CJ, Fang Y, Freeman GJ, Secrist H, Marshall JD, Hwang PT, Frankel LR, DeKruyff RH, Umetsu DT. Human alveolar macrophages present antigen ineffectively due to defective expression of B7 costimulatory cell surface molecules. J Clin Invest. 1995;95(3):1415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Blumenthal RL, Campbell DE, Hwang P, DeKruyff RH, Frankel LR, Umetsu DT. Human alveolar macrophages induce functional inactivation in antigen-specific CD4 T cells. J Allergy Clin Immunol. 2001;107(2):258–64. [DOI] [PubMed] [Google Scholar]
- 153.van Furth R, Cohn ZA, Hirsch JG, Humphrey JH, Spector WG, Langevoort HL. The mononuclear phagocyte system: a new classification of macrophages, monocytes, and their precursor cells. Bull World Health Organ. 1972;46(6):845–52. [PMC free article] [PubMed] [Google Scholar]
- 154.Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, Cumano A, Geissmann F. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311(5757):83–7. [DOI] [PubMed] [Google Scholar]
- 155.Bruno L, Seidl T, Lanzavecchia A. Mouse pre-immunocytes as non-proliferating multipotent precursors of macrophages, interferon-producing cells, CD8alpha(+) and CD8alpha(−) dendritic cells. Eur J Immunol. 2001;31(11): 3403–12. [DOI] [PubMed] [Google Scholar]
- 156.Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, Greter M, Mortha A, Boyer SW, Forsberg EC, Tanaka M, van Rooijen N, García-Sastre A, Stanley ER, Ginhoux F, Frenette PS, Merad M. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38(4):792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, Lambrecht BN. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210(10):1977–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330(6005):841–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sawai CM, Babovic S, Upadhaya S, Knapp DJHF, Lavin Y, Lau CM, Goloborodko A, Feng J, Fujisaki J, Ding L, Mirny LA, Merad M, Eaves CJ, Reizis B. Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity. 2016;45(3):597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SEW, Pollard JW, Frampton J, Liu KJ, Geissmann F. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336(6077):86–90. [DOI] [PubMed] [Google Scholar]
- 161.Nayak DK, Zhou F, Xu M, Huang J, Tsuji M, Hachem R, Mohanakumar T. Long-term persistence of donor alveolar macrophages in human lung transplant recipients that influences donor specific immune responses. Am J Transplant. 2016;16(8):2300–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Eguíluz-Gracia I, Schultz HHL, Sikkeland LIB, Danilova E, Holm AM, Pronk CJH, Agace WW, Iversen M, Andersen C, Jahnsen FL, Baekkevold ES. Long-term persistence of human donor alveolar macrophages in lung transplant recipients. Thorax. 2016;71(11):1006–11. [DOI] [PubMed] [Google Scholar]
- 163.Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, Ivanov S, Duan Q, Bala S, Condon T, van Rooijen N, Grainger JR, Belkaid Y, Ma’ayan A, Riches DWH, Yokoyama WM, Ginhoux F, Henson PM, Randolph GJ. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity. 2013;39(3):599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Guilliams M, Scott CL. Does niche competition determine the origin of tissue-resident macrophages? Nat Rev Immunol. 2017;17(7):451–60. [DOI] [PubMed] [Google Scholar]
- 165.Eidsmo L, Allan R, Caminschi I, van Rooijen N, Heath WR, Carbone FR. Differential migration of epidermal and dermal dendritic cells during skin infection. J Immunol. 2009;182(5):3165–72. [DOI] [PubMed] [Google Scholar]
- 166.Molawi K, Wolf Y, Kandalla PK, Favret J, Hagemeyer N, Frenzel K, Pinto AR, Klapproth K, Henri S, Malissen B, Rodewald H-R, Rosenthal NA, Bajenoff M, Prinz M, Jung S, Sieweke MH. Progressive replacement of embryo-derived cardiac macrophages with age. J Exp Med. 2014;211(11):2151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tamoutounour S, Guilliams M, Montanana Sanchis F, Liu H, Terhorst D, Malosse C, Pollet E, Ardouin L, Luche H, Sanchez C, Dalod M, Malissen B, Henri S. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity. 2013;39(5):925–38. [DOI] [PubMed] [Google Scholar]
- 168.Mitchell AJ, Pradel LC, Chasson L, van Rooijen N, Grau GE, Hunt NH, Chimini G. Technical advance: autofluorescence as a tool for myeloid cell analysis. J Leukoc Biol. 2010;88(3):597–603. [DOI] [PubMed] [Google Scholar]
- 169.Vermaelen K, Pauwels R. Accurate and simple discrimination of mouse pulmonary dendritic cell and macrophage populations by flow cytometry: methodology and new insights. Cytom A. 2004;61(2):170–7. [DOI] [PubMed] [Google Scholar]
- 170.Yu Y-RA, Hotten DF, Malakhau Y, Volker E, Ghio AJ, Noble PW, Kraft M, Hollingsworth JW, Gunn MD, Tighe RM. Flow cytometric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. Am J Respir Cell Mol Biol. 2016;54(1):13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bharat A, Bhorade SM, Morales-Nebreda L, McQuattie-Pimentel AC, Soberanes S, Ridge K, DeCamp MM, Mestan KK, Perlman H, Budinger GRS, Misharin AV. Flow cytometry reveals similarities between lung macrophages in humans and mice. Am J Respir Cell Mol Biol. 2016;54(1):147–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Duan M, Steinfort DP, Smallwood D, Hew M, Chen W, Ernst M, Irving LB, Anderson GP, Hibbs ML. CD11b immunophenotyping identifies inflammatory profiles in the mouse and human lungs. Mucosal Immunol. 2016;9(2): 550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ohnishi K, Komohara Y, Saito Y, Miyamoto Y, Watanabe M, Baba H, Takeya M. CD169-positive macrophages in regional lymph nodes are associated with a favorable prognosis in patients with colorectal carcinoma. Cancer Sci. 2013;104(9):1237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jakubzick CV, Randolph GJ, Henson PM. Monocyte differentiation and antigen-presenting functions. Nat Rev Immunol. 2017;17(6):349–62. [DOI] [PubMed] [Google Scholar]
- 175.Juarez E, Nuñez C, Sada E, Ellner JJ, Schwander SK, Torres M. Differential expression of Toll-like receptors on human alveolar macrophages and autologous peripheral monocytes. Respir Res. 2010;11:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Asada K, Sasaki S, Suda T, Chida K, Nakamura H. Anti-inflammatory roles of peroxisome proliferator-activated receptor gamma in human alveolar macrophages. Am J Respir Crit Care Med. 2004;169(2):195–200. [DOI] [PubMed] [Google Scholar]


