Abstract
Bacterial recombinational repair of double-strand breaks often begins with creation of initiating 3′ single-stranded DNA (ssDNA) tails on each side of a double-strand break (DSB). Importantly, if the RecBCD pathway is followed, RecBCD creates a gap between the sequences at 3′ ends of the initiating strands. The gap flanks the DSB and extends at least to the nearest Chi site on each strand. Once the initiating strands form ssDNA–RecA filaments, each ssDNA–RecA filament searches for homologous double-stranded DNA (dsDNA) to use as a template for the DNA synthesis needed to fill the gap created by RecBCD. Our experimental results show that the DNA synthesis requires formation of a heteroduplex dsDNA that pairs >20 contiguous bases in the initiating strand with sequence matched bases in a strand from the original dsDNA. To trigger synthesis, the heteroduplex must be near the 3′ end of the initiating strand. Those experimentally determined requirements for synthesis combined with the Chi site dependence of the function of RecBCD and the distribution of Chi sites in bacterial genomes could allow the RecBCD pathway to avoid some genomic rearrangements arising from directly induced DSBs; however, the same three factors could promote other rearrangements.
INTRODUCTION
Rearrangements between repetitive sequence elements underlie many examples of genomic instability in both prokaryotes and eukaryotes (1). These rearrangements include potentially deleterious gene deletion events (1–3). Eukaryotes use complex strategies (4,5) to avoid dangerous rearrangements that can result when repeated sequences interfere with double-strand break (DSB) repair (4–6). Bacterial strategies have largely remained mysterious.
It is known that repeated ribosomal RNA (rrn) loci do not contain Chi sites (7). If Chi sites were randomly positioned, then there is only a 0.5% probability that the loci would not contain any Chi sites. Thus, placement of Chi sites in rrn loci is suppressed (7). The authors propose that the suppression of Chi sites within rrn loci prevents initiation of duplication within rrn loci because RecBCD degrades all of the DNA within the repeat (7).
When a DSB occurs in bacteria, it can be repaired using RecA-mediated homologous recombination. Homologous recombination dominates replication fork-associated DSBs in both prokaryotes (8) and eukaryotes (9); however, in this work we will focus on the repair of directly induced double-strand breaks.
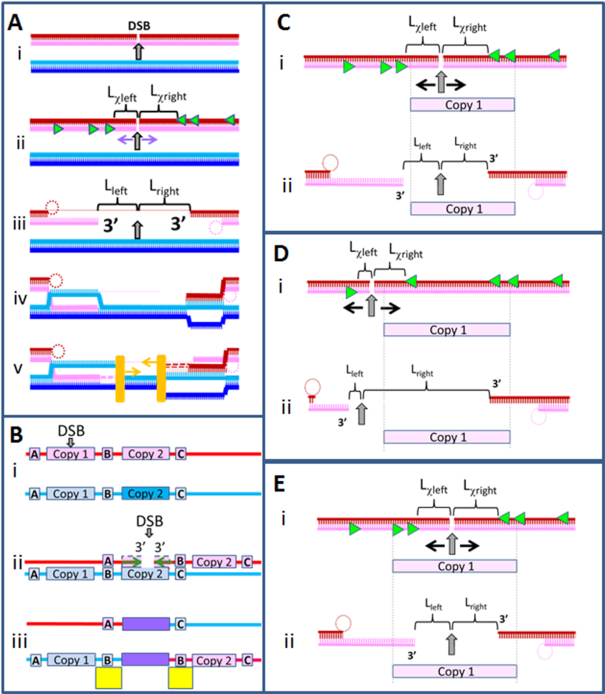
It has long been supposed that after a directly induced DSB, repair follows the process illustrated in Figure 1A. Figure 1Ai shows a DSB in one of two identical DNA sequences. On each side of the DSB, RecBCD interacts with the double-stranded DNA (dsDNA) end. RecBCD then progresses along the dsDNA until it recognizes a Chi site. Figure 1Aii shows the Chi sites that could be recognized by RecBCD. Recognition of a Chi site triggers a functional change in RecBCD (10–16). The change in function either stops RecBCD degradation of an initiating strand (17) or causes RecBCD to nick an initiating strand that was already separated from the complementary strand (14). In either case, after recognizing a Chi site, RecBCD creates two 3′ single-stranded DNA (ssDNA) tails (14,15,17–20) (Figure 1Aiii). As indicated in Figure 1Aiii and Supplementary Table S1, the left tail is separated from the DSB by Lleft ≥ Lχleft bases and the right tail is separated from the DSB by Lright ≥ Lχright bases. As illustrated in Figure 1A, these inequalities are true even if RecBCD does not recognize the first Chi site it encounters, or if the 3′ ends of the initiating ssDNA are not occupied by Chi sites. The inequalities simply depend on RecBCD not changing function until it recognizes a Chi site.
Figure 1.
Schematic of a RecBCD-dependent repair of a double-strand break and a genomic rearrangement that might result. (A) i. The dsDNA sequences are identical. The DSB is indicated by the gray arrow. ii. The bright green triangles indicate the Chi sites on each strand that are oriented so that they would be recognized by RecBCD as it proceeds along the dsDNA from the DSB. The Lχleft and Lχright are the separations between the position of the DSB and the nearest appropriately oriented Chi site on each initiating strand. iii. RecBCD creates the two initiating ssDNAs, while the complementary strands are degraded or looped (dotted circles). Lleft and Lright are the distances separating the DSB from the 3′ end of each initiating strand. Note that Lleft ≥ Lχleft and Lright ≥ Lχright. iv. RecA mediated strand exchange creates heteroduplex products that reach the 3′ ends of the filaments. v. DNA polymerase (orange rectangles) extends both initiating ssDNAs by copying the complementary strands beginning at the 3′ end of an initiating strand in a RecA filament. (B) The red and blue lines represent two identical dsDNAs that include two copies of a repeated sequence, Copy 1 and Copy 2 separated by a non-repeated region. i. A DSB occurs near the center of Copy 1.ii. After the DSB, RecBCD creates two ssDNA–RecA filaments by loading RecA onto the initiating ssDNA created by RecBCD. The two ssDNA–RecA filaments are indicated by the green horizontal arrows pointing toward the 3′ end. iii. Both filaments pair with the sequence matched regions in Copy 2 and form sequence matched heteroduplex products that extend to the 3′ ends of the initiating strands. DNA synthesis then completes two dsDNAs. The sequence region that includes the heteroduplex products and the newly synthesized DNA is shown in purple. The completion of the dsDNA is followed by Holliday junction resolution that is a crossover on the right side. As a result, the upper dsDNA lacks the sequence region between the two copies, and the lower dsDNA has two copies of the region between the two repeats. The duplicated regions are highlighted by the yellow rectangles. (C) Schematic of a DSB that occurs in Copy 1 of a repeated sequence. The appropriately oriented Chi sites are indicated by the bright green triangles, and the direction of RecBCD is shown by the black arrows. Copy 1 only includes appropriately oriented Chi sites in the dark red strand, so only the dark red strand can terminate in a region of Copy 1. (D) Same as C), but the DSB occurs to the left of Copy 1, which changes the number of correctly oriented Chi site in this region. The light pink filament cannot include a region of Copy 1 because RecBCD moves away from the DSB. (E) In a different sequence than C), Copy 1 includes appropriately oriented Chi sites on both strands, allowing the rearrangement shown in B); however, this never occurs in any of the 12 enteric bacteria we studied in this work.
Two ssDNA–RecA filaments are formed when RecA binds to the two 3′ ssDNA tails (14,15,17–20). The two ssDNA–RecA filaments then search for homologous regions in the dsDNA. Importantly, the bases in the gap between the 3′ ends of the initiating strands do not participate in the homology search because they are not included in the ssDNA–RecA filaments.
To determine whether a region of dsDNA is homologous to the initiating strand in the ssDNA–RecA filament, strand exchange is attempted. Strand exchange establishes Watson-Crick pairing between ∼ 8 bases in the initiating strand and 8 bases in one of the strands in the dsDNA (21–23). If the pairing is successful, the initiating and complementary strands form a heteroduplex. Since the complementary strand was originally base paired with the outgoing strand, formation of the heteroduplex leaves the outgoing strand unpaired. After formation of a short heteroduplex product, strand exchange can extend the heteroduplex product in a 5′ to 3′ direction with respect to the initiating ssDNA (18,24,25) (Figure1Aiv). In vitro experiments have probed the stability of strand exchange products as a function of N, the number of contiguous bases in the initiating strand that are sequence matched with the corresponding bases in the dsDNA. The stability of sequence matched strand exchange products in vitro increases strongly as N increases from 8 to 20 bp (22,23,26,27). In the absence of ATP hydrolysis, 20 bp products are nearly irreversible (22,23,26,27). In the presence of ATP hydrolysis, 20 bp products are quite unstable, and heteroduplex stability in vitro increases only slightly as the product extends from 20 to 75 bp (27). In vitro, products remain unstable even if N > 80 bp as long as the outgoing strand remains single stranded (27,28).
Figure 1B provides an example that illustrates how genomic rearrangement could result from a DSB repair that joins different copies of long repeated sequences. Importantly, strand exchange products remain reversible (27,28) unless two complete dsDNA strands are formed and DNA synthesis replaces bases in the gap between the 3′ ends of the initiating ssDNA. Thus, the genomic rearrangement illustrated in Figure 1B is unlikely unless DNA synthesis begins at the terminal 3′ OH on each initiating strand and uses the complementary strand as a template (Figure 1Aiv) (29).
Figure 1C shows that if a DSB occurs in a repeat, RecBCD can create a filament that terminates in a region of that repeat. Figure 1D shows that if a DSB occurs outside of a repeat, then RecBCD can still form a filament that terminates in a region of a repeat. Importantly, a DSB outside of a repeat cannot create two filaments that terminate in regions of the same copy of the repeat because RecBCD progresses away from the DSB along the dsDNA, as illustrated in Figure 1D.
In contrast, Figure 1E shows that if a DSB occurs within a repeat, RecBCD can create two filaments that terminate in regions of the same copy of the repeat, if the repeat contains appropriately positioned Chi sites on each strand. The results we will present in this work suggest that a combination of the action of RecBCD and the positioning of Chi sites in the genomes of enteric bacteria implies that no DSB can create two filaments that both include regions of a copy of a repeat that extends over more than 20 bp. We also present experimental results indicating that DNA synthesis by Pol IV is not significant unless a heteroduplex product extending over >20 contiguous homologous bp is located within ∼8 bp of the 3′ end of the filament. Thus, the experimental results imply that since RecBCD cannot form two filaments that terminate in regions of the same copy of a repeat longer than 20 bp, DSB repair that follows the RecBCD pathway illustrated in Figure 1A cannot create genomic rearrangement by joining both filaments to the analogous regions in another copy of the repeat, as illustrated in Figure 1B.
MATERIALS AND METHODS
Escherichia coli Pol IV was purified from the TMCΔT strain (BL21-AIΔdinB, ΔumuDC, ΔrecA) by ion exchange chromatography and hydrophobic interaction chromatography as previously described (30,31). Oligonucleotides were obtained from Integrated DNA Technologies (IDT) and are listed in the Supplementary Materials and Methods section.
The dsDNA containing 90 bp with internal labels was obtained by heating and cooling down slowly the corresponding oligonucleotides from 90 to 40°C with 1°C steps equilibrated for 1 min; the emission at 518 nm was acquired (excitation at 493 nm) at each temperature step. The dsDNA containing 180 bp was prepared by initially annealing a 90 nt ssDNA containing an internal rhodamine label on base 58 from the 5′ end and a 5′-end phosphorylated oligonucleotide (82 bases) containing an internal fluorescein label (position 57 from the 3′ end). Another dsDNA without labels was annealed using two oligonucleotides containing 90 and 98 bases; the former was 5′-end phosphorylated. Finally, the two dsDNAs were annealed and ligated overnight at 16°C in the presence of T4 DNA ligase in ligase reaction buffer (50 mM Tris, 10 mM MgCl2, 1 mM ATP, and 10 mM dithiothreitol, pH 7.5, NEB). The 180 bp construct was further purified by running a 3% agarose gel in TBE (Tris/borate/EDTA) buffer for 2 h (6 V/cm). The 180 bp band was visualized with a midrange UV trans-illuminator and cut. Finally the dsDNA was extracted from the agarose using a Nucleospin kit (Machery and Nagel, Bethlehem, PA) and concentrated on a YM-100 centrifugal filter (Millipore). The sample containing 98 bp dsDNA was prepared by annealing the complementary oligonucleotides from 90 to 40°C with 1°C steps equilibrated for 1 min; the emission at 518 nm was acquired (excitation at 493 nm) at each temperature step.
FRET measurements
Strand exchange reactions were performed by mixing an aliquot of 0.06 μM 98 nt ssDNA/RecA filament, 0.06 μM labeled dsDNA, and 1 μM Escherichia coli DNA Polymerase IV (Pol IV) or 5 units Bacillus subtilis DNA polymerase, Large fragment (LF-Bsu) (New England Biolabs (NEB), 5000 units/ml) and rapidly transferring the solution to a quartz cuvette. For DNA Pol IV measurements, the RecA buffer contained 0.1 mg/ml BSA, 2 mM dATP and 0.4 mM dNTPs. Measurements in the presence of Bsu polymerase were performed in RecA buffer containing 1 mM ATP and 0.1 mM dNTPs. The filaments were initially prepared by incubating 0.06 μM ssDNA (final concentration ∼6 μM in bases) with 2 μM RecA (NEB) in the presence of 1 mM cofactor (ATP or dATP), 10 U/ml of pyruvate kinase, 3 mM phosphoenolpyruvate, and 0.2 μM single-stranded binding protein (SSB) (Epicentre) in RecA buffer (70 mM Tris–HCl, 10 mM MgCl2 and 5 mM dithiothreitol, pH 7.6) at 37°C for 10 min.
FRET experiments followed the emission of the fluorescein label by using 493-nm excitation during 30 min; the emission was read as counts per second (cps) at 518 nm every one second. The integration was 0.5 s and the band width 2 nm. The sample was kept at all times at 37°C.
RESULTS
Studies of the positioning of Chi sites within repeats in bacterial genomes
As illustrated in Figure 1B, genomic rearrangement can result if both ssDNA–RecA filaments include sub-regions of a repeated sequence that trigger DNA synthesis after pairing with adjacent regions of another copy of the repeat.
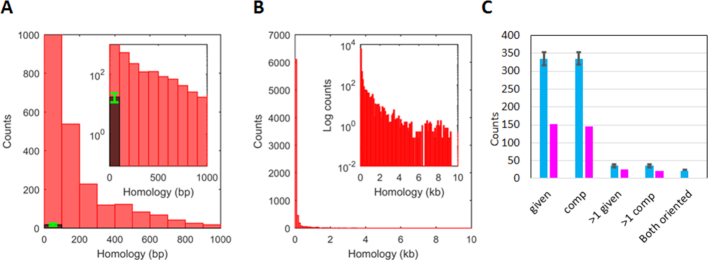
The challenge posed by repeated sequences is illustrated in Figure 2A and B. The red histograms show the number of repeats of a given length averaged over four ∼5 Mb long E. coli genomes (Figure 2A). The darker histogram shows the average number of repeats that occur in one hundred 5 Mb sequences composed of randomly chosen bases (random sequences) (Figure 2A, green error bars indicate the standard deviation). Importantly, though no randomly chosen sequence contains a repeat longer than 25 bp, real genomes contain thousands of repeats that extend beyond 25 bp. Those longer repeats include the ∼5 kb rDNA operon, as well as repeats that are even longer (32).
Figure 2.
Statistical distributions from bacterial genomes showing repeats >20 bp and distributions of Chi sites or randomly positioned markers in repeats >20 bp. (A) The red bars in the histogram represent repeats with length between 20 and 1000 bp averaged over the four E. coli genomes using a 100 bp bin width. The dark bar shows an analogous result obtained for 100 5-Mbp sequences composed of randomly chosen bases. The green error bar shows the standard deviation for those 100 sequences. (B) Same as A), but including all repeat lengths. (C) The magenta bars show the total number of Chi sites positioned within repeats summed over 12 enteric bacteria, whereas the cyan bars show the results for the same sequences when random markers are positioned in the sequence. The error bars correspond to the results for 100 realizations of the randomly placed markers. Importantly, for each genome the number of markers is the same as the number of Chi sites. The label below each pair of bars indicates the strand to which the results apply. The given strand is the strand whose sequence is given in the sequence database, and the comp strand is the strand that is complementary to that sequence. Thus, the first two pairs of bars correspond to the number of repeats that contain at least one Chi site on the given and comp strands, respectively. Similarly, the second two pairs of bars indicate the number of repeats that contain more than one Chi site on the given and comp strands, respectively. The final cyan bar shows the number of repeats that would contain properly oriented Chi sites on both strands. The corresponding magenta bar is zero.
Figure 1B illustrates a DSB that occurs in a repeat that leads to rearrangement resulting from both filaments pairing with adjacent regions of another copy of the repeat. That rearrangement requires the repeat to include a Chi site on each strand that could be recognized by RecBCD after the DSB, as illustrated in Figure 1E. To determine whether any long repeat contains two Chi sites that meet this criterion, we studied the positioning of Chi sites within repeats longer than 20 bp in 12 enteric bacteria (Supplementary Material and Methods). The results shown in the figure represent the sum over all 12 of the enteric bacteria that we considered. The results for each individual bacterial genome are shown in Supplementary Table S2.
We wanted to consider the sequence of each strand in the genome separately, so we assigned names to each strand. For each bacteria, the ‘given strand’ is the strand whose sequence is given in the referenced database, and the ‘comp strand’ is the sequence of the strand that is complementary to the given strand. The magenta bars at the left of Figure 2C show the total number of repeats that contain at least one Chi site. The figure shows that 153 repeats longer than 20 bp contain at least one Chi site in the given strand, and that 146 repeats longer than 20 bp contain at least one Chi site in the comp strand. We also considered the number of repeats that contain more than one Chi site on each strand. The two sets of bars near the center of Figure 2C show that 25 repeats in the given strand contain >1 Chi site, and 22 repeats in the comp strand contain >1 Chi site. The set of bars at the far right in Figure 2C that is labeled ‘both oriented’ shows that no repeat longer than 20 bp contains Chi sites on both strands that could be recognized by RecBCD after a DSB. Thus, for repeats that are 20 bp or longer, the condition shown in Figure 1E never occurs.
To determine whether the placement of Chi sites in repeats longer than 20 bp is suppressed, we compared the actual results for Chi sites to the results for markers that we randomly positioned in the same bacterial genomes. Importantly, for each genome the number of randomly placed markers was equal to the number of Chi sites actually found in that genome. For each genome, we ran 100 realizations of those random marker placements and calculated the average value for those 100 realizations. We then summed those average values over the 12 enteric bacteria that we considered. The cyan bars indicate those sums, and the accompanying error bars represent the standard deviations (Figure 2C).
The two sets of bars at the left side of Figure 2C show that on average 334.8 repeats in the given strand contain at least one randomly placed marker, and 335.4 repeats in the comp strand contain at least one randomly placed marker. On average 35.6 and 35.9 repeats contain >1 randomly placed markers on the given and comp strands, respectively. Finally, on average 45 repeats contain randomly placed markers on each strand. In half of those cases, Chi sites in the same positions could both be recognized by RecBCD because the 3′ sides of the markers face toward each other. Thus, as indicated by the cyan bar at the far right side of Figure 2C, if Chi sites were randomly positioned in genomes, on average 22.5 repeats would contain Chi sites on each strand that could both be recognized by RecBCD after a DSB. In contrast, no repeat that is 20 bp or longer contains Chi sites on each strand that could both be recognized by RecBCD after a DSB, so the condition shown in Figure 1E never occurs.
Since Figure 2C shows that no repeat that is 20 bp or longer contains appropriately positioned Chi sites on each strand, RecBCD cannot create two filaments that could trigger the genomic rearrangement illustrated in Figure 1B by joining both filaments to adjacent regions in another copy of the repeat. Figure 2C only considers repeats longer than 20 bp. Thus, the rearrangement illustrated in Figure 1B could still occur if repeats shorter than 20 bp have a significant probability of triggering synthesis. We therefore performed experiments to test the N dependence of synthesis. If synthesis is unlikely unless N > 20, then Figure 2C implies that if the RecBCD pathway is followed, the positioning of Chi sites in the genomes of enteric bacteria completely suppresses genomic rearrangements that require two filaments that contain regions of the same copy of a repeat.
Homology influences DNA synthesis
In these experiments, we study the dependence of DNA synthesis by Pol IV on length of homology (N). Previous work suggested that extension of the initiating strand by E. coli DNA Polymerase IV (Pol IV) stabilizes D-loops prior to re-establishment of a DNA polymerase III-dependent replication (33). Even in eukaryotic cells, translesion polymerases may aid DSB repair by stabilizing strand invasion intermediates (33). The expression level of the chromosomal Pol IV gene is upregulated during the SOS response, which may allow Pol IV to effectively compete against other DNA polymerases for this function (34,35). Additionally, in vivo studies indicate that RecA recruits Pol IV onto regions of DNA damage (36), and in vitro Pol IV can do synthesis that extends the initiating strand following RecA mediated homologous recombination, whereas Pol V cannot (37). Furthermore, new work indicates that most Pol IV molecules carry out DNA synthesis outside replisomes (38). Thus, we studied DNA synthesis by Pol IV. To test whether the results involve some very special property of Pol IV or whether they apply more generally to DNA polymerases, we also studied synthesis by the large fragment of Bacillus subtilis DNA polymerase I (LF-Bsu) which has been modified to remove the exonuclease activity that Pol IV intrinsically lacks.
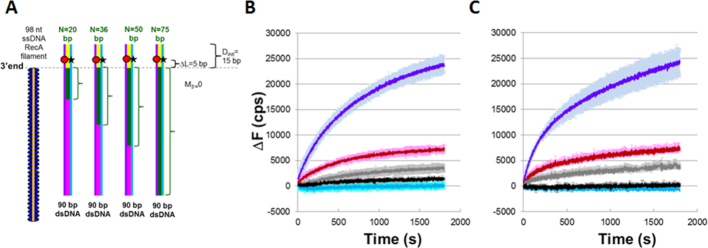
We first formed ssDNA–RecA filaments, and then allowed these filaments to interact with the dsDNA. If a sufficiently stable heteroduplex forms, a DNA polymerase can bind and extend the initiating strand. Extension begins at the terminal 3′ OH of the initiating strand and proceeds in the 5′ to 3′ direction with respect to the initiating strand, using the complementary strand as a template. We monitored the base pairing between the two strands in the dsDNA by measuring the fluorescence emission due to a fluorescein label on the outgoing strand of the dsDNA (Figure 3A). Initially, the rhodamine label on the complementary strand quenches the fluorescein emission on the outgoing strand because the complementary strand is annealed with the outgoing strand. If the complementary and outgoing strands separate due to strand exchange or DNA synthesis, the fluorescence emission will increase. To study effects due to the DNA polymerases, we positioned the fluorescent labels ΔL base pairs beyond the 3′ end of the filament. ΔL was chosen to be long enough that long strand exchange products do not produce large fluorescence increases even in the absence of a DNA polymerase.
Figure 3.
DNA synthesis stabilizes DSB repair occurring at a repeat. (A) Experimental schematic showing a typical ssDNA–RecA filament (orange line with blue ellipses) and 90 bp dsDNA. ΔL = Dlabel– Dinit = 5 bp. The labeled dsDNA used in all of the experiments was the same, so each N value corresponds to a different filament sequence. For each N value, the green brackets highlight the green regions of the dsDNA that are homologous to the N bases at the 3′ end of the ssDNA. The other bases in the dsDNA are heterologous to the initiating ssDNA. The yellow region indicates Dinit = 15 bp. The remaining dsDNA is shown in magenta. The red circle and black star represent the rhodamine and fluorescein labels, respectively. They are positioned on the complementary (purple line) and outgoing (blue line) strands, respectively. (B) Graph representing the average over three trials of the change in fluorescence (ΔF) versus time curves in experiments with dATP–ssDNA–RecA filaments and DNA Pol IV represented in A) for N = 75 (dark blue), 50 (red), 36 (gray), 20 (black), and heterologous filament (light blue). ΔF in counts per second (cps) is calculated as the difference between the measured fluorescence and the average initial fluorescence for heterologous dsDNA. The error bars show the standard deviation based on three trials. (C) Same as B) in the presence of ATP–ssDNA–RecA filaments and LF-Bsu polymerase.
We will specify positions in the dsDNA using D, their separation from the end of the dsDNA toward which synthesis is directed. In the first set of experiments, the 3′ end of the initiating strand was positioned 15 bp from the end of the dsDNA, so Dinit = 15 bp. Thus, if the polymerase adds 15 bases to the initiating strand, then the polymerase will reach the end of the dsDNA, which is also the end of the complementary strand. Furthermore, the fluorescent labels were located ∼10 bp from the end of the dsDNA, so Dlabel ∼10 bp and ΔL = Dinit – Dlabel ∼ 5 bp (Figure 3A).
The same 90 bp labeled dsDNA target was used in all of the experiments illustrated in Figure 3A. We varied the homology between the dsDNA and the ssDNA–RecA filaments by changing the sequence of the initiating ssDNA. The different 98 nt ssDNA sequences (Supplementary Materials and Methods) were designed to be heterologous to the dsDNA except for N contiguous bases at the 3′ end of the filament that match the corresponding N bases in the dsDNA (green brackets in Figure 3A encompassing 20, 36, 50 and 75 bp).
Figure 3B shows graphs of ΔF, the difference between the measured fluorescence as a function of time and the average initial fluorescence value for a heterologous ssDNA–RecA filament. These experiments were carried out with DNA Pol IV, ssDNA–RecA filaments in dATP, and dNTPs. Figure 3C shows the analogous results with LF-Bsu. In Figure 3B, C each of the curves represents results for different N values. Results obtained without DNA polymerase are shown in Supplementary Figure S1, along with results obtained with DNA, Pol IV and RecA, but without dNTPs. That figure also shows results with ssDNA, Pol IV and dNTPs, but no RecA. Comparison of Figure 3B and C with Supplementary Figure S1A–E suggests that for either cofactor, the observed fluorescence increase is dominated by DNA synthesis triggered by RecA mediated strand exchange.
Additional results for LF-Bsu are shown in Supplementary Figure S2. In these experiments Dinit is also 15 bp, but the fluorescent labels are positioned at the end of the dsDNA (Dlabel = 0, Dinit = 0 and ΔL = 15), whereas in Figure 3, Dlabel = 10 and ΔL = 5. These results also indicate that fluorescence increase due to DNA synthesis is small unless N > ∼36 bp. Supplementary Figure S1 also shows results for DNA Pol IV, ssDNA–RecA filaments in ATP, and dNTPs. The results obtained when ATP is the cofactor are very similar to those obtained in the presence of dATP, except that the magnitudes of the fluorescence signals obtained using ATP are approximately a factor of 2 smaller than those obtained using dATP. In sum, the similarity between Figures 3B, C and Supplementary Figure S2 suggests that the N dependence of the results represents general features of DNA synthesis triggered by the formation of heteroduplex strand exchange products, at least for DNA polymerases lacking 3′ to 5′ exonuclease activity.
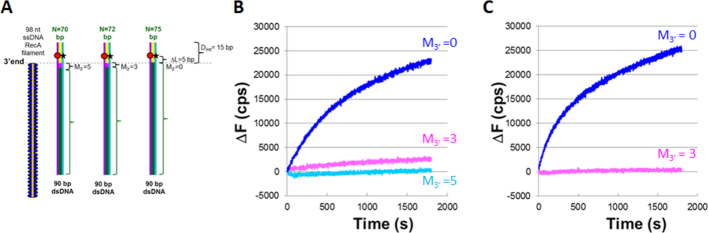
Synthesis is blocked by mismatches at the 3′ ends of ssDNA
In vivo, heterologous bases will always separate the 3′ ends of ssDNA–RecA filaments from sequence matched strand exchange products formed by joining different copies of the repeat, unless the repeat extends to the 3′ ends of the initiating ssDNA. Like eukaryotic recombinases, RecA can create strand exchange products that include some mismatches (39,40); however, there is also evidence indicating that the efficiency of strand exchange decreases in the presence of mismatches (22,23). To test whether mismatches at the 3′ end of the filament can inhibit the DNA synthesis required to make recombination irreversible, we designed the experiments illustrated schematically in Figure 4A. These experiments are identical to those performed in Figure 3, except that the number of mismatched bases at the 3′ end of the filament (M3′) is varied between 0 and 8 bp. Figure 4B shows the ΔF curve obtained in the presence of DNA Pol IV and indicates that even M3′ = 3 strongly suppresses the fluorescence increase, suggesting there is very little strand separation due to DNA synthesis. Furthermore, with Pol IV, the result for M3′ = 5 is indistinguishable from the results for heterologous controls, which is also true for LF-Bsu when M3′ = 3 (Figure 4C). Controls for these experiments are shown in Supplementary Figure S3. In sum, even heteroduplex products that are very stable cannot trigger synthesis unless they are very close (<5 bp) to the 3′ ends of the filaments.
Figure 4.
DNA synthesis is required for a DNA polymerase to stabilize strand exchange products. (A) Schematic for experiments with M3′ values of 0, 3, 5. (B) Graphic representation of the change in fluorescence (ΔF) versus time curves from single trial experiments performed with dATP–ssDNA–RecA filaments and DNA Pol IV in which the blue, pink, and light-blue curves correspond to M3′ values of 0, 3 and 5 base mismatches, respectively. ΔF is calculated as the difference between the measured fluorescence and the average initial fluorescence for heterologous dsDNA. (C) Analogous experiments as (B), but with LF-Bsu polymerase instead of DNA Pol IV.
Adjacent homoduplex dsDNA decreases the fluorescence signal associated with DNA synthesis
In vivo, heteroduplex products resulting from the pairing of the initiating and complementary strands are almost always flanked by homoduplex dsDNA in which complementary and outgoing strands remain base paired. Previous work has suggested that this homoduplex dsDNA drives reversal of adjacent heteroduplex products (27). As the experimental schematic shown in Figure 3A indicates, if a polymerase extends the initiating strand by 15 bases, the polymerase will reach the end of the template strand. If synthesis extends to the end of the dsDNA, then no homoduplex tail will extend beyond the 3′ side of the initiating strand.
To probe the influence of the homoduplex dsDNA on DNA synthesis, we increased Dinit from 15 bp to 66 bp. When Dinit = 66 bp, the DNA polymerase must synthesize 66 bases in order to remove the homoduplex tail at the 3′ end of the filament (Figure 5). If during our observation time strand displacement synthesis by a DNA polymerase is rapid enough to reach the end of the dsDNA when Dinit = 15, but not rapid enough to reach the end when Dinit = 66, then comparison of results from experiments with the two different Dinit values may provide insight into the influence of homoduplex dsDNA adjacent to heteroduplex products.
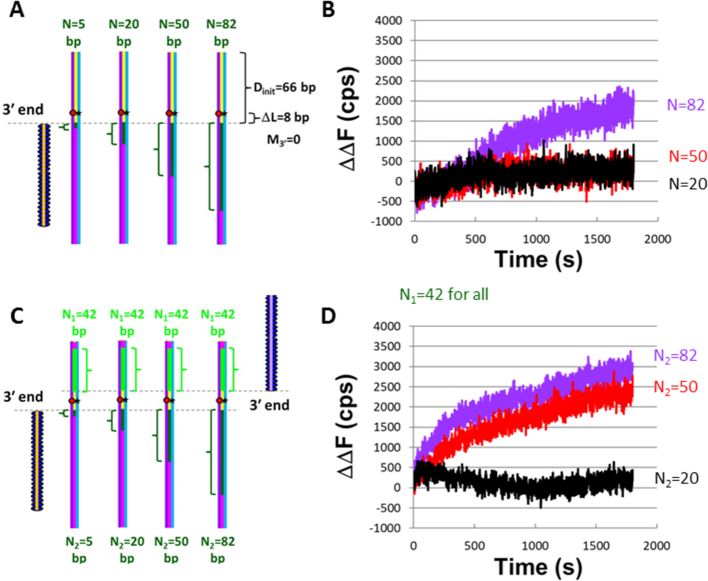
Figure 5.
The presence of a second filament rescues instability caused by an ssDNA outgoing strand and a long homoduplex dsDNA (180 bp) that extends beyond the 3′ end of a filament. (A) Schematic of experiments with 66 bp of homoduplex dsDNA on the 3′ end of the filament and N values of 5, 20, 50 and 82 bp. (B) Graphic representation of the change in fluorescence (ΔΔF) versus time curves of the experiment represented in (A) with dATP–ssDNA–RecA filaments and DNA Pol IV. ΔΔF is calculated as the difference between the measured fluorescence and the fluorescence for N = 5. The purple, red, and black curves correspond to N2 = 82, 50 and 20 nt, respectively. (C) Schematic for experiments performed involving two filaments. In all of these experiments N1 = 42. (D) Graphic representation of the change in fluorescence ΔΔF versus time curves for single trials of the experiment represented in (C) with dATP–ssDNA–RecA filaments and DNA Pol IV. ΔΔF was calculated as indicated above. Different N2 values are represented with different curve colors, where purple, red and black correspond to N2 = 82, 50 and 20 nt, respectively.
The same dsDNA with Dlabel = 58 bp was used in all of the experiments illustrated in Figure 5A, and N was controlled by varying the 98-nt sequence of the initiating strands. For this construct, even for N = 82, we see no increase in fluorescence in the absence of DNA synthesis. The raw fluorescence curves obtained with dATP–ssDNA–RecA filaments, DNA Pol IV, and dNTPs are shown in Supplementary Figure S4, and Figure 5B shows the corresponding ΔΔF versus time curves, where ΔΔF is the difference between the observed fluorescence and the fluorescence for N = 5 at each time. Notably, in Figure 3 and Supplementary Figure S2, the ΔF signal for the heterologous ssDNA–RecA filament is much smaller than the signal for the homologous filaments, so for Figure 3 and Supplementary Figure S2 ΔF(t) = ΔΔF(t). Thus, the results in these figures can be compared to the results in Figures 5 and 6. In Figures 5, 6 and Supplementary Figures S4 and S5, the purple, red, and black curves represent results for N = 82, 50 and 20, respectively.
Figure 6.
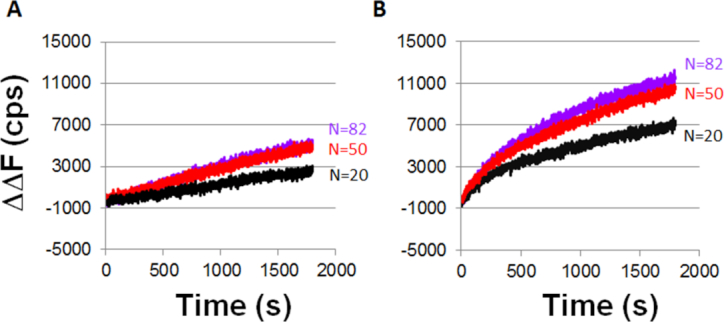
ΔΔF versus time curves obtained with ATP–ssDNA–RecA filaments, LF-Bsu polymerase, and 180 bp labeled dsDNA construct. The purple, red and black curves correspond to N = 82, 50 and 20, respectively. (A) ΔΔF versus time curves for one filament experiments with LF-Bsu, represented by the schematic shown in Figure 5A and after subtracting the heterologous DNA curve shown in Supplementary Figure S5A. (B) ΔΔF versus time curves for two filament experiments in the presence of LF-Bsu, represented by the schematic shown in Figure 5C and after subtracting the heterologous DNA curve shown in Supplementary Figure S5B.
Figure 5B shows that when Dinit = 66 bp, the increase in fluorescence due to Pol IV synthesis is only significant if N > 50; however, Figure 3B shows that both N = 50 and N = 36 produce significant fluorescence signals when Dinit = 15 bp. In contrast, Figure 6A shows results for LF-Bsu synthesis indicating that when Dinit = 66 even N = 20 produces significant fluorescence, and the fluorescence signals for N = 50 and N = 82 are nearly indistinguishable. We note that Supplementary Figure S6 shows that in these longer constructs no significant fluorescence increase is observed when M3′ = 8 bp, so the M3′ requirement applies when DNA synthesis cannot reach the end of the dsDNA.
In sum, the presence of homoduplex dsDNA does influence synthesis, and for Pol IV the results shown in Figure 5B and Supplementary Figure S6 may be closer to the in vivo results than those shown in Figure 3B and Figure 4B. The results in Figure 5B may better approximate in vivo conditions because Dinit = 66 is so long that Pol IV synthesis does not reach the end of the dsDNA during our observation time, whereas Dinit = 15 bp is short enough that DNA synthesis could reach the end of the dsDNA during our observation time.
Pol IV synthesis triggered by two filaments enhances the fluorescence
Finally, we wanted to probe synthesis in cases in which the action of RecBCD positions a repeat at the 3′ end of one of the filaments, as illustrated in Figure 1C and D. As discussed above, if both filaments must trigger synthesis, then the conditions illustrated in Figures 1C and D will not produce genomic rearrangement even though they include one filament that meets the N and M3′ requirements for synthesis.
To study synthesis triggered by the initiating ssDNA formed at both sides of a DSB, we performed the experiments illustrated in Figure 5C. All of the experiments illustrated in Figure 5C included one filament with N1 = 42 contiguous bases that are homologous to the corresponding bases in the dsDNA. The sequence of the second filament was varied so that N2, the number of contiguous bases that are homologous to the corresponding bases in the other strand of the dsDNA, varied from 0 to 82 bases. As in previous experiments, the fluorescence monitors the separation between the outgoing and complementary strands in the region near the fluorescent probes. In these experiments, we compare the fluorescence signal obtained when only one filament can trigger synthesis to that obtained in experiments in which both filaments can trigger synthesis that can complete two dsDNAs.
The ΔΔF results shown in Figure 5D indicate that the fluorescence change for N2 = 50 is quite significant, even though no detectable fluorescence change was observed in one-filament experiments with N = 50 (Figure 5B). Since a second filament with N1 = 42 significantly increased the fluorescence shift observed for filaments with 50 contiguous homologous bases, it is reasonable to suggest that one filament with N = 50 does trigger some synthesis, but the synthesis only produces a detectable fluorescence increase when a second filament also triggers synthesis. For N = 82, Figure 5 shows that the ratio of the total change in the ΔΔF values for the one filament case to the analogous values for the two filament case is 0.55 ± 0.08. For lower N values, the one filament signal did not exceed the noise, so no ratio can be obtained.
The analogous ΔΔF results for LF-Bsu are shown in Figure 6B, while Supplementary Figure S5A and B shows the corresponding ΔF(t) raw curves. Figure 6 shows that the ratio of the total change in the ΔΔF values for the one-filament case to the analogous values for the two-filament case is 0.47 ± 0.04, 0.66 ± 0.05, 0.3 ± 0.05 for N = 82, 50 and 20, respectively.
In sum, when the dsDNA is sufficiently long that synthesis is unlikely to reach the end of the dsDNA, if two RecA filaments trigger synthesis by Pol IV, the fluorescence signal is detectable for N ≥ 50, whereas N = 82 was required to produce detectable fluorescence in the one filament experiments. Comparison of the results observed when only one filament triggers synthesis suggests that even in the long dsDNA N = 50 can trigger synthesis by one filament, but the base pairing between the complementary and outgoing strands quenches the fluorescence effectively unless a second filament also triggers synthesis.
DISCUSSION
Figure 1A shows RecBCD creating a gap that extends Lright ≥ Lχvright bases on the right side of the DSB and Lleft ≥ Lχleft bases on the left side of the DSB (14,17,41,42). In vivo the gap can extend over tens of kb (13). It has been unclear why RecBCD would create a gap since energy is required to remove the bases and to replace the bases after they are removed.
Figure 2 indicates that the positioning of Chi sites in bacterial genomes implies that RecBCD cannot create two filaments that include sub-regions of the same copy of a repeat that extends over 20 or more bp. Importantly, if Chi sites were positioned randomly in genomes, then ∼20 repeats longer than 20 bp would include Chi sites that would allow RecBCD to create filaments that both include regions of the repeat. The complete absence of such occurrences in the 12 enteric bacteria we considered suggests that the strong suppression of such occurrences has an important role in vivo.
The experimental results shown in Figures 3–6 and Supplementary Figure S2 suggest that synthesis is unlikely unless an ssDNA-filament includes N > 20 bp within 8 bp of the 3′ end of the filament. If synthesis is required to create an irreversible strand exchange product, the results shown in Figure 2 indicate that RecBCD cannot create two ssDNA–RecA filaments that could produce genomic rearrangement by triggering synthesis after pairing with adjacent regions in another copy of a repeat. Figure 1B shows an example of a rearrangement that could occur if Chi sites were randomly positioned in bacterial genomes but is eliminated by the positioning of Chi sites in genomes.
If initiating strands do not terminate at or near Chi sites, then one cannot make any conclusions about how Chi sites influence rearrangements that require only one filament to trigger synthesis. In what follows we will consider the possibility that all filaments do terminate in Chi sites and assume that rearrangement can be triggered if only one of the filaments terminates in a repeat. Consistent with previous proposals, repeats that do not contain Chi sites will not generate rearrangements because RecBCD will remove the entire repeat (7). In contrast, breaks that occur outside of repeats can generate filaments that terminate in a repeat if RecBCD recognizes a Chi site in the repeat, as illustrated in Figure 1D. Thus, RecBCD could enhance the number of locations in the genome at which a DSB could lead to an initiating ssDNA that terminates in a repeat that contains a Chi site, even if RecBCD reduces the rearrangement rate for repeats that do not include Chi sites. In sum, if filaments always terminate in Chi sites then rearrangement associated with repeats that do not contain Chi sites is suppressed, but rearrangement associated with repeats that do contain Chi sites could be enhanced. This is consistent with Chi sites having a dual role that promotes some rearrangements at targeted locations near Chi sites and inhibits other rearrangements.
The gap created by RecBCD creates N and M3′ requirements for the synthesis that underlies Chi site dependent suppression of rearrangement, but those requirements may also promote RecBCD dependent rejection of interactions in which Chi sites do not play a direct role. In particular, these requirements for synthesis can reject interactions involving short regions of accidental homology positioned anywhere in the ssDNA–RecA filament or long regions of accidental homology that are separated from the 3′ end of the filament by >8 bp, as illustrated in Figure 7A.
Figure 7.
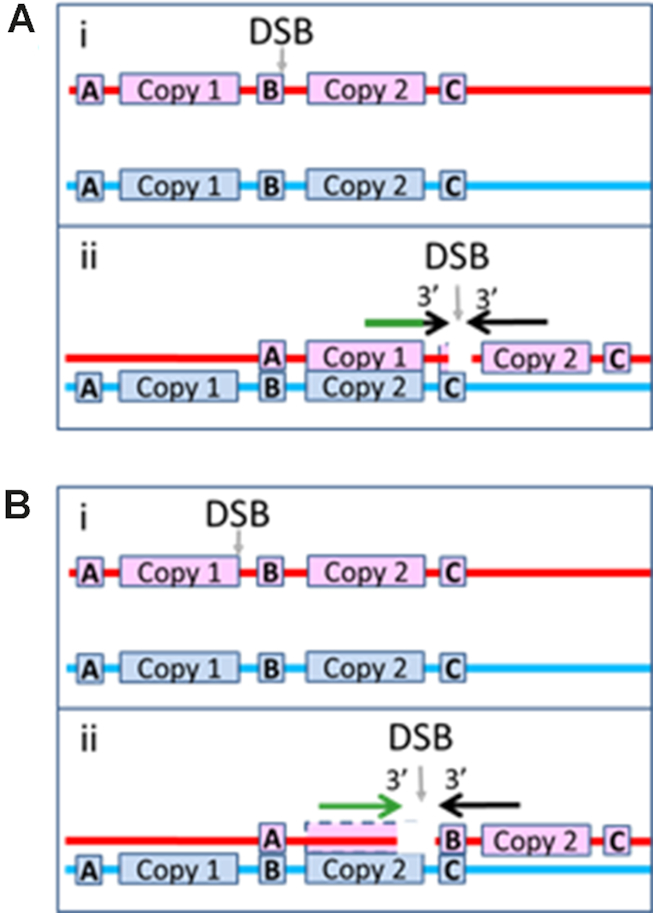
Illustration of how genomic rearrangement can be reduced for DSBs that occur outside of repeats. Same as Figure 1B, but the DSB occurs outside of a repeat. (A) The right filament is heterologous to the sequence to the right of Copy 2, so it cannot form a heteroduplex product by pairing with the dsDNA to the right of Copy 2. The left filament includes enough of Copy 1 to form a very stable N bp heteroduplex product when paired with the corresponding bases in Copy 2, but the black region near the tip of the arrow shows that the N homologous bp are separated from the 3′ of the filament by M3′ bases that are not homologous to the corresponding bases in Copy 2. Those M3′ mismatched bases could inhibit the progress of strand exchange beyond the sequence matched N bp heteroduplex because strand exchange proceeds less effectively through regions that contain mismatches. (B) i. A DSB occurs just before the right edge of Copy 1. ii. The left filament can form a long sequence matched heteroduplex product at the 3′ end of the initiating strand, but the right filament cannot form a stable heteroduplex in the region to the right of Copy 2. If synthesis triggered by one filament can produce an irreversible strand exchange product, then this geometry can produce genomic rearrangement; however, if both filaments must trigger synthesis that completes two dsDNAs, this interaction would not produce genomic rearrangement, even though the right filament could trigger synthesis.
Finally, the results in Figures 5 and 6 show that the base pairing between the complementary and outgoing strands is decreased if a second filament triggers synthesis. This is consistent with the base pairing between the complementary strand and the newly synthesized extension of the initiating strand being more stable if both filaments trigger synthesis that completes two dsDNAs. That increase in stability could result from synthesis removing the ssDNA regions that experiments suggest can reverse strand exchange (27,28). Thus, since RecBCD creates a gap that produces greater product stability when two filaments trigger synthesis, the increase in stability due to a second filament provides RecBCD discrimination between truly homologous interactions and interactions that involve regions of accidental homology. This discrimination could suppress rearrangement even if Chi sites were randomly distributed in bacterial genomes, but the positioning of Chi sites in genomes enhances effectiveness of this suppression mechanism.
In vivo, >10 kb of synthesis will frequently be required to fill the gap created by RecBCD, so the <20 bp gaps used in our experiments do not accurately capture the in vivo conditions (13). In vivo, synthesis by two filaments could still increase product stability by removing the ssDNA regions that experiments suggest can reverse strand exchange (27,28). If irreversible product formation is unlikely unless two filaments trigger synthesis in vivo, then rearrangement will rarely be triggered by DSBs that create only one filament that includes a repeat. Figures 1C and D shows situations in which RecBCD will create one filament that meets the criterion for synthesis, but Figure 7B illustrates why genomic rearrangement will be suppressed if both filaments are required to trigger synthesis.
Overall, even if filaments do not terminate in Chi sites, these results are consistent with RecBCD suppressing rearrangements that require both filaments to trigger synthesis after binding with adjacent regions in a different copy of a repeat. In contrast, if filaments always terminate in Chi sites and only one filament is required to trigger synthesis, the RecBCD pathway may enhance rearrangements involving repeats that contain Chi sites, while suppressing rearrangements associated with repeats that do not contain Chi sites. This dual role for the RecBCD pathway is consistent with homologous recombination enhancing cell survival by maintaining genome integrity, while also promoting genome rearrangement that leads to diversity, evolution, and speciation (43).
Supplementary Material
ACKNOWLEDGEMENTS
T.F.T. and V.G. would like to thank members of the Godoy Lab for their help, especially Margaret Downs. M.P. would also like to acknowledge useful conversations with Prof. Phillip Sharp.
Author contributions: C.L. performed almost all analysis of bacterial sequences; C.D. performed all experiments, and contributed to the experimental design and data analysis; T.F.T. and V.G provided the DNA Pol IV protein, contributed to the experimental design, and offered structural insight; C.P. contributed to the understanding of the interaction between Pol IV and RecA; and M.P. conceived the study, contributed to the analysis of bacterial sequences, experimental design, and data analysis. All of the authors discussed the results, contributed to the manuscript preparation, and approved the final version of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Northeastern University and NIGMS [RO1GM088230 to V.G.]; ′Initiative d′ Excellence′ program of the French State [′DYNAMO′, ANR-11-LABX-0011-01 to C.P.]; HCRP (Harvard College) (to C.L.); Harvard University. Funding for open access charge: Harvard University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bzymek M., Lovett S.T.. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8319–8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Farabaugh P.J., Schmeissner U., Hofer M., Miller J.H.. Genetic studies of the lac repressor: VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J. Mol. Biol. 1978; 126:847–863. [DOI] [PubMed] [Google Scholar]
- 3. Krawczak M., Cooper D.N.. Gene deletions causing human genetic-disease-mechanisms of mutagenesis and the role of the local DNA-sequence environment. Hum. Genet. 1991; 86:425–441. [DOI] [PubMed] [Google Scholar]
- 4. Ryu T., Bonner M.R., Chiolo I.. Cervantes and Quijote protect heterochromatin from aberrant recombination and lead the way to the nuclear periphery. Nucleus. 2016; 7:485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amaral N., Ryu T., Li X., Chiolo I.. Nuclear dynamics of heterochromatin repair. Trends Genet. 2017; 33:86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bao W., Kojima K.K., Kohany O.. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA. 2015; 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reams A.B., Kofoid E., Duleba N., Roth J.R.. Recombination and annealing pathways compete for substrates in making rrn duplications in Salmonella enterica. Genetics. 2014; 196:119–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cox M.M. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 2001; 35:53–82. [DOI] [PubMed] [Google Scholar]
- 9. Arnaudeau C., Lundin C., Helleday T.. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001; 307:1235–1245. [DOI] [PubMed] [Google Scholar]
- 10. Taylor A.F., Smith G.R.. RecBCD enzyme is altered upon cutting DNA at a Chi recombination hotspot. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:5226–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Handa N., Bianco P.R., Baskin R.J., Kowalczykowski S.C.. Direct visualization of RecBCD movement reveals cotranslocation of the RecD motor after Chi recognition. Mol. Cell. 2005; 17:745–750. [DOI] [PubMed] [Google Scholar]
- 12. Dillingham M.S., Kowalczykowski S.C.. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008; 72:642–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cockram C.A., Filatenkova M., Danos V., El Karoui M., Leach D.R.F.. Quantitative genomic analysis of RecA protein binding during DNA double-strand break repair reveals RecBCD action in vivo. Proc. Natl Acad. Sci. U.S.A. 2015; 112:E4735–E4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith G.R. How RecBCD enzyme and Chi promote DNA break repair and recombination: a molecular biologist's view. Microbiol. Mol. Biol. Rev. 2012; 76:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Symington L.S. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb. Perspect. Biol. 2014; 6:a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wiktor J., van der Does M., Büller L., Sherratt D.J., Dekker C.. Direct observation of end resection by RecBCD during double-stranded DNA break repair in vivo. Nucleic Acids Res. 2018; 46:1821–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kowalczykowski S.C. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2015; 7:a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mawer J.S.P., Leach D.R.F.. Branch migration prevents DNA loss during double-strand break repair. PLoS Genet. 2014; 10:e1004485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Azeroglu B., Mawer J.S.P., Cockram C.A., White M.A., Hasan A.M.M., Filatenkova M., Leach D.R.F.. RecG directs DNA synthesis during double-strand break repair. PLoS Genet. 2016; 12:e1005799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith G.R. Conjugational recombination in Escherichia-coli - myths and mechanisms. Cell. 1991; 64:19–27. [DOI] [PubMed] [Google Scholar]
- 21. Yang D.R., Boyer B., Prévost C., Danilowicz C., Prentiss M.. Integrating multi-scale data on homologous recombination into a new recognition mechanism based on simulations of the RecA-ssDNA/dsDNA structure. Nucleic Acids Res. 2015; 43:10251–10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qi Z., Redding S., Lee J.Y., Gibb B., Kwon Y., Niu H., Gaines W.A., Sung P., Greene E.C.. DNA Sequence alignment by microhomology sampling during homologous recombination. Cell. 2015; 160:856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Danilowicz C., Yang D., Kelley C., Prévost C., Prentiss M.. The poor homology stringency in the heteroduplex allows strand exchange to incorporate desirable mismatches without sacrificing recognition in vivo. Nucleic Acids Res. 2015; 43:6473–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cox M.M. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 2007; 8:127–138. [DOI] [PubMed] [Google Scholar]
- 25. Gupta R.C., Golub E.I., Wold M.S., Radding C.M.. Polarity of DNA strand exchange promoted by recombination proteins of the RecA family. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:9843–9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hsieh P., Camerini-Otero C.S., Camerini-Otero R.D.. The synapsis event in the homologous pairing of DNAs: RecA recognizes and pairs less than one helical repeat of DNA. Proc. Natl Acad. Sci. U.S.A. 1992; 89:6492–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Danilowicz C., Hermans L., Coljee V., Prévost C., Prentiss M.. ATP hydrolysis provides functions that promote rejection of pairings between different copies of long repeated sequences. Nucleic Acids Res. 2017; 45:8448–8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rosselli W., Stasiak A.. The ATPase activity of RecA is needed to push the DNA strand exchange through heterologous regions. EMBO J. 1991; 10:4391–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu J., Sneeden J., Heyer W.D.. Tsubouchi H. In vitro assays for DNA pairing and recombination-associated DNA synthesis. DNA Recombination: Methods and Protocols. 2011; 745:363–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cafarelli T.M., Rands T.J., Benson R.W., Rudnicki P.A., Lin I., Godoy V.G.. A single residue unique to DinB-like proteins limits formation of the polymerase IV multiprotein complex in Escherichia coli. J. Bacteriol. 2013; 195:1179–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tashjian T.F., Lin I., Belt V., Cafarelli T.M., Godoy V.G.. RNA primer extension hinders DNA synthesis by Escherichia coli mutagenic DNA Polymerase IV. Front. Microbiol. 2017; 8:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koren S., Harhay G.P., Smith T.P., Bono J.L., Harhay D.M., McVey S.D., Radune D., Bergman N.H., Phillippy A.M.. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013; 14:R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lovett S. Replication arrest-stimulated recombination: Dependence on the RecA paralog, RadA/Sms and translesion polymerase, DinB. DNA Repair. 2006; 5:1421–1427. [DOI] [PubMed] [Google Scholar]
- 34. Kim S.R., Matsui K., Yamada M., Gruz P., Nohmi T.. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics. 2001; 266:207–215. [DOI] [PubMed] [Google Scholar]
- 35. Hastings P.J., Hersh M.N., Thornton P.C., Fonville N.C., Slack A., Frisch R.L., Ray M.P., Harris R.S., Leal S.M., Rosenberg S.M.. Competition of Escherichia coli DNA Polymerases I, II and III with DNA Pol IV in stressed cells. PLoS One. 2010; 5:e10862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mallik S., Popodi E.M., Hanson A.J., Foster P.L.. Interactions and localization of Escherichia coli error-Prone DNA polymerase IV after DNA damage. J. Bacteriol. 2015; 197:2792–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pomerantz R.T., Kurth I., Goodman M.F., O’Donnell M.E.. Preferential D-loop extension by a translesion DNA polymerase underlies error-prone recombination. Nat. Struct. Mol. Biol. 2013; 20:748–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Henrikus S.S., Wood E.A., McDonald J.P., Cox M.M., Woodgate R., Goodman M.F., van Oijen A.M., Robinson A.. DNA polymerase IV primarily operates outside of DNA replication forks in Escherichia coli. PLOS Genet. 2018; 14:e1007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Volodin A.A., Bocharova T.N., Smirnova E.A., Camerini-Otero R.D.. Reversibility, equilibration, and fidelity of strand exchange reaction between short oligonucleotides promoted by RecA protein from Escherichia coli and human Rad51 and Dmc1 proteins. J. Biol. Chem. 2009; 284:1495–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sagi D., Tlusty T., Stavans J.. High fidelity of RecA-catalyzed recombination: a watchdog of genetic diversity. Nucleic Acids Res. 2006; 34:5021–5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Singleton M.R., Dillingham M.S., Gaudier M., Kowalczykowski S.C., Wigley D.B.. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004; 432:187–193. [DOI] [PubMed] [Google Scholar]
- 42. Taylor A.F., Amundsen S.K., Guttman M., Lee K.K., Luo J., Ranish J., Smith G.R.. Control of RecBCD enzyme activity by DNA binding- and chi hotspot-dependent conformational changes. J. Mol. Biol. 2014; 426:3479–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Darmon E., Leach D.R.F.. Bacterial genome instability. Microbiol. Mol. Biol. Rev. 2014; 78:1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.