Abstract
High levels of salinity induce serious oxidative damage in plants. Flavonoids, as antioxidants, have important roles in reactive oxygen species (ROS) scavenging. In the present study, the tobacco R2R3 MYB type repressor, NtMYB4, was isolated and characterized. The expression of NtMYB4 was suppressed by salinity. Overexpression of NtMYB4 reduced the salt tolerance in transgenic tobacco plants. NtMYB4 repressed the promoter activity of NtCHS1 and negatively regulated its expression. Rutin accumulation was significantly decreased in NtMYB4 overexpressing transgenic plants and NtCHS1 RNAi silenced transgenic plants. Moreover, high H2O2 and contents were detected in both types of rutin-reduced transgenic plants under high salt stress. In addition, exogenous rutin supplementation effectively scavenged ROS (H2O2 and ) and improved the salt tolerance of the rutin-reduced transgenic plants. In contrast, NtCHS1 overexpressing plants had increased rutin accumulation, lower H2O2 and contents, and higher tolerance to salinity. These results suggested that tobacco NtMYB4 acts as a salinity response repressor and negatively regulates NtCHS1 expression, which results in the reduced flavonoid accumulation and weakened ROS-scavenging ability under salt stress.
Keywords: NtMYB4, NtCHS1, flavonoid pathway, ROS level, salt stress
Introduction
Salinity is one of the most significant abiotic stresses affecting plant growth and agricultural productivity. The response of plants to salinity is a complex set of traits that involves signaling and metabolic processes at molecular, cellular, and whole-plant levels. Extensive research has been carried out on the mechanisms of plant salt tolerance, including ion homeostasis, osmotic adjustment, reactive oxygen species (ROS) scavenging, detoxification, signal transduction, transcription, and crosstalk with other stresses (Tuteja, 2007; Zhang et al., 2012; Deinlein et al., 2014). However, the detailed mechanism underlying plant salt tolerance is unclear.
The exposure of plants to salinity results in massive changes in gene expression. Transcription factors (TFs) are initially vital in sensing salt and stimulating tolerance responses (Deinlein et al., 2014). TFs, including MYB, WRKY, bHLH, bZIP, and NAC families, are differentially expressed in response to salinity (Jiang and Deyholos, 2009; Jiang et al., 2009; Yang et al., 2009; He et al., 2012; Cui et al., 2013; Deinlein et al., 2014). These TFs regulate the expression levels of various genes that may function in salt tolerance of plants. Studies have demonstrated that gene manipulation could alter the salt tolerance of plants. For example, the ectopic expression of TaMYB73 from wheat in Arabidopsis thaliana activates the expression of stress signaling genes such as AtCBF3 and AtABF3, and enhances the tolerance to ionic and salinity stress (He et al., 2012). In turn, the TF AtbZIP24, which down-regulates the expression of stress inducible-genes such as the Na+ transporter AtHKT1;1, is required during salt stress in plants. In this trend, AtbZIP24 silenced transgenic Arabidopsis shows increased salt tolerance compared to wild type (WT) plants (Yang et al., 2009). In the same line, the ethylene responsive factor BpERF11 down-regulates the expression of important genes involved in abiotic stress tolerance such as LEA and DHN, and leads to reduced levels of proline and the accumulation of ROS. Thus, BpERF11 negatively regulates plant salt and osmotic tolerance (Zhang et al., 2016).
Flavonoids are important secondary metabolites in plants and have important roles in the resistance to oxidative damage caused by ROS during plant growth and abiotic stresses due to their antioxidant activity (Moore et al., 2005; Yu and Jez, 2008; Agati et al., 2012; Pandey et al., 2015). It has been reported that flavonols modulate ROS levels to control the stomata aperture (Watkins et al., 2017) and the formation of root hairs (Maloney et al., 2014). Accumulation of flavonoids, such as flavonols, anthocyanins, and proanthocyanidins in plants could enhance salt tolerance (Liu et al., 2014; Li P. et al., 2017). In this trend, moderate salt stress was shown to induce the accumulation of flavonoids and promote the quality of agricultural crops and medicinal plants (Lim et al., 2012; Colla et al., 2013). Rutin is a glycoside flavonoid that is abundant in a wide variety of plants. Rutin has received much attention due to its health benefits, such as activity against diabetes and inflammation, prevention of hypertension, and antioxidant properties (Kerdudo et al., 2014; Yoo et al., 2014; Kaur and Muthuraman, 2016; Ghorbani, 2017). Rutin biosynthesis is regulated by flavonoid structural genes and TFs. Six structural genes, PAL, C4H, 4CL, CHS, CHI, and FLS, are the major genes involved in the rutin biosynthetic pathway (Winkel-Shirley, 2001). Rutin is one of the most abundant flavonoids, accounting for more than 90% of total flavonoids in tobacco (Li et al., 2014). For this reason, tobacco is the proper model plant to explore the function of rutin in salt tolerance.
Increasing evidence has indicated that R2R3 MYB TFs play important roles in regulating the flavonoid biosynthetic pathway in plants (Dubos et al., 2010; Zhang et al., 2014; Zhou et al., 2017a). R2R3 MYB subgroup 4 TFs, AtMYB3, AtMYB4, AtMYB7, and AtMYB32 are involved in repression of the phenylpropanoid and flavonoid pathways in Arabidopsis (Jin et al., 2000; Dubos et al., 2010; Fornalé et al., 2014; Zhou et al., 2017b). Recently, many genes orthologous to AtMYB4, including ZmMYB31, ZmMYB42, EgMYB1, BrMYB4, CsMYB4a, and FtMYB11, were identified from different plants, all of which negatively regulate phenylpropanoid biosynthesis (Fornalé et al., 2006; Legay et al., 2010; Zhang et al., 2014; Li M. et al., 2017; Zhou et al., 2017a). To the best of our knowledge, tobacco MYB repressors have not been studied yet.
In the present study, we identified an R2R3 MYB TF, NtMYB4, which is orthologous to AtMYB4. We determined that the expression of the NtMYB4 gene was significantly repressed by high salinity. In addition overexpression of NtMYB4 resulted in reduced rutin accumulation, which affects the antioxidant ability of plants under salt stress. Our results indicate that the reduced rutin contents might be a consequence of the repression of the biosynthetic gene NtCHS1 by NtMYB4. As a whole, these data indicate that NtMYB4 plays an important function during the responses of plant to saltine stress.
Materials and Methods
NtMYB4 Gene Isolation
A search for sequences orthologous to AtMYB4 was conducted using the Blastp tool in China Tobacco Genome Database1. The isolated MYB protein sequences were aligned and MYB proteins containing the conserved LLsrGIDPxT/SHRxI/L, EAR repression, zinc-finger (CX1–2CX7–12CX2C), and GY/FDFLGL motifs in the C-termini were selected for further analysis (Jin et al., 2000; Zhou et al., 2015). PCR was used to clone the full length cDNA of NtMYB4 with the primers NtMYB4-F/NtMYB4-R (Supplementary Table S1). The PCR products were purified and cloned with a pEASY-Blunt Cloning Kit (TransGen Biotech, China) and then sequenced. RNA isolation from tobacco leaves, cDNA synthesis, and PCR were performed as described in Chen et al. (2017). Protein sequences were aligned in DNAMAN software (Lynnon Biosoft, United States). A phylogenetic tree was constructed with the neighbor-joining (NJ) algorithm in the MEGA6.0 program (Kumar et al., 2008). The robustness of the tree topology was assessed using 1,000 bootstrap replicates.
Genetic Construction and Plant Transformation
For the preparation of cassettes for the overexpression of NtMYB4 and NtCHS1 and for the silencing of NtCHS1, each gene-specific amplicon was amplified from tobacco leaf cDNA, synthetized as reported in the following paragraph. In order to construct the overexpression vector NtMYB4-pCHF1, the entire coding sequence (CDS) of NtMYB4 was amplified using primers NtMYB4OE-F/NtMYB4OE-R (Supplementary Table S1). The PCR product was digested with XbaI/SacI and cloned into the XbaI/SacI digested pCHF1 plasmid under the control of the CaMV 35S promoter. To construct the overexpression vector NtCHS1-pC3301-ZDS, coding region (CDS) of NtCHS1 (Chen et al., 2017) was amplified using primers NtCHS1OE-F/NtCHS1OE-R (Supplementary Table S1). The PCR product was digested with XbaI/SacI and cloned into the XbaI/SacI digested pC3301-ZDS plasmid under the control of the CaMV 35S promoter. A 293 bp long fragment of NtCHS1 was amplified using the gene-specific primers NtCHS1RNAi-F/NtCHS1RNAi-R, which contain attB sites (underlined) (Supplementary Table S1). Using BP and LR reactions, the target fragment was then ligated into the destination vector pH7GWIWG2(I) (Xu et al., 2011) according to the GatewayTM manufacturer’s instructions (Invitrogen, Carlsbad, CA, United States), to yield the NtCHS1-RNAi vector. The pCHF1-NtMYB4, pC3301-ZDS-NtCHS1 and NtCHS1-RNAi vectors were introduced into Agrobacterium tumefaciens strain LBA4404. Transformation of tobacco was performed using the leaf disc method as reported by Horsch et al. (1985).
In order to construct the subcellular localization vector NtMYB4-CPB-YFP, NtMYB4-YFP-F/NtMYB4-YFP-R primers (Supplementary Table S1) were used to amplify the NtMYB4 coding region and the PCR product was cloned into the BamHI digested pCPB-YFP plasmid using the ClonExpress® Entry One Step Cloning Kit (Vazyme Biotech, China).
With the purpose of constructing the reporters and effectors for UAS/GAL4-based transcriptional repression assay. NtMYB4BD-F/NtMYB4BD-R primers (Supplementary Table S1) were used to amplify the NtMYB4 coding region. For constructing the pGBKT7-NtMYB4 vector, the PCR product was cloned into the BamHI digested pGBKT7 plasmid using the ClonExpress® Entry One Step Cloning Kit (Vazyme Biotech, China). To construct the effector vectors, two primer pairs GAL4BD-F/GAL4BD-R and GAL4BD-F/PMDC32-NtMYB4-R (Supplementary Table S1) were used to amplify the fusion sequence from pGBKT7-NtMYB4. Then the fusion sequences were cloned into the BamHI digested pMDC32 plasmid using the ClonExpress® Entry One Step Cloning Kit. The reporters 35S-UAS::GUS were constructed according to the method of Chen et al. (2014).
With the purpose of constructing the reporters and effectors for the dual luciferase assay, seven primer pairs pPAL-F/pPAL-R, pC4H-F/pC4H-R, p4CL-F/p4CL-R, pCHS1-F/pCHS1-R, pCHI-F/pCHI-R, pFLS-F/pFLS-R, and pANS-F/pANS-R (Supplementary Table S1) were designed to amplify the promoter sequences of the genes NtPAL, NtC4H, Nt4CL, NtCHS1, NtCHI, NtFLS, and NtANS, respectively. For pGreen vectors construction, the promoter sequences of seven genes were cloned into the BamHI digested pGreen plasmid using the ClonExpress® Entry One Step Cloning Kit.
Quantitative RT-PCR Analysis (qRT-PCR)
Total RNA was extracted from WT tobacco tissue, such as leaves, roots, stems, buds and flowers, and from transgenic tobacco leaves according to the method described in Chen et al. (2017). First-strand cDNA was synthesized using a PrimeScriptTM RT Master Mix (Perfect Real Time) (Takara, Clontech, Japan) according to the manufacturer’s instructions. The qRT-PCR was performed using SYBR® Premix Ex TaqTM (Tli RNaseH Plus) (Takara, Clontech, Japan), and primers listed in Supplementary Table S1. The Tob103 gene (GenBank accession no. U60495) served as an internal control. The reaction mixtures consisted in 10 μl SYBR Green mix, 0.4 μl forward qRT-PCR primer, 0.4 μl reverse primer, 1 μl template cDNA, and 7.8 μl sterile water. The thermal cycling parameters were 95°C for 30 s, followed by 40 cycles of 95°C for 15 s and 58°C for 34 s. The relative expression levels were normalized to the expression of the Tob103 gene. The comparative cycle threshold (ΔΔCT) method was used to calculate the relative expression levels of the target genes. Data were expressed as the mean ± SD as determined from three independent biological replicates.
Subcellular Localization
NtMYB4-CPB-YFP construct was transformed into A. tumefaciens EHA105 strain. A single bacterial clone was grown at 28°C overnight, centrifuged for 10 min at 4,200 rpm, re-suspended in 10 mM MgCl2/10 mM MES to a final OD = 1.0 and cultured for 2–3 h with 2 μl 100 mM acetosyringone in the dark. Transformed bacteria were infiltrated into leaves of Nicotiana benthamiana (Chen et al., 2014). The leaves were collected between 48 and 72 h after infiltration and examined by confocal laser scanning microscopy. Images of triplicate infiltrated leaves were acquired with a Fluo ViewTM FV1000 microscope equipped with an argon laser line of 515 nm (excitation) for YFP signal.
Dual Luciferase Assay
The binding to promoters of flavonoid structural genes was assessed to test the transcriptional regulation activity of NtMYB4. Five promoter-pGreen vectors, pCHF1-NtMYB4, pCHF1 empty vector, and P19 were transformed into A. tumefaciens AH105. The bacteria were grown with 2 μl 100 mM acetosyringone at 28°C overnight. To prepare the suspension used for infiltration, bacteria were re-suspended in 10 mM MgCl2/10 mM MES to a final OD = 1.0. Transformed bacteria containing pGreen, pCHF1 and P19 vectors were combined at a 3:3:1 volume ratio, cultured for 1–2 h in the dark and then infiltrated into leaves of N. benthamiana as reported above. The leaves were harvested for dual luciferase assay analysis at 72 h after infiltration according to the instructions of the Dual-Luciferase® Reporter Assay System (Promega, United States).
UAS/GAL4-Based Transcriptional Repression Assay
A transient assay was carried out to test NtMYB4 transcriptional repression activity. To prepare the suspension used for infiltration, bacteria were re-suspended in 10 mM MgCl2/10 mM MES to a final OD = 1.0. Transformed bacteria containing 35S-UAS::GUS, GAL4BD/GAL4BD-NtMYB4 and P19 vectors were combined at a 2:2:1 volume ratio and cultured for 2–3 h in the dark. Transformed bacteria were infiltrated into leaves of N. benthamiana and cultured under weak light conditions. The leaves were harvested for GUS staining at 72 h after infiltration (Liu et al., 2010).
Plant Growth and Salt Treatment
Seeds of WT and transgenic plants of Nicotiana tabacum cultivar Honghuadajinyuan (HD) were sterilized in 75% ethanol for 30 s and then washed three times in sterile water. After that, they were further sterilized in 15% (w/v) H2O2 solution for 8 min and then washed three times in sterile water. For the germination test, the sterilized seeds were cultured on MS solid medium with 0 mM NaCl, 200 mM NaCl, or 200 mM NaCl + 100 μM rutin, respectively. The germination rate was measured in the next 0–11 days after sowing. For the root length test, the sterilized seeds were cultured on 1/2 MS solid medium. Ten day-old seedlings were then transferred to MS solid medium with 0 mM NaCl, 200 mM NaCl, or 200 mM NaCl + 100 μM rutin. The plates were cultured in growth chambers under a 16 h light/8 h dark photoperiod at 28°C. Root length was measured at 20 days after transfer. Forty seeds were used per experiment, and all experiments were repeated three times independently. For the analysis of NtMYB4 expression under salt stress, seedlings with three or four leaves grown on basal MS medium were transferred to a liquid MS medium containing 200 mM NaCl for 12 or 24 h. The corresponding control was prepared by incubating seedlings in MS liquid medium for the same time. Leaves were harvested after 0, 12, 24 h, and 30 days of treatments according to the method of Chen et al. (2017). All collected samples were frozen in liquid nitrogen and stored at −80°C.
Measurements of H2O2 and Content
Collected leaves of 1-month-old plants treated with high salt stress (200 mM NaCl) were used to measure ROS levels. The measurement of H2O2 content was based on the method described by Libik et al. (2005). The content was determined by measuring the formation of red azo compound, which has specific absorption peak at 530 nm, according to the Detection Kit (SA-1-G, Keming, China). In addition, H2O2 and were detected visually using the 3’3’-diaminobenzidine (DAB) and nitro blue tetrazolium (NBT) histochemical staining method as described by Zhao et al. (2016). For measurements of each treatment, leaves were collected from at least three plants.
Measurement of Rutin by High-Performance Liquid Chromatography
Rutin was extracted according to the method described by Li et al. (2015), with some modifications. Dried leaves samples (0.1 g) were extracted with 1 ml of 80% ethanol by 20 min at 40°C with ultrasound, followed by a new extraction with 95% ethanol for 10 min, and then three washes with 80% ethanol 10 min. Supernatants were collected and the volume brought to 50 ml by distilled water and filtered in a 0.45 μm, 13 mm Millex Syringe Filter (Merck Millipore, Carrigtwohill Co., Cork, Ireland). Rutin contents in extracts were analyzed by HPLC as described previously (Huang et al., 2016). Analysis was carried out on three independent biological replicates each containing three technical replicates.
Measurement of Anthocyanin Content
The flowers were collected in the full-blossom period of WT and NtCHS1 transgenic tobacco. Anthocyanin from flowers were measured by spectrophotometry according to the method of Pattanaik et al. (2010). All samples were measured as triplicates in three independent biological replicates.
Statistical Analysis
Data were analyzed by Duncan’s multiple range tests in the ANOVA program of SPSS (IBM SPSS 22). The P-value less than 0.05 and 0.001 was considered statistically significant.
Results
Characterization of NtMYB4, a Nuclear Localized Transcription Repressor
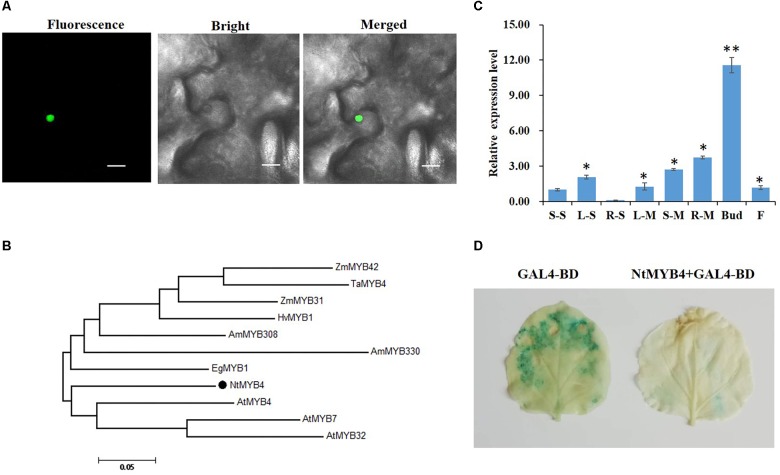
By blasting the tobacco genome database with AtMYB4 amino acid sequence, we identified a R2R3 MYB, NtMYB4, as orthologous to AtMYB4. An alignment of amino acid sequences of NtMYB4 and three other orthologous sequences revealed that it contained the R2 and R3 conserved motifs in the N-terminal region, and the conserved LLsrGIDPxT/SHRxI/L motif, EAR repression motif, zinc-finger domain (CX1–2CX7–12CX2C), and GY/FDFLGL motif in the C-terminal region (Supplementary Figure S1). A phylogenetic analysis was performed using the amino acid sequences of NtMYB4 and 10 other orthologous sequences. The result indicated that NtMYB4 clustered with AtMYB4, AtMYB7, and AtMYB32 (Figure 1B), which are members of subgroup 4 and repressors of the phenylpropanoid pathway. Subcellular localization analysis showed that NtMYB4 was a nuclear localized protein (Figure 1A). Spatial transcript accumulation analysis showed that in seedlings NtMYB4 was more expressed in leaves, whereas in mature plants in all organs assayed, especially the floral bud (Figure 1C). A UAS/GAL4-based transcriptional repression assay showed that NtMYB4 had strong repression activity in tobacco leaves (Figure 1D). These results suggested that NtMYB4 might act as a nuclear localized transcription repressor in tobacco.
FIGURE 1.
NtMYB4 is a nuclear localized transcription repressor in tobacco. (A) Subcellular localization analysis of NtMYB4. An NtMYB4-GFP construct was transformed into leaves of N. benthamiana and examined by confocal laser scanning microscopy. A confocal micrograph is shown at the left (green fluorescent protein, GFP), the corresponding differential interference contrast (bright) image is in the middle, and the merged image is at the right. Bars, 20 μm. (B) Phylogenetic tree based on the amino acid sequences of NtMYB4 and 10 other R2R3 MYB subgroup 4 members. The NtMYB4 isolated in this study is highlighted by a black dot. Accession numbers for MYB subgroup 4 sequences are: AmMYB308 (JQ0960), AmMYB330 (P81395), AtMYB4 (AY519615), AtMYB7 (AEC06531), AtMYB32 (NP_195225), EgMYB1 (CAE09058), HvMYB1 (P20026), TaMYB4 (AAT37167), ZmMYB31 (CAJ42202), and ZmMYB42 (CAJ42204). (C) The expression pattern of NtMYB4 in tobacco root, stem, leaf flower and bud was detected by qRT-PCR. Expression data were detected in two representative growth stages of common tobacco, including the seedling (S) and mature (M) stages. Organs sampled included leaf (L), stem (S), root (R), flower (F). Asterisks in (C) indicate differences between tissues (∗P < 0.05, ∗∗P < 0.01). (D) UAS/GAL4-based transcriptional repression assay of NtMYB4 in N. benthamiana leaves. NtMYB4 repressed the 35S-driven GUS activity. Pairwise combinations of constructs as indicated were co-infiltrated into tobacco leaves and stained for GUS activity. The left leaf marked GAL4-BD is 35S-driven GAL4 DNA binding domain (BD) as control; the right leaf marked NtMYB4+GAL4-BD was 35S-driven full-length NtMYB4 fused to the Gal4-BD domain.
NtMYB4 Overexpression Significantly Reduced Salt Tolerance
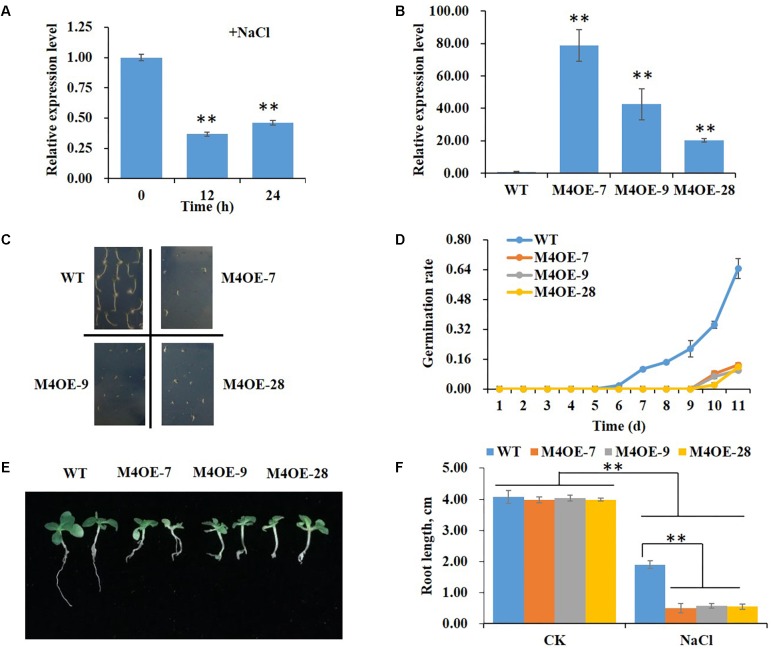
Real-time PCR was performed to determine changes in expression levels of NtMYB4 in WT tobacco in response to high salt stress. The result showed that the level of NtMYB4 mRNA in leaves was significantly lower after 12 and 24 h of seedling treatment with 200 mM NaCl (Figure 2A).
FIGURE 2.
Overexpression of NtMYB4 significantly reduces salt tolerance. The relative expression levels were normalized to the expression of the Tob103 gene (internal control). The relative expression of NtMYB4 in leaves of WT plants was set to 1. (A) The expression of NtMYB4 in leaves of WT plants was significantly suppressed by the addition of 200 mM NaCl in the medium. Asterisks in (A) indicate significant difference (∗∗P < 0.01) between control condition and salt treatment. (B) The expression level of NtMYB4 in leaves was significantly increased in overexpressing transgenic tobacco compared to the WT. (C,D) The seed germination of WT and three NtMYB4 overexpressing transgenic tobacco lines under 200 mM NaCl. (E,F) The root elongation of WT and three NtMYB4 overexpressing transgenic tobacco lines under 200 mM NaCl. Data are expressed as the mean ± SD as determined from three independent biological replicates. Values in (F) that are significantly different between treatments and among lines within each treatment are marked with ∗∗P < 0.01 and ∗P < 0.05.
To verify the function of NtMYB4 in salt tolerance, we overexpressed the NtMYB4 gene in tobacco. Three transgenic lines M4OE-7, M4OE-9, and M4OE-28 were obtained (Figure 2B) and analyzed in detail. The germination rate and root length of WT and transgenic seeds under high salt stress treatment were measured. Even though there were no obvious differences in germination rates between WT and transgenic seeds when grown on basal MS medium (0 mM NaCl), severe suppression was observed in NtMYB4 overexpressing transgenic seeds under 200 mM NaCl as the germination rate was decreased 92% (Figure 2C). Moreover, we also observed that salt stress provoked a delay in germination, which normally occurs at the fourth day. Thus, WT seeds started to germinate on the sixth day in MS amended with 200 mM NaCl, while the NtMYB4 overexpressing transgenic seeds on the ninth day (Figure 2D). On basal medium, no obvious differences in seedling growth and root length were observed between WT and transgenic plants (Figure 2F). Conversely, 200 mM NaCl treatment caused a significant inhibition in root elongation in both WT and transgenic lines, being 53, 87, 86, and 86% the inhibition observed in WT, M4OE-7, M4OE-9, and M4OE-28, respectively. Interestingly, under salt treatment, the root length was significantly reduced in all transgenic lines with respect to WT plants (Figure 2E,F). These results showed that overexpression of NtMYB4 reduced the seed germination rate and root elongation under 200 mM NaCl compared to WT, which indicated that NtMYB4 plays a negative role in plant salt tolerance.
NtMYB4 Overexpression Leads to Reduced Flavonoid Accumulation
Numerous reports have demonstrated that salt stress can induce the accumulation of phenolic compounds in plant tissues (Ksouri et al., 2007; Baâtour et al., 2013). The key structural genes of the phenylpropanoid pathway PAL, CHS, CHI, DFR, FLS, and ANS were induced by salt stress in leaves of WT tobacco (Supplementary Figure S2A). Moreover, rutin accumulation increased significantly under 200 mM NaCl in tobacco (Supplementary Figure S2B). In order to further determine the effects of salt stress in flavonoid biosynthesis, we investigated rutin accumulation in NtMYB4 overexpressing transgenic plants. Rutin content significantly decreased in NtMYB4 overexpressing transgenic tobacco compared to WT (Figure 3A). We speculated that rutin might play a significant role in plant salt tolerance. In order to verify this hypothesis, we supplemented culture media with 100 μmol exogenous rutin. Under these experimental conditions, the inhibition of root length elongation was partially counterbalanced (Figure 3B,C), and plants were more robust overall (Figure 3B). Rutin supplementation in fact significantly reduced the inhibition of root length elongation observed both in WT and transgenic lines on MS with 200 mM NaCl. Only one, M4OE-9, of the three transgenic lines showed root length similar to WT plants on MS supplemented with 200 mM NaCl and 100 μmol rutin (Figure 3C). These result showed that supplementation with rutin enhances the salt tolerance of both WT and NtMYB4 overexpressing transgenic tobacco seedlings.
FIGURE 3.
Supplementation with rutin increases salt tolerance of tobacco. (A) Rutin accumulation was significantly decreased in the leaves of NtMYB4 overexpressing transgenic tobacco lines compared to WT. DW, dry weight. Asterisks in (A) indicate significant difference (∗∗P < 0.01) between NtMYB4-OE and WT. (B,C) The root length of WT and three NtMYB4 overexpressing transgenic lines under 200 mM NaCl with or without 100 μM rutin. Data are expressed as the mean ± SD as determined from three independent biological replicates. Values in (C) that are significantly different between treatments and among lines within each treatment are marked with ∗∗P < 0.01 and ∗P < 0.05.
Rutin Effectively Scavenges ROS Under Salt Stress
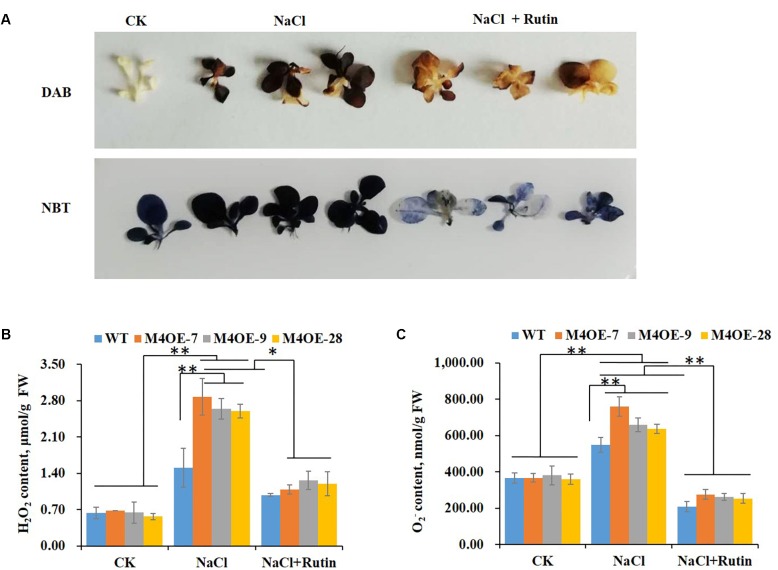
It is well documented that salt stress usually results in the excessive accumulation of ROS, which have deleterious effects on plant cells (Ashraf and Harris, 2004; Torun, 2018). Thanks to their antioxidant capacity, flavonoids play an important role against oxidative injury caused by salt stress (Dixon and Paiva, 1995). To determine the ROS scavenging capability of rutin, seedlings of WT plants with or without 200 mM NaCl treatment were stained with DAB or NBT. The histochemical staining suggested that salt stress dramatically increased the H2O2 or content compared to control. Conversely, supplementation with 100 μmol exogenous rutin during salt stress appeared to markedly decrease the levels of ROS (Figure 4A). The content of H2O2 or was measured using visible spectrophotometry. NtMYB4 overexpressing transgenic plants showed a higher level of ROS (H2O2 or content) than WT plants under salt stress, while exogenous rutin effectively scavenged the ROS (Figure 4B,C). These results showed that rutin plays important roles in scavenging ROS.
FIGURE 4.
Rutin effectively scavenges the ROS levels under salt stress. (A) DAB or NBT staining of WT plants under control (CK) and salt (NaCl) treatment with or without rutin supplementation. (B,C) H2O2 or content of WT plants and NtMYB4 overexpressing transgenic lines under control (CK) and salt (NaCl) treatment with or without rutin supplementation, measured by visible spectrophotometry. FW, fresh weight. Data are expressed as the mean ± SD as determined from three independent biological replicates. Values in (B,C) that are significantly different between treatments and among lines within each treatment are marked with ∗∗P < 0.01 and ∗P < 0.05.
NtMYB4 Negatively Regulates the NtCHS1 Activity
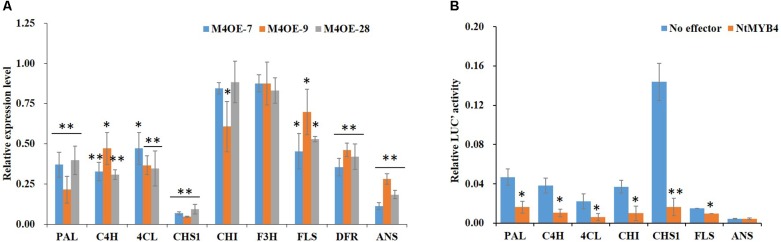
In order to reveal the molecular mechanism mediated by NtMYB4 in regulating flavonoid biosynthesis, we determined the expression levels of flavonoid pathway genes and flavonoid accumulation in WT and NtMYB4 overexpressing transgenic lines. qRT-PCR result showed that transcript levels of PAL, C4H, 4CL, CHS1, FLS, DFR, and ANS were reduced in NtMYB4 overexpressing transgenic plants. Notably, CHS1 and ANS expression was significantly reduced by 94 and 89%, respectively (Figure 5A). Dual-luciferase assay was used to demonstrate whether or not NtMYB4 represses the promoter activity of PAL, C4H, 4CL, CHS1, CHI, FLS, and ANS genes. Results showed that NtMYB4 significantly reduced the luciferase signal controlled by each promoter, except for that of the ANS gene. In particular, the luciferase signal controlled by the promoter of NtCHS1 were reduced by 88.66% (Figure 5B). These results indicated that NtMYB4 regulates negatively the transcription of NtCHS1 and thus represses the flavonoid pathway.
FIGURE 5.
NtMYB4 negatively regulates the expression of flavonoid pathway genes. (A) The expression level of structural genes in NtMYB4 overexpressing transgenic plants were significant inhibited compared to WT. The relative expression of structural genes for the WT control were set to 1. (B) Dual luciferase (LUC) assays of NtMYB4 (effector) and the promoters fused to pGreen luciferase plasmid (reporter). Empty vector was used as the effector in the control assay. The promoters of PAL, C4H, 4CL, CHI, CHS1, FLS, and ANS genes were used in dual luciferase assays (n = 3). Data are expressed as the mean ± SD as determined from three independent biological replicates. Asterisks indicate that the value is significantly different from that of the control (∗P < 0.05, ∗∗P < 0.01).
NtCHS1 RNAi Silenced Transgenic Plants Showed a Similar Phenotype to NtMYB4 Overexpressing Transgenic Plants Under Salt Stress
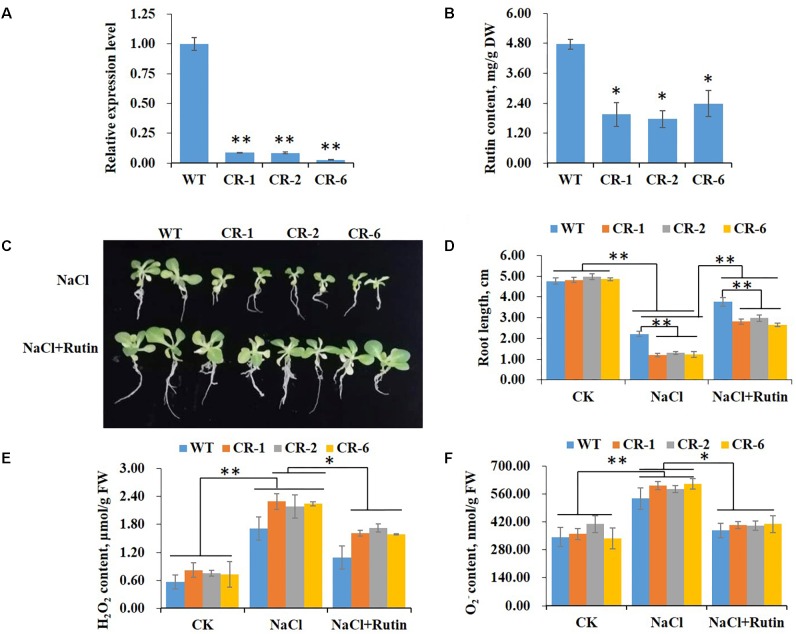
Our previous study revealed that the expression level of NtCHS1 was significantly induced by 200 mM NaCl treatment (Chen et al., 2017). To validate the function of NtCHS1 in salt stress tolerance, we silenced the gene by RNA interference. The relative expression level of NtCHS1 in the leaves of silenced transgenic tobacco lines was then analyzed using qRT-PCR. Three RNAi silenced lines (CR-1, CR-2, CR-6), out of the five independent NtCHS1-RNAi lines produced, were selected for further analyses, as they showed a significant decrease in the expression level of NtCHS1 compared to WT plants (Figure 6A).
FIGURE 6.
NtCHS1 RNAi silenced transgenic plants show a similar phenotype to NtMYB4 overexpressing transgenic plants in salt stress. (A) The expression level of NtCHS1 decreased in RNAi silenced T0 transgenic tobacco leaves compared to WT. Asterisks in (A) indicate remarkable difference (∗∗P < 0.01) between NtCHS1 RNAi transgenic lines and WT. (B) Rutin content decreased in NtCHS1 RNAi silenced transgenic tobacco lines compared to WT. DW, dry weight. Asterisks in (B) indicate remarkable difference (∗P < 0.05) between NtCHS1 RNAi transgenic lines and WT. (C,D) Root length of WT and NtCHS1 RNAi transgenic plants treated with salt stress with or without exogenous rutin. Values in (D) that are significantly different between treatments and among lines within each treatment are marked with ∗∗P < 0.01 and ∗P < 0.05. (E,F) H2O2 or content of WT plants and NtCHS1 RNAi silenced transgenic lines under control (CK) and salt treatment (NaCl) with or without rutin supplementation, measured by visible spectrophotometry. FW, fresh weight. Data are expressed as the mean ± SD as determined from three independent biological replicates. Values in (E,F) that are significantly different between treatments and among lines within each treatment are marked with ∗∗P < 0.01 and ∗P < 0.05.
As shown in Supplementary Figure S3B, except for PAL and DFR, the expression of 4CL, CHI, F3H, FLS, and ANS decreased significantly in NtCHS1 RNAi lines. These lines also displayed a significant decrease in the levels of anthocyanins in the flowers and rutin in the leaves with respect to WT plants (Supplementary Figure S4B and Figure 6B). The results indicated that regulating the expression level of NtCHS1 might alter the expression of major metabolic structural genes and the production of flavonoids in the rutin biosynthesis pathway.
To determine the role of NtCHS1 in salt tolerance, seed germination rate and root length of WT and transgenic tobacco plants cultured on MS medium with or without the addition of 200 mM NaCl were measured. Severe suppression was observed in RNAi-silenced transgenic seeds, as the germination rate was decreased 95% under 200 mM NaCl. Moreover, the germination of NtCHS1 RNAi-silenced transgenic seeds was delayed and it was almost abolished in the presence of 200 mM NaCl (Supplementary Figure S5). For root elongation, there was no obvious difference in seedling growth and root length between WT and transgenic lines under controlled condition (Figure 6D). In the 200 mM NaCl treatment, significant inhibition of root elongation was observed both in WT and RNAi transgenic lines. In addition, under the above condition RNAi transgenic lines displayed a significant reduction in root length with respect to the WT (Figure 6C,D). Supplementation with 100 μmol rutin in 200 mM NaCl MS medium enhanced root elongation of both WT and NtCHS1 RNAi-silenced transgenic tobacco seedlings significantly (Figure 6C,D). Supplementation with rutin also reduced the ROS levels in the leaves of NtCHS1 RNAi silenced transgenic plants (Figure 6E,F), as in NtMYB4 overexpressing transgenic plants.
NtCHS1 Overexpressing Transgenic Plants Enhanced Salt Tolerance
To further determine the function of NtCHS1 in tolerance to high salt stress, we overexpressed the NtCHS1 gene in tobacco. Three overexpression lines (CO-2, CO-3, and CO-5) were obtained. The results showed that the expression level of NtCHS1 was significantly increased in the leaves of overexpressing lines compared to WT plants (Figure 7A). We also analyzed the expression levels of the structural genes involved in the flavonoid biosynthetic pathway in NtCHS1 overexpressing lines. As shown in Supplementary Figure S3A, except for PAL with no obvious expression change, the expression of 4CL, CHI, F3H, DFR, FLS, and ANS were significantly up-regulated in NtCHS1 overexpressing lines. In addition, rutin content was significantly increased in overexpressing lines (Figure 7B). However, no obvious difference in anthocyanin content was detected between the three overexpression lines and WT (Supplementary Figure S4A).
FIGURE 7.
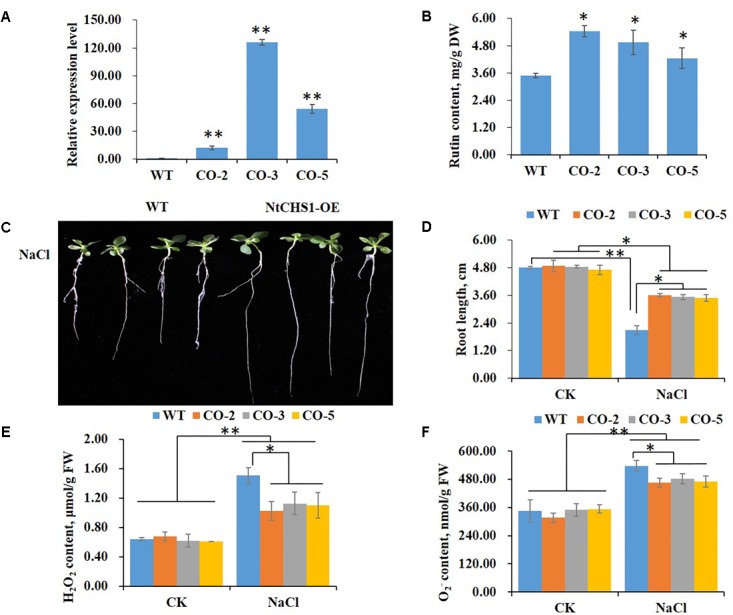
NtCHS1 overexpressing transgenic plants significantly increase salt tolerance. (A) The expression level of NtCHS1 in leaves was significantly increased in overexpressing T0 transgenic tobacco leaves compared to WT. Asterisks in (A) indicate remarkable difference (∗∗P < 0.01) between NtCHS1 overexpressing transgenic lines and WT. (B) Rutin content increased in the leaves of typical NtCHS1 overexpressing transgenic tobacco lines compared to WT. Asterisks in (B) indicate remarkable difference (∗P < 0.05) between NtCHS1 overexpressing transgenic lines and WT. (C,D) Root length of WT and NtCHS1 overexpressing transgenic plants (CO-3 line) treated with salt stress. Values in (D) that are significantly different between treatments and among lines within each treatment are marked with ∗∗P < 0.01 and ∗P < 0.05. (E,F) H2O2 or content of WT plants and NtCHS1 overexpressing transgenic lines under control (CK) and salt treatment (NaCl), measured by visible spectrophotometry. FW, fresh weight. Data are expressed as the mean ± SD as determined from three independent biological replicates. Values in (E,F) that are significantly different between treatments and among lines within each treatment are marked with ∗∗P < 0.01 and ∗P < 0.05.
The seed germination rate and root length of WT and NtCHS1 overexpressing transgenic tobacco plants were also measured. These parameters did not change between WT and transgenic lines under controlled condition. Under 200 mM NaCl, the germination delayed with respect to the controlled condition in all plants, but the germination rate increased in NtCHS1 overexpressing lines with respect to the control (Supplementary Figure S5). Notably, salt stress induced a significant inhibition of root elongation in both WT (55%), and, although less markedly (26%) in transgenic lines as well (Figure 7C,D). Furthermore, salt stress induced a significant increased in H2O2 or content in both WT and transgenic tobacco leaves. However, NtCHS1 overexpressing transgenic lines had significant lower H2O2 or content than WT plants under salt stress (Figure 7E,F). These results showed that NtCHS1 overexpression caused flavonoid accumulation and affected the rate of ROS scavenging.
Discussion
In this study, we identified and characterized the NtMYB4 gene from tobacco. Sequence analysis revealed that NtMYB4 contains the R2R3 domain, the zinc-finger domain as well as the EAR and LLsrGIDPxT/SHRxI/L motifs (Supplementary Figure S1), which are typical of sub-group 4 R2R3 MYB repressors (Jin et al., 2000; Zhou et al., 2015). 35S-UAS::GUS leaf transient assay and subcellular analysis showed that NtMYB4 is a nuclear localized transcription repressor in tobacco (Figure 1). Furthermore, dual-luciferase assay verified that NtMYB4 significantly decreased the promoting activity of most of the key genes in flavonoid biosynthesis, including PAL, C4H, 4CL, CHS, CHI, and FLS (Figure 5B). qRT-PCR results confirmed that NtMYB4 overexpression significantly reduced the expression level of the six key genes (Figure 5A).
The expression of CHS was negatively regulated by subgroup 4 R2R3 TFs, including AtMYB4, AtMYB7, CsMYB4a, and FtMYB11 (Jin et al., 2000; Fornalé et al., 2014; Li M. et al., 2017; Zhou et al., 2017a). Our results indicated that NtMYB4 directly repressed NtCHS1 gene expression to negatively regulate rutin biosynthesis in tobacco.
Transcription factors are initially vital in sensing salt and their expression levels are changed by salinity, leading to many tolerance responses (Deinlein et al., 2014). Little is known about how subgroup 4 R2R3 MYB TFs function in salt stress. The expression of AtMYB7 was reported to be induced by salinity in A. thaliana, while AtMYB4 expression was not induced (Fornalé et al., 2014; Kim et al., 2015). Here, the expression level of NtMYB4 was found to be highly repressed by salinity (Figure 2A), indicating that NtMYB4 is a salinity-responsive TF. In order to demonstrate whether NtMYB4 functions in salinity tolerance, we overexpressed NtMYB4 (Figure 2B) and investigated plant response to salinity. The results showed that NtMYB4 overexpressing transgenic lines exhibited a much lower seed germination rate and shorter root length than WT plants (Figure 2C–F). In addition, the rutin content was significantly reduced in NtMYB4 overexpressing transgenic lines. Supplementation with exogenous rutin in saline solution improved the salt tolerance of the NtMYB4 overexpressing transgenic lines. These results indicated that inadequate content of rutin might directly result in the salt sensitivity of NtMYB4 overexpressing transgenic lines.
It is well known that CHS expression is induced by various stimuli, including abiotic and biotic stress (Richard et al., 2000; Zabala et al., 2006; Dao et al., 2011; Chen et al., 2015). In our previous study, we found that NtCHS1 was induced by high salt stress (Chen et al., 2017). Here, we further investigated the functions of NtCHS1 transgenic plants under high salt stress, including overexpression or RNAi-silenced transgenic tobacco lines. Our data showed that NtCHS1 RNAi-silenced transgenic tobacco lines, as NtMYB4 overexpressing transgenic tobacco lines, exhibited reduced rutin contents and salt sensitivity (Figure 6). In addition, NtCHS1 overexpressing transgenic tobacco lines exhibited increased rutin accumulation and higher salt tolerance than WT tobacco (Figure 7). These results further suggest that NtMYB4, by repressing CHS, decreases rutin biosynthesis and, in turn, salt tolerance.
Salinity leads to the overproduction of ROS in plants. ROS are highly reactive and toxic and result in oxidative stress (Gill and Tuteja, 2010). Evidence has accumulated that flavonoids are an important class of antioxidants, and have important roles in plant abiotic stress (Yang et al., 2001; Agati et al., 2012). AtMYB12 has been reported to confer salt and drought tolerance by increasing the levels of flavonoids in transgenic A. thaliana (Wang et al., 2016). Moreover, an extensive integrated analysis of single overexpression of AtMYB75 or AtMYB12, or double overexpression of AtMYB12 and AtMYB75, and tt4 as a flavonoid-deficient mutant, demonstrated that flavonoid over-accumulation was key to enhanced abiotic stress tolerance in Arabidopsis (Nakabayashi et al., 2014). The accumulation of flavonoids is enhanced by salinity (Colla et al., 2013), and the expression of the biosynthetic genes, including PAL, CHS, CHI, DFR, FLS and ANS, were shown to be increased in the present study (Supplementary Figure S3A). Similarly moderate salt stress increased rutin accumulation in buckwheat sprout (Lim et al., 2012), which indicated its function in plant salt tolerance. In the present study, higher ROS levels (H2O2 and activity) were detected in NtMYB4 overexpressing (Figure 3A, 4) and NtCHS1 RNAi-silenced transgenic tobacco, with reduced rutin content (Figure 6). Supplementation with exogenous rutin reduced H2O2 and activity in leaves. Moreover, NtCHS1 overexpressing transgenic tobacco increased rutin accumulation and enhanced scavenging ability (Figure 7) under salinity. These results indicated that rutin exhibits strong ROS scavenging ability.
Conclusion
We characterized NtMYB4 functions as a repressor in salt responsiveness. The expression of NtMYB4 was inhibited by salinity, resulting in the activation of NtCHS1 transcription, followed by the accumulation of rutin to scavenge ROS. The results show that rutin is an important antioxidant and is effective in maintaining the balance of ROS in tobacco under salt stress. These results add more light to the roles played by flavonoids in the plant responses to saline stress.
Author Contributions
SC and AY conceived and designed the research. SC, FW, YL, YQ, XP, CF, FL, and ZW performed the experiments and analyzed the data. SC and FL wrote the manuscript. YW, AY, and HL support for the research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by The Agricultural Science and Technology Innovation Program (ASTIP-TRIC01) and China Postdoctoral Science Foundation (2018M630808).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00178/full#supplementary-material
References
- Agati G., Azzarello E., Pollastri S., Tattini M. (2012). Flavonoids as antioxidants in plants: location and functional significance. Plant Sci. 196 67–76. 10.1016/j.plantsci.2012.07.014 [DOI] [PubMed] [Google Scholar]
- Ashraf M., Harris P. J. C. (2004). Potential biochemical indicators of salinity tolerance in plants. Plant Sci. 166 3–16. 10.1016/j.plantsci.2003.10.024 [DOI] [Google Scholar]
- Baâtour O., Mahmoudi H., Tarchoun I., Nasri N., Trabelsi N., Kaddour R., et al. (2013). Salt effect on phenolics and antioxidant activities of Tunisian and Canadian sweet marjoram (Origanum majorana L.) shoots. J. Sci. Food Agric. 93 134–141. 10.1002/jsfa.5740 [DOI] [PubMed] [Google Scholar]
- Chen G. H., Sun J. Y., Liu M., Liu J., Yang W. C. (2014). SPOROCYTELESS is a novel embryophyte-specific transcription repressor that interacts with TPL and TCP proteins in Arabidopsis. J. Genet. Genomics 41 617–625. 10.1016/j.jgg.2014.08.009 [DOI] [PubMed] [Google Scholar]
- Chen L. J., Guo H. M., Lin Y., Cheng H. M. (2015). Chalcone synthase EaCHS1 from Eupatorium adenophorum functions in salt stress tolerance in tobacco. Plant Cell Rep. 34 885–894. 10.1007/s00299-015-1751-7 [DOI] [PubMed] [Google Scholar]
- Chen S., Pan X. H., Li Y. T., Cui L. J., Zhang Y. C., Zhang Z. M., et al. (2017). Identification and characterization of chalcone synthase gene family members in Nicotiana tabacum. J. Plant Growth Regul. 36 374–384. 10.3389/fpls.2016.01089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colla G., Rouphael Y., Cardarelli M., Svecova E., Rea E., Lucini L. (2013). Effects of saline stress on mineral composition, phenolic acids and flavonoids in leaves of artichoke and cardoon genotypes grown in floating system. J. Sci. Food Agric. 93 1119–1127. 10.1002/jsfa.5861 [DOI] [PubMed] [Google Scholar]
- Cui M. H., Yoo K. S., Hyoung S., Nguyen H. T., Kim Y. Y., Kim H. J., et al. (2013). An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 587 1773–1778. 10.1016/j.febslet.2013.04.028 [DOI] [PubMed] [Google Scholar]
- Dao T. T., Linthorst H. J., Verpoorte R. (2011). Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10 397–412. 10.1007/s11101-011-9211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinlein U., Stephan A. B., Horie T., Luo W., Xu G. H., Schroeder J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19 371–379. 10.1016/j.tplants.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. A., Paiva N. L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7 1085–1097. 10.1105/tpc.7.7.1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaa B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15 573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Fornalé S., Lopez E., Salazar-Henao J. E., Fernández-Nohales P., Rigau J., Caparros-Ruiz D. (2014). AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 55 507–516. 10.1093/pcp/pct187 [DOI] [PubMed] [Google Scholar]
- Fornalé S., Sonbol F. M., Maes T., Capellades M., Puigdomènech P., Rigau J., et al. (2006). Down-regulation of the maize and Arabidopsis thaliana caffeic acid O-methyl-transferase genes by two new maize R2R3-MYB transcription factors. Plant Mol. Biol. 62 809–823. 10.1007/s11103-006-9058-2 [DOI] [PubMed] [Google Scholar]
- Ghorbani A. (2017). Mechanisms of antidiabetic effects of flavonoid rutin. Biomed. Pharmacother. 96 305–312. 10.1016/j.biopha.2017.10.001 [DOI] [PubMed] [Google Scholar]
- Gill S. S., Tuteja N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48 909–930. 10.1016/j.plaphy.2010.08.016 [DOI] [PubMed] [Google Scholar]
- He Y., Li W., Lv J., Jia Y., Wang M., Xia G. (2012). Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 63 1511–1522. 10.1093/jxb/err389 [DOI] [PubMed] [Google Scholar]
- Horsch R. F., Fry J. E., Hoffmann N. L., Eichholtz D., Rogers S. G., Fraley R. T. (1985). A simple and general method for transferring genes into plants. Science 227 1229–1231. 10.1126/science.227.4691.1229 [DOI] [PubMed] [Google Scholar]
- Huang X., Yao J., Zhao Y., Xie D., Jiang X., Xu Z. (2016). Efficient rutin and quercetin biosynthesis through flavonoids-related gene expression in Fagopyrum tataricum Gaertn. Hairy root cultures with UV-B irradiation. Front. Plant Sci. 7:63. 10.3389/fpls.2016.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Deyholos M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69 91–105. 10.1007/s11103-008-9408-3 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Yang B., Deyholos M. K. (2009). Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol. Genet. Genomics 282 503–516. 10.1007/s00438-009-0481-3 [DOI] [PubMed] [Google Scholar]
- Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19 6150–6161. 10.1093/emboj/19.22.6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Muthuraman A. (2016). Therapeutic evaluation of rutin in two-kidney one-clip model of renovascular hypertension in rat. Life Sci. 150 89–94. 10.1016/j.lfs.2016.02.080 [DOI] [PubMed] [Google Scholar]
- Kerdudo A., Dingas A., Fernandez X., Faure C. (2014). Encapsulation of rutin and naringenin in multilamellar vesicles for optimum antioxidant activity. Food Chem. 159 12–19. 10.1016/j.foodchem.2014.03.005 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Hyun W. Y., Nguyen H. N., Jeong C. Y., Xiong L., Hong S. W., et al. (2015). AtMyb7, a subgroup 4 R2R3 Myb, negatively regulates ABA-induced inhibition of seed germination by blocking the expression of the bZIP transcription factor ABI5. Plant Cell Environ. 38 559–571. 10.1111/pce.12415 [DOI] [PubMed] [Google Scholar]
- Ksouri R., Megdiche W., Debez A., Falleh H., Grignon C., Abdelly C. (2007). Salinity effects on polyphenol content and antioxidant activities in leaves of the halophyte Cakile maritima. Plant Physiol. Biochem. 45 244–249. 10.1016/j.plaphy.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J., Tamura K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9 299–306. 10.1093/bib/bbn017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legay S., Sivadon P., Blervacq A. S., Pavy N., Baghdady A., Tremblay L., et al. (2010). EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytol. 188 774–786. 10.1111/j.1469-8137.2010.03432.x [DOI] [PubMed] [Google Scholar]
- Li M., Li Y., Guo L., Gong N., Pang Y., Jiang W., et al. (2017). Functional characterization of tea (Camellia sinensis) MYB4a transcription factor using an integrative approach. Front. Plant Sci. 8:943. 10.3389/fpls.2017.00943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Li Y. J., Zhang F. J., Zhang G. Z., Jiang X. Y., Yu H. M., et al. (2017). The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 89 85–103. 10.1111/tpj.13324 [DOI] [PubMed] [Google Scholar]
- Li X., Du Y., Zhang H., Kuai Y., Liu C., Li D. (2015). Extracting chlorogenic acid, rutin, nicotine, and solanesol from tobacco. Chin. Tobacco Sci. 36 1–4. [Google Scholar]
- Li Y., Pang T., Shi J., Lu X., Deng J., Lin Q. (2014). Liquid chromatography with mass spectrometry method based two-step precursor ion scanning for the structural elucidation of flavonoids. J. Sep. Sci. 37 3067–3073. 10.1002/jssc.201400720 [DOI] [PubMed] [Google Scholar]
- Libik M., Konieczny R., Pater B., Slesak I., Miszalski Z. (2005). Differences in the activities of some antioxidant enzymes and in H2O2 content during rhizogenesis and somatic embryogenesis in callus cultures of the ice plant. Plant Cell Rep. 23 834–841. 10.1007/s00299-004-0886-8 [DOI] [PubMed] [Google Scholar]
- Lim J. H., Park K. J., Kim B. K., Jeong J. W., Kim H. J. (2012). Effect of salinity stress on phenolic compounds and carotenoids in buckwheat (Fagopyrum esculentum M.) sprout. Food Chem. 135 1065–1070. 10.1016/j.foodchem.2012.05.068 [DOI] [PubMed] [Google Scholar]
- Liu L. J., Zhang Y. Y., Tang S. Y., Zhao Q. Z., Zhang Z. H., Zhang H. W., et al. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61 893–903. 10.1111/j.1365-313X.2009.04109.x [DOI] [PubMed] [Google Scholar]
- Liu S., Ju J., Xia G. (2014). Identification of the flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes from Antarctic moss and their regulation during abiotic stress. Gene 543 145–152. 10.1016/j.gene.2014.03.026 [DOI] [PubMed] [Google Scholar]
- Maloney G. S., DiNapoli K. T., Muday G. K. (2014). The anthocyanin reduced tomato mutant demonstrates the role of flavonols in tomato lateral root and root hair development. Plant Physiol. 166 614–631. 10.1104/pp.114.240507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. P., Westall K. L., Ravenscroft N., Farrant J. M., Lindsey G. G., Brandt W. F. (2005). The predominant polyphenol in the leaves of the resurrection plant Myrothamnus flabellifolius, 3,4,5 tri-O-galloylquinic acid, protects membranes against desiccation and free radical-induced oxidation. Biochem. J. 385 301–308. 10.1042/BJ20040499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi R., Yonekura-Sakakibara K., Urano K., Suzuki M., Yamada Y., Nishizawa T., et al. (2014). Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77 367–379. 10.1111/tpj.12388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Misra P., Choudhary D., Yadav R., Goel R., Bhambhani S., et al. (2015). AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues. Sci. Rep. 5:12412. 10.1038/srep12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanaik S., Kong Q., Zaitlin D., Werkman J. R., Xie C. H., Patra B., et al. (2010). Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta 231 1061–1076. 10.1007/s00425-010-1108-y [DOI] [PubMed] [Google Scholar]
- Richard S., Lapointe G., Rutledge R. G., Séguin A. (2000). Induction of chalcone synthase expression in white spruce by wounding and jasmonate. Plant Cell Physiol. 41 982–987. 10.1093/pcp/pcd017 [DOI] [PubMed] [Google Scholar]
- Torun H. (2018). Time-course analysis of salicylic acid effects on ROS regulation and antioxidant defense in roots of hulled and hulless barley under combined stress of drought, heat and salinity. Physiol. Plant. 10.1111/ppl.12798 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Tuteja N. (2007). Chapter twenty-four-mechanisms of high salinity tolerance in plants. Methods Enzymol. 428 419–438. 10.1016/S0076-6879(07)28024-3 [DOI] [PubMed] [Google Scholar]
- Wang F., Kong W., Wong G., Fu L., Peng R., Li Z., et al. (2016). AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genomics 291 1545–1559. 10.1007/s00438-016-1203-2 [DOI] [PubMed] [Google Scholar]
- Watkins J. M., Chapman J. M., Muday G. K. (2017). Abscisic acid-induced reactive oxygen species are modulated by flavonols to control stomata aperture. Plant Physiol. 175 1807–1825. 10.1104/pp.17.01010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126 485–493. 10.1104/pp.126.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P. S., Bai J. F., Liu J. W., Li H. G. (2011). Construction of LSV and LMoV binary virus resistant RNAi vector using gateway technology. Chin. Agric. Sci. Bull. 27 144–147. [Google Scholar]
- Yang B., Kotani A., Arai K., Kusu F. (2001). Estimation of the antioxidant activities of flavonoids from their oxidation potentials. Anal. Sci. 17 599–604. 10.2116/analsci.17.599 [DOI] [PubMed] [Google Scholar]
- Yang O., Popova O. V., Süthoff U., Lüking I., Dietz K. J., Golldack D. (2009). The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 436 45–55. 10.1016/j.gene.2009.02.010 [DOI] [PubMed] [Google Scholar]
- Yoo H., Ku S. K., Baek Y. D., Bae J. S. (2014). Anti-inflammatory effects of rutin on HMGB1-induced inflammatory responses in vitro and in vivo. Inflamm. Res. 63 197–206. 10.1007/s00011-013-0689-x [DOI] [PubMed] [Google Scholar]
- Yu O., Jez J. M. (2008). Nature’s assembly line: biosynthesis of simple phenylpropanoids and polyketides. Plant J. 54 750–762. 10.1111/j.1365-313X.2008.03436.x [DOI] [PubMed] [Google Scholar]
- Zabala G., Zou J., Tuteja J., Gonzalez D. O., Clough S. J., Vodkin L. O. (2006). Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection. BMC Plant Biol. 6:26. 10.1186/1471-2229-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Han B., Wang T., Chen S., Li H., Zhang Y., et al. (2012). Mechanisms of plant salt response: insights from proteomics. J. Proteome Res. 11 49–67. 10.1021/pr200861w [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang Y., Sun M., Wang J., Kawabata S., Li Y. (2014). BrMYB4, a suppressor of genes for phenylpropanoid and anthocyanin biosynthesis, is down-regulated by UV-B but not by pigment-inducing sunlight in turnip cv. Tsuda. Plant Cell Physiol. 55 2092–2101. 10.1093/pcp/pcu137 [DOI] [PubMed] [Google Scholar]
- Zhang W., Yang G., Mu D., Li H., Zang D., Xu H., et al. (2016). An ethylene-responsive factor BpERF11 negatively modulates salt and osmotic tolerance in Betula platyphylla. Sci. Rep. 6:23085. 10.1038/srep23085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Ren Y. R., Wang Q. J., Wang X. F., You C. X., Hao Y. J. (2016). Ubiquitination-related MdBT scaffold proteins target a bHLH transcription factor for iron homeostasis. Plant Physiol. 172 1973–1988. 10.1104/pp.16.01323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Sun Z., Ding M., Logacheva M. D., Kreft I., Wang D., et al. (2017a). FtSAD2 and FtJAZ1 regulate activity of the FtMYB11 transcription repressor of the phenylpropanoid pathway in Fagopyrum tataricum. New Phytol. 216 814–828. 10.1111/nph.14692 [DOI] [PubMed] [Google Scholar]
- Zhou M., Zhang K., Sun Z., Yan M., Chen C., Zhang X., et al. (2017b). LNK1 and LNK2 corepressors interact with the MYB3 transcription factor in phenylpropanoid biosynthesis. Plant Physiol. 174 1348–1358. 10.1104/pp.17.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Sun Z., Wang C., Zhang X., Tang Y., Zhu X., et al. (2015). Changing a conserved amino acid in R2R3-MYB transcription repressors results in their cytoplasmic accumulation and abolishes their repressive activity in Arabidopsis. Plant J. 84 395–403. 10.1111/tpj.13008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.