Abstract
The ability to persist in the absence of growth triggered by low oxygen levels is a critical process for the survival of mycobacterial species in many environmental niches. MSMEG_5243 (fsq), a gene of unknown function in Mycobacterium smegmatis, is up-regulated in response to hypoxia and regulated by DosRDosS/DosT, an oxygen- and redox-sensing two-component system that is highly conserved in mycobacteria. In this communication, we demonstrate that MSMEG_5243 is a flavin-sequestering protein and henceforth refer to it as Fsq. Using an array of biochemical and structural analyses, we show that Fsq is a member of the diverse superfamily of flavin- and deazaflavin-dependent oxidoreductases (FDORs) and is widely distributed in mycobacterial species. We created a markerless deletion mutant of fsq and demonstrate that fsq is required for cell survival during hypoxia. Using fsq deletion and overexpression, we found that fsq enhances cellular resistance to hydrogen peroxide treatment. The X-ray crystal structure of Fsq, solved to 2.7 Å, revealed a homodimeric organization with FAD bound noncovalently. The Fsq structure also uncovered no potential substrate-binding cavities, as the FAD is fully enclosed, and electrochemical studies indicated that the Fsq:FAD complex is relatively inert and does not share common properties with electron-transfer proteins. Taken together, our results suggest that Fsq reduces the formation of reactive oxygen species (ROS) by sequestering free FAD during recovery from hypoxia, thereby protecting the cofactor from undergoing autoxidation to produce ROS. This finding represents a new paradigm in mycobacterial adaptation to hypoxia.
Keywords: reactive oxygen species (ROS), mycobacteria, Mycobacterium smegmatis, hypoxia, flavin, flavin adenine dinucleotide (FAD), bacterial genetics, oxidative stress, anoxia, dos regulon, DosR, oxidoreductase
Introduction
Mycobacteria are obligately aerobic bacteria that are able to survive prolonged periods of hypoxia and nutrient limitation. One of the key challenges for mycobacterial persistence during hypoxia is combating the generation of reactive oxygen species (ROS)4 during re-entry into aerobic conditions and ensuing growth. The majority of ROS production in mycobacterial cells is generated through the autoxidation of molecules with sufficient electrochemical potential to form superoxide (1). Flavins, which include FAD, are one class of molecules that have sufficient potential to autoxidize (in the presence of light or metal ions) (1, 2). In mycobacteria, autoxidation of flavins generates superoxide, which is subsequently converted into hydrogen peroxide by superoxide dismutase. Catalase then in turn catalyzes the decomposition of hydrogen peroxide to water and oxygen (3, 4). Mycobacteria also have a range of other strategies to detoxify ROS, for example using a phenolic glycolipid present in their cell wall to scavenge ROS and mycothiol to reduce disulfide bonds (5–7).
In mycobacteria, a conserved mechanism in the adaptation to low oxygen is the activation of the two-component regulatory system DosRDosS/DosT. The histidine kinases DosS and DosT use a heme group to sense the redox state and oxygen concentration of the cell (8, 9). Both kinases in turn phosphorylate DosR, a LuxR-type helix-turn-helix transcriptional activator that binds 18-bp palindromic sequences in promoter regions (10–12). In the fast-growing soil bacterium Mycobacterium smegmatis, the dos system controls 11 operons and 49 genes in response to oxygen deprivation (13). Many of the genes under Dos control remain uncharacterized.
Fsq is among the mycobacterial proteins of the dos regulon and is highly up-regulated under hypoxia (13). Fsq has recently been identified as belonging to a diverse and largely uncharacterized superfamily of flavin/deazaflavin oxidoreductases (FDORs) that are abundant in mycobacteria and other Actinobacteria (14, 15). This superfamily is characterized by a split β barrel structural fold that generally binds flavin/deazaflavin cofactors. The FDOR superfamily can be further split into two families: the FDOR-A family in which each has F420 as a cofactor and the more diverse FDOR-B family that can bind F420, FAD, or FMN as cofactors, or heme as a substrate. The FDOR-B family exhibits diverse functions, including pyridoxine 5′-phosphate oxidases (PnPOxs), biliverdin reductases, and heme oxygenases (14).
Understanding of physiological roles of the uncharacterized genes of the dos regulon remains an important goal as they may provide insight into the unique mechanisms of adaptation these bacteria have evolved to survive both the stress of entry into hypoxia and re-entry into aerobic growth conditions during their normal life cycle. In this communication, we determined the role of Fsq in adaptation to hypoxia through a combination of bioinformatic, genetic, biochemical, and physiological approaches.
Results and discussion
Homologues of Fsq are highly conserved among mycobacteria
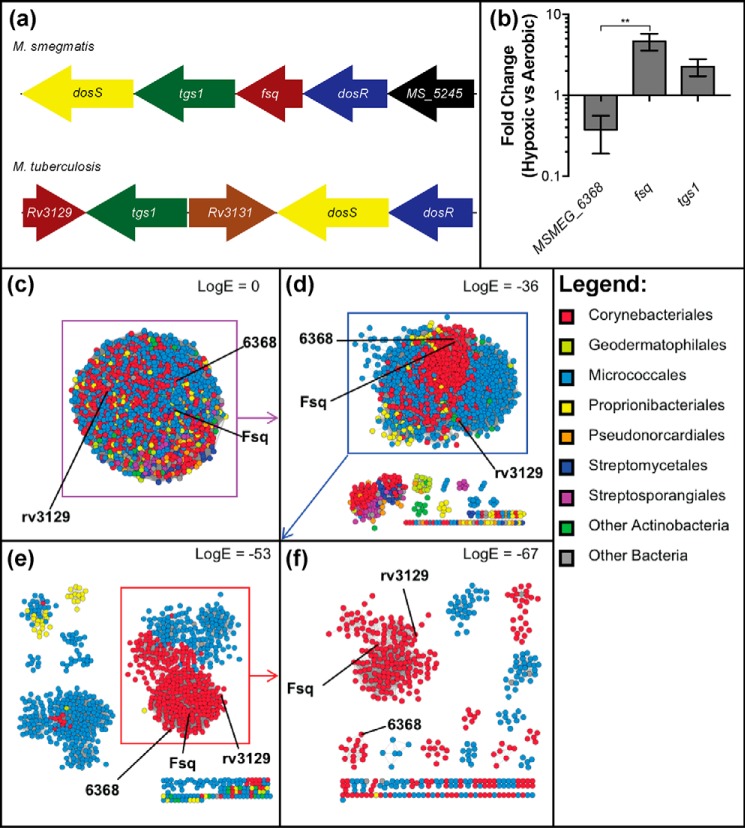
A previous bioinformatics analysis of the diverse superfamily of FDORs placed Fsq within a discrete FAD-binding subfamily alongside MSMEG_6368 and rv3129 from Mycobacterium tuberculosis (14). We compared the genetic organization, expression levels, and primary sequences of Fsq and MSMEG_6368 to gain insight into their possible cellular roles. Based on pairwise alignment, Fsq shares 64% amino acid identity with rv3129 and 54% identity with MSMEG_6368. The fsq and rv3129 genes are both part of the dosR regulon and share similar genomic contexts, with both being in the same region as dosR (Fig. 1a). Conversely, MSMEG_6368 is not part of the dosR regulon and does not share the same genomic context as fsq (13). Quantitative RT-PCR confirmed that fsq, but not MSMEG_6368, is up-regulated by 4.7-fold under hypoxia (p = 0.003) (Fig. 1b). The tgs1 gene encoding the TAG synthase 1, a gene known to be controlled by DosR, was up-regulated 2.3-fold under hypoxia (Fig. 1b).
Figure 1.
Organization, regulation, and abundance of Fsq. a, genomic context of fsq in M. smegmatis and its homolog rv3129 in M. tuberculosis. Matching colors between M. smegmatis and M. tuberculosis represent homologous genes. Gene sizes are not to scale. b, comparison of expression ratio of MSMEG_6368 and fsq, with tgs1 as a positive control, from cells harvested at 6 days' growth in aerobic and hypoxic conditions. The gene sigA was used as the reference. Error bars represent S.D. of biological triplicates. Statistical testing was performed using an unpaired Student's t test, where ** = p < 0.01. c–f, SSNs of Fsq-related proteins are shown with different cutoffs. In an SSN, protein sequences are represented as nodes (dots) that are joined by edges (lines) which represent a similarity metric such as Basic Local Alignment Search Tool (BLAST) E-value (48). Nodes are colored according to taxonomic order. c, SSN with no logE cutoff applied. d, SSN of Fsq-related proteins with a logE cutoff of −36. A large heterogeneous cluster containing Fsq, MSMEG_6368, and rv3129 separates from several small clusters. e, the large heterogenous cluster examined with a cutoff of −53. A large compact cluster of mycobacterial sequences separates from other clusters. f, the large cluster from previous panel with a cutoff of −67. A cluster containing MSMEG_6368 separates from one containing Fsq and rv3129, suggesting that they may have distinct functions.
A sequence similarity network (SSN) was used to examine the FAD-binding subfamily that includes Fsq and MSMEG_6368 in greater detail than in our previous study (14). We initially truncated the dataset such that sequences with less than 70% coverage and less than 30% identity to Fsq were excluded from analysis. This gave an initial network of 1124 nodes, which formed a large group without application of a logE filter (Fig. 1c). Several small groups (primarily consisting of sequences from orders other than Corynebacteriales and Micrococcales) separate out when only edge scores with logE <−36 were considered (Fig. 1d). The main cluster contained nodes including FSQ, MSMEG_6368, and rv3129. When edges were further filtered to logE <−53, a large cluster containing only sequences from Corynebacteriales becomes apparent (Fig. 1e). The mycobacterial sequences represented include slow-growing pathogenic species such as M. tuberculosis, Mycobacterium kansasii, and Mycobacterium canettii, as well as fast-growing species such as Mycobacterium wolinskyi, Mycobacterium phlei, and Mycobacterium fortuitum, suggesting that Fsq homologs are highly conserved throughout mycobacterial species. Finally, when edge scores with logE <−67 were considered, a cluster containing MSMEG_6368 separates from Fsq and rv3129. The high level of sequence conservation among this final group of homologs, across such a large number of mycobacterial species, suggests that Fsq and rv3129 are under strong selective pressure to maintain a physiological function (Fig. 1f), and therefore are proposed to be flavin-sequestering family proteins (FSFP). In contrast, MSMEG_6368 appears to be less conserved among mycobacteria.
fsq contributes to the survival of Mycobacterium smegmatis under hypoxic growth conditions
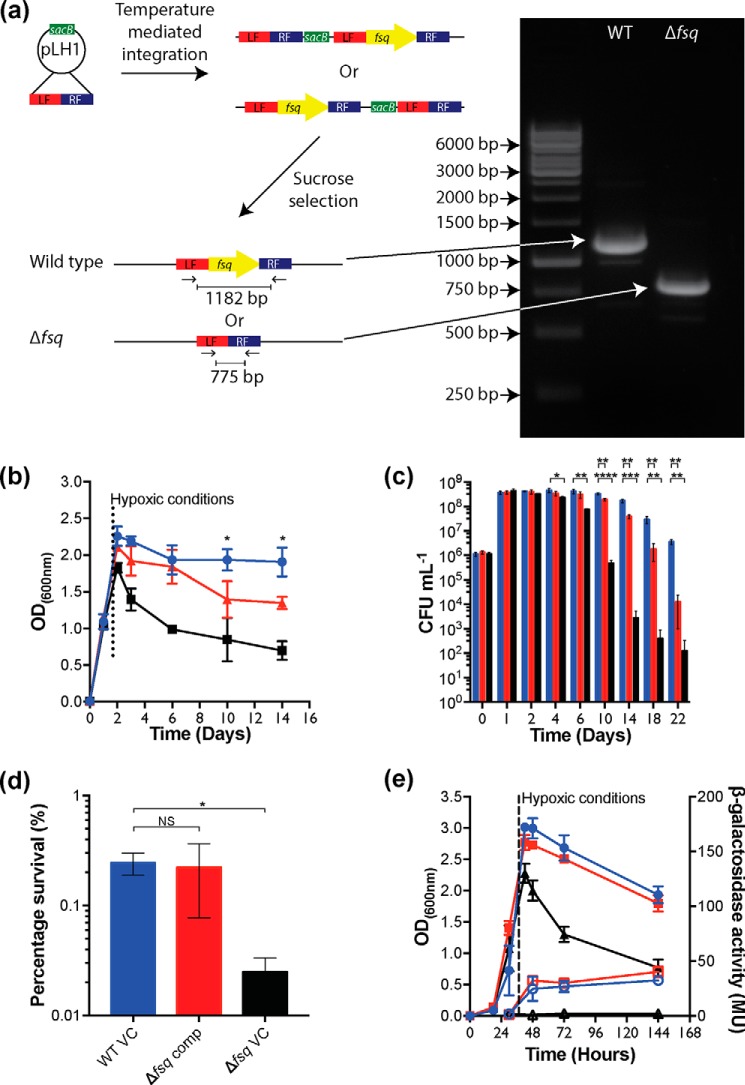
To determine the function of fsq, we constructed a markerless deletion mutant strain Δfsq (Fig. 2a) (Table S1). The Δfsq deletion mutant was confirmed by PCR and whole genome sequencing. Strain Δfsq and the isogenic WT parent strain were grown under aerobic conditions on a range of different carbon sources (fermentable and nonfermentable) and no differences in growth rate or final cell yield were observed between the strains (Fig. S1). However, when strains were grown in sealed serum vials that became hypoxic as cells grew (∼37 h), we observed a difference in the final optical density (600 nm) between the Δfsq mutant and the WT (Fig. 2b). To investigate this effect in more detail, we compared the cell viability of the Δfsq mutant to the isogenic WT and a ΔdosR mutant during prolonged incubation under hypoxia (Fig. 2c). The Δfsq mutant exhibited a survival defect compared with WT strain from day 10 post inoculation onwards, with the maximum difference observed being a 2-log difference at 22 days post inoculation (Fig. 2c). Consistent with previous findings (13), the ΔdosR mutant showed a 3-log decrease in viability after 10 days compared with WT. We were able to complement the Δfsq defect under hypoxia using complementing vector (pLH2, fsq+) induced with 20 ng/ml tetracycline (Fig. 2d). After 18 days, there was 0.24% survival for the WT with an empty vector, 0.22% survival for complemented Δfsq, and 0.02% survival for Δfsq mutant with an empty vector (Fig. 2d).
Figure 2.
Creation and hypoxic survival of Δfsq markerless deletion mutant. a, diagram of fsq markerless deletion construction. b and c, long-term survival of M. smegmatis WT (blue) compared with ΔdosR (black) and Δfsq (red) mutants under hypoxic conditions. Growth was in HdB minimal medium supplemented with 22 mm glycerol. Entry of the vials into a hypoxic state was estimated by the decolorization of methylene blue (1.5 μg/ml) which occurred at 27 h. b, growth measured using optical density at 600 nm wavelength with an average of three biological replicates shown with error bars representing S.D. Unpaired Student's t test of WT compared with Δfsq. *, p < 0.05 at each time point. c, survival measured using colony-forming units (CFU) with an average of three biological replicates shown with error bars representing S.D. Unpaired Student's t test of WT compared with Δfsq and WT compared with ΔdosR at each time point. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. d, complementation of survival defect of Δfsq mutant using pLH2 (fsq+) (Δfsq comp) (red) compared with an empty vector control in wild type (WT VC) (blue) and an empty vector in Δfsq mutant (Δfsq VC) (black). Vectors were induced using 20 ng/ml tetracycline just prior to entry into hypoxia. Survival percentage of day 18 with an average of three biological replicates shown with error bars representing S.D. Unpaired Student's t test of WT compared with Δfsq comp and WT compared with Δfsq VC. *, p < 0.05. e, measurement of DosR activity through hyd3 LacZ expression in M. smegmatis WT compared with ΔdosR and Δfsq mutants under hypoxic conditions. Strains were grown into hypoxia in HdB with 22 mm glycerol. Entry into hypoxia was indicated by methylene blue (1.5 μg/ml) control vials. Samples (2 ml) were taken at 27, 48, 72, and 144 h and used to preform β-gal assays to assess the level of dosR activity. Key: WT, blue circles; ΔdosR mutant, black triangles; Δfsq mutant, red squares; β-gal activity, open symbols; optical density (600 nm), closed symbols. Each sample shows average of a biological triplicate with error bars representing S.D.
The loss of viability observed for Δfsq mutant cells was initially hypothesized to be caused by Fsq playing a role in the complete activation of DosR given the two genes are in close proximity (Fig. 1a). To address this hypothesis, we determined the effect of the Δfsq mutation on the expression of a dos-regulated gene (hyd3-lacZ) in M. smegmatis (16). When the expression of hyd3-lacZ was measured in the WT background, hyd3-lacZ was up-regulated in response to hypoxia (Fig. 2e). No up-regulation of hyd3-lacZ was observed in the ΔdosR mutant, but WT levels of hyd3-lacZ expression were observed in the Δfsq mutant, confirming that fsq did not play a role in DosR activation.
MSMEG_6368 shares 54% identity with Fsq but is not part of the dosR regulon, suggesting a distinct functional role. To investigate the functional role of MSMEG_6368 and rule out MSMEG_6368 compensating for the loss of fsq in the Δfsq mutant, we created a markerless deletion mutant of MSMEG_6368 (strain Δ6368) (Table S1). We then grew the Δ6368 mutant and the isogenic WT strain on a range of carbon sources (fermentable and nonfermentable) under aerobic conditions and found that there was no detectable difference in the growth rate or final cell yield (Fig. S2, a and b). No phenotypic differences in growth or cell viability between the Δ6368 mutant and the isogenic WT strain were observed under hypoxia (Fig. S2, c and d). We then used qPCR to monitor the expression of MSMEG_6368 in the WT and Δfsq mutant (Fig. S2e). During hypoxia, the expression of MSMEG_6368 was not different in the Δfsq mutant compared with the WT. These data suggest that MSMEG_6368 does not provide compensation for fsq in the Δfsq mutant during hypoxia.
Biochemical, structural, and electrochemical analysis suggests Fsq is an FAD-binding protein
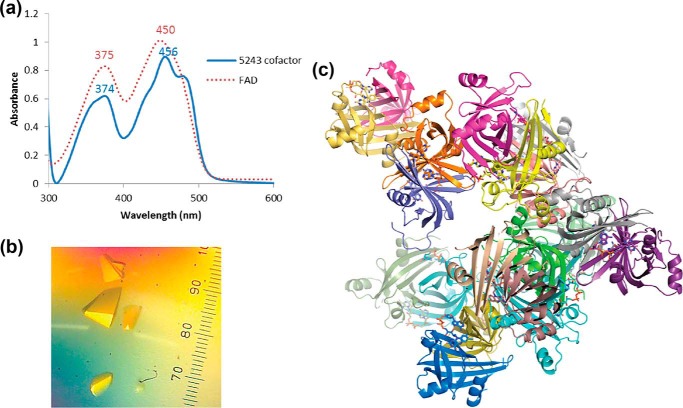
To determine the biochemical function of Fsq, we expressed Fsq in Escherichia coli and purified the protein to homogeneity. Fsq had previously been demonstrated to preferentially bind to FAD over other structurally similar cofactors FMN and F420, with a Kd value of 3.6 μm (14). We set about testing whether Fsq could function as a flavin-sequestering protein by solving and analyzing its X-ray crystal structure and testing its electrochemical properties. Fsq heterologously produced in E. coli co-purified with a cofactor that has a UV-visible absorbance spectrum with peaks at 374 nm and 456 nm, which is consistent with FAD (Fig. 3a). To confirm the identity of this cofactor, we crystallized the purified protein (Fig. 3b), obtaining 2.7 Å resolution diffraction (Table 1). The crystal contained 20 molecules in the asymmetric unit, arranged as homodimers (Fig. 3c). Omit electron density (mFo − DFc) consistent with FAD was observed at the dimer interfaces/cofactor-binding sites for 17 of these chains (Fig. 4, a and b). The remaining three chains had no FAD bound at this site, although they were part of dimers in which the other monomer was bound to FAD. There was no electron density to support covalent attachment of FAD to the protein, which is consistent with previous work showing that bound FAD can be readily removed from Fsq through partial unfolding-refolding (14).
Figure 3.
a, UV-visible absorbance spectra comparing the cofactor co-purified with Fsq (blue) to FAD (red). b, crystals of Fsq bound to the co-purified FAD, with no cofactor stripping or additional reconstitution (40× magnification). c, the 20 chains of apo- and holo-Fsq found in the asymmetric unit. 17 chains were bound to FAD.
Table 1.
Crystallography statistics for Fsq
| Data collection | |
| Space group | P2 21 21 |
| Unit cell parameters | |
| a (Å) | 80.27 |
| b (Å) | 186.04 |
| c (Å) | 195.39 |
| α, β, γ (°) | 90, 90, 90 |
| Wavelength (Å) | 1.5418 |
| Resolution range (Å)a | 29.6–2.69 (2.74–2.69) |
| Unique reflections | 82,043 |
| Completeness (%)a | 99.9 (99.9) |
| Multiplicitya | 7.4 (7.3) |
| Rmergea,b | 0.215 (1.444) |
| Rpima,c | 0.084 (0.565) |
| Mean 〈I/σ(I)〉a | 10.5 (1.7) |
| CC1/2a,d | 0.993 (0.551) |
| Solvent content (%, v/v) | 45.7 |
| Molecules per asymmetric unit | 20 |
| Refinement | |
| Reflections used | 81,939 |
| Resolution range (Å)a | 29.6–2.69 (2.72–2.69) |
| Rwork/Rfreea,e | 0.209/0.258 |
| No. of atoms (all) | 20515 |
| Water molecules | 362 |
| Average B-factor (Å2) | |
| Main chains | 44.7 |
| Side chains | 47.6 |
| Water molecules | 34.1 |
| FAD molecules | 46.4 |
| Root mean square deviations | |
| Bond length (Å) | 0.003 |
| Bond angles (°) | 0.682 |
| Ramachandran plot regions (%) | |
| Favored | 94.23 |
| Allowed | 4.62 |
| Outliers | 1.14 |
| PDB ID | 6ECI |
a Values in parentheses are for the highest-resolution shell.
bRmerge = (Σh Σi |Ih,i − 〈Ih〉|)/(Σh Σi 〈Ih〉), where 〈Ih〉 is the average intensity of i symmetry-related observations of the unique reflection h.
cRpim = (Σh Σi (1/(nh − 1))1/2 |Ih,i − 〈Ih〉|)/(Σh Σi 〈Ih〉).
dCC1/2 = linear correlation coefficient between intensities from random half-datasets.
eRwork = Σh |F(obs) − F(calc)|/Σh |F(obs)| and 5% of the data that were excluded from the refinement were used to calculate Rfree.
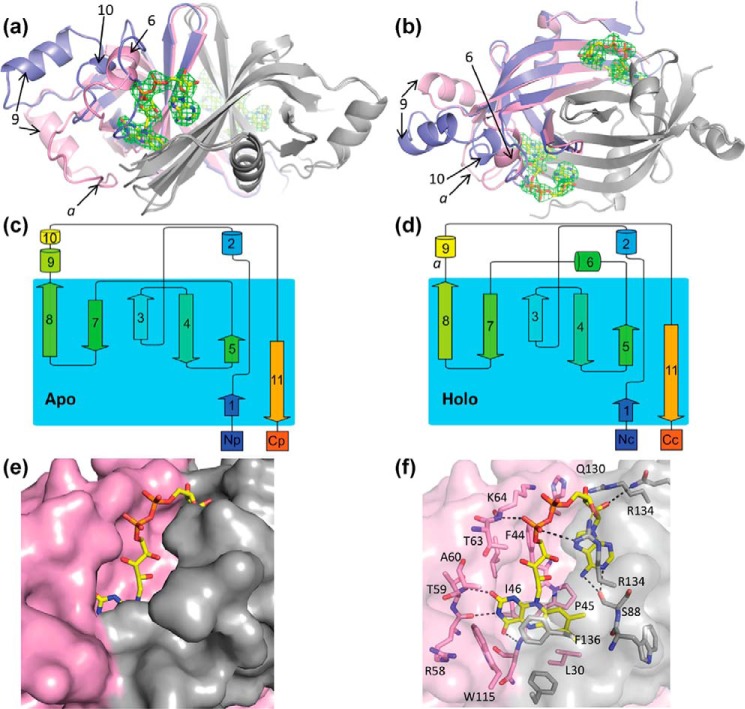
Figure 4.
Structure of apo- and holo-FAD Fsq. a and b, side (a) and top (b) views of the overlaid structures of apo (blue, chain P) and holo (pink, chain B) Fsq. The secondary chain of the dimer for both structures is shown in gray. FAD bound to the holo-structure is shown in yellow and the electron density for the cofactor on both binding sites of the dimer is shown, representing the Fo − Fc omit map contoured at 3 σ. Key loops and α-helixes are labeled to reference parts c and d. c and d, topology maps of the apo- and holo-structures, respectively, adapted from the cartoons generated by Pro-origami. e, solvent-accessible surface of Fsq showing the buried flavin moiety in the holo-structure. f, residues involved in FAD binding.
Comparison of the apo- and holo- forms of Fsq revealed a large conformational change in the holo-structure that allows for the accommodation of FAD (Fig. 4, a and b). Most notably, a loop region in the apo-protein forms α-helix-6 in the holo-protein, which interacts with the phosphate groups of FAD (Fig. 4, c and d). Although the apo-protein contains both α-helix-9 and α-helix-10, the position of α-helix-9 is shifted in the holo-protein, causing α-helix-10 to unravel. This lengthens loop-a between α-helix-9 and β-sheet-7, allowing it to accommodate the isoalloxazine group of FAD (Fig. 4, b and d). This substantial change upon cofactor binding is unusual for proteins of the FDOR family, as structurally characterized FMN- and F420-binding proteins show only slight topological differences between their apo- and holo- forms (Fig. S3) (14, 15, 17, 18). This structural rearrangement results in essentially complete enclosure of the cofactor within the interior of the protein. From the pairwise alignment of Fsq and rv3129 we found that the majority of the key residues were maintained except for a proline 45 and leucine 30, which were deleted in rv3129.
Flavin/deazaflavin-binding proteins can typically carry out three broad roles: enzymes that catalyze chemical reactions (19), electron transport proteins that transfer electrons to partner enzymes for catalysis (20), or cofactor-binding proteins that bind to and sequester or transport cofactors (21–23). We therefore sought to test each of these hypotheses. In FDORs with known enzymatic activity, catalysis occurs on the Re-face of the isoalloxazine rings of their flavin cofactors, which is solvent accessible to allow substrate binding (15, 17). In contrast, for Fsq, we found that the isoalloxazine ring of FAD is fully enclosed by the protein, making the Re-face completely inaccessible as a substrate-binding site (Fig. 4e). This is achieved by an offset parallel stacking interaction between the xylene and pyrazine rings of the isoalloxazine and Phe-136 of the secondary chain in the dimer, and loop-a (which undergoes a conformational change upon FAD binding) covers the pyrimidine ring with hydrophobic interactions from Trp-115 from the primary chain (Fig. 4f). The lack of any substrate-binding site will prevent binding or catalysis of small molecule substrates, making it unlikely that Fsq is an enzyme.
Protein film voltammetry was employed to determine the effect of Fsq on the electrochemical properties of the cofactor, which could also provide insight into its physiological role. Both Fsq and FAD displayed a single reversible redox peak at all pH values that we tested, with no evidence of one-electron reductions (Fig. 5a), as is seen in some flavin-dependent enzymes such as fumarate reductase (24). At pH 7.3, the redox potential of FAD was found to be −260 ± 3 mV (versus normal hydrogen electrode), whereas that of Fsq:FAD was −246 ± 9 mV (Fig. 5a). Thus, the redox potential of FAD is not substantially changed by Fsq. This is in contrast to many enzymes and electron-transfer proteins that can alter the redox potentials more than 100 mV in either direction to carry out catalysis (25, 26). There was relatively little difference in the effect of pH on the redox potential of free and complexed FAD (−45 mV/pH unit and −39 mV/pH unit, respectively) with no redox-linked pKa observed in the range of 3.6–7.3. This contrasts again with electron-transfer proteins, such as flavodoxins, that have a redox-linked pKa between pH 4.6 and 6.8 (25, 27) (Fig. 5b). The values for free FAD are consistent with previous studies of free FAD on carbon nanotube–modified electrodes (28). In summary, FAD appears to be relatively stable and unreactive when bound to Fsq, making it unlikely that this protein:flavin complex is involved in catalysis or electron-transfer reactions, leaving the possibility that it is involved in FAD sequestration.
Figure 5.
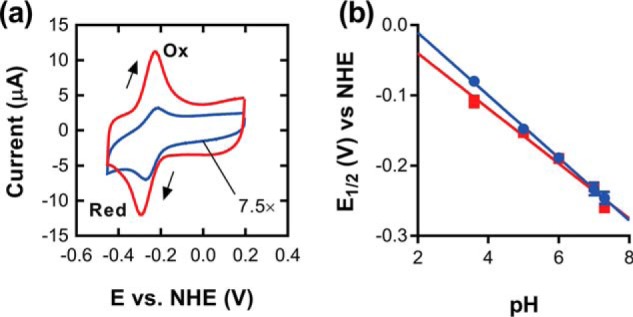
Electrochemical properties of Fsq. a, cyclic voltammograms of Fsq (blue) and FAD (red) adsorbed onto modified glassy carbon electrodes. Scans were conducted at 50 mV/s in the direction indicated by the arrows in 100 mm sodium phosphate, pH 7.3. Oxidative and reductive peaks are labeled. The cyclic voltammogram of Fsq is scaled by a factor of 7.5 for comparison. b, effect of pH on the redox potential of Fsq and FAD. Over the range tested Fsq and FAD varied by −45 mV/pH unit and −39 mV/pH unit, respectively. NHE, normal hydrogen electrode; E, electrochemical potential.
Fsq promotes resistance to oxidative stress
Based on the structural and biochemical data that suggest Fsq is unlikely to have either a catalytic or electron-transfer role in mycobacterial physiology, and the reduced fitness of the Δfsq strain in hypoxic conditions, we hypothesized one benefit of FAD sequestration by Fsq could be the prevention of FAD autoxidation to avert superoxide formation and cellular damage. One of the key challenges that mycobacterial species must overcome to survive hypoxia is mitigating oxidative damage upon entry to and re-aeration from hypoxia. To test this hypothesis, we challenged the Δfsq mutant and isogenic WT grown in either hypoxia (inducing conditions) or aerobic conditions (noninducing) with hydrogen peroxide (Fig. 6). Under aerobic conditions, there was no significant difference in cell viability between the WT and Δfsq mutant in response to hydrogen peroxide challenge (Fig. 6a). However, under hypoxic conditions, the Δfsq mutant was significantly (p = 0.0082) more sensitive to hydrogen peroxide challenge compared with the WT (Fig. 6b). When fsq was overexpressed in cells using the tetracycline-inducible vector pLH2 (fsq+), we could protect cells from killing by hydrogen peroxide stress compared with the empty vector control (VC) (Fig. 6c). These results establish the molecular process by which Fsq confers a fitness advantage to M. smegmatis during recovery from hypoxia. However, they do not explain the fitness advantage of Fsq to M. smegmatis during long-term hypoxic growth. Thus, although at least one physiological function of Fsq is now defined, other specific functions related to hypoxic survival could also exist.
Figure 6.
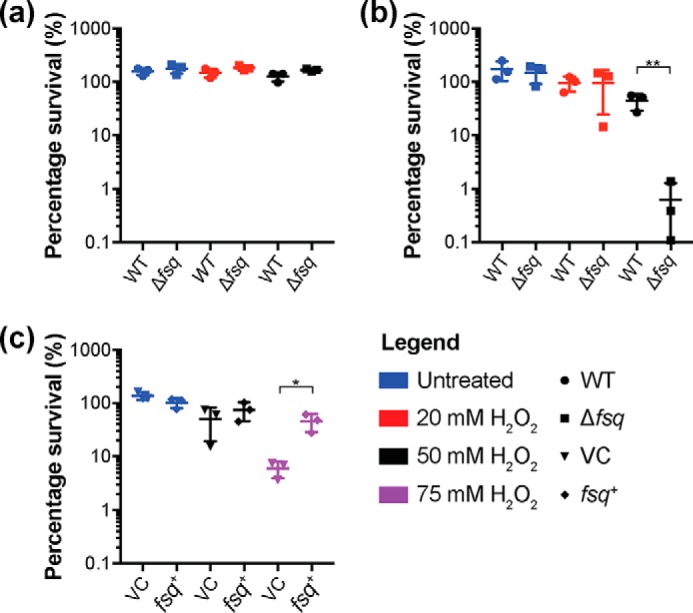
Fsq heightens resistance to hydrogen peroxide. a, challenge of WT compared with Δfsq with 20 or 50 mm hydrogen peroxide for 3 h. Cells were grown aerobically in HdB with 22 mm glycerol for 10 days before harvest and resuspension in phosphate buffer saline with 0.5% tyloxapol. Cells were normalized to 1 optical density (600 nm) before treatment. b, challenge of WT compared with Δfsq with 20 or 50 mm hydrogen peroxide for 3 h. Cells were grown hypoxically in HdB with 22 mm glycerol for 10 days before harvest and resuspension in phosphate buffer saline with 0.5% tyloxapol. Cells were normalized to 1 optical density (600 nm) before treatment. c, challenge of WT either expressing fsq (fsq+) (WT pLH2) or a vector control (VC) (WT pMind) control induced with 20 ng−1 ml tetracycline, challenged with 50 or 75 mm hydrogen peroxide for 3 h. Cells were grown aerobically to exponential phase n HdB with 22 mm glycerol before harvest and resuspension in phosphate buffer saline with 0.5% tyloxapol. Cells were normalized to 1 optical density (600 nm) before treatment. a–c, error bars represent S.D. of three biological replicates. Unpaired Student's t test compared with WT for each treatment, where *, p < 0.05; **, p < 0.01.
Comparison to other cofactor-binding proteins in mycobacteria
Having confirmed that Fsq protects M. smegmatis against oxidative damage when emerging from hypoxic conditions by sequestering free FAD, we examined whether other examples of mycobacterial cofactor-sequestering proteins have been reported in the literature. M. tuberculosis and M. smegmatis have both been shown to harbor a nonfunctional nitroreductase designated Acg that has been shown to function as a flavin mononucleotide (FMN) storage protein or used to sequester FMN from other biochemical pathways (29). Acg is not related in sequence or structure to Fsq. However, as we have observed with Fsq, the Acg structure encloses (“caps”) FMN in a way to sterically prevent reactivity with NAD(P)H. Thus, like Fsq, Acg also appears to have evolved from a superfamily in which most proteins are enzymes and, under selective pressure, have lost the ability to bind substrate to develop a cofactor-sequestration function. Notably, the acg gene is also regulated as part of the dosR regulon (29). The observation that mycobacteria has evolved two different proteins that each sequester flavin cofactors to prevent deleterious interactions, both of which are under control of the DosR regulon, highlights the biochemical challenges imposed by the descent into, and emergence from, hypoxia. These examples provide us with deeper understanding of the mechanisms that mycobacteria have evolved to adapt to the pressures associated with hypoxic conditions.
Conclusions
Mycobacteria utilize a diverse array of flavins in a wide variety of essential electron-transfer reactions, including FAD, FMN, and F420 (30). Previously, it has been shown that mycobacteria possess proteins such as Acg to sequester FMN in hypoxic conditions. In this study we infer, using a combination of biochemical, structural, and electrochemical data, that Fsq from M. smegmatis functions to sequester FAD under hypoxia. The expression of fsq is controlled by DosR and mutants defective in fsq are hypersensitive to oxidative stress when recovering from hypoxia. These data suggest that the ability of Fsq to sequester FAD protects it from autoxidation and the production of ROS under hypoxia.
Experimental procedures
Strains and growth conditions
M. smegmatis mc2155 was grown in either lysogeny broth with 0.05% Tween 80 (LBT) or Hartmans de Bont (HdB) minimal medium (31) supplemented with 0.05% tyloxapol and either 22 mm glycerol or 30 mm succinate as the sole carbon and energy source. When required, media were supplemented with 20 μg/ml kanamycin, 50 μg/ml hygromycin, or 5 μg/ml gentamicin. If solid media were required, 1.5% agar LBT was used. Aerobic growth was carried out in flasks of 125 ml with 25 ml medium or 250 ml with 50 ml medium in rotary incubator shaking at 200 rpm, 37 °C. E. coli strain DH10β was grown in lysogeny broth or M9 minimal medium and, when required, 50 μg/ml kanamycin, 50 μg/ml hygromycin, or 20 μg/ml gentamicin. M. smegmatis cells were grown into hypoxia using 30 ml of medium in 120-ml serum vials using the Berney-Cook model of hypoxia (32) where depletion of oxygen was confirmed using a control vial that contained methylene blue (1.5 μg/ml) that becomes colorless when low oxygen is present. To count colony-forming units (cfu/ml), each culture was serially diluted in PBS (pH 7.0) and spotted onto agar plates. All survival studies were performed with biological triplicates, and plates were spotted in technical triplicate. Optical density to monitor growth of bacteria were done at 600 nm wavelength using a Jenway 6300 spectrophotometer. Once optical density of cells was above 0.5, the cells were diluted 1 in 10 in 0.85% saline buffer to bring the optical density (600 nm) below 0.5. β-gal assays were performed as described previously (33).
Sequence similarity network
Sequences were obtained via BLAST searches using MSMEG_5243 as the query and were curated with CD-Hit (34) to remove sequences with more than 90% identity. Sequence similarity networks were obtained using an all-versus-all BLAST of the curated sequences as described previously (14) using BLAST+ (35). Networks were visualized in Cytoscape 3.6.1 (36) with nodes colored according to taxonomic order and BLAST E-values used as the edges.
Markerless deletion mutant construction
In brief, to generate the deletion of fsq, two homologous regions either side of target gene, we generated and fused together using overlap extension PCR (Primers LH1, LH2, LH3, LH4) (Table S2) and cloned into pX33 using the SpeI restriction enzyme (New England Biolabs). For the deletion of MSMEG_6368, following the initial PCR that generated the two homologous regions either side of target gene (Primers LH7, LH8, LH9, LH10) (Table S2), we used Gibson cloning (New England Biolabs) as per the manufacturer's instructions to join the homologous regions to pX33. The pX33 constructs with homologous regions were then electroporated into M. smegmatis and grown at 28 °C to ensure permissibility of the pX33 temperature-sensitive plasmid. Confirmation of the plasmid in the cell was achieved by treating cell colonies with 250 mm catechol, which causes any colonies containing the plasmid to turn yellow because of the catechol dioxygenase produced by pX33. Following confirmation of the plasmid presence, the cells were grown on liquid media at 28 °C and then plated 40 °C to force integration based on the homology of the gene flanking regions. Integrants were confirmed through catechol screening (as above). Integrants were then spread-plated on sucrose (10% w/v) containing media to force recombination to either the WT genotype or the markerless deletion mutant. All mutants were confirmed using whole genome sequencing (WGS) and aligned using Geneious to an isogenic WT to confirm that the gene was deleted.
Expression and purification of Fsq
The vector encoding Fsq includes an N-terminal His-tag followed by a TEV protease cleavage site, as described previously (14). E. coli BL21(DE3) (Invitrogen) cells were transformed with the pETMCSIII vector containing the Fsq construct using electroporation. Colonies were picked and grown in 5-ml starter cultures before being transferred to 1 liter modified autoinduction medium (5 g of yeast extract, 20 g of tryptone, 85.5 mm NaCl, 22 mm KH2PO4, 42 mm Na2HPO4, 0.6% glycerol, 0.05% glucose, 0.2% lactose, and 100 μg/ml of ampicillin) and grown for 16 h at 30 °C before centrifugation at 4000 × g for 15 min and removal of supernatant. Cell pellets were frozen and resuspended in 40 ml of buffer A (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, 25 mm imidazole) before being ruptured by sonication on ice. Turbonuclease (0.1 μl) (Sigma-Aldrich) was added prior to pelleting cell debris by centrifugation at 20,000 × g for 60 min. The resulting supernatant was then loaded onto a 5-ml HisTrap nickel affinity column (GE Healthcare) pre-equilibrated with buffer A. The column was washed with 25 ml of buffer A containing 25 mm imidazole, 25 ml of 8% buffer B (43 mm imidazole) and eluted in 100% buffer B (250 mm imidazole). Buffer B has the same composition as buffer A except for the addition of 250 mm imidazole. The eluted protein was then passed over a GE HiPrep 26/10 desalting column to buffer exchange into 50 mm Tris-HCl, 200 mm NaCl, pH 8, and His-tag was cleaved by incubating the eluent overnight with TEV protease (purified in-house) (37), 1 mm DTT, and 0.5 mm EDTA. The cleaved His-tag and TEV protease were then removed by passing the reaction mixture over a 5-ml HisTrap nickel affinity column and collecting the flow-through. The sample was concentrated by centrifugation in an Amicon Ultra-15 10 MWCO spin filter at 4000 × g to a final volume of 2 ml and further purified by size-exclusion chromatography (SEC) using a GE HiLoad 16/600 Superdex 75-pg column with buffer containing 20 mm HEPES, 150 mm NaCl, pH 7.5. The final sample was diluted to adjust buffer concentration to 20 mm HEPES, 50 mm NaCl, pH 7.5, and concentrated to 1.3 mm for storage at 4 °C.
X-ray crystallography
High-throughput screens from Hampton Research were used to identify initial crystal-forming conditions for Fsq using the 1.3 mm protein solution. Crystals grew in 20% PEG 1500, 4% 2-methyl-2,4-pentanediol, and 0.1 m citric acid, pH 3.5. Using the same conditions as a cryo-buffer, data were collected in-house with a Xenocs GeniX 3D Cu HF microbeam X-ray generator and a MAR345 plate detector. The diffraction data were integrated using XDS (38) and AIMLESS (39) as part of the CCP4 suite (40) was used for scaling. This was followed by molecular replacement with Phaser (41) using the structure of a related protein of unknown function from Corynebacterium glutamicum (PDB ID: 3FKH), which has 45% amino acid sequence identity to Fsq. Initial model building was performed using Buccaneer (42), followed by manual model building in Coot (43). Refinement was performed with Refmac 5.5 (44) and PHENIX 1.13_2998 (45). Omit mFo − DFc electron density was used to model the position of bound FAD molecules.
DNA manipulation and cloning
All molecular biology techniques were carried out according to standard procedures. All restriction enzymes and DNA modifying enzymes used were from New England Biolabs and were in their appropriate buffers. All constructs were confirmed using either PCR or restriction digests followed by sequencing. PCR for construct creation was done using Thermo Fisher Phusion polymerase following the recommended conditions with appropriate annealing temperatures and extension time. The construct for fsq tetracycline (pLH2)–inducible expression was made using the pMIND vector backbone as described previously (46) with an artificial ribosome-binding site (47) using the primers LH5 and LH6 to amplify fsq for cloning (Table S2).
Quantitative RT-PCR
Total RNA was extracted using TRIzol (Ambion) as per the manufacturer's instructions and then purified using the RNA Clean and Concentrator kit (Zymo) as per the manufacturer's instructions. The cells were lysed by three cycles in a mini-Beadbeater (Biospec Products) at 4800 rpm for 30 s and on ice between each of the cycles. DNA was removed by treatment with 3 units RNase-free DNase using the TURBO DNA-free kit (Ambion) as per the manufacturer's instructions. The RNA concentration was determined using a NanoDrop One spectrophotometer. Reverse transcriptase PCR was performed using SuperScript III (Invitrogen), according to the manufacturer's instructions for cDNA synthesis. After cDNA synthesis, qPCR was carried out using Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen). The qPCR reactions were carried out on a QuantStudio 6 (Applied Biosystems). As an internal control and for the normalization of results the gene sigA was used. Error bars are representative of biological triplicate. Primers used are listed in Table S2 (MSMEG_6368 = LH11, LH12; fsq = LH13, LH14; tgs1 = tgsfw, tgsrev).
Hydrogen peroxide challenges
Cells were grown in HdB minimal media with glycerol as the sole carbon and energy source. Cell were then harvested and resuspended in PBS (pH 7.5) with 0.05% tyloxapol normalized to an A600 of 1.0 and then treated with 20, 50, or 75 mm hydrogen peroxide and left shaking at 180 rpm at 37 °C for 3 h. Cell were then plated using serial dilutions 5 μl spots on LBT solid media to determine cell viability.
Electrochemistry
Cyclic voltammetry was carried out using a BASi Epsilon Potentiostat with a C3 cell stand. A three-electrode system was employed comprising a modified glassy carbon electrode as working electrode, Pt wire counter and Ag/AgCl-saturated NaCl reference electrode. All potentials are quoted versus the normal hydrogen electrode, calculated as Ag/AgCl + 196 mV. The working electrode was prepared by polishing according to the manufacturer's instructions using 0.05 mm alumina polish, followed by sonication for 5–10 min in Milli-Q water. Carbon nanotubes (single-walled COOH-functionalized 1–2 nm, nanostructured and amorphous materials, outer diameter 1–2 nm) were dispersed in Milli-Q water using sonication to give a concentration of 1 mg/ml. SEC-purified Fsq (∼1 μm) was mixed with an equal volume of nanotube suspension and applied directly to the freshly polished electrode surface and allowed to dry at ambient temperature. Films prepared with protein-free FAD were used as controls. Oxygen was removed from the buffer through purging with argon.
The accession number for MSMEG_5243 is PDB ID: 6ECI.
Author contributions
L. K. H., J. A., and F. H. A. conceptualization; L. K. H., J. A., and F. H. A. data curation; L. K. H., J. A., F. H. A., and K. H. formal analysis; L. K. H. and J. A. investigation; L. K. H., J. A., F. H. A., K. H., P. D. C., and T. R. methodology; L. K. H., J. A., and F. H. A. writing-original draft; L. K. H., C. G., C. J. J., and G. M. C. project administration; L. K. H., J. A., F. H. A., K. H., C. G., C. J. J., and G. M. C. writing-review and editing; F. H. A., P. D. C., T. R., C. G., C. J. J., and G. M. C. supervision; C. G., C. J. J., and G. M. C. funding acquisition.
Supplementary Material
Acknowledgments
We thank Dr. Rhys Grinter and the two reviewers for their helpful comments.
This work was supported by the Maurice Wilkins Centre for Molecular Biodiscovery and the Marsden Fund, Royal Society, New Zealand. This work was also supported by a University of Otago Doctoral Scholarship and a Sandy Smith Scholarship (to L. K. H.) and by a CSIRO OCE Postdoctoral Fellowship, ARC DECRA Fellowship DE170100310, and NHMRC New Investigator Grant APP1139832 (to C. G.). In addition, this work was supported by NHMRC Project Grant APP1128929 (to C. J. J., C. G., and G. M. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and Tables S1 and S2.
The atomic coordinates and structure factors (code 6ECI) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ROS
- reactive oxygen species
- FDOR
- flavin/deazaflavin oxidoreductase
- SSN
- sequence similarity network
- qPCR
- quantitative PCR
- VC
- vector control
- FMN
- flavin mononucleotide
- LBT
- lysogeny broth with 0.05% Tween 80
- HdB
- Hartmans de Bont
- TEV
- tobacco etch virus
- SEC
- size-exclusion chromatography.
References
- 1. Miller D. M., Buettner G. R., and Aust S. D. (1990) Transition-metals as catalysts of autoxidation reactions. Free Radic. Biol. Med. 8, 95–108 10.1016/0891-5849(90)90148-C [DOI] [PubMed] [Google Scholar]
- 2. Massey V. (1994) Activation of molecular-oxygen by flavins and flavoproteins. J. Biol. Chem. 269, 22459–22462 [PubMed] [Google Scholar]
- 3. Piddington D. L., Fang F. C., Laessig T., Cooper A. M., Orme I. M., and Buchmeier N. A. (2001) Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect. Immun. 69, 4980–4987 10.1128/IAI.69.8.4980-4987.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ng V. H., Cox J. S., Sousa A. O., MacMicking J. D., and McKinney J. D. (2004) Role of KatG catalase-peroxidase in mycobacterial pathogenesis: Countering the phagocyte oxidative burst. Mol. Microbiol. 52, 1291–1302 10.1111/j.1365-2958.2004.04078.x [DOI] [PubMed] [Google Scholar]
- 5. Flynn J. L., and Chan J. (2001) Immunology of tuberculosis. Annu. Rev. Immunol. 19, 93–129 10.1146/annurev.immunol.19.1.93 [DOI] [PubMed] [Google Scholar]
- 6. Buchmeier N., and Fahey R. C. (2006) The msha gene encoding the glycosyltransferase of mycothiol biosynthesis is essential in Mycobacterium tuberculosis Erdman. FEMS Microbiol. Lett. 264, 74–79 10.1111/j.1574-6968.2006.00441.x [DOI] [PubMed] [Google Scholar]
- 7. Neill M. A., and Klebanoff S. J. (1988) The effect of phenolic glycolipid-1 from Mycobacterium leprae on the antimicrobial activity of human macrophages. J. Exp. Med. 167, 30–42 10.1084/jem.167.1.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar A., Toledo J. C., Patel R. P., Lancaster J. R. Jr., and Steyn A. J. (2007) Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U.S.A. 104, 11568–11573 10.1073/pnas.0705054104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sousa E. H., Tuckerman J. R., Gonzalez G., and Gilles-Gonzalez M. A. (2007) DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci. 16, 1708–1719 10.1110/ps.072897707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park H. D., Guinn K. M., Harrell M. I., Liao R., Voskuil M. I., Tompa M., Schoolnik G. K., and Sherman D. R. (2003) Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48, 833–843 10.1046/j.1365-2958.2003.03474.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wisedchaisri G., Wu M., Sherman D. R., and Hol W. G. J. (2008) Crystal structures of the response regulator DosR from Mycobacterium tuberculosis suggest a helix rearrangement mechanism for phosphorylation activation. J. Mol. Biol. 378, 227–242 10.1016/j.jmb.2008.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Florczyk M. A., McCue L. A., Purkayastha A., Currenti E., Wolin M. J., and McDonough K. A. (2003) A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires rv3133c (dosR or devR) for expression. Infect. Immun. 71, 5332–5343 10.1128/IAI.71.9.5332-5343.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berney M., Greening C., Conrad R., Jacobs W. R. Jr., and Cook G. M. (2014) An obligately aerobic soil bacterium activates fermentative hydrogen production to survive reductive stress during hypoxia. Proc. Natl. Acad. Sci. U.S.A. 111, 11479–11484 10.1073/pnas.1407034111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahmed F. H., Carr P. D., Lee B. M., Afriat-Jurnou L., Mohamed A. E., Hong N. S., Flanagan J., Taylor M. C., Greening C., and Jackson C. J. (2015) Sequence-structure-function classification of a catalytically diverse oxidoreductase superfamily in mycobacteria. J. Mol. Biol. 427, 3554–3571 10.1016/j.jmb.2015.09.021 [DOI] [PubMed] [Google Scholar]
- 15. Ahmed F. H., Mohamed A. E., Carr P. D., Lee B. M., Condic-Jurkic K., O'Mara M. L., and Jackson C. J. (2016) Rv2074 is a novel F420 H2-dependent biliverdin reductase in Mycobacterium tuberculosis. Protein Sci. 25, 1692–1709 10.1002/pro.2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berney M., Greening C., Hards K., Collins D., and Cook G. M. (2014) Three different [NiFe] hydrogenases confer metabolic flexibility in the obligate aerobe Mycobacterium smegmatis. Environ. Microbiol. 16, 318–330 10.1111/1462-2920.12320 [DOI] [PubMed] [Google Scholar]
- 17. Mashalidis E. H., Gittis A. G., Tomczak A., Abell C., Barry C. E. 3rd, Garboczi D. N. (2015) Molecular insights into the binding of coenzyme F420 to the conserved protein Rv1155 from Mycobacterium tuberculosis. Protein Sci. 24, 729–740 10.1002/pro.2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Safo M. K., Mathews I., Musayev F. N., di Salvo M. L., Thiel D. J., Abraham D. J., and Schirch V. (2000) X-ray structure of Escherichia coli pyridoxine 5′-phosphate oxidase complexed with FMN at 1.8 Å resolution. Structure 8, 751–762 10.1016/S0969-2126(00)00162-3 [DOI] [PubMed] [Google Scholar]
- 19. Walsh C. (1980) Flavin coenzymes—at the crossroads of biological redox chemistry. Acc. Chem. Res. 13, 148–155 10.1021/ar50149a004 [DOI] [Google Scholar]
- 20. Buckel W., and Thauer R. K. (2018) Flavin-based electron bifurcation, ferredoxin, flavodoxin, and anaerobic respiration with protons (Ech) or NAD+ (Rnf) as electron acceptors: A historical review. Front. Microbiol. 9, 401 10.3389/fmicb.2018.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bourdeaux F., Hammer C. A., Vogt S., Schweighöfer F., Nöll G., Wachtveitl J., and Grininger M. (2018) Flavin storage and sequestration by Mycobacterium tuberculosis dodecin. ACS Infect. Dis. 4, 1082–1092 10.1021/acsinfecdis.7b00237 [DOI] [PubMed] [Google Scholar]
- 22. Grininger M., Staudt H., Johansson P., Wachtveitl J., and Oesterhelt D. (2009) Dodecin is the key player in flavin homeostasis of archaea. J. Biol. Chem. 284, 13068–13076 10.1074/jbc.M808063200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu F., Xiong J., Kumar S., Yang C., Ge S., Li S., Xia N., and Swaminathan K. (2011) Structural and biophysical characterization of Mycobacterium tuberculosis dodecin Rv1498a. J. Struct. Biol. 175, 31–38 10.1016/j.jsb.2011.04.013 [DOI] [PubMed] [Google Scholar]
- 24. Jeuken L. J. C., Jones A. K., Chapman S. K., Cecchini G., and Armstrong F. A. (2002) Electron-transfer mechanisms through biological redox chains in multicenter enzymes. J. Am. Chem. Soc. 124, 5702–5713 10.1021/ja012638w [DOI] [PubMed] [Google Scholar]
- 25. Segal H. M., Spatzal T., Hill M. G., Udit A. K., and Rees D. C. (2017) Electrochemical and structural characterization of Azotobacter vinelandii flavodoxin II. Protein Sci. 26, 1984–1993 10.1002/pro.3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heuts D. P., Scrutton N. S., McIntire W. S., and Fraaije M. W. (2009) What's in a covalent bond? On the role and formation of covalently bound flavin cofactors. FEBS J. 276, 3405–3427 10.1111/j.1742-4658.2009.07053.x [DOI] [PubMed] [Google Scholar]
- 27. Heering H. A., and Hagen W. R. (1996) Complex electrochemistry of flavodoxin at carbon-based electrodes: Results from a combination of direct electron transfer, flavin-mediated electron transfer and comproportionation. J. Electroanal. Chem. 404, 249–260 10.1016/0022-0728(95)04248-2 [DOI] [Google Scholar]
- 28. Goran J. M., and Stevenson K. J. (2013) Electrochemical behavior of flavin adenine dinucleotide adsorbed onto carbon nanotube and nitrogen-doped carbon nanotube electrodes. Langmuir 29, 13605–13613 10.1021/la403020y [DOI] [PubMed] [Google Scholar]
- 29. Chauviac F. X., Bommer M., Yan J., Parkin G., Daviter T., Lowden P., Raven E. L., Thalassinos K., and Keep N. H. (2012) Crystal structure of reduced MsAcg, a putative nitroreductase from Mycobacterium smegmatis and a close homologue of Mycobacterium tuberculosis Acg. J. Biol. Chem. 287, 44372–44383 10.1074/jbc.M112.406264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Macheroux P., Kappes B., and Ealick S. E. (2011) Flavogenomics—a genomic and structural view of flavin-dependent proteins. FEBS J. 278, 2625–2634 10.1111/j.1742-4658.2011.08202.x [DOI] [PubMed] [Google Scholar]
- 31. Smeulders M. J., Keer J., Speight R. A., and Williams H. D. (1999) Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181, 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Berney M., Weimar M. R., Heikal A., and Cook G. M. (2012) Regulation of proline metabolism in mycobacteria and its role in carbon metabolism under hypoxia. Mol. Microbiol. 84, 664–681 10.1111/j.1365-2958.2012.08053.x [DOI] [PubMed] [Google Scholar]
- 33. Gebhard S., Tran S. L., and Cook G. M. (2006) The Phn system of Mycobacterium smegmatis: A second high-affinity ABC-transporter for phosphate. Microbiology 152, 3453–3465 10.1099/mic.0.29201-0 [DOI] [PubMed] [Google Scholar]
- 34. Fu L., Niu B., Zhu Z., Wu S., and Li W. (2012) CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T. L. (2009) Blast+: Architecture and applications. BMC Bioinformatics 10, 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Doerks T., Copley R. R., Schultz J., Ponting C. P., and Bork P. (2002) Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 12, 47–56 10.1101/gr.203201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cabrita L. D., Gilis D., Robertson A. L., Dehouck Y., Rooman M., and Bottomley S. P. (2007) Enhancing the stability and solubility of TEV protease using in silico design. Protein Sci. 16, 2360–2367 10.1110/ps.072822507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 41. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cowtan K. (2008) Fitting molecular fragments into electron density. Acta Crystallogr. D Biol. Crystallogr. 64, 83–89 10.1107/S0907444907033938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blokpoel M. C., Murphy H. N., O'Toole R., Wiles S., Runn E. S., Stewart G. R., Young D. B., and Robertson B. D. (2005) Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res. 33, e22 10.1093/nar/gni023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robson J., McKenzie J. L., Cursons R., Cook G. M., and Arcus V. L. (2009) The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin–antitoxin module that controls growth via inhibition of translation. J. Mol. Biol. 390, 353–367 10.1016/j.jmb.2009.05.006 [DOI] [PubMed] [Google Scholar]
- 48. Atkinson H. J., Morris J. H., Ferrin T. E., and Babbitt P. C. (2009) Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One 4, e4345 10.1371/journal.pone.0004345 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.