Abstract
Accumulating evidence indicates that a wide range of E3 ubiquitin ligases are involved in the development of many human diseases. Searching for small-molecule modulators of these E3 ubiquitin ligases is emerging as a promising drug discovery strategy. Here, we report the development of a cell-based high-throughput screening method to identify modulators of E3 ubiquitin ligases by integrating the ubiquitin-reference technique (URT), based on a fusion protein of ubiquitin located between a protein of interest and a reference protein moiety, with a Dual-Luciferase system. Using this method, we screened for small-molecule modulators of SMAD ubiquitin regulatory factor 1 (SMURF1), which belongs to the NEDD4 family of E3 ubiquitin ligases and is an attractive therapeutic target because of its roles in tumorigenesis. Using RAS homolog family member B (RHOB) as a SMURF1 substrate in this screen, we identified a potent SMURF1 inhibitor and confirmed that it also blocks SMURF1-dependent degradation of SMAD family member 1 (SMAD1) and RHOA. An in vitro auto-ubiquitination assay indicated that this compound inhibits both SMURF1 and SMURF2 activities, indicating that it may be an antagonist of the catalytic activity of the HECT domain in SMURF1/2. Moreover, cell functional assays revealed that this compound effectively inhibits protrusive activity in HEK293T cells and blocks transforming growth factor β (TGFβ)-induced epithelial-mesenchymal transition (EMT) in MDCK cells, similar to the effects on these processes caused by SMURF1 loss. In summary, the screening approach presented here may have great practical potential for identifying modulators of E3 ubiquitin ligases.
Keywords: high-throughput screening (HTS), E3 ubiquitin ligase, ubiquitylation (ubiquitination), inhibitor, cellular regulation, protein degradation, epithelial-mesenchymal transition (EMT), auto-ubiquitination, chemical screen, HECT domain, SMAD ubiquitin regulatory factor 1 (SMURF1), ubiquitin-reference technique (URT)
Introduction
The ubiquitin-dependent pathway is a key regulatory mechanism that controls the degradation of proteins important in various biological processes such as cell cycle, DNA repair, signal transduction, transcriptional regulation, endocytosis, and stress responses (1–3). Ubiquitination of proteins requires a multienzyme system comprised of ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase enzyme (E3). Ubiquitin is first activated by binding to E1 and then transferred to an E2 before being covalently linked to a protein substrate in a reaction catalyzed by E3 ubiquitin ligase (1). Compared with only one E1 enzyme and limited number of E2s in most organisms, there is a much larger number of E3 ubiquitin ligases. Each E3 ubiquitin ligase recognizes a set of substrates and controls the specificity in ubiquitin-mediated protein degradation (2). Therefore, targeting specific E3 ubiquitin ligases using small molecules is a promising strategy to regulate degradation of specific proteins.
SMURF1 and SMURF2 are two closely related members of the NEDD4 family of E3 ubiquitin ligases (4). SMURFs were first identified as negative regulators of TGFβ4 signaling that either directly target receptor-regulated SMADs for degradation (5–7) or target the TGFβ receptors via interactions with inhibitory SMADs (8–10). SMURF1 also plays a key role in regulating cell polarity, protrusive activity, and dissolution of tight junctions in TGFβ-induced EMT by targeting the small GTPase RHOA for degradation (11, 12). Thus, SMURF1 functions in the TGFβ signaling both as an antagonist of SMAD signaling and an effector of the TGFβ/PAR6 pathway during EMT. As the SMAD pathway is a tumor suppressor pathway, and the TGFβ/PAR6-regulated EMT plays an important role in cancer progression by promoting an invasive, metastatic phenotype (13), SMURF1 might be a key player in controlling both cancer cell proliferation and metastasis during tumorigenesis. Moreover, our recent study found that SMURF1 targets RHOB, a close family member of RHOA, for degradation to control its abundance in cells (14). It is noteworthy that RHOB is recognized as a tumor suppressor by promoting death of transformed cells (15), suggesting that SMURF1 may also prevent cancer cell death by targeting RHOB for degradation. Hence, these distinct roles of SMURF1 make it a good candidate as a target for modulation by small molecules, which will be greatly beneficial for both a mechanistic understanding of HECT family ligase catalytic mechanism and cancer drug development.
Present high-throughput screening methods for identifying small-molecule modulators of E3 ubiquitin ligases are mainly limited to in vitro systems, which are complicated and inconvenient. Here we report a general cell-based high-throughput screening method by integrating the Dual-Luciferase system with the ubiquitin-reference technique (URT) (16, 17) to identify small-molecule modulators of E3 ubiquitin ligases. URT uses a linear fusion in which a ubiquitin is located between a protein of interest and a reference protein moiety. The fusion protein is co-translationally (or nearly so) cleaved by ubiquitin-specific processing proteases (Ubps) after ubiquitin to produce equimolar amounts of the protein of interest and the reference protein bearing the C-terminal ubiquitin moiety (17). By introducing the internal reference, URT can compensate for sample-to-sample variation inherent in cell-based screens. In this study, we applied the URT system to a high-throughput screen for SMURF1 modulators and identified a novel SMURF1 small-molecule inhibitor.
Results
Construction of URT-luciferase high-throughput screening system
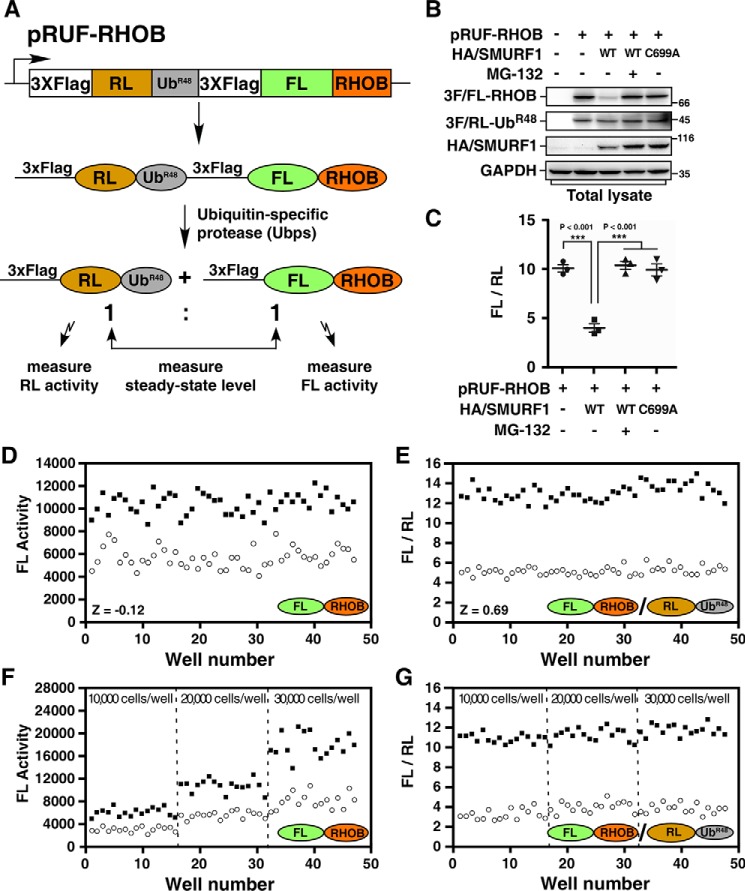
To screen for small-molecule inhibitors of SMURF1, we used the SMURF1 substrate RHOB as a target protein to develop a cell-based high-throughput screening system using Dual-Luciferase and URT. In this procedure, N-terminally triple FLAG-tagged Renilla luciferase (RL) is linked to the N terminus of a ubiquitin K48R mutant (UbR48) moiety that is in turn linked to triple FLAG-tagged firefly luciferase (FL). The resulting 3×FLAG-RL-UbR48-3×FLAG-FL was then fused to the target substrate RHOB to generate the final fusion protein 3×FLAG-RL-UbR48-3×FLAG-FL-RHOB. The expression plasmid is designated as pRUF(RL-UbR48-FL)-RHOB as diagrammed in Fig. 1A. The very end amino acid residue (glycine) in UbR48 provides a proteolytic cleavage site for the endogenous Ubps, yielding equimolar amount of the triple FLAG-tagged target protein FL-RHOB and reference protein RL-UbR48. The UbR48 mutant was used instead of WT ubiquitin (Ub) to prevent a potential K48 ubiquitin conjugation on RL-Ub, which may function as a degradation signal of RL-Ub. The steady-state levels of FL-RHOB and RL-UbR48 can then be quantified by measuring the activities of FL and RL, respectively, using the Dual-Glo Luciferase Assay System (Promega). The RL-UbR48 is stable and acts as an internal reference, whereas FL-RHOB is a substrate of SMURF1 and its degradation inversely correlates with the activity of FL; therefore, the activity of SMURF1 can be revealed by the ratio of FL activity to RL activity (FL/RL). The presence of SMURF1 inhibitor will block SMURF1-mediated FL-RHOB degradation and increase the FL/RL ratio. Conversely, the FL/RL ratio will be reduced in the presence of SMURF1 activator.
Figure 1.
Cell-based high-throughput URT-Dual-Luciferase Assay system for SMURF1. A, a schematic of the pRUF-RHOB construct. A fusion protein comprised of triple FLAG-tagged RL, UbR48 moiety, triple FLAG-tagged FL, and RHOB is cleaved in vivo by Ubps at the UbR48-RHOB junction to yield equimolars of triple FLAG-tagged RL-UbR48 and triple FLAG-tagged FL-RHOB. The triple FLAG-tagged FL-RHOB is a substrate of SMURF1 and will be degraded in the presence of SMURF1. B, analysis of the steady-state levels of FL-RHOB by immunoblotting. HEK293T cells were transfected with pRUF-RHOB and HA-tagged SMURF1 (HA/SMURF1), WT, or catalytically inactive mutant C699A, as indicated. After overnight treatment with or without 5 μm MG-132, total cell lysates were subjected to immunoblotting using indicated antibodies to determine the steady-state protein levels. C, luciferase assay of FL-RHOB. HEK293T cells were transfected with pRUF-RHOB and HA/SMURF1 WT or C699A, and treated with or without 5 μm MG-132 as in (B) and then applied to Dual-Glo Luciferase Assay to measure the activities of FL and RL. Results were plotted as the ratio of FL activity to RL activity (FL/RL). D and E, evaluation of the screening system. HEK293T cells co-transfected with pRUF-RHOB and WT HA/SMURF1 were treated overnight with DMSO (open circles) and 5 μm MG-132 (filled squares) as negative or positive controls, respectively. Luciferase activities were measured and plotted using either FL activity alone (D) or the FL/RL ratio (E). F and G, the URT system effectively corrects variation of cell numbers. HEK293T cells co-transfected with pRUF-RHOB and WT HA/SMURF1 were seeded with varying number of cells as indicated and treated with DMSO or MG-132 as in (D). Luciferase activities were measured and plotted as FL alone (F) or FL/RL (G).
To test the feasibility of this system, we first examined the steady-state levels of FL-RHOB fusion protein by immunoblotting. As expected, the steady-state level of FL-RHOB was reduced by co-expression of WT SMURF1, but not by the catalytically inactive form SMURF1-C699A, whereas the steady-state level of RL-UbR48 was not affected by either of them (Fig. 1B). Furthermore, the proteasome inhibitor MG-132 prevented SMURF1-dependent loss of FL-RHOB, indicating that SMURF1-mediated RHOB degradation is through the ubiquitin-proteasome pathway (Fig. 1B). Next, we calculated the ratio of the two luciferase reporter proteins by measuring their activities. Quantification of the FL/RL ratio was in good agreement with our analysis by immunoblotting, showing that co-expression of WT SMURF1 reduced the FL/RL ratio from 10 to 4, whereas co-expression of SMURF1-C699A had no effect on the FL/RL ratio. Treatment with MG-132, however, prevented the decrease of FL/RL ratio caused by SMURF1 (Fig. 1C). Thus, analysis of protein levels using URT–Dual-Luciferase Assay provides an accurate measurement of SMURF1-dependent RHOB turnover.
To explore the capacity of the URT–Dual-Luciferase method in a high-throughput setting, pRUF-RHOB was co-transfected with SMURF1 into HEK293T cells, which were subsequently seeded in 96-well plates and then treated with either the solvent DMSO as negative control or MG-132 as positive control. After overnight treatment, the activities of FL and RL were measured using the Dual-Glo Luciferase Assay System with a Varioskan Flash instrument (Thermo Scientific) in 96-well format. Assay performance was then assessed by measuring the value of Z-factor, which is a coefficient for the screening window (18). Z-factor is calculated using the formula Z = 1 − (3σs+3σc)/|μs-μc|, in which σs and σc are the standard deviations (S.D.s) of samples and controls, μs and μc are the means of samples and controls, respectively. As shown in Fig. 1D, using FL activity alone, the assay yielded a very poor Z value (−0.12). However, the assay quality was dramatically improved when the FL/RL ratio was used, with the Z-factor rising to 0.69 (Fig. 1E), converting a poor method into an excellent assay (18). In addition, URT normalization provided an excellent correction of variation because of differences in cell-seeding densities. Seeding with different amount of cells drastically changed the FL activity in the screen (Fig. 1F); however, the FL/RL ratio was not significantly affected by the cell seeding (Fig. 1G).
High-throughput screening for SMURF1 inhibitors
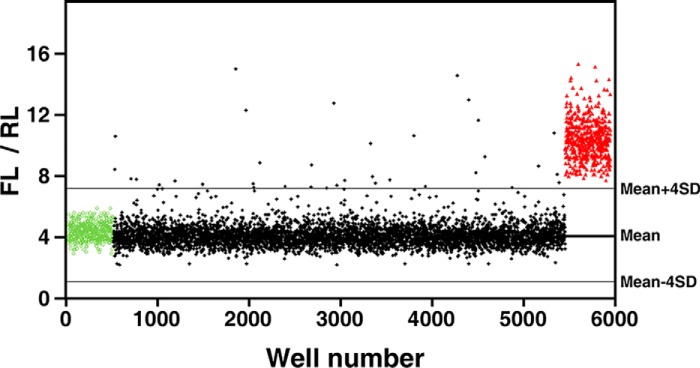
Next, the high-throughput URT–Dual-Luciferase screening system was applied to screen for SMURF1 inhibitors using one of our in-house compound libraries, which contains a total of 5000 compounds. For this, HEK293T cells were co-transfected with pRUF-RHOB and SMURF1 in a large batch and subsequently seeded in 96-well plates. Individual wells were then treated overnight with 10 μm of each compound from this library, and the FL and RL activities were then measured and analyzed. In each plate, the first column was treated with DMSO as a negative control, whereas the last column received MG-132 as a positive control. A cutoff corresponding to the assay mean ± 4 × S.D. was used for hit selection, which yielded 37 out of 5000 compounds as potential SMURF1 antagonists (0.74% hit rate) (Fig. 2).
Figure 2.
High-throughput screening for SMURF1 inhibitors. HEK293T cells co-transfected with pRUF-RHOB and WT HA/SMURF1 were treated overnight with 10 μm of each compound from the compound library. DMSO (green circles) and 5 μm MG-132 (red triangles) were used as negative or positive controls, respectively. Activities of FL and RL were measured and results plotted as FL/RL. Solid lines represent the mean and the mean ± 4 × S.D. of all assay points excluding MG-132–treated wells.
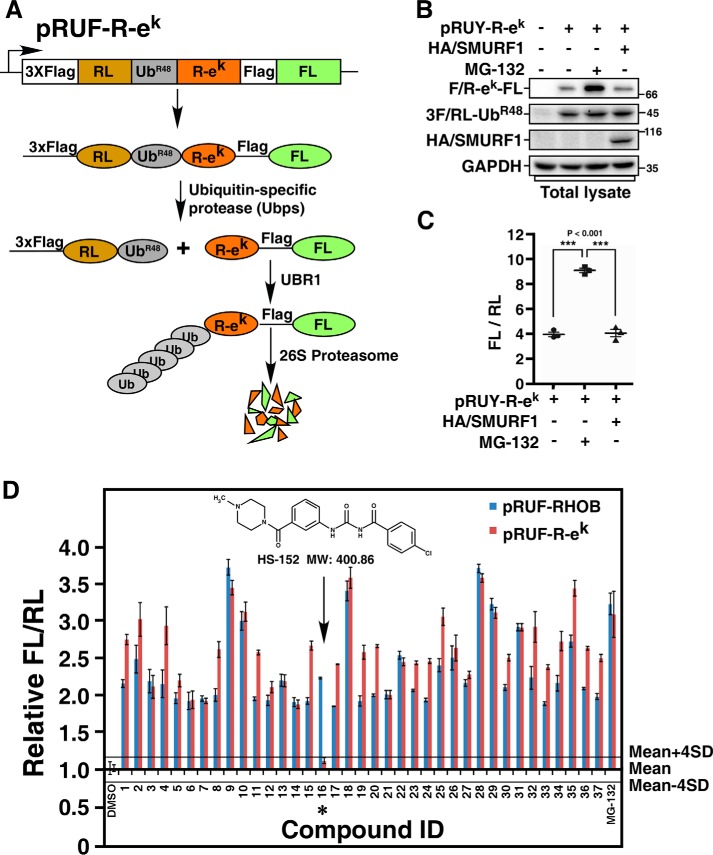
Because nonspecific inhibitors that target E1, E2, or proteasome could also block SMURF1-dependent degradation of FL-RHOB, we designed a secondary screen to rule out the nonspecific inhibitors by applying these compounds to a SMURF1-unrelated ubiquitin-dependent proteasome pathway. For this purpose, the N-end rule pathway, which determines the in vivo half-life of a protein depending on the identity of its N-terminal residues (19, 20), was employed. The ek sequence, which encodes a 45-residue segment of the Escherichia coli Lac repressor and was used in previous studies of N-end rule pathway (17, 21), was inserted into pRUF vector to generate pRUF–R-ek (Fig. 3A). Expression of pRUF–R-ek in cells will generate the fusion protein 3×FLAG-RL-UbR48–R-ek–FLAG-FL, which will subsequently yield an equimolar amount of the reference protein RL-UbR48 and the target protein R-ek–FL with the arginine (R) residue at the N-terminal (Fig. 3A). According to the N-end rule pathway, short-lived R-ek–FL generated by pRUF–R-ek will be targeted by endogenous E3 ubiquitin ligase UBR1 that belongs to the family of RING-type E3 ubiquitin ligases (19, 22), which differs from the HECT family E3s in structure and catalytic mechanism. As expected, the steady-state level of R-ek–FL was significantly increased by MG-132 treatment, whereas overexpression of SMURF1 did not significantly affect the levels of R-ek–FL, confirming that R-ek–FL is not a substrate for SMURF1 (Fig. 3, B and C). pRUF–R-ek was thus used in the secondary screen to eliminate nonspecific inhibitors from the 37 selected compounds from the primary screen. The major difference between primary screen and secondary screen is the two different types of E3s that were used in the screens. Therefore, compounds affecting both FL-RHOB degradation and R-ek–FL degradation are likely false-positive hits that target common components in these two screens, such as E1, E2, or proteasome. The compound HS-152, 4-chloro-N-((3-(4-methylpiperazine-1-carbonyl) phenyl) carbamoyl)-benzamide, with molecular weight of 400.86, was the only one affecting the steady-state level of FL-RHOB, but not the level of R-ek–FL, and therefore was identified as a potential specific inhibitor of SMURF1 (Fig. 3D). The compound HS-152 was resynthesized for further characterization. The detailed synthetic procedure and chemical characterization of HS-152 are provided in the supporting data.
Figure 3.
Counterscreen using the N-end rule pathway. A, a schematic of the pRUF–R-ek construct. The pRUF–R-ek is designed similarly to pRUF-RHOB (Fig. 1A) except that triple FLAG-tagged FL-RHOB is replaced by R-ek–FLAG-FL. B, immunoblotting assay of the steady-state levels of R-ek–FL. HEK293T cells were transfected with pRUF–R-ek with or without HA/SMURF1 as indicated. After 3 h treatment with or without 10 μm MG-132, steady-state protein levels were determined by immunoblotting total cell lysates using indicated antibody. C, luciferase assay of R-ek–FL. HEK293T cells were transfected with or without HA/SMURF1 and pRUF–R-ek, and treated with or without MG-132 as in (B). The activities of FL and RL were then measured and plotted as FL/RL. D, counterscreen of selected compounds from the primary screen. HEK293T cells transfected with pRUF–R-ek were treated 6 h with 10 μm of each of the 37 compounds identified in the primary screen. DMSO and MG-132 were used as negative and positive controls, respectively. The effect of each compound on the FL-RHOB/RL or R-ek–FL/RL ratio was plotted relative to the mean of the eight DMSO-treated control wells, respectively. Solid lines represent the mean ± 4 × S.D. of the DMSO controls, as indicated. Arrow and asterisk show the compound that had minimal effect on R-ek–FL/RL ratio.
Characterization of the SMURF1 inhibitor
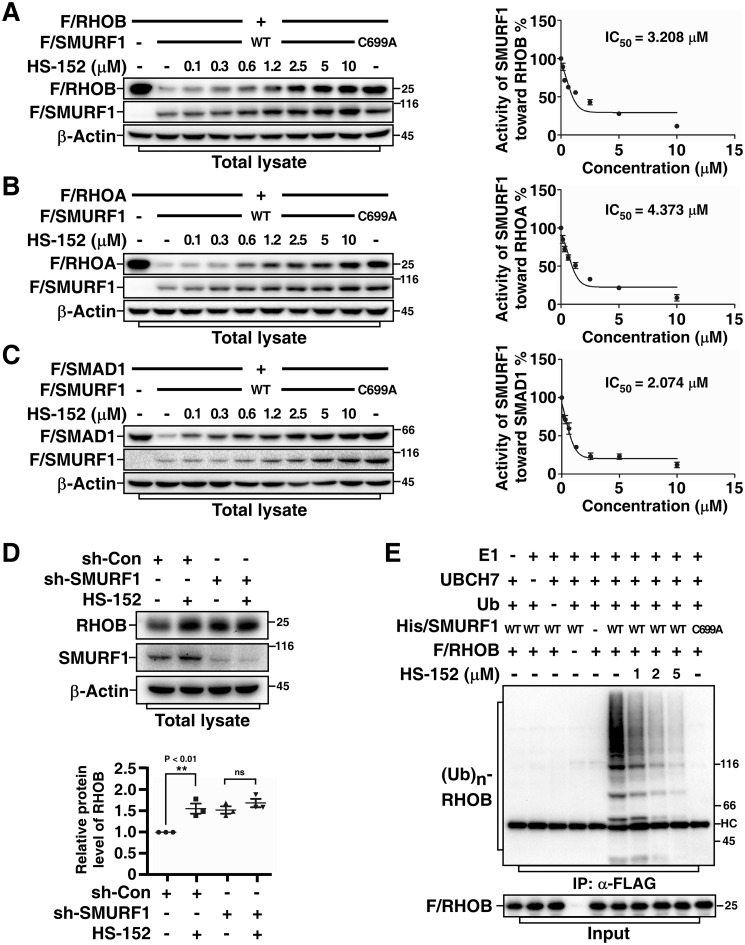
To further characterize the capability of compound HS-152 to inhibit SMURF1 activity, we first examined its efficacy in preventing SMURF1-dependent protein degradation. Indeed, HS-152 not only potently inhibited SMURF1-mediated RHOB degradation with an IC50 of 3.2 μm (50% inhibitory concentration) (Fig. 4A), but also strongly blocked SMURF1-mediated RHOA and SMAD1 degradation (IC50 4.4 μm and 2.1 μm, respectively) (Fig. 4, B and C). We investigated whether HS-152–caused up-regulation of SMURF1 substrates is indeed through affecting SMURF1. To this end, we examined the effects of HS-152 on endogenous RHOB levels in control and SMURF1 knockdown cells. As shown in Fig. 4D, treatment of HS-152 significantly increased endogenous RHOB levels in control cells. Knockdown of SMURF1 also increased RHOB levels; however, HS-152 could not further increase RHOB levels in SMURF1 knockdown cells, indicating that HS-152–mediated increase of RHOB levels is through SMURF1. We next performed in vitro ubiquitination assay to investigate whether HS-152 stabilizes RHOB via inhibiting SMURF1-mediated RHOB ubiquitination. As expected, HS-152 significantly blocked SMURF1-mediated RHOB ubiquitination in a dose-dependent manner (Fig. 4E).
Figure 4.
HS-152 inhibits SMURF1-mediated ubiquitination and degradation. A, HS-152 inhibits SMURF1-mediated RHOB degradation in a dose-dependent manner. HEK293T cells were transfected with FLAG-tagged RHOB (F/RHOB) and WT or C699A FLAG-tagged SMURF1 (F/SMURF1) as indicated. After overnight treatment with or without different doses of HS-152, total cell lysates were subjected to immunoblotting using indicated antibodies. The steady-state protein levels were quantified using Image Lab software (Bio-Rad) with β-actin as a loading control. Results were plotted in right panel as the levels of F/RHOB in cells co-transfected with WT F/SMURF1 and F/RHOB at each dose of HS-152 treatment relative to the level of F/RHOB in cells transfected with F/RHOB alone and without HS-152 treatment. B, HS-152 inhibits SMURF1-mediated RHOA degradation in a dose-dependent manner. HEK293T cells transfected with indicated FLAG-tagged RHOA (F/RHOA) and F/SMURF1 (WT or C699A) were treated with different doses of HS-152 and subjected to immunoblotting and then quantified and plotted as in (A). C, HS-152 inhibits SMURF1-mediated SMAD1 degradation in a dose-dependent manner. HEK293T cells transfected with indicated FLAG-tagged SMAD1 (F/SMAD1) and F/SMURF1 (WT or C699A) were treated with a different dose of HS-152 and subjected to immunoblotting and then quantified and plotted as in (A). D, HS-152 up-regulates endogenous RHOB levels through SMURF1. HEK293T cells transfected with control shRNA (sh-Con) or shRNA against SMURF1 (sh-SMURF1) and treated 4 h with or without 2 μm HS-152 and then subjected to immunoblotting assay. The lower panel presents quantitative analysis of Western blotting results (mean ± S.D. of three independent experiments). E, HS-152 inhibits SMURF1-mediated RHOB ubiquitination in vitro. FLAG-tagged RHOB (F/RHOB) and His-tagged SMURF1 (His/SMURF1) expressed and purified from bacteria were subjected to an in vitro ubiquitination assay in the absence or presence of different doses of HS-152 as indicated. The reaction products were then subjected to anti-FLAG immunoprecipitation (IP) followed by immunoblotting assay to detect ubiquitin-conjugated RHOB ((Ub)n-RHOB) using an anti-ubiquitin antibody.
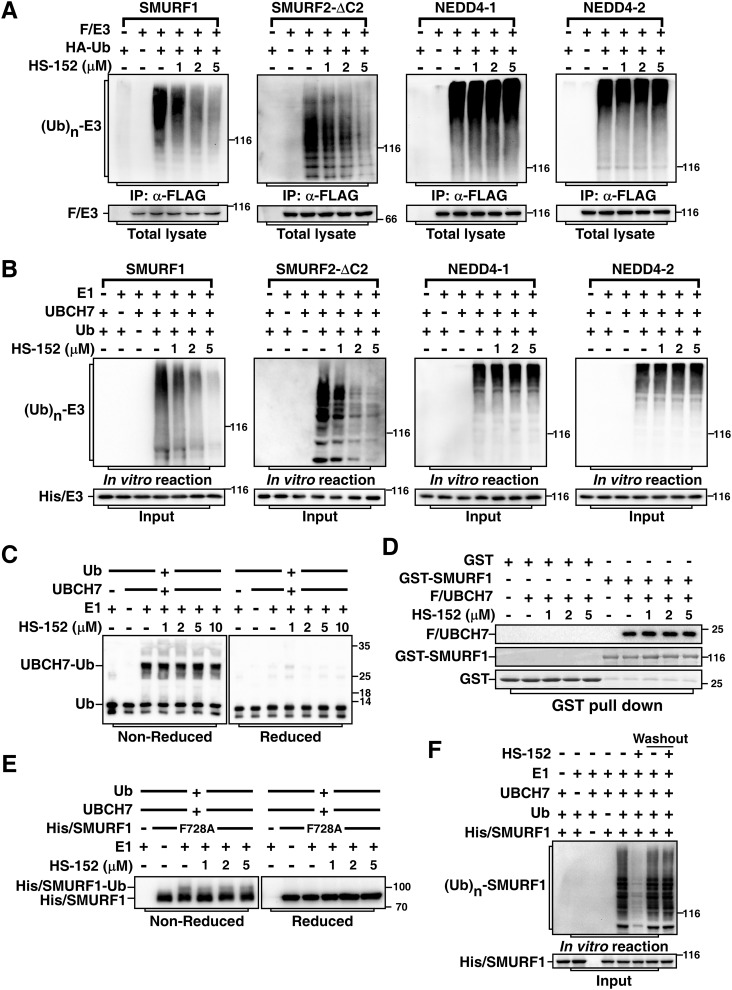
We further examined the specificity of HS-152 by comparing its capability on blocking catalytic activities of SMURF1 and its family members SMURF2, NEDD4–1, and NEDD4–2. Unlike SMURF1, SMURF2 contains an extra WW domain and is subject to auto-inhibiting its catalytic activity via its C2 domain (23). Therefore, we assessed HS-152 activity toward SMURF2 using SMURF2-ΔC2, which is a hyperactive form of SMURF2 that lacks of the N-terminal C2 domain. Treatment with HS-152 significantly blocked auto-ubiquitination of SMURF1 and SMURF2-ΔC2, but not NEDD4–1 and NEDD4–2, in cells and in vitro (Fig. 5, A and B), indicating that HS-152 is a relatively specific inhibitor of SMURFs. Furthermore, to confirm that the inhibitory activity of HS-152 toward SMURFs did not reflect activity on upstream steps in the ubiquitination cascade, we examined E1-dependent ubiquitin charging of E2 UBCH7, the preferred E2 for SMURF1 (24). Even at 10 μm, a concentration at which HS-152 strongly inhibited SMURF1 and SMURF2 activity, UBCH7 was still charged efficiently (Fig. 5C), indicating that HS-152 works indeed at the E3 step. Meanwhile, UBCH7 showed similar affinity to SMURF1 in the presence or absence of HS-152, indicating that HS-152 does not affect binding of E2 to SMURF1 (Fig. 5D). Previous study showed that the phenylalanine residue located four amino acids from the C-terminal of HECT domain (−4F) is essential for transferring ubiquitin from HECT domain to substrate, but not critical for the formation of ubiquitin-thioester intermediate with the catalytic cysteine residue of HECT domain (25). We next further used the −4F to alanine mutant SMURF1 (SMURF1-F728A) to investigate in which step HS-152 interrupts SMURF1 catalysis. Indeed, SMURF1-F728A formed thioester bond with ubiquitin. Interestingly, treatment of HS-152 did not affect the formation of the ubiquitin-thioester intermediate (Fig. 5E), suggesting that the mechanism by which HS-152 inhibits SMURF1 activity is through blocking the transfer of ubiquitin from HECT domain to the substrates to form isopeptide bond. In addition, we verified that HS-152 functions as a reversible inhibitor of SMURF1 by washout experiment (Fig. 5F). Altogether, these data suggest that HS-152 directly targets the catalytic activity of SMURF HECT domains.
Figure 5.
HS-152 inhibits catalytic activities of SMURFs. A, HS-152 inhibits auto-ubiquitination of SMURF1 and SMURF2-ΔC2 in cells. HEK293T cells were transfected with HA-tagged ubiquitin (HA/Ub) and FLAG-tagged SMURF1, SMURF2-ΔC2, NEDD4–1, or NEDD4–2 as indicated. The cells were treated 3 h with 40 μm MG-132 and then subjected to anti-FLAG immunoprecipitation (IP) followed by immunoblotting assay to detect ubiquitin-conjugated E3s ((Ub)n-E3) using anti-HA antibody. B, HS-152 inhibits auto-ubiquitination of SMURF1 and SMURF2-ΔC2 in vitro. Purified SMURF1, SMURF2-ΔC2, NEDD4–1, and NEDD4–2 were subjected to an in vitro auto-ubiquitination assay in the absence or presence of HS-152 at the indicated concentrations and ubiquitin-conjugated E3s ((Ub)n-E3) were detected by immunoblotting with anti-ubiquitin antibody. C, HS-152 does not inhibit E2 UBCH7 ubiquitin conjugation. E2 UBCH7 conjugation by E1 enzyme was conducted with varying concentrations of HS-152 as indicated. Reactions were stopped in nonreducing SDS sample buffer and then were applied to immunoblotting under nonreducing condition (left panel), or were reduced using DTT prior to SDS-PAGE (right panel). Ubiquitin and ubiquitin conjugated to UBCH7 via the reductant-sensitive thioester bond are indicated. D, HS-152 does not affect binding of E2 to Smurf1. Purified Smurf1 and UBCH7 were subjected to GST pulldown assay in the absence or presence of indicated amount of HS-152. E, HS-152 does not affect formation of ubiquitin-thioester intermediate in vitro. Purified Smurf1-F728A mutant was subjected to an in vitro auto-ubiquitination assay in the absence or presence of indicated amount HS-152. Reactions were stopped as in (C) to examine the thioester bond formation. F, HS-152 is a reversible inhibitor of SMURF1. Purified SMURF1 was incubated with 5 μm HS-152 for 20 min and then carried out with or without washing three times, as indicated, before subjected to an in vitro auto-ubiquitination assay.
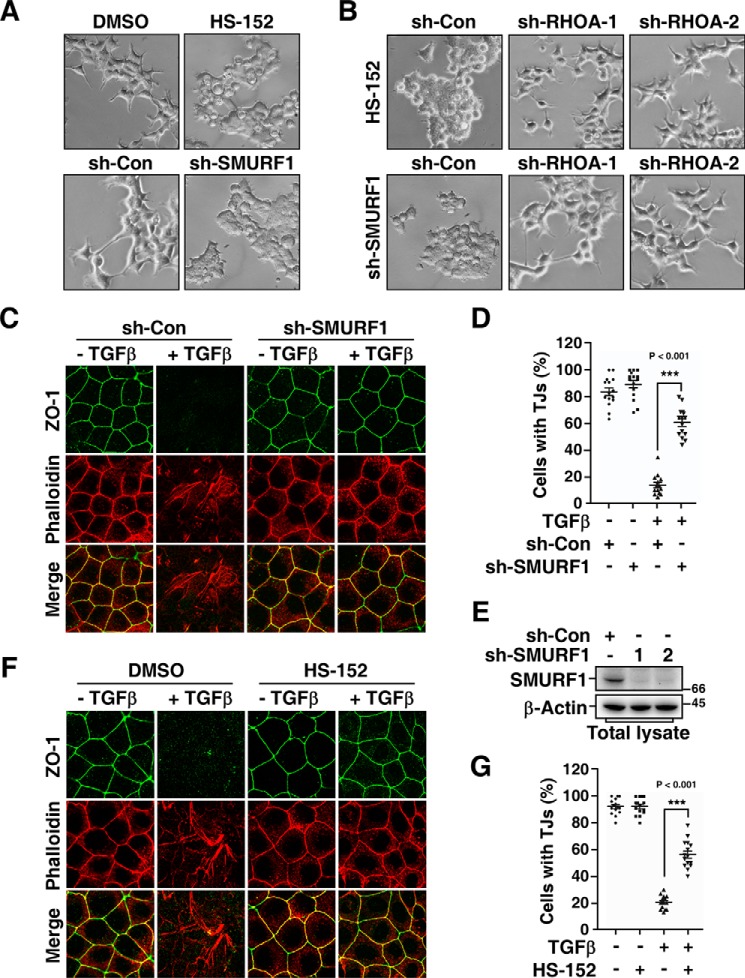
Our previous studies showed that SMURF1 functions as a downstream effector of PAR6, a component of the polarity complex and is important for cell protrusive activity and TGFβ-induced EMT (11, 12). Therefore, we sought to examine whether HS-152 is able to inhibit these SMURF1-dependent pathways. We first examined the effect of HS-152 on the protrusive activity of HEK293T cells. Similar to the phenotype observed when SMURF1 expression was knocked down (12), overnight treatment with HS-152 dramatically blocked the formation of cell protrusions (Fig. 6A). Because SMURF1-mediated RHOA degradation is a key step in the SMURF1-regulated protrusive activity, knockdown of SMURF1 results in an accumulation of RHOA, which subsequently inhibits the cell protrusive activity. Therefore, concomitantly knocking down RHOA levels suppresses the effect of SMURF1 knockdown and restores the protrusive activity (12). Similarly, knockdown of RHOA significantly rescued the protrusion formation of HEK293T cells treated with HS-152 (Fig. 6B), indicating that the loss of protrusion caused by HS-152 treatment is also because of accumulation of RHOA.
Figure 6.
HS-152 inhibits cell protrusive activity and TGFβ-induced EMT. A, HS-152 has a similar effect with SMURF1 shRNA on inhibiting protrusive activity of HEK293T cells. Twenty-four h after being transfected with control shRNA (sh-Con) or shRNA against SMURF1 (sh-SMURF1) (lower panel), or overnight after being treated with or without 1 μm HS-152 (upper panel) as indicated, HEK293T cell morphology was imaged by phase contrast microscopy. B, HS-152–caused loss of protrusion depends on RHOA. HEK293T cell morphology was imaged by phase contrast microscopy 24 h after co-transfection with the indicated combination of sh-SMURF1 and sh-Con or shRNA against RHOA (sh-RHOA) (lower panel), or treated another 4 h with DMSO or 1 μm HS-152 20 h after transfection with sh-Con or sh-RHOA (upper panel) as indicated. C and D, knockdown of SMURF1 blocks TGFβ-induced EMT. MDCK cells transduced with lentivirus encoding sh-Con or sh-SMURF1 were treated 20 h with or without 100 pm TGFβ, and then subjected to immunofluorescence assay. ZO-1 staining was detected with an anti-ZO-1 antibody (green), and F-actin was visualized with Texas red-conjugated phalloidin (red) (C). The percentages of cells with tight junctions (TJs) were plotted in (D). Five random areas were counted for each experiment and data of three independent experiments were assessed and represented as mean ± S.D. (D). E, knockdown efficiency of SMURF1. MDCK cells transduced with lentivirus encoding sh-Con or sh-SMURF1 were subjected to immunoblotting assay to examine the knockdown efficiency of SMURF1. F and G, HS-152 inhibits TGFβ-induced EMT. MDCK cells pretreated 12 h with DMSO or 0.05 μm HS-152 were treated another 20 h with or without 100 pm TGFβ and then subjected to immunofluorescence assay (F). Quantification of cells with tight junctions was as in (D).
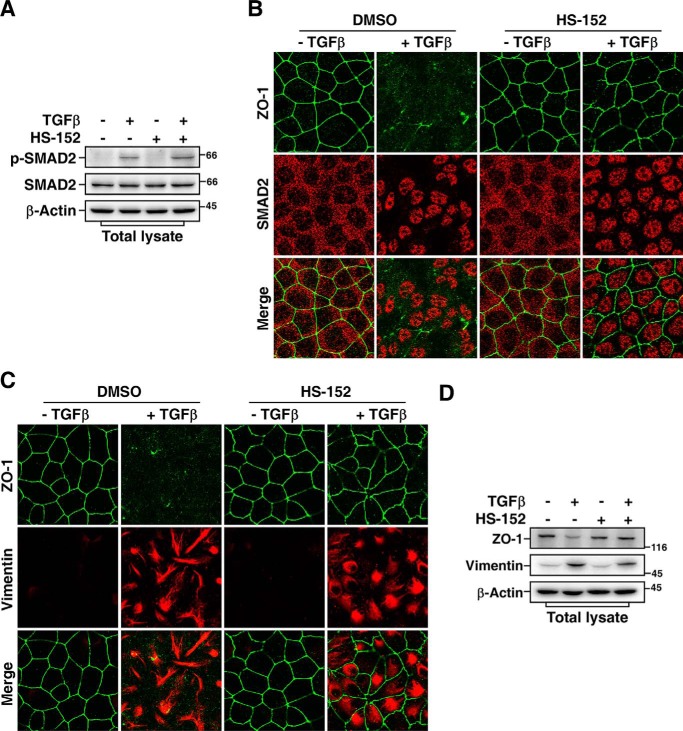
Previous study showed that SMURF1 activity is required for TGFβ-induced EMT (11). The Madin-Darby canine kidney (MDCK) cells displayed apical-basal polarity and formed tight junctions in the absence of TGFβ. After 24 h treatment with TGFβ, MDCK cells underwent EMT that was characterized by loss of tight junctions and rearrangement of cortical actin cytoskeleton into stress fibers. Indeed, knockdown of SMURF1 dramatically blocked TGFβ-induced EMT in MDCK cells (Fig. 6, C and D). The efficiency of knockdown of SMURF1 was examined by immunoblotting assay (Fig. 6E). Similarly, HS-152 treatment significantly blocked TGFβ-induced dissociation of tight junctions even after 24 h treatment with TGFβ (Fig. 6, F and G). Furthermore, despite blocking dissolution of tight junctions, HS-152 treatment did not block TGFβ-dependent phosphorylation and nuclear accumulation of SMAD2 (Fig. 7, A and B), and subsequent expression of mesenchymal marker protein vimentin (Fig. 7, C and D), indicating that HS-152 does not affect TGFβ-dependent SMAD2 activation, which is in good agreement with previous study showing that blocking TGFβ-dependent EMT by interfering with the PAR6/SMURF1/RHOA pathway is independent of SMAD activation (11).
Figure 7.
HS-152 does not interfere with TGFβ/SMAD pathway. A, HS-152 does not inhibit TGFβ-induced phosphorylation of SMAD2. MDCK cells pretreated 12 h with DMSO or 0.05 μm HS-152 were treated 20 h with or without 100 pm TGFβ and then subjected to immunoblotting of total cell lysates with indicated antibodies. B, HS-152 does not hinder the nuclear accumulation of SMAD2 in response to TGFβ treatment. MDCK cells pretreated 12 h with or without 0.05 μm HS-152 were treated another 20 h with or without 100 pm TGFβ, and then subjected to immunofluorescence assay. ZO-1 staining was detected with an anti-ZO-1 antibody (green), and SMAD2 was detected with an anti-SMAD2 antibody (red). C, HS-152 does not block TGFβ-induced vimentin expression. MDCK cells treated as in (B) were subjected to immunofluorescence assay to detect ZO-1 (green) and vimentin (red) staining using anti–ZO-1 and anti-vimentin antibodies, respectively. D, HS-152 blocks TGFβ-induced down-regulation of ZO-1 but not up-regulation of vimentin. MDCK cells treated as in (B) were subjected to immunoblotting assay with indicated antibodies.
Discussion
Dysregulation of protein degradation plays pivotal roles in development of many human diseases like cancer (26, 27). Inhibition of proteasome would induce multiple events that lead to cell death or growth inhibition of malignant cells (27). As a matter of fact, bortezomib is the first proteasomal inhibitor that was approved by the Food and Drug Administration for treatment of relapsed and refractory multiple myeloma (28, 29). However, because of their nonspecific effects on all ubiquitin-proteasome pathways, proteasomal inhibitors inevitably involve the risk of side effects and the administered dose needs to be carefully monitored (27, 29). Continually increasing evidence shows that a number of E3 ubiquitin ligases are closely related to human cancer (30, 31). Unlike proteasome, E1, or E2, E3 ubiquitin ligases have much higher substrate specificity. Specifically targeting desired E3 ubiquitin ligases would be an alternative way with less associated toxicity than proteasomal inhibitors. Hence, high-throughput screening for inhibitors of E3 ubiquitin ligases is drawing much attention for cancer drug development from the pharmaceutical industry and academic studies.
Present general high-throughput screening methods for screening E3 ubiquitin ligase inhibitors have mainly relied on in vitro strategies including UbFluor, fluorescence resonance energy transfer (FRET), homogenous time-resolved fluorescence (HTRF), electrochemiluminescence, scintillation proximity assay (SPA), and dissociation-enhanced lanthanide fluoroimmunoassay (DELFIA) (32, 33). However, all these screening assays require purification of proteins (ubiquitin, E1, E2, E3 and often a substrate) and in vitro fluorescence or isotope labeling, which make the assays complicated and expensive. Moreover, compounds identified in vitro entail a verification or validation by cell-based assays for their specificity, permeability, and efficacy. In this work, we have developed a cell-based high-throughput screening method for E3 ubiquitin ligases using the URT–Dual-Luciferase. In this system, FL was used as target protein reporter, whereas RL was used as reference reporter to normalize the variation resulting from cell seeding, transfection, expression level, toxicity effect and other sample-to-sample variation. Moreover, URT significantly improved the assay quality of the screen, allowing us to fulfill the screen that was hard to execute without the URT normalization. It must be addressed that co-transfection of an RL vector and a substrate-linked FL vector cannot fully serve to replace our single fusion protein approach, as the two individually transcribed mRNAs may be affected or translated differently in the presence of added drugs. Through adaptation of proper substrates, this method allows high-throughput screening for both inhibitors and activators of other E3 ubiquitin ligases, for example, the E3 of N-end rule pathway that was used in our secondary screen. Hence, this cell-based in vivo high-throughput screening system is a highly sensitive, cost effective, and convenient system for screening modulators of E3 ubiquitin ligases. Alternatively, fluorescent proteins such as YFP and CFP might also be used as reporters in our high-throughput screening system, which will further cut down the cost and even will be able to provide a real-time monitoring system for living cell assays. In addition, we observed some conjugation products of RL-UbR48, although these conjugations had no significant effect on RL activity so that did not affect the screen, suggesting that RL-UbR48 can still be used as a source of ubiquitin for conjugation reactions and/or be conjugated with Ub. Therefore, a Ub mutant with all K to R substitutions and mutation(s) that may preclude it as a substrate of E3 Ub ligases for conjugation reactions but not affect the cleavage by Ubps might be a better choice in the future application of this method.
Recent studies indicate that SMURF1 and SMURF2 are also involved in cancer progress (34). SMURFs negatively regulate TGFβ signaling by targeting TGFβ receptors and R-SMADs. Intriguingly, TGFβ signaling plays a complex role during tumor progress. In normal tissue TGFβ is a tumor suppressor, but in later stages of cancer progression TGFβ can often stimulate a metastatic phenotype (35). Therefore, the dual role of TGFβ signaling in tumorigenesis leads to a dilemma for therapeutic purpose through either stimulating or suppressing TGFβ signaling. Our previous studies showed that SMURF1 regulates cell polarity, protrusive activity, and TGFβ-dependent EMT by targeting small GTPase RHOA for degradation, suggesting that SMURF1 plays a key role at late stages of tumorigenesis through promoting invasiveness and metastasis of tumor cells (11, 12). A potential way for cancer treatment we proposed could be through inhibiting SMURF1 activity, which will block metastasis of tumor cells without impairing TGFβ-induced cell growth inhibition.
Previous study using a virtual screening strategy identified small-molecule compounds that block the interaction of SMURF1 WW1 domain and SMAD1/5 therefore inhibit SMURF1-mediated SMAD1/5 ubiquitination (36). Using the same strategy, Zhang et al. (37) identified small compounds that target Ub-binding region of SMURF1 HECT domain to inhibit SMURF1 ligase activity. However, all these compounds only showed effects on blocking SMURF1-mediated SMAD1/5 degradation but not RHOA degradation; therefore, it is not clear whether they are able to prevent tumor cell migration and EMT. Through our screen, we successfully identified a potent SMURF1 small-molecule inhibitor, which not only blocks SMURF1-mediated SMAD1 degradation, but also blocks SMURF1-mediated RHOA or RHOB degradation. Indeed, this compound dramatically blocked SMURF1-regulated protrusive activity and TGFβ-induced EMT, but did not inhibit TGFβ/SMAD pathway. Thus, the SMURF1 inhibitor we obtained in this screen could be a potential lead compound for cancer therapy to block TGFβ-promoted tumor metastasis.
Experimental procedures
DNA constructs and reagents
Constructs for expression of human SMURF1 (WT and C699A), SMURF2 (WT and ΔC2), RHOA, RHOB, and SMAD1 have been described previously (7, 12, 14, 23, 38). SMURF1-F728A mutant was generated by PCR-based site-directed mutagenesis. pRUF-RHOB was generated by PCR FL from pGL2 vector (Promega), RL from pRL-CMV (Promega), RHOB from pCMV5-RHOB (14), Ub-K48R from pcDNA3-URT construct (gift from Dr. A. Varshavsky), and then sequentially cloned into pCMV5 vector. One triple FLAG tag was added to the N-terminal of RL, and another triple FLAG tag was inserted between Ub and FL (Fig. 1A). The pRUF–R-ek was modified from pRUF-RHOB using Arg-ek (R-ek)-FL to replace FL-RHOB. The arginine (R) residue of R-ek was directly after Ub. The ek was generated by PCR from pGEX-4T-1 vector (GE Healthcare). A FLAG tag was inserted between ek and FL to allow for easy detection of the fusion protein (Fig. 3A). Short hairpin RNAs (shRNAs) against SMURF1 and RHOA were described previously (14). Dual-Glo assay reagent is from Promega. MG-132 is from Sigma-Aldrich.
Cell culture and transfection
HEK293T and MDCK cells were grown in high-glucose Dulbecco's modified Eagle's medium (HyClone) containing 10% (v/v) fetal calf serum (Gibco), 100 units/ml streptomycin and penicillin (EMD Millipore) at 37 °C in a humidified 5% CO2 incubator. HEK293T cells were transiently transfected using the calcium-phosphate method as described previously (39).
URT–Dual-Luciferase assay
HEK293T cells were trypsinized 16 h after transient transfection with the desired constructs, then seeded at a density of 20,000 cells per well in 96-well plates and allowed to attach for 8–10 h. Subsequently, the cells were treated with different compounds from the compound library for 12–16 h incubation before applied to measure activities of firefly luciferase and Renilla luciferase using the Dual-Glo Luciferase Assay System (Promega) according to the company's protocol using a Varioskan Flash Multimode Microplate Reader (Thermo Scientific).
Immunoblotting, in vitro ubiquitination, and immunofluorescence assays
Immunoblotting and in vitro ubiquitination assay were performed as described previously except RHOB was used instead of RHOA (39). Immunofluorescence was performed as described previously (14) and fluorescence imaging was done by a Leica TCS SP8 confocal microscope. The antibodies used were anti-FLAG M2 mAb (Sigma-Aldrich), anti-HA mAb (Santa Cruz Biotechnology, SC7392), anti-ubiquitin P4D1 mAb (Santa Cruz Biotechnology, SC8017), anti-His antibody (Santa Cruz Biotechnology, SC8036), anti-SMURF1 antibody (Abcam, ab57573), anti-RHOB antibody (Santa Cruz Biotechnology, SC180), anti–ZO-1 antibody (EMD Millipore, MABT11), anti-vimentin antibody (BD Biosciences, 550513), anti-SMAD2 mAb (Cell Signaling Technology, 3122S), anti–p-SMAD2 mAb (Cell Signaling Technology, 3108S), anti–β-actin mAb (Santa Cruz Biotechnology, SC47778), and anti-GAPDH mAb (Santa Cruz Biotechnology, SC32233). Phalloidin conjugated to Texas Red (Invitrogen) was used to visualize F-actin.
Statistical analysis
Two-tailed Student's t tests were performed to evaluate statistical significance. p < 0.05 was considered a statistically significant change. *, p < 0.05; **, p < 0.01; ***, p < 0.001. All the values were presented as mean ± S.D. of at least triplicate experiments.
Author contributions
M. T. and T. Zeng data curation; M. T. and T. Zeng formal analysis; M. T. and T. Zeng validation; M. T., T. Zeng, M. L., Q. L., L. L., T. J., and G. L. investigation; M. T. and T. Zeng visualization; T. Zeng, X. D., and H.-R. W. funding acquisition; T. Zeng, Huayue Lin, and H.-R. W. methodology; T. Zeng project administration; S. H., Hong Lin, T. Zhang, Q. K., and X. D. resources; H.-R. W. supervision; H.-R. W. writing-original draft; H.-R. W. writing-review and editing.
Supplementary Material
Acknowledgment
We thank Dr. A. Varshavsky for generously providing reagents.
This work was supported by National Natural Science Foundation of China U1605222, 81472459, and 31070771 (to H.-R. W.); 31601132 (to T. Z.); and U1405223 (to X. D.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supporting data.
- TGFβ
- transforming growth factor β
- EMT
- epithelial-mesenchymal transition
- URT
- ubiquitin-reference technique
- Ubps
- ubiquitin-specific processing proteases
- RL
- Renilla luciferase
- FL
- firefly luciferase
- Ub
- ubiquitin
- MDCK
- Madin-Darby canine kidney
- R-ek
- Arg-ek.
References
- 1. Hershko A., and Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 2. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- 3. Varshavsky A. (2017) The ubiquitin system, autophagy, and regulated protein degradation. Annu. Rev. Biochem. 86, 123–128 10.1146/annurev-biochem-061516-044859 [DOI] [PubMed] [Google Scholar]
- 4. Zou X., Levy-Cohen G., and Blank M. (2015) Molecular functions of NEDD4 E3 ubiquitin ligases in cancer. Biochim. Biophys. Acta 1856, 91–106 10.1016/j.bbcan.2015.06.005 [DOI] [PubMed] [Google Scholar]
- 5. Lin X., Liang M., and Feng X. H. (2000) Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-β signaling. J. Biol. Chem. 275, 36818–36822 10.1074/jbc.C000580200 [DOI] [PubMed] [Google Scholar]
- 6. Zhang Y., Chang C., Gehling D. J., Hemmati-Brivanlou A., and Derynck R. (2001) Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 98, 974–979 10.1073/pnas.98.3.974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu H., Kavsak P., Abdollah S., Wrana J. L., and Thomsen G. H. (1999) A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400, 687–693 10.1038/23293 [DOI] [PubMed] [Google Scholar]
- 8. Ebisawa T., Fukuchi M., Murakami G., Chiba T., Tanaka K., Imamura T., and Miyazono K. (2001) Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 276, 12477–12480 10.1074/jbc.C100008200 [DOI] [PubMed] [Google Scholar]
- 9. Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., and Wrana J. L. (2000) Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell 6, 1365–1375 10.1016/S1097-2765(00)00134-9 [DOI] [PubMed] [Google Scholar]
- 10. Murakami G., Watabe T., Takaoka K., Miyazono K., and Imamura T. (2003) Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol. Biol. Cell 14, 2809–2817 10.1091/mbc.e02-07-0441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ozdamar B., Bose R., Barrios-Rodiles M., Wang H. R., Zhang Y., and Wrana J. L. (2005) Regulation of the polarity protein Par6 by TGFβ receptors controls epithelial cell plasticity. Science 307, 1603–1609 10.1126/science.1105718 [DOI] [PubMed] [Google Scholar]
- 12. Wang H. R., Zhang Y., Ozdamar B., Ogunjimi A. A., Alexandrova E., Thomsen G. H., and Wrana J. L. (2003) Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 302, 1775–1779 10.1126/science.1090772 [DOI] [PubMed] [Google Scholar]
- 13. Zavadil J., and Böttinger E. P. (2005) TGF-β and epithelial-to-mesenchymal transitions. Oncogene 24, 5764–5774 10.1038/sj.onc.1208927 [DOI] [PubMed] [Google Scholar]
- 14. Wang M., Guo L., Wu Q., Zeng T., Lin Q., Qiao Y., Wang Q., Liu M., Zhang X., Ren L., Zhang S., Pei Y., Yin Z., Ding F., and Wang H. R. (2014) ATR/Chk1/Smurf1 pathway determines cell fate after DNA damage by controlling RhoB abundance. Nat. Commun. 5, 4901 10.1038/ncomms5901 [DOI] [PubMed] [Google Scholar]
- 15. Karlsson R., Pedersen E. D., Wang Z., and Brakebusch C. (2009) Rho GTPase function in tumorigenesis. Biochim. Biophys. Acta 1796, 91–98 10.1016/j.bbcan.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 16. Lévy F., Johnsson N., Rümenapf T., and Varshavsky A. (1996) Using ubiquitin to follow the metabolic fate of a protein. Proc. Natl. Acad. Sci. U.S.A. 93, 4907–4912 10.1073/pnas.93.10.4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Varshavsky A. (2005) Ubiquitin fusion technique and related methods. Methods Enzymol. 399, 777–799 10.1016/S0076-6879(05)99051-4 [DOI] [PubMed] [Google Scholar]
- 18. Zhang J. H., Chung T. D., and Oldenburg K. R. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen 4, 67–73 10.1177/108705719900400206 [DOI] [PubMed] [Google Scholar]
- 19. Varshavsky A. (1996) The N-end rule: Functions, mysteries, uses. Proc. Natl. Acad. Sci. U.S.A. 93, 12142–12149 10.1073/pnas.93.22.12142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Varshavsky A. (2005) Regulated protein degradation. Trends Biochem. Sci. 30, 283–286 10.1016/j.tibs.2005.04.005 [DOI] [PubMed] [Google Scholar]
- 21. Bachmair A., and Varshavsky A. (1989) The degradation signal in a short-lived protein. Cell 56, 1019–1032 10.1016/0092-8674(89)90635-1 [DOI] [PubMed] [Google Scholar]
- 22. Tasaki T., Mulder L. C., Iwamatsu A., Lee M. J., Davydov I. V., Varshavsky A., Muesing M., and Kwon Y. T. (2005) A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell Biol. 25, 7120–7136 10.1128/MCB.25.16.7120-7136.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wiesner S., Ogunjimi A. A., Wang H. R., Rotin D., Sicheri F., Wrana J. L., and Forman-Kay J. D. (2007) Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130, 651–662 10.1016/j.cell.2007.06.050 [DOI] [PubMed] [Google Scholar]
- 24. Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., and Wrana J. L. (2005) Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol. Cell 19, 297–308 10.1016/j.molcel.2005.06.028 [DOI] [PubMed] [Google Scholar]
- 25. Salvat C., Wang G., Dastur A., Lyon N., and Huibregtse J. M. (2004) The –4 phenylalanine is required for substrate ubiquitination catalyzed by HECT ubiquitin ligases. J. Biol. Chem. 279, 18935–18943 10.1074/jbc.M312201200 [DOI] [PubMed] [Google Scholar]
- 26. Mani A., and Gelmann E. P. (2005) The ubiquitin-proteasome pathway and its role in cancer. J. Clin. Oncol. 23, 4776–4789 10.1200/JCO.2005.05.081 [DOI] [PubMed] [Google Scholar]
- 27. Roeten M. S. F., Cloos J., and Jansen G. (2018) Positioning of proteasome inhibitors in therapy of solid malignancies. Cancer Chemother. Pharmacol. 81, 227–243 10.1007/s00280-017-3489-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams J., and Kauffman M. (2004) Development of the proteasome inhibitor Velcade (bortezomib). Cancer Invest. 22, 304–311 10.1081/CNV-120030218 [DOI] [PubMed] [Google Scholar]
- 29. Gandolfi S., Laubach J. P., Hideshima T., Chauhan D., Anderson K. C., and Richardson P. G. (2017) The proteasome and proteasome inhibitors in multiple myeloma. Cancer Metastasis Rev. 36, 561–584 10.1007/s10555-017-9707-8 [DOI] [PubMed] [Google Scholar]
- 30. Senft D., Qi J., and Ronai Z. A. (2018) Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 18, 69–88 10.1038/nrc.2017.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang D., Ma L., Wang B., Liu J., and Wei W. (2017) E3 ubiquitin ligases in cancer and implications for therapies. Cancer Metastasis Rev. 36, 683–702 10.1007/s10555-017-9703-z [DOI] [PubMed] [Google Scholar]
- 32. Foote P. K., Krist D. T., and Statsyuk A. V. (2017) High-throughput screening of HECT E3 ubiquitin ligases using UbFluor. Curr. Protoc. Chem. Biol. 9, 174–195 10.1002/cpch.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun Y. (2005) Overview of approaches for screening for ubiquitin ligase inhibitors. Methods Enzymol. 399, 654–663 10.1016/S0076-6879(05)99043-5 [DOI] [PubMed] [Google Scholar]
- 34. David D., Nair S. A., and Pillai M. R. (2013) Smurf E3 ubiquitin ligases at the cross roads of oncogenesis and tumor suppression. Biochim. Biophys. Acta 1835, 119–128 10.1016/j.bbcan.2012.11.003 [DOI] [PubMed] [Google Scholar]
- 35. Massagué J. (2008) TGFβ in cancer. Cell 134, 215–230 10.1016/j.cell.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cao Y., Wang C., Zhang X., Xing G., Lu K., Gu Y., He F., and Zhang L. (2014) Selective small molecule compounds increase BMP-2 responsiveness by inhibiting Smurf1-mediated Smad1/5 degradation. Sci. Rep. 4, 4965 10.1038/srep04965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y., Wang C., Cao Y., Gu Y., and Zhang L. (2017) Selective compounds enhance osteoblastic activity by targeting HECT domain of ubiquitin ligase Smurf1. Oncotarget 8, 50521–50533 10.18632/oncotarget.10648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoodless P. A., Haerry T., Abdollah S., Stapleton M., O'Connor M. B., Attisano L., and Wrana J. L. (1996) MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell 85, 489–500 10.1016/S0092-8674(00)81250-7 [DOI] [PubMed] [Google Scholar]
- 39. Wang H. R., Ogunjimi A. A., Zhang Y., Ozdamar B., Bose R., and Wrana J. L. (2006) Degradation of RhoA by Smurf1 ubiquitin ligase. Methods Enzymol. 406, 437–447 10.1016/S0076-6879(06)06032-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.