Abstract
The transcription elongation and pre-mRNA splicing factor Tat-SF1 associates with the U2 small nuclear ribonucleoprotein (snRNP) of the spliceosome. However, the direct binding partner and underlying interactions mediating the Tat-SF1–U2 snRNP association remain unknown. Here, we identified SF3b1 as a Tat-SF1–interacting subunit of the U2 snRNP. Our 1.1 Å resolution crystal structure revealed that Tat-SF1 contains a U2AF homology motif (UHM) protein–protein interaction module. We demonstrated that Tat-SF1 preferentially and directly binds the SF3b1 subunit compared with other U2AF ligand motif (ULM)-containing splicing factors, and further established that SF3b1 association depends on the integrity of the Tat-SF1 UHM. We next compared the Tat-SF1–binding affinities for each of the five known SF3b1 ULMs and then determined the structures of representative high- and low-affinity SF3b1 ULM complexes with the Tat-SF1 UHM at 1.9 Å and 2.1 Å resolutions, respectively. These structures revealed a canonical UHM–ULM interface, comprising a Tat-SF1 binding pocket for a ULM tryptophan (SF3b1 Trp338) and electrostatic interactions with a basic ULM tail. Importantly, we found that SF3b1 regulates Tat-SF1 levels and that these two factors influence expression of overlapping representative transcripts, consistent with a functional partnership of Tat-SF1 and SF3b1. Altogether, these results define a new molecular interface of the Tat-SF1–U2 snRNP complex for gene regulation.
Keywords: X-ray crystallography, protein–protein interaction, isothermal titration calorimetry (ITC), RNA splicing, protein structure, RNA-binding protein, protein domain, gene regulation
Introduction
Tat stimulatory factor 1 (Tat-SF1)4 was originally identified as a host cofactor that activates Tat-directed HIV-1 transcription (1, 2). Subsequent studies revealed that Tat-SF1 normally stimulates human transcription elongation (3, 4) as a complex with P-TEFb, SPT5, and RAP30 (2, 5–7). The Saccharomyces cerevisiae Tat-SF1 homologue, CUS2, promotes transcription elongation in combination with the RNA unwindases PRP5 and PRP11 (8). Beyond transcription, CUS2 is well-established as an assembly factor for the U2 small nuclear (sn)RNA in the early stages of pre-mRNA splicing (9–13). Like CUS2, human Tat-SF1 influences pre-mRNA splicing (14–18). Reduced Tat-SF1 levels typically promote intron retention rather than exon-skipping or alternative splice sites. Most recently, critical Tat-SF1 functions in pre-mRNA splicing have emerged for embryonic stem cell differentiation (17, 18).
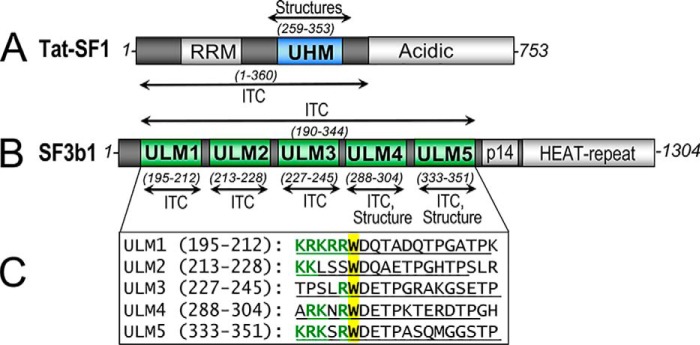
Tat-SF1 co-immunoprecipitates with the U2 small nuclear ribonucleoprotein particle (17–20), which anneals with the pre-mRNA branch point site in the early steps of spliceosome assembly. Yet, the direct interaction partner(s) of Tat-SF1 in the human spliceosome remains unknown. Clues arise from experimental observations coupled with the primary sequences of Tat-SF1 and the U2 snRNP components. Primary sequence analysis suggests that Tat-SF1 contains two modular domains, an RNA recognition motif (RRM) and a U2AF homology motif (UHM) (Fig. 1A). The UHM is a protein-interaction module with specialized features for recognizing short “U2AF ligand motifs” (ULM) of target proteins (21). Among U2 snRNP subunits, SF3b1 singularly comprises ULMs that could mediate Tat-SF1 UHM association; namely, five motifs near the N-terminal region of the SF3b1 protein (Fig. 1, B and C).
Figure 1.
SF3b1 and Tat-SF1 splicing factors contain ULM and UHM motifs. Human Tat-SF1 (A, blue) and SF3b1 (B, green) domain organization are shown. Residue ranges of constructs used for crystallization (structure) or isothermal titration calorimetry (ITC) are given in parentheses. p14, p14a binding site. C, the aligned sequences of five ULM repeats from human SF3b1 are shown in the inset below. The boundaries of synthetic peptides (in parentheses and underlined) terminate at the second TP motif, which for ULM1/ULM2 and ULM2/ULM3 also corresponds to the start of the next ULM. The central tryptophan (yellow) is preceded by characteristic basic (R/K) amino acids (green).
Here, we explore the hypothesis that a UHM–ULM interface couples Tat-SF1 to the SF3b1 subunit of the U2 snRNP. We determined a 1.1 Å resolution structure that details the Tat-SF1 UHM-fold. We show that Tat-SF1 directly and specifically recognizes the ULM region of SF3b1, and determine high resolution structures of Tat-SF1 UHM complexes comparing complexes with high- and low-affinity SF3b1 ULMs. The Tat-SF1–SF3b1 ULM interactions are important for association of the proteins in cell extracts. We demonstrate that SF3b1 regulates Tat-SF1 expression levels, and that these splicing factors both regulate representative transcripts. Altogether these results elucidate a functional UHM–ULM-mediated partnership of Tat-SF1 with the SF3b1 subunit of the U2 snRNP spliceosome particle.
Results
Tat-SF1 contains a U2AF homology motif
To confirm that Tat-SF1 contains a UHM and detail its fold, we determined the crystal structure of this region at 1.1 Å resolution (Fig. 2, Table 1). Analysis using the DALI 3D comparison server (22) shows that the structure most closely matches established members of the UHM family, including CAPERα (PDB ID 4OZ1, Z-score 13.3) and U2AF65 (PDB ID 4Z2X, Z-score 13.2). The core βαββαβ-fold also resembles an RRM (e.g. the U1A prototype, PDB ID 3K0J, Z-score 12.8), which shares a common topology and appears to be an ancestor of the UHM. A 310 helix at the N terminus of the Tat-SF1 UHM is stabilized by crystal packing, yet may have relevance in the context of the full-length protein-fold.
Figure 2.
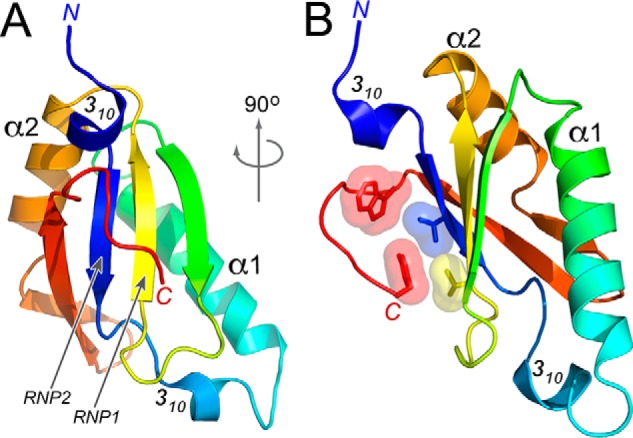
Structure of the Tat-SF1 UHM at 1.13 Å resolution. A, view into the β-sheet. Arrows indicate the β-strands corresponding to RNP1 and RNP2 of the RRM-like fold. B, view rotated 90° about the y axis. Interactions between the C-terminal Tat-SF1 UHM extension and degenerate RNP residues are shown in surface and stick representation. The UHM is colored in a rainbow gradient from blue to red at the N and C termini.
Table 1.
X-ray data collection and refinement statistics
Values in parentheses refer to the highest resolution shell: apo 1.17–1.13 Å; ULM4 1.96–1.89 Å; ULM5 2.17–2.10 Å. For Rfree, 7% of the reflections were randomly-selected then excluded from all stages of refinement.
| Tat-SF1 UHM ligand | Apo | ULM4 | ULM5 |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.83 | 1.13 | 0.98 |
| Resolution range (Å) | 30.20–1.13 | 35.05–1.89 | 37.31–2.10 |
| Space group | P 43 | P 32 21 | P 21 21 21 |
| Unit cell lengths (Å) | 30.20, 30.20, 98.74 | 58.37, 58.37, 97.31 | 46.32, 58.71, 125.96 |
| Unit cell angles (°) | 90 90 90 | 90 90 120 | 90 90 90 |
| Total reflections | 178,395 | 90,661 | 101,323 |
| Unique reflections | 32,586 | 15,748 | 20,337 |
| Multiplicity | 8.7 (1.9) | 5.7 (1.8) | 5.0 (5.0) |
| Completeness (%) | 98.8 (99.6) | 99.3 (94.5) | 98.4 (97.0) |
| Mean I/(I) | 13.6 (2.9) | 24.3 (2.7) | 10.1 (1.8) |
| Rmerge (%) | 5.4 (25.2) | 4.4 (45.2) | 8.7 (65.0) |
| Rp.i.m. (%) | 3.3 (25.2) | 2.0 (24.3) | 4.3 (36.1) |
| CC1/2 | 0.99 (0.84) | 0.99 (0.81) | 0.98 (0.99) |
| Refinement | |||
| Rwork/Rfree (%) | 12.7/16.0 | 17.1/19.8 | 17.5/21.9 |
| Tat-SF1 UHM in ASU | 1 | 1 | 2 |
| SF3b1 ULM in ASU | – | 1 | 1 |
| No. non-hydrogen atoms | |||
| Tat-SF1 UHM | 899 | 799 | 1573 |
| SF3b1 ULM | – | 72 | 39 |
| PEG/Glycerol | 13 | 9 | 13 |
| Ions | 5 | – | – |
| Waters | 114 | 105 | 151 |
| R.m.s.d. bonds (Å) | 0.007 | 0.010 | 0.013 |
| R.m.s.d. angles (°) | 0.83 | 0.97 | 1.16 |
| Ramachandran (%) | 98.9/1.1/0 | 100/0/0 | 97.3/2.7/0 |
| favored/allowed/outlier | |||
| Clashscore (percentile) | 2.76 (93) | 2.93 (99) | 0.94 (100) |
| 〈B-factor〉 (Å2) | 19.3 | 44.8 | 43.8 |
| Tat-SF1 UHM | 16.8 | 41.6 | 42.1 |
| SF3b1 ULM | – | 69.5 | 89.9 |
| PEG/glycerol | 45.2 | 61.0 | 80.1 |
| Ions | 20.7 | – | – |
| Water | 36.4 | 51.3 | 46.5 |
| PDB code | 6N3D | 6N3E | 6N3F |
The canonical RNA interface of the RRM family involves nucleobase stacking with aromatic residues in two ribonucleoprotein consensus motifs (RNP1 and RNP2) of the β-sheet surface. In all prior UHM structures (23–28), the RNP motifs are degenerate and masked by C-terminal extensions of the core-fold. Accordingly, a C-terminal tail of the Tat-SF1 UHM (namely Trp347 and Tyr353) packs against the putative RNP surface (Val314 and Ile267). These key RNP residues have diverged from the typical aromatic residues of RRMs, consistent with specialized roles for the Tat-SF1 UHM in protein rather than RNA interactions.
Tat-SF1 preferentially and directly binds SF3b1 compared with other ULM-containing proteins
We next tested whether the Tat-SF1 UHM binds a ULM-containing partner (Fig. 3). Most ULM-containing proteins are pre-mRNA splicing factors, including U2AF65 (29), SF1 (30), and SF3b1 (31–33). The inner nuclear membrane protein MAN1 (34) and the poly(Q)-expanded protein ATX1 (35) also contain ULMs. Considering the influence of Tat-SF1 on pre-mRNA splicing (14–18) and its co-immunoprecipitation with the SF3b1-containing U2 snRNP (17–20), we focused on testing Tat-SF1 association with SF3b1 compared with other ULM-containing splicing factors.
Figure 3.
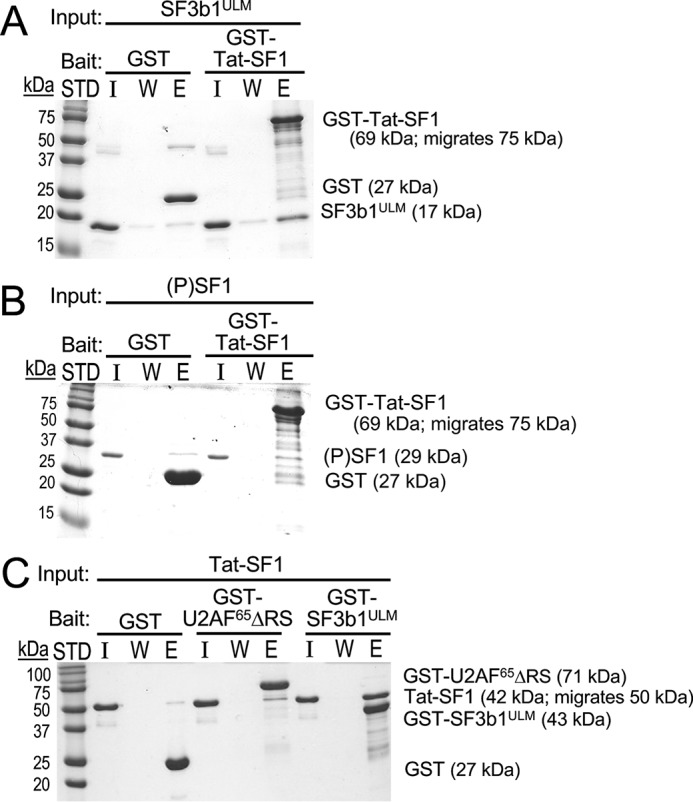
Tat-SF1 directly and preferentially associates with the ULM region of SF3b1. GST-pulldowns of purified recombinant proteins were resolved by SDS-PAGE and stained with Coomassie® Brilliant Blue. Three lanes were loaded for each experiment: I, input (1 μg); W, final wash of bound resin; E, elution. The sizes of molecular weight standards (STD) and subunits are indicated. GST is a control for nonspecific binding. A, GST-fused Tat-SF1 (residues 1–360) retains the SF3b1 ULM region (“SF3b1ULM”, residues 190–344) and B, shows little retention of UHMK1-phosphorylated (P) SF1. Likewise, C, GST-U2AF65ΔRS (residues 85–475) shows little Tat-SF1 retention, whereas GST-SF3b1ULM significantly retains Tat-SF1.
To ensure direct interactions, we used GST pulldown assays of purified recombinant proteins (Fig. 3). Our Tat-SF1 construct comprises the RRM and UHM domains that are sufficient for snRNP co-immunoprecipitation (19) (residues 1–360, Fig. 1A). We compared Tat-SF1 retention by the ULM-containing region of SF3b1 (SF3b1ULM, residues 190–344, Fig. 1B) with nearly full-length U2AF65 that lacks an aggregation prone arginine-serine repeat region (ΔRS), each N-terminally tagged with GST. To avoid overlapping molecular weights, we conversely compared GST–Tat-SF1 retention of SF3b1ULM with SF1. The SF1 protein was phosphorylated (P) in vitro with UHMK1 as described (25, 36), which subtly enhances SF1-U2AF65 association (36, 37) and is the major form of SF1 in proliferating cells (36, 38).
GST–Tat-SF1 retains a substantial fraction of the input SF3b1ULM and has little detectable association with (P)SF1 (Fig. 3, A and B). Although GST–U2AF65ΔRS retains a slight fraction of untagged Tat-SF1, this interaction is likely to be nonspecific considering the nanomolar affinity of the constitutive U2AF65-2AF35 heterodimer (29). Instead, the amount of Tat-SF1 retained by GST–SF3b1ULM is significant (Fig. 3C). This result demonstrates that Tat-SF1 specifically and directly associates with the ULM region of SF3b1.
Tat-SF1 preferentially binds the fifth SF3b1 ULM
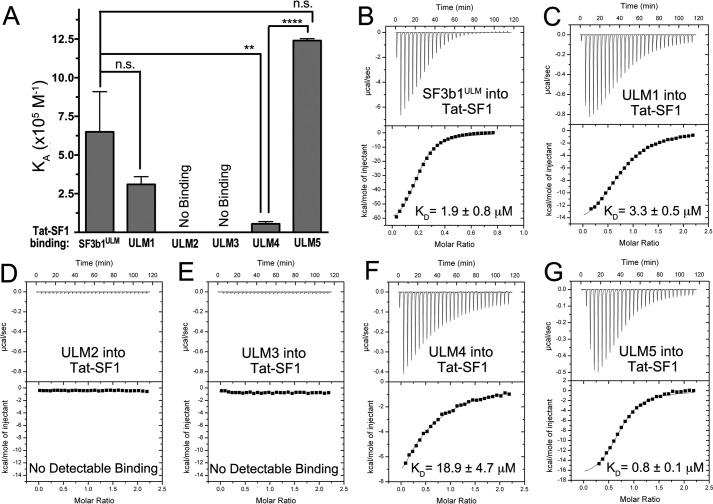
To quantitatively characterize Tat-SF1–SF3b1 interactions, we used isothermal titration calorimetry (ITC) to measure the binding affinities of Tat-SF1 for SF3b1ULM and each of the individual SF3b1 ULMs (Fig. 4, Table 2). Our Tat-SF1 construct for ITC experiments included residues 1–360, which supports snRNP co-immunoprecipitation (19) and avoids precipitation of the UHM fragment during titration. Unlike CAPERα (23) but similar to U2AF65 (31), the Tat-SF1–SF3b1ULM isotherms show no evidence of cooperative association. We chose boundaries of peptides corresponding to the SF3b1 ULMs to minimize overlap among consecutive motifs (Fig. 1B). Tat-SF1 detectably binds three of the five documented SF3b1 ULMs (ULM1, ULM4, and ULM5). The binding affinity of Tat-SF1 for ULM5 is >20-fold greater than for ULM4 and slightly greater than ULM1 (4-fold). No heats were detected during titrations of Tat-SF1 with ULM2 or ULM3. The apparent binding affinity of Tat-SF1 for the entire SF3b1ULM region agrees with the average values of the highest affinity ULM5 and ULM1 sites (Table 2). We conclude that Tat-SF1 is capable of recognizing three local SF3b1 ULM sites, yet has lower affinity for ULM4 compared with either ULM5 or ULM1.
Figure 4.
Tat-SF1 prefers SF3b1 ULM5 by isothermal titration calorimetry analysis of binding. A, apparent equilibrium affinities (KA) of the indicated SF3b1 ULMs binding Tat-SF1 (residues 1–360). SF3b1ULM, SF3b1 residues 190–344. Unpaired, two tailed t-tests with Welch's correction: **, p < 0.005; ****, p < 0.0001. B–G, representative isotherms of the titrations.
Table 2.
Thermodynamics of Tat-SF1 binding SF3b1 ULMs
Average values and standard deviations of three independent titrations.
| Interaction | KD | ΔH | −TΔSa |
|---|---|---|---|
| μm | kcal mol−1 | ||
| Wildtype Tat-SF1titrated with ULM regions | |||
| SF3b1ULM | 1.8 ± 0.9 | −66.1 ± 3.0 | 58.0 ± 3.0 |
| ULM1 | 3.0 ± 0.5 | −11.8 ± 0.3 | 4.1 ± 0.3 |
| ULM2b | No detectable binding | ||
| ULM3b | No detectable binding | ||
| ULM4c | 18.9 ± 4.7 | −34.1 ± 25.3 | 27.5 ± 25.5 |
| ULM5 | 0.8 ± 0.1 | −18.7 ± 1.0 | 10.2 ± 1.2 |
| E286K/D290K mutant Tat-SF1 titrated with ULM5 | |||
| ULM5b | No detectable binding | ||
| E286K/F337A mutant Tat-SF1 titrated with ULM5 | |||
| ULM5b | No detectable binding | ||
a Calculated using −TΔS = ΔG-ΔH, with ΔG = −RT ln (KD−1) and T = 303 K.
b No heats detected during titration of 200 μm ULM into 20 μm Tat-SF1 protein.
c Values are estimates due to low binding affinity.
Structures of Tat-SF1 bound to high and low affinity SF3b1 ULMs
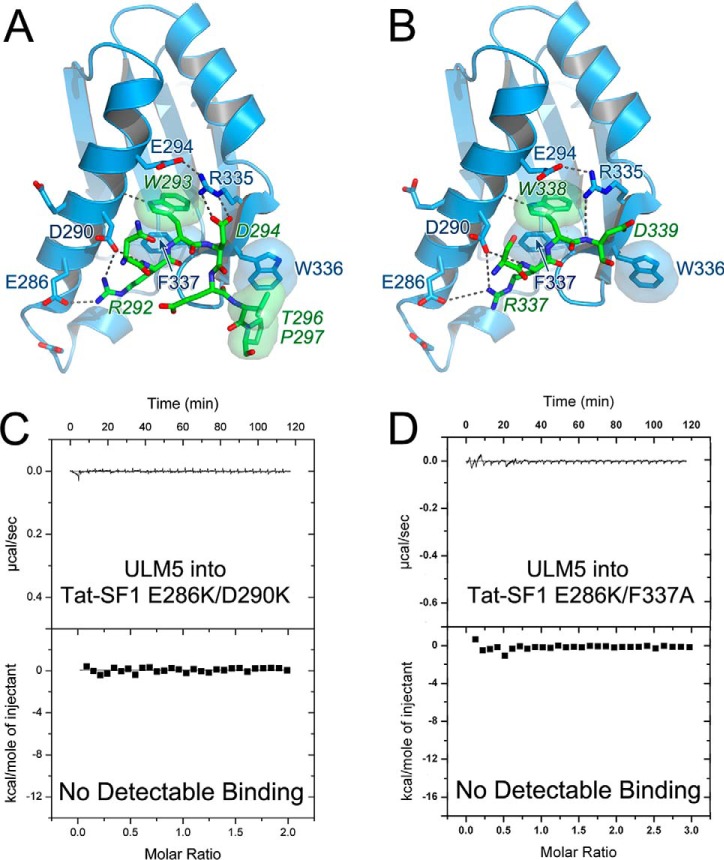
To identify and compare the interactions of the Tat-SF1 UHM with high versus low affinity SF3b1 ULMs, we determined the crystal structures of its complexes with ULM4 and ULM5 at 1.9 Å and 2.1 Å resolutions (Fig. 5, Table 1). Seven residues (residues 291–297) of a 17-residue ULM4 peptide were observed bound to a single Tat-SF1 UHM in the crystallographic asymmetric unit (Fig. 5A). Fewer ULM5 residues are evident (four ordered residues) and one of the two Tat-SF1 UHM copies in the asymmetric unit lacks bound ligand (Fig. 5B). This difference is most likely due to the distinct packing environments of the crystal structures.
Figure 5.
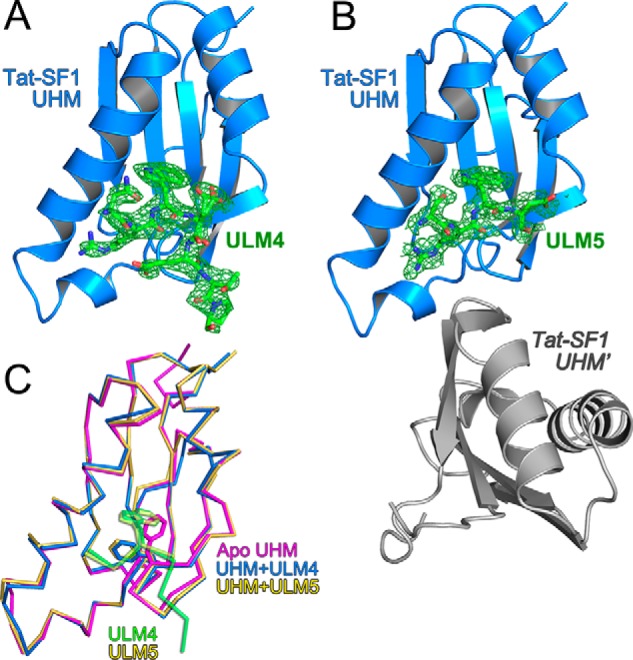
Structures of Tat-SF1 UHM bound to SF3b1 ULM. A, feature-enhanced electron density maps (65) (green, 1.5 σ contour level) for SF3b1 ULM4 or B, ULM5 bound to Tat-SF1 UHM (blue). A second Tat-SF1 UHM copy that lacks bound ULM5 (Tat-SF1 UHM) is colored gray. View is rotated 180° relative to the left view of Fig. 2. C, superposed backbone traces of unliganded (magenta) Tat-SF1 UHM with ULM4-bound (blue) or ULM5-bound (yellow) UHM structures. The SF3b1 ULM4 (green) and ULM5 (yellow) are overlaid for reference. Stick diagrams show SF3b1 Trp293/Trp338 and compare Tat-SF1 Phe337 positions.
The core-fold of the apo-Tat-SF1 UHM is similar to its SF3b1 ULM-bound complexes (r.m.s.d. n 0.35–0.45 Å between 86 matching Cα atoms) (Fig. 5C). However, the empty ULM-binding site of the apo-Tat-SF1 structure is filled by a phenylalanine (Phe337) donated by the so-called “RXF” loop of the UHM (21). In the Tat-SF1–ULM complexes, this Phe337 side chain rearranges to enclose the consensus ULM tryptophan.
Tat-SF1 displays characteristic features of UHM-type interactions with the SF3b1 ULMs (Fig. 6, A and B). The ULM tryptophan (ULM4 Trp293 or ULM5 Trp338) inserts between the α-helices of the Tat-SF1 UHM. There, the ULM tryptophan is sandwiched between a salt bridge (Tat-SF1 Glu294–Arg335) and a T-shaped aromatic interaction (39) with the Phe337 side chain. The Tat-SF1-bound ULM4 and ULM5 peptides both adopt a U-shape, which is shared by the two other known structures of SF3b1 ULMs bound to UHMs (ULM5 bound to CAPERα (23) or SPF45 (26)). In contrast, the SF3b1 ULM trajectories differ from that of the SF1 ULM, which binds to the U2AF65 UHM in a distinct, linear conformation that is facilitated by a C-terminal coiled-coil extension of the SF1 ULM (24, 25).
Figure 6.
Tat-SF1 UHM-SF3b1 ULM interactions. A, ULM4 and B, ULM5. Hydrogen bonds are indicated by dashed lines. Key interacting residues are labeled; SF3b1 residues are italicized. Packing interactions are indicated by semitransparent surfaces. Isothermal titration calorimetry confirms structure-guided mutations of the Tat-SF1 interface abolish detectable binding to SF3b1 ULMs. Representative isotherms for: C, E286K/D290K or D, E286K/F337A mutant Tat-SF1 (residues 1–360) titrated with SF3b1 ULM5 are shown.
At the C terminus of SF3b1 ULM4, a threonine-proline (TP) motif packs against an exposed Tat-SF1 tryptophan (Trp336) at the X position of the UHM RXF loop (Fig. 6A). Although the surrounding crystal contacts prevent an analogous interaction of Tat-SF1 Trp336 with the ULM5 TP motif, the SF3b1 ULM5 and ULM4 share identical C-terminal sequences (Fig. 1C). As such, ULM5 is likely to contact Tat-SF1 Trp336 in a solution environment. The SF3b1ULM region contains >20 TP motifs near the ULMs. These SF3b1 sites are highly phosphorylated (phospho.ELM (40)), which could provide means to regulate SF3b1 association with UHM-containing proteins.
Positively-charged residues at the N terminus of each ULM orient toward Glu294, Glu289, Asp290, and Glu286 in an acidic α-helix of the Tat-SF1 UHM (Fig. 6, A and B). Although several N-terminal ULM residues are disordered in the electron density, an ion pair between a conserved ULM arginine (ULM4 Arg292 or ULM5 Arg337) and Tat-SF1 Glu286 is well-defined. Considering that the Tat-SF1–interacting residues of the SF3b1 ULM3, ULM4, and ULM5 C termini are identical, the different number of basic residues in the N-terminal tails are likely to dominate the range of binding affinities. For example, the different Tat-SF1 binding energies of ULM4 versus ULM5 (ΔΔG 1.9 kcal mol−1) are consistent with the loss of an ion pair between the acidic Tat-SF1 α-helix and SF3b1 ULM5 Lys333 (41). A prior study showed that basic residues substituted in the normally acidic U2AF65 α-helix eliminated detectable binding to the SF1 ULM in pulldown assays (30). To investigate the importance of corresponding charge–charge interactions for the Tat-SF1–ULM complex, we introduced analogous Tat-SF1 E286K and D290K mutations, and found that these substitutions abolished detectable binding to SF3b1 ULM5 by ITC (Fig. 6C).
Integrity of the Tat-SF1 UHM is critical for co-immunoprecipitation with SF3b1
We evaluated the contribution of UHM–ULM interactions to the association of full-length Tat-SF1 and SF3b1 in human cell extracts (Fig. 7). Plasmids expressing Tat-SF1 variants or N-terminally FLAG-tagged SF3b1 were co-transfected into HEK293T cells. The associated protein complexes were immunoprecipitated using Tat-SF1–specific agarose beads. We found that wildtype (WT) Tat-SF1 retained significant levels of FLAG-SF3b1 in lysates from transfected but not mock-transfected cells. This result confirms the existence of a Tat-SF1–SF3b1 complex, in agreement with prior observations in distinct contexts (17–20). We next compared FLAG-SF3b1 co-immunoprecipitates of a Tat-SF1 variant with E286K/F337A mutations, which prevent detectable binding of Tat-SF1 to SF3b1 ULM5 by ITC (Fig. 6D). The mutant Tat-SF1 variant lacked detectable retention of FLAG-SF3b1, which supports the importance of an intact UHM for association of Tat-SF1 with SF3b1 in human cell extracts.
Figure 7.
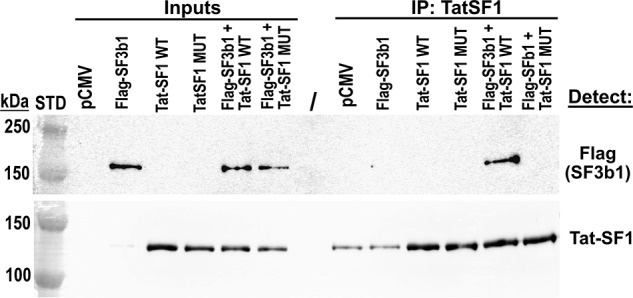
Tat-SF1 UHM mediates association with SF3b1 in cell extracts. The amount of transiently expressed FLAG-SF3b1 (143 kDa) retained by immobilized Tat-SF1 proteins (86 kDa; migrates 135 kDa) was detected. Cell lysates (0.5%) were loaded as input controls (left lanes). The sizes of Bio-Rad Precision Plus Dual Color pre-stained markers are indicated (STD). Tat-SF1 (WT) interacts with SF3b1, whereas E286K/F337A mutations (MUT) of the UHM abolished detectable Tat-SF1 association with SF3b1.
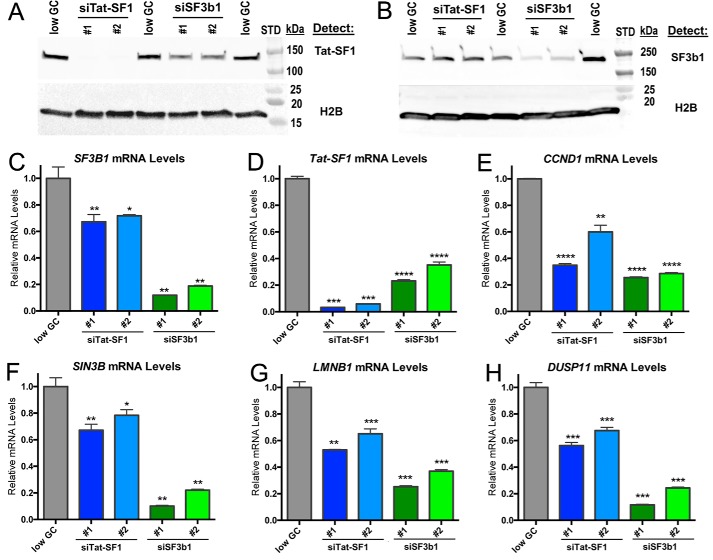
Tat-SF1 expression is sensitive to SF3b1 levels
The association of Tat-SF1 with SF3b1 suggested that its expression level would be mutually regulated to ensure formation of a functional macromolecular complex. To test this hypothesis, we used siRNAs to reduce Tat-SF1 or SF3b1 expression levels in HEK293T cells, and evaluated changes in expression of the other factor by RT-qPCR and immunoblotting (Fig. 8, A–D). Tat-SF1 knockdown had little effect on SF3b1 levels, consistent with prior observations (18). Nevertheless, SF3b1 knockdown significantly reduced Tat-SF1 expression levels. We conclude that whereas SF3b1 serves Tat-SF1-independent functions as a major spliceosome subunit (42), a feedback mechanism modulates Tat-SF1 levels in response to SF3b1.
Figure 8.
Tat-SF1 expression levels decline following SF3b1 depletion and in turn decrease expression of similar representative transcripts as SF3b1. A and B, immunoblots demonstrating Tat-SF1 or SF3b1 knockdown following transfection of HEK293T cells with the indicated StealthTM siRNAs or a low G/C control siRNA (“low GC”). Whole cell lysates were analyzed by SDS-PAGE and immunoblotted for either: A, Tat-SF1 or B, SF3b1. H2B is encoded by an intronless gene and hence an appropriate loading control. C–H, relative levels of TAT-SF1, SF3B1, or the indicated transcripts were determined by RT-qPCR of total RNA isolated from HEK 293T cells following transfection with the indicated siRNAs. Amplification of the LMNB1 exon 5/6 junction is shown. The LMNB1 exon 7/8 junction gave similar results as shown in Fig. S1. Primer locations and sequences are given in Table S1. Unpaired, two-tailed t tests with Welch's correction: *, p < 0.05; **, p < 0.005; ***, p < 0.0005; ****, p < 0.0001.
Tat-SF1 and SF3b1 regulate overlapping gene transcripts
To further explore the functional outcomes of a Tat-SF1 partnership with SF3b1, we evaluated whether SF3b1 regulates expression of Tat-SF1 responsive genes. We focused on three representative transcripts (CCND1, SIN3B, and LMNB1) that have documented expression changes in response to Tat-SF1 levels (16). As expected, levels of these transcripts decreased following siRNA-mediated reduction of Tat-SF1 (Fig. 8, E–G). Notably, SF3b1 knockdown also decreased the levels of all three Tat-SF1–sensitive transcripts (by 3–5-fold). These effects are likely to be direct, because SF3b1 expression is only subtly affected by Tat-SF1 levels (Fig. 8, B and C) (18). Despite numerous studies (e.g. Refs. 14, 16, and 18), to our knowledge neither we nor others have succeeded in rescuing gene expression by reintroducing Tat-SF1, which suggests that Tat-SF1 activities are delicately balanced by localization and/or post-translational modifications. To reinforce the functional convergence of Tat-SF1 and SF3b1, we compared an additional transcript (DUSP11) that is known to be sensitive to an SF3b1-targeted small molecule, spliceostatin-A (43). Both SF3b1 and Tat-SF1 knockdown decreased levels of the DUSP11 transcript (Fig. 8H).
As a preliminary check for alternative splicing, we compared the sensitivity of a distinct splice site from LMNB1 to Tat-SF1 and SF3b1 knockdown (Fig. S1). We observed similar changes in transcript levels when probing either LMNB1 splice site. Consistently, Tat-SF1 reduction typically leads to intron retention rather than alternatively splicing (17, 18), and most intron-retained transcripts would be removed by quality control pathways (44). As such, our experiments lack discrimination between altered transcription and splicing. Regardless, the significant impact of SF3b1 knockdown for all Tat-SF1–sensitive transcripts supports altered pre-mRNA splicing, considering the well-established roles of SF3b1 in the spliceosome (42). Altogether, our results establish convergent roles for Tat-SF1 and SF3b1 in human gene expression.
Discussion
Here, we identify direct and specific coupling of the Tat-SF1 transcriptional and pre-mRNA splicing factor with the SF3b1 subunit of the U2 snRNP. Underscoring the importance of the Tat-SF1–SF3b1 complex, associated snRNPs are required for Tat-SF1 to stimulate transcription (19). Following clues from structural homology, we pinpoint a critical UHM–ULM interface for Tat-SF1–SF3b1 association in cell extracts or purified proteins. Our high resolution structures reveal that the Tat-SF1 UHM exhibits a characteristic RRM-like topology with an extended C terminus that masks degenerate ribonucleoprotein motifs. The Tat-SF1 UHM encloses a central tryptophan of the SF3b1 ULM between two α-helices, which is typically essential for UHM–ULM binding (e.g. Refs. 29–31 and 36). Tat-SF1 and SF3b1 association further depends on electrostatic interactions between an acidic UHM α-helix and a basic ULM tail.
The Tat-SF1 UHM–SF3b1 ULM structures expand panoptic views of this protein interaction motif (21). A recurrent theme of tryptophan-mediated and charge–charge interactions underscores the question of how cognate UHM–ULM pairs are determined. Although we show that the Tat-SF1 UHM is required for detectable SF3b1 association, it remains likely that auxiliary interfaces mediated by distinct protein or RNA regions could enhance the binding specificity and affinity of the Tat-SF1–SF3b1 complex in a physiological context. The binding affinities of the Tat-SF1–SF3b1 protein domains are micromolar (Table 2), consistent with other SF3b1 ULM–UHM protein complexes (21). Such relatively weak interactions could be important for dynamic associations (45), yet do not rule out enhancement by transient interfaces during spliceosome assembly. For example, the UHM-mediated association of Tat-SF1 with the SF3b1 ULMs may synergize with the U2 snRNA, which Tat-SF1 and SF3b1 bind in the context of the intact U2 snRNP. Indeed, an RRM mutation of the yeast homologue Cus2 prevents its U2 snRNA association and rescue of U2 snRNA misfolding (9, 19).
Macromolecular complexes such as spliceosome particles often employ feedback mechanisms to coordinate subunit expression (e.g. Refs. 46–48). Our finding that Tat-SF1 expression decreases following SF3b1 depletion (Fig. 8, A and D) supports functional coordination of Tat-SF1 and SF3b1. Conversely, SF3b1 levels are relatively insensitive to reduced Tat-SF1, which agrees with SF3b1 ULM associations with a variety of alternative splicing factors (23, 26, 27, 31). The SF3b1-dependent Tat-SF1 expression is analogous to a previously characterized precedent, the U2AF heterodimer (46). U2AF65 knockdown reduces U2AF35 protein levels, whereas reduced U2AF35 levels are insensitive to U2AF65. Functionally, in vitro splicing can be reconstituted by the U2AF65 subunit alone (49–51), and moreover, Schizosaccharomyces pombe introns that are affected by U2AF65 depletion are affected by U2AF35 depletion (52). Accordingly, we here find that the levels of representative downstream transcripts respond to both Tat-SF1 and SF3b1 reductions. These effects are likely to be specific, because SF3b1 (43, 53, 54) and Tat-SF1 (16, 17, 55) knockdowns alter expression and splicing of only a subset of genes. We suggest that a feedback mechanism promotes the formation of Tat-SF1 complexes with SF3b1.
Altogether, we have identified SF3b1 as a directly interacting partner of Tat-SF1, and detailed a requisite ULM–UHM interface of this functional complex at atomic resolution. Beyond our study of Tat-SF1, the SF3b1 ULM-binding preferences have been comprehensively determined for one other UHM-containing protein, U2AF65 (31). Both Tat-SF1 and U2AF65 bind the SF3b1 ULM region with similar apparent affinities (approximate KD values 2 μm/3 μm for Tat-SF1/U2AF65 binding to SF3b1ULM), and both show a subtle binding preference for ULM5 among the SF3b1 ULMs. Immunoaffinity purification followed by MS has shown that Tat-SF1 associates with U2 snRNP particles, but is absent from assembled, “A”-stage spliceosomes (20). In contrast, U2AF65 is an essential splicing factor that stabilizes U2 snRNP association with pre-mRNA splice sites (56, 57), and is detected only in the A- and later “B”-stages of spliceosome assembly (20). The similar SF3b1 binding preferences of Tat-SF1 and U2AF65, coupled with sequential Tat-SF1 and U2AF65 functions during transcription and spliceosome assembly, call for future studies to explore whether Tat-SF1 serves as a checkpoint factor for handoff of functional U2 snRNP to U2AF65 during spliceosome assembly.
Experimental procedures
Recombinant protein expression and purification
Human Tat-SF1 (residues 1–360 of NCBI Refseq NP_055315), Tat-SF1 UHM (residues 260–353), SF3b1ULM (residues 190–344 of NCBI Refseq NP_036565), U2AF65 (residues 85–475 of NCBI Refseq NP_009210), or SF1 (residues 1–255 of NCBI RefSeq NP_004621) were expressed in Escherichia coli as GST fusions using either pGEX-6p or an in-house pGEX-6p variant encoding a tobacco etch virus protease site. The splicing factor proteins were purified by GST affinity followed by anion exchange or heparin affinity chromatography. For crystallization experiments, the Tat-SF1 UHM was separated from aggregates by size exclusion chromatography in 50 mm NaCl, 15 mm HEPES, pH 7.4, 0.2 mm tris(2-carboxyethyl)phosphine. Purified SF1 proteins were phosphorylated by UHMK1 as described (25) and then separated from UHMK1 by size exclusion chromatography. Synthetic peptides corresponding to SF3b1 ULMs (with residue ranges shown in Fig. 1) were purchased at 98% purity (Biomatik or Bio-Synthesis, Inc.).
GST pulldown assays
The GST fusion “bait” protein (100 μg) was incubated for 1 h on pre-packaged GSH-agarose spin columns with 0.2-ml bed volume (Thermo Scientific PierceTM) that were equilibrated three times with binding buffer (100 mm NaCl, 25 mm HEPES, pH 7.4, 3% glycerol, 1 mm EDTA, 10 mm DTT), and supplemented with protease inhibitor. The immobilized GST fusions were incubated with purified protein (500 μg, 1 mg/ml) in binding buffer for 45 min at 4 °C. The resin was then washed five times with binding buffer. Retained proteins were eluted by heating in Laemmli loading buffer supplemented with 10 mm GSH, and analyzed by SDS-PAGE with Coomassie® Blue staining.
Isothermal titration calorimetry
Protein samples were dialyzed into 25 mm HEPES, pH 7.4, 50 mm NaCl, 0.2 mm tris(2-carboxyethyl)phosphine, and peptides were diluted into >200-fold greater volume of dialysis buffer, prior to ITC. The thermodynamics of Tat-SF1 titrated with SF3b1ULM or ULM peptides were measured using a VP-ITC and fit using Origin 7.0 (MicroCal, LLC). Control titrations of ULM peptides into buffer were linear and showed negligible heats of dilution, and were subtracted to correct the relatively low affinity isotherms of ULM1 and ULM4. Other isotherms were corrected for the heats of dilution using an average of three data points at the saturated plateau of the titration. The apparent stoichiometries are consistent with 1:1 binding except SF3b1ULM, which is underdetermined and appears to bind between two and five Tat-SF1 molecules considering χ2 values of different fits.
Crystallization and structure determination
For crystallization of the Tat-SF1 UHM–SF3b155 ULM4 and ULM5 complexes, 0.1 mm Tat-SF1 UHM was mixed with a 1.2-fold molar excess of SF3b155 ULM and concentrated in a 2-kDa MWCO Vivaspin® Hydrosart (Sartorius) concentrator. The Tat-SF1 UHM and its ULM complexes were crystallized by vapor diffusion using the JCSG+TM screen (Molecular Dimensions) at 277 K. The apo-Tat-SF1 UHM crystallization conditions were optimized in hanging drop format to 0.4 m MgCl2, 0.1 m Tris, pH 7.5, 15% PEG 8000. The respective crystallization solutions of ULM4 and ULM5 complexes contained 0.2 m magnesium formate, 20% PEG 3350, and 0.1 m Tris, pH 8.5, 15% PEG 8000. The crystals were sequentially transferred to 20% glycerol in synthetic mother liquor then flash-cooled in liquid nitrogen. Crystallographic data sets were collected remotely at Stanford Synchrotron Radiation Lightsource beamlines 12-2, 7-1, and 9-2 (58). Data were processed using the SSRL AUTOXDS script (A. Gonzalez and Y. Tsai) implementation of XDS (59) and CCP4 packages (60).
The apo-Tat-SF1 UHM structure was determined with an ensemble search model comprising the U2AF65, SPF45, and CAPERα UHM structures generated using Ensembler (61). A molecular replacement solution using Phaser (61) via the Phenix interface (62) (TF Z-score 5.8, LLG 304) was rebuilt in Coot (63), and refined using Phenix (62). Alternative strategies to determine the apo-Tat-SF1 UHM structure were unsuccessful, including direct methods at 1.13 Å resolution and molecular replacement searches with either individual UHM crystal structures stripped of ligand coordinates (U2AF65, PDB ID 4FXW; SPF45, PDB ID 2PEH; CAPERα, PDB ID 4OZ0) or an unpublished apo-NMR structure (PDB ID 2DIT). The ULM-bound structures were determined by molecular replacement starting from the apo-Tat-SF1 UHM structure. Crystallographic data and refinement statistics are given in Table 1.
Cell culture and transfections
HEK 293T cells (ATCC® CRL-3216TM) were cultured in GibcoTM minimum essential media supplemented with 10% fetal calf serum (Atlanta Biologicals) and 2 mm l-glutamine and maintained at 37 °C in a humidified atmosphere containing 5% CO2. Cells were transfected using JetPrime® (Polyplus-transfection®) as instructed by the manufacturer.
Co-immunoprecipitation
HEK 293T cells were transfected with plasmids encoding human Tat-SF1 (modified from Origene #SC114990), N-terminally FLAG-tagged SF3b1 (modified from a gift of Benjamin Ebert (64)), or empty control vector (pCMV). After 24 h, cells were harvested and lysed in a IP buffer (50 mm Tris, pH 8.0, 75 mm NaCl, 5% glycerol, 10 mm CaCl2, cOmpleteTM EDTA-free protease inhibitor (Sigma), 0.5 mm DTT) + 0.5% Triton X-100. Lysates were clarified by centrifugation and a fraction of each sample was set aside as input controls. The remaining lysates were diluted 1:5 with IP buffer + 0.1% Triton X-100, then incubated for 1 h on a rocking platform at 4 °C protein G-Sepharose (GE Healthcare number 17061801) pre-bound to Tat-SF1-specific antibody (Santa Cruz Biotechnology number sc-514351). Beads were collected by centrifugation and washed extensively with IP buffer. Final washes included RNase A (Sigma).
Immunoblotting
The immunoprecipitated complexes on beads were resuspended in SDS-PAGE loading buffer. For total protein analysis, cells were lysed in a buffer containing 50 mm Tris, pH 8.0, 10 mm EDTA, 1% SDS, 1 mm DTT, and protease inhibitors. Proteins were separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and immunoblotted with antibodies specific for Tat-SF1 (Santa Cruz Biotechnology number sc-514351), FLAG (Cell Signaling Technology number 2368), SF3b1 (MBL number D221-3), or Histone H2B (Cell Signaling Technology number 12354). Secondary antibodies included anti-mouse IgG horseradish peroxidase or anti-rabbit IgG horseradish peroxidase (GE Healthcare numbers NA931 or NA934). Chemiluminescent signal from SuperSignal WestPico chemiluminescent substrate (ThermoFisher Scientific) was detected on a ChemidocTM Touch Imaging System (Bio-Rad).
Quantitative-reverse transcriptase real time PCR
Cells were harvested 3 days following transfection with StealthTM siRNA duplexes (ThermoFisher Scientific) specific for HTATSF1 (assay IDs: HSS120650, HSS120651), SF3B1 (assay IDs: HSS146413, HSS146415), or a negative control (12935200). Total RNA was isolated and DNase I-treated using the RNeasy Kit (Qiagen). The cDNAs were synthesized by Moloney murine leukemia virus RT with random primers (Invitrogen), and analyzed by quantitative PCR with SYBRTM Green in triplicate on a Bio-Rad CFX thermal cycler. The RT-qPCR products were quantified by the relative standard curve method and normalized to the level of 18S rRNA. Results in Figs. 7 and 8 represent at least three biological replicates. Primers are listed in Table S1.
Author contributions
S. L. and C. L. K. conceptualization; S. L., J. L. J., M. P., and C. L. K. validation; S. L., J. R. L., S. W. H., D. M., J. L. J., M. P., and C. L. K. investigation; S. L. and C. L. K. writing-original draft; S. L., D. M., J. L. J., M. P., and C. L. K. writing-review and editing; J. L. J., M. P., and C. L. K. methodology; C. L. K. supervision; C. L. K. funding acquisition.
Supplementary Material
Acknowledgments
We thank Manuel Ares (University of California, Santa Cruz) and Aaron Hoskins (University of Wisconsin, Madison, WI) for stimulating discussions and sharing unpublished data. We appreciate the skilled technical assistance of Nazish Jeffery and Eliezra Glasser. SSRL is supported by the United States Dept. of Energy under Contract No. DE-AC02-76SF00515 and NIH Grant P41 GM103393.
This work was supported by National Institutes of Health (NIH) Grants R01 GM070503 and GM117005. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Fig. S1.
The atomic coordinates and structure factors (codes 6N3D, 6N3E, and 6N3F) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Tat-SF1
- Tat stimulatory factor 1
- GST
- glutathione S-transferase
- ITC
- isothermal titration calorimetry
- RT-qPCR
- quantitative real time reverse transcriptase PCR
- snRNP
- small nuclear ribonucleoprotein particle
- UHM
- U2AF homology motif
- ULM
- U2AF ligand motif
- RRM
- RNA recognition motif
- IP
- immunoprecipitation
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation.
References
- 1. Zhou Q., and Sharp P. A. (1996) Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science 274, 605–610 10.1126/science.274.5287.605 [DOI] [PubMed] [Google Scholar]
- 2. Parada C. A., and Roeder R. G. (1999) A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 18, 3688–3701 10.1093/emboj/18.13.3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li X. Y., and Green M. R. (1998) The HIV-1 Tat cellular coactivator Tat-SF1 is a general transcription elongation factor. Genes Dev. 12, 2992–2996 10.1101/gad.12.19.2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Y., Yamaguchi Y., Tsugeno Y., Yamamoto J., Yamada T., Nakamura M., Hisatake K., and Handa H. (2009) DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 23, 2765–2777 10.1101/gad.1834709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou Q., Chen D., Pierstorff E., and Luo K. (1998) Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 17, 3681–3691 10.1093/emboj/17.13.3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim J. B., Yamaguchi Y., Wada T., Handa H., and Sharp P. A. (1999) Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol. Cell. Biol. 19, 5960–5968 10.1128/MCB.19.9.5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fong Y. W., and Zhou Q. (2000) Relief of two built-in autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 20, 5897–5907 10.1128/MCB.20.16.5897-5907.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chathoth K. T., Barrass J. D., Webb S., and Beggs J. D. (2014) A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol. Cell 53, 779–790 10.1016/j.molcel.2014.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yan D., Perriman R., Igel H., Howe K. J., Neville M., and Ares M. (1998) CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol. Cell. Biol. 18, 5000–5009 10.1128/MCB.18.9.5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perriman R., and Ares M. Jr. (2000) ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 14, 97–107 [PMC free article] [PubMed] [Google Scholar]
- 11. Perriman R., Barta I., Voeltz G. K., Abelson J., and Ares M. Jr. (2003) ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl. Acad. Sci. U.S.A. 100, 13857–13862 10.1073/pnas.2036312100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perriman R. J., and Ares M. Jr. (2007) Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 21, 811–820 10.1101/gad.1524307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rodgers M. L., Tretbar U. S., Dehaven A., Alwan A. A., Luo G., Mast H. M., and Hoskins A. A. (2016) Conformational dynamics of stem II of the U2 snRNA. RNA 22, 225–236 10.1261/rna.052233.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miller H. B., Saunders K. O., Tomaras G. D., and Garcia-Blanco M. A. (2009) Tat-SF1 is not required for Tat transactivation but does regulate the relative levels of unspliced and spliced HIV-1 RNAs. PLoS ONE 4, e5710 10.1371/journal.pone.0005710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jablonski J. A., Amelio A. L., Giacca M., and Caputi M. (2010) The transcriptional transactivator Tat selectively regulates viral splicing. Nucleic Acids Res. 38, 1249–1260 10.1093/nar/gkp1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller H. B., Robinson T. J., Gordân R., Hartemink A. J., and Garcia-Blanco M. A. (2011) Identification of Tat-SF1 cellular targets by exon array analysis reveals dual roles in transcription and splicing. RNA 17, 665–674 10.1261/rna.2462011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abramczuk M. K., Burkard T. R., Rolland V., Steinmann V., Duchek P., Jiang Y., Wissel S., Reichert H., and Knoblich J. A. (2017) The splicing co-factor barricade/Tat-SF1 is required for cell cycle and lineage progression in Drosophila neural stem cells. Development 144, 3932–3945 10.1242/dev.152199 [DOI] [PubMed] [Google Scholar]
- 18. Corsini N. S., Peer A. M., Moeseneder P., Roiuk M., Burkard T. R., Theussl H. C., Moll I., and Knoblich J. A. (2018) Coordinated control of mRNA and rRNA processing controls embryonic stem cell pluripotency and differentiation. Cell Stem Cell 22, 543–558 e512 10.1016/j.stem.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 19. Fong Y. W., and Zhou Q. (2001) Stimulatory effect of splicing factors on transcriptional elongation. Nature 414, 929–933 10.1038/414929a [DOI] [PubMed] [Google Scholar]
- 20. Agafonov D. E., Deckert J., Wolf E., Odenwälder P., Bessonov S., Will C. L., Urlaub H., and Lührmann R. (2011) Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol. Cell. Biol. 31, 2667–2682 10.1128/MCB.05266-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Loerch S., and Kielkopf C. L. (2016) Unmasking the U2AF homology motif family: a bona fide protein-protein interaction motif in disguise. RNA 22, 1795–1807 10.1261/rna.057950.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holm L., and Laakso L. M. (2016) Dali server update. Nucleic Acids Res. 44, W351–355 10.1093/nar/gkw357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loerch S., Maucuer A., Manceau V., Green M. R., and Kielkopf C. L. (2014) Cancer-relevant splicing factor CAPERα engages the essential splicing factor SF3b155 in a specific ternary complex. J. Biol. Chem. 289, 17325–17337 10.1074/jbc.M114.558825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Y., Madl T., Bagdiul I., Kern T., Kang H. S., Zou P., Mäusbacher N., Sieber S. A., Krvmer A., and Sattler M. (2013) Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3′-splice site recognition. Nucleic Acids Res. 41, 1343–1354 10.1093/nar/gks1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang W., Maucuer A., Gupta A., Manceau V., Thickman K. R., Bauer W. J., Kennedy S. D., Wedekind J. E., Green M. R., and Kielkopf C. L. (2013) Structure of phosphorylated SF1 bound to U2AF65 in an essential splicing factor complex. Structure 21, 197–208 10.1016/j.str.2012.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Corsini L., Bonna S., Basquin J., Hothorn M., Scheffzek K., Valcárcel J., and Sattler M. (2007) U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat. Struct. Mol. Biol. 14, 620–629 10.1038/nsmb1260 [DOI] [PubMed] [Google Scholar]
- 27. Corsini L., Hothorn M., Stier G., Rybin V., Scheffzek K., Gibson T. J., and Sattler M. (2009) Dimerization and protein binding specificity of the U2AF homology motif of the splicing factor Puf60. J. Biol. Chem. 284, 630–639 10.1074/jbc.M805395200 [DOI] [PubMed] [Google Scholar]
- 28. Yoshida H., Park S. Y., Oda T., Akiyoshi T., Sato M., Shirouzu M., Tsuda K., Kuwasako K., Unzai S., Muto Y., Urano T., and Obayashi E. (2015) A novel 3′ splice site recognition by the two zinc fingers in the U2AF small subunit. Genes Dev. 29, 1649–1660 10.1101/gad.267104.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kielkopf C. L., Rodionova N. A., Green M. R., and Burley S. K. (2001) A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106, 595–605 10.1016/S0092-8674(01)00480-9 [DOI] [PubMed] [Google Scholar]
- 30. Selenko P., Gregorovic G., Sprangers R., Stier G., Rhani Z., Krämer A., and Sattler M. (2003) Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell 11, 965–976 10.1016/S1097-2765(03)00115-1 [DOI] [PubMed] [Google Scholar]
- 31. Thickman K. R., Swenson M. C., Kabogo J. M., Gryczynski Z., and Kielkopf C. L. (2006) Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein-protein interactions among pre-mRNA splicing factors. J. Mol. Biol. 356, 664–683 10.1016/j.jmb.2005.11.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cass D. M., and Berglund J. A. (2006) The SF3b155 N-terminal domain is a scaffold important for splicing. Biochemistry 45, 10092–10101 10.1021/bi060429o [DOI] [PubMed] [Google Scholar]
- 33. Spadaccini R., Reidt U., Dybkov O., Will C., Frank R., Stier G., Corsini L., Wahl M. C., Lührmann R., and Sattler M. (2006) Biochemical and NMR analyses of an SF3b155–p14-U2AF–RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA 12, 410–425 10.1261/rna.2271406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kondé E., Bourgeois B., Tellier-Lebegue C., Wu W., Pérez J., Caputo S., Attanda W., Gasparini S., Charbonnier J. B., Gilquin B., Worman H. J., and Zinn-Justin S. (2010) Structural analysis of the Smad2–MAN1 interaction that regulates transforming growth factor-β signaling at the inner nuclear membrane. Biochemistry 49, 8020–8032 10.1021/bi101153w [DOI] [PubMed] [Google Scholar]
- 35. de Chiara C., Menon R. P., Strom M., Gibson T. J., and Pastore A. (2009) Phosphorylation of S776 and 14-3-3 binding modulate ataxin-1 interaction with splicing factors. PLoS ONE 4, e8372 10.1371/journal.pone.0008372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manceau V., Swenson M., Le Caer J. P., Sobel A., Kielkopf C. L., and Maucuer A. (2006) Major phosphorylation of SF1 on adjacent Ser-Pro motifs enhances interaction with U2AF65. FEBS J. 273, 577–587 10.1111/j.1742-4658.2005.05091.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chatrikhi R., Wang W., Gupta A., Loerch S., Maucuer A., and Kielkopf C. L. (2016) SF1 phosphorylation enhances specific binding to U2AF65 and reduces binding to 3′-splice-site RNA. Biophys. J. 111, 2570–2586 10.1016/j.bpj.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Manceau V., Kremmer E., Nabel E. G., and Maucuer A. (2012) The protein kinase KIS impacts gene expression during development and fear conditioning in adult mice. PLoS ONE 7, e43946 10.1371/journal.pone.0043946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burley S. K., and Petsko G. A. (1985) Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science 229, 23–28 10.1126/science.3892686 [DOI] [PubMed] [Google Scholar]
- 40. Diella F., Gould C. M., Chica C., Via A., and Gibson T. J. (2008) Phospho.ELM: a database of phosphorylation sites: update 2008. Nucleic Acids Res. 36, D240–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Horovitz A., Serrano L., Avron B., Bycroft M., and Fersht A. R. (1990) Strength and co-operativity of contributions of surface salt bridges to protein stability. J. Mol. Biol. 216, 1031–1044 10.1016/S0022-2836(99)80018-7 [DOI] [PubMed] [Google Scholar]
- 42. Lee Y., and Rio D. C. (2015) Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323 10.1146/annurev-biochem-060614-034316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Corrionero A., Miñana B., and Valcárcel J. (2011) Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 25, 445–459 10.1101/gad.2014311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Popp M. W., and Maquat L. E. (2013) Organizing principles of mammalian nonsense-mediated mRNA decay. Annu. Rev. Genet. 47, 139–165 10.1146/annurev-genet-111212-133424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Acuner Ozbabacan S. E., Engin H. B., Gursoy A., and Keskin O. (2011) Transient protein-protein interactions. Protein Eng. Des. Sel. 24, 635–648 10.1093/protein/gzr025 [DOI] [PubMed] [Google Scholar]
- 46. Pacheco T. R., Coelho M. B., Desterro J. M., Mollet I., and Carmo-Fonseca M. (2006) In vivo requirement of the small subunit of U2AF for recognition of a weak 3′ splice site. Mol. Cell. Biol. 26, 8183–8190 10.1128/MCB.00350-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rösel-Hillgartner T. D., Hung L. H., Khrameeva E., Le Querrec P., Gelfand M. S., and Bindereif A. (2013) A novel intra-U1 snRNP cross-regulation mechanism: alternative splicing switch links U1C and U1–70K expression. PLoS Genet. 9, e1003856 10.1371/journal.pgen.1003856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Turunen J. J., Verma B., Nyman T. A., and Frilander M. J. (2013) HnRNPH1/H2, U1 snRNP, and U11 snRNP cooperate to regulate the stability of the U11–48K pre-mRNA. RNA 19, 380–389 10.1261/rna.036715.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guth S., Martínez C., Gaur R. K., and Valcárcel J. (1999) Evidence for substrate-specific requirement of the splicing factor U2AF35 and for its function after polypyrimidine tract recognition by U2AF65. Mol. Cell. Biol. 19, 8263–8271 10.1128/MCB.19.12.8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zamore P. D., and Green M. R. (1991) Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 10, 207–214 10.1002/j.1460-2075.1991.tb07937.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zamore P. D., Patton J. G., and Green M. R. (1992) Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355, 609–614 10.1038/355609a0 [DOI] [PubMed] [Google Scholar]
- 52. Webb C. J., Lakhe-Reddy S., Romfo C. M., and Wise J. A. (2005) Analysis of mutant phenotypes and splicing defects demonstrates functional collaboration between the large and small subunits of the essential splicing factor U2AF in vivo. Mol. Biol. Cell 16, 584–596 10.1091/mbc.e04-09-0768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dolatshad H., Pellagatti A., Fernandez-Mercado M., Yip B. H., Malcovati L., Attwood M., Przychodzen B., Sahgal N., Kanapin A. A., Lockstone H., Scifo L., Vandenberghe P., Papaemmanuil E., Smith C. W., Campbell P. J., et al. (2015) Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia 29, 1798 10.1038/leu.2015.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang Y., Hale J., Wang Y., Li W., Zhang S., Zhang J., Zhao H., Guo X., Liu J., Yan H., Yazdanbakhsh K., Huang G., Hillyer C. D., Mohandas N., et al. (2018) SF3B1 deficiency impairs human erythropoiesis via activation of p53 pathway: implications for understanding of ineffective erythropoiesis in MDS. J. Hematol. Oncol. 11, 19 10.1186/s13045-018-0558-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Finci L. I., Zhang X., Huang X., Zhou Q., Tsai J., Teng T., Agrawal A., Chan B., Irwin S., Karr C., Cook A., Zhu P., Reynolds D., Smith P. G., et al. (2018) The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action. Genes Dev. 32, 309–320 10.1101/gad.311043.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Valcárcel J., Gaur R. K., Singh R., and Green M. R. (1996) Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science 273, 1706–1709 10.1126/science.273.5282.1706 [DOI] [PubMed] [Google Scholar]
- 57. Ruskin B., Zamore P. D., and Green M. R. (1988) A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52, 207–219 10.1016/0092-8674(88)90509-0 [DOI] [PubMed] [Google Scholar]
- 58. Soltis S. M., Cohen A. E., Deacon A., Eriksson T., González A., McPhillips S., Chui H., Dunten P., Hollenbeck M., Mathews I., Miller M., Moorhead P., Phizackerley R. P., Smith C., Song J., et al. (2008) New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta Crystallogr. D Biol. Crystallogr. 64, 1210–1221 10.1107/S0907444908030564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 10.1107/S0907444909047374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 64. Paolella B. R., Gibson W. J., Urbanski L. M., Alberta J. A., Zack T. I., Bandopadhayay P., Nichols C. A., Agarwalla P. K., Brown M. S., Lamothe R., Yu Y., Choi P. S., Obeng E. A., Heckl D., Wei G., et al. (2017) Copy-number and gene dependency analysis reveals partial copy loss of wild-type SF3B1 as a novel cancer vulnerability. Elife 6, e23268 10.7554/eLife.23268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Afonine P. V., Moriarty N. W., Mustyakimov M., Sobolev O. V., Terwilliger T. C., Turk D., Urzhumtsev A., and Adams P. D. (2015) FEM: feature-enhanced map. Acta Crystallogr. D Biol. Crystallogr. 71, 646–666 10.1107/S1399004714028132 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.