Abstract
A hexanucleotide GGGGCC expansion in intron 1 of chromosome 9 open reading frame 72 (C9orf72) gene is the most frequent cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). The corresponding repeat-containing sense and antisense transcripts cause a gain of toxicity through the accumulation of RNA foci in the nucleus and deposition of dipeptide-repeat (DPR) proteins in the cytoplasm of the affected cells. We have previously reported on the potential of engineered artificial anti-C9orf72-targeting miRNAs (miC) targeting C9orf72 to reduce the gain of toxicity caused by the repeat-containing transcripts. In the current study, we tested the silencing efficacy of adeno-associated virus (AAV)5-miC in human-derived induced pluripotent stem cell (iPSC) neurons and in an ALS mouse model. We demonstrated that AAV5-miC transduces different types of neuronal cells and can reduce the accumulation of repeat-containing C9orf72 transcripts. Additionally, we demonstrated silencing of C9orf72 in both the nucleus and cytoplasm, which has an added value for the treatment of ALS and/or FTD patients. A proof of concept in an ALS mouse model demonstrated the significant reduction in repeat-containing C9orf72 transcripts and RNA foci after treatment. Taken together, these findings support the feasibility of a gene therapy for ALS and FTD based on the reduction in toxicity caused by the repeat-containing C9orf72 transcripts.
Keywords: C9orf72, ALS, FTD, miRNA, gene therapy, AAV
Introduction
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are two severe neurodegenerative diseases, with overlapping pathologic and genetic features but distinct clinical features. ALS is the most common adult-onset motor neuron degenerative disorder that affects mainly the upper and lower motor neurons in the brain and corticospinal tract.1, 2 FTD is a presenile dementia characterized by the degeneration of neurons in the frontal and temporal lobes of the brain.2, 3 A significant number of patients develop both diseases (ALS-FTD).4
The most common genetic mutation in both ALS and FTD is a hexanucleotide GGGGCC (G4C2) repeat expansion in the first intron of the chromosome 9 open reading frame 72 (C9orf72) gene.4, 5, 6 ALS and FTD patients display hundreds to a few thousand copies of the G4C2 repeat in the C9orf72 gene.5 The contribution of this mutation to the pathogenesis of both diseases has been debated for several years, with the loss of C9orf72 function (haploinsufficiency), a gain of toxicity, or a combination of both being implicated.7, 8 Reduced C9orf72 mRNA levels in patients due to hypermethylation of the G4C2 repeat supports haploinsufficiency.6, 9 On the other hand, a causal role for RNA-mediated toxicity is supported by accumulation of the repeat-containing transcripts that fold into stable structures, forming RNA foci enriched with RNA-binding proteins in the nucleus.5, 10, 11, 12, 13 RNA foci are detected in several repeat expansion diseases and can sequester RNA-binding proteins. RNA foci produced from both the sense (G4C2) and antisense (G2C4) repeat transcripts are detected in tissues and in induced pluripotent stem cell (iPSC)-derived neurons from ALS and FTD patients, proving that the repeat region is bidirectionally transcribed.12, 14
Gain of toxicity is also supported by repeat-associated non-ATG (RAN) translation of the sense and antisense repeat transcripts, resulting in the accumulation of five aberrant dipeptide-repeat (DPR) proteins (poly(GA), poly(GR), poly(GP), poly(PA), and poly(PR)) in the cytoplasm, all with different toxicity profiles shown in vitro and in vivo.12, 15, 16 In addition, DPRs disrupt the nucleocytoplasmic transport system of cells.17, 18 Furthermore, autopsy studies revealed that ∼90% of C9orf72 ALS and ∼50% of FTD patients have cytoplasmic aggregation of the transactive response DNA-binding protein of 43 kDa (TDP-43; encoded by TAR DNA binding protein [TARDBP]).8, 19 Abnormal aggregation of P62 and ubiquitin have also been described in C9orf72-related ALS and FTD patients.20, 21, 22 A recently developed bacterial artificial chromosome (BAC) transgenic mouse model expressing the human C9orf72, including the expanded G4C2 repeat, showed gain-of-toxicity features such as RNA foci, DPRs, TDP-43, and p62 inclusions.23 These mice also develop neurodegeneration and ALS and/or FTD-like phenotypes, suggesting that these inclusions result in a gain of toxicity and contribute to the pathology observed in ALS and FTD.23
Regardless of the contribution of either RNA-mediated toxicity or haploinsufficiency, a therapy reducing the repeat-containing transcripts could potentially translate into a reduction of RNA foci and DPR proteins, slowing down the disease progression. Silencing of C9orf72 transcripts by RNAi strategies, such as duplex and single-stranded small interfering RNAs (siRNAs), or by the RNase H-mediated antisense oligonucleotides (ASOs) indeed resulted in the reduction of RNA foci and DPR proteins in patient-derived iPSC neurons and in mouse models.14, 24, 25, 26, 27 Interestingly, targeting only the sense strand of C9orf72 with ASOs not only reduced RNA foci but also rescued the disrupted nucleocytoplasmic transport in patient-derived iPSC neurons.18
Another promising strategy to achieve similar outcomes is by adeno-associated virus (AAV)-delivered artificial microRNAs (miRNAs) engineered to target C9orf72. miRNAs are short non-coding RNAs that bind to a complementary mRNA through specific base pairing, inducing its degradation and/or translational repression. As AAVs express stable extrachromosomal nuclear episomes, the primary miRNA transcripts can be continually produced, resulting in a longer-lasting therapeutic effect. The primary miRNA transcripts are processed into precursor miRNAs that are transported to the cytoplasm for further processing and incorporation into the RNA-induced silencing complex (RISC).28 Thus, one major challenge to overcome for a miRNA-based gene therapy approach is targeting of the repeat-containing C9orf72 transcripts within the cell nucleus. In one study, siRNAs, which are also processed by RISC, demonstrated the silencing of C9orf72 mRNA in patient-derived iPSC neurons, but the nuclear repeat-containing transcripts and RNA foci were unaffected, indicating a predominant efficacy in the cytoplasm.14 We previously reported on miC that target the sense, antisense, or both transcripts of C9orf72, and we demonstrated in vitro using luciferase reporter systems that all three approaches are feasible.29 In addition, we used cell models and showed crucial evidence that miC can be functional in the cell nucleus, where the repeat-containing transcripts accumulate and form RNA foci.
In the current study, we investigated the efficacy of AAV-delivered miC on the lowering of C9orf72 in human-derived iPSC neurons and in the Tg(C9orf72_3) line 112 mice as a proof of concept for an AAV-based gene therapy.30 Different human neuronal cell types were transduced, and the C9orf72-lowering efficacy in the nucleus and cytoplasm was investigated. In addition, we showed evidence that miC targeting C9orf72 in the mouse brain causes the reduction in nuclear RNA foci. Our study provides strong evidence that AAV-delivered miC can target C9orf72 in the cell nucleus and may be promising to alleviate the RNA-mediated toxicity in ALS and FTD patients.
Results
AAV5 Can Efficiently Transduce Neuronal and Non-neuronal Cells
The neuronal cells affected in ALS and FTD deviate. The main affected cells in ALS patients are motor neurons in the brain and spinal cord, whereas neurons in the frontal and temporal lobes of the brain are mainly affected in patients with FTD. About 15% of patients develop both ALS and FTD, where different types of neurons in the brain and spinal cord are affected.31 Besides motor neurons, other CNS cell types, such as astrocytes, microglia, and oligodendrocytes, may contribute to the progression of the diseases.32, 33, 34, 35 For example, it has been shown that astrocytes carrying the C9orf72 hexanucleotide expansion are toxic to motor neurons.32, 34 Although the underlying mechanisms remain unclear, intercellular seeding and transmission of DPRs between the two cell types could be a contributing factor.32, 36 Thus, ideally, a therapeutic drug for ALS and/or FTD should target a large variety of neuronal and non-neuronal cell types.
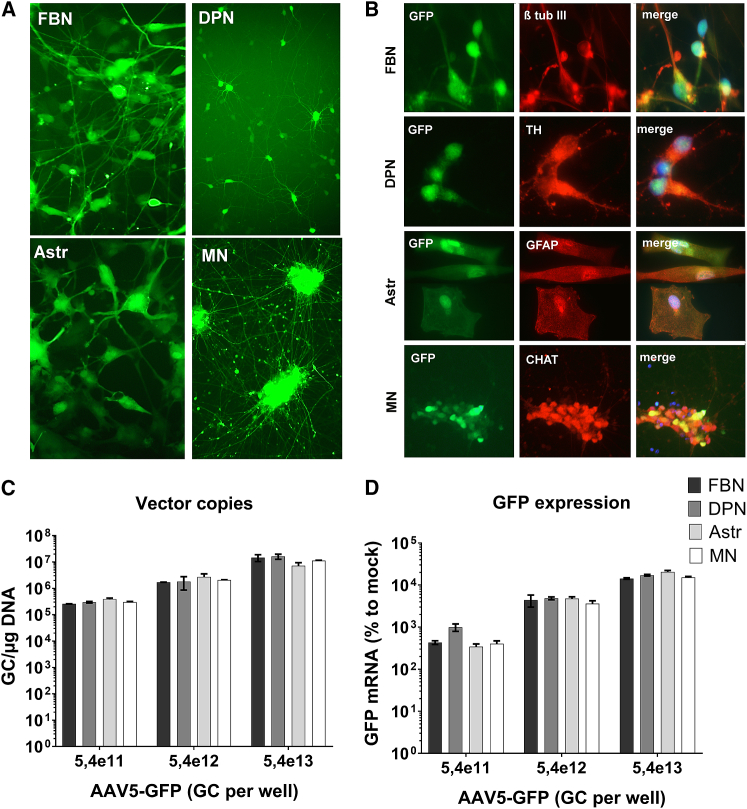
We generated and characterized different human-derived iPSC neurons and astrocytes to validate the transduction of AAV5 in different CNS cell types (Figure 1). iPSCs were induced into a neural progenitor state and differentiated into frontal brain-like neurons (FBNs) or astrocytes (Figure S1). In addition, commercially available mature dopaminergic neurons and motor neurons from a healthy person were obtained. Immunohistochemistry was performed and about 60% of FBNs were β-tubulin III positive and glial fibrillary acidic protein (GFAP) negative, implicating a successful differentiation rate of iPSCs into mature neurons. Similarly, mature astrocytes were ∼90% GFAP positive, confirming a successful differentiation of iPSCs into astrocytes. Mature dopaminergic neurons were ∼90% tyroxine hydroxylase (TH) positive, confirming successful differentiation. Mature motor neurons were also successfully differentiated as ∼85% were choline acetyltransferase (CHAT) positive.
Figure 1.
Transduction of Different iPSC-Derived Cells by AAV5
(A) Human iPSCs were differentiated into mature frontal brain-like neurons (FBNs), dopaminergic neurons (DPNs), astrocytes (Astrs), and motor neurons (MNs). The cells were transduced with 5e12 genomic copies (GC) AAV5-CAG-GFP, and live-cell imaging was performed at 2 weeks post-transduction. (B) Characterization of iPSC-derived cells. FBNs, DPNs, Astrs, and MNs were transduced with AAV5-CAG-GFP and fixed at 2 weeks post-transduction. Immunohistochemistry was performed with antibodies detecting β-tubulin III (β tub III) for mature FBNs, tyrosine hydroxylase (TH) for mature DPNs, glial fibrillary acidic protein (GFAP) for mature astrocytes, and choline acetyltransferase (CHAT) for mature MNs. (C and D) Transduction efficiency of AAV5 in iPSC-derived neurons. FBNs, DPNs, Astrs, and MNs were transduced with increasing doses of AAV5-CAG-GFP. The vector copy distribution (C) and GFP mRNA expression (D) were evaluated at 2 weeks post-transduction. Vector copies were calculated using a standard curve. For GFP mRNA expression, the input of RNA was corrected for GAPDH, and expression was calculated relative to cells treated with the formulation buffer (mock). Each bar represent the mean and standard deviation of one experiment performed in triplicate.
Following transduction with AAV5-GFP, ∼90% of all the different cell types expressed GFP (Figure 1A). Immunohistochemistry for GFP combined with either β-tubulin III, TH, GFAP, or CHAT antibodies confirmed that all four cell types were transduced by AAV5 (Figure 1B). To compare the AAV transduction tropism of the different cell types, we isolated DNA and RNA of transduced cells and quantitated vector copies and GFP mRNA expression in the cells (Figures 1C and 1D). A similar dose-dependent transduction efficiency was observed in all cell types, and transduction correlated with GFP expression. Thus, AAV5 efficiently transduces different human CNS-specific cell types, including FBNs, dopaminergic neurons, motor neurons, and astrocytes, and, hence, it is a promising vector to deliver therapeutic genes to the CNS to treat neurogenerative diseases such as ALS and FTD.
C9orf72 Expression Is Reduced in Neuronal Cells Derived from an FTD Patient
iPSCs from an FTD patient (ND42765) and a healthy non-diseased person (ND42245) were differentiated into FBNs (FTD-FBN) and astrocytes to compare the levels of C9orf72 mRNA and repeat-containing transcripts (Figure S1). qRT-PCR was performed 2 weeks after maturation for total C9orf72 mRNA (detecting all transcript variants) and the sense intronic transcripts (detecting sense transcripts containing the G4C2 repeat) to compare the expression levels in these cells. Primers amplifying a region spanning exon 2 to exon 4 were used to detect total C9orf72 mRNA (Figure 2A).10 The sense intronic transcripts were detected with primers amplifying a region in intron 1.10 The levels of total C9orf72 mRNA were significantly reduced in the FTD patient-derived cells: a reduction of ∼60% was observed in FBNs and ∼25% in astrocytes from the FTD patient as compared to healthy cells (Figure 2B). Interestingly, although at a low level, sense intronic transcript levels were increased by ∼30% in FBNs and ∼20% in astrocytes of the FTD patient as compared to healthy cells (Figure 2C). Thus, while total C9orf72 mRNA levels were reduced, sense intronic transcripts seem to accumulate in iPSC-derived FBNs and astrocytes from the FTD patient.
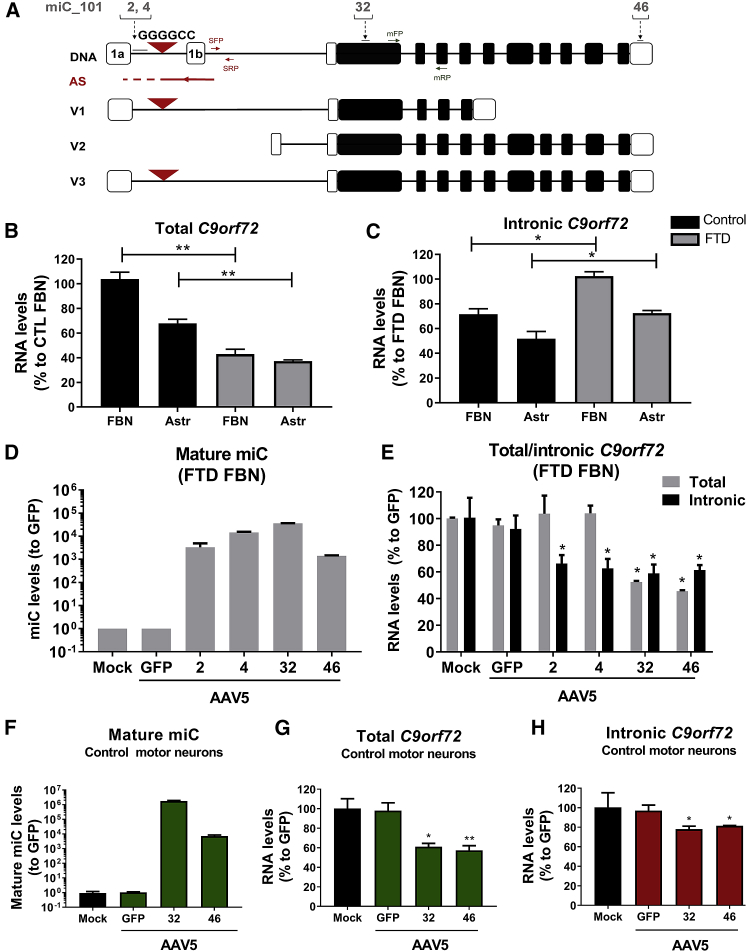
Figure 2.
Silencing of C9orf72 in iPSC Neurons by AAV5-miC
(A) Schematic of C9orf72 gene and location of the miC-binding sites. The C9orf72 gene consists of 12 exons, including the two alternatively spliced exon 1a and exon 1b. The G4C2 expansion is in the first intron between exon 1a and 1b. The gene produces three sense transcripts (V1, V2, and V3) and an antisense transcript. miC candidates were designed with binding sites in intron 1 (miC2 and miC4), exon 2 (miC32), and exon 11 (miC46). Primer sets in intron 1 were used to detect the sense intronic transcripts of C9orf72 (primerset for C9orf72 sense intronic transcript [SFP-SRP]). Total C9orf72 mRNA was detected with primers spanning exon 2 and exon 4 (primerset for total C9orf72 mRNA [mFP-mRP]), as described by others.10 (B and C) Expression of C9orf72 mRNA (B) and sense intronic transcripts (C) in FBNs and astrocytes. iPSCs were differentiated into FBNs and astrocytes. RNA was isolated from cells after 2 weeks of maturation, and qRT-PCR was performed to detect the endogenously expressed total C9orf72 mRNA and sense intronic transcripts (intronic C9orf72). The RNA input levels were corrected to GAPDH and calculated relative to the cell line with the highest expression of C9orf72 (FBNs). Error bars indicate the mean of two independent experiments. Data were evaluated using Student’s t test (*p < 0.05 and **p < 0.01). (D) Expression of the mature miC guide strands in FBNs after transduction with AAV5. Mature FTD-FBNs were transduced with 2e12 GC AAV5-miC2, AAV5-miC4, AAV5-miC32, and AAV5-miC46. Cells treated with the formulation buffer (mock) or AAV5-GFP served as controls. RNA was isolated 7 days post-transduction, and expressions of the mature miC2, miC4, miC32, and miC46 were determined by TaqMan. MicroRNA input levels were normalized to U6 small nuclear RNA and set relative to cells treated with AAV5-GFP. (E) Silencing of C9orf72 mRNA and sense intronic transcripts in iPSC-derived FBNs. Mature FTD-FBNs were transduced with 2e12 GC AAV5-miC2, AAV5-miC4, AAV5-miC32, and AAV5-miC46. RNA was isolated 7 days post-transduction. The levels of total C9orf72 mRNA and the sense intronic transcripts were determined by qRT-PCR. mRNA input was normalized to GAPDH and set relative to cells treated with AAV5-GFP. Data were evaluated using a one-way ANOVA with Dunnett’s multiple comparison test (*p < 0.05) to compare cells treated with AAV5-miC to AAV5-GFP. (F) miC32 and miC46 expressions in transduced motor neurons. Healthy motor neurons differentiated from human iPSCs were transduced with AAV5-GFP, AAV5-miC32, and AAV5-miC46 for 2 weeks. Total RNA was isolated, and small RNA TaqMan was performed to detect the mature miC32 and miC46, as described in (D). (G and H) C9orf72 reduction in motor neurons by AAV5-miC. RNA was isolated from transduced motor neurons 2 weeks post-transduction, and qRT-PCR was performed to detect the total C9orf72 mRNA (G) and sense intronic transcripts (H), as described in (E). Error bars represent the mean of two independent experiments. Data were evaluated using a one-way ANOVA with Dunnett’s multiple comparison test (*p < 0.05 and **p < 0.01) to compare cells treated with AAV5-miC to AAV5-GFP.
AAV5-miC Can Lower the Repeat-Containing Transcripts of C9orf72 in iPSC Neurons
Sequences on the human C9orf72 were previously selected to design miC.29 The miC sequences were embedded in the primary miR-101 and/or miR-451 scaffold by replacing the naturally expressed guide strand sequences. Four lead miC candidates were selected in the miR-101 scaffold based on their efficacy on reporter genes and their ability to reduce the endogenously expressed C9orf72 mRNA and sense intronic transcripts in cells.29 miC32 and miC46 were designed to target C9orf72 exon 2 and exon 11, respectively, targeting all sense C9orf72 transcripts (Figure 2A). miC2 and miC4 were designed in intron 1 to selectively silence the sense G4C2 sense intronic transcripts.
To determine whether miC delivered by AAV5 is functional in patient-derived cells, FTD-FBNs were transduced with AAV5-miC2, AAV5-miC4, AAV5-miC32, and AAV5-miC46. At 2 weeks following transduction, all four mature miC were expressed, suggesting a successful transduction by AAV5-miC and efficient processing into a mature miC (Figure 2D). Sense intronic transcript levels were reduced by ∼40% in FBNs transduced with miC2 and miC4, while the C9orf72 mRNA levels were not affected (Figure 2E). Thus, both candidates exclusively target the sense intronic transcripts while preserving normal levels of C9orf72 mRNA. Of the candidates targeting the total C9orf72 mRNA, both miC32 and miC46 reduced the levels of C9orf72 mRNA (∼50%) and the sense intronic transcript (∼40%). Thus, the sense intronic transcripts could be also targeted by silencing total C9orf72 mRNA.
Additionally, we investigated silencing of C9orf72 in a healthy motor neuron cell line, as motor neurons are highly affected in ALS. The expression of C9orf72 was first evaluated in control (non-transduced) motor neurons, and both total and intronic C9orf72 were detected. However, the intronic C9orf72 expression in this cell line, which lacks the G4C2 expansion, was very low and slightly above the detection limit (data not shown). Having established that C9orf72 can be detected in healthy motor neurons, the cells were transduced with AAV5-miC32 and AAV5-miC46 for 2 weeks. We found expression of miC32 and miC46, confirming that AAV5 efficiently transduces human motor neurons (Figure 2F). Consistently, we observed an ∼40% reduction in total C9orf72 mRNA by both miC candidates and a mild reduction in the intronic C9orf72 (∼20%) (Figures 2G and 2H). Altogether, we demonstrated the reductions in total and intronic C9orf72 levels in FBNs and motor neurons, confirming that both neuronal cell types are transduced and that the miC candidates are effective at lowering C9orf72 in these cells.
Efficient Silencing of C9orf72 in the Nucleus of iPSC-Derived Neurons by AAV-miC
The accumulation of the G4C2 repeat-containing transcripts in the cell nucleus seems to highly contribute to the progression of both ALS and FTD. These transcripts form RNA foci in the cell nucleus that sequester RNA-binding proteins and inhibit their function or are transported to the cytoplasm for RAN translation into toxic DPRs.12, 16, 24, 37 Thus, for a therapeutic approach, efficacy within the cell nucleus is required to effectively target the RNA-mediated toxicity in ALS and FTD. The processing of miRNAs occurs through a multi-step process involving nuclear and cytoplasmic phases, but the mature miRNA product is produced in the cytoplasm.28, 38, 39, 40 Therefore, miRNAs were initially thought to be predominantly expressed and active in the cytoplasm. We previously demonstrated that active mature miC is also detected in the nucleus of cells transfected with miC constructs, but at lower levels than in the cytoplasm.29 Here we evaluated whether the transduction of iPSC neurons by AAV5-miC is sufficient to express the mature miC and reduce C9orf72 levels in the nucleus (Figure 3).
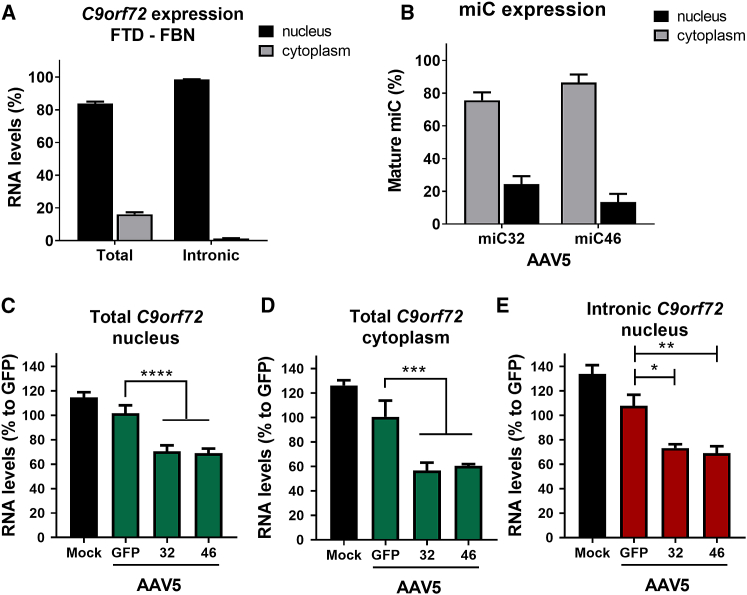
Figure 3.
Reduction of C9orf72 in the Nucleus by miC
(A) Nuclear and cytoplasmic expressions of C9orf72 in FTD-FBNs. RNA was isolated from nuclear and cytoplasmic fractions of mature FTD-FBNs, and qRT-PCR was performed to detect the total C9orf72 mRNA (total C9orf72) and sense intronic transcripts (intronic C9orf72). Total and intronic C9orf72 mRNA levels were normalized to GAPDH (n = 4). The sum of nuclear and cytoplasmic C9orf72 expression values was set at 100%. (2−Δct nuclear C9orf72 RNA + 2−Δct cytoplasmic C9orf72 RNA = 100%). (B) Mature miC expression in nucleus and cytoplasm. FTD-FBNs were transduced with mock, AAV5-GFP, AAV5-miC31, and AAV5-miC46 for 7 days (n = 4). RNA was isolated from nucleus and cytoplasm, and expressions of mature miC31 and miC46 were determined by small RNA TaqMan. mRNA input levels were normalized to GAPDH. The sum of nuclear and cytoplasmic miC expression values was set at 100%. (C and D) Silencing of total C9orf72 in nucleus and cytoplasm. RNA was isolated from the nucleus (C) and cytoplasm (D) of FTD-FBNs transduced for 7 days with mock, AAV5-GFP, AAV5-miC31, and AAV5-miC46. mRNA levels were normalized to GAPDH, and total C9orf72 expression was determined relative to AAV5-GFP-treated cells. (E) Reduction of intronic C9orf72 in nucleus of FTD-FBNs. Performed as described in (C) and (D), intronic C9orf72 expression was determined relative to AAV5-GFP-treated cells. Data were evaluated using a one-way ANOVA with Dunnett’s multiple comparison test (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001) to compare cells treated with AAV5-miC to AAV5-GFP (n = 4).
FTD-FBNs were transduced with AAV5-miC32 and AAV5-miC46, and after a week RNA was isolated from nuclear and cytoplasmic fractions to calculate the percentage of RNA transcripts in both cellular compartments. In control FBNs, ∼80% of total C9orf72 mRNA was detected in the nucleus and ∼20% was measured in the cytoplasm, whereas sense intronic transcripts were predominantly (∼95%) found in the nucleus of FTD-FBNs (Figure 3A). Thus, both C9orf72 mRNA and sense intronic transcript levels were significantly higher in the nucleus of FTD-FBNs. Next, the percentage of the mature miC and the silencing of C9orf72 were determined in nucleus and cytoplasm after transducing FTD-FBNs with AAV5-miC32 and AAV5-miC46. About 20% of the mature miC32 was detected in the nucleus while ∼80% was measured in the cytoplasm (Figure 3B). In cells treated with AAV5-miC46, ∼10% of the mature miC was expressed in the nucleus and ∼90% in the cytoplasm. Interestingly, both AAV5-miC32 and AAV5-miC46 resulted in an ∼30% reduction in C9orf72 mRNA in the nucleus and an ∼40% reduction in the cytoplasm (Figures 3C and 3D). Consistently, an ∼25% reduction in the sense intronic transcripts was observed in the nucleus (Figure 3E). Our data show that the mature miC32 and miC46 can both shuttle from the cytoplasm to the cell nucleus and can reduce levels of both C9orf72 mRNA and the sense intronic transcripts in the nucleus as well as in the cytoplasm.
AAV5-miC32 and AAV5-miC46 Can Both Reduce C9orf72 in Tg(C9orf72_3) Line 112 Mice
Having established the efficacy of AAV5-miCs in different human neuronal cell types, we next evaluated their efficacy in vivo in Tg(C9orf72_3) line 112 mice.30 This mouse model is based on several tandem copies of the human C9orf72, with repeat sizes ranging from 100–1,000 repeats. Although the progressive neurodegeneration seen in ALS and FTD patients is not observed in these mice, they do exhibit some of the pathological features seen in patients, such as RNA foci (starting at ∼3 months of age) and poly(GP) protein (starting at ∼6–20 months of age).
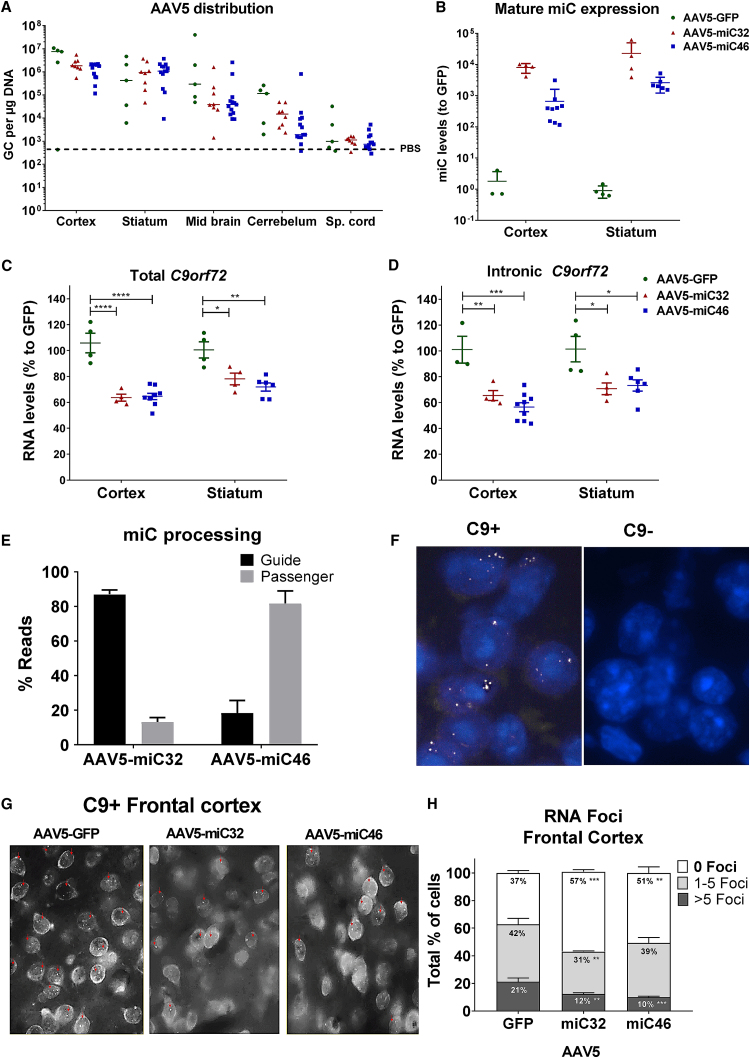
The 3-month-old mice were injected bilaterally in the striatum with AAV5-GFP, AAV5-miC32, and AAV5-miC46. Mice were sacrificed 6 weeks post-injection to determine the distribution of AAV5, mature miC expression, C9orf72 lowering, and the effect of miC on RNA foci formation. A widespread distribution of AAV5 to the cortex, striatum, and midbrain was observed after administration in the striatum (Figure 4A). A weak transduction of the cerebellum was observed while the spinal cord was not transduced. Consistent with the AAV5 distribution, small RNA TaqMan showed high expression of miC32 and miC46 in the cortex and striatum, which resulted in a 20%–40% lowering of C9orf72 mRNA and the sense intronic transcripts (Figures 4B–4D). Both AAV5-miC32 and AAV5-miC46 also target the mouse C9orf72 ortholog (3110043O21 Rik), and indeed they lowered the target 3110043O21 Rik. No behavioral and/or phenotypic changes were observed in mice treated with AAV5-miC32 or AAV5-miC46 (Figure S3).
Figure 4.
Reduction of C9orf72 in C9BAC Mice
(A) Vector copy distribution of AAV5 upon intrastriatal injection. The 3-month-old Tg(C9orf72_3) line 112 mice were injected with AAV5-GFP (5e10 GC), AAV5-miC32 (5e10 GC), and AAV5-miC46 (1e10 GC) bilaterally in the striatum. All mice were sacrificed 6 weeks after surgeries, and frontal cortex, striatum, midbrain, cerebellum, and spinal cord were collected. DNA was isolated from the tissues, and qPCR was performed with primers amplifying a 95-bp fragment from the CAG promoter region. The genome copies per tissue were calculated using a standard curve. (B) Expressions of mature miC32 and miC46 guide strands in the cortex and striatum of Tg(C9orf72_3) line 112 mice. Performed as described in (A), total RNA was isolated from the cortex and striatum for small RNA TaqMan. MicroRNA input levels were normalized to U6 small nuclear RNA and set relative to AAV-GFP-treated mice. (C and D) Lowering of total (C) and intronic C9orf72 (D) by miC in Tg(C9orf72_3) line 112 mice. Performed as described in (A), total RNA was isolated from the cortex and striatum, and qRT-PCR was performed using primers for total C9ORF72 mRNA and sense intronic transcripts. RNA input levels were normalized to GAPDH and set relative to AAV-GFP mice. (E) Processing of miC32 and miC46 in mice. Small RNA NGS was performed on RNA isolated from the striatum to determine the length and ratio of guide and passenger strands. (F) detection of RNA foci in the cortex of Tg(C9orf72_3) line 112 mice. Mouse brain was frozen and sectioned in OCT. RNA FISH was performed using a TYE563-(CCCCGG)3 LNA probe to detect the sense foci. Sense foci (shown as white spots) were detected in Tg(C9orf72_3) line 112 mice (C9+), but not in control littermates (C9−). (G and H) Reduction of RNA foci in frontal cortex. Cells with 0, 1–5, or >5 foci were counted in control (AAV5-GFP) and treated groups (AAV5-miC32 and AAV5-miC46). Red arrows show the cells that contain RNA foci (G). The percentages of cells containing 0, 1–5, or >5 foci were calculated from 6 different images per treatment group (H) (n = 3). Data were evaluated using a one-way ANOVA with Dunnett’s multiple comparison test (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
AAV5-miC32 and AAV5-miC46 Are Processed Differently in the Mouse Brain
We further investigated the fidelity of miC processing in the mouse brain. Following transcription of the miC construct, the primary miR-101 is processed by Drosha cleavage and then by Dicer cleavage into a miRNA duplex. The miRNA duplex is then separated, and the guide strand is usually incorporated into the RISC while in most cases the passenger strand is degraded. The processing of the miC32 and miC46 was analyzed by small RNA sequencing to determine the ratio of guide and passenger strands that are produced. Small transcriptome analysis was performed on RNA isolated from the striatum of four mice that were injected with AAV5-miC32 or AAV5-miC46. For each sample, we obtained between 15 and 30 million small RNA reads that were subsequently adaptor trimmed and aligned against the corresponding reference sequence. All reads shorter than 10 nt, longer than 45 nt, or represented less than 10 times were excluded from the analysis. miC32 was processed into predominantly guide strands (∼87%), 19–20 nt long, with a low percentage of the passenger strand (∼13%). However, miC46 processing yielded more passenger strands (∼82%) of between 19 and 22 nt long and low amounts (∼18%) of guide strands (Figure 4E; Table S1).
AAV5-miC Reduces RNA Foci in Tg(C9orf72_3) Line 112 Mice
RNA foci formation by the repeat-containing transcripts is considered a hallmark of the RNA-mediated toxicity in ALS and/or FTD.4, 11, 12, 13 Fluorescence in situ hybridization (FISH) using a TYE563-(C4G2)3 locked nucleic acid (LNA) probe showed that ∼60%–80% of cells in cortex, hippocampus, and cerebellum of the Tg(C9orf72_3) line 112 mice contained RNA foci (Figure 4F).30 After confirming the presence of sense and antisense RNA foci in the cortex, hippocampus, and cerebellum, the efficacy of AAV5-miC32 and AAV5-miC46 to reduce RNA foci was determined. Both miC candidates caused a significant drop of sense RNA foci in cortex and hippocampus and the number of cells containing RNA foci, as well in as the total amount of RNA foci per cell (Figures 4G and 4H; Figure S2). AAV5-miC32 resulted in a 20% drop of cells containing RNA foci, and the number of cells containing 1–5 RNA foci and >5 RNA foci were reduced by 11% and 9%, respectively. Similarly, AAV5-miC46 treatment resulted in a 14% reduction in foci-containing cells, a 3% reduction in cells containing 1–5 foci, and an 11% reduction in cells containing >5 RNA foci. Hence, these data confirm that AAV5-delivered miC candidates against total C9orf72 mRNA are functional in reducing nuclear sense RNA foci in brain tissues of the Tg(C9orf72_3) line 112 mice.
Overall, we demonstrated that AAV5 can transduce different CNS cell types relevant for ALS and/or FTD treatment and that miC candidates targeting C9orf72 are successfully delivered and are functional in the mouse brain. Furthermore, we showed that total C9orf72 mRNA and sense intronic C9orf72 transcripts can be lowered in both the nucleus and cytoplasm of cells, increasing the potential for achieving therapeutic benefit in patients.
Discussion
The intronic G4C2 repeat of C9orf72 produces repeat-containing transcripts that cause RNA foci and DPR proteins, which contribute to ALS and/or FTD pathology. Thus, a therapy reducing these gain-of-toxicity features could slow down disease progression in ALS and/or FTD patients. In this study, we provide evidence that AAV5-delivered miRNAs targeting C9orf72 reduce gain-of-toxicity features caused by the G4C2 repeat in a murine model.
Initially described as a pure motor neuron disease, it is now thought that other cell types, including resident glial cells, are involved in ALS.41, 42 Abnormal neuropsychological testing is observed in ∼50% of ALS patients, indicating that, besides motor neurons, other neuronal cell types in the brain are affected.31 In addition, some patients develop both ALS and FTD, affecting different types of neurons in the brain and spinal cord. Delivery of therapeutics to the affected cell types is a major challenge for ALS and/or FTD therapy. We demonstrated that AAV5 can transduce various cell types of the CNS that are relevant to both diseases.
We observed a reduction of C9orf72 mRNA in astrocytes and FBNs derived from an FTD patient compared to healthy cells. This finding was consistent with several other studies that reported a reduction of C9orf72 mRNA and protein in iPSC neurons and in brain and spinal cord tissues from C9orf72-related ALS and/or FTD patients.6, 9, 43, 44, 45, 46 This reduction is caused by methylation of the repeat region of C9orf72, which is located in or near the promoter region, leading to transcription inhibition.47, 48 However, C9orf72 haploinsufficiency alone is most likely not sufficient to cause neurodegeneration. The reduction of C9orf72 in mice was tolerable, while complete elimination of C9orf72 caused splenomegaly and enlarged lymph nodes, but not neurodegeneration or mis-localization of TDP-43.49, 50, 51
Compared to C9orf72 mRNA levels, the expression of sense intronic transcripts was low in FTD and control cells. Yet, sense intronic transcripts were increased in FTD cells as compared to healthy cells, consistent with what has been reported by others.10 The elevated sense intronic transcript levels detected in the FTD cells seem to be caused by defective splicing of intron 1 due to the presence of the G4C2 repeat.5, 52 The corresponding transcripts containing the G4C2 repeat may be protected from degradation, which allows them to accumulate in the cell.44, 52, 53, 54 Despite a relatively low abundance as compared to C9orf72 mRNA, the intronic transcripts could still be sufficient to accumulate in RNA foci over time.29, 47, 52 Furthermore, it has been estimated that hundreds of protein products could be produced by a single mRNA, suggesting that even low levels of intronic transcripts are sufficient to cause the accumulation of toxic DPR proteins in the cell.9, 55
Several approaches, such as ASOs, duplex and single-stranded siRNAs, and small compounds, have been tested and proved promising to reduce gain-of-toxicity features caused by the G4C2 repeat.24, 26, 56 In this study, we investigated the feasibility of a miRNA-based gene therapy to obtain long-term silencing of the repeat-containing transcripts of C9orf72. Four AAV5-miC candidates were tested on human-derived iPSC neurons, and all four resulted in sufficient transduction to express therapeutically relevant levels of the corresponding mature miC. Selective reduction of the sense intronic transcripts was achieved with the miC targeting intron 1, without affecting normal C9orf72 mRNA levels. This approach could prevent RNA-mediated toxicity without further reducing the C9orf72 protein. However, as sequence variations within intron 1 have been observed, genomic screening of patients for selection could be necessary when targeting this region.57 Thus, sequence conservation of miC2 and miC4 targets should be determined in larger cohort studies.
We previously used a publicly available RNA sequencing (RNA-seq) database of patients to investigate the conservation of miC2 and miC4 target sites in intron 1, but the intronic transcript levels detected were too low to determine their conservation.29, 58 The target sequences of miC32 and miC46, targeting either exon 2 or exon 11, were well conserved between the patients, and both candidates also reduced levels of the sense intronic transcripts in iPSC neurons. The reduction of sense intronic transcripts by miC32 and miC46 supports previous findings that intron 1 is still present in the mature C9orf72 mRNA, suggesting defective splicing.52 Although miC32 and miC46 also reduced the levels of normal C9orf72 mRNA, its expression was not completely eliminated. Additionally, we observed transduction, mature miC expression, and the reduction of C9orf72 mRNA in a healthy motor neuron cell line. As expected, the expression of the sense intronic transcripts in this cell line was very low, as the repeat expansion is absent in healthy individuals.
Having established that miCs delivered by AAV5 are effective in human-derived iPSC neurons, we specifically studied their efficacy in the cell nucleus where C9orf72 mRNA and intronic transcripts are predominantly expressed. Although the primary and precursor miRNAs originate from the cell nucleus, their transport to the cytoplasm to exert post-transcriptional gene silencing via the RISC has been well described.28, 38, 39, 59 Indeed, most studies have initially focused on post-transcriptional gene silencing of miRNAs in the cytoplasm. However, the discovery of several mature miRNA and RISC components enriched in the nucleus indicate that nuclear miRNAs do exist.60 Additionally, several proteins mediating nucleus-cytoplasm shuttling of small RNAs have been identified.60, 61 We found ∼5 times lower levels of mature miC in the nucleus compared to cytoplasm of transduced iPSC neurons, but the nuclear miC levels were still sufficient to reduce the levels of C9orf72 mRNA and the sense intronic transcripts in the nucleus. Thus, AAV-delivered miC can lower the repeat-containing transcripts that accumulate in the nucleus of ALS and/or FTD patients.
Moving forward to an in vivo proof-of-concept study, we tested the delivery and efficacy of AAV5-miC in the BAC transgenic Tg(C9orf72_3) line 112 mouse model.30 These mice exhibit pathologic features such as sense and antisense RNA foci and poly(GP) protein but no TDP-43 or P62 inclusions. These mice also do not develop the ALS and/or FTD-like phenotype in their lifespan, possibly due to the lack of 5′ and 3′ regulatory elements needed to control the sufficient expression of sense and antisense transcripts.23, 30 Intrastriatal injection of AAV5 resulted in a strong localized transduction on the injection site and surrounding areas, including frontal cortex and midbrain area, but it was not sufficient to transduce the spinal cord. Thus, further studies in larger animals are required to predict the best routes of injection into the cerebrospinal fluid (CSF) to transduce the brain and spinal cord of patients.
Intrastriatal delivery of AAV5-miC caused the high expression of mature miC and significant lowering of C9orf72 mRNA and the sense intronic transcripts in the transduced areas. The efficacy of AAV5-miC in the nucleus was further confirmed by the finding of a significant reduction of nuclear sense RNA foci in the cortex and hippocampus, suggesting that the mature miC is functional in the nucleus of transduced neurons in vivo. Moreover, a reduction in the mouse C9orf72 ortholog was also observed and was well tolerated in mice (3110043O21Rik). The processing of the miC in the mouse brain revealed that miC32 has a low amount of passenger strand, decreasing the risk for off-target effects in comparison to miC46. In this study, all mice were sacrificed at 5 months of age; future studies in older mice that have more accumulation of poly(GP) proteins would be needed to investigate the effect of miC on the DPR protein accumulation. However, based on the observed reduction of the sense intronic transcripts, a reduction of poly(GP) protein would be expected, as fewer repeat-containing transcripts are available to undergo RAN translation.
Taken together, these data provide a proof of concept for the silencing of C9orf72 by AAV5-miC in relevant cell types as a potential treatment approach for ALS and/or FTD. miC32 offers the superior profile as a candidate to reduce gain of toxicity in ALS and/or FTD due to the sense intronic transcripts. We showed nuclear and cytoplasmic silencing, increasing the potential for a therapeutic effect of a miC32-based gene therapy that silences C9orf72.
Materials and Methods
Cell Culture
Human control (ND42245) and Frontotemporal Dementia (ND42765) iPSCs derived from fibroblasts were ordered from Coriell Biorepository and cultured on Matrigel (Corning)-coated 6-well plates in mTeSR1 (STEMCELL Technologies). For embryoid body-based neural induction, iPSCs were seeded on AggreWell800 plates and cultured in STEMdiff Neural Induction Medium (STEMCELL Technologies) for 5 days with daily medium changes. Embryoid bodies were harvested and plated on 6-well plates coated with poly-D-lysine (Sigma-Aldrich) and laminin (Sigma-Aldrich) in STEMdiff Neural Induction Medium for 7 days with daily medium changes. Rosettes were harvested with STEMdiff Neural Rosette Selection Reagent (STEMCELL Technologies) and plated on poly-D-lysine- and laminin-coated 6-well plates in STEMdiff Neural Induction Medium for 24 h.
For differentiation into FBNs, STEMdiff Neural Induction Medium was replaced with STEMdiff Neuron Differentiation Medium (STEMCELL Technologies), and neuroprogenitor cells were differentiated for 5 days. For differentiation into astrocytes, neuroprogenitor cells were differentiated in STEMdiff Astrocyte Differentiation Medium (STEMCELL Technologies). The neuroprogenitor cells were then plated on poly-D-lysine- and laminin-coated plates in STEMdiff Neuron Maturation Medium (STEMCELL Technologies) for 1 week or STEMdiff Astrocytes Maturation Medium for 3 weeks. The mature FBNs and astrocytes were stored in liquid nitrogen in Neuroprogenitor Freezing Medium (STEMCELL Technologies).
Cryopreserved non-diseased mature dopaminergic neurons (iCELL Dopaneurons, 01279, C1028, lot 102477) were ordered at FUJIFILM Cellular Dynamics. Cryopreserved non-diseased mature motor neurons (40HU-005, lot 400089) were ordered at iXCells Biotechnologies.
Generation of AAV5-miC Vectors and Transductions
The design and cloning of the miC constructs were performed as described previously.29, 62 The miC constructs were all expressed by the synthetic cytomegalovirus (CMV) early enhancer and chicken β-actin (CAG) promoter. To produce AAV5, the CAG-miC2, CAG-miC4, CAG-miC32, and CAG-miC46 cassettes were obtained by digestion with restriction enzymes HindIII and PvuI and cloned in a uniQure transfer plasmid in order to generate an entry plasmid. The presence of the two inverted terminal repeats (ITRs) was confirmed by restriction digestion with SmaI. The ITR-CAG-miC cassettes were inserted into a recombinant baculovirus vector by homologous recombination in Spodoptera frugiperda Sf9 cells, and clones were selected by plague purification and insert PCR. The recombinant baculovirus containing the ITR-CAG-miC was further amplified until passage 6 (P6) in Sf+ cells and screened for the best production and stability by PCR and qRT-PCR. To generate AAV5, Sf+ cells were triple infected with three different recombinant baculoviruses expressing the ITRs-CAG-miC, the replicon enzyme, and the capsid protein. The cells were lysed 72 h after the triple infection, and the crude lysate was treated with 50 U/mL Benzonase (Merck, Darmstadt, Germany) for 1 h at 37°C. AAV5 was purified on an AVB Sepharose column (GE Healthcare, Little Chalfont, UK) and eluted in a formulation buffer consisting of 1× PBS and 4% sucrose. The final titer was determined by qRT-PCR with primers amplifying a 95-bp fragment from the CAG promoter region.
For transductions with AAV, FBNs, dopaminergic neurons (DPNs), and motor neurons (MNs) were plated in 24-well plates at 0.3*106 cells/well. Astrocytes were plated at 0.1*106 cells/well in STEMdiff Astrocyte Maturation Medium (STEMCELL Technologies) on Matrigel-coated plates. FBNs were plated in STEMdiff Neuron Maturation Medium (STEMCELL Technologies) on poly-D-lysine- and laminin-coated plates. Dopaminergic neurons were plated in iCell Neural Base Medium (FUJIFILM Cellular Dynamics), according to the manufacturer’s description on poly-D-lysine- and laminin-coated plates. Motor neurons were plated in Motor Neuron Maintenance Medium, according to the manufacturer’s description on Matrigel-coated plates. After 1 week of acclimation, cells were transduced with AAV5 for 1–2 weeks.
RNA and DNA Isolation
For RNA, plated cells and tissues were lysed in 300 μL TRizol. RNA was isolated from TRizol using the DIRECT-ZOL miniprep kit (R2050, Zymo Research). DNA was isolated using the DNeasy Blood & Tissue Kit (69506, QIAGEN), according to the manufacturer’s protocol.
Next-Generation Sequencing
Small RNA sequencing libraries for the Illumina sequencing platform were generated using high-quality total RNA as input and the NEXTflex Small RNA Sequencing kit (Bioo Scientific, Austin, TX, USA). Briefly, the small RNA species were subjected to ligation with 3′ and 5′ RNA adapters, first-strand reverse transcription, and PCR amplification. Sample-specific barcodes were introduced in the PCR step. The PCR products were separated on Tris/Borate/EDTA (TBE)-PAGE, and the expected band around 30 bp was recovered for each sample. The resulting sequencing libraries were quantified on a BioAnalyzer (Agilent Technologies, Santa Clara, CA). The libraries were multiplexed, clustered, and sequenced on an Illumina HiSeq 2000 (TruSeq version [v.]3 chemistry), with a single-read 36-cycle sequencing protocol and indexing. The sequencing run was analyzed with the Illumina CASAVA pipeline (v.1.8.2), with demultiplexing based on sample-specific barcodes. The raw sequencing data produced were processed, removing the sequence reads that were of too low quality (only “passing filter” reads were selected). In total, we generated between 15 and 35 million reads per sample.
NGS Data Analysis
Next-generation sequencing (NGS) small RNA raw datasets were analyzed using the CLC Genomics Workbench 8 (QIAGEN). The obtained reads were adaptor trimmed, which decreased the average read size from ∼50 to ∼25 bp. All reads containing ambiguity N symbols, reads shorter than 10 nt, longer than 45 nt, and reads represented less than 10 times were discarded. Next, the obtained unique small RNA reads were aligned to the reference sequences of the pre-miC9 constructs, with a maximum of 3-nt mismatches allowed. The percentages of reads based on the total number of reads matching the reference sequence were calculated (Table S1).
qRT-PCR and miRNA TaqMan Assay
To determine C9orf72 mRNA knockdown in cells, RNA was isolated, and first-strand complementary DNA was reverse transcribed using random hexamer primers with the Dynamo kit (Finnzymes, Espoo, Finland). Real-time PCR amplification was performed with primers to detect total C9orf72 mRNA and the sense intronic transcripts of human. Total C9orf72 mRNA was detected with the following primers: mFP 5′-CGGAAAGGAAGAATATGGATGC-3′, mRP 5′-CCATTACAGGAATCACTTCTCCA-3′, and probe mPRB 5′-AGCATTGGAATAATACTCTGACCCTGATCTTC-3′. The sense intronic transcripts were detected with the following primers: SFP 5′-ACGCCTGCACAATTTCAGCCCAA-3′, SRP 5′-CAAGTCTGTGTCATCTCGGAGCTG-3′, and probe SPRB 5′-TGAGGGCAGCAATGCAAGTCGGTGTG-3′. The mRNA expression levels were normalized to human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (forward 5′-GAAGGTGAAGGTCGGAGTC-3′, reverse 5′-GAAGATGGTGATGGGATTTC-3′, and probe 5′-CAAGCTTCCCGTTCTCAGCC-3′) as an internal control.
PCR reaction conditions were as follows: 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The assays were performed on an ABI 7000 or ABI 7500 (Applied Biosystems, Foster City, CA, USA). Gene expression levels were normalized to GAPDH as an internal control, and the level of gene expression was calculated relative to control cells. To determine the expressions of miC2, miC5, miC32, and miC46, Custom TaqMan Small RNA Assay (Themo Fisher Scientific) was used (miC2, assay ID CTEPR3R; miC4, assay ID CTFVKNN; miC32, assay ID CSGJPRB; and miC46, assay ID CSHSNXJ). The RT reaction and TaqMan were performed according to the manufacturer’s protocol.
Animals
Two breeding couples were ordered at The Jackson Laboratory (stock number 023099) and were kept and bred at the Neurosciences Division, Center for Applied Medical Research, CIMA, University of Navarra, Pamplona, Spain. The animals were housed 4–5 per cage with ad libitum access to food and water and maintained in a temperature-controlled environment on a 12-h dark-light cycle. All procedures were carried out in accordance with the current European and Spanish regulations (86/609/EEC; RD1201/2005). This study was approved by the Ethical Committee of the University of Navarra (137/010).
Intrastriatal Injection in Tg(C9ORF72_3) Line 112 mice
Surgeries were performed as described previously.63 In brief, 3-month-old mice were anesthetized with ketamine and xylazine (80/10 mg/kg, intraperitoneally [i.p.]) and placed in a stereotactic frame. The scalp was shaved, and a longitudinal incision was made along the midline of the skull. The dorsal surface of the skull was then exposed, and two burr holes were drilled above the infusion sites. 2 or 5 μL virus suspension or PBS solution (sham mice) was infused bilaterally to striatum (+0.8 mm anterior-posterior [AP], ±2 mm medial-lateral [ML], −4.0 mm dorsal-ventral [DV] to bregma), according to the Paxinos and Watson.64 A 5-μL Hamilton syringe (or 10 for ventricle surgeries) was used for the infusion (Hamilton, Reno, NV, USA). The infusion rate was 0.2 μL/min, and the needle remained in place for 5 min after the infusion for vector absorption. Finally, the site was stitched closed.
RNA Foci FISH
RNA FISH was performed as described previously with some adjustments.23, 30, 56 In brief, whole mouse brains were fixed in 4% paraformaldehyde (PFA) for 1 week at 4°C. Brains were transferred into 15-mL tubes containing 10 mL 30% PBS and sucrose and left at 4°C until brains sunk to the bottom. The brains were then frozen in optimal cutting temperature (OCT) compound and 25-μM-thick cryostat sections were prepared. Brain sections were permeabilized in 0.2% Triton and 1× PBS for 10 min and incubated for 1 h in hybridization buffer (50% formamide, 10% dextran sulfate, 0.1 mg/mL yeast tRNA, 2× saline-sodium citrate (SSC), and 50 mM sodium phosphate) at 55°C. Hybridization was performed overnight with 40 nm TYE563-(C4G2)3 LNA probe in hybridization buffer at 55°C. Brain sections were then washed once with 40% formamide and 1× SSC for 30 min at 55°C, twice in 2× SSC and 0.1% Tween-20 at room temperature for 5 min, and 3 times in 0.1× SSC for 10 min at room temperature. The slides were mounted with ProLong Gold Antifade Mountant with DAPI (Invitrogen) and visualized using a Leica DM2500 fluorescence microscope.
Statistical Analysis
Data were analyzed using Student’s t test or ordinary one-way ANOVA to determine statistical significances. The p values are represented by the following: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Author Contributions
Conceptualization, P.K and R.M.; Investigation, R.M., J.M.L., A.G.-O., and J.M.; Resources, A.G.-O., M.C.-T., M.E., and S.U.; Supervision, Formal Analysis, Visualization, and Writing – Initial Draft, P.K., R.M., M.M.E., and S.J.v.D.; Project Administration and Writing – Review and Editing, P.K., S.J.v.D., and H.P.; Funding Acquisition, P.K.
Acknowledgments
The authors would like to thank Ellen Broug and Eileen Sawyer for reviewing the manuscript.
Footnotes
Supplemental Information includes three figures and one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2019.02.001.
Contributor Information
Raygene Martier, Email: r.martier@uniqure.com.
Pavlina Konstantinova, Email: p.konstantinova@uniqure.com.
Supplemental Information
References
- 1.Geser F., Lee V.M.Y., Trojanowski J.Q. Amyotrophic lateral sclerosis and frontotemporal lobar degeneration: a spectrum of TDP-43 proteinopathies. Neuropathology. 2010;30:103–112. doi: 10.1111/j.1440-1789.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrari R., Kapogiannis D., Huey E.D., Momeni P. FTD and ALS: a tale of two diseases. Curr. Alzheimer Res. 2011;8:273–294. doi: 10.2174/156720511795563700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liscic R.M., Grinberg L.T., Zidar J., Gitcho M.A., Cairns N.J. ALS and FTLD: two faces of TDP-43 proteinopathy. Eur. J. Neurol. 2008;15:772–780. doi: 10.1111/j.1468-1331.2008.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renton A.E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., ITALSGEN Consortium A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heutink P., Jansen I.E., Lynes E.M. C9orf72; abnormal RNA expression is the key. Exp. Neurol. 2014;262(Pt B):102–110. doi: 10.1016/j.expneurol.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 7.Gendron T.F., Belzil V.V., Zhang Y.J., Petrucelli L. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014;127:359–376. doi: 10.1007/s00401-013-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling S.C., Polymenidou M., Cleveland D.W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y., Lin S., Staats K.A., Li Y., Chang W.-H., Hung S.-T., Hendricks E., Linares G.R., Wang Y., Son E.Y. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat. Med. 2018;24:313–325. doi: 10.1038/nm.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J., Hu J., Ludlow A.T., Pham J.T., Shay J.W., Rothstein J.D., Corey D.R. c9orf72 Disease-Related Foci Are Each Composed of One Mutant Expanded Repeat RNA. Cell Chem. Biol. 2017;24:141–148. doi: 10.1016/j.chembiol.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper-Knock J., Walsh M.J., Higginbottom A., Robin Highley J., Dickman M.J., Edbauer D., Ince P.G., Wharton S.B., Wilson S.A., Kirby J. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain. 2014;137:2040–2051. doi: 10.1093/brain/awu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zu T., Liu Y., Bañez-Coronel M., Reid T., Pletnikova O., Lewis J., Miller T.M., Harms M.B., Falchook A.E., Subramony S.H. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. USA. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wojciechowska M., Krzyzosiak W.J. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagier-Tourenne C., Baughn M., Rigo F., Sun S., Liu P., Li H.-R., Jiang J., Watt A.T., Chun S., Katz M. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. USA. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freibaum B.D., Taylor J.P. The Role of Dipeptide Repeats in C9ORF72-Related ALS-FTD. Front. Mol. Neurosci. 2017;10:35. doi: 10.3389/fnmol.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ash P.E.A., Bieniek K.F., Gendron T.F., Caulfield T., Lin W.L., Dejesus-Hernandez M., van Blitterswijk M.M., Jansen-West K., Paul J.W., 3rd, Rademakers R. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jovičić A., Mertens J., Boeynaems S., Bogaert E., Chai N., Yamada S.B., Paul J.W., 3rd, Sun S., Herdy J.R., Bieri G. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K., Donnelly C.J., Haeusler A.R., Grima J.C., Machamer J.B., Steinwald P., Daley E.L., Miller S.J., Cunningham K.M., Vidensky S. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scaber J., Talbot K. What is the role of TDP-43 in C9orf72-related amyotrophic lateral sclerosis and frontemporal dementia? Brain. 2016;139:3057–3059. doi: 10.1093/brain/aww264. [DOI] [PubMed] [Google Scholar]
- 20.Gijselinck I., Van Langenhove T., van der Zee J., Sleegers K., Philtjens S., Kleinberger G., Janssens J., Bettens K., Van Cauwenberghe C., Pereson S. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 21.Al-Sarraj S., King A., Troakes C., Smith B., Maekawa S., Bodi I., Rogelj B., Al-Chalabi A., Hortobágyi T., Shaw C.E. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- 22.Brettschneider J., Van Deerlin V.M., Robinson J.L., Kwong L., Lee E.B., Ali Y.O., Safren N., Monteiro M.J., Toledo J.B., Elman L. Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol. 2012;123:825–839. doi: 10.1007/s00401-012-0970-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y., Pattamatta A., Zu T., Reid T., Bardhi O., Borchelt D.R., Yachnis A.T., Ranum L.P. C9orf72 BAC Mouse Model with Motor Deficits and Neurodegenerative Features of ALS/FTD. Neuron. 2016;90:521–534. doi: 10.1016/j.neuron.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Jiang J., Zhu Q., Gendron T.F., Saberi S., McAlonis-Downes M., Seelman A., Stauffer J.E., Jafar-Nejad P., Drenner K., Schulte D. Gain of Toxicity from ALS/FTD-Linked Repeat Expansions in C9ORF72 Is Alleviated by Antisense Oligonucleotides Targeting GGGGCC-Containing RNAs. Neuron. 2016;90:535–550. doi: 10.1016/j.neuron.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gendron T.F., Chew J., Stankowski J.N., Hayes L.R., Zhang Y.J., Prudencio M., Carlomagno Y., Daughrity L.M., Jansen-West K., Perkerson E.A. Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Sci. Transl. Med. 2017;9:eaai7866. doi: 10.1126/scitranslmed.aai7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu J., Rigo F., Prakash T.P., Corey D.R. Recognition of c9orf72 Mutant RNA by Single-Stranded Silencing RNAs. Nucleic Acid Ther. 2017;27:87–94. doi: 10.1089/nat.2016.0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu J., Liu J., Li L., Gagnon K.T., Corey D.R. Engineering Duplex RNAs for Challenging Targets: Recognition of GGGGCC/CCCCGG Repeats at the ALS/FTD C9orf72 Locus. Chem. Biol. 2015;22:1505–1511. doi: 10.1016/j.chembiol.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graves P., Zeng Y. Biogenesis of mammalian microRNAs: a global view. Genomics Proteomics Bioinformatics. 2012;10:239–245. doi: 10.1016/j.gpb.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martier R., Liefhebber J.M., Miniarikova J., van der Zon T., Snapper J., Kolder I., Petry H., van Deventer S.J., Evers M.M., Konstantinova P. Artificial microRNAs targeting C9ORF72 have the potential to reduce accumulation of the intra-nuclear transcripts in ALS and FTD patients. Mol. Ther. Nucleic Acids. 2019 doi: 10.1016/j.omtn.2019.01.010. Published online January 30, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Rourke J.G., Bogdanik L., Muhammad A.K.M.G., Gendron T.F., Kim K.J., Austin A., Cady J., Liu E.Y., Zarrow J., Grant S. C9orf72 BAC Transgenic Mice Display Typical Pathologic Features of ALS/FTD. Neuron. 2015;88:892–901. doi: 10.1016/j.neuron.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S., Huang E.J. Modeling ALS and FTD with iPSC-derived neurons. Brain Res. 2017;1656:88–97. doi: 10.1016/j.brainres.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer K., Ferraiuolo L., Miranda C.J., Likhite S., McElroy S., Renusch S., Ditsworth D., Lagier-Tourenne C., Smith R.A., Ravits J. Direct conversion of patient fibroblasts demonstrates non-cell autonomous toxicity of astrocytes to motor neurons in familial and sporadic ALS. Proc. Natl. Acad. Sci. USA. 2014;111:829–832. doi: 10.1073/pnas.1314085111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Giorgio F.P., Boulting G.L., Bobrowicz S., Eggan K.C. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 34.Haidet-Phillips A.M., Hester M.E., Miranda C.J., Meyer K., Braun L., Frakes A., Song S., Likhite S., Murtha M.J., Foust K.D. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat. Biotechnol. 2011;29:824–828. doi: 10.1038/nbt.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ilieva H., Polymenidou M., Cleveland D.W. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J. Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selvaraj B.T., Livesey M.R., Chandran S. Modeling the C9ORF72 repeat expansion mutation using human induced pluripotent stem cells. Brain Pathol. 2017;27:518–524. doi: 10.1111/bpa.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters O.M., Cabrera G.T., Tran H., Gendron T.F., McKeon J.E., Metterville J., Weiss A., Wightman N., Salameh J., Kim J. Human C9ORF72 Hexanucleotide Expansion Reproduces RNA Foci and Dipeptide Repeat Proteins but Not Neurodegeneration in BAC Transgenic Mice. Neuron. 2015;88:902–909. doi: 10.1016/j.neuron.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y., Kim M., Han J., Yeom K.-H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim V.N., Nam J.W. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 41.Tyzack G., Lakatos A., Patani R. Human Stem Cell-Derived Astrocytes: Specification and Relevance for Neurological Disorders. Curr. Stem Cell Rep. 2016;2:236–247. doi: 10.1007/s40778-016-0049-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madill M., McDonagh K., Ma J., Vajda A., McLoughlin P., O’Brien T., Hardiman O., Shen S. Amyotrophic lateral sclerosis patient iPSC-derived astrocytes impair autophagy via non-cell autonomous mechanisms. Mol. Brain. 2017;10:22. doi: 10.1186/s13041-017-0300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Z., Poidevin M., Li X., Li Y., Shu L., Nelson D.L., Li H., Hales C.M., Gearing M., Wingo T.S., Jin P. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc. Natl. Acad. Sci. USA. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A.J., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Blitterswijk M., Gendron T.F., Baker M.C., DeJesus-Hernandez M., Finch N.A., Brown P.H., Daughrity L.M., Murray M.E., Heckman M.G., Jiang J. Novel clinical associations with specific C9ORF72 transcripts in patients with repeat expansions in C9ORF72. Acta Neuropathol. 2015;130:863–876. doi: 10.1007/s00401-015-1480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donnelly C.J., Zhang P.W., Pham J.T., Haeusler A.R., Mistry N.A., Vidensky S., Daley E.L., Poth E.M., Hoover B., Fines D.M. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rizzu P., Blauwendraat C., Heetveld S., Lynes E.M., Castillo-Lizardo M., Dhingra A., Pyz E., Hobert M., Synofzik M., Simón-Sánchez J. C9orf72 is differentially expressed in the central nervous system and myeloid cells and consistently reduced in C9orf72, MAPT and GRN mutation carriers. Acta Neuropathol. Commun. 2016;4:37. doi: 10.1186/s40478-016-0306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dodge J.C., Treleaven C.M., Fidler J.A., Hester M., Haidet A., Handy C., Rao M., Eagle A., Matthews J.C., Taksir T.V. AAV4-mediated expression of IGF-1 and VEGF within cellular components of the ventricular system improves survival outcome in familial ALS mice. Mol. Ther. 2010;18:2075–2084. doi: 10.1038/mt.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koppers M., Blokhuis A.M., Westeneng H.J., Terpstra M.L., Zundel C.A.C., Vieira de Sá R., Schellevis R.D., Waite A.J., Blake D.J., Veldink J.H. C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann. Neurol. 2015;78:426–438. doi: 10.1002/ana.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sudria-Lopez E., Koppers M., de Wit M., van der Meer C., Westeneng H.J., Zundel C.A.C., Youssef S.A., Harkema L., de Bruin A., Veldink J.H. Full ablation of C9orf72 in mice causes immune system-related pathology and neoplastic events but no motor neuron defects. Acta Neuropathol. 2016;132:145–147. doi: 10.1007/s00401-016-1581-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Rourke J.G., Bogdanik L., Yáñez A., Lall D., Wolf A.J., Muhammad A.K., Ho R., Carmona S., Vit J.P., Zarrow J. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351:1324–1329. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niblock M., Smith B.N., Lee Y.-B., Sardone V., Topp S., Troakes C., Al-Sarraj S., Leblond C.S., Dion P.A., Rouleau G.A. Retention of hexanucleotide repeat-containing intron in C9orf72 mRNA: implications for the pathogenesis of ALS/FTD. Acta Neuropathol. Commun. 2016;4:18. doi: 10.1186/s40478-016-0289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fratta P., Mizielinska S., Nicoll A.J., Zloh M., Fisher E.M.C., Parkinson G., Isaacs A.M. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reddy K., Zamiri B., Stanley S.Y.R., Macgregor R.B., Jr., Pearson C.E. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 2013;288:9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 56.Su Z., Zhang Y., Gendron T.F., Bauer P.O., Chew J., Yang W.Y., Fostvedt E., Jansen-West K., Belzil V.V., Desaro P. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83:1043–1050. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nordin A., Akimoto C., Wuolikainen A., Alstermark H., Forsberg K., Baumann P., Pinto S., de Carvalho M., Hübers A., Nordin F. Sequence variations in C9orf72 downstream of the hexanucleotide repeat region and its effect on repeat-primed PCR interpretation: a large multinational screening study. Amyotroph. Lateral Scler. Frontotemporal Degener. 2017;18:256–264. doi: 10.1080/21678421.2016.1262423. [DOI] [PubMed] [Google Scholar]
- 58.Prudencio M., Belzil V.V., Batra R., Ross C.A., Gendron T.F., Pregent L.J., Murray M.E., Overstreet K.K., Piazza-Johnston A.E., Desaro P. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 2015;18:1175–1182. doi: 10.1038/nn.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu H., Lei C., He Q., Pan Z., Xiao D., Tao Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer. 2018;17:64. doi: 10.1186/s12943-018-0765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schraivogel D., Meister G. Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem. Sci. 2014;39:420–431. doi: 10.1016/j.tibs.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 62.Miniarikova J., Zanella I., Huseinovic A., van der Zon T., Hanemaaijer E., Martier R., Koornneef A., Southwell A.L., Hayden M.R., van Deventer S.J. Design, Characterization, and Lead Selection of Therapeutic miRNAs Targeting Huntingtin for Development of Gene Therapy for Huntington’s Disease. Mol. Ther. Nucleic Acids. 2016;5:e297. doi: 10.1038/mtna.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pascual-Lucas M., Viana da Silva S., Di Scala M., Garcia-Barroso C., González-Aseguinolaza G., Mulle C., Alberini C.M., Cuadrado-Tejedor M., Garcia-Osta A. Insulin-like growth factor 2 reverses memory and synaptic deficits in APP transgenic mice. EMBO Mol. Med. 2014;6:1246–1262. doi: 10.15252/emmm.201404228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paxinos G., Watson C. Academic Press; 1998. The Rat Brain in Stereotaxic Coordinates. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.