Abstract
The diverse structures and multifaceted roles of carotenoids make these colorful pigments attractive targets for synthetic biology.
Carotenoids are structurally diverse pigments and related derivatives that mediate photosynthetic function, responses to biotic and abiotic signals, and control plant architecture. It is these multifaceted roles that make carotenoids uniquely attractive as synthetic biology targets when considering ways to alter plant form and function to meet the needs of food security or new agricultural and industrial applications. This Update seeks to explore how synthetic biology might capitalize on the recent advances made in the carotenoid field. Opportunities are discussed along with the research needed to drive the carotenoid synthetic biology era forward.
Functional Diversity of Carotenoids
Carotenoids, a large class of lipophilic compounds, bestow fruits and flowers with beautiful yellow, orange, and red hues attributable to the conjugated double bond chromophore of these pigments (Britton et al., 2004). Carotenoids include β-carotene, which colors carrots orange and is a natural food colorant and provitamin, lycopene which colors tomato fruits red, astaxanthin, the red pigment found in flowers and algae which serves as a valuable food additive for the fish industry, and the commercially valuable red carotenoid pigments of saffron, to name a few. Plant carotenoids and their derivatives are also important to other species that consume plants. Certain carotenoids are enzymatically converted into visual pigments and signals needed to control development while other carotenoids and their derivatives (apocarotenoids) contribute to eye health, cardiovascular health, function of the immune system, cognitive function, antioxidant activity, general fitness and interspecies communication (Thomas et al., 2014; Murillo and Fernandez, 2016; Rivera-Madrid et al., 2016; de Carvalho and Caramujo, 2017; Rubin et al., 2017; Toews et al., 2017; Zhao and Nabity, 2017; Eggersdorfer and Wyss, 2018; Harrison and Quadro, 2018; Saint et al., 2018). However, carotenoids in plants are far more than colorful pigments. A major role of carotenoids in plants is the contribution to light capture in photosynthesis and photoprotection under high light conditions (Polívka and Frank, 2010; Dall’Osto et al., 2012; Qin et al., 2015; Hashimoto et al., 2016; Leonelli et al., 2017; Emiliani et al., 2018). Without carotenoids in photosynthetic tissue, plants do not survive outside of the laboratory (Robertson, 1955; Anderson and Robertson, 1960; Matthews et al., 2003). Carotenoids are also chemical signals that attract pollinators that are essential for plant reproduction (Bradshaw and Schemske, 2003). Specific apocarotenoids (products of enzymatic cleavage of carotenoids) serve as signaling molecules (Hou et al., 2016). These include internal signals that mediate stress responses and developmental programs and external signals for communication with other organisms.
The study of carotenoids from chemical structure elucidation to molecular biology has been ongoing for over 120 years (Britton et al., 2017). The chemistry and biochemistry of carotenoids and carotenoid enzymes has been recently reviewed (Moise et al., 2014; Rodriguez-Concepcion et al., 2018). Chemical structural variation is the foundation for carotenoid functional diversity in plants and in other species that respond to plant carotenoids and their derivatives for nutritional or symbiotic benefit. It is the manifold roles of carotenoids that make them uniquely attractive for synthetic biology when considering ways to alter plant form and function to meet the needs of food security or new agricultural and industrial applications. Here, we will focus on novel ideas, open questions and research needed to drive opportunities (Box 1) in synthetic biology.
Redesigning Plant Membranes
Typical carotenoid structures are forty carbons in length with a central conjugated double bond system (Britton et al., 2004). The conjugated double bonds absorb light and are responsible for the unique colors, spectral properties, and biochemical activities. The Plant Kingdom is known to produce hundreds of different carotenoid structures including geometrical isomers (cis/trans or Z/E), carotenoids with different end-groups and modifications, and altered positions of double bonds. Carotenoid chemical structures differ widely in terms of shape, biophysical properties, and how the molecules also interact and alter the physical properties of plant membranes where hydrophobic carotenoids are localized. For example, various carotenoid structures uniquely impact membrane permeability, membrane fluidity, and diffusion of small molecules across artificial membranes (Gruszecki and Strzałka, 2005). It is possible to create many new types of carotenoids given the wealth of available carotenoid genes and encoded enzymes coupled with the ability to create novel carotenoids both with and without further added moieties (e.g. esters, sugars, etc.). Therefore, a better understanding of how specific carotenoids affect membrane biophysical properties could lead to novel synthetic plant membranes for a multitude of purposes, including the improvement of nutrient and oxygen uptake in roots or creation of more efficient barriers against phytopathogens.
Synthetic Photoprotection Systems
Variation in carotenoid structures not only influences their spectral properties or color but can also influence biological activities including energy transfer, photoprotection and quenching of damaging singlet oxygen in plants and bioactivities in humans and other organisms. Binding of a carotenoid to a protein will also alter the carotenoid spectral properties and bioactivity. A classic example is the astaxanthin-binding carotenoprotein, alpha-crustacyanin, which imparts a blue color to lobster (Homarus gammarus). A boiled lobster turns red as the red carotenoid is released upon denaturation of the carotenoprotein (Weesie et al., 1995). The cyanobacterial orange carotenoid protein mediates photoprotection by relying on protein-carotenoid interactions that exhibit unique spectral states associated with the photoprotection mechanism (Maksimov et al., 2017). Depending upon light intensity, photosynthetic bacteria are known to control the carotenoid pigment-protein composition to alter the spectral characteristics and efficiency of energy transfer (Magdaong et al., 2014). These observations point out the prospects of improving energy transfer or photoprotection in plants by engineering bioenergetic changes in carotenoids and caroteno-protein complexes. Simply changing the suite of photoprotective carotenoids can improve plant responses to high light stress, as seen when the xanthophyll cycle pigment composition was altered (Leonelli et al., 2017). The unusual features of carotenoids represent an opportunity for creating synthetic systems based on novel carotenoids to further improve photoprotection and efficiency of energy capture. Molecular dynamics simulations currently being applied to investigate protein-carotenoid interactions (Daskalakis, 2018) will be valuable in designing optimal interactions between photosystem proteins and nonnative carotenoids. Photosynthetic bacteria are good test beds to build and test features of this system (Nagashima et al., 2017; Yukihira et al., 2017).
Enroute to “Plug and Play” Metabolic Engineering of Carotenoids
Carotenoid biosynthetic enzymes and genes have been identified over the last thirty years or so (Moise et al., 2014; Rodriguez-Concepcion et al., 2018) and earlier genetic studies and mutants from the 1940s and later provide an important resource for gene discovery (Wurtzel et al., 2012). Today, there is an enormous collection of genes for biosynthesis of a wide array of carotenoids and carotenoid cleavage products. This extensive toolbox is comprised of skeleton enzymes and genes derived from diverse organisms that biosynthesize carotenoids, including plants, bacteria, fungi, and aphids (Zhao and Nabity, 2017). Another novel source of enzymes can be found in organisms that do not biosynthesize carotenoids but instead process dietary carotenoids for health and species fitness (Toews et al., 2017). The first efforts to metabolically engineer the pathway in plants began with the humanitarian effort to alleviate vitamin A deficiency with the creation of high provitamin A crops (Burkhardt et al., 1997; Ye et al., 2000). The accumulation of the provitamin A β-carotene in rice (Oryza sativa) seed endosperm was not predicted given the three genes introduced. This surprising result could be explained by unanticipated endogenous enzyme activities (Schaub et al., 2005). In fact, any synthetic biology approach will be limited by natural genetic variation, as described for carotenoids (Harjes et al., 2008; Vallabhaneni et al., 2009, 2010; Vallabhaneni and Wurtzel, 2009; Lee et al., 2012; Schulz et al., 2016; Sulli et al., 2017; Price et al., 2018; Teixeira da Silva et al., 2018). Nonetheless, metabolic engineering of the pathway has been demonstrated for many crops (Giuliano, 2017; Majer et al., 2017; Nogueira et al., 2017, 2018; Camagna et al., 2018; McQuinn et al., 2018; Sankari et al., 2018). These strategies have also included the successful application of gene editing technology (Dahan-Meir et al., 2018) which in the future could be used to incorporate favorable allelic variation from wild species. There is also the possibility of generating new plant sources of carotenoids. For example, cotton (Gossypium sp.) is well known for its valuable fiber. Cotton seed is also a good potential source of protein and oil, but at present the seed is not suitable for human and monogastric animal consumption because of the toxic gossypol present in the seed. Recent engineering of carotenoid accumulation in cotton seed (Yao et al., 2018) can now be combined with the engineered seed where gossypol biosynthesis has been inhibited (Zhou et al., 2013), thus making the cotton seed a valuable and new food source in a crop that is already supported by an active commercial pipeline.
Current metabolic engineering strategies to modify plant carotenoids rely on controlling the overexpression of known enzymes/components by use of constitutive promoters to control expression. What synthetic biology can bring are tunable promoters that activate/deactivate in response to external signals to control the expression of individual rate-controlling pathway enzymes (Kassaw et al., 2018). Multiplexed pathways could be combined in a synthetic biological circuit with other needed parts for tuning responses to biotic and abiotic cues and incorporating other emerging technologies (Wurtzel and Kutchan, 2016). “Toggle switches” could be used to optimize expression of all the rate-controlling enzyme and nonenzyme components needed for pathway function. If a plant was channeling isoprenoid substrates to competing pathways because stress programs were induced, then reduced isoprenoid pools could be sensed through artificial sensors that would signal control circuits to ramp up isoprenoid precursors. Synthetic biology could be used to replace native metabolons with new enzymes and metabolons built to be more efficient, structurally unique from the endogenous “parts,” and not restricted by associations with native complexes. Such artificial pathway components could then be usable as “parts” across genotypes and species, thus eliminating the interference caused by genetic diversity. Other options for synthetic biology might include the engineering in limited tissues or cell types so that the altered pathway does not interfere with photosynthetic function and requirements for carotenoids. “Toggle switches” could be introduced for manufacturing certain carotenoids needed to attract beneficial organisms; once the associated organism was no longer needed, the switch would turn off and the specific carotenoid production would cease. These examples necessitate synthetic biology parts and bring metabolic engineering to a new level of nuanced control that could serve diverse plants and crops. However, further research at many levels is needed to accomplish “plug and play” predictability for simultaneous engineering of multiple metabolic targets in an unlimited array of plant genotypes and species. With deeper understanding of the multilevel regulation of carotenoid biosynthesis, sequestration and trafficking, it will be feasible to imagine novel and synthetic systems that build on carotenoids.
Carotenoid Biosynthesis in Brief
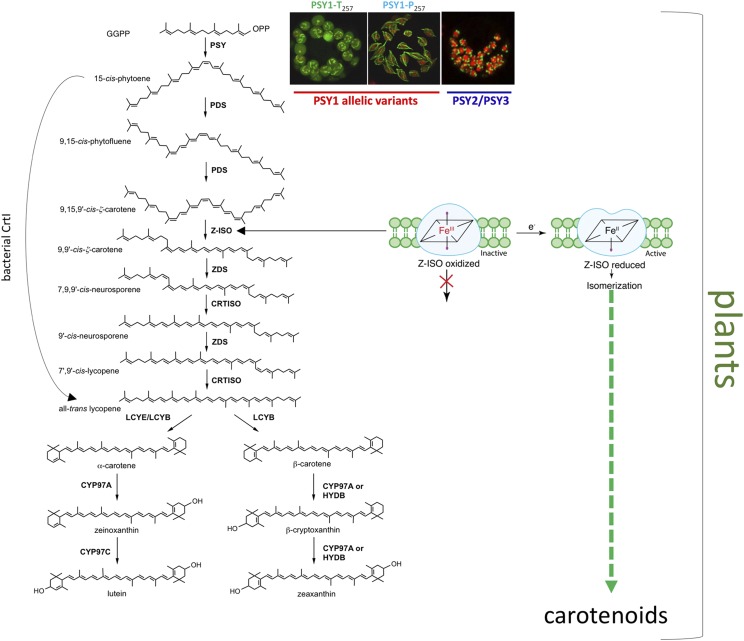
The carotenoid biosynthetic pathway in plants is mediated by nuclear-encoded enzymes which are imported into plastids of varying types (Moise et al., 2014; Rodriguez-Concepcion et al., 2018; Sun et al., 2018). Carotenoids are biosynthesized from five-carbon isoprenoids which are combined to create the first pathway compound, the 40-carbon, 15-cis-phytoene (Fig. 1). With only three conjugated double bonds, phytoene is colorless, but as in all carotenoids, the compound possesses a unique absorption spectrum which is characteristic for identification and contributes to bioactivity (Britton et al., 2004). The later biosynthetic steps leading from 15-cis-phytoene include catalysis of multiple desaturations and isomerizations to extend the conjugated double bond system which confers color and unique spectra to the various intermediates and pathway products. The core portion of the pathway leading to all-translycopene is common to all plants. Later steps include ring closure to produce carotenes, and further modifications to create xanthophylls, keto-carotenoids, and so on.
Figure 1.
Simplified plant carotenoid biosynthetic pathway to zeaxanthin and lutein. The major question of metabolon organization and assembly is highlighted by the inset showing varying localization of maize (Zea mays) phytoene synthase (PSY) allelic variants and isozymes fused to GFP and introduced into chloroplasts marked by red chlorophyll autofluorescence (adapted from Shumskaya et al., 2012). Formation of lutein actually requires the synergistic interactions of CYP97A and CYP97C (Quinlan et al., 2012). Steps catalyzed by the bacterial enzyme, CrtI, are also indicated and correspond to the four steps catalyzed by the plant enzymes PDS, Z-ISO, ZDS, and CRTISO. These reactions also require an electron transfer chain and plastoquinones to channel electrons/protons produced during desaturation steps. Redox states of the Z-ISO heme iron depict posttranslational regulation of the pathway. Redox changes cause conformational changes in Z-ISO leading to a switch of the heme ligands and activation/deactivation of Z-ISO. When the heme iron is oxidized, the pathway is interrupted whereas when the heme iron is reduced, Z-ISO is active and the pathway progresses. “Z-ISO oxidized,” refers to the Z-ISO heme iron in the oxidized Fe+3 state; “Z-ISO reduced,” refers to the Z-ISO heme iron in the reduced Fe+2 state. CrtI, bacterial phytoene desaturase. Plant enzymes: PSY, phytoene synthase; PDS, phytoene desaturase; Z-ISO, 15-cis-ζ-carotene isomerase; ZDS, ζ-carotene desaturase; CRTISO, carotenoid isomerase; LCYE, lycopene ε-cyclase; LCYB, lycopene β-cyclase; CYP97A, cytochrome P450 97A (P450 type β-ring hydroxylase); CYP97C, cytochrome P450 97C (P450 type ε-ring hydroxylase); HYDB, β-ring hydroxylase (nonheme diiron type). For more details related to this figure, see Moise et al. (2014) and Beltran et al. (2015).
Regulation of the Biosynthetic Pathway at Multiple Levels
The structural genes of the biosynthetic pathway are under transcriptional control as evidenced by the correlation of transcripts with carotenoid content or composition, whether by natural genetic variation (Harjes et al., 2008; Vallabhaneni et al., 2009; Vallabhaneni and Wurtzel, 2009), or by transgenic overexpression (Burkhardt et al., 1997; Ye et al., 2000; Giuliano, 2017). Thus, the pathway is amenable to metabolic engineering by overexpression of genes controlling flux into the pathway (Giuliano, 2014, 2017) or by controlling the transcript levels of other genes that lead to specific pathway intermediates (Harjes et al., 2008; Vallabhaneni et al., 2009). These successes have inspired efforts to discover transcription factors that efficiently coordinate expression of the entire pathway. However, an important point to consider in searching for carotenoid-specific transcription factors is that many plants cultivated today have been bred with mutations that are not representative of the ancestral state. For example, although the yellow endosperm allele of maize (Zea mays) is generally considered dominant to the white allele, the ancestral state is white endosperm as found in teosinte, the wild progenitor (Palaisa et al., 2003, 2004). Another example is that of carrot (Daucus carota) root for which commercial varieties are typically orange, whereas the wild progenitor has a white root. Recent genome analysis suggests that the orange phenotype is due to a photomorphogenic mutation that has been widely bred into carrots (Iorizzo et al., 2016; Llorente et al., 2017). Therefore, these breeding alleles may actually conceal unknown physiological roles of carotenoids that link developmental cues with other complex metabolic processes. In addition, the primary allele used for breeding the carotenoid trait may have undergone regulatory changes as in the case of yellow endosperm (Li et al., 2009). There are other examples where crop domestication has led to loss of certain biochemical features, such as apocarotenoids no longer detected in tomato (Solanum lycopersicum) as compared to wild species (Tieman et al., 2017). A number of transcription factors that modulate transcriptional levels of the pathway structural genes have been identified (Welsch et al., 2007; Ampomah-Dwamena et al., 2018; Lu et al., 2018; Xiong et al., 2019). However, these transcription factors coregulate many other noncarotenogenic genes to coordinate networks related to carotenoid function (e.g. tomato fruit development, photosynthesis, abscisic acid responses; Meier et al., 2011; Su et al., 2015). It is therefore no surprise that carotenoid-specific regulatory mutants were never identified among the extensive collection of maize carotenoid mutants (Wurtzel et al., 2012). The complex coregulation of carotenoids with other metabolic processes explains in part why engineering through transcriptional over-expression leads to unexpected off-target results that were revealed when transgenic plants were subjected to nontargeted metabolomic analyses (Decourcelle et al., 2015) or global transcriptomic analysis (Lu et al., 2018). Therefore, more effective targeted control of the carotenoid pathway might necessitate creation of artificial transcription factors and cognate promoters.
In addition to transcriptional activation, there likely exists more nuanced posttranscriptional and posttranslational control to accommodate the constantly changing environment of plants which demands varied carotenoid resources. Pathway engineering might alternatively utilize knowledge related to these posttranscriptional phenomena, for which evidence is growing. Phytoene synthase (PSY) is well-recognized for controlling pathway flux and as such has been a major target for metabolic engineering through transcriptional overexpression (Giuliano, 2017). PSY expression is also controlled through feedback regulation derived from carotenoid pathway products (Kachanovsky et al., 2012; Enfissi et al., 2017). While PSY is under transcriptional regulation, there is mounting evidence for its posttranscriptional regulation. PSY, the first committed step of the pathway, is encoded by a multigene family in all of the major food crops of the Poaceae (grass family; Li et al., 2008a, 2008b). The different isozymes were shown to target to unique locations within the chloroplast (Shumskaya et al., 2012), either to plastoglobules known to be associated with thylakoid membranes (van Wijk and Kessler, 2017) or to the envelope membrane and stroma (Fig. 1). PSY in apple (Malus x domestica) is also represented by multiple isozymes which were targeted to different suborganellar locations (Ampomah-Dwamena et al., 2015). Variant localization may alter enzyme activity as suggested by several studies on PSY. Daffodil (Narcissus sp.) PSY was shown to be active only when associated with membranes created with plastid-type galactolipids but inactive as a soluble protein or if the membranes were formed of phospholipids (Schledz et al., 1996). Etiolated seedlings of Arabidopsis thaliana mutants with reduced digalactosyldiacylglycerol showed a reduction of total carotenoids, with little effect on carotenoid composition, which would be consistent with the requirement of galactolipids for PSY activity (Fujii et al., 2018). When PSY was overexpressed in Arabidopsis, enzyme activity was only associated with the membrane-bound form and not the stromal-localized form (Lätari et al., 2015). In white mustard (Synapis alba) seedlings, as light-induced thylakoid membrane formation occurred concomitantly with photomorphogenesis, PSY relocalized from the stroma, where the enzyme was inactive, to thylakoid membranes, where PSY became activated (Welsch et al., 2000). These observations reveal the dynamic state of pathway organization and raise the question of how PSY intraplastidial targeting is controlled to alter the activity of PSY and pathway function. It is conceivable that nonenzyme factors are involved in controlling enzyme localization and stability within the plastid. For example, stability of Arabidopsis PSY, encoded by a single copy gene, was shown to be controlled through opposite actions of the stabilizing Or protein (Zhou et al., 2015) and degradative Clp protease (Welsch et al., 2018). Plants with favorable alleles of the Or gene, exhibited increased levels of carotenoids (Chayut et al., 2017; Morikawa et al., 2018; Yazdani et al., 2019). The example of engineering with the Or protein shows that the metabolic engineering of carotenoids can be achieved without modifying expression of the pathway structural genes. Elucidation of the complete suite of factors that control pathway assembly and function are needed to advance the predictability of carotenoid metabolic engineering.
Insight into Post-Translational Control
The complexity of carotenoid pathway regulation transcends the first biosynthetic step which becomes obvious in evolutionary analysis of the following steps that occur both in plants and in carotenogenic bacteria (see Fig. 1). What is intriguing, is that bacteria biosynthesize all-translycopene with a single enzyme. In contrast, plants utilize two desaturases and two isomerases which yield a complex array of geometrical isomers postulated to play a role in feed-back regulation, retrograde signaling from plastid to nucleus (Chan et al., 2016), and serving as putative substrates for signaling apocarotenoids (Avendaño-Vázquez et al., 2014). Early attempts to modify the pathway in rice and other species utilized the bacterial enzyme, CrtI, which is a FAD (FAD)-dependent desaturase (Schaub et al., 2012). However, it is unknown how CrI integrates into the multienzyme plant pathway, nor whether engineering with CrtI is potentially less efficient because of some unknown interference with the endogenous signaling network (Enfissi et al., 2010, 2017; Nogueira et al., 2013). The four plant enzymes include three FAD proteins, phytoene desaturase (PDS), zetacarotene desaturase (ZDS), and carotenoid isomerase (CRTISO), and a newly identified heme protein,15-cis-zetacarotene isomerase (Z-ISO; Li et al., 2007; Chen et al., 2010; Beltrán et al., 2015). Z-ISO shares ancestry with an obscure protein in noncarotenogenic denitrifying bacteria and utilizes the heme to mediate double-bond isomerization within a membrane. Z-ISO catalyzes isomerization of the central 15-15’ double-bond from cis to trans by using a novel mechanism yet to be described for any heme protein (Fig. 1; Beltrán et al., 2015). Most notably, this heme is regulated through the redox state of the heme iron, suggesting that Z-ISO activity in plants is posttranslationally regulated through plastid redox status. The transient inhibition of Z-ISO activity is predicted to lead to accumulation of 9,15’,9’-tricis-zetacarotene, which might be involved in signaling and the bent shape of this geometric isomer would be expected to behave differently within the membrane as compared to the linear-shaped Z-ISO product, 9,9’-di-cis-zetacarotene. Lastly, the Z-ISO heme cofactor may be the missing link that connects stress responses to nitric oxide by controlling flux through novel geometrical isomers in the carotenoid biosynthetic pathway (Beltrán et al., 2015). Although understanding of posttranslational regulation of the pathway is limited, this is an area of research that deserves further attention with a potential for pathway redesign.
Solving Structural Mysteries of the Biosynthetic Machinery
Engineering carotenoids through better understanding of posttranslational control, including pathway assembly, may be a more viable approach to manipulation of the pathway for strategic endpoints. It is this latter approach that will require much further research in understanding the mechanisms controlling posttranslational regulation of the biosynthetic pathway, carotenoid trafficking and carotenoid sequestration. To date, we know only that the biosynthetic pathway enzymes are widely distributed within plastids and that the distribution of enzymes depends on plastid type (e.g. chloroplast, chromoplast; Shumskaya and Wurtzel, 2013; Sun et al., 2018). We have an incomplete picture of how the enzymes might interact to create a fully functioning metabolic pathway multienzyme complex. Clearly, there are oligomeric carotenoid protein complexes as these have been observed, and there is evidence for substrate channeling as would be expected of a multienzyme complex (Quinlan et al., 2012; Shumskaya and Wurtzel, 2013; Beltrán et al., 2015; Lundquist et al., 2017). How is this oligomeric complex assembled? Z-ISO is encoded by a single copy gene, and the gene product is an integral membrane protein as compared to PSY, PDS, and ZDS, which are peripheral membrane proteins. How are pathway components recruited when PSY might be located in multiple sites and Z-ISO is in a fixed position? These reactions also require an electron transfer chain and plastoquinones to channel electrons/protons produced during the desaturation steps. These auxiliary components must be physically integrated into the pathway scheme, with the anticipation that different plastid forms (Sun et al., 2018) will have unique membrane architectures available for this purpose. Any efforts for metabolic engineering of the carotenoid pathway or new and improved versions will require a detailed understanding of enzyme structures and organization of the fully functional complex. It is reasonable to assume that this complex may change form and location depending upon the plastid type and carotenoid endpoint function. For example, in chloroplasts, carotenoids are directed to thylakoid-localized photosystems, or alternatively trafficked by an unknown mechanism passing through the double membrane to reach the exterior of the plastid for cleavage into apocarotenoids (Demurtas et al., 2018). These disparate processes likely require distinct topological conformations of the pathway given the variant endpoints of carotenoids. In addition, the function of the carotenoid pathway is linked to other pathways, such as chlorophyll biosynthesis and plastoquinone biosynthesis (Meier et al., 2011; Hunter et al., 2018), suggesting that enzyme components could be physically linked in particular tissues or under certain physiological conditions. Identification of otherwise elusive pathway controlling components could be facilitated by developing robust computational algorithms based on machine learning (Tzfadia et al., 2012). Discovery of the auxiliary nonstructural genes whose products specifically coordinate the function of the carotenoid pathway, would be valuable in developing metabolic engineering strategies with more predictable “plug and play” features.
Elucidating the functional organization of the multienzyme complex in plants is challenging. Fortunately, reconstruction of the plant carotenoid biosynthetic pathway can be achieved in Escherichia coli or yeast, which both synthesize the isoprenoid precursors (Chen et al., 2010; Pollmann et al., 2017; Ren et al., 2017; Schwartz et al., 2017; Zhang et al., 2018). These heterologous systems are excellent test beds for rapid characterization of carotenoid biosynthetic enzymes of the present and synthetic future, as well as for study of enzyme interactions and biosynthetic complex organization. Carotenogenic bacteria and marine algae have also been used to engineer carotenoid biosynthesis (Wang et al., 2014) and there is also the option of utilizing plant tissue culture systems (Oleszkiewicz et al., 2018; Schaub et al., 2018). One of the challenges for elucidating structures of the pathway enzymes and complexes is that the enzymes are membrane proteins and thus are inherently difficult to obtain ordered crystals which are needed for structure elucidation. However, there are emerging tools, such as cryo-electron microscopy (Cryo-EM), which does not depend upon protein crystallization. Cryo-EM has been used to exquisitely elucidate the photosystem II light harvesting II supercomplex from spinach (Spinacia oleracea) containing proteins, chlorophylls, and carotenoids (Wei et al., 2016). As imaging technology is improving, the current size limitations of Cryo-EM will be less of a problem (Cheng, 2018). This imaging method can computationally discern structural variants to advance our understanding of the structures of the enzymes as well as structures of the functional complexes. Purified proteins, protein complexes, or proteins assembled onto near-native lipid nanodiscs (Efremov et al., 2017; Matthies et al., 2018) can be imaged for structure elucidation. The recent crystal structure determination (Brausemann et al., 2017) for the second enzyme of the pathway, phytoene desaturase, which was found to be active as a homotetramer (Gemmecker et al., 2015), will facilitate structure elucidation of the higher-order oligomeric complexes of the pathway.
Novel Ideas for Carotenoid Sequestration
Metabolic engineering of carotenoids must also take into account mechanisms that promote carotenoid accumulation and sequestration. For example, the transition of the green chloroplast to the red-colored and highly carotenogenic chromoplast is accompanied by de novo biosynthesis and accumulation of carotenoids. After carotenoids are biosynthesized, these hydrophobic pigments generally accumulate in membranes or other lipophilic structures such as plastoglobules (van Wijk and Kessler, 2017). Regulated control of carotenoid sequestration could be used to manipulate downstream bioactivities in the plant. For example, apocarotenoid glycosylation is postulated to inhibit apocarotenoid bioactivity in plants (Lätari et al., 2015). Xanthophylls in pepper (Capsicum annuum) fruit chromoplasts are primarily of the esterified class (Hornero-Méndez and Mínguez-Mosquera, 2000). That is, fatty acids are covalently attached to the terminal hydroxyl groups of xanthophylls. The lipophilic tail appears to facilitate both carotenoid sequestration and chromoplast development. The importance of esterification in carotenoid sequestration and plant development is reflected by the PALE YELLOW PETAL 1 mutant of tomato, which exhibits reduced esterified carotenoids and aberrant chromoplast development due to a lesion in a gene encoding an acyl transferase (Ariizumi et al., 2014). Esterification similarly enhances astaxanthin accumulation in algae (Chen et al., 2015). Carotenoid modification also occurs in nonphotosynthetic organisms, where the esterified moiety may be chemically different from a fatty acid. In naturally carotenogenic bacteria, the covalent attachment of sugars to carotenoids is thought to stabilize carotenoid accumulation by decreasing lipophilicity (e.g. zeaxanthin-p-diglucoside in Erwinia uredovora, known also as Pantoea ananas). Bacterial genes that encode enzymes to catalyze carotenoid glycosylation are known (Misawa et al., 1990) and could be utilized to alter carotenoid deposition in plants. Monitoring such metabolic changes at the single cell level is possible utilizing emerging methods such as confocal raman spectroscopy (Timlin et al., 2017) and MALDI mass spectrometry imaging (MALDI-MSI; Horn et al., 2012; Horn and Chapman, 2014; Dalisay et al., 2015). Further elucidation of the process that links carotenoid modification, sequestration, bioactivity and plastid development in plants, would be valuable in creating new synthetic paradigms for directing and sequestering carotenoids or carotenoid-like structures for nutritional engineering or other novel purposes.
Metabolite Repair
Some carotenoids are interconverted through epoxidase/de-epoxidase activities, such as activities which control the formation of energetically optimal pigments for photoprotection (Leonelli et al., 2017; Leuenberger et al., 2017). Carotenoid epoxides also form nonenzymatically, as a consequence of saturating light intensities during photosynthesis. For example, excess light at the reaction center leads to nonenzymatic formation of β-carotene epoxides and consequential harmful reactive oxygen species. The repair of β-carotene epoxide has been postulated to be controlled by CruP, an enzyme that likely evolved from a carotenoid biosynthetic enzyme, lycopene cyclase (Bradbury et al., 2012; Li et al., 2016). Overexpression of maize CruP in Arabidopsis reduced the formation of β-carotene epoxides and reactive oxygen species, and produced cold-tolerant and anoxia-tolerant plants (Bradbury et al., 2012). Therefore, a better understanding of the mechanisms controlling epoxidase/de-epoxidase activities, especially those related to metabolite repair, could contribute to efforts at creating more resilient plants.
The Bursting Field of Apocarotenoids
Carotenoids are also enzymatically cleaved to apocarotenoids which are gaining attention for bioactivities related to growth, development, plant architecture, stress responses and signaling with other organisms (Tian, 2015; Hou et al., 2016). The wonderful scent of roses (Rosa sp.) or aroma of tomatoes, key factors for consumer appeal, are due in part to volatile apocarotenoids which are likely biosynthesized for ecological purposes.
The first apocarotenoid discovered in plants was abscisic acid (ABA), well known for mediating stress responses. The plastid-made zeaxanthin is enzymatically converted outside of the plastid to ABA via 9-cis-epoxycarotenoid dioxygenase (NCED; Qin and Zeevaart, 1999). The enzymes for biosynthesis and degradation of ABA are well described and easily identified (Li et al., 2008b; Wasilewska et al., 2008; Vallabhaneni and Wurtzel, 2010; Finkelstein, 2013). It is now known that plants biosynthesize many more apocarotenoids beyond ABA. However, mechanisms of action, associated enzymes, regulation, and transporters are largely unknown (Ahrazem et al., 2016). Candidate carotenoid cleavage dioxygenases (CCDs) can be identified through bioinformatics and chemical screens, although not all have been associated with specific apocarotenoids (Vallabhaneni et al., 2010; Van Norman et al., 2014; Chen et al., 2018; Dickinson et al., 2018). Plant apocarotenoid research is bursting with discoveries that will transform synthetic biology applications in agriculture.
The role of apocarotenoids in rhizosphere signaling can be important for synthetic biology approaches to alter plant yield. Unique apocarotenoids are released into the rhizosphere as signals with other organisms. For example, strigolactones are apocarotenoids derived from β-carotene that regulate plant architectural changes (Baz et al., 2018; Gao et al., 2018; Jia et al., 2018). These apocarotenoids inhibit shoot branching above ground and signal below ground with rhizosphere organisms to mediate root architectural changes necessary for symbiosis with beneficial mycorrhizal fungi and mediate resistance to harmful fungi (Al-Babili and Bouwmeester, 2015; Borghi et al., 2016; Decker et al., 2017; Stauder et al., 2018). Biosynthesis of strigolactones is regulated through PSY expression, subjected to feedback regulation and regulated in response to other plant hormones (e.g. ABA, gibberellic acid, auxin) and to inorganic phosphate (Stauder et al., 2018; Jia et al., 2018). Strigolactone transport was shown to be mediated by an ABC transporter (Kretzschmar et al., 2012; Sasse et al., 2015). Modifying strigolactone transport improved the mycorrhizal associations needed to enhance plant yield (Borghi et al., 2016; Liu et al., 2018b).
While strigolactones attract beneficial organisms to promote plant growth, these apocarotenoids also facilitate the germination of harmful parasitic plants (Gomez-Roldan et al., 2008; Umehara et al., 2008). Striga is a devastating weed prevalent in Africa that destroys crops as a consequence of germinating in response to strigolactone produced in host plant roots. Synthetic biology could be used to build a tunable biological circuit (Liu et al., 2018a) that delivers a novel strigolactone structure that functions in plants but is no longer recognizable by harmful rhizosphere organisms and parasitic plants.
New Growth Regulator is an Apocarotenoid
The full range of apocarotenoid structures and associated bioactivities is unknown. There are yet to be described apocarotenoid signals that control plant architecture as suggested by the architectural abnormalities that result when carotenoid biosynthesis is blocked at certain steps or expression of certain carotenoid cleavage dioxygenase enzymes is eliminated (for review, see Hou et al., 2016). The volatile apocarotenoid, β-cyclocitral, was recently discovered to be a new growth regulator, or the unique precursor to a new regulator, that controls root architecture by stimulating cell division and root growth in both monocots and dicots (Dickinson et al., 2018). β-cyclocitral could also stimulate growth of root systems which would otherwise be inhibited by salt stress. The discovery was made through a biochemical screen where lateral root formation was inhibited by D15, a chemical designed to inhibit CCD1, an enzyme with broad carotenoid substrate specificity (Vogel et al., 2008; Van Norman et al., 2014). Determination that β-cyclocitral is the active compound, and not a direct precursor, will require elucidation of the receptor.
β-Cyclocitral is a volatile apocarotenoid already known to modulate gene expression related to stress and defense responses (Ramel et al., 2012; Havaux, 2014; Lv et al., 2015). It has been widely thought that β-cyclocitral is only produced nonenzymatically by oxidative cleavage of β-carotene that occurs in the presence of excess light at the photosynthetic reaction center. It would be surprising that β-cyclocitral formation would not be regulated through some biosynthetic route since β-cyclocitral has an important regulatory role. The possibility of a biosynthetic route is suggested in the biogenesis of the chemically similar 3-OH-β-cyclocitral, which is a precursor of safranal found in Autumn crocus (Crocus sativus) stigmas used to make the costly saffron spice (Frusciante et al., 2014). 3-OH-β-cyclocitral is enzymatically produced upon cleavage of zeaxanthin (but not β-carotene) by CCD2 which simultaneously forms the nonvolatile, crocetindialdehyde (Frusciante et al., 2014). Further evidence supporting the biosynthetic formation of β-cyclocitral has been observed in cyanobacteria associated with toxic algal blooms. These photosynthetic organisms emit β-cyclocitral, a survival allelochemical that negatively affects the odor of drinking water and deters feeding by other species (Jüttner et al., 2010). It is thought that such cyanobacteria make β-cyclocitral enzymatically from β-carotene along with crocetindialdehyde, as occurs with the formation of 3-OH-β-cyclocitral from zeaxanthin in plants. A cleavage enzyme of the CCD class has been suspected. Taken together with the observations in plants, it would not be surprising that the volatile β-cyclocitral also functions in rhizosphere signaling, a yet to be explored hypothesis.
The finding of β-cyclocitral as a new growth regulator (or its precursor) argues strongly that there is a yet to be defined cadre of enzymes dedicated to biosynthesis and degradation, comparable to the regulatory suite of enzymes controlling pools of ABA (Finkelstein, 2013). Whether there exists an in vivo biosynthetic route in plants to form β-cyclocitral, one could be created to better regulate deployment of a network of stress-responsive programs. A synthetic biosynthetic route could build on the framework of the known metabolic enzymes for carotenoid biosynthesis in combination with a synthetic cleavage dioxygenase created to specifically direct the formation of β-cyclocitral. However, tinkering with carotenoid and apocarotenoid networks could potentially interfere with the poorly understood signaling that facilitates normal plant growth under changing environmental conditions. Thus, further research on the physiological role, biogenesis and transport of β-cyclocitral and other apocarotenoids is essential.
Redesigning Plant Structures
The discovery of bioactive apocarotenoids can potentially lead to remodeled plant architecture and synthetic plant parts. The future might include design of robust root systems to promote growth under poor environmental conditions, introduction of mycorrhizal symbioses where none presently exists, or creation of plant parts as chemical factories for biosynthesis of novel phytochemicals that could be rapidly harvested from exudates. Alternatively, volatile apocarotenoids could be created as new chemical scents or developed into a volatile chemical arsenal to create a protective barrier against pathogenic organisms. Such goals would clearly require the elucidation of the current biosynthetic range of apocarotenoids and how those apocarotenoids function and interact ecologically with favorable and harmful organisms. Interestingly, many apocarotenoids have been shown to have biological activity in animals (Harrison and Quadro, 2018), which might provide a means to create novel apocarotenoids or to at least discover the elusive apocarotenoid signals of plants. The reporters used to test responses in animal cells (Linnewiel et al., 2009) could be re-engineered for plants as sensor beacons to elucidate the novel signaling apocarotenoids that control plant architecture.
Conclusions
Carotenoids have diverse bioactivities and potential for retooling to meet emerging needs of agriculture. There has been great progress made in understanding mechanisms controlling carotenoid biosynthesis, modification, degradation, and properties related to energy transfer. An extensive collection of genes and enzymes is available and already much progress has been made in metabolic engineering of carotenoids in multiple species. The recent discoveries of apocarotenoids and their associated bioactivities suggest an untapped potential for creating new toolkits for modifying plant form to address agricultural and industrial needs. With these resources, many opportunities await application of synthetic biology (Box 1). However, true “plug and play” predictability will require deeper investigations into the biology and biochemistry of carotenoids (see Outstanding Questions).
Acknowledgments
I thank Dr. Jesús Beltrán (Penn State University) for his thoughtful suggestions on this review and extensive contributions to research on Z-ISO as a graduate student in my lab. I thank Dr. Alexandra Dickinson (Duke University) for providing early data on the new growth regulator and for helpful comments on this review. I also thank the many members of my laboratory, my collaborators, and my colleagues in the carotenoid community for their extensive contributions to the research described herein.
Footnotes
Research in the Wurtzel laboratory has been funded by the National Institutes of Health (grant no. GM081160), National Science Foundation, American Cancer Society, Rockefeller Foundation International Rice Biotechnology Program, McKnight Foundation, USDA, DOD, New York State, CUNY, and Lehman College.
Articles can be viewed without a subscription.
References
- Ahrazem O, Gómez-Gómez L, Rodrigo MJ, Avalos J, Limón MC (2016) Carotenoid cleavage oxygenases from microbes and photosynthetic organisms: features and functions. Int J Mol Sci 17: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Babili S, Bouwmeester HJ (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol 66: 161–186 [DOI] [PubMed] [Google Scholar]
- Ampomah-Dwamena C, Driedonks N, Lewis D, Shumskaya M, Chen X, Wurtzel ET, Espley RV, Allan AC (2015) The Phytoene synthase gene family of apple (Malus x domestica) and its role in controlling fruit carotenoid content. BMC Plant Biol 15: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampomah-Dwamena C, Thrimawithana AH, Dejnoprat S, Lewis D, Espley RV, Allan AC (2018) A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson IC, Robertson DS (1960) Role of carotenoids in protecting chlorophyll from photodestruction. Plant Physiol 35: 531–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Kishimoto S, Kakami R, Maoka T, Hirakawa H, Suzuki Y, Ozeki Y, Shirasawa K, Bernillon S, Okabe Y, Moing A, Asamizu E, et al. (2014) Identification of the carotenoid modifying gene PALE YELLOW PETAL 1 as an essential factor in xanthophyll esterification and yellow flower pigmentation in tomato (Solanum lycopersicum). Plant J 79: 453–465 [DOI] [PubMed] [Google Scholar]
- Avendaño-Vázquez AO, Cordoba E, Llamas E, San Román C, Nisar N, De la Torre S, Ramos-Vega M, Gutiérrez-Nava MD, Cazzonelli CI, Pogson BJ, León P (2014) An uncharacterized apocarotenoid-derived signal generated in zeta-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. Plant Cell 26: 2524–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baz L, Mori N, Mi J, Jamil M, Kountche BA, Guo X, Balakrishna A, Jia KP, Vermathen M, Akiyama K, Al-Babili S (2018) Mol Plant 11: 1312–1314 [DOI] [PubMed] [Google Scholar]
- Beltrán J, Kloss B, Hosler JP, Geng J, Liu A, Modi A, Dawson JH, Sono M, Shumskaya M, Ampomah-Dwamena C, Love JD, Wurtzel ET (2015) Control of carotenoid biosynthesis through a heme-based cis-trans isomerase. Nat Chem Biol 11: 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi L, Liu GW, Emonet A, Kretzschmar T, Martinoia E (2016) The importance of strigolactone transport regulation for symbiotic signaling and shoot branching. Planta 243: 1351–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury LMT, Shumskaya M, Tzfadia O, Wu S-B, Kennelly EJ, Wurtzel ET (2012) Lycopene cyclase paralog CruP protects against reactive oxygen species in oxygenic photosynthetic organisms. Proc Natl Acad Sci USA 109: E1888–E1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw HD, Schemske DW (2003) Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 426: 176–178 [DOI] [PubMed] [Google Scholar]
- Brausemann A, Gemmecker S, Koschmieder J, Ghisla S, Beyer P, Einsle O (2017) Structure of phytoene desaturase provides insights into herbicide binding and reaction mechanisms involved in carotene desaturation. Structure 25: 1222-1232 [DOI] [PubMed] [Google Scholar]
- Britton G, Liaaen-Jensen S, Pfander H, eds, (2004) Carotenoids Handbook. Birkhäuser Verlag, Basel [Google Scholar]
- Britton G, Liaaen-Jensen S, Pfander H (2017) Carotenoids: A Colourful History. Fagtrykk Trondheim AS, Norway [Google Scholar]
- Burkhardt PK, Beyer P, Wünn J, Klöti A, Armstrong GA, Schledz M, von Lintig J, Potrykus I (1997) Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin A biosynthesis. Plant J 11: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Camagna M, Grundmann A, Bär C, Koschmieder J, Beyer P, Welsch R (2018) Enzyme fusion removes competition for geranylgeranyl diphosphate in carotenogenesis. Plant Physiol pp.01026.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Chayut N, Yuan H, Ohali S, Meir A, Sa’ar U, Tzuri G, Zheng Y, Mazourek M, Gepstein S, Zhou X, Portnoy V, Lewinsohn E, et al. (2017) Distinct mechanisms of the ORANGE protein in controlling carotenoid flux. Plant Physiol 173: 376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Wang B, Han D, Sommerfeld M, Lu Y, Chen F, Hu Q (2015) Molecular mechanisms of the coordination between astaxanthin and fatty acid biosynthesis in Haematococcus pluvialis (Chlorophyceae). Plant J 81: 95–107 [DOI] [PubMed] [Google Scholar]
- Chen H, Zuo X, Shao H, Fan S, Ma J, Zhang D, Zhao C, Yan X, Liu X, Han M (2018) Genome-wide analysis of carotenoid cleavage oxygenase genes and their responses to various phytohormones and abiotic stresses in apple (Malus domestica). Plant Physiol Biochem 123: 81–93 [DOI] [PubMed] [Google Scholar]
- Chen Y, Li F, Wurtzel ET (2010) Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiol 153: 66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. (2018) Single-particle cryo-EM-How did it get here and where will it go. Science 361: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C, Bocobza S, Czosnek H, Aharoni A, Levy AA (2018) Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J 95: 5–16 [DOI] [PubMed] [Google Scholar]
- Dalisay DS, Kim KW, Lee C, Yang H, Rübel O, Bowen BP, Davin LB, Lewis NG (2015) Dirigent protein-mediated lignan and cyanogenic glucoside formation in flax seed: integrated omics and MALDI mass spectrometry imaging. J Nat Prod 78: 1231–1242 [DOI] [PubMed] [Google Scholar]
- Dall’Osto L, Holt NE, Kaligotla S, Fuciman M, Cazzaniga S, Carbonera D, Frank HA, Alric J, Bassi R (2012) Zeaxanthin protects plant photosynthesis by modulating chlorophyll triplet yield in specific light-harvesting antenna subunits. J Biol Chem 287: 41820–41834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalakis V. (2018) Protein-protein interactions within photosystem II under photoprotection: the synergy between CP29 minor antenna, subunit S (PsbS) and zeaxanthin at all-atom resolution. Phys Chem Chem Phys 20: 11843–11855 [DOI] [PubMed] [Google Scholar]
- de Carvalho CCCR, Caramujo MJ (2017) Carotenoids in aquatic ecosystems and aquaculture: a colorful business with implications for human health. Front Mar Sci 4: 93 [Google Scholar]
- Decker EL, Alder A, Hunn S, Ferguson J, Lehtonen MT, Scheler B, Kerres KL, Wiedemann G, Safavi-Rizi V, Nordzieke S, Balakrishna A, Baz L, et al. (2017) Strigolactone biosynthesis is evolutionarily conserved, regulated by phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss, Physcomitrella patens. New Phytol 216: 455–468 [DOI] [PubMed] [Google Scholar]
- Decourcelle M, Perez-Fons L, Baulande S, Steiger S, Couvelard L, Hem S, Zhu C, Capell T, Christou P, Fraser P, Sandmann G (2015) Combined transcript, proteome, and metabolite analysis of transgenic maize seeds engineered for enhanced carotenoid synthesis reveals pleotropic effects in core metabolism. J Exp Bot 66: 3141–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demurtas OC, Frusciante S, Ferrante P, Diretto G, Azad NH, Pietrella M, Aprea G, Taddei AR, Romano E, Mi J, Al-Babili S, Frigerio L, et al. (2018) Candidate enzymes for saffron crocin biosynthesis are localized in multiple cellular compartments. Plant Physiol 177: 990–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson AJ, Lehner K, Mijar M, Benfey PN (2018) β-cyclocitral is a natural root growth regulator. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov RG, Gatsogiannis C, Raunser S (2017) Chapter One - Lipid nanodiscs as a tool for high-resolution structure determination of membrane proteins by single-particle Cryo-EM. In Ziegler C, ed, Methods in Enzymology, Vol 594 Academic Press, pp 1–30 [DOI] [PubMed] [Google Scholar]
- Eggersdorfer M, Wyss A (2018) Carotenoids in human nutrition and health. Arch Biochem Biophys 652: 18–26 [DOI] [PubMed] [Google Scholar]
- Emiliani J, D’Andrea L, Lorena Falcone Ferreyra M, Maulión E, Rodriguez E, Rodriguez-Concepción M, Casati P (2018) A role for β,β-xanthophylls in Arabidopsis UV-B photoprotection. J Exp Bot 69: 4921–4933 [DOI] [PubMed] [Google Scholar]
- Enfissi EMA, Barneche F, Ahmed I, Lichtlé C, Gerrish C, McQuinn RP, Giovannoni JJ, Lopez-Juez E, Bowler C, Bramley PM, Fraser PD (2010) Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 22: 1190–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enfissi EM, Nogueira M, Bramley PM, Fraser PD (2017) The regulation of carotenoid formation in tomato fruit. Plant J 89: 774–788 [DOI] [PubMed] [Google Scholar]
- Finkelstein R. (2013) Abscisic Acid synthesis and response. Arabidopsis Book 11: e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frusciante S, Diretto G, Bruno M, Ferrante P, Pietrella M, Prado-Cabrero A, Rubio-Moraga A, Beyer P, Gomez-Gomez L, Al-Babili S, Giuliano G (2014) Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc Natl Acad Sci USA 111: 12246–12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Kobayashi K, Nagata N, Masuda T, Wada H (2018) Digalactosyldiacylglycerol is essential for organization of the membrane structure in etioplasts. Plant Physiol 177: 1487–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Zhang T, Xu B, Jia L, Xiao B, Liu H, Liu L, Yan H, Xia Q (2018) CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 8 (CCD8) in tobacco affects shoot and root architecture. Int J Mol Sci 19: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmecker S, Schaub P, Koschmieder J, Brausemann A, Drepper F, Rodriguez-Franco M, Ghisla S, Warscheid B, Einsle O, Beyer P (2015) Phytoene desaturase from Oryza sativa: oligomeric assembly, membrane association and preliminary 3D-analysis. PLoS One 10: e0131717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G. (2014) Plant carotenoids: genomics meets multi-gene engineering. Curr Opin Plant Biol 19: 111–117 [DOI] [PubMed] [Google Scholar]
- Giuliano G. (2017) Provitamin A biofortification of crop plants: a gold rush with many miners. Curr Opin Biotechnol 44: 169–180 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, Bouwmeester H, Bécard G, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Gruszecki WI, Strzałka K (2005) Carotenoids as modulators of lipid membrane physical properties. Biochimica et Biophysica Acta (BBA) -. Molecular Basis of Disease 1740: 108–115 [DOI] [PubMed] [Google Scholar]
- Harjes CE, Rocheford TR, Bai L, Brutnell TP, Kandianis CB, Sowinski SG, Stapleton AE, Vallabhaneni R, Williams M, Wurtzel ET, Yan J, Buckler ES (2008) Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319: 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison EH, Quadro L (2018) Apocarotenoids: emerging roles in mammals. Annu Rev Nutr 38: 153–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Uragami C, Cogdell RJ (2016) Carotenoids and Photosynthesis. Subcell Biochem 79: 111–139 [DOI] [PubMed] [Google Scholar]
- Havaux M. (2014) Carotenoid oxidation products as stress signals in plants. Plant J 79: 597–606 [DOI] [PubMed] [Google Scholar]
- Horn PJ, Chapman KD (2014) Lipidomics in situ: insights into plant lipid metabolism from high resolution spatial maps of metabolites. Prog Lipid Res 54: 32–52 [DOI] [PubMed] [Google Scholar]
- Horn PJ, Korte AR, Neogi PB, Love E, Fuchs J, Strupat K, Borisjuk L, Shulaev V, Lee Y-J, Chapman KD (2012) Spatial mapping of lipids at cellular resolution in embryos of cotton. Plant Cell 24: 622–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornero-Méndez D, Mínguez-Mosquera MI (2000) Xanthophyll esterification accompanying carotenoid overaccumulation in chromoplast of Capsicum annuum ripening fruits is a constitutive process and useful for ripeness index. J Agric Food Chem 48: 1617–1622 [DOI] [PubMed] [Google Scholar]
- Hou X, Rivers J, León P, McQuinn RP, Pogson BJ (2016) Synthesis and function of apocarotenoid signals in plants. Trends Plant Sci 21: 792–803 [DOI] [PubMed] [Google Scholar]
- Hunter CT, Saunders JW, Magallanes-Lundback M, Christensen SA, Willett D, Stinard PS, Li QB, Lee K, DellaPenna D, Koch KE (2018) Maize w3 disrupts homogentisate solanesyl transferase (ZmHst) and reveals a plastoquinone-9 independent path for phytoene desaturation and tocopherol accumulation in kernels. Plant J 93: 799–813 [DOI] [PubMed] [Google Scholar]
- Iorizzo M, Ellison S, Senalik D, Zeng P, Satapoomin P, Huang J, Bowman M, Iovene M, Sanseverino W, Cavagnaro P, Yildiz M, Macko-Podgórni A, et al. (2016) A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat Genet 48: 657–666 [DOI] [PubMed] [Google Scholar]
- Jia KP, Baz L, Al-Babili S (2018) From carotenoids to strigolactones. J Exp Bot 69: 2189–2204 [DOI] [PubMed] [Google Scholar]
- Jüttner F, Watson SB, von Elert E, Köster O (2010) β-cyclocitral, a grazer defence signal unique to the cyanobacterium Microcystis. J Chem Ecol 36: 1387–1397 [DOI] [PubMed] [Google Scholar]
- Kachanovsky DE, Filler S, Isaacson T, Hirschberg J (2012) Epistasis in tomato color mutations involves regulation of phytoene synthase 1 expression by cis-carotenoids. Proc Natl Acad Sci USA 109: 19021–19026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassaw TK, Donayre-Torres AJ, Antunes MS, Morey KJ, Medford JI (2018) Engineering synthetic regulatory circuits in plants. Plant Sci 273: 13–22 [DOI] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483: 341–344 [DOI] [PubMed] [Google Scholar]
- Lätari K, Wüst F, Hübner M, Schaub P, Beisel KG, Matsubara S, Beyer P, Welsch R (2015) Tissue-specific apocarotenoid glycosylation contributes to carotenoid homeostasis in Arabidopsis leaves. Plant Physiol 168: 1550–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Joung J-G, McQuinn R, Chung M-Y, Fei Z, Tieman D, Klee H, Giovannoni J (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Leonelli L, Brooks MD, Niyogi KK (2017) Engineering the lutein epoxide cycle into Arabidopsis thaliana. Proc Natl Acad Sci USA 114: E7002–E7008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger M, Morris JM, Chan AM, Leonelli L, Niyogi KK, Fleming GR (2017) Dissecting and modeling zeaxanthin- and lutein-dependent nonphotochemical quenching in Arabidopsis thaliana. Proc Natl Acad Sci USA 114: E7009–E7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Murillo C, Wurtzel ET (2007) Maize Y9 encodes a product essential for 15-cis-zeta-carotene isomerization. Plant Physiol 144: 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vallabhaneni R, Wurtzel ET (2008b) PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiol 146: 1333–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vallabhaneni R, Yu J, Rocheford T, Wurtzel ET (2008a) The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol 147: 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Tsfadia O, Wurtzel ET (2009) The phytoene synthase gene family in the Grasses: subfunctionalization provides tissue-specific control of carotenogenesis. Plant Signal Behav 4: 208–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Peers G, Dent RM, Bai Y, Yang SY, Apel W, Leonelli L, Niyogi KK (2016) Evolution of an atypical de-epoxidase for photoprotection in the green lineage. Nat Plants 2: 16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnewiel K, Ernst H, Caris-Veyrat C, Ben-Dor A, Kampf A, Salman H, Danilenko M, Levy J, Sharoni Y (2009) Structure activity relationship of carotenoid derivatives in activation of the electrophile/antioxidant response element transcription system. Free Radic Biol Med 47: 659–667 [DOI] [PubMed] [Google Scholar]
- Liu D, Mannan AA, Han Y, Oyarzún DA, Zhang F (2018a) Dynamic metabolic control: towards precision engineering of metabolism. J Ind Microbiol Biotechnol 45: 535–543 [DOI] [PubMed] [Google Scholar]
- Liu G, Pfeifer J, de Brito Francisco R, Emonet A, Stirnemann M, Gübeli C, Hutter O, Sasse J, Mattheyer C, Stelzer E, Walter A, Martinoia E, et al. (2018b) Changes in the allocation of endogenous strigolactone improve plant biomass production on phosphate-poor soils. New Phytol 217: 784–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Martinez-Garcia JF, Stange C, Rodriguez-Concepcion M (2017) Illuminating colors: regulation of carotenoid biosynthesis and accumulation by light. Curr Opin Plant Biol 37: 49–55 [DOI] [PubMed] [Google Scholar]
- Lu S, Zhang Y, Zhu K, Yang W, Ye J, Chai L, Xu Q, Deng X (2018) The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol 176: 2657–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist PK, Mantegazza O, Stefanski A, Stühler K, Weber APM (2017) Surveying the oligomeric state of Arabidopsis thaliana chloroplasts. Mol Plant 10: 197–211 [DOI] [PubMed] [Google Scholar]
- Lv F, Zhou J, Zeng L, Xing D (2015) β-cyclocitral upregulates salicylic acid signalling to enhance excess light acclimation in Arabidopsis. J Exp Bot 66: 4719–4732 [DOI] [PubMed] [Google Scholar]
- Magdaong NM, LaFountain AM, Greco JA, Gardiner AT, Carey AM, Cogdell RJ, Gibson GN, Birge RR, Frank HA (2014) High efficiency light harvesting by carotenoids in the LH2 complex from photosynthetic bacteria: unique adaptation to growth under low-light conditions. J Phys Chem B 118: 11172–11189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer E, Llorente B, Rodríguez-Concepción M, Daròs J-A (2017) Rewiring carotenoid biosynthesis in plants using a viral vector. Sci Rep 7: 41645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimov EG, Sluchanko NN, Slonimskiy YB, Slutskaya EA, Stepanov AV, Argentova-Stevens AM, Shirshin EA, Tsoraev GV, Klementiev KE, Slatinskaya OV, Lukashev EP, Friedrich T, et al. (2017) The photocycle of orange carotenoid protein conceals distinct intermediates and asynchronous changes in the carotenoid and protein components. Sci Rep 7: 15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews PD, Luo R, Wurtzel ET (2003) Maize phytoene desaturase and zeta-carotene desaturase catalyse a poly-Z desaturation pathway: implications for genetic engineering of carotenoid content among cereal crops. J Exp Bot 54: 2215–2230 [DOI] [PubMed] [Google Scholar]
- Matthies D, Bae C, Toombes GE, Fox T, Bartesaghi A, Subramaniam S, Swartz KJ (2018) Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs. eLife 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuinn RP, Wong B, Giovannoni JJ (2018) AtPDS overexpression in tomato: exposing unique patterns of carotenoid self-regulation and an alternative strategy for the enhancement of fruit carotenoid content. Plant Biotechnol J 16: 482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier S, Tzfadia O, Vallabhaneni R, Gehring C, Wurtzel ET (2011) A transcriptional analysis of carotenoid, chlorophyll and plastidial isoprenoid biosynthesis genes during development and osmotic stress responses in Arabidopsis thaliana. BMC Syst Biol 5: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K (1990) Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol 172: 6704–6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moise AR, Al-Babili S, Wurtzel ET (2014) Mechanistic aspects of carotenoid biosynthesis. Chem Rev 114: 164–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa T, Uraguchi Y, Sanda S, Nakagawa S, Sawayama S (2018) Overexpression of DnaJ-like chaperone enhances carotenoid synthesis in Chlamydomonas reinhardtii. Appl Biochem Biotechnol 184: 80–91 [DOI] [PubMed] [Google Scholar]
- Murillo AG, Fernandez ML (2016) Potential of dietary non-provitamin a carotenoids in the prevention and treatment of diabetic microvascular complications. Adv Nutr 7: 14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima KVP, Sasaki M, Hashimoto K, Takaichi S, Nagashima S, Yu LJ, Abe Y, Gotou K, Kawakami T, Takenouchi M, Shibuya Y, Yamaguchi A, et al. (2017) Probing structure-function relationships in early events in photosynthesis using a chimeric photocomplex. Proc Natl Acad Sci USA 114: 10906–10911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira M, Mora L, Enfissi EM, Bramley PM, Fraser PD (2013) Subchromoplast sequestration of carotenoids affects regulatory mechanisms in tomato lines expressing different carotenoid gene combinations. Plant Cell 25: 4560–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira M, Enfissi EMA, Martínez Valenzuela ME, Menard GN, Driller RL, Eastmond PJ, Schuch W, Sandmann G, Fraser PD (2017) Engineering of tomato for the sustainable production of ketocarotenoids and its evaluation in aquaculture feed. Proc Natl Acad Sci USA 114: 10876–10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira M, Enfissi EM, Almeida J, Fraser PD (2018) Creating plant molecular factories for industrial and nutritional isoprenoid production. Curr Opin Biotechnol 49: 80–87 [DOI] [PubMed] [Google Scholar]
- Oleszkiewicz T, Klimek-Chodacka M, Milewska-Hendel A, Zubko M, Stróż D, Kurczyńska E, Boba A, Szopa J, Baranski R (2018) Unique chromoplast organisation and carotenoid gene expression in carotenoid-rich carrot callus. Planta [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaisa KA, Morgante M, Williams M, Rafalski A (2003) Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 15: 1795–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaisa K, Morgante M, Tingey S, Rafalski A (2004) Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proc Natl Acad Sci USA 101: 9885–9890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polívka T, Frank HA (2010) Molecular factors controlling photosynthetic light harvesting by carotenoids. Acc Chem Res 43: 1125–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollmann H, Breitenbach J, Wolff H, Bode HB, Sandmann G (2017) Combinatorial biosynthesis of novel multi-hydroxy carotenoids in the red yeast Xanthophyllomyces dendrorhous. J Fungi (Basel) 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price EJ, Bhattacharjee R, Lopez-Montes A, Fraser PD (2018) Carotenoid profiling of yams: Clarity, comparisons and diversity. Food Chem 259: 130–138 [DOI] [PubMed] [Google Scholar]
- Qin X, Zeevaart JA (1999) The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci USA 96: 15354–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Suga M, Kuang T, Shen JR (2015) Photosynthesis. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 348: 989–995 [DOI] [PubMed] [Google Scholar]
- Quinlan RF, Shumskaya M, Bradbury LMT, Beltrán J, Ma C, Kennelly EJ, Wurtzel ET (2012) Synergistic interactions between carotene ring hydroxylases drive lutein formation in plant carotenoid biosynthesis. Plant Physiol 160: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Hu P, Zhao H (2017) A plug-and-play pathway refactoring workflow for natural product research in Escherichia coli and Saccharomyces cerevisiae. Biotechnol Bioeng 114: 1847–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Madrid R, Aguilar-Espinosa M, Cárdenas-Conejo Y, Garza-Caligaris LE (2016) Carotenoid derivates in achiote (Bixa orellana) seeds: synthesis and health promoting properties. Front Plant Sci 7: 1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DS. (1955) The genetics of vivipary in maize. Genetics 40: 745–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M, Avalos J, Bonet ML, Boronat A, Gomez-Gomez L, Hornero-Mendez D, Limon MC, Meléndez-Martínez AJ, Olmedilla-Alonso B, Palou A, Ribot J, Rodrigo MJ, et al. (2018) A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Prog Lipid Res 70: 62–93 [DOI] [PubMed] [Google Scholar]
- Rubin LP, Ross AC, Stephensen CB, Bohn T, Tanumihardjo SA (2017) Metabolic effects of inflammation on vitamin a and carotenoids in humans and animal models. Adv Nutr 8: 197–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint SE, Renzi-Hammond LM, Khan NA, Hillman CH, Frick JE, Hammond BR (2018) The macular carotenoids are associated with cognitive function in preadolescent hildren. Nutrients 10: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankari M, Rao PR, Hemachandran H, Pullela PK, Doss C GP, Tayubi IA, Subramanian B, Gothandam KM, Singh P, Ramamoorthy S (2018) Prospects and progress in the production of valuable carotenoids: Insights from metabolic engineering, synthetic biology, and computational approaches. J Biotechnol 266: 89–101 [DOI] [PubMed] [Google Scholar]
- Sasse J, Simon S, Gübeli C, Liu GW, Cheng X, Friml J, Bouwmeester H, Martinoia E, Borghi L (2015) Asymmetric localizations of the ABC transporter PaPDR1 trace paths of directional strigolactone transport. Curr Biol 25: 647–655 [DOI] [PubMed] [Google Scholar]
- Schaub P, Al-Babili S, Drake R, Beyer P (2005) Why is golden rice golden (yellow) instead of red? Plant Physiol 138: 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub P, Yu Q, Gemmecker S, Poussin-Courmontagne P, Mailliot J, McEwen AG, Ghisla S, Al-Babili S, Cavarelli J, Beyer P (2012) On the structure and function of the phytoene desaturase CRTI from Pantoea ananatis, a membrane-peripheral and FAD-dependent oxidase/isomerase. PLoS One 7: e39550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub P, Rodriguez-Franco M, Cazzonelli CI, Álvarez D, Wüst F, Welsch R (2018) Establishment of an Arabidopsis callus system to study the interrelations of biosynthesis, degradation and accumulation of carotenoids. PLoS One 13: e0192158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schledz M, al-Babili S, von Lintig J, Haubruck H, Rabbani S, Kleinig H, Beyer P (1996) Phytoene synthase from Narcissus pseudonarcissus: functional expression, galactolipid requirement, topological distribution in chromoplasts and induction during flowering. Plant J 10: 781–792 [DOI] [PubMed] [Google Scholar]
- Schulz DF, Schott RT, Voorrips RE, Smulders MJ, Linde M, Debener T (2016) Genome-wide association analysis of the anthocyanin and carotenoid contents of rose petals. Front Plant Sci 7: 1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, Frogue K, Misa J, Wheeldon I (2017) Host and pathway engineering for enhanced lycopene biosynthesis in Yarrowia lipolytica. Front Microbiol 8: 2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumskaya M, Wurtzel ET (2013) The carotenoid biosynthetic pathway: thinking in all dimensions. Plant Sci 208: 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumskaya M, Bradbury LMT, Monaco RR, Wurtzel ET (2012) Plastid localization of the key carotenoid enzyme phytoene synthase is altered by isozyme, allelic variation, and activity. Plant Cell 24: 3725–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauder R, Welsch R, Camagna M, Kohlen W, Balcke GU, Tissier A, Walter MH (2018) Strigolactone levels in dicot roots are determined by an ancestral symbiosis-regulated clade of the PHYTOENE SYNTHASE gene family. Front Plant Sci 9: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, Roustan JP, Bouzayen M, Giuliano G, Chervin C (2015) Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biol 15: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulli M, Mandolino G, Sturaro M, Onofri C, Diretto G, Parisi B, Giuliano G (2017) Molecular and biochemical characterization of a potato collection with contrasting tuber carotenoid content. PLoS One 12: e0184143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Yuan H, Cao H, Yazdani M, Tadmor Y, Li L (2018) Carotenoid metabolism in plants: the role of plastids. Mol Plant 11: 58–74 [DOI] [PubMed] [Google Scholar]
- Teixeira da Silva JA, Dobránszki J, Rivera-Madrid R (2018) The biotechnology (genetic transformation and molecular biology) of Bixa orellana L. (achiote). Planta 248: 267–277 [DOI] [PubMed] [Google Scholar]
- Thomas DB, McGraw KJ, Butler MW, Carrano MT, Madden O, James HF (2014) Ancient origins and multiple appearances of carotenoid-pigmented feathers in birds. Proceedings of the Royal Society B: Biological Sciences 281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L. (2015) Recent advances in understanding carotenoid-derived signaling molecules in regulating plant growth and development. Front Plant Sci 6: 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D, Zhu G, Resende MF Jr., Lin T, Nguyen C, Bies D, Rambla JL, Beltran KS, Taylor M, Zhang B, Ikeda H, Liu Z, et al. (2017) A chemical genetic roadmap to improved tomato flavor. Science 355: 391–394 [DOI] [PubMed] [Google Scholar]
- Timlin JA, Collins AM, Beechem TA, Shumskaya M, Wurtzel ET (2017) Localizing and quantifying carotenoids in intact cells and tissues. In Cvetkovic Dragan and Nikolic Goran, eds, Carotenoids, 3. IntechOpen Limited, London, UK [Google Scholar]
- Toews DPL, Hofmeister NR, Taylor SA (2017) The evolution and genetics of carotenoid processing in animals. Trends Genet 33: 171–182 [DOI] [PubMed] [Google Scholar]
- Tzfadia O, Amar D, Bradbury LMT, Wurtzel ET, Shamir R (2012) The MORPH algorithm: ranking candidate genes for membership in Arabidopsis and tomato pathways. Plant Cell 24: 4389–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, Kyozuka J, Yamaguchi S (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Vallabhaneni R, Wurtzel ET (2009) Timing and biosynthetic potential for carotenoid accumulation in genetically diverse germplasm of maize. Plant Physiol 150: 562–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhaneni R, Wurtzel ET (2010) From epoxycarotenoids to ABA: the role of ABA 8′-hydroxylases in drought-stressed maize roots. Arch Biochem Biophys 504: 112–117 (Carotenoids Highlight edition) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhaneni R, Gallagher CE, Licciardello N, Cuttriss AJ, Quinlan RF, Wurtzel ET (2009) Metabolite sorting of a germplasm collection reveals the hydroxylase3 locus as a new target for maize provitamin A biofortification. Plant Physiol 151: 1635–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhaneni R, Bradbury LM, Wurtzel ET (2010) The carotenoid dioxygenase gene family in maize, sorghum, and rice. Arch Biochem Biophys 504: 104–111 (Carotenoids Highlight edition) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman JM, Zhang J, Cazzonelli CI, Pogson BJ, Harrison PJ, Bugg TD, Chan KX, Thompson AJ, Benfey PN (2014) Periodic root branching in Arabidopsis requires synthesis of an uncharacterized carotenoid derivative. Proc Natl Acad Sci USA 111: E1300–E1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk KJ, Kessler F (2017) Plastoglobuli: plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Annu Rev Plant Biol 68: 253–289 [DOI] [PubMed] [Google Scholar]
- Vogel JT, Tan B-C, McCarty DR, Klee HJ (2008) The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J Biol Chem 283: 11364–11373 [DOI] [PubMed] [Google Scholar]
- Wang C, Kim JH, Kim SW (2014) Synthetic biology and metabolic engineering for marine carotenoids: new opportunities and future prospects. Mar Drugs 12: 4810–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frei dit Frey N, Leung J (2008) An update on abscisic acid signaling in plants and more... Mol Plant 1: 198–217 [DOI] [PubMed] [Google Scholar]
- Weesie RJ, Askin D, Jansen FJ, de Groot HJ, Lugtenburg J, Britton G (1995) Protein-chromophore interactions in alpha-crustacyanin, the major blue carotenoprotein from the carapace of the lobster, Homarus gammarus. A study by 13C magic angle spinning NMR. FEBS Lett 362: 34–38 [DOI] [PubMed] [Google Scholar]
- Wei X, Su X, Cao P, Liu X, Chang W, Li M, Zhang X, Liu Z (2016) Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature 534: 69–74 [DOI] [PubMed] [Google Scholar]
- Welsch R, Beyer P, Hugueney P, Kleinig H, von Lintig J (2000) Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 211: 846–854 [DOI] [PubMed] [Google Scholar]
- Welsch R, Maass D, Voegel T, Dellapenna D, Beyer P (2007) Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol 145: 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R, Zhou X, Yuan H, Álvarez D, Sun T, Schlossarek D, Yang Y, Shen G, Zhang H, Rodriguez-Concepcion M, Thannhauser TW, Li L (2018) Clp protease and OR directly control the proteostasis of phytoene synthase, the crucial enzyme for carotenoid biosynthesis in Arabidopsis. Mol Plant 11: 149–162 [DOI] [PubMed] [Google Scholar]
- Wurtzel ET, Kutchan TM (2016) Plant metabolism, the diverse chemistry set of the future. Science 353: 1232–1236 [DOI] [PubMed] [Google Scholar]
- Wurtzel ET, Cuttriss A, Vallabhaneni R (2012) Maize provitamin a carotenoids, current resources, and future metabolic engineering challenges. Front Plant Sci 3: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong C, Luo D, Lin A, Zhang C, Shan L, He P, Li B, Zhang Q, Hua B, Yuan Z, Li H, Zhang J, et al. (2019) A tomato B-box protein SlBBX20 modulates carotenoid biosynthesis by directly activating PHYTOENE SYNTHASE 1, and is targeted for 26S proteasome-mediated degradation. New Phytol 221: 279–294 [DOI] [PubMed] [Google Scholar]
- Yao D, Wang Y, Li Q, Ouyang X, Li Y, Wang C, Ding L, Hou L, Luo M, Xiao Y (2018) Specific upregulation of a cotton phytoene synthase gene produces golden cottonseeds with enhanced provitamin A. Sci Rep 8: 1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani M, Sun Z, Yuan H, Zeng S, Thannhauser TW, Vrebalov J, Ma Q, Xu Y, Fei Z, Van Eck J, Tian S, Tadmor Y, et al. (2019) Ectopic expression of ORANGE promotes carotenoid accumulation and fruit development in tomato. Plant Biotechnol J 17: 33–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287: 303–305 [DOI] [PubMed] [Google Scholar]
- Yukihira N, Sugai Y, Fujiwara M, Kosumi D, Iha M, Sakaguchi K, Katsumura S, Gardiner AT, Cogdell RJ, Hashimoto H (2017) Strategies to enhance the excitation energy-transfer efficiency in a light-harvesting system using the intra-molecular charge transfer character of carotenoids. Faraday Discuss 198: 59–71 [DOI] [PubMed] [Google Scholar]
- Zhang C, Chen X, Lindley ND, Too H-P (2018) A “plug-n-play” modular metabolic system for the production of apocarotenoids. Biotechnol Bioeng 115: 174–183 [DOI] [PubMed] [Google Scholar]
- Zhao C, Nabity PD (2017) Phylloxerids share ancestral carotenoid biosynthesis genes of fungal origin with aphids and adelgids. PLoS One 12: e0185484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Zhang C, Wu Y, Tang Y (2013) Metabolic engineering of gossypol in cotton. Appl Microbiol Biotechnol 97: 6159–6165 [DOI] [PubMed] [Google Scholar]
- Zhou X, Welsch R, Yang Y, Álvarez D, Riediger M, Yuan H, Fish T, Liu J, Thannhauser TW, Li L (2015) Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc Natl Acad Sci USA 112: 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]