Abstract
Myelin is composed primarily of lipids and diseases affecting myelin are associated with alterations in its lipid composition. However, correlation of the spatial (in situ) distribution of lipids with the disease-associated compositional and morphological changes is not well defined. Herein we applied high resolution matrix-assisted laser desorption ionization imaging mass spectrometry (MALDI-IMS), immunohistochemistry (IHC), and liquid chromatography–electrospray ionization–mass spectrometry (LC-ESI-MS) to evaluate brain lipid alterations in the dysmyelinating shiverer (Shi) mouse and cuprizone (Cz) mouse model of reversible demyelination. MALDI-IMS revealed a decrease in the spatial distribution of sulfatide (SHexCer) species, SHexCer (d42:2), and a phosphatidylcholine (PC) species, PC (36:1), in white matter regions like corpus callosum (CC) both in the Shi mouse and Cz mouse model. Changes in these lipid species were restored albeit not entirely upon spontaneous remyelination after demyelination in the Cz mouse model. Lipid distribution changes correlated with the local morphological changes as confirmed by IHC. LC-ESI-MS analyses of CC extracts confirmed the MALDI-IMS derived reductions in SHexCer and PC species. These findings highlight the role of SHexCer and PC in preserving the normal myelin architecture and our experimental approaches provide a morphological basis to define lipid abnormalities relevant to myelin diseases.
Keywords: brain, cuprizone, electrospray ionization, mass spectrometry, matrix-assisted laser desorption ionization, myelin, shiverer
Introduction
The central nervous system (CNS) is composed primarily of lipids and the majority of these lipids are present in myelin. Myelin is a mixture of lipids and proteins arranged as lamellar membranes that wrap around axons thus enabling high velocity neuronal conduction. Myelin is composed of 70% to 85% lipid based on the dry weight1 and is constituted mainly of the lipid classes: phospholipids, cholesterols, and complex sphingolipids in an approximately equal proportion.2 The structural integrity of myelin largely depends on the interaction between lipids and membrane proteins such as myelin basic protein (MBP) and proteolipid protein (PLP).3–5 Even subtle alterations in lipid or protein composition can disrupt the normal myelin structure and function.
Lipid-rich myelin membranes are the targets in many CNS diseases, the most notable of which is multiple sclerosis, a disease-associated with damage and loss of myelin and neuronal axons.6,7 In human patients as well as rodent models of multiple sclerosis, myelin damage is associated with alterations in lipid composition.7–10 A decrease in the tissue levels and a rise in cerebrospinal fluid levels of sulfatides/sulfated hexosyl ceramides (SHexCer) was previously reported.11,12 In addition, in multiple sclerosis patients, SHexCer antibodies were detected in serum,13,14 and serum levels of phospholipids such as phosphatidylcholine (PC) and lysophosphatidylcholine were also elevated.13,15,16 Other neurodegenerative and neurological lysosomal storage disorders including Alzheimer’s,17,18 Niemann-Pick’s diseases,19 and metachromatic leukodystrophy20 are associated with changes in composition and turnover of sphingolipids such as SHexCer that make up nearly 5% of the myelin lipid content.21,22 These findings implicate that a widespread disruption in lipid composition occurs in diseases affecting myelin. However, these studies all employed classic “bind and grind” methods and conventional mass spectrometry requiring tissue extracts for lipid analysis; thus, tissue morphology is not preserved. To fully understand the role of lipids in myelin and neuronal function, it is crucial not only to identify but also map the disease-associated in situ lipid distribution in the context of tissue anatomy.
Lipids are diverse biomolecules and detection of lipids using probes or antibodies is confined to the lipid type and individual lipid species that differ in fatty acid chain length, degree of saturation, and other modifications like hydroxylation cannot be distinguished. Newly emerging technologies, such as imaging mass spectrometry (IMS), enable simultaneous in situ detection of individual molecular species of lipids while preserving the tissue morphology.23–25 The three major sample ionization methods of IMS include secondary ion mass spectrometry (SIMS), matrix-assisted laser desorption ionization (MALDI), and desorption electrospray ionization (DESI). Although SIMS is the first introduced method, MALDI is the most widely used sample ionization method.26 SIMS yields highest resolution (subcellular) and can also be used to map elemental distribution on the tissue surface.26 It is, however, a time-consuming method from perspectives of data collection and analysis, due to the prevalence of fragment ions in the individual mass spectra and is also limited to a mass range below m/z 500. DESI is a simpler process that does not require specific tissue preparation steps, has good sensitivity, and can identify a wide range of species from low to high m/z27; however, it yields a spatial resolution that is quite poor. MALDI offers a wide range of molecular size detection, medium to high spatial resolution, and high sensitivity. Thus, the three ionization methods complement each other, and the choice of ionization method depends on the user’s preferred requirements such as spatial resolution, m/z detection range, area to be imaged, and the time required for sample preparation, data acquisition, and analysis.
Recently, multiple studies have reported IMS derived CNS profiles of lipids, proteins, and neuropeptides in rodent and human tissues.26–32 IMS has been increasingly used as a promising tool to study distribution of small molecules, drugs, and metabolites in various disease conditions including tumors, chronic inflammation, and neurodegenerative diseases, and for biomarker discovery and validation studies.26,33–35 However, IMS-based methods have not been utilized to study lipids in myelin disorders. In this study, we used Fourier-transform ion cyclotron resonance (FTICR)-based MALDI-IMS to demonstrate in situ brain lipid profiles associated with dysmyelination (loss of biochemically abnormal myelin) in the shiverer (Shi) mouse and in a mouse model of cuprizone (Cz) induced reversible demyelination (loss of normally formed myelin).
The Shi mouse is a deletion mutant of the gene encoding MBP, an integral myelin membrane protein that maintains myelin stability through its interaction with lipid head groups.4–6 Lack of MBP in Shi mice causes dysmyelination as a result of improper myelin compaction. Cz is a copper-chelating agent when fed at a concentration of 0.2% to 0.3% in diet selectively induces oligodendrocyte apoptosis.36 Loss of oligodendrocytes results in demyelination that accompanies reactive responses by the CNS-resident microglial and astroglial cells. Demyelination resolves and remyelination occurs after removal of Cz from the diet. The Shi and Cz mouse models thus provided the opportunity to explore changes in the situ distribution and composition of brain lipids associated with myelin lesions. As the most extensive dynamics of lipid regulation is expected in the myelin-rich white matter (WM), we focused our analyses to the prototypical WM region, the corpus callosum (CC). In situ lipid profiles of both the mouse models were correlated with local morphology by immunohistochemistry (IHC) and verified by liquid chromatography–electrospray ionization–mass spectrometry (LC-ESI-MS) analysis of lipid extracts prepared from affected brain regions.
Materials and Methods
Chemicals and Reagents
Methanol, acetonitrile, trifluoroacetic acid (TFA), sodium trifluoroacetate (NaTFA), 50 mM triethylammonium bicarbonate (TEAB), bicinchoninic acid (BCA) protein assay kit, Optimal Cutting Temperature (OCT) embedding medium, 10% neutral-buffered formaldehyde, and hematoxylin and eosin (H&E) were purchased from Thermo Fisher Scientific (Waltham, MA). The internal standards for LC-ESI-MS including galactosyl(ß) ceramide (d18:1/12:0) and mono-sulfo galactosyl(ß) ceramide (d18:1/17:0) were purchased from Avanti Polar Lipids (Alabaster, AL). Bis-cyclohexanone oxaldihydrazone (Cz), ammonium bicarbonate, dichloromethane, 1, 5-diaminonapthalene (DAN), Urea, and luxol fast blue (LFB) were obtained from Sigma-Aldrich (St. Louis, MO). The following rabbit polyclonal primary antibodies were used: glial fibrillary acidic protein (GFAP; Agilent Technologies, Carpinteria, CA), ionized calcium-binding adapter molecule 1 (IBA1; Wako Chemicals, Richmond, VA), and PLP (Sigma-Aldrich, St. Louis, MO). Antirabbit horseradish peroxidase (HRP) secondary antibody was purchased from Sigma-Aldrich (St. Louis, MO).
Animals
All animal experiments were performed in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited vivarium at Biogen according to the Institutional Animal Care and Use Committee (IACUC) approved protocols. Six to 8 weeks old, male C3HeB/FeJ-shiverer (also known as C3Fe.SWV-Mbpshi/J; referred here as “Shi”) mice and age-matched wild type (WT) controls (C3HeB/FeJ) were obtained from The Jackson Laboratory. To induce demyelination, 8 weeks old, male C57BL/six mice (obtained from The Jackson Laboratory) were fed with standard chow diet (CD) mixed with 0.2% Cz (Research Diets, Inc, NJ) for 6 weeks. Age-matched control mice were fed with CD. After 6 weeks, Cz was removed from the diet and replaced with CD. The study was continued for additional 6 weeks to allow remyelination. Cz- and CD-fed mice were sacrificed at 6- and 12-week study time points.
Tissue Preparation
Following carbon dioxide euthanasia, mouse brains were harvested and immediately dissected into two equal halves along the mid sagittal plane. One half was snap-frozen on dry ice and stored at −80C. The other half was fixed in 10% neutral-buffered formalin. Cryosections (10 μm thick) were prepared at −20C lateral to the midline at approximately sagittal level 69 (1.5 mm distal to Bregma) based on Allen’s reference atlas for mouse brain.37 Sections were thaw-mounted on indium tinoxide (ITO)-coated glass slides (Bruker Daltonics, Billerica, MA). OCT was used to hold the tissue block on the cryostat chuck with only a minimal contact between the tissue section and OCT was confined to the periphery to avoid interference in the MS signal. Sections were vacuum dried in a desiccator for 10 min before washing or stored at −80C for later use. For washing, sections were carefully dipped in ice-cold 25 mM ammonium bicarbonate (pH 7.2) for 15 sec. The optimal wash conditions were chosen based on previously reported recommendations.38 Excess wash buffer was wiped off the slides and allowed to air-dry.
Matrix Application and MALDI-IMS
An optical image (4800 dpi) of the tissue section was obtained after washing using an Epson Perfection V500 photo scanner. DAN matrix, that has been reported to facilitate excellent detection sensitivity for lipid analysis in both positive and negative polarities,39 was applied as matrix using a custom-built sublimation apparatus based on previously described devices in the literature.40,41 Matrix slurry was prepared in methanol and allowed to sublime at 110C to 120C in a vacuum chamber below 50 mTorr for 5 min resulting in a uniform matrix deposition on the tissue surface. The deposited matrix was allowed to recrystallize with vapors of solvent containing 95% acetonitrile and 0.3% TFA at 37C for 30 min.40,41 The estimated coating density of the matrix was 300 µg/cm2.
The glass slide was then mounted onto the MALDI adapter plate and a wite-out correction pen was used to mark three teach points for aligning the slide and tissue images. IMS was performed with a 7 Tesla SolariX XR FTICR mass spectrometer (Bruker Daltonics, Billerica, MA). Before scanning images, the instrument was calibrated through masses of NaTFA clusters. The laser beam was set to raster randomly across the entire tissue surface to be imaged. During the acquisition process mass spectra are generated from each laser spot (pixel) covering the entire area of interest. Mass spectra within the m/z range of 200 to 2000 were acquired in negative and positive modes in separate experiments from two serial sections of the respective samples both with spatial resolution of 80 µm and mass resolution of 200,000. Each mass spectrum was obtained as the sum of 20 laser shots with the laser beam set to small or minimum. The following software tools (versions 4.0) from Bruker Daltonics (Billerica, MA) were used: FlexControl for acquiring mass spectra, FlexImaging for visualizing and processing MS images, and Data Analysis for processing mass spectra and generating the peak list. The list of identified lipid species, mass spectra of negative and positive lipid ions, and the normalized MS images of various lipid species revealing anatomical features of the WT mouse brain WM and gray matter regions are provided in supplementary information (Table S1 and Figs. S1–S3).
Histology and Immunohistochemistry
After MALDI-IMS, matrix was removed from the slide by washing in absolute ethanol for 2 min followed by clearing in 70% ethanol an additional 30 sec. Sections were then stained with H&E using ST5020–CV5030 Stainer Integrated Workstation from Leica Biosystems Inc. (Buffalo Grove, IL). Tissue slices were trimmed from the formalin-fixed mouse brain halves approximately 1.5 mm lateral to the midline, processed and embedded in paraffin. Sagittal sections (5 μm thick) were collected at the same approximate level as the frozen samples analyzed by MALDI-IMS. After deparaffinizing and rehydrating, serial sections were used for LFB/H&E co-staining and IHC. Deparaffinization and rehydration steps for were done as a part of automated H&E staining protocol on the ST5020–CV5030 Stainer Integrated Workstation from Leica Biosystems Inc. (Buffalo Grove, IL). Briefly, slides were placed in three different containers with xylene for 5 min each. This was followed by rehydrating steps: first, slides were kept in two different containers with 100% ethanol for 10 min each; second, in two different containers with 95% ethanol for 10 min each; and last, in two different containers with deionized water for 5 min each. This was followed by automated H&E staining. For LFB staining, sections were stained with 0.1% LFB (prepared in 95% ethanol) for 24 hr at 60C. Excess stain was washed with 95% ethanol and distilled water. Sections were differentiated in 0.05% lithium carbonate for 30 sec and in 70% ethanol for 30 sec followed by automated H&E staining as described above. IHC was performed using the XT/Ultra-automated IHC research platform (Ventana Medical Systems, Tucson, AZ) with the manufacturer provided buffers and reagents. The following polyclonal rabbit primary antibodies were used: GFAP (1:2000), IBA1 (1:2000), and PLP (1:1000); and antirabbit HRP (1:5000) was used as the secondary antibody. After IHC, slides were scanned using Aperio Scanscope (Leica Biosystems, Buffalo Grove, IL) or VS-120 (Olympus Life Science, Waltham, MA) scanners for whole slide imaging.
Lipid Extraction and LC-ESI-MS Analysis
The CC region from the frozen WT and Shi mouse and from the Cz-fed and CD-fed mouse brain tissue samples were dissected with a razor blade inside the cryostat chamber at −20C and homogenized in a buffer composed of 4 M urea and 50 mM TEAB. The total protein concentration of the cleared tissue lysates was measured by BCA assay. Equal amount of total protein (3 µg and 10 µg for positive and negative ion analysis, respectively) per sample were reconstituted with 30 µL of dichloromethane/methanol (2:1), vortexed and centrifuged. The water phase was discarded and the organic phase was speed vacuum dried. The dried samples were resuspended in 15 µL (for negative mode) or 20 µL (for positive mode) of methanol containing an internal standard. The internal standards: galactosyl(ß) ceramide (d18:1/12:0) and mono-sulfo galactosyl(ß) ceramide (d18:1/17:0) were used for analysis in the positive mode (4 pmol/µL) and negative mode (6 pmol/µL), respectively. In all, 1 to 2 µL per sample was loaded into a high-performance liquid chromatography (HPLC) Scherzo SM-C18 column (Imtakt, Portland, OR) for lipid separation using the gradient of 65% to 100% B (20 min) at 3.5 µl/min flow rate. Eluting solutions were composed of (1) water (70%)/methanol (30%) and 0.1% formic acid; and (2) water (10%)/90% 2-propanol/acetonitrile (3:2), 0.1% formic acid, and 100 mM ammonium acetate. Chromatography was performed on an Acquity UPLC System (Waters Corp., Milford, MA) in-line with a 7 Tesla SolariX XR FTICR mass spectrometer (Bruker Daltonics, Billerica, MA). Mass spectra were collected in the range of m/z 150 to 3000 in the positive and negative modes with a mass resolution of 200,000.
Lipid Identification
Identification of lipid species observed in MS images and LC-ESI-MS spectra was based on accurate mass matching using the Lipid Maps database42,43 taking advantage of high mass accuracy of the FTICR-instrument with less than three parts per million (ppm) mass errors. The ion forms [M+H]+, [M+NH4]+, [M+Na]+, and [M+K]+ were considered for mass matching in the positive mode, and ion forms [M-H]− and [M+Cl]− were used in the negative mode. Identities of selected lipids were confirmed by tandem mass spectrometry (see Table S2), wherein precursor ions were fragmented by Quadrupole collision-induced dissociation (QCID). Mass spectra (m/z range of 50–3000) of generated fragments were evaluated with a mass resolution of 70,000. Additional confirmation of lipid identities was based on comparisons with published reports.28,44,45
Data Analysis and Statistics
For visualizing spatial distribution of lipids across the tissue sections, MS images were root mean squared (RMS; vector) normalized, which is the default of the flexImaging software (Bruker Daltonics) for FTICR MALDI-IMS data. We used a total ion current (TIC) and mass window-based normalization procedure for semiquantitative comparison of different phenotypes with both MALDI-IMS and LC-MS generated data. Normalized peak intensities were calculated as integrated values across either the region of interest (MALDI-IMS) or the LC elution space (LC-ESI MS). For this purpose, integrated peak intensities were extracted by the Data Analysis software (Bruker Daltonics, Billerica, MA) or by an in-house developed script (for LC-MS peak extraction) enforcing peak matching across pixels or LC-MS scans within a mass tolerance of 3 ppm in a retention time window of 1.5 min (LC-MS only). Subsequently the peak list and peak intensities were exported to Microsoft Excel. Ion intensities of individual lipid species were then normalized to TIC calculated as the sum of intensities of all lipid species ranging from 400 to 920 m/z. Normalized intensities of individual lipid species obtained from biological replicates were averaged to evaluate differences between WT and Shi mice (n=5), and between Cz-fed and CD-fed mice (n=4). For LC-MS data, ratios of individual peak intensities to the intensity of internal standard were calculated to correct for signal fluctuation during data acquisition. A detailed comparison of the (pixel-by-pixel) RMS and TIC-based normalization demonstrated convincingly that the impact of different numerical procedures had no significant effect on quantification and conclusions from this study (see Fig. S4).
To analyze the IHC data, regions of interest (ROI’s) within the CC and cerebral cortex (CerC) were annotated and positive staining was quantified by applying computer-generated algorithms (Visiopharm, Hoersholm, Denmark). Unpaired Student’s t-test with Welch’s correction and two-way ANOVA with Bonferroni post hoc test were used for statistical analysis. Data were expressed as mean ± SD. Differences were considered statistically significant at p<0.05. The p values, bar charts, correlation coefficients, and regression best-fit lines were generated with GraphPad Prism version 6 (GraphPad Software Inc., La Jolla, CA).
Results
Altered In Situ Distribution of Sulfatide and Phosphatidylcholine Species in the CC of Shi Mouse and Cz-Fed Mouse
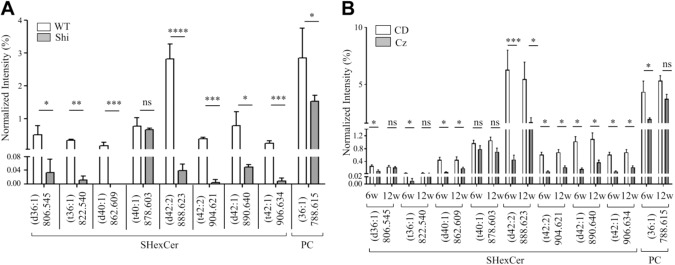
MALDI-IMS revealed differences in the spatial distribution of individual SHexCer and PC species in the CC and other regions including mid brain, thalamus, hypothalamus, pons, and cerebellar WM of the Shi mouse brain compared with the WT mouse (Fig. 1). The spatial distribution of SHexCer (d42:2) (m/z 888.623) and PC (36:1) (m/z 788.615) was decreased; whereas, spatial localization of SHexCer (d36:1) (m/z 806.545) was increased within the affected regions (Fig. 1A–C). For a semiquantitative comparison of their tissue levels, TIC normalized ion intensities were calculated from selected ROI in the CC (Fig. 1A–C; dashed outlines) of the Shi and WT mouse. Consistent with the decreased spatial distribution, the normalized intensities of SHexCer (d42:2) and PC (36:1) were lower in the CC of the Shi mouse indicating their reduced tissue levels compared with the WT (Fig. 1A and C). Furthermore, in agreement with the increased spatial signals, the normalized intensity of SHexCer (d36:1) was elevated in the CC of the Shi mouse compared with WT (Fig. 1B). Changes were statistically significant for SHexCer (d42:2) and SHexCer (d36:1) with an average 4-fold decrease and 2-fold increase in their intensity levels, respectively (Fig. 1A and B).
Figure 1.
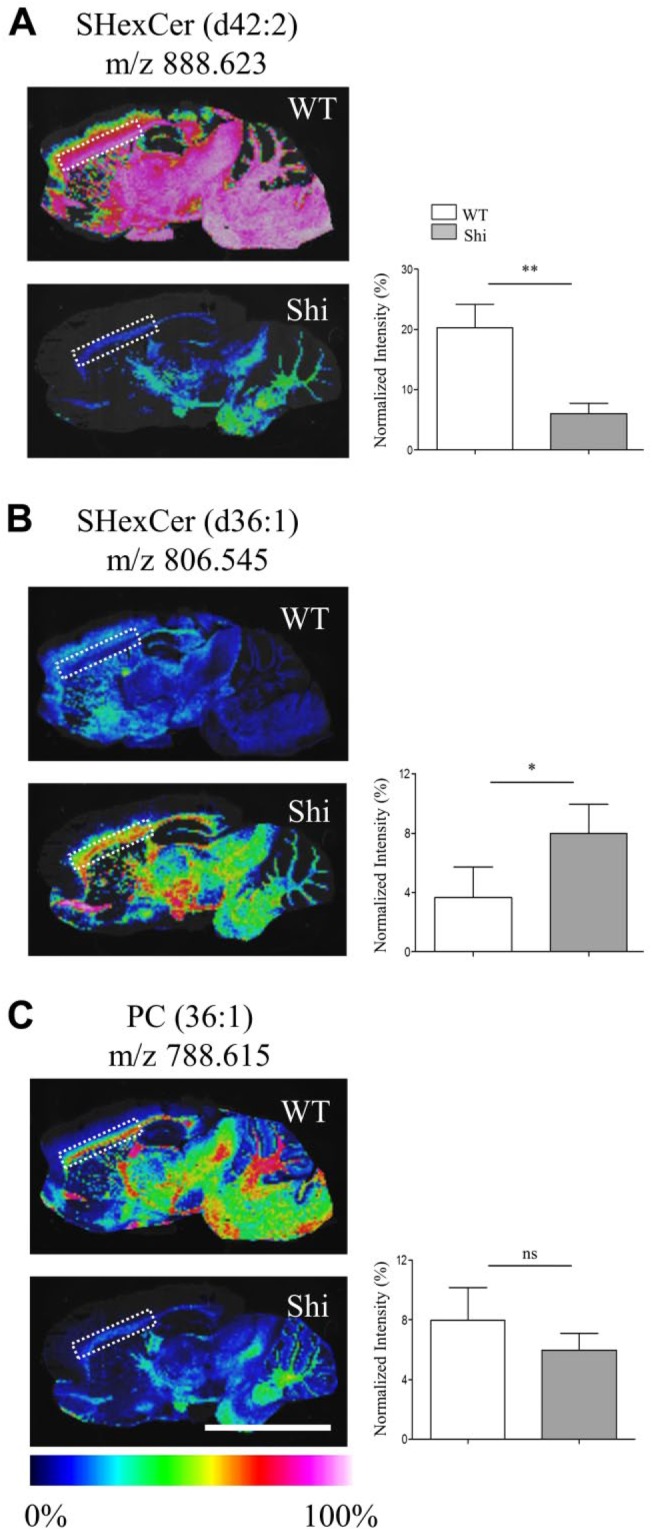
Changes in the spatial distribution of SHexCer and PC species within white matter regions like the CC of Shi mouse brain. (A–C) Normalized MS images of SHexCer (d42:2), SHexCer (d36:1), and PC (36:1), respectively. The spatial distribution of SHexCer (d42:2) and PC (36:1) was reduced in the Shi mouse brain compared with the WT in myelin enriched white matter regions like the CC; whereas, spatial distribution of SHexCer (d36:1) was increased within the affected regions. Lipid annotation and the corresponding m/z are listed above each image. Charts next to each image show normalized intensities (plotted as %) calculated from the selected ROI in the CC (dashed outline in each image). The normalized intensities of SHexCer (d42:2) were significantly decreased; whereas, there was a nonsignificant reduction in the normalized intensities of PC (36:1) in the CC of the Shi mouse compared with WT mouse. Normalized intensities of SHexCer (d36:1) were significantly increased in the Shi mouse compared with WT mouse (n=5, five Shi mice and five WT mice animals were evaluated; unpaired two-tailed t-test; spatial resolution, 80 μm; and scale bar, 5 mm). Abbreviations: CC, corpus callosum; Shi, shiverer; MS, mass spectrometry; SHexCer, sulfatide; PC, phosphatidylcholine; WT, wild type; ROI, region of interest. *p<0.05, **p<0.01.
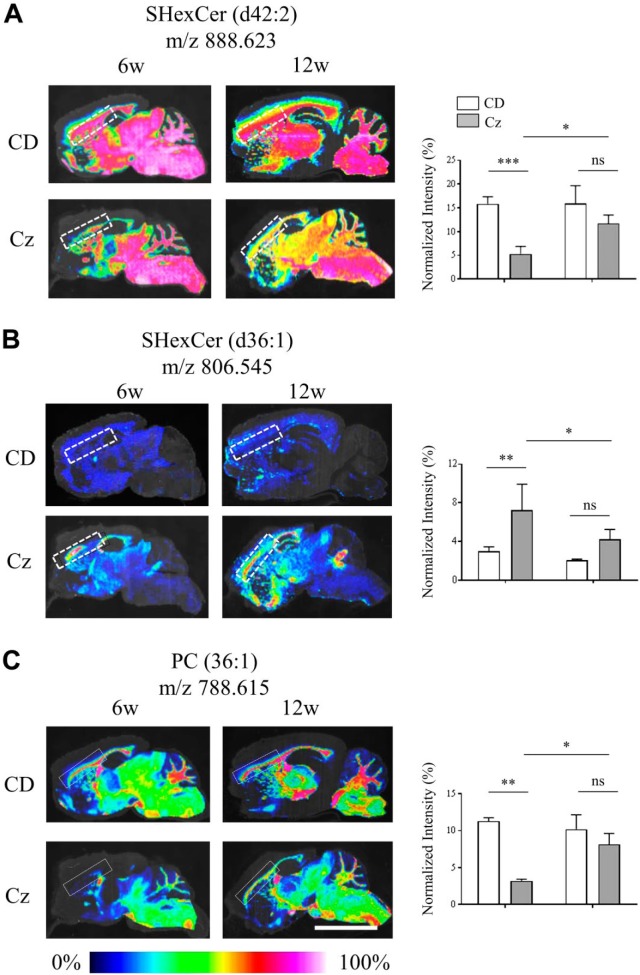
MALDI-IMS also depicted temporal changes in the spatial distribution pattern of SHexCer and PC species in the CC of Cz-fed mouse compared with the CD-fed mouse. Similar to what was observed in the Shi mouse, there was a decrease in the spatial distribution of SHexCer (d42:2) and PC (36:1) and a rise in distribution of SHexCer (d36:1) upon demyelination in the CC of Cz mouse at 6 weeks (Fig. 2A–C; dashed outlines). Spatial distribution of both lipid species was restored albeit not completely back to baseline during remyelination after the removal of Cz at 12 weeks (Fig. 2A–C). Normalized intensities of SHexCer (d42:2) and PC (36:1) significantly lowered after 6-week Cz-feeding indicating their reduced tissue levels (Fig. 2B and C). Alternatively, normalized intensity of SHexCer (d36:1) was significantly elevated after 6-week Cz-feeding (Fig. 2A). The ion intensities restored at 12 weeks, although not completely but significantly compared with the 6-week Cz-fed mouse (Fig. 2A–C). Intensities of the Cz-fed mouse were not significantly different compared with the CD-fed mouse at 12 weeks.
Figure 2.
Altered spatial and temporal distribution of SHexCer and PC species in the CC of the Cz-fed mouse. (A–C) Normalized MS images of SHexCer (d42:2), SHexCer (d36:1), and PC (36:1), respectively. Shown in each panel are the MS images acquired from sagittal brain sections collected from CD-fed mouse (upper row) and Cz-fed mouse (bottom row) at 6w (demyelination) and 12w (remyelination) study time points. Compared with the CD-fed mouse, the spatial distribution of SHexCer (d42:2) and PC (36:1) was reduced within the CC (dashed outline) of Cz-fed mouse after 6w demyelination; whereas, the spatial distribution SHexCer (d36:1) was increased within the affected areas of the CC. Spatial distribution changes of all three lipid species restored albeit not completely during remyelination (12w) in the CC of Cz-fed mouse. Lipid annotation and the corresponding m/z are listed above each image. Charts next to each image show normalized intensities (plotted as %) calculated from the selected ROI in the CC (dashed outline in each image). Normalized intensities of SHexCer (d42:2) and PC (36:1) were significantly lower after the 6w Cz-feeding indicating their reduced tissue levels compared with the CD-fed mouse. Alternatively, normalized intensities of SHexCer (d36:1) were significantly elevated after the 6w Cz-feeding indicating a rise in their tissue levels compared with the CD-fed mouse. The ion intensities restored at 12w although not completely but significantly compared with the 6w Cz-fed mouse. Intensities of the Cz-fed mouse were not significantly different compared with the CD-fed mouse at 12w (n=4, four Cz-fed mice and four CD-fed mice were evaluated per time point; two-way ANOVA; spatial resolution, 80 μm; and scale bar, 5 mm). Abbreviations: CC, corpus callosum; Cz, cuprizone; MS, mass spectrometry; SHexCer, sulfatide; PC, phosphatidylcholine; ROI, region of interest; CD, chow diet; w, weeks. *p<0.05, **p<0.01, ***p<0.001.
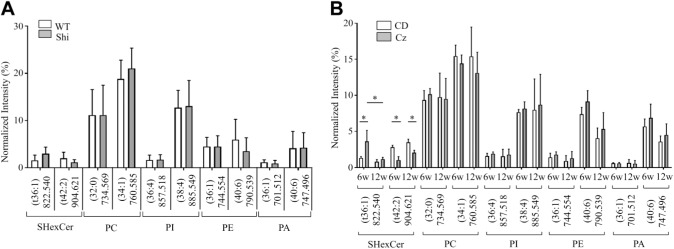
In both, the Shi and Cz-fed mice, changes occurred in a few other SHexCer species located in the WM. The normalized intensity of SHexCer (t36:1) was elevated; whereas, normalized intensity of SHexCer (t42:2) was reduced in the CC of Shi mouse compared with the WT (Fig. 3A). However, these differences were not statistically significant. Cz-fed mouse exhibited a significant rise in the normalized intensity of SHexCer (t36:1) and significant decrease in the normalized intensity of SHexCer (t42:2) within the CC at 6 weeks (Fig. 3B). By 12 weeks, the intensities of both lipid species restored (Fig. 3B). However, the intensity of SHexCer (t42:2) remained significantly lower compared with the CD-fed mouse.
Figure 3.
Changes occurred in other SHexCer species located in the white matter but phospholipid species localized in gray matter regions did not differ significantly either in the Shi or Cz-fed mouse. (A) The normalized intensity of SHexCer (t36:1) was elevated; whereas, normalized intensity of SHexCer (t42:2) was reduced in the CC of Shi mouse compared with the WT. However, these differences were not statistically significant. PC shown here as well as phospholipid species that belong to PI, PE, and PA did not differ significantly between the Shi and WT mouse. Additional species belonging to these lipid types are shown in the supplementary information. (B) There was a significant rise in the normalized intensity of SHexCer (t36:1) and significant decrease in the normalized intensity of SHexCer (t42:2) in the CC of Cz-fed mouse at 6w. By 12w, the intensities of both lipid species restored. However, the intensity of SHexCer (t42:2) remained significantly lower compared with the CD-fed mouse. Like observed in the Shi mouse, the normalized intensities of PC, PI, PE, and PA species did not differ significantly between the Cz-fed and CD-fed mouse at 6w and 12w. Additional species of the above listed lipid types are shown in the supplementary information. Unpaired two-tailed t-test or two-way ANOVA. Differences were considered as significant when p<0.05. Abbreviations: Shi, shiverer; Cz, cuprizone; SHexCer, sulfatide; CC, corpus callosum; WT, wild type; PC, phosphatidylcholine; PI, phosphatidylinositol; PE, phosphatidylethanolamine; PA, phosphatidic acid; CD, chow diet; w, weeks.
Other PC as well as phospholipids belonging to phosphatidylinositol (PI), phosphatidic acid (PA), and phosphatidylethanolamine (PE) did not differ significantly between the Shi and WT mouse as well as between the Cz-fed and CD-fed mouse (Fig. 3A and B). Normalized intensities for these lipids were calculated from the CerC because they were localized in the gray matter regions. Profiles of additional phospholipid and SHexCer species are shown in the supplementary information (Fig. S5 and S6).
MALDI-IMS Derived Changes in Sulfatide and Phosphatidylcholine Species in the CC of Shi Mouse and Cz-Fed Mouse Correlated With the Local Morphological Changes
It is known that the severity and progression of dysmyelinating phenotype of the Shi mouse correlates with the myelin and axon pathology.46 Also, many studies have reported demyelination and the associated microglial and astroglial responses in the CC of Cz-fed mouse that were restored during remyelination after the removal of Cz in diet.36,47–49 To determine if the MALDI-IMS derived distribution changes in SHexCer and PC correlate with the regional brain histopathology in the Shi and Cz-fed mouse, IHC of sagittal brain sections was performed using established myelin and glial cell markers.
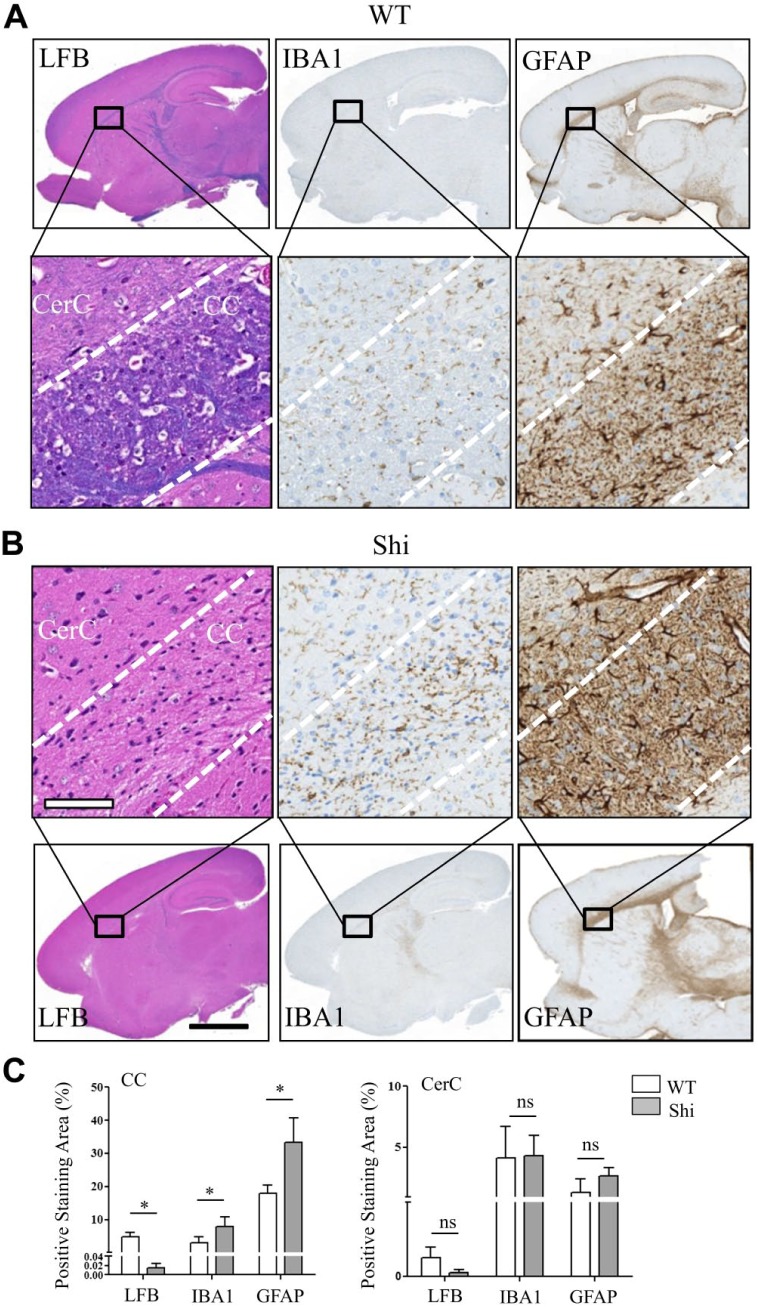
There was a decrease in the LFB positive staining within the CC, mid brain, thalamus, hypothalamus, pons, and cerebellar WM confirming hypomyelination in the Shi mouse compared with the WT (Fig. 4A and B). Also within these regions, there were increased number of microglia and astrocytes in the Shi mouse as indicated by the increased IBA1 and GFAP positive staining, respectively (Fig. 4A and B). Quantitative comparison of staining levels in the CC revealed significant hypomyelination and a significant reactive microgliosis and astrogliosis in the Shi mouse compared with the WT (Fig. 4C). No significant difference was observed between the WT and Shi mouse in the staining levels of the tested markers in the gray matter regions such as CerC (Fig. 4C). These morphological changes correlated with the changes in spatial distribution and intensities of SHexCer and PC species in the Shi mouse (Fig. 1A–C).
Figure 4.
Altered spatial distribution patterns of sulfatide and phosphatidylcholine species correlated with reduced myelin staining, reactive macrogliosis and reactive astrocytosis in the CC of Shi mouse brain. A full view of digital scanned images from the WT and Shi mouse sagittal brain sections stained with LFB, IBA1, and GFAP (A, top row, and B, bottom row). (A) Bottom row and (B) top row magnified views of CC and CerC of the corresponding image shown in the column. (C) Whereas the LFB positive staining was significantly decreased, the IBA1 and GFAP positive staining was significantly increased in the CC of Shi mouse. There was no significant difference between the mice of either genotype in the levels of all markers in the CerC. Unpaired two-tailed t-test; n=5; spatial resolution, 80 μm; white scale bar, 90 μm; and black scale bar, 3 mm. Abbreviations: CC, corpus callosum; Shi, shiverer; WT, wild type; LFB, luxol fast blue; IBA, ionized calcium-binding adapter molecule; GFAP, glial fibrillary acidic protein; CerC, cerebral cortex. *p<0.05.
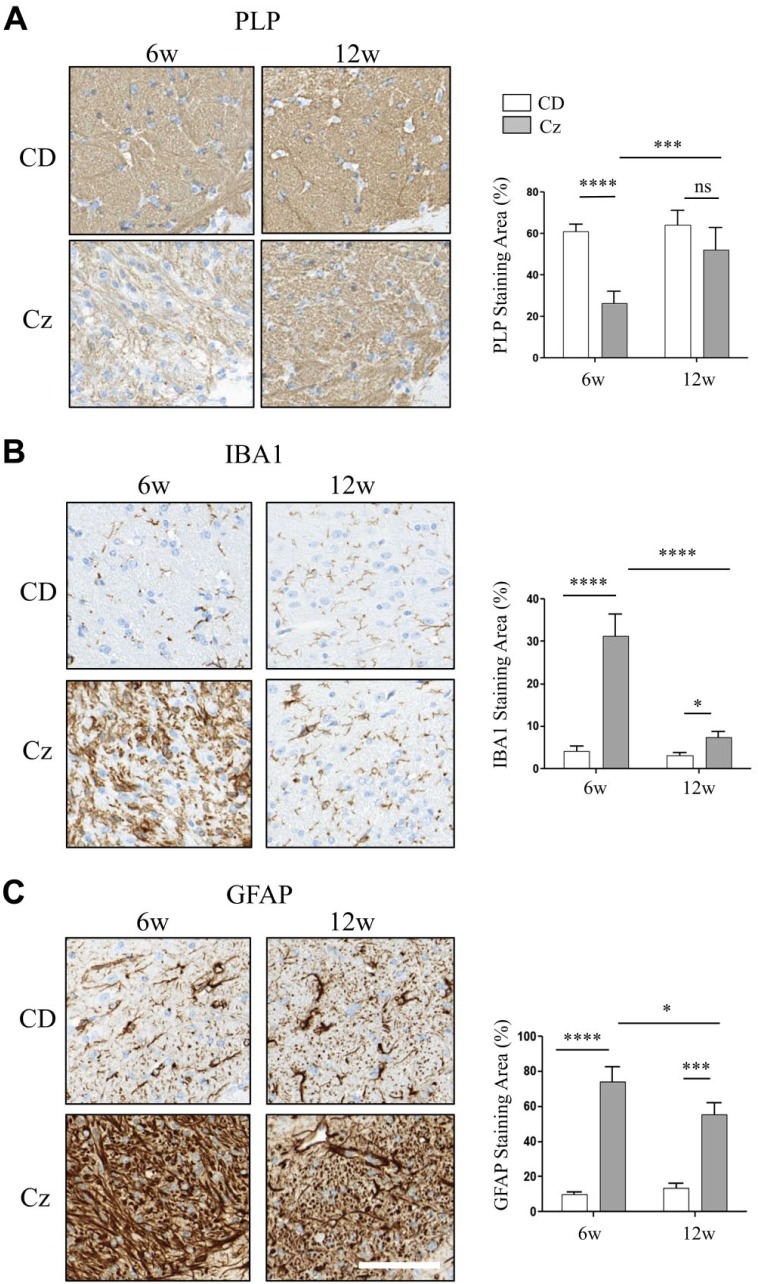
A significantly lower positive staining for PLP, the major myelin-specific protein, was noticed in the CC of Cz-fed mouse at 6 weeks confirming demyelination (Fig. 5A). In addition, within these foci, there were increased numbers of microglia and astrocytes as indicated by a significant rise in the IBA1 and GFAP positive staining, respectively (Fig. 5B and C). After Cz removal in diet, PLP staining was significantly restored at 12 weeks compared with 6 weeks and did not significantly differ compared with the CD-fed mouse (Fig. 5A). Cz removal significantly reduced the microglial and astroglial response at 12 weeks compared with 6 weeks, but IBA1 and GFAP staining levels remained significantly higher than the CD-fed mouse (Fig. 5B and C). These morphological changes overlapped with the temporal changes in spatial distribution of SHexCer and PC species in the Cz-fed mouse (Fig. 2A–C).
Figure 5.
Spatial and temporal distribution changes of sulfatide and phosphatidylcholine species correlated with reduced myelin staining, reactive macrogliosis, and reactive astrocytosis in the CC of Cz-fed mouse brain. (A–C) CC region of brain sections collected from the CD-fed (upper row) and Cz-fed mouse (bottom row) at 6w and 12w study time points and stained with PLP, IBA1, and GFAP, respectively. Chart next to each image show positive staining area (plotted as %). There was a significant decrease in the PLP-positive staining; whereas, IBA1 and GFAP positive staining was significantly increased in the CC of Cz-fed mouse after 6w demyelination. Consistent with remyelination, PLP staining was restored at 12w and there was no significant difference in the staining levels compared with the age-matched CD-fed mouse. By 12w, IBA and GFAP positive staining also resolved significantly compared with the 6w Cz-fed mouse. However, staining levels remain significantly elevated compared with the age-matched CD-fed mouse. Two-way ANOVA, n ranged from five to eight animals per time point, and scale bar, 90 μm. Abbreviations: CC, corpus callosum; Cz, cuprizone; CD, chow diet; PLP, proteolipid protein; IBA, ionized calcium-binding adapter molecule; GFAP, glial fibrillary acidic protein; w, weeks. *p<0.05, ***p<0.001, ****p<0.0001.
LC-ESI-MS Analysis Confirmed the Reduced Abundance of Many Sulfatide and Phosphatidylcholine Species in the CC of Shi Mouse and Cz-Induced Demyelination but No Species With Elevated Abundance Were Found
To qualify whether the spatial distribution changes in SHexCer and PC species occurring in the Shi mouse and Cz-fed mouse correspond to their actual local tissue levels, LC-ESI-MS analysis was performed using the dissected brain tissue. As the in situ lipid profiles in the CC correlated with the associated histopathological changes, segments of brain tissue were meticulously dissected from CC of the Shi and WT mouse, and from the Cz-fed and CD-fed mouse. Lipid extracts were prepared from the tissue homogenate and lipid levels were measured using an internal standard (see methods). Consistent with the MALDI-IMS results, LC-ESI-MS derived normalized intensities for SHexCer (d42:2) and PC (36:1) were significantly decreased in the Shi mouse compared with the WT mouse (Fig. 6A). The intensities of SHexCer (t42:2), SHexCer (d40:1), SHexCer (d42:1), and SHexCer (t42:1) were also significantly decreased in the Shi mouse (Fig. 6A). However, in contrast to the MALDI-IMS derived elevations (Figs. 1B and 2B), LC-ESI-MS reported a significant decrease in the normalized intensities of SHexCer (d36:1) and SHexCer (t36:1) in the Shi mouse (Fig. 6A). The levels of SHexCer species were reduced by a greater fold change compared with PC (36:1), and the SHexCer (d42:2) and SHexCer (t42:2) exhibited a greater fold reduction compared with other SHexCer species (Fig. 6A). Similar changes in the profiles of above described lipid species were observed upon demyelination at 6 weeks in the Cz-fed mouse (Fig. 6B). In addition, lipid levels were partially restored at 12 weeks after removal of Cz from the diet (Fig. 6B).
Figure 6.
LC-ESI-MS analysis confirmed the reduced abundance of SHexCer and PC species in the CC of Shi mouse brain and upon Cz-induced demyelination but no lipid species were elevated. (A) LC-ESI-MS-derived normalized intensities (plotted as %) for SHexCer (d40:1), SHexCer (d42:2), SHexCer (t42:2), SHexCer (d42:1), SHexCer (t42:1), and PC (36:1) were significantly decreased in the Shi mouse. Normalized intensities of SHexCer (d36:1) and SHexCer (t36:1) were also significantly decreased in the Shi mouse. There was no significant difference in intensities of SHexCer (t40:1) between the Shi and WT mouse. Unpaired two-tailed t-test, five animals of each genotype were evaluated. (B) LC-ESI-MS-derived normalized intensities (plotted as %) for SHexCer (d40:1), SHexCer (d42:2), SHexCer (t42:2), SHexCer (d42:1), SHexCer (t42:1), and PC (36:1) were significantly decreased after 6w in Cz-treatment. Intensities of all these lipid species partially restored at 12 weeks after removal of Cz from the diet. Normalized intensities of SHexCer (d36:1) and SHexCer (t36:1) were reduced after 6w of Cz-feeding but the intensities these lipid species completely restored by 12w. There was a small reduction in the intensities of SHexCer (t40:1) in the Cz-fed mouse at both 6w and 12w but the intensities were not significantly different compared with the age-matched CD-fed mouse. Two-way ANOVA, five to eight animals of each type were analyzed per time point. Abbreviations: LC-ESI-MS, liquid chromatography–electrospray ionization–mass spectrometry; CC, corpus callosum; Shi, shiverer; Cz, cuprizone; SHexCer, sulfatide; PC, phosphatidylcholine; WT, wild type; CD, chow diet; w, weeks. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Discussion
The purpose of this study was to define myelin damage associated changes in lipid distribution and composition combining FTICR-based MALDI-IMS, IHC, and LC-ESI-MS analyses. In both the Shi mouse and Cz mouse models, most notable lipid distribution changes occurred in well-myelinated WM regions such as the CC. The spatial distribution and abundance of SHexCer (d42:2) and PC (36:1) were reduced in the CC of Shi mouse compared with WT mouse (Fig. 1A and C and Fig. 6A), and upon Cz-induced demyelination compared with the CD-fed mouse (Fig. 2A and C and Fig. 6B). The abundance of additional SHexCer species was also reduced in the Shi mouse CC (Fig. 6A) as well as upon Cz-induced demyelination (Fig. 6B). These changes resolved but not entirely after removal of Cz from the diet. Consistent with the changes in SHexCer and PC (36:1), IHC confirmed reduced myelin staining and reactive gliosis in the CC (Figs. 4 and 5). No differences were observed in PI, PE, PA, and PC lipid species that predominantly distributed in the gray matter, in both the Shi and Cz mouse models (Fig. 3A and B; and Fig. S1 and 2).
It is noteworthy that both the Shi and Cz mouse models suffered a significant decline in the CC levels of SHexCer (d42:2), the most abundant SHexCer species in myelin sheaths. Myelinating oligodendrocytes are the major sources of SHexCer in the CNS.50 SHexCer-s are crucial for maintaining the myelin structure,51 regulating oligodendrocyte development and survival, and for axon-glial signaling.52,53 They are sulfated-glycosphingolipids synthesized downstream of ceramides and cerebrosides by the enzymes UDP-galactose:ceramide galactosyltransferase (CGT) and cerebroside sulfotransferase (CST).54,55 Three structural variants of SHexCer species include chain length, hydroxylation, and degree of unsaturation of the fatty acid chain.56 Fatty acid chains containing at least 40 carbons are considered as “very long” (VLCFA) and fatty acid chains with 36 to 38 carbons are referred as “long” (LCFA). For example, SHexCer (42:2), which contains a total of 42 carbons (monounsaturated fatty acid chain 24C:1 and sphingosine group 18:1) is a SHexCer-VLCFA; whereas, SHexCer (36:1) is a SHexCer-LCFA that contains 36 carbons (18C:0 fatty acid chain and 18:1 sphingosine group). The annotations d and t represent hydroxylation and nonhydroxylation of the fatty acid chain, respectively. Following synthesis, the SHexCer-VLCFA mainly SHexCer (42:2), are incorporated into the myelin membranes where they stabilize myelin structure in association with MBP and PLP.4,5 SHexCer are degraded in lysosomes by the enzyme arylsulfatase A (ASA).57 Disturbances in the synthesis of SHexCer secondary to abnormal expression of CGT or CST; defective assembly of SHexCer into the myelin membranes, or their degradation results in alterations in their levels thereby leading to impaired conduction of nerve impulses.58,59
Prior studies have reported a decrease in the total SHexCer content in CNS myelin fractions evaluated using traditional lipid analysis methods.60,61 Here, we showed decreased local composition of individual SHexCer species including the myelin-specific SHexCer-VLCFA. No associated changes in the expression levels of SHexCer metabolizing enzymes like CGT, CST, or ASA were reported in the Shi mouse brain62 indicating that loss of MBP does not impact SHexCer synthesis or degradation. Therefore, we presume that lack of MBP in Shi mice results in paucity of compact myelin and subsequent defective assembly into myelin membranes eventually reducing their amounts. Reduced amounts of SHexCer (d42:2) and other SHexCer-VLCFA has also been reported in the brain tissue from rodent models and human patients with multiple sclerosis.7 Conversely, there was a rise in their levels in the cerebrospinal fluid as well as presence of antibodies to SHexCer in serum from multiple sclerosis patients indicating that myelin loss results in loss of SHexCer from tissue and a subsequent increase in their outflow into the extracellular milieu.11
Studies with Cz model have shown a decrease in the expression of CGT as well as of the cholesterol-synthesizing enzyme, HMG-CoA reductase in mouse brain upon demyelination.9,63 As a result, the levels of cerebrosides and cholesterol were reduced. Cz-treatment also upregulated the expression of serine palmitoyl transferase (SPT), the rate limiting enzyme required for de novo synthesis of ceramides, in reactive-astrocytes.64 However, in our study, cholesterol, ceramide, or cerebroside species were not detected by MALDI-IMS probably due to their weak ionization efficiency. Cholesterol ions were detected by LC-ESI-MS and there was a small reduction in the ion intensities of 6-week Cz-fed mouse compared with CD-fed mouse (Fig. S7) but the changes were not statistically significant. Therefore, these were not evaluated further, and we limited semiquantitative analysis to lipid species detected in common by both MALDI-IMS and LC-ESI-MS. However, cholesterol being one of three major lipid species in myelin plays an important role in myelin formation and myelin stability; mapping of cholesterol species in future studies using other ionization methods like SIMS or specific sample preparation optimized for the detection of cholesterol65 may provide more insights into understanding lipid alterations associated with myelin damage.
For most lipid species, MALDI-IMS and LC-ESI-MS results were congruent in both the mouse models. However, opposite fold differences were observed with a few lipid species. For example, in contrast to the MALDI-IMS derived rise in the spatial distribution and normalized intensities of SHexCer-LCFA like SHexCer (d36:1) and SHexCer (t36:1) (Fig. 1B and 2B and Fig. 3A and B), LC-ESI-MS reported a significant decrease in the abundance of SHexCer (d36:1) and SHexCer (t36:1) in both the Shi mouse and Cz-induced demyelination (Fig. 6A and B). The distribution and abundance of both species recovered after removal of Cz from the diet (Figs. 2B, 3B, and 6B). In another instance, MALDI-IMS did not yield difference in signal intensities of SHexCer-VLCFA like SHexCer (d40:1), SHexCer (d42:1), and SHexCer (t42:1) in the Shi mouse (Fig. S5A) as well as SHexCer (d40:1) in Cz model (Fig. S6A). However, LC-ESI-MS inferred a significant reduction in the levels of these SHexCer species in both mouse models (Fig. 6A and B). We speculate these differences are an outcome of the differences in matrix effects and dynamic range of detection between the two methods.30,32,66,67
Normally, CC is mainly composed of myelinating oligodendrocytes that are enriched in very-long chain fatty acid containing sulfatides (SHexCer-VLCFA) compared with long chain fatty acid containing sulfatides (SHexCer-LCFA). We presume that highly abundant SHexCer-VLCFA might have minimized the signal from or suppressed detection of less abundant SHexCer-LCFA in the control mouse. However, upon Cz-induced demyelination or in dysmyelinating Shi mouse, SHexCer levels were reduced in CC. It is noteworthy that the levels of SHexCer-VLCFA were reduced to a greater degree than the levels of SHexCer-LCFA (per LC-ESI-MS). As a result, it is possible that IMS detection of SHexCer-LCFA improved thereby giving a false impression of an increase in their spatial distribution. It is also likely that SHexCer-LCFA were better exposed to MALDI laser beam thus allowing improved detection compared with otherwise complex (SHexCer-VLCFA enriched myelin) tissue environment in control tissue.
During remyelination in the Cz model, myelin and tissue complexity as well as SHexCer-VLCFA levels were restored although not to the condition of normal myelin. Therefore, this poses a greater interference to the detection of SHexCer-LCFA by MALDI-IMS compared with their detection from demyelinated samples. LC-ESI-MS analysis revealed that the levels of above listed both lipid species were reduced upon myelin loss. This was probably because, both the lipid species were better separated and detected when lipid extracts prepared from normal or diseased myelin were evaluated.
Alternatively, it is possible that other dominant lipid species in myelin like cholesterols may impact the detection of other lipid species. Previous reports using SIMS have shown that cholesterol migrates to surface and form crystals during the process of sublimation thereby decreasing their levels within the tissue.68 Consequently, this decreases the suppression of yield of other lipid ions including the less abundant SHexCer-LCFA thus increasing their detection. Quenching effect on IMS signals of certain lipid species by other abundant lipid species within the ROI has been previously reported.69 SHexCer in kidney medulla and papilla quenched the IMS signal of PI species such as PI (38:4). The quenching effect was minimized when lipid extracts from these regions were analyzed by LC-ESI-MS. Future development and availability of improved analytical approaches combining use of deuterated or isotope-labeled reference lipid standards70–72 should distinguish the effects of ion suppression and signal variations due to local tissue environment.
In summary, we demonstrated that lack of MBP in the Shi mouse and demyelination in Cz-fed mouse mainly impacts the SHexCer composition and to a lesser extent the PC species. Among the SHexCer species, the levels of SHexCer-VLCFA were reduced to a greater extent than SHexCer-LCFA. These alterations were restored upon remyelination following removal of Cz from the diet. By applying MALDI-IMS for the first time in myelin disease mouse models, we could convincingly demonstrate the representative spatial as well as temporal distribution of brain lipids with histological correlation. A broader application of this technique complementing with LC-ESI-MS and other morphology-based assays to study lipids in other myelin diseases might provide new insights in understanding disease mechanisms.
Supplemental Material
Supplemental material, DS_10.1369_0022155418815860 for Defining Changes in the Spatial Distribution and Composition of Brain Lipids in the Shiverer and Cuprizone Mouse Models of Myelin Disease by Rajanikanth J. Maganti, Xiaoping L. Hronowski, Robert W. Dunstan, Brian T. Wipke, Xueli Zhang, Luke Jandreski, Stefan Hamann and Peter Juhasz in Journal of Histochemistry & Cytochemistry
Footnotes
Competing Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: RJM is an employee of and holds stock options in Moderna Therapeutics. XLH, BTW, XZ, LJ, SH, and PJ are/were employees of and hold stock/options in Biogen, Inc. RWD is an employee of and holds stock/options in AbbVie, Inc.
Author Contributions: RJM conceived the experimental design, executed the in vivo and ex vivo studies including data analysis and interpretation, and drafted the manuscript; XLH assisted in matrix-assisted laser desorption ionization imaging mass spectrometry (MALDI-IMS) preliminary method development, performed liquid chromatography–electrospray ionization–mass spectrometry (LC-ESI-MS) and assisted in data analysis, and drafted portion of LC-ESI-MS methods section; RWD assisted with review and interpretation of histology, immunohistochemistry (IHC) and MALDI-IMS data; BTW assisted with design and execution of animal studies, data interpretation and manuscript review; XZ performed histology and IHC; SH and LJ performed digital scanning of histology and IHC slides, ran the image analysis algorithms for the IHC data quantification; and PJ assisted with review and interpretation of MALDI-IMS and LC-ESI-MS data.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The financial support for the study was provided by Biogen (Cambridge, MA), which is a commercial entity.
Contributor Information
Rajanikanth J. Maganti, Biogen, Cambridge, Massachusetts Moderna Therapeutics, Cambridge, Massachusetts.
Xiaoping L. Hronowski, Biogen, Cambridge, Massachusetts
Robert W. Dunstan, Biogen, Cambridge, Massachusetts AbbVie, Worcester, Massachusetts.
Brian T. Wipke, Biogen, Cambridge, Massachusetts
Xueli Zhang, Biogen, Cambridge, Massachusetts.
Luke Jandreski, Biogen, Cambridge, Massachusetts.
Stefan Hamann, Biogen, Cambridge, Massachusetts.
Peter Juhasz, Biogen, Cambridge, Massachusetts.
Literature Cited
- 1. Quarles RH, Macklin WB, Morell P. Myelin formation, structure and biochemistry. In: Brady S, Siegel G, Albers RW, Price D, editors. Basic neurochemistry: molecular, cellular and medical aspects. 7th edition. Burlington: Elsevier Academic Press; 2005. p. 51–71. [Google Scholar]
- 2. Norton WT, Poduso SE. Myelination in rat brain: changes in myelin composition during brain maturation. J Neurochem. 1973;21(4):759–73. [DOI] [PubMed] [Google Scholar]
- 3. Bradl M. Myelin dysfunction/degradation in the central nervous system: why are myelin sheaths susceptible to damage? J Neural Transm Suppl. 1999;55:9–17. [DOI] [PubMed] [Google Scholar]
- 4. Min Y, Kristiansen K, Boggs JM, Husted C, Zasadzinski JA, Israelachvili J. Interaction forces and adhesion of supported myelin lipid bilayers modulated by myelin basic protein. Proc Natl Acad Sci U S A. 2009;106(9):3154–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ohler B, Graf K, Bragg R, Lemons T, Coe R, Genain C, Isrealachvili J, Husted C. Role of lipid interactions in autoimmune demyelination. Biochim Biophys Acta. 2004;1688(1):10–7. [DOI] [PubMed] [Google Scholar]
- 6. Duncan ID, Kondo Y, Zhang SC. The myelin mutants as models to study myelin repair in the leukodystrophies. Neurotherapeutics. 2011;8(4):607–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wheeler D, Bandaru VV, Calabresi PA, Nath A, Haughey NJ. A defect of sphingolipid metabolism modifies the properties of normal appearing white matter in multiple sclerosis. Brain. 2008;131(Pt 11):3092–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gregson NA. Lysolipids and membrane damage: lysolecithin and its interaction with myelin. Biochem Soc Trans. 1989;17(2):280–3. [DOI] [PubMed] [Google Scholar]
- 9. Jurevics H, Hostettler J, Muse ED, Sammond DW, Matsushima GK, Toews AD, Morell P. Cerebroside synthesis as a measure of the rate of remyelination following cuprizone-induced demyelination in brain. J Neurochem. 2001;77(4):1067–76. [DOI] [PubMed] [Google Scholar]
- 10. Muse ED, Jurevics H, Toews AD, Matsushima GK, Morell P. Parameters related to lipid metabolism as markers of myelination in mouse brain. J Neurochem. 2001;76(1):77–86. [DOI] [PubMed] [Google Scholar]
- 11. Haghighi S, Lekman A, Nilsson S, Blomqvist M, Andersen O. Myelin glycosphingolipid immunoreactivity and CSF levels in multiple sclerosis. Acta Neurol Scand. 2012;125(1):64–70. [DOI] [PubMed] [Google Scholar]
- 12. Ilyas AA, Chen ZW, Cook SD. Antibodies to sulfatide in cerebrospinal fluid of patients with multiple sclerosis. J Neuroimmunol. 2003;139(1–2):76–80. [DOI] [PubMed] [Google Scholar]
- 13. Del Boccio P, Pieragostino D, Di Ioia M, Petrucci F, Lugaresi A, De Luca G, Gambi D, Onofrj M, Di Ilio C, Sacchetta P, Urbani A. Lipidomic investigations for the characterization of circulating serum lipids in multiple sclerosis. J Proteomics. 2011;74(12):2826–36. [DOI] [PubMed] [Google Scholar]
- 14. Marbois BN, Faull KF, Fluharty AL, Raval-Fernandes S, Rome LH. Analysis of sulfatide from rat cerebellum and multiple sclerosis white matter by negative ion electrospray mass spectrometry. Biochim Biophys Acta. 2000;1484(1):59–70. [DOI] [PubMed] [Google Scholar]
- 15. Balood M, Zahednasab H, Siroos B, Mesbah-Namin SA, Torbati S, Harirchian MH. Elevated serum levels of lysophosphatidic acid in patients with multiple sclerosis. Hum Immunol. 2014;75(5):411–3. [DOI] [PubMed] [Google Scholar]
- 16. Tong XW, Xue QM. Alterations of serum phospholipids in patients with multiple sclerosis. Chin Med J (Engl). 1993;106(9):650–4. [PubMed] [Google Scholar]
- 17. Han X, Holtzman DM, McKeel DW, Jr, Kelley J, Morris JC. Substantial sulfatide deficiency and ceramide elevation in very early Alzheimer’s disease: potential role in disease pathogenesis. J Neurochem. 2002;82(4):809–18. [DOI] [PubMed] [Google Scholar]
- 18. Irizarry MC. A turn of the sulfatide in Alzheimer’s disease. Ann Neurol. 2003;54(1):7–8. doi: 10.1002/ana.10642. [DOI] [PubMed] [Google Scholar]
- 19. He X, Chen F, McGovern MM, Schuchman EH. A fluorescence-based, high-throughput sphingomyelin assay for the analysis of Niemann-Pick disease and other disorders of sphingomyelin metabolism. Anal Biochem. 2002;306(1):115–23. [DOI] [PubMed] [Google Scholar]
- 20. Ramakrishnan H, Hedayati KK, Lullmann-Rauch R, Wessig C, Fewou SN, Maier H, Goebel HH, Gieselmann V, Eckhardt M. Increasing sulfatide synthesis in myelin-forming cells of arylsulfatase A-deficient mice causes demyelination and neurological symptoms reminiscent of human metachromatic leukodystrophy. J Neurosci. 2007;27(35):9482–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ishizuka I. Chemistry and functional distribution of sulfoglycolipids. Prog Lipid Res. 1997;36(4):245–319. [DOI] [PubMed] [Google Scholar]
- 22. Norton WT, Autilio LA. The chemical composition of bovine CNS myelin. Ann N Y Acad Sci. 1965;122:77–85. [DOI] [PubMed] [Google Scholar]
- 23. Barcelo-Coblijn G, Fernandez JA. Mass spectrometry coupled to imaging techniques: the better the view the greater the challenge. Front Physiol. 2015;6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chaurand P, Schwartz SA, Billheimer D, Xu BJ, Crecelius A, Caprioli RM. Integrating histology and imaging mass spectrometry. Anal Chem. 2004;76(4):1145–55. [DOI] [PubMed] [Google Scholar]
- 25. Hanrieder J, Phan NT, Kurczy ME, Ewing AG. Imaging mass spectrometry in neuroscience. ACS Chem Neurosci. 2013;4(5):666–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nilsson A, Goodwin RJ, Shariatgorji M, Vallianatou T, Webborn PJ, Andren PE. Mass spectrometry imaging in drug development. Anal Chem. 2015;87(3):1437–55. [DOI] [PubMed] [Google Scholar]
- 27. Takats Z, Wiseman JM, Cooks RG. Ambient mass spectrometry using desorption electrospray ionization (DESI): instrumentation, mechanisms and applications in forensics, chemistry, and biology. J Mass Spectrom. 2005;40(10):1261–75. [DOI] [PubMed] [Google Scholar]
- 28. Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, Murphy RC. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem Rev. 2011;111(10):6491–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–60. [DOI] [PubMed] [Google Scholar]
- 30. Heeren RM, Smith DF, Stauber J, Kukrer-Kaletas B, MacAleese L. Imaging mass spectrometry: hype or hope? J Am Soc Mass Spectrom. 2009;20(6):1006–14. [DOI] [PubMed] [Google Scholar]
- 31. Nimesh S, Mohottalage S, Vincent R, Kumarathasan P. Current status and future perspectives of mass spectrometry imaging. Int J Mol Sci. 2013;14(6):11277–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sparvero LJ, Amoscato AA, Dixon CE, Long JB, Kochanek PM, Pitt BR, Bayir H, Kagan VE. Mapping of phospholipids by MALDI imaging (MALDI-MSI): realities and expectations. Chem Phys Lipids. 2012;165(5):545–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andersson M, Andren P, Caprioli RM. MALDI imaging and profiling mass spectrometry in neuroproteomics. In: Alzate O, editor. Neuroproteomics. Frontiers in neuroscience. Boca Raton: CRC Press; 2010. p. 115–34. [PubMed] [Google Scholar]
- 34. Hanada M, Sugiura Y, Shinjo R, Masaki N, Imagama S, Ishiguro N, Matsuyama Y, Setou M. Spatiotemporal alteration of phospholipids and prostaglandins in a rat model of spinal cord injury. Anal Bioanal Chem. 2012;403(7):1873–84. [DOI] [PubMed] [Google Scholar]
- 35. Shariatgorji M, Nilsson A, Goodwin RJ, Kallback P, Schintu N, Zhang X, Crossman AR, Bezard E, Svenningsson P, Andren PE. Direct targeted quantitative molecular imaging of neurotransmitters in brain tissue sections. Neuron. 2014;84(4):697–707. doi: 10.1016/j.neuron.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 36. Praet J, Guglielmetti C, Berneman Z, Van der Linden A, Ponsaerts P. Cellular and molecular neuropathology of the cuprizone mouse model: clinical relevance for multiple sclerosis. Neurosci Biobehav Rev. 2014;47:485–505. [DOI] [PubMed] [Google Scholar]
- 37. Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–76. [DOI] [PubMed] [Google Scholar]
- 38. Angel PM, Spraggins JM, Baldwin HS, Caprioli R. Enhanced sensitivity for high spatial resolution lipid analysis by negative ion mode matrix assisted laser desorption ionization imaging mass spectrometry. Anal Chem. 2012;84(3):1557–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thomas Al, Charbonneau JL, Fournaise E, Chaurand PJ. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1, 5-diaminonapthalene deposition. Anal Chem. 2012;84(4):2048–54. [DOI] [PubMed] [Google Scholar]
- 40. Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom. 2007;18(9):1646–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murphy RC, Hankin JA, Barkley RM, Zemski Berry KA. MALDI imaging of lipids after matrix sublimation/deposition. Biochim Biophys Acta. 2011;1811(11):970–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Subramaniam S. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35(Database issue):D527–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sud M, Fahy E, Cotter D, Dennis EA, Subramaniam S. LIPID MAPS-nature lipidomics gateway: an online resource for students and educators interested in lipids. J Chem Educ. 2012;89(2):291–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gode D, Volmer DA. Lipid imaging by mass spectrometry: a review. Analyst. 2013;138(5):1289–315. [DOI] [PubMed] [Google Scholar]
- 45. Gopalakrishnan G, Awasthi A, Belkaid W, De Faria O, Jr, Liazoghli D, Colman DR, Dhaunchak AS. Lipidome and proteome map of myelin membranes. J Neurosci Res. 2013;91(3):321–34. [DOI] [PubMed] [Google Scholar]
- 46. Loers G, Aboul-Enein F, Bartsch U, Lassmann H, Schachner M. Comparison of myelin, axon, lipid, and immunopathology in the central nervous system of differentially myelin-compromised mutant mice: a morphological and biochemical study. Mol Cell Neurosci. 2004;27(2):175–89. [DOI] [PubMed] [Google Scholar]
- 47. Blakemore WF, Franklin RJ. Remyelination in experimental models of toxin-induced demyelination. Curr Top Microbiol Immunol. 2008;318:193–212. [DOI] [PubMed] [Google Scholar]
- 48. Kipp M, Clarner T, Dang J, Copray S, Beyer C. The cuprizone animal model: new insights into an old story. Acta Neuropathol. 2009;118(6):723–36. [DOI] [PubMed] [Google Scholar]
- 49. Matsushima GK, Morell P. The neurotoxicant, cuprizone, as a model to study demyelination and remyelination in the central nervous system. Brain Pathol. 2001;11(1):107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marcus J, Popko B. Galactolipids are molecular determinants of myelin development and axo-glial organization. Biochim Biophys Acta. 2002;1573(3):406–13. [DOI] [PubMed] [Google Scholar]
- 51. Ishibashi T, Dupree JL, Ikenaka K, Hirahara Y, Honke K, Peles E, Popko B, Suzuki K, Nishino H, Baba H. A myelin galactolipid, sulfatide, is essential for maintenance of ion channels on myelinated axon but not essential for initial cluster formation. J Neurosci. 2002;22(15):6507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bansal R, Pfeiffer SE. Reversible inhibition of oligodendrocyte progenitor differentiation by a monoclonal antibody against surface galactolipids. Proc Natl Acad Sci U S A. 1989;86(16):6181–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shroff SM, Pomicter AD, Chow WN, Fox MA, Colello RJ, Henderson SC, Dupree JL. Adult CST-null mice maintain an increased number of oligodendrocytes. J Neurosci Res. 2009;87(15):3403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bosio A, Binczek E, Le Beau MM, Fernald AA, Stoffel W. The human gene CGT encoding the UDP-galactose ceramide galactosyl transferase (cerebroside synthase): cloning, characterization, and assignment to human chromosome 4, band q26. Genomics. 1996;34(1):69–75. [DOI] [PubMed] [Google Scholar]
- 55. Honke K, Tsuda M, Hirahara Y, Ishii A, Makita A, Wada Y. Molecular cloning and expression of cDNA encoding human 3’-phosphoadenylylsulfate:galactosylceramide 3’-sulfotransferase. J Biol Chem. 1997;272(8):4864–8. [DOI] [PubMed] [Google Scholar]
- 56. Takahashi T, Suzuki T. Role of sulfatide in normal and pathological cells and tissues. J Lipid Res. 2012;53(8):1437–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Eckhardt M. The role and metabolism of sulfatide in the nervous system. Molecular neurobiology. 2008;37(2–3):93–103. [DOI] [PubMed] [Google Scholar]
- 58. Bosio A, Binczek E, Stoffel W. Functional breakdown of the lipid bilayer of the myelin membrane in central and peripheral nervous system by disrupted galactocerebroside synthesis. Proc Natl Acad Sci U S A. 1996;93(23):13280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Honke K, Taniguchi N. Sulfotransferases and sulfated oligosaccharides. Med Res Rev. 2002;22(6):637–54. [DOI] [PubMed] [Google Scholar]
- 60. Cammer W, Kahn S, Zimmerman T. Biochemical abnormalities in spinal cord myelin and CNS homogenates in heterozygotes affected by the shiverer mutation. J Neurochem. 1984;42(5):1372–8. [DOI] [PubMed] [Google Scholar]
- 61. Inouye H, Ganser AL, Kirschner DA. Shiverer and normal peripheral myelin compared: basic protein localization, membrane interactions, and lipid composition. J Neurochem. 1985;45(6):1911–22. [DOI] [PubMed] [Google Scholar]
- 62. Bird TD, Farrell DF, Stranahan S, Austin E. Developmental dissociation of myelin synthesis and “myelin-associated” enzyme activities in the shiverer mouse. Neurochem Res. 1980;5(8):885–95. [DOI] [PubMed] [Google Scholar]
- 63. Jurevics H, Largent C, Hostettler J, Sammond DW, Matsushima GK, Kleindienst A, Toews AD, Morell P. Alterations in metabolism and gene expression in brain regions during cuprizone-induced demyelination and remyelination. J Neurochem. 2002;82(1):126–36. [DOI] [PubMed] [Google Scholar]
- 64. Kim S, Steelman AJ, Zhang Y, Kinney HC, Li J. Aberrant upregulation of astroglial ceramide potentiates oligodendrocyte injury. Brain Pathol. 2012;22(1):41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dufresne M, Thomas A, Breault-Turcot J, Masson JF, Chaurand P. Silver-assisted laser desorption ionization for high spatial resolution imaging mass spectrometry of olefins from thin tissue sections. Anal Chem. 2013;85(6):3318–24. [DOI] [PubMed] [Google Scholar]
- 66. Taylor PJ. Matrix effects: the Achilles heel of quantitative high-performance liquid chromatography–electrospray–tandem mass spectrometry. Clin Biochem. 2005;38(4):328–34. [DOI] [PubMed] [Google Scholar]
- 67. Trufelli H, Palma P, Famiglini G, Cappiello A. An overview of matrix effects in liquid chromatography–mass spectrometry. Mass Spectrom Rev. 2011;30(3):491–509. [DOI] [PubMed] [Google Scholar]
- 68. Dowlatshahi Pour MD, Malmberg P, Ewing A. An investigation on the mechanism of sublimed DHB matrix on molecular ion yields in SIMS imaging of brain tissue. Anal Bioanal Chem. 2016;408(12):3071–81. [DOI] [PubMed] [Google Scholar]
- 69. Marsching C, Jennemann R, Heilig R, Grone HJ, Hopf C, Sandhoff R. Quantitative imaging mass spectrometry of renal sulfatides: validation by classical mass spectrometric methods. J Lipid Res. 2014;55(11):2343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pirman DA, Kiss A, Heeren RM, Yost RA. Identifying tissue-specific signal variation in MALDI mass spectrometric imaging by use of an internal standard. Anal Chem. 2013;85(2):1090–6. [DOI] [PubMed] [Google Scholar]
- 71. Pirman DA, Yost RA. Quantitative tandem mass spectrometric imaging of endogenous acetyl-L-carnitine from piglet brain tissue using an internal standard. Anal Chem. 2011;83(22):8575–81. [DOI] [PubMed] [Google Scholar]
- 72. Prideaux B, Dartois V, Staab D, Weiner DM, Goh A, Via LE, Barry CE, Stoeckli M. High-sensitivity MALDI-MRM-MS imaging of moxifloxacin distribution in tuberculosis-infected rabbit lungs and granulomatous lesions. Anal Chem. 2011;83(6):2112–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1369_0022155418815860 for Defining Changes in the Spatial Distribution and Composition of Brain Lipids in the Shiverer and Cuprizone Mouse Models of Myelin Disease by Rajanikanth J. Maganti, Xiaoping L. Hronowski, Robert W. Dunstan, Brian T. Wipke, Xueli Zhang, Luke Jandreski, Stefan Hamann and Peter Juhasz in Journal of Histochemistry & Cytochemistry