Abstract
Although cytarabine has been widely considered as one of the chemotherapy drugs for high-risk myelodysplastic syndromes (MDS), the overall response rate is only approximately 20-30%. Nuclear factor erythroid 2-related factor 2 (NRF2, also called NFE2L2) has been shown to play a pivotal role in preventing cancer cells from being affected by chemotherapy. However, it is not yet known whether NRF2 can be used as a prognostic biomarker in MDS, or whether elevated NRF2 levels are associated with cytarabine resistance. Here, we found that NRF2 expression levels in bone marrow from high-risk patients exceeded that of low-risk MDS patients. Importantly, high NRF2 levels are correlated with inferior outcomes in MDS patients (n=137). Downregulation of NRF2 by the inhibitor Luteolin, or lentiviral shRNA knockdown, enhanced the chemotherapeutic efficacy of cytarabine, while MDS cells treated by NRF2 agonist Sulforaphane showed increased resistance to cytarabine. More importantly, pharmacological inhibition of NRF2 could sensitize primary high-risk MDS cells to cytarabine treatment. Mechanistically, downregulation of dual specificity protein phosphatase 1, an NRF2 direct target gene, could abrogate cytarabine resistance in NRF2 elevated MDS cells. Silencing NRF2 or dual specificity protein phosphatase 1 also significantly sensitized cytarabine treatment and inhibited tumors in MDS cells transplanted mouse models in vivo. Our study suggests that targeting NRF2 in combination with conventional chemotherapy could pave the way for future therapy for high-risk MDS.
Introduction
Myelodysplastic syndromes (MDS) are a heterogeneous disease of clonal hematopoietic stem cell neoplasms characterized by ineffective hematopoiesis, cytopenia, dysplasia of the myeloid cells, and an inherent risk of progression to acute myeloid leukemia (AML).1 There are several prognostic scores, and the International Prognostic Scoring System (IPSS) and its revised vision (IPSS-R) are commonly used to stratify patients into two risk groups, defining lower and higher risk patients.2,3 Higher risk MDS patients have an increased risk of developing AML and are associated with poor clinical outcomes. Treatment strategies were made according to the risk categories of MDS. Cytarabine (Ara-C) is a pyrimidine nucleoside analog that interferes with the synthesis of DNA when the cycle holds in the S phase. Over the last decades, Ara-C-based therapies have been widely used to manage MDS patients, especially those at higher risk.4 However, the overall response rate of single Ara-C treatment was only approximately 20-30%.5,6 Clinically, MDS patients who remained unresponsive to the routine treatment of Ara-C (100mg/m2) were defined as Ara-C-resistant MDS patients.
Nuclear factor erythroid 2-related factor 2 (NRF2, also called NFE2L2) is a transcription factor that protects cells from oxidative damage.7,8 Under oxidative stress, NRF2 is released from its cytosolic inhibitor Kelch-like ECH-associated protein 1 (KEAP1) and translocates to the nucleus.9 It has recently been shown that NRF2 underlies drug resistance in acute myeloid leukemia (AML), chronic myeloid leukemia (CML), and chronic lymphocytic leukemia (CLL).10–12 NRF2 binding to antioxidant responsive element (ARE) allows induction of a number of cytoprotective and detoxification genes, such as NAD(P)H: quinone oxidoreductase 1 (NQO1), heme oxygenase-1 (HO-1), and glutamate-cysteine ligase (GCL).7,13,14 Few studies have shown the mechanisms of NRF2 in drug resistance. NRF2 target genes, such as HO-1, have been reported to facilitate resistance of tumor cells to chemotherapy in AML cells.15,16
Here, we aimed to correlate NRF2 expression and its clinical outcome in a large cohort of MDS patients (n=137). We also performed in vitro and in vivo experiments to validate our findings regarding the function of NRF2 in chemo-resistance in MDS. We found that NRF2 expressions were elevated in higher risk MDS and correlated with inferior clinical outcomes. High levels of NRF2 reduced MDS cell sensitivity to Ara-C treatment partly through its direct target gene DUSP1.
Methods
Immunohistochemistry
Immunohistochemistry (IHC) was performed on 4 μm thick bone marrow (BM) sections. BM samples were stained for NRF2 expression (dilution 1:200; Abcam, UK) or DUSP1 expression (dilution 1:100; Abcam, UK). Samples were incubated using primary antibody for 30 minutes (min) at 37°C. Secondary antibody (dilution 1:50; Dako, Denmark) was applied for 15 min. Binding was visualized by the horseradish peroxidase (HRP)/3,3′-diaminobenzidine (DAB) kit (ZSGB-BIO, China). Staining results were semi-quantified using an arbitrary score as follows: No staining, 0; Pale yellow, 1; Tan, 2; Brown, 3; Nuclear staining in 0-25% of cells, 0; Nuclear staining in 25-50% of cells, 1; Nuclear staining in 50-75% of cells, 3; Nuclear staining in 75-100% of cells, 4. The stain color score multiplied by the nuclear staining proportion score is the final IHC score.
In vitro cytotoxicity assay
Myelodysplastic syndrome cell lines (5×105/mL) and primary MDS cells (1×106/mL) were seeded in 96-well flat bottom plates and treated with increasing concentrations of Ara-C. Cell proliferation was determined using the MTS proliferation assay. 20 μl of MTS (Promega, USA) was added to 100 μl of cell suspension, and cells were further incubated in 5% CO2 for 3-4 hours at 37 °C. The plates were then analyzed on an enzyme immunoassay plate reader at 490 nm. The half inhibitory concentration (IC50) values of Ara-C were calculated by Prism Graphpad software. All experiments were performed in triplicate.
Mice models
NOD/SCID-IL2Rγnull-SGM3 (NSGS) mice were bred and maintained in Cincinnati Children’s Hospital Medical Center (CCHMC).17 Mice were randomized into six groups. NRF2 shRNA SKM-1 (transfected with shRNA targeting NRF2), DUSP1 shRNA SKM-1 (transfected with shRNA targeting DUSP1), and scramble shRNA SKM-1 were resuspended in 300 μl phosphate buffer saline (PBS) and then injected intravenously into the non-irradiated mice (1 million cells per mouse). Ten days after cell inoculation (Day 0), the mice received 50 milligram/kilogram (mg/kg) of Ara-C or PBS once a day for five consecutive days (Day 10-15). Ara-C and PBS were injected intraperitoneally. All experiments were performed in accordance with protocols approved by the Institutional Review Board of CCHMC.
Statistical analysis
Data were analyzed using SPSS 16.0 and GraphPad Prism 6. Statistical analyses were performed using Student t-test or one-/two-way ANOVA with multiple comparisons correction. P<0.05 was considered statistically significant.
Results
NRF2 is elevated in higher risk MDS and correlates with inferior overall survival
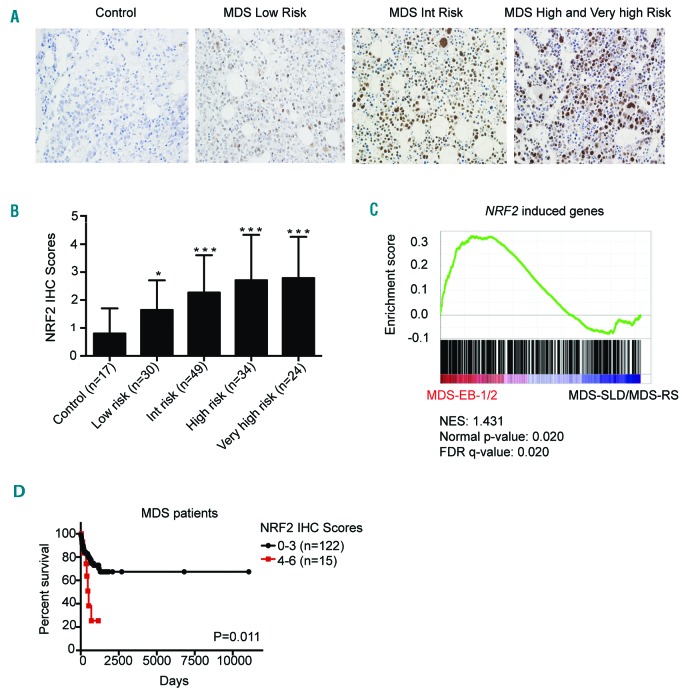
To explore NRF2 expression and its clinical outcome in MDS, we performed immunohistochemistry (IHC) on 137 MDS patients and 17 controls (Figure 1A). IHC staining results showed that NRF2 was over-expressed in the BM biopsies from MDS patients (P<0.01) (Figure 1B). The NRF2 levels of intermediate-, high-, and very high-risk IPSS-R patients exceeded that of low-risk IPSS-R patients (P=0.004) (Online Supplementary Table S1). To validate the expression of NRF2 with its downstream target signature in MDS, we analyzed published gene expression profiles of CD34+ BM cells purified from samples obtained from 183 MDS patients and 17 healthy controls.18 We performed gene set enrichment analysis (GSEA) to explore the downstream targets’ signature of NRF2 in this cohort.19 Although the expression of NRF2 is not significantly activated in MDS patients compared to the healthy controls (P=0.225) (Online Supplementary Figure S1A), NRF2 expression is significantly enriched in higher risk MDS patients (MDS-EB-1/2) when compared to those with lower risk MDS (MDS-SLD/MDS-RS) (P=0.020) (Figure 1C). Leading edge genes are shown in Online Supplementary Appendix Lists 1 and 2. It is worth noting that MDS patients with higher NRF2 expression levels (IHC scores, 4-6) displayed worse overall survival (OS) than patients with lower NRF2 levels (IHC scores, 0-3) (median, 391 vs. 554 days; P=0.011) (Figure 1D). We further performed CD34 and NRF2 double staining by immunofluorescence with MDS patient BM aspiration samples (Online Supplementary Figure S1B). CD34 and NRF2 double staining in particular cells demonstrated that NRF2 was also expressed at protein levels in CD34+ cells (Online Supplementary Figure S1C). High NRF2 expression levels were closely associated with higher risk according to the 2016 WHO subtype (P=0.022), IPSS cytogenetics (P=0.001), and IPSS categories (P=0.001). There were no significant differences in other clinical features between MDS patients with higher and lower NRF2 levels (Online Supplementary Table S2).
Figure 1.
Expression and clinical outcomes of NRF2 in myelodysplastic syndrome (MDS) patients. (A) NRF2 immunohistochemistry (IHC) staining of bone marrow biopsy samples (magnification ×400). (B) MDS patients had higher NRF2 IHC scores compared to controls. (C) Gene set enrichment plot showed that NRF2 target genes were enriched in higher-risk MDS patients. (D) MDS patients with higher NRF2 levels displayed worse overall survival (OS). *P<0.05; **P<0.01; ***P≤0.001. Int: intermediate; MDS-SLD: myelodysplastic syndrome single-lineage dysplasia; MDS-RS: myelodysplastic syndrome with ring sinderoblasts.
Pharmacological modulations of NRF2 regulate chemotherapeutic efficacy of Ara-C in MDS cells
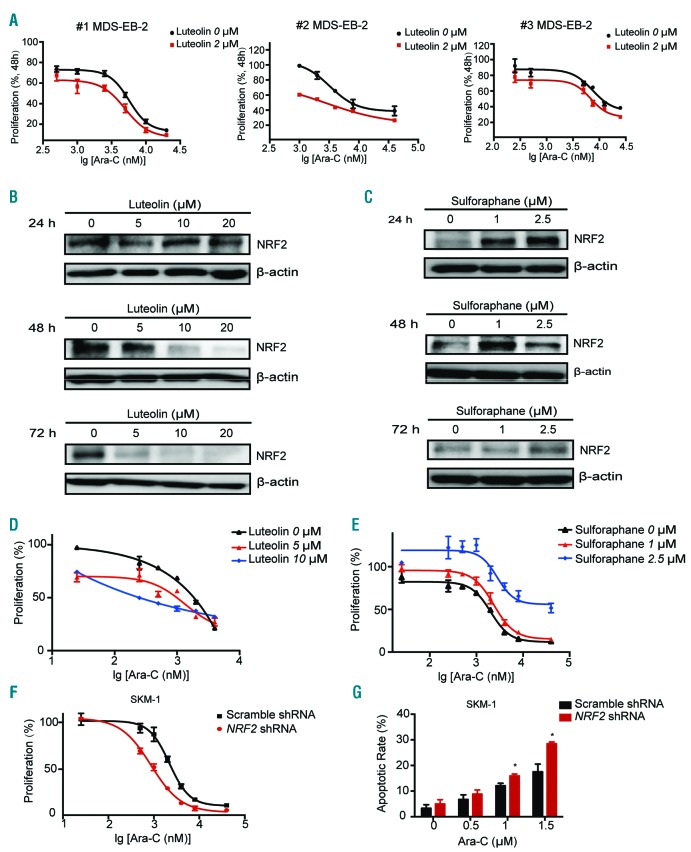
The effect of NRF2 on Ara-C resistance was first evaluated in primary MDS cells. After 72 h exposure to increasing doses of Ara-C, proliferation of primary MDS cells with 2 μM NRF2 inhibitor Luteolin was significantly reduced compared to vehicle control-treated MDS cells (# 1 MDS-EB-2 IC50: 5.7 μM vs. 4.8 μM, P=0.04; # 2 MDS-EB-2 IC50: 3.2 μM vs. 1.9 μM, P=0.006; # 3 MDS-EB-2 IC50: 7.5 μM vs. 6.9 μM; P=0.03) (Figure 2A).
Figure 2.
NRF2 inhibitor and activator regulate the sensitivity of myelodysplastic syndrome (MDS) cells to cytarabine (Ara-C) treatment. (A) Ara-C IC50 was significantly decreased in primary MDS cells treated with the NRF2 inhibitor Luteolin. (B) Luteolin decreased NRF2 protein levels in SKM-1. (C) The NRF2 agonist Sulforaphane increased NRF2 protein levels in SKM-1. (D) Ara-C IC50 was significantly decreased by Luteolin in SKM-1. (E) Ara-C IC50 was significantly increased by Sulforaphane in SKM-1. (F) NRF2 silencing significantly decreased IC50 of Ara-C in SKM-1. (G) NRF2 shRNA enhanced apoptosis induced by Ara-C in SKM-1 cell lines. *P<0.05; **P<0.01; ***P≤0.001. h: hours.
To further identify the function of NRF2 in MDS, we examined the pharmacological effects of NRF2 inhibitor and activator in MDS-patient-derived SKM-1 and murine MDS model cells MLLPTD/WT/RUNX1-S291fs cells.20 The highest dose of Ara-C resulted in approximately 90% inhibition of SKM-1 at 72 h (IC50, 1.72 μM) and MLLPTD/WT/RUNX1-S291fs cells at 48 h (IC50, 0.17 μM) (Online Supplementary Figure S2A and B). In agreement with previous reports on human lung carcinoma and colorectal cancer cell lines,21,22 we found that the NRF2 inhibitor Luteolin (3, 4, 5, 7-tetrahydroxy flavone) suppressed the protein expression of NRF2 in SKM-1 (Figure 2B). Sulforaphane (SFN) has been shown to be a potent NRF2 activator.23 SFN treatments in SKM-1 cells increased the protein expression of NRF2 (Figure 2C). NRF2 mRNA levels in MDS cells treated with the NRF2 inhibitor or agonist were measured. There was little change at mRNA levels, but obvious changes of NRF2 were seen at protein levels (Online Supplementary Figure S2C-F). Lower doses of Luteolin treatment had little effect on cell proliferation (Online Supplementary Figure S3A), but significantly enhanced the cytotoxicity of Ara-C (0-4 μM) to SKM-1. The IC50 values of Ara-C in SKM-1 cells were 1.41 μM and 0.93 μM with 5 μM and 10 μM Luteolin treatment, respectively (P<0.001) (Figure 2D). Similar results were also found in MLLPTD/WT/RUNX1-S291fs cells (Online Supplementary Figure S3B-D). The IC50 was reduced from 0.17 μM to 0.11 μM by 1 μM Luteolin in MLLPTD/WT/RUNX1-S291fs (P=0.007) (Online Supplementary Figure S3D). 1μM SFN treatments had little effect on the proliferation of SKM-1 cells (Online Supplementary Figure S3E). Interestingly, SFN treatment can decrease the chemotherapeutic effect of Ara-C (0-40 μM) in SKM-1. The IC50 was raised from 1.72 μM to 5.73 μM in SKM-1 by 2.5 μM SFN treatment (P=0.008) (Figure 2E). A similar effect of SFN could also be found in MLLPTD/WT/RUNX1-S291fs (Online Supplementary Figure S3F-H). 1 μM SFN treatment increased the Ara-C IC50 from 0.17 μM to 0.26 μM compared to vehicle treatment (P=0.001) (Online Supplementary Figure S3H).
Re-sensitizing MDS cells to Ara-C treatment in vitro by knockdown of NRF2
Transduction of NRF2 shRNA plasmid in human and mouse MDS cell lines repressed NRF2 mRNA levels by approximately 40-60%, compared with scramble shRNA plasmid transduction (Online Supplementary Figure S4A and B). Immunoblot analysis revealed that NRF2 shRNA robustly reduced the expression of NRF2 protein (Online Supplementary Figure S4C and D). Knockdown of NRF2 resulted in a significant reduction of Ara-C IC50 in SKM-1 (72 h Ara-C IC50, 2.20 μM vs. 0.87 μM; P=0.001) (Figure 2F) and MLLPTD/WT/RUNX1-S291fs (48 h Ara-C IC50, 0.37 μM vs. 0.25 μM; P=0.049) (Online Supplementary Figure S4E). Knockdown of NRF2 enhanced apoptosis induced by Ara-C in MDS cell lines (Figure 2G and Online Supplementary Figure S4F). We also found that NRF2 silenced MDS cell lines after Ara-C treatment tended to be arrested in the S phase (Online Supplementary Figure S4G-J).
DUSP1 is an NRF2 direct target gene in MDS
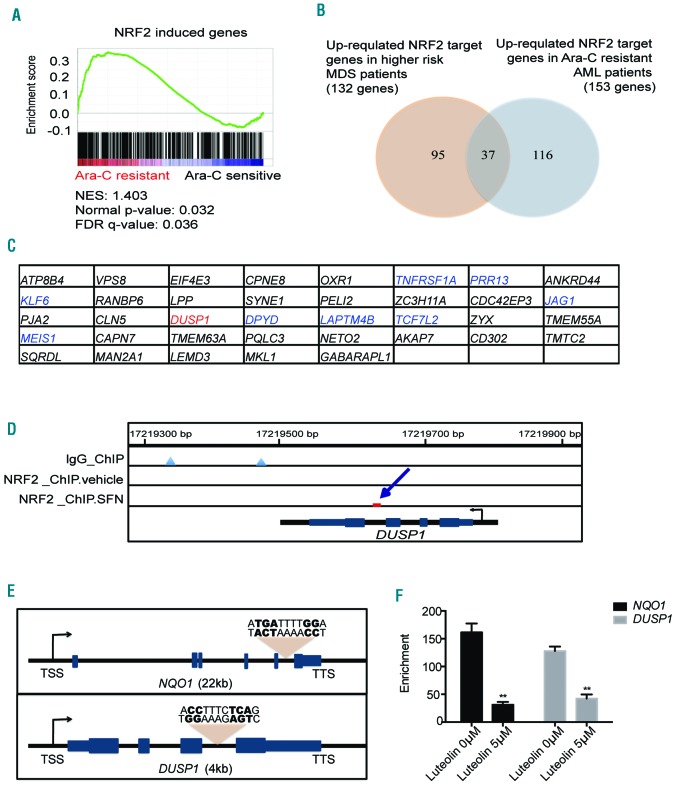
To further investigate the mechanisms involved in NRF2-mediated Ara-C resistance, we also analyzed published gene expression profiles of Ara-C-sensitive and Ara-C-resistant AML patient samples. Our analysis indicated that a group of NRF2 target genes might be responsible for Ara-C resistance in AML (P=0.032) (Figure 3A). Leading edge genes are shown in Online Supplementary Appendix List 3. After overlapping the up-regulated NRF2 target genes in high-risk MDS patients (total 132 genes) and Ara-C resistant AML patients (total 153 genes), we found a list of common up-regulated NRF2 target genes (n=37) (Figure 3B). Interestingly, dual-specificity protein phosphatase 1 (DUSP1) was one of the genes up-regulated in both high-risk MDS patients and Ara-C-resistant AML patients (Figure 3C). We then performed NRF2 ChIP-seq analysis based on a published dataset from human lymphoblastoid cell lines.24 In cells treated with NRF2 agonist, ChIP analysis vaildated the NRF2 binding site in the region of DUSP1 gene loci (Figure 3D). The NRF2 binding regions proximal to NQO1 and DUSP1 genes contained a conserved NRF2 binding TGAnnnnGG motif, as previously reported (Figure 3E).25 ChIP q-PCR analysis revealed that the NRF2 binding signals in the NQO1 and DUSP1 genes were significantly higher than the negative control loci. Lower NRF2 signals were detected in SKM-1 with 5 μM NRF2 inhibitor treatment (48 h, P<0.01) (Figure 3F).
Figure 3.
DUSP1 is an NRF2 target gene in myelodysplastic syndrome (MDS). (A) Gene set enrichment plot showed that NRF2 target genes were enriched in cytarabine (Ara-C)--resistant acute myeloid leukemia (AML) patients. (B) Overlap of up-regulated NRF2 target genes in higher-risk MDS patients and Ara-C-resistant AML patients. (C) The gene list of 37 overlapped genes. (D) ChIP sequence analysis of published data24 indicated the NRF2 binding site in the region of DUSP1 gene. (E) NRF2 binding sites in the regions of NQO1 and DUSP1 genes. TSS: transcription start site; TTS: transcription termination site. (F) NRF2 ChIP q-PCR analysis of SKM-1 cells.
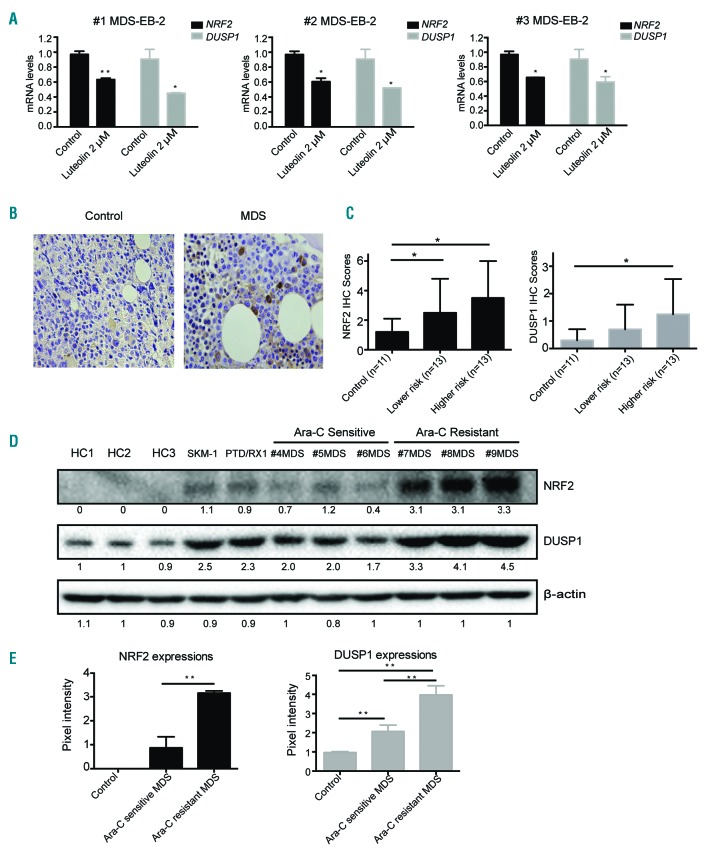
Consistent with the mRNA expression of NRF2, mRNA expression of DUSP1 could also be inhibited by 2 μM NRF2 inhibitor Luteolin treatment in primary MDS cells (Figure 4A). Our q-PCR results confirmed that DUSP1 was an NRF2 direct target gene in SKM-1 and MLLPTD/WT/RUNX1-S291fs cells (Online Supplementary Figure S5A-D).We also performed immunohistochemistry assay to detect DUSP1 expression in 11 controls and 26 MDS patients (Figure 4B). NRF2 and DUSP1 IHC scores were both significantly increased in the higher-risk MDS group (high-risk and very high-risk by IPSS-R) compared to the control group (P<0.05) (Figure 4C). To better detect the small differences at the protein levels, we further compared the levels of NRF2 and DUSP1 with the BM mononuclear cells from Ara-C sensitive and Ara-C-resistant MDS patients by immunoblotting (Figure 4D). Samples from MDS patients who were responsive to Ara-C treatment were selected as Ara-C-sensitive MDS samples. MDS patients in whom Ara-C treatment was seen to be ineffective were chosen as Ara-C-resistant cases. Elevated levels of NRF2 and DUSP1 were seen in the BM samples of Ara-C-resistant MDS patients by immunoblotting analysis; this was statistically significant based on intensities (Figure 4E).
Figure 4.
NRF2 and DUSP1 expressions were elevated in higher-risk myelodysplastic syndrome (MDS) or cytarabine (Ara-C)--resistant MDS patients. (A) NRF2 and DUSP1 mRNA levels were both repressed by Luteolin in primary MDS cells. (B) DUSP1 immunohistochemistry (IHC) staining of bone marrow (BM) biopsy samples (magnification ×400). (C) NRF2 and DUSP1 IHC scores in controls and MDS. (D) Immunoblotting analysis was conducted for NRF2 and DUSP1 protein levels in healthy controls, MDS cell lines, and primary MDS cells. (E) Elevations of NRF2 and DUSP1 were confirmed in the BM samples of Ara-C-resistant MDS by immunoblotting analysis. *P<0.05; **P<0.01.
NRF2 mediates Ara-C resistance partly through its direct target gene DUSP1
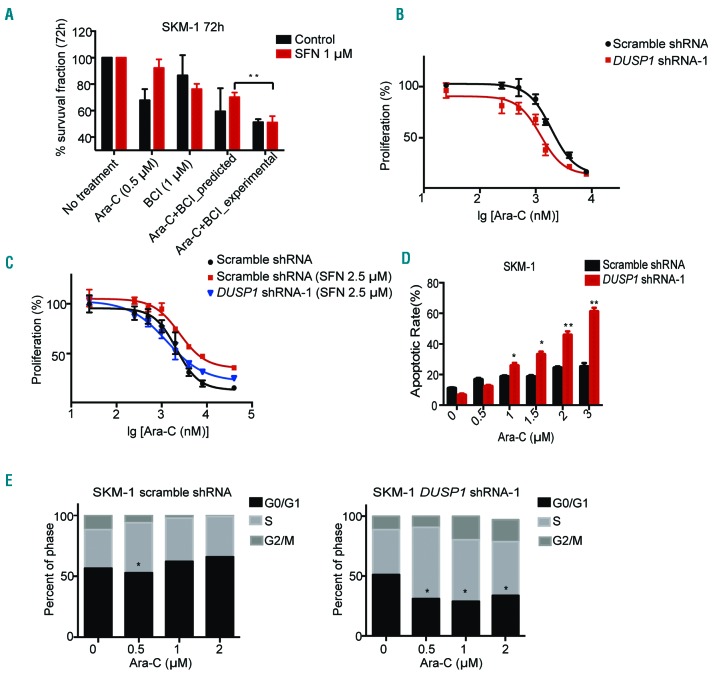
A Dusp1 and Dusp6 inhibitor,26,27 was used to test the potential of targeting DUSP1 for Ara-C therapy. The combined inhibitory effect was predicted using the Bliss independent model.28,29 Our data indicate that BCI and Ara-C have statistically significant synergistic effects on NRF2 activated SKM-1 cells (%survival, predicted 70.1% vs. experimental 51.0%; P=0.005) and MLLPTD/WT/RUNX1-S291fs cells (%survival, predicted 57.0% vs. experimental 41.0%; P=0.048) (Figure 5A and Online Supplementary Figure S5E). To further demonstrate that the therapeutic synergistic effect is due to the inhibition of DUSP1, we conducted DUSP1 shRNA on SKM-1 cells. DUSP1 shRNA-1 and Dusp1 shRNA-1 were chosen because the data indicated that both of them mediated the best knockdown efficiency in human and mouse MDS cell lines, respectively (Online Supplementary Figure S6A-D). Downregulation of DUSP1 by lentivirus shRNA could sensitize SKM-1 cells to Ara-C treatment (72 h Ara-C IC50, scramble shRNA 1.91 μM vs. DUSP1 shRNA-1 1.23 μM; P=0.002) (Figure 5B). Treatment of the NRF2 agonist SFN significantly mitigated Ara-C toxicity in scramble shRNA SKM-1 cells (72 h Ara-C IC50, control treatment 2.05 μM vs. SFN treatment 2.50 μM; P=0.044). Significantly, DUSP1 shRNA on NRF2 elevated SKM-1 cell lines abrogate Ara-C resistance (72 h Ara-C with SFN treatment IC50, scramble shRNA 2.50 μM vs. DUSP1 shRNA-1 1.35 μM; P=0.020) (Figure 5C). In MLLPTD/WT/RUNX1-S291fs cells, Dusp1 downregulation could sensitize MDS mouse cells to Ara-C treatment (48 h Ara-C IC50, scramble shRNA 0.20 μM vs. Dusp1 shRNA-1 0.15 μM; P=0.001) (Online Supplementary Figure S6E). Consistent with the data in SKM-1 cells, Dusp1 shRNA could also re-sensitize Ara-C-resistant MDS mouse cells [48 h Ara-C IC50, scramble shRNA with control treatment 0.20 μM vs. scramble shRNA with SFN treatment 0.22 μM (P=0.025) scramble shRNA with SFN treatment 0.22 μM vs. Dusp1 shRNA-1 with SFN treatment 0.16 μM (P=0.001)] (Online Supplementary Figure S6F). Knockdown of DUSP1 led to an increase in the apoptotic rate of SKM-1 treated with Ara-C compared to scramble shRNA SKM-1 cells (Figure 5D). DUSP1 silenced SKM-1 cell lines with Ara-C treatment also tended to be arrested in the S phase (Figure 5E). These results indicated that DUSP1 was a downstream gene of NRF2 and partially responsible for NRF2-mediated Ara-C resistance. To determine whether silencing of NRF2 or DUSP1 compromised the reactive oxygen specis (ROS) levels in MDS cells, we analyzed ROS production by flow cytometry. No significant difference in ROS levels was observed between NRF2 or DUSP1 knockdown MDS cells and control cells (Online Supplementary Figure S7A-D). To further investigate other possible pathways involved in Ara-C resistance, we adjusted the fold change value to 1.1 and the rawp value to 0.05 so that we obtained two larger cohorts of up-regulated genes in high-risk MDS (total 5477 genes) or Ara-C-resistant AML (total 3074 genes). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed on 331 overlapped common genes (Online Supplementary Figure S7E), which revealed potential involvement of genes in a number of pathways (Online Supplementary Figure S7F).
Figure 5.
NRF2 confers cytarabine (Ara-C) resistance partly through the activation of DUSP1 in myelodysplastic syndrome (MDS). (A) Ara-C and (E)-2-benzylidene-3-(cyclohexylamino)-2,3-dihydro-1H-inden-1-one (BCI) have synergistic effects in NRF2 agonist-treated SKM-1 cells. (B) DUSP1 shRNA-1 sensitized SKM-1 cells to Ara-C treatment. (C) DUSP1 shRNA-1 re-sensitized NRF2 agonist treated SKM-1 cells to Ara-C treatment. (D) DUSP1 shRNA-1 enhanced apoptosis induced by Ara-C in SKM-1 cell lines. (E) DUSP1 shRNA-1 induced S phrase arrest in SKM-1. *P<0.05; **P<0.01; ***P≤0.001.
Re-sensitizing MDS cells to Ara-C treatment in vivo by knockdown of NRF2 or DUSP1
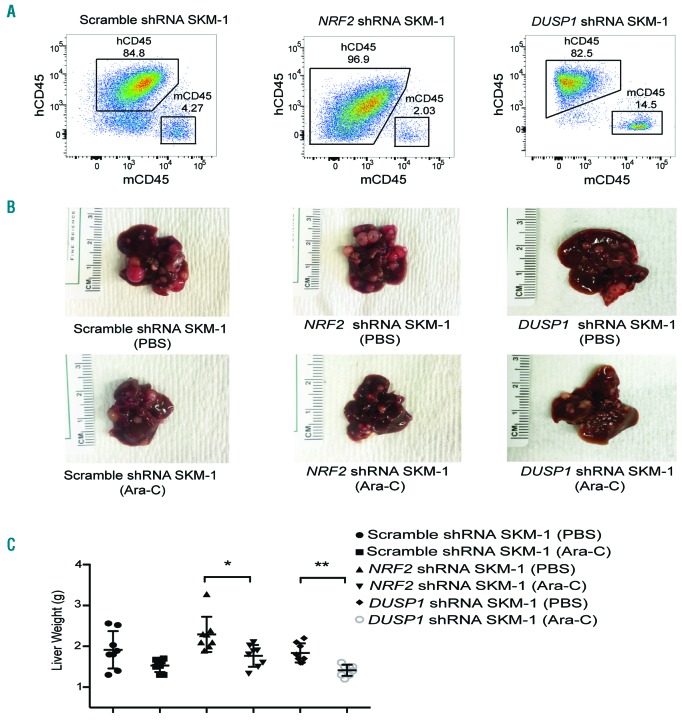
To determine the effect of NRF2 and DUSP1 on chemoresistance in vivo, we established xenograft mouse models through intravenously injecting NRF2 shRNA, DUSP1 shRNA, or scramble shRNA SKM-1 cells into NOD/SCID-IL2Rγnull-SGM3 (NSGS) mice. The ratios of human CD45+ cells to mouse CD45+ cells were more than 80% in tumors (Figure 6A) but less than 5% in bone marrow (Online Supplementary Figure S8A) or peripheral blood (Online Supplementary Figure S8B). However, no significant differences were observed in the survival of transplanted NSGS mice (Online Supplementary Figure S8C-E). In scramble shRNA MDS mice, the tumor weight showed a trend to decrease, but did not reach a significant change in the Ara-C treatment group (1.91 g vs. 1.53 g with PBS vs. Ara-C treatment group, respectively; P=0.062) (Figure 6B and C). Treatment of NRF2 or DUSP1 silencing MDS mice with Ara-C resulted in significantly smaller tumors in the liver (NRF2 silencing MDS mice, 2.30 g vs. 1.77 g with PBS vs. Ara-C treatment group, respectively, P=0.010; DUSP1 silencing MDS mice, 1.84 g vs. 1.41 g, respectively, P=0.001).
Figure 6.
Knockdown of NRF2 significantly sensitizes myelodysplastic syndrome (MDS) cells to cytarabine (Ara-C) in vivo. (A) Flow cytometry showed appearance of human CD45+ cells in liver tumors of scramble, NRF2 and DUSP1 shRNA SKM-1 transplanted NSGS mice. (B) Liver tumors in scramble, NRF2 and DUSP1 shRNA SKM-1 transplanted NSGS mice with phosphate buffer saline (PBS) or Ara-C treatment. (C) Liver tumor volumes were significantly smaller in NRF2 or DUSP1 shRNA SKM-1 transplanted MDS mice treated with Ara-C compared with PBS. *P<0.05; **P<0.01; ***P≤0.001. g: grams.
Discussion
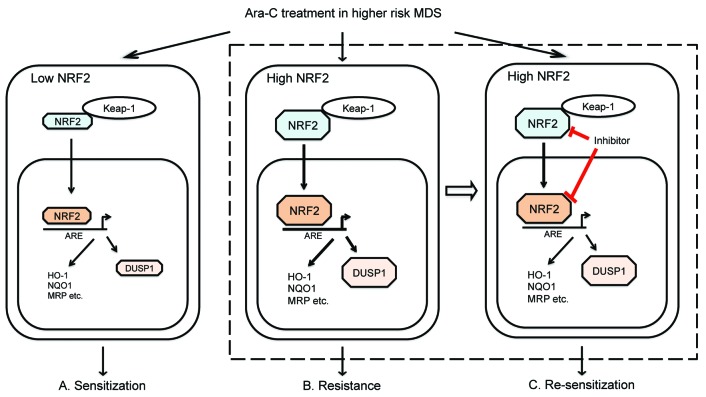
The present study was aimed to investigate the role of NRF2 in MDS and its molecular mechanism involved in chemoresistance, particularly in Ara-C-based therapy. Using IHC and unbiased analysis, we identified the predictive role of NRF2 in clinical outcomes amongst MDS patients. Based on our data and published evidence, we proposed a model of NRF2 in high-risk MDS with Ara-C treatment (Figure 7). Activation of NRF2-DUSP1 signaling and other pathways might lead to Ara-C resistance in high-risk MDS. Inhibition of NRF2 could re-sensitize MDS cells to Ara-C treatment.
Figure 7.
A proposed model of NRF2 in higher-risk myelodysplastic syndrome (MDS) with cytarabine (Ara-C) treatment. (A) Ara-C treatment inhibits the cell viability of the MDS cells with low NRF2 levels. (B) NRF2 confers Ara-C resistance partly through its downstream target gene DUSP1 in MDS cells. (C) The inhibition of NRF2 re-sensitizes MDS cells to Ara-C treatment.
Garcia-Manero et al. had previously measured NRF2 mRNA levels in peripheral blood mononuclear cells of AML or MDS cases (n=31) and reported that higher mRNA levels of NRF2 were associated with longer survival.30 Here, we explored the prognostic impact of NRF2 in a larger cohort of MDS patients (n=137). Our IHC data indicated that higher risk MDS patients had higher NRF2 expression levels in BM samples compared to lower risk patients by IPSS-R (P=0.004). GSEA results of CD34+ BM cell gene expressions from published MDS patient cohort data (n=183) further confirmed NRF2 was elevated in higher-risk MDS patients (MDS-EB-1/2) compared to lower-risk patients (MDS-SLD/RS) (Figure 1C). We speculate that activated NF-κB signaling may drive the overexpression of NRF2 in high-risk MDS.10,31 It has also been reported that mitochondrial dynamics could regulate neural stem cell fate by modifying ROS signaling to activate NRF2-dependent pathways.32,33 Future studies may help to elucidate the possible mechanisms involved in high NRF2 expression levels in higher-risk MDS patients. Importantly, our survival analysis indicated that MDS patients with higher NRF2 levels in BM cells correlated with worse OS than patients with lower NRF2 levels (P=0.011) (Figure 1D). The discrepancy between our results and the previously published data may be related to the methods, cells studied (mononuclear cells of PB vs. BM), or sample size (31 vs. 137). This needs to be further investigated in larger independent cohorts.
Ara-C is widely used as a treatment approach in higher-risk MDS patients; however, single Ara-C treatment has limited therapeutic effect. Drug resistance is the major cause of treatment failure. High NRF2 proteins in human primary AML cells have been shown to be driven by NF-κB and knockdown of NRF2 reduced colony formation of AML cells in response to treatment of Ara-C and daunorubine.10 However, there are no research reports on the role of NRF2 in mediating drug resistance in MDS. The SKM-1 cell line was established from a Japanese male patient in 1985 who was initially diagnosed as higher-risk MDS (MDS-EB-2).34 It has been reported that SKM-1 cells had a higher IC50 of Ara-C than other myeloid leukemia cell lines, indicating that SKM-1 cells were more Ara-C resistant.35,36 The incidences of MLL-PTD and RUNX1 mutations showed an increase in higher-risk MDS compared to lower-risk MDS.37 Thus, SKM-1 cells and MDS mouse model cell line RUNX1 mutant-transduced MllPTD/WT BM cells (MllPTD/WT/RUNX1-S291fs) were used in our study. BM mononuclear cells from Ara-C-sensitive and Ara-C-resistant MDS patients were also studied. Our results indicated that NRF2-mediated drug resistance in MDS was similar to other conditions.10,38
Luteolin is a potential NRF2 inhibitor that can promote the degradation of NRF2 mRNA. Our results revealed that NRF2 downregulation in primary MDS cells, by inhibitor Luteolin, decreased IC50 of Ara-C. As primary MDS cells were mostly composed of non-transformed cells, we also validated our results in human and mouse MDS cell lines. Downregulation of NRF2 by Luteolin could also enhance the chemotherapeutic efficacy of Ara-C in MDS cell lines. SFN is a well-known NRF2 agonist. SFN is an isothiocyanate that forms a KEAP1– Sulforaphane thionoacyl adduct to stabilize NRF2.39 We found that upregulation of NRF2, mediated by agonist SFN, induced resistance of MDS cells to Ara-C. Previous reports in AML indicated the protective role of NRF2 against apoptosis.15 NRF2 regulates homologs miR-125B1 and miR-29B1 to repress the apoptosis induced by the front-line AML chemotherapy agent daunorubicin.40 To better define how suppression of NRF2 sensitized MDS cells to Ara-C, we used lentivirus-mediated shRNA for knockdown of NRF2. Knockdown of NRF2 markedly enhanced apoptosis and triggered S-phrase arrest in MDS cell lines treated with Ara-C. Taken together, NRF2 levels regulate MDS cells’ sensitivity to Ara-C therapy.
There is increasing evidence to suggest that NRF2 target genes, such as HO-1, NQO1, and multidrug resistance-associated protein (MRP), are involved in cytoprotection and detoxification, thus providing drug resistance in anti-cancer therapy.12,41,42 To determine the mechanisms of NRF2-mediated Ara-C resistance in MDS, we performed GSEA analysis on published data and then discovered a list of the genes (n=37) up-regulated in both high-risk MDS patients and Ara-C-resistant AML patients. One of the genes on the list is DUSP1 (also known as MKP1), which regulates mitogen-activated protein kinase (MAPKinase) by dephosphorylation of threonine and tyrosine residues.43 DUSP1 may play an important role in the cellular response to environmental oxidative stress and agents that damage DNA.44 However, little is known about the relationship between DUSP1 and NRF2 or the effect of DUSP1 on chemo-resistance in MDS. Our ChIP q-PCR and q-PCR data indicated DUSP1 was an NRF2 direct target gene. Our IHC and immunoblotting data showed that DUSP1 expressions were elevated in higher-risk or Ara-C-resistant MDS. Given the small number of MDS patients studied, future validation with larger cohorts is needed.
Interestingly, downregulation of DUSP1 by inhibitor or lentivirus shRNA could abrogate Ara-C resistance in NRF2-elevated MDS cells. There is growing evidence to demonstrate that NRF2 activation by antioxidant interventions increased cancer cell migration and induced tumor metastasis by decreasing ROS levels.45,46 It has been suggested that NRF2 improved sensitivity of AML cells to chemotherapy by compromising the ability of the AML cell to scavenge the ROS.38 Our results suggested that ROS signaling pathways may play limited roles in NRF2-mediated Ara-C resistance in MDS cells. We identified a larger cohort of the genes (n=331) up-regulated in both high-risk MDS patients and Ara-C-resistant AML patients. We found significant enrichment of the up-regulated genes in 12 KEGG pathways pertaining to cell signaling (e.g. MAPK and JAK-STAT signaling), immune responses (e.g. chemokine signaling and lyso-some signaling), and cell death (e.g. apoptosis and FoxO signaling). It has been reported that alterations of SETD2 (encoding the histone 3 lysine 36 trimethyltransferase) and EZH2 (catalyzing the trimethylation of lysine 27 of histone H3) also led to resistance to DNA damaging-chemotherapy such as Ara-C in leukemia via different mechanisms.47,48 Our current study indicated that NRF2 conferred Ara-C resistance partly through DUSP1 in MDS. Other signaling pathways identified in this study warrant further investigation in the future.
Our data showed that silencing NRF2 or DUSP1 significantly sensitized tumors to Ara-C by measuring tumor size in the livers of our SKM-1-transplanted mouse models. SKM-1 is a cell line carrying a large number of mutations, affecting ASXL1, BCORL1, EZH2, SF1, STAG2, TET2, TP53, and WT1, which are close to the characteristics of high-risk MDS.49 Thus, SKM-1 cell lines were used in this study. To better establish stable NRF2 or DUSP1 knockdown MDS mouse models, SKM-1 cells were transfected and then injected into NSGS mice. SKM-1 cells mainly generated tumors in the livers but not in BM or peripheral blood of NSGS mice. Although not the ideal model, our in vivo results indicated that silencing NRF2 or DUSP1 increased the sensitivity of SKM-1 cells to Ara-C treatment. Future MDS patient-derived xenograft models are needed to validate our findings.50
In conclusion, our clinical and experimental results revealed that NRF2 expression levels are elevated in high-risk MDS patients and serve as a statistically significant prognostic variable for OS in MDS patients. Pharmacological inhibition of NRF2 re-sensitizes MDS cells to Ara-C treatment while activation of NRF2 by agonist resulted in the reduced sensitivity to Ara-C. NRF2 mediates Ara-C resistance partly through its direct target gene DUSP1. Taken together, our findings suggest that silencing NRF2 re-sensitizes high-risk MDS cells to Ara-C treatment. Targeting NRF2 in combination with conventional chemotherapy could overcome drug resistance in high-risk MDS patients.
Supplementary Material
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/3/485
Funding
This work was supported by grants from the National Key Technology R&D Program (2014BAI09B13), National Natural Science Foundation of China Grants (81270582, 81470290, 81700121, 81800121), Major Program Fund of the Science Technology Department of Zhejiang Province (2013c03043-2), the Taub Foundation (to GH), and National Institutes of Health (NIH) (R01DK105014 to GH).
References
- 1.Cazzola M, Malcovati L. Myelodysplastic syndromes--coping with ineffective hematopoiesis. N Engl J Med. 2005; 352(6):536–538. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg P, Cox C, LeBeau MM, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89(6):2079–2088. [PubMed] [Google Scholar]
- 3.Greenberg PL, Tuechler H, Schanz J, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120(12):2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Killick SB, Carter C, Culligan D, et al. Guidelines for the diagnosis and management of adult myelodysplastic syndromes. Br J Haematol. 2014;164(4):503–525. [DOI] [PubMed] [Google Scholar]
- 5.Burnett AK, Milligan D, Prentice AG, et al. A comparison of low-dose cytarabine and hydroxyurea with or without all-trans retinoic acid for acute myeloid leukemia and high-risk myelodysplastic syndrome in patients not considered fit for intensive treatment. Cancer. 2007;109(6):1114–1124. [DOI] [PubMed] [Google Scholar]
- 6.Miller KB, Kim K, Morrison FS, et al. The evaluation of low-dose cytarabine in the treatment of myelodysplastic syndromes: a phase-III intergroup study. Ann Hematol. 1992;65(4):162–168. [DOI] [PubMed] [Google Scholar]
- 7.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39(4):199–218. [DOI] [PubMed] [Google Scholar]
- 8.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27(20):2179–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rushworth SA, Zaitseva L, Murray MY, Shah NM, Bowles KM, MacEwan DJ. The high Nrf2 expression in human acute myeloid leukemia is driven by NF-kappaB and underlies its chemo-resistance. Blood. 2012;120(26):5188–5198. [DOI] [PubMed] [Google Scholar]
- 11.Zhang BP, Zhao J, Li SS, et al. Mangiferin activates Nrf2-antioxidant response element signaling without reducing the sensitivity to etoposide of human myeloid leukemia cells in vitro. Acta Pharmacol Sin. 2014;35(2):257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu RP, Hayashi T, Cottam HB, et al. Nrf2 responses and the therapeutic selectivity of electrophilic compounds in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2010;107(16):7479–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niture SK, Khatri R, Jaiswal AK. Regulation of Nrf2-an update. Free Radic Biol Med. 2014;66:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rushworth SA, Macewan DJ. The role of nrf2 and cytoprotection in regulating chemotherapy resistance of human leukemia cells. Cancers (Basel). 2011; 3(2):1605–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rushworth SA, Bowles KM, MacEwan DJ. High basal nuclear levels of Nrf2 in acute myeloid leukemia reduces sensitivity to proteasome inhibitors. Cancer Res. 2011; 71(5):1999–2009. [DOI] [PubMed] [Google Scholar]
- 16.Cho JM, Manandhar S, Lee HR, Park HM, Kwak MK. Role of the Nrf2-antioxidant system in cytotoxicity mediated by anticancer cisplatin: implication to cancer cell resistance. Cancer Lett. 2008;260(1-2):96–108. [DOI] [PubMed] [Google Scholar]
- 17.Wunderlich M, Stockman C, Devarajan M, et al. A xenograft model of macrophage activation syndrome amenable to anti-CD33 and anti-IL-6R treatment. JCI Insight. 2016;1(15):e88181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellagatti A, Cazzola M, Giagounidis A, et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia. 2010;24(4):756–764. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi Y, Zhang Y, Yokota A, Yan XM, Liu JQ, et al. Pathobiologic pseudohypoxia as a putative mechanism underlying myelodysplastic syndromes. Cancer Discov. 2018. August 23 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang X, Wang H, Fan L, et al. Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radic Biol Med. 2011;50(11):1599–1609. [DOI] [PubMed] [Google Scholar]
- 22.Chian S, Li YY, Wang XJ, Tang XW. Luteolin sensitizes two oxaliplatin-resistant colorectal cancer cell lines to chemotherapeutic drugs via inhibition of the Nrf2 pathway. Asian Pac J Cancer Prev. 2014;15(6):2911–2916. [DOI] [PubMed] [Google Scholar]
- 23.Jazwa A, Rojo AI, Innamorato NG, Hesse M, Fernandez-Ruiz J, Cuadrado A. Pharmacological targeting of the transcription factor Nrf2 at the basal ganglia provides disease modifying therapy for experimental parkinsonism. Antioxid Redox Signal. 2011;14(12):2347–2360. [DOI] [PubMed] [Google Scholar]
- 24.Chorley BN, Campbell MR, Wang X, et al. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40(15):7416–7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi EH, Suzuki T, Funayama R, et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun. 2016;7:11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molina G, Vogt A, Bakan A, et al. Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat Chem Biol. 2009;5(9):680–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kesarwani M, Kincaid Z, Gomaa A, et al. Targeting c-FOS and DUSP1 abrogates intrinsic resistance to tyrosine-kinase inhibitor therapy in BCR-ABL-induced leukemia. Nat Med. 2017;23(4):472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greco WR, Bravo G, Parsons JC. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev. 1995;47(2):331–385. [PubMed] [Google Scholar]
- 29.Zhao W, Sachsenmeier K, Zhang L, Sult E, Hollingsworth RE, Yang H. A New Bliss Independence Model to Analyze Drug Combination Data. J Biomol Screen. 2014;19(5):817–821. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Manero G, Tambaro FP, Bekele NB, et al. Phase II trial of vorinostat with idarubicin and cytarabine for patients with newly diagnosed acute myelogenous leukemia or myelodysplastic syndrome. J Clin Oncol. 2012;30(18):2204–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun T, Carvalho G, Coquelle A, et al. NF-kappaB constitutes a potential therapeutic target in high-risk myelodysplastic syndrome. Blood. 2006;107(3):1156–1165. [DOI] [PubMed] [Google Scholar]
- 32.Khacho M, Clark A, Svoboda DS, et al. Mitochondrial Dynamics Impacts Stem Cell Identity and Fate Decisions by Regulating a Nuclear Transcriptional Program. Cell Stem Cell. 2016;19(2):232–247. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Li M, Sun C, et al. Altered hematopoietic cell gene expression precedes development of therapy-related myelodysplasia/acute myeloid leukemia and identifies patients at risk. Cancer Cell. 2011; 20(5):591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakagawa T, Matozaki S, Murayama T, et al. Establishment of a leukaemic cell line from a patient with acquisition of chromosomal abnormalities during disease progression in myelodysplastic syndrome. Br J Haematol. 1993;85(3):469–476. [DOI] [PubMed] [Google Scholar]
- 35.Liu Z, Chen J, Wang H, et al. Chidamide shows synergistic cytotoxicity with cytarabine via inducing G0/G1 arrest and apoptosis in myelodysplastic syndromes. Am J Transl Res. 2017;9(12):5631–5642. [PMC free article] [PubMed] [Google Scholar]
- 36.Murase M, Iwamura H, Komatsu K, et al. Lack of cross-resistance to FF-10501, an inhibitor of inosine-5′-monophosphate dehydrogenase, in azacitidine-resistant cell lines selected from SKM-1 and MOLM-13 leukemia cell lines. Pharmacol Res Perspect. 2016;4(1):e00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dicker F, Haferlach C, Sundermann J, et al. Mutation analysis for RUNX1, MLL-PTD, FLT3-ITD, NPM1 and NRAS in 269 patients with MDS or secondary AML. Leukemia. 2010;24(8):1528–1532. [DOI] [PubMed] [Google Scholar]
- 38.Karathedath S, Rajamani BM, Musheer Aalam SM, et al. Role of NF-E2 related factor 2 (Nrf2) on chemotherapy resistance in acute myeloid leukemia (AML) and the effect of pharmacological inhibition of Nrf2. PLoS One. 2017;12(5):e0177227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong F, Freeman ML, Liebler DC. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem Res Toxicol. 2005;18(12):1917–1926. [DOI] [PubMed] [Google Scholar]
- 40.Shah NM, Zaitseva L, Bowles KM, MacEwan DJ, Rushworth SA. NRF2-driven miR-125B1 and miR-29B1 transcriptional regulation controls a novel anti-apoptotic miRNA regulatory network for AML survival. Cell Death Differ. 2015;22(4):654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai H, Scott E, Kholghi A, et al. Cancer chemoprevention: Evidence of a nonlinear dose response for the protective effects of resveratrol in humans and mice. Sci Transl Med. 2015;7(298):298ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole SP. Targeting multidrug resistance protein 1 (MRP1, ABCC1): past, present, and future. Annu Rev Pharmacol Toxicol. 2014;54:95–117. [DOI] [PubMed] [Google Scholar]
- 43.Checker R, Gambhir L, Sharma D, Kumar M, Sandur SK. Plumbagin induces apoptosis in lymphoma cells via oxidative stress mediated glutathionylation and inhibition of mitogen-activated protein kinase phosphatases (MKP1/2). Cancer Lett. 2015;357(1):265–278. [DOI] [PubMed] [Google Scholar]
- 44.Keyse SM, Emslie EA. Oxidative stress and heat shock induce a human gene encoding a protein-tyrosine phosphatase. Nature. 1992;359(6396):644–647. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, Liu X, Long M, et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci Transl Med. 2016;8(334):334ra351. [DOI] [PubMed] [Google Scholar]
- 46.Tschop MH, Stumvoll M, Ristow M. Opposing Effects of Antidiabetic Interventions on Malignant Growth and Metastasis. Cell Metab. 2016;23(6):959–960. [DOI] [PubMed] [Google Scholar]
- 47.Mar BG, Chu SH, Kahn JD, et al. SETD2 alterations impair DNA damage recognition and lead to resistance to chemotherapy in leukemia. Blood. 2017;130(24):2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gollner S, Oellerich T, Agrawal-Singh S, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med. 2017;23(1):69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palau A, Mallo M, Palomo L, et al. Immunophenotypic, cytogenetic, and mutational characterization of cell lines derived from myelodysplastic syndrome patients after progression to acute myeloid leukemia. Genes Chromosomes Cancer. 2017;56(3):243–252. [DOI] [PubMed] [Google Scholar]
- 50.Medyouf H, Mossner M, Jann JC, et al. Myelodysplastic cells in patients reprogram mesenchymal stromal cells to establish a transplantable stem cell niche disease unit. Cell Stem Cell. 2014;14(6):824–837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.