Although the relationship between Acinetobacter baumannii efflux pumps and antimicrobial resistance is well documented, less is known about the involvement of these proteins in the pathogenicity of this nosocomial pathogen. In previous work, we identified the AbaQ major facilitator superfamily (MFS) efflux pump and demonstrated its participation in the motility and virulence of A. baumannii.
KEYWORDS: Acinetobacter, efflux pumps, motility, virulence
ABSTRACT
Although the relationship between Acinetobacter baumannii efflux pumps and antimicrobial resistance is well documented, less is known about the involvement of these proteins in the pathogenicity of this nosocomial pathogen. In previous work, we identified the AbaQ major facilitator superfamily (MFS) efflux pump and demonstrated its participation in the motility and virulence of A. baumannii. In the present study, we examined the role in these processes of A. baumannii transporters belonging to different superfamilies of efflux pumps. Genes encoding known or putative permeases belonging to efflux pump superfamilies other than the MFS were selected, and the corresponding knockouts were constructed. The antimicrobial susceptibilities of these mutants were consistent with previously reported data. In mutants of A. baumannii strain ATCC 17978 carrying inactivated genes encoding the efflux pumps A1S_2736 (resistance nodulation division [RND]), A1S_3371 (multidrug and toxic compound extrusion [MATE]), and A1S_0710 (small multidrug resistance [SMR]), as well as the newly described ATP-binding cassette (ABC) permeases A1S_1242 and A1S_2622, both surface-associated motility and virulence were reduced compared to the parental strain. However, inactivation of the genes encoding the known ABC permeases A1S_0536 and A1S_1535, the newly identified putative ABC permeases A1S_0027 and A1S_1057, or the proteobacterial antimicrobial compound efflux (PACE) transporters A1S_1503 and A1S_2063 had no effects on bacterial motility or virulence. Our results demonstrate the involvement of antimicrobial transporters belonging at least to five of the six known efflux pump superfamilies in both surface-associated motility and virulence in A. baumannii ATCC 17978.
INTRODUCTION
Acinetobacter baumannii is an opportunistic microorganism that causes nosocomial infections of the central nervous system, skin, soft tissues, and bone, as well as pneumonia (1). The health threat posed by this bacterium is related to its enormous potential to develop antimicrobial resistance, its ability to survive in the hospital environment, and its virulence factors (2). Among the resistance mechanisms of A. baumannii are enzymatic modification of antimicrobial agents, changes in membrane permeability, alternative metabolic processes, alterations of the site of antimicrobial action, and reduction of the intracellular drug concentration through drug extrusion (3). The last mechanism is carried out by efflux pumps. However, the demonstrated involvement of these antimicrobial determinants in multidrug resistance and thus in the pathogenicity of diverse bacterial species implies their role not only in drug extrusion but also in virulence (4, 5). Efflux pumps can be classified into six superfamilies according to their amino acid similarity, energy source, number of components, number of transmembrane regions, and substrate types (4): major facilitator superfamily (MFS), resistance nodulation division (RND), multidrug and toxic compound extrusion (MATE), small multidrug resistance (SMR), ATP-binding cassette (ABC), and proteobacterial antimicrobial compound efflux (PACE) (6, 7). In A. baumannii, transporters belonging to all six superfamilies have been identified.
Although A. baumannii lacks flagella, this bacterium is able to move through surface-associated motility (8, 9). In previous reports we identified the MFS efflux pump AbaQ, which in A. baumannii is involved not only in quinolone extrusion but also in surface-associated motility and virulence (10, 11). Additional evidence implicating efflux pumps in the pathogenicity of A. baumannii, and therefore its virulence has been observed in several studies. For instance, overexpression of the RND efflux pump AdeABC increased bacterial virulence in a murine model (12), while inactivation of the MFS efflux pump AbaF lowered bacterial virulence in a Caenorhabditis elegans model (13). However, to our knowledge, whether transporters belonging to the other efflux pump superfamilies participate in the motility and virulence of A. baumannii has yet to be determined. Thus, the aim of this work was to investigate the role of transporters belonging to different efflux pumps superfamilies in the surface-associated motility and virulence of A. baumannii.
RESULTS
Selection and mutant construction of different A. baumannii efflux pumps belonging to the RND, MATE, SMR, ABC, and PACE superfamilies.
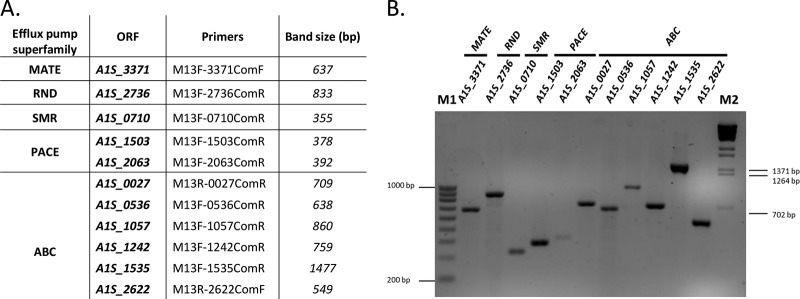
As noted above, we previously described the role of the MFS AbaQ quinolone-efflux pump in the surface-associated motility and virulence of A. baumannii (10, 11). To examine the role of other efflux pumps in these processes, genes encoding transporters belonging to the other efflux pump superfamilies were selected (Table 1). From the RND efflux pump superfamily, the gene A1S_2736 was chosen. This gene encodes a permease homologous (99% identity) to the AdeJ component corresponding to the AdeIJK RND efflux pump described in A. baumannii strain BM4454 (14). The AbeM3 transporter, encoded by the gene A1S_3371, and the transporter encoded by the gene A1S_0710, both identified in A. baumannii ATCC 17978, were chosen as members of the MATE and SMR efflux pump superfamilies, respectively (15, 16). From the ABC efflux pump superfamily, the genes A1S_0536 and A1S_1535 were selected. These genes encode permeases from the only two thus-far-identified ABC transporters involved in antimicrobial resistance, and both have been identified in strain ATCC 17978 (17, 18). In addition, by analyzing the annotated genome of the same A. baumannii strain, several genes annotated as possible permeases of ABC transporters were identified. Based on their permease-like structure, determined by using the Protter web platform (http://wlab.ethz.ch/protter) to analyze both the amino acid sequence and transmembrane characteristics of the putative proteins, the genes A1S_0027, A1S_1057, A1S_1242, and A1S_2622 were selected as putative members of the ABC superfamily. Finally, from the PACE superfamily, we selected the only two members of this recently discovered superfamily: the AceI transporter encoded by the A1S_2063 gene and the A1S_1503 gene, both identified in A. baumannii ATCC 17978 (19). No other PACE members were found in the annotation of the genome of this strain. Mutants of all these genes were obtained using a suicide vector, as described in Materials and Methods. All of the resulting mutants were confirmed by PCR (Fig. 1) and sequencing (Macrogen) using the appropriate primers (Table 2). All of the constructed A. baumannii mutants were stable, as confirmed after ten passages without selective pressure (data not shown).
TABLE 1.
Efflux pump transporters of A. baumannii ATCC 17978 (CP000521.1) analyzed in this studya
| Transporter | Accession no. | Efflux pump superfamily | Participation in motility | Participation in virulence |
|---|---|---|---|---|
| A1S_2736 | WP_000046679.1 | RND | + | + |
| A1S_3371 | WP_001135124.1 | MATE | + | + |
| A1S_0710 | WP_000312237.1 | SMR | + | + |
| A1S_0536 | WP_000165905.1 | ABC | – | – |
| A1S_1535 | WP_000922240.1 | ABC | – | – |
| A1S_0027 | WP_000031022.1 | ABC | – | – |
| A1S_1057 | WP_000536102.1 | ABC | – | – |
| A1S_1242 | WP_000988396.1 | ABC | + | + |
| A1S_2622 | WP_000657706.1 | ABC | + | + |
| A1S_1503 | WP_001161758.1 | PACE | – | – |
| A1S_2063 | WP_005135057.1 | PACE | – | – |
The accession number, superfamily of each protein, and its participation (+) or nonparticipation (–) in motility and virulence are indicated.
FIG 1.
(A) Primers used to confirm each of the indicated A. baumannii mutants. The lengths of the resulting PCR products are shown in base pairs (bp). (B) PCR verification of each A. baumannii mutant generated by gene disruption. V Ladder NzyTech (lane M1) and ʎ BsteII-digested DNA (lane M2) were used as molecular size markers.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′–3′) | Application |
|---|---|---|
| MUT0027F | AGTCGAATTCGTGGAACGAAGCAAGTCG | Mutant construction |
| MUT0027R | AGTCGAATTCTTTCATCAATACCGAACC | Mutant construction |
| COM0027R | GCTTGTGCTGAAATAGTCTCAGC | Mutant verification |
| MUT0710F | AGTCGAATTCGCAGGTGTATTTGAAGTG | Mutant construction |
| MUT0710R | AGTCGAATTCCAATGCCTGTCCAAACCG | Mutant construction |
| COM0710R | CAGTCCTGATACAATCAGTACAGCG | Mutant verification |
| MUT0536F | AGTCGAATTCTGAAGGTGAGCTGGTTGC | Mutant construction |
| MUT0536R | AGTCGAATTCATCGTATGACCAGCCGCG | Mutant construction |
| COM0536R | TTCGATAATACGCGTTGC | Mutant verification |
| MUT1057F | AGTCGAATTCGTGGTCGTGCAAATTGCG | Mutant construction |
| MUT1057R | AGTCGAATTCCACTAGGTTTTGTAATGG | Mutant construction |
| COM1057R | GGATAGTCTCGCTGAACAATTGC | Mutant verification |
| MUT1242F | AGTCGAATTCCAGCCTTGGCTACAGGCG | Mutant construction |
| MUT1242R | AGTCGAATTCAACCACGCGGTCATACAC | Mutant construction |
| COM1242R | CACGTAAATCAGCACGTTTACC | Mutant verification |
| MUT1535F | AGTCGAATTCCCTGAAAGTTTCTTGAGC | Mutant construction |
| MUT1535R | AGTCGAATTCGTAGGCGTCAGTTACACGG | Mutant construction |
| COM1535R | GCAGTAGTAGCAGCAATTACGC | Mutant verification |
| MUT2063F | AGTCGAATTCGGTCATCATTGCGATTGC | Mutant construction |
| MUT2063R | AGTCGAATTCGCAATCAGCAAACCACCC | Mutant construction |
| COM2063R | GGTATAAACCAAAATACATAAGG | Mutant verification |
| MUT2622F | AGTCGAATTCCCAAGTAATTTAACGC | Mutant construction |
| MUT2622R | AGTCGAATTCCTGCAAAACGAATAGG | Mutant construction |
| COM2622F | GTCATTGGCCAAACAGCCAATAGC | Mutant verification |
| MUT2736F | AGTCGAATTCCGTCTTTCAATTGAAAGTG | Mutant construction |
| MUT2736R | AGTCGAATTCGTAGCTATGCGACATGCG | Mutant construction |
| COM2736R | TCACGATTTATGCTCCTGAG | Mutant verification |
| MUT3371F | AGTCGAATTCGTCCGGTACCATTAACAC | Mutant construction |
| MUT3371R | AGTCGAATTCTGATGCTATAGCCGGTGC | Mutant construction |
| COM3371R | ACGGTTCGGCCGAAGTGG | Mutant verification |
| CMPL0710F | ATCGTCTAGAATGAAAATGTCTGAAGG | Mutant complementation |
| CMPL0710R | ATCGCCATGGTTAGGATGGAGACGAA | Mutant complementation |
| CMPL1242F | ATCGTCTAGAATGATGACAAAAATAAATTAC | Mutant complementation |
| CMPL1242R | ATCGTCTAGATCATGAATTGCCCCCTTGG | Mutant complementation |
| CMPL2736F | ATCGTCTAGAATGCCTATTGCACAATATC | Mutant complementation |
| CMPL2736R | ATCGTCTAGATCACGATTTATGCTCCTGAG | Mutant complementation |
| CMPL3371F | ATCGTCTAGAATGCCACCAAGACAGTC | Mutant complementation |
| CMPL3371R | ATCGTCTAGACTAAATAAGAACTTTG | Mutant complementation |
| M13FpUC | GTTTTCCCAGTCACGAC | Sequencing primer for pCR-BluntII-TOPO and pVRL Gmr vectors |
| M13RpUC | CAGGAAACAGCTATGAC | Sequencing primer for pCR-BluntII-TOPO and pVRL Gmr vectors |
Determination of the substrate specificity of the different efflux pumps.
The antimicrobial susceptibility of the A. baumannii ATCC 17978 wild-type (WT) strain and its derivative mutants was determined by the microdilution method (Table 3). The results confirmed the susceptibility of the RND efflux pump mutant A1S_2736 to a wide range of antimicrobial agents (such as β-lactams and quinolones) and to other compounds (such as sodium dodecyl sulfate [SDS]), as previously described (14). Moreover, this mutant was highly susceptible to previously untested compounds, including deoxycholate (DC). The A1S_3371 MATE efflux pump mutant only showed a slight difference in tetracycline even though no antimicrobial resistances were previously associated with this transporter (15). The A1S_0710 SMR efflux pump mutant exhibited greater susceptibility than the wild type to compounds such as SDS and DC. Accordingly, overexpression of this transporter in Escherichia coli increased its resistance to both compounds (16). In addition, this same mutant was slightly more susceptible than the wild-type to several antimicrobials, including quinolones, tetracycline, and trimethoprim. Among the ABC efflux pump mutants, the genes A1S_0536 and A1S_1535 exhibited resistance to erythromycin and to gentamicin and chloramphenicol, respectively, as previously described (17, 18). Mutants of two newly described ABC permeases, encoded by the genes A1S_1242 and A1S_2622, showed modest differences in their resistance profiles compared to the wild-type strain (Table 3). However, inactivation of the putative ABC permeases encoded by the genes A1S_0027 and A1S_1057 did not result in differences in susceptibility to any of the compounds tested. Finally, among members of the PACE superfamily, the AceI (A1S_2063) mutant was more susceptible to chlorhexidine and the A1S_1503 mutant to acriflavin, as previously reported (7, 19). The latter mutant also showed a slight susceptibility to meropenem, amikacin, and minocycline (Table 3).
TABLE 3.
MICs of the tested antimicrobials for wild-type A. baumannii ATCC 17978 (WT) and the derivatives carrying mutant efflux pumpsa
| Antimicrobial agent | MIC (mg/liter) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | Superfamily |
|||||||||
| RND (A1S_2736) | MATE (A1S_3371) | SMR (A1S_0710) | PACE |
ABC |
||||||
| A1S_1503 | A1S_2063 | A1S_0536 | A1S_1535 | A1S_1242 | A1S_2622 | |||||
| Ciprofloxacin | 0.25 | 0.031 | 0.25 | 0.125 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| Levofloxacin | 0.125 | 0.0156 | 0.125 | 0.062 | 0.125 | 0.125 | 0.125 | 0.125 | 0.062 | 0.125 |
| Nalidixic acid | 8 | 2 | 8 | 4 | 8 | 8 | 8 | 8 | 4 | 8 |
| Trimethoprim | 16 | 4 | 16 | 8 | 16 | 16 | 16 | 16 | 8 | 16 |
| Novobiocin | 8 | 0.062 | 8 | 8 | 8 | 8 | 2 | 8 | 4 | 8 |
| Meropenem | 2 | 0.5 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 |
| Ampicillin | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 |
| Amikacin | 1 | 0.5 | 1 | 1 | 0.5 | 1 | 0.5 | 1 | 1 | 1 |
| Gentamicin | 0.5 | 0.25 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.25 | 0.5 | 0.25 |
| Erythromycin | 4 | 2 | 4 | 4 | 4 | 4 | 2 | 4 | 4 | 4 |
| Colistin | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Chloramphenicol | 16 | 8 | 16 | 16 | 16 | 16 | 16 | 8 | 16 | 16 |
| Tetracycline | 4 | 1 | 2 | 2 | 4 | 4 | 4 | 4 | 2 | 4 |
| Minocycline | 0.062 | 0.0038 | 0.062 | 0.031 | 0.031 | 0.062 | 0.062 | 0.062 | 0.031 | 0.031 |
| Rifampin | 4 | 1 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| DC | 5,000 | 62.5 | 5,000 | 1,250 | 5,000 | 5,000 | 5,000 | 2,500 | 5,000 | 5,000 |
| SDS | 2,500 | 156 | 2,500 | 1,250 | 2,500 | 2,500 | 2,500 | 2,500 | 2,500 | 2,500 |
| Acriflavin | 12.5 | 12.5 | 12.5 | 12.5 | 6.25 | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 |
| Chlorhexidine | 6 | 6 | 6 | 6 | 6 | 3 | 6 | 6 | 6 | 6 |
Mutants that did not at all differ from the wild type are not included (A1S_0027 and A1S_1057). Differences compared to the WT strain are indicated in boldface.
Surface-associated motility of the A. baumannii efflux pump mutants.
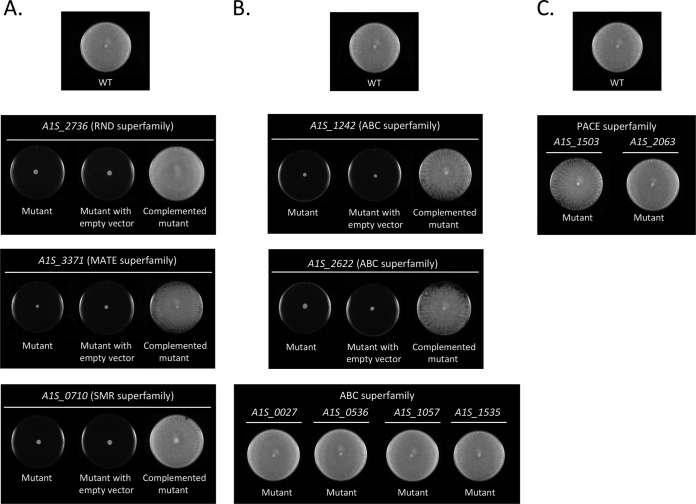
To examine whether the selected efflux pumps play important roles in the surface-associated motility of A. baumannii, the motility of the knockout mutants on agar motility plates was investigated. A loss of motility was determined in mutants of the RND (A1S_2736), MATE (A1S_3371), and SMR (A1S_0710) efflux pumps (Fig. 2A). Introduction of plasmid pVRL1 Gmr, carrying the corresponding wild-type gene, restored the parental surface-associated motility phenotype in all cases (Fig. 2A). In the only two A. baumannii ABC efflux pumps described so far (A1S_0536 and A1S_1535), there were no changes in the motility profile compared to the respective wild-type strain (Fig. 2B). However, mutants of the ABC permeases identified here and encoded by the genes A1S_1242 and A1S_2622 showed a loss of surface-associated motility, while the motility of mutants A1S_0027 and A1S_1057 was similar to that of the wild-type strain (Fig. 2B). Introduction of the plasmid pVRL1 Gmr, carrying the corresponding wild-type gene, restored motility in both A1S_1242 and A1S_2622 mutants (Fig. 2B). Finally, the motility of both mutants of the PACE superfamily transporters (A1S_1503 and A1S_2063) was similar to that of the wild-type strain (Fig. 2C). Since the growth curves of all mutants grown in liquid medium (Luria-Bertani [LB]) were comparable to that of the respective wild-type strain (data not shown), the observed defects in surface-associated motility cannot be attributed to reductions in the bacterial growth rate.
FIG 2.
Surface-associated motility assays of wild-type A. baumannii strain ATCC 17978 (WT) and the indicated derivative mutants belonging to the RND, MATE, and SMR (A), ABC (B), and PACE (C) superfamilies. The surface-associated motility of the indicated mutants carrying either the empty expression vector pVRL1 Gmr (mutant with empty vector) or the same plasmid containing the corresponding wild-type (WT) gene (complemented mutants) is also shown.
Effect of the inactivation of genes encoding efflux pumps on the virulence of A. baumannii.
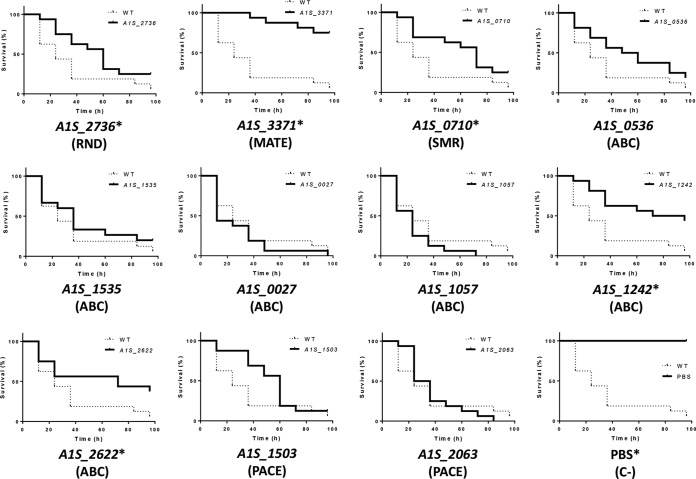
The nematode C. elegans has been used as a model organism to study the virulence of A. baumannii by counting the progeny of worms fed with the strain of interest (20). In the present study, the virulence of mutants of the efflux pumps encoded by the genes A1S_2736 (RND), A1S_3371 (MATE), and A1S_0710 (SMR), as well as that of mutants of the newly described ABC permeases (A1S_1242 and A1S_2662), was attenuated compared to the wild-type strain (Fig. 3). However, in the mutants A1S_0536, A1S_1535, A1S_0027, and A1S_1057 (ABC), as well as mutants A1S_1503 and A1S_2063 (PACE), the level of virulence was similar to that of the wild-type strain (Fig. 3). The validity of the Galleria mellonella killing in studies of in vivo host-pathogen interactions in A. baumannii has been reported (21). Accordingly, we used this animal model to verify the results obtained in the nematode fertility assay. As expected, the virulence of the same A. baumannii mutants, lacking the genes encoding the permeases A1S_2736 (RND), A1S_3371 (MATE), A1S_0710 (SMR), A1S_1242 (ABC), and A1S_2662 (ABC), was significantly decreased compared to the wild-type strain (Fig. 4).
FIG 3.
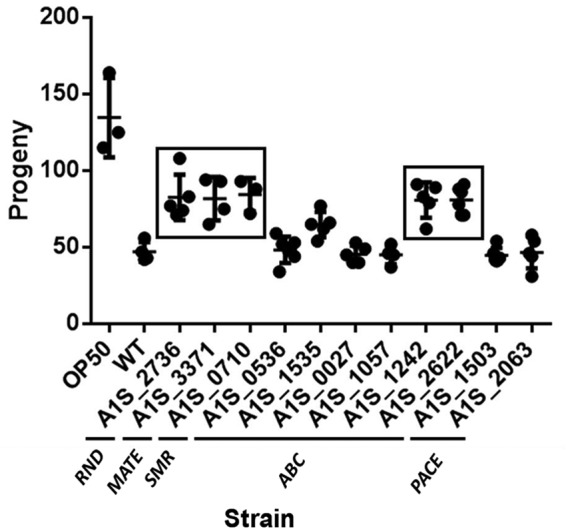
Representative C. elegans fertility assay results for wild-type A. baumannii strain ATCC 17978 (WT) and derivative mutants. Boxes indicate strains that differed significantly (P < 0.05) compared to the WT strain. E. coli OP50 was included as a low-virulence control.
FIG 4.
Survival of G. mellonella larvae (n = 16 per group) infected with approximately 106 CFU of the indicated A. baumannii strains or PBS. The concentrations of all mutant strains differed by <0.5 log CFU/larva with respect to the wild-type (WT; strain ATCC 17978). In the negative-control group (C–) the caterpillars were inoculated with PBS to take into account killing due to physical trauma. All assays were carried out simultaneously. For clarity, the results obtained from cultures inoculated with the different mutants or PBS are presented separately and compared to the group inoculated with the WT strain. In those comparisons, a P value of <0.05, indicated by an asterisk (*), was considered to indicate significant decreased virulence relative to the WT strain. The results of a representative assay are shown.
DISCUSSION
Bacterial efflux pumps actively prevent the intracellular accumulation of toxic compounds and thus, play an important role in antimicrobial resistance (6). In previous studies, we identified a quinolone efflux pump, AbaQ, belonging to the MFS superfamily and involved in the motility and virulence of A. baumannii (10, 11). This led us to examine the participation of other transporters, belonging to different efflux pump superfamilies, in the motility and virulence of A. baumannii. Mutants of genes encoding the selected transporters of each of the other five remaining superfamilies (Table 1) were therefore constructed using A. baumannii ATCC 17978 (Fig. 1). Their MICs to different compounds were determined using the microdilution method (Table 3).
Knowledge of the role of efflux pumps in the motility and virulence of A. baumannii is limited. We therefore examined the motility and virulence of mutants lacking representative transporters belonging to all superfamilies of efflux pumps using motility plates and two in vivo animal models, respectively. The mutant lacking the RND efflux pump A1S_2736 had an altered motility pattern and an attenuated virulence compared to the wild-type strain (Fig. 2A, 3, and 4). In A. baumannii, some members of the RND superfamily have been implicated in the pathogenicity of the bacterium. For example, in A. baumannii strain AB5075, the attenuated virulence of two mutants (arpA and arpB) lacking components of an RND system was demonstrated in the G. mellonella model (22). Furthermore, inactivation of the gene A1S_0114, involved in the synthesis of a lipopeptide, reduces the motility and virulence of A. baumannii. A1S_0114 is located on the same operon as A1S_0116, which encodes an RND transporter possibly involved in the secretion of this lipopeptide (23).
During the preparation of the manuscript, Knight et al. reported that the protein AdeJ, the permease component of an RND system, is involved in the motility of Acinetobacter nosocomialis whereas the permease AdeB, which also belongs to the same efflux pump superfamily, is not (24). Although our study does not include an analysis of the AdeB protein, we analyzed the permease encoded by the gene A1S_2736, which has 99% identity with AdeJ of A. nosocomialis, and obtained the same results. Previous studies showed that the absence of the gene encoding the AdeRS two-component system, which regulates the RND efflux pump AdeABC in A. baumannii, causes a loss of motility but not of virulence (25). Further alterations in the motility and virulence of A. nosocomialis resulting from the deregulation of efflux pumps have also been demonstrated. Specifically, deletion of the repressor of the RND efflux pump, AcrAB (AcrR), increased both the motility and the virulence of the mutant (26).
The role of members of the RND superfamily in motility and virulence has also been studied in other pathogens. Thus, in Pseudomonas aeruginosa, the RND transporter MexEF-OprN was shown to be involved in the transport of a signal molecule that mediates quorum sensing and therefore swarming motility, biofilm formation, and the production of virulence factors (27). In the opportunistic pathogen Stenotrophomonas maltophilia, mutants lacking the RND transporter SmeYZ have an increased susceptibility to human serum and neutrophils, exhibit a decreased swimming motility, and are less virulent (28). The absence of the RND AcrAB efflux system in the respiratory pathogen Klebsiella pneumoniae results in a lower capacity of infection (29), while in Enterobacter cloacae a transporter homologous to the AcrAB-TolC system contributes to environmental adaptation, fitness, and virulence (30). There are also several examples linking other RND efflux pumps to virulence in the pathogens Borrelia burgdorferi (31), Salmonella enterica (32), Vibrio cholerae (33), and Erwinia amylovora (34).
Mutants in the MATE efflux pump A1S_3371 and the SMR efflux pump A1S_0710 also show a reduced motility and an attenuated virulence in A. baumannii (Fig. 2A, 3, and 4). In Ralstonia solanacearum the absence of the MATE DinF efflux pump causes a decreased virulence of the bacterium in tomato plants (35), and in K. pneumoniae the SMR KpnEF efflux pump participates in capsule synthesis and therefore, presumably, virulence as well (36).
While the involvement of ABC efflux pumps in the virulence of A. baumannii has not been demonstrated, we previously reported a gene encoding a putative taurine ABC transporter component involved in both motility and virulence of this bacterial species (10). Inactivation of the ABC transporters encoded by the previously described genes A1S_0536 and A1S_1535 had no effects on the motility or virulence of A. baumannii. However, in this work we newly identified two genes encoding ABC permeases (A1S_1242 and A1S_2622) that seem to be involved in both functions (Fig. 2B, 3, and 4). In contrast, mutants of other genes encoding putative components of ABC transporters (A1S_0027 and A1S_1057) did not show changes in motility or virulence (Fig. 2B, 3, and 4). Genes encoding ABC efflux pump components in other pathogens have also been implicated in virulence, such as the ABC efflux pumps MacAB in S. enterica (37), MrtAB in Yersinia pseudotuberculosis (38), YejABEF in Brucella melitensis (39), and Ssu in Xanthomonas citri (40).
The motility and virulence profiles of A. baumannii mutants belonging to the PACE superfamily (A1S_1503 and A1S_2063) were very similar to those of the wild-type strain (Fig. 2C, 3, and 4). To our knowledge, the role of the recently discovered PACE superfamily in virulence has not been studied in any other microorganism. However, since these protein are highly conserved in other pathogenic bacteria, while their physiological role remains to be determined, their participation in drug resistance seems to be accidental (41).
The mechanisms by which efflux pumps participate in bacterial pathogenicity have yet to be fully elucidated. However, it is clear that the loss of an efflux pump may in itself, have secondary consequences, by altering the composition and structure of the cell envelope. Furthermore, efflux pumps also may contribute to the secretion of virulence factors, as observed for the RND-type transporter VexB of V. cholerae and the ABC-type transporter MacAB of E. coli (42, 43). In addition, efflux pumps may translocate host-derived antimicrobial molecules, allowing survival of the pathogen. This is the case of the MFS efflux pump FarAB of Neisseria gonorrhoeae, which protects the bacterium against long-chained antibacterial fatty acids produced by the host (44). Other examples are the RND efflux pumps AcrAB and CmeABC (belonging to E. coli and Campylobacter jejuni, respectively), both of which translocate the bactericidal bile encountered in the intestinal tracts of their host animals (45, 46). Likewise, efflux pumps may affect motility and virulence, by translocating molecules that modulate processes such as quorum sensing, as shown for the RND efflux pump MexEF-OprN of P. aeruginosa (27). In A. baumannii, the function of these efflux systems might be the translocation of surfactants or molecules related to surface-associated motility or quorum sensing, since alterations in molecules mediating bacterial population responses have been related to the inhibition of motility (47).
Overall, our study provides new insights into the efflux pumps of A. baumannii, especially their involvement in the motility and virulence of the bacterium. Although not all efflux pumps contribute to motility and virulence, our results demonstrate that in A. baumannii ATCC 17978, all superfamilies of efflux pumps, except the recently discovered PACE superfamily, include transporters involved in surface-associated motility. Moreover, all mutants lacking an efflux transporter involved in this kind of motility were also less virulent. While the mechanisms underlying the participation of these efflux pumps in motility and virulence remain unclear, further studies of these proteins will broaden current understanding of the pathogenicity of A. baumannii. In addition, the results reported here suggest that inhibitors against these transporters will be valuable tools with which to fight multiresistant pathogens, by targeting not only antimicrobial resistance but also bacterial motility, thereby reducing the capacity for host colonization.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
A. baumannii ATCC 17978 and Escherichia coli (strains DH5α and OP50) were grown at 37°C in LB agar or in LB broth with shaking at 180 rpm. Plasmids were maintained and selected in E. coli and A. baumannii hosts using the same medium but supplemented as needed with kanamycin (50 mg/liter) or gentamicin (20 mg/liter for E. coli or 40 mg/liter for A. baumannii). Bacterial growth was monitored as follows. A bacterial culture in LB broth was inoculated with a 1:100 dilution of an overnight culture and incubated with shaking (180 rpm) at 37°C. The optical density at 600 nm (OD600) was measured hourly. Each test was performed in triplicate. The plasmids pCR-BluntII-TOPO (Invitrogen) and pVRL1 Gmr (48) were used for mutant construction and complementation, respectively.
Construction of A. baumannii knockouts and complementation.
Gene inactivation was carried out as previously described (49). Briefly, an internal fragment of the target gene was PCR amplified with the corresponding primers (Table 2) using genomic DNA from A. baumannii ATCC 17978 as the template. This fragment was cloned into the kanamycin and zeocin resistance plasmid pCR-BluntII-TOPO, which is unable to replicate in A. baumannii, and then propagated in E. coli DH5α. The recombinant plasmids (0.1 μg) were electroporated into A. baumannii ATCC 17978. Candidate transformant clones were selected on kanamycin-containing plates. Mutants were confirmed by sequencing (Macrogen) of the PCR products. To determine the stability of the obtained mutants, the cultures were passaged 10 times (every 24 h) without selective pressure and the colonies that developed on LB agar plates with or without kanamycin were counted. For complementation, all genes were cloned into the XbaI-NcoI site or, when necessary, into the XbaI site of the pVRL1 Gmr vector. The recombinant plasmids were propagated in E. coli DH5α and then electroporated into the corresponding A. baumannii mutant. The complemented mutant was selected on plates containing gentamicin. The resulting recombinant plasmids were verified by PCR and sequencing. The primers used in this study are shown in Table 2.
In vitro antimicrobial susceptibility assay.
MICs for all antimicrobials and disinfectants (Sigma) were determined on 96-well plates by the broth microdilution method, using Mueller-Hinton medium (Merck), according to CLSI guidelines (50). All MIC assays were repeated a minimum of three times.
Motility assays.
Motility assays were conducted on fresh LB agar motility plates (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.5% glucose, and 0.5% Difco agar) used on the day of their preparation. To test the motility of the different A. baumannii strains, a 5-μl drop of an exponential-phase culture of each strain was placed in the center of the plate. The inoculated plates were incubated at 30°C, usually for 16 to 20 h, until growth of the wild-type strain reached to the plate border. All assays were carried out in triplicate in a minimum of three independent experiments. The representative images shown in the figures were obtained using the ChemiDoc XRS+ System (Bio-Rad).
In vivo nematode model of virulence.
The virulence of the different A. baumannii strains was determined in a nematode fertility assay performed as previously described (20). Briefly, C. elegans strain N2 was fed with low-virulence E. coli strain OP50 grown as a lawn on nematode growth medium (NGM) plates. To physiologically synchronize the worms, the nematode eggs were hatched in M9 medium (0.02 M KH2PO4, 0.04 M Na2HPO4, 0.08 M NaCl, and 0.001 M MgSO4) and worms in the first larval (L1) stage were growth arrested overnight at 16°C. The L1 worms were then grown to the L4 stage on NGM plates seeded with the bacterial strains investigated in this study. One nematode in the L4 state was placed on a peptone-glucose-sorbitol plate with the same strain and incubated at 25°C. During the next 3 days, adult nematodes were transferred to new plates seeded with the same bacterial strain. To determine the fertility of the worms, their progeny was counted daily, 48 h after removal of the parent, using a stereomicroscope (Olympus SZ51). Six independent replicates were established for each strain and each fertility assay was performed in triplicate.
G. mellonella killing assay.
The wax worm model of virulence was used following the methodology described by Peleg et al. (21) with some modifications. Briefly, 16 caterpillars (approximately 300 mg in weight) were used for each A. baumannii strain. Cells from bacterial inocula prepared from exponential cultures (OD600 = 0.5, approximately 108 CFU/ml) were washed with phosphate-buffered saline (PBS) and adjusted again to the same OD600. The concentrations of the inocula were confirmed by bacterial colony counts on LB agar. A 10-μl inoculum of the corresponding strain or 10 μl of PBS (as a negative control) was injected into the hemocoel of each worm through the last left proleg. The caterpillars were incubated at 37°C in the dark, and their survival was checked every 12 h for a total of 96 h. All G. mellonella killing experiments were performed at least twice.
Statistics.
Data for the nematode model of virulence are presented as means ± the standard deviations and were analyzed using two-tailed, one-way analysis of variance, followed by the Tukey test for post hoc multiple group comparisons. In the G. mellonella killing assay, survival curves were plotted using the Kaplan-Meier method. Differences in survival were calculated using the log-rank test. In all analyses, a P value of <0.05 was considered to indicate statistical significance.
ACKNOWLEDGMENTS
We thank Joan Ruiz (Universitat Autònoma de Barcelona [UAB]) and Susana Escribano (UAB) for excellent technical assistance, as well as our UAB student, Paula García, for helpful support. We also thank Paolo Visca (University Roma Tre, Rome, Italy) for kindly providing plasmid pVRL1 Gmr and Ana Morton (Departament de Biologia Animal, de Biologia Vegetal i d’Ecologia, UAB) for guidance during the G. mellonella experiments.
This study was supported by grant BIO2016-77011-R from the Ministerio de Economía y Competitividad. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. J.A. is a Serra Húnter Fellow, Generalitat de Catalunya, Barcelona, Spain.
REFERENCES
- 1.Howard A, O’Donoghue M, Feeney A, Sleator RD. 2012. Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 3:243–250. doi: 10.4161/viru.19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee C-R, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha C-J, Jeong BC, Lee SH. 2017. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol 7:55. doi: 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piddock LJV. 2006. Multidrug-resistance efflux pumps? not just for resistance. Nat Rev Microbiol 4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 5.Dijun D, Wang-Kan X, Neuberger A, van Veen HW, Pos KM, Piddock LJV, Luisi BF. 2018. Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol 16:523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 6.Coyne S, Courvalin P, Perichon B. 2011. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 55:947–953. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassan KA, Liu Q, Henderson PJF, Paulsen IT. 2015. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio 6:e01982-14. doi: 10.1128/mBio.01982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemmer KM, Bonomo RA, Rather PN. 2011. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157:2534–2544. doi: 10.1099/mic.0.049791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skiebe E, de Berardinis V, Morczinek P, Kerrinnes T, Faber F, Lepka D, Hammer B, Zimmermann O, Ziesing S, Wichelhaus TA, Hunfeld KP, Borgmann S, Gröbner S, Higgins PG, Seifert H, Busse HJ, Witte W, Pfeifer Y, Wilharm G. 2012. Surface-associated motility, a common trait of clinical isolates of Acinetobacter baumannii, depends on 1,3-diaminopropane. Int J Med Microbiol 302:117–128. doi: 10.1016/j.ijmm.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Pérez-Varela M, Corral J, Vallejo JA, Rumbo-Feal S, Bou G, Aranda J, Barbé J. 2017. Mutations in the β-subunit of the RNA polymerase impair the surface-associated motility and virulence of Acinetobacter baumannii. Infect Immun 85:e00327-17. doi: 10.1128/IAI.00327-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pérez-Varela M, Corral J, Aranda J, Barbé J. 2018. Functional characterization of AbaQ, a novel efflux pump mediating quinolone resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 62:e00906-18. doi: 10.1128/AAC.00906-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon EJ, Balloy V, Fiette L, Chignard M, Courvalin P, Grillot-Courvalin C. 2016. Contribution of the Ade resistance-nodulation-cell division-type efflux pumps to fitness and pathogenesis of Acinetobacter baumannii. mBio 7:e00697-16. doi: 10.1128/mBio.00697-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma A, Sharma R, Bhattacharyya T, Bhando T, Pathania R. 2017. Fosfomycin resistance in Acinetobacter baumannii is mediated by efflux through a major facilitator superfamily (MFS) transporter-AbaF. J Antimicrob Chemother 72:68–74. doi: 10.1093/jac/dkw382. [DOI] [PubMed] [Google Scholar]
- 14.Damier-Piolle L, Magnet S, Brémont S, Lambert T, Courvalin P. 2008. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents Chemother 52:557–562. doi: 10.1128/AAC.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. 2011. Development of a high-throughput cloning strategy for characterization of Acinetobacter baumannii drug transporter proteins. J Mol Microbiol Biotechnol 20:211–219. doi: 10.1159/000329836. [DOI] [PubMed] [Google Scholar]
- 16.Rajamohan G, Stevenson K, Marcon M, Pancholi P, Srinivasan V, Gebreyes WA. 2008. Novel secondary active transporters conferring antimicrobial resistance in Acinetobacter baumannii with broad substrate specificity. URL: https://idsa.confex.com/idsa/2008/webprogram/Paper27595.html.
- 17.Okada U, Yamashita E, Neuberger A, Morimoto M, Van Veen HW, Murakami S. 2017. Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat Commun 8:1336. doi: 10.1038/s41467-017-01399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Hassan KA, Brown MH, Paulsen IT. 2016. Rapid multiplexed phenotypic screening identifies drug resistance functions for three novel efflux pumps in Acinetobacter baumannii. J Antimicrob Chemother 71:1223–1232. doi: 10.1093/jac/dkv460. [DOI] [PubMed] [Google Scholar]
- 19.Hassan KA, Jackson SM, Penesyan A, Patching SG, Tetu SG, Eijkelkamp BA, Brown MH, Henderson PJF, Paulsen IT. 2013. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc Natl Acad Sci U S A 110:20254–20259. doi: 10.1073/pnas.1317052110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vallejo JA, Beceiro A, Rumbo-Feal S, Rodríguez-Palero MJ, Russo TA, Bou G. 2015. Optimization of the Caenorhabditis elegans model for studying the pathogenesis of opportunistic Acinetobacter baumannii. Int J Antimicrob Agents pii:S0924-857900241-S0921. doi: 10.1016/j.ijantimicag.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 21.Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother 53:2605–2609. doi: 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tipton KA, Farokhyfar M, Rather PN. 2017. Multiple roles for a novel RND-type efflux system in Acinetobacter baumannii AB5075. Microbiologyopen 6. doi: 10.1002/mbo3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rumbo-Feal S, Pérez A, Ramelot TA, Álvarez-Fraga L, Vallejo JA, Beceiro A, Ohneck EJ, Arivett BA, Merino M, Fiester SE, Kennedy MA, Actis LA, Bou G, Poza M. 2017. Contribution of the A. baumannii A1S_0114 gene to the interaction with eukaryotic cells and virulence. Front Cell Infect Microbiol 7:108. doi: 10.3389/fcimb.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knight DB, Rudin SD, Bonomo RARP. 2018. Acinetobacter nosocomialis: Defining the role of efflux pumps in resistance to antimicrobial therapy, surface motility, and biofilm formation. Front Microbiol 9:1902. doi: 10.3389/fmicb.2018.01902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Silva PM, Kumar A. 2018. Effect of sodium chloride on surface-associated motility of Acinetobacter baumannii and the role of AdeRS two-component system. J Membr Biol 251:5–13. doi: 10.1007/s00232-017-9985-7. [DOI] [PubMed] [Google Scholar]
- 26.Subhadra B, Kim J, Kim DH, Woo K, Oh Mh CC. 2018. Local repressor AcrR regulates AcrAB efflux pump required for biofilm formation and virulence in Acinetobacter nosocomialis. Front Microbiol 8:270. doi: 10.3389/fcimb.2018.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamarche MG, Déziel E. 2011. MexEF-oprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline). PLoS One 6:e24310. doi: 10.1371/journal.pone.0024310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin YT, Huang YW, Chen SJ, Chang CW, Yang TC. 2015. The SmeYZ efflux pump of Stenotrophomonas maltophilia contributes to drug resistance, virulence-related characteristics, and virulence in mice. Antimicrob Agents Chemother 59:4067–4073. doi: 10.1128/AAC.00372-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez A, Poza M, Fernández A, Del Carmen Fernández M, Mallo S, Merino M, Rumbo-Feal S, Cabral MP, Bou G. 2012. Involvement of the AcrAB-TolC efflux pump in the resistance, fitness, and virulence of Enterobacter cloacae. Antimicrob Agents Chemother 56:2084–2090. doi: 10.1128/AAC.05509-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bunikis I, Denker K, Östberg Y, Andersen C, Benz R, Bergström S. 2008. An RND-type efflux system in Borrelia burgdorferi is involved in virulence and resistance to antimicrobial compounds. PLoS Pathog 4:e1000009. doi: 10.1371/journal.ppat.1000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishino K, Latifi T, Groisman EA. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol 59:126–141. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 33.Nishiyama S, Takahashi Y, Yamamoto K, Suzuki D, Itoh Y, Sumita K, Uchida Y, Homma M, Imada K, Kawagishi I. 2016. Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants. Sci Rep 6:20866. doi: 10.1038/srep20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burse A, Weingart H, Ullrich MS. 2004. The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol Plant Microbe Interact 17:43–54. doi: 10.1094/MPMI.2004.17.1.43. [DOI] [PubMed] [Google Scholar]
- 35.Brown DG, Swanson JK, Allen C. 2007. Two host-induced Ralstonia solanacearum genes, acrA and dinF, encode multidrug efflux pumps and contribute to bacterial wilt virulence. Appl Environ Microbiol 73:2777–2786. doi: 10.1128/AEM.00984-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srinivasan VB, Rajamohan G. 2013. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob Agents Chemother 57:4449–4462. doi: 10.1128/AAC.02284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogomolnaya LM, Andrews KD, Talamantes M, Maple A, Ragoza Y, Vazquez-Torres A, Andrews-Polymenis H. 2013. The ABC-type efflux pump MacAB protects Salmonella enterica serovar Typhimurium from oxidative stress. mBio 4:e00630-13. doi: 10.1128/mBio.00630-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crimmins GT, Mohammadi S, Green ER, Bergman MA, Isberg RR, Mecsas J. 2012. Identification of MrtAB, an ABC transporter specifically required for Yersinia pseudotuberculosis to colonize the mesenteric lymph nodes. PLoS Pathog 8:e1002828. doi: 10.1371/journal.ppat.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Bie P, Cheng J, Lu L, Cui B, Wu Q. 2016. The ABC transporter YejABEF is required for resistance to antimicrobial peptides and the virulence of Brucella melitensis. Sci Rep 6:31876. doi: 10.1038/srep31876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tófoli De Araújo F, Bolanos-Garcia VM, Pereira CT, Sanches M, Oshiro EE, Ferreira RCC, Chigardze DY, Gonçalves Barbosa JA, Ferreira LCDS, Benedetti CE, Blundell TL, Balan A. 2013. Structural and physiological analyses of the alkanesulphonate-binding protein (SsuA) of the citrus pathogen Xanthomonas citri. PLoS One 8:e80083. doi: 10.1371/journal.pone.0080083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hassan KA, Liu Q, Elbourne LDH, Ahmad I, Sharples D, Naidu V, Chan CL, Li L, Harborne SPD, Pokhrel A, Postis VLG, Goldman A, Henderson PJF, Paulsen IT. 2018. Pacing across the membrane: the novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res Microbiol 169:450–454. doi: 10.1016/j.resmic.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bina XR, Provenzano D, Nguyen N, Bina JE. 2008. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect Immun 76:3595–3605. doi: 10.1128/IAI.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamanaka H, Kobayashi H, Takahashi E, Okamoto K. 2008. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J Bacteriol 190:7693–7698. doi: 10.1128/JB.00853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee EH, Shafer WM. 1999. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol Microbiol 33:839–845. [DOI] [PubMed] [Google Scholar]
- 45.Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE. 1995. Genes acrA and acrB encode a stress‐induced efflux system of Escherichia coli. Mol Microbiol 16:45–55. [DOI] [PubMed] [Google Scholar]
- 46.Lin J, Michel LO, Zhang Q. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother 46:2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicol M, Alexandre S, Luizet JB, Skogman M, Jouenne T, Salcedo SP, Dé E. 2018. Unsaturated fatty acids affect quorum sensing communication system and inhibit motility and biofilm formation of Acinetobacter baumannii. Int J Mol Sci 19:214. doi: 10.3390/ijms19010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lucidi M, Runci F, Rampioni G, Frangipani E, Leoni L, Visca P. 2018. New shuttle vectors for gene cloning and expression in multidrug-resistant Acinetobacter species. Antimicrob Agents Chemother 62:e02480-17. doi: 10.1128/AAC.02480-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, Parreira JR, Rodriguez-Velo P, Bou G. 2010. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol 10:279. doi: 10.1186/1471-2180-10-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]