Abstract
K562 is widely used in biomedical research. It is one of three tier-one cell lines of ENCODE and also most commonly used for large-scale CRISPR/Cas9 screens. Although its functional genomic and epigenomic characteristics have been extensively studied, its genome sequence and genomic structural features have never been comprehensively analyzed. Such information is essential for the correct interpretation and understanding of the vast troves of existing functional genomics and epigenomics data for K562. We performed and integrated deep-coverage whole-genome (short-insert), mate-pair, and linked-read sequencing as well as karyotyping and array CGH analysis to identify a wide spectrum of genome characteristics in K562: copy numbers (CN) of aneuploid chromosome segments at high-resolution, SNVs and indels (both corrected for CN in aneuploid regions), loss of heterozygosity, megabase-scale phased haplotypes often spanning entire chromosome arms, structural variants (SVs), including small and large-scale complex SVs and nonreference retrotransposon insertions. Many SVs were phased, assembled, and experimentally validated. We identified multiple allele-specific deletions and duplications within the tumor suppressor gene FHIT. Taking aneuploidy into account, we reanalyzed K562 RNA-seq and whole-genome bisulfite sequencing data for allele-specific expression and allele-specific DNA methylation. We also show examples of how deeper insights into regulatory complexity are gained by integrating genomic variant information and structural context with functional genomics and epigenomics data. Furthermore, using K562 haplotype information, we produced an allele-specific CRISPR targeting map. This comprehensive whole-genome analysis serves as a resource for future studies that utilize K562 as well as a framework for the analysis of other cancer genomes.
K562 is an immortalized chronic myelogenous leukemia (CML) cell line derived from a 53-yr-old Caucasian female in 1970 (Lozzio and Lozzio 1975). Since being established, K562 has been widely used in biomedical research as a “workhorse” cell line, contributing to the understanding of fundamental human biological processes as well as to basic and translational cancer research (Grzanka et al. 2003; Drexler et al. 2004; Butler and Hirano 2014). Along with the H1 human embryonic stem cell line and the GM12878 lymphoblastoid cell line, K562 is one of the three tier-one cell lines of the ENCyclopedia Of DNA Elements Project (ENCODE) (The ENCODE Project Consortium 2012), forming the basis of more than 1300 ENCODE data sets to date. Furthermore, it is also one of the few cell lines most commonly used for large-scale CRISPR/Cas9 gene-targeting screens (Wang et al. 2015; Adamson et al. 2016; Arroyo et al. 2016; Morgens et al. 2016; Han et al. 2017; Liu et al. 2017).
Although the functional genomic characteristics of K562 have been extensively studied and documented, reflected in close to 600 ChIP-seq, 400 RNA-seq, 50 DNase-seq, and 30 RIP-seq data sets available through the ENCODE portal (Sloan et al. 2016), the sequence and structural features of the K562 genome have never been comprehensively characterized, even though past cytogenetic studies using G-banding, fluorescence in situ hybridization (FISH), multiplex-FISH, and comparative genomic hybridization (CGH) showed that K562 cells contain pervasive aneuploidy and multiple gross structural abnormalities (Selden et al. 1983; Wu et al. 1995; Gribble et al. 2000; Naumann et al. 2001), not unexpected for a cancer cell line. In other words, the rich amount of K562 functional genomics and epigenomics work conducted to date—in particular, integrative analyses that have been carried out using the vast troves of K562 ENCODE data—were done without taking into account the many differences of the K562 genome relative to the human reference genome. This leads to skewed interpretations and reduces the amount of knowledge that can be gained from the rich, multilayered ENCODE data sets that continue to accumulate.
Here, we report for the first time a comprehensive characterization of the K562 genome (Fig. 1; Supplemental Fig. S1A) that includes copy numbers (CNs) of chromosome segments at high-resolution; single-nucleotide variants (SNVs, also including single-nucleotide polymorphisms [SNPs]); and small insertions and deletions (indels) with allele frequencies corrected by CN in aneuploid regions, loss of heterozygosity, megabase-scale phased haplotypes often spanning entire chromosome arms, and structural variants (SVs), including small and large-scale complex SVs with phasing. We then took first steps into exploring how knowledge about genome sequence and structural features can influence the interpretation of functional genomics and epigenomics data and show examples of how deeper insights into genome regulatory complexity can be obtained by integrating genomic context. These insights also shed light on important questions regarding cancer evolution.
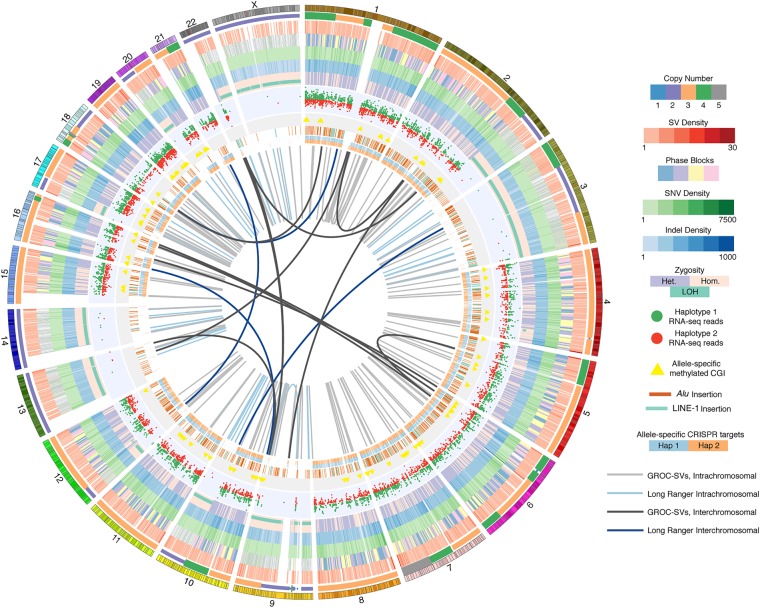
Figure 1.
Comprehensive overview of the K562 genome. Circos (Krzywinski et al. 2009) visualization of the K562 genome with the following tracks in inward concentric order: chromosomes; CN, i.e., ploidy by chromosome segment; merged SV density in 1.5-Mb windows of deletions, duplications, and inversions identified using ARC-SV (Arthur et al. 2018), BreakDancer (Chen et al. 2009), BreakSeq (Lam et al. 2010), LUMPY (Layer et al. 2014), Pindel (Ye et al. 2009), and Long Ranger (Zheng et al. 2016; Marks et al. 2018); phased haplotype blocks (demarcated with four colors for clearer visualization); SNV density in 1-Mb windows; indel density in 1-Mb windows; dominant zygosity in 1-Mb windows (heterozygous or homozygous >50%) with regions exhibiting loss of heterozygosity (LOH) indicated; RNA-seq reads for loci exhibiting allele-specific expression; CpG islands (CGI) exhibiting allele-specific methylation; histogram (log-scale) of allele-specifically methylated CpGs in 50-kb windows; nonreference Alu and LINE-1 insertions; allele-specific CRISPR target sites; large-scale rearrangements detected by Long Ranger (Zheng et al. 2016; Marks et al. 2018) (light blue: intrachromosomal; dark blue: interchromosomal); and by GROC-SVs (Spies et al. 2017) (light gray: intrachromsomal; dark gray: interchromosomal).
Results
Karyotyping
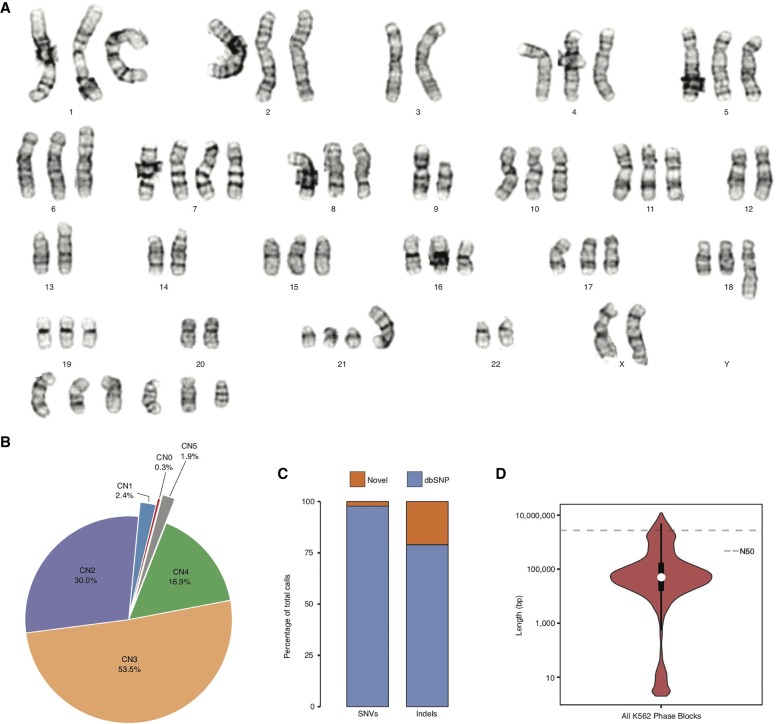
The K562 cell line exhibits pervasive aneuploidy (Fig. 2A). Analysis of 20 individual K562 cells using GTW banding showed that all cells demonstrated a near-triploid karyotype and are characterized by multiple structural abnormalities. The karyotype of our line of K562 cells is overall consistent (although not identical) with previously published karyotypes (Selden et al. 1983; Wu et al. 1995; Gribble et al. 2000; Naumann et al. 2001), suggesting that its near-triploid state arose during leukemogenesis or early in the establishment of the cell line. It also suggests that different K562 cell lines kept and passaged in different laboratories may exhibit some additional karyotypic differences. Although the karyotype for all chromosomes in our K562 cell line was supported by previous karyotype analyses, slight variations do exist (Supplemental Table S1) with Chromosomes 10, 12, and 21 showing the most variability.
Figure 2.
K562 ploidy and haplotypes. (A) Representative karyogram of K562 cells produced by GTW banding showing multiple numerical and structural chromosomal abnormalities and an overall near-triploid karyotype. ISCN 2013 description in relationship to a triploid karyotype [<3n>]: 53∼70<3n>,XX,-X or Y,-3,?dup(6)(p21p25),+7,?inv(7)(p13p22),add(7)(q32),-9,add(9)(p24),del(9)(p13),add(10)(q22),-13,add(13)(p11),-14,add(17)(p11.2)x2,add(18)(q23),-20,der(21)t(1;21)(q21;p11),-22,+4∼7mar[cp20]. (B) CN (i.e., ploidy) by percentage across the K562 genome. (C) Percentage of K562 SNVs and indels that are novel and known in dbSNP (Sherry et al. 2001). (D) Violin plot, with overlaid box plot, of phased haplotype block sizes (y-axis, log-scaled) in which the dashed line represents the N50 value (2,721,866 bp).
Identification of copy number (CN) by chromosome segments
We used read-depth analysis (Abyzov et al. 2011) to assign a CN, that is, ploidy to all chromosome segments at 10-kb resolution or entire chromosomes in the K562 genome (Fig. 1; Supplemental Table S2). We first calculated WGS coverage in 10-kb bins and plotted it against %GC content where five distinct clusters were clearly observed (Supplemental Fig. S2). Clusters were designated as corresponding to particular CNs based on the mean coverage of each cluster (Supplemental Methods). Such designations confirm that the triploid state is the most common in the K562 genome. The CNs assigned to all chromosome segments using this approach are consistent with array CGH (Supplemental Fig. S3; Supplemental Data) and also with previous CGH analyses (Gribble et al. 2000; Naumann et al. 2001) with minor differences on Chromosomes 7, 10, 11, and 20 (Supplemental Table S3). Although on a general level, the CNs identified based on read-depth analysis track the findings from karyotyping, read-depth analysis reveals the CNs of many chromosome segments that would not have been apparent from karyotyping alone (Supplemental Fig. S3; Supplemental Data; Supplemental Table S2). We see that 53.5% of the K562 genome has a baseline CN of three (consistent with the karyotype) (Fig. 2A), 16.9% CN of four, 1.9% CN of five, 2.4% CN of one, and only 30.0% has remained in a diploid state (Fig. 2B). In addition, two large regions (5.8 and 3.1 Mb in size) on Chromosome 9 (20,750,000–26,590,000 and 28,560,000–31,620,000, respectively) were lost entirely (Supplemental Table S2).
SNVs and indels
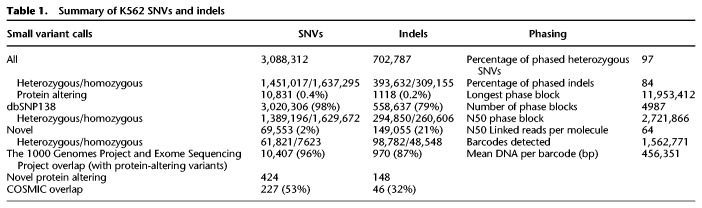
We identified SNVs and indels in the K562 genome. We assigned heterozygous allele frequencies to these variants by taking into account ploidy in which nonconventional frequencies are included (e.g., 0.33 and 0.67 in triploid regions; 0.25, 0.50, and 0.75 in tetraploid regions). Using this approach, we detected and genotyped a total of 3.09 million SNVs (1.45 million heterozygous, 1.64 million homozygous) and 0.70 million indels (0.39 million heterozygous, 0.31 million homozygous) (Table 1; Supplemental Data Set S1). There are 13,471 heterozygous SNVs and indels that have more than two haplotypes in aneuploid regions where CN is >2 (Supplemental Data Set S1). Furthermore, Chromosomes 3, 9, 13, 14, and X along with large stretches of Chromosomes 2, 10, 12, 17, 20, and 22 show loss of heterozygosity (LOH) (Fig. 1; Supplemental Table S4). Although a normal tissue sample corresponding to K562 is not available for comparative analysis, we overlapped these SNVs and indels with those in dbSNP138 (Sherry et al. 2001) and found the overlap to be 98% and 79%, respectively (Fig. 2C; Supplemental Data Set S1), suggesting an accumulation of a significant number of K562-specific SNVs and indels relative to germline variants present in the population. After filtering for protein-altering SNVs and indels in K562 that overlap with those identified from the 1000 Genomes Project or from the Exome Sequencing Project (http://evs.gs.washington.edu/EVS/), we found that 424 SNVs and 148 indels are private protein-altering (PPA) (Table 1; Supplemental Table S5). Furthermore, the overlap between the PPA variants and the Catalogue of Somatic Mutations in Cancer (COSMIC) is 53% and 31% for SNVs and indels, respectively (Supplemental Table S6). Eighteen genes that acquired PPA variants overlap with the Sanger Cancer Gene Census; canonical tumor suppressor genes and oncogenes such as RAD51B, TP53, PDGFRA, RABEP1, EPAS1, and WHISC1 are notably present among them (Supplemental Table S7).
Table 1.
Summary of K562 SNVs and indels
Haplotype phasing
We performed haplotype phasing for the K562 genome by performing 10x Genomics linked-read library preparation and sequencing (Zheng et al. 2016; Marks et al. 2018). This library was sequenced (2 × 151 bp) to 59× genome coverage. Post-sequencing quality-control analysis showed that 1.06 ng, or about 320 genome equivalents, of high molecular weight (HMW) K562 genomic DNA fragments (average fragment size = 59 kb, 95.3% >20 kb, 11.9% >100 kb) were partitioned into 1.56 million oil droplets for unique barcoding. Half of all reads come from HMW DNA molecules with at least 64 linked reads (N50 Linked Reads per Molecule or LPM) (Table 1). We estimate the actual physical coverage (CF) to be 191× (Supplemental Methods). Using Long Ranger (Marks et al. 2018), 1.41 million (97.2%) of heterozygous SNVs and 0.58 million (83.7%) of indels (previously identified) (Supplemental Data Set S1) were successfully phased into 4987 haplotype blocks (Fig. 1; Table 1; Supplemental Data Set S2). The longest is 11.95 Mb (N50 = 2.72 Mb) (Fig. 2D; Table 1; Supplemental Data Set S2); however, haplotype block lengths vary widely across different chromosomes (Supplemental Fig. S4; Fig. 1) with poorly phased regions corresponding to regions with LOH (Fig. 1; Supplemental Table S4; Supplemental Data Set S2).
Mega-haplotypes encompassing entire chromosome arms
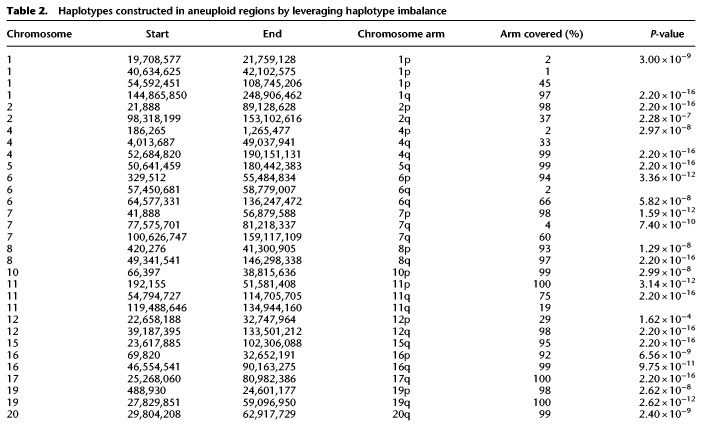
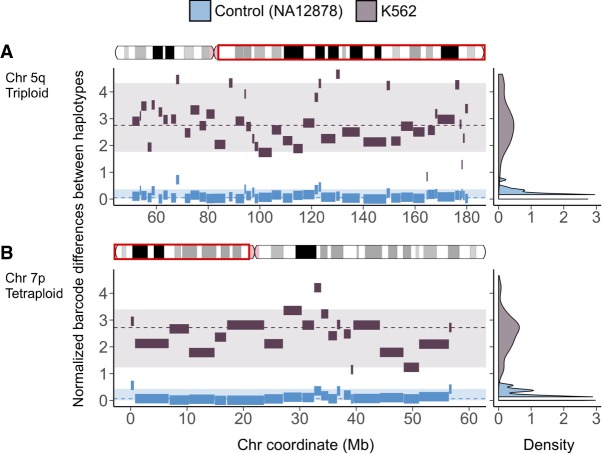
Leveraging the haplotype imbalance in aneuploid regions, we constructed mega-haplotypes (Table 2; Fig. 3; Supplemental Data), often encompassing entire K562 chromosome arms, by “stitching” the phased haplotype blocks obtained from Long Ranger (Supplemental Data Set S2) that contain ≥100 phased heterozygous SNVs derived from linked reads using a recently published method (Supplemental Methods; Bell et al. 2017). Using this approach, a total of 31 autosomal mega-haplotypes were constructed (Table 2; Supplemental Data), 15 of which encompass entire (or >95%) chromosome arms such as 19p, 19q, 10p, 7p, and 5q (Fig. 3). The average mega-haplotype is 50.7 Mb or roughly four times longer than the longest phased haplotype block from Long Ranger (Fig. 2D; Tables 1, 2; Supplemental Data Set S2). The longest mega-haplotype is approximately 137 Mb long (4q). In this approach, smaller phase blocks (less than 100 SNVs) from Long Ranger are not included in the mega-haplotype assembly. Thus, these mega-haplotypes (referred to as such hereafter) do not directly supplant the Long Ranger phase blocks (Supplemental Data Set S2) in terms of detailed local phasing information.
Table 2.
Haplotypes constructed in aneuploid regions by leveraging haplotype imbalance
Figure 3.
Mega-haplotypes of entire K562 chromosome arms: (x-axis) chromosome coordinate (Mb); (y-axis) difference in unique linked-read barcode counts between major and minor haplotypes, normalized for SNV density. Haplotype blocks from of normal control sample (NA12878) in blue and from K562 in dark gray. Density plots on the right reflect the distribution of the differences in haplotype-specific barcode counts for the control sample (blue) and K562 (dark gray). These density distributions are used for testing of significant difference (P < 0.001) using a one-sided t-test. Significant difference in haplotype-specific barcode counts indicates aneuploidy and haplotype imbalance. Haplotype blocks (with 100 or more phased SNVs) generated from Long Ranger (Supplemental Data Set S2) for the major and minor haplotypes were then “stitched” to mega-haplotypes encompassing the entire chromosome arm: (A) 5q (triploid); (B) 7p (tetraploid).
Identification and reconstruction of structural variants (SVs) from linked reads
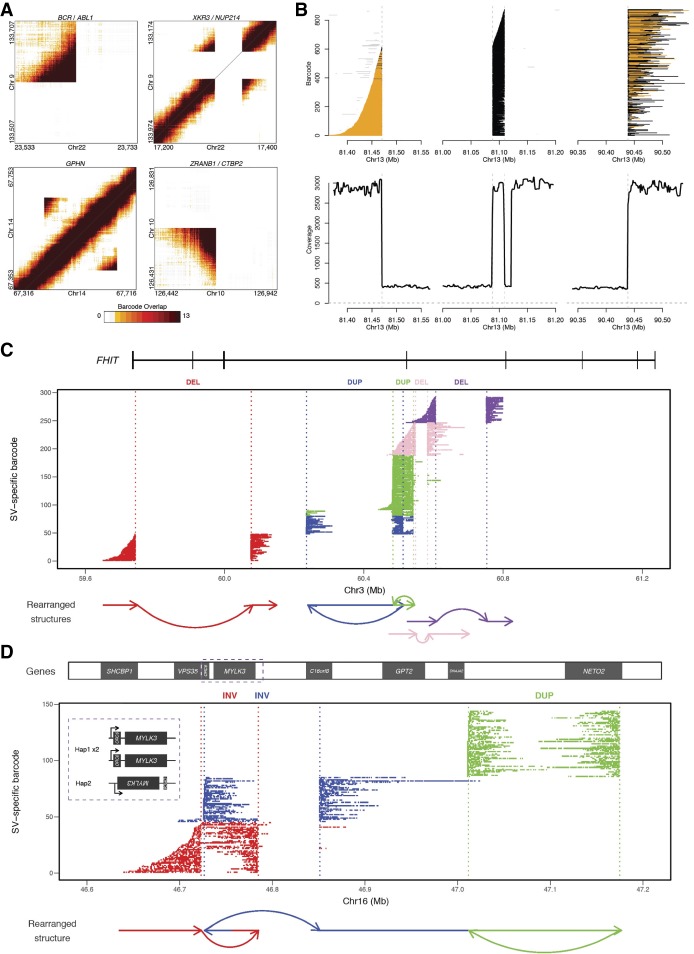
In addition to phasing, another use for the linked-read sequencing data is to identify breakpoints of large-scale SVs by searching for the discordant mapping of clusters of linked reads carrying the same barcodes. The identified SVs can then also be assigned to specific haplotypes if the breakpoint-supporting reads contain phased SNVs or indels (Zheng et al. 2016). Using this approach, which is also implemented by the Long Ranger software from 10x Genomics, we identified 186 large SVs >30 kb (98% phased) (Supplemental Data Set S3) and 3541 deletions between 50 bp and 30 kb (79% phased) (Supplemental Data Set S4). The large SVs include deletions, inversions, duplications, and inter- and intrachromosomal rearrangements (Supplemental Data Set S3; Fig. 4A). As expected, we detected the BCR/ABL1 gene fusion, a hallmark of K562, as one of the SV calls with highest quality score (Fig. 4A; Supplemental Data Set S3), along with two other known gene fusions in K562 (Engreitz et al. 2012): XKR3/NUP214 between Chromosomes 9 and 22 (Fig. 4A), and CDC25A/GRID1 between Chromosomes 3 and 10 (Supplemental Data Set S5; Supplemental Data).
Figure 4.
K562 SVs including large complex rearrangements resolved using linked-read sequencing. (A) Heat maps of overlapping barcodes for SVs in K562 resolved from linked-read sequencing using Long Ranger (Zheng et al. 2016; Marks et al. 2018). BCR/ABL1 translocation between Chromosomes 9 and 22. XKR3/NUP214 translocation between Chromosomes 9 and 22. Duplication within GPHN on Chromosome 14. Deletion that partially overlaps ZRANB1 and CTB2 on Chromosome 10. (B) Large complex rearrangement occurring on Chromosome 13 with informative reads from only one haplotype (region with loss of heterozygosity). Each line depicts a fragment inferred from linked reads based on clustering of identical barcodes (y-axis) using GROC-SVs (Spies et al. 2017). Abrupt endings (vertical dashed lines) of fragments indicate locations of breakpoints of this complex rearrangement. Fragments are phased locally with respect to surrounding SNVs (colored orange for same haplotype and black when no informative SNVs are found nearby). Gray lines indicate portions of fragments that do not support the current breakpoint. Fragments end abruptly at 81.47 Mb, indicating a breakpoint, picking up again at 81.09 Mb and continuing to 81.11 Mb where they end abruptly, then picking up again at 90.44 Mb. Coverage from 81.12 to 81.20 Mb are from reads with different sets of linked-read barcodes and thus are not part of this fragment set. (C,D) Complex rearrangements involving multiple haplotype-resolved SVs. Using gemtools (Greer et al. 2017), each SV is identified from linked reads grouped by identical barcodes (i.e., SV-specific barcodes, y-axis) indicative of single HMW DNA molecules (depicted by each row) that span the breakpoints. SVs are represented in different colors. The x-axis shows the hg19 genomic coordinate. Dotted lines represent individual breakpoints with schematic diagram of the rearranged structures drawn below the plot. (C) Multiple SVs within FHIT on 3p14.2. Deletion (DEL; red) (59.74–60.08 Mb) results in the loss of multiple exons. Two overlapping duplications (DUP; blue and green) in cis orientation (same allele of FHIT) indicated by the presence of HMW molecules spanning both DUPs. Two adjacent DELs (pink and purple) in trans (different alleles of FHIT), indicated by the absence of shared SV-specific barcodes for the HWM molecules spanning each DEL. SV haplotypes analyzed using SV-specific barcodes (not enough informative SNVs due to LOH). (D) Complex, intrachromosomal rearrangement spanning approximately 0.5 Mb on 16q11.2 and 16q12.1 that involve two overlapping inversions, 63 kb (red) and 125 kb (blue), and a 163-kb tandem duplication (green). This rearrangement resides on the nonduplicated haplotype of this triploid region. ORC6 is located entirely within the 63-kb inversion on 16q11.2 and is “deleted” by the left breakpoint of the 125-kb inversion, which also inverts MYLK3. C16orf8 on the same haplotype is also partially “deleted” by the 125-kb inversion (blue); NETO2 is duplicated by the 163-kb tandem duplication (green). (Inset) MYLK3 and ORC6 show allele-specific expression (Supplemental Table S12). MYLK3 is only expressed from this rearranged allele (Haplotype 2); ORC6 is expressed from the non-rearranged “diploid” allele (Haplotype 1).
We also leveraged the long-range information derived from the linked reads to identify, assemble, and reconstruct SV-spanning breakpoints (including those of large-scale complex rearrangements) in the K562 genome using the recently established method Genome-wide Reconstruction of Complex Structural Variants (GROC-SVs) (Spies et al. 2017). In this method, long DNA fragments that span breakpoints are statistically inferred and refined by quantifying barcode similarity between pairs of genomic regions, similar to Long Ranger (Marks et al. 2018). Sequence reconstruction is then performed by assembling the relevant linked reads around the identified breakpoints from which complex SVs are then automatically reconstructed. The breakpoints that have supporting evidence from the K562 3-kb mate-pair data set (Supplemental Methods) were determined as high-confidence events (Supplemental Data Set S5). GROC-SVs identified a total of 161 high-confidence breakpoints including 12 interchromosomal events (Figs. 1, 4B; Supplemental Data Set S5); each event is accompanied with visualization (Supplemental Data); and 138 of the breakpoints were successfully sequence-assembled with nucleotide-level resolution of breakpoints as well the exact sequence in the cases where nucleotides have been added or deleted (Supplemental Data Set S5). A notable example of assembly by GROC-SVs is a complex intrachromosomal rearrangement on Chromosome 13 (Fig. 4B).
Using gemtools as described (Greer et al. 2017), we identified phased structural rearrangements (multiple deletions and tandem duplications) within the tumor suppressor gene FHIT on 3p14.2 (Fig. 4C; Waters et al. 2014). Because K562 exhibits LOH on Chromosome 3, the SVs within FHIT were phased using linked-read barcodes instead of heterozygous SNVs. The hemizygous deletion between 59.74 and 60.08 Mb of 3p14.2 results in the loss of FHIT exons 6, 7, and 8. For the two phased tandem duplications on the same allele, one is intronic, and the other duplicates exon 5 (Fig. 4C). The two deletions downstream from the phased duplications are on two different alleles of FHIT. Another allele-specific, complex, intrachromosomal rearrangement in K562 spans ∼0.5 Mb on 16q11.2 and 16q12.1 (Fig. 4D), involving two overlapping inversions (62 and 125 kb) and a tandem duplication (163 kb). These events affect ORC6, MYLK3, RHBDF1 (previously known as C16orf8), and NETO2, which has recently been identified as a cancer marker gene (Oparina et al. 2012; Hu et al. 2015). This rearrangement resides on the nonduplicated haplotype of this triploid region. ORC6 is located entirely within the more centromeric inversion of this locus on 16q11.2 and is “deleted” by the left breakpoint of the more telemetric inversion, which also “deletes” RHBDF1 and inverts MYLK3, possibly disrupting its promoter region or proximal enhancers or disconnecting MYLK3 from their regulation (Fig. 4D).
Small-scale complex SVs from deep-coverage WGS
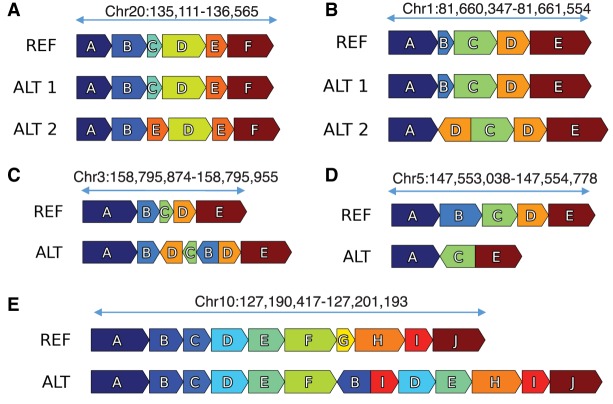
Small-scale complex SVs (Fig. 5A–E) as well as noncomplex SVs were identified using a novel algorithm called Automated Reconstruction of Complex Structural Variants (ARC-SV) (Arthur et al. 2018) from deep-coverage WGS data (Supplemental Data Set S6; Supplemental Data). These small-scale complex SVs are defined as genomic rearrangements with multiple breakpoints that cannot be explained by one well-defined (noncomplex) SV type such as deletions, insertions, tandem duplications, or inversions. After filtering out SVs <50 bp or with breakpoints that reside in simple repeats, low complexity regions, satellite repeats, or segmental duplications, we identified 122 complex SVs (accompanied with schematic visualizations), 2235 deletions, 320 tandem duplications, and 6 inversions (Supplemental Data Set S6). Examples of complex SVs include dispersed duplications in which duplicated sequences are inserted elsewhere in the genome in a nontandem fashion (Fig. 5A). These dispersed duplications sometimes involve inversions of the inserted sequence and deletions at the insertion site (Fig. 5B,C). Other examples include inversions flanked on one or both sides by deletions (Fig. 5D), duplications that involve multiple nonexact copies, as well as deletion, inversion, and multiple duplications residing at the same locus (Fig. 5E). Eight of 10 breakpoints from five complex SVs were successfully validated by PCR and Sanger sequencing (Supplemental Table S8).
Figure 5.
Small-scale complex SVs in K562 resolved using ARC-SV. Examples of small-scale complex SVs resolved using ARC-SV (Arthur et al. 2018) from the K562 WGS data set. (A) Deletion of Block C and duplication of Block E between Blocks B and D on Chromosome 20 (135,111–136,565). This variant has been validated by PCR. (B) Deletion of Block B and inverted duplication of Block D between Blocks A and C on Chromosome 1 (81,660,347–81,661,554). (C) Duplication and inversion of Blocks B, C, and D between Blocks B and D on Chromosome 3 (158,795,874–158,795,955) overlapping IQCJ-SCHIP1. (D) Inversion of Block C flanked by deletions of Blocks C and D on Chromosome 5 (147,553,038–147,554,778) inside SPINK14 (coding for a serine peptidase inhibitor). (E) Deletion of Block G, duplications of blocks I, D, and E, and inverted duplication of Block B between Blocks F and H on Chromosome 10 (127,190,417–127,201,193).
SVs from mate-pair sequencing analysis
To increase the sensitivity of detecting medium-sized SVs (1–100 kb) in K562, we constructed a 3-kb mate-pair library and sequenced (2 × 151 bp) to 6.9× nonduplicate coverage. The sequence coverage (CR) of each 3-kb insert is 302 bp or 10%, which translates to a physical coverage (CF) of 68.5×. From the mate-pair library, SVs (deletions, inversions, and tandem duplications) were identified by clustering discordant read-pairs and split-reads using LUMPY (Layer et al. 2014). Only SVs that have both discordant read-pair and split-read support were retained. Overall, we identified 270 deletions, 35 inversions, and 124 tandem duplications using this approach (Supplemental Data Set S7). Approximately 83% of these SVs are between 1 and 10 kb, and 88% are between 1 and 100 kb (Supplemental Data Set S7). Twelve deletions and five tandem duplications were randomly selected for PCR and Sanger sequencing validation (Supplemental Table S8). The validation rates were 83% and 80%, respectively.
Noncomplex SVs from deep-coverage WGS
Noncomplex SVs (deletions, inversions, insertions, and tandem duplications) in K562 were called from deep-coverage WGS data using a combination of established methods, namely Pindel (Ye et al. 2009), BreakDancer (Chen et al. 2009), and BreakSeq (Lam et al. 2010). These SVs were combined with those of the same SV type that were identified using ARC-SV, LUMPY, and Long Ranger, in which SVs (n = 2665) with support from multiple methods by ≥50% reciprocal overlap were merged. Through this combination of methods, a total of 9082 noncomplex SVs were identified in the K562 genome, including 5490 deletions, 531 duplications, 436 inversions, and 2602 insertions (Supplemental Data). (We note that only BreakDancer [Chen et al. 2009] was designed to call insertions.) Consistent with previous analyses (e.g., Lam et al. 2012), deletions show the highest number of concordant calls across the various methods compared to duplications and inversions (Supplemental Fig. S5; Supplemental Data). Eighteen deletions (>1 kb) and 18 tandem duplications, both with split-read support, were randomly chosen for experimental validation using PCR and Sanger sequencing. The validation rates were 89% and 72%, respectively (Supplemental Table S8).
LINE-1 and Alu insertions
We identified nonreference LINE-1 and Alu retrotransposon insertions (REIs) in the K562 genome from our deep-coverage short-insert WGS data using a modified RetroSeq (Keane et al. 2013) approach (Supplemental Methods). Nonreference REIs were identified from paired-end reads that have one of the paired reads mapping to the human reference genome and the other read mapping to either the Alu or LINE-1 consensus sequence in a full or split-read fashion (Methods). We identified 1147 nonreference Alu insertions and 85 nonreference LINE-1 insertions in K562 (Supplemental Table S9; Fig. 1). Nine Alu and 10 LINE-1 insertions with split-read support were randomly chosen for validation using PCR and Sanger sequencing. The validation rates were 88% and 100%, respectively (Supplemental Table S10).
Allele-specific gene expression
Integrating CN information (i.e., allele frequencies) of the heterozygous SNVs (Supplemental Data Set S1), we reanalyzed two replicates of ENCODE poly(A)-mRNA RNA-seq data to identify allele-specific gene expression in K562. We identified 5053 and 5149 genes that show allele-specific expression (P < 0.05) in replicates one and two, respectively (Fig. 1; Supplemental Table S11). We also identified 2342 and 2176 genes that would have been falsely identified to have allele-specific expression and 1641 and 1710 genes that would not have been identified to have allele-specific expression in replicates one and two, respectively, if the allele frequencies of heterozygous SNVs in aneuploid regions were not taken into consideration (Supplemental Table S12).
Allele-specific DNA methylation
By integrating CN and phase information of heterozygous SNVs of K562, we identified 110 CpG islands (CGIs) that exhibit allele-specific DNA methylation (Fig. 1; Supplemental Table S13). We obtained K562 whole-genome bisulfite sequencing (WGBS) reads from ENCODE (Sloan et al. 2016) and aligned the reads to hg19 using Bismark (Krueger and Andrews 2011), in which 76.9% of reads were uniquely mapped and 26.2% of cytosines were methylated in a CpG context (Supplemental Methods). We then used reads that overlap both phased heterozygous SNVs (Supplemental Data Set S2) and CpGs to phase the methylated and unmethylated CpGs to their respective haplotypes. We then grouped the phased individual CpGs into CGIs. Fisher's exact test (taking the CN of a given CGI locus into account) was used to evaluate allele-specific methylation (P < 0.05), and significant results were selected using a target false discovery rate of 10%. Of these 110 CGIs, 35 reside within promoter regions (here defined as 1 kb upstream of a gene), 83 are intragenic, and 28 lie within 1 kb downstream from 113 different genes. The following six genes are within 1 kb of a differentially methylated CGI and overlap with the Sanger Cancer Gene Census: ABL1, AXIN2, CCND1, HOXD11, KDR, and PRDM16.
Allele-specific CRISPR targets
We identified a total of 28,511 targets in the K562 genome suitable for allele-specific CRISPR targeting (Fig. 1; Supplemental Table S14). Sequences (including reverse complement) of phased variants that differ by more than 1 bp between the alleles were extracted to find all possible CRISPR targets by searching for the pattern [G, C, or A]N20GG. Using a selection method previously described and validated (Sunagawa et al. 2016), only conserved high-quality targets were retained (Supplemental Methods). Of the 28,511 allele-specific CRISPR target sites, 15,488 are within an annotated protein-coding or noncoding RNA transcript, 705 within an exon, and 13 targets are within an experimentally validated enhancer (Supplemental Table S14; Visel et al. 2007).
Genomic structural context provides insight into regulatory complexity
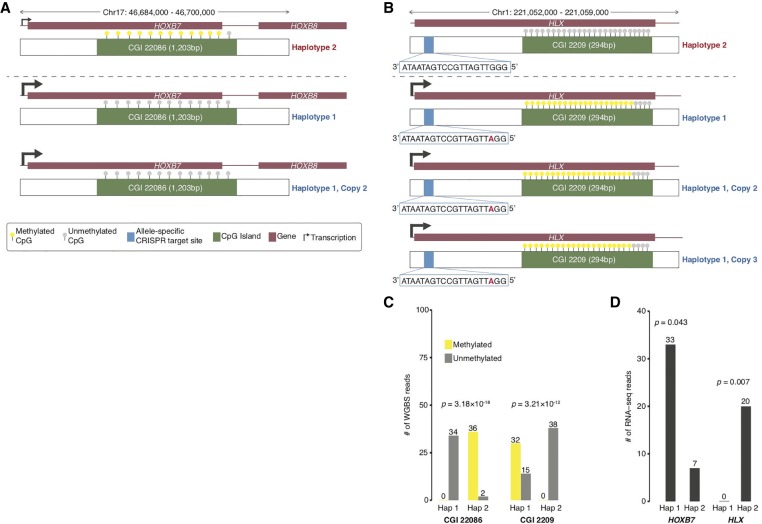
We show examples of how deeper insights into gene regulation and regulatory complexity can be obtained by integrating genomic structural contexts with functional genomics and epigenomics data (Fig. 6A–D). One example is the allele-specific RNA expression and allele-specific DNA methylation in K562 at the HOXB7 locus on Chromosome 17 (Fig. 6A). By incorporating the genomic context of HOXB7 in K562, we see that HOXB7 exhibits highly preferential RNA expression from the two copies of Haplotype 1 (P = 0.007) in which the CGI near its promoter is completely unmethylated (P = 3.18 × 10−18) (Fig. 6A,C). The second example is allele-specific RNA expression and allele-specific DNA methylation of the HLX gene in K562 (Fig. 6B). The HLX locus on Chromosome 1 is tetraploid, and we see that HLX is only expressed from Haplotype 1, which has three copies, and is not expressed in Haplotype 2 (P = 0.043) (Fig. 6B,D). The CGI of the HLX locus is unmethylated in Haplotype 2 but highly methylated on Haplotype 1 (P = 5.14 × 10−15) (Fig. 6B,C). There is also an allele-specific CRISPR targeting site for both haplotypes within HLX (Fig. 6B). In addition, we performed Pearson correlation analysis between our deep-coverage K562 WGS data and K562 POLR2A ChIP-seq data (previously released on the ENCODE data portal) to determine whether changes in K562 genome CN or ploidy affected binding of the polymerase molecule to genomic DNA in a large-scale fashion (Supplemental Fig. S6). The two sets of data are very well correlated (r = 0.51, P < 2.2 × 10−16) suggesting that RNA polymerase activity is generally influenced by ploidy in the K562 genome. In addition, we also correlated the K562 POLR2A ChIP-seq data with the FPKM values from four independent K562 poly(A) RNA-seq experiments (also previously released on the ENCODE portal) and find that these data sets are also very well correlated consistently (r = 0.46, P < 2.2 × 10−16; r = 0.58, P < 2.2 × 10−16; r = 0.47, P < 2.2 × 10−16; r = 0.46, P < 2.2 × 10−16) (Supplemental Fig. S7A–D).
Figure 6.
Genomic structural contexts provide insights into regulatory complexity. (A) Chr 17: 46,687,000–46,700,000 locus (triploid in K562) containing HOXB7 and HOXB8 and CpG Island (CGI) 22086 (1203 bp) where phased Haplotype 1 has two copies and Haplotype 2 has one copy. Allele-specific expression of HOXB7 from Haplotype 1. CpGs in CGI 22086 are unmethylated in Haplotype 1 and methylated in Haplotype 2. (B) Chr 1: 221,052,000–221,059,000 locus (tetraploid in K562) containing HLX and CGI 2209 (294 bp) where phased Haplotype 1 has three copies and Haplotype 2 has one copy. Allele-specific expression of HLX from Haplotype 1. CpGs in CGI 2209 are unmethylated in Haplotype 2 and highly methylated in Haplotype 1. Allele-specific CRISPR targeting site 797 bp inside the 5′ end of the HLX for both Haplotypes. (C) Number of methylated and unmethylated phased WGBS reads for Haplotypes 1 and 2 in CGI 22086 and CGI 2209 in which both CGIs exhibit allele-specific DNA methylation. (D) Number of RNA-seq reads for Haplotypes 1 and 2 of HLX and HOXB7 in which both genes exhibit allele-specific RNA expression.
Furthermore, we also find allele-specific RNA expression for the rearranged copy of MYLK3 (P < 1.93 × 10−17) and the normal, non-rearranged copies (CN = 2) of ORC6 (P < 1.58 × 10−8) in which expression from rearranged allele (CN = 1) of ORC6 is “depleted” in K562 (Supplemental Table S11; Fig. 4C,D). These observations made by integrating our K562 linked-read data and ENCODE RNA expression data provide novel insights into gene regulatory mechanisms in terms of ectopic expression and dosage compensation, which also raises important questions regarding the history of the K562 cell line in terms of mutations, selective pressures, and adaption.
Discussion
Despite its wide usage and impact on biomedical research, K562's genomic sequence and structural features have never been comprehensively characterized, beyond its karyotype (Selden et al. 1983; Wu et al. 1995; Gribble et al. 2000; Naumann et al. 2001) and SNPs called from 30×-coverage WGS but without taking aneuploidy or CN into consideration (Cavalli et al. 2016). Analysis, integration, and interpretation of the extensive collection of functional genomics and epigenomics data sets for K562 had so far relied solely on the human reference genome. Here, we present the first detailed and comprehensive characterization of the K562 genome. In summary, by performing deep-coverage short-insert WGS, 3-kb-insert mate-pair sequencing, deep-coverage linked reads sequencing, array CGH, karyotyping, and integrating a compendium of novel and established analysis methods (Supplemental Fig. S1A), we produced a comprehensive spectrum of genomic structural features (Fig. 1) for K562 that includes SNVs (Supplemental Data Set S1), indels (Supplemental Data Set S1), ploidy by chromosome segments at 10-kb resolution (Supplemental Table S2), phased haplotypes (Supplemental Data Set S2; Supplemental Data)—often of entire chromosome arms (Table 2; Supplemental Data)—phased CRISPR targets (Supplemental Table S14), nonreference REIs (Supplemental Table S9), and SVs (Supplemental Data) including deletions, duplications, and inversions, and complex SVs (Supplemental Data Sets S6, S7). Many SVs were also phased, assembled, and experimentally verified (Supplemental Data Sets S2–S5; Supplemental Tables S8, S10). Of the 3,784,863 variants that were haplotype-phased in the K562 genome (Supplemental Data Sets S2–S5), 3,088,185 (81.6%) are SNVs; 692,998 (18.31%) are indels; 3451 are deletion SVs (51 bp to 30 kb; 0.1%); and 229 are large SVs. We used the hg19 genome build for this study in order to keep the data sets consistent for analysis and integration since the data generation phase started before the GRCh38 genome release. Due to the genome-wide nature of the study, realigning the sequencing reads to GRCh38 will not significantly affect the results.
Pervasive aneuploidy is a characteristic of many cancers. Previous studies have confirmed the near-triploid karyotype of K562 (Selden et al. 1983; Wu et al. 1995; Gribble et al. 2000; Naumann et al. 2001). In our analysis, however, we also found considerable portions of the K562 genome to be much more varied than what had previously been reported. This is because by leveraging deep-coverage WGS, the CN across different chromosome segments (Fig. 1; Supplemental Table S2; Supplemental Data), as determined by our read-depth analysis, is of much higher resolution than karyotyping. Furthermore, the identified chromosome segments with aneuploidy (CN > 2) and orthogonally supported by karyotyping and array CGH were further validated (Supplemental Fig. S3; Supplemental Data), also orthogonally, from a statistical approach in which significant differences in unique linked-read barcode counts between the major and minor haplotypes were determined using a one-sided t-test (P < 0.001) (Fig. 3; Supplemental Data; Bell et al. 2017). In addition, it has to be taken into consideration that for a widely used cell line with decades of history such as K562, additional genome variation is expected (Supplemental Discussion).
Sensitive and accurate identification of SNVs and indels requires relatively deep WGS coverage (>33× and >60×, respectively) (Bentley et al. 2008; Fang et al. 2014). From our greater than 70× nonduplicate coverage WGS data, we identified large numbers of SNVs and indels that we could subsequently correct for their allele frequencies according to ploidy. In addition to being essential for correct haplotype identification, these ploidy-corrected variants are also needed for functional genomics or epigenomics analyses such as the determination of allele-specific gene expression or of allele-specific transcription factor binding in K562 (Cavalli et al. 2016). From RNA-seq or ChIP-seq data analysis, a statistically significant increase in transcription or transcription factor binding signal in one allele compared to the other at a heterozygous locus, may be identified as a case of allele-specific expression or allele-specific transcription-factor binding, which usually suggests allele-specific gene regulation at this locus. However, if aneuploidy can be taken into consideration and the signals normalized by ploidy, the case identified might be a result of increased CN rather than the preferential activation of one allele over the other on the epigenomic level. Indeed, in our reanalysis of two replicates of ENCODE K562 RNA-seq data, we identified 2359 and 2643 genes that would have been falsely identified to have allele-specific expression in addition to 1808 and 2063 genes that would not have been identified to have allele-specific expression in replicates one and two, respectively, if ploidy was not taken into consideration (Supplemental Table S12).
It was previously shown that integrating orthogonal methods and signals improves SV-calling sensitivity and accuracy (Layer et al. 2014; Mohiyuddin et al. 2015). Here, we combined deep-coverage short-insert WGS, mate-pair sequencing, linked-read sequencing, and several SV-calling methods to identify many noncomplex SVs. To obtain the union set of noncomplex SV calls from the various methods, the SVs identified by multiple methods were merged and indicated accordingly (Supplemental Data). For deletions (Supplemental Fig. S5A), we see strong overlap for the various methods, but this overlap is less pronounced for duplications (Supplemental Fig. S5B) and inversions (Supplemental Fig. S5C). This is consistent with previous analysis (Lam et al. 2012) as inversions and duplications are more difficult in principle to accurately resolve (Lin et al. 2015; Sudmant et al. 2015). We also expect the detection of many SVs to be method-specific, since each method is designed to utilize different types of signals and also optimized to identify different classes of SVs (Pabinger et al. 2014; Lin et al. 2015). Again, if particular SVs are of interest for follow-up studies, they should first be experimentally validated.
The complex rearrangements identified by using ARC-SV from short-insert WGS (Fig. 5A–E; Supplemental Data Set S6; Supplemental Data) and by using GROC-SVs from linked reads (Fig. 4B; Supplemental Data Set S5; Supplemental Data) are classes of SVs that could not be easily identified and automatically reconstructed using previously existing methods. The small-scale complex SVs that were identified by using ARC-SV and experimentally validated (Fig. 5; Supplemental Table S8; Supplemental Data) describe a subtle class of complex rearrangements in cancer genomes that have been relatively understudied (Perry et al. 2008; Quinlan and Hall 2012; Collins et al. 2017). Detecting and automatically reconstructing these small-scale complex SVs, especially in a “hay” of canonical SVs and in highly rearranged cancer genomes, has remained an unsolved problem for many years. In other words, our results reveal a class of previously overlooked complex SVs in cancer that can now be identified from standard short-insert WGS data and elucidated further. They have clear implications for the conventional models of cancer evolution which often assume gradual, step-by-step mutations; however, these complex SVs support a form of punctuated genome evolution (Davis et al. 2017). A major unsolved question still is how complex SVs arise mechanistically for which there are general models: template switching during replication (Lee et al. 2007; Hastings et al. 2009) and chromothripsis (Stephens et al. 2011). Furthermore, the functional consequences of these small-scale complex SVs are also unknown. These important questions remain unsolved mainly due to the lack of data and examples. It is possible that this mutational complexity contributes to genome innovation, at least in cancer, or is just a curious sideshow (Quinlan and Hall 2012). Only the accumulation of such examples and data will allow researchers in fields such as cancer evolution to begin to address these important questions.
Before the existence of linked-read sequencing, haplotype phasing and resolving large SVs (>30 kb) relied heavily on fosmid libraries (Kitzman et al. 2011; Williams et al. 2012; Adey et al. 2013; Cao et al. 2015; Snyder et al. 2015), which were laborious, costly, time consuming, and much less efficient. Using linked-read sequencing and gemtools (Greer et al. 2017), we phased and resolved complex SVs that are especially compelling on 3p14.2 (within the tumor suppressor gene FHIT) and on 16q11.2 and 16q12.1 of the K562 genome (Fig. 4C,D; Supplemental Discussion).
Data generated from this comprehensive whole-genome analysis of K562 is available through the ENCODE portal (Supplemental Fig. S1B,C; Sloan et al. 2016). We envision that this analysis will serve as a valuable resource for further understanding the vast troves of ENCODE data available for K562, such as determining whether a potential or known regulatory sequence element has been altered by SNVs or SNPs, indels, retrotransposon insertions, a gain or loss of copies of that given element, or allele-specific regulation. As additional examples of how integrating genomic context can yield further understanding of existing ENCODE data, we showed, as examples, the complex gene regulatory scenarios uncovered at the HOXB7 and HLX loci in K562 (Fig. 6; Supplemental Discussion). In addition, we also observed that the K562 POLR2A ChIP-seq signals in both replicates are very well correlated with poly(A) RNA-seq signal and with WGS coverage, suggesting an association between polymerase binding and active transcription and between polymerase binding and ploidy (Supplemental Figs. S6, S7).
Our work here serves to guide future studies that utilize the K562 “workhorse” cell line, such as CRISPR screens where knowledge of the sequence variants can extend or modify the number of editing targets (Supplemental Table S13) and knowledge of aberrant CN will allow for much more confident data interpretation. To give an example, in a recent study that uses CRISPRi to screen and elucidate the function of long noncoding RNAs in human cells, of the seven cell types studied, the number of gRNA hits varied considerably among the various cell types, with 89.4% of hits unique to only one cell type and none in more than five cell types (Liu et al. 2017). Although a large portion of the phenomenon are very likely explained by cell-specific effects, it is still quite possible that many of the gRNA hit differences were the result of differences in genome sequence or ploidy. Our list of allele-specific CRISPR targets (Supplemental Table S13) will allow for the discernment between these two potential reasons for differences in CRISPR effects and should be particularly valuable for future large-scale CRISPR screens that utilize K562. Lastly, this study also serves as a technical example for the advanced, integrated, and comprehensive analyses of other heavily utilized cell lines and genomes in biomedical research such as HepG2.
Methods
Genomic DNA extraction and karyotyping
K562 cells were obtained from the Stanford ENCODE Product Center (NHGRI Project 1U54HG006996). Genomic DNA was extracted using the QIAGEN DNeasy Blood and Tissue Kit (Catalog No. 69504) and quantified using the Qubit dsDNA HS Assay Kit (Invitrogen). DNA was then verified to be pure (OD260/280 > 1.8; OD260/230 > 1.5) using NanoDrop (Thermo Fisher Scientific) and high molecular weight (mean >30 kb) using field-inversion gel electrophoresis on the Pippin Pulse System (Sage Science). K562 cells were sent to the Cytogenetics Laboratory at Stanford University (http://cytogenetics.stanford.edu) for karyotyping where 20 metaphase cells were analyzed using GTW banding.
Data generation and analysis
Illumina short-insert WGS, mate-pair WGS, 10x Genomics linked-read WGS, and array CGH were all performed using standard experimental techniques. Genomic sequence and structural features of the K562 genome were plotted using Circos (Krzywinski et al. 2009). For full descriptions of experimental and computational procedures (including analysis code), see Supplemental Methods.
Experimental validation
Random sets of SV calls from short-insert WGS and mate-pair sequencing were selected from PCR validation. These sets include complex SVs (from ARC-SV), deletions (>1 kb), and tandem duplications. PCR primers were designed such that the amplicons span the breakpoints and produce products between 200 and 500 bp. In the case of complex SVs, pairs of PCR primers were designed to validate multiple breakpoints. Ten Alu and 10 LINE-1 events with split-read support were randomly chosen for validation. PCR primers were designed to amplify products ranging from 65 to 150 bp in which one primer anneals to unique sequence and the other anneals to the retrotransposon sequence. All PCR amplicons were gel purified and Sanger sequenced.
Data access
All resources generated for K562 in this study are listed with detailed descriptions in Supplemental Figure S1. All data (raw sequences, processed data, and Supplemental Data Sets S1–S7) generated in this study have been submitted to ENCODE (https://www.encodeproject.org) under accession number ENCBS806UYV. Analysis code is available as Supplemental Material. Accession numbers for individual data files such as Supplemental Data Sets S1–S7 and experiments are listed in Supplemental Figure S1B,C.
Supplementary Material
Acknowledgments
We thank Ms. Aditi Narayanan, Dr. Idan Gabdank, Mr. Nathaniel Watson, Dr. Carrie Davis, Ms. Kathrina Onate, Mr. Zachary Myers, Ms. Khine Z. Lin, and Dr. Cricket Sloan for assistance with data release on the ENCODE portal. We thank Dr. Athena Cherry and the Stanford Cytogenetics Laboratory for karyotype analysis and Ms. Arineh Khechaduri for performing genomic DNA preparation. We thank Dr. Minyi Shi for providing K562 cells. A.E.U. is a Tashia and John Morgridge Faculty Scholar of the Stanford Child Health Research Institute. A.E.U. was supported by National Institutes of Health (NIH), National Human Genome Research Institute (NHGRI) grant P50-HG007735 and the Stanford Medicine Faculty Innovation Program. B.Z. was additionally supported by NIH training grant T32-HL110952. W.H.W. received support from NIH/NHGRI grants R01-HG007834 and P50-HG007735. J.G.A. received funding from NIH training grant T32-GM096982 and National Science Foundation Graduate Fellowship DGE-114747. A.A. was funded by NIH grant U24CA220242. H.P.J. was funded by the Intermountain Healthcare Research Award and NIH/NHGRI grants R01-HG006137 and P01-HG00205.
Author contributions: B.Z. and A.E.U. conceived and designed the study. B.Z., R.P., N.B.-E., M.S.H., and R.R.H. performed experiments. B.Z., S.S.H., S.U.G., J.M.B., J.G.A., N.S., X. Zhu, X. Zhang, S.B., G.S., D.P., and A.A. performed data analysis. A.E.U. contributed to data interpretation. H.P.J., W.H.W., and A.E.U. contributed resources and supervised the study. B.Z., S.S.H., and A.E.U. wrote the manuscript with input from S.U.G., J.M.B., J.G.A., N.S., D.P., and A.A.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.234948.118.
References
- Abyzov A, Urban AE, Snyder M, Gerstein M. 2011. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21: 974–984. 10.1101/gr.114876.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. 2016. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167: 1867–1882.e21. 10.1016/j.cell.2016.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adey A, Burton JN, Kitzman JO, Hiatt JB, Lewis AP, Martin BK, Qiu R, Lee C, Shendure J. 2013. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature 500: 207–211. 10.1038/nature12064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo JD, Jourdain AA, Calvo SE, Ballarano CA, Doench JG, Root DE, Mootha VK. 2016. A genome-wide CRISPR death screen identifies genes essential for oxidative phosphorylation. Cell Metab 24: 875–885. 10.1016/j.cmet.2016.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JG, Chen X, Zhou B, Urban AE, Wong WH. 2018. Detection of complex structural variation from paired-end sequencing data. bioRxiv 10.1101/200170 [DOI]
- Bell JM, Lau BT, Greer SU, Wood-Bouwens C, Xia LC, Connolly ID, Gephart MH, Ji HP. 2017. Chromosome-scale mega-haplotypes enable digital karyotyping of cancer aneuploidy. Nucleic Acids Res 45: e162 10.1093/nar/gkx712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59. 10.1038/nature07517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MO, Hirano N. 2014. Human cell-based artificial antigen-presenting cells for cancer immunotherapy. Immunol Rev 257: 191–209. 10.1111/imr.12129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Wu H, Luo R, Huang S, Sun Y, Tong X, Xie Y, Liu B, Yang H, Zheng H, et al. 2015. De novo assembly of a haplotype-resolved human genome. Nat Biotechnol 33: 617–622. 10.1038/nbt.3200 [DOI] [PubMed] [Google Scholar]
- Cavalli M, Pan G, Nord H, Wallerman O, Wallén Arzt E, Berggren O, Elvers I, Eloranta ML, Rönnblom L, Lindblad Toh K, et al. 2016. Allele-specific transcription factor binding to common and rare variants associated with disease and gene expression. Hum Genet 135: 485–497. 10.1007/s00439-016-1654-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. 2009. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 6: 677–681. 10.1038/nmeth.1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Redin CE, Hanscom C, Antolik C, Stone MR, Glessner JT, Mason T, Pregno G, Dorrani N, et al. 2017. Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol 18: 36 10.1186/s13059-017-1158-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A, Gao R, Navin N. 2017. Tumor evolution: linear, branching, neutral or punctuated? Biochim Biophys Acta 1867: 151–161. 10.1016/j.bbcan.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler HG, Matsuo Y, MacLeod RAF. 2004. Malignant hematopoietic cell lines: in vitro models for the study of erythroleukemia. Leuk Res 28: 1243–1251. 10.1016/j.leukres.2004.03.022 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Agarwala V, Mirny LA. 2012. Three-dimensional genome architecture influences partner selection for chromosomal translocations in human disease. PLoS One 7: e44196 10.1371/journal.pone.0044196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Wu Y, Narzisi G, O'Rawe JA, Barrón LTJ, Rosenbaum J, Ronemus M, Iossifov I, Schatz MC, Lyon GJ. 2014. Reducing INDEL calling errors in whole genome and exome sequencing data. Genome Med 6: 89 10.1186/s13073-014-0089-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer SU, Nadauld LD, Lau BT, Chen J, Wood-Bouwens C, Ford JM, Kuo CJ, Ji HP. 2017. Linked read sequencing resolves complex genomic rearrangements in gastric cancer metastases. Genome Med 9: 57 10.1186/s13073-017-0447-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble SM, Roberts I, Grace C, Andrews KM, Green AR, Nacheva EP. 2000. Cytogenetics of the chronic myeloid leukemia-derived cell line K562. Cancer Genet Cytogenet 118: 1–8. 10.1016/S0165-4608(99)00169-7 [DOI] [PubMed] [Google Scholar]
- Grzanka A, Grzanka D, Orlikowska M. 2003. Cytoskeletal reorganization during process of apoptosis induced by cytostatic drugs in K-562 and HL-60 leukemia cell lines. Biochem Pharmacol 66: 1611–1617. 10.1016/S0006-2952(03)00532-X [DOI] [PubMed] [Google Scholar]
- Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC. 2017. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol 35: 463–474. 10.1038/nbt.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327 10.1371/journal.pgen.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Chen HY, Cai J, Yang GZ, Feng D, Zhai YX, Gong H, Qi CY, Zhang Y, Fu H, et al. 2015. Upregulation of NETO2 expression correlates with tumor progression and poor prognosis in colorectal carcinoma. BMC Cancer 15: 1006 10.1186/s12885-015-2018-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Wong K, Adams DJ. 2013. RetroSeq: transposable element discovery from next-generation sequencing data. Bioinformatics 29: 389–390. 10.1093/bioinformatics/bts697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzman JO, MacKenzie AP, Adey A, Hiatt JB, Patwardhan RP, Sudmant PH, Ng SB, Alkan C, Qiu R, Eichler EE, et al. 2011. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat Biotechnol 29: 59–63. 10.1038/nbt.1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. 10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HY, Mu XJ, Stütz AM, Tanzer A, Cayting PD, Snyder M, Kim PM, Korbel JO, Gerstein MB. 2010. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat Biotechnol 28: 47–55. 10.1038/nbt.1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HY, Pan C, Clark MJ, Lacroute P, Chen R, Haraksingh R, O'Huallachain M, Gerstein MB, Kidd JM, Bustamante CD, et al. 2012. Detecting and annotating genetic variations using the HugeSeq pipeline. Nat Biotechnol 30: 226–229. 10.1038/nbt.2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, Hall IM. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15: R84 10.1186/gb-2014-15-6-r84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131: 1235–1247. 10.1016/j.cell.2007.11.037 [DOI] [PubMed] [Google Scholar]
- Lin K, Smit S, Bonnema G, Sanchez-Perez G, de Ridder D. 2015. Making the difference: integrating structural variation detection tools. Brief Bioinform 16: 852–864. 10.1093/bib/bbu047 [DOI] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et al. 2017. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355: eaah7111 10.1126/science.aah7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozzio CB, Lozzio BB. 1975. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood 45: 321–334. [PubMed] [Google Scholar]
- Marks P, Garcia S, Barrio AM, Belhocine K, Bernate J, Bharadwaj R, Bjornson K, Catalanotti C, Delaney J, Fehr A, et al. 2018. Resolving the full spectrum of human genome variation using linked-reads. bioRxiv 10.1101/230946 [DOI] [PMC free article] [PubMed]
- Mohiyuddin M, Mu JC, Li J, Bani Asadi N, Gerstein MB, Abyzov A, Wong WH, Lam HY. 2015. MetaSV: an accurate and integrative structural-variant caller for next generation sequencing. Bioinformatics 31: 2741–2744. 10.1093/bioinformatics/btv204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens DW, Deans RM, Li A, Bassik MC. 2016. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol 34: 634–636. 10.1038/nbt.3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann S, Reutzel D, Speicher M, Decker HJ. 2001. Complete karyotype characterization of the K562 cell line by combined application of G-banding, multiplex-fluorescence in situ hybridization, fluorescence in situ hybridization, and comparative genomic hybridization. Leuk Res 25: 313–322. 10.1016/S0145-2126(00)00125-9 [DOI] [PubMed] [Google Scholar]
- Oparina NY, Sadritdinova AF, Snezhkina AV, Dmitriev AA, Krasnov GS, Senchenko VN, Melnikova NV, Belenikin MS, Lakunina VA, Veselovsky VA, et al. 2012. Increase in NETO2 gene expression is a potential molecular genetic marker in renal and lung cancers. Russ J Genet 48: 506–512. 10.1134/S1022795412050171 [DOI] [PubMed] [Google Scholar]
- Pabinger S, Dander A, Fischer M, Snajder R, Sperk M, Efremova M, Krabichler B, Speicher MR, Zschocke J, Trajanoski Z. 2014. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform 15: 256–278. 10.1093/bib/bbs086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Ben-Dor A, Tsalenko A, Sampas N, Rodriguez-Revenga L, Tran CW, Scheffer A, Steinfeld I, Tsang P, Yamada NA, et al. 2008. The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet 82: 685–695. 10.1016/j.ajhg.2007.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2012. Characterizing complex structural variation in germline and somatic genomes. Trends Genet 28: 43–53. 10.1016/j.tig.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selden JR, Emanuel BS, Wang E, Cannizzaro L, Palumbo A, Erikson J, Nowell PC, Rovera G, Croce CM. 1983. Amplified Cλ and c-abl genes are on the same marker chromosome in K562 leukemia cells. Proc Natl Acad Sci 80: 7289–7292. 10.1073/pnas.80.23.7289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29: 308–311. 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, Gabdank I, Narayanan AK, Ho M, Lee BT, et al. 2016. ENCODE data at the ENCODE portal. Nucleic Acids Res 44: D726–D732. 10.1093/nar/gkv1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder MW, Adey A, Kitzman JO, Shendure J. 2015. Haplotype-resolved genome sequencing: experimental methods and applications. Nat Rev Genet 16: 344–358. 10.1038/nrg3903 [DOI] [PubMed] [Google Scholar]
- Spies N, Weng Z, Bishara A, McDaniel J, Catoe D, Zook JM, Salit M, West RB, Batzoglou S, Sidow A. 2017. Genome-wide reconstruction of complex structural variants using read clouds. Nat Methods 14: 915–920. 10.1038/nmeth.4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40. 10.1016/j.cell.2010.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa GA, Sumiyama K, Ukai-Tadenuma M, Perrin D, Fujishima H, Ukai H, Nishimura O, Shi S, Ohno R-I, Narumi R, et al. 2016. Mammalian reverse genetics without crossing reveals Nr3a as a short-sleeper gene. Cell Rep 14: 662–677. 10.1016/j.celrep.2015.12.052 [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. 2007. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–D92. 10.1093/nar/gkl822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. 2015. Identification and characterization of essential genes in the human genome. Science 350: 1096–1101. 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CE, Saldivar JC, Hosseini SA, Huebner K. 2014. The FHIT gene product: tumor suppressor and genome “caretaker”. Cell Mol Life Sci 71: 4577–4587. 10.1007/s00018-014-1722-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LJS, Tabbaa DG, Li N, Berlin AM, Shea TP, MacCallum I, Lawrence MS, Drier Y, Getz G, Young SK, et al. 2012. Paired-end sequencing of Fosmid libraries by Illumina. Genome Res 22: 2241–2249. 10.1101/gr.138925.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SQ, Voelkerding K V, Sabatini L, Chen XR, Huang J, Meisner LF. 1995. Extensive amplification of bcr/abl fusion genes clustered on three marker chromosomes in human leukemic cell line K-562. Leukemia 9: 858–862. [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. 10.1093/bioinformatics/btp394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Lau BT, Schnall-Levin M, Jarosz M, Bell JM, Hindson CM, Kyriazopoulou-Panagiotopoulou S, Masquelier DA, Merrill L, Terry JM, et al. 2016. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol 34: 303–311. 10.1038/nbt.3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.