Abstract
The zebrafish embryo is transcriptionally mostly quiescent during the first 10 cell cycles, until the main wave of zygotic genome activation (ZGA) occurs, accompanied by fast chromatin remodeling. At ZGA, homologs of the mammalian stem cell transcription factors (TFs) Pou5f3, Nanog, and Sox19b bind to thousands of developmental enhancers to initiate transcription. So far, how these TFs influence chromatin dynamics at ZGA has remained unresolved. To address this question, we analyzed nucleosome positions in wild-type and maternal-zygotic (MZ) mutants for pou5f3 and nanog by MNase-seq. We show that Nanog, Sox19b, and Pou5f3 bind to the high nucleosome affinity regions (HNARs). HNARs are spanning over 600 bp, featuring high in vivo and predicted in vitro nucleosome occupancy and high predicted propeller twist DNA shape value. We suggest a two-step nucleosome destabilization-depletion model, in which the same intrinsic DNA properties of HNAR promote both high nucleosome occupancy and differential binding of TFs. In the first step, already before ZGA, Pou5f3 and Nanog destabilize nucleosomes at HNAR centers genome-wide. In the second step, post-ZGA, Nanog, Pou5f3, and SoxB1 maintain open chromatin state on the subset of HNARs, acting synergistically. Nanog binds to the HNAR center, whereas the Pou5f3 stabilizes the flanks. The HNAR model will provide a useful tool for genome regulatory studies in a variety of biological systems.
The development of multicellular organisms is first driven by maternal products and occurs in the absence of transcription (Tadros and Lipshitz 2009). The main wave of the embryo's own transcription, zygotic genome activation (ZGA), occurs several hours after the egg is fertilized. The mechanistic reasons for this transcriptional delay are not completely understood. In Drosophila and zebrafish Danio rerio, transcription after ZGA depends on a small number of maternal enhancer-binding transcription factors (TFs). Zygotic genome activators include Zelda in Drosophila (Liang et al. 2008), as well as three homologs of pluripotency TFs, Pou5f3, Nanog, and Sox19b in zebrafish (Lee et al. 2013; Leichsenring et al. 2013). Recent analysis of chromatin accessibility and gene expression also implicated TFs POU5F1 and NFYA as ZGA activators in humans (Gao et al. 2018) and mice (Lu et al. 2016), respectively. The interactions of ZGA activators with chromatin are only starting to be understood, and it is unclear if the same ZGA mechanisms are implemented in different animals.
The basic units of chromatin structure are the nucleosomes, which restrict the in vivo access of most TFs to their target sites (Beato and Eisfeld 1997; Luo et al. 2014). Although many TFs cannot bind their target site in the context of nucleosome DNA in vitro, cooperative interactions among multiple factors allow binding, even in the absence of defined orientation of their recognition motifs in DNA (Adams and Workman 1993; Zinzen et al. 2009). In steady-state cell culture systems, most of the TF binding sites (TFBSs) fall within nucleosome-free DNA regions (Thurman et al. 2012), and it is difficult to conclude if the chromatin accessibility is a cause or a consequence of TF binding. Studies of hematopoietic cell fate transitions suggested a hierarchical model, in which a relatively small set of “pioneer” TFs collaboratively compete with nucleosomes to bind DNA in a cell-type–specific manner. The binding of pioneer TFs, also called lineage-determining factors and master regulators, is hypothesized to prime DNA by moving nucleosomes and inducing the deposition of epigenetic enhancer marks. This enables concurrent or subsequent binding of signal-dependent TFs that direct regulated gene expression (Heinz et al. 2010; Trompouki et al. 2011; Li et al. 2018). Two alternative scenarios of TF-guided chromatin opening at enhancers during cell-fate transitions have been suggested: the nucleosome-mediated cooperativity model and the model based on specialized properties of pioneer TFs (Calo and Wysocka 2013; Slattery et al. 2014). The nucleosome-mediated cooperativity model postulates that if the recognition motifs for multiple cooperating TFs are located within the length of one nucleosome, the nucleosome will be removed (Mirny 2010; Moyle-Heyrman et al. 2011). The alternative model evokes special properties of pioneer factors to bind their DNA sites directly on nucleosomes (Iwafuchi-Doi and Zaret 2016). The pioneer factor model predicts that pioneer TFs will engage into the silent chromatin, whereas the majority of nonpioneer TFs will occupy nucleosome-depleted regions.
In Drosophila, the ZGA activator Zelda binds to regions of high predicted nucleosome occupancy and creates competency for other factors to bind the DNA, thus providing an example of a pioneer factor (Schulz et al. 2015; Sun et al. 2015). Genomic binding of zebrafish ZGA activator Pou5f3 is detectable at the 512-cell stage, just before the main ZGA wave (Leichsenring et al. 2013). Binding of Pou5f3 to the enhancers of its early regulated target genes could be significantly outcompeted by increasing histone concentration at ZGA (3 hpf, 10th cell cycle) (Joseph et al. 2017). In zebrafish, nucleosome positioning signals guide the transcription start site (TSS) selection after ZGA but not before, suggesting that the rules of nucleosome positioning may change over ZGA (Haberle et al. 2014).
Genetic ablation of maternal and zygotic expression of Pou5f3 and Nanog results in severe pleiotropic phenotypes, global deregulation of transcription, and gastrulation arrest (Lunde et al. 2004; Onichtchouk et al. 2010; Gagnon et al. 2018; Veil et al. 2018), suggesting that these factors act at the genome-wide level. However, it is not clear whether Pou5f3 or Nanog preferentially target the regions occupied by nucleosomes before ZGA or are able to remove nucleosomes from the regions to which they bind. We address these questions below.
Results
The properties of TFBSs bound by Pou5f3, SoxB1, and Nanog individually or in combination
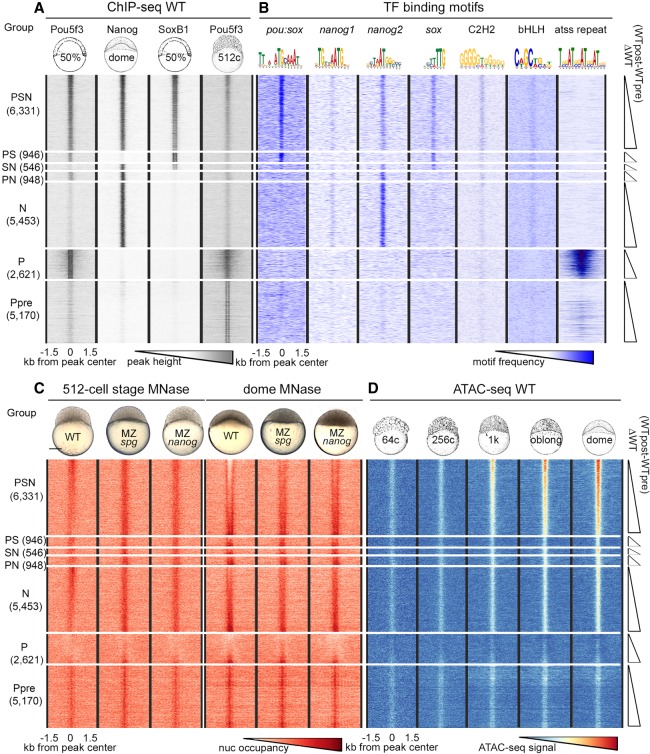
Our analysis strategy was to categorize the genomic regions by their binding of Pou5f3 (P), SoxB1 (S), and Nanog (N), individually or in combination. For this analysis, we focused on the TFBSs within ChIP-seq peaks from our own and others’ previous work (Xu et al. 2012; Leichsenring et al. 2013) and on a group of randomly chosen control regions of similar size. Based on TFs binding data, genomic sites were classified into the following groups: P and N groups (Pou5f3 and Nanog bind individually); PS, PN, and SN groups (TFs bind pair-wise); and the PSN group (all three TFs bind). The group of genomic regions occupied by Pou5f3 alone and only pre-ZGA, termed Ppre, served as an additional control. We set to compare the relative changes in nucleosome occupancy using MNase-seq method for each of these groups between MZspg, MZnanog mutants, and the wild-type (WT). In the cases in which we found statistically significant differences in nucleosome occupancy between the groups, we evaluated to which extent these differences could be explained by sequence-specific binding of the TFs to their consensus recognition motifs. We use the terms “consensus binding” and “nonconsensus binding,” as suggested by Afek et al. (2015), to distinguish between TF binding to its cognate consensus motifs on DNA (“consensus binding”) and TF binding at the absence of motifs (“nonconsensus binding”). The heat maps of TF occupancy for the seven groups are shown in Figure 1A, summary profiles in Supplemental Figure S1, and the genomic regions in the Supplemental Table S1. We performed de novo motif search to estimate the relative abundance of the sequence-specific recognition consensus motifs for each TF in each group. In addition to the sequences closely matching known Pou5f3, SoxB1, and Nanog consensus motifs (pou:sox, sox, nanog1, and nanog2), we detected enrichment for generic motifs for C2H2-type zinc-finger proteins and β-HLH domain TFs, dinucleotide repeats, and tetranucleotide ATSS repeats on the regions bound by Pou5f3 alone. Figure 1B shows distribution of the motifs within the TFBS groups (for all logos, see Supplemental Fig. S2; for matrices, see Supplemental Table S2).
Figure 1.
Nucleosomes cover all TFBSs pre-ZGA and are depleted from PSN triple-bound regions post-ZGA. Heat maps of six groups of genomic regions defined by combinatorial binding of Pou5f3, Nanog, and SoxB1 post-ZGA, as indicated at the left, and Ppre group bound by Pou5f3 only pre-ZGA. Three-kilobase genomic regions were aligned at the ChIP-seq peak centers. Within each group, the data were sorted by ascending difference between wild-type (WT) post-ZGA and pre-ZGA nucleosome occupancy (ΔWT post-pre). (A) TF binding (Pou5f3 50% epiboly, Nanog and SoxB1 dome stage, Pou5f3 512-cell stage). (B) Occurrence of TF binding motifs. (C) Nucleosome occupancy in the embryos of indicated genotypes. (D) Accessible chromatin signals in pre-ZGA (64c, 256c; c indicates cell stage), ZGA (1K), and post-ZGA stages (oblong, dome). (C) Scale bar, 200 µm.
Pou5f3, SoxB1, and Nanog preferentially bind to the enhancers of developmental genes (Xu et al. 2012; Leichsenring et al. 2013). To compare the enrichment in developmental enhancers in the groups defined above, we used GREAT analysis (McLean et al. 2010). The enrichment in enhancers within PSN group was an order of magnitude higher than that in the N, PS, PN, SN, and Ppre groups (Supplemental Fig. S3). We found no enrichment for the P group, indicating that nonconsensus binding of Pou5f3 alone post-ZGA does not mark enhancers.
All TFBSs are occupied by nucleosomes before ZGA; nucleosomes are depleted only from the triple-bound Pou5f3, Nanog, and SoxB1 regions after ZGA
We isolated chromatin from the WT, the MZspg and MZnanog mutants at pre-ZGA (512-cell stage) and post-ZGA (dome stage) and performed micrococcal nuclease treatment followed by deep sequencing of the resulting fragments (MNase-seq) as described (Supplemental Fig. S4; Supplemental Table S3; Zhang et al. 2014). At the 512-cell stage, all TFBSs had higher nucleosome occupancy than surrounding sequences (Fig. 1C, 512-cell stage). At the dome stage, a nucleosome-depleted region appeared in the PSN group, with these depletions being strongly reduced in the MZnanog and MZspg mutants compared with the WT (Fig. 1C, dome). Single or dual Pou5f3, SoxB1, or Nanog occupation sites remained covered by nucleosomes. We compared our data with recently published ATAC-seq–based chromatin accessibility data (Liu et al. 2018). Weak ATAC-seq signals that did not meet the standard cutoffs in Liu et al. (2018) and Meier et al. (2018) were detectable when aligned on all TFBSs (Fig. 1D). Before ZGA, the elevated nucleosome occupancy (MNase-seq) and accessible chromatin (ATAC-seq) signals colocalized on all TFBSs irrespectively to their regulatory potential (Fig. 1C, 512-cell stage; Fig. 1D, 64- and 256-cell stage). We interpreted mixed signals of opposite direction as local destabilization of nucleosomes on TFBSs. Mixed high MNase-seq and high ATAC-seq signals could reflect heterogeneity of the cells in the embryo: In a few cells, the TFBS was nucleosome-free, whereas in most of the cells, the TFBS was covered by nucleosome. Low MNase-seq and high ATAC-seq signals colocalized only on the PSN group, indicating nucleosome depletion from only PSN regions post-ZGA. All other TFBSs were covered by destabilized nucleosomes, pre- and post-ZGA.
Pou5f3 and Nanog nonspecifically reduce nucleosome occupancy before ZGA and act sequence-specifically after ZGA
We tested whether the differences in pre- and post-ZGA nucleosome occupancy between each of the two mutants, MZspg and MZnanog, and the WT were significantly different from zero in each TFBS group and significantly different among the TFBS groups. This was the case for both stages (one-way ANOVA for all comparisons) (Supplemental Fig. S5; Supplemental Table S4). At the 512-cell stage, MNase-seq signal in both mutants was higher than in the WT for all TFBSs (except for P group in MZnanog) (Supplemental Fig. S5B,C). At the dome stage, MNase-seq signal in both mutants mostly increased in the PSN group compared with the WT (Supplemental Fig. S5D–F). This indicated that Pou5f3 and Nanog displace nucleosomes at both stages. To test if nucleosome displacement by TFs correlates with gene expression, we linked each TFBS to the nearest promoter defined by Haberle et al. (2014) and divided them into four categories (early zygotic, late zygotic, maternal-zygotic, and nonexpressed) (Supplemental Fig. S6A). Nucleosome displacement by TFs pre-ZGA and in double- and single-TF-bound TFBSs post-ZGA did not differ between gene expression categories. On the PSN (triple-bound) TFBSs linked to the early zygotic genes, nucleosome displacement post-ZGA was stronger and more dependent on Pou5f3 and Nanog than on the PSN TFBSs linked to the other categories (Supplemental Fig. S6B–G).
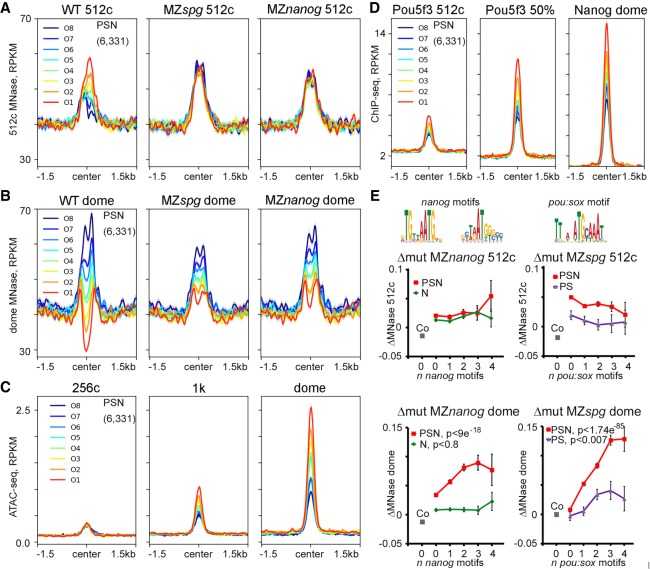
To test whether the same regions within the PSN group are affected at the 512-cell and dome stages, we ranked 6331 PSN regions by descending difference between pre- and post-ZGA nucleosome occupancy (ΔWT = WTpost − WTpre) (Fig. 1C,D) and divided them to octiles O1–O8, O1 being the most open at dome stage. The order of octiles by nucleosome occupancy did not match between the 512-cell and dome stages (Fig. 2A,B); neither did the mutant effects (Supplemental Fig. S7A,B). The ranking of octiles at the dome stage correlated with the ATAC-signal strength starting from ZGA (1K) (Fig. 2C) and with the occupancy by Pou5f3 and Nanog TFs (Fig. 2D). The reduction in nucleosome occupancy by Pou5f3 and Nanog assayed by the MNase-seq method at the post-ZGA dome stage corresponded to the increase of the accessible chromatin signals assayed by ATAC-seq at the post-ZGA oblong stage (Pou5f3 and Nanog morpholino knockdown experiments of Liu et al. 2018; Supplemental Fig. S7B,C).
Figure 2.
Pou5f3 and Nanog displace nucleosomes from different regions before and after ZGA. (A–D) PSN group regions ranked into octiles by ascending difference between WT post-ZGA and pre-ZGA MNase-seq signal (ΔWT post-pre). Three-kilobase genomic regions were aligned at the ChIP-seq peak centers. Summary plots per octile: (A) nucleosome occupancy 512-cell stage, (B) nucleosome occupancy dome, (C) ATAC-seq, and (D) TF occupancy. (E) TF effects on nucleosome density were estimated as MNase-seq signal difference between the WT and indicated mutants (Δmut) and related to the number of specific motifs (zero, one, two, three, or four or more motifs) per 320-bp binding region. Motifs are indicated on top. Pou5f3 and Nanog act nonsequence-specifically at the 512-cell stage (upper row). At dome stage (lower row), Pou5f3 and Nanog act by sequence-specific binding to their motifs on triple-occupancy PSN regions but not on single and double TFBSs. PSN indicated by red; N, green; and PS, violet. (Co) Random control regions (zero motifs; gray); (y-axis) Δmut.
To evaluate the contribution of TF consensus binding to the nucleosome displacement, we compared MNase-seq signal changes between each mutant and the WT (Δmut MZspg and Δmut MZnanog) within the PSN, N, or PS groups on the regions with zero, one, two, three, or four or more cognate Pou5f3/SoxB1 or Nanog motifs (Fig. 2E; for one-way ANOVA, see Supplemental Table S5). Δmut MZspg and Δmut MZnanog did not depend on the number of motifs at the 512-cell stage (Fig. 2E, 512c). However, Δmut MZspg and Δmut MZnanog on TFBSs without motifs were higher than in randomly chosen control sequences, suggesting either indirect or nonspecific effects of the TFs (Fig. 2E, 512-cell stage, gray Co data point; for Student's t-test, see Supplemental Table S6). At the dome stage, Δmut MZspg and Δmut MZnanog depended on the number of respective motifs in the PSN group regions, but not in the N group and only marginally in the PS group (Fig. 2E, dome). These results paralleled accessible chromatin changes assayed by morpholino knockdown experiments and ATAC-seq (Supplemental Fig. S7D). Thus, in contrast to nonspecific pre-ZGA events, post-ZGA nucleosome depletion relied on specific binding of Pou5f3, Nanog, and SoxB1 (PSN) and required synergistic action of all three TFs.
Pou5f3 and Nanog bind chromatin independently from preexisting chromatin modifications and DNA methylation
The mixed elevated MNase and ATAC-seq signals observed on all TFBSs could reflect nucleosome destabilization by either distinct sequence composition or epigenetic premarking. Murphy et al. (2018) have described “placeholder” H2AZ/H3K4me1 containing nucleosomes, which occupy the regions lacking DNA methylation in cleavage embryos. Only 5.3% of TFBS defined in our study overlapped with placeholder nucleosome locations (Supplemental Fig. S8A), and these 5.3% TFBSs were bound by Pou5f3 and Nanog similarly to the rest of the sites (Supplemental Fig. S8B–D). Liu et al. (2018) suggested that Pou5f3 and Nanog specifically recognize methylated motifs; we did not find a support for this statement (see Supplemental Fig. S9 and legend). As we could not find strong evidence for preexisting epigenetic marking, we focused on sequence composition of TFBS regions in order to explain nucleosome destabilization effects.
Pou5f3, SoxB1, and Nanog bind to 600-bp-long asymmetric high nucleosome affinity regions
The reasons for high nucleosome occupancy of pre-ZGA TFBS could be intrinsic nucleosome-favoring DNA features. Nucleosomes cover 75%–90% of the genome (Kornberg 1974) and can form virtually anywhere, but the properties of the underlying DNA may change nucleosome affinity up to several orders of magnitude (Field et al. 2008). Increased GC content and 10-bp periodic fluctuations of dinucleotides underlie the high ability of DNA to wrap around histone octamers, whereas AT-rich sequences strongly disfavor nucleosome formation (Satchwell et al. 1986; Field et al. 2008; Chung and Vingron 2009; Kaplan et al. 2009; Tillo and Hughes 2009). To evaluate if Pou5f3, SoxB1, and Nanog preferentially recognize nucleosome-positioning signals, we used nucleosome predictions based on the in vitro nucleosome DNA sequence preferences mentioned above (Kaplan et al. 2009). The predicted nucleosome occupancy is scored by a probability for each nucleotide position to be located in the nucleosome center (or dyad). We selected the maximal [nucmax] score (for the most likely predicted dyad position) within the mean length of ChIP-seq peak (320 bp) for each TFBS and control region and compared them across the groups. [Nucmax] predicted scores of all (individual, double-bound, and triple-bound) TFBSs were higher than respective scores for the random control regions (Supplemental Fig. S10A). Notably, the [nucmax] scores of TFBS groups in which Nanog binds (i.e., N, PN, SN, and PSN groups) were significantly higher than of those of the groups in which Nanog does not bind (Supplemental Fig. S10B; Supplemental Table S7).
The minimum nucleosome prediction scores within the 320-bp interval [nucmin] most likely corresponded to the predicted inter-nucleosomal sequences within the region (Segal et al. 2006). In all TFBSs [nucmin], scores were also significantly higher than the respective scores for random control regions (Supplemental Fig. S10C,D; Supplemental Table S8). The difference in [nucmin] predictions agreed with experimental data: Experimental nucleosome occupancy of random control regions aligned on [nucmin] positions was below the genomic average, as predicted, but it was above the genomic average in [nucmin] positions of all TFBSs (Supplemental Fig. S10E–L). These data implied that high nucleosome affinity regions (HNARs) underlying TFBSs span at least 320 bp, that is, more than two nucleosome lengths.
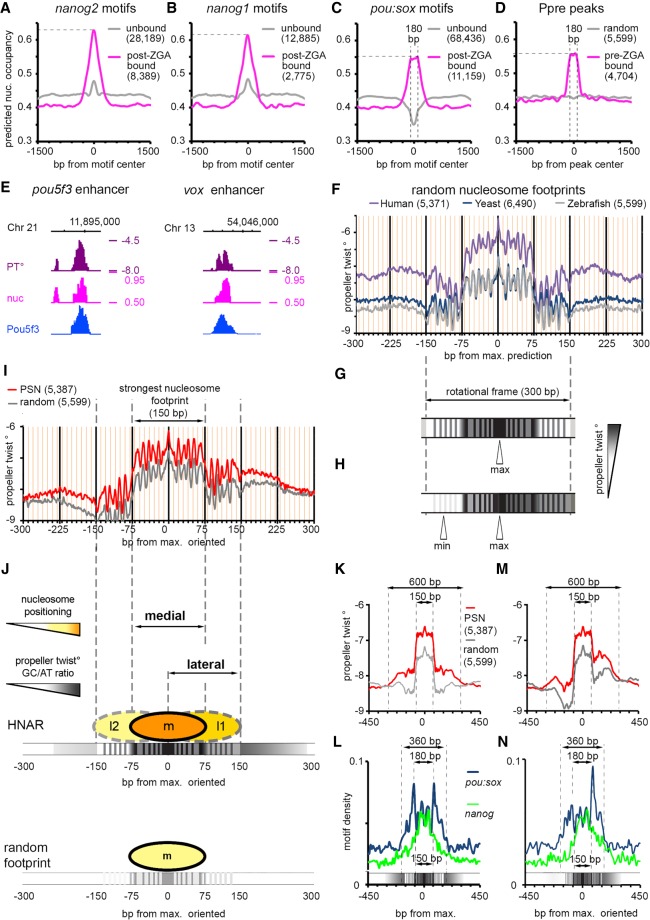
TF recognition motif sequences may themselves contain high predicted nucleosome occupancy features, or alternatively, the high predicted nucleosome occupancy may be encoded by sequences neighboring the recognition motifs. To distinguish between these possibilities, we compared predicted nucleosome occupancy of the motifs that are actually bound by Nanog or Pou5f3/SoxB1 (within 320 bp of respective TFBS), with predicted nucleosome occupancy of randomly occurring unbound motifs (within the interval from 1 to 1.5 kb away from the TFBS center). All bound motifs could be discriminated from the unbound ones by higher predicted nucleosome occupancy of neighboring sequences (Fig. 3A–C). Nucleosome prediction profiles around the nanog1 and nanog2 motifs were higher and different in shape from the profiles on pou:sox motifs (Fig. 3, cf. A,B and C), whereas pou:sox motif profiles (Fig. 3C) were similar to the flattened profiles on the regions bound by Pou5f3 pre-ZGA (Ppre, Fig. 3D).
Figure 3.
Pou5f3, SoxB1, and Nanog bind to the high nucleosome affinity regions (HNARs). (A–C) Mean predicted nucleosome occupancy plots on the indicated motifs bound by respective TF (magenta) versus unbound random motif matches (gray). (D) Mean predicted nucleosome occupancy plot centered on Pou5f3 pre-MBT TF binding peaks (magenta) versus random sequences (gray). (E) Pou5f3 binds to the regions of high predicted nucleosome occupancy and propeller twist on the enhancers of pou5f3 and vox. (PT°) Propeller twist; (nuc) in vitro nucleosome occupancy prediction, tick marks every 1 kb. (F) Propeller twist summary plot of random zebrafish, yeast, and human genomic regions aligned at the base pair with maximal predicted nucleosome occupancy within 320 bp [nucmax]. Yellow x-axis gridlines 10 bp apart. (G) Scheme of the PT periodic oscillations in 300 bp around [nucmax]. (H) Scheme of the oriented PT plot aligned at [nucmax]. (I) Oriented PT plot of PSN group (red) and control genomic regions (gray). (J) Scheme of medial and lateral strong nucleosome footprints at HNAR. Central 300-bp periodic frame supports alternative medial, lateral one, and lateral two nucleosome positions with decreasing strength (m > l1 > l2 = random footprint); in random sequence, only medial footprint is supported (bottom). (K,M) Symmetric (K) and oriented (M) smoothened PT plots (80-bp moving average) of PSN and control genomic regions, aligned at [nucmax]. (L,N) Distribution of nanog and pou:sox motifs in the PSN group for K and M, respectively. Motif density in base-pair motif per base-pair sequence.
There is no agreement in the literature on the intrinsic nucleosome occupancy features within regulatory sequences. Underrepresentation of nucleosome positioning sequences in enhancers has been reported (Daenen et al. 2008; Papatsenko et al. 2009; Khoueiry et al. 2010; He et al. 2010), whereas other studies reported their overrepresentation (Tillo et al. 2010; Barozzi et al. 2014; Sun et al. 2015). Our data strongly support the notion that intrinsic nucleosome occupancy in zebrafish early enhancers is high, but in the view of the contradictory literature, we aimed to confirm this conclusion by using an independent approach. Propeller twist (PT), the angle between the heterocycles in the two complementary bases in a base pair, is a DNA shape parameter that is not directly encoded by primary sequence. PT values have been related to the flexibility of DNA bending around proteins (el Hassan and Calladine 1996) and strongly correlate with the ability of DNA to wrap around nucleosomes in yeast (Lee et al. 2007; Gan et al. 2012). Intrinsic nucleosome-forming preferences of the DNA on the known PSN-bound enhancers of pou5f3 (Parvin et al. 2008), vox, and vent genes (Belting et al. 2011) were independently captured by the primary sequence–derived model of Kaplan et al. (2009) and high values of PT (Fig. 3E; Supplemental Fig. S11).
We aimed to further describe the length and structure of putative enhancers that underlie TFBSs by combining PT and the nucleosome predictions based on the Kaplan et al. (2009) model. Because it is unknown how the features captured by sequence-based nucleosome positioning programs are reflected in PT values, we started from the control sequences. We randomly selected 5000–6000 genomic sequences from human and yeast genomes as 320-bp control sequences and found [nucmax] predicted nucleosome scores (dyad) within each region by the model of Kaplan et al. (2009). Then we plotted the average PT values for zebrafish, human, and yeast aligned on the [nucmax] position. As shown in Figure 3F, a sharp increase in PT marked the central 150-bp nucleosome footprint, which was embedded into a 300-bp periodic shape. The differences in the height of PT graphs roughly corresponded to the median GC content in zebrafish, yeast, and human genomes (36.8%, 38.4%, and 40.9%, respectively) as provided by the NCBI.
The ∼10-bp periodicity for PT prediction within the central 300 bp of DNA (Fig. 3G) represents a single rotational frame, which was reported to stabilize nucleosome positioning on DNA (see Supplemental Fig. S12 and legend). To uncover possible hidden asymmetry of the nucleosome positioning signals, we oriented each 320-bp genomic region centered on [nucmax] so that the minimum nucleosome prediction value within ±160 bp around [nucmax] is positioned at the left (5′) side of the interval, as shown in Figure 3H. We compared summary PT heatmaps of TFBS groups aligned on [nucmax] or [nucmin] with their nucleotide composition and found that higher PT values corresponded to increased GC content (Supplemental Fig. S13A,B and the legend). Thus, PT sensitively reflected two attributes of DNA affinity to nucleosomes: GC content and dinucleotide rotational periodicity. We next used the [nucmax] and [nucmin] positions as viewpoints to estimate the differences between TFBSs and random nucleosome footprints using symmetric and asymmetric PT plots. All TFBSs except the P and PS groups differed from the control group by higher PT values over ∼600 bp around [nucmax], as shown in Figure 3I for PSN group regions (for all groups, see Supplemental Figs. S14, S15). We define here the 600-bp DNA region with high predicted nucleosome occupancy/PT/GC content, which is centered on a 300-bp periodic frame, as a HNAR.
HNAR structure supports two strong nucleosome positioning sequences, medial and lateral
The random nucleosome footprint and HNAR, both oriented from low to high nucleosome prediction/PT, are shown in Figure 3J. PT values/relative GC content is higher in HNAR, which should theoretically result in stronger nucleosome positioning. The graph above HNAR shows three positions within the 300-bp frame, given that high PT and periodic PT signal represent two independent factors that stabilize nucleosome on DNA. The central (medial), lateral one (l1), and lateral two (l2) positions are supported by 10 periodic peaks each. The medial position is the most stable; the l1 position is weaker because of the overall lower PT but is still stronger than a random nucleosome footprint. The l2 position is the weakest, being supported by an even lower PT value compared with the medial and l1 positions. In the random nucleosome footprint, only the medial position is stabilized by underlying DNA features (Fig. 3J, bottom). TF binding motifs for Nanog and Pou5f3/SoxB1 were differentially distributed around HNAR center. When HNAR was oriented symmetrically (as shown in Fig. 3K), most nanog motifs localized within the medial 150-bp nucleosome footprint, whereas pou:sox motifs were present over the whole 300-bp central periodic shape, with two peaks 180 bp apart from each other (Fig. 3L). In the oriented projection of HNAR (Fig. 3M), the pou:sox motifs added up in one peak within the higher PT flank (lateral footprint), whereas the nanog motifs were mostly central (Fig. 3N). The nucleosome distribution in the model on Figure 3J is theoretical, and we next addressed if it could help us to interpret in vivo data.
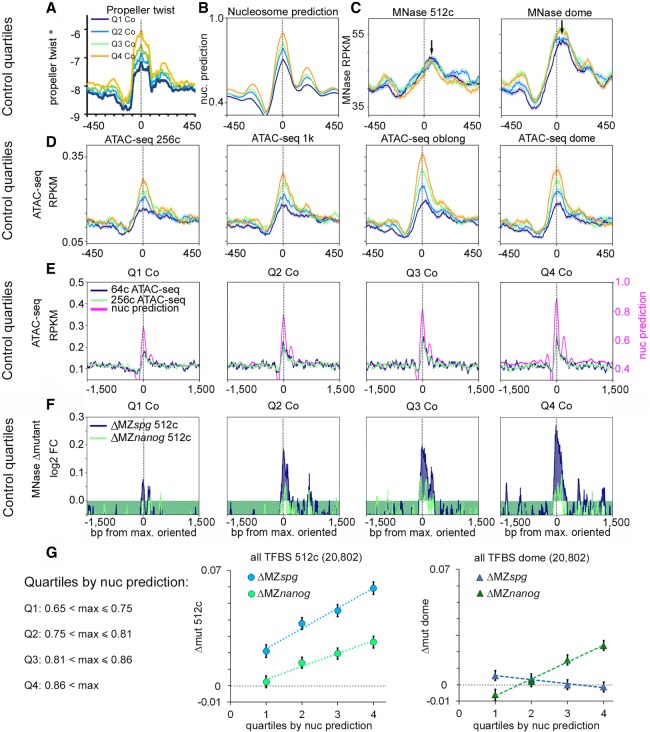
Pou5f3 and Nanog nonspecifically destabilize nucleosomes at HNAR centers pre-ZGA
Before ZGA, Pou5f3 and Nanog destabilized nucleosomes on all TFBS groups independently from the presence of their consensus motifs. Could structural properties of HNARs explain this effect? To find out if this is the case, we divided the TFBS and random control regions, matched by [nucmax] value, into quartiles Q1 through Q4, Q1 having the lowest [nucmax] value. The PT and nucleosome prediction graphs for control oriented nucleosome footprints are shown in Figure 4, A and B (for the TFBS groups, see Supplemental Fig. S16A,B). High MNase-seq and high ATAC-seq signals colocalized within random strong nucleosome footprint (Fig. 4C,D), indicating that footprint features alone caused destabilization of nucleosomes. On random nucleosome footprints and on TFBSs, the ATAC-seq signal peaked on the predicted dyad (Fig. 4D), whereas nucleosomes were shifted to the lateral positions (black arrows on Fig. 4C). The strength of the ATAC-seq signal positively correlated with the predicted nucleosome occupancy value in all stages and groups (Fig. 4D; for TFBS groups, see Supplemental Fig. S16C,D; for statistics, see Supplemental Table S9). Thus, HNAR structure itself promoted nucleosome destabilization using a mechanism that relies on predicted nucleosome occupancy. We tested if the nucleosome displacement effects of Pou5f3 and Nanog pre-ZGA contribute to this mechanism. Pre-ZGA, Pou5f3 and Nanog displaced nucleosomes from the random nucleosome footprints and HNAR centers, and the strength of displacement increased with the predicted nucleosome occupancy (Fig. 4E,F; Supplemental Fig. S17A–D). Pou5f3 ChIP-seq signals at the 512-cell stage were present at all HNAR centers and even on a control footprint (Supplemental Fig. S17E), indicating that Pou5f3 may act by direct binding. At the 512-cell stage, the nucleosome displacement by Pou5f3 and Nanog increased with the predicted nucleosome footprint strength in all TFBS groups and the control (for the whole data set, see Fig. 4G; for TFBS groups, see Supplemental Fig. S18; for statistics, see Supplemental Table S10). Post-ZGA, nucleosome displacement only by Nanog depended on nucleosome prediction strength (Fig. 4G), suggesting that Pou5f3 acts differently before and after ZGA. We concluded that nonspecific nucleosome destabilization by Pou5f3 and Nanog pre-ZGA depends on HNAR structural features, is not restricted to enhancers, and occurs genome-wide.
Figure 4.
Pou5f3 and Nanog randomly displace nucleosomes from HNAR centers pre-ZGA. (A–D) Random control genomic regions were ranked by ascending nucleosome prediction score into quartiles Q1 through Q4, aligned at HNAR centers ([nucmax]; gray dotted lines), and oriented as in Figure 3H. (A) Propeller twist (PT°); (B) nucleosome prediction; (C) MNase-seq (black arrows show the lateral shift from [nucmax]); and (D,E) ATAC-seq signal colocalizes with [nucmax] and increases with nucleosome prediction value. (F) Random control quartiles: Nucleosome displacement from HNAR centers by Pou5f3 and Nanog at the 512-cell stage increases with nucleosome prediction value. Gray dotted lines indicate [nucmax]. (G) Dependencies of Δmut (nucleosome occupancy) from nucleosome prediction strength in the whole TFBS data set: (left) 512-cell stage; (right) dome.
Sequence-specific Nanog binding at the HNAR center and Pou5f3 binding on +90-bp lateral position deplete nucleosomes post-ZGA
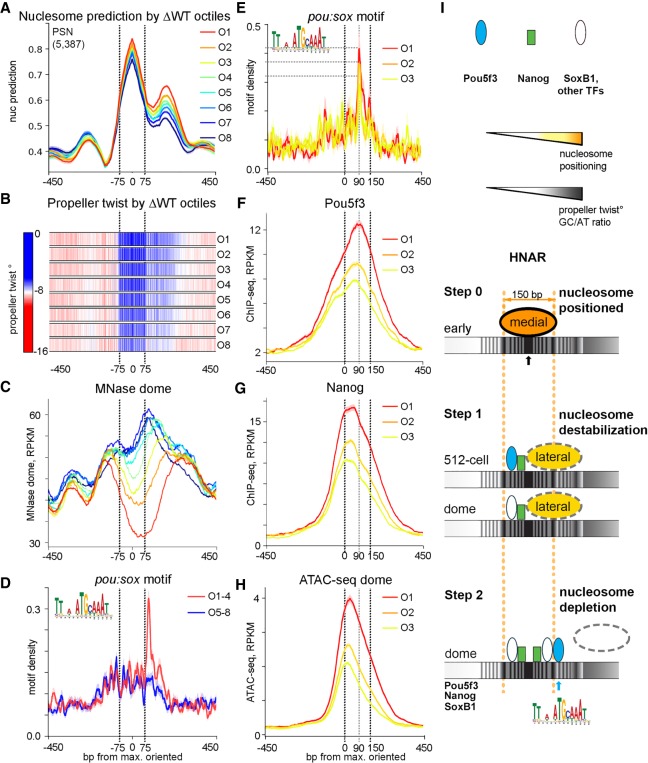
Widespread nucleosome destabilization may facilitate specific binding of TFs if their motifs are present at HNAR centers. Nucleosome-mediated cooperativity model (also referred as “assisted loading” or “collaborative binding”) (Spitz and Furlong 2012) postulates that nucleosome depletion occurs when multiple TFs specifically bind their motifs on DNA within one strong nucleosome positioning sequence. HNAR contains at least two overlapping strong nucleosome positioning sequences, medial and lateral footprints (Fig. 3J). Our next question was if synergistic nucleosome depletion by Pou5f3, SoxB1, and Nanog after ZGA depends on the amount and distribution of the specific TF motifs on medial (−75 to 75 bp from HNAR center) or lateral (0–150 bp from HNAR center) footprints. By using the octiles defined above (O1 being the most open; O8, most closed at dome stage), we centered the genomic regions of each octile on [nucmax], oriented as in Figure 3H, and compared nucleosome prediction values, PT, MNase-seq signal, and localization of specific motifs between the octiles (Fig. 5A–D). Nucleosome prediction values and PT decreased from O1 to O8 (Fig. 5A,B). The abundance of nanog, sox, C2H2, and bHLH motifs, localized within the medial nucleosome footprint, did not significantly change among octiles (Supplemental Fig. S19). Thus, specific TF binding within the medial footprint displaces nucleosomes laterally, but it is not sufficient to deplete them (as in octiles O6 through O8) (Fig. 5C). In contrast, pou:sox motif on the lateral footprint was enriched in open octiles (O1 through O3) (Fig. 5D,E), suggesting that this motif in the +90-bp position is critical for nucleosome depletion. Post-ZGA, Pou5f3 bound at +90 bp (Fig. 5F), whereas Nanog and ATAC-seq peaks were localized more centrally (Fig. 5G,H). We concluded that post-ZGA nucleosome depletion involves two overlapping nucleosome footprints and is therefore distinct from pure nucleosome-mediated cooperativity mechanism. Specific Nanog and Pou5f3 binding to the medial and lateral footprints, respectively, blocks nucleosome assembly on both nucleosome positioning sequences and leads to nucleosome depletion.
Figure 5.
Post-ZGA nucleosome depletion requires Nanog central binding and Pou5f3 binding to the +90-bp position at HNAR. PSN group regions ranked into octiles as in Figure 2, aligned on [nucmax] and oriented as in Figure 3H. Black dotted lines mark the borders of the medial nucleosome footprint in A through D and of the lateral nucleosome footprint in E through H. (A) Nucleosome prediction profiles per octile. (B) Propeller twist heat maps per octile. (C) MNase-seq (dome) per octile; note nucleosome displacement to the lateral footprint in O6 through O8. (D,E) Pou:sox motif density, base-pair motif per base-pair sequence. (D) Pou:sox motif peak at +90 bp is present in the open O1 through O4 but not in the O5 through O8 octiles. (E) Pou:sox motif abundance at the +90-bp position decreases O1 > O2 > O3. (F) Pou5f3 binds at the +90-bp position in octiles O1 through O3. (G) Nanog binds centrally in O1 through O3. (H) ATAC-seq in O1 through O3. (I) Two-step nucleosome destabilization-depletion model (see main text). Black and blue arrows show 0 (center) and +90-bp position Pou5f3 binding at HNAR.
Two-step nucleosome destabilization-depletion model
We summarize our findings in a two-step HNAR-centered nucleosome destabilization-depletion model (Fig. 5I). Initially, in the early pre-ZGA cell cycles, nucleosomes assemble on the strongest medial nucleosome positioning sequence of HNAR (Fig. 5I, Step 0). At 512-cell stage, Pou5f3, Nanog, and possibly other nonhistone DNA binding proteins nonspecifically destabilize nucleosomes on central nucleosome footprints of HNARs and shift them laterally (Fig. 5I, Step 1, 512-cell). At dome stage, nucleosomes are destabilized by Nanog but not by Pou5f3 (Fig. 5I, Step 1, dome). In the second step, which occurs only after ZGA, simultaneous specific Nanog binding within the medial and Pou5f3 binding within the lateral nucleosome positioning sequence deplete nucleosomes from HNAR (Fig. 5I, step 2, dome).
HNAR model is applicable to the mammalian system
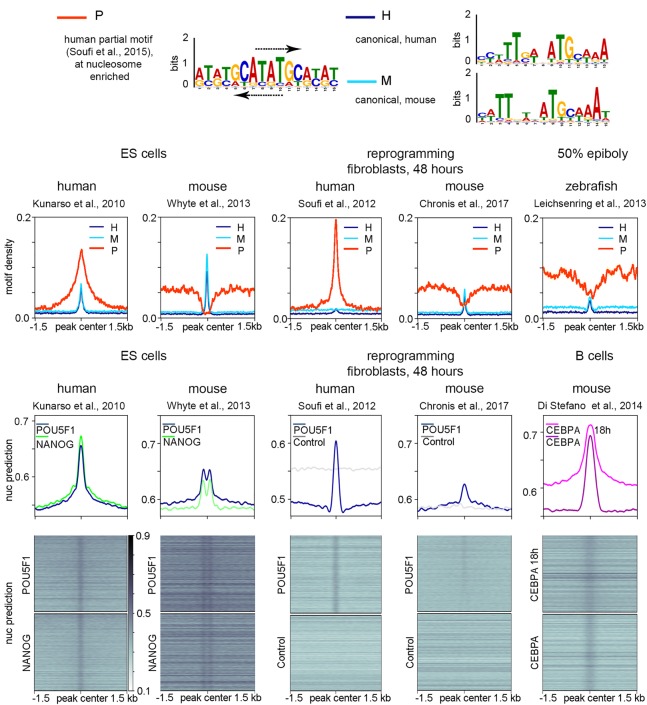
We tested if the HNAR model is applicable to the mammalian homologs of Pou5f3 and Nanog. Experiments using POU5F1, SOX2, KLF, and MYC (OSKM mix) (Takahashi and Yamanaka 2006) for reprogramming of human or mouse fibroblasts to induced pluripotent stem (iPS) cells suggested different modes of human POU5F1 and mouse POU5F1 interactions with chromatin. Soufi et al. (2015) showed that at the initial stage of human fibroblast reprogramming, POU5F1 targets its partial motifs directly on nucleosomal DNA. Chronis et al. (2017), on the other hand, did not find evidence for direct nucleosome targeting by POU5F1 in the reprogramming experiment in mouse cells. We addressed if the differences between species can be explained by the sequence composition of POU5F1/Pou5f3 TFBSs and the location of their motifs at HNARs. We recovered human partial nucleosomal motif (Soufi et al. 2015) and canonical pou:sox motifs using mouse and human ES cell ChIP-seq data for POU5F1 (for logos, see Fig. 6A; for matrices, see Supplemental Table S2; Kunarso et al. 2010; Whyte et al. 2013). We then compared the relative abundances of human partial motif (P), mouse canonical (M), and human canonical (H) motifs across the POU5F1/Pou5f3 ChIP-seq data sets from human and mouse ES cells (Kunarso et al. 2010; Whyte et al. 2013), reprogramming experiments (Soufi et al. 2012; Chronis et al. 2017), and zebrafish embryos. The results indicated that only human POU5F1 can bind the partial nucleosomal motif, whereas canonical motifs are recognized by all POU5F1 homologs (Fig. 6B–D).
Figure 6.
Species-specific chromatin recognition features of POU5F1 homologs. (A) Logos of motifs recognized by human POU5F1 (P indicates partial; H, canonical), and by mouse POU5F1 (M indicates canonical). Arrows show the matches to the partial nucleosomal motif of Soufi et al. (2015) on the forward and reverse strands of human palindromic motif. (B–D) Densities of partial human motif (P), human canonical motif (H), and mouse canonical motif (M) in the POU5F1/Pou5f3 ChIP-seq data from the sources indicated at the top. (E–G) Nucleosome predictions for the ChIP-seq genomic regions from the sources indicated at the top.
Nucleosome-binding preferences of POU5F1 protein could indicate that it binds to the HNAR center in humans, and their absence mouse POU5F1 could indicate that it binds outside of the HNAR center. To test this, we examined the distribution of the predicted nucleosome occupancy around the POU5F1 ChIP-seq data sets (Fig. 6E,F). Single peaks of predicted nucleosome occupancy in human ES cells indicated that POU5F1 is located at the HNAR center, whereas bimodal peaks in mouse ES cells indicated that POU5F1 binds HNAR, but it is located at a distance from the HNAR center (Fig. 6E). In reprogramming experiments, human POU5F1, but not mouse POU5F1, could bind HNAR centers in a way similar to ES cells (Fig. 6F). The HNAR destabilization-depletion model plausibly explains the differences in pioneer properties of mouse and human POU5F1 orthologs: POU5F1, but not mouse POU5F1, specifically binds to the palindromic motif at the HNAR center, which may destabilize and shift strongly positioned nucleosomes. CEBPA TF poises mouse B cells for rapid reprogramming into iPS cells by OSKM (Di Stefano et al. 2014). CEBPA was strongly bound to HNAR centers in two experimental conditions (Fig. 6G; data from Di Stefano et al. 2016), suggesting that it may destabilize strongly positioned nucleosomes. We concluded that the HNAR model of zebrafish ZGA is applicable to other systems.
Discussion
We show that the affinity of both the TFs Pou5f3 and Nanog, and nucleosomes to the same locations within the genome is supported by the intrinsic DNA features favoring high in vitro nucleosome occupancy. We define the regions spanning 600 bp of high predicted nucleosome occupancy/PT/GC content around the central periodic structure as “high nucleosome affinity regions” (HNARs) to distinguish them from strong nucleosome positioning sequences of one nucleosome length (Fig. 3J). We show that Pou5f3 and Nanog binding is the cause, not a consequence, of chromatin opening at HNARs, which defines them as pioneer factors. ZGA TFs are involved in two steps of nucleosome displacement: nonspecific competition with histones on strong nucleosome footprints before ZGA and maintenance of open chromatin state by synergistic binding to HNARs after ZGA (Fig. 5I). We think that although the same TFs are involved in both steps, the two chromatin remodeling steps are driven by separate processes. Connection between the two steps can be described as a weak regulatory linkage (sensu Gerhart and Kirschner 2007).
Step 1: destabilization of nucleosomes by nonconsensus TF binding to periodic sequences at the HNAR centers
At the ninth cell cycle and before ZGA commences, Pou5f3 and Nanog bind to HNARs and reduce the nucleosome occupancy within the central nucleosome footprint in a nonspecific manner. We find unlikely that Pou5f3 acts by direct binding to nucleosomes, as it does not recognize human nucleosomal motifs (Fig. 6A–D). Our MNase-seq protocol digested the chromatin to ∼80% mononucleosomes. This method depletes “fragile” nucleosomes on the regulatory regions (Iwafuchi-Doi et al. 2016; Mieczkowski et al. 2016). Despite this, we see mixed ATAC-seq and MNase-seq signals on TFBS: This indicates that the step 1 process is not restricted to regulatory regions and is stochastic. Mixed signals presumably come from different cells: In some cells, HNARs are nucleosome-free, but in most cells, they are closed by nucleosomes. Pre-ZGA, the Pou5f3 ChIP-seq signal is enriched at all HNAR centers (Supplemental Fig. S17E). A single-cell, single-molecule imaging study of POU5F1 and SOX2 showed that nonconsensus interactions with chromatin are central to the in vivo search for functional binding sites (Chen et al. 2014). These nonconsensus interactions provide a measurable ChIP signal in a population of cells (Chen et al. 2014). We assume that the high sequencing depth of pre-ZGA Pou5f3 ChIP-seq (Leichsenring et al. 2013) allows us to detect nonconsensus Pou5f3 binding pre-ZGA. It was previously shown that multiple TFs outcompete nucleosomes from certain genomic locations (Afek et al. 2015). Nonconsensus binding is enhanced at genomic locations with periodic and symmetric DNA sequences in which nucleotides of different types alternate (Sela and Lukatsky 2011). HNAR centers are symmetric (Fig. 3J) and are underlain by alternating periodic nucleotide stretches (Kaplan et al. 2009), which make them likely templates for enhanced nonconsensus binding of multiple TFs. The theoretical model of TF–nucleosome competition on a single nucleosome footprint was suggested by Mirny (2010). One nontrivial prediction of his model is that stabilization of nucleosomes by strong nucleosome-positioning sequence will increase the nucleosome displacement effect of multiple cooperating TFs. This is exactly what we observed: Pou5f3 and Nanog pre-ZGA effects increase with nucleosome prediction strength, and so does ATAC-signal (Fig. 4). Given a nonspecific nature of the process, there is no reason to assume that Pou5f3 or Nanog are indispensable for it: Other TFs may be also involved. In summary, the simplest explanation for pre-ZGA nucleosome destabilization by Pou5f3 and Nanog is nonconsensus binding to periodic sequences at HNAR centers, where they compete with histones thus preventing nucleosome formation.
Step 2: post-ZGA nucleosome depletion by synergistic action of Pou5f3 and Nanog in the presence of SoxB1
We detected widespread alignment of nucleosomes on their DNA-encoded nucleosome positioning signals from pre- to post-ZGA (Fig. 4C, black arrows). Increase of nucleosome positioning strength was previously documented for promoters: Zygotic TSS selection grammar is characterized by nucleosome positioning signals, precisely aligned with the first downstream (+1) nucleosome (Haberle et al. 2014). Strengthening of nucleosome attachment to their DNA sites over ZGA is independent of transcription (Haberle et al. 2014; Zhang et al. 2014) but may depend on the elongation of the cell cycle or on the rapid exchange of maternal nucleosome-associated proteins, such as of conserved maternal linker histones B4/H1M to their zygotic variants (Müller et al. 2002). We speculate that after an initial short phase during the ninth to 10th cell cycle when histones may be nonspecifically outcompeted with TFs and vice versa (Joseph et al. 2017), temporally “open” chromatin sites are closed again unless stabilized by PSN-specific binding (step 2 in our model). Stabilization occurs at HNARs, which are bound by all three TFs (PSN group) and, in addition, carry specific pou:sox motif outside of the central nucleosome footprint (+90 bp position at Fig. 5I).
HNAR model and working hypotheses
The length of HNARs well matches the average size of developmental enhancers (Levine and Tjian 2003). It has been suggested that strong nucleosome positioning signals overlap with the enhancers (Tillo et al. 2010) and that pioneer TFs may recognize these signals (Barozzi et al. 2014; Sun et al. 2015). However, it is currently unclear how ubiquitously occurring strong nucleosome positioning signals differ from those contributing to the enhancers. Dinucleotide repeats that we detected on all TFBSs (Supplemental Fig. S2) and that had been previously characterized as general enhancer features (Yanez-Cuna et al. 2014) contribute to the periodic nucleosome positioning signal underlying HNAR center (Kaplan et al. 2009) and, at the same time, serve as a template for nonconsensus recognition by multiple TFs (Sela and Lukatsky 2011). We hypothesize that most of the TFs present in the cell transiently bind to HNAR centers by default. Further TF-specific DNA shape cues may be decoded as differential specific binding positions of the TFs within the HNAR relatively to its center. We suggest two testable hypotheses:
If TF destabilizes nucleosomes in a manner dependent on nucleosome prediction strength of the underlying sequence, this TF is involved in nonspecific nucleosome-mediated cooperativity at HNAR center (as in step 1 in Fig. 5I).
Functional TF binding motifs involved in transcriptional regulation reside in HNAR outside of medial nucleosome footprint (as pou:sox motif in step 2 in Fig. 5I).
The 300-bp PT periodic regions, similar in shape, can be recovered from zebrafish, yeast, and human genomes (Fig. 3F). HNARs can be distinguished from random regions by elevated PT and GC/AT ratio (Fig. 3I). These features promote nucleosome assembly and nonconsensus binding of various TFs: Thus, HNAR provides a platform for competition between histones and TFs. In a way, HNARs can be viewed as naturally occurring enhancer templates: Adding specific TF binding motifs to HNAR will make an enhancer. We envision that the HNAR concept will be instrumental for future studies of genome regulatory biology of eukaryotes.
Methods
Zebrafish maintenance and embryo collection
WT fish of AB/TL strain were raised, maintained, and crossed under standard conditions as described by Westerfield (2000). The mutant spgm793 and nanogm1435 lines were maintained as described previously (Lunde et al. 2004; Veil et al. 2018). Embryos obtained from crosses were collected within 10 min and raised in egg water at 28.5°C. Staging was performed following the 1995 Kimmel staging series (Kimmel et al. 1995). Stages of MZspgm793 and MZnanogm1435 embryos were indirectly determined by observation of WT embryos born at the same time and incubated under identical conditions. All experiments were performed in accordance with German Animal Protection Law (TierSchG).
MNase digestion and sequencing
Around 200–400 WT, MZspg and MZnanog embryos were staged to the 512-cell (2.75 h postfertilization) or dome (4.3 h postfertilization) stage. Embryo fixation and the MNase digestion was performed as previously described (Zhang et al. 2014). The yield of and degree of digestion were controlled using Agilent high sensitivity DNA kit on Agilent Bioanalyzer, according to the manufacturer's instructions. Chromatin was digested so that it contained 80% mononucleosomes (Supplemental Fig. S4). Libraries were prepared using the Illumina sequencing library preparation protocol and single-end sequenced on an Illumina HiSeq 2500 by Eurofins. Mapping, data processing, and statistical analysis details are in the Supplemental Material.
Data access
MNase-seq raw and processed data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE109410. Other processed data are available in the main text or the Supplemental Materials.
Supplementary Material
Acknowledgments
We thank Nadine Vastenhouw for sharing MNase-seq protocol; Wolfgang Driever, Sebastian Arnold, Patrick Lemaire, Meijiang Gao, and Marco Ell for commenting on the manuscript; Sabine Goetter for excellent fish care; and Andrea Buderer for administrative support. We especially thank Remo Rohs for maintaining the community access to TFBShape server. This work was supported by the Deutsche Forschungsgemeinschaft (DFG) DFG-ON86/4-1, DFG-ON86/4-2, and DFG-EXC294 A7-2 for D.O. The Freiburg Galaxy Team is funded by DFG grant SFB 992/1 2012 and BMBF grant 031 A538A RBC.
Author contributions: D.O. conceived the project; M.V. performed all the experiments; D.O., L.Y., and B.G. performed the data analysis; and D.O. and M.V. wrote the manuscript. All authors edited the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.240572.118.
References
- Adams CC, Workman JL. 1993. Nucleosome displacement in transcription. Cell 72: 305–308. 10.1016/0092-8674(93)90109-4 [DOI] [PubMed] [Google Scholar]
- Afek A, Cohen H, Barber-Zucker S, Gordân R, Lukatsky DB. 2015. Nonconsensus protein binding to repetitive DNA sequence elements significantly affects eukaryotic genomes. PLoS Comput Biol 11: e1004429 10.1371/journal.pcbi.1004429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. 2014. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell 54: 844–857. 10.1016/j.molcel.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M, Eisfeld K. 1997. Transcription factor access to chromatin. Nucleic Acids Res 25: 3559–3563. 10.1093/nar/25.18.3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belting HG, Wendik B, Lunde K, Leichsenring M, Mössner R, Driever W, Onichtchouk D. 2011. Pou5f1 contributes to dorsoventral patterning by positive regulation of vox and modulation of fgf8a expression. Dev Biol 356: 323–336. 10.1016/j.ydbio.2011.05.660 [DOI] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49: 825–837. 10.1016/j.molcel.2013.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. 2014. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156: 1274–1285. 10.1016/j.cell.2014.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, Plath K. 2017. Cooperative binding of transcription factors orchestrates reprogramming. Cell 168: 442–459 e420. 10.1016/j.cell.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HR, Vingron M. 2009. Sequence-dependent nucleosome positioning. J Mol Biol 386: 1411–1422. 10.1016/j.jmb.2008.11.049 [DOI] [PubMed] [Google Scholar]
- Daenen F, van Roy F, De Bleser PJ. 2008. Low nucleosome occupancy is encoded around functional human transcription factor binding sites. BMC Genomics 9: 332 10.1186/1471-2164-9-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano B, Sardina JL, van Oevelen C, Collombet S, Kallin EM, Vicent GP, Lu J, Thieffry D, Beato M, Graf T. 2014. C/EBPα poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 506: 235–239. 10.1038/nature12885 [DOI] [PubMed] [Google Scholar]
- Di Stefano B, Collombet S, Jakobsen JS, Wierer M, Sardina JL, Lackner A, Stadhouders R, Segura-Morales C, Francesconi M, Limone F, et al. 2016. C/EBPα creates elite cells for iPSC reprogramming by upregulating Klf4 and increasing the levels of Lsd1 and Brd4. Nat Cell Biol 18: 371–381. 10.1038/ncb3326 [DOI] [PubMed] [Google Scholar]
- el Hassan MA, Calladine CR. 1996. Propeller-twisting of base-pairs and the conformational mobility of dinucleotide steps in DNA. J Mol Biol 259: 95–103. 10.1006/jmbi.1996.0304 [DOI] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216 10.1371/journal.pcbi.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Obbad K, Schier AF. 2018. The primary role of zebrafish nanog is in extra-embryonic tissue. Development 145: dev147793 10.1242/dev.147793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Guan J, Zhou S, Zhang W. 2012. Structural features based genome-wide characterization and prediction of nucleosome organization. BMC Bioinformatics 13: 49 10.1186/1471-2105-13-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Wu K, Liu Z, Yao X, Yuan S, Tao W, Yi L, Yu G, Hou Z, Fan D, et al. 2018. Chromatin accessibility landscape in human early embryos and its association with evolution. Cell 173: 248–259 e215. 10.1016/j.cell.2018.02.028 [DOI] [PubMed] [Google Scholar]
- Gerhart J, Kirschner M. 2007. The theory of facilitated variation. Proc Natl Acad Sci 104: 8582–8589. 10.1073/pnas.0701035104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Li N, Hadzhiev Y, Plessy C, Previti C, Nepal C, Gehrig J, Dong X, Akalin A, Suzuki AM, et al. 2014. Two independent transcription initiation codes overlap on vertebrate core promoters. Nature 507: 381–385. 10.1038/nature12974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. 2010. Nucleosome dynamics define transcriptional enhancers. Nat Genet 42: 343–347. 10.1038/ng.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. 2016. Cell fate control by pioneer transcription factors. Development 143: 1833–1837. 10.1242/dev.133900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. 2016. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol Cell 62: 79–91. 10.1016/j.molcel.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SR, Pálfy M, Hilbert L, Kumar M, Karschau J, Zaburdaev V, Shevchenko A, Vastenhouw NL. 2017. Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. eLife 6: e23326 10.7554/eLife.23326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366. 10.1038/nature07667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoueiry P, Rothbächer U, Ohtsuka Y, Daian F, Frangulian E, Roure A, Dubchak I, Lemaire P. 2010. A cis-regulatory signature in ascidians and flies, independent of transcription factor binding sites. Curr Biol 20: 792–802. 10.1016/j.cub.2010.03.063 [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kornberg RD. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184: 868–871. 10.1126/science.184.4139.868 [DOI] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. 2010. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634. 10.1038/ng.600 [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244. 10.1038/ng2117 [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ. 2013. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503: 360–364. 10.1038/nature12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M, Maes J, Mossner R, Driever W, Onichtchouk D. 2013. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science 341: 1005–1009. 10.1126/science.1242527 [DOI] [PubMed] [Google Scholar]
- Levine M, Tjian R. 2003. Transcription regulation and animal diversity. Nature 424: 147–151. 10.1038/nature01763 [DOI] [PubMed] [Google Scholar]
- Li R, Cauchy P, Ramamoorthy S, Boller S, Chavez L, Grosschedl R. 2018. Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming. Genes Dev 32: 96–111. 10.1101/gad.309583.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. 2008. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456: 400–403. 10.1038/nature07388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Wang W, Hu S, Wang X, Zhang Y. 2018. Inherited DNA methylation primes the establishment of accessible chromatin during genome activation. Genome Res 28: 998–1007. 10.1101/gr.228833.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, Zhang Y. 2016. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 165: 1375–1388. 10.1016/j.cell.2016.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde K, Belting HG, Driever W. 2004. Zebrafish pou5f1/pou2, homolog of mammalian Oct4, functions in the endoderm specification cascade. Curr Biol 14: 48–55. 10.1016/j.cub.2003.11.022 [DOI] [PubMed] [Google Scholar]
- Luo Y, North JA, Rose SD, Poirier MG. 2014. Nucleosomes accelerate transcription factor dissociation. Nucleic Acids Res 42: 3017–3027. 10.1093/nar/gkt1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier M, Grant J, Dowdle A, Thomas A, Gerton J, Collas P, O'Sullivan JM, Horsfield JA. 2018. Cohesin facilitates zygotic genome activation in zebrafish. Development 145: dev156521 10.1242/dev.156521 [DOI] [PubMed] [Google Scholar]
- Mieczkowski J, Cook A, Bowman SK, Mueller B, Alver BH, Kundu S, Deaton AM, Urban JA, Larschan E, Park PJ, et al. 2016. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat Commun 7: 11485 10.1038/ncomms11485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA. 2010. Nucleosome-mediated cooperativity between transcription factors. Proc Natl Acad Sci 107: 22534–22539. 10.1073/pnas.0913805107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle-Heyrman G, Tims HS, Widom J. 2011. Structural constraints in collaborative competition of transcription factors against the nucleosome. J Mol Biol 412: 634–646. 10.1016/j.jmb.2011.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Thisse C, Thisse B, Raz E. 2002. Expression of a linker histone-like gene in the primordial germ cells in zebrafish. Mech Dev 117: 253–257. 10.1016/S0925-4773(02)00174-0 [DOI] [PubMed] [Google Scholar]
- Murphy PJ, Wu SF, James CR, Wike CL, Cairns BR. 2018. Placeholder nucleosomes underlie germline-to-embryo DNA methylation reprogramming. Cell 172: 993–1006 e1013. 10.1016/j.cell.2018.01.022 [DOI] [PubMed] [Google Scholar]
- Onichtchouk D, Geier F, Polok B, Messerschmidt DM, Mössner R, Wendik B, Song S, Taylor V, Timmer J, Driever W. 2010. Zebrafish Pou5f1-dependent transcriptional networks in temporal control of early development. Mol Syst Biol 6: 354 10.1038/msb.2010.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatsenko D, Goltsev Y, Levine M. 2009. Organization of developmental enhancers in the Drosophila embryo. Nucleic Acids Res 37: 5665–5677. 10.1093/nar/gkp619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvin MS, Okuyama N, Inoue F, Islam ME, Kawakami A, Takeda H, Yamasu K. 2008. Autoregulatory loop and retinoic acid repression regulate pou2/pou5f1 gene expression in the zebrafish embryonic brain. Dev Dyn 237: 1373–1388. 10.1002/dvdy.21539 [DOI] [PubMed] [Google Scholar]
- Satchwell SC, Drew HR, Travers AA. 1986. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol 191: 659–675. 10.1016/0022-2836(86)90452-3 [DOI] [PubMed] [Google Scholar]
- Schulz KN, Bondra ER, Moshe A, Villalta JE, Lieb JD, Kaplan T, McKay DJ, Harrison MM. 2015. Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res 25: 1715–1726. 10.1101/gr.192682.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang JP, Widom J. 2006. A genomic code for nucleosome positioning. Nature 442: 772–778. 10.1038/nature04979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela I, Lukatsky DB. 2011. DNA sequence correlations shape nonspecific transcription factor-DNA binding affinity. Biophys J 101: 160–166. 10.1016/j.bpj.2011.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordan R, Rohs R. 2014. Absence of a simple code: how transcription factors read the genome. Trends Biochem Sci 39: 381–399. 10.1016/j.tibs.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151: 994–1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. 2015. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161: 555–568. 10.1016/j.cell.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13: 613–626. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- Sun Y, Nien CY, Chen K, Liu HY, Johnston J, Zeitlinger J, Rushlow C. 2015. Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res 25: 1703–1714. 10.1101/gr.192542.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042. 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. 10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Hughes TR. 2009. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10: 442 10.1186/1471-2105-10-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR. 2010. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One 5: e9129 10.1371/journal.pone.0009129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. 2011. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147: 577–589. 10.1016/j.cell.2011.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veil M, Schaechtle MA, Gao M, Kirner V, Buryanova L, Grethen R, Onichtchouk D. 2018. Maternal Nanog is required for zebrafish embryo architecture and for cell viability during gastrulation. Development 145: dev155366 10.1242/dev.155366 [DOI] [PubMed] [Google Scholar]
- Westerfield M. 2000. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). University of Oregon Press, Eugene, OR. [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan ZP, Müller P, Fogley R, Dibiase A, Trompouki E, Unternaehrer J, Xiong F, Torregroza I, Evans T, et al. 2012. Nanog-like regulates endoderm formation through the Mxtx2-nodal pathway. Dev Cell 22: 625–638. 10.1016/j.devcel.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Cuna JO, Arnold CD, Stampfel G, Boryn LM, Gerlach D, Rath M, Stark A. 2014. Dissection of thousands of cell type-specific enhancers identifies dinucleotide repeat motifs as general enhancer features. Genome Res 24: 1147–1156. 10.1101/gr.169243.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Vastenhouw NL, Feng J, Fu K, Wang C, Ge Y, Pauli A, van Hummelen P, Schier AF, Liu XS. 2014. Canonical nucleosome organization at promoters forms during genome activation. Genome Res 24: 260–266. 10.1101/gr.157750.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. 2009. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462: 65–70. 10.1038/nature08531 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.