Abstract
There continues to be interest in targeting epigenetic ‘readers, writers and erasers’ for the treatment of cancer and other pathologies. A mechanistic understanding is frequently lacking, however, for the synergy observed when combining deacetylase and bromodomain inhibitors. Here we identify cell cycle and apoptosis regulator 2 (CCAR2) as an early target for acetylation in colon cancer cells treated with sulforaphane (SFN). N-terminal acetylation of CCAR2 diminished its interactions with histone deacetylase 3 (HDAC3) and β-catenin, interfering with Wnt coactivator functions of CCAR2 including in cells harboring genetically encoded CCAR2 acetylation. Protein domain arrays and pull-down assays identified acetyl ‘reader’ proteins that recognized CCAR2 acetylation sites, including BRD9 and members of the bromodomain and extraterminal domain (BET) family. Treatment with the BET inhibitor JQ1 synergized with SFN in colon cancer cells and suppressed tumor development effectively in a preclinical model of colorectal cancer. Studies with SFN+JQ1 in combination implicated a BET/BRD9 acetyl switch and a shift in the pool of acetyl ‘reader’ proteins in favor of BRD9-regulated target genes.
Introduction
Cell cycle and apoptosis regulator 2 (CCAR2), also known as DBC1/KIAA1967, has gained attention as a ‘master regulator’ of metabolism, aging, and cancer (1–4). This designation derives from the interactions of CCAR2 with protein partners that exert critical roles in physiology and pathophysiology, including Sirtuin 1 (SIRT1) and CHK2, linking CCAR2 to p53 function and DNA repair (1–7). Less is known about the N-terminal region of CCAR2 that associates with, and inhibits, histone deacetylase 3 (HDAC3), while also interacting with β-catenin to stabilize β-catenin/Tcf complexes in the nucleus (6,7). In so doing, CCAR2 serves as a coactivator of Wnt signaling, a well-studied pathway in disease and development (8).
Our attention was drawn to CCAR2 based on two converging observations. First, when CCAR2 is overexpressed in colon tumors, the corresponding patients exhibit significantly reduced survival (7). Second, as reported here, CCAR2 was identified as an early target for acetylation by sulforaphane (SFN), an agent that causes inhibition and turnover of HDAC3 in colon cancer cells (9–12). Notably, when SFN was combined with JQ1, an inhibitor of the bromodomain and extraterminal domain (BET) family (13–15), CCAR2 no longer served as an effective coactivator of Wnt/β-catenin signaling in vitro and in vivo.
There is growing interest in targeting epigenetic ‘readers, writers, and erasers’ deregulated in cancer and other pathologies (13–16). This investigation combined SFN+JQ1 to affect CCAR2 acetylation, and in so doing provided new mechanistic insights into the competition that exists among the ‘readers’ of acetylated histone and non-histone proteins that are regulated during epigenetic combination therapies.
Materials and Methods
Cells and treatments
HCT116, SW480 (human colon cancer cells) and CCD841 (non-transformed colonic epithelial cells) were from ATCC (Manassas, VA, USA), and used within 10–15 passages from receipt. Each cell line was confirmed independently to be of human origin, with no mammalian inter-species contamination, and with the correct genetic profile based on allele-specific markers (Idexx Radil, Columbia, MO (17,18)). Cells were cultured in McCoy’s 5A media (Invitrogen) or EMEM (Invitrogen), supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin, at 370C in a humidified chamber with 5% CO2. All cells were tested routinely for mycoplasma by DAPI staining, and by using a PCR-based methodology (19).
JQ1 was purchased from MedChem Express (Monmouth Junction, NJ), whereas the other test agents were from the sources noted elsewhere (11). Nominal concentrations were as follows: 15 μM SFN, 6-methylsulfinylhexyl isothiocyanate (6-SFN), 9-methylsulfinylnonyl isothiocyanate (9-SFN), and allyl isothiocyanate (AITC); 1 μM trichostatin A (TSA); 10 mM sodium butyrate (NaB); and 1 mM valproic acid (VPA). For combination index (CI) experiments, SFN, JQ1, and suberoylanilide hydroxamic acid (SAHA) were tested in the range 2–17 μM, 1–60 μM and 0.1–2 μM, respectively, with dimethylsulfoxide (DMSO) as vehicle. In most experiments, cells were treated with test agents 24 h after seeding (10,11), except in HDAC3 siRNA knockdown assays, which were conducted according to a published methodology (11,12). HDAC3 siRNA (Trilencer-27) and control siRNA were procured from Origene, and cells were transfected with RNAiMAX reagent (Invitrogen) for 24–48 h, using the manufacturer’s protocol. Two of the target siRNAs, designated as siRNA(1) and siRNA(3), produced the most efficient knockdown of HDAC3, and the data are shown in the corresponding figures. Unless indicated otherwise, whole cell lysates or nuclear and cytoplasmic fractions (10) were harvested 6 h after treatment with test agents, followed by RNA or protein expression analyses.
Additional experiments involved CCAR2 deletion from colon cancer cells via CRISPR/Cas9 genome editing (20,21). The PX459 Vector control (Addgene) included a non-targeting gRNA sequence integrated into the vector. For re-introduction of CCAR2 into CCAR2-null cells, transient transfection was conducted using expression constructs for WT protein or acetylation mutants. In the latter case, a Q5 Site-Directed Mutagenesis kit (New England BioLabs, Ipswich, MA) was used to convert Lys to Arg, starting with CCAR2 plasmid pcDNA Myc DBC1 (Addgene plasmid #35096) (22), with confirmation by direct sequencing.
Genetically encoded acetylation of CCAR2
A system for genetically encoded Lys acetylation on histones (23) was adopted for CCAR2. In brief, CCAR2 and 3xHA were PCR amplified and sub-cloned into pGEM-9Zf(−) (Promega # P2391) to generate HA-CCAR2. HA-CCAR2 was restriction cloned into pE337, replacing H3.3-HA. Q5 Site-Directed Mutagenesis was used to convert Lys to TAG stop codons at defined sites in CCAR2. Plasmids pE312 and pE337-HA-CCAR2 WT and Lys mutants were stably expressed in CCAR2 null HCT116 cells using Super PiggyBac Transposase vector (SBI #PB210PA-1) and selected with puromycin and neomycin. Cells were treated for 24 h with 10 mM N-acetyl-L-lysine to express acetylated CCAR2 (Sigma #A4021).
Immunoblotting (IB) and immunohistochemistry (IHC)
IB used published procedures for whole cell lysates, nuclear/cytoplasmic fractions, and tissue lysates of colon tumors or normal colon biopsies (9–11,17,24). Antibody to CCAR2 was from Bethyl Labs (Montgomery, TX), whereas acetyl-lysine (Ac-Lys), histone H3, histone H4, histone H4K12-acetylated (H4K12ac), 14–3-3, RAD54, HDAC3, β-catenin, c-Myc, cyclin D1, matrix metalloproteinase 7 (MMP7), poly(ADP-ribose)polymerase (PARP), Caspase-3, Pin1, Lamin, and β-actin primary antibodies were from sources reported (9–11,17,24). IHC followed the general procedures described elsewhere (17,24).
Proximity ligation assays (PLA)
Protein-protein interactions were examined in situ, in cell-based assays and tissue sections, using the Duolink PLA Fluorescence Protocol (Sigma-Aldrich, St. Louis, MO), according to the manufacturer’s recommendations.
Pulldown assays
Immunoprecipitation (IP) methodologies were as reported for endogenous proteins (10,11), or Myc-, GST- and HA-tagged proteins (5,25–27).
Mass spectrometry
Acetylation sites on CCAR2 were identified following the general approach reported (5). In brief, 24 h after seeding, HCT116 cells were treated with SFN or DMSO, and 6 h later the cell lysates were subjected to IP using CCAR2 antibody. Following SDS-PAGE separation, the CCAR2 band was excised from the gel and digested overnight with trypsin prior to extraction and analysis on an Eksigent cHiPLC™ with nanoLC linked via a nanoflex to an ABSCIEX TripleTOF 5600™ mass spectrometer (Mass Spectrometry-Proteomics Core, Baylor College of Medicine, Houston, TX). Peaks Studio version 7.0 (Bioinformatics Solutions Inc.) was used to match spectra to peptides using the NCBI non-redundant database, including consideration of lysine acetylation. Modified peptides were verified by manual inspection of MS/MS data.
RNA analyses
RNA-seq sequencing (RNA-seq) and bioinformatics analyses were as reported (28) for adenomatous colon polyps from familial adenomatous polyposis (FAP) patients (GSE88945, GSE106500) and the polyposis in rat colon (Pirc) preclinical model (29). Library preparation via a NEBNext® Ultra™ Directional RNA Library Prep Kit was followed by Illumina® sequencing on a NextSeq 500/550 instrument (Illumina, La Jolla, CA). Real-time reverse transcription quantitative PCR (RT-qPCR) used a reported methodology (28).
Docking in silico
After multiple sequence alignment (30), docking of BRD2, BRD3, BRD4 and BRD9 was performed using AutoDock Vina (31), on CCAR2 structures predicted via SWISS-MODEL (32). Ligand-protein interactions were analyzed using PDBePISA (33,34) and LPC/CSU (35). Initial work-up confirmed that the docking of JQ1 with BRD2, BRD3, and BRD4 corresponded favorably with the reported orientations (13).
Chromatin immunoprecipitation (ChIP)
The ChIP-IT Express Enzymatic kit (Active Motif, Carlsbad, CA) was used, as reported (12). Following drug treatment, HCT116 cells were cross-linked with formaldehyde and homogenized in order to isolate the nuclear fraction. DNA fragmentation was performed using a Biorupter for 15 cycles of 20 secs each. Ten microliters of fragmented chromatin was kept as input, while the remainder was subjected to IP with anti-CCAR2 (Cell Signaling), BRD9 (Active Motif), or BRD3 (Active Motif) antibodies. After reversing the cross-linking, and proteinase treatment, DNA was purified using the QIAquick PCR Purification kit (Qiagen). PCR was run on a Roche Light Cycler 480 II with pre-incubation for 5 min at 95 °C, then 55 cycles at 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 10 s. Each experiment was repeated at least twice. Primer sequences were as follows:
E1 (Forward Primer) TTGTCGCAGGTATGCTGAGTC
E1 (Reverse Primer) TGTGATTACCCAGGCACACT
E2 (Forward Primer) TCCTGAGTCACGGAGTTGTCT
E2 (Reverse Primer) TGCGATCTTCAGAGGGCCTA
E3 (Forward Primer) GAGATTACAGGGAGTGGCAGTG
E3 (Reverse Primer) TGGAAACTCAGATACTCCTGGG
E4 (Forward Primer) CTCCCGAGGGCGATAAAAGG
E4 (Reverse Primer) GGATGTTTGCTGGAACGCTG
Promoter (Forward Primer) TGCATGACCGCATTTCCAATA
Promoter (Reverse Primer) CGGACAAACCGGACGTTTAATTC
Preclinical experiments
All studies were approved by the Institutional Animal Care and Use Committee. For xenograft experiments, 5×106 cells (SW480 CCAR2 CRISPR/Cas9 knockout or vector controls) were injected into either flank of male athymic nude mice (Envigo, Somerset, NJ). After 10 d, animals were randomized as follows (n=5 mice/group): SFN, 100 mg/kg body weight (BW) via daily oral gavage; JQ1, 50 mg/kg BW, twice weekly i.p. injection; SFN+JQ1, at doses of the individual compounds, or vehicle. Tumor volumes were measured twice/week using calipers. In rat experiments, Pirc males (29) at 5 months of age were assigned to study groups (3–4/group), and 2 months later occluding colon polyps were resected (36). Rats were then treated for 5 weeks with test agents, as follows: SFN, 400 parts per million (p.p.m.) in AIN93 diet; JQ1, 12.5 mg/kg BW via twice weekly i.p. injection; SFN+JQ1, at the doses of the individual compounds, or vehicle. The study was terminated 2 months after polypectomy, and GI lesions were enumerated prior to IB and RNA-seq, as reported (28). To our knowledge, this is the first report to examine secondary prevention in a murine model of FAP, following surgical intervention.
Statistics
Results are representative of findings from at least three independent experiments, expressed as mean±SE, unless indicated otherwise. Student’s t-test was used for paired comparisons, whereas multiple groups were subjected to analysis of variance and Bonferroni’s test (GraphPad Prism™ v5.04, La Jolla, CA, USA). Statistical significance was shown in the corresponding figures, as follows: *P<0.05, **P<0.01, ***P<001, ****P<0.0001.
Results
Novel acetylation sites are produced on CCAR2 by SFN
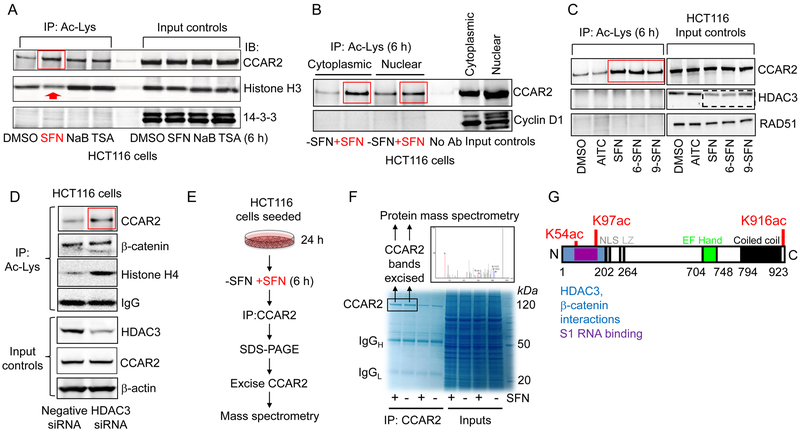
Whole cell lysates were prepared from HCT116 colon cancer cells incubated with SFN for 6 h, and anti-acetyl-lysine (Ac-Lys) antibody was used to IP endogenous acetylated proteins. IB confirmed the increased histone acetylation after treatment with pan-HDAC inhibitors NaB and TSA, but not by SFN at 6 h (Fig. 1A, red arrow). Under the same conditions, robust acetylation of CCAR2 was detected in SFN-treated cells (Fig. 1A, red box), comparable to that of NaB and TSA. None of the test agents caused 14–3-3 acetylation. Similar observations were made when SW480 cells were treated with SFN, TSA, SAHA, NaB, or VPA (Supplementary Fig. S1); all of the test agents except SFN (arrow) caused increased histone acetylation at 6 h, and all of the compounds – including SFN (box) – produced a marked increase in CCAR2 acetylation without affecting the acetylation status of its paralog, CCAR1. Acetylation of CCAR2 at 6 h occurred in both the cytoplasmic and nuclear compartments (Fig. 1B), whereas SFN had no effect on the acetylation status of cyclin D1. Structural analogs of SFN that also were reported to inhibit HDAC activity and to turnover HDAC3 protein (11), namely 6-SFN and 9-SFN, similarly increased the acetylation status of CCAR2 without affecting a negative control, RAD51 (Fig. 1C). Increased CCAR2 acetylation was not observed for allyl isothiocyanate (AITC), which lacks HDAC inhibitory activity in colon cancer cells (11). No HDAC3 acetylation was detected under conditions in which HDAC3 protein levels were reduced by SFN/6-SFN/9-SFN at 6 h (Fig. 1C, dashed box). SiRNA-mediated knockdown of HDAC3 recapitulated the induction of CCAR2 acetylation in colon cancer cells (Fig. 1D and Supplementary Fig. S2), without changing the acetylation status of β-catenin (Fig. 1D). Thus, CCAR2 acetylation can occur in colon cancer cells in the absence of similar changes to other non-histone proteins, including β-catenin and HDAC3.
Figure 1.
CCAR2 is an early target for acetylation in SFN-treated colon cancer cells. A, HCT116 cells were treated with test agents, and 6 h later cell lysates were subjected to IP with Ac-Lys antibody. B, Protocol from A, applied to nuclear and cytoplasmic extracts. C, Protocol from A, repeated with SFN analogs 6-SFN, 9-SFN and AITC. D, siRNA-mediated knockdown of HDAC3 and IP/IB of cell lysates, as indicated. For HDAC3 knockdown in SW480 cells, refer Fig. S2. E,F, After IP and SDS-PAGE, CCAR2 was excised from the gel, digested with trypsin, and analyzed by protein mass spectrometry. G, Positions of SFN-induced acetylation sites. NLS, nuclear localization signal; LZ, leucine zipper. The IP and IB data shown in each figure panel are from a single experiment in each case, and are representative of the findings from three or more independent experiments.
CCAR2 was pulled down from colon cancer cells (Fig. 1E), and after protein separation the CCAR2 band was excised from the gel (Fig. 1F) and subjected to tandem mass spectrometry. Following SFN treatment, three novel acetylation sites were identified on CCAR2 at K54, K97, and K916 (Fig. 1G and Supplementary Fig. S3). N-terminal acetylation sites were within a region that interacts with HDAC3 and β-catenin, whereas C-terminal acetylation was adjacent to a coiled coil domain (Fig. 1G).
CCAR2 acetylation interferes with Wnt coactivator functions
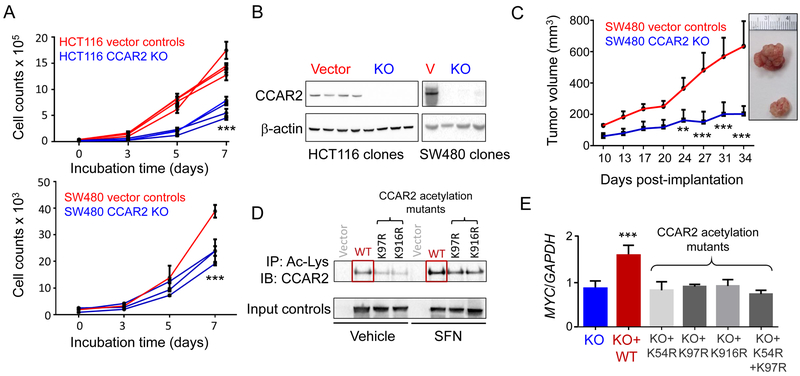
To examine the functional consequences of SFN-induced CCAR2 acetylation, we first deleted CCAR2 from colon cancer cells using CRISPR/Cas9 (Fig. 2A and 2B). Clones that lacked CCAR2 protein had reduced growth rates compared with the vector controls in vitro (Fig. 2A), and when injected into nude mice (Fig. 2C). CCAR2 was then re-introduced back into CCAR2-null cells via transient transfection of the corresponding expression constructs, either as wild type (WT) CCAR2 or as acetylation mutants. IP with Ac-Lys antibody revealed low basal acetylation for WT CCAR2 in the vehicle controls, which was increased after SFN treatment, but was less marked for the acetylation mutants K97R and K916R (Fig. 2D). Starting with CCAR2 null cells, re-introduction of acetylation mutants K54R, K97R, and K916R, or the double-mutant K54R/K97R, had no effect on MYC expression, whereas reintroduction of WT CCAR2 increased MYC levels significantly (Fig. 2E). Similar results were obtained for MMP7 (Supplementary Fig. S4A), and while this was reversed by SFN treatment following transient transfection of WT CCAR2, acetylation mutants such as K54R were resistant to SFN (Supplementary Fig. S4B).
Figure 2.
CCAR2 acetylation lowers oncogene expression in colon cancer cells. A, Deletion of CCAR2, with each line signifying a different clone, and each data-point representing mean±SD (n=3). B, Confirmation by IB of CCAR2 loss after CRISPR/ Cas9 genome editing. V, vector; KO, knockout. C, Xenograft studies in mice. Each data-point represents mean±SD (n=5). D, CCAR2 null colon cancer cells transiently transfected with vector control, or expression constructs for wild type (WT) or acetylation mutants of CCAR2. After 24 h, cells were treated with SFN or vehicle, and 6 h later cell lysates were subjected to IP/IB, as indicted. E, After treatment of colon cancer cells as indicated in D, total RNA was isolated and RT-qPCR was performed for MYC normalized to GAPDH. Data bars = mean±SD (n=3). **P<0.01 and ***P<0.001 significant difference from the corresponding vector/KO control. The data shown in each figure panel are from a single experiment in each case, and are representative of the findings from two or more independent experiments.
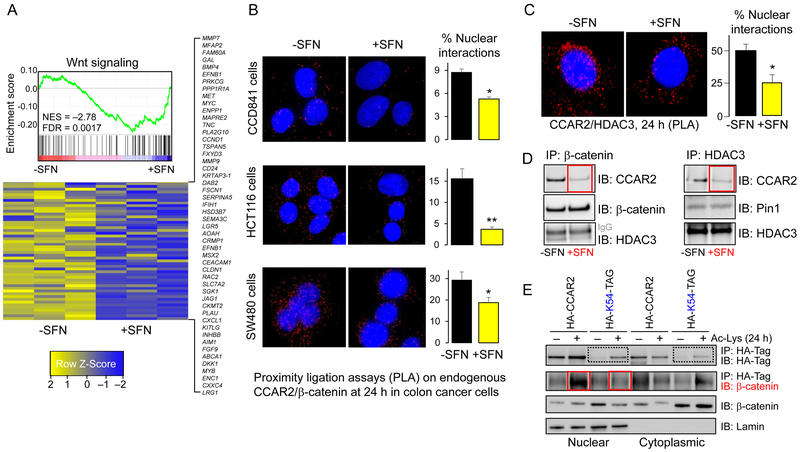
In addition to MYC and MMP7, RNA-seq revealed a suit of Wnt/β-catenin target genes downregulated in SFN-treated colon cancer cells (Fig. 3A). Next, we examined direct interactions between endogenous CCAR2 and β-catenin proteins via PLA (37,38). In CCD841 non-transformed colonic epithelial cells, CCAR2/β-catenin interactions were detected at low levels, whereas numerous interactions were observed in HCT116 and SW480 cells (Fig. 3B, red dots). After SFN treatment, fewer CCAR2/β-catenin interactions were detected, especially in the nucleus. Using the same approach, we also detected diminished CCAR2/HDAC3 nuclear interactions (Fig. 3C).
Figure 3.
CCAR2 protein interactions are disrupted by SFN treatment, and genetically encoded acetylation of CCAR2 Lys 54 blocks β-catenin interactions. A, RNA-seq data from HCT116 colon cancer cells, 6 h after treatment with SFN or vehicle. Each column is a biological replicate (n=3). Wnt signaling was among the top five cancer-related pathways altered by SFN (49), and gene set enrichment analysis (GSEA) prioritized 118 Wnt-related genes among 22727 genes in the dataset. B, PLA identified endogenous interactions of CCAR2 and β-catenin proteins. Cells were imaged 24 h after treatment with SFN or vehicle. C, The approach in B, used to examine endogenous CCAR2/HDAC3 interactions. Data bars designate mean±SD (n=3); *P<0.05, **P<0.01, compared with vehicle. D, 24 h after treating HCT116 cells with SFN or vehicle, nuclear extracts were subjected to IP/IB. E, A system for genetically encoding lysine modifications on histones (23) was used to engineer K54 acetylation on CCAR2. Nuclear and cytoplasmic extracts were subjected to IP/IB with the antibodies shown, 24 h after addition of Ac-Lys, to trigger the designed acetylation on CCAR2. The data shown in each figure panel are from a single experiment in each case, and are representative of the findings from two or more independent experiments.
To corroborate these findings, we pulled-down endogenous β-catenin or HDAC3 from nuclear extracts of colon cancer cells (Fig. 3D), and confirmed that interactions with CCAR2 were reduced markedly after SFN treatment (red boxes). Peptidyl-prolyl cis/trans isomerase 1 (Pin1), which interacts with HDAC3 in the nuclear compartment (10), was used as a control in some experiments. We conclude that CCAR2/HDAC3/β-catenin interactions are disrupted in SFN-treated colon cancer cells, interfering with the Wnt coactivator role of CCAR2 (Supplementary Fig. S5).
Next, a system for genetically encoding lysine modifications on histones (23) was used, for the first time, to engineer acetylation sites on a non-histone protein, CCAR2. Starting with CCAR2 null colon cancer cells, stable clones were generated containing HA-tagged CCAR2 or HA-tagged CCAR2-K54-TAG (abbreviated hereafter as HA-CCAR2 and HA-K54-TAG) – the ‘TAG’ premature stop codon preventing protein expression in the absence of acetyl-lysine (Ac-Lys) reagent (23). Twenty-four hours after the addition of Ac-Lys, nuclear and cytoplasmic extracts were subjected to IP with an antibody to the HA-tag on CCAR2, followed by IB with the same antibody (Fig. 3E). In cells stably transfected with HA-K54-TAG, no band was detected until the addition of Ac-Lys reagent, consistent with the formation of genetically encoded CCAR2-K54 acetylated protein (Fig. 3E, dotted boxes). In the presence of Ac-Lys reagent, IP with HA antibody followed by IB for β-catenin revealed a strong band in the nuclear compartment of cells stably transfected with HA-CCAR2 but not HA-K54-TAG (Fig. 3E, solid boxes). Thus, acetylation of Lys 54 on CCAR2 was sufficient to block its interactions with β-catenin in the nuclear compartment. Colon cancer cells also were generated containing stably transfected HA-K97-TAG; no CCAR2-K97ac band was detected after Ac-Lys treatment (Supplementary Fig. S6). The K97ac site may be destabilizing to CCAR2 under the conditions used, in the absence of K54ac.
CCAR2 acetyl ‘readers’ include BET family members and BRD9
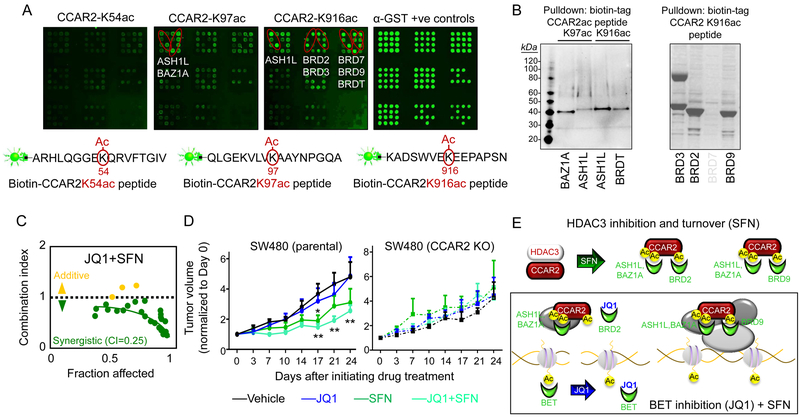
Based on the CCAR2 acetylation sites observed after SFN treatment (Fig. 1G), we turned our attention to the acetyl ‘reader’ proteins. Biotin-tagged peptide mimetics of CCAR2 were screened (25–27) via protein arrays comprising all known acetyl readers (Fig. 4A), followed by GST-pulldown assays for validation (Fig. 4B). CCAR2-K97ac peptide was recognized by bromodomains of ASH1L and BAZ1A, whereas CCAR2-K916ac peptide interacted with bromodomains of ASH1L, BRDT, BRD2, BRD3 and BRD9. The arrays also implicated BRD7 interacting with CCAR2-K916ac peptide, but this was not corroborated in pulldown experiments (Fig. 4B, right panel). Protein arrays did not recognize a peptide mimetic for CCAR2-K54ac (Fig. 4A, left panel).
Figure 4.
Acetylation sites on CCAR2 are recognized by acetyl ‘reader’ proteins. A, Protein arrays comprising all BROMO/ PHD/PWWP/YEATS domains were screened using peptide mimetics of acetylated CCAR2, followed B, by GST-pulldown assays for validation. C, Viability of colon cancer cells; CI, combination index. D, Mice were injected in either flank with parental SW480 cells or SW480 CCAR2 KO cells, and 10 d later animals were treated with test agents (see Methods). Data-points, mean±SD (n=5); **P<0.01, *P<0.05 compared with vehicle time-point. E, Working model of acetyl readers and CCAR2 interactions. The data shown in each figure panel are from a single experiment in each case, and are representative of the findings from two or more independent experiments.
The BET members BRD2, BRD3, and BRDT, along with BRD4, interact with high specificity on the arrays with JQ1 (M.T. Bedford, manuscript in preparation). However, CCAR2 peptide mimetics did not interact with BRD4 on the arrays, despite favorable docking scores in silico (Supplementary Fig. S7). Docking scores supported the preferred interactions of BET members and BRD9 with CCAR2K916ac versus H4K16ac, suggesting a scenario in which acetylated CCAR2 competes with acetylated histones for the binding of acetyl readers and their inhibitors, such as JQ1.
JQ1 exhibited strong synergy with SFN in colon cancer cells, the combination index of 0.25 (Fig. 4C) being comparable to 0.13 and 0.33, respectively, for JQ1 plus SAHA or 6-SFN (Supplementary Fig. S8A). Co-treatment with JQ1 plus SFN or 6-SFN increased the levels of cleaved PARP and cleaved Caspase-3, indicating enhanced apoptosis in colon cancer cells (Supplementary Fig. S8B, dotted boxes), consistent with prior studies using SFN and 6-SFN alone (10–12). Next, we took SW480 cells that are reported to be resistant to JQ1 (39), but also exhibited reduced CCAR2/β-catenin interactions after SFN treatment (Fig. 3B), and examined their growth in nude mice. As expected for a JQ1 resistant cell line (39), JQ1 alone had no effect, but JQ1 enhanced the tumor suppressive actions of SFN in vivo (Fig. 4D, left panel, JQ1+SFN, **P<0.01) despite an apparent lack of synergy under the conditions employed. When mice were injected with SW480 CCAR2-null cells, as before (Fig. 2C), no inhibition was observed for SFN, JQ1, or SFN+JQ1 (Fig. 4D, right panel). Thus, SFN required the presence of CCAR2, and circumvented resistance mechanisms in JQ1 resistant colon cancer cells (39,40). Our working model (Fig. 4E) proposes a shift to increased BRD9/CCAR2-containing chromatin complexes as a basis for the synergistic interactions of SFN+JQ1 in colon cancer cells, supported by bioinformatics data – see below.
Cooperative inhibition by SFN+JQ1 in a genetic model of colorectal cancer
We next re-examined RNA-seq data from a recent study (28) and observed the stratification of CCAR2 in colon adenomas of FAP patients (Fig. 5A). Subjects with high CCAR2 levels in colon adenomas also had high CCAR2 expression in normal-looking tissues, whereas patients with lower CCAR2 levels in colon adenomas had reduced CCAR2 expression in normal-looking tissues. Normal-looking colon in FAP patients, and in preclinical models of FAP, is rarely ‘normal’ due to the presence of microadenomas and other preneoplastic lesions.
Figure 5.
SFN+JQ1 protect in a murine model of FAP. A, RNA-seq data (28) mined for CCAR2 levels in adenomatous colon polyps from FAP patients and the Pirc rat (29). Each data-point designates an individual polyp or a normal-looking colonic mucosa sample. B, Polypectomy in the Pirc model (36). C, At 5 months of age, Pirc males (3–4/ group) were assigned to different arms of the study, and two months thereafter occluding colon polyps were resected. Rats were then treated for 5 wks with SFN, JQ1, SFN+JQ1, or vehicle (see Methods). The study was terminated 2 months after polypectomy, and duplicate lesions in each group provided data for D,E, RNA-seq and F IB. The IB data are from a single experiment, and are representative of the findings from two independent experiments. D, Principle component analysis of 1436 differentially-expressed genes (DEGs) identified E, Wnt targets downregulated by JQ1+SFN. G, Compared with vehicle, JQ1+SFN inhibited the growth of colon polyps significantly (**P<0.01).
Interestingly, the ‘CCAR2 high’ molecular phenotype also was detected (Fig. 5A) in the Pirc model of FAP (29). We resected occluding polyps in the rat (Fig. 5B), as reported (36), and animals were then treated with SFN, JQ1, SFN+JQ1, or vehicle (Fig. 5C). When the study was terminated, 2 months after polypectomy, paired colon polyps in each group were subjected to RNA-seq and IB. RNA-seq segregated the groups based on principle component analyses of 1436 genes in the dataset (Fig. 5D). Notably, RNA-seq recapitulated findings from cell-based assays with respect to Wnt genes downregulated by SFN (Fig. 3A), and these observations were extended to JQ1 and SFN+JQ1 groups (Fig. 5E). IB of tissue lysates from Pirc colon tumors also showed reduced expression of target proteins such as CCAR2, cyclin D1, and MMP7, especially for SFN+JQ1 in combination (Fig. 5F, red box). Consistent with these molecular changes, SFN+JQ1 suppressed colon tumor growth significantly, exceeding the inhibition observed for SFN or JQ1 alone (Fig. 5G, **P<0.01).
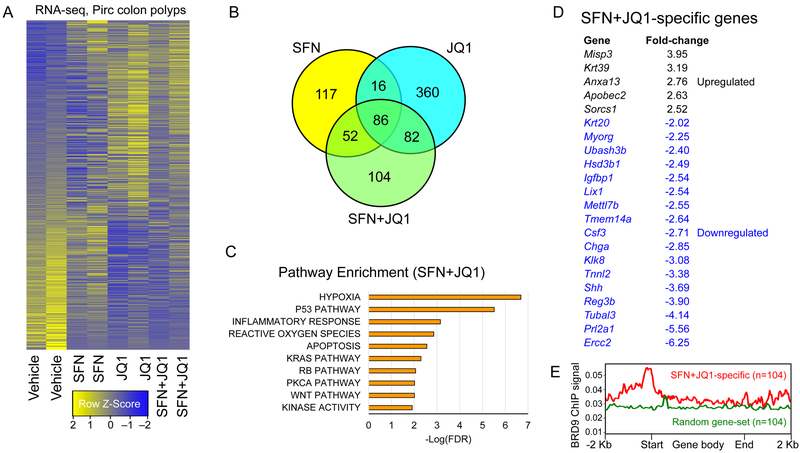
Based on the working model (Fig. 4E), we took the entire RNA-seq dataset (Fig. 6A), and prioritized 104 combination-specific ‘cooperativity/synergy’ candidates among 324 genes in the SFN+JQ1 group (green circle, Fig. 6B). In addition to Wnt, top cancer-specific pathways included hypoxia, p53, inflammation, reactive oxygen species (ROS), KRAS, RB, and apoptosis (Fig. 6C), and the most upregulated and downregulated genes were identified (Fig. 6D). Notably, when all 104 SFN+JQ1 ‘cooperativity/synergy’ genes were interrogated together with available ChIP-seq data for BRD9 (GSM2092891), BRD9 was localized at the corresponding transcription start sites (Fig. 6E, red line). No corresponding BRD9 signal was detected at transcription start sites of 104 randomly selected genes (Fig. 6E, green line), implicating BRD9 enrichment on the promoters of SFN+JQ1 cooperativity/synergy genes as being mechanistically relevant.
Figure 6.
RNA-seq prioritizes HDAC+BET ‘cooperativity’ genes in Pirc colon tumors. A, Heatmaps for groups analyzed in duplicate (1436 DEGs, no cutoff applied). B, Number of DEGs compared to vehicle controls. C, Pathway enrichment analysis for SFN+JQ1 DEGs. D, Highly upregulated and downregulated SFN+JQ1-specific genes. E, Human genome reference GRCh38 was interrogated using BRD9 ChIP-seq data downloaded from GSM2092891. The profile was plotted using Mmint, with the red line representing the average BRD9 signal for SFN+JQ1 ‘synergy/cooperativity’ genes. The corresponding BRD9 signal also was examined for a set of 104 randomly selected genes, not among the ‘synergy/cooperativity’ candidates (green line). Start, transcription start sites; End, transcription stop sites (n=104).
Discussion
Acetylation of CCAR2 by hMOF at K112/K215 sites is known to displace SIRT1 (5), and we speculated that novel N-terminal acetylation sites identified here might similarly interfere with β-catenin interactions. Consistent with this idea, we observed reduced nuclear CCAR2/β-catenin interactions coinciding with downregulation of multiple Wnt targets. Expression of a genetically encoded K54ac site on CCAR2 was sufficient to block its interactions with β-catenin, indicating a key role for this post-translational modification in regulating β-catenin associations. Further, the K54 acetylation mutant interfered with the ability of SFN to reduce downstream targets of β-catenin, such as MMP7 (Supplementary Fig. S4B). Xenograft studies in mice indicated that CCAR2 was required for tumor growth inhibition by SFN+JQ1 in vivo, and we extended these observations to the Pirc model, showing suppression of adenomatous colon polyps by SFN+JQ1 in the rat. In Pirc colon polyps and in a subset of adenomas from FAP patients (Fig. 5A), as well as in adenomatous polyps from a screening colonoscopy trial (Supplementary Fig. S9), a ‘CCAR2 high’ molecular phenotype was observed, which is noteworthy given that CCAR2 overexpression is associated with poor prognosis in colorectal cancer patients (7). ‘CCAR2 high’ adenomatous polyps from the screening colonoscopy trial had elevated expression of β-catenin and its downstream targets, such as, MMP7, c-Myc, and cyclin D1, and increased β-catenin/CCAR2 interactions were detected by PLA (Supplementary Fig. S9).
The C-terminal K916 CCAR2 acetylation site appears to be distant from N-terminal K54/K97 acetylation sites that overlap with HDAC3/β-catenin interacting domains (Fig. 1G). However, as a protein with structural flexibility (41), circumstances might dictate that the ends become aligned, for example after binding lysine methyltransferase ASH1L, which interacted with acetylated peptides from N- and C-terminal regions of CCAR2 (Fig. 4B), or BAZ1A. BAZ1A is a non-catalytic ISWI subunit that associates relatively weakly with acetylated histones, but is critical for DNA damage recovery (42), which is a key function of CCAR2 (43,44). An intriguing question is whether BAZ1A and ASH1L interact preferentially with acetylated non-histone proteins such as CCAR2, affecting gene expression changes as members of specific chromatin remodeling complexes in response to SFN+JQ1 treatment. In this context, competition between BET members and BRD9 for the K916ac site on CCAR2 would shift in favor of CCAR2/BRD9 complexes after SFN+JQ1 treatment (Fig. 4E).
The latter working model derives from three interrelated observations: (i) like BRD2 and BRD3, BRD9 interacts favorably with CCAR2-K916ac (Fig. 4B); (ii) unlike BRD2 and BRD3, BRD9 is not subject to inhibition by JQ1; and (iii) BRD9 is a required subunit of SWI-SNF complexes (45). We speculate that SFN-induced acetylation sites on CCAR2 might exert distinct functions, with K54ac for β-catenin displacement, K97ac for ASH1L/BAZ1A-mediated chromatin interactions, and K916ac as an acetyl switch between BET vs. BRD9 functions. This does not preclude JQ1 also inhibiting BET acetyl readers on histones (Fig. 4E) to affect changes in gene expression (13–16). The possibility that SFN and JQ1 might interact synergistically at the level of MYC transcription was investigated via ChIP assays, with the following observations: (i) CCAR2 interactions were confirmed on promoter and superenhancer regions, (ii) these interactions were almost completely inhibited by the combination of SFN+JQ1 (Supplementary Fig. S10A), and (iii) BRD3 interactions on superenhancer regions also were reduced, to a lesser degree, by JQ1 alone (Supplementary Fig. S10B).
Finally, as an HDAC3-interacting protein, CCAR2 might be targeted using HDAC3-selective inhibitors (46,47), although these agents have yet to enter clinical trials. One approach to enhancing efficacy might involve modifying SFN as a lead compound (48,49), and combining with improved, second-generation bromodomain inhibitors (50,51). This strategy could provide further insights into the ‘cooperativity/synergy’ candidate genes prioritized here, and the associated regulatory pathways to be targeted in future clinical trials. We conclude that JQ1+SFN interferes with the Wnt coactivator role of CCAR2, and shifts the pool of acetyl readers in favor of BRD9-regulated genes, providing a mechanistic basis for new therapeutic avenues combining HDAC3+BET inhibition.
Supplementary Material
Significance:These results highlight the competition that exists among the ‘readers’ of acetylated histone and non-histone proteins and provide a mechanistic basis for potential new therapeutic avenues involving epigenetic combination treatments.
Acknowledgments
We thank R. Jaimes, L. Chew and A. Khan for technical assistance. Dr. O. Hiraike (University of Tokyo, Japan) provided a Myc-DBC expression construct, whereas plasmids pE312 (pPB 4xU25C EF1 AcKRS-TAGT2A-Dendra2 IRES Puro) and pE337 (pPB 4xU25C EF1 H33 3xHA IRES Neo) were from Dr. J. Chin (MRC Laboratory of Molecular Biology, Cambridge, UK). Protein arrays were run by C. Sagum in the Protein Array and Analysis Core, supported by Cancer Prevention & Research Institute of Texas grant RP130432. L.M. Lui provided technical help with mass spectrometry (Protein Mass Spectrometry Core, Baylor College of Medicine), and N. Otto performed IHC in the MD Anderson Pathology & Imaging Core. Initial RNA-seq was conducted at the Center for Genome Research and Biocomputing at Oregon State University. This work was supported by grants CA090890 and CA122959 from the National Cancer Institute, by the John S. Dunn Foundation, and by a Chancellor’s Research Initiative. Funding also was provided by grants R25TCA057730, CA208461 and CA016672, and a gift from the Feinberg Family to E. Vilar.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Kim J-E, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. [DOI] [PubMed] [Google Scholar]
- 2.Escande C, Chini CCS, Nin V, Dykhouse KM, Novak CM, Levine J, et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J Clin Invest. 2010;120:545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Bonkowski MS, Moniot S, Zhang D, Hubbard BP, Ling AJY, et al. A conserved NAD+ binding pocket that regulates protein-protein interactions during aging. Science. 2017;355:1312–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giguère SSB, Guise AJ, Jean Beltran PM, Joshi PM, Greco TM, Quach OL, et al. The proteomic profile of deleted in breast cancer 1 (DBC1) interactions points to a multifaceted regulation of gene expression. Mol Cell Proteomics. 2016;15:791–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng H, Yang L, Peng L, Izumi V, Koomen J, Seto E, et al. hMOF acetylation of DBC1/CCAR2 prevents binding and inhibition of SirT1. Mol Cell Biol. 2013;33:4960–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chini CCS, Escande C, Nin V, Chini EN. HDAC3 is negatively regulated by the nuclear protein DBC1. J Biol Chem. 2010;285:40830–40837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu EJ, Kim SH, Kim HJ, Heo K, Ou CY, Stallcup MR, et al. Positive regulation of β-catenin-PROX1 signaling axis by DBC1 in colon cancer progression. Oncogene. 2016;35:3410–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nusse R, Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985–999. [DOI] [PubMed] [Google Scholar]
- 9.Myzak MC, Karplus PA, Chung F-L, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. [DOI] [PubMed] [Google Scholar]
- 10.Rajendran P, Delage B, Dashwood WM, Yu T-W, Wuth B, Williams DE, et al. Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: competing actions of 14–3-3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly. Mol Cancer. 2011;10:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajendran P, Kidane AI, Yu T-W, Dashwood W-M, Bisson WH, Löhr CV, et al. HDAC turnover, CtIP acetylation and dysregulated DNA damage signaling in colon cancer cells treated with sulforaphane and related dietary isothiocyanates. Epigenetics. 2013;8:612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajendran P, Dashwood W-M, Li L, Kang Y, Kim E, Johnson G, et al. Nrf2 status affects tumor growth, HDAC3 gene promoter associations, and the response to sulforaphane in the colon. Clin Epigenetics. 2015;7:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazur PK, Herner A, Mello SS, Wirth M, Hausmann S, Sánchez-Rivera FJ, et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med. 2015;21:1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujisawa T, Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat Rev Mol Cell Biol. 2017;18:246–262. [DOI] [PubMed] [Google Scholar]
- 16.Kleppe M, Koche R, Zou L, van Galen P, Hill CE, Dong L, et al. Dual targeting of oncogenic activation and inflammatory signaling increases therapeutic efficacy in myeloproliferative neoplasms. Cancer Cell. 2018;33:29–43.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R, Dashwood W-M, Nian H, Löhr CV, Fischer KA, Tsuchiya N, et al. NADPH oxidase overexpression in human colon cancers and rat colon tumors induced by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). Int J Cancer. 2011;128:2581–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parasramka MA, Dashwood WM, Wang R, Saeed HH, Williams DE, Ho E, et al. A role for low-abundance miRNAs in colon cancer: the miR-206/Krüppel-like factor 4 (KLF4) axis. Clin Epigenetics. 2012;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young L, Sung J, Stacey G, Masters JR. Detection of Mycoplasma in cell cultures. Nat Protoc. 2010;5:929–934. [DOI] [PubMed] [Google Scholar]
- 20.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiraike H, Wada-Hiraike O, Nakagawa S, Koyama S, Miyamoto Y, Sone K, et al. Identification of DBC1 as a transcriptional repressor for BRCA1. Br J Cancer. 2010;102:1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elsässer SJ, Ernst RJ, Walker OS, Chin JW. Genetic code expansion in stable cell lines enables encoded chromatin modification. Nat Methods. 2016;13:158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang R, Löhr CV, Fischer K, Dashwood WM, Greenwood JA, Ho E, et al. Epigenetic inactivation of endothelin-2 and endothelin-3 in colon cancer. Int J Cancer. 2013;132:1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gates LA, Shi J, Rohira AD, Feng Q, Zhu B, Bedford MT, et al. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J Biol Chem. 2017;292:14456–14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espejo A, Bedford MT. Protein-domain microarrays. Methods Mol Biol. 2004;264:173–181. [DOI] [PubMed] [Google Scholar]
- 28.Ertem FU, Zhang W, Chang K, Mohaiza Dashwood W, Rajendran P, Sun D, et al. Oncogenic targets Mmp7, S100a9, Nppb and Aldh1a3 from transcriptome profiling of FAP and Pirc adenomas are downregulated in response to tumor suppression by Clotam. Int J Cancer. 2017;140:460–468. [DOI] [PubMed] [Google Scholar]
- 29.Amos-Landgraf JM, Kwong LN, Kendziorski CM, Reichelderfer M, Torrealba J, Weichert J, et al. A target-selected Apc-mutant rat kindred enhances the modeling of familial human colon cancer. Proc Natl Acad Sci USA. 2007;104:4036–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gille C, Fähling M, Weyand B, Wieland T, Gille A. Alignment-Annotator web server: rendering and annotating sequence alignments. Nucleic Acids Res. 2014;42:W3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paxman JJ, Heras B. Bioinformatics tools and resources for analyzing protein structures. Methods Mol Biol. 2017;1549:209–220. [DOI] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. [DOI] [PubMed] [Google Scholar]
- 35.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. [DOI] [PubMed] [Google Scholar]
- 36.Ertem F, Dashwood W-M, Rajendran P, Raju G, Rashid A, Dashwood R. Development of a murine colonoscopic polypectomy model (with videos). Gastrointest Endosc. 2016;83:1272–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius K-J, Jarvius J, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. [DOI] [PubMed] [Google Scholar]
- 38.Blokzijl A, Nong R, Darmanis S, Hertz E, Landegren U, Kamali-Moghaddam M. Protein biomarker validation via proximity ligation assays. Biochim Biophys Acta. 2014;1844:933–939. [DOI] [PubMed] [Google Scholar]
- 39.Tögel L, Nightingale R, Chueh AC, Jayachandran A, Tran H, Phesse T, et al. Dual Targeting of Bromodomain and Extraterminal Domain Proteins, and WNT or MAPK Signaling, Inhibits c-MYC Expression and Proliferation of Colorectal Cancer Cells. Mol Cancer Ther. 2016;15:1217–1226. [DOI] [PubMed] [Google Scholar]
- 40.Engelke CG, Chinnaiyan AM. aBETting therapeutic resistance by Wnt signaling. Cell Res. 2015;25:1187–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunquell J, Yuan J, Erwin A, Westerheide SD, Xue B. DBC1/CCAR2 and CCAR1 Are Largely Disordered Proteins that Have Evolved from One Common Ancestor. Biomed Res Int. 2014;2014:418458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oppikofer M, Sagolla M, Haley B, Zhang H-M, Kummerfeld SK, Sudhamsu J, et al. Non-canonical reader modules of BAZ1A promote recovery from DNA damage. Nat Commun. 2017;8:862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magni M, Ruscica V, Restelli M, Fontanella E, Buscemi G, Zannini L. CCAR2/DBC1 is required for Chk2-dependent KAP1 phosphorylation and repair of DNA damage. Oncotarget. 2015;6:17817–17831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.López-Saavedra A, Gómez-Cabello D, Domínguez-Sánchez MS, Mejías-Navarro F, Fernández-Ávila MJ, Dinant C, et al. A genome-wide screening uncovers the role of CCAR2 as an antagonist of DNA end resection. Nat Commun. 2016;7:12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hohmann AF, Martin LJ, Minder JL, Roe J-S, Shi J, Steurer S, et al. Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nat Chem Biol. 2016;12:672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLeod AB, Stice JP, Wardell SE, Alley HM, Chang C-Y, McDonnell DP. Validation of histone deacetylase 3 as a therapeutic target in castration-resistant prostate cancer. Prostate. 2018;78:266–277. [DOI] [PubMed] [Google Scholar]
- 47.Harada T, Ohguchi H, Grondin Y, Kikuchi S, Sagawa M, Tai YT, et al. HDAC3 regulates DNMT1 expression in multiple myeloma: therapeutic implications. Leukemia. 2017;31:2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okonkwo A, Mitra J, Johnson GS, Li L, Dashwood WM, Hegde M, et al. Heterocyclic analogs of sulforaphane trigger DNA damage and impede DNA repair in colon cancer cells: interplay of HATs and HDACs. Mol Nutr Food Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson GS, Li J, Beaver LM, Dashwood WM, Sun D, Rajendran P, et al. A functional pseudogene, NMRAL2P, is regulated by Nrf2 and serves as a coactivator of NQO1 in sulforaphane-treated colon cancer cells. Mol Nutr Food Res. 2017;61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheung K, Lu G, Sharma R, Vincek A, Zhang R, Plotnikov AN, et al. BET N-terminal bromodomain inhibition selectively blocks Th17 cell differentiation and ameliorates colitis in mice. Proc Natl Acad Sci USA. 2017;114:2952–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu L, Chen Y, Mayakonda A, Koh L, Chong YK, Buckley DL, et al. Targetable BET proteins- and E2F1-dependent transcriptional program maintains the malignancy of glioblastoma. Proc Natl Acad Sci USA. 2018;115:E5086–E5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.