Abstract
Despite the presence of cytotoxic T lymphocytes (CTLs) in the tumor microenvironment, the majority of immunogenic human colon cancer does not respond to immune checkpoint inhibitor immunotherapy, and microsatellite instable (MSI) tumors are not naturally eliminated. The molecular mechanism underlying the inactivity of tumor-infiltrating CTLs is unknown. We report here that CTLs were present in both MSI and microsatellite stable (MSS) colon tumors. The expression of the H3K9me3-specific histone methyltransferase SUV39H1 was significantly elevated in human colon carcinoma compared to normal colon tissues. Using a mouse colon carcinoma model, we further determined that tumor-infiltrating CTLs in the colon tumor microenvironment have high expression of SUV39H1. To target SUV39H1 in the tumor microenvironment, a virtual chemical library was screened based on the SET (suppressor of variegation 3–9, enhancer of zeste and trithorax) domain structure of the human SUV39H1 protein. Functional enzymatic activity assays identified a small molecule that inhibits SUV39H1 enzymatic activity. Based on the structure of this small molecule, we modified it and chemically synthesized a small molecule, termed F5446, which has an EC50 of 0.496 μM for SUV39H1 enzymatic activity. H3K9me3 was enriched in the promoters of GZMB, PRF1, FASLG, and IFNG in quiescent T cells. F5446 inhibited H3K9me3, thereby, upregulating expression of these effectors in tumor-infiltrating CTLs and suppressing colon carcinoma growth in a CD8+ CTL-dependent manner in vivo. Our data indicate that SUV39H1 represses CTL effector gene expression and, in doing so, confers colon cancer immune escape.
Keywords: SUV39H1, H3K9me3, T-cell effector, immune evasion, epigenetics
Introduction
Colon cancer was the first human neoplasia that was found to be under immunosurveillance by the adaptive immune response (1–3). The type, density, and location of tumor-infiltrating immune cells within the tumor microenvironment correlates with tumor growth, progression and recurrence, and predicts patient clinical outcomes, and therefore, human colon tumors are considered immunogenic (1–8). Given the presence of helper T cells (Th1s) and cytotoxic T lymphocytes (CTLs) in the tumor microenvironment, it might be expected that the host immune system would naturally eliminate these immunogenic colon tumors. However, colon cancer is the third most commonly diagnosed cancer and the second leading cause of cancer death in men and women combined, suggesting that this is not the case. Why this host defense mechanism fails to occur is not yet known. One possibility is that the microsatellite instable (MSI) colon tumors selectively upregulate multiple counter-inhibitory checkpoint molecules affording immune escape (9). Consistent with this notion, PD-1 blocking antibody immunotherapy yields high response rates in MSI colon cancer patients (10).
However, despite breakthroughs in immune checkpoint inhibitor (ICI) immunotherapy for many types of human cancers, colon cancer stands out as one of the few human cancers that does not respond to ICI immunotherapy (except for the small subset of MSI positive patients, and these represent only ~4% of all colon cancer cases)(10). Why the majority of human immunogenic colon cancer cases do not respond to ICI immunotherapy is unknown (11).
ICI immunotherapy, such as anti–PD-1 and anti–PD-L1, block interactions between PD-L1 and PD-1 to reverse immune suppression to unleash CTLs that kill tumor cells in the tumor microenvironment. CTLs kill tumor cells through inducing tumor cell apoptosis, and for this to occur, CTLs must be activated and the target tumor cells must be sensitive to apoptosis induction. Resistance to apoptosis is a hallmark of human cancer (12), particularly in human colon cancer (13–15). If tumor cells are not sensitive to apoptosis induction, then regardless of how potent the CTLs are, the tumor cells will not be eliminated. Other PD-L1–independent mechanisms of immune suppression, such as myeloid-derived suppressive cells (MDSCs), may suppress ICI-activated CTLs in the tumor microenvironment (16) in addition to PD-L1–mediated immune suppression. The fact that CTL infiltration is observed in both MSS and MSI human colon cancer suggests that a lack of CTL infiltration is unlikely to be the sole mechanism underlying human colon cancer non-response to ICI immunotherapy (1,3,4,16,17). Therefore, targeting the effector gene expression and function of tumor-infiltrating CTLs can potentially bypass immune suppression to directly augment their effector function to control colon carcinoma development. To test this hypothesis, we developed a novel small molecule inhibitor (termed F5446) specific to histone methyltransferase SUV39H1 that decreased H3K9me3 at T-cell effector promoters to increase effector gene expression in tumor-infiltrating CTLs, thereby, suppressing colon carcinoma growth in vivo.
Materials and Methods
Mice and cells.
BALB/c and C57BL/6 mice were obtained from the Charles River Frederick mouse facility. B6.129S7-Rag1tm1Mom/J mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Six to eight–week old female mice were used. All mice were housed, maintained, and studied in accordance with approved protocols by Augusta University and the Ohio State University Institutional Animal Use and Care Committees. Mouse colon carcinoma CT26 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA) in 2013 and stored in liquid nitrogen as aliquots. Cells were used within 30 passages. ATCC characterized these cells by morphology, immunology, DNA fingerprint, and cytogenetics. Murine colon carcinoma MC38 cells were provided by Dr. Jeffrey Schlom (National Cancer Institute, Bethesda, MD) and characterized as previously described (18). Cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) plus 10% HyClone fetal bovine serum (FBS)(Cat# SH30396.03, GE Healthcare, Chicago, IL). Cells are tested for mycoplasma every two months, and all cells used in this study were mycoplasma-negative.
Database assessment.
Human dataset of SUV39H1 expression in human primary tumor (n=380) and normal solid tissue (n=51) was extracted from TCGA Colon and Rectal Cancer (COADREAD) ploy A+ IlluminaHiSeq pancan normalized RNA-seq datasets using UCSC Xena Cancer Genomics Browser (University of California, Santa Cruz, CA). H3K9me3 ChIP-Seq data set were extracted from the Gene Expression Omnibus (GEO) Database (accession# GSM1058783) and analyzed with IGV 2.4x program (Broad Institute, Cambridge, MA and University of California, San Diego, CA).
Synthesis of F5446.
Synthesis of F5446 was conducted by Leadgen Labs LLC (Orange, CT) using reagents purchased from Sigma-Aldrich (St Luis, MO) and methods as previously described (19). 1H NMR spectra were recorded at Brucker 400 MHz, and 13C NMR spectra were recorded at 100 MHz at room temperature using CDCl3 as solvent and tetramethylsilane (TMS) as an internal standard. Chemical shifts were reported in ppm relative to TMS as the internal standard. The following abbreviations are used to describe spin multiplicity: s=singlet, d=doublet, t=triplet, q=quartet, m=multiplet, brs=broad singlet, dd=doublet of doublets.
The LC/MS was recorded on an Agilent 1100 with LC/MSD SLMobile Phase: A-H2O+0.1%HCOOH; B-MeCN+0.1HCOOH. 5: 6-(4-Chloro-benzenesulfonyl)-4,5-dioxo-3,4,5,6-tetrahydro-3,6-diaza-asindacene-1,7-dicarboxylic acid 1-benzyl ester 7-methyl ester: beige crystalline powder; m.p. 141–152°C; 1H NMR (400 MHz, CDCl3, δ) 11.05–11.02 (m, 1H), 8.56 (d, J = 0.8 Hz, 1H), 8.54 (d, J = 0.8 Hz, 1H), 7.82–7.81 (m, 2H), 7.53–7.51 (m, 2H), 7.45–7.30 (m, 5H), 5.37 (s, 2H), 4.00 (s, 3H); 13C NMR (100 MHz, CDCl3, δ) 169.3, 169.2, 141.1, 136.7, 135.0, 133.9, 133.9, 133.9, 131.4, 131.2, 128.7, 128.3, 128.1, 128.0, 127.2, 125.9, 117.4, 114.8, 66.5, 53.1. LC/MS for C26H17ClN2O8S [M+H]+ 552.95, found 553.0. Step A: Methyl 1-(4-chlorobenzenesulfonyl)-4-formyl-1H-pyrrole-2-carboxylate. Solution of pyrrole 1 (1.0 g, 6.5 mmol) in tetrahydrofuran (THF) (15 mL) was slowly added to a suspension of NaH (0.36 g, 60% in mineral oil, 9.0 mmol) in THF (15 mL). The reaction mixture was then stirred for 1 hour at room temperature, followed by slow addition of p-ClPhSO2Cl (1.65 g, 7.8 mmol) in 15 mL of THF. The reaction mixture was then stirred at room temperature overnight followed by quenching with CH3COOH.
After solvent was evaporated, the crude product was purified by column chromatography using hexane/EtOAc=7/3. Product 2 was isolated with the yield of 1.7 g, 70%. Step B: Methyl 4-[(1E)-3-(benzyloxy)-3-oxoprop-1-en-1-yl]-1-(4-chlorobenzenesulfonyl)-1H-pyrrole-2-carboxylate. (EtO)2P(O)CH2CO2Bn (1.69 g, 5.9 mmol) was added dropwise at −30°C to a suspension of NaH (0.29 g 60% in mineral oil, 6.7 mmol) in THF (12 mL) and the mixture was then warmed up to 0°C. After the mixture was cooled to –30°C, the pyrrole 2 (1.7 g, 5.2 mmol) in THF (10 mL) was added at –10°C. After 5 minutes of stirring the mixture was quenched with 10% aqueous NH4Cl (7 mL), and the THF was evaporated under reduced pressure. The residue was extracted with CH2Cl2 (3 × 50 mL), dried (Na2SO4), and evaporated to give a tan solid (2.1 g). Recrystallization from acetone gave product 3: 1.56 g, 74%; Step C: Methyl 4-{4-[(benzyloxy)carbonyl]-1H-pyrrol-3-yl}−1-(4-chlorobenzenesulfonyl)-1H-pyrrole-2-carboxylate. Freshly prepared LiHMDS [prepared from HMDS (0.88 g, 5.4 mmol) and 2.5 M n-BuLi (1.56 mL, 3.9 mmol) in THF (8 mL)] was added at –78°C to a solution of TosMIC (0.64 g, 3.3 mmol) and product 3 (1.13 g, 2.5 mmol) in THF (25 mL). After stirring for 15 minutes at –70°C, the reaction mixture was quenched with 10% aqueous NH4Cl (8 mL) and evaporated under reduced pressure to remove THF. The residue was then extracted with CH2Cl2 (4 × 20 mL), dried (Na2SO4), and filtered.
After evaporation, the residue was chromatographed over silica gel using 5% EtOAc/25% petroleum ether/70% CH2Cl2 to give product 4: yield 0.55g, 45%. Step D: 1-Benzyl 7-methyl 6-(4-chlorobenzenesulfonyl)-4,5-dioxo-3H,4H,5H,6H-pyrrolo[3,2-e]indole-1,7-dicarboxylate. Oxalyl chloride (0.11 mL, 1.3 mmol) was added dropwise to a solution of the bipyrrole 4 (0.52 g, 1.0mmol) in CH2Cl2 (20 mL) at 0°C. More oxalyl chloride (0.06 mL) was slowly added in an hour, and the mixture was left at 0°C for 24 hours. The mixture was cooled to –78°C, and SnCl4 (0.118 mL, 1.0 mmol) was added dropwise. After 1.5 hours, the mixture was quenched by dropwise addition of water (4.0 mL). The CH2Cl2 layer was separated, and the aqueous phase was extracted with CH2Cl2 (3 × 10 mL). Combined organic phases were dried (Na2SO4) and evaporated under reduced pressure. The residue was then chromatographed on silica gel using CHCl3/MeOH=25/1.
Molecular docking.
Structures in the NCI Diversity Set V were supplied in 2D form (https://wiki.nci.nih.gov/display/NCIDTPdata/Compound+Sets). Three-dimensional representations were generated with computer program obprep from the Openbabel suite (20). Docking of these structures was done with computer program Autodock Vina 2. 0 (21)(The Scripps Research Institute, San Diego, CA) to predict binding modes of a drug candidate against the protein target using a box centered on the ligand binding site. The figure-of-merit in the scoring output of Autodock Vina is expressed as a binding affinity in kcal/mol, with a standard error of 2.85 kcal/mol. Suppressor of variegation 3–9, enhancer of zeste and trithorax (SET) domain structure of human SUV39H1 protein was used for molecular docking (22).
Histone methyltransferase activity assay.
Recombinant SUV39H1 protein (250 nM, Cat# SUV39H1–406H; Reaction Biology Corp, Malvern, PA) was incubated with human histone 3 peptide (amino acids 1–21, 5 μM), S-Adenosyl-L-[methyl-3H] methionine (1 μM, Cat# NET15500, PerkinElmer, Boston, MA), and the test compounds (0–50 μM) in reaction buffer [50 mM Tris-HCl pH8.5, 50 mM NaCl, 5 mg MgCl2, 1 mM dithiothreitol (Sigma-Aldrich), and 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich)] at 30°C for 60 minutes. The reaction was stopped by adding EDTA to 50 mM. the reaction mixture (6 μL) from each reaction was spotted onto a 1.2 × 1.2cm Whatman P81 ion exchange paper (Cat# 3698–915, obtained from Reaction Biology Corp) disc. The filter discs were air-dried at room temperature for 30 minutes, washed 3 times in washing buffer (50 mM Na2CO3 and NaHCO3 pH 9.2) for 10 minutes each time, and air-dried again. The filters were transferred to 20 mL scintillation vials containing 1 mL water and 10 mL Ultima Gold scintillation cocktail (Cat# 6013329, PerkinElmer) and counted in a LS6500 Multi-Purpose Scintillation Counter (Beckman, Indianapolis, IN). Enzymatic inhibition was calculated using Prism GraphPad 6.0 (San Diego, CA). F5446 inhibitory activity was determined in a 10-dose EC50 mode with 3-fold serial dilution starting at 10 μM essentially as described above in Reaction Biology Corp (Malvern, PA).
F5446 tolerability in vivo.
C57BL/6 mice (7–8 weeks of age) were injected intraperitoneally with vehicle [10% Cremophor EL (Cat# C5135, Sigma-Aldrich) in PBS] or F5446 at doses of 10 and 20 mg/kg body weight (n=5 per group), respectively, every 2 days for a total of 7 doses. Mouse weights were measured every 2 days. Mice were euthanized approximately 24 hours after the last dose. Blood was collected from mice immediately after CO2 euthanasia via cardiac puncture. Whole blood from each mouse was dispensed into a microvette 100 K3E tube (Cat# 20.1278.100, Sarstedt, Numbrecht, Germany) and a 1.5 mL microcentrifuge tube, which were then submitted to the Comparative Pathology and Mouse Phenotyping Shared Resource at The Ohio State University Comprehensive Cancer Center (https://cancer.osu.edu/research-and-education/shared-resources/comparative-pathology-and-mouse-phenotyping) for determination of complete blood counts and serum biochemical panel, respectively. Complete blood counts were performed on a FORCYTE Autosampler 10 Hematology Analyzer (Oxford Science, Inc., Oxford, CT), followed by manual confirmation of white blood cell differential counts. Serum biochemical panel was determined on a VetAce Chemistry Analyzer (Alfa Wassermann, West Caldwell, NJ) using undiluted serum collected manually as supernatant after removal of blood clot by centrifugation.
Mouse tumor models.
MC38 tumor cells (1.5×105 cells/mouse) were injected into C57BL/6 or Rag1 KO mice, and CT26 cells (2×105 cells/mouse) were injected into BALB/c mice. MC38 tumor-bearing mice were treated eight days later, and CT26 tumor-bearing mice were treated ten days later, respectively, with vehicle (10% cremophor in PBS) and IgG (200 μg/mouse; Control), F5446 only (10 mg/kg body weight), anti–PD-1 only (200 μg/mouse, clone 29F.1A12; Bio X cell, West Lebanon, NH), or F5446 plus anti–PD-1 every 2 days for a total of 7 times. To determine CD8+ CTL function, tumor-bearing mice were treated with vehicle and IgG, F5446, or F5446 plus a CD8-specific neutralization monoclonal antibody (200μg/mouse, clone BE0061; Bio X cell, NH) at same concentrations. Tumor size was measured in two dimensions with a digital micrometer caliper at the indicated time points. Tumor volume was calculated by the formula: (tumor length x tumor width2)/2. MC38 tumor-bearing mice were sacrificed 22 days after tumor injection, and CT26 tumor-bearing mice were sacrificed 24 days after tumor injection.
Isolation of tumor cells and tumor-infiltrating CTLs.
MC38 tumor cells (1.5 ×105 cells/mouse) were injected to C57BL/6 mice subcutanouesly (s.c.). Tumors were excised 22 days after tumor cell injection and digested in collagenase solution – collagenase (1 mg/mL; Sigma-Aldrich), hyaluronidase (0.1 mg/mL; Sigma-Aldrich), and DNaseI (30 U/mL; Roche Diagnostics, Indianapolis, IN) – in RPMI 1640 medium plus 10% FBS at 37°C on a magnet stirrer for 30 minutes to obtain single cells. The digested tumor mixtures were centrifuged in a Sorvall Legend RT centrifuge (Kendro Laboratory Products, Germany) at 1500 RPM for 5 minutes. The cell pellet was resuspended in red cell lysis buffer (0.15 mole/L NH4Cl, 10 mmole/L KHCO3, and 0.1 mM EDTA pH7.4), incubated at room temperature for 5 minutes, centrifuged, and washed with PBS once. The cells were then resuspended in PBS and the MojoSort mouse CD45 nanobeads (Cat# 480028, Biolegend) were added to the cell suspension. The cell and beads mixtures were incubated in 4°C with rotation for 30 minutes, and then placed on DYNAL MPC-S magnet stand (Invitrogen) for 2 minutes. The supernatant was collected and used as the CD45– (>95% CD45- cells) tumor cells. To isolate tumor-infiltrating CTLs, tumor mixtures were incubated with mouse CD8 Dynabeads (Cat# 114.47D, clone lyt-2, Invitrogen Dynal AS, Oslo, Norway) in PBS and incubated at 4°C with rotation for 30 minutes. The bead and cell mixtures were placed on a DYNAL MPC-S magnet stand (Invitrogen) for 2 minutes. The supernatant was removed and the beads were resuspended in 1 mL PBS, and placed on the magnet stand for 2 minutes. This washing step was repeated once. The bead-bound CD8+ cells were directly used for analysis.
T cell purification and activation.
Spleens were collected from C57BL/6 mice and broken into single cells in 10 mL PBS by using two sterile frosted microscope slides (Cat#12–552-3, Fisher Scientific, Hampton, NH). The cell suspension was passed through a 100 μM cell strainer (Cat # 22363549, Fisher Scientific) and centrifuged. The cell pellet was resuspended in PBS and washed twice. The cell pellet was then resuspended in PBS, and CD3+ T cells were purified using the MojoSort mouse CD3 T cell isolation kit (Cat #480031, Biolegend) according to the manufacturer’s instructions. The CD3+ T cell purity was above 90%. For T-cell activation, purified T cells (1.5×106 cells/well in 24-well plate) were cultured in RPMI 1640 medium plus 10% FBS in tissue culture plates coated with anti-mouse CD3 (8 μg/mL, clone 145–2C11; Bio X cell) and anti-mouse CD28 (10 μg/mL, clone 37.51; Bio X cell) for 2 days.
mRNA analysis.
Total RNA was isolated from CD3+ T cells, purified tumor-infiltrating CD8+ T cells, and fresh whole tumor tissues using the GeneJET RNA isolation kit according to the manufacturer’s instructions (Fisher Scientific). RNA concentrations were measured using a NanoDrop spectrophotometer (Thermo Scientific, Mansfield, TX). RNA purity was determined based on OD260/OD280 ratio, and only samples with ratio >1.90 were used. cDNA was synthesized from 500 ng total RNA in a 20 μL reaction volume using the ThermoScript RT-PCR kit (Cat#11146–016, Invitrogen) with the substitution of ThermoScript reverse transcriptase by M-MLV reverse transcriptase (Cat #M170B, Promega, Madison, WI). PCR was performed in triplicates with 1 μL cDNA mixture using gene-specific primers (Supplementary Table S1) in the StepOne Plus Real-Time PCR System (Applied Biosystems). Gene expression of Gzmb, Prf1, Faslg, and Ifng was determined as ΔΔCT with β-actin as internal normalization control.
Western blots.
Western blotting analysis was performed as previously described (23). Briefly, resting and in vitro-activated spleen CD3+ T cells and tumor-infiltrating CD8+ T cells were lysed in total cell lysate buffer (20 mM Hepes pH 7.4, 20 mM NaCl, 10% glycerol, 1% Triton x100) plus protease inhibitor cocktail (Cat#535140, Millipore, Burlington, MA) on ice for 60 minutes. The cell lysate was centrifuged at 13,000 RPM in 4°C for 10 minutes in a Eppendorf 5415R microcentrifuge (Eppendorf, Hauppauge, NY). The supernatant was collected and measured for protein concentration using Protein Assay Dye Reagent (Cat#500–0006, Bio-Rad Laboratories, Hercules, CA) according to the manufacturer’s instructions using bovine serum albumin (Cat#BP1605, Fisher Scientific) as concentration standard. Total lysates (20 μg per sample) were loaded to a 4–20% mini-protean TGX precast gel (Cat#456–1096, Bio-Rad Laboratories), and the proteins were transferred to the Immun-Blot PVDF membrane (Cat#162–0177, Bio-Rad Laboratories) after electrophoresis using a Pierce power blot cassette. The membrane was incubated in 5% blotting-grade milk (Bio-Rad Laboratories) in PBS plus 1% Tween-20 (Cat#P1379, Sigma-Aldrich) at room temperature for 1 hour. The membrane was then washed 3 times in PBS plus 1% Tween 20 and incubated with anti-mouse CD8 (BD Pharmingen, clone 53–6.7) at 1:1000, anti-mouse CD11b (Abcam, clone M1/70) at 1:1000, anti-mouse perforin (clone H315, Santa Cruz, Dallas, TX) at 1:500, and anti-mouse β-actin (clone: AC-74, Sigma-Aldrich) at 1:10000, respectively, in PBS plus 1% Tween 20 and 5% blotting-grade milk (Bio-Rad Laboratories) at 4°C on a shaker overnight. The membrane was then washed 3 times with PBS plus 1% tween 20 and incubated with anti-rabbit IgG at 1:3000, anti-mouse IgG at 1:3000, and anti-Rat IgG at 1:3000 dilution, respectively, for 45 minutes. The membrane was developed using the Western Lightning Plus-ECL kit (PerkinElmer, Waltham, MA) for 1 minute, exposed to x-ray film (Cat#E3012, Denville Scientific, Holliston, MA), and processed in the SRX-101A medical file processor (Konica Monolta Medical and Graphic Inc., Hino-shi, Japan).
Flow cytometry analysis:
Resting and activated CD3+ T cells (approximately 1.5 ×106 cells) were blocked with TruStain fcX™ (Cat#101320, Biolegend), stained with monoclonal antibody specific for Zombie Violet (Cat#423108, Biolegend) and FasL (clone MFL3, Biolegend), fixed and permeabilized using the Cytofix/Cytoperm Plus Kit (BD Bioscience, San Diego, CA) according to the manufacturer’s instructions, and stained with monoclonal antibodies specific for granzyme B (clone GB11, Biolegend), perforin (clone eBioOMAK-D, eBiosciences) and IFNγ (clone XMG1.2, Biolegend). Aliquots of fresh tumor digestion mixtures were incubated with monoclonal antibodies specific for Zombie Violet, CD45.2 (Clone 104, Biolegend), and CD8 (Biolegend, clone 53–6.7). Zombie– live cells and CD45+ cells were gated and analyzed for CD8+ T cells. The stained cells were analyzed using LSR Fortessa cytometer and FACSDiva software (BD Biosciences). Data were analyzed with FlowJo V10 program (Flow Jo, Ashland, OR).
Immunohistochemistry.
De-identified human MSS (n=9) and MSI (n=8) colon carcinoma specimens were obtained from the Cooperative Human Tissue Network (CHTN) Southern Division (University of Alabama, Birmingham, AL). Human tonsil tissue was obtained from Georgia Cancer Center Biorepository (Augusta, GA) and used as a positive control for CD8+ T cell staining. Immunohistochemical staining was carried out by the Georgia Esoteric Molecular Labs (Augusta, GA), and the CD8 antibody used was obtained from Dako (clone C8/144B, Dako, Santa Clara, CA). The tissues were then probed with mouse specific IHC polymer detection kit (Cat# ab209101, Abcam) according to the manufacturer’s instructions. The stained tissues were counterstained with hematoxylin (Richard-Allan Scientific, Kalamazoo, MI). Tumor tissues stained without the primary antibody were used as negative controls. CD8+ T cell infiltration was graded by a board-certified Pathologist (N.M.S) as low, medium, and high, and was also quantified by double-blind counting of CD8+ T cells per view field for each stained specimen.
Chromatin immunoprecipitation (ChIP).
Purified CD3+ T cells and the bead-bound isolated tumor-infiltrating CD8+ T cells, as described above, were fixed with 1% formaldehyde (Cat#BP531, Fisher Scientific) in RPMI 1640 medium plus 10% FBS for 10 minutes. ChIP analyses were performed using anti-H3K9me3 (Cat#ab8898, Abcam) and the Simple ChIP Plus Enzymatic Chromatin IP Kit (Cat# 9004, Cell Signaling Technologies, Danvers, MA) according to the manufacturer’s instructions. The H3K9me3-bound DNA of Gzmb, Prf1, Faslg, Ifng, and Nxf2 gene promoters was quantified by qPCR in triplicates using gene promoter-specific primers (Supplementary Table S1).
Statistical analysis.
All statistical analysis was performed using SAS 9.4 (SAS Institute Inc., Cary, NC), and statistical significance was assessed using an alpha level of 0.05. A repeated-measures mixed model with fixed effects of treatment groups and day, as well as all two- and three-factor interactions between these effects, was used to examine changes in tumor size over time. A one- or two-factor ANOVA with fixed effects of F5446 and anti–PD-1 groups and the two-factor interaction of these effects was used to examine differences in mean tumor size and tumor weight. A Tukey-Kramer multiple comparison procedure was used to examine post hoc pairwise differences between the groups. Statistical analysis was also performed using two-tailed t test. EC50 was calculated using Prism GraphPad 6.0 (GraphPad Software, San Diego, CA).
Results
CTL tumor-infiltration in MSS and MSI human colon carcinoma
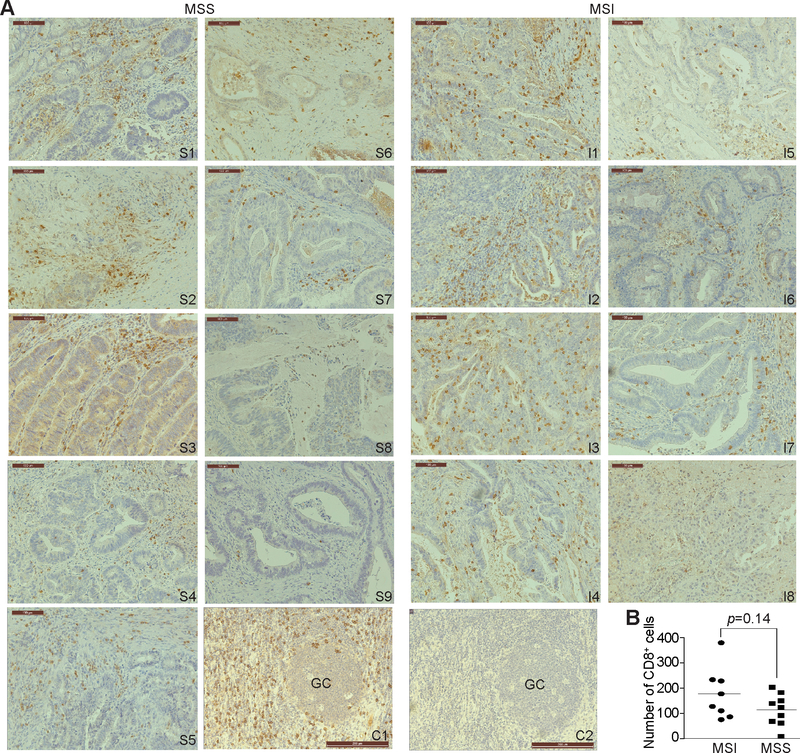
Human MSS and MSI colon carcinoma specimens were analyzed by immunohistochemical staining of CD8+ T cells. As expected, all eight MSI colon carcinoma specimens exhibited medium to high infiltration of CTLs. However, eight of the nine MSS colon carcinoma specimens also exhibited medium infiltration by CTLs (Fig. 1A). No statistically significant difference in CTL tumor infiltration was observed between MSI and MSS colon carcinoma specimens (Fig. 1B).
Figure 1. Tumor-infiltrating CTLs in human MSS and MSI colon carcinoma.
A. Human colon carcinoma MSS (S1-S9, n=9) and MSI (I1-I8, n=8) tumor specimens were stained with CD8-specific antibody. Shown are representative images of tumor-infiltrating CD8+ CTLs. Human tonsil tissue was stained with the CD8-specific antibody (positive control, C1) or no primary antibody (negative control, C2) as in A. GC: germinal center. Scale bar=100 μM. B. The CD8+ CTLs were quantified in each tumor specimen and presented. Each dot represents number of CD8+ CTLs in one view area of a tumor specimen. Two-tail t tests were used to determine differences in CD8+ CTL infiltration between MSS and MSI tumor specimens with p<0.05 as being statistically significant. Bar: Mean.
SUV39H1 is expressed in tumor cells and tumor-infiltrating CTLs in tumors
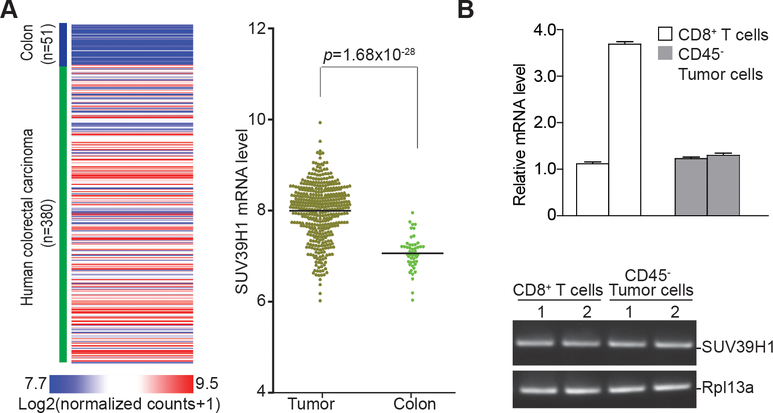
The above observation that no significant difference in CTL infiltration exists between MSS and MSI colon carcinomas suggests that a lack of CTL infiltration might not be the sole mechanism underlying colon cancer immune escape. It is, therefore, possible that the functional status of the tumor-infiltrating CTLs is also responsible for colon cancer immune escape. We screened the TCGA database for altered gene expression between human colon carcinoma tissues and normal colon tissues to identify genes with known function in regulating CTL functions. We found that the expression of SUV39H1 which encodes H3K9me3-specific histone methyltransferase recently shown to regulate CTL effector expression (24), was significantly elevated in the tumor tissue (Fig. 2A).
Figure 2. SUV39H1 expression is elevated in both colon carcinoma cells and tumor-infiltrating CTLs.
A. Data sets of SUV39H1 mRNA in human colon carcinoma (n=380) and normal colon tissues (n=51) extracted from TCGA database and plotted as a heatmap shown in the left panel. The SUV39H1 mRNA expression range is represented by red and blue color gradient as indicated at the left bottom panel. The relative mRNA expression is represented as Log2 (normalized counts+1). The SUV39H1 mRNA levels was compared between colon carcinoma and normal colon tissues and presented in the right panel. Two-tail t tests were used to determine differences in SUV39H1 mRNA expression between tumor tissues and normal colon tissues with p<0.05 as being statistically significant. Bar: Mean. B. Tumors were excised from two MC38 tumor-bearing mice and digested in a collagenase solution. Portion of the tumor cells were incubated with anti-CD45–conjugated magnetic beads to deplete leukocytes from tumor cells. Another portion of the tumor cells was incubated with anti-CD8–conjugated magnetic beads to isolated CD8+ tumor-infiltrating CTLs. RNA was prepared and analyzed by qPCR using SUV39H1-specific primers. The two open bars indicate SUV39H1 mRNA in CD8+ tumor-infiltrating CTLs from two tumor-bearing mice. The two grey filled bars represent SUV39H1 mRNA in CD45-depleted tumor cells from two tumor-bearing mice. Bottom panel: agarose gel showing the PCR-amplified Suv39h1 DNA fragment. Rpl13a was used as normalization control.
Next, we sought to determine Suv39h1 expression in tumor-infiltrating CTLs in the MC38 colon carcinoma mouse model. CD45– tumor cells and CD8+ CTLs were isolated from fresh MC38 tumor tissues and analyzed for Suv39h1 mRNA levels. As in the human colon carcinoma tissues, tumor-infiltrating CTLs, as well as tumor cells, had high expression of Suv39h1 in the MC38 tumor model (Fig. 2B).
H3K9me3 is enriched in the promoter region of effectors in quiescent T cells
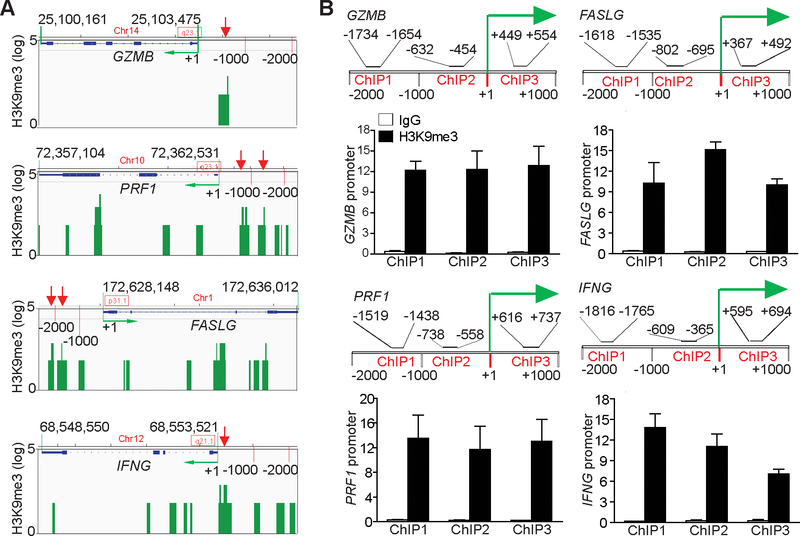
SUV39H1 catalyzes H3K9me3 (25,26). Analysis of H3K9me3 ChIP-Seq data in the GEO Database revealed that H3K9me3 is enriched in the promoter regions of GZMB, PRF1, FASLG, and IFNG in human T cells (Fig. 3A). To validate this finding in the mouse T-cell genome, we purified CD3+ T cells from C57BL/6 mouse spleens and performed ChIP analysis using an H3K9me3-specific antibody and PCR primers that covered the promoters approximately from −2000 to +1000 relative to the transcription start sites of these four effector genes (Fig. 3B). ChIP analysis of these four individual gene promoter regions indicated that H3K9me3 was enriched in the promoter regions of all 4 effector genes in resting mouse T cells (Fig. 3B). These observations indicate that the expression of T-cell effector genes might be silenced by an H3K9me3-mediated epigenetic mechanism in resting T cells.
Figure 3. The effector promoter chromatin is enriched with H3K9me3 in T cells.
A. Genome-wide ChIP-Sequencing data set of human CD3+ T cells was extracted from GEO (accession #GSM 1058783). The genome regions of genes encoding T-cell effector genes GZMB, PRF1, FASLG, and IFNG were visualized using program IGV. The gene locations are indicated. The H3K9me3 peaks are presented as green bars. The green arrow indicates transcription direction. The H3K9me3 peaks in the gene promoter regions are indicated by red arrows. B. CD3+ T cells were purified from spleens of C57BL/6 mice and analyzed by ChIP with anti-H3K9me3 and gene-specific PCR primers in triplicates as indicated. The promoter structures of each gene are shown at the top panel of each bar graph. ChIP1–3 indicate the three ChIP PCR-amplified regions at the respective gene promoter. The number above the bar indicates the ChIP PCR-amplified region as defined by nucleotide locations relative to each gene transcription initiation site. The bottom bar graphs show ChIP H3K9me3. Column: Mean; Bar: SD.
Development of an SUV39H1-specific small molecule inhibitor
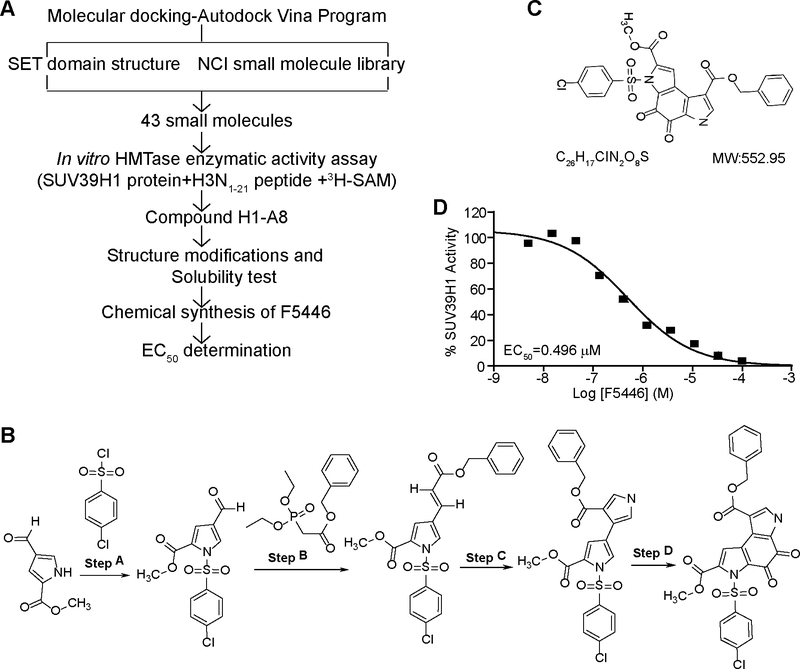
The above observations that tumor-infiltrating CTLs expressed high SUV39H1 and that H3K9me3 was enriched in the effector gene promoters in quiescent T cells suggest that effector gene expression may be repressed by the SUV39H1-H3K9me3 axis in tumor-infiltrating CTLs. Therefore, targeting the SUV39H1-H3K9me3 axis may increase effector gene expression and thereby, suppress tumors without the need to activate CTLs in the tumor microenvironment. To develop a SUV39H1-specific inhibitor, the molecular docking program Autodock Vina (21) was used to screen the NCI small chemical virtual library based on the SET domain structure of human SUV39H1 protein. The top 43 hits were selected and tested for inhibition of human SUV39H1 enzyme activity in vitro (Fig. 4A). Compound A8 and H1 exhibited inhibitory activity against recombinant SUV39H1. These two compounds were further tested at Reaction Biology Corp. Compound A8 [8-O-Benzyl 2-O-methyl 3-(4-methylphenyl) sulfonyl-4,5-dioxo-6H-pyrrolo(3,2-e) indole-2,8-dicarboxylate, C27H20N2O8S] was then selected and structurally modified. A chemical synthesis procedure was developed for this A8 derivative, termed F5446 (Fig. 4B). F5446 has a molecular weight of 552.95 g/mol (Fig. 4C) and exhibited an EC50 of 0.496 μM in an in vitro enzymatic activity assay (Fig. 4D). The 1H and 13C spectra (Supplementary Fig. S1 and S2) and HPLC/MS data of F5446 (Supplementary Fig. S3) confirmed the structure and composition.
Figure 4. Development of the SUV39H1-specific small molecule inhibitor F5446.
A. Scheme of F5446 development. B. Chemical synthesis procedures of F5446. The detailed procedures are described in the methods. C. F5446 chemical structure. D. F5446 was tested in a 10-dose EC50 mode with 3-fold serial dilutions using recombinant human SUV39H1 protein as the methyltransferase, S-(methyl-3H) adenosyl-L-methionine as the substrate, and Histone 3 peptide (N1–21) as the template in the presence of indicated doses of F5446. The EC50 was calculated with GraphPad 6.0 as described in the methods.
In vivo tolerability of F5446
To determine the in vivo tolerability of F5446, mice were dosed every two days with vehicle or 10 or 20 mg/kg body weight F5446 for a total of 7 doses. Observations of body weights during the 2-week dosing period indicated mice in the 20 mg/kg group lost weight on average (<10%). The 10 mg/kg body weight group maintained a stable body weight throughout the dosing period, whereas the vehicle-treated control group increased average body weight by ~10%. These data are summarized in Supplementary Fig. S4 and Supplementary Table S2. Complete blood counts indicated no significant differences between vehicle and the F5446 treatment groups (Supplementary Table S3). Serum chemistry data suggested no difference between vehicle and drug-treated groups with all markers, except amylase and lipase, which were reduced, although still within normal levels in the F5446 treatment groups (Supplementary Table S4).
SUV39H1 suppresses the expression of cytotoxic effector genes through H3K9me3
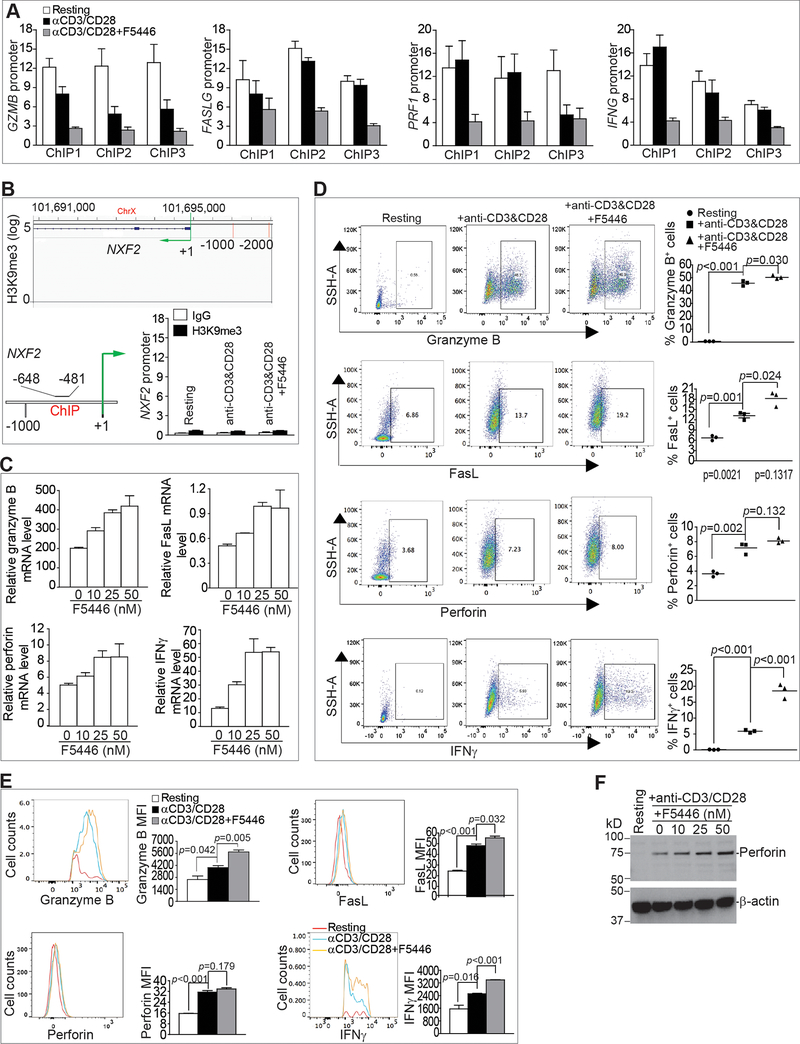
H3K9me3 is a repressive chromatin marker and often silences gene expression (24,25,27). We, therefore, reasoned that inhibition of SUV39H1 by F5446 would decrease H3K9me3 at these effector gene promoters, thereby, un-silencing the gene expression. CD3+ T cells were cultured in anti-CD3/CD28-coated plates in the absence or presence of F5446 and analyzed for H3K9me3. Stimulation of T cells decreased H3K9me3 at the promoter regions of Gzmb and Prf1 (Fig. 5A). F5446 treatment further reduced H3K9me3 in the promoter regions of Gzmb, Prf1, Faslg, and Ifng in activated T cells (Fig. 5A). As a negative control, we analyzed the effect of F5446 on H3K9me3 in the Nxf2 promoter in T cells because the Nxf2 gene promoter has no H3K9me3 deposition in resting human T cells (Fig. 5B). No H3K9me3 deposition was detected in Nxf2 gene promoter region in resting and activated mouse T cells, and F5446 did not alter H3K9me3 in the Nxf2 gene promoter in activated T cells (Fig. 5B). To determine whether reduced H3K9me3 leads to increased gene expression, the expression of the four effector genes was analyzed. Consistent with decreased H3K9me3 in their promoter regions, F5446 treatment increased the expression of all four effector genes in a dose-dependent manner (Fig. 5C). F5446 significantly increased the percentage of granzyme B-, IFNγ-, and FasL-positive T cells (Fig. 5D). F5446 treatment also significantly increased the protein levels (MFI) of granzyme B, IFNγ, and FasL in the T cells (Fig. 5E). Lastly, F5446 treatment increased perforin protein in activated T cells, albeit to a small degree (Fig. 5F). Taken together, we conclude that H3K9me3 directly represses Gzmb, Prf1, Faslg, and Ifng transcription in T cells.
Figure 5. The SUV39H1-H3K9me3 pathway represses effector gene expression in T cells.
A. CD3+ T cells (1.5 × 106 cells/well) were stimulated in a 24-well plate coated with anti-CD3 (8 μg/mL) and anti-CD28 (10 μg/mL) for 2 days in the absence or presence of F5446 (25 nM). The resting, stimulated, and F5446-treated T cells were analyzed by ChIP with anti-H3K9me3 and gene promoter-specific PCR primers in triplicates. ChIP 1–3 represents the three regions amplified by ChIP PCR at the respective gene promoter as shown in Fig. 3B. Column: Mean, Bar: SEM. B. H3K9me3 deposition at the Nxf2 locus was visualized and analyzed as in Fig. 3. C. Purified CD3+ T cells (1.5 × 106 cells/well) were stimulated in a 24-well plate coated with anti-CD3 (8 μg/mL) and anti-CD28 (10 μg/mL) in the presence of F5446 at the indicated doses for 2 days. The treated T cells were then analyzed for mRNA expression of the four indicated effectors. D. Purified CD3+ T cells (1.5 × 106 cells/well) were stimulated in a 24-well plate coated with anti-CD3 (8 μg/mL) and anti-CD28 (10 μg/mL) in the presence of F5446 (25 nM) for 2 days. Cells were then intracellularly stained for granzyme B, perforin, and IFNγ. Cells were also stained for cell surface FasL. Shown on the left are representative dot plots. Cells were gated as shown and quantified as the percentage of positive cells for each of the four proteins (right panel). Bar: mean. Differences in the % respective cell subsets between the indicated groups were determined by two-tail t test with p<0.05 as being statistically significant. E. The gated granzyme B- and IFNγ-positive cells, as shown in D, were further analyzed for MFI of granzyme B and IFNγ. For perforin and FasL, total cell population was analyzed for MFI of perforin and FasL. F. Purified CD3+ T cells (1.5 × 106 cells/well) were stimulated in a 24-well plate coated with anti-CD3 (8 μg/mL) and anti-CD28 (10 μg/mL) in the presence of F5446 at the indicated doses for 2 days. Cells were then analyzed for perforin protein by Western blotting. β-actin was used as a normalization control.
F5446 increases T-cell effector expression to suppress colon carcinoma growth in vivo
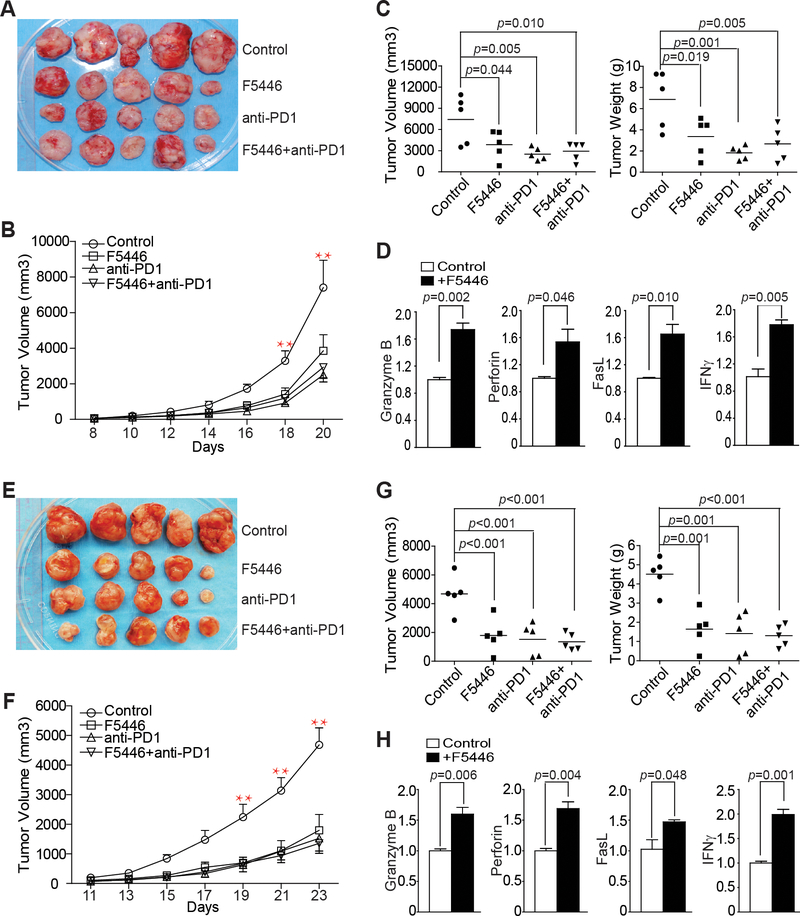
IFNγ and lymphocytes are the two primary components of the host cancer immunosurveillance system (28). The perforin-granzyme and FasL-Fas cytotoxic pathways are the two primary effector mechanisms by which T lymphocytes induce apoptosis of tumor cells (29–31). Colon tumor-infiltrating CTLs expressed high SUV39H1 in the tumor microenvironment (Fig. 2B). We, therefore, hypothesized that targeting SUV39H1 should increase effector expression in the tumor-infiltrating CTLs to suppress tumor growth in an immune competent host. To test this hypothesis, we used the MC38 colon tumor mouse model. MC38 is immunogenic and responds to anti–PD-1 immunotherapy (32). We, therefore, included anti–PD-1 immunotherapy as a positive control. MC38 tumor-bearing mice were treated with IgG, F5446, anti-PD-1, or F5446+anti–PD-l. Both F5446 and anti–PD-1 treatments suppressed the established MC38 tumor growth in a time-dependent manner (Fig. 6A and 6B). Both F5446 and anti–PD-1 treatments significantly reduced tumor growth as measured by tumor size and tumor weight (Fig. 6C). However, F5446 and anti–PD-1 did not show additive or synergistic effects (Fig. 6C). To determine whether inhibition of SUV39H1 increases T-cell effector gene expression in vivo, tumor tissues were analyzed for the expression of the four effector genes. qPCR analysis revealed that F5446 treatment significantly increased the expression of granzyme B, perforin, FasL, and IFNγ (Fig. 6D).
Figure 6. F5446 increases T-cell effector gene expression to suppress colon carcinoma growth in vivo.
A. MC38 cells (1.5×105 cells/mouse) were injected to thirty C57BL/6 mice subcutaneously. Twenty mice with similar sized tumors were randomly assigned into four groups (n=5/group) at day 8 after tumor cell injection. The four groups of tumor-bearing mice were treated with vehicle and IgG (control), F5446 (10 mg/kg), anti-PD-1 (200 μg/mouse), and F5446+anti-PD-1 every two days for 14 days. Shown are tumor images. B. Tumor growth rates in control and treatment groups were measured as tumor volume over time. Differences in tumor volume between control and treatment groups were analyzed statistically as described in the methods. **p<0.01. Bar: SEM. C. Mice were sacrificed at day 22 after tumor cell injection. Tumors were dissected and measured for volume (left panel) and weight (right panel). Differences in tumor volume and weight between control and treatment groups were determined by two-tail t test with p<0.05 as being statistically significant. Bar: mean. D. RNA was prepared from the tumor tissues of the control and F5446 treatment groups. RNA from three mice in each group was pooled and analyzed by qPCR in triplicates for the expression of indicated T-cell effector genes. β-actin was used as an internal normalization control. Differences in each gene expression between control and the treatment groups were determined by two-tail t test with p<0.05 as being statistically significant. Column: mean; Bar, SEM. E. CT26 cells (2×105 cells/mouse) were injected to thirty BALB/c mice subcutaneously. Twenty mice with similar sized tumors were randomly assigned into four groups and treated as in A. Shown are tumor images. F. Tumor growth rates in the control and treatment groups were measured as tumor volume over time and differences in tumor volume between control and treatment groups were determined statistically as described in methods.** p<0.01. Bar: SEM. G. Mice were sacrificed at day 24 after tumor cell injection. Tumors were dissected and measured for volume (left panel) and weight (right panel). Differences in tumor volume and weight between control and treatment groups were determined by two-tail t test. Bar: mean. H. RNA was prepared from total tumor tissues of the control and F5446 treatment groups. RNA from three mice in each group was pooled and analyzed by qPCR in triplicates for the expression of the indicated T-cell effector genes. β-actin was used as an internal normalization control. Differences in each gene expression between control and the treatment groups were determined by two-tail t test with p<0.05 as being statistically significant. Column: mean; Bar, SEM.
To validate the above finding, we then tested F5446 in the complimentary CT26 colon tumor mouse model. CT26 tumor-bearing mice were treated as the MC38 tumor-bearing mice. Similar to observations in the MC38 tumor model, both F5446 and anti–PD-1 treatments suppressed the established CT26 tumor growth in a time-dependent manner (Fig. 6E and6F). Both F5446 and anti–PD-1 treatments significantly reduced tumor growth as measured by tumor size and tumor weight (Fig. 6G). However, F5446 and anti–PD-1 did not show additive or synergistic effects (Fig. 6G). Analysis of total tumor tissues revealed that F5446 treatment also significantly increased the expression of granzyme B, perforin, FasL, and IFNγ (Fig. 6H).
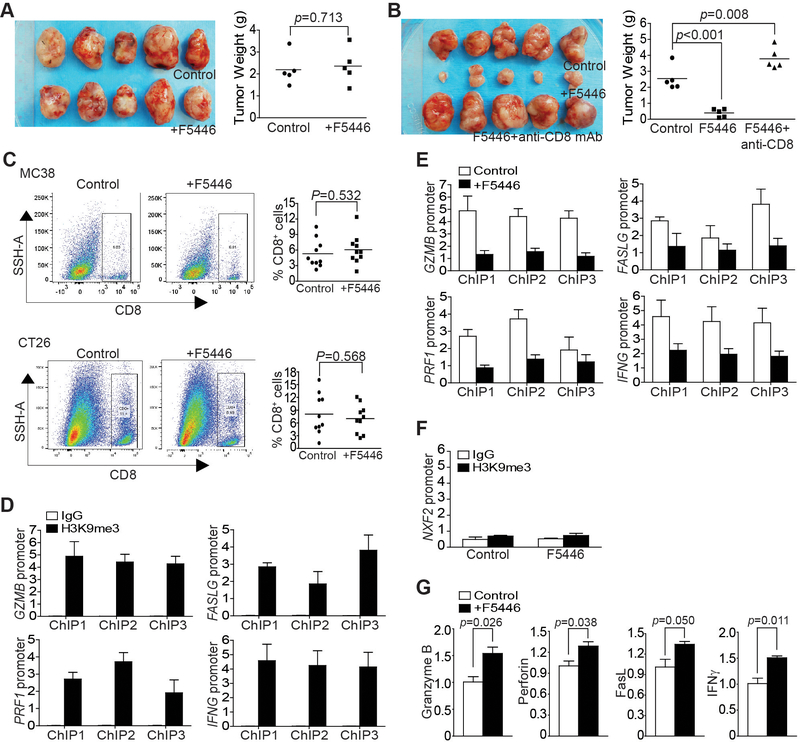
The above observations indicate that F5446 inhibits SUV39H1 to increase T-cell effector gene expression to suppress tumor growth. To validate this, MC38 tumor cells were injected to Rag1 KO mice, and the tumor-bearing mice were treated with F5446. The tumor suppression efficacy of F5446 disappeared in the Rag1 KO mice (Fig. 7A). A complimentary approach was then used to further determine the role of CTLs in F5446-mediated tumor suppression. MC38 tumor-bearing mice were treated with F5446 in the absence or presence of CD8 neutralization. Again, the efficacy of F5446 in tumor suppression was abolished when CD8+ CTLs were neutralized in the tumor-bearing mice (Fig. 7B). F5446 did not alter the level of tumor-infiltrating CTLs in both MC38 and CT26 tumors (Fig. 7C).
Figure 7. The SUV39H1-H3K9me3 pathway represses T-cell effector gene expression in tumor-infiltrating CTLs in vivo.
A. MC38 cells were injected to twelve Rag1 KO mice subcutaneously. Ten mice with similar sized tumors were randomly assigned into two groups at day 8, and treated with vehicle (control, n=5) or F5446 (10 mg/kg, n=5) every two days for 14 days. Left panel shows tumor images. Tumors were excised from sacrificed mice 22 days after tumor cell injection, weighed and presented at the right panel. Each dot represents tumor weight of tumor from one mouse. Differences in tumor weights between control and the treatment groups were determined by two-tail t test with p<0.05 as being statistically significant. B. CT26 cells (2×105 cells/mouse) were injected to twenty BALB/c mice subcutaneously. Fifteen mice with similar sized tumors were randomly assigned into three groups at day 10 after tumor cell injection and treated with vehicle (control, n=5), F5446 (10 mg/kg, n=5), and F5446+anti-CD8 (n=5) every two days for 14 days. Left panel shows tumor images. Tumors were excised from sacrificed mice 24 days after tumor cell injection, weighed, and presented at the right panel. Each dot represents tumor weight of tumor from one mouse. Differences in tumor weights between control and the treatment group were determined by two-tail t test with p<0.05 as being statistically significant. C. Tumor tissues were collected from the control (two independent experiments, n=5 each) and F5446-treated (two independent experiments, n=5 each) MC38 and CT26 tumor-bearing mice, respectively, digested with a collagenase solution and stained with Zombie Violet, anti-CD45 and anti-CD8. CD45+ cells were gated from live cells (Zombie Violet- cells) and were further gated for CD8+ cells. Shown are representative dot plots (left panel). The % CD8+ T cells was quantified and pooled from the two independent experiments and presented in the right panel. Each dot represents data from one mouse. Bar: mean. Differences in % CD8+ cells between control and the treatment groups were determined by two-tail t test with p<0.05 as being statistically significant. D. CD8+ CTLs were isolated from pooled tumor tissues of five CT26 tumor-bearing mice as shown in Fig. 6E and analyzed by ChIP with anti-H3K9me3 and gene-specific PCR primers in triplicates. ChIP 1–3 represents the three regions of the respective gene promoter amplified by ChIP PCR as shown in Fig. 3B. Column: mean; bar: SEM. E. CD8+ CTLs were isolated from pooled tumor tissues of five CT26 tumor-bearing mice from the control and F5446 treatment groups as shown in Fig. 6E. The isolated CTLs were then analyzed by ChIP with anti-H3K9me3 and gene-specific PCR primers in triplicates as indicated in Fig. 3B. The H3K9me3 was normalized to input. ChIP 1–3 indicates the ChIP PCR amplified three regions of the respective gene promoter as in Fig. 3B. Column: mean; Bar: SEM. F. CD8+ CTLs were isolated from pooled tumor tissues of five CT26 tumor-bearing mice from the control and F5446 treatment groups as shown in Fig. 6E. The isolated CTLs were then analyzed by ChIP with IgG and anti-H3K9me3, respectively, and nxf2 promoter-specific PCR primers in triplicates as indicated in Fig. 5B. The H3K9me3 was normalized to input. G. CD8+ CTLs were isolated from pooled tumor tissues of five CT26 tumor-bearing mice from the control and F5446 treatment groups as shown in Fig. 6E. The isolated CTLs were also analyzed by qPCR in triplicates for the expression of genes for granzyme B, perforin, FasL, and IFNγ. Differences in each gene expression between control and the treatment groups were determined by two-tail t test with p<0.05 as being statistically significant. Column: mean; Bar, SEM.
We then isolated CD8+ CTLs from tumor tissues of the control and the F5446-treated groups. ChIP analysis of H3K9me3 in the promoter regions of Gzmb, Prf1, Faslg, and Ifng indicated that H3K9me3 is enriched in the promoter regions of these four T-cell effector genes (Fig. 7D). Treatment with F5446 decreased H3K9me3 in the promoter regions of all four effector genes in the tumor-infiltrating CTLs (Fig. 7E). As a negative control, F5446 did not alter H3K9me3 deposition at the Nxf2 promoter region in T cells (Fig. 7F). qPCR analysis determined that the expression of granzyme B, perforin, FasL, and IFNγ were significantly increased by F5446 treatment (Fig. 7G). As a quality control, the purified CD8+ CTLs were analyzed by Western blot, and the purified cells confirmed to be CD8+ T cells (Supplementary Fig. S5). Taken together, these data indicate that F5446 inhibits SUV39H1 to increase the expression of granzyme B, perforin, FasL, and IFNγ in tumor-infiltrating CTLs, resulting in enhanced CTL lytic function to suppress tumor growth.
Discussion
The H3K9me3-specific histone methyltransferase SUV39H1 plays an essential role in epigenetic regulation of gene silencing in T cells under physiological conditions (24,33,34), as it regulates constitutive heterochromatin (25,27,35). We observed significantly elevated SUV39H1 expression in human colon carcinoma, suggesting that the SUV39H1-H3K9me3 pathway may play a critical role in silencing gene expression in tumor-infiltrating T cells and tumor cells in the tumor microenvironment under pathological conditions (36–40). To target SUV39H1, we screened the NCI small molecule virtual library and developed an SUV39H1-specific small molecule inhibitor F5446. Chaeotocin and verticillin A are natural products that inhibit the enzymatic activity of several histone methyltransferases including SUV39H1 (37,41). Although very potent as histone methyltransferase inhibitors, chaetocin and verticillin A have multiple targets. Due to their structural complexity, synthetic procedures have not been developed for either of them. We chemically synthesized F5446 and confirmed its composition using an array of analytical methods. F5446 is a small molecule inhibitor of SUV39H1and exhibited potent and specific tumor suppressor activity in vivo. Our in vivo tolerability and efficacy studies demonstrated F5446 can be administered at an efficacious dose of 10 mg/kg body weight without severe toxicities in mice.
SUV39H1 expression in tumor-infiltrating CTLs was as high as in the tumor cells. It is known that SUV39H1-H3K9me3-mediated gene silencing is critical for CD4+ T cell lineage differentiation under physiological conditions (33,34). SUV39H1 represses cytokine gene expression through H3K9me3-dependent transcription silencing to control Th1/2 cell lineage commitment and Th17 cell differentiation (33,34). It is also reported that SUV39H1-dependent H3K9me3 controls the expression of a set of stem cell-related genes in CD8+ T cells and that IFNγ+ and granzyme B+ cells decrease in SUV39H1-deficient mice compared to wild-type mice (24). In this study, we observed H3K9me3 enrichment in the promoter regions of genes encoding granzyme B, perforin, FasL and IFNγ in resting human and mouse T cells, which is consistent with the inactivation of transcription of these four effector genes in resting T cells. However, the repressive epigenetic silencing marks are diminished to create a transcriptionally active chromatin conformation in activated T cells, thereby, allowing activation of these effectors under physiological conditions (42). F5446 treatment did not alter the tumor-infiltrating CTL levels in both MC38 and CT26 colon tumor models. Further studies are needed to determine whether SUV39H1regulates survival and differentiation of subsets of CTLs in the tumor microenvironment (24). However, it is clear that SUV39H1 regulates expression of CTL effector genes in the tumor microenvironment. Our observations that tumor-infiltrating CTLs expressed high SUV39H1 indicates that these tumor-infiltrating CTLs were not functionally activated, or at least only partially activated in their lytic function. The tumor microenvironment consists of PD-L1+ tumor cells, myeloid-derived suppressor cells (MDSCs), immunosuppressive cytokines, chemokines, metabolic factors, and other immune suppressive cells. These immunosuppressive cells and factors can directly suppress T-cell activation and function through PD-L1–independent mechanisms (11,43–45), thereby, reducing tumor-infiltrating CTL activity and preventing suppression of immunogenic colon cancers. Therefore, targeting tumor-infiltrating CTL effector gene expression is a potentially effective approach to enhance the antitumor immune response.
In summary, H3K9me3 was enriched in the promoter regions of T-cell effector genes for granzyme B, perforin, FasL, and IFNγ in resting T cells and tumor-infiltrating CTLs. IFNγ and lymphocytes are the two key components of the host cancer immunosurveillance system (28). T cells, particularly CD8+ CTLs, are the major lymphocytes that suppress tumor development (46). CD8+ CTLs use the perforin-granzyme and FasL-Fas cytotoxic pathways as the two primary effector mechanisms to induce tumor cell apoptosis (15,29–31,47–49). F5446 treatment decreased H3K9me3 at the promoter regions of all four effector genes and increased their expression in activated T cells in vitro and in vivo. Treatment of mice bearing colon tumors with F5446 significantly increased the expression of granzyme B, perforin, FasL, and IFNγ and suppressed established tumor growth in a CD8+ CTL-dependent manner to a degree comparable to the anti–PD-1 immunotherapy (32). However, F5446 and anti–PD-1 showed no additive or synergistic effects on suppression of colon tumor growth in vivo, suggesting that F5446 and anti–PD-1 may use similar mechanisms to inhibit tumor growth. Nevertheless, our data document that colon tumor-infiltrating CTLs are partially inactivated by a PD-L1–independent mechanism in the tumor microenvironment and that targeting the SUV39H1-H3K9me3 pathway is potentially an effective mechanism to augment effector gene expression in tumor-infiltrating CTLs to suppress tumor growth.
Supplementary Material
Acknowledgements
We thank Dr. Kimya Jones at Georgia Esoteric Molecular Labs for immunohistochemical staining of tumor tissues and The Ohio State University Pharmacoanalytical and Comparative Pathology and Mouse Phenotyping Shared Resources for F5446 mouse tolerability studies. Grant support from NIH/NCI CA182518, CA133085 (K.L.), CA016058 (M.A.P.), and CA227433 (M.W.G., N.H.O., K.L.).
Grant support: NIH/NCI CA182518, CA133085 (K.L.) and CA227433 (M.W.G., N.H.O., K.L.) and VA Merit Review BX001962 (K.L.)
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists
Disclosure
The authors declare no conflict of interest.
References
- 1.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960–4 [DOI] [PubMed] [Google Scholar]
- 2.Galon J, Fridman WH, Pages F. The adaptive immunologic microenvironment in colorectal cancer: a novel perspective. Cancer Res 2007;67:1883–6 [DOI] [PubMed] [Google Scholar]
- 3.Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med 2005;353:2654–66 [DOI] [PubMed] [Google Scholar]
- 4.Camus M, Tosolini M, Mlecnik B, Pages F, Kirilovsky A, Berger A, et al. Coordination of intratumoral immune reaction and human colorectal cancer recurrence. Cancer Res 2009;69:2685–93 [DOI] [PubMed] [Google Scholar]
- 5.Kroemer G, Galluzzi L, Zitvogel L, Fridman WH. Colorectal cancer: the first neoplasia found to be under immunosurveillance and the last one to respond to immunotherapy? Oncoimmunology 2015;4:e1058597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fridman WH, Galon J, Pages F, Tartour E, Sautes-Fridman C, Kroemer G. Prognostic and predictive impact of intra- and peritumoral immune infiltrates. Cancer Res 2011;71:5601–5 [DOI] [PubMed] [Google Scholar]
- 7.Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res 2011;71:1263–71 [DOI] [PubMed] [Google Scholar]
- 8.Mlecnik B, Van den Eynde M, Bindea G, Church SE, Vasaturo A, Fredriksen T, et al. Comprehensive Intrametastatic Immune Quantification and Major Impact of Immunoscore on Survival. J Natl Cancer Inst 2018;110 [DOI] [PubMed] [Google Scholar]
- 9.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le DT, Hubbard-Lucey VM, Morse MA, Heery CR, Dwyer A, Marsilje TH, et al. A Blueprint to Advance Colorectal Cancer Immunotherapies. Cancer Immunol Res 2017;5:942–9 [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74 [DOI] [PubMed] [Google Scholar]
- 13.Moller P, Koretz K, Leithauser F, Bruderlein S, Henne C, Quentmeier A, et al. Expression of APO-1 (CD95), a member of the NGF/TNF receptor superfamily, in normal and neoplastic colon epithelium. Int J Cancer 1994;57:371–7 [DOI] [PubMed] [Google Scholar]
- 14.von Reyher U, Strater J, Kittstein W, Gschwendt M, Krammer PH, Moller P. Colon carcinoma cells use different mechanisms to escape CD95-mediated apoptosis. Cancer Res 1998;58:526–34. [PubMed] [Google Scholar]
- 15.Strater J, Hinz U, Hasel C, Bhanot U, Mechtersheimer G, Lehnert T, et al. Impaired CD95 expression predisposes for recurrence in curatively resected colon carcinoma: clinical evidence for immunoselection and CD95L mediated control of minimal residual disease. Gut 2005;54:661–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masugi Y, Nishihara R, Yang J, Mima K, da Silva A, Shi Y, et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut 2017;66:1463–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marisa L, Svrcek M, Collura A, Becht E, Cervera P, Wanherdrick K, et al. The Balance Between Cytotoxic T-cell Lymphocytes and Immune Checkpoint Expression in the Prognosis of Colon Tumors. J Natl Cancer Inst 2018;110. [DOI] [PubMed] [Google Scholar]
- 18.Hodge JW, Schlom J. Comparative studies of a retrovirus versus a poxvirus vector in whole tumor-cell vaccines. Cancer Res 1999;59:5106–11 [PubMed] [Google Scholar]
- 19.Carter P, Fitzjohn S, Halazy S, Magnus P. Studies on the synthesis of the antitumor agent CC-1065. Synthesis of PDE I and PDE II, inhibitors of cyclic adenosine-3’,5’-monophosphate phosphodiesterase using the 3,3’-bipyrrole strategy. J Ameri Chem Soc 1987;109:2711–7 [Google Scholar]
- 20.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. Journal of cheminformatics 2011;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T, Xu C, Liu Y, Fan K, Li Z, Sun X, et al. Crystal structure of the human SUV39H1 chromodomain and its recognition of histone H3K9me2/3. PLoS One 2012;7:e52977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C, Yang D, Sabbatini ME, Colby AH, Grinstaff MW, Oberlies NH, et al. Contrasting roles of H3K4me3 and H3K9me3 in regulation of apoptosis and gemcitabine resistance in human pancreatic cancer cells. BMC Cancer 2018;18:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pace L, Goudot C, Zueva E, Gueguen P, Burgdorf N, Waterfall JJ, et al. The epigenetic control of stemness in CD8(+) T cell fate commitment. Science 2018;359:177–86 [DOI] [PubMed] [Google Scholar]
- 25.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000;406:593–9 [DOI] [PubMed] [Google Scholar]
- 26.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 2003;12:1591–8 [DOI] [PubMed] [Google Scholar]
- 27.Fodor BD, Shukeir N, Reuter G, Jenuwein T. Mammalian Su(var) genes in chromatin control. Annu Rev Cell Dev Biol 2010;26:471–501 [DOI] [PubMed] [Google Scholar]
- 28.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 2001;410:1107–11 [DOI] [PubMed] [Google Scholar]
- 29.Kagi D, Vignaux F, Ledermann B, Burki K, Depraetere V, Nagata S, et al. Fas and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science 1994;265:528–30 [DOI] [PubMed] [Google Scholar]
- 30.Afshar-Sterle S, Zotos D, Bernard NJ, Scherger AK, Rodling L, Alsop AE, et al. Fas ligand-mediated immune surveillance by T cells is essential for the control of spontaneous B cell lymphomas. Nat Med 2014;20:283–90 [DOI] [PubMed] [Google Scholar]
- 31.LA O, Tai L, Lee L, Kruse EA, Grabow S, Fairlie WD, et al. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature 2009;461:659–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juneja VR, McGuire KA, Manguso RT, LaFleur MW, Collins N, Haining WN, et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med 2017;214:895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quintana FJ, Jin H, Burns EJ, Nadeau M, Yeste A, Kumar D, et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat Immunol 2012;13:770–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allan RS, Zueva E, Cammas F, Schreiber HA, Masson V, Belz GT, et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature 2012;487:249–53 [DOI] [PubMed] [Google Scholar]
- 35.Muller MM, Fierz B, Bittova L, Liszczak G, Muir TW. A two-state activation mechanism controls the histone methyltransferase Suv39h1. Nat Chem Biol 2016;12:188–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olcina MM, Leszczynska KB, Senra JM, Isa NF, Harada H, Hammond EM. H3K9me3 facilitates hypoxia-induced p53-dependent apoptosis through repression of APAK. Oncogene 2016;35:793–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paschall AV, Yang D, Lu C, Choi JH, Li X, Liu F, et al. H3K9 Trimethylation Silences Fas Expression To Confer Colon Carcinoma Immune Escape and 5-Fluorouracil Chemoresistance. J Immunol 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nizialek EA, Sankunny M, Niazi F, Eng C. Cancer-predisposition gene KLLN maintains pericentric H3K9 trimethylation protecting genomic stability. Nucleic Acids Res 2016;44:3586–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suryo Rahmanto Y, Jung JG, Wu RC, Kobayashi Y, Heaphy CM, Meeker AK, et al. Inactivating ARID1A Tumor Suppressor Enhances TERT Transcription and Maintains Telomere Length in Cancer Cells. J Biol Chem 2016;291:9690–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Z, Wang L, Sun Y, Jiang Z, Domsic J, An C, et al. Menin and Daxx Interact to Suppress Neuroendocrine Tumors through Epigenetic Control of the Membrane Metallo-Endopeptidase. Cancer Res 2017;77:401–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat Chem Biol 2005;1:143–5 [DOI] [PubMed] [Google Scholar]
- 42.Russ BE, Olshanksy M, Smallwood HS, Li J, Denton AE, Prier JE, et al. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity 2014;41:853–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ries CH, Cannarile MA, Hoves S, Benz J, Wartha K, Runza V, et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell 2014;25:846–59 [DOI] [PubMed] [Google Scholar]
- 44.Holmgaard RB, Brachfeld A, Gasmi B, Jones DR, Mattar M, Doman T, et al. Timing of CSF-1/CSF-1R signaling blockade is critical to improving responses to CTLA-4 based immunotherapy. Oncoimmunology 2016;5:e1151595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol 2012;12:253–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanson HL, Donermeyer DL, Ikeda H, White JM, Shankaran V, Old LJ, et al. Eradication of established tumors by CD8+ T cell adoptive immunotherapy. Immunity 2000;13:265–76 [DOI] [PubMed] [Google Scholar]
- 47.Russell JH, Ley TJ. Lymphocyte-mediated cytotoxicity. Annu Rev Immunol 2002;20:323–70 [DOI] [PubMed] [Google Scholar]
- 48.Strater J, Wellisch I, Riedl S, Walczak H, Koretz K, Tandara A, et al. CD95 (APO-1/Fas)-mediated apoptosis in colon epithelial cells: a possible role in ulcerative colitis. Gastroenterology 1997;113:160–7 [DOI] [PubMed] [Google Scholar]
- 49.Strater J, Koretz K, Gunthert AR, Moller P. In situ detection of enterocytic apoptosis in normal colonic mucosa and in familial adenomatous polyposis. Gut 1995;37:819–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.