Abstract
In recent years, growing research interest has focused on the so-called liquid biopsy. A simple blood test offers access to a plethora of information, which might be extremely helpful in understanding or characterizing specific diseases. Blood contains different molecules, of which circulating free DNA (cfDNA), circulating tumor DNA (ctDNA), circulating tumor cells (CTCs) and extracellular vesicles (EVs) are the most relevant. Conceivably, these molecules have the potential for tumor diagnosis, monitoring tumor evolution, and evaluating treatment response and pharmacological resistance. This review aims to present a state-of-the-art of recent advances in circulating DNA and circulating RNA in gastrointestinal stromal tumors (GISTs). To date, progress in liquid biopsy has been scarce in GISTs due to several issues correlated with the nature of the pathology. Namely, heterogeneity in primary and secondary mutations in key driver genes has greatly slowed the development and application in GISTs, unlike in other tumor types in which liquid biopsy has already been translated into clinical practice. However, meaningful novel data have shown in recent years a significant clinical potential of ctDNA, CTCs, EVs and circulating RNA in GISTs.
Keywords: circulating tumor cell, ctDNA, CTCs, epigenetics, gastrointestinal stromal tumor, GIST, liquid biopsy, personalized medicine, precision medicine, soft tissue sarcoma
Introduction
Technological advances, particularly next-generation sequencing (NGS), have paved the way to personalized medicine by drastically reducing the time and costs required to assess an individual’s and disease’s genetic make-up. Nowadays, it is indisputable that NGS technology provides the opportunity to look with unprecedented depth into biological samples, identifying low and ultralow frequency DNA variants.1–3 One promising application of NGS technology is liquid biopsy for cancer detection and monitorization towards a personalized cancer-medicine strategy. In recent years, not surprisingly, we have witnessed a growing research interest in liquid biopsy. The term ‘liquid biopsy’, according to the NCI Dictionary of Cancer Terms (www.cancer.gov), is defined as ‘a test done on a sample of blood to look for cancer cells from a tumor that are circulating in the blood or for pieces of DNA from tumor cells that are in the blood’. Indeed, a simple blood test offers access to a plethora of information, which might be helpful in understanding or characterizing a broad spectrum of diseases, including cancer.4,5 Blood contains different molecules, including circulating free DNA (cfDNA), circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), circulating RNA (cRNA) and extracellular vesicles (EVs).6–11 All these molecules together potentially permit the diagnosis of tumors, monitoring their evolution, and evaluating treatment response and drug resistance.6,8,12–14 Consequently, liquid biopsy offers pivotal implications in clinical management, promising to revolutionize the standard management of oncological patients (Figure 1). Specifically, the variety of liquid biopsy applications includes:
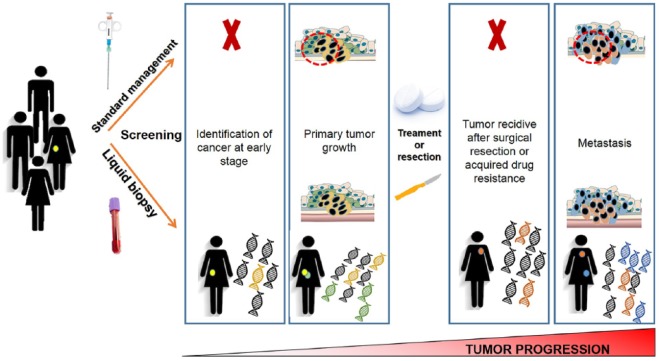
Figure 1.
Patients’ management: standard biopsy versus liquid biopsy. Potentially, a simple blood test may promote the identification of tumors at an early stage, in contrast with standard biopsy, which can be done only with advanced disease. Moreover, liquid biopsy has the advantage of providing a dynamic picture of the tumor, whereas standard biopsy may give only a static image, resulting from the small tumor tissue collected. Finally, liquid biopsy can be helpful to monitor the therapy response, due to the fact that it can detect novel resistance mutations which suggest the tumor is no longer responding to the treatment.
- Biological and clinical understanding of the disease
- Risk-based stratification of tumor patients
- Personalization of therapy
- Evaluation of clinical outcome, including therapeutic efficacy assessment.
cfDNA, ctDNA, CTCs, cRNA and EVs
cfDNA and ctDNA
Current evidence points to cfDNA being released during physiological cell functions and refers to DNA fragments outside of cells in different body fluids, including the plasma, serum, urine, and saliva.15,16 The major part of extracellular DNA is adsorbed to the surface of leukocytes or erythrocytes (cell-bound DNA) in the bloodstream.17,18 A portion can be identified in the plasma and it is known as cfDNA. cfDNA has a short half-life and is often heavily damaged, mainly due to its easy degradation by nucleases.15,19–21 cfDNA also includes ctDNA, which is DNA-derived from tumors.6 The exact mechanism through which ctDNA reaches body fluids is still unclear, although it has been proposed that apoptosis or necrosis of tumor cells, or active secretion from macrophages that have phagocytized necrotic cells, may have a prominent role in this process.9,18,21 DNA concentration in plasma varies greatly from one individual to another; for example, the cfDNA concentration is lower in healthy people than in cancer patients (10–20 mg/ml versus >1000 mg/ml),11,22,23 suggesting that the major contribution is given by ctDNA, while normal DNA only represents a small portion. As a result, ctDNA has emerged during the last decade as a novel and key source of information, profoundly diverse from tissue biopsy. Some key studies, across several cancer types, have also shown that mutations leading to treatment resistance can be detected in ctDNA several months before detection by imaging, suggesting its potential in monitoring drug response.24,25 Finally, liquid biopsy offers repeatability due to its minimally invasive nature, which in turn leads to better acceptance by patients.19
CTCs
Recently, CTC analysis has become a significant field of study in biomedical research. In particular, CTC detection has emerged as an early marker of tumor recurrence, occurring before clinical symptoms manifestation, in various tumor types.26,27 CTCs are tumor cells that may be released by early tumor lesions or metastases, generating expectations by the research community for the development of a blood-based cancer test. However, CTCs detected in blood are usually in low numbers, being estimated that ~1–10 CTCs per ml of blood released by primary tumors or metastases may be detected in peripheral blood.28–31 Therefore, the development of a reliable CTC-based test for early cancer detection or monitoring cancer progression remains challenging. In addition, CTCs are heterogeneous and may circulate as single cells or clusters of cells, making their use in the clinical setting even more complex. For example, it has been observed that CTC clusters may have a higher metastatic potential and a shorter half-life in circulation.32,33 The majority of CTCs die in the bloodstream due to different causes, including physical and oxidative stress and paucity of growth factors and cytokines. However, the cancer cells that survive can exit the bloodstream and reach the surrounding tissues, where they start to divide and grow.34,35
cRNA
More recently, research in the liquid biopsy field has also focused on cRNAs, which includes mRNA and noncoding RNA (ncRNA). To date, it is well established that cRNAs are crucial mediators in cell-to-cell communication and in the regulation of gene expression and biological functions in recipient cells, thereby acting like hormones.36,37 Similarly to cfDNA, cRNAs are highly vulnerable to degrading enzymes in the bloodstream, such as RNases. Given the critical role of these messengers, cRNA is preserved enclosed in EVs, including microvesicles, exosomes and apoptotic bodies, or complexed with specific RNA binding proteins, such as Argonaute 2 (AGO2), high-density lipoprotein and low-density lipoprotein.7,38–41
According to a length cut-off of 200 nucleotides (nts), ncRNAs encompass two super families: small ncRNAs and long ncRNAs (lncRNAs). Small ncRNAs comprise (1) microRNA (miRNAs) and small interfering RNAs (siRNAs), mediating RNA-silencing at the post-transcriptional level; (2) small nuclear RNAs (snRNAs), regulating splicing; (3) small nucleolar RNAs (snoRNAs), which may affect ribosomal RNA, transfer RNA, and snRNA processing; and (4) P-element-induced-wimpy testis (piwi)-interacting RNAs (piwiRNAs), which regulate chromatin modification and have transposon-silencing capabilities.36 The lncRNA family comprises ncRNAs that are heterogeneous in both size, from 200 to 10,000 nts, and role. Specifically, lncRNAs may act as regulators of gene expression, as scaffolds for protein binding and as decoys for different RNA molecules, including miRNAs.36,42–44 Additionally, deregulated lncRNA expression has been associated with the development of diseases, including cancer.45–47 Among all these circulating ncRNAs, miRNAs are the most known and well-characterized. miRNAs, at approximately 20–22 nts in length, exert their action as modulators by binding specific seed sequences on the 3′UTR of specific target genes. It has been widely reported that circulating miRNAs are extraordinarily stable in body fluids48 and the amount and composition of exosomal miRNAs differ between cancer patients and healthy controls, suggesting these miRNAs may represent potential non-invasive biomarkers.49–51 The most recently discovered class are circular RNAs (circRNA), functioning as sponges for miRNAs or proteins.52 circRNAs generally formed by the alternative splicing of pre-miRNAs, with 3′ and 5′ ends covalently linked,53 are relatively abundant in exosomes and represent a new frontier in cancer research.54–58
EVs
EVs are cell-derived submicron membranous vesicles released into extracellular space.59–61 They are small, lipid-bound particles packaging diverse nucleic acids and protein cargo, which are secreted from cells within normal and pathological conditions.62 These can be potentially released in all body fluids, including saliva, blood, urine, breast milk and tears. Exosomes are the smallest vesicles, ranging in size from 30 to 100 nm and are generated by exocytosis of multivesicular bodies (MVBs) (Figure 2).63 Microvesicles (MVs) are larger vesicles with a size spanning between 100 and 1000 nm, formed through a finely regulated budding/blebbing of the plasmatic membrane. MV production is generally low under physiological conditions, whereas tumors excrete them in a high constitutive manner.63–68
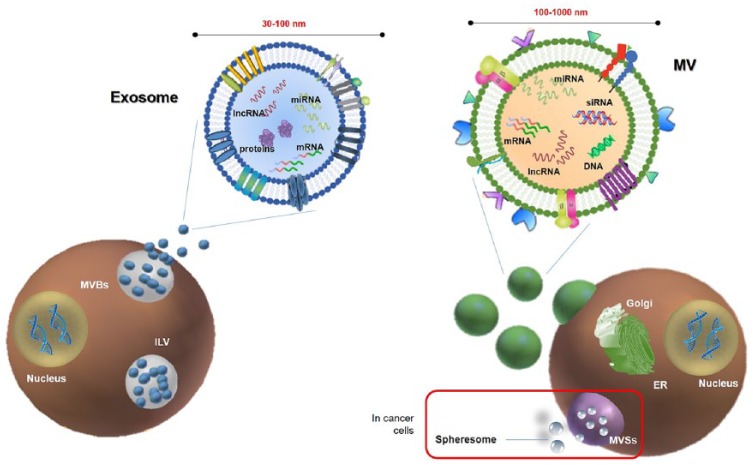
Figure 2.
Schematic representation of extracellular vesicle (EV) release. Left panel: Exosomes (30–100 nm in size) are released in extracellular space from multivesicular bodies (MVBs) through exocytosis. MVBs contain various intraluminal vesicles (ILVs) which are generated by the inward budding of the endosome membrane. Exosome cargo may include different kind of RNAs, such as miRNA, lncRNAs, mRNAs, otherwise quickly degraded if free. Right panel: Microvesicles (100–1000 nm in size) originate through a finely regulated budding/blebbing of the plasmatic membrane involving the Golgi apparatus. According to the classical secretory pathway, vesicles with their protein cargo, are sorted and packed in the Golgi apparatus, and then transported to the plasma membrane. In cancer, it has been proposed that there is an additional mechanism of EV release. Specifically, cancer cells may produce multivesicular spheres (MVSs), which contain many spheresomes.
Taken together, ctDNA and cRNA offer the chance to gain a time-dynamic picture of the tumor, allowing the following of the eventual progression, pharmacological response or appearance of drug resistance.13,69
This review aims to present a state-of-the-art of the current progresses in ctDNA and RNA findings in GISTs.
Table 1 summarizes the main features of cfDNA, ctDNA, CTCs, cRNA and EVs
Table 1.
Summary of the main features of circulating biomaterials.
| Biomaterial | Type | Molecular size | Physiology | Analytical technique (I: isolation; A: analysis approaches) |
Advantages | Disadvantages |
|---|---|---|---|---|---|---|
| cDNA | cfDNA | Highly fragmented; usually 166–320 bp in lenght | Released through necrosis, apoptosis, and active secretion. In healthy controls cfDNA levels are usually on average 30 ng/ml; in tumor patients cfDNA is usually 30% higher than healthy controls but can be up to 1000 ng/ml |
I: Automated extraction method with MyOne Dynabeads;20 polymer mediated enrichment or a phenol-chloroform based extraction procedure;11 magnetic bead-based method A: NGS methods, ranging from WGS or WES to targeted sequencing of a limited gene panel |
• Non-invasive; • Usable as prognostic and predictive biomarker; • Monitoring tumor progression |
• Integrity of cfDNA may be compromised in the transportation, storage, and handling of samples; plasma cfDNA degrades by 30% for each year of storage;21
• inability to specifically quantitate ctDNA fraction in cfDNA; • concentration of cfDNA did not seem to be associated with OS or PFS11 |
| ctDNA | Shorter fragment than cfDNA, highly fragmented DNA | 0.01–90% of the total cfDNA;11 secreted by tumor cells as a signaling molecule to drive tumor metastasis | I: ctDNA isolation follows the same approaches of cfDNA extraction; A: targeted approaches in which few tumor-specific mutations known from the primary tumor are used for monitoring residual disease (qRT-PCR, BEAMing, Safe-SeqS, CAPP-Seq, and TAm-Seq); genome-wide analysis for CNAs or point mutations by WGS or WES | • Monitoring therapy efficiency by detecting mutation-driven resistance; • Non-invasive |
• Limited half-life: from 16 minutes to 2.5 hours;11 The rate depends on the location, size, and vascularity of the tumor | |
| CTCs | 5–30 µm | EpCAM+/cytokeratin+/DAPI+/CD45 cell31,35 |
I: Centrifugation or high-resolution imaging combined with immunocyto-fluorescent staining;7 specific detection kit (i.e Cellsearch® system) A: CTCs can theoretically be characterized by all of the ‘omic’ assays that are usually performed on tissue biopsies. (i.e RNA-seq, CNAs, mutations’ analysis, epigenetic changes evaluation) |
• CTCs allow all the ‘omic’ analyses, as DNA sequence analysis, and study of mRNA and proteins expressed by tumor cells35 | • Very rare (~1 CTC per ml of blood) | |
| cRNA | ncRNA in complex with specific RNA binding proteins | Sizes varies upon RNA types (i.e. miRNA: ~20–22 nt, lncRNA: >200 nt) | Released mostly by through active secretion (i.e. role of messenger between cells); cRNAs are highly vulnerable due to degrading enzymes in the bloodstream; preserved enclosed in EVs or complexed with specific RNA binding proteins | Specific kits (i.e. Mirvana Paris or Trizol LS); qRT-PCR, microarray, and NGS58 | • Potential non-invasive diagnostic biomarkers; • More stable than other circulating molecules |
• Difficult measurement due to the low concentration and high RNase activities in the extracellular space |
| EVs | Exosomes | 30–100 nm | Released during reticulocyte differentiation as a consequence of multivesicular endosome fusion with the plasma membrane |
I: Ultracentrifugation, Specific kit, novel high-resolution flow cytometry-based method. Electron microscopy, flow cytometry, DLS, and nanoparticle tracking analysis61
A: analysis depend upon exosome cargo to evaluate (i.e. protein, RNA or DNA) |
• Content of exosomes is a reflection of what the cell is experiencing; dynamic mediators of intercellular communication;57
• Sensitive and non-invasive method, allowing the detection of tumors at an early stage; • Efficient tissue-specific, non-immunogenic carrier to deliver therapeutic drugs68 |
• Exosomes are potentially released by all the type of cells, including the nontumoral cells. • Some proteins are consistently associated with exosomes, not specific for cancer |
| MVs | 100–1000 nm | Budding/ blebbing of the plasmatic membrane, from lipid rafts |
I: same methods used for exosomes A: same used for exosomes |
• Similar to exosomes | • Similar to exosomes |
CAPP-Seq: cancer personalized profiling by deep sequencing; CNA: copy number aberration; CTC: circulating tumor DNA; DLS: dynamic light scattering; EV, extracellular vesicle; GIST, gastrointestinal stromal tumor; MVs: microvesicles; NGS: next-generation sequencing; OS: overall survival; PCR, polymerase chain reaction; PFS, progression-free survival; qRT-PCR: quantitative-real-time reverse transcription PCR; Safe-SeqS: safe-sequencing system; TAm-Seq: tagged-amplicon deep sequencing; WGS: whole genome sequencing; WES: whole exome sequencing.
Clinical epidemiology of GISTs
GISTs are rare sarcoma with an incidence in Europe of 15–20 cases per million per year. Although GISTs account for fewer than 1% of all gastrointestinal tumors, they constitute the most common sarcoma subtype.70 KIT or PDGFRA gain-of-function mutations are the key drivers of this neoplasm and are found in approximately 85–90% of GISTs, whereas 10–15% do not present any alteration on these genes and are referred to as KIT/PDGFRA wild-type (WT) GISTs.71–76 KIT/PDGFRA mutant GISTs include a plethora of different primary mutations in well-known regions. Primary mutations in KIT involve exons 9, 11, 13 or 17, whereas exons 12, 14 or 18 are the major hotspots in the PDGFRA gene. Single amino-acid substitutions, in-frame deletions or insertions are the predominant mutations types regardless of the gene and the exons. The development of tyrosine-kinase inhibitors (TKIs), such as first-line imatinib, revolutionized GIST management in the early 2000s through specific targeting of KIT/PDGFRA molecular drivers. However, the majority of GIST patients experience disease progression associated with the acquisition of secondary KIT/PDGFRA alterations; alternatively, the involvement of pharmacogenetic and epigenetic mechanisms on imatinib resistance has been investigated, without conclusive results.75–80 To overcome the loss of imatinib response, the multikinase inhibitors sunitinib and regorafenib have been introduced successively as standard second- and third-line treatments, adding clinical benefits to GIST patients.81–85 The emergence of new studies, together with the implementation of cutting-edge technologies in the field, highlights that the ever-increasing complexity in GIST molecular biology challenges the success of consecutive lines of treatment.86 In the light of this complexity, liquid biopsy could emerge as a windfall in the near future. To date, progress in liquid biopsy in GISTs have been hampered mainly by the heterogeneity of primary and secondary mutations in KIT and PDGFRA receptor tyrosine kinases.87,88 Indeed, this heterogeneity in mutational hotspots has slowed down liquid biopsy development and clinical use in GIST patients, unlike in other tumors such as lung cancer, in which it is being successfully implemented.89 Given this assumption, in the following sections, we will analyze the most recent findings in liquid biopsy, considering both cDNA and cRNA in GISTs.
ctDNA and cfDNA in GISTs
The majority of reports are focused on primary and secondary KIT alterations, and aim to develop ctDNA as a novel biomarker to help clinicians in management of GIST patients. The first reported evidence for liquid biopsy in GISTs was presented at the 2013 ASCO Annual Meeting.90 In this study, Demetri and colleagues analyzed both plasma and tissue DNA from a subgroup of GIST patients with advanced disease following imatinib and sunitinib failure, enrolled in the phase III GRID study. Specifically, the authors compared DNA mutational status (performed by Sanger sequencing) from archival tumor tissues with plasma samples (analyzed with bead emulsion amplification and magnetics; BEAMing). The analysis showed an 84% overall concordance in detection of primary KIT exon 9 and 11 mutations between tumor tissue and plasma. In particular, the concordance was 100% for KIT exon 9, but only 79% for KIT exon 11. Remarkably, BEAMing detects more frequently KIT secondary mutations in plasma (47%) compared with tumor tissue (12%). Overall, this represents the first report validating the feasibility of plasma-based ctDNA analysis in GISTs. In the same year, Maier and colleagues reported the results from patients enrolled in the NCT01462994 trial.91 This study aimed to detect KIT and PDGFRA mutations in cfDNA in plasma from GIST patients with known activating KIT/PDGFRA mutations in tumor tissue, regardless of disease status and planned treatment.92 Specifically, the authors evaluated 291 plasma samples from 38 GIST patients using 25 different allele-specific ligation (L)-polymerase chain reaction (PCR) assays covering common KIT/PDGFRA mutations. Mutations were detected in 15 patients, 9 having active disease (i.e. having at least one progressing lesion or responding to treatment) while the remaining 6 were without evidence of residual disease after surgery. Interestingly, the authors observed dynamic changes in mutant/wt allele ratios correlated with the course of the disease.92 For instance, they showed a decrease in mutant cfDNA or negativization in patients responding to TKIs. Of note, all six patients with no evidence of disease had a high (n = 5) or moderated (n = 1) risk of relapse (Miettinen criteria), emphasizing that mutant cfDNA might also serve as tumor-specific biomarkers for the early prediction of recurrence in localized GISTs. Despite these promising data, reports on liquid biopsy in GISTs are still relatively limited (Table 2).
Table 2.
Summary of the studies performed in GIST patients and included in the review.
| Author, year | Aim | Patients |
Technique | Result | |
|---|---|---|---|---|---|
|
N
of tumor samples |
N
of plasma samples |
||||
| Demetri and colleagues90 | To consider circulating DNA in plasma as a source of tumor DNA. | 102 tissue | 163 | Sanger sequencing (tissue) and BEAMing (plasma) | Demonstrated utility of plasma-based circulating DNA analysis of target oncogenes. |
| Maier and colleagues92 | To detect tumor DNA carrying KIT or PDGFRA mutation in plasma and correlate its discovery with the disease clinical course. | FFPE from 38 GISTs |
291 (from 38 GISTs) |
Allele-specific L-PCR assay | - Confirmed presence of KIT/PDGFRA mutant cfDNA. - The amount of mutant cfDNA correlates with the disease clinical course, being significantly higher in patients with active disease compared with those in complete remission. |
| Yoo and colleagues93 | To assess the relevance of soluble serum proteins and ctDNA as biomarkers for TKI-refractory GISTs. | Archival tissue from 28 GIST |
58 (from 30 GISTs: n = 30 day 1 of cycle 1; n = 28 day 1 of cycle 2) |
BEAMing | Demonstrated usefulness of serum ctDNA for the identification of TKI-resistant mutations. |
| Bauer and colleagues94 | To evaluate plasma sequencing to detect or monitor the spectrum of resistance mutations in GISTs. | Tumor tissue from 15 GISTs |
30 (from 22 GISTs) |
Illumina MiSeq platform | Showed that plasma sequencing detects a multitude of resistance mutation in KIT and other genes. |
| Kang and colleagues95 | To analyze ctDNA from the plasma of GIST patients on TKI therapy. | Tumor tissue from 3 GISTs |
Not specified (from 3 GISTs) |
NGS | - Demonstrated detection of primary and secondary mutations in ctDNA. - Resistant mutations in ctDNA may represent early biomarkers for treatment response. |
| Wada and colleagues96 | To investigate if secondary KIT mutations can be detected in ctDNA. | Primary tumor and imatinib-resistant lesion from 4 GISTs | 8 (from 4 GISTs: samples taken before and after the treatment of imatinib-resistant lesions with sunitinib) |
NGS | - Confirmed detection of KIT secondary mutation in ctDNA. - Secondary mutation in plasma were the same identified in imatinib-resistant tumor tissue. - The fraction of ctDNA changed along with tumor status. |
| Kang and colleagues97 | To validate the use of ctDNA as a biomarker for determining KIT and PDGFRA mutations. |
FFPE from 25 GISTs |
25 (from 25 GISTs, taken before surgery) |
Sanger sequencing (tissue), NGS (tissue and plasma) | Demonstrated the feasibility of using ctDNA as a surrogate tissue for the presence of KIT/PDGFRA mutations prior to resection of primary tumor. |
| Boonstra and colleagues98 | To develop a ddPCR assay to detect common KIT exon 11 mutations in both tumor tissue and ctDNA. | Archival FFPE from 27 GISTs |
22 (from 22 GIST, taken before start of TKI treatment) |
Sanger sequencing (FFPE), NGS (FFPE), ddPCR (FFPE and plasma) | Demonstrated the feasibility of a single ddPCR assay for the detection of multiple KIT exon 11 mutations in ctDNA. |
| Namløs and colleagues99 | To detect KIT and PDGFRA mutations with high sensitivity in ctDNA from patients with GISTs through NGS. | Tissue from 50 GIST | 44 blood samples from treatment-naïve patients and 6 from GISTs under TKIs | NGS (tissue and plasma) | - Plasma from high-risk patients or with metastatic disease showed more frequently detectable mutations in ctDNA compared with patients with localized or intermediate to low-risk GISTs. - Detection of ctDNA in patients undergoing TKI treatment can be related to the disease development. |
| Li and colleagues100 | To investigate feasibility of detecting ANO1 in CTCs in GISTs and association between ANO1 expression and clinical outcome of GIST. | Blood samples from 121 GISTs (of whom, 52 were high-risk GISTs, 42 intermediate risk, 18 low or very low risk), 21 gastric cancer, 23 colorectal cancer patients and 10 healthy controls | qRT-PCR | - ANO1 is a specific marker of CTCs in GISTs -High ANO1 correlated with high risk, large tumor size and high mitotic count - ANO1 positive expression correlated with poor disease-free survival. - In the neoadjuvant setting, reduction of ANO1 expression correlated with the response to imatinib. |
|
| Atay and colleagues101 | To provide a comprehensive proteome analysis and characterization of GIST-derived exosomes that might be used as a resource for the discovery of new diagnostic biomarkers and therapeutic targets. | 30 (from 18 GISTs and 12 healthy donors) |
Mass spectrometry | Showed proteomic analysis of circulating exosomes is suitable for diagnosis, prognosis and monitoring of treatment response. | |
BEAMing: bead emulsion amplification and magnetics; cfDNA: circulating free DNA; ctDNA: circulating tumor DNA; ddPCR: digital droplet PCR; FFPE: formalin-fixed paraffin-embedded; GIST, gastrointestinal stromal tumor; PCR, polymerase chain reaction; qRT-PCR: quantitative-real-time reverse transcription PCR; NGS: next-generation sequencing; TKI, tyrosine-kinase inhibitor.
Following the work by Maier and colleagues, Yoo and collaborators assessed circulating biomarkers in TKI-refractory GIST patients recruited in a single-arm phase II trial using dovitinib.93 BEAMing analysis of ctDNA identified primary kinase mutations in 16.7% of the patients; these mutations were 100% concordant with the results observed in the corresponding tumor tissue.51 The detection of primary mutations was relatively low, compared with secondary KIT/PDGFRA mutations. This result is not surprising considering that BEAMing better detects predesigned point mutations, common as secondary mutations, rather than the complex KIT primary mutations in exon 11. Subsequently, Bauer and collaborators reported additional results on liquid biopsy in GISTs at the 2015 ASCO Annual Meeting. In particular, they prospectively collected 30 plasma and 15 matched tumor samples from 22 metastatic GIST patients.94 Using a custom-designed targeted sequencing panel in an Illumina Miseq platform, they detected a total of 87 nonsynonymous KIT mutations in plasma samples. Primary mutations, all matching tumor analysis, were identified in 41% of GIST patients; resistance mutations were observed in 86% of GIST patients, although they were also observed in patients responding to imatinib.94 Recently, Kang and collaborators, using NGS, provided more data from plasma samples through the monitorization of three GIST patients under TKI treatment. The authors analyzed tumor mutational status in baseline tumor biopsies and plasma samples collected during the follow up.95 Additional mutations in plasma emerged in those patients who had a partial response or progressive disease, whereas they kept detecting only the primary mutation in the patient with stable disease.95 The study had important limitations, such as the lack of plasma samples with matched biopsies and the absence of NGS data confirmation with a different technique. Particularly, considering that KIT and PDGFRA mutations are mutually exclusive, we may assume that the presence of a secondary PDGFRA mutation on a patient harboring a clonal KIT primary mutation is an artefact derived from the NGS methodology. Nevertheless, NGS applications hold the pivotal advantage to detect novel secondary mutations conferring resistance. Therefore, it may represent an enrichment for future studies or clinical trials of novel/repositioned/existing drugs specifically targeting secondary mutations. In the wake of this idea, Wada and coworkers investigated four imatinib-resistant GIST patients, who underwent surgical resection.96 In particular, the authors analyzed, through an NGS approach, mutations in tumor tissue from resected primary and imatinib-resistant lesions and in ctDNA isolated before and after imatinib treatment. All the four patients had a primary KIT exon 11 lesion with deletions involving codons 550 to 559. Patients with imatinib-resistant lesions had resistance mutations in the KIT exon 13 (n = 3) and exon 18 (n = 1); the same genetic alterations were measured in ctDNA.
The non-invasive detection of mutations is pivotal for the selection process of target agents. Indeed, the efficacy of sunitinib correlates with the secondary mutation genotype; specifically, sunitinib is more effective in KIT exon 13 or 14-mutant GISTs.95,102 With regard to the third-line treatment, regorafenib, the GRID study reported the same benefits for patients harboring the most common primary KIT mutations. More recently, the study by Ben-Ami and coworkers suggested regorafenib provides long-term benefit in metastatic GIST patients with KIT exon 11 primary mutations and WT for KIT/PDGFRA.80,83 Overall, it is clearly important to know the tumor secondary mutational status to predict the efficacy of TKIs in imatinib-resistant GISTs. Wada and collaborators evaluated also the cfDNA as a surrogate biomarker of response. Indeed, in the literature, there are different reports evaluating its feasibility in this context.103 The study by Wada and colleagues, also reported that cfDNA decreased marginally with treatment in two patients, while another patient with stable disease exhibited a substantial increment in the cfDNA concentration. These data pinpoint that the concentration of cfDNA might not accurately reflect tumor evolution.96 However, we should not under-evaluate that specific tumor markers may be predominantly present in the cell-bound rather than in the cell-free fraction.104 This issue should be considered with caution as tumor treatment often influences leukocytes or erythrocytes apoptosis, with consequent release of cell-bound DNA into plasma. Therefore, an increase in some markers will be a clue to blood cell death, rather than reflecting tumor growth. Actually, the work by Wada and colleagues in one patient following surgery of primitive lesion, showed that cfDNA increased substantially, while ctDNA was below the threshold of detection before recurrence. After progression of the imatinib-resistant lesion, ctDNA increased and then returned to the value below the threshold following sunitinib treatment, while cfDNA was constantly at high levels. In this regard, ctDNA may be a better biomarker compared with cfDNA.96 Nonetheless, this has to be taken with caution as ctDNA may reflect a mixed population: it can derive from dying tumor cells responding to therapy or from tumor cells resistant to therapy.18 Unfortunately, to date, no further reports focusing on the impact of resistance mutations have been published. Taken together, the data led us to speculate on the importance of liquid biopsy to follow the tumor evolution under TKI treatment. However, the available literature is still too scarce, and additional prospective investigations recruiting a major number of patients are critical before its translation into clinical practice in GISTs.
More recently, three papers have focused their attention on the detection of primary mutations in GISTs. In the first of these works, Kang and colleagues97 analyzed plasma samples, collected before surgery, from 25 patients with localized gastric GISTs. This is one of the few studies addressing the role of ctDNA detection in localized GISTs. The standard treatment for localized GISTs is complete surgical resection; however, mutational status is important for the indication of adjuvant or neoadjuvant imatinib therapy, and occasionally helps in the diagnosis of GISTs. However, tumor tissue samples before surgery can be inadequate for standard mutation analysis. In this context, liquid biopsy may have the potential to detect primary mutations prior to resection. In addition, presence of mutant ctDNA after surgery might allow assessing microscopic residual disease, possibly responsible of recurrence, and guiding adjuvant therapy recommendation. Mutational status of the paired plasma-tissue samples were investigated through Sanger sequencing (tissue) and an NGS panel covering KIT exons 9, 11, 13, 17 and PDGFRA exon 18 (plasma). A total of 18 out of 25 GISTs were KIT exon 11 mutants, and the remaining were KIT/PDGFRA WT. The reported concordance between plasma and tissue samples was 72%, with 13 patients identified as KIT exon 11 mutants in plasma. None of the seven KIT/PDGFRA WT patients had measurable mutations in the plasma DNA.97 In a subsequent study, Boonstra and collaborators showed digital droplet PCR (ddPCR) may be useful in the detection of common KIT exon 11 mutations in both GIST tumor tissue and ctDNA.98 In particular, the authors used an in-house designed single ddPCR assay covering two hotspots in exon 11. According to COSMIC, around 80% of the mutations in this exon cluster in two hotspot regions of approximately 25 bp within a 100 bp range from each other.105 The authors first validated ddPCR in 36 pretreatment biopsies of GIST patients previously tested via Sanger sequencing or NGS. A total of 27 patients were KIT exon 11 mutants, whereas 9 had no KIT exon 11 mutations and served as negative controls. ddPCR resulted in 100% of specificity since all controls turned out to be negative, and in 77% of sensitivity, detecting 21/27 mutations. However, five mutations were located within the annealing sequence of the primers; one of the five samples had a duplication that was considered negative even characterized by a typical pattern of droplet distribution. Considering the remaining 22 samples covered by ddPCR, only 1 with a single nucleotide variant located within the detection range of probe 2 was a true false-negative tumor, and therefore the assay showed an overall sensibility of 95% for the regions covered in KIT exon 11. Subsequently, the ddPCR assay was tested on plasma samples available before and at multiple time points during imatinib therapy for 14 GIST patients with metastatic disease and 8 with localized GISTs. All the 22 patients had measurable disease before collection of the first (baseline) plasma sample. Analysis of the baseline plasma sample highlighted the presence of a KIT exon 11 mutation in 13 of 14 metastasized patients, and only in 1 of 8 with localized disease. The authors also used ddPCR to monitor the treatment response in serial plasma samples from 11 metastasized GISTs under TKI treatment. They showed a decrease in KIT exon 11 mutant ctDNA during treatment, which was in agreement with radiological treatment response or stable disease, evaluated according to RECIST criteria.98
More lately, Namløs and colleagues applied an NGS approach to analyze ctDNA samples from 44 treatment-naïve GIST patients (n = 35 KIT and n = 9 PDGFRA mutants).99 Somatic mutations in ctDNA were found in 36% of the plasma samples (n = 16 patients). The ctDNA detection rate was higher for KIT mutants (42.8% mutants detected) compared with PDGFRA mutant GISTs (11.1%). In addition, plasma from high-risk patients or with metastatic disease showed more frequently detectable mutations in ctDNA compared with patients with localized or intermediate to low-risk GISTs. Furthermore, the authors showed that ctDNA detection in patients undergoing TKI treatment might be related to disease development. Indeed, analysis on six KIT mutant GISTs receiving TKIs at the time of blood collection, revealed the presence of ctDNA in patients with progressive disease; no mutations were observed in patients with stable disease.
CTCs in GIST
Research data on CTCs in GISTs are very scarce, and currently, only one study has been published in the literature.100 In particular, Li and collaborators investigated the feasibility of detecting ANO1 (known as DOG1) expression in peripheral blood mononuclear cells (PBMCs) of GIST patients. ANO1 is, together with KIT, a diagnostic biomarker in GISTs. A total of 54% of the patients analyzed were ANO1-positive and a higher expression was significantly associated with a larger tumor size, high mitotic count and risk. The authors investigated also the prognostic role of ANO1. In particular, ANO1 expression was tested in 112 before and 4 weeks after surgical resection. A total of 51.8% of patients were ANO1-positive pre-resection, and only 12.1% (n = 7) of them turned out positive after surgery; these patients were characterized by liver metastasis. Afterwards, ANO1-positive status emerged in 21 GISTs experiencing recurrence after surgery. Finally, the authors evaluated imatinib efficacy after 3 months of neoadjuvant treatment in 26 GIST patients, preoperatively treated with TKIs. ANO1 expression was tested in PBMCs pre and post imatinib treatment. The 17 patients with disease control (partial response or stable disease) showed a reduction trend of ANO1 expression and 10 patients became negative, whereas the expression level did not change in the 9 patients with progressive disease. Despite the limitations of the study, including the sample size, these results showed that CTC detection in PBMCs by quantifying ANO1 could be taken into account and may offer an interesting opportunity to monitor the disease course as well as the clinical response to imatinib.100
Circulating vesicles and RNA in GIST
If the literature on circulating DNA in GISTs is quite scarce, reports on the different molecules are anecdotal, representing a new, valid and largely unexplored field of investigation. Up to now, the majority of the studies focused on circulating vesicles. The first evidence of exosome release in GISTs dates back to 2014, when Atay and collaborators investigated the role of exosomes in mediating the complex interplay between the tumor and stroma during disease progression.101 In particular, they selected the human cell line GIST-T1 as an in vitro disease model, expressing the most common type of mutation involved in GIST pathogenesis (i.e. KIT exon 11). The authors showed GIST cells secreted high number of exosomes, or ‘oncosomes’, carrying the activated oncogenic KIT receptor. Interestingly, the authors reported that the invasion of stromal cells, through these specific exosomes, led to the production of interstitial cells of Cajal (ICC)-like cells. Indeed, these oncosomes act like phenotypic modifiers of their microenvironment, promoting tumor progression through the regulation of downstream KIT-signaling pathways in stromal cells, which differentiate to ICC-like cells. Moreover, conditioning with GIST-T1-derived exosomes promotes enhanced secretion of the matrix metalloproteinase (MMP) 1, which is recognized to dynamically contribute to tumor cell invasion.101 Even if the authors could not prove the direct contribution of KIT in this process due to methodological issues, these preliminary data indicate that tumor transformation in not solely driven by oncogenes but other factors are involved. Indeed, the selective blocking of MMP exosome-mediated, MMP-1 secretion abrogated tumor invasiveness. In other words, this first report highlighted the existence of a feedback loop between a signaling mediated by the exosomes and matrix MMPs and suggests a potential role for exosomes as stroma-modifiers.101 Subsequently, a work by Junquera and collaborators, described for the first time multivesicular sphere (MVS) production in GIST cells in vivo.106 Specifically, MVSs are spherical membrane structures produced through a budding process from the plasma membrane, containing many MVs referred to as spheresomes. Interestingly, spheresomes are different from exosomes, and represent a novel mechanism coming from a spherical membrane structure. Analyzing eight gastric biopsies from GIST patients, MVSs containing spheresomes were observed establishing interactions with cytoskeleton filaments and the extracellular matrix. In particular, once in the extracellular matrix, medium MVSs can release the spheresomes (remaining empty) or cross the wall of blood vessels near cancer cells, entering the circulation. This last observation contributes to strengthening the evidence that tumor-derived EVs, besides stimulating cells at distant sites in the organism, play a key role in the initiation of the metastatic niche. The idea is that tumor-derived EVs give rise to the receptive microenvironment supporting the cell arrival, engraftment and survival in the metastatic site.106,107 In addition, Junquera and colleagues observed a considerable variability depending on the tumor sample; in particular, early stage tumors (<4.5 cm and low mitotic activity index) secrete a high number of spheresomes, while tumor with high mitotic activity do not show a presence of spheresomes. This suggests that exacerbation of a specific communication process between mesenchymal cells within tumors could occur, facilitating growth or metastases.106 Therefore, MVSs may represent a novel and alternative approach to cancer treatment in which MVSs are important therapeutic targets, in a strategy aimed at neutralizing or trapping, thus preventing the signaling process they initiate. After these preliminary studies, the most recent data in GISTs were presented, this year, by Atay and collaborators.108 In this work, they performed a comprehensive vesicular proteome profiling of GIST-derived exosomes (GDEs), from two GIST cell lines (GIST-T1 and GIST-882), providing important information on the content, biological role and therapeutic value of these vesicles. Specifically, authors showed that GIST cell lines are characterized by an inherent overactive exosome production mechanism, leading to their release and accumulation. Proteomic analysis showed that total exosomal protein content was significantly higher in GDEs compared with the non-transformed primary myometrial smooth muscle cells, representing the host healthy cells surrounding the tumor in vivo. In particular, the authors identified a core of 1060 proteins supporting the exosomal origin (e.g. features shared with exosomes-derived from other cells types), while maintaining the tumor identity. Specifically, the core protein was enriched in diagnostic markers and other features related to GISTs as well as novel kinases, phosphatases and tumor-associated antigens, previously unreported in GISTs. Interestingly, among the markers, the authors showed an enrichment of the markers of autophagy, which is involved in GIST survival and progression (for a review see Ravegnini and colleagues109). An added value of this work is the evaluation of selected GDE-associated core protein in clinical specimens, with the capture and isolation, of KIT positive (KIT+) exosomes from the plasma of GIST patients (n = 18) and healthy donors (n = 12). In this preliminary analysis, Atay and colleagues showed that the number of KIT+ vesicles in controls was small, suggesting the majority of them in GIST patients originate from the tumor.101 This concept was further supported by the correlation between circulating KIT+ levels with tumor burden and treatment response. Indeed, the authors observed that accumulation of circulating KIT+ exosomes was: (1) enhanced in the peripheral blood of patients with metastatic GISTs compared with primary disease, and (2) decreased in patients responding to treatment. In view of these considerations, quantitative changes in exosomes might represent a tool to predict malignant capabilities (e.g. recurrence or metastasis) and response to therapy. Overall, the results of this comprehensive proteomic analysis of exosomes secreted by GIST cells have unveiled different clinically relevant candidates to be circulating diagnostic and monitoring-disease biomarkers. Nevertheless, despite an in-depth examination of the proteome of isolated KIT+ exosomes holding great promise, the feasibility of the analysis remains to be elucidated.
To date, no additional studies in GISTs described these or additional kinds of circulating molecules. In particular, the literature on circulating miRNAs or lncRNAs is missing, leaving many questions unanswered on their potential role in tumor progression and metastasis, as well as in TKIs response.
A small number of studies on miRNAs and lncRNAs, limited to tissue GISTs (for a review see Nannini and colleagues110 and Kupcinskas111) have showed a few miRNAs and lncRNAs involved in regulating several genes and biological processes in GIST pathogenesis; however, up to now, none of these have been translated to the clinic or obtained from the bloodstream. Nevertheless, in the near future, molecular investigation of miRNA and lncRNA could represent interesting circulating candidates as prognostic, diagnostic and disease monitoring biomarkers.
Liquid biopsy in GISTs: clinical utility and challenging issues
Liquid biopsy has demonstrated to be a valuable tool, and recent technological innovations are generating promising clinical results, suggesting that liquid biopsy might be incorporated into clinical practice in the near future. This is the case of ctDNA analysis in non-small cell lung carcinoma and CTCs analysis in breast and colon cancer.112–115 However, for other tumors, liquid biopsy is still a wish, highlighting a great lack of homogeneity between the various types of cancer patients. In this context, GISTs, despite the attractive biology based on a few driver mutations, are among the tumors in which the advances in liquid biopsy are very limited. Certainly, even if the studies so far reported do not permit drawing any definitive conclusion, some individual cases underscore that liquid biopsy may be useful in monitoring the clinical response to TKIs in GISTs. The development of resistance mutations is the main mechanism of acquired resistance to TKIs, found in 80–90% of patients experiencing disease progression.87,116,117 Therefore, the prompt identification of these mutations might be clinically relevant to drive therapeutic decision-making. On the other hand, the application of liquid biopsy to improve GIST early diagnosis appears to be challenging. In particular, the heterogeneity of primary mutations in KIT or PDGFRA and the relative paucity of circulating elements in the bloodstream clearly impairs the sensitivity. Moreover, the lack of standardized methods of analysis realizes that we are still far behind in the application of liquid biopsy in GISTs. Therefore, in order to broaden the knowledge and application of liquid biopsy in early diagnosis and prognosis, methodological issues need to be addressed. In this regard, KIT exon 11 mutations, accounting for approximately 90% of KIT mutations, vary remarkably in length and location; consequently, their detection in plasma, through PCR-based methods is quite problematic.94 Great enthusiasm has emerged regarding NGS applications, and their ability to identify low and ultralow frequency mutations, although this approach involves inherent experimental errors. Indeed, even for methods with the lowest reported error rate, thousands of false positive variants are possible in a fully sequenced human genome.118 Errors can result from bioinformatics analysis and experimental process (e.g. sample or library preparation and sequencing chemistries). Experimental errors can be reduced through confirmatory sequencing studies, in a manner independent of the algorithms and chemistries used. Nevertheless, NGS applications hold the pivotal advantage of reducing false-negative results, compared with BEAMing and allele-specific L-PCR, which are certainly limited by the identification of predesigned mutations conferring resistance. In view of this consideration, overcoming this limitation with novel NGS techniques is mandatory. Actually, optimizing target mutation profiling is beneficial for patients, as the identification of known and novel resistance mutations may help in selecting optimal responders for molecular therapies.
Conclusion
The concept of ‘oncogene addiction’ was first introduced in the late 20th century to describe the constitutive activity of specific activated/overexpressed oncogenes needed for the continuous maintenance of the malignant phenotype.119 Afterwards, it did not take long to realize that drugs, specifically targeting hyperactivated oncogenes, could selectively kill cancer cells. Overall, this finding paved the way to the era of precision medicine and targeted therapy, which are based on correct patient selection. The study and understanding of biological processes underlying tumor development and progression have deeply changed cancer treatment, as witnessed in GISTs. In the early 2000s, imatinib revolutionized the field of targeted treatment, particularly in a disease in which no effective treatments were available at that time. The identification of a specific gene status (KIT and PDGFRA mutations) in a precise tumor type (GISTs) enables the selection of patients for targeted therapies. In this panorama, it is extremely important to have tools available for early diagnosis, improving the prognosis, for real-time monitoring of the disease, and ultimately the survival rate.
Currently, tissue biopsy represents the gold standard for a precise diagnosis of cancer; however, this approach suffers from several limitations, summarized in Figure 3. In view of these limits, there is an urgent need of minimally invasive techniques allowing a strict patient follow up at different time points; here originates the concept of liquid biopsy. The development of a ‘liquid biomarker’, which can be easily isolated from any body fluids, represents a great opportunity for early diagnosis and drug response monitoring. Despite the promising expectation, the research is still in its embryonic phase. Indeed, we can list a series of pros and cons (Figure 3) related to liquid biopsy, whilst for some circulating components, such as exosomes, we are even far from clinical applications.
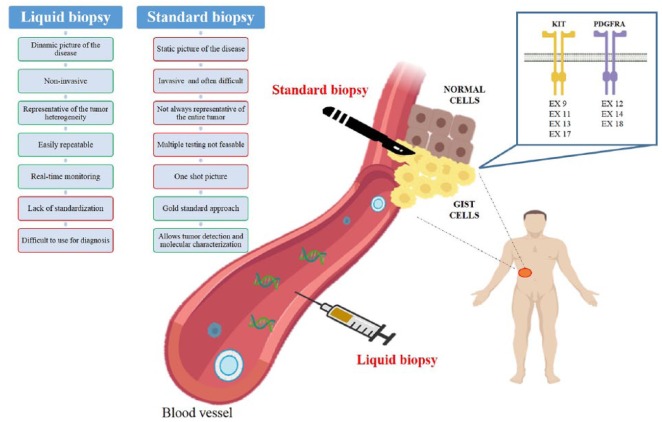
Figure 3.
Pros and cons of liquid and standard biopsy in GIST.
GIST, gastrointestinal stromal tumor.
In conclusion, liquid biopsy has entered the scene of the era of personalized medicine, representing a key tool to complement the other available techniques routinely used in the clinic. With regard to GISTs, a global effort should be considered as mandatory to translate the use of liquid biopsy into the clinic.
Footnotes
Funding: Gloria Ravegnini is supported by an MSD Italia fellowship granted by and on behalf of Merck Sharp & Dohme Corporation and L’Oréal-UNESCO for Women in Science. Giulia Sammarini is supported by Fondazione Famiglia Parmiani. This work was supported by the Ministry of Education, University and Research of Italy (MIUR, grant number 2015Y3C5KP_002 to SA).
Conflict of interest statement: The authors declare that there is no conflict of interest.
ORCID iD: Gloria Ravegnini  https://orcid.org/0000-0002-7774-402X
https://orcid.org/0000-0002-7774-402X
Contributor Information
Gloria Ravegnini, Department of Pharmacy and Biotechnology, University of Bologna, Bologna, Italy.
Giulia Sammarini, Department of Pharmacy and Biotechnology, University of Bologna, Bologna, Italy.
César Serrano, Vall d’Hebron Institute of Oncology, Vall d’Hebron University Hospital, Barcelona, Spain.
Margherita Nannini, Department of Specialized, Experimental and Diagnostic Medicine, Sant’Orsola-Malpighi Hospital, University of Bologna, Bologna, Italy.
Maria A. Pantaleo, Department of Specialized, Experimental and Diagnostic Medicine, Sant’Orsola-Malpighi Hospital, University of Bologna, Bologna, Italy
Patrizia Hrelia, Department of Pharmacy and Biotechnology, University of Bologna, Bologna, Italy.
Sabrina Angelini, Department of Pharmacy and Biotechnology, Via Irnerio 48, 40126 Bologna, Italy.
References
- 1. Rehm HL. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet 2013; 14: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kamps-Hughes N, McUsic A, Kurihara L, et al. ERASE-Seq: leveraging replicate measurements to enhance ultralow frequency variant detection in NGS data. PLoS One 2018; 13: e0195272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rabbani B, Nakaoka H, Akhondzadeh S, et al. Next generation sequencing: implications in personalized medicine and pharmacogenomics. Mol Biosyst 2016; 12: 1818–1830. [DOI] [PubMed] [Google Scholar]
- 4. Alix-Panabieres C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov 2016; 6: 479–491. [DOI] [PubMed] [Google Scholar]
- 5. Bardelli A, Pantel K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017; 31: 172–179. [DOI] [PubMed] [Google Scholar]
- 6. Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017; 14: 531–48. [DOI] [PubMed] [Google Scholar]
- 7. Bayraktar R, Van Roosbroeck K, Calin GA. Cell-to-cell communication: microRNAs as hormones. Mol Oncol 2017; 11: 1673–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murtaza M, Dawson S-J, Pogrebniak K, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 2015; 6: 8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013; 10: 472–484. [DOI] [PubMed] [Google Scholar]
- 10. Tseng J-Y, Yang C-Y, Liang S-C, et al. Dynamic changes in numbers and properties of circulating tumor cells and their potential applications. Cancers (Basel) 2014; 6: 2369–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elazezy M, Joosse SA. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J 2018; 16: 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reinert T, Schøler L V, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016; 65: 625–634. [DOI] [PubMed] [Google Scholar]
- 13. Dawson S-J, Tsui DWY, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013; 368: 1199–1209. [DOI] [PubMed] [Google Scholar]
- 14. Murtaza M, Dawson S-J, Tsui DWY, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013; 497: 108–112. [DOI] [PubMed] [Google Scholar]
- 15. Stewart CM, Tsui DWY. Circulating cell-free DNA for non-invasive cancer management. Cancer Genet. 2018; 228–229: 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stewart CM, Kothari PD, Mouliere F, et al. The value of cell-free DNA for molecular pathology. J Pathol 2018; 244: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heitzer E, Haque IS, Roberts CES, et al. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. Epub ahead of print 8 November 2018. DOI: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 18. Kidess E, Jeffrey SS. Circulating tumor cells versus tumor-derived cell-free DNA: rivals or partners in cancer care in the era of single-cell analysis? Genome Med 2013; 5: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017; 17: 223–238. [DOI] [PubMed] [Google Scholar]
- 20. Ehrich M, Tynan J, Mazloom A, et al. Genome-wide cfDNA screening: clinical laboratory experience with the first 10,000 cases. Genet Med 2017; 19: 1332–1337. [DOI] [PubMed] [Google Scholar]
- 21. Mehra N, Dolling D, Sumanasuriya S, et al. Plasma cell-free DNA concentration and outcomes from taxane therapy in metastatic castration-resistant prostate cancer from two phase III trials (FIRSTANA and PROSELICA). Eur Urol 2018; 74: 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001; 61: 1659–1665. [PubMed] [Google Scholar]
- 23. Fiala C, Diamandis EP. Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC Med 2018; 16: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012; 486: 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Diaz LA, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012; 486: 537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang ZF, Cristofanilli M, Shao ZM, et al. Circulating tumor cells predict progression-free and overall survival in Chinese patients with metastatic breast cancer, HER2-positive or triple-negative (CBCSG004): a multicenter, double-blind, prospective trial†. Ann Oncol 2013; 24: 2766–2772. [DOI] [PubMed] [Google Scholar]
- 27. Hiltermann TJN, Pore MM, van den Berg A, et al. Circulating tumor cells in small-cell lung cancer: a predictive and prognostic factor. Ann Oncol Off J Eur Soc Med Oncol 2012; 23: 2937–2942. [DOI] [PubMed] [Google Scholar]
- 28. Lianidou ES, Markou A, Strati A. The role of CTCs as tumor biomarkers. Adv Exp Med Biol 2015; 867: 341–367. [DOI] [PubMed] [Google Scholar]
- 29. Zhang C, Guan Y, Sun Y, et al. Tumor heterogeneity and circulating tumor cells. Cancer Lett 2016; 374: 216–23. [DOI] [PubMed] [Google Scholar]
- 30. Gwak H, Kim J, Kashefi-Kheyrabadi L, et al. Progress in circulating tumor cell research using microfluidic devices. Micromachines 2018; 9: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ferreira MM, Ramani VC, Jeffrey SS. Circulating tumor cell technologies. Mol Oncol 2016; 10: 374–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aceto N, Bardia A, Miyamoto DT, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014; 158: 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mitra A, Mishra L, Li S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget 2015; 6: 10697–10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer 2007; 7: 834–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cabel L, Proudhon C, Gortais H, et al. Circulating tumor cells: clinical validity and utility. Int J Clin Oncol 2017; 22: 421–430. [DOI] [PubMed] [Google Scholar]
- 36. Anfossi S, Babayan A, Pantel K, et al. Clinical utility of circulating non-coding RNAs - an update. Nat Rev Clin Oncol 2018; 15: 541–563 [DOI] [PubMed] [Google Scholar]
- 37. Cortez MA, Bueso-Ramos C, Ferdin J, et al. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol 2011; 8: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011; 108: 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011; 13: 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pucci M, Reclusa Asiáin P, Duréndez Sáez E, et al. Extracellular vesicles As miRNA nano-shuttles: dual role in tumor progression. Target Oncol 2018; 13: 175–187. [DOI] [PubMed] [Google Scholar]
- 41. Kim KM, Abdelmohsen K, Mustapic M, et al. RNA in extracellular vesicles. Wiley Interdiscip Rev RNA 2017; 8: e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10: 155–159. [DOI] [PubMed] [Google Scholar]
- 43. Ørom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 2013; 154: 1190–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ulitsky I. Interactions between short and long noncoding RNAs. FEBS Lett 2018; 592: 2874–2883. [DOI] [PubMed] [Google Scholar]
- 45. Shen X, Qi P, Du X. Long non-coding RNAs in cancer invasion and metastasis. Mod Pathol 2015; 28: 4–13. [DOI] [PubMed] [Google Scholar]
- 46. Qi P, Zhou X, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer 2016; 15: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grossi E, Sánchez Y, Huarte M. Expanding the p53 regulatory network: LncRNAs take up the challenge. Biochim Biophys Acta 2016; 1859: 200–208. [DOI] [PubMed] [Google Scholar]
- 48. Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci 2008; 105: 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cheng G. Circulating miRNAs: Roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev 2015; 81: 75–93. [DOI] [PubMed] [Google Scholar]
- 50. Berindan-Neagoe I, Monroig P, del C, Pasculli B, et al. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin 2014; 64: 311–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Redis RS, Calin S, Yang Y, et al. Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol Ther 2012; 136: 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495: 384–388. [DOI] [PubMed] [Google Scholar]
- 53. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016; 143: 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015; 25: 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fanale D, Taverna S, Russo A, et al. Circular RNA in exosomes. Adv Exp Med Biol 2018; 1087: 109–117. [DOI] [PubMed] [Google Scholar]
- 56. Greene J, Baird A-M, Brady L, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci 2017; 4: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest 2016; 126: 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee I, Baxter D, Lee MY, et al. The importance of standardization on analyzing circulating RNA. Mol Diagn Ther 2017; 21: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lane RE, Korbie D, Hill MM, et al. Extracellular vesicles as circulating cancer biomarkers: opportunities and challenges. Clin Transl Med 2018; 7: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Becker A, Thakur BK, Weiss JM, et al. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell 2016; 30: 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kastelowitz N, Yin H. Exosomes and microvesicles: identification and targeting by particle size and lipid chemical probes. Chembiochem 2014; 15: 923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang W, Xia W, Lv Z, et al. Liquid biopsy for cancer: circulating tumor cells, circulating free DNA or exosomes? Cell Physiol Biochem 2017; 41: 755–768. [DOI] [PubMed] [Google Scholar]
- 63. György B, Szabó TG, Pásztói M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci 2011; 68: 2667–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen Y, Li G, Liu M-L. Microvesicles as emerging biomarkers and therapeutic targets in cardiometabolic diseases. Genomics Proteomics Bioinformatics 2018; 16: 50–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 2009; 9: 581–593. [DOI] [PubMed] [Google Scholar]
- 66. Latifkar A, Cerione RA, Antonyak MA. Probing the mechanisms of extracellular vesicle biogenesis and function in cancer. Biochem Soc Trans 2018; 46: 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013; 200: 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. De Toro J, Herschlik L, Waldner C, et al. Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front Immunol 2015; 6: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pantel K, Alix-Panabieres C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res 2013; 73: 6384–6388. [DOI] [PubMed] [Google Scholar]
- 70. Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer 2011; 11: 865–878. [DOI] [PubMed] [Google Scholar]
- 71. Maleddu A, Pantaleo MA, Nannini M, et al. The role of mutational analysis of KIT and PDGFRA in gastrointestinal stromal tumors in a clinical setting. J Transl Med 2011; 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pantaleo MA, Astolfi A, Di Battista M, et al. Insulin-like growth factor 1 receptor expression in wild-type GISTs: a potential novel therapeutic target. Int J cancer 2009; 125: 2991–2994. [DOI] [PubMed] [Google Scholar]
- 73. Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol 2004; 22: 3813–3825. [DOI] [PubMed] [Google Scholar]
- 74. Pantaleo MA, Urbini M, Indio V, et al. Genome-wide analysis identifies MEN1 and MAX mutations and a neuroendocrine-like molecular heterogeneity in quadruple WT GIST. Mol Cancer Res 2017; 15: 553–562. [DOI] [PubMed] [Google Scholar]
- 75. Pantaleo MA, Ravegnini G, Astolfi A, et al. Integrating miRNA and gene expression profiling analysis revealed regulatory networks in gastrointestinal stromal tumors. Epigenomics 2016; 8: 1347–1366. [DOI] [PubMed] [Google Scholar]
- 76. Angelini S, Ravegnini G, Fletcher JA, et al. Clinical relevance of pharmacogenetics in gastrointestinal stromal tumor treatment in the era of personalized therapy. Pharmacogenomics 2013; 14: 941–956. [DOI] [PubMed] [Google Scholar]
- 77. Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med 2002; 347: 472–480. [DOI] [PubMed] [Google Scholar]
- 78. Serrano C, Wang Y, Mariño-Enríquez A, et al. KRAS and KIT gatekeeper mutations confer polyclonal primary imatinib resistance in GI stromal tumors: relevance of concomitant phosphatidylinositol 3-kinase/AKT dysregulation. J Clin Oncol 2015; 33: e93–e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ravegnini G, Urbini M, Simeon V, et al. An exploratory study by DMET array identifies a germline signature associated with imatinib response in gastrointestinal stromal tumor. Pharmacogenomics J. Epub ahead of print 20 September 2018. DOI: 10.1038/s41397-018-0050-4. [DOI] [PubMed] [Google Scholar]
- 80. Angelini S, Ravegnini G, Nannini M, et al. Folate-related polymorphisms in gastrointestinal stromal tumours: susceptibility and correlation with tumour characteristics and clinical outcome. Eur J Hum Genet 2015; 23: 817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Demetri GD, van Oosterom AT, Garrett CR, et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. Lancet (London, England) 2006; 368: 1329–1338. [DOI] [PubMed] [Google Scholar]
- 82. Ravegnini G, Nannini M, Zenesini C, et al. An exploratory association of polymorphisms in angiogenesis-related genes with susceptibility, clinical response and toxicity in gastrointestinal stromal tumors receiving sunitinib after imatinib failure. Angiogenesis 2017; 20:139–148. [DOI] [PubMed] [Google Scholar]
- 83. George S, Wang Q, Heinrich MC, et al. Efficacy and safety of regorafenib in patients with metastatic and/or unresectable GI stromal tumor after failure of imatinib and sunitinib: a multicenter phase II trial. J Clin Oncol 2012; 30: 2401–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ravegnini G, Sammarini G, Angelini S, et al. Pharmacogenetics of tyrosine kinase inhibitors in gastrointestinal stromal tumor and chronic myeloid leukemia. Expert Opin Drug Metab Toxicol 2016; 12: 733–742. [DOI] [PubMed] [Google Scholar]
- 85. Ravegnini G, Nannini M, Sammarini G, et al. Personalized medicine in Gastrointestinal Stromal Tumor (GIST): clinical implications of the somatic and germline DNA analysis. Int J Mol Sci 2015; 16: 15592–15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Serrano C, George S, Valverde C, et al. Novel insights into the treatment of imatinib-resistant gastrointestinal stromal tumors. Target Oncol 2017; 12: 277–288. [DOI] [PubMed] [Google Scholar]
- 87. Liegl B, Kepten I, Le C, et al. Heterogeneity of kinase inhibitor resistance mechanisms in GIST. J Pathol 2008; 216: 64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wardelmann E, Merkelbach-Bruse S, Pauls K, et al. Polyclonal evolution of multiple secondary kit mutations in gastrointestinal stromal tumors under treatment with imatinib mesylate. Clin Cancer Res 2006; 12: 1743–1749. [DOI] [PubMed] [Google Scholar]
- 89. Castellanos-Rizaldos E, Grimm DG, Tadigotla V, et al. Exosome-based detection of EGFR T790M in plasma from non-small cell lung cancer patients. Clin Cancer Res 2018; 24: 2944–2950. [DOI] [PubMed] [Google Scholar]
- 90. Demetri GD, Jeffers M, Reichardt P, et al. Mutational analysis of plasma DNA from patients (pts) in the phase III GRID study of regorafenib (REG) versus placebo (PL) in tyrosine kinase inhibitor (TKI)-refractory GIST: correlating genotype with clinical outcomes. J Clin Oncol 2013; 31: (15 Suppl.) ASCO abstracts 10503. [Google Scholar]
- 91.www.clinicaltrial.gov.
- 92. Maier J, Lange T, Kerle I, et al. Detection of mutant free circulating tumor DNA in the plasma of patients with gastrointestinal stromal tumor harboring activating mutations of CKIT or PDGFRA. Clin Cancer Res 2013; 19: 4854–4867. [DOI] [PubMed] [Google Scholar]
- 93. Yoo C, Ryu M-H, Na YS, et al. Analysis of serum protein biomarkers, circulating tumor DNA, and dovitinib activity in patients with tyrosine kinase inhibitor-refractory gastrointestinal stromal tumors. Ann Oncol Off J Eur Soc Med Oncol 2014; 25: 2272–2277. [DOI] [PubMed] [Google Scholar]
- 94. Bauer S, Herold T, Muhlenberg T, et al. Plasma sequencing to detect a multitude of secondary KIT resistance mutations in metastatic gastrointestinal stromal tumors (GIST). In: Annual Meeting of the American-Society-of-Clinical-Oncology (ASCO), 2015. [Google Scholar]
- 95. Kang G, Bae BN, Sohn BS, et al. Detection of KIT and PDGFRA mutations in the plasma of patients with gastrointestinal stromal tumor. Target Oncol 2015; 10: 597–601. [DOI] [PubMed] [Google Scholar]
- 96. Wada N, Kurokawa Y, Takahashi T, et al. Detecting secondary C-KIT mutations in the peripheral blood of patients with imatinib-resistant gastrointestinal stromal tumor. Oncology 2016; 90: 112–117. [DOI] [PubMed] [Google Scholar]
- 97. Kang G, Sohn BS, Pyo J-S, et al. Detecting primary KIT mutations in presurgical plasma of patients with gastrointestinal stromal tumor. Mol Diagn Ther 2016; 20: 347–351. [DOI] [PubMed] [Google Scholar]
- 98. Boonstra PA, ter Elst A, Tibbesma M, et al. A single digital droplet PCR assay to detect multiple KIT exon 11 mutations in tumor and plasma from patients with gastrointestinal stromal tumors. Oncotarget 2018; 9: 13870–13883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Namløs HM, Boye K, Mishkin SJ, et al. Noninvasive detection of ctDNA reveals intratumor heterogeneity and is associated with tumor burden in gastrointestinal stromal tumor. Mol Cancer Ther 2018; 17: 2473–2480. [DOI] [PubMed] [Google Scholar]
- 100. Li Q, Zhi X, Zhou J, et al. Circulating tumor cells as a prognostic and predictive marker in gastrointestinal stromal tumors: a prospective study. Oncotarget 2016; 7: 36645–36654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Atay S, Banskota S, Crow J, et al. Oncogenic KIT-containing exosomes increase gastrointestinal stromal tumor cell invasion. Proc Natl Acad Sci 2014; 111: 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Heinrich MC, Owzar K, Corless CL, et al. Correlation of kinase genotype and clinical outcome in the North American Intergroup Phase III Trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor: CALGB 150105 Study by Cancer and Leukemia Group B and Southwest Oncology Gr. J Clin Oncol 2008; 26: 5360–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wimberger P, Roth C, Pantel K, et al. Impact of platinum-based chemotherapy on circulating nucleic acid levels, protease activities in blood and disseminated tumor cells in bone marrow of ovarian cancer patients. Int J cancer 2011; 128: 2572–2580. [DOI] [PubMed] [Google Scholar]
- 104. Skvortsova TE, Rykova EY, Tamkovich SN, et al. Cell-free and cell-bound circulating DNA in breast tumours: DNA quantification and analysis of tumour-related gene methylation. Br J Cancer 2006; 94: 1492–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015; 43: D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Junquera C, Castiella T, Muñoz G, et al. Biogenesis of a new type of extracellular vesicles in gastrointestinal stromal tumors: ultrastructural profiles of spheresomes. Histochem Cell Biol 2016; 146: 557–567. [DOI] [PubMed] [Google Scholar]
- 107. Graves LE, Ariztia E V, Navari JR, et al. Proinvasive properties of ovarian cancer ascites-derived membrane vesicles. Cancer Res 2004; 64: 7045–7049. [DOI] [PubMed] [Google Scholar]
- 108. Atay S, Wilkey DW, Milhem M, et al. Insights into the proteome of gastrointestinal stromal tumors-derived exosomes reveals new potential diagnostic biomarkers. Mol Cell Proteomics 2018; 17: 495–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ravegnini G, Sammarini G, Nannini M, et al. Gastrointestinal stromal tumors (GIST): facing cell death between autophagy and apoptosis. Autophagy 2017; 13: 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Nannini M, Ravegnini G, Angelini S, et al. MiRNA profiling in gastrointestinal stromal tumors: implication as diagnostic and prognostic markers. Epigenomics; 72015; 7: 1033–1049. [DOI] [PubMed] [Google Scholar]
- 111. Kupcinskas J. Small molecules in rare tumors: emerging role of MicroRNAs in GIST. Int J Mol Sci 2018; 19: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hardingham JE, Grover P, Winter M, et al. Detection and clinical significance of circulating tumor cells in colorectal cancer–20 years of progress. Mol Med 2015; 21(Suppl. 1): S25–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ignatiadis M, Rack B, Rothé F, et al. Liquid biopsy-based clinical research in early breast cancer: the EORTC 90091–10093 treat CTC trial. Eur J Cancer 2016; 63: 97–104. [DOI] [PubMed] [Google Scholar]
- 114. Manicone M, Poggiana C, Facchinetti A, et al. Critical issues in the clinical application of liquid biopsy in non-small cell lung cancer. J Thorac Dis 2017; 9: S1346–S1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lim SY, Lee JH, Diefenbach RJ, et al. Liquid biomarkers in melanoma: detection and discovery. Mol Cancer 2018; 17: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Maleddu A, Pantaleo MA, Nannini M, et al. Mechanisms of secondary resistance to tyrosine kinase inhibitors in gastrointestinal stromal tumours (Review). Oncol Rep 2009; 21: 1359–1366. [DOI] [PubMed] [Google Scholar]
- 117. Gounder MM, Maki RG. Molecular basis for primary and secondary tyrosine kinase inhibitor resistance in gastrointestinal stromal tumor. Cancer Chemother Pharmacol 2011; 67: 25–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Robasky K, Lewis NE, Church GM. The role of replicates for error mitigation in next-generation sequencing. Nat Rev Genet 2014; 15: 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Weinstein IB. Cancer. Addiction to oncogenes–the Achilles heal of cancer. Science 2002; 297: 63–64. [DOI] [PubMed] [Google Scholar]