ASCT2 is a transporter that exchanges neutral amino acids such as glutamine across the cell membrane, an essential process for rapidly growing cells, including cancerous ones. Ndaru et al. report the synthesis and characterization of a novel class of inhibitors useful for studying this transporter.
Abstract
The neutral amino acid transporter alanine serine cysteine transporter 2 (ASCT2) belongs to the solute carrier 1 (SLC1) family of transport proteins and transports neutral amino acids, such as alanine and glutamine, into the cell in exchange with intracellular amino acids. This amino acid transport is sodium dependent, but not driven by the transmembrane Na+ concentration gradient. Glutamine transport by ASCT2 is proposed to be important for glutamine homoeostasis in rapidly growing cancer cells to fulfill the energy and nitrogen demands of these cells. Thus, ASCT2 is thought to be a potential anticancer drug target. However, the pharmacology of the amino acid binding site is not well established. Here, we report on the synthesis and characterization of a novel class of ASCT2 inhibitors based on an amino acid scaffold with a sulfonamide/sulfonic acid ester linker to a hydrophobic group. The compounds were designed based on an improved ASCT2 homology model using the human glutamate transporter hEAAT1 crystal structure as a modeling template. The compounds were shown to inhibit with a competitive mechanism and a potency that scales with the hydrophobicity of the side chain. The most potent compound binds with an apparent affinity, Ki, of 8 ± 4 µM and can block the alanine response with a Ki of 40 ± 23 µM at 200 µM alanine concentration. Computational analysis predicts inhibitor interactions with the binding site through molecular docking. In conclusion, the sulfonamide/sulfonic acid ester scaffold provides facile synthetic access to ASCT2 inhibitors with a potentially large variability in chemical space of the hydrophobic side chain. These inhibitors will be useful chemical tools to further characterize the role of ASCT2 in disease as well as improve our understanding of inhibition mechanisms of this transporter.
Introduction
The solute carrier 1 (SLC1) family of transporters consists of acidic amino acid transporters, excitatory amino acid transporters (EAATs; Arriza et al., 1994), and two neutral amino acid transporters, the alanine serine cysteine transporters 1 (ASCT1; Zerangue and Kavanaugh, 1996) and ASCT2 (Kanai, 1996). These neutral amino acid transporters were shown to transport amino acids with small, hydrophilic side chains, such as serine, cysteine, asparagine, and glutamine (Bröer et al., 1999), but also alanine with the nonpolar methyl side chain (Pinilla et al., 2001). Interestingly, amino acids are not transported by a regular sodium symport mechanism but are rather imported into the cell by obligate exchange with an internal, neutral amino acid (Bussolati et al., 1992; Bröer et al., 2000; Zander et al., 2013). While this exchange process is not driven by the transmembrane Na+ concentration gradient, it is modulated by sodium, which binds to the transporter with high affinity (Grewer and Grabsch, 2004; Zander et al., 2013). Therefore, exchange, while electroneutral (Bussolati et al., 1992), was also proposed to be dependent on the transmembrane potential (Zander et al., 2013).
Recently, ASCT2 has received increasing attention in the literature because of its proposed involvement in nutrient transport in cancer cells (Fuchs and Bode, 2005; Nicklin et al., 2009; Schulte et al., 2018; van Geldermalsen et al., 2018). These rapidly growing cells have a high nitrogen and energy demand, which can be fulfilled by increased import of glutamine as a source of nitrogen and energy. ASCT2 is thought to play a pivotal role in this process in synergy with another neutral amino acid transporter, LAT1 (Large Neutral Amino Acids Transporter 1; Fuchs and Bode, 2005; Nicklin et al., 2009). Together, these transporters also supply leucine to the cell in a glutamine-dependent mechanism, which in turn activates intracellular signaling cascades important for cell growth. Therefore, it was suggested that specifically inhibiting glutamine transport by ASCT2 provides a strategy for cancer therapeutics (Schulte et al., 2018). However, the pharmacology of the transporter binding site is not very well developed at present.
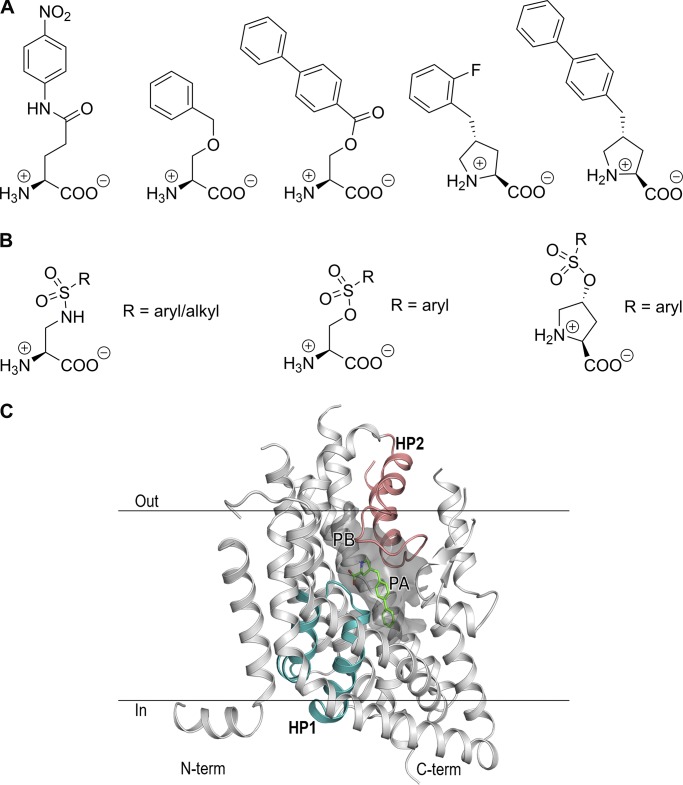
In 2004, we identified the first competitive inhibitors for ASCT2 based on structural similarity with competitive EAAT inhibitors (Grewer and Grabsch, 2004). One of these blockers is benzylserine, which blocks transport with poor apparent affinity in the 1–2 mM range, depending on alanine concentration, and was later shown to reduce cell proliferation in melanoma cells (Wang et al., 2014). Subsequently, we and others have developed blockers with higher affinities based on a variety of different scaffolds (Esslinger et al., 2005; Albers et al., 2012; Colas et al., 2015; Schulte et al., 2015, 2016; Singh et al., 2017; Garibsingh et al., 2018). For example, L-γ-glutamyl-p-nitroanilide is a commercially available ASCT2 inhibitor with micromolar affinity from a series of glutamyl anilide derivatives (Esslinger et al., 2005). Our laboratory has published results from inhibitors based on serine esters (Albers et al., 2012), as well as a structure-function analysis of benzylproline derivatives, with the best compound demonstrating an apparent Ki value of 3 µM. (Singh et al., 2017). Furthermore, a recent study demonstrated low micromolar Ki values for compounds with increased aromatic bulk in the side chain based on diaminobutanoic acid as a scaffold (Schulte et al., 2016). Some of the previously developed classes of ASCT2 inhibitors are shown in Fig. 1 A.
Figure 1.
Structures of ASCT2 inhibitors and predicted binding pockets. (A) Representative structures of previously published series of ASCT2 inhibitors. (B) Inhibitors based on amino acid scaffolds and sulfonamide and sulfonic acid ester linkages. (C) Homology model of ASCT2, illustrating P and pocket B (PB). A previously characterized inhibitor, (R)-γ-(4-biphenylmethyl)-L-proline (Singh et al., 2017), is shown in green. HP1 and HP2, bordering the binding site, are highlighted in blue and magenta, respectively.
ASCT2 inhibitors have also been developed based on in silico screening methods (Colas et al., 2015; Singh et al., 2017; Garibsingh et al., 2018). Our groups, for example, have identified competitive inhibitors and activators based on screening of databases containing close to 600,000 compounds, including the ZINC database (Colas et al., 2015). This screen resulted in γ-2-fluorobenzyl proline as a top hit, which was later used as a scaffold to identify benzylproline derivatives with higher affinity. The recently published EAAT1 crystal structure (Canul-Tec et al., 2017) allowed further refinement of the ASCT2 homology models and predicted several experimentally verified hits, which are not based on amino acid scaffolds (Garibsingh et al., 2018), thus further increasing the chemical space to be explored in the substrate binding site.
In the present study, we use computational modeling, medicinal chemistry, and electrophysiology assays to characterize a novel series of ASCT2 inhibitors, including sulfonamide and sulfonic acid ester linkers, in an effort to explore specific regions in the ASCT2 substrate binding site. These compounds are easily accessible in a two-step synthetic pathway and display apparent affinities up to 8 µM for the most potent sulfonic acid ester. These compounds add to the current library of ASCT2 inhibitors, increasing our understanding of ASCT2–ligand interactions and pharmacology of the binding site.
Materials and methods
Cell culture and transfection
Rat ASCT2 (rASCT2; Bröer et al., 1999), EAAT1 and YFP complementary DNAs were used to transiently cotransfect subconfluent human embryonic kidney 293 (HEK293) cells with POLYPLUS Jet-prime transfection reagent, according to the instructions from the supplier. Human ASCT2 (hASCT2) complementary DNA was obtained from Genecopoeia. Cells were analyzed using electrophysiological techniques 24–30 h after transfection.
Electrophysiological techniques
Electrophysiological experiments were performed as described previously (Albers et al., 2012). Except for compounds 1–3b (dissolved directly in buffer), all other compounds were first dissolved in DMSO in varying stock concentrations. Subsequently, the DMSO stock solution was diluted to various concentrations with external buffer, depending on the compounds’ solubility in water. For compounds with a single phenyl ring, concentrations of up to 4–5 mM could be achieved. In contrast, the biphenyl derivatives were only soluble up to 0.5 mM. The maximum DMSO concentration was 3%, which did not affect electrophysiological recording, as shown in control experiments. The external buffer used to dissolve the compounds and applied to cells in electrophysiological experiments contained 140 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, and 10 mM HEPES, pH 7.40. The compounds were applied to HEK293 cells expressing rat ASCT2 suspended from a current recording electrode in whole-cell configuration (Hille, 2001), immersed in the external buffer used to dissolve the compound through a rapid solution exchange device, as described previously (Grewer et al., 2000). Intracellular buffer contained 130 mM NaSCN, 2 mM MgCl2, 10 mM EGTA, and 10 mM HEPES, pH 7.40. The open pipette resistance was between 3 and 6 MΩ. Due to the relatively small currents, series resistance was not compensated. Currents traces were recorded using an Adams and List EPC7 amplifier and digitized using a Molecular Devices Digidata A/D converter.
Data analysis
Linear and nonlinear curve fittings of the experimental data were performed using MicroCal Origin software. Linear plots were fitted using the standard equation (y = a + bx) obtaining R2 and Pearson’s r values. Nonlinear dose–response relationships were fitted with a Michaelis–Menten-like equation to obtain Km, Ki, and Imax (current at saturating substrate or inhibitor concentrations) values. Each electrophysiological experiment was performed at least four times with at least three different cells at different times. Unless stated otherwise, the error bars in all our graphs represent mean ± SD.
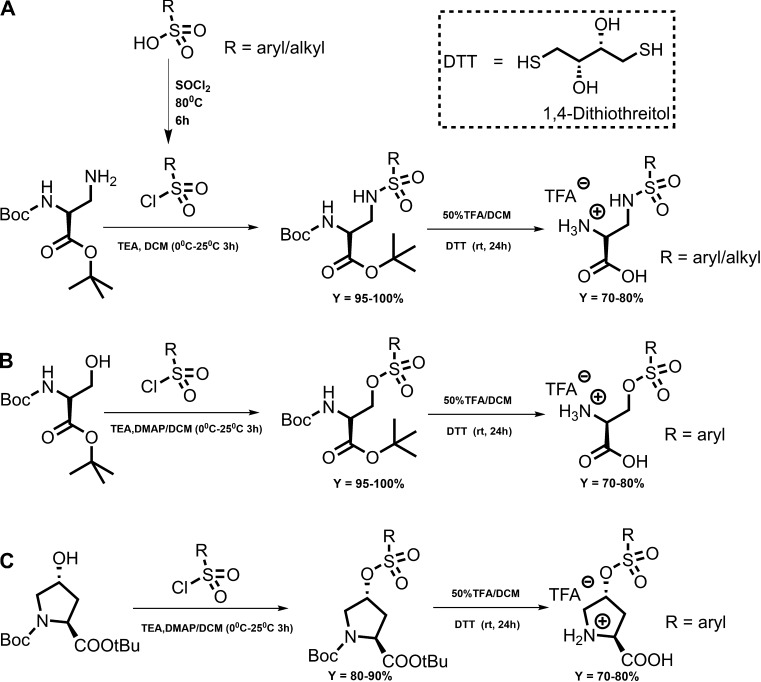
Synthesis
Chemicals were purchased from VWR or Sigma-Aldrich. Unless otherwise stated, all synthesized sulfonamides and sulfonic acid esters followed a general procedure as shown by the scheme below. The protected compounds are indicated by the suffix “a” and the deprotected compounds by the suffix “b.”
General procedure for the synthesis of sulfonamides and sulfonic acid esters through coupling with sulfonyl chlorides
Step 1 (a): Sulfonamides
100.0 mg (0.337 mmol) Boc-DAP-Otbu hydrochloride (tert-butyl-(2S)-3-amino-2-[(2-methylpropan-2-yl)oxycarbonylamino]propanoate) was charged into a previously oven-dried 10-mL round-bottomed flask fitted with a stir bar and dissolved in 3.5 ml dry dichloromethane after purging the flask with N2 gas for at least 5 min. The flask was stirred and cooled to 0°C (ice bath), and triethylamine (2 equivalent) was added via syringe under N2 gas. The mixture was stirred at 0°C for 30 min, after which the respective sulfonyl chloride (1 equivalent) dissolved in 1.5 ml of dry dichloromethane was added dropwise at 0°C under N2 gas atmosphere. The reaction was then allowed to warm up to room temperature and monitored with TLC. After 2 h, TLC showed full disappearance of starting materials. The solvent was removed by evaporation under reduced pressure and the residue suspended in 2 ml of 25% EtOAc in hexanes. The precipitate was filtered off, and the filtrate, concentrated in vacuo, was then purified by flash silica gel column chromatography (5–20% EtOAc in hexanes) to yield a pure product (>90% yield).
Step 1 (b): Sulfonic acid esters
100.0 mg (0.337 mmol) Boc-L-trans-4-hydroxyproline tert-butyl ester and 4-dimethylaminopyridine (0.1 equivalent) was charged into a previously oven-dried 10-ml round-bottomed flask fitted with a stir bar and dissolved in 3.5 ml dry dichloromethane after purging the flask with N2 gas for at least 5 min. The flask was stirred and cooled to 0°C (ice bath) and triethylamine (2 equivalent) added via syringe under N2 gas. The mixture was stirred at 0°C for 10 min, after which the respective sulfonyl chloride (1 equivalent), dissolved in 1.5 ml dry dichloromethane, was added dropwise at 0°C under N2 gas atmosphere. The reaction was then allowed to warm up to room temperature and monitored with TLC (25% EtOAc in hexanes). After 20 h, TLC showed full disappearance of starting materials. The solvent was removed by evaporation under reduced pressure and the residue suspended in 2 ml of 25% EtOAc in hexanes. The white precipitate formed was filtered off, the filtrate was concentrated in vacuo, and the product was purified by flash silica gel column chromatography (5–20% EtOAc in hexanes) to yield a white pure product (>90% yield).
For synthesis of compound 4a, the sulfonylchloride was first synthesized. Benzene sulfonic acid (500.0 mg, 3.161 mmol) was dissolved in 9 ml SOCl2 and a catalytic amount of dimethylformamide (0.1 ml) added. The reaction mixture was refluxed at 75–80 0°C for 3 h to form a yellow mixture. TLC on 3:1 hexane/ethyl acetate showed 100% conversion. Excess SOCl2 was removed under reduced pressure by successive addition of dichloromethane (DCM) and concentration in vacuo to yield a dark-brown crude sulfonyl chloride (547 mg, 98% yield) used in the next step without further purification.
The structures of the synthesized compounds were confirmed using TLC and NMR spectroscopy. The exact synthetic procedures for each compound, including NMR characterization, are shown in the Supplemental materials and methods.
General deprotection procedure for sulfonic acid esters and sulfonamides
To a cooled, oven-dried, round-bottom flask (2 or 10 mL) fitted with a stir bar, 20–100 mg pure N-Boc, t-butyl protected compound, and DL-dithiothreitol (2 equivalent) were charged, and the flask was purged with N2 for 10 min. 0.7 ml dry DCM was then added under N2 to dissolve the flask’s contents. After 10 min, 0.4 ml dry trifluoroacetic acid (TFA; 32 equivalent) was added dropwise at 0°C. The reaction mixture was left to warm up to room temperature and then stirred for 24 h monitored by TLC (5–20% MeOH in DCM). After the reaction was complete, excess TFA was removed under reduced pressure by successive addition of DCM and toluene and finally methanol. The final light-pink/white product was obtained after trituration with ether at 0°C to give pure product (confirmed by TLC). This precipitate (TFA salt) was dried overnight under vacuum and then used without further purification in electrophysiological experiments.
Molecular docking
We used our previously published model of ASCT2 (Garibsingh et al., 2018), which was based on the x-ray structure of the human EAAT1 in the outward-open conformation (PDB accession no. 5MJU; Canul-Tec et al., 2017). Docking was performed using Glide from the Schrödinger suite (Schrödinger, 2017). Specifically, the ASCT2 model was prepared with the Maestro Protein Preparation Wizard under default parameters. We then generated a grid with the Receptor Grid Generation Panel to define the binding site. The box that outlined the binding site was derived from the coordinates of the inhibitor, (3S)-3-[[3-[[4-(trifluoromethyl)benzoyl]amino]phenyl]methoxy]-L-aspartic acid (TFB-TBOA), bound to the template structure (EAAT1). Atoms of the ligand UCPH101 were removed before assigning the box. Based on our previous analysis (Garibsingh et al., 2018), docking was set up to satisfy at least one of the following three constraints: hydrogen bond of the ligand with (1) S351 or (2) S353 and (3) the nuclear Overhauser effect (NOE) constraint that requires that one or more of the ligand atoms occupy a specified region within a given distance range from the constraint center. This constraint allowed us to select ligands based on their potential to specifically dock in pocket A (PA). The NOE constraint center was defined as the carbon atom at the center of the CF3 group on TFB-TBOA with the default minimum and maximum distance parameters. This constraint suggested that the smaller molecules were not large enough to dock in PA and therefore allowed us to select ligands based on their potential to specifically dock in PA. Finally, we used induced fit docking from the Schrödinger suite (Sherman et al., 2006) to redock the ligands and evaluate the binding site flexibility. No significant movement was observed in the side chains of the residues constituting the binding site, increasing our confidence in the docking with Glide.
LigPrep was used to prepare the ligands for docking with Glide. We used the default parameters for Glide docking and specified NOE SMARTS pattern as “Donor Including Aromatic H + Halogens” based on our ligand types. PyMOL was used to visualize the docking results (Schrodinger, 2018).
Relative binding affinity estimation
To estimate the relative binding affinity between compounds and ASCT2, we used molecular mechanics generalized with born surface area solvation (MM-GBSA) with Prime in the Schrödinger suite (2017-2). This calculation provides the approximate free energies of binding, where a more negative value indicates higher binding affinity. We used the docked ligands and the prepared model (as above) without ligands as input. The distance from ligand selection was set to 5 Å to perform the calculation.
rASCT2 versus hASCT2
If not stated otherwise, electrophysiology experiments were performed using rASCT2. rASCT2 shares sequence identity of 79% (88% similarity) with hASCT2, including a highly conserved binding site (Fig. S1). For example, the residues of PA and PB in rASCT2 and hASCT2 are identical (Garibsingh et al., 2018). In addition, we have previously shown that rASCT2 used in our electrophysiology experiments yielded ranges of Ki comparable to those obtained with ASCT2 in cell-based glutamine uptake assays (Garibsingh et al., 2018). Therefore, we conclude that rASCT2 provides a valid model for investigating inhibitors targeting the substrate binding site of the hASCT2.
Online supplemental material
The online supplemental material contains five additional figures, as well as the NMR data for the synthesized compounds. Fig. S1 shows the sequence alignment; Fig. S2 shows the dose–response curve of inhibition of EAAT1 by compound 16b; Fig. S3 shows a plot of Ki versus the MM-GBSA score; Fig. S4 shows a plot of log(P) versus the MM-GBSA score; and Fig. S5 shows the predicted binding poses of all tested compounds.
Results
In this paper, the objective was to synthesize and characterize a novel class of ASCT2 inhibitors as chemical tools to characterize ASCT2. We focused on 2,3-di-aminopropionic acid and 4-hydroxyproline amino acid with sulfonamide and sulfonic acid ester linkages to hydrophobic side chains to explore the newly proposed PA in the ASCT2 substrate binding site (Colas et al., 2015), as illustrated in Fig. 1 C. Based on previous work, we hypothesized that addition of a linker region to the amino acid scaffold would allow the compounds to occupy PA, which is available to small molecules when the transporter is in an outward-open conformation (Fig. 1 C). Sulfonamides and sulfonic acid esters allow facile synthesis based on commercially available sulfonyl chlorides, providing access to a potentially large pool of compounds to be tested. The two-/three-step synthetic procedure involves the initial coupling reaction between di-protected L-2,3-diaminopropionic acid (protected at α – NH2 and α – COOH) and the respective alkly/aryl sulfonyl chloride followed by deprotection using TFA to yield the final compound. The synthetic scheme is shown in Fig. 2.
Figure 2.
Two-step synthetic scheme. (A) Sulfonamides based on the 1,2-DAP scaffold. (B) Sulfonic acid esters based on the serine scaffold. (C) Sulfonic acid esters based on hydroxyproline. DTT, DL-dithiothreitol.
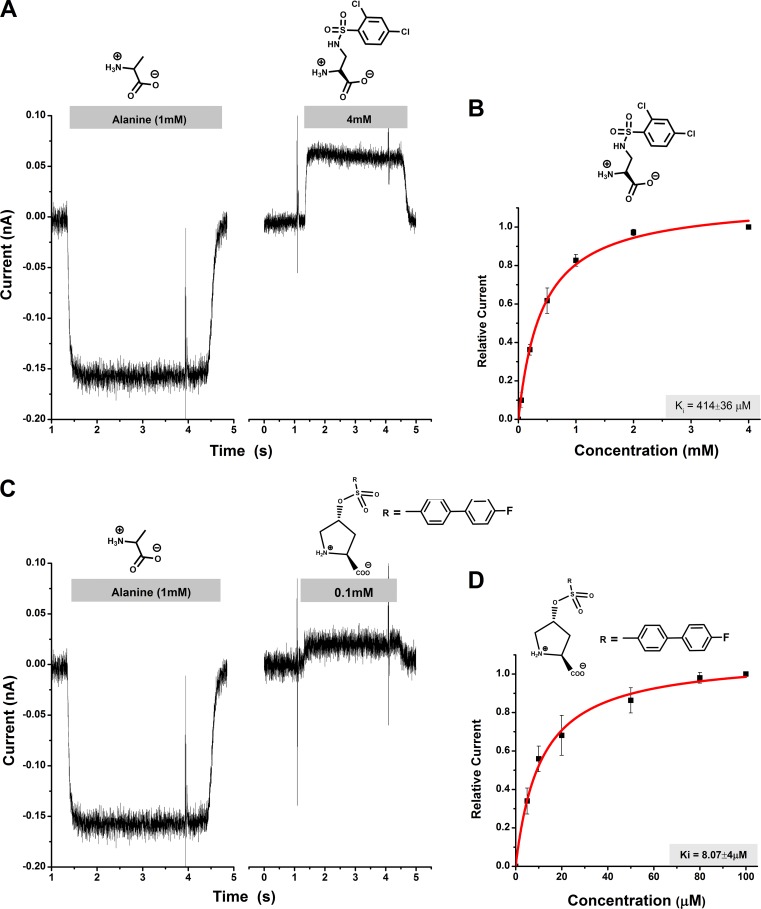
These compounds were then tested to analyze their ability to inhibit leak anion current in HEK293 cells expressing rASCT2. HEK293 cells do not express detectable levels of ASCT2 before transfection (Grewer and Grabsch, 2004), thus making them a suitable system for analysis of ASCT2 function. It was previously shown by others (Bröer et al., 2000) and us (Grewer and Grabsch, 2004) that a transporter-mediated anion current can be used to measure ASCT2 function and is a reliable measure of transport activity. This fact is also well established for the homologous glutamate transporters (Watzke et al., 2001), which have a much better established pharmacology (Shimamoto et al., 1998; Koch et al., 1999). Transport current cannot be used to test the function of ASCT2 substrates/inhibitors, because ASCT2 is an amino acid exchanger, and, thus, steady-state transport current is not observed (Bröer et al., 2000; Zander et al., 2013). However, competitive inhibitors block charge movement induced by voltage-dependent movement of transported substrates across the membrane (Zander et al., 2013), suggesting that the anion current assay reflects on exchange activity (Albers et al., 2012). Consistent with these previous observations, anion current was induced upon application of a transported substrate, such as alanine, which was inwardly directed in the presence of permeable intracellular anion (SCN−), as shown in Fig. 3 (A and C). Therefore, in all subsequent experiments, 1 mM L-alanine was used as a control.
Figure 3.
Characterization of ASCT2 inhibitors through electrophysiology. (A and C) Representative current responses to the application of 1 mM alanine (left) and selected DAP and hydroxyproline derivatives (right; time of application and concentration indicated by the gray bar) to ASCT2-expressing HEK293 cells. (B and D) Dose–response relationships for the inhibitors shown in A and C. The red lines represent the best fit to a Michaelis–Menten-like equation (apparent Ki stated in graph). Experiments were performed at 0 mV transmembrane potential in the presence of 140 mM external NaCl, 135 mM internal NaSCN, and 10 mM internal alanine. Error bars represent ±SD.
In contrast to transported substrates, competitive inhibitors were shown to block a tonic leak anion current (Albers et al., 2012; Singh et al., 2017), resulting in apparent outward direction of the current. This apparent outward current, however, is caused by the inhibition of the tonic outward flow of intracellular thiocyanate. The inhibitor-induced apparent outward current is illustrated for two representative inhibitors of the sulfonamide and sulfonic acid ester classes in Fig. 3 (A and C). The outward current was dose dependent and could be fitted to a Michaelis–Menten-type equation to yield an apparent Ki. The Ki values are summarized in Table 1. Our results show that all of the synthesized compounds block the leak anion current, indicating that they all show characteristics of competitive ASCT2 inhibitors. One of the sulfonic acid ester-based derivatives, compound 16b, blocked ASCT2 anion leak current with lower micromolar affinity (8.1 ± 4.0 µM), in the same range as the best serine-ester and benzyl proline derivatives reported so far (Albers et al., 2012; Singh et al., 2017). This was not unexpected, as this compound includes a side chain with the largest log(P) value of the compounds tested here, which shows that the side chain has the highest hydrophobicity. These results are consistent with previously reported ASCT2 inhibitors (Albers et al., 2012; Singh et al., 2017) and is well explained by our structural ASCT2 model (see below).
Table 1. Summary of inhibition constants and structures of the compounds synthesized and tested as well as docking MM-GBSA scores.
| Structure | Ki (µM) | MM-GBSA |
|---|---|---|
| 1 | 553 ± 50 | −16.06 |
| 2b | 919 ± 174 | −27.81 |
| 3b | 1,480 ± 146 | −21.05 |
| 4b | 771 ± 213 | −22.05 |
| 5b | 647 ± 87 | −33.93 |
| 6b | NA | −30.82 |
| 7b | NA | −29.44 |
| 8b | 889 ± 226 | −31.65 |
| 9b | 841 ± 80 | −31.09 |
| 10b | 642 ± 64 | −35.44 |
| 11b | 414 ± 40 | −41.85 |
| 12b | 309 ± 82 | −38.13 |
| 13b | 410 ± 356 | −34.29 |
| 14b | 418 ± 187 | −39.95 |
| 15b | 267 ± 82 | −44.47 |
| 16b | 8.07 ± 4 | −45.70 |
NA, not assessed.
Another interesting finding is that the fluoro-substituent at the 4′-position of the biphenyl ring increased apparent affinity by more than 30-fold over the nonsubstituted biphenyl derivative, as shown in Table 1. One of the sulfonamide derivatives, 2,4-dichlorobenzene, was shown to have a higher affinity compared with other derivatives at the 2 position of the ring. This fact is consistent with previous structure–function analysis of benzylproline derivatives (Singh et al., 2017). It was not possible to obtain reliable apparent binding affinities for compounds 6b and 7b due to their limited solubility in buffer and DMSO; however, the outward current induced by these compounds clearly demonstrates their inhibitory behavior.
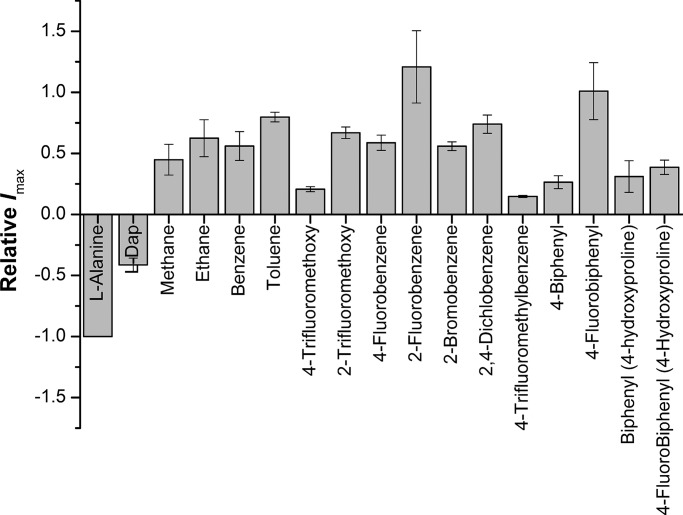
The maximum currents induced by the tested compounds at saturating concentrations (Imax) varied from one compound to another, with 4-fluorobenzene sulfonamide (compound 9b) exhibiting the highest outward current, greater in absolute magnitude than the inward current induced by the transported substrate alanine (Fig. 4). This is not consistent with previously reported results of serine esters and benzylproline derivatives (Albers et al., 2012; Singh et al., 2017), which showed the inhibitor-induced outward current to be ∼25–30% of the substrate-induced anion conductance at saturating concentrations. This deviation from previously observed behavior is likely caused by the different chemistries of the linker. The varying magnitude of Imax for the compounds reported here indicates that the ability to block the leak anion conductance is a function of the exact structure of the side chain. In addition, the Imax and Ki values did not show any significant correlation, suggesting that the two properties are caused by differing structural aspects of the inhibitors. We also tested L-2,3-diaminopropionic acid, which we used as scaffold for the sulfonamides. Interestingly, DAP (compound 1) showed properties of a transported substrate (Fig. 4 and Table 1).
Figure 4.
Current responses, Imax, induced by 1 mM alanine or saturating concentrations of sulfonamides and sulfonic acid esters. Saturating current was calculated from the response at the tested concentration (1 mM) and the known Km value, I/Imax = c/(c + Km), where c is the concentration. Apparent outward (positive) current (inhibition of anion leak current) reflects inhibitory behavior. Error bars represent ±SD.
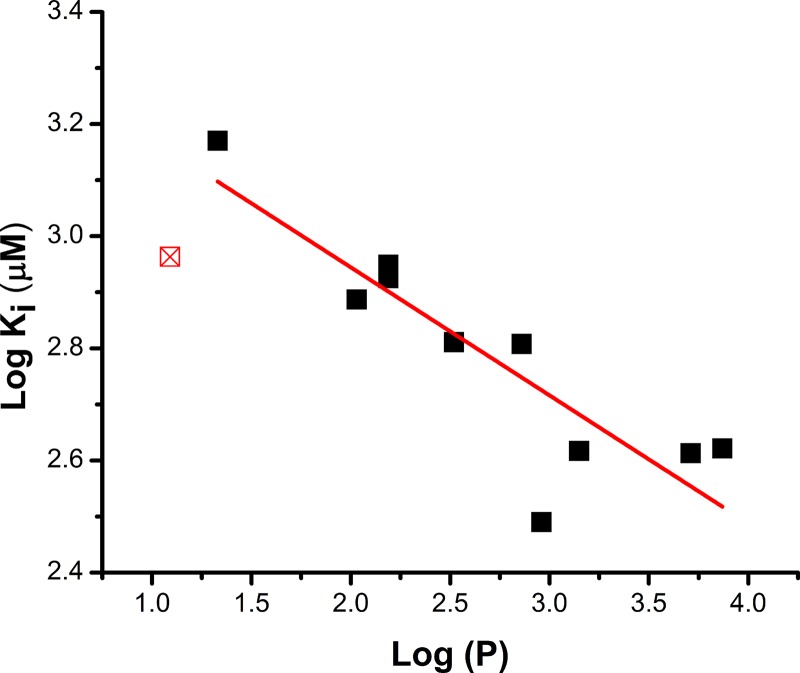
To test this hypothesis further, we looked for correlations of the Ki with physical properties of the compounds. We plotted log(Ki) versus the log(P) of the hydrophobic side chain (Fig. 5), where P is the octanol/water partition coefficient of the side chain. The data show reasonable correlation of these quantities, with an R2 for the linear regression of 0.74 and a Pearson’s r value of 0.88, indicating a good linear fit. A similar correlation with side-chain hydrophobicity has been observed previously (Singh et al., 2017) for another series of compounds, supporting the idea of a hydrophobic binding pocket accepting the side chain of the compounds. Interestingly, the methanesulfonamide (compound 2b) does not fit well with the log(Ki) versus log(P) relationship of the other compounds (Fig. 5). A possible explanation could be that sulfonamides with very short side chains start displaying substrate-like behavior, as was previously proposed for serine esters (Albers et al., 2012) and consistent with our computational analysis (see below).
Figure 5.
Inhibitor affinity correlates with the hydrophobicity of the hydrocarbon side chain. Log(Ki) is plotted as a function of log(P) of the side chain. Log(P) was calculated according to (Petrauskas and Kolovanov, 2000). R2 for the linear regression (solid line) is 0.74. Pearson’s r value is −0.88, indicating good correlation. Compound 2b was excluded from the fit (red square) because of borderline substrate-like properties.
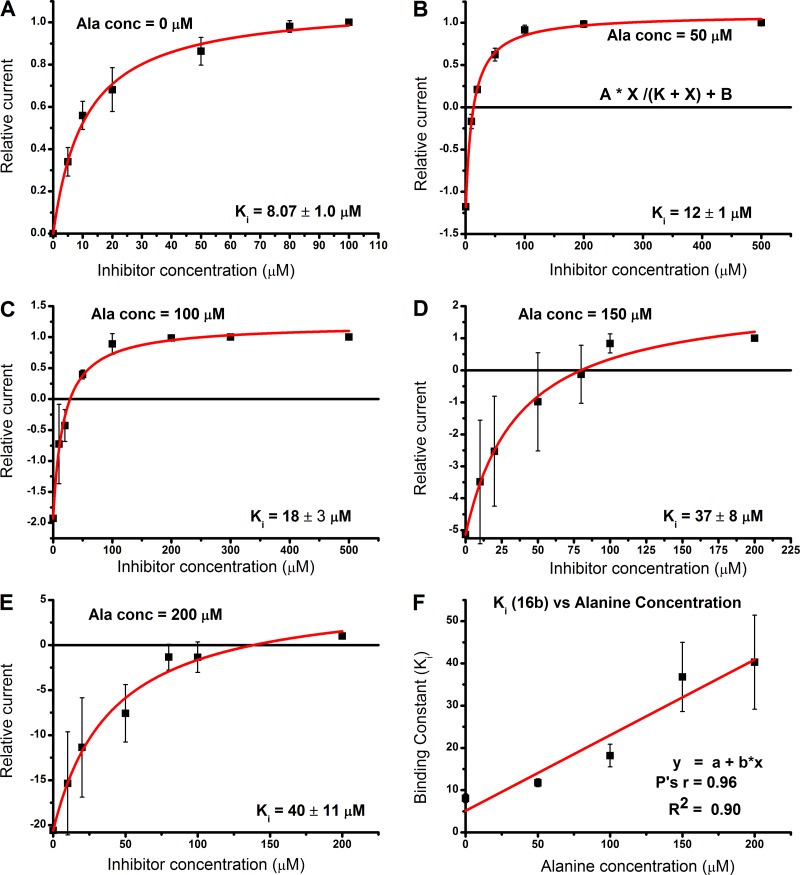
To ascertain that these compounds are competitive inhibitors, we performed competition experiments in the presence of varying alanine concentrations using the compound with the highest apparent affinity in this class (16b). As expected, at higher concentrations of alanine and low concentrations of 16b, an inward current was observed due to the inability of the compound to displace alanine, while the current reversed direction to an outward current as inhibitor concentrations were increased (Fig. 6, A–E). We also investigated the relationship of inhibitor binding constants at different alanine concentrations, which, consistent with expectations, demonstrated a linear relationship, increasing in Ki value at increasing alanine concentration (Fig. 6 F). This behavior has been reported previously for other ASCT2 blockers (Albers et al., 2012). Together, these data demonstrate that compound 16b shows the hallmark properties of a competitive inhibitor, consistent with the idea that amino acid analogues bind to the substrate binding site. Importantly, compound 16b remains a potent inhibitor with a Ki of 40 µM at alanine concentrations as high as 0.2 mM (Fig. 6 E).
Figure 6.
The inhibition mechanism is competitive. (A–E) Determination of inhibition constant Ki for compound 16b at different concentrations of alanine. The red lines are fits of the currents according to the following equation: I = I1 + I2 [Inh]/(Ki + [Inh]), where I1 is the alanine-induced current without inhibitor, and I2 is the maximum current in the presence of saturating inhibitor concentration, [Inh]max (Albers et al., 2012). (F) KI plotted as a function of the alanine concentration, showing a linear relationship according to the equation Ki(Ala) = Ki(0) + [Ala] Ki(0)/Km(Alanine), where Ki(Ala) and Ki(0) are the KI values in the presence and absence of alanine and Km(Alanine) is the apparent Michaelis–Menten constant for alanine activation of anion current. Error bars represent ±SD.
In contrast to rASCT2, hASCT2 does not transport cysteine, which rather plays the role of a modulator (Scalise et al., 2015). Therefore, inhibitor interaction may also vary between the rat and human transporters. To test this possibility, currents were recorded in response to application of compound 16b. While these currents were outward (8 ± 2 pA), indicating that the compound also acts as an inhibitor in hASCT2 at a concentration of 100 µM, we were unable to reliably determine the apparent Ki due to the poor expression of hASCT2 in the HEK293 system and, thus, small currents (<20 pA for saturating [alanine]). In addition, we tested compound 16b for interaction with rat EAAT1. As shown in Fig. S2, EAAT1 was inhibited with an apparent Ki of 7 ± 2 µM, indicating that the sulfonamide compounds are not specific for ASCT2 within the SLC1 family. This result is not unexpected, because β-sulfonamido functionalized aspartates have been previously found to inhibit glutamate transporters (Hansen et al., 2016).
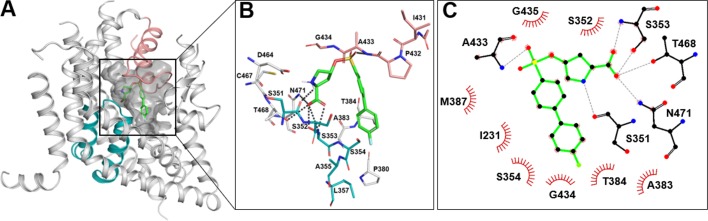
We docked the 16 compounds to our recently published model of ASCT2 in the outward-open conformation (Garibsingh et al., 2018; see Materials and methods). This model was based on the EAAT1 crystal structure that was solved with TFB-TBOA bound to PA. In agreement with previous studies (Colas et al., 2015; Garibsingh et al., 2018), the carboxy and amino group of the ligands are predicted to make polar contacts with the backbone of S351 and S353 (hairpin loop 1 [HP1]) as well as the side chain amine of N471 and the hydroxyl group of T468 (transmembrane domain 8). In addition, the oxygen of the linker region is also predicted to make hydrogen bonds with HP2. For example, the oxygen in the sulfonic acid ester linker of compound 16b makes a hydrogen bond with A433 (Fig. 7). Finally, the hydrophobic side chains of the ligands form hydrophobic interactions with PA (Fig. 7).
Figure 7.
Compound 16b predicted binding mode. (A) Compound 16b (green sticks) docked in ASCT2 outward-open binding site; HP1 is shown in teal and HP2 in salmon. (B) Docking pose of compound 16b with hydrogen bonds shown as gray dashes. Oxygen, nitrogen, sulfur and fluorine are represented in red, blue, yellow, and cyan, respectively. (C) Compound 16b–ASCT2 interactions are visualized in 2D with LigPlot (Wallace et al., 1995). Residues predicted to form hydrogen bonds are shown in black, and residues making hydrophobic contacts are shown as red arcs.
Compounds without an aromatic group in the side chain did not protrude into PA. We also observed that it was necessary to have an attachment on the aromatic ring itself if the compound was to begin angling downward toward PA. We saw a significant change in the MM-GBSA score and Ki between compounds 4b and 5b that is explained by the docking pose (Fig. S3); compound 5b is the first compound in the series with a group attached to the phenyl ring, which tilts the compound down toward PA, whereas compound 4b is too short.
Next, to relate the docking pose with predicted binding affinity, we estimated the binding affinity score with MM-GBSA and compared the data with the experimental results. While these scores are not expected to correctly predict the absolute binding energy values, they correlated well with the experimental data (Figs. 5 and S4 and Table 1). For example, compound 2b showed a substrate-like docking pose and had a more negative MM-GBSA score than compounds 3b and 4b, which had larger hydrophobic side chains, in agreement with the experimental results (Fig. S5).
We observed that the size of the hydrophobic side chain attached to the linker affected the pose and binding affinity score. Only the longer, biphenyl-substituted compounds were able to successfully dock to PA, (Fig. S5 and Table 1), with the exception of compounds 10b and 11b with their two-substituted halogen atoms, which agrees with our previous study, showing that halogen substitution on the phenyl ring increase affinity (Singh et al., 2017). It should be noted that the smallest compound (compound 1) is an outlier in the MM-GBSA/Ki correlation (Fig. S3). This result suggests that our structural model and MM-GBSA calculations capture the activity of the larger compounds that bind PA and are not optimal for identifying smaller ligands (substrates or inhibitors). In summary, from the docking poses, predicted ligand interactions, and the accompanying MM-GBSA, we predicted that compounds 15b and 16b would be the most potent inhibitors. This was confirmed with the experimental data (Table 1).
Discussion
The development of novel chemical probes to study ASCT2 is important for the further characterization of this transporter. For example, ASCT2 inhibitors can be used to stabilize uncharacterized conformations for structure determination with cryo-EM or x-ray crystallography. Here, we report on a new class of competitive ASCT2 inhibitors based on two amino acid scaffolds and the sulfonic acid/sulfonamide linker in the side chain. We selected serine, 1,2-diamino-propionic acid, and proline as amino acids, because they successfully served as scaffolds for inhibitors with other linkers to the hydrophobic side chain in the past (Albers et al., 2012; Singh et al., 2017). In addition, the inhibitors are synthetically easily accessible, with a two-step reaction scheme based on coupling and deprotection, using mostly commercially available sulfonyl chlorides. Apparent affinity of ASCT2 for the inhibitors is in the millimolar to midmicromolar range, with the strongest inhibitor showing a Ki value of ∼8 µM. These values are in a similar range of those previously found for hydrophobic moieties connected to amino acid scaffolds with carbon or ester linkages (Albers et al., 2012; Singh et al., 2017). Thus, the sulfonylamides/esters provide a useful scaffold for straightforward synthetic access to ASCT2 inhibitors. However, the sulfonyl group does not increase affinity over carboxylic esters with the same hydrophobic side chain. Inhibition of ASCT2 by the most potent compound is consistent with a competitive mechanism, with the Ki proportionally increasing with the concentration of the coapplied transported substrate alanine. The inhibitors exhibit competitive inhibition binding mode, which is expected because they are amino acid–like inhibitors that are predicted to bind to the substrate binding site (Albers et al., 2012; Singh et al., 2017). The physiological concentration of alanine and glutamine are in the 500 µM range (Cantor et al., 2017). At this concentration, compound 16b is predicted to be still quite potent, with a Ki of 94 µM, a ∼2.5-times higher affinity compared with L-γ-glutamyl-p-nitroanilide at comparable substrate concentrations (Esslinger et al., 2005).
Like other members of the SLC1 family of transporters (Wadiche et al., 1995; Ryan et al., 2004), ASCT2 catalyzes passive anion movement across the membrane through the kinetically coupled anion conductance (Bröer et al., 2000; Grewer and Grabsch, 2004; Zander et al., 2013). This anion conductance is activated by transported substrates (Bröer et al., 2000). A leak component of the anion conductance is blocked by competitive inhibitors (Grewer and Grabsch, 2004). It was previously shown for a series of serine esters that compounds with small side-chain volume, such as the ethyl and isopropyl esters, exhibited properties of transported substrates, increasing the anion conductance (Albers et al., 2012). None of the compounds reported here demonstrate activating behavior, even the compounds with the small methane- and ethane-sulfonamide groups. Therefore, it can be concluded that the sulfonyl linker is incompatible with transported substrate behavior, most likely because of molecular properties other than size. Interestingly, the potency of the inhibitors to block the ASCT2 leak anion conductance at saturating concentrations was variable (Fig. 4). This is an effect that has not been observed for previous series of inhibitory compounds (Singh et al., 2017), which all blocked the anion conductance to a similar extent. It is, therefore, possible that some of the compounds still allow the leak anion conductance to operate, even when all inhibitor binding sites are saturated.
The glutamine transporter ASCT2 has recently been proposed as a potential drug target in several studies. This proposal was based on the observation that rapidly growing cancer cells become addicted to glutamine (Wise and Thompson, 2010; Schulze and Harris, 2012; Altman et al., 2016) and that limiting the glutamine supply to these cells can lead to suppression of cancer cell growth (Wang et al., 2015; van Geldermalsen et al., 2016). Since ASCT2 is overexpressed in several cancer cells and cell lines (Kim et al., 2013; van Geldermalsen et al., 2016), a potential strategy is to target ASCT2 either through knockdown using RNA silencing methods (Fuchs et al., 2004; Wang et al., 2015), or by inhibition with small, organic molecules (Wang et al., 2014). Currently, however, no clinical drugs or even potent tool compounds are available that inhibit ASCT2 glutamine transport in vivo. One issue is that current inhibitors have a low potency, with the best inhibitors having apparent affinities in the low micromolar range. It will be crucial to further develop structure–function relationships to achieve affinities in the nanomolar range. The sulfonamide/sulfonic acid ester linkage may provide an opportunity for further structure–function analysis, because a wide variety of side chains with diverse functional properties will be available for testing through a simple two-step synthetic approach.
Another challenge for the development of ASCT2 inhibitors is the lack of an atomic-resolution experimentally determined structure in the outward-facing conformation. Recently, a cryo-EM crystal structure of hASCT2 was published (Garaeva et al., 2018). While this structure generally confirms sequence alignments used for previous homology models generated based on GltPh (Boudker et al., 2007; Colas et al., 2015) and EAAT1 (Canul-Tec et al., 2017; Garibsingh et al., 2018), it is in the inward-facing configuration and substrate bound. Therefore, this structure is less suitable for in silico docking of potential inhibitors from the extracellular side of the membrane. Notably, the potency of the new ASCT2 inhibitors correlated with our docking and MM-GBSA calculations, which is a highly nontrivial finding, suggesting that this approach may provide a framework for the design of more potent ASCT2 inhibitors. However, the structure will be useful to determine differences between intracellular and extracellular-facing binding sites, as well as the differentiation between one-gate and two-gate transport mechanisms (Garaeva et al., 2018). In this respect, it may be interesting to test for binding of current competitive inhibitors from the intracellular side, because in the potential one-gate mechanism (Garaeva et al., 2018), the intracellular binding site may not be large enough to accept inhibitors with large, hydrophobic side chains.
Another structural model for ASCT2 inhibitor binding was recently published by Scopelliti and colleagues (Scopelliti et al., 2018). This model is based on a crystal structure of the prokaryotic ASCT2 homologue GltPh with mutations to two nonconserved amino acid residues in the binding site, aiming to replicate the native ASCT2 binding site. The mutant GltPh was crystallized with bound cysteine as a transported substrate, as well as the inhibitor benzylcysteine. Interestingly, benzylcysteine was found to bind in a pose, in which the hydrophobic benzyl group in the side chain occupies PB, consistent with inhibitor binding predicted from our previous homology models built on GltPh. The absence of the GltPh-R397 arginine side chain, which is replaced by shorter cysteine side chain in ASCT2, creates a void into which hydrophobic ligands can bind (Garibsingh et al., 2018). It may be possible to exploit pocket B in future iterations of our ASCT2 inhibitors.
The ASCT2 model used in the present work was based on the human EAAT1 structure in the outward-open conformation, which is highly conserved between these proteins, both of which are divergent from GltPh’s HP2 used in previous models. Similarly to the EAAT1 structure, the ASCT2 model has a pronounced opening of HP2 that leads to a larger binding site. Additionally, docking of our previously characterized (Singh et al., 2017) and newly discovered inhibitors suggests that the hydrophobic side chain of the inhibitors binds in PA instead of pocket B. These docking poses of the current ASCT2 inhibitors are similar to the binding pose of TFB-TBOA in the EAAT1 crystal structure, increasing our confidence in our model. It should be noted, however, that the relatively low resolution (3.7 Å) of the EAAT1 structure may result in uncertainty in both the EAAT1 structure and the ASCT2 model. This weakness in the model is mitigated by the (a) high enrichment of known ASCT2 ligands and (b) high correlation between MM-GBSA score of the new inhibitors and their Ki (Fig. S3).
Remarkably, a comparison between HP2 of the newly determined cryo-EM inward-facing hASCT2 structure and our outward-open model (used here) and outward-occluded model (not shown; Garibsingh et al., 2018) reveals a highly similar HP2 conformation (RMSDs of 1.06 Å and 1 Å, respectively). Moreover, our outward-open model exhibits better ligand enrichment scores than those of the inward-facing hASCT2 (logAUC of 47.9 and 61.7, respectively). These results are further suggestive of the model’s accuracy and relevance of rational design.
We anticipate that the differentiation of these binding modes is expected to be of critical importance for developing future ASCT2 inhibitors. Therefore, we propose that using the EAAT1-based ASCT2 model is better for ASCT2 blocker development. The compounds we have experimentally tested in this study dock well to this improved structural model and allow us to focus on hydrophobic interaction with PA.
In conclusion, the sulfonamides/sulfonic ester series of compounds characterized here as ASCT2 inhibitors add to our repertoire of scaffolds on which future structure–function studies can be based. Our improved ASCT2 homology model, based on the hEAAT1 crystal structure, allows refined prediction of binding poses, agreeing well with the experimental data. While specificity for ASCTs over EAATs has yet to be tested for most inhibitors and/or to be achieved, our results are encouraging for the development of future higher-affinity ASCT2 inhibitors, which will be necessary to further establish ASCT2 as an anticancer drug target.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health (grant R01 GM108911 to A. Schlessinger and C. Grewer and grant T32 CA078207 to R.-A.A. Garibsingh) and the National Science Foundation (grant 1515028 awarded to C. Grewer).
The authors declare no competing financial interests.
Author contributions: E. Ndaru and R.A. Garibsingh designed and performed experiments and wrote the manuscript. Y. Shi, E. Wallace, P. Zakrepine, and J. Wang performed experiments. A. Schlessinger and C. Grewer designed the strategy and experiments and wrote the manuscript.
José D. Faraldo-Gómez served as editor.
Footnotes
This work is part of the special collection entitled "Molecular Physiology of the Cell Membrane: An Integrative Perspective from Experiment and Computation."
References
- Albers T., Marsiglia W., Thomas T., Gameiro A., and Grewer C.. 2012. Defining Substrate and Blocker Activity of Alanine Serine Cysteine Transporter 2 (ASCT2) Ligands with Novel Serine Analogs. Mol. Pharmacol. 81:356–365. 10.1124/mol.111.075648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman B.J., Stine Z.E., and Dang C.V.. 2016. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer. 16:619–634. 10.1038/nrc.2016.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriza J.L., Fairman W.A., Wadiche J.I., Murdoch G.H., Kavanaugh M.P., and Amara S.G.. 1994. Functional comparisons of three glutamate transporter subtypes cloned from human motor cortex. J. Neurosci. 14:5559–5569. 10.1523/JNEUROSCI.14-09-05559.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudker O., Ryan R.M., Yernool D., Shimamoto K., and Gouaux E.. 2007. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 445:387–393. 10.1038/nature05455 [DOI] [PubMed] [Google Scholar]
- Bröer A., Brookes N., Ganapathy V., Dimmer K.S., Wagner C.A., Lang F., and Bröer S.. 1999. The astroglial ASCT2 amino acid transporter as a mediator of glutamine efflux. J. Neurochem. 73:2184–2194. [PubMed] [Google Scholar]
- Bröer A., Wagner C., Lang F., and Bröer S.. 2000. Neutral amino acid transporter ASCT2 displays substrate-induced Na+ exchange and a substrate-gated anion conductance. Biochem. J. 346:705–710. 10.1042/bj3460705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussolati O., Laris P.C., Rotoli B.M., Dall’Asta V., and Gazzola G.C.. 1992. Transport system ASC for neutral amino acids. An electroneutral sodium/amino acid cotransport sensitive to the membrane potential. J. Biol. Chem. 267:8330–8335. [PubMed] [Google Scholar]
- Cantor J.R., Abu-Remaileh M., Kanarek N., Freinkman E., Gao X., Louissaint A. Jr., Lewis C.A., and Sabatini D.M.. 2017. Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell. 169:258–272.e17. 10.1016/j.cell.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canul-Tec J.C., Assal R., Cirri E., Legrand P., Brier S., Chamot-Rooke J., and Reyes N.. 2017. Structure and allosteric inhibition of excitatory amino acid transporter 1. Nature. 544:446–451. 10.1038/nature22064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas C., Grewer C., Otte N.J., Gameiro A., Albers T., Singh K., Shere H., Bonomi M., Holst J., and Schlessinger A.. 2015. Ligand Discovery for the Alanine-Serine-Cysteine Transporter (ASCT2, SLC1A5) from Homology Modeling and Virtual Screening. PLOS Comput. Biol. 11:e1004477 10.1371/journal.pcbi.1004477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esslinger C.S., Cybulski K.A., and Rhoderick J.F.. 2005. Ngamma-aryl glutamine analogues as probes of the ASCT2 neutral amino acid transporter binding site. Bioorg. Med. Chem. 13:1111–1118. 10.1016/j.bmc.2004.11.028 [DOI] [PubMed] [Google Scholar]
- Fuchs B.C., and Bode B.P.. 2005. Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? Semin. Cancer Biol. 15:254–266. 10.1016/j.semcancer.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Fuchs B.C., Perez J.C., Suetterlin J.E., Chaudhry S.B., and Bode B.P.. 2004. Inducible antisense RNA targeting amino acid transporter ATB0/ASCT2 elicits apoptosis in human hepatoma cells. Am. J. Physiol. Gastrointest. Liver Physiol. 286:G467–G478. 10.1152/ajpgi.00344.2003 [DOI] [PubMed] [Google Scholar]
- Garaeva A.A., Oostergetel G.T., Gati C., Guskov A., Paulino C., and Slotboom D.J.. 2018. Cryo-EM structure of the human neutral amino acid transporter ASCT2. Nat. Struct. Mol. Biol. 25:515–521. 10.1038/s41594-018-0076-y [DOI] [PubMed] [Google Scholar]
- Garibsingh R.A., Otte N.J., Ndaru E., Colas C., Grewer C., Holst J., and Schlessinger A.. 2018. Homology Modeling Informs Ligand Discovery for the Glutamine Transporter ASCT2. Front Chem. 6:279 10.3389/fchem.2018.00279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewer C., and Grabsch E.. 2004. New inhibitors for the neutral amino acid transporter ASCT2 reveal its Na+-dependent anion leak. J. Physiol. 557:747–759. 10.1113/jphysiol.2004.062521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewer C., Watzke N., Wiessner M., and Rauen T.. 2000. Glutamate translocation of the neuronal glutamate transporter EAAC1 occurs within milliseconds. Proc. Natl. Acad. Sci. USA. 97:9706–9711. 10.1073/pnas.160170397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J.C., Bjørn-Yoshimoto W.E., Bisballe N., Nielsen B., Jensen A.A., and Bunch L.. 2016. β-Sulfonamido Functionalized Aspartate Analogues as Excitatory Amino Acid Transporter Inhibitors: Distinct Subtype Selectivity Profiles Arising from Subtle Structural Differences. J. Med. Chem. 59:8771–8786. 10.1021/acs.jmedchem.6b01066 [DOI] [PubMed] [Google Scholar]
- Hille B. 2001. Ion channels of excitable membranes. 3rd. Sinauer Associates, Inc., Sunderland, MA. [Google Scholar]
- Kanai Y. 1996. [Na+ -dependent amino acid transporters: their structure and function]. Nihon Rinsho. 54:638–645. [PubMed] [Google Scholar]
- Kim S., Jung W.H., and Koo J.S.. 2013. The expression of glutamine-metabolism-related proteins in breast phyllodes tumors. Tumour Biol. 34:2683–2689. 10.1007/s13277-013-0819-7 [DOI] [PubMed] [Google Scholar]
- Koch H.P., Kavanaugh M.P., Esslinger C.S., Zerangue N., Humphrey J.M., Amara S.G., Chamberlin A.R., and Bridges R.J.. 1999. Differentiation of substrate and nonsubstrate inhibitors of the high-affinity, sodium-dependent glutamate transporters. Mol. Pharmacol. 56:1095–1104. 10.1124/mol.56.6.1095 [DOI] [PubMed] [Google Scholar]
- Nicklin P., Bergman P., Zhang B., Triantafellow E., Wang H., Nyfeler B., Yang H., Hild M., Kung C., Wilson C., et al. 2009. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 136:521–534. 10.1016/j.cell.2008.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrauskas A., and Kolovanov E.. 2000. ACD/Log P method description. Perspect. Drug Discov. Des. 19:99–116. 10.1023/A:1008719622770 [DOI] [Google Scholar]
- Pinilla J., Barber A., and Lostao M.P.. 2001. Active transport of alanine by the neutral amino-acid exchanger ASCT1. Can. J. Physiol. Pharmacol. 79:1023–1029. 10.1139/y01-087 [DOI] [PubMed] [Google Scholar]
- Ryan R.M., Mitrovic A.D., and Vandenberg R.J.. 2004. The chloride permeation pathway of a glutamate transporter and its proximity to the glutamate translocation pathway. J. Biol. Chem. 279:20742–20751. 10.1074/jbc.M304433200 [DOI] [PubMed] [Google Scholar]
- Scalise M., Pochini L., Pingitore P., Hedfalk K., and Indiveri C.. 2015. Cysteine is not a substrate but a specific modulator of human ASCT2 (SLC1A5) transporter. FEBS Lett. 589:3617–3623. 10.1016/j.febslet.2015.10.011 [DOI] [PubMed] [Google Scholar]
- Schrodinger L. 2018. The PyMOL Molecular Graphics System, Version 2.0. [Google Scholar]
- Schrödinger L. 2017. Schrödinger Release 2017-2: Glide. New York, NY. [Google Scholar]
- Schulte M.L., Dawson E.S., Saleh S.A., Cuthbertson M.L., and Manning H.C.. 2015. 2-Substituted Nγ-glutamylanilides as novel probes of ASCT2 with improved potency. Bioorg. Med. Chem. Lett. 25:113–116. 10.1016/j.bmcl.2014.10.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte M.L., Khodadadi A.B., Cuthbertson M.L., Smith J.A., and Manning H.C.. 2016. 2-Amino-4-bis(aryloxybenzyl)aminobutanoic acids: A novel scaffold for inhibition of ASCT2-mediated glutamine transport. Bioorg. Med. Chem. Lett. 26:1044–1047. 10.1016/j.bmcl.2015.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte M.L., Fu A., Zhao P., Li J., Geng L., Smith S.T., Kondo J., Coffey R.J., Johnson M.O., Rathmell J.C., et al. 2018. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat. Med. 24:194–202. 10.1038/nm.4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A., and Harris A.L.. 2012. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 491:364–373. 10.1038/nature11706 [DOI] [PubMed] [Google Scholar]
- Scopelliti A.J., Font J., Vandenberg R.J., Boudker O., and Ryan R.M.. 2018. Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5. Nat. Commun. 9:38 10.1038/s41467-017-02444-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman W., Beard H.S., and Farid R.. 2006. Use of an induced fit receptor structure in virtual screening. Chem. Biol. Drug Des. 67:83–84. 10.1111/j.1747-0285.2005.00327.x [DOI] [PubMed] [Google Scholar]
- Shimamoto K., Lebrun B., Yasuda-Kamatani Y., Sakaitani M., Shigeri Y., Yumoto N., and Nakajima T.. 1998. DL-threo-beta-benzyloxyaspartate, a potent blocker of excitatory amino acid transporters. Mol. Pharmacol. 53:195–201. 10.1124/mol.53.2.195 [DOI] [PubMed] [Google Scholar]
- Singh K., Tanui R., Gameiro A., Eisenberg G., Colas C., Schlessinger A., and Grewer C.. 2017. Structure activity relationships of benzylproline-derived inhibitors of the glutamine transporter ASCT2. Bioorg. Med. Chem. Lett. 27:398–402. 10.1016/j.bmcl.2016.12.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Geldermalsen M., Wang Q., Nagarajah R., Marshall A.D., Thoeng A., Gao D., Ritchie W., Feng Y., Bailey C.G., Deng N., et al. 2016. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene. 35:3201–3208. 10.1038/onc.2015.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Geldermalsen M., Quek L.E., Turner N., Freidman N., Pang A., Guan Y.F., Krycer J.R., Ryan R., Wang Q., and Holst J.. 2018. Benzylserine inhibits breast cancer cell growth by disrupting intracellular amino acid homeostasis and triggering amino acid response pathways. BMC Cancer. 18:689 10.1186/s12885-018-4599-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadiche J.I., Amara S.G., and Kavanaugh M.P.. 1995. Ion fluxes associated with excitatory amino acid transport. Neuron. 15:721–728. 10.1016/0896-6273(95)90159-0 [DOI] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski R.A., and Thornton J.M.. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127–134. 10.1093/protein/8.2.127 [DOI] [PubMed] [Google Scholar]
- Wang Q., Beaumont K.A., Otte N.J., Font J., Bailey C.G., van Geldermalsen M., Sharp D.M., Tiffen J.C., Ryan R.M., Jormakka M., et al. 2014. Targeting glutamine transport to suppress melanoma cell growth. Int. J. Cancer. 135:1060–1071. 10.1002/ijc.28749 [DOI] [PubMed] [Google Scholar]
- Wang Q., Hardie R.A., Hoy A.J., van Geldermalsen M., Gao D., Fazli L., Sadowski M.C., Balaban S., Schreuder M., Nagarajah R., et al. 2015. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J. Pathol. 236:278–289. 10.1002/path.4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watzke N., Bamberg E., and Grewer C.. 2001. Early intermediates in the transport cycle of the neuronal excitatory amino acid carrier EAAC1. J. Gen. Physiol. 117:547–562. 10.1085/jgp.117.6.547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise D.R., and Thompson C.B.. 2010. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 35:427–433. 10.1016/j.tibs.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander C.B., Albers T., and Grewer C.. 2013. Voltage-dependent processes in the electroneutral amino acid exchanger ASCT2. J. Gen. Physiol. 141:659–672. 10.1085/jgp.201210948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerangue N., and Kavanaugh M.P.. 1996. ASCT-1 is a neutral amino acid exchanger with chloride channel activity. J. Biol. Chem. 271:27991–27994. 10.1074/jbc.271.45.27991 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.