How resident inner nuclear membrane (INM) proteins are turned over is unclear. Koch et al. identify an APC/C-dependent mechanism controlling the degradation of Mps3, a conserved integral protein of the INM.
Abstract
The nucleus is enclosed by the inner nuclear membrane (INM) and the outer nuclear membrane (ONM). While the ONM is continuous with the endoplasmic reticulum (ER), the INM is independent and separates the nucleoplasm from the ER lumen. Turnover of ER proteins has been well characterized by the ER-associated protein degradation (ERAD) pathway, but very little is known about turnover of resident INM proteins. Here we show that the anaphase-promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase, regulates the degradation of Mps3, a conserved integral protein of the INM. Turnover of Mps3 requires the ubiquitin-conjugating enzyme Ubc7, but was independent of the known ERAD ubiquitin ligases Doa10 and Hrd1 as well as the recently discovered Asi1–Asi3 complex. Using a genetic approach, we have found that Cdh1, a coactivator of APC/C, modulates Mps3 stability. APC/C controls Mps3 degradation through Mps3’s N terminus, which resides in the nucleoplasm and possesses two putative APC/C-dependent destruction motifs. Accumulation of Mps3 at the INM impairs nuclear morphological changes and cell division. Our findings therefore reveal an unexpected mechanism of APC/C-mediated protein degradation at the INM that coordinates nuclear morphogenesis and cell cycle progression.
Introduction
The nucleus is enclosed by two membranes that demarcate the nucleoplasm from the cytoplasm. The outer nuclear membrane (ONM) is continuous with the ER, whereas the inner nuclear membrane (INM), which harbors hundreds of proteins (Ungricht and Kutay, 2015; Smoyer et al., 2016), interacts with the nucleoplasm. INM-localized proteins regulate a diverse range of nuclear activities that include chromosome movement, gene expression, and signal transduction. Nascent INM proteins are synthesized at the ER, transported through the nuclear pore complex, and then anchored at the INM (Katta et al., 2014; Ungricht and Kutay, 2015). Abnormal accumulation of INM proteins, such as the integral membrane protein SUN1, has been linked to the pathogenesis of progeric and dystrophic laminopathies in mammals (Chen et al., 2012; Burke and Stewart, 2014). But how homeostasis of resident INM proteins is achieved to maintain proper INM function remains to be further elucidated.
The ER-associated protein degradation (ERAD) pathway regulates the turnover of many ER proteins by marking them for proteasome degradation (Vembar and Brodsky, 2008; Zattas and Hochstrasser, 2015). ERAD acts in a step-wise manner, which involves the target protein being polyubiquitylated by the joint actions of an E2 ubiquitin-conjugating enzyme and an E3 ubiquitin ligase. Two partially redundant E2 enzymes, Ubc6 and Ubc7, function with one or both ERAD E3 ligases, Doa10 and Hrd1, to mediate substrate turnover (Bordallo and Wolf, 1999; Swanson et al., 2001; Carvalho et al., 2006). In addition, Doa10 also regulates protein degradation at the INM (Deng and Hochstrasser, 2006).
Recent work from budding yeast has shown that the Asi1–Asi3 protein complex in particular acts in concert with Ubc6 and Ubc7 to polyubiquitinate INM proteins that are sorted for proteasome degradation (Foresti et al., 2014; Khmelinskii et al., 2014), thereby defining the first known dedicated INM protein quality control pathway, which has been called INMAD for INM-associated protein degradation (Pantazopoulou et al., 2016). However, the Asi proteins, including Asi1, Asi2, and Asi3, are not conserved in mammals, and genome-wide proteomic analysis in budding yeast has shown that they are mostly responsible for the degradation of proteins mislocalized at the INM (Foresti et al., 2014; Khmelinskii et al., 2014). Noticeably, being a resident INM protein itself, Asi1 is highly unstable and subject to proteasome degradation, but the responsible E3 ligase for Asi1 turnover remains unknown (Pantazopoulou et al., 2016). These observations indicate that additional E3 ligases function at the INM to regulate protein turnover.
We show here that the anaphase-promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase best known for its role in controlling cell cycle progression (Irniger et al., 1995; King et al., 1995; Sudakin et al., 1995), regulates the degradation of the SUN-domain protein Mps3, an integral INM protein and an essential component of the linker of the nucleoskeleton to cytoskeleton complex in budding yeast (Jaspersen et al., 2002; Conrad et al., 2007). Using a genetic approach, we show that APC/C and its cofactor Cdh1 mediate Mps3 degradation through Mps3’s N terminus, which resides in the nucleoplasm and possesses two putative APC/C-dependent degradation motifs. Accumulation of Mps3 at the INM impairs nuclear morphological changes and cell division. Our work reveals, for the first time to our knowledge, that APC/C regulates the degradation of an integral INM protein and thereby defines a new pathway for protein turnover at the INM.
Results
Degradation of Mps3 is regulated by the ubiquitin-proteasome system
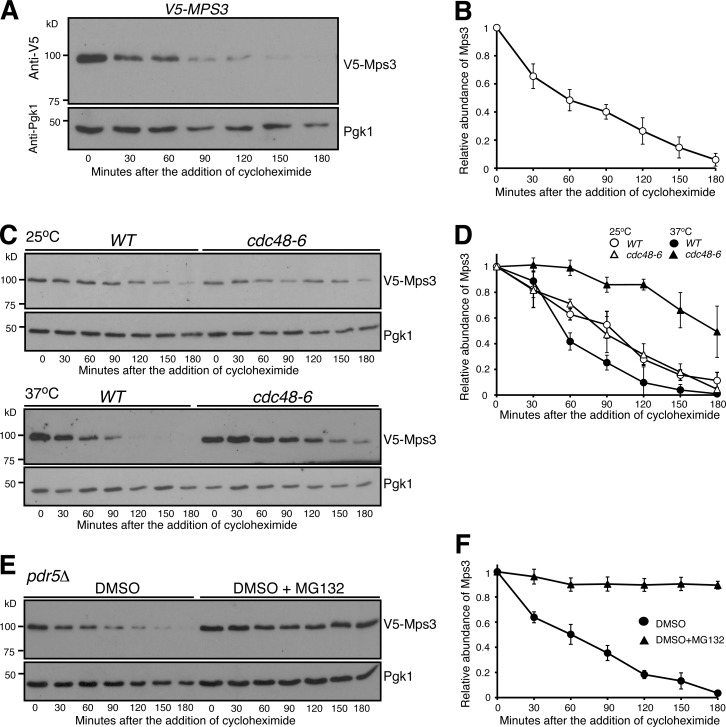
We and others have shown previously that the INM-localized protein Mps3 regulates centrosome duplication and separation in budding yeast (Jaspersen et al., 2002; Friederichs et al., 2011; Li et al., 2017). Here we seek to determine how Mps3 is subject to protein degradation during the cell cycle. We generated an N-terminal V5-tagged allele, PMPS3-V5-MPS3 (Fig. 1 A), which is under the control of its endogenous promoter and serves as the only functional copy of MPS3 in the yeast genome. PMPS3-V5-MPS3 cells displayed a similar doubling time to that of untagged yeast cells (see below). To determine the half-life of Mps3, we used cycloheximide (CHX), a potent inhibitor of protein biosynthesis, and performed CHX-chase experiments (Fig. 1 A). By Western blotting, we found that V5-Mps3 exhibited a half-life of ∼45 min when cells proliferated in rich medium (Fig. 1 B). We note that, when probed by a polyclonal antibody that recognizes amino acids 65 to 76 from the N terminus of Mps3 (Li et al., 2017), the untagged Mps3 showed a similar half-life of ∼45 min (Fig. S1, A and B), suggesting that V5-Mps3 acts just like the endogenous Mps3. Because the critical mutations investigated in this study lie in this region, we used the V5-tagged alleles as described below. Taken together, our data demonstrate that INM-localized Mps3 is a short-lived protein.
Figure 1.
Proteasome-dependent degradation of Mps3. (A and B) CHX-chase experiment showing Mps3 half-life. Yeast cells were grown to the exponential phase, then CHX was added to the culture media. Cell aliquots were withdrawn at indicated times, and protein extracts were prepared for Western blotting. An anti-V5 antibody was used to probe V5-Mps3. The level of Pgk1 serves as a loading control. Time zero is the point of CHX addition. Quantification of Mps3 protein abundance is shown in B. Error bars represent the standard deviation from the mean of seven biological replicates. (C and D) Cdc48 regulates Mps3 degradation. Cells were grown at 25°C to the exponential phase, and CHX was then added as shown in A. To inactivate cdc48-6, cells were shifted to 37°C for 1 h before the addition of CHX. Quantification of Mps3 protein abundance in shown in D. Samples were analyzed as in B; error bars represent the standard deviation from the mean of biological replicates (n = 3). Notably, the half-life of Mps3 at the nonpermissive temperature increased more than twofold. (E and F) The proteasome is responsible for Mps3 turnover. Protein extracts and Western blot were prepared as in A. The use of the pdr5Δ allele allows yeast to uptake MG132, which is dissolved in DMSO. Quantification of Mps3 protein abundance in shown in F. Samples were analyzed as in B. Error bars represent the standard deviation from the mean of biological replicates (n = 3). Note that Mps3 is stabilized in the presence of MG132.
Because Mps3 is an integral membrane protein (Jaspersen et al., 2002) synthesized at the ER before being transported to the INM, we determined whether Mps3 is subject to ERAD regulation. For integral ER membrane protein substrates of ERAD, the AAA-ATPase Cdc48 facilitates their extraction from the ER membrane and is critical for transporting ubiquitinated substrates to the proteasome (Rabinovich et al., 2002; Schuberth and Buchberger, 2005). We therefore determined whether Mps3 degradation is regulated by Cdc48 (Fig. 1 C). We used a temperature-sensitive allele, cdc48-6, to inactivate Cdc48 at 37°C. At the permissive temperature (25°C), the half-life of Mps3 was comparable in wild-type and cdc48-6 cells (Fig. 1, C and D). In contrast, the Mps3 protein level stabilized in cdc48-6 cells at the restrictive temperature (37°C), with a half-life of ∼150 min (Fig. 1 D), a threefold increase. This finding suggests that Mps3 is potentially subject to ERAD regulation. To further confirm that Mps3 is degraded by the proteasome, we used the cell-permeable drug MG132 to inhibit proteasome activity. For yeast cells to retain MG132 and thereby keep the proteasome at an inactive state, the plasma membrane ABC transporter, encoded by PDR5, was removed (Leppert et al., 1990). Noticeably, in pdr5Δ cells treated with MG132, Mps3 became highly stable, showing a half-life of >180 min (Fig. 1, E and F). We therefore conclude that the ubiquitin-proteasome system regulates Mps3 degradation.
Mps3 is degraded by nucleus-localized proteasomes
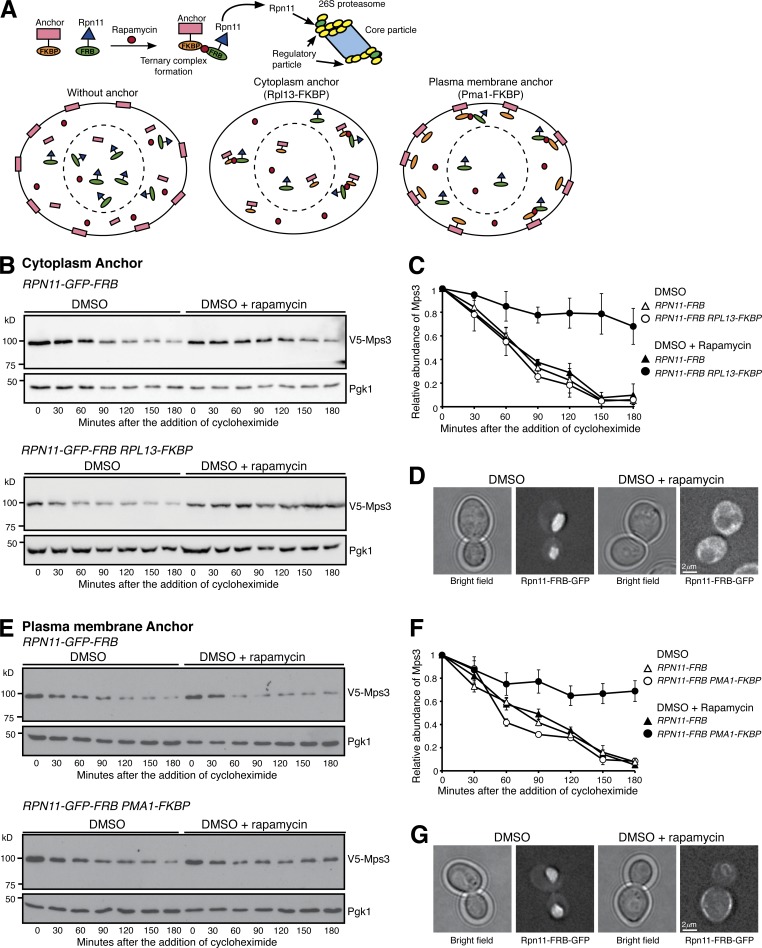
The N-terminal domain of Mps3 is positioned in the nucleoplasm (Jaspersen et al., 2002; Li et al., 2017); we therefore asked whether Mps3 is degraded by nucleus-localized proteasomes. To deplete proteasomes from the yeast nucleus, we used the anchor-away system to force proteasomes to relocate to either the cytoplasm or the plasma membrane (Haruki et al., 2008; Nemec et al., 2017; Fig. 2 A). Of note, tethering the proteasome to the anchor protein, which is mediated by rapamycin-triggered dimerization of FKBP12-rapamycin binding domain (FRB) and human FK506 binding protein (FKBP12), is conditional (Haruki et al., 2008; Fig. 2 A), and the TOR1-1 mutation renders yeast cells resistant to rapamycin (Helliwell et al., 1994). We found that the half-life of Mps3 was not altered upon the addition of rapamycin, which served as a control in our experiments (Fig. 2, B and C). However, upon the addition of rapamycin and by way of Rpn11-FRB and Rpl13-FKBP interaction, proteasomes were sequestered to the cytoplasm (Fig. 2 D). As a result, the half-life of Mps3 reached >180 min, a fourfold increase over the wild type (Fig. 2, B and C), demonstrating that degradation of Mps3 was impaired when proteasomes were depleted from the nucleus. Similarly, anchoring proteasomes to the plasma membrane also increased the half-life of Mps3 to >180 min (Fig. 2, E–G). These findings indicate that nuclear proteasomes are required for adequate turnover of Mps3.
Figure 2.
Nucleus-localized proteasome regulates Mps3 degradation. (A) A schematic representation of forced proteasome localization using the anchor-away system. Briefly, Rpn11, a subunit of the 26S proteasome, was tagged with FRB, which in the presence of rapamycin induces a ternary complex formation between Rpn11-FRB and the FKBP-tagged anchor, either at the cytoplasm (Rpl13-FKBP) or at the plasma membrane (Pma1-FKBP). (B) Forced localization of proteasomes to the cytoplasm. Yeast cells were prepared for CHX chase, and rapamycin dissolved in DMSO was added to relocate the proteasome to the cytoplasm. (C) Quantification of Mps3 degradation as shown in B. Error bars represent the standard deviation from the mean of biological replicates (n = 3). (D) Representative images showing forced localization of the proteasome to the cytoplasm 180 min after the addition of rapamycin. Note that Rpn11-GFP-FRB became dispersed in the cytoplasm in the presence of rapamycin. (E) Forced localization of the proteasome to the plasma membrane. Samples were analyzed as in B. (F) Quantification of Mps3 degradation as shown in E. Error bars represent the standard deviation from the mean of biological replicates (n = 3). (G) Representative images showing forced localization of the proteasome to the plasma membrane 180 min after the addition of rapamycin. Note that Rpn11-GFP-FRB was enriched at the cell periphery in the presence of rapamycin.
To further determine the effect of nuclear localization on Mps3 turnover, we generated two mps3 deletion alleles. One expressed only the first 150 amino acids (mps3(1–150)), the other an Mps3 fragment that lacked amino acids 1 to 93 (mps3(Δ1-93)), and both were under the control of either the endogenous MPS3 or the galactose promoter (Fig. S1, C–I). Both the Mps3-150 and Mps3(Δ1–93) fragments were localized to the nucleus (Fig. S1, F and I), with Mps3(Δ1–93), which retained the transmembrane domain, concentrated at the nuclear periphery (Fig. S1 I). Coinciding with these observations, Mps3-150 demonstrated a half-life comparable to the full-length Mps3 (Fig. S1, D and E). In contrast, deletion of the first 93 amino acids of Mps3 significantly affected the protein half-life despite its localizing to the nuclear periphery (Fig. S1, G–I). Taken together, our findings demonstrate that the bulk of Mps3 is degraded within the nucleus by nuclear proteasomes, but the N terminus of Mps3 plays an important role in its stability.
The E2 enzymes Ubc7 and Ubc6 regulate Mps3 degradation
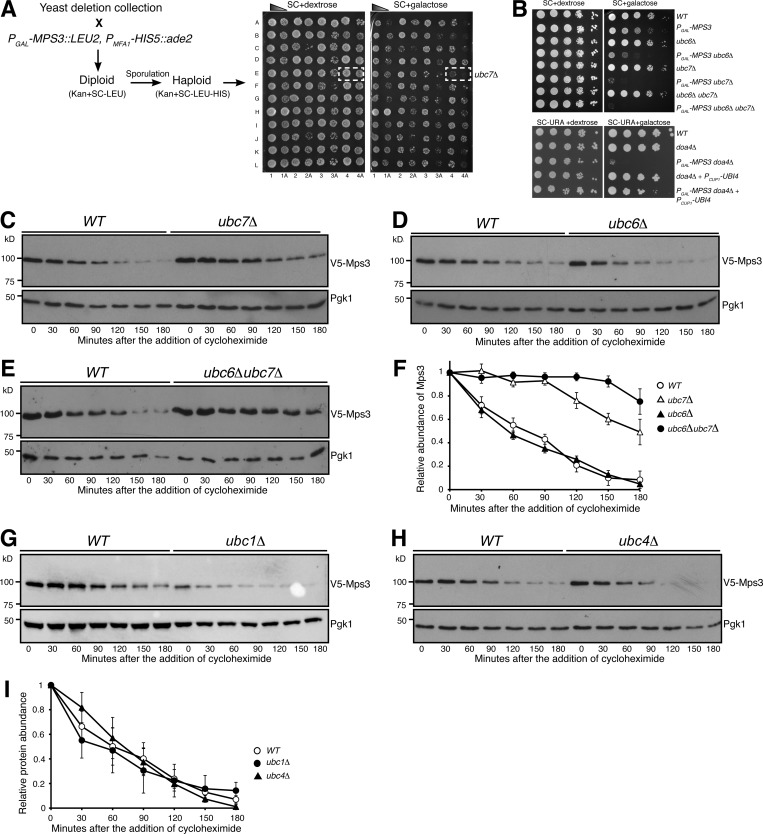
We sought to identify the E2 ubiquitin conjugating enzyme and the E3 ubiquitin ligase that are responsible for Mps3 degradation. We have reported previously that overproduction of Mps3 by way of PGAL-MPS3 leads to a slight growth defect in vegetative yeast cells (Li et al., 2017). Because Mps3 is an essential component of the INM and the yeast spindle pole body (SPB), we reasoned that impairing the degradation system specific to Mps3 would render a lethal phenotype when Mps3 is overproduced. From the yeast deletion collection library (ATCC-GSA-5), we pooled together 93 deletions of nonessential yeast genes that encode either an E2 or an E3 or their interacting factors (Table S1, modified from Xie et al., 2010) and screened their genetic interactions with PGAL-MPS3, which overproduced Mps3 in the galactose medium (Fig. 3 A as an example). Of note, in the glucose medium, the GAL promoter is repressed. We found that deletion of UBC7, which encodes an E2 for ERAD (Gilon et al., 1998) and INMAD (Foresti et al., 2014; Khmelinskii et al., 2014; Pantazopoulou et al., 2016), showed synthetic lethality with PGAL-MPS3 when yeast cells were cultured in the galactose medium (Fig. 3, A and B). In addition, DOA4, which encodes the ubiquitin hydrolase that is responsible for recycling ubiquitin from proteasome-bound intermediates (Papa and Hochstrasser, 1993), also genetically interacted with PGAL-MPS3 (Fig. 3 B). Importantly, the synthetic lethality observed in PGAL-MPS3 doa4Δ cells was suppressed upon the introduction of a high-copy-number plasmid that expressed ubiquitin (Fig. 3 B). These findings indicate that Ubc7 is a putative E2 enzyme for Mps3 degradation and further support the idea that degradation of Mps3 is mediated by the ubiquitin-proteasome system.
Figure 3.
Ubc7 is a key regulator of Mps3 degradation. (A) A representation of the genetic screen performed using the yeast deletion collection. Briefly, a deletion collection specific for genes encoding known ubiquitin conjugating enzymes, ubiquitin ligases, and their regulators (a total of 93 gene deletions) was crossed to a strain containing an MPS3 overexpression plasmid under the galactose inducible promoter. Selective media allowed for the selection of haploid yeast cells containing both PGAL-MPS3 and a gene deletion. This method enabled the identification of putative genetic interactors of MPS3. The synthetic lethality observed with ubc7Δ, doa4Δ, and others was further tested to confirm the genetic interaction. (B) MPS3 genetically interacts with UBC7 and DOA4. To determine cell viability, yeast cells were grown overnight in YPD liquid medium to reach saturation, 10-fold diluted, spotted onto SC plates with either 2% dextrose or 2% galactose, and then incubated at 30°C for ∼2 d. Overexpression of MPS3 yielded a sick phenotype. The overexpression of MPS3 coupled with ubc6Δ cells showed a stronger sick phenotype, while the ubc7Δ and ubc6Δubc7Δ had a lethal phenotype. Notably, the similar near-lethal phenotype observed when coupled with a doa4Δ can be suppressed by the overexpression of ubiquitin (PCUP1-UBI4, one of the ubiquitin-coding repeats from UBI4, was under the control of the CUP1 promoter). (C–E) Ubc7 regulates degradation of Mps3. WT, ubc6Δ, ubc7Δ, and ubc6Δubc7Δ yeast cells were prepared for CHX chase as in Fig. 1 A. (F) Quantification of the relative abundance of Mps3 as shown in C–E. Error bars represent the standard deviation from the mean of biological replicates (n = 3). (G–I) The E2 enzymes Ubc1 and Ubc4 do not regulate Mps3 degradation. WT, ubc1Δ, and ubc4Δ yeast cells were prepared for CHX chase as in Fig. 1 A. Quantification of the relative abundance of Mps3 is shown in I. Error bars represent the standard deviation from the mean of biological replicates (n = 3).
To determine whether Ubc7 governs Mps3 protein stability, we determined the half-life of Mps3 in ubc7Δ cells (Fig. 3 C). Indeed, in the absence of Ubc7, the half-life of Mps3 increased to more than 150 min (Fig. 3 C and see below). Another E2 enzyme involved in ERAD and INMAD, Ubc6, is partially redundant to Ubc7 (Chen et al., 1993). We therefore determined Mps3’s half-life in ubc6Δ cells (Fig. 3 D and see below). Deletion of UBC6 alone did not change Mps3’s stability when compared with wild type (Fig. 3 D). However, removal of both Ubc6 and Ubc7 (ubc6Δ ubc7Δ) drastically increased Mps3’s half-life compared with the ubc7Δ single mutant (Fig. 3, E and F). This finding indicates that in the absence of Ubc7, Ubc6 can partially serve as an alternative E2 for Mps3 degradation. We therefore conclude that the E2 enzymes Ubc7 and, to a lesser degree, Ubc6 regulate Mps3 protein turnover.
Mps3 is not regulated by the known ERAD and INMAD E3 ubiquitin ligases
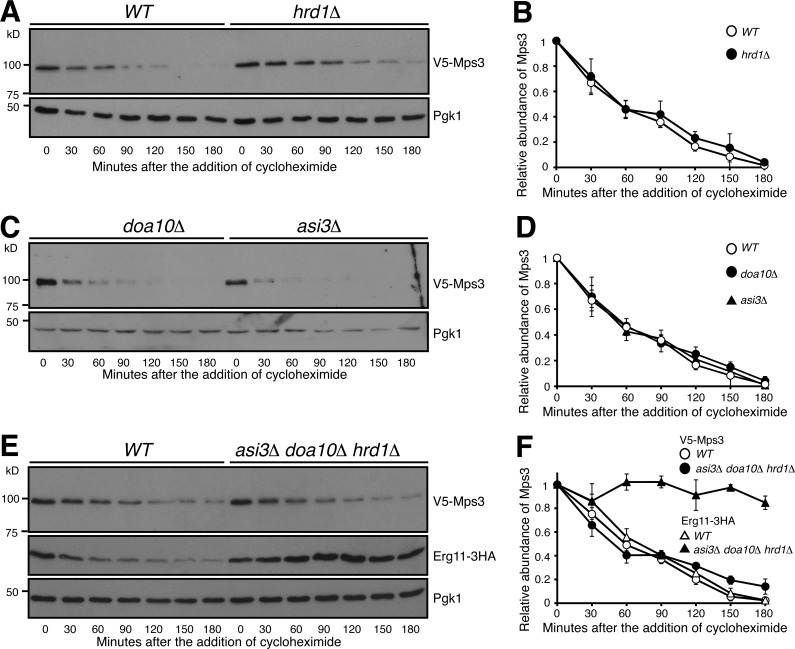
Both Ubc6 and Ubc7 are involved in ERAD- and INMAD-mediated protein degradation; we therefore hypothesized that the known ERAD E3 ubiquitin ligases, Hrd1 and Doa10, or the INMAD-specific Asi1–Asi3 complex (Bordallo and Wolf, 1999; Swanson et al., 2001; Carvalho et al., 2006; Foresti et al., 2014; Khmelinskii et al., 2014), are required for Mps3 degradation. To test our hypothesis, we used gene deletions of HRD1, DOA10, and ASI3 to abolish their corresponding E3 enzymatic activities (Fig. 4). Using CHX chase, we observed that the degradation of Mps3 is independent of Hrd1 (Fig. 4, A and B), an E3 ubiquitin ligase responsible for retrotranslocating substrates from the ER to cytosol for degradation (Bordallo and Wolf, 1999; Baldridge and Rapoport, 2016). This result supports our finding that turnover of Mps3 takes place inside the yeast nucleus (Fig. 2). The other two E3 enzymes, Doa10 and the Asi1–Asi3 complex, both have been implicated in degradation of certain INM-localized proteins (Swanson et al., 2001; Deng and Hochstrasser, 2006; Foresti et al., 2014; Khmelinskii et al., 2014). However, the half-life of Mps3 remained unchanged in doa10Δ or asi3Δ, or doa10Δasi3Δ double mutant cells (Fig. 4, C and D; and not depicted). More importantly, removal of all the three E3 enzyme activities had no noticeable impact on the half-life of Mps3 (Fig. 4, E and F). In contrast, a previously identified INM protein and substrate for the Asi1–Asi3 complex, Erg11 (Foresti et al., 2014), was stabilized when the activities of the three E3 ligases were absent (Fig. 4, E and F). Taken together, our findings demonstrate that degradation of Mps3 is not governed by the known ERAD or INMAD E3 ubiquitin ligases and indicate that a different E3 ligase is responsible for regulating Mps3 protein stability at the INM.
Figure 4.
Mps3 degradation is independent of the known ERAD and INMAD E3 ligases. (A and B) Canonical E3 ligase Hrd1 does not regulate Mps3 degradation. WT and hrd1Δ yeast cells were prepared for CHX chase as in Fig. 1 A. Quantification of Mps3 protein abundance is shown in B. Error bars represent the standard deviation from the mean of biological replicates (n = 3). (C and D) Canonical E3 ligases Doa10 and Asi3 do not regulate Mps3 degradation. Yeast cells were prepared for CHX chase in a doa10Δ or asi3Δ background and were analyzed as in Fig. 1 A. Quantification of Mps3 protein abundance as shown in C. Error bars represent the standard deviation from the mean of biological replicates (n = 3). (E and F) The canonical E3 ligases involved in ERAD do not regulate Mps3 degradation through redundancy. WT and asi3Δdoa10Δhrd1Δ yeast cells were prepared for CHX chase and analyzed as in Fig. 1 A. Erg11-3HA was used as a positive control, and Pgk1 was probed for a loading control. Quantification of Mps3 relative protein abundance is shown in F. Error bars represent the standard deviation from the mean of biological replicates (n = 3). Note there is no change in Mps3 half-life between the WT and asi3Δdoa10Δhrd1Δ cells.
APC/C regulates Mps3 degradation
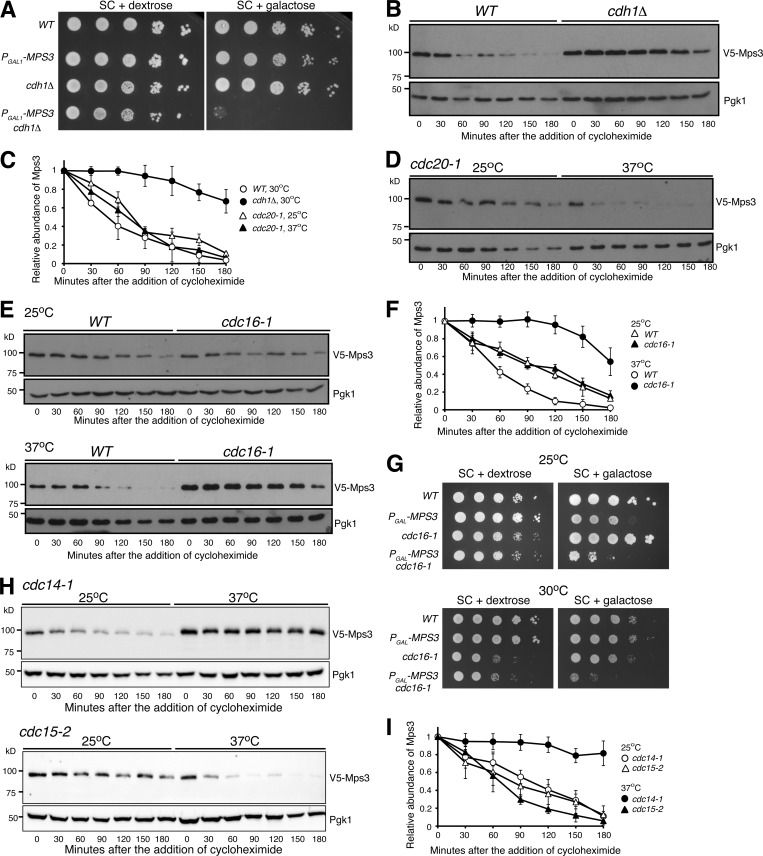
To search for the E3 ubiquitin ligase that is responsible for Mps3 degradation, we determined other gene deletions that genetically interacted with PGAL-MPS3 in our screen (Table S1). One of them, CDH1, which encodes an activator of the APC/C (King et al., 1995; Visintin et al., 1997; Zachariae et al., 1998), genetically interacted with PGAL-MPS3; deletion of CDH1 caused a lethal phenotype when MPS3 was overexpressed (Fig. 5 A). Cdh1 acts during late mitosis and at the G1 phase (Visintin et al., 1997); we therefore enriched yeast cells in G1 with α factor and performed CHX-chase experiments to determine Mps3’s half-life (Fig. 5 B). Consistent with their genetic interaction, the half-life of Mps3 increased to >3 h in the absence of Cdh1 (Fig. 5, B and C). As a control, we found that the kinetics of Erg11 degradation in cdh1Δ cells remained similar to the wild type (Fig. S2 A). Cdh1 has a paralog called Cdc20, which activates APC/C at the metaphase-to-anaphase transition during mitosis (Visintin et al., 1997). Because CDC20 is an essential gene, we used a temperature-sensitive cdc20-1 allele to inactivate Cdc20 at 37°C (Fig. 5, C and D). The half-life of Mps3 remained unchanged in cdc20-1 cells at the restrictive temperature as in wild-type cells (Fig. 5, C and D). We therefore conclude that Cdh1, but not Cdc20, specifically regulates Mps3 turnover.
Figure 5.
APC/CCdh1-dependent regulation of Mps3 degradation. (A) MPS3 genetically interacts with CDH1. To determine cell viability, yeast cells were grown overnight in YPD liquid medium to reach saturation, 10-fold diluted, spotted onto SC plates with either 2% dextrose or 2% galactose, and then incubated at 30°C for ∼2 d. Overexpression of MPS3 is lethal in cdh1Δ cells. (B) The APC/C activator, Cdh1, regulates Mps3 degradation. The relative protein abundance of Mps3 in WT and cdh1Δ cells was analyzed after arresting cells in G1 using α factor (10 µg/ml) for 2 h, and then CHX chase was performed as described in Fig. 1 A. (C) Quantification of the relative abundance of Mps3 as shown in B and D. Representative blots from three biological replicates of each experiment are shown. (D) Mps3 turnover is independent of Cdc20. Cells were grown at 25°C to the exponential phase, and CHX was then added as shown in Fig. 1 A. To inactivate cdc20-1, cells were shifted to 37°C for 1 h before the addition of CHX. (E and F) The APC/C subunit Cdc16 regulates Mps3 degradation. Cells were grown at 25°C to the exponential phase, and CHX was then added as shown in Fig. 1 A. To inactivate cdc16-1, cells were shifted to 37°C for 1 h before the addition of CHX. Quantification of Mps3 protein abundance is shown in F. Error bars represent the standard deviation from the mean of biological replicates (n = 3). Notably, inactivation of the APC through the cdc16-1 allele at the nonpermissive temperature resulted in a threefold increase in Mps3 half-life. (G and H) MPS3 genetically interacts with cdc16-1. Overexpression of MPS3 in cdc16-1 cells showed a sick phenotype at a semipermissive temperature. Yeast cells were grown overnight in YPD liquid medium to reach saturation, 10-fold diluted, spotted onto SC plates with either 2% dextrose or 2% galactose, and then incubated at 30°C for ∼2 d. (H and I) Cdc14, but not Cdc15, regulates Mps3 degradation. Yeast cells were grown at 25°C to the exponential phase, and CHX was then added as shown in Fig. 1 A. To inactivate either cdc14-1 or cdc15-2, cells were shifted to 37°C for 1 h before the addition of CHX. Quantification of Mps3 protein abundance is shown in I. Error bars represent the standard deviation from the mean of biological replicates (n = 3). Note that inactivation of Cdc14, but not Cdc15, stabilized Mps3.
We hypothesized that APC/C serves as the E3 ligase for Mps3 degradation. To test this hypothesis, we inactivated the APC/C using the temperature-sensitive cdc16-1 allele, the wild type of which encodes an essential subunit of the APC/C (Zachariae et al., 1996). By CHX chase and Western blotting, we found that Mps3 was stabilized when cdc16-1 was inactivated at 37°C, reaching a half-life of 160 min, an approximately fourfold increase over that of wild type (Fig. 5, E and F). As a control, we found that Asi1, another INM-localized protein whose corresponding E3 is currently unknown, was degraded at a similar kinetics in both cdh1Δ and cdc16-1 cells compared with wild-type cells (Fig. S2), demonstrating the specificity of APC/CCdh1 for Mps3 degradation. On the basis of this observation, we hypothesized that a genetic interaction would exist between MPS3 and CDC16. To test this idea, we overproduced Mps3 in cdc16-1 cells (Fig. 5 G). Even at the permissive temperature (25°C), we observed a severe growth defect when PGAL-MPS3 cdc16-1 cells were cultured in the galactose medium (Fig. 5 G). It became lethal at the semipermissive temperature of 30°C (Fig. 5 G). To address whether Ubc1 and Ubc4, two E2 enzymes previously known to act in concert with APC/C (Rodrigo-Brenni and Morgan, 2007), regulate Mps3 turnover, we determined Mps3 stability in ubc1Δ and ubc4Δ cells (Fig. 3, G–I). In the absence of either Ubc1 or Ubc4, Mps3 displayed a half-life similar to that of wild-type cells (Fig. 3 I). Taken together, our findings indicate that APC/CCdh1, likely in conjunction with the E2 enzyme Ubc7, regulates Mps3 degradation.
Because Cdh1, and therefore the activity of APC/CCdh1, is activated by the protein phosphatase Cdc14 after anaphase onset (Visintin et al., 1998; Jaspersen et al., 1999), we hypothesized that the degradation of Mps3 is regulated during the cell cycle. In the absence of the phosphatase activity of Cdc14 by way of cdc14-1 at the restrictive temperature (37°C), Mps3 was stabilized (Fig. 5, H and I), indicating that Cdc14 is required for APC/CCdh1 activation and therefore Mps3 degradation. In contrast, in cdc15-2 mutant cells, which were blocked at anaphase and prevented exit from mitosis (Hartwell et al., 1973), Mps3 was degraded in a similar kinetics as in wild-type cells (Fig. 5, H and I). These findings support the idea that APC/CCdh1 regulates Mps3 turnover during the cell cycle.
The N terminus of Mps3 possesses two putative APC/C-dependent destruction motifs
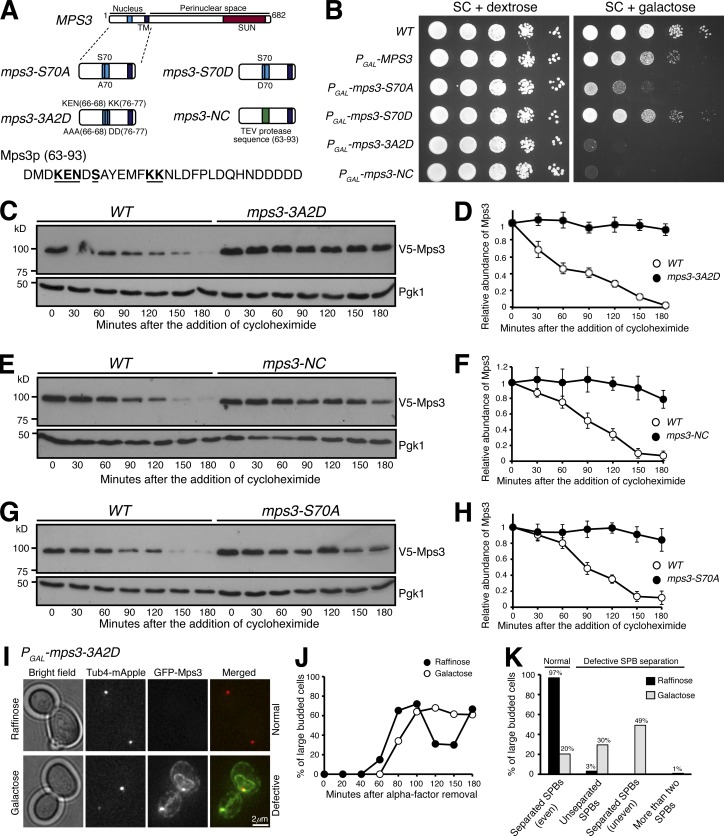
To mediate protein degradation, APC/C recognizes two well-defined destruction motifs, the D and KEN boxes, on its substrates (Glotzer et al., 1991; Pfleger and Kirschner, 2000). Deletion of amino acids from 63 to 93 (mps3-NC) drastically increased Mps3 protein stability in yeast meiosis (Li et al., 2017). This region harbors a KEN sequence from amino acids 66 to 68, whereas the amino acids from 76 to 84 loosely fit the consensus sequence of the D box (Fig. 6 A). To determine whether the putative KEN or D box, or both, regulate Mps3 protein stability, we generated individual and double mutants: mps3-3A, in which the KEN sequence was replaced by three alanines; mps3-2D, in which two lysine residues at positions 76 and 77 were replaced by two glutamates; and mps3-3A2D, a double mutation (see diagram in Fig. 6 A). Crucially, overexpression of mps3-3A2D, but not mps3-3A or mps3-2D, rendered a lethal phenotype (Figs. 6 A and S3). Similarly, in either mps3-3A or mps3-2D mutant cells, whose expressions were under the control of the endogenous promoter, the half-life of Mps3 remained at or close to the wild-type level (Fig. S3, A–F). In contrast, in the double mutant mps3-3A2D, the half-life of Mps3 increased to >3 h (Fig. 6, C and D). In addition, we found that the half-life of Mps3 dramatically increased in mps3-NC cells, in which the two putative destruction motifs both are absent (Fig. 6, E and F). These findings indicate that Mps3 possesses two putative destruction motifs, and the KEN and D boxes are somehow redundant in regulating Mps3 degradation.
Figure 6.
The N terminus of Mps3 possesses two putative destruction motifs. (A) A schematic representation of Mps3 protein domains and subsequent mutations introduced. TM, transmembrane domain. (B) The N terminus of Mps3 is crucial for cell viability. Yeast cells were grown overnight in YPD liquid medium to reach saturation, 10-fold diluted, spotted onto SC plates with either 2% dextrose or 2% galactose, and then incubated at 30°C for ∼2 d. Overexpression of a phosphorylation mutant (S70A) produces a very sick phenotype that can be reverted to wild type by the phosphomimetic (S70D) version. Amino acids 63–93 appear to be a key region of Mps3, where removal and specific point mutations result in a complete synthetic lethality. (C–H) The N terminus of Mps3 regulates protein stability. WT, mps3-3A2D (C and D), mps3-NC (E and F), and mps3-S70A (G and H) yeast cells were prepared for CHX chase and analyzed as described in Fig. 1 A. Quantification of Mps3 protein abundance is shown in D, F, and H. Error bars represent the standard deviation from the mean of biological replicates (n = 3). Note that each of these mutations results in a severe impairment of Mps3 degradation, with observed half-lives well over 180 min. (I–K) Accumulation of PGAL1-mps3-3A2D results in failed SPB separation or misseparation. Yeast cells were grown in raffinose medium and arrested at G1 with α-factor. Galactose was added 30 min before removal of α-factor to induce the GAL1 promoter. Aliquots were withdrawn and prepared for live-cell fluorescence microscopy. Tub4-RFP marks the SPB. Representative images of mps3-3A2D cells grown in raffinose or galactose are shown in I (t = 180 min). Budding index is shown in J. Cell aliquots were withdrawn at indicated times, and budding morphology was determined by phase-contrast microscopy. Quantification of SPB separation is shown in K. More than 500 cells were counted in both raffinose and galactose experiments. Four categories of SPB separation in large-budded cells were classified, the first being normal SPB separation (even) and the remaining three being types of defective SPB separation: unseparated SPBs, uneven separation of SPBs, and more than two SPBs. Tub4-RFP marks the SPB.
We have reported previously that Mps3 is phosphorylated at S70 (Li et al., 2017). Because protein phosphorylation regulates ubiquitination (Hunter, 2007), we addressed whether S70 phosphorylation regulates Mps3 stability using the phosphomutant MPS3-S70A (Li et al., 2017). Overproduction of Mps3-S70A, but not Mps3-S70D, which is a phophosmimetic, caused a severe growth defect (Figs. 6 B and S3). In the absence of S70 phosphorylation, Mps3-S70A became highly stable, with a half-life of >3 h (Fig. 6, G and H), whereas the turnover of Mps3-S70D resembled that of the wild type (Fig. S3, G and H). Taken together, the above findings provide further evidence that posttranslational modification regulates Mps3 degradation.
The mps3-3A2D mutant exhibited a discernible cell proliferation defect compared with the wild type (Fig. S3 J). To further determine the biological significance of Mps3 accumulation at the INM, we used α factor to synchronize yeast cells at G1 and determined nuclear morphology and cell cycle progression in cells with excessive Mps3-3A2D (Fig. 6, I–K). Upon G1 release by removal of the α factor, we overexpressed PGAL-MPS3-3A2D in the galactose medium (Figs. 6 I and S4). Overproduced Mps3-3A2D localized to the SPB and accumulated at the nuclear periphery (Figs. 6 I and S4), forming extended membrane structures, a phenotype similar to that we reported previously of the mps3-NC allele (Li et al., 2017). On the basis of budding index and the dynamics of Tub4-marked SPBs, we determined that mutant cells overproducing Mps3-3A2D showed delayed cell cycle progression (Fig. 6, J and K). In large budded cells at mitosis, 80% of them displayed SPB separation defects (Fig. 6 K). These findings explain the cell lethality caused by Mps3-3A2D overproduction.
The N terminus of Mps3 is both necessary and sufficient for APC/CCdh1-mediated protein degradation
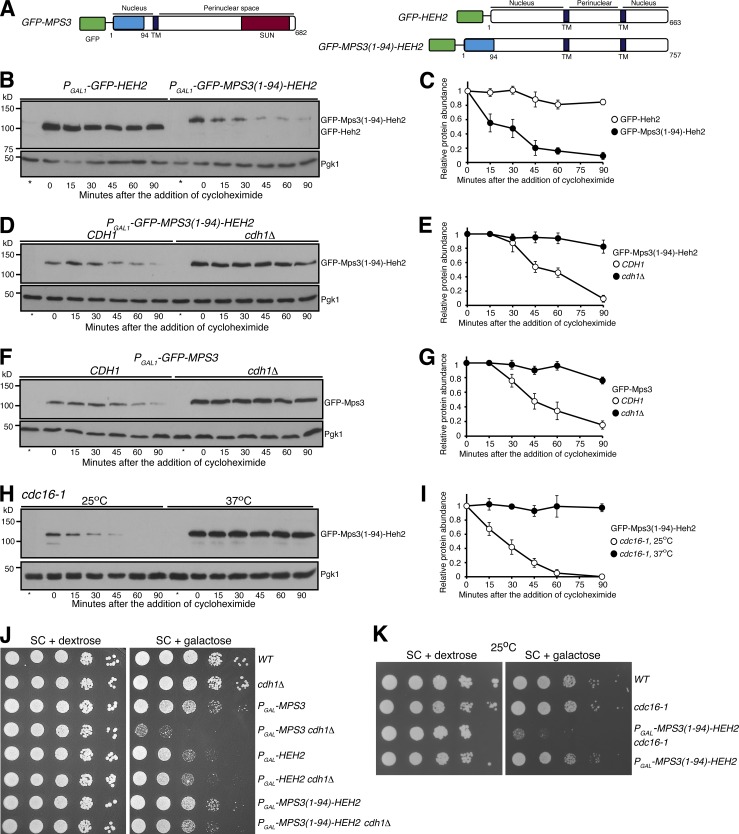
To determine if the N terminus of Mps3 is sufficient for APC/C-mediated protein turnover, we engineered a Mps3(1–94)-Heh2 hybrid protein by grafting the N terminus of Mps3 to Heh2 (Fig. 7 A), another INM-localized protein (King et al., 2006). We used the galactose medium to induce the expression of PGAL-HEH2 and PGAL-MPS3(1–94)-HEH2 and determined protein stability by CHX chase (Fig. 7, B–I). Heh2 was a relatively stable protein with a half-life of >180 min, whereas Mps3(1–94)-Heh2 displayed a half-life of <30 min (Fig. 7, B and C), demonstrating that the N terminus of Mps3 is sufficient to mediate the degradation of the hybrid protein. Crucially, in the absence of Cdh1, Mps3(1–94)-Heh2 restored protein stability to a level similar to that of Heh2 (Fig. 7, D and E). As a control, overproduced Mps3 by way of PGAL-GFP-MPS3 responded to Cdh1 regulation similarly to the endogenous Mps3 (Fig. 7, F and G; and see above). To determine whether APC/C regulates the degradation of the fusion protein, we used the cdc16-1 allele to inactivate APC/C as shown in Fig. 5 E. At the restrictive temperature and in the absence of APC/C activity, Mps3(1–94)-Heh2 became highly stable (Fig. 7, H and I). These findings support our conclusion that the N terminus of Mps3 specifically responds to APC/CCdh1-mediated protein degradation.
Figure 7.
The N terminus of Mps3 is necessary and sufficient for APC/CCdh1-dependent protein degradation. (A) A schematic representation of GFP-Mps3, GFP-Heh2, and the fusion protein GFP-Mps3(1–94)-Heh2 protein structure and membrane topology. TM, transmembrane domain. (B and C) The N terminus of Mps3 is sufficient for triggering the degradation of Mps3(1–94)-Heh2. CHX chase is as described in Fig. 1 A. To induce protein production, yeast cells were grown in galactose medium for ∼3 h before the addition of CHX. Quantification of protein abundance is shown in C. Error bars represent the standard deviation from the mean of biological replicates (n = 3). Note that the fusion protein Mps3(1–94)-Heh2 has a half-life of ∼30 min. (D and E) Cdh1 regulates Mps3(1–94)-Heh2 degradation. CHX-chase experiments were performed as in B. Quantification of protein abundance is shown in E. Error bars represent the standard deviation from the mean of biological replicates (n = 3). (F and G) Cdh1 regulates the degradation of overproduced Mps3. CHX-chase experiments were performed as in B. Quantification of protein abundance is shown in G. Error bars represent the standard deviation from the mean of biological replicates (n = 3). (H and I) Cdc16 regulates the degradation of the fusion protein Mps3(1–94)-Heh2. CHX-chase experiments were performed as in Fig. 5 E. To inactivate cdc16-1, cells were shifted to 37°C for 1 h before the addition of CHX. Quantification of Mps3 protein abundance is shown in I. Error bars represent the standard deviation from the mean of biological replicates (n = 3). (J) Genetic evidence showing the N terminus of Mps3 regulates protein stability. Yeast cells were grown overnight in YPD liquid medium to reach saturation, 10-fold diluted, spotted onto SC plates with either 2% glucose or 2% galactose, and then incubated at 30°C for ∼2 d. (K) Genetic interaction between MPS3(1–94)-HEH2 and cdc16-1. Overexpression of MPS3(1–94)-HEH2 in cdc16-1 cells showed a sick phenotype at the permissive temperature 25°C. Yeast cells were grown overnight in YPD liquid medium to reach saturation, 10-fold diluted, spotted onto SC plates with either 2% dextrose or 2% galactose, and then incubated at 25°C for ∼2 d.
Finally, overproduction of Heh2 by means of PGAL-HEH2 caused a slow growth defect in yeast cells plated on the galactose medium (Fig. 7 J). In contrast, PGAL-MPS3(1–94)-HEH2 cells displayed close to normal growth (Fig. 7 J), supporting the idea that the Mps3 N terminus promotes the degradation of the fusion protein Mps3(1–94)-Heh2. Importantly, in the absence of Cdh1, the fusion protein was stabilized (Fig. 7 C), and PGAL-MPS3(1–94)-HEH2 cells grew slower in the galactose medium (Fig. 7 J). Similarly, overproduction of Mps3(1–94)-Heh2 is lethal even at the permissive temperature of cdc16-1 (Fig. 7 K). Taken together, our findings demonstrate that the N terminus of Mps3 is both necessary and sufficient for targeted protein degradation mediated by APC/CCdh1.
Discussion
In this study, we have shown that the degradation of the resident INM protein Mps3 is regulated by the E3 ubiquitin ligase APC/C in concert with the E2 ubiquitin-conjugating enzymes Ubc7 and Ubc6, thereby defining a new pathway for protein turnover at the nuclear envelope. In contrast to the two known ERAD E3 ligases, Doa10 and Hrd1, and the Asi1–Asi3 complex, all of which contain integral membrane proteins (Vembar and Brodsky, 2008; Zattas and Hochstrasser, 2015), APC/C does not possess membrane components and is best known for its role in degrading soluble proteins and regulating cell cycle progression (Pines, 2011; Davey and Morgan, 2016). Our work therefore extends the role of APC/C in modulating the turnover of an integral membrane protein. It also expands the known pathways for INM-associated protein degradation. Accumulation of Mps3 leads to nuclear membrane expansion and impairs cell division (Friederichs et al., 2011; Li et al., 2017), thus demonstrating the significance of timely turnover of resident INM proteins, such as Mps3, at the nuclear envelope.
APC/C regulates Mps3 degradation at the INM
Four lines of evidence that we have obtained support the idea that APC/CCdh1 modulates Mps3 turnover by targeting the N terminus of Mps3. First, our combined genetic, cytological, and biochemical observations demonstrate that Cdh1, an APC/C regulator, controls Mps3 degradation. In the absence of Cdh1, Mps3 is stabilized and accumulates at the nuclear periphery. Second, inactivation of APC/C leads to increased Mps3 stability. Third, at its N terminus, which is located in the nucleoplasm, Mps3 harbors two redundant degradation motifs that resemble the KEN and D boxes that are subject to APC/C regulation. Finally, by grafting the N terminus of Mps3 onto an unrelated INM protein, we show that the chimeric Mps3(1–94)-Heh2 behaves in a similar manner to Mps3, responding to APC/CCdh1 regulation. APC/CCdh1 modulates cell cycle progression by controlling the degradation of mitotic factors to ensure proper exit of mitosis (Visintin et al., 1997; Zachariae et al., 1998). In addition, APC/CCdh1 is active during the G1 phase in preparation for mitosis (Pines, 2011). That Mps3 is subject to APC/CCdh1 regulation indicates that turnover of Mps3 is coupled to cell cycle progression, consistent with Mps3’s mode of action in SPB duplication and separation in budding yeast (Jaspersen et al., 2002; Friederichs et al., 2011; Li et al., 2017). Our observation of Mps3 being degraded by nucleus-localized proteasomes further supports the idea that Mps3 is ubiquitinated by APC/C, which is concentrated inside the nucleus in budding yeast. To our knowledge, ours is the first example demonstrating that APC/CCdh1 controls the degradation of an INM-localized protein.
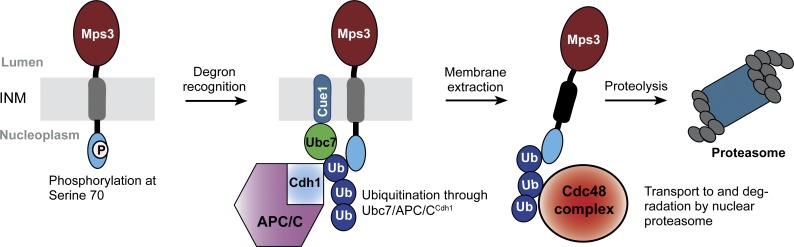
How, then, does APC/CCdh1 regulate the degradation of a membrane protein at the INM? We have shown that the N-terminal domain of Mps3, which is located in the nucleus, harbors two putative destruction motifs recognized by APC/C. We speculate that APC/CCdh1 is recruited to the nuclear periphery to ubiquitinate Mps3, the process of which is further enhanced by protein phosphorylation at S70, as crosstalk between ubiquitination and protein phosphorylation is common for APC/C substrates (Hall et al., 2008; He et al., 2013; Davey and Morgan, 2016). Ubiquitinated Mps3 could then serve as a substrate for the Cdc48 protein complex, which extracts Mps3 from the INM and delivers it to the nuclear proteasome for degradation (Fig. 8). Alternatively, we have shown previously that Mps3 is cleaved near the destruction motifs at its N terminus (Li et al., 2017). Cleavage of Mps3 perhaps signals the critical first step in Mps3 turnover, followed by extraction of the remaining Mps3 from the INM for proteasome degradation. We note that the above two possibilities are not mutually exclusive, and future studies will clarify how membrane-bound Mps3 is transported to the proteasome.
Figure 8.
Model for APC/C-dependent Mps3 degradation. Mps3 is oligomerized at the INM; for simplicity, only one copy of Mps3 is shown. Ub, ubiquitin.
APC/C works in concert with the E2 enzymes Ubc7 and Ubc6
Turnover of Mps3 requires the E2 conjugating enzymes Ubc7, and to some degree Ubc6, indicating that APC/CCdh1 acts in concert with a set of ERAD- and INMAD-specific E2 ubiquitin-conjugating enzymes for proteolysis. The cognizant E2 enzymes pairing with APC/C are Ubc1 and Ubc4 (Finley et al., 2012). Neither Ubc1 nor Ubc4 appears to be necessary for Mps3 turnover (this study). Nevertheless, our findings have expanded the repertoire of the E3 ligases involved in membrane protein degradation but also revealed the possibility that APC/C may pair with previously unrecognized E2 enzymes to regulate protein degradation at specialized cellular compartments, such as the INM.
Previous works have revealed that pathways governed by the E3 ligases Doa10 and the Asi1–Asi3 complex regulate INM protein degradation (Swanson et al., 2001; Foresti et al., 2014; Khmelinskii et al., 2014), in particular for degradation of membrane proteins mislocalized to the INM (Khmelinskii et al., 2014). However, the specificity of resident INM protein degradation is unknown. In contrast, APC/C recognizes its substrates primarily through the D and KEN boxes (Davey and Morgan, 2016). Although it is currently unknown whether APC/C regulates resident INM proteins other than Mps3, our findings suggest that the destruction motifs that reside on INM proteins play a critical role in modulating their stability.
The significance of protein turnover at the INM
The SUN-domain protein Mps3 is a resident INM protein, whose anchoring at the INM is enhanced by its binding partners in the nucleus as well as the KASH-domain/like proteins bound to the ONM (Tapley and Starr, 2013). Mps3 is homologous to mammalian SUN1, whose accumulation at the INM has been implicated in progeric and dystrophic laminopathies in a mouse model (Chen et al., 2012). Similarly, accumulation of Mps3 in budding yeast leads to nuclear envelope expansion and nuclear tubule formation (Friederichs et al., 2011) and impairs cell cycle progression (Li et al., 2017; this study). Currently, little is known of how SUN1 is degraded in mammalian cells. Our finding that APC/C mediates Mps3 turnover during the cell cycle has implications for understanding the clearance of SUN1 and its associated proteins in animal cells, and therefore is relevant to the understanding of laminopathies in humans.
Materials and methods
Yeast strains and plasmids used in this study
Yeast strains used in this study and their genotypes are shown in Table S2. To tag the N terminus of Mps3 with V5, we used a homologous recombination-based gene replacement method as follows. We linearized plasmid pHG553 (see below) with AflII and integrated it at the endogenous MPS3 locus by yeast transformation. URA3-positive colonies were then counterselected on a 5-fluoroorotic acid plate to remove the untagged MPS3 copy, thereby leaving V5-MPS3 as the only functional MPS3 gene in the yeast genome. A similar method was used to generate V5-tagged Mps3-S70A, Mps3-S70D, Mps3-3A, Mp3-2D, Mps3-3A2D, and Mps3-NC using the derivatives of plasmid pHG553. All these MPS3 alleles were verified by DNA sequencing before use. For C-terminal tagging of Asi1 and Erg11 with 3xHA and V5, we used a standard PCR-based homologous recombination method (Longtine et al., 1998). Tub4-mApple and others have been reported previously (Shirk et al., 2011; Li et al., 2015). Gene deletion strains of cdh1Δ, asi3Δ, doa10Δ, hrd1Δ, ubc4Δ, ubc6Δ, and ubc7Δ were obtained from the ATCC deletion collection (ATCC-GSA-5). The gene deletion of ubc1Δ was a gift from the laboratory of Mark Hochstrasser (Yale University, New Haven, CT). To generate the PGAL-GFP-MPS3(1–94)-HEH2 fusion allele, we transformed plasmid pHG479 (see below). The alleles cdc16-1, cdc20-1, cdc48-6, and PGAL-GFP-MPS3 have been reported previously (Sethi et al., 1991; Yamamoto et al., 1996; Li et al., 2017). The anchor-away strains have been described previously (Nemec et al., 2017). Briefly, rapamycin was used to force the formation of a ternary complex between an anchor protein fused with FKBP12, and the target protein fused with GFP and FRB; Haruki et al., 2008). In this case, the target protein was the proteasomal lid subunit RPN11 and the anchor proteins were either RPL13 or PMA1, to force localization of RPN11 to the cytoplasm or plasma membrane, respectively (Nemec et al., 2017). Because of the toxic nature of rapamycin to wild-type yeast cells, rapamycin-resistant strains containing mutated TOR1 were used. The yeast homologue to FKBP12 is FPR1, so to reduce competition for FRB binding between FPR1 and the anchor protein, the FPR1 gene was deleted using standard PCR-based homologous recombination (Nemec et al., 2017).
Plasmids used in this study and their main features are shown in Table S3. We used pRS306 as the backbone to generate PMPS3-V5-MPS3 (pHG553). The MPS3 promoter was obtained from pHG454 (Li et al., 2017). PCR-based site-directed mutagenesis was used to generate mps3 mutant alleles (Table S3). To generate PGAL-GFP-HEH2 (plasmid pHG479), the ORF of HEH2 was amplified from the S288C yeast strain background and cloned into the BamHI and SacI sites of pHG302 (Li et al., 2015). To create PGAL-GFP-MPS3(1–94)-HEH2 (pHG572), the first 282 bp of the MPS3 gene was cloned into the BamHI site of pHG479. All plasmids were confirmed by DNA sequencing before use. PCR primers used in this study are listed in Table S4.
Yeast culture methods and cell viability assay
For CHX-chase experiments, yeast cells were grown overnight in YPD (1% yeast extract, 2% peptone, and 2% dextrose) liquid medium to saturation at 30°C. Cell cultures were diluted with YPD to reach an OD (optical density, λ = 600 nm) of 0.2 and incubated at 30°C until the OD reached 0.7. CHX was then added to a final concentration of 200 µg/ml, the point of which was defined as time zero. Cell aliquots were withdrawn at indicated times for protein extraction and prepared for Western blotting.
To chemically inactivate the proteasome activity (Fig. 1 E), yeast cells with pdr5Δ were prepared as described above to reach an OD of 0.7; the culture was then split into three fractions: DMSO only, DMSO with CHX (200 µg/ml final concentration), and DMSO with CHX (200 µg/ml final concentration) and MG132 (50 µM final concentration). Cell aliquots were withdrawn at indicated times, and protein extracts were prepared for Western blotting.
To relocate the proteasomes from the nucleus to the cytoplasm or the plasma membrane (Fig. 2), yeast cells containing the anchor-away system were prepared as described above to reach an OD of 0.7. The culture was then split into two fractions: DMSO with CHX (200 µg/ml final concentration) and DMSO with CHX (200 µg/ml final concentration) and rapamycin (1 µg/ml final concentration). Cell aliquots were then withdrawn at the indicated times for microscopy and protein extraction.
To arrest yeast cells at G1 (Fig. 5 B), strains were grown in YPD liquid medium to an OD of 0.5, then 10 µg/ml of α factor was added. After 2 h of incubation, CHX (200 µg/ml final concentration) was added, and cell aliquots were collected at the indicated times for protein extraction and Western blotting.
Strains carrying temperature-sensitive alleles (cdc48-6, cdc16-1, cdc20-1, cdc14-1, and cdc15-2) were grown overnight at 25°C to saturation in YPD liquid medium and diluted as described above, then incubated at 25°C until the OD reached 0.6. The culture was then split into two fractions and allowed to incubate for 1 h at either 25°C or 37°C. CHX was then added to a final concentration of 200 µg/ml, and cell aliquots were withdrawn at the indicated times for protein extraction and subsequent Western blotting (Figs. 1, 5, and 7).
For experiments using the GAL1-10 promoter (Fig. 7), synthetic complete (SC) medium with 2% dextrose was used to grow cells to an OD of 0.5, and an aliquot was collected for protein extraction and Western blotting. Then, yeast cells were washed twice in water and incubated for ∼3 h in SC medium with 2% galactose to induce expression of the GAL1-10 promoter. CHX (200 µg/ml final concentration) was added, and cell aliquots were collected at the indicated times for protein extraction and Western blotting. To determine cell viability, yeast cells were grown overnight in YPD liquid medium to reach saturation, 10-fold diluted, spotted onto SC plates with either 2% dextrose or 2% galactose, and then incubated at 30°C for ∼2 d (Figs. 3, 5, 6, and 7).
Cell proliferation experiments were performed in triplet in each indicated strain background (Fig. S3 J). Yeast cells were grown overnight in YPD liquid medium to reach saturation, diluted to an OD (λ = 600 nm) of 0.2, and continued to grow with vigorous shaking at 30°C. Cell proliferation was determined by optical density every hour for 10 h (Fig. S3 J). All time-course experiments were repeated at least three times.
Genetic screen to identify MPS3 interacting factors
From the yeast deletion collection (ATCC-GSA-5), a pooled Mata library containing deletions of individual ORFs that encode the known E2 and E3 enzymes and their regulators was used to screen MPS3 interacting genes. This library was crossed to the yeast strain HY4671, which contains a galactose-inducible MPS3 construct, PGAL-MPS3, along with PMFA1-HIS5::ade2, which is specifically expressed in Mata cells (Tong et al., 2001). After mating, diploids were selected, grown to saturation, and incubated in sporulation medium (1% potassium acetate, 0.1% yeast extract, and 0.05% dextrose) for ∼4 d at 30°C. To select for haploids, sporulated cells were then transferred to SC-leucine and -histidine double dropout medium with G418 in the presence of either 2% dextrose or 2% galactose, and then incubated at 30°C for ∼2 d to determine cell viability (Fig. 3 A).
Protein extraction and Western blotting
Yeast aliquots were withdrawn at the indicated times for protein extraction by precipitation in the presence of 20 mM NaOH, and standard SDS-PAGE and Western blotting were performed. V5-Mps3, its derivatives, and Asi1-V5 were detected by a mouse monoclonal anti-V5 antibody (1:5,000, R960-25; Thermo Fisher Scientific). Erg11-3HA was detected by an anti-HA mouse monoclonal antibody (1:1,000, 12CA5; Sigma). GFP-tagged proteins were detected by an anti-GFP mouse monoclonal antibody (1:10,000 dilution, GF28R; Thermo Fisher Scientific). A polyclonal antibody (1:10,000 dilution) against the amino acids 65–77 of Mps3 (Li et al., 2017) was used to probe the protein level of untagged and V5-tagged Mps3 (Fig. S1 A). The level of Pgk1 was probed by a Pgk1 antibody (PA5-28612; Thermo Fisher Scientific) to serve as a loading control. HRP-conjugated secondary antibodies, goat anti-mouse (1706516; Bio-Rad), were used to probe the proteins of interest by an ECL kit (1705060; Bio-Rad).
Two ECL-based Western blot detection methods were used, x-ray film (Figs. 1, 2, 3, 4, 5, 6, 7, and S2) and the ChemiDoc MP Imaging System (17001402; Bio-Rad; Figs. 2 B, 3 G, 5 H, S1, S3, and S4). To calculate the relative protein abundance at each time point, individual band intensities were measured using IPLab Imaging Software in conjunction with an in-house GelAnalyzer script and exported to Microsoft Excel. Target protein band intensities were made relative to those of the loading control (Pgk1). The mean value and standard deviation from the mean of the relative protein band intensities were determined for each time point between biological replicates. The mean protein band intensity was plotted in Microsoft Excel, with error bars representing the standard deviation.
Fluorescence microscopy
A DeltaVision microscope system (GE Healthcare) was used to acquire live-cell fluorescence images as described previously (Li et al., 2015). Briefly, an agarose pad with 2% raffinose was prepared on a concave slide, and an aliquot of yeast cells was placed on the agarose. The slide was then sealed with LVP (Li et al., 2015), and images were acquired at each of the indicated time points at 30°C. Images were acquired with a 63× (NA = 1.40) objective lens on an inverted microscope (IX-71; Olympus) equipped with a CoolSNAP HQ2 CCD camera (Photometrics). Pixel size was set at 0.10700 µm. For images attained during time course experiments, 10 optical sections with a 0.5-µm thickness were acquired at each time point. To reduce phototoxicity, a neutral density filter was used to diminish the intensity of the excitation light to ∼50% or less of the equipment output. For GFP, the excitation spectrum was set at 470/40 nm, and emission spectrum at 525/50 nm; for mApple, excitation was at 572/35, and emission at 632/60 nm. Acquired images were deconvolved using the SoftWorx package (GE Healthcare). Projected images are used for display (Fig. 2, D and G; and Figs. 6 I, S1, and S4).
Online supplemental material
Fig. S1 shows the extended data on Mps3 localization and its degradation profile. Fig. S2 shows the degradation profile of Erg11 and Asi1. Fig. S3 shows the redundant destruction motifs located at the N terminus of Mps3. Fig. S4 shows the mutant phenotypes of mps3-3A2D overexpression. Table S1 shows yeast strains included in the targeted genetic screen. Table S2 shows yeast strains used in this study. Table S3 shows plasmids used. Table S4 shows DNA primers used.
Supplementary Material
Acknowledgments
We thank Dr. Yanchang Wang for insightful discussion and for providing plasmids and yeast strains. Elizabeth Staley provided technical assistance and Jen Kennedy assisted with text editing.
This work is supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number GM117102.
The authors declare no competing financial interests.
Author contributions: Conceptualization: B.A. Koch, H.-G. Yu; Data curation: B.A. Koch, H. Jin; Formal analysis: B.A. Koch, H. Jin, H.-G. Yu; Funding acquisition: H.-G. Yu; Investigation: B.A. Koch, H. Jin; Methodology: B.A. Koch, H. Jin, R.J. Tomko, H.-G. Yu; Project administration: H.-G. Yu; Supervision: H.-G. Yu; Validation: B.A. Koch, H. Jin; Visualization: B.A. Koch, H. Jin; Writing – original draft: B.A. Koch, H.-G. Yu; Writing – review and editing: B.A. Koch, R.J. Tomko, H.-G. Yu.
References
- Baldridge R.D., and Rapoport T.A.. 2016. Autoubiquitination of the Hrd1 Ligase Triggers Protein Retrotranslocation in ERAD. Cell. 166:394–407. 10.1016/j.cell.2016.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo J., and Wolf D.H.. 1999. A RING-H2 finger motif is essential for the function of Der3/Hrd1 in endoplasmic reticulum associated protein degradation in the yeast Saccharomyces cerevisiae. FEBS Lett. 448:244–248. 10.1016/S0014-5793(99)00362-2 [DOI] [PubMed] [Google Scholar]
- Burke B., and Stewart C.L.. 2014. Functional architecture of the cell’s nucleus in development, aging, and disease. Curr. Top. Dev. Biol. 109:1–52. 10.1016/B978-0-12-397920-9.00006-8 [DOI] [PubMed] [Google Scholar]
- Carvalho P., Goder V., and Rapoport T.A.. 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 126:361–373. 10.1016/j.cell.2006.05.043 [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Chi Y.H., Mutalif R.A., Starost M.F., Myers T.G., Anderson S.A., Stewart C.L., and Jeang K.T.. 2012. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell. 149:565–577. 10.1016/j.cell.2012.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Johnson P., Sommer T., Jentsch S., and Hochstrasser M.. 1993. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 74:357–369. 10.1016/0092-8674(93)90426-Q [DOI] [PubMed] [Google Scholar]
- Conrad M.N., Lee C.Y., Wilkerson J.L., and Dresser M.E.. 2007. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 104:8863–8868. 10.1073/pnas.0606165104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey N.E., and Morgan D.O.. 2016. Building a Regulatory Network with Short Linear Sequence Motifs: Lessons from the Degrons of the Anaphase-Promoting Complex. Mol. Cell. 64:12–23. 10.1016/j.molcel.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., and Hochstrasser M.. 2006. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 443:827–831. 10.1038/nature05170 [DOI] [PubMed] [Google Scholar]
- Finley D., Ulrich H.D., Sommer T., and Kaiser P.. 2012. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics. 192:319–360. 10.1534/genetics.112.140467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti O., Rodriguez-Vaello V., Funaya C., and Carvalho P.. 2014. Quality control of inner nuclear membrane proteins by the Asi complex. Science. 346:751–755. 10.1126/science.1255638 [DOI] [PubMed] [Google Scholar]
- Friederichs J.M., Ghosh S., Smoyer C.J., McCroskey S., Miller B.D., Weaver K.J., Delventhal K.M., Unruh J., Slaughter B.D., and Jaspersen S.L.. 2011. The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet. 7:e1002365 10.1371/journal.pgen.1002365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilon T., Chomsky O., and Kulka R.G.. 1998. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 17:2759–2766. 10.1093/emboj/17.10.2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M., Murray A.W., and Kirschner M.W.. 1991. Cyclin is degraded by the ubiquitin pathway. Nature. 349:132–138. 10.1038/349132a0 [DOI] [PubMed] [Google Scholar]
- Hall M.C., Jeong D.E., Henderson J.T., Choi E., Bremmer S.C., Iliuk A.B., and Charbonneau H.. 2008. Cdc28 and Cdc14 control stability of the anaphase-promoting complex inhibitor Acm1. J. Biol. Chem. 283:10396–10407. 10.1074/jbc.M710011200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Mortimer R.K., Culotti J., and Culotti M.. 1973. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 74:267–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H., Nishikawa J., and Laemmli U.K.. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell. 31:925–932. 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- He J., Chao W.C., Zhang Z., Yang J., Cronin N., and Barford D.. 2013. Insights into degron recognition by APC/C coactivators from the structure of an Acm1-Cdh1 complex. Mol. Cell. 50:649–660. 10.1016/j.molcel.2013.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell S.B., Wagner P., Kunz J., Deuter-Reinhard M., Henriquez R., and Hall M.N.. 1994. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell. 5:105–118. 10.1091/mbc.5.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. 2007. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell. 28:730–738. 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Irniger S., Piatti S., Michaelis C., and Nasmyth K.. 1995. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 81:269–278. 10.1016/0092-8674(95)90337-2 [DOI] [PubMed] [Google Scholar]
- Jaspersen S.L., Charles J.F., and Morgan D.O.. 1999. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol. 9:227–236. 10.1016/S0960-9822(99)80111-0 [DOI] [PubMed] [Google Scholar]
- Jaspersen S.L., Giddings T.H. Jr., and Winey M.. 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159:945–956. 10.1083/jcb.200208169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katta S.S., Smoyer C.J., and Jaspersen S.L.. 2014. Destination: inner nuclear membrane. Trends Cell Biol. 24:221–229. 10.1016/j.tcb.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Blaszczak E., Pantazopoulou M., Fischer B., Omnus D.J., Le Dez G., Brossard A., Gunnarsson A., Barry J.D., Meurer M., et al. 2014. Protein quality control at the inner nuclear membrane. Nature. 516:410–413. 10.1038/nature14096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M.C., Lusk C.P., and Blobel G.. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 442:1003–1007. 10.1038/nature05075 [DOI] [PubMed] [Google Scholar]
- King R.W., Peters J.M., Tugendreich S., Rolfe M., Hieter P., and Kirschner M.W.. 1995. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 81:279–288. 10.1016/0092-8674(95)90338-0 [DOI] [PubMed] [Google Scholar]
- Leppert G., McDevitt R., Falco S.C., Van Dyk T.K., Ficke M.B., and Golin J.. 1990. Cloning by gene amplification of two loci conferring multiple drug resistance in Saccharomyces. Genetics. 125:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Shao Y., Jin H., and Yu H.G.. 2015. Ndj1, a telomere-associated protein, regulates centrosome separation in budding yeast meiosis. J. Cell Biol. 209:247–259. 10.1083/jcb.201408118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Jin H., Koch B.A., Abblett R.L., Han X., Yates J.R. III, and Yu H.G.. 2017. Cleavage of the SUN-domain protein Mps3 at its N-terminus regulates centrosome disjunction in budding yeast meiosis. PLoS Genet. 13:e1006830 10.1371/journal.pgen.1006830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Nemec A.A., Howell L.A., Peterson A.K., Murray M.A., and Tomko R.J. Jr. 2017. Autophagic clearance of proteasomes in yeast requires the conserved sorting nexin Snx4. J. Biol. Chem. 292:21466–21480. 10.1074/jbc.M117.817999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulou M., Boban M., Foisner R., and Ljungdahl P.O.. 2016. Cdc48 and Ubx1 participate in a pathway associated with the inner nuclear membrane that governs Asi1 degradation. J. Cell Sci. 129:3770–3780. 10.1242/jcs.189332 [DOI] [PubMed] [Google Scholar]
- Papa F.R., and Hochstrasser M.. 1993. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 366:313–319. 10.1038/366313a0 [DOI] [PubMed] [Google Scholar]
- Pfleger C.M., and Kirschner M.W.. 2000. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pines J. 2011. Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 12:427–438. 10.1038/nrm3132 [DOI] [PubMed] [Google Scholar]
- Rabinovich E., Kerem A., Fröhlich K.U., Diamant N., and Bar-Nun S.. 2002. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22:626–634. 10.1128/MCB.22.2.626-634.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni M.C., and Morgan D.O.. 2007. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 130:127–139. 10.1016/j.cell.2007.05.027 [DOI] [PubMed] [Google Scholar]
- Schuberth C., and Buchberger A.. 2005. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat. Cell Biol. 7:999–1006. 10.1038/ncb1299 [DOI] [PubMed] [Google Scholar]
- Sethi N., Monteagudo M.C., Koshland D., Hogan E., and Burke D.J.. 1991. The CDC20 gene product of Saccharomyces cerevisiae, a beta-transducin homolog, is required for a subset of microtubule-dependent cellular processes. Mol. Cell. Biol. 11:5592–5602. 10.1128/MCB.11.11.5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirk K., Jin H., Giddings T.H. Jr., Winey M., and Yu H.G.. 2011. The Aurora kinase Ipl1 is necessary for spindle pole body cohesion during budding yeast meiosis. J. Cell Sci. 124:2891–2896. 10.1242/jcs.086652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoyer C.J., Katta S.S., Gardner J.M., Stoltz L., McCroskey S., Bradford W.D., McClain M., Smith S.E., Slaughter B.D., Unruh J.R., and Jaspersen S.L.. 2016. Analysis of membrane proteins localizing to the inner nuclear envelope in living cells. J. Cell Biol. 215:575–590. 10.1083/jcb.201607043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Ganoth D., Dahan A., Heller H., Hershko J., Luca F.C., Ruderman J.V., and Hershko A.. 1995. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell. 6:185–197. 10.1091/mbc.6.2.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R., Locher M., and Hochstrasser M.. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 15:2660–2674. 10.1101/gad.933301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley E.C., and Starr D.A.. 2013. Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr. Opin. Cell Biol. 25:57–62. 10.1016/j.ceb.2012.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Pagé N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294:2364–2368. 10.1126/science.1065810 [DOI] [PubMed] [Google Scholar]
- Ungricht R., and Kutay U.. 2015. Establishment of NE asymmetry—targeting of membrane proteins to the inner nuclear membrane. Curr. Opin. Cell Biol. 34:135–141. 10.1016/j.ceb.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Vembar S.S., and Brodsky J.L.. 2008. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9:944–957. 10.1038/nrm2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Prinz S., and Amon A.. 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 278:460–463. 10.1126/science.278.5337.460 [DOI] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E.S., Prinz S., Tyers M., and Amon A.. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 2:709–718. 10.1016/S1097-2765(00)80286-5 [DOI] [PubMed] [Google Scholar]
- Xie Y., Rubenstein E.M., Matt T., and Hochstrasser M.. 2010. SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev. 24:893–903. 10.1101/gad.1906510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Guacci V., and Koshland D.. 1996. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s). J. Cell Biol. 133:99–110. 10.1083/jcb.133.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W., Shin T.H., Galova M., Obermaier B., and Nasmyth K.. 1996. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science. 274:1201–1204. 10.1126/science.274.5290.1201 [DOI] [PubMed] [Google Scholar]
- Zachariae W., Schwab M., Nasmyth K., and Seufert W.. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 282:1721–1724. 10.1126/science.282.5394.1721 [DOI] [PubMed] [Google Scholar]
- Zattas D., and Hochstrasser M.. 2015. Ubiquitin-dependent protein degradation at the yeast endoplasmic reticulum and nuclear envelope. Crit. Rev. Biochem. Mol. Biol. 50:1–17. 10.3109/10409238.2014.959889 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.