Abstract
After years of continuous exposure to HIV envelope antigens, a minority of HIV-infected individuals develop a cognate polyclonal humoral response comprising very potent and extremely cross-reactive neutralizing antibodies (broadly neutralizing antibodies, or bNAbs). Isolated bNAbs derived from memory B cell pools have been the focus of intense studies over the past decade. However, it is not yet known how to translate the features of bNAbs into practical HIV prevention methods. In this review, we attempt to seek insights from emerging information about the human broadly neutralizing plasma response; its frequency, clonal composition, specificity, potency and commonality among infected subjects. We also consider how this information points to selecting and prioritizing certain epitope targets and strategies for HIV vaccine design.
Keywords: bNAbs, HIV, gp120, vaccine, antibody, neutralization
The broadly neutralizing antibody (bNAb) response in HIV infection
The prevention of HIV/AIDS is a tall order for the human immune system. HIV is an integrating retrovirus that rapidly establishes permanent infection and cannot be naturally cleared thereafter. Once infection is established, HIV has an extreme capacity to evade immune responses by genetic variation [1]. Viral evolution translates to worldwide envelope sequence diversity, which clusters into genetic subtypes, or clades, at the population level. Therefore, an anti-HIV immune response must be very potent, sterilizing, and highly cross-reactive to have protective efficacy.
In theory, cross-reactive, neutralizing anti-HIV envelope antibodies (commonly termed broadly neutralizing antibodies, or bNAbs) could be used to prevent or treat HIV in a myriad of ways (Figure 1). It has been repeatedly shown that some HIV-infected subjects develop highly cross-reactive or near “pan-neutralizing” antibody responses against various epitopes on the HIV envelope [2–6]. Further, a variety of monoclonal antibodies (mAbs) isolated from these individuals targeting different epitopes (Figure 2) exhibit impressive breadth and potency in vitro [5, 7–10] and confer sterilizing protection in animal models of HIV infection [11–16]. Accordingly, bNAbs have been used as guides in attempts to engineer immunogens that might raise qualitatively similar humoral responses via vaccination (approaches termed “reverse vaccinology” or “rational design”; see [17–19]). Unfortunately, these approaches have not succeeded in reliably generating broadly neutralizing antibody responses, or in producing an effective neutralization-based HIV vaccine.
Figure 1.
Potential uses for HIV-1 bNAbs
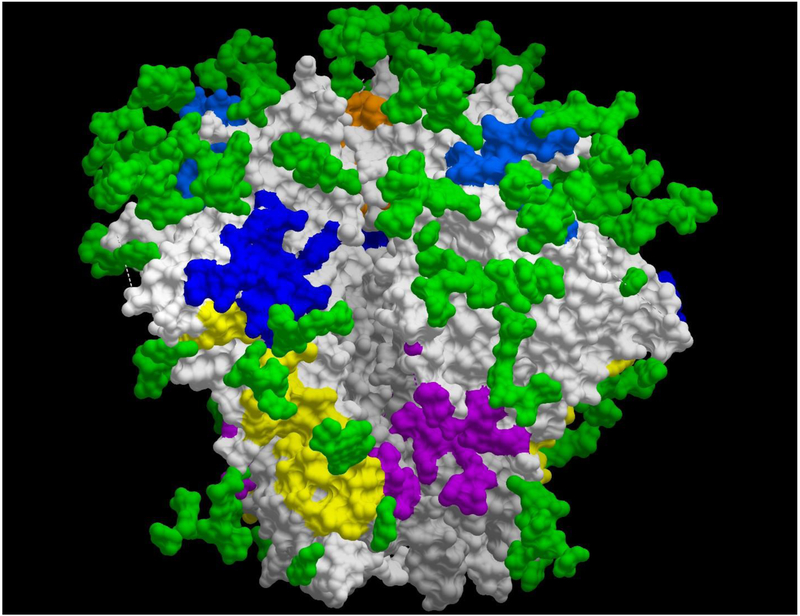
Figure 2.
Epitopes targeted by bNAbs on the HIV BG505 trimer. The HIV-1 gp120 is shown in light grey, and HIV-1 gp41 in grey. The glycosylation throughout the trimer (glycan shield) is represented by green residues. The following colors were used for the epitopes: orange (V3-glycan), light blue (V2-glycan), dark blue (CD4bs), yellow (subunit interface), and purple (fusion peptide). Epitope residues for PGT145 (V2-glycan), PGT 121 (V3-glycan), 8ANC19 (subunit interface), and PGT 151 (fusion peptide) obtained from [128], and N49P7 (CD4bs) was obtained from [7].
Despite tremendous progress, more information may be needed to effectively use the features and targets of broadly neutralizing responses for prevention. Molecular information concerning the structure and function of isolated bNAbs abounds. However, knowledge of the conditions that surround the evolution and persistence of broadly neutralizing antibodies in HIV-infected subjects will also inform the feasibility of vaccine design [20, 21]. For example, rare bNAbs that demand aberrant circumstances to evolve and persist may be less practical design templates versus ones that are recurring across the spectrum of HIV-infected humans. Accordingly, in this review we address the knowns and unknowns surrounding HIV bNAb responses that are detected in circulation.
The basic view of HIV neutralizing activity
Direct neutralizing activity amounts to the inactivation of an HIV virion via the binding of an anti-envelope antibody, preventing viral attachment/entry. Thus, an antibody’s direct neutralizing activity does not require Fc-mediated functions, although engagement with cell surface Fc receptor is probably unavoidable and even beneficial in vivo. All known antibodies that neutralize HIV bind to the viral envelope trimer at either the surface antigen, gp120, or the transmembrane protein, gp41 [22, 23]. Both proteins can be targeted for neutralization before and after attachment to host cells [5, 24–29].
Historically, HIV neutralizing activity has been assessed in a wide variety of in vitro assays. Primary T cells [30–33], primary macrophages [34] and various immortalized cell lines [35, 36] have been used as target cells for either primary viruses grown in activated human CD4+ T cells or cell line-adapted viruses. Other assay formats mix infected and uninfected cells to measure neutralization of cell-cell virus transmission [37]. More recently developed assays use pseudotyped viruses that express the envelopes of various HIV strains. Currently, the dominant assay of this sort mixes pseudotyped viruses with engineered HeLa cells (TZM-bl) expressing the HIV receptor (CD4) and coreceptors (CXCR4 and CCR5). The target cells express luciferase signals after a single round of infection. This assay format is highly reproducible, relatively high-throughput, and amenable to validation [38, 39]. It helped identify certain bNAbs as candidates for clinical development [8, 9, 40, 41]. Overall, it must be recognized that the potencies of neutralizing antibodies may vary significantly according to assay format. For example, neutralizing antibodies generally show lower potencies in assays using wild type viruses and/or primary CD4+ T cells as targets versus cell line-based assays with pseudoviruses [42].
Neutralization assay development has also involved a strategy of sorting viruses into “Tiers” based on their sensitivity to selected monoclonal neutralizing antibodies or reference from HIV-infected plasma samples [43]. The Tier 1 class comprises the greatest and most frequent sensitivity to neutralization. Tier 2 viruses exhibit intermediate sensitivity; Tier 3 viruses the least sensitivity. These distinctions are somewhat arbitrary, as neutralization sensitivity follows a continuum among viruses [43], and there have been recent attempts to account for this [44]. Nevertheless, the Tier system has been used for evaluating neutralizing antibody breadth and potency. Antibodies that cover all tiers in vitro are rare and highly cross-reactive. As such they are typically deemed to be superior members of their class.
Broadly neutralizing human antibodies: what’s out there?
Over the years a host of neutralizing mAbs have been recovered from HIV-infected subjects using a variety of methods [45–50]. The advent circa 2009 of single cell sorting with HIV envelope antigens, coupled with powerful new tools for sequencing immunoglobulin genes, significantly expanded the array of available antibodies and identified a collection of new epitopes and/or binding site details [5, 8–10, 41, 51, 52].
The properties of bNAbs inform the identification of vulnerable domains on the HIV envelope and reveal what humans are capable of producing. But faced with so many choices, which epitopes and antibodies do we really care about? The simple answer is that the most desirable antibodies will be the most potent and broadly reactive ones. But there are some important nuances to consider.
Gauging neutralization breadth demands some sort of rubric. A simple tally of neutralized strains is insufficient: one could simply line up and test a wide array of neutralization sensitive “Tier 1” viruses (see above) to secure a large total. Therefore, neutralization breath is currently assessed using panels of viruses representing multiple clades and neutralization Tiers (see above). Antibodies are typically judged as broadly neutralizing if they exhibit some capacity to reach across clades and more difficult to neutralize Tier 2 and 3 viruses.
As it stands, mAbs directed against the CD4 binding site (CD4bs) on gp120 and the 10E8 epitope in the membrane proximal region of gp41 exhibit superior cross-clade, cross-tier breadth and potency (Table 1). Several anti-CD4bs mAbs neutralize all viruses in routine test panels and assays [7]. The least (albeit still impressive) breadth is observed with bNAbs against epitopes containing glycan arrays on the gp120 V1/V2 loop, gp120 V3 loop, and the gp120-gp41 interface; although these bNAbs tend to have more potency than those with more breadth (Table 1).
Table 1:
Potency and neutralization of bNAbs
| mAb | Epitope | % neutralization breadth |
Potency (IC50 μg/ml) |
Potency (IC80 μg/ml) |
Reference |
|---|---|---|---|---|---|
| N49P6* | CD4bs | 100 | 0.31 | 0.71 | [7] |
| N49P7* | CD4bs | 100 | 0.10 | 0.39 | [7] |
| N49P11* | CD4bs | 100 | 1.98 | 10.64 | [7] |
| N6* | CD4bs | 98 | 0.09 | 0.32 | [7] |
| DH511–2 | gp41 | 98 | 1.20 | 6.60 | [52] |
| 10E8* | gp41 | 97 | 0.72 | 4.20 | [7] |
| VRC07* | CD4bs | 92 | 0.11 | 0.39 | [7] |
| 12A12 | CD4bs | 92 | 0.24 | 1.13 | [8] |
| 3BNC117* | CD4bs | 90 | 0.09 | 0.31 | [7] |
| N49P9* | CD4bs | 89 | 0.12 | 0.42 | [7] |
| VRC01* | CD4bs | 88 | 0.25 | 0.95 | [7] |
| NIH 45–46* | CD4bs | 86 | 0.09 | 0.38 | [7] |
| PG9* | VlV2-glycan | 85 | 0.11 | 0.30 | [7] |
| VRC-CH33 | CD4bs | 83 | 0.07 | 0.34 | [58] |
| PG16* | VlV2-glycan | 82 | 0.03 | 0.15 | [7] |
| PGDM1400* | VlV2-glycan | 80 | 0.02 | 0.14 | [7] |
| VRC-PG04 | CD4bs | 76 | 0.17 | 0.74 | [58] |
| PGT145* | VlV2-glycan | 74 | 0.11 | 0.40 | [7] |
| PGT151* | Fusion peptide | 66 | 0.01 | 0.02 | [7] |
| 8ANC195* | Subunit interface | 65 | 0.91 | 3.54 | [7] |
| PGT121* | V3-glycan | 66 | 0.03 | 0.08 | [7] |
| BG18 | V3-glycan | 64 | 0.01 | 0.04 | [87] |
| 3BNC55 | CD4bs | 63 | 0.57 | 2.28 | [8] |
| 35022 | Subunit interface | 62 | .03 | ND | [88] |
| PGT128* | V3-glycan | 60 | 0.02 | 0.05 | [7] |
| N60P1.1* | CD4bs | 59 | 1.71 | 6.88 | [7] |
| CH103 | CD4bs | 59 | 7.08 | 5.15 | [89] |
| 10–1074* | V3-glycan | 57 | 0.05 | 0.16 | [7] |
Neutralization tested in same panel in same lab
ND: Not Determined
Inter-epitope differences in neutralization breadth and potency reveal some intriguing details. Notably, the breadth of certain bNAbs are restricted by clade-specific holes in coverage. Anti-V3-glycan neutralizing antibodies tend to be resistant to CRF01_AE viruses [53]. The anti-V1V2-glycan mAbs demonstrate poor activity against Clade B viruses [54]. Furthermore, the potencies of certain anti-V1V2-glycan bNAbs, exemplified by PG9, PG16, and PGT145 [55], suffer from “incomplete neutralization,” a phenomenon where maximal neutralization plateaus without ever reaching 100%. This phenomenon occurs due to existence of some virions that are already resistant to antibody neutralization, regardless of the concentration, leading to non-sigmoidal dose-response neutralization curves. In addition to the anti-glycan antibodies, this was noted to be a problem for some anti-gp41 and anti-gp41–120 interface antibodies. Incomplete neutralization is thought to be due glycan heterogeneity, though it remains incompletely understood [55]. Perhaps because of this, bNAbs targeting the CD4 binding site seem to be less likely to show this phenomenon (an exception being b12) [55]. In any case, these characteristics are stark reminders of the capacity for HIV to evade the humoral immune system at all levels.
More encouraging data suggest that bNAbs share similar characteristics across multiple HIV-infected subjects, and thus restricted germline immunoglobulin gene usage across individuals could be targeted by a vaccine. Such commonality lowers the chances that the antibodies are products of rare, peculiar and/or donor-specific circumstances. For example, anti-CD4bs bNAbs arise from VH1–2 (can be found in 95% of populations) [5, 8, 9] or VH1–46 germline Ig heavy genes [7, 9, 56] across source subjects. Although the light chain of VRC01-like bNAbs makes fewer contacts with gp120, they also arise from a limited set of germ-line genes [8]. The recent finding of public antibody clonotypes (antibodies from different individuals that have an identical or near-identical CDRH3) makes this all the more relevant [57], but it remains to be seen if this will hold true for anti-CD4bs antibodies.
Almost all anti-CD4bs bNAbs exhibit a high degree of somatic hypermutation, which is essential for their breadth and potency [7, 8, 58, 59]. VRC01-like antibodies contact gp120 through the D loop, the CD4-binding loop, and V5 region [10]. VH1–2*02-derived antibodies are conserved in Arg71HC, Trp50HC, Asn58HC, and Trp100BHC [60] and the unusual short 5 length amino acid CDRL3 which is needed to stabilize and contact to gp120 (V5 and loop D) [61, 62]. This short CDRL3 is necessary to avoid the glycan at position N267 in the D loop of gp120 [63, 64]. Anti-CD4bs mAbs with the most extensive breadth (N49P6, N49P7 and NIH45–46) lack the deletion in CDRH3, distinguishing them from VRC01 and related bNAbs. This feature allows mAbs N49P6 and N49P7 to bypass Phe43 pocket in the CD4 binding site of gp120 and access the highly conserved inner domain of the envelope, thus allowing extraordinary breath [7].
About 50% of gp120 trimer mass consist of glycan which most of them are in form of high-mannose glucans (incomplete processed glycan) [65, 66]. Most but not all of the anti-glycan bNAbs that bind to gp120 to/or around N332 supersite, using their long CDRH3 to reach to the protein surface of gp120 at V3 region [40, 41, 67, 68]. PGT121, PGT128, and 10–1074 are among the V3-glycan bNAbs that are highly dependent on the glycan at positions 301 and 332 [40, 41]. Anti-V1V2-glycan bNAbs are well known for their very long CDRH3 and their acidic amino acids at the tip of the CDRH3 for their binding to the lysine-rich region of the V2 loop. Their binding to gp120 is dependent on the conformation of the trimeric form of gp120 [5, 54].
The frequencies and complexities of the broadly neutralizing plasma responses
Most if not all HIV-infected humans mount a neutralizing response against their homologous viruses; infected subjects frequently exhibit responses that neutralize a limited range of Tier 1 viruses; responses that neutralize across clades and cover more neutralization-resistant viruses are less prevalent [6, 69, 70]. In a cross-sectional study, Doria-Rose found that plasma responses in roughly 38% of subjects neutralized 4 out of 5 selected test viruses; roughly 8% could neutralize Tier 2 viruses in a cross-clade panel [70]. A cross-sectional study by Hraber et al observed that roughly 50% of infected subjects had circulating responses capable of neutralizing 50% of strains across clades in the TZM-bl format, along a continuum of potency [71]. Within this continuum are “elite neutralizers,” first defined by potent plasma activity against more than one pseudovirus within a clade group and across at least four clade groups [72]. In a global cross-sectional study, Simek et al. found that only 1% of subjects had plasma responses that met these criteria [72]. Among such persons, a smaller subset has plasma responses with near pan-neutralizing breadth [7, 10, 52, 73]. There is no clear evidence, however, that such responses control autologous viral replication in the host. Further, every known “elite neutralizer” from which a broadly neutralizing antibody has been derived harbors HIV replication. Such resistant strains may be rather common in the general HIV-infected population [74, 75].
The above studies have logically progressed toward attempts to understand what types of circulating antibodies are responsible for “elite” or pan-neutralizing plasma activity. To be sure, extremely broad and potent bNAbs with multiple utilities have been isolated from the memory B cell pools of elite neutralizers [8–10, 41, 73]. However, it has been shown that memory B cell and circulating anti-envelope antibody repertoires are discordant in HIV infection [76]. Accordingly, significant efforts have been applied toward direct deconvolution of plasma neutralizing responses. Approaches toward this end have included neutralization assays with mutant viruses selectively sensitive or resistant to specific types of neutralizing antibodies [5, 77–79]; antigen-specific depletion of plasma [4, 77, 80]; or bulk immunoglobulin fractionation [7, 81–83]. Recently, Williams et al. deconvoluted an anti-gp41 bNAb lineage from an African HIV-1 clade C chronically infected individual from both memory B cell and plasma [52]. Sajadi et al combined single-cell sequencing of bone marrow plasma cells with protein sequencing of gp120-specific plasma antibodies [7] to provide a comprehensive picture of the circulating repertoire. The approach yielded a family of closely related, near pan-neutralizing plasma antibodies (N49P6 and N49P7 in Table 1) from a subject with equally broad and potent plasma activity.
In view of the above studies, we may step back and ask: do broadly neutralizing plasma responses in one person stem from monoclonal, pauciclonal or polyclonal (one, several, or many) antibodies? So far, the answer seems to be “all of the above.” In some cases broadly neutralizing plasma activity tracks to one or two lineages of highly related antibodies. In a study of plasma from 13 HIV-infected patients using a panel of mutant pseudoviruses, Walker et al. demonstrated that broadly neutralizing plasma activity typically tracks to 1 or 2 epitope specificities [84]. Similarly, Bonsignori et al. found that 2 distinct specificities isolated from a single subject seemed to explain >95% of the serum neutralization [85]. Thus, very broad and potent plasma neutralizing responses are unlikely to be monoclonal although they may be monospecific; i.e., comprising a swarm of related antibodies sharing a major epitope specificity [7].
More intriguing are cases where the overall plasma neutralizing profile cannot be explained by the limited number of specificities as the examples given above [7, 80, 81]. Scheid et al reported that diverse mixtures of anti-HIV envelope mAbs derived from memory B cells approached but could not recapitulate the full spectrum of plasma neutralizing activity of the source subject [86]. In one Elite Neutralizer, a case was identified where no single bNAb found in plasma could explain more than 60% of the neutralization breadth in circulation [7]. Simple mixing of multiple broad and potent plasma bNAbs from this subject achieved no more than ~90% of the circulating neutralization breadth. Collectively, these findings present a mixed bag for HIV vaccine development. Observations linking broadly neutralizing plasma activity to monoclonal or pauciclonal specificity are a potential boon, as practical translation is conceptually uncomplicated. Data indicating that a more complicated plasma milieu explains “elite” neutralizing activity warrants further exploration.
A related question concerns whether circulating “non-neutralizing” or poorly neutralizing antibodies abrogate the persistence and/or activity of bNAbs. Deconvolution of a near pan-neutralizing plasma response showed that the anti-CD4 binding site antibody family responsible for plasma activity comprised a minority of the anti-gp120 repertoire, co-circulating along with a much larger population of non-neutralizing anti-gp120 antibodies [7]. The same situation was observed in this donor at multiple sampling times and also in another donor [7].
What aspects of HIV infection promote broad and potent HIV neutralizing antibodies in circulation?
Multiple studies [71, 90–92] have shown duration of infection and chronic antigen exposure to be key factors for bNAb development. For unknown reasons, anti-gp120 responses tend to arise later than those against other HIV antigens, including gp41 (the median time for appearance of anti-gp41 and anti-gp120 IgG antibodies are 14 and 28 days, respectively) [93]. Anti-gp41 antibodies are thought to arise earlier than anti-gp120 because of engagement of pre-existing memory B cells that have been generated through contact with commensal microbiota Nevertheless, neutralizing antibodies take several months to develop [36, 94], neutralization breadth takes a median of 2.5 years to first be detected [95]. The most broad and potent bNAbs manifest with unusually high levels of somatic hypermutation [96, 97], reflecting the impact of prolonged antigenic stimulation. The prolonged time it takes to develop the bNAb response has profound implications for HIV vaccine development (potential number/amount of antigen exposure, as well as high hypermutation requirement).
Early germinal centers (GCs) are made up of highly diverse B cell clones [98], but clonal selection leads to the more homogeneous GCs, with affinity maturation constantly eliminating low-affinity antibodies and replacing them with higher-affinity antibodies. In the germinal center, other factors can be important as well. B cells reactive to gp41 antigen have been found to present in the memory B cell pool of HIV-negative persons [99]. In certain other situations, different B cell lineages can work synergistically (viral escapes mutants against one antibody have been shown to higher binding affinity to a second lineage) [100].
The magnitude of viral replication is an intriguing variable in bNAb development. Several studies indicated an association between high HIV-1 viral load and bNAb production [69, 101–103]. However, no such relationship was reported elsewhere [6, 102]. Some of this discrepancy may be due to varying definitions of bNAb activity between studies. Remarkably, Clade B “elite neutralizers,” reflect sustained, low-level viremia between 102 and 104 copies/ml [6]. In other words, many will also fall within the category of “elite controllers” or “viremic controllers.” This trend is evident in many of the subjects from which bNAbs of different specificities have been derived: Donor 45 (VRC01, VRC07, NIH45–46), Donor N49 (N49P6, N49P7, N49P9), Donor N60 (N60P1.1, N60P2.1), Donor Z258 (N6), Patient 3 (3BNC117), N152 (10E8), EB354 (BG18), Patient 8 (8ANC195), and Donor Ch0210 (DH511–2) [7–10, 104] (Table 2). In addition to low viral loads, these subjects (whose data is available) have relatively intact CD4 counts and prolonged duration of infection (Table 2). It may be that beginning of breadth noted with individuals with higher viral loads is due to an acceleration of the process initially (from higher viral loads) but that the bNAb production cannot be sustained due to CD4 cell loss and B cell dysfunction with disease progression. In those with extreme bNAb activity, sustained viral control and CD4 preservation over many years seem to be needed.
Table 2.
Donor and antibody characteristics of bNAbs
| MAb | Clade | Donor | Years after diagnosis |
HIV-1 RNA copies/ml |
CD4 (cells/ul) |
Epitope | Ig gene family | Ref |
|---|---|---|---|---|---|---|---|---|
| N49P6 N49P7 N49P9 |
B | N49 | 20 | 7,854 | 447 | CD4bs | IGHV 1–2*02 IGLV2–11*01/2 IGLV2–23*01/2/3 |
[7] |
| N6 | B | Z258 | 21 | 996 | 733 | CD4bs | IGHV 1–2*02 IGKV1–33 |
[10] |
| 3BNC117 3BNC55 |
B | Patient 3 |
Unknown (dx 2002) |
880 | 427 | CD4bs | IGHV 1–2*02 IGKV1D-33 | [8] |
| VRC01 VRC07 NIH 45–46 |
B | Donor 45 |
18 | <15,000 | >500 | CD4bs | IGHV 1–2*02 IGKV3–11*01 |
[8, 9, 104] |
| BG18 | B | EB354 | 28 | 454 (65 to 2,520) |
697 (554–1323) |
V3-glycan | IGHV4–4 IGLV3–25 |
[87] |
| N60P1.1 | B | N60 | 5 | <400 | 888 | CD4bs | IGHV 1–2*02 IGKV 1–5*01 |
[7] |
| 10E8 | B | N152 | >20 | 3,811 | 325 | gp41 | IGHV3–15*05 IGLV3–19*01 |
[73] |
| 35022 | B | N152 | >20 | 3,811 | 325 | Subunit interface |
IGHV1–18*02 IGLV2–14*02, or IGLV2–23*01/2/3 |
[73] |
| 8ANC195 | B | Patient 8 |
Unknown (dx 1989) |
<50 | 580 | Subunit interface |
IGHV 1–69 IGKV 1–5 |
[8] |
| PGT151 | C | Donor 31 |
>3 | Unknown | Unknown | Fusion peptide |
IGHV3–30*04 IGKV2D-29*02 |
[114] |
| DH511–2 | C | CH021 0 |
Unknown | 5,180 | Unknown | gp41 | IGHV3–15 IGKV1–39 |
[52] |
| CHI03 | C | CH505 | 2.6 | ~104–105 | 69–431 | CD4bs | IGH4–59*01 IGL3-l*01 |
[89] |
| PGT145 PGDM1400 |
C | Donor 84 |
>3 | Unknown | Unknown | VlV2-glycan | IGHV 1–8*01 IGKV2–28*01/ IGKV2D-28*01 |
[40,51] |
| PGT121 10–1074 |
A | Donor 17 |
>3 | Unknown | Unknown | V3-glycan | IGHV4–59*01 IGLV3–21*02 |
[40, 41, 115] |
| PG9 PG16 |
A | Donor 24 |
>3 | Unknown | Unknown | VlV2-glycan | IGHV3–33*05 IGLV2–14*01 |
[5] |
| VRC-CH33 | A | CH021 9 |
>3 | Unknown | Unknown | CD4bs | IGHV1–2*02 IGK1–33*01 |
[58] |
| VRC-PG04 | A/D | Donor 74 |
Unknown | Unknown | Unknown | CD4bs | IGHV1–2*02 IGK1–33*01 |
[58] |
| 12A12 | CRF02_AG | Patient 12 |
>3 | Unknown | Unknown | CD4bs | IGHV1–2 IGKV1D-33 |
[8,84, 116] |
| PGT128 | CRF02_AG | Donor 36 |
>3 | Unknown | Unknown | V3-glycan | IGHV4–39*07 IGLV2–8*01 |
[40] |
The infecting clade of a patient seems to have a role in the types of bNAbs they can generate. In a study of patients from Malawi, South Africa, Holland, and the United States, broadly neutralizing anti-CD4bs antibodies (not necessarily bNAbs) were detected in 88% of all sera collected between 99 and 258 weeks post-HIV-1 infection subjects [105]. However, the most broad and potent CD4bs bNAbs have been isolated from Clade B infected patients (Table 2). In comparison, anti-V1V2-glycan and anti-V3-glycan bNAbs have been isolated from non-Clade B subjects (Table 2). This finding has also been validated on the population level. In a study with over 4,000 patients, Rusert et al. found that antibodies targeting the CD4bs were associated with subtype B infection, whereas anti-V2-glycan antibodies were associated with non-B subtypes [90]. Thus, the infecting HIV clade can have implications on the type of bNAb produced, and this should be a factor when considering the design of an HIV vaccine that will generate bNAbs.
The qualitative aspects of the viral envelopes being presented over time should also influence whether or not anti-envelope responses become broadly neutralizing. Exposure to envelope diversity is a contributing factor, during early infection [103] or with superinfection with heterologous HIV strains [54, 106] On the other hand, a recent report argues that certain transmitted viruses possess unique phenotypic and/or genotypic properties that imprint broadly neutralizing antibody lineages into the humoral immune system at the time of exposure [107].
If bNAbs regularly arise in certain HIV-infected populations, HIV envelope evolution should manifest a temporal drift toward neutralization resistance in the same groups of people [1]. Longitudinal analyses of clade B and clade C infected cohorts indicate this may indeed be the case. Bunnik et al examined the neutralization sensitivities of viruses isolated from over 30 clade B HIV-infected men having sex with men (MSM) ≤ 4 months after seroconversion between 1985–1989 and 2003–2006 [108]. Neutralization assays indicated an overall trend toward decreased neutralization sensitivity over time. Bouvin-Pley et al examined the neutralization sensitivities of viruses isolated from early/transmitted clade B HIV envelope variants from 40 men having sex with men during the period from 1997 to 2010 [109]. This study revealed a continuous, temporal drift toward decreased neutralization sensitivity to a number of anti-gp120 neutralizing mAbs, including ones directed against the CD4bs and variable loop glycans. In another study done in a clade C infected population, the envelopes showed increasing resistance over a 13 year time period [110]. On a sobering note, the more recent, neutralization-resistant variants were not only fully replication-competent but also less immunogenic for generating plasma neutralizing responses [108, 109]. It will be interesting to follow such population dynamics as efforts to use anti-CD4bs broadly neutralizing antibodies for prevention and therapy progress worldwide.
Host factors could further contribute to bNAb development in HIV infection. Landais et al. investigated the effects of age, geographic origin, gender, viral load, CD4 T cell count, virus subtypes, and HLA types on the development of neutralization breadth in patients from Eastern and Southern Africa. They found no detectable influence of gender, age, and/or geographic origin on the development of bNAb breadth [102]. However, in several studies in infants, bNAbs appear to develop differently than adults. In study, over 75% of children less than 6 years old were able to neutralize a panel of 16 viruses, compared to only 19% of adults [111]. In another study, 20 of 28 infants less than 1 year of age developed cross-clade neutralizing antibody, some within 1 year of becoming infected [112]. Interestingly, children appear to routinely develop bNAbs against more than one epitope [113]. Whether this may be due to a function of a more intact immune system [111], or to the something specific in the developing immune system are still not known.
What impedes the generation and circulation of broad and potent HIV neutralizing antibodies?
HIV bNAbs have been frustratingly difficult to raise in most mammals. Circa 2005, Haynes et al recognized that the broadly neutralizing mAbs 2F5 and 4E10, reactive with the membrane proximal region (MPER) of gp41, were auto/polyreactive with cardiolipin and other mammalian antigens [117]. Based on such findings, these investigators posited that immunization strategies routinely fail to raise circulating titers of neutralizing antibodies against MPER or other neutralizing epitopes because the parental B cells are auto/polyreactive. As such, they are deleted via natural immune tolerance mechanisms [118]. This concept was initially met with strong skepticism and resistance [119]. Nevertheless, it was ultimately shown that B cell development in 2F5 VDJ knock-in mice is blocked at the stage of pre–B to immature B cells, similar to what is observed in mice expressing known autoreactive B cell receptors against self-antigens [120]. Because of these elegant studies, the concept that bNAbs are linked to polyreactivity is a cornerstone for understanding both natural anti-HIV envelope responses and also why such immunity is so difficult to translate into a vaccine setting.
Ironically, some of the broadest and most potent HIV bNAbs are defined by characteristics that are typical of polyreactive antibodies such as long and hydrophobic CDRH3 domains. The 4E10 CDRH1 and CDRH3 domains can bind a variety of lipids (e.g. cardiolipin) potentially located in the vicinity of the gp41 MPER peptide. The near pan-neutralizing anti-MPER mAb 10E8 is predicted to interact with membrane lipids via its CDRL1, CDRL2 and/or CDRH3 domains [121]. 10E8 was originally believed to not bind lipids [73]; however, it was later demonstrated that it binds cholesterol-rich bilayers [122]. Thus, the most broadly neutralizing anti-MPER neutralizing antibodies bind hybrid epitopes comprising multiple “self” components. Anti-V1V2-glycan and anti-V3-glycan bNAbs also exhibit the connection between a long CDRH3 domain and polyreactivity. Fortunately, very broad and potent CD4bs bNAbs have been identified that are not apparently auto/polyreactive, according to standard measures [123].
Concluding remarks:
The translation of broadly neutralizing antibody responses into clinical applications currently follows two paths, each with its own hurdles. The first path involves the rational optimization of bNAbs for direct use as antiviral agents. This exercise is largely technical, success mainly hinges on the nature and performance of the antibodies themselves. Passive immunization of HIV-infected humans with single bNAbs has delivered only modest and transient clinical benefit, in part because of the emergence of resistant variants, some of which were present in subjects prior to antibody treatment [74, 75, 124]. Major efforts to broaden virus coverage, including the use of bispecific and mixtures of bNAbs, are underway [125]. The second path involves the use of bNAbs as templates for vaccine design. Here, success depends in part on whether the immune systems of most healthy humans are capable of generating the template bNAbs in an effective manner. Studies of circulating bNAb responses in HIV-infected subjects are helping to inform this issue, yielding contextual information not easily divulged from interrogations of memory B cell repertoires.
Two key lessons are emerging from studies of plasma neutralizing responses. The first is that persistent, near pan-neutralizing plasma activity can track to one or a few related antibody species comprised by the circulating polyclonal anti-Env response. These observations lend further support to the development of vaccines built around targeted epitopes. Notably, near pan-neutralizing antibodies actively persist in plasma even as a minor part of a much larger repertoire of non-neutralizing anti-envelope antibodies. Specifically, circulating bNAbs are not “swamped out” by a higher prevalence of other anti-Env specificities. This situation is additional good news for vaccine development, as it obviates concerns that near-pristine epitope specificity and selectivity is needed to elicit a bNAb response. A third lesson is that bNAb activities seen as most relevant to vaccine development (e.g. ones with near pan-neutralizing activity) are infrequently detected in plasma from HIV-infected subjects. As these activities require immunoglobulins with atypical characteristics [5, 7, 9, 51, 52, 86], emergence and persistence in plasma could depend on peculiar characteristics of the host; the nature of the infecting HIV strain; coincident opportunistic infections; or other variables [54, 96, 97, 99, 100, 123, 126, 127]; [107]. On the other hand, it is promising that broadly neutralizing antibodies with similar specificities and biochemical properties are repeatedly observed in diverse subjects [7, 9, 10]. These findings could mean that certain types of broadly neutralizing responses are not strongly dependent on rare, subject-specific immune stimulation/evolution events and may be achieved in healthy humans via vaccination.
Highlights.
A minority of HIV-infected subjects develop plasma neutralizing responses with near pan-breadth against heterologous virus strains. The development of such responses, depending on epitope specificity, is linked to infecting clade, viral load, duration of infection, and virus and host genetic factors.
New approaches allow extremely broad and potent bNabs to be derived directly from human plasma responses.
In some cases, broadly neutralizing plasma activity traces to a single lineage of a few related bNabs, representing a minority of the polyclonal anti-HIV envelope response. The co-circulation of poorly neutralizing/non-neutralizing anti-envelope antibodies does not abrogate the persistence and activities of the bNabs.
Each class of known bNAb exhibits at least one “atypical” trait; e.g., high somatic hypermutation, long HCDR3, and/or auto/polyreactivity.
Among bNAbs types, those against the CD4bs are seen more frequently; share common features among diverse subjects; and exhibit extreme neutralization breadth and potency.
Box 1. Clinician’s Corner.
Most individuals with HIV infection make type-specific neutralizing antibody, capable of only neutralizing their own infecting clade of virus. Although a substantial portion of HIV-infected individuals do make some cross-reactive antibodies (to different clades) by several years after infection, only a minority of HIV infected individuals (1%) make broadly neutralizing antibodies that are potent enough to neutralize most strains from different clades around the world.
The study of the bNAb response as it occurs in circulation, and the factors that lead to the development of such a response are intensive areas of study. Some factors that affect bNAb development are duration of HIV infection, amount of circulating virus, infecting clade, and viral diversity.
Currently, bNAbs are being used as templates from which to design an HIV vaccine, and being evaluated as a potential therapy that can prevent, treat, or “cure” HIV infection.
bNAbs are known to target several different epitopes on the HIV envelope, including CD4bs, V1V2-glycan, V3-glycan, gp41, and the gp41-gp120 interface. Besides the difference in epitopes, bNAbs have different properties (gene family use, degree of somatic hypermutation, autoreactivity, polyreactivity, potency, breadth, ability to completely neutralize virus, etc.), some of which appear among to be specific to the classes of bNAbs. The CD4bs bNAbs appear to several characteristics that are favorable for clinical development compared to the others.
Outstanding questions.
Despite tremendous progress, efforts to develop bNAb-based HIV countermeasures remain hindered by several hurdles and gaps in knowledge. Questions addressable by studies of plasma neutralizing responses include:
What host/virus conditions and characteristics support the generation and persistence of bNAb responses in plasma? Are they rare or recurring?
What explains broadly neutralizing plasma activity when it is not linked to one or few related bNAbs? Why do combinations of bNAbs from a single subject fail to replicate the full spectrum of plasma neutralizing activity? What is the missing factor?
In elite neutralizers, what virus/host relationships are in play when ongoing, low-level plasma viral replication coexists with broad and potent plasma bNAbs? Can epitope-paratope relationships in these situations inform the design of engineered bNAbs with truly pan-neutralizing activity and/or inform improved envelope-based vaccine designs?
An expanded database of deconvoluted plasma neutralizing responses should help resolve these questions, thus elucidating immune landscapes most likely to promote the development of pan-neutralizing antibody responses. Such information should provide valuable guideposts for developing optimized HIV countermeasures based on humoral immunity.
ACKNOWLEDGMENTS
The work on this article was supported by NIH 1R01AI110259–01A1 and VA Merit Award 1I01BX002358–01A1 to M.M.S.
Glossary
- Autoreactive
antibody having this trait are those with the ability to bind to self antigens
- bNAbs
Broadly neutralizing antibodies. A type of antibody (directed at different epitopes) that arises in some HIV patients that are able to neutralize the majority of known HIV strains. These antibodies harbor unusual traits such as high somatic hypermutation, long HCDR3s, and auto/polyreactivity
- Clade
subtype; HIV-1 is made up of 3 groups (M, N, and O), with group M representing the vast majority of HIV strains worldwide. Group M is made up of many clades, with clades A, B, and C being the most predominant, followed by other clades and between-clade recombinants
- CD4 binding site
The part of HIV-1 envelope that binds to the CD4 receptor, which initiates viral entry. Antibodies which bind to the CD4 binding site of gp120 are termed anti-CD4bs antibodies
- Elite Controller
Rare HIV-infected individuals who can naturally suppress viral replication of HIV in their bodies to extremely low levels
- Elite Neutralizer
Rare HIV-infected individuals whose serum or plasma immunoglobulin can neutralize a majority of known HIV strains
- Fc receptor
receptor on the surface of certain cells (such as B cells, NK cells, macrophages, neutrophils, and mast cells) that recognize the Fc portion (constant region) of an antibody
- gp120
glycoprotein 120, one of the proteins (along with gp41) making up the HIV envelope. gp120 is made up of 5 constant domains (labelled C1–C5) and variable domains or loops (labeled V1–V5)
- HIV
Human immunodeficiency virus, a positive-sense single-stranded retrovirus covered with a viral envelope composed of trimers composed of gp120 and gp41 proteins
- HIV envelope
The viral envelope of HIV, which is punctuated by spikes, each one made of three gp120s and gp41s forming a heterodimer
- Neutralization
The ability of an antibody to prevent prevent viral attachment, entry, and/or infection of otherwise susceptible cells without engagement of Fc-mediated effector functions. In HIV this occurs with inactivation of a virion via the binding of an anti-envelope antibody. The outcome of the interaction is direct abrogation of envelope function (e.g. host receptor attachment)
- Polyreactive
antibody having this trait are those with the ability to bind to multiple antigens
- Somatic hypermutation (SHM)
The process of mutation affecting the variable region of B-cell receptors (BCR) in germinal center B cells undergoing affinity maturation, leading to higher affinity BCR and antibodies
- Viremia
the amount of virus in the blood. In HIV-1 infection, this can vary approximately 7 logs (100-107 HIV-1 RNA copies/ml), with the mean level in chronic infection (post acute infection and pre-AIDS) between 104-105 HIV-1 RNA copies/ml
- Virion
A complete virus article (infective form of a virus outside a host cell, with a core of RNA or DNA and a capsid)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lewis GK et al. (2017) Survivors Remorse: antibody-mediated protection against HIV-1. Immunol Rev 275 (1), 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sather DN and Stamatatos L (2010) Epitope specificities of broadly neutralizing plasmas from HIV-1 infected subjects. Vaccine 28 Suppl 2, B8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y et al. (2009) Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J Virol 85 (17), 8954–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhillon AK et al. (2007) Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J Virol 81 (12), 6548–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker LM et al. (2009) Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326 (5950), 285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sajadi MM et al. (2011) Correlation between circulating HIV-1 RNA and broad HIV-1 neutralizing antibody activity. J Acquir Immune Defic Syndr 57 (1), 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sajadi MM et al. (2018) Identification of Near-Pan-neutralizing Antibodies against HIV-1 by Deconvolution of Plasma Humoral Responses. Cell 173 (7), 1783–1795 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheid JF et al. (2011) Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333 (6049), 1633–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X et al. (2010) Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329 (5993), 856–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J et al. (2016) Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth. Immunity 45 (5), 1108–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moldt B et al. (2012) Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proceedings of the National Academy of Sciences of the United States of America 109 (46), 18921–18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shingai M et al. (2014) Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med 211 (10), 2061–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pegu A et al. (2014) Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci Transl Med 6 (243), 243ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saunders KO et al. (2015) Sustained Delivery of a Broadly Neutralizing Antibody in Nonhuman Primates Confers Long-Term Protection against Simian/Human Immunodeficiency Virus Infection. J Virol 89 (11), 5895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudicell RS KY, Ko SY, Pegu A, Louder MK, et al. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. . J Virol 88, 12669–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saunders KO et al. (2015) Broadly Neutralizing Human Immunodeficiency Virus Type 1 Antibody Gene Transfer Protects Nonhuman Primates from Mucosal Simian-Human Immunodeficiency Virus Infection. J Virol 89 (16), 8334–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medina-Ramirez M et al. (2017) Stabilized HIV-1 envelope glycoprotein trimers for vaccine use. Curr Opin HIV AIDS 12 (3), 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Taeye SW et al. (2016) HIV-1 Envelope Trimer Design and Immunization Strategies To Induce Broadly Neutralizing Antibodies. Trends Immunol 37 (3), 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders RW and Moore JP (2017) Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol Rev 275 (1), 161–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Regenmortel MH (2012) Basic research in HIV vaccinology is hampered by reductionist thinking. Front Immunol 3, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Regenmortel MHV (2017) Immune systems rather than antigenic epitopes elicit and produce protective antibodies against HIV. Vaccine 35 (16), 1985–1986. [DOI] [PubMed] [Google Scholar]
- 22.Wibmer CK et al. (2015) HIV broadly neutralizing antibody targets. Curr Opin HIV AIDS 10 (3), 135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrabi R et al. (2015) Identification of Common Features in Prototype Broadly Neutralizing Antibodies to HIV Envelope V2 Apex to Facilitate Vaccine Design. Immunity 43 (5), 959–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorny MK et al. (2005) Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J Virol 79 (8), 5232–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu L et al. (1996) CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384 (6605), 179–83. [DOI] [PubMed] [Google Scholar]
- 26.Trkola A et al. (1996) CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384 (6605), 184–7. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan N et al. (1998) CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol 72 (6), 4694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burton D et al. (1994) Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266 (5187), 1024–27. [DOI] [PubMed] [Google Scholar]
- 29.Ray K et al. (2014) Antigenic properties of the HIV envelope on virions in solution. J Virol 88 (3), 1795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown BK et al. (2008) Cross-clade neutralization patterns among HIV-1 strains from the six major clades of the pandemic evaluated and compared in two different models. Virology 375 (2), 529–38. [DOI] [PubMed] [Google Scholar]
- 31.D’Souza MP et al. (1991) Evaluation of monoclonal antibodies to HIV-1 by neutralization and serological assays: an international collaboration. Collaborating Investigators. AIDS 5 (9), 1061–70. [DOI] [PubMed] [Google Scholar]
- 32.D’Souza MP et al. (1997) Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. AIDS Clinical Trials Group Antibody Selection Working Group. J Infect Dis 175 (5), 1056–62. [DOI] [PubMed] [Google Scholar]
- 33.Nara PL et al. (1987) Simple, rapid, quantitative, syncytium-forming microassay for the detection of human immunodeficiency virus neutralizing antibody. AIDS Res Hum Retroviruses 3 (3), 283–302. [DOI] [PubMed] [Google Scholar]
- 34.Ruppach H et al. (2000) Human immunodeficiency virus (HIV)-positive sera obtained shortly after seroconversion neutralize autologous HIV type 1 isolates on primary macrophages but not on lymphocytes. J Virol 74 (12), 5403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polonis VR et al. (2008) Recent advances in the characterization of HIV-1 neutralization assays for standardized evaluation of the antibody response to infection and vaccination. Virology 375 (2), 315–20. [DOI] [PubMed] [Google Scholar]
- 36.Wei X et al. (2003) Antibody neutralization and escape by HIV-1. Nature 422 (6929), 307–12. [DOI] [PubMed] [Google Scholar]
- 37.Reh L et al. (2015) Capacity of Broadly Neutralizing Antibodies to Inhibit HIV-1 Cell-Cell Transmission Is Strain- and Epitope-Dependent. PLoS Pathog 11 (7), e1004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montefiori DC (2009) Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 485, 395–405. [DOI] [PubMed] [Google Scholar]
- 39.Sarzotti-Kelsoe M et al. (2014) Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods 409, 131–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker LM et al. (2011) Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477 (7365), 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mouquet H et al. (2012) Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109 (47), E3268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen YZ et al. (2018) Neutralizing Activity of Broadly Neutralizing Anti-HIV-1 Antibodies against Clade B Clinical Isolates Produced in Peripheral Blood Mononuclear Cells. J Virol 92 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seaman MS et al. (2010) Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84 (3), 1439–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hraber P et al. (2018) A single, continuous metric to define tiered serum neutralization potency against HIV. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robert-Guroff M et al. (1985) HTLV-III-neutralizing antibodies in patients with AIDS and AIDS-related complex. Nature 316 (6023), 72–4. [DOI] [PubMed] [Google Scholar]
- 46.Weiss RA et al. (1985) Neutralization of human T-lymphotropic virus type III by sera of AIDS and AIDS-risk patients. Nature 316 (6023), 69–72. [DOI] [PubMed] [Google Scholar]
- 47.Purtscher M et al. (1994) A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses 10 (12), 1651–8. [DOI] [PubMed] [Google Scholar]
- 48.Zwick MB et al. (2001) Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol 75 (22), 10892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammond PW (2010) Accessing the human repertoire for broadly neutralizing HIV antibodies. mAbs 2 (2), 157–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roben P et al. (1994) Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol 68 (8), 4821–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sok D et al. (2014) Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci U S A 111 (49), 17624–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams LD et al. (2017) Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Science Immunology 2 (7), eaal2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H et al. (2018) Evaluation of susceptibility of HIV-1 CRF01_AE variants to neutralization by a panel of broadly neutralizing antibodies. Arch Virol 163 (12), 3303–3315. [DOI] [PubMed] [Google Scholar]
- 54.Doria-Rose NA et al. (2014) Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509 (7498), 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCoy LE et al. (2015) Incomplete Neutralization and Deviation from Sigmoidal Neutralization Curves for HIV Broadly Neutralizing Monoclonal Antibodies. PLoS Pathog 11 (8), e1005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.West AP Jr. et al. (2012) Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci U S A 109 (30), E2083–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Setliff I et al. (2018) Multi-Donor Longitudinal Antibody Repertoire Sequencing Reveals the Existence of Public Antibody Clonotypes in HIV-1 Infection. Cell Host Microbe 23 (6), 845–854 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu X et al. (2011) Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333 (6049), 1593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou T et al. (2010) Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329 (5993), 811–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scharf L et al. (2013) Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proc Natl Acad Sci USA 110 (15), 6049–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jardine JG et al. (2015) HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349 (6244), 156–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burton DR and Hangartner L (2016) Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu Rev Immunol 34, 635–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou T et al. (2007) Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445 (7129), 732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jardine J et al. (2013) Rational HIV immunogen design to target specific germline B cell receptors. Science 340 (6133), 711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korber B et al. (2001) Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull 58, 19–42. [DOI] [PubMed] [Google Scholar]
- 66.Doores KJ et al. (2010) Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci USA 107 (31), 13800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Calarese DA et al. (2003) Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300 (5628), 2065–71. [DOI] [PubMed] [Google Scholar]
- 68.Kong L et al. (2013) Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 20 (7), 796–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doria-Rose NA et al. (2010) Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol 84 (3), 1631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doria-Rose NA et al. (2009) Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol 83 (1), 188–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hraber P et al. (2014) Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28 (2), 163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simek MD et al. (2009) Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol 83 (14), 7337–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang J et al. (2012) Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491 (7424), 406–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scheid JF et al. (2016) HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535 (7613), 556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bar KJ et al. (2016) Effect of HIV Antibody VRC01 on Viral Rebound after Treatment Interruption. N Engl J Med 375 (21), 2037–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guan Y et al. (2009) Discordant memory B cell and circulating anti-Env antibody responses in HIV-1 infection. Proc Natl Acad Sci USA 106 (10), 3952–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gray ES et al. (2009) Antibody specificities associated with neutralization breadth in plasma from HIV-1 subtype C infected blood donors. J Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davis KL et al. (2009) High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology 387 (2), 414–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Decker JM et al. (2005) Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201 (9), 1407–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Y et al. (2009) Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J Virol 83 (2), 1045–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sajadi MM et al. (2012) Signature biochemical properties of broadly cross-reactive HIV-1 neutralizing antibodies in human plasma. Journal of Virology 86 (9), 5014–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sajadi MM et al. (2015) lambda Light Chain Bias Associated With Enhanced Binding and Function of Anti-HIV Env Glycoprotein Antibodies. J Infect Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tomaras GD et al. (2011) Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J Virol 85 (21), 11502–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Walker LM et al. (2010) A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6 (8), e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bonsignori M et al. (2012) Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol 86 (8), 4688–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scheid JF et al. (2009) Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458 (7238), 636–40. [DOI] [PubMed] [Google Scholar]
- 87.Barnes CO et al. (2018) Structural characterization of a highly-potent V3-glycan broadly neutralizing antibody bound to natively-glycosylated HIV-1 envelope. Nat Commun 9 (1), 1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang J et al. (2014) Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515 (7525), 138–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liao HX et al. (2013) Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496 (7446), 469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rusert P et al. (2016) Determinants of HIV-1 broadly neutralizing antibody induction. Nat Med 22 (11), 1260–1267. [DOI] [PubMed] [Google Scholar]
- 91.Subbaraman H et al. (2018) Broadly neutralizing antibodies: What is needed to move from a rare event in HIV-1 infection to vaccine efficacy? Retrovirology 15 (1), 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dugast AS et al. (2017) Virus-driven Inflammation Is Associated With the Development of bNAbs in Spontaneous Controllers of HIV. Clin Infect Dis 64 (8), 1098–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tomaras GD et al. (2008) Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol 82 (24), 12449–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aasa-Chapman MM et al. (2004) Development of the antibody response in acute HIV-1 infection. AIDS 18 (3), 371–81. [DOI] [PubMed] [Google Scholar]
- 95.Mikell I et al. (2011) Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog 7 (1), e1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kwong PD and Mascola JR (2012) Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37 (3), 412–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klein F et al. (2013) Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153 (1), 126–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tas JM et al. (2016) Visualizing antibody affinity maturation in germinal centers. Science 351 (6277), 1048–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liao HX et al. (2011) Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J Exp Med 208 (11), 2237–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gao F et al. (2014) Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell 158 (3), 481–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pereyra F et al. (2008) Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis 197 (4), 563–71. [DOI] [PubMed] [Google Scholar]
- 102.Landais E et al. (2016) Broadly Neutralizing Antibody Responses in a Large Longitudinal Sub-Saharan HIV Primary Infection Cohort. PLoS Pathog 12 (1), e1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Piantadosi A et al. (2009) Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J Virol 83 (19), 10269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rudicell RS et al. (2014) Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J Virol 88 (21), 12669–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lynch RM et al. (2012) The development of CD4 binding site antibodies during HIV-1 infection. J Virol 86 (14), 7588–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Williams KL et al. (2018) Superinfection Drives HIV Neutralizing Antibody Responses from Several B Cell Lineages that Contribute to a Polyclonal Repertoire. Cell Rep 23 (3), 682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kouyos RD et al. (2018) Tracing HIV-1 strains that imprint broadly neutralizing antibody responses. Nature 561 (7723), 406–410. [DOI] [PubMed] [Google Scholar]
- 108.Bunnik EM et al. (2010) Adaptation of HIV-1 envelope gp120 to humoral immunity at a population level. Nat Med 16 (9), 995–7. [DOI] [PubMed] [Google Scholar]
- 109.Bouvin-Pley M et al. (2013) Evidence for a continuous drift of the HIV-1 species towards higher resistance to neutralizing antibodies over the course of the epidemic. PLoS Pathog 9 (7), e1003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rademeyer C et al. (2016) Features of Recently Transmitted HIV-1 Clade C Viruses that Impact Antibody Recognition: Implications for Active and Passive Immunization. PLoS Pathog 12 (7), e1005742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Muenchhoff M et al. (2016) Nonprogressing HIV-infected children share fundamental immunological features of nonpathogenic SIV infection. Sci Transl Med 8 (358), 358ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goo L et al. (2014) Early development of broadly neutralizing antibodies in HIV-1-infected infants. Nat Med 20 (6), 655–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ditse Z et al. (2018) HIV-1 Subtype C-Infected Children with Exceptional Neutralization Breadth Exhibit Polyclonal Responses Targeting Known Epitopes. J Virol 92 (17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Falkowska E et al. (2014) Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40 (5), 657–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mouquet H et al. (2011) Memory B Cell Antibodies to HIV-1 gp140 Cloned from Individuals Infected with Clade A and B Viruses. PLoS One 6 (9), e24078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhou T et al. (2013) Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39 (2), 245–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haynes BF et al. (2005) Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308 (5730), 1906–8. [DOI] [PubMed] [Google Scholar]
- 118.Haynes BF et al. (2005) Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Hum Antibodies 14 (3–4), 59–67. [PMC free article] [PubMed] [Google Scholar]
- 119.Scherer EM et al. (2007) Difficulties in eliciting broadly neutralizing anti-HIV antibodies are not explained by cardiolipin autoreactivity. AIDS 21 (16), 2131–9. [DOI] [PubMed] [Google Scholar]
- 120.Verkoczy L et al. (2010) Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci U S A 107 (1), 181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Irimia A et al. (2016) Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10. Immunity 44 (1), 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen J et al. (2014) Mechanism of HIV-1 neutralization by antibodies targeting a membrane-proximal region of gp41. J Virol 88 (2), 1249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bonsignori M et al. (2016) Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody. Cell 165 (2), 449–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Caskey M et al. (2015) Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522 (7557), 487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Padte NN et al. (2018) Engineering multi-specific antibodies against HIV-1. Retrovirology 15 (1), 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mouquet H and Nussenzweig MC (2012) Polyreactive antibodies in adaptive immune responses to viruses. Cell Mol Life Sci 69 (9), 1435–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu M et al. (2015) Polyreactivity and autoreactivity among HIV-1 antibodies. J Virol 89 (1), 784–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chuang GY et al. (2019) Structural Survey of Broadly Neutralizing Antibodies Targeting the HIV-1 Env Trimer Delineates Epitope Categories and Characteristics of Recognition. Structure 27 (1), 196–206 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]