Abstract
Bats host diverse viruses due to their unique ecology, behavior, and immunology. However, the role of other organisms with which bats interact in nature is understudied as a contributor to bat viral diversity. We discovered five viruses in the blood of fruit bats (Hypsignathus monstrosus) from the Republic of Congo. Of these five viruses, four have phylogenetic and genomic features suggesting an arthropod origin (a dicistrovirus, a nodavirus, and two tombus-like viruses), while the fifth (a hepadnavirus) is clearly of mammalian origin. We also report the parallel discovery of related tombus-like viruses in fig wasps and primitive crane flies from bat habitats, as well as high infection rates of bats with haemosporidian parasites (Hepatocystis sp.). These findings suggest transmission between arthropods and bats, perhaps through ingestion or hyperparasitism (viral infection of bat parasites). Some “bat-associated” viruses may be epidemiologically linked to bats through their ecological associations with invertebrates.
Keywords: bats, arthropods, virus, ecology, evolution, dicistrovirus, nodavirus, tombusvirus, hepadnavirus, next-generation sequencing
1. Introduction
Many emerging viral pathogens have their origins in the fruit bats of the Old World (family Pteropidae). Pteropid bats are natural reservoir hosts of highly virulent pathogens such as Marburg virus, Nipah virus, and Hendra virus, and they are often presumed to host the ebolaviruses. Despite extensive studies of bats and their zoonotic or potentially zoonotic infections, little is knows about the broader ecology of bat viruses. To date, much emphasis has been placed on understanding direct spillover from bats to humans (Brierley et al., 2016; Marí Saéz et al., 2015; Plowright et al., 2015). Less effort has been placed on understanding the origins of viral diversity in bats or the maintenance of bat-associated viruses in nature. In particular, few studies have considered bat viruses in an ecological context that includes not only bats but also the diverse other taxa with which bats interact (Kuno and Chang, 2005; Malmlov et al., 2017).
Bats (order Chiroptera) are the second largest order of mammals after rodents (order Rodentia). Overall, bats host more emerging viruses per species than any other mammalian clade (Luis et al., 2013). The underlying ecological and phylogenetic diversity of bats contributes to the diversity and richness of viruses that they host (Luis et al., 2013; Webber et al., 2017; Willoughby et al., 2017). Other drivers of this viral diversity are varied (Luis et al., 2013), including factors related to viral exposure and persistence, such as long lifespans relative to body size (Munshi-South and Wilkinson, 2010), population connectivity (Plowright et al., 2011), the impacts of prolonged torpor (in some species) on immune function (Dempster et al., 1966), and diverse, densely aggregated multispecies roosting that can sustain chains of viral transmission (Willoughby et al., 2017).
Bats have close ecological relationships with arthropods (Allen et al., 1956; Goldberg et al., 2017; Kalka et al., 2008; Kuno and Chang, 2005; Palmer and Gunier, 1975; Sulkin et al., 1965). Most widely appreciated among these relationships is predation by insectivorous bats. Insectivorous bats consume insect biomass from 25% to greater than 100% of their body weight each night (Coutts et al., 1973; Kunz et al., 2011, 1995). Insectivorous bats are thereby likely exposed to a diversity of arthropod viruses through ingestion (Kuno and Chang, 2005), and a broad range of insect-origin viruses have been discovered in the guano of insectivorous bats (Kuno and Chang, 2005; Li et al., 2010; Reuter et al., 2014). These “dietary viruses” (Li et al., 2010; Reuter et al., 2014; Sulkin et al., 1965) result from passive transport through the gastrointestinal tract; however it is unknown whether viral nucleic acid or viral particles can cross the gut wall or infect bats in some cases.
Fruit bats, as their common name suggests, are dietary specialists on fruits. Fruit bats are among the most important seed dispersers in tropical forests, playing a critical role in forest regeneration and other ecosystem services (Kunz et al., 2011; Oleksy et al., 2017, 2015). Fruits, however, are themselves complex “ecosystems,” with a diversity of closely associated and often co-evolved arthropods in and on them. For example, one of the most specific fruit-arthropod associations is the obligate relationship between figs (family Moraceae, genus Ficus) and their highly specialized pollinating wasps (families Agaonidae, Chalcidoidea)(Yang et al., 2015). The “fruit” of the fig tree (syconium) is in fact the flower of the plant. The female wasp lays eggs in the syconium and dies. Her hatching male offspring tunnel through the “fruit” and die without exiting, while the female offspring exit through these tunnels, carrying pollen to another tree (Cook and West, 2005). Hammer-headed fruit bats (Hypsignathus monstrosus), the focus of our study, consume figs as a primary food source, although their diet also includes the juice and pulp of mangoes, bananas, guavas, and soursops (Bradbury, 1977; Langevin and Barclay, 1990). Fruit bats therefore consume insects not merely incidentally, but rather as a significant component of their nutritional intake (Barclay et al., 2006; Clulow and Blundell, 2011; Herrera et al., 2002). Although opportunities for oral exposure of fruit bats to fruit-associated arthropod viruses are plentiful, this type of exposure is generally not considered in studies of viral ecology and evolution (Plowright et al., 2015).
Oral exposure of fruit bats to fruit-associated arthropod viruses is notably distinct from the transmission of vector-borne viruses by hematophagous arthropods. Vector-borne viruses transmitted by mosquitoes, including Chikungunya virus, West Nile virus, and Rift Valley fever virus have been detected in Pteropid bats (Boiro et al., 1987; Bunde et al., 2006; Diallo et al., 1999). Bat specialist ectoparasites such as bat flies (families Nycteribiidae, Streblidae) may also play a role as vectors (Dick and Dittmar, 2014). We recently described a novel rhabdovirus (family Rhabdovirus, genus Ledantevirus) of nycteribiid bat flies of fruit bats in Uganda with phylogenetic and genomic features that ally it with both zoonotic rhabdoviruses and insect-adapted viruses (Goldberg et al., 2017).
Here we describe a study of hammer-headed fruit bats in Republic of Congo in which we tested blood for viruses by next-generation sequencing (NGS). Our results reveal diverse viruses circulating in the blood of these fruit bats, four out of five of which are phylogenetically and genomically allied with viruses of arthropods. The new arthropod-origin viruses include a nodavirus (family Nodaviridae), a dicistrovirus (family Dicistroviridae), and two distinct variants of the recently recognized tombus-like viruses (family Tombusviridae). Furthermore, we surveyed the virome of Ceratosolen sp. fig wasps and primitive crane flies from bat habitats in Uganda and report novel tumbus-like viruses in these insects that are close relatives of the tombus-like virus from the blood of fruit bats, offering parallel evidence for the presence of such viruses in bat-associated arthropods. We also report high prevalence of a novel hepatitis B-like virus (Family: hepadnaviridae), a clearly mammalian virus, circulating in the same population of bats, as well as a high prevalence of infection with haemosporidian parasites of the genus Hepatocystis.
2. Results:
From 23 February 2015 to 1 March 2015, we captured 44 hammerheaded fruit bats (10 adult females, 29 adult males, 5 juvenile males) from a lekking site (described below). Analysis of NGS data from the serum of these bats revealed nucleic acid sequences from five previously unknown viruses, with near full genome coverage at sequencing depths of 531 reads per base on average. Of the 44 bats sequenced, 1 (2.3%) contained sequences of a novel dicistrovirus (family Dicistroviridae), 1 (2.3%) contained sequences of and a novel nodavirus (family Nodaviridae), 3 (6.8%) contained sequences of one of two novel tombus-like viruses (family Tombusviridae), and 10 (22.7%) contained sequences of a hepatitis B-like virus (family Hepadnaviridae). Forty-two bats (95.5%) contained sequences of only one of these viruses, but 2 coinfected bats (4.5%) contained sequences from two of these viruses. Frequencies of infection with each virus are shown in Table 1.
Table 1:
Novel viruses in Hammerhead fruit bats
| Arthropod Origin | Vertebrate Origin | ||||
|---|---|---|---|---|---|
| Dicistrovirus | Nodavirus | Tombus-like virus | Hepadnavirus | ||
| closest relative: | Drosophila C Virus |
Nodamura Virus RNA 1 |
Wuhan Insect Virus 35 |
Beihai tombus-like virus 7 |
Woodchuck Hepatitis Virus |
| Accession | MH310078 | MH324435 | MH324433 | MH324432 | MH324435 |
| Percent identity | 73% | 54% | 69% | 39% | 72% |
| Genome Coverage | 80% | 74% | 100% | 90% | 20% |
| Prevalence (n=44) | 2.3% | 2.3% | 2.3% | 4.5% | 22.7% |
| CpG Frequency | 0.7 | 1.0 | 0.9 | 0.8 | 0.7 |
2.1. Dicistrovirus
Dicistroviruses are dicistronic, picorna-like viruses, with two open reading frames (ORF) each accessed by independent internal ribosome entry sites (IRES). ORF1 encodes non-structural elements including a helicase, protease, and polymerase. ORF2 translation is mediated through the intergenic region (IGR) IRES and encodes the 4 capsid proteins (Nakashima and Uchiumi, 2009). The dicistrovirus we identified was of the genus Cripavirus. Cripaviruses are typically pathogenic in invertebrates, with the type strain, cricket paralysis virus (CrPV) causing paralysis and death in its natural host (Valles et al., 2017). Previously, a dicistrovirus was detected in insectivorous bat guano (Li et al., 2010; Reuter et al., 2014), presumably having passed through the gut.
1.9% of 728,482 trimmed reads from bat RML-1502254 mapped to what we provisionally named hypsignathus dicistrovirus, with deep coverage of both non-overlapping open reading frames (ORF1 and ORF2). Contiguous sequences (contigs) were assembled and trimmed reads were remapped iteratively to fill gaps and produce a viral consensus sequence of ~9300 nucleotides in length. The nucleotide identity of the capsid gene of the new virus to its closest known relative (drosophila C virus) is 73%. The species demarcation for dicistroviruses is accepted to be 90% identity in the capsid protein genes (Valles et al., 2017), making hypsignathus dicistrovirus a putative new species within the Cripavirus genus (Fig. 1A).
Figure 1.
Maximum likelihood phylogenies (1,000 bootstrap replicates) of polymerase genes sequences of A. dicistroviruses (~2700 bp), B. nodaviruses (~2000 bp), C. tombus-like viruses (~1200 bp), and D. hepadnaviruses (~570 bp). Maximum likelihood phylogenetic analyses were performed with codon-based alignments of polymerase genes, with poorly aligned regions removed (see text for full description).
RT-PCR using hypsignathus dicistrovirus specific primers (ORF1) confirmed the presence of hypsignathus dicistrovirus in the sample identified as positive by NGS. RNA extract from water blanks were negative, confirming that the virus was not present as a contaminant of the silica columns. Retrospective analysis of samples and blanks run in our lab in the past showed no evidence of these viruses, further confirming that they did not result from contamination or cross-contamination.
2.2. Nodavirus
Nodavirus genomes are bipartite, with one segment coding for RNA replicase (RNA1: ~3.2 kb) and one segment (RNA2: ~1.2 kb) coding for the viral capsid precursor protein. Nodaviruses are separated into two genera, Alphanodavirus (arthropod infecting) and Betanodavirus (fish infecting). The alphanodaviruses, including the type strain Nodamura virus, have been detected in a range of insects including Drosophila fruit flies (Aguiar et al., 2015), hematophageous insects such as mosquitoes (Schuster et al., 2014; Tesh, 1980), and phlebotomine sandflies (Aguiar et al., 2015).
We obtained ~2kb of the RNA replicase of segment RNA1 of a novel alphanodavirus. The sequence was most closely related to the type strain, Nodamura virus (Johnson et al., 2004)., and clusters with Nodamura virus phylogenetically (Fig. 1B), but with a percent amino acid identity of only 54%. For convenience, we refer to this virus as hypnovirus (“hypsignathus nodavirus”), indicating its discovery in H. monstrosus and its being a putative new genus (Hypnovirus, provisionally), following naming conventions for nodaviruses discovered in the feces of canines (Caninovirus) (Conceição-Neto et al., 2017) and in mosquitos (Mosinovirus) (Schuster et al., 2014). The next most closely related nodavirus, lutzomyia nodavirus, infects phlebotamine sandflies and is considered potentially vector-borne (Aguiar et al., 2015).
2.3. Tombus-like viruses
Tombus viruses, named for the type strain tomato bushy stunt virus, are positive sense, single stranded RNA viruses that typically infect plants (Stuart et al., 2004). However, recently discovered tombus-like viruses have diverse hosts, including marine invertebrates, terrestrial arthropods, and potentially free-living or parasitic protists (Dolja and Koonin, 2018; Shi et al., 2016). Tombus-like viruses have diverse genome architectures, utilizing both segmented and non-segmented strategies (Shi et al., 2016).
We detected RNA-dependent RNA polymerase (RDRP) sequences of two distinct clades of tombus-like viruses in H. monstrosus (Fig. 1C). The first, tentatively named hypsignathus monstrosus tombus-like virus 1, most closely resembled Wuhan insect virus 35 (2221 bp genome with two overlapping ORFs), a tombus-like virus discovered in an insect pool from Wuhan, China (Shi et al., 2016), but was highly divergent (54% amino acid identity). In two other H. monstrosus, we detected similar sequences (96% nucleotide identity) representing a second tombus-like virus, tentatively named hypsignathus monstrosus tombus-like virus 2.Hypsignathus monstrosus tombus-like virus 2 was a ~90% complete genome (3600 bp), and by BlastX (Altschul et al., 1990) was most closely related by to Beihai tombus-like virus 7 (3939 bp genome with 3 ORFS, 2 overlapping), originally detected in panaeid shrimp from Beihai, China. However amino acid percent identity was only 39% between hypsignathus monstrosus tombuslike virus 2 and Beihai tombus-like virus 7.
In 1 pool of 20 Ceratosolen sp. fig wasps (order Hymenoptera, family Agaonidae) collected from a Ficus brachylepis syconium and in 3 pools of two primitive crane flies each (order Diptera, family Tanyderidae) from fruit bat habitats in Uganda (hollow roosting trees of the genera and species Pterygota mildbraedii, Olea witchii, and Parinari excelsa), we also detected tombus-like virus sequences representing partial RDRP genes. These viruses cluster with Wuhan insect virus 35 and hypsignathus monstrosus tombus-like virus 1. In particular, fig wasp tombus-like virus shared 52.5% identity with hypsignathus monstrosus tombus-like virus 1, and the primitive crane fly tombus-like virus shared 60% identity with hypsignathus monstrosus tombus-like virus 1, making crane fly tombus-like virus the closest known relative of hypsignathus monstrosus tombus-like virus 1.
2.4. Hepadnaviruses
22.7% of H. monstrosus sampled (10/44) contained a novel hepatitis B-like virus provisionally named hypsignathus monstrosus hepatitis B virus (HMHBV). Phylogenetic analysis based on ~1000 nt of the hepatitis B polymerase gene (pol) indicates that HMHBV clusters with rodent orthohepadnaviruses, sharing 72% nucleotide identity with Woodchuck Hepatitis virus (Fig. 1D). In contrast, HMHBV pol shares only 50% nucleotide identity with the nearest bat hepadnavirus, tent-making bat hepatitis virus (Drexler et al., 2013).
2.5. Haemosporidian parasites
In addition to the viruses described above, we detected small subunit ribosomal RNA gene sequences of the haemosporidian parasite genus Hepatocystis in 43/44 (97.7%) bats (GenBank accession numbers MK234751-MK234793). These sequences were very similar to each other on average (98.5% ± 0.3% SE nucleotide identity) and were most closely related (mean 92.3% ± 1.2% SE nucleotide identity) to Hepatocystis sp. isolate CRNP11 (GenBank accession number DQ396536) detected in a Horsfield’s fruit bat (Cynopterus horsfieldii) in Malaysia. Of the 43 Hepatocystis-infected bats, 3 were also infected with hypsignathus monstrosus tombus-like viruses (1 and 2), 1 with hypnovirus and 1 with hypsignathus dicistrovirus.
2.6. Analysis of dinucleotide frequencies
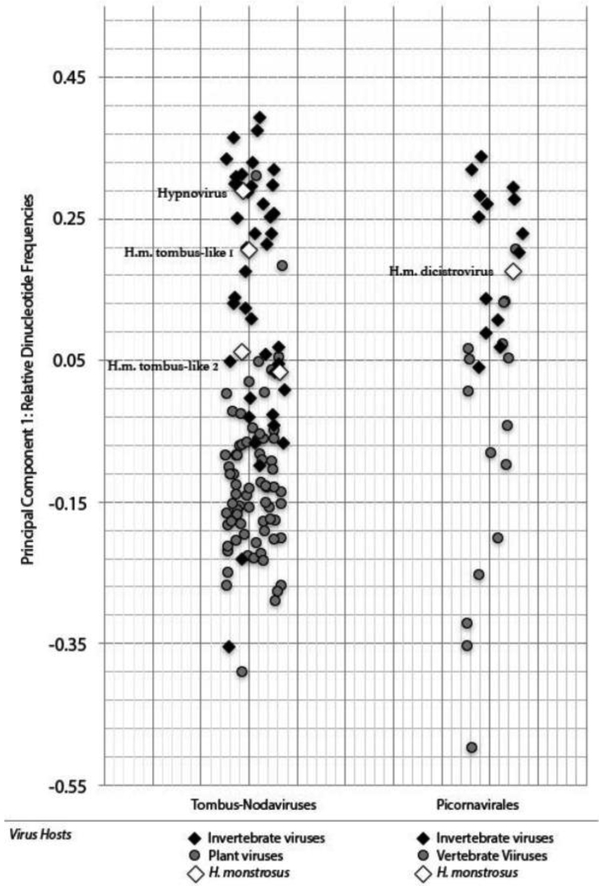
In order to explore virus-host associations, we calculated dinucleotide frequencies of the novel viral sequences we discovered. The new tombus-noda viruses detected in H. monstrosus do not exhibit a depletion of CpG dinucleotides nor an excess of dinucleotide TpA, as is typical of mammalian and plant RNA viruses (CpG: Avg 0.94 StDev: 0.09; TpA: Avg 0.82, StDev 0.23) (Glass et al., 2007; Upadhyay et al., 2013). Rather, values for both CpG and TpA of arthropod tombus-like viruses were statistically indistinguishable from Tombus-Noda viruses identified in bats and other mammals (CpG: Two-Tailed P-Value 0.86; TpA: 0.5752). Hypsignathus dicistrovirus showed limited CpG depletion and no excess of TpA (CpG: 0.67, TpA: 0.77), with both values falling within have a standard deviation of average for dicistroviruses (Average CpG: 0.75, StDev: 0.15; Average TpA: 0.78, StDev: 0.08). To compare these values directly, we conducted two principal components analyses of dinucleotide frequencies of the Picornavirales (the picornaviruses and dicistroviruses) and the Tombus-Noda clade of viruses (Fig. 2). This analysis shows that variations in dinucleotide frequencies can discriminate between related viruses of vertebrates and invertebrates, as well those of invertebrates and plants. In both analyses, the viruses we detected in H. monstrosus cluster with viruses of arthropods.
Figure 2.
Principal components analysis of relative dinucleotide frequencies of Tombus-Noda viruses and Picornavirus-dicistroviruses, from a diversity of hosts. The plot shows separation of groups by the first principal component, which can be interpreted as the direction, uncorrelated to other components, which maximizes the variance of the samples when projected onto the component. Points represent individual sequences.
3. Discussion
Many bats are “keystone” species, in that their absence would have cascading effects across ecosystems (Kunz et al., 2011). This high ecological connectivity may predispose bats to exposure to viruses of the species with which they interact in nature, including a diversity of arthropod viruses. Our results suggest that, in at least some cases, arthropod viruses can be found in bat blood. Although bites by vectors are a well-established route by which arthropods transmit viruses to bats (Klimpel and Mehlhom, 2013), our results suggest that other mechanisms may also be important. Oral exposure may be one such mechanism. “Fruit” bats (despite their common name) consume a great biomass and diversity of arthropods on fruits, not simply incidentally but as an important nutritional component of their diets (Barclay et al., 2006; Clulow and Blundell, 2011; Herrera et al., 2002). Our discoveries of diverse, novel, arthropod origin viruses in the blood of frugivorous bats suggests that such interactions might create an “ecological link” between arthropod and bat viromes.
Dicistroviruses have been identified in the feces of mammals, including insectivorous bats and even humans, but the fecal virome includes food-associated viruses that may simply have passed through the gut (Janowski et al., 2017; Kapoor et al., 2010; Reuter et al., 2014). Viruses of plants or insects detected in feces are frequently present due to dietary exposures, but may not indicate replicative infection (Balique et al., 2015; Zhang et al., 2006). It remains unclear whether arthropod dicistroviruses can replicate in mammalian cells. Conflicting reports have been published regarding the culture of taura syndrome virus in primate cell lines (Audelo-del-Valle et al., 2003; Pantoja et al., 2004). Our finding of a dicistrovirus circulating in the blood of a fruit bat could indicate passage of viral material from the gastrointestinal to the circulatory system. We speculate that such a process might occur through micro-abrasions during mastication and swallowing of rough food matter (Soave, 1966). Given its high level of similarity to drosophila C virus, hypsignathus dicistrovirus may have been acquired through ingestion of a dipteran insect associated with fruit.
The diversity of tombusviruses, nodaviruses, and the intermediary “tombus-like” viruses such as those in the blood of hammer-headed fruit bats has only recently become elucidated (Shi et al., 2016). Tombus-noda viruses are a super-family of single stranded positive sense RNA viruses with diverse hosts (Longdon et al., 2014). Tombus-noda virus clades contain phylogenetic admixtures of viruses of nematodes, marine invertebrates, terrestrial invertebrates, plants and mammals. Host-switches have been common over evolutionary time, although the current cross-species transmission potential of tombus-noda viruses requires further study (Ball et al., 1992; Johnson et al., 2004; Tesh, 1980).
Our parallel discoveries of tombus-like viruses in fig wasps and primitive crane flies associated with bat habitats strengthens the argument for a link between bat and arthropod viromes. Hypsignathus monstrosus tombus-like virus 1 is most closely related to the new tombus-like virus from primitive crane flies, which are commonly associated with habitats frequented by forest roosting bats. Also closely related to the tombus-like viruses of hammer-headed fruit bats are the new viruses from fig wasps of F. brachylepis figs. These wasps have evolved mutualistic relationships with fig trees, laying eggs inside figs and serving as their pollinators (Wang et al., 2010). Figs are important foods for many frugivorous animals across the tropics, including bats (Shanahan et al., 2001). Hypsignathus monstrosus tombus-like virus 2 falls within a clade with Beihai tombus-like virus 8 and other marine invertebrate-hosted tombus-like viruses, suggesting the existence of unknown tombus-like viruses in invertebrates of terrestrial origin with which fruit bats interact.
Alternatively, such viruses may be hosted by bat ecto- or endoparasites and thus be “hyperparasites” (Dolja and Koonin, 2018; Goldberg et al., 2017; Grybchuk et al., 2018; Jansen van Vuren et al., 2017, 2016). For example, a recent study detected fungus-associated partitiviruses in human sera and on that basis inferred viral hyperparasitism of a fungal pathogen (Phan et al., 2018). In this light, we note the high prevalence of infection of the sampled bats with haemosporidian parasites of the genus Hepatocystis, which are common in African fruit bats (Perkins and Schaer, 2016; Schaer et al., 2017) The tombus-like viruses we detected could be viruses of this or another parasite. Such hierarchical ecological associations complicate traditional definitiions of “host” but recall the notion of the “holobiont” virome, which considers the assemblage of a host and its parasites as an ecological unit (Richardson 2017).
By contrast, hypnovirus, the novel nodavirus we detected in hammer-headed fruit bat sera, is likely vector-borne. The type strain, Nodamura virus, was first detected in a mosquito, but nodaviruses have been shown to cause lethal infections in insects, mammals, and fish (Bailey and Scott, 1973; Shetty et al., 2012; Tesh, 1980). Transfection of Nodamura virus genomic RNAs has resulted in replication in a wide variety of cultured cells, showing intracellular competence for cross species transmission (Ball et al., 1992). The close evolutionary relationships of hypnovirus to the vector-borne alphanodavirus Nodamura virus and the more recently discovered phlebotomine sandfly lutzomyia nodavirus (Aguiar et al., 2015) suggests that this virus was detected at a time of transient viremia following transmission by a hematophagous arthropod. Sampling of potential vectors at the lekking site would be necessary to identify the vector.
Our detection of a novel hepatitis B virus, hypsignathus monstrosus hepatitis B virus (HMHBV), re-enforces the notion that our data accurately reflect the true blood virome of this bat population. Hepadnaviruses are small, partially double-stranded DNA viruses with circular genomes that infect a diversity of mammals, and the closest relatives of HMHBV are hosted by ground squirrels (Testut et al., 1996) and woodchucks (Tyler et al., 1981). Rodent hepadnaviruses have been discovered from the Arctic to the temperate regions, and bat hepadnaviruses have been detected in the Americas, Asia, and West African insectivorous bats (Drexler et al., 2013; Rasche et al., 2016; Wang et al., 2017), but not previously, to our knowledge, in African pteropids. HMHBV shared the greatest nucleotide identity (72%) in the polymerase gene with woodchuck hepatitis virus, and was only distantly related to the most similar known bat hepatitis B virus (Drexler et al., 2013). This places HMHBV in what was previously considered the rodent clade of hepatitis B viruses (Testut et al., 1996).
Dinucleotide frequency analysis offers further insights into the patterns observed above. Dinucleotide frequency can offer insight into viral host adaptation, at least across widely divergent host biologies (e.g. vertebrates versus invertebrates, and plants versus invertebrates; Babayan et al., 2018; Kapoor et al., 2010; Upadhyay et al., 2014). RNA viruses of vertebrates and plants typically show a depletion of CpG dinucleotide, and an excess of TpA dinucleotide. By contrast, RNA viruses of invertebrates do not exhibit extreme CpG and TpA frequencies (Kapoor et al., 2010; Karlin et al., 1994; Rima and McFerran, 1997). The CpG and TpA frequencies of the Tombus-Noda viruses and dicistroviruses that we detected in hammer-headed fruit bats were statistically indistinguishable from frequencies in related viruses adapted to the cellular replication machinery of arthropods.
Our results bear on a long-standing problem in infectious disease ecology. Some viruses that are considered “bat-associated” have not yet been isolated from bats. Most famous among these are the ebolaviruses which have yet to be isolated from bats (Leendertz et al., 2016) but have been detected in bats by PCR (Goldstein et al., 2018; Leroy et al., 2005) and antibody tests (Hayman et al., 2012, 2010; Ogawa et al., 2015; Pourrut et al., 2009, 2007). Over evolutionary time, many medically important viral taxa appear to have originated in invertebrates, with viruses of mammals in general, and bats in particular, interspersed among those of arthropod hosts (Shi et al., 2016). Our results provide a mechanism by which such a pattern might occur – namely frequent transmission of invertebrate viruses to bats via non-vector-borne modes of transmission such as ingestion or hyperparasitism.
For example, we recently documented an unusual rhabdovirus in nycteribiid bat flies (obligate hematophagous ectoparasites of bats) on pteropids in Uganda (Goldberg et al., 2017). This virus is a member of the genus Ledantevirus, which contains the zoonotic Le Dantec virus (Woodruff et al., 1977). Members of this viral genus are considered “bat associated” despite some never having been found in bats themselves (Blasdell et al., 2015; Goldberg et al., 2017). Bats may be associated ecologically and evolutionarily with these “bat associated” viruses not as reservoirs, but rather as intermediary hosts of arthropod viruses. The discovery of Bombali ebolavirus in Sierra Leone validates the consensus view that bats play a role in the ecology of ebolaviruses, but because it was found in two sympatric insectivorous bat species the exact nature of its association with bats remains unclear (Goldstein et al., 2018). Since the initial discovery of Sudan ebolavirus, it has been speculated based on circumstantial evidence that the reservoirs of certain ebolaviruses might non-hematophagous arthropods (Leendertz, 2016; Monath, 1999; Preston, 2012). Our results support the plausibility of this notion and suggest that occasional arthropod-bat transmission under ecologically favorable conditions might account for the sporadic appearance of such “bat associated” viruses.
Virus discoveries in apparently discordant hosts and sample types must be carefully scrutinized. In rare cases such discoveries have been linked to contaminated laboratory reagents (Naccache et al., 2013; Simmons et al., 2011). For example, marine-sourced silica in nucleic acid extraction columns has previously been linked to the unlikely presence of a marine hybrid parvovirus-like virus in samples from human patients (Naccache et al., 2013). Although marine dicistroviruses exist (Bonning and Miller, 2010), hypsignathus dicistrovirus is phylogenetically allied with terrestrial dicistroviruses (and absent from other samples analyzed previously, concurrently, and subsequently in our lab). Furthermore we were able to detect hysignathus dicistrovirus by NGS and RT-PCR amplification only in some bat serum samples and not in other bat serum samples or in extraction blanks. Hypsignathus dicistrovirus is not therefore a misassigned marine dicistrovirus contaminant of the silica columns, but rather a virus in the blood of the bats.
We also caution that our inferences are based on the detection of viral nucleic acid in the serum of bats, and not on detection of live viruses. Although complete genes of naked, dietary double-stranded DNA can pass from gut into the serum (Spisák et al., 2013), the half-life of unprotected viral RNA is significantly shorter (Chen et al., 2008; Dickson and Wilusz, 2011). Unfortunately, biosafety considerations required inactivation of all samples prior to processing. Nevertheless, we do not believe the viruses we detected are incidental. The coverage and depth of sequencing varied across the viruses discovered, ranging from 80-100% genome coverage, and up to 500x average coverage depth. Canonical ORFs did not contain premature stop codons. Bat and insect samples were sequenced on different days, and we have never before sequenced viruses similar to those reported herein.
Overall, our discovery of diverse arthropod origin viruses circulating in the blood of hammer-headed fruit bats of the Republic of Congo, as well as our discovery of a new mammalian virus related to rodent orthohepadnaviruses, are suggestive of diverse modes of viral transmission, all of which may all contribute to the observed diversity of viruses in bats. We suspect that cross-species transmission of viruses to bats and other mammals from invertebrates may occur with more regularity than has been appreciated, and that arthropods may host many “bat associated” viruses that have defied detection in bats themselves. Further analysis of these and other samples of arthropods from bat habitats is warranted. Future efforts to explore alternative reservoirs of “bat associated” viruses should include sampling of invertebrates that are ecologically associated with bats and that may host viruses capable of infecting bats.
4. Methods:
4.1. Collection of samples
The study site lies in the buffer corridor of Odzala-Kokoua National Park (OKNP), along National Route 2 roughly 100 km southwest of Ouesso in the Republic of the Congo (00° 54' 33" N and 15° 36' 01" E). Hammer-headed fruit bats (Hypsignathus monstrosus) were captured at night using canopy-level mist nets at a lekking site and transported to the sampling set-up in cloth bags. After sampling, the bats were released at the location of capture. Blood samples were obtained from the cephalic vein and centrifuged in MiniCollect Z serum tubes (Greiner Bio-One). Sera was collected and frozen in liquid nitrogen in the field and stored at −80°C in Brazzaville, Republic of the Congo, until shipment to the USA. All methods approved by Institutional Animal Care and Use Committee of the Rocky Mountain Laboratories (NIH ASP #2015-010).
In a related effort, in January 2016, 80 samples of fig wasps (4 pools) from 4 species of figs (F. brachylepis, F. spongii, F. mucuso, and F. capensis) were collected from locations in and near Kibale and Semliki National Parks, in western Uganda. Briefly, ripe, intact synconia were collected from the ground underneath the trees and were opened using sterile instruments. Adult fig wasps and larvae (encased in galls) were collected into sterile tubes containing DNA/RNA Shield buffer (Zymo Research Corporation, Irvine, CA, USA). Primitive crane flies were collected from 4 hollow tree bat roosts (Pterygota mildbraedii, Olea witchii, Parinari excelsa, Strombosia scheffleri) using sterile forceps and also placed into sterile tubes containing DNA/RNA Shield buffer. Insect samples were then processed using previously described methods (Goldberg et al., 2017).
4.2. Virus detection and characterization
RNA was extracted from samples (200-300 uL of bat serum or homogenized insects), using QIAamp MinElute Virus Spin kit (Qiagen Inc., Valencia, CA), without carrier RNA and stored at −80°C until shipment on dry ice to the University of Wisconsin-Madison for NGS. NGS was performed using methods previously described (Bennett et al., 2016; Toohey-Kurth et al., 2017). Briefly, RNA was DNAse treated by using the Turbo DNA-free Kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Extracted, DNAse treated, RNA was then converted to double-stranded cDNA (dscDNA) using the Superscript double-stranded cDNA Synthesis kit (Invitrogen, Carlsbad, CA, USA) with random hexamer priming of first-strand synthesis. Double stranded cDNA was purified using Agencourt Ampure XP beads (Beckman Coulter, Brea, CA, USA). DscDNA was prepared for paired-end NGS by Illumina MiSeq (MiSeq Reagent Kit v3, 300 cycle, Illumina, San Diego, CA, USA) using the Nextera XT DNA sample prep kit (Illumina, San Diego, CA, USA). NGS reads were analyzed as previously described (Goldberg et al., 2017).
4.3. Nucleotide composition analyses
The relative frequency of CpG dinucleotide pairs was calculated for CDS of each virus consensus sequence using the R Biostrings package (Pages et al., 2017). Relative dinucleotide frequency is the ratio of the observed to the expected frequency of a particular dinucleotide. A dinucleotide is considered underrepresented when the relative dinucleotide frequency is <0.78, and overrepresented when >1.22. Statistical analyses were performed in the computer package R (R Core Team, 2013). Principal components analysis of relative dinucleotide frequencies of all 16 dinucleotides was performed in R using the Stats package.
4.4. Phylogenetics
Maximum likelihood phylogenetic analyses were performed with codon-based alignments of polymerase genes. Alignments were created using the MAFFT algorithm (Katoh et al., 2002) in TranslatorX (Abascal et al., 2010), using Gblocks (Talavera et al., 2007) to exclude poorly aligned regions. Maximum likelihood phylogenies were constructed in PhyML (Guindon et al., 2010) and displayed utilizing FigTree (Rambaut, Andrew, 2016).
4.5. Quality control RT-PCR
RT-PCR primers were developed for hypsignathus dicistrovirus ORF1 (HDorf1F: 5’ – TTG CAG CAA AAC AGT TGA GG – 3’; HDorflR: 5’ - TGA GAC CAC AAA CCC AGA CA – 3’) to confirm NGS-based results and to test laboratory reagents as a potential source of contamination. RNA from NGS positive sera and water blank negative controls were extracted using both trizol and Qiagen column based methods (QIAamp MinElute Virus Spin kit; Qiagen Inc., Valencia, CA). RT-PCR was performed using New England Biolabs OneTaq One-Step RT-PCR kit (New England Biolabs, Ipswich, Mass.) under standard conditions, with the following thermocycling parameters: 48°C for 30 min; 94°C for 60 sec; 40 cycles at 94°C for 15 sec, 51°C for 30 sec, 68°C for 45 sec; 68°C for 5 min. PCR products were visualized under ultraviolet light on 1.5% agarose gels stained with ethidium bromide.
Supplementary Material
Highlights:
Fruit bats from Republic of Congo host diverse RNA viruses of arthropod origin.
Four arthropod viruses were found in the blood of bats at a forest breeding site.
Bats may acquire invertebrate viruses through their diets or from bat parasites.
“Bat associated” viruses may reside in invertebrates with ecological links to bats.
Acknowledgements:
We thank the Ministre de la Recherche Scientifique et de l’Innovation Technologique Republique du Congo and the Uganda Wildlife Authority for permission to conduct this research. This research was supported by the Intramural Research Programs of the National Institute of Allergy and Infectious Diseases (NIAID), the United States Fish and Wildlife Service (F13AC00558), The Jon and Wendy Neu Family Foundation, the University of Wisconsin-Madison’s Virology Training Grant (NIH T32 AI078985), and the University of Wisconsin-Madison’s John D. MacArthur Fellowship Program. The contents are the responsibility of the authors and do not necessarily reflect the views of the United States Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers of viruses:
Hypsignathus dicistrovirus (MH310078), Hypnovirus (MH324435), Hypsignathus monstrosus tombus-like virus 1 (MH324433), Hypsignathus monstrosus tombus-like virus 2 (MH324432), crane fly tombus-like virus 1-3 (MH324428-MH324430), fig wasp tombus-like virus (MH324431), Hypsignathus monstrosus hepatitis B virus (MH324435-MH324444).
Conflict of Interest:
The authors declare no conflict of interest.
References
- Abascal F, Zardoya R, Telford MJ, 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38, W7–W13. 10.1093/nar/gkq291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguiar ERGR, Olmo RP, Paro S, Ferreira FV, de Faria IJ da S, Todjro YMH, Lobo FP, Kroon EG, Meignin C, Gatherer D, Imler J-L, Marques JT, 2015. Sequence-independent characterization of viruses based on the pattern of viral small RNAs produced by the host. Nucleic Acids Res. 43, 6191–6206. 10.1093/nar/gkv587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R, Sulkin SE, Wallis C, 1956. Relationship of bat salivary gland virus to St. Louis encephalitis groups of viruses. Proc. Soc. Exp. Biol. Med. Soc. Exp. Biol. Med. N. Y. N 93, 79–81. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Audelo-del-Valle J, Clement-Mellado O, Magaña-Hernández A, Flisser A, Montiel-Aguirre F, Briseño-García B, 2003. Infection of Cultured Human and Monkey Cell Lines with Extract of Penaeid Shrimp Infected with Taura Syndrome Virus. Emerg. Infect. Dis. 9, 265–266. 10.3201/eid0902.020181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babayan SA, Orton RJ, Streicker DG, 2018. Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science 362, 577–580. 10.1126/science.aap9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey L, Scott HA, 1973. The Pathogenicity of Nodamura Virus for Insects. Nature 241, 545 10.1038/241545a0 [DOI] [PubMed] [Google Scholar]
- Balique F, Lecoq H, Raoult D, Colson P, 2015. Can Plant Viruses Cross the Kingdom Border and Be Pathogenic to Humans? Viruses 7, 2074–2098. 10.3390/v7042074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball LA, Amann JM, Garrett BK, 1992. Replication of nodamura virus after transfection of viral RNA into mammalian cells in culture. J. Virol. 66, 2326–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay RMR, Barclay LE, Jacobs DS, 2006. Deliberate insectivory by the fruit bat Rousettus aegyptiacus. Acta Chiropterologica 8, 549–553. 10.3161/1733-5329(2006)8[549:DIBTFB]2.0.CO;2 [DOI] [Google Scholar]
- Bennett AJ, Sibley SD, Lauck M, Weny G, Hyeroba D, Tumukunde A, Friedrich TC, O’Connor DH, Johnson CA, Rothman JM, Goldberg TL, 2016. Naturally Circulating Hepatitis A Virus in Olive Baboons, Uganda. Emerg. Infect. Dis. 22, 1308–1310. 10.3201/eid2207.151837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasdell KR, Guzman H, Widen SG, Firth C, Wood TG, Holmes EC, Tesh RB, Vasilakis N, Walker PJ, 2015. Ledantevirus: A Proposed New Genus in the Rhabdoviridae Has A Strong Ecological Association with Bats. Am. J. Trop. Med. Hyg. 92, 405–410. 10.4269/ajtmh.14-0606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiro I, Konstaninov OK, Numerov AD, 1987. [Isolation of Rift Valley fever virus from bats in the Republic of Guinea]. Bull. Soc. Pathol. Exot. Filiales 80, 62–67. [PubMed] [Google Scholar]
- Bonning BC, Miller WA, 2010. Dicistroviruses. Annu. Rev. Entomol. 55, 129–150. 10.1146/annurev-ento-112408-085457 [DOI] [PubMed] [Google Scholar]
- Bradbury JW, 1977. Lek Mating Behavior in the Hammer-headed Bat. Z. Für Tierpsychol. 45, 225–255. 10.1111/j.1439-0310.1977.tb02120.x [DOI] [Google Scholar]
- Brierley L, Vonhof MJ, Olival KJ, Daszak P, Jones KE, 2016. Quantifying Global Drivers of Zoonotic Bat Viruses: A Process-Based Perspective. Am. Nat. 187, E53–64. 10.1086/684391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunde JM, Heske EJ, Mateus-Pinilla NE, Hofmann JE, Novak RJ, 2006. A survey for West Nile virus in bats from Illinois. J. Wildl. Dis. 42, 455–458. 10.7589/0090-3558-42.2.455 [DOI] [PubMed] [Google Scholar]
- Chen C-YA, Ezzeddine N, Shyu A-B, 2008. Messenger RNA Half-Life Measurements in Mammalian Cells. Methods Enzymol. 448, 335–357. 10.1016/S0076-6879(08)02617-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clulow S, Blundell AT, 2011. Deliberate Insectivory by the Fruit Bat Pteropus poliocephalus by Aerial Hunting. Acta Chiropterologica 13, 201–205. 10.3161/150811011X578750 [DOI] [Google Scholar]
- Conceição-Neto N, Godinho R, Álvares F, Yinda CK, Deboutte W, Zeller M, Laenen L, Heylen E, Roque S, Petrucci-Fonseca F, Santos N, Van Ranst M, Mesquita JR, Matthijnssens J, 2017. Viral gut metagenomics of sympatric wild and domestic canids, and monitoring of viruses: Insights from an endangered wolf population. Ecol. Evol 7, 4135–4146. 10.1002/ece3.2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JM, West SA, 2005. Figs and fig wasps. Curr. Biol. 15, R978–R980. 10.1016/j.cub.2005.11.057 [DOI] [PubMed] [Google Scholar]
- Coutts RA, Fenton MB, Glen E, 1973. Food Intake by Captive Myotis lucifugus and Eptesicus fuscus (Chiroptera: Vespertilionidae). J. Mammal. 54, 985–990. 10.2307/1379098 [DOI] [Google Scholar]
- Dempster G, Grodums EI, Spencer WA, 1966. Experimental Coxsackie B-3 virus infection in Citellus lateralis. J. Cell. Physiol. 67, 443–453. 10.1002/jcp.1040670309 [DOI] [PubMed] [Google Scholar]
- Diallo M, Thonnon J, Traore-Lamizana M, Fontenille D, 1999. Vectors of Chikungunya virus in Senegal: current data and transmission cycles. Am. J. Trop. Med. Hyg. 60, 281–286. [DOI] [PubMed] [Google Scholar]
- Dick CW, Dittmar K, 2014. Parasitic Bat Flies (Diptera: Streblidae and Nycteribiidae): Host Specificity and Potential as Vectors, in: Bats (Chiroptera) as Vectors of Diseases and Parasites, Parasitology Research Monographs. Springer, Berlin, Heidelberg, pp. 131–155. 10.1007/978-3-642-39333-4_6 [DOI] [Google Scholar]
- Dickson AM, Wilusz J, 2011. Strategies for viral RNA stability: live long and prosper. Trends Genet. TIG 27, 286–293. 10.1016/j.tig.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja VV, Koonin EV, 2018. Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer. Virus Res. 244, 36–52. 10.1016/j.virusres.2017.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Geipel A, König A, Corman VM, van Riel D, Leijten LM, Bremer CM, Rasche A, Cottontail VM, Maganga GD, Schlegel M, Müller MA, Adam A, Klose SM, Carneiro AJB, Stöcker A, Franke CR, Gloza-Rausch F, Geyer J, Annan A, Adu-Sarkodie Y, Oppong S, Binger T, Vallo P, Tschapka M, Ulrich RG, Gerlich WH, Leroy E, Kuiken T, Glebe D, Drosten C, 2013. Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc. Natl. Acad. Sci. U. S. A. 110, 16151–16156. 10.1073/pnas.1308049110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JL, Thompson RF, Khulan B, Figueroa ME, Olivier EN, Oakley EJ, Van Zant G, Bouhassira EE, Melnick A, Golden A, Fazzari MJ, Greally JM, 2007. CG dinucleotide clustering is a species-specific property of the genome. Nucleic Acids Res. 35, 6798–6807. 10.1093/nar/gkm489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg TL, Bennett AJ, Kityo R, Kuhn JH, Chapman CA, 2017. Kanyawara Virus: A Novel Rhabdovirus Infecting Newly Discovered Nycteribiid Bat Flies Infesting Previously Unknown Pteropodid Bats in Uganda. Sci. Rep 7 10.1038/s41598-017-05236-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein T, Anthony SJ, Gbakima A, Bird BH, Bangura J, Tremeau-Bravard A, Belaganahalli MN, Wells HL, Dhanota JK, Liang E, Grodus M, Jangra RK, DeJesus VA, Lasso G, Smith BR, Jambai A, Kamara BO, Kamara S, Bangura W, Monagin C, Shapira S, Johnson CK, Saylors K, Rubin EM, Chandran K, Lipkin WI, Mazet JAK, 2018. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat. Microbiol 3, 1084 10.1038/s41564-018-0227-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grybchuk D, Akopyants NS, Kostygov AY, Konovalovas A, Lye L-F, Dobson DE, Zangger H, Fasel N, Butenko A, Frolov AO, Votýpka J, d’Avila-Levy CM, Kulich P, Moravcová J, Plevka P, Rogozin IB, Serva S, Lukeš J, Beverley SM, Yurchenko V, 2018. Viral discovery and diversity in trypanosomatid protozoa with a focus on relatives of the human parasite Leishmania. Proc. Natl. Acad. Sci. 115, E506–E515. 10.1073/pnas.1717806115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O, 2010. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 59, 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hayman DTS, Emmerich P, Yu M, Wang L-F, Suu-Ire R, Fooks AR, Cunningham AA, Wood JLN, 2010. Long-Term Survival of an Urban Fruit Bat Seropositive for Ebola and Lagos Bat Viruses. PLOS ONE 5, e11978 10.1371/journal.pone.0011978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman DTS, Yu M, Crameri G, Wang L-F, Suu-Ire R, Wood JLN, Cunningham AA, 2012. Ebola Virus Antibodies in Fruit Bats, Ghana, West Africa. Emerg. Infect. Dis. 18, 1207–1209. 10.3201/eid1807.111654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera GL, Gutierrez E, Hobson KA, Altube B, Díaz WG, Sánchez-Cordero V, 2002. Sources of assimilated protein in five species of New World frugivorous bats. Oecologia 133, 280–287. 10.1007/s00442-002-1036-z [DOI] [PubMed] [Google Scholar]
- Janowski AB, Krishnamurthy SR, Lim ES, Zhao G, Brenchley JM, Barouch DH, Thakwalakwa C, Manary MJ, Holtz LR, Wang D, 2017. Statoviruses, A novel taxon of RNA viruses present in the gastrointestinal tracts of diverse mammals. Virology 504, 36–44. 10.1016/j.virol.2017.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen van Vuren P, Wiley M, Palacios G, Storm N, McCulloch S, Markotter W, Birkhead M, Kemp A, Paweska JT, 2016. Isolation of a Novel Fusogenic Orthoreovirus from Eucampsipoda africana Bat Flies in South Africa. Viruses 8 10.3390/v8030065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen van Vuren P, Wiley MR, Palacios G, Storm N, Markotter W, Birkhead M, Kemp A, Paweska JT, 2017. Isolation of a novel orthobunyavirus from bat flies (Eucampsipoda africana). J. Gen. Virol. 98, 935–945. 10.1099/jgv.0.000753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL, Price BD, Eckerle LD, Ball LA, 2004. Nodamura Virus Nonstructural Protein B2 Can Enhance Viral RNA Accumulation in both Mammalian and Insect Cells. J. Virol. 78, 6698–6704. 10.1128/JVI.78.12.6698-6704.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalka MB, Smith AR, Kalko EKV, 2008. Bats Limit Arthropods and Herbivory in a Tropical Forest. Science 320, 71–71. 10.1126/science.1153352 [DOI] [PubMed] [Google Scholar]
- Kapoor A, Simmonds P, Lipkin WI, Zaidi S, Delwart E, 2010. Use of nucleotide composition analysis to infer hosts for three novel picorna-like viruses. J. Virol. 84, 10322–10328. 10.1128/JVI.00601-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Doerfler W, Cardon LR, 1994. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J. Virol. 68, 2889–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T, 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimpel S, Mehlhorn H, 2013. Bats (Chiroptera) as Vectors of Diseases and Parasites: Facts and Myths. Springer Science & Business Media. [Google Scholar]
- Kuno G, Chang G-JJ, 2005. Biological Transmission of Arboviruses: Reexamination of and New Insights into Components, Mechanisms, and Unique Traits as Well as Their Evolutionary Trends. Clin. Microbiol. Rev. 18, 608–637. 10.1128/CMR.18.4.608-637.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz TH, Braun de Torrez E, Bauer D, Lobova T, Fleming TH, 2011. Ecosystem services provided by bats. Ann. N. Y. Acad. Sci. 1223, 1–38. 10.1111/j.1749-6632.2011.06004.x [DOI] [PubMed] [Google Scholar]
- Kunz TH, Whitaker JO, Wadanoli MD, 1995. Dietary energetics of the insectivorous Mexican free-tailed bat Tadarida brasiliensis during pregnancy and lactation. Oecologia 101, 407–415. 10.1007/BF00329419 [DOI] [PubMed] [Google Scholar]
- Langevin P, Barclay RMR, 1990. Hypsignathus monstrosus. Mamm. Species 1–4. 10.2307/3504110 [DOI] [Google Scholar]
- Leendertz SAJ, 2016. Testing New Hypotheses Regarding Ebolavirus Reservoirs. Viruses 8 10.3390/v8020030 [DOI] [Google Scholar]
- Leendertz SAJ, Gogarten JF, Düx A, Calvignac-Spencer S, Leendertz FH, 2016. Assessing the Evidence Supporting Fruit Bats as the Primary Reservoirs for Ebola Viruses. EcoHealth 13, 18–25. 10.1007/s10393-015-1053-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez J-P, Swanepoel R, 2005. Fruit bats as reservoirs of Ebola virus. Nature 438, 575 10.1038/438575a [DOI] [PubMed] [Google Scholar]
- Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH, Delwart E, 2010. Bat Guano Virome: Predominance of Dietary Viruses from Insects and Plants plus Novel Mammalian Viruses. J. Virol. 84, 6955–6965. 10.1128/JVI.00501-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B, Brockhurst MA, Russell CA, Welch JJ, Jiggins FM, 2014. The Evolution and Genetics of Virus Host Shifts. PLoS Pathog. 10 10.1371/journal.ppat.1004395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis AD, Hayman DTS, O’Shea TJ, Cryan PM, Gilbert AT, Pulliam JRC, Mills JN, Timonin ME, Willis CKR, Cunningham AA, Fooks AR, Rupprecht CE, Wood JLN, Webb CT, 2013. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. R. Soc. Lond. B Biol. Sci 280, 20122753. 10.1098/rspb.2012.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmlov A, Seetahal J, Carrington C, Ramkisson V, Foster J, Miazgowicz KL, Quackenbush S, Rovnak J, Negrete O, Munster V, Schountz T, 2017. Serological evidence of arenavirus circulation among fruit bats in Trinidad. PLOS ONE 12, e0185308 10.1371/journal.pone.0185308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí Saéz A, Weiss S, Nowak K, Lapeyre V, Zimmermann F, Düx A, Kühl HS, Kaba M, Regnaut S, Merkel K, Sachse A, Thiesen U, Villányi L, Boesch C, Dabrowski PW, Radonić A, Nitsche A, Leendertz SAJ, Petterson S, Becker S, Krähling V, Couacy-Hymann E, Akoua-Koffi C, Weber N, Schaade L, Fahr J, Borchert M, Gogarten JF, Calvignac-Spencer S, Leendertz FH, 2015. Investigating the zoonotic origin of the West African Ebola epidemic. EMBO Mol. Med. 7, 17–23. 10.15252/emmm.201404792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monath TP, 1999. Ecology of Marburg and Ebola Viruses: Speculations and Directions for Future Research. J. Infect. Dis. 179, S127–S138. 10.1086/514281 [DOI] [PubMed] [Google Scholar]
- Munshi-South J, Wilkinson GS, 2010. Bats and birds: Exceptional longevity despite high metabolic rates. Ageing Res. Rev. 9, 12–19. 10.1016/j.arr.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Naccache SN, Greninger AL, Lee D, Coffey LL, Phan T, Rein-Weston A, Aronsohn A, Hackett J, Delwart EL, Chiu CY, 2013. The Perils of Pathogen Discovery: Origin of a Novel Parvovirus-Like Hybrid Genome Traced to Nucleic Acid Extraction Spin Columns. J. Virol. 87, 11966–11977. 10.1128/JVI.02323-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N, Uchiumi T, 2009. Functional analysis of structural motifs in dicistroviruses. Virus Res. 139, 137–147. 10.1016/j.virusres.2008.06.006 [DOI] [PubMed] [Google Scholar]
- Ogawa H, Miyamoto H, Nakayama E, Yoshida R, Nakamura I, Sawa H, Ishii A, Thomas Y, Nakagawa E, Matsuno K, Kajihara M, Maruyama J, Nao N, Muramatsu M, Kuroda M, Simulundu E, Changula K, Hang’ombe B, Namangala B, Nambota A, Katampi J, Igarashi M, Ito K, Feldmann H, Sugimoto C, Moonga L, Mweene A, Takada A, 2015. Seroepidemiological Prevalence of Multiple Species of Filoviruses in Fruit Bats (Eidolon helvum) Migrating in Africa. J. Infect. Dis. 212 Suppl 2, S101–108. 10.1093/infdis/jiv063 [DOI] [PubMed] [Google Scholar]
- Oleksy R, Giuggioli L, McKetterick TJ, Racey PA, Jones G, 2017. Flying foxes create extensive seed shadows and enhance germination success of pioneer plant species in deforested Madagascan landscapes. PLOS ONE 12, e0184023 10.1371/journal.pone.0184023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksy R, Racey PA, Jones G, 2015. High-resolution GPS tracking reveals habitat selection and the potential for long-distance seed dispersal by Madagascan flying foxes Pteropus rufus. Glob. Ecol. Conserv. 3, 678–692. 10.1016/j.gecco.2015.02.012 [DOI] [Google Scholar]
- Pages H, Aboyoun P, Gentleman R, DebRoy S, 2017. Biostrings: String objects representing biological sequences, and matching algorithms [WWW Document]. R Package. [Google Scholar]
- Palmer DB, Gunier WJ, 1975. A Preliminary Survey of Arthropods Associated with Bats and Bat Caves in Missouri and Two Counties of Oklahoma. J. Kans. Entomol. Soc. 48, 524–531. [Google Scholar]
- Pantoja CR, Navarro SA, Naranjo J, Lightner DV, Gerba CP, 2004. Nonsusceptibility of Primate Cells to Taura Syndrome Virus. Emerg. Infect. Dis. 10, 2106–2112. 10.3201/eid1012.040419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins SL, Schaer J, 2016. A Modern Menagerie of Mammalian Malaria. Trends Parasitol. 32, 772–782. 10.1016/j.pt.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Phan TG, del Valle Mendoza J, Sadeghi M, Altan E, Deng X, Delwart E, 2018. Sera of Peruvians with fever of unknown origins include viral nucleic acids from nonvertebrate hosts. Virus Genes 54, 33–40. 10.1007/s11262-017-1514-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, Eby P, Hudson PJ, Smith IL, Westcott D, Bryden WL, Middleton D, Reid PA, McFarlane RA, Martin G, Tabor GM, Skerratt LF, Anderson DL, Crameri G, Quammen D, Jordan D, Freeman P Wang L-F, Epstein JH, Marsh GA, Kung NY, McCallum H, 2015. Ecological dynamics of emerging bat virus spillover. Proc. R. Soc. B Biol. Sci. 282 10.1098/rspb.2014.2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, Foley P, Field HE, Dobson AP, Foley JE, Eby P, Daszak P, 2011. Urban habituation, ecological connectivity and epidemic dampening: the emergence of Hendra virus from flying foxes (Pteropus spp.). Proc. R. Soc. Lond. B Biol. Sci. 278, 3703–3712. 10.1098/rspb.2011.0522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourrut X, Délicat A, Rollin PE, Ksiazek TG, Gonzalez J-P, Leroy EM, 2007. Spatial and temporal patterns of Zaire ebolavirus antibody prevalence in the possible reservoir bat species. J. Infect. Dis. 196 Suppl 2, S176–183. 10.1086/520541 [DOI] [PubMed] [Google Scholar]
- Pourrut X, Souris M, Towner JS, Rollin PE, Nichol ST, Gonzalez J-P, Leroy E, 2009. Large serological survey showing cocirculation of Ebola and Marburg viruses in Gabonese bat populations, and a high seroprevalence of both viruses in Rousettus aegyptiacus. BMC Infect. Dis 9, 159 10.1186/1471-2334-9-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston R, 2012. The Hot Zone: The Terrifying True Story of the Origins of the Ebola Virus. Knopf Doubleday Publishing Group. [Google Scholar]
- R Core Team, 2013. R: A language environment for statistical computing. R Foundation for Statistical Computing. [Google Scholar]
- Rambaut Andrew, 2016. FigTree, v 1.4.3. [Google Scholar]
- Rasche A, Souza BF de CD, Drexler JF, 2016. Bat hepadnaviruses and the origins of primate hepatitis B viruses. Curr. Opin. Virol., Emerging viruses• Viral immunology 16, 86–94. 10.1016/j.coviro.2016.01.015 [DOI] [PubMed] [Google Scholar]
- Reuter G, Pankovics P, Gyöngyi Z, Delwart E, Boros Á, 2014. Novel dicistrovirus from bat guano. Arch. Virol. 159, 3453–3456. 10.1007/s00705-014-2212-2 [DOI] [PubMed] [Google Scholar]
- Rima BK, McFerran NV, 1997. Dinucleotide and stop codon frequencies in single-stranded RNA viruses. J. Gen. Virol. 78 (Pt 11), 2859–2870. 10.1099/0022-1317-78-11-2859 [DOI] [PubMed] [Google Scholar]
- Schaer J, Perkins SL, Ejotre I, Vodzak ME, Matuschewski K, Reeder DM, 2017. Epauletted fruit bats display exceptionally high infections with a Hepatocystis species complex in South Sudan. Sci. Rep. 7, 6928 10.1038/s41598-017-07093-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S, Zirkel F, Kurth A, van Cleef KWR, Drosten C, van Rij RP, Junglen S, 2014. A unique nodavirus with novel features: mosinovirus expresses two subgenomic RNAs, a capsid gene of unknown origin, and a suppressor of the antiviral RNA interference pathway. J. Virol. 88, 13447–13459. 10.1128/JVI.02144-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan M, So S, Compton SG, Corlett R, 2001. Fig-eating by vertebrate frugivores: a global review. Biol. Rev. Camb. Philos. Soc. 76, 529–572. [DOI] [PubMed] [Google Scholar]
- Shetty M, Maiti B, Shivakumar Santhosh K, Venugopal MN, Karunasagar I, 2012. Betanodavirus of Marine and Freshwater Fish: Distribution, Genomic Organization, Diagnosis and Control Measures. Indian J. Virol. Off. Organ Indian Virol. Soc. 23, 114–123. 10.1007/s13337-012-0088-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Lin X-D, Tian J-H, Chen L-J, Chen X, Li C-X, Qin X-C, Li J, Cao J-P, Eden J-S, Buchmann J, Wang W, Xu J, Holmes EC, Zhang Y-Z, 2016. Redefining the invertebrate RNA virosphere. Nature 540, 539–543. 10.1038/nature20167 [DOI] [PubMed] [Google Scholar]
- Simmons G, Glynn SA, Komaroff AL, Mikovits JA, Tobler LH, Hackett J, Tang N, Switzer WM, Heneine W, Hewlett IK, Zhao J, Lo S-C, Alter HJ, Linnen JM, Gao K, Coffin JM, Kearney MF, Ruscetti FW, Pfost MA, Bethel J, Kleinman S, Holmberg JA, Busch MP, Group (SRWG), for the B.X.S.R.W., 2011. Failure to Confirm XMRV/MLVs in the Blood of Patients with Chronic Fatigue Syndrome: A Multi-Laboratory Study. Science 334, 814–817. 10.1126/science.1213841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soave OA, 1966. Transmission of rabies to mice by ingestion of infected tissue. Am. J. Vet. Res. 27, 44–46. [PubMed] [Google Scholar]
- Spisák S, Solymosi N, Ittzés P, Bodor A, Kondor D, Vattay G, Barták BK, Sipos F, Galamb O, Tulassay Z, Szállási Z, Rasmussen S, Sicheritz-Ponten T, Brunak S, Molnár B, Csabai I, 2013. Complete Genes May Pass from Food to Human Blood. PLOS ONE 8, e69805 10.1371/journal.pone.0069805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart G, Moffett K, Bozarth RF, 2004. A whole genome perspective on the phylogeny of the plant virus family Tombusviridae. Arch. Virol. 149, 1595–1610. 10.1007/s00705-004-0298-7 [DOI] [PubMed] [Google Scholar]
- Sulkin SE, Allen R, Sims R, Taylor SK, 1965. Bats in Relation to Arthropod-Borne Viruses: An Experimental Approach with Speculations. Am. J. Public Health Nations Health 55, 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J, Kjer K, Page R, Sullivan J, 2007. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 56, 564–577. 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Tesh RB, 1980. Infectivity and pathogenicity of Nodamura virus for mosquitoes. J. Gen. Virol. 48, 177–182. [Google Scholar]
- Testut P, Renard CA, Terradillos O, Vitvitski-Trepo L, Tekaia F, Degott C, Blake J, Boyer B, Buendia MA, 1996. A new hepadnavirus endemic in arctic ground squirrels in Alaska. J. Virol. 70, 4210–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toohey-Kurth K, Sibley SD, Goldberg TL, 2017. Metagenomic assessment of adventitious viruses in commercial bovine sera. Biol. J. Int. Assoc. Biol. Stand. 47, 64–68. 10.1016/j.biologicals.2016.10.009 [DOI] [PubMed] [Google Scholar]
- Tyler GV, Summers JW, Snyder RL, 1981. Woodchuck hepatitis virus in natural woodchuck populations. J. Wildl. Dis. 17, 297–301. 10.7589/0090-3558-17.2.297 [DOI] [PubMed] [Google Scholar]
- Upadhyay M, Samal J, Kandpal M, Vasaikar S, Biswas B, Gomes J, Vivekanandan P, 2013. CpG dinucleotide frequencies reveal the role of host methylation capabilities in parvovirus evolution. J. Virol. 87, 13816–13824. 10.1128/JVI.02515-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay M, Sharma N, Vivekanandan P, 2014. Systematic CpT (ApG) Depletion and CpG Excess Are Unique Genomic Signatures of Large DNA Viruses Infecting Invertebrates. PLOS ONE 9, e111793. 10.1371/journal.pone.0111793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles SM, Chen Y, Firth AE, Guérin DMA, Hashimoto Y, Herrero S, de Miranda JR, Ryabov E, ICTV Report Consortium, 2017. ICTV Virus Taxonomy Profile: Dicistroviridae. J. Gen. Virol. 98, 355–356. 10.1099/jgv.0.000756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yang X-L, Li W, Zhu Y, Ge X-Y, Zhang L-B, Zhang Y-Z, Bock C-T, Shi Z-L, 2017. Detection and genome characterization of four novel bat hepadnaviruses and a hepevirus in China. Virol. J. 14, 40 10.1186/s12985-017-0706-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R-W, Sun B-F, Zheng Q, 2010. Diffusive coevolution and mutualism maintenance mechanisms in a fig-fig wasp system. Ecology 91, 1308–1316. [DOI] [PubMed] [Google Scholar]
- Webber QMR, Fletcher QE, Willis CKR, 2017. Viral Richness is Positively Related to Group Size, but Not Mating System, in Bats. EcoHealth 14, 652–661. 10.1007/s10393-017-1276-3 [DOI] [PubMed] [Google Scholar]
- Willoughby AR, Phelps KL, PREDICT Consortium, Olival KJ, 2017. A Comparative Analysis of Viral Richness and Viral Sharing in Cave-Roosting Bats. Diversity 9, 35 10.3390/d9030035 [DOI] [Google Scholar]
- Woodruff AW, Ansdell VE, Bowen ET, 1977. Le Dantec virus infection in a patient who had not been to West Africa. Br. Med. J. 2, 1632–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L-Y, Machado CA, Dang X-D, Peng Y-Q, Yang D-R, Zhang D-Y, Liao W-J, 2015. The incidence and pattern of copollinator diversification in dioecious and monoecious figs. Evolution 69, 294–304. 10.1111/evo.12584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Breitbart M, Lee WH, Run J-Q, Wei CL, Soh SWL, Hibberd ML, Liu ET, Rohwer F, Ruan Y, 2006. RNA Viral Community in Human Feces: Prevalence of Plant Pathogenic Viruses. PLoS Biol. 4 10.1371/journal.pbio.0040003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.