Abstract
The most frequent malignancy of the pancreas is the pancreatic ductal adenocarcinoma (PDAC). Despite many efforts PDAC has still a dismal prognosis. Biomarkers for early disease stage diagnosis as a prerequisite for a potentially curative treatment are still missing. Novel blood-based markers may help to overcome this limitation.
Methods: Prior to surgery plasma levels of thrombospondin-2 (THBS2), which was recently published as a novel biomarker, and CA19-9 from 52 patients with histologically proven PDAC were determined, circulating cell-free (cfDNA) was quantified. 15 patients with side-branch IPMNs without worrisome features and 32 patients with chronic pancreatitis served for comparison. Logit (logistic regression) models were used to test the performance of single biomarkers and biomarker combinations.
Results: CA19-9 and THBS2 alone showed comparable c-statistics of 0.80 and 0.73, respectively, improving to 0.87 when combining these two markers. The c-statistic was further increased to 0.94 when combining CA19-9 and THBS2 with cfDNA quantification. This marker combination performed best for all PDAC stages but also for PDACs grouped by stage. The greatest improvement over CA19-9 was seen in the group of stage I PDAC, from 0.69 to 0.90 for the three marker combination.
Conclusion:These data establish the combination of CA19-9, THBS2 and cfDNA as a composite liquid biomarker for non-invasive diagnosis of early-stage PDAC.
Keywords: liquid biopsy, pancreatic cancer, circulating tumor DNA, CA19-9, thrombospondin-2
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most frequent malignant tumor of the pancreas and is likely to become the second leading cause of cancer mortality in the near future due to a global increase in prevalence and still insufficient progress in therapy 1. Diagnosis of PDAC is usually made only at an advanced stage and prognosis is correspondingly poor with a 5-year overall survival rate of only 5 % 2. About 20 % of the patients diagnosed with PDAC are eligible for pancreatic surgery 3. Overall, even upon resection in curative intent and adjuvant chemotherapy the median disease-free survival is at best about 22 months 4. Prognosis is far better at early stages 5. Therefore, early diagnosis of PDAC has a decisive impact on patient survival 4. Neoadjuvant chemo- or radiochemotherapy of resectable PDAC could improve patient survival 6 but are currently only recommended in the framework of clinical trials. However, histological confirmation of PDAC is necessary before any cytotoxic therapy is initiated 7-9. Despite the fact that histological confirmation of a pancreatic lesion is not required for pancreatic surgery according to current guidelines, it is also important to confirm a suspected diagnosis of PDAC e.g. in case of unclear lesions in an inflammatory background of chronic pancreatitis. Biopsy of a pancreatic lesions is preferentially done by endoscopic ultrasound (EUS)-guided fine needle aspiration (FNA). But pancreatic tumors are at times challenging for EUS-guided FNA biopsies. Type, size and localization of the lesion in the pancreas, experience of the endoscopist, and number of passes of the needle through the lesion and the dense tumor stroma impact on outcome of EUS-guided FNA of pancreatic lesions. Thus, inconclusive pathological results in tissue or cells obtained for diagnosis are reported in up to 30 % of case 10. Accordingly, the diagnostic accuracy of EUS-guided FNA of PDAC is 76 % - 90 %, the false negative rate is about 15 % 11.
Serum carbohydrate antigen 19-9 (CA19-9) is an established tumor marker for PDAC and is elevated in up to 85 % of the patients with PDAC 12. CA19-9 is not regarded as useful to establish diagnosis of the disease, but to monitor the course of a given disease in PDAC patients 13, 14. The limitations of CA19-9 measurements must be appreciated as they are critical for using this biomarker for patients with suspected PDAC. CA19-9 is a sialylated Lewis (a) antigen that is found on the erythrocyte surface. About 10 % of the caucasian population do not show detectable levels of CA19-9 due to the disruption of its biosynthesis by the absence of the enzyme fucosyltransferase 9, 15. There is also a lack of specificity of CA19‑9 for PDAC, since it is also moderately elevated by chronic pancreatitis, cholestasis or cholangitis 16. Therefore, CA19-9 is not recommended as a screening tool for the detection of PDAC 17, 18 and new diagnostic approaches are urgently needed. A promising tool for this purpose could be the analysis of circulating cell-free DNA (cfDNA). CfDNA is isolated from blood plasma and allows a non-invasive, tumor-specific genotyping in malignancies 19. Recently we have shown, that patients with metastatic PDAC harbor higher cfDNA levels than those patients with benign pancreatic lesions or healthy controls 20. Several alterations in key oncogenes and tumor suppressors have been identified in PDAC with KRAS being the most frequently mutated oncogene 21, 22. Recently it was shown, that KRAS mutations are detectable through a liquid biopsy of patients with early as well as late stage malignancies 23, namely in PDAC 20, 24, 25. The tumor derived fraction of cfDNA that is carrying cancer specific mutations is called circulating tumor DNA (ctDNA) and was shown to be highly representative for the respective tumor tissue and suitable for cancer genotyping 26 as long as the method used is sensitive enough. This actually opens the avenue for a molecular monitoring of PDAC during systemic treatment. However, ctDNA analysis is limited in cases where the primary tumours release only small amounts of or even no ctDNA into the circulation. There are other secreted markers that have been reported to be diagnostic for PDAC, such as circulating amino-acids 27, cancer exosomes 28, proteome based analytes 29, epigenetic biomarkers such as nucleosomes 30 and miRNAs 31. A recently published study developed a fluorescence nanobiosensor for ultrasensitive arginase and protease detection and identified a PDAC-specific enzymatic signature in serum, consisting of arginase, matrix metalloproteinase-1, -3, and - 9, cathepsin-B and -E, urokinase plasminogen activator and neutrophil elastase 32. However, all these approaches are still under evaluation and have not reached the clinic.
Obviously, proteins released from precursor lesions during tumor progression such as pancreatic intraepithelial neoplasias (PanIN) might serve as innovative and effective diagnostic biomarkers. Thrombospondin-2 (THBS2) is a disulfide-linked homotrimeric glycoprotein that mediates cell-to-cell and cell-to-matrix interactions. THBS2 probably inhibits angiogenesis and depletion of the THBS2 gene in a mouse model increases the susceptibility to cancer 33. THBS2 is secreted or released from human precursor PanIN organoids and may hence serve as a biomarker for early PDAC 34. Preclinical data were recently validated in a large cohort with PDAC patients at various disease stages compared to healthy controls and patients with cystic tumors or chronic pancreatitis 35. Normal pancreatic cells express the THBS2 antigen but under physiological conditions the plasma concentration is low. In contrary, it is highly expressed by PDAC tumor cells and the plasma of PDAC patients shows elevated THBS2 levels 35. The concentration of THBS2 in plasma is reported to allow the discrimination between resectable PDAC stage I cancer and advanced stage III/IV. However, the mechanism of THBS2 release into the blood stream remains elusive 35.
Here, we examined whether integrating established and novel blood-based markers i. e. CA19‑9, THBS2 and cfDNA, will facilitate and further improve diagnosis of PDAC in case of a resectable lesion in the pancreas detected by imaging.
Methods
Plasma separation
Whole venous blood was collected in 7.5 ml EDTA tubes (S-Monovette® K3E, Sarstedt) by peripheral blood draw and kept at 4 °C until separation, which was carried out within one hour after collection. For separation whole blood was centrifuged for 10 minutes at 820 x g and 4 °C, plasma fraction was then transferred to cold 2 ml tubes (RNA/DNA LoBind micro-centrifuge tubes, Eppendorf) and subsequently centrifuged again for 10 min at 20.000 x g and 4 °C. Pure plasma was then transferred to fresh 2 ml tubes for immediate storage at ‑80 °C until further use.
Measurement of CA19-9 and THBS2 in plasma samples
Plasma levels of CA19-9 were determined routinely by ELISA (Roche). Plasma levels of THBS2 were determined by Quantikine® ELISA Human Thrombospondin-2 (R&D Systems) following the instructions of the manufacturer. Plasma samples were diluted 4-fold and analyzed in duplicate. A standard curve of the positive control proteins provided in the kit was used to produce reference concentrations using a 4-parameter logistic nonlinear regression model in SoftMax Pro 5.3 Software (Molecular Device).
Isolation and characterization of cfDNA
cfDNA was isolated from 2 ml of plasma using the QIAamp® MinElute® ccfDNA Mini Kit (Qiagen) according to the instructions of the manufacturer and recovered in 50 µl ultra-clean water. Concentration of cfDNA was then measured using the Qubit™ dsDNA HS Assay Kit with a Qubit™ 2.0 fluorometer (Invitrogen).
Statistical analysis
Spearman correlation coefficients were calculated for continuous variables and Mann-Whitney U test was used to compare the medians of categorical variables. Univariate and multivariable logit (logistic regression) models were applied to test the performance of single biomarkers and biomarker combinations in differentiating PDAC from control samples (IPMN or pancreatitis). The sample set was randomly divided into a training and a validation set. The logistic regression models were fitted to the training set and test performance was subsequently verified on a user-blinded validation sample set 36, 37. The binary dependent variable was coded 1 for PDAC and 0 for IPMN. CA19-9 and THBS2 were dichotomized as 0 when negative (˂ 55 U/ml and ˂ 42 ng/ml, respectively) and 1 when positive (≥ 55 U/ml and ≥ 42 ng/ml, respectively). cfDNA level was considered as continuous variable. The area under the curve (AUC) was calculated for each model and a bootstrap test with 2000 replicates was used to test if there is a statistically significant difference between the AUC of the marker combination (CA19-9, THBS2 and cfDNA) and the AUC of CA19-9 alone. The 95 % confidence intervals were also estimated based on 2000 stratified bootstrap replicates. R version 3.4.2 (R Foundation for statistical computing) and GraphPad Prism version 7.00 (GraphPad Software) were used for statistical analyses.
Results
Study cohort and patient characteristics
Plasma samples from 52 patients with resectable PDAC were prospectively collected and underwent retrospective translational analysis. The PDAC diagnosis was confirmed by pathological assessment of each resected sample. The control group comprised 15 individuals with side-branch IPMNs without worrisome features as determined by MRI imaging and/or EUS and 32 individuals with chronic pancreatitis of different origin. Plasma samples were collected with an approval of the institutional review board of Ulm University (approval numbers: 317/12, 230/14, 128/15) and at the Department of Surgery at the TUM, Munich, with the assurance of ethical and data protection standards by supervision of the ethics committee of the Faculty of Medicine at the TUM (#1926/07, #5428/12). After informed consent, venous blood samples were taken from the participants. The patients were all therapy naïve and samples were taken prior the initiation of any disease-specific treatment. Plasma samples were stored at -80 °C until further use. Clinical characteristics of the study cohort are shown in Table 1.
Table 1.
Clinical characteristics
| PDAC | IPMN | Pancreatitis | |
|---|---|---|---|
| N | 52 | 15 | 32 |
| Age (years) | 68 (42-81) | 70 (44-82) | 57 (21-81) |
| Male / Female | 26 / 26 | 5 / 10 | 23 / 9 |
| Ethnic origin | 51 caucasian / 1 asian | 15 caucasian | 32 caucasian |
| History of diabetes | 13 (25 %) | 0 (0 %) | 3 (9 %) |
| CA19-9 positive | 31 (60 %) | 0 (0 %) | 3 (9 %) |
| Histological / cytological diagnosis | 51 (98 %) | 0 (0 %) | |
| Imaging based diagnosis | 1 (3 %) | 15 (100 %) | |
| Pancreatic mass | 52 (100 %) | 15 (100 %) | 7 (22 %) |
| Genesis of pancreatitis | |||
| Hereditary / autoimmune / idiopathic / alcohol-induced | 4 / 9 / 10 / 9 | ||
| Disease stage | |||
| I | 14 (27 %) | ||
| II | 17 (33 %) | ||
| III | 21 (40 %) | ||
| Grading X / 1 / 2 / 3 | 16 / 3 / 16 / 17 | ||
| Localization h / b / t | 39 / 9 / 4 | 10 / 13 / 9 | |
| Sum of diameters (mm) | 30 | 11 | 5 |
X=unknown, h=head, b=body, t=tail
Total cfDNA quantification
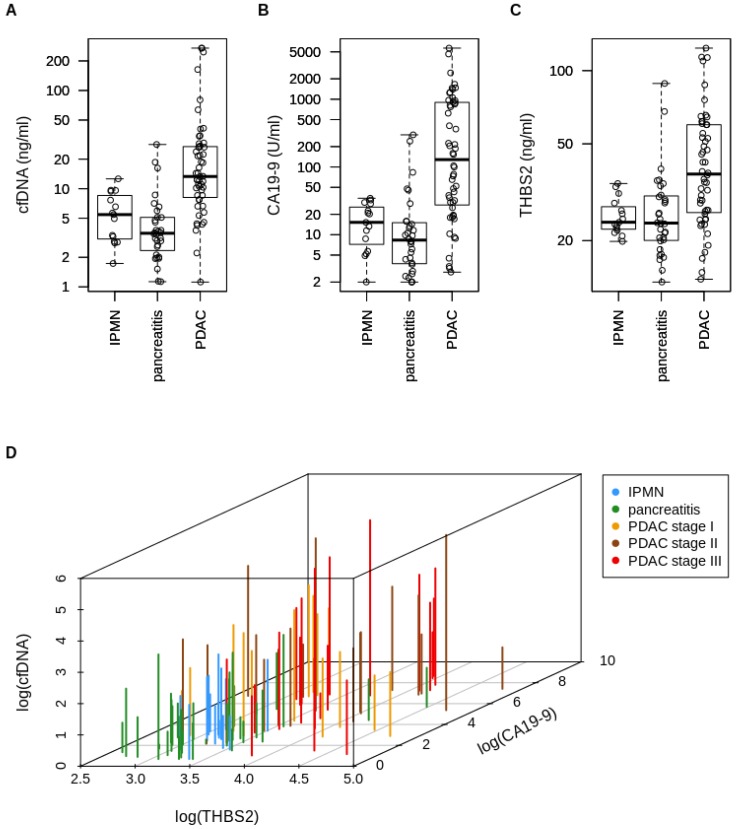
CfDNA amount in the plasma of PDAC patients (13.3 ng/ml) was increased by 2.4 fold compared to that of individuals with IPMNs (5.5 ng/ml, p < 0.0001) and by 3.8 fold compared to individuals with chronic pancreatitis (3.5 ng/ml, p < 0.0001). A scatter plot of cfDNA concentrations is shown in Figure 1A. Age had a minor effect on the cfDNA amount in the PDAC group (r² = 0.17) and in the IPMN group (r² = 0.31) but not in the chronic pancreatitis group (r² = 0.05). The PDAC and control group were not age matched with a median age of 68 years in PDAC group versus 61 years in the control group (p = 0.0038). There was no association between cfDNA concentration and history of diabetes (data not shown).
Figure 1.
Scatter plot of (A) cfDNA concentrations (IPMN vs. PDAC: p<0.0001; pancreatitis vs. PDAC: p<0.0001), (B) CA19-9 concentrations (IPMN vs. PDAC: p<0.0001; pancreatitis vs. PDAC: p<0.0001), (C) THBS2 concentrations (IPMN vs. PDAC: p=0.0007; pancreatitis vs. PDAC: p=0.0001), and (D) three-dimensional plot of log-transformed cfDNA, THBS2 and CA19-9 concentrations of patients with PDAC (n=52), IPMN (n=15) and chronic pancreatitis (n=32). cfDNA=Circulating cell-free DNA, IPMN=Intraductal papillary mucinous neoplasia, PDAC=Pancreatic ductal adenocarcinoma, CA19-9=Carbohydrate antigen 19-9, THBS2=Thrombospondin-2.
Median CA19-9 and THBS2 levels are increased in early PDAC
The median CA19-9 level in PDAC plasma samples (128.5 U/ml) was increased by 8-fold compared to that of IPMN patients (15.2 U/ml, p < 0.0001) and by 15-fold compared to that of patients with chronic pancreatitis (8.33 ng/ml, p < 0.0001). A scatter plot of CA19-9 concentrations is shown in Figure 1B. CA19-9 was considered “positive" when reaching the conventional cut-off at 55 U/ml. Median THBS2 concentration in our PDAC plasma samples (37.57 ng/ml) was increased by 1.6-fold compared to that of IPMN patients (23.81 ng/ml, p = 0.0007) and again by 1.6-fold compared to that of the chronic pancreatitis group (23.61 ng/ml, p = 0.0001). A scatter plot of THBS2 concentrations is shown in Figure 1C. The recently published 35 cut-off for THBS2 (≥ 42 ng/ml) was used to dichotomize the cohort into “THBS2 positive” and “negative”. 46 % (24/52) of PDAC patients, none of the IPMN patients and 6 % (2/32) of the chronic pancreatitis patients were considered “THBS2 positive” according to this threshold. Figure 1D shows a three-dimensional plot of total cfDNA amount, THBS2 concentration and CA19-9 plasma levels for each patient to visualize the levels of all three markers per patient.
Test characteristics of single markers and marker combinations
We established a cut-off for cfDNA concentration in plasma based on the validation set to discriminate between PDAC and IPMN or pancreatitis, considering a range of 0 to 5 false positives. A threshold for cfDNA concentration at 16.2 ng/ml was able to detect 70 % of PDAC cases with 96 % specificity in the validation set. By combining the conventional cut-off for CA19-9 (≥ 55 U/ml), the recently reported 35 cut-off for THBS2 (≥ 42 ng/ml) and a cut-off at 16.2 ng/ml for cfDNA we reached a specificity of 92 % with a sensitivity of 87 % in the validation dataset (Table 2).
Table 2.
Test performance of single markers and marker combinations
| Training set | Validation set | ||||
|---|---|---|---|---|---|
| Marker | Cut-off | Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) |
| CA19-9 (≥ 55 U/ml) | |||||
| 55 | 91 | 63 | 96 | ||
| THBS2 (≥ 42 ng/ml) | |||||
| 41 | 96 | 50 | 96 | ||
| cfDNA (ng/ml) | |||||
| 5 FP | 12.4 | 86 | 70 | 80 | 79 |
| 4 FP | 16.0 | 68 | 87 | 70 | 96 |
| 3 FP | 16.2 | 68 | 87 | 70 | 96 |
| 2 FP | 18.8 | 65 | 65 | 57 | 96 |
| 1 FP | 19.0 | 45 | 91 | 57 | 96 |
| 0 FP | 28.0 | 32 | 100 | 43 | 96 |
| CA19-9 (≥ 55 U/ml) + THBS2 (≥ 42 ng/ml) | |||||
| 73 | 91 | 80 | 96 | ||
| CA19-9 (≥ 55 U/ml) + THBS2 (≥ 42 ng/ml) + cfDNA (ng/ml) | |||||
| 5 FP | 12.4 | 86 | 78 | 93 | 92 |
| 4 FP | 16.0 | 86 | 83 | 87 | 92 |
| 3 FP | 16.2 | 86 | 87 | 87 | 92 |
| 2 FP | 18.8 | 73 | 91 | 80 | 92 |
| 1 FP | 19.0 | 73 | 96 | 80 | 92 |
| 0 FP | 28.0 | 41 | 96 | 50 | 96 |
FP=false-positives
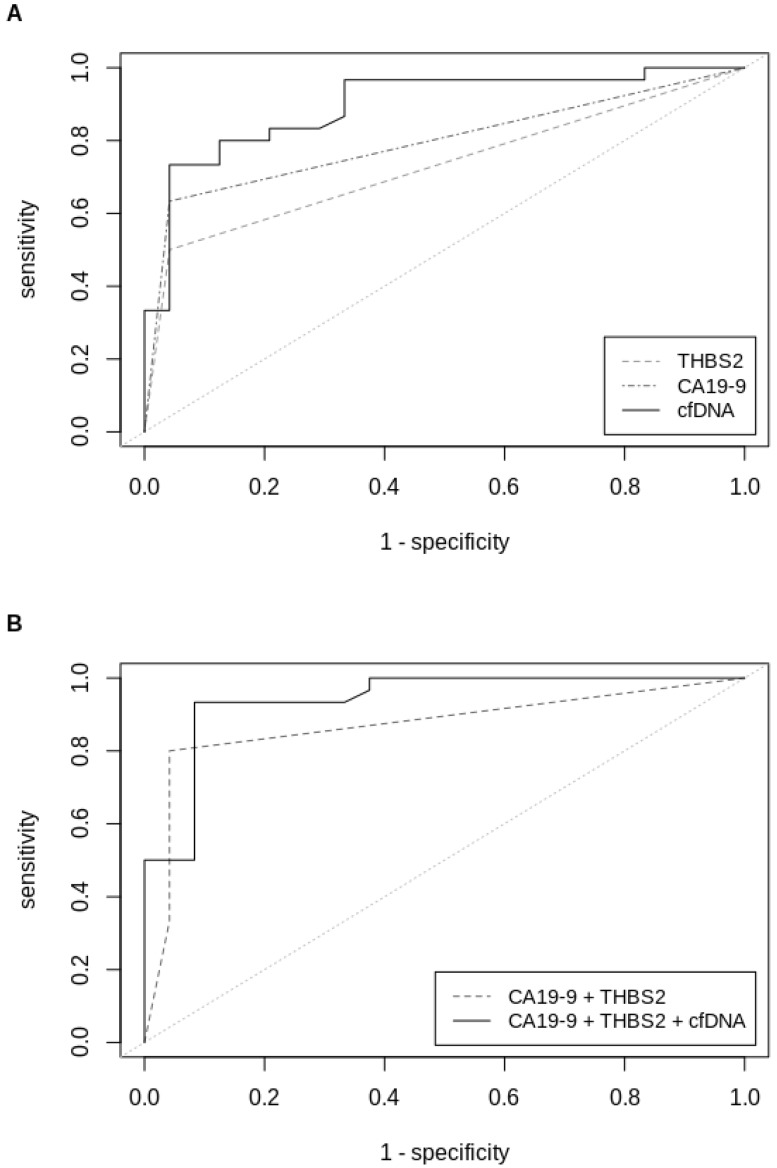
THBS2 alone had a c-statistic of 0.73 for all stages of PDACs in the validation set, which was comparable to that of CA19-9 alone (0.80). The combination of both, CA19-9 and THBS2, showed a superior c-statistic of 0.87 (Table 3). The c-statistic was further improved to 0.94 by combining the categorical variables CA19-9 and THBS2 with the continuous measurement of the cfDNA amount, determined by fluorometric quantification. This marker combination performed best for all stages but also when separating PDAC patients by stage. C-statistics of PDAC stage I, stage II and stage III were 0.90, 0.96 and 0.94, respectively. The biggest improvement in diagnosis compared to the established marker CA1 9-9 was seen for PDAC stage I, for which c-statistic could be improved from 0.69 to 0.90 (Table 3). ROC curves for single markers and marker combinations are shown in Figure 2.
Table 3.
Receiver-operating characteristics of validation set
| All Stages | Stage I | Stage II | Stage III | ||
|---|---|---|---|---|---|
| N | 30/24 | 7/24 | 9/24 | 14/24 | |
| CA19-9 (≥ 55 U/ml) | AUC | 0.80 | 0.69 | 0.87 | 0.83 |
| 95 % CI | 0.70 | 0.49 | 0.72 | 0.69 | |
| 0.89 | 0.90 | 1.00 | 0.96 | ||
| THBS2 (≥ 42 ng/ml) | AUC | 0.73 | 0.69 | 0.76 | 0.71 |
| 95 % CI | 0.63 | 0.49 | 0.58 | 0.56 | |
| 0.83 | 0.90 | 0.93 | 0.86 | ||
| cfDNA (ng/ml) | AUC | 0.90 | 0.89 | 0.97 | 0.84 |
| 95 % CI | 0.81 | 0.77 | 0.92 | 0.69 | |
| 0.98 | 1.00 | 1.00 | 0.99 | ||
| CA19-9 (≥ 55 U/ml) + THBS2 (≥ 42 ng/ml) | AUC | 0.87 | 0.89 | 0.86 | 0.86 |
| 95 % CI | 0.78 | 0.74 | 0.71 | 0.73 | |
| 0.96 | 1.00 | 1.00 | 0.99 | ||
| CA19-9 (≥ 55 U/ml) + THBS2 (≥ 42 ng/ml) + cfDNA (ng/ml) | AUC | 0.94 | 0.90 | 0.96 | 0.94 |
| 95 % CI | 0.88 | 0.78 | 0.91 | 0.87 | |
| 1.00 | 1.00 | 1.00 | 1.00 | ||
| p-value | 0.0013 | 0.0143 | 0.1424 | 0.0549 |
AUC=Area under curve, CI=Confidence interval, p-values are calculated for CA19-9 (≥ 55 U/ml) + THBS2 (≥ 42 ng/ml) + cfDNA (ng/ml) model versus established marker CA19-9.
Figure 2.
ROC-curves (validation sample set) of (A) univariate analysis of THBS2, CA19-9 and cfDNA concentrations and (B) multivariable analysis of CA19-9+THBS2 and CA19‑9+THBS2+cfDNA concentrations in plasma samples from patients with all stages of PDAC versus patients with IPMN or pancreatitis. ROC=Receiver operating characteristics, THBS2=Thrombospondin-2, CA19-9=Carbohydrate antigen 19-9, cfDNA=Circulating cell-free DNA.
Biomarkers CA19-9, THBS2 and cfDNA are complementary
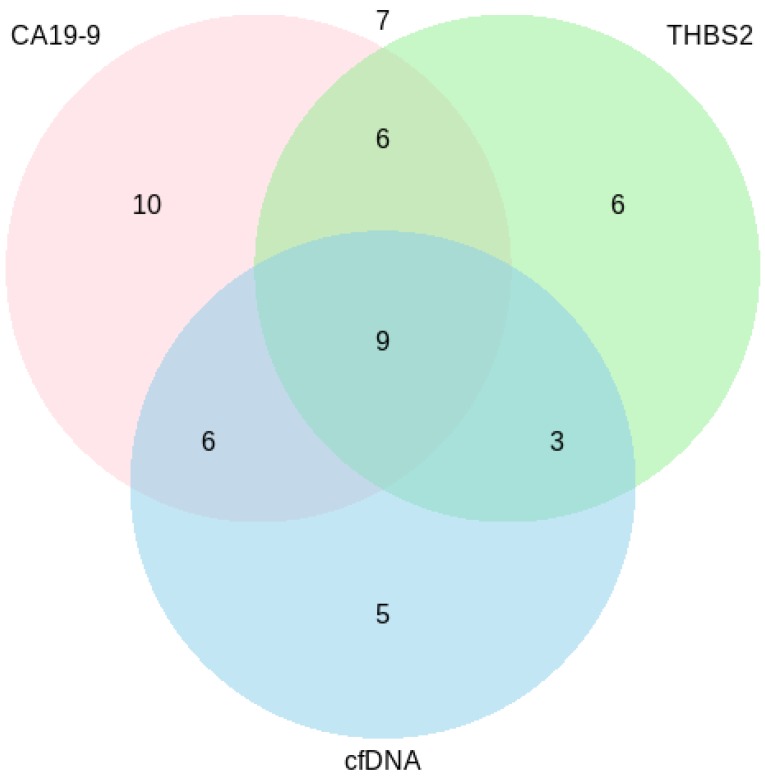
Figure 3 illustrates the number of PDAC patients that were tested positive for each marker and marker combination. 19 % (10) of 52 samples were only positive for CA19-9, 12 % (6/52) were only positive for THBS2 and 10 % (5/52) were only positive for cfDNA. 12 % (6/52) of the samples were positive for CA19‑9 and cfDNA, another 12 % (6/52) for CA19-9 and THBS2 and 6 % (3/52) for cfDNA and THBS2. 17 % (9/52) were positive for all three markers. 13 % (7/52) of the samples were negative for all of the analyzed markers. Two of these negatives were stage I, another two were stage II and another three were stage III, respectively.
Figure 3.
Venn diagram showing the number of PDAC patients (n=52) tested positive for each marker/marker combination. CA19-9=Carbohydrate antigen 19-9, cfDNA=Circulating cell-free DNA, THBS2=Thrombospondin-2.
Discussion
Despite recent advances in the clinical management of resectable PDAC, the survival of PDAC patients is mainly determined by the stage, at which the disease is diagnosed. New strategies for diagnosis of PDAC at an early stage of the disease are therefore urgently needed.
Neoadjuvant chemo- or chemo-radiotherapy 38, 39, followed by surgery and intensive adjuvant treatment 4, 5 could further improve the prognosis of patients with resectable PDAC. These treatment modalities require preoperative tissue diagnostics to establish diagnosis. However, even the analysis of tissue is sometimes not informative in case of extensive fibrosis and low cellular yield in FNAs. Indeed, as shown in Table 1, it is frequently difficult to obtain sufficient amounts of high quality tumor material for further histological examination, resulting in a high number of samples for which grading is impossible (16/52 cases, 31 %). National and international guidelines recommend a histological diagnosis prior to the initiation of any cytotoxic treatment 7-9. EUS guided FNA is currently widely accepted to obtain material for histological or at least cytological assessment of a pancreatic lesion, but the sensitivity of EUS-guided FNA of PDAC ranks only between 64 % 40 and 93 % 41 at best. In addition, the false negative rate is reported to be about 15 % 8, being even higher in a daily practice setting. A review covering 191 patients with pancreatic lesions showed a diagnostic accuracy of EUS-FNA cytology (EUS-FNAC) of only 78.4 %. Alterations in the specimen occurring after sampling can be mimicking states like necrosis, chromatin clearing, mitosis, macronucleoli and hyperchromasia and can further lead to false positive diagnosis 42. The poorly accessible anatomic localization of the pancreas and the difficulty in distinguishing inflammatory changes from a malignant lesion make it at times difficult to secure a preoperative diagnosis of early PDAC. New biomarkers that allow an accurate diagnosis of an early PDAC are therefore urgently needed and biomarkers that can be obtained in a minimally invasive fashion would be desirable.
CA19-9 is the only validated serum biomarker for PDAC so far. It is reported to be elevated in up to 85 % of patients with PDAC 11 but shows a disappointing sensitivity of 63 % for all PDAC stages in our validation set. While CA19-9 serum levels under 100 U/ml likely imply a resectable disease state, levels above 100 U/ml suggest poor resectability or metastatic disease 43. However, as a single biomarker for non-invasive diagnosis of resectable PDAC CA19-9 alone will not suffice. The prognostic information of pre-operative CA19-9 serum levels is relevant, since patients with normal levels (< 37 U/ml) have a prolonged median survival compared to patients with elevated levels. A decrease of CA19-9 serum levels by ≥ 20 - 50 % compared to baseline or even normalization after surgical resection or chemotherapy is associated with prolonged survival 43. Of note, up to 10 % of the caucasian population do not show detectable levels of CA19-9 due to an impaired biosynthesis by the absence of fructosyltransferase. This also limits the value of CA19-9 as a diagnostic biomarker.
It has been shown that cfDNA concentrations are significantly increased in patients with PDAC compared to healthy individuals or patients with pancreatic cysts 20. In line with this finding, cfDNA can be used as a monitoring marker and thereby help to detect therapy response or disease progression 44-46. However, the use of the total cfDNA amount is challenging, because it has a high range and elevated levels are observed in several conditions, such as simple muscle training, end-stage renal failure, pregnancy, stroke, surgery, trauma or myocardial infarction 47. Mutations detectable in cfDNA are highly representative for the respective tumor tissue, resulting in a blood-tissue concordance between 82 % 48 and 95 % 24. The tumor-derived fraction of cfDNA, termed ctDNA, was analyzed in several studies, mostly with a focus on KRAS mutations. Patients with no detectable ctDNA (e.g. no tumor-specific mutations detectable) in blood samples after surgery had a longer disease-free survival and a longer overall survival than the group of PDAC patients with detectable ctDNA 24. While high levels of mutated KRAS alleles were detectable in the blood of patients with metastatic PDAC, the frequency of KRAS mutations detectable in patients with resectable PDAC ranges between 19 % and 43 % 24. Therefore, we focused on the three-marker combination, consisting of cfDNA, THBS2 and CA19-9 and did not include KRAS genotyping of cfDNA.
THBS2 has been suggested as a new promising biomarker for non-invasive early PDAC diagnosis 35. However, as for cfDNA or CA19-9, each of the individual biomarkers has certain limitations. In our validation set THBS2 alone showed a c-statistic of 0.73 considering all PDAC stages versus controls and a c-statistic of 0.69 or 0.76 when considering the prognostically favorable stage I and stage II, respectively. Thus, as in case of CA19-9 and cfDNA, THBS2 alone also is not appropriate for the noninvasive detection of early stages of pancreatic cancer. Our data however support the notion that a combined assessment of CA19-9 and THBS2 levels greatly improves the test performance. Sensitivity and specificity previously reported for THBS2 were based on a comparison with healthy controls. We used benign pancreatic lesions as controls to simulate the clinical situation, when there is a suspicious lesion detected by imaging and it is unclear whether the lesion is malignant or benign. For the combination of THBS2 and CA19-9 Kim et al. reported an AUC of 0.95 when discriminating IPMN from PDAC and an AUC of 0.87 when discriminating pancreatitis from PDAC. We also report an AUC of 0.87 for THBS2 + CA19-9 in our validation set which comprises PDAC patients versus pancreatitis and IPMN patients, respectively. The AUC for the three-marker combination including the cfDNA concentration in this set is increased to 0.94. The median concentration of THBS2 reported by Kim et al. is lower than that reported in this manuscript (29.7 vs. 37.9 ng/ml). This is probably caused by a difference in sample size (278 vs. 52). We also detected about 50 % of our PDAC cases with 96 % specificity using the cutoff reported by Kim et al. (42 ng/ml). The most promising approach for a non-invasive differential diagnosis of PDAC is a combination of several markers maximizing the sensitivity and specificity of the approach, as previously reported 49, 50. Our proposed composite liquid biomarker panel, consisting of the combination of total cfDNA, THBS2 and CA19-9 plasma levels may indeed fulfill the clinical requirements for such a diagnostic test. This panel improved the sensitivity to 87 % with 92 % specificity in discriminating PDAC from IPMN or chronic pancreatitis. This marker combination performed best for all PDAC stages but also for PDACs grouped by stage. The greatest improvement over CA19-9 was seen in the group of stage I PDAC, from 0.69 to 0.90 for the three-marker combination. The almost even distribution of the 7 PDAC patients negative for all three markers across the disease stages (two times stage I, two times stage II and three times stage III) suggests that this detection failure is not linked to a generally low marker expression in very early PDAC. It is also not linked to the lesion diameter. Given these data this approach may be also useful for screening high risk populations. Longitudinal prospective studies in larger groups of patients are now needed to further examine the power of this approach.
Acknowledgments
The authors wish to thank all recruiting and participating centers of the NEONAX trial. For excellent technical assistance the authors also would like to thank Mrs. Magdalena Bienek-Ziolkowski and Mrs. Rosina Sing (Biobank, Department of Internal Medicine I, Ulm University).
Financial Support
This work was supported by an unrestricted research grant from CELGENE within the framework of the NEONAX trial to T.S.
References
- 1.Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV. et al. Pancreatic cancer. Nature reviews Disease primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 2.Oettle H, Neuhaus P, Hochhaus A, Hartmann JT, Gellert K, Ridwelski K. et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the CONKO-001 randomized trial. Jama. 2013;310:1473–81. doi: 10.1001/jama.2013.279201. [DOI] [PubMed] [Google Scholar]
- 3.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet (London, England) 2011;378:607–20. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conroy T, Hammel P, Hebbar M, Abdelghani MB, Wei AC-c, Raoul J-L. et al. Unicancer GI PRODIGE 24/CCTG PA.6 trial: A multicenter international randomized phase III trial of adjuvant mFOLFIRINOX versus gemcitabine (gem) in patients with resected pancreatic ductal adenocarcinomas. Journal of Clinical Oncology. 2018;36:LBA4001–LBA. [Google Scholar]
- 5.Neoptolemos JP, Palmer DH, Ghaneh P, Psarelli EE, Valle JW, Halloran CM. et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. The Lancet. 2017;389:1011–24. doi: 10.1016/S0140-6736(16)32409-6. [DOI] [PubMed] [Google Scholar]
- 6.Mokdad AA, Minter RM, Zhu H, Augustine MM, Porembka MR, Wang SC, Neoadjuvant Therapy Followed by Resection Versus Upfront Resection for Resectable Pancreatic Cancer: A Propensity Score Matched Analysis. J Clin Oncol; 2016. [DOI] [PubMed] [Google Scholar]
- 7.Ducreux M, Cuhna AS, Caramella C, Hollebecque A, Burtin P, Goere D. et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(Suppl 5):v56–68. doi: 10.1093/annonc/mdv295. [DOI] [PubMed] [Google Scholar]
- 8.Seufferlein T, Porzner M, Becker T, Budach V, Ceyhan G, Esposito I. et al. [S3-guideline exocrine pancreatic cancer] Z Gastroenterol. 2013;51:1395–440. doi: 10.1055/s-0033-1356220. [DOI] [PubMed] [Google Scholar]
- 9.Tempero MA, Malafa MP, Behrman SW, Benson AB 3rd, Casper ES, Chiorean EG. et al. Pancreatic adenocarcinoma, version 2.2014: featured updates to the NCCN guidelines. J Natl Compr Canc Netw. 2014;12:1083–93. doi: 10.6004/jnccn.2014.0106. [DOI] [PubMed] [Google Scholar]
- 10.Tellez-Avila FI, Martinez-Lozano JA, Rosales-Salinas A, Bernal-Mendez AR, Guerrero-Velasquez C, Ramirez-Luna MA. et al. Repeat endoscopic ultrasound fine needle aspiration after a first negative procedure is useful in pancreatic lesions. Endoscopic ultrasound. 2016;5:258–62. doi: 10.4103/2303-9027.187889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gimeno-Garcia AZ, Elwassief A. How to improve the success of endoscopic ultrasound guided fine needle aspiration cytology in the diagnosis of pancreatic lesions. Journal of interventional gastroenterology. 2012;2:31–6. doi: 10.4161/jig.20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartwig W, Strobel O, Hinz U, Fritz S, Hackert T, Roth C. et al. CA19-9 in potentially resectable pancreatic cancer: perspective to adjust surgical and perioperative therapy. Ann Surg Oncol. 2013;20:2188–96. doi: 10.1245/s10434-012-2809-1. [DOI] [PubMed] [Google Scholar]
- 13.Satake K, Kanazawa G, Kho I, Chung YS, Umeyama K. A clinical evaluation of carbohydrate antigen 19-9 and carcinoembryonic antigen in patients with pancreatic carcinoma. J Surg Oncol. 1985;29:15–21. doi: 10.1002/jso.2930290106. [DOI] [PubMed] [Google Scholar]
- 14.Duffy MJ, Sturgeon C, Lamerz R, Haglund C, Holubec VL, Klapdor R. et al. Tumor markers in pancreatic cancer: a European Group on Tumor Markers (EGTM) status report. Ann Oncol. 2010;21:441–7. doi: 10.1093/annonc/mdp332. [DOI] [PubMed] [Google Scholar]
- 15.Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143:725–9. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- 16.Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol. 2007;33:266–70. doi: 10.1016/j.ejso.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Krokowski M, Bodalski J, Bratek A, Machejko P, Caillat-Zucman S. CTLA-4 gene polymorphism is associated with predisposition to IDDM in a population from central Poland. Diabetes Metab. 1998;24:241–3. [PubMed] [Google Scholar]
- 18.Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS. et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–27. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 19.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–12. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 20.Berger AW, Schwerdel D, Costa IG, Hackert T, Strobel O, Lam S. et al. Detection of Hot-Spot Mutations in Circulating Cell-Free DNA From Patients With Intraductal Papillary Mucinous Neoplasms of the Pancreas. Gastroenterology. 2016;151:267–70. doi: 10.1053/j.gastro.2016.04.034. [DOI] [PubMed] [Google Scholar]
- 21.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J. et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nature communications. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Science translational medicine. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pietrasz D, Pecuchet N, Garlan F, Didelot A, Dubreuil O, Doat S. et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23:116–23. doi: 10.1158/1078-0432.CCR-16-0806. [DOI] [PubMed] [Google Scholar]
- 25.Dabritz J, Preston R, Hanfler J, Oettle H. Follow-up study of K-ras mutations in the plasma of patients with pancreatic cancer: correlation with clinical features and carbohydrate antigen 19-9. Pancreas. 2009;38:534–41. doi: 10.1097/MPA.0b013e31819f6376. [DOI] [PubMed] [Google Scholar]
- 26.Odegaard JI, Vincent JJ, Mortimer S, Vowles JV, Ulrich BC, Banks KC, Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clinical Cancer Research; 2018. [DOI] [PubMed] [Google Scholar]
- 27.Mayers JR, Wu C, Clish CB, Kraft P, Torrence ME, Fiske BP. et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med. 2014;20:1193–8. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–82. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu KH, Barry CG, Austin D, Busch CM, Sangar V, Rustgi AK. et al. Stable isotope dilution multidimensional liquid chromatography-tandem mass spectrometry for pancreatic cancer serum biomarker discovery. J Proteome Res. 2009;8:1565–76. doi: 10.1021/pr800904z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauden M, Pamart D, Ansari D, Herzog M, Eccleston M, Micallef J. et al. Circulating nucleosomes as epigenetic biomarkers in pancreatic cancer. Clin Epigenetics. 2015;7:106. doi: 10.1186/s13148-015-0139-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE. et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311:392–404. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- 32.Kalubowilage M, Covarrubias-Zambrano O, Malalasekera AP, Wendel SO, Wang H, Yapa AS. et al. Early detection of pancreatic cancers in liquid biopsies by ultrasensitive fluorescence nanobiosensors. Nanomedicine. 2018;14:1823–32. doi: 10.1016/j.nano.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Hoffman JP, Alpaugh RK, Rhim AD, Reichert M, Stanger BZ. et al. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep. 2013;3:2088–99. doi: 10.1016/j.celrep.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawighorst T, Velasco P, Streit M, Hong YK, Kyriakides TR, Brown LF. et al. Thrombospondin-2 plays a protective role in multistep carcinogenesis: a novel host anti-tumor defense mechanism. EMBO J. 2001;20:2631–40. doi: 10.1093/emboj/20.11.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Bamlet WR, Oberg AL, Chaffee KG, Donahue G, Cao XJ, Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers. Science translational medicine; 2017. p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ko J, Bhagwat N, Yee SS, Ortiz N, Sahmoud A, Black T. et al. Combining Machine Learning and Nanofluidic Technology To Diagnose Pancreatic Cancer Using Exosomes. ACS Nano. 2017;11:11182–93. doi: 10.1021/acsnano.7b05503. [DOI] [PubMed] [Google Scholar]
- 37.Cao Z, Liu C, Xu J, You L, Wang C, Lou W. et al. Plasma microRNA panels to diagnose pancreatic cancer: Results from a multicenter study. Oncotarget. 2016;7:41575–83. doi: 10.18632/oncotarget.9491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy JE, Wo JY, Ryan DP, Jiang W, Yeap BY, Drapek LC, Total Neoadjuvant Therapy With FOLFIRINOX Followed by Individualized Chemoradiotherapy for Borderline Resectable Pancreatic Adenocarcinoma: A Phase 2 Clinical Trial. JAMA Oncol; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reni M, Balzano G, Zanon S, Zerbi A, Rimassa L, Castoldi R. et al. Safety and efficacy of preoperative or postoperative chemotherapy for resectable pancreatic adenocarcinoma (PACT-15): a randomised, open-label, phase 2-3 trial. Lancet Gastroenterol Hepatol. 2018;3:413–23. doi: 10.1016/S2468-1253(18)30081-5. [DOI] [PubMed] [Google Scholar]
- 40.Bhutani MS, Hawes RH, Baron PL, Sanders-Cliette A, van Velse A, Osborne JF. et al. Endoscopic ultrasound guided fine needle aspiration of malignant pancreatic lesions. Endoscopy. 1997;29:854–8. doi: 10.1055/s-2007-1004321. [DOI] [PubMed] [Google Scholar]
- 41.Gress FG, Hawes RH, Savides TJ, Ikenberry SO, Lehman GA. Endoscopic ultrasound-guided fine-needle aspiration biopsy using linear array and radial scanning endosonography. Gastrointestinal endoscopy. 1997;45:243–50. doi: 10.1016/s0016-5107(97)70266-9. [DOI] [PubMed] [Google Scholar]
- 42.Clarke DL, Clarke BA, Thomson SR, Garden OJ, Lazarus NG. The role of preoperative biopsy in pancreatic cancer. HPB (Oxford) 2004;6:144–53. doi: 10.1080/13651820410030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. Journal of gastrointestinal oncology. 2012;3:105–19. doi: 10.3978/j.issn.2078-6891.2011.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berger AW, Schwerdel D, Welz H, Marienfeld R, Schmidt SA, Kleger A. et al. Treatment monitoring in metastatic colorectal cancer patients by quantification and KRAS genotyping of circulating cell-free DNA. PloS one. 2017;12:e0174308. doi: 10.1371/journal.pone.0174308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J. et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M. et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–86. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berger AW, Schwerdel D, Ettrich TJ, Hann A, Schmidt SA, Kleger A. et al. Targeted deep sequencing of circulating tumor DNA in metastatic pancreatic cancer. Oncotarget. 2018;9:2076–85. doi: 10.18632/oncotarget.23330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P. et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:10202–7. doi: 10.1073/pnas.1704961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–30. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]