Abstract
Background and objective
Transcriptional coactivator with PDZ-binding motif (TAZ) has been found to be associated with tumor progression. Mitochondrial homeostasis regulates cancer cell viability and metastasis. However, the roles of TAZ and mitochondrial homeostasis in liver cancer viability have not been explored. The aim of our study was to investigate the influence of TAZ on HepG2 liver cancer cell apoptosis.
Materials and methods
HepG2 liver cancer cell was used in the present study, and shRNA against TAZ was transfected into HepG2 cell to knockdown TAZ expression. Mitochondrial function was analyzed using Western blotting, immunofluorescence assay, and flow cytometry. Pathway blocker was used to confirm the role of CaMKII pathway in TAZ-mediated cancer cell death.
Results
Our results indicated that TAZ deletion induced death in HepG2 cell via apoptosis. Biological analysis demonstrated that mitochondrial stress, including mitochondrial bioenergetics disorder, mitochondrial oxidative stress, and mitochondrial apoptosis, were activated by TAZ deletion. Furthermore, we found that TAZ affected mitochondrial stress by triggering mitochondrial elongation factor 1 (MIEF1)-related mitochondrial dysfunction. The loss of MIEF1 sustained mitochondrial function and promoted cancer cell survival. Molecular investigation illustrated that TAZ regulated MIEF1 expression via the CaMKII signaling pathway. Blockade of the CaMKII pathway prevented TAZ-mediated MIEF1 upregulation and improved cancer cell survival.
Conclusion
Taken together, our results highlight the key role of TAZ as a master regulator of HepG2 liver cancer cell viability via the modulation of MIEF1-related mitochondrial stress and the CaMKII signaling pathway. These findings define TAZ and MIEF1-related mitochondrial dysfunction as tumor suppressors that act by promoting cancer apoptosis via the CaMKII signaling pathway, with potential implications for new approaches to liver cancer therapy.
Keywords: TAZ, liver cancer, death, MIEF1, CaMKII signaling pathway
Introduction
Liver cancer is the second leading cause of cancer-related death worldwide. Although young patients have an improved prognosis, older patients (generally considered >45 years) have an increased mortality rate after being diagnosed with liver cancer. At the molecular level, reducing the survival ratio of cancer cells is vital to treat liver cancer. Cancer cell apoptosis is regulated via caspase-independent and caspase-dependent death pathways, such as the caspase-9-involved endogenous mitochondrial apoptotic and Fas receptor-related exogenous apoptotic pathway.1 Notably, ample evidence has indicated a correlation between mitochondrial stress and liver cancer death. Liver tissue contains abundant mitochondria, and several liver biological processes are closely regulated by mitochondria such as protein synthesis, redox balance, fat metabolization, and vitamin storage. In an in vitro study, the mitochondrial reactive oxygen species levels (mROS) are highly correlated with the therapeutic sensitivity of liver cancer to sulforaphane. Moreover, the mitochondrial energy metabolism and mitochondrial calcium homeostasis drastically control the viability of liver cancer.2 These findings indicate that mitochondria seem to be the therapeutic target to control the development of liver cancer.
Recently, mitochondrial stress has been reported as the upstream mediator to trigger mitochondrial dysfunction.3–5 Mitochondrial stress could promote cell death in liver cancer, colorectal cancer, pancreatic cancer, cervical cancer, and lung cancer.6 Mechanistically, mitochondrial stress is regulated by an array of mitochondrial fission adaptors, including Drp1, Mff, and Fis1.7 Notably, mitochondrial elongation factor 1 (MIEF1) is a novel mitochondrial fission mediator.8 Once activated by the JNK and/or CaMKII signaling pathway,9,10 MIEF1 expression is upregulated, and increased MIEF1 promotes Drp1 recruitment onto the surface of mitochondria. Subsequently, active Drp1 interacts with mitochondria to form a constriction ring around the mitochondria.11 Ultimately, with the help of ATP, mitochondria are divided into several fragmentations. MIEF1-related mitochondrial stress has been observed in acute myocardial infarction,12 diabetes,13 and ultraviolet irradiation-mediated cell apoptosis.14 However, limited insights have been gained to understand the role of the CaMKII pathway and MIEF1-related mitochondrial fission in liver cancer.
Transcriptional coactivator with PDZ-binding motif (TAZ), a coactivator of the Hippo-pathways, promotes the transcription of multiple tumorigenesis genes, including Cyclin D1 and connective tissue growth factor.15 Accumulative evidence has established the necessary role of TAZ in the development and progression of various types of cancers, such as liver cancer, colon adenocarcinoma,16 osteosarcoma, and cervical cancer.17 At the molecular level, mitochondrial oxidative stress is also modulated by TAZ. Recently, increased TAZ, which functions to attenuate mitochondrial fission and maintain the mitochondrial dynamics balance, has been found in the heart, suggesting that TAZ seems to be the upstream mediator of mitochondrial homeostasis. More importantly, TAZ activation has emerged as a growth advantage for cancer cells. Increased TAZ expression works together with Yes-associated protein to promote the epithelial–mesenchymal transition (EMT), cell cycle transition, and lamellipodia formation, ultimately contributing to tumor growth, survival, and metastasis. However, the molecular mechanism by which TAZ promotes the survival of liver cancer is not elucidated. In light of the regulatory effects of TAZ on mitochondrial homeostasis, we questioned whether TAZ promotes liver cancer survival via the modulation of mitochondrial stress. Thus, the aim of our study was to observe the biological alterations of liver cancer in response to TAZ deletion and validate whether liver cancer viability was controlled by TAZ via MIEF1-related mitochondrial stress in a manner dependent on the CaMKII signaling pathway.
Materials and methods
Cell culture and reagents
The HepG2 cell line (ATCC, Cat. no #HB-8065™) was purchased from American Type Culture Collection (Manassas, VA, USA). Cells were grown to 70% confluence in DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (v) under 37°C/5% CO2. To observe the role of TAZ in cancer cell viability, two independent shRNAs against Taffazin and control shRNA were transfected into HepG2 cells. To explore the role of CaMKII in cancer cell death, KN93 (25 µM, Selleck Chemicals, Houston, TX, USA),18 an inhibitor of CaMKII, was added into the cell medium for 45 minutes.
TUNEL staining and MTT assay
Cell death was measured via the TUNEL assay using an In Situ Cell Death Detection Kit (Hoffman-La Roche Ltd., Basel, Switzerland). The TUNEL kit stains nuclei that contain fragmented DNA. After treatment, the cells were fixed with 3.7% paraformaldehyde for 30 minutes at room temperature.19 An equilibration buffer, nucleotide mix, and rTdT enzyme were subsequently incubated with the samples at 37°C for 60 minutes. A saline–sodium citrate buffer was then used to stop the reaction. After loading with DAPI, the samples were visualized via fluorescence microscopy (BX-61; Olympus Corporation, Tokyo, Japan). In addition, the MTT assay was performed to analyze the cell viability according to the methods described in a previous study.3 The absorbance at 570 nm was determined. The relative cell viability was recorded as a ratio with the control group. The experiments were performed in triplicate and repeated three times with similar results.20
Western blotting
Cytosolic and mitochondrial fractions were used for the Western blotting assays. Proteins (40–60 µg) were loaded for immunodetection. The samples were resolved by 10% SDS-PAGE and then transferred to PVDF membranes (85 V for 60 minutes).21 Then, 5% nonfat dried milk in Tris-buffered saline was used to block the membranes, which were incubated with primary antibodies overnight at 4°C. The membranes were subsequently incubated with a secondary antibody for 45 minutes at room temperature.22 The membranes were washed with TBST at least three times. The immunoblots were then detected using an enhanced chemiluminescence substrate (Applygen Technologies, Inc. Beijing, China).23 The primary antibodies used in the present study were as follows: cyt-c (1:500; Abcam Cambridge, UK; #ab90529), caspase-9 (1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA; #9504), pro-caspase-3 (1:1,000, Abcam, #ab13847), cleaved caspase-3 (1:1,000, Abcam, #ab49822), complex III subunit core (CIII-core2, 1:1,000, Thermo Fisher Scientific, #459220), complex II (CII-30, 1:1,000, Abcam, #ab110410), complex IV subunit II (CIV-II, 1:1,000, Abcam, #ab110268), Cyclin D1 (1:1,000, Abcam, #ab134175) Drp1 (1:1,000, Abcam, #ab56788), Fis1 (1:1,000, Abcam, #ab71498), Opa1 (1:1,000, Abcam, #ab42364), Mfn1 (1:1,000, Abcam, #ab57602), Mfn2 (1:1,000, Abcam, #ab56889), c-IAP (1:1,000, Cell Signaling Technology, #4952), MIEF1 (1:1,000, Abcam, #ab89944), Tom20 (1:1,000, Abcam, #ab186735), CDK4 (1:1,000, Abcam, #ab137675), Bcl2 (1:1,000, Cell Signaling Technology, #3498), Bax (1:1,000, Cell Signaling Technology, #2772), CaMKII (1:1,000, Cell Signaling Technology, #3362), p-CaMKII (1:1,000, Cell Signaling Technology, #12716), PARP (1:1,000, Abcam, #ab32064), cleaved PARP (1:1,000, Abcam, #ab110315), and TAZ (1:1,000, Abcam, #ab84927). The experiments were performed in triplicate and repeated three times with similar results.
Transwell assay
Transwell assays were performed using 24-well transwell chambers that contain an insert with an 8-µm pore size.24 Approximately 2.5×104 cells suspended in 50 µL of L-DMEM were seeded in the upper chamber, and L-DMEM supplemented with 5% FBS was added to the lower chamber. After 24 hours, nonmigrated cells on the upper surface were removed.25 The membranes were then stained with crystal violet staining solution (Sigma-Aldrich Co., St Louis, MO, USA) for 1 hour at room temperature. The membranes were then photographed, and the number of migrated cells was counted.26
EdU staining
Cellular proliferation was detected via EdU staining. Cells were fixed with 4% paraformaldehyde for 15 minutes. The cells were subsequently incubated with EdU staining solution (FluoProbes®, Montlucon Cedex, France, catalog number FP-MM9829)27 for 20 minutes in the dark. After being washed with PBS, the cells were labeled by DAPI. Images were obtained using fluorescence microscopy.28
Immunofluorescent staining
The samples were washed with cold-PBS three times and then permeabilized using 0.1% Triton X-100, followed by neutralization with NH4Cl buffer for 45 minutes. The samples were subsequently incubated overnight with the following primary antibodies: cyt-c (1:500; Abcam; #ab90529), p-CaMKII (1:1,000, Cell Signaling Technology, #12716), MIEF1 (1:1,000, Abcam, #ab89944), and Tom-20 (1:1,000, Abcam, #ab186735). Confocal immunofluorescence images were collected using FV10-ASW 1.7 software and an Olympus IX81 microscope (Olympus Corporation). The fluorescence intensity was calculated using Image-Pro Plus 6.0 software. First, fluorescence pictures were converted to grayscale with Image-Pro Plus 6.0 software. The fluorescence intensities were then separately recorded as grayscale intensities.29 Mitochondria were observed in at least 100 cells, and the average length of the mitochondria was measured under an inverted microscope to quantify mitochondrial fission (BX51; Olympus Corporation), according to a previous study.30 The experiments were performed in triplicate and repeated three times with similar results.31
Mitochondrial potential observation and LDH release assay
The Mitochondrial Membrane Potential Detection Kit (JC-1; Beyotime Institute of Biotechnology, Shanghai, China) was used to observe changes in the mitochondrial potential.32 Briefly, 5 mg/mL JC-1 working solution was added to the medium and incubated for 30 minutes at 37°C with CO2. The cells were subsequently washed with PBS to remove the JC-1 probe, and images were obtained via fluorescence microscopy (BX-61; Olympus Corporation). The ratio of red to green fluorescence was analyzed using Image-Pro Plus version 4.5 (Media Cybernetics, Inc., Rockville, MD, USA).33 The LDH release assay was used to observe cell death according to the manufacturer’s guidelines.34 The relative LDH release was recorded as the ratio to that of the control group. The experiments were performed in triplicate and repeated three times with similar results.
Flow cytometry for mROS
Flow cytometry was used to analyze mROS production. After treatment, the cells were washed three times with PBS and were then resuspended in PBS using 0.25% trypsin. The cells were subsequently incubated with the MitoSOX red mitochondrial superoxide indicator (Molecular Probes, Eugene, OR, USA) for 15 minutes at 37°C in the dark.35 After three washes with PBS, the mROS production was analyzed via flow cytometry (Sysmex Partec GmbH, Görlitz, Germany), and the data were analyzed using Flowmax software (Sysmex Partec GmbH, Version 2.3, Germany).36 The experiments were performed in triplicate and repeated three times with similar results.
Caspase activity detection and ELISA
The caspase-3 and caspase-9 activities were determined using commercial kits (Beyotime Institute of Biotechnology). The levels of antioxidant factors, including GPX, SOD, and GSH, were measured with ELISA kits purchased from the Beyotime Institute of Biotechnology.37 The experiments were performed in triplicate and repeated three times with similar results.38
RNA isolation and qPCR
Total RNA was extracted with RIPA lysis buffer (Beyotime Institute of Biotechnology). cDNA was then reverse-transcribed according to the methods described in a previous study. The mRNA expression was measured via qRT-PCR (SYBR Green method).39 The SYBR Green reagent was obtained from Solarbio Life Sciences (Beijing, China). The relative mRNA levels were normalized to GAPDH and calculated using the 2−ΔΔCt method as described in a previous study.40 The following primers were used in the present study: ROCK-1 (Forward: 5′-ACCTGTAACCCAAGGAGATGTG-3′, Reverse 5′-CACAATTGGCAGGAAAGTGG-3′), and Rac1 (Forward 5′-ATGCAGGCCATCAAGTGTGTGG-3′, Reverse: 5′-TTACAACAGCAGGCATTTTCTC-3′). The experiments were performed in triplicate and repeated three times with similar results.
Transfection
Transfection with shRNA was used to inhibit TAZ expression. Two independent shRNAs (shRNA1-TAZ and shRNA2-TAZ, Yangzhou Ruibo Biotech Co., Ltd., Yangzhou, China) were used to infect cells using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s protocol.41,42 To inhibit the MIEF1-related mitochondrial fission, shRNA against MIEF1 was trisected into cancer cells. The negative control group was transfected with a negative control shRNA. Transfection was performed for approximately 48 hours. Western blotting was subsequently used to verify the knockdown efficiency after harvesting the transfected cells.43
Statistical analyses
All results presented in this study were acquired from at least three independent experiments. The statistical analyses were performed using SPSS 16.0 (SPSS, Inc., Chicago, IL, USA). All results in the present study were analyzed with one-way analysis of variance, followed by Tukey’s test. P<0.05 was considered statistically significant.
Results
TAZ deletion promotes HepG2 liver cancer cell apoptosis
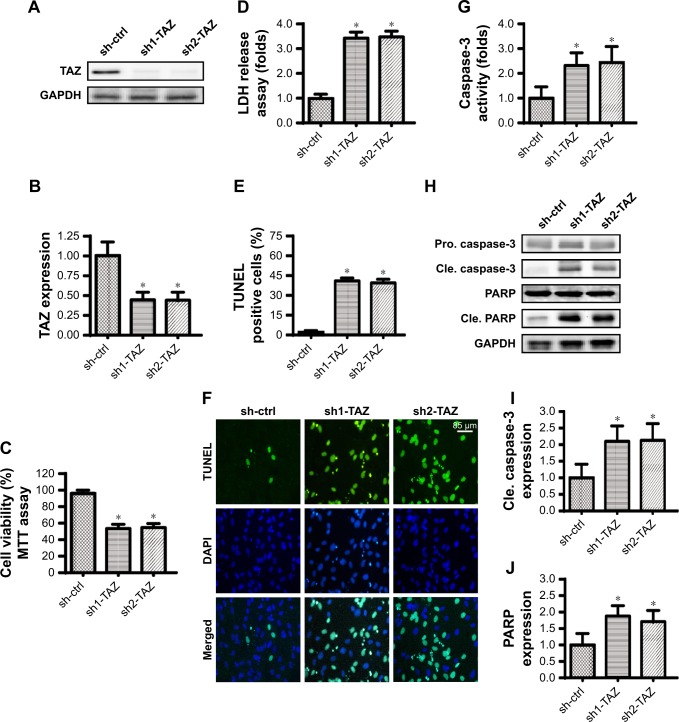
Two independent shRNAs against TAZ were used to silence the expression of TAZ in the HepG2 liver cancer cell line. The knockdown efficiency was confirmed via Western blotting (Figure 1A and B). The cell viability was subsequently measured via MTT assay. Compared with the control shRNA group, the transfection of TAZ shRNA significantly reduced the cell viability in HepG2 cells (Figure 1C). To confirm whether the cell viability reduction resulted from increased cell death, an LDH release assay was used. As shown in Figure 1D, the loss of TAZ in HepG2 cells promoted LDH release in the medium compared with that in the control group. This information was further supported via TUNEL staining. Compared with the control group, the number of TUNEL positive cells was rapidly increased in HepG2 cells transfected with TAZ shRNA (Figure 1E and F). These data illustrate that TAZ deletion seems to activate cell death in liver cancer cells in vitro. It was subsequently verified whether cell death was executed via apoptosis. The caspase-3 activity was measured to reflect the activation of the caspase family. As shown in Figure 1G, compared with the control group, TAZ shRNA transfection significantly elevated the caspase-3 activity. This result was further supported by measuring the expression of cleaved caspase-3 and its substrate cleaved PARP. Western blot analysis demonstrated that the levels of cleaved caspase-3 and cleaved PARP were drastically upregulated in response to TAZ shRNA transfection (Figure 1H–J), compared with the control shRNA transfection. Overall, this information indicated that the loss of TAZ induced death in HepG2 liver cancer cells via the activation of apoptosis.
Figure 1.
TAZ deletion induces apoptosis in liver cancer cells in vitro.
Notes: (A, B) Two independent TAZ shRNAs were transfected into HepG2 liver cancer cells. Western blotting was used to observe the knockdown efficiency. (C) The MTT assay was used for cell viability detection. Two independent TAZ shRNAs were transfected into HepG2 liver cancer cells. (D) LDH release assay for cell death. (E, F) TUNEL staining for apoptotic cells. Green dots were recorded, and the ratio of TUNEL-positive cells was evaluated to reflect cell apoptosis. (G) Caspase-3 activity was measured via ELISA. Two independent shRNAs were transfected into HepG2 liver cancer cells. (H–J) Western blotting was performed to analyze the expression of pro-apoptotic proteins, such as cleaved caspase-3 and cleaved PARP. *P<0.05 vs sh-ctrl.
Abbreviations: Pro. caspase-3, pro-apoptotic caspase-3; Cle. caspase-3, cleaved caspase-3; LDH, lactate dehydrogenase; sh-ctrl, control shRNA; TAZ, transcriptional co-activator with PDZ-binding motif.
TAZ knockdown impairs HepG2 liver cancer cell proliferation and migration
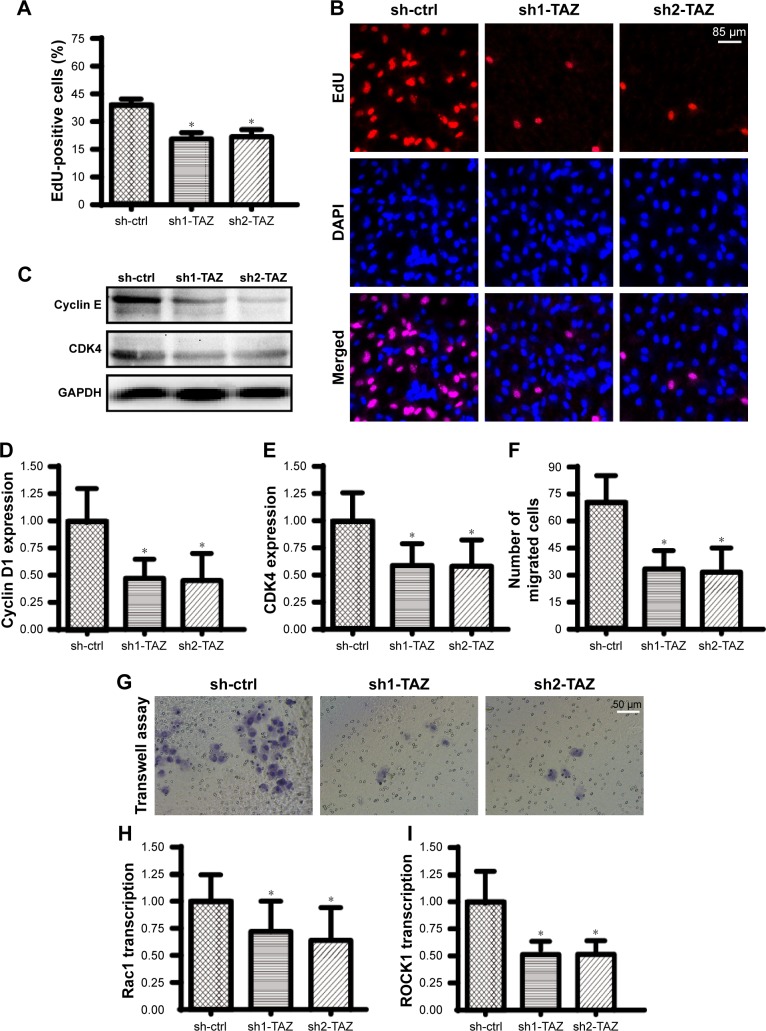
In addition to cell death, we also observed the influence of TAZ on cell migration and proliferation. EdU staining was used to observe the cell number at the S-phase. As shown in Figure 2A and B, compared with the control group, the number of EdU-positive cells was rapidly reduced after transfection with TAZ shRNA, suggesting that TAZ deletion attenuated the proliferation in HepG2 liver cancer cells in vitro. This conclusion was further validated by measuring the cell cycle protein expressions. Western blot analyses indicated that the expressions of Cyclin D1 and CDK4 were abundant in normal HepG2 liver cancer cells (Figure 2C–E). However, following exposure to TAZ shRNA, the levels of Cyclin D1 and CDK4 were rapidly downregulated. This information indicates that HepG2 liver cancer cell proliferation is controlled by TAZ by affecting the cell cycle protein stabilization.
Figure 2.
TAZ deletion impairs HepG2 liver cancer cell proliferation and migration.
Notes: (A, B) EdU assays were performed to verify the functional role of TAZ in HepG2 cell growth. The number of EdU-positive cells was recorded. (C–E) To assess the cellular response to TAZ deletion, protein was collected, and Western blotting was used to analyze the expression of Cyclin D1 and CDK4. (F, G) Transwell assays were used to observe HepG2 cell migration. Z-VAD-FMK was used to exclude the influence of apoptosis on cell migration. (H, I) RNA was isolated from HepG2 cells, and qPCR was conducted to analyze the expression of ROCK1 and Rac1, which are key factors involved in the cancer cell migratory response. *P<0.05 vs sh-ctrl (control shRNA).
Abbreviations: sh-ctrl, control shRNA; TAZ, transcriptional co-activator with PDZ-binding motif.
Furthermore, a transwell assay was used to observe the mobilization of HepG2 liver cancer cells in response to TAZ deletion. To exclude the influence of apoptosis on the transwell assay results, a caspase inhibitor (Z-VAD-FMK) was used in the TAZ-deleted cells, and these cells were used as the control group to verify the influence of apoptosis on the transwell assay. As shown in Figure 2F and G, compared with the control group, the loss of TAZ reduced the number of migrated cells. Interestingly, although treatment with Z-VAD-FMK could partially reverse the cell mobility, the ratio of migrated cells remained lower in the TAZ knockdown group than that in the control group (Figure 2F and G). These data indicated that cell migration was closely regulated by TAZ. At the molecular level, the transcription of metastatic genes, such as Rac1 and ROCK1, was rapidly repressed in response to TAZ (Figure 2H and I). Overall, these findings indicate that HepG2 cell migration and proliferation are also modulated by TAZ.
TAZ repression impairs mitochondrial bioenergetics
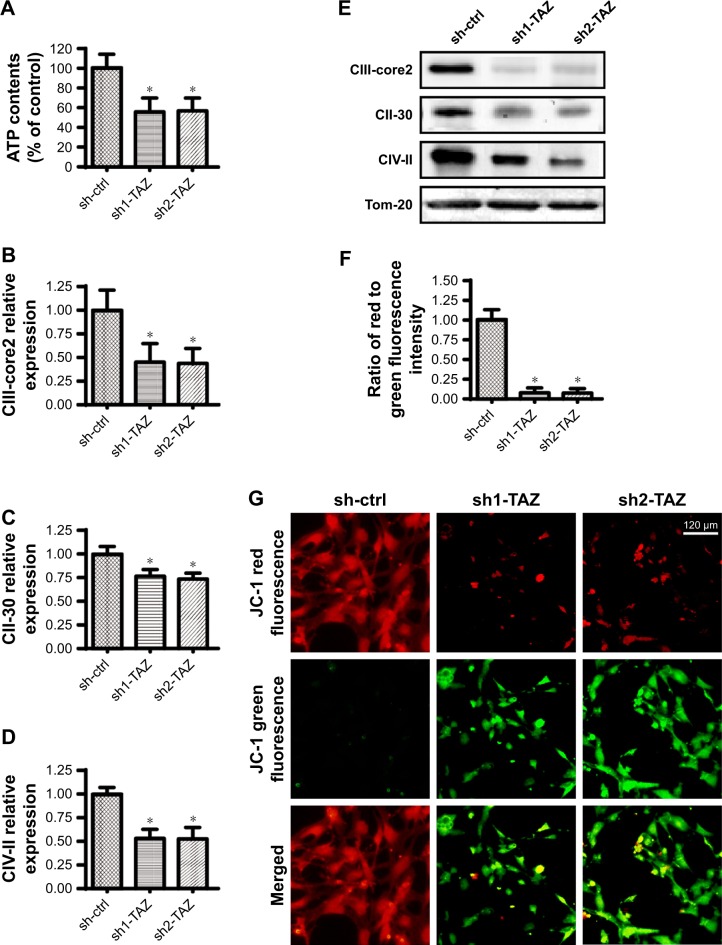
Cancer viability, growth, and metastasis are strongly regulated by mitochondrial bioenergetics, which provide sufficient energy to sustain cell proliferation, metabolism, differentiation, and invasion.44,45 In light of the central role played by mitochondrial bioenergetics in various cancer biological functions,46,47 we questioned whether TAZ affected mitochondrial bioenergetics. First, the total ATP production was rapidly reduced in response to TAZ deletion compared with that in the control group (Figure 3A). Previous studies have reported that ATP is primarily generated by mitochondria via the mitochondrial respiratory function,48,49 which converts the mitochondrial membrane potential into chemical energy.50 Interestingly, the expression of the mitochondrial respiratory complex was clearly downregulated once TAZ was silenced (Figure 3B–E). In response to the mitochondrial respiratory dysfunction, the mitochondrial membrane potential, assessed via JC-1 staining, was also reduced in TAZ-deleted cells (Figure 3F–G). Taken together, these data illustrate the important function of TAZ to sustain mitochondrial respiratory function and cancer bioenergetics in HepG2 cells.
Figure 3.
TAZ governs mitochondrial respiratory function and energy metabolism.
Notes: (A) Cellular ATP production was determined in an HepG2 cell that was transfected with TAZ shRNA. (B–E) After treatment, proteins were isolated from the HepG2 cell, and Western blotting was subsequently applied to analyze the expression of the mitochondrial respiratory complex. (F–G) Mitochondrial membrane potential was observed using JC-1 probe. The red-to-green fluorescence intensity was used to quantify the mitochondrial membrane potential. *P<0.05 vs sh-ctrl.
Abbreviations: sh-ctrl, control shRNA; TAZ, transcriptional co-activator with PDZ-binding motif.
Loss of TAZ activates MIEF1-related mitochondrial fission
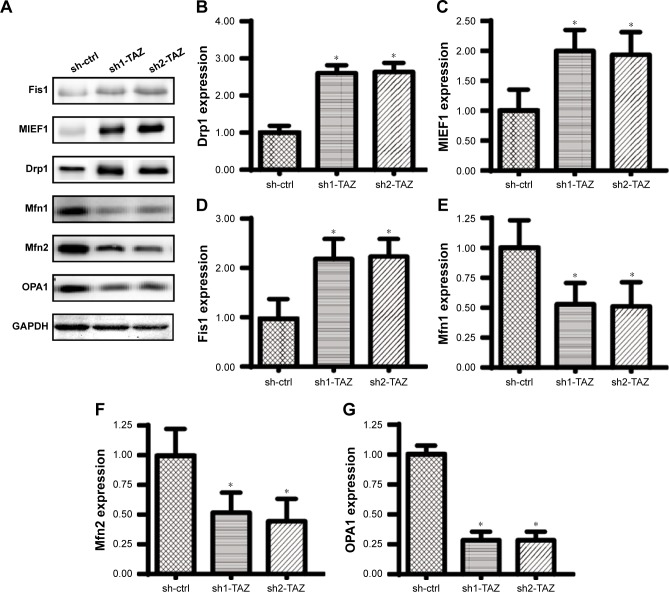
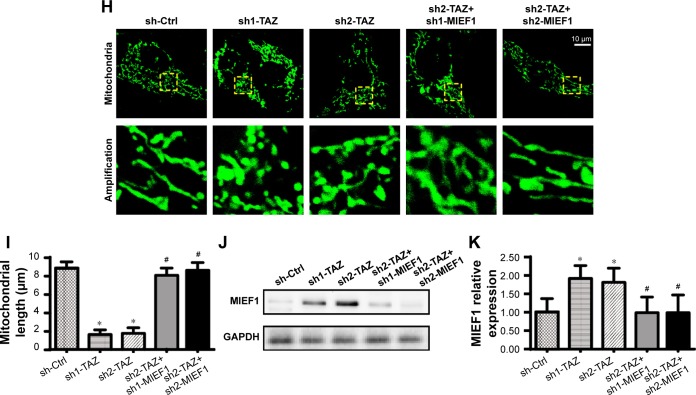
Recent studies have identified MIEF1-related mitochondrial fission as the upstream mediator of mitochondrial homeostasis.51 Increased MIEF1 promotes mitochondrial fission, and the latter activates mitochondrial stress, including mitochondrial dysfunction, mitochondrial apoptosis, mitochondrial oxidation, and mitochondrial calcium overloading. Thus, we wondered whether MIEF1-related mitochondrial fission was activated by TAZ deletion and contributed to the mitochondrial dysfunction and cell injury in HepG2 cells. First, Western blot analyses demonstrated that the parameters related to mitochondrial fission were rapidly increased in the TAZ-deleted cells, including Drp1, MIEF1, and Fis1 (Figure 4A–G). In contrast, the expressions of mitochondrial fusion markers, such as Mfn1, Mfn2, and Opa1 (Figure 4A–G), were downregulated in response to TAZ deletion. This information indicated that the loss of TAZ activated mitochondrial fission and repressed mitochondrial fusion in HepG2 cells. Mitochondrial fission was subsequently observed using an immunofluorescence assay according to previous studies.3,44 The average length of the mitochondria in cells was recorded to reflect the degree of mitochondrial fission. As shown in Figure 4H and I, compared with the control group, TAZ deletion promoted the formation of mitochondrial fragmentation with a shorter length. Furthermore, to verify whether MIEF1 was involved in TAZ-induced mitochondrial fission, shRNA against MIEF1 was used to neutralize the promotive effects of TAZ deletion on MIEF1 activation. The knockdown efficiency was confirmed via Western blotting as shown in Figure 4J and K. The mitochondrial fission and mitochondrial length were subsequently measured again. Compared with the control group, TAZ deletion increased the levels of MIEF1 (Figure 4J and K), an effect that was accompanied by a decrease in the mitochondria length (Figure 4H and I), indicative of mitochondrial fission activation. However, the loss of MIEF1 abolished the TAZ-mediated MIEF1 upregulation (Figure 4J and K), and this result also occurred with an increase in the mitochondrial length (Figure 4H and I), indicative of mitochondrial fission inhibition. Overall, these findings confirm that mitochondrial fission is effectively activated by TAZ deletion via MIEF1 upregulation.
Figure 4.
TAZ deletion triggers mitochondrial fission in a manner dependent on MIEF1.
Notes: (A–G) Western blotting was applied to evaluate the expression of pro-fission proteins, including Drp1, MIEF1 and Fis1. Pro-fusion factors, such as Mfn1, Mfn2 and Opa1, were also evaluated via Western blotting. (H, I) Immunofluorescence assay for mitochondria using a mitochondrial-specific antibody Tom-20. The average length of the mitochondria was measured, and this parameter was used to quantify mitochondrial fission. (J, K) The expression of MIEF1 was also evaluated via Western blotting. shRNA against MIEF1 was transfected into HepG2 cells, and the expression of MIEF1 was subsequently measured. *P<0.05 vs sh-ctrl, #P<0.05 vs sh-TAZ.
Abbreviations: MIEF1, mitochondrial elongation factor 1; sh-ctrl, control shRNA; TAZ, transcriptional co-activator with PDZ-binding motif.
MIEF1-related mitochondrial fission promotes mitochondrial stress
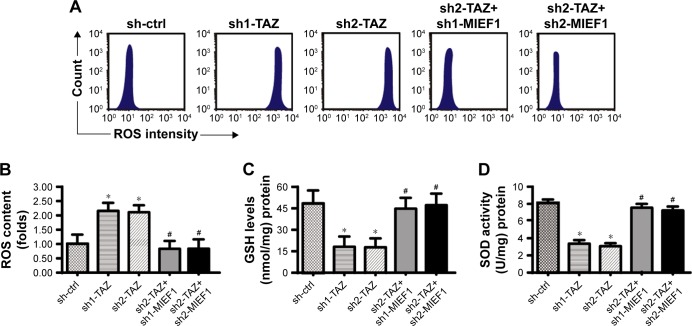
To explain the role of MIEF1-related mitochondrial fission in HepG2 cells, mitochondrial stress was detected. First, the mROS production was determined via flow cytometry. Compared with the control group, TAZ deletion augmented the production of mROS (Figure 5A and B), and this effect could be negated by MIEF1 shRNA transfection. As a consequence of the mROS overloading, the concentrations of SOD, GSH, and GPX were substantially downregulated in the TAZ-deleted cells (Figure 5C–E), indicative of the redox imbalance achieved by TAZ deletion. Interestingly, the loss of MIEF1 reversed the levels of SOD, GSH, and GPX (Figure 5C–E), suggesting that the cell antioxidant homeostasis was regulated by MIEF1-related mitochondrial fission.
Figure 5.
MIEF1-related mitochondrial fission modulates mitochondrial stress.
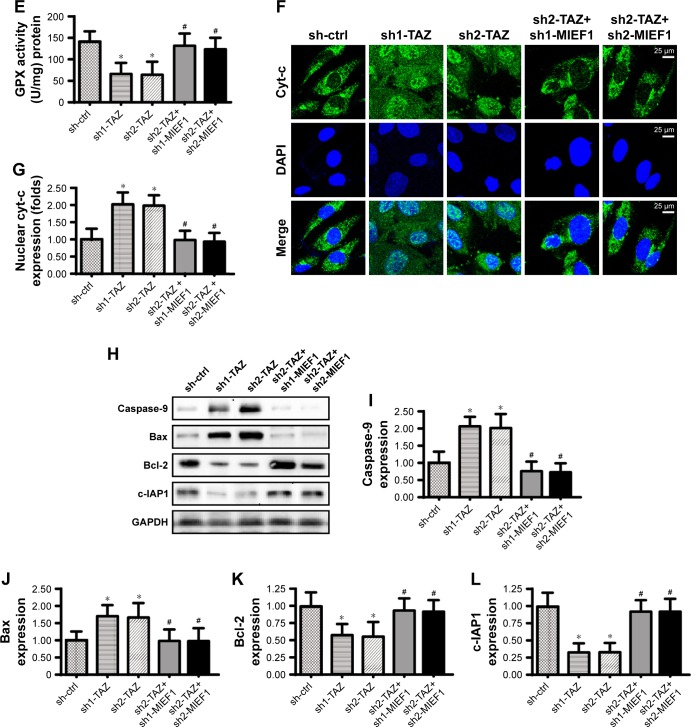
Notes: (A, B) Mitochondrial ROS production was analyzed using flow cytometry. The relative ROS content was recorded as a ratio to that of the control group. shRNAs against TAZ and MIEF1 were transfected into HepG2 cells. (C–E) The concentrations of cellular antioxidants, such as GSH, GPX, and SOD, were measured using ELISA. (F, G) Immunofluorescence assay for cyt-c liberation. Relative nuclear cyt-c expression was recorded as a ratio to that of the control group. (H–L) Western blotting analysis was used to analyze the alterations of mitochondrial apoptotic proteins, such as caspase-9, Bax, Bcl-2, and c-IAP1. shRNAs against TAZ and MIEF1 were transfected into HepG2 cells. *P<0.05 vs sh-ctrl, #P<0.05 vs sh-TAZ.
Abbreviations: MIEF1, mitochondrial elongation factor 1; ROS, reactive oxygen species; sh-ctrl, control shRNA; TAZ, transcriptional co-activator with PDZ-binding motif.
Furthermore, mitochondrial apoptosis was measured. Mitochondrial cyt-c liberation into cytoplasm is the key feature of mitochondrial injury.52 With the assistance of immunofluorescence assay, we demonstrated that TAZ deletion contributed to the cyt-c liberation into the cytoplasm (Figure 5F and G), and this effect was neutralized by MIEF1 shRNA transfection. Excessive cyt-c liberation would facilitate caspase-9 activation, which was accompanied by a decrease in mitochondrial anti-apoptotic proteins. As shown in Figure 5H–L, compared with the control group, TAZ deletion elevated the expression of caspase-9/Bax and reduced the levels of Bcl-2/c-IAP. Interestingly, the depression of MIEF1 reversed the content of Bcl-2/c-IAP and prevented the upregulation of caspase-9/Bax (Figure 5H–L). Overall, this information highlighted the necessary role played by MIEF1-related mitochondrial fission in TAZ-induced mitochondrial stress in HepG2 liver cancer cells.
TAZ modulates MIEF1 expression via CaMKII signaling pathway
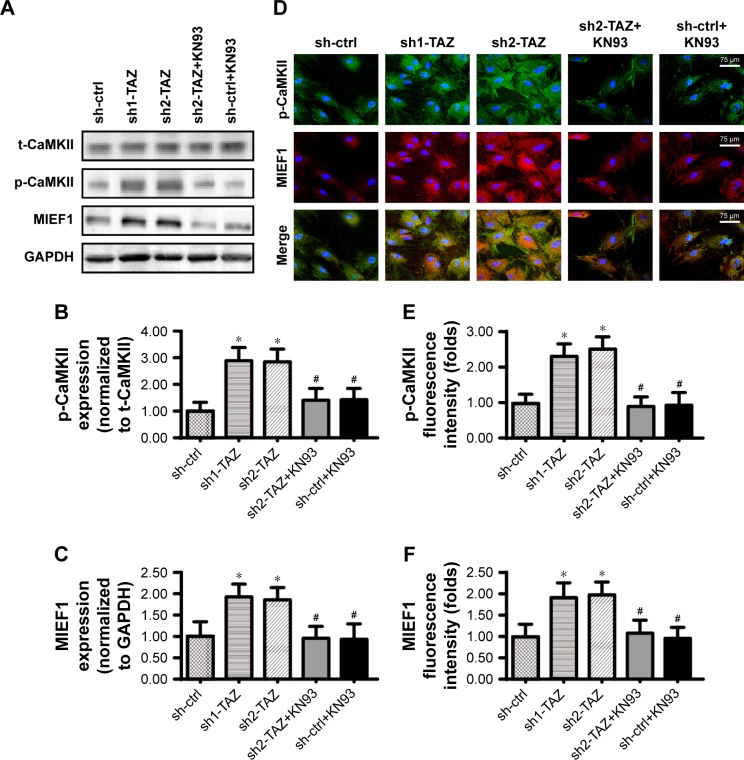
Recent studies have indicated the links between CaMKII activation and mitochondrial fission initiation.53 In the present study, we aimed to determine whether TAZ increases MIEF1 expression via the CaMKII signaling pathway. Western blot analyses demonstrated that the CaMKII pathway was activated by TAZ deletion, as indicated by increased p-CaMKII expression (Figure 6A–C). Interestingly, blockade of the CaMKII pathway using KN93 not only repressed the p-CaMKII expression but also reduced the MIEF1 expression in TAZ-deleted cells (Figure 6A–C). These data suggested that the CaMKII pathway was required for TAZ-mediated MIEF1 upregulation. This conclusion was further supported via an immunofluorescence assay using p-CaMKII and MIEF1 antibodies. Low fluorescence intensities of p-CaMKII and MIEF1 were noted in normal HepG2 cells (Figure 6D–F). Interestingly, TAZ deletion rapidly upregulated the levels of p-CaMKII and MIEF1, and this effect could be abolished by KN93 (Figure 6D–F). Overall, we provide evidence to support that TAZ regulates MIEF1 expression via the CaMKII signaling pathway.
Figure 6.
TAZ regulates MIEF1 expression via the CaMKII signaling pathway.
Notes: (A–C) Western blotting was used to analyze p-CaMKII and MIEF1 expressions. The CaMKII blocker KN93 was used to prevent CaMKII activation in TAZ-deleted cells. (D–F) Immunofluorescence assay for MIEF1 and p-CaMKII. The relative fluorescence intensities of MIEF1 and p-CaMKII were measured. *P<0.05 vs sh-ctrl; #P<0.05 vs sh-TAZ.
Abbreviations: MIEF1, mitochondrial elongation factor 1; sh-ctrl, control shRNA; TAZ, transcriptional co-activator with PDZ-binding motif.
Discussion
TAZ has been found to be overexpressed in several kinds of tumors.54 However, the functional role of TAZ in liver cancer development and progression has not been adequately explored. Our results provided an answer to this issue, and we found that TAZ was associated with liver cancer survival by sustaining mitochondrial homeostasis. The knockdown of TAZ promoted HepG2 liver cancer death via the activation of mitochondrial apoptosis. Moreover, the loss of TAZ also impaired cancer cell proliferation and migration. Biological analysis demonstrated that TAZ repression enhanced mitochondrial fission by upregulating MIEF1. Activated MIEF1-related mitochondrial stress induced mitochondrial stress, including mitochondrial bioenergetics disorder, mitochondrial oxidative injury, and mitochondrial apoptosis activation. The knockdown of MIEF1 sustained mitochondrial function and structure in TAZ-deleted cells. Moreover, we found that TAZ modulated MIEF1 via the CaMKII signaling pathway; the blockade of the CaMKII axis favored cancer survival and maintained mitochondrial function. Overall, these data help us to further understand the molecular mechanisms of TAZ that underlie the biological characterization of liver cancer. Moreover, our results also indicate that TAZ and MIEF1-related mitochondrial fission are potential targets to treat liver cancer via the induction of cancer cell apoptosis and mitochondrial dysfunction.
TAZ is the core component in the Hippo pathway that controls the tissue size and oncogenesis. TAZ has been demonstrated to be involved in the development of multiple organs, including the lungs and the heart, as well as in numerous cellular processes, including stem cell differentiation, cell proliferation, and the EMT.55 Recent studies have reported the links between TAZ and cancer development. TAZ is significantly increased in patients with colon cancer, and its expression was downregulated by radiation.16 Moreover, in cervical cancer, TAZ promotes the tumorigenicity and inhibits cancer apoptosis.17 Similar to the previous findings, the present study indicated that the loss of TAZ via transfection shRNA reduces the viability of the liver cancer HepG2 cell line via the activation of apoptosis. These results, combined with the previous studies, substantiated the sufficiency and necessity of pathogenically relevant degrees of TAZ to promote cancer survival.
Notably, there is strong evidence supporting the regulatory role of TAZ in inducing mitochondrial damage in different types of cells. In cardioskeletal myopathy, TAZ depression reduces mitochondrial ATP production and consequently induces muscle weakness.56 In fatty liver disease, TAZ downregulation promotes lipid accumulation and suppresses mitochondrial respiratory function.57 Moreover, TAZ is also involved in mitophagy regulation,58 mitochondrial cardiolipin metabolism,59 and the mitochondrial redox balance.60 In the present study, we found that TAZ deletion evoked mitochondrial bioenergetics disorder, mitochondrial oxidative injury, mitochondrial potential reduction, mitochondrial fission, and mitochondrial apoptosis. These results lay the foundation for detailed investigations of the molecular mechanisms of TAZ repression and its downstream events in liver cancer development and progression.
Furthermore, we found that TAZ modulated mitochondrial homeostasis by activating MIEF1-related mitochondrial fission. MIEF1 is a protein expressed on the outer membrane of the mitochondria. Ample MIEF1 expression promotes the accumulation of Drp1 on the surface of mitochondria, and the latter interacts with mitochondria to form the contractile ring61 to proceed with mitochondrial division. Increased MIEF1 is required for successive mitochondrial fission in various disease models, such as cardiac IR injury,12 cell reprogramming,62 and diabetes.13 The present study further demonstrated that MIEF1-related mitochondrial fission was also activated by TAZ and contributed to mitochondrial stress. This information fills in the gap regarding how TAZ handles mitochondrial homeostasis, and this process is employed with MIEF1-related mitochondrial fission. To this end, we found that TAZ controlled MIEF1 expression via the CaMKII signaling pathway. Furthermore, more robust data concerning the relationship of CaMKII and mitochondrial fission have been provided by genetic loss- and gain-of-function studies in vivo and in vitro. In cardiac hypertrophy63 and neuronal apoptosis,64 CaMKII has been acknowledged as the upstream trigger of mitochondrial fission. Similar to our finding, a recent study in liver cancer also demonstrates that mitochondrial fission is primarily regulated by CaMKII.10 These findings illustrate the functional importance of CaMKII in regulating mitochondrial stress in liver cancer.
There are several limitations in the present study. First, only one liver cancer cell line was used in the present study. Additional experiments using other liver cancer cell lines are necessary to support our findings. Animal studies are also required to validate our results. At the molecular level, shRNA was used to knockdown the expression of TAZ in HepG2 cells and more studies using adenovirus-mediated TAZ overexpression assay are also vital to further verify the role of TAZ in liver cancer cell death. To the end, we cannot exclude the influence of extrinsic apoptosis pathway on liver cancer cell death. Accordingly, it is necessary to figure out the relationship between TAZ and extrinsic apoptosis pathway in liver cancer.
Conclusion
Our results explained the central role of TAZ deletion and the subsequent MIEF1-related mitochondrial fission that leads to apoptosis execution in liver cancer. Based on our results, strategies to repress TAZ expression and enhance MIEF1-related mitochondrial fission may bring a new entry point for the treatment of liver cancer. However, additional research using animal models and/or human samples is required in the future to validate our findings.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Xie LL, Shi F, Tan Z, Li Y, Bode AM, Cao Y. Mitochondrial network structure homeostasis and cell death. Cancer Sci. 2018;109(12):3686–3694. doi: 10.1111/cas.13830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu L, Jiang B, Chen Z, et al. Cytisine induces endoplasmic reticulum stress caused by calcium overload in HepG2 cells. Oncol Rep. 2018;39(3):1475–1484. doi: 10.3892/or.2018.6200. [DOI] [PubMed] [Google Scholar]
- 3.Zhou H, Zhu P, Wang J, Zhu H, Ren J, Chen Y. Pathogenesis of cardiac ischemia reperfusion injury is associated with CK2α-disturbed mitochondrial homeostasis via suppression of FUNDC1-related mitophagy. Cell Death Differ. 2018;25(6):1080–1093. doi: 10.1038/s41418-018-0086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou H, Wang J, Zhu P, et al. NR4A1 aggravates the cardiac microvascular ischemia reperfusion injury through suppressing FUNDC1-mediated mitophagy and promoting Mff-required mitochondrial fission by CK2α. Basic Res Cardiol. 2018;113(4):23. doi: 10.1007/s00395-018-0682-1. [DOI] [PubMed] [Google Scholar]
- 5.Jin Q, Li R, Hu N, et al. DUSP1 alleviates cardiac ischemia/reperfusion injury by suppressing the Mff-required mitochondrial fission and Bnip3-related mitophagy via the JNK pathways. Redox Biol. 2018;14:576–587. doi: 10.1016/j.redox.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srinivasan S, Guha M, Kashina A, Avadhani NG. Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection. Biochim Biophys Acta Bioenerg. 2017;1858(8):602–614. doi: 10.1016/j.bbabio.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H, Wang S, Zhu P, Hu S, Chen Y, Ren J. Empagliflozin rescues diabetic myocardial microvascular injury via AMPK-mediated inhibition of mitochondrial fission. Redox Biol. 2018;15:335–346. doi: 10.1016/j.redox.2017.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagerweij T, Dusoswa SA, Negrean A, et al. Optical clearing and fluorescence deep-tissue imaging for 3D quantitative analysis of the brain tumor microenvironment. Angiogenesis. 2017;20(4):533–546. doi: 10.1007/s10456-017-9565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu P, Zhang G, Sha L, Hou S. DUSP1 alleviates cerebral ischaemia reperfusion injury via inactivating JNK-Mff pathways and repressing mitochondrial fission. Life Sci. 2018;210:251–262. doi: 10.1016/j.lfs.2018.08.049. [DOI] [PubMed] [Google Scholar]
- 10.Sun X, Cao H, Zhan L, et al. Mitochondrial fission promotes cell migration by Ca2+/CaMKII/ERK/FAK pathway in hepatocellular carcinoma. Liver Int. 2018;38(7):1263–1272. doi: 10.1111/liv.13660. [DOI] [PubMed] [Google Scholar]
- 11.Henderson D, Huebner C, Markowitz M, et al. Do developmental temperatures affect redox level and lifespan in C. elegans through upregulation of peroxiredoxin? Redox Biol. 2018;14:386–390. doi: 10.1016/j.redox.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samangouei P, Crespo-Avilan GE, Cabrera-Fuentes H, et al. MiD49 and MiD51: new mediators of mitochondrial fission and novel targets for cardioprotection. Cond Med. 2018;1(5):239–246. [PMC free article] [PubMed] [Google Scholar]
- 13.Wada J, Nakatsuka A. Mitochondrial dynamics and mitochondrial dysfunction in diabetes. Acta Med Okayama. 2016;70(3):151–158. doi: 10.18926/AMO/54413. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Liu L, Wu S, Xing D. Drp1, Mff, Fis1, and MiD51 are coordinated to mediate mitochondrial fission during UV irradiation-induced apoptosis. FASEB J. 2016;30(1):466–476. doi: 10.1096/fj.15-274258. [DOI] [PubMed] [Google Scholar]
- 15.Kriz V, Korinek V. Wnt, RSPO and Hippo signalling in the intestine and intestinal stem cells. Genes. 2018;9(1):20. doi: 10.3390/genes9010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pathak S, Banerjee A, Meng WJ, Kumar Nandy S, Gopinath M, Sun XF. Significant expression of tafazzin (TAZ) protein in colon cancer cells and its downregulation by radiation. Int J Radiat Biol. 2018;94(1):79–87. doi: 10.1080/09553002.2018.1400191. [DOI] [PubMed] [Google Scholar]
- 17.Chen M, Zhang Y, Zheng PS. Tafazzin (TAZ) promotes the tumorigenicity of cervical cancer cells and inhibits apoptosis. PLoS One. 2017;12(5):e0177171. doi: 10.1371/journal.pone.0177171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong W, Gao X, Wang S, et al. Prox1-GFP/Flt1-DsRed transgenic mice: an animal model for simultaneous live imaging of angiogenesis and lymphangiogenesis. Angiogenesis. 2017;20(4):581–598. doi: 10.1007/s10456-017-9572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antoniou C, Chatzimichail G, Xenofontos R, et al. Melatonin systemically ameliorates drought stress-induced damage in Medicago sativa plants by modulating nitro-oxidative homeostasis and proline metabolism. J Pineal Res. 2017;62(4):e12401. doi: 10.1111/jpi.12401. [DOI] [PubMed] [Google Scholar]
- 20.Antunes F, Brito PM. Quantitative biology of hydrogen peroxide signaling. Redox Biol. 2017;13:1–7. doi: 10.1016/j.redox.2017.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan T, Pi H, Li M, et al. Inhibiting MT2-TFE3-dependent autophagy enhances melatonin-induced apoptosis in tongue squamous cell carcinoma. J Pineal Res. 2018;64(2):e12457. doi: 10.1111/jpi.12457. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Li X, Chen C, Chan MTV, Wu WKK, Shen J. Melatonin inhibits nucleus pulposus (NP) cell proliferation and extracellular matrix (ECM) remodeling via the melatonin membrane receptors mediated PI3K-Akt pathway. J Pineal Res. 2017;63(3):e12435. doi: 10.1111/jpi.12435. [DOI] [PubMed] [Google Scholar]
- 23.Armartmuntree N, Murata M, Techasen A, et al. Prolonged oxidative stress down-regulates early B cell factor 1 with inhibition of its tumor suppressive function against cholangiocarcinoma genesis. Redox Biol. 2018;14:637–644. doi: 10.1016/j.redox.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sajib S, Zahra FT, Lionakis MS, German NA, Mikelis CM. Mechanisms of angiogenesis in microbe-regulated inflammatory and neoplastic conditions. Angiogenesis. 2018;21(1):1–14. doi: 10.1007/s10456-017-9583-4. [DOI] [PubMed] [Google Scholar]
- 25.Koentges C, Pepin ME, Müsse C, et al. Gene expression analysis to identify mechanisms underlying heart failure susceptibility in mice and humans. Basic Res Cardiol. 2018;113(1):8. doi: 10.1007/s00395-017-0666-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu D, Zeng X, Li X, Mehta JL, Wang X. Role of NLRP3 inflammasome in the pathogenesis of cardiovascular diseases. Basic Res Cardiol. 2018;113(1):5. doi: 10.1007/s00395-017-0663-9. [DOI] [PubMed] [Google Scholar]
- 27.Landry NM, Cohen S, Dixon IMC. Periostin in cardiovascular disease and development: a tale of two distinct roles. Basic Res Cardiol. 2018;113(1):1. doi: 10.1007/s00395-017-0659-5. [DOI] [PubMed] [Google Scholar]
- 28.Koopman CD, Zimmermann WH, Knöpfel T, de Boer TP. Cardiac optogenetics: using light to monitor cardiac physiology. Basic Res Cardiol. 2017;112(5):56. doi: 10.1007/s00395-017-0645-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong H, Tao T, Chen S, et al. MicroRNA-143 promotes cardiac ischemia-mediated mitochondrial impairment by the inhibition of protein kinase Cepsilon. Basic Res Cardiol. 2017;112(6):60. doi: 10.1007/s00395-017-0649-7. [DOI] [PubMed] [Google Scholar]
- 30.Zhou H, Zhu P, Guo J, et al. Ripk3 induces mitochondrial apoptosis via inhibition of FUNDC1 mitophagy in cardiac IR injury. Redox Biol. 2017;13:498–507. doi: 10.1016/j.redox.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou H, Zhang Y, Hu S, et al. Melatonin protects cardiac microvasculature against ischemia/reperfusion injury via suppression of mitochondrial fission-VDAC1-HK2-mPTP-mitophagy axis. J Pineal Res. 2017;63(1):e12413. doi: 10.1111/jpi.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conradi LC, Brajic A, Cantelmo AR, et al. Tumor vessel disintegration by maximum tolerable PFKFB3 blockade. Angiogenesis. 2017;20(4):599–613. doi: 10.1007/s10456-017-9573-6. [DOI] [PubMed] [Google Scholar]
- 33.Zhou H, Ma Q, Zhu P, Ren J, Reiter RJ, Chen Y. Protective role of melatonin in cardiac ischemia-reperfusion injury: from pathogenesis to targeted therapy. J Pineal Res. 2018;64(3):e12471. doi: 10.1111/jpi.12471. [DOI] [PubMed] [Google Scholar]
- 34.Casadonte L, Verhoeff BJ, Piek JJ, Vanbavel E, Spaan JAE, Siebes M. Influence of increased heart rate and aortic pressure on resting indices of functional coronary stenosis severity. Basic Res Cardiol. 2017;112(6):61. doi: 10.1007/s00395-017-0651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou H, Li D, Zhu P, et al. Inhibitory effect of melatonin on necroptosis via repressing the Ripk3-PGAM5-CypD-mPTP pathway attenuates cardiac microvascular ischemia-reperfusion injury. J Pineal Res. 2018;65(3):e12503. doi: 10.1111/jpi.12503. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez NR, Liou R, Kurth F, Jiang H, Saver J. Antiangiogenesis and medical therapy failure in intracranial atherosclerosis. Angiogenesis. 2018;21(1):23–35. doi: 10.1007/s10456-017-9578-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H, Li D, Zhu P, et al. Melatonin suppresses platelet activation and function against cardiac ischemia/reperfusion injury via PPARγ/FUNDC1/mitophagy pathways. J Pineal Res. 2017;63(4):e12438. doi: 10.1111/jpi.12438. [DOI] [PubMed] [Google Scholar]
- 38.Espinosa-Díez C, Miguel V, Vallejo S, et al. Role of glutathione biosynthesis in endothelial dysfunction and fibrosis. Redox Biol. 2018;14:88–99. doi: 10.1016/j.redox.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crooke A, Huete-Toral F, Colligris B, Pintor J. The role and therapeutic potential of melatonin in age-related ocular diseases. J Pineal Res. 2017;63(2):e12430. doi: 10.1111/jpi.12430. [DOI] [PubMed] [Google Scholar]
- 40.Zhou HAO, Yang J, Xin T, et al. Exendin-4 enhances the migration of adipose-derived stem cells to neonatal rat ventricular cardiomyocyte-derived conditioned medium via the phosphoinositide 3-kinase/Akt-stromal cell-derived factor-1α/CXC chemokine receptor 4 pathway. Mol Med Rep. 2015;11(6):4063–4072. doi: 10.3892/mmr.2015.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou H, Hu S, Jin Q, et al. Mff-dependent mitochondrial fission contributes to the pathogenesis of cardiac microvasculature ischemia/reperfusion injury via induction of mROS-mediated cardiolipin oxidation and HK2/VDAC1 disassociation-involved mPTP opening. J Am Heart Assoc. 2017;6(3):e005328. doi: 10.1161/JAHA.116.005328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou H, Du W, Li Y, et al. Effects of melatonin on fatty liver disease: the role of NR4A1/DNA-PKcs/p53 pathway, mitochondrial fission, and mitophagy. J Pineal Res. 2018;64(1):e12450. doi: 10.1111/jpi.12450. [DOI] [PubMed] [Google Scholar]
- 43.Erland LAE, Yasunaga A, Li ITS, Murch SJ, Saxena PK. Direct visualization of location and uptake of applied melatonin and serotonin in living tissues and their redistribution in plants in response to thermal stress. J Pineal Res. 2019;66(1):e12527. doi: 10.1111/jpi.12527. [DOI] [PubMed] [Google Scholar]
- 44.Zhou H, Shi C, Hu S, Zhu H, Ren J, Chen Y. BI1 is associated with microvascular protection in cardiac ischemia reperfusion injury via repressing Syk-Nox2-Drp1-mitochondrial fission pathways. Angiogenesis. 2018;21(3):599–615. doi: 10.1007/s10456-018-9611-z. [DOI] [PubMed] [Google Scholar]
- 45.Zhou H, Wang J, Zhu P, Hu S, Ren J. Ripk3 regulates cardiac microvascular reperfusion injury: the role of IP3R-dependent calcium overload, XO-mediated oxidative stress and F-action/filopodia-based cellular migration. Cell Signal. 2018;45:12–22. doi: 10.1016/j.cellsig.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 46.Zhou H, Yue Y, Wang J, Ma Q, Chen Y. Melatonin therapy for diabetic cardiomyopathy: a mechanism involving Syk-mitochondrial complex I-SERCA pathway. Cell Signal. 2018;47:88–100. doi: 10.1016/j.cellsig.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 47.Zhou H, Wang S, Hu S, Chen Y, Ren J. ER-Mitochondria microdomains in cardiac ischemia-reperfusion injury: a fresh perspective. Front Physiol. 2018;9:755. doi: 10.3389/fphys.2018.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu P, Hu S, Jin Q, et al. Ripk3 promotes ER stress-induced necroptosis in cardiac IR injury: a mechanism involving calcium overload/XO/ROS/mPTP pathway. Redox Biol. 2018;16:157–168. doi: 10.1016/j.redox.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu H, Jin Q, Li Y, et al. Melatonin protected cardiac microvascular endothelial cells against oxidative stress injury via suppression of IP3R-[Ca2+]c/VDAC-[Ca2+]m axis by activation of MAPK/ERK signaling pathway. Cell Stress Chaperones. 2018;23(1):101–113. doi: 10.1007/s12192-017-0827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Souza LEB, Beckenkamp LR, Sobral LM, et al. Pre-culture in endothelial growth medium enhances the angiogenic properties of adipose-derived stem/stromal cells. Angiogenesis. 2018;21(1):15–22. doi: 10.1007/s10456-017-9579-0. [DOI] [PubMed] [Google Scholar]
- 51.Losón OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24(5):659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou H, Wang J, Hu S, Zhu H, Toan S, Ren J. BI1 alleviates cardiac microvascular ischemia-reperfusion injury via modifying mitochondrial fission and inhibiting XO/ROS/F-actin pathways. J Cell Physiol. 2019;234(4):5056–5069. doi: 10.1002/jcp.27308. [DOI] [PubMed] [Google Scholar]
- 53.Huang CY, Lai CH, Kuo CH, et al. Inhibition of ERK-Drp1 signaling and mitochondria fragmentation alleviates IGF-IIR-induced mitochondria dysfunction during heart failure. J Mol Cell Cardiol. 2018;122:58–68. doi: 10.1016/j.yjmcc.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 54.de Cristofaro T, Di Palma T, Ferraro A, et al. TAZ/WWTR1 is overexpressed in papillary thyroid carcinoma. Eur J Cancer. 2011;47(6):926–933. doi: 10.1016/j.ejca.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 55.Liu CY, Lv X, Li T, et al. PP1 cooperates with ASPP2 to dephosphorylate and activate TAZ. J Biol Chem. 2011;286(7):5558–5566. doi: 10.1074/jbc.M110.194019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson JM, Ferrara PJ, Verkerke ARP, et al. Targeted overexpression of catalase to mitochondria does not prevent cardioskeletal myopathy in Barth syndrome. J Mol Cell Cardiol. 2018;121:94–102. doi: 10.1016/j.yjmcc.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan EM, Fix A, Crouch MJ, et al. Murine diet-induced obesity remodels cardiac and liver mitochondrial phospholipid acyl chains with differential effects on respiratory enzyme activity. J Nutr Biochem. 2017;45:94–103. doi: 10.1016/j.jnutbio.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsu P, Liu X, Zhang J, Wang HG, Ye JM, Shi Y. Cardiolipin remodeling by TAZ/tafazzin is selectively required for the initiation of mitophagy. Autophagy. 2015;11(4):643–652. doi: 10.1080/15548627.2015.1023984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Angelini R, Lobasso S, Gorgoglione R, Bowron A, Steward CG, Corcelli A. Cardiolipin fingerprinting of leukocytes by MALDI-TOF/MS as a screening tool for Barth syndrome. J Lipid Res. 2015;56(9):1787–1794. doi: 10.1194/jlr.D059824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He Q, Harris N, Ren J, Han X. Mitochondria-targeted antioxidant prevents cardiac dysfunction induced by tafazzin gene knockdown in cardiac myocytes. Oxid Med Cell Longev. 2014;2014(4):1–12. doi: 10.1155/2014/654198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ji WK, Hatch AL, Merrill RA, Strack S, Higgs HN. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. Elife. 2015;4:e11553. doi: 10.7554/eLife.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prieto J, León M, Ponsoda X, et al. Dysfunctional mitochondrial fission impairs cell reprogramming. Cell Cycle. 2016;15(23):3240–3250. doi: 10.1080/15384101.2016.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hobson SR, Gurusinghe S, Lim R, et al. Melatonin improves endothelial function in vitro and prolongs pregnancy in women with early-onset preeclampsia. J Pineal Res. 2018;65(3):e12508. doi: 10.1111/jpi.12508. [DOI] [PubMed] [Google Scholar]
- 64.Kim DI, Lee KH, Gabr AA, et al. Aβ-Induced Drp1 phosphorylation through Akt activation promotes excessive mitochondrial fission leading to neuronal apoptosis. Biochim Biophys Acta. 2016;1863(11):2820–2834. doi: 10.1016/j.bbamcr.2016.09.003. [DOI] [PubMed] [Google Scholar]