Abstract
SgrAI is a type IIF restriction endonuclease that cuts an unusually long recognition sequence and exhibits self-modulation of DNA cleavage activity and sequence specificity. Previous studies have shown that SgrAI forms large oligomers when bound to particular DNA sequences and under the same conditions where SgrAI exhibits accelerated DNA cleavage kinetics. However, the detailed structure and stoichiometry of SgrAI:DNA as well as the basic building block of the oligomers, has not been fully characterized. Ion mobility mass spectrometry (IM-MS) was employed to analyze SgrAI/DNA complexes and show that the basic building block of the oligomers is the DNA-bound SgrAI dimer (DBD) with one SgrAI dimer bound to two pre-cleaved duplex DNA molecules each containing one half of the SgrAI primary recognition sequence. The oligomers contain variable numbers of DBDs with as many as 19 DBDs. Observation of the large oligomers shows that nano-electrospray ionization (nano-ESI) can preserve the proposed activated form of an enzyme. Finally, the collision cross section (CCS) of the SgrAI/DNA oligomers measured by IM-MS was found to have a linear relationship with the number of DBD in each oligomer suggesting a regular, repeating structure.
Keywords: SgrAI, protein-DNA complex, ion mobility, collision cross section
Graphical Abstract
Restriction endonucleases (ENases) are bacterial enzymes that bind to specific DNA sequences (recognition sites) in duplex DNA and cleave at or near this sequence.1–4 Most Type II ENases recognize and cleave 4–6 bp sites. ENases with long and specific recognition sites are useful in genomic technology due to the lower frequency of occurrence of the longer sequences producing larger DNA fragments.5, 6 SgrAI is a Type IIF ENase from Streptomyces griseus that recognizes an 8 bp DNA sequence 5’ CR|CCGGYG 3’ (where R = A or G, Y = C or T, and | indicates the site of cleavage), known as the cognate or primary site.7 SgrAI also cleaves at secondary sites — 5’ CR|CCGGY(A,C,T) 3’ and 5’ CR|CCGGGG 3’ under particular reaction conditions. SgrAI cleaves plasmids with two copies of the primary site faster than plasmids with only one copy. In addition, when both primary and secondary sites are on the same plasmid, SgrAI cleaves both sites although cleavage of the secondary site is slower (approximately 26-fold) than that of the primary site. However, in the absence of the primary site, the cleavage rate of the secondary site is significantly slower (200-fold).8–10 It has also been shown that the pre-cleaved primary site DNA sequence will stimulate the cleavage of primary and secondary sites by SgrAI. Such a self-activation with expansion of sequence specificity is unusual in Type II ENases. 8, 9, 11
The x-ray crystallographic structures of the homologous (as evidenced by their common core fold and similar DNA recognition sequences) Type IIF ENases SfiI, NgoMIV and Cfr10I show homotetramers composed of two dimers in a tail-to-tail orientation.12–14 Two duplex DNA molecules containing the recognition sequence bind to opposite sides of the tetramer.12–16 However, SgrAI is a dimer in solution without DNA.6, 11 Both Mg2+ or Ca2+ stimulate DNA binding by most ENases, with Mg2+ conferring cleavage activity while Ca2+ inhibits DNA cleavage but maintains site specific DNA binding activity.17–23 SgrAI has been shown to form large and heterogeneous species in the presence of primary site DNA containing sufficient flanking sequences, and under the same conditions where activation of DNA cleavage by SgrAI occurs.6, 11 Native gel electrophoresis and analytical ultracentrifugation show that the oligomer is heterogeneous and composed of possibly 4–12 DNA-bound SgrAI dimers (DBDs). However, the exact size, stoichiometry and basic building block of these species (inferred as DBD 6) has not been measured directly.
Structural information of non-covalent protein complexes is important in understanding the mechanism of protein function.24–29 X-ray crystallography and nuclear magnetic resonance (NMR) can provide high resolution structural information, however, these techniques require a large amount of pure and homogeneous protein. Because the SgrAI/DNA oligomers are heterogeneous in size based on analytical ultracentrifugation results,6 analysis via x-ray crystallography is challenging. Nano-electrospray ionization (nano-ESI) mass spectrometry (MS) is widely used to determine stoichiometry, and with specialized tandem MS (MS/MS) and IM experiments the spatial organization – topology – of supramolecular complexes in gas phase can also be investigated.29 Considerable evidence exists to support that protein assemblies remain intact and folded in vacuum.24–30 However, nano-ESI MS cannot provide atomic level information for protein complexes.
Ion mobility mass spectrometry (IM-MS) has been coupled with nano-ESI to reveal geometric information for protein complexes.24 The collision cross section (CCS) includes information about physical size and shape of the ion. Under the acceleration of electric fields, ions with larger CCS and lower charge state migrate slower in the drift tube filled with bath gas, analogous to the concept of electrophoresis. Therefore the drift times of different ions reflect differences in charge state and CCS.31, 32 Protocols have been developed to calculate ion-bath gas CCS based on x-ray crystallograpic or NMR structures. The Jarrold group has developed an open source software program – MOBCAL – to calculate helium based CCSs of biological molecules with three approaches33, 34 – projection approximation (PA), exact hard sphere scattering (EHSS) and the trajectory method (TM).35, 36 PA determines CCS by averaging all possible geometric projection areas. The calculation is simpler than the other two methods because it does not consider scattering processes and long-range interactions between the ions and the bath gas. Consequently, PA underestimates the CCSs.37, 38 EHSS calculates CCS by averaging the momentum transfer cross section that is related to the scattering angle over the relative velocity and the collision geometry. However, EHSS ignores long-range interactions.33, 34, 38 TM takes into account scattering, long-range interactions and the effect of multiple collisions and it is considered to be the most reliable and accurate CCS calculation approach. In this method, interatomic potentials are defined and then the trajectories are calculated within the potentials to obtain the scattering angles. The disadvantage of TM is that the calculation is the most computationally intense of the three methods.36, 39 PA and EHSS are widely used for large molecule CCS calculations.40–45 But the deviation between PA and EHSS can be as large as 20% for some geometries with grossly concave surfaces.33 In the current study, an empirical method, named scaled PA and developed by the Robinson group, was used, because the calculated CCS using scaled PA agree with the experimentally measured CCS.46, 47
Different charge states of the same protein usually have different CCSs. In most cases, as the number of charges increases, the CCS increases. This is believed to be due to unfolding or other expansion of the protein structure due to repulsion between the greater number of charges, leading to a larger CCS.35, 48, 49 However, the situation has been found to be different for some complexes. The TRAP 11-mer ring, for example, collapses (CCS decreases) with an increasing number of charges.40 Generally, the lower charge states of proteins are used to calculate CCSs because they are more likely to be those of the native state. 35, 48, 49
For this article, SgrAI/DNA complexes were analyzed by IM-MS under native-like conditions. SgrAI/DNA oligomers of different sizes were observed, and their masses indicate that the DBD is the basic building block of these oligomers, with one SgrAI dimer bound to two copies of the pre-cleaved SgrAI primary recognition site (PC DNA) in each DBD. The experimentally determined CCSs of the SgrAI/DNA oligomers were found to have a linear relationship with the number of DBD per oligomer, suggesting a regular, repeating structure in the oligomers. By comparing the CCSs of SgrAI dimer bound to DNA of different lengths, the conformation of the DNA in the unoligomerized DBD IM-MS is proposed.
Materials and Methods
Protein preparation.
Wild type SgrAI ENase was expressed and purified as described previously6, 50 and estimated at 99% purity by Coomassie stained SDS-PAGE. Before analysis, the SgrAI was dialyzed into a buffer containing 300 mM ammonium acetate and 1 mM dithiothreitol (DTT) using a Slide-A-Lyzer MINI dialysis device, with a 3.5K molecular weight cut off (Thermo Fisher Scientific, Rockford, IL) at 4°C. The final concentration of SgrAI was measured based on absorbance at 280 nm with Thermo Scientific NanoDrop 2000 micro-volume UV-visible spectrophotometer (Wilmington, DE), and the extinction coefficient of the SgrAI derived from the protein sequence.51
DNA preparation.
The oligonucleotides were made synthetically and purified using C18 reverse phase HPLC by Sigma-Aldrich (St. Louis, MO). The concentration of each single strand was then measured spectrophotometrically with extinction coefficients calculated from standard values for the nucleotides.52 DNA samples were annealed and aliquoted into small amounts, flash frozen in liquid nitrogen, and stored at −80°C. Each aliquot was used only once after being removed from the freezer.6, 50 The sequences of the 40 bp secondary site (40–2) DNA , PC DNA, and PC minus 9 flanking bp (PC-9) DNA are shown below, with recognition sites indicated in red:
| 40–2 (2° site) | 5'-GATGCGTGGGTCTTCACACCGGGGTGAAGACCCACGCATC-3' |
| 3' CTACGCACCCAGAAGTGTGGCCCCACTTCTGGGTGCGTAG-5' | |
| PC | 5'GATGCGTGGGTCTTCACA –3' |
| 3'-CTACGCACCCAGAAGTGTGGCC-5' | |
| PC-9 | 5'-GTCTTCACA -3' |
| 3'CAGAAGTGTGGCC -P5' |
IM-MS.
SgrAI (10–12 μM) and DNA are in a buffer with 100 mM ammonium acetate, 5 mM calcium tartrate and 1 mM DTT to give a mole ratio between SgrAI dimer and DNA of 1:1 to 1:4. Nano-ESI MS was used to analyze the SgrAI/DNA solution on a Synapt G2 HDMS (Waters MS Technologies, Manchester, UK) as described in the literature.53–56 Briefly, each sample was loaded into a tapered glass capillary pulled in-house using a Sutter Instruments P-97 micropipette puller (Novato, CA). A platinum wire was inserted into the non-tapered end of the capillary and a voltage of 1.1–1.6 kV was applied until optimal ion transmission and protein desolvation were achieved. The cone voltage was optimized at 50 V and the ion source temperature was ~30°C to minimize denaturation of protein or DNA. Pressure in the source region was raised to about 7 mbar by partially restricting the vacuum line to the rotary pump to optimize ion collisional cooling and transmission.
All samples were analyzed with argon as the collision gas and nitrogen as the ion mobility gas. The pressure of argon in the transfer ion guide was 3.0×10−2 mbar and the pressure of argon in the trap was 4.6×10−2 mbar. Collision induced dissociation (CID) was performed over a range of collision voltages in the trap collision cell, with the “quadrupole profile” set to 10,000 to transmit ions with m/z from 8,000 to 30,000 efficiently. The flow rate of the helium cell was 180 mL/min. The pressure of nitrogen in the ion mobility cell was 3.2 mbar. The ion mobility spectrometry (IMS) wave height and velocity were optimized at 16.0 V and 200 ms−1, respectively. The time-of-flight (TOF) analyzer pressure was 9.0×10−7 mbar. The IM-MS conditions were optimized based on the literature and research group experience with large complexes.41
Calibration of the CCS was performed as described.35 Native serum amyloid P (EMD Millipore Corporation, Chicago, IL) pentamer and decamer and concanavalin A (Sigma-Aldrich, St. Louis, MO) tetramer were used as standard proteins to create a CCS calibration curve because their sizes are similar to those of the analytes. All mass spectra were analyzed by MassLynx v4.1 and all mobiligrams were visualized using DriftScope v2.1 provided by Waters MS Technologies (Manchester, UK).
CCS calculation and model building.
Theoretical CCSs were calculated using the PA and EHSS methods in MOBCAL using models prepared from x-ray crystallographic structures.41 Scaled PA was used to calculate the theoretical CCSs46, 47. The DBD models with different DNA conformations were prepared manually using the PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.57 B-form DNA was manually added to the ends of the DNA in the x-ray crystal structure of SgrAI bound to cleaved primary site DNA (PDB code 3MQY58). Bends in the modeled DNA were introduced manually using PyMOL, taking care to maintain proper bond lengths and angles. Computations of CCSs with MOBCAL were performed on a 2.83 GHz quad-core Xeon SGI Altix ICE 8200 (Fremont, CA) server at the University of Arizona.
Results
Spectra of SgrAI alone and with 40–2 and PC DNA
Nano-ESI-MS was used to detect species in solutions of SgrAI alone (Fig. 1A), and with DNA (Fig. 1B-C). The spectrum of SgrAI without DNA (Fig. 1A) showed mostly SgrAI dimer, as expected6, 11 with a small amount of the total signal corresponding to SgrAI tetramer. Three different DNA constructs were also examined for their effects on oligomerization of SgrAI. The sequence 40–2 is a 40 bp duplex DNA containing a secondary site SgrAI recognition sequence (Materials and Methods). Secondary site sequences differ from primary site sequences by one bp, and are cleaved slowly by SgrAI in the absence of primary site sequences.6, 8, 9 SgrAI presented as largely dimeric, bound to one copy of 40–2, in the spectrum (Fig. 1B), with some SgrAI tetramer bound to two copies of 40–2, and a very small amount of hexamer bound to three copies of 40–2 (Fig. 1B). Next, the primary site sequence PC DNA, which mimicks the product of cleavage by SgrAI, was analyzed (Fig. 1C). The spectrum of SgrAI with PC DNA, at a molar ratio of 1:2 SgrAI dimer to PC DNA, shows a dimer of SgrAI bound to two copies of PC DNA, as well as a significant amount of larger species including a SgrAI tetramer+4PC DNA, and SgrAI hexamer+6PC DNA. In addition, unresolved species are found at m/z of 9,000–13,000 (Fig. 1C). The masses of all of the identifiable species are consistent with two copies of PC DNA binding per SgrAI dimer.
Figure 1.
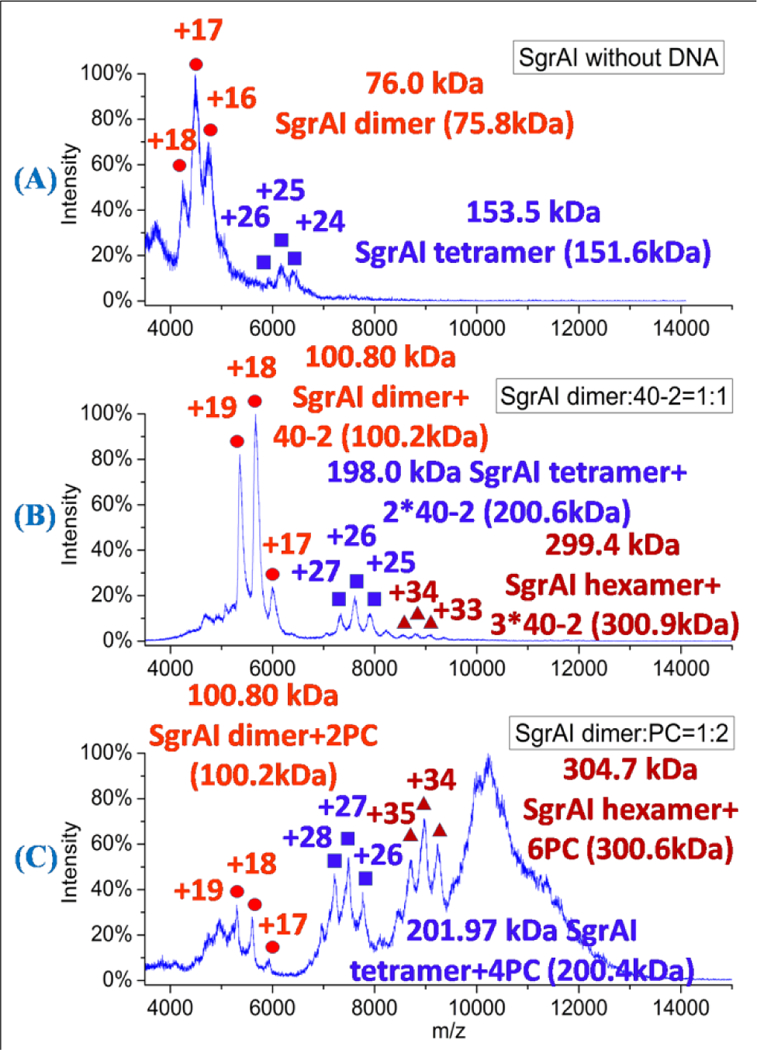
Spectra of SgrAI and SgrAI with different DNA. (A) SgrAI without DNA. Peaks corresponding to the SgrAI dimer (red circles) and SgrAI tetramer (purple squares) are visible. (B) SgrAI with 40–2 DNA (molar ratio 1:1). Peaks corresponding to the SgrAI dimer bound to one duplex of 40–2 DNA (red circles), SgrAI tetramer bound to two 40–2 duplexes (purple squares), and SgrAI hexamer bound to three 40–2 duplexes (red triangles) are visible. (C) SgrAI with PC DNA (molar ratio 1:2). Peaks corresponding to the SgrAI dimer bound to two PC DNA duplexes (red circles), SgrAI tetramer bound to four PC DNA duplexes (purple squares), and SgrAI hexamer bound to 6 PC DNA duplexes (red triangles) are visible, along with a large unresolved peak (900–1200 m/z). The theoretical masses were listed in the parentheses.
The effect of varying the molar ratio of SgrAI dimer to PC DNA was examined using nano-ESI mass spectrometry (Fig. S1). At a molar ratio of 1:1, SgrAI dimer+2PC DNA (1×DBD) and SgrAI tetramer+2PC DNA (2×DBD) complexes were observed (Fig. S1A and Table S1). Some species with higher m/z were also found, however their abundances were relatively small. A large unresolved species occurs at 4,000–5,000 m/z that overlaps with the peaks for the DBD (SgrAI dimer bound to two copies of PC DNA, Fig. S1B-D). At a molar ratio of 1:2, SgrAI dimer+2PC DNA (1×DBD), SgrAI tetramer+4PC DNA (2×DBD), SgrAI hexamer+6PC DNA (3×DBD) complexes are observed (Fig. 1C and Fig. S1B). In addition, large m/z species are also observed but with unresolvable peaks (Fig. 1C, Fig. S1B and Table S1). At the two higher ratios (1:2.6 and 1:4, Fig. S1C-D), the SgrAI hexamer+6PC DNA species is no longer resolvable, and the relative intensities of species above 8,000 m/z diminishes.
Collision induced dissociation (CID) to resolve species
CID was used in order to attempt to resolve peaks from the broad features in some spectra. Figure S2 shows the effect of increasing collision voltage in the MS/MS quadrupole-selected high m/z range in radio frequency profile mode, m/z 8,000–30,000 selected, of the 1:2.6 molar ratio mixture of SgrAI dimer with PC DNA. Low energy CID may result in larger SgrAI/DNA oligomers dissociating to smaller species that can then be distinguished. In the experiment, the “quadrupole profile” was set to transport ions with m/z from 8,000 to 30,000 efficiently. The ions were then dissociated by CID at different collision energies. With low collision voltage (4 V), no obvious peaks were observed in the spectrum of SgrAI:PC DNA with a molar ratio of 1:2.6 (Fig. S2A). At a collision voltage above 30 V, some peaks of SgrAI hexamer+6PC DNA complex (3×DBD) (Table S2) appeared at m/z 8,400 and their relative abundance increased as well (Fig. S2B). In addition, the broad peak at m/z 10,700 diminished. At collision voltages of 50 V and 70V, the spectrum showed no obvious additional changes (Fig. S2C-D). Thus, the peaks corresponding to SgrAI hexamer+6PC DNA can be clearly resolved at 50 V (Fig. S2C and Table S2), and are nearly resolved at 30 V (Fig. S2B), and remain similar at 70 V (Fig. S2D) as they are at 50 V (Fig. S2C). The broad peak at 9,000–10,000 remained unresolvable even at 70 V (Fig. S2D).
Low energy CID was also used to investigate unresolved features of the spectrum of SgrAI with PC-9 DNA (Fig. S3 and Table S3). PC-9 has fewer (7 bp vs. 16 bp) flanking bp than PC DNA. The length of DNA flanking the recognition sequence has been shown to influence activation and oligomerization of SgrAI.51 The spectrum of SgrAI with PC-9 (at a 1:2 SgrAI dimer:PC DNA) with CID of 4 V shows three broad, unresolved peaks (Fig. S3A). At 30 V, peaks corresponding to SgrAI dimer+2PC-9 and SgrAI tetramer+4PC-9 are resolvable (Fig. S3B). The features in general appear to be diminished with CID, and the feature at 12,000–14,000 m/z nearly disappears altogether.
Ion mobility-mass spectrometry (IM-MS) to resolve species
IM-MS was utilized to resolve species with overlapping m/z (Fig. 1) in a 1:2 molar ratio mixture of SgrAI dimer with PC DNA. Species are separated by drift time as well as m/z, and drift time varies with CCSs and charge states. Six clusters can be discerned, labeled I-VI (Fig. 2A). Cluster I, II, and III contain peaks in m/z corresponding to SgrAI dimer+2PC DNA, SgrAI tetramer+4PC DNA, and SgrAI hexamer+6PC DNA, respectively. No interpretable m/z peaks are immediately discernible in clusters IV and V. However close inspection of cluster VI (Fig. 1B-C) shows peaks that appear to correspond to SgrAI dodecamer+12PC DNA. Species in clusters IV and V were revealed using IM-MS of a 1:2.6 mixture of SgrAI dimer and PC DNA (Fig. S4-S8) and region selection. The highest intensity region of cluster IV (Fig. S4B and S7A-C) showed m/z peaks (Fig. S4C and S6) corresponding to SgrAI eicosamer+20PC DNA, and the highest intensity region of cluster V (Fig. S5B and S7D-E) revealed m/z peaks (Fig. S5C and S7) corresponding to SgrAI 38mer+38PC DNA. The identified peaks are reproducible (Fig. S6-S7). Because the highest intensity regions of the IM-MS spectra are difficult to discern in the spectral plots with log scales (Figs. S4B and S5B), Figure S8 shows the plots in log, square root, and linear scales. The widths of the identified m/z peaks are narrower (relative to those of the smaller oligomers, (Fig. 1) due to region selection (see Supporting Information and Figs. S9-S10).
Figure 2.
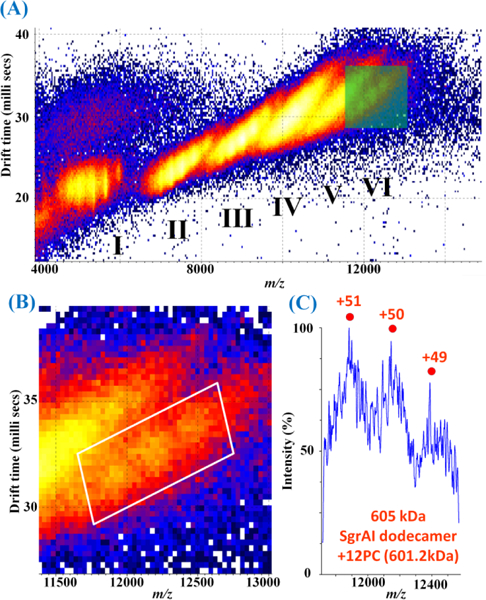
Mobiligram of SgrAI dimer:PC DNA at a molar ratio of 1:2. (A) Six distinct regions are visible (I-VI). Regions I-III correspond to SgrAI dimer+2PC DNA, SgrAI tetramer+4PC DNA, and SgrAI hexamer+6PC DNA, respectively. Regions IV-VI overlap in the mass spectrum (Figure 1C) but are distinct regions separated by drift time in IMS. (B) Enlargement of the green boxed region in (A). (C) Spectrum extracted from the region in the white boxed region in (B) with peaks corresponding to SgrAI dodecamer+12PC DNA. These peaks are resolvable only after IMS which separates species that otherwise overlap in the mass spectrum.
From IM-MS, the CCSs were measured for each of the identified SgrAI/PC DNA species (Fig. 3 and Table S4). The CCS of the lowest discernible charge state of each oligomer was used to represent the CCS of the oligomer. A linear relationship was found between CCSs and the number of SgrAI dimer+2PC DNA complexes (Fig. 3).
Figure 3.
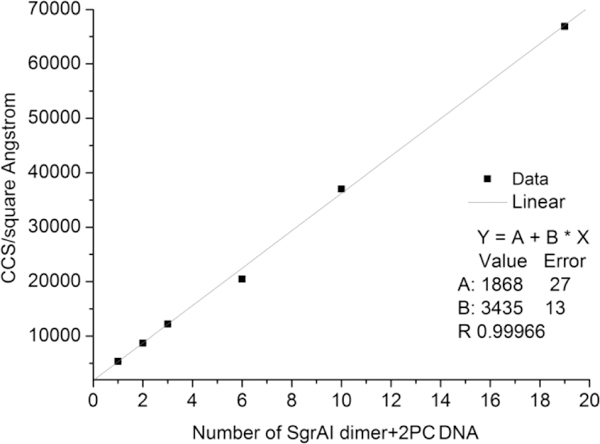
Linear relationship between experimental CCSs of SgrAI, PC DNA complexes and the number of DBD.
Model building and CCS comparison
The IM-MS CCSs of the SgrAI dimer, the SgrAI dimer+2PC-9, the SgrAI dimer+40–2 DNA, and the SgrAI dimer+2PC DNA complexes were used to compare with those calculated CCSs from models derived from x-ray crystal structures (Fig. 4 and Table S5). The charge state for all of these complexes is +17. The CCSs of SgrAI bound to 40–2 DNA and SgrAI bound to PC DNA (blue and purple triangles, respectively, Fig. 4B) were found by IM-MS to be approximately 11% larger than the CCS of SgrAI with PC-9 DNA (red circles, Fig. 4B) complex. In other words, the 18 bp of added flanking DNA (9 bp on either side of the recognition sequence) present in the SgrAI dimer+2PC DNA complex relative to the SgrAI dimer+2PC-9 complex increases the CCS by ~11%.
Figure 4.
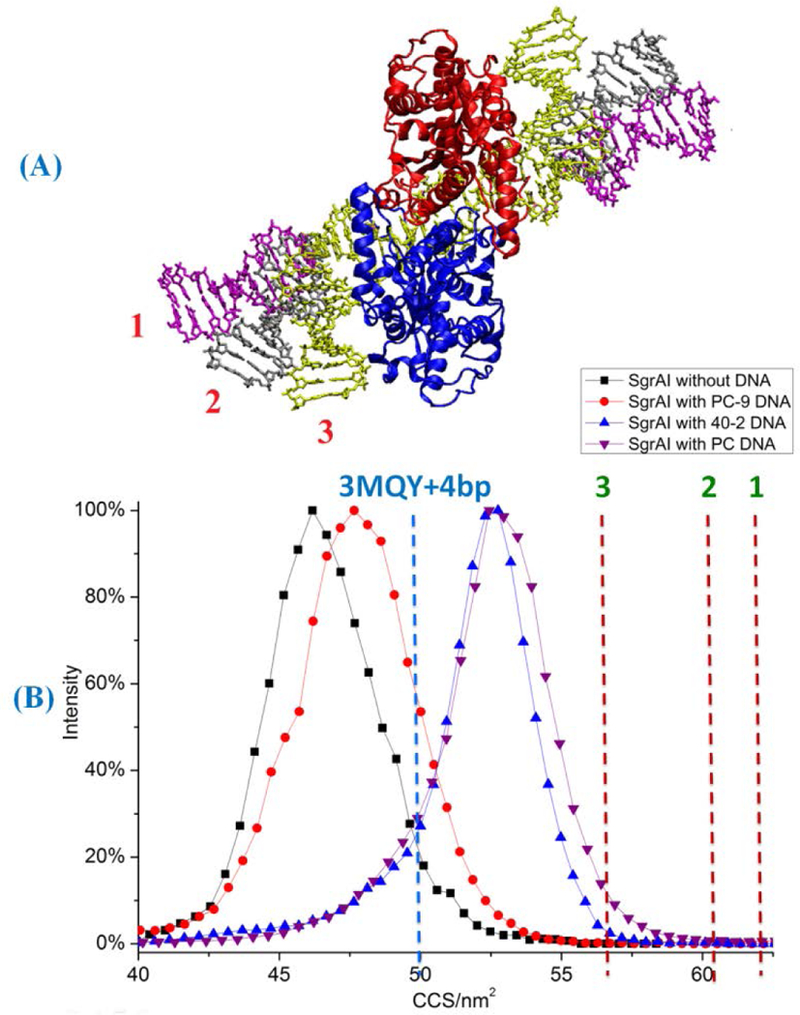
CCSs of SgrAI dimer with different DNA complexes. (A) Three different conformations of flanking DNA (labeled with different colors). 11 bp of B-DNAs were added on both sides of x-ray crystallographic structure 3MQY as flanking DNA. Models with three different flanking DNA conformations were built. In the first model, the flanking DNA is modeled as straight B-form DNA elongated straightly outward into space. In the second model, the flanking DNA bends towards SgrAI by ~30° based on the first model. In the third model, the flanking DNA bends towards SgrAI by ~80°. (B) CCSs of experimental and modeled complexes: black squares, SgrAI dimer without DNA, red circles, SgrAI with PC-9 DNA, blue triangles, SgrAI with 40–2 DNA, purple triangles, SgrAI with PC DNA complex. Dashed lines are calculated CCS of 3MQY with 4 bp of modeled flanking DNA (2 each site of recognition sequence, blue dash line), and calculated CCSs (red dash lines) of models 1–3 (A).
The observed CCS was used to model the conformation of the DNA bound to SgrAI. The crystallographic structure of SgrAI bound to DNA, 3MQY, contains only 5 bp flanking the 8 bp SgrAI recognition site, therefore the conformations of the additional 2 bp or 11 bp flanking this site that is present in PC-9 or both 40–2 and PC DNA, respectively, are not known. The flanking DNA influences formation of the large oligomers, and activation of DNA cleavage by SgrAI.6, 10 Models for the complexes of SgrAI bound to PC DNA and PC-9 DNA were prepared by the addition of flanking DNA (11 bp B-form on either side of the recognition sequence for PC DNA, 2 bp for PC-9 DNA) to the structure found in 3MQY.
The DNA conformation of the modeled flanking DNA in the model of SgrAI bound to PC DNA was varied giving rise to three different models (1–3, Fig. 4A). In the first model, the flanking DNA extends with no bends (1 of Fig. 4A). In the second model, the flanking DNA bends by 30° toward the SgrAI protein (2 of Fig. 4A). In the third model, the flanking DNA bends as in the second model but by an additional 50° (80° total) (3 of Fig. 4A). The CCSs were calculated for the model of the SgrAI dimer + 2PC-9 DNA complex (3MQY with 4 bp DNA, blue dashed line, Fig. 3B and Table S6), and the three models of SgrAI dimer + 2PC DNA complexes (1–3, red dashed lines, Fig. 4B and Table S6) using a linear combination method.59
Discussion
The SgrAI dimer+2PC DNA (DBD) is the basic building block of the oligomers.
Previous studies have shown that SgrAI forms a homodimer in the absence of DNA.6, 11 Nano-ESI-MS was employed to analyze SgrAI without DNA to confirm that the native dimer can be preserved under the experimental conditions. The major species observed in the mass spectrum is the SgrAI dimer (Fig. 1A and Table S1). The SgrAI tetramer was also observed in a relatively smaller amount consistent with dimer as the dominant species in solution at the given concentration. No remarkable larger oligomers were observed in these experimental conditions. The results show that the non-covalent interaction to form the SgrAI dimer can be preserved in nano-ESI-MS.
Secondary site DNA can bind to SgrAI but will not stimulate the DNA cleavage activity.68 A 40 bp DNA containing the secondary site sequence, 40–2 (Materials and Methods) was used to test the binding of secondary site DNA to SgrAI. SgrAI dimer with one 40–2 DNA, tetramer with two 40–2 DNA and a small amount of hexamer with three 40–2 DNA complexes were observed (Fig. 1B and Table S1). The relative ratio of the DNA bound dimer and DNA bound tetramer is similar to the ratio of dimer and tetramer in the absence of DNA (Fig. 1A-B). No other obvious larger species were observed in this experiment. The mass spectra of SgrAI without DNA and SgrAI with 40–2 DNA reveal that 40–2 DNA does not stimulate formation of the larger oligomers, as found previously by other techniques.6 In addition, DNA bound SgrAI can be observed under these experimental conditions. The charge states of SgrAI without DNAare +16, +17, and +18, which are similar to those of 40–2 DNA bound SgrAI (Fig. 1A-B), suggesting that the negative charges of 40–2 DNA were neutralized under the experimental conditions. The pKa of the phosphodiester bonds in DNA is about 3,60 therefore not expected to be protonated at pH 8.0. It is likely that cations such as calcium and ammonium are also attached, neutralizing the negative charge of the DNA. The number of these cations may vary in different experiments so the theoretical masses do not include cation or neutral species attachment. As a result, most experimental masses (in parentheses in Fig. 1) are larger than the theoretical masses by 1–2%. Furthermore, the attachment leads to peak broadening.58
To explore the influence of pre-cleaved primary site DNA which can stimulate formation of the large oligomers,6 different amounts of PC DNA were mixed with SgrAI resulting in different molar ratios of SgrAI dimer to PC DNA. When SgrAI dimer:PC DNA was 1:1, only the SgrAI dimer+2PC DNA (1×DBD) complex was observed, and not a complex with only 1 bound PC DNA. Two PC DNAs can form a DNA duplex structure that is similar to the uncleaved DNA substrate of the enzyme that binds tightly to the SgrAI dimer. If this interpretation is true, then the amount of SgrAI dimer+2PC DNA complex should increase when the SgrAI dimer:PC DNA ratio is 1:2.
When the SgrAI dimer:PC DNA ratio is 1:2, the relative amounts of large m/z species are greater (Fig. S1B) than that in the 1:1 SgrAI dimer:PC DNA mixture (Fig. S1A) suggesting that the 1:1 SgrAI dimer:PC DNA mixture molar ratio is not optimal for formation of the large m/z species. In the 1:2 molar ratio mixture, the observation of SgrAI tetramer+4PC DNA (2×DBD) and SgrAI hexamer+6PC DNA (3×DBD) supports the DBD complex as the basic building block of the larger oligomeric species. The peaks at m/z of 10,000–13,000 were not interpretable, possibly because of overlap from multiple species in the same m/z range derived from either neutral species attachment binding and/or oligomers with different masses. The complex of the SgrAI dimer with 40–2 DNA was found to contain mostly DBD (Fig. 1B), consistent with previous work6,11 showing that the secondary site DNA does not stimulate SgrAI in oligomer formation. The large oligomers are found to occur only with DNA capable of activating the DNA cleavage activity of SgrAI, hence occur only when the enzyme is activated.6 Thus observation of the larger oligomers shows that nano-ESI can preserve not only native protein but also the activated form of an enzyme.
At SgrAI:PC DNA molar ratios of 1:2.6 and 1:4, the spectra are similar (Fig. S1C-D), but reduced in total relative intensity of larger m/z species (above 8,000) and the SgrAI hexamer+6PC DNA species is no longer resolved. Both SgrAI dimer+2PC DNA (1×DBD) and SgrAI tetramer+4PC DNA (2×DBD) complexes are observed (Fig. S1C-D and Table S1). These results also support the 1:2 molar ratio as optimal for forming larger oligomers of SgrAI/DNA.
Low energy CID was used to reduce the influence of non-specific attachment of small molecules/salts to the protein and to investigate the composition of the SgrAI/DNA complexes, particularly in the unresolved peaks at m/z of 8,000–13,000 in various spectra. The results suggest that CID either dissociated the larger m/z species to the SgrAI hexamer+6PC DNA complex (3×DBD), or dissociated non-specific attachment from this species reducing its m/z complexity. If derived from the larger SgrAI/DNA oligomers, the results suggest that these large oligomers are less stable since they require relatively low energy to dissociate to the SgrAI hexamer+6PC DNA (3×DBD) complex. The SgrAI hexamer+6PC DNA (3×DBD) complex was found to be stable at the highest collision voltage of the instrument and did not appear to dissociate into smaller species such as SgrAI dimer+2PC DNA (1×DBD) complex or SgrAI tetramer+4PC DNA complex (2×DBD) (Fig. S1D). These results also show that when the concentration of PC DNA is equal to or higher than two times the SgrAI concentration, SgrAI/DNA oligomers can form.
CID was also used to distinguish peaks in the spectra of SgrAI bound to PC-9 DNA (Fig. S3). PC-9 DNA has fewer base pairs (7 bp) flanking the SgrAI recognition sequences than PC DNA (with 16 flanking bp, Materials and Methods). Previous studies have shown that a primary site with more flanking DNA binds tighter to SgrAI.6 Also, sufficient flanking DNA is required for stimulation of the DNA cleavage activity of SgrAI. 6, 10 At a SgrAI dimer:PC-9 DNA ratio of 1:2, no distinct peaks were observed and only unresolved broad features were present (Fig. S3A). CID was then applied to dissociate non-specific attachment of small molecules/salts or larger SgrAI/DNA oligomers overlapping in m/z. Distinct peaks of SgrAI dimer+2PC-9 DNA (1×DBD) and SgrAI tetramer+4PC-9 DNA (2×DBD) complexes are observed at a CID of 30 V (Fig. S3B and Table S3), and much of the larger m/z species was diminished. The results show that large oligomers may form when PC-9 DNA is mixed with SgrAI, although the relative amount appears to be less than with PC DNA (Fig. S2). These results also show that the length of flanking DNA does not affect the ratio between SgrAI dimer and PC DNA in the complexes.
Previous analytical ultracentrifugation results have shown that oligomers of DNA bound SgrAI may contain as many as twelve DBD.6 The mass spectra of SgrAI/DNA complexes provide direct evidence that the DBD is the basic building block of these oligomers. However, the oligomers are heterogeneous and their peaks overlap so the exact masses of these oligomers are still unclear from mass spectra alone, suggesting that ion mobility coupled to MS might help clarify the oligomeric states.
Linear relationship between CCSs and the number of DBD.
IM-MS was used in this study to separate species with different CCSs, including those with the same m/z but different overall mass and charge, and to characterize the number of DBD in the different oligomers. The mobiligram of SgrAI dimer:PC DNA at a molar ratio of 1:2 is shown in Fig. 2. Peaks of SgrAI dimer+2PC DNA (1×DBD), SgrAI tetramer+4PC DNA (2×DBD) and SgrAI hexamer+6PC DNA (3×DBD) complexes were observed in clusters I-III, respectively. A series of features were also observed in the boxed region of Fig. 2B, in cluster VI, corresponding to SgrAI dodecamer+12PC DNA (6×DBD). Because the intensities of these peaks are weaker than the intensities of the non-resolvable broad peaks in the 9000–12000 m/z range, they were not observed directly in Fig. 1. IMS therefore facilitated the extraction of these “buried” peaks based on differences in drift time. The result shows the power of IMS to separate peaks of protein complexes or DNA/protein complexes having different drift times; even though these species have similar m/z. Because clusters I-III correspond to oligomers with 1×, 2×, and 3×DBD, and cluster VI corresponds to oligomers with 6×DBD, it follows that clusters IV and V would correspond to oligomers of SgrAI+2PC DNA with 4× and 5×DBD, however no peaks in m/z could be resolved for confirmation. The inability to distinguish these directly may be due to the fact that larger species (ie. larger than 6×DBD) occur with m/z similar to the peaks of 4×DBD and 5×DBD, or that neutral species binding to the complexes results in heterogeneity and m/z overlap.
To identify the composition in clusters IV-V, the highest intensity regions in these clusters in the 1:2.6 SgrAI dimer:PC DNA mobiligram were selected (Fig. S4-S5, and S8). SgrAI eicosamer+20 PC DNA (10×DBD) and SgrAI 38mer+38 PC DNA (19×DBD) complexes were observed in the clusters IV and V of the mobiligram. The masses derived from the peaks agree well with the theoretical masses of SgrAI eicosamer+20 PC DNA (10×DBD) and SgrAI 38mer+38 PC DNA (19×DBD) complexes (Table S1), and the observed peaks are reproducible (Fig. S6-S7). The m/z peaks for these species are narrower than for the smaller oligomers due to region selection in IMS (see Supporting Information and Figs. S9-S10). The discovery of large complexes, as well as the complexity of the spectrum hence requiring region selection, are both consistent with the formation of a heterogeneous mixture of oligomers with different numbers of DBD.6 It is interesting that the total charge of these large species is approximately double that of globular proteins with similar mass.61 This is likely due to a non-globular shape of the SgrAI dimer+2PC DNA oligomers, because m/z is related to the surface area to volume ratio of the macromolecule.61, 62,63 It is possible that these were discerned only in the spectra with a higher ratio of PC DNA to SgrAI due to the formation of larger oligomers under these conditions. However, the diminished intensity in these regions with higher ratios of PC DNA to SgrAI suggests an effect by the DNA to reduce the amount of the larger oligomers, perhaps by competing with a second DNA binding site on SgrAI within the oligomer. Such a binding site is suggested by the dependence of oligomer formation on flanking DNA length.6, 10
To further confirm the observation of SgrAI eicosamer+20 PC DNA (10×DBD) and SgrAI 38mer+38 PC DNA (19×DBD) complexes, their CCSs were compared with the CCSs of the smaller complexes. Fig. 3 shows that the CCSs of the larger SgrAI PC DNA complexes are on the same straight line as smaller SgrAI PC DNA complexes. The CCSs suggest that the larger species observed have similar structures to smaller complexes. Further, the linear relationship suggests a regular, repeating structure, since each added DBD results in roughly the same increase in CCS. Because some oligomers differ by 1 DBD, this must be the basic building block of the oligomer. Both the regular, repeating nature of the structure and its heterogeneity in length are consistent with a run-on oligomer.
Model building and CCS calculations of SgrAI/DNA complexes.
The size of the 40–2 DNA, or that of two PC DNA annealed at the recognition sequence, is 40 bp, about twice the size of two PC-9 DNA (20 bp, Materials and Methods). However, the experimentally observed increase in CCS when adding PC-9 DNA to SgrAI dimer alone (black squares, Fig. 4B) was much smaller than the CCS increase when adding 40–2 DNA or two PC DNAs (each with 16 bp flanking each side of the recognition site) to the same (compare blue and purple triangles to red circles, Fig. 4B). This is because the DNA inserts into the DNA binding site of SgrAI. To a first approximation, the overall size and shape of the SgrAI dimer is likely to be similar to that of the SgrAI dimer+2PC-9 DNA complex. When longer DNA is used, the flanking DNA is expected to extend away from the protein, consistent with the greater increase in observed CCS.
The calculated CCS of the complex between SgrAI dimer+2PC-9 DNA (blue dashed line, Fig. 4B and Table S6) is close to (4% larger than) the experimental CCS determined for this complex (red circles, Fig. 4B). The calculated CCSs of the three different models of SgrAI dimer+2PC DNA complex are ~62.0 nm2 (1, red dashed line, Fig. 4B), ~60.0 nm2 (2, red dashed line, Fig. 4B), and ~57.0 nm2 (3, red dashed line, Fig. 4B), which are 17%, 13%, and 7% larger than the experimentally determined value for this complex (purple triangles, Fig. 4B and Table S6). Therefore, the third conformation of flanking DNA for the SgrAI dimer+2PC DNA complex, with an 80° bend (3 of Fig. 4A) is most consistent with the experimental CCS. The bend in DNA may be the result of the gas phase medium; with no solvent to shield charge, the DNA bridges back to the protein.
The experimental CCS of the oligomer allows for comparison to possible models built from assemblies of the DBD. However, several different models were found to agree with the experimental CCS (data not shown). In addition, the likelihood of conformational changes that are expected to occur with activation of SgrAI in the oligomeric complex which are currently unknown, adds further complexity to the modeling process. Therefore, a description of the SgrAI/DNA oligomer structure must await determination by higher resolution structural methods, such as cryoelectron microscopy or x-ray crystallography. However, the CCS determined herein will provide an independent measure to compare to any such model.
Conclusions
The masses of SgrAI:PC DNA complexes determined by IM-MS reveal that the basic building block of the SgrAI/DNA oligomer is the DBD with two copies of PC DNA per SgrAI dimer. The IM-MS data also show that this oligomer is heterogeneous and contains species with 1–6 DBD, as well as some as large as 10 and 19 DBD. This finding is similar to that previously reported using analytical ultracentrifugation.11, 51 Observation of the oligomers shows that nano-ESI can preserve the noncovalent stoichiometries of both the native and activated forms of an enzyme. Finally, the CCSs of the oligomers of DBD show a linear relationship with the number of the DBD, showing that oligomers differing in the number of DBD had similar changes in CCS and therefore a similar, regular structure.
Restriction endonucleases such as SgrAI are proposed to function as a defense against invading phage DNA. Cleavage of the host genome is prevented by methylation of SgrAI recognition sequences by the cognate SgrAI methyltransferase. SgrAI derives from Streptomyces griseus, a prokaryote with an unusually large genome (8.5 Mb, nearly twice the size of E. coli). The relatively long (8 bp vs. 4–6 bp) SgrAI recognition sequence may have evolved to reduce the number of recognition sequences in the genome that would otherwise occur, which reduces pressure in the form of sites required for methylation by the methyltransferase, as well as sites that could be potentially cleaved by SgrAI. Similarly, the relatively low DNA cleavage activity of unstimulated SgrAI may also have evolved to reduce this pressure. However, the low intrinsic cleavage activity, and the reduced number of cleavage sites would both contribute to a less efficient defense against any invading phage DNA. Hence the activation of SgrAI upon encounter of at least two unmethylated SgrAI recognition sequences, combined with the expansion of its sequence specificity, result in more cleavages occurring more rapidly to inactivate this invading DNA. But this activation and expansion of sequence specificity could in principle also harm the host genome. Therefore the oligomerization of activated SgrAI may have evolved to sequester SgrAI on the phage DNA and away from the host genome.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation DBI grant 0923551 to V.H.W.
Abbreviations and Textual Footnotes
- CCS
collision cross section
- CID
collision induced dissociation
- DBD
DNA-bound SgrAI dimer
- DTT
dithiothreitol
- EHSS
exact hard sphere scattering
- ENase
restriction endonucleases
- IM-MS
Ion mobility mass spectrometry
- IMS
ion mobility spectrometry
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- nano-ESI
nano-electrospray ionization
- NMR
nuclear magnetic resonance
- PA
projection approximation
- PC DNA
pre-cleaved SgrAI primary recognition site
- PC-9
PC minus 9 flanking bpTM, trajectory method
- TOF
time-of-flight
Footnotes
Supporting Information Available including spectra of different molar ratios of SgrAI dimer to PC DNA, spectra with CID of SgrAI dimer:PC DNA 1:2.6 and 1:2. In addition, tables containing the masses of SgrAI/DNA complexes, CID data, IM-MS CCSs of SgrAI PC DNA complexes, and calculated CCSs of SgrAI dimer with different DNA complexes. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bickle TA, and Kruger DH (1993) Biology of DNA restriction, Microbiol. Rev. 57, 434–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kruger DH, and Bickle TA (1983) Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts, Microbiol. Rev. 47, 345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer HW (1971) DNA restriction and modification mechanisms in bacteria, Annu. Rev. Microbiol. 25, 153–176. [DOI] [PubMed] [Google Scholar]
- 4.McClelland M (1981) The effect of sequence specific DNA methylation on restriction endonuclease cleavage, Nucleic Acids Res. 9, 5859–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilcock DT, Daniels LE, Bath AJ, and Halford SE (1999) Reactions of type II restriction endonucleases with 8-base pair recognition sites, J. Biol. Chem. 274, 36379–36386. [DOI] [PubMed] [Google Scholar]
- 6.Park CK, Stiteler AP, Shah S, Ghare MI, Bitinaite J, and Horton NC (2010) Activation of DNA cleavage by oligomerization of DNA-bound SgrAI, Biochemistry 49, 8818–8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tautz N, Kaluza K, Frey B, Jarsch M, Schmitz GG, and Kessler C (1990) SgrAI, a novel class-II restriction endonuclease from Streptomyces griseus recognizing the octanucleotide sequence 5’-CR/CCGGYG-3’ [corrected], Nucleic Acids Res. 18, 3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bitinaite J, and Schildkraut I (2002) Self-generated DNA termini relax the specificity of SgrAI restriction endonuclease, Proceedings of the National Academy of Sciences of the United States of America 99, 1164–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hingorani-Varma K, and Bitinaite J (2003) Kinetic analysis of the coordinated interaction of SgrAI restriction endonuclease with different DNA targets, J Biol Chem 278, 40392–40399. [DOI] [PubMed] [Google Scholar]
- 10.Wood KM, Daniels LE, and Halford SE (2005) Long-range communications between DNA sites by the dimeric restriction endonuclease SgrAI, J Mol Biol 350, 240–253. [DOI] [PubMed] [Google Scholar]
- 11.Daniels LE, Wood KM, Scott DJ, and Halford SE (2003) Subunit assembly for DNA cleavage by restriction endonuclease SgrAI, J Mol Biol 327, 579–591. [DOI] [PubMed] [Google Scholar]
- 12.Siksnys V, Skirgaila R, Sasnauskas G, Urbanke C, Cherny D, Grazulis S, and Huber R (1999) The Cfr10I restriction enzyme is functional as a tetramer, J. Mol. Biol. 291, 1105–1118. [DOI] [PubMed] [Google Scholar]
- 13.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, and Huber R (2000) Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA, Nat. Struct. Biol. 7, 792–799. [DOI] [PubMed] [Google Scholar]
- 14.Vanamee ES, Viadiu H, Kucera R, Dorner L, Picone S, Schildkraut I, and Aggarwal AK (2005) A view of consecutive binding events from structures of tetrameric endonuclease SfiI bound to DNA, EMBO J. 24, 4198–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nobbs TJ, Szczelkun MD, Wentzell LM, and Halford SE (1998) DNA excision by the Sfi I restriction endonuclease, J. Mol. Biol. 281, 419–432. [DOI] [PubMed] [Google Scholar]
- 16.Bozic D, Grazulis S, Siksnys V, and Huber R (1996) Crystal structure of Citrobacter freundii restriction endonuclease Cfr10I at 2.15 A resolution, J. Mol. Biol. 255, 176–186. [DOI] [PubMed] [Google Scholar]
- 17.Vipond IB, and Halford SE (1995) Specific DNA recognition by EcoRV restriction endonuclease induced by calcium ions, Biochemistry 34, 1113–1119. [DOI] [PubMed] [Google Scholar]
- 18.Engler LE, Welch KK, and Jen-Jacobson L (1997) Specific binding by EcoRV endonuclease to its DNA recognition site GATATC, J. Mol. Biol. 269, 82–101. [DOI] [PubMed] [Google Scholar]
- 19.Martin AM, Horton NC, Lusetti S, Reich NO, and Perona JJ (1999) Divalent metal dependence of site-specific DNA binding by EcoRV endonuclease, Biochemistry 38, 8430–8439. [DOI] [PubMed] [Google Scholar]
- 20.Reid SL, Parry D, Liu HH, and Connolly BA (2001) Binding and recognition of GATATC target sequences by the EcoRV restriction endonuclease: a study using fluorescent oligonucleotides and fluorescence polarization, Biochemistry 40, 2484–2494. [DOI] [PubMed] [Google Scholar]
- 21.Nastri HG, Evans PD, Walker IH, and Riggs PD (1997) Catalytic and DNA binding properties of PvuII restriction endonuclease mutants, J. Biol. Chem. 272, 25761–25767. [DOI] [PubMed] [Google Scholar]
- 22.Etzkorn C, and Horton NC (2004) Ca2+ binding in the active site of HincII: implications for the catalytic mechanism, Biochemistry 43, 13256–13270. [DOI] [PubMed] [Google Scholar]
- 23.Joshi HK, Etzkorn C, Chatwell L, Bitinaite J, and Horton NC (2006) Alteration of sequence specificity of the type II restriction endonuclease HincII through an indirect readout mechanism, J. Biol. Chem. 281, 23852–23869. [DOI] [PubMed] [Google Scholar]
- 24.Heck AJ (2008) Native mass spectrometry: a bridge between interactomics and structural biology, Nat. Methods 5, 927–933. [DOI] [PubMed] [Google Scholar]
- 25.Benesch JL, Ruotolo BT, Simmons DA, and Robinson CV (2007) Protein complexes in the gas phase: technology for structural genomics and proteomics, Chem. Rev. 107, 3544–3567. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez H, and Robinson CV (2007) Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry, Nat. Protoc. 2, 715–726. [DOI] [PubMed] [Google Scholar]
- 27.Veenstra TD (1999) Electrospray ionization mass spectrometry: a promising new technique in the study of protein/DNA noncovalent complexes, Biochem. Biophys. Res. Commun. 257, 1–5. [DOI] [PubMed] [Google Scholar]
- 28.Loo JA (1997) Studying noncovalent protein complexes by electrospray ionization mass spectrometry, Mass Spectrom Rev. 16, 1–23. [DOI] [PubMed] [Google Scholar]
- 29.Rusconi F, Guillonneau F, and Praseuth D (2002) Contributions of mass spectrometry in the study of nucleic acid-binding proteins and of nucleic acid-protein interactions, Mass Spectrom Rev. 21, 305–348. [DOI] [PubMed] [Google Scholar]
- 30.Pace CN, Trevino S, Prabhakaran E, and Scholtz JM (2004) Protein structure, stability and solubility in water and other solvents, Philos Trans R Soc Lond B Biol Sci 359, 1225–1234; discussion 1234–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borsdorf H, and Eiceman GA (2006) Ion Mobility Spectrometry: Principles and Applications, Appl. Spectrosc. Rev. 41, 323–375. [Google Scholar]
- 32.Borsdorf H, Mayer T, Zarejousheghani M, and Eiceman GA (2011) Recent Developments in Ion Mobility Spectrometry, Appl. Spectrosc. Rev. 46, 472–521. [Google Scholar]
- 33.Shvartsburg AA, and Jarrold MF (1996) An exact hard-spheres scattering model for the mobilities of polyatomic ions, Chem. Phys. Lett. 261, 86–91. [Google Scholar]
- 34.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, and Jarrold MF (1996) Structural information from ion mobility measurements: effects of the long-range potential, J. Phys. Chem. 100, 16082–16086. [Google Scholar]
- 35.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, and Ruotolo BT (2010) Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology, Anal. Chem. 82, 9557–9565. [DOI] [PubMed] [Google Scholar]
- 36.Jurneczko E, and Barran PE (2011) How useful is ion mobility mass spectrometry for structural biology? The relationship between protein crystal structures and their collision cross sections in the gas phase, Analyst 136, 20–28. [DOI] [PubMed] [Google Scholar]
- 37.von Heiden G, Hsu M, Kemper PR, and Bowers MT (1991) Structures of carbon cluster ions from 3 to 60 atoms: linears to rings to fullerenes, J. Chem. Phys. 95, 3835–3837. [Google Scholar]
- 38.Wyttenbach T, von Helden G, Batka JJ, Carlat JD, and Bowers MT (1996) Effect of the long-range potential on ion mobility measurements, J. Am. Soc. Mass Spectrom. 8, 275–282. [Google Scholar]
- 39.Shvartsburg AA, Schatz GC, and Jarrold MF (1998) Mobilities of carbon cluster ions: critical importance of the molecular attractive potential, J. Chem. Phys. 108, 2416–2423. [Google Scholar]
- 40.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, and Robinson CV (2005) Evidence for macromolecular protein rings in the absence of bulk water, Science 310, 1658–1661. [DOI] [PubMed] [Google Scholar]
- 41.Ruotolo BT, Benesch JL, Sandercock AM, Hyung SJ, and Robinson CV (2008) Ion mobility-mass spectrometry analysis of large protein complexes, Nat Protoc 3, 1139–1152. [DOI] [PubMed] [Google Scholar]
- 42.Uetrecht C, Barbu IM, Shoemaker GK, van Duijn E, and Heck AJ (2011) Interrogating viral capsid assembly with ion mobility-mass spectrometry, Nat Chem 3, 126–132. [DOI] [PubMed] [Google Scholar]
- 43.van Duijn E, Barendregt A, Synowsky S, Versluis C, and Heck AJ (2009) Chaperonin complexes monitored by ion mobility mass spectrometry, J. Am. Chem. Soc. 131, 1452–1459. [DOI] [PubMed] [Google Scholar]
- 44.Knapman TW, Morton VL, Stonehouse NJ, Stockley PG, and Ashcroft AE (2010) Determining the topology of virus assembly intermediates using ion mobility spectrometry-mass spectrometry, Rapid Commun. Mass Spectrom. 24, 3033–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Politis A, Park AY, Hyung SJ, Barsky D, Ruotolo BT, and Robinson CV (2010) Integrating ion mobility mass spectrometry with molecular modelling to determine the architecture of multiprotein complexes, PLoS One 5, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall Z, Politis A, Bush MF, Smith LJ, and Robinson CV (2012) Charge-state dependent compaction and dissociation of protein complexes: insights from ion mobility and molecular dynamics, J Am Chem Soc 134, 3429–3438. [DOI] [PubMed] [Google Scholar]
- 47.Benesch JL, and Ruotolo BT (2011) Mass spectrometry: come of age for structural and dynamical biology, Curr Opin Struct Biol 21, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scarff CA, Thalassinos K, Hilton GR, and Scrivens JH (2008) Travelling wave ion mobility mass spectrometry studies of protein structure: biological significance and comparison with X-ray crystallography and nuclear magnetic resonance spectroscopy measurements, Rapid Commun. Mass Spectrom. 22, 3297–3304. [DOI] [PubMed] [Google Scholar]
- 49.Fernandez-Lima FA, Blase RC, and Russell DH (2010) A study of ion-neutral collision cross section values for low charge states of peptides, proteins, and peptide/protein complexes, Int. J. Mass Spectrom. 298, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunten PW, Little EJ, Gregory MT, Manohar VM, Dalton M, Hough D, Bitinaite J, and Horton NC (2008) The structure of SgrAI bound to DNA; recognition of an 8 base pair target, Nucleic Acids Res. 36, 5405–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park CK, Joshi HK, Agrawal A, Ghare MI, Little EJ, Dunten PW, Bitinaite J, and Horton NC (2010) Domain swapping in allosteric modulation of DNA specificity, PLoS biology 8, e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cavaluzzi MJ, and Borer PN (2004) Revised UV extinction coefficients for nucleoside-5’-monophosphates and unpaired DNA and RNA, Nucleic Acids Res. 32, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, and Bateman RH (2004) Applications of a travelling wave-based radio-frequency-only stacked ring ion guide, Rapid Commun Mass Spectrom 18, 2401–2414. [DOI] [PubMed] [Google Scholar]
- 54.Shvartsburg AA, and Smith RD (2008) Fundamentals of traveling wave ion mobility spectrometry, Anal Chem 80, 9689–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giles K, Williams JP, and Campuzano I (2011) Enhancements in travelling wave ion mobility resolution, Rapid Commun Mass Spectrom 25, 1559–1566. [DOI] [PubMed] [Google Scholar]
- 56.Zhou M, Dagan S, and Wysocki VH (2012) Protein subunits released by surface collisions of noncovalent complexes: nativelike compact structures revealed by ion mobility mass spectrometry, Angew Chem Int Ed Engl 51, 4336–4339. [DOI] [PubMed] [Google Scholar]
- 57.DeLano WL (2002) The PyMOL User’s Manual DeLano Scientific, Palo Alto, CA. [Google Scholar]
- 58.Little EJ, Dunten PW, Bitinaite J, and Horton NC (2011) New clues in the allosteric activation of DNA cleavage by SgrAI: structures of SgrAI bound to cleaved primary-site DNA and uncleaved secondary-site DNA, Acta Crystallogr D Biol Crystallogr 67, 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bush MF, Hall Z, Politis A, Barsky D, and Robinson CV (2011) Interpreting collision cross section of protein complexes: models, approximations, errors, and best practices, 59th American Society for Mass Spectrometry annual conference, Section ion mobility mass spectrometry: integration into structural biology. [Google Scholar]
- 60.Schooling SR, Hubley A, and Beveridge TJ (2009) Interactions of DNA with biofilm-derived membrane vesicles, J. Bacteriol. 191, 4097–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernandez de la Mora J (2000) Electrospray ionization of large multiply charged species proceeds via Dole’s charged residue mechanism, Analytica Chimica Acta 406, 93–104. [Google Scholar]
- 62.Fernandez de la Mora J (2012) Why do GroEL ions exhibit two gas phase conformers?, J Am Soc Mass Spectrom 23, 2115–2121. [DOI] [PubMed] [Google Scholar]
- 63.Kaltashov IA, and Mohimen A (2005) Estimates of protein surface areas in solution by electrospray ionization mass spectrometry, Anal Chem 77, 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


