HERV loci transcribed lncRNA TROJAN promotes TNBC progression through ZMYND8 degradation by the ubiquitin-proteasome pathway.
Abstract
Human endogenous retroviruses (HERVs) play pivotal roles in the development of breast cancer. However, the detailed mechanisms of noncoding HERVs remain elusive. Here, our genome-wide transcriptome analysis of HERVs revealed that a primate long noncoding RNA, which we dubbed TROJAN, was highly expressed in human triple-negative breast cancer (TNBC). TROJAN promoted TNBC proliferation and invasion and indicated poor patient outcomes. We further confirmed that TROJAN could bind to ZMYND8, a metastasis-repressing factor, and increase its degradation through the ubiquitin-proteasome pathway by repelling ZNF592. TROJAN also epigenetically up-regulated metastasis-related genes in multiple cell lines. Correlations between TROJAN and ZMYND8 were subsequently confirmed in clinical samples. Furthermore, our study verified that antisense oligonucleotide therapy targeting TROJAN substantially suppressed TNBC progression in vivo. In conclusion, the long noncoding RNA TROJAN promotes TNBC progression and serves as a potential therapeutic target.
INTRODUCTION
Breast cancer is the most common malignancy and the second leading cause of cancer death among females (1). Since the definition of intrinsic molecular subtypes in 2000 (2), breast cancer has been divided into different subtypes and treated accordingly. Anti-estrogen and/or anti–human epidermal growth factor receptor 2 (HER2) therapies are standard targeted treatments for luminal-type and/or HER2-positive breast cancer (3). However, triple-negative breast cancer (TNBC), a subtype with highly aggressive biological behavior and a higher risk of recurrence and distant metastasis, still lacks an effective targeted therapy (4). Thus, screening for TNBC therapeutic targets is of great significance and urgent need.
Human endogenous retroviruses (HERVs) are formed by exogenous retroviruses that integrate into host cell genomes (5); they cover approximately 8% of the human genomic sequence and are distributed in approximately 700,000 genomic loci (6). HERVs play an important role in a diverse range of biological processes (7), including tumor progression (8), and HERV-K may be a target for immunotherapy or a biomarker of tumor imitation or progression (9). Unfortunately, as HERVs cover several hundred thousand genome loci, very few studies have focused on unraveling the expression profile of each HERV locus, and even less knowledge exists regarding the correlation and molecular mechanisms of HERVs and breast cancer progression.
Previously, we developed a prognostic mRNA–long noncoding RNA (lncRNA) signature in TNBC through the transcriptome profiling of TNBC samples. This signature, consisting of three mRNAs and two lncRNAs (10), accurately predicted the relapse-free survival of patients with TNBC. In the preliminary study that followed, one lncRNA, AK124454, was demonstrated to promote TNBC cell proliferation and invasion. This AK124454 sequence (later dubbed TROJAN) highly overlaps that of a long terminal repeat (LTR) belonging to HERVs. These factors inspired us to perform an in-depth study of long noncoding HERVs.
Here, we used RNA sequencing (RNA-seq)–based transcriptome analysis to examine TNBC-related HERV species, uncover the detailed mechanism of TROJAN in promoting breast cancer progression, and explore the potential of TROJAN as an anti-TNBC therapeutic target.
RESULTS
Transcriptome profiling of HERVs reveals TROJAN, a HERV-derived lncRNA that is highly expressed in TNBC
We first used RNA-seq to perform transcriptional analysis of all the HERVs in eight pairs of TNBC and normal breast tissues. In total, 523,655 HERVs were subjected to further analysis (table S1). We identified a set of 51 HERVs as being highly expressed in TNBC tissues (Fig. 1A). One LTR70 was identified as a potential TNBC-related HERV based on its enrichment fold change in TNBC tissues (Fig. 1, B and C). This LTR70 sequence showed high overlap with that of our previously described lncRNA, AK124454 (10). This transcript is also annotated as ENST00000624228.1 or CTC-260E6.4 and is primate specific (fig. S1A). To identify the full-length sequence of this transcript, we performed rapid amplification of cDNA (complementary DNA) ends (RACE) assays and found that the full-length transcript was approximately 1300 nucleotides (Fig. 1D). We named our newly identified full transcript TROJAN (a reference to the hidden enemies inside the Trojan Horse). According to Repbase, the 3′ end of TROJAN contains a complete LTR70 sequence of several mosaic LTRs flanked by MER67C and LTR56 (fig. S1, B and C). The estimated number of TROJAN copies per cell was approximately 361.5 in MDA-MB-231 LM2 cells and 246 in BT549 cells (fig. S1D). We revealed that TROJAN expression was independent of genomic sequence contamination (fig. S1E). In The Cancer Genome Atlas (TCGA) database, TROJAN (we used the CTC-260E6.4 transcript as a substitution) was associated with reduced disease-free survival in patients with breast cancer (fig. S1F). In an independent cohort of 53 paired TNBC tissues and adjacent normal breast tissues, TROJAN expression was substantially higher in TNBC tissues (Fig. 1E and table S1), and TROJAN was associated with reduced relapse-free survival in FUSCC (Fudan University Shanghai Cancer Center) cohort 1 (Fig. 1F). Through RNA in situ hybridization (ISH), we further confirmed that TROJAN was highly enriched in TNBC samples, correlated with the poor survival of patients with TNBC in FUSCC cohort 2, and subcellularly localized to the nucleus (Fig. 1, G and H, and fig. S1G). TROJAN was also identified as being polyadenylated (fig. S1H). Together, these findings demonstrate that TROJAN is an lncRNA containing a HERV sequence that is highly expressed in TNBC and correlates with poor patient survival.
Fig. 1. HERV transcriptome profile in human breast cancer.
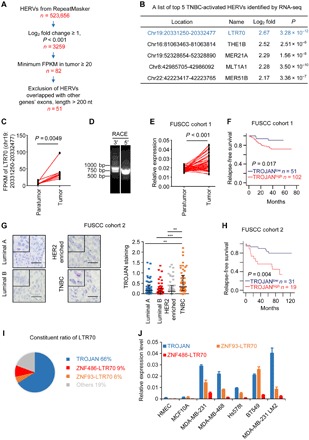
(A) Schematic diagram depicting the screening of TNBC-related HERVs. RNA-seq detected HERV expression across eight paired TNBC tissues and adjacent normal tissues for HERVs derived from RepeatMasker. FPKM, fragments per kilobase of transcript per million mapped reads. (B) The top 5 TNBC-related HERVs identified by RNA-seq. (C) The expression of LTR70 [chromosome (chr) 19: 20331250-20332477] in eight paired TNBC tissues and adjacent normal tissues. P value was determined using two-tailed paired Student’s t test. (D) Polymerase chain reaction (PCR) products generated in the 3′ (left) and 5′ (right) RACE assay covering the 5′ and 3′ ends of the TROJAN transcript. (E) The quantitative PCR (qPCR) analysis of the relative TROJAN transcription levels in TNBC tissues (n = 53) versus the adjacent normal breast tissues (n = 53) in FUSCC cohort 1. P value was determined using two-tailed paired Student’s t test. (F) Kaplan-Meier analysis of the relapse-free survival of 153 patients with TNBC in FUSCC cohort 1. A log-rank test was used to determine the statistical significance between the low TROJAN expression group (n = 51) and the high TROJAN expression group (n = 102). (G) RNA ISH of TROJAN in breast cancer tissues with different subtypes (n = 50 each) (FUSCC cohort 2). Scale bars, 50 μm. The data are presented as the median with interquartile range; two-tailed unpaired Student’s t test. **P < 0.01 and ***P < 0.001. (H) Kaplan-Meier analysis of the relapse-free survival of 50 patients with TNBC in FUSCC cohort 2. The log-rank test was used to determine statistical significance between the low TROJAN expression group (n = 31) and the high TROJAN expression group (n = 19). (I) The constituent ratio of LTR70. The assay was performed by RNA-seq. (J) The qPCR analysis of the expression of TROJAN and two other LTR70s in multiple cell lines. The data are presented as the mean ± SD; n = 3 independent experiments. See also figs. S1 and S2.
TROJAN is a predominantly expressed LTR70 transcript in TNBC
Because the LTR sequences were highly homologous, we explored whether TROJAN was the only TNBC-related, LTR70-containing transcript. The LTR70 RNA-seq data highlighted that these transcripts were expressed differently in tumors and normal tissues (table S1). Hence, we reasoned that LTR70s could not be researched in their entirety. We screened out eight expressed LTR70s (named according to their intronic genes, such as ZNF93-LTR70) from among the other 20 highly homologous transcripts from RNA-seq data on MDA-MB-231 LM2 cells (GSE104910) and public RNA-seq data of multiple cell lines in the University of California, Santa Cruz (UCSC) database [long RNA-seqs from the Encyclopedia of DNA Elements (ENCODE)/Cold Spring Harbor Laboratory]. We next performed polymerase chain reaction (PCR)–enriched RNA-seq in MDA-MB-231 LM2 cells based on a pair of primers compatible with all eight of the expressed LTR70s to further find the relative abundance of each transcript (fig. S2A). The read counts of the RNA-seq data showed that TROJAN had the highest expression; the other two major expressed LTR70s were located in chromosome 19 at 20020176-20021457 and 20289348-20290288 (Fig. 1I). These LTR70s were located in the first introns of ZNF93 and ZNF486 and were therefore named ZNF93-LTR70 and ZNF486-LTR70 according to their genome locations. To further exclude other transcripts, we designed four random pairs of primers to amplify the TROJAN sequence. The PCR products were cloned in a pGEM-T Easy Vector and sequenced. The proportion of TROJAN transcripts ranged from 71.4 to 100% (fig. S2B). TROJAN was highly expressed in the TNBC cell lines (MDA-MB-231, MDA-MB-468, Hs578t, and BT549) compared with two normal cell lines (HMEC and MCF10A human mammary epithelial cells; Fig. 1J). Through the UCSC genome browser database, we found that TROJAN was also highly expressed in H1-hESC embryonic stem cells (fig. S2C). These results demonstrate that TROJAN is the dominant transcript of LTR70 and is highly expressed in TNBC.
TROJAN promotes TNBC proliferation and metastasis and is a potential therapeutic target of TNBC
To explore the TROJAN phenotype in breast cancer, we knocked down its expression using short hairpin RNAs (shRNAs) and overexpressed its entire transcript (fig. S3, A and B). TROJAN down-regulation impaired the proliferative potential of breast cancer cell lines in vitro, while TROJAN overexpression promoted this ability (Fig. 2, A and B). We used the mammary fat pad injection model of female nonobese diabetic (NOD)/severe combined immunodeficient (SCID) mice to investigate the effects of TROJAN on breast cancer proliferation in vivo. Down-regulation of TROJAN reduced tumor volume (Fig. 2C). We performed an in vitro proliferation assay using HMEC and MCF10A cells and found that their proliferation was not impaired (fig. S3C). TROJAN down-regulation also suppressed cell migration and invasion abilities, while its up-regulation promoted migration and invasion abilities in vitro (Fig. 2, D to F, and fig. S3, D to G). Moreover, we established a tail vein injection model with female BALB/c nude mice to investigate the effects of TROJAN on metastasis in vivo. In both TROJAN knockdown groups (n = 5), the bioluminescence imaging signal indicating disseminated metastasis in the lung tissue was weaker, and there were fewer lung nodules than in the control group (n = 5; Fig. 2G and fig. S3H). We constructed an in vivo bone metastasis model through the intracardiac injection of SCP2 cells (with or without TROJAN knockdown). We found that TROJAN knockdown cells had a reduced bone metastasis ability (Fig. 2H and fig. S3I). We also observed that TROJAN knockdown cells had reduced liver metastasis incidence (fig. S3J). We generated a modified expression vector using two guide RNAs (gRNAs) flanking TROJAN (fig. S4, A to D), which significantly impaired TROJAN expression. We found that the deletion of TROJAN caused decreased tumor volume and lung metastasis ability in vivo, which were restored by the reintroduction of full-length TROJAN (fig. S4, E and F). Together, these results demonstrate that TROJAN is a driver of breast cancer progression, especially for TNBC metastasis.
Fig. 2. The oncogenic role of TROJAN in breast cancer.
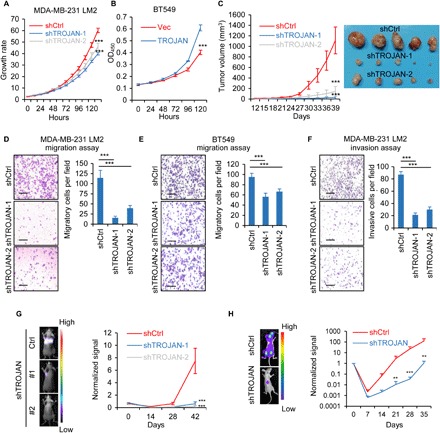
(A) In vitro growth curves of the breast cancer cell line MDA-MB-231 LM2 expressing control (Ctrl) or TROJAN shRNAs. The data are presented as the mean ± SD; n = 3 independent experiments; two-tailed unpaired Student’s t test. (B) In vitro growth curves of the breast cancer cell line BT549 expressing a control empty vector (Vec) or TROJAN. The data are presented as the mean ± SD; n = 3 independent experiments; two-tailed unpaired Student’s t test. OD450, optical density at 450 nm. (C) In vivo growth of MDA-MB-231 LM2 (39 days; n = 5) expressing control or TROJAN shRNAs. Tumor volume quantification (left) and representative tumor images (right) are shown. The data are presented as the mean ± SD; two-tailed unpaired Student’s t test. (D and E) In vitro transwell migration assay of MDA-MB-231 LM2 (D) and BT549 (E) cells expressing control or TROJAN shRNAs. The data are presented as the mean ± SD; n = 3 independent experiments; two-tailed unpaired Student’s t test. Scale bars, 200 μm. (F) In vitro transwell invasion assay of MDA-MB-231 LM2 cells expressing control or TROJAN shRNAs. The data are presented as the mean ± SD; n = 3 independent experiments; two-tailed unpaired Student’s t test. Scale bars, 200 μm. (G) In vivo intravenous xenograft mouse model of the MDA-MB-231 LM2 (42 days; n = 5) cells expressing control or TROJAN shRNAs. The relative bioluminescence intensities (BLIs) indicate lung metastasis. The data are presented as the mean ± SD; two-tailed unpaired Student’s t test. (H) BLI of bone metastasis of SCP2 cells expressing control or TROJAN shRNA. The data are presented as the mean ± SE; Wilcoxon signed-rank test. **P < 0.01 and ***P < 0.001. See also figs. S3 and S4.
Given the lack of effective targeted therapies for patients with TNBC (4, 11–13), we explored the potential role of TROJAN as a TNBC therapeutic target using antisense oligonucleotides (ASOs). We designed eight ASOs and confirmed that they all decreased TROJAN expression (Fig. 3A). The three ASOs with the highest interference efficiencies (ASO-1, ASO-4, and ASO-6) significantly impaired the proliferation of MDA-MB-231 LM2 cells (Fig. 3B). ASO-4 also impaired the proliferation of BT549 cells, while its impact on Hs578t cells, a cell line with lower TROJAN expression, was mild (fig. S5A). The proliferative ability of the ASO-transfected murine TNBC cell line 4 T1 was not reduced, which indicated that the phenotype was not caused by off-target effects (fig. S5B). We transfected the cells with ASOs without using transfection reagents to simulate in vivo conditions, and we demonstrated that ASO-4 was the most effective (Fig. 3C). However, truncation of ASO-4 did not improve its interference efficiency (fig. S5C); thus, the full 20-mer ASO-4 was selected for in vivo assays. To evaluate the efficacy of anti-TROJAN targeted therapy and the therapeutic potential of TROJAN inhibition, we created an intravenous xenograft mouse model (Fig. 3D). During the treatment process, we observed that the lung metastasis signal was significantly reduced in ASO-treated mice (Fig. 3E) and that the number of lung metastasis nodules was significantly lower in ASO-treated mice than in phosphate-buffered saline (PBS)–treated control mice (Fig. 3F). Moreover, TROJAN expression was significantly lower in the lung metastasis nodules of ASO-treated mice than in those of PBS-treated mice (fig. S5D). Previous research has suggested that ASOs may accumulate in liver, kidney, and spleen tissues (14). We measured biochemical parameters and organ weights and found that the liver, kidney, and spleen weights were similar between the PBS-treated and ASO-treated mice (fig. S5E). The murine blood chemistry analysis revealed that the alanine aminotransferase, aspartate aminotransferase, total bilirubin, and blood urea levels were similar between the treatment and control groups (fig. S5F), which suggests that ASO toxicity was limited. We also created a mammary fat pad xenograft mouse model (Fig. 3G) and observed that the tumor volume was significantly reduced in ASO-4–treated mice (Fig. 3H). Together, these results demonstrate that TROJAN is a potential therapeutic target that can be modified by ASO treatment in TNBC.
Fig. 3. Potential therapeutic role of TROJAN in breast cancer progression.
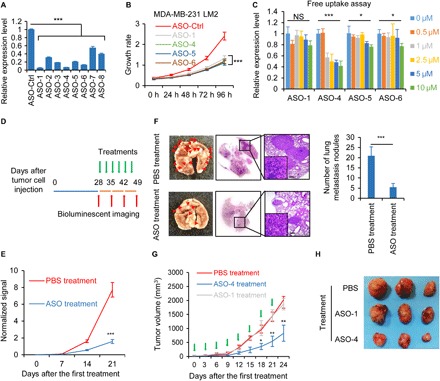
(A) Quantitative reverse transcription PCR (qRT-PCR) analysis of the relative TROJAN transcription in the MDA-MB-231 LM2 cells transfected with ASOs targeting TROJAN or control. The data are presented as the mean ± SD; n = 3 independent experiments; two-tailed unpaired Student’s t test. (B) In vitro growth assay of MDA-MB-231 LM2 cells transfected with ASOs targeting TROJAN or control. The data are presented as the mean ± SD; n = 3 independent experiments; two-tailed unpaired Student’s t test. h, hours. (C) In vitro free uptake assay in the MDA-MB-231 LM2 cells. Six different concentrations of ASO were assessed (0, 0.5, 1, 2.5, 5, and 10 μM). The 0 μM group was set as the control. qRT-PCR detection of relative TROJAN expression. The data are presented as the mean ± SD; n = 3 independent experiments. (D) Illustration of the TROJAN-targeted ASO treatment timeline. Briefly, we injected MDA-MB-231 LM2 cells into the tail veins of female BALB/c nude mice to construct a lung metastasis model. Approximately 28 days after injection, we randomly assigned eight mice with similar lung BLI signals to two groups, the PBS and ASO treatment groups. Both groups received treatment (PBS or ASO) twice per week immediately after randomization, and BLI imaging was performed once per week. The arrows indicate the different events (green, treatment; red, bioluminescence imaging). (E and F) Lung metastasis quantification (E), gross anatomy, hematoxylin and eosin (H&E) lung staining, and number of metastatic nodules in the lungs (F) are shown. The data are presented as the mean ± SD; two-tailed unpaired Student’s t test. Scale bars, 200 μm. (G) The TROJAN-targeted ASO treatment timeline and the tumor volumes of the three treatment groups (PBS, ASO-1, and ASO-4) are shown. Briefly, we injected MDA-MB-231 LM2 cells into the mammary fat pads of female NOD/SCID mice to construct an orthotopic xenograft model. Approximately 21 days after injection, we randomly assigned nine mice with similar tumor volumes to three treatment groups: PBS, ASO-1, and ASO-4 groups. All groups received treatment (PBS, ASO-1, or ASO-4) twice per week immediately after randomization. The green arrows indicate the treatments. The data are presented as the mean ± SD; two-tailed unpaired Student’s t test. (H) Representative tumor images are shown. *P < 0.05, **P < 0.01, and ***P < 0.001. NS, not significant. See also fig. S5.
TROJAN interacts with ZMYND8 protein
To explore the underlying molecular mechanisms of TROJAN, we performed RNA pull-down assays combined with stable isotope labeling with amino acids in cell culture (SILAC)–based quantitative proteomics to identify proteins associated with TROJAN. Full-length sense and antisense TROJAN strands were transcribed in vitro, purified, and labeled with biotin at their 3′ ends. We incubated TROJAN sense strands, TROJAN antisense strands, and blank beads with stable isotopes of Lys8 and Arg10–, Lys4 and Arg6–, and Lys0 and Arg0–labeled MDA-MB-231 LM2 cell lysates, respectively (fig. 6A). One irrelevant transcript, SYSL1, served as the mass spectrometry (MS) control to exclude general RNA binding proteins (table S1). After excluding the general RNA binding proteins, we identified 10 potential TROJAN-specific binding proteins (fig. S6B). We were most interested in one chromatin binding protein, zinc finger MYND-type containing 8 (ZMYND8), which reportedly plays an important role in breast cancer progression (15, 16). Because ZMYND8 was shown to repress the expression of several metastasis-related oncogenes (16), we reasoned that it might be the most important TROJAN binding protein. However, the ZMYND8 regulators remained unclear. To confirm the interaction between TROJAN and ZMYND8, we performed RNA immunoprecipitation (RIP). Compared with immunoglobulin G (IgG)–combined RNA, ZMYND8 was significantly enriched for TROJAN (Fig. 4A). Confocal microscopy of TROJAN fluorescent ISH and ZMYND8 immunofluorescence showed the colocalization of TROJAN and ZMYND8 in the nucleus (Fig. 4B).
Fig. 4. TROJAN associates with the ZMYND8 protein.
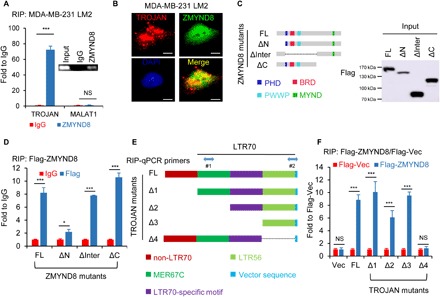
(A) RIP assay and the subsequent qRT-PCR assay and semi–RT-PCR (inset). Relative quantification of TROJAN and MALAT1 expression in MDA-MB-231 LM2 RNA-protein complexes immunoprecipitated with IgG or ZMYND8 antibodies. MALAT1 was used as a negative control. The data are presented as the mean ± SD; n = 3 independent experiments; two-tailed unpaired Student’s t test. (B) Confocal RNA fluorescence ISH and immunofluorescence images showing the colocalization of TROJAN and ZMYND8. Scale bars, 10 μm. DAPI, 4′,6-diamidino-2-phenylindole. (C) Schematic representation of Flag-tagged full-length human ZMYND8 and its deletion mutants. The ΔN, ΔInter, and ΔC constructs are depicted. The anti-Flag Western blot image showing ZMYND8 full-length or deletion mutant expression in human embryonic kidney (HEK) 293T cells. (D) RIP assay and the subsequent qRT-PCR assay performed in HEK293T cells ectopically expressing full-length Flag-tagged ZMYND8 and its deletion mutants. Relative quantification of TROJAN expression in RNA-protein complexes immunoprecipitated with IgG or Flag antibodies. The data are presented as the mean ± SD; n = 3 independent experiments; two-tailed unpaired Student’s t test. (E) TROJAN motif and schematic representation of full-length TROJAN and its deletion mutants. (F) RIP assay and the subsequent qRT-PCR assay. HEK293T cells were cotransfected with full-length TROJAN (FL)/mutants (Δ1 to Δ4)/control vector as well as Flag-ZMYND8/Flag-vector (Flag-Vec). After 48 hours, the cell lysates were immunoprecipitated with beads coated with a Flag antibody. Two primers (#1 for FL, Δ1, and Δ4 and #2 for Δ2 and Δ3) were used in the qRT-PCR assay. Fold enrichment was adjusted by each input and Flag-Vec. The data are presented as the mean ± SD; n = 3 independent experiments; two-tailed unpaired Student’s t test. *P < 0.05 and ***P < 0.001. See also fig. S6.
To identify the binding regions between TROJAN and ZMYND8, we first ectopically expressed full-length Flag-ZMYND8 and its three mutants: N-terminal truncated (ΔN), interregional truncated (ΔInter), and C-terminal truncated (ΔC) (Fig. 4C). Using the RIP assay, we demonstrated that deleting the N terminus abolished the TROJAN-ZMYND8 interaction (Fig. 4D), which indicated that TROJAN binds to the ZMYND8 N terminus. Next, we constructed a series of TROJAN mutants to screen the ZMYND8 binding motif. We truncated the length of TROJAN according to this motif (Fig. 4E). We used the anti-Flag RIP assay to demonstrate that deleting the 3′ end of TROJAN (TROJAN-Δ4) reduced its binding capacity for ZMYND8 (Fig. 4F), which indicated that this region contains the ZMYND8 binding motif. The MDA-MB-231 LM2 cells with TROJAN-Δ4 expression had an impaired migration ability in vitro compared with those with full-length TROJAN (fig. S6, C to E). Moreover, we established a tail vein injection model with female nude mice to investigate the effects of TROJAN-Δ4 on metastasis in vivo. Similar to the in vitro results, TROJAN-Δ4 had impaired lung metastasis formation ability compared with full-length TROJAN in vivo (fig. S6F). As the sequence of the TROJAN-Δ3 is highly homologous with that of several LTR56s (listed in fig. S6G, left), we performed RIP assays of cells ectopically expressing Flag-ZMYND8 and found that only three LTR56s (LTR56-2: 1.6-fold, LTR56-3: 2.1-fold, and LTR56-5: 1.8-fold) were significantly enriched in ZMYND8 immunoprecipitates (fig. S6G, right). As their fold changes compared with IgG were relatively low, we speculated that their binding capacities to ZMYND8 were weak. Together, these results indicate that TROJAN can bind the ZMYND8 N terminus at its 3′ end.
TROJAN promotes ZMYND8 protein degradation by repelling ZNF592
To explore the mechanism underlying the association between TROJAN and ZMYND8, we immunoblotted ZMYND8 in TROJAN–down-regulated cells and demonstrated that TROJAN knockdown significantly increased ZMYND8 protein levels in MDA-MB-231 LM2 cells (Fig. 5A). After treatment with the proteasomal inhibitor MG132, the control group cells expressed higher ZMYND8 levels than did the TROJAN–down-regulated cells (Fig. 5B), and similar results were observed in BT549 cells (Fig. 5, C and D). TROJAN overexpression significantly decreased ZMYND8 protein levels, and MG132 treatment antagonized this effect (Fig. 5E). To further explore the underlying mechanisms, we treated MDA-MB-231 LM2 cells with the protein synthesis inhibitor cycloheximide (CHX) and analyzed ZMYND8 stability upon TROJAN down-regulation. We found that the ZMYND8 protein half-life was substantially longer in TROJAN–down-regulated cells (Fig. 5F), suggesting that TROJAN regulates the stability of ZMYND8. In addition, ZMYND8 expression was increased in TROJAN–down-regulated lung metastasis xenografts (fig. S7A), and TROJAN knockdown did not affect the transcriptional level of ZMYND8 (fig. S7B). We next performed a ubiquitination assay to confirm whether TROJAN promotes ZMYND8 degradation through the ubiquitin-proteasome system and demonstrated that ZMYND8 ubiquitination levels were reduced upon TROJAN knockdown (Fig. 5G). Treatment with the autophagy/lysosome inhibitor chloroquine diphosphate did not alter the increase in ZMYND8 expression caused by TROJAN knockdown (fig. S7C). Together, these findings suggest that TROJAN promotes ZMYND8 degradation via ubiquitination.
Fig. 5. TROJAN regulates ZMYND8 protein stability via ubiquitination.
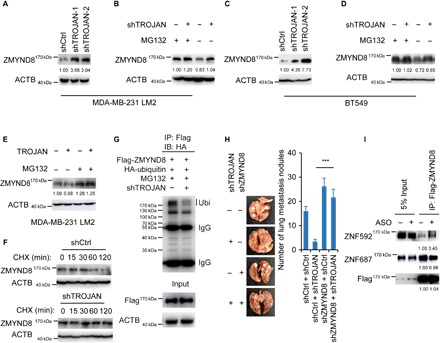
(A) Western blot images showing ZMYND8 expression in MDA-MB-231 LM2 cells expressing control or TROJAN shRNAs. (B) Western blot images showing ZMYND8 expression in MDA-MB-231 LM2 cells expressing control or TROJAN shRNA treated with dimethyl sulfoxide (DMSO) (control) or MG132. (C) Western blot images showing ZMYND8 expression in BT549 cells expressing control or TROJAN shRNAs. (D) Western blot images of ZMYND8 expression in BT549 cells expressing control or TROJAN shRNAs treated with DMSO or MG132. (E) Western blot images showing ZMYND8 expression in MDA-MB-231 LM2 cells overexpressing control or TROJAN treated with DMSO or MG132. (F) Western blot images of ZMYND8 expression in MDA-MB-231 LM2 cells expressing control or TROJAN shRNAs treated with DMSO or CHX. (G) Western blot images showing ZMYND8-associated ubiquitination (Ubi) in control and TROJAN–down-regulated HEK293T cells ectopically expressing full-length Flag-tagged ZMYND8 treated with MG132. HA, hemagglutinin. (H) In vivo intravenous xenograft mouse model in MDA-MB-231 LM2 cells expressing TROJAN and/or ZMYND8 shRNAs (n = 6 for each group). The data are presented as the mean ± SD; two-tailed unpaired Student’s t test. ***P < 0.001. (I) Western blot images showing the interaction of ZNF592 with ZMYND8 under TROJAN knockdown. See also figs. S7 and S8.
To explore whether the tumor-promoting function of TROJAN is dependent on its interaction with ZMYND8, we performed an in vivo lung metastasis assay in MDA-MB-231 LM2 cells expressing TROJAN and/or ZMYND8 shRNAs. ZMYND8 knockdown promoted their metastatic ability, which could be partially rescued by simultaneously knocking down TROJAN and ZMYND8 (fig. S7D and Fig. 5H). We performed a migration assay with MDA-MB-231 LM2 cells treated with ASO-4 under ZMYND8 knockdown (fig. S7E). The inhibition of migration by ASO-4 treatment was weakened by the additional knockdown of ZMYND8. ZMYND8 down-regulation did not affect TROJAN expression (fig. S7F). Furthermore, we evaluated the interactions between ZMYND8 and its two strong binding partners, ZNF592 and ZNF687 (15, 17). The binding capacity of ZNF592 with ZMYND8 was strengthened under TROJAN knockdown (Fig. 5I). We immunoblotted for ZMYND8 in ZNF592–down-regulated cells and demonstrated that ZNF592 knockdown significantly decreased ZMYND8 protein levels in MDA-MB-231 LM2 cells (fig. S7G). After treatment with the proteasome inhibitor MG132, the expression of ZMYND8 was recovered. We next performed a ubiquitination assay and demonstrated that ZMYND8 ubiquitination levels were increased under ZNF592 knockdown (fig. S7H), indicating that ZNF592 prevented ZMYND8 degradation and that TROJAN might have hindered their interaction, resulting in ZMYND8 instability. These data suggest that TROJAN promotes breast cancer progression mainly by degrading ZMYND8 via ubiquitination.
To identify the regulatory role of TROJAN in comprehensive gene expression, we first performed transcriptional gene microarray analysis using both TROJAN–down-regulated and control cells. Pathway analysis revealed that the genes altered by TROJAN down-regulation were mainly enriched in pathways associated with cancer (fig. S8A). Therefore, we selected the top 3 down-regulated oncogenes, EGFR (epidermal growth factor receptor), VEGFA (vascular endothelial growth factor A), and MDM2, as potential targets of TROJAN. EGFR, VEGFA, and MDM2 expression levels were decreased in the TROJAN–down-regulated TNBC cell lines (fig. S8B). We confirmed that the expression of these genes could be rescued in the TROJAN and ZMYND8 double knockdown/knockout cells (fig. S8, C and D). Previous research has demonstrated that ZMYND8 suppresses tumor progression by transcriptionally down-regulating several metastasis-related genes (16). We performed a chromatin immunoprecipitation (ChIP) assay and found that ZMYND8 occupied the EGFR, VEGFA, and MDM2 promoters (fig. S8E). Through chromatin isolation by RNA purification (ChIRP), we confirmed that TROJAN directly bound to the promoter regions of EGFR, VEGFA, and MDM2 (fig. 8F). We revealed that EGFR, VEGFA, and MDM2 were directly coregulated by TROJAN and ZMYND8.
Recent studies have also shown that ZMYND8 suppresses the expression of target genes through the recruitment of histone demethylases (such as KDM5C or KDM5D) to chromatin, causing the demethylation of transcriptional activation–related histones [such as histone H3 lysine 4 trimethylation (H3K4me3)]. We first examined the occupancy of ZMYND8 and KDM5C at the promoter of ZMYND8 target genes upon TROJAN knockdown through a ChIP assay and found that the occupancies of ZMYND8 and KDM5C were significantly increased (fig. S8G). We then examined H3K4me3 levels at the EGFR, VEGFA, and MDM2 promoters in TROJAN–down-regulated cells. H3K4me3 levels were significantly reduced in the promoter regions of target genes in TROJAN–down-regulated cells (fig. S8H). Together, these findings demonstrate that EGFR, VEGFA, and MDM2 are directly regulated by both TROJAN and ZMYND8.
TROJAN and ZMYND8 correlation in clinical samples
To investigate the clinical correlation between TROJAN and ZMYND8, we performed immunohistochemistry (IHC) analysis to detect ZMYND8 expression in breast cancer specimens (FUSCC cohorts 1 and 2, Fig. 6A). TROJAN staining negatively correlated with ZMYND8 translation but not transcription (Fig. 6, B and C). TROJAN was also positively correlated with EGFR, VEGFA, and MDM2 (fig. S9A). Patients with low ZMYND8 expression tended to have poorer prognoses (this finding was not significant in FUSCC cohort 1, and the difference in relapse-free survival was not significant in FUSCC cohort 2), as demonstrated by log-rank tests of the Kaplan-Meier curves (fig. S9, B and C). However, the TROJAN-ZMYND8 combined expression signature was able to predict the survival of patients with TNBC (Fig. 6, D and E). Together, these data support the model that TROJAN negatively regulates ZMYND8 in a clinical context as a tumor promoter.
Fig. 6. Correlation among TROJAN, ZMYND8, and tumor progression in clinical samples.
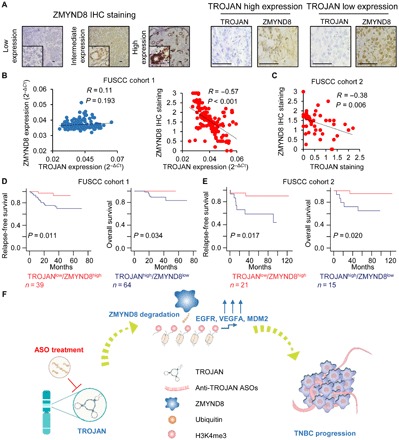
(A) IHC detection of ZMYND8 in FUSCC cohorts 1 and 2. Scale bars, 100 μm. (B and C) Correlation between TROJAN and ZMYND8 expression in FUSCC cohorts 1 (B) and 2 (C), as determined by Pearson correlation analysis. (D and E) Kaplan-Meier analysis of the relapse-free survival and overall survival of TROJANlow/ZMYND8high and TROJANhigh/ZMYND8low patients in FUSCC cohorts 1 (D) and 2 (E). (F) Schematic diagram of TROJAN regulating breast cancer progression via ZMYND8. See also fig. S9.
DISCUSSION
In this study, TROJAN was identified as both a tumor promoter and a potential therapeutic target for breast cancer, especially for TNBC, by screening breast cancer–related ERVs based on their genome-wide transcriptome profiles. TROJAN partially regulates TNBC progression via the ZMYND8 pathway (Fig. 6F).
HERVs occupy approximately 8% of the human genome (18) and are activated in a cell type–specific manner (19). Because a large number of HERVs exist, and those within the same families are highly homologous, identifying their functionality is challenging. Although 66 ERV members have been identified according to the GenBank database (6), most of the reported HERVs have protein-coding capacities (i.e., contain gag, pol, and env open reading frames) (5). In addition, the relationship between HERV transcripts and their genomic loci is complicated. One recent study classified HERV transcription patterns according to four modes: initiation in the proviral LTR, read-through transcription, part of an lncRNA, and intronic transcription (9). TROJAN is an lncRNA transcribed from a genomic locus containing an LTR70 sequence. According to Montesion et al.’s research (9), the LTR70 sequence is part of the lncRNA TROJAN.
Only one previous study has demonstrated an association of noncoding HERVs with hepatocellular carcinoma (20). Several other lines of evidence support the function of lncRNAs in TNBC (21). The lncRNA BCAR4 is highly expressed in TNBC and promotes TNBC metastasis through the hedgehog pathway (22). The lncRNA LINK-A promotes the tumorigenesis of TNBC through activation of the normoxic hypoxia-inducible factor–1α signaling pathway (23). The lncRNA LINP1 regulates the radiotherapy sensitivity of TNBC through enhancing the repair of DNA double-strand breaks (24). The functionality of most HERV regions, especially the noncoding regions, in breast cancer remains unclear. Thus, we performed genome-wide transcriptome analysis to identify additional functional long noncoding HERVs. On the basis of the 523,655 HERVs included in the RepeatMasker database, we identified one LTR70 (named TROJAN) as being highly activated in TNBC. Notably, the expression patterns of the LTR70 members differ from each other, indicating that HERV expression cannot be evaluated at the family level. As the functional noncoding HERV identified in breast cancer, TROJAN promoted breast cancer progression and was related to poor survival.
We also found that TROJAN lies upstream of ZMYND8. According to recent, emerging researches, ZMYND8 acts as a transcriptional repressor and as a tumor suppressor in different cancer types, such as breast cancer and prostate cancer (25). Shen et al. (15) reported that ZMYND8 could interact with KDM5C to suppress the transcription of enhancer RNAs, which led to the suppression of breast cancer metastasis. Li et al. (16) found that ZMYND8 could repress the metastasis but not the proliferation of cancer. Another group published several studies that also showed that ZMYND8 is a tumor suppressor (26, 27). However, one recent study claimed that ZMYND8 could be an oncogene under hypoxic conditions (28), possibly due to differences in experimental context or cell type. Given our findings and those of previous studies, we reasoned that ZMYND8 might be a metastasis repressor. Previous studies have also suggested that lncRNAs could regulate protein stability via ubiquitination. For example, LINC00673 promotes PTPN11 degradation by reinforcing the interaction between PTPN11 and PRPF19, an E3 ubiquitin ligase (29), and Hu et al. (30) described that FAL1 could stabilize the BMI1 protein by inhibiting ubiquitination. While our findings uncovered ZMYND8 ubiquitination, similar to Hu et al.’s (30) research, the detailed mechanisms of how TROJAN regulates ZMYND8 stability through the ubiquitination process require further study. TROJAN could promote breast cancer progression through multiple targets, of which ZMYND8 may be the most vital. Among the several potential target genes of TROJAN identified by microarray analysis, we found that EGFR, VEGFA, and MDM2 were coregulated by TROJAN and ZMYND8. Therefore, we conclude that TROJAN can regulate breast cancer progression via the ZMYND8-dependent pathway.
Patients with TNBC have poor outcomes and lack effective targeted therapies (4, 11–13). Although some therapeutic targets, such as immune checkpoint inhibitors (targeting programmed cell death protein 1/programmed cell death protein ligand 1), poly(adenosine 5′-diphosphate–ribose) polymerase inhibitors, and androgen receptor inhibitors, have been found (4), their efficacy in treating patients with TNBC is still uncertain (31). Furthermore, transcriptional products as therapeutic targets are more extensive because some protein products (such as MYC) are difficult to target (32, 33). Hence, we attempted to investigate the therapeutic potential of TROJAN in TNBC. Compared with ASOs, traditional RNA interference technologies, such as small interfering RNAs (siRNAs), have several drawbacks when used in the clinic: (i) short half-lives (34), (ii) generally poor target cell uptake ability (34, 35), and (iii) weak nuclear transcriptional silencing effects (36, 37). Several ASOs have been successfully used in the clinic, most notably fomivirsen and mipomersen, and have been approved by the U.S. Food and Drug Administration. Fomivirsen was registered in 1998 for the treatment of cytomegalovirus (CMV)–induced retinitis in immunocompromised patients (33, 34), and mipomersen (also known as ISIS 301012) has been approved for the treatment of homozygous familial hypercholesterolemia. Mipomersen targets and degrades apolipoprotein B100 mRNA, which is synthesized by the liver through ribonuclease H activity, and thus, reduces the concentrations of both apolipoprotein B and low-density lipoprotein cholesterol in the plasma (38). Several other ASOs have been applied in clinical trials, especially for cancer treatment (33, 39). Thus, we applied ASOs that target TROJAN as a therapeutic approach. Our current research demonstrated that all the ASOs tested could impair TROJAN expression, and we identified the one that exerted the strongest uptake ability. In the in vivo assay, the tested ASO successfully inhibited the progression of breast cancer. Our current research provides a TNBC-related transcript as a promising therapeutic target that is also critical for breast cancer progression. Our findings warrant future development to improve the efficacy and uptake ability and reduce the toxicity of ASOs. In conclusion, the breast cancer–related noncoding HERV TROJAN is not only a trigger for tumor progression but also a potential candidate for future preclinical validation for treating breast cancer, especially TNBC.
MATERIALS AND METHODS
Clinical samples
For the HERV RNA-seq analysis (Fig. 1, B and C), eight paired tissues and adjacent normal tissues were collected from the primary sites of patients with TNBC. The samples were immediately snap-frozen and stored at −80°C. To validate TROJAN expression (Fig. 1E), 53 TNBC tumors paired with adjacent normal breast tissue were included. For IHC and quantitative PCR (qPCR) analysis, 153 patients with TNBC (FUSCC cohort 1) who underwent surgery at the FUSCC from 2010 to 2013 were included. For IHC and RNAscope analyses, 200 patients (FUSCC cohort 2) who underwent surgery at the FUSCC from 2001 to 2010 were included. The survival analysis for the TCGA cohort of breast cancer was obtained from GEPIA (Gene Expression Profiling Interactive Analysis; http://gepia.cancer-pku.cn/).
RNA isolation and real-time quantitative reverse transcription–PCR
Total RNA was isolated from patient specimens or cells using the RNeasy Plus Mini Kit (QIAGEN) following the manufacturer’s protocol. First-strand cDNA synthesis from total RNA was performed using the PrimeScript RT Reagent Kit with gDNA (genomic DNA) Eraser (TAKARA). Real-time qPCR was performed using SYBR Premix Ex Taq (TAKARA) on an ABI 7900HT Fast Real-Time PCR system (Applied Biosystems). The results were analyzed with SDS v2.1 software and the 2−ΔΔCt method.
RNA-seq data, sources, and HERV analysis
RNA library preparation was performed as described in the Illumina TruSeq Stranded Total RNA LT Sample Preparation Kit with Ribo-Zero Gold (Illumina Inc.). Briefly, ribosomal RNA was depleted from 0.3 to 1 μg of total RNA. Following depletion, mRNA was fragmented to an average size of 200 to 400 bp at 94°C for 4 min. The cleaved RNA fragments were copied into first-strand cDNA using a reverse transcriptase (Invitrogen) and random primers. The first-strand cDNA was converted into double-stranded DNA in the presence of deoxyuridine triphosphate (dUTP). The incorporation of dUTP in second-strand synthesis quenched the second strand during amplification and thus improved the strand specificity of the library. These cDNA fragments then had the addition of a single “A” base and subsequent ligation of the adapter. The products were purified and enriched with PCR to create the final library. After qualification with the Agilent 2100 Bioanalyzer (Agilent Technologies) with the DNA chip and quantification with the Qubit 3.0 Fluorometer (Invitrogen), the samples were subjected to 150-bp paired-end sequencing using an Illumina HiSeq X Ten platform (Illumina Inc.). The raw data were subjected to quality control analyses using Seqtk. The RNA-seq data were then mapped to the human reference genome hg19 with TopHat2 (version: 2.0.9). The HERVs were obtained from RepeatMasker (www.repeatmasker.org/species/hg.html) for expression analyses by HTSeq-count. In total, 553 HERV species (distributed across 523,656 genomic loci) were included. The HERV differential analysis was performed using edgeR.
IHC analysis
Paraffin-embedded tissue sections were deparaffinized at 60°C for 20 min, cleared in xylene, and subjected to a graded series of alcohol. For hematoxylin and eosin (H&E) staining, slides were stained with Mayer’s hematoxylin (Sigma-Aldrich), blued in 0.1% sodium bicarbonate, and counterstained with Eosin Y solution (Sigma-Aldrich). For IHC, the slides were heated with saline sodium citrate (SSC) buffer at 95° to 100°C. After cooling, the slides were blocked with blocking solution (2% goat serum, 2% bovine serum albumin, and 0.05% Tween 20 in PBS) at room temperature and incubated with a primary antibody diluted in blocking solution at 4°C. Endogenous peroxidase activity was quenched with 0.3% H2O2. Slides were incubated with a horseradish peroxidase (HRP)–conjugated secondary antibody (GeneTech) at room temperature, developed with 3,3′-diaminobenzidine substrate (GeneTech), counterstained and blued with hematoxylin, and dehydrated with a graded series of alcohol. The positive staining density was measured using a computerized imaging system composed of a Leica CCD camera DFC420 connected to a Leica DM IRE2 microscope (Leica Microsystems Imaging Solutions Ltd.). The H-score scoring system was used, which evaluated staining intensity (0 to 3) and the percentage of positively stained cells (0 to 1), with a final score ranging from 0 to 3.
RNA ISH
RNA ISH was performed on tissue microarrays using an RNAscope 2.5 HD Detection Kit (RED, Advanced Cell Diagnostics) or an RNAscope Multiplex Fluorescent Reagent Kit according to the manufacturer’s instructions. For ISH, tissue sections were deparaffinized with xylene and 100% ethanol, incubated in a H2O2 solution for 10 min, heated in target retrieval buffer at 95° to 99°C for 20 min, and digested in protease solution for 30 min. The slides were then hybridized with a custom probe, Hs-LOC105372310-O2, in a HybEZ oven (Advanced Cell Diagnostics) at 40°C for 2 hours. After signal amplification and detection, the slides were dried in a 60°C dry oven for 15 min and mounted with EcoMount. Positive staining density was measured using a computerized imaging system composed of a Leica CCD DFC420 camera connected to a Leica DM IRE2 microscope (Leica Microsystems Imaging Solutions Ltd.). The H-score scoring system was used, which evaluates staining intensity (0 to 3) and the percentage of positively stained cells (0 to 1), with a final score ranging from 0 to 3. For fluorescent ISH, cells were rinsed in PBS twice, fixed in 4% formaldehyde in PBS (pH 7.4) for 30 min at room temperature, incubated in a H2O2 solution for 10 min, and digested in protease solution for 10 min. The slides were then hybridized with a custom probe, Hs-LOC105372310-O2, in a HybEZ oven (Advanced Cell Diagnostics) at 40°C for 2 hours. After signal amplification, the slides were conjugated with TSA Plus Cy3 (PerkinElmer). The slides were then incubated with anti-ZMYND8 antibody overnight at 4°C. After incubation with the fluorescein-conjugated secondary antibody, the slides were mounted with ProLong Gold Antifade Reagent with 4′,6-diamidino-2-phenylindole.
Microarray
Total RNA (300 ng) was amplified using the WT Expression Kit (Ambion) according to the manufacturer’s instructions. cRNA (10 μg) was used for reverse transcription, and 5.5 μg of cDNA was hybridized to GeneChip Human Transcriptome Array 2.0 (Affymetrix) according to the manufacturer’s protocol. After overnight hybridization, the chips were terminally labeled and scanned using an Affymetrix GeneChip 3000 7G scanner.
Cell lines, transfection, and lentiviral shRNA vectors
Immortalized HMEC and MCF-10A cells; human breast cancer cells MDA-MB-231, MDA-MB-468, Hs578t, and BT549; and human embryonic kidney (HEK) 293T cells were obtained from the American Type Culture Collection and cultured under standard conditions. The standard Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) protocol was followed for all transient plasmid transfections.
Lentiviruses were generated using the pLKO.1 vector and packaging plasmids (psPAX2 and pMD2.G) in the HEK293T cell line. Annealing oligonucleotides targeting TROJAN were synthesized (Sangon Biotech) and cloned into the pLKO.1-Puro vector. Supernatants were collected and filtered through a 0.45-μm syringe filter. Target cells were infected with lentivirus in the presence of polybrene (6 mg/ml; Sigma-Aldrich) at a multiplicity of infection of 0.7. Stably transduced cells were generated by puromycin selection for 3 days beginning 24 hours after infection. The knockdown efficiency and specificity of all the shRNAs were validated with qPCR or immunoblotting. The two shRNAs producing the best knockdown efficiency were used in subsequent studies (shTROJAN-1, 5′-GCAGTCTCTTAAGCAGATTGA-3′; shTROJAN-2, 5′-GCAACTGCTGTTAATGAAAGT-3′; shZMYND8, 5′-CGGAGTAATAAATCCAGTT-3′).
The siRNA transfections were conducted with the Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The target sequence for siZNF592 was 5′-GCCAGATCCTGATGATCCA-3′.
Establishment of knockout strains
Two gRNA sequences spanning the TROJAN genomic locus were designed with the CRISPR Design Tool (https://zlab.bio/guide-design-resources). A one-step cloning method was used to generate two single gRNAs in LentiCRISPRv2 as previously described (40), with the exception that one of the U6 promoters was replaced with an H1 promoter to avoid potential homologous recombination. The H1 promoter sequence was amplified from pUC-H1-gRNA (Addgene plasmid 61089). The TROJAN deletion efficiency was measured by PCR. The following gRNA was used for ZMYND8 knockout: 5′-GTGATGTGTCCTGCGGCGAG-3′. Single clones were established by dilution cloning.
5′ and 3′ RACE
5′-RACE and 3′-RACE analyses were performed using the SMARTer RACE cDNA Amplification Kit (Clontech Laboratories) according to the manufacturer’s instructions. Briefly, total RNA from MDA-MB-231 cells was extracted with an RNeasy Mini Kit (QIAGEN), and gDNA was removed by on-column deoxyribonuclease I [ribonuclease (RNase) free; New England Biolabs] digestion. After a poly(A) tail was added using poly(A) polymerase (TAKARA), 5′-RACE and 3′-RACE products were amplified with their respective lncRNA-specific primers and cloned into the pGEM-T Easy Vector (Promega) for sequencing, and the spliced full-length lncRNAs were obtained using a new pair of primers. The poly(A) tail detection assay was performed using total RNA from MDA-MB-231 cells, which were either treated or not treated with poly(A) polymerase (TAKARA). The relative abundance of TROJAN in poly(A) polymerase–treated or untreated RNA was determined by qPCR. Glyceraldehyde-3-phosphate dehydrogenase [containing poly(A)] and U6 [without poly(A)] were used as reference genes.
Cloning procedures
Full-length TROJAN and its truncated mutants were delivered in pcDNA3.1(+) (Thermo Fisher Scientific) or pCDH-CMV-MCS-EF1-blast (modified from pCDH-CMV-MCS-EF1-puro from System Biosciences). Full-length ZMYND8 (cloned into pCMV-TAG2B) was a gift from F. Lan. The truncated ZMYND8 mutant cDNAs (ΔInter and ΔC) were gifts from K. M. Miller and were lastly cloned into pCMV-TAG2B.
In vitro cell growth assay
Cells were seeded in 96-well plates and imaged using the IncuCyte ZOOM System (Essen BioScience). Frames were captured at 12-hour intervals from four separate regions per well. Cultures were maintained at 37°C in an incubation chamber, and the growth rate was analyzed using IncuCyte software (2013A Rev2).
Transwell migration and invasion assays
For the migration assay, 5 × 104 cells in serum-free medium were seeded in the top chamber of the 24-well insert (pore size, 8 μm; BD Biosciences). Medium containing 10% serum was used as a chemoattractant in the lower chamber. After incubation for 6 to 8 hours, the cells that had migrated to the opposite side of the membrane were fixed with 4% paraformaldehyde and stained with crystal violet. For the invasion assays, 1 × 105 MDA-MB-231 LM2 cells in serum-free medium were plated in the top chamber containing a Matrigel-coated membrane with a 24-well insert (pore size, 8 μm; BD Biosciences). Medium supplemented with 10% serum was used as a chemoattractant in the lower chamber. After incubation for 24 hours, cells that had invaded the opposite side of the membrane were fixed with 4% paraformaldehyde and stained with crystal violet.
Antibodies
The following primary antibodies were used: anti-ZMYND8 (Proteintech), anti-ACTB (Actin Beta) (Proteintech), anti-hemagglutinin (HA; Proteintech), anti-Flag (Proteintech), anti-ubiquitin (Proteintech), anti-H3K4me3 (Cell Signaling Technology), anti-KDM5C (Bethyl), anti-ZNF592 (Bethyl), and anti-ZNF687 (Bethyl). The HRP-linked anti-mouse (1/5000) and anti-rabbit antibodies (1/5000) were purchased from Cell Signaling Technology.
Western blotting
Cells were lysed in SDS lysis buffer [50 mM tris (pH 8.1), 1 mM EDTA, 1% SDS, 1 mM fresh dithiothreitol, sodium fluoride, and leupeptin]. The lysates were centrifuged at 10,000g for 20 min, supernatants were collected, and protein concentrations were determined with the bicinchoninic acid (BCA) Protein Assay Kit (Pierce). A total of 30 to 60 μg of protein was separated by SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore). The primary and secondary antibodies were described above, and the SuperSignal West Femto Substrate Trial Kit (Pierce) was used as the enhanced chemiluminescence substrate. ImageJ was used to quantify the Western blotting results by densitometry.
Cytoplasmic/nuclear RNA purification
Cytoplasmic and nuclear RNAs were purified using the Cytoplasmic and Nuclear RNA Purification Kit (Norgen) according to the manufacturer’s instructions. Briefly, 3 × 106 MDA-MB-231 LM2 cells were harvested and incubated with lysis solution for 5 min. After centrifugation, cytoplasmic RNA was isolated from the supernatants, and nuclear RNA was isolated from the pellets.
Xenograft in vivo model
For the in vivo mammary fat pad xenograft model illustrated in Fig. 2C and fig. S4E, 6-week-old female NOD/SCID mice were used. MDA-MB-231 LM2 cells (1 × 106) were harvested and resuspended in 50 μl of PBS and 50 μl of Matrigel. Cells were injected directly into the mammary fat pads of the mice, and tumor volumes were calculated by V = L × (W × ½)2, where L is length (longest dimension) and W is width (shortest dimension). For the in vivo lung metastasis xenograft model illustrated in Fig. 2G and figs. S4F and S6F, 6-week-old female BALB/c nude mice were used. MDA-MB-231 LM2 cells (1 × 105; stably expressing shTROJAN or shCtrl) were harvested in PBS and injected into the lateral tail vein in a total volume of 100 μl. For the in vivo bone metastasis xenograft model illustrated in Fig. 2H, 6-week-old female BALB/c nude mice were used. SCP2 cells (1 × 105; stably expressing shTROJAN or shCtrl) were harvested in PBS and injected into the left ventricle in a total volume of 100 μl. The mice were imaged for luciferase activity immediately after injection to exclude baseline bias. After the mice were euthanized, their lungs or livers were removed, imaged, and placed into 4% paraformaldehyde for paraffin embedding. For the bone metastasis xenograft models, the forelimb and hindlimb long bones of the mice were removed, fixed in 4% paraformaldehyde after 24 hours, decalcified (10% EDTA, 2 weeks), dehydrated through a graded alcohol series, embedded in paraffin, and stained with H&E.
The bioluminescence imaging shown in Fig. 2G was performed using the Lumazone imaging system (MAG BioSystems), and the bioluminescence imaging shown in Fig. 2H and figs. S4F and S6F was performed using the NightOWL LB 983 in vivo imaging system (Berthold Technologies). Relative bioluminescence signal quantitation was calculated by the respective imaging system software packages.
For biochemical analysis, the eyeballs of the mice were extracted, and blood samples were collected, placed at room temperature for 30 min, and centrifuged at 4500g for 15 min. The serum (supernatants) samples were collected for biochemical analysis using the automatic biochemical analyzer 8021A (URIT).
ASO delivery
Eight phosphorothioate-modified ASOs targeting TROJAN were designed and purchased from BioSune in Shanghai, China: ASO-1, 5′-mUmUmAmGmCAGCTTGAAGCmCmAmGmGmU-3′; ASO-2, mCmAmAmUmCTGCTTAAGAGmAmCmUmGmC; ASO-3, mUmGmAmAmCAGTTTCAAGAmGmAmUmGmA; ASO-4, mAmCmUmUmGTGGCAATAGTmGmAmAmCmA; ASO-5, mGmGmAmCmAGAGAGTGGGCmUmGmAmUmA; ASO-6, mAmGmGmAmCAGAGAGTGGGmCmUmGmAmU; ASO-7, mGmGmAmUmTATAACCTGGGmAmGmCmAmA; ASO-8, mCmUmGmCmATCCTTTGGTGmUmUmAmCmA; ASO-4 (18-mer), mCmUmUmGTGGCAATAGTmGmAmAmC; ASO-4 (16-mer), mUmUmGTGGCAATAGTmGmAmA; ASO-4 (14-mer), mUmGTGGCAATAGTmGmA; ASO-4 (13-mer), mUmGTGGCAATAGmUmG.
An ASO not matching any known human transcripts (Ionis 141923) was used as the negative control (ASO-Ctrl, mCmCmUmUmCCCTGAAGGTTmCmCmUmCmC). For in vitro ASO delivery, 1 × 105 MDA-MB-231 LM2 cells were seeded in six-well plates and transfected with 1.4 μg of ASO premixed with 5.6 μl of Lipofectamine 2000, and RNA was isolated after 24 hours. For the in vitro proliferation assay, 1 × 103 MDA-MB-231 LM2 cells were seeded in 96-well plates and transfected with 0.35 μg of ASO premixed with 0.35 μl of Lipofectamine 2000. For the in vitro free uptake assay, 1 × 105 MDA-MB-231 LM2 cells were seeded in six-well plates and transfected with ASOs at various concentrations (0, 0.5, 1, 2.5, 5, and 10 μM) without a transfection reagent, and RNA was isolated after 24 hours. For in vivo ASO delivery, ASOs were delivered by intraperitoneal injection at a dose of 25 mg/kg (lung metastasis xenograft model) or 50 mg/kg (mammary fat pad xenograft model). PBS was injected as the control.
Preparation of triple SILAC
SILAC Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific) was supplemented with 10% dialyzed fetal bovine serum (Thermo Fisher Scientific) and 1% streptomycin/penicillin. SILAC medium containing lysine (42 mg/liter), arginine (73 mg/liter), and proline (200 mg/liter) was added to prevent the arginine-to-proline conversion. The heavy culture medium was supplemented with lysine (U-13C6, 99%; U-15 N2, 99%) (Lys8) and arginine (U-13C6, 99%; U-15 N4, 99%) (Arg10). The culture medium was supplemented with lysine (4,4,5,5-D4, 96 to 98%) (Lys4) and arginine (U-13C6, 99%) (Arg6). All isotope amino acids were synthesized by Cambridge Isotope Laboratories, and unlabeled amino acids were purchased from Sigma-Aldrich. Cells were grown in parallel for at least five generations.
RNA pull-down assay
Full-length sense and antisense TROJAN strands were cloned into pGEM-T Easy Vector (Promega). In vitro transcription was performed using the HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs), and RNA was purified using the RNeasy MinElute Cleanup Kit (QIAGEN). TROJAN was labeled using the Biotin 3′ End DNA Labeling Kit (Thermo Fisher Scientific). Cells were harvested and resuspended in freshly prepared radioimmunoprecipitation assay lysis buffer supplemented with RNaseOUT recombinant ribonuclease inhibitor (50 U/ml; Thermo Fisher Scientific), SUPERase In RNase inhibitor (50 U/ml; Thermo Fisher Scientific), and a protease/phosphatase inhibitor cocktail (Roche). RNA secondary structure formation was assessed as follows. Briefly, 20 μg of total biotin-labeled RNA was first restructured in RNA structure buffer [10 mM tris (pH 7.0), 0.1 M KCl, and 10 mM MgCl2] at 90°C for 2 min, immediately placed on ice for 2 min, and then incubated at room temperature for 20 min. Labeled RNA was incubated with streptavidin magnetic beads (Pierce) for 30 min at room temperature with agitation. The RNA-captured magnetic beads were washed twice with wash buffer (Thermo Fisher Scientific). In total, 10 mg of SILAC cell lysates was added to yeast transfer RNA (0.1 μg/μl) and 5 mM MgCl2. Sense and antisense RNA captured on the magnetic beads, as well as blank beads, were incubated with Lys0 and Arg0, Lys4 and Arg6, and Lys8 and Arg10 cell lysates, respectively, in protein-RNA binding buffer (Thermo Fisher Scientific) overnight at 4°C with agitation. RNA binding protein complexes were washed five times with ice-cold wash buffer and boiled in SDS lysis buffer.
Mass spectrometry
Liquid chromatography tandem MS (LC-MS/MS) experiments were performed on a high-performance LC system comprising a nanoACQUITY Binary Solvent Manager LC pump and a nanoACQUITY Sample Manager (all from Waters Corporation, Milford, MA, USA) connected to an linear trap quadropole (LTQ) Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Samples were injected onto an Acclaim PepMap precolumn (0.1 mm by 20 mm; Thermo Fisher Scientific) for 2 min at a flow rate of 8 μl/min and subsequently separated on an Acclaim PepMap RSLC column (0.075 mm by 250 mm; Thermo Fisher Scientific) at a flow rate of 300 μl/min. The mobile phases comprised 0.1% formic acid (phase A and the loading phase) and 99.9% acetonitrile with 0.1% formic acid (phase B). A 90-min linear phase B gradient from 3 to 35% was used. The separated samples were introduced into the mass spectrometer via a nanoelectrospray source (Thermo Electron Corporation). The spray voltage and heating capillary were set at 1.6 kV and 200°C, respectively. The mass spectrometer was operated in data-dependent mode, and each duty cycle consisted of one full MS survey scan in the 350- to 1600-Da mass range with a resolution power of 60,000 using an Orbitrap, followed by MS2 experiments to identify the 10 strongest peaks with the LTQ. Peptides were fragmented in the LTQ instrument using collision-induced dissociation with helium. Normalized collision energy values were set at 35%, and previously fragmented peptides were excluded for 60 s.
Database search
Protein searches were performed with MaxQuant 1.5.3.8 software against a database comprising 246 common contaminant proteins and the SWISS-PROT database for humans (version 2014_07; downloaded from www.uniprot.org). Triple multiplicities for Lys0 and Arg0, Lys4 and Arg6, and Lys8 and Arg10 were used for SILAC quantitation, and the variable modifications were methionine (M) oxidation, protein N-terminal acetylation, and lysine (K) acetylation. The maximum number of missed cleavage sites was set to 2, and the false discovery rate identification acceptance criteria were less than 1% for peptides, proteins, and modification sites.
RIP assay
RIP was performed using the Magna RIP Kit according to the manufacturer’s instructions (Millipore). Briefly, 2 × 107 MDA-MB 231 LM2 cells were trypsinized, rinsed twice with ice-cold PBS, resuspended in an equal pellet volume of RIP lysis buffer supplemented with protease inhibitor cocktail and an RNase inhibitor, and subjected to a single freeze-thaw cycle for gentle lysis. A total of 5 μg of antibody was added to the magnetic beads, and the mixture was incubated for 30 min at room temperature in RIP wash buffer with agitation. The beads were washed three times in RIP wash buffer, and the cell lysates were added and incubated at 4°C overnight. Each immunoprecipitant was resuspended in proteinase K buffer [proteinase K (1.2 μg/μl) and 1% SDS] and incubated at 55°C for 30 min. RNA was isolated by phenol, chloroform, and isoamyl alcohol according to the manufacturer’s instructions and detected by quantitative reverse transcription (qRT)–PCR.
IP and co-IP assays
For ubiquitination analysis, 2 × 106 HEK293T cells were transiently transfected with 5 μg of HA-ubiquitin and 5 μg of Flag-ZMYND8. After 48 hours, the cells were treated with MG132 (10 mg/ml; Sigma-Aldrich) for 4 hours before harvesting. Cells were lysed in lysis buffer [50 mM tris (pH 7.4), 150 mM NaCl, 1% NP-40, and 0.25% sodium deoxycholate] and immunoprecipitated with anti-Flag M2 magnetic beads (Sigma-Aldrich) at 4°C overnight. The immunoprecipitants were extensively washed with lysis buffer five times, eluted with SDS loading buffer by boiling for 5 min, and immunoblotted with an anti-HA antibody. For the co-IP assay, 1 × 107 cells were lysed in lysis buffer and immunoprecipitated with anti-Flag M2 magnetic beads at 4°C overnight. The immunoprecipitants were thoroughly washed with lysis buffer five times, eluted by boiling with SDS loading buffer for 5 min, and immunoblotted with anti-ZNF592, anti-ZNF687, and anti-Flag antibodies.
Chromatin immunoprecipitation
Briefly, 1 × 107 cells were cross-linked with 1% formaldehyde, sonicated to create 200- to 500-bp fragments in ChIP lysis buffer [50 mM Hepes (pH 7.5), 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% Na-deoxycholate, supplemented with protease inhibitor cocktail], and incubated with 3 μg of specific antibodies with protein A/G magnetic beads (Millipore). The beads were washed three times with lysis buffer, three times with wash buffer (50 mM Hepes, 300 mM LiCl, 1 mM EDTA, 0.5% NP-40, and 0.7% Na-deoxycholate), and once with tris-EDTA buffer (TE). Each immunoprecipitate was eluted and reverse cross-linked in elution buffer [50 mM tris-HCl (pH 8.0), 10 mM EDTA, and 1% SDS] at 65°C for 6 hours. After RNase A and proteinase K digestion, DNA samples were isolated with phenol, chloroform, and isoamyl alcohol according to the manufacturer’s instructions and analyzed by qRT-PCR.
Chromatin isolation by RNA purification
Briefly, 1 × 107 cells were cross-linked with 1% glutaraldehyde, sonicated to create 200- to 500-bp fragments in ChIRP lysis buffer [50 mM tris-Cl (pH 7.0), 10 mM EDTA, and 1% SDS, supplemented with protease inhibitor cocktail and an RNase inhibitor], diluted in a double volume of hybridization buffer [750 mM NaCl, 1% SDS, 50 mM tris-Cl (pH 7.0), 1 mM EDTA, and 15% formamide, supplemented with protease inhibitor cocktail and an RNase inhibitor], and incubated with 100 pmol of 3′-biotin–labeled probes (RiboBio) with streptavidin magnetic beads (Pierce) at 37°C for 4 hours. The beads were washed five times in wash buffer (2× SSC and 0.5% SDS, supplemented with protease inhibitor cocktail and an RNase inhibitor) at 37°C. RNA samples were digested by proteinase K; isolated with phenol, chloroform, and isoamyl alcohol according to the manufacturer’s instructions; and analyzed by qRT-PCR. DNA samples were digested by RNase A, RNase H, and proteinase K; isolated with phenol, chloroform, and isoamyl alcohol according to the manufacturer’s instructions; and analyzed by qRT-PCR.
Ethical approval
All the procedures involving patients were in accordance with the Declaration of Helsinki (1964, amended in 1975, 1983, 1989, 1996, and 2000) of the World Medical Association. This study was approved by the Ethics Committee of FUSCC, and each participant signed an informed consent document. The animal protocols were approved by the Animal Welfare Committee of Shanghai Medical College at Fudan University.
Statistical analysis
Statistical analysis was performed using SPSS and R software. P < 0.05 indicates significance. The survival curves were constructed according to the Kaplan-Meier method and compared with the log-rank test. Relapse-free survival was assessed from the date of surgery to the date of local relapse or distant relapse. Overall survival was assessed from the date of surgery to the date of death or last follow-up. Patients without events or death were censored at the last follow-up.
Supplementary Material
Acknowledgments
We are grateful to J. Wu, G.-Y. Liu, and Z.-Z. Shen for excellent data management. We also thank K. M. Miller for providing the cDNA of the ZMYND8-truncated mutants, Q.-Y. Lei for the helpful comments, and P.-Y. Zhang and M. Yao for providing technical assistance with the animal experiments and bioluminescence imaging. Funding: This work was supported by the National Natural Science Foundation of China (81572583, 81874113, 81874112, 81502278, 81372848, 81370075, 81530075, 81773155, 81672600, and 81722032), the Municipal Project for Developing Emerging and Frontier Technology in Shanghai Hospitals (SHDC12010116), the Cooperation Project of Conquering Major Diseases in the Shanghai Municipality Health System (2013ZYJB0302), the Innovation Team of the Ministry of Education (IRT1223), the Shanghai Key Laboratory of Breast Cancer (12DZ2260100), the Training Plan of Excellent Talents in Shanghai Municipality Health System (2017YQ038), the Training Plan of Excellent Talents in FUSCC (YJJQ201801), the “Chen Guang” project supported by the Shanghai Municipal Education Commission and Shanghai Education Development Foundation (17CG01), the Shanghai Pujiang Program (18PJD007), and the Training Plan of Excellent Talents of FUSCC (YJYQ201602). Author contributions: X.J. and X.-E.X. conducted and analyzed most of the experiments. X.-E.X., Y.-Z.J., K.-D.Y., and Z.-M.S. designed the study. Y.-R.L., W.S., Y.-X.R., and Y.-J.G. conducted the remaining experiments. Y.-Z.J. contributed to the tissue preparation. X.H., X.-H.H., S.-L.H., D.-Q.L., H.-J.S., F.L., S.L., Y.-F.H., G.-H.H., and G.-H.D. provided crucial reagents and discussion. X.J., X.-E.X., W.-J.Z., and Y.-Z.J. prepared the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the corresponding authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/3/eaat9820/DC1
Fig. S1. TROJAN properties.
Fig. S2. Expression pattern of TROJAN.
Fig. S3. TROJAN phenotype.
Fig. S4. Construction of the TROJAN knockout cell line.
Fig. S5. Potential therapeutic role of TROJAN in breast cancer progression.
Fig. S6. TROJAN associates with the ZMYND8 protein.
Fig. S7. TROJAN degrades the ZMYND8 protein.
Fig. S8. TROJAN and ZMYND8 regulate the transcription of diverse target genes.
Fig. S9. TROJAN regulates breast cancer progression via ZMYND8.
Table S1. TNBC-related HERVs, basic patient information, primers, and MS data for proteins pulled down by TROJAN and SYSL1.
REFERENCES AND NOTES
- 1.DeSantis C. E., Fedewa S. A., Sauer A. G., Kramer J. L., Smith R. A., Jemal A., Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J. Clin. 66, 31–42 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Perou C. M., Sørlie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Rees C. A., Pollack J. R., Ross D. T., Johnsen H., Akslen L. A., Fluge Ø., Pergamenschikov A., Williams C., Zhu S. X., Lønning P. E., Børresen-Dale A.-L., Brown P. O., Botstein D., Molecular portraits of human breast tumours. Nature 406, 747–752 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Zardavas D., Piccart M., Neoadjuvant therapy for breast cancer. Annu. Rev. Med. 66, 31–48 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Denkert C., Liedtke C., Tutt A., von Minckwitz G., Molecular alterations in triple-negative breast cancer—The road to new treatment strategies. Lancet 389, 2430–2442 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Thompson P. J., Macfarlan T. S., Lorincz M. C., Long terminal repeats: From parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol. Cell 62, 766–776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer J., Blomberg J., Seal R. L., A revised nomenclature for transcribed human endogenous retroviral loci. Mobile DNA 2, 7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pi W., Yang Z., Wang J., Ruan L., Yu X., Ling J., Krantz S., Isales C., Conway S. J., Lin S., Tuan D., The LTR enhancer of ERV-9 human endogenous retrovirus is active in oocytes and progenitor cells in transgenic zebrafish and humans. Proc. Natl. Acad. Sci. U.S.A. 101, 805–810 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherkasova E., Scrivani C., Doh S., Weisman Q., Takahashi Y., Harashima N., Yokoyama H., Srinivasan R., Linehan W. M., Lerman M. I., Childs R. W., Detection of an immunogenic HERV-E envelope with selective expression in clear cell kidney cancer. Cancer Res. 76, 2177–2185 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montesion M., Bhardwaj N., Williams Z. H., Kuperwasser C., Coffin J. M., Mechanisms of HERV-K (HML-2) transcription during human mammary epithelial cell transformation. J. Virol. 92, e01258-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Y.-Z., Liu Y.-R., Xu X.-E., Jin X., Hu X., Yu K.-D., Shao Z.-M., Transcriptome analysis of triple-negative breast cancer reveals an integrated mRNA-lncRNA signature with predictive and prognostic value. Cancer Res. 76, 2105–2114 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Dent R., Trudeau M., Pritchard K. I., Hanna W. M., Kahn H. K., Sawka C. A., Lickley L. A., Rawlinson E., Sun P., Narod S. A., Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 13, 4429–4434 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Xu H., Eirew P., Mullaly S. C., Aparicio S., The omics of triple-negative breast cancers. Clin. Chem. 60, 122–133 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Lehmann B. D., Pietenpol J. A., Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J. Pathol. 232, 142–150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu R. Z., Grundy J. S., Geary R. S., Clinical pharmacokinetics of second generation antisense oligonucleotides. Expert Opin. Drug Metab. Toxicol. 9, 169–182 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Shen H., Xu W., Guo R., Rong B., Gu L., Wang Z., He C., Zheng L., Hu X., Hu Z., Shao Z.-M., Yang P., Wu F., Shi Y. G., Shi Y., Lan F., Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell 165, 331–342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li N., Li Y., Lv J., Zheng X., Wen H., Shen H., Zhu G., Chen T.-Y., Dhar S. S., Kan P.-Y., Wang Z., Shiekhattar R., Shi X., Lan F., Chen K., Li W., Li H., Lee M. G., ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Mol. Cell 63, 470–484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong F., Chiu L.-Y., Cox B., Aymard F., Clouaire T., Leung J. W., Cammarata M., Perez M., Agarwal P., Brodbelt J. S., Legube G., Miller K. M., Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 29, 197–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordaux R., Batzer M. A., The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10, 691–703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacques P.-É., Jeyakani J., Bourque G., The majority of primate-specific regulatory sequences are derived from transposable elements. PLOS Genet. 9, e1003504 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimoto K., Suzuki A. M., Dos Santos A., Desterke C., Collino A., Ghisletti S., Braun E., Bonetti A., Fort A., Qin X.-Y., Radaelli E., Kaczkowski B., Forrest A. R. R., Kojima S., Samuel D., Natoli G., Buendia M. A., Faivre J., Carninci P., CAGE profiling of ncRNAs in hepatocellular carcinoma reveals widespread activation of retroviral LTR promoters in virus-induced tumors. Genome Res. 25, 1812–1824 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q., Gao S., Li H., Lv M., Lu C., Long noncoding RNAs (lncRNAs) in triple negative breast cancer. J. Cell. Physiol. 232, 3226–3233 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Xing Z., Lin A., Li C., Liang K., Wang S., Liu Y., Park P. K., Qin L., Wei Y., Hawke D., Hung M.-C., Lin C., Yang L., lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159, 1110–1125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin A., Li C., Xing Z., Hu Q., Liang K., Han L., Wang C., Hawke D. H., Wang S., Zhang Y., Wei Y., Ma G., Park P. K., Zhou J., Zhou Y., Hu Z., Zhou Y., Marks J. R., Liang H., Hung M.-C., Lin C., Yang L., The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat. Cell Biol. 18, 213–224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., He Q., Hu Z., Feng Y., Fan L., Tang Z., Yuan J., Shan W., Li C., Hu X., Tanyi J. L., Fan Y., Huang Q., Montone K., Dang C. V., Zhang L., Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat. Struct. Mol. Biol. 23, 522–530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong F., Miller K. M., Double duty: ZMYND8 in the DNA damage response and cancer. Cell Cycle 17, 414–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basu M., Khan M. W., Chakrabarti P., Das C., Chromatin reader ZMYND8 is a key target of all trans retinoic acid-mediated inhibition of cancer cell proliferation. Biochim. Biophys. Acta 1860, 450–459 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Basu M., Sengupta I., Khan M. W., Srivastava D. K., Chakrabarti P., Roy S., Das C., Dual histone reader ZMYND8 inhibits cancer cell invasion by positively regulating epithelial genes. Biochem. J. 474, 1919–1934 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Chen Y., Zhang B., Bao L., Jin L., Yang M., Peng Y., Kumar A., Wang J. E., Wang C., Zou X., Xing C., Wang Y., Luo W., ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J. Clin. Invest. 128, 1937–1955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng J., Huang X., Tan W., Yu D., Du Z., Chang J., Wei L., Han Y., Wang C., Che X., Zhou Y., Miao X., Jiang G., Yu X., Yang X., Cao G., Zuo C., Li Z., Wang C., Cheung S. T., Jia Y., Zheng X., Shen H., Wu C., Lin D., Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat. Genet. 48, 747–757 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Hu X., Feng Y., Zhang D., Zhao S. D., Hu Z., Greshock J., Zhang Y., Yang L., Zhong X., Wang L.-P., Jean S., Li C., Huang Q., Katsaros D., Montone K. T., Tanyi J. L., Lu Y., Boyd J., Nathanson K. L., Li H., Mills G. B., Zhang L., A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 26, 344–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lord C. J., Tutt A. N. J., Ashworth A., Synthetic lethality and cancer therapy: Lessons learned from the development of PARP inhibitors. Annu. Rev. Med. 66, 455–470 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Gustafson W. C., Meyerowitz J. G., Nekritz E. A., Chen J., Benes C., Charron E., Simonds E. F., Seeger R., Matthay K. K., Hertz N. T., Eilers M., Shokat K. M., Weiss W. A., Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell 26, 414–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams B. D., Parsons C., Walker L., Zhang W. C., Slack F. J., Targeting noncoding RNAs in disease. J. Clin. Invest. 127, 761–771 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kole R., Krainer A. R., Altman S., RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 11, 125–140 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fox R. G., Lytle N. K., Jaquish D. V., Park F. D., Ito T., Bajaj J., Koechlein C. S., Zimdahl B., Yano M., Kopp J. L., Kritzik M., Sicklick J. K., Sander M., Grandgenett P. M., Hollingsworth M. A., Shibata S., Pizzo D., Valasek M. A., Sasik R., Scadeng M., Okano H., Kim Y., MacLeod A. R., Lowy A. M., Reya T., Image-based detection and targeting of therapy resistance in pancreatic adenocarcinoma. Nature 534, 407–411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutschner T., Hämmerle M., Eißmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Groß M., Zörnig M., MacLeod A. R., Spector D. L., Diederichs S., The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73, 1180–1189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lennox K. A., Behlke M. A., Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 44, 863–877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raal F. J., Santos R. D., Blom D. J., Marais A. D., Charng M.-J., Cromwell W. C., Lachmann R. H., Gaudet D., Tan J. L., Chasan-Taber S., Tribble D. L., Flaim J. D., Crooke S. T., Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: A randomised, double-blind, placebo-controlled trial. Lancet 375, 998–1006 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Viney N. J., van Capelleveen J. C., Geary R. S., Xia S., Tami J. A., Yu R. Z., Marcovina S. M., Hughes S. G., Graham M. J., Crooke R. M., Crooke S. T., Witztum J. L., Stroes E. S., Tsimikas S., Antisense oligonucleotides targeting apolipoprotein(a) in people with raised lipoprotein(a): Two randomised, double-blind, placebo-controlled, dose-ranging trials. Lancet 388, 2239–2253 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Cao J., Wu L., Zhang S.-M., Lu M., Cheung W. K. C., Cai W., Gale M., Xu Q., Yan Q., An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 44, e149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/3/eaat9820/DC1
Fig. S1. TROJAN properties.
Fig. S2. Expression pattern of TROJAN.
Fig. S3. TROJAN phenotype.
Fig. S4. Construction of the TROJAN knockout cell line.
Fig. S5. Potential therapeutic role of TROJAN in breast cancer progression.
Fig. S6. TROJAN associates with the ZMYND8 protein.
Fig. S7. TROJAN degrades the ZMYND8 protein.
Fig. S8. TROJAN and ZMYND8 regulate the transcription of diverse target genes.
Fig. S9. TROJAN regulates breast cancer progression via ZMYND8.
Table S1. TNBC-related HERVs, basic patient information, primers, and MS data for proteins pulled down by TROJAN and SYSL1.


