Abstract
The hydroxyornithine transformylase from Pseudomonas aeruginosa is known by the gene name pvdF, and has been hypothesized to use N10-formyltetrahydrofolate (N10-fTHF) as a co-substrate formyl donor to convert N5-hydroxyornithine (OHOrn) to N5-formyl- N5-hydroxyornithine (fOHOrn). PvdF is in the biosynthetic pathway for pyoverdin biosynthesis, a siderophore generated under iron-limiting conditions that has been linked to virulence, quorum sensing and biofilm formation. The structure of PvdF was determined by X-ray crystallography to 2.3 Å, revealing a formyltransferase fold consistent with N10-formyltetrahydrofolate dependent enzymes, such as the glycinamide ribonucleotide transformylases, N-sugar transformylases and methionyl-tRNA transformylases. Whereas the core structure, including the catalytic triad, is conserved, PvdF has three insertions of 18 or more amino acids, which we hypothesize are key to binding the OHOrn substrate. Steady state kinetics revealed a non-hyperbolic rate curve, promoting the hypothesis that PvdF uses a random-sequential mechanism, and favors folate binding over OHOrn.
Keywords: PvdF, N5-hydroxyornithine, N5-formyl-N5-hydroxyornithine, transformylase, formyltransferase, pyoverdin
INTRODUCTION
Iron is required for major metabolic processes such as cellular respiration and nucleotide biosynthesis. Due to insolubility and toxicity, iron is sequestered and highly regulated in human cells and is thus unavailable to bacterial pathogens, a phenomenon that has been called nutritional immunity1. Pathogens have developed elaborate mechanisms to overcome the paucity of available iron in the human host, including producing high affinity chelators called siderophores. Once secreted, siderophores bind iron, and are taken up in the iron-loaded form to provide the pathogen with the required iron2, 3.
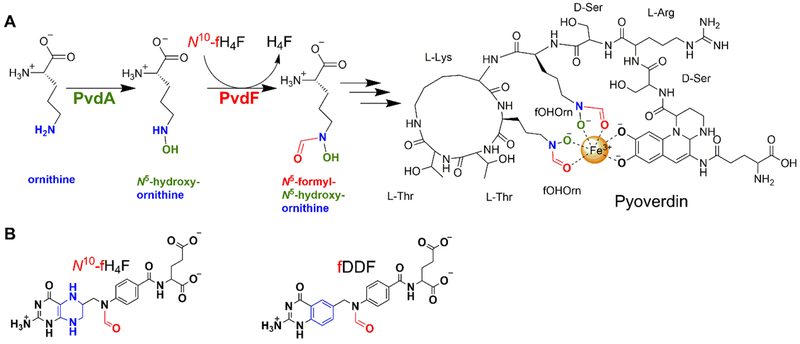
The focus of this study is the second step in the biosynthesis of the siderophore pyoverdin, which is linked to virulence, quorum sensing, and biofilm development2 in the ESKAPE pathogen4, 5 Pseudomonas aeruginosa. Pyoverdins, whose structure and composition is dependent on bacterial strain, are composed of a dihydroxyquinoline chromophore core with an α-ketoacid sidechain attached to a 6–14 amino acid peptide that is assembled by nonribosomal peptide synthethase (NRPS) enzymes (Figure 1)3. Along with the NRPS enzymes, there are also enzymes in pyoverdin biosynthesis that are required for the production of precursors, maturation and tailoring of the peptide, and chromophore synthesis2, 3, 6, 7 . All pyoverdins from P. aeruginosa strains include N5-formyl-N5-hydroxyornithine, a nonproteinogenic amino acid derived from ornithine that has been hydroxylated and formylated on the sidechain amine resulting in a hydroxamate moiety of the siderophore. The biosynthetic operons for production of pyoverdin include proteins for conversion of L-ornithine (Orn) to N5-hydroxyornithine (OHOrn) by the ornithine hydroxylase PvdA, and for subsequent formation of N5-formyl-N5-hydroxyornithine (fOHOrn) by the hydroxyornithine transformylase PvdF (Figure 1)2. PvdA has been structurally and biochemically characterized8–10, but little is known about PvdF. Deletion strains of PvdF do not produce pyoverdin and are avirulent, and cell extracts from those strains showed formation of ornithine hydroxylamine without conversion to the hydroxamate form11.
Figure 1.
(A) Role of PvdF in pyoverdin biosynthesis. PvdA and PvdF derivatize ornithine to generate the precursor N5-formyl-N5-hydroxyornithine (fOHOrn). This molecule is subsequently incorporated into pyoverdin by nonribosomal peptide synthetases PvdF catalyzes formyl group transfer from N10-formyltetrahydrofolate (N10-fTHF) to N5 of N5-hydroxyornithine. (B) Chemical representation of N10-formyl-THF (right) and cofactor analogue fDDF (left). The ring altered between the cofactor and the analogue is highlighted in blue.
N5-formyl-N5-hydroxyornithine is a component of other siderophores, including rhodochelin (Rhodococcuss jostii RHA1)12, coelichelin (Streptomyces coelicolor)13 and amychelin (Amycolatopsis sp.AA4)14. These chelators are similarly constructed by NRPS assembly lines that are dependent on accessory enzymes to generate fOHOrn. The enzyme characterizations for these pathways are at the initial stages, with activities confirmed and some steady state analyses performed12–14. Rhodochelin formyltransferase (Rft) has been definitively shown to perform a N10-formyltetrahydrofolate (N10-fTHF) dependent formylation reaction to convert OHOrn to fOHOrn12.
Here we report structural characterization of a hydroxyornithine transformylase, the PvdF enzyme from Pseudomonas aeruginosa. The structure reveals a core fold common among N10-fTHF dependent transformylases, including the glycinamide ribonucleotide transformylases (GART)15–19, the methionyl-tRNA transformylase (MTF)20, 21, and N-sugar transformylases of O-antigen formation22–26. However, the structure reveals large, unique insertions that we propose are important for binding the substrate OHOrn, and that place PvdF as the first documented member of a new structural subclass. This work includes a steady state kinetic analysis that indicates a partially ordered, formally random-sequential bireactant system that favors folate binding.
MATERIALS AND METHODS
Preparation of PvdF Overexpression Plasmid.
The pvdF gene was cloned from Pseudomonas aeruginosa (PAO1) genomic DNA, using polymerase chain reaction (PCR) with Herculase polymerase (Stratagene). The reaction was supplemented with 8% (v/v) DMSO as per manufacturer instructions due to high G-C content (61%). The forward primer (5’-AAT TAT ATA CAT ATG ACG AAA AGG AAA CTG GCC TA −3’) contains an NdeI restriction site (underlined), and the reverse primer (5’-AAT ATA ATA CAG ATC TGG GAG CTT CTC GGC GAG CAG C-3’) contains an BglII restriction site (underlined). The amplified DNA fragment was ligated into the correspondingly digested pET29b vector (Novagen) with T4 DNA ligase (New England BioLabs). The overexpression plasmid generates the PvdF protein with a C-terminal thrombin cleavage site followed by a histidine tag. This construct was further modified by site-directed mutagenesis to incorporate two stop codons at the C-terminus of the PvdF sequence so that the native PvdF protein, without purification tags, could be expressed. The Quik-Change® site-directed mutagenesis kit (Stratagene) was used with the forward primer (5’-CTG CTG GCC GAG AAG CTC TGA TGA CTG GGT ACC CTG GTG-3’) and reverse complement primer (stop codons underlined).
Preparation of K72A,K74A-PvdF expression plasmid.
The PvdF K72A,K74A expression plasmid was prepared by the Genscript plasmid preparation and mutagenesis services. The pvdF gene was cloned into the pET29b expression plasmid at the HindIII restriction site on the 3’ end and the NdeI restriction site on the 5’ end following the stop codon. The gene was synthesized such that the codon for K72 (AAA) was changed to encode alanine (GCA) and the codon for K74 (AAG) to encode alanine (GCG).
Wildtype PvdF expression and purification.
The PvdF plasmid was transformed into BL21(DE3) E.coli (New England BioLabs) for expression. Baffled flasks containing 1 L of LB Miller media containing 50 μg/ml of kanamycin were inoculated with 10 mL of overnight culture and grown at 37 °C in a shaker incubator (225rpm). When the OD600 reached 0.5, the temperature was lowered to 25 °C and allowed to equilibrate for 15 min. Expression was induced with isopropyl-β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.2 mM with shaking incubation for 16 hours. The cells were harvested by centrifugation (6000 × g, 5 min, 4 °C). The cell pellets were resuspended in 20 mL of 50 mM Tris-HCl pH 8.5 and lysed by three passes through a French press apparatus (35,000 psi). The lysate was centrifuged (12000 × g, 30 min, 4 °C) and the supernatant was injected onto a Source 30Q affinity column (GE Healthcare) pre-equilibrated with 50 mM Tris-HCl pH 8.5. The protein was eluted with a linear gradient of increasing NaCl to 500 mM. Protein fractions containing PvdF were confirmed by 15% SDS-PAGE and pooled. The salt concentration was adjusted to 1 M final concentration by slow addition of solid NaCl with gentle mixing. The protein was injected onto a Source Phenyl Sepharose (GE Healthcare) column pre-equilibrated in 50 mM Tris-HCl pH 8.5, 1 M NaCl. The PvdF protein was eluted from the column using a gradient to a buffer with no NaCl (50 mM Tris-HCl pH 8.5). Fractions containing PvdF were concentrated and injected onto a Superdex 200 gel filtration column (GE Healthcare) pre-equilibrated in 50 mM potassium phosphate pH 7.4. PvdF eluted at a molecular weight consistent with monomeric protein. The protein was concentrated with an Amicon® Ultracell® 30K centrifugal filter to 70 mg/mL as determined by Bradford assay, and stored at −80 °C. The purification protocol yielded 148 mg per liter of culture.
Expression and purification of K72A,K74A-PvdF.
The K72A,K74A-PvdF expression plasmid was transformed into BL21(DE3) E.coli (New England BioLabs). The variant protein was expressed and purified in a similar manner to wildtype PvdF, except that the phenyl sepharose column was not required to attain high purity. Therefore, the protein eluted from the Source 30Q affinity column was directly injected onto the Superdex 75 gel filtration column. This preparation yielded 55 mg of protein per liter of cell culture.
Selenomethionine substituted PvdF expression and purification.
Se-Met PvdF was produced according to the protocol by Van Duyne et al.27 with some modifications. M9 minimal media was augmented with 2 mM MgCl2, 0.1 mM CaCl2, 0.4% (w/v) glucose and 50 μg/mL kanamycin. Growth cultures (1L) were inoculated with 10 mL of overnight culture and incubated at 37 °C in a shaker incubator (225 rpm) until an OD600 of 0.5 was reached. The temperature was lowered to 25 °C and an amino acid mixture was added to inhibit methionine production and allow for selenomethionine incorporation (the amino acid mixture included: 100 mg each of lysine, phenylalanine, threonine; 50 mg each of isoleucine, leucine, valine; 60 mg of selenomethionine, per liter of culture). When the OD600 of the culture reached 1.0, IPTG was added to a final concentration of 0.2 mM and the culture was incubated for a further 16 hours with shaking. The SeMet protein purification was performed as for the native protein, with the exception that all buffers were supplemented with 2 mM dithiothreitol (DTT). The purified protein was concentrated with an Amicon® Ultracell® 30K centrifugal filter to 80 mg/mL as determined by Bradford assay and stored in −80 °C. The purification protocol yielded 100 mg per liter of culture.
PvdA protein expression and purification.
The PvdA enzyme was expressed and purified as previously reported8, 10.
Preparation of hydroxyornithine (OHOrn).
N5-hydroxyornithine was prepared by Garrett Moraski (Montana State University) according to the published protocol.28
Preparation of 10-formyl-5,8 dideazafolate (fDDF) and 5,8 dideazafolate (DDF).
Both fDDF and DDF were a generous gift from Dr. Carol Caperelli (University of Cincinnati). Both substrates were prepared according to the published protocols.29
Steady state activity assays.
The assay buffer contained 50 mM potassium phosphate buffer pH 7.4. The deformylation of fDDF is followed by the change in extinction coefficient at 295 nm (Δε=18.9 mM−1cm−1)30. The assay was performed using a TgK scientific stopped-flow instrument at 25 °C equipped with a mercury-xenon lamp. Enzyme (200 nM) with 11 mM N5-hydroxyornithine was mixed at 1:1 ratio with varied fDDF concentration (5 μM to 642 μM). The rate was measured for 30 sec and the rate dependence was fit to the Michaelis-Menten equation. When hydoxyornithine was varied, the reaction was performed in 96 well flat–bottom plate (Corning cat # 9107) using a Varian 50MPR Microplate Reader, with a total reaction mixture per well of 300 μL. Each well contained a final enzyme concentration of 100 nM. The hydroxyornithine concentration was varied from 5.8 mM to 100 mM. The reaction was initiated with addition of 150 μM fDDF; the highest concentration possible at the fDDF λmax, within the linear range of the instrument. Reaction progress was monitored at 295 nm in 1 sec cycle reads for 90 seconds. Data were fit to the Michaelis-Menten equation.
Coupled steady state activity assay.
Pseudomonas aeruginosa ornithine hydroxylase (PvdA) was used to generate the substrate OHOrn. The standard assay buffer contained 50 mM potassium phosphate buffer pH 7.4. The reaction was performed in a 1.5 mL quartz cuvette using a Cary 50 Bio UV-visible spectrophotometer. The initial reaction (600 μL) contained 1 μM PvdA, 150 μM FAD, 10 mM ornithine. Varying concentrations of NADPH were added (133 μM to 1 mM) for PvdA to generate defined OHOrn concentrations. The progress of the reaction was monitored at 300 nm as a measure of NADPH turnover. When the change in absorbance at 300 nm ceased, 1 mM fDDF (final concentration) was added to the cuvette and the spectrometer was blanked. The transformylase reaction was initiated by the addition of 200 nM PvdF and monitored at 295 nm for 30 sec using a 0.5 cm pathlength. The initial rates were plotted versus [OHOrn], assuming that each NADPH consumed by PvdA produced one OHOrn molecule. The plot showed a non-hyperbolic velocity curve, with a decreasing rate at concentrations of OHOrn above 400 μM. Points lower than 133 μM were not obtained due to insufficient signal-to-noise.
Wildtype and K72A,K74A-PvdF progress curves.
Progress curves were measured using the same buffer as the steady state assays, generating OHOrn with 1 μM PvdA, 150 μM FAD, 10 mM ornithine, 500 μM NADPH. After the reaction ceased to change at 300 nm, the PvdF reaction was initiated by the addition of fDDF (46–183 μM) and 200 nM PvdF. The reaction was monitored at 295 nm for 60 min using 0.5 cm pathlength quartz cuvette.
Mass Spectrometry.
Samples from the PvdA-PvdF reaction were diluted 1000-fold with LC-MS grade water (Sigma-Aldrich), and 10 μL of each dilution was analyzed by LC-MS over 65 minutes on an LCMS-IT-TOF (Shimadzu Scientific Instruments) with a Shim-pack XR-ODS column. The mobile phase consisted of 95% of an aqueous 0.1% formic acid solution and 5% acetonitrile (Sigma Aldrich), with a total flow rate of 0.2 mL/min. An ESI source was used, and acquisition was performed in scan mode from 120–550 m/z for both positive and negative ion modes. A 10 msec ion accumulation time was used, and event time was set to 100 msec. A three stage gradient was run as follows: 5% acetonitrile for 5 minutes, a linear gradient from 5% to 95% acetonitrile over 20 minutes, and 95% acetonitrile for another 20 minutes.
PvdF Crystallization.
Purified SeMet protein was exchanged into 50 mM potassium phosphate buffer pH 7.4, 2 mM DTT and diluted to 40 mg/mL. A few flakes of powdered DDF were added to the protein solution, and the mixture was incubated on ice for 15 min. The protein solution was centrifuged (12000 × g, 30 sec, 4 °C). The protein was crystallized using the hanging-drop vapor diffusion method. Crystallization drops were prepared by mixing 1.5 μL protein solution with 1.5 μL precipitant solution containing 0.55 M sodium citrate, 0.1 M Tris-HCl pH 8.5. Rectangular-prism shaped crystals with dimensions 0.15 μm × 0.15 μm × 0.04 μm grew within 2 weeks. For data collection, crystals were soaked in a cryoprotectant solution containing the precipitant solution augmented with 20% ethylene glycol and flash cooled in liquid nitrogen.
Single crystal X-ray diffraction data collection and processing.
A single wavelength anomalous dispersion (SAD) dataset was collected at the Stanford Synchrotron Radiation Laboratory (SSRL, Stanford, CA) beamline 12–2 using a wavelength of 0.9795 at 100 K. This wavelength was based on a selenium fluorescence scan which showed a strong signal, with an inflection point at 0.9795 Å. The software package Blu-Ice31 was used to collect 847 oscillation images (0.15 º per image). The exposure time per frame was 0.2 sec with a transmission of 3%, and the crystal-to-detector distance set at 400 mm. Diffraction data were processed using XDS32 to 2.3 Å with anomalous signal to 2.73 Å in the space group P21 with cell dimensions of a=128 Å, b=92.7 Å, c=128 Å, β=90.1 º. While the crystals appeared single, the diffraction pattern showed twinning (overlapping lattices) making space group determination problematic. Data were frequently auto-processed as P422, but had to be manually re-processed in P21 in order to obtain the solution. Despite a strong anomalous signal, the SAD data did not lead to a solution using PHENIX. Crank233 in the CCP4 program suite34 was used to determine the location of 24 Se atoms, providing initial phases to build eight monomers in the asymmetric unit. This solution had a figure of merit (FOM) of 0.782 and Rcomb of 0.351. XTRIAGE35 identified the twin fractions (-l, k, h; -h, -k, l; l, -k, h) with (-h, -k, l) showing the highest twin fraction of 0.49. This twin fraction was applied in subsequent rounds of model building and refinement using Coot36 and Phenix.Refine37. Water molecules were added automatically and inspected manually using Coot. Citrate molecules, derived from the crystallization conditions, were modeled manually using Coot. DDF molecules were built using LigandFit38, 39 with restraints generated using eLBOW40 and REEL41. Statistics for data refinement and analysis can be found in Table 1.
Table 1.
PvdF SeMet data collection and refinement statistics
| SeMet | |
|---|---|
| Data collection | |
| Beamline | SSRL 12–2 |
| Wavelength (Å) | 0.9795 |
| Space group | P21 |
| Cell dimensions; a, b, c (Å), (°) | 127.9 92.7 127.9 90 90.11 90 |
| Resolution (Å) | 38.74 – 2.3 (2.38 – 2.3) |
| Rmergea | 0.089 (0.381) |
| Total observations | 324216 (14816) |
| Total unique observations | 121662 (5715) |
| Mean ((I) / sd(I)) | 9.2 (2.2) |
| Completeness (%) | 91.8 (87.4) |
| CC(l/2) | 0.909 (0.856) |
| Redundancy | 2.7 (2.6) |
| Refinement | |
| Resolution (Å) | 38.67 – 2.3 (2.38 – 2.3) |
| Rcrystb | 0.2270 (0.2930) |
| Rfreec | 0.2494 (0.3019) |
| Total unique observations | 121938 (12169) |
| No. of non-hydrogen atoms | 17617 |
| Protein | 16963 |
| Ligand | 328 |
| Water | 326 |
| rms deviation bonds (Å) | 0.037 |
| rms deviation angles (°) | 2.31 |
| Overall mean B-factor (Å2) | 39.3 |
| Ramachandran plot analysise | |
| Favored region | 96.53 |
| Allowed region | 3.47 |
| DDF | |
| Chain A; occupancy, B-factor (Å2) | 1.0, 30.5 |
| Chain B; occupancy, B-factor (Å2) | none |
| Chain C; occupancy, B-factor (Å2) | 0.69, 25.8 |
| Chain D; occupancy, B-factor (Å2) | 1.0, 31.5 |
| Chain E; occupancy, B-factor (Å2) | 0.72, 25.8 |
| Chain F; occupancy, B-factor (Å2) | 0.66, 25.3 |
| Chain G; occupancy, B-factor (Å2) | 1.0, 21.5 |
| Chain H; occupancy, B-factor (Å2) | 1.0, 29.6 |
Rmerge = Σh|Ih - <I>|/ΣhIh, where Ih is the intensity of reflection h, and <I> is the mean intensity of all symmetry-related reflections
Rcryst = Σ||Fo| - |Fc||/Σ|Fo|, Fo and Fc are observed and calculated structure factor amplitudes.
Five percent of the reflections were initially reserved to create an Rfree test set used during each subsequent round of refinement.
PvdF crystallographic model.
The final PvdF model contains eight monomers; however, the model is discontinuous with several chain breaks per monomer due to disorder. The amino acid summary can be found in Table 2. The model contains 336 waters, seven DDF molecules and eight citrate molecules. The DDF molecules are located at an interface between monomers. Four DDF molecules are present in 100% occupancy, whereas the remaining three were refined to 66–73% occupancy. Final Ramachandran analysis has been calculated with MolProbity42 with 96.5% in the favored regions and one outlier in an area of poor density. Root mean square deviation values were calculated using PDBeFold43 and protein interaction interfaces were calculated using PDBePISA44. Structures figures were prepared using Pymol45. Atomic coordinates and structure factors for SeMet PvdF were deposited into the Protein Data Bank, with the accession code 6CUL.
Table 2.
Ordered amino acids PvdF monomers
| Residues | |
|---|---|
| Monomer A | 3 – 225, 231 – 274 |
| Monomer B | 3 – 225, 231 – 274 |
| Monomer C | 3 – 26, 30 – 80, 85 – 224, 231 – 272 |
| Monomer D | 2 – 225, 232 – 274 |
| Monomer E | 3 – 224, 231 – 272 |
| Monomer F | 3 – 158, 163 – 225, 230 – 273 |
| Monomer G | 2 – 27, 32 – 225, 229 – 275 |
| Monomer H | 3 – 80, 84 – 225, 230 – 274 |
RESULTS
Preparation of PvdF.
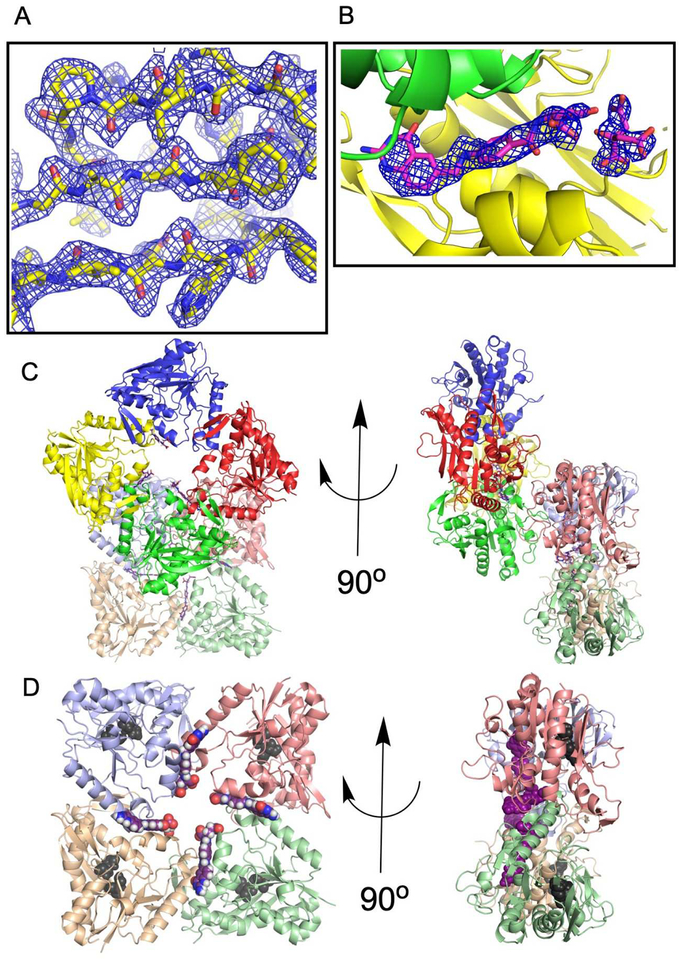
PvdF protein was heterologously produced in E. coli and purification was completed in three steps, using anion exchange, phenyl sepharose and gel filtration chromatography. The 31 kDa protein eluted from the gel filtration column at a molecular weight consistent with monomeric protein in solution (Supplemental Figure S1). The SeMet protein was purified using a similar protocol, with the addition of a reducing agent to all buffers. The SeMet protein was crystallized using citrate as the precipitant, and crystals only formed in the presence of the product analogue 5,8-dideazafolate (DDF). As defined in the Materials and Methods section, the structure determination was complicated by twinning. Despite the technical difficulties, initial phase estimates for PvdF were determined by single wavelength anomalous dispersion phasing using the selenomethionine-substituted form of the protein to 2.34 Å (Table 1). A representative electron density map of the refined structure is depicted in Figure 2A, with example density for the DDF in Figure 2B. The asymmetric unit contains eight monomers, arranged in two rings with 4-fold rotational symmetry (Figure 2C). Consistent with the gel filtration data for the protein in solution, the average interface area between monomers calculated by PDBePISA44 was 719 Å2, indicating that the four-fold symmetry is the result of the arrangement of the monomers in the crystal lattice and not indicative of an oligomeric state. Unexpectedly contacts within this monomer-monomer interface are mediated by the bound product analogue, 5,8 dideazafolate (DDF) which is not observed to bind in the putative active site, defined by the conserved catalytic triad (Figure 2D).
Figure 2.
The asymmetric unit. (A) PvdF crystals appeared single in the drop; however, they showed evident twinning in the diffraction images. Refinement required implementation of the twin law (-h,-k,l). Representative electron density for a 2Fo-Fc simulated annealing map (residues 166–174; 215–243) contoured at 1.5σ is shown. (B) PvdF crystals did not form without the cofactor analogue DDF. Electron density at the interfaces between seven of eight monomers is assigned to DDF and citrate. Electron density is displayed as a 2Fo-Fc simulated annealing omit map contoured at 2σ. (C) PvdF crystallized with eight monomers in the asymmetric unit, as two rings with four-fold symmetry. Each monomer is a distinct color. (D) DDF (magenta) was observed at an interface between PvdF monomers. PvdF monomers E,F,G,H are shown and the location of the active site is highlighted with two of the three residues of the catalytic triad shown in black (the remaining residue is part of a disordered loop). If this were a productive catalytic binding mode, the formate would have to travel ~22 Å. In all other transformylases, the folate binds with the formate directly adjacent to the catalytic triad (within 5 Å).
Monomer architecture.
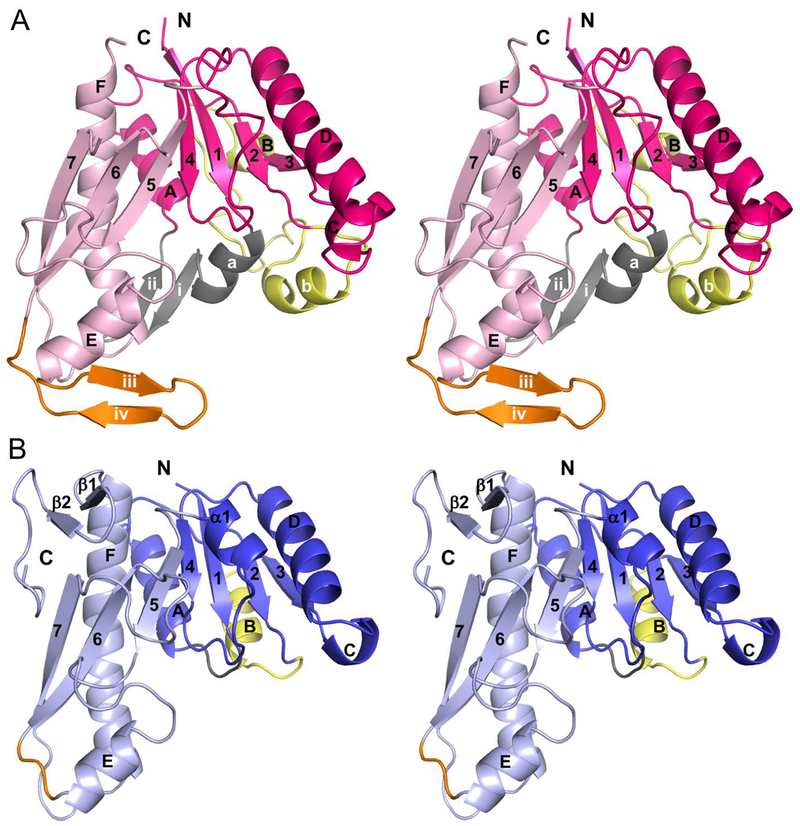
The core of the PvdF monomer shows the standard formyltransferase fold found in the N10-formyltetrahydrofolate dependent enzymes, with a central 7-stranded sheet surrounded by helices and loops (Figure 3A). The fold has been previously divided into two subdomains: an N-terminal subdomain for binding the folate substrate and a C-terminal subdomain for binding the substrate to be formylated18, 19. Glycinamide ribonucleotide transformylase (GART) is one of the best studied formyltransferases15, 17, and E. coli GART (EcGART) will serve as a frame of reference for our discussion (Figure 3B). The core of PvdF and EcGART are structurally conserved; however, PvdF has three major structural insertions. In the N-terminal subdomain, EcGART has a short loop between strand 1 and helix A. In PvdF, strand 1 is followed by a short helix, which is labeled helix a, and then an antiparallel β-sheet (strands i and ii), before helix A (grey in Figure 3). This is a total insertion of 23 amino acids (residues 12 to 34 in PvdF). In EcGART, the connection between the second β-strand and helix B is also a short loop. Helix B is followed by another short loop that connects to strand 3. PvdF has a large insertion at this connection (40 amino acids), beginning at residue 63. Helix B of the Rossman fold is replaced with helix b, which is roughly parallel to helix a (not packed against the central sheet). Helix b is followed by a long loop that does pack against the central sheet, structurally replacing helix B of EcGART. PvdF does have a short turn of a helix B before it rejoins the standard fold at residue 102 in strand 3. These changes are highlighted in yellow in Figure 3. Finally, in the C-terminal subdomain, PvdF has an insertion between helix E and strand 6, shown in orange in Figure 3. This insertion, residues 197 to 214 (18 amino acids), forms an antiparallel β-sheet with strands labeled iii and iv. EcGART continues after the F helix, with 2 additional strands forming a small sheet, structural elements not within the formyltransferase fold and not found in PvdF.
Figure 3.
Walleye stereoimages showing monomer topology comparison between PvdF and EcGART. (A) While the formyltransferase fold and core of these proteins is similar, PvdF contains three major insertions sites respectively colored in gray (residues 12–34, helix a, strands i and ii), yellow (residues 63–102, loop and helix b) and orange (197–214, strands iii and iv). The N-terminal domain of PvdF (residues 1–162) in shown in magenta and the C terminal domain is in light pink (residues 163–275). (B) The N-terminal domain of EcGART (residues 1–100) is shown in dark blue and the C terminal domain is in light blue (residues 101–209). Insertion sites are represented in same colors as in panel A. EcGART contains structural elements that are not seen in PvdF, helix α1 and terminal sheet β1- β2. Helices are labeled with letters and sheets are indicated in numbers.
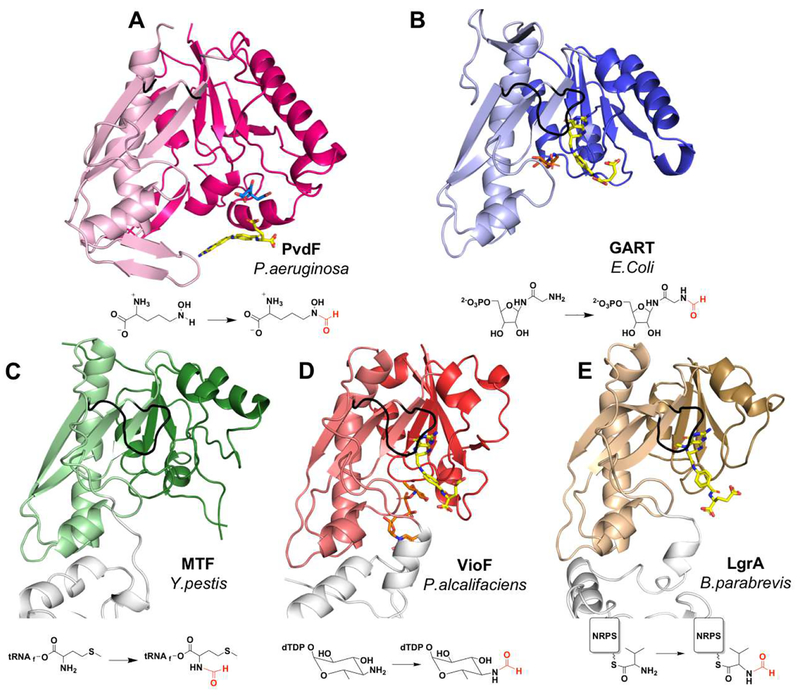
Structural homologues.
The closest structural homologues to PvdF are the GART proteins, which are found in the pathway for the de novo biosynthesis of purines. The root mean square deviation (rmsd) calculated for the comparison of PvdF to EcGART is 2.2 Å for 165 Cα residues (Figure 4A and 4B). PvdF is 275 amino acids in length, whereas EcGART is 209 amino acids. The three insertions listed above account for most of the differences in Cα comparison, with other more subtle changes found within other loops. Unlike PvdF, GART proteins are dimeric, though the interfaces for dimerization within the GART family are not conserved. The methionyl-tRNA formyltransferase (MTF), which formylates the primary amine of the methionine attached to the initiator methionyl-tRNA, is monomeric and also shares a similar fold with PvdF and EcGART. The enzyme from Yersinia pestis is shown in Figure 4C, which when compared to PvdF has an rmsd of 2.3 Å over 150 Cα residues. The sugar N-transformylases involved in production of modified sugars for incorporation into O-antigens also fall into this structural and functional class. Of these, VioF from P. alcalifaciens O30 showed the closest structural similarity to PvdF with an rmsd of 2.5 Å over 145 Cα residues and is dimeric using a structural feature not found in PvdF (Figure 4D). Many of the sugar N-transformylases include C-terminal domains with other catalytic activities or regulatory roles22–26. Recently, the structure of the gramicidin initiation module (LgrA) was determined, which includes a formyltransferase domain (Figure 4E). Interestingly, this formyltransferase domain has been proposed to be incorporated into the NRPS assembly line as the result of a gene duplication and horizontal transfer of a sugar N-transformylase46, potentially an evolutionary precursor of VioF. At the C-terminal end of the LgrA transformylase domain is a new structural element, a loop that includes an α-helix, that serves as a linker to the adenylation domain of the NRPS module46. Note that the secondary structure insertions of PvdF are unique among these enzymes (Figure 4), placing PvdF in a new structural subclass of N10-fTHF dependent transformylase enzymes.
Figure 4.
Comparison of PvdF with structural and functional homologues. (A) PvdF (B) GART from E.Coli (PDB:1CDE; sequence identity when compared to PvdF: 27%, calculated in LALIGN); (C) methionyl t-RNA transformylase from Y.pestis (PDB:3R8X; seq id: 26%); (D) VioF, sugar N-transformylase from P.alcalifaciens (PDB:4YFY; seq id: 28%); and (E) the NRPS formyltransferase domain of LgrA from B.parabrevis (PDB:5ES7; seq id: 25%). In each case the darker shade is the N-terminal and lighter shade is the C-terminal domain. In the bottom row the transformylase is part of a large multi-functional enzyme and domains without transformylase activity are white. The active site loop is shown in black. Folate analogues are shown in yellow with respective substrates in orange. The citrate molecule from crystallization in PvdF is shown in cyan. The reaction catalyzed by each enzyme is represented under the structure.
Folate binding pocket.
Enzymes dependent on N10-formyltetrahydrofolate are sometimes identified by a folate binding motif, HxSLLPxxxG where x is any residue, in the C-terminal subdomain (despite the N-terminal domain being labeled the folate-binding domain) (Figure 5)22. The histidine in this sequence is one of three residues in the catalytic triad, discussed later. This sequence starts in strand 5 and continues through the loop that connects to helix E, forming a portion of the folate binding pocket. For many of the N-sugar transformylases and for PvdF, this sequence is not conserved. The initial histidine and final glycine residues are conserved in three dimensions in PvdF. However, the remaining residues are not conserved, and the loop is 5 amino acids longer. The resulting sequence is: 170-HxGVTRyyyyyxxxG-184, where y is any residue in the extended PvdF loop. The primary contacts with the folate as determined in the GART and N-sugar transformylases are through hydrophobic interactions and hydrogen bonds between the methylpterin rings and the protein backbone found in the loop connecting strands 4 and 5, and in the loop connecting strands 6 and 7. The loop connecting strands 4 and 5 in EcGART and most of the other transformylases includes a helix (labeled α1 in Figure 3B). In PvdF, this loop is similarly coiled, but does not make the hydrogen bonds requisite to define this as a helix. While the residues that interact with the methylpterin of the folates are not conserved between the GART and N-sugar transformylases, the shape, hydrophobicity of the pocket, and hydrogen bonding interactions are. In PvdF, the loop connecting strands 4 and 5 maintains the proper shape for interaction with the methylpterin rings; however, the loop connecting strands 6 and 7 is disordered, potentially because no folate is bound in the active site. This loop has been noted in other transformylases to be mobile18, 47, 48. We hypothesize that for catalysis, the folate binds in the analogous location in PvdF when compared to both the GART and N-sugar transformylases. PvdF crystals would not grow without inclusion of a folate analogue. However, the folates evident in the electron density map are found forming crystal contacts, as was described previously, rather than in the active site.
Figure 5.
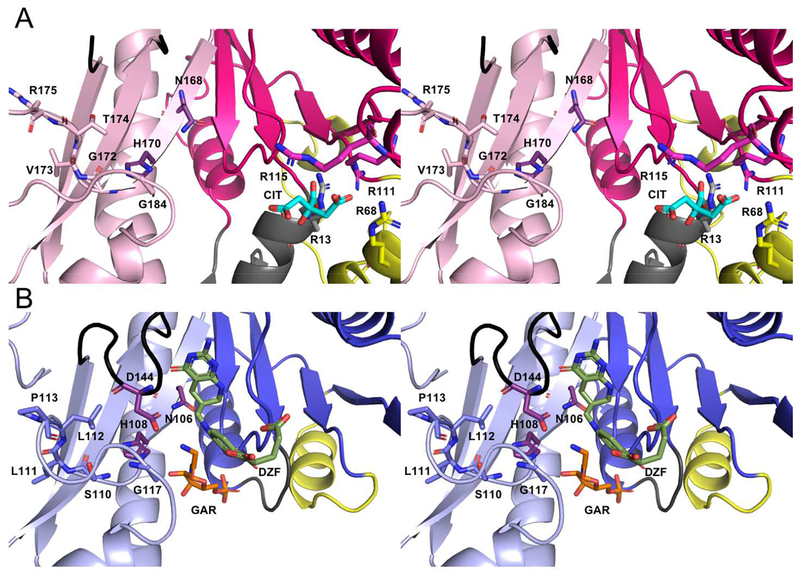
Walleye stereo comparison of the active sites of PvdF (A) and EcGART (B). EcGART contains the folate binding motif (S110-P113, light blue), that is not conserved in PvdF (G172-R175, light pink). The catalytic triad of Asn, His and Asp (shown in purple) is conserved in both PvdF and EcGART with Asp being part of the folate binding loop (shown in black). This loop was mobile in PvdF and not resolved. Because of unique insertions (gray and yellow, Panel A), PvdF contains an arginine binding pocket that contains a citrate molecule (cyan) derived from the crystallization mother liquor. By comparison to the binding orientation of the THF analogue in GART (dark green, Panel B), we propose that this arginine pocket may stabilize the mobile glutamate tail of the folate analogue in PvdF during catalysis.
OHOrn binding.
The substrates differ widely between N-transformylase groups, and include nucleotide precursors, sugar-nucleotides, tRNA, amino acids, and amino acids attached to a NRPS carrier domain through a phosphopantetheinyl linker, as documented in Figure 4. It is not surprising that the individual substrate binding interactions are specific, with the ultimate goal of presenting the amine group undergoing formylation within range of the formyl group of the N10-formyltetrahydrofolate. While there is no substrate bound in the PvdF structure, comparison to the holo-structures of transformylases previously determined suggests that the OHOrn substrate will bind in an analogous location (Figure 5A). The loop between strand 1 and helix A forms important hydrogen bonding and ionic interactions with the phosphates of the substrates in the GART and N-sugar transformylases (grey in Figure 5B)15, 18, 23, 24. In PvdF, the loop that contains these amino acids is not present, and this is instead the location of the first major insertion. Indeed, helix a, which is in the same three-dimensional space, has two sidechains, from Asn14 and Asp18, that point into the cavity and may form interactions with the backbone of the OHOrn substrate. The PvdF helix a is placed more interior in the active site than the GART or N-sugar transformylase 1-A loop, and the Asn14 and Asp18 sidechains would place the substrate deeper in the active site, potentially accounting for the considerably shorter OHOrn substrate.
The loop in the tRNA transformylase connecting β-strand 2 and helix B has been considered important for binding of the tRNA substrate20, 21. This loop corresponds with the second major insertion in PvdF; however helix b is in the comparable three dimensional location (yellow in Figure 5). Helix b is unlikely to play a direct role in hydroxyornithine binding in PvdF, being too distant from the putative substrate binding site. Finally, helix F has a proline forming a kink in these homologues, not necessarily at the same turn in the helix, but still suitable to promote a conformation in which the N-terminus of the helix is bent toward the substrate binding cavity. In the GART and N-sugar transformylases, charged and polar residues of helix F form hydrogen bonds with the substrates. The analogous residue in PvdF is Arg252, which may serve a similar role. In all, the location for substrate binding is likely analogous to that seen in the GART and sugar transformylases, but the residues that promote binding may be contributed at least in part by the structural features that are unique to PvdF.
Citrate.
The model of PvdF has a well-ordered citrate bound by three arginine residues (13, 68, and 111) from helix a, helix b, and helix C in every monomer (Figure 5A). The citrate molecule is derived from the mother liquor, which required greater than 0.5 M citrate for protein crystal formation. The binding of citrate in this site is undoubtedly a crystallization artifact. Nevertheless, the citrate is in close proximity of N10-fTHF binding site, and is bound with a free carboxylate less than 3 Å from the putative location for the folate glutamate tail (Figure 5B). This suggests that the citrate from the crystallization conditions, in large molar excess, prevented binding of folate in the active site. If this is correct, Arg 111 or potentially Arg 115 (nearby but not bound to citrate) may be involved in binding the glutamate tail of N10-fTHF; however, a new crystal form with folate bound in the active site would be necessary to establish this. It is important to note that there are no comparable binding interactions for binding the glutamate tail within the previously determined N10-fTHF dependent transformylase domains: the tail is frequently found to be disordered or having high B-factors in structures where folate is bound15, 19, 23, 24.
Catalytic Triad.
The loop connecting strands 6 and 7 (black in Figure 4 and Figure 5) has been named both the active site loop18 and the folate binding loop19. As noted before, this loop has been documented in other transformylases to be mobile, so it is not surprising that this loop is disordered in PvdF in the absence of folate. This loop harbors an aspartic acid that is one of three residues in a proposed catalytic triad. The other two residues, a histidine and an asparagine, are located in strand 5. These three residues are conserved in PvdF: Asn168, His170, and Asp229 (purple in Figure 5). In the GART and sugar transformylases, the binding pose of the folate is such that the formyl group on N10-fTHF is positioned at the center of the triad17, 22, 23. The proposed mechanism for the GART enzymes, and by extension all enzymes of this class, suggests that the amino group of the substrate performs a nucleophilic attack on the carbonyl of the formyl group of N10-fTHF, generating a tetrahedral intermediate. The catalytic triad residues are proposed to serve as general acid-general base residues to promote intermediate formation and resolution of the catalytic cycle17, 22.
PvdF steady state kinetics.
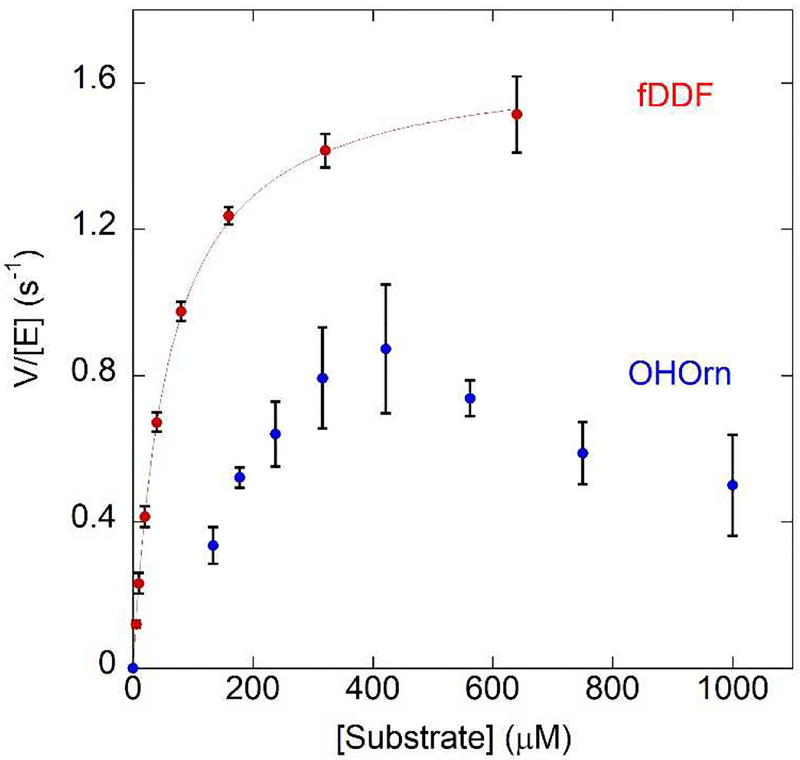
N10-fTHF dependent transformylases, such as those from purine biosynthesis, have been successfully assayed using the analogue 10-formyl-5,8-dideazafolate (fDDF)30, 49. When the formyl group is removed from fDDF, there is an increase in absorbance at 295 nm, allowing for a convenient continuous spectroscopic assay. When OHOrn is held in excess, and the varied substrate is fDDF, kinetic parameters are readily determined: Km = 60 ± 10 μM, kcat = 1.7 ± 0.1 sec−1 (Figure 6, red curve). However, the converse reaction, with fDDF in excess and OHOrn as the varied substrate, yielded a physiologically improbable kinetic constants (Supplemental Figure S2). This is likely due to difficulties with the assay: the experiment was not repeatable with each subsequent experiment showing an increase in Km and a decrease in kcat. The Km effect can be rationalized as OHOrn is known to be unstable50–52, and so the effective substrate concentration was diminishing with time. We hypothesized that a solution to this problem was to have the preceding enzyme of the biosynthetic pathway, the ornithine hydroxylase PvdA, generate the necessary substrate in situ. PvdA is a flavin-dependent enzyme that must be reduced by NADH with each catalytic cycle8–10. The appropriate concentration of OHOrn was produced from PvdA by varying and limiting the concentration of NADH. The steady state plot produced in this manner, showed a non-hyperbolic velocity curve (blue in Figure 6). The curve does not fit well to a Michealis-Menten or a substrate inhibition model. Instead, the data suggest a random-sequential bireactant mechanism in which the pathway for fDDF binding first is preferred for product generation, a model that as has been previously described for other enzymes53, 54.
Figure 6.
In the presence of fDDF as the varied substrate PvdF shows typical Michaelis-Menten kinetics (red trace, Km = 60 ± 10 μM, kcat = 1.7 ± 0.1 sec−1). In this experiment, the synthesized OHOrn was used. Higher concentrations of fDDF were not possible due to the highly absorbant nature of the compound exceeding the linear range of the spectrometer. Using hydroxyornithine as the varied substrate generated by PvdA in a coupled assay, PvdF initially exhibits a sigmoidal curve (blue dotted trace) until 400 uM, after which the rate decreases. The model that best describes this behavior is a random bireactant mechanism in which the binding of fDDF is preferred first step
Product detection.
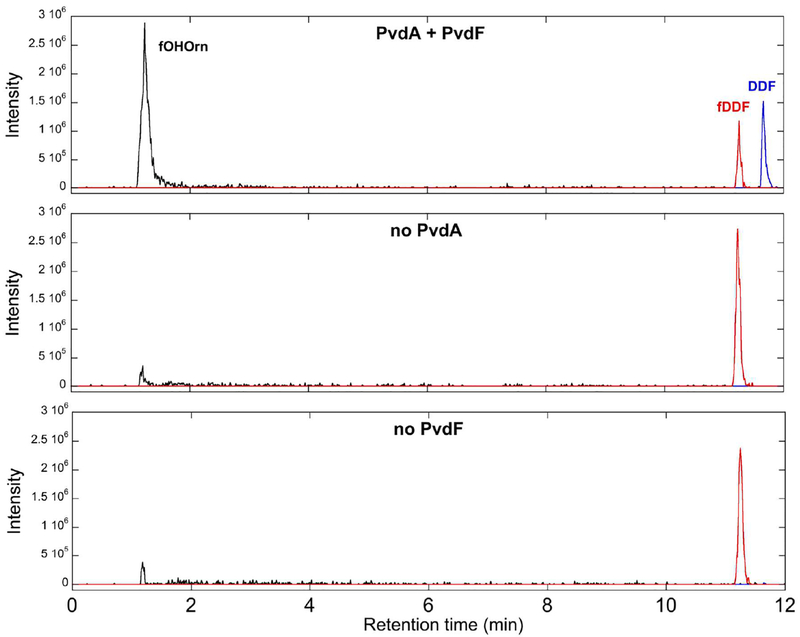
The absorbance assay described above indicates the loss of the formyl group from fDDF, but not necessarily for formation of the product fOHOrn. To confirm fOHOrn production, the PvdA-PvdF reaction was analyzed by LCMS, monitoring for fOHOrn (m/z=177.1), fDDF (m/z=466.2) and DDF (m/z=438.1) (Figure 7). fOHOrn and DDF were observed, and the fDDF decreased in reactions containing both enzymes and all necessary substrates (flavin, NADH, Orn, fDDF), whereas controls that did not contain one of the enzymes (PvdA or PvdF) did not show production of the fOHOrn or DDF products.
Figure 7.
The formation of the product fOHOrn was monitored by LCMS (black trace, m/z=177.0), along with DDF (blue trace, m/z=438.1) and fDDF (red trace, m/z=466.2). Unlike in the control samples (lower two panels), the complete reaction (top panel) shows clear formation of the product by catalytic formyl group transfer.
The observed DDF binding mode is a crystallization artifact.
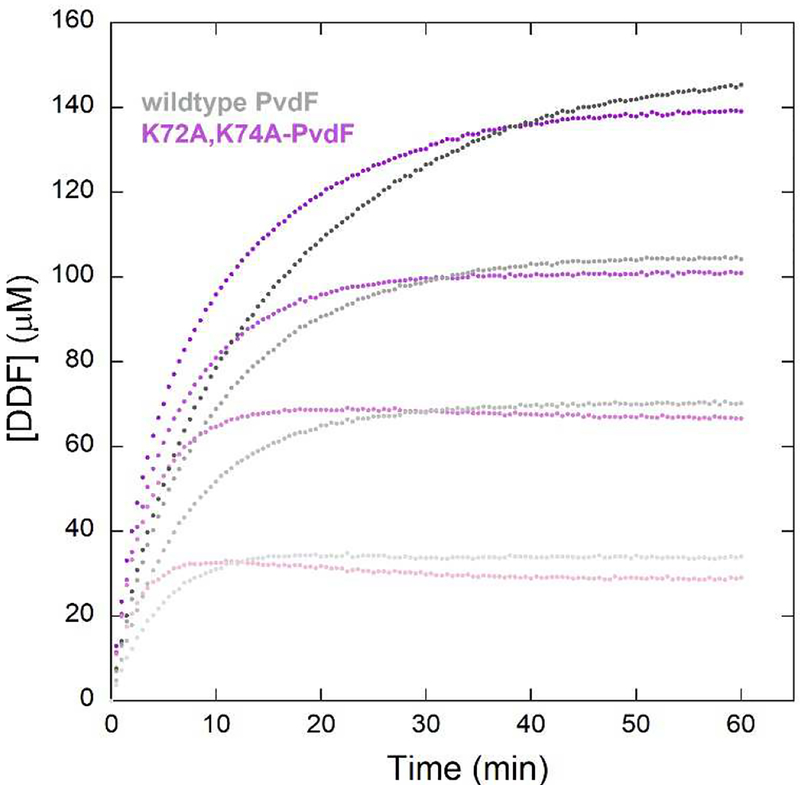
We hypothesized that the observed binding site for DDF, outside the active site and 22 Å distant from the catalytic triad, is a crystallization artifact (Figure 2D). As mentioned previously, the crystals only grew in the presence of DDF, so potentially this binding promoted the formation of an oligomerization interface that promoted crystallization. Despite >10 years of effort, these twinned crystals were the best to date and the only ones that produced a refined structure. However, we now have the benefit of a refined structure in which we can analyze crystallization contacts. We generated a variant, K72A,K74A-PvdF. These two lysine residues flank the DDF binding site. In monomers C and D, K72 directly hydrogen bond with the glutamate tail of DDF. K74 of one monomer is proximity of E65 of the next monomer in the ring, and in two of the eight cases, these residues form a hydrogen bond. The double K→A variant did not crystallize, and the protein was active as shown in full progress curves (Figure 8). Therefore, the catalytically relevant binding of the folate is not seen in this PvdF structure. Instead, we hypothesize that in the catalytic complex, the folate will bind such that the formate to be transferred (attached to N10) will be adjacent to the catalytic triad, as seen in all other homologues of this family.
Figure 8.
Progress curve comparison between the wild type (shade of grey) and K72A, K74A-PvdF variant (shades of purple/pink). The curves represent four concentrations of fDDF (from light to dark shades). The K72A,K74A-PvdF does not crystallize, suggesting that the crystallographic binding site for DDF is disrupted. The variant is more active than the wildtype, suggesting that the catalytic binding site for the folate is not what is observed in the structure.
DISCUSSION
PvdF is the formyltransferase that converts N5-hydroxyornithine (OHOrn) to N5-formyl-N5-hydroxyornithine (fOHOrn) so that fOHOrn can be incorporated into the siderophore pyoverdin by a nonribosomal peptide synthetase assembly line (Figure 1). An N10-fTHF-dependent hydroxyornithine transformylase involved in siderophore biosynthesis has been functionally characterized previously12. The enzyme, rhodochelin formyltransferase, or Rft, is involved in the biosynthesis of the mixed catecholate-hydroxamate siderophore rhodochelin by Rhodococcus jostii RHA1, a gram positive soil bacteria. Like pyoverdin, this siderophore includes two formylhydroxyornithine residues for iron chelation, and is assembled by a nonribosomal peptide synthetase. Using an HPLC-MS assay, the authors showed conversion of OHOrn to fOHOrn, but there are no structural data for Rft. Sequence comparisons indicate that Rft is not a close structural homologue of PvdF. Instead, Rft is likely to be structurally similar to either the tRNA transformylase (FMT) or the N-sugar transformylase ArnA, an E. coli enzyme involved in lipid A modification that transformylates UDP-4-amino-4-deoxy-L-arabinose12. Rft, FMT, and ArnA all have the conserved HxSLLPxxxG motif for binding the folate co-substrate that PvdF lacks, and they all lack the major insertions highlighted in Figure 3 that are specific to PvdF. Also unlike PvdF, they all have a C-terminal domain that provides additional functionality (to enhance substrate binding or to provide an additional catalytic activity). Finally, Rft is an allosteric enzyme showing positive cooperativity, and is proposed to be a tetramer in solution12. PvdF is a monomer in solution and in the crystals. The 4-fold ring structure seen in Figure 2B is the result of crystal packing. Therefore, Rft is more functionally and likely structurally similar to the N-sugar transformylases and the tRNA transformylases than to PvdF.
The initiation module of the NRPS for the biosynthesis of the antibiotic gramicidin (LgrA) includes a formyltransferase that has been structurally characterized. The LgrA N10-fTHF-dependent formyltransferase domain transfers a formate to the backbone amine of a valine while the amino acid is covalently attached to the peptidyl carrier domain by a phosphopantethienyl tether46. This is in contrast to PvdF, which is a stand-alone accessory enzyme that formylates the sidechain amine of the free OHOrn substrate before the product fOHOrn is activated by an NRPS adenylation domain and attached to the carrier domain of the PvdI or PvdJ proteins2. LgrA has been hypothesized to be an evolutionary descendent of an N-sugar transformylase46. Both LgrA and N-sugar transformylases are considerably shorter in length, and lack the secondary structure insertions of PvdF, again suggesting that PvdF belongs to a distinct structural class of N10-fTHF dependent formyltransferase.
PvdF maintains the 7-stranded β-sheet core in a formyltransferase fold common to the N10-fTHF dependent transformylases, and the catalytic triad characteristic for this class of enzymes is conserved. The structurally unique features of PvdF are likely responsible for interaction with the smaller amino acid substrate. The mechanism previously defined for the GART enzymes and hypothesized to be conserved in the class likely holds for PvdF. A detailed steady state kinetic analysis of human GART demonstrated an ordered-sequential kinetic mechanism in which the folate binds first55. Subsequent, pre-steady state kinetic experiments for the E. coli GART defined a random sequential kinetic mechanism in which folate and GAR bind in no obligatory order, but for which the apoenzyme has higher affinity for fDDF than GAR49. For PvdF, varying the concentration of the substrate analogue fDDF (OHOrn in excess) generated Michaelis-Menten kinetics with kcat and Km values in keeping with an enzyme from secondary metabolism. N5-hydroxyornithine was a difficult substrate with which to work, and when used as the varied substrate, the data were not reproducible and provided kinetic values that were not physiologically relevant. However, when the ornithine hydroxylase (PvdA) of the same biosynthetic pathway was used to generate the OHOrn in situ, reproducible data could be generated. Interestingly, the curve was nonhyperbolic and not well fit by Michealis-Menten nor a substrate inhibition model (Figure 6). Instead, these data represent a model defined by Ferdinand53 and later echoed by Segel54 in which the bireactant system shows random binding of the two substrates, but favors binding of the folate over the OHOrn, very similar to the kinetic models for the GART proteins.
Supplementary Material
ACKNOWLEDGEMENTS
This publication was made possible by funds from NSF Grant CHE-1403293 (ALL), NSF Grant CHE-1402475 (GRM), NIH Grant GM127655 (ALL), and the University of Kansas General Research Fund #2301642 (ALL). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02–76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the NIH and NIGMS (including P41 GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIH or NIGMS. Thank you to the staff at the SSRL for their generous assistance. We are grateful to Dr. Kathy Meneely, Dr. Jose Olucha, Annemarie Chilton, and Anindita Basu for their assistance with the PvdF project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Hood MI, and Skaar EP (2012) Nutritional immunity: transition metals at the pathogen-host interface, Nat Rev Microbiol 10, 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Visca P, Imperi F, and Lamont IL (2007) Pyoverdine siderophores: from biogenesis to biosignificance, Trends Microbiol 15, 22–30. [DOI] [PubMed] [Google Scholar]
- [3].Meyer JM (2000) Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species, Arch Microbiol 174, 135–142. [DOI] [PubMed] [Google Scholar]
- [4].Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, and Bartlett J (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America, Clin Infect Dis 48, 1–12. [DOI] [PubMed] [Google Scholar]
- [5].Rice LB (2008) Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE, J Infect Dis 197, 1079–1081. [DOI] [PubMed] [Google Scholar]
- [6].Mossialos D, Ochsner U, Baysse C, Chablain P, Pirnay JP, Koedam N, Budzikiewicz H, Fernandez DU, Schafer M, Ravel J, and Cornelis P (2002) Identification of new, conserved, non-ribosomal peptide synthetases from fluorescent pseudomonads involved in the biosynthesis of the siderophore pyoverdine, Mol Microbiol 45, 1673–1685. [DOI] [PubMed] [Google Scholar]
- [7].Gulick AM (2017) Nonribosomal peptide synthetase biosynthetic clusters of ESKAPE pathogens, Natural Product Reports 34, 981–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Meneely KM, and Lamb AL (2007) Biochemical characterization of a flavin adenine dinucleotide-dependent monooxygenase, ornithine hydroxylase from Pseudomonas aeruginosa, suggests a novel reaction mechanism, Biochemistry 46, 11930–11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Olucha J, and Lamb AL (2011) Mechanistic and structural studies of the N-hydroxylating flavoprotein monooxygenases, Bioorg Chem 39, 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Olucha J, Meneely KM, Chilton AS, and Lamb AL (2011) Two structures of an N-hydroxylating flavoprotein monooxygenase: ornithine hydroxylase from Pseudomonas aeruginosa, J Biol Chem 286, 31789–31798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].McMorran BJ, Shanta Kumara HM, Sullivan K, and Lamont IL (2001) Involvement of a transformylase enzyme in siderophore synthesis in Pseudomonas aeruginosa, Microbiology 147, 1517–1524. [DOI] [PubMed] [Google Scholar]
- [12].Bosello M, Mielcarek A, Giessen TW, and Marahiel MA (2012) An enzymatic pathway for the biosynthesis of the formylhydroxyornithine required for rhodochelin iron coordination, Biochemistry 51, 3059–3066. [DOI] [PubMed] [Google Scholar]
- [13].Pohlmann V, and Marahiel MA (2008) Delta-amino group hydroxylation of L-ornithine during coelichelin biosynthesis, Org Biomol Chem 6, 1843–1848. [DOI] [PubMed] [Google Scholar]
- [14].Seyedsayamdost MR, Traxler MF, Zheng SL, Kolter R, and Clardy J (2011) Structure and biosynthesis of amychelin, an unusual mixed-ligand siderophore from Amycolatopsis sp. AA4, J Am Chem Soc 133, 11434–11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Almassy RJ, Janson CA, Kan CC, and Hostomska Z (1992) Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase, Proc Natl Acad Sci U S A 89, 6114–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Deis SM, Doshi A, Hou Z, Matherly LH, Gangjee A, and Dann CE 3rd. (2016) Structural and Enzymatic Analysis of Tumor-Targeted Antifolates That Inhibit Glycinamide Ribonucleotide Formyltransferase, Biochemistry 55, 4574–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Klein C, Chen P, Arevalo JH, Stura EA, Marolewski A, Warren MS, Benkovic SJ, and Wilson IA (1995) Towards structure-based drug design: crystal structure of a multisubstrate adduct complex of glycinamide ribonucleotide transformylase at 1.96 A resolution, J Mol Biol 249, 153–175. [DOI] [PubMed] [Google Scholar]
- [18].Zhang Y, Desharnais J, Greasley SE, Beardsley GP, Boger DL, and Wilson IA (2002) Crystal structures of human GAR Tfase at low and high pH and with substrate beta-GAR, Biochemistry 41, 14206–14215. [DOI] [PubMed] [Google Scholar]
- [19].Zhang Z, Caradoc-Davies TT, Dickson JM, Baker EN, and Squire CJ (2009) Structures of glycinamide ribonucleotide transformylase (PurN) from Mycobacterium tuberculosisreveal a novel dimer with relevance to drug discovery, J Mol Biol 389, 722–733. [DOI] [PubMed] [Google Scholar]
- [20].Schmitt E, Blanquet S, and Mechulam Y (1996) Structure of crystalline Escherichia coli methionyl-tRNA(f)Met formyltransferase: comparison with glycinamide ribonucleotide formyltransferase, EMBO J 15, 4749–4758. [PMC free article] [PubMed] [Google Scholar]
- [21].Schmitt E, Panvert M, Blanquet S, and Mechulam Y (1998) Crystal structure of methionyltRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet, EMBO J 17, 6819–6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gatzeva-Topalova PZ, May AP, and Sousa MC (2005) Crystal structure and mechanism of the Escherichia coli ArnA (PmrI) transformylase domain. An enzyme for lipid A modification with 4-amino-4-deoxy-L-arabinose and polymyxin resistance, Biochemistry 44, 5328–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Genthe NA, Thoden JB, Benning MM, and Holden HM (2015) Molecular structure of an N-formyltransferase from Providencia alcalifaciens O30, Protein Sci 24, 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Thoden JB, Goneau MF, Gilbert M, and Holden HM (2013) Structure of a sugar N-formyltransferase from Campylobacter jejuni, Biochemistry 52, 6114–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Woodford CR, Thoden JB, and Holden HM (2015) New role for the ankyrin repeat revealed by a study of the N-formyltransferase from Providencia alcalifaciens, Biochemistry 54, 631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zimmer AL, Thoden JB, and Holden HM (2014) Three-dimensional structure of a sugar N-formyltransferase from Francisella tularensis, Protein Sci 23, 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, and Clardy J (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin, J Mol Biol 229, 105–124. [DOI] [PubMed] [Google Scholar]
- [28].Mayfield JA, Frederick RE, Streit BR, Wencewicz TA, Ballou DP, and DuBois JL (2010) Comprehensive spectroscopic, steady state, and transient kinetic studies of a representative siderophore-associated flavin monooxygenase, J Biol Chem 285, 30375–30388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Caperelli CA, and Conigliaro J (1986) Synthesis of 10-Acetyl-5,8-Dideazafolic Acid - a Potent Inhibitor of Glycinamide Ribonucleotide Transformylase, Journal of Medicinal Chemistry 29, 2117–2119. [DOI] [PubMed] [Google Scholar]
- [30].Smith GK, Mueller WT, Benkovic PA, and Benkovic SJ (1981) On the cofactor specificity of glycinamide ribonucleotide and 5-aminoimidazole-4-carboxamide ribonucleotide transformylase from chicken liver, Biochemistry 20, 1241–1245. [DOI] [PubMed] [Google Scholar]
- [31].McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, and Kuhn P (2002) Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines, Journal of Synchrotron Radiation 9, 401–406. [DOI] [PubMed] [Google Scholar]
- [32].Kabsch W (2010) Integration, scaling, space-group assignment and post-refinement, Acta Crystallogr D Biol Crystallogr 66, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pannu NS, Waterreus WJ, Skubak P, Sikharulidze I, Abrahams JP, and de Graaff RA (2011) Recent advances in the CRANK software suite for experimental phasing, Acta Crystallogr D Biol Crystallogr 67, 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, and Wilson KS (2011) Overview of the CCP4 suite and current developments, Acta Crystallographica Section D-Biological Crystallography 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot, Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine, Acta Crystallogr D Biol Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Terwilliger TC, Adams PD, Moriarty NW, and Cohn JD (2007) Ligand identification using electron-density map correlations, Acta Crystallogr D Biol Crystallogr 63, 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Terwilliger TC, Klei H, Adams PD, Moriarty NW, and Cohn JD (2006) Automated ligand fitting by core-fragment fitting and extension into density, Acta Crystallogr D Biol Crystallogr 62, 915–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Moriarty NW, Grosse-Kunstleve RW, and Adams PD (2009) electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation, Acta Crystallogr D Biol Crystallogr 65, 1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Moriarty NW, Draizen EJ, and Adams PD (2017) An editor for the generation and customization of geometry restraints, Acta Crystallogr D Struct Biol 73, 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography, Acta Crystallogr D Biol Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Krissinel E, and Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions, Acta Crystallogr D Biol Crystallogr 60, 2256–2268. [DOI] [PubMed] [Google Scholar]
- [44].Krissinel E, and Henrick K (2007) Inference of macromolecular assemblies from crystalline state, Journal of Molecular Biology 372, 774–797. [DOI] [PubMed] [Google Scholar]
- [45].The PyMOL Molecular Graphics Systems, Version 2.0 Schrödinger, LLC. [Google Scholar]
- [46].Reimer JM, Aloise MN, Harrison PM, and Schmeing TM (2016) Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase, Nature 529, 239–242. [DOI] [PubMed] [Google Scholar]
- [47].Chen P, Schulze-Gahmen U, Stura EA, Inglese J, Johnson DL, Marolewski A, Benkovic SJ, and Wilson IA (1992) Crystal structure of glycinamide ribonucleotide transformylase from Escherichia coli at 3.0 A resolution. A target enzyme for chemotherapy, J Mol Biol 227, 283–292. [DOI] [PubMed] [Google Scholar]
- [48].Su Y, Yamashita MM, Greasley SE, Mullen CA, Shim JH, Jennings PA, Benkovic SJ, and Wilson IA (1998) A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A, J Mol Biol 281, 485–499. [DOI] [PubMed] [Google Scholar]
- [49].Shim JH, and Benkovic SJ (1998) Evaluation of the kinetic mechanism of Escherichia coli glycinamide ribonucleotide transformylase, Biochemistry 37, 8776–8782. [DOI] [PubMed] [Google Scholar]
- [50].Franke J, Ishida K, Ishida-Ito M, and Hertweck C (2013) Nitro versus hydroxamate in siderophores of pathogenic bacteria: effect of missing hydroxylamine protection in malleobactin biosynthesis, Angew Chem Int Ed Engl 52, 8271–8275. [DOI] [PubMed] [Google Scholar]
- [51].Neumann CS, Jiang W, Heemstra JR, Gontang EA, Kolter R, and Walsh CT (2012) Biosynthesis of Piperazic Acid via N5-Hydroxy-Ornithine in Kutzneria spp. 744, Chembiochem 13, 972–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tomlinson G, and Viswanatha T (1973) Synthesis and properties of -N-hydroxyornithine, Can J Biochem 51, 754–763. [DOI] [PubMed] [Google Scholar]
- [53].Ferdinand W (1966) The interpretation of non-hyperbolic rate curves for two-substrate enzymes. A possible mechanism for phosphofructokinase, Biochem J 98, 278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Segel IH (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady - State Enzyme Systems John Wiley & Sons, Inc. [Google Scholar]
- [55].Caperelli CA (1989) Mammalian glycinamide ribonucleotide transformylase. Kinetic mechanism and associated de novo purine biosynthetic activities, J Biol Chem 264, 5053–5057. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.