Abstract
In meiotic prophase, chromosomes are organized into compacted loop arrays to promote homolog pairing and recombination. Here, we probe the architecture of the mouse spermatocyte genome in early and late meiotic prophase using Hi-C. Our data support the established loop-array model of meiotic chromosomes, and infer loops averaging 0.8–1 Mb in early prophase and extending to 1.5–2 Mb in late prophase as chromosomes compact and homologs undergo synapsis. Topologically associating domains (TADs) are lost in meiotic prophase, suggesting that assembly of the meiotic chromosome axis alters the activity of chromosome-associated cohesin complexes. While TADs are lost, physically-separated A and B compartments are maintained in meiotic prophase. Moreover, meiotic DNA breaks and inter-homolog crossovers preferentially form in the gene-dense A compartment, revealing a role for chromatin organization in meiotic recombination. Finally, direct detection of inter-homolog contacts genome-wide reveals the structural basis for homolog alignment and juxtaposition by the synaptonemal complex.
In the specialized meiotic cell division program, homologs must identify one another, pair along their lengths, and physically link to ensure their accurate segregation in the meiosis I division. Inter-homolog links are formed by homologous recombination, in which DNA double-strand breaks (DSBs) are first introduced along each chromosome, and are then repaired using the homolog as a template 1. A subset of DSBs are repaired as inter-homolog crossovers, reciprocal exchanges of genetic material that drive eukaryotic evolution by shuffling alleles along chromosomes in each generation, and also constitute specific physical links between each pair of homologs 2. Failure to form inter-homolog crossovers can cause chromosome mis-segregation in the meiosis I division. In humans, aneuploidy resulting from meiotic chromosome mis-segregation is a major cause of miscarriage and the source of developmental disorders including Down Syndrome 3.
To promote the formation of accurate inter-homolog crossovers, chromosomes undergo dramatic morphological changes during meiotic prophase 2. In leptonema (Latin for “thin threads”), chromosomes become individualized and compacted as linear loop arrays around the proteinaceous chromosome axis. The axis comprises cohesin complexes with meiosis-specific subunits 4–6 plus filamentous axis “core” proteins 7, that together aid chromosome compaction and serve as a platform for recombination 8,9. Later, in zygonema (“paired threads”), telomeres cluster on the nuclear envelope and form a distinctive “bouquet” arrangement, and homologs begin to undergo synapsis. Synapsis, mediated by assembly of the synaptonemal complex (SC) between paired chromosome axes 2,10, is completed in pachynema (“thick threads”) along with further linear compaction of chromosomes. Meiotic recombination occurs alongside these morphological changes, with DSBs introduced in leptonema, and inter-homolog recombination driving pairing and synapsis of homologs in zygonema and pachynema. Finally, the SC is disassembled in diplonema (“two threads”), followed by further compaction and homolog segregation in meiosis I.
In mice, meiotic prophase occurs over the course of ~10 days, during which time the chromosomes are also highly transcriptionally active. Overall transcription levels are low in early prophase, then massively increase in mid-pachynema to support sperm development 11–13. Thus, meiotic prophase chromosomes must achieve a balance between two seemingly-conflicting needs: first, overall compaction and organization around the meiotic chromosome axis to support homolog pairing and synapsis; and second, high-level transcription at many loci. This balance between compaction and transcriptional activity contrasts with mitosis, where transcription is largely shut down as chromosomes become tightly compacted in mitotic prophase 11–13.
While recent technological advances have driven a fundamental rethinking of the forces driving mammalian chromosome organization in interphase and mitosis, the organization of the meiotic genome and how it relates to somatic-cell genome organization is largely unknown. Here, we performed chromosome conformation capture (Hi-C) 14,15 on synchronized mouse spermatocytes in both early and late meiotic prophase, revealing how chromosomes are reorganized to meet the needs of this unique developmental stage. We find that meiotic chromosomes show a near-complete loss of long-range contacts as they are reorganized around the meiotic chromosome axis. We show that topologically associating domains (TADs), a key organizational feature of interphase chromosomes, are lost as cohesin complexes become integrated into the chromosome axis to form a stable loop array. At the same time, transcriptional activity in pachynema drives spatial clustering of highly-transcribed loci into transcription “hubs” that manifest as long-range Hi-C contacts. Separate detection of intra- vs. inter-homolog contacts in a high polymorphism density hybrid allows us to define the physical parameters of homolog pairing by the synaptonemal complex as cells progress from zygonema to pachynema. Finally, we show that chromosome compartments are maintained in meiotic prophase, and that both DSBs and crossovers show a strong bias toward the gene-dense A compartment, revealing a key role for chromatin state in meiotic recombination.
Results
Hi-C analysis of mouse spermatogenesis
While chromosome conformation capture methods (Hi-C) 14,15 have recently enabled an unprecedented exploration of eukaryotic genome structure and regulation, analysis of mammalian meiotic prophase by Hi-C has been limited by an inability to isolate pure populations of meiotic prophase cells. To overcome this challenge, we developed methods to purify large numbers of highly-synchronized mouse spermatocytes 16,17 (R.K. and F.C., unpublished) (Fig. S1a-c) and performed Hi-C in both early prophase (zygonema) and late prophase (late pachynema/diplonema) (Fig. 1a, Table S1). To capture inter-homolog contacts during recombination and synapsis, we isolated spermatocytes from C57BL6/J (B6) x M. castaneus (CAST/EiJ; CAST) F1 hybrid mice, which possess 0.83% overall single-nucleotide polymorphism (SNP) density between haplotypes. We performed Hi-C using 100-base paired-end sequencing reads, theoretically allowing us to unambiguously assign B6 vs. CAST haplotype for over half of individual reads, and over a quarter of paired-end reads. We generated 351 million Hi-C contacts for zygonema from two independent samples, and 487 million contacts for pachynema from three independent samples (Fig. S1e-g, Table S2). The resulting Hi-C contact maps from the two prophase stages were visually distinct, yet maps from biological replicates showed high reproducibility (Fig. S2), demonstrating the robustness of our synchronization and purification method. We could assign 3.3% of zygonema read pairs (11.7 million) and 3.6% of pachynema read pairs (17.7 million) as unambiguous inter-homolog contacts (Methods, Table S2). As a control, we used a recent Hi-C dataset from unsynchronized cultured mouse embryonic stem cells (hereafter termed “interphase”) 18. Overall, our data provide an unprecedented picture of dynamic genome reorganization in mammalian meiotic prophase.
Figure 1. Hi-C analysis of the meiotic prophase genome.
(a) Experimental workflow. C57BL/6 x CAST/EiJ male F1 hybrid mice were treated with WIN18,446 followed by retinoic acid to synchronize spermatogenesis, then spermatocytes were isolated and purified by FACS (Fig. S1). Samples of 600,000–800,000 cells were analyzed by Hi-C using 100-base paired end sequencing followed by a haplotype-aware analysis pipeline (Table S1, S2). (b) Hi-C contact maps for cells in interphase (E14 cell culture cells), early zygonema, and late pachynema. Compared to interphase cells, meiotic prophase chromosomes lose all long-range (> 10 Mb) contacts. Color scale for all panels is white (zero Hi-C contacts per bin) to red (indicated CPKM (contacts per kb per billion mapped contacts; see Methods) or higher Hi-C contacts per bin. See Fig. S1e-g for genome-wide Hi-C contact maps. (c) Pearson correlation matrices for chromosome 3 in interphase, zygonema, and pachynema. These matrices graphically illustrate the correlation between different chromosomal regions’ Hi-C contact patterns. Red indicates strongly-correlated contacts, and blue indicates strongly anti-correlated contacts. The observed red-blue checkerboard patterns strongly indicate the presence of compartments in all datasets. (d) Eigenvector analysis of chromosome 3 in interphase, zygonema, and pachynema. Correlations were calculated using a two-tailed non-parametric Spearman correlation coefficient. See Fig. S4 for Eigenvector analysis of additional chromosomes.
Meiotic prophase chromosomes maintain compartment structure but lose topologically associating domains
The eukaryotic genome is organized in all developmental and cell-cycle stages to achieve the particular needs of each cell. In interphase, chromosomes occupy individual “territories” in the nucleus, and also show multiple levels of internal organization. Dynamic DNA binding, loop extrusion modulated by chromosome-bound CTCF, and dissociation from DNA by cohesin complexes gives rise to megabase-sized topologically associating domains (TADs) with high local interaction propensity 22–31. Interphase chromosomes are also arranged into “compartments,” with the gene-dense and transcriptionally-active “A” compartment physically separated from the gene-poor, heterochromatic “B” compartment 15,19. In contrast to TADs, compartments are not formed through dynamic loop extrusion and do not depend on cohesin 20–22, rather they likely form through the tendency of heterochromatin to self-associate through a phase separation-like mechanism 23–25.
In meiotic prophase, we observe a near-complete loss of very long-range contacts (over ~5–10 Mb) consistent with the known organization of meiotic chromosomes as linear arrays of loops anchored to the meiotic chromosome axis (Fig. 1b). We also observe “X”-shaped inter-chromosomal contact patterns consistent with the alignment of chromosomes into the prophase bouquet, which are particularly strong in zygonema but also detectable in pachynema (Fig. S2, S3a-b). Despite the reorganization of chromosomes into loop arrays, we find that meiotic prophase chromosomes maintain strong A/B compartment identity, observable in Hi-C contact maps as a checkerboard pattern near the diagonal axis (Fig. 1b). A/B compartments are also clearly visible in chromosome-wide Pearson correlation matrices (Fig. 1c), and are remarkably consistent with interphase compartments (Fig. 1d, S4a-c). Thus, despite the reorganization of chromosomes into loop arrays in meiotic prophase, the fundamental organization of chromatin into A/B compartments is maintained.
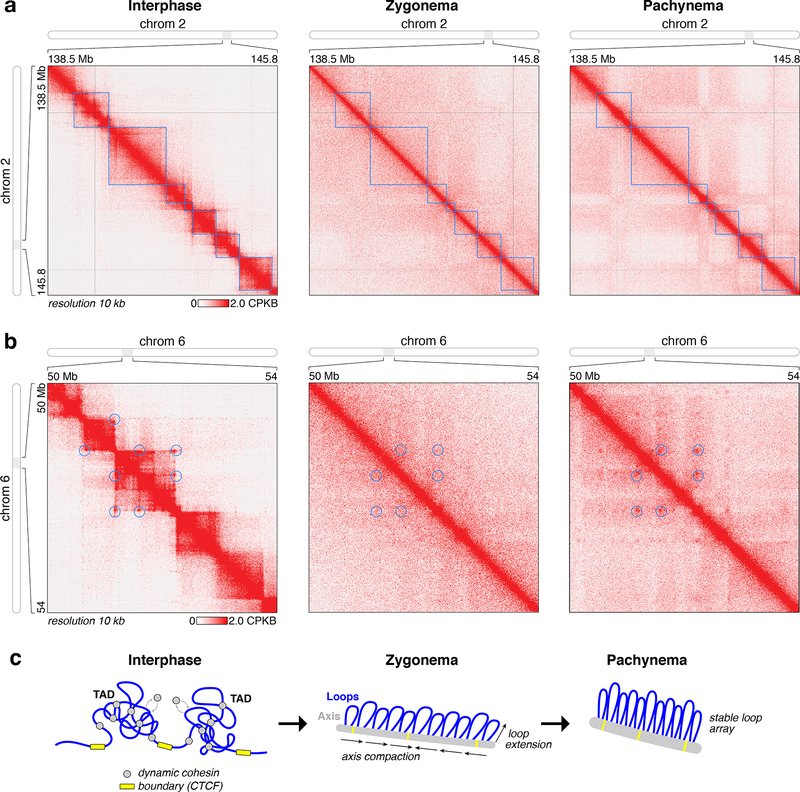
We next examined TADs, which are visible in Hi-C contact maps as squares with high contact propensity, often with strong corner signals that result from looping interactions between TAD boundaries 26. We find that in meiosis, TADs are mostly lost despite the continued presence of cohesin on chromosomes (Fig. 2a) 2,27. A few loci show evidence of looping interactions between TAD boundaries (Fig. 2b), but most loci show a complete loss of both the square and corner TAD signals. These data suggest that if cohesin-constrained loops are present in meiotic chromosomes, as ample cytological and electron microscopy data suggest 2, the locations of these loops most likely vary from cell to cell. This may arise from a reduction in CTCF’s influence on loop positioning, or from modulation of cohesin activity upon association with the filamentous chromosome axis “core” proteins. We propose that association with the chromosome axis reduces the dynamics of chromosome association and dissociation by cohesin, leading to the formation of a stable loop array 7. Our data do not reveal whether cohesin-mediated loop extrusion activity is reduced upon axis association, though the increase in average loop size as cells progress from zygonema to pachynema (see below) suggests that loop extrusion continues through prophase (Fig. 2c). Our data indicating a lack of reproducible loop positions in meiosis contrasts with recent Hi-C analyses of S. cerevisiae meiosis, which showed strong looping interactions between cohesin binding sites across the genome in pachynema 28,29. While binding sites for S. cerevisiae meiotic cohesin complexes are highly reproducible 8,30, likely leading to these strong looping signals, there is so far no evidence of reproducible cohesin binding along chromosomes in mouse spermatocytes.
Figure 2. Loss of TADs in meiotic chromosomes.
(a) High-resolution view of a region of chromosome 2, showing loss of topologically-associating domains (blue boxes) in meiotic prophase. (b) High-resolution view of a region of chromosome 6, showing establishment of a stable loop array in pachynema with loop bases (blue circles) corresponding to interphase TAD boundaries. (c) Model for assembly of meiotic chromosomes. Association of dynamic cohesin complexes (grey) with chromosome axis core proteins mediates assembly of the axis and reduces cohesin dynamics (chromatin association and dissociation, and loop extension) as cells enter leptonema/zygonema, then loops further extend in coordination with axis compaction as cells enter pachynema. At some loci, the bases of stable loops in pachynema coincide with interphase TAD boundaries (yellow).
Formation of transcription “hubs” on meiotic chromosomes
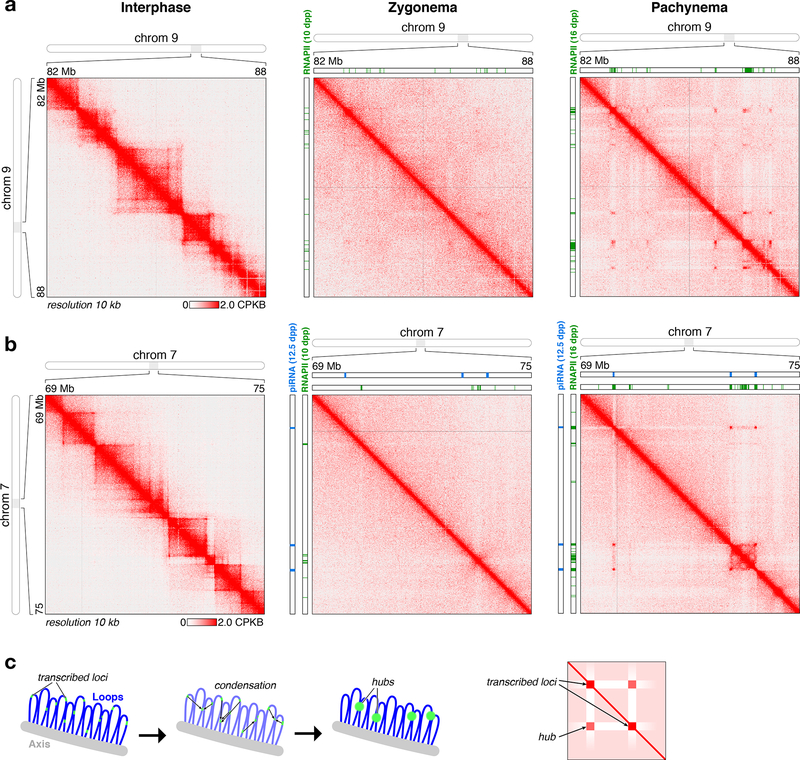
While we observe a near-complete loss of TAD signal in meiotic chromosomes, a large fraction of the genome shows looping or clustering interactions at the 1–10 Mb scale, which are present in zygonema but very pronounced in pachynema (Fig. 3a-b, S5). When we overlaid Hi-C contact maps with RNA Polymerase II-bound loci in both prophase stages 31, we found that the clustered loci correspond to loci undergoing active transcription in both meiotic stages (Fig. 3a). Some clusters also correspond to highly-transcribed clusters of piRNAs, short RNAs with specialized roles in transposon silencing and sperm development (Fig. 3b) 32,33. These data suggest that transcribed loci self-associate or condense within the meiotic chromosome structure to form clusters or “hubs” (Fig. 3c). While prior studies have shown that transcription machinery can localize to “transcription factories” 34 and form phase-separated condensates within the nucleus 35–38, the strong interactions evident in our Hi-C contact maps suggest that meiotic prophase chromosomes are particularly susceptible to these influences. Supporting the idea of transcription hub formation in meiotic prophase, several prior studies have shown that RNA polymerase II 39–41 and nascent RNA transcripts 41,42 form highly punctate localization patterns in mouse and human spermatocytes.
Figure 3. Transcription-mediated interaction hubs in meiotic chromosomes.
(a) High-resolution view of a region of chromosome 9 in interphase, zygonema, and pachynema. Shown in green are RNA Polymerase II peaks detected at 10 dpp (zygonema) or 16 dpp (pachynema) 31. (b) High-resolution view of a region of chromosome 7 in interphase, zygonema, and pachynema. Shown in blue are piRNA clusters transcribed at 12.5 days post-partum (dpp), during pachynema of the first wave of spermatogenesis (piRNA clusters measured at 14.5 dpp, later in pachynema, were nearly identical) 69, and shown in green are RNA Polymerase II peaks detected at 10 dpp (zygonema of the first wave of spermatogenesis) or 16 dpp (pachynema). (c) Left: Model for assembly of transcription-mediated interaction hubs. In the absence of dynamic cohesin complexes disrupting chromatin-chromatin interactions, highly-transcribed loci (green) will condense through cooperative self-interactions into interaction hubs. Right: Schematic of Hi-C contact maps resulting from assembly of interaction hubs. Highly-transcribed regions show depletion of short-range contacts with non-transcribed regions, and increased interactions with highly-transcribed regions up to several Mb away. See Fig. S6 for additional examples of transcription-mediated interaction hubs.
Global organization of meiotic chromosomes
To characterize the global organization of meiotic chromosomes, we next analyzed genome-wide Hi-C contact probability (P) as a function of genomic distance (s). We find that for genomic distances less than ~5 Mb, contact probability P(s) follows a power-law scaling proportional to s−0.5, dramatically different from the typical scaling of interphase chromosomes (between s−1 and s−1.5) 15,43 (Fig. 4a, S6). The P(s)~s−0.5 scaling we observe in meiosis is similar to prior findings on mitotic chromosomes, which are organized as helical arrays of loops by cohesin-related condensin complexes 12,13,44. Meiotic chromosomes are also morphologically similar to early mitotic prophase chromosomes, being individualized and compacted, but much longer than mitotic prometaphase or metaphase chromosomes 45. In agreement with this idea, the P(s) curves of meiotic prophase cells are most similar to those of chromosomes in early mitotic prophase, which have lost detectable TADs and are organized as linear arrays of loops, but have not yet formed the highly compacted helical arrays characteristic of metaphase chromosomes 13. While contact probability in mitotic prophase chromosomes drops sharply beyond ~2 Mb 13, meiotic chromosomes retain a P(s)~s−0.5 scaling relationship up to ~5 Mb (Fig. 4a). Chromosomes in pachynema show high contact probability at slightly longer distances than in zygonema, suggesting that cohesin-constrained loops may continue to extend through zygonema until final stabilization of the loop array in pachynema. This model agrees with prior reports of axis compaction as cells progress from zygonema to pachynema, and the more general inverse relationship between loop size and axis length in mutants of both meiosis-specific cohesin subunits (e.g. Smc1β) and chromosome axis core proteins (SYCP3) 46–48. To estimate average loop length genome-wide, we examined plots of the slope, or derivative, of the P(s) function, maxima in which have been shown to correlate with average loop lengths inferred from polymer simulations 49. This analysis suggests that average loop lengths are 0.8–1 Mb in zygonema, and extend to 1.5–2 Mb in pachynema (Fig. 4a, lower panel). To estimate average loop density along chromosomes, we measured the total length of synapsed chromosome axis in B6 x CAST pachynema spermatocytes at 215 +/− 33 μm (Fig. S1d). If the entire 2.8 Gb (haploid) genome is contained within loops averaging 1.5 Mb in length, this suggests an average loop density of ~10 loops per micron of chromosome axis in pachynema.
Figure 4. Global organization of chromosomes and detection of inter-homolog contacts in meiotic prophase.
(a) Genome-wide contact probability versus genomic distance (P(s)) in zygonema (green) and pachynema (purple) versus interphase (black), with dotted lines corresponding to P(s)~s−0.5 and P(s)~s-1.5. The observed scaling is consistent across all chromosomes (Fig. S6a,b,d) and for both B6 and CAST chromosomes (Fig. S6f-g). Lower panel: Plot of the slope of the P(s) curves shown above reveals average loop size in zygonema (0.8–1.0 Mb) and pachynema (1.5–2.0 Mb). (b) Schematic model of chromosome organization and homolog synapsis in meiotic prophase, with the chromosome axis (gray line) constraining sister chromosomes as aligned loop arrays (two shades of blue in inset). Notably, loops likely extend in all directions from each axis (see cross-section view at right), resulting in the interdigitation of loops from homologous chromosomes (blue and yellow). Synaptonemal complex (SC) transverse filaments are shown in gray. (c) Inter-homolog Hi-C contact maps for chromosome 5 in zygonema. Close-up views of boxed regions 1 (middle of chromosome) and 2 (end of chromosome) are shown in lower panels. (d) Inter-homolog Hi-C contact maps for chromosome 5 in pachynema, as in (c). See Fig. S7 for additional examples. (e) P(s) curves for inter-homolog contacts (zygonema green, pachynema purple), with dotted line corresponding to P(s)~s-0.18. Modeling the convolution of two P(s)~s−0.5 functions, representing the physical alignment and interdigitation of two loop arrays in pachynema, gives a function proportional to s−0.206 (Fig. S8). Intra- versus Inter-homolog P(s) functions are illustrated in magenta in panel (b).
Hi-C captures homolog pairing in meiotic prophase
Meiotic prophase is the only developmental stage in mammals where homologous chromosomes are physically associated along their lengths. The 0.83% single-nucleotide polymorphism (SNP) density between B6 and CAST haplotypes in our F1 hybrid mice allowed us to assign 3.3% of zygonema read pairs (11.7 million) and 3.6% of pachynema read pairs (17.7 million) as unambiguous inter-homolog contacts, enabling analysis of inter-homolog contacts genome-wide (Methods, Table S2). Hi-C contact maps constructed using only inter-homolog contacts showed strong diagonal signal in all intra-chromosomal maps, clearly indicating that homologs are aligned along their lengths (Fig. 4c-d, S7). This general relationship was true in both zygonema and pachynema, despite the fact that chromosomes are only partially synapsed in zygonema. Preferential association within A/B compartments, visible as a checkerboard pattern in the inter-homolog Hi-C maps, was also evident along the entire lengths of most chromosomes (Fig. 4c-d, S7a-b). This finding supports a model in which the chromatin loops of paired homologs are extensively interdigitated (Fig. 4b), allowing preferential self-association of the A and B compartments between these chromosomes. In agreement with this idea, we also observe evidence of transcription-mediated interactions between homologs (Fig. S7c-d).
We next plotted contact probability versus genomic distance specifically for inter-homolog contacts (Fig. 4e). The inter-homolog P(s) function shows a significantly shallower slope than the intra-homolog P(s) function, with a power-law scaling roughly proportional to s−0.18 (Fig. 4e). When considering the structure of a synapsed homolog pair, we envision that two factors may contribute to this shallower slope. First, synapsed homologs are aligned and juxtaposed arrays of chromatin loops, whose bases are held apart by the SC but which can likely extensively interdigitate (Fig. 4b). The effect of this loop interdigitation can be modeled mathematically as a convolution of two P(s)=s−0.5 functions, which results in a power-law scaling function proportional to P(s)=s−0.2 (Fig. S8). Second, chromosomes are unlikely to be held in perfect juxtaposition by the synaptonemal complex. Local variation in packing density due to differences in loop size and positioning, plus variations in axis structure, likely give rise to small displacements of aligned homologs relative to one another. The effect on interhomolog P(s) would be to increase long-range contacts relative to short-range contacts, as we observe (Fig. 4e). Overall, our data support a model in which synapsed homologs are closely aligned along their length, while individual loci within the aligned loop arrays retain significant freedom to access sequences on the homologous chromosome within a +/− 5–8 Mb region.
Meiotic recombination frequency is strongly correlated with compartment structure
Spo11-catalyzed DSBs, which initiate meiotic recombination, occur preferentially in “hotspots” whose locations are dictated by a combination of chromatin structure and protein factors, and in yeast correlate with high-GC content regions 8,50–52. In most mammals, hotspot locations are controlled by PRDM9, a histone methyltransferase that generates trimethylated histone H3 lysine 4 and 36 (H3K4me3, H3K36me3) marks in chromatin near its binding sites 53. PRDM9 has been shown to direct recombination away from functional elements like promoters at the fine scale 54, but control of DSB formation at larger scales is not well understood in mammals. Prior reports that the recombinase RAD51 preferentially localizes to R-band (A compartment) chromatin in meiotic prophase 55, and also that meiotic chiasma appear more frequently in R bands in mouse spermatocytes 56, have hinted that compartment identity may play a role in mammalian meiotic recombination. More recently, PRDM9 was shown to bind and promote DSB formation more effectively in euchromatin than in heterochromatin or lamin-associated regions 57, suggesting that chromatin accessibility may directly affect meiotic recombination rates through differential PRDM9 binding. Finally, genome-wide maps of meiotic DSBs have shown a bias toward nucleosome-depleted regions flanked by H3K4me3- and H3K36me3 nucleosomes in euchromatin 58.
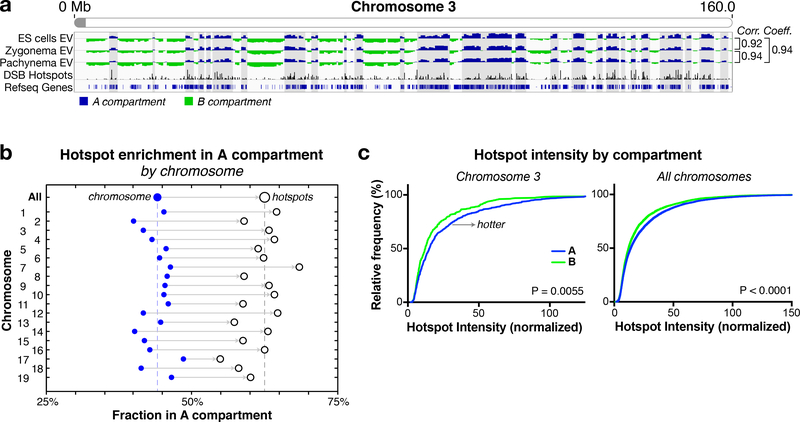
To further explore the connection between chromosome compartments and meiotic recombination, we overlaid the chromosome compartment structure with a previously-reported map of meiotic DSB hotspots in B6 x CAST F1 hybrids 59. We found that both hotspot density (Fig. 5a-b, S9a) and relative intensity (Fig. 5c, S9e) are significantly higher in the A compartment compared to the B compartment. The A compartment is also enriched in both PRDM9-bound sites (Fig. S9b) and H3K4me3 peaks (Fig. S9c) in B6 x CAST spermatocytes 60. Finally, a set of ~800 crossovers between B6 and CAST chromosomes in the multi-species Collaborative Cross 61 also shows a strong bias toward the A compartment (Figure S9d). Overall, these data indicate that the meiotic recombination landscape, while controlled at the fine scale by the location of PRDM9 binding sites, is strongly correlated at the megabase scale with compartment identity and chromatin state.
Figure 5. Meiotic DSB hotspots show strong compartment bias.
(a) Eigenvector analysis of chromosome 3 in interphase, zygonema, and pachynema, as in Fig. 1d, with the addition of DSB hotspot locations and intensities 59. (b) Fraction of each chromosome in the A compartment (blue circles, genome-wide data at top) and the fraction of DSB hotspots in that chromosome that are located in the A compartment (open circles). Every chromosome shows a strong bias toward the A compartment (Fig. S9a). (c) Cumulative distribution of hotspot intensity by compartment (A blue, B green) in chromosome 3 (left) and genome-wide (right). P values calculated using a Kolmogorov-Smirnov test. See Fig. S9e for graphs of each individual chromosome.
Isolation and silencing of the X-chromosome in pachynema
In mammalian meiosis, chromosomes that fail to pair and synapse are subject to a pathway termed meiotic silencing of unsynapsed chromatin (MSUC), in which these regions obtain repressive chromatin marks and are transcriptionally silenced 62–65. In male mice, the X and Y chromosomes pair, synapse, and form crossovers in a ~1 Mb “pseudo-autosomal region”, but the bulk of these chromosomes remain unpaired. As spermatocytes enter pachynema, the unsynapsed regions of the X and Y are silenced by MSUC, also termed “meiotic sex chromosome inactivation” (MSCI) 66, and become isolated from other chromosomes as they are packaged into the “sex body” or XY body.
Our Hi-C contact maps clearly illustrate the reorganization of the X chromosome in pachynema. In zygonema, the X chromosome behaves equivalently to autosomes, showing strong “X”-shaped inter-chromosomal interaction patterns (Fig. S3c) and maintaining compartment structure while losing visible TADs (Fig. 6a-b). While these features are maintained through pachynema on autosomes, however, the X chromosome shows dramatic changes. First, the X chromosome becomes strongly isolated from all autosomes in pachynema, completely losing the “X”-shaped inter-chromosomal contact pattern observed in zygonema (Fig. S3c). Second, the X chromosome’s compartment structure is completely lost in pachynema (Fig. 6b, S4d). Third, consistent with the idea that the looping or clustering interactions we observe on autosomes are linked to transcription, we observe a near-complete loss of this clustering on the X chromosome as it becomes transcriptionally silenced in pachynema (Fig. 6b).
Figure 6. X chromosome organization in pachynema.
(a) Hi-C contact maps for the X chromosome in interphase, zygonema, and pachynema. Dotted boxes indicate the area shown in close-up in insets. (b) Closeup view of a region of chromosome X that shows transcription-mediated clustering of loci in zygonema, which is largely lost in pachynema. Shown in green are RNA Polymerase II binding peaks at 10 dpp (zygonema) or 16 dpp (pachynema) 31. (c) Pearson correlation matrices for the X chromosome in interphase, zygonema, and pachynema. See Figure S4d for Eigenvector analysis of the X chromosome.
While these data reveal significant reorganization of the X chromosome in pachynema, its underlying structure as a linear array of loops appears mostly unaffected. We plotted P(s) for the X chromosome in both zygonema and pachynema, and found that while the X chromosome shows a subtly different contact probability curve in pachynema compared to autosomes, the overall shape and slope of the curve is largely unchanged from zygonema (Fig. S6h,i). Thus, the pachynema X chromosome can be considered to represent a “basal state” of meiotic chromosome organization, in which the axis-associated loop structure is unperturbed by either transcription-mediated clustering of loci or A/B compartment structure. In agreement with the idea that meiotic chromosome axis-associated chromatin loop locations are mostly stochastic, we observe no evidence of reproducibly-located loops along the pachynema X chromosome in our Hi-C contact maps (Fig 6a-b).
Discussion
In meiotic prophase, chromosomes are highly organized by the meiotic chromosome axis and synaptonemal complex to promote homolog recognition and recombination, yet these chromosomes must also be transcriptionally active to support later stages of spermatogenesis. Here, we use Hi-C to directly visualize chromosome reorganization in meiotic prophase, revealing the physical parameters of chromosome organization by the meiotic chromosome axis and of homolog juxtaposition by the synaptonemal complex. We find that meiotic chromosomes lose TADs, retain strong A/B compartment structure, and form transcription “hubs” through clustering of highly-transcribed loci. These changes can be explained by a model in which association of cohesin complexes with the meiotic chromosome axis stabilizes their association with chromatin, reduces the influence of CTCF on the positioning of cohesin-constrained loops, and may also affect loop extrusion rate or processivity. As cohesin complexes coalesce on the chromosome axis, they mediate the assembly of a stable array of loops, and our data suggests that loop lengths continue to increase as the axis undergoes linear compaction through leptonema/zygonema and into pachynema.
The loss of TADs, retention of A/B compartment structure, and formation of transcription “hubs” are all strongly reminiscent of recent reports of the effects of cohesin depletion in somatic cells. Cohesin depletion causes an almost immediate loss of TADs in aggregate Hi-C data, strengthening and in some cases fragmentation of the A/B compartment structure, and the formation of multivalent enhancer clusters, or “superenhancer hubs” 20–22. The similarity of meiotic chromosomes to chromosomes that have lost cohesin entirely supports our model in which cohesin dynamics are strongly suppressed as they are repurposed for assembly of a stable chromatin loop array. The loops themselves, which exceed 1 Mb in length in pachynema, are essentially free of dynamic cohesin complexes which would otherwise counteract the tendency of both heterochromatin and transcription machinery to self-associate.
The overall structure of meiotic chromosomes as linearly-compacted loop arrays organized by cohesin complexes and other axis components is highly conserved throughout eukaryotes 2. When we compare our data from M. musculus spermatocytes with two recent Hi-C analyses of S. cerevisiae meiotic chromosomes 28,29, we find some clear differences, but overall a striking level of agreement. The most obvious difference is that while Hi-C contact maps of S. cerevisiae chromosomes show strong evidence of looping between known cohesin binding sites, M. musculus chromosomes show little evidence for reproducible loop locations across the cell population. This difference is likely due to the known preferential binding of S. cerevisiae meiotic cohesin complexes near the 3’ ends of open reading frames, and particularly between convergent gene pairs 9,30,67; there is so far no evidence of preferred cohesin binding sites in mouse meiocytes. Another major difference between S. cerevisiae and M. musculus is the length of chromatin loops: our data suggest that average chromatin loops in M. musculus chromosomes extend from 0.8–1 Mb in zygonema to 1.5–2 Mb in pachynema, while Hi-C and polymer simulations of S. cerevisiae chromosomes indicate an average loop length of ~26 kb in this organism. Loop lengths are shorter, ~20 kb, in the absence of interhomolog synapsis (zip1Δ), in agreement with our finding that loops extend as chromosomes undergo synapsis during the zygonema-pachynema transition. The extremely short loops in S. cerevisiae meiotic chromosomes, combined with the preferential cohesin binding sites and the transient nature of pachynema in S. cerevisiae, probably precludes any clustering of transcribed loci as we observe in M. musculus spermatocytes. While loop lengths are ~50-fold different between S. cerevisiae and M. musculus, the density of loops along the chromosome axis is remarkably similar in the two organisms. With a total pachytene axis length of ~36 μm in S. cerevisiae (based on measurements in 68), and 65% of the genome packaged into 26 kb loops as estimated by Schalbetter et. al 29, we estimate a loop density of ~8.5 per micron of axis, very close to our estimate of 10 loops per micron in M. musculus. This finding agrees with prior proposals that while loop lengths vary widely between eukaryotes, scaling roughly with overall genome size, the architecture of the chromosome axis and its looping structure is highly conserved 2. Thus, while the details of meiotic chromosome structure vary between organisms, the fundamental architecture of the chromosome axis-constrained loop array is extremely consistent.
Prior studies have pointed out strong morphological similarities between chromosomes in meiotic prophase and very early mitotic prophase 45, in which chromosomes are individualized and partially compacted, but have not yet become the highly-compacted helical loop arrays found in mitotic prometaphase and metaphase 12,13. Whereas mitotic prophase is a transient state characterized by the replacement of cohesins with condensins and eviction of transcriptional machinery, meiotic prophase is a highly stable state mediated by meiosis-specific cohesin complexes and characterized by high transcriptional activity. Nonetheless, in keeping with chromosomes’ overall morphological similarity, we find that global genome organization in these two states is similar, with P(s) curves showing power-law scaling proportional to s-0.5. Meiotic chromosomes retain a P(s)~s−0.5 scaling relationship over longer distances than mitotic prophase chromosomes (5 Mb versus 2 Mb 13), consistent with significantly longer loops in meiotic prophase (1.5 Mb in pachynema, compared to 60–80 kb in mitotic prophase 13).
The high transcriptional activity of meiotic chromosomes impacts chromosome organization in several ways. First, meiotic chromosomes retain physically-separated A and B compartments, in contrast to mitotic chromosomes which lose compartment separation entirely 12,13. Second, we observe strong clustering of highly-transcribed loci, suggesting that these loci tend to self-associate or condense during the extended meiotic prophase. This condensation is likely a consequence of the loss of dynamic cohesin complexes on extended chromatin loops, which would otherwise act to dissociate these clusters much as they counteract the self-association of A and B compartments in somatic cells 21,22. Due to meiotic chromosomes’ organization as linear loop arrays, clustering of transcribed loci can only occur locally, within a 5–10 Mb range, but apparently can occur between loci on homologs due to interdigitation of the paired loop arrays.
The maintenance of A/B compartments and high-level transcription in meiotic prophase shows that the genome retains many chromatin-structure features of interphase chromosomes, despite reorganization by the meiotic chromosome axis and in stark contrast to chromosomes entering mitosis. Chromatin structure in turn strongly affects the overall distribution of meiotic DSBs and eventual inter-homolog crossovers, strongly biasing recombination toward the gene-dense A compartment over the more heterochromatic B compartment. This effect is likely due to differential chromatin accessibility, with PRDM9 able to more easily access its binding sites and generate H3K4me3 marks in the A compartment 57.
Newly-developed methods for the synchronization and purification of mouse spermatocytes, and their analysis by Hi-C, can provide a new window into the organization and function of meiotic chromosomes. These advances will be critical to advance our understanding of the roles of structural proteins including cohesins and chromosome axis components, the interplay between chromosome organization and transcription, and homolog interactions during recombination and synapsis.
Online Methods
Mouse husbandry and spermatocyte isolation
We mated female C57BL/6J and male CAST/EiJ mice (obtained from The Jackson Laboratory). We treated male F1 neonates with the retinoic acid inhibitor WIN 18,446 at two days post-partum to block spermatogonial differentiation, then injected retinoic acid at 9 days post-partum as previously described 16 (Fig. S1a). Treated animals were allowed to recover for 25–47 days to enable isolation of synchronized cell populations in zygonema and pachynema of the second through fifth waves, with harvesting times calculated as previously described 70 (Table S1). Spermatocytes from synchronized testes were isolated and stained with Hoechst 33342 to allow isolation of cells with 4C DNA content (prophase I) by flow cytometry 17. Compared to unsynchronized animals, cells isolated from synchronized testes showed a significantly different cell profile with only a few densely populated 4C regions (Fig. S1b). Purity and prophase stage were determined by chromosome spreads of sorted cells, stained with antibodies to SYCP3 (sc-74569, mouse monoclonal clone D-1 from Santa Cruz) and H1t (a gift from Dr. Mary Ann Handel) (Fig. S1c). Final cell numbers and purity for each sample are noted in Table S1.
Hi-C library preparation and sequencing
Hi-C experiments were performed largely as previously described 15,26,71. Briefly, 600,000–800,000 cells (Table S1) were cross-linked with 2% formaldehyde for 10 min at room temperature, then the reaction was quenched using 200 mM glycine for 5 min at room temperature, then 15 minutes on ice, then samples were frozen in liquid nitrogen. Nuclei were isolated and directly applied for digestion using the 4-base cutter restriction enzyme MboI (NEB) at 37°C overnight. The single strand overhang was filled with biotin-14-dATP (Life Technologies) using Klenow DNA polymerase (New England Biolabs). In contrast to traditional Hi-C, the ligation was performed when the nuclear membrane was still intact (in situ protocol). DNA was ligated for 4 hours at 16°C using T4 ligase (New England Biolabs). Protein was degraded by proteinase K (New England Biolabs) treatment at 55°C for 30 min. The crosslinking was reversed with addition of 500 mM NaCl and incubation at 65°C overnight. DNA was purified by ethanol precipitation, sonicated to 300–700 bp fragments, and size-selected using SPRI magnetic beads as described 26. Biotinylated DNA was selected with Dynabeads MyOne T1 Streptavidin beads (Life Technologies). Sequencing libraries were prepared on beads, checked using an Agilent Bioanalyzer 2100 and quantified using a Qubit (Life Technologies). Libraries were sequenced on an Illumina Hiseq 4000 with 100 cycles of paired-end reads.
Hi-C data analysis and bioinformatics
Hi-C data pre-processing and analysis was performed largely as previously described 72, with modifications for assignment of haplotype of each read. We aligned each read to the mm10 genome assembly using BWA-MEM 73 with default parameters except the clipping penalty (-L flag) was set to 13. Next, WASP 74 was adapted to identify reads containing one or more SNPs, then the read was re-aligned after flipping each allele to the value in the CAST genome. For SNP identification, we used data from the Wellcome Sanger Institute Mouse Genomes Project 75, accession code ERS076381. Dividing 226,138,14 SNPs by a total genome length of 2,725,521,370 (one copy of each chromosome including X and Y) gives 0.83% SNP density, or one SNP in 120 bp on average. SNP-containing reads were kept for further analysis only if the read mapped to the same genomic location in both mapping steps. Next, the haplotype at each SNP location was identified, and the haplotype of the read classified as either ambiguous (no SNPs), B6 (all SNPs mapping to B6), or CAST (all SNPs mapping to CAST) (Table S2). On average, only 0.5% of reads containing multiple SNPs showed a mixture of B6 and CAST alleles, and these reads were discarded (these reads could arise from multiple sources, including an inter-homolog ligation junction within the read, capture of a meiotic or pre-meiotic recombination event, or sequencing errors). Read pairs corresponding to B6-B6, CAST-CAST, or B6-CAST interactions were then separated for later analysis. For construction of Hi-C contact maps and contact probability analysis, all read pairs including those without SNPs were used. BAM files were further processed using pairtools (https://github.com/mirnylab/pairtools) to identify ligation junctions and produce Hi-C. pairs files. Hi-C contact maps in .hic format were constructed using the pre function in Juicer 76. Hi-C contact maps were visualized in Juicebox 77 with balanced normalization applied 78. For assignment of compartments, we used the eigenvector function in Juicer 76. Eigenvectors were calculated for control E14 cells, zygonema (combined dataset), and pachynema (sample #1). Eigenvectors and additional genomic features were visualized using Integrated Genomics Viewer v. 2.4.10 79. Custom scripts are available at Github (https://github.com/lucaspatel/nsmb_mousehic).
For comparisons of our Hi-C contact maps with interphase Hi-C contact maps, we used a previously-published dataset obtained using the above library preparation and sequencing methodology from mouse E14 embryonic stem cells (strain background 12910la; GEO sample GSM1908921) 18. We re-mapped this dataset to the mm10 genome assembly using the same procedure as above, but without considering haplotype. All figures showing Hi-C contact maps are displayed with a linear white-to-red gradient, and report maximum contrast (red) in terms of CPKB, “Hi-C Contacts Per Kilobase per Billion mapped contacts”, calculated as follows:
Contrast levels for each panel in a given figure are adjusted to equivalent CPKB values.
For correlation of compartments with other genomic features, we used A/B compartment calls from the control interphase dataset as these were more robust than from zygotene or pachytene, but agreed closely with meiotic datasets across most of the genome. For DSB hotspots, we used hotspot locations and normalized intensity assignments from a previously-published ssDNA map from spermatocytes of a C57BL/6J x CAST/EiJ F1 hybrid mouse (GEO sample GSM1954839) 59. For crossovers, we used the B6xCAST and CASTxB6 crossover locations from a previously-published multi-species cross (797 crossovers total; re-mapped from mm9 to mm10 reference genome) 61. For PRDM9 binding sites and H3K4me3 sites, we used PRDM9 and H3K3me3 peaks called from ChIP-Seq data on B6xCAST F1 hybrid mice (GEO sample GSE60906) 60. Peak locations (6,955 PRDM9 peaks and 80,940 H3K4me3 peaks) were re-mapped from mm9 to mm10 reference genome.
For DSB hotspot analysis, we first calculated the center of each DSB hotspot and assigned the hotspot to either the A or B compartment. We then plotted the cumulative hotspot intensity distribution for each compartment (genome-wide or per chromosome) in Prism 7, and calculated P-values using a Kolmogorov-Smirnov test (Figure S9e). We next used regioneR 80 to calculate the significance of enrichment of DSB hotspots overlapping the A compartment, compared to a random distribution (Figure S9a). We used a similar analysis to calculate the significance of enrichment of crossovers, PRDM9 binding sites, and H3K4me3 peaks (Figure S9b-d). Other statistical tests were performed using Prism v. 7 (GraphPad Software).
For comparison of Hi-C contacts with transcription data, we used a published RNA Polymerase II ChIP-Seq dataset for 16-dpp C57BL/6J mouse testes (GEO Sample GSM1083638), remapped to mm10 31. For comparison with piRNA clusters, we used data from the piRNA cluster database 81,82, specifically 12.5-dpp (SRR772029/GEO GSM1096583) and 14.5-dpp (SRR7720230/GEO GSM1096584) C57BL/6J mouse testes samples 69.
Contact Probability Calculation
Contact probability versus genomic distance (P(s)) curves were calculated as previously described 12,83. Briefly, we divided all genomic separations into logarithmically-sized bins, starting at 10 kb and increasing by a factor of 1.12 per bin. We first calculated the number of Hi-C contacts in each dataset that fell into each bin. We next calculated the number of possible Hi-C contacts at each distance across the genome or within an individual chromosome, using a fragment size of 250 bp to approximate the ~256 bp size of MboI-generated restriction fragments. Dividing contact number by potential contacts in each bin yielded contact probability P(s), which we then normalized by setting the value of P(s) at a distance of 100 kb to 1. Due to their distinctive organization in meiotic prophase, the X and Y chromosomes were considered separately in this analysis.
Supplementary Material
Acknowledgements
We thank M. Handel for the kind gift of H1T antibodies; members of the Corbett, Cole, and Ren labs; and A. Desai for helpful discussions. We thank S. Aigner, M. Neale, G. Fudenberg, and L. Mirny for helpful suggestions on Hi-C data interpretation, and M. Griswold and C. Hogarth for assistance with synchronization of spermatogenesis. K.D.C. acknowledges support from the Ludwig Institute for Cancer Research and the National Institutes of Health grant #R01GM104141. F.C. acknowledges support from National Institutes of Health grant #DP2HD087943. B.R. acknowledges support from the Ludwig Institute for Cancer Research. We acknowledge the National Institutes of Health grant #CA16672 for support of the Research Animal Support Facility Smithville and the CPRIT grant #RP170628 for support of the Flow Cytometry and Cellular Imaging Core. R.K. is supported by a CPRIT Research Training Award, #RP170067. R.R. was supported by a Ruth L. Kirschstein National Research Service Award NIH/NCI T32 CA009523.
Footnotes
Competing interests
The authors declare no competing interests.
Accession Codes
All sequencing data have been deposited in the NCBI Gene Expression Omnibus database under accession number GSE122622.
Data and Code Availability
All custom scripts and code are available at Github (https://github.com/lucaspatel/nsmb_mousehic) or from the authors (K.D.C.) upon request.
All sequencing data have been deposited in the NCBI Gene Expression Omnibus database under accession number GSE122622: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122622
All previously-published data used in our analysis is available at the links below:
GSM1908921: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM1908921
GSM1954839: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM1954839
ERS076381: ftp://ftp-mouse.sanger.ac.uk/current_snps/strain_specific_vcfs/CAST_EiJ.mgp.v5.snps.dbSNP142.vcf.gz
GSE101406: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60906
GSM1083638: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM1083638
SRR772029/SRR7720230 (processed): http://www.smallrnagroup.uni-mainz.de/piRNAclusterDB.htm
SRR772029/GSM1096583 (raw data): https://www.ncbi.nlm.nih.gov/sra/SRX248863
SRR7720230/GSM1096584 (raw data): https://www.ncbi.nlm.nih.gov/sra/SRX248864
References
- 1.Keeney S, Lange J & Mohibullah N Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu Rev Genet 48, 187–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zickler D & Kleckner N Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33, 603–754 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Hassold T, Hall H & Hunt P The origin of human aneuploidy: where we have been, where we are going. Human Molecular Genetics 16 Spec No. 2, R203–8 (2007). [DOI] [PubMed] [Google Scholar]
- 4.McNicoll F, Stevense M & Jessberger R Cohesin in gametogenesis. Curr Top Dev Biol 102, 1–34 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Lee J Roles of cohesin and condensin in chromosome dynamics during mammalian meiosis. J. Reprod. Dev 59, 431–436 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rankin S Complex elaboration: making sense of meiotic cohesin dynamics. FEBS J. 282, 2426–2443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West AMV et al. A conserved mechanism for meiotic chromosome organization through self-assembly of a filamentous chromosome axis core. bioRxiv 375220 (2018). [Google Scholar]
- 8.Blat Y, Protacio RU, Hunter N & Kleckner N Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111, 791–802 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Panizza S et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146, 372–383 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Page SL & Hawley RS The Genetics and Molecular Biology of the Synaptonemal Complex. Annu Rev Cell Dev Biol 20, 525–558 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Parsons GG & Spencer CA Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol Cell Biol 17, 5791–5802 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naumova N et al. Organization of the mitotic chromosome. Science 342, 948–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibcus JH et al. A pathway for mitotic chromosome formation. Science 359, eaao6135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekker J, Rippe K, Dekker M & Kleckner N Capturing chromosome conformation. Science 295, 1306–1311 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Lieberman-Aiden E et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogarth CA et al. Turning a spermatogenic wave into a tsunami: synchronizing murine spermatogenesis using WIN 18,446. Biol. Reprod 88, 40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole F et al. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat Genet 46, 1072–1080 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan J et al. Histone H3 lysine 4 monomethylation modulates long-range chromatin interactions at enhancers. Cell Res. 28, 204–220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shopland LS et al. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol 174, 27–38 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarzer W et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao SSP et al. Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wutz G et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36, 3573–3599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jost D, Carrivain P, Cavalli G & Vaillant C Modeling epigenome folding: formation and dynamics of topologically associated chromatin domains. Nucleic Acids Res 42, 9553–9561 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Pierro M, Zhang B, Aiden EL, Wolynes PG & Onuchic JN Transferable model for chromosome architecture. Proc. Natl. Acad. Sci. USA 113, 12168–12173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falk M et al. Heterochromatin drives organization of conventional and inverted nuclei. bioRxiv 244038 (2018). doi: 10.1101/244038 [DOI] [Google Scholar]
- 26.Rao SSP et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biswas U, Hempel K, Llano E, Pendas A & Jessberger R Distinct Roles of Meiosis-Specific Cohesin Complexes in Mammalian Spermatogenesis. PLoS Genet 12, e1006389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller H et al. Characterizing meiotic chromosomes’ structure and pairing using a designer sequence optimized for Hi-C. Mol. Syst. Biol 14, e8293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schalbetter SA, Fudenberg G, Baxter J, Pollard KS & Neale MJ Principles of Meiotic Chromosome Assembly. bioRxiv 442038 (2018). doi: 10.1101/442038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun X et al. Transcription dynamically patterns the meiotic chromosome-axis interface. Elife 4, 8522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margolin G, Khil PP, Kim J, Bellani MA & Camerini-Otero RD Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics 15, 39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vagin VV et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Siomi MC, Sato K, Pezic D & Aravin AA PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12, 246–258 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Buckley MS & Lis JT Imaging RNA Polymerase II transcription sites in living cells. Curr Opin Genet Dev 25, 126–130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hnisz D, Shrinivas K, Young RA, Chakraborty AK & Sharp PA A Phase Separation Model for Transcriptional Control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chong S et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabari BR et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho W-K et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modzelewski AJ et al. Dgcr8 and Dicer are essential for sex chromosome integrity during meiosis in males. J Cell Sci 128, 2314–2327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page J et al. Inactivation or non-reactivation: what accounts better for the silence of sex chromosomes during mammalian male meiosis? Chromosoma 121, 307–326 (2012). [DOI] [PubMed] [Google Scholar]
- 41.de Vries M et al. Human male meiotic sex chromosome inactivation. PLoS ONE 7, e31485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner JMA et al. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr. Biol 14, 2135–2142 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Flyamer IM et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marsden MP & Laemmli UK Metaphase chromosome structure: evidence for a radial loop model. Cell 17, 849–858 (1979). [DOI] [PubMed] [Google Scholar]
- 45.Liang Z et al. Chromosomes Progress to Metaphase in Multiple Discrete Steps via Global Compaction/Expansion Cycles. Cell 161, 1124–1137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novak I et al. Cohesin Smc1beta determines meiotic chromatin axis loop organization. J Cell Biol 180, 83–90 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Revenkova E et al. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol 6, 555–562 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Yuan L et al. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science 296, 1115–1118 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Gassler J et al. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J 36, 3600–3618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan J et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerton JL et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA 97, 11383–11390 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baudat F & Nicolas A Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci USA 94, 5213–5218 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paigen K & Petkov PM PRDM9 and Its Role in Genetic Recombination. Trends Genet 34, 291–300 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brick K, Smagulova F, Khil P, Camerini-Otero RD & Petukhova GV Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plug AW, Xu J, Reddy G, Golub EI & Ashley T Presynaptic association of Rad51 protein with selected sites in meiotic chromatin. Proc Natl Acad Sci USA 93, 5920–5924 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holmquist GP Chromosome bands, their chromatin flavors, and their functional features. Am. J. Hum. Genet 51, 17–37 (1992). [PMC free article] [PubMed] [Google Scholar]
- 57.Walker M et al. Affinity-seq detects genome-wide PRDM9 binding sites and reveals the impact of prior chromatin modifications on mammalian recombination hotspot usage. Epigenetics Chromatin 8, 31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lange J et al. The Landscape of Mouse Meiotic Double-Strand Break Formation, Processing, and Repair. Cell 167, 695–708.e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smagulova F, Brick K, Pu Y, Camerini-Otero RD & Petukhova GV The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev. 30, 266–280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baker CL et al. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet 11, e1004916 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu EY et al. High-resolution sex-specific linkage maps of the mouse reveal polarized distribution of crossovers in male germline. Genetics 197, 91–106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turner JMA Meiotic Silencing in Mammals. Annu Rev Genet 49, 395–412 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Turner JMA et al. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 37, 41–47 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Baarends WM et al. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol Cell Biol 25, 1041–1053 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Homolka D, Ivanek R, Capkova J, Jansa P & Forejt J Chromosomal rearrangement interferes with meiotic X chromosome inactivation. Genome Res 17, 1431–1437 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKee BD & Handel MA Sex chromosomes, recombination, and chromatin conformation. Chromosoma 102, 71–80 (1993). [DOI] [PubMed] [Google Scholar]
- 67.Blat Y & Kleckner N Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98, 249–259 (1999). [DOI] [PubMed] [Google Scholar]
- 68.Zhang L et al. Topoisomerase II mediates meiotic crossover interference. Nature 511, 551–556 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li XZ et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50, 67–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hogarth CA & Griswold MD The key role of vitamin A in spermatogenesis. J. Clin. Invest 120, 956–962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dixon JR et al. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van de Geijn B, McVicker G, Gilad Y & Pritchard JK WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat Methods 12, 1061–1063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keane TM et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Durand NC et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Durand NC et al. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst 3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Knight PA & Ruiz D A fast algorithm for matrix balancing. IMA J Numer Anal 33, 1029–1047 (2013). [Google Scholar]
- 79.Thorvaldsdóttir H, Robinson JT & Mesirov JP Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinformatics 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gel B et al. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32, 289–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rosenkranz D piRNA cluster database: a web resource for piRNA producing loci. Nucleic Acids Res 44, D223–30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rosenkranz D & Zischler H proTRAC--a software for probabilistic piRNA cluster detection, visualization and analysis. BMC Bioinformatics 13, 5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Imakaev M et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods 9, 999–1003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.