Abstract
Aberrant activation of the Wnt signal transduction pathway triggers the development of colorectal cancer. The ADP-ribose polymerase Tankyrase (TNKS) mediates proteolysis of Axin—a negative regulator of Wnt signaling—and provides a promising therapeutic target for Wnt-driven diseases. Proteolysis of TNKS substrates is mediated through their ubiquitination by the poly-ADP-ribose (pADPr)-dependent RING-domain E3 ubiquitin ligase RNF146/Iduna. Like TNKS, RNF146 promotes Axin proteolysis and Wnt pathway activation in some cultured cell lines, but in contrast with TNKS, RNF146 is dispensable for Axin degradation in colorectal carcinoma cells. Thus, the contexts in which RNF146 is essential for TNKS-mediated Axin destabilization and Wnt signaling remain uncertain. Herein, we tested the requirement for RNF146 in TNKS-mediated Axin proteolysis and Wnt pathway activation in a range of in vivo settings. Using null mutants in Drosophila, we provide genetic and biochemical evidence that Rnf146 and Tnks function in the same proteolysis pathway in vivo. Furthermore, like Tnks, Drosophila Rnf146 promotes Wingless signaling in multiple developmental contexts by buffering Axin levels to ensure they remain below the threshold at which Wingless signaling is inhibited. However, in contrast with Tnks, Rnf146 is dispensable for Wingless target gene activation and the Wingless-dependent control of intestinal stem cell proliferation in the adult midgut during homeostasis. Together, these findings demonstrate that the requirement for Rnf146 in Tnks-mediated Axin proteolysis and Wingless pathway activation is dependent on physiological context, and suggest that, in some cell types, functionally redundant pADPr-dependent E3 ligases or other compensatory mechanisms promote the Tnks-dependent proteolysis of Axin in both mammalian and Drosophila cells.
Keywords: ADP-ribosylation, Axin, beta-catenin, Iduna, intestinal stem cell, midgut, RNF146, Tankyrase, Wingless, Wnt
THE Wnt/Wingless signal transduction pathway directs fundamental processes during metazoan development (Nusse and Clevers 2017) . Inhibition of Wnt signaling underlies numerous birth defects, and inappropriate activation of Wnt signaling is associated with several cancers, including the vast majority of colorectal cancers. Thus, understanding the basic mechanisms that underlie activation of the Wnt pathway will facilitate the design of innovative strategies for the treatment of several human diseases. Regulation of the stability of β-catenin/Armadillo, the central transcriptional activator in the pathway, dictates the level of Wnt pathway activity. A “destruction complex,” which includes the concentration-limiting scaffold protein Axin, the tumor suppressor Adenomatous polyposis coli (APC), and glycogen synthase kinase 3, targets β-catenin for proteasomal degradation. Wnt stimulation inhibits the destruction complex and induces the recruitment of Axin to an activation complex termed the “signalosome,” thus stabilizing β-catenin and promoting activation of the pathway. The regulation of Axin holds significant clinical relevance, as the level of Wnt signaling is modulated by Axin activity (Liu and He 2010).
Therapeutic agents that selectively target the Wnt pathway are a priority for the field, but have yet to enter the clinic (Lum and Clevers 2012). A promising lead was suggested by previous small molecule screens that identified Tankyrase (TNKS)—the enzyme that mediates the poly-ADP-ribosylation of Axin—as a novel therapeutic target for Wnt-driven diseases (Huang et al. 2009). Poly-ADP-ribosylation mediated by TNKS promotes Wnt signaling by targeting Axin for proteasomal degradation, and thereby stabilizing β-catenin. TNKS inhibitors impede the Wnt-dependent proliferation of cultured cells, and, in mice with conditional targeted deletion of Apc, TNKS inhibitors disrupt the growth of colonic adenomas, raising the possibility that these agents hold clinical promise (Waaler et al. 2012; Lau et al. 2013).
In mammalian cultured cells, the targeting of Axin, and of TNKS itself, for proteasomal degradation through TNKS-dependent poly-ADP-ribosylation requires their subsequent ubiquitination by the poly-ADP-ribose (pADPr)-dependent RING-domain E3 ubiquitin ligase RNF146/Iduna (Callow et al. 2011; Kang et al. 2011; Zhang et al. 2011; DaRosa et al. 2015). Furthermore, RNF146 also promotes Axin degradation in Xenopus embryos (Zhu et al. 2018). Thus, in principle, RNF146 could potentially provide another therapeutic target for Wnt-driven cancer. However, in contrast with the effects of TNKS inhibition, depletion of RNF146 neither stabilized Axin nor inhibited the transcriptional activation of Wnt target genes in colorectal carcinoma cell lines harboring truncations in APC (Callow et al. 2011). These findings raised the question of whether RNF146 is indeed essential for all TNKS-mediated Axin degradation in vivo, and whether RNF146 is a viable therapeutic target for Wnt-driven disease. An in vivo mouse model for RNF146 inactivation to address this question has not yet been reported.
Herein, we sought to test the extent to which RNF146 is essential for TNKS-mediated Axin proteolysis and Wnt signaling in a range of in vivo contexts. We built upon a previously established genetic model that demonstrated evolutionary conservation in Tnks function in Drosophila. These studies revealed that Tnks-mediated ADP-ribosylation targets Axin for proteolysis under basal conditions throughout development, and that the in vivo requirement for Tnks is context-dependent (Wang et al. 2016b,c). Specifically, in the adult Drosophila intestine, where gradients of Wingless signaling exist at high levels at each compartment boundary, and decrease as a function of distance from these boundaries (Buchon et al. 2013; Tian et al. 2016), Tnks is essential for transcriptional activation of target genes in regions where Wingless is present at low concentration and controls the Wingless-dependent regulation of intestinal stem cell (ISC) proliferation (Tian et al. 2016; Wang et al. 2016c). Furthermore, Tnks also serves to buffer Axin activity in other in vivo contexts, by ensuring that Axin levels remain below the threshold at which Wingless pathway activation is inhibited (Wang et al. 2016b; Yang et al. 2016). For example, Tnks is required for the Wingless-dependent specification of cell fate in the embryonic epidermis when endogenous Axin levels are increased by only twofold (Yang et al. 2016), and also serves this function in Wingless-dependent cell fate specification in the larval wing imaginal disc and in the pupal abdomen.
In this report, we demonstrate that Drosophila Rnf146/Iduna mediates the pADPr-dependent degradation of Tnks substrates, including Axin and Tnks itself, under basal conditions throughout development. We provide genetic and biochemical evidence that Tnks and Rnf146 function in the same pADPr-dependent proteolytic pathway, indicating that RNF146 function is evolutionarily conserved. Furthermore, like Tnks, Rnf146 promotes Wingless signaling in multiple in vivo contexts by buffering Axin levels such that they remain below the threshold that inhibits Wingless signaling. Surprisingly, however, and in contrast to Tnks, Rnf146 is dispensable in the adult midgut for both promoting Wingless target gene activation and for regulating the Wingless-dependent control of ISC proliferation. Together, these findings reveal a context-dependent role for RNF146 in Tnks-mediated Axin proteolysis and Wingless signaling in vivo, and support the hypothesis that, in specific contexts, functionally redundant pADPr-dependent E3 ligase(s) may mediate the proteolysis of Axin not only in mammalian cells, but also in Drosophila cells.
Materials and Methods
Drosophila stocks and transgenes
To generate deletions in Rnf146, P{Δ2-3}99B was used to mobilize the P element EY09040 (Bloomington Drosophila Stock Center, BDSC) (Bellen et al. 2004), which is inserted in the first intron of Rnf146. By PCR screening, seven lines with deletions in Rnf146 were identified, including Rnf14636 and Rnf146157. Rnf14636 has a deletion of 5970 nucleotides (+774 to +6743 with reference to the Rnf146 transcriptional start site); 14 nucleotides from EY09040 remain at this site. Rnf146157 has a deletion of 3540 nucleotides (+774 to +4313 with reference to the Rnf146 transcriptional start site). Approximately 1.2K nucleotides from EY09040 remain at this site. The Rnf146 mutant alleles were recombined with FRT2A for clonal analysis.
Other stocks:
Tnks19 (Wang et al. 2016c), Tnks503 (Wang et al. 2016c), C765-Gal4 (BDSC) (Brand and Perrimon 1993), 71B-Gal4 (BDSC) (Brand and Perrimon 1993), UAS Axin-V5 (Yang et al. 2016), BAC Axin-V5 integrated at the VK30 site (PBac{y[+]-attP-9A}VK00030) (Gerlach et al. 2014), UAS-AxinΔTBD-V5, UAS-AxinΔRGS-V5, UAS-AxinΔArm-V5, UAS-AxinΔPP2A-V5, UAS-AxinΔDIX-V5 were integrated at the attP33 site (Tacchelly-Benites et al. 2018), esg-Gal4 UAS-GFP (Micchelli and Perrimon 2006), fz3-RFP (Olson et al. 2011), FRT82B pygoS123 (Thompson et al. 2002); hsFLP1 (Golic and Lindquist 1989), FRT82B arm-lacZ (Vincent et al. 1994) (provided by J. Treisman, Skirball Institute, New York, NY), and FRT2A ubi-GFPnls (BDSC#5825).
MARCM lines:
MARCM 82B: hsflp, UAS-mCD8:GFP; tub-Gal4, FRT82B, tub-Gal80 (Li et al. 2013).
MARCM 2A: y w hs-flp; tub-Gal4 UAS-mCD8::GFPLL5/CyO act-GFPJMR1; FRT2A tub-Gal80LL9I (Jiang and Reichert 2012).
Canton-S flies were used as wild-type controls. All crosses were performed at 25° unless otherwise indicated.
Clonal analysis
Mitotic mutant clones were generated by FLP-mediated recombination (Xu and Rubin 1993) in the larval wing imaginal discs using hsFLP1. Clones were induced by subjecting first and second instar larvae to a 37° heat shock for 2 hr, and were detected by the loss of expression of an arm-lacZ or ubi-GFP transgene in third-instar larval wing imaginal discs.
Genotypes for generating mitotic clones in the wing were as follows:
Tnks mutant wing disc clones expressing Axin-V5 or Axin-V5 deletion mutants with the 71B driver: hsFLP1/+; UASAxin-V5/+; FRT82B Tnks19/71B-Gal4 FRT82B arm-lacZ.
Rnf146 mutant wing disc clones expressing Axin-V5 or Axin-V5 deletion mutants with the C765 driver: hsFLP1/+; UAS-Axin-V5/+; Rnf146 FRT2A, C765-Gal4/ubi-GFP FRT2A.
Mitotic clones were generated in the intestine using the MARCM system (Lee and Luo 2001). For analysis of fz3-RFP expression, clones were induced in third-instar larvae by a single 2 hr heat shock and examined at 1–2 days after eclosion, as fz3-RFP expression becomes less homogenous with age.
Antibodies
The primary antibodies used for immunostaining were mouse anti-V5 (1:5000; Invitrogen), mouse anti-Wingless (1:200, 4D4 concentrated antibody; Developmental Studies Hybridoma Bank, DSHB), guinea pig anti-Senseless (1:1000) (Nolo et al. 2000), rabbit anti-β-gal (1:1000; MP Biomedicals), mouse anti-pADPr (1:1000, clone 10H; Tulip Biolabs), rabbit anti-GFP (1:200; Invitrogen). The primary antibodies used for immunoblotting were guinea pig anti-Axin (1:1000) (Wang et al. 2016c), guinea pig anti-Tnks (1:1000) (Wang et al. 2016c), rabbit anti-Kinesin Heavy Chain (1:10,000; Cytoskeleton), rabbit anti-Gluthathione-S-Transferase (1:10,000; Invitrogen) and rabbit anti-pADPr 96-10 (1:10,000) (Affar et al. 1999).
The secondary antibodies used for immunostaining were goat or donkey Alexa Fluor 488 or 555 conjugates (1:400; Invitrogen). The secondary antibodies used for immunoblotting were: goat anti-rabbit HRP conjugate (1:10,000; Bio-Rad), goat anti-mouse HRP conjugate (1:10,000; Bio-Rad), and goat anti-guinea pig HRP conjugate (1:10,000; Jackson ImmunoResearch).
Immunostaining and immunoblotting
For immunostaining, third-instar larval wing imaginal discs were dissected in PBS, fixed in 4% paraformaldehyde in PBS for 20 min, and washed with PBS with 0.1% Triton X-100, followed by incubation in PBS with 0.5% Triton X-100 and 10% BSA for 1 hr at room temperature. Incubation with primary antibodies was performed at 4° overnight in PBS with 0.5% Triton X-100. Incubation with secondary antibodies was for 2 hr at room temperature. Fluorescent images were obtained on a Nikon A1RSi confocal microscope and processed using Adobe Photoshop software.
For larval lysates, third-instar larvae were dissected to remove salivary glands, fat body, and gut tissues in cold PBS. After removal of PBS, 4× Laemmli loading buffer supplemented with 1 M DTT was added and the lysates were vortexed briefly. For lysates from embryos, embryos were lysed in lysis buffer [50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1% NP-40, 10% glycerol, 1.5 mM EDTA (pH 8.0)], supplemented with phosphatase and protease inhibitor cocktail (1:100; Thermo Scientific) and 1 μM of the poly (ADP-ribose) glycohydrolase inhibitor ADP-HPD (Enzo Life Sciences). All lysates were incubated for 5 min at 100° before SDS-PAGE analysis. For detection of pADPr in immunoblots: third-instar larvae of the indicated phenotypes were dissected in cold 1× PBS supplemented with protease inhibitor cocktail ProteaseArrest (1:100; GBiosciences) and 1 μM of the poly(ADP-ribose) glycohydrolase inhibitor ADP-HPD (Enzo Life Sciences). Tissues were lysed as described above and samples were resolved by SDS-PAGE using 4–15% gradient gels (Bio-Rad). Quantification of immunoblots was performed with ImageJ (Wayne Rasband, National Institutes of Health).
Quantification and statistics
Progenitor cells were identified by esg > GFP. For quantification, the number of total progenitor cells was counted in a field of 0.032 mm2 within the R5a midgut region (Buchon et al. 2013). For quantification of pH 3-positive cells, the total number of pH 3+ cells in the posterior midgut of the indicated genotypes was counted. All t-tests were performed using Prism (GraphPad Software).
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article and figures. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7523393.
Results
Drosophila Rnf146/Iduna targets Tnks substrates for degradation in vivo
In vitro studies have revealed that human RNF146/Iduna, a pADPr-dependent RING-domain E3 ubiquitin ligase, promotes Wnt signaling in some cultured cell lines by targeting Axin for degradation, whereas in colonic carcinoma cells, RNF146 is dispensable for this process (Callow et al. 2011; Zhang et al. 2011). We sought to determine whether RNF146 is essential for Wnt signaling in distinct in vivo contexts. We hypothesized that, if the ability of Tnks to promote Wingless signaling is fully dependent on RNF146, the in vivo requirements for Rnf146 would be similar to those of Tnks. The single Drosophila Rnf146 homolog shares sequence homology with mammalian RNF146 in their two major domains: the RING domain, which is predicted to interact with an E2 ubiquitin conjugating enzyme, and the poly-ADP-ribose (pADPr)-binding WWE domain (Figure 1A). Indeed, all amino acids within the human RNF146 WWE domain that bind pADPr (Wang et al. 2012) are identical in Drosophila Rnf146 (Figure 1A).
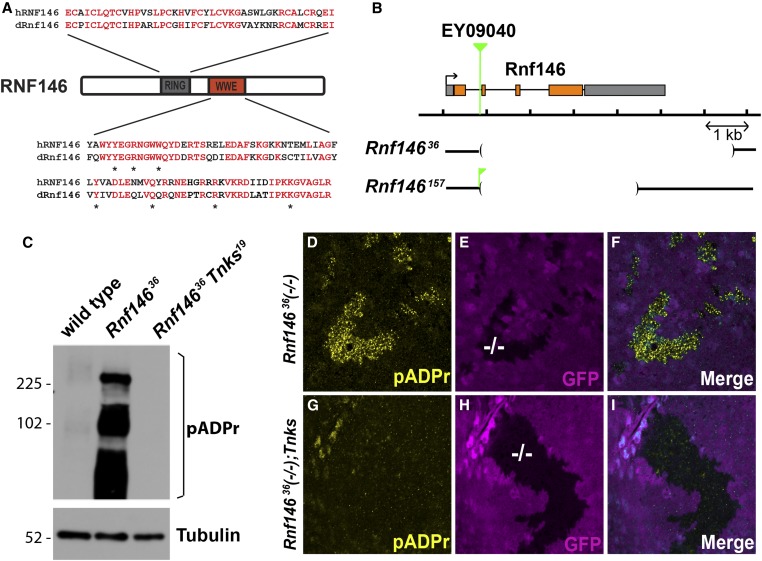
Figure 1.
Drosophila Rnf146/Iduna targets Tnks substrates for degradation in vivo. (A) Schematic representation of the domain structure of RNF146. Residues in human RNF146 and the Drosophila homolog that are identical are shown in red. All residues in the human RNF146 WWE domain that bind poly-ADP-ribose are identical in Drosophila Rnf146 and indicated with asterisks. (B) Schematic representation of Drosophila Rnf146 genomic region and deletions in two Rnf146 mutants. Insertion site of the P element EY09040 is indicated (green). A fragment of the P element EY09040 remains in the Rnf146157 mutant. (C) Lysates from wild-type and Rnf146 mutant larvae were analyzed by immunoblot using pADPr antibody. Tubulin was used as a loading control. The levels of poly-ADP-ribosylated proteins are increased in Rnf146 null mutant third-instar larvae, but revert to baseline in Rnf146 Tnks double null mutants. (D–I) Third-instar larval wing imaginal discs with Rnf14636 null mutant clones marked by the absence of GFP (−/−; magenta) were stained with indicated antibodies. Proteins modified by poly-ADP-ribose (D) accumulate cell autonomously in Rnf14636 mutant clones but revert to baseline upon concomitant inactivation of Tnks (Tnks19) (G–I). Blue is the merge of magenta and yellow.
To study Rnf146 function in vivo, we generated Drosophila Rnf146 mutants by inducing imprecise excision of a transposon inserted within the first intron of the Rnf146 gene. We isolated several deletions that eliminate most of the open reading frame, including Rnf14636 and Rnf146157 (Figure 1B). Like Tnks null mutants, Rnf146 null mutants (lacking both maternal and zygotic Rnf146) are viable and display no overt external patterning defects.
We tested the hypothesis that proteins normally targeted for pADPr-directed degradation would accumulate after their ADP-ribosylation in Rnf146 mutants using an antibody directed against pADPr in immunoblots. We found a marked increase in levels of proteins modified by pADPr in Rnf146 mutant larval lysates, but that these levels were restored to wild-type levels by concomitant inactivation of Rnf146 and Tnks in Rnf146 Tnks double null mutants (Figure 1C). These results support the conclusion that Tnks and Rnf146 function in the same pADPr-dependent proteolysis pathway in Drosophila. To determine whether the pADPr-dependent protein turnover mediated by Rnf146 is a cell autonomous function, we examined clones of Rnf146 null mutant cells in third-instar larval imaginal discs by immunostaining with a pADPr antibody. We observed an increase in pADPr staining intensity that is confined to all Rnf146 mutant cells in all discs examined and eliminated by concomitant inactivation of Tnks (Figure 1, D–I and Figure S1). These findings suggested that Drosophila Rnf146, like human RNF146, is a pADPr-directed ubiquitin ligase that promotes turnover of proteins ADP-ribosylated by Tnks, and that Rnf146 activity is cell autonomous and ubiquitous.
Rnf146 mediates the pADPr-directed proteolysis of Axin and Tnks in vivo
Given that human RNF146 regulates Wnt signaling in cultured human cells through Tnks-dependent Axin degradation, and that human RNF146 promotes the turnover of both Tnks and Axin, we sought to determine whether this activity is functionally conserved in Drosophila Rnf146. In comparison with wild type, Tnks levels are markedly increased in lysates from Rnf146 mutant larvae and embryos (Figure 2A and Figure S2A). Furthermore, immunostaining of third-instar larval imaginal discs with a Tnks antibody revealed a cell autonomous increase in Tnks staining in clones of Rnf146 null mutant cells (Figure2, C–E and Figure S2, B–D). These findings provide in vivo evidence that the essential role of RNF146 in Tnks degradation is evolutionarily conserved, demonstrate that this requirement for Rnf146 is cell-intrinsic, and indicate that Rnf146 destabilizes Tnks at multiple developmental stages.
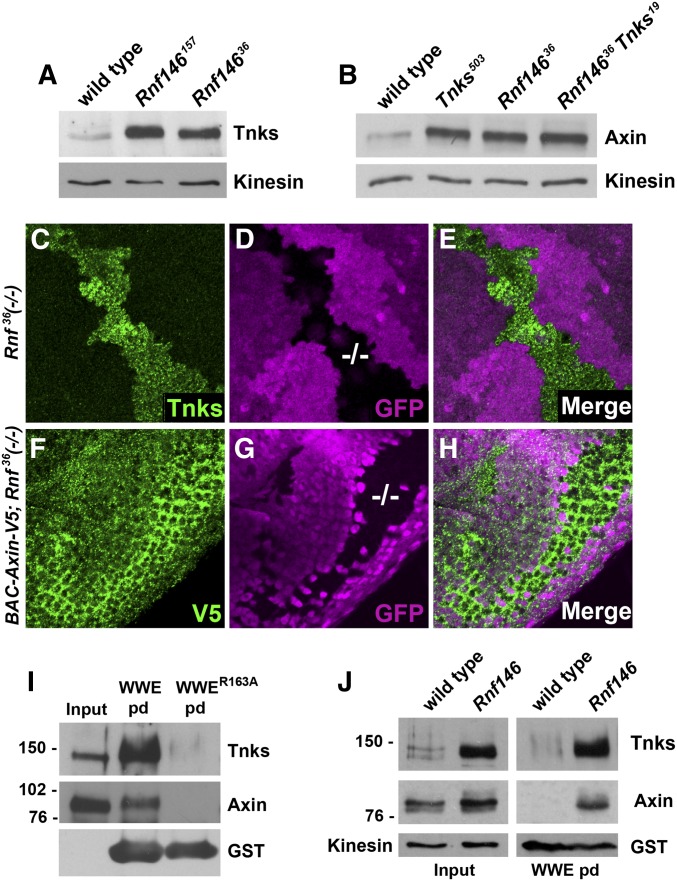
Figure 2.
Drosophila Rnf146 mediates ADP-ribose-directed destabilization of Axin and Tnks in vivo. (A) Immunoblot of lysates from wild-type and Rnf146 null mutant larvae probed with Tnks and Axin antibody. Tnks and Axin protein levels are increased in Rnf146 mutant larvae. Kinesin was used as a loading control. (B) Immunoblot of lysates from wild type, Tnks null mutant, Rnf146 null mutant, and Rnf146 Tnks double null mutant larvae probed with Axin antibody. Axin protein levels are increased in Tnks and Rnf146 mutants, but not further increased in Rnf146 Tnks double mutants. Kinesin was used as a loading control. (C–E) Third-instar larval wing imaginal discs with Rnf14636 null mutant clones marked by the absence of GFP (−/−; magenta) were stained with indicated antibodies. Tnks accumulates cell autonomously in Rnf14636 mutant clones. (F–H) Eye imaginal discs from third-instar larvae expressing a BAC Axin-V5 transgene with Rnf146 null mutant clones were stained with indicated antibodies. Rnf146 mutant clones were marked by the absence of GFP (−/−; magenta). (I) Detection of poly-ADP-ribosylated Tnks and ADP-ribosylated Axin using the GST-WWE pulldown assay. (J) poly-ADP-ribosylated Tnks and poly-ADP-ribosylated Axin accumulated in Rnf146 null mutants.
To determine whether Drosophila Rnf146 also promotes the turnover of Axin in vivo, we examined lysates from Rnf146 mutant larvae in immunoblots with an Axin antibody. By comparison with wild type, Axin levels are increased in Rnf146 mutant larvae and embryos (Figure 2B and Figure S2A). The magnitude of this increase in Axin levels is comparable upon Tnks loss, Rnf146 loss, or concomitant loss of Tnks and Rnf146, as revealed in lysates from Rnf146 Tnks double mutant larvae (Figure 2B). These in vivo findings also support the conclusion that Tnks and Rnf146 function in the same pADPr-dependent degradation pathway. Importantly, Rnf146 targets Axin for degradation even prior to 3 hr of embryonic development (Figure S2A), when wingless expression is first detectable. Therefore, like Tnks, Rnf146 also targets Axin for proteolysis independently of Wingless pathway activation. To determine whether Rnf146 promotes Axin degradation in a cell autonomous manner, we generated Rnf146 mutant clones in transgenic flies that express a V5-tagged Axin under the control of its endogenous promoter/enhancer (BAC Axin-V5) (Gerlach et al. 2014). As anticipated, Axin-V5 levels were increased in a cell-autonomous manner within Rnf146 mutant clones (Figure 2, F–H). Together, these findings indicate that Drosophila Rnf146 is required for the turnover of both Tnks and Axin at multiple developmental stages, and that human and Drosophila Rnf146 share evolutionarily conserved roles in Tnks-dependent Axin destabilization.
We hypothesized that the Tnks and Axin that accumulates in Rnf146 mutants would be ADP-ribosylated. To test this hypothesis, we performed pull down assays with the Trp-Trp-Glu (WWE) domain of Rnf146, which binds robustly to pADPr in Tnks substrates (Zhang et al. 2011; Wang et al. 2012). We previously performed three distinct controls that verified the specificity of the GST-WWE pull downs for detection of Tnks-dependent Axin ADP-ribosylation in vivo: Axin pull down was abolished in Tnks null mutants (Wang et al. 2016a; Yang et al. 2016), or when the Tnks binding domain within Axin was deleted (Yang et al. 2016), or by the GST-WWER164A negative control (Yang et al. 2016), in which an arginine-to-alanine substitution in the RNF146 WWE domain abolishes interaction with poly-ADP-ribose (Zhang et al. 2011). In lysates from Rnf146 mutant larvae, ADP-ribosylated Tnks and Axin were pulled down by WWE, but not the WWER163A mutant control (Figure 2I). Mobility shifts in ADP-ribosylated Axin were small, consistent with previous findings that ADP-ribosylation does not alter the electrophoretic mobility of Axin (Huang et al. 2009; Zhang et al. 2011) (Figure 2I). To determine the extent to which ADP-ribosylated Axin accumulates in Rnf146 mutants, we performed WWE pulldowns with lysates from wild-type vs. Rnf146 mutant larvae. ADP-ribosylated Axin was not detected in wild-type larvae, indicating that ADP-ribosylated Axin is turned over rapidly in vivo. By comparison, ADP-ribosylated Axin levels were markedly increased in Rnf146 mutants, despite the fact that elimination of Rnf146 resulted in only a moderate increase in total levels of Axin protein (Figure 2J). Taken together, these data provide evidence that like human RNF146, Drosophila Rnf146 targets both ADP-ribosylated Tnks and ADP-ribosylated Axin for degradation in vivo.
Rnf146 promotes Wingless signaling by buffering Axin activity
Tnks inactivation increases Axin levels by only twofold to threefold, which is below the threshold at which Axin levels inhibit activation of the Wingless pathway. However, an increase in Axin to levels that do not inhibit Wingless signaling in wild type are sufficient to inhibit Wingless-dependent cell fate specification in Tnks mutants, suggesting that Tnks serves to buffer Axin activity (Wang et al. 2016b). As our findings indicated that Tnks and Rnf146 likely function in the same proteolysis pathway, we postulated that Rnf146 would also promote Wingless signaling by maintaining Axin levels below this in vivo threshold. To test this hypothesis, we first expressed an Axin-V5 transgene near physiological levels using the 71B-Gal4 driver, which drives expression in the dorsal and ventral regions of the pouch of third-instar larval wing imaginal discs (Wang et al. 2016b). As demonstrated previously, under these conditions, the expression of both wingless and the Wingless target gene senseless at the dorsoventral boundary of the third-instar larval wing imaginal discs are indistinguishable from wild type (Figure 3, A and B), as is patterning of the adult wing margin (Figure 3, C and D). However, under the same conditions, inactivation of Rnf146 resulted in the loss of senseless expression and aberrant expansion of wingless expression at the dorsoventral boundary of larval wing discs, loss of sensory bristles at the adult wing margin, notched wings, and ectopic bristles within the wing blade (Figure 3, E–H, 100% penetrance, n = 27); each of these defects is indicative of the inhibition of Wingless signaling. These findings indicate that increases in Axin levels that are within the physiological threshold in wild-type flies inhibited Wingless signaling if Rnf146 activity is lost.
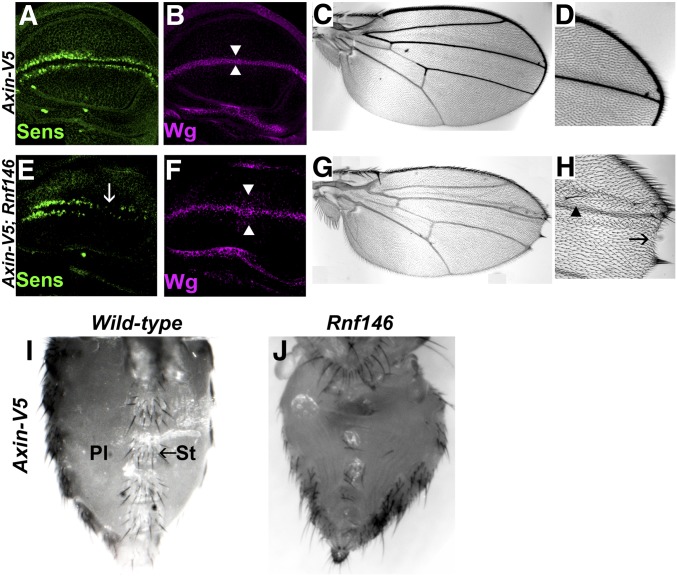
Figure 3.
Rnf146 prevents the accumulation of excess Axin levels and promotes Wingless signaling. (A and B) Confocal images of third-instar larval wing imaginal discs expressing Axin-V5 with the 71B-Gal4 driver, which drives expression in the dorsal and ventral regions of the pouch of third-instar larval wing imaginal discs. Expression of Axin-V5 does not disrupt the Wingless target gene senseless (A), wingless expression (B) or cell fate in the adult wing (C and D). (E–H) Expression of Axin-V5 in Rnf14636/Rnf146157 null mutants causes disruption of Senseless [(E), arrow] and a slight increase in the number of cells expressing Wingless [(F), arrowheads]. In the adult wing, expression of Axin-V5 in Rnf146 null mutants results in the loss of wing blade tissue (G), loss of sensory bristles at the wing margin [(H), arrow] and ectopic bristles in the wing blade [(H), arrowhead]; 100% of Rnf146 mutants had wing margin defects, n = 13. (I) Ventral abdomen of wild-type adult female expressing Axin-V5 with the 71B-Gal4 driver. Sternites and sternal bristles (St) marked by arrow; pleura (Pl). Some sternites and sternal bristles are lost in Rnf14636/Rnf146157 mutants (J), indicating that Rnf146 promotes Wingless-dependent cell fate specification.
To determine if Rnf146 regulates Wingless signaling in other physiological contexts, we examined a different developmental stage and tissue. As demonstrated previously, when an Axin-V5 transgene is expressed in the pupal abdomen with the 71B-Gal4 driver, Wingless-dependent cell fate specification is indistinguishable from wild-type, as revealed by the size, morphology, spacing, and number of sternites in adults (Wang et al. 2016b) (Figure 3I). In contrast, under the same conditions, the abdomens of Rnf146 mutant adults displayed a reduced number of sternites and sternal bristles, a decreased size in the sternites that remained, and expansion of the pleura (Figure 3J, 100% penetrance, n = 27). These phenotypes indicate the loss of Wingless signaling, as observed previously in wingless mutants, or upon inactivation of the Wingless pathway transcriptional activators dTCF/Pangolin and Legless/BCL9 (Baker 1988; Brunner et al. 1997; Kramps et al. 2002). Thus, as observed in the wing, increases in Axin transcription that are compatible with normal development in the wild-type abdomen inhibit Wingless-dependent developmental processes upon Rnf146 inactivation. These findings suggested that by targeting ADP-ribosylated Axin for degradation, Rnf146 serves to buffer Axin activity in multiple in vivo contexts.
An in vivo analysis of domains required for Tnks- and Rnf146-dependent degradation of Axin
To further test the hypothesis that Tnks and Rnf146 act in the same pathway to mediate Axin proteolysis, we performed a structure-function analysis to compare the domains required for Axin degradation by Tnks or Rnf146. We previously generated a series of Axin transgenes with deletions in domains required for Axin’s interaction with other Wingless pathway components, including Tnks (AxinΔTBD-V5), the tumor suppressor Apc (AxinΔRGS-V5), the transcriptional activator Armadillo (AxinΔArm-V5), the phosphatase PP2A (AxinΔPP2A-V5), and the signalosome component Dishevelled (AxinΔDIX-V5) (Tacchelly-Benites et al. 2018). As demonstrated previously, immunostaining of wild-type Axin-V5 expressed in larval wing discs using the 71B-Gal4 driver, which drives expression in the dorsal and ventral wing pouch, revealed a marked increase in Axin-V5 levels in Tnks mutant clones (Figure 4, A–C) (Wang et al. 2016b), indicating that V5-tagged Axin is subject to Tnks-dependent degradation in vivo. A similarly marked increase in Axin-V5 levels was present in Rnf146 null mutant clones (Figure 4, G–I). Consistent with previous work (Wang et al. 2016b), deletion of the TBD, but not the RGS domain, abolished the ability of Tnks to promote Axin degradation (Figure 4, D–F and Figure S3, A–C). Similarly, deletion of the TBD, but not the RGS domain, also abolished the ability of Rnf146 to promote Axin degradation (Figure 4, J–L and Figure S4, A–C). Together, these in vivo results support that hypothesis that Tnks and Rnf146 act in the same Axin proteolysis pathway.
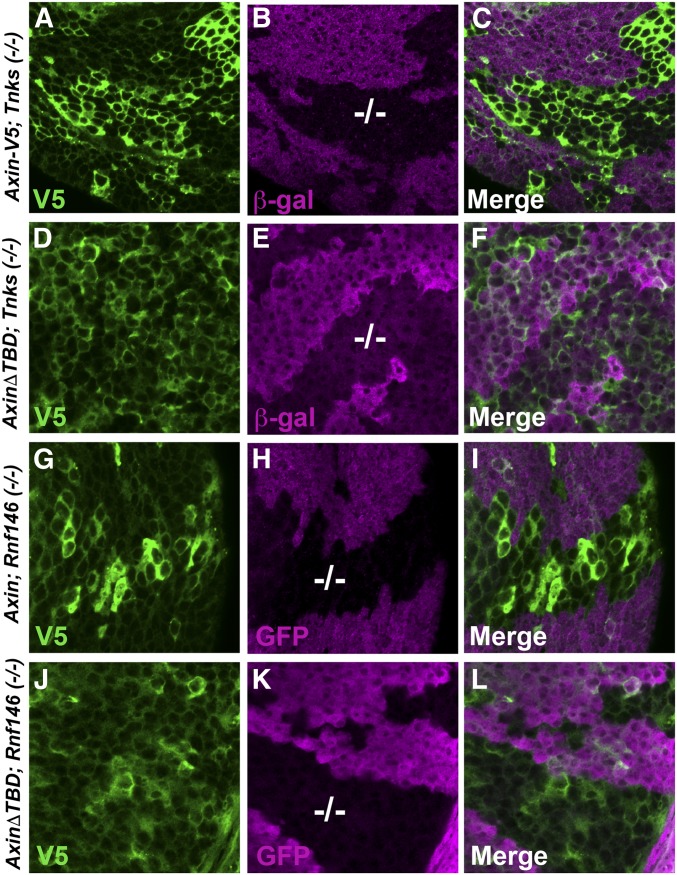
Figure 4.
The Tankyrase binding domain in Axin is required for Tnks- and Rnf146-dependent Axin degradation. Confocal images of third-instar larval wing imaginal discs expressing indicated Axin transgenes with the 71B-Gal4 driver, which drives expression in the dorsal and ventral regions of the pouch of third-instar larval wing imaginal discs. Axin-V5 or an Axin deletion mutant that lacks the Tankyrase binding domain (AxinΔTBD-V5) were stained with V5 antibody (green). Tnks mutant clones were marked by the lack of β-gal staining (−/−; magenta), and Rnf146 mutant clones were marked by the lack of GFP staining (−/−; magenta). The levels of wild-type Axin are increased in Tnks mutant clones (A–C) and in Rnf146 mutant clones (G–I) whereas the levels of AxinΔTBD are indistinguishable inside and outside both Tnks mutant clones (D–F) and Rnf146 mutant clones (J–L).
Similar to Axin-V5, the levels of AxinΔArm-V5 were increased in both Tnks mutant and Rnf146 mutant wing disc clones (Figures S3, D–F and S4, D–F), indicating that the Arm-binding domain is dispensable for Tnks-dependent Axin degradation. Analysis of AxinΔPP2A-V5 and AxinΔDIX-V5 in the same assay revealed some disruption of Axin regulation by Tnks or Rnf146 in the majority of wing disc clones. In many cells, AxinΔPP2A-V5 and AxinΔDIX-V5 were present at the same levels in Tnks null mutant clones or Rnf146 null mutant clones vs. in the neighboring cells outside these clones, suggesting loss of Axin destabilization in most clones (Figures S3, G–I and M–O and S4, G–I and M–O); however, we observed that, occasionally, the levels of AxinΔPP2A-V5 and AxinΔDIX-V5 were increased within some cells in Tnks mutant or Rnf146 mutant wing disc clones, suggesting that Tnks- and Rnf146-mediated degradation remained at least partially intact (Figures S3, J–L and P–R and S4, J–L and P–R). Together, our results indicate that the same domains are required for Axin degradation by Tnks or Rnf146, with not only the Tnks-binding domain, but also the PP2A-binding and DIX domains important in this process. These results provide further in vivo evidence for the existence of a Tnks-Rnf46 degradation pathway.
Rnf146 is dispensable for Wingless-dependent regulation of ISC proliferation
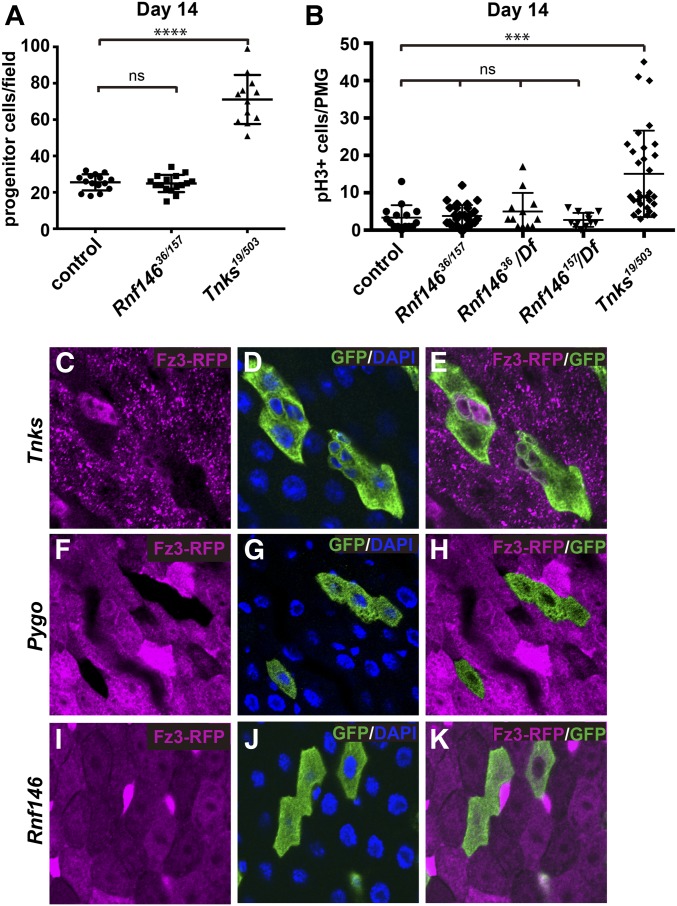
Tnks is dispensable for the majority of Wingless-dependent developmental processes under standard laboratory conditions, but is essential for the Wingless-dependent control of ISC proliferation during homeostasis in the adult midgut (Wang et al. 2016c). In this physiological context, the role of Tnks in targeting Axin for degradation is essential to regulate Wingless signaling and thus ISC proliferation. To determine whether Rnf146 similarly regulates ISC proliferation during homeostasis, we compared the number of midgut progenitor cells, which we identified by their expression of the transcription factor escargot (esg) (Micchelli and Perrimon 2006), in controls vs. Rnf146 mutants or Tnks mutants. In contrast with Tnks mutants, Rnf146 mutants did not exhibit an increased number of progenitor cells during homeostasis at 14 days after eclosion (Figure 5A and Figure S5, A–C). To directly compare the proliferation rate of ISCs, we identified dividing ISCs with phospho-histone H3 (pH3)—a marker for mitosis. Quantification indicated that in contrast with Tnks mutants, no significant difference in the ISC proliferation rate existed between controls and homozygous Rnf146 mutants (Figure 5B). To rule out genetic background or the accumulation of suppressors that could mask an increased ISC proliferation rate, we tested the Rnf146 null mutants in trans to a deficiency that deletes the entire Rnf146 locus. Analysis of these transheterozygotes also revealed no significant increase in the ISC proliferation rate (Figure 5B). Therefore, we conclude that, in contrast with Tnks, Rnf146 is not required to regulate ISC proliferation during homeostasis in the adult midgut, suggesting that Rnf146 is dispensable for the regulation of Axin and Wingless signaling in this context.
Figure 5.
Rnf146 is dispensable for Wingless-dependent regulation of ISC proliferation and Wingless target gene activation in the adult midgut. (A) Quantification of the relative number of progenitor cells in wild-type (n = 15), Rnf146 mutants (n = 16), or Tnks mutants (n = 12) at 14 days posteclosion. Values are presented as mean ± SD **** P < 0.0001 (t-test), ns: nonsignificant. (B) Quantification of pH 3-positive cells in posterior midguts (PMG) of wild-type, Rnf146 null mutants, or Tnks null mutants at 14 days post-eclosion. Values are presented as mean ± SD *** P < 0.001, (t-test), ns: nonsignificant. Numbers of flies examined for each genotype (from left to right) are 15, 27, 12, 12 and 31 respectively. (C–E) fz3-RFP expression in ECs is dependent on Wingless signaling activation. Fz3-RFP (magenta) is absent in Tnks19 null mutant clones that are at a distance from the midgut–hindgut boundary (marked by GFP). (F–H) Fz3-RFP (magenta) is absent in all pygoS123 null mutant clones (marked by GFP). (I–K) Rnf146 is dispensable for fz3-RFP expression in all regions examined, regardless of distance from the compartment boundary. Mutant clones (marked by GFP) were induced in third-instar larvae and examined 1–2 days after eclosion.
To further test this conclusion, we analyzed the effects of Rnf146 loss on the activation of Wingless target gene expression in the adult midgut. In the adult intestine, Wingless pathway activity peaks at major compartment boundaries, decreases as a function of distance from these boundaries, and is present at a low levels within compartments (Buchon et al. 2013; Tian et al. 2016). Previous work revealed that during adult midgut homeostasis, Tnks is critical for the activation of Wingless target gene expression in compartments, where Wingless pathway activity is relatively low, but dispensable for pathway activation at the compartment boundaries, where Wingless pathway activity is high (Wang et al. 2016c) (Figure 5, A–C). We sought to determine if Rnf146 similarly promotes Wingless target gene activation in the adult midgut by examining the expression of fz3-RFP, a Wingless target gene reporter (Sato et al. 1999; Sivasankaran et al. 2000). As reported previously, the expression of fz3-RFP in differentiated enterocytes (ECs) requires Wingless signaling (Tian et al. 2016; Wang et al. 2016c), and is completely lost upon inactivation of the essential Wingless signaling transcription cofactor Pygopus (Pygo) (Kramps et al. 2002; Parker et al. 2002; Thompson et al. 2002; Tian et al. 2016; Wang et al. 2016c) (Figure 5, F–H). However, fz3-RFP expression was unchanged in Rnf146 mutant clones, even within the compartment where Wingless signaling activity is relatively low (Figure 5, I–K). Taken together, these findings indicate that, in contrast with Tnks, Rnf146 is dispensable for regulating Wingless signaling and ISC proliferation in the adult midgut during homeostasis.
Discussion
In principle, as the pADPr-directed E3 ligase RNF146/Iduna is known to target Axin for TNKS-mediated proteasomal degradation in mammalian cells, it could provide an effective therapeutic target to inhibit Wnt signaling (Huang et al. 2009; Callow et al. 2011; Zhang et al. 2011). However, previous studies revealed that RNF146 is essential for this function in only some cultured cell lines, whereas in others, including colorectal cancer cells with truncations in APC, RNF146 is dispensable for Axin destabilization and Wnt signaling. Therefore, the extent to which RNF146 is required to regulate Wnt-dependent processes in vivo has remained unclear, as is the extent to which RNF146 could provide a viable therapeutic target for Wnt-driven diseases. Herein, we discovered that RNF146/Iduna function is conserved in Drosophila, and we evaluated the extent to which RNF146 is required for TNKS-dependent Wingless pathway regulation in vivo. Following the isolation of null mutants, we provide multiple lines of in vivo evidence that Drosophila Rnf146/Iduna mediates the pADPr-directed proteolysis of Tnks substrates. We demonstrate by both immunostaining and immunoblotting of null mutant larval imaginal discs that the level of pADPr-modified proteins is markedly increased in vivo following Rnf146 inactivation, but reverts to baseline upon concomitant inactivation of Tnks. These findings suggest that most, if not all Rnf146 substrates in larval imaginal discs require poly-ADP-ribosylation that is catalyzed specifically by Tnks, rather than by other ADP-ribose polymerases.
Our findings demonstrate that, as in cultured mammalian cells, Drosophila Axin and Tnks both serve as Rnf146 substrates in vivo. We show that following Rnf146 inactivation in larvae, both Axin and Tnks levels increase in vivo, with even larger relative increases in the levels of ADP-ribosylated Axin and ADP-ribosylated Tnks. Like Tnks, Rnf146 promotes Axin degradation ubiquitously and prior to the initiation of Wingless signaling, suggesting that Tnks and Rnf146 participate in a constitutive pathway for Axin proteolysis that acts under basal conditions but is necessary to promote signaling following Wingless stimulation. Our findings demonstrate that the same domains in Axin are required for Tnks- and Rnf146-dependent degradation, providing additional in vivo evidence supporting the conclusion that Tnks and Rnf146 function in the same pADPr-dependent pathway for Axin proteolysis. Although the ADP-ribosylation sites and the pADPr-dependent ubiquitination sites that target Axin for degradation are not yet known, our findings indicate that these sites are likely to be found in the domains that we identified herein as essential for Tnks- and Rnf146-mediated Axin degradation.
Our discovery that the function of mammalian RNF146 in Axin degradation is evolutionary conserved in Drosophila allowed us to investigate whether Rnf146 promotes Wingless signaling in vivo, and whether Tnks and Rnf146 possess identical roles in Wingless-dependent processes. We achieved this by comparing the consequences of their inactivation in vivo in multiple physiological contexts, and by determining whether there exist differences that may underlie their disparate requirements in Axin degradation vs. Wnt signaling in cultured mammalian cells. The threshold at which Axin levels inhibit Wingless signaling is between threefold and ninefold above the endogenous Axin level in wing discs (Peterson-Nedry et al. 2008; Wang et al. 2016b) and approximately fourfold to eightfold in embryos (Yang et al. 2016; Schaefer et al. 2018). Like inactivation of Tnks (Feng et al. 2014; Wang et al. 2016b), inactivation of Rnf146 has no overt effects on Wingless-dependent patterning of imaginal discs or the expression of Wingless target genes in larval wing discs at endogenous Axin levels. Herein, we demonstrate that an increase in Axin to levels that are still compatible with normal development in wild-type flies inhibit Wingless signaling in Rnf146 null mutants. These Wingless signaling defects in cell fate specification in larval wing imaginal discs and the pupal abdomen in Rnf146 mutants phenocopy Tnks mutants. Thus, Rnf146 is essential for the role of Tnks in preventing Axin accumulation above a threshold level, and, thereby, serves to buffer the negative regulation of Wingless signaling by Axin in several physiological contexts.
However, in one physiological context, the consequences of Tnks inactivation differ markedly from that of Rnf146 inactivation. During homeostasis in the adult intestine, the regulation of Axin by Tnks is essential for the Wingless-dependent control of ISC proliferation in the epithelium (Wang et al. 2016c). In this context, Tnks is essential for the expression of Wingless target genes in the midgut compartments, where Wingless pathway activity is relatively low, but dispensable at the compartment boundaries, where Wingless pathway activity is high. Tnks inactivation results in phenotypes that are also observed upon inactivation of positive regulatory Wingless pathway components, and results in ISC overproliferation driven by the nonautonomous activation of JAK/STAT signaling in the epithelium. Importantly, reduction in the gene dosage of Axin by one-half suppresses this overproliferation phenotype, providing evidence that aberrantly increased Axin levels underlie the deregulation of ISC proliferation in Tnks mutants. In sharp contrast with Tnks loss, inactivation of Rnf146 neither inhibits the transcriptional activation of a Wingless target gene nor results in increased ISC proliferation. Together, these findings indicate that in many, but not all, physiological contexts, Rnf146 serves as an essential E3 ubiquitin ligase for Tnks-mediated Axin proteolysis and thus for promoting Wingless signaling.
These results, together with previous studies in cultured mammalian cell lines, suggest that Rnf146 is dispensable for Tnks-dependent regulation of Wnt signaling in both Drosophila and mammalian intestinal cells. Our findings provide additional evidence to support the hypothesis that in specific cell types, functionally redundant pADPr-dependent E3 ligase(s) promote Tnks-dependent Axin proteolysis not only in mammalian cells (Callow et al. 2011), but also in Drosophila cells. Alternatively, other mechanisms may promote Axin degradation to compensate for the loss of Rnf146. If similar modes of Axin regulation exist in human cells, our results suggest that inhibition of RNF146 may indeed suffice to attenuate the aberrant activation of Wnt signaling in certain Wnt-driven disease contexts but not others, and that differences will likely be determined by the existence of underlying compensatory mechanisms for Axin proteolysis in specific cell types.
Acknowledgments
We thank Victoria Marlar for technical support, Ann Lavanway for microscopy advice, and the investigators listed in Materials and Methods for generously providing reagents. This work was funded by the National Institutes of Health (R01GM121421 and R01GM122222 to Y.A., P30CA023108 to the Norris Cotton Cancer Center, P40OD018537 to the BDSC), and the Canadian Institutes of Health Research (MO-178013 to G.G.P.).
Author contributions: Z.W., O.T.-B., and Y.A. conceived the study. Z.W., O.T.-B., G.P.N., M.K.J., and Y.A designed and performed the experiments. J.-P.G. and G.G.P. provided reagents and technical advice. Z.W., O.T.-B., and Y.A. wrote the article.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7523393.
Communicating editor: K. O’Connor-Giles
Literature Cited
- Affar E. B., Duriez P. J., Shah R. G., Winstall E., Germain M., et al. , 1999. Immunological determination and size characterization of poly(ADP-ribose) synthesized in vitro and in vivo. Biochim. Biophys. Acta 1428: 137–146. 10.1016/S0304-4165(99)00054-9 [DOI] [PubMed] [Google Scholar]
- Baker N. E., 1988. Transcription of the segment-polarity gene wingless in the imaginal discs of Drosophila, and the phenotype of a pupal-lethal wg mutation. Development 102: 489–497. [DOI] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., et al. , 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. 10.1534/genetics.104.026427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brunner E., Peter O., Schweizer L., Basler K., 1997. Pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature 385: 829–833. 10.1038/385829a0 [DOI] [PubMed] [Google Scholar]
- Buchon N., Osman D., David F. P., Fang H. Y., Boquete J. P., et al. , 2013. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 3: 1725–1738 (erratum: Cell Rep. 3: 1755) 10.1016/j.celrep.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Callow M. G., Tran H., Phu L., Lau T., Lee J., et al. , 2011. Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS One 6: e22595 10.1371/journal.pone.0022595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaRosa P. A., Wang Z., Jiang X., Pruneda J. N., Cong F., et al. , 2015. Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature 517: 223–226. 10.1038/nature13826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Li X., Ray L., Song H., Qu J., et al. , 2014. The Drosophila tankyrase regulates Wg signaling depending on the concentration of Daxin. Cell. Signal. 26: 1717–1724. 10.1016/j.cellsig.2014.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach J. P., Emmink B. L., Nojima H., Kranenburg O., Maurice M. M., 2014. Wnt signalling induces accumulation of phosphorylated beta-catenin in two distinct cytosolic complexes. Open Biol. 4: 140120 10.1098/rsob.140120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic K. G., Lindquist S., 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59: 499–509. 10.1016/0092-8674(89)90033-0 [DOI] [PubMed] [Google Scholar]
- Huang S. M., Mishina Y. M., Liu S., Cheung A., Stegmeier F., et al. , 2009. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461: 614–620. 10.1038/nature08356 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Reichert H., 2012. Programmed cell death in type II neuroblast lineages is required for central complex development in the Drosophila brain. Neural Dev. 7: 3 10.1186/1749-8104-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. C., Lee Y. I., Shin J. H., Andrabi S. A., Chi Z., et al. , 2011. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. USA 108: 14103–14108. 10.1073/pnas.1108799108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramps T., Peter O., Brunner E., Nellen D., Froesch B., et al. , 2002. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell 109: 47–60. 10.1016/S0092-8674(02)00679-7 [DOI] [PubMed] [Google Scholar]
- Lau T., Chan E., Callow M., Waaler J., Boggs J., et al. , 2013. A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res. 73: 3132–3144. 10.1158/0008-5472.CAN-12-4562 [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L., 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24: 251–254. 10.1016/S0166-2236(00)01791-4 [DOI] [PubMed] [Google Scholar]
- Li X., Erclik T., Bertet C., Chen Z., Voutev R., et al. , 2013. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 498: 456–462. 10.1038/nature12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., He X., 2010. Destruction of a destructor: a new avenue for cancer therapeutics targeting the Wnt pathway. J. Mol. Cell Biol. 2: 70–73. 10.1093/jmcb/mjp040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L., Clevers H., 2012. Cell biology. The unusual case of Porcupine. Science 337: 922–923. 10.1126/science.1228179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli C. A., Perrimon N., 2006. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439: 475–479. 10.1038/nature04371 [DOI] [PubMed] [Google Scholar]
- Nolo R., Abbott L. A., Bellen H. J., 2000. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102: 349–362. 10.1016/S0092-8674(00)00040-4 [DOI] [PubMed] [Google Scholar]
- Nusse R., Clevers H., 2017. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell 169: 985–999. 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- Olson E. R., Pancratov R., Chatterjee S. S., Changkakoty B., Pervaiz Z., et al. , 2011. Yan, an ETS-domain transcription factor, negatively modulates the Wingless pathway in the Drosophila eye. EMBO Rep. 12: 1047–1054. 10.1038/embor.2011.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D. S., Jemison J., Cadigan K. M., 2002. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development 129: 2565–2576. [DOI] [PubMed] [Google Scholar]
- Peterson-Nedry W., Erdeniz N., Kremer S., Yu J., Baig-Lewis S., et al. , 2008. Unexpectedly robust assembly of the Axin destruction complex regulates Wnt/Wg signaling in Drosophila as revealed by analysis in vivo. Dev. Biol. 320: 226–241. 10.1016/j.ydbio.2008.05.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Kojima T., Ui-Tei K., Miyata Y., Saigo K., 1999. Dfrizzled-3, a new Drosophila Wnt receptor, acting as an attenuator of Wingless signaling in wingless hypomorphic mutants. Development 126: 4421–4430. [DOI] [PubMed] [Google Scholar]
- Schaefer K. N., Bonello T. T., Zhang S., Williams C. E., Roberts D. M., et al. , 2018. Supramolecular assembly of the beta-catenin destruction complex and the effect of Wnt signaling on its localization, molecular size, and activity in vivo. PLoS Genet. 14: e1007339 10.1371/journal.pgen.1007339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankaran R., Calleja M., Morata G., Basler K., 2000. The Wingless target gene Dfz3 encodes a new member of the Drosophila Frizzled family. Mech. Dev. 91: 427–431. 10.1016/S0925-4773(99)00313-5 [DOI] [PubMed] [Google Scholar]
- Tacchelly-Benites O., Wang Z., Yang E., Benchabane H., Tian A., et al. , 2018. Axin phosphorylation in both Wnt-off and Wnt-on states requires the tumor suppressor APC. PLoS Genet. 14: e1007178 10.1371/journal.pgen.1007178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B., Townsley F., Rosin-Arbesfeld R., Musisi H., Bienz M., 2002. A new nuclear component of the Wnt signalling pathway. Nat. Cell Biol. 4: 367–373. 10.1038/ncb786 [DOI] [PubMed] [Google Scholar]
- Tian A., Benchabane H., Wang Z., Ahmed Y., 2016. Regulation of stem cell proliferation and cell fate specification by Wingless/Wnt signaling gradients enriched at adult intestinal compartment boundaries. PLoS Genet. 12: e1005822 10.1371/journal.pgen.1005822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J. P., Girdham C. H., O’Farrell P. H., 1994. A cell-autonomous, ubiquitous marker for the analysis of Drosophila genetic mosaics. Dev. Biol. 164: 328–331. 10.1006/dbio.1994.1203 [DOI] [PubMed] [Google Scholar]
- Waaler J., Machon O., Tumova L., Dinh H., Korinek V., et al. , 2012. A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res. 72: 2822–2832. 10.1158/0008-5472.CAN-11-3336 [DOI] [PubMed] [Google Scholar]
- Wang Z., Michaud G. A., Cheng Z., Zhang Y., Hinds T. R., et al. , 2012. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 26: 235–240. 10.1101/gad.182618.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Tacchelly-Benites O., Yang E., Ahmed Y., 2016a. Dual roles for membrane association of Drosophila Axin in Wnt signaling. PLoS Genet. 12: e1006494 10.1371/journal.pgen.1006494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Tacchelly-Benites O., Yang E., Thorne C. A., Nojima H., et al. , 2016b. Wnt/Wingless pathway activation is promoted by a critical threshold of Axin maintained by the tumor suppressor APC and the ADP-ribose polymerase tankyrase. Genetics 203: 269–281. 10.1534/genetics.115.183244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Tian A., Benchabane H., Tacchelly-Benites O., Yang E., et al. , 2016c. The ADP-ribose polymerase Tankyrase regulates adult intestinal stem cell proliferation during homeostasis in Drosophila. Development 143: 1710–1720. 10.1242/dev.127647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Rubin G. M., 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117: 1223–1237. [DOI] [PubMed] [Google Scholar]
- Yang E., Tacchelly-Benites O., Wang Z., Randall M. P., Tian A., et al. , 2016. Wnt pathway activation by ADP-ribosylation. Nat. Commun. 7: 11430 10.1038/ncomms11430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu S., Mickanin C., Feng Y., Charlat O., et al. , 2011. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 13: 623–629. 10.1038/ncb2222 [DOI] [PubMed] [Google Scholar]
- Zhu X., Xing R., Tan R., Dai R., Tao Q. 2011. The RNF146 E3 ubiquitin ligase is required for the control of Wnt signalling and body pattern formation in Xenopus. Mech. Dev. 147: 28–36. 10.1038/ncb2222 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article and figures. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7523393.