Sorghum (Sorghum bicolor (L.) Moench) is a major staple food cereal for millions of people worldwide. Valluru et al. identify putative deleterious mutations among ∼5.5M segregating variants of 229 diverse sorghum...
Keywords: deleterious mutations, genetic load, genome-wide predictions, mutation burden, sorghum
Abstract
Sorghum (Sorghum bicolor L.) is a major food cereal for millions of people worldwide. The sorghum genome, like other species, accumulates deleterious mutations, likely impacting its fitness. The lack of recombination, drift, and the coupling with favorable loci impede the removal of deleterious mutations from the genome by selection. To study how deleterious variants impact phenotypes, we identified putative deleterious mutations among ∼5.5 M segregating variants of 229 diverse biomass sorghum lines. We provide the whole-genome estimate of the deleterious burden in sorghum, showing that ∼33% of nonsynonymous substitutions are putatively deleterious. The pattern of mutation burden varies appreciably among racial groups. Across racial groups, the mutation burden correlated negatively with biomass, plant height, specific leaf area (SLA), and tissue starch content (TSC), suggesting that deleterious burden decreases trait fitness. Putatively deleterious variants explain roughly one-half of the genetic variance. However, there is only moderate improvement in total heritable variance explained for biomass (7.6%) and plant height (average of 3.1% across all stages). There is no advantage in total heritable variance for SLA and TSC. The contribution of putatively deleterious variants to phenotypic diversity therefore appears to be dependent on the genetic architecture of traits. Overall, these results suggest that incorporating putatively deleterious variants into genomic models slightly improves prediction accuracy because of extensive linkage. Knowledge of deleterious variants could be leveraged for sorghum breeding through either genome editing and/or conventional breeding that focuses on the selection of progeny with fewer deleterious alleles.
Plant genomes continually accumulate new mutations due to population demographichistory (Brandvain et al. 2013), random drift (Lynch and Gabriel 1990), the mating system (Hartfield and Glémin 2014), domestication (Lu et al. 2006; Ramu et al. 2017), and linked selection due to genetic interactions (Felsenstein 1974). While a sizeable portion of such new mutations are neutral (Shaw et al. 2002; Covert et al. 2013), a small portion of new mutations are likely to be deleterious because they disrupt evolutionarily conserved sites, protein function (Yampolsky et al. 2005; Doniger et al. 2008), or gene expression (Kremling et al. 2018) in a way that results in negative impacts on fitness. The elimination of deleterious mutations from breeding populations has therefore been suggested as a prospective avenue for crop improvement (Morrell et al. 2012; Moyers et al. 2018).
Sorghum (Sorghum bicolor L., 2n = 20) is an important and versatile crop that is grown for food, forage, and fuel. It was domesticated from its wild ancestor ∼8000 years ago in Africa (Wendorf et al. 1992). Five major morphological forms have traditionally been recognized: bicolor, caudatum, durra, guinea, and kafir. While these races are widespread in distinct regions of Africa, reflecting the diverse agro-ecological environments (Dillon et al. 2007; Evans et al. 2013), sorghum has maintained minimal genome redundancy due to the absence of any whole-genome duplication for > 70 MY (Paterson et al. 2004, 2009). However, inbreeding sorghum is likely to accumulate more weakly deleterious mutations when compared to an outcrossing species, which accumulates strong recessive deleterious mutations that reduce the mean fitness of the species over time (Moyers et al. 2018). Nonetheless, there is accumulating evidence showing that enhanced homozygosity (Kumaravadivel and Rangasamy 1994), relaxed selection (Arunkumar et al. 2015), and low levels of outcrossing (Pamilo et al. 1987; Nakayama et al. 2012) can act to purge deleterious mutations leading to lower mutation burden in selfing populations. Though the relative contributions of these processes to mutation burden has long been debated, both theoretical and experimental evidence suggests that reduced population size effects usually outcompete processes that enhance the purging of deleterious mutations caused by selfing (Bustamante et al. 2002; Slotte et al. 2010, 2013; Arunkumar et al. 2015), leading to an influx of deleterious mutations into selfing species.
Modern breeding and domestication results in an increased mutation burden in domesticates when compared to their wild progenitors, and a decreased mutation burden in elite cultivars when compared to landraces (Gaut et al. 2015; Ramu et al. 2017; Yang et al. 2017). The demographic history and inbreeding allow deleterious variants of weaker effect to reach appreciable frequencies owing to random drift, which can contribute significantly to mutation burden and affect fitness-related traits (Kono et al. 2016). An estimated 20–30% of nonsynonymous variants are deleterious in rice (Lu et al. 2006), Arabidopsis (Günther and Schmid 2010), maize (Mezmouk and Ross-Ibarra 2014), and cassava (Ramu et al. 2017). Renaut and Rieseberg (2015) identified an excess of nonsynonymous single-nucleotide polymorphisms (SNPs) segregating in domesticated sunflower and globe artichoke relative to natural populations. Similarly, ∼20–40% of protein-coding SNPs are predicted to have a deleterious allele in maize (Mezmouk and Ross-Ibarra 2014). Indeed, deleterious mutations are predicted to be enriched near regions of strong selection (Chun and Fay 2011; Gaut et al. 2015; Kono et al. 2016), pointing to a potentially important role for deleterious variants in shaping agronomic phenotypes.
Genomic selection (GS) can help to accelerate crop breeding when compared to conventional phenotype-based selection approaches. In genome-wide prediction (GWP) models employed in GS, the genetic variance is modeled by accounting for either the biological additive or dominant effects of the markers that can potentially improve the prediction accuracy of phenotypic traits (Vitezica et al. 2013, 2016). Genes associated with complex traits carry an uncertain number of deleterious mutations distributed across the genome, and such a mutation burden contributes significantly to the total phenotypic variation of traits (Yang et al. 2017). Because deleterious mutations can occur in both homozygous and heterozygous states depending on the genetic context, trait-specific and genetic-context based GWP models could be expected to capture the phenotypic effects of deleterious mutations. Therefore, GWP models encompassing deleterious variants are expected to account for the total genetic contribution to and improve the prediction accuracy of complex traits (Yang et al. 2017). However, the improvement of GWP will depend on how strongly correlated deleterious variants are to all other variants.
In this study, we examine the contribution of putatively deleterious variants to phenotypic variation in sorghum. We used a racially, geographically, and phenotypically diverse biomass sorghum population that represents the ancestry of four major sorghum types (Brenton et al. 2016). All accessions were phenotyped for two agronomic traits, dry biomass (DBM) and plant height (PH), and for two physiological traits, specific leaf area (SLA) and tissue starch content (TSC), under field conditions. We performed whole-genome resequencing (WGS) on 229 sorghum lines and identified putative deleterious mutations in the genome. The main objectives of this study were to determine (1) whether empirical patterns of deleterious mutation burden differ among sorghum racial groups, and (2) whether deleterious variants improve prediction accuracy of complex traits and, if so, whether such accuracy differs among phenotypic traits that have different genetic architecture. To address these questions, we first identified the putative deleterious mutations and their biological effect sizes, and then estimated an individual mutation burden and its relationship with phenotypic traits. Taking advantage of a Bayesian GS framework (Habier et al. 2011), we tested the biological significance of deleterious variants on prediction of DBM, PH, SLA, and TSC.
Materials and Methods
Plant material, field experiments, and phenotypic data
A biomass sorghum diversity panel assembled for the Transportation Energy Resource from Renewable Agriculture-Mobile Energy-Crop Phenotyping Platform (TERRAMEPP) and Transportation Energy Resource from Renewable Agriculture-Water Efficient Sorghum Technologies (TERRA-WEST) projects was used in this study. This panel was composed of 869 lines: 339 lines coming from Fernandes et al. (2018), 117 lines coming from Brenton et al. (2016), 273 lines coming from Yu et al. (2016), and 140 additional lines obtained from John Burke (United States Department of Agriculture, Lubbock, TX). Although phenotypic data for the entire panel were collected, only a subset of 229 lines for which WGS data were available were used in the study. These 229 lines belong to four major races of sorghum (caudatum, durra, guinea, and kafir) with representatives from the African continent, Asia, and the Americas (Supplemental Material, Figure S1).
Field experiments were conducted in Illinois during 2016 in an augmented block design that consisted of 960 four-row plots with a row length of 3 m, 1.5 m alleys and 0.76 m row spacing. All plots were arranged in 40 rows and 24 columns. Target density of the plant population was ∼270,368 plants ha−1, and experiments were planted in late May and harvested in early October. PH was measured from the ground to the uppermost leaf whorl at seven developmental stages starting 4 weeks after planting (WAP) up to 16 WAP, with an interval of 2 weeks (seven stages), and averaged across the plot. Biomass data were collected at harvest using a four-row Kemper head attached to a John Deere 5830 tractor. A plot sampler equipment with a near infrared sensor (model 130S, RCI Engineering) was used to measure the wet weight of total biomass (lb), and to quantify biomass moisture (%) and starch (%) contents of plants (Li et al. 2015) in the two middle rows of each four-row plot. Biomass yield in dry U.S. tons per acre was calculated as: dry U.S. tons per acre = total plot wet weight (lb) × (1−plot moisture) / (plot area in acre) × 0.0005. Because some accessions had flowered (38 accessions), flowering data were recorded in 2018 (flowering data were not available for 2016). We conducted an additional set of analyses that had excluded these 38 accessions to assess the potential confounding effect of flowering time on PH.
To estimate SLA, the youngest fully expanded leaves from two randomly selected plants of the middle two rows of each plot were excised just above the ligule 60–70 days after planting. Damaged leaves were avoided. Excised leaves were then recut under water and the cut surface kept immersed. In the laboratory, three 1.6-cm leaf discs were collected from the middle of each leaf while avoiding the midrib. Leaf discs were immediately transferred to an oven set at 60° for 2 weeks. The dry mass of leaf discs was determined and SLA was expressed as the ratio of fresh leaf area to dry leaf mass (cm2 g−1). Considering a 10-day interval among the SLA sampling, we used “date of sampling” as a term in the model to generate best linear unbiased predictors (BLUPs).
Statistical analysis of phenotypic data
Phenotypic data analysis was conducted according to experimental design, which consisted of a series of incomplete blocks connected through common checks. The following model was used to generate BLUPs for all genotypes included in the field trial:
where μ is the overall mean, gi is the random effect of the ith genotype, ej is the random effect of the jth environment, bk(j) is the random effect of the kth incomplete block nested within the jth location, geij represents the effect of genotype-by-environment interaction, and εijk is the residual error for the ith genotype in the kth incomplete block in the jth location.
For SLA, we fitted another model that accounted for the sampling date:
where μ is the overall mean, gi is the random effect of the ith genotype, ej is the random effect of the jth environment, bk(j) is the random effect of the kth incomplete block nested within the jth environment, dl(j) is the random effect of the lth sampling date nested within kth incomplete block and the jth location, geij represents the effect of genotype-by-environment interaction, and εijkl is the residual error for the ith genotype in the kth incomplete block and lth sampling date in the jth environment.
For the purpose of estimating the broad-sense heritability (H2) of each phenotype, we estimated variance components using the restricted maximum likelihood. All effects were assumed to be random. Broad-sense heritability on an entry-mean basis was calculated as H2 = σ2G/(σ2G + σ2GXE/number of locations + σ2e/number of environments × number of replicates), where σ2G is the variance among accessions, σ2GXE is the accession-by-environment variance, and σ2e is the error variance. All analyses were conducted in R software (R Development Core Team 2015).
Genotyping
Genomic DNA (gDNA) was extracted using the cetyl trimethylammonium bromide (CTA) method and quantified using picogreen (Molecular Probes, Eugene, OR) on a microplate reader of Synergy HT (BioTek, Winooski, VT). After preprocessing steps of the gDNA samples, 10 libraries were prepared (24 samples in each library) and sequenced on HiSeq 4000 (PE_2x150) using sequencing kit version 1. Fastq files were demultiplexed with the bcl2fastq v2.17.1.14 conversion software of Illumina. We used Sentieon Genomics Pipeline DNA sequencing (Freed et al. 2017) and a series of custom bash scripts to process the raw reads. Briefly, fastq files were aligned to the Sorghum bicolor reference genome version 3.1 (https://phytozome.jgi.doe.gov). PCR duplicates were removed, base quality was recalibrated based on a “known SNPs” file, and recalibrated files were processed through the Haplotype Caller (HC). No realignment around insertions/deletions was performed. The data set therefore contained 239 samples, corresponding to 229 unique accessions, of which seven had one or two replicates.
To create a list of known SNPs for the recalibration step, the HC pipeline was run without recalibration on the list of 239 BAM files. The output was filtered removing SNPs that had a number of heterozygote genotypes across all accessions > 10% and/or a number of heterozygote genotypes more than two times the number of minor alleles [hereafter referred to as “homozygosity-based filter” (Chia et al. 2012)]. In addition, “SNP clusters,” defined as three or more SNPs located within 5 bp were also filtered out. Clusters of SNPs are often generated by misalignment and were conservatively considered as spurious. The filtered list of SNPs was used as known SNPs to recalibrate the BAM files and to generate a final list of SNPs. The vcf file generated by the HC contained biallelic SNPs (n = 22,359,733) and was further filtered to only retain SNPs with at least 4× coverage (n = 21,865,512), and with a nonmissing genotype in ≤ 40% of the samples (n = 14,535,156). After removing SNP clusters and applying homozygosity-based filters, the final data set contained 5,512,653 SNPs that were used for further analyses.
Identifying putatively deleterious mutations
The substitution of amino acid effect on protein function was predicted with the Sorting Intolerant From Tolerant (SIFT) algorithm (Vaser et al. 2016). A nonsynonymous mutation with a SIFT score < 0.05 was defined as a putative deleterious mutation. To identify a higher-confidence set of deleterious mutations, we used genomic evolutionary rate profiling (GERP > 2) (Davydov et al. 2010) estimated from a multi-species whole-genome alignment of six species including Zea mays, Oryza sativa, Setaria italica, Brachypodium distachyon, Hordeum vulgare, and Musa acuminate. We therefore used both an estimate of sequence conservation (GERP > 2) and protein conservation (SIFT < 0.05) to identify more conservative deleterious mutations (hereafter H(high-confidence)GERPDEL-SNPs) in constrained portions of the genome. Using these HGERPDEL-SNPs, we estimated the mutation burden, which was defined as the number of derived deleterious alleles carried by an individual divided by the total number of nonmissing alleles (Vitezica et al. 2016), based on a putative derived deleterious allele that was defined as a minor allele in the multi-species alignment (Yang et al. 2017). First, we counted the total number of deleterious alleles in a given genotype. Here, each allele was given a score of 0.5. If both were deleterious alleles at a given position, we counted them as 1 (0.5 for each allele). If only one allele was deleterious, then it was counted as 0.5. We summed all these homozygous (1’s) and heterozygous (0.5’s) deleterious alleles. Second, we counted the total number of alleles used to score deleterious alleles in a given genotype. Finally, the total number of deleterious alleles was divided by the total number of scored alleles, and the resulting ratio was defined as the mutation burden.
To account for the effects of linkage, we calculated linkage disequilibrium (LD) between SNPs and identified random variants (nondeleterious) to be used as a control set to compare with deleterious variants. A subset of 100,000 random SNP markers were selected and all possible pairwise r2 values were calculated using Plink 1.9 (Chang et al. 2015). Using the 1% of all the possible pairwise calculations, we calculated the relationship of distance between markers and r2. To define local LD structure across each chromosome, we also calculated the mean LD score (Bulik-Sullivan et al. 2015) for each marker. LD scores were calculated with a window of 1 Mb using the software GCTA (Yang et al. 2011; Bulik-Sullivan et al. 2015). Each LD score was divided by the total number of SNPs within each window (Figure S2). To identify SNPs in high LD with deleterious variants, we first explored the effect of window size and r2 threshold on the number of SNPs selected (Figure S3). Given the LD pattern observed, we used a window size of 250 kb and an r2 threshold of 0.9, meaning that if any marker within 250 kb of a deleterious variants has an r2 ≥ 0.9, it would be excluded from further analysis. This yielded a list of ∼1 million SNPs that were in LD with deleterious SNPs, which were excluded from all SNPs. An equal proportion of 100 sets of random variants with the similar allele frequency range of deleterious variants were selected (Figure S4).
Estimating effect sizes of deleterious and nondeleterious variants
Despite the different assumptions in genetic architecture made by the different models, and the fact that the QTL effects are not of equal size and have different genetic architectures, the simplest model ridge regression (RR)-BLUP often performs just as well in extensive cross-validation and empirical studies. Unless indicated otherwise, effect sizes were estimated using the RR-BLUP model implemented in the R-package rrBLUP version 4.2 (Endelman 2011). We fitted the model y = μ + Zu + e, where y is a vector of BLUPs of phenotype; μ is an intercept vector; and Z is an n × p incidence matrix (either deleterious or random variants) containing the allelic states of the p marker loci (z = {−1, 0, 1}), where −1 represents the minor allele; u is the p × 1 vector of marker effects; and e is a n × 1 vector of residuals. Under RR-BLUP, u ∼ MVN (0, Iσ2u) where σ2u is the variance of the common distribution of marker effects and was estimated using restricted maximum likelihood.
Partitioning of genetic variance and GWP
We compared the variance explained by deleterious variants to that of an equal proportion of randomly sampled variants from the distribution of nondeleterious variants. Following the method of Brenton et al. (2016), we used a two-dimensional sampling approach to create 100 equal-sized data sets of randomly sampled variants matched for minor allele frequency. For each trait, we fitted the model separately for each variant set (either deleterious variant or nondeleterious variant) and estimated the phenotypic variance explained.
For each variant set (deleterious variant vs. nondeleterious set), we fitted a standard genomic (G)BLUP model including only additive effects by fitting a linear mixed model of the following form: y = Zg + e, where y is a vector of BLUPs for the phenotype, the vector g is a random effect, the BLUP represents the genomic estimated breeding values (GEBV) for each individual, Z is a design matrix indicating observations of genotype identities, and e is a vector of residuals. The GEBV were obtained by assuming g ∼ MVN (0, Kσ2g), where σ2g is the additive genetic variance and K is the square genomic relationship matrix based on SNP data, implemented in TASSEL (Bradbury et al. 2007). Predictive abilities for all traits were evaluated using a fivefold cross-validation approach repeated 100 times and were implemented in the R statistical software.
Data availability
Genotypic data is available in CyVerse (doi: https://doi.org/10.25739/6yts-xq12). Phenotypic data is available at bitbucket (https://bitbucket.org/bucklerlab/sorghum_geneticload/src/master). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7638122.
Results
Around 33% of nonsynonymous substitutions are putatively deleterious
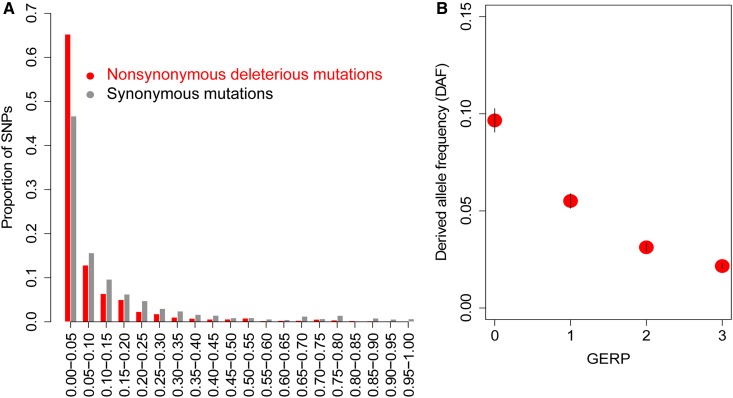
We resequenced the whole genome of 229 diverse biomass sorghum accessions, belonging to four racial groups that were selected to be representative of diverse geographical regions (Figure S1) (Brown et al. 2011; Thurber et al. 2013). The mean sequencing depth was 5.8×, resulting in a data set consisting of ∼5.5 M SNPs. Out of 5.5 M SNPs, ∼6.3% of SNPs are located in coding regions. To determine the distribution of putatively deleterious SNPs in coding regions of the sorghum genome, we first annotated deleterious SNPs using a SIFT score (SIFT < 0.05) that predicts an amino acid substitution effect on protein function (Vaser et al. 2016). Based on SIFT score < 0.05, we find that ∼33% of the total nonsynonymous substitutions are putatively deleterious (average SIFT score of 0.08), while 67% are predicted as tolerated mutations (average SIFT score of 0.47). We estimated the “derived allele” frequency (DAF) spectrum, with the derived allele defined as a minor allele in the multi-species sequence alignment (Yang et al. 2017). Our results reveal that a large proportion of deleterious SNPs have a lower DAF (< 0.05; Figure 1a). While DAF shows a negative association with GERP scores (Figure 1b) (Yang et al. 2017), it has a positively associated pattern with SIFT scores (Figure S5).
Figure 1.
Deleterious mutations in the sorghum genome. (a) Site allele-frequency spectrum of nonsynonymous deleterious mutations and synonymous mutations in the sorghum genome. The derived allele frequency (DAF) distribution of alleles is shown where a minor allele in the multi-species alignment was considered as a derived deleterious allele (Yang et al. 2017). (b) The allele frequency of the derived alleles in bins of different genomic evolutionary rate profiling (GERP) scores. The vertical bars in (b) indicate SE.
We then combined GERP (> 2) and SIFT (< 0.05) scores to identify a higher-confidence set of deleterious SNPs (HGERPDEL-SNPs; Figure S6). Unless otherwise indicated, all further analyses were performed using HGERPDEL-SNPs. While the majority of HGERPDEL-SNPs had an average SIFT score of < 0.01 (Figure S6a), they also showed a low overall allele frequency (average minor allele frequency = 0.07, Figure S6c) that is consistent with population genetic expectations. All identified HGERPDEL-SNPs show comparably similar distributions among all chromosomes (P = 0.34; Figure S6b) and arise from noncentromeric regions of the chromosomes (Figure S7). Our results corroborate previous studies showing that selection acts on deleterious variants to keep them rare (Mezmouk and Ross-Ibarra 2014), and support a combined use of SIFT and GERP scores (Figure 1) as effective quantitative measures of an observed variant for its long-term fitness consequences (Yang et al. 2017).
Both deleterious and nondeleterious variants exhibit different effect size distributions
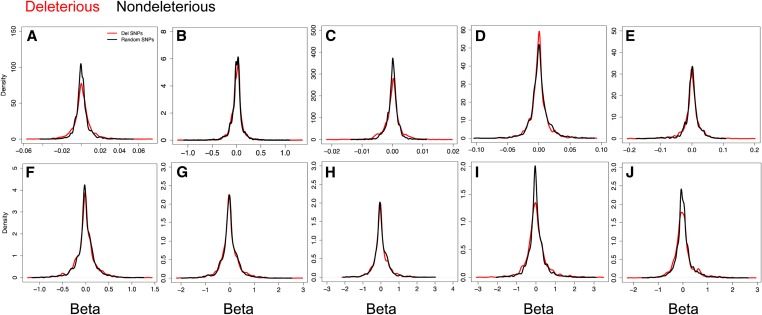
We estimated the additive effect sizes explained by HGERPDEL-SNPs for all phenotypic traits. An equal number of nondeleterious variants were used as a control, which are not in LD but have a similar minor allele frequency spectrum of HGERPDEL-SNPs across the genome (Figure S4). We compared the full density distribution of the effect sizes of both HGERPDEL-SNPs and nondeleterious variants to avoid the winner’s curse (Zöllner and Pritchard 2007; Jun et al. 2018) and examined whether HGERPDEL-SNPs effect sizes are overall higher in magnitude compared to nondeleterious variants (Figure 2).
Figure 2.
Smoothed estimate of density distribution of regression coefficients associated with highly conserved Del variants (HGERPDEL-SNPs) and nondeleterious variants for 10 phenotypic traits [(a) biomass, (b) specific leaf area, (c) tissue starch content, and (d–j) plant height 4, 6, 8, 10, 12, 14, and 16 weeks after planting]. Del, deleterious; GERP, genomic evolutionary rate profiling.
Our results show that the density distribution of the effect sizes of both HGERPDEL-SNPs and nondeleterious variants follow a similar pattern, albeit showing some subtle differences in the density peak and distribution. The density distribution of HGERPDEL-SNPs extends much farther than the distribution of nondeleterious variants, both at the highest and lowest range of distribution (Figure 2), which is similar to the results of previous studies (Zöllner and Pritchard 2007; Jun et al. 2018). While such density distributions are consistent across all traits, HGERPDEL-SNPs show different density peaks compared to nondeleterious variants. For some traits, HGERPDEL-SNPs show reduced-density peaks while for height at 4WAP, HGERPDEL-SNPs show higher-density peaks compared to nondeleterious variants (Figure 2, a–j).
We then compared the empirical cumulative distribution of effect sizes of HGERPDEL-SNPs and nondeleterious variants. Using the two-sample Kolmogorov–Smirnov test, we demonstrate that the effect sizes of both HGERPDEL-SNPs and nondeleterious variants show different density patterns for all phenotypes studied (Figure S8). This suggests that HGERPDEL-SNPs have more variable effect sizes compared to nondeleterious variants for all phenotypic traits. Indeed, the observed variance for estimated effects across all traits was twofold higher for HGERPDEL-SNPs, suggesting that HGERPDEL-SNPs have substantially larger and more subtle effects overall.
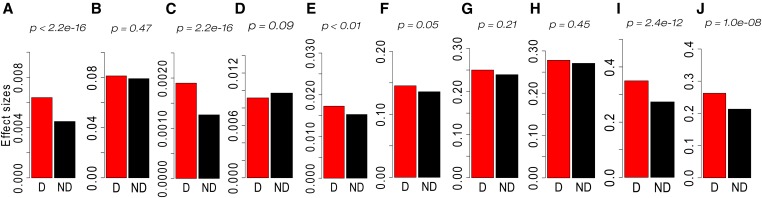
We also compared the means of folded distributions of both HGERPDEL-SNPs and nondeleterious variants. Across all phenotypes, HGERPDEL-SNPs have on average 30.14% (ranging 0–42.34%) higher effects than those observed for nondeleterious variants (Figure 3 and Figure S9). The average effect sizes captured by HGERPDEL-SNPs therefore appear to have greater effect sizes than the average effect sizes explained by nondeleterious variants, which are consistent with the previous results observed in maize (Yang et al. 2017), humans (Marouli et al. 2017; Jun et al. 2018), and mice (Ji et al. 2016).
Figure 3.
Bar plots of means of folded distributions of effect sizes of highly conserved deleterious variants (HGERPDEL-SNPs) and nondeleterious variants for 10 phenotypic traits [(a) biomass, (b) specific leaf area, (c) tissue starch content, and (d–j) plant height 4, 6, 8, 10, 12, 14, and 16 weeks after planting]. GERP, genomic evolutionary rate profiling. D, deleterious; ND, nondeleterious.
Deleterious mutation burden varies among racial groups and negatively correlates with phenotypes
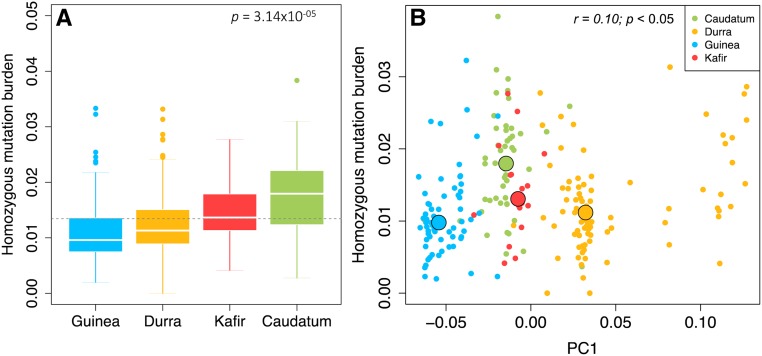
We estimated the mutation burden based on HGERPDEL-SNPs as the count of derived deleterious alleles carried by an individual divided by the total number of scored (nonmissing) alleles (see Materials and Methods; Figure 4). This reveals a substantial variation for mutation burden among racial groups (P = 3.14 × 10−05) based on the HGERPDEL-SNPs (Figure 4a). We observed that the caudatum group is significantly higher, with an average of 36%, for homozygous mutation burden as compared to other racial groups. Compared to the median burden across all racial groups, the guinea group has a proportionately lower burden (−20%), while the caudatum group has a proportionately higher burden (+49%). On average, an individual typically carries 0.0112 (SD 0.006), 0.0124 (SD 0.006), 0.0140 (SD 0.006), and 0.0178 (SD 0.007) mutation burden in the homozygous state in the guinea, durra, kafir, and caudatum groups, respectively. Across all racial groups, individual mutation burden ranges from 0.001 to 0.038 based on the HGERPDEL-SNPs, suggesting that all racial groups showed variable mutation burden.
Figure 4.
Homozygous mutation burden in sorghum. (a) Homozygous mutation burden estimated for different racial groups of sorghum based on highly conserved deleterious variants (HGERPDEL-SNPs). The derived allele is defined as a minor allele from multi-species sequence alignments (Yang et al. 2017). The mutation burden was estimated as the count of derived deleterious alleles carried by an individual divided by the total number of scored (nonmissing) alleles. The horizontal broken line indicates the mean of homozygous mutation burden across all racial groups. (b) Scatter plots of homozygous mutation burden and PC1 derived from genome-wide SNP markers. The black circles indicate the median values for each group. GERP, genomic evolutionary rate profiling; PC, principal coordinate.
Given that there is a considerable amount of admixture present in sorghum lines, we checked if admixture influenced the mutation burden estimation among racial groups. We plotted the relationship between the homozygous mutation burden and the principal components derived from genome-wide SNP markers (Figure 4b and Figure S10). This shows that although there are admixed lines, a tendency toward a higher and lower mutation burden was observed for the caudatum and guinea groups, respectively (Figure 4, a and b). These results indicate that the deleterious mutation burden estimated based on the derived deleterious allele is largely due to the genomic architecture of racial groups, while it is less biased with admixture.
We further evaluated the underlying relationship of mutation burden with phenotypic traits. Four putative phenotypic fitness traits were selected for this study: DBM, PH (seven developmental stages), SLA, and TSC. We selected these traits because total biomass has been explicitly used as an index of fitness in several species, as it can integrate the overall capacity for survival and reproduction (Donovan et al. 2009; Younginger et al. 2017). PH is an ecological fitness trait that incorporates processes for coexistence along spectra of light gradients (Falster and Westoby 2003). SLA is generally regarded as a useful summary ecological trait that often strongly correlates with many key plant attributes of ecological interest (Westoby 1998; Meziane and Shipley 1999). Starch production and its utilization on the diurnal basis, and its role under diverse growth conditions, is regarded as a major integrator in the regulation of plant growth and hence can be considered as a determinant of plant fitness (Sulpice et al. 2009; Thalmann and Santelia 2017).
We observed a substantial phenotypic variation for all traits among racial groups [Figure S11, biomass: P < 0.001; SLA: P < 0.001; starch: P < 0.05; height: P = 5.9e−5 (4WAP), P = 0.04 (6WAP), P = 3.1e−6 (8WAP), P = 3.9e−6 (10WAP), P = 7.5e−5 (12WAP), P = 0.001 (14WAP), and P < 0.05 (16WAP)], with highly heritable variation observed for PH [H2 = 0.87 (at 10 WAP)] and biomass (H2 = 0.73), consistent with previous studies (Brenton et al. 2016). We also found strong correlations among traits (Figure S12).
Using a simple linear regression model between mutation burden and phenotypic traits across all racial groups, we consistently found a negative relationship of mutation burden with all phenotypic traits (Table S1). We also performed a grouped regression combining racial groups that show parallel response, and show that the combined slopes further confirmed the negative correlations between mutation burden and phenotypes (Table S1). These results suggest that deleterious variants decrease trait fitness. However, the majority of these correlations are not significant except for PH (in case of grouped regression only), indicating that the deleterious mutation burden can be strongly linked to the variation in PH in the biomass sorghum lines studied.
Deleterious variants contribute considerably to phenotypic variation but vary substantially among traits
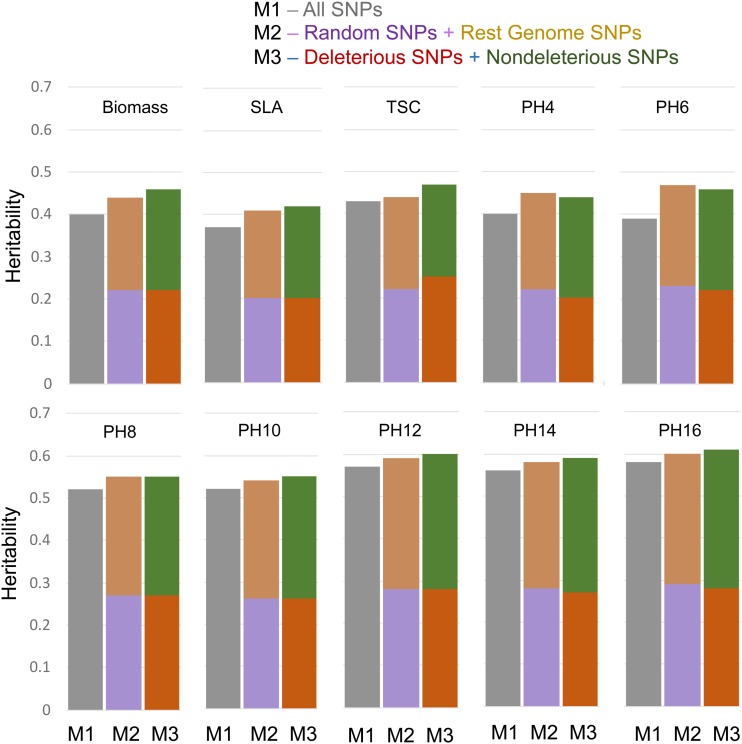
We tested whether incorporating putatively deleterious variants could inform GS models and improve the GWP of phenotypes. HGERPDEL-SNPs identified from WGS were used as priors and integrated into a genomic prediction framework (Figure 5). We quantified the amount of genetic variance, heritability, and model improvement by deleterious variants, and compared them with those of random variants. Based on a variance partitioning approach with a two-kernel model (see Materials and Methods), the model with putatively HGERPDEL-SNPs explained roughly half of the genetic variance [biomass: 52%, SLA: 48%, starch: 46%, and PH: 45–49% (across all stages)] (Figure 5). There was a modest improvement in total heritable variance explained for biomass (7.6%, h2 = 0.24 against 0.22 for random variants) and PH (3.1%, h2 = 0.33 against 0.32 for random variants across seven developmental stages). However, there was no advantage regarding heritable variance for SLA and TSC (Figure 6, a and b) for HGERPDEL-SNPs as compared to random variants.
Figure 5.
Heritability estimates for all traits using a two-kernel model. M, model; PH4–16, plant height at 4, 6, 8, 10, 12, 14, and 16 weeks after planting; SLA, specific leaf area; TSC, tissue starch content.
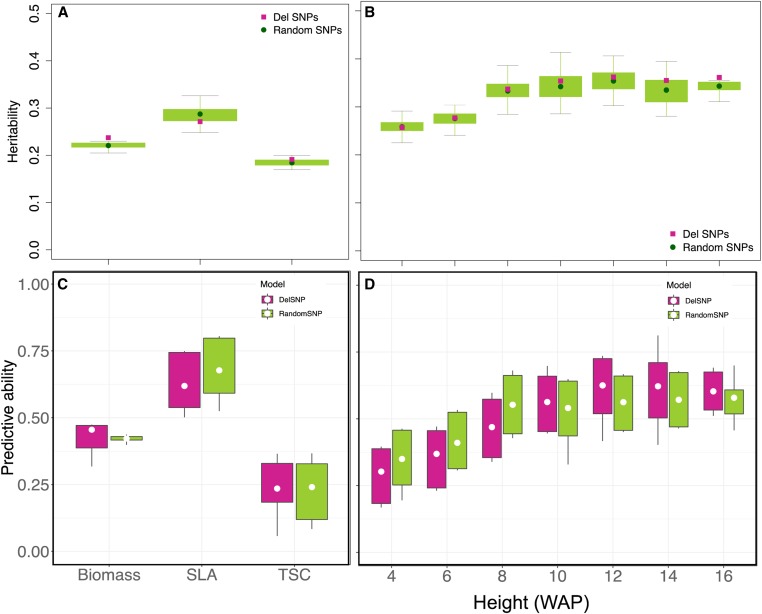
Figure 6.
Genome-wide prediction models incorporating putatively deleterious variants. (a and b) Heritability estimates for all traits using a single-kernel model. Heritability estimates for nondeleterious variants are derived based on 100 independent sets that are randomly chosen across the genome from variants that are not in linkage disequilibrium with deleterious variants. (c and d) Boxplots showing a fivefold cross-validation prediction ability estimation for deleterious variants and random variants. Del, deleterious; SLA, specific leaf area; TSC, tissue starch content; WAP, weeks after planting.
We addressed the potential confounding effects of flowering on PH. We performed heritability estimates based on nonflowered lines (all flowered lines were excluded) within and across racial groups. We observed only minor nonsignificant differences on heritability and these model results are complementary to the model results obtained using all genotypes (Figure S13).
To evaluate the predictive ability, we performed a fivefold cross-validation, repeated 100 times, which was implemented in a GBLUP model with either the HGERPDEL-SNPs or the nondeleterious SNP data sets. Consistent with the results of heritability, we observed 8.1 and 7.0% improvements on predictive ability for biomass and PH (at 10–16WAP only), respectively, while there was no improvement for SLA, TSC, or PH at early stages (at 4–8WAP, Figure 6, c and d). These results suggest that the contribution of putative HGERPDEL-SNPs to phenotypic variation varies considerably among traits.
Discussion
Sorghum, a genus that evolved across diverse environments in Africa, exhibits a wide range of phenotypic diversity (Wright 1931; Doggett 1970; Dillon et al. 2007). This raises the question of whether sorghum racial groups carry variable deleterious mutation burdens, allowing the mutation consequences to be tested for phenotypic diversity. In this study, we whole-genome resequenced 229 biomass sorghum lines and defined a high-confidence set of putative deleterious mutations using SIFT (< 0.05) and GERP (> 2) scores. All racial groups of sorghum showed variable mutation burdens (ranging from 0.001 to 0.038) that correlated negatively with phenotypic traits. We observed that an average deleterious variant had larger biological effects than an average nondeleterious variant. We further noticed that the prediction ability of the GWP models encompassing deleterious variants are largely trait-dependent.
Combining the criteria of SIFT (< 0.05) and GERP (> 2) scores, we first show that sorghum racial groups accumulate appreciable amounts of deleterious mutations in the genome, estimated to be ∼33% of total nonsynonymous substitutions (Figure 1). Although the number and frequency of such mutations within a population largely depends on effective population size, our results match well with previous studies that estimate 20–30% of nonsynonymous variants to be deleterious in several crop species, including model plant species (Lu et al. 2006; Günther and Schmid 2010; Mezmouk and Ross-Ibarra 2014; Ramu et al. 2017). Considering highly frequent (DAF > 0.9) mutations, there are 63 nonsynonymous deleterious mutations across racial groups, which are distributed across all chromosomes. These mutations could likely be a combination of variants of important domestication targets, recent pseudogenes, and some truly deleterious variants that are the product of drift (de Alencar Figueiredo et al. 2008; Smith et al. 2018).
We next estimated an individual mutation burden as the count of derived deleterious alleles carried by an individual divided by the total number of scored (nonmissing) alleles, which differed considerably among individuals and racial groups (Figure 4). It is notable but expected, given that different racial groups have had varying patterns of population dynamics, selection intensities, and domestication histories that could detectably alter the influx of deleterious mutations (Wendorf et al. 1992; Dillon et al. 2007; Paterson et al. 2009). Contrasting deleterious burden has previously been reported in different populations of crop species (Lu et al. 2006; Renaut and Rieseberg 2015; Ramu et al. 2017) and humans (Lohmueller et al. 2008; Fu et al. 2014; Simons et al. 2014). Comparatively, the caudatum group appears to have a higher mutation burden than the guinea group, the oldest of the specialized sorghum races (Stemler et al. 1975; Harlan et al. 1976). We propose that the higher mutation burden of the caudatum group might be potentially related to the population bottleneck, resulting in a smaller population size that increases the chances of inbreeding, genetic homogeneity, and an increased influx of deleterious mutations (Renaut and Rieseberg 2015; Yang et al. 2017; Moyers et al. 2018). On the other hand, a lower mutation burden in the guinea group might be due partly to its higher outcrossing rates, which can reach up to 20% when compared to other races (Barro-Kondombo et al. 2010; Ranwez et al. 2017). Therefore, our results suggest that, first, negative selection is less effective at removing weakly deleterious mutations, yielding variable mutation burden among racial groups. Second, the combined effects of a bottleneck and directional selection during domestication (Hamblin et al. 2006; Lohmueller et al. 2008) can have an important impact on the deleterious mutation burden even in smaller racial groups of sorghum in which founder events can be more frequent (Charlesworth and Wright 2001; Szövényi et al. 2014).
Although informative, our estimation of mutation burden has some important limitations. First, the deleterious mutations identified in the population were based on the degree of sequence conservation, which is often poorly constructed. Second, our derivation of deleterious mutations does not include noncoding or structural variants, which can contribute substantially to the total load of deleterious mutations (Huang et al. 2017; Bastarache et al. 2018). Third, our burden estimation assumes equal fitness effects for all mutations, which is unlikely, as mutations can have different fitness effects that can vary with environments (Henn et al. 2016). Fourth, we consider the same sign of the effect when estimating the burden, which would be misestimated, as some deleterious mutations may be locally adaptive or neutral (Vikram et al. 2015; Bastarache et al. 2018). Nonetheless, despite these caveats, our findings revealed a substantial genomic burden of deleterious mutations in sorghum.
We investigated the phenotypic effects of deleterious mutations (Table S1). We found negative correlations between mutation burden and phenotypic traits, suggesting a considerable cost of deleterious mutations on phenotypic traits (Yang et al. 2017) in a species that has been subjected to recent demographic expansion (Hamblin et al. 2006). Consistently, we find that an average deleterious variant has demonstrably larger biological effect, which could likely have an important impact contributing to heritable phenotypic variation (Figure 2 and Figure 3). In grasses, it has been previously shown that heritable phenotypic variation can be increased as much as 0.1–1% by new mutations (Sprague et al. 1960; Houle et al. 1996; Bataillon 2000). However, the fate of such large-effect mutations on phenotypes is unclear, and whether such mutations are attributable to unconditional deleteriousness or can grant adaptable heritable variation to diverse growing conditions has been actively debated (Glémin and Bataillon 2009). Nonetheless, previous studies have revealed novel variations of genes resulting from postdomestication mutations in sorghum and suggest that neodiversity contributed to new adaptations (de Alencar Figueiredo et al. 2008; Glémin and Bataillon 2009).
Across four traits, we find that putatively deleterious alleles explain roughly one-half of the genetic variance (46–49%), but that there is only a moderate improvement in total heritable variance explained for biomass (7.6%) and PH (3.1%). Additionally, there is no advantage for SLA and TSC (Figure 5 and Figure 6). Such a difference in the contribution of deleterious variants to phenotypic traits was recently observed in maize where dominance contributed substantially to grain yield, while phenology traits appeared to be largely additive (Yang et al. 2017). Though the effects of mutations being deleterious or compensatory depends greatly upon the genetic background into which that mutation is incorporated (Moyers et al. 2018), the trivial contributions of mutations to SLA and TSC indicate that such mutations could be either nearly neutral or negatively synergistic. Therefore, our results support the proposition that deleterious mutational effects vary with phenotypic traits and appear to be often larger for fitness-related quantitative traits, while they are unclear for traits that are not directly linked to fitness (Park et al. 2011). Fitness-related quantitative traits, which are expected to have a more complex genetic architecture, could potentially carry a higher polygenic mutation burden that could considerably affect phenotypes (Purcell et al. 2014). Also, such expectations are in line with the longstanding understanding that fitness-linked quantitative traits showing directional dominance generally exhibit inbreeding depression (Wright 1931; Kelly 1999; Charlesworth and Charlesworth 1999), which indeed is strongly linked to the degree of deleterious burden in the genome (Mezmouk and Ross-Ibarra 2014).
Finally, although our study did not account for sampling error while estimating an individual deleterious variant effect, which is generally greater for rare variants (Jun et al. 2018), our heritability estimates are consistent with the prediction abilities of phenotypic traits. Therefore, our work adds to ongoing GWP efforts to explore the cumulative effects of deleterious mutations on phenotypic diversity (Yang et al. 2017; Moyers et al. 2018). However, since rare deleterious variants are less correlated with each other and their associations greatly suffer from low statistical power (Park et al. 2011; Auer and Lettre 2015), employing either gene- and/or family-based approaches (Auer and Lettre 2015; Ji et al. 2016; Jun et al. 2018), or leveraging the phenotypic patterns (Bastarache et al. 2018), in which deleterious mutations have detectable phenotypic consequences would assist in examining how rare deleterious mutations shape an individual phenotype.
Conclusions
We used phenotypic and genomic data from different racial groups of sorghum to show that sorghum accumulates an appreciable number of deleterious mutations in the genome. Mutation burden differs substantially among racial groups that negatively correlate with phenotypes. GS models encompassing deleterious mutations show variable predictive ability across traits and, given the relatively high level of population structure in sorghum, disentangling deleterious effects at the single-variant level would take a tremendous amount of effort and recombination. Deleterious variants could be prioritized through work with intermediate phenotypes or with more extensive evolutionary analysis among closely related species. Both of these avenues, if combined with high-throughput genome editing and conventional breeding approaches involving parental lines with fewer deleterious variants, could be used to systematically start removing deleterious variants from elite sorghum lines.
Acknowledgments
We thank Robert Bukowski for assistance with the SIFT pipeline, Sara Miller for editorial assistance, and two reviewers and the Editor for their constructive comments on the earlier version of the manuscript. The information, data, or work presented herein was funded in part by the Advanced Research Projects Agency-Energy, United States Department of Energy, under award numbers DE-AR0-000598 and DE-AR0-000661. Support from the United States Department of Agriculture, Agricultural Research Service is greatly acknowledged. The views and opinions of the authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7638122.
Communicating editor: T. Juenger
Literature Cited
- Arunkumar R., Ness R. W., Wright S. I., Barrett S. C. H., 2015. The evolution of selfing is accompanied by reduced efficacy of selection and purging of deleterious mutations. Genetics 199: 817–829. 10.1534/genetics.114.172809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer P. L., Lettre G., 2015. Rare variant association studies: considerations, challenges and opportunities. Genome Med. 7: 16 10.1186/s13073-015-0138-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barro-Kondombo C., Sagnard F., Chantereau J., Deu M., Vom Brocke K., et al. , 2010. Genetic structure among sorghum landraces as revealed by morphological variation and microsatellite markers in three agroclimatic regions of Burkina Faso. TAG Theor. Appl. Genet. Theor. Angew. Genet. 120: 1511–1523. 10.1007/s00122-010-1272-2 [DOI] [PubMed] [Google Scholar]
- Bastarache L., Hughey J. J., Hebbring S., Marlo J., Zhao W., et al. , 2018. Phenotype risk scores identify patients with unrecognized Mendelian disease patterns. Science 359: 1233–1239. 10.1126/science.aal4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataillon T., 2000. Estimation of spontaneous genome-wide mutation rate parameters: whither beneficial mutations? Heredity 84: 497–501. 10.1046/j.1365-2540.2000.00727.x [DOI] [PubMed] [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y., et al. , 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- Brandvain Y., Slotte T., Hazzouri K. M., Wright S. I., Coop G., 2013. Genomic identification of founding haplotypes reveals the history of the selfing species Capsella rubella. PLoS Genet. 9: e1003754 10.1371/journal.pgen.1003754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenton Z. W., Cooper E. A., Myers M. T., Boyles R. E., Shakoor N., et al. , 2016. A genomic resource for the development, improvement, and exploitation of sorghum for bioenergy. Genetics 204: 21–33. 10.1534/genetics.115.183947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. J., Myles S., Kresovich S., 2011. Genetic support for phenotype-based racial classification in sorghum. Crop Sci. 51: 224–230. 10.2135/cropsci2010.03.0179 [DOI] [Google Scholar]
- Bulik-Sullivan B. K., Loh P.-R., Finucane H. K., Ripke S., Yang J., et al. , 2015. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47: 291–295. 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C. D., Nielsen R., Sawyer S. A., Olsen K. M., Purugganan M. D., et al. , 2002. The cost of inbreeding in Arabidopsis. Nature 416: 531–534. 10.1038/416531a [DOI] [PubMed] [Google Scholar]
- Chang C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M., et al. , 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4: 7 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 1999. The genetic basis of inbreeding depression. Genet. Res. 74: 329–340. 10.1017/S0016672399004152 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Wright S. I., 2001. Breeding systems and genome evolution. Curr. Opin. Genet. Dev. 11: 685–690. 10.1016/S0959-437X(00)00254-9 [DOI] [PubMed] [Google Scholar]
- Chia J.-M., Song C., Bradbury P. J., Costich D., de Leon N., et al. , 2012. Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44: 803–807. 10.1038/ng.2313 [DOI] [PubMed] [Google Scholar]
- Chun S., Fay J. C., 2011. Evidence for hitchhiking of deleterious mutations within the human genome. PLoS Genet. 7: e1002240 10.1371/journal.pgen.1002240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert A. W., Lenski R. E., Wilke C. O., Ofria C., 2013. Experiments on the role of deleterious mutations as stepping stones in adaptive evolution. Proc. Natl. Acad. Sci. USA 110: E3171–E3178. 10.1073/pnas.1313424110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov E. V., Goode D. L., Sirota M., Cooper G. M., Sidow A., et al. , 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLOS Comput. Biol. 6: e1001025 10.1371/journal.pcbi.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alencar Figueiredo L. F., Calatayud C., Dupuits C., Billot C., Rami J.-F., et al. , 2008. Phylogeographic evidence of crop neodiversity in sorghum. Genetics 179: 997–1008. 10.1534/genetics.108.087312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon S. L., Shapter F. M., Henry R. J., Cordeiro G., Izquierdo L., et al. , 2007. Domestication to crop improvement: genetic resources for sorghum and saccharum (Andropogoneae). Ann. Bot. 100: 975–989. 10.1093/aob/mcm192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett H., 1970. Sorghum. Longmans, London. [Google Scholar]
- Doniger S. W., Kim H. S., Swain D., Corcuera D., Williams M., et al. , 2008. A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet. 4: e1000183 10.1371/journal.pgen.1000183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan L. A., Ludwig F., Rosenthal D. M., Rieseberg L. H., Dudley S. A., 2009. Phenotypic selection on leaf ecophysiological traits in Helianthus. New Phytol. 183: 868–879. 10.1111/j.1469-8137.2009.02916.x [DOI] [PubMed] [Google Scholar]
- Endelman J. B., 2011. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4: 250–255. 10.3835/plantgenome2011.08.0024 [DOI] [Google Scholar]
- Evans J., McCormick R. F., Morishige D., Olson S. N., Weers B., et al. , 2013. Extensive variation in the density and distribution of DNA polymorphism in sorghum genomes. PLoS One 8: e79192 10.1371/journal.pone.0079192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falster D. S., Westoby M., 2003. Plant height and evolutionary games. Trends Ecol. Evol. 18: 337–343. 10.1016/S0169-5347(03)00061-2 [DOI] [Google Scholar]
- Felsenstein J., 1974. The evolutionary advantage of recombination. Genetics 78: 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes S. B., Dias K. O. G., Ferreira D. F., Brown P. J., 2018. Efficiency of multi-trait, indirect, and trait-assisted genomic selection for improvement of biomass sorghum. Theor. Appl. Genet. 131: 747–755. 10.1007/s00122-017-3033-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed D. N., Aldana R., Weber J. A., Edwards J. S., 2017. The sentieon genomics tools - a fast and accurate solution to variant calling from next-generation sequence data. bioRxiv. Available at: 10.1101/115717 [DOI] [Google Scholar]
- Fu W., Gittelman R. M., Bamshad M. J., Akey J. M., 2014. Characteristics of neutral and deleterious protein-coding variation among individuals and populations. Am. J. Hum. Genet. 95: 421–436. 10.1016/j.ajhg.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B. S., Díez C. M., Morrell P. L., 2015. Genomics and the contrasting dynamics of annual and perennial domestication. Trends Genet. 31: 709–719. 10.1016/j.tig.2015.10.002 [DOI] [PubMed] [Google Scholar]
- Glémin S., Bataillon T., 2009. A comparative view of the evolution of grasses under domestication. New Phytol. 183: 273–290. 10.1111/j.1469-8137.2009.02884.x [DOI] [PubMed] [Google Scholar]
- Günther T., Schmid K. J., 2010. Deleterious amino acid polymorphisms in Arabidopsis thaliana and rice. Theor. Appl. Genet. 121: 157–168. 10.1007/s00122-010-1299-4 [DOI] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Kizilkaya K., Garrick D. J., 2011. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics 12: 186 10.1186/1471-2105-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin M. T., Casa A. M., Sun H., Murray S. C., Paterson A. H., et al. , 2006. Challenges of detecting directional selection after a bottleneck: lessons from sorghum bicolor. Genetics 173: 953–964. 10.1534/genetics.105.054312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan J. R., De Wet J. M. J., Stemler A. B. L., 1976. Origins of African Plant Domestication. De Gruyter, Berlin. [Google Scholar]
- Hartfield M., Glémin S., 2014. Hitchhiking of deleterious alleles and the cost of adaptation in partially selfing species. Genetics 196: 281–293. 10.1534/genetics.113.158196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn B. M., Botigué L. R., Peischl S., Dupanloup I., Lipatov M., et al. , 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl. Acad. Sci. USA 113: E440–E449. 10.1073/pnas.1510805112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D., Morikawa B., Lynch M., 1996. Comparing mutational variabilities. Genetics 143: 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-F., Gulko B., Siepel A., 2017. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat. Genet. 49: 618–624. 10.1038/ng.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X., Kember R. L., Brown C. D., Bućan M., 2016. Increased burden of deleterious variants in essential genes in autism spectrum disorder. Proc. Natl. Acad. Sci. USA 113: 15054–15059. 10.1073/pnas.1613195113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G., Manning A., Almeida M., Zawistowski M., Wood A. R., et al. , 2018. Evaluating the contribution of rare variants to type 2 diabetes and related traits using pedigrees. Proc. Natl. Acad. Sci. USA 115: 379–384. 10.1073/pnas.1705859115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. K., 1999. An experimental method for evaluating the contribution of deleterious mutations to quantitative trait variation. Genet. Res. 73: 263–273. 10.1017/S0016672399003766 [DOI] [PubMed] [Google Scholar]
- Kono T. J. Y., Fu F., Mohammadi M., Hoffman P. J., Liu C., et al. , 2016. The role of deleterious substitutions in crop genomes. Mol. Biol. Evol. 33: 2307–2317. 10.1093/molbev/msw102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremling K. A. G., Chen S.-Y., Su M.-H., Lepak N. K., Romay M. C., et al. , 2018. Dysregulation of expression correlates with rare-allele burden and fitness loss in maize. Nature 555: 520–523. 10.1038/nature25966 [DOI] [PubMed] [Google Scholar]
- Kumaravadivel N., Rangasamy S. R. S., 1994. Plant regeneration from sorghum anther cultures and field evaluation of progeny. Plant Cell Rep. 13: 286–290. 10.1007/BF00233321 [DOI] [PubMed] [Google Scholar]
- Li J., Danao M.-G. C., Chen S.-F., Li S., Singh V., et al. , 2015. Prediction of starch content and ethanol yields of sorghum grain using near infrared spectroscopy. J. Infrared Spectrosc. 23: 85–92. 10.1255/jnirs.1146 [DOI] [Google Scholar]
- Lohmueller K. E., Indap A. R., Schmidt S., Boyko A. R., Hernandez R. D., et al. , 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature 451: 994–997. 10.1038/nature06611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Tang T., Tang H., Huang J., Shi S., et al. , 2006. The accumulation of deleterious mutations in rice genomes: a hypothesis on the cost of domestication. Trends Genet. TIG 22: 126–131. 10.1016/j.tig.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Lynch M., Gabriel W., 1990. Mutation load and the survival of small populations. Evolution 44: 1725–1737. 10.1111/j.1558-5646.1990.tb05244.x [DOI] [PubMed] [Google Scholar]
- Marouli E., Graff M., Medina-Gomez C., Lo K. S., Wood A. R., et al. , 2017. Rare and low-frequency coding variants alter human adult height. Nature 542: 186–190. 10.1038/nature21039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meziane D., Shipley B., 1999. Interacting determinants of specific leaf area in 22 herbaceous species: effects of irradiance and nutrient availability. Plant Cell Environ. 22: 447–459. 10.1046/j.1365-3040.1999.00423.x [DOI] [Google Scholar]
- Mezmouk S., Ross-Ibarra J., 2014. The pattern and distribution of deleterious mutations in maize. G3 (Bethesda) 4: 163–171. 10.1534/g3.113.008870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell P. L., Buckler E. S., Ross-Ibarra J., 2012. Crop genomics: advances and applications. Nat. Rev. Genet. 13: 85–96. 10.1038/nrg3097 [DOI] [PubMed] [Google Scholar]
- Moyers B. T., Morrell P. L., McKay J. K., 2018. Genetic costs of domestication and improvement. J. Hered. 109: 103–116. 10.1093/jhered/esx069 [DOI] [PubMed] [Google Scholar]
- Nakayama S.-I., Shi S., Tateno M., Shimada M., Takahasi K. R., 2012. Mutation accumulation in a selfing population: consequences of different mutation rates between selfers and outcrossers. PLoS One 7: e33541 10.1371/journal.pone.0033541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamilo P., Nei M., Li W.-H., 1987. Accumulation of mutations in sexual and asexual populations. Genet. Res. 49: 135–146. 10.1017/S0016672300026938 [DOI] [PubMed] [Google Scholar]
- Park J.-H., Gail M. H., Weinberg C. R., Carroll R. J., Chung C. C., et al. , 2011. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc. Natl. Acad. Sci. USA 108: 18026–18031. 10.1073/pnas.1114759108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Chapman B. A., 2004. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101: 9903–9908. 10.1073/pnas.0307901101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., et al. , 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. 10.1038/nature07723 [DOI] [PubMed] [Google Scholar]
- Purcell S. M., Moran J. L., Fromer M., Ruderfer D., Solovieff N., et al. , 2014. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506: 185–190. 10.1038/nature12975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu P., Esuma W., Kawuki R., Rabbi I. Y., Egesi C., et al. , 2017. Cassava haplotype map highlights fixation of deleterious mutations during clonal propagation. Nat. Genet. 49: 959–963. 10.1038/ng.3845 [DOI] [PubMed] [Google Scholar]
- Ranwez V., Serra A., Pot D., Chantret N., 2017. Domestication reduces alternative splicing expression variations in sorghum. PLoS One 12: e0183454 10.1371/journal.pone.0183454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2015. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org.
- Renaut S., Rieseberg L. H., 2015. The accumulation of deleterious mutations as a consequence of domestication and improvement in sunflowers and other composite crops. Mol. Biol. Evol. 32: 2273–2283. 10.1093/molbev/msv106 [DOI] [PubMed] [Google Scholar]
- Shaw F. H., Geyer C. J., Shaw R. G., 2002. A comprehensive model of mutations affecting fitness and inferences for Arabidopsis thaliana. Evolution 56: 453–463. 10.1111/j.0014-3820.2002.tb01358.x [DOI] [PubMed] [Google Scholar]
- Simons Y. B., Turchin M. C., Pritchard J. K., Sella G., 2014. The deleterious mutation load is insensitive to recent population history. Nat. Genet. 46: 220–224. 10.1038/ng.2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Foxe J. P., Hazzouri K. M., Wright S. I., 2010. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol. Biol. Evol. 27: 1813–1821. 10.1093/molbev/msq062 [DOI] [PubMed] [Google Scholar]
- Slotte T., Hazzouri K. M., Ågren J. A., Koenig D., Maumus F., et al. , 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45: 831–835. 10.1038/ng.2669 [DOI] [PubMed] [Google Scholar]
- Smith O., Nicholson W. V., Kistler L., Mace E., Clapham A., et al. , 2018. A domestication history of dynamic adaptation and genomic deterioration in sorghum. bioRxiv. Available at: 10.1101/336503 [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Russell W. A., Penny L. H., 1960. Mutations affecting quantitative traits in the selfed progeny of doubled monoploid maize stocks. Genetics 45: 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemler A. B. L., Harlan J. R., de Wet J. M. J., 1975. Evolutionary history of cultivated sorghums (Sorghum bicolor [Linn.] Moench) of Ethiopia. Bull. Torrey Bot. Club 102: 325–333. 10.2307/2484758 [DOI] [Google Scholar]
- Sulpice R., Pyl E.-T., Ishihara H., Trenkamp S., Steinfath M., et al. , 2009. Starch as a major integrator in the regulation of plant growth. Proc. Natl. Acad. Sci. USA 106: 10348–10353. 10.1073/pnas.0903478106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szövényi P., Devos N., Weston D. J., Yang X., Hock Z., et al. , 2014. Efficient purging of deleterious mutations in plants with haploid selfing. Genome Biol. Evol. 6: 1238–1252. 10.1093/gbe/evu099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann M., Santelia D., 2017. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 214: 943–951. 10.1111/nph.14491 [DOI] [PubMed] [Google Scholar]
- Thurber C. S., Ma J. M., Higgins R. H., Brown P. J., 2013. Retrospective genomic analysis of sorghum adaptation to temperate-zone grain production. Genome Biol. 14: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R., Adusumalli S., Leng S. N., Sikic M., Ng P. C., 2016. SIFT missense predictions for genomes. Nat. Protoc. 11: 1–9. 10.1038/nprot.2015.123 [DOI] [PubMed] [Google Scholar]
- Vikram P., Swamy B. P. M., Dixit S., Singh R., Singh B. P., et al. , 2015. Drought susceptibility of modern rice varieties: an effect of linkage of drought tolerance with undesirable traits. Sci. Rep. 5: 14799 10.1038/srep14799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitezica Z. G., Varona L., Legarra A., 2013. On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics 195: 1223–1230. 10.1534/genetics.113.155176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitezica Z. G., Varona L., Elsen J.-M., Misztal I., Herring W., et al. , 2016. Genomic BLUP including additive and dominant variation in purebreds and F1 crossbreds, with an application in pigs. Genet. Sel. Evol. 48: 6 10.1186/s12711-016-0185-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendorf F., Close A. E., Schild R., Wasylikowa K., Housley R. A., et al. , 1992. Saharan exploitation of plants 8,000 years BP. Nature 359: 721–724. 10.1038/359721a0 [DOI] [Google Scholar]
- Westoby M., 1998. A leaf-height-seed (LHS) plant ecology strategy scheme. Plant Soil 199: 213–227. 10.1023/A:1004327224729 [DOI] [Google Scholar]
- Wright S., 1931. Evolution in mendelian populations. Genetics 16: 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yampolsky L. Y., Kondrashov F. A., Kondrashov A. S., 2005. Distribution of the strength of selection against amino acid replacements in human proteins. Hum. Mol. Genet. 14: 3191–3201. 10.1093/hmg/ddi350 [DOI] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., Visscher P. M., 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88: 76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Mezmouk S., Baumgarten A., Buckler E. S., Guill K. E., et al. , 2017. Incomplete dominance of deleterious alleles contributes substantially to trait variation and heterosis in maize. PLoS Genet. 13: e1007019 10.1371/journal.pgen.1007019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younginger B. S., Sirová D., Cruzan M. B., Ballhorn D. J., 2017. Is biomass a reliable estimate of plant fitness?1. Appl. Plant Sci. 5: 1600094 10.3732/apps.1600094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li X., Guo T., Zhu C., Wu Y., et al. , 2016. Genomic prediction contributing to a promising global strategy to turbocharge gene banks. Nat. Plants 2: 1–7. [DOI] [PubMed] [Google Scholar]
- Zöllner S., Pritchard J. K., 2007. Overcoming the winner’s curse: estimating penetrance parameters from case-control data. Am. J. Hum. Genet. 80: 605–615. 10.1086/512821 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genotypic data is available in CyVerse (doi: https://doi.org/10.25739/6yts-xq12). Phenotypic data is available at bitbucket (https://bitbucket.org/bucklerlab/sorghum_geneticload/src/master). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7638122.