Abstract
Calorie restriction (CR) delays aging and affects the circadian clocks by reprogramming circadian rhythms in gene expression. To expand on the circadian mechanisms in CR, we assayed rhythms in the protein translation by analyzing polysome-associated mRNAs in the liver of mice fed ad libitum (AL) and CR diets. Global comparison of the diets revealed that <1% of transcripts were differentially abundant in the polysomes. In contrast, the large differential, up to 10%, was detected when CR and AL diets were compared at individual times throughout the day. Most transcripts that were rhythmic under AL lost their rhythms, and many new transcripts gained rhythms under CR. Only a small fraction of transcripts, including the circadian clock genes, were rhythmic under both diets. Thus, CR strongly reprograms translation. CR affected translation of enzymes regulating long-chain acetyl-coenzyme A (Acyl-CoA) metabolism. The expression of the Acyl-CoA thioesterase (ACOT) family was induced upon CR, leading to the increased transcriptional activity of peroxisome proliferator–activated receptor α, the transcriptional factor regulated by the ACOT products. We propose that the differential translation induced by CR leads to a temporal partition and reprogramming of metabolic processes and provides a link between CR, lipid metabolism, and the circadian clock.—Makwana, K., Gosai, N., Poe, A., Kondratov, R. V. Calorie restriction reprograms diurnal rhythms in protein translation to regulate metabolism.
Keywords: ACOTs, lipid metabolism, mRNA-sequencing
There is growing evidence on the interplay between feeding, aging process, and the circadian system (1–5). In mammals, the circadian system is a hierarchal network of circadian clocks that operates in many organs and tissues under the control of the central clock in the suprachiasmatic nuclei. In a cell, clocks operate as interconnected loops formed by several transcriptional factors that regulate the activity and expression of each other (6). There are a dozen core clock genes that are essential for the circadian clock to operate. The circadian rhythms generated by the clocks orchestrate gene expression, metabolic processes, cell signaling, and synchronize the physiologic processes inside the organism that allow better adaptation to the periodic environment. Disruption of these rhythms in animal models significantly affects many metabolic processes, such as glucose, fat, and redox homeostasis, which may lead to metabolic syndromes and later to the development of diabetes, cardiometabolic pathology, and cancer. Disruption of circadian rhythms in humans through environmental disturbances, such as shift work, increases the risk of similar diseases supporting the importance of the circadian clocks and rhythms (7–10).
Aging is accompanied by changes in both the central and peripheral clocks. These changes are manifested by alterations of circadian rhythms in behavior and metabolism. Calorie restriction (CR) delays aging and increases longevity in a variety of organisms (11, 12) and affects circadian clocks. CR changes circadian rhythms in behavior and gene expression, thus regulating both central and peripheral circadian clocks, and the effect is conserved between flies and rodents (13–17). Circadian clocks are master regulators of metabolism (18), and it was speculated that at least some of the beneficial outcomes of CR are due to the effect on the clock (2, 3). Recent analysis of circadian rhythms of mRNA abundance in the liver and skeletal muscles of young and old mice fed ad libitum (AL) or CR diets identifies a significant reprogramming of the circadian transcriptome by aging and CR (16, 19), supporting the importance of the interaction between biologic clocks, diet, and aging.
Multiple high-throughput analyses, such as transcriptomic, proteomic, acetylomic, and metabolomic analysis, have been performed to study the effects of CR on metabolism in different tissues and organisms, including primates (20–22). Importantly, the large-scale comparisons found little congruency between transcriptome and proteome (23), which suggests the importance of posttranscriptional regulation such as RNA processing (20) or control of translation in the CR mechanism. To further unravel the molecular pathways that contribute to the adaptation of organism physiology to CR and establish a mechanistic connection between the CR-induced changes in the expression and metabolism, we studied the effect of CR on differential translation. Polysome association of mRNAs is a good reflection of active translation (24–26), and here, we assayed the polysome-associated mRNAs (P-mRNAs) in the livers of mice fed AL and CR diets across the day using RNA sequencing (RNA-seq). We found significant changes in the translational for many transcripts whose products are known to be associated with CR, such as components of insulin, AMPK, and forkhead box signaling pathways (27). Our analysis also revealed several genes that were not previously implicated in CR mechanisms, such as enzymes involved in long-chain acetyl-coenzyme (Acyl-CoA) metabolism. Together these data provide an explanation for CR-induced effects on physiology and suggest the importance of the interaction between the circadian clock and translational machinery as a mechanism of organismal adaption to dietary challenges.
MATERIALS AND METHODS
Animal experiments
All experiments with animals were performed according to protocol 21141-KON-AS, approved by Cleveland State University Institutional Animal Care and Use Committee. We used carbon dioxide as the euthanizing agent followed by cervical dislocation to confirm death. This method of euthanasia is consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association. C57B6/J male littermates were randomly grouped into AL or CR groups and were kept at alternate 12:12 h light and dark phase. Animals were housed in groups. At the start of the experiment, all the animals were 3 mo old. Control group was fed AL, whereas CR animals were fed a restricted diet with 30% less calories without malnutrition. Both groups received 18% Teklad (Envigo, Huntington, United Kingdom) (composition: protein 18.6%, fat 5%, and carbohydrate 44.2%). CR treatment was implimented gradually with 10% reduction of calories in the first wk, 20% in the second wk, and 30% reduction began in the third wk. CR animals received food once per day, 3 h after the lights were turned off [zeitgeber time (ZT)14]. Body weight was taken after 2 mo on the diet (10 animals for AL and 17 for CR). Liver tissues were collected across a 24-h period, every 4 h, and immediately frozen and kept at −80°C for subsequent processing. At time point ZT14, tissues were harvested before feeding.
Polysome profiling
Liver tissues were lysed and homogenized using mortar and pestle in a buffer containing KCL (2 M), HEPES (pH 7.4), Mgcl2 (1 M), DTT (1 M), cyclohexamide (20 mg/ml), 2% Triton X-100, and RNAsin (N2115; Promega, Madison, WI, USA). For polyribosome separation, the liver lysates were loaded on 10–50% sucrose gradient containing KCL (2 M), HEPES (pH 7.4), Mgcl2 (1 M), DTT (1 M), cyclohexamide (20 mg/ml). The gradient was centrifuged at 4°C and 17,000 rpm for 14 h using an SW 32.1 rotor (Beckman Coulter, Brea, CA, USA). An Isco Programmable Density Gradient System (Teledyne Isco, Lincoln, USA) was used to collect the gradients at 254 nm using an Isco UA-6 absorbance detector (Teledyne Isco).
RNA isolation
RNA from the polysomal fractions was isolated using Trizol LS Reagent (9,852,301; Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Equal concentration of RNA from individual polysomal fractions were pooled together in equal concentration for mRNA-sequencing (mRNA-seq). Two biologic samples per circadian time point for AL and CR animals were sent for mRNA-seq. Total RNA from liver tissues of 3 or more biologic replicates for each circadian time was isolated for both AL and CR using Trizole (Ambion 157,708; Thermo Fisher Scientific) according to the manufacturer’s protocol. Concentration was determined by NanoDrop 2000 (Thermo Fisher Scientific).
mRNA-seq
RNA was isolated from polysome fractions. RNA samples for two biologic replicates for each time point for both AL and CR groups were prepared and sent to Genewiz (South Plainfield, NJ, USA). Libraries were prepared using poly A tail selection and were paired end and nonstrand specific.
Western blotting
Lysis of frozen liver was done in cell signaling buffer with a protease/phosphatase inhibitor cocktail (Cell Signaling Technology, Danvers, MA, USA). Bradford Protein Assay Kit (500-0006; Bio-Rad, Hercules, CA, USA) was used for concentration determination. A total of 30 μg protein was loaded equally. Proteins were transferred on PVDF membrane at 110 mA for 70 min. Ponceau staining was done to check for the uniform loading of the lysates. Anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (5174, RRID:AB_10622025; Cell Signaling Technology) was using as loading control. To assay the total levels of Acyl-CoA thioesterase 1 (ACOT1) and ACOT4 proteins following anti-ACOT1 antibodies (ab133948; Abcam, Cambridge, United Kingdom) and anti-ACOT4 antibodies (PA551453, RRID:AB_2637603; Thermo Fisher Scientific).
Real-time quantitative PCR
RNA (1 μg) was used to make cDNA using Superscript IV reverse transcriptase (Thermo Fisher Scientific). Real-time quantitative PCR was done using 50 μg of cDNA using universal SYBR Green mix (1725124; Bio-Rad). Gene 18S was used for normalization of PCR for total mRNA, and β-actin was used for normalization of PCR for samples extracted from polysomes. See Table 1 for primer sequences.
TABLE 1.
Primer sequences
| Primer sequence, 5′–3′ |
||
|---|---|---|
| Primer | Forward | Reverse |
| Acot1 | CTTGGATAGCTCCAGTTTCCA | CTTGGATAGCTCCAGTTTCCA |
| Acot3 | CACCGCTACCTGGAATGTAAT | CCTTCCAAGCCTCTTTCTAGTC |
| Acot4 | CATCCTGGAACTTGCCATGTA | GGCCGAGCCTTTAATCCTATC |
| Fmo3 | CACCACCATCCAGACAGATTAC | CCTTGAGAAACAGCCATAGGAG |
| Serpina 12 | CTGGACCCACTGATAAT | CCTGACTGGAGAATCATA |
| 18S | GCTTAATTTGACTCAACACGGGA | AGCTATCAATCTGTCAATCCTGTC |
| β-actin | CCTCTGAACCCTAAGGCCAA | AGCCTGGATGGCTACGTACA |
Fasting blood glucose measurement
Plasma glucose concentration measurement was performed on 5 AL and 11 CR mice. Mice in both groups were 5 mo old. CR mice were fed a diet for 2 mo. The food was removed at ZT0 (7 am). Animals were unfed for 14 h and glucose levels were measured at ZT14 (9 pm). The blood was collected from a tail vein, and glucose concentration was assayed with a glucometer (Truetrack; Home Diagnostics, Fort Lauderdale, FL, USA).
Kyoto Encyclopedia of Genes and Genomes pathway analysis
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) 6.8 (https://david.ncifcrf.gov/summary.jsp) was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Stable gene IDs from Ensembl (https://useast.ensembl.org/index.html) was used for analysis. Rhythmically and differentially abundant transcripts in the polysome fractions were analyzed. Pathways were sorted according to the cutoff value of P < 0.01.
Analysis of mRNA-seq library
Fastq files were checked for quality before and after the removal of adapters using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc). Removal of adapters were performed using AfterQC (28). Reads were aligned to the mouse reference genome (GRCm38.p5 Release M15) using STAR aligner (29). All these steps were done using Owens cluster at Ohio Supercomputer Center. Read counts were done using HTSeq (30) with nonstranded option, and differential expression was analyzed using DESeq2 (31) using default parameters at https://usegalaxy.org/. Transcripts were sorted out with a false discovery value of P < 0.049, mean base higher than 249, and log2 ≥ ±0.7.
Statistical analysis
Three animals for every time point for each diet were used for PCR and Western blot experiments (mRNA-seq data, n = 2 for every time point for each diet). Data are shown as means ± sd. SPSS Statistics 20 (IBM, Armonk, NY, USA) and Prism software packages (GraphPad, La Jolla, CA, USA) were used for analysis. The AL vs. CR effect and time of day effect were tested for significance with 2-way repeated ANOVA corrected for multiple comparison using Bonferroni. A value of P < 0.05 was considered as a statistically significant difference.
JTK_Cycle analysis
Rhythmic association of transcripts with polysomes was analyzed using JTK_Cycle software (Hughes Lab, Toronto, ON, Canada) (32). Normalized read counts generated from DESeq2 (31) were used for this analysis. A value of < 0.05 was set as a cutoff for rhythmicity. Heatmap for rhythmic transcripts was generated using gplots package in Rstudio (https://www.rstudio.com/).
RESULTS
Effect of CR on differential mRNA translation in the liver
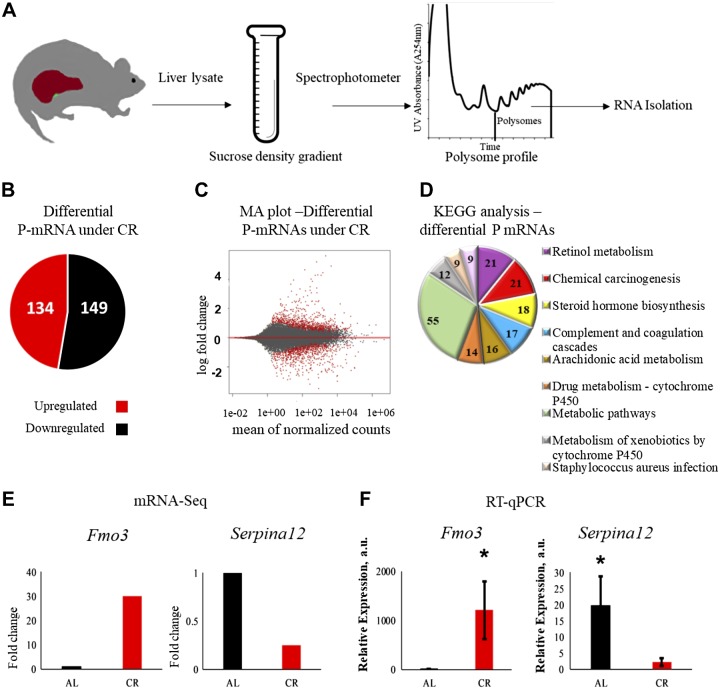
To study the effect of CR on the temporal protein translation, we decided to isolate mRNA associated with ribosomes. There are several methods for isolation of ribosome-loaded mRNA (24, 25, 33), and we analyzed polysome-associated transcripts. It is generally accepted, with few exceptions, that mRNAs associated with polysomes are actively translated (26). While some of the transcripts might escape the analysis because of some limitations of this approach, but generally changes in the transcript abundance in polysomes provide the information on the state of the cellular translatome (25). We isolated polyribosomes from livers of wild-type mice on AL and CR diets. Effects of CR on physiology and metabolism are sex dependent (34); therefore, the study was performed with male mice only. At 3 mo old, C57B6/J male mice were randomly assigned to AL and 30% CR groups. The food for the CR group was provided once per day at ZT14 (2 h after the light was off). Mice on the CR diet were healthy and active, their body weight was significantly smaller compared with mice on AL feeding, and their fasting blood glucose was also significantly reduced (Table 2) in agreement with well-documented effects of CR diet on body weight and glucose homeostasis. After 2 mo on the AL and CR diets, when the animals were 5 mo old, the samples (3 animals/time point) were collected at 6 time points across a 24 h cycle at intervals of 4 h. Ribosomes were fractionated by sucrose density gradient (Fig. 1A). RNA extracted from polysomes (P-mRNAs) were subjected to mRNA-seq (Fig. 1A). P-mRNA sequence reads mapped mainly to mRNA coding DNA regions, and biologic reproducibility of mRNA-seq data was high. P-mRNAs for 26,913 genes were found in polysomes. To check for the overall effect of the CR diet, we drew comparison between AL and CR animals after combining all the biologic replicates of all 6 time points across the day for each group. Interestingly, only about 0.1% of transcripts (the cutoff was set to 1.6-fold change) showed differential association with polysomes in the liver of mice on CR and AL diets (Fig. 1B, C): abundance of 134 P-mRNAs were up-regulated and 149 P- mRNAs were down-regulated under CR (Fig. 1B). To validate our RNA-seq data, we selected 2 genes, Fmo3 and Serpina12, and performed quantitative real-time PCR on the same mRNA samples isolated from polysomal fractions that were used for RNA-seq. In agreement with RNA-seq data (Fig. 1E), we found increased polysome association for Fmo3 and decreased for Serpina12 (Fig. 1F). We also confirmed the agreement between RNA-seq and real-time quantitative PCR for several other transcripts. Therefore, we are confident in our RNA-seq data. We applied KEGG pathway analysis using the publicly available DAVID information and found that metabolic, retinol, and steroid metabolism were among the top pathways affected by CR (Fig. 1D).
TABLE 2.
Body weight and fasting glucose measurement of AL and CR animals
| Diet (mean ± sem) |
|||
|---|---|---|---|
| Measurement | AL | CR | P |
| Body weight (g) | 32.731 ± 2.64 | 24.631 ± 1.82 | <0.0001 |
| Fasting blood glucose (mg/dl) | 166.8 ± 11.77 | 116.8 ± 9.15 | <0.0001 |
Figure 1.
CR mediated differential translation in the liver. A) The experimental workflow. Mouse liver lysates were subjected to fractionation using sucrose gradient. Total RNA was extracted from polysome fractions and mRNA-seq was performed. B) The effect of CR on the P-mRNA abundance. P-mRNAs, whose abundance was increased (red), and P-mRNAs, whose abundance were decreased (black), in the liver polysomes in response to CR. C) Log ratio/mean average (MA plot) of log-fold change for mean of normalized read counts, every dot represents individual gene, gene for P-mRNAs with differential abundance between AL and CR samples are shown by red dots. D) Enriched pathways for P-mRNAs differentially abundant under CR treatment. Pathways were sorted in decreasing order of significance. Values on the pie chart represent the number of the P-mRNAs involved in the pathways. Cutoff value was set to P ≤ 0.01. E, F) Validation of mRNA-seq data: mRNA-seq data for Fmo3 and Serpina12 (n = 12) (E) and real-time quantitative PCR on RNA extracted from polysomal fraction for Fmo3 and Serpina12 (n = 12) (F). *P < 0.05.
Rhythms in translation of circadian clock transcripts under AL and CR diets
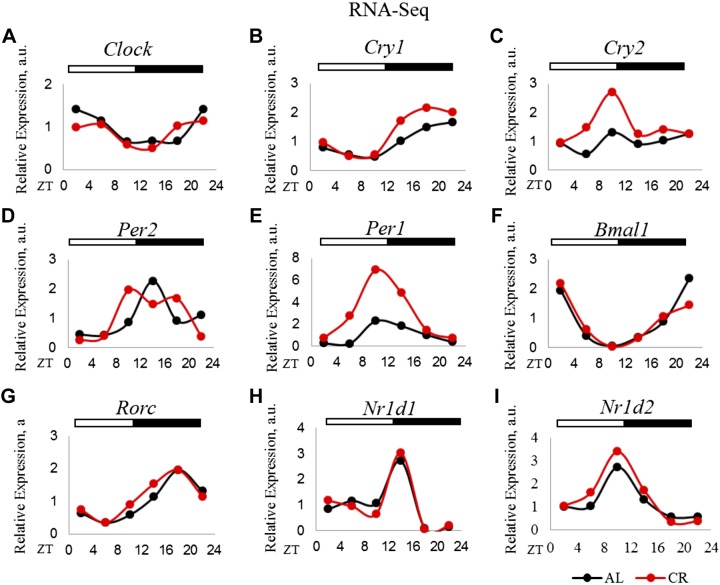
We, and others, have found that mRNAs for core circadian clock genes oscillate under both AL and CR conditions, and CR increases the expression of several clock genes in the mouse liver (13, 14) and Drosophila fat-body and head (15). We analyzed the rhythmic abundance of P-mRNAs for several core circadian clock and clock-controlled genes using the JTK_Cycle algorithm (32). Most of the clock genes (Clock, Bmal1, Per1, Per2, Cry1, Nr1d1, Nr1D2, and Rorc) were associated with polysomes in a rhythmic manner under both diets (Fig. 2). P-mRNA for Cry2 was arrhythmic under AL and CR. P-mRNAs for Per1, Per2, Cry1, and Cry2 were induced in a time-dependent manner. The phase of P-mRNAs was in good agreement with the phases of the rhythms for the appropriate mRNAs [Astafev et al. (14); Table 3]. Thus, for most of the clock gene transcripts, the changes in polysome association under CR diet were driven by CR-induced changes in mRNA abundance. Importantly, CR did not affect the phases of clock gene expression, confirming that the food for the CR group was provided at a physiologically appropriate time for feeding.
Figure 2.
Translation of core clock transcripts under AL and CR diets. P-mRNA abundance of circadian clock genes in the liver polysomes for AL and CR animals: Clock (A), Cry1 (B), Cry2 (C), Per2 (D), Per1 (E), Bmal1 (F), Rorc (G), Nrld1 (H), Nrld2 (I). Two biologic replicates for each time point of AL animals were compared to those of CR for differential P-mRNA abundance using Deseq2 software. Log2 values for the fold change thus obtained were used to plot graphs (n = 2 for each time point for each diet). The light/dark bars on the top represent light and dark phases of the day (light is on at ZT0, and off at ZT12).
TABLE 3.
Phases of rhythmicity in translation and transcription for circadian clock genes
| Translation phase |
Transcription phase |
||||||
|---|---|---|---|---|---|---|---|
| Gene ID | Gene name | AL | CR | Circadian | AL | CR | Circadian |
| ENSMUSG00000055116.8 | BMAL1 | 22 | 22 | AL/CR | 22.64 | 22.8 | AL/CR |
| ENSMUSG00000020893.17 | Per1 | 12 | 10 | AL/CR | 12.12 | 11.22 | AL/CR |
| ENSMUSG00000055866.9 | Per2 | 16 | 12 | AL/CR | 14.36 | 15.51 | AL/CR |
| ENSMUSG00000020038.10 | Cry1 | 18 | 18 | AL/CR | 18.7 | 19.7 | AL/CR |
| ENSMUSG00000068742.11 | Cry2 | 22 | 10 | None | 13.51 | 18.86 | AL |
Rhythmic translation in the liver of mice on AL and CR diets
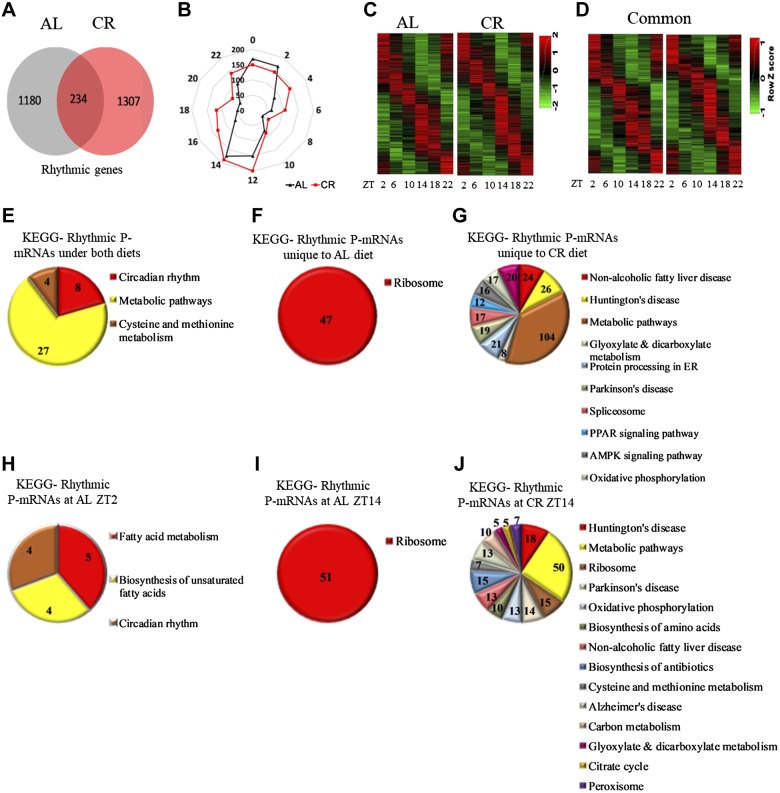
We used the JTK_Cycle algorithm to identify rhythmically abundant P-mRNAs in the liver polysomes of AL and CR animals. We found that under AL diet, 1414 P-mRNAs were rhythmically abundant, and 1541 P-mRNAs were rhythmic under CR diet (Fig. 3A–C). Interestingly, while about the same percentage of transcripts were rhythmically abundant under both diets—4.7% P-mRNA on AL and 5.7% P-mRNA on CR—the lists of these rhythmically abundant P-mRNAs under AL and CR diets were very different. Only 234 transcripts [about 1% of the total number (26,913) and about 20% of all rhythmic transcripts under AL] were rhythmic under both diets (Fig. 3A, D). Thus, for most of the rhythmically abundant P-mRNAs under AL diet, the rhythms ceased under CR, and an equally large and different group of P-mRNAs acquired rhythms. Interestingly, it was recently reported that CR reprograms the circadian transcriptome, and the list of genes rhythmic under AL only partially overlap with the list of genes rhythmic under CR (16).Thus, like the effect of CR on the transcriptome, the circadian output was also significantly reprogrammed.
Figure 3.
Rhythms in the abundance of P-mRNA in the liver of mice under AL and CR diets. A) The number of transcripts found to be circadian in the liver polysomes of AL and CR animals by JTK_cycle analysis. Cutoff value was set at P < 0.05. B) Radar diagram for the distribution pattern of circadian P-mRNAs in the liver polysomes of AL and CR animals. The numbers at the periphery of radar (0–22) represent time of the day (light is on at ZT0, and off at ZT12). The numbers on the circle represent number of P-mRNAs whose abundance peaked at that time of the day. C) Heatmap for P-mRNA abundance and distribution of circadian P-mRNAs in the liver polysomes of AL and CR animals. D) Heat map displays the peak in P-mRNA abundance and distribution of circadian transcripts rhythmic under both AL and CR diets. E) Enriched pathways for P-mRNAs rhythmic under both AL and CR diets. F) Enriched pathways for P-mRNAs rhythmic only in the liver polysomes of AL animals. G) The enriched pathways for P-mRNAs rhythmic only in the liver polysomes of CR animals. H) The enriched pathways for rhythmic P-mRNAs peaked at ZT2 in the livers of AL animals. I) Enriched pathways for rhythmic P-mRNAs peaked at ZT14 in livers of the AL animals. J) Enriched pathways for rhythmic P-mRNAs peaked at ZT14 for CR animals. Pathways are sorted in decreasing order of significance. Values on the pie chart represent transcripts involved in the pathways. Cutoff value was set at P < 0.01.
KEGG pathway analysis revealed a significant shift in rhythmic biologic processes induced by CR. As it is illustrated on Fig. 3E, the circadian rhythm was one of the main pathways that was enriched for P-mRNAs that were rhythmic under AL and CR diets. A significant number of P-mRNAs rhythmically abundant exclusively in liver polysomes of AL animals were found to be involved in the ribosomal protein synthesis pathway (Fig. 3F), which is in the agreement with previously reported circadian rhythms in ribosome biogenesis (35) and translation (36). AMPK signaling was enriched for P-mRNAs that were exclusively rhythmic under CR (Fig. 3G) and may contribute to well-documented CR-induced improvement in glucose homeostasis (37). Another pathway enriched under CR was nonalcoholic fat liver disease pathway (Fig. 3G), which is in agreement with well-documented changes in fat metabolism upon CR (4, 38). Thus, the data on changes in rhythmic translation of various transcripts under AL and CR diets suggest that circadian output was reprogramed to fit metabolic needs of the organism. The phase distribution for the rhythmic P-mRNAs is shown on Fig. 3B. For both diets, rhythms in polysomal abundance for most P-mRNAs peaked around light/dark and dark/light transitions (Fig. 3B, C). For AL animals, the pathways enriched at ZT2 were circadian rhythm and fat metabolism (Fig. 3H), whereas, at ZT14, the ribosomal protein synthesis pathway was enriched (Fig. 3I). For CR animals, at ZT14, metabolic pathways, oxidative phosphorylation, and TCA cycle were among the significantly enriched pathways (Fig. 3J). Endocytosis and protein processing pathways were enriched at ZT2 upon CR but are not shown, as they fall above the cutoff P value of 0.01.
For P-mRNAs rhythmic under both diets (Fig. 3A, D), we investigated the effect of CR on their phase, amplitude, and daily average. We found that for most of the rhythmic transcripts CR did not change the phase: peaks for 178 (76%) transcripts were not shifted or shifted by only 2 h, peaks for 38 transcripts were phase shifted by 4 h, and for 18 transcripts by 6 or more hours (Supplemental Fig. S1). For 11 transcripts, the daily average abundance was increased upon CR by 1.5-fold or more, and for 37 transcripts, the daily average abundance was reduced by 1.5-fold or more. A 1.5-fold increase and decrease in rhythm amplitude was observed for 47 and for 30 transcripts, respectively.
CR significantly changed temporal translation in the liver
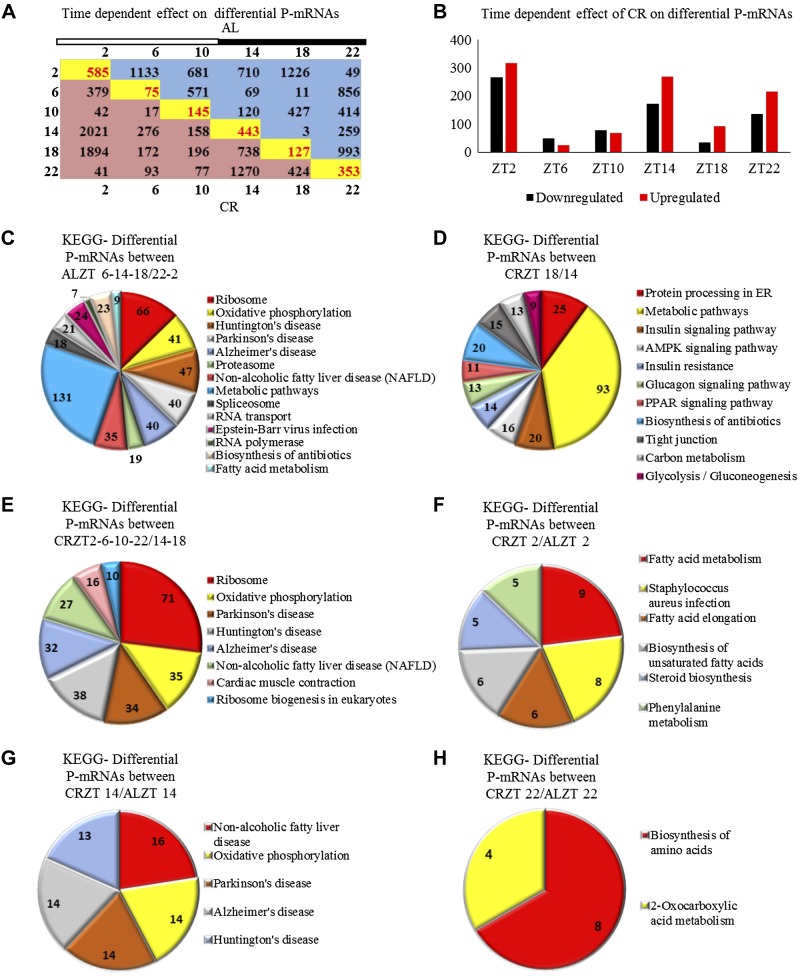
The transcripts translated during the active and resting phase of the day might be different. Rhythmically translated transcripts have been analyzed in the previous section. Arrhythmic transcripts might also be affected by CR, but these changes could be missed through the global comparison presented in Fig. 1B, C. To understand the effect of time and diet on differential translation, we compared the abundance of P-mRNAs in AL and CR animals at all 6 individual time points across the day. In this analysis, we did not differentiate between rhythmic and arrhythmic P-mRNAs.
First, we compared the effect of time of the day on P-mRNA abundance within the AL and CR groups (Fig. 4A). For both diets, different pathways were enriched at every time point of the day. Interestingly, the circadian times can be clustered based on their difference/similarity. For the AL diet, based on the number of P-mRNAs differentially associated with polysomes, ZT2 and ZT22 formed a close group with a low number of P-mRNAs different (only 49 transcripts are different) between these times (Fig. 4A); another group was formed by ZT6, ZT14, and ZT18 (the numbers of differential transcripts are 69, 11, or 3 as shown in Fig. 4A). The number of differential P-mRNAs between these 2 groups were high (from 259 for ZT14 vs. ZT22 and 993 for ZT22 vs. ZT18 (as shown in Fig. 4A). Differential abundance comparison between these groups was performed followed by KEGG analysis. Ribosomal protein synthesis, oxidative phosphorylation, nonalcoholic fatty liver disease, and spliceosome were among the pathways that were revealed to be enriched between these 2 clustered groups (Fig. 4C). ZT10 was moderately different from both the above mentioned groups (no significant pathways were enriched upon comparing ZT10 with the group formed of ZT2, ZT6, ZT14, ZT18, and ZT22 clusters). Similarly, for CR animals, ZT2, ZT6, ZT10, and ZT22 formed a group with low numbers of P-mRNAs different between these times (except for the difference between ZT2 and ZT6—379 as shown in Fig. 4A). The second group was formed by ZT14 and ZT18. The time of feeding, ZT14, and the time just after feeding, ZT18, showed high numbers of P-mRNAs different from ZT2 (>1500) and ZT22 (several hundred as shown in Fig. 4A). ZT14 and ZT18 were moderately different from ZT6 and ZT10 groups (Fig. 4A). Differential abundance comparison between these groups revealed ribosomal subunit protein synthesis, oxidative phosphorylation, and enzymes involved in ribosomal biogenesis as the enriched pathways (Fig. 4E). Apparently, the difference in P-mRNA abundance was also high between ZT14 and ZT18 (738 transcripts Fig. 4A). Interestingly, differentially abundant P-mRNA’s between ZT14 and ZT18 were found to be involved in insulin signaling, insulin resistance, and AMPK signaling (Fig. 4D).
Figure 4.
CR significantly changed temporal translation in the liver. A) Time-of-day–dependent differential abundance of P-mRNAs in the liver polysomes of AL (blue) and CR (red) animals. Numbers of transcripts were obtained by comparing time points on the x axis with the y axis. Time-dependent effect of CR diet is shown in yellow boxes where AL was compared with CR for the respective time point. Light/dark bar at the top represents light and dark phases of the day. B) Time-dependent effect of CR on P-mRNA abundance in polysomes. Black bars show number of P-mRNA, whose abundance was decreased, and red bars show number of P-mRNAs, whose abundance was increased at a particular time of the day in liver polysome when CR animals were compared with AL animals (n = 2 for every time point for each diet). C) Enriched pathways between AL ZT6-14-18 and AL ZT2-22. D) The enriched pathways between CR ZT18 and CR ZT14. E) Enriched pathways between CR ZT2-6-10-22 and CR ZT14-18. F) Unique enriched pathways between CR and AL at ZT2. G) Unique enriched pathways between CR and AL at ZT14. H) Unique enriched pathways between CR and AL at ZT22. Light is on at ZT0, and off at ZT12.
We also compared AL and CR animals at every time followed by KEGG pathway analysis to find unique pathways enriched at every time point across the day as a result of CR treatment. We found that there is a significant effect of diet on translation at every time point. Abundance for many P-mRNAs was found to be increased or decreased (Fig. 4B). The largest difference in P-mRNA abundance between CR and AL group was found to be at ZT2; these differentially abundant transcripts are involved in fatty acid metabolism and biosynthesis of unsaturated fatty acids (Fig. 4F). Differentially abundant P-mRNA numbers were also high at ZT14, and genes of these P-mRNAs were involved in oxidative phosphorylation and nonalcoholic fatty liver disease (Fig. 4G). For ZT22, pathways enriched were biosynthesis of amino acids and 2-oxocarboxylic acid metabolism (Fig. 4H). A minimal difference between the diets was observed at ZT6 (Fig. 4B).
CR regulated the translation of enzymes regulating long-chain Acyl-CoA metabolism
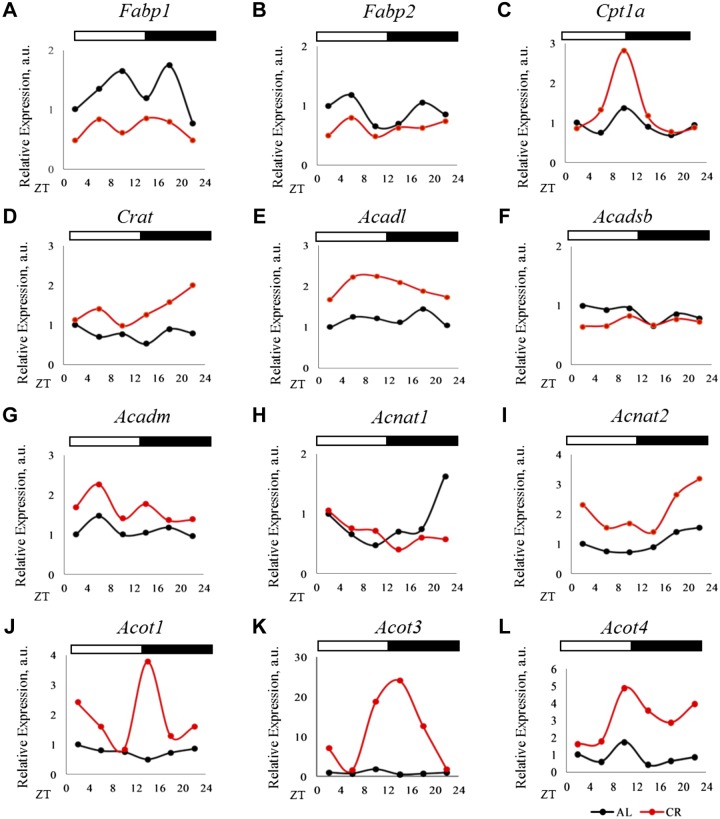
Changes in the expression of many lipid metabolic genes under CR were reported (39, 40) and we observed similar effects of CR on their P-mRNAs. We found enrichment of transcripts that were involved in long-chain Acyl-CoA metabolism. Importantly, many of these transcripts were not previously associated with CR. Lipid chaperons known as fatty acid binding proteins (FABPs) regulate lipid transport and availability (41). Under CR, we observed reduction in P-mRNA for the liver-specific Fabp1 and no change for Fabp2 P-mRNA (Fig. 5A, B). Carnitine acetyltransferases regulate the transport of fatty acids into the mitochondria. P-mRNA for carnitine palmitoyltransferase 1A (Cpt1a), which preferentially transports long-chain fatty acids into the mitochondria, was increased during the late stage of the dietary restriction period (Fig. 5C), whereas increased P-mRNA for carnitine acetyltransferase (42) (preferentially transport short chain) was observed after feeding (Fig. 5D). Enzymes from Acyl-CoA dehydrogenases family control the rate-limiting step in β-oxidation of fatty acids in mitochondria (43). We observed induction of P-mRNA for long-chain Acyl-CoA dehydrogenases (Acadl) (Fig. 5E), but not for Acyl-CoA dehydrogenase, short/branch chain (Fig. 5F). Time-dependent induction was observed for Acyl-CoA dehydrogenase, medium chain P-mRNA (Fig. 5G). N-acyltransferases [bile acyl-CoA: amino acid N-acyltransferase (BAAT), Acyl-CoA amino acid N-acyltransferase 1 (ACNAT1), and ACNAT2] conjugate Acyl-CoA molecules with glycine and taurine and release CoA inside the peroxisomes. Substrates for ACNAT1 are long chain and very long chain Acyl-CoA molecules, whereas, substrate for ACNAT2 is not yet known (44). We observed induction of Acnats at several time points (Fig. 5H, I). ACOT enzymes hydrolyze Acyl-CoA molecules to free fatty acids (FFAs) and CoA-SH (45–47). We observed strong time-dependent induction in the P-mRNA for Acot1, 3, and 4 (Fig. 5J–L), and these enzymes act on long-chain Acyl-CoAs, succinyl-CoA, and glutaryl-CoA (44). The P-mRNAs of other family members were not significantly affected. Interestingly, the peak induction in the P-mRNA for ACOTs was at the end of the rest/dietary restriction period, and for ACNAT1 at the end of the active period. Therefore, CR significantly changed the translation for several rate-limiting enzymes involved in very long and long-chain Acyl-CoA metabolism, and the effect of CR on enzymes involved in short-chain Acyl-CoA metabolism was minimal.
Figure 5.
CR regulated the translation of enzymes regulating long-chain Acyl-CoA metabolism. A, B) P-mRNA abundance of FABPs. Fabp1 and Fabp2 in the liver polysomes of AL and CR animals. C, D) P-mRNA abundance for acyltransferases, Cpt1a and carnitine acetyltransferase (Crat) in the liver polysomes of AL and CR animals. E–G) P-mRNA abundance for Acyl-CoA dehydrogenase family proteins—Acadl, Acyl-CoA dehydrogenase, short/branch chain, and Acyl-CoA dehydrogenase, medium chain—in the liver polysomes of AL and CR animals. H, I) P-mRNA abundance for N-acyltransferases—Acnat1 and Acnat2—in the liver polysomes of AL and CR animals. J–L) P-mRNA abundance for Acot1, Acot3, and Acot4 in the liver polysomes of AL and CR animals. Light/dark bar at the top represents light and dark phases of the day; light is on at ZT0, and off at ZT12. Two biologic replicates for each time point of AL animals were compared to those of CR for differential P-mRNA abundance using Deseq2 software. Log2 values for the fold change thus obtained were used to plot graphs (n = 2 for each time point for each diet).
CR induced the translation of proteins from ACOT family
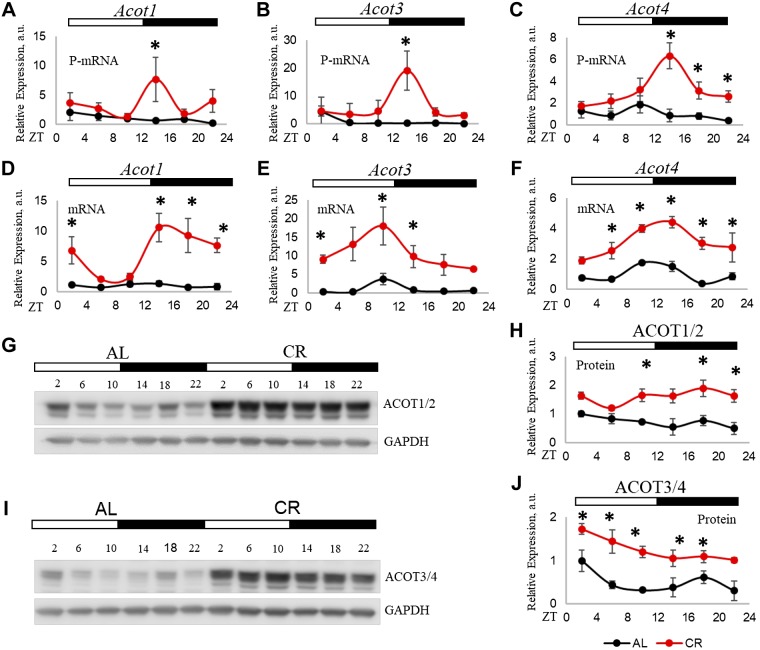
We decided to study the effect of CR on the expression of ACOT family. First, we performed PCR on polysomal fraction and confirmed the results of RNA-seq. In the AL group, P-mRNA abundance for Acot3, Acot4, and Acot1 P-mRNA was arrhythmic. Under CR treatment, Acot4 became rhythmic. CR has 2 main effects on the Acot P-mRNAs: the induction at several times, with the strongest effect at ZT14, and phase shift (Fig. 6A–C). Under CR, all 3 Acots peaked at ZT14, while under AL, Acot1 and Acot3 peaked at ZT2, and Acot4 peaked at ZT10. The food for the CR group was provided at ZT14 during the experiment, and on the day of sample collection, the samples at ZT14 time point were collected before the food was provided. Thus, the peaks in Acot P-mRNAs were not an acute response to the feeding but, rather, represented the entrainment and food anticipation. mRNA expressions for Acots were affected by CR (Fig. 6D–F); CR did not change the peaks but significantly induced mRNA expression for all 3 genes. CR-induced P-mRNAs to some extent correlates with the changes in their corresponding mRNA levels. However, P-mRNAs for all 3, Acot1, Acot3, and Acot4, peaked at the same time, at ZT14 under CR (Fig. 6A–C), whereas, mRNAs reached peaks at different times: ZT14 for Acot1 and Acot4 and ZT10 for Acot3 under CR (Fig. 6D–F). To further investigate if changes in P-mRNA abundance would result in changes in protein abundance, we performed Western blotting for ACOT1 and ACOT4. We found that in agreement with the increased translation, the expression of ACOT proteins was significantly induced by CR (Fig. 6G–J). Some difference between daily translation and protein abundance might be due to the different half-lives of the proteins. In addition, it is important to mention that, because of high-sequence homology and almost identical MW between ACOT1 and ACOT2 and between ACOT3 and ACOT4, it is possible that the antibodies cross-reacted, and the bands might represent the mixed signals of respective closely identical proteins.
Figure 6.
CR induced the translation of proteins from ACOT family. A–C) P-mRNA abundance of Acot1, Acot3, and Acot4 as assayed by real-time quantitative PCR in the liver polysome of CR and AL animals. The β-actin gene was used as control. D–F) mRNA expression of Acot1, Acot3, and Acot4 in total RNA from the liver of CR and AL animals. The 18S gene was used as a control. G) Total ACOT1 protein level, assayed by Western blot, in the liver of CR and AL animals. GAPDH protein was used as a loading control. H) Quantification of ACOT1 protein levels. I) Total ACOT4 protein level, assayed by Western blot, in the liver of CR and AL animals. GAPDH protein was used as a loading control. J) Quantification of ACOT3/4 protein levels. Three biologic replicates (n = 3 for each time point for each diet) were analyzed. Two-way ANOVA was performed using GraphPad Prism for statistical analysis. *P < 0.05.
CR-induced daily rhythms in peroxisome proliferator-activated receptor α transcriptional activity
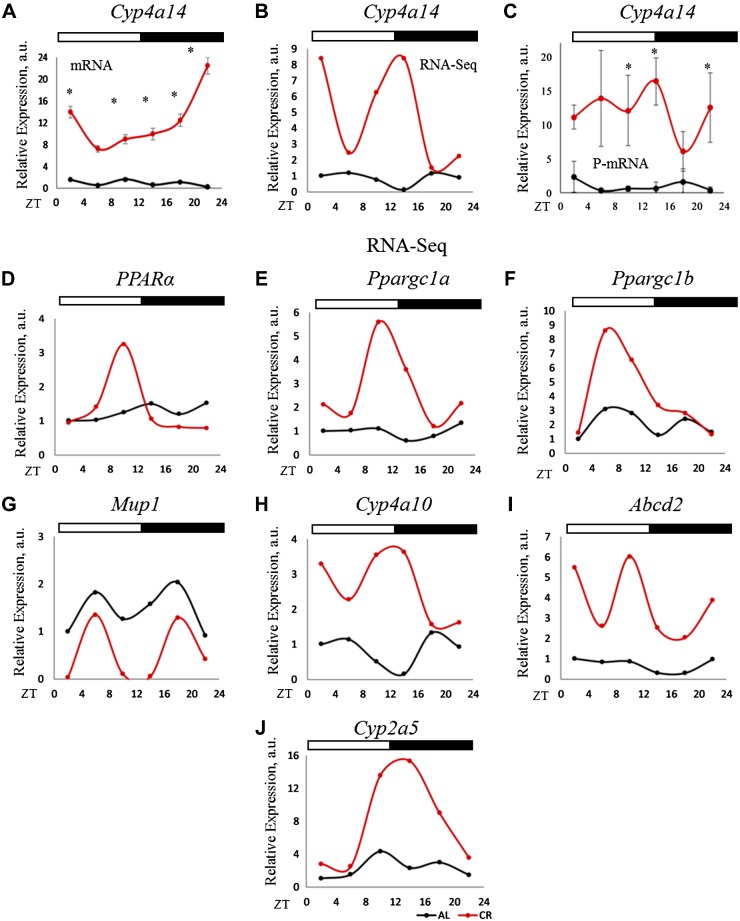
Cytoplasmic ACOT1 removes CoA enzyme from Acyl-CoA molecule to generate FFAs. Some of these FFAs serve as ligands to peroxisome proliferator-activated receptor α (PPARα) and regulate its transcriptional activity. We predicted that the increased ACOT1 expression will result in increased PPARα transcriptional activity. Acots are transcriptional targets of PPARα, and the increased mRNA of Acots expression under CR (Fig. 6D–F) is also in agreement with increased PPARα activation. We assayed the expression of another PPARα target gene Cyp4a14, which is a member of the cytochrome P450 superfamily. PCR assay confirms the up-regulation of Cyp4a14 mRNA level (Fig. 7A). Cyp4a14 expression was significantly up-regulated under CR according to mRNA-seq data (Fig. 7B), and PCR for Cyp4a14 P-mRNA level in the polysomal fraction (Fig. 7C) confirmed the findings of the RNA-seq data (Fig. 7B). Changes in P-mRNAs abundance for several other known PPARα targets were assayed. As it was expected, the P-mRNAs for Cyp4a10, Abcd2, and Cyp2a5 (Fig. 7H–J) were induced and for Mup1 (Fig. 7G) was suppressed under CR (Supplemental Fig. S2) (48–50). We also investigated polysome association of PPARγ coactivators: Ppargc1a and Ppargc1b transcripts. Polysome association for these transcripts were induced at ZT10 for Ppargc1a, and ZT6 for Ppargc1b by CR (Fig. 7E, F). Thus, CR significantly induced the activity of transcriptional factor PPARα and increased ACOT1 expression, which might be one of the contributing factors.
Figure 7.
P-mRNA and mRNA abundance for Cyp4a14, a PPARα-target gene. A) mRNA abundance for Cyp4a14 in the total RNA from the liver of CR and AL animals (n = 3 for each time point for each diet). B) mRNA-seq data for P-mRNA of Cyp4a14, a target of PPARα transcription factor, in the liver polysomes of CR and AL animals (n = 2 for every time point for each diet). C) Validation of mRNA-seq data of Cyp4a14 P-mRNA (n = 3 for every time point for each diet). The 18S gene was used as control for total RNA, whereas β-actin was used for P-mRNA expression analysis. Two-way ANOVA was performed using GraphPad Prism, and a value of P < 0.05 was considered significant. D) P-mRNA of PPARα in the liver polysomes of AL and CR and animals. E–J) P-mRNA of PPARα target genes in the liver polysomes of AL and CR and animals. Two biologic replicates for each time point of AL animals were compared to that of CR for differential P-mRNA abundance using Deseq2 software. Log2 values for the fold change thus obtained were used to plot graphs (n = 2 for each time point for each diet). Light/dark bars at the top represents light and dark phases of the day (light on at ZT0 and off at ZT12). *P < 0.05.
DISCUSSION
To extend the knowledge on the molecular mechanisms of CR and circadian clock interaction, we performed polysome profiling in the liver of mice on AL and CR diets across the day. The analysis revealed several major impacts of CR on liver metabolism. First, CR did not significantly affect the translation of the core circadian clock proteins. Some changes in the absolute level of translation for most of clock transcripts were in agreement with changes in the abundance of the appropriate mRNAs with exception for Clock and Bmal1. Thus, we did not observe any disruption of the rhythms in circadian clock genes, most likely, because the food was provided during physiologically active periods. In contrast to the effect on the core clock components, CR significantly reprogramed the circadian output in translation. Under AL and CR, there were similar fractions of transcripts that were rhythmically associated with polysomes but only a small fraction of genes was rhythmic on both diets. Atger et al. (36) recently compared ribosome associated transcripts for the night time restricted feeding and AL diets in the mouse liver. They observed an increased amplitude in rhythm but did not report significant change in the pattern of rhythms, which is a big contrast to our data. Thus, the changes in the temporal rhythms of the polysomal transcripts induced by CR are very different from the effect of time-restricted feeding. The difference in the approaches might also contribute to the difference in results between the studies: Atger et al. used ribosomal footprint and we used polysome profiling.
What is the driving force for the observed rhythms in P-mRNAs? It was recently reported that under AL conditions about 70% of rhythmically transcribed mRNAs are also rhythmically associated with ribosomes (36). The recent analysis of circadian hepatic transcriptome revealed that about 1900 genes oscillate exclusively under AL, 4000 genes oscillate exclusively under CR conditions, and 2300 genes oscillate at both conditions (16). Thus, more than 50% of transcripts rhythmic under AL diets are rhythmic under both diets. In our study, <20% of polysomal transcripts rhythmic under AL diets were still rhythmic under CR. We expect that at least some of the changes in polysomal transcripts induced by CR are a direct consequence of the changes in transcript abundance, and other mechanisms might exist as well. For example, under CR, the mRNA for Acot3 peaked at ZT10 and P-mRNA peaked at ZT14. Of note, we cannot directly compare our data with Sato et al. (16) because of the difference in age of mice (22 wk in our study vs. 46–56 wk in Sato et al.), duration of the CR (2 mo in our study vs. 6 mo in Sato et al.), and time of feeding (ZT14 in our study vs. ZT12 in Sato et al.).
Another important change induced by CR is the effect on the translation of proteins involved in different metabolic pathways. These changes correlate with known changes in physiology induced by CR: for example, changes in AMPK signaling correlate with improved glucose metabolism (37, 51–53). One of the noticeable pathways associated was ribosomal protein synthesis. This pathway was rhythmic under AL, which supports previously reported circadian rhythms in ribosome biogenesis (35), these rhythms were proposed to be an important driving force for the rhythms in translation (54). CR is known to change protein homeostasis, but there is some controversy on the CR-induced changes in protein synthesis (55–57). Ribosomal biogenesis was previously linked to CR (58). We found that under CR rhythms in ribosomal protein, synthesis was ceased. The ribosome biogenesis is an energy-consuming process, and ceased rhythms in ribosomal protein synthesis suggest that under conditions of limited resources cells select different strategies for rhythmic control of translation, which warrants further investigation. Interestingly, the recent study on the effect of CR on transcriptome and proteome in the liver of rhesus monkey revealed significant changes in ribosomal gene expression, but transcript and protein data are not in agreement (20). Our data suggest that CR also regulates translation of ribosomal proteins, thus, providing a potential explanation for the discrepancy discussed above. Importantly, while only a small number of transcripts were affected by CR across the day, a significantly higher magnitude of CR-induced changes in differential translation were detected at every circadian time. The circadian times also affected the polysome association for many transcripts for both AL and CR diets. Therefore, CR caused temporal partition of metabolic processes through differential translation of transcripts involved in various metabolic pathways across the day. For example, P-mRNAs linked with oxidative phosphorylation and peroxisomes were differentially associated with polysomes between ZT2 and ZT14 for CR, when the main sources of energy for liver is fat, whereas P-mRNAs for insulin signaling are enriched at ZT18 when carbohydrates are utilized as a prime source to produce energy. The effect of CR on energy metabolism is well documented, and multiple transcriptome analyses demonstrated that CR causes changes in transcription of the components of TCA cycle and mitochondria electron transport chain (27). Our results revealed an additional level of regulation and provided further mechanistic links through temporal control of differential translation.
Previous studies in Drosophila (15) and mammals (59) revealed strong effects of CR on fat metabolism. Our study revealed a strong effect on the expression of enzymes involved in long-chain Acyl-CoA metabolism. We identified type I ACOTs as potential targets of CR. ACOTs are enzymes that hydrolyze Acyl-CoA to produce CoA and FFAs. The expression of Acots is regulated by different diets: high-fat diet, ketogenic diet, and dietary restriction (47, 60). Acot1-6 genes form a cluster on mouse chromosome 12, and it was proposed that they are expressed in synchrony and transcription of the whole cluster is regulated by PPARα and by bZip transcriptional factors (49). We observed in our study an increased mRNA expression for Acots, which may be a consequence of the increased PPARα activity under CR, but it was reported recently that Acot1 expression in the liver can be induced in PPARα independent manner (61). It was reported that the Acot1 mRNA expression increases during dietary restriction and decreases in response to refeeding (61). CR animals were at least 20 h postprandial, and therefore, we cannot exclude the possibility that increased expression was an adaptation to prolonged dietary restriction. In our study, under CR diet, the level of ACOT1 protein did not decrease at ZT18 (after the feeding) and it was lower at ZT6-10 (unfed state). Thus, there is a difference in the regulation of Acot1 expression between CR and dietary restriction. The changes in Acot1, Acot3, and Acot4 polysome association can be partially explained through the changes in mRNA abundance; at the same time, the daily profiles of P-mRNA and mRNA abundances are not the same, suggesting that CR might regulate the expression of some of these genes at the level of translation. We also cannot exclude the regulation at the level of protein stability because there is no absolute correlation between changes in ACOT protein level and appropriate P-mRNA. Thus, we propose that Acot expression is regulated by CR on several levels: transcription, translation, and protein stability.
The circadian clock is a master regulator of metabolism (18). Recent data suggest that the circadian clock plays an important role in mitochondrial energy production. Under dietary restriction conditions, mitochondrial fat oxidation is under strong circadian control; mechanistically, the clock was linked with oxidative phosphorylation through NAD+/SIRTUIN-3–dependent deacetylation of mitochondria oxidative enzymes (62). CR is periodic dietary restriction, and it was previously reported that CR recruits the circadian clock. In the current study, the phases of clock gene translation were not affected by CR, but the amplitudes were significantly enhanced in agreement with previously reported mRNA data (14). We speculate that CR might enhance the amplitude of circadian clock output in gene expression and through that might facilitate mitochondrial oxidative functions.
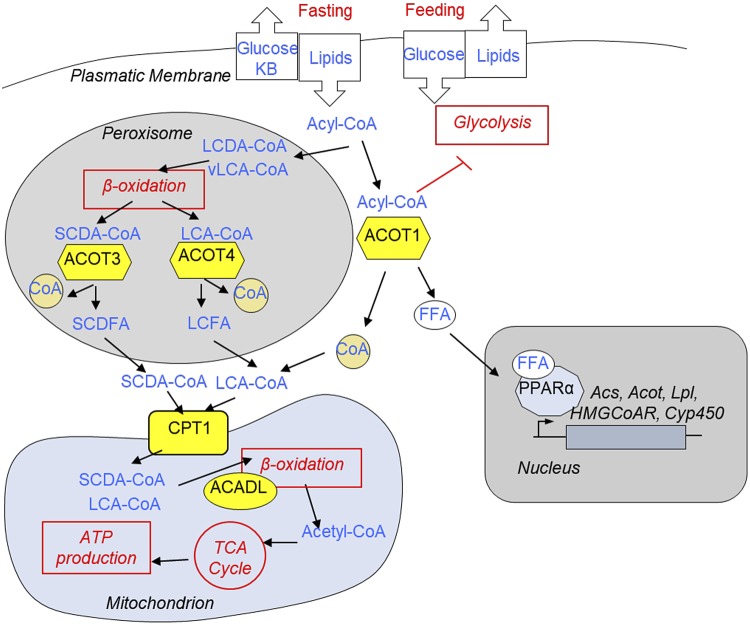
What is the physiologic importance of the CR-induced changes in ACOT expression? We have proposed the following model (Fig. 8). The expression of enzymes involved in fat metabolism is a subject for circadian regulation. The effect of CR on translation of many enzymes involved in fat metabolism was also time-of-day dependent. We hypothesized that this temporal regulation is important for the transition between feeding and dietary restriction under CR diet. ACOT3 and ACOT4 are localized in the peroxisomes. Peroxisomes oxidize very long-chain Acyl-CoA (vLCA-CoA) and long-chain dicarboxylic Acyl-CoA (LCDA-CoA) (44, 63). This β-oxidation is not coupled with ATP production. vLCA-CoA and LCDA-CoA get oxidized in peroxisomes to generate long-chain Acyl-CoA (LCA-CoA) and short-chain dicarboxylic Acyl-CoA. It was proposed that these Acyl-CoAs are further hydrolyzed inside peroxisomes by ACOT3 (LCA-CoA) and ACOT4 (short-chain dicarboxylic Acyl-CoA) to their inactive forms and produce FFAs and CoA enzyme (44). In agreement with this, the translation of peroxisome-localized ACOTs occurred between ZT14 and ZT18, and the maximum of their protein levels was observed at ZT2-ZT6, when energy production is switched from carbohydrate to fat sources. These FFAs are exported from peroxisomes, get activated in the cytoplasm, and these activated long Acyl-CoA molecules are transported into the mitochondria by long-chain Acyl-CoA–specific Cpt1, an abundance of P-mRNA for Cpt1 was also induced by CR at ZT10 when maximum transport of fatty acids to mitochondria might be expected. In mitochondria, the β-oxidation is coupled with ATP production, and in agreement with that, the translation of rate-limiting Acadl has also been increased under CR across the day with maximum at ZT6-10. Thus, under nutrient-limited conditions, coordinated increased expression of peroxisomal ACOTs and mitochondrial enzymes help to utilize vLCA-CoA and LCDA-CoA for ATP production. Importantly, peroxisomal enzymes from Acnat family use similar Acyl-CoA molecules as substrates to produce Acyl-taurines (44). Thus, ACOTs and ACNAT enzymes compete for similar substrate, and their translation occurs in antiphase: Acots peaked at ZT10-ZT14, and Acnats peaked at ZT22-ZT2. Thus, their translation is timely partitioned to minimize the competition for similar substrate.
Figure 8.
Reprogramming of fatty acid metabolism by CR (hypothesis). CR is a state of periodic feeding and dietary restriction. During dietary restriction, there is significant influx of fatty acids in the liver. Activated fatty acids—Acyl-CoA—can be used to produce ketone bodies or oxidized in mitochondria to generate ATP for hepatocyte homeostasis and to fuel gluconeogenesis. vLCA-CoA and LCDA-CoA molecules are oxidized in the peroxisomes. It was proposed that after few rounds of carbon chain shortening of vLCA-CoA and LCDA-CoA, ACOTs would localize in peroxisome (i.e., ACOT3 and ACOT4) hydrolyze LCA-CoA and SCDFA-CoA. Generated short-chain and long-chain FFA molecules leave the peroxisome, get activated in the cytoplasm and through carnitine dependent shuttle (catalyzed by CPT1), and enter into the mitochondria. In mitochondria, Acyl-CoA molecules undergo β-oxidation (ACADL catalyzes the rate-limiting step), and generated Acyl-CoA enters the TCA cycle to produce ATPs. Cytoplasmic ACOT1 hydrolyzes Acyl-CoA into FFAs and CoA. It was proposed that this can serve several purposes: limit the supply of Acyl-CoA to mitochondria and reduce the level of cytoplasmic acyl-CoA molecules known to inhibit glycolysis, thus contributing to metabolic flexibility. Finally, it will provide FFAs that serve as ligands for nuclear receptor PPARα, master regulator of fat metabolism. Only the enzymes whose translation has been affected by CR in our study (yellow) are presented in the figure.
ACOT1 is localized in the cytoplasm and known to regulate fat metabolism (61). Under CR diet, mice consume the provided food in 2–3 h; therefore, the unfed period is about 21 h. During the unfed period in the liver, fat molecules are used for β-oxidation and to generate ketone bodies. For these processes, fatty acids must be activated (converted to Acyl-CoA), but hydrolysis of Acyl-CoA into inactive FFA by CR-induced ACOT1 seems counterintuitive. We propose the following explanation to this observation: It was proposed that during dietary restriction, significant influx of fatty acids may result in the excessive accumulation of Acyl-CoA esters and intermediate metabolites of fatty acid oxidation (FAO), compromising the FAO (64, 65). ACOT1 limits the supply of Acyl-CoA molecules available for β-oxidation and keep this process in synchrony with the demands of the tricarboxylic acid cycle and give enough time for the oxidative phosphorylation complex to adjust accordingly. In agreement, the translation of lipid chaperon FABP1 (41), which is involved in fatty acid transport, is down-regulated under CR (Fig. 6A). At the same time when an animal is about to receive food, high level of Acyl-CoA could lead to the inhibition of glycolysis (66) and interfere with glucose uptake and limit metabolic flexibility at the time when liver must prepare to switch from fat to carbohydrate metabolism. In agreement with that, the translation of ACOT1 peaked at ZT14, and the expression of ACOT1 protein peaked at ZT18.Therefore, ACOT1 helps to prepare the liver for the consumption of carbohydrates when animals are about to expect food. Finally, products of ACOT1 enzymatic activity (i.e., FFAs) serve as ligands for PPARα, regulating its transcriptional activity. In support of this, we observed increased mRNA levels and increased polysome association of mRNAs for several well-known targets of PPARα (Fig. 7). Further studies will help to understand if any of these hypothetical scenarios occur. Potential importance of ACOTs in CR mechanisms is coming from the observation that mice with significantly reduced expression of Acot1-6 genes (mice triple deficient for transcriptional factors in bZIp family) fail to adapt to CR (49).
Our study has several limitations that need to be addressed in the future. First, as it was mentioned above, on CR diet, mice consume the provided food in 2–3 h following prolonged dietary restriction; therefore, some of the observed changes might be an adaption to periodic feeding while others could be a specific response to the reduced calorie intake. The experiments with time-restricted feeding might help to address this problem. At the same time, most of published data on metabolic effects of CR were done for CR vs. AL comparison; therefore, the data presented in our study are highly relevant to previous works. Second, the experiments were performed only in wild-type mice and it is unclear if the circadian clock is involved in the observed effects of CR on the rhythms of P-mRNA. The experiments with circadian clock mutants will help to understand the contribution of the circadian clock. Finally, the study was done only with male mice because of the clear sex dependence of many of CR effects (14, 67), including the effects on the rhythms, so a similar study with female mice is important to conduct.
There is more evidence that protein translation is not a simple transition from mRNA to protein but an important regulatory step. In response to the external and internal signals, cells change both global translation and translation of selective proteins. Adaptation to CR requires significant rewiring of many physiologic processes, including chromatin organization, transcription, and, as our data demonstrates, translation. In addition to the previously reported changes in global protein translation, we found that CR-induced changes in the translation of proteins involved in metabolic pathways are known to be affected by CR. Importantly, these changes in translation are time-of-day dependent and link the temporal rhythms with liver response to nutrients.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
This work was supported by U.S. National Institues of Health, National Institute on Aging Grant 5RO1-AG039547 (to R.V.K.), and internal support by the Gene Regulation in Health and Diseases Department at Cleveland State University (to R.V.K.). The authors declare no conflicts of interest.
Glossary
- Acadl
Acyl-CoA dehydrogenases, long-chain
- ACNAT1
Acyl-CoA amino acid N-acyltransferase 1
- ACNAT2
Acyl-CoA amino acid N-acyltransferase 2
- Acyl-CoA
acetyl-coenzyme A
- ACOT
Acyl-CoA thioesterase
- AL
ad libitum
- Cpt1
carnitine palmitoyltransferase 1
- CR
calorie restriction
- FABP
fatty acid binding protein
- FFA
free fatty acid
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LCA-CoA
long-chain Acyl-CoA
- LCDA-CoA
long-chain dicarboxylic acid Acyl-CoA
- mRNA-seq
mRNA sequencing
- PPARα
peroxisome proliferator-activated receptor α
- P-mRNA
polysome-associated mRNA
- RNA-seq
RNA sequencing
- vLCA-CoA
very long-chain Acyl-CoA
- ZT
zeitgeber time
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
K. Makwana and R. V. Kondratov designed experiments and wrote the manuscript; K. Makwana performed all the experiments and data analysis; K. Makwana and N. Gosai prepared all the figures and tables; and K. Makwana, A. Poe, and R. Kondratov reviewed the manuscript.
REFERENCES
- 1.Asher G., Sassone-Corsi P. (2015) Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92 [DOI] [PubMed] [Google Scholar]
- 2.Chaudhari A., Gupta R., Makwana K., Kondratov R. (2017) Circadian clocks, diets and aging. Nutr. Healthy Aging 4, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Froy O. (2013) Circadian aspects of energy metabolism and aging. Ageing Res. Rev. 12, 931–940 [DOI] [PubMed] [Google Scholar]
- 4.Chalasani N., Younossi Z., Lavine J. E., Diehl A. M., Brunt E. M., Cusi K., Charlton M., Sanyal A. J. (2012) The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 55, 2005–2023 [DOI] [PubMed] [Google Scholar]
- 5.Manoogian E. N. C., Panda S. (2017) Circadian rhythms, time-restricted feeding, and healthy aging. Ageing Res. Rev. 39, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ko C. H., Takahashi J. S. (2006) Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 15, R271–R277 [DOI] [PubMed] [Google Scholar]
- 7.Potter G. D. M., Skene D. J., Arendt J., Cade J. E., Grant P. J., Hardie L. J. (2016) Circadian rhythm and sleep disruption: causes, metabolic consequences, and countermeasures. Endocr. Rev. 37, 584–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M.-D., Li C.-M., Wang Z. (2012) The role of circadian clocks in metabolic disease. Yale J. Biol. Med. 85, 387–401 [PMC free article] [PubMed] [Google Scholar]
- 9.Scheer F. A. J. L., Hilton M. F., Mantzoros C. S., Shea S. A. (2009) Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl. Acad. Sci. USA 106, 4453–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savvidis C., Koutsilieris M. (2012) Circadian rhythm disruption in cancer biology. Mol. Med. 18, 1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heilbronn L. K., Ravussin E. (2003) Calorie restriction and aging: review of the literature and implications for studies in humans. Am. J. Clin. Nutr. 78, 361–369 [DOI] [PubMed] [Google Scholar]
- 12.Lee C., Longo V. (2016) Dietary restriction with and without caloric restriction for healthy aging. F1000 Res. 5, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel S. A., Velingkaar N., Makwana K., Chaudhari A., Kondratov R. (2016) Calorie restriction regulates circadian clock gene expression through BMAL1 dependent and independent mechanisms. Sci. Rep. 6, 25970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Astafev A. A., Patel S. A., Kondratov R. V. (2017) Calorie restriction effects on circadian rhythms in gene expression are sex dependent. Sci. Rep. 7, 9716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katewa S. D., Akagi K., Bose N., Rakshit K., Camarella T., Zheng X., Hall D., Davis S., Nelson C. S., Brem R. B., Ramanathan A., Sehgal A., Giebultowicz J. M., Kapahi P. (2016) Peripheral circadian clocks mediate dietary restriction-dependent changes in lifespan and fat metabolism in Drosophila. Cell Metab. 23, 143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato S., Solanas G., Peixoto F. O., Bee L., Symeonidi A., Schmidt M. S., Brenner C., Masri S., Benitah S. A., Sassone-Corsi P. (2017) Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell 170, 664–677.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acosta-Rodríguez V. A., de Groot M. H. M., Rijo-Ferreira F., Green C. B., Takahashi J. S. (2017) Mice under caloric restriction self-impose a temporal restriction of food intake as revealed by an automated feeder system. Cell Metab. 26, 267–277.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bass J. (2012) Circadian topology of metabolism. Nature 491, 348–356 [DOI] [PubMed] [Google Scholar]
- 19.Solanas G., Peixoto F. O., Perdiguero E., Jardí M., Ruiz-Bonilla V., Datta D., Symeonidi A., Castellanos A., Welz P. S., Caballero J. M., Sassone-Corsi P., Muñoz-Cánoves P., Benitah S. A. (2017) Aged stem cells reprogram their daily rhythmic functions to adapt to stress. Cell 170, 678–692.e20 [DOI] [PubMed] [Google Scholar]
- 20.Rhoads T. W., Burhans M. S., Chen V. B., Hutchins P. D., Rush M. J. P., Clark J. P., Stark J. L., McIlwain S. J., Eghbalnia H. R., Pavelec D. M., Ong I. M., Denu J. M., Markley J. L., Coon J. J., Colman R. J., Anderson R. M. (2018) Caloric restriction engages hepatic RNA processing mechanisms in rhesus monkeys. Cell Metab. 27, 677–688.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swindell W. R. (2009) Genes and gene expression modules associated with caloric restriction and aging in the laboratory mouse. BMC Genomics 10, 585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jové M., Naudí A., Ramírez-Núñez O., Portero-Otín M., Selman C., Withers D. J., Pamplona R. (2014) Caloric restriction reveals a metabolomic and lipidomic signature in liver of male mice. Aging Cell 13, 828–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maier T., Güell M., Serrano L. (2009) Correlation of mRNA and protein in complex biological samples. FEBS Lett. 583, 3966–3973 [DOI] [PubMed] [Google Scholar]
- 24.Iwasaki S., Ingolia N. T. (2017) The growing toolbox for protein synthesis studies. Trends Biochem. Sci. 42, 612–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dermit M., Dodel M., Mardakheh F. K. (2017) Methods for monitoring and measurement of protein translation in time and space. Mol. Biosyst. 13, 2477–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chassé H., Boulben S., Costache V., Cormier P., Morales J. (2017) Analysis of translation using polysome profiling. Nucleic Acids Res. 45, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson R. M., Weindruch R. (2010) Metabolic reprogramming, caloric restriction and aging. Trends Endocrinol. Metab. 21, 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S., Huang T., Zhou Y., Han Y., Xu M., Gu J. (2017) AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinformatics 18 (Suppl 3), 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S., Pyl P. T., Huber W. (2015) HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love M. I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes M. E., Hogenesch J. B., Kornacker K. (2010) JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms 25, 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melamed D., Arava Y. (2007) Genome-wide analysis of mRNA polysomal profiles with spotted DNA microarrays. Methods Enzymol. 431, 177–201 [DOI] [PubMed] [Google Scholar]
- 34.Mitchell S. J., Madrigal-Matute J., Scheibye-Knudsen M., Fang E., Aon M., González-Reyes J. A., Cortassa S., Kaushik S., Gonzalez-Freire M., Patel B., Wahl D., Ali A., Calvo-Rubio M., Burón M. I., Guiterrez V., Ward T. M., Palacios H. H., Cai H., Frederick D. W., Hine C., Broeskamp F., Habering L., Dawson J., Beasley T. M., Wan J., Ikeno Y., Hubbard G., Becker K. G., Zhang Y., Bohr V. A., Longo D. L., Navas P., Ferrucci L., Sinclair D. A., Cohen P., Egan J. M., Mitchell J. R., Baur J. A., Allison D. B., Anson R. M., Villalba J. M., Madeo F., Cuervo A. M., Pearson K. J., Ingram D. K., Bernier M., de Cabo R. (2016) Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab. 23, 1093–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jouffe C., Cretenet G., Symul L., Martin E., Atger F., Naef F., Gachon F. (2013) The circadian clock coordinates ribosome biogenesis. PLoS Biol. 11, e1001455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atger F., Gobet C., Marquis J., Martin E., Wang J., Weger B., Lefebvre G., Descombes P., Naef F., Gachon F. (2015) Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc. Natl. Acad. Sci. USA 112, E6579–E6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cantó C., Auwerx J. (2011) Calorie restriction: is AMPK a key sensor and effector? Physiology (Bethesda) 26, 214–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hernandez-Rodas M. C., Valenzuela R., Videla L. A. (2015) Relevant aspects of nutritional and dietary interventions in non-alcoholic fatty liver disease. Int. J. Mol. Sci. 16, 25168–25198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulkarni S. R., Armstrong L. E., Slitt A. L. (2013) Caloric restriction-mediated induction of lipid metabolism gene expression in liver is enhanced by Keap1-knockdown. Pharm. Res. 30, 2221–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park C. Y., Park S., Kim M. S., Kim H.-K., Han S. N. (2017) Effects of mild calorie restriction on lipid metabolism and inflammation in liver and adipose tissue. Biochem. Biophys. Res. Commun. 490, 636–642 [DOI] [PubMed] [Google Scholar]
- 41.Furuhashi M., Hotamisligil G. S. (2008) Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 7, 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsiao Y. S., Jogl G., Tong L. (2004) Structural and biochemical studies of the substrate selectivity of carnitine acetyltransferase. J. Biol. Chem. 279, 31584–31589 [DOI] [PubMed] [Google Scholar]
- 43.Swigonová Z., Mohsen A. W., Vockley J. (2009) Acyl-CoA dehydrogenases: dynamic history of protein family evolution. J. Mol. Evol. 69, 176–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunt M. C., Siponen M. I., Alexson S. E. (2012) The emerging role of acyl-CoA thioesterases and acyltransferases in regulating peroxisomal lipid metabolism. Biochim. Biophys. Acta 1822, 1397–1410 [DOI] [PubMed] [Google Scholar]
- 45.Hunt M. C., Rautanen A., Westin M. A., Svensson L. T., Alexson S. E. (2006) Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional evolution results in a reduced number of human peroxisomal ACOTs. FASEB J. 20, 1855–1864 [DOI] [PubMed] [Google Scholar]
- 46.Brocker C., Carpenter C., Nebert D. W., Vasiliou V. (2010) Evolutionary divergence and functions of the human acyl-CoA thioesterase gene (ACOT) family. Hum. Genomics 4, 411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellis J. M., Bowman C. E., Wolfgang M. J. (2015) Metabolic and tissue-specific regulation of acyl-CoA metabolism. PLoS One 10, e0116587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rakhshandehroo M., Knoch B., Michael M., Kersten S. (2010) Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010, 612089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gachon F., Leuenberger N., Claudel T., Gos P., Jouffe C., Fleury Olela F., de Mollerat du Jeu X., Wahli W., Schibler U. (2011) Proline- and acidic amino acid-rich basic leucine zipper proteins modulate peroxisome proliferator-activated receptor alpha (PPARalpha) activity. Proc. Natl. Acad. Sci. USA 108, 4794–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mandard S., Müller M., Kersten S. (2004) Peroxisome proliferator-activated receptor alpha target genes. Cell. Mol. Life Sci. 61, 393–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss E. P., Racette S. B., Villareal D. T., Fontana L., Steger-May K., Schechtman K. B., Klein S., Holloszy J. O.; Washington University School of Medicine CALERIE Group (2006) Improvements in glucose tolerance and insulin action induced by increasing energy expenditure or decreasing energy intake: a randomized controlled trial. Am. J. Clin. Nutr. 84, 1033–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larson-Meyer D. E., Heilbronn L. K., Redman L. M., Newcomer B. R., Frisard M. I., Anton S., Smith S. R., Alfonso A., Ravussin E. (2006) Effect of calorie restriction with or without exercise on insulin sensitivity, beta-cell function, fat cell size, and ectopic lipid in overweight subjects. Diabetes Care 29, 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Long Y. C., Zierath J. R. (2006) AMP-activated protein kinase signaling in metabolic regulation. J. Clin. Invest. 116, 1776–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinturel F., Gerber A., Mauvoisin D., Wang J., Gatfield D., Stubblefield J. J., Green C. B., Gachon F., Schibler U. (2017) Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell 169, 651–663.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D’Costa A. P., Lenham J. E., Ingram R. L., Sonntag W. E. (1993) Moderate caloric restriction increases type 1 IGF receptors and protein synthesis in aging rats. Mech. Ageing Dev. 71, 59–71 [DOI] [PubMed] [Google Scholar]
- 56.Miller B. F., Robinson M. M., Reuland D. J., Drake J. C., Peelor F. F., III, Bruss M. D., Hellerstein M. K., Hamilton K. L. (2013) Calorie restriction does not increase short-term or long-term protein synthesis. J. Gerontol. A Biol. Sci. Med. Sci. 68, 530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karunadharma P. P., Basisty N., Dai D. F., Chiao Y. A., Quarles E. K., Hsieh E. J., Crispin D., Bielas J. H., Ericson N. G., Beyer R. P., MacKay V. L., MacCoss M. J., Rabinovitch P. S. (2015) Subacute calorie restriction and rapamycin discordantly alter mouse liver proteome homeostasis and reverse aging effects. Aging Cell 14, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barger J. L., Anderson R. M., Newton M. A., Da Silva C., Vann J. A., Pugh T. D., Someya S., Prolla T. A., Weindruch R. (2015) A conserved transcriptional signature of delayed aging and reduced disease vulnerability is partially mediated by SIRT3. PLoS One 10, e0120738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bruss M. D., Khambatta C. F., Ruby M. A., Aggarwal I., Hellerstein M. K. (2010) Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. Am. J. Physiol. Endocrinol. Metab. 298, E108–E116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tognini P., Murakami M., Liu Y., Eckel-Mahan K. L., Newman J. C., Verdin E., Baldi P., Sassone-Corsi P. (2017) Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab. 26, 523–538.e5 [DOI] [PubMed] [Google Scholar]
- 61.Franklin M. P., Sathyanarayan A., Mashek D. G. (2017) Acyl-CoA thioesterase 1 (ACOT1) regulates PPARα to couple fatty acid flux with oxidative capacity during fasting. Diabetes 66, 2112–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peek C. B., Affinati A. H., Ramsey K. M., Kuo H. Y., Yu W., Sena L. A., Ilkayeva O., Marcheva B., Kobayashi Y., Omura C., Levine D. C., Bacsik D. J., Gius D., Newgard C. B., Goetzman E., Chandel N. S., Denu J. M., Mrksich M., Bass J. (2013) Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342, 1243417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hunt M. C., Solaas K., Kase B. F., Alexson S. E. H. (2002) Characterization of an acyl-coA thioesterase that functions as a major regulator of peroxisomal lipid metabolism. J. Biol. Chem. 277, 1128–1138 [DOI] [PubMed] [Google Scholar]
- 64.Van Eunen K., Simons S. M. J., Gerding A., Bleeker A., den Besten G., Touw C. M. L., Houten S. M., Groen B. K., Krab K., Reijngoud D. J., Bakker B. M. (2013) Biochemical competition makes fatty-acid β-oxidation vulnerable to substrate overload. PLOS Comput. Biol. 9, e1003186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moffat C., Bhatia L., Nguyen T., Lynch P., Wang M., Wang D., Ilkayeva O. R., Han X., Hirschey M. D., Claypool S. M., Seifert E. L. (2014) Acyl-CoA thioesterase-2 facilitates mitochondrial fatty acid oxidation in the liver. J. Lipid Res. 55, 2458–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parvin R., Dakshinamurti K. (1970) Inhibition of gluconeogenic enzymes by free fatty acids and palmitoyl coenzyme A. J. Biol. Chem. 245, 5773–5778 [PubMed] [Google Scholar]
- 67.Chou S. H., Lee Y. C., Huang C. F., Wang Y. R., Yu H. P., Lau Y. T. (2010) Gender-specific effects of caloric restriction on the balance of vascular nitric oxide and superoxide radical. Cardiovasc. Res. 87, 751–759 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.