Abstract
RNA editing occurs in the endosymbiont organelles of higher plants as C-to-U conversions of defined nucleotides. The availability of large quantities of RNA sequencing data makes it possible to identify RNA editing sites and to quantify their editing extent. We have investigated RNA editing in 34 protein-coding mitochondrial transcripts of four Populus species, a genus noteworthy for its remarkably small number of RNA editing sites compared to other angiosperms. 27 of these transcripts were subject to RNA editing in at least one species. In total, 355 RNA editing sites were identified with high confidence, their editing extents ranging from 10 to 100%. The most heavily edited transcripts were ccmB with the highest density of RNA editing sites (53.7 sites / kb) and ccmFn with the highest number of sites (39 sites). Most of the editing events are at position 1 or 2 of the codons, usually altering the encoded amino acid, and are highly conserved among the species, also with regard to their editing extent. However, one SNP was found in the newly sequenced and annotated mitochondrial genome of P. alba resulting in the loss of an RNA editing site compared to P. tremula and P. davidiana. This SNP causes a C-to-T transition and an amino acid exchange from Ser to Phe, highlighting the widely discussed role of RNA editing in compensating mutations.
Keywords: RNA editing, mitochondria, poplar, Populus
RNA editing is the term for a post-transcriptional process by which the RNA is altered resulting in a sequence deviating from its corresponding genomic template (Benne et al. 1986). The alterations encompass insertions, deletions, or chemical modification of single bases. RNA editing sites refer to specific RNA positions affected by RNA editing, and also to the corresponding DNA positions.
In land plants, RNA editing was first discovered in plant mitochondria in 1989 (Covello and Gray 1989; Gualberto et al. 1989; Hiesel et al. 1989), and somewhat later also in chloroplasts (Hoch et al. 1991). In nuclear encoded transcripts, RNA editing was also described, but not extensively analyzed (Meng et al. 2010).
In endosymbiont organelles of higher plants, the only RNA editing mechanism is the conversion from C to U by deamination (Takenaka et al. 2013), while U-to-C conversion occurs in lycopods, ferns, and hornworts. Insertions and deletions have not been observed in plants, but are present in kinetoplastids, a group of flagellated protists, where the phenomenon was first described (Benne et al. 1986).
In some instances, RNA editing occurs with an extent of virtually 100% (i.e., the affected C is edited to U in all transcripts), compensating mutations in the genomic sequence that would otherwise lead to the exchange of highly conserved amino acids in the encoded proteins by restoring the original transcript sequence (Gualberto et al. 1989). This view is supported by the circumstance that most RNA editing events occur at position 1 or 2 of a codon, usually altering the encoded amino acid (Takenaka et al. 2013). These RNA editing sites are highly conserved across plant species and are efficiently edited as shown recently in a comparison of RNA editing sites in 17 angiosperm species (Edera et al. 2018). Another line of evidence for the mutational compensatory mechanism outside of higher plants has been recently provided in dinoflagellates, a photoautotrophic group with extensively edited mRNAs in their organelles and high conservation of editing sites (Klinger et al. 2018).
In other instances, RNA editing is a regulated process, meaning that a given editing site may only be edited to an extent of less than 100%, sometimes even to less than 10% (Benne 1989; Simpson and Shaw 1989; Bentolila et al. 2013; Takenaka et al. 2014). Therefore, RNA editing may serve as a transcriptional control mechanism. This view is supported by the introduction of translational initiation or termination codons by RNA editing (Hoch et al. 1991).
Apart from mature mRNA, RNA editing can be found in untranslated regions and introns, where it is in some instances a prerequisite for splicing (Börner et al. 1995). It is also thought to be involved in trans-splicing (Binder et al. 1992). In non-protein-coding RNA species, editing events were identified in tRNAs (Binder et al. 1992), whereas editing in rRNAs is rare, if it happens at all (Takenaka et al. 2013).
Edited C nucleotides cannot be recognized by a common sequence motif in the vicinity. Thus, editing sites are individually recognized. For a number of editing sites, 20 to 25 bp long cis elements have been identified, localized 5′ of the edited C, the crucial residues being 5 to 15 nucleotides upstream (Bock et al. 1996; Chaudhuri and Maliga 1996; Verbitskiy et al. 2008). In other instances, nucleotides further upstream or downstream of the editing site appear to have influence on editing (Verbitskiy et al. 2008). The great variability of both sequence and location of the cis elements relative to the edited nucleotide imply that different site-specific trans factors individually recognizing single editing sites direct RNA editing (Takenaka et al. 2013). The proteins of the PLS-class of pentatricopeptide repeat (PPR) proteins have been identified as the trans factors in RNA editing (Kotera et al. 2005; Hammani et al. 2009; Zehrmann et al. 2009; Barkan and Small 2014; Takenaka et al. 2014). The PPR proteins are encoded in the nuclear genome, but the translated proteins are almost exclusively targeted to plastids and mitochondria (Colcombet et al. 2013). As summarized in several reviews (Barkan and Small 2014; Takenaka et al. 2014), the PPR proteins are characterized by a number of consecutive tandem modules, each of which binds to a specific upstream nucleotide (Kindgren et al. 2015). PPR proteins may contain a DYW element which is expected to act as the deaminase enzyme (Barkan and Small 2014; Takenaka et al. 2014). When a DYW element is missing, a second PPR protein contributing the DYW function may be recruited with the support of the MORF/RIP proteins. Additional proteins unrelated to PPRs are also involved in organellar RNA editing, suggesting that the process is mediated by complex editosoms (Barkan and Small 2014; Takenaka et al. 2014).
A straightforward way to detect RNA editing sites is to compare RNAs with their corresponding DNA templates. As an alternative approach to Sanger sequencing of cDNAs (Gualberto et al. 1989; Giegé and Brennicke 1999), next-generation sequencing of transcriptomes (RNA-seq) is increasingly being used for the identification of C-to-U RNA editing sites in recent years (Picardi et al. 2010; Fang et al. 2012; Grimes et al. 2014; Wu et al. 2015; Sahraeian et al. 2017; Edera et al. 2018). Although poly(A)+ RNA is usually not (rarely) present in the organelles (Schuster and Stern 2009), poly(A)+ RNA in combination with oligo-dT priming for reverse transcription was successfully used for assessing RNA editing in many studies (e.g., Picardi et al. 2010; Shearman et al. 2014). However, quantitative analysis by such an approach should be handled with care (Stone and Storchova 2015).
In this approach, RNA-seq reads are mapped to genomic sequences (ideally of the same genotype) to identify editing sites and to quantify their editing extent. This strategy is challenging because RNA editing site detection can be distorted by genomic reads that might still be present in RNA-seq data and by RNA-seq reads that may originate from nuclear loci in case of dual transcription of homologs (Choi et al. 2006) and map unspecific to the mitochondrial genome sequence. Especially the adjustment of mapping parameters is difficult because stringent mapping settings may lead to false negatives, while more relaxed settings may increase the number of false positives (Guo et al. 2015; Edera et al. 2018). Nevertheless, this strategy allows a transcriptome-wide fast detection of editing sites and has enormous potential to deepen our knowledge of transcriptional processes in plant mitochondria (Edera et al. 2018).
This study focused on the identification of RNA editing sites in the coding sequences of mitochondrial genes in four different Populus species to deepen our understanding of RNA editing in this genus. Because RNA-seq data are still rare for Populus, RNA-seq data sets from different tissues have been used in this study, taking into consideration that tissue-specific RNA editing events cannot be excluded (Picardi et al. 2010; Tseng et al. 2013; Chen et al. 2017; Ichinose and Sugita 2017; Rodrigues et al. 2017) which potentially could restrict species comparisons for some editing sites.
Materials and Methods
Detection and plotting of RNA editing sites
Detection of RNA editing sites relied on SNP detection comparing sequencing reads of transcriptomic experiments (RNA-seq) with a genomic template. The sequencing runs used for this study (RNA-seq runs downloaded from SRA at NCBI or newly generated runs available at the SRA of NCBI: PRJNA514029) are listed in Table S1. The genomic template was a FASTA file containing all 78 potentially transcribed RNAs including hypothetical genes, rRNAs, and tRNAs derived from the annotated mitochondrial genome of P. tremula W52 (Genbank accession KT337313; Kersten et al. 2016). The NGS reads were mapped to the set of 78 transcripts using CLC Genomics Workbench (CLC-GWB) Version 11.0 (QIAGEN, Venlo, The Netherlands), which provided all tools mentioned below. The detailed parameters are listed in Table S2. In brief, read data (QC controlled and – if necessary – trimmed using the Trim Reads tool) were used as the input for the Map Reads to Reference tool. The resulting read mappings were used as the input for the Local Realignment tool. The Reads Track output was then used by the Low Frequency Variant Detection tool to produce a Variant Track. The SNP tables contained within the Variant Track output files and the detailed mapping coverage reports were exported from CLC-GWB. Both mappings for single reads and mappings combining multiple reads from species, accessions, etc. were carried out this way. At this stage, coverage and count filters were kept deliberately relaxed in order to investigate as much of the dataset as possible. More stringent filtering was applied at later steps of the analysis (see below).
The SNP tables were filtered for C-to-T polymorphisms. These filtered tables were analyzed using a custom R script (File S1) alongside with the mapping coverage report and the FASTA file containing the genomic information in order to produce graphical representations of editing sites, frequency, and coverage. Stringent filtering for coverage, count, and frequency was performed here using the following para-meters: Minimum coverage ≥ 10, Minimum count ≥ 3, Minimum Frequency ≥ 10, Probability ≥ 0.95.
A summarizing table including all RNA editing sites identified in the four species analyzed was generated using a modified version of Variant Tools (File S2). The original version of the software Variant Tools is available on https://github.com/ThuenenFG/varianttools (Schroeder et al. 2016).
Codon position affected by RNA editing and amino acid changes produced by RNA editing
Codon positions affected by RNA editing were identified based on the position of a related RNA editing site in the CDS using simple equations. The following equations are true for the different codon positions: position mod 3 = 1 → codon position 1; position mod 3 = 2 → codon position 2; position mod 3 = 0 → codon position 3.
Codon changes and amino acid changes produced by RNA editing were identified using an in-house Ruby script where the following exceptions from the standard genetic code were considered: UGA→Trp and CGG→Trp (Table S3b).
Sequencing, assembly and annotation of the complete mitochondrial genome of Populus alba clone Monrepos
Total genomic DNA was isolated from leaves of the male P. alba clone Monrepos (original provenance: Germany, Baden-Wuerttemberg) according to a published protocol (Dumolin et al. 1995). Genomic library generation and sequencing on the Illumina MiSeq v3 (2x300 bp paired-end reads; 25x coverage) and on the PacBio RS (10x coverage) was done by GATC Biotech AG (Konstanz, Germany).
Initial quality control of the NGS reads was done with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). If not otherwise stated, CLC-GWB (v. 10.0.1; CLC bio, A QIAGEN company; Aarhus, Denmark) was used for data processing. Using the Trim Reads tool, all reads were trimmed including adapter trimming, quality trimming (quality limit of 0.01), trimming of ambiguous nucleotides (maximal two nucleotides allowed), trimming of 10 nucleotides at the 5′-end and 1 nucleotide at the 3′-end and removing reads of less than 80 bp in length. All other options were set to default. In total 218,626 contigs of a length of at least 200 bp were generated by de novo assembly of all trimmed reads, using the De novo Assembly tool. The mapping mode was set to “Map reads back to contigs” (using a length fraction of 0.9 and a similarity fraction of 0.95). All other parameters were set to default. Duplicates and containments (>=98% identity) were removed using Dedupe included in BBMap (https://sourceforge.net/projects/bbmap/). The remaining 207,725 contigs were subjected to the Join Contigs tool of the Genome Finishing Module (plugin in CLC-GWB). Contig analysis type was set to “use long reads”, where all PacBio subreads were used as long reads. This step was repeated three times. The longest scaffold representing the entire mitochondrial genome was selected from the scaffolds. Overlapping sequence ends were removed from this scaffold and a N-strech inside the sequence was replaced by a related sequence obtained from one of the original MiSeq contigs.
The entire mtDNA sequences of P. alba clone Monrepos (838,420 bp; Genbank accession MK034705) was annotated based on the annotations of the mtDNA sequence of P. tremula W52 (KT337313; Kersten et al. 2016). Briefly, the related GenBank file (KT337313.1) was transferred to a draft SQN-file using the CHLOROBOX-GenBank2Sequin-tool (https://chlorobox.mpimp-golm.mpg.de/GenBank2Sequin.html). This SQN-file was edited using the Sequin tool (v13.05; https://www.ncbi.nlm.nih.gov/Sequin/) by importing the new mtDNA sequence of P. alba (clone Monrepos) with “update sequence”. A GenBank file of the P. alba mtDNA was exported from Sequin and served as input to create the related circular gene map using the OrganellarGenomeDRAW software (OGDRAW v1.2, https://chlorobox.mpimp-golm.mpg.de/OGDraw.html; Lohse et al. 2013).
Detection of DNA polymorphisms in Populus CDS and analyses of nad6-146 in different individuals
The reference CDS sequences of P. tremula W52 (Genbank accession KT337313) were used as query in a BlastN analysis vs. the CDS sequences of P. davidiana (KY216145.1) and P. alba clone Monrepos (MK034705) extracted from the related GB accessions. All SNPs vs. the P. tremula reference identified in the alignments were listed (Table S4).
Data availability statement
The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article, its tables, figures, and supplemental material deposited at figshare. RNA-seq data of P. tremula and P. trichocarpa generated in this study are publicly available at the SRA of NCBI (PRJNA514029). The annotated complete mtDNA sequence of P. alba clone Monrepos is available at GenBank (MK034705). Supplemental material available at Figshare: https://doi.org/10.25387/g3.7166141.
Results
Identification of C-to-U RNA editing sites in mitochondrial CDSs of four Populus species
RNA editing sites (C-to-U) were identified based on mappings of RNA-seq data of P. tremula, P. alba, P. davidiana and P. trichocarpa (Table 1; Table S1; newly generated RNA-seq data at the SRA of NCBI: PRJNA514029) to coding sequences (CDSs) of 34 mitochondrial genes previously annotated in P. tremula W52 (Genbank accession KT337313; Kersten et al. 2016) including putative CDS of rpl16 and mttb, both annotated as potential pseudogenes (CDS sequences of the genes analyzed are given in File S3).
Table 1. RNA-seq data sets from four Populus species used in the study.
| Species (Section) | Genotypes | Total number of reads | Total amount of data (Gb) |
|---|---|---|---|
| P. tremula (Populus) | W52, W100, Asp201a | 1,763,130,526 | 178.08 |
| P. davidiana (Populus) | Palgong2a, Seogwang9a, Seogwang15a | 1,680,315,108 | 169.71 |
| P. alba (Populus) | P. alba var. pyramidalisa (no genotype information) | 362,749,552 | 54.41 |
| P. trichocarpa (Tacamahaca) | Muhle_Larsen, NW7_17C, Weser4, Weser6 | 1,680,315,108 | 211.72 |
Data downloaded from NCBI (SRA). Details on the data sets are provided in Table S1. RNA-seq data of P. tremula (W52 and W100) and P. trichocarpa generated in this study are publicly available at the SRA of NCBI (PRJNA514029).
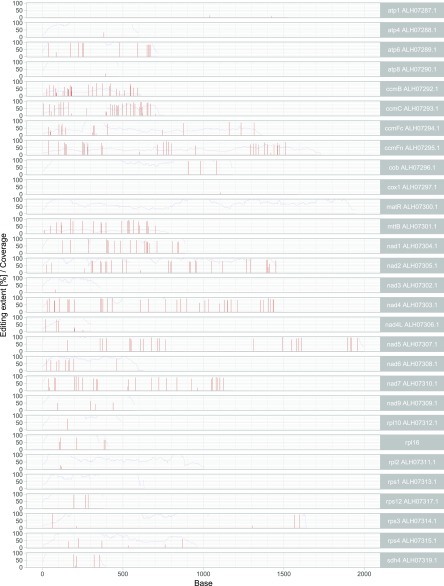
In total, 377 potential RNA editing sites (Table S3a) were identified in the CDS of 29 of the 34 mitochondrial genes analyzed, when considering all editing sites detected in at least one of the four Populus species. For all 29 CDS with RNA editing sites, the sites were plotted as red bars to their related nucleotide position as presented for P. tremula in Figure 1 (Figure S1 for the other three Populus species). The editing extent as given by the height of the red bars was in the range of 10–100%. A value of 10% was set as threshold for the SNP frequency (equivalent to editing extent) when filtering original SNP data according to SNP frequency values. The coverage value at each position is plotted as a blue line allowing to check if there are regions in the CDS escaping RNA editing site detection by an insufficient coverage value (coverage threshold was set to 10 reads in SNP filtering).
Figure 1.
Potential RNA editing sites in 29 mitochondrial CDS of P. tremula. In total, 377 potential RNA editing sites identified in combined RNA-seq data sets of three P. tremula genotypes (Table 1) were plotted to the nucleotide positions (Base) of the related CDS annotated in P. tremula W52 (Genbank accession KT337313; Kersten et al. 2016). Bars in red indicate edited bases (editing sites), their height shows the editing extent in percent. Blue lines show the coverage at each base as long as it is 100 or below. All 29 CDS that are potentially affected by RNA editing in at least one of the four Populus species investigated are shown in individual rows.
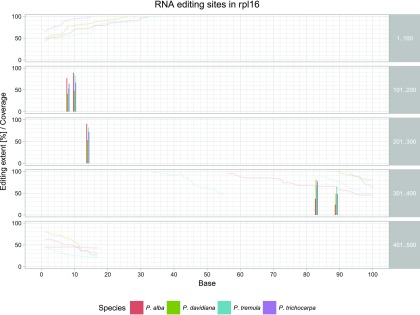
To make comparisons between species easier, the potential RNA editing sites of all species were plotted together in individual graphical representations of the 29 genes (Figure S2) as shown for rpl16 as an example in Figure 2. In the CDS of this gene, five RNA editing sites were identified occurring in all four Populus species.
Figure 2.
RNA editing in rpl16 in four Populus species. The mitochondrial rpl16 gene (417 bp) shows five editing sites at positions 108, 110, 214, 383 and 389 bp, which are conserved in the four Populus species investigated. Bars indicate edited bases, their height shows the editing extent in percent. Lines show the coverage at each base as long as it is 100 or below. The four Populus species are color-coded in both bars and lines.
The annotation of the complete DNA sequence of the P. tremula W52 mitochondrial genome (Genbank accession KT337313; Kersten et al. 2016) was checked for overlapping CDS to avoid false-positive/negative detection caused by overlaps. The CDS of cox3 (246,064 to 246,861 bp) and sdh4 (246,789 to 247,184 bp) – both annotated in forward direction – show a 72-bp overlap. No potential RNA editing site was identified in the overlapping region of both genes (Table S3a).
The 377 potential RNA editing sites identified in at least one of the four Populus species in this study (Table S3a) were compared with RNA editing sites recently identified for P. tremula in another study (Edera et al. 2018).
All sites identified in only one Populus species in our study and not identified by Edera et al. (2018), were manually validated in the related mappings. In case of sdh4 and rps4, all sites were validated because nucleotide polymorphisms others than C-to-U were detected in some of the mapped reads. These reads mapped unspecifically to the mitochondrial genome and originated from the nuclear genome as proven by BlastN of related P. tremula read sequences vs. P. tremula scaffolds at PopGenIE (http://popgenie.org/; Sundell et al. 2015) and vs. nuclear P. trichocarpa scaffolds at Phytozome (https://phytozome.jgi.doe.gov/; Tuskan et al. 2006). The selected P. tremula sdh4 reads showed 100% identity to a nuclear P. tremula scaffold (Potra000847) and 96% identity to P. trichocarpa chromosome 4 in a region where the gene Potri.004g049600 is annotated as “similar to sdh4”. BlastN analyses of the selected P. tremula rps4-reads provided hits with 99–100% identity to Potra185431, a nuclear P. tremula scaffold and with 96% identity to P. trichocarpa chromosome 18 in the genic region of Potri.018G031500 annotated as “rps4, mitochondrial”. These results indicate dual transcription of mitochondrial genes and their nuclear orthologs in the case of sdh4 and rps4 in P. tremula. Dual transcription of homologs in the nuclear and mitochondrial genomes has been previously reported for sdh4 in the Populus lineage (Choi et al. 2006).
After this manual validation, 355 RNA editing sites in 27 genes remained (Table S3b). Figure 3 shows the numbers and densities of these RNA editing sites in the related CDS. No editing sites were identified in atp9, cox1, cox2, cox3, rpl2, rps7 and rps14. In case of rps7, the mean coverage of the CDS sequence was below the detection threshold for editing sites in all species except for P. trichocarpa.
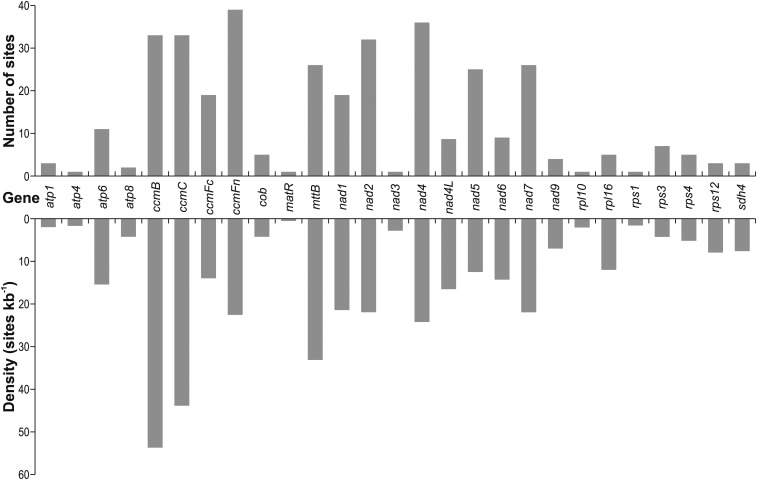
Figure 3.
Number (top) and density (bottom) of RNA editing sites in the CDS of 27 mitochondrial protein-coding genes across four Populus species. All RNA editing sites detected in at least one of the four Populus species investigated are counted. Exact values for the numbers and densities of sites in the related CDS are given in Table S3 (“number and density”).
The highest number of editing sites was identified in the CDS of ccmFn (39 sites) and the highest density in the CDS of ccmB (53.7 sites/kb; Figure 3).
Most RNA editing sites identified in this study are located at codon position 1 (33%; 118 sites) or 2 (53%; 189 sites). Only 14% of the editing sites (48 sites) are at the third codon position (Table S3b).
Comparison of the mitochondrial RNA editing sites in the four Populus species
Protein-coding sequences of mitochondrial Populus genes were analyzed for DNA polymorphisms to check if there is any overlap with RNA editing site positions identified in this study. Since entire mtDNA sequences with annotated genes were only available for P. tremula W52 (Genbank accession KT337313; Kersten et al. 2016) and P. davidiana Odae19 (KY216145.1; Choi et al. 2017), the complete mitochondrial genome sequence of P. alba (clone Monrepos) was assembled and annotated in addition (Genbank accession MK034705; Figure S3).
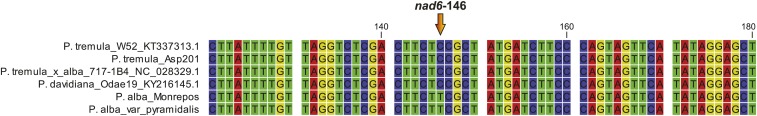
In total, 16 SNPs were identified in the CDS of the P. davidiana and/or the P. alba individual when compared with the P. tremula individual (Table S3). Only C-to-N or N-to-C SNPs were further considered because only such SNPs may result in a loss or gain of an RNA editing site depending on the location. Only one of these SNPs is located at an RNA editing site, namely a C-to-T SNP identified in nad6 at position 146 in P. alba clone Monrepos (Table S4). The nad6-CDS sequences of more Populus individuals were compared (Figure 4; Figure S4). In all individuals with a P. tremula or P. davidiana mitochondrial genome, a C-allele was detected at position nad6-146, whereas a T-allele was identified in the two P. alba genotypes including a P. alba var. pyramidalis individual which was analyzed in a recent whole genome assembly (Ma et al. 2018) and which was the source individual of the RNA-seq data used in this study (Table S1). The C-to-T SNP at nad6-146 results in a loss of the related RNA editing site via replacement to thymidine in P. alba. The SNP is at codon position 2 and results in a codon change (TCC to TTC) and in an amino acid exchange (Ser to Phe; Table S4). RNA editing at C-146 detected in P. tremula and P. davidiana results in the same amino acid exchange.
Figure 4.
Replacement of an editable cytidine by thymidine at the genomic level in the CDS of nad6 at position 146 in two P. alba genotypes. The nad6 CDS of P. alba clone Monrepos is according to GenBank accession MK034705. The nad6 CDS of P. alba var. pyramidalis was extracted from scaffold GWHAAEP00000188 (105625-106254 bp) of a recent whole genome assembly (Ma et al. 2018). The related CDS of other Populus species were taken from GenBank accession KY216145.1 (P. davidiana Odae19) and KT337313 (P. tremula W52). The nad6 CDS of P. tremula Asp201 was extracted from the scaffold Potra197846 (19887-20516 bp) of the P. tremula v1.1 whole genome assembly at PopGenIE (http://popgenie.org/; Sundell et al. 2015). For P. trichocarpa, a related genomic reference sequence is missing. The complete nad6 alignment is presented in Figure S4.
For a comparison of the RNA editing sites in the four Populus species, 343 sites identified in at least one species were considered, which are covered by at least 10 reads in all four species analyzed (Table S3d). Among these 343 RNA editing sites, 238 sites were identified in all four Populus species (Table S3e), indicating that most of the RNA editing sites are highly conserved between the individuals analyzed and probably between the four related Populus species. The individual differences at the other RNA editing sites, especially differences observed in the P. alba individual compared to the other individuals are expected to be mainly due to a too low coverage at these positions as discussed in more detail below. In case of P. alba, only 54 GB of RNA-seq data were available, whereas for the other individuals more than 160 GB of RNA-seq data were included in the study (Table 1).
Discussion
In our study, 73 new RNA editing sites were detected (355 sites in total) in at least one of the four Populus species analyzed. These new sites included 26 sites in mttb and 4 sites in rpl16 (Figure 2) previously annotated as potential pseudogenes because of lacking identification of related start codons (NC_028096.1). No RNA editing sites were reported for these genes by Edera et al. (2018) because they did not include these genes in the analysis. The identification of RNA editing sites in mttb and rpl16 in our study suggests both genes are functional genes in Populus mitochondria. The expression of mitochondrial-encoded mttb has been previously demonstrated on the RNA level in Nicotiana tabacum (van der Merwe and Dubery 2007) as well as on the protein level in Arabidopsis thaliana (Carrie et al. 2016). There are also indications for the expression of rpl16 in plant mitochondria, where probably a GTG codon acts as translation initiation codon (Bock et al. 1994).
Some of the 329 editing sites identified by Edera et al. (2018) for P. tremula were not identified in our study, among them one editing site in cox3 and one in rps14; both are editing sites with very low RNA editing extent. The detection of RNA editing sites with low editing extent, which is dependent on the threshold set for SNP detection, is difficult and requires sufficient coverage at the related position. Often, a very large amount of RNA-seq data is needed for obtaining enough coverage for mitochondrial genes, as most of the RNA-seq data publicly available are derived from oligo-dT-primed cDNA-libraries and include only a small fraction of mitochondrial RNA molecules (see Introduction). Moreover, contamination of genomic DNA in RNA preparations used for RNA-seq can distort (“dilute”) the values for editing extents.
In general, some differences in identified RNA editing sites and related editing extents are not unexpected between different studies of a species especially if different RNA-seq data sets from different individuals and tissues as well as different methods for the identification of editing sites, especially different mapping parameters are used as in our and Edera’s study (Edera et al. 2018). Different strategies have been developed and discussed to improve the detection of RNA editing sites (Guo et al. 2015; Stone and Storchova 2015; Edera et al. 2018; Wu et al. 2018). In mappings of RNA-seq data to CDS, the addition of flanking regions to the CDS may help to increase the read coverage and thus the detection of RNA editing sites at the ends of the CDS (Edera et al. 2018).
False-positive RNA editing site detection may also arise from unspecific mapping of nuclear expressed transcripts to mitochondrial reference sequences in cases of dual transcription of nuclear and mitochondrial transcripts as known for sdh4 in Salicaceae and Lupinus (Choi et al. 2006; Havird and Sloan 2016) and suggested for sdh4 and rps4 in P. tremula based on our study. One might circumvent this problem by using genomic sequences of all cellular compartments (nucleus, chloroplast, mitochondrion) of the individual of interest as reference sequences for mappings of RNA-seq data.
False–positive results may also arise when C-to-U RNA editing is “mimicked” by a genomic C-to-T DNA polymorphism at an editable cytidine, which may happen if the genomic reference used for mapping RNA-seq data is from another individual/species than the RNA-seq data.
Substitutions of editable cytidines with thymidines are the main cause of losses of editing sites along angiosperm evolution as shown in 17 genera (Edera et al. 2018). The authors expect that consecutive and highly conserved editing sites had been replaced by thymidines (thymidine footprints) as result of retroprocessing, by which edited transcripts are reverse transcribed to cDNA and integrated into the genome by homologous recombination. However, point mutations have also been proposed for the loss of editing sites favored by natural selection (Mower 2008).
Even within one genus, replacements of editable cytidines by thymidine may occur as shown for the loss of the Populus RNA editing site nad6-146 in two P. alba genotypes (Figure 4). Our study indicated that this loss of an RNA editing site could be P. alba-specific within the Populus genus, however more data are needed to confirm this conclusion. A loss of the nad6-146 RNA editing site has also been described in other genera (Cucumis, Malus, Arabidopsis and some Asterids; Edera et al. 2018). In general, Populus showed the largest number of thymidine footprints and the lowest number of mitochondrial RNA editing sites in the comparison of 17 genera (Edera et al. 2018). Early-diverging lineages, such as Liriodendron – in contrast – show the highest numbers of editing sites among angiosperms (Richardson et al. 2013; Edera et al. 2018).
Considering the proportion of RNA editing sites at the different codon positions (33% at position 1; 53% at position 2; 14% at position 3), our results are in agreement with numerous other studies showing that editing sites are predominantly found at non-synonymous positions in protein-coding genes, most frequently at the second position (Giegé and Brennicke 1999; Mulligan et al. 2007; Yura and Go 2008; Cuenca et al. 2010; Picardi et al. 2010; Sloan and Taylor 2010; Edera et al. 2018). Recently, it has been shown that conservation levels varied among codon positions across 17 angiosperm genera with lowest conservation at the third positions, as expected for synonymous sites with no obvious impact in the resulting protein (Edera et al. 2018).
Among the 355 RNA editing sites identified in this study, 238 sites were identified in all four Populus species analyzed (Table S3; “sites_all_species”) indicating that most of the RNA editing sites are highly conserved between the individuals analyzed and probably between the related species. In a recent study in the genus Leucaena, 607 conserved RNA editing positions have been identified in the mitochondrial genome when considering all three genome groups in this genus (Kovar et al. 2018).
As RNA-seq data sets from various tissues have been used in our study, individual differences due to tissue-specific RNA editing events may not be excluded (Picardi et al. 2010; Tseng et al. 2013; Chen et al. 2017; Ichinose and Sugita 2017; Rodrigues et al. 2017). It will be exciting to test in the future whether some of the non-conserved editing sites represent real differences between species and may even have functional implications.
Conclusions
In this study, the previous finding of Edera et al. (2018) that the number of RNA editing sites in poplar mitochondria is the smallest among all angiosperm genera has not only been confirmed, but also expanded from one species to four species within the genus Populus. Furthermore, a high level of conservation has been found throughout all poplar species investigated. Interestingly, the loss of an RNA editing site by genomic substitution of an editable cytidine with thymidine was observed in two P. alba genotypes. If this finding reflects an ongoing reduction of RNA editing sites within the genus Populus cannot be clarified without deeper phylogenetic analyses in the future.
Acknowledgments
We are grateful to Katrin Groppe for technical assistance.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7166141.
Communicating editor: D. Threadgill
Literature Cited
- Barkan A., Small I., 2014. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65: 415–442. 10.1146/annurev-arplant-050213-040159 [DOI] [PubMed] [Google Scholar]
- Benne R., 1989. RNA-editing in trypanosome mitochondria. Biochim. Biophys. Acta 1007: 131–139. 10.1016/0167-4781(89)90031-6 [DOI] [PubMed] [Google Scholar]
- Benne R., Van den Burg J., Brakenhoff J. P., Sloof P., Van Boom J. H., et al. , 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46: 819–826. 10.1016/0092-8674(86)90063-2 [DOI] [PubMed] [Google Scholar]
- Bentolila S., Oh J., Hanson M. R., Bukowski R., 2013. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 9: e1003584 10.1371/journal.pgen.1003584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S., Marchfelder A., Brennicke A., Wissinger B., 1992. RNA editing in trans-splicing intron sequences of nad2 mRNAs in Oenothera mitochondria. J. Biol. Chem. 267: 7615–7623. [PubMed] [Google Scholar]
- Bock H., Brennicke A., Schuster W., 1994. Rps3 and rpl16 genes do not overlap in Oenothera mitochondria: GTG as a potential translation initiation codon in plant mitochondria? Plant Mol. Biol. 24: 811–818. 10.1007/BF00029863 [DOI] [PubMed] [Google Scholar]
- Bock R., Hermann M., Kössel H., 1996. In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J. 15: 5052–5059. 10.1002/j.1460-2075.1996.tb00885.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner G. V., Mörl M., Wissinger B., Brennicke A., Schmelzer C., 1995. RNA editing of a group II intron in Oenothera as a prerequisite for splicing. Mol. Gen. Genet. 246: 739–744. 10.1007/BF00290721 [DOI] [PubMed] [Google Scholar]
- Carrie C., Weißenberger S., Soll J., 2016. Plant mitochondria contain the protein translocase subunits TatB and TatC. J. Cell Sci. 129: 3935–3947. 10.1242/jcs.190975 [DOI] [PubMed] [Google Scholar]
- Chaudhuri S., Maliga P., 1996. Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J. 15: 5958–5964. 10.1002/j.1460-2075.1996.tb00982.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.-C., Liu Y.-C., Wang X., Wu C.-H., Huang C.-H., et al. , 2017. Whole plastid transcriptomes reveal abundant RNA editing sites and differential editing status in Phalaenopsis aphrodite subsp. formosana. Bot. Stud. (Taipei, Taiwan) 58: 38 10.1186/s40529-017-0193-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C., Liu Z., Adams K. L., 2006. Evolutionary transfers of mitochondrial genes to the nucleus in the Populus lineage and coexpression of nuclear and mitochondrial Sdh4 genes. New Phytol. 172: 429–439. 10.1111/j.1469-8137.2006.01821.x [DOI] [PubMed] [Google Scholar]
- Choi M. N., Han M., Lee H., Park H.-S., Kim M.-Y., et al. , 2017. The complete mitochondrial genome sequence of Populus davidiana Dode. Mitochondrial DNA B Resour. 2: 113–114. 10.1080/23802359.2017.1289346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J., Lopez-Obando M., Heurtevin L., Bernard C., Martin K., et al. , 2013. Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biol. 10: 1557–1575. 10.4161/rna.26128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W., 1989. RNA editing in plant mitochondria. Nature 341: 662–666. 10.1038/341662a0 [DOI] [PubMed] [Google Scholar]
- Cuenca A., Petersen G., Seberg O., Davis J. I., Stevenson D. W., 2010. Are substitution rates and RNA editing correlated? BMC Evol. Biol. 10: 349 10.1186/1471-2148-10-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumolin S., Demesure B., Petit R. J., 1995. Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor. Appl. Genet. 91: 1253–1256. 10.1007/BF00220937 [DOI] [PubMed] [Google Scholar]
- Edera A. A., Gandini C. L., Sanchez-Puerta M. V., 2018. Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol. Biol. 97: 215–231. 10.1007/s11103-018-0734-9 [DOI] [PubMed] [Google Scholar]
- Fang Y., Wu H., Zhang T., Yang M., Yin Y., et al. , 2012. A complete sequence and transcriptomic analyses of date palm (Phoenix dactylifera L.) Mitochondrial Genome. PLoS One 7: e37164 10.1371/journal.pone.0037164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé P., Brennicke A., 1999. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA 96: 15324–15329. 10.1073/pnas.96.26.15324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes B. T., Sisay A. K., Carroll H. D., Cahoon A. B., 2014. Deep sequencing of the tobacco mitochondrial transcriptome reveals expressed ORFs and numerous editing sites outside coding regions. BMC Genomics 15: 31 10.1186/1471-2164-15-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto J. M., Lamattina L., Bonnard G., Weil J.-H., Grienenberger J.-M., 1989. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 341: 660–662. 10.1038/341660a0 [DOI] [PubMed] [Google Scholar]
- Guo W., Grewe F., Mower J. P., 2015. Variable frequency of plastid RNA editing among ferns and repeated loss of Uridine-to-Cytidine editing from vascular plants. PLoS One 10: e0117075 10.1371/journal.pone.0117075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K., Okuda K., Tanz S. K., Chateigner-Boutin A.-L., Shikanai T., et al. , 2009. A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21: 3686–3699. 10.1105/tpc.109.071472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havird J. C., Sloan D. B., 2016. The roles of mutation, selection, and expression in determining relative rates of evolution in mitochondrial vs. nuclear genomes. Mol. Biol. Evol. 33: 3042–3053. 10.1093/molbev/msw185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesel R., Wissinger B., Schuster W., Brennicke A., 1989. RNA editing in plant mitochondria. Science 246: 1632–1634. 10.1126/science.2480644 [DOI] [PubMed] [Google Scholar]
- Hoch B., Maier R. M., Appel K., Igloi G. L., Kössel H., 1991. Editing of a chloroplast mRNA by creation of an initiation codon. Nature 353: 178–180. 10.1038/353178a0 [DOI] [PubMed] [Google Scholar]
- Ichinose M., Sugita M., 2017. RNA editing and its molecular mechanism in plant organelles. Genes (Basel) 8: 5 10.3390/genes8010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten B., Faivre Rampant P., Mader M., Le Paslier M.-C., Bounon R., et al. , 2016. Genome sequences of Populus tremula chloroplast and mitochondrion: Implications for holistic poplar breeding. PLoS One 11: e0147209 10.1371/journal.pone.0147209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P., Yap A., Bond C. S., Small I., 2015. Predictable alteration of sequence recognition by RNA editing factors from Arabidopsis. Plant Cell 27: 403–416. 10.1105/tpc.114.134189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger C. M., Paoli L., Newby R. J., Wang M. Y., Carroll H. D., et al. , 2018. Plastid transcript editing across dinoflagellate lineages shows lineage-specific application but conserved trends. Genome Biol. Evol. 10: 1019–1038. 10.1093/gbe/evy057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera E., Tasaka M., Shikanai T., 2005. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330. 10.1038/nature03229 [DOI] [PubMed] [Google Scholar]
- Kovar L., Nageswara-Rao M., Ortega-Rodriguez S., Dugas D. V., Straub S., et al. , 2018. PacBio-based mitochondrial genome assembly of Leucaena trichandra (Leguminosae) and an intrageneric assessment of mitochondrial RNA Editing. Genome Biol. Evol. 10: 2501–2517. 10.1093/gbe/evy179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Drechsel O., Kahlau S., Bock R., 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41: W575–W581. 10.1093/nar/gkt289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Wan D., Duan B., Bai X., Bai Q., et al. , 2018. Genome sequence and genetic transformation of a widely distributed and cultivated poplar. Plant Biotechnol. J. 10.1111/pbi.12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Chen D., Jin Y., Mao C., Wu P., et al. , 2010. RNA editing of nuclear transcripts in Arabidopsis thaliana. BMC Genomics 11: S12 10.1186/1471-2164-11-S4-S12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower J. P., 2008. Modeling Sites of RNA Editing as a fifth nucleotide state reveals progressive loss of edited sites from angiosperm mitochondria. Mol. Biol. Evol. 25: 52–61. 10.1093/molbev/msm226 [DOI] [PubMed] [Google Scholar]
- Mulligan R. M., Chang K. L. C., Chou C. C., 2007. Computational analysis of RNA editing sites in plant mitochondrial genomes reveals similar information content and a sporadic distribution of editing sites. Mol. Biol. Evol. 24: 1971–1981. 10.1093/molbev/msm125 [DOI] [PubMed] [Google Scholar]
- Picardi E., Horner D. S., Chiara M., Schiavon R., Valle G., et al. , 2010. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucleic Acids Res. 38: 4755–4767. 10.1093/nar/gkq202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A. O., Rice D. W., Young G. J., Alverson A. J., Palmer J. D., 2013. The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 11: 29 10.1186/1741-7007-11-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues N. F., Fonseca G. C., Kulcheski F. R., Margis R., 2017. Salt stress affects mRNA editing in soybean chloroplasts. Genet. Mol. Biol. 40: 200–208. 10.1590/1678-4685-gmb-2016-0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahraeian S. M. E., Mohiyuddin M., Sebra R., Tilgner H., Afshar P. T., et al. , 2017. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat. Commun. 8: 59 10.1038/s41467-017-00050-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder H., Cronn R., Yanbaev Y., Jennings T., Mader M., et al. , 2016. Development of molecular markers for determining continental origin of wood from white oaks (Quercus L. sect. Quercus). PLoS One 11: e0158221 10.1371/journal.pone.0158221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G., Stern D., 2009. RNA polyadenylation and decay in mitochondria and chloroplasts. Prog. Mol. Biol. Transl. Sci. 85: 393–422. 10.1016/S0079-6603(08)00810-6 [DOI] [PubMed] [Google Scholar]
- Shearman J. R., Sangsrakru D., Ruang-Areerate P., Sonthirod C., Uthaipaisanwong P., et al. , 2014. Assembly and analysis of a male sterile rubber tree mitochondrial genome reveals DNA rearrangement events and a novel transcript. BMC Plant Biol. 14: 45 10.1186/1471-2229-14-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L., Shaw J., 1989. RNA editing and the mitochondrial cryptogenes of kinetoplastid protozoa. Cell 57: 355–366. 10.1016/0092-8674(89)90911-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan D. B., Taylor D. R., 2010. Testing for selection on synonymous sites in plant mitochondrial DNA: The role of codon bias and RNA editing. J. Mol. Evol. 70: 479–491. 10.1007/s00239-010-9346-y [DOI] [PubMed] [Google Scholar]
- Stone J. D., Storchova H., 2015. The application of RNA-seq to the comprehensive analysis of plant mitochondrial transcriptomes. Mol. Genet. Genomics 290: 1–9. 10.1007/s00438-014-0905-6 [DOI] [PubMed] [Google Scholar]
- Sundell D., Mannapperuma C., Netotea S., Delhomme N., Lin Y.-C., et al. , 2015. The Plant Genome Integrative Explorer Resource: PlantGenIE.org. New Phytol. 208: 1149–1156. 10.1111/nph.13557 [DOI] [PubMed] [Google Scholar]
- Takenaka M., Verbitskiy D., Zehrmann A., Härtel B., Bayer-Császár E., et al. , 2014. RNA editing in plant mitochondria - Connecting RNA target sequences and acting proteins. Mitochondrion 19: 191–197. 10.1016/j.mito.2014.04.005 [DOI] [PubMed] [Google Scholar]
- Takenaka M., Zehrmann A., Verbitskiy D., Härtel B., Brennicke A., 2013. RNA editing in plants and its evolution. Annu. Rev. Genet. 47: 335–352. 10.1146/annurev-genet-111212-133519 [DOI] [PubMed] [Google Scholar]
- Tseng C.-C., Lee C.-J., Chung Y.-T., Sung T.-Y., Hsieh M.-H., 2013. Differential regulation of Arabidopsis plastid gene expression and RNA editing in non-photosynthetic tissues. Plant Mol. Biol. 82: 375–392. 10.1007/s11103-013-0069-5 [DOI] [PubMed] [Google Scholar]
- Tuskan G. A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., et al. , 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604. 10.1126/science.1128691 [DOI] [PubMed] [Google Scholar]
- van der Merwe J. A., Dubery I. A., 2007. Expression of mitochondrial tatC in Nicotiana tabacum is responsive to benzothiadiazole and salicylic acid. J. Plant Physiol. 164: 1231–1234. 10.1016/j.jplph.2006.11.009 [DOI] [PubMed] [Google Scholar]
- Verbitskiy D., van der Merwe J. A., Zehrmann A., Brennicke A., Takenaka M., 2008. Multiple specificity recognition motifs enhance plant mitochondrial RNA editing in vitro. J. Biol. Chem. 283: 24374–24381. 10.1074/jbc.M803292200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Liu W., Aljohi H. A., Alromaih S. A., Alanazi I. O., et al. , 2018. REDO: RNA editing detection in plant organelles based on variant calling results. J. Comput. Biol. 25: 509–516. 10.1089/cmb.2017.0214 [DOI] [PubMed] [Google Scholar]
- Wu Z., Stone J. D., Štorchová H., Sloan D. B., 2015. High transcript abundance, RNA editing, and small RNAs in intergenic regions within the massive mitochondrial genome of the angiosperm Silene noctiflora. BMC Genomics 16: 938 10.1186/s12864-015-2155-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura K., Go M., 2008. Correlation between amino acid residues converted by RNA editing and functional residues in protein three-dimensional structures in plant organelles. BMC Plant Biol. 8: 79 10.1186/1471-2229-8-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehrmann A., Verbitskiy D., van der Merwe J. A., Brennicke A., Takenaka M., 2009. A DYW domain–containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell 21: 558–567. 10.1105/tpc.108.064535 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article, its tables, figures, and supplemental material deposited at figshare. RNA-seq data of P. tremula and P. trichocarpa generated in this study are publicly available at the SRA of NCBI (PRJNA514029). The annotated complete mtDNA sequence of P. alba clone Monrepos is available at GenBank (MK034705). Supplemental material available at Figshare: https://doi.org/10.25387/g3.7166141.