Abstract
Non-homologous end joining (NHEJ) repairs the majority of DNA double-strand breaks in human cells, yet the detailed order of events in this process has remained obscure. Here, we describe how to employ Xenopus laevis egg extract for the study of NHEJ. The egg extract is easy to prepare in large quantities, and it performs efficient end joining that requires the core end joining proteins Ku, DNA-PKcs, XLF, XRCC4, and DNA ligase IV. These factors, along with the rest of the soluble proteome, are present at endogenous concentrations, allowing mechanistic analysis in a system that begins to approximate the complexity of cellular end joining. We describe an ensemble assay that monitors covalent joining of DNA ends and fluorescence assays that detect joining of single pairs of DNA ends. The latter assay discerns at least two discrete intermediates in the bridging of DNA ends.
1. INTRODUCTION
DNA double-strand breaks (DSBs) are common and extremely toxic DNA lesions that must be repaired to maintain genomic integrity. The major pathway for DSB repair in human cells is nonhomologous end joining (NHEJ), which rejoins DNA ends by direct ligation. NHEJ is a versatile mechanism that can ligate incompatible and chemically damaged ends. It does this by using a variety of different enzymes, including polymerases and exonucleases, to make DNA ends compatible for joining. How end processing is regulated to minimize mutations is poorly understood.
During NHEJ, ends are first bound by a heterodimer of the Ku70 and Ku80 proteins (Ku), which encircles the broken DNA end like a ring (Walker, Corpina, & Goldberg, 2001). Ku recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), creating the DNA-PK holo-enzyme (Carter, Vancurová, Sun, Lou, & DeLeon, 1990; Dvir, Peterson, Knuth, Lu, & Dynan, 1992; Dvir, Stein, Calore, & Dynan, 1993; Gottlieb & Jackson, 1993; Lees-Miller, Chen, & Anderson, 1990). DNA-PK phosphorylates a number of proteins, including itself, and DNA-PKcs autophosphorylation is important for its function (Dobbs, Tainer, & Lees-Miller, 2010; Jette & Lees-Miller, 2015; Jiang et al., 2015). A variety of different enzymes, including DNA polymerases λ and μ, poly-nucleotide kinase/phosphatase (PNKP), aprataxin, aprataxin and PNKP-like factor (APLF), and Artemis, are used to process damaged or mismatched ends and make them compatible for joining (Menon & Povirk, 2016; Waters, Strande, Wyatt, Pryor, & Ramsden, 2014). Ends are ligated by DNA ligase IV (LIG4), which resides in a complex with its essential accessory factor XRCC4 (Critchlow, Bowater, & Jackson, 1997; Grawunder et al., 1997; Li et al., 1995). XRCC4 in turn binds to its homolog XRCC4-like factor (XLF), which stimulates the activity of the LIG4:XRCC4 complex (Ahnesorg, Smith, & Jackson, 2006; Buck et al., 2006; Gu, Lu, Tsai, Schwarz, & Lieber, 2007; Hentges et al., 2006; Lu, Pannicke, Schwarz, & Lieber, 2007; Tsai, Kim, & Chu, 2007). XLF also interacts with the Ku-DNA complex (Yano, Morotomi-Yano, Lee, & Chen, 2011; Yano et al., 2008). Recently, a new paralog of XRCC4 and XLF (PAXX) has been discovered, which is redundant with XLF in some but not all contexts (Balmus et al., 2016; Craxton et al., 2015; Kumar, Alt, & Frock, 2016; Lescale et al., 2016; Liu, Shao, Jiang, Lee, & Zha, 2017; Ochi et al., 2015; Roy et al., 2015; Tadi et al., 2016; Xing et al., 2015). Unlike XLF, PAXX does not interact with XRCC4, although it does interact with Ku.
Cell-free systems provide a powerful tool to study biochemical mechanisms. Cell-free extracts derived from human cells recapitulate NHEJ, although the efficiency of end joining is generally low (Akopiants et al., 2009; Baumann & West, 1998; Chappell, Hanakahi, Karimi-Busheri, Weinfeld, & West, 2002; Cortes et al., 1996; Feldmann, Schmiemann, Goedecke, Reichenberger, & Pfeiffer, 2000; Hanakahi, Bartlet-Jones, Chappell, Pappin, & West, 2000; Jayaram, Ketner, Adachi, & Hanakahi, 2008; Labhart, 1999a; Lee et al., 2004; Lee, Yannone, Chen, & Povirk, 2003; Pfeiffer, Feldmann, Odersky, Kuhfittig-Kulle, & Goedecke, 2005; Pfeiffer, Odersky, Goedecke, & Kuhfittig-Kulle, 2014; Smeaton, Miller, Ketner, & Hanakahi, 2007; Weis-Garcia et al., 1997). Similarly, defined systems using purified NHEJ proteins join DNA ends less efficiently than in vivo NHEJ, possibly because they do not contain all of the proteins that participate in and regulate NHEJ in the cell (Chang et al., 2016; Ma & Lieber, 2006; Ma et al., 2004; Nick McElhinny et al., 2005; Tsai et al., 2007). For these reasons, a whole egg lysate from the frog Xenopus laevis is an attractive alternative. These extracts contain the entire proteome of the egg and can be produced in large quantities. Early experiments showed that egg extracts join DNA ends rapidly at room temperature dependent on Ku and DNA-PK (Chen et al., 2001; Di Virgilio & Gautier, 2005; Labhart, 1999b; Postow et al., 2008; Taylor et al., 2010). More recently, we showed that end joining additionally requires XLF, XRCC4-LIG4, and LIG4 catalytic activity (Graham, Walter, & Loparo, 2016).a In addition to joining compatible DNA ends, egg extract joins incompatible ends by filling in or degrading incompatible overhangs (Chen et al., 2001; Daza et al., 1996; Di Virgilio & Gautier, 2005; Gu, Bennett, & Povirk, 1996; Labhart, 1999b; Pfeiffer & Vielmetter, 1988; Sandoval & Labhart, 2002; Thode, Schäfer, Pfeiffer, & Vielmetter, 1990; Zhu & Peng, 2016). These processing events depend on the factors mentioned earlier (our unpublished results), indicating that egg extract will be useful to elucidate how end processing is regulated during NHEJ.
Here, we describe ensemble and single-molecule NHEJ assays in egg extract. Ensemble assays report on the processing and ligation of DNA ends by monitoring DNA products, while single-molecule assays reveal transient intermediates in end joining. We describe two variations of a single-molecule fluorescence assay in which DNA substrates are labeled near their ends with the fluorescent dyes Cy3 and Cy5 and bridging of ends is indicated by colocalization and Förster resonance energy transfer (FRET) between these two fluorophores.
2. ENSEMBLE END JOINING ASSAYS
In this section, we describe ensemble biochemical assays for monitoring end joining in egg extract. We typically use a high-speed supernatant (HSS) of unfertilized eggs arrested in interphase (Lebofsky, Takahashi, & Walter, 2009). However, end joining also occurs in low-speed egg lysates that contain membranes and in “CSF-arrested” extract that is arrested in metaphase II of meiosis (Di Virgilio & Gautier, 2005; Postow et al., 2008). Highly concentrated nucleoplasmic extracts derived from either in vitro-reconstituted nuclei or germinal vesicles can join DNA ends under certain conditions, although they primarily tend to resect DNA ends and may join ends through alternative end joining (Alt-EJ) mechanisms (Budzowska, Graham, Sobeck, Waga, & Walter, 2015; Lehman & Carroll, 1991; Lehman, Clemens, Worthylake, Trautman, & Carroll, 1993; Liao, Toczylowski, & Yan, 2008; Toczylowski & Yan, 2006; Yan, McCane, Toczylowski, & Chen, 2005; B. Stinson & R. Amunugama, personal communication).
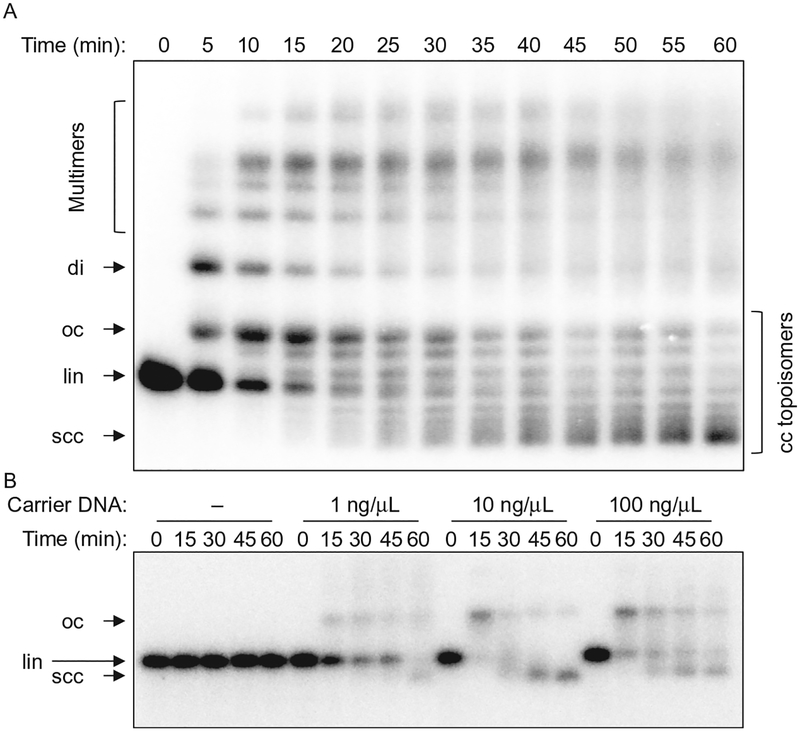
Linear DNA fragments introduced into egg extract are joined end to end by the NHEJ machinery. This yields circular or linear end joining products (Fig. 1A), the ratio of which varies depending on substrate length and concentration. Egg extracts including HSS are also capable of performing the 5′–3′ end resection step that initiates homologous recombination or Alt-EJ (Liao, Guay, Toczylowski, & Yan, 2012; Liao, Toczylowski, & Yan, 2011). We find that the overall DNA concentration added to extract influences the types of products formed and the balance between end joining and resection. We include 10–100ng/μL of closed-circular plasmid DNA as a “carrier” in our reactions, as this suppresses resection and promotes end joining (through an unknown mechanism; see Fig. 1B). The efficiency of DNA replication in egg extract has similarly been shown to depend on the total DNA concentration (Lebofsky, van Oijen, & Walter, 2011). The optimal concentration of carrier DNA may vary between different preparations of extract. Importantly, the effect of carrier DNA does not depend on the presence of homology between the substrate and the carrier DNA, as both heterologous and homologous carrier plasmids support efficient end joining. We typically incubate end joining reactions at room temperature; however, joining at 13–15°C has also been reported (Gu et al., 1996; Labhart, 1999b).
Fig. 1.
Ensemble end joining assay. (A) Time course of an end joining reaction, showing conversion of linear substrate (lin) into open circular (oc), supercoiled closed-circular (scc), dimeric (di), and multimeric products. Supercoiling of closed-circular DNA in extract arises from nucleosome assembly. Closed-circular (cc) topoisomers are visible between the oc and scc bands. The substrate DNA band is slightly overexposed at the 0-min timepoint. (B) Carrier DNA dependence of end joining. A very low concentration of substrate DNA (~0.05ng/μL) was incubated with an extract containing different amounts of closed-circular carrier DNA. End joining does not occur at all if the overall DNA concentration is too low.
End joining reactions may be performed with unlabeled DNA, which is then separated on an agarose gel and stained with an intercalating dye such as SYBR Gold. Alternatively, substrate DNA may be site-specifically labeled with a fluorescent dye or a radioactive nucleotide. Below, we describe how to perform end joining reactions using radiolabeled substrate DNA, which provides high sensitivity and good signal to noise for reactions with small amounts of substrate. At low substrate DNA concentrations (~1ng/μL), the products formed are predominantly circles, which result from joining of the two ends of the same molecule of DNA. We commonly use linearized 3–5kb plasmid DNA to permit efficient circularization.
2.1. Substrate Preparation
2.1.1. Materials
Maxiprepped plasmid DNA containing a single EcoRI site
EcoRI-HF® (New England Biolabs, Cat. #R3101). Any restriction enzyme may be used, provided that it cuts the plasmid to leave 5′ over-hangs that can be filled in with a suitable radiolabeled dNTP
Molecular biology grade agarose
Horizontal gel electrophoresis rig, casting tray (~10cm × 10cm), and comb
Heavy-duty plastic packing tape
DNA ladder (e.g., 2-log ladder from New England Biolabs, Cat. #N3200)
Klenow fragment of DNA polymerase I, 5000U/mL (New England Biolabs, Cat. #M0210L). Note: Do not use the 3′–5′ exonuclease-deficient version of the Klenow fragment, as this enzyme can add nontemplated nucleotides to blunt-ended DNA
[α−32P]-dATP (EasyTides, Perkin Elmer)
Unlabeled dCTP, dGTP, and dTTP
Ethidium bromide, 10mg/mL solution
6 × gel loading dye (New England Biolabs, Cat. #B7024S)
Tris–borate–EDTA (TBE) buffer: Dissolve the following per liter to make a 10 × stock: 108g of Tris base, 55g of boric acid, and 40mL of 0.5M EDTA solution, pH 8.0. Use EDTA, not EDTA disodium salt, and adjust the pH of the EDTA stock solution to 8.0 with sodium hydroxide.
Long-wavelength (365nm) UV light box
Dialysis tubing (SpectraPor #132650, 23mm, 6–8kDa MWCO) and clips
Cooled microcentrifuge
Isobutanol, equilibrated with aqueous buffer at pH 8
3 M sodium acetate, pH 5.2
100% and 70% ethanol
Clean razor blade or scalpel
Broad-tipped forceps
1 × TE buffer: 10mM Tris–HCl, pH 8, 1mM EDTA
10mM Tris–HCl, pH 7.5
PCR purification kit (Qiagen)
Nanodrop spectrophotometer
2.1.2. Protocol
Digest 100μg of plasmid DNA with 5μL of 20U/μL EcoRI in a 100μL volume of CutSmart buffer at 37°C for 3h.
Tape all wells except one of a gel loading comb together with packing tape, and cast a 1 × TBE agarose gel containing 1μg/mL ethidium bromide. Add 20μL of loading dye to the DNA digestion reaction and load in the large well. Load a DNA ladder in the small well. Separate by electrophoresis at 7.5V/cm.
On a UV light box, cut out the band containing the DNA with a sharp razor blade or a scalpel. To prevent photodamage, avoid excessive exposure of the DNA to UV light. Alternatively, the band may be visible under room light when viewed against a white background.
- Electroelute the DNA from the gel slice:
- Cut a piece of dialysis tubing several centimeters longer than the gel slice. Rinse the tubing with water and open the ends by rubbing them between your thumb and forefinger. Clamp one end with a plastic clip.
- Submerge the dialysis tubing in a gel box containing 1 × TBE to fill it with buffer, pushing out any bubbles.
- Slide the product band (preferably as a single piece of agarose) into the dialysis bag, squeeze out any bubbles and excess buffer, and clamp the other end.
- Add ethidium bromide to the 1 × TBE running buffer to a final concentration of 1μg/mL.
- Orient the tubing perpendicular to the direction of the electric field and push the gel slice to the edge of the tubing nearest the cathode.
- Apply voltage at 7.5V/cm. The DNA will migrate out of the gel slice and accumulate on the opposite edge of the dialysis tubing. The stained DNA is often visible in room light, and it can also be visualized more clearly under ultraviolet light.
- After all of the DNA has exited the gel (about 30min to 1h), remove one of the dialysis clips, and pull out the gel slice with a pair of forceps, being careful not to disturb the DNA that has accumulated on the opposite side of the dialysis tubing.
- Carefully rub the dialysis tubing between your thumb and forefinger to free DNA bound to the sides, being careful not to spill the solution from the tubing. Pipette out the DNA.
- To remove ethidium bromide, extract the DNA solution twice with 1 volume of isobutanol. Mix well at each extraction step by vortexing, and centrifuge briefly to separate the phases. Isobutanol forms the top phase.
- Precipitate DNA by adding 0.1 volumes of 3 M sodium acetate, pH 5.2 and 2.5 volumes of 100% ethanol. Incubate at −20°C for ≥10min and spin at 16,000 × g for 30min at 4°C in a microcentrifuge. Large volumes can be divided between microcentrifuge tubes or spun at ≥4000 × g in 15mL conical tubes in a swinging bucket rotor. Wash the pellet briefly with 70% ethanol and centrifuge again for 1min. For samples divided between multiple tubes or large samples in 15mL conical tubes, disrupt all pellets by pipetting up and down with 70% ethanol, transfer to a single microcentrifuge tube, and centrifuge for 10min at 16,000 × g. Thoroughly aspirate the 70% ethanol and dissolve the pellet in 100 μL of 10mM Tris–HCl pH 7.5. Determine the concentration by 280nm absorbance on a low-volume UV–Vis spectrophotometer (e.g., Nanodrop).
- Set up the following radiolabeling reaction:
- 1.0μg of gel purified linear DNA fragment
- 2μL 10 × NEBuffer 2 (New England Biolabs; supplied with Klenow fragment)
- 1μL [α−32P]-dATP
- 0.67μL each 1mM dTTP, dCTP, and dGTP. All dNTPs are included in the reaction to prevent resection by the 3′–5′ exonuclease activity of the Klenow fragment
- 0.5μL of 5000U/mL Klenow fragment
- Water to 20μL
Incubate at 25°C for 15min.
Stop the reaction by adding 100μL of PCR purification kit buffer PB (Qiagen). Purify the DNA using a PCR purification kit (Qiagen) following the manufacturer’s instructions. Elute the DNA from the spin column with 50μL of 10mM Tris–HCl, pH 7.5. Full recovery of the DNA would yield a final concentration of 20ng/μL.
2.2. End Joining Reactions
2.2.1. Materials
HSS of unfertilized X. laevis eggs (Lebofsky et al., 2009). We typically flash-freeze 33μL aliquots in liquid nitrogen and store them at −80°C.
Nocodazole (0.5mg/mL solution in dimethylsulfoxide)
Creatine phosphokinase (CPK; Sigma, #C-3755, 35,000U; Type I from rabbit muscle), 5mg/mL solution in 10mM HEPES, pH 7.5, 50% glycerol, 50mM NaCl. Store at −20°C.
Adenosine triphosphate (ATP; Sigma, #A-5394), 200mM solution. Adjust the pH to approximately 7 with NaOH using pH indicator paper, and store aliquots at −20°C.
Phosphocreatine (PC; Sigma, #P-6502), 1 M solution in 10mM KH2PO4, pH 7.0. Store at −20°C.
Closed circular plasmid DNA, 1μg/μL in 1 × TE buffer
Stop solution/loading dye: 8mM EDTA, 0.13% phosphoric acid, 10% Ficoll, 5% SDS, 0.2% bromophenol blue, 80mM Tris, pH 8
Proteinase K, 1mg/mL solution
Hybond-XL membrane (GE Healthcare Life Sciences)
Heavy Whatman filter paper
C-fold or multifold paper towels
Plastic wrap
Large book or other flat, heavy object
Vacuum gel dryer
Storage phosphorscreen and phosphorimager
2.2.2. Protocol
- Prepare an ATP regeneration system by combining the following:
- 0.5μL of 5mg/mL CPK
- 5μL of 200mM ATP
- 10μL of 1 M PC
Add 0.5μL of 0.5mg/mL nocodazole to a single 33μL aliquot of egg extract, and mix thoroughly by pipetting, being careful to avoid bubbles. Also add 1μL of ATP regeneration system and 3.3μL of 1μg/μL closed-circular carrier plasmid. Mix thoroughly.
For each end joining reaction, combine 10μL of this extract mixture with 0.5μL of radiolabeled substrate DNA on ice.
Withdraw an initial 2μL sample into 5μL of stop solution/loading dye. Transfer the reactions to room temperature and withdraw additional 2μL samples at desired time points, e.g., 20, 40, and 60min.
Treat the stopped reactions with proteinase K (1μg per sample) for 30min at 37°C or overnight at room temperature.
Load the samples on a 0.8% 1 × TBE agarose gel and separate by electrophoresis for 1.5–2h at 7.5V/cm.
Sandwich the gel between two pieces of the Hybond-XL membrane to trap the DNA and then sandwich this between two pieces of a heavy Whatman filter paper. Place the gel sandwich on a stack of paper towels, cover with a piece of plastic wrap, and compress under a large book or other heavy, flat object for at least 15min to remove most of the buffer from the gel.
Remove the paper towels and dry the gel for at least 1h on a vacuum gel dryer set to 80°C.
Remove the Whatman filter paper, cover the dried gel/Hybond-XL sandwich with a single layer of plastic wrap, and expose to a storage phosphorscreen for several hours to overnight. Image exposed screen on a phosphorimager.
2.2.3. Notes
For imaging nonradiolabeled DNA by intercalating dye staining, treat the samples with RNase A (2μg per sample for 30min at 37°C) prior to PK treatment to degrade endogenous egg RNA in the extract. Separate DNA on a gel without intercalating dye and stain the gel after electrophoresis.
The amount of DNA in different bands may be quantified using image analysis software such as ImageJ. The intensity of radiolabeled DNA summed over the entire lane usually decreases over the course of the reaction, likely due to some fraction of the substrate DNA being resected. End joining efficiency varies between batches of extract, but the majority of labeled DNA remaining at 30min is typically found in joined products (Fig. 1).
2.3. Immunodepletion Analysis
Immunodepletion of proteins from egg extract can be used to study their functions in end joining, both in ensemble and in single-molecule assays. Custom antibodies are raised against the desired factor by immunizing rabbits with the purified full-length protein, a fragment of the protein, or a synthetic peptide. We have had good success with antibodies directed against 12- to 16-amino acid synthetic peptides derived from the C-termini of proteins of interest. The antibody is typically affinity-purified from serum by coupling the antigen to a solid support (e.g., SulfoLink resin [Thermo Fisher]), passing crude serum over the resin, eluting bound antibody with low pH, and rapidly neutralizing the eluate with concentrated high-pH Tris buffer (Harlow & Lane, 1988). Commercial services are available for immunization and bleeding of rabbits and affinity purification of antibody.
2.3.1. Materials
Affinity-purified antibody (1mg/mL)
Nonspecific IgG antibody (see Note 1)
Phosphate-buffered saline (PBS; Teknova): 135mM NaCl, 2.7mM KCl, 4.3mM Na2HPO4, 1.4mM KH2PO4
PBS + 0.02% sodium azide (Warning: Sodium azide is toxic!)
Protein A sepharose CL-4B beads (GE Healthcare Life Sciences), stored as a 33% slurry in PBS + 0.02% sodium azide
Low-binding 0.65mL and 1.7mL microcentrifuge tubes (Corning Costar® #3206 and Sigma Aldrich #T3406)
Swinging bucket or horizontal microcentrifuge
Vacuum aspirator
Gel loading pipette tips
Ultrafine gel loading pipette tips (USA Scientific, 0.25mm OD orifice, #1022–8950)
Egg lysis buffer: 10mM HEPES, pH 7.7, 50mM KCl, 2.5mM MgCl2, 250mM sucrose. Filter sterilize and store at 4°C.
Clean scissors
2.3.2. Protocol
- Bead preparation:
- To prepare enough beads for several depletion experiments, dispense 300μL of a 33% protein A sepharose bead slurry (i.e., 100μL bed volume) to a 1.7-mL low-binding microcentrifuge tube. Centrifuge for 30s at 2000 × g to pellet the beads. Aspirate most of the supernatant with a vacuum aspirator fitted with a gel loading tip.
- Add 300μg of 1mg/mL affinity-purified antibody and incubate on a rotary mixer for 1h at room temperature. A larger quantity of antibody is necessary in some cases for proteins that are difficult to deplete. Crude serum can sometimes be used without affinity purification.
- Wash beads three times with 1mL of PBS, centrifuging 30s at 2000 × g between washes. Aspirate thoroughly with a gel loading tip followed by an ultrafine gel loading tip, and resuspend with 2 bed volumes (200μL) of PBS + 0.02% sodium azide to make a 33% slurry.
- Immunodepletion:
- Dispense 0.1 bed volumes of beads for each volume of extract you wish to deplete in two 0.65mL low-binding microcentrifuge tubes (see Note 2). For instance, to deplete 33μL of extract, aliquot 10μL of 33% bead slurry to each tube.
- Wash beads twice with 500μL of 1 × ELB, centrifuging for 1min at 2000 × g after each wash.
- Supplement extract with 0.5mg/mL nocodazole in DMSO to a final concentration of 8ng/μL to prevent microtubule formation. Mix well.
- Thoroughly aspirate buffer from the first tube of beads using an ultrafine gel loading tip and add extract to the beads. Inject a small (~1μL) bubble to facilitate mixing.
- Place the tube on a rotary mixer and rotate at a moderate rate (~0.1–0.2 revolutions per second) for 20min at room temperature.
- Pellet the beads by centrifuging for 1min at 2000 × g. Transfer the supernatant to the second tube of beads. To avoid transferring beads from one tube to another, it helps to first remove the bulk of the extract using a regular pipette tip and then remove the remainder with an ultrafine gel loading tip immersed in the beads. Cut off the end of the tip with a pair of clean scissors to avoid transferring beads and to make it easier to dispense the extract from the tip.
- Repeat steps e and f. After the final round of immunodepletion, the extract may be stored on ice for over an hour without loss of activity.
2.3.3. Notes
A mock depletion control with nonspecific IgG bound to protein A sepharose beads is typically included. Nonspecific IgG can be purchased or purified from the serum of nonimmunized rabbits (normal rabbit serum) using protein A sepharose (Harlow & Lane, 1988). The most important control is to rescue any defects resulting from immunodepletion with the addition of recombinant protein added at or near endogenous concentration.
Immunodepletion conditions are optimized for each protein. For each round, we use 0.1–0.2 bed volumes of beads per 1 volume of extract. A 20-min incubation time at room temperature has been effective for all NHEJ factors tested so far; however, some proteins may require a longer incubation at 4°C.
3. SINGLE-MOLECULE END JOINING ASSAY
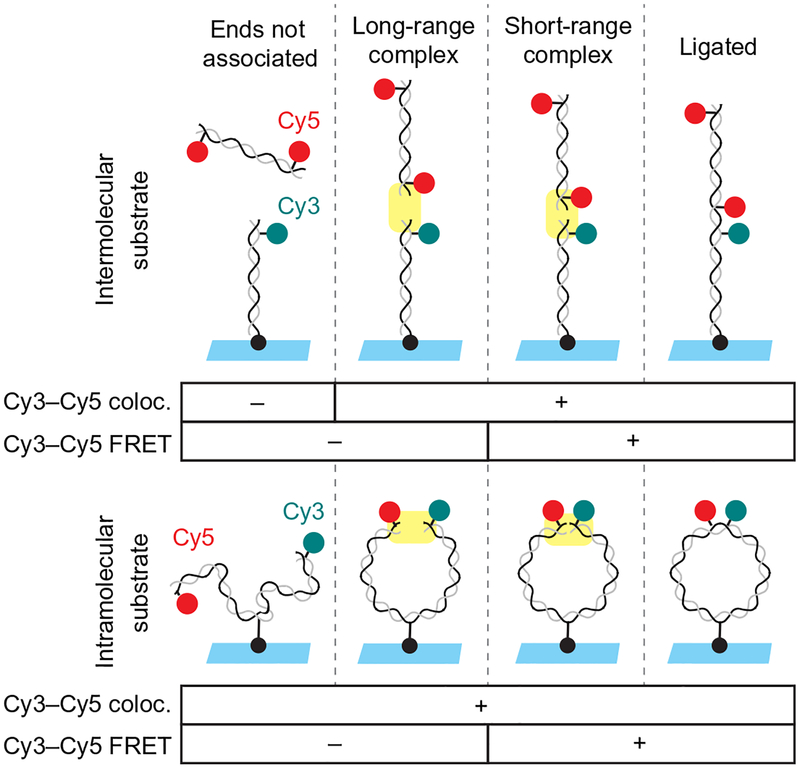
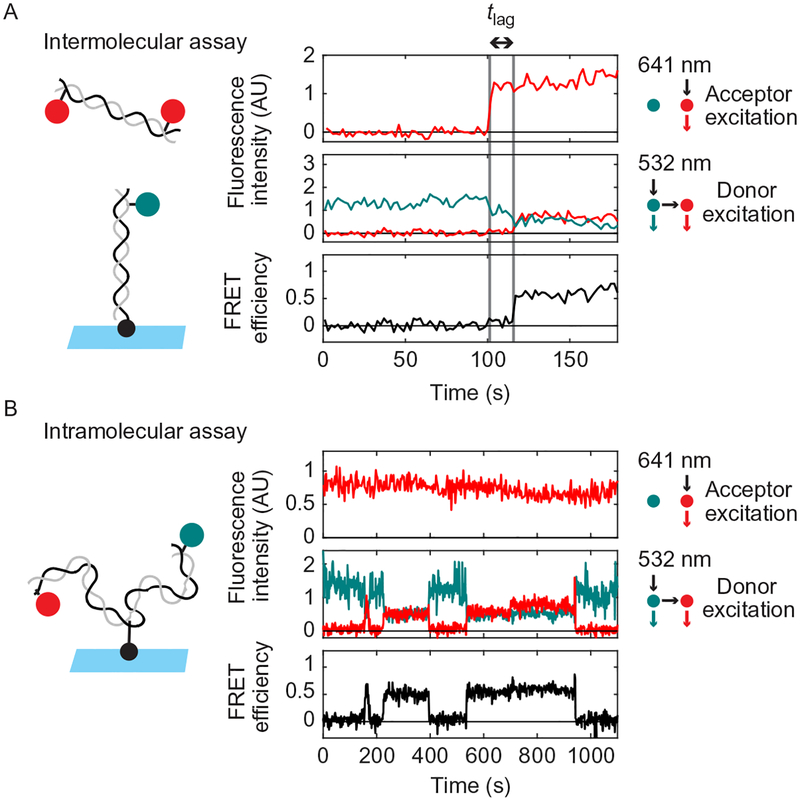
While the ensemble assay described earlier allows covalently joined NHEJ products to be observed, it does not permit the detection of transient, noncovalent reaction intermediates. Single-molecule fluorescence colocalization and FRET can be used to monitor bridging of DNA ends. We have developed two assay configurations (Fig. 2): In the “intermolecular” version of the assay, a short DNA duplex is tethered to a glass surface at one end and labeled at the other end with Cy3 (Fig. 2, upper panel). This tethered DNA duplex is incubated with egg extract containing a second DNA duplex that is labeled with Cy5 near both ends. This assay revealed two stages in DNA bridging (Graham et al., 2016): the Cy3- and Cy5-labeled DNA duplexes are first tethered in a “long-range” complex in which Cy3 and Cy5 colocalize, but the dyes are not sufficiently close for detection of FRET. The long-range synaptic complex is subsequently converted into a “short-range” complex in which DNA ends are aligned, as indicated by an increase in FRET signal. The short-range complex can also be studied using an “intramolecular” assay in which the two ends of a longer, internally tethered DNA are labeled with Cy3 and Cy5 (Fig. 2, lower panel). Though formation of the long-range complex cannot be detected using this substrate, it is useful for characterizing the high-FRET short-range complex, which forms at a high rate due to the increased local concentration of DNA ends. Using these two assays, we previously showed that formation of the long-range synaptic complex requires Ku and DNA-PKcs but not DNA-PK catalytic activity, XLF, or XRCC4-LIG4, while formation of the short-range synaptic complex requires DNA-PK catalytic activity, XLF, and XRCC4-LIG4, but not LIG4 catalytic activity.
Fig. 2.
Schematic of intramolecular and intermolecular single-molecule DNA bridging assays. The intermolecular assay (upper panel) relies on a 100-bp DNA substrate labeled near one end with Cy3 (cyan circle) and tethered to the coverslip at the other end by a biotin–streptavidin attachment (black circle). This is incubated with egg extract containing a second 100-bp DNA labeled near both ends with Cy5 (red circles). Formation of the long-range synaptic complex (Graham et al., 2016) is detected in the inter-molecular assay based on the appearance of a discrete Cy5 spot that colocalizes with a Cy3-labeled DNA on the surface. Subsequent formation of the short-range synaptic complex is indicated by the appearance of FRET between Cy3 and Cy5. The intramolecular DNA substrate (lower panel) consists of a single, 2-kb DNA labeled near one end with Cy3 and near the other end with Cy5 and tethered to a streptavidin-coated coverslip via an internal biotin. Formation of the short-range synaptic complex (Graham et al., 2016) is indicated by appearance of FRET between Cy3 and Cy5.
3.1. Substrate Preparation
3.1.1. Short Duplexes for Intermolecular Assay
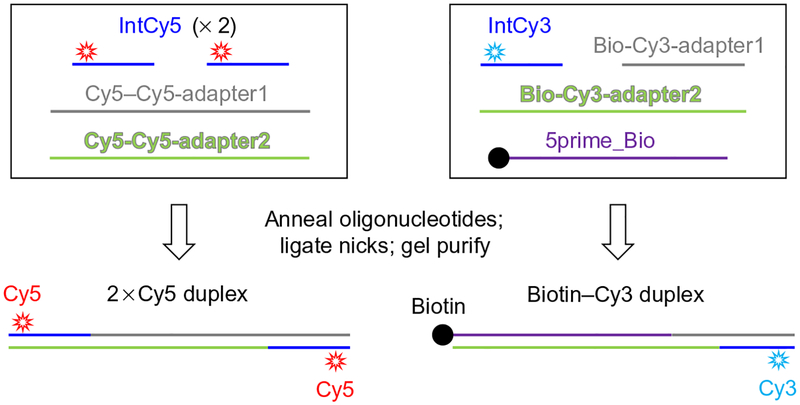
This protocol describes the preparation of labeled 100bp DNA duplexes suitable for the intermolecular single-molecule NHEJ assay (Figs. 2, upper panel, and 3). Similar results have been obtained for longer (~1kb) DNA fragments; however, these longer DNAs are susceptible to aggregation in the extract, probably as a result of chromatinization.
Fig. 3.
Preparation of substrates for intermolecular single-molecule assay.
3.1.1.1. Materials
Fluorescently labeled oligonucleotides. Oligonucleotides labeled internally 7nt from the 5′ end permit end joining while allowing FRET to be observed. We used the following sequences:
IntCy3 5′ GGATCT/iCy3N/ACCGCTGTTGAGATC 3′. IntCy5 5′ AACTCT/iCy5N/TTTCCGAAGGTAACTGG 3′.
/iCy3N/ and /iCy5N/ represent C6-amino-deoxyuridine residues labeled with Cy3 and Cy5 NHS esters (off-catalog modification from Integrated DNA Technologies).
The following long oligonucleotides:
2xCy5 substrate
Cy5–Cy5-adapter1 5′/5Phos/acatttactctctaacatcacgcctagatagaaacagatagcttgaacagatCC AGTTACCTTCGGAAAAAGAGTT 3′ Cy5–Cy5-adapter2 5′/5Phos/atctgttcaagctatctgtttctatctaggcgtgatgttagagagtaaatgtCCA GTTACCTTCGGAAAAAGAGTT 3′
Biotin-Cy3 substrate
Bio-Cy3-adapter1 5′/5Phos/GTTACATCGAACTGGATCTCAACAGCGGTAAGATCC 3′ Bio-Cy3-adapter2 5′/5Phos/CAGTTCGATGTAACTTAGAACCGCATCTTT CACAGGTTCTTTTTGCTCTCATTGTTAGTCATTTGT CAGATTCAACTG 3′ 5prime_Bio 5′/5Biosg/CAGTTGAATCTGACAAATGACTAACAATGA GAGCAAAAAGAACCTGTGAAAGATGCGGTTCTAA 3′
/5Phos/ represents 5′ phosphorylation and /5Biosg/ represents a 5′ biotin modification (Integrated DNA Technologies)
5% 19:1 bis-acrylamide 0.5 × TBE-PAGE gel
Low-binding 0.65 and 1.7mL microcentrifuge tubes (Corning Costar® #3206 and Sigma Aldrich #T3406)
27-gauge needle
Isopropanol
NEBuffer 3 (New England Biolabs; 100mM NaCl, 50mM Tris–HCl, 10mM MgCl2, 1mM DTT, pH 7.9)
20mM ATP
T4 DNA ligase (New England Biolabs, #M0202M)
Cellulose acetate spin filters (Corning Costar Spin-X)
10-bp DNA ladder (Invitrogen, #10821–015)
3.1.1.2. Procedure
- Mix equimolar quantities of the following oligonucleotides in 25μL of 1 × NEBuffer 3 (New England Biolabs; 100mM NaCl, 50mM Tris–HCl, 10mM MgCl2, 1mM DTT, pH 7.9):
- Cy5 duplex: Cy5–Cy5-adapter1, Cy5–Cy5-adapter2, and IntCy5 (add two molar equivalents of IntCy5, which anneals on both sides of the duplex)
- Biotin-Cy3 duplex: Bio-Cy3-adapter1, Bio-Cy3-adapter2, 5prime_Bio, and IntCy3
Heat to 95°C for 1min in a PCR machine and then cool at 0.1°C/s to 25°C.
Add 1.3μL of 20mM ATP and 1μL of T4 DNA ligase and incubate for 1h at 37°C to seal nicks in the DNA.
Separate products on a 5% 19:1 bis-acrylamide 0.5 × TBE-PAGE gel, dividing the reaction between several lanes. Run a DNA ladder and the original oligonucleotides in separate lanes to verify the size of the final product and to confirm that it is well separated from the reactants.
Place the gel against a light background. Using a sharp scalpel or a razor blade, excise the product, which should be visible without staining as a bright pink (Cy3) or blue (Cy5) band. Excise as narrow a slice of polyacrylamide as possible while recovering the bulk of the product. Stain the remainder of the gel for about 2–5min with 1μg/mL ethidium bromide in 0.5 × TBE to visualize the molecular weight standards. Image promptly to minimize diffusion of the DNA out of the gel.
Place the gel band in a low-binding 0.65mL microcentrifuge tube that is clean on the outside. Regular tubes may also be used. Use a 27-gauge needle to poke a hole in the bottom of the 0.65mL tube and place it in a 1.7-mL low-binding microcentrifuge tube. Spin at maximum speed for 1min in a microcentrifuge, which will force the gel through the needle hole, grinding it into small pieces. Use a pipette tip or clean, fine-tipped forceps to transfer residual gel pieces from the small tube to the large tube. Add 500μL of 1 × TE buffer (10mM Tris–HCl, pH 8, 1mM EDTA) to the gel fragments in the large tube and rotate for several hours to overnight on a rotary mixer at room temperature.
Using a P1000 pipette tip with the end cut off, transfer the polyacryl-amide slurry to a cellulose acetate spin filter and spin at 16,000 × g in a microcentrifuge. Add 50μL of 3 M sodium acetate, pH 5.2 and 1mL of isopropanol to the filtrate. Incubate at −20°C for ≥10min and then spin at maximum speed in a 4°C microcentrifuge for 30min. A brightly colored pellet should be visible. Aspirate the supernatant and wash the pellet with 70% ethanol. Respin for 1min and dissolve in 1 × ELB salts (10mM HEPES, pH 7.7, 50mM KCl, 2.5mM MgCl2). Determine the concentration of the DNA and the fluorescent dyes using a Nanodrop spectrophotometer.
3.1.2. Blunt-Ended Intramolecular (Circularization) Substrate
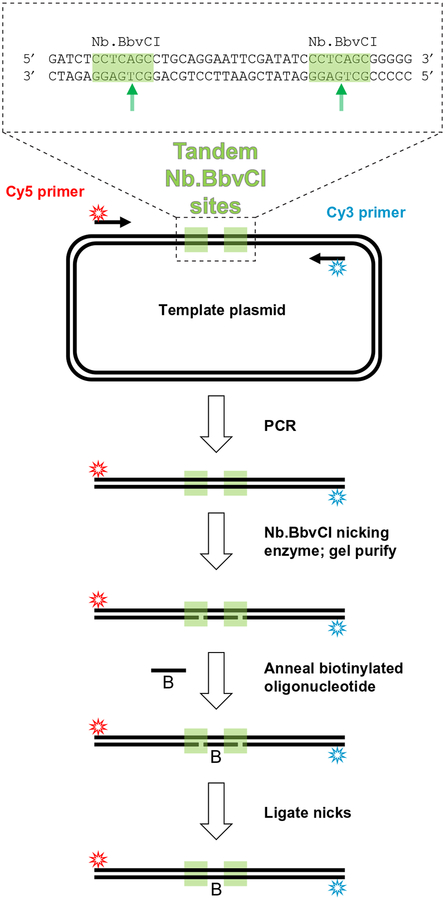
This section describes how to prepare a single-molecule FRET reporter for monitoring bridging of the two ends of a single linear DNA molecule (Figs. 2, lower panel, and 4). This reporter is a 2-kbp long, internally biotinylated DNA fragment labeled 7nt from one end with Cy3 and 7nt from the other end with Cy5. A PCR product is first generated with internally Cy3- and Cy5-labeled primers using a template that contains tandem sites for the nicking restriction endonuclease Nb.BbvCI (see Note 1). After digestion of the PCR product with the nicking enzyme, the short oligonucleotide between these restriction sites is replaced with an excess of internally biotinylated, phosphorylated oligonucleotide by heat denaturation followed by annealing. T4 DNA ligase is used to seal the nicks in the DNA, and the final product is gel purified, yielding an internally biotinylated, double-stranded DNA molecule with Cy3 and Cy5 labels near each end.
Fig. 4.
Intramolecular FRET substrate preparation.
3.1.2.1. Materials
The same fluorescently labeled oligonucleotides used for making the 100bp duplex substrates (see above).
2 × Q5 polymerase master mix (NEB) or similar PCR master mix. Taq polymerase should not be used as it will add a nontemplated dA nucleotide to the 3′ end of the PCR product.
Template DNA. The template contains tandem recognition sites for the nicking restriction endonuclease Nb.BbvCI (Fig. 4). Our full-length template plasmid is available upon request.
Internally biotinylated oligonucleotide; identical to the intervening sequence (lower strand) between the nicking sites of the PCR template:
5′ /5Phos/TGAGGGATATCGAA/iBiodUK/TCCTGCAGGC 3′
/5Phos/ represents a 5′ phosphate modification and /iBiodUK/ represents an internal biotinylated deoxyuridine (Integrated DNA Technologies).
Dialysis tubing (SpectraPor #132650, 23mm, 6–8kDa MWCO) and clips
4°C microcentrifuge
Nb.BbvCI restriction enzyme (New England Biolabs, Cat. #R0631S)
T4 DNA ligase (New England Biolabs, #M0202M)
Molecular biology grade agarose
TBE buffer (see above)
1 × TE buffer (see above)
Phenol:chloroform:isoamyl alcohol (25:24:1) mixture, equilibrated with aqueous buffer at pH 8
Isobutanol, equilibrated with aqueous buffer at pH 8
3 M sodium acetate, pH 5.2
100% and 70% ethanol
Clean razor blade or scalpel
Ethidium bromide 10mg/mL solution
Nanodrop spectrophotometer
3.1.2.2. Protocol
In a 400μL total reaction volume, PCR amplify a 2-kbp fragment using the above primer and template combination. We use Q5 polymerase 2 × master mix (New England Biolabs). Note: Prior to ordering expensive fluorescently labeled primers, verify that the PCR works with unlabeled primers.
Extract the PCR product once with 1 volume of 25:24:1 phenol:chloroform:isoamyl alcohol and once with 1 volume of chloroform. Centrifuge briefly at maximum speed in a microcentrifuge after each extraction step. Retain the aqueous fraction, which will be on top.
Precipitate the DNA by adding 40μL of 3 M sodium acetate, pH 5.2 and 800μL of 100% ethanol. Incubate on ice for ≥10min and then spin in a 4°C microcentrifuge for 30min at maximum speed. Wash the pellet with 70% ethanol, spin briefly, and aspirate the ethanol thoroughly with an ultrafine gel loading tip.
Redissolve the pellet in 200μL of 1 × CutSmart buffer (New England Biolabs; 50mM potassium acetate, 20mM Tris–acetate, 10mM magnesium acetate, 100μg/mL bovine serum albumin, pH 7.9).
Digest for 1h at 37°C with 4μL (40U) of Nb.BbvCI.
Separate the reaction products on a 1 × TBE 0.8% agarose gel containing 1μg/mL ethidium bromide.
Cut out the product band with a clean razor blade or a scalpel.
Extract and purify DNA from the gel slice using the electroelution protocol given earlier for preparation of the ensemble end joining substrate. DNA may also be purified by splitting the product between several spin columns of a commercial gel purification kit; however, this typically gives a lower yield.
Determine the concentration of the purified DNA using a spectrophotometer that can accommodate small volumes (e.g., Nanodrop). Add 10 molar equivalents of an internally biotinylated oligonucleotide and 0.1 volumes of 10 × T4 DNA ligase buffer (New England Biolabs). Heat to 80°C for 5min and slowly cool to room temperature.
Add 1μL of T4 DNA ligase and incubate either at 37°C for 1h or overnight at room temperature to seal the nicks in the DNA. (Note: Because the DNA ends are not phosphorylated, they will not be ligated in this step.)
Heat-inactivate T4 DNA ligase at 65°C for 10min and store the ligated product at −20°C.
3.1.2.3. Notes
Nb.BbvCI retains residual activity toward the top strand, meaning that this enzyme can generate some double-strand breaks in DNA as well as nicks. Nt.BbvCI, the BbvCI mutant that nicks the top strand rather than the bottom strand, has higher fidelity (New England Biolabs, personal communication). Using Nt.BbvCI, in combination with a top-strand biotinylated oligonucleotide, may help to limit unwanted double-strand cleavage of the PCR product.
Our protocol does not remove free biotinylated oligonucleotide, which does not seem to interfere with the single-molecule assay.
3.2. Preparation of Flowcells
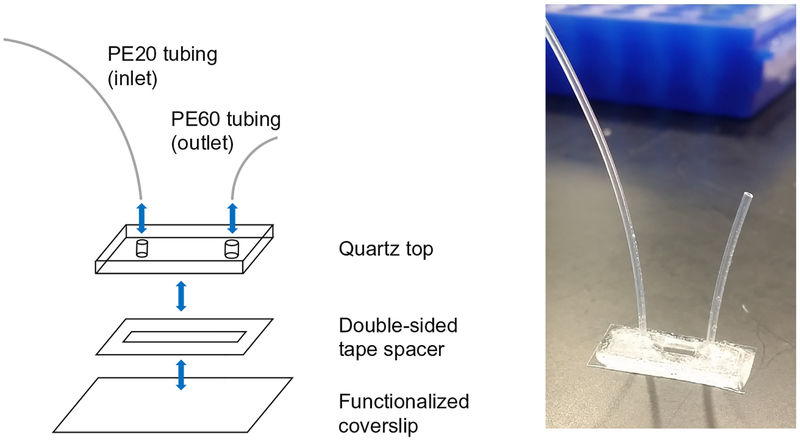
Glass coverslips are passivated and functionalized with a mixture of polyethylene glycol (PEG) and biotin-PEG, as described previously (Tanner & van Oijen, 2010). A simple flowcell is constructed by cutting a small channel in a piece of double-sided tape and sandwiching this between the functionalized coverslip and a quartz top containing two drilled holes. Plastic tubing is inserted into the holes, and the assembly is sealed with epoxy (Fig. 5).
Fig. 5.
Schematic of flowcell assembly (left) and completed flowcell (right).
3.2.1. Materials
For coverslip functionalization (see Tanner & van Oijen, 2010 for protocol):
1 M potassium hydroxide
100% ethanol
Acetone, ACS reagent grade
Aminopropyltriethoxysilane
Sodium bicarbonate
mPEG-SVA and Biotin-mPEG-SVA (Laysan)
Water bath sonicator
Coverslips (VWR Micro Cover Glasses, No. 1.5)
For flowcell construction:
PE20 and PE60 polyethylene tubing (BD Intramedic, #427416 and 427406)
Quartz tops (OZ grade clear fused quartz plate, 20 × 7 × 1mm custom size)
Epoxy (Devcon 5 Minute Epoxy #14250)
Diamond-tipped scribe
Double-sided tape sheet (7” × 10”, 0.12-mm thick, double-sided SecureSeal™ Adhesive Sheet, Grace Biolabs #620001)
Flat plastic coverslip forceps
Flat metal forceps
No. 5 scalpel handle and stainless steel blades
Scissors
Plastic cutting board
Dremel tool with a diamond-tipped drill bit (A&M Instruments; 1.2mm Flame Medium 3/32” Shank, #HP863–012)
3.2.2. Protocol
- Prepare functionalized coverslips essentially as described before (Tanner & van Oijen, 2010), with the following modifications (step numbers refer to the protocol provided in the reference):
- Do not quench the silanization reaction by flooding staining jars with a large volume of water (step 3). Rather, discard the silanization mixture in a hazardous waste bottle and rinse the coverslips five to six times with ultrapure water.
- In step 5, use mPEG-SVA and Biotin-mPEG-SVA from Laysan Bio, Inc.
- Place silanized coverslips directly on top of each other during PEGylation in step 6 instead of using spacer coverslips.
- Do not use compressed gas to dry the coverslips in step 7. Instead, extensively wash the coverslip with ultrapure water until it forms tight beads on the surface. Blot remaining beads of water from the edges using a lint-free wipe.
- Prepare quartz tops:
- Drill two holes in a 20 × 6 × 1-mm piece of quartz using a Dremel rotary tool with a diamond-tipped bit. Support the piece of quartz on a stack of paper towels submerged in water. A relatively small amount of force is required to drill through the quartz. Pressing too hard can easily break the quartz. Continue drilling each hole until it is just wide enough to accommodate a piece of PE20 (inlet) or PE60 (outlet) tubing.
- If you are recycling an already-used quartz top, wash it thoroughly with acetone, scrape off remnants of double-sided tape and epoxy with a razor blade and remove debris from the holes by pushing through a piece of PE20/PE60 tubing and cutting it off on the other side with a sharp razor blade.
Cut a 1-mm wide, 1-cm long channel out of double-sided tape using a razor blade or a sharp scalpel.
Peel off the transparent side of the double-sided tape, and stick it to a quartz top, making sure that the holes are aligned with the channel.
Using the end of a pair of blunt forceps or the bottom of a microcentrifuge tube, press against the double-sided tape to eliminate any air-filled spaces between it and the quartz.
Use a diamond-tipped scribe to cut a piece of a functionalized coverslip slightly larger than the quartz top. When cutting coverslips, hold the scribe vertically and do not press very hard. Practice with nonfunctionalized coverslips before using functionalized coverslips.
Peel off the other side of the double-sided tape on the quartz top and place the quartz top in the center of the coverslip.
With the quartz top flat on the benchtop, carefully press on the cover-slip with the tip of a pair of blunt plastic forceps or the bottom of a microcentrifuge tube to eliminate any air-filled spaces between the coverslip and the tape. It is important to press gently, as the coverslip can easily crack.
Cut a 2-cm piece of PE60 tubing for the outlet and a 10-cm piece of PE20 tubing for the inlet. To avoid clogging, angle each cut slightly so that the end of the tubing will not be pressed flat against the coverslip.
Turn the flowcell over and insert the PE20 and PE60 tubing into the appropriate holes. It helps to grasp the tubing near the end using a pair of flat metal forceps.
Use epoxy to glue the tubing in place. Touch the tubing on each side with a drop of epoxy using a gel loading pipette tip and then drag the tip to connect the two drops. Then, use epoxy to seal the edges of the quartz top. The epoxy should extend from the top surface of the quartz down to the coverslip.
3.3. Single-Molecule NHEJ Assay
This assay monitors the bridging of single pairs of DNA ends by smFRET. The intramolecular and intermolecular assays are performed in essentially the same way, except that in the latter case, a second labeled DNA fragment is included in the extract mixture. Alternating laser excitation (ALEX) is used to visualize both the donor (Cy3) and the acceptor (Cy5) dyes. This is important, as it allows loss of FRET signal due to dye separation to be distinguished from loss of FRET signal due to bleaching or blinking of Cy5.
3.3.1. Materials
Labeled DNA substrate(s) (see above)
1 × egg lysis buffer (ELB) salts—10mM HEPES, pH 7.7, 50mM KCl, 2.5mM MgCl2
Bell jar connected to a vacuum line
Streptavidin—1mg/mL in PBS
Laser power meter (LaserMate Q, Coherent)
- Control sample for aligning channels (see Section 4.3). We use one of the two samples:
- A “nanogrid” of 150nm holes in a thin metal film, generated by electron beam lithography, which is imaged with transillumination to provide an array of calibration points (Baday et al., 2012).
- TetraSpeck fluorescent microspheres (Thermo Fisher) deposited on a glass coverslip, which are imaged using total internal reflection fluorescence (TIRF).
Protocatechuic acid (PCA), 250mM solution in 1 × ELB salts. Adjust the stock solution to pH 7.7 with NaOH. Store large aliquots at −80°C and a small working aliquot at −20°C. Make a new working aliquot when the old one begins yellowing, which indicates oxidation.
Protocatechuate 3,4-dioxygenase (PCD), 5μM solution in 10mM HEPES, pH 7.5, 50mM KCl, 1.25mM MgCl2, 50% glycerol. Store aliquots at −20°C.
Creatine phosphokinase (CPK), adenosine triphosphate (ATP), and phosphocreatine (PC) stock solutions; see ensemble end joining reaction protocol earlier.
Maxiprepped plasmid DNA to use as carrier, 1μg/μL in 1 × TE (10mM Tris–HCl, pH 8, 1mM EDTA)
3.3.2. Microscope Setup
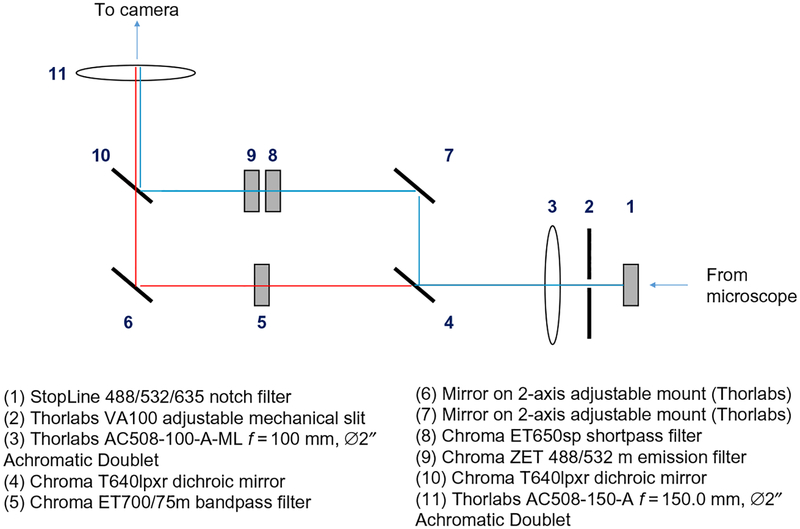
For our single-molecule experiments, we use a home-built, through-objective TIRF microscope with laser illumination. A home-built dual view device (Fig. 6) permits simultaneous wide-field imaging of Cy3 and Cy5 emission. Our instrument is constructed around an Olympus IX-71 upright microscope fitted with a ZT532/638rpc filter cube (Chroma). Beams from a Coherent Sapphire 532-nm laser and a Coherent CUBE 641-nm laser are expanded with telescopes made of pairs of converging lenses, combined using dichroic mirrors, expanded again with another two-lens telescope, and directed into the back port of the microscope. The laser beam is focused at the back focal plane of the objective using a lens on a vertical translation mount, whose position can be adjusted to set the TIRF angle. Fluorescence emission is directed through the side port of the microscope into a homemade dual view (Fig. 6), which images Cy3 and Cy5 emission on the two halves of an EMCCD camera (ImageEM, Hamamatsu). Lasers are switched on and off with Uniblitz VS14 shutters controlled by a Uniblitz VMM-D3 3-channel shutter controller. Shutter timing is synchronized with the output trigger signal of the camera using an NI USB-6009 DAQ card (National Instruments) controlled by custom software written in LabView. A motorized microstage (Mad City Labs) is used to position the sample in the x–y plane and is controlled with LabView software provided by the manufacturer. A Cordless Rumble Pad 2 game controller (Logitech) is used for manual stage control, and custom LabView software (available upon request) is used to coordinate automated movement sequences with shutter timing.
Fig. 6.
Schematic of home-built dual view for separating Cy3 and Cy5 emission signals.
3.3.3. Protocol
Turn on the EMCCD camera of the microscope to allow the camera chip to cool down, and turn on the lasers.
Place 10mL of 1 × ELB salts in a conical tube with its lid loosened; allow to sit under vacuum in the bell jar for at least 10min to degas. Degassing will reduce the amount of oxygen in the solution and prevent the formation of bubbles in the flowcell.
While the 1 × ELB salts is degassing, use a gel loading tip to inject 15μL of 1mg/mL streptavidin in PBS into the outlet PE60 tubing of a micro-fluidic flowcell. Ensure that streptavidin gets all the way through the channel and comes out into the PE20 tubing on the other side. Allow the streptavidin to sit in the flowcell for ≥5min.
Set the power of each laser to the desired value using a laser power meter. For simplicity, we typically measure the laser power at one of its focal points on the table and later determine the corresponding power density at the sample.
Place a drop of immersion oil on the objective lens, being very careful to avoid air bubbles, which can skew the angle of the TIRF beam. Moving fringes of light and dark intensity or a large dark patch that creeps into the image are symptomatic of immersion oil bubbles.
Mount the flowcell securely on the microscope and raise the objective until a circle of immersion oil touches the coverslip.
Using brightfield illumination (by eye or with EM gain set to 0 on the camera), focus on the double-sided tape on the edge of the channel. This will bring the objective near the correct focal plane.
Wash the streptavidin out of the flowcell with ELB salts. To this end, place the inlet tubing in the bottom of a 0.65-mL microcentrifuge tube containing 0.5mL of ELB salts. Insert a gel loading tip attached to a P200 pipette into the outlet and draw 200μL of the buffer through the flowcell. When removing the gel loading tip from the tubing, hold on to the tubing so as not to accidentally rip it out of the flowcell. Alternatively, a syringe pump connected to the outlet may be used to draw solutions into the flowcell.
Draw ~30–50μL of a dilution of the appropriate biotinylated DNA substrate into the flowcell. We do not generally determine the absolute concentration of the substrate, but instead titrate the concentration empirically for each new substrate preparation and batch of coverslips until the desired density of substrates on the surface is achieved. Substrates should be tethered at a low enough density that they are mostly nonoverlapping (~300 substrates per field of view (FOV) on our microscope).
Open the shutter on the 532-nm laser, and focus on the substrate DNA, imaging at about 100ms per frame. Focus slowly using the fine focus knob. Near the correct plane of focus, the relative background intensity in the two channels changes in a characteristic way that becomes recognizable with experience. One common mistake is focusing on the wrong (bottom) side of the coverslip. For the intramolecular substrate, check that the microscope is focused on the top side by switching to the 641-nm laser; a similar density of spots in the Cy5 channel should be visible. If there is any ambiguity, focus downward to the bottom surface of the coverslip (where surface-bound fluorescent contaminants should be visible) and then focus back up to the top surface.
Wash the channel again with ELB salts.
Supplement a 33-μL aliquot of HSS with 0.5μL of 0.5mg/mL nocodazole in DMSO. Mix thoroughly.
- Prepare an ATP regeneration system by combining the following:
- 0.5μL of 5mg/mL creatine phosphokinase
- 5μL of 200mM ATP
- 10μL of 1 M phosphocreatine
- Make a reaction mix with HSS of egg cytosol:
- 25μL HSS supplemented with nocodazole
- 2.5μL of 1μg/μL closed-circular plasmid DNA (“carrier DNA”). A lower concentration of carrier DNA may be optimal for some preparations of extract. This can be optimized in ensemble end joining experiments (see above)
- 0.8μL ATP regeneration system
- 0.6μL of 250mM PCA
- 0.6μL of 5μM PCD
- 0.6μL of 50mM Trolox. Mix well after adding the Trolox.
- 1μL of 100nM 2xCy5-labeled 100bp duplex (for intermolecular assay only)
Pop any bubbles by holding the tube firmly against one index finger and flicking sharply with the nail of the other index finger. Spin 30s at maximum speed in a microcentrifuge.
Prepare shutter control software (we use custom LabView code, which is available upon request) and image acquisition software to begin recording as soon as extract is pulled into the flowcell. We typically excite Cy3 with one or two frames of 532nm light for every one frame of Cy5 excitation with 641nm light. Illumination may be continuous or stroboscopic. Continuous illumination at a lower laser power is generally preferable. Typical laser powers through the objective for a 500-ms exposure are 0.1mW for the 647nm laser and 0.5mW for the 532nm laser.
Pull the extract into the flowcell, turn off the room lights, and begin recording a movie. Manual refocusing may be necessary if a single FOV is imaged for an extended period of time. Switch to fresh FOVs as desired. To collect long trajectories, we sometimes record in a single FOV for 30min with intermittent illumination. To obtain overall rates of long- and short-range synaptic complex formation, each FOV may be sampled for a shorter period of time—typically 18–30s for the intra-molecular assay or 3min for the intermolecular assay.
At the end of the experiment, acquire several images of a nanogrid of subdiffraction-limited holes under transillumination (with camera EM gain turned off) or a slide with TetraSpeck fluorescent beads under TIRF illumination to serve as a standard for registering the two channels. A new set of images should be acquired for every day of experiments, as the optics may shift (or be deliberately realigned) from day to day. For bead samples, data can be combined from several FOVs to achieve uniform coverage of the visible area.
Quartz tops can be recycled by plucking the tubing out of the flowcell and submerging the flowcell in acetone overnight. This will soften the epoxy and double-sided tape, allowing the coverslip and quartz top to be peeled apart.
4. ANALYSIS OF SINGLE-MOLECULE DATA
4.1. Opening Data in MATLAB
We analyze our imaging data using custom MATLAB scripts, which are available upon request. Data are recorded using HCImage software and are saved as “.cxd” files. This file format, and a variety of other imaging file formats, can be imported into MATLAB using the BioFormats package. To use this plugin, download the MATLAB Toolbox from the BioFormats website (http://downloads.openmicroscopy.org/bio-formats/5.2.4/), extract the downloaded archive, and add the extracted folder to the MATLAB search path using the addpath command.
4.2. Field of View Segmentation
A single movie file may contain data from many different FOVs. Movie segments from different FOVs are divided using an automated function that calculates the correlation between pixel intensities in successive frames. A change of FOV is indicated by a drop in this correlation below a user-defined threshold, which may need to be manually adjusted between movies, depending on the number of tethered substrates and the amount of background noise in the images.
4.3. Channel Alignment
The Cy3 and Cy5 channels of the microscope are realigned to within a few pixels prior to each experiment. However, because of slight imperfections in the alignment and distortions in the optics, it is necessary to register the channels more precisely during data analysis. We do this in one of the two ways.
For the intramolecular FRET reporter, for which the same molecules are reliably labeled with Cy3 and Cy5, the substrates themselves may be used for channel alignment. Each channel is divided into eight sectors, and the optimal translation for aligning each sector is determined by image correlation between the Cy3 and Cy5 channels across several FOVs.
Alternatively, a reference sample containing point sources that appear in both the Cy3 and Cy5 channels may be used for alignment. A coverslip with nonspecifically adsorbed TetraSpeck fluorescent microspheres (Thermo Fisher) works for this purpose. However, we typically image a trans-illuminated “nanogrid” consisting of a thin film of aluminum containing 150nm holes generated by electron beam lithography. The channels are first roughly aligned using image correlation to find an approximate translation over the entire visible area. Small defects in the array facilitate unambiguous alignment. Points are detected in one channel using a peak-finding function, and approximate positions in the other channel are found by applying the translation. The points in the two channels are then localized more precisely by two-dimensional Gaussian fitting. The MATLAB function cp2tform is used to generate a spatial transformation function between the fitted points in the two channels.
4.4. Drift Correction
Within each single-FOV movie segment, drift is corrected by calculating the spatial cross-correlation function between the initial frame in the segment and each subsequent frame. This, and other operations involving image correlation, can be done efficiently by Fourier transforming each image (MATLAB function fft2) multiplying in the frequency domain, and inverse transforming (MATLAB function ifft2). The peak of the cross-correlation function, rounded to the nearest integer, is taken to be the offset of the frame in pixels from the initial frame in the segment.
4.5. Extraction of Integrated Intensities
For locating single DNA substrates, the first several frames in each FOV are averaged and processed with a spatial bandpass filter to extract features the size of a diffraction-limited spot. Points of interest are identified in one of the two channels using a peak-finding function that identifies local maxima that are not closer together than some minimum distance (typically 7 pixels). For the intermolecular substrate, statically bound DNAs are located in the Cy3 channel. For the intramolecular FRET reporter, which is labeled with both Cy3 and Cy5, points are located in the Cy5 channel to avoid bias against high-FRET spots. A transformation function between the two channels (see above) is used to find corresponding points in the other channel.
The fluorescence intensity of each substrate is determined by summing the pixel intensities in a disc of radius 4 pixels centered on the peak. Local background is determined as the mean intensity of a circle of pixels surrounding this disc. The background-subtracted intensity is given by: (summed intensity of the disc) – (average background intensity) * (area of the disc).
4.6. Analysis and Interpretation of Single-Molecule Traces
The earlier analysis yields single-substrate traces of Cy3 and Cy5 emission with 532nm excitation as well as Cy5 emission with 641nm excitation. FRET efficiency (EFRET) is calculated after applying a correction for Cy5 direct excitation by 532nm light, Cy3 bleedthrough into the Cy5 channel, and differences in quantum yield and detection efficiency of Cy3 and Cy5 emission, as described previously (Lee et al., 2005).
4.6.1. Analysis of Traces From the Intermolecular Assay
For the intermolecular assay, we manually inspect traces using a homemade browser function written in MATLAB. The times of Cy5-DNA binding (long-range complex formation) and transition to FRET (short-range complex formation) are manually annotated within the browser function. An example of a Cy5-DNA-binding event followed by a transition to high FRET is shown in Fig. 7A. A waiting time histogram can be compiled for the transition between long- and short-range complexes.
Fig. 7.
Example single-molecule traces from the intermolecular (A) and intramolecular (B) NHEJ assays. The upper panel in (A) and (B) shows Cy5 emission with direct excitation by 641nm light. The middle panel shows Cy3 and Cy5 emission with excitation of Cy3 by 532nm light. The lower panel shows calculated FRET efficiency. Substrate schematics are shown to the left. Note that 0s corresponds to the first frame in the particular field of view being imaged, not the time of extract addition. (A) In the intermolecular assay, binding of a Cy5-DNA to a Cy3-DNA on the surface is detected by appearance of Cy5 signal with 641nm excitation (first dashed gray line). Formation of the short-range synaptic complex is indicated by the appearance of a high-FRET signal (second dashed gray line) after a time delay (“tlag”). (B) A sample trace from the intramolecular assay showing three rounds of short-range complex formation and dissolution, as indicated by increases and decreases in FRET. This trace was acquired in extract immunodepleted of XRCC4-LIG4 and supplemented with catalytically inactive XRCC4-LIG4K278R complex, which supports short-range complex formation but not ligation.
A survival curve for the long-range complex and the overall rate of long-range complex formation are determined by detecting Cy5-DNA-binding events in an automated fashion: A histogram of background-subtracted Cy5 intensity is first compiled for all particles over all frames of the movie. Because most Cy3-DNAs are not bound by a Cy5-DNA, the highest peak of this histogram represents zero Cy5 molecules. This peak is fit locally to a Gaussian function, and a threshold is chosen two standard deviations above the mean. Cy5-binding events are assigned whenever the Cy5 intensity of a particular spot crosses this threshold. A survival time distribution is then calculated from the durations of these binding events.
Calculating the survival time distribution of the long-range complex from Cy5 traces is complicated by two considerations: (1) some Cy5-positive spots may transition to a short-range complex, and (2) some Cy5-binding events may be cut off by the end of a movie. To deal with this issue, we calculate survival curves using the Kaplan–Meier estimator, which takes into account the fact that survival times may be known incompletely if individuals (in this case, molecules) are lost to follow-up measurements, a condition known as “right censorship.” We consider Cy5-binding events to be right-censored by transitions to high FRET or by the end of a movie. Events are also considered to be right-censored when Cy3 photobleaches, which makes it impossible (by measuring FRET) to determine whether the complex is in a long-range or short-range/ligated state. Our criterion for transitions to high FRET for the purpose of censoring is that the calculated FRET efficiency exceeds 0.4 for four consecutive frames (see Note 1). Our criterion for Cy3 photobleaching is that the Cy3 intensity drops two standard deviations below the center of the single-Cy3 peak in the Cy3 intensity histogram.
The rate of long-range complex formation is calculated by dividing the number of observed binding events by the number of frames in which Cy3-DNAs are “available” to be bound (i.e., not already bound and not photobleached). Cy5-DNA-binding events are not counted after Cy3 has photobleached, as this makes it impossible to determine whether the complex is in a low-FRET or a high-FRET state.
4.6.2. Analysis of Traces From the Intramolecular Assay
Similar to the intermolecular assay, traces from the intramolecular assay are inspected using a homemade browser function written in MATLAB. Photobleaching events and transitions between low- and high-FRET states are manually annotated. Events are considered right-censored for Kaplan–Meier survival curve analysis if they are terminated by the end of the movie or by photobleaching of Cy3 or Cy5.
To obtain the kinetics of high-FRET complex formation (including both short-range complex and ligated product), we use an automated stage control program to switch to a new FOV every 15–30s. This provides more statistically independent data points than imaging a single FOV, and it avoids loss of signal due to photobleaching. The FRET efficiency histogram can be obtained for the substrates within each FOV, and the fraction of FRET-positive spots (EFRET > 0.25) can be plotted as a function of time (e.g., Fig. 4 in Graham et al., 2016).
4.7. Notes
The requirement that EFRET > 0.4 over four consecutive frames helps to avoid spurious detection of FRET transitions due to measurement noise. This requirement is reasonable, given that high-FRET short-range complexes generally persist much longer than this.
5. CONCLUSIONS AND OUTLOOK
X. laevis egg extract provides a powerful approach for detailed mechanistic studies of nonhomologous end joining. The system is unique because it supports highly efficient, cell-free end joining that depends on the core end joining factors (Ku, DNA-PKcs, DNA ligase IV-XRCC4, and XLF). This property, together with the presence of other NHEJ regulators in extract, both known (e.g., DNA polymerase λ, DNA polymerase μ, PNKP, Artemis) and unknown, makes it likely that any mechanistic insights gained reflect the complexity of NHEJ as it occurs in the cell. The use of single-molecule approaches within this system makes it possible to probe transient end joining intermediates. Measuring smFRET between labeled DNA ends, while a useful readout of end proximity, is only one possible configuration of the assay. Many variations can be envisioned, including multiwavelength imaging of fluorescently labeled proteins together with FRET between DNA ends. The extract-based approach could potentially be extended to study the competition between NHEJ and other repair pathways, such as homologous recombination (in particular, the initial 5′–3′ resection step) or Alt-EJ. Furthermore, replication-competent extracts could be used to investigate how NHEJ is suppressed at double-strand breaks arising from replication fork collapse or at DNA interstrand cross-links (Long, Räschle, Joukov, & Walter, 2011; Räschle et al., 2008; Zhang & Walter, 2014).
Footnotes
End joining independent of Ku and DNA-PKcs may also occur in egg extract (Di Virgilio & Gautier, 2005; Graham et al., 2016; Labhart, 1999b; Sandoval & Labhart, 2002). The relative contributions of classical Ku/DNA-PKcs-dependent and alternative Ku/DNA-PKcs-independent end joining pathways seem to depend on the exact reaction conditions and DNA substrates employed. Under the conditions described here, end joining occurs primarily through the classical pathway (Graham et al., 2016).
REFERENCES
- Ahnesorg P, Smith P, & Jackson SP (2006). XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell, 124, 301–313. [DOI] [PubMed] [Google Scholar]
- Akopiants K, Zhou R-Z, Mohapatra S, Valerie K, Lees-Miller SP, Lee K-J, et al. (2009). Requirement for XLF/Cernunnos in alignment-based gap filling by DNA polymerases lambda and mu for nonhomologous end joining in human whole-cell extracts. Nucleic Acids Research, 37, 4055–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baday M, Cravens A, Hastie A, Kim H, Kudeki DE, Kwok P-Y, et al. (2012). Multicolor super-resolution DNA imaging for genetic analysis. Nano Letters, 12, 3861–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmus G, Barros AC, Wijnhoven PWG, Lescale C, Hasse HL, Boroviak K, et al. (2016). Synthetic lethality between PAXX and XLF in mammalian development. Genes & Development, 30, 2152–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, & West SC (1998). DNA end-joining catalyzed by human cell-free extracts. Proceedings of the National Academy of Sciences of the United States of America, 95, 14066–14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondanèche M-C, Sanal O, et al. (2006). Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell, 124, 287–299. [DOI] [PubMed] [Google Scholar]
- Budzowska M, Graham TGW, Sobeck A, Waga S, & Walter JC (2015). Regulation of the Rev1-pol ζ complex during bypass of a DNA interstrand cross-link. The EMBO Journal, 34, 1971–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter T, Vancurová I, Sun I, Lou W, & DeLeon S (1990). A DNA-activated protein kinase from HeLa cell nuclei. Molecular and Cellular Biology, 10, 6460–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HHY, Watanabe G, Gerodimos CA, Ochi T, Blundell TL, Jackson SP, et al. (2016). Different DNA end configurations dictate which NHEJ components are most important for joining efficiency. The Journal of Biological Chemistry, 291, 24377–24389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, & West SC (2002). Involvement of human polynucleotide kinase in double-strand break repair by nonhomologous end joining. The EMBO Journal, 21, 2827–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Inamdar KV, Pfeiffer P, Feldmann E, Hannah MF, Yu Y, et al. (2001). Accurate in vitro end joining of a DNA double strand break with partially cohesive 3′ overhangs and 3′-phosphoglycolate termini: Effect of Ku on repair fidelity. The Journal of Biological Chemistry, 276, 24323–24330. [DOI] [PubMed] [Google Scholar]
- Cortes P, Weis-Garcia F, Misulovin Z, Nussenzweig A, Lai JS, Li G, et al. (1996). In vitro V(D)J recombination: Signal joint formation. Proceedings of the National Academy of Sciences of the United States of America, 93, 14008–14013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craxton A, Somers J, Munnur D, Jukes-Jones R, Cain K, & Malewicz M (2015). XLS (c9orf142) is a new component of mammalian DNA double-stranded break repair. Cell Death and Differentiation, 22, 890–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchlow SE, Bowater RP, & Jackson SP (1997). Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Current Biology, 7, 588–598. [DOI] [PubMed] [Google Scholar]
- Daza P, Reichenberger S, Göttlich B, Hagmann M, Feldmann E, & Pfeiffer P (1996). Mechanisms of nonhomologous DNA end-joining in frogs, mice and men. Biological Chemistry, 377, 775–786. [DOI] [PubMed] [Google Scholar]
- Di Virgilio M, & Gautier J (2005). Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. The Journal of Cell Biology, 171, 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs TA, Tainer JA, & Lees-Miller SP (2010). A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst), 9, 1307–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir A, Peterson SR, Knuth MW, Lu H, & Dynan WS (1992). Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America, 89, 11920–11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir A, Stein LY, Calore BL, & Dynan WS (1993). Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. The Journal of Biological Chemistry, 268, 10440–10447. [PubMed] [Google Scholar]
- Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, & Pfeiffer P (2000). DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: Implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Research, 28, 2585–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb TM, & Jackson SP (1993). The DNA-dependent protein kinase: Requirement for DNA ends and association with Ku antigen. Cell, 72, 131–142. [DOI] [PubMed] [Google Scholar]
- Graham TGW, Walter JC, & Loparo JJ (2016). Two-stage synapsis of DNA ends during non-homologous end joining. Molecular Cell, 61, 850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, et al. (1997). Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature, 388, 492–495. [DOI] [PubMed] [Google Scholar]
- Gu XY, Bennett RA, & Povirk LF (1996). End-joining of free radical-mediated DNA double-strand breaks in vitro is blocked by the kinase inhibitor wortmannin at a step preceding removal of damaged 3′ termini. The Journal of Biological Chemistry, 271, 19660–19663. [DOI] [PubMed] [Google Scholar]
- Gu J, Lu H, Tsai AG, Schwarz K, & Lieber MR (2007). Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: Influence of terminal DNA sequence. Nucleic Acids Research, 35, 5755–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanakahi LA, Bartlet-Jones M, Chappell C, Pappin D, & West SC (2000). Binding of inositol phosphate to DNA-PK and stimulation of double-strand break repair. Cell, 102, 721–729. [DOI] [PubMed] [Google Scholar]
- Harlow E, & Lane D (1988). Antibodies: A laboratory manual. Woodbury, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Hentges P, Ahnesorg P, Pitcher RS, Bruce CK, Kysela B, Green AJ, et al. (2006). Evolutionary and functional conservation of the DNA non-homologous end-joining protein, XLF/Cernunnos. The Journal of Biological Chemistry, 281, 37517–37526. [DOI] [PubMed] [Google Scholar]
- Jayaram S, Ketner G, Adachi N, & Hanakahi LA (2008). Loss of DNA ligase IV prevents recognition of DNA by double-strand break repair proteins XRCC4 and XLF. Nucleic Acids Research, 36, 5773–5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jette N, & Lees-Miller SP (2015). The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Progress in Biophysics and Molecular Biology, 117, 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Crowe JL, Liu X, Nakajima S, Wang Y, Li C, et al. (2015). Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and endligation during nonhomologous end-joining. Molecular Cell, 58, 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Alt FW, & Frock RL (2016). PAXX and XLF DNA repair factors are functionally redundant in joining DNA breaks in a G1-arrested progenitor B-cell line. Proceedings of the National Academy of Sciences of the United States of America, 113, 10619–10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P (1999a). Nonhomologous DNA end joining in cell-free systems. European Journal of Biochemistry, 265, 849–861. [DOI] [PubMed] [Google Scholar]
- Labhart P (1999b). Ku-dependent nonhomologous DNA end joining in Xenopus egg extracts. Molecular and Cellular Biology, 19, 2585–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, Takahashi T, & Walter JC (2009). DNA replication in nucleus-free Xenopus egg extracts. Methods in Molecular Biology, 521, 229–252. [DOI] [PubMed] [Google Scholar]
- Lebofsky R, van Oijen AM, & Walter JC (2011). DNA is a co-factor for its own replication in Xenopus egg extracts. Nucleic Acids Research, 39, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, et al. (2004). Implication of DNA polymerase lambda in alignment-based gap filling for Nonhomologous DNA end joining in human nuclear extracts. The Journal of Biological Chemistry, 279, 805–811. [DOI] [PubMed] [Google Scholar]
- Lee NK, Kapanidis AN, Wang Y, Michalet X, Mukhopadhyay J, Ebright RH, et al. (2005). Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophysical Journal, 88, 2939–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Yannone SM, Chen DJ, & Povirk LF (2003). Requirement for XRCC4 and DNA ligase IV in alignment-based gap filling for nonhomologous DNA end joining in vitro. Cancer Research, 63, 22–24. [PubMed] [Google Scholar]
- Lees-Miller SP, Chen YR, & Anderson CW (1990). Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Molecular and Cellular Biology, 10, 6472–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman CW, & Carroll D (1991). Homologous recombination catalyzed by a nuclear extract from Xenopus oocytes. Proceedings of the National Academy of Sciences of the United States of America, 88, 10840–10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman CW, Clemens M, Worthylake DK, Trautman JK, & Carroll D (1993). Homologous and illegitimate recombination in developing Xenopus oocytes and eggs. Molecular and Cellular Biology, 13, 6897–6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescale C, Lenden Hasse H, Blackford AN, Balmus G, Bianchi JJ, Yu W, et al. (2016). Specific roles of XRCC4 paralogs PAXX and XLF during V(D)J recombination. Cell Reports, 16, 2967–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Otevrel T, Gao Y, Cheng HL, Seed B, Stamato TD, et al. (1995). The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell, 83, 1079–1089. [DOI] [PubMed] [Google Scholar]
- Liao S, Guay C, Toczylowski T, & Yan H (2012). Analysis of MRE11’s function in the 5′ → 3′ processing of DNA double-strand breaks. Nucleic Acids Research, 40, 4496–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Toczylowski T, & Yan H (2008). Identification of the Xenopus DNA2 protein as a major nuclease for the 5′ → 3′ strand-specific processing of DNA ends. Nucleic Acids Research, 36, 6091–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Toczylowski T, & Yan H (2011). Mechanistic analysis of Xenopus EXO1’s function in 5′-strand resection at DNA double-strand breaks. Nucleic Acids Research, 39, 5967–5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Shao Z, Jiang W, Lee BJ, & Zha S (2017). PAXX promotes KU accumulation at DNA breaks and is essential for end-joining in XLF-deficient mice. Nature Communications, 8, 13816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, R€aschle M, Joukov V, & Walter JC (2011). Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science, 333, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Pannicke U, Schwarz K, & Lieber MR (2007). Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity. The Journal of Biological Chemistry, 282, 11155–11162. [DOI] [PubMed] [Google Scholar]
- Ma Y, & Lieber MR (2006). In vitro nonhomologous DNA end joining system. Methods in Enzymology, 408, 502–510. [DOI] [PubMed] [Google Scholar]
- Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, et al. (2004). A biochemically defined system for mammalian nonhomologous DNA end joining. Molecular Cell, 16, 701–713. [DOI] [PubMed] [Google Scholar]
- Menon V, & Povirk LF (2016). End-processing nucleases and phosphodiesterases: An elite supporting cast for the non-homologous end joining pathway of DNA double-strand break repair. DNA Repair (Amst), 43, 57–68. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juárez R, Bebenek K, Kee BL, et al. (2005). A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Molecular Cell, 19, 357–366. [DOI] [PubMed] [Google Scholar]
- Ochi T, Blackford AN, Coates J, Jhujh S, Mehmood S, Tamura N, et al. (2015). DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science, 347, 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P, Feldmann E, Odersky A, Kuhfittig-Kulle S, & Goedecke W (2005). Analysis of DNA double-strand break repair by nonhomologous end joining in cell-free extracts from mammalian cells. Methods in Molecular Biology, 291, 351–371. [DOI] [PubMed] [Google Scholar]
- Pfeiffer P, Odersky A, Goedecke W, & Kuhfittig-Kulle S (2014). Analysis of double-strand break repair by nonhomologous DNA end joining in cell-free extracts from mammalian cells. Methods in Molecular Biology, 1105, 565–585. [DOI] [PubMed] [Google Scholar]
- Pfeiffer P, & Vielmetter W (1988). Joining of nonhomologous DNA double strand breaks in vitro. Nucleic Acids Research, 16, 907–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, and Funabiki H (2008). Ku80 removal from DNA through double strand break-induced ubiquitylation. The Journal of Cell Biology 182, 467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räschle M, Knipscheer P, Knipsheer P, Enoiu M, Angelov T, Sun J, et al. (2008). Mechanism of replication-coupled DNA interstrand crosslink repair. Cell, 134, 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, de Melo AJ, Xu Y, Tadi SK, N egrel A, Hendrickson E, et al. (2015). XRCC4/XLF interaction is variably required for DNA repair, and is not required for ligase IV stimulation. Molecular and Cellular Biology, 35, 3017–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval A, & Labhart P (2002). Joining of DNA ends bearing non-matching 3′-overhangs. DNA Repair (Amst), 1, 397–410. [DOI] [PubMed] [Google Scholar]
- Smeaton MB, Miller PS, Ketner G, & Hanakahi LA (2007). Small-scale extracts for the study of nucleotide excision repair and non-homologous end joining. Nucleic Acids Research, 35, e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadi SK, Tellier-Lebègue C, Nemoz C, Drevet P, Audebert S, Roy S, et al. (2016). PAXX is an accessory c-NHEJ factor that associates with Ku70 and has overlapping functions with XLF. Cell Reports, 17, 541–555. [DOI] [PubMed] [Google Scholar]
- Tanner NA, & van Oijen AM (2010). Visualizing DNA replication at the single-molecule level. Methods in Enzymology, 475, 259–278. [DOI] [PubMed] [Google Scholar]
- Taylor EM, Cecillon SM, Bonis A, Chapman JR, Povirk LF, & Lindsay HD (2010). The Mre11/Rad50/Nbs1 complex functions in resection-based DNA end joining in Xenopus laevis. Nucleic Acids Research, 38, 441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thode S, Schäfer A, Pfeiffer P, & Vielmetter W (1990). A novel pathway of DNA endto-end joining. Cell, 60, 921–928. [DOI] [PubMed] [Google Scholar]
- Toczylowski T, & Yan H (2006). Mechanistic analysis of a DNA end processing pathway mediated by the Xenopus Werner syndrome protein. The Journal of Biological Chemistry, 281, 33198–33205. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Kim SA, & Chu G (2007). Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proceedings of the National Academy of Sciences of the United States of America, 104, 7851–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, & Goldberg J (2001). Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature, 412, 607–614. [DOI] [PubMed] [Google Scholar]
- Waters CA, Strande NT, Wyatt DW, Pryor JM, & Ramsden DA (2014). Nonhomologous end joining: A good solution for bad ends. DNA Repair (Amst), 17, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis-Garcia F, Besmer E, Sawchuk DJ, Yu W, Hu Y, Cassard S, et al. (1997). V(D)J recombination: In vitro coding joint formation. Molecular and Cellular Biology, 17, 6379–6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M, Yang M, Huo W, Feng F, Wei L, Jiang W, et al. (2015). Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nature Communications, 6, 6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, McCane J, Toczylowski T, & Chen C (2005). Analysis of the Xenopus Werner syndrome protein in DNA double-strand break repair. The Journal of Cell Biology, 171, 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Morotomi-Yano K, Lee K-J, & Chen DJ (2011). Functional significance of the interaction with Ku in DNA double-strand break recognition of XLF. FEBS Letters, 585, 841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Morotomi-Yano K, Wang S-Y, Uematsu N, Lee K-J, Asaithamby A, et al. (2008). Ku recruits XLF to DNA double-strand breaks. EMBO Reports, 9, 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, & Walter JC (2014). Mechanism and regulation of incisions during DNA inter-strand cross-link repair. DNA Repair (Amst), 19, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, & Peng A (2016). Non-homologous end joining repair in Xenopus egg extract. Scientific Reports, 6, 27797. [DOI] [PMC free article] [PubMed] [Google Scholar]