Abstract
An ion concentration polarization (ICP)-based electrokinetic concentration device is used for accelerating the surface hybridization reaction between exosomal microRNAs (miRNAs) and morpholinos (MOs) as a synthetic oligo capture probe in the nanomolar concentration range in a microfluidic channel. Compared with standard hybridization at the same concentration, the hybridization time of the miRNA target on MO capture probes could be reduced from ∼24 h to 30 min, with an increase in detection speed by 48 times. This ICP-enhanced hybridization method not only significantly decreases the detection time but also makes workflow simple to use, circumventing use of quantitative reverse transcription polymerase chain reaction or other conventional enzyme-based amplification methods that can cause artifacts.
I. INTRODUCTION
Exosomes have gained immense research interest in recent years owing to their promising diagnostic and therapeutic potential.1 They contain ample proteomic and genetic information for disease diagnostics, monitoring of cancer progression, metastasis, and drug efficacy.2 In particular, “exosomal microRNAs” could provide a rich source of biomarker molecules for early detection and monitoring of diseases.3 MicroRNAs (miRNAs) are short, single-stranded non-coding RNA molecules containing between 18 and 22 nucleotides in length which play a crucial role in gene expression regulation with atypical expression closely linked with pathologies, most notably cancer.4 Therefore, there is significant interest in detecting and quantifying these important regulators. To date, no technology can accurately quantify and profile these biomarkers with accuracy, ease, low cost, and high throughput.4 Current mainstream technologies such as quantitative reverse transcription polymerase chain reaction (qRT-PCR), microarray hybridization, and next-generation sequencing (NGS) face challenges in nucleic acid profiling.5 In qRT-PCR, the golden standard for the detection and quantification of RNA targets, the transcription step can generate variability because the efficiency of reverse transcriptases can be dependent on the sequence, length, and structure.6 In multiplexed qRT-PCR amplification, biases can be introduced by varying the GC content, length, starting concentration of cDNAs, and buffer composition.7–9 To overcome limitations of mainstream technologies and build a nucleic acid profiling platform that is accurate, simple to use, sensitive, and selective, research efforts are being undertaken based on micro- and nanofluidic technologies.10
In recent years, several promising biosensing technologies have been presented to overcome limitations of mainstream technologies. These technologies include but not limited to the rolling-circle isothermal amplification within hydrogel microparticles to compartmentalize multiple reactions,11 free-solution electrophoretic detection with drag-tags and single stranded DNA binding proteins to increase the differences in the electrophoretic mobility of target-probe complexes,12,13 and a strategy whereby gating a nonselective microfluidic channel with an ion-selective membrane to establish a quasistationary depletion front, which enable high-throughput processing of multiple samples in various medical applications.14
To detect exosomal miRNAs without performing qRT-PCR, we propose integration of an ion concentration polarization (ICP)-based microfluidic concentrator with a morpholino (MO) microarray to significantly enhance the MO-miRNA hybridization assay speed and sensitivity. This is the same concentrator device platform we have used in our earlier publication15 in Biosensors. However, the key differences are as follows: miRNA target instead of DNA, redesigned morpholino capture probes, and adjusted several operational parameters as shown in Table II. MOs are synthetic nucleic acid analogs with a non-charged backbone of morpholine rings16 which facilitate surface hybridization-based detection at low and moderate ionic strengths down to ∼10 nM17 synergistic to ICP. However, MO-based assays suffered from low detection sensitivity and long hybridization periods, especially at low concentrations.18,19 The ICP concentration reduces the analyte diffusion length and increases the hybridization rate due to the locally enhanced concentration of nucleic acids. Using this ICP-based approach, our group has already demonstrated rapid DNA detection.15
TABLE II.
Comparison between miRNA and DNA in ICP hybridization.
| miRNA (miRNA 21) | DNA15 | |
|---|---|---|
| MO concentration (capture probe) | 100 μM | 40 μM |
| MO capture probe | 5′ NH2-CAG TCA ACA TCA GTC TGA TAA GCT A 3′ | 5′ NH2-GTA GCT AAT GAT GTG GCA TCG GTT 3′ |
| Voltage during ICP hybridization | 15 V | 75 V |
| Heat applied to sample before experiment | 50 °C for 7 min | No heat |
| Plug stability | 1 h | 30 min |
| Concentration factor | 90× (Co = 100 nM) | 1000× (Co = 10 nM) |
| Concentration time | 30 min | 5 min |
| Limit of detection | 25 nM | 1 nM |
II. MATERIALS AND METHODS
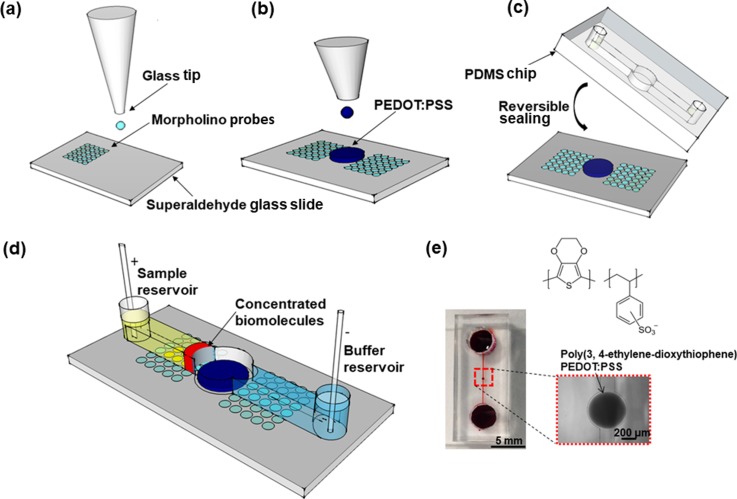
We followed the standard soft lithography protocol to fabricate a polydimethylsiloxane (PDMS) microfluidic device.15 As shown in Fig. 1(a), we built a single concentrator device by printing an array of MO capture probes using a fluid GIX microplotter II (Sonoplot Inc.) on a superaldehyde-functionalized microarray slide from Arrayit and the conductive polymer poly(3,4-ethylenedioxythiophene)-poly(styrenesulfonate) (PEDOT: PSS) as a cation-selective membrane next to an array of MO capture probes Fig. 1(b) and reversibly sealed the substrate with a PDMS microchannel from the top Fig. 1(c). As target analytes, we selected miRNA 21 and miRNA 155 for the study since they are expressed in a wide variety of cancers including breast and lung cancer cells.20 Once the analyte and buffer solutions were loaded, an electrical voltage was applied to induce ICP as shown in Fig. 1(d). A photograph of a fully assembled PDMS concentrator with a PEDOT: PSS circular membrane is shown in Fig. 1(e). Unlabeled MO probe sequence MO 21, MO 155 and fluorescein isothiocyanate-labeled MO 21 FITC, MO 155 FITC in Table I were purchased from Gene Tools. FITC-labeled MOs were used to verify the printing quality of MO probes on glass slides. The 22 and 23 nucleotides long miRNA's 21 and 155 targets, respectively, were labeled at the 3′-end with Cyanine 5 (Cy5) (Integrated DNA Technologies). Conductive polymer PEDOT: PSS 3.0%–4.0% in H2O (high conductivity grade) was obtained from Sigma-Aldrich. The MO concentration was set to 100 μM offering sufficient hybridization sites between MO and miRNA on the surface. However, due to electrostatic repulsion between hybridized miRNA targets, further increasing MO sites per area did not lead to more targets per area.21
FIG. 1.
Fabrication of a microfluidic concentrator chip. (a) Printing of morpholino capture probes on a superaldehyde-functionalized glass slide. (b) Deposition of the conductive polymer next to an array of MO capture probes. (c) Reversible sealing of the pre-patterned substrate with a PDMS chip from the top. (d) 3D schematic microfluidic concentrator—electrical voltage was applied to induce ICP after the analyte and buffer solutions were loaded. (e) Photograph of a fully assembled PDMS concentrator with the PEDOT: PSS circular layer. The PEDOT: PSS circular layer is ∼3.0 μm high, the microchannel is 17 μm high, and the distance between printed PEDOT:PSS and the top surface of the microchannel is ∼14 μm, 200 μm wide, and 1 cm long with reservoir (4 mm diameter) at each end.
TABLE I.
miRNA target sequences and complementary MO capture probes.
| RNA/MO | Sequence | Comments |
|---|---|---|
| miRNA 21 | 5′ UAG CUU AUC AGA CUG AUG UUG A-CY5 3′ | Target to MO 21 |
| MO 21 | 5′ NH2-CAG TCA ACA TCA GTC TGA TAA GCT A 3′ | Capture probe miRNA 21 |
| MO 21 FITC | 5′ NH2-CAG TCA ACA TCA ATC TGA TAA GTA-FITC 3′ | Immobilization control |
| miRNA 155 | 5′ UUA AUG CUA AUC GUG AUA GGG GU-CY5 3′ | Target to MO 155 |
| MO 155 | 5′ NH2-AAA ACC CCT ATC ACG ATT AGC ATT AAC 3′ | Capture probe miRNA 155 |
| MO 155 FITC | 5′ NH2-TCA ACA TCA GTC TGA TAA GCT A-FITC 3′ | Immobilization control |
To test the operation of the device in hybridization assays, the cathodic reservoir was first loaded with 50 μl of 0.1× PBS buffer to fill the microchannel up to the edge of the anodic reservoir by capillarity. It takes approximately 20 min to equilibrate the conductive polymer layer. Prior to loading the miRNA target solution in 16 mM phosphate buffer solution, it was heated for 7 min at 50 °C before dispensing into the opposite (anodic) reservoir. Finally, another 50 μl of 0.1× PBS buffer was loaded into the cathodic reservoir to form a column height difference between the reservoirs. Finally, to activate the electroosmotic flow, 15 V was then applied along the microchannel through the Pt wires (0.5 mm in diameter, Goodfellow) in each reservoir, using a DC source meter (Keithley 2400). The source meter was controlled using a LabVIEW program (National Instruments). We estimated the total throughput of the device is ∼2.4 μl (see S3 in the supplementary material for the calculation).
The voltage difference initiated the formation of a miRNA plug in front of PEDOT: PSS. During a concentration period of 30 min, the miRNA target molecules in the plug hybridized with the immobilized MO probes located at the bottom of the microchannel. At the end of the surface hybridization assay, the voltage was switched off and the plug was quickly washed away from the detection site by the pressure-driven flow created by the column height difference between the reservoirs.
To quantify the miRNA concentration in the plug, we measured the fluorescence intensity of the microchannel filled with miRNA samples, miRNA 21 and miRNA 155, of known concentrations (1, 10, and 100 μM), as shown in Fig. S2(a) (supplementary material) for miRNA 21, and used them as reference/calibration lines in Fig. S2(b) (supplementary material). We then plotted the fluorescence intensity data as a function of concentration time during ICP electrokinetic preconcentration. To calculate the concentration factor, we estimated the final concentration of miRNA obtained after 30 min by comparing the fluorescence intensity of miRNA plug against the reference lines that correspond to the fluorescence intensity values of different miRNA concentrations. Finally, we simply calculated the ratio of the miRNA concentrations before and after as a measure of the concentration factor. Similar to our previous results with DNA preconcentration, the electrokinetic preconcentration of miRNA exhibited a non-linear behavior, resulting in different preconcentration factors depending on the starting miRNA concentration. The exact reason for this nonlinear behavior is still under investigation.
III. RESULTS AND DISCUSSION
A. Denaturing miRNAs for MO surface hybridization
Hairpin structures are a common feature of RNA molecules.22 The overall structure and physical properties of small single-stranded hairpin molecules and the effects of the sequence and loop size on hairpin stability have been widely reported. It is known that sufficiently low ionic strength combined with temperature in the range of 59 °C to 80 °C destabilizes the hairpin structure independent of the molecular concentration.23
Since miRNA 21 and 155 have a predicted and stable hairpin, we destabilized pure mir21 and mir155's internal base pairing by heating the sample to ∼50 °C for 7 min with a buffer ionic strength of 16 mM to better enhance the denaturing process. In standard hybridization assays for ∼21–24 h, heating the miRNA sample solution increased the fluorescent intensity by ∼2.5-fold (Fig. S1, supplementary material). For this reason, we used preheated miRNA samples for the subsequent MO-miRNA hybridization experiment.
B. Characterization of the electrokinetic miRNA concentration
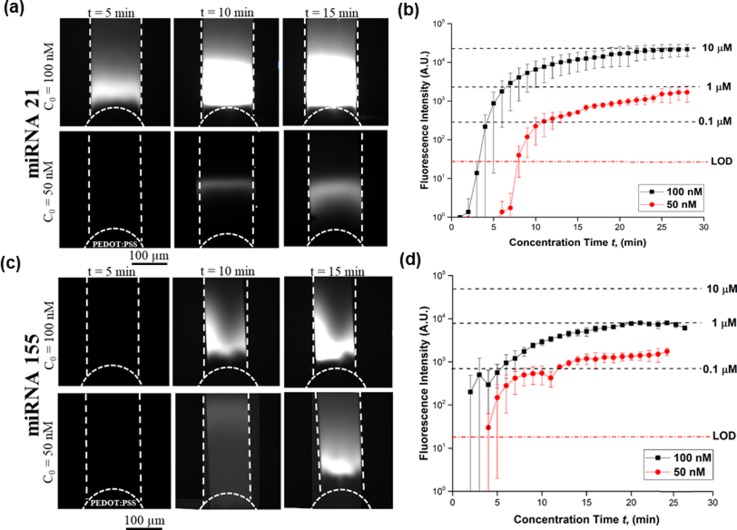
To characterize the concentration performance of our device, we conducted experiments at 50 nM and 100 nM under various voltages of 15 V, 30 V, and 50 V applied between the Pt electrodes. At higher voltages, the miRNA plug was less stable due to strong vortex formation in the depletion zone. At 15 V, approximately 0.8 μA flows through the entire cross section of the microchannel, a fluorescent plug started forming in front of the conductive polymer. After the concentration period of 5 min, a bright fluorescent plug was visible inside the channel, indicating the miRNA near the conductive polymer membrane and its size continued to increase as a function of concentration time, as shown in Fig. 2(a). Within 30 min, the concentration for miRNA 21 increased by ∼90-fold for 100 nM and ∼20-fold for 50 nM as shown in Fig. 2(b) (see Movie S1 in the supplementary material for the plug formation during the first 5 min of the miRNA electrokinetic concentration). The fluorescence intensity observed for miRNA 155 was lower than that for miRNA 21 as shown in Fig. 2(c). This difference in fluorescence intensity can be explained by the poly G-sequence (4 G bases in a row) of miRNA 155, which results in strong secondary and potentially tetraplex formations,24 causing lower purities and yields. It was confirmed by the manufacturer that the miRNA 21 sample has 90% purity, whereas miRNA 155 had lower purity than miRNA 21; however, exact purity could not be determined. For miRNA 155, the concentration increased by ∼10-fold for 100 nM and ∼4-fold for 50 nM as shown in Fig. 2(d).
FIG. 2.
Characterization of miRNA concentrations 50 nM and 100 nM at 15 V. (a) Bright fluorescent plug inside the channel indicating miRNA 21 near the conductive polymer membrane—size continued to increase as a function of concentration time. (b) Fluorescence intensity for ND 1 filter and 1 s exposure time for miRNA 21. Within 30 min, the concentration of miRNA 21 increased by ∼90-fold for 100 nM and ∼20-fold for 50 nM. (c) Fluorescent plug inside the channel indicating miRNA 155 near the conductive polymer membrane. (d) Its concentration increased by ∼10-fold for 100 nM and ∼4-fold for 50 nM.
The concentration plug is in direct contact and spatially above the MO capture probes. In our previous publication,25 we investigated the DNA concentration plug three-dimensional concentration profile with confocal microscopy along the channel depth in the z-direction. Our finding shows that the fluorescence signal intensity of the DNA plug was maximal near the glass surface and decreased almost linearly as the focal plane moved upward to the top of the channel, where the fluorescence signal intensity value decreased to about one-fourth of the maximal value at the top of the PDMS channel. We hypothesize that the reason for this concentration distribution along the channel depth could be linked to the different electroosmotic mobilities of glass and PDMS.
C. Enhanced MO-miRNA surface hybridization
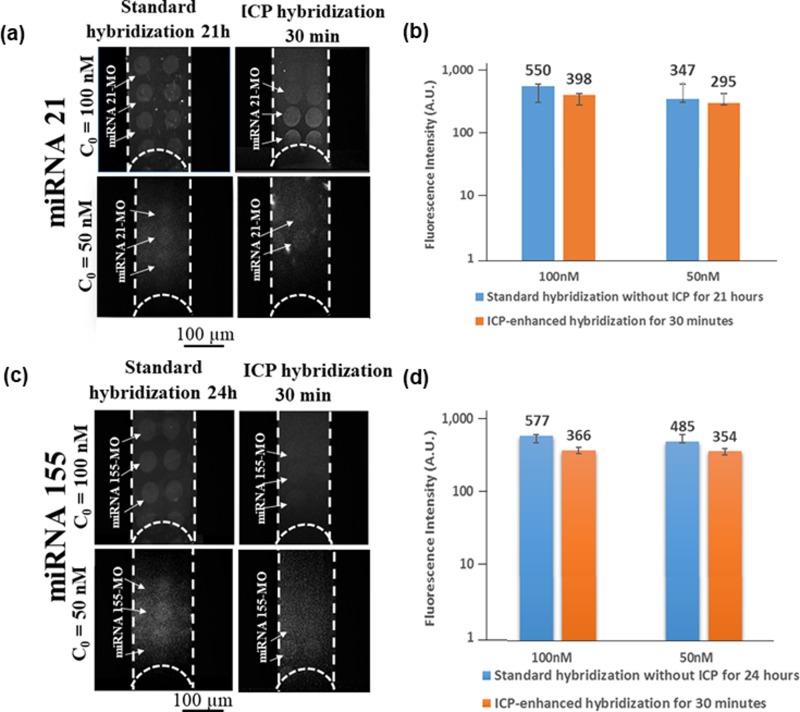
After concentrating the target miRNA on MO capture probes for 30 min, we could achieve fluorescence signal intensity comparable to that of standard diffusion-based incubation after approximately 24 h as evidenced in Figs. 3(a) and 3(b). We determined the limit of detection (LOD) for miRNA 21 at ∼25 nM with ICP over 30 min and for miRNA 155 at ∼50 nM after 30 min (see Movie S2 in the supplementary material showing the initial plug formation and the last 3 min of MO-miRNA 21 surface hybridization with the ICP concentration starting from an initial miRNA concentration of 25 nM).
FIG. 3.
Comparison of fluorescence intensity with and without ICP enhancement for miRNA 21 and 155 at 50 nM and 100 nM. (a) Fluorescence micrographs for MO-miRNA 21 hybridization. Left column: hybridization without ICP and right column: hybridization with ICP. (b) Bar graph quantifying fluorescence intensity with and without ICP. After concentrating the target miRNA 21 on MO spots for 30 min, we could achieve a comparable fluorescence intensity to that realized without ICP after ∼21 h. (c) Fluorescence micrographs for MO-miRNA 155 hybridization under standard incubation and ICP-enhanced hybridization. (d) Quantification of fluorescence signal intensity between standard and ICP hybridization.
Compared to the detection of DNA samples, miRNA posed more challenges for detection at low concentrations, as indicated in Table II. Several operational parameters such as preheating of miRNA samples and increasing surface density of MO capture probes were necessary to achieve a lower detection limit. Whilst the purpose of this study was not to conduct an exhaustive analysis of differences between results of ICP hybridization of miRNA and DNA, it is important to note challenges for miRNA hybridization assays. Since the heated miRNA solution cools down quickly inside the microchannel, one way to improve detection sensitivity is to create an ITO heater underneath the glass substrate to keep a consistent temperature of 50 °C during the electrokinetic concentration. Alternative to MO, more sensitive capture probes for miRNA such as locked-nucleic acids (LNAs) and peptide nucleic acids (PNAs) can also be used.26
IV. CONCLUSIONS
We have demonstrated an enhancement of the miRNA 21 target concentration by ∼90 times in 30 min, resulting in a ∼48-fold increase in the MO-miRNA reaction rate relative to hybridization without ICP enhancement inside a microfluidic channel. In the case of miRNA 155, the concentration increase was by ∼10 folds within 30 min. In both cases, the ICP-enhanced hybridization assays resulted in an effective reduction of required incubation time from 24 h to 30 min to detect a hybridization between 100 nM miRNA solution and MO capture probes inside a microfluidic channel. Hence, our focus for this research was to use ICP to overcome the diffusion-limited transport of miRNA onto the MO capture probes, thereby reaching comparable fluorescence signal intensity for detection with a significantly short amount of time as opposed to the lengthy standard incubation-based detection at lower miRNA concentrations. Further optimization of the ICP-based concentrator device such as incorporating an integrated ITO heater in the same design platform during the ICP concentration or using a more sensitive capture probe could achieve a lower LOD within the same amount of concentration time and potentially applied to miRNAs extracted directly from cancer cell lines at lower sample concentration levels.
V. SUPPLEMENTARY MATERIAL
See supplementary material for the standard hybridization of miRNA 21 at 100 nM with heat and without heat (Fig. S1), the method used to quantify miRNA in the plug (Fig. S2), the calculation for the total throughput of the device (Fig. S3), a video showing the plug formation during the first 5 min of the miRNA electrokinetic concentration for miRNA 21 at 100 nM (Movie S1), and a video showing the initial plug formation and the last 3 min of MO-miRNA 21 surface hybridization with the ICP concentration starting from an initial miRNA concentration of 25 nM (Movie S2).
ACKNOWLEDGMENTS
We gratefully acknowledge the financial support from the New York University Abu Dhabi (NYUAD) through the NYUAD Research Enhancement Fund and Abu Dhabi Education Council Award for Research Excellence 2015. The device fabrication was conducted in the microfabrication core facility of NYUAD. We would also like to thank Professor Rastislav Levicky, Department of Chemical and Biomolecular Engineering, New York University Tandon School of Engineering, Brooklyn, New York, US, for his discussion on MO-miRNA hybridization.
References
- 1. Hon K. W., Abu N., Ab Mutalib N.-S., and Jamal R., Front. Pharmacol. 8, 583 (2017). 10.3389/fphar.2017.00583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vlassov A. V., Magdaleno S., Setterquist R., and Conrad R., Biochim. Biophys. Acta - Gen. Subj. 1820, 940 (2012). 10.1016/j.bbagen.2012.03.017 [DOI] [PubMed] [Google Scholar]
- 3. Habbe N., Koorstra J. B. M., Mendell J. T., Offerhaus G. J., Ji K. R., Feldmann G., Mullendore M. E., Goggins M. G., Hong S. M., and Maitra A., Cancer Biol. Ther. 8, 340 (2009). 10.4161/cbt.8.4.7338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Egatz-Gomez A., Wang C., Klacsmann F., Pan Z., Marczak S., Wang Y., Sun G., Senapati S., and Chang H. C., Biomicrofluidics 10, 032902 (2016) 10.1063/1.4948525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Senapati S., Basuray S., Slouka Z., Cheng L. J., and Chang H. C., Top. Curr. Chem. 304, 153 (2011). 10.1007/978-3-642-23050-9 [DOI] [PubMed] [Google Scholar]
- 6. Lorenz T. C., J. Visualized Exp. 63, 4–14 (2012). [Google Scholar]
- 7. Raabe C. A., Tang T.-H., Brosius J., and Rozhdestvensky T. S., Nucl. Acids Res. 42, 1414 (2014). 10.1093/nar/gkt1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dabney J. and Meyer M., Biotechniques 52, 87 (2012) 10.2144/000113809. [DOI] [PubMed] [Google Scholar]
- 9. Swindell W. R., Xing X., Voorhees J. J., Elder J. T., Johnston A., and Gudjonsson J. E., Physiol. Genomics 46, 533 (2014). 10.1152/physiolgenomics.00022.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taller D., Richards K., Slouka Z., Senapati S., Hill R., Go D. B., and Chang H.-C., Lab Chip 15, 1656 (2015). 10.1039/C5LC00036J [DOI] [PubMed] [Google Scholar]
- 11. Chapin S. C. and Doyle P. S., Anal. Chem. 83, 7179 (2011). 10.1021/ac201618k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wegman D. W. and Krylov S. N., Angew. Chem., Int. Ed. 50, 10335 (2011). 10.1002/anie.201104693 [DOI] [PubMed] [Google Scholar]
- 13. Goldman J. M., Zhang L. A., Manna A., Armitage B. A., Ly D. H., and Schneider J. W., Biomacromolecules 14, 2253 (2013). 10.1021/bm400388a [DOI] [PubMed] [Google Scholar]
- 14. Sun G., Pan Z., Senapati S., and Chang H. C., Phys. Rev. Appl. 7, 064024 (2017) 10.1103/PhysRevApplied.7.064024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martins D., Levicky R., and Song Y. A., Biosens. Bioelectron. 72, 87 (2015). 10.1016/j.bios.2015.04.063 [DOI] [PubMed] [Google Scholar]
- 16. Tercero N., Wang K., Gong P., and Levicky R., J. Am. Chem. Soc. 131, 4953 (2009). 10.1021/ja810051q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qiao W., Kalachikov S., Liu Y., and Levicky R., Anal. Biochem. 434, 207 (2013). 10.1016/j.ab.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karkare S. and Bhatnagar D., Appl. Microbiol. Biotechnol. 71, 575 (2006). 10.1007/s00253-006-0434-2 [DOI] [PubMed] [Google Scholar]
- 19. Wang X. and Smirnov S., ACS Nano 3, 1004 (2009). 10.1021/nn900113x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang R., Liao Y., Zhou X., Fu Y., and Xing D., Sens. Actuators, B 247, 505 (2017). 10.1016/j.snb.2017.03.055 [DOI] [Google Scholar]
- 21. Gong P., Wang K., and Liu Y., J. Am. Chem. Soc. 132, 9663 (2010). 10.1021/ja100881a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chastain M. and Tinoco I., Prog. Nucl. Acid Res. Mol. Biol. 41, 131 (1991). 10.1016/S0079-6603(08)60008-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rentzeperis D., Alessi K., and Marky L. A., Nucl. Acids Res. 21, 2683 (1993). 10.1093/nar/21.11.2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yafe A., Etzioni S., Weisman-Shomer P., and Fry M., Nucl. Acids Res. 33, 2887 (2005). 10.1093/nar/gki606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martins D., Wei X., Levicky R., and Song Y. A., Anal. Chem. 88, 3539 (2016). 10.1021/acs.analchem.5b03875 [DOI] [PubMed] [Google Scholar]
- 26. Lautner G. and Gyurcsányi R. E., Electroanalysis 26, 1224 (2014). 10.1002/elan.201400055 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for the standard hybridization of miRNA 21 at 100 nM with heat and without heat (Fig. S1), the method used to quantify miRNA in the plug (Fig. S2), the calculation for the total throughput of the device (Fig. S3), a video showing the plug formation during the first 5 min of the miRNA electrokinetic concentration for miRNA 21 at 100 nM (Movie S1), and a video showing the initial plug formation and the last 3 min of MO-miRNA 21 surface hybridization with the ICP concentration starting from an initial miRNA concentration of 25 nM (Movie S2).