Abstract
The human colonic microbiota is a dense ecosystem comprised of numerous microbes including bacteria, phage, fungi, archaea, and protozoa that compete for nutrients and space. Studies are beginning to reveal the antagonistic mechanisms that gut bacteria use to compete with other members of this ecosystem. In the healthy human colon, the majority of the Gram negative bacteria are of the order Bacteroidales. Proteobacteria, such as E. coli, are numerically fewer, but confer important properties to the host, such as colonization resistance. Several enteric pathogens use T6SSs to antagonize symbiotic gut E. coli facilitating colonization and disease progression. T6SSs loci are also widely distributed in human gut Bacteroidales, which includes three predominant genera: Bacteroides, Parabacteroides, and Prevotella. There are three distinct genetic architectures of T6SS loci among the gut Bacteroidales, termed GA1, GA2, and GA3. GA1 and GA2 T6SS loci are contained on integrative and conjugative elements and are the first T6SS loci shown to be readily transferred in the human gut between numerous species and families of Bacteroidales. In contrast, the GA3 T6SSs are present exclusively in Bacteroides fragilis. There are divergent regions in all three T6SS GAs that contain genes encoding effector and immunity proteins, many of which function by unknown mechanisms. To date, only the GA3 T6SSs have been shown to antagonize bacteria, and they target nearly all gut Bacteroidales species analyzed. This chapter will delve more deeply into properties of the T6SSs of these human gut bacteria and the ecological outcomes of their synthesis in vivo. Type VI secretion systems were first identified and characterized in pathogenic bacteria of the Proteobacterial phylum (1, 2). The discovery in 2010 that these secretion systems can target and intoxicate not only eukaryotic cells, but also other bacteria (3), revealed that some T6SSs help bacteria compete with other bacteria in a community setting. Indeed, many Proteobacterial symbionts, including the plant symbiont Pseudomonas putida, the bumble bee gut symbiont Snodgrassella alvi, and the squid symbiont Vibrio fischeri, all have T6SSs that provide a competitive advantage in their natural ecosystems (4–6). An early in silico analysis using COG models of Proteobacterial T6SS proteins against primary sequence databases suggested that T6SSs are largely confined to Proteobacterial species, which are minor members of some human-associated microbial communities such as the gut microbiota (7, 8)
In the dense microbial ecosystem of the human colon, contact-dependent mechanisms of antagonism, such as a T6SS, should be an effective means of thwarting competitors. In this microbial ecosystem, the predominant Gram negative bacteria are of the phylum Bacteroidetes, comprising approximately half of the colonic bacteria in most people, and vastly outnumbering commensal Proteobacteria. Most human gut Bacteroidetes species are contained within the order Bacteroidales, which includes several different families, each with one predominant genus in the human gut: Bacteroides (family Bacteroidaceae), the Parabacteroides (family Tannerellaceae), Prevotella (family Prevotellaceae), and Alistipes (family Rickenellaceae), as well as families with more minor representatives. Humans are colonized at high density with numerous Bacteroidales species simultaneously (9–11) and the abundant species in one individual may not be the same as in another (9, 10). T6SSs were not discovered in species of the phylum Bacteroidetes until recently (12–14), mostly due to the lack of primary sequence or profile-sequence similarity of the 13 core Proteobacterial T6SS proteins with Bacteroidetes proteins. The use of profile-profile analyses revealed that Bacteroidales species of the human gut are a rich source of diverse T6SSs (15). As structural and mechanistic properties of T6SSs are discussed in the preceding chapter, here we will focus on unique properties of the T6SS of the human gut Bacteroidales, the ecological and functional properties of these T6SSs that are known to date, and the ecological advantages conferred by the T6SSs of human gut Proteobacterial strains in vivo.
GENETIC CHARACTERISTICS AND PROPERTIES OF THE T6SS OF GUT BACTEROIDALES
An analysis of 205 human gut Bacteroidales genomes including 35 different species of four genera, Bacteroides, Parabacteroides, Prevotella, and Alistipes, revealed the presence of 130 T6SS loci in 115 of these strains with 15 strains containing two different T6SS loci (16). These T6SS loci were found in the genomes of 19 different species of Bacteroides, Parabacteroides, and Prevotella, but were not found in any of the nine Alistipes genomes analyzed. A notable feature is that these T6SSs segregated into three very distinct genetic architectures (GA), termed GA1, GA2 and GA3 (16) (Fig 1A). Within a given GA, the majority of the T6SS genes are of high DNA identify, interspersed with small regions that are variable (Fig 1A). The variable regions of all three T6SS GAs contain genes encoding effector and immunity proteins, some of which are similar to those previously described in other organisms (16, 17). Most of the GA3 T6SS effector and immunity proteins are unlike those previously described and function by mechanisms currently unknown. By comparing the divergent regions within a GA, there are predicted to be at least 30 different variable regions in the 48 GA1 loci analyzed, 21 in the 9 GA2 regions analyzed, and 17 in the 56 GA3 regions analyzed, with each divergent region likely encoding at least one effector immunity pair. Therefore, the T6SSs of gut Bacteroidales encode numerous distinct toxins, many of which operate via unknown mechanisms.
Figure 1.
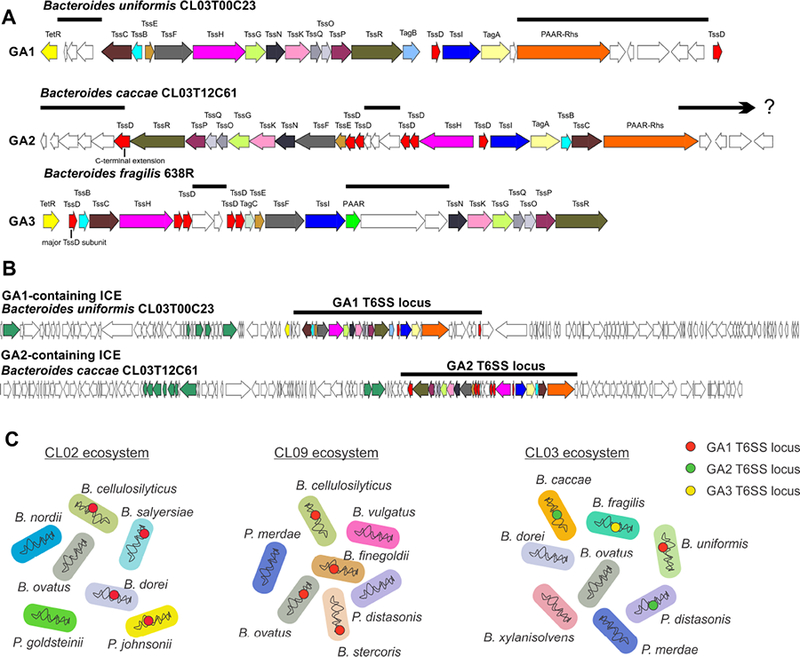
A. ORF maps of one representative locus of each of the three genetic architectures (GA) of T6SS loci of gut Bacteroidales. T6SS loci of GA1 and GA2 are present in diverse Bacteroidales species whereas GA3 T6SS loci are confined to B. fragilis. T6SS loci of a given GA are extremely similar to each other except for the divergent regions noted by lines above the genes, which encode known or putative effector and immunity proteins. The major TssD protein of GA3 is noted as is the TssD protein of the GA2 loci that have C-terminal extensions likely conferring toxin activity. The ends of the GA1 and GA2 loci have not been precisely determined. B. ORF maps of ICE containing GA1 and GA2 T6SS loci of two Bacteroides species. The T6SS loci are designated by a line above the map. Genes involved in conjugative transfer (tra genes) are colored green (15). C. The abundant fecal gut Bacteroidales from three different healthy humans (CL02, CL09, and CL03) were analyzed for the presence of T6SSs. Seven Bacteroidales species were isolated and sequenced from subject CL02 and from subject CL09. In each subject, four of the seven species harbor nearly identical GA1 T6SSs loci within a subject, demonstrating transfer of these ICE between these strains in their gut (12, 15). In contrast, of the eight species isolated and sequenced from human subject CL03, two contain GA2 T6SS loci, albeit with different divergent regions. Therefore, these GA2 ICE were not transferred between these species. In addition, one species contains a GA1 T6SS locus and the B. fragilis strain from this individual contains a GA3 T6SS locus (15). Red, green and yellow dots represent the GA1, GA2, and GA3 T6SS loci.
An analysis of the predicted proteins produced by these loci showed that four of the conserved T6SS proteins of Proteobacterial species are missing in gut Bacteroidales T6SS: TssA, TssJ, TssL, and TssM. TssJ, L, and M are components of the transmembrane complex (18) and TssA binds this complex and likely recruits the baseplate assemblage and coordinates tail tube and sheath biogenesis (19, 20). Instead of encoding these proteins, all three GAs of Bacteroidales T6SS loci encode four conserved proteins not present in Proteobacterial T6SS loci, namely TssO-TssR (13, 16). These likely serve as functional orthologs of the Proteobacterial proteins comprising the transmembrane complex.
All three T6SS GAs encode multiple TssD (Hcp) needle proteins, with GA2 and GA3 loci encoding six and five distinct TssD proteins, respectively. In the GA2 loci, one of the six TssD proteins has a C-terminal extension, likely conferring toxin activity to this protein (Fig 1A). In the GA3 locus, the main structural TssD protein was identified among the five TssD proteins (Fig 1A)(21). The function of the accessory TssD proteins may be to bind effector proteins and incorporate them into the needle structure, as in both the GA2 and GA3 loci, four of the tssD genes flank the effector and immunity gene variable regions (Fig 1A).
An important feature of the three different GAs is their distribution among gut Bacteroidales species. GA1 and GA2 T6SS loci are present in numerous species of Bacteroides, Parabacteroides, and Prevotella, whereas GA3 T6SSs are present exclusively in B. fragilis. The wide distribution of GA1 and GA2 in at least three distinct families of Bacteroidales is due to their presence on integrative and conjugative elements (ICE) (12, 15) (Fig 1B). ICE containing GA1 T6SSs are in the range of 120 kb and are extremely similar along their length to ICE containing distinct GA1 T6SS. Excluding the T6SS divergent regions, the DNA of ICE containing distinct GA1 T6SS loci are approximately 95% similar over their entire length. Although very dissimilar to the GA1 ICE, GA2 containing ICE are on the order of 100 kb and share approximately 75% - 99% DNA identify over their length with other GA2 containing ICE. As ICE are chromosomal self-transmissible mobile elements (22, 23), they have the ability to move between species. In fact, the GA1 containing ICE have been found to be transferred extensively between Bacteroidales species that are co-resident in the human intestine (12, 16)(Fig 1C). Examination of hundreds of genomes of gut Bacteroidales has indicated that GA2 T6SS are not present in a strain that has either a GA1 or GA3 T6SS locus. However, B. fragilis strains can have both GA1 and GA3 T6SS loci in their genome. The reason for the apparent lack of GA2 T6SS loci in the same chromosome with GA1 or GA3 T6SS loci is currently unknown. Bacteroidales species present in the gut of an individual can, however, collectively contain all three different T6SS GAs (16) (Fig 1C).
ECOLOGICAL CONSEQUENCES OF GUT BACTEROIDALES T6SS ANTAGONISM
The fitness benefits of theT6SSs of the gut Bacteroidales are still incompletely understood. The best studied are the GA3 T6SSs of B. fragilis. GA3 T6SSs were found to in approximately 86% of B. fragilis strains based on genome or metagenome analyses including hundreds of strains (16, 24), making them widely distributed in the species. The GA3 T6SSs of two different B. fragilis strains were found to antagonize all gut Bacteroidales species tested including Bacteroides and Parabacteroides species, but not the one Prevotella copri strain analyzed nor other B. fragilis with the same GA3 T6SS region or immunity genes (21, 25). In addition, no killing was evident against any gut Proteobacterial species analyzed (21), suggesting specificity for Bacteroidales. The TssD needle protein of the B. fragilis 638R GA3 T6SS is present in the supernatant of actively growing cells in vitro as well as in the feces of monoassociated gnotobiotic mice (21), suggesting this antagonistic system is constitutively synthesized and firing, rather than only responding to specific external signals or threats. The number of GA3 T6SS transmission events in a human gut colonized at typical levels with B. fragilis was predicted to be on the order of 6 × 1010-1011 per day (25).
Effector and immunity proteins were identified in three different GA3 T6SS loci (21, 25, 26). These toxic effector proteins are not similar to other known proteins and therefore intoxicate by as yet unidentified mechanisms. The toxicity of two of these effectors requires an added N-terminal periplasmic targeting sequence when they are produced inside a sensitive cell from an inducible promoter (27), suggesting that they may need to be localized to the periplasm for toxicity. In a few strains, genes encoding immunity proteins to GA3 effectors were found outside of the T6SS regions, in some cases in strains that did not have a GA3 T6SS locus (25). These immunity genes were found to confer protection from attack by a B. fragilis strain synthesizing the T6SS effectors to which the immunity proteins are directed, suggesting that acquisition of these immunity genes confers an advantage to an organisms by protecting it from GA3 T6SS-mediated antagonism (25). A recent study analyzing human gut metageonimc data revealed arrays of orphan immunity genes, which were termed Acquired Interbacterial Defense (AID) gene clusters (28). These orphan immunity islands reside on predicted mobile elements and include immunity genes likely derived from B. fragilis GA3 T6SS loci and disseminated by lateral transfer.
Although the GA3 T6SSs are very effective at antagonizing Bacteroidales species in vitro, the effects in vivo are more variable. As B. fragilis are known to co-exist in the human gut with numerous other Bacteroidales species that are susceptible to their GA3 T6SS, the spatial organization of the microbiota likely dictates the effectiveness of this weapon. In a gnotobiotic mouse competitive colonization model, an isogenic T6SS+ wild type strain outcompetes an isogenic mutant strain lacking the T6SS effector and immunity genes (21, 25). As isogenic strains should share the same spatial and nutritional niche and therefore should make frequent contacts, a strong antagonistic effect is expected. The ability of GA3 T6SSs of B. fragilis to antagonize other wild type non-isogenic B. fragilis strains in vivo was also demonstrated (25, 26). However, the effectiveness of the GA3 T6SS was less when analyzing antagonism of different Bacteroides species (Fig 2B). A significant effect was observed with a B. vulgatus strain (25), but not with B. thetiotaomicron (21, 25). It may be that in this model, where the mice are provided ample nutrients and likely utilize different nutrients in the gut (29, 30), B. fragilis and B. thetaiotaomicron would make infrequent contacts (Fig 2B).
Figure 2.
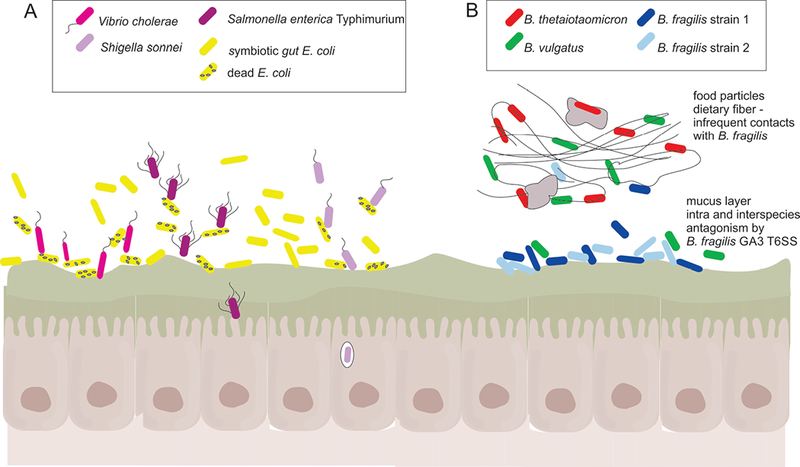
T6SS mediated antagonism in the mammalian gut. A. Three different Proteobacterial enteric pathogens, Vibrio cholerae, Salmonella enterica Typhimurium, and Shigella sonnei use T6SSs to target resident gut E. coli to overcome colonization resistance and cause disease in animal models (33–37). In the case of V. cholerae, the lysed E. coli initiate innate immune responses that upregulate virulence factors and increase dissemination (33). B. Bacteroides fragilis GA3 T6SS antagonize nearly all gut Bacteroidales species in vitro. In vivo, strong antagonistic effects are seen between two distinct B. fragilis strains likely due to their localization at the mucosal surface where they will make frequent contacts. This intraspecies antagonism may lead to the dominance of one strain. B. vulgatus was also significantly antagonized by a B. fragilis GA3 T6SS, possibly due to overlapping nutritional niches. In contrast, a significant antagonistic effect by the GA3 T6SS of B. fragilis was not observed when co-inoculated with B. thetaiotaomicron. These varied effects may be due to the substrate preferences of these species that may spatially segregate them under normal dietary conditions.
Analysis of metagenomic datasets of human gut samples revealed a link between the presence of Bacteroidales T6SSs and the composition of the microbiota, especially in regard to the GA3 T6SSs of B. fragilis (24). The presence of GA3 T6SS genes correlated significantly with an abundance of Bacteroides and a decrease in specific Firmicutes. Moreover, the microbiota of the developing infant gut are significantly more likely to contain GA3 T6SSs than those of adults. Also, B. fragilis strain replacement in the infant microbiota is more pronounced than in adults, which appear to be dominated by a single strain of B. fragilis. These findings suggest that competition amongst B. fragilis strains for dominance is fiercest early in life, and that the ultimate microbiota composition may be influenced by GA3 T6SSs (24).
The functions of the GA1 and GA2 T6SSs have not yet been elucidated. There has been no demonstration that these T6SSs target bacteria as do the GA3 T6SSs. Many of the identifiable effectors are predicted nucleases or other nucleic acid targeting enzymes (16) that could function in bacteria, archaea, or eukaryotes. The fact that the ICE containing these T6SSs are shared between different co-resident species of Bacteroidales in the human gut suggests they likely do not have a Bacteroidales target and may instead provide defense against a common enemy. It is also possible that these T6SSs may allow for nutrient acquisition, or protection from environmental stressors, as has been shown for T6SSs in other organisms (31, 32). The prevalence and transfer of these systems among human gut Bacteroidales species makes them intriguing secretion systems for continued analysis.
T6SS EFFECTS OF ENTERIC PROTEOBACTERIA IN THE MAMMALIAN GUT
Although less abundant in the healthy human gut microbiota than Bacteroidales, E. coli gut symbionts play a crucial role in colonization resistance against enteric pathogens of the Proteobacterial phylum. Enteric pathogens such as Vibrio cholerae, Shigella sonnei, and Salmonella enterica, have all been shown to utilize T6SSs to overcome colonization resistance by antagonizing resident gut E. coli (33–37) (Fig 2A). What is less studied is whether the symbiotic gut E. coli have T6SSs that function in colonization resistance against enteric pathogens. Antagonism by the diffusible microcin toxin produced by E. coli Nissle was shown to function as a colonization barrier to enteric pathogens (38) providing precedent for such an effect. Queries of a set of 1267 human gut metagenomes consolidated into the “three cohorts gene catalog (3CGC)” (39) for matches to Pfam models identifying the four conserved Proteobacterial T6SS proteins absent in Bacterodales T6SSs (TssA, J, L, and M) revealed that 174 of these metagenomes encoded proteins with motifs that met or exceeded the gathering threshold of all four of the Proteobacterial models used (Coyne and Comstock, unpublished). These findings suggest that resident gut E. coli strains likely harbor T6SSs, the in vivo effects of which remain to be determined.
CONCLUDING REMARKS
The discovery of T6SS loci and their prevalence in diverse human gut Bacteroidales species has revealed that the composition of the human gut microbiota is likely significantly influenced by these secretion systems. We still know very little about these secretion systems, including the targets of the GA1 and GA2 T6SSs, the advantage of transfer of these regions to co-resident Bacteroidales species, and the mechanisms of action of many of the toxic effectors. In addition, it will be interesting to determine the prevalence of T6SSs in human gut E. coli strains and whether these antagonistic systems contribute to their ability to affect colonization by enteric pathogens.
Acknowledgements
The Comstock lab is funded to study T6SSs of gut Bacteroidales by Public Health Service grant 01AI120633 from the NIH/National Institute of Allergy and Infectious Diseases.
References
- 1.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernal P, Allsopp LP, Filloux A, Llamas MA. 2017. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J 11:972–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steele MI, Kwong WK, Whiteley M, Moran NA. 2017. Diversification of Type VI Secretion System Toxins Reveals Ancient Antagonism among Bee Gut Microbes. MBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speare L, Cecere AG, Guckes KR, Smith S, Wollenberg MS, Mandel MJ, Miyashiro T, Septer AN. 2018. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1808302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Human Microbiome Project C. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grice EA, Segre JA. 2012. The human microbiome: our second genome. Annu Rev Genomics Hum Genet 13:151–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi H, Sakamoto M, Benno Y. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol 46:535–548. [DOI] [PubMed] [Google Scholar]
- 10.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M. 2005. Diversity of the human intestinal microbial flora. Science 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zitomersky NL, Coyne MJ, Comstock LE. 2011. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect Immun 79:2012–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coyne M, Zitomersky N, McGuire A, Earl A, Comstock L. 2014. Evidence of extensive DNA transfer between Bacteroidales species within the human gut. mBio 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD. 2014. A Type VI Secretion-Related Pathway in Bacteroidetes Mediates Interbacterial Antagonism. Cell Host Microbe 16:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson MM, Anderson DE, Bernstein HD. 2015. Analysis of the outer membrane proteome and secretome of Bacteroides fragilis reveals a multiplicity of secretion mechanisms. PLoS One 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coyne MJ, Roelofs KG, Comstock LE. 2016. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coyne M, Roelofs KG, Comstock LE. 2016. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. 2012. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durand E, Nguyen VS, Zoued A, Logger L, Pehau-Arnaudet G, Aschtgen MS, Spinelli S, Desmyter A, Bardiaux B, Dujeancourt A, Roussel A, Cambillau C, Cascales E, Fronzes R. 2015. Biogenesis and structure of a type VI secretion membrane core complex. Nature 523:555–560. [DOI] [PubMed] [Google Scholar]
- 19.Zoued A, Durand E, Brunet YR, Spinelli S, Douzi B, Guzzo M, Flaugnatti N, Legrand P, Journet L, Fronzes R, Mignot T, Cambillau C, Cascales E. 2016. Priming and polymerization of a bacterial contractile tail structure. Nature 531:59–63. [DOI] [PubMed] [Google Scholar]
- 20.Zoued A, Durand E, Santin YG, Journet L, Roussel A, Cambillau C, Cascales E. 2017. TssA: The cap protein of the Type VI secretion system tail. Bioessays 39. [DOI] [PubMed] [Google Scholar]
- 21.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. 2016. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci U S A 113:3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waters JL, Salyers AA. 2013. Regulation of CTnDOT conjugative transfer is a complex and highly coordinated series of events. MBio 4:e00569–00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8:552–563. [DOI] [PubMed] [Google Scholar]
- 24.Verster AJ, Ross BD, Radey MC, Bao Y, Goodman AL, Mougous JD, Borenstein E. 2017. The Landscape of Type VI Secretion across Human Gut Microbiomes Reveals Its Role in Community Composition. Cell Host Microbe 22:411–419 e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wexler AG, Bao Y, Whitney JC, Bobay LM, Xavier JB, Schofield WB, Barry NA, Russell AB, Tran BQ, Goo YA, Goodlett DR, Ochman H, Mougous JD, Goodman AL. 2016. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci U S A 113:3639–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hecht AL, Casterline BW, Earley ZM, Goo YA, Goodlett DR, Bubeck Wardenburg J. 2016. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep 17:1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim B, Zimmermann M, Barry NA, Goodman AL. 2017. Engineered Regulatory Systems Modulate Gene Expression of Human Commensals in the Gut. Cell 169:547–558 e515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross BD, Verster AJ, Radey MC, Schmidtke DT, Pope CE, Hoffman LR, Hajjar A, Peterson SB, Borenstein E, Mougous J. 2018. Acquired interbacterial defense systems protect against interspecies antagonism in the human gut microbiome. bioRxiv doi: 10.1101/471110:471110. [DOI] [Google Scholar]
- 29.Pudlo NA, Urs K, Kumar SS, German JB, Mills DA, Martens EC. 2015. Symbiotic Human Gut Bacteria with Variable Metabolic Priorities for Host Mucosal Glycans. MBio 6:e01282–01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tuncil YE, Xiao Y, Porter NT, Reuhs BL, Martens EC, Hamaker BR. 2017. Reciprocal Prioritization to Dietary Glycans by Gut Bacteria in a Competitive Environment Promotes Stable Coexistence. MBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Si M, Zhao C, Burkinshaw B, Zhang B, Wei D, Wang Y, Dong TG, Shen X. 2017. Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc Natl Acad Sci U S A 114:E2233–E2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, Wei G, Qian PY, Luo ZQ, Shen X. 2017. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun 8:14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao W, Caro F, Robins W, Mekalanos JJ. 2018. Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 359:210–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fast D, Kostiuk B, Foley E, Pukatzki S. 2018. Commensal pathogen competition impacts host viability. Proc Natl Acad Sci U S A 115:7099–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson MC, Vonaesch P, Saffarian A, Marteyn BS, Sansonetti PJ. 2017. Shigella sonnei Encodes a Functional T6SS Used for Interbacterial Competition and Niche Occupancy. Cell Host Microbe 21:769–776 e763. [DOI] [PubMed] [Google Scholar]
- 36.Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, Durand E, Journet L, Cascales E, Monack DM. 2016. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A 113:E5044–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pezoa D, Blondel CJ, Silva CA, Yang HJ, Andrews-Polymenis H, Santiviago CA, Contreras I. 2014. Only one of the two type VI secretion systems encoded in the Salmonella enterica serotype Dublin genome is involved in colonization of the avian and murine hosts. Vet Res 45:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. 2016. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540:280–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, Juncker AS, Manichanh C, Chen B, Zhang W, Levenez F, Wang J, Xu X, Xiao L, Liang S, Zhang D, Zhang Z, Chen W, Zhao H, Al-Aama JY, Edris S, Yang H, Wang J, Hansen T, Nielsen HB, Brunak S, Kristiansen K, Guarner F, Pedersen O, Dore J, Ehrlich SD, Bork P, Wang J, Meta HITC. 2014. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 32:834–841. [DOI] [PubMed] [Google Scholar]


