Abstract
Gestational exposure to air pollution is associated with negative outcomes in newborns and children. In a previous study, we demonstrated a synergistic negative effect of pre- and postnatal exposure to PM2.5 on lung development in mice. However, the means by which air pollution affects development of the lung have not yet been identified.
In this study, we exposed pregnant BALB/c mice and their offspring to concentrated urban PM2.5 (from São Paulo, Brazil; target dose 600 μg/m3 for 1 h daily). Exposure was started on embryonic day 5.5 (E5.5, time of placental implantation). Lung tissue of fetuses and offspring was submitted to stereological and transcriptomic analyses at E14.5 (pseudoglandular stage of lung development), E18.5 (saccular stage) and P40 (postnatal day 40, alveolarized lung). Additionally, lung function and cellularity of bronchoalveolar lavage (BAL) fluid were studied in offspring animals at P40.
Compared to control animals that were exposed to filtered air throughout gestation and postnatal life, PM-exposed mice exhibited higher lung elastance and a lower alveolar number at P40 whilst the total lung volume and cellularity of BAL fluid were not affected. Glandular and saccular structures of fetal lungs were not altered upon gestational exposure; transcriptomic signatures, however, showed changes related to DNA damage and its regulation, inflammation and regulation of cell proliferation. A differential expression was validated at E14.5 for the candidates Sox8, Angptl4 and Gas1.
Our data substantiate the in utero biomolecular effect of gestational exposure to air pollution and provide first-time stereological evidence that pre- and early life-postnatal exposure compromise lung development, leading to a reduced number of alveoli and an impairment of lung function in the adult mouse.
Keywords: Air pollution, Transcriptomics, Lung development, Particulate matter, Alveolarization
1. Introduction
According to the World Health Organization, about 3 million deaths were attributable to effects of ambient air pollution worldwide in 2012. With growing population, urbanization and industrialization, pollution of ambient air constitutes an increasing health risk, particularly in developing and low-income regions (World Health Organization, 2016).
Amongst other air pollutants such as ozone and NO2, particulate matter (PM) is the pollutant whose ambient air levels are most commonly used to assess air quality and air pollution-related health risks (Burnett et al., 2014; World Health Organization, 2016). Its composition is highly heterogeneous and variable, depending on the location and sources (both natural and human-made) of particle emission. PM deposits along the respiratory system dependent on the aerodynamic diameter of contained particles (Kelly and Fussell, 2012). The fraction with an aerodynamic diameter of 2.5 μm and lower (PM2.5) is considered small enough to reach alveolar spaces, with ultrafine particles (<0.1 μm) being able to cross the air/blood barrier.
Exposure to PM has frequently been linked to respiratory and cardiovascular morbidity and mortality (Feng et al., 2016), alongside a possible involvement in the development of diabetes mellitus and neurological diseases (Calderon-Garciduenas et al., 2016). Moreover, maternal exposure to PM during pregnancy has been linked to adverse birth outcomes (Feng et al., 2016; Li et al., 2017; Siddika et al., 2016; Teng et al., 2016; Veras et al., 2016) and was demonstrated to increase susceptibility to respiratory infections and cause lung function deficits in early childhood (Jedrychowski et al., 2010; Jedrychowski et al., 2013; Korten et al., 2016; Vieira, 2015), as shown for other pollutants (Morales et al., 2015). The rapid growth during organ development, involving morphological changes with complex regulation of proliferation and differentiation, is thought to predispose for susceptibility to environmental influences during the intrauterine and early-life period (Veras et al., 2016; Wright and Brunst, 2013).
During pregnancy, air pollutants might cause a systemic effect in the mother that subsequently impacts the fetus or could, once blood-borne, act directly on placenta and fetus (Soto et al., 2017; Veras et al., 2008). Plausible biological mechanisms include oxidative stress, inflammation, DNA damage as well as coagulative and hemodynamic changes in the mother (Hertz-Picciotto et al., 2008; Kannan et al., 2006; Veras et al., 2016). However, animal model studies on this field are scarce and the developmental effect of intrauterine exposure remains experimentally poorly defined. Similarly, the mechanisms by which prenatal exposure to air pollution impairs lung development are not yet understood.
Based on previous findings indicating that pre- and postnatal exposure of mice to urban-level air pollution synergistically affect lung development (Mauad et al., 2008), we hypothesized that prenatal exposure to PM2.5 causes changes in the development of the mouse lung that are already perceptive in utero, leading to altered lung structure and function during later life. We presumed that analysis of the fetal lung transcriptome provides novel insights into the underlying processes mediating PM-associated developmental changes and damage.
In this study, we exposed pregnant mice and their offspring to concentrated urban PM2.5 (concentrated ambient particles, CAP). In a comprehensive approach, we studied lung stereology and the lung transcriptome at pre- and postnatal time points. In addition, we assessed lung function and cellularity at an early-life stage and characterized the composition of particulate matter at our site.
2. Methods
2.1. Animals
Pregnant BALB/c mice were provided by the animal facility of the University of São Paulo School of Medicine, where breeding of BALB/c mice is conducted on a continuous basis. Pregnant mice were continuously sourced throughout the study period. Using the presence of a vaginal plug as indicator, the first light cycle after nightly mating was defined as embryonic day 0.5 (E0.5). Apart from the daily periods of exposure, mice were housed in polypropylene cages in an environment with constant room temperature (22 °C), filtered air and a 12 h/12 h light/dark cycle. Free access to diet (Nuvital, Colombo, PR, Brazil) and water was granted. This study was approved by the Institutional Review Board for Experimental Studies of the School of Medicine of São Paulo, Brazil (approval number 072/12). All animals received care in accordance with guidelines published by the National Research Council (USA) (National Research Council, 2011) with appropriate attention given to the alleviation of animals’ distress and discomfort.
2.2. Study design
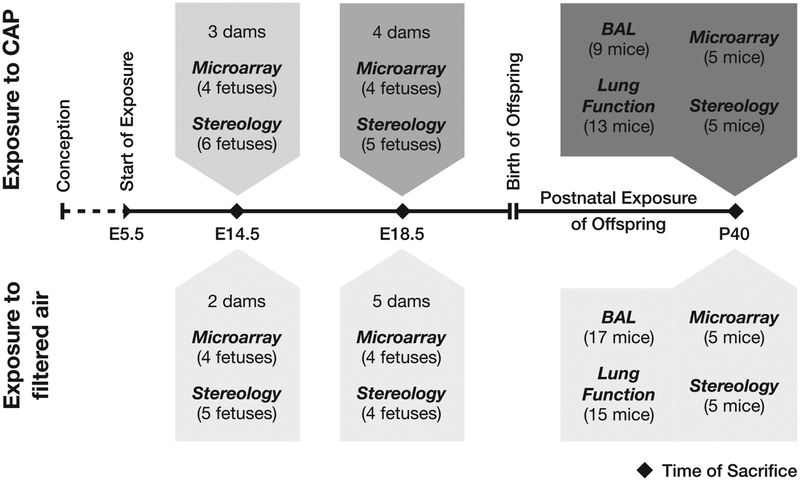
A total of 22 pregnant mice was randomly assigned to a CAP-exposed and a control group (exposed to filtered air). Exposure to CAP was started at embryonic day 5.5 (E5.5), after implantation of the embryo (Cross et al., 1994), and was continued in offspring after birth (Fig. 1). In part, pregnant mice were sacrificed and fetal lung tissue was submitted to stereological and microarray analysis at E14.5 (pseudoglandular stage of lung development) and E18.5 (saccular stage). Naturally-born offspring (14 exposed, 19 control mice) was kept until P40 (postnatal day 40, terminated lung alveolarization) and underwent lung function testing as well as bronchoalveolar lavage. As at prenatal time points, lung tissue of the offspring was submitted to stereological and microarray analysis. Detailed information about the number of fetuses retrieved at prenatal time points and about litter sizes of offspring can be found in Supplementary Tables S1 and S2.
Fig. 1.
Study design. The number of animals submitted to the respective analyses is specified in parentheses.
2.3. Exposure setup
The animal exposure took place during the dry season from August to November 2012.
As previously described, we used an ambient particle concentrator developed at the Harvard School of Public Health (HAPC) (Sioutas et al., 1995; Yoshizaki et al., 2016). This system uses a series of virtual impactors to concentrate fine ambient particles with an aerodynamic diameter <2.5 μm (PM2.5) from ambient air and is located on the campus of the University of São Paulo School of Medicine (23°33′18.1”S 46°40′15.0”W, http://www.inaira.org/br/intoxicacao/concentrador.shtml). Air pollution at this site has previously been characterized as emission from mainly vehicular sources (Andrade et al., 2012; CETESB, 2012; de Miranda et al., 2012). During the period of animal exposure, we assessed the metal elemental composition and black carbon (BC) concentration of ambient air PM2.5 (for methods and results, please refer to the supplement). The polycyclic aromatic hydrocarbon content of PM2.5 at our site was studied in July 2012, results were previously published elsewhere (Yoshizaki et al., 2016).
Animals were placed in temperature- and humidity-controlled exposure chambers, either connected to the CAP-stream derived from the HAPC (exposed group) or to a clean air supply provided using a high-efficiency particulate air filter (control group). The impactor setup of the HAPC creates a very light vacuum in the exposure chamber (Sioutas et al., 1995) (in our facility about −18 mmHg in relation to the surrounding atmospheric pressure) that itself could possibly influence lung physiology in mice. To account for this bias, the atmospheric pressure inside the CAP-exposure chamber and the clean-air chamber were regulated to be identical.
Mass concentrations of generated CAP were measured using an airborne particulate monitor (two-wavelength nephelometer DataRam DR-4000, Thermo Fisher Scientific, Waltham, MA, USA). Our target dose of CAP-exposure was 600 μg•m−3 for 1 h daily, equivalent to the WHO guideline value for 24-h mean PM2.5 concentration (25 μg•m−3) (World Health Organization, 2005). To ensure the best possible constancy in delivered CAP dose from day to day, the exposure time was proportionally adjusted to the HAPC-derived CAP concentration at the start of each exposure. For alleviation of animals’ discomfort, the maximum daily exposure time was limited to 120 min. Daily lengths of stay in the exposure chambers were the same for exposed and control mice. The animals received their last exposure on the day before their respective sacrifice.
The concentration of CAP derived from the HAPC is subject to continuous fluctuations and short-term alterations based on changes in environmental conditions. To approximate the effective daily CAP delivery, we calculated P, the mean of the CAP concentrations measured at the start and end of each exposure. Knowing the respective exposure time t, we calculated the proportion of our target dose and the CAP dose every mouse i received on their respective exposure day d. We calculated the exposure efficacy E over the respective number of exposure days n with the following formula:
A hypothetical exposure perfectly matching our target CAP concentration PT = 600 μg•m−3 and target exposure time tT = 1 h would herewith result in an exposure efficacy of 1.
2.4. Lung function testing
At P40, offspring mice (n = 13 in exposure, n = 15 in control group) were deeply anesthetized by intraperitoneal injection of thiopental (70 mg•kg−1), tracheotomized and connected to a flexiVent small animal ventilation device (SCIREQ, Montral, QC, Canada). Before starting mechanical ventilation, they were paralyzed by intraperitoneal injection of pancuronium bromide (1 mg•kg−1). Animals were ventilated with a tidal volume of 10 mL•kg−1 at a breathing frequency of 120 breaths•min−1. With single frequency forced oscillation maneuvers at a sinusoidal frequency of 2.5 Hz, we measured the dynamic resistance R and the dynamic elastance E (single compartment model). Input impedance of the respiratory system was measured applying oscillatory volume perturbations composed of the sum of 13 sinusoidal frequencies from 1 to 20.5 Hz (broadband forced oscillation technique). Based on the impedance, the Newtonian resistance (Rn, reflecting the resistance of airways), tissue damping (G) and tissue elastance (H) were calculated using the constant phase model (Bates, 2009; Gomes et al., 2000; Hantos et al., 1992).
2.5. Bronchoalveolar lavage (BAL)
Following lung function measurements, offspring animals were exsanguinated via the inferior vena cava. The right main bronchus was ligated and before BAL, the right lung was removed and stored for later RNA preparation (see below). Remaining left lungs were washed with 0.5 mL of sterile physiological saline for three times. Collected BAL fluids were centrifuged at 900 g for 8 min at 5 °C, supernatants were discarded and cell pellets were resuspended in 1 mL of physiological saline. A total cell count was performed using a Neubauer chamber (Carl Roth, Karlsruhe, Germany). Subsequently, cytocentrifuge slides were prepared and stained with Diff-Quik® (Medion Diagnostics, Dündingen, Switzerland). We performed differential cell counts by microscopic slide examination considering macrophages, lymphocytes, neutrophils, eosinophils and respiratory epithelial cells according to standard morphological criteria; 300 cells per slide were counted (n = 9 in exposure, n = 17 in control group).
2.6. Lung tissue collection
At E14.5 and E18.5, pregnant dams were anesthetized by isoflurane and subsequently euthanized by intraperitoneal injection of thiopental (200 mg•kg−1). Following exsanguination via the inferior vena cava, fetuses were retrieved. For microarray analysis, fetal lungs were dissected under stereomicroscopic view, snap frozen in liquid nitrogen and subsequently stored at −80 °C. Right lungs of P40 offspring (see above) were frozen and stored similarly.
2.7. Stereology
Whole E14.5 fetuses and dissected E18.5 lungs were fixed in 4% phosphate-buffered paraformaldehyde (PFA) for 24 h and embedded in paraffin. One 5 μm thick section was prepared and stained with H.E. for every 200 μm of tissue. We used digital scans of the slides (scanner: Pannoramic SCAN, 3DHistech, Budapest, Hungary) and the publically available software ImageJ (developed at the National Institute of Mental Health, Bethesda, MD, USA) for stereological analysis (exposure group: n = 6 at E14.5, n = 5 at E18.5; control group: n = 5 at E14.5, n = 4 at E18.5). Total lung volume was determined according to the Cavalieri principle (Hsia et al., 2010) superimposing a point grid on the digitized slides. We measured the relative volumes of lung compartments (E14.5: epithelial tubes, blood vessels, mesenchymal stroma; E18.5: airways, blood vessels, mesenchymal stroma, saccules) by point counting. We further used a cycloid grid to determine the surface area of saccules at E18.5. Absolute volumes of lung compartments were calculated multiplying relative volumes with total lung volumes.
Left lungs of offspring mice were inflated with 4 mL of PFA (equivalent to a pressure of ca. 20 cmH2O) after BAL, ligated and placed in PFA for 24 h. Fixed lungs were embedded in agar and cut into 2 mm thick slices in a vertical, uniform and random manner (Knust et al., 2009). A point grid was used to determine lung volumes (Cavalieri principle). Subsequently, agar slices were embedded in paraffin and 5 μm H.E.-stained sections were prepared. We used the newCAST stereology software (Visiopharm, Hoersholm, Denmark), coupled to an optical microscope (Eclipse Ni, Nikon, Tokyo, Japan), for determination of relative lung volumes and surface areas (airways, blood vessels, alveolar septa/spaces). Central airways and vessels were not considered in our analysis; for volume fractions, we did not differentiate between alveolar ducts and alveoli. Digitized scans of pairs of consecutive slices were used to perform an alveolar count based on the Euler characteristic (Hyde et al., 2004; Knust et al., 2009) (n = 5 per group: 3 males, 2 females in exposed; 1 male and 4 females in control group). In short, this method is based on counting fusions or separations of alveolar septal segments between a reference and a consecutive look-up section as a surrogate to count alveolar openings within a certain lung volume defined by the slice thickness and the analyzed frame area (for more information see Knust et al., 2009). Per animal, 3 pairs of consecutive slices were examined. In turn, a total of 4–6 disectors (frame size 11410.1 μm2) per slice pair was analyzed. Mean alveolar volumes and alveolar number densities were calculated with the derived alveolar numbers and the previously determined total alveolar/whole lung volumes.
2.8. Statistical analysis
For statistical evaluation of results (except microarray and qPCR analysis) as well as data visualization, SPSS® version 15.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA) were used. Data was found to be normally distributed. Results were considered statistically significant at p < 0.05 (Student′s t-test). Data is reported as mean ± standard deviation (SD).
2.9. Extraction of total RNA
Total RNA was extracted from tissue samples (n = 4 in E14.5 and E18.5 subgroups, n = 5 in P40 subgroups) using the RNeasy Mini Kit (Qiagen, Hilden, Germany), following manufacturer′s instructions. RNA purity and integrity were determined by spectrophotometry (NanoDrop 1000, Thermo Fisher Scientific) and capillary electrophoresis (Bioanalyzer 2100, RNA 6000 Nano Kit, Agilent Technologies, Santa Clara, CA, USA). An A260/A280 ratio between 1.8 and 2.1 and a RNA Integrity Number (RIN) > 7 were our RNA quality thresholds for subsequent microarray analysis.
2.10. Microarray analysis
100 ng of total RNA from each sample was amplified with the Ambion WT Expression Kit (Life Technologies, Carlsbad, CA, USA) and hybridized to the GeneChip Mouse Gene 2.0 ST array (Affymetrix, Santa Clara, CA, USA), following manufacturer’s standard protocols. Hybridized arrays were washed and stained using the GeneChip Hybridization Wash and Stain Kit with a Fluidics Station 450 (standard protocols, using the AGCC Fluidics Control) and scanned with a GeneChip Scanner 3000 7G (standard settings, AGCC Scan Control, Affymetrix). The expression data of this experiment have been deposited in NCBI’s Gene Expression Omnibus, accessible through GEO series accession number GSE104656 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE104656).
Output CEL-files were imported to the Affymetrix Expression Console software for summarization, background correction and normalization of expression data. The default Robust Multi-Array Analysis (RMA) algorithm was applied at gene level. Annotation of transcript clusters within the software was based on the most recent annotation data available (release 36). On-chip controls were removed from the data set, only main design transcript clusters were included in subsequent analyses. Processed expression data was imported to the TM4 MultiExperiment Viewer (MeV) version 4.9 (Saeed et al., 2003) (available at http://mev.tm4.org), differentially expressed genes (DEGs) were identified at a cutoff p = 0.01 (t-test).
2.11. Interpretation of results
For Venn diagram creation we used an online tool provided by the group of Bioinformatics & Evolutionary Genomics at the Ghent University (http://bioinformatics.psb.ugent.be/webtools/Venn). Enrichment analyses were carried out through publically available online tools using official gene symbols as provided by the Affymetrix annotation data as gene identifiers for data upload.
With Enrichr (http://amp.pharm.mssm.edu/Enrichr) (Kuleshov et al., 2016), we used the following library builds as reference libraries: KEGG 2016, BioCarta 2016, WikiPathways 2016, Reactome 2016, Panther 2016, GO (Biological Process, Cellular Component and Molecular Function) 2017, MGI Mammalian Phenotype 2017 and Jensen DISEASES. Enrichment results were explored using output tables ranked by Enrichr’s combined score, considering pathways/terms enriched at p < 0.05 and to a lesser extent those in the upper list that were ranked higher than at least one other pathway/term enriched at p < 0.05.
We used the Functional Annotation Tool of DAVID (https://david.ncifcrf.gov, version 6.8) (Huang da et al., 2009) to retrieve comparative information about enrichment of KEGG and BioCarta pathways at standard settings (cutoff EASE p = 0.1, minimum count = 2, standard background for Mus musculus).
Reactome pathway analysis was also performed via the official online platform (http://reactome.org, database version 60) (Fabregat et al., 2016) using the analysis tool with the projection to human option enabled. Results were explored focusing on over-represented pathways with p < 0.05. The PANTHER pathway database (version 3.4.1, PANTHER version 11.1) was used for analysis of DEG lists through the Gene List Analysis tool (PANTHER Over-representation Test) on the official website (http://pantherdb.org) (Mi et al., 2017) (cutoff p = 0.05). We supplied a custom background gene list for analysis, containing all genes included in the Mouse Gene 2.0 ST microarray′s main design. This custom background was further used in GO term enrichment with GORILLA (http://cbl-gorilla.cs.technion.ac.il, database update 06/17/17) (Eden et al., 2009) at a significance cutoff p = 0.001.
2.12. qPCR validation
We used quantitative real-time PCR (qPCR) to validate the differential expression of six possible candidate genes at E14.5. Per sample, 1000 ng of total RNA underwent reverse transcription to cDNA using the High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific), according to manufacturer′s instructions. PCR was performed using TaqMan Gene Expression Assays (FAM-labeled MGB probes) for the following genes: Gas1 (growth arrest specific 1, Mm01700206_g1), Sox8 (SRY-box 8, Mm00803422_m1), Muc4 (mucin 4, Mm00466886_m1), Cdkn1c (cyclin-dependent kinase inhibitor 1C, Mm01272135_g1), Csnk1d (casein kinase 1, delta, Mm00503623_m1) and Angptl4 (angiopoetin-like 4, Mm00480431_m1). The StepOnePlus RT-PCR system and the Taq-Man Fast Advanced Master Mix (Thermo Fisher Scientific) were used according to manufacturer’s protocols. Each sample was run in triplicate, Actb (actin, beta, Mm00607939_s1) and Rpl13a (ribosomal protein L13A, Mm01612986_gH) were used as endogenous reference genes.
Fold changes were calculated with the comparative Ct method (2−ΔΔCt). We tested for statistical significance with Mann-Whitney U tests using SPSS version 18.0 (IBM Corp., Armonk, NY, USA). Results were considered statistically significant at p < 0.05.
3. Results
3.1. Animal exposure
The calculated mean exposure efficacy during the exposure period (78 days) was 1.10 ± 0.22. This shows that the CAP-exposed animals effectively received a slightly higher dose than targeted, equivalent to a mean 24-h PM2.5 exposure to 27.5 μg•m−3. On 11 days within the whole exposure period (78 days in total), it was not possible to perform an exposure to CAP due to high relative air humidity and the consequential insufficient CAP concentration derived from the HAPC. The metal element composition and black carbon content of PM2.5 specified at our site are shown in Supplementary Table S3. Data about the content of polycyclic aromatic hydrocarbons has been published previously (Yoshizaki et al., 2016).
3.2. Lung function testing
We observed a statistically significant increase of tissue elastance (32.44 ± 8.10 vs. 27.08 ± 5.20 cmH2O•mL−1, p = 0.04) in mice pre- and postnatally exposed to CAP, compared to control animals (Table 1). The dynamic lung elastance was also increased (34.31 ± 7.77 vs. 29.68 ± 4.16 cmH2O•mL−1) with a strong statistical tendency (p = 0.06). The effect of exposure to CAP on elastance parameters was observed in both male and female animals (see Supplementary Tables S4 and S5). Parameters reflecting the resistance (dynamic resistance, Newtonian resistance and tissue damping) were not affected by exposure to CAP. Though not a primary endpoint of our study and not observed in female animals, exposed male mice showed a lower body weight at P40 than male controls (20.1 ± 1.6 vs. 23.1 ± 1.5 g, p = 0.006).
Table 1.
Lung function (flexiVent) of exposed and control mice at P40.
| Parameter | Mean ± SD | p | ||
|---|---|---|---|---|
| Exposed | Control | |||
| Weight | (g) | 19.9 ± 1.6 | 20.7 ± 2.5 | 0.34 |
| R | (cmH2O·s·mL−1) | 0.83 ± 0.15 | 0.76 ± 0.14 | 0.20 |
| E | (cmH2O·mL−1) | 34.31 ± 7.77 | 29.68 ± 4.16 | 0.06 |
| Rn | (cmH2O·s·mL−1) | 0.30 ± 0.10 | 0.28 ± 0.09 | 0.61 |
| G | (cmH2O·mL−1) | 5.98 ± 1.91 | 5.96 ± 1.06 | 0.97 |
| H | (cmH2O·mL−1) | 32.44 ± 8.10 | 27.08 ± 5.20 | 0.04* |
R: dynamic resistance, E: dynamic elastance, Rn: Newtonian resistance, G: tissue damping, H: tissue elastance.
3.3. Bronchoalveolar lavage
We observed no significant differences between the exposed and the control group animals in cellularity of BAL fluid (see Supplementary Table S6).
3.4. Stereology
We did not observe a difference in stereological parameters of the developing lung between the CAP-exposed and the control group at E14.5 and E18.5 (Supplementary Tables S7 and S8).
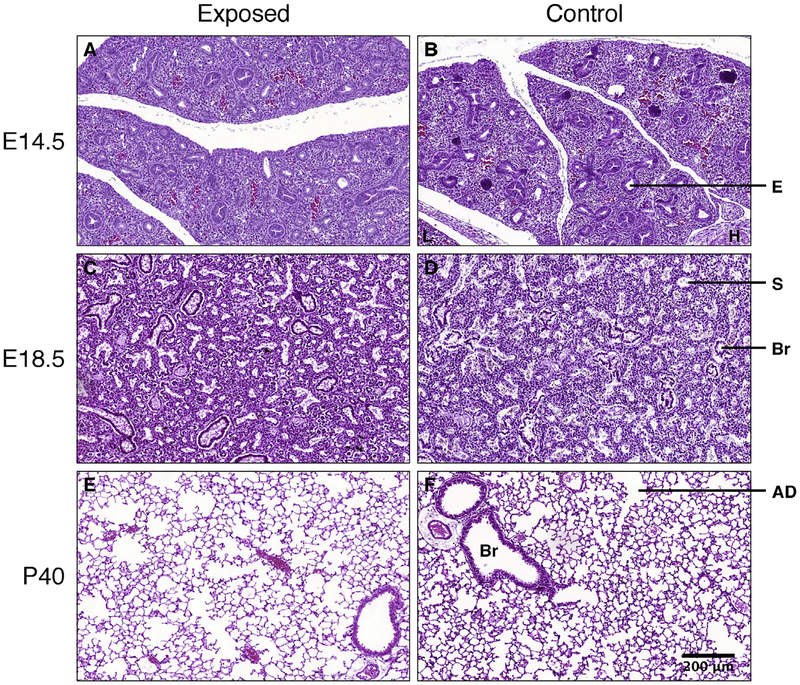
At P40 (alveolarized lung), however, exposed animals had a significantly decreased number of alveoli (in 106 alveoli: 0.91 ± 0.38 vs. 1.51 ± 0.22, p = 0.02). Accordingly, the alveolar number density (in 103 alveoli per mm3 lung: 9.73 ± 2.09 vs. 13.73 ± 1.08, p = 0.02) was reduced and the mean alveolar volume (in 103 μm3: 53.7 ± 10.0 vs. 39.2 ± 4.2, p = 0.02) was increased. Total lung volumes, alveolar surface areas and surface densities were not significantly decreased in the exposed group (Table 2, Fig. 2A–F).
Table 2.
Stereological measures of lungs of pre-/postnatally exposed mice and controls at P40.
| Parameter | Mean ± SD | p | |
|---|---|---|---|
| Exposed | Control | ||
| Animal weight (g) | 19.4 ± 1.6 | 18.5 ± 1.0 | 0.32 |
| Lung volume (mm3) | 92 ± 28 | 109 ± 10 | 0.22 |
| vol/wt ratio (mm3·g−1) | 4.7 ± 1.3 | 5.9 ± 0.7 | 0.11 |
| ND of alveoli (103 mm−3) | 9.73 ± 2.09 | 13.73 ± 1.08 | 0.005** |
| N of alveoli (106) | 0.91 ± 0.38 | 1.51 ± 0.22 | 0.02* |
| of alveoli (103 μm3) | 53.7 ± 10.0 | 39.2 ± 4.2 | 0.02* |
| Vf of alveoli (%) | 50.8 ± 5.0 | 53.5 ± 2.6 | 0.33 |
| Vf of alveolar septa (%) | 39.5 ± 2.3 | 36.3 ± 3.2 | 0.11 |
| Vf of airways (%) | 6.7 ± 2.5 | 7.4 ± 2.9 | 0.68 |
| Vf of vessels (%) | 2.9 ± 1.6 | 2.8 ± 1.2 | 0.88 |
| Vt of alveoli (mm3) | 47.5 ± 17.1 | 58.5 ± 6.6 | 0.22 |
| Vt of alveolar septa (mm3) | 35.9 ± 9.6 | 39.8 ± 5.5 | 0.46 |
| Vt of airways (mm3) | 5.9 ± 2.4 | 8.1 ± 3.4 | 0.26 |
| Vt of vessels (mm3) | 2.5 ± 1.3 | 3.0 ± 1.3 | 0.56 |
| SD of alveolar septa (mm−1) | 192.5 ± 40.3 | 219.8 ± 24.3 | 0.23 |
| SD of airways (mm−1) | 18.5 ± 7.1 | 16.7 ± 1.5 | 0.59 |
| SD of vessels (mm−1) | 45.6 ± 13.4 | 61.1 ± 45.6 | 0.49 |
| SA of alveolar septa (cm2) | 184.5 ± 86.7 | 241.2 ± 40.1 | 0.22 |
| SA of airways (cm2) | 15.8 ± 5.4 | 18.3 ± 3.0 | 0.39 |
| SA of vessels (cm2) | 42.8 ± 21.5 | 67.7 ± 52.7 | 0.36 |
ND: number density, N: total number, : mean volume, Vf: volume fraction, Vt: total volume, SD: surface density, SA: surface area.
Fig. 2.
Lung tissue from CAP-exposed (A, C, E) and control animals (B, D, F) at E14.5 (pseudoglandular stage, A, B), E18.5 (saccular stage, C, D) and P40 (alveolarized lung, E, F). H.E.-staining, all images taken at same magnification. E: budding epithelial tube, L: liver, H: heart (myocardium), Br: bronchiole, S: saccule, AD: alveolar duct. Alveoli appear enlarged in the CAP-exposed group at P40.
3.5. Transcription microarray analysis
At E14.5, 329 genes were differentially expressed (112 down-regulated, 217 up-regulated). At E18.5 and P40, the numbers of DEGs were 319 (209 down, 110 up) and 249 (151 down, 98 up), respectively. The overall expression changes were found to be comparatively low, only 1.8% of all DEGs amongst the three time points exhibited log2 fold changes of an absolute value > 1. A total of 6 genes were found to be regulated both at E14.5 and E18.5 but exhibited opposite fold changes at the respective time points. Three genes were differentially expressed at both E14.5 and P40, but not at E18.5 (Supplementary Fig. S1, Supplementary Tables S9 and S10).
Enrichment analyses of E14.5 DEGs resulted in terms related to DNA metabolism and damage repair, siRNA and miRNA metabolism, chemokine and G protein signaling, nuclear receptor transcription pathway as well as Wnt and cadherin signaling. “Respiratory distress” was the second most enriched term in the Enrichr MGI Phenotype reference library and “lung disease” appeared on top of the list of enriched terms in the Jensen DISEASES library.
At E18.5, enrichment results included terms related to MAP kinase and phosphatidylinositol signaling, cytokine/interleukin signaling, T cell and Toll-like receptor signaling as well as regulation of cell proliferation/apoptosis. Enriched MGI Phenotype terms were highly related to inflammation and immune response.
DEGs at P40 were found to be enriched in terms related to steroid/cholesterol biosynthesis and metabolism, phosphatidylinositol and calcium signaling, amino acid and lipid metabolism, SREBP (sterol regulatory element-binding protein) signaling, actin skeleton regulation and oxidative stress. Full lists of enrichment results can be found in the data supplement.
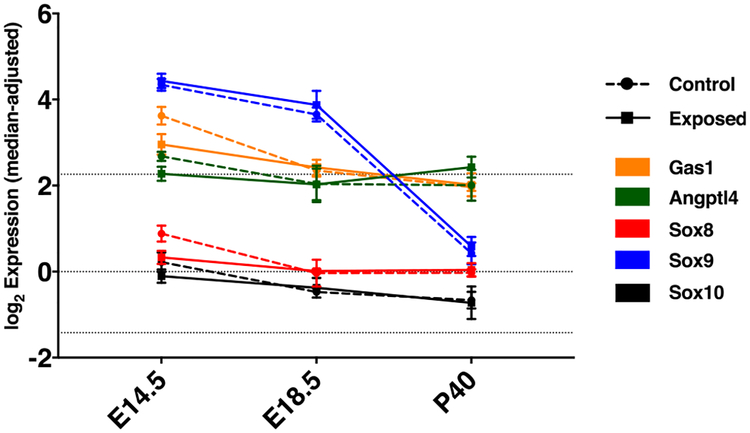
The six DEGs at E14.5 that had been chosen for validation (Gas1, Sox8, Muc4, Cdkn1c, Csnk1d, Angptl4) showed qPCR fold changes concordant to the microarray results (Table 3). Whereas Csnk1d had an even smaller FC in qPCR (close to 0), the other five genes showed larger fold changes than in the microarray analysis. All six genes underwent qPCR in E18.5 and P40 samples as well, confirming that differential expression was observed at E14.5 only (data not shown). However, the qPCR results showed a statistically significant expression difference (down-regulation) only for Sox8 (log2 FC = −1.84, p = 0.02) and Angptl4 (log2 FC = −0.49, p = 0.02). Gas1 exhibited a statistical tendency for being down-regulated (log2 FC = −0.84, p = 0.08). The time course of expression (based on microarray data) of Gas1, Sox8 and Angptl4 is depicted in Fig. 3.
Table 3.
Microarray and qPCR fold changes of candidate genes at E14.5.
| DEG | Microarray | qPCR | ||
|---|---|---|---|---|
| log2 FC | p | log2 FC | p | |
| Gas1 | −0.67 | 0.005 | −0.84 | 0.08 |
| Sox8 | −0.55 | 0.004 | −1.84 | 0.02* |
| Muc4 | 0.43 | 0.006 | 0.86 | 0.25 |
| Cdkn1c | −0.48 | 0.005 | −0.60 | 0.15 |
| Csnk1d | 0.12 | 0.004 | 0.03 | 0.56 |
| Angptl4 | −0.41 | 0.006 | −0.49 | 0.02* |
Fig. 3.
Median-adjusted and log2-transformed expression (transcription microarray) of Gas1, Angptl4 and Sox 8 as well as the SoxE family members Sox9 and Sox10 in lungs of CAP-exposed and control mice. Error bars show SD, dotted lines represent quartiles of the overall distribution of expression values in the experiment.
4. Discussion
In this study, we present for the first time stereological evidence that pre- and early postnatal exposure to urban, vehicle-derived PM2.5 leads to a decrease in alveolar number, resulting in enlarged alveolar spaces and increased lung elastance at an early stage of life. We further demonstrated transcriptional changes in the fetal lung linked to DNA damage and regulation, inflammation and cell proliferation upon gestational exposure to PM2.5. These findings imply not only that exposure to PM2.5 during lung development disturbs the alveolarization process and alters physiological properties, but also that the lung in utero is susceptible to gestational exposure at the molecular level, possibly causing or promoting PM-associated effects during later life.
In contrast to our previous study (Mauad et al., 2008), here we implemented an experimental setup exposing animals to concentrated ambient particles for a specified time per day (pulsed exposure), rather than keeping them exposed to ambient pollution levels for 24 h. This allowed us to better control the delivered pollutant dose since the concentration of ambient particles could partly compensate for environmental conditions that lowered ambient particle levels. Thereby, we established an exposure setup with more constancy and higher delivered particle doses. At the same time, by keeping the mice in a dedicated environment separated from the surroundings, we could ensure that the animals were not open to disturbing environmental influences such as noise or temperature fluctuations that were present in the former 24 h-exposition setup (Mauad et al., 2008).
To which extent pulsed and continuous exposures are equivalent to each other regarding the elicited biological responses, cannot be evaluated definitively at this point. A pulsed (experimental) exposure to high pollutant levels over a short period of time may lead to physiological changes different from those caused by a long-time (e.g. life-long) exposure to continuous, lower levels of pollutants. This could potentially explain the lack of change in BAL-cellularity upon CAP-exposure observed in our study. Dependent on the pattern of exposure, differential molecular mechanisms may be involved in causing developmental damage. However, the increase in lung elastance at P40 upon pre- and postnatal pulsed exposure to PM2.5 found in this study matches the alteration of pressure/volume curves upon continuous 24 h-exposure shown in our previous work (Mauad et al., 2008). So far, other groups had investigated air pollutant-related lung elastance changes in adult mice only (Mazzoli-Rocha et al., 2016; Thevenot et al., 2013).
There were no significant stereological alterations in the saccular structures of distal fetal lungs. In young adult mice, however, PM2.5-exposure induced an important decrease in the absolute number of alveoli, with increased alveolar sizes, without significant changes of the alveolar surface density. There were no changes in airways or vessel volumes. These data indicate that PM2.5-exposure affects mainly alveolarization. For ozone, it had been previously demonstrated that exposure of infant Rhesus monkeys during the early postnatal period affects the development of distal airways and the appearance and timing of alveolar growth and maturation (Fanucchi et al., 2006).
At P40, mice lungs are regarded to as having just finished secondary septation and microvascular maturation (Schittny, 2017), entering adulthood. It has been shown that the overall number of alveoli physiologically continues to increase together with the lung volume beyond P40, without an alteration of alveolar size and density (Pozarska et al., 2017). Hence, the observed change in alveolar structure at P40 could possibly persist during later life since the alveolarization process itself had been terminated. However, evidence from our data is not conclusive as we did not investigate later time points. A recent study demonstrated that exposure to PM2.5 and ozone increases the risk of development of an Asthma-COPD overlap syndrome in adult asthmatics (To et al., 2016). Whether (prenatal) exposure to air pollution of children causes changes in alveolar structure and thereby impacts their adult respiratory health remains to be investigated.
A study in the ENVIRONAGE birth cohort analyzed the transcriptome of cord blood cells of newborns and showed that chronic exposure to PM2.5 during gestation is associated with altered expression of genes related to DNA damage in newborns of both sexes, amongst other abnormalities (Winckelmans et al., 2017). So far, several mouse studies have focused on effects of maternal exposure to cigarette smoke (CS) on the fetus′ and the offspring′s lung. Adverse effects on lung function (Drummond et al., 2017) and morphology (Noel et al., 2017) have been shown and synergistic effects between pre- and postnatal exposure have been identified. A recent study provided first insights into CS-related transcriptional alterations in the fetal lung (Unachukwu et al., 2017). Regarding ambient air pollution, however, we yet remain the first to provide evidence of an effect of prenatal and early-life postnatal exposure to particulate matter on lung development.
Our results indicate that exposure to PM2.5 alters gene transcription differentially in the developing lung at distinct developmental stages. The developmental stages themselves, each requiring a unique “program” for cell proliferation, differentiation and function, as well as the different ways of exposure before and after birth (transplacental and transpulmonary) possibly contribute to this finding. DEGs during the prenatal period were linked to DNA damage and regulation, inflammation as well as cell proliferation. This is consistent with the hypotheses that the effect exerted by maternal exposure to PM2.5 on the developing fetus may be mediated by both a systemic (inflammatory) response of the dam as well as a blood/placenta-mediated effect of particles on the fetus possibly leading to DNA damage in a fetus-specific response (Veras et al., 2016). DEGs at P40, on the other hand, can be seen in the picture of an alveolarized lung that was exposed to PM2.5 throughout development and might be affected by means of oxidative stress and inflammation.
Upon exposure to PM2.5, we found altered expression levels of genes that might be of interest in the setting of lung development. Particularly in the case of Sox8, but also in most of the other candidates studied, we observed a larger fold change in qPCR results than in microarray analysis. Compression of fold changes, especially for lower expressed genes, is a common feature of microarray experiments (Choe et al., 2005; Dallas et al., 2005).
Sox8, SRY (sex determining region Y)-box 8, has been shown to play a role in primary sex determination and neural development (She and Yang, 2015; Weider and Wegner, 2017) and has also been linked to non-small cell lung cancer (Xie et al., 2014). At E14.5, it is expressed in the epithelial buds of distal airways (Eurexpress atlas) (Diez-Roux et al., 2011). Information about its role during lung development remain highly scarce compared to the better studied role of the related SoxE group member Sox9 which has been shown to be a regulator in branching lung morphogenesis and differentiation of respiratory epithelial progenitors (Chang et al., 2013; Rockich et al., 2013; Ustiyan et al., 2016), being linked to Wnt/β-catenin as well as Kras signaling (Caprioli et al., 2015; Rockich et al., 2013; Ustiyan et al., 2016). Sox8-deficient mice exhibit reduced body weight starting at postnatal week 3 (Sock et al., 2001). Our data indicate a lower, but nevertheless present expression of Sox8 (and Sox10) in fetal lung tissue with less over-time regulation than Sox9 (Fig. 3). This matches information retrievable from recent expression databases such as the Lung Gene Expression Analysis (LGEA) Web Portal (Du et al., 2017), the LungMAP database (Ardini-Poleske et al., 2017) and the EMBL-EBI Expression Atlas (Petryszak et al., 2016).
The role in developmental processes of Angptl4, angiopoietin-like 4, is poorly studied. Angptl4 has been shown to exhibit a plethora of different capacities, being involved in lipid metabolism, wound healing, regulation of reactive oxygen species, inflammation, tumorigenesis and regulation of vascular integrity/leakiness (La Paglia et al., 2017; Zhu et al., 2012; Zhu et al., 2016). The Angptl4 protein is subject to proteolytic cleavage with different functions attributed to the N- and C-terminal regions and possible differential processing between tissues (Dijk and Kersten, 2014). ANGPTL4 is expressed in human airway smooth muscle cells and its induction has been shown to be regulated through PKC and MAPK pathways (Stapleton et al., 2010). In the E14.5 lung, it is predominantly expressed in the subepithelial mesenchyme (Eurexpress atlas). Its role in angiogenesis still remains controversial and seems to depend on the tissue in question (Guo et al., 2014).
Growth-arrest-specific 1 (Gas1) is known to be a regulator of cell proliferation, inducing cell arrest and mediating apoptosis. It plays a role in embryonal differentiation and has been shown to be a positive regulator of the Sonic hedgehog pathway (Rosti et al., 2015; Segovia and Zarco, 2014). During lung development, it is strongly expressed in the developing bronchioles (Lee et al., 2001) and, analogue to our microarray data, shows a slight downregulation over the course of development in the aforementioned databases.
A limitation to our study is the restricted number of animals that could be exposed in parallel due to the complex and time-intensive exposure setup. This did neither allow us to implement a cross-over between study groups at birth of the offspring, nor to expose the dams (and males) before mating. Furthermore, it limited the number of biological samples per group which particularly requires a careful interpretation of the transcriptomic data.
Generally speaking, the small fold changes might further implicate a low biological impact of the found differential gene expression. Assuming that air pollution does not exert its effects via a single specific pathway but rather via a complex interplay of alterations at several biological levels (genetic, epigenetic, transcriptional, translational and posttranslational changes to the point of alteration and regulation of proteins steering complex processes such as cell growth, differentiation and intra- or intercellular signaling) this seems explicable. Similarly, transcriptional micro-array analysis can only reflect a part of the whole picture of cellular processes and regulation. Besides, our whole-lung transcriptomic approach might have missed detecting transcriptional alterations only present in single non-abundant cell types due to a dilution effect on the RNA level. Whilst the time point P40 was essential in this study to assess the consequences of developmental exposure at an early-life stage, microarray data from P40 does not allow to draw conclusions about foregone biomolecular processes during alveolarization.
We did not separately analyze male and female animals for stereology. Male mice are known to have a higher number of alveoli than females, most likely based on a higher total lung volume rather than higher alveolar density (Pozarska et al., 2017). In fact, more lungs of male animals were analyzed in the exposed group than in the control group. However, at this early-life stage we did not observe a difference in lung volumes between groups and, taking into account that the alveolar number densities were significantly altered upon CAP-exposure, this suggests that the found differences in alveolar parameters are not caused by differences in male-to-female ratios between the groups.
Previous data indicates that pre- and postnatal exposure to PM2.5 account for developmental changes synergistically (Mauad et al., 2008). In addition to the mild transcriptomic changes found in fetal lungs upon maternal exposure, epigenetic alterations could further be responsible for the demonstrated impairment of lung development in early postnatal life (Traboulsi et al., 2017). Future larger-scale animal studies will be required to further decipher the pathophysiology of pollution-related developmental damage.
5. Conclusion
Intrauterine and early postnatal exposure to urban, vehicle-derived PM2.5 decreases the number of alveoli and affects lung function, associated with development- and inflammation-related transcriptomic changes in fetal lung tissue. Our findings emphasize the importance of making efforts to reduce the global burden of ambient air pollution in order to improve lung health of generations to come.
Supplementary Material
Acknowledgements
We thank Regiani Oliveira, Luis Amato and Rosana Astolfo for their contribution to the characterization of particulate matter. Furthermore, we would like to thank Henrique Moriya for his scientific support.
Funding: This study was funded by the Brazilian National Institute of Science and Technology – National Institute of Integrated Analysis of Environmental Risk (INCT-INAIRA), the Brazilian National Council for Scientific and Technological Development (CNPq, grant number 573813/2006–6) and the São Paulo Research Foundation (FAPESP, grant number 08/577717–6).
This paper has been recommended for acceptance by Payam Dadvand.
Footnotes
Conflicts of interest: None.
Appendix A. Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.envpol.2018.05.055.
References
- Andrade MD, de Miranda RM, Fornaro A, Kerr A, Oyama B, de Andre PA, Saldiva P, 2012. Vehicle emissions and PM(2.5) mass concentrations in six Brazilian cities. Air Qual. Atmos. Health 5, 79–88. 10.1007/s11869-010-0104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardini-Poleske ME, Clark RF, Ansong C, Carson JP, Corley RA, Deutsch GH, Hagood JS, Kaminski N, Mariani TJ, Potter SS, Pryhuber GS, Warburton D, Whitsett JA, Palmer SM, Ambalavanan N, Lung MAPCT, 2017. LungMAP: the molecular atlas of lung development program. Am. J. Physiol. Lung Cell Mol. Physiol ajplung.00139.02017 10.1152/ajplung.00139.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates JHT, 2009. Lung Mechanics An Inverse Modeling Approach. Cambridge University Press, Cambridge, NY, USA. [Google Scholar]
- Burnett RT, Pope CA 3rd, Ezzati M, Olives C, Lim SS, Mehta S, Shin HH, Singh G, Hubbell B, Brauer M, Anderson HR, Smith KR, Balmes JR, Bruce NG, Kan H, Laden F, Pruss-Ustun A, Turner MC, Gapstur SM, Diver WR, Cohen A, 2014. An integrated risk function for estimating the global burden of disease attributable to ambient fine particulate matter exposure. Environ. Health Perspect 122, 397–403. 10.1289/ehp.1307049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Garciduenas L, Leray E, Heydarpour P, Torres-Jardon R, Reis J, 2016. Air pollution, a rising environmental risk factor for cognition, neuroinflammation and neurodegeneration: the clinical impact on children and beyond. Rev. Neurol. (Paris) 172, 69–80. 10.1016/j.neurol.2015.10.008. [DOI] [PubMed] [Google Scholar]
- Caprioli A, Villasenor A, Wylie LA, Braitsch C, Marty-Santos L, Barry D, Karner CM, Fu S, Meadows SM, Carroll TJ, Cleaver O, 2015. Wnt4 is essential to normal mammalian lung development. Dev. Biol 406, 222–234. 10.1016/j.ydbio.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CETESB, 2012. Qualidade do Ar no Estado de São Paulo 2012 (in Portuguese). Available on. http://cetesb.sp.gov.br/ar/publicacoes-relatorios/. (Accessed 9 April 2018).
- Chang DR, Martinez Alanis D, Miller RK, Ji H, Akiyama H, McCrea PD, Chen J, 2013. Lung epithelial branching program antagonizes alveolar differentiation. Proc. Natl. Acad. Sci. U. S. A 110, 18042–18051. 10.1073/pnas.1311760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe SE, Boutros M, Michelson AM, Church GM, Halfon MS, 2005. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 6, R16 10.1186/gb-2005-6-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross JC, Werb Z, Fisher SJ, 1994. Implantation and the placenta: key pieces of the development puzzle. Science 266, 1508–1518. [DOI] [PubMed] [Google Scholar]
- Dallas PB, Gottardo NG, Firth MJ, Beesley AH, Hoffmann K, Terry PA, Freitas JR, Boag JM, Cummings AJ, Kees UR, 2005. Gene expression levels assessed by oligonucleotide microarray analysis and quantitative real-time RTPCR – how well do they correlate? BMC Genom. 6, 59 10.1186/1471-2164-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Miranda RM, de Fatima Andrade M, Fornaro A, Astolfo R, de Andre PA, Saldiva P, 2012. Urban air pollution: a representative survey of PM(2.5) mass concentrations in six Brazilian cities. Air Qual. Atmos. Health 5, 63–77. 10.1007/s11869-010-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nurnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia-Lopez R, Echevarria D, Puelles E, Garcia-Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dolle P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G, Ballabio A, 2011. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 9, e1000582 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijk W, Kersten S, 2014. Regulation of lipoprotein lipase by Angptl4. Trends Endocrinol. Metabol 25, 146–155. 10.1016/j.tem.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Drummond D, Baravalle-Einaudi M, Lezmi G, Vibhushan S, Franco-Montoya ML, Hadchouel A, Boczkowski J, Delacourt C, 2017. Combined effects of in utero and adolescent tobacco smoke exposure on lung function in C57Bl/6J mice. Environ. Health Perspect 125, 392–399. 10.1289/ehp54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Kitzmiller JA, Sridharan A, Perl AK, Bridges JP, Misra RS, Pryhuber GS, Mariani TJ, Bhattacharya S, Guo M, Potter SS, Dexheimer P, Aronow B, Jobe AH, Whitsett JA, Xu Y, 2017. Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax 72, 481–484. 10.1136/thoraxjnl-2016-209598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z, 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinf. 10, 48 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMBL-EBI Expression Atlas: https://www.ebi.ac.uk/gxa/home/, accessed on June 14, 2017.
- Eurexpress atlas: http://www.eurexpress.org/, accessed on October 21, 2017.
- Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F, McKay S, Matthews L, May B, Milacic M, Rothfels K, Shamovsky V, Webber M, Weiser J, Williams M, Wu G, Stein L, Hermjakob H, D’Eustachio P, 2016. The reactome pathway knowledgebase. Nucleic Acids Res. 44, D481–D487. 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanucchi MV, Plopper CG, Evans MJ, Hyde DM, Van Winkle LS, Gershwin LJ, Schelegle ES, 2006. Cyclic exposure to ozone alters distal airway development in infant rhesus monkeys. Am. J. Physiol. Lung Cell Mol. Physiol 291, L644–L650. 10.1152/ajplung.00027.2006. [DOI] [PubMed] [Google Scholar]
- Feng S, Gao D, Liao F, Zhou F, Wang X, 2016. The health effects of ambient PM2.5 and potential mechanisms. Ecotoxicol. Environ. Saf 128, 67–74. 10.1016/j.ecoenv.2016.01.030. [DOI] [PubMed] [Google Scholar]
- Gomes RF, Shen X, Ramchandani R, Tepper RS, Bates JH, 2000. Comparative respiratory system mechanics in rodents. J. Appl. Physiol 89, 908–916. [DOI] [PubMed] [Google Scholar]
- Guo L, Li SY, Ji FY, Zhao YF, Zhong Y, Lv XJ, Wu XL, Qian GS, 2014. Role of Angptl4 in vascular permeability and inflammation. Inflamm. Res 63, 13–22. 10.1007/s00011-013-0678-0. [DOI] [PubMed] [Google Scholar]
- Hantos Z, Daroczy B, Suki B, Nagy S, Fredberg JJ, 1992. Input impedance and peripheral inhomogeneity of dog lungs. J. Appl. Physiol 72, 168–178. [DOI] [PubMed] [Google Scholar]
- Hertz-Picciotto I, Park HY, Dostal M, Kocan A, Trnovec T, Sram R, 2008. Prenatal exposures to persistent and non-persistent organic compounds and effects on immune system development. Basic Clin. Pharmacol. Toxicol 102, 146–154. 10.1111/j.1742-7843.2007.00190.x. [DOI] [PubMed] [Google Scholar]
- Hsia CC, Hyde DM, Ochs M, Weibel ER, 2010. An official research policy statement of the American Thoracic Society/European Respiratory Society: standards for quantitative assessment of lung structure. Am. J. Respir. Crit. Care Med. 181, 394–418. 10.1164/rccm.200809-1522ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA, 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hyde DM, Tyler NK, Putney LF, Singh P, Gundersen HJ, 2004. Total number and mean size of alveoli in mammalian lung estimated using fractionator sampling and unbiased estimates of the Euler characteristic of alveolar openings. Anat. Rec. Part A Discov. Mol. Cell. Evol. Biol 277, 216–226. 10.1002/ar.a.20012. [DOI] [PubMed] [Google Scholar]
- Jedrychowski WA, Perera FP, Maugeri U, Mroz E, Klimaszewska-Rembiasz M, Flak E, Edwards S, Spengler JD, 2010. Effect of prenatal exposure to fine particulate matter on ventilatory lung function of preschool children of non-smoking mothers. Paediatr. Perinat. Epidemiol 24, 492–501. 10.1111/j.1365-3016.2010.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrychowski WA, Perera FP, Spengler JD, Mroz E, Stigter L, Flak E, Majewska R, Klimaszewska-Rembiasz M, Jacek R, 2013. Intrauterine exposure to fine particulate matter as a risk factor for increased susceptibility to acute broncho-pulmonary infections in early childhood. Int. J. Hyg Environ. Health 216, 395–401. 10.1016/j.ijheh.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S, Misra DP, Dvonch JT, Krishnakumar A, 2006. Exposures to airborne particulate matter and adverse perinatal outcomes: a biologically plausible mechanistic framework for exploring potential effect modification by nutrition. Environ. Health Perspect 114, 1636–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly FJ, Fussell JC, 2012. Size, source and chemical composition as determinants of toxicity attributable to ambient particulate matter. Atmos. Environ 60, 504–526. 10.1016/j.atmosenv.2012.06.039. [DOI] [Google Scholar]
- Knust J, Ochs M, Gundersen HJ, Nyengaard JR, 2009. Stereological estimates of alveolar number and size and capillary length and surface area in mice lungs. Anat. Rec 292, 113–122. 10.1002/ar.20747. [DOI] [PubMed] [Google Scholar]
- Korten I, Ramsey K, Latzin P, 2016. Air pollution during pregnancy and lung development in the child. Paediatr. Respir. Rev 10.1016/j.prrv.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A, 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Paglia L, Listi A, Caruso S, Amodeo V, Passiglia F, Bazan V, Fanale D, 2017. Potential role of ANGPTL4 in the cross talk between metabolism and cancer through PPAR signaling pathway. PPAR Res. 2017, 8187235. 10.1155/2017/8187235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Leung AK, Tang MK, Cai DQ, Schneider C, Brancolini C, Chow PH, 2001. Functions of the growth arrest specific 1 gene in the development of the mouse embryo. Dev. Biol 234, 188–203. 10.1006/dbio.2001.0249. [DOI] [PubMed] [Google Scholar]
- LGEA Web Portal: https://research.cchmc.org/pbge/lunggens/mainportal.html, accessed on June 14, 2017.
- Li X, Huang S, Jiao A, Yang X, Yun J, Wang Y, Xue X, Chu Y, Liu F, Liu Y, Ren M, Chen X, Li N, Lu Y, Mao Z, Tian L, Xiang H, 2017. Association between ambient fine particulate matter and preterm birth or term low birth weight: an updated systematic review and meta-analysis. Environ. Pollut 227, 596–605. 10.1016/j.envpol.2017.03.055. [DOI] [PubMed] [Google Scholar]
- LungMAP database: http://www.lungmap.net/, accessed on June 14, 2017.
- Mauad T, Rivero DH, de Oliveira RC, Lichtenfels AJ, Guimaraes ET, de Andre PA, Kasahara DI, Bueno HM, Saldiva PH, 2008. Chronic exposure to ambient levels of urban particles affects mouse lung development. Am. J. Respir. Crit. Care Med 178, 721–728. 10.1164/rccm.200803-436OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoli-Rocha F, Oliveira VR, Barcellos BC, Moreira DK, Saldiva PH, Faffe DS, Zin WA, 2016. Time-dependency of mice lung recovery after a 4-week exposure to traffic or biomass air pollutants. Respir. Physiol. Neurobiol 230, 16–21. 10.1016/j.resp.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD, 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189. 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales E, Garcia-Esteban R, de la Cruz OA, Basterrechea M, Lertxundi A, de Dicastillo MD, Zabaleta C, Sunyer J, 2015. Intrauterine and early postnatal exposure to outdoor air pollution and lung function at preschool age. Thorax 70, 64–73. 10.1136/thoraxjnl-2014-205413. [DOI] [PubMed] [Google Scholar]
- National Research Council, 2011. Guide for the Care and Use of Laboratory Animals, eighth ed. The National Academies Press, Washington, DC: Available on https://www.nap.edu/catalog/12910/guide-for-the-care-and-use-of-laboratory-animals-eighth. [Google Scholar]
- Noel A, Xiao R, Perveen Z, Zaman H, Le Donne V, Penn A, 2017. Sex-specific lung functional changes in adult mice exposed only to second-hand smoke in utero. Respir. Res 18, 104 10.1186/s12931-017-0591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T, Fullgrabe A, Fuentes AM, Jupp S, Koskinen S, Mannion O, Huerta L, Megy K, Snow C, Williams E, Barzine M, Hastings E, Weisser H, Wright J, Jaiswal P, Huber W, Choudhary J, Parkinson HE, Brazma A, 2016. Expression Atlas update–an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 44, D746–D752. 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozarska A, Rodriguez-Castillo JA, Surate Solaligue DE, Ntokou A, Rath P, Mizikova I, Madurga A, Mayer K, Vadasz I, Herold S, Ahlbrecht K, Seeger W, Morty RE, 2017. Stereological monitoring of mouse lung alveolarization from the early postnatal period to adulthood. Am. J. Physiol. Lung Cell Mol. Physiol 312, L882–L895. 10.1152/ajplung.00492.2016. [DOI] [PubMed] [Google Scholar]
- Rockich BE, Hrycaj SM, Shih HP, Nagy MS, Ferguson MA, Kopp JL, Sander M, Wellik DM, Spence JR, 2013. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc. Natl. Acad. Sci. U. S. A 110, E4456–E4464. 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosti K, Goldman A, Kajander T, 2015. Solution structure and biophysical characterization of the multifaceted signalling effector protein growth arrest specific-1. BMC Biochem. 16, 8 10.1186/s12858-015-0037-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J, 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378. [DOI] [PubMed] [Google Scholar]
- Schittny JC, 2017. Development of the lung. Cell Tissue Res. 367, 427–444. 10.1007/s00441-016-2545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segovia J, Zarco N, 2014. Gas1 is a pleiotropic regulator of cellular functions: from embryonic development to molecular actions in cancer gene therapy. Mini Rev. Med. Chem 14, 1139–1147. [DOI] [PubMed] [Google Scholar]
- She ZY, Yang WX, 2015. SOX family transcription factors involved in diverse cellular events during development. Eur. J. Cell Biol 94, 547–563. 10.1016/j.ejcb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Siddika N, Balogun HA, Amegah AK, Jaakkola JJ, 2016. Prenatal ambient air pollution exposure and the risk of stillbirth: systematic review and meta-analysis of the empirical evidence. Occup. Environ. Med 73, 573–581. 10.1136/oemed-2015-103086. [DOI] [PubMed] [Google Scholar]
- Sioutas C, Koutrakis P, Burton RM, 1995. A technique to expose animals to concentrated fine ambient aerosols. Environ. Health Perspect 103, 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock E, Schmidt K, Hermanns-Borgmeyer I, Bosl MR, Wegner M, 2001. Idiopathic weight reduction in mice deficient in the high-mobility-group transcription factor Sox8. Mol. Cell Biol 21, 6951–6959. 10.1128/mcb.21.20.6951-6959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto SF, Melo JO, Marchesi GD, Lopes KL, Veras MM, Oliveira IB, Souza RM, de Castro I, Furukawa LNS, Saldiva PHN, Heimann JC, 2017. Exposure to fine particulate matter in the air alters placental structure and the renin-angiotensin system. PLoS One 12, e0183314 10.1371/journal.pone.0183314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton CM, Joo JH, Kim YS, Liao G, Panettieri RA Jr., Jetten AM, 2010. Induction of ANGPTL4 expression in human airway smooth muscle cells by PMA through activation of PKC and MAPK pathways. Exp. Cell Res 316, 507–516. 10.1016/j.yexcr.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng C, Wang Z, Yan B, 2016. Fine particle-induced birth defects: impacts of size, payload, and beyond. Birth Defects Res. Part C Embryo Today 108, 196–206. 10.1002/bdrc.21136. [DOI] [PubMed] [Google Scholar]
- Thevenot P, Saravia J, Giaimo J, Happel KI, Dugas TR, Cormier SA, 2013. Chronic alcohol induces M2 polarization enhancing pulmonary disease caused by exposure to particulate air pollution. Alcohol Clin. Exp. Res 37, 1910–1919. 10.1111/acer.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To T, Zhu J, Larsen K, Simatovic J, Feldman L, Ryckman K, Gershon A, Lougheed MD, Licskai C, Chen H, Villeneuve PJ, Crighton E, Su Y, Sadatsafavi M, Williams D, Carlsten C, 2016. Progression from Asthma to chronic obstructive pulmonary disease. Is air pollution a risk factor? Am. J. Respir. Crit. Care Med 194, 429–438. 10.1164/rccm.201510-1932OC. [DOI] [PubMed] [Google Scholar]
- Traboulsi H, Guerrina N, Iu M, Maysinger D, Ariya P, Baglole CJ, 2017. Inhaled pollutants: the molecular scene behind respiratory and systemic diseases associated with ultrafine particulate matter. Int. J. Mol. Sci 18 10.3390/ijms18020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unachukwu U, Trischler J, Goldklang M, Xiao R, D’Armiento J, 2017. Maternal smoke exposure decreases mesenchymal proliferation and modulates Rho-GTPase-dependent actin cytoskeletal signaling in fetal lungs. Faseb. J 31, 2340–2351. 10.1096/fj.201601063R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustiyan V, Zhang Y, Perl AK, Whitsett JA, Kalin TV, Kalinichenko VV, 2016. beta-catenin and Kras/Foxm1 signaling pathway are critical to restrict Sox9 in basal cells during pulmonary branching morphogenesis. Dev. Dynam 245, 590–604. 10.1002/dvdy.24393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veras MM, Damaceno-Rodrigues NR, Caldini EG, Maciel Ribeiro AA, Mayhew TM, Saldiva PH, Dolhnikoff M, 2008. Particulate urban air pollution affects the functional morphology of mouse placenta. Biol. Reprod 79, 578–584. 10.1095/biolreprod.108.069591. [DOI] [PubMed] [Google Scholar]
- Veras MM, de Oliveira Alves N, Fajersztajn L, Saldiva P, 2016. Before the first breath: prenatal exposures to air pollution and lung development. Cell Tissue Res. 10.1007/s00441-016-2509-4. [DOI] [PubMed] [Google Scholar]
- Vieira SE, 2015. The health burden of pollution: the impact of prenatal exposure to air pollutants. Int. J. Chronic Obstr. Pulm. Dis 10, 1111–1121. 10.2147/copd.s40214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weider M, Wegner M, 2017. SoxE factors: transcriptional regulators of neural differentiation and nervous system development. Semin. Cell Dev. Biol 63, 35–42. 10.1016/j.semcdb.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Winckelmans E, Vrijens K, Tsamou M, Janssen BG, Saenen ND, Roels HA, Kleinjans J, Lefebvre W, Vanpoucke C, de Kok TM, Nawrot TS, 2017. Newborn sex-specific transcriptome signatures and gestational exposure to fine particles: findings from the ENVIRONAGE birth cohort. Environ. Health 16 (52). 10.1186/s12940-017-0264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 2005. Air Quality Guidelines: Global Update 2005. WHO Regional Office for Europe, Copenhagen: Available on: http://www.who.int/. [Google Scholar]
- World Health Organization, 2016. Ambient Air Pollution: a Global Assessment of Exposure and Burden of Disease. World Health Organization, Geneva: Available on: http://www.who.int/. [Google Scholar]
- Wright RJ, Brunst KJ, 2013. Programming of respiratory health in childhood: influence of outdoor air pollution. Curr. Opin. Pediatr 25, 232e239 10.1097/MOP.0b013e32835e78cc. [DOI] [PubMed] [Google Scholar]
- Xie C, Han Y, Liu Y, Han L, Liu J, 2014. miRNA-124 down-regulates SOX8 expression and suppresses cell proliferation in non-small cell lung cancer. Int. J. Clin. Exp. Pathol 7, 7518–7526. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yoshizaki K, Fuziwara CS, Brito JM, Santos TM, Kimura ET, Correia AT, Amato-Lourenco LF, Vasconcellos P, Silva LF, Brentani MM, Mauad T, Saldiva PH, Macchione M, 2016. The effects of urban particulate matter on the nasal epithelium by gender: an experimental study in mice. Environ. Pollut 213, 359–369. 10.1016/j.envpol.2016.02.044. [DOI] [PubMed] [Google Scholar]
- Zhu P, Goh YY, Chin HF, Kersten S, Tan NS, 2012. Angiopoietin-like 4: a decade of research. Biosci. Rep 32, 211–219. 10.1042/bsr20110102. [DOI] [PubMed] [Google Scholar]
- Zhu X, Guo X, Wu S, Wei L, 2016. ANGPTL4 correlates with NSCLC progression and regulates epithelial-mesenchymal transition via ERK pathway. Lung 194, 637–646. 10.1007/s00408-016-9895-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.