Abstract
Gene-panel and whole-exome analyses are now standard methodologies for mutation detection in Mendelian disease. However, the diagnostic yield achieved is at best 50%, leaving the genetic basis for disease unsolved in many individuals. New approaches are thus needed to narrow the diagnostic gap. Whole-genome sequencing is one potential strategy, but it currently has variant-interpretation challenges, particularly for non-coding changes. In this study we focus on transcriptome analysis, specifically total RNA sequencing (RNA-seq), by using monogenetic neuromuscular disorders as proof of principle. We examined a cohort of 25 exome and/or panel “negative” cases and provided genetic resolution in 36% (9/25). Causative mutations were identified in coding and non-coding exons, as well as in intronic regions, and the mutational pathomechanisms included transcriptional repression, exon skipping, and intron inclusion. We address a key barrier of transcriptome-based diagnostics: the need for source material with disease-representative expression patterns. We establish that blood-based RNA-seq is not adequate for neuromuscular diagnostics, whereas myotubes generated by transdifferentiation from an individual’s fibroblasts accurately reflect the muscle transcriptome and faithfully reveal disease-causing mutations. Our work confirms that RNA-seq can greatly improve diagnostic yield in genetically unresolved cases of Mendelian disease, defines strengths and challenges of the technology, and demonstrates the suitability of cell models for RNA-based diagnostics. Our data set the stage for development of RNA-seq as a powerful clinical diagnostic tool that can be applied to the large population of individuals with undiagnosed, rare diseases and provide a framework for establishing minimally invasive strategies for doing so.
Keywords: RNA-seq, transcriptomics, diagnostics, Mendelian disease, transdifferentiation, myotubes, muscular dystrophy
Introduction
Identifying a causative genetic variant (mutation) in an individual with Mendelian disease is a critical event that ends the diagnostic odyssey, enabling accurate anticipatory care and prognostic guidance and providing the opportunity for individualized treatment. In the case of neuromuscular disorders, the diagnostic algorithm historically has included muscle biopsy, muscle imaging, and targeted genetic testing.1, 2, 3, 4 Recently, gene panels and whole-exome sequencing (WES) have dramatically improved the diagnostic yield, enabling genetic resolution in 35%–50% of cases (as compared to 10% resolved with chromosomal microarray), and have been associated with reducing health care costs and improving outcomes.2, 5, 6, 7, 8, 9 Despite these advantages, a substantial percentage of individuals remain undiagnosed and thus have not derived the many benefits that genetic diagnosis provides.10, 11, 12, 13
There are several potential explanations to account for the large fraction of unresolved cases. These include (1) the challenge of interpreting variants of unknown significance (VUSs),14, 15 (2) the fact that WES cannot delineate certain changes (e.g., structural rearrangements, copy-number variants (CNVs), and tandem-repeat expansions), (3) the uncertainty of evaluating variants in genes not yet linked to diseases, and (4) the fact that WES does not adequately capture intronic and regulatory regions.16 The exact number of mutations residing in the non-coding genome is debatable.17 Evidence suggests that as many as 9%18 to 30% of disease-causing variants impact RNA expression and/or processing and are found in non-coding regions.16
Whole-genome sequencing (WGS) enumerates variants in both coding and non-coding regions, and it can provide information related to CNVs.19 However, WGS presents important (and currently unresolved) challenges to data interpretation for the more than 3 million SNVs per sample.20, 21 Moreover, the validation of non-coding variants (as well as coding changes that impact RNA expression and splicing) usually requires additional functional studies at the transcriptional level.3, 22
The number of discovered pathogenic splicing mechanisms related to human disease is rapidly growing.23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 For example, aberrant splicing events in LMNA (MIM: 150330), DMD (MIM: 300377), SMN1 (MIM: 600354), TTN (MIM: 188840), and HNRPDL (MIM: 607137) have been linked to several neuromuscular disorders, and 185 deep intronic mutations have been identified across 77 genes associated with neuromuscular disease.24, 25, 26, 27 Transcriptome sequencing (i.e., RNA-seq) is ideal for detecting such changes because it allows for the detection of both coding and non-coding variants and provides transcript-level information for interpreting splice changes. Moreover, RNA-seq enables the comparison of expression levels in an individual sample with the levels in controls, and it can reveal expression outliers and imbalances in allele expression, changes that can then be used to prioritize DNA variants.35 In a cohort of rare, undiagnosed muscle disorders, RNA-seq analysis from muscle biopsies provided a diagnosis in 35% of cases.33
Of note, each cell type is known to have a unique gene-expression profile that includes expression of tissue-specific isoforms and alternative splicing. Hence, a key consideration for RNA-seq analysis is the requirement for disease-relevant source material. As an example, RNA sampled from blood does not adequately represent the transcriptome necessary for the analysis of many rare disorders.33, 36, 37 Diagnostic biopsies are most likely the best tissue source for RNA-seq but are not available in many cases.
In this study we focus on transcriptome analysis as a means of validating the utility of RNA-seq for mutation detection (Figure 1). In support of previous data,33 we demonstrate that RNA-seq from muscle biopsies can resolve a substantial portion of panel and exome “negative” cases (36% in our cohort). Because biopsies are invasive and not available in many cases, we additionally tested the suitability of derived cell lines for RNA-seq-based diagnostics. We demonstrate that, in contrast to blood, primary skin fibroblasts and myotubes created by transdifferentiating fibroblasts (t-myotubes) share significant aspects of the expression profile of skeletal muscle and can be used for accurate identification of mutations.34, 38, 39, 40, 41 Lastly, we developed PAGE (Panel Analysis of Gene Expression), a web-based tool that (1) enables the comparison of gene expression across multiple tissues and the identification of the optimal tissues to study and (2) allows for the exploration of variants and splicing changes identified in our analysis. In total, we describe the strengths and challenges of transcriptome analysis and establish a minimally invasive strategy for RNA-seq-based diagnostics.
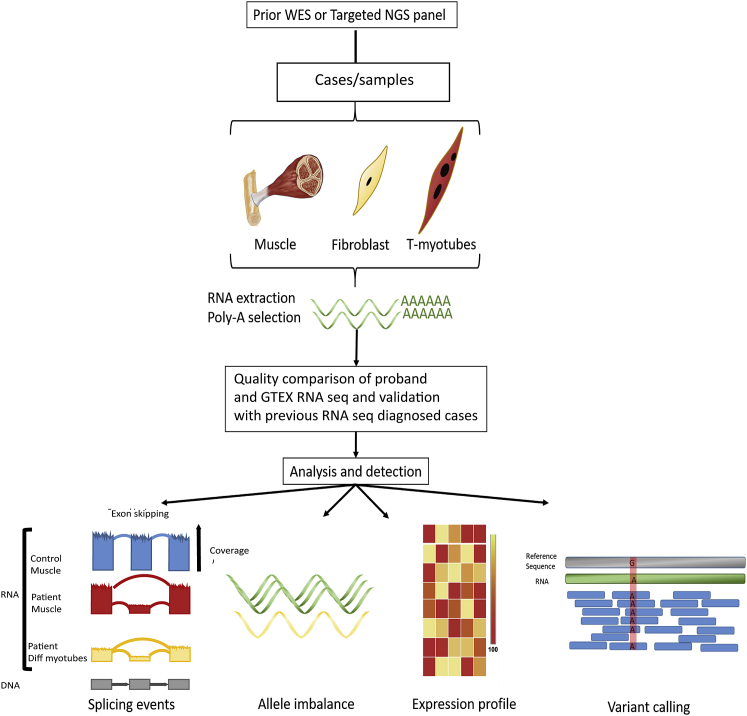
Figure 1.
Overview of the Diagnostic Pipeline
70 samples were processed through our pipeline. Total RNA was extracted from muscle biopsies, fibroblasts, and t-myotubes, was polyA selected, and then sequenced at a depth of 50–100 paired-end reads. Our RNA-seq diagnostic algorithm compares undiagnosed individuals with our in-house database and with control transcriptome data obtained from GTEx. We worked first from a panel of genes known to be mutated in neuromuscular disorders (n = 132), and we focused our analysis in parallel on (1) novel splicing events (far left), (2) imbalances in allelic expression (middle left), (3) statistically significant differences in expression (middle right), and (4) rare sequence variants of clinical relevance (far right). Using this strategy, we were able to solve 36% of cases that were “negative” according to the gene panel and/or exome sequencing (n = 25 total unknown cases).
Material and Methods
Study Design
We analyzed 70 samples from 29 families (Table 1 and Table S1). The genetic diagnosis was undetermined in 25 families; in nine of these it was still undetermined after testing by gene panel (“negative”; 162-gene panel), 12 lacked a diagnosis (“negative”) after WES, and four were “negative” for both. The four families wherein causative mutation(s) had previously been established (families 4, 7, 9, and 10) served as our positive controls. In 25 families, only samples from the proband were available, but in four families we had access to samples from additional family members. For reference controls, we used healthy skeletal muscle samples from the Common Fund (CF) Genotype-Tissue Expression Project (GTEx)36, 37 (50 controls for expression analysis and 184 for junction analysis); the controls were selected according to quality metrics established by Cummings et al.33 (Table S2). Additionally, we used data from 10 immortalized fibroblast controls from GTEx and 10 blood controls characterized as “fast death – natural causes” (Table S2).
Table 1.
Families Studied with RNA-Seq
| Family | Sample | Phenotype | Diagnosis | Splice Classification | Transcript levels | Allele Imbalance |
|---|---|---|---|---|---|---|
| 4a | f; m; myo | CM | RYR1 (GenBank: NM_001042723.1) c.2862G>A (p.Pro894Pro); RYR1 c.9859C>T (p. Arg3287>Cys) | exon extension | down 2× | no |
| 5 | f; m; myosingleton | CMD | GMPPB heterozygous (GenBank: NM_013334.3) c.94C>T (p.Pro32Ser); GMPPB heterozygous, 5′ UTR variant, g.349761246 G>A, new start codon | new start codon | down 4,1× | no |
| 6 | f; m; myosingleton | CMD | POMT2 (GenBank: NM_013382.5) c.1502A>C (p.Glu501Ala) homozygous | N/A | no | yes |
| 7a | f | CMD | LAMA2 (GenBank: NM_000426.3) c.9212–1G>A homozygous | exon extension | down 2,1× | no |
| 8 | msingleton | CM | TTN (GenBank: NM_001267550.2) c.74837_74840dupTTAG (p.Arg24947Ser∗2);TTN c.28114 +1G>A | exon extension | no | no |
| 9a | f; m; myo | DMD | DMD g.chrX: 32,366,860 A>C dbSNP: r.5326_5327ins51bp | novel exon | down 4,8× | X-linked disorder |
| 10a | m | CM | MTM1 (GenBanK: NM_000252.2) c.1262G>A (p.Arg421Gln) | N/A | no | X-linked disorder |
| 12 | f; m; myosingleton | CM | N/A | N/A | N/A | N/A |
| 13 | msingleton | CM | N/A | N/A | N/A | N/A |
| 14 | f; m; myo siblings | multiple pterygium syndrome | N/A | N/A | N/A | N/A |
| 15 | ftrio | MDC1A | LAMA2 (GenBank: NM_000426.3.) c.4860G>A; LAMA2 (GenBank: NM_000426.3) c.2584T>C (p.Cys862Arg) | exon skipping | down 2× | yes |
| 16 | f; myosingleton | restrictive cardiomyopathy | N/A | N/A | N/A | N/A |
| 17 | f; m; myosingleton | Pompe | GAA (NM_000152.5) c.−32–13T>G (intron 1); GAA c.1927G>A (p.Gly643Arg) | splice polypyrimidine tract variant | down 2× | yes |
| 18 | f; m; myosingleton | CMD | N/A | N/A | N/A | N/A |
| 21 | f; myosingleton | LGMD | N/A | N/A | N/A | N/A |
| 26 | f; m; myosingleton | DMD | UI for specific mutation | UI | down 7× | X-linked disorder |
| 27 | f; myosingleton | myopathy | N/A | N/A | N/A | N/A |
| 28 | f; m; myosingleton | CMD | N/A | N/A | N/A | N/A |
| 29 | fsingleton | myopathy | N/A | N/A | N/A | N/A |
| 30 | f; myosingleton | CMD | N/A | N/A | N/A | N/A |
| 31 | msingleton | arthrogryposis multiplex congenita | N/A | N/A | N/A | N/A |
| 32 | msingleton | CM | N/A | N/A | N/A | N/A |
| 33 | msingleton | distal myopathy | N/A | N/A | N/A | N/A |
| 34 | msingleton | DMD | DMD X: 33192302–33192451; UI for point mutation | novel exon | down 3,8× | X-linked disorder |
| 35 | msingleton | DMD | DMD (GenBank: NM_000109.3); c.93+1G>C exon 2 | exon extension | down 18,5x | X-linked disorder |
| 36 | msingleton | muscular dystrophy | N/A | N/A | N/A | N/A |
| 38 | msingleton | LGMD | DYSF (GenBank: NM_003494.3) c.4060_4062del (p.Ser1354del); UI for second mutation | novel exon | no | no |
| 39 | mproband & father | muscular dystrophy | N/A | N/A | N/A | N/A |
| 40 | f; m; myosiblings | recurrentrhabdomyolysis | N/A | N/A | N/A | N/A |
Abbreviations are as follows: Myo = myotubes; UI = under investigation; CM = congenital myopathy; CMD = congenital muscular dystrophy; LGMD = limb girdle muscle dystrophy; DMD = Duchenne muscular dystrophy; MDC1A = congenital muscular dystrophy 1A; m = male; and f = female. An extended version of the table is available in the Supplemental Data (Table S1).
Control samples.
Individuals were recruited through an REB (research ethics board)-approved protocol and were identified at the Hospital for Sick Children (HSC) and at different centers across Canada, the USA, and Europe. All participating individuals signed an REB-approved consent form. Except for four positive controls, all selected families lacked a conclusive genetic diagnosis. Muscle biopsies were obtained from the Department of Pathology and Laboratory Medicine (DPLM) at HSC or were shipped frozen from referring centers. Skin-derived fibroblasts were obtained at HSC, were shipped frozen, or were submitted in culture media. We used 15–25 mg of muscle per muscle biopsy. RNA was extracted from the muscle biopsies with the miRNeasy fibrous tissue mini kit (QIAGEN, Cat No./ID: 74704) according to the manufacturer’s instructions, and a minimum of 200 ng of RNA for library preparation. The cutoffs for the RNA integrity number (RIN) and the DV200 value were 8 and ≥92%, respectively.
Generation of T-Myotubes via the Transdifferentiation of Fibroblasts Derived from Affected Individuals
Fibroblast cultures were obtained from skin biopsies of selected individuals according to previously described protocols.41, 42 The fibroblasts were cultured in DMEM with 10% fetal bovine serum (FBS) without the addition of antibiotics. All of the fibroblasts we used for RNA extraction were 70%–80% confluent at the time of extraction. We employed early-passage (<10) fibroblast lines for transdifferentiation. They were seeded at 50% confluence in 10 cm dishes coated with 20% matrigel (Corning Matrigel Matrix). At 70% confluence, the cells were infected with ad-MyoD (Vector Biolab cat. no.1492), with a MOI of 100, that was in infection medium (skeletal-muscle cell-growth medium; Promocel cat. no. 23060). Infected dishes were swirled every 20 min and incubated at 37°C for 3 h. Then, the medium infected with the virus was replaced with fresh skeletal-muscle cell-growth medium and left for 24 h, after which the medium was changed to differentiation medium (DMEM plus 2% horse serum and 0.1% insulin). Differentiation was carried out for 21 days, and 50% of the medium was replaced every other day. On the 5th day of differentiation, the dishes were coated again with 20% matrigel according to the manufacturer’s recommendations.
To evaluate the differentiation process, we extracted RNA on days 1, 3, 5, 14, and 21. qRT-PCR quantification was carried out for selected transcripts (RYR1 [MIM:180901], DMD, DES [MIM: 125660], MYOD1 [MIM: 159970], MYH3 [MIM: 160720], and MYOG [MIM: 159980]) known to be present at different stages of t-myotube maturation (Figure S1). The results were compared against qRT-PCR carried out on RNA from control human muscle. To corroborate the expression of proteins unique to skeletal muscle, we performed immunofluorescence (IF) for dystrophin, alpha actinin, and ryanodine receptor type I (antibodies were mouse dystrophin, MANDRA 1[7A10 DSHB], mouse musculoskeletal α-actinic, sarcomeric [A7811 Sigma], and rabbit RyR1 [HPA056416 Sigma]; secondary antibodies were Alexa Fluor 555 goat anti-rabbit and Alexa Fluor 555 goat anti-mouse [Life Technology]).
Generating Sequencing Data
Sequencing was done at the Centre for Applied Genomics (TCAG) at the HSC and involved poly-A selection of mRNA (Illumina TruSeq) or total RNA extraction followed by ribosomal RNA depletion (in family 4 only). Paired-end 126+126 bp sequencing was performed with Illumina HiSeq 2000 instruments at a sequencing depth of 50–100 million paired reads per sample. Raw sequencing reads for GTEx control samples were downloaded from dbGAP (accession no. phs000424.v6.p1) with the Sequence Read Archive (SRA) toolkit.43, 44
Gene Panels
We created eight virtual neuromuscular-disease-associated gene panels containing 132 genes in total (Table S3); the panels were created on the basis of the 2017 gene table of monogenic neuromuscular disorders.45 We focused our initial analysis on the genes in these panels and extended the analysis, when necessary, to all OMIM genes or all protein-coding genes according to ENSEMBL annotation.46, 47 In family 40 we also used a mitochondrial Mitocarta gene panel (1,158 genes).48
Read Alignment, Quality Control, and Read Quantitation
Alignment, variant calling, and quality control were carried out via the RNA-seq workflow from the bcbio-nextgen bioinformatics framework (version 1.1.0). In brief, raw reads were aligned to the GRCh37 (hg19) version of the human reference genome with the splice-aware aligner STAR in two-pass mode (the first pass discovers new splice junctions and inserts them into the junction database, and the second pass calls junctions and calculates their counts).49 Multiqc was then used for quality control and assurance analysis of the resulting bam file by comparing to metrics gathered from bcbio-nextgen, samtools, and fastqc.50, 51 Finally, we quantitated reads by assigning them to genes (features) annotated in Ensembl (release 75) and counting them with the featureCounts tool.47, 52
Expression Analysis
Gene-level and exon-level expression values were calculated with the edgeR package (R version 3.5.0, edgeR version 3.22.3, bioconductor version [Biobase package] 2.40.0).53 Heatmaps of the expression of the genes in the gene panels were plotted with the pheatmap R package (version 1.0.10) (Figure S2). To prioritize splicing events and genomic variants, we selected genes expressed at ≥1 RPKM and identified expression outliers. We carried out outlier detection at the gene level to detect expression outliers. We calculated Z scores for the RPKM expression values of every gene in every skeletal-muscle sample and compared these to the mean and standard deviation of the 50 GTEx controls, as well as the mean and standard deviation of the 25 skeletal-muscle samples from the cohort. We filtered genes with an average expression in GTEx of ≥0.1 RPKM and |Z score| ≥ 1.5 and reported them as potential outliers for the set of 132 neuromuscular genes, 3,739 OMIM genes, and 1,158 Mitocarta genes in family 40. We focused on downregulated genes and ordered them according to n-fold decrease in expression. We confirmed the outlier status with OUTRIDER (version 0.99.31) and reported genes whose expression had changed by 2-fold versus their mean expression in the control GTEx cohort. Detailed expression information is provided in Tables S4–S15.
Variant Calling and Annotation
The coverage across reference nucleotides in RNA-seq is highly variable because of the differential expression of genes, isoforms, and alleles. These factors, plus post-transcriptional modifications, result in lower variant-calling precision when RNA-seq data are used. On the basis of previous data, RNA-seq-specific filters enable the calling of ≥95% (compared to >99% in WES or WGS) pathogenic genomic variants.54
We measured the precision of small-variant calling of the Genome Analysis Toolkit (GATK) HaplotypeCaller (version 4.0.1.2) using the RNA-seq data of the GM12878 cell line, sequenced by ENCODE (SRA accession number in SRR307898), and the National Institute of Standards and Technology (NIST) Genome-in-a-Bottle variant calls as a true positive set.55, 56, 57 We found that the GATK HaplotypeCaller 4.0.1.2 had lower precision than the GATK HaplotypeCaller 3.6: 85% versus 98% for SNVs and 52% versus 70% for indels, and thus we chose the latest version of the GATK HaplotypeCaller branch 3.6×.
Variants were called with the GATK HaplotypeCaller (version 3.8) according to the GATK best practices for variant calling in RNA-seq as implemented in the bcbio-nextgen RNA-seq pipeline. Namely, we used the GATK SplitNCigarReads command to prepare bam files, and we set the RNA-seq specific filters (–cluster-window-size 35–cluster-size 3–filter-expression ‘FS > 30.0’–filter-name FS–filter-expression ‘QD < 2.0’–filter-name QD). We filtered out sites of RNA editing according to the RADAR database (version 2-20180202).58 Variants were annotated with Ensembl VEP (version 93.3), vcfanno (version 0.3.0), and CRE with data sources from GEMINI (version 0.20.1), bcbio-nextgen (version 1.1.0), gnomAD (version 2.0.1), dbNSFP (version 3.5a), and the Human Gene Mutation Database (HGMD) (version 20180411_v2018.1).59, 60, 61, 62, 63 Rare variants (allele frequency [AF] < 0.01 in gnomAD WES and WGS) that had significant impact (MED or HIGH according to GEMINI terms) and that were covered by at least 5 reads were selected. We combined the variant reports within a family to prioritize variants according to the inheritance pattern of the particular disease and to check for the consistency of variant calling across the tissues.
Identification and Annotation of Splice Variants
To compare methods of genomic variant calling and to choose the best one, we used standard benchmarks (Genome in a Bottle, Illumina Platinum Genomes) to assess the specificity and sensitivity of different tools.57, 64 The evidence of splicing abnormalities that have been confirmed as true positives is growing; however, this evidence is not sufficient for the creation of a benchmark. Most splicing analysis methods are compared through the use of synthetic tests.27 Thus, we chose the method on the basis of its ability to call pathogenic splicing changes in several published cases.
Several tools that predict the pathogenicity of splice-affecting variants have been developed.65, 66, 67 In our splicing analysis, we chose the Mendelian RNA-seq method described in Cummings et al.33 Unlike other splicing analysis methods, which were developed by expanding on the idea of the differential expression analysis of genes, Mendelian RNA-seq treats splice junctions similarly to how they are treated in genomic variant analyses: it calls a junction in a large cohort of skeletal muscle controls from GTEx and then annotates it with the frequency.68 Junctions with a frequency of zero are considered as candidates for pathogenic events, and then additional evidence (genomic variants, phenotype, expression status, functional studies, and Sanger or RT-PCR validation) is explored to assess the pathogenicity. This approach significantly simplifies the analysis, reduces computing time, and allows the identification of a limited set of candidate pathogenic events for further investigation.
We made several improvements to Mendelian RNA-seq. First, we built a reusable database of junctions generated from the 184 controls, and we utilized this database for comparison, thus speeding up our analysis. Second, we opined that the requirement for a novel junction to be absent from all controls might lead to false negatives because a random read originating from the transcriptional noise in a few of the controls might mask the well-covered novel junction in a sample. We therefore reported junctions for further analysis if they were present in ≤5/184 (2.7%) controls. This allowed us to detect pathogenic splice events in families 38 and 34, where aberrant junctions were present in 1 or 2 GTEx samples with very low coverage (1 or 2 reads).
We annotated rare (absent or appearing ≤5 times in GTEx control samples) splicing junctions with the following statuses in GENCODE: (1) both donor and acceptor sites are present in GENCODE, i.e., the junction is not new; (2) only the donor site is present in GENCODE, and the acceptor site (junction end) is absent, i.e., there is a new acceptor site; (3) only the acceptor site is present in GENCODE, and the donor site (junction start) is absent, i.e., there is a new donor site; (4) both the donor and acceptor sites of the junction are present in GENCODE annotation, but separately (i.e., not as one junction), i.e., there is a new incidence of exon skipping; (5) neither the donor or acceptor sites are present in the GENCODE annotation. We report rare junctions having at least one site absent in GENCODE (Table S16).
Our system of filters was designed to remove splicing junctions that fit the following criteria: (1) they are present in ≥5 of the 184 GTEx skeletal muscle controls; (2) they are annotated in GENCODE with both sites as a single junction; (3) they are covered by <5 reads; (4) they have a normalized coverage score ≤0.05 (normalization accounts for coverage in other junctions of the same gene and removes sequencing noise; see the definition of the normalization score in Figure S5B from Cummings et al.14); and (5) they are shared between >2 samples in a cohort. Finally, we annotated novel splicing events with information about gene expression (e.g., the expression outlier status and whether it is up- or down-regulated) for better filtering. For splicing junctions in genes that are not part of the gene panels, we added two additional filters to remove (1) junctions with read coverage <30, (2) junctions with normalized coverage <0.5, and (3) junctions with a frequency in the cohort of <7 (Table S17).
Coverage Analysis
The coverage of a gene in RNA-seq data depends on both the level of expression of the gene, as well as the depth of sequencing. The read coverage of a gene determines the ability to call variants and splice junctions in a gene. In our work we set the minimum coverage threshold at five reads.
We measured average per-nucleotide coverage in our muscular, fibroblast, and t-myotubes samples for the genes in our gene panels. We split the genes into bins according to average coverage: <10×, 10×–100×, 100×–1,000×, 1,000×–10,000×, and >10,000× (Figure S4). Genes in the lowest coverage bin were deemed not suitable for variant detection by RNA-seq.
Allelic Imbalance Analysis
We filtered variants in heterozygous sites with a coverage depth ≥20 by using bcftools, and we calculated minor-allele read-count ratios by using information from the AD (allelic depth) format field of a vcf file produced by GATK HaplotypeCaller.51 Then, we calculated a median of these ratios for genes having ≥5 variants; we used this value to characterize the allelic imbalance of a gene by computing a Z score and comparing sample data with data from 50 GTEx controls. We prioritized genes according to absolute Z scores (Table S22). This approach is a modification of allele imbalance analysis used in Cummings et al.33.
PAGE—A Web Interface for Exploring the Data
We developed PAGE, a web interface for exploring the data and the results generated in this project. The PAGE web application is built with React and Redux JavaScript libraries for front-end visualization and a Python FLASK framework for the back end API. PAGE uses a SQLITE3 database to store gene information such as the gene name, Ensembl ID, and HGNC synonyms. This database also stores gene-panel information such as the panel name and the list of genes included in each panel. PAGE uses a MongoDB database to store gene expression data such as RPKM and RAW expression counts for every exon in the gene across various tissues in GTEx and our samples. Finally, the webpage interfaces with a JBrowse instance—a genome browser for visualizing expression and splice-junction data in our sample cohort, along with the aggregated data from 184 GTEX controls. User inputs to the PAGE application are handled by the React library, which communicates with the Python FLASK API end points to fetch query-specific data from the SQLITE3 and MongoDB databases in JSON format; the data are then parsed and displayed on the webpage.
Results
In this study, we applied our transcriptome analysis pipeline to 65 samples from individuals with monogenetic neuromuscular disorders (Figure 1); we performed RNA-seq on 26 skeletal-muscle biopsies, 22 skin-fibroblast cultures, 17 transdifferentiated-myotube cultures (t-myotubes; Table 1 and Table S1), and 5 fibroblast control samples, for a total of 70 samples.
Variability of Expression in Muscle Biopsies
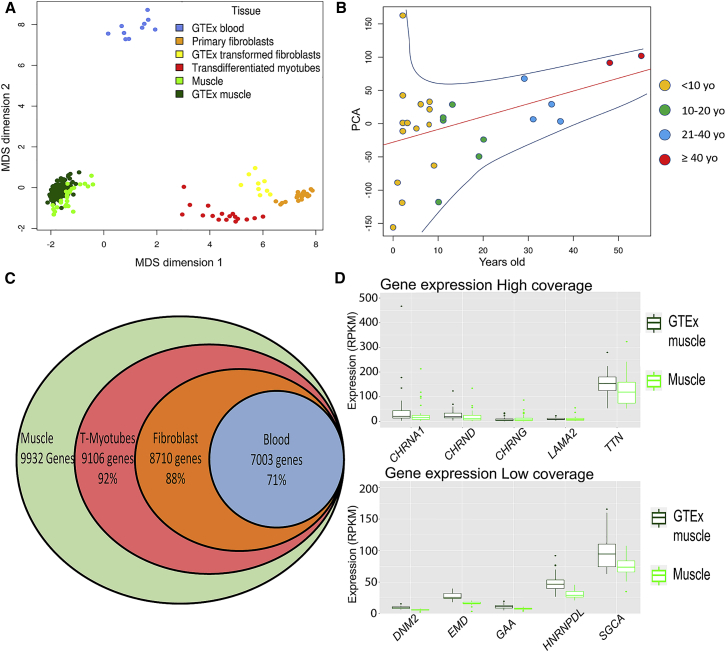
We assessed the quality and variability of our muscle samples by comparing them with the transcriptomes from GTEx skeletal muscle controls. On the basis of the multidimensional scaling plot of expression profiles (Figure 2A), our samples clustered with GTEx skeletal muscle, indicating that our cohort had a transcriptome profile similar to that of GTEx controls. However, more than half of our samples came from pediatric individuals, 60% of them ≤10 years old. When our cohort is stratified by age, we see that the expression profile of affected pediatric individuals differs from that of affected adults, and we found that the transcriptome tends to adopt an “adult profile” at ∼10 years of age (Figure 2B).
Figure 2.
Sample Distribution and Gene-Expression Profile
(A) A multi-dimensional scaling (MDS) plot of our cohort of 70 samples (primary fibroblasts, t-myotubes, and muscle) compared with tissue-matched sets from GTEx (blood, transformed fibroblasts, and muscle). There is significant overlap between the muscle samples from our cohort and GTEx. Samples from blood, muscle, and fibroblasts formed distinct clusters, but transdifferentiated myotubes cluster as a group between the fibroblasts and muscle samples.
(B) A principal-component analysis (PCA) of only our muscle samples shows a clear clustering of samples on the basis of age (x axis; 1st PCA). There is increased variability in expression in the younger samples; this variability diminishes with increased age of the samples.
(C) We identified 9,932 genes expressed at >1 RPKM. Of these genes, ∼92% were expressed at ≥1 RPKM in t-myotubes, 88% in fibroblasts, and 71% in blood.
(D) Comparison of expression between our muscle samples and GTEx muscle for the five highest- and lowest-coverage genes. There is no significant difference between our samples and those from GTEx.
In our cohort of muscle biopsies, 9,932 protein-coding genes were expressed, on average, at >1 RPKM (Figure 2C). Out of 132 genes previously associated with neuromuscular disease (Figure S3 and Table S5), 15 genes (ALG14 [MIM: 612866], CACNA1A [MIM: 601011], CCDC78 [MIM: 614666], CHAT [MIM: 118490], CHKB [MIM: 612395], CHRNE [MIM: 100725], CLN3 [MIM: 607042], CNTN1 [MIM: 600016], DUX4 [MIM: 606009], GMPPB [MIM: 615320], ISPD [MIM: 614361], KCNA1 [MIM: 176260], KCNE3 [MIM: 176263], POMK [MIM: 615247], and SYT2 [MIM: 600104]) were expressed at <1 RPKM in our cohort and in GTEx skeletal-muscle controls. Among these 15 genes with low expression, seven (CACNA1A, CCDC78, CHAT, DUX4, KCNA1, KCNE3, and SYT2) had an average per-base coverage below 10×. Given the overall high sequencing depths in our experiment (50–100 M paired reads), we predict that detecting variants in these seven genes with RNA-seq will be challenging. An additional three genes (AGRN [MIM: 103320], MEGF10 [MIM: 612453], and MSTN [MIM: 601788]) were expressed below 1 RPKM in GTEx but not in our samples. We thus estimate that 95% of genes in a comprehensive neuromuscular gene panel can be studied with an RNA-seq analysis of skeletal-muscle biopsies.
Each gene in our 132-gene panel presented some degree of expression variability. 33.8% of the genes in our muscle biopsies have a coefficient of variation (CofV; defined as the ratio of the standard deviation to the mean) >50% (Table S11), a value similar to the CofV of these genes in GTEx. To examine this further, we chose five genes (CHRNA1 [MIM: 100690], CHRNG [MIM: 100730], CHRND [MIM: 100720], TTN, and LAMA2 [MIM: 156225]) with high variability and five genes (GAA [MIM: 606800], HNRNPDL [MIM: 607137], DNM2 [MIM: 602378], EMD [MIM: 300384], and SGCA [MIM :600119]) with low variability, and we showed that their CofV is comparable between our samples and GTEx controls (Figure 2D).
Splice Changes
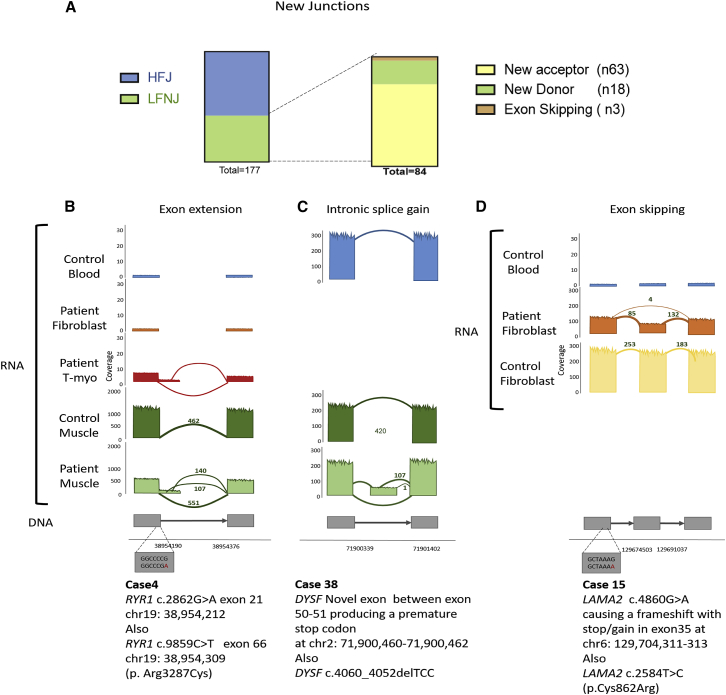
Abnormalities in RNA processing are a potential mutational pathomechanism that can be captured by RNA-seq. We hypothesized that these should be uncovered by examining novel junctions. In our 25 polyA purified muscle-biopsy transcriptomes, we identified a total of 166 novel junctions that were present in ≤10 GTEx controls (Table S16); 84 of them were present at a low frequency (≤2 samples) within our cohort (we designated them as low-frequency novel junctions [LFNJs]). The median number of LFNJs per muscle sample was 5 (min 1, max 20). The LFNJs we encountered (including four pseudo-exons) were as follows: new acceptors (n = 63), new donors (n = 18), and exon-skipping events (n = 3). We discovered no rare junctions where neither end site was annotated in GENCODE (Figure 3A). Using more stringent quality filters, but not restricting analysis to our gene panels, we discovered 50 novel junctions: 23 new donor sites, 20 new acceptor sites, and seven exon-skipping events (Table S16). LFNJs were considered disease-causing changes in eight of our affected families. Examples are described in Figure 3 and explained below.
Figure 3.
Analysis of Low-Frequency Novel Junctions
(A) 177 novel junctions were detected in the transcriptomes of our muscle samples, and we divided them into high-frequency junctions (HFJs) and low-frequency novel junctions (LFNJs). The LFNJs were subdivided into new acceptor (n = 63), new donor (n = 18), and exon-skipping (n = 3) events. Pseudo exons (n = 4) are counted within new acceptor or donor events.
(B) Example of a novel donor site detected in t-myotubes and muscle biopsy (family 4). The canonical donor splice site of exon 21 of RYR1 was reduced (551 reads), and two alternative, cryptic donor splice sites were activated. The first (chr19: 38,954,212) (new donor with 107 reads) extends exon 21 by 44 bp, causing a +1 frameshift and stop-gain at chr19: 38,955,338341. The second (chr19: 38,954,309) (new donor with 140 reads), extends exon 21 by 141 bp, causing a 1 nt frameshift with stop-gain at chr19: 38,954,395397. Overall, RYR1 expression is decreased 2.1-fold in affected cells compared to in controls.
(C) Example of a pseudo exon (family 38). A novel exon was found in the muscle between exons 50 and 51 in DYSF (supported by 107 reads), creating a premature stop codon at chr2: 71,900,460–71,900,462.
(D) Example of exon skipping (family 15). An exon-skipping event was detected in fibroblasts in LAMA2 (supported by four reads), causing a frameshift with stop-gain in exon 35 at chr6: 129,704,311–313. Overall, LAMA2 expression was decreased (see also Figure 4C).
We identified four cases of exonic extensions that were pathogenic (Table 1). One example is found in individual 4, an individual with centronuclear myopathy, where we found an exonic extension caused by a synonymous point mutation in the last nucleotide of exon 21 of RYR1. The canonical donor splice site of exon 21 was suppressed, and two alternative cryptic donor splice sites were activated (Figure 3B). The first (GenBank: NM_001042723.1; chr19: 38,954,212) extends exon 21 by 44 bp, causing a +1 frameshift and the occurrence of a premature stop codon TGA at chr19: 38,955,338–341. The second (GenBank: NM_001042723.1; chr19: 38,954,309) extends exon 21 by 141 bp and causes a −1 frameshift in exon 22 and a resulting premature stop codon at chr19: 38,954,395–397. Correspondingly, there is a 2.1-fold drop in RYR1 expression compared to that in controls.
We identified three cases of novel pathogenic pseudo-exons caused by deep intronic mutations. In family 38, the presence of new start and stop junctions (Figure 3C) helped us to identify an intronic splice gain (resulting in a new exon) in DYSF [MIM: 603009]. In proband 34, who presented a clear phenotype for Duchenne muscular dystrophy but for whom no diagnosis was reached after multiplex ligation-dependent probe amplification (MLPA) and sequencing for DMD, we found an intronic inclusion (pseudo-exon) between exons 1 and 2. In sample 9 (a positive control), we previously reported a reduced expression of DMD (10× lower) and a deep intronic mutation that created a pseudo-exon with a premature stop codon; we now confirm the discovery of this new junction in t-myotubes as well (see below).32
We also uncovered one exon-skipping event (in family 15) caused by a synonymous variant (GenBank: NM_000426.3: c.4860G>A) in the splice region of exon 33 of LAMA2 (Figure 3D).
Genomic Variants Identified by RNA-Seq
Identifying coding variants from RNA-seq data is complicated by multiple factors, including uneven coverage stemming from the variable expression of genes, exons, isoforms, and alleles.26 In our cohort, we observed that, on average, 80% of the nucleotides found in the 132 genes in the neuromuscular panel are covered by at least 20 reads (Table S19). In family 14, we had both skeletal-muscle RNA-seq and blood WGS data for two samples, enabling us to compare the variants identified from each of the datasets (Table S20). Excluding KCNJ12 [MIM: 602323] variants (excluded because of misassembly in the reference genome), RNA-seq correctly identified 90% (259/287 and 272/305 variants) of variants identified by WGS. The remaining 10% of variants not detected by RNA-seq came from three sources: sites of good coverage where a variant was not called by a variant-calling program (GATK false negatives), sites of low or zero coverage (e.g., exons that are not expressed in muscle but are included in the canonical isoform), and variants filtered out by an RNA-seq-specific variant filter (for example, when many variants are detected in a small window). 2% of variants detected by RNA-seq across the neuromuscular gene panel of 132 genes were not found in the WGS. These might represent somatic variants and/or false discoveries.
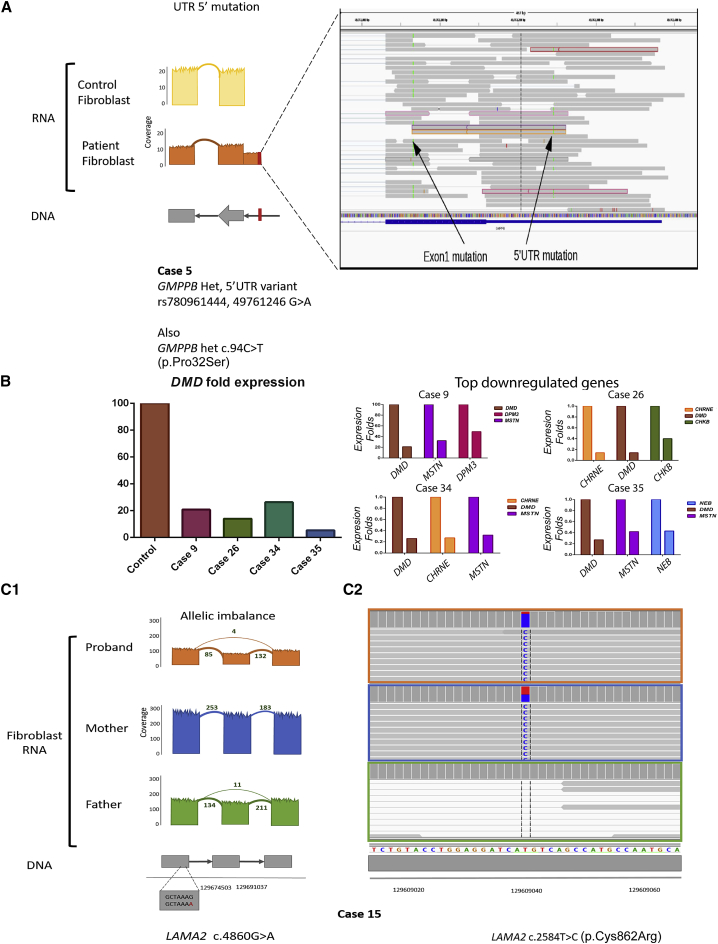
RNA-seq identified pathogenic variants in nine of our cases (Table 1) and additionally provided resolution on a rare variant found in a 5′ untranslated region (UTR). Family 5 presented with a phenotype compatible with congenital muscular dystrophy, and WES identified a heterozygous missense variant in GMPPB (GenBank: NM_013334.3) c.94C>T [p.Pro32Ser]). RNA-seq analysis additionally uncovered a heterozygous variant in the 5′ UTR of GMPPB (g.349761246 G>A) in trans with c.94C>T (Figure 4A).69 This SNV is predicted to create an alternative ATG start codon that adds 29 new amino acids to the protein. Expression of GMPPB in this case was decreased by 4× (compared to in our cohort) and by 57× (compared to in GTEx skeletal muscle) (Table S6).
Figure 4.
Analysis of Variants, Expression Profiles, and Allele-Specific Expression
(A) Detection of a 5′ UTR mutation with RNA-seq. WES for family 5 reported a heterozygous missense variant in GMPPB c.94C>T (p.Pro32Ser). We identified this variant in RNA from the fibroblasts (left arrow; labeled “Exon1 mutation”), along with a heterozygous 5′ UTR variant, g.349761246G>A (right arrow; labeled “5′UTR mutation”), that results in a new start codon. This variant was seen only in the transcriptome data and not by WES. It produces an in-frame insertion of 116 bp, potentially adding 29 new amino acids to the protein. Of note, both variants were absent from control fibroblasts.
(B) The left panel depicts the n-fold difference in DMD RNA levels between GTEx control muscle and 4 cases of Duchenne muscular dystrophy (in family 9 = intronic splice gain, family 26 = mutation still elusive, family 34 = intronic splice gain, and family 35 = disruption of exon 1 ATG). The right panel shows the top 3 most downregulated genes (from a panel of 132 neuromuscular genes) for each of these cases.
(C) Transcript expression ([C1], in RPKM) of LAMA2 from fibroblasts (in orange) from an individual with congenital muscular dystrophy and from the parents (father in green and mother in blue) (family 15). The proband had a greater-than-2-fold reduction in transcript expression as compared to his mother. (C2) He also had a maternally inherited pathogenic missense variant in LAMA2 (c.2548T>G [p.(Cys862Arg)]). This variant demonstrated mono-allelic expression, and it was found in 85% of the reads in the proband, 50% of the reads in the mother, and not found in the father.
Expression Outliers and Allele Imbalance
We next used comparative gene expression and allelic imbalance as ways to prioritize genes and variants for subsequent analysis. After multiple testing correction with the Bonferroni method, the mean number of abnormally expressed genes in our muscle gene panel (|Z score| > 1.5) per muscle sample was 17 (Table S10). In seven of the 10 previously undiagnosed families, we found that the candidate gene was within the group of expression outliers. In six of these seven cases, the causative gene was one of the top 5 outlier genes (it was the 31st outlier gene in the other). The following cases, where the causative gene was downregulated by more than 2-fold, are examples of the utility of expression analysis: family 4, RYR1 (Figure 3B); family 5, GMPPB (Figure 4A); and families 9, 26, 34, and 35, DMD (Figure 4B).
An examination of the balance of allele expression, particularly in combination with expression level analysis, might also reveal causative mutations. Allele imbalance was observed in instances of mutations in RYR1 (abnormal junctions, Figure 3B) and GMPPB (5′ UTR variant, Figure 4A), but not in the case of the DYSF pseudo exon, which occurs between exons 50 and 51 out of 55 exons total and thus probably evades the nonsense mediated decay pathway (NMD), Figure 3B. Allele imbalance also directed us to the cause of disease in three additional cases (described below).
Family 15 included the proband, diagnosed with congenital muscular dystrophy, and samples from both parents. We examined RNA from fibroblasts; in the proband, we observed a missense variant (GenBank: NM_000426.3: c.2584T>C [p.Cys862Arg]) in exon 19 of LAMA2 in 85% of reads. In the father, we found an exon 33 skipping event, resulting from c.4860G>A (GenBank: NM_000426.3), in LAMA2 (Figure 3D). This exon 33 skipping was also supported at very low coverage (four reads) in the proband. In the mother we found the p.Cys862Arg variant in 50% of reads (Figure 4C). Taken together, we concluded that the individual had compound-heterozygous mutations in LAMA2 that resulted in almost complete loss of expression from the paternal (exon-skipping) allele (Table S21).
A similar scenario was present in an individual 17, who had a phenotype consistent with Pompe disease. RNA-seq identified an exonic missense variant in GAA (GenBank: NM_000152.5) c.1927G>A (p.Gly643Arg) in 74% of reads. It also uncovered a second variant, (GenBank: NM_000152.5) c.32–13T>G (intron 1), associated with intronic inclusion and the introduction of a premature stop codon. This variant was represented in only 16% of reads, consistent with allele-specific reduction, which together with the missense mutation in trans revealed compound-heterozygous GAA mutations as the cause of the disease in this case.
Lastly, we clarified with RNA-seq one case of an individual with congenital muscular dystrophy and suspected dystroglycanopathy (family 6). RNA-seq identified a homozygous POMT2 mutation, (MIM: 607439) (GenBank: NM_013382.5.) c.1502A>C (p.Glu501Ala), that resulted in no reduction of expression of the POMT2 transcript. This homozygous variant, which originally was poorly delineated by exome sequencing, was subsequently identified in the individual’s DNA by exome reanalysis. Maternal DNA was heterozygous for the same variant. Further analysis of the transcriptome showed that adjacent genes also had mono-allelic expression. A chromosomal microarray confirmed a large region, approximately 15 Mb in length, of homozygosity on chromosome 14 (chr14: 75,699,438–90,713,574) but no other regions of homozygosity throughout the remainder of the genome, thus supporting an argument in favor of uniparental isodisomy.70
Blood Is an Inadequate Source Material for the Analysis of the Muscle Transcriptome
A major limitation of RNA-seq is the need to have source material with a disease-relevant transcriptome. In most cases this requires biopsied material. In addition, as with WES, trio analysis greatly improves data interpretation, as exemplified in family 15 earlier. For neuromuscular diseases, biopsies are invasive and limited in amount and/or availability, and parental samples are rarely (if ever) available. Thus, we looked for alternate, minimally invasive sources for study.
First, we compared the expression profiles of blood and skeletal muscle from the GTEx database. Of the 9,932 genes expressed in muscle at >1 RPKM, only 71% were expressed in blood at >1 RPKM (Figure 2C and Table S4). Moreover, the genes most commonly associated with muscle disease have low expression in blood. 89% of 132 genes from the neuromuscular panel are expressed at >1 RPKM in skeletal muscle, but only 53% of the 132 genes reach this level in blood (Table S5). Thus, blood is an inadequate source for transcriptome analysis for most neuromuscular diseases.
Proband-Derived Fibroblasts Recapitulate Aspects of the Muscle Transcriptome
We next examined skin fibroblasts because these can be obtained via minimally invasive means and can be reprogrammed into other cell types.41, 71, 72, 73 We obtained transcriptomes from fibroblasts from 26 individuals, and we additionally considered the GTEx control data for immortalized fibroblasts. Our skin fibroblasts (passage 1–4) clustered together in a similar way as immortalized fibroblasts, though their expression profiles overlapped incompletely (Figure 2A). When we compared our cohort of fibroblasts to skeletal muscle, we found an increased coverage of known neuromuscular genes as compared to blood, and there was particularly high coverage of genes associated with congenital muscular dystrophy (Figure 2D). In total, 88% of all genes, including 61% of genes from the neuromuscular gene panel (Table S5), expressed in muscle were also expressed in fibroblasts at >1 RPKM (Figure 2C and Table S4). In terms of specific disease-gene coverage, the transcripts that were poorly expressed in fibroblasts were components of the excitation-contraction coupling apparatus (e.g., RYR1, CACNA1S [MIM: 114208]), and some components of the sarcomere (e.g., NEB [MIM: 161650] and TTN) (Table S5).
Transcriptomes from Transdifferentiated Myotubes Match Those from Skeletal Muscle
Given that transcriptomes from blood and fibroblasts did not adequately represent skeletal muscle, we lastly turned to an ex vivo model of muscle (skeletal myotubes derived by myoD overexpression in fibroblasts; see Material and Methods). We first verified transdifferentiation and measured myotube maturation at different time points by monitoring several markers of myotube differentiation (using qRT-PCR and immunofluorescence) and by evaluating cell morphology (Figure S1). Between 50% and 85% of fibroblasts were converted to t-myotubes with our technique. Resulting t-myotubes continually progressed in differentiation status throughout the time course; 21 days after differentiation marked the point at which cultures were most mature and still fully viable. Of note, despite the variable conversion and differentiation rates, for simplicity we did not remove fibroblasts from the t-myotubes for subsequent RNA sequencing.
In t-myotubes, 11,185 protein-coding genes were expressed at >1 RPKM. Among the 9,932 genes expressed at >1 RPKM in skeletal-muscle samples, 9,106 (92%) were expressed in t-myotubes at >1 RPKM (Figure 2C and Table S4). Importantly, 87% of the genes in the neuromuscular panel were expressed in t-myotubes at >1 RPKM; only ANO5 [MIM: 608662], CAPN3 [MIM: 114240], CLCN1 [MIM: 118425], COLQ [MIM: 603033], KBTBD13 [MIM: 613727], SCN4A [MIM: 603967], and SYNE2 [MIM: 608442] were expressed at <1 RPKM in t-myotubes and at >1 RPKM in skeletal muscle (Figures 5 and S3 and Table S5). Cluster analysis showed that t-myotubes have an expression profile intermediate between fibroblasts and skeletal muscle, and they have much more variability compared to either (Figure 2A). From the 117 (out of 132) genes that were expressed at >1 RPKM in muscle, we detected a coefficient of variation >0.5 in 34% (40/117) in muscle, 49% (57/117) in t-myotubes, and 9% (10/117) in fibroblasts (Tables S6–S8). This variability is likely driven by the variable percentage of fibroblasts and t-myotubes in our cultures, as well as by the variable differentiation state of the t-myotubes. The muscle-disease-associated genes with the highest variability in t-myotubes are ACTA1 (MIM: 12610), KLHL1 (MIM: 605332), DES, TNNT1 (MIM: 191041), and CHRNA1.
Figure 5.
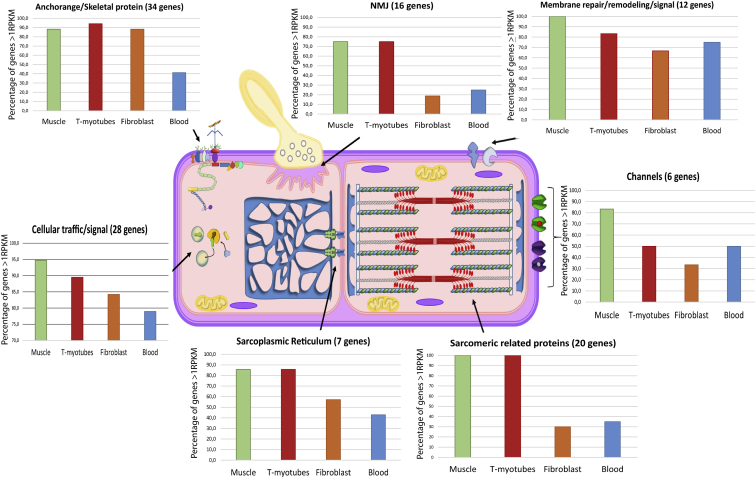
Most Common Muscle-Disease-Causing Genes, Classified by Their Main Function, That Are Expressed at >1 RPKM
We organized 123 of the 132 genes in our panel into seven distinct groups associated with muscle function, and we determined their expression in four different tissues (skeletal muscle, t-myotubes, fibroblasts, and blood). We calculated the percentage of genes in each category that were expressed at ≥1 RPKM in each of the tissues, and we saw that most of the muscle genes are poorly expressed in blood and fibroblasts and sufficiently expressed in t-myotubes (adapted from Dowling et al., 201779).
For data analysis, we began by looking at sequence variants present in our neuromuscular panel. In t-myotubes, we could detect 75% of the total exonic variants identified in matched skeletal muscle biopsies. Correspondingly, t-myotubes had a median of 74% of base pairs covered at 20×, whereas 83% of base pairs were covered in skeletal muscle. Of note, fibroblasts only had 50% median coverage of neuromuscular gene variants (Tables S19 andS20).
We next looked at splicing events in our 13 matched sample sets (muscle biopsy, t-myotubes, and fibroblasts) (11 unknowns and two positive controls, Table S18). Out of 49 novel junctions detected in muscle, we detected six in t-myotubes and two in fibroblasts. Interestingly, 55.3% of the muscle novel junctions not found in t-myotubes belonged to NEB, TTN, or TPM3 (MIM: 191030) (27.7%; 19.1%; and 8.5%, respectively), although 13% belonged to genes with low expression in the reprogrammed cells (mainly SYNE2 and ANO5). In t-myotubes we detected a median of 15 novel splice junctions unique to t-myotubes (these were novel junctions that were absent in primary fibroblasts and muscle). The genes with a higher number of splicing events in the t-myotubes are mainly involved in the extracellular matrix (COL6A3 [MIM: 120250] and COL6A1 [120220]), membrane trafficking (SYT2 and CAV3 [MIM: 601253]), and the sarcolemmal membrane structure (SGCA and SGCB [MIM: 600900).
Lastly, we analyzed the suitability of t-myotubes for identifying disease-causing genetic variants. Importantly, in our cohort of 11 unknown matched samples, in all five cases in which pathogenic disease-causing changes were found in skeletal muscle and/or fibroblasts, the disease-causing changes were also found in t-myotubes (Table 1).
PAGE—An Interface for Data Exploration
As exemplified by our t-myotube data and by the comparisons between blood and skeletal muscle, one of the key considerations for RNA-seq is source material. In an effort to provide a simple platform for interrogating RNA-seq data to determine the most suitable tissue for study, we created a web-based, searchable data portal (PAGE). This portal enables the user to search for genes or gene panels of interest and then visualize the coverage of those genes in different tissues. This platform incorporates the new data we generated for fibroblasts and t-myotubes and additionally includes the major tissue types from GTEx.
In PAGE, we enable comparisons at the exon level—the user picks genes (or a pre-loaded panel of genes) and two (or more) tissues of interest to compare. For each gene in the panel, PAGE counts the number of exons that are expressed in the primary tissue and checks if the same exon(s) are also expressed in the other tissues of interest. On the basis of the percentage overlap in the number of exons expressed in the tissues being compared, the tissues are ranked as in descending order of similarity to the primary tissue of interest. The user also has the ability to view the expression of the exons across all the samples as box plots by clicking on the genes of interest. Additionally, the user can explore the identified variants, expression levels, and splicing changes of the gene through a link to JBrowse.74 We have precomputed the similarity of all protein-coding genes across 31 tissues in GTEx and ranked the tissues on the basis of the similarity in expression of the exons. PAGE is freely available (see Web Resources).
Discussion
Taken together, our results validate RNA-seq as a tool for the detection and discovery of rare disease mutations. We demonstrate that it provides a considerable improvement in the overall diagnostic rate over exome sequencing alone and provides clarity for the interpretation and validation of non-coding variants. Our pipeline allowed us to arrive at a diagnosis in 36% (9 of 25) of instances previously unsolved by gene panels or WES. Furthermore, we provide extensive data supporting the suitability of using cell lines derived from individuals with the disease in RNA-seq-based diagnostics. Specifically, we show that transdifferentiated myotubes accurately reflect the transcriptome of skeletal muscle; these data open the door to minimally invasive strategies for transcriptome-based diagnostics.
In terms of primary-mutation discovery, all of our solved cases represent examples of mutations either that would not have been discovered by exomic analysis or that would have required validation by secondary methods. Our data thus broaden the range of mutations that can be discovered via RNA-seq. In addition to identifying exonic, splice-site, and deep intronic mutations, we validate the capability of transcriptome analysis to identify variants in regulatory upstream regions (promoters, enhancers, and UTRs) and variants that impact allele-specific expression. Allele imbalance is a particularly useful phenomenon to observe because it can point to more complex disease mechanisms, such as imprinting, uniparental disomy, X inactivation, and chromosomal rearrangements.35 Moreover, the findings from our families demonstrate the utility of RNA-seq for providing clarity to some variants in large genes, such as TTN and RYR1, with frequently encountered VUSs; such genes are notoriously challenging to evaluate.
Our study identified several additional points related to the use of RNA-seq for mutation discovery. Somewhat unsurprisingly, and in keeping with WES and WGS, the yield of proband analysis was improved through the use of sibling and parental samples.75 Specifically, in those families in which we had samples available from the proband and a relative, the difficulties of filtering and identifying posttranscriptional defects were significantly reduced. This not only impacted the accuracy of detecting aberrant splicing and/or variant calling, but also enabled better interpretation of potential allelic imbalance. The challenge to this, of course, is that trio analysis necessitates obtaining disease-relevant tissue from the proband as well as the parents; in the case of myopathies, these tissue samples would historically be skeletal muscle biopsies. We show, however, as discussed below, that cells derived from skin biopsies provide a suitable alternative that makes trio studies feasible.
Another important consideration that emerged from our study (and is also consistent with exome and genome sequencing) is the importance of accurate clinical phenotyping for data interpretation. In our cohort, we arrived at a diagnosis primarily for the individuals or families where the phenotype was well delineated. This was particularly true for families with a clinical or pathological phenotype, such as Duchenne muscular dystrophy, Pompe disease, and merosin-deficient congenital muscular dystrophy, associated with mutations in a single gene.
A further consideration from the overall data is the age of the sample vis-a-vis the GTEx control database. The GTEx consortium provides an invaluable reference database for transcriptome analysis and the identification of posttranscriptional abnormalities. However, these data are limited to samples from adults >18 years old. We observed a wider variability in the expression profile in children under the age of 10 years old and noticed a trend of clustering after this age was reached. We did not do a deeper pediatric transcriptome analysis because it was not the primary aim of our project, and we currently lack sufficient control samples to fully understand and define the critical differences between pediatric and adult cases. We believe, however, that that pediatric controls and an analysis of the differences in pediatric and adult samples are necessary to better understand the changes in the transcriptome during the maturation of the skeletal muscle, especially in the pediatric population and, eventually, during the aging process, and thus advocate for efforts related to developing control datasets for these age ranges.
Lastly, we have made advances in terms of the pipeline and analysis of RNA-seq data for mutation detection. In particular, we have focused on novel junctions, as well as on the combination of outlier expression sets and allele balance. Interestingly, our approach identified in our samples, as compared to in GTEx, a median of 5 LFNJs. Although a handful of these junctions proved to be pathogenic, there were several additional and equally rare junctions that were identified in our samples. Some of these “novel” junctions could be due to samples’ biological variability, caused by known and yet-to-be-identified splicing mechanisms. Because all of our samples are pediatric, this could result in biased usage of some junctions. We classify these rare, novel junctions as “junctions of unknown significance (JUSs)” and feel their biological role and pathogenicity will become clearer with the analysis of a much larger number of RNA-seq samples. Additionally, the focus of our splicing analysis was to identify novel junctions and not to look for changes in splice-site usage and shifts in isoform levels. In the future, we would like to extend the pipeline to study changes in splicing, as well as novel splicing, as more mature algorithms become available for tackling this problem.
One of the important developments from our study is the description of the suitability of fibroblasts and transdifferentiated myotubes for muscle transcriptomics. Although blood is the easiest sample to obtain, it only expresses ∼75% of muscle genes at >1 RPKM and only half of the most common genes used in clinical panels of muscle disorders. In particular, genes involved in the sarcomere, sarcolemmal membrane, and the neuromuscular junction are poorly represented in the blood transcriptome. Transcriptomes from skin fibroblasts offer an improvement over those from blood, particularly for genes associated with congenital muscular dystrophies (CMDs). This corroborates previous studies showing the utility of fibroblasts for studying CMDs and particularly for mutation discovery for dystroglycanopathies.38, 76, 77 RNA-seq analysis of fibroblasts has previously been explored for individuals with mitochondrial disease, and it was shown to improve diagnostic accuracy in this setting by 10%.34 However, primary fibroblasts lack the expression of most of the genes involved in the sarcomere (30%) and neuromuscular junction (18.8%) (Figure 5). This limitation is greatly compensated for by their potential to be reprogrammed into different cell types, such as myotubes.
Our study reports a thorough and extensive transcriptomic analysis of transdifferentiated myotubes. We show that most of muscle-disease-related genes, including all components of the sarcomere, the sarcolemmal membrane, and the neuromuscular junction, are expressed in t-myotubes (Figure S1). Variants found in both exonic and intronic regions can be reliably uncovered in t-myotubes. Critically, in our matched cohort of biopsies and t-myotubes from the same individuals, we successfully identified all causative mutations that were present in the muscle biopsies. We conclude therefore that t-myotubes can effectively be used for RNA-seq-based diagnostics when a biopsy is not available, and they can offer additional clarity for variant evaluation by providing easy access to parental samples.
There are also a few limitations of RNA-seq performed on t-myotubes as compared to that performed on muscle biopsies. Although we achieved >1 RPKM expression in nearly all disease-relevant muscle transcripts, the expression of these transcripts in t-myotubes is lower for most genes. This might hinder the identification of expression outliers and allele imbalance. Expression analysis is also complicated by the variability of the cultures (which contain a mix of fibroblasts and myotubes at different stages of maturity), by the lack of a control dataset, and by the influence of forced overexpression of MyoD.78 In terms of specific sequence evaluation, we could identify only 75% of genomic SNVs seen in blood (as compared to 90% in biopsies) and uncover only a portion of the novel junctions seen in muscle RNA-seq. These last two challenges most likely relate, at least in part, to reduced gene expression (providing lower coverage for SNVs) and to splice changes that accompany myotube maturation (causing some exons found in muscle biopsies not to be expressed in our myotubes). To address these limitations, we are aiming our future work at increasing our cohorts of controls and t-myotubes derived from affected individuals and on developing improved culture settings to promote more uniformity and maturation.
Our study underscores the importance of source material for RNA-seq-based diagnostics. Although we have convincingly demonstrated that t-myotubes derived from affected individuals can be used in diagnostics for muscle diseases when biopsies are not available, we appreciate that these particular cell lines are not suitable for other subsets of rare disease. In these settings, biopsies of an affected tissue will likely remain the gold standard for transcriptomics. However, there is an opportunity to employ similar strategies of fibroblast transdifferentiation to generate other disease-relevant cell models (neurons, cardiomyocytes, etc.). Future studies will focus on the development of these cell models and thus will hopefully expand the breadth of diseases that can be evaluated by RNA-seq.
Of note, in order to simplify the choice of suitable source material for a mutation-discovery study with RNA-seq, we developed our PAGE interface. PAGE currently contains transcriptome data from the tissues present in GTex and from our fibroblasts and t-myotubes, and it will incorporate information from new cell derivatives as they are developed. The goal of the PAGE portal is to provide users with the ability to quickly and easily evaluate the expression levels of a gene or a set of genes in tissue(s) of interest. For example, a clinician working on neuromuscular disorders could evaluate whether genes expressed in skeletal muscle are also expressed at “sufficient” levels in fibroblasts. This will give the clinician the ability to decide whether they can use fibroblasts for transcriptome analysis instead of a muscle biopsy.
In addition to the advantages of using RNA-seq for diagnostics, some caveats need to be considered. Even though we identify many disease-relevant mutations, the use of RNA-seq for variant discovery is mainly restricted to gene-coding regions of the genome (and particularly to highly expressed genes that have typically high coverage). Furthermore, as has been shown here, because expression is tissue specific, analysis requires the sequencing of disease-relevant tissues. The ability to identify variants and splicing changes of interest is dependent on the gene of interest, tissue of expression, and high depth of coverage; because of this, the cost associated with RNA sequencing can be a major factor in utilizing the technology in a clinical diagnostic setting. Differential expression analysis requires many replicates for sample comparison, and this is especially true when one is looking to identify subtle (yet significant) changes in gene expression. Finally, gene expression is a highly regulated process and therefore requires matched “normal” samples; this will require the sequencing of samples from multiple tissues over multiple developmental stages, and all of these have to be analyzed in a consistent manner.
In summary, we provide convincing support for the utility of RNA-seq for the detection of rare, disease-causing mutations associated with neuromuscular disorders, and we show that RNA-seq can identify mutations in settings where gene panels and exome sequencing do not. Furthermore, we demonstrate the feasibility of using minimally invasive material derived from individuals for transcriptome-based diagnostics, and we thus establish the groundwork for using easily obtainable cell models for trio studies and in settings where biopsies are not available. We also fully describe RNA-seq’s strengths (providing clarity on some coding variants, identifying deep intronic variants, and illuminating UTR mutations) and point out some of the challenges that remain with its interpretation (age dependent variation and the presence of novel junctions of unclear significance).
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was supported by funding from a Genome Canada Disruptive Innovations in Genomics grant (J.J.D. and M.B.), as well as by seed funding to J.J.D. from the Chase-an-Idea grant mechanism (Centre for Brain and Mental Health, the Hospital for Sick Children) and by funding from the Muscular Dystrophy Association (J.J.D., MDA 380397). Computational analysis was conducted by the Canadian Centre for Computational Genomics (C3G), funded by Genome Canada and Genome Quebec. H.D.G. was supported by a Restracomp grant (the Hospital for Sick Children). K.D.M. and S.A.M. are partially supported by a National Institutes of Health (NIH) grant, U54, NS053672, that funds the Iowa Wellstone Muscular Dystrophy Cooperative Research Center (MDCRC). Carrie Stephan and Mary Cox provided extensive clinical or laboratory support for the Iowa MDCRC. A.N.O. would like to acknowledge l’Hospital Infantil Sant Joan de Déu per a la Investigació, integrated as part of the biobank (BioNER) of the Carlos III Health Institute (ISCIII), for samples and data procurement. We thank Vandana Shashi and Kelly Schoch for contributing samples. They are partially supported by NIH grant U01, HG007672.
Published: February 28, 2019
Footnotes
Supplemental Data can be found with this article online at https://doi.org/10.1016/j.ajhg.2019.01.012.
Contributor Information
Michael Brudno, Email: brudno@cs.toronto.edu.
James J. Dowling, Email: james.dowling@sickkids.ca.
Web Resources
bcbio-nextgen, https://github.com/chapmanb/bcbio-nextgen
The Centre for Applied Genomics, http://www.tcag.ca
Common Fund (CF) Genotype-Tissue Expression Project (GTEx), https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000424.v7.p2
CRE, https://github.com/ccmbioinfo/cre, https://github.com/naumenko-sa/cre
CRT, https://github.com/ccmbioinfo/crt, https://github.com/naumenko-sa/crt
Ensembl, http://www.ensembl.org
fastqc, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
GATK Best Practices. Calling Variants in RNA-seq, https://software.broadinstitute.org/gatk/documentation/article.php?id=3891
GENCODE, https://www.gencodegenes.org/
MITOCARTA: An Inventory of Mammalian Mitochondrial Genes, https://www.broadinstitute.org/scientific-community/science/programs/metabolic-disease-program/publications/mitocarta/mitocarta-in-0
OMIM, https://www.omim.org
OUTRIDER, https://github.com/gagneurlab/OUTRIDER
Portal, https://gtexportal.org/home/
The R Project for Statistical Computing, https://www.r-project.org/
R Studio, https://www.rstudio.com/
RNA-seq of GM12878, SRR307898, https://www.ncbi.nlm.nih.gov/sra/?term=SRR307898GTEx
vcfanno, https://github.com/brentp/vcfanno
Supplemental Data
References
- 1.North K.N., Wang C.H., Clarke N., Jungbluth H., Vainzof M., Dowling J.J., Amburgey K., Quijano-Roy S., Beggs A.H., Sewry C., International Standard of Care Committee for Congenital Myopathies Approach to the diagnosis of congenital myopathies. Neuromuscul. Disord. 2014;24:97–116. doi: 10.1016/j.nmd.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kress W., Rost S., Kolokotronis K., Meng G., Pluta N., Müller-Reible C. The genetic approach: Next-generation sequencing-based diagnosis of congenital and infantile myopathies/muscle dystrophies. Neuropediatrics. 2017;48:242–246. doi: 10.1055/s-0037-1602660. [DOI] [PubMed] [Google Scholar]
- 3.Bönnemann C.G., Wang C.H., Quijano-Roy S., Deconinck N., Bertini E., Ferreiro A., Muntoni F., Sewry C., Béroud C., Mathews K.D., Members of International Standard of Care Committee for Congenital Muscular Dystrophies Diagnostic approach to the congenital muscular dystrophies. Neuromuscul. Disord. 2014;24:289–311. doi: 10.1016/j.nmd.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonald C.M. Clinical approach to the diagnostic evaluation of hereditary and acquired neuromuscular diseases. Phys. Med. Rehabil. Clin. N. Am. 2012;23:495–563. doi: 10.1016/j.pmr.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasli N., Böhm J., Le Gras S., Muller J., Pizot C., Jost B., Echaniz-Laguna A., Laugel V., Tranchant C., Bernard R. Next generation sequencing for molecular diagnosis of neuromuscular diseases. Acta Neuropathol. 2012;124:273–283. doi: 10.1007/s00401-012-0982-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., Centers for Mendelian Genomics The genetic basis of mendelian phenotypes: Discoveries, challenges, and opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark M.M., Stark Z., Farnaes L., Tan T.Y., White S.M., Dimmock D., Kingsmore S.F. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genomic Med. 2018;3:16. doi: 10.1038/s41525-018-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan T.Y., Dillon O.J., Stark Z., Schofield D., Alam K., Shrestha R., Chong B., Phelan D., Brett G.R., Creed E. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr. 2017;171:855–862. doi: 10.1001/jamapediatrics.2017.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stark Z., Tan T.Y., Chong B., Brett G.R., Yap P., Walsh M., Yeung A., Peters H., Mordaunt D., Cowie S. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet. Med. 2016;18:1090–1096. doi: 10.1038/gim.2016.1. [DOI] [PubMed] [Google Scholar]
- 10.Todd E.J., Yau K.S., Ong R., Slee J., McGillivray G., Barnett C.P., Haliloglu G., Talim B., Akcoren Z., Kariminejad A. Next generation sequencing in a large cohort of patients presenting with neuromuscular disease before or at birth. Orphanet J. Rare Dis. 2015;10:148. doi: 10.1186/s13023-015-0364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chae J.H., Vasta V., Cho A., Lim B.C., Zhang Q., Eun S.H., Hahn S.H. Utility of next generation sequencing in genetic diagnosis of early onset neuromuscular disorders. J. Med. Genet. 2015;52:208–216. doi: 10.1136/jmedgenet-2014-102819. [DOI] [PubMed] [Google Scholar]
- 12.Schofield D., Alam K., Douglas L., Shrestha R., MacArthur D.G., Davis M., Laing N.G., Clarke N.F., Burns J., Cooper S.T. Cost-effectiveness of massively parallel sequencing for diagnosis of paediatric muscle diseases. NPJ Genom. Med. 2017;2:4. doi: 10.1038/s41525-017-0006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boycott K.M., Vanstone M.R., Bulman D.E., MacKenzie A.E. Rare-disease genetics in the era of next-generation sequencing: Discovery to translation. Nat. Rev. Genet. 2013;14:681–691. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
- 14.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper G.M. Parlez-vous VUS? Genome Res. 2015;25:1423–1426. doi: 10.1101/gr.190116.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma M., Ru Y., Chuang L.S., Hsu N.Y., Shi L.S., Hakenberg J., Cheng W.Y., Uzilov A., Ding W., Glicksberg B.S., Chen R. Disease-associated variants in different categories of disease located in distinct regulatory elements. BMC Genomics. 2015;16(Suppl 8):S3. doi: 10.1186/1471-2164-16-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper G.M., Shendure J. Needles in stacks of needles: Finding disease-causal variants in a wealth of genomic data. Nat. Rev. Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 18.Stenson P.D., Mort M., Ball E.V., Evans K., Hayden M., Heywood S., Hussain M., Phillips A.D., Cooper D.N. The Human Gene Mutation Database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volk A.E., Kubisch C. The rapid evolution of molecular genetic diagnostics in neuromuscular diseases. Curr. Opin. Neurol. 2017;30:523–528. doi: 10.1097/WCO.0000000000000478. [DOI] [PubMed] [Google Scholar]
- 20.Schuelke M., Øien N.C., Oldfors A. Myopathology in the times of modern genetics. Neuropathol. Appl. Neurobiol. 2017;43:44–61. doi: 10.1111/nan.12374. [DOI] [PubMed] [Google Scholar]
- 21.Lohmann K., Klein C. Next generation sequencing and the future of genetic diagnosis. Neurotherapeutics. 2014;11:699–707. doi: 10.1007/s13311-014-0288-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darras B.T., Jones H.R. Diagnosis of pediatric neuromuscular disorders in the era of DNA analysis. Pediatr. Neurol. 2000;23:289–300. doi: 10.1016/s0887-8994(00)00202-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang G.S., Cooper T.A. Splicing in disease: Disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 24.Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sibley C.R., Blazquez L., Ule J. Lessons from non-canonical splicing. Nat. Rev. Genet. 2016;17:407–421. doi: 10.1038/nrg.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byron S.A., Van Keuren-Jensen K.R., Engelthaler D.M., Carpten J.D., Craig D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016;17:257–271. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaz-Drago R., Custódio N., Carmo-Fonseca M. Deep intronic mutations and human disease. Hum. Genet. 2017;136:1093–1111. doi: 10.1007/s00439-017-1809-4. [DOI] [PubMed] [Google Scholar]
- 28.Pagliarini V., La Rosa P., Sette C. Faulty RNA splicing: Consequences and therapeutic opportunities in brain and muscle disorders. Hum. Genet. 2017;136:1215–1235. doi: 10.1007/s00439-017-1802-y. [DOI] [PubMed] [Google Scholar]
- 29.Gallego-Paez L.M., Bordone M.C., Leote A.C., Saraiva-Agostinho N., Ascensão-Ferreira M., Barbosa-Morais N.L. Alternative splicing: The pledge, the turn, and the prestige : The key role of alternative splicing in human biological systems. Hum. Genet. 2017;136:1015–1042. doi: 10.1007/s00439-017-1790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anna A., Monika G. Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 2018;59:253–268. doi: 10.1007/s13353-018-0444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Hashim A., Gonorazky H.D., Amburgey K., Das S., Dowling J.J. A novel intronic mutation in MTM1 detected by RNA analysis in a case of X-linked myotubular myopathy. Neurol Genet. 2017;3:e182. doi: 10.1212/NXG.0000000000000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonorazky H., Liang M., Cummings B., Lek M., Micallef J., Hawkins C., Basran R., Cohn R., Wilson M.D., MacArthur D. RNAseq analysis for the diagnosis of muscular dystrophy. Ann. Clin. Transl. Neurol. 2015;3:55–60. doi: 10.1002/acn3.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cummings B.B., Marshall J.L., Tukiainen T., Lek M., Donkervoort S., Foley A.R., Bolduc V., Waddell L.B., Sandaradura S.A., O’Grady G.L., Genotype-Tissue Expression Consortium Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med. 2017;9:eaal5209. doi: 10.1126/scitranslmed.aal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kremer L.S., Bader D.M., Mertes C., Kopajtich R., Pichler G., Iuso A., Haack T.B., Graf E., Schwarzmayr T., Terrile C. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun. 2017;8:15824. doi: 10.1038/ncomms15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith R.M., Webb A., Papp A.C., Newman L.C., Handelman S.K., Suhy A., Mascarenhas R., Oberdick J., Sadee W. Whole transcriptome RNA-Seq allelic expression in human brain. BMC Genomics. 2013;14:571. doi: 10.1186/1471-2164-14-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium G.T., GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 38.Stevens E., Torelli S., Feng L., Phadke R., Walter M.C., Schneiderat P., Eddaoudi A., Sewry C.A., Muntoni F. Flow cytometry for the analysis of α-dystroglycan glycosylation in fibroblasts from patients with dystroglycanopathies. PLoS ONE. 2013;8:e68958. doi: 10.1371/journal.pone.0068958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barateau A., Vadrot N., Vicart P., Ferreiro A., Mayer M., Héron D., Vigouroux C., Buendia B. A novel lamin A mutant responsible for congenital muscular dystrophy causes distinct abnormalities of the cell nucleus. PLoS ONE. 2017;12:e0169189. doi: 10.1371/journal.pone.0169189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butterfield R.J., Dunn D.M., Hu Y., Johnson K., Bönnemann C.G., Weiss R.B. Transcriptome profiling identifies regulators of pathogenesis in collagen VI related muscular dystrophy. PLoS ONE. 2017;12:e0189664. doi: 10.1371/journal.pone.0189664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandez-Fuente M., Martin-Duque P., Vassaux G., Brown S.C., Muntoni F., Terracciano C.M., Piercy R.J. Adenovirus-mediated expression of myogenic differentiation factor 1 (MyoD) in equine and human dermal fibroblasts enables their conversion to caffeine-sensitive myotubes. Neuromuscul. Disord. 2014;24:250–258. doi: 10.1016/j.nmd.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 42.Normand J., Karasek M.A. A method for the isolation and serial propagation of keratinocytes, endothelial cells, and fibroblasts from a single punch biopsy of human skin. In Vitro Cell. Dev. Biol. Anim. 1995;31:447–455. doi: 10.1007/BF02634257. [DOI] [PubMed] [Google Scholar]
- 43.Mailman M.D., Feolo M., Jin Y., Kimura M., Tryka K., Bagoutdinov R., Hao L., Kiang A., Paschall J., Phan L. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leinonen R., Sugawara H., Shumway M., International Nucleotide Sequence Database Collaboration The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonne G., Rivier F., Hamroun D. The 2018 version of the gene table of monogenic neuromuscular disorders (nuclear genome) Neuromuscul. Disord. 2017;27:1152–1183. doi: 10.1016/j.nmd.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 46.McKusick V.A. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C.G. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calvo S.E., Clauser K.R., Mootha V.K. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44(D1):D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobin A., Gingeras T.R. Mapping RNA-seq Reads with STAR. Curr Protoc Bioinformatics. 2015;51:1–19. doi: 10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y., Smyth G.K., Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 53.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piskol R., Ramaswami G., Li J.B. Reliable identification of genomic variants from RNA-seq data. Am. J. Hum. Genet. 2013;93:641–651. doi: 10.1016/j.ajhg.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Current Protoc Bioinformatics. 2013;43:1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Consortium E.P., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zook J.M., Catoe D., McDaniel J., Vang L., Spies N., Sidow A., Weng Z., Liu Y., Mason C.E., Alexander N. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci. Data. 2016;3:160025. doi: 10.1038/sdata.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramaswami G., Li J.B. RADAR: A rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014;42:D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pedersen B.S., Layer R.M., Quinlan A.R. Vcfanno: Fast, flexible annotation of genetic variants. Genome Biol. 2016;17:118. doi: 10.1186/s13059-016-0973-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paila U., Chapman B.A., Kirchner R., Quinlan A.R. GEMINI: Integrative exploration of genetic variation and genome annotations. PLoS Comput. Biol. 2013;9:e1003153. doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: A one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eberle M.A., Fritzilas E., Krusche P., Källberg M., Moore B.L., Bekritsky M.A., Iqbal Z., Chuang H.Y., Humphray S.J., Halpern A.L. A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res. 2017;27:157–164. doi: 10.1101/gr.210500.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohno K., Takeda J.I., Masuda A. Rules and tools to predict the splicing effects of exonic and intronic mutations. Wiley Interdiscip. Rev. RNA. 2018;9 doi: 10.1002/wrna.1451. [DOI] [PubMed] [Google Scholar]
- 66.Shibata A., Okuno T., Rahman M.A., Azuma Y., Takeda J., Masuda A., Selcen D., Engel A.G., Ohno K. IntSplice: Prediction of the splicing consequences of intronic single-nucleotide variations in the human genome. J. Hum. Genet. 2016;61:633–640. doi: 10.1038/jhg.2016.23. [DOI] [PubMed] [Google Scholar]
- 67.Leman R., Gaildrat P., Gac G.L., Ka C., Fichou Y., Audrezet M.P., Caux-Moncoutier V., Caputo S.M., Boutry-Kryza N., Leone M. Novel diagnostic tool for prediction of variant spliceogenicity derived from a set of 395 combined in silico/in vitro studies: an international collaborative effort. Nucleic Acids Res. 2018;46:11656–11657. doi: 10.1093/nar/gky979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hartley S.W., Mullikin J.C. Detection and visualization of differential splicing in RNA-seq data with JunctionSeq. Nucleic Acids Res. 2016;44:e127. doi: 10.1093/nar/gkw501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jensen B.S., Willer T., Saade D.N., Cox M.O., Mozaffar T., Scavina M., Stefans V.A., Winder T.L., Campbell K.P., Moore S.A., Mathews K.D. GMPPB-associated dystroglycanopathy: Emerging common variants with phenotype correlation. Hum. Mutat. 2015;36:1159–1163. doi: 10.1002/humu.22898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brun B.N., Willer T., Darbro B.W., Gonorazky H.D., Naumenko S., Dowling J.J., Campbell K.P., Moore S.A., Mathews K.D. Uniparental disomy unveils a novel recessive mutation in POMT2. Neuromuscul. Disord. 2018;28:592–596. doi: 10.1016/j.nmd.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang P., Zhang L., Gao Y., He Z., Yao D., Wu Z., Cen J., Chen X., Liu C., Hu Y. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell. 2014;14:370–384. doi: 10.1016/j.stem.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 72.Engel J.L., Ardehali R. Direct cardiac reprogramming: Progress and promise. Stem Cells Int. 2018;2018:1435746. doi: 10.1155/2018/1435746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gopalakrishnan S., Hor P., Ichida J.K. New approaches for direct conversion of patient fibroblasts into neural cells. Brain Res. 2017;1656:2–13. doi: 10.1016/j.brainres.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H. JBrowse: A next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ghaoui R., Cooper S.T., Lek M., Jones K., Corbett A., Reddel S.W., Needham M., Liang C., Waddell L.B., Nicholson G. Use of whole-exome sequencing for diagnosis of limb-girdle muscular dystrophy: Outcomes and lessons learned. JAMA Neurol. 2015;72:1424–1432. doi: 10.1001/jamaneurol.2015.2274. [DOI] [PubMed] [Google Scholar]
- 76.Herzog C., Has C., Franzke C.W., Echtermeyer F.G., Schlötzer-Schrehardt U., Kröger S., Gustafsson E., Fässler R., Bruckner-Tuderman L. Dystroglycan in skin and cutaneous cells: Beta-subunit is shed from the cell surface. J. Invest. Dermatol. 2004;122:1372–1380. doi: 10.1111/j.0022-202X.2004.22605.x. [DOI] [PubMed] [Google Scholar]
- 77.Willer T., Lee H., Lommel M., Yoshida-Moriguchi T., de Bernabe D.B., Venzke D., Cirak S., Schachter H., Vajsar J., Voit T. ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome. Nat. Genet. 2012;44:575–580. doi: 10.1038/ng.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cacchiarelli D., Qiu X., Srivatsan S., Manfredi A., Ziller M., Overbey E., Grimaldi A., Grimsby J., Pokharel P., Livak K.J. Aligning single-cell developmental and reprogramming trajectories identifies molecular determinants of myogenic reprogramming outcome. Cell Syst. 2018;7:258–268. doi: 10.1016/j.cels.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 79.Dowling J.J., Gonorazky H., Cohn R., Campbell C. Treating pediatric neuromuscular disorders: The future is now. Am. J. Med. Genet. 2018;176:804–841. doi: 10.1002/ajmg.a.38418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.