Abstract
Alzheimer’s disease (AD) is considered a polygenic disorder. This view is clouded, however, by lingering uncertainty over how to treat the quasi “monogenic” role of apolipoprotein E (APOE). The APOE4 allele is not only the strongest genetic risk factor for AD, it also affects risk for cardiovascular disease, stroke, and other neurodegenerative disorders. This review, based mostly on data from human studies, ranges across a variety of APOE-related pathologies, touching on evolutionary genetics and risk mitigation by ethnicity and sex. The authors also address one of the most fundamental question pertaining to APOE4 and AD: Does APOE4 increase AD risk via a loss- or gain-of-function? The answer will be of the utmost importance in guiding future research in AD.
Keywords: Apolipoprotein E (APOE), Alzheimer’s disease (AD), neurodegenerative disease, cardiovascular disease, pleiotropy, evolutionary genetics, ethnicity, sex, gene-based therapy, anti-sense oligonucleotide (ASO)
In brief
Does APOE4 increase risk for Alzheimer’s disease via a gain- or loss-of-function? Belloy, et al., focusing on human data, examine critical issues like pleiotropy, sex, and ancestral background, to address this fundamental question.
Introduction
Recent advances in gene-based therapeutics have led to startling responses in neurologic disorders that have previously proven resistant to treatment. Starting in the periphery, patients with neuropathy caused by transthyretin amyloidosis (OMIM#105210) have responded well to treatment with either RNA interference (RNAi) or an antisense oligonucleotide (ASO) directed against the mutated TTR gene (Adams et al., 2018; Benson et al., 2018). ASO treatment has been FDA-approved [though not without controversy (Aartsma-Rus and Krieg, 2017)] for Duchenne muscular dystrophy (OMIM#310200), using an exon-skipping strategy (wherein the ASO prevents the mutated exon from being incorporated into the protein resulting in a shortened, but functional, form of dystrophin). Moving into the central nervous system (CNS), where treatment efforts are further complicated by the blood-brain barrier, neurology has entered a revolutionary new phase in our approach to fatal, neurodegenerative diseases. Spinal muscular atrophy (OMIM#253300), a previously, universally fatal motor neuron disorder of childhood, has recently proven amenable to intrathecal ASO treatment (Mercuri et al., 2018b, 2018a). Additional ASO studies are underway in Huntington’s disease (OMIM#143100) and amyotrophic lateral sclerosis (ALS, OMIM#105400) due to superoxide dismutase mutations (Miller et al., 2013; Wild and Tabrizi, 2017), two diseases believed to result from a toxic gain of function in the mutated protein. Thus, ASOs have already proven highly versatile in their potential for treatment using disparate mechanisms including eliminating a toxic protein; excising a mutated, interior exon to spare the rest of the protein; and, critically, preventing transcription of the upstream open-reading frame that typically inhibits translation, thereby resulting in increased translation of a protein (Rinaldi and Wood, 2018). Across this remarkably broad spectrum of ASO-based strategies, the common theme in these initial gene-based treatment efforts is the targeting of a relatively rare, monogenic disorder.
Late-onset Alzheimer’s disease (referred to in this review as AD, OMIM#104300) is generally considered a polygenic disorder (Escott-Price et al., 2015; Lambert et al., 2013). This view is clouded, however, by lingering uncertainty over how to treat the apolipoprotein E4 (APOE4) allele. What is a neurologist to make of a 56 year-old woman with AD and two copies of APOE4? Does she have a monogenic version of Alzheimer’s disease? If so, should the physician consider an ASO-based approach to treating her, knowing that this has the potential for serious side effects (Shugart, 2017)? Most critically, would the physician want to increase or decrease the availability of the APOE4 protein? We will use this thought experiment as a point of departure for considering the critical question of whether APOE4-related AD risk reflects a loss or a gain of APOE function. Several reviews, centered on rodent and cell work, have already critically advanced this dialogue (Huang, 2010; Huynh et al., 2017a; Kim et al., 2009; Liu et al., 2013; Michaelson, 2014). The current review will focus mainly on human studies—ranging across a variety of topics from coronary artery disease to evolutionary genetics—in the hopes of settling on an evidence-supported answer to this fundamental question pertaining to APOE4 and AD.
A Brief History of APOE in Human Genetics
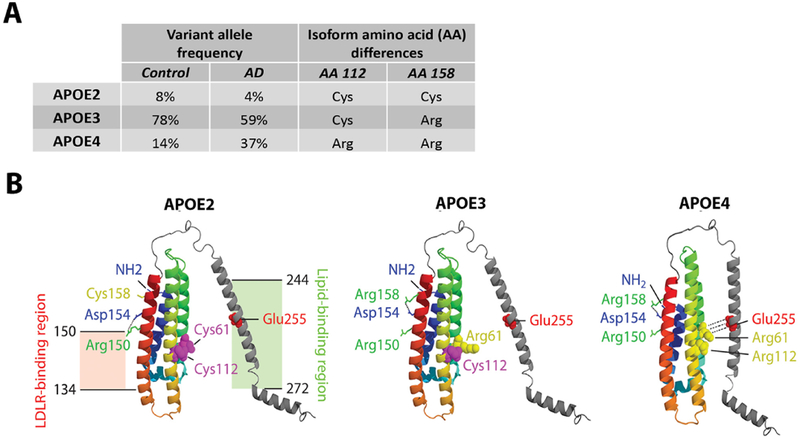
In 1973, Havel and Kane described a mysterious “arginine-rich” protein in the very low-density lipoprotein (VLDL) of patients with a rare disorder of lipid metabolism called familial hypercholesterolemia type III (OMIM#143890) (Havel and Kane, 1973). Utermann and colleagues further characterized this protein and dubbed it APOE (Utermann et al., 1975), with subsequent work characterizing the three main isoforms of the protein: APOE2, APOE3, and APOE4 (Zannis et al., 1982). These initial findings unveiled a role for APOE and its variants in lipid metabolism and cardiovascular disease (Mahley, 1998; Mahley and Rall, 2000). The three isoforms result from common polymorphisms at two nearby loci on the APOE gene. The genetic polymorphisms are labeled rs429358 (C>T) and rs7412 (C>T) and result in amino acid changes at position 112 and 158, respectively, of the APOE protein (Rall et al., 1982; Weisgraber et al., 1981). As shown in Figure 1A, the haplotype combination at the two SNPs defines the three APOE protein isoforms, where the most common isoform, APOE3, has a cysteine at position 112 and an arginine at position 158. APOE2, the least common isoform, has a cysteine at both positions, and APOE4 has an arginine at both positions. These amino acid substitutions have been shown to drive a domain interaction in the protein such that the N-terminal and C-terminal domains, which are normally separated in APOE2 and APOE3, are joined by a salt bridge in APOE4 (Figure 1B).
Figure 1. APOE isoforms, allele frequencies, and protein structures.
A) The three main APOE isoforms APOE2, APOE3, and APOE4, respectively encoded by the Apolipoprotein E2, E3, and E4 alleles, are the result of non-synonymous polymorphisms that cause amino acid changes at position 112 and 158 of the APOE protein (Rall et al., 1982; Weisgraber et al., 1981). APOE3 is the most common variant in the general population. The APOE4 variant is a major genetic risk factor for AD, while APOE2 is protective (Farrer et al., 1997). B) Structural models of lipid-free APOE are shown for each major isoform, based on X-ray crystallography, structure prediction, and circular dichroism spectroscopy (Zhong and Weisgraber, 2009). The N-terminal domain contains APOE’s low-density lipoprotein receptor (LDLR) region at amino acid residues 134 to 150, while the C-terminal holds the lipid-binding region at residues 244 to 272. Amino acid substitutions in APOE4 promote a salt-bridge between Arg61 and Glu255, which, compared to the APOE2 and APOE3 variants, drives increased domain interaction between the N- and C-terminal domains. In figure panel A, APOE allele frequencies are obtained, with permission, from American Medical Association © Farrer, L. et al. JAMA. 278, 1349–1356 (1997). Figure panel B is a reprint, with permission, from Annual Reviews © Yu, J. et al. Annu. Rev. Neurosci. 37, 79–100 (2014).
The diligent reader will, in looking carefully at Figure 1A, wonder why there is not a genotype corresponding to an arginine at 112 and a cysteine at 158. It seems that the rs429358 and rs7412 SNPs are in almost perfect linkage equilibrium (The 1000 Genomes Project Consortium, 2015), so that the combination of an arginine at position 112 and a cysteine at position 158 is extremely rare and defines the APOE1 (also called APOE3r) isoform (Seripa et al., 2007). Only four subjects carrying the APOE1 variant have been reported in the literature: one healthy 70 year-old African Yoruba and three Italians (a 76 year-old with motor neuron disease and, from a separate family, a healthy mother and her autistic son) (Murrell et al., 2006; Persico et al., 2004; Seripa et al., 2007). The frequency (and potential clinical relevance) of this rare APOE1 variant on human diseases may have been underestimated, since it cannot be genotyped by standard sequencing or SNP-arrays but requires restriction-fragment length polymorphism (RFLP)-PCR.
These critical genetic and structural protein insights were already well-established when the link between APOE4 and AD risk was made by Allen Roses and colleagues at Duke University in a pair of landmark studies published in 1993 (Corder et al., 1993; Strittmatter et al., 1993). The next year the same group demonstrated that the APOE2 allele was protective against AD (Corder et al., 1994). In the ensuing quarter century, roughly ten thousand papers have been published pertaining to some aspect of the relationship between AD and APOE. Several critical findings have been replicated and are now essentially indisputable. The risk of AD increases and the age-at-onset decreases with the number of APOE4 alleles (Corder et al., 1993; Farrer et al., 1997). The increase in risk varies substantially, as we will consider later, depending on ancestral background and sex, but as a rough estimate, having a single APOE4 allele increases risk 2–4 fold and having two APOE4 alleles increases risk about 8–12 fold (Farrer et al., 1997). The APOE4 allele also drives the age-at-onset down such that APOE4 carriers are, on average, about 12 years younger than non-carriers (Corder et al., 1993; Roses, 1996). The risk of AD decreases and the age-at-onset increases with the number of APOE2 alleles (Chartier-Harlin et al., 1994; Corder et al., 1994; Farrer et al., 1997). These associations of APOE alleles with age-at-onset have also been reported for early-onset AD due to mutations in the amyloid precursor protein (APP) and presenilin (PSEN) genes, although the size of the APOE effect tends to be diminished some on this autosomal dominant background (Van Duijn et al., 1994; Sorbi et al., 1995; Vélez et al., 2016; Wijsman et al., 2005). Similar observations have been made in subjects with Down Syndrome (OMIM#190685), who, due to trisomy of chromosome 21, carry three copies of APP and develop early-onset AD pathology (Coppus et al., 2008; Royston et al., 1996).
Before leaving the relative safety of solid findings about APOE4 and AD for shakier ground, it is worth making the point that the association between APOE4 and AD is highly likely to be due to the APOE4 variant itself. This is never a foregone conclusion in genetic association studies. Many common SNPs that are associated with a given disease are merely markers that indicate that the actual, causal genetic change is nearby [and in linkage disequilibrium (LD) with the common SNP]. This has been a particularly controversial issue in the APOE literature because the senior author from the landmark 1993 APOE studies, made a case that APOE4’s link to AD was actually mediated by a causal variation in a nearby gene called TOMM40 (Roses et al., 2010). Roses and colleagues developed this hypothesis across a series of papers (Roses et al., 2016, 2014; Zeitlow et al., 2017), but two, much larger studies by independent groups failed to replicate the main finding (Cruchaga et al., 2011; Jun et al., 2012). While some groups continue to pursue this hypothesis, the general consensus in the field is that the association of APOE4 with AD is, in fact, mediated by the corresponding amino acid change in the APOE protein itself. The TOMM40 saga serves, nonetheless, as an important reminder that a SNP identified in a genome-wide association study (GWAS) is rarely the causal variant itself but, instead, a nearby genetic co-traveler in linkage disequilibrium with the causal variant. With APOE and AD, we have the rare good fortune of the association locus being a relatively common, non-synonymous exonic SNP, which decades of basic science in cell and animal models have all but proven to be the causal variant.
Role of APOE in Alzheimer’s Disease Pathogenesis
Several outstanding reviews have covered the molecular and cell biology related to APOE’s role in cardiovascular disease and AD (Huang, 2010; Huang and Mahley, 2014; Kim et al., 2009). A few consistent findings are worth pointing out here to lay the groundwork for interpreting the human data in this review. In the periphery, APOE is produced mainly by the liver, but also by the adrenal gland and macrophages (Elshourbagy et al., 1985; Kockx et al., 2018). In the CNS, APOE is expressed mainly by astrocytes and microglia, but also, to a less clear extent, by neurons under stress conditions (Huang et al., 2004b; Kockx et al., 2018; Saura et al., 2003; Stone et al., 1997; Uchihara et al., 1995; Xu et al., 2006). APOE does not appear to cross the blood-brain barrier and so the peripheral pool of APOE and the CNS pool of APOE are considered to be largely independent of one another (Linton et al., 1991; Liu et al., 2012). In the periphery and the CNS, APOE is a protein that shuttles cholesterol and other lipids between cells (Dietschy and Turley, 2011; Mahley, 1998). To do so, APOE interacts with other proteins (like ABCA1) that can add lipid moieties to APOE, transforming it into a lipoprotein (Koldamova et al., 2005; Wahrle et al., 2004). APOE then delivers its cholesterol cargo to cells via one of several receptors belonging to the low density lipoprotein receptor (LDLR) family (Herz and Bock, 2002). Much research on APOE in the CNS has focused on its critical role in shuttling cholesterol to neurons for the maintenance of cell membranes and synapses, and for their repair after injury (Lane-Donovan et al., 2014; Pfrieger, 2003).
Quality?
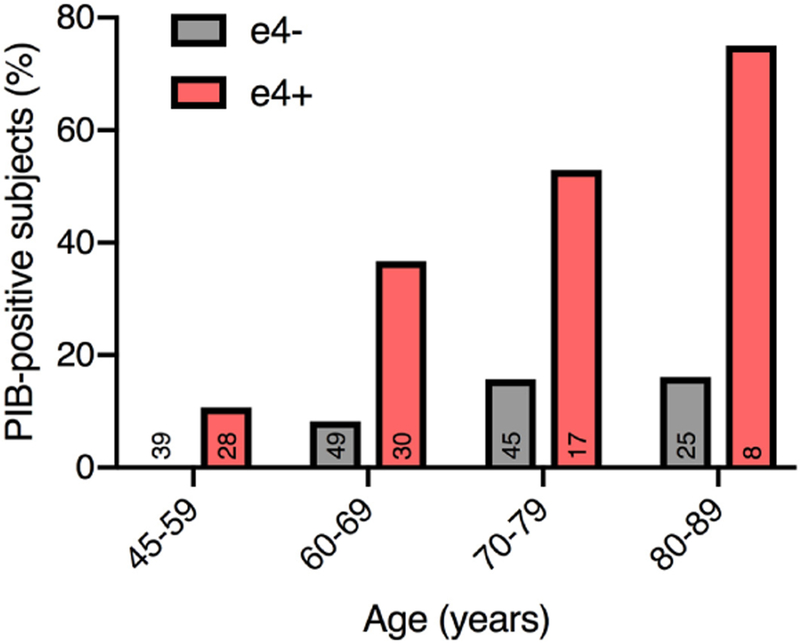
With the ability to measure beta-amyloid peptides in the spinal fluid and plaques in the brain using positron emission tomography (PET), it is now well-established that even among cognitively healthy, older controls, APOE4 carriers are more likely to have abnormally high amyloid levels (Figure 2) (Fleisher et al., 2013; Morris et al., 2010). Conversely, while there is a bit less evidence, given the relative rarity of APOE2 carriers, it appears that the APOE2 allele is associated with a reduced amyloid burden in healthy older controls (Fleisher et al., 2013; Morris et al., 2010). Given the primary role of beta-amyloid in AD, it is presumed that the effect of APOE4 on beta-amyloid accumulation is the chief, though perhaps not the sole (Conejero-Goldberg et al., 2011; Giles et al., 2017; Reiman et al., 2001; Yu et al., 2014), mechanism linking APOE4 to AD risk. While it is clear that APOE4 is associated with accumulation of beta-amyloid, it is not clear how APOE4 does this. A landmark study [now widely replicated (Dickson et al., 1997; Wisniewski and Frangione, 1992)] established early on that APOE is co-deposited with Aβ in amyloid plaques, suggesting that a direct protein-protein interaction might link APOE to amyloid aggregation (Namba et al., 1991). This was later expanded by observations in mice that the APOE isoforms differentially affect Aβ clearance from the brain (Castellano et al., 2011; Verghese et al., 2013). For many years, the leading hypothesis, derived mainly from mouse models, was thus that APOE4 results in impaired processing and clearance of amyloid (Huynh et al., 2017a; Kim et al., 2009). Recent work in human stem-cell derived astrocytes and microglia supports this view (Lin et al., 2018). Conversely, studies of human stem-cell derived neurons also found a direct increase in beta-amyloid production and secretion by APOE4 neurons (Huang et al., 2017; Lin et al., 2018; Wang et al., 2018). Another recent discovery is that microglia appear to upregulate APOE expression as part of a unique phenotype that develops in the context of neurodegeneration and amyloid pathology (Keren-shaul et al., 2017; Krasemann et al., 2017). This phenotype in turn is critically regulated by signaling of TREM2, a microglial surface receptor, that interacts with Aβ oligomers and APOE to enable amyloid clearance and APOE-mediated immune regulation (Li et al., 2018; Yeh et al., 2016). These insights are particularly compelling, given that many microglia-associated genes, such as TREM2, are significantly associated with risk for AD (Pimenova et al., 2018).
Figure 2. APOE4 drives amyloid accumulation.
Binding of Pittsburgh Compound-B (PIB) reflects cerebral amyloid accumulation. Graph displays, per age group and stratified by presence of APOE4 allele (number of subjects indicated at bottom of bars), frequency of PIB-positive subjects in a cohort of cognitively normal individuals. APOE4 carriers more frequently display amyloid accumulation, an effect that becomes more pronounced with age. Figure is adapted, with permission, from John Wiley and Sons © Morris, J. C. et al. Ann. Neurol. 67, 122–131 (2010).
Or Quantity?
While the basic mechanisms remain ill-defined, one contributing factor of the APOE effect on amyloid and AD risk may relate to the availability of the protein. Measuring APOE, whether in plasma or CSF, can be challenging. Most approaches make use of enzyme-linked immunosorbent assay (ELISA)-based strategies for APOE detection, but these are hobbled by intrinsic assay variability, bias from antibody isoform-specific binding preference, and a lack of isoform specificity (Baker-nigh et al., 2016; Mart et al., 2014). In response to these concerns, a few studies have used mass-spectrometry approaches to decrease measurement bias (Baker-nigh et al., 2016; Mart et al., 2014; Martínez and Oskar, 2014). The mass-spectrometry approaches tend to show similar general trends, but with smaller effect sizes. That is, APOE genotype-dependent differences in APOE protein levels appear less pronounced in mass-spectrometry studies compared to ELISA-based methods, supporting concerns about isoform-specific binding preference in ELISA (Baker-nigh et al., 2016; Rasmussen et al., 2015). Given the limited sample sizes, differing strategies in lipoprotein extraction, and varying analytical standards of these mass-spectrometry studies, future large-scale studies are needed to determine the optimal measurement technique. Ultimately, the goal would be to have reliable, reproducible, and isoform-specific assays, so that, in a person with the APOE (3/4) genotype, one could measure the amount of plasma or CSF APOE3 and APOE4, rather than just the total amount of APOE.
Bearing these caveats in mind, several large studies have now shown that plasma APOE levels vary, consistently, as a function of APOE genotype (Corsetti et al., 2016; Cruchaga et al., 2012; Khan et al., 2013; Rasmussen et al., 2015; Schiele et al., 2000). From APOE2 to APOE3 to APOE4, there tends to be a linear decline in APOE levels. The Copenhagen study group in Denmark has also shown that plasma APOE levels tend to be lower in patients with AD compared to age-matched controls, even after controlling for genotype (Rasmussen et al., 2015, 2018). That is, even among APOE4 carriers, lower plasma APOE levels are associated with greater risk of AD. As noted above, the plasma and CSF pools of APOE appear to be independent of one another, with plasma APOE derived mainly from hepatocytes, CSF and brain APOE derived mainly from astrocytes, and little to no capacity for APOE to move across the blood-brain barrier (Linton et al., 1991; Liu et al., 2012). As such, it may not be surprising that the consistent story of APOE genotype impacting APOE levels in the plasma is not replicated in human CSF studies. Some have shown a similar APOE2>APOE3>APOE4 effect on CSF APOE levels (Cruchaga et al., 2012; Toledo et al., 2014), but others have not (Bekris et al., 2008; Talwar et al., 2016). CSF APOE measurements also appear to vary with age, sex, and with the amount of beta-amyloid in the CSF (Baker-nigh et al., 2016; Bekris et al., 2008; Cruchaga et al., 2012; Darreh-shori et al., 2011; Toledo et al., 2014). A step closer to the pathological substrate, findings on brain APOE levels have also been marked by heterogeneity. As with CSF, some studies of APOE levels in brain have found the plasma-like pattern of APOE2>APOE3>APOE4 (Beffert et al., 1999; Bertrand and Maria, 1995), while others have not (Conejero-Goldberg et al., 2011; Glöckner et al., 2002). An inverse correlation between brain APOE levels and beta-amyloid has also repeatedly been observed (Beffert et al., 1999; Glöckner et al., 2002; Lambert et al., 2005; Shinohara et al., 2013). Across these studies, age, degree of pathology, and brain region studied vary considerably, which prohibits making a solid conclusion about the effect of APOE genotype on APOE levels in the brain. In summary, plasma APOE levels have consistently been reported to depend on APOE genotype, while levels in the CSF and brain tissue, likely in part due to small sample sizes, measurement issues, and other covariates, do not yet abide by a conclusive and replicable pattern across genotypes. Some factors that may contribute to APOE genotype affecting APOE levels (as in plasma) are the differential effects of APOE isoforms on receptor binding preferences, instability due to N- and C-terminal domain interaction, lipidation status, beta-amyloid sequestration, and differential clearance efficiency [for reviews, cfr. (Huang and Mahley, 2014; Huang et al., 2004b)].
APOE Pleiotropy
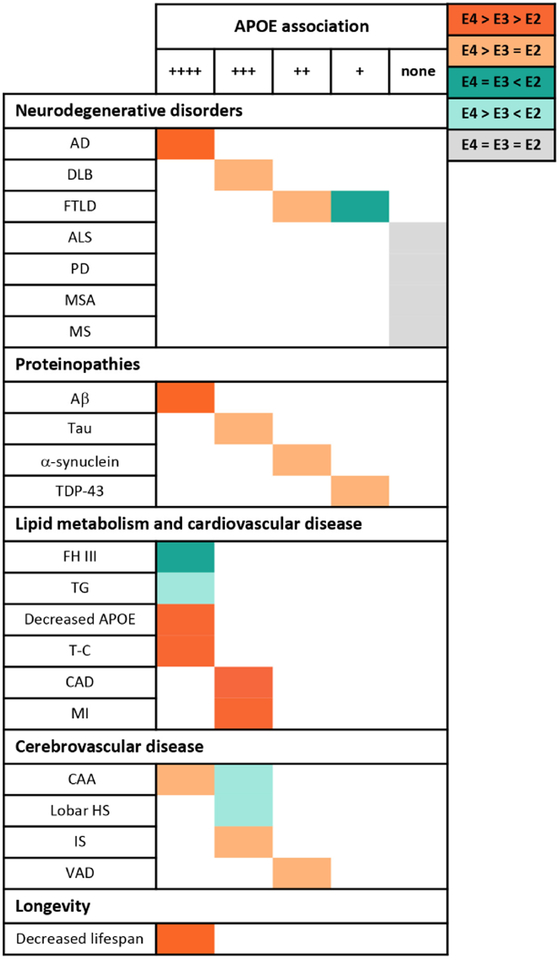
As is clear from the brief history above, APOE is not just an AD-relevant gene, but has been strongly implicated in cardiovascular disease as well. Its role in other disorders of the nervous system is less clear-cut, but association studies have been done in nearly every neurologic disorder known to humankind. The various implications of APOE isoforms across disorders is relevant to shed light on its gain or loss of function properties and to guide therapy development. In this section we will review both the firm and the less firm associations between APOE and diseases other than AD. An overview of the main association between APOE genotypes and these different pathologies is summarized in Figure 3.
Figure 3. APOE is a pleiotropic gene.
The table illustrates the relationship between the three major APOE genotypes and their association with various diseases and pathologies. Position on the matrix, from left to right, indicates the strength of association, while color marks the APOE genotype relationship. Abbreviations: AD, Alzheimer’s Disease; DLB, Dementia with Lewy Bodies; FTLD, Frontotemporal Lobar Degeneration; ALS, Amyotrophic Lateral Sclerosis; PD, Parkinson’s Disease; MSA, Multiple Systems Atrophy; MS, Multiple Sclerosis; FH III, familial hypolipoproteinemia type III; TG, Triglycerides; CAD, Coronary Artery Disease; T-C; Total Cholesterol; MI, Myocardial Infarction; CAA, Cerebral Amyloid Angiopathy; HS, Hemorrhagic Stroke; IS, Ischemic Stroke; VAD, Vascular Dementia.
Cardiovascular Disease
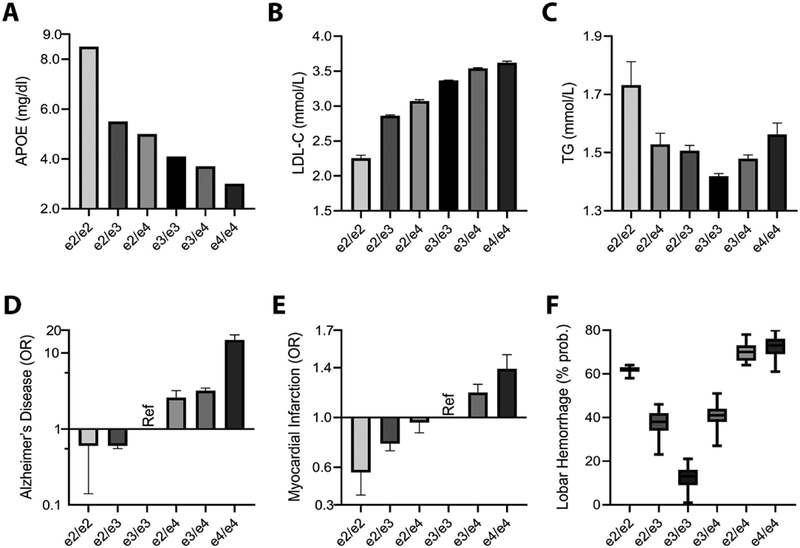
The APOE genotypes show a pronounced, step-wise effect on several, interrelated cardiovascular phenotypes. Levels of plasma APOE and, generally also, high-density lipoprotein cholesterol (HDL-C) decrease in a perfect parametric fashion across the six APOE genotypes (2/2 > 2/3 > 2/4 > 3/3 > 3/4 > 4/4; Figure 4A), and the reverse is true for levels of low-density lipoprotein cholesterol (LDL-C; Figure 4B) (Bennet et al., 2007; Khan et al., 2013; Rasmussen, 2016). A similar step-wise pattern is seen in the risk for coronary artery disease (CAD) and myocardial infarction (MI), where APOE2 carriers have the least risk, APOE4 carriers have the greatest risk, and APOE3 carriers fall in the middle [(Anand et al., 2009; Bennet et al., 2007; Wang et al., 2015; Xu et al., 2016); Figure 4E]. For comparison, the risk for AD across genotypes is shown in Figure 4D. The impact of APOE genotype on CAD is undoubtedly linked, in large part, to the effect of APOE on LDL-C and HDL-C (Goldstein and Brown, 2015). Some studies have suggested that CAD risk may be above and beyond that attributable solely to the cholesterol level changes, while others have not (Corsetti et al., 2012; Rasmussen and Tybjaerg-hansen, 2016; Ward et al., 2009). These effects are relatively strong with a single APOE4 allele increasing LDL-C by roughly 10 % and increasing the risk of CAD by 6 % (Bennet et al., 2007; Khan et al., 2013; Rasmussen, 2016). Two copies of APOE4 increase LDL-C by 20 % and the risk of CAD by 20 %. In contrast to these linear trends in HDL-C, LDL-C, and CAD, the effect of APOE genotype in triglycerides (TG) is quite distinct (Figure 4C). APOE2 carriers and APOE4 carriers are both at increased risk of hypertriglyceridemia (Bennet et al., 2007; Dallongeville et al., 1992; Huang, 2010; Marais et al., 2014). A small portion of APOE2 homozygotes, already the rarest of the six genotypes, are prone to a severe disorder of lipid metabolism called Type III hyperlipoproteinemia (OMIM#617347), characterized by extreme elevations in triglycerides and total cholesterol. These patients, in addition to their risk for early CAD, can develop changes such as palmar xanthomata (orange creases in the palms) and tuberous xanthomas (nodular lipid deposits) typically appearing on skin over the knees and elbows (Marais et al., 2014).
Figure 4. Parametric associations of APOE alleles with Alzheimer’s disease, cardiovascular traits, and cerebrovascular pathology.
Across all six combination of APOE alleles, clear parametric decreases can be observed for plasma APOE levels (A) and increases for low density lipoprotein cholesterol (LDL-C) levels [mean ± standard error of mean (SEM)] (B), while triglyceride (TG) levels are marked by a U-shape (geometric mean ± SEM) (C), indicating risk for hypertriglyceridemia in APOE2 and APOE4 carriers (Rasmussen, 2016). The parametric association of APOE genotypes with lipid traits is also reflected in the association with risk, indicated by the odds ratio (OR ± SE), for AD (Caucasians, pathology-confirmed; Farrer et al., 1997) (D) and Myocardial Infarction (MI) (Wang et al., 2015) (E). While the APOE-related risk for AD follows APOE2 > APOE3 > APOE4, cerebral amyloid angiopathy (CAA) and related risk for intracerebral lobar hemorrhage display a U-shape (% probability of cases versus controls; median ± min/max) (F), indicating increased risk for both APOE2 and APOE4 carriers (Biffi et al., 2010). In panels C, D, and F, the position of the APOE (2/4) genotype is shifted to better indicate the combined effect of the two detrimental APOE alleles. Figure panel A is a graphical representation, and B&C are adaptations, with permission, from Elsevier © Rasmussen, K. Atherosceloris. 255, 145–155 (2016). Figure panel D is a visual adaptation of data, with permission, from American Medical Association © Farrer, L. et al. JAMA. 278, 1349–1356 (1997). Figure panel E is a visual adaptation of data, under CC BY, from Wang, Y. et al. FEBS open bio. 5, 852–858 (2015). Figure panel F is a visual adaptation, with permission, from John Wiley and Sons © Biffi, A. et al. Ann. Neurol. 68, 934–943 (2010).
Cerebrovascular Disease
Given the prominent effects of APOE genotypes on cholesterol and CAD, it is not surprising that there would be parallel effects in the CNS. Indeed, the risk of ischemic stroke is roughly 30 % greater for APOE4 carriers (Wei et al., 2017). It is unclear if this increased risk is solely attributable to the risks of hypercholesterolemia and CAD (both of which predispose to stroke), or if there are additional brain-specific effects at play, for example, the role of APOE in astrocytes and pericytes at the blood-brain barrier (Bell et al., 2012; Halliday et al., 2016). The APOE2 allele does not appear to be protective against ischemic stroke. In terms of hemorrhagic stroke, the role of the APOE genotype is likely mediated in part by cerebral amyloid angiopathy (CAA, resulting from amyloid deposition in blood vessel walls). CAA predisposes to lobar hemorrhages, which are more common in APOE2 carriers and APOE4 carriers, resulting in a U-shaped curve when looking at hemorrhage risk across the six APOE genotypes [(Viswanathan and Greenberg, 2011) and Figure 4F]. While APOE2 and APOE4 both increase risk of CAA and related hemorrhage compared to APOE3, the risks are qualitatively distinct. APOE4 carriers tend to have a greater burden of amyloid in vessels, leading to microbleeds, but APOE2 carriers appear to be more prone to such vessels rupturing and causing larger hemorrhages (Biffi et al., 2011; Charidimou et al., 2015; Yu et al., 2015). This increased propensity of APOE2 carriers to hemorrhage from CAA is also supported by a recent paper demonstrating that they are much more likely to show superficial siderosis on MRI scans (Pichler et al., 2017). The importance of beta-amyloid deposits in vessels has been highlighted recently in the replicated finding that APOE4-positive AD patients treated with anti-amyloid antibodies are at increased risk for amyloid-related imaging abnormalities (ARIA). APOE4 carriers are more prone both to the edematous and hemorrhagic forms of ARIA, prompting some pharmaceutical companies to adjust the antibody dose for APOE4 carriers in these studies (Sevigny et al., 2016; Sperling et al., 2012).
Vascular Dementia
Vascular dementia (VAD) is, for lack of a better term, a squirrelly diagnosis that lacks standard pathological criteria and can be difficult to operationalize for research purposes (O’Brien and Thomas, 2015). It comes in several flavors, including post-stroke dementia, multi-infarct dementia, and small vessel ischemic disease (also called subcortical vascular dementia or, to use an older term, Binswanger’s disease). To complicate matters further, cerebrovascular disease and AD often co-occur, particularly in older patients (Attems and Jellinger, 2014; Tenenholz et al., 2010). Because APOE impacts the risk of both ischemic and hemorrhagic stroke, it is bound to have an impact on the risk for VAD. Indeed, earlier reviews and meta-analyses suggest an increased risk in APOE4 carriers (Dwyer et al., 2013; Rohn, 2014; Sun et al., 2015; Verghese et al., 2011). A more interesting question is whether, when controlling for stroke burden and other potential confounds, an APOE4 stroke patient is more likely to develop post-stroke dementia than an APOE3 stroke patient. These are challenging studies to undertake and there is a good deal of methodological variability across studies. For now, the jury is still out with several studies suggesting that APOE4 carriers are more prone to post-stroke cognitive decline and dementia (Ballard et al., 2004; Wagle et al., 2009, 2010) and several other studies suggesting no effect of APOE4 (Bour et al., 2010; Qian et al., 2012; Rowan et al., 2005).
Other Neurodegenerative Disorders
The reader will forgive us if we lump multiple sclerosis (MS) in with the neurodegenerative disorders, but this is an increasing defensible position (Calabrese et al., 2015; Eshaghi et al., 2018). Numerous studies have looked at whether the APOE4 allele increases risk for MS and it appears, on balance, that it does not (Burwick et al., 2006; Pinholt et al., 2006). This conclusion is best reflected in a very large study aptly titled “Closing the case of APOE in multiple sclerosis: No association with disease risk in over 29 000 subjects.” (Lill et al., 2012a). As with post-stroke cognitive decline, it is also reasonable to ask whether APOE4 could affect the likelihood of developing dementia due to MS. This has been looked at by a number of investigators, but these study sample sizes are uniformly small (ranging from 50 to 500 patients) and the results are fairly heterogeneous (Carmona et al., 2011; Ghaffar and Feinstein, 2010; Ghaffar et al., 2010; Koutsis et al., 2007; Shi et al., 2011). This question of cognitive decline in MS is particularly compelling in some ways because this patient population skews considerably younger than the other neurodegenerative disorders and any APOE4 effects in this age range would likely be wholly unrelated to concomitant AD or vascular pathology. Cognitive impairment in MS is a growing sub-field and, as MS-specific cognitive assessments become finer-tuned, the potential role of APOE4 will be worth revisiting in a large n study (Sumowski et al., 2018).
The genetic advances in frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS) over the last decade have been astounding (DeJesus-Hernandez et al., 2011; Renton et al., 2011). On the fringe of these major advances, there is waning interest in the role of APOE in these two often, but not always, related disorders. The verdict is in for ALS. A large meta-analysis (4k cases and 10k controls) in 2014 found no association between ALS risk and APOE genotypes (Govone et al., 2014). The story in FTLD is less straight-forward. This is due, in part, to FTLD being more challenging to diagnose accurately in-vivo compared to ALS. In addition, FTLD is now commonly sub-divided into the two main neuropathological subtypes of tau-positive or TDP-43 positive (with still more esoteric branching points below that), whereas ALS is, with the exception of rare SOD1 and FUS mutations, almost always due to TDP-43 pathology (van Es et al., 2017; Olney et al., 2017; Taylor et al., 2016). Furthermore, a clinical diagnosis of FTLD will not uncommonly be corrected at autopsy with a gold-standard pathologic diagnosis of AD. This confound, in combination with the younger age-at-onset in FTLD patients, will bias studies towards finding an APOE effect if the FTLD phenotype is not defined pathologically. A recent, multisite study of FTLD, for example, found that roughly 12% of clinically diagnosed behavioral variant frontotemporal dementia cases have AD pathology (Perry et al., 2017). Keeping these caveats in mind, three meta-analyses done over the last 20 years have come up with some divergent findings. The first found that APOE2 increased the risk of FTLD but found no effect of APOE4 while the two more recent (and larger) studies found no effect of APOE2, but did show that APOE4 increased risk of FTLD (Rubino et al., 2013; Su et al., 2017; Verpillat et al., 2002). The effect size, reflected in the odds ratio (OR), for a single APOE4 allele is considerably smaller in these two positive studies of FTLD (varying from 1.6 to 1.8) than in AD (ranging from 2 to 4). The studies collated across these meta-analyses have a mixture of clinically-defined and autopsy-confirmed phenotypes and co-mingle TDP-43 and tau pathology. The international frontotemporal dementia consortium recently completed a study that examined the role of APOE (and other loci) in the three main FTLD clinical subtypes (Mishra et al., 2017). Only 3% of cases were autopsy-confirmed. They found that APOE4 increased the risk of the behavioral, progressive non-fluent aphasia, and semantic dementia variants. The finding was strongest in the behavioral variant group (which has the largest sample size), but across all three clinical subtypes, the odds ratios here were, again, modest (from 1.3 to 1.4). It should be noted that the two language variants of FTLD are probably more likely than the behavioral variant to be caused by AD pathology at autopsy. Importantly, they also looked at frontotemporal dementia with motor neuron disease (the most reliable diagnosis given that these patients have ALS, which allows for a highly accurate clinical diagnosis) and found no association with APOE4 even in the homozygous state. A recent autopsy study of roughly 1000 subjects from the Religious Orders Study and Memory and Aging Project (ROSMAP) has made a compelling case that APOE4 drives TDP-43 pathology even after covarying for co-morbid AD or alpha-synuclein pathology, though with a moderate odds ratio hovering around 1.5 in different models that were run (Yang et al., 2018). This suggests that the finding of an APOE4 link to FTLD might be driven by the TDP-43 cases, but this runs counter to the lack of an association in ALS and frontotemporal dementia with ALS (known TDP-43opathies). The potential role of APOE4 in FTLD remains an open question to be settled soon, we suspect, once the question is asked in the growing samples of pathology-confirmed and pathology-stratified FTLD cases. As in MS, even if APOE does not impact the risk of developing FTLD it is worth considering whether it might affect the subsequent course of disease as has been suggested in both human and mouse studies (Agosta et al., 2009; Giles et al., 2017).
The strongest case for a role of APOE in non-AD neurodegenerative diseases can be made in dementia with Lewy bodies (DLB). Even more so than FTLD, the clinical diagnosis of DLB is often confounded with AD (McKeith et al., 2017). More challenging still, there is a high degree of overlap between AD and DLB pathology at post-mortem (Robinson et al., 2018; Spires-Jones et al., 2017). This is due, in part, to the strong correlation of age with each disease such that dual AD/DLB pathology is particularly prevalent in patients who die in their 80s and 90s (Spires-Jones et al., 2017). Owing to these challenges, the only means of determining a clear link between DLB and APOE would be a study done with autopsy-confirmed diagnoses. Tsuang and colleagues undertook such a daunting task and characterized subjects as pure AD (n = 244), mixed AD/DLB (n=224), pure DLB (n=91), pure Parkinson’s disease dementia (n=81) and 269 controls without significant AD or Lewy body pathology (Tsuang et al., 2013). In this multisite, tour-de-force the authors showed, convincingly, that APOE4 increases risk not just for AD (OR=9.9), but also for mixed AD/DLB (OR=12.6), pure DLB (OR=6.1), and Parkinson’s disease dementia (PDD) (OR=3.1). Another subsequent study showed that patients with both AD and Lewy Body pathology (n=215) had a higher frequency of at least one APOE4 allele compared to just AD pathology (n=316) (Chung et al., 2015). Despite this strong link to DLB, APOE has not clearly been associated with other synucleinopathies such as multiple system atrophy (MSA) or Parkinson’s disease (PD). MSA is relatively rare and to date studies examining a link between APOE and disease risk or age-at-onset have been negative (Cairns et al., 1997; Morris et al., 2000, 2001). In PD, an earlier meta-analysis suggested that APOE2 may be associated with increased risk (Huang et al., 2004a), but subsequent work further updated this meta-analysis and suggested that no clear significant associations could be determined (Williams-Gray et al., 2009). Since then, as is the case for other neurodegenerative disorders, PD has been marked by advances in genetic association analyses, culminating in the creation of the PDGene database and two large-scale meta-analyses that could not establish APOE as a significant risk locus for PD (Lill et al., 2012b; Nalls et al., 2014). As in MS, there remains the important possibility that while APOE may not increase risk for PD, it may increase the risk for dementia in PD (Aarsland et al., 2017; Collins and Williams-Gray, 2016; Monsell et al., 2014). This touches on the ongoing controversy of whether PDD and DLB are two distinct entities or one in the same thing. The authors of the current review are firmly among the lumpers, as opposed to the splitters, in viewing PDD and DLB as slightly distinct clinical disorders that merge into a single phenomenon at pathology (Jellinger and Korczyn, 2018; Langston et al., 2015). Viewed in this light, it seems likely that future studies with pathology-confirmed diagnoses will expand on the work of Tsuang et al. to show that APOE4 increases risk for PDD/DLB, in part via AD-related co-pathology, but independently as well. This association has solid basic science support as well with work in a PD mouse model showing a pronounced upregulation of APOE in response to alpha-synuclein pathology (Gallardo et al., 2008).
Longevity
Unsurprisingly—perhaps, given the strong effect on cardiovascular disease, stroke, and at least two age-related neurodegenerative disorders—APOE has consistently been reported as one of the two most robust genetic loci that associate with longevity across populations (Shadyab and Lacroix, 2015; Slagboom et al., 2018). The other locus contains the transcription factor FOXO3, a member of the forkhead box O family, which is an important regulator of homeostasis and response to cellular stress (Eijkelenboom and Burgering, 2013). Clearly, longevity is a highly pleiotropic trait, influenced by many lifestyle and environmental factors that contribute to the aging process (Christenen et al., 2006). In addition to these environmental factors, there appears to be a substantial genetic component to longevity as well, with heritability in twin-studies estimated at about 25 % (Maria et al., 1996). It did not take long after the discoveries of APOE as a risk factor for cardiovascular disease and AD, before the research community established a link between APOE and aging (Smith, 2002). Subsequent work on a Danish cohort of centenarians suggested that APOE4 exerts increasing influence on mortality across aging (Jacobsen et al., 2010). A meta-analysis of centenarian studies (4k centenarians and 7k younger controls) determined that APOE2 increased (OR=1.3) and APOE4 decreased (OR=0.6) the probability of longevity (Deelen et al., 2011). On the other hand, a large meta-analysis of GWAS of aging in the general population, from the CHARGE consortium (n = 25k), did not find any significant effects at the APOE locus (Walter et al., 2011). The mean age at death in this study was lower, however, compared to the prior two studies, due to the focus on participants from the general population rather than centenarians. This suggested that the analysis by Walter and colleagues lacked power to determine an association with longevity or that APOE may play a more important role particularly for very old age. Follow-up analysis on particularly long-lived subjects (>90 years of age, 6k cases versus 4k younger controls) of the CHARGE consortium did, indeed, identify a significant association with the APOE locus (Broer et al., 2015). The largest, to date, meta-GWAS on longevity (>90 years old, 21k cases and 77k younger controls) also established the APOE LD block as the most strongly associated genetic locus. Several other meta-analyses have consistently reproduced this finding and established the association of APOE2 with increased, and APOE4 with decreased, longevity (Garatachea et al., 2014, 2015; Revelas et al., 2018; Sebastiani et al., 2013). In terms of the size of these effects, the most recent meta-analysis found an OR of 0.4 for APOE4 and of 1.4 for APOE2 (Revelas et al., 2018). Using parental lifespan as a proxy for longevity, several recent, very large studies from the UK Biobank and Lifespan projects have also found a significant effect at the APOE locus (Joshi et al., 2016, 2017, Pilling et al., 2016, 2017).
A Spectrum of APOE Function?
Across the disorders described above, where there is a clear association with APOE, a few distinct patterns emerge (Figure 3). The first is that in AD (as in CAD, LDL-C levels, longevity, etc.), there tends to be a step-wise increase in risk from APOE2 to APOE3 to APOE4. While this pattern cannot definitively support a toxic gain- versus loss-of-function mechanism for APOE4 in AD, in our view, it tips the balance somewhat towards a loss of function. Combined with data from biomarker studies in humans, it seems that APOE4 carriers are more prone to amyloid deposition and AD than APOE3 carriers, who in turn are more prone than APOE2 carriers. If the increased risk of AD in APOE4 carriers were due to a gain of function in APOE4, we would need a separate explanation for the protective effect of APOE2 compared to APOE3. It seems more straight-forward, instead, to invoke a spectrum of decreasing protein function from APOE2 to APOE3 to APOE4. This spectrum perspective is not universally applicable, however, as illustrated by APOE’s binding to the LDL receptor, which is roughly equivalent between APOE3 and APOE4 but severely defective for APOE2 (Huang, 2010; Huang and Mahley, 2014). A second point to make is that there are at least a couple, replicable examples in non-AD disorders of a U-shaped curve across the six genotypes in which APOE2 and APOE4 carriers are both at increased risk for disease. This serves as a reminder that the general pattern of APOE2 “better” than APOE3 “better” than APOE4 does not always hold. A final point to make is that the step up in AD risk from the APOE (3/4) to (4/4) genotype is probably not linear but, rather, quadratic. This is important when considering the essentially required step of co-varying for APOE4 dose in AD genetic and biomarker studies. It is arguable that rather than simply co-varying for the number of APOE4 alleles, as is commonly done in genetic association analyses, it may be better to use all six genotypes weighted by their specific ORs, as derived from a large-sample AD meta-analysis (Darst et al., 2017).
Gleaning Biological Insights into APOE from Statistical Interactions
Cell and animal model experiments in APOE biology have, inconsistencies notwithstanding, provided some crucial insights into potential mechanisms of AD pathogenesis. These approaches have several obvious advantages pertaining to experimental control, precision, and reproducibility that we cannot hope to enjoy in human studies of APOE and AD (which are inherently noisy, time-consuming, and costly). On the other hand, human studies have the decided advantage of real-world applicability in that we are studying the disease we want to cure in the organism in which we want to cure it. While our capacity to alter biological variables in human studies is limited, we can profit from looking at natural independent variables such as ancestral background, sex, education, medications, and environmental exposures and how they affect the dependent variable of APOE-related risk for AD. Here we will focus specifically on two such independent variables: ancestral background and sex, which appear to interact strongly with APOE genotype to affect AD risk. Leveraging these interactions has the potential to reveal critical new biological insights into APOE-related AD pathogenesis.
APOE and Ancestral Background
Genetic association studies provide essential information about the association of APOE polymorphisms with a given trait. Examining the evolutionary genetics and geographical distribution of APOE alleles may provide further insights into the diverse molecular functions of this protein. The evolutionary shifts in the APOE genotypes are reflected in the wide range of genotype frequencies encountered today across different ethnic backgrounds. Considering the evolutionary changes in APOE, together with the variance in genotypes across modern-day ethnicities, will help make sense of the remarkable differences in APOE-related AD risk between different ancestral backgrounds.
Evolutionarily speaking, ancient predecessors of all apolipoproteins already existed in very early eukaryotes and have individually evolved with increasing phyla, orders and species (Babin et al., 1999). In vertebrates, APOE is widely expressed in fish, reptiles and mammals (Duggan and Callard, 2001). Consistent evidence from primates and early human DNA sequencing indicates that APOE4 is the ancestral allele, and was later substituted by APOE3 during human evolution through selective adaptation (Hanlon and Rubinsztein, 1995; Hixson et al., 1988; Mcintosh et al., 2012). APOE2 represents the most recent variation at the locus, with an approximate age of 80,000 years (Fullerton et al., 2000). As reviewed in detail by Huebbe and Rimbach, several hypotheses have been formulated about the functional reasons and potential selective pressures contributing to the evolution and the global distribution of human APOE alleles (Huebbe and Rimbach, 2017). Adaptations in APOE function likely reflect selection for early-life survival and effective reproduction within a particular environment [e.g. protection against high infection pressure (Van Exel et al., 2017)], rather than for late-life traits like AD that are more common to modern-day affluent societies. Although these selective pressures have acted over the course of millennia of human evolution, by shedding light on ethnogeographically specific, early-life survival demands, they may provide useful information to understand and establish research lines aiming to counteract the deleterious effect of APOE4 in late-onset traits. Notably, the ancestral background of APOE is also relevant to provide context for translational research in rodent models. APOE is strongly conserved between rodents and humans (78% and 70% homology at the cDNA and protein level, respectively). Both rats and mice have the same amino acid as the human APOE4 allele at position 112, confirming the ancestral origin of this allele (Rajavashisth et al., 1985). Despite this, mouse APOE functionally behaves more like human APOE3 in that it does not display the APOE4 domain interaction (as mice have a threonine rather than an arginine at the critical salt bridge position 61) (Raffaı et al., 2001). Both in vitro and in vivo, humanized mouse apoE (Arg-61), like human APOE4, binds preferentially to VLDL. In contrast, wild-type mouse apoE (Thr-61) displays an APOE3-like preference for HDL (Raffaı et al., 2001). Clearly, these differences will be critical when studying AD pathology on a mouse APOE background compared to human APOE knock-in models (which is why the latter approach is generally preferred).
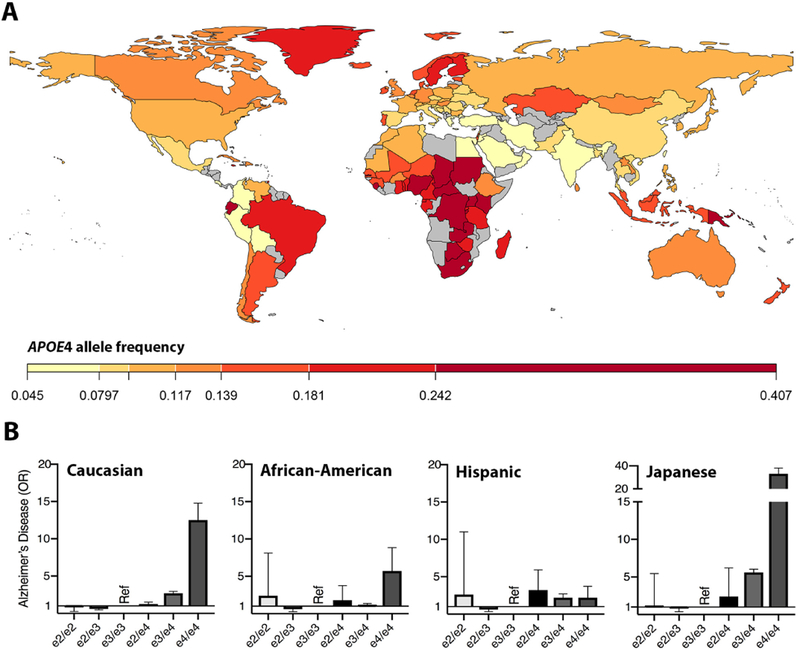
The world-wide distribution of human APOE alleles varies considerably. APOE3 is the most common in all the human populations, but at frequencies ranging from 85 % (Asia) to 69 % (Africa) (Corbo and Scacchi, 1999; Singh et al., 2006). APOE4 allele frequency is negatively correlated with APOE3 allele frequency (r=−0.97) and it is enriched in indigenous populations of Central Africa (40 %), Oceania (37 %), and Australia (26 %) (Corbo and Scacchi, 1999). The distribution across Europe and Asia follows an apparent north-to-south gradient, with the low APOE4 frequency in the Mediterranean area or South China (<10 %) and increasing in more northern regions (up to 25 %) (Egert et al., 2012; Hu et al., 2011). APOE2 is the least common major allele with a general worldwide frequency of 7.3 % and it is absent in many indigenous populations, without showing any apparent geographical trend (Corbo and Scacchi, 1999; Singh et al., 2006). Some factors that may contribute to these geographical distributions are adaptation to climate extremes and infection load [for review, cfr. (Huebbe and Rimbach, 2017)]. The wide geographical variability of APOE4 distribution can be appreciated in Figure 5A.
Figure 5. Geographical differences in APOE4 frequency and ethnic risk mitigation.
A) Worldwide APOE4 allele frequency (For methods and references, see Supplementary text). B) APOE-related risk for Alzheimer’s disease (clinically defined) across all six APOE genotypes, for Caucasians (left), African-Americans (second from left), Hispanics (second from right), and Japanese (right) patient groups (Farrer et al., 1997). Figure panel B is a visual adaptation of data, with permission, from American Medical Association © Farrer, L. et al. JAMA. 278, 1349–1356 (1997).
The three APOE polymorphisms demonstrate remarkable, ethnicity-specific differences in the risk for AD and other late-onset diseases. This point can be illustrated by examining the role of APOE genotypes in three common diseases—AD, lobar hemorrhage and CAD—across the three most studied populations (African, East-Asian and European). East-Asians seem to be the most vulnerable to the APOE4 effect in all three of these diseases (Bennet et al., 2007; Farrer et al., 1997; Tzourio et al., 2008; Zhang et al., 2014). A similar trend has been reported in regard to longevity in East-Asians versus Europeans (Garatachea et al., 2014). Conversely, despite the increased risk for AD in African-Americans (Hendrie et al., 2014) compared to European-Americans, the association of APOE4 with AD is relatively weak in African-Americans and only observable at homozygosity in Africans (Farrer et al., 1997; Hendrie et al., 2014). An early illustration of AD risk differences based on APOE genotype and ethnicity is shown in Figure 5B (Farrer et al., 1997). Similarly, APOE alleles appear to have no effect on lobar hemorrhage in African-Americans (Sawyer et al., 2018) or on CAD in Afro-Carribeans (Larifla et al., 2017). Recent efforts further demonstrate that the continuum of African ancestry in admixed African-Americans impacts the effect of APOE4 on AD and cognitive decline (Deters et al., 2018; Rajabli et al., 2018), such that APOE4 confers greater risk with decreasing percent of African ancestry. Rajalbi et al. (2018) point out that this ethnicity-specific effect may be due to local ancestry at the APOE locus. The controversial debate on AD risk due to APOE cis elements was initiated shortly after the link between APOE4 and AD was established in 1993 (Lambert et al., 1997; Templeton, 1995). While three main promoter SNPs have since been identified, they have inconsistently been associated with AD risk (Rasmussen et al., 2015; Xiao et al., 2017; Xin et al., 2010). These discrepancies may in part be due to differing ancestries across study cohorts.
The evolutionary shifts in APOE genotype provide a window on to the pleiotropic role of this protein and may shed light on its potential role not just in cholesterol metabolism, but in other functions as disparate as bone metabolism and innate immunity (Dieckmann et al., 2013; Gale et al., 2014). Elucidating the role of the three APOE genotypes in these other physiological functions should, in turn, enhance our understanding of potential mechanisms related to AD risk. The wide range of APOE-related risk across ancestral backgrounds is a potentially very rich biological vein to mine, The APOE-by-ancestral background interaction is likely hiding critical gene-gene interactions that will reveal novel protein interactions and potential drug targets. Aside from the biological insights to be gleaned, with the move towards personalized medicine, the strong variability in APOE-related risk across ancestral backgrounds is critical for clinicians to factor into their assessment of a patient’s risk of developing AD.
APOE and Sex
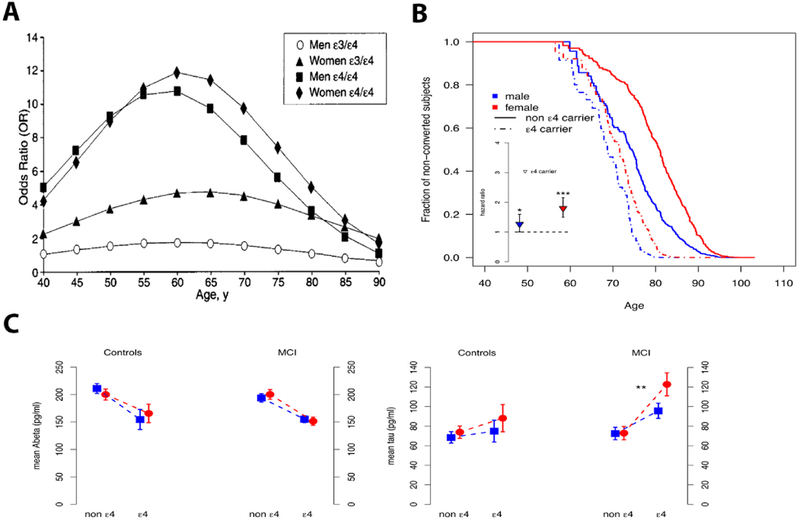
Just one year after the initial studies linking APOE4 to AD, a paper by Payami and colleagues found that the APOE4-asociated increased risk for AD was seen mainly in women (Payami et al., 1994). The plot thickened over the next two years when the Duke group could not replicate this increased risk in women (Corder et al., 1995a), and in a separate paper, pointed out, rightly, that the question is complicated by the competing risks in APOE4 carriers of death and AD (Corder et al., 1995b). Payami and colleagues returned the following year with a follow-up paper redemonstrating the APOE-by-sex interaction in a large sample of cases and controls (Payami et al., 1996). The dust settled in 1997 when Farrer and colleagues published the first, definitive meta-analysis on the role of APOE in AD, examining the APOE2 and APOE4 effects in 5k+ cases and 8k+ controls (Farrer et al., 1997). This landmark 1997 paper not only replicated the main effects of APOE4 and APOE2, but strengthened, considerably, the case for the APOE-by-sex interaction. Figure 2 in the Farrer meta-analysis is reprinted here as Figure 6A and demonstrates that among Caucasians, the OR of AD in APOE (3/4) heterozygotes (compared to APOE (3/3) homozygotes) hovers around 3–4-fold in women between the ages of 50–80 whereas the OR in APOE (3/4) men hardly moves above 1. The OR in APOE (4/4) leaps to 10 and above for men and women, but even among the homozygotes there appears to be a slightly greater effect in women. For reasons that remain obscure (to these authors in any case), this prominent APOE-by-sex interaction did not initially receive a great deal of research attention and, to this day, is still not considered very often in the clinical setting. Some small studies in the late 90s did replicate this APOE-by-sex interaction (Breitner et al., 1999; Bretsky et al., 1999), but overall the importance of this interaction seemed not to take hold for another 10–15 years.
Figure 6. Sex interacts with APOE to affect risk of Alzheimer’s disease, clinical decline and biomarker levels.
A) Risk for Alzheimer’s disease based on APOE genotype, stratified by male and female sex (Farrer et al., 1997). B) Risk of clinical decline, defined as conversion from healthy controls to mild cognitive impairment (MCI) or Alzheimer’s disease (AD), across the age range, stratified by sex. Inset shows the hazard ratio for conversion as determined for each sex independently, marking higher risk in women (Altmann et al., 2014). * p<0.05; *** p<0.001. C) In MCI patients, the APOE4 allele’s effect on increasing tau levels was significantly greater in women than in men. [Analysis was adjusted for age and education; blue squares correspond to men and red circles to women]. Panels display levels of CSF Abeta (left) and tau (right). ** p<0.01. Figure panel A is a reprint, with permission, from American Medical Association © Farrer, L. et al. JAMA. 278, 1349–1356 (1997). Figure panel B&C are reprints, with permission, from John Wiley and Sons © Altmann, A. et al. Ann. Neurol. 75, 563–573 (2014).
Interest in sex-based differences in disease has grown considerably over the last several years as reflected by, and, in part due to, the National Institute of Health’s insistence on considering the role of sex as a biological variable (National Institutes of health, 2015). We undertook a review of the interaction between sex and APOE 5 years ago, that covered animal model work and human studies (Ungar et al., 2014). One of the most notable findings, expanded upon in that review, pertained to mouse models of AD. Historically, mouse model studies in AD and most disorders, have tended to ignore female mice so as not to have to contend with the prominent effects of the estrus cycle. ApoE4 mouse models stood out in this regard in that, from the earliest studies, investigators found that the AD-relevant effects of these apoE mouse models were more obvious in female mice and so tended to focus on them (Raber et al., 1998). In other words, the APOE-by-sex interaction was found to play a prominent role in apoE mouse models and, happily, in the same direction (increasing APOE4 effects in females).
Regarding work in humans, the last 5 years have seen a number of studies published supporting the hypothesis that the APOE4 risk of AD is greater in women than in men. Our group, moving beyond the case-control analysis approach, used longitudinal data to examine the APOE-by-sex interaction on the risk of converting from healthy aging to cognitive impairment. As shown in Figure 6B, the APOE4-related increase in risk of conversion was significantly greater in women than in men (Altmann et al., 2014). This study then examined the same interaction on spinal fluid levels of beta-amyloid and tau (biomarkers of AD) and found that the APOE4 allele increased the risk for abnormal amyloid in men and women equally, but an APOE4 effect on tau was more prominent in women (Figure 6C). Revisiting the case-control approach, Neu et al. recently undertook a massive meta-analysis (combined cases and controls > 50k) and found that, in the 65–75 age range, the APOE4 effect was significantly stronger in women than in men (Neu et al., 2017). A recent study by Hohman and colleagues has replicated, in a larger sample, our spinal fluid biomarker findings showing that the APOE4 effect on beta-amyloid is strong and similar in men and women, but that the APOE4 effect on tau is greater in women (Hohman et al., 2018). The spinal fluid data from our study and that of Hohman and colleagues can be interpreted in (at least) two ways. It is possible that the APOE4 effect on amyloid is equivalent between men and women and that, for a given amount of amyloid pathology, APOE4 has a greater effect on tau pathology in women. Alternatively, it may be that the greater APOE4 effect on tau pathology in women is indirect and due to the fact that APOE4 drives amyloid pathology at an earlier age in women than in men. This latter possibility strikes us as more likely and cannot be resolved until more biomarker data is acquired in men and women in their early 50s when, as shown in Figure 2, amyloid pathology first becomes detectable in APOE4 carriers (Morris et al., 2010).
Two decades of research in humans (and mouse models) support the hypothesis that the APOE4 effect on AD risk is greater in women than in men. Investigators have only begun to scratch the surface of why this might be the case. Some evidence suggests that APOE expression is impacted by estradiol (Lambert et al., 2004; Levin-allerhand et al., 2001; Srivastava et al., 2008; Stone et al., 1997; Struble et al., 2003). In this age of enormous AD GWAS meta-analyses, the role of the X-chromosome variants remains essentially unexplored (note that the Manhattan plots in all the large AD GWAS papers only show chromosomes 1–22) (Lambert et al., 2013; Marioni et al., 2018). In addition to these hormonal and genetic possibilities, there are myriad environmental factors that likely play into the APOE-by-sex interaction, including, but not limited to, sex-based differences in education, head trauma, and all-cause mortality.
Gain or loss of function?
A quarter century after the initial studies linking APOE4 to AD, the most fundamental question remains unanswered: is APOE4 intrinsically bad or just not as good as APOE3 (which is not as good as APOE2)? This question looms particularly large now that clinical neuroscience is making remarkable headway with gene-targeting therapies. Prior animal and cell work has contributed substantially to this issue and stimulated the development of various therapies aimed at APOE (Huang and Mucke, 2012; Michaelson, 2014; Yu et al., 2014; Zhao et al., 2018). Some of these studies pointed out that the mere presence of APOE, regardless of isoform, may drive AD pathology, suggesting it may even be better to not have APOE at all (Bien-Ly et al., 2012; Giles et al., 2017; Huynh et al., 2017b; Kim et al., 2011). Clearly, investigating this condition in human studies is highly complex due to the rarity of APOE-deficient patients and the lack of critical experimental controls in human genetic research. Nonetheless, in the first descriptions of APOE-deficient patients, no clinical symptoms referable to the nervous system were evident (Ghiselli et al., 1981; Lohse et al., 1992a), which motivated the authors’ conclusion that APOE may be of minimal physiological importance beyond its role in peripheral lipid metabolism (Lohse et al., 1992b). Critical limitations to these early case reports include the fact that they appeared before the link between APOE and AD had been formed and the patients described are generally quite young when considering the possibility of AD-relevant phenotypes.
A more recent and far more compelling case report that touches on this critical question also highlights the many challenges inherent in human APOE studies. Mak and colleagues have described a 40 year-old African-American patient with severe hyperlipidemia due to compound heterozygosity for ablative, frameshift mutations in APOE (Mak et al., 2014). In addition to a cardiovascular assessment, the patient agreed to spinal fluid analysis which showed normal levels of amyloid and tau. Structural brain imaging was reportedly normal and without evidence of vascular disease. The patient’s cognitive assessment was complicated in that he had a history of presumed dyslexia, but also showed substantial deficits across multiple non-language domains, including memory and visuospatial skills, though these deficits were felt to be longstanding and stable. In a single, potentially invaluable case study, with the capability to inform the field as to whether the lack of APOE increases the risk of AD, we are faced with all the challenges related to APOE research. The patient is about 10 years younger than when one begins to see amyloid abnormalities in healthy controls with APOE4 [(Morris et al., 2010), Figure 2]. He has an ancestral background which likely mitigates the effects of APOE dysfunction on AD risk [(Deters et al., 2018; Farrer et al., 1997; Hendrie et al., 2014), Figure 5B]. His cognitive impairment could reflect an important role of APOE in neural development or could reflect amyloid-independent effects of APOE and prodromal AD pathogenesis (Conejero-Goldberg et al., 2011; Shi and Holtzman, 2018; Yu et al., 2014). It is also pointed out, in subsequent scientific correspondence related to the cognitive impairment (Malloy et al., 2015), that the patient was “from a socially deprived environment and had profound disadvantages during childhood”, raising the complex issue of gene-environment interactions. It would be most informative, of course, to repeat this assessment when the patient is 80, but he is at high risk of premature coronary artery disease and, in fact, already has some suggestive findings on stress testing, which will likely limit his longevity (Bennet et al., 2007; Joshi et al., 2017; Slagboom et al., 2018). If he were assessed at 75 or 80, were cognitively stable, and showed no evidence for abnormal amyloid, one could conclude that APOE deficiency does not drive AD pathogenesis. This would be concordant with recent human IPSC-derived neuron work showing that APOE-null cell phenotypes were similar to APOE (3/3) cell phenotypes (Wang et al., 2018). Such an outcome would provide good, albeit indirect, evidence that it would be safe, from a brain standpoint at least, to try knocking down APOE4 to prevent or slow AD in APOE4 carriers. From a systemic standpoint, however, given that peripheral APOE deficiency causes a profound dyslipidemia and increases risk for CAD (Marais et al., 2014), one would need either to consider CSF-specific approaches (intrathecal ASO-based therapy for example) or to monitor and correct any treatment-associated dyslipidemia.. On the other hand, it is, in our view, equally likely that if he were reassessed at 50, he might then have evidence of amyloid abnormalities on spinal fluid assessment and perhaps some worsening of his cognitive impairment. This outcome would, of course, suggest that APOE deficiency is as bad, or worse, than APOE4 homozygosity and that, perhaps, APOE4’s AD-related risk is due to reduced function that could be mitigated by increasing its expression.
The Path forward
We do not intend to end this review, however, on a note of hopeless complexity. On the contrary, in many ways, the case study described above points the way forward and showcases the remarkable tools now at our disposal to solve the APOE/AD puzzle. APOE is pleiotropic and as neurologists and neuroscientists we will do well to collaborate closely with colleagues in cardiovascular research and lipid metabolism who have an additional decade of experience studying APOE. The ever-widening availability of next-generation sequencing and AD biomarkers means that these sorts of rare case studies will slowly expand into rare case series. A quick look at the exome data available in the Alzheimer’s disease sequencing project, for example, revealed two healthy heterozygous carriers of APOE loss-of-function mutations both above 85 years old and one with the APOE (3/4) genotype. At least two centers, including ours, are building a cohort of deeply-phenotyped and whole-genome-sequenced, “protected” or “resilient” APOE4 carriers who remain cognitively healthy into their 80s and beyond. The expectation is that people with this extreme phenotype harbor rare, protective genetic variants that will offer critical insights into APOE4-related pathogenesis and, in turn, present novel targets for drug development. Additional promising approaches will explore the biology underlying the prominent APOE-by-ancestry and APOE-by-sex interactions on AD risk. Further, while not the focus of this review, there is no question that increasingly sophisticated cell-based and animal-model APOE research will continue to complement human studies. The best approaches will continue to be those that move back and forth between human findings and experimental models as the latter are critical in helping to validate and characterize (or refute and discard) candidate genetic variants. There is, undoubtedly, work to be done, but the tools are now at hand to take full advantage of the APOE lever and create a drug that eradicates AD.
Supplementary Material
Acknowledgements
This work was supported in part by the National Institutes of Health (R01 AG060747; P50 AG047366) and the Asad Jamal Center for Cognitive Health in Aging. We thank Katherine Laura Rasmussen, Alessandro Biffi, and Jonathan Rosand for sharing data used in figures. We thank Yongha Kim and Grace Tam for their careful review of this article.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
The authors declare no competing interests.
References
- Aarsland D, Creese B, Politis M, Chaudhuri KR, Ffytche DH, Weintraub D, and Ballard C (2017). Cognitive decline in Parkinson disease. Nat. Rev. Neurol 13, 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, and Krieg AM (2017). FDA Approves Eteplirsen for Duchenne Muscular Dystrophy: The Next Chapter in the Eteplirsen Saga. Nucleic Acid Ther. 27, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D, Gonzalez-Duarte A, O’Riordan WD, Yang C-C, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk JL, et al. (2018). Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med 379, 11–21. [DOI] [PubMed] [Google Scholar]
- Agosta F, Vossel KA, Miller BL, Migliaccio R, Bonasera SJ, Filippi M, Boxer AL, Karydas A, Possin KL, and Gorno-Tempini ML (2009). Apolipoprotein E ε4 is associated with disease-specific effects on brain atrophy in Alzheimer’s disease and frontotemporal dementia. Proc. Natl. Acad. Sci 106, 2018–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann A, Tian L, and Henderson VW (2014). Sex Modifies the APOE-Related Risk of Developing Alzheimer Disease. Am. Neurol. Assoc 75, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand SS, Xie C, Paré G, Montpetit A, Rangarajan S, McQueen MJ, Cordell HJ, Keavney B, Yusuf S, Hudson TJ, et al. (2009). Genetic variants associated with myocardial infarction risk factors in over 8000 individuals from five ethnic groups the INTERHEART genetics study. Circ. Cardiovasc. Genet 2, 16–25. [DOI] [PubMed] [Google Scholar]
- Attems J, and Jellinger KA (2014). The overlap between vascular disease and Alzheimer’s disease - lessons from pathology. BMC Med. 12, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babin PJ, Bogerd J, Kooiman FP, Van Marrewijk WJA, and Van Der Horst DJ (1999). Apolipophorin II / I, Apolipoprotein B, Vitellogenin, and Microsomal Triglyceride Transfer Protein Genes Are Derived from a Common Ancestor. J Mol Evol 49, 150–160. [DOI] [PubMed] [Google Scholar]
- Baker-nigh AT, Mawuenyega KG, Bollinger JG, Ovod V, and Kasten T (2016). Human Central Nervous System (CNS) ApoE Isoforms are Increased by Age, Differentially Altered by Amyloidosis, and Relative Amounts Reversed in the CNS Compared to Plasma. J. Biol. Chem 291, 27204–27218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard CG, Morris CM, Rao H, O’Brien JT, Barber R, Stephens S, Rowan E, Gibson A, Kalaria RN, and Kenny RA (2004). APOE ε4 and cognitive decline in older stroke patients with early cognitive impairment. Neurology 63, 1399–1402. [DOI] [PubMed] [Google Scholar]
- Beffert U, Cohn JS, Petit-Turcotte C, Tremblay M, Aumont N, Ramassamy C, Davignon J, and Poirier J (1999). Apolipoprotein E and b-amyloid levels in the hippocampus and frontal cortex of Alzheimer’s disease subjects are disease-related and apolipoprotein E genotype dependent. Brain Res. 843, 87–94. [DOI] [PubMed] [Google Scholar]
- Bekris LM, Millard SP, Galloway NM, Vuletic S, Albers JJ, Li G, Galasko DR, DeCarli C, Farlow MR, Clark CM, et al. (2008). Multiple SNPs within and surrounding the apolipoprotein E gene influence cerebrospinal fluid apolipoprotein E protein levels. J. Alzheimer’s Dis 13, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RD, Winkler EA, Singh I, Sagare AP, Deane R, Wu Z, Holtzman DM, Betsholtz C, Armulik A, Sallstorm J, et al. (2012). Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature 485, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennet AM, Angelantonio E. Di, Ahlbom A, Keavney B, Collins R, Wiman B, de Faire U, and Danesh J (2007). Association of Apolipoprotein E Genotypes With Lipid Levels and Coronary Risk. JAMA 298, 1300–1311. [DOI] [PubMed] [Google Scholar]
- Benson MD, Waddington-Cruz M, Berk JL, Polydefkis M, Dyck PJ, Wang AK, Planté-Bordeneuve V, Barroso FA, Merlini G, Obici L, et al. (2018). Inotersen Treatment for Patients with Hereditary Transthyretin Amyloidosis. N. Engl. J. Med 379, 22–31. [DOI] [PubMed] [Google Scholar]
- Bertrand P, and Maria G (1995). Association of apolipoprotein E genotype with brain levels of apolipoprotein E and apolipoprotein J (clusterin) in Alzheimer disease. Mol. Brain Res 33, 174–178. [DOI] [PubMed] [Google Scholar]
- Bien-Ly N, Gillespie AK, Walker D, Yoon SY, and Huang Y (2012). Reducing Human Apolipoprotein E Levels Attenuates Age-Dependent Aβ Accumulation in Mutant Human Amyloid Precursor Protein Transgenic Mice. J. Neurosci 32, 4803–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A, Sonni A, Anderson CD, Kissela B, Jagiella JM, Schmidt H, Jimenez-Conde J, Hansen BM, Fernandez-Cadenas I, Cortellini L, et al. (2010). Variants at APOE influence risk of deep and lobar intracerebral hemorrhage. Ann. Neurol 68, 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A, Anderson CD, Jagiella JM, Schmidt H, Kissela B, Hansen BM, and Jimenez-Conde J (2011). APOE Genotype Predicts Extent of Bleeding and Outcome in Lobar Intracerebral Hemorrhage. Lancet Neurol. 10, 702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour AMJJ, Rasquin SMC, Baars L, Van Boxtel MPJ, Visser PJ, Limburg M, and Verhey FRJ (2010). The effect of the APOE-e4 allele and ACE-I/D polymorphism on cognition during a two-year follow-up in first-ever stroke patients. Dement. Geriatr. Cogn. Disord 29, 534–542. [DOI] [PubMed] [Google Scholar]
- Breitner JCS, Wyse BW, Anthony JC, Welsh-Bohmer KA, Steffens DC, Norton MC, Tschanz JT, Plassman BL, Meyer MR, Skoog I, et al. (1999). APOE-e4 count predicts age when prevalence of AD increases, then declines: The Cache County Study. Neurology 53, 321–336. [DOI] [PubMed] [Google Scholar]
- Bretsky PM, Buckwalter JG, Seeman TE, Miller CA, Poirier J, Schellenberg GD, Finch CE, and Henderson VW (1999). Evidence for an interaction between apolipoprotein E Genotype, Gender, and Alzheimer disease. Alzheimer Dis. Assoc. Disord 13, 216–221. [DOI] [PubMed] [Google Scholar]
- Broer L, Buchman AS, Deelen J, Evans DS, Faul JD, Lunetta KL, Sebastiani P, Smith JA, Smith AV, Tanaka T, et al. (2015). GWAS of Longevity in CHARGE Consortium Confirms APOE and FOX3 Candidacy. J. Gerontol. Med. Sci 70, 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwick RM, Ramsay PP, Haines JL, Hauser SL, Oksenberg JR, Pericak-Vance MA, Schmidt S, Compston A, Sawcer S, Cittadella R, et al. (2006). APOE epsilon variation in multiple sclerosis susceptibility and disease severity: Some answers. Neurology 66, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Cairns NJ, Atkinson PF, Kovács T, Lees AJ, Daniel SE, and Lantos PL (1997). Apolipoprotein E e4 allele frequency in patients with multiple system atrophy. Neurosci. Lett 221, 161–164. [DOI] [PubMed] [Google Scholar]
- Calabrese M, Magliozzi R, Ciccarelli O, Geurts JJG, Reynolds R, and Martin R (2015). Exploring the origins of grey matter damage in multiple sclerosis. Nat. Rev. Neurosci 16, 147–158. [DOI] [PubMed] [Google Scholar]
- Carmona O, Masuet C, Santiago O, Alía P, Moral E, Alonso-Magdalena L, Casado V, and Arbizu T (2011). Multiple sclerosis and cognitive decline: Is ApoE-4 a surrogate marker? Acta Neurol. Scand 124, 258–263. [DOI] [PubMed] [Google Scholar]
- Castellano JM, Kim J, Stewart FR, Jiang H, Demattos RB, Patterson BW, Fagan AM, Morris JC, Kwasi G, Cruchaga C, et al. (2011). Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci. Transl. Med 3, 89ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charidimou A, Martinez-Ramirez S, Shoamanesh A, Oliveira-Filho J, Frosch M, Vashkevich A, Ayres A, Rosand J, Gurol ME, Greenberg SM, et al. (2015). Cerebral amyloid angiopathy with and without hemorrhage. Neurology 84, 1206–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier-Harlin M-C, Parfitt M, Legrain S, Pérez-tur J, Brousseau T, Evans A, Berr C, Vldal O, Roques P, Gourlet V, et al. (1994). Apolipoprotein E, e4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer’s disease: analysis of the 19q13.2 chromosomal region. Hum. Mol. Genet 3, 569–574. [DOI] [PubMed] [Google Scholar]
- Christenen K, Johnson TE, and Vaupel JW (2006). The quest for genetic determinants of human longevity: challenges and insights. Nat. Rev. Genet 7, 436–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EJ, Babulal GM, Monsell SE, Cairns NJ, Roe CM, and Morris JC (2015). Clinical features of Alzheimer disease with and without Lewy bodies. JAMA Neurol. 72, 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins LM, and Williams-Gray CH (2016). The genetic basis of cognitive impairment and dementia in parkinson’s disease. Front. Psychiatry 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conejero-Goldberg C, Hyde T, Chen S, Dreses-Werringloer U, Herman M, Kleinman J, Davies P, and Goldberg T (2011). Molecular signatures in post-mortem brain tissue of younger individuals at high risk for Alzheimer’s disease as based on APOE genotype. Mol. Psychiatry 16, 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppus AMW, Evenhuis HM, Verberne GJ, Visser FE, Arias-Vasquez A, Sayed-Tabatabaei FA, Vergeer-Drop J, Eikelenboom P, van Gool WA, and van Duijn CM (2008). The impact of apolipoprotein E on dementia in persons with Down’s syndrome. Neurobiol. Aging 29, 828–835. [DOI] [PubMed] [Google Scholar]
- Corbo RM, and Scacchi R (1999). Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a ‘thrifty’ allele? Ann Hum Genet 63, 301–310. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, and Pericak-Vance MA (1993). Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s Disease in Late Onset Families. Science (80-.). 261, 921–923. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC Jr., Rimmler JB, Locke PA, Conneally PM, Schmader KE, et al. (1994). Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet 7, 180–184. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Rloses AD, Pericak-Vance MA, Small GW, Haines JL, Payami H, et al. (1995a). The Apolipoprotein E E4 allele and sex-specific risk of Alzheimer’s Disease. JAMA Lett. to Ed 273, 373–374. [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Rimmler JB, Locke PA, Conneally PM, Schmader KE, Tanzi RE, et al. (1995b). Apolipoprotein E, survival in Alzheimer’s disease patients, and the competing risks of death and Alzheimer’s disease. Neurology 45, 1323–1328. [DOI] [PubMed] [Google Scholar]
- Corsetti JP, Gansevoort RT, Bakker SJL, Navis G, Sparks CE, and Dullaart RPF (2012). Apolipoprotein e predicts incident cardiovascular disease risk in women but not in men with concurrently high levels of high-density lipoprotein cholesterol and C-reactive protein. Metabolism. 61, 996–1002. [DOI] [PubMed] [Google Scholar]
- Corsetti JP, Gansevoort RT, Bakker SJL, and Dullaart RPF (2016). Apolipoprotein E levels and apolipoprotein E genotypes in incident cardiovascular disease risk in subjects of the Prevention of Renal and Vascular End-stage disease study. J. Clin. Lipidol 10, 842–850. [DOI] [PubMed] [Google Scholar]
- Cruchaga C, Nowotny P, Kauwe JSK, Ridge PG, Mayo K, Bertelsen S, Hinrichs A, Fagan AM, Holtzman DM, Morris JC, et al. (2011). Association and Expression Analyses With Single-Nucleotide Polymorphisms in TOMM40 in Alzheimer Disease. Arch Neurol 68, 1013–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C, Kauwe JSK, Nowotny P, Bales K, Pickering EH, Mayo K, Bertelsen S, Hinrichs A, Initiative TADN, Fagan AM, et al. (2012). Cerebrospinal fluid APOE levels: an endophenotype for genetic studies for Alzheimer’s disease. Hum. Mol. Genet 21, 4558–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallongeville J, Lussier-Cacan S, and Davignon J (1992). Modulation of plasma triglyceride levels by apoE phenotype: a meta-analysis. J. Lipid Res. 33, 447–454. [PubMed] [Google Scholar]
- Darreh-shori T, Modiri N, Blennow K, Baza S, Kamil C, Ahmed H, Andreasen N, and Nordberg A (2011). The apolipoprotein E e4 allele plays pathological roles in AD through high protein expression and interaction with butyrylcholinesterase. Neurobiogy of Aging 32, 1236–1248. [DOI] [PubMed] [Google Scholar]
- Darst BF, Koscik RL, Racine AM, Oh JM, Krause RA, Carlsson CM, Zetterberg H, Blennow K, Christian BT, Bendlin BB, et al. (2017). Pathway-specific polygenic risk scores as predictors of β-amyloid deposition and cognitive function in a sample at increased risk for Alzheimer’s disease. J. Alzheimer’s Dis 55, 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen J, Beekman M, Uh H, Helmer Q, Kuningas M, Kremer D, van der Bergen R, Suchiman HED, Lakenberg N, Van Den Akker EB, et al. (2011). Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell 10, 686–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Gilmer F, Adamson J, et al. (2011). Expanded GGGGCC hexanucleotide repeat in non-coding region of C9ORF72 causes chromosome 9p-linked frontotemporal dementia and amyotrophic lateral sclerosis. Neuron 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deters K, Napolioni V, Greicius MD, and Mormino BC (2018). African ancestry moderates the effect of APOE4 on cognitive decline. In Alzheimer’s & Dementia, pp. P1027–P1028. [Google Scholar]
- Dickson T, Saunders H, and Vickers J (1997). Relationship between apolipoprotein E and the amyloid deposits and dystrophic neurites in Alzheimer’s disease. Neuropathol. Appl. Neurobiol 23. [DOI] [PubMed] [Google Scholar]
- Dieckmann M, Beil FT, Muelle B, Bartelt A, Marshall RP, Koehne T, Amling M, Ruether W, Cooper JA, Humphries SE, et al. (2013). Human Apolipoprotein E Isoforms Differentially affect Bone Mass and Turnover in Vivo. J. Bone Miner. Res 28, 234–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietschy J, and Turley S (2011). Cholesterol metabolism in the brain. Curr. Opin. Lipidol 12, 105–112. [DOI] [PubMed] [Google Scholar]
- Duggan AE, and Callard IANP (2001). Phylogenetic Distribution of Apolipoproteins A-I and E in Vertebrates as Determined by Western Blot Analysis. J. Exp. Zool 290, 255–264. [DOI] [PubMed] [Google Scholar]
- Van Duijn CM, de Knijff P, Cruts M, Wehnert A, Havekes LM, Hofman A, and Van Broeckhoven C (1994). Apolipoprotein E4 allele in a population–based study of early–onset Alzheimer’s disease. Nat. Genet 7, 74–78. [DOI] [PubMed] [Google Scholar]
- Dwyer R, Skrobot OA, Dwyer J, Munafo M, and Kehoe PG (2013). Using Alzgene-like approaches to investigate susceptibility genes for vascular cognitive impairment. J. Alzheimer’s Dis 34, 145–154. [DOI] [PubMed] [Google Scholar]
- Egert S, Rimbach G, and Huebbe P (2012). ApoE genotype: from geographic distribution to function and responsiveness to dietary factors. Proc. Nutr. Soc 71, 410–424. [DOI] [PubMed] [Google Scholar]
- Eijkelenboom A, and Burgering BMT (2013). FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol 14, 83–97. [DOI] [PubMed] [Google Scholar]
- Elshourbagy NA, Liao WS, Mahley RW, and Taylor JM (1985). Apolipoprotein E mRNA is abundant in the brain and adrenals, as well as in the liver, and is present in other peripheral tissues of rats and marmosets. Proc. Natl. Acad. Sci. USA 82, 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es MA, Hardiman O, Chio A, Al-Chalabi A, Pasterkamp RJ, Veldink JH, and van den Berg LH (2017). Amyotrophic lateral sclerosis. Lancet 390, 2084–2098. [DOI] [PubMed] [Google Scholar]
- Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, Badarinarayan N, Morgan K, Passmore P, Holmes C, et al. (2015). Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain 138, 3673–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi A, Marinescu RV, Young AL, Firth NC, Prados F, Jorge Cardoso M, Tur C, De Angelis F, Cawley N, Brownlee WJ, et al. (2018). Progression of regional grey matter atrophy in multiple sclerosis. Brain 141, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Exel E, Koopman JJE, Van Bodegom D, Meij JJ, De Knijff P, Ziem JB, Finch CE, and Westendorp RGJ (2017). Effect of APOE ε 4 allele on survival and fertility in an adverse environment. PLoS One 12, e017949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cuppples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-vance MA, Risch N, van Duijn CM, et al. (1997). Effects of Age, Sex, and Ethnicity on the Association Between Apolipoprotein E Genotype and Alzheimer Disease. JAMA 278, 1349–1356. [PubMed] [Google Scholar]
- Fleisher AS, Chen K, Liu X, Ayutyanont N, Roontiva A, Thiyyagura P, Protas H, Joshi AD, Sabbagh M, Sadowsky CH, et al. (2013). Apolipoprotein E ε4 and age effects on florbetapir positron emission tomography in healthy aging and Alzheimer disease. Neurobiol. Aging 34, 1–12. [DOI] [PubMed] [Google Scholar]
- Fullerton SM, Clark AG, Weiss KM, Nickerson DA, Taylor SL, Stenga JH, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, et al. (2000). Apolipoprotein E Variation at the Sequence Haplotype Level: Implications for the Origin and Maintenance of a Major Human Polymorphism. Am J Hum Genet 67, 881–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale SC, Gao L, Mikacenic C, Coyle SM, Rafaels N, Murray Dudenkov T, Madenspacher JH, Draper DW, Ge W, Aloor JJ, et al. (2014). APOε4 is associated with enhanced in vivo innate immune responses in human subjects. J. Allergy Clin. Immunol 134, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo G, Schlüter OM, and Südhof TC (2008). A molecular pathway of neurodegeneration linking α-synuclein to ApoE and Aβ peptides. Nat. Neurosci 11, 301–308. [DOI] [PubMed] [Google Scholar]
- Garatachea N, Emanuele E, Calero M, Fuku N, Arai Y, Abe Y, Murakami H, Miyachi M, Yvert T, Verde Z, et al. (2014). ApoE gene and exceptional longevity: Insights from three independent cohorts. Exp. Gerontol 53, 16–23. [DOI] [PubMed] [Google Scholar]
- Garatachea N, Marín PJ, Santos-Lozano A, Sanchis-Gomar F, Emanuele E, and Lucia A (2015). The ApoE Gene Is Related with Exceptional Longevity: A Systematic Review and Meta-Analysis. Rejuvenation Res. 18, 3–13. [DOI] [PubMed] [Google Scholar]
- Ghaffar O, and Feinstein A (2010). APOE epsilon4 and cognitive dysfunction in multiple sclerosis: a review. J Neuropsychiatry Clin Neurosci 22, 155–165. [DOI] [PubMed] [Google Scholar]
- Ghaffar O, Reis M, Pennell N, O’Connor P, and Feinstein A (2010). APOE ε4 and the cognitive genetics of multiple sclerosis. Neurology 74, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli G, Schaefer EJ, Gascon P, and Brewer HB (1981). Type III Hyperlipoproteinemia Associated with Apolipoprotein E Deficiency. Science (80-). 214, 1239–1241. [DOI] [PubMed] [Google Scholar]
- Giles DA, Moreno-fernandez ME, Stankiewicz TE, Graspeuntner S, Cappelletti M, Wu D, Mukherjee R, Chan CC, Lawson MJ, Klarquist J, et al. (2017). APOE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner F, Meske V, and Ohm TG (2002). Genotype-related differences of hippocampal apolipoprotein E levels only in early stages of neuropathological changes in Alzheimer’s disease. Neuroscience 114, 1103–1114. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, and Brown MS (2015). A century of cholesterol and coronaries: From plaques to genes to statins. Cell 161, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govone F, Vacca A, Rubino E, Gai A, Boschi S, Gentile S, Orsi L, Pinessi L, and Rainero I (2014). Lack of association between APOE gene polymorphisms and amyotrophic lateral sclerosis: A comprehensive meta-analysis. Amyotroph. Lateral Scler. Front. Degener 15, 551–556. [DOI] [PubMed] [Google Scholar]
- Halliday MR, Rege SV, Ma Q, Zhao Z, Miller CA, Winkler EA, and Zlokovic BV (2016). Accelerated pericyte degeneration and blood – brain barrier breakdown in apolipoprotein E4 carriers with Alzheimer’s disease. J. Cereb. Blood Flow Metab 36, 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon CS, and Rubinsztein DC (1995). Arginine residues at codons 112 and 158 in the apolipoprotein E gene correspond to the ancestral state in humans. Atherosclerosis 112, 85–90. [DOI] [PubMed] [Google Scholar]
- Havel RJ, and Kane JP (1973). Primary dysbetalipoproteinemia: predominance of a specific apoprotein species in triglyceride-rich lipoproteins. Proc Natl Acad Sci U S A 70, 2015–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrie HC, Murrell J, Baiyewu O, Lane KA, Purnell C, Ogunniyi A, Unverzagt FW, Hall K, Callahan CM, Saykin AJ, et al. (2014). APOE ε4 and the risk for Alzheimer disease and cognitive decline in African American and Yoruba. Int. Psychogeriatrics 26, 977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J, and Bock HH (2002). Lipoprotein Receptors in the Nervous System. Annu. Rev. Biochem 71, 405–434. [DOI] [PubMed] [Google Scholar]
- Hixson JE, Cox LA, and Borenstein S (1988). The Baboon Apolipoprotein E Gene: Structure, Expression, Linkage with the Gene for Apolipoprotein C-I. Genomics 2, 315–323. [DOI] [PubMed] [Google Scholar]
- Hohman TJ, Dumitrescu L, Barnes LL, Thambisetty M, Beecham G, Kunkle B, Gifford KA, Bush WS, Chibnik LB, Mukherjee S, et al. (2018). Sex-Specific Assocation of Apolipoprotein E With Cerebrospinal Fluid Levels of Tau. JAMA Neurol. 75, 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Qin YH, Jing CX, Lu L, Hu B, and Du PF (2011). Does the geographical gradient of ApoE4 allele exist in China? A systemic comparison among multiple Chinese populations. Mol Biol Rep 38, 489–494. [DOI] [PubMed] [Google Scholar]
- Huang Y (2010). Mechanisms linking apolipoprotein E isoforms with cardiovascular and neurological diseases. Curr. Opin. Lipidol 21, 337–345. [DOI] [PubMed] [Google Scholar]
- Huang Y, and Mahley RW (2014). Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol. Dis 72, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, and Mucke L (2012). Alzheimer mechanisms and therapeutic strategies. Cell 148, 1204–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Chen PC, and Poole C (2004a). APOE-ε2 allele associated with higher pevalence of sporadic Parkinson disease. Neurology 62, 2198–2202. [DOI] [PubMed] [Google Scholar]
- Huang Y, Weisgraber KH, Mucke L, and Mahley RW (2004b). Apolipoprotein E: Diversity of cellular origins, structural and biophysical properties, and effects in Alzheimer’s disease. J. Mol. Neurosci 23, 189–204. [DOI] [PubMed] [Google Scholar]
- Huang YA, Zhou B, Wernig M, Su TC, Huang YA, Zhou B, Wernig M, and Su TC (2017). ApoE2, ApoE3 and ApoE4 Differentially Stimulate APP Transcription and A-beta Secretion. Cell 168, 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebbe P, and Rimbach G (2017). Evolution of human apolipoprotein E (APOE) isoforms: Gene structure, protein function and interaction with dietary factors. Ageing Res. Rev 37, 146–161. [DOI] [PubMed] [Google Scholar]
- Huynh T-PV, Davis AA, Ulrich JD, and Holtzman DM (2017a). Apolipoprotein E and Alzheimer’s disease: the influence of apolipoprotein E on amyloid-β and other amyloidogenic proteins. J. Lipid Res 58, 824–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TV, Liao F, Francis CM, Ulrich JD, Cole T, Holtzman DM, Huynh TV, Liao F, Francis CM, Robinson GO, et al. (2017b). Age-Dependent Effects of apoE Reduction Using Antisense Oligonucleotides in a Model of beta-amyloidosis. Neuron 96, 1013–1023.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen R, Martinussen T, Christiansen L, Jeune B, Vaupel JW, and Christensen K (2010). Increased effect of the ApoE gene on survival at advanced age in healthy and long-lived Danes: two nationwide cohort studies. Aging Cell 9, 1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellinger KA, and Korczyn AD (2018). Are dementia with Lewy bodies and Parkinson’s disease dementia the same disease? BMC Med. 16, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PK, Fischer K, Schraut KE, Campbell H, Esko T, and Wilson JF (2016). Variants near CHRNA3/5 and APOE have age- and sex-related effects on human lifespan. Nat. Commun 7, 11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PK, Pirastu N, Kentistou KA, Fischer K, Hofer E, Schraut KE, Clark DW, Nutile T, Barnes CLK, Timmers PRHJ, et al. (2017). Genome-wide meta-analysis associates HLA-DQA1/DRB1 and LPA and lifestyle factors with human longevity. Nat. Commun 8, 910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Vardarajan BN, Buros J, Yu CE, Hawk MV, Dombroski BA, Crane PK, Larson EB, Mayeux R, Haines JL, et al. (2012). Comprehensive search for Alzheimer disease susceptibility loci in the APOE region. Arch. Neurol 69, 1270–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-shaul H, Spinrad A, Weiner A, Colonna M, Schwartz M, Amit I, Keren-shaul H, Spinrad A, Weiner A, Matcovitch-natan O, et al. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- Khan TA, Shah T, Prieto D, Zhang W, Price J, Fowkes GR, Cooper J, Talmud PJ, Humphries SE, Sundstrom J, et al. (2013). Apolipoprotein E genotype, cardiovascular biomarkers and risk of stroke: Systematic review and meta-analysis of 14015 stroke cases and pooled analysis of primary biomarker data from up to 60883 individuals. Int. J. Epidemiol 42, 475–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Basak JM, and Holtzman DM (2009). The Role of Apolipoprotein E in Alzheimer’s Disease. Neuron 63, 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jiang H, Park S, Eltorai AEM, Stewart FR, Yoon H, Basak JM, Finn MB, and Holtzman DM (2011). Haploinsufficiency of Human APOE Reduces Amyloid Deposition in a Mouse Model of Amyloid-  Amyloidosis. J. Neurosci 31, 18007–18012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kockx M, Traini M, and Kritharides L (2018). Cell-specific production, secretion, and function of apolipoprotein E. J. Mol. Med 96, 361–371. [DOI] [PubMed] [Google Scholar]
- Koldamova R, Staufenbiel M, and Lefterov I (2005). Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice. J. Biol. Chem 280, 43224–43235. [DOI] [PubMed] [Google Scholar]
- Koutsis G, Panas M, Giogkaraki E, Potagas C, Karadima G, Sfagos C, and Vassilopoulos D (2007). APOE ε4 is associated with impaired verbal learning in patients with MS. Neurology 68, 546–549. [DOI] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, et al. (2017). The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566–581.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J-C, Pérez-Tur J, Dupire MJ, Galasko D, Mann D, Amouyel P, Hardy J, Delacourte A, and Chartier-Harlin MC (1997). Distortion of allelic expression of apolipoprotein E in Alzheimer’s disease. Hum. Mol. Genet 6, 2151–2154. [DOI] [PubMed] [Google Scholar]
- Lambert J, Coyle N, Lendon C, and Elizabeth Q (2004). The allelic modulation of apolipoprotein E expression by oestrogen: potential relevance for Alzheimer’s disease. J Med Genet 41, 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J, Mann D, Richard F, Tian J, Shi J, Thaker U, Merrot S, Harris J, Frigard B, Iwatsubo T, et al. (2005). Is there a relation between APOE expression and brain amyloid load in Alzheimer’s disease ? J Neurol Neurosurg Psychiatry 76, 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, DeStefano AL, Bis JC, Beecham GW, et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane-Donovan C, Philips GT, and Herz J (2014). More than Cholesterol Transporters: Lipoprotein Receptors in CNS Function and Neurodegeneration. Neuron 83, 771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston JW, Schüle B, Rees L, Nichols RJ, and Barlow C (2015). Multisystem Lewy body disease and the other parkinsonian disorders. Nat. Genet 47, 1378–1384. [DOI] [PubMed] [Google Scholar]
- Larifla L, Armand C, Bangou J, Blanchet-Deverly A, Numeric P, Fonteau C, Michel CT, Ferdinand S, Bourrhis V, and Vélayoudom-Céphise FL (2017). Association of APOE gene polymorphism with lipid profile and coronary artery disease in Afro-Caribbeans. PLoS One 12, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-allerhand J, Mcewen BS, Lominska CE, Lubahn DB, Korach KS, and Smith JD (2001). Brain region-specific up-regulation of mouse apolipoprotein E by pharmacological estrogen treatments. J. Neurochem 79, 796–803. [DOI] [PubMed] [Google Scholar]
- Li X, An Z, Zhao Y, Wu X, Li X, Jiang L, Gui X, Liu Y, Sun Y, and Zhu B (2018). TREM2 Is a Receptor for beta-Amyloid that Mediates Microglial Function. Neuron 97, 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill CM, Liu T, Schjeide BMM, Roehr JT, Akkad DA, Damotte V, Alcina A, Ortiz MA, Arroyo R, de Lapuente AL, et al. (2012a). Closing the case of APOE in multiple sclerosis: No association with disease risk in over 29 000 subjects. J. Med. Genet 49, 558–562. [DOI] [PubMed] [Google Scholar]
- Lill CM, Roehr JT, McQueen MB, Kavvoura FK, Bagade S, Schjeide BMM, Schjeide LM, Meissner E, Zauft U, Allen NC, et al. (2012b). Comprehensive research synopsis and systematic meta-analyses in Parkinson’s disease genetics: The PDgene database. PLoS Genet. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Seo J, Gao F, Feldman HM, Wen H, Penney J, Cam HP, Gjonseka E, Raja WK, Cheng J, et al. (2018). APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron 98, 1141–1154.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton MF, Gish R, Hubl ST, Butler E, Esquivel C, Bry W, Janet K, Wardell MR, and Young SG (1991). Phenotypes of Apolipoprotein B and Apolipoprotein E after Liver Transplantation. J. Clin. Invest 88, 270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kanekiyo T, Xu H, and Bu G (2013). Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Kuhel DG, Shen L, Hui DY, and Woods SC (2012). Apolipoprotein E does not cross the blood-cerebrospinal fluid barrier, as revealed by an improved technique for sampling CSF from mice. Am J Physiol Regul Integr Comp Physiol 303, 903–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse P, Brewer HB, Meng MS, Skarlatos SI, Larosa JC, and Brewer HB (1992a). Familial apolipoprotein E deficiency and type Ill hyperlipoproteinemia due to a premature stop codon in the apolipoprotein E gene. J. Lipid Res 33, 1583–1590. [PubMed] [Google Scholar]
- Lohse P, Brewer HB, Meng MS, Skarlatos SI, Larosa JC, and Jr HBB (1992b). Familial apolipoprotein E deficiency and type Ill hyperlipoproteinemia due to a premature stop codon in the apolipoprotein E gene. J. Lipid Res 33, 1583–1590. [PubMed] [Google Scholar]
- Mahley RW (1998). Apolipoprotein E: Cholesterol Transport Protein with Expanding Role in Cell Biology. Science (80-.). 240, 622–630. [DOI] [PubMed] [Google Scholar]
- Mahley RW, and Rall SC (2000). Apolipoprotein E: Far More Than a Lipid Transport Protein. Annu. Rev. Genomics Hum. Genet 2000 01, 507–537. [DOI] [PubMed] [Google Scholar]
- Mak ACY, Pullinger CR, Tang LF, Wong JS, Deo RC, Schwarz JM, Gugliucci A, Movsesyan I, Ishida BY, Chu C, et al. (2014). Effects of the absence of apolipoprotein E on lipoproteins, neurocognitive function, and retinal function. JAMA Neurol. 71, 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malloy M, Miller B, and Kane J (2015). Apolipoprotein E and neurocognitive function: In reply. JAMA Neurol. 72, 479. [DOI] [PubMed] [Google Scholar]
- Marais AD, Solomon GAE, and Blom DJ (2014). Dysbetalipoproteinaemia: A mixed hyperlipidaemia of remnant lipoproteins due to mutations in apolipoprotein E. Crit. Rev. Clin. Lab. Sci 51, 46–62. [DOI] [PubMed] [Google Scholar]
- Maria A, Matthew H, Holm NV, Sørensen TIA, Harvald B, and Vaupel JW (1996). The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870 – 1900.Hum. Genet 97, 319–323. [DOI] [PubMed] [Google Scholar]
- Marioni RE, Harris SE, Zhang Q, Mcrae AF, Hagenaars SP, Hill WD, Davies G, Ritchie CW, Gale CR, Starr JM, et al. (2018). GWAS on family history of Alzheimer’s disease. Transl. Psychiatry 8, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mart E, Nielsen HM, Batruch I, Drabovich AP, Begcevic I, Lopez MF, Minthon L, Bu G, Mattsson N, Portelius E, et al. (2014). Assessment of Peptide Chemical Modifications on the Development of an Accurate and Precise Multiplex Selected Reaction Monitoring Assay for Apolipoprotein E Isoforms. J. Proteome Res 13, 1077–1087. [DOI] [PubMed] [Google Scholar]
- Martínez E, and Oskar M (2014). Total apolipoprotein E levels and specific isoform composition in cerebrospinal fluid and plasma from Alzheimer’s disease patients and controls. Acta Neuropathol. 127, 633–643. [DOI] [PubMed] [Google Scholar]
- Mcintosh AM, Bennett C, Dickson D, Anestis SF, Watts DP, Webster TH, Fontenot MB, and Bradley BJ (2012). The Apolipoprotein E (APOE) Gene Appears Functionally Monomorphic in Chimpanzees (Pan troglodytes). PLoS One 7, e47760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeith IG, Boeve BF, Dickson DW, Halliday G, Taylor J-P, Weintraub D, Aarsland D, Calvin J, Attems J, Ballard CG, et al. (2017). Diagnosis and management of dementia with Lewy bodies Fourth consensus report of the DLB Consortium. Neurology 89, 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, Connolly AM, Iannaccone ST, Kirschner J, Kuntz NL, Saito K, et al. (2018b). Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N. Engl. J. Med 378, 625–635. [DOI] [PubMed] [Google Scholar]
- Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, Connolly AM, Iannaccone ST, Kirschner J, Kuntz NL, Saito K, et al. (2018a). Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy. N. Engl. J. Med 378, 625–635. [DOI] [PubMed] [Google Scholar]
- Michaelson DM (2014). APOE ε 4: The most prevalent yet understudied risk factor for Alzheimer’s disease. Alzheimer’s Dement. 10, 861–868. [DOI] [PubMed] [Google Scholar]
- Miller TM, Pestronk A, David W, Rothstein J, Simpson E, Appel SH, Andres PL, Mahoney K, Allred P, Alexander K, et al. (2013). An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: A phase 1, randomised, first-in-man study. Lancet Neurol. 12, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Ferrari R, Heutink P, Hardy J, and Pijnenburg Y (2017). Gene-based association studies report genetic links for clinical subtypes of frontotemporal dementia. Brain 1–10. [DOI] [PubMed] [Google Scholar]
- Monsell SE, Besser LM, Heller KB, Checkoway H, Litvan I, and Kukull WA (2014). Clinical and pathologic presentation in Parkinson’s disease by Apolipoprotein e4 allele status. Park. Relat Disord 20, 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris HR, Vaughan JR, and Datta SR (2000). Multiple System Atrophy and Progressive Supranuclear Palsy: α-Synuclein, synphilin, tau, and APOE. Neurology 55, 1918–1920. [DOI] [PubMed] [Google Scholar]
- Morris HR, Schrag A, Nath U, Burn D, Quinn NP, Daniel S, Wood NW, and Lees AJ (2001). Effect of ApoE and tau on age of onset of progressive supranuclear palsy and multiple system atrophy. Neurosci. Lett 312, 118–120. [DOI] [PubMed] [Google Scholar]
- Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, and Mintun MA (2010). APOE predicts Amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann. Neurol 67, 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell JR, Price BM, Baiyewu O, Gureje O, Deeg M, Hendrie H, Ogunniyi A, and Hall K (2006). The fourth Apolipoprotein E haplotype found in the Yoruba of Ibadan. Am J Med Genet B Neuropsychiatr Genet 141B, 426–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, et al. (2014). Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet 46, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba Y, Tomonaga M, Kawasaki H, Otomo E, and Ikeda K (1991). Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer’s disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 541, 163–166. [DOI] [PubMed] [Google Scholar]
- National Institutes of health (2015). Consideration of sex as a biological variable in NIH-funded research. [Google Scholar]
- Neu SC, Pa J, Kukull W, Beekly D, Kuzma A, Gangadharan P, Wang L, Romero K, Arneric SP, Redolfi A, et al. (2017). Apolipoprotein E Genotype and Sex Risk Factors for Alzheimer Disease A Meta-analysis. JAMA Neurol. 74, 1178–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien JT, and Thomas A (2015). Vascular dementia. Lancet 386, 1698–1706. [DOI] [PubMed] [Google Scholar]
- Olney NT, Spina S, and Miller BL (2017). Frontotemporal Dementia. Lancet 4, 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payami H, Montee KR, Kaye JA, Bird TD, Yu C-E, Wijsman EM, and Schellenberg GD (1994). Alzheimer’s Disease, Apolipoprotein E4, and Gender. JAMA Lett. to Ed 271, 1316–1317. [PubMed] [Google Scholar]
- Payami H, Zareparsi S, Montee KR, Sexton GJ, Kaye JA, Bird TD, Yu C, Wijsman EM, Heston LL, Litt M, et al. (1996). Gender Difference in Apolipoprotein E-Associated Risk for Familial Alzheimer Disease: A Possible Clue to the Higher Incidence of Alzheimer Disease in Women. Am J Hum Genet 58, 803–811. [PMC free article] [PubMed] [Google Scholar]
- Perry DC, Brown JA, Possin KL, Datta S, Trujillo A, Radke A, Karydas A, Kornak J, Sias AC, Rabinovici GD, et al. (2017). Clinicopathological correlations in behavioural variant frontotemporal dementia. Brain 140, 3329–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persico AM, D’Agruma L, Zelante L, Militerni R, Bravaccio C, Schneider C, Melmed R, Trillo S, Montecchi F, Elia M, et al. (2004). Enhanced APOE2 transmission rates in families with autistic probands. Psychiatr. Genet 14, 73–82. [DOI] [PubMed] [Google Scholar]
- Pfrieger FW (2003). Role of cholesterol in synapse formation and function. Biochim. Biophys. Acta - Biomembr. 1610, 271–280. [DOI] [PubMed] [Google Scholar]
- Pichler M, Vemuri P, Rabinstein AA, Aakre J, Flemming KD, Brown RD, Kumar N, Kantarci K, Kremers W, Mielke MM, et al. (2017). Prevalence and Natural History of Superficial Siderosis: A Population-Based Study. Stroke 48, 3210–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling LC, Atkins JL, Bowman K, Jones SE, Tyrrell J, Beaumont RN, Ruth KS, Tuke MA, Yaghootkar H, Wood AR, et al. (2016). Human longevity is influenced by many genetic variants: evidence from 75000 UK Biobank participants. Aging (Albany. NY). 8, 547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling LC, Kuo CL, Sicinski K, Tamosauskaite J, Kuchel GA, Harries W, Herd P, Wallace R, Ferrucci L, and Melzer D (2017). Human longevity: 25 genetic loci associated in 389,166 UK biobank participants. Aging (Albany. NY). 9, 2504–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenova AA, Ray T, and Goate AM (2018). Untangling Genetic Risk for Alzheimer’s Disease. Biol. Psychiatry 83, 300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinholt M, Frederiksen JL, and Christiansen M (2006). The association between apolipoprotein E and multiple sclerosis. Eur. J. Neurol 13, 573–580. [DOI] [PubMed] [Google Scholar]
- Qian L, Ding L, Cheng L, Zhu X, Zhao H, Jin J, Guan D, Zhang B, Chen X, and Xu Y (2012). Early biomarkers for post-stroke cognitive impairment. J. Neurol 259, 2111–2118. [DOI] [PubMed] [Google Scholar]
- Raber J, Wong D, Buttini M, Orth M, Bellosta S, Pitas RE, Mahley RW, and Mucke L (1998). Isoform-specific effects of human apolipoprotein E on brain function revealed in ApoE knockout mice: Increased susceptibility of females. Proc. Natl. Acad. Sci. USA 95, 10914–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaı RL, Dong L, Farese RV, and Weisgraber KH (2001). Introduction of human apolipoprotein E4 ‘“domain interaction”‘ into mouse apolipoprotein E. Proc. Natl. Acad. Sci. U. S. A 98, 11587–11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabli F, Vance JM, Feliciano-Astacio BE, Celis K, Adams LD, Hamilton-Nelson KL, Wang L, Nuytemans K, Whitehead PL, Hofmann NK, et al. (2018). Identifying a protective variant that lowers the risk for developing AD in APOE-E4 carriers. In Alzheimer’s & Dementia, p. P1028. [Google Scholar]
- Rajavashisth TB, Kaprein JS, Reue KL, and Lusis AJ (1985). Evolution of apolipoprotein E: Mouse sequence and evidence for an 11-nucleotide ancestral unit. Proc. Natl. Acad. Sci. USA 82, 8085–8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rall SC, Weisgraber KH, and Mahley RW (1982). Human apolipoprotein E the complete amino acid sequence. J. Biol. Chem 257, 4171–4178. [PubMed] [Google Scholar]
- Rasmussen KL (2016). Plasma levels of apolipoprotein E, APOE genotype and risk of dementia and ischemic heart disease: A review. Atherosclerosis 255, 145–155. [DOI] [PubMed] [Google Scholar]
- Rasmussen KL, and Tybjaerg-hansen A (2016). Data on plasma levels of apolipoprotein E, correlations with lipids and lipoproteins stratified by APOE genotype, and risk of ischemic heart disease. Data Br. 6, 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KL, Tybjaerg-Hansen A, Nordestgaard BG, and Frikke-Schmidt R (2015). Plasma levels of apolipoprotein E and risk of dementia in the general population. Ann. Neurol 77, 301–311. [DOI] [PubMed] [Google Scholar]
- Rasmussen KL, Tybjaerg-hansen A, and Nordestgaard BG (2018). Plasma apolipoprotein E levels and risk of dementia: A Mendelian randomization study of 106,562 individuals. Alzheimer’s Dement. 14, 71–80. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Caselli RJ, Chen K, Alexander GE, Bandy D, and Frost J (2001). Declining brain activity in cognitively normal apolipoprotein E e4 heterozygotes: A foundation for using positron emission tomography to efficiently test treatments to prevent Alzheimer’s disease. Proc. Natl. Acad. Sci 98, 3334–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revelas M, Thalamuthu A, Oldmeadow C, Evans TJ, Armstrong NJ, Kwok JB, Brodaty H, Schofield PR, Scott RJ, Sachdev PS, et al. (2018). Review and meta-analysis of genetic polymorphisms associated with exceptional human longevity. Mech. Ageing Dev 175, 24–34. [DOI] [PubMed] [Google Scholar]
- Rinaldi C, and Wood MJA (2018). Antisense oligonucleotides: The next frontier for treatment of neurological disorders. Nat. Rev. Neurol 14, 9–22. [DOI] [PubMed] [Google Scholar]
- Robinson JL, Lee EB, Xie SX, Rennert L, Suh E, Bredenberg C, Caswell C, Van Deerlin VM, Yan N, Yousef A, et al. (2018). Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain 141, 2181–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohn TT (2014). Is apolipoprotein E4 an important risk factor for vascular dementia? Int. J. Clin. Exp. Pathol 7, 3504–3511. [PMC free article] [PubMed] [Google Scholar]
- Roses AD (1996). Apolipoprotein E Alleles As Risk Factors in Alzheimer’s Disease. Annu. Rev. Med 47, 387–400. [DOI] [PubMed] [Google Scholar]
- Roses A, Sundseth S, Saunders A, Gottschalk W, Burns D, and Lutz M (2016). Understanding the genetics of APOE and TOMM40 and role of mitochondrial structure and function in clinical pharmacology of Alzheimer’s disease. Alzheimer’s Dement. 12, 687–694. [DOI] [PubMed] [Google Scholar]
- Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS, Huentelman MJ, Welsh-Bohmer KA, and Reiman EM (2010). A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease. Pharmacogenomics J. 10, 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roses AD, Lutz MW, Saunders AM, Goldgaber D, Saul R, Sundseth SS, Akkari PA, Roses SM, Gottschalk WK, Whitfield KE, et al. (2014). African-American TOMM40’523-APOE haplotypes are admixture of West African and Caucasian alleles. Alzheimer’s Dement. 10, 592–601. [DOI] [PubMed] [Google Scholar]
- Rowan E, Morris CM, Stephens S, Ballard C, Dickinson H, Rao H, Saxby BK, McLaren AT, Kalaria RN, and Kenny RA (2005). Impact of hypertension and apolipoprotein E4 on poststroke cognition in subjects >75 years of age. Stroke 36, 1864–1868. [DOI] [PubMed] [Google Scholar]
- Royston MC, Mann D, Pickering-Brown S, Owen F, Perry R, Ragbavan R, Khin-Nu C, Tyner S, Day K, Crook R, et al. (1996). ApoE2 allele, Down’s syndrome, and dementia. Ann. N. Y. Acad. Sci 777, 255–259. [DOI] [PubMed] [Google Scholar]
- Rubino E, Vacca A, Govone F, De Martino P, Pinessi L, and Rainero I (2013). Apolipoprotein e polymorphisms in frontotemporal lobar degeneration: A meta-analysis. Alzheimer’s Dement. 9, 706–713. [DOI] [PubMed] [Google Scholar]
- Saura J, Petegnief V, Wu X, Liang Y, and Paul SM (2003). Microglial apolipoprotein E and astroglial apolipoprotein J expression in vitro: Opposite effects of lipopolysaccharide. J. Neurochem 85, 1455–1467. [DOI] [PubMed] [Google Scholar]
- Sawyer RP, Sekar P, Osborne J, Kittner SJ, Moomaw CJ, Flaherty ML, Langefeld CD, Anderson CD, Rosand J, and Woo D (2018). Racial/ethnic variation of APOE alleles for lobar intracerebral hemorrhage. Neurology 0, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiele F, Bacquer D. De, Vincent-viry M, Beisiegel U, and Ehnholm C (2000). Apolipoprotein E serum concentration and polymorphism in six European countries: the ApoEurope Project. Atherosclerosis 152, 475–488. [DOI] [PubMed] [Google Scholar]
- Sebastiani P, Bae H, Sun FX, Andersen SL, Daw EW, Kojima T, Hirose N, Schupf N, Puca A, and Perls TT (2013). Meta - analysis of genetic variants associated with human exceptional longevity. Aging (Albany. NY). 5, 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seripa D, Matera MG, Daniele A, Bizzarro A, Rinaldi M, Gravina C, Bisceglia L, Corbo RM, Panza F, Solfrizzi V, et al. (2007). The missing ApoE allele. Ann. Hum. Genet 71, 496–500. [DOI] [PubMed] [Google Scholar]
- Sevigny J, Chiao P, Bussière T, Weinreb PH, Williams L, Maier M, Dunstan R, Salloway S, Chen T, Ling Y, et al. (2016). The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nat. Publ. Gr 537, 50–56. [DOI] [PubMed] [Google Scholar]
- Shadyab AH, and Lacroix AZ (2015). Genetic factors associated with longevity: A review of recent findings. Ageing Res. Rev 19, 1–7. [DOI] [PubMed] [Google Scholar]
- Shi Y, and Holtzman DM (2018). Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat. Rev. Immunol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Tu JL, Gale SD, Baxter L, Vollmer TL, Campagnolo DI, Tyry TM, Zhuang Y, and Kuniyoshi SM (2011). APOE ε4 is associated with exacerbation of cognitive decline in patients with multiple sclerosis. Cogn. Behav. Neurol 24, 128–133. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Petersen RC, Dickson DW, and Bu G (2013). Brain regional correlation of amyloid-β with synapses and apolipoprotein E in non-demented individuals: potential mechanisms underlying regional vulnerability to amyloid-β accumulation. Acta Neuropathol. 125, 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugart J (2017). What Price Success? Ionis Drug Worked in Phase 3 but Had Serious Side Effects. [Google Scholar]
- Singh PP, Singh M, and Mastana SS (2006). APOE distribution in world populations with new data from India and the UK. Ann. Hum. Biol 33, 279–308. [DOI] [PubMed] [Google Scholar]
- Slagboom PE, Van Den Berg, N., and Deelen J (2018). Phenome and genome based studies into human ageing and longevity: An overview. BBA - Mol. Basis Dis 1864, 2742–2751. [DOI] [PubMed] [Google Scholar]
- Smith JD (2002). Apolipoproteins and aging: emerging mechanisms. Ageing Res. Rev 1, 345–365. [DOI] [PubMed] [Google Scholar]
- Sorbi S, Nacmias B, Forleo P, Piacentini S, Latorraca S, and Amaducci L (1995). Epistatic effect of APP717 mutation and apolipoprotein E genotype in familial Alzheimer’s disease. Ann. Neurol 38, 124–127. [DOI] [PubMed] [Google Scholar]
- Sperling R, Salloway S, Brooks DJ, Tampieri D, Barakos J, Fox NC, Raskind M, Sabbagh M, Honig LS, Porsteinsson AP, et al. (2012). Amyloid-related imaging abnormalities in patients with Alzheimer’s disease treated with bapineuzumab: a retrospective analysis. Lancet Neurol. 11, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spires-Jones TL, Attems J, and Thal DR (2017). Interactions of pathological proteins in neurodegenerative diseases. Acta Neuropathol. 134, 187–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava N, Averna M, and Srivastava R (2008). Dietary cholesterol and estrogen administration elevate brain apolipoprotein E in mice by different mechanisms. Indian J Biochem Biophys. 45, 410–415. [PubMed] [Google Scholar]
- Stone DJ, Rozovsky I, Morgan TE, Anderson CP, Hajian H, and Finch CE (1997). Astrocytes and microglia respond to estrogen with increased apoE mRNA in vivo and in vitro. Exp. Neurol 143, 313–318. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, and Roses AD (1993). Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci 90, 1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struble RG, Rosario ER, Kircher ML, Ludwig SM, Mcadamis PJ, Watabe K, Mcasey ME, Cady C, and Nathan BP (2003). Regionally specific modulation of brain apolipoprotein E in the mouse during the estrous cycle and by exogenous 17β estradiol. Exp. Neurol 183, 638–644. [DOI] [PubMed] [Google Scholar]
- Su W-H, Shi Z-H, Liu S-L, Wang X-D, Liu S, and Ji Y (2017). Updated meta-analysis of the role of APOE ε2/ε3/ε4 alleles in frontotemporal lobar degeneration. Oncotarget 8, 43721–43732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumowski JF, Benedict R, Enzinger C, Filippi M, Geurts JJ, Hamalainen P, Hulst H, Inglese M, Leavitt VM, Rocca MA, et al. (2018). Cognition in multiple sclerosis: State of the field and priorities for the future. Neurology 90, 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JH, Tan L, Wang HF, Tan MS, Tan L, Li JQ, Xu W, Zhu XC, Jiang T, and Yu JT (2015). Genetics of vascular dementia: Systematic review and meta-analysis. J. Alzheimer’s Dis 46, 611–629. [DOI] [PubMed] [Google Scholar]
- Talwar P, Sinha J, Grover S, Agarwal R, Kushwaha S, Srivastava MVP, and Kukreti R (2016). Meta-analysis of apolipoprotein E levels in the cerebrospinal fluid of patients with Alzheimer’s disease. J. Neurol. Sci 360, 179–187. [DOI] [PubMed] [Google Scholar]
- Taylor JP, Jr RHB, and Cleveland DW (2016). Decoding ALS: from genes to mechanism. Nature 539, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR (1995). A Cladistic Analysis of Phenotypic Associations with Haplotypes Inferred from Restriction Endonuclease Mapping or DNA Sequencing. V. Analysis of Case/Control Sampling Designs: Alzheimer’s Disease and the Apoprotein E Locus. Genetics 140, 403–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenholz L, Dietmar G, and Thal R (2010). Vascular pathology in the aged human brain. Acta Neuropathol. 119, 277–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium (2015). A global reference for human genetic variation. Nature 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo JB, Da X, Weiner MW, Wolk DA, Xie SX, Arnold SE, Davatzikos C, Shaw LM, Trojanowski JQ, and For the Alzheimer’s Disease Neuroimaging Initiative (2014). CSF Apo-E levels associate with cognitive decline and MRI changes. Acta Neuropathol. 127, 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang D, Leverenz JB, Lopez OL, Hamilton RL, Bennett DA, Schneider JA, Buchman AS, Larson EB, Crane PK, Kaye JA, et al. (2013). APOE ε4 increases risk for dementia in pure synucleinopathies. JAMA Neurol. 70, 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio C, Arima H, Harrap S, Anderson C, Godin O, Woodward M, Neal B, Bousser MG, Chalmers J, Cambien F, et al. (2008). APOE genotype, ethnicity, and the risk of cerebral hemorrhage. Neurology 70, 1322–1328. [DOI] [PubMed] [Google Scholar]
- Uchihara T, Duyckaerts C, He Y, Kobayashi K, Seilhean D, Amouyel P, and Hauw JJ (1995). ApoE immunoreactivity and microglial cells in Alzheimer’s disease brain. Neurosci. Lett 195, 5–8. [DOI] [PubMed] [Google Scholar]
- Ungar L, Altmann A, and Greicius MD (2014). Apolipoprotein E, gender, and Alzheimer’s disease: an overlooked, but potent and promising interaction. Brain Imaging Behav. 8, 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utermann G, Jaeschke M, and Menzel J (1975). Familial Hyperlipproteinemia type III: Deficiency of a specific apolipoprotein (APO E-III) in the very-low density lipoproteins. FEBS Lett. 56, 352–355. [DOI] [PubMed] [Google Scholar]
- Vélez JI, Lopera F, Sepulveda-Falla D, Patel HR, Johar AS, Chuah A, Tobón C, Rivera D, Villegas A, Cai Y, et al. (2016). APOE*E2 allele delays age of onset in PSEN1 E280A Alzheimer’s disease. Mol. Psychiatry 21, 916–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese PB, Castellano JM, and Holtzman DM (2011). Roles of Apolipoprotein E in Alzheimer’s Disease and Other Neurological Disorders. Lancet Neurol. 10, 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese PB, Castellano JM, Garai K, Wang Y, Jiang H, Shah A, Bu G, Frieden C, and Holtzman DM (2013). ApoE influences amyloid-β (Aβ) clearance despite minimal apoE/Aβ association in physiological conditions. Proc Natl Acad Sci U. S. A 110, E1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpillat P, Camuzat A, Hannequin D, Thomas-Anterion C, Puel M, Belliard S, Dubois B, Didic M, Lacomblez L, Moreaud O, et al. (2002). Apolipoprotein E gene in frontotemporal dementia: An association study and meta-analysis. Eur. J. Hum. Genet 10, 399–405. [DOI] [PubMed] [Google Scholar]
- Viswanathan A, and Greenberg SM (2011). Cerebral Amyloid Angiopathy in the Elderly. Ann. Neurol 70, 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle J, Farner L, Flekkøy K, Wyller TB, Sandvik L, Eiklid KL, Fure B, Stensrød B, and Engedal K (2009). Association between apoe e4 and cognitive impairment after stroke. Dement. Geriatr. Cogn. Disord 27, 525–533. [DOI] [PubMed] [Google Scholar]
- Wagle J, Farner L, Flekkøy K, Wyller TB, Sandvik L, Eiklid KL, Fure B, Stensrød B, and Engedal K (2010). Cognitive impairment and the role of the ApoE e4-allele after stroke - A 13 months follow-up study. Int. J. Geriatr. Psychiatry 25, 833–842. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, and Holtzman DM (2004). ABCA1 is required for normal central nervous system apoE levels and for lipidation of astrocyte-secreted apoE. J. Biol. Chem 279, 40987–40993. [DOI] [PubMed] [Google Scholar]
- Walter S, Atzmon G, Demerath EW, Garcia ME, Kaplan RC, Kumari M, Lunetta KL, Milaneschi Y, Tanaka T, Tranah GJ, et al. (2011). A Genome-Wide Association Study of Aging. Neurobiol. Aging 32, 2109.e15–2109.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Najm R, Xu Q, Jeong D, Walker D, Balestra ME, Yoon SY, Yuan H, Li G, Miller ZA, et al. (2018). Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat. Med 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Sun LM, Zhang L, Xu HT, Dong Z, Wang LQ, and Wang ML (2015). Association between Apolipoprotein E polymorphism and myocardial infarction risk: A systematic review and meta-analysis. FEBS Open Bio 5, 852–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward H, Mitrou PN, Bowman R, Luben R, Wareham NJ, Khaw K-T, and Bingham S (2009). Genotype, Lipids, and Coronary Heart Disease Risk a prospective population study. Arch Intern Med 169, 1424–1430. [DOI] [PubMed] [Google Scholar]
- Wei LK, Au A, Menon S, Griffiths LR, Kooi CW, Irene L, Zhao J, Lee C, Alekseevna AM, Hassan MRA, et al. (2017). Polymorphisms of MTHFR, eNOS, ACE, AGT, ApoE, PON1, PDE4D, and Ischemic Stroke: Meta-Analysis. J. Stroke Cerebrovasc. Dis 26, 2482–2493. [DOI] [PubMed] [Google Scholar]
- Weisgraber KH, Rall SC, and Mahley RW (1981). Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J. Biol. Chem 256, 9077–9083. [PubMed] [Google Scholar]
- Wijsman EM, Daw EW, Yu X, Steinbart EJ, Nochlin D, Bird TB, and Schellenberg GD (2005). APOE and Other Loci Affect Age-At-Onset in Alzheimer’s Disease Families With PS2 Mutation. Am. J. Med. Genet. - Neuropsychiatr. Genet 132B, 14–20. [DOI] [PubMed] [Google Scholar]
- Wild EJ, and Tabrizi S (2017). Therapies targeting DNA and RNA in Huntington’s disease. Lancet Neurol. 16, 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Gray CH, Goris A, Saiki M, Foltynie T, Compston DAS, Sawcer SJ, and Barker RA (2009). Apolipoprotein e genotype as a risk factor for susceptibility to and dementia in Parkinson’s Disease. J. Neurol 256, 493–498. [DOI] [PubMed] [Google Scholar]
- Wisniewski T, and Frangione B (1992). Apolipoprotein E: a pathological chaperone protein in patients with cerebral and systemic amyloid. Neurosci. Lett 135, 235–238. [DOI] [PubMed] [Google Scholar]
- Xiao H, Gao Y, Liu L, and Li Y (2017). Association between polymorphisms in the promotor region of the apolipoprotein E (APOE) gene and Alzheimer’s disease: a meta-analysis. EXCLI J. 16, 921–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XY, Ding JQ, and Chen S. Di (2010). Apolipoprotein e promoter polymorphisms and risk of Alzheimer’s disease: Evidence from meta-analysis. J. Alzheimer’s Dis 19, 1283–1294. [DOI] [PubMed] [Google Scholar]
- Xu M, Zhao J, Zhang Y, Ma X, Dai Q, Zhi H, Wang B, and Wang L (2016). Apolipoprotein e Gene Variants and Risk of Coronary Heart Disease: A Meta-Analysis. Biomed Res. Int 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Bernardo A, Walker D, Kanegawa T, Mahley RW, and Huang Y (2006). Profile and Regulation of Apolipoprotein E (ApoE) Expression in the CNS in Mice with Targeting of Green Fluorescent Protein Gene to the ApoE Locus. Neurobiol. Dis. Profile 26, 4985–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Yu L, White CC, Chibnik LB, Chhatwal JP, Sperling RA, Bennett DA, Schneider JA, and De Jager PL (2018). Evaluation of TDP-43 proteinopathy and hippocampal sclerosis in relation to APOE ε4 haplotype status: a community-based cohort study. Lancet Neurol. 17, 773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh FL, Wang Y, Tom I, Gonzalez LC, and Sheng M (2016). TREM2 Binds to Apolipoproteins, Including APOE and Amyloid-beta by Microglia. Neuron 91, 328–340. [DOI] [PubMed] [Google Scholar]
- Yu J-T, Tan L, and Hardy J (2014). Apolipoprotein E in Alzheimer’s Disease: An Update. Annu. Rev. Neurosci 37, 79–100. [DOI] [PubMed] [Google Scholar]
- Yu L, Boyle PA, Nag S, Leurgans S, Buchman AS, Wilson RS, Arvanitakis Z, Farfel JF, De Jager PL, Bennett DA, et al. (2015). APOE and Cerebral Amyloid Angiopathy in Community Dwelling Older Persons. Neurobiol. Aging 36, 2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis VI, Breslow JL, Utermann G, Mahley RW, Weisgraber KH, Havel RJ, Goldstein JL, Brown MS, Schonfeld G, Hazzard WR, et al. (1982). Proposed nomenclature of apoE isoproteins, apoE genotypes, and phenotypes. J. Lipid Res 23, 911–914. [PubMed] [Google Scholar]
- Zeitlow K, Charlambous L, Ng I, Gagrani S, Mihovilovic M, Luo S, Rock DL, Saunders A, Roses AD, and Gottschalk WK (2017). The Biological Foundation of the Genetic Association of TOMM40 with Late-onset Alzheimer’s disease. Biochim Biophys Acta 1863, 2973–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wang X, Tang Z, Liu J, Yang S, Zhang Y, Wei Y, Luo W, Wang J, Li J, et al. (2014). Apolipoprotein E gene polymorphism and the risk of subarachnoid hemorrhage: a meta-analysis of case–control studies. Lipids Heal. Dis. 2014, 13, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Liu C, Qiao W, and Bu G (2018). Apolipoprotein E, Receptors, and Modulation of Alzheimer’s Disease. Biol. Psychiatry 83, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N, and Weisgraber KH (2009). Understanding the association of apolipoprotein E4 with Alzheimer disease: Clues from its structure. J. Biol. Chem 284, 6027–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.