Abstract
The risk stratification of prostate cancer and breast cancer tumours from patients relies on histopathology, selective genomic testing, or on other methods employing fixed formalin tissue samples. However, static biomarker measurements from bulk fixed-tissue samples provide limited accuracy and actionability. Here, we report the development of a live-primary-cell phenotypic-biomarker assay with single-cell resolution, and its validation with prostate cancer and breast cancer tissue samples for the prediction of post-surgical adverse pathology. The assay includes a collagen-I/fibronectin extracellular-matrix formulation, dynamic live-cell biomarkers, a microfluidic device, machine-vision analysis and machine-learning algorithms, and generates predictive scores of adverse pathology at the time of surgery. Predictive scores for the risk stratification of 59 prostate cancer patients and 47 breast cancer patients, with values for area under the curve in receiver-operating-characteristic curves surpassing 80%, support the validation of the assay and its potential clinical applicability for the risk stratification of cancer patients.
One in six men will be diagnosed with prostate cancer during their lifetime, accounting for over 28% of total cancer cases in the United States1. Most newly diagnosed prostate cancer cases represent low-risk disease with less than a 4% chance of death2. The use of currently available screening and diagnostic methods in prostate cancer has resulted in the significant over-diagnosis and over-treatment of patients with Gleason 6 prostate cancer, as well as in the under-treatment of more aggressive cancers2–4. Similarly, in the United States, approximately one in eight women will be diagnosed with breast cancer during their lifetime5. Of the breast cancer subtypes, ductal carcinoma in situ (DCIS) is recognized as a large, low-risk breast cancer in need of diagnostic methods that help identify women who require aggressive treatment4. The gold standard for prognosis is adverse pathology (formalin-fixed tissue histology) in surgical specimens6,7. Predicting surgical adverse-pathology features with strong accuracy (>80 %)—currently not available in prostate cancer and breast cancer treatment planning—would provide oncologists with important information necessary for precision medicine. Furthermore, the assessment of physiologically relevant biomarkers of an individual’s tumour aggressiveness to categorize low-risk or indolent disease versus aggressive high-risk disease could improve diagnostic risk assessment in prostate cancer and breast cancer, and provide clinically actionable performance metrics8–14. Phenotypic biomarkers can be used in cancer diagnosis and in risk stratification because of the inherent genetic heterogeneity of cancer15–17. Direct evaluation of the dynamic phenotypic behaviour of single, living tumour cells grown in a controlled microenvironment could provide deeper insights into multiple and coordinated signalling pathways, and offer an improved risk stratification and diagnostic tool18,19. Previous attempts to analyse dynamic biomarkers from single cells derived from primary biopsy tissue have been limited by inherent difficulties in culturing primary tumour cells (particularly prostate cells). Hence, biomarkers for the prediction of cancer incidence and progression and of the risk of local growth, aggressiveness and metastasis have remained inaccessible.
Here, we describe a microfluidic-based high-content assay for the analysis (with single-cell resolution) of cell cultures derived from solid prostate tumour tissue or breast tumour tissue (Fig. 1). The assay uses live-cell phenotypic biomarkers—including protein localization, protein dynamics, protein modification state, cytoskeletal dynamics, membrane dynamics, cell morphology and cell motility—and leverages machine vision and machine learning to overcome the limitations of traditional, static, formalin-fixed histochemical biomarker analysis and also genomic tests that measure a small number of selected genes19–23 from bulk and static formalin-fixed tissue samples. The assay requires an extracellular-matrix formulation (ECMf; ref.19 and Supplementary Methods) that enables rapid culture (<72 h) of primary cancer cells and the measurement of previously inaccessible live-cell phenotypic biomarkers, as well as custom machine-vision software and machine-learning algorithms (Supplementary Methods) that quantify both live-cell and fixed-cell molecular and cellular phenotypic biomarkers from single cells to generate predictive scores via specific machine-learning algorithms for a given prediction18.
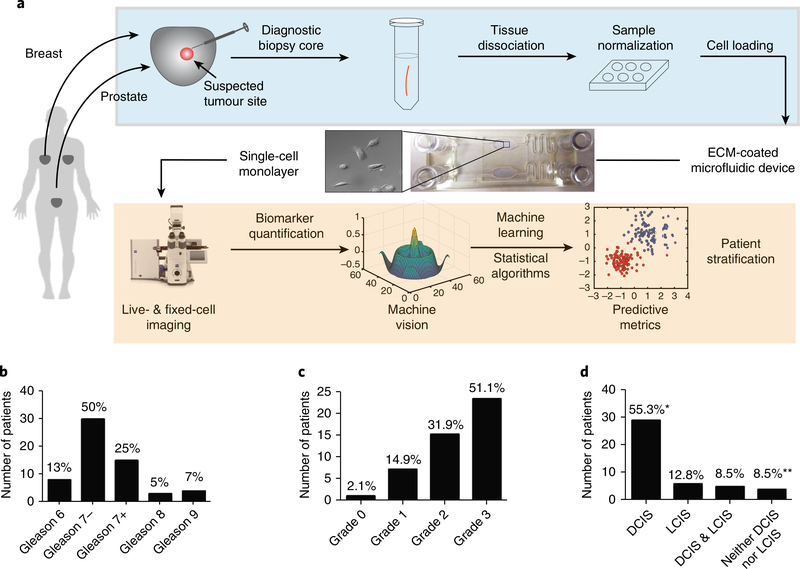
Fig. 1 |. Workflow for the risk stratification of patients via surgical adverse-pathology features using the live-primary-cell phenotypic-biomarker assay (STRAT-AP) and patient-sample characteristics of the clinical study.
a, Post-radical prostatectomy or mastectomy or lumpectomy biopsy cores were taken from tumour lesions at clinical collaborator sites. Cores were shipped overnight on cold packs to the central processing laboratory, and enzymatically dissociated. Cells were then cultured to normalize to in vitro conditions. Cells were imaged for a suite of phenotypic biomarkers via automated live-cell and fixed-cell microscopy on a microfluidic device. Images were analysed by machine-vision algorithms (processes in the orange shaded box are automated). The resulting data were objectively analysed by machine-learning statistical algorithms. b, Distribution of samples on the basis of Gleason score: 7−(Gleason 3+ 4) and 7+ (Gleason 4+ 3) samples (percentages of each Gleason score in the sample population are noted). The prostate sample set used in this study is representative of the naturally occurring distribution of Gleason scores found in the US population, according to initial biopsy reports. c, Grade distribution for the breast sample set. d, DCIS and LCIS distribution of the breast sample set. *Four samples were positive for DCIS yet did not have LCIS data reported. **Three samples were negative for DCIS yet did not have LCIS data reported. Image of microscope reproduced with permission from ZEISS Microscopy.
We evaluated the assay—which we name stratification of adverse pathology (STRAT-AP) because of its ability to risk-stratify patients on the basis of surgical adverse pathology features—as a personalized tool for the prediction of a patient’s risk of locally aggressive (invasive or metastatic) cancer. Specifically, we derived predictive risk scores for individual patients from over 300 molecular and cellular phenotypic primary and aggregate biomarkers (Supplementary Tables 2 and 3). The scores were derived and selected on the basis of the ability to predict true indolent disease or latent or occult aggressive disease with metastatic potential from a pathologically defined low-risk biopsy. The predictive scores are the general adverse pathology potential (GAPP) for assessing the risk of any adverse pathology, the local adverse pathology potential (LAPP) for assessing local tumour extension, and the metastatic adverse pathology potential (MAPP) for assessing the risk of metastatic disease (Table 1). The GAPP, LAPP and MAPP scores generated by STRAT-AP showed strong performance at individual and group levels, validating the performance of the assay for the risk stratification of prostate cancer and breast cancer patients.
Table 1 |.
Machine-learning-derived GAPP, LAPP and MAPP clinical scores
| GAPP | LAPP | MAPP | |
|---|---|---|---|
| Prostate | Any of the six adverse pathologies (SVI, PSM, EPE, PNI, LNP, LVI) | Seminal vesicle invasion (SVI) Positive surgical margins (PSM) | Perineural invasion (PNI) Lymph node positive (LNP) |
| Extra prostatic extension (EPE) | Lympho-vascular invasion (LVI) | ||
| Breast | Any of the four adverse pathologies (ENE, PSM, LVI, LI) | Extranodal extension (ENE) Positive surgical margins (PSM) | Lympho-vascular invasion (LVI) Lymph invasion (LI) |
Results
Overview of the assay.
A successful risk stratification test needs to mitigate the complexities of tumour heterogeneity and of the tumour microenvironment, and be able to predict post-surgical adverse pathologies. STRAT-AP includes five distinct components: a defined extracellular-matrix formulation designed for primary-cell adhesion, survival and for the measurement of relevant biomarkers19 (Supplementary Methods), a suite of dynamic and static molecular and cellular phenotypic biomarkers (Supplementary Methods), a microfluidic device for high-throughput live-cell and fixed-cell imaging (Methods and Supplementary Methods), machine-vision software to objectively measure biomarkers (Supplementary Methods) and machine-learning algorithms (Supplementary Methods) to generate clinically relevant scores that predict postsurgery adverse pathology states related to the local growth and metastatic behaviour of single, tumour-derived, primary biopsied cells19–23, and ultimately to the patient tumour samples.
Figure 1a provides an overview of the STRAT-AP assay and summarizes its main components. The assay has been designed as a clinically relevant and actionable laboratory-developed test24. Sample-handling and rapid-culturing conditions were established to develop, from tumour samples, single-cell suspensions and short-term cell cultures (<72 h) enriched with epithelial cells19 (see Methods). Cells derived from patient biopsies were placed on an ECMf-coated microfluidic device (Fig. 1a) and imaged using a standard high-throughput format (Fig. 1a; see Methods). More than 70% of the primary live-biopsy cells survived during transport, and more than 80% of those cells adhered and spread within the ECMf-coated imaging chamber of the microfluidic device19.
The characteristics of the patient population for the samples used in the validation of the STRAT-AP platform are detailed in Fig. 1b–d. The specific post-surgical adverse pathology features (pathologies) for each patient after radical prostatectomy or breast lumpectomy and mastectomy were determined by pathologists located at the site of surgery and blinded to the predictions (see Methods).
Biomarker quantification and generation of aggregate biomarkers.
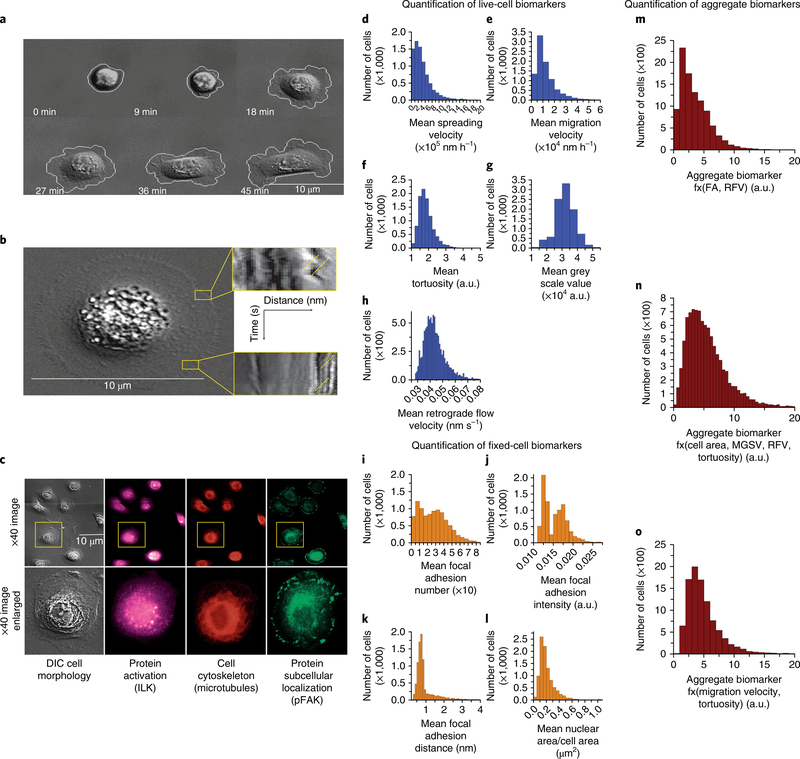
Oncoproteins localize in focal adhesions (FAs) in two-dimensional environments regulate cytoskeletal and signalling dynamics, and are implicated in the genesis and ontogeny of tumour cells21–23. We thus hypothesized that a biomarker suite that focuses on FAs and cytoskeletal cancer markers could be used in the STRAT-AP assay. Cell-spreading velocity (CSV; Fig. 2d), tortuosity (Fig. 2f), mean square grey value (MSGV; used as a measure of cell height and adhesion; Fig. 2g) and actin retrograde flow velocity (RFV) measured by membrane fluctuations at the cell leading edge (Fig. 2b,h) cumulatively represent dynamic live-cell, label-free biomarkers quantified during its live-cell imaging.
Fig. 2 |. Phenotypic (cellular and molecular) biomarkers measured via sequential live-cell imaging and fixed-cell imaging in a standardized microfluidic environment.
a–h, Representative live-cell biomarkers include cell spreading (a) and tortuosity, cell adhesion rate to the device substrate, cell area change during adhesion, and tortuosity of cell membrane as a measure of morphology. Rapid dynamics of the membrane surface are measured as retrograde flow through kymographs (b). The yellow lines indicate examples of where the retrograde flow measurements are made. The expression, localization and phosphorylation state of subcellular protein complexes (phospho-focal adhesion kinase, pFAK) and individual proteins (integrin-linked kinase, ILK) as well as microtubules are measured on corresponding fixed cells and matched to respective live-cell images (c). Quantification of the total cell population for mean cell-spreading velocity (d), mean cell-migration velocity (e), mean cell tortuosity (f), mean greyscale value (g) and mean actin-retrograde-flow velocity (h). i–l, Representative fixed-cell biomarkers are mean cell FA (i), mean cell FA intensity (j), mean cell FA distance from the membrane edge (k) and mean cell nuclear area/cell area (l). m–o, LAPP4 = f(FA, RFV) aggregate biomarker (m), MAPP10 = f(Area, MGSV, RFV, Tortuosity) aggregate biomarker (n), MAPP17 = f(Migration velocity, Tortuosity) aggregate biomarker (o).
On completion of the live-cell imaging protocol, primary biopsy-derived single-cell cultures were fixed and stained with antibodies to measure static biomarkers, such as protein localization, cytoskeletal architecture and protein activation (Fig. 2c). Because FAs harbour proteins implicated in both prostate and breast tumorigenesis—for example, integrin-linked kinase (ILK), FA kinase (FAK; ref. 25), and breast cancer anti-oestrogen resistance protein 1 (BCAR1) or protein product p130Cas—mean FA number (FA-N; Fig. 2I), mean FA intensity (FA-I) and mean FA distance (FA-D) were also quantified for each cell from the leading membrane edge (Fig. 2k) to assess FA activation as well as force generation and motility from the coupling of FA and actin. FA measurements are representative of static biomarkers measured in the fixed-cell imaging regime. We also quantified cellular-nucleus area (CAN; Fig. 2l), a traditionally important histological feature, from measurements of dynamic live single cells interacting with the ECMf.
Before biomarker quantification, each single cell from a sample was assigned a unique identification number in order to track individual cells over the course of the live-cell and fixed-cell imaging sequences (Fig. 3a). Molecular and cellular phenotypic biomarkers, selected for their potential predictive power, such as FA-N (Fig. 2j), CSV and tortuosity (measurement of the curves in the cell surface) were quantified (Fig. 2d,f) to indirectly measure FA activity, actin cytoskeletal activation and myosin activation, respectively, by using fluorescent and differential interference contrast (DIC) microscopy in concert with machine-vision software. Additionally, MSGV via DIC illumination was quantified to assess cell height, adhesion and the cytoskeletal activation necessary for cell spreading (Fig. 2g). Similarly, RFV was measured by membrane fluctuations at the cell leading edge (Fig. 2b,h) to assess the extent of FA–actin coupling as its levels have been correlated with the downstream signalling important for survival, growth and motility23. Cell-migration velocity, cell area and cell perimeter were also measured over time. RFV of the cellular membrane was monitored by the creation of kymographs, which were scored for slope of the membrane above background. From the slope, we derived the velocity of the membrane flow. Each FA measurement was monitored by identifying the highest greyscale values around the perimeter of the cell membrane above background, and then by scoring them for total size on an individual FA. Antibody control images for prostate and breast cancer primary cells and cell lines are shown in Supplementary Figs. 6 and 7.
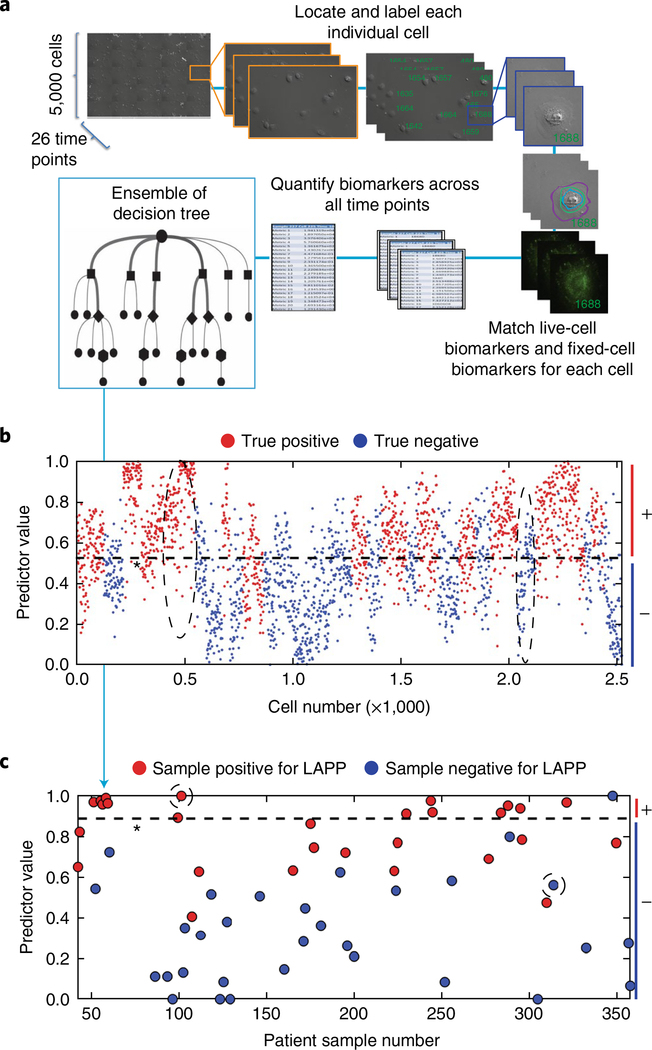
Fig. 3 |. Quantification of automated machine-vision biomarkers informs random-forest decision trees for the stratification of single cells and the prediction of surgical pathology features.
a, Live-cell images were collected on approximately 5,000 cells across 26 time points. Cells were fixed and stained and a 27th image was taken. Each cell was assigned a unique identifier and measured for respective live-cell and fixed-cell cellular and molecular phenotypic biomarkers, leading to an average of 42 million measurements per sample. These measurements were consolidated across time points to ~328 measurements per cell. The set of biomarkers measured for each cell were fed to a statistical-analysis algorithm that generates multiple decision trees to stratify negative cells and positive cells for a given pathological outcome. Decision trees were weighted to optimize algorithm accuracy. b, Characterization of individual normal and potiential cancer cells. A representative plot showing the stratification of negative cells and positive cells through combinations of biomarkers determined by random-forest decision tree analysis. Patient-level results were obtained by summarizing the cell-level results. c, Stratification and prediction of whether patients are positive or negative for surgical adverse pathological features of interest. A representative plot shows the stratification of patients for a given predicted pathology feature. Dashed ovals in b show groups of cells predicted as true positives and true negatives for LAPP, and correspond to the circled patient-level predictions in c. Dashed lines indicate the machine-learning-derived thresholds for the discrimination of negative (−) and positive (+ ) cells (b) and patients (c).
Importantly, from the quantification of both dynamic and static biomarkers for individual cells, aggregate biomarkers can be generated for each cell (Fig. 2m–o and Supplementary Table 3). Supplementary Fig. 1 shows the quantification of primary and aggregate biomarkers for breast cancer. Aggregate biomarkers were calculated to garner insights into relationships, cooperative or antagonistic, between ensembles of signalling pathways or between subcellular protein–protein interaction networks (Fig. 2m–o). By assigning each cell an individual number, live-cell cellular biomarkers and fixed-cell molecular and cellular biomarkers18 could be measured on the same cell and then integrated into the GAPP, LAPP and MAPP scores (Table 1).
Determination of adverse pathology states.
After a suite of both primary and aggregate single-cell biomarkers have been quantified by machine vision and mathematical processing, the machine-learning algorithms integrate biomarker values to generate both cell-level and patient-level predictive scores for individual adverse pathological features (Fig. 3). Single-cell analyses from heterogeneous tumour tissue samples require additional mathematical transformations to translate single-cell-level biomarker measurements for the prediction of patient-level adverse pathology. To achieve this, we used ~5,000 images of live single cells per patient sample over 26 time points (Fig. 3a) to obtain longitudinal biomarker data (live-cell imaging regime). Live-cell imaging is accomplished using label-free DIC to image cell-edge and cell-membrane contours and dynamics. Importantly, no antibodies or probes were used in the live-cell imaging regime, to ensure minimal perturbation of primary-cell behaviour. STRAT-AP then records a 27th image of the same ~5,000 single cells after cells have been fixed and stained with relevant antibodies for four FA-associated proteins (fixed-cell imaging regime). When quantifying fluorescence from antibodies in the fixed-cell imaging regime, the local background (immediately adjacent area outside the cell) is subtracted from each individual cell.
Primary biomarkers from the tracked single cells throughout both the live-cell and fixed-cell imaging regimes were tabulated for each individual cell (Fig. 3a). Aggregate biomarker quantities derived from functions of primary biomarkers are then calculated on the basis of mathematical functions26 (Supplementary Table 3) such that both primary and aggregate biomarker quantifications can be input to a machine-learning algorithm (see Methods and Supplementary Methods) to generate a plot that predicts if a specific cell exhibits a biomarker profile indicative of having positive or negative potential for a given adverse pathology (Fig. 3b).
The machine-learning algorithm was trained on 70% of the single cells from the entire patient population to derive statistical algorithms that were then tested on the remaining 30% of patient cells in a blinded fashion (see Methods). The resulting cell-level predictions were synthesized into patient-level predictions on the basis of the percentage of cells that were predicted to be positive or negative (Fig. 3c).
Prediction of post-surgical adverse pathology features and patientrisk stratification.
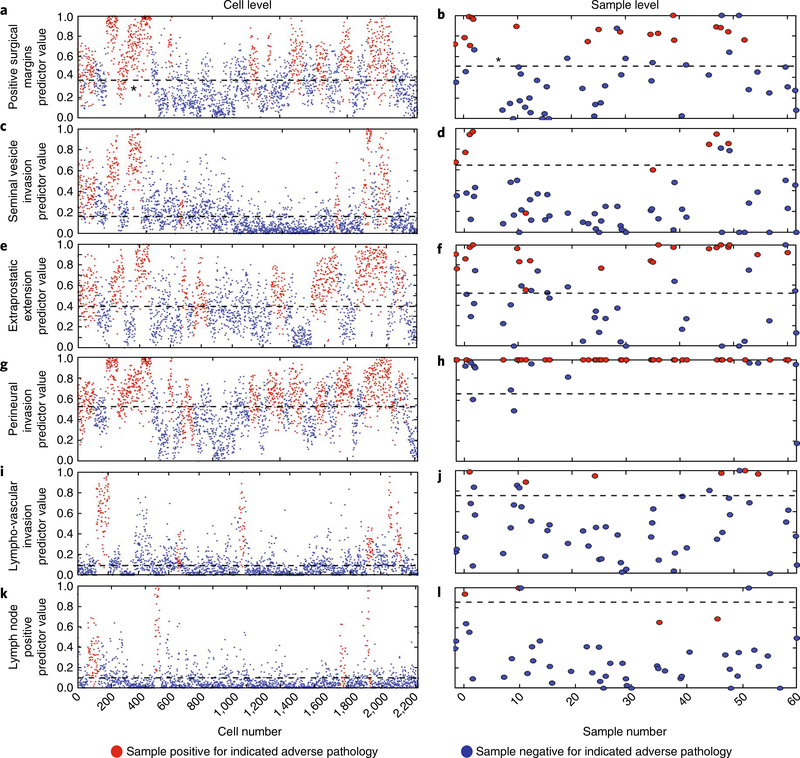
The machine-learning model was designed to maximize both sensitivity and specificity, and to ensure an optimal level of detection of true positives and true negatives while minimizing false-positive and false-negative predictions. Figure 4a,c,e–g,i,k displays output predictions for adverse-pathology features of individual prostate cancer patients: positive surgical margins (PSMs), seminal vesical invasion (SVI), extra-prostatic extension (EPE), perineural invasion (PNI), lympho-vascular invasion (LVI) and lymphnode positive (LNP). These plots separate each patient’s cells into positive (above the determined threshold value) or negative (below the threshold value) for a given adverse-pathology feature.
Fig. 4 |. Machine-learning statistical algorithms predict specific surgical adverse-pathology features in blinded test-sample sets.
a, A cell-level plot stratifying negative (blue circles) and positive (red circles) cells for PSMs. The black dashed line indicates the threshold, determined by machine-learning-derived statistical algorithms. b, Corresponding patient-level plot stratifying positive (red circles) and negative (blue circles) for PSMs. Results were obtained by summarizing the cell-level results. The black dashed line indicates the threshold set by the machine-learning-derived statistical algorithm. c–l, Cell-level plots stratifying negative and positive cells for SVI (c), EPE (e), PNI (g), LVI (i) and LNP (k), and patient-level plots stratifying patients for SVI (d), EPE (f), PNI (h), LVI (j) and LNP (l).
To identify positive cells and subsequently positive patients, STRAT-AP undertakes the following process (Supplementary Fig. 7). Primary biomarkers are first measured and quantified using machine-vision software. Primary biomarkers are then input into algorithms that generate aggregate biomarkers. Both primary and aggregate biomarkers are input into a machine-learning decision tree. Having been trained on 70% of the cells across the patient population, the decision tree sets an optimal threshold value to determine if a cell is positive or negative for a given group of specific adverse-pathology features. Then a second threshold value is established to decide how many cells need to be positive to accurately predict if a patient is positive for a specific set of adverse-pathology features. Those thresholds are then applied to the remaining 30% of cells in a blinded fashion to generate predictions on patient adverse pathology, and the performance of those predictions is assessed after unblinding the pathology data. Importantly, the thresholds are set by the decision tree, with no operator input (see Methods).
Figure 4b,d,f,h,j,l displays predictions for individual adverse-pathology features of prostate cancer patients. Briefly, patient-level predictions are a function of the number of single cells that STRAT-AP determines to be positive or negative for any given adverse-pathology state (see Methods). Supplementary Fig. 2 shows that the transformation from cell-based to patient-based predictions for prostate cancer samples is also applicable to breast cancer. The predictions were then compared with the surgical pathology reports to calculate true positives, false positives, true negatives and false negatives. Finally, the full data set was analysed by the machine-learning algorithm (random-forest classifier), and the outcome compared with the results from the validation process to determine the robustness of the developed algorithm. The sensitivities and specificities were calculated for each adverse-pathologic category. Receiver operating characteristic (ROC) curves were generated and the associated area under the curve (AUC) calculated to verify the accuracy and discrimination ability of the classification algorithm.
The sensitivities and specificities of the patient-based predictions were calculated to determine the performance of the STRAT-AP algorithms in predicting adverse pathology after radical prostatectomy (Table 2). For the prediction of individual adverse pathology features, AUCs were greater than 0.80 (Fig. 5a–f). We also performed a similar analysis for the predictions of post-surgery adverse pathology in breast cancer (Fig. 5g–n).
Table 2 |.
Predictive performance results for adverse pathologies from prostate tissue and breast tissue samples
| Predicted adverse pathology | Sensitivity | Specificity | AuC | N | True positive | True negative | Predicted positive | Predicted negative |
|---|---|---|---|---|---|---|---|---|
| Prostate tissue | ||||||||
| Seminal vesicle invasion | 0.89 | 0.96 | 0.93 | 57 | 9 | 48 | 8 | 46 |
| Positive surgical margin | 0.99 | 0.93 | 0.94 | 59 | 18 | 41 | 18 | 38 |
| Extra-prostatic extension | 0.95 | 0.97 | 0.96 | 53 | 21 | 32 | 20 | 31 |
| Perineural invasion | 0.99 | 0.99 | 0.99 | 50 | 37 | 13 | 37 | 13 |
| Lymph node positive | 0.95 | 0.96 | 0.81 | 47 | 4 | 43 | 4 | 41 |
| Lymph vascular invasion | 0.99 | 0.98 | 0.98 | 54 | 6 | 48 | 6 | 47 |
| GAPP | 0.91 | 0.93 | 0.88 | 59 | 45 | 14 | 41 | 13 |
| LAPP | 0.93 | 0.90 | 0.93 | 59 | 28 | 31 | 26 | 28 |
| MAPP | 0.95 | 0.84 | 0.89 | 59 | 40 | 19 | 38 | 16 |
| Breast tissue | ||||||||
| Extra-nodal extension | 0.99 | 0.73 | 0.84 | 37 | 14 | 23 | 13 | 19 |
| Positive surgical margin | 0.99 | 0.95 | 0.98 | 45 | 3 | 42 | 3 | 39 |
| Lympho-vascular invasion | 0.90 | 0.87 | 0.87 | 44 | 21 | 23 | 19 | 19 |
| Lymph invasion | 0.96 | 0.79 | 0.91 | 46 | 27 | 19 | 20 | 18 |
| GAPPa | 0.81 | 0.93 | 0.85 | 47 | 32 | 15 | 26 | 14 |
| LAPPa | 0.99 | 0.72 | 0.81 | 47 | 15 | 32 | 15 | 23 |
| MAPPa | 0.84 | 0.88 | 0.85 | 47 | 31 | 16 | 26 | 14 |
| MAPPLIb | 0.90 | 0.85 | 0.83 | 32 | 19 | 13 | 15 | 12 |
| MAPPLVIb | 0.83 | 0.87 | 0.86 | 32 | 18 | 14 | 15 | 12 |
Sensitivity, specificity, AUC, total number of samples, true-positive and true-negative numbers, and number of samples predicted positive or negative were obtained from machine-learning-derived statistical algorithms.
For breast samples, GAPP, LAPP and MAPP scores are derived from algorithms trained on all breast samples.
For breast samples, MAPPLI and MAPPLVI scores are derived from algorithms trained on DCIS positive samples only.
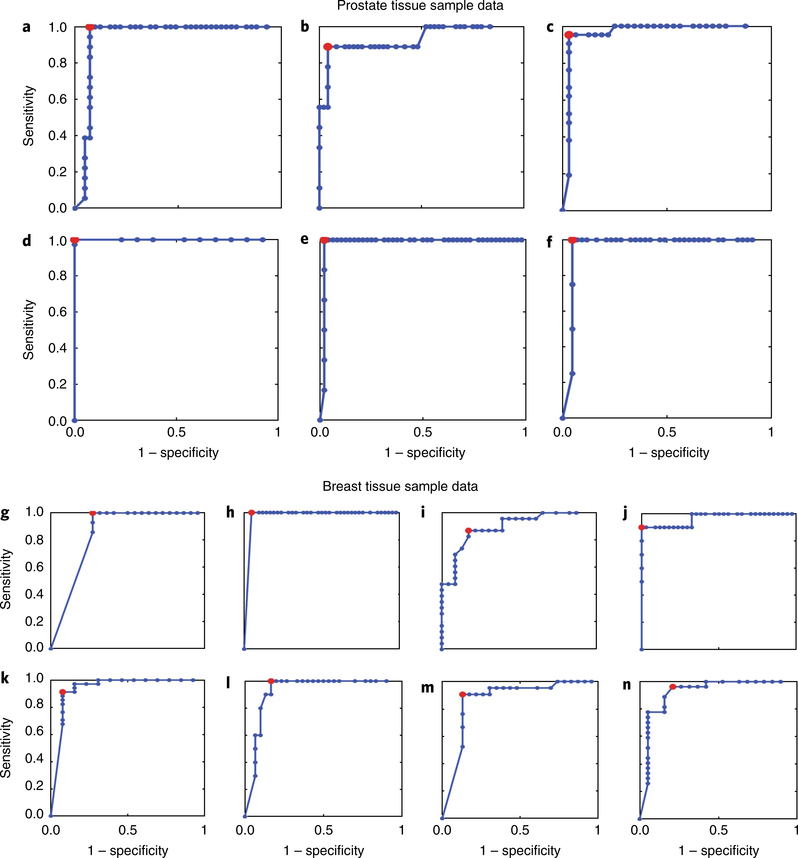
Fig. 5 |. RoC curve analysis of predictions of surgical adverse-pathology features.
The optimal sensitivity and specificity for each surgical adverse-pathology feature was calculated on the basis of machine-learning-derived ROC curve analysis. a–f, Scores for predicting adverse-pathology features for samples from prostate cancer patients predict PSMs (a), SVI (b), EPE (c), PNI (d), LVI (e) and LNP (f) with high levels of sensitivity and specificity. g–n, Scores for predicting adverse-pathology features for samples from breast cancer patients predict ENE (g), PSMs (h), grade (Gr, i), Her2/Neu positive (H/NP, j), DCIS (k), LCIS (l), LVI (m) and LI (n) with high levels of sensitivity and specificity.
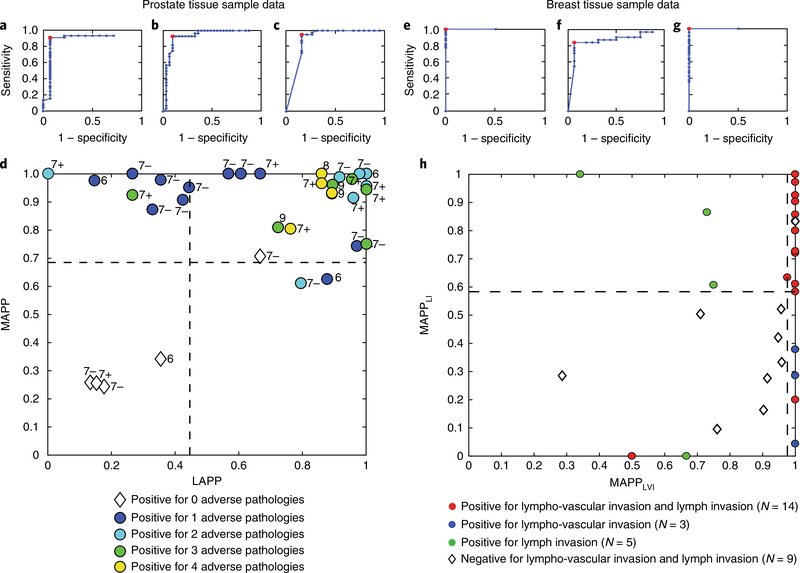
The calculated GAPP, LAPP and MAPP scores (defined for both prostate cancer and breast cancer in Table 1) for the risk stratification of patients were predicted with AUCs > 0.80 (Fig. 6a–c,e–g). With LAPP, MAPP and individual predictor values, risk stratification plots can be generated (Fig. 6d,h) to separate prostate and breast DCIS cancer patients on the basis of the number and type of adverse-pathology states and metastatic potential. Importantly, the machine-learning predictions were robust and reproducible. Training the algorithm with bigger sample sizes should increase the AUC values for risk stratification. ROC curves show that GAPP, LAPP and MAPP scores derived from quantified biomarkers were able to predict adverse pathologies with high sensitivity and specificity (>0.80) and to separate or stratify patients into distinct, quantifiable groups on the basis of predicted surgical adverse-pathology features.
Fig. 6 |. RoC curve analysis for predicting groups of specific surgical adverse-pathology features generates scores that predict cancer severity and risk-stratify patients with high levels of sensitivity and specificity.
a, GAPP scores predict if prostate patient samples have any adverse pathologies. b, LAPP scores predict locally aggressive behaviour. c, MAPP scores predict metastatic potential. d, Risk stratification plot using MAPP and LAPP scores for prostate cancer patients shows that patients with different adverse pathologies cluster in centralized quadrants. Dotted lines indicate machine-learning-derived thresholds. e, GAPP scores predict if breast patient samples have any adverse pathologies. f, Breast cancer LAPP scores predict locally aggressive behaviour. g, Breast cancer MAPP scores predict metastatic potential. h, Plot of MAPPLI (MAPP-lymph invasion) versus MAPP LVI (MAPP-lympho-vascular invasion) scores for breast cancer DCIS patients, showing that these patients can be risk-stratified via machine-learning-derived thresholds, with aggressive cancer patients and indolent cancer patients correctly clustering in separate quadrants.
Discussion
The ability to predict adverse pathology before radical prostatectomy in low-risk, indolent prostate cancer (Gleason 6 and Gleason 3+ 4) and before lumpectomy or mastectomy in DCIS breast cancer would significantly improve the risk stratification of patients. Immunohistochemistry-based nomograms and molecular and genomic approaches have been developed to address the need for precise diagnoses and risk stratification in prostate cancers and breast cancers. There are multiple tests for prostate cancer and breast cancer risk stratification, including molecular and genomic testing, imaging (MRI) and risk nomograms27–34. However, they all have limited application, clinical actionability and performance (in particular, low sensitivity or specificity in the prediction of post-surgical adverse-pathological features). By way of example, standard histopathological methods for prostate cancer—the Gleason score—have AUCs of 0.60–0.70 (ref. 35) when predicting clinical outcomes for low-risk and intermediate-risk patients. This level of predictive performance of the Gleason scoring system has led to notable over-diagnosis and over-treatment of prostate cancer patients36. By enabling predictive-performance levels closer to an AUC of 0.90 when used in conjunction with the Gleason score, the STRAT-AP assay has the potential to dramatically improve the ability to predict if a low-risk or intermediate-risk cancer will progress to an aggressive cancer or remain non-aggressive or indolent. This performance level will enable additional and more personalized risk stratification than the Gleason score.
The STRAT-AP assay was developed to analyse live cells instead of fixed tissue. Importantly, STRAT-AP uses a significant body of meaningful and previously inaccessible phenotypic biomarkers, and overcomes the limitations of bulk, formalin-fixed, paraffin-embedded tissue sample analysis and of static pre-selected biomarkers characteristic of existing histopathological and genomic risk stratification tests. The approach taken by the STRAT-AP assay is backed by evidence in several cancers that deregulation of key signalling pathways (for example, PI3K, FAK, Rho-ROCK and MAPK) is responsible for disease-relevant molecular and cellular phenotypes, including changes in cytoskeletal dynamics, migration velocity and membrane fluctuations37,38, which are central to tumorigenesis and metastasis.
The rationale for biomarker selection is based on the observations that numerous proteins involved in oncogenesis localize to focal adhesion sites and that actin polymerization is physically and biochemically coupled to FA sites when cells are cultured in 2D in vitro environments23. For example, localization and activation of the FAK protein has been implicated in multiple epithelial cancers23. Similarly, GTPase-mediated actin polymerization, as measured by actin retrograde-flow velocity, has also been implicated in metastatic progression of epithelial cancers. Furthermore, as oncogenesis is linked with FA formation and stabilization as well as with enhanced actin polymerization events in 2D in vitro environments23, STRAT-AP’s biomarkers were selected to measure FA and actin-activation states. A list of STRAT-AP’s biomarkers and their relative importance is presented and discussed in Supplementary Tables 2–5. The high performance of STRAT-AP can be attributed to the use of live cells to measure dynamic biomarkers with single-cell resolution objectively through machine-vision algorithms. Additionally, machine-learning algorithms objectively rank and maximize the predictive value of a diverse set of phenotypic live-cell and fixed-cell biomarkers. STRAT-AP was also designed to be easily and seamlessly integrated into the clinical workflow. Such a workflow would involve the following steps. Physicians simply place a biopsy in a tube pre-filled with transport medium instead of a tube filled with formalin (as they would in current clinical workflows). Similarly, that tube is sent to the pathology laboratory and placed in a box containing ice packs and shipped overnight to a central laboratory. The sample is stable for up to 72 h at 4 °C, thus allowing time for transport. In the central laboratory, the sample is removed and placed in a dissociation buffer and allowed to normalize to the ECMf before the single-cell suspension is seeded on a microfluidic device for imaging. Image acquisition and data analysis are automated, and patient scores (GAPP, LAPP and MAPP) are ready to be returned back to the physician within 72 h. Because the GAPP score predicts whether the patient is experiencing any adverse pathology, it is a simple to interpret whether a patient has low-risk indolent disease or low-risk aggressive disease. The LAPP and MAPP scores provide further information about whether the patient is experiencing local or metastatic adverse pathology, guiding the clinician through personalized treatment options for the patient (Fig. 6d,h). With automated microscopy and machine vision, STRAT-AP is user-friendly for both the clinician ordering the test and the technician conducting the test, and provides clinically useful prediction metrics (Table 2).
The concept of live-cell phenotypic biomarkers is not unprecedented. High-throughput, live-cell imaging of cellular phenotypic biomarkers has been used to screen for drug responses39, but has not been used to risk-stratify patients by predicting adverse pathologies, as the timelines of previous methods are too long to be relevant to treatment guidance. In addition, methods that are unable to culture patient samples to measure and analyse relevant phenotypic biomarkers would be ineffective. Therefore, STRAT-AP fills an important gap in current treatment planning, as it allows for personalized risk assessments to guide treatment. Also, STRAT-AP is a clinically applicable test with single-cell resolution using live-primary-cell phenomics—rapid analysis of the behaviour of live primary biopsied cells via dynamic molecular and cellular biomarkers18,19,22,23,26.
The machine-vision software for image analysis in STRAT-AP was developed to objectively measure biomarkers in individual live cells across space and time to achieve the necessary spatiotemporal resolution to overcome the challenges of tumour heterogeneity. The combination of automated live-cell imaging microscopy and machine-vision software allows for molecular and cellular phenotypic biomarkers to be objectively measured on individual cells. The ECMf was designed not just to keep cells alive in vitro, but to provide a reproducible reference microenvironment to compare single-cell behaviour and quantify biomarkers, and is not considered to represent a complete recapitulation of native ECM in vivo. A powerful feature of the STRAT-AP assay is its use of machine learning to generate predictions on both individual adverse-pathology features and aggregate groupings of biologically related pathology features. The machine-learning algorithm, which uses ~300 individual and aggregate biomarker inputs, provides cell-level and patient-level tissue scores. The GAPP, LAPP and MAPP scores generated by the machine-learning algorithms differentiate cell populations and cancer patients with low-grade and intermediate-grade disease and tumour behaviour. The scores predict adverse pathologic findings in surgical specimens (Fig. 5 and Table 2), upgrading the Gleason score, and biochemical recurrence in prostate cancer (Supplementary Figs. 3 and 4). GAPP predicts both overall tumour aggressiveness and metastatic potential, with high sensitivity and specificity for prostate cancer and breast cancer (Fig. 6a,e and Table 2). LAPP predicts positive surgical margins, seminal vesicle invasion and extra-prostatic extension for prostate cancer, and extra-nodal extension and positive surgical margins for breast cancer, also with high sensitivity and specificity (Fig. 6b,f and Table 2). Moreover, pathological findings such as DCIS can be independently predicted, to then predict subsets of GAPP, LAPP and MAPP scores so that DCIS patients can be effectively stratified on the basis of the potential of their respective DCIS lesions to aggressively grow locally or distally (Fig. 6h and Table 2). MAPP predicts PNI, LVI and LNP disease for prostate cancer, and LVI and LI in breast cancer, again with high sensitivity and specificity (Fig. 5g and Table 2). Each of these adverse pathologies has implications for individual and personalized patient prognosis.
The STRAT-AP assay supplements both the traditional formalin-fixed paraffin-embedded tissue-scoring systems used in prostate and breast cancer as well as recently introduced genomic tests, and improves tumour risk assessment and treatment decisions without interrupting the current oncological and pathological workflows. Using live-cell phenotypic-biomarker measurements from individual cells directly encompasses cellular heterogeneity40 and single-cell behaviour such as cell–ECM interactions, cytoskeletal and signalling dynamics, subcellular protein localization, protein modification, functional protein-complex formation, membrane fluctuations and motility parameters implicated in cancer aggressiveness and metastasis41. Furthermore, STRAT-AP as a live-primary-cell assay could be applied as a companion diagnostic tool for personalized therapy selection and, by making it into an automated high-throughput system, as a biomarker and lead-compound discovery tool. Although this validation study with prostate cancer and breast cancer shows the versatility and performance of the assay, further risk stratification validation studies in these and other solid tumours are warranted.
Methods
Study design.
Annually, over one million men undergo prostate biopsies and over one million women undergo breast biopsies in the United States1. Using these numbers as a guide, we developed a clinical validation study for the STRAT-AP assay. The study involves growing the sample size in two stages: from 60 (stage I) to 300 (stage II) biopsies. These numbers were determined mathematically using the following equation:
where N is the required study size for assumed sensitivity, sN is the sensitivity, and TP and FN the true-positive and false-negative probabilities. P is the percentage of the total population that has the characteristic of interest, W = 0.05 is the confidence interval, and Z = 1.96 is the z score for the desired confidence interval42–44. All 59 prostate tissue samples and 47 breast tissue samples were de-identified and collected under Institutional Review Board (IRB) approval at the following institutions: Lahey Hospital and Medical Center, Department of Cancer Research, American Medical Professionals of New York, Urology Place of San Antonio and the National Cancer Institute Cooperative Human Tissue Network (CHTN). All procedures performed in the studies involving human participants were carried out in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or with comparable ethical standards. Inclusion and exclusions were determined as follows. The conditions for the inclusion of prostate cancer samples were male patients from 40 to 80 years of age; all races; the patient is willing to participate and has signed the written informed-consent form; patients with Gleason score 6, 7,8, 9 or 10; patients undergoing radical prostatectomy; patients with known clinical outcomes. The conditions for the exclusion of prostate cancer samples were patients that have received prior chemotherapy and patients that have tested positive for infectious blood-borne pathogens. The conditions for the inclusion of breast cancer samples were female patients from 40 to 90 years of age; all races; the patient is willing to participate and has signed the written informed-consent form; basal-cell and squamous-cell malignancies of the skin may be included. The conditions for the inclusion of breast cancer samples were non-treated patients (without previous hormonal therapy, chemotherapy or radiotherapy before surgery); pre-menopausal and post-menopausal women; patient primary treatment plan is surgery (lumpectomy or mastectomy); patients diagnosed with metastatic cancer or other malignancy apart from basal cell and squamous cell. The conditions for the exclusion of breast cancer samples were patients diagnosed with positive infectious disease such as HIV/AIDs, hepatitis or syphilis; patients with known breast disease undergoing treatment for the disease; pregnant or lactating women. Consenting patients that met inclusion and exclusion criteria were selected by clinical collaborators. Patient samples were randomized, and the investigators were blind to patient selection and surgical pathological data before testing the performance of the machine-learning algorithm. Following biomarker data analysis, surgical–pathology findings were unblinded, and the predictive power and statistics of STRAT-AP were assessed and validated.
Pathology.
The specific data collected from the pathology reports were Gleason primary and secondary pattern and score, tumour stage, percentage of tumour involvement, SVI, PSMs, EPE, LVI, PNI, pelvic lymph-node involvement and biochemical recurrence. Typically, aggressive prostate cancer presents with one, or several, of the following six specific adverse pathologies: (1) SVI, (2) PSMs, (3) EPE, (4) LVI, (5) PNI and (6) LNP6. In breast cancer, patients typically present with one, or several, of the following specific adverse pathologies: (1) ENE, (2) PSMs, (3) H/NP, (4) DCIS, (5) LCIS, (6) LVI and (7) LI.
Tissue handling and culture of primary prostate cells.
Cold-chain or temperature-controlled transport ensured that fresh samples from surgical specimens could be maintained alive while being sent to the central laboratory facility. Samples were collected from areas of suspected tumour on the basis of presurgery biopsy data, collected in transport medium supplemented with purified extracellular protein factors to support cell and tissue survival in suspension and with Dulbecco’s modified Eagle’s medium (DMEM)22, and shipped from the hospital site to the core laboratory overnight at 4 °C. The transport medium used for the transportation of tissue samples from the operating room to the central laboratory was developed by Cellanyx (cat. nos. TM-PR001 (prostate) and TM-BR001 (breast)). Albeit with less cell-survival efficiency, DMEM can also be used to transport samples. Transport time can be up to 72 h (to maintain the viability of cells19) (Supplementary Fig. 11). On arrival at the laboratory, samples were mechanically disrupted and then treated with collagenase in Cellanyx’s dissociation medium supplemented with Lonza prostate epithelial basal medium (PrEBM). After overnight digestion, samples were resuspended in fresh culturing medium free of collagenase, and seeded on tissue-culture plates coated with ECMf19(Supplementary Fig. 10). Dissociation medium used to dissociate tissue samples into single-cell suspensions was developed by Cellanyx (cat. nos. DM-PR001 (prostate) and DM-BR001 (breast)). Alternatively, DMEM supplemented with 10 μg ml−1 collagenase I (Millipore Sigma, cat. no. 9001–12-1) can also be used to dissociate the cells (Supplementary Methods). Cells were allowed to acclimate to the culture conditions for 24 h before image analysis. Specifically, a collagen-type-I/ECMf19 was used to coat culturing substrates containing Cellanyx’s culturing medium (cat. nos. CM-PR001 (prostate) and CM-BR001 (breast)). ECMf was designed to support cell adhesion and survival as well as to create a reference environment for measuring cellular and molecular phenotypic biomarkers under well-defined, robust and reproducible conditions. Cells were removed from culture plates via standard trypsin treatment, and an average of 5,000 cells were seeded on a given microfluidic device for image analysis. Alternatively, laminin may also be used as a protein coating to promote adhesion and survival, albeit less efficiently, as the engagement of pro-growth integrins facilitates the adhesion and survival of primary biopsy cells in 2D culture systems. Culturing medium used for in vitro culturing of primary biopsy cells was developed by Cellanyx (cat. nos. CM-PR001 (prostate) and CM-BR001 (breast)). DMEM supplemented with 10 mM glutamine (Life Technologies) may also be used as the medium for culturing primary cells in vitro (Supplementary Methods).
A biopsy core sample typically contains a variety of cell types, such as epithelial cells (luminal and apical), fibroblasts, mesenchymal cells, immune cells and endothelial cells45,46. The ECMf and media formulations select for epithelial cells (~70%, Supplementary Fig. 5) while maintaining stromal cells (~15%) and other cell types (~15%) in the population.
Integration into clinical workflows and platform–operator interface.
Designed as a laboratory-developed test (LDT), STRAT-AP can fit seamlessly into an on-site pathology laboratory or a centralized laboratory. Its key elements, as with any LDT, are its process and equipment. STRAT-AP uses media, reagents and disposables that are cost-effective (<$100 per sample) and easily managed with traditional culturing techniques. Furthermore, the assay uses a commercially available microscope to acquire images that are digitized using standard imaging software and analysed by custom machine-vision software. Once biomarker measurements are quantified by the software, the values are input into machine-learning algorithms that return the predictive GAPP, LAPP and MAAP scores. Image acquisition is currently automated, and other processes such as cell dissociation and antibody staining for the fixed-cell imaging regime can also be automated.
ECM and cell-response tests.
Freshly dissociated primary prostate samples and breast samples were distributed evenly among the three substrate-coated wells. After 24 h of incubation at 37 °C and 5% CO2, substrate wells were washed of loose cells and cellular debris. Wells were imaged and percentage of confluence calculated. For growth curves of cells on varied substrates, 15,000 cells were seeded per condition, and the cells were incubated at 37 °C and 5% CO2. Images were collected at 24 h intervals for three days.
Microfluidic device.
A microfluidic device was developed as a high-throughput, standardized, cell-measurement environment (Supplementary Fig. 9). The microfluidic device consisted of luer-lock inlet and outlet ports for automated cell loading and fluid exchange. The imaging chamber could be modulated to accommodate one or more substrates for exposing cells to multiple environments, thus increasing the total possible biomarkers measured. The physical dimensions of the device are similar to those of a standard microscope slide. The microfluidic device acts as a self-contained environmental chamber that standardizes biomarker measurements as the biopsied cells interact with the ECMf-coated glass surface and culturing medium. After live-cell imaging, cells were fixed with paraformaldehyde and stained with antibodies on the microfluidic device, enabling the live-cell biomarkers to be coordinated with the fixed-cell biomarkers, given cell location cataloguing. Cell viability was measured by adhesion and cell spreading on the microfluidic device. Viability levels were greater than >98% for the duration of live-cell culturing (2 h) on the device. Alternative glass-bottom imaging methods may be used with less high-throughput capabilities given that the fluid exchange of media, and of fixative and antibody solutions, may not be automated.
Cell imaging.
STRAT-AP relies on the label-free evaluation of dynamic live-cell phenotypic biomarkers, followed by the antibody-labelled evaluation of static molecular phenotypic biomarkers in fixed cells (Fig. 2 and Supplementary Figs. 1, 3 and 5). Live-cell imaging was carried out for 2 h after cells were seeded onto the microfluidic device in order to observe adhesion dynamics, cell spreading and early cell-motility events. During this time, cell location was tracked and stored to correlate it with cell location after fixation. Cells were imaged using label-free DIC microscopy, and visible membrane features such as adhesion cell-nucleus size, membrane ruffling and motility were quantified. Cells were then permeabilized, fixed and stained with antibodies to probe for protein localization and modification state. During live-cell imaging, cells were tracked and their location stored via automated software. After fixation, cells were imaged and biomarkers from tracked live cells were merged, for each corresponding cell. All images were captured using a Nikon TE-2000 system and Nikon Elements software with a Nikon CFI Plan Apo Lambda ×20, 0.75 numerical aperture, 1.0 mm working-distance DIC objective (MRD00205) and exposure time of 100 ms for live-cell imaging, and a Nikon CFI Plan Fluor ×40, 0.75 numerical aperture Eco glass objective (MRH00401), exposure times of 500 ms and Nikon filter sets, C-FL GFP HC HISN zero shift filter set (96362), C-FL Texas Red HC HISN zero shift filter set (96365), C-FL CY5 HC HISN zero shift filter set (96366) for fluorescence imaging. All image analyses were performed using custom Matlab software (Mathworks), as described in Supplementary Methods and ref. 26. Cells were identified using a combination of centre of mass and greyscale values. The custom machine-vision software for performing biomarker quantification was developed by Cellanyx (cat. nos. SS-MV-PR001 (prostate analysis) and SS-MV-BR001 (breast analysis)). Machine-vision software may also be built with the open-source software ImageJ.
Immunofluorescence.
Cells were fixed using 4% paraformaldehyde (Thermo Fisher Scientific product no. FB002) for 20 min and permeabilized using 0.1% Triton X-100 (Millipore Sigma cat. no. T8787). Cells were treated with mouse phosphorylated FA kinase (pFAK) antibody (BD Transduction Labs, product no. 611806) for active FA staining, rabbit integrin linked kinase (ILK) antibody (Abcam product no. ab74336) and rat α -tubulin antibody (Thermo Fisher Scientific product no. MA1–80189). Secondary Alexa-Fluor 488 anti-mouse (Thermo Fisher Scientific Probes product no. A11029), Alexa-Fluor 647 anti-rabbit (Thermo Fisher Scientific Probes product no. A21245) and Alexa-Fluor 594 anti-rat antibodies (Thermo Fisher Scientific product no. A11007) were used to stain the pFAK, ILK and α-tubulin primary antibodies, respectively. Supplementary Figs. 6 and 7 display staining controls in primary cancer cells and cancer cell lines, respectively.
Statistical analyses.
Custom machine-learning software (Supplementary Fig. 8) to generate thresholds and predictive clinical scores was developed by Cellanyx (cat. nos. SS-ML-PR001 (prostate analysis) and SS-ML-BR001 (breast analysis)). Machine-learning algorithms may also be built with MatLab software (Mathworks). The algorithms are based on random-forest decision trees47. Random forests are trained collections of decision trees with random selection to average outcomes or predictions. Individual random-forest-based algorithms are used for each specific prediction. For example, a specific adverse pathology (for example, SVI) or specific grouping of adverse pathologies (for example, LAPP) is predicted by a unique random-forest algorithm. The variance is reduced by averaging the ensemble of prediction with the advantage of keeping bias low from decision trees. Individual cells were analysed thereafter. The whole population of analysed cells was used to develop a sample score. On the basis of ROC curve analysis, decision thresholds (positive or negative for a given prediction) were set to maximize both sensitivity and specificity. The sensitivity and specificity of the assay were determined by comparing the algorithm-generated predictions with post-surgical pathology reports.
The following procedure was used to develop the machine-learning algorithm to predict and classify a specific adverse pathology (Supplementary Fig. 8). A random-forest classifier is a meta-estimator that uses multiple and distinct subsamples of a data set to fit a number of decision trees using randomly selected subsets of the biomarkers and end points, and that then uses averaging to improve the predictive accuracy of the machine-learning algorithms48,49. Because it uses random selection and averaging, over-fitting is controlled for. The performance reported is based on the predictions of the blinded validation data set. The 70% subsample was used in conjunction with the post-surgery pathology reports associated with each sample for the development of the ‘learned’ classification algorithm. Each objectively quantified biomarker was input into the machine-learning algorithms, which evaluate mathematical permutations of biomarkers along with individual biomarker scores to output the predictive GAPP, LAPP and MAPP scores40. The machine-learning algorithms were implemented using custom Matlab software26.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability.
The machine-vision and machine-learning MATLAB codes are available upon request from the corresponding author.
Supplementary Material
Acknowledgements
Tissue samples were provided by Lahey Hospital and Medical Center, Department of Cancer Research, American Medical Professionals of New York, by Urology Place of San Antonio and by the NCI Cooperative Human Tissue Network (CHTN). The authors thank S.K. Sia for a thoughtful review of the manuscript and P. Chaturvedi, R. Gottileb and M.B. Lisman for discussions, as well as M. Foroohar, S. Zappala, H. Rashid, V. Mouraviev, K. Christ, T.B. Sullivan and N. Kella. M.S.M. thanks J.A. Manak, C.A. Manak, G.H. Manak, P.L. Manak, L.L. Manak and P.W. Manak for support. J.S.V. thanks S.H. Varsanik for support. A.C.C. thanks C.S. Chandersekaran, A.C. Chandersekaran, A.B. Cravens Chander and I.E.A. Chander for support.
Footnotes
Competing interests
The authors declare the following competing financial interests: M.S.M., J.S.V., M.J.W., W.R.S., N.J., N.S., A.M., D.B., R.J.S., G.D., T.M., H.M.C., K.B.K., G.R.S. and A.C.C.: Cellanyx Diagnostics, stock options. B.J.H.: Cellanyx Diagnostics, consultant stock options. D.M.A.: Genomic Health, speaker; Myriad Genetics, speaker; Cellanyx Diagnostics, stock options; Applied Medical, stock options. The following patents WO2016138041, WO2013075145 and WO201204826921,22,26 cover aspects of the technology presented in this paper.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. Anonymized biomarker quantifications are available upon request from the corresponding author.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41551-018-0285-z.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel R, Ma J, Zou Z & Jemal A Cancer statistics, 2014. CA Cancer J. Clin 64, 9–29 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Bell N et al. Recommendations on screening for prostate cancer with the prostate-specific antigen test. CMAJ 186, 1225–1234 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mariotto AB et al. Cancer survival: an overview of measures, uses, and interpretation. J. Natl Cancer Inst. Monogr 2014, 145–186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shieh Y et al. Population-based screening for cancer: hope and hype. Nat. Rev. Clin. Oncol 13, 550–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.U.S. Cancer Statistics Working Group United States Cancer Statistics: 1999–2013 Incidence and Mortality Web-based Report http://www.cdc.gov/uscs (U.S. Department of Health and Human Services, 2016). [Google Scholar]
- 6.Canfield SE et al. A guide for clinicians in the evaluation of emerging molecular diagnostics for newly diagnosed prostate cancer. Rev. Urol 16, 172–180 (2014). [PMC free article] [PubMed] [Google Scholar]
- 7.Dahabreh IJ et al. Core Needle and Open Surgical Biopsy for Diagnosis of Breast Lesions: An Update to the 2009 Report AHRQ Comparative Effectiveness Reviews (Agency for Healthcare Research and Quality, 2014). [PubMed] [Google Scholar]
- 8.Alvarado M, Ozanne E & Esserman L Overdiagnosis and overtreatment of breast cancer. Am. Soc. Clin. Oncol. Educ. Book 2012, e40–e45 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Evans A & Vinnicombe S Overdiagnosis in breast imaging. Breast 31, 270–273 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Hugosson J & Carlsson S Overdetection in screening for prostate cancer. Curr. Opin. Urol 24, 256–263 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Klotz L Cancer overdiagnosis and overtreatment. Curr. Opin. Urol 22, 203–209 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Lebeau A & Kuhn T Updates in the treatment of ductal carcinoma in situ of the breast. Curr. Opin. Obstet. Gynecol 28, 49–58 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Strope SA & Andriole GL Prostate cancer screening: current status and future perspectives. Nat. Rev. Urol 7, 487–493 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Verma M, Patel P & Verma M Biomarkers in prostate cancer epidemiology. Cancers 3, 3773–3798 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koren S & Bentires-Alj M Breast tumor heterogeneity: source of fitness, hurdle for therapy. Mol. Cell 60, 537–546 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Marusyk A, Almendro V & Polyak K Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer 12, 323–334 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Zbuk KM & Eng C Cancer phenomics: RET and PTEN as illustrative models. Nat. Rev. Cancer 7, 35–45 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Albala D, Knopf K & Sant G Phenotypic cancer biomarkers-future role in precision oncology?. NPJ Precision Oncology 1, 21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chander A et al. Rapid and short-term extra-cellular matrix-mediated in vitro culturing of tumor and non-tumor human primary prostate cells from fresh radical prostatectomy tissue. Urology 105, 91–100 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Chander AC et al. Systems, devices and methods for microfluidic culturing, manipulation and analysis of tissues and cells. US patent US20130149724A1 (2013).
- 21.Chander AC et al. Systems, devices and methods for microfluidic culturing, manipulation and analysis of tissues and cells. US patent US20160272934A1 (2014).
- 22.Chander AC Systems, methods and devices for measuring growth/oncogenic and migration/metastatic potential. US patent US20130237453A1 (2011).
- 23.Chander AC Integrin-Linked Kinase, ECM Compostion and Substrate Rigidity Regulate Focal Adhesion—Actin Coupling, Modulating Survival, Proliferation andMigration: Towards a Biophysical Cancer Biomarker. PhD thesis, Columbia Univ; (2012). [Google Scholar]
- 24.Abdeen AA, Lee J & Kilian KA Capturing extracellular matrix properties in vitro: microengineering materials to decipher cell and tissue level processes. Exp. Biol. Med 241, 930–938 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kacsinta AD et al. Intracellular modifiers of integrin alpha 6p production in aggressive prostate and breast cancer cell lines. Biochem. Biophys. Res. Commun 454, 335–340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chander AC, Su WR & Varsanik JS Cell imaging and analysis to differentiate clinically relevant sub-populations of cells. Patent WO2016138041A2 (2015).
- 27.Cuzick J et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 12, 245–255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein EA et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur. Urol 66, 550–560 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Little J et al. Multigene panels in prostate cancer risk assessment: a systematic review. Genet. Med 18, 535–544 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Mendhiratta N, Meng X & Taneja SS Using multiparametric MRI to ‘personalize’ biopsy for men. Curr. Opin. Urol 25, 498–503 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Ross AE et al. A genomic classifier predicting metastatic disease progression in men with biochemical recurrence after prostatectomy. Prostate Cancer Prostatic. Dis 17, 64–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scarpato KR & Barocas DA Use of mpMRI in active surveillance for localized prostate cancer. Urol. Oncol 34, 320–325 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Sternberg IA, Vela I & Scardino PT Molecular profiles of prostate cancer: to treat or not to treat. Annu. Rev. Med 67, 119–135 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Zhuang L & Johnson MT How precisely can prostate cancer be managed? Int. Neurourol. J 20, S120–S130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong LM et al. Evaluation of models predicting insignificant prostate cancer to select men for active surveillance of prostate cancer. Prostate Cancer Prostatic Dis. 18, 137–143 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Tosoian JJ, Carter HB, Lepor A & Loeb S Active surveillance for prostate cancer: current evidence and contemporary state of practice. Nat. Rev. Urol 13, 205–215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Street CA & Bryan BA Rho kinase proteins—pleiotropic modulators of cell survival and apoptosis. Anticancer Res. 31, 3645–3657 (2011). [PMC free article] [PubMed] [Google Scholar]
- 38.Wirtz D, Konstantopoulos K & Searson PC The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer 11, 512–522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang J et al. Improving drug discovery with high-content phenotypic screens by systematic selection of reporter cell lines. Nat. Biotechnol 34, 70–77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Almendro V, Marusyk A & Polyak K Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol 8, 277–302 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Cross SE, Jin YS, Rao J & Gimzewski JK Nanomechanical analysis of cells from cancer patients. Nat. Nanotech 2, 780–783 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Akobeng AK Understanding diagnostic tests 1: sensitivity, specificity and predictive values. Acta Paediatr. 96, 338–341 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Akobeng AK Understanding diagnostic tests 3: receiver operating characteristic curves. Acta Paediatr. 96, 644–647 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Akobeng AK Understanding diagnostic tests 2: likelihood ratios, pre- and post-test probabilities and their use in clinical practice. Acta Paediatr. 96, 487–491 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Liu AY & True LD Characterization of prostate cell types by CD cell surface molecules. Am. J. Pathol 160, 37–43 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Signoretti S & Loda M Defining cell lineages in the prostate epithelium. Cell Cycle 5, 138–141 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Criminisi A & Shotton J Decision Forests for Computer Vision and Medical Image Analysis (Springer, New York, NY, 2013). [Google Scholar]
- 48.Ho TK Proceedings of 3rd International Conference on Document Analysis and Recognition Vol. 2, A1–A7 (IEEE, Montreal, 1995).. [Google Scholar]
- 49.Ho TK The random subspace method for constructing decision forests. IEEE. Trans. Pattern. Anal. Mach. Intell 20, 832–844 (1998). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.