Abstract
The mosquito, Aedes baisasi, which inhabits brackish mangrove swamps, is known to feed on fish. However, its host assemblage has not been investigated at the species level. We amplified and sequenced the cytochrome oxidase subunit I barcoding regions as well as some other regions from blood-fed females to identify host assemblages in the natural populations from four islands in the Ryukyu Archipelago. Hosts were identified from 230 females. We identified 15 host fish species belonging to eight families and four orders. Contrary to expectations from previous observations, mudskippers were detected from only 3% of blood-engorged females. The dominant host was a four-eyed sleeper, Bostrychus sinensis (Butidae, Gobiiformes), in Iriomote-jima Island (61%), while it was a snake eel, Pisodonophis boro (Ophichthidae, Anguilliformes), in Amami-oshima and Okinawa-jima islands (78% and 79%, respectively). Most of the identified hosts were known as air-breathing or amphibious fishes that inhabit mangroves or lagoons. Our results suggest that A. baisasi females locate the bloodmeal hosts within the mangrove forests and sometimes in the adjacent lagoons and land on the surface of available amphibious or other air-breathing fishes exposed in the air to feed on their blood.
Introduction
For most mosquito species, blood proteins are essential nutrients for egg production and thus, for reproductive fitness1. Therefore mosquito species have evolved to utilize the vertebrates as hosts (bloodmeals)2. When a number of possible hosts exist in their habitats, host preferences may develop. This may be the case, especially if blood quality affects reproductive output1.
The range of host vertebrates and host preferences has been investigated intensively for species that feed on humans1,2 as they can be vectors of pathogens that cause a variety of infectious diseases, including malaria, dengue fever, Japanese encephalitis, West Nile fever, etc. In the 1980s to 1990s, the enzyme-linked immunosorbent assay (ELISA) was mainly used for host identification3,4. However, this technique requires anti-sera preparation, for which the researchers need to identify candidate hosts before species-level identification. Otherwise, they can first apply anti-sera with broad activity (e.g., anti-bird and anti-mammal), but additional steps are needed for identification at the species level5.
DNA sequencing-based identification has been popular for the last decade. Development of vertebrate-specific primers for the regions frequently used for phylogeny analysis represented a breakthrough using this approach (e.g., mitochondrial 12S rRNA and 16S rRNA6, cytochrome b4 and cytochrome oxidase subunit I4,7). Studies on bloodmeal hosts from human-fed mosquito species using this approach have identified the broad range of host taxa including mammals, birds, reptiles8–10 and amphibians4,9 to the species level.
Some mosquito species, such as Culex peccator, C. erraticus, Aedes albopictus, A. togoi, and several Uranotaenia spp., are known to feed mainly on ectothermic hosts9,11,12. However, there are few species reported to feed on fish. Slooff and Marks13 observed A. longiforceps feeding on a mudskipper, Periophthalmus musgravei, in the Solomon Islands. Okudo et al.14 observed Aedes baisasi feeding on another mudskipper, Pe. argentilineatus in a cage and confirmed by ELISA using anti-fish antibodies that mosquitoes collected in the field had fed on fish. These authors suggested that mudskippers should be most readily accessible hosts because they are very common and are out of water for long periods. Tamashiro et al.9 further investigated the host range of wild A. baisasi using DNA sequencing of the 16S rRNA region and found DNA sequences of fish origin in 94% of blood-engorged A. baisasi females (the remaining 6% had fed on frogs). BLAST searches resulted in either a goby (Gobiiformes) or a snake eel (Anguilliformes) with relatively low similarities (91–92% and 89–95%, respectively) due to a deficiency of 16S rRNA data of potential candidates in the NCBI database, and they could not identify the host species.
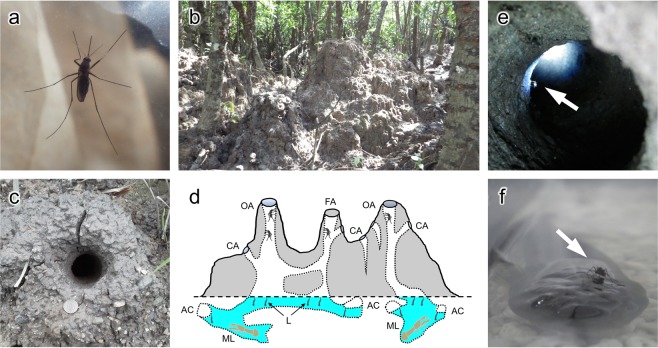
Aedes baisasi15 (Fig. 1a) inhabits burrows made by a mud lobster Thalassina anomala or land crabs such as Cardisoma carnifex, Discoplax hirtipes, and Episesarma lafondii, which are often found in the upper intertidal zone of mangrove forests in the Ryukyu Archipelago14,16 (Fig. 1). Larvae grow in brackish water (salinity 0–31) within burrows16. Adults are nocturnal; they rest in the upper parts of the holes in the daytime and fly out at night. Physiological states of females collected in the burrows suggest that adult females have a short flight range and that their mating and feeding activities are restricted to mangroves close to the burrows16. Therefore, it is expected that most candidate hosts also inhabit mangroves.
Figure 1.
The life-history of Aedes baisasi. (a) An adult female. (b) Mounds made by a mud lobster Thalassina anomala in a mangrove forest. (c) Aperture of a burrow in the mound. The coin is 22.6 mm in diameter. (d) Hypothesized structure of a mound, modified with permission from Miyagi & Toma57. Apertures of lobster burrows are basically open (OA), but are sometimes filled with mud (FA). Apertures of burrows made by crabs on the mounds (CA) are often connected to those made by the lobster. The lower part of the burrow is filled with brackish water, where A. baisasi larvae (L) live. Mud lobsters (ML) usually stay in brackish water and in an air chamber (AC) in the burrow. (e) An adult resting on the wall of the burrow (arrow). (f) A female feeding on the exposed surface of Bostrychus sinensis (arrow) in an enclosed cage during a preliminary experiment. Photo credits: Takashi Miyake.
Accordingly, we determined mitochondrial genome sequences of 11 species from the orders Gobiiformes and Anguilliformes that are expected to appear near mangroves on Okinawa-jima and Iriomote-jima to enhance the reference database (Supplementary Table S1). We also determined 15,400 bp of the mitochondrial DNA sequence of a snake eel Pisodonophis boro (Anguilliformes; An2 in Table S1). Although we could not complete the entire circular mitochondrial genome, we used it as a reference, as it contains sequences of all regions used for host identification (cytochrome oxidase subunit I, cytochrome b, 12S rRNA, and 16S rRNA). In this study, specimens of the mosquito, A. baisasi, were collected from four islands in the Ryukyu Archipelago. With the enhanced database, we identified their hosts to the species level. In addition to the 16S r RNA, used in Tamashiro et al.9, we used three regions; (1) cytochrome oxidase subunit I, since data for this region has become increasingly available due to its use in ‘DNA barcoding’17, (2) cytochrome b, as this region has been used for phylogenetic analysis in the order Gobiiformes18,19, and (3) a hypervariable region of the 12S rRNA gene using the MiFish primer set developed for identification of fish species20. We demonstrate that A. baisasi uses a variety of host fishes and that host usage patterns vary among islands of the Ryukyu Archipelago. We discuss whether the broad host usage of A. baisasi and its inter-island variation reflect host preference.
Results
Host identification
We collected 758 adult females of A. baisasi from the four islands. Proportions of blood-engorged individuals among the females on each island ranged from 27.3 to 75.0% (Table 1). The highest proportion (75.0%) may have been due to small sample size (N = 8). We performed PCR amplification for 263 blood-engorged females and successfully amplified host DNA for 87.8% of them (Table 1).
Table 1.
Numbers of specimens of Aedes baisasi collected, their status, and success rate of PCR amplification from their bloodmeals.
| Population | Year | Male | Female | PCR | ||||
|---|---|---|---|---|---|---|---|---|
| Blood-present | Blood-absent | % Blood-present | Female no. | Succeeded | % success | |||
| Iriomote-jima-1 | 2015 | NA | 52 | 83 | 38.5 | 52 | 50 | 96.2 |
| Iriomote-jima-2 | 2015 | NA | 19 | 28 | 40.4 | 19 | 19 | 100.0 |
| 2016 | 83 | 58 | 43 | 57.4 | 40 | 28 | 70.0 | |
| Ishigaki-jima | 2015 | NA | 6 | 2 | 75.0 | 6 | 6 | 100.0 |
| Okinawa-jima | 2016 | 27 | 21 | 56 | 27.3 | 21 | 20 | 95.2 |
| 2017 | 372 | 70 | 177 | 28.3 | 70 | 61 | 87.1 | |
| Amami-oshima | 2017 | 129 | 60 | 83 | 42.0 | 55 | 47 | 85.5 |
| Total | 286 | 472 | 37.7 | 231 | 263 | 87.8 | ||
Of the COI sequences from 211 samples for which we conducted BLASTn searches against the GenBank database, only those from 96 samples achieved ≥99% similarity (Supplementary Table S2). Samples with 92–94% similarity to four-eyed sleeper Bostrychus sinensis (Butidae, Gobiiformes) sequences in the GenBank database, reached ≥99% similarity with B. sinensis in our supplementary database. All samples that resulted in ≥99% similarity to a conger eel Uroconger lepturus (Congridae, Anguilliformes) in the GenBank achieved ≥99% similarity to a snake eel Pisodonophis boro (Ophichthidae, Anguilliformes) in our database. We confirmed that BLASTn searches for the 12S rRNA sequences from some of these samples against the GenBank database resulted in same-species identification (Pi. boro) with ≥98% similarity (AMA9 and AMA10, for example, Supplementary Table S2). Accordingly, we concluded that Pi. boro was the correct host. We found considerable discrepancies among the sequences of both COI and 12S rRNA of a spaghetti eel, Moringua microchir (Moringuidae, Anguilliformes) within the GenBank database. Many of the 12S rRNA sequences from our samples matched one of them (accession no. LC020870) closely; thus, they were identified as M. microchir.
Combined with the GenBank database and ours (Supplementary Table S1), we could identify host fish species with ≥99% similarity from all 230 female mosquitoes analyzed (Table 2, Supplementary Fig. S1), but for one sample, intermingled DNA sequences were obtained for both COI and cytochrome b regions, which we considered as originated from two different blood sources (Supplementary Table S2), most likely from Pi. boro and B. sinensis. This sample was excluded from further analysis.
Table 2.
Numbers of host species identified from abdomens of blood-engorged mosquitoes.
| Order | Family | Species | Islands | |||
|---|---|---|---|---|---|---|
| Amami-oshima | Okinawa-jima | Ishigaki-jima | Iriomote-jima | |||
| Anguilliformes | Moringuidae | Moringua microchir | 0 | 4 | 3 | 17 |
| Muraenidae | Gymnothorax pictus | 0 | 0 | 0 | 1 | |
| Uropterygius concolor | 0 | 0 | 0 | 1 | ||
| Ophichthidae | Pisodonophis boro | 36 | 64 | 0 | 5 | |
| Gobiiformes | Butidae | Bostrychus sinensis | 9 | 11 | 0 | 60 |
| Oxudercidae | Mugilogobius sp. ‘Izumi-haze' | 1 | 0 | 0 | 0 | |
| Periophthalmus argentilineatus | 0 | 1 | 0 | 6 | ||
| Trypauchenopsis intermedia | 0 | 1 | 0 | 0 | ||
| Gobiidae | Myersina macrostoma | 0 | 0 | 0 | 1 | |
| Blenniiformes | Blenniidae | Blenniella bilitonensis | 0 | 0 | 1 | 0 |
| Entomacrodus striatus | 0 | 0 | 0 | 1 | ||
| Istiblennius edentulus | 0 | 0 | 2 | 0 | ||
| Salarias fasciatus | 0 | 0 | 0 | 2 | ||
| Salarias luctuosus | 0 | 0 | 0 | 2 | ||
| Tetraodontiformes | Balistidae | Rhinecanthus verrucosus | 0 | 0 | 0 | 1 |
| Total | 46 | 81 | 6 | 97 | ||
Host assemblage
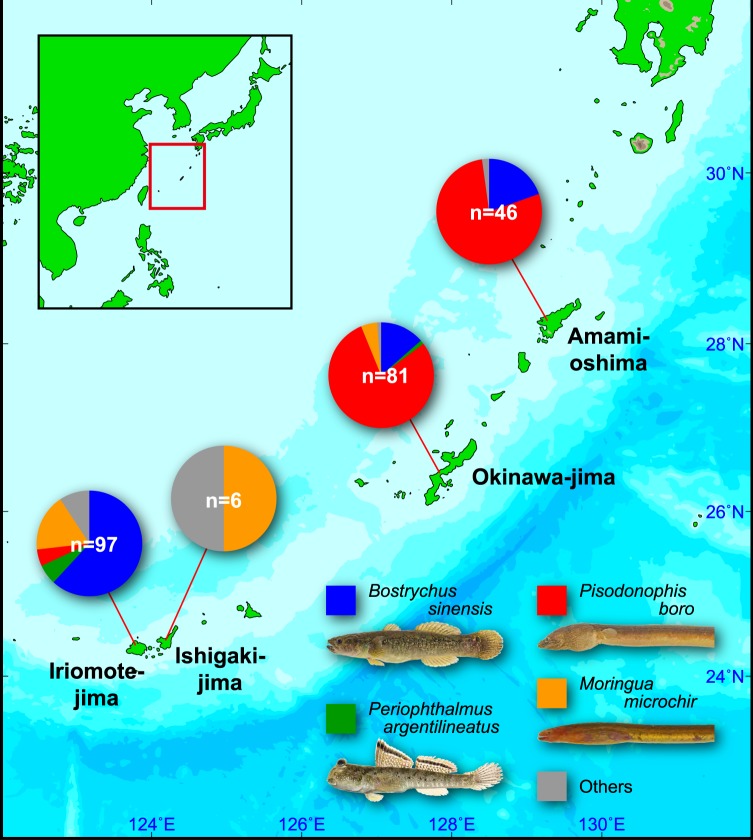
Although we identified 15 host species from four islands, host assemblages were quite different among the islands, except for between Amami-oshima and Okinawa-jima (Fig. 2, Table 2). We identified bloodmeal hosts from 46 female mosquitoes in Amami-oshima, and only three host species were found, with Pi. boro as the most frequent host (78%). About 20% of all females fed on B. sinensis, and the blood from a goby Mugilogobius sp. ‘Izumi-haze’ was detected in one female. A similar pattern was observed in Okinawa-jima; five host species were found, again with Pi. boro as the most frequent host (79%) and B. sinensis as the second most frequent (14%). Some fed on M. microchir (5%) and the blood from a mudskipper, Periophthalmus argentilineatus, was detected in one female and from a bearded eel goby, Trypauchenopsis intermedia, in another.
Figure 2.
A map of study areas in the Ryukyu Archipelago, Japan and host assemblages of Aedes baisasi in each area. The main map was generated by GMT ver. 5.4.2 (http://gmt.soest.hawaii.edu/) and a template of the small scale map is from http://www.freemap.jp/. Photo credits: Ken Maeda.
On the other hand, we identified 11 host species from 97 female mosquitoes in Iriomote-jima. Bostrychus sinensis was the most frequent host (61%), followed by M. microchir (18%), Pe. argentilineatus (6%) and Pi. boro (5%). Some host species were found only in Iriomote-jima, but they are detected from only one or two female mosquitoes. These included three blennies Entomacrodus striatus, Salarias fasciatus and S. luctuosus (Blenniidae, Blenniiformes), a goby Myersina macrostoma (Gobiidae, Gobiiformes), two moray eels Gymnothorax pictus and Uropterygius concolor (Muraenidae, Anguilliformes), and a triggerfish Rhinecanthus verrucosus (Balistidae, Tetraodontiformes).
Although we collected only 6 blood-engorged females on Ishigaki-jima, host composition was quite different from the other islands; M. microchir was the most frequent (50%), followed by two blennies Blenniella bilitonensis and Istiblennius edentulous.
Discussion
Host identification based on DNA sequences
The DNA-sequence-based technique has brought great progress in bloodmeal host identification of mosquitoes and other arthropods (e.g., biting midges21–23 and ticks7,24). In our study, using a combination of some DNA regions used for phylogenetic studies and DNA barcoding, host identification for fish-fed A. baisasi was much improved. Tamashiro et al.9 aimed at revealing feeding habits of 35 mosquito species from 11 genera with broad spectra of potential host animals. They also used the mitochondrial 16S rRNA region for this species and this enabled identification at higher taxonomic levels (i.e., at the order level). In our study, we applied the COI region to host identification4,7 and also benefited from recently developed universal PCR primers, which were originally designed for metabarcoding environmental DNA from fishes20. This resulted in host identification at the species level. Therefore, this study was the first to demonstrate host assemblages of mosquito species that are parasitic upon fish in natural populations.
While the COI region is well established for DNA barcoding and the amount of available data has been increasing, many of our samples failed to surpass 99% similarity to any taxa in GenBank. There are several possible reasons for this. First, the GenBank database does not include sequence diversity among localities. For example, B. sinensis is widely distributed and regional differentiation may account for the low similarity (92–94%). Second, taxonomy has often not been clearly established and that makes it difficult to complete the database. Regardless, we developed a database including more species from more localities with reliable identification and the genomic data. The sequence database of candidate fishes from the sampling areas greatly improved the similarity, and the short fragment of 12S rRNA (using a “MiFish” primer set)20 helped to further resolve these problems (Supplementary Table S2).
Feeding habits of Aedes baisasi
Host identification at the species level has some interesting implications. First, contrary to our expectations, based on the laboratory observation by Okudo et al.14, mudskippers were not the main source of bloodmeals in A. baisasi; DNA sequences that originated with a mudskipper Periophthalmus argentilineatus were detected in only 7 out of 229 (3%) host-identified females. Second, bloodmeal hosts fed frequently on fish species which inhabit mangrove swamps, such as B. sinensis, Pi. boro and Pe. argentilineatus25,26, confirming that feeding activities of A. baisasi are restricted to the mangroves16. Some other species documented less frequently included Mugilogobius sp., Myersina macrostoma, Trypauchenopsis intermedia, and Uropterygius concolor, which also inhabit mangroves26–28. Some species, such as Entomacrodus striatus, Salarias fasciatus, Istiblennius edentulus, and Gymnothorax pictus inhabit tidepools along rocky shore and/or coral reefs29, implying that A. baisasi may sometimes search for hosts out of mangrove forests and may visit nearby lagoons. Third, many bloodmeal hosts can remain out of water for prolonged periods. Bostrychus sinensis is known as a facultative air-breathing fish30, and can survive out of water for more than a day31. Pisodonophis boro also has air-breathing habits32. Some blennies are amphibious, e.g., E. striatus33–35, I. edentulus35,36 (but see Platt et al.33) and also B. bilitonensis, sometimes clings to rocks out of water37. The moray eel G. pictus also leaves water and wriggles across dry places38. Given the behavioral attributes of these fishes, it is likely that A. baisasi searches for and lands on fishes either when they leave the water or when parts of their bodies are exposed to the air.
Many of these air-breathing or amphibious fish are reported to be more active out of the water at night than during the day. Most amphibious blennies emerge from water mainly at night to avoid the risk of desiccation39,40. Bostrychus sinensis usually hides in hollows on mud, beneath rocks, or gaps in mangrove roots, and in caves during the daytime and emerges at night41. We have seen many individuals feeding in shallow waters at night, which may often expose themselves to the air. Pisodonophis boro is also active at night at shallows42, foraging mainly for sesarmid crabs43, and is also an accessible host for mosquitoes. These habits match the feeding activity of A. baisasi.
Our results, with support from other observations14,16, suggest that females rest in lobster holes during the daytime and leave at night to search for bloodmeal hosts within the mangroves and sometimes in adjacent lagoons. They locate and land on the surfaces of exposed amphibious or other air-breathing fishes and feed there, but are not attracted to humans or warm-blooded animals14. However, some ecological factors remain unclear regarding seasonal variation in host selection, the stage of fishes on which A. baisasi feed (e.g., juveniles, immatures or adults) and the cues A. baisasi uses to locate bloodmeal hosts. Seasonal variation is expected because fish communities vary seasonally in mangroves25,42. Tamashiro et al.9 found three A. baisasi females with blood from frogs. These were sampled on Iriomote-jima in May (Ichiro Miyagi and Takako Toma, Laboratory of Mosquito Systematics of Southeast Asia and South Pacific, personal communication). Olfaction is the likely cue for the mosquitoes to locate a host, which is implied by the fact that air-breathing fishes have special excretion systems in their skin44–46, but laboratory and/or field assays are needed to confirm this hypothesis.
Host preference of Aedes baisasi
Host assemblages were quite different among the islands. This may reflect differences in abundance of available fishes among the islands, considering that B. sinensis is abundant on Iriomote-jima, but less on Okinawa-jima and Amami-oshima41,47.
On the other hand, it is puzzling that A. baisasi do not feed frequently on the mudskipper, Pe. argentilineatus, which is apparently the most accessible fish host in this region. Periophthalmus argentilineatus seems less active at night48, and we often saw it still staying out of water. The mudskipper may have some kind of mechanism that keeps off A. baisasi. It is also the puzzling that significant blood feeding from spaghetti eels, Moringua microchir, occurred on three of the four islands. Little information is available on the life history and ecology of moringuid eels, including M. microchir29,49. Moringua microchir is recognized as a rather rare species in Japan, and it has not reported from mangroves. Keith et al.50 reported that the juveniles inhabit estuaries and lower reaches of rivers, while adult females stay on shallow marine bottoms. However, frequent predation by mosquitoes implies that spaghetti eels are actually common around mangroves. Our data also suggest that A. baisasi may preferentially feed on it. Although A. baisasi chooses bloodmeal hosts according to their availability (abundance and nocturnal activity out of water), host preferences may exist.
The subgenus Geoskusea, to which A. baisasi belongs, includes 10 species, all of which also inhabit brackish water51. In addition to A. baisasi and A. longiforceps13, other species also probably use air-breathing fishes as bloodmeal hosts. Further studies of host identification in these species will help our understanding of the evolution in host preference and exploitation in niche adaptation.
Methods
Sampling
Adult mosquitoes were collected during the daytime (between 9:00 a.m. and 6:00 p.m.) in mangrove forests on four islands in the Ryukyu Archipelago, Japan (Fig. 2): Iriomote-jima Island (Iriomote-jima-1: Uehara, 24° 23′ N, 123° 49′ E, and Iriomote-jima-2: Komi, 24° 19′ N, 123° 54′ E) in November, 2015 and October, 2016, Ishigaki-jima Island (24° 25′ N, 124° 14′ E) in November, 2015, Okinawa-jima Island (24° 19′ N, 123° 54′ E) in October, 2016 and September, 2017, and Amami-oshima Island (28° 15′ N, 129° 24′ E) in September, 2017. Mosquitoes resting in crab or mangrove lobster burrows during the daytime16 were forced out by disturbing them with a twig and trapped with a sweep net. We found from preliminary surveys that it was more effective than using a handheld vacuum16. Collected mosquitoes were preserved in 99.5% ethanol and brought to the laboratory.
Bloodmeal host identification
Each mosquito was individually examined to determine species, sex, and blood-feeding status under a microscope. For blood-engorged females, their abdomens were isolated using forceps. Genomic DNA was extracted from the abdomen either using a DNeasy Blood & Tissue Kit (Qiagen) or by the HotSHOT method52.
We conducted PCR to amplify either mitochondrial COI (cytochrome oxidase subunit I) with primers M13BC-FW (5′-TGT AAA ACG ACG GCC AGT HAA YCA YAA RGA YAT YGG NAC-3′) and COI_long (r) (5′-AAG AAT CAG AAT ARG TGT TG-3′) designed to amplify exclusively from vertebrate DNA4,7, or mitochondrial 12S rRNA region with M13F-attached MiFish-U-F (5′-GTA AAA CGA CGG CCA GGT CGG TAA AAC TCG TGC CAG C-3′) and MiFish-U-R (5′-CAT AGT GGG GTA TCT AAT CCC AGT TTG-3′)20. PCR was performed using KOD FX Neo DNA polymerase kit (Toyobo, Japan) in a 20 µL total volume: 5 µL of 2x Buffer, 0.3 µL of 10 µM each primer, 2 µL of 2 mM dNTPs, 0.2 µL of KOD FX Neo, 0.4 µl of template DNA and 1.8 µL of dH2O. PCR cycling conditions were as follows: initial denaturation at 94 °C for 2 min, then 35 cycles of 98 °C for 10 s, 48 °C for 30 s, and 68 °C for 1 min for COI, and 35 cycles of 98 °C for 10 s, 50 °C for 10 s, and 68 °C for 15 s for 12S rRNA. PCR products were visualized on 1.5% agarose gels to confirm amplification. Amplified products were purified using Exo-SAP-IT (USB corp., Cleveland, OH, USA). For both regions, sequence reactions were conducted with M13F primer (5′-GTA AAA CGA CGG CCA G-3′) using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosytems). Sequencing products were analyzed on an ABI PRISM 3130 capillary DNA sequencer (Applied Biosystems).
All sequenced specimens were identified using BLASTn searches against the GenBank nucleotic acid sequence database (NCBI website, http://www.ncbi.nlm.nih.gov/BLAST/) and/or a similar search using BLAST+ ver. 2.3.0+ against the additional reference sequences stated below. The most similar fish species (≥99% sequence identity) based upon blood from engorged female mosquitoes was considered to be the parasitized host.
When we could not find any data that matched our query sequences at ≥99%, we conducted PCR using the same DNA polymerase kit for other regions with primers designed to amplify exclusively from vertebrate or fish DNA: the mitochondrial cytochrome B region with CytB(f)(5′-GAG GMC AAA TAT CMT TCT GAG G-3′) and CytB(r)(5′-TAG GGC VAG KAC TCC TCC TAG T-3′)4, or the mitochondrial 16S rDNA region with 16Sa-L (5′-CGC CTG TTT ACC AAA AAC ATC GCC T-3′) and 16Sb-H (5′-CCG GTC TGA ACT CAG ATC ACG T-3′)53. PCR cycling conditions were as follows: initial denaturation at 94 °C for 2 min, then 35 cycles of 98 °C for 10 s, 55 °C for 40 s, and 68 °C for 1 min for cytochrome B, and 35 cycles of 98 °C for 10 s, 50 °C for 40 s, and 68 °C for 1 min for 16S rDNA. Positive amplicons were sequenced with one of the primers used in PCR reactions. Identities of all sequenced specimens were determined as described above.
Reference sequences of fishes
Total genomic DNA of 15 specimens belonging to six anguilliform species and six gobiiform species collected from Okinawa-jima and Iriomote-jima was extracted from the right pectoral fins (Gobiiformes) or muscle pieces (Anguilliformes) preserved in 99.5% ethanol, using a DNeasy Blood & Tissue Kit (Quiagen, Hilden, Germany) or a Maxwell RSC Blood DNA Kit (Promega, Fitchburg, Wisconsin, USA).
Whole genome shotgun sequencing libraries were prepared using a KAPA HyperPlus Kit, PCR-free (KAPA Biosystems, Wilmington, Massachusetts, USA). Extracted genomic DNA was enzymatically fragmented into pieces of 200–1000 bp. After repairing the protruding ends and A-tailing, sequencing adaptors were ligated onto both ends of the DNA fragments. Shotgun libraries were then sequenced on either an Illumina MiSeq sequencer (Illumina, San Diego, California, USA) with MiSeq V3 600 cycle kit (Illumina) or an Illumina HiSeq 2500 sequencer in Rapid Run mode version 2 using a HiSeq Rapid Cluster Kit v2-Paired-End (Illumina) and HiSeq Rapid SBS Kit v2 (Illumina) or an Illumina HiSeq 4000 sequencer with HiSeq 3000/4000 PE Cluster Kits and HiSeq 3000/4000 SBS kit (300 cycles, Illumina) following manufacturer instructions.
Sequencing data from each library were assembled with the IDBA_UD assembler version 1.1.154 with different kmer lengths (60, 80, 100). Identification of complete mitochondrial genomes from assembled contigs was performed by (1) comparing them with the complete Stiphodon alcedo mitochondrial genome (accession: AB613000.1) (BLASTN e-value B 1e-100), and by (2) confirming that 100 bp of both head and tail DNA sequences of a contig were identical, indicating that the sequence was circular. Complete mitochondrial genomes were aligned using MAFFT v7.24455 and all positions with gaps were removed using trimAl56. All sequenced raw data are available in the DDBJ Sequence Read Archive under BioProject accession number PRJDB5763. Assembled mitochondrial genome sequences with gene annotations are available in the DDBJ database under accession numbers: AP019348–AP019362. Accession numbers for each individual are shown in Supplementary Table S1.
Procedures used to handle fish specimens in this study were approved by the Animal Care and Use Committees of both Okinawa Institute of Science and technology Graduate University and Gifu University. All experiments and samplings were performed in accordance with relevant guidelines and regulations of the committees.
Supplementary information
Acknowledgements
We are grateful to Norikazu Shikatani, Tohru Naruse, Motohiro Kawanishi, Haruo Okudo, Ichiro Miyagi and Takako Toma for field data, and to Saki Tsujita, Kosei Yamada, Yasutaka Kuroi for assistance in the field, and to the Amami Ranger Office (Amami Wildlife Conservation Center) and the Municipal Board of Education at Nago for permission to collect samples in Amamigunto National Park and Oura Mangrove Forest, respectively. We thank Katsunori Tachihara (University of the Ryukyus) for providing permission to use his photos of Gymnothorax pictus and Rhinecanthus verrucosus, Atsushi Tawa (National Research Institute of Far Seas Fisheries, Fisheries Research Agency) and Toshifumi Saeki (Rivus, Okinawa) for providing fish specimens, and Steven D. Aird (OIST) for editing the manuscript. This study was supported partially by Collaborative Research of the Tropical Biosphere Research Center, University of the Ryukyus to T.M.
Author Contributions
T.M. and N.A. conceived and designed the experiments. All authors performed data curation. T.M., N.A., K.M., C.S. and R.K. analyzed the data. T.M. and N.A. led the writing, with substantial contributions from all authors.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40509-6.
References
- 1.Takken W, Verhulst NO. Host preferences of blood-feeding mosquitoes. Annu. Rev. Entomol. 2013;58:433–453. doi: 10.1146/annurev-ento-120811-153618. [DOI] [PubMed] [Google Scholar]
- 2.Lyimo IN, Ferguson HM. Ecological and evolutionary determinants of host species choice in mosquito vectors. Trends Parasitol. 2009;25:189–196. doi: 10.1016/j.pt.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Silver, J. B. Mosquito ecology: field sampling methods. (Springer, 2008).
- 4.Townzen JS, Brower AVZ, Judd DD. Identification of mosquito bloodmeals using mitochondrial cytochrome oxidase subunit I and cytochrome b gene sequences. Med. Vet. Entomol. 2008;22:386–393. doi: 10.1111/j.1365-2915.2008.00760.x. [DOI] [PubMed] [Google Scholar]
- 5.Apperson CS, et al. Host-feeding habits of Culex and other mosquitoes (Diptera: Culicidae) in the Borough of Queens in New York City, with characters and techniques for identification of Culex mosquitoes. J. Med. Entomol. 2002;39:777–785. doi: 10.1603/0022-2585-39.5.777. [DOI] [PubMed] [Google Scholar]
- 6.Kitano T, Umetsu K, Tian W, Osawa M. Two universal primer sets for species identification among vertebrates. Int. J. Legal Med. 2007;121:423–427. doi: 10.1007/s00414-006-0113-y. [DOI] [PubMed] [Google Scholar]
- 7.Alcaide M, et al. Disentangling vector-borne transmission networks: A universal DNA barcoding method to identify vertebrate hosts from arthropod bloodmeals. PLoS One. 2009;4:1–6. doi: 10.1371/journal.pone.0007092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roiz D, et al. Blood meal analysis, flavivirus screening, and influence of meteorological variables on the dynamics of potential mosquito vectors of West Nile virus in northern Italy. J. Vector Ecol. 2012;37:20–28. doi: 10.1111/j.1948-7134.2012.00196.x. [DOI] [PubMed] [Google Scholar]
- 9.Tamashiro M, Toma T, Mannen K, Higa Y, Miyagi I. Bloodmeal identification and feeding habits of mosquitoes (Diptera: Culicidae) collected at five islands in the Ryukyu Archipelago, Japan. Med. Entomol. Zool. 2011;62:53–70. doi: 10.7601/mez.62.53. [DOI] [Google Scholar]
- 10.Martínez-de la Puente J, Ruiz S, Soriguer R, Figuerola J. Effect of blood meal digestion and DNA extraction protocol on the success of blood meal source determination in the malaria vector Anopheles atroparvus. Malar. J. 2013;12:109. doi: 10.1186/1475-2875-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyagi I. Feeding habits of some Japanese mosquitoes on cold-blooded animals in laboratory. (in Japanese) Trop. Med. 1972;14:203–217. [Google Scholar]
- 12.Cupp EW, et al. Identification of reptilian and amphibian blood meals from mosquitoes in an eastern equine encephalomyelitis virus focus in central Alabama. Am. J. Trop. Med. Hyg. 2004;71:272–276. doi: 10.4269/ajtmh.2004.71.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slooff R, Marks EN. Mosquitoes (Culicidae) biting a fish (Periophthalmidae) J. Med. Entomol. 1965;2:16. doi: 10.1093/jmedent/2.1.16. [DOI] [PubMed] [Google Scholar]
- 14.Okudo H, et al. Crab-hole mosquito, Ochlerotatus baisasi, feeding on mudskipper (Gobiidae: Oxudercinae) in the Ryukyu Islands, Japan. J. Am. Mosq. Control Assoc. 2004;20:134–137. [PubMed] [Google Scholar]
- 15.Knight KL, Hull WB. Three new species of Aedes from the Philippines (Diptera, Culicidae) Pacific Sci. 1951;5:197–203. [Google Scholar]
- 16.Toma T, et al. Bionomics of the mud lobster–hole mosquito Aedes (Geoskusea) baisasi in the mangrove swamps of the Ryukyu Archipelago, Japan. J. Am. Mosq. Control Assoc. 2011;27:207–216. doi: 10.2987/11-6123.1. [DOI] [PubMed] [Google Scholar]
- 17.Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agorreta A, et al. Molecular phylogenetics of Gobioidei and phylogenetic placement of European gobies. Mol. Phylogenet. Evol. 2013;69:619–633. doi: 10.1016/j.ympev.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 19.Thacker CE, Hardman MA. Molecular phylogeny of basal gobioid fishes: Rhyacichthyidae, Odontobutidae, Xenisthmidae, Eleotridae (Teleostei: Perciformes: Gobioidei) Mol. Phylogenet. Evol. 2005;37:858–871. doi: 10.1016/j.ympev.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Miya M, et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015;2:150088. doi: 10.1098/rsos.150088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bobeva A, et al. Host preferences of ornithophilic biting midges of the genus Culicoides in the Eastern Balkans. Med. Vet. Entomol. 2015;29:290–296. doi: 10.1111/mve.12108. [DOI] [PubMed] [Google Scholar]
- 22.Lassen SB, Nielsen SA, Skovgård H, Kristensen M. Molecular identification of bloodmeals from biting midges (Diptera: Ceratopogonidae: Culicoides Latreille) in Denmark. Parasitol. Res. 2011;108:823–829. doi: 10.1007/s00436-010-2123-4. [DOI] [PubMed] [Google Scholar]
- 23.M-D L Puente J, et al. Genetic characterization and molecular identification of the bloodmeal sources of the potential bluetongue vector Culicoides obsoletus in the Canary Islands, Spain. Parasites and Vectors. 2012;5:1–7. doi: 10.1186/1756-3305-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humair P-F, et al. Molecular identification of bloodmeal source in Ixodes ricinus ticks using 12S rDNA as a genetic marker. J. Med. Entomol. 2007;44:869–880. doi: 10.1093/jmedent/44.5.869. [DOI] [PubMed] [Google Scholar]
- 25.Kuo S-R, Lin H-J, Shao K-T. Fish assemblages in the mangrove creeks of Northern and Southern Taiwan. Estuaries. 1999;22:1004. doi: 10.2307/1353079. [DOI] [Google Scholar]
- 26.Tachihara K, et al. Ichthyofauna in mangrove estuaries of the Okinawa, Miyako, Ishigaki and Iriomote Islands during August from 2000 to 2002. Bull. Soc. Sea Water Sci. Japan. 2003;57:481–490. [Google Scholar]
- 27.Akihito P, Meguro K. First record of the goby Myersina macrostoma from Japan. (in Japanese) Japanese J. Ichthyol. 1978;24:295–299. [Google Scholar]
- 28.McCosker JE, Hatooka K, Sasaki K. Japanese moray eels of the genus. Uropterygius. Japanese J. Ichthyol. 1984;31:261–267. [Google Scholar]
- 29.Nakabo, T. Fishes of Japan with Pictorial Keys to the Species. (Tokai University Press, 2013).
- 30.Peh WYX, Chew SF, Wilson JM, Ip YK. Branchial and intestinal osmoregulatory acclimation in the four-eyed sleeper, Bostrychus sinensis (Lacepède), exposed to seawater. Mar. Biol. 2009;156:1751–1764. doi: 10.1007/s00227-009-1210-5. [DOI] [Google Scholar]
- 31.Ip YK, et al. The sleeper Bostrichthys sinensis (Family Eleotridae) stores glutamine and reduces ammonia production during aerial exposure. J. Comp. Physiol. B. 2001;171:357–367. doi: 10.1007/s003600100184. [DOI] [PubMed] [Google Scholar]
- 32.Hora SL. A note on the biology of the precipitating action of the mucus of Boro fish, Pisodonophis boro (Ham. Buch.) J. Asiat. Soc. Bengal. 1933;29:271–274. [Google Scholar]
- 33.Platt ERM, Fowler AM, Ord TJ. Land colonisation by fish is associated with predictable changes in life history. Oecologia. 2016;181:769–781. doi: 10.1007/s00442-016-3593-6. [DOI] [PubMed] [Google Scholar]
- 34.Ord TJ, Cooke GM. Repeated evolution of amphibious behavior in fish and its implications for the colonization of novel environments. Evolution. 2016;70:1747–1759. doi: 10.1111/evo.12971. [DOI] [PubMed] [Google Scholar]
- 35.Ord TJ, Summers TC, Noble MM, Fulton CJ. Ecological release from aquatic predation is associated with the emergence of marine blenny fishes onto land. Am. Nat. 2017;189:570–579. doi: 10.1086/691155. [DOI] [PubMed] [Google Scholar]
- 36.Ba-Omar T, Al-Riyami MM. Integumentary histology of the amphibious blenny, Isteblennius edentulus (Forester and Schneider, 1801) Sultan Qaboos Univ. J. Sci. 2009;14:9–15. [Google Scholar]
- 37.Froese, R. & Pauly, D. FishBase. http://www.fishbase.org (2018).
- 38.Chave EHN, Randall HA. Feeding behavior of the moray eel. Gymnothorax pictus. Copeia. 1971;3:570–574. doi: 10.2307/1442464. [DOI] [Google Scholar]
- 39.Luck AS, Martin KLM. Tolerance of forced air emergence by a fish with a broad vertical distribution, the rockpool blenny, Hypsoblennius gilberti (Blenniidae) Environ. Biol. Fishes. 1999;54:295–301. doi: 10.1023/A:1007584406324. [DOI] [Google Scholar]
- 40.Dabruzzi TF, Wygoda ML, Wright JE, Eme J, Bennett WA. Direct evidence of cutaneous resistance to evaporative water loss in amphibious mudskipper (family Gobiidae) and rockskipper (family Blenniidae) fishes from Pulau Hoga, southeast Sulawesi, Indonesia. J. Exp. Mar. Bio. Ecol. 2011;406:125–129. doi: 10.1016/j.jembe.2011.05.032. [DOI] [Google Scholar]
- 41.Ministry of Environment Japan. Red Data Book2014 – Threatened Wildlife of Japan. (in Japanese)(Gyosei Corporation, 2015).
- 42.Lin HJ, Shao KT. Seasonal and diel changes in a subtropical mangrove fish assemblage. Bull. Mar. Sci. 1999;65:775–794. [Google Scholar]
- 43.Lampang PN, Palasai A, Kettratad J. Gut content analysis of the snake eel Pisodonophis boro (Hamilton, 1822) from estuary of Pranburi River, Thailand. Proc. 3rd Natl. Meet. Biodivers. Manag. Thail. 2016;3:246–249. [Google Scholar]
- 44.Chew SF, Ip YK. Excretory nitrogen metabolism and defence against ammonia toxicity in air-breathing fishes. J. Fish Biol. 2014;84:603–638. doi: 10.1111/jfb.12279. [DOI] [PubMed] [Google Scholar]
- 45.Park JY. Structure of the skin of an air-breathing mudskipper. Periophthalmus magnuspinnatus. J. Fish Biol. 2002;60:1543–1550. doi: 10.1111/j.1095-8649.2002.tb02446.x. [DOI] [Google Scholar]
- 46.Randall DJ, Ip YK. Ammonia as a respiratory gas in water and air-breathing fishes. Respir. Physiol. Neurobiol. 2006;154:216–225. doi: 10.1016/j.resp.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Nature Conservation Division, Kagoshima Prefecture. Revision of Endangered Animals and Plants in Kagoshima Prefecture: Red Data Book. (in Japanese)(Kagoshima Prefectural Environmental Technology Association, 2016).
- 48.Colombini I, Berti R, Nocita A, Chelazzi L. Foraging strategy of the mudskipper Periophthalmus sobrinus Eggert in a Kenyan mangrove. J. Exp. Mar. Bio. Ecol. 1996;197:219–235. doi: 10.1016/0022-0981(95)00160-3. [DOI] [Google Scholar]
- 49.Castle PHJ. Early life-history of the eel Moringua edwardsi (Pisces, Moringuidae) in the Western North Atlantic. Bull. Mar. Sci. 1979;29:1–18. [Google Scholar]
- 50.Keith, P., Marquet, G., Valade, P., Bosc, P. & Vigneux, E. Atlas des poissons et des crustacés d’eau douce des Comores, Mascareignes et Seychelles. (Patrimoines Naturels, v. 65. Publications scientifiques du Muséum National d’Histoire Naturelle, 2006).
- 51.Miyagi I, Toma T, Lien JC. Ochlerotatus (Geoskusea) timorensis (Culicidae: Diptera), a new species from crab-holes, West Timor, Indonesia. Med. Entomol. Zool. 2004;55:107–114. doi: 10.7601/mez.55.107. [DOI] [Google Scholar]
- 52.Truett GE, et al. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- 53.Tang KL, Fielitz C. Phylogeny of moray eels (Anguilliformes: Muraenidae), with a revised classification of true eels (Teleostei: Elopomorpha: Anguilliformes) Mitochondrial DNA. 2013;24:55–66. doi: 10.3109/19401736.2012.710226. [DOI] [PubMed] [Google Scholar]
- 54.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 55.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyagi, I. & Toma, T. The National History of Mosquitoes in the Ryukyu Islnads. (in Japanese)(Tokai University Press, 2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.